- Department of Oceanography, Chonnam National University, Gwangju, South Korea
Dinoflagellates in the family Suessiaceae, so-called suessiacean dinoflagellates, play diverse roles in aquatic ecosystems, being distributed from tropical to polar waters and from marine to freshwater habitats and encompassing free-living forms, symbionts, and parasites. Despite their importance due to the variety of ecological roles and biodiversity, very few studies have characterized small suessiacean species. Recent advances in molecular techniques could provide insights into the yet unexplored ecological roles they play in aquatic environments. Using a global DNA metabarcoding dataset, this study elucidated the hidden biodiversity, global distribution, and ecological characteristics of suessiacean dinoflagellates. The results of this study indicated that the family Suessiaceae was the sixth highest in terms of read count and the ninth highest in terms of amplicon sequence variant (ASV) richness from a total of 42 categorized dinoflagellate families, suggesting that their global abundance has been greatly underestimated. Furthermore, metabarcodes of suessiacean dinoflagellates were found to be cosmopolitan in distribution, although the ecological niche of each taxon was distinctly different within the group based on their latitudinal and vertical distribution patterns. Moreover, phylogenetic analysis discovered at least five new phylogenetic groups and three new individual species within the family. Collectively, the findings of this study highlight the significance of suessiacean dinoflagellates in global aquatic ecosystems and reveal the importance of big data obtained from environmental DNA in exploring the ecological functions of understudied species.
Introduction
Dinoflagellates (Alveolata: Dinophyceae) are among the most diverse and abundant groups of unicellular protists commonly found in aquatic environments. They are both evolutionarily and ecologically important since they form diverse lineages and have a variety of ecological roles such as primary producers, consumers, mutualists, and parasites; they can also cause harmful algal blooms (Coats, 1999; Jeong et al., 2010; Murray et al., 2016; Suggett et al., 2017; Jang et al., 2022). The dinoflagellate group currently consists of 18 orders, including Suessiales, and more than 2,000 extant species have so far been described (Gómez, 2012; Guiry and Guiry, 2022).
The family Suessiaceae belongs to the order Suessiales, which was first described by Fensome et al. (1993). The order Suessiales originally contained two families: Suessiaceae (fossil dinosporin cysts) and Symbiodiniaceae (extant symbionts). Although both families were merged into Suessiaceae by Moestrup et al. (2009a), they were later separated again along with the rapidly increasing biodiversity found within the groups (LaJeunesse et al., 2018). Since the first report of an extant species in the family Suessiaceae in 1999, numerous extant species in the group have been discovered and described in aquatic environments around the world (e.g., Montresor et al., 1999; Moestrup et al., 2009a,b; Jang et al., 2017b; Sampedro et al., 2022). Suessiacean species have been known to take various life forms including typical free-living cells and parasitic and symbiotic cells, thereby playing a variety of complex ecological roles in aquatic ecosystems (Levy et al., 2007; Siano et al., 2010; Gómez, 2012; Jang et al., 2017a). Although they all belong to the same phylogenetic clade, suessiacean species are found in marine and freshwater ecosystems at all latitudes because of their highly diversified ecophysiological characteristics (Montresor et al., 1999; Takahashi et al., 2015; Sampedro et al., 2022). There have been several records of blooms that were caused by members of this family and/or presumably suessiacean species based on morphological observations (e.g., Stoecker et al., 2000; Moestrup et al., 2009a; Moreira-González et al., 2021). However, understanding of these phenomena has been difficult due to the small size of the culprits (i.e., generally 7–15 μm in length) and the lack of effective species identification methods (Kang et al., 2019).
With the advent of both electron microscopy and genetic validation methods in recent decades, a sophisticated taxonomic classification and identification system has been established for the members of this family (Moestrup et al., 2009a). Moreover, with the growing popularity of molecular techniques (e.g., DNA metabarcoding, which allows simultaneous identification of many taxa within the environmental samples through DNA barcoding), the prevalence and dominance of suessiacean dinoflagellates within diverse protistan assemblages is gradually being revealed (Smith et al., 2017; Kellogg et al., 2019; Liu et al., 2020; Salmaso et al., 2020). The number of published environmental sequencing datasets has rapidly increased in recent years. In the past, such datasets existed primarily in an unprocessed state and thus were not easily accessible. However, the recent creation of metaPR2, a platform containing large, standardized datasets of 18S environmental rRNA sequences for protist groups, has dramatically improved data accessibility (Vaulot et al., 2022). This platform combines samples obtained from small-scale expeditions and standardizes methodological differences (e.g., bioinformatic workflows), thereby enabling meaningful data comparison on a global scale.
The objective of the current study was to provide insights into the hidden biodiversity, global distribution, and putative ecological niches of suessiacean dinoflagellates using a DNA metabarcoding dataset. The obtained results contributed to the better understanding of the ecological importance of the suessiacean group and further reveal the ecological niches these organisms occupy within aquatic ecosystems worldwide.
Materials and methods
Defining taxonomic groups within suessiacean dinoflagellates
Nine genera (Ansanella, Asulcocephalium, Biecheleria, Biecheleriopsis, Leiocephalium, Pelagodinium, Piscinoodinium, Polarella, and Yihiella) have so far been assigned to the family Suessiaceae based on both morphology and molecular phylogeny (Table 1). In this study, all amplicon sequence variants (ASVs)—which are single DNA sequences recovered from a high-throughput marker-gene data analysis (Callahan et al., 2017)—belonging to the family Suessiaceae were initially categorized at the genus level before further analyses were conducted. This is because many suessiacean species are belong to monospecific genera. Moreover, in some genera within the Suessiaceae group, the 18S rRNA gene does not have sufficient resolution (Jang et al., 2017b; Raho et al., 2018; Jang et al., 2022). Due to the above, the data were classified as “Ansanella, Asulcocephalium, Biecheleria, Biecheleriopsis, Leiocephalium, Pelagodinium, Piscinoodinium, Polarella, and Yihiella,” and sequences that did not match these genera but belonged to the Suessiaceae family were categorized as “Unidentified.” The ASVs that had initially been incorrectly named “Protodinium,” were integrated into the “Biecheleriopsis” category according to the current taxonomic classification system (Takahashi et al., 2014; Jang et al., 2015). To date, no 18S gene sequence has been reported for true Protodinium (Lohmann) Kofoid & Swezy, although its ultrastructure had been investigated by Dodge (1974). Nevertheless, phylogenetic analysis was performed on all ASVs belonging to the “Protodinium” category, thereby confirming that all the ASVs belonged to the Biecheleriopsis clade (Supplementary Figure S1).
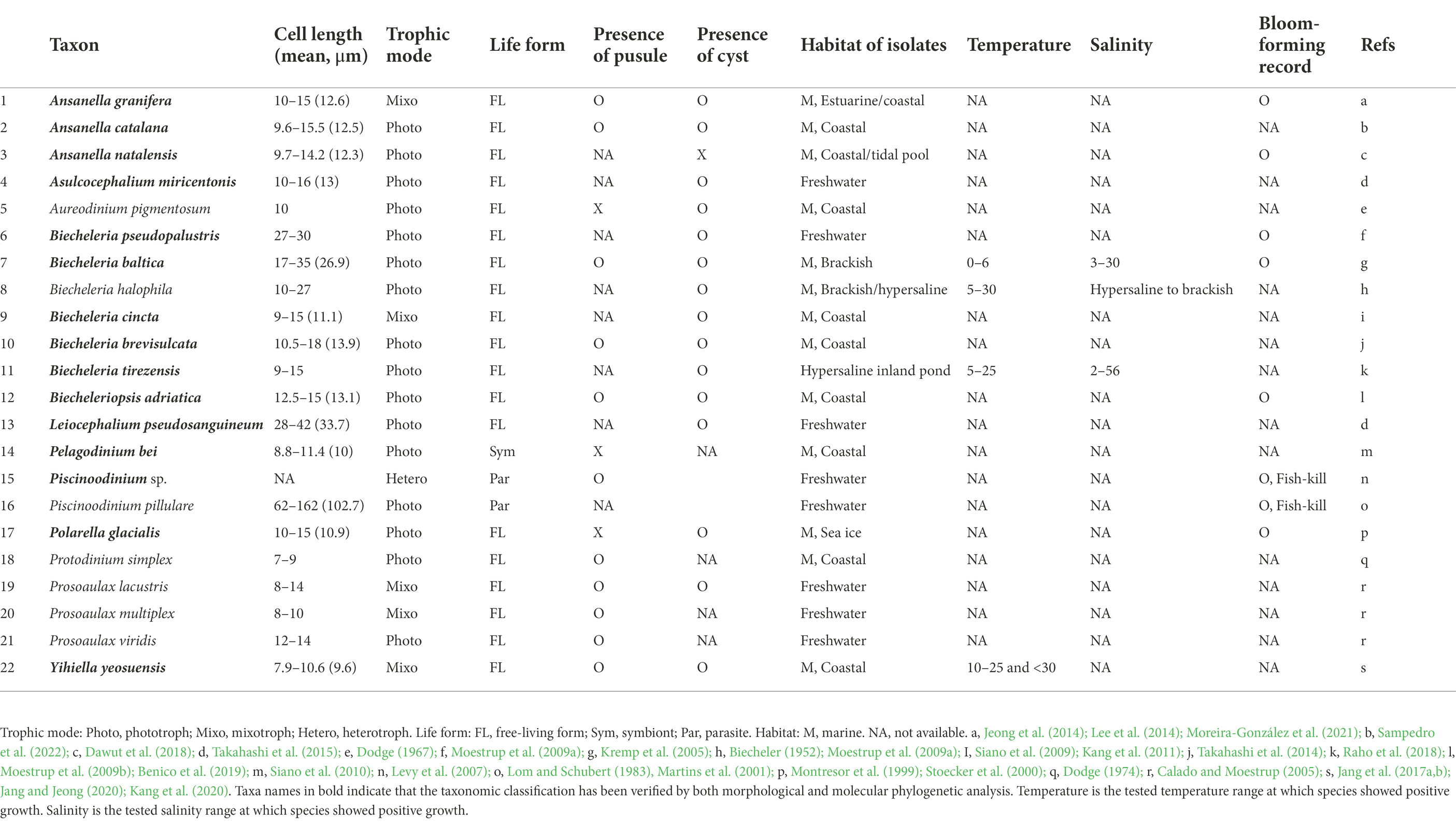
Table 1. Comparison of the taxonomic and ecophysiological characteristics and records of extant species presumably belonging the family Suessiaceae.
Dataset selection and processing
All of the Dinophyceae metabarcoding datasets gathered from worldwide sampling projects were obtained from the newly created metaPR2 v.1.0.3 database,1 which is a database of 18S rRNA metabarcodes that have been reprocessed with a pipeline based on the dada2 R package (Callahan et al., 2016). Taxonomic classifications were assigned using the Protist Ribosomal Reference database (PR2; Guillou et al., 2012). The detailed bioinformatic workflow is described by Vaulot et al. (2022).
For the current analysis, 38 datasets including 3,284 samples collected from oceanic, coastal, and freshwater lakes and rivers (terrestrial ecosystems were excluded) were investigated out of 41 datasets available from the metaPR2 v.1.0.3 database. The data analyzed in this study are summarized in Supplementary Table S1. Both the V4 and V9 regions of the 18S ribosomal sequences containing at least 100 total reads per ASV were selected for the analysis (Supplementary Figure S2). The total read number per sample was normalized to 100 so that the value displayed in the different panels corresponded to the percentage of total protist reads (i.e., the number of reads for each ASV in a given sample was divided by the total number of reads in the sample and then multiplied by 100). The sample types included in the analysis were water, ice, epibiota, and sediment, and the data were retrieved without a specific screen for cell size fractions. Taxa selection was performed according to the classification criterion of the metaPR2 database: “Alveolata” for Supergroup, “Dinoflagellata” for Division, “Dinophyceae” for Class, “Suessiales” for Order, and “Suessiaceae” for Family. The physical and chemical environmental conditions during sampling and other available metadata associated with samples were obtained from the metaPR2 website and are summarized in Supplementary Table S2. For further statistical verification, the read distributions of different genera associated with various factors (i.e., water temperature, salinity, latitude, and depth) were compared using Chi-squared homogeneity tests of contingency tables, which use observed vs. expected values (generated according to standard formulas) to calculate the Chi-squared statistics (Sokal and Rohlf, 1987).
Multiple sequence alignment and phylogenetic analysis of 18S rRNA genes
18S rRNA sequences belonging to the family Suessiaceae were downloaded from the PR2 database to construct a phylogenetic tree. The database includes sequences of strains in which taxonomic identification was verified along with numerous environmental sequences. Only sequences of the 18S rRNA region that were longer than 800 bp were selected from the database, while sequences from new species that had not been registered in the database (e.g., Ansanella natalensis and A. catalana) were added manually. Additionally, five sequences belonging to a different family but within order Suessiales were selected as an outgroup. A total of 234 sequences were aligned using the ClustalW algorithm implemented in MEGA v.11 (Tamura et al., 2021), of which 146 were uncultured environmental sequences while the other 88 were from cultured strains. The final alignment contained 1,680 aligned nucleotide positions of 18S rRNA gene.
The aligned dataset was used to construct a phylogenetic tree via the maximum-likelihood (ML) statistical method. The dataset was tested for the best-fitting substitution model in MEGA v.11 (Tamura et al., 2021), with the model indicated by the Akaike Information Criterion (AIC) as the best-fitting one (the TN93 + G model) being chosen (Tamura et al., 2021). The tree was visualized and edited using iTOL v.6 (Letunic and Bork, 2021).
Results and discussion
Taxonomic composition and relative abundance based on metabarcoding data
Most of the ASVs were taxonomically assigned to one of the 41 taxa at the family level, with the remaining sequences assigned to the “Unidentified” category (Figure 1A). The family Gymnodiniaceae contained the highest read abundances in the Dinophyceae metabarcoding dataset, which was consistent with previous findings, as Gymnodiniaceae is one of the most common families worldwide, both in terms of frequency of occurrence and abundance (Kudela and Gobler, 2012; Thessen et al., 2012; Kodama et al., 2021). Of the 42 family groups of dinoflagellates, Suessiaceae was the sixth highest in terms of read abundance and the ninth highest in terms of ASV richness (Supplementary Table S3). The amount of DNA in dinoflagellates is known to be directly correlated with cell size (LaJeunesse et al., 2005; Murray et al., 2016; Liu et al., 2021). Although suessiacean species are among the smallest cells in the dinoflagellate group, the high read abundances observed in this study suggest that their abundance in nature may have been heavily underestimated.

Figure 1. Treemaps displaying taxonomic composition and relative abundance of dinoflagellates based on 18S rRNA gene metabarcoding at (A) the family level and (B) the genus level within the family Suessiaceae. The left side represents the number of reads while the right side is the number of amplicon sequence variants (ASVs). The taxon Suessiaceae in (A) is marked with a blue box. Detailed data on the taxa and values used in this analysis are separately provided in Supplementary Table S3.
In the metabarcoding datasets of the family Suessiaceae, the highest read abundances were found for Biecheleria and Biecheleriopsis (=Protodinium; Figure 1B). Although relatively recently Moestrup et al. (2009a,b) re-established the two genera based on both ultrastructure and phylogenetic relationships, some species in those genera have been reported as Gymnodinium for approximately half a century, with several records of blooms (Biecheler, 1952; Moestrup et al., 2009a).
When compared with the high read abundance in the genus Polarella, ASV richness was surprisingly low, suggesting that Polarella might have evolved to dominate certain environments instead of increasing species diversity to survive in multiple environments (Figure 1B). In contrast, the genus Pelagodinium had a higher ASV richness and lower read abundance, which may have resulted from symbiosis and diverse genetic pools being created through relationships with its host organisms (Spero, 1987; Siano et al., 2010). Additionally, the large number of both reads and ASVs in the “Unidentified” category suggests that the biodiversity of the family Suessiaceae remains largely unexplored. A further analysis of 20 ASVs belonging to the “Unidentified” category showed that only two of them contributed greatly to the total read abundance those ASVs belonged to phylogenetically unknown groups (Supplementary Figure S3).
Global distribution and habitat
Members of the family Suessiaceae have been identified in both marine and freshwater environments (Table 1). According to the metabarcoding analysis results, reads of all genera in Suessiaceae were found in marine environmental samples, whereas the majority of reads collected in freshwater ecosystems were limited to Asulcocephalium, Biecheleria, and Biecheleriopsis (Figures 2A,B). The two monospecific genera Asulcocephalium and Leiocephalium were established based on isolates from freshwater samples, while the genus Biecheleria has been reported to contain both marine and freshwater species (Moestrup et al., 2009a; Takahashi et al., 2015). However, the discovery of large numbers of reads belonging to Biecheleriopsis in freshwater samples suggests a hidden ecophysiological diversity of the species within this genus.
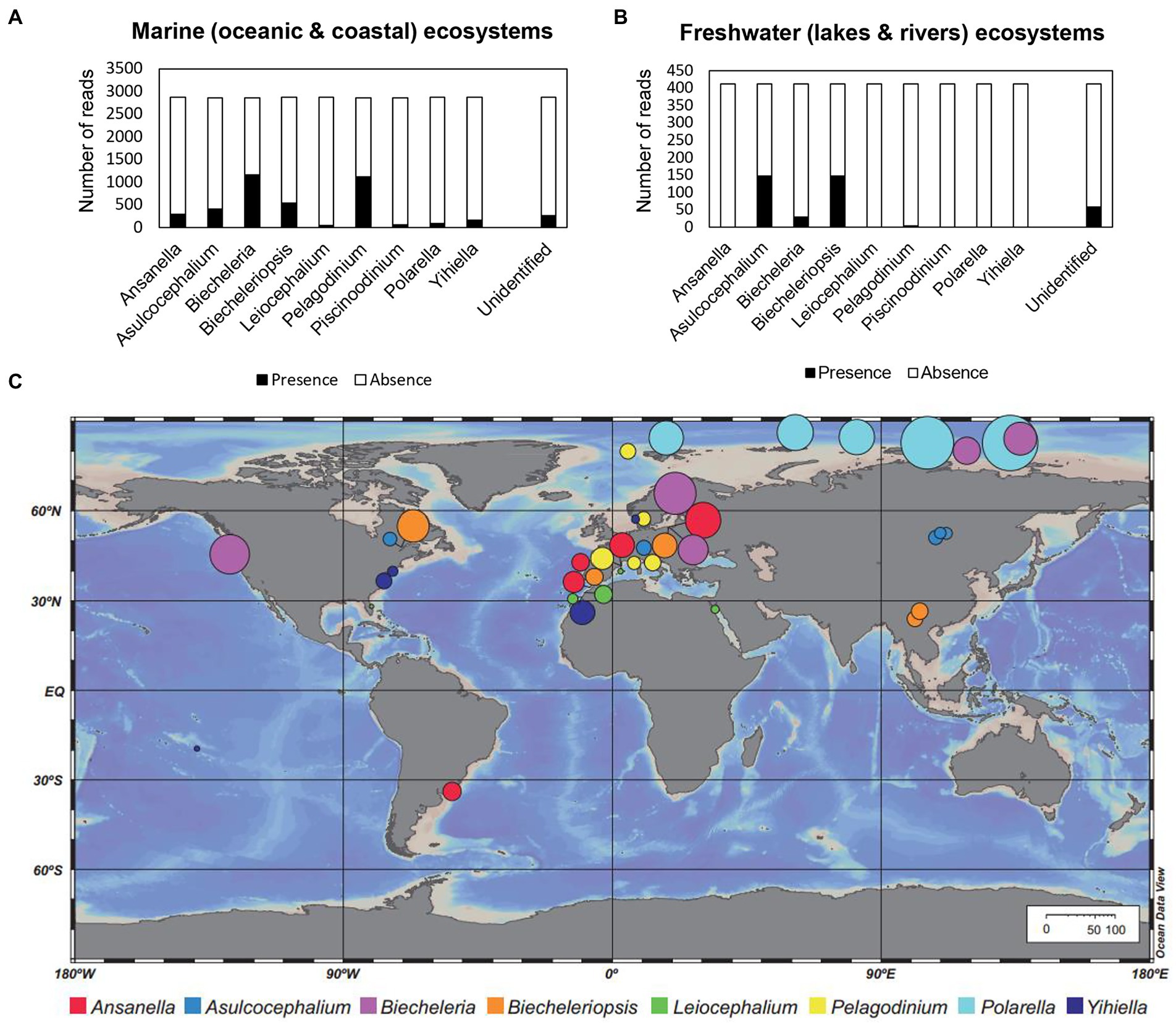
Figure 2. Number of reads in the metaPR2 dataset annotated to each genus (family Suessiaceae) among the samples collected from (A) marine (oceanic and coastal) and (B) freshwater (lakes and rivers) ecosystems. (C) Distribution map showing the five samples with the highest abundance of each genus among the surveyed samples. The diameter of each circle (based on the scale bars) represents the percentage (%) of reads belonging to each genus within the entire protist community.
A few dinoflagellate groups that have colonised freshwater habitats play a key role in understanding the evolution of microbial lineages (Logares et al., 2007). Despite the overall lack of knowledge regarding the marine-freshwater transition, scientists have provided phylogenetic data supporting these relationships (Logares et al., 2007; Annenkova et al., 2015). Salinity is a key factor in determining the distribution of the marine-freshwater boundary. Thus, the fact that many suessiacean species possess osmoregulatory organelles (e.g., pusular systems) may explain the frequent marine-freshwater transitions among lineages within this family (Table 1; Dodge, 1972). It is also worth noting that most suessiacean species have a known cyst stage, which may further facilitate the evolutionary transition.
The five samples with the highest abundance of the Polarella genus were all taken from the Arctic region (Figure 2C). Specifically, the reads of Polarella accounted for 25–65% of the total eukaryotic reads in these five samples, indicating that this genus may be a common bloom-forming taxon in polar regions. Unlike Polarella, the genus Biecheleria tends to predominate in a wide range of aquatic environments without clear environmental restrictions. Biecheleria baltica is known to live in cold waters, whereas other Biecheleria species are mainly found in temperate waters (Kremp et al., 2005; Siano et al., 2009; Takahashi et al., 2014; Raho et al., 2018). For example, most isolates of Biecheleria cincta were collected from temperate waters (Siano et al., 2009; Kang et al., 2011; Luo et al., 2013). However, the morphology of a cold-water dwelling strain of B. cincta, RCC2013, perfectly matched that of the type strains of other temperate strains, while its 18S and 28S rRNA sequences were identical to those of its warmer water counterparts (Siano et al., 2009; Balzano et al., 2012). On the other hand, the highly variable internal transcribed spacer (ITS) region exhibited a faster rate of evolution among strains of B. cincta, suggesting that this species has evolved ability to rapidly adapt to new environments (Luo et al., 2013).
Ecophysiological diversity and differentiated ecological niches
This study investigated the relative read abundance of each genus within the family Suessiaceae in relation to water temperature and salinity (Figures 3A,B). Similar to the temperature dependence of the growth rates of common dinoflagellate species (Paerl, 2014), for many genera, high proportions of read counts were distributed between water temperatures of 15 to 30°C (Figure 3A). However, reads attributed to the genus Leiocephalium generally occupied high-temperature intervals, with a significant proportion of the reads occupying the 30–35°C interval (Chi-square homogeneity test, p < 0.01). This suggests that the taxon was likely a warm-water species. The read distribution of Polarella indicated that the members of this genus were cold-water species, which is consistent with the observed field data (Stoecker et al., 2000). Moreover, given that Yihiella showed positive growth rates at 10–30°C in lab-based experiments (Kang et al., 2020), the fact that more than 97% of Yihiella reads were distributed precisely within a 10–30°C temperature interval demonstrates that combining metabarcoding data sets provides a promising means of understanding taxon-specific ecophysiology. When the reads of each genus were classified by salinity, considerable proportions of the reads attributed to the genera Asulcocephalium, Biecheleriopsis, and Polarella were observed at a 0–5 psu interval, including freshwater samples (Chi-square homogeneity test, p < 0.01; Figure 3B). Polarella is commonly distributed in and around ice samples, and its cyst stage might improve osmotic resistance. Considering the above, it is proposed that metabarcoding analyses may provide novel insights into the ecophysiological diversity of genera whose distributions have previously been recorded exclusively in either freshwater or marine environments. For example, through the current analysis it was possible to detect the probable existence of marine-dwelling species in the genera Asulcocephalium and Leiocephalium and freshwater species in the genus Biecheleriopsis.
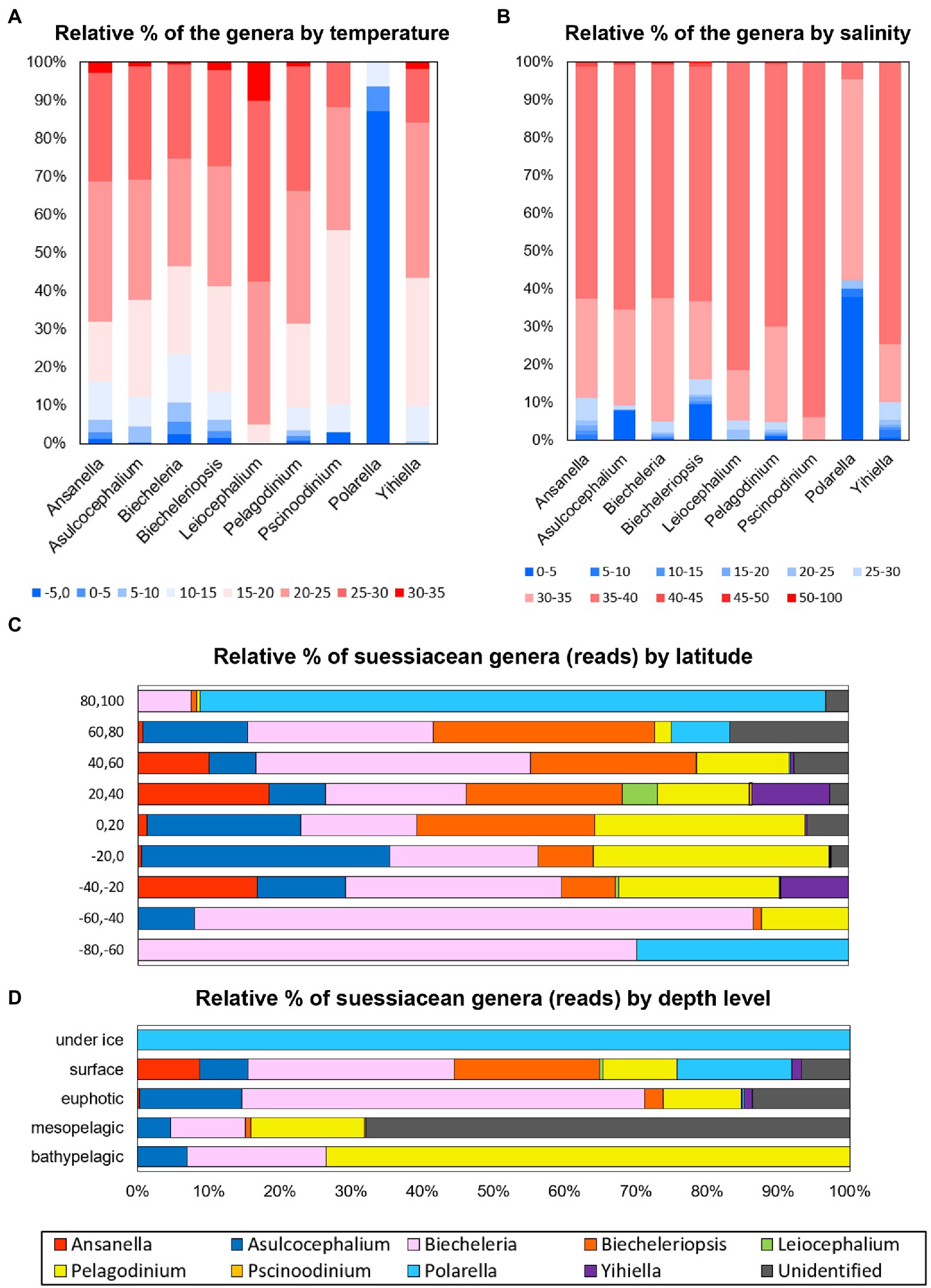
Figure 3. Relative read abundance of each genus (family Suessiaceae) according to (A) water temperature and (B) salinity of the surveyed samples. The color scheme represents the water temperature (-5–35°C) and salinity (0–100 psu). Relative read abundance of each genus classified by (C) horizontal (i.e., latitude) and (D) vertical (i.e., water depth) criteria.
To understand the ecological niches of suessiacean species in the context of their known ecophysiological characteristics, the distribution of each genus within its natural environment was classified by horizontal and vertical criteria (i.e., latitude and water depth; Figures 3C,D). The read distributions of genera in accordance with latitude and water depth were significantly different, as supported by the Chi-square test for homogeneity (both p < 0.01). Polarella and Biecheleria comprised most of the suessiacean species in Arctic and Antarctic waters (Figure 3C). Unlike other genera, the read abundances of Ansanella and Yihiella were low in tropical waters. The ecological niches of the suessiacean genera according to water depth showed a more pronounced difference (Figure 3D). The reads of genus Pelagodinium and “Unidentified” category dominated the deep-water layers (i.e., mesopelagic and bathypelagic). The vertical distribution of Pelagodinium, which is only 10 μm in long, could reach the deep-water layer because these microorganisms live as symbionts in several hosts such as foraminifera (Siano et al., 2010). Moreover, the high number of “Unidentified” reads in the mesopelagic zone suggests that this environment may be inhabited by a yet unexplored diversity of Suessiaceae. Two ASVs, 536fcef820 and caffe54124, accounted for a high proportion of the “Unidentified” category in the mesopelagic group (236–562 m) collected from various regions in the Arctic and Antarctic (Supplementary Table S4). Information about their taxonomy could be obtained through phylogenetic study (See next section for details).
Biodiversity estimations based on phylogenetic analysis
A phylogenetic analysis based on the 18S rRNA sequences obtained from the PR2 database was performed to estimate the phylogenetic biodiversity of the family Suessiaceae (Figure 4). Although there were slight variations in the phylogenetic position depending on the method the trees were analyzed, at least five new phylogenetic groups and three phylogenetically novel sequences were commonly identified among the analyzed phylogenetic trees (Figure 4; Supplementary Figures S3B, S4). The 18S rRNA sequences belonging to Group A differed by up to 4% from those of the Asulcocephalium genus. Considering the level of differences in the 18S rRNA sequence among different genera in family Suessiaceae (i.e., 1.2–5.3%), this group is now believed to be a new genus (Jeong et al., 2014; Jang et al., 2017b). The environmental sequences belonging to Unknown Group B differed by approximately 1–3% from those of Asulcocephalium miricentonis, suggesting that these sequences may belong to new species within the genus. The sequences in this group were globally distributed, being found in lakes in Luxembourg and France, as well as in the Columbia River estuary in the United States (Lefranc et al., 2005; Kahn et al., 2014). Group C contained a variety of sub-clades with sequence differences ranging from 2 to 4% relative to other known species, suggesting that this group have a high species diversity. The sequences belonging to Group D were collected from colder regions such as the North Pole and Sweden (Bachy et al., 2011; Majaneva et al., 2012), and the members of this clade are therefore expected to share similar ecophysiological features with Polarella glacialis. However, in addition to a 1–2% sequence difference between the two groups, morphological analyses are necessary in order to confirm that Group D represents a new species. Moreover, the sequences in Group E are presumed to be those of freshwater species, as they originated from high mountain lakes (Triadó-Margarit and Casamayor, 2012). However, this clade did not appear to belong to any other genus based on its phylogenetic position. In addition, three individual sequences (GQ483664, HQ438122, and EU087256) that do not form clades but diverged from the near groups could also represent new species. Finally, although the resolution of the 18S rRNA gene for intrageneric classification of the genus Biecheleria was too low, the high number of environmental sequences obtained from various aquatic waters suggests that the genus Biecheleria may have diverse ecological roles in global aquatic ecosystems.
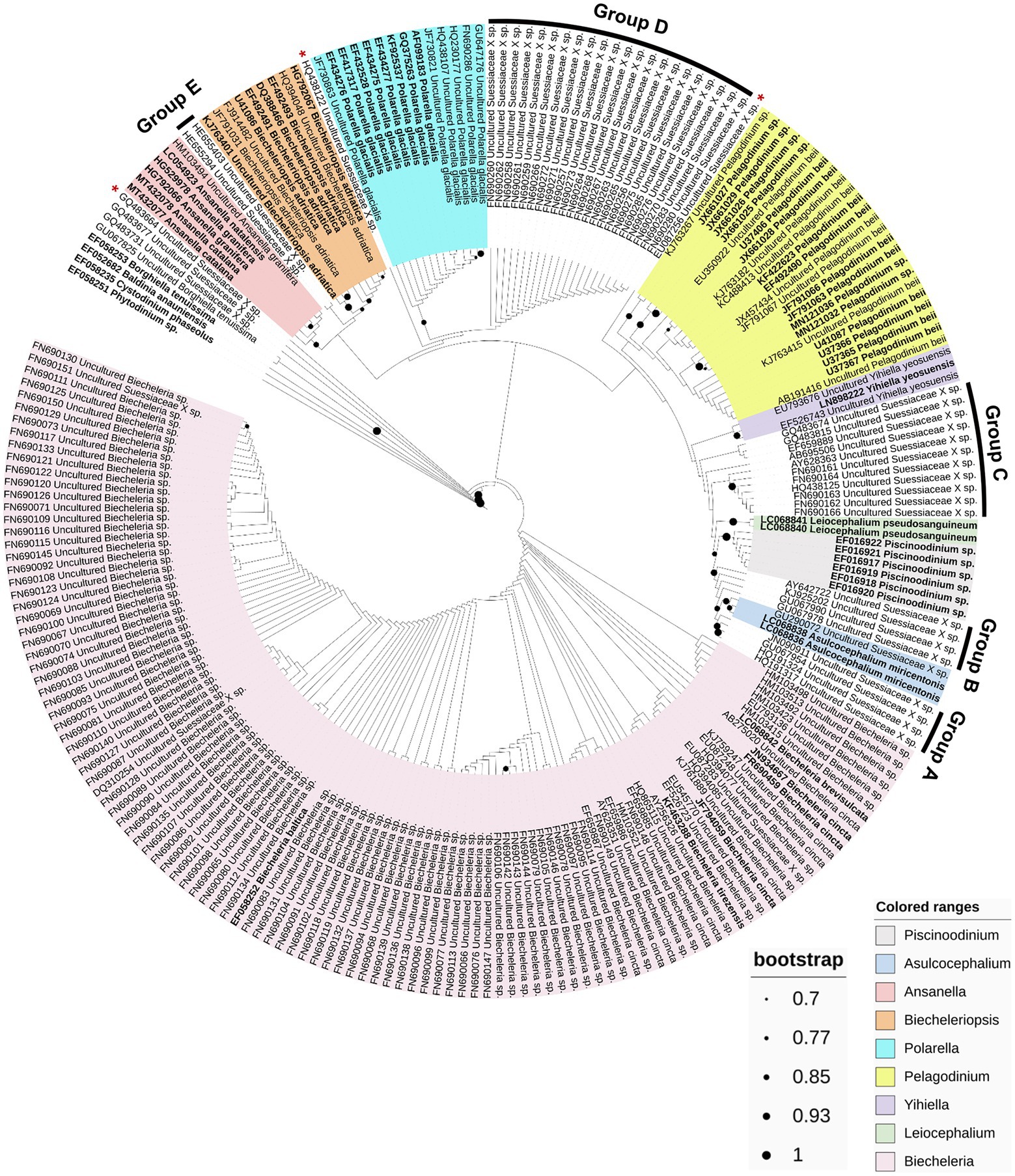
Figure 4. Maximum-likelihood phylogenetic tree based on 234 partial 18S rRNA gene sequences within the family Suessiaceae. The potential new taxonomic groups and sole sequences are denoted as A–E, and red asterisks (*), respectively. The strains sequenced based on the isolates are indicated in bold, and the others are environmental sequences. The colored ranges represent each genus. Bootstrap support values over 70% (0.7) are shown as dots on the interior nodes.
Metabarcoding approach for suessiacean dinoflagellate species
Because the average cell length of species in the family Suessiaceae is only 14.5 μm (except for Piscinoodinium species, which vary in length depending on their life cycle; Table 1), metabarcoding approach is essential for distribution and ecophysiological studies of these small species in the field. By using the recent and well-established taxonomic classification system of suessiacean dinoflagellates, the metabarcoding data is beginning to reveal the ecology and diversity of individual species. As an example, two environmental sequences (JQ516748 and EF526743) with high homologies to the genus Yihiella were found in the coral microbiome and anoxic water in fjords, respectively, allowing assumption of the potential ecological niches of Yihiella-like species (Behnke et al., 2010; Kimes et al., 2013). In addition, the metabarcoding analysis is also expected to provide more information about blooms caused by small suessiacean species. The results of this study have also confirmed that the metabarcoding approach could be used as a tool to provide new information to other multidisciplinary studies of the family Suessiaceae. For access to a variety of species, however, genetic information about species that have not yet been barcoded, such as Aureodinium and Prosoaulax, should be revealed in the future.
Conclusion
This study is the first to reveal the global species diversity, distribution, and ecological niches of the family Suessiaceae in aquatic ecosystems through the analysis of a large metabarcoding dataset. Out of the 42 family groups of dinoflagellates, Suessiaceae were the sixth in terms of read abundance and the ninth in terms of ASV richness, suggesting that their abundance in the natural environment has been significantly underestimated. Furthermore, at least five new phylogenetic groups and three individual novel sequences within the family were discovered based on 18S rRNA environmental sequences. These sequences are likely to indicate the presence of new species. Metabarcodes and associated environmental information from the surveyed samples provided novel insights into the ecophysiological characteristics of individual taxa, and even their putative ecological niches. The findings of this study highlight the importance of suessiacean dinoflagellates in aquatic ecosystems worldwide and demonstrate that big data obtained from environmental DNA constitute a promising tool for characterizing the ecological functions of understudied species.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT; nos. 2022R1C1C1004755 and 2022M3I6A1085991 awarded to SJ).
Acknowledgments
Many thanks to Daniel Vaulot and collaborators for developing the publicly available database metaPR2, as well as to Wordvice (https://wordvice.com) for English language editing. Two reviewers and editor made valuable comments that improved an earlier draft of this manuscript.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1010854/full#supplementary-material
Footnotes
References
Annenkova, N. V., Hansen, G., Moestrup, Ø., and Rengefors, K. (2015). Recent radiation in a marine and freshwater dinoflagellate species flock. The ISME J. 9, 1821–1834. doi: 10.1038/ismej.2014.267
Bachy, C., López-García, P., Vereshchaka, A., and Moreira, D. (2011). Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the north pole at the end of the polar night. Front. Microbiol. 2:106. doi: 10.3389/fmicb.2011.00106
Balzano, S., Gourvil, P., Siano, R., Chanoine, M., Marie, D., Lessard, S., et al. (2012). Diversity of cultured photosynthetic flagellates in the Northeast Pacific and Arctic oceans in summer. Biogeosciences 9, 4553–4571. doi: 10.5194/bg-9-4553-2012
Behnke, A., Barger, K. J., Bunge, J., and Stoeck, T. (2010). Spatio-temporal variations in protistan communities along an O2/H2S gradient in the anoxic Framvaren Fjord (Norway). FEMS Microbiol. Ecol. 72, 89–102. doi: 10.1111/j.1574-6941.2010.00836.x
Benico, G. A., Takahashi, K., Lum, W. M., Yñiguez, A. T., Azanza, R. V., Leong, S. C. Y., et al. (2019). First report of Biecheleriopsis adriatica in Bolinao, northwestern Philippines and its wide distribution in Southeast Asia and adjacent waters. Philipp. J. Nat. Sci. 24, 34–41.
Calado, A. J., and Moestrup, Ø. (2005). On the freshwater dinoflagellates presently included in the genus Amphidinium, with a description of Prosoaulax gen. Nov. Phycologia 44, 112–119. doi: 10.2216/0031-8884(2005)44[112,OTFDPI]2.0.CO;2
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. The ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/NMETH.3869
Coats, D. W. (1999). Parasitic life styles of marine dinoflagellates. J. Eukaryot. Microbiol. 46, 402–409. doi: 10.1111/j.1550-7408.1999.tb04620.x
Dawut, M., Sym, S. D., and Horiguchi, T. (2018). Re-investigation of Gymnodinium natalense (Dinophyceae), a tidal pool dinoflagellate from South Africa and the proposal of a new combination Ansanella natalensis. Phycol. Res. 66, 300–309. doi: 10.1111/pre.12329
Dodge, J. D. (1967). Fine structure of the dinoflagellate Aureodinium pigmentosum gen. et sp. nov. Br. Phycol. Bul. 3, 327–336. doi: 10.1080/00071616700650211
Dodge, J. D. (1972). The ultrastructure of the dinoflagellate pusule: a unique osmo-regulatory organelle. Protoplasma 75, 285–302. doi: 10.1007/BF01279820
Dodge, J. D. (1974). A redescription of the dinoflagellate Gymnodinium simplex with the aid of electron microscopy. J. Mar. Biol. Assoc. U. K. 54, 171–177. doi: 10.1017/S0025315400022141
Fensome, R. A., Taylor, F. J. R., Norris, G., Sarjeant, W. A. S., Wharton, D. I., and William, G. L. (1993). A Classification of Living and Fossil Dinoflagellates: Micropaleontology. Hanover, PA: Sheridan Press.
Gómez, F. (2012). A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata). Syst. Biodivers. 10, 267–275. doi: 10.1080/14772000.2012.721021
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., et al. (2012). The Protist ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Guiry, M. D., and Guiry, G. M. (2022). AlgaeBase. Available at: https://www.algaebase.org (Accessed December 02, 2022).
Jang, S. H., and Jeong, H. J. (2020). Spatio-temporal distributions of the newly described mixotrophic dinoflagellate Yihiella yeosuensis (Suessiaceae) in Korean coastal waters and its grazing impact on prey populations. Algae 35, 45–59. doi: 10.4490/algae.2020.35.2.24
Jang, S. H., Jeong, H. J., Kwon, J. E., and Lee, K. H. (2017a). Mixotrophy in the newly described dinoflagellate Yihiella yeosuensis: a small, fast dinoflagellate predator that grows mixotrophically, but not autotrophically. Harmful Algae 62, 94–103. doi: 10.1016/j.hal.2016.12.007
Jang, S. H., Jeong, H. J., Moestrup, Ø., Kang, N. S., Lee, S. Y., Lee, K. H., et al. (2015). Morphological, molecular and ecophysiological characterization of the phototrophic dinoflagellate Biecheleriopsis adriatica from Korean coastal waters. Eur. J. Phycol. 50, 301–317. doi: 10.1080/09670262.2015.1054892
Jang, S. H., Jeong, H. J., Moestrup, Ø., Kang, N. S., Lee, S. Y., Lee, K. H., et al. (2017b). Yihiella yeosuensis gen. et sp. nov. (Suessiaceae, Dinophyceae), a novel dinoflagellate isolated from the coastal waters of Korea. J. Phycol. 53, 131–145. doi: 10.1111/jpy.12486
Jang, S. H., Na, S. I., Lee, M. J., and Yoo, Y. D. (2022). Assessing the utility of mitochondrial gene markers in the family Suessiaceae (Dinophyta) with phylogenomic validation. Mol. Phylogenet. Evol. 177:107625. doi: 10.1016/j.ympev.2022.107625
Jeong, H. J., Jang, S. H., Moestrup, O., Kang, N. S., Lee, S. Y., Potvin, E., et al. (2014). Ansanella granifera gen. et sp. nov. (Dinophyceae), a new dinoflagellate from the coastal waters of Korea. Algae 29, 75–99. doi: 10.4490/algae.2014.29.2.075
Jeong, H. J., Yoo, Y. D., Kim, J. S., Seong, K. A., Kang, N. S., and Kim, T. H. (2010). Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci. J. 45, 65–91. doi: 10.1007/s12601-010-0007-2
Kahn, P., Herfort, L., Peterson, T. D., and Zuber, P. (2014). Discovery of a Katablepharis sp. in the Columbia River estuary that is abundant during the spring and bears a unique large ribosomal subunit sequence element. Microbiol. Open 3, 764–776. doi: 10.1002/mbo3.206
Kang, H. C., Jeong, H. J., Lim, A. S., Ok, J. H., You, J. H., Park, S. A., et al. (2020). Effects of temperature on the growth and ingestion rates of the newly described mixotrophic dinoflagellate Yihiella yeosuensis and its two optimal prey species. Algae 35, 263–275. doi: 10.4490/algae.2020.35.8.20
Kang, H. C., Jeong, H. J., Ok, J. H., You, J. H., Jang, S. H., Lee, S. Y., et al. (2019). Spatial and seasonal distributions of the phototrophic dinoflagellate Biecheleriopsis adriatica (Suessiaceae) in Korea: quantification using qPCR. Algae 34, 111–126. doi: 10.4490/algae.2019.34.5.25
Kang, N. S., Jeong, H. J., Yoo, Y. D., Yoon, E. Y., Lee, K. H., Lee, K., et al. (2011). Mixotrophy in the newly described phototrophic dinoflagellate Woloszynskia cincta from western Korean waters: feeding mechanism, prey species and effect of prey concentration. J. Eukaryot. Microbiol. 58, 152–170. doi: 10.1111/j.1550-7408.2011.00531.x
Kellogg, C. T., McClelland, J. W., Dunton, K. H., and Crump, B. C. (2019). Strong seasonality in arctic estuarine microbial food webs. Front. Microbiol. 10:2628. doi: 10.3389/fmicb.2
Kimes, N. E., Johnson, W. R., Torralba, M., Nelson, K. E., Weil, E., and Morris, P. J. (2013). The Montastraea faveolata microbiome: ecological and temporal influences on a Caribbean reef-building coral in decline. Environ. Microbiol. 15, 2082–2094. doi: 10.1111/1462-2920.12130
Kodama, T., Watanabe, T., Taniuchi, Y., Kuwata, A., and Hasegawa, D. (2021). Micro-size plankton abundance and assemblages in the western North Pacific subtropical gyre under microscopic observation. PLoS One 16:e0250604. doi: 10.1371/journal.pone.0250604
Kremp, A., Elbrachter, M., Schweikert, M., Wolny, J. L., and Gottschling, M. (2005). Woloszynskia Halophila (Biecheler) comb. Nov.: a bloom-forming cold-water Dinoflagellate co-occurring with Scrippsiella hangoei (Dinophyceae) in the Baltic Sea. J. Phycol. 41, 629–642. doi: 10.1111/j.1529-8817.2005.00070.x
Kudela, R. M., and Gobler, C. J. (2012). Harmful dinoflagellate blooms caused by Cochlodinium sp.: global expansion and ecological strategies facilitating bloom formation. Harmful Algae 14, 71–86. doi: 10.1016/j.hal.2011.10.015
LaJeunesse, T. C., Lambert, G., Andersen, R. A., Coffroth, M. A., and Galbraith, D. W. (2005). Symbiodinium (pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 41, 880–886. doi: 10.1111/j.0022-3646.2005.04231.x
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e6. doi: 10.1016/j.cub.2018.07.008
Lee, S. K., Jeong, H. J., Jang, S. H., Lee, K. H., Kang, N. S., Lee, M. J., et al. (2014). Mixotrophy in the newly described dinoflagellate Ansanella granifera: feeding mechanism, prey species, and effect of prey concentration. Algae 29, 137–152. doi: 10.4490/algae.2014.29.2.137
Lefranc, M., Thénot, A., Lepere, C., and Debroas, D. (2005). Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71, 5935–5942. doi: 10.1128/AEM.71.10.5935-5942.2005
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Levy, M. G., Litaker, R. W., Goldstein, R. J., Dykstra, M. J., Vandersea, M. W., and Noga, E. J. (2007). Piscinoodinium, a fish-ectoparasitic dinoflagellate, is a member of the class Dinophyceae, subclass Gymnodiniphycidae: convergent evolution with Amyloodinium. J. Parasitol. 93, 1006–1015. doi: 10.1645/GE-3585.1
Liu, Y., Hu, Z., Deng, Y., Shang, L., Gobler, C. J., and Tang, Y. Z. (2021). Dependence of genome size and copy number of rRNA gene on cell volume in dinoflagellates. Harmful Algae 109:102108. doi: 10.1016/j.hal.2021.102108
Liu, L., Wang, Z., and Lu, S. (2020). Diversity and geographical distribution of resting stages of eukaryotic algae in the surface sediments from the southern Chinese coastline based on metabarcoding partial 18S rDNA sequences. Mar. Ecol. 41, 1–17. doi: 10.1111/maec.12585
Logares, R., Shalchian-Tabrizi, K., Boltovskoy, A., and Rengefors, K. (2007). Extensive dinoflagellate phylogenies indicate infrequent marine–freshwater transitions. Mol. Phylogenet. Evol. 45, 887–903. doi: 10.1016/j.ympev.2007.08.005
Lom, J., and Schubert, G. (1983). Ultrastructural study of Piscinoodinium pillulare (Schäperclaus, 1954) Lom, 1981 with special emphasis on its attachment to the fish host. J. Fish Dis. 6, 411–428. doi: 10.1111/j.1365-2761.1983.tb00096.x
Luo, Z., Yang, W., Xu, B., and Gu, H. (2013). First record of Biecheleria cincta (Dinophyceae) from Chinese coasts, with morphological and molecular characterization of the strains. Chin. J. Oceanol. Limnol. 31, 835–845. doi: 10.1007/s00343-013-2315-8
Majaneva, M., Rintala, J. M., Piisilä, M., Fewer, D. P., and Blomster, J. (2012). Comparison of wintertime eukaryotic community from sea ice and open water in the Baltic Sea, based on sequencing of the 18S rRNA gene. Polar Biol. 35, 875–889. doi: 10.1007/s00300-011-1132-9
Martins, M. L., Moraes, J. R. E., Andrade, P. M., Schalch, S. H. C., and Moraes, F. R. D. (2001). Piscinoodinium pillulare (Schäperclaus, 1954) Lom, 1981 (Dinoflagellida) infection in cultivated freshwater fish from the northeast region of São Paulo state, Brazil: parasitological and pathological aspects. Braz. J. Biol. 61, 639–644. doi: 10.1590/S1519-69842001000400013
Moestrup, Ø., Lindberg, K., and Daugbjerg, N. (2009a). Studies on woloszynskioid dinoflagellates IV: the genus Biecheleria gen. Nov. Phycol. Res. 57, 203–220. doi: 10.1111/j.1440-1835.2009.00540.x
Moestrup, Ø., Lindberg, K., and Daugbjerg, N. (2009b). Studies on woloszynskioid dinoflagellates V. ultrastructure of Biecheleriopsis gen. Nov., with description of Biecheleriopsis adriatica sp. nov. Phycol. Res. 57, 221–237. doi: 10.1111/j.1440-1835.2009.00541.x
Montresor, M., Procaccini, G., and Stoecker, D. K. (1999). Polarella glacialis, gen. Nov., sp. nov. (Dinophyceae): Suessiaceae are still alive! J. Phycol. 35, 186–197. doi: 10.1046/j.1529-8817.1999.3510186.x
Moreira-González, A. R., Alonso-Hernández, C. M., Arencibia-Carballo, G., Betanzos-Vega, A., Morton, S. L., and Richlen, M. L. (2021). First report of an Ansanella granifera bloom associated with eutrophication in cuban waters, caribbean region. Harmful Algae News 67, 10–11.
Murray, S. A., Suggett, D. J., Doblin, M. A., Kohli, G. S., Seymour, J. R., Fabris, M., et al. (2016). Unravelling the functional genetics of dinoflagellates: a review of approaches and opportunities. Perspect. Phycol. 3, 37–52. doi: 10.1127/pip/2016/0039
Paerl, H. W. (2014). Mitigating harmful cyanobacterial blooms in a human-and climatically-impacted world. Life 4, 988–1012. doi: 10.3390/life4040988
Raho, N., Fraga, S., Abad, J. P., and Marín, I. (2018). Biecheleria tirezensis sp. nov. (Dinophyceae, Suessiales), a new halotolerant dinoflagellate species isolated from the athalassohaline Tirez natural pond in Spain. Eur. J. Phycol. 53, 99–113. doi: 10.1080/09670262.2017.1386328
Salmaso, N., Boscaini, A., and Pindo, M. (2020). Unraveling the diversity of eukaryotic microplankton in a large and deep perialpine lake using a high throughput sequencing approach. Front. Microbiol. 11:789. doi: 10.3389/fmicb.2020.00789
Sampedro, N., Reñé, A., Matos, J., Fortuño, J. M., and Garcés, E. (2022). Detection of the widespread presence of the genus Ansanella along the Catalan coast (NW Mediterranean Sea) and the description of Ansanella catalana sp. nov. (Dinophyceae). Eur. J. Phycol. 57, 125–142. doi: 10.1080/09670262.2021.1914861
Siano, R., Kooistra, W. H., Montresor, M., and Zingone, A. (2009). Unarmoured and thin-walled dinoflagellates from the Gulf of Naples, with the description of Woloszynskia cincta sp. nov. (Dinophyceae, Suessiales). Phycologia 48, 44–65. doi: 10.2216/08-61.1
Siano, R., Montresor, M., Probert, I., Not, F., and de Vargas, C. (2010). Pelagodinium gen. Nov. and P. béii comb. nov., a dinoflagellate symbiont of planktonic foraminifera. Protist 161, 385–399. doi: 10.1016/j.protis.2010.01.002
Smith, K. F., Kohli, G. S., Murray, S. A., and Rhodes, L. L. (2017). Assessment of the metabarcoding approach for community analysis of benthic-epiphytic dinoflagellates using mock communities. N. Z. J. Mar. Freshwater Res. 51, 555–576. doi: 10.1080/00288330.2017.1298632
Spero, H. J. (1987). Symbiosis in the planktonic foraminifer, Orbulina universa, and the isolation of its symbiotic dinoflagellate, Gymnodinium béii sp. nov. J. Phycol. 23, 307–317. doi: 10.1111/j.1529-8817.1987.tb04139.x
Stoecker, D. K., Gustafson, D. E., Baier, C. T., and Black, M. M. (2000). Primary production in the upper sea ice. Aquat. Microb. Ecol. 21, 275–287. doi: 10.3354/ame021275
Suggett, D. J., Warner, M. E., and Leggat, W. (2017). Symbiotic dinoflagellate functional diversity mediates coral survival under ecological crisis. Trends Ecol. Evol. 32, 735–745. doi: 10.1016/j.tree.2017.07.013
Takahashi, K., Moestrup, Ø., Jordan, R. W., and Iwataki, M. (2015). Two new freshwater woloszynskioids Asulcocephalium miricentonis gen. et sp. nov. and Leiocephalium pseudosanguineum gen. et sp. nov. (Suessiaceae, Dinophyceae) lacking an apical furrow apparatus. Protist 166, 638–658. doi: 10.1016/j.protis.2015.10.003
Takahashi, K., Sarai, C., and Iwataki, M. (2014). Morphology of two marine woloszynskioid dinoflagellates, Biecheleria brevisulcata sp. nov. and Biecheleriopsis adriatica (Suessiaceae, Dinophyceae), from Japanese coasts. Phycologia 53, 52–65. doi: 10.2216/13-192.1
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thessen, A. E., Patterson, D. J., and Murray, S. A. (2012). The taxonomic significance of species that have only been observed once: the genus Gymnodinium (Dinoflagellata) as an example. PLoS One 7:e44015. doi: 10.1371/journal.pone.0044015
Triadó-Margarit, X., and Casamayor, E. O. (2012). Genetic diversity of planktonic eukaryotes in high mountain lakes (Central Pyrenees, Spain). Environ. Microbiol. 14, 2445–2456. doi: 10.1111/j.1462-2920.2012.02797.x
Keywords: 18S rRNA, amplicon sequencing, bloom, dinoflagellate, metagenomics, metaPR2, Suessiaceae, metabarcoding
Citation: Jang SH (2022) Assessment of biodiversity, global distribution, and putative ecological niches of suessiacean dinoflagellates by DNA metabarcoding. Front. Ecol. Evol. 10:1010854. doi: 10.3389/fevo.2022.1010854
Edited by:
Roksana Majewska, North-West University, South AfricaReviewed by:
Elianne Egge, Norwegian Institute for Water Research (NIVA), NorwayAlberto G. Sáez, Complutense University of Madrid, Spain
Copyright © 2022 Jang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Se Hyeon Jang, shjang@jnu.ac.kr
 Se Hyeon Jang
Se Hyeon Jang