- 1Department of Ecology and Evolutionary Biology, Cornell University, Ithaca, NY, United States
- 2Biodiversity Unit, Department of Biology, Lund University, Lund, Sweden
- 3Department of Ecology and Evolutionary Biology, University of California, Irvine, Irvine, CA, United States
- 4Rocky Mountain Biological Laboratory, Crested Butte, CO, United States
- 5Department of Neurobiology and Behavior, Cornell University, Ithaca, NY, United States
Research on floral volatiles has grown substantially in the last 20 years, which has generated insights into their diversity and prevalence. These studies have paved the way for new research that explores the evolutionary origins and ecological consequences of different types of variation in floral scent, including community-level, functional, and environmentally induced variation. However, to address these types of questions, novel approaches are needed that can handle large sample sizes, provide quality control measures, and make volatile research more transparent and accessible, particularly for scientists without prior experience in this field. Drawing upon a literature review and our own experiences, we present a set of best practices for next-generation research in floral scent. We outline methods for data collection (experimental designs, methods for conducting field collections, analytical chemistry, compound identification) and data analysis (statistical analysis, database integration) that will facilitate the generation and interpretation of quality data. For the intermediate step of data processing, we created the R package bouquet, which provides a data analysis pipeline. The package contains functions that enable users to convert chromatographic peak integrations to a filtered data table that can be used in subsequent statistical analyses. This package includes default settings for filtering out non-floral compounds, including background contamination, based on our best-practice guidelines, but functions and workflows can be easily customized as necessary. Next-generation research into the ecology and evolution of floral scent has the potential to generate broadly relevant insights into how complex traits evolve, their genomic architecture, and their consequences for ecological interactions. In order to fulfill this potential, the methodology of floral scent studies needs to become more transparent and reproducible. By outlining best practices throughout the lifecycle of a project, from experimental design to statistical analysis, and providing an R package that standardizes the data processing pipeline, we provide a resource for new and seasoned researchers in this field and in adjacent fields, where high-throughput and multi-dimensional datasets are common.
Introduction
Volatile organic compounds (VOCs) are studied in many subfields of chemical ecology, in both aquatic and terrestrial environments. Over the last 20 years, research on floral VOCs has increased dramatically and moved beyond descriptive studies. This progression in the field has led to the emergence of insights in three key areas, which has laid the groundwork for next-generation research in floral scent. First, despite the existence of nearly 2,000 described floral scent compounds (Knudsen et al., 2006), most studied plants share a core set of “greatest hits,” ubiquitous compounds (e.g., limonene, linalool, β-caryophyllene, benzaldehyde) that are produced by a small number of pathways (Pichersky and Dudareva, 2006) and in some cases, have multiple demonstrated functions (Kessler et al., 2013; Raguso, 2016). Second, despite an initial bias that floral scent was exclusively relevant to specialized pollination systems, it is now clear that floral scent is critical for pollinator attraction in more generalized systems (e.g., Johnson and Hobbhahn, 2010; Lemaitre et al., 2014; Larue et al., 2016; Schiestl et al., 2018). Third, despite an early expectation that floral scent would be a reliable phylogenetic marker, some phylogenetic studies have found evidence for convergent evolution and homoplasy across groups of flowering plants, in addition to substantial population-level variation (see below). In light of these insights, which suggest that floral scent composition may be less dauntingly complex, more broadly relevant, and more evolutionarily labile than was thought previously, we need to deepen our understanding of what determines variation in floral scent, what consequences this variation has for species interactions, and how such variation evolves.
Sources of variation in floral scent include genetics, plant morphology and phenology, the abiotic environment, and the biotic community context (across and within trophic levels). In the small number of systems studied so far, single genes can allow for emission of a particular compound (Box 1, section 1A), but the extent to which scent is regulated by a small number of genes of large effect that could be candidate “speciation genes” (Schlüter et al., 2011; Byers et al., 2014) across taxa with different pollinators is just beginning to be explored. Furthermore, recent studies suggest that genetic variation and heritability in emission rate can take on a wide range of values (Box 1, section 1B). Floral scent is often considered an inflorescence- or plant-level trait, but significant variation in emissions exists both within flowers on a plant (due to differential emission across flower parts; Box 1, section 2A) and among flowers on a plant (due to changes in emissions following visitation from pollinators or herbivores or to changes in environmental conditions at the time of flowering; Box 1, section 2B). While the putative roles of floral scents often are assigned to biotic interactions, environmental factors may have significant effects on the expression and evolution of floral scent, by generating patterns of intraspecific variation along environmental gradients (Box 1, section 3A), or by generating plastic change in scent “chemotypes” (=chemical phenotypes, often dominated by one or a few compounds; Box 1, section 3B; rev. by Farré-Armengol et al., 2020). In addition, variation in floral scent can be influenced by or be a direct product of associations with other organisms, such as epiphytic bacteria on plant tissues, microbes in nectar, plant pathogens, or soil and soil nutrients (Box 1, section 4A). Finally, although variation in floral scent at the plant community level is well-documented (Owen et al., 2001; Courtois et al., 2009; Junker et al., 2011; Junker, 2016), the extent to which such variation in scent affects ecological interactions is only beginning to emerge through both correlative (Kantsa et al., 2018) and manipulative (Larue et al., 2016) studies (Raguso, 2012; Junker, 2016). A small number of studies provide proof-of-concept that floral scent variation at the community level can contribute to plant-pollinator network structure (Box 1, section 5A) and ecological function (Box 1, section 5B).
| BOX 1 Sources, functions, and evolution of variation in floral scent. | ||||
| Broad category | Narrow category and insights | Systems | References | |
| Sources of variation | (1) Genetic variation | (A) The molecular basis for floral scent is determined primarily by a relatively small number of loci of large effect | Petunia sp. | Hoballah et al., 2007; Klahre et al., 2011 |
| Orchids | Schlüter et al., 2011; Xu et al., 2012 | |||
| Mimulus sp. | Byers et al., 2014; Peng et al., 2017 | |||
| (B) The extent of genetic variation in scent emission rate ranges from low to high. | Brassica rapa Ipomopsis sp. Sorbus sp. | Zu et al., 2016 Campbell et al., 2022b Feulner et al., 2014 | ||
| (2) Within plant variation | (A) Volatile emissions vary within flowers (e.g., among flower parts) | Review | García et al., 2021 | |
| (B) Volatile emissions vary among flowers on a plant | Nicotiana sp. Oenothera sp. | Euler and Baldwin, 1996; Baldwin et al., 1997 Eisen et al., 2022 | ||
| (3) Abiotic environment variation | (A) Intraspecific variation in scent may occur along environmental gradients | Latitude Cypripedium sp. Encephalartos sp. Echinopsis sp. | Braunschmid et al., 2021 Suinyuy et al., 2015 Schlumpberger and Raguso, 2008 | |
| Elevation Cypripedium sp. Gymnadenia sp. Ficus sp. | Braunschmid et al., 2017 Gross et al., 2016 Souto-Vilarós et al., 2018 | |||
| (B) Scent chemotypes may change plastically in response to environmental factors | Water availability Ipomopsis sp. Four species found in Rocky Mountains, USA | Campbell et al., 2019 Burkle and Runyon, 2016 | ||
| Nutrients Arabis sp. | Luizzi et al., 2021 | |||
| Ozone | ||||
| Fagopyrum sp. | Rering et al., 2020 | |||
| Brassicaceae sp. | Saunier and Blande, 2019 | |||
| Multiple environmental factors | ||||
| Hesperis sp. | Majetic et al., 2009 | |||
| Four species found in Rocky Mountains, USA | Glenny et al., 2018 | |||
| (4) Associations with other organisms | (A) Microorganisms affe ct volatile emissions, or are the source of “floral” volatiles | Epiphytic bacteria | Peñuelas et al., 2014; Helletsgruber et al., 2017; Burdon et al., 2018 | |
| Nectar microbes | Rering et al., 2018; Yang et al., 2019 | |||
| Plant pathogens | Majetic et al., 2017; Cellini et al., 2019 | |||
| (5) Plant community variation | (A) Variation can affect network structure in communities | Two species in a German grassland | Larue et al., 2016 | |
| Mediterranean scrubland communities in Greece | Kantsa et al., 2018 | |||
| Montane meadow in Northern Rocky Mountains | Burkle and Runyon, 2019 | |||
| (B) Variation can affect ecological function in communities | Mediterranean scrubland community in Spain | Filella et al., 2013 | ||
| Functional consequences of variation | (6) Volatiles that have functional consequences are often a subset of all of the compounds produced by the plant | Bat pollinated sp. in Costa Rica | Von Helversen et al., 2000 | |
| Datura and Agave sp. | Riffell et al., 2009 | |||
| Chiloglottis sp. | Peakall et al., 2010 | |||
| Echium sp. | Burger et al., 2012 | |||
| Mimulus sp. | Byers et al., 2014 | |||
| (7) Floral volatiles can serve additional functions beyond pollinator attraction, such as repelling antagonists | Rafflesia sp. Nicotiana sp. Penstemon sp. Petunia sp. Polemonium sp. | Lev-Yadun et al., 2009 Kessler et al., 2008, 2019 Burdon et al., 2018 Kessler et al., 2013 Galen et al., 2011 | ||
| (8) Different pollinators can have different behavioral responses to the same compounds | Review | Junker and Blüthgen, 2010 | ||
| Evolution of variation | (9) Macroevolutionary processes | (A) Scent is phylogenetically conserved | ||
| Chiloglottis sp. | Mant et al., 2002; Peakall et al., 2010 | |||
| Nicotiana sp. | Raguso et al., 2006 | |||
| Oil-secreting orchids in South Africa | Steiner et al., 2011 | |||
| Ophrys sp. | Joffard et al., 2020 | |||
| Streptanthoid sp. | Weber et al., 2018 | |||
| (B) Scent displays convergent evolution | Bat pollinated sp. Lysimachia sp. Ophrys sp. Passiflora sp. Seven fly pollinated sp. with fetid odors | Knudsen and Tollsten, 1995 Schäffler et al., 2012 Stökl et al., 2005 Clifford, 2017 Johnson and Jürgens, 2010 | ||
| (C) Scent displays homoplastic variation | Stanhopea and Embreea sp. | Williams and Whitten, 1999 | ||
| Acleisanthes and Mirabilis sp. | Levin et al., 2003 | |||
| (10) Microevolutionary processes | (A) Volatiles can be the targets of different forms of selection (e.g., directional or stabilizing) | Gymnadenia sp. | Schiestl et al., 2011; Gross et al., 2016; Chapurlat et al., 2019 | |
| Brassica sp. | Gervasi and Schiestl, 2017 | |||
| Knauer and Schiestl, 2017; Schiestl et al., 2018 | ||||
| Anacamptis sp. | Joffard et al., 2020 | |||
| Ipomopsis sp. | Campbell et al., 2022a | |||
| (B) Volatiles can be under selection from different agents of selection | Arum sp. Brassica sp. Biscutella sp. Penstemon sp. Primula sp. Solanum sp. | Gfrerer et al., 2021 Gervasi and Schiestl, 2017; Knauer and Schiestl, 2017 Knauer et al., 2018 Parachnowitsch et al., 2012 Ehrlén et al., 2012 Kessler and Halitschke, 2009 | ||
| (C) Differences in scent are linked to reproductive isolation or changes in mating system, while similarities are linked to hybridization | Arabis sp. Calendula sp. Capsella sp. Chiloglottis and Ophrys sp. | Petrén et al., 2021 Zito et al., 2018 Sas et al., 2016 Ayasse et al., 2010; Peakall and Whitehead, 2014; Whitehead and Peakall, 2014; Whitehead et al., 2015; Gervasi et al., 2017 | ||
| Ipomopsis sp. | Bischoff et al., 2015 | |||
| Mandevilla sp. | Rubini Pisano et al., 2019 | |||
| Phlox sp. | Majetic et al., 2018 | |||
| Platanthera sp. | Esposito et al., 2018 | |||
| Spiranthes sp. | Tao et al., 2018 | |||
Floral scent can vary at many scales, from within-plant to patch, population, species and phylogenetic scales, but the functional consequences of variation at different levels are less well-documented or understood. There are several ways in which functional variation in scent may differ from observed or measured variation: only a subset of compounds in a volatile profile may have functional consequences (Box 1, section 6), volatiles may play roles in more than one type of species interaction (Box 1, section 7), and functional responses may vary across pollinator taxa (Box 1, section 8) or geographically across the distribution of a plant. A major remaining challenge is to understand the full spectrum of selective forces acting upon variation in floral scent, and thereby to establish the functional significance of different volatile compounds constituting floral scent blends. This task is daunting, given our underestimation of the variety of functions involved. For example, it is challenging enough to demonstrate pollinator-related functions, either directly through electrophysiological and behavioral assays (Schiestl and Marion-Poll, 2002; rev. by Raguso, 2020) or indirectly through studies of selection gradients informed by such assays (Schiestl et al., 2011). However, there is growing evidence that natural enemies are just as likely as mutualists to shape floral scent as selective agents (Galen et al., 2011; Kessler et al., 2013; Campbell et al., 2022a), even in cases where flowers are small, scent emissions are miniscule and natural enemies are microbial (Huang et al., 2012). Thus, there are very few documented cases in which floral scent composition is simple and the functions of all components are known (e.g., Wiemer et al., 2009).
There are open questions about the evolution of floral scent on both macro- and micro-evolutionary scales. The results of phylogenetic studies suggest that scent can be evolutionarily labile (Box 1, section 9), but they do not resolve the extent to which macroevolutionary patterns result from phylogenetic constraints and natural selection across many systems (Knudsen et al., 2006; Raguso et al., 2015). This insight, in combination with evidence for rapid microevolutionary change in scent (e.g., Gervasi and Schiestl, 2017; Schiestl et al., 2018), leads to a number of areas for next-generation research on the microevolution of floral volatiles, including determining the most common forms of natural selection on floral volatiles (e.g., directional, stabilizing; Box 1, section 10A), the most relevant agents of selection (Box 1, section 10B), and the relative contribution(s) of floral scent to reproductive isolation, population structure, and mating system evolution (Box 1, section 10C).
Although conceptual frameworks exist for addressing these questions, the development of suitable methods has lagged in three key areas. First, these types of studies require larger sample sizes to characterize intraspecific variation in floral scent and link this variation to differences in community dynamics, functional responses, environmental variation, or evolutionary processes (Friberg et al., 2019). As such, computational tools that make processing large numbers of chromatograms feasible (Box 2) will be critical, but these types of tools have not yet been widely adopted in the field. Second, studies that expand the field into new plant systems or that address relatively understudied sources of variation will require analytical chemical methods as well as data processing methods that provide rigorous quality control. In particular, the abilities to identify compounds correctly, to retain minor products consistently, and to remove artifact and contaminant compounds appropriately in large datasets are all critical for expanding our knowledge reputably. Third, research on plant volatiles has been historically somewhat isolated from work occurring in related fields including pollination ecology (Raguso, 2008a,b) and community ecology (Raguso, 2014). However, the number of publications on floral volatiles has increased since 2006, when a landmark book on the field was published (Dudareva and Pichersky, 2006; for additional reviews, see Stashenko and Martínez, 2008, 2012). This pattern indicates the beginning of a long-desired integration of research on floral volatiles into the fields of community ecology, behavior, and evolution (Raguso, 2001, 2008a). As such, new studies need to adopt methodological approaches that are transparent, can be understood by non-chemists, and can be accessible for meta-analyses.
BOX 2 Software for pre-processing GC-MS datasets.
Whereas many investigators will rely upon proprietary instrument software to pre-process a GC-MS data set, a large array of open-source software is also available and can provide additional capabilities. Key features to look for in these software packages are broad interoperability with equipment and other packages, ease of batch processing, scripting interfaces that enable reproducible analyses by consolidating parameters, and actively updated, well-documented, open-source algorithms (Bartlett et al., 2011). Comprehensive solutions that provide the majority of steps for high throughput analyses include: XCMS (Smith et al., 2006) adapted for identifying compounds in GC-MS datasets with either metaMS (Wehrens et al., 2014) or PyMS (O’Callaghan et al., 2012), eRah (Domingo-Almenara et al., 2016), and ADAP-GC (Jiang et al., 2010; Ni et al., 2012, 2016; Smirnov et al., 2019) featured in the MZmine software (Katajamaa and Orešič, 2007; Pluskal et al., 2010). Many other tools exist that implement some but not all of the pre-processing steps: deconvolution in AMDIS (Stein, 1999) followed by tracking conserved unknowns in SpectConnect (Styczynski et al., 2007); or peak alignment in MetAlign (Lommen, 2009) followed by grouping mass peaks in MSClust (Tikunov et al., 2012). A detailed comparative listing of 29 packages for GC-MS pre-processing is provided by Spicer et al. (2017).
Literature review
We conducted a literature review of studies that measured floral scent between 2005 and 2019 (Sections “Methods” and “Results” in Supplementary material 1, list of studies in Supplementary material 2) to address two questions: (1) Are floral scent studies being published in more mainstream journals at an increasing rate? (2) How many studies provide detailed data processing protocols? Our review revealed that the number of publications on floral volatiles has increased at a similar rate in more general, mainstream journals as in journals that are specific to the subfields of chemistry, biochemistry, and plant sciences. At the same time, very few of the studies report how datasets were cleaned and processed–for instance, under 30% of studies (140 studies out of 509 studies that measured scent between 2014 and 2019) report collecting ambient control samples, which are important to compare to biological samples to determine the extent of background contamination (see Section “Compound filtering”). Of those studies that did collect ambient control samples, two-thirds of them (93 studies) did not identify how ambient control data were used in the study. Fewer than five percent of studies reported using other methods of cleaning data, such as removing known contaminants or removing rare compounds from datasets, although these are commonly performed practices. Similarly, while most studies reported using authentic standards or Kovats retention indices (see Section “Analytical chemistry”) to identify the detected compounds, about 15% of studies did not provide any information on how compounds were identified. In addition, less than 10% of studies published between 2014 and 2019 include archived raw data, which vastly limits our ability to make advances via meta-analyses (e.g., Junker et al., 2018).
These three pressing methodological issues are pertinent to the accessibility of new research in the ecology, evolution, and function of floral scent, and, more broadly, to cutting-edge, high-throughput studies of any type of VOC. These issues are especially important as incentives for beginning researchers to add volatile data to their research programs. To address these issues, we present recommendations for best practices and new computational tools that form an analytical pipeline, given that most recent studies in the field do not report their data processing and cleaning practices in a reproducible way (but see Clifford, 2017; Lahondère et al., 2020). While various aspects of methods in the field have been reviewed previously (e.g., Tholl and Röse, 2006; Stashenko and Martínez, 2008; Tholl et al., 2020), here we aim to increase the accessibility of this expanding field through two primary aims. First, we blend a synthesis of the existing literature on methods in this and related fields with our own experiences into a guide for data collection (experimental design, collection techniques, analytical chemistry, and compound identification), and for data analysis (statistics and database integration). Second, for the intermediate step of data processing, we introduce a new R package, bouquet, that facilitates transparent, consistent, and reproducible data filtering and manipulation.
Steps in the study of plant volatiles, from experimental design to statistical analysis
Experimental design
We present recommendations for aspects of experimental design that are particularly important when analyzing variation in floral volatiles: sampling effort, the use of controls, and the standardization or measurement of environmental conditions. Sampling effort varies widely across published studies–for instance, some studies have used a sample from one plant to characterize the floral scent of a species, while other studies have analyzed hundreds of samples from a species. Specifically, in the subset of papers that we reviewed (see Supplementary materials 1, 2) that were published in 2018 and 2019, the number of samples collected per group (e.g., populations, species, experimental treatments, etc.) ranged from 1 to 154, with a median of only 3 samples per group. Some of this variation stems from the inherent trade-off between sampling within and across species (or any other type of group), but the variation also results from a lack of agreement in the field as to what constitutes a suitable sample size for a group (e.g., species, population) of interest. While a minimum sample size of ten individuals per species was arbitrarily proposed nearly 25 years ago by Raguso and Pellmyr (1998), this convention has not been adopted in the field (only 12% of papers published in 2018–2019 met this minimum number of samples per group). Much larger samples sizes than typically used (median = 3) are needed to detect compounds found at low to moderate frequencies and to properly characterize intraspecific variation. For example, 100 samples were needed to detect 28 out of 34 VOCs emitted by Oenothera harringtonii (Onagraceae) across their geographic range [see accumulation curve in Skogen et al. (2022)]. A similar accumulation curve constructed for 68 plants of Ipomopsis aggregata (Polemoniaceae) from a single population revealed that 20 samples were needed to detect 26 of 29 VOCs (data from Campbell et al., 2019). Given the rise of analytical methods that can handle larger sample sizes (see Section “Analytical chemistry”), we encourage researchers to increase their sampling effort at levels appropriate to their research questions, simply to be able to capture potential polymorphism, plasticity, and other sources of variation (Delle-Vedove et al., 2017). Appropriate sample sizes for other questions will depend upon levels of variation and how they affect statistical power, but significantly larger sample sizes may be required for some studies, including those that estimate heritable variation in and natural selection on scent production (e.g., Gross et al., 2016), or that map finer spatial and temporal resolution of variation in scent composition (Gfrerer et al., 2021; Skogen et al., 2022).
For any method of scent collection (see Section “Sample collection,” where we review methods briefly and refer to prior, more detailed reviews), ambient control samples must be collected from empty headspace chambers as well as from samples that contain only vegetation, in order to rule out these potential sources of volatiles. The relative merits of headspace chambers are reviewed by Stewart-Jones and Poppy (2006) and Tholl et al. (2020). At least a few ambient and vegetative control samples should be collected concurrently with each replicate set (e.g., each day’s sample) of floral samples. A set of floral samples might consist of all the samples collected on a given day if plants are close to each other (e.g., within a growth chamber or on a single greenhouse bench), however, if samples that are collected concurrently are more dispersed in space (e.g., across one or more field sites), then one or more ambient controls should be collected in each sampling location. One or more vegetative control samples should be collected from each group that will factor into the analyses. For instance, in a study comparing scent profiles of species within the same community, vegetative controls should be collected from each species, while in a study comparing scent profiles of several species across several populations, vegetative controls should be collected from each species at each population. This is important for plant lineages bearing complex vegetative trichomes (e.g., Lamiaceae, Asteraceae, Polemoniaceae) and when induced whole plant volatile responses to herbivores or pathogens are anticipated. Protocols for filtering out compounds detected in the ambient and vegetative controls are described below (see Section “Chromatogram to data table pipeline”).
Because environmental conditions can generate plastic variation in floral scent emissions, floral scent collections should be made under standardized environmental conditions unless the natural variation is specifically of interest, as in studies of plasticity or heritability in the field. When possible, we advise measuring environmental variables such as temperature and soil moisture, during each sampling period, as these variables can influence the composition of floral emissions (e.g., Farré-Armengol et al., 2014; Glenny et al., 2018).
Sample collection
The suitability of VOC sampling methods for different kinds of scientific questions has been reviewed comprehensively by Tholl and Röse (2006), Stashenko and Martínez (2008) and more recently by Tholl et al. (2020) and is summarized in Figure 1. Readers are referred to these reviews for detailed methodological comparisons and practical considerations. Here we present a guide to selecting and optimizing a method and extracting and interpreting the data.
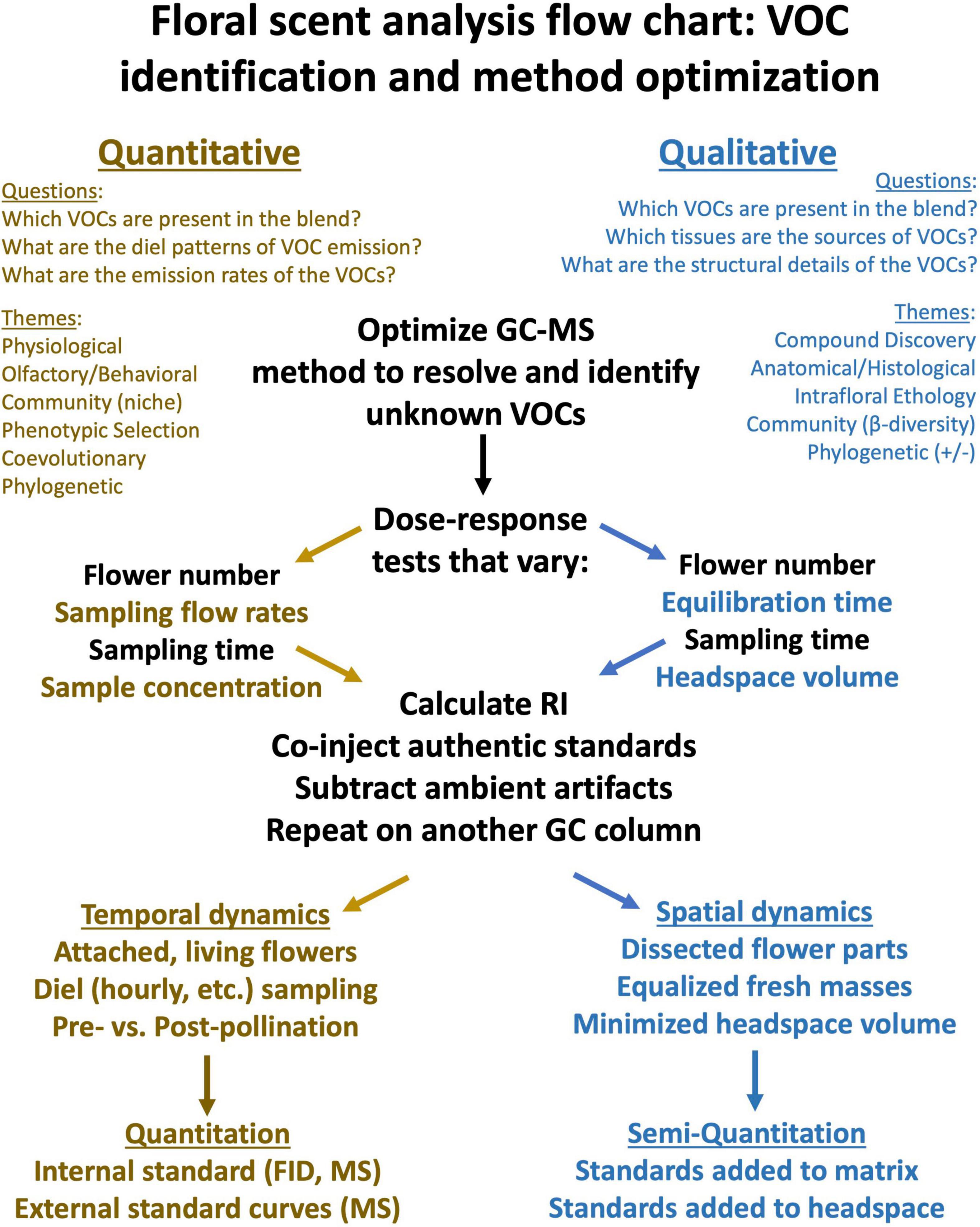
Figure 1. A graphical illustration of how to develop and optimize methods for the analysis of floral VOCs. Brown text indicates questions and themes that are primarily quantitative in nature, whereas blue text is relevant for approaches that are often qualitative, and black text applies more generally. Note that these categories are not exclusive, but are meant to provide guidance to new investigators in this field.
Scent collection methods can be generally grouped into two categories: “static headspace” methods and “dynamic headspace” methods, although below we describe an additional method that can be hybrid (micro-SPE or TD-GC-MS). Static headspace approaches are fully equilibration based and involve the equilibration of volatile samples within a standardized headspace volume and passive adsorption onto a matrix that can be directly thermally desorbed for coupled gas chromatography–mass spectrometry (GC-MS). Benefits of static headspace approaches include increasing the range and sensitivity of volatile trapping, providing better representation of small, highly volatile compounds often masked by solvents and, conversely, more complete mass spectra for identification of large, less volatile compounds due to equilibration (Tholl et al., 2021). These benefits are weighed against the costs of losing the entire sample in one injection and additional challenges associated with sample quantification, specifically that it is difficult to quantify emission rates (reviewed by Ruiz-Hernández et al., 2018). Three common methods for sampling static headspace involve the use of solid phase microextraction (SPME) fibers, twister, and silicone tube-based traps for thermal desorption; these methods have all been reviewed extensively (Tholl et al., 2020). Because the entire sample is lost to injection, additional replicate samples should be collected when using these methods.
In contrast, dynamic headspace methods involve the replacement of headspace air over time. As such, they provide insight into quantitative aspects of floral scent production by enabling the estimation of emission rates of each compound of interest, which can be standardized across samples by the number or mass of flowers sampled (Tholl et al., 2020). With solvent-based dynamic headspace methods, volatiles are collected using an adsorbent material that is packed within a glass or steel trap, and traps are eluted with solvent following the collection period for subsequent analysis via GC-MS. Aspects of the protocol that need to be tailored to the particulars of each system include the length of the collection period, the amount of adsorbent in the traps, the volume of eluate, and the amount of floral material in each sample (Dobson et al., 2005). In addition to facilitating quantitative analysis, this method also produces samples that can be frozen and stored for subsequent (re)analysis (e.g., using different GC columns or detector systems).
An additional method of dynamic headspace collection involves the use of volatile adsorbent traps for shorter collection times, followed by direct thermal desorption of trapped compounds for GC-MS analysis on an instrument that is set up for that form of injection (TD-GC-MS). The traps can be small (e.g., containing only 2–5 mg of Tenax and/or charcoal) or larger to fit some automated thermal desorption systems. With small traps, the method is sometimes referred to as micro-solid phase extraction (micro-SPE) (Amirav and Dagan, 1997; Dötterl et al., 2005). Advantages of this method include reduced sample collection times (minutes, instead of hours) and stable sample storage in the absence of refrigeration. Furthermore, some thermal desorption systems can recollect some of the sample prior to entering the GC column, which is generally not an option when SPME or stir-bar methods are used. Because collections made using this method are not diluted by a solvent, thermal desorption-based dynamic headspace sampling is particularly well suited for trapping compounds emitted in small amounts (Bischoff et al., 2014), such as volatiles emitted by pollen or dissected flower parts (Jürgens and Dötterl, 2004) as well as small compounds (e.g., ethyl acetate) whose evaluation by GC-MS would be impaired by co-elution with a large solvent peak (Arguello et al., 2013).
Regardless of the method used, preparing and collecting samples in a way that minimizes contamination and background noise is essential, because a clean baseline is critical for compound identification and quantification (described below). For all methods, collection vials (e.g., glass scintillation vials, PET cups or bottles) should be cleaned prior to use via washing with non-scented detergent and baked in a clean oven that is not used to dry plant material. Similarly, sampling apparatuses (e.g., SPME fibers, adsorbent traps) should also be cleaned via the injection port of the GC (for SPME fibers), heating (for thermal desorption traps) or via rinsing with (or soaking in) copious volumes of solvents (solvent traps) and then heating (silicone tubing). Note that specific temperatures and solvents used will vary with materials and applications. These steps increase the time required to collect samples, but, when used in combination with ambient and vegetative control samples (see Section “Experimental design”), can greatly reduce the background noise and thus increase the power of a dataset, while also reducing time wasted during analysis (i.e., screening out artifacts).
Analytical chemistry
The combination of gas chromatography (GC) as a means of separating complex chemical blends, with mass spectrometry (MS) as a detector system used to identify unknowns, has played a crucial role in the study of plant volatiles, including essential oils and floral scent (Adams, 2001). Although many analytical methods can be applied to the study of floral and plant volatiles (Stashenko and Martínez, 2012; Ruiz-Hernández et al., 2018; Tholl et al., 2020), here we focus on the use of GC-MS to identify floral scent constituents. In Figure 1, we outline how GC-MS-based analytical approaches might differ for researchers using floral scent to address quantitative vs. qualitative questions. It should be clear from this figure that complementary analytical methods are used to address different kinds of research questions exploring floral scent at different levels of biological organization.
Selecting a suitable analytical approach involves several iterations of data collection and fine-tuning. First, the researcher must obtain proof-of-concept data confirming that volatiles can be collected, analyzed and identified from their system. Such pilot studies can be accomplished through collaboration with analytical chemists or chemical ecologists with suitable equipment (Raguso et al., 2015), or through core facilities at research institutions, usually for a fee. One consideration at this stage is the choice of GC capillary column dimensions and stationary phase polarity, which can be tailored to efficiently separate different chemical mixtures. The most commonly available all-purpose GC column, non-polar (5%-phenyl)-methylpolysiloxane (DB-5, HP-5 or similar product names) offers the benefits of high stability, a large published literature for compound retention indices (see Section “Adding biochemical context”), and an appealing logic for beginners (retention times generally track compound size and boiling points). Polar GC columns (DB-WAX, etc.) have polyethylene glycol as a stationary phase, through which blends are separated according to relative polarity as well as boiling point differences. Thus, compound retention order is less intuitive, and the polar columns have lower maximum temperature, a noisier baseline and a shorter working life, but they are superb for separating complex blends of similar compounds (e.g., sesquiterpene hydrocarbons) or small, polar volatiles (e.g., related to fermentation) that would elute too early on a non-polar GC column.
When planning projects that involve scent sampling in new systems or using new methods, we suggest that investigators dedicate time for developing and revising their analytical methods (Figure 1)–pilot studies rarely reveal the full complexity of a volatile bouquet, due to the exploratory nature of the methodology. For example, the unusual yeast-like scent of Asimina triloba (Annonaceae) flowers was not adequately captured using dynamic headspace trapping, solvent elution with hexane and separation on a conventional non-polar DB-5 GC column, due to the masking of small, highly volatile fermentation compounds by the solvent injection peak on that column (Goodrich et al., 2006). Even after switching to a polar DB-WAX GC column and a solvent-free, static headspace approach, the common, all-purpose SPME fiber (100 μm PDMS) was insufficient, necessitating the use of a hybrid SPME fiber (65 μm DVB-PDMS) (Goodrich et al., 2006). The researcher should optimize methods for their system and question, both to enhance the sensitivity and consistency of data analysis and to minimize costs, especially when high replication is needed. Method optimization may involve tradeoffs between slower GC runs needed to resolve all peaks to baseline (Adams, 2001) and faster runs promoting more data replication per unit of analytical time (Eisen et al., 2022). Truncated GC programs that accelerate oven temperature ramps during intervals when few peaks are eluting can shave run times and costs (Doubleday et al., 2013).
Compound identification and removal of contaminants
Chemical analysis using GC-MS provides a wealth of information that can be used to identify individual compounds in complex floral scents (Box 3). We recommend a combination of approaches to provide improved specificity and confidence in identifications, recognizing that some approaches may not be possible for some compounds. Tentative identification of a compound generally begins with performing an automated search of an unknown mass spectrum against a mass spectral library that generates a list of candidate compounds ranked by similarity. However, many reference compounds have similar mass spectra, and not all compounds are present in MS libraries, so the top result should only be treated as a tentative identification, and compounds with library match scores below 90% (or some other justified level) should be treated as unknowns (Box 3; criterion 3). For compounds emitted at low levels relative to the chromatographic background, or that nearly co-elute with other compounds, it is often helpful to perform deconvolution or subtract the mass spectrum adjacent to the peak of interest before a library search. Considering price and comprehensiveness, users may select from large universal libraries such as NIST, Wiley, MassBank (free and open source, Horai et al., 2010), and the Golm Metabolome Database (free, Hummel et al., 2013), or libraries specific to plant compounds such as the Adams Essential Oils Library (Adams, 2001), the MassFinder Terpenoids Library, the Flavor and Fragrance Natural and Synthetic Compounds Library (FFNSC), or the Mass Spectra for Chemical Ecology database (MACE, free, Schulz and Möllerke, 2022).
BOX 3 Criteria used to identify unknown compounds resulting from GC-MS analysis.
Criterion 1: co-chromatography and MS-match with an authentic standard, preferably on two different stationary phases (GC columns). Criterion 1 should be sufficient for compounds that are common to floral scent blends and have straightforward chemistry (e.g., methyl salicylate), but may not be sufficient for compounds whose stereochemistry is unknown (due to chiral centers, positional isomers or stereoisomers with potential differences in biological function). Criterion 1 would be sufficient in the case of co-chromatography with authentic enantiomers on a chiral GC column.
Criterion 2: in the absence of an authentic standard, through the combination of standard retention indices (RI) and MS libraries, with reference to other published studies on an appropriate GC column. Criterion 2 may be necessary when standards are not easily obtained but may be deduced through co-chromatography with essential oils for which high quality analyses have been published (e.g., zingiberene in ginger essential oil; Millar, 1998).
Criterion 3: absence of published RI value, but MS library fit is high (>90%) or confidence can be drawn from key mass spectral ion fragments. Criterion 3 is not definitive but is relevant when VOCs appear in metabolic series, such as phenylethyl esters with a common base peak of m/z 104, or aliphatic ethyl esters with m/z 88 (Arguello et al., 2013), or when the nitrogen rule is used to evaluate putative pyrroles and piperidines (Moré et al., 2019).
Absent Criteria 1, 2 or 3, compound identity remains unknown. In such cases it is best to either present the full mass spectrum or to list it in descending order (for consistency, we suggest listing the top 10 m/z fragments in descending order of% abundance, including [by definition] the base peak and, when possible, the molecular ion) with fragment abundance expressed as% relative to the base peak (most abundant MS ion = 100%).
To improve on the tentative identification provided by a library search, a researcher can compare the Kovats retention index (RI, a standardized retention time calculated by running a series of n-alkanes with the same GC method) of an unknown compound to values in the literature provided in databases such as The PheroBase (El-Sayed, 2021), the NIST Chemistry WebBook (Linstrom and Mallard, 2001), Adams (2001), and FFNSC (Box 3, criterion 2). While this approach can narrow the list of candidate compounds, there are multiple ways in which RIs may be calculated, which could generate some degree of noise when comparing calculated RIs to published values (discussed in Stashenko and Martínez, 2012). Researchers using temperature-programed (rather than isothermal) GC-MS separation of volatile blends will need to calculate RIs (and compare with published data) using the equation of Van Den Dool and Kratz (1963). One challenging aspect of compound identification using RIs is that published values can vary due to factors reviewed by Babushok (2015), who provides standard deviations and confidence intervals for essential oil components identified on the most commonly used polar and non-polar GC columns (see Babushok et al., 2011). Calculated RIs falling outside of these distributions for published values on the same GC column should be considered false positives.
The most rigorous way to confirm a tentative identification of a compound is to run a reference standard of the known compound on the same GC column and program (Box 3, criterion 1). This approach is often unavailable to investigators who do not have chemical supplies, may not be affordable when a large number of compounds need to be purchased and may not be practical when a compound is not commercially available. However, some researchers amass collections of authentic standards and often are willing to share aliquots with colleagues for peak identification.
Volatile blends include structural variants such as positional isomers, cis/trans isomers and enantiomers, which often require additional analytical methods to determine absolute configuration. For example, GC analysis with a chiral column was needed to separate enantiomers of lilac aldehydes and alcohols (Dötterl et al., 2007), whereas derivatization reactions were required to determine the double bond positions and cis/trans conformations of medium chain alkenes in yuccas (Tröger et al., 2019). Thus, mass spectral library matches alone are insufficient to determine whether a given flower emits cis- or trans-β-ocimene, or the (+) or (−) enantiomers of β-pinene, compounds that are common floral scent constituents (Knudsen and Gershenzon, 2006). Thus, when the NIST mass spectral library returns “D-limonene” as a best fit to a sample spectrum, it is inappropriate in the absence of a chiral GC column.
A challenge of interpreting GC-MS data is the presence of chromatographic artifacts and impurities, which can arise at several stages of collection (see Section “Sample collection” above) and analysis. Supplementary material 3 summarizes the most common ambient contaminants in GC-MS analyses, derived from the apparatus (oven bags, plastic tubing, glassware), growth chamber or greenhouse air, dirty solvents, or fume hoods. Many of these sources are eliminated in natural settings, but in such cases other contaminants arise related to sample transport and storage, community complexity (e.g., ants within headspace chamber, strongly scented neighboring plants) or the researchers themselves (lotions, dirty hands, breath lozenges, insect repellent and sunscreen). Usually, contaminants can be identified due to their presence in ambient controls (see Section “Compound filtering”), but contamination may be specific to a faulty tube, pump or vial. Although novel floral volatiles are identified each year (e.g., Maia et al., 2019; Milet-Pinheiro et al., 2021), by now there is a clear sense of what kinds of compounds can be produced by flowers. Investigators new to floral scent analysis should be skeptical of phthalates, silanes or squalene (in tubing), xylenes or halogenated phenols (in solvents), as well as nonanal and decanal (human skin contaminants; see Supplementary material 3). Compounds such as limonene and benzaldehyde constitute special cases, as they are nearly ubiquitous floral scent components (Knudsen et al., 2006) but also are commonly found as low abundance ambient contaminants. When these compounds are genuinely floral, the collection of headspace samples from larger numbers of flowers or for longer collection times should increase GC peak areas significantly over those of accompanying ambient controls.
Chromatogram to data table pipeline
After volatiles are collected and analyzed by GC, there are several data processing steps that create a table of emissions of each confirmed floral volatile compound in each sample that can be used for analysis. These steps include peak picking, alignment, and integration, compound identification, filtering compounds by comparison to ambient controls, and quantification of emission rates from peak areas. As the accuracy of the filtered emissions table affects all downstream analyses, we advocate adopting data processing protocols that are robust, reproducible, and clearly presented. Our survey of 509 floral scent studies (see Supplementary material 2) conducted from 2005 to 2019 found that while 85% of studies described how compounds were identified, only 28% collected ambient controls and 9% reported if or how those controls were used in filtering the dataset (detailed methods and results in Supplementary material 1). Therefore, many studies are not reporting methods in sufficient detail to make studies accessible to a broad audience. We argue that this gap in methodological and reporting practices is a serious problem for this growing field, but one that can be solved by using a well-documented data processing pipeline. As the steps in each experiment will depend on the analytical method, software, and dataset size, we offer the following flexible guidelines for implementing each step and encourage authors to report all relevant parameters and procedures. We present a new R package, bouquet, that enables easy implementation and reporting of the compound filtering steps (see Section “Compound filtering”).
Data pre-processing
Analyses that rely on detecting, identifying, and quantifying hundreds of previously unknown compounds consistently in tens to hundreds of biological samples require data pre-processing steps that overcome some of the unique challenges of raw GC-MS datasets. First, compounds that co-elute from the GC column form overlapping chromatographic peaks that are difficult to integrate and identify. Various signal processing algorithms have been devised to deconvolute overlapping peaks into separated pure mass spectra that can be better matched to reference databases and quantified (reviewed in Du and Zeisel, 2013; Domingo-Almenara et al., 2016). Second, retention times drift slightly among batches of samples, so peaks must be aligned across the dataset before comparison. Performance of numerous peak alignment algorithms has been compared (Koh et al., 2010; Coble and Fraga, 2014; Niu et al., 2014). Third, compounds present at low amounts are often not recognized initially by peak detection algorithms in all samples, so to avoid coding these small amounts as zero, they can be filled in by integrating ions present in other samples at that retention time (Katajamaa and Orešič, 2007; Domingo-Almenara et al., 2016; Müller et al., 2020). Modern implementations of these three algorithms and others for pre-processing of large GC-MS datasets are given in Box 2.
Quantification with standards
There are several techniques available for quantifying volatiles by conversion of integrated peak areas into relative abundances (percentage analysis) or absolute emission rates (e.g., mass emitted per hour per g fresh weight) using internal or external standards (reviewed in Qualley and Dudareva, 2014; Ruiz-Hernández et al., 2018). Presenting data as emission rates, which can be presented in different units, rather than peak areas enables comparisons across study systems that may require different sampling methods (for instance, different numbers of flowers enclosed, or different collection windows). For both liquid extraction-based dynamic headspace and stir-bar static headspace methods, an internal standard can be added (to the sample after elution, or to the stir bar before collection) to facilitate quantification in ng of the internal standard (Qualley and Dudareva, 2014; Ruiz-Hernández et al., 2018). Alternatively, calibration curves of external standards can be calculated by running replicated (three or more times) dilution series of standards with known concentrations spanning several orders of magnitude across the linear range of the detector. For thermal desorption methods, internal standards and dilutions can be pipetted directly onto the trapping material, while dilutions can be injected in solvent for solvent-based methods. Due to differences in mass spectral fragmentation patterns, response factors differ among and within compound classes (terpenoids, benzenoids, aliphatics, etc.), so at least one standard of each compound class should be included in the mixture.
Compound filtering
An important part of the chromatogram to data table pipeline is determining which compounds will be included in the final dataset; compounds could be excluded if they are present in the control samples, if they are rare in the dataset, or if they are known contaminants (see Compound identification above). As mentioned above, there are many sources of undesired chemical background in floral scent chromatograms: compounds that originate from the environment, vegetative tissue, or sampling apparatus (e.g., nylon bags, storage vials), compounds produced by impurities or breakdown of the trapping material, and compounds originating from the instrument (the autosampler, inlet, or GC column) (see Supplementary material 3). This background can be effectively reduced during data processing by contrasting floral samples with control samples of ambient air and/or vegetative tissue (see Section “Experimental design”). Our survey of volatile processing methods revealed four strategies for comparing samples with controls that each have advantages and drawbacks. First, one may exclude any compounds present in the controls; this is straightforward but may be too stringent if floral volatiles occur at low levels in vegetative tissue or the ambient air. Second, one may subtract the average amounts of each compound found in controls from each sample; this strategy isolates contributions from floral tissues from those in leaves or the environment but may result in negative values or contaminants that are reduced but not entirely eliminated from the dataset. Third, one may exclude compounds in the controls unless they occur at several-fold (e.g., four times) greater amounts in samples on average or on the particular date of sampling. Fourth, one may exclude compounds in the controls unless they pass a statistical test (amounts in samples significantly greater than those in controls). Due to the large number of comparisons involved in conducting statistical tests on the levels of hundreds of compounds in floral and control samples, we recommend controlling the number of false positives using the false discovery rate method. Because the third and fourth strategies do not rely on a uniform operation as the exclusion or subtraction methods do, these methods allow for custom thresholds for inclusion depending on the background level of noise but may exclude rare floral compounds.
After background compounds have been removed, manual filters can focus the final dataset on common or high-emission volatiles, although these should be applied with caution if volatile diversity or differences in minor compounds are of interest. Note that such an approach can miss entirely a compound that is responsible for pollinator attraction, as seen for example in the case of a minute quantity of species-specific indole emission attracting hawkmoths (Bischoff et al., 2015). Authors may choose to exclude compounds that only occur rarely, using a threshold of the count or frequency of occurrence in each species, population, or treatment group. Compounds with low emissions may also be excluded with a minimum threshold for the largest peak encountered across samples. Finally, known contaminants (Supplementary material 3) or compounds that elute outside the retention time range of most floral volatiles may be excluded (e.g., early eluting small molecules such as CO2 or late-eluting phthalates or other contaminants).
To perform all these filtering steps in an easily documented and customizable data processing pipeline, we developed the R package bouquet, available for download at https://github.com/jmpowers/bouquet. This package enables users to start with a dataset of integrated and identified peaks, and associated sample metadata, and perform filtering steps of their choosing (described above). As such, this package differs from the recently developed gcProfileMakeR (Perez-Sanz et al., 2021), which enables users to identify metabolites that are produced constitutively or non-constitutively. Supplementary material 4 provides an introductory vignette with an example dataset, and complete documentation of each function is included in the package. Users first load a table of integrated and identified peaks from a batch of floral samples and ambient and/or vegetative controls. The format of this initial table is flexible to allow inputs from many instrument-specific software packages as well as freely available tools like AMDIS and OpenChrom. Next, the user loads the experimental metadata with information on the type of sample, experimental treatments or groups, and some measure of biomass or flower number used to standardize emission rates. The user can then perform the filtering steps of their choosing (all filtering steps described above are implemented), and combine the filters using logical operators. For example, to be included in the dataset, a compound may need to be present in floral samples at four times the average of ambient controls and occur in at least 5% of floral samples. The resulting filtered dataset is then ready for downstream analyses. The package includes a plot_filters function to visualize which compounds passed the filtering steps by color and position (Supplementary material 3). One plot shows the rarity and maximum amount of each compound, with points sized by the average amount. Another plot shows how mean amounts compare in samples vs. controls. These visual diagnostics can show whether likely floral compounds are erroneously filtered out, or if contaminants are being kept in the current filtering scheme. Whereas this software automates the process of filtering volatiles for large numbers of chromatograms, it is best combined with some manual inspection of chromatograms to ensure that key floral volatiles have not been missed, for example due to overlap of peaks, and with consideration of previously known sources of volatiles (e.g., Chapurlat et al., 2019).
Statistical analysis
Even after filtering out contaminants, floral scent bouquets often contain a large number of compounds. Scent data, like other forms of metabolomics (Hall, 2006), are thus inherently multivariate (van Dam and Poppy, 2008). The high dimensionality presents challenges for statistical analysis, but these are not unlike those in some other biological fields such as species composition of communities (Anderson and Willis, 2003) and gene expression (Allison et al., 2006). The appropriate method of analysis can vary as much as the ecological or evolutionary question being asked, so our approach here is to focus on some key points rather than attempting a comprehensive review of methods for analysis.
A very common goal is to determine the compounds that differ between some groups, such as different plant species, different sites, or different experimental treatments. This is often accomplished through some form of ordination, a way of collapsing the multiple dimensions that differentiate the groups into a smaller number of interpretable dimensions that can be easily visualized in a 2 (or 3) dimensional graph. An important decision to make in choosing an ordination analysis is between using an unconstrained (i.e., unsupervised) method or a constrained (supervised) method. Unconstrained methods find axes that explain variation in the entire data set, whereas constrained methods find axes that explain variation among pre-defined groups (Anderson and Willis, 2003). Published studies of floral volatiles have tended to emphasize unconstrained methods such as principal components analysis and non-metric dimensional scaling (NMDS), which may be more familiar to some practitioners. These unconstrained methods are most suited to exploratory analyses without a few specific groups to compare. It is important to realize that the axes that explain variation in a full data set may differ greatly from the axes that actually separate the groups of interest. For example, consider the case of comparing two species, in which case constrained ordination is most appropriate. If variation reflects largely variation in total emissions, then an unconstrained ordination will give relatively equal weights to all compounds, even if the primary difference between species is in a specific compound. In the example in Figure 2, variation within species is similar for both volatiles (covariance is equal in both dimensions), yielding a principal component that reflects total emissions and weights VOC 1 and 2 similarly. It does a poor job describing the differences between the species. But the canonical discriminant function correctly captures the species difference (larger mean for VOC 2 and smaller mean for VOC 1 for species 2 compared to species 1), and separates the two species well. Such a situation will occur whenever within-species and between-species variation show different patterns and illustrates the importance of performing constrained ordination if the intent is to compare specific groups.
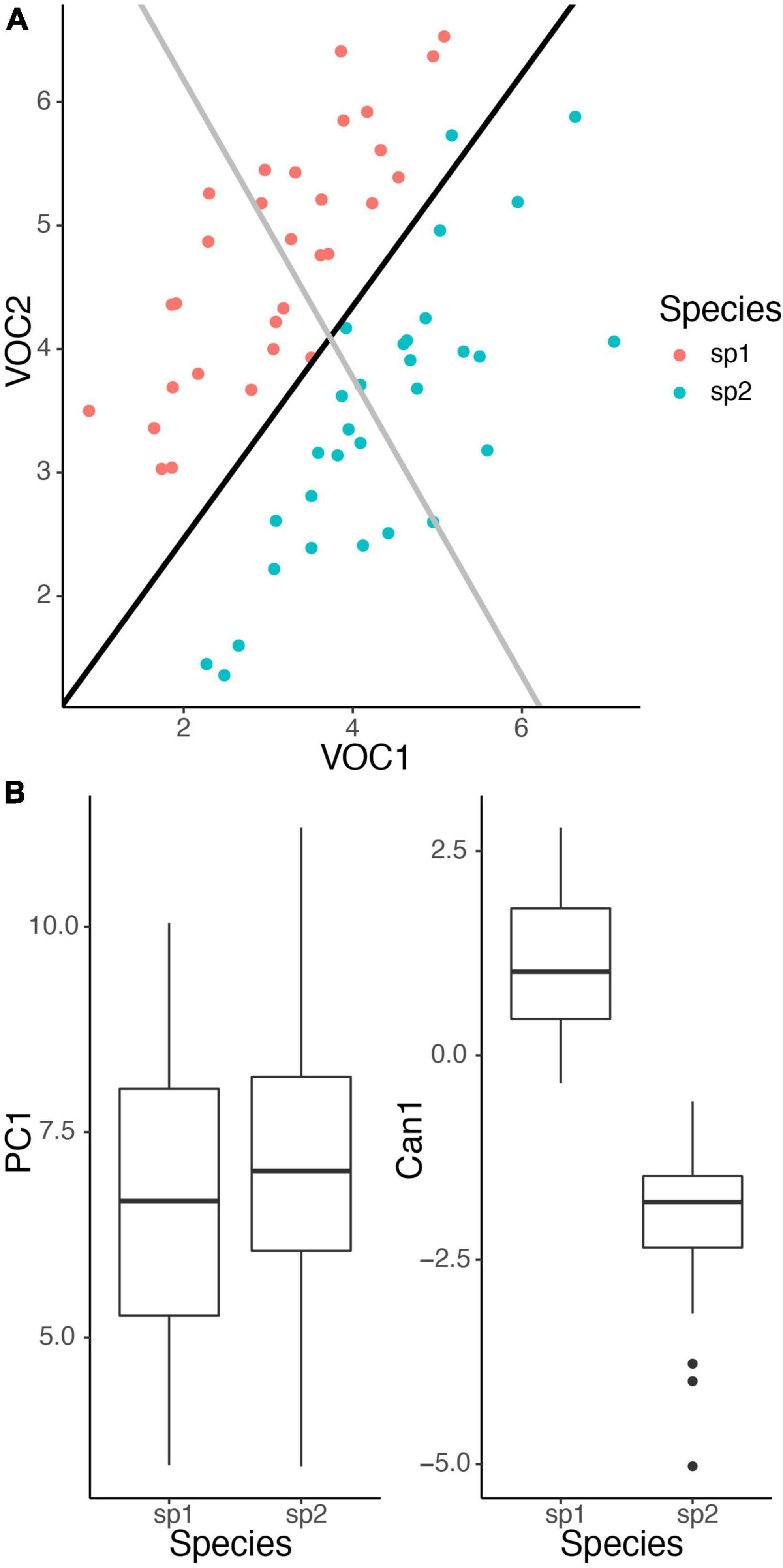
Figure 2. Illustration of unconstrained ordination vs. constrained ordination. Species 1 (sp1) has lower average emissions of VOC 1 (mean = 3) and higher average emissions of VOC 2 (mean = 4.5) than species 2 (sp2) (means = 4.5 and 3.5). Within both species emissions of the two volatiles follow a multivariate normal distribution with covariance between the volatiles equal to 0.75. (A) Unconstrained ordination with principal components analysis (black line) reflects total emissions, whereas constrained ordination with canonical discriminant analysis (CDA) (gray line) correctly captures the difference between the species. (B) Boxplots for ordination scores. Projecting the points onto the two axes shows that only CDA separates the two species well. All analyses performed in R.
Both unconstrained and constrained methods of ordination can be divided further into methods that are based on traditional parametric statistics vs. non-parametric methods (Table 1). The parametric methods of principal component analysis (PCA) and canonical discriminant analysis (CDA) are based on eigenvector analysis using Euclidean distances to define separation. For these parametric methods, log or square root transformation of the data may make distributions of residuals closer to fitting the assumption of multivariate normality. In contrast, the methods of NMDS and canonical analysis of principal coordinates (CAP or distance-based redundancy analysis) do not make such rigid distributional assumptions, as they typically use permutation tests for significance testing (e.g., in the R package “vegan”; Legendre et al., 2011). Another potential benefit of these methods is that they can also use other distance measures to define dissimilarity of two samples, such as Bray-Curtis distance (Legendre and Anderson, 1999). For both reasons, they are often more suitable for situations in which a large number of samples have zero for some volatiles, which is typical for GC-MS data. The CAP method is especially valuable for comparing groups because it combines the features of a constrained analysis with lack of distributional assumptions and lack of the need for high sample sizes of traditional CDA (in which the sample size has to exceed the number of volatiles). An alternative to ordination is machine learning methods, such as random forests (Cutler et al., 2007; Ranganathan and Borges, 2010). Random forests takes bootstrap samples and fits a classification tree using binary partitioning based on a small number of variables. Then the observations not included in the original bootstrapping are predicted from the trees to assess a misclassification rate. The importance of a particular variable can be assessed by the difference made to the misclassification rate if it is randomly shuffled. This method is well suited to situations with a very large number of variables (i.e., VOCs), but does not provide the interpretability of methods like PCA and CDA in which axes are linear combinations of variables. Some examples of application to floral volatiles are provided by Parachnowitsch et al. (2012) and Bischoff et al. (2014).
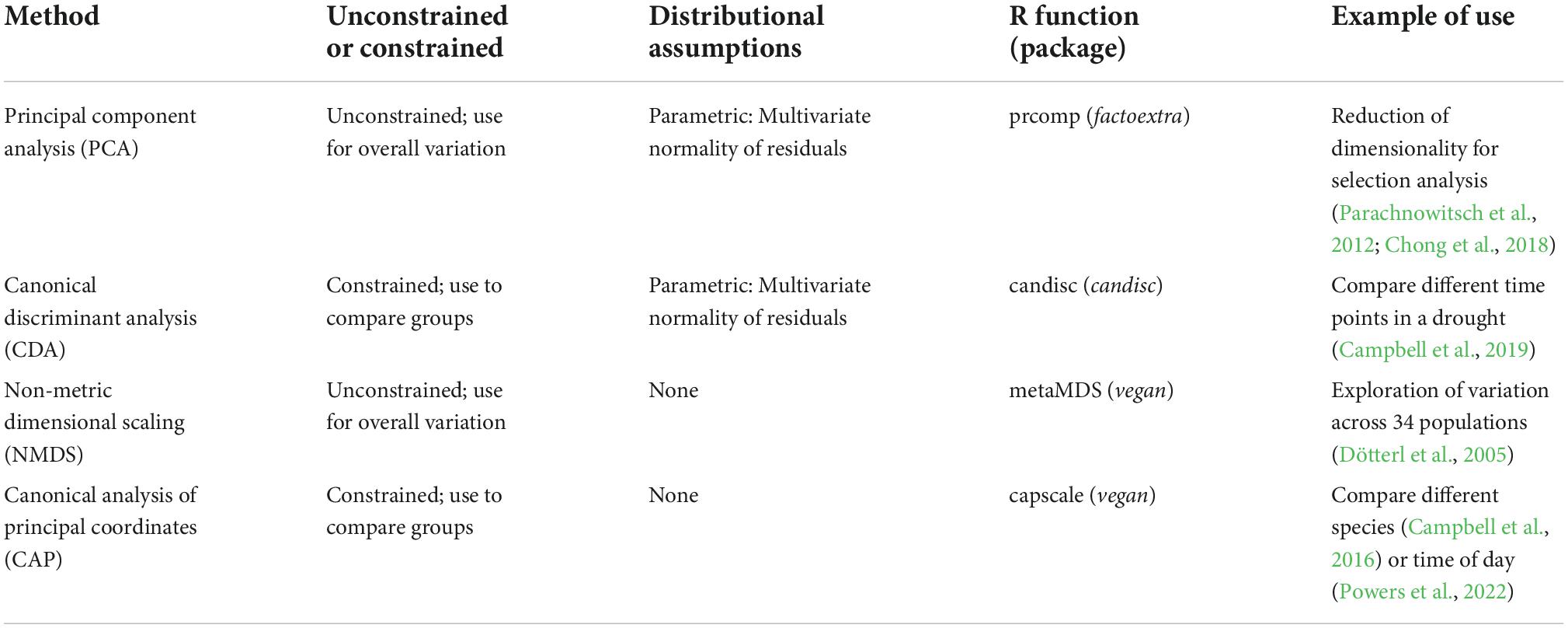
Table 1. Four categories of ordination methods for analyzing multivariate data, along with descriptions of whether the method is unconstrained or constrained, when to use it, distributional assumptions, R functions and packages, and examples of use.
The problem of many variables becomes more acute when the intent is to combine the information on floral volatiles with other complex data, such as plant fitness to investigate natural selection, a phylogeny to investigate macroevolution, DNA markers to investigate genomic architecture, or a plant-pollinator network to investigate impacts on species interactions. Phenotypic selection analysis has long wrestled with the problem of regressing fitness on a high dimensional set of multivariate traits. Some solutions include projection pursuit regression (Schluter and Nychka, 1994) and canonical analysis (Blows and Brooks, 2003), as applied to floral traits in Campbell et al. (2022a), penalized multivariate regression (Gfrerer et al., 2021), and Bayesian reduced rank regression (Opedal et al., 2022), all of which reduce dimensionality by finding new axes that explain variation in fitness. The latter method allows back-transforming a selection gradient on the constructed axis to estimate selection gradients on the original VOCs. A dimensionality reduction method for unconstrained analysis back-transforms selection gradients onto principal components (Chong et al., 2018). Similar issues arise with testing for phylogenetic signature of floral scents. Several recent papers used phylogenetic principal components analysis to identify the global and local phylogenetic structure of floral scents (Prieto-Benítez et al., 2016; Joffard et al., 2020). Multivariate comparative methods are now available to test evolutionary hypotheses about scent composition in a phylogenetic framework (Adams, 2014; Goolsby, 2017). Studies of genomic architecture have largely focused on one or a few volatile compounds (Box 1, section 1A) and so have not yet dealt with integrating extensive molecular marker data with the whole suite of VOC emissions.
Examination of the role of scent in generating links in plant-pollinator networks is particularly challenging because both scent and the links between plants and pollinators are of high dimension. Few investigators have taken on this challenge (Junker et al., 2010). One remarkable study of a Mediterranean scrubland had 41 plant species and 168 species of insect visitors to flowers (Kantsa et al., 2018), with 10,400 links between individual VOCs and insect visitation (Kantsa et al., 2019). Analysis shows two general ways that such a large number of links can be handled. The first approach is to collapse floral volatiles into groups chosen on the basis of prior knowledge, which could reflect biochemical pathways (Junker, 2017) or prior knowledge of perception on the part of the visitors, such as the use of electroantennogram responses to identify volatiles to keep in the analysis (Schiestl et al., 2011; Chapurlat et al., 2019). In this case, the authors grouped VOCs into the chemical classes of aliphatics, benzenoids, phenylpropanoids, monoterpenes, and sesquiterpenes (Kantsa et al., 2018). They then treated visitation by different insect species as a multivariate response vector, using plant traits (including the chemical classes of scent) of different plant species as predictors in a multivariate generalized linear model. The second approach is to use statistical methods to group links, in this case illustrated by using network methods to identify modules in the bipartite network of VOCs and insects (Guimera and Amaral, 2005). By looking at both the volatile composition and the insect composition of each module it was possible to detect associations, with one module, for example, containing both a high proportion of benzenoids and a high proportion of visits by bees (Kantsa et al., 2019). Incorporation of floral volatiles into networks is still in its infancy. In general for analysis of floral volatiles, there is room for development and consideration of other statistical categorization methods suitable for sparse data in which there are fewer samples than parameters (Cao et al., 2011).
Adding biochemical context
After an analysis has identified volatiles of interest, modern chemical databases can be leveraged to add information on structures, biochemical pathways, and ecological and phylogenetic context. Basic chemical information including structures, nomenclature, physical data, and links to relevant studies is accessible through PubChem, ChEBI, and ChemSpider, which all have scripting interfaces for bulk queries (Szöcs et al., 2020). Metabolic pathways for thousands of plant volatiles can be cross-referenced in databases such as MetaCyc (Caspi et al., 2020) and KEGG (Kanehisa and Goto, 2000), which cover thousands of pathways across a large taxonomic breadth. This information is useful in identifying the general pathway that synthesizes a volatile (terpenoid, fatty acid, shikimate, etc.), the specific enzymes involved in its synthesis, and other volatiles that originate from the same pathway. This knowledge can be leveraged in biosynthetic distance measures between samples or species (Junker, 2017) that take into account the non-independent origin of volatiles from shared pathways. Databases such as SCENTbase (Knudsen et al., 2006), SuperScent (Dunkel et al., 2009), and AromaDb (Kumar et al., 2018) provide curated occurrence data across plant species, or in the case of mVOC (Lemfack et al., 2018), from microbes that may be found on plant surfaces. The PheroBase (El-Sayed, 2021) aggregates information on plant occurrences as well as documented use of volatiles as behavioral signals in insects. We encourage authors to weave this contextual data into their analyses and interpretation to highlight connections among the diversity of volatiles emitted by plants.
Conclusion
Over the last 25 years, methodological and conceptual advances have led to the rapid expansion of the field of floral volatiles research. While studies were previously focused on identifying and describing variation in scent in isolation, the current challenge for the field is to determine what contributes to variation in floral scent, what consequences this variation has for species interactions, and how such variation evolves. These frontiers in floral scent research have the potential to generate broadly relevant insights into the evolution of complex traits and the genetic architecture and ecological implications of trait variation. However, in order for the next-generation of floral scent research to achieve these goals, studies must become more reproducible and transparent to facilitate comparisons across systems, and to enable non-specialists to contribute to the field. Here we present a set of best practices for next-generation research in floral volatiles, from experimental design to statistical analysis and interpretation. In particular, we highlight the need for more reproducible workflows in the data processing stage, and introduce an R package, bouquet, to systematize the data cleaning steps that lead to the production of a final dataset. While our guide is focused on floral volatiles research, the considerations and recommendations presented here are also relevant to many other fields that generate high-throughput, multi-dimensional data.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://github.com/jmpowers/bouquet.
Author contributions
KE and RR conceived of the study. KE led the literature review with contributions from JP. JP created the R package and vignette with contributions from KE and DC. All authors contributed to the design of the study, drafted sections of the manuscript, contributed to the manuscript revision, read, and approved the submitted version.
Funding
This manuscript was inspired by our collective experiences reading the literature of this growing field, and by RR and DC’s experiences teaching the Volatile Analysis at RMBL–Workshop and Lab-to-Field Training Course, funded by NSF DBI-1624073. KE was partially supported by an NSF Graduate Research Fellowship (DGE-1650441) and an NSF Postdoctoral Fellowship in Biology (DBI-2007075). JP and DC were partially supported by funding from NSF-DEB-1654655 and NSF-DEB-2135270. Open access publication costs were covered by Lund University APC Fund (20220204).
Acknowledgments
We thank SD and two reviewers for their constructive feedback on the manuscript. We also thank Valerie Martin, David Hopp, and members of the 2022 RMBL Volatiles Analysis Course for testing bouquet and providing feedback.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with one of the authors, RR.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1006416/full#supplementary-material
Supplementary material 1 | The rationale, methods, and results of a literature review of the transparency of floral scent methods in studies published between 2005 and 2019.
Supplementary material 2 | The studies included in the literature review described in Supplementary material 1.
Supplementary material 3 | Typical non-biological contaminants common to GC-MS studies of plant volatiles.
Supplementary material 4 | Introductory vignette for bouquet package.
References
Adams, D. C. (2014). A method for assessing phylogenetic least squares models for shape and other high-dimensional multivariate data. Evolution 68, 2675–2688. doi: 10.1111/evo.12463
Adams, R. P. (2001). Identification of Essential Oil Components by Gas Chromatography/Quadrupole Mass Spectroscopy. Carol Stream, IL: Allured Publishing Corporation.
Allison, D. B., Cui, X., Page, G. P., and Sabripour, M. (2006). Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 7, 55–65. doi: 10.1038/nrg1749
Amirav, A., and Dagan, S. (1997). A direct sample introduction device for mass spectrometry studies and gas chromatography mass spectrometry analyses. Eur. J. Mass Spectrom. 3:105. doi: 10.1255/ejms.27
Anderson, M. J., and Willis, T. J. (2003). Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 84, 511–525. doi: 10.1890/0012-96582003084[0511:CAOPCA]2.0.CO;2
Arguello, J. R., Sellanes, C., Lou, Y. R., and Raguso, R. A. (2013). Can yeast (S. cerevisiae) metabolic volatiles provide polymorphic signaling? PLoS One 8:e70219. doi: 10.1371/journal.pone.0070219
Ayasse, M., Gögler, J., and Stökl, J. (2010). “Pollinator-driven speciation in sexually deceptive orchids of the genus Ophrys,” in Evolution in Action: Case studies in Adaptive Radiation, Speciation and the Origin of Biodiversity, ed. M. Glaubrecht (Berlin: Springer-Verlag), 101–118.
Babushok, V. I. (2015). Chromatographic retention indices in identification of chemical compounds. Trends Analyt. Chem. 69, 98–104. doi: 10.1016/j.trac.2015.04.001
Babushok, V. I., Linstrom, P. J., and Zenkevich, I. G. (2011). Retention indices for frequently reported compounds of plant essential oils. J. Phys. Chem. Ref. Data 40, 043101. doi: 10.1063/1.3653552
Baldwin, I. T., Preston, C., Euler, M., and Gorham, D. (1997). Patterns and consequences of benzyl acetone floral emissions from Nicotiana attenuata plants. J. Chem. Ecol. 23, 2327–2343. doi: 10.1023/B:JOEC.0000006677.56380.cd
Bartlett, J. C., Ishimura, Y., and Kloda, L. A. (2011). Why choose this one? Factors in scientists’ selection of bioinformatics tools. Inf. Res. 16, 1.
Bischoff, M., Jürgens, A., and Campbell, D. R. (2014). Floral scent in natural hybrids of Ipomopsis (Polemoniaceae) and their parental species. Ann. Bot. 113, 533–544. doi: 10.1093/aob/mct279
Bischoff, M., Raguso, R. A., Jurgens, A., and Campbell, D. R. (2015). Context-dependent reproductive isolation mediated by floral scent and color. Evolution 69, 1–13. doi: 10.1111/evo.12558
Blows, M. W., and Brooks, R. (2003). Measuring nonlinear selection. Am. Natural. 162, 815–820. doi: 10.1086/378905
Braunschmid, H., Guilhot, R., and Dötterl, S. (2021). Floral scent and pollinators of Cypripedium calceolus L. at different latitudes. Diversity 13:5. doi: 10.3390/d13010005
Braunschmid, H., Mükisch, B., Rupp, T., Schäffler, I., Zito, P., Birtele, D., et al. (2017). Interpopulation variation in pollinators and floral scent of the lady’s-slipper orchid Cypripedium calceolus L. Arthropod-Plant Interact. 11, 363–379. doi: 10.1007/s11829-017-9512-x
Burdon, R. C. F., Junker, R. R., Scofield, D. G., and Parachnowitsch, A. L. (2018). Bacteria colonising Penstemon digitalis show volatile and tissue-specific responses to a natural concentration range of the floral volatile linalool. Chemoecology 28, 11–19. doi: 10.1007/s00049-018-0252-x
Burger, H., Dötterl, S., Häberlein, C. M., Schulz, S., and Ayasse, M. (2012). An arthropod deterrent attracts specialised bees to their host plants. Oecologia 168, 727–736. doi: 10.1007/s00442-011-2136-4
Burkle, L. A., and Runyon, J. B. (2016). Drought and leaf herbivory influence floral volatiles and pollinator attraction. Glob. Change Biol. 22, 1644–1654. doi: 10.1111/gcb.13149
Burkle, L. A., and Runyon, J. B. (2019). Floral volatiles structure plant–pollinator interactions in a diverse community across the growing season. Funct. Ecol. 33, 2116–2129. doi: 10.1111/1365-2435.13424
Byers, K. J. R. P., Vela, J. P., Peng, F., Riffell, J. A., and Bradshaw, H. D. (2014). Floral volatile alleles can contribute to pollinator-mediated reproductive isolation in monkeyflowers (Mimulus). Plant J. Cell Mol. Biol. 80, 1031–1042. doi: 10.1111/tpj.12702
Campbell, D. R., Bischoff, M., Raguso, R. A., Briggs, H. M., and Sosenski, P. (2022a). Selection of floral traits by pollinators and seed predators during sequential life history stages. Am. Natural. 199, 808–823. doi: 10.1086/716740
Campbell, D. R., Jürgens, A., and Johnson, S. D. (2016). Reproductive isolation between Zaluzianskya species: The influence of volatiles and flower orientation on hawkmoth foraging choices. N. Phytol. 210, 333–342. doi: 10.1111/nph.13746
Campbell, D. R., Raguso, R. A., Midzik, M., Bischoff, M., and Broadhead, G. T. (2022b). Genetic and spatial variation in vegetative and floral traits across a hybrid zone. Am. J. Bot. doi: 10.1002/ajb2.16067
Campbell, D. R., Sosenski, P., and Raguso, R. A. (2019). Phenotypic plasticity of floral volatiles in response to increasing drought stress. Ann. Bot. 123, 601–610. doi: 10.1093/aob/mcy193
Cao, K. L., Boitard, S., and Besse, P. (2011). Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12:253. doi: 10.1186/1471-2105-12-253
Caspi, R., Billington, R., Keseler, I. M., Kothari, A., Krummenacker, M., Midford, P. E., et al. (2020). The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 48, D445–D453. doi: 10.1093/nar/gkz862
Cellini, A., Giacomuzzi, V., Donati, I., Farneti, B., Rodriguez-Estrada, M. T., Savioli, S., et al. (2019). Pathogen-induced changes in floral scent may increase honeybee-mediated dispersal of Erwinia amylovora. ISME J. 13, 847–859. doi: 10.1038/s41396-018-0319-2
Chapurlat, E., Ågren, J., Anderson, J., Friberg, M., and Sletvold, N. (2019). Conflicting selection on floral scent emission in the orchid Gymnadenia conopsea. N. Phytol. 222, 2009–2022. doi: 10.1111/nph.15747
Chong, V. K., Fung, H. F., and Stinchcombe, J. R. (2018). A note on measuring natural selection on principal component scores. Evol. Lett. 2, 272–280. doi: 10.1002/evl3.63
Clifford, M. R. (2017). Scents and Sense Ability: The Evolution and Role of Chemical Cues in the Pollination and Herbivory of Passiflora. Available online at: https://digital.lib.washington.edu:443/researchworks/handle/1773/39959 (accessed January 8, 2019).
Coble, J. B., and Fraga, C. G. (2014). Comparative evaluation of preprocessing freeware on chromatography/mass spectrometry data for signature discovery. J. Chromatogr. A 1358, 155–164. doi: 10.1016/j.chroma.2014.06.100
Courtois, E. A., Paine, C. E. T., Blandinieres, P.-A., Stien, D., Bessiere, J.-M., Houel, E., et al. (2009). Diversity of the volatile organic compounds emitted by 55 species of tropical trees: A survey in French Guiana. J. Chem. Ecol. 35:1349. doi: 10.1007/s10886-009-9718-1
Cutler, D. R., Edwards, T. C., Beard, K. H., Cutler, A., Hess, K. T., Gibson, J., et al. (2007). Random forests for classification in ecology. Ecology 88, 2783–2792. doi: 10.1890/07-0539.1
Delle-Vedove, R., Schatz, B., and Dufay, M. (2017). Understanding intraspecific variation of floral scent in light of evolutionary ecology. Ann. Bot. 120, 1–20. doi: 10.1093/aob/mcx055
Dobson, H. E. M., Raguso, R. A., Knudsen, J. T., and Ayasse, M. (2005). “Scent as an attractant,” in Practical Pollination Biology, eds A. Dafni, P. G. Kevan, and B. C. Husband (Cambridge: Enviroquest Ltd), 230.
Domingo-Almenara, X., Brezmes, J., Vinaixa, M., Samino, S., Ramirez, N., Ramon-Krauel, M., et al. (2016). eRah: A computational tool integrating spectral deconvolution and alignment with quantification and identification of metabolites in GC/MS-based metabolomics. Anal. Chem. 88, 9821–9829. doi: 10.1021/acs.analchem.6b02927
Dötterl, S., Burkhardt, D., Jürgens, A., and Mosandl, A. (2007). Stereoisomeric pattern of lilac aldehyde in Silene latifolia, a plant involved in a nursery pollination system. Phytochemistry 68, 499–504. doi: 10.1016/j.phytochem.2006.11.013
Dötterl, S., Wolfe, L. M., and Jürgens, A. (2005). Qualitative and quantitative analyses of flower scent in Silene latifolia. Phytochemistry 66, 203–213. doi: 10.1016/j.phytochem.2004.12.002
Doubleday, L. A. D., Raguso, R. A., and Eckert, C. G. (2013). Dramatic vestigialization of floral fragrance across a transition from outcrossing to selfing in Abronia umbellata (Nyctaginaceae). Am. J. Bot. 100, 2280–2292. doi: 10.3732/ajb.1300159
Du, X., and Zeisel, S. H. (2013). Spectral deconvolution for gas chromatography mass spectrometry-based metabolomics: Current status and future perspectives. Comput. Struct. Biotechnol. J. 4:e201301013. doi: 10.5936/csbj.201301013
Dunkel, M., Schmidt, U., Struck, S., Berger, L., Gruening, B., Hossbach, J., et al. (2009). SuperScent—a database of flavors and scents. Nucleic Acids Res. 37, D291–D294. doi: 10.1093/nar/gkn695
Ehrlén, J., Borg-Karlson, A.-K., and Kolb, A. (2012). Selection on plant optical traits and floral scent: Effects via seed development and antagonistic interactions. Basic Appl. Ecol. 13, 509–515. doi: 10.1016/j.baae.2012.08.001
Eisen, K. E., Geber, M. A., and Raguso, R. A. (2022). Emission rates of species-specific volatiles vary across communities of Clarkia species: Evidence for multimodal character displacement. Am. Natural. 199, 824–840. doi: 10.1086/715501
El-Sayed, A. M. (2021). The PheroBase: Database of Insect Pheromones and Semiochemicals. Available online at: http://www.pherobase.com (accessed September 10, 2021).
Esposito, F., Vereecken, N. J., Gammella, M., Rinaldi, R., Laurent, P., and Tyteca, D. (2018). Characterization of sympatric Platanthera bifolia and Platanthera chlorantha (Orchidaceae) populations with intermediate plants. PeerJ 6:e4256. doi: 10.7717/peerj.4256
Euler, M., and Baldwin, I. T. (1996). The chemistry of defense and apparency in the corollas of Nicotiana attenuata. Oecologia 107, 102–112. doi: 10.1007/BF00582240
Farré-Armengol, G., Fernández-Martínez, M., Filella, I., Junker, R. R., and Peñuelas, J. (2020). Deciphering the biotic and climatic factors that influence floral scents: A systematic review of floral volatile emissions. Front. Plant Sci. 11:1154. doi: 10.3389/fpls.2020.01154
Farré-Armengol, G., Filella, I., Llusià, J., Niinemets, Ü, and Peñuelas, J. (2014). Changes in floral bouquets from compound-specific responses to increasing temperatures. Glob. Change Biol. 20, 3660–3669. doi: 10.1111/gcb.12628
Feulner, M., Pointner, S., Heuss, L., Aas, G., Paule, J., and Dötterl, S. (2014). Floral scent and its correlation with AFLP data in Sorbus. Organ. Divers. Evol. 14, 339–348. doi: 10.1007/s13127-014-0180-8
Filella, I., Primante, C., Llusià, J., Martín González, A. M., Seco, R., Farré-Armengol, G., et al. (2013). Floral advertisement scent in a changing plant-pollinators market. Sci. Rep. 3:3434. doi: 10.1038/srep03434
Friberg, M., Schwind, C., Guimarães, P. R., Raguso, R. A., and Thompson, J. N. (2019). Extreme diversification of floral volatiles within and among species of Lithophragma (Saxifragaceae). Proc. Natl. Acad. Sci. U.S.A. 116, 4406–4415. doi: 10.1073/pnas.1809007116
Galen, C., Kaczorowski, R., Todd, S. L., Geib, J., and Raguso, R. A. (2011). Dosage-dependent impacts of a floral volatile compound on pollinators, larcenists, and the potential for floral evolution in the alpine skypilot Polemonium viscosum. Am. Natural. 177, 258–272. doi: 10.1086/657993
García, Y., Friberg, M., and Parachnowitsch, A. L. (2021). Spatial variation in scent emission within flowers. Nordic J. Bot. 39, e3014. doi: 10.1111/njb.03014
Gervasi, D. D. L., and Schiestl, F. P. (2017). Real-time divergent evolution in plants driven by pollinators. Nat. Commun. 8:14691. doi: 10.1038/ncomms14691
Gervasi, D. D. L., Selosse, M. A., Sauve, M., Francke, W., Vereecken, N. J., Cozzolino, S., et al. (2017). Floral scent and species divergence in a pair of sexually deceptive orchids. Ecol. Evol. 7, 6023–6034. doi: 10.1002/ece3.3147
Gfrerer, E., Laina, D., Gibernau, M., Fuchs, R., Happ, M., Tolasch, T., et al. (2021). Floral scents of a deceptive plant are hyperdiverse and under population-specific phenotypic selection. Front. Plant Sci. 12:1910. doi: 10.3389/fpls.2021.719092
Glenny, W. R., Runyon, J. B., and Burkle, L. A. (2018). Drought and increased CO2 alter floral visual and olfactory traits with context-dependent effects on pollinator visitation. N. Phytol. 220, 785–798. doi: 10.1111/nph.15081
Goodrich, K. R., Zjhra, M. L., Ley, C. A., and Raguso, R. A. (2006). When flowers smell fermented: The chemistry and ontogeny of yeasty floral scent in pawpaw (Asimina triloba: Annonaceae). Int. J. Plant Sci. 167, 33–46. doi: 10.1086/498351
Goolsby, E. W. (2017). Rapid maximum likelihood ancestral state reconstruction of continuous characters: A rerooting-free algorithm. Ecol. Evol. 7, 2791–2797. doi: 10.1002/ece3.2837
Gross, K., Sun, M., and Schiestl, F. P. (2016). Why do floral perfumes become different? Region-specific selection on floral scent in a terrestrial orchid. PLoS One 11:e0147975. doi: 10.1371/journal.pone.0147975
Guimera, R., and Amaral, L. A. N. (2005). Functional cartography of complex metabolic networks. Nature 433, 895–900. doi: 10.1038/nature03288
Hall, R. D. (2006). Plant metabolomics: From holistic hope, to hype, to hot topic. N. Phytol. 169, 453–468. doi: 10.1111/j.1469-8137.2005.01632.x
Helletsgruber, C., Dötterl, S., Ruprecht, U., and Junker, R. R. (2017). Epiphytic bacteria alter floral scent emissions. J. Chem. Ecol. 43, 1073–1077. doi: 10.1007/s10886-017-0898-9
Hoballah, M. E., Gubitz, T., Stuurman, J., Broger, L., Barone, M., Mandel, T., et al. (2007). Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19, 779–790. doi: 10.1105/tpc.106.048694
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., et al. (2010). MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714. doi: 10.1002/jms.1777
Huang, M., Sanchez-Moreiras, A. M., Abel, C., Sohrabi, R., Lee, S., Gershenzon, J., et al. (2012). The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. N. Phytol. 193, 997–1008. doi: 10.1111/j.1469-8137.2011.04001.x
Hummel, J., Strehmel, N., Bölling, C., Schmidt, S., Walther, D., and Kopka, J. (2013). “Mass spectral search and analysis using the Golm metabolome database,” in The Handbook of Plant Metabolomics, eds W. Weckwerth and G. Kahl (Hoboken, NJ: John Wiley and Sons, Ltd), 321–343. doi: 10.1002/9783527669882.ch18
Jiang, W., Qiu, Y., Ni, Y., Su, M., Jia, W., and Du, X. (2010). An automated data analysis pipeline for GC-TOF-MS metabonomics studies. J. Proteom. Res. 9, 5974–5981. doi: 10.1021/pr1007703
Joffard, N., Arnal, V., Buatois, B., Schatz, B., and Montgelard, C. (2020). Floral scent evolution in the section Pseudophrys: Pollinator-mediated selection or phylogenetic constraints? Plant Biol. 22, 881–889. doi: 10.1111/plb.13104
Johnson, S. D., and Hobbhahn, N. (2010). Generalized pollination, floral scent chemistry, and a possible case of hybridization in the African orchid Disa fragrans. S. Afr. J. Bot. 76, 739–748. doi: 10.1016/j.sajb.2010.07.008
Johnson, S. D., and Jürgens, A. (2010). Convergent evolution of carrion and faecal scent mimicry in fly-pollinated angiosperm flowers and a stinkhorn fungus. S. Afr. J. Bot. 76, 796–807. doi: 10.1016/j.sajb.2010.07.012
Junker, R. R. (2016). “Multifunctional and diverse floral scents mediate biotic interactions embedded in communities,” in Deciphering chemical language of plant communication, signaling and communication in plants, eds J. D. Blande and R. Glinwood (New York, NY: Springer International), 257–282. doi: 10.1007/978-3-319-33498-1
Junker, R. R. (2017). A biosynthetically informed distance measure to compare secondary metabolite profiles. Chemoecology 28, 1–9. doi: 10.1007/s00049-017-0250-4
Junker, R. R., Kuppler, J., Amo, L., Blande, J. D., Borges, R. M., van Dam, N. M., et al. (2018). Covariation and phenotypic integration in chemical communication displays: Biosynthetic constraints and eco-evolutionary implications. New Phytol. 220, 739–749. doi: 10.1111/nph.14505
Junker, R. R., and Blüthgen, N. (2010). Floral scents repel facultative flower visitors, but attract obligate ones. Ann. Bot. 105, 777–782. doi: 10.1093/aob/mcq045
Junker, R. R., Daehler, C. C., Dötterl, S., Keller, A., and Blüthgen, N. (2011). Hawaiian ant–flower networks: Nectar-thieving ants prefer undefended native over introduced plants with floral defenses. Ecological Monographs 81, 295–311. doi: 10.1890/10-1367.1
Junker, R. R., Höcherl, N., and Blüthgen, N. (2010). Responses to olfactory signals reflect network structure of flower-visitor interactions. J. Anim. Ecol. 79, 818–823. doi: 10.1111/j.1365-2656.2010.01698.x
Jürgens, A., and Dötterl, S. (2004). Chemical composition of anther volatiles in Ranunculaceae: Genera-specific profiles in Anemone, Aquilegia, Caltha, Pulsatilla, Ranunculus, and Trollius species. Am. J. Bot. 91, 1969–1980. doi: 10.3732/ajb.91.12.1969
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kantsa, A., Raguso, R. A., Dyer, A. G., Olesen, J. M., Tscheulin, T., and Petanidou, T. (2018). Disentangling the role of floral sensory stimuli in pollination networks. Nat. Commun. 9:1041. doi: 10.1038/s41467-018-03448-w
Kantsa, A., Raguso, R. A., Lekkas, T., Kalantzi, O.-I., and Petanidou, T. (2019). Floral volatiles and visitors: A meta-network of associations in a natural community. J. Ecol. 107, 2574–2586. doi: 10.1111/1365-2745.13197
Katajamaa, M., and Orešič, M. (2007). Data processing for mass spectrometry-based metabolomics. J. Chromatogr. A 1158, 318–328. doi: 10.1016/j.chroma.2007.04.021
Kessler, A., and Halitschke, R. (2009). Testing the potential for conflicting selection on floral chemical traits by pollinators and herbivores: Predictions and case study. Funct. Ecol. 23, 901–912. doi: 10.1111/j.1365-2435.2009.01639.x
Kessler, D., Bing, J., Haverkamp, A., and Baldwin, I. T. (2019). The defensive function of a pollinator-attracting floral volatile. Funct. Ecol. 33, 1223–1232. doi: 10.1111/1365-2435.13332
Kessler, D., Diezel, C., Clark, D. G., Colquhoun, T. A., and Baldwin, I. T. (2013). Petunia flowers solve the defence/apparency dilemma of pollinator attraction by deploying complex floral blends. Ecol. Lett. 16, 299–306. doi: 10.1111/ele.12038
Kessler, D., Gase, K., and Baldwin, I. T. (2008). Field experiments with transformed plants reveal the sense of floral scents. Science 321, 1200–1202. doi: 10.1126/science.1160072
Klahre, U., Gurba, A., Hermann, K., Saxenhofer, M., Bossolini, E., Guerin, P. M., et al. (2011). Pollinator choice in Petunia depends on two major genetic loci for floral scent production. Curr. Biol. 21, 730–739. doi: 10.1016/j.cub.2011.03.059
Knauer, A. C., and Schiestl, F. P. (2017). The effect of pollinators and herbivores on selection for floral signals: A case study in Brassica rapa. Evol. Ecol. 31, 285–304. doi: 10.1007/s10682-016-9878-8
Knauer, A. C., Bakhtiari, M., and Schiestl, F. P. (2018). Crab spiders impact floral-signal evolution indirectly through removal of florivores. Nat. Commun. 9:1367. doi: 10.1038/s41467-018-03792-x
Knudsen, J. T., and Tollsten, L. (1995). Floral scent in bat-pollinated plants: A case of convergent evolution. Botanic. J. Linnean Soc. 119, 45–57. doi: 10.1111/j.1095-8339.1995.tb00728.x
Knudsen, J. T., and Gershenzon, J. (2006). “The chemical diversity of floral scent,” in Biology of floral scent, eds N. Dudareva and E. Pichersky (Boca Raton, FL: CRC Press), 27–52.
Knudsen, J. T., Eriksson, R., Gershenzon, J., and Ståhl, B. (2006). Diversity and distribution of floral scent. Bot. Rev 72:1. doi: 10.1663/0006-8101200672[1:DADOFS]2.0.CO;2
Koh, Y., Pasikanti, K. K., Yap, C. W., and Chan, E. C. Y. (2010). Comparative evaluation of software for retention time alignment of gas chromatography/time-of-flight mass spectrometry-based metabonomic data. J. Chromatogr. A 1217, 8308–8316. doi: 10.1016/j.chroma.2010.10.101
Kumar, Y., Prakash, O., Tripathi, H., Tandon, S., Gupta, M. M., Rahman, L.-U., et al. (2018). AromaDb: A database of medicinal and aromatic plant’s aroma molecules with phytochemistry and therapeutic potentials. Front. Plant Sci. 9:1081. doi: 10.3389/fpls.2018.01081
Lahondère, C., Vinauger, C., Okubo, R. P., Wolff, G. H., Chan, J. K., Akbari, O. S., et al. (2020). The olfactory basis of orchid pollination by mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 117, 708–716. doi: 10.1073/pnas.1910589117
Larue, A.-A. C., Raguso, R. A., and Junker, R. R. (2016). Experimental manipulation of floral scent bouquets restructures flower–visitor interactions in the field. J. Anim. Ecol. 85, 396–408. doi: 10.1111/1365-2656.12441
Legendre, P., and Anderson, M. J. (1999). Distance-based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecol. Monogr. 69, 1–24. doi: 10.1890/0012-96151999069[0001:DBRATM]2.0.CO;2
Legendre, P., Oksanen, J., and ter Braak, C. J. F. (2011). Testing the significance of canonical axes in redundancy analysis. Methods Ecol. Evol. 2, 269–277. doi: 10.1111/j.2041-210X.2010.00078.x
Lemaitre, A. B., Pinto, C. F., and Niemeyer, H. M. (2014). Generalized pollination system: Are floral traits adapted to different pollinators? Arthropod-Plant Interact. 8, 261–272. doi: 10.1007/s11829-014-9308-1
Lemfack, M. C., Gohlke, B.-O., Toguem, S. M. T., Preissner, S., Piechulla, B., and Preissner, R. (2018). mVOC 2.0: A database of microbial volatiles. Nucleic Acids Res. 46, D1261–D1265. doi: 10.1093/nar/gkx1016
Levin, R. A., McDade, L. A., and Raguso, R. A. (2003). The systematic utility of floral and vegetative fragrance in two genera of Nyctaginaceae. Syst. Biol. 52, 334–351. doi: 10.1080/10635150390196975
Lev-Yadun, S., Ne’eman, G., and Shanas, U. (2009). A sheep in wolf’s clothing: Do carrion and dung odours of flowers not only attract pollinators but also deter herbivores? BioEssays 31, 84–88. doi: 10.1002/bies.070191
Linstrom, P. J., and Mallard, W. G. (2001). The NIST chemistry WebBook:? A chemical data resource on the internet. J. Chem. Eng. Data 46, 1059–1063. doi: 10.1021/je000236
Lommen, A. (2009). MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Analyt. Chem. 81, 3079–3086. doi: 10.1021/ac900036d
Luizzi, V. J., Friberg, M., and Petrén, H. (2021). Phenotypic plasticity in floral scent in response to nutrient, but not water, availability in the perennial plant Arabis alpina (Brassicaceae). Funct. Ecol. 35, 1655-1665. doi: 10.1111/1365-2435.13866
Maia, A. C. D., Grimm, C., Schubert, M., Etl, F., Gonçalves, E. G., Do Amaral Ferraz Navarro, D. M., et al. (2019). Novel floral scent compounds from night-blooming Araceae pollinated by cyclocephaline scarabs (Melolonthidae, Cyclocephalini). J. Chem. Ecol. 45, 204–213. doi: 10.1007/s10886-018-1018-1
Majetic, C. J., Castilla, A. R., and Levin, D. A. (2018). Losing a scent of one’s self: Is there a reduction in floral scent emission in self-pollinating Phlox cuspidata versus outcrossing Phlox drummondii? Int. J. Plant Sci. 180, 86–92. doi: 10.1086/701102
Majetic, C. J., Fetters, A. M., Beck, O. M., Stachnik, E. F., and Beam, K. M. (2017). Petunia floral trait plasticity in response to soil nitrogen content and subsequent impacts on insect visitation. Flora 232, 183–193. doi: 10.1016/j.flora.2016.08.002
Majetic, C. J., Raguso, R. A., and Ashman, T.-L. (2009). Sources of floral scent variation: Can environment define floral scent phenotype? Plant Signal. Behav. 4, 129–131. doi: 10.4161/psb.4.2.7628
Mant, J. G., Schiestl, F. P., Peakall, R., and Weston, P. H. (2002). A phylogenetic study of pollinator conservatism among sexually deceptive orchids. Evolution 56, 888–898. doi: 10.1111/j.0014-3820.2002.tb01402.x
Milet-Pinheiro, P., Domingos-Melo, A., Olivera, J. B., Albuquerque, N. S. L., Costa, A. C. G., Albuquerque-Lima, S., et al. (2021). A semivolatile floral scent marks the shift to a novel pollination system in bromeliads. Curr. Biol. 31, 860–868.e4. doi: 10.1016/j.cub.2020.11.012
Millar, J. G. (1998). Rapid and simple isolation of zingiberene from ginger essential oil. J. Natural Prod. 61, 1025–1026. doi: 10.1021/np9800699
Moré, M., Mulieri, P., Battán-Horenstein, M., Cocucci, A. A., and Raguso, R. A. (2019). The role of fetid olfactory signals in the shift to saprophilous fly pollination in Jaborosa (Solanaceae). Arthropod-Plant Interact. 13, 375–386. doi: 10.1007/s11829-018-9640-y
Müller, E., Huber, C. E., Brack, W., Krauss, M., and Schulze, T. (2020). Symbolic aggregate approximation improves gap filling in high-resolution mass spectrometry data processing. Anal. Chem. 92, 10425–10432. doi: 10.1021/acs.analchem.0c00899
Ni, Y., Qiu, Y., Jiang, W., Suttlemyre, K., Su, M., Zhang, W., et al. (2012). ADAP-GC 2.0: Deconvolution of coeluting metabolites from GC/TOF-MS data for metabolomics studies. Analyt. Chem. 84, 6619–6629. doi: 10.1021/ac300898h
Ni, Y., Su, M., Qiu, Y., Jia, W., and Du, X. (2016). ADAP-GC 3.0: Improved peak detection and deconvolution of co-eluting metabolites from GC/TOF-MS data for metabolomics studies. Analyt. Chem. 88, 8802–8811. doi: 10.1021/acs.analchem.6b02222
Niu, W., Knight, E., Xia, Q., and McGarvey, B. D. (2014). Comparative evaluation of eight software programs for alignment of gas chromatography-mass spectrometry chromatograms in metabolomics experiments. J. Chromatogr. A 1374, 199–206. doi: 10.1016/j.chroma.2014.11.005
O’Callaghan, S., De Souza, D. P., Isaac, A., Wang, Q., Hodkinson, L., Olshansky, M., et al. (2012). PyMS: A Python toolkit for processing of gas chromatography-mass spectrometry (GC-MS) data. Application and comparative study of selected tools. BMC Bioinform. 13:115. doi: 10.1186/1471-2105-13-115
Opedal, ØH., Gross, K., Chapurlat, E., Parachnowitsch, A., Joffard, N., Sletvold, N., et al. (2022). Measuring, comparing, and interpreting phenotypic selection on floral scent. J. Evol. Biol. doi: 10.1111/jeb.14103
Owen, S. M., Boissard, C., and Hewitt, C. N. (2001). Volatile organic compounds (VOCs) emitted from 40 Mediterranean plant species: VOC speciation and extrapolation to habitat scale. Atmospheric Environ. 35, 5393–5409. doi: 10.1016/S1352-2310(01)00302-8
Parachnowitsch, A. L., Raguso, R. A., and Kessler, A. (2012). Phenotypic selection to increase floral scent emission, but not flower size or colour in bee-pollinated Penstemon digitalis. N. Phytol. 195, 667–675. doi: 10.1111/j.1469-8137.2012.04188.x
Peakall, R., and Whitehead, M. R. (2014). Floral odour chemistry defines species boundaries and underpins strong reproductive isolation in sexually deceptive orchids. Ann. Bot. 113, 341–355. doi: 10.1093/aob/mct199
Peakall, R., Ebert, D., Poldy, J., Barrow, R. A., Francke, W., Bower, C. C., et al. (2010). Pollinator specificity, floral odour chemistry and the phylogeny of Australian sexually deceptive Chiloglottis orchids: Implications for pollinator-driven speciation. N. Phytol. 188, 437–450. doi: 10.1111/j.1469-8137.2010.03308.x
Peng, F., Byers, K. J. R. P., and Bradshaw, H. D. (2017). Less is more: Independent loss-of-function OCIMENE SYNTHASE alleles parallel pollination syndrome diversification in monkeyflowers (Mimulus). Am. J. Bot. 104, 1055–1059. doi: 10.3732/ajb.1700104
Peñuelas, J., Farré-Armengol, G., Llusia, J., Gargallo-Garriga, A., Rico, L., Sardans, J., et al. (2014). Removal of floral microbiota reduces floral terpene emissions. Sci. Rep. 4:6727. doi: 10.1038/srep06727
Perez-Sanz, F., Ruiz-Hernández, V., Terry, M. I., Arce-Gallego, S., Weiss, J., Navarro, P. J., et al. (2021). gcProfileMakeR: An R package for automatic classification of constitutive and non-constitutive metabolites. Metabolites 11:211. doi: 10.3390/metabo11040211
Petrén, H., Toräng, P., Ågren, J., and Friberg, M. (2021). Evolution of floral scent in relation to self-incompatibility and capacity for autonomous self-pollination in the perennial herb Arabis alpina. Ann. Bot. 127, 737–747. doi: 10.1093/aob/mcab007
Pichersky, E., and Dudareva, N. (2006). “Floral scent metabolic pathways: Their regulation and evolution,” in Biology of Floral Scent, eds E. Pichersky and N. Dudareva (Boca Raton, FL: CRC Press), 55–78.
Pluskal, T., Castillo, S., Villar-Briones, A., and Orešiè, M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 11:395. doi: 10.1186/1471-2105-11-395
Powers, J. M., Sakai, A. K., Weller, S. G., and Campbell, D. R. (2022). Variation in floral volatiles across time, sexes, and populations of wind-pollinated Schiedea globosa. Am. J. Bot. 109, 345–360. doi: 10.1002/ajb2.1820
Prieto-Benítez, S., Millanes, A. M., Dötterl, S., and Giménez-Benavides, L. (2016). Comparative analyses of flower scent in Sileneae reveal a contrasting phylogenetic signal between night and day emissions. Ecol. Evol. 6, 7869–7881. doi: 10.1002/ece3.2377
Qualley, A. V., and Dudareva, N. (2014). “Quantification of plant volatiles,” in Plant Metabolism: Methods and Protocols Methods in Molecular Biology, ed. G. Sriram (Totowa, NJ: Humana Press), 41–53. doi: 10.1007/978-1-62703-661-0_4
Raguso, R. A. (2001). “Floral scent, olfaction, and scent-driven foraging behavior,” in Cognitive Ecology of Pollination: Animal Behaviour and Floral Evolution, eds J. D. Thomson and L. Chittka (Cambridge: Cambridge University Press), 83–105. doi: 10.1017/CBO9780511542268.006
Raguso, R. A. (2008a). Start making scents: The challenge of integrating chemistry into pollination ecology. Entomol. Exp. Appl. 128, 196–207. doi: 10.1111/j.1570-7458.2008.00683.x
Raguso, R. A. (2008b). Wake up and smell the roses: The ecology and evolution of floral scent. Annu. Rev. Ecol. Evol. Syst. 39, 549–569. doi: 10.1146/annurev.ecolsys.38.091206.095601
Raguso, R. A. (2012). New synthesis: Exploring the chemical links in ecological food webs. J. Chem. Ecol. 38, 441–441. doi: 10.1007/s10886-012-0128-4
Raguso, R. A. (2014). A wrapped bouquet: The untapped potential of floral chemistry. J. Chem. Ecol. 40, 412–413. doi: 10.1007/s10886-014-0440-2
Raguso, R. A. (2016). More lessons from linalool: Insights gained from a ubiquitous floral volatile. Curr. Opin. Plant Biol. 32, 31–36. doi: 10.1016/j.pbi.2016.05.007
Raguso, R. A. (2020). “Behavioral responses to floral scent: Experimental manipulations and multimodal plant–pollinator communication,” in Biology of Plant Volatiles, eds E. Pichersky and N. Dudareva (Boca Raton, FL: CRC Press).
Raguso, R. A., and Pellmyr, O. (1998). Dynamic headspace analysis of floral volatiles: A comparison of methods. Oikos 81, 238–254. doi: 10.2307/3547045
Raguso, R. A., Schlumpberger, B. O., Kaczorowski, R. L., and Holtsford, T. P. (2006). Phylogenetic fragrance patterns in Nicotiana sections Alatae and Suaveolentes. Phytochemistry 67, 1931–1942. doi: 10.1016/j.phytochem.2006.05.038
Raguso, R. A., Thompson, J. N., and Campbell, D. R. (2015). Improving our chemistry: Challenges and opportunities in the interdisciplinary study of floral volatiles. Nat. Prod. Rep. 32, 893–903. doi: 10.1039/C4NP00159A
Ranganathan, Y., and Borges, R. M. (2010). Reducing the babel in plant volatile communication: Using the forest to see the trees. Plant Biol. 12, 735–742. doi: 10.1111/j.1438-8677.2009.00278.x
Rering, C. C., Beck, J. J., Hall, G. W., McCartney, M. M., and Vannette, R. L. (2018). Nectar-inhabiting microorganisms influence nectar volatile composition and attractiveness to a generalist pollinator. N. Phytol. 220, 750–759. doi: 10.1111/nph.14809
Rering, C. C., Franco, J. G., Yeater, K. M., and Mallinger, R. E. (2020). Drought stress alters floral volatiles and reduces floral rewards, pollinator activity, and seed set in a global plant. Ecosphere 11:e03254. doi: 10.1002/ecs2.3254
Riffell, J. A., Lei, H., and Hildebrand, J. G. (2009). Neural correlates of behavior in the moth Manduca sexta in response to complex odors. Proc. Natl. Acad. Sci. U.S.A. 106, 19219–19226. doi: 10.1073/pnas.0910592106
Rubini Pisano, A., Moré, M., Cisternas, M. A., Raguso, R. A., and Benitez-Vieyra, S. (2019). Breakdown of species boundaries in Mandevilla: Floral morphological intermediacy, novel fragrances and asymmetric pollen flow. Plant Biol. 21, 206–215. doi: 10.1111/plb.12924
Ruiz-Hernández, V., Roca, M. J., Egea-Cortines, M., and Weiss, J. (2018). A comparison of semi-quantitative methods suitable for establishing volatile profiles. Plant Methods 14:67. doi: 10.1186/s13007-018-0335-2
Sas, C., Müller, F., Kappel, C., Kent, T. V., Wright, S. I., Hilker, M., et al. (2016). Repeated inactivation of the first committed enzyme underlies the loss of benzaldehyde emission after the selfing transition in Capsella. Curr. Biol. 26, 3313–3319. doi: 10.1016/j.cub.2016.10.026
Saunier, A., and Blande, J. D. (2019). The effect of elevated ozone on floral chemistry of Brassicaceae species. Environ. Pollut. 255:113257. doi: 10.1016/j.envpol.2019.113257
Schäffler, I., Balao, F., and Dötterl, S. (2012). Floral and vegetative cues in oil-secreting and non-oil-secreting Lysimachia species. Ann. Bot. 110, 125–138. doi: 10.1093/aob/mcs101
Schiestl, F. P., and Marion-Poll, F. (2002). “Detection of physiologically active flower volatiles using gas chromatography coupled with electroantennography,” in Analysis of Taste and Aroma Molecular Methods of Plant Analysis, eds J. F. Jackson and H. F. Linskens (Berlin: Springer), 173–198. doi: 10.1007/978-3-662-04857-3_9
Schiestl, F. P., Balmer, A., and Gervasi, D. D. (2018). Real-time evolution supports a unique trajectory for generalized pollination. Evolution 72, 2653–2668. doi: 10.1111/evo.13611
Schiestl, F. P., Huber, F. K., and Gomez, J. M. (2011). Phenotypic selection on floral scent: Trade-off between attraction and deterrence? Evol. Ecol. 25, 237–248. doi: 10.1007/s10682-010-9409-y
Schlumpberger, B. O., and Raguso, R. A. (2008). Geographic variation in floral scent of Echinopsis ancistrophora (Cactaceae); evidence for constraints on hawkmoth attraction. Oikos 117, 801–814. doi: 10.1111/j.0030-1299.2008.16211.x
Schluter, D., and Nychka, D. (1994). Exploring fitness surfaces. Am. Natural. 143, 597–616. doi: 10.1086/285622
Schlüter, P. M., Xu, S., Gagliardini, V., Whittle, E., Shanklin, J., Grossniklaus, U., et al. (2011). Stearoyl-acyl carrier protein desaturases are associated with floral isolation in sexually deceptive orchids. Proc. Natl. Acad. Sci. U.S.A. 108, 5696–5701. doi: 10.1073/pnas.1013313108
Schulz, S., and Möllerke, A. (2022). MACE – an open access data repository of mass spectra for chemical ecology. J. Chem. Ecol. 48, 589–597. doi: 10.1007/s10886-022-01364-4
Skogen, K. A., Jogesh, T., Hilpman, E. T., Todd, S. L., and Raguso, R. A. (2022). Extensive population-level sampling reveals clinal variation in (R)-(-)-linalool produced by the flowers of an endemic evening primrose, Oenothera harringtonii. Phytochemistry 200:113185. doi: 10.1016/j.phytochem.2022.113185
Smirnov, A., Qiu, Y., Jia, W., Walker, D. I., Jones, D. P., and Du, X. (2019). ADAP-GC 4.0: Application of clustering-assisted multivariate curve resolution to spectral deconvolution of gas chromatography–mass spectrometry metabolomics data. Analyt. Chem. 91, 9069–9077. doi: 10.1021/acs.analchem.9b01424
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R., and Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analyt. Chem. 78, 779–787. doi: 10.1021/ac051437y
Souto-Vilarós, D., Proffit, M., Buatois, B., Rindos, M., Sisol, M., Kuyaiva, T., et al. (2018). Pollination along an elevational gradient mediated both by floral scent and pollinator compatibility in the fig and fig-wasp mutualism. J. Ecol. 106, 2256–2273. doi: 10.1111/1365-2745.12995
Spicer, R., Salek, R. M., Moreno, P., Cañueto, D., and Steinbeck, C. (2017). Navigating freely-available software tools for metabolomics analysis. Metabolomics 13:106. doi: 10.1007/s11306-017-1242-7
Stashenko, E. E., and Martínez, J. R. (2008). Sampling flower scent for chromatographic analysis. J. Sep. Sci. 31, 2022–2031. doi: 10.1002/jssc.200800151
Stashenko, E. E., and Martínez, J. R. (2012). “GC-MS analysis of volatile plant secondary metabolites,” in Gas Chromatography in Plant Science, Wine Technology, Toxicology and Some Specific Applications, eds B. Salih and Ö Çelikbıçak (Rijeka: InTech).
Stein, S. E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10, 770–781. doi: 10.1016/S1044-0305(99)00047-1
Steiner, K. E., Kaiser, R., and Dötterl, S. (2011). Strong phylogenetic effects on floral scent variation of oil-secreting orchids in South Africa. Am. J. Bot. 98, 1663–1679. doi: 10.3732/ajb.1100141
Stewart-Jones, A., and Poppy, G. M. (2006). Comparison of glass vessels and plastic bags for enclosing living plant parts for headspace analysis. J. Chem. Ecol. 32:845. doi: 10.1007/s10886-006-9039-6
Stökl, J., Paulus, H., Dafni, A., Schulz, C., Francke, W., and Ayasse, M. (2005). Pollinator attracting odour signals in sexually deceptive orchids of the Ophrys fusca group. Plant Syst. Evol. 254, 105–120. doi: 10.1007/s00606-005-0330-8
Styczynski, M. P., Moxley, J. F., Tong, L. V., Walther, J. L., Jensen, K. L., and Stephanopoulos, G. N. (2007). Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Analyt. Chem. 79, 966–973. doi: 10.1021/ac0614846
Suinyuy, T. N., Donaldson, J. S., and Johnson, S. D. (2015). Geographical matching of volatile signals and pollinator olfactory responses in a cycad brood-site mutualism. Proc. R. Soc. B Biol. Sci. 282:20152053. doi: 10.1098/rspb.2015.2053
Szöcs, E., Stirling, T., Scott, E. R., Scharmüller, A., and Schäfer, R. B. (2020). webchem: An R package to retrieve chemical information from the web. J. Stat. Softw. 93, 1–17. doi: 10.18637/jss.v093.i13
Tao, Z.-B., Ren, Z. X., Bernhardt, P., Liang, H., Li, H. D., Zhao, Y. H., et al. (2018). Does reproductive isolation reflect the segregation of color forms in Spiranthes sinensis (Pers.) Ames complex (Orchidaceae) in the Chinese Himalayas? Ecol. Evol. 8, 5455–5469. doi: 10.1002/ece3.4067
Tholl, D., and Röse, U. S. R. (2006). “Detection and identification of floral scent compounds,” in Biology of floral scent, eds E. Pichersky and N. Dudareva (Boca Raton, FL: CRC Press), 3–26.
Tholl, D., Hossain, O., Weinhold, A., Röse, U. S. R., and Wei, Q. (2021). Trends and applications in plant volatile sampling and analysis. Plant J. 106, 314–325. doi: 10.1111/tpj.15176
Tholl, D., Weinhold, A., and Röse, U. S. R. (2020). “Practical approaches to plant volatile collection and analysis,” in Biology of Plant Volatiles, eds E. Pichersky and N. Dudareva (Boca Raton, FL: CRC Press).
Tikunov, Y. M., Laptenok, S., Hall, R. D., Bovy, A., and de Vos, R. C. H. (2012). MSClust: A tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8, 714–718. doi: 10.1007/s11306-011-0368-2
Tröger, A., Svensson, G. P., Althoff, D. M., Segraves, K. A., Raguso, R. A., and Francke, W. (2019). The pattern of straight chain hydrocarbons released by Yucca flowers (Asparagaceae). J. Chem. Ecol. 45, 46–49. doi: 10.1007/s10886-018-1037-y
van Dam, N. M., and Poppy, G. M. (2008). Why plant volatile analysis needs bioinformatics: Detecting signal from noise in increasingly complex profiles. Plant Biol. 10, 29–37. doi: 10.1055/s-2007-964961
Van Den Dool, H., and Kratz, P. D. (eds) (1963). A generalization of the retention index system including linear temperature programmed gas-liquid partition chromatography. J. Chromatogr. 11, 463–471. doi: 10.1016/S0021-9673(01)80947-X
Von Helversen, O., Winkler, L., and Bestmann, H. J. (2000). Sulphur-containing “perfumes” attract flower-visiting bats. J. Comp. Physiol. A 186, 143–153. doi: 10.1007/s003590050014
Weber, M. G., Cacho, N. I., Phan, M. J. Q., Disbrow, C., Ramírez, S. R., and Strauss, S. Y. (2018). The evolution of floral signals in relation to range overlap in a clade of California Jewelflowers (Streptanthus s.l.). Evolution 72, 798–807. doi: 10.1111/evo.13456
Wehrens, R., Weingart, G., and Mattivi, F. (2014). metaMS: An open-source pipeline for GC–MS-based untargeted metabolomics. J. Chromatogr. B 966, 109–116. doi: 10.1016/j.jchromb.2014.02.051
Whitehead, M. R., and Peakall, R. (2014). Pollinator specificity drives strong prepollination reproductive isolation in sympatric sexually deceptive orchids. Evolution 68, 1561–1575. doi: 10.1111/evo.12382
Whitehead, M. R., Linde, C. C., and Peakall, R. (2015). Pollination by sexual deception promotes outcrossing and mate diversity in self-compatible clonal orchids. J. Evol. Biol. 28, 1526–1541. doi: 10.1111/jeb.12673
Wiemer, A. P., Moré, M., Benitez-Vieyra, S., Cocucci, A. A., Raguso, R. A., and Sérsic, A. N. (2009). A simple floral fragrance and unusual osmophore structure in Cyclopogon elatus (Orchidaceae). Plant Biol. 11, 506–514. doi: 10.1111/j.1438-8677.2008.00140.x
Williams, N. H., and Whitten, W. M. (1999). Molecular phylogeny and floral fragrances of male euglossine bee-pollinated orchids: A study of Stanhopea (Orchidaceae). Plant Spec. Biol. 14, 129–136. doi: 10.1046/j.1442-1984.1999.00016.x
Xu, S., Schlüter, P. M., Grossniklaus, U., and Schiestl, F. P. (2012). The genetic basis of pollinator adaptation in a sexually deceptive orchid. PLoS Genet. 8:e1002889. doi: 10.1371/journal.pgen.1002889
Yang, M., Deng, G. C., Gong, Y. B., and Huang, S. Q. (2019). Nectar yeasts enhance the interaction between Clematis akebioides and its bumblebee pollinator. Plant Biol. 21, 732–737. doi: 10.1111/plb.12957
Zito, P., Tavella, F., Sajeva, M., Carimi, F., and Dötterl, S. (2018). Inflorescence scents of Calendula maritima, Calendula suffruticosa subsp. fulgida, and their hybrid. Int. J. Plant Sci. 179, 415–421. doi: 10.1086/697240
Keywords: analytical chemistry, chemical ordination, GC-MS, floral scent, data processing, reproducible research, high-dimensional traits
Citation: Eisen KE, Powers JM, Raguso RA and Campbell DR (2022) An analytical pipeline to support robust research on the ecology, evolution, and function of floral volatiles. Front. Ecol. Evol. 10:1006416. doi: 10.3389/fevo.2022.1006416
Received: 29 July 2022; Accepted: 20 September 2022;
Published: 20 October 2022.
Edited by:
Stefan Dötterl, University of Salzburg, AustriaReviewed by:
Adam Shuttleworth, University of KwaZulu-Natal, South AfricaEva Gfrerer, University of Salzburg, Austria
Copyright © 2022 Eisen, Powers, Raguso and Campbell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherine E. Eisen, a2F0aGVyaW5lLmVpc2VuQGJpb2wubHUuc2U=
†These authors have contributed equally to this work
 Katherine E. Eisen
Katherine E. Eisen John M. Powers
John M. Powers Robert A. Raguso
Robert A. Raguso Diane R. Campbell
Diane R. Campbell