- 1Department of Plant Physiology, Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University Jena, Jena, Germany
- 2School of Ecology and Conservation, University of Agricultural Sciences, Gandhi Krishi Vigyana Kendra (GKVK), Bengaluru, India
- 3Department of Crop Physiology, University of Agricultural Sciences, Gandhi Krishi Vigyana Kendra (GKVK), Bengaluru, India
- 4Molecular and Applied Microbiology, Leibniz Institute for Natural Product Research and Infection Biology - Hans Knöll Institute (HKI), Jena, Germany
- 5Institute of Microbiology, Friedrich Schiller University Jena, Jena, Germany
Numerous Trichoderma strains are beneficial for plants, promote their growth, and confer stress tolerance. A recently described novel Trichoderma strain strongly promotes the growth of Arabidopsis thaliana seedlings on media with 50 mM NaCl, while 150 mM NaCl strongly stimulated root colonization and induced salt-stress tolerance in the host without growth promotion. To understand the dynamics of plant-fungus interaction, we examined the secretome from both sides and revealed a substantial change under different salt regimes, and during co-cultivation. Stress-related proteins, such as a fungal cysteine-rich Kp4 domain-containing protein which inhibits plant cell growth, fungal WSC- and CFEM-domain-containing proteins, the plant calreticulin, and cell-wall modifying enzymes, disappear when the two symbionts are co-cultured under high salt concentrations. In contrast, the number of lytic polysaccharide monooxygenases increases, which indicates that the fungus degrades more plant lignocellulose under salt stress and its lifestyle becomes more saprophytic. Several plant proteins involved in plant and fungal cell wall modifications and root colonization are only found in the co-cultures under salt stress, while the number of plant antioxidant proteins decreased. We identified symbiosis- and salt concentration-specific proteins for both partners. The Arabidopsis PYK10 and a fungal prenylcysteine lyase are only found in the co-culture which promoted plant growth. The comparative analysis of the secretomes supports antioxidant enzyme assays and suggests that both partners profit from the interaction under salt stress but have to invest more in balancing the symbiosis. We discuss the role of the identified stage- and symbiosis-specific fungal and plant proteins for salt stress, and conditions promoting root colonization and plant growth.
Introduction
Symbiotic Trichoderma species colonize host plants, often promote their growth and confer tolerance to various (a)biotic stresses (Shoresh et al., 2010; Contreras-Cornejo et al., 2011, 2021; Brotman et al., 2012; Mukherjee et al., 2012; Mangiest, 2020; Tseng et al., 2020), which make them attractive for agricultural applications. We recently described a novel salt-tolerant Trichoderma strain isolated from a stress-exposed Leucas aspera (Wild.) tree which is closely related to T. confertum (Tseng et al., 2020). The fungus also colonized the roots of Arabidopsis thaliana and Nicotiana attenuata, and the interaction of the Trichoderma spp. strain with Arabidopsis seedlings was strongly dependent on the salt concentration in the medium. The fungus promoted the growth of the host plants in soil and media with 50 mM NaCl, while no growth stimulation was observed without salt or concentrations >100 mM. Colonized plants were better protected against pathogen attacks and tolerated higher salt concentrations (Tseng et al., 2020).
The secretomes of beneficial fungi in association with host plants contain stress-, symbiosis-, and defense-specific proteins which orchestrate the diverse responses (Hermosa et al., 2013; Lamdan et al., 2015; Morán-Diez et al., 2015; Druzhinina et al., 2018; Nogueira-Lopez et al., 2018). Likewise, colonized roots respond to stress such as salt, e.g., by activating abscisic acid (ABA)-dependent responses (Kaushal, 2020; van Zelm et al., 2020) and have to balance them with appropriate responses which control root colonization. Overcolonization or aggressive behavior of the microbes activates defense-related responses such as the hormones jasmonic acid or salicylic acid, depending on the mode of interaction between the two partners (Mengistu, 2020). The fungi counteract the plant’s defense by secretion of e.g., carbohydrate-binding enzymes for cell wall degradation, proteases, and mainly uncharacterized small cysteine-rich proteins. They weaken the plant and generate degradation products of plant polymers for the fungal primary metabolism. Secreted plant proteins, which are involved in defense to restrict hyphal propagation in the host, are activated by damage/microbe-associated molecular patterns (D/MAMPs) (Hermosa et al., 2013), such as breakdown products of chitin or β-glucans of the Trichoderma cell wall. Likewise, fungal (chitinases, β-1,3-glucanases, and proteases) and plant (pectinases, cellulases, and xylanases) cell wall hydrolases generate DAMPs by the degradation of their own cell wall or that of the symbiont (Sharon et al., 1993; Seidl et al., 2006; Djonovic et al., 2007; Martinez et al., 2008; Hermosa et al., 2013; Lamdan et al., 2015; Alkooranee et al., 2017; Sarrocco et al., 2017; Silva et al., 2019). In return, fungi secrete effector proteins which inhibit plant defense (Kubicek et al., 2011; Guzmán-Guzmán et al., 2017; Mendoza-Mendoza et al., 2018).
Apart from the battle for establishing conditions for a stable symbiosis, many Trichoderma strains stimulate the host’s growth. They can produce or stimulate the production of plant growth hormones, in particular auxin or auxin-precursors, or promote the nutrient supply from the rhizosphere to the host, or induce alterations in the root architecture which allows better access of the host to nutrients in the soil (Contreras-Cornejo et al., 2009; Hermosa et al., 2012; Samolski et al., 2012; Ruocco et al., 2015; Nieto-Jacobo et al., 2017).
Very little is known about how plant and fungal secreted proteins are involved in these processes. Therefore, we investigated which proteins are secreted by the two partners under the different salt conditions to identify which of them are specific for the beneficial phase (50 mM NaCl), and for the phase when both partners suffer under salt stress (150 mM NaCl). Our results demonstrate that less stress-related proteins are secreted under co-culture conditions whereas the investment in proteins involved in colonization increases.
Results and Discussion
Secretome Analysis of Trichoderma spp. and Arabidopsis thaliana Exposed to Different Salt Concentrations
Previously, we showed that co-cultivation of a Trichoderma strain with Arabidopsis seedlings resulted in growth promotion on media with 50 mM salt whereas, under higher salt concentrations, the fungus conferred salt tolerance to the host (Tseng et al., 2020). To identify plant and fungal proteins with specific functions in the interaction, we co-cultivated the symbionts, either alone or together, in a liquid plant nutrient medium (PNM) with 0, 50, 100, or 150 mM NaCl for 5 days. After centrifugation, the remaining culture media were filtered and proteins were isolated by solid-phase extraction followed by LC-MS/MS (liquid chromatography-tandem mass spectrometry) analysis (cf. section “Materials and Methods”). In total, 722 and 605 proteins from Arabidopsis and Trichoderma spp., respectively, were uniquely identified in all salt concentrations and three independent biological replicates. In the Trichoderma spp. cultures, 236, 114, 102, and 311 proteins were observed in at least 2 of 3 biological replicates in 0, 50, 100, and 150 mM NaCl, respectively, whereas 19, 107, 81, and 119 proteins were observed in the plant cultures. In the co-culture, 82, 135, 160, and 116 different plant proteins and 241, 248, 278, and 170 fungal proteins were observed, respectively, in each salt concentration. Just ten Arabidopsis and 49 Trichoderma spp. proteins were common to all salt concentrations and replicates (cf. data submission).
Classification of Secreted Fungal Proteins
The fungal proteins were classified according to their biological functions. The vast majority of them were uncharacterized (Figure 1). Furthermore, in many cases, a pathway is only represented by a single protein. Therefore, it is difficult to draw general conclusions from the pie diagrams shown in Figure 1 without a more detailed analysis of the individual proteins (cf. below). However, it is obvious that the percentage of proteins with known functions is almost twice as high in the exudate preparation from fungi grown in the co-culture with 50 mM NaCl compared to all other exudate preparations. More importantly, 9% of all identified fungal proteins in the exudate of the co-culture with 150 mM NaCl are lytic polysaccharide monooxygenases (Andlar et al., 2018; Forsberg et al., 2019; Almeida et al., 2021; Calderaro et al., 2021; Jagadeeswaran et al., 2021). These enzymes are major contributors to the recycling of carbon in nature, mainly by degradation of plant lignocellulose (Sagarika et al., 2021; Vuong and Master, 2021; Zerva et al., 2021). They catalyze the oxidative cleavage of glycosidic bonds in polysaccharides such as chitin and cellulose, hemicellulose or starch and their discovery has revolutionized our understanding of enzymatic biomass conversion (Manavalan et al., 2021; Wang et al., 2021). Lytic polysaccharide monooxygenases require oxygen and partners to donate electrons during catalytic action on the cellulose backbone which must be present in the cell wall and released into the apoplastic space (Rani Singhania et al., 2021). Due to their enormous potential for application in agricultural waste decomposition, the current research focusses on biochemical application. However, also a few studies showed that they are involved in plant/microbe interactions (reviewed in Jagadeeswaran et al., 2021). The diagram in Figure 1 shows that lytic polysaccharide monooxygenases increase with increasing salt concentrations in the co-culture. This indicates that the fungus degrades more plant cell wall material under salt stress which might be an indication of a more aggressive behavior of the fungus under stress, associated with a shift toward a more saprophytic lifestyle.
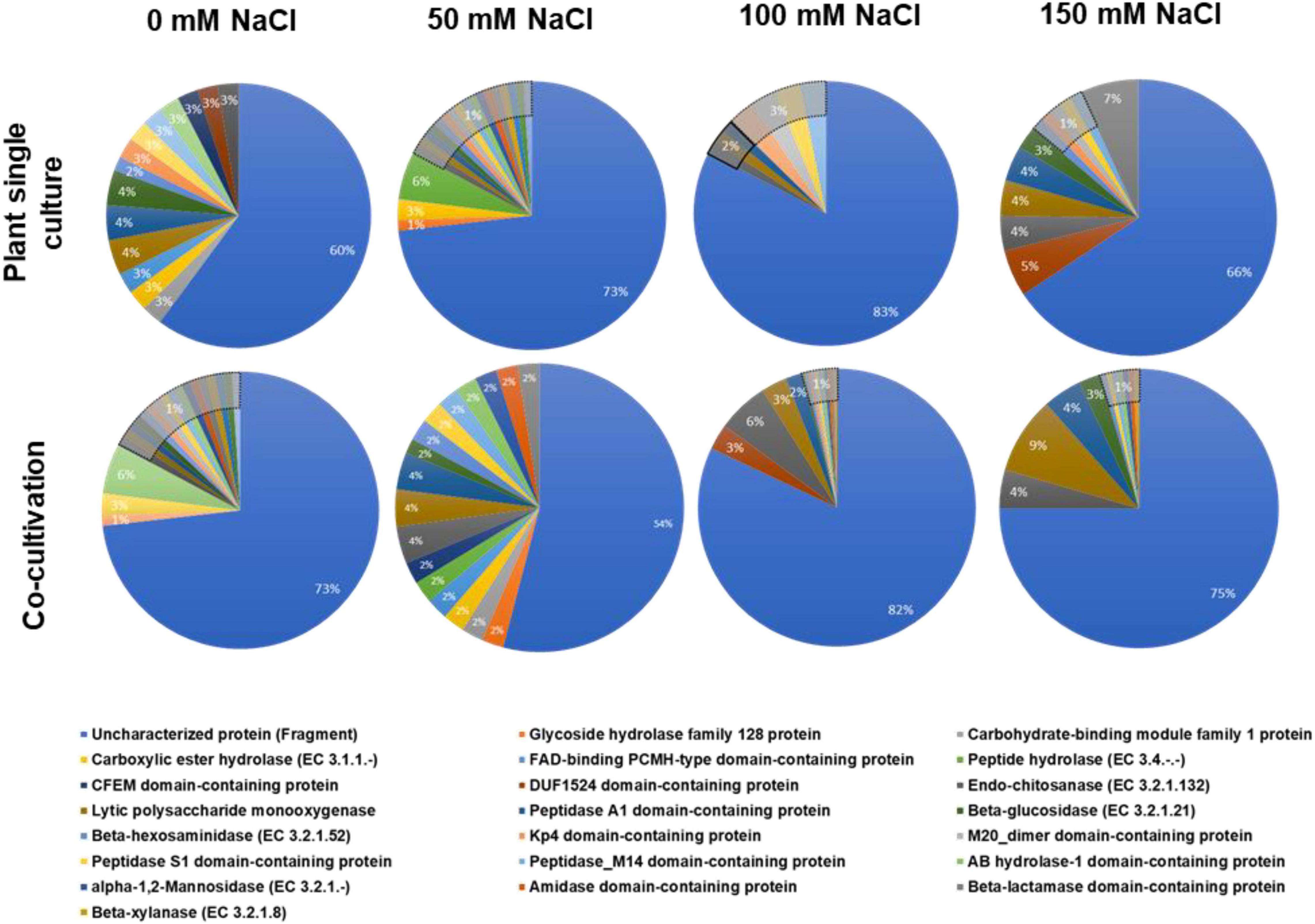
Figure 1. Annotation of the identified fungal proteins according to the biological pathways (GO annotation). The fungus was either grown alone or in co-culture with the plants in media supplemented with 0, 50, 100, or 150 mM NaCl.
Classification of the Secreted Plant Proteins
All identified plant proteins were classified according to their biological functions (Figure 2). Overall inspection of the generated pie diagrams for the plant proteins under the different salt conditions demonstrated that the diversity of the protein functions is much higher in the co-cultures with 0–100 mM salt compared to the diversity of the proteins from plants grown without the fungus (Figure 2). For instance, in media with 50 mM NaCl which favors growth promotion of the host, the identified proteins of plants growing alone belong to 4 categories, whereas those in the co-culture to 17 categories. In media with 150 mM NaCl, the complexity of the identified proteins of the plant growing without the fungus becomes more complex (cf. below), which is consistent with many observations that 150 mM salt induces massive salt stress responses. Overall, the result demonstrates that the fungus has a strong influence on the plant protein pattern, in particular in media with 0–100 mM salt.
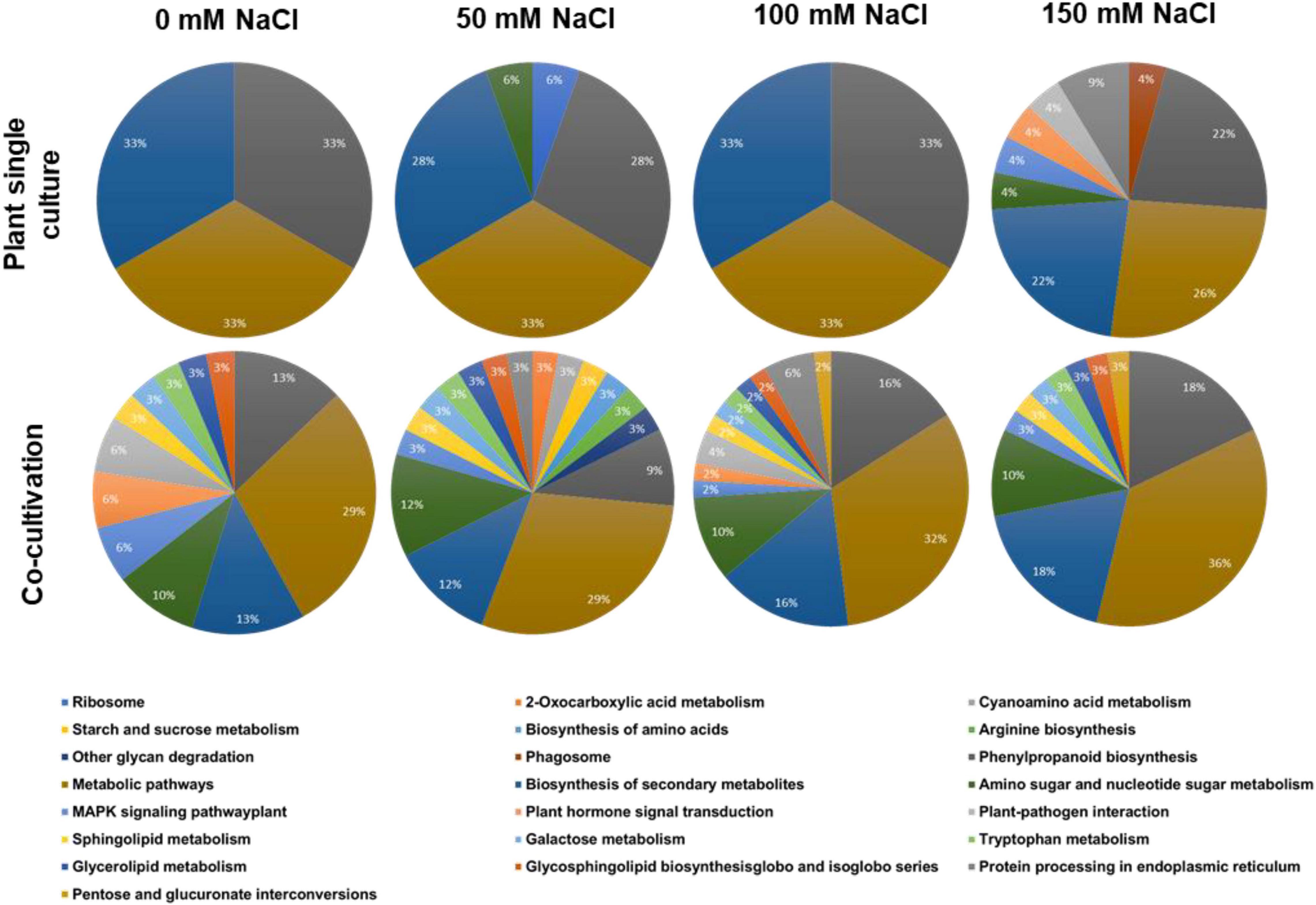
Figure 2. Annotation of the identified plant proteins according to the biological pathways (GO annotation). The plants were either grown alone or in co-culture with the fungus in media supplemented with 0, 50, 100, or 150 mM NaCl.
Among all identified proteins, 193 plant and 199 fungal proteins were predicted to possess an N-terminal secretion sequence. After annotation according to the GO terms “molecular functions,” “biological processes,” and “cellular components,” the 20 most abundant proteins for each salt concentration were further analyzed. The number of proteins annotated as “Response to stress” in Arabidopsis cultures was reduced when Trichoderma was present (Figure 3B). Only 4% of the secreted proteins in the co-culture without salt were annotated as “Response to stress” and none of them were present in the single cultures. In particular, the number of secreted plant proteins of the categories “Antioxidant activity,” “Peroxidase activity,” and “Oxidoreductase activity, acting on peroxide as acceptor” was highly reduced in the presence of Trichoderma spp. in media with 50, 100, or 150 mM NaCl (Figure 3C). This suggests that the fungus reduces oxidative stress to the host under salt stress. In contrast, proteins with predicted “Hydrolase activity” showed elevated abundances in the co-cultures with salt (Figure 3C).
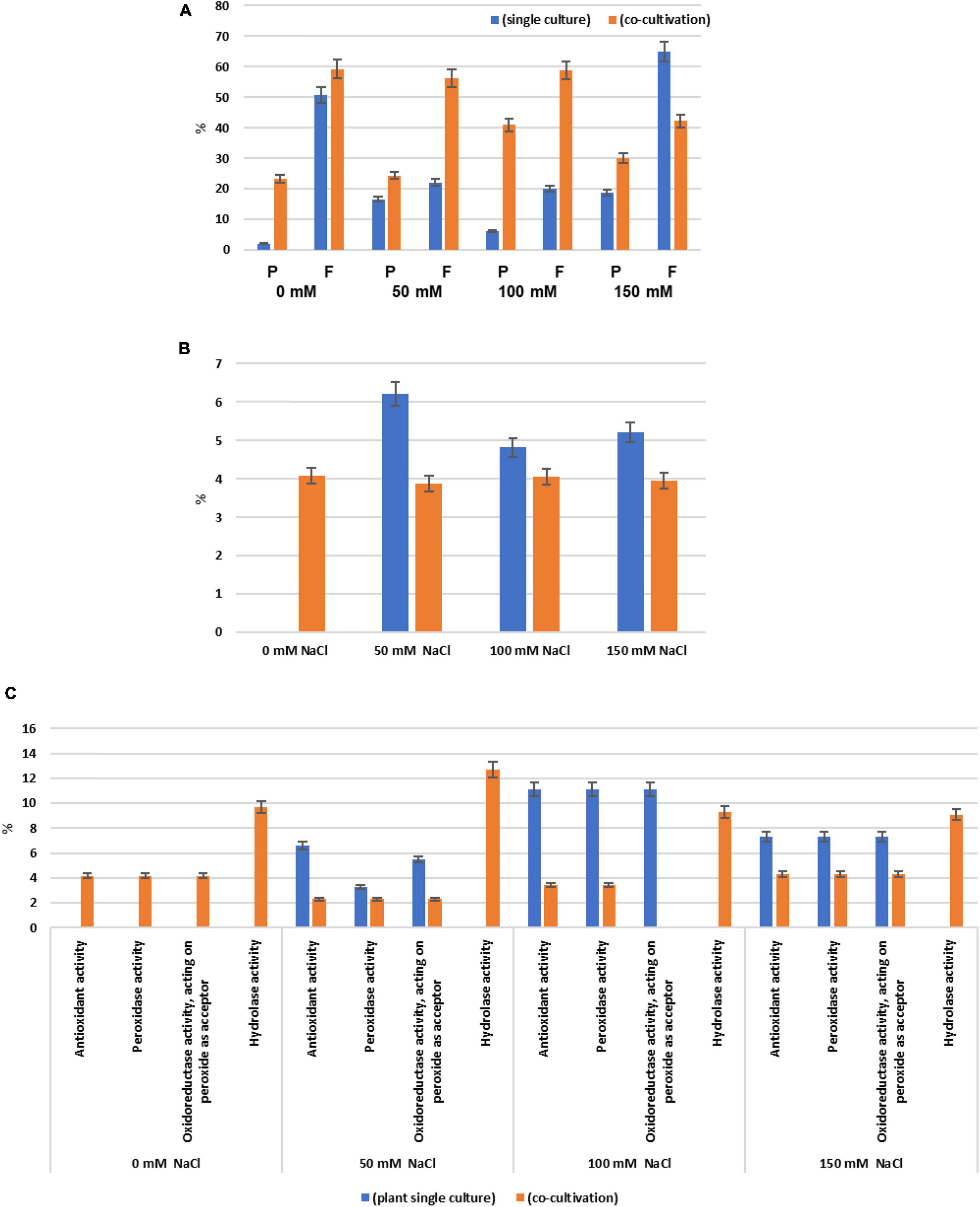
Figure 3. (A) Percentage of the secreted plant (P) and fungal (F) proteins of all identified proteins when the two symbionts are either growing alone or in co-culture, in the presence of 0 mM, 50 mM, 100 mM, or 150 mM NaCl, respectively. Secreted proteins are those with predicted N-terminal secretion sequences. (B) Percentage of the plant proteins annotated as “Response to stress” among all secreted plant proteins. (C) Percentage of plant proteins predicted to contain “Antioxidant activity,” “Peroxidase activity,” “Oxidoreductase activity, acting on peroxide as acceptor,” and “Hydrolase activity” among all secreted plant proteins. Error bars (SEs) refer to the differences between the three replicates.
Plant Secretome
We classified the secreted plant proteins into those which are only detectable in the presence of the fungus under low (i) or high (ii) salt concentrations, (iii) those which are detectable in the presence of the fungus under all salt conditions, and (iv) those which disappear in the presence of the fungus.
(i) Plant proteins which are only secreted in the presence of the fungus under 50 mM salt
Three plant proteins were only detected in co-cultures which stimulated plant growth (50 mM NaCl; Table 1; Tseng et al., 2020). PYK10 (Q9SR37) is the most abundant β-glucosidase in Brassicae roots and a major constituent of endoplasmic reticulum (ER) bodies. Several studies showed that PYK10 is involved in the mutualistic interaction between Arabidopsis and the endophytic fungus Piriformospora indica (Thürich et al., 2018, and ref. therein). The protein is released from the ER bodies to the extracellular space after cell disruption. The detectability of this protein only in co-cultures with 50 mM NaCl suggests that it has a specific function under these conditions. Since one would expect more cell injury or disruption under higher salt conditions (cf. below), an uncontrolled accumulation of PYK10 in the exudate fraction under 50 mM salt conditions is unlikely. The two other plant proteins specific for this co-cultivation condition are the uncharacterized GDSL-motif-containing esterase Q9C7N5 and glycosyl hydroxylase P94078.
(ii) Plant proteins which are only secreted in co-cultures with high salt
Plant proteins that are specific for exudates with high salt are mainly involved in stress responses (Table 1). DEGRADATION OF PERIPLASMIC PROTEIN 8 (DEG8; Q9FMA9), RESPONSIVE TO DEHYDRATION 19 (P43296) and the lectins Q9LJR2 and Q9LK72 are related to salt and/or microbial stress (Bernier and Berna, 2001; Lyou et al., 2009). Q9LV60 is a stress-inducible receptor-like kinase and the lipid recognition protein F4J7G5 is involved in sterol transport under stress (TAIR; arabidopsis.org). The mannose-binding lectin Q9ZVA4 has anti-fungal activities (Wu and Bao, 2013; Barre et al., 2019; Ghahremani et al., 2019), and the EPITHIOLSPECIFIER MODIFIER 1 Q9LJG3 participates in isothiocyanate production during glucosinolate hydrolysis (Burow et al., 2008; cf. Sato et al., 2019). Finally, the oligogalacturonidase oxidases Q9SA85/7 generate oligomers which function as DAMPs for the activation of the plant immune system (Benedetti et al., 2018). Besides proteins involved in salt stress responses, the appearance of defense-related proteins indicates that the fungus becomes more aggressive (cf. below) and the plant activates its defense machinery to restrict fungal growth. Consistent with this interpretation, we observe a strong increase in root colonization when the salt concentration increases to 150 mM (Tseng et al., 2020). Hyphae are not only found in roots but also around and in the stem tissue as well as on leaves (Figure 4).
(iii) Plant proteins from co-cultures under all salt conditions
Plant proteins with known functions which were detected in the presence of the fungus under all salt conditions (Table 1) are α-galactosidase 3 (Q8VXZ7) which hydrolzses β-L-arabinofuranose in cell wall-localized arabinogalactan-proteins (Imaizumi et al., 2017), the β-xylosidase Q9FLG1 involved in systemic acquired resistance (Breitenbach et al., 2014), the cysteine proteinase RESPONSIVE TO DEHYDRATION 21(A) (P43297) which is involved in immunity under drought stress (Ormancey et al., 2019; Pogorelko et al., 2019; Liu et al., 2020) and the chitinase Q9M2U5 which participates in cell wall remodeling and degradation of Trichoderma hyphae (Kappel and Gruber, 2020). All these proteins have protective functions in plant/microbe interactions, with a focus on the cell wall, and accumulate independently of salt stress.
(iv) Plant proteins that disappear in the presence of the fungus
Proteins that are only found in the absence of the fungus are either not synthesized/released by the host, because they are not required under symbiotic conditions, or they are degraded by fungal enzymes, e.g., for weakening the plant to allow better colonization (Table 2). The lipid-transfer proteins (LTPs, Q9FJ65, Q9LV65) respond to drought and salt stress (Yeats and Rose, 2008; Golldack et al., 2014). Q9FJ65 increases the glutathione content and enhances pathogen resistance (McLaughlin et al., 2015). DIR-1 like (Q9LV65) has an overlapping function with DIR1 involved in systemic acquired resistance (Champigny et al., 2013). The absence of this protein in the co-culture suggests that the fungus restricts systemic signaling under salt stress. The β-glucosidase 1 Q9SCW1 and pectin methylesterase inhibitor O49297 are involved in cell wall remodeling (Moneo-Sánchez et al., 2019). The disappearance of these two enzymes in the co-culture indicates that the fungus reduces the pressure on the plant to adapt the cell wall to salt stress. Calreticulin (O04151) accumulation indicates ER stress, which appears to be reduced in the co-culture as well. β-glucosides and lipid-transfer proteins are involved in membrane stabilization under biotic (Q9LLR6) and abiotic stress (Q9LLR6, Q9LLR7 Q9FF10) (cf.TAIR)1. PELPK1 (Q9LXB8) might be an important target of fungal signals to restrict root growth under salt stress (Rashid and Deyholos, 2011; Guo et al., 2013). In summary, the functional analysis of the proteins which disappear in the co-culture under salt stress, is in line with the overall analysis that the fungus reduces salt stress for the plant (e.g., results in Figure 1). On the other hand, in spite of the strong increase in root colonization in media with 150 mM NaCl (Figure 3), stress-related proteins which are found in uncolonized roots disappear in the presence of the fungus (Table 1). A detailed analysis of the identified proteins might help to understand how the balance between beneficial features in the symbiotic interaction and restriction of fungal growth is achieved.
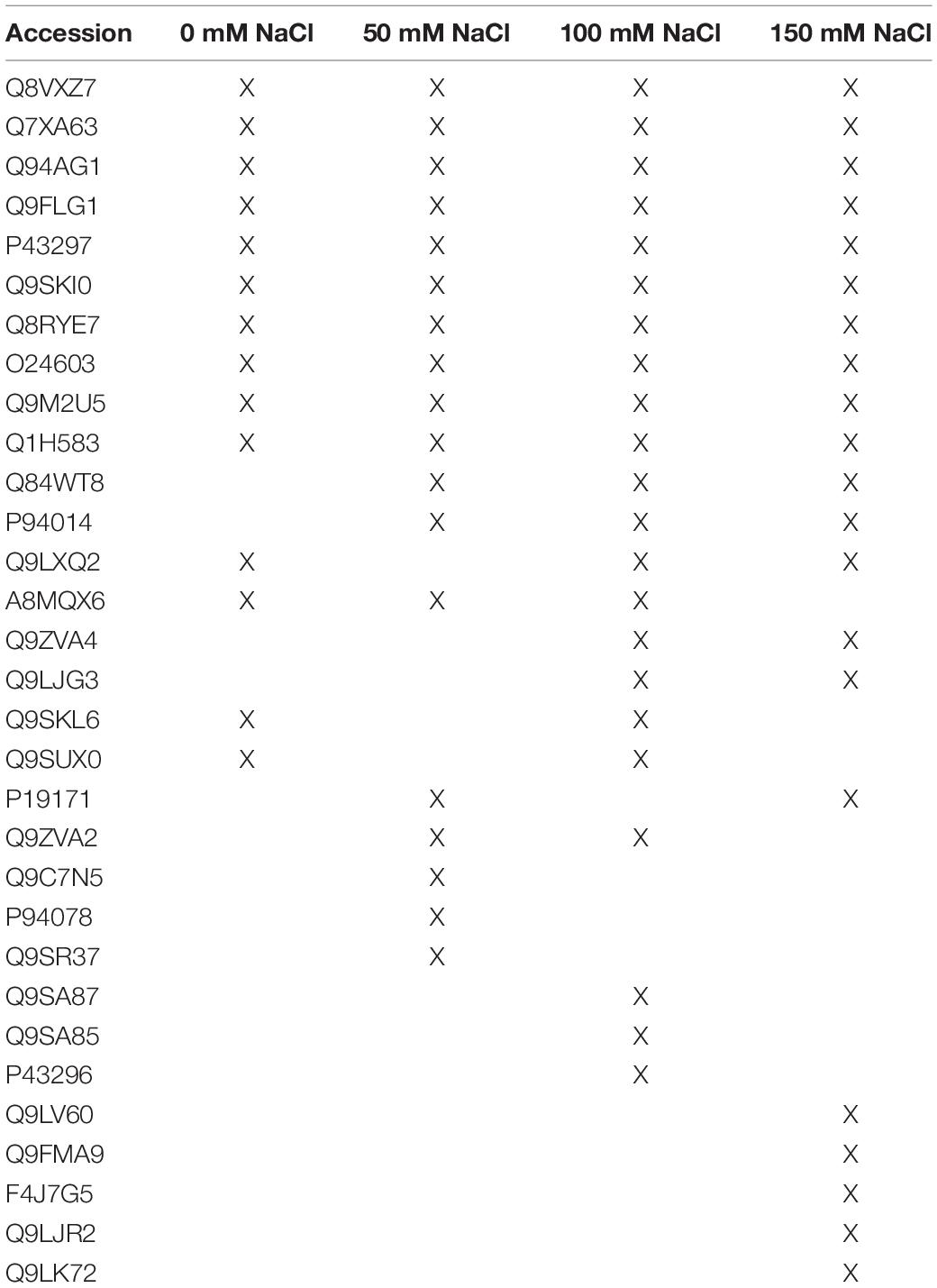
Table 1. Secreted plant proteins in the presence of the fungus under the different salt concentrations.
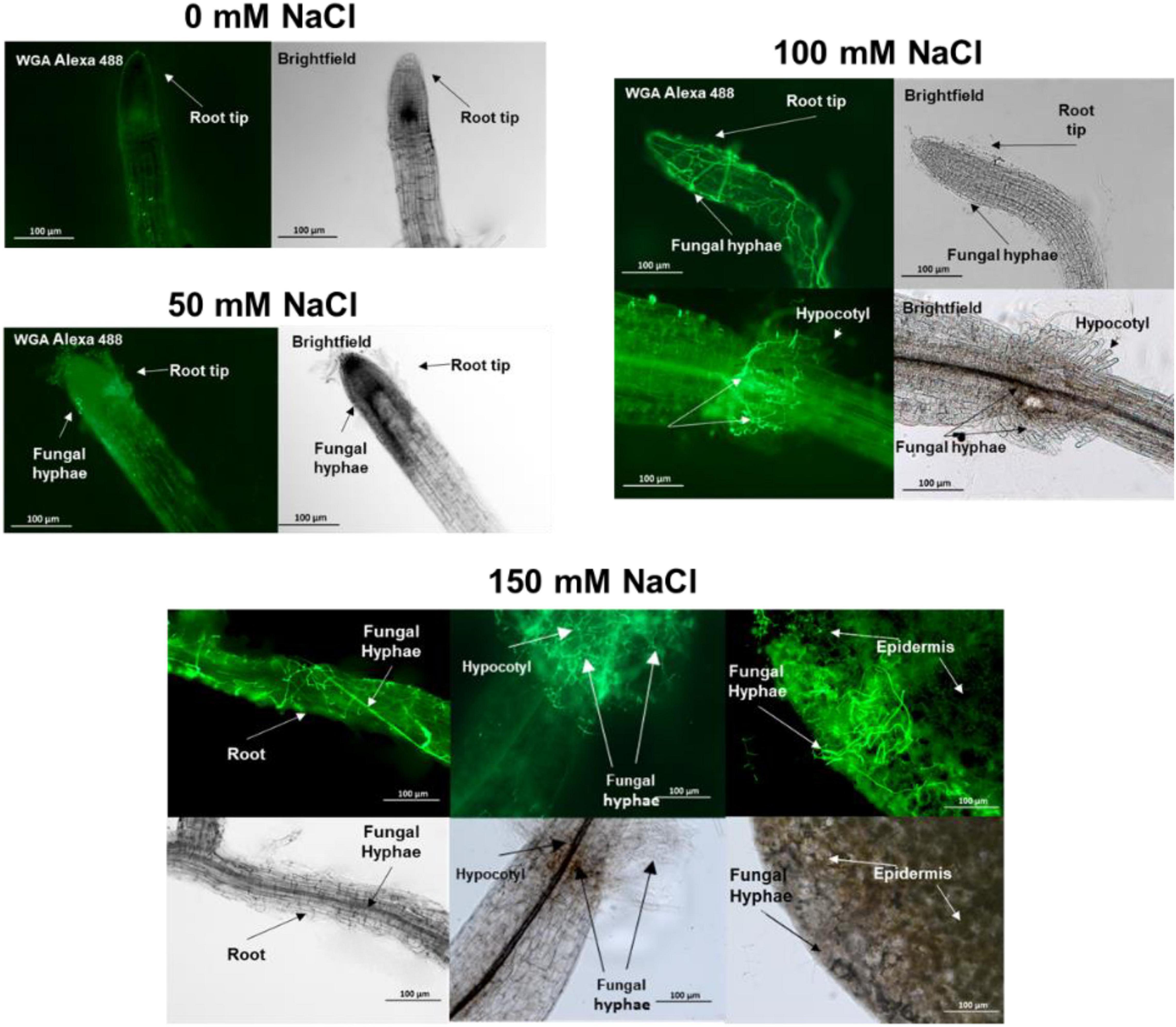
Figure 4. Trichoderma spp. hyphae in the root, hypocotyl and leaf of 15 day-old Arabidopsis seedlings colonized by the fungus since 10 days. Cocultivation occurred on media with 0–150 mM NaCl.

Table 2. Plant proteins with predicted N-terminal secretion sequences in media with/without salt in the absence of the fungus.
The comparative analysis identified secreted plant proteins that are specific for the described conditions. While some of them are well characterized (e.g., PYK10), barely anything is known for others. Their appearance in the plant secretome under the specific conditions and the context of other proteins should be investigated in the future in more detail.
Fungal Secretome
Fungal Proteins in the Co-culture
Without salt in the co-cultivation medium, only three fungal enzymes with known functions were found: the glucosaminidase A0A2T4AMN0 removes glucosamine from chitosans, the monooxygenase A0A2T3ZSA1 and the glucosidase A0A2T4A915 are involved in polysaccharide catabolism. The absence of these proteins in salt-containing media suggests specific functions of these enzymes in cell wall degradation.
In media with 50 mM salt, the number of secreted fungal enzymes involved in catabolism is higher: we found the β-xylanase A0A2T4A4K2 and β-1,3-glucanase A0A2T4ADC1 involved in polysaccharide catabolism, the peptidases A0A2T3ZS57, A0A2T4A515, and A0A2T4AAN8 and three uncharacterized proteins (A0A1P8ATL2, A0A2T4A7S8, A0A2T4A7H4). The catabolic enzymes might be important for the generation of metabolites required for the stimulation of the growth of the host. A prenylcysteine lyase (A0A2T4AFC0) generates cysteine, H2O2, and an isoprenoid aldehyde. The H2O2 generated by this enzyme might participate in promoting lateral root and root hair development and thus root growth of the host. Furthermore, the release of a farnesyl or geranyl-geranyl residue from the cysteine of a lipoprotein suggests that the fungus might interfere with the isoprenoid metabolism of the plant.
Trichoderma-Secreted Proteins That Disappear in the Co-culture Under Salt Stress
The fungus secretes 11 proteins in a medium with 150 mM salt which disappear in the co-culture (Table 3). A0A2T4AS91 is a cysteine-rich Kp4 domain-containing protein, which inhibits cell growth and division by blocking calcium uptake and signaling (Gu et al., 1995; Gage et al., 2002) and responds to environmental stress (Skamnioti et al., 2008; Brown, 2011; Zapparata et al., 2021). Two CFEM domain-containing proteins (A0A2T4ATW6 and A0A2T4APB9) with predicted GPI anchors (Kulkarni et al., 2003; Zhang et al., 2015; Kou et al., 2017) are proposed to function in surface sensing and stress-tolerance (Sabnam and Barman, 2017; Zhu et al., 2017), heme-iron acquisition (Nasser et al., 2016), cell wall stability and biofilm formation (Vaknin et al., 2014). Recent studies also showed the involvement of the CFEM motif in redox regulation in rice blast (Kou et al., 2017). Furthermore, the cell wall stress-responsive (WSC) domain in A0A2T4AAU9 function as a sensor of multiple stresses at the surface of the plasma membrane (Tong et al., 2016, 2019; Oide et al., 2019) and might be involved in β-glucan remodeling in the fungal cell wall (Wawra et al., 2019). The disappearance of these proteins in the presence of the host indicates that the fungus suffers less under salt stress in the symbiosis which might be the driving force for the intensified root colonization.
Malondialdehyde Levels and Antioxidant Enzyme Activities
The malondialdehyde (MDA) level is an indicator of oxidative stress. In the absence of salt in the culture medium, the MDA level was identical in seedlings with and without Trichoderma. With increasing salt concentrations, the MDA level increased and was always lower in the presence of the fungus. The fungal inhibition in media with up to 100 mM NaCl was<10%, whereas 21% reduction was observed with 150 mM NaCl (Figure 5). Furthermore, the activities of the antioxidant enzymes superoxide dismutase (SOD), catalase (CAT) and glutathione reductase (GR) increased with increasing salt concentrations in the medium, and the activity levels were always lower in the presence of the fungus (Figure 5). These results are consistent with the interpretation of the proteomic data that the fungus reduces salt stress for the plants, despite the massive increase in root colonization in 150 mM NaCl. Apparently, interaction is still better than living alone. Furthermore, without salt in the medium, the roots do not profit from the presence of the fungus. Consistent with the microscopic data (Figure 4), and the observation that without salt, no growth-promoting effect of the fungus on Arabidopsis seedlings can be detected (Tseng et al., 2020), it appears that there is barely any interaction between the two partners without salt. The sharp difference in the symbiotic interaction between 0 mM and 50 mM NaCl needs to be investigated in more detail.
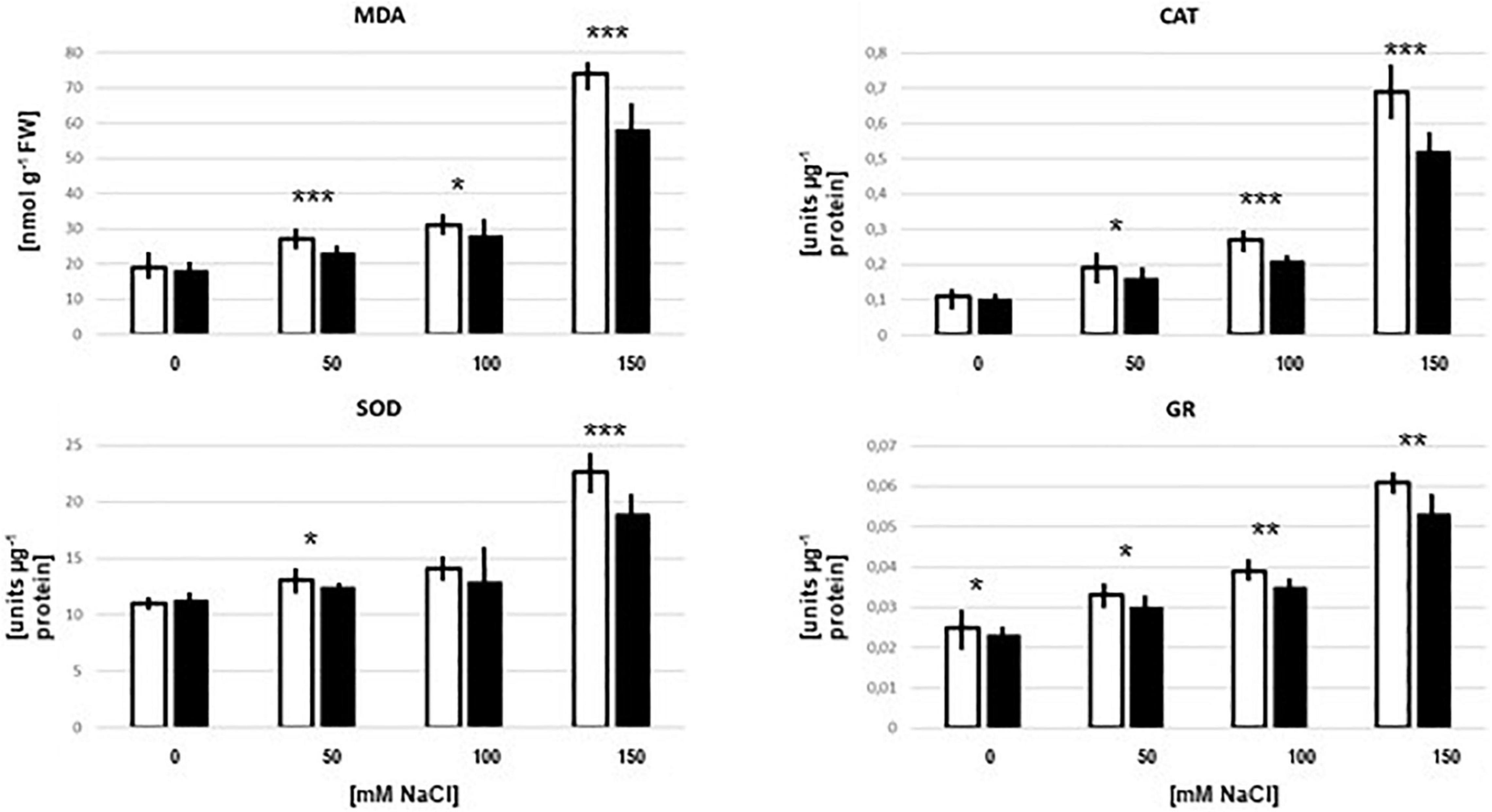
Figure 5. MDA levels and CAT, SOD, and GR enzyme activities in the root of Arabidopsis seedlings which were either colonized (black bars) or mock-treated (white bars) with Trichoderma on media with 0–150 mM NaCl. The treatment was identical to that for the peptide extraction. The results are based on 3 independent experiments, bars represent SEs. Asterisks above the bar pairs indicate significant differences by Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001).
Conclusion
The plant and fungal secretome patterns differ substantially under different salt conditions and when the two organisms are co-cultured. Overall, the identified proteins support the idea that the symbiotic interaction between the two organisms has an advantage for both partners under salt stress. The number of plant enzymes involved in oxidative stress is lower (Figure 3) and fungal proteins with known functions in stress sensing are reduced in co-cultures with 150 mM NaCl. The high salt concentration promotes root colonization by the fungus and the higher demand for nutrients by the microbe is visible by the appearance of fungal catabolic enzymes. Not surprisingly, the root counteracts the increased root colonization by releasing proteins with antifungal activities. However, comparison of the number of apoplastic plant proteins in co-cultures with high salt (ii) with fungal proteins which attack the plant cell wall (Figure 1 and Table 3) indicates that the plant apoplastic defense machinery is probably too weak to restrict fungal propagation in the host under salt stress. It would be interesting to see how the protein composition within the plant cell responds to the hyphal propagation and whether the host accumulates proteins which restrict fungal entry into the host cell or reduce loss of nutrients to the microbe.
Only a few proteins are exclusively found under co-cultivation conditions which resulted in the promotion of plant growth. Among them is the well-characterized plant PYK10. Since this abundant root protein is not detectable in exudate preparation from co-cultures with no or higher NaCl concentrations, nor in the plant cultures without the fungus, it is tempting to speculate that PYK10 is involved in beneficial features of the symbiosis resulting in plant growth promotion. Moreover, the appearance of this protein in this specific exudate fraction suggests the existence of a secretion mechanism that operates under the growth-stimulating conditions. Among the fungal proteins which are specific for co-cultures with 50 mM NaCl, the H2O2-generating prenylcysteine lyase has the potential to promote root growth.
Besides the general reduction of plant enzymes with antioxidant activities in co-cultures with 150 mM salt, also proteins with other functions confirm that symbiotic roots are less stressed. Examples are calreticulin for ER stress, cell wall remodeling and membrane-stabilizing enzymes, but also PELPK1 involved in root growth inhibition. The majority of fungal proteins which disappear in the co-culture with 150 mM salt stabilize the plasma membrane or senses stress.
Numerous studies investigated the Trichoderma secretome in symbiotic interactions with hosts (e.g., Gómez-Mendoza et al., 2014; Lamdan et al., 2015; González-López et al., 2021; Zhao et al., 2021). Besides similarities in the protein patterns and their interpretation (cf. Lamdan et al., 2015; Nogueira-Lopez et al., 2018) several Trichoderma proteins proposed to be involved in root colonization are not found in our study, such as the secreted cysteine-rich proteins swollenins, hydrophobins, and SM1 or SM2 (Viterbo and Chet, 2006; Brotman et al., 2008; Crutcher et al., 2015; Guzmán-Guzmán et al., 2017). We provide a list of proteins that are secreted under specific salt conditions. Since these conditions also influence the symbiotic interaction, the role of these proteins should be investigated in more detail. Finally, the role of the lytic polysaccharide monooxygeneases for symbiotic plant/fungus interactions requires more attention.
Materials and Methods
Cultivation of the Symbionts Alone and in Co-culture
Growth Conditions of Plants and Fungus
Arabidopsis thaliana (ecotype Columbia-0) seeds were surface-sterilized with a solution containing sterile distilled water (dH2O), sodium lauroyl sarcosinate, and DanKlorix (GP GABA GmBH Hamburg, Germany; 64%, 4%, 32%; v/v) for 8 min under constant shaking, followed by six rinses with dH2O. Surface-sterile seeds were sown on MS media supplemented with 0.3% gelrite (Murashige and Skoog, 1962). After cold treatment at 4°C for 48 h, plates were incubated for 10 days at 22°C under continuous illumination (100 μmol m–2 s–1). The Trichoderma strain was propagated on KM medium (pH 6.5) for a week at 25°C in the dark (Tseng et al., 2020).
Co-cultivation of A. thaliana and Trichoderma strain was performed under in vitro culture conditions. Ten-day-old A. thaliana seedlings of equal sizes were directly transferred from MS medium to the plant nutrient medium (PNM) medium. The fungus was grown on PNM plates. The co-cultivation of Arabidopsis plants and Trichoderma strain were performed on PNM medium with the salt concentrations mentioned in the text. A 5 mm plug of the Trichoderma strain was placed at the center of co-cultivation’s plate, as control only a plaque of the medium without hyphae was transferred. Microscopic analyses occurred after 5 day-incubation at 22°C under continuous illumination (100 μmol m–2 s–1) (Peškan-Berghöfer et al., 2004; Tseng et al., 2020). For the secretome analysis, the co-cultivation (or growth of the partners alone) was performed in liquid PNM with 0, 50, 100, or 150 mM NaCl for 5 days. After centrifugation, the remaining culture media were filtered and proteins were isolated by solid-phase extraction followed by LC-MS/MS (liquid chromatography-tandem mass spectrometry) analysis.
In-Solution Digest
The culture supernatant was filtered through 3 kDa MWCO spin filters (VWR) at 16,000 × g for 10 min. The protein retentate was resolubilized in 25 μl 100 mM TEAB and the solution was mixed with 25 μl 2,2,2-trifluoroethanol. Reduction and alkylation of cysteine thiols were performed by the addition of each 1 μL of 500 mM TCEP (tris(2-carboxyethyl) phosphine) and 625 mM 2-chloroacetamide incubated at 70°C for 30 min. The proteins were further cleaned up using C4 spin columns (Thermo Fisher Scientific): Acetonitrile (ACN) for reconstitution, 0.1% trifluoroacetic acid (TFA) for equilibration, loading, and washing, 0.1% TFA in 80:20 ACN/H2O (v/v) for elution. Proteins were dried in a vacuum evaporator (Eppendorf), resolubilized in 50 μl of 100 mM TEAB, and digested overnight (18 h) with a Trypsin + LysC mixture (Promega) at a protein to protease ratio of 25:1. Samples were vacuum-evaporated, resolubilized in 25 μL of 0.05% TFA in H2O/ACN 98/2 (v/v), and filtered through Ultrafree-MC 0.2 μm PTFE membrane spin filters (Merck-Millipore) at 14 000 × g for 15 min. The filtrate was transferred to HPLC vials and injected into the LC-MS/MS instrument. Each sample was measured in triplicate (3 analytical replicates of 3 biological replicates of 16 different conditions equals 144 LC-MS/MS runs).
Liquid Chromatography-Tandem Mass Spectrometry Analysis
LC-MS/MS analysis was performed on an Ultimate 3,000 nano RSLC system connected to a QExactive Plus mass spectrometer (both Thermo Fisher Scientific, Waltham, MA, United States). Peptide trapping for 5 min on an Acclaim Pep Map 100 column (2 cm × 75 μm, 3 μm) at 5 μL/min was followed by separation on an analytical Acclaim Pep Map RSLC nano column (50 cm × 75 μm, 2μm). Mobile phase gradient elution of eluent A (0.1% (v/v) formic acid in water) mixed with eluent B (0.1% (v/v) formic acid in 90/10 acetonitrile/water) was performed as follows: 0 min at 4% B, 4 min at 5% B, 6 min at 6% B, 14 min at 10% B, 20 min at 14% B, 35 min at 20% B, 42 min at 26%B, 46 min at 32% B, 52 min at 42% B, 55 min at 50% B 58 min at 65% B, 60–64.9 min at 96% B, 65–90 min at 4% B.
Positively charged ions were generated at a spray voltage of 2.2 kV using a stainless steel emitter attached to the Nanospray Flex Ion Source (Thermo Fisher Scientific). The quadrupole/orbitrap instrument was operated in Full MS/data-dependent MS2 (Top10) mode. Precursor ions were monitored at m/z 300–1,500 at a resolution of 70,000 FWHM (full width at half maximum) using a maximum injection time (ITmax) of 120 ms and an AGC (automatic gain control) target of 1 ⋅ 106. Precursor ions with a charge state of z = 2–5 were filtered at an isolation width of m/z 1.6 amu for further HCD fragmentation at 30% normalized collision energy (NCE). MS2 ions were scanned at 17,500 FWHM (ITmax = 120 ms, AGC = 2 ⋅ 105) using a fixed first mass of m/z 120 amu. Dynamic exclusion of precursor ions was set to 30 s and the underfill ratio was set to 1.0%. The LC-MS/MS instrument was controlled by Chromeleon 6.8, QExactive Tune 2.8 and Xcalibur 4.0 software.
Protein Database Search
Tandem mass spectra were searched against the UniProt databases of A. thaliana and T. harzianum by using Proteome Discoverer (PD) 2.2 (Thermo) and the algorithms of Mascot 2.4.1 (Matrix Science, United Kingdom), Sequest HT (version of PD2.2) and MS Amanda 2.0. Two missed cleavages were allowed for the tryptic digestion. The precursor mass tolerance was set to 10 ppm and the fragment mass tolerance was set to 0.02 Da. Modifications were defined as dynamic Met oxidation and protein N-term acetylation as well as static Cys carbamidomethylation. A strict false discovery rate (FDR) < 1% (peptide and protein level) was required for positive protein hits. If only 1 peptide per protein has been identified the hit was accepted if the Mascot score was >30 or the MS Amanda score was >300 or the Sequest score was >4. The Percolator node of PD2.2 and a reverse decoy database was used for q-value validation of spectral matches. Only rank 1 proteins and peptides of the top-scored proteins were counted. Label-free protein quantification was based on the Minora algorithm of PD2.2 using a signal-to-noise ratio > 5. Imputation of missing quan values was applied by setting the abundance to 75% of the lowest abundance identified for each sample. Significantly different abundances were based on a fold change >4, a ratio-adjusted significance level<0.05, which is the p-value calculated by the 2s t-test divided by the log2 ratio, and the proteins needed to be identified at least in 2 of 3 biological replicates of the sample group with the highest abundance.
Databank Analyses
All identified proteins were examined for an N-terminal secretion sequence, using the TargetP 1.1,2 SignalP 4.1,3 and Predotar 1.34 software (Small et al., 2004; Emanuelsson et al., 2007). The secreted protein candidates were classified according to their Gene ontology (GO) terms using ShinyGO (Ashburner et al., 2000; Gene Ontology Consortium, 2015), or the classification occurred according to their functional description on UniProt Database.
Root Colonization Analyzed by Microscopy
The entire Arabidopsis roots, root tips, hypocotyls and leaves were imaged using an AXIO Imager.M2 (Zeiss Microscopy GmbH, Jena, Germany) equipped with a 10x objective (N-Achroplan 10x/0.3). The bright field and fluorescence images (EX 545/25 and EM 605/70) were recorded with a color camera (AXIOCAM 503 color Zeiss, Jena, Germany) by use of an EGFP (EM 525/50 nm) and DsRED filter (EM 605/70 nm). Digital images were processed with the ZEN software (Zeiss, Jena, Germany), treated with Adobe R Photoshop to optimize brightness, contrast, and coloring and to overlay the photomicrographs to confirm fluorescence information.
Malondialdehyde Levels and Antioxidant Enzyme Activities
The assays for malondialdehyde levels and antioxidant enzyme activities were described in detail in Tsai et al. (2020). About 50 seedlings were collected from the co-cultivation (or growth of the plant partner alone) after 5 days from liquid PNM with 0, 50, 100, or 150 mM NaCl, as outlined above. Data are based on 3 repetitions, bars represent SEs.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD028957.
Author Contributions
HR and Y-HT designed and performed the experiments. TK, OK, and AB performed the LC-MS/MS analysis. KN, RU, and RO supervised and coordinated the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the DFG (Deutsche Forschungsgemeinschaft) within the scope of CRC1127 ChemBioSys (project number 239748522, projects A3 and B2) and CRC/TR 124 FungiNet (project number 210879364, project Z2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ arabidopsis.org
- ^ http://www.cbs.dtu.dk/services/TargetP/
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ https://urgi.versailles.inra.fr/Tools/Predotar
References
Alkooranee, J. T., Aledan, T. R., Ali, A. K., Lu, G., Zhang, X., Wu, J., et al. (2017). Detecting the hormonal pathways in oilseed rape behind induced systemic resistance by trichoderma harzianum TH12 to Sclerotinia sclerotiorum. PLoS One 12:e0168850. doi: 10.1371/journal.pone.0168850
Almeida, D. A., Horta, M. A. C., Ferreira Filho, J. A., Murad, N. F., and de Souza, A. P. (2021). The synergistic actions of hydrolytic genes reveal the mechanism of Trichoderma harzianum for cellulose degradation. J. Biotechnol. 334, 1–10. doi: 10.1016/j.jbiotec.2021.05.001
Andlar, M., Rezić, T., Marđetko, N., Kracher, D., Ludwig, R., and Šantek, B. (2018). Lignocellulose degradation: an overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 18, 768–778. doi: 10.1002/elsc.201800039
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Barre, A., Simplicien, M., Benoist, H., Van Damme, E. J. M., and Rougé, P. (2019). Mannose-specific lectins from marine algae: diverse structural scaffolds associated to common virucidal and anti-cancer properties. Mar. Drugs 17:440. doi: 10.3390/md17080440
Benedetti, M., Verrascina, I., Pontiggia, D., Locci, F., Mattei, B., De Lorenzo, G., et al. (2018). Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 94, 260–273. doi: 10.1111/tpj.13852
Bernier, F., and Berna, A. (2001). Germins and germin-like proteins: plant do-all proteins. But what do they do exactly? Plant Physiol. Biochem. 39, 545–554. doi: 10.1016/s0981-9428(01)01285-2
Breitenbach, H. H., Wenig, M., Wittek, F., Jordá, L., Maldonado-Alconada, A. M., Sarioglu, H., et al. (2014). Contrasting roles of the apoplastic aspartyl protease apoplastic, enhanced disease susceptibility1-dependent1 and legume lectin-like Protein1 in arabidopsis systemic acquired resistance. Plant Physiol. 165, 791–809. doi: 10.1104/pp.114.239665
Brotman, Y., Briff, E., Viterbo, A., and Chet, I. (2008). Role of swollenin, an expansin-like protein from trichoderma, in plant root colonization. Plant Physiol. 147, 779–789. doi: 10.1104/pp.108.116293
Brotman, Y., Lisec, J., Méret, M., Chet, I., Willmitzer, L., and Viterbo, A. (2012). Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology 158, 139–146. doi: 10.1099/mic.0.052621-0
Brown, D. W. (2011). The KP4 killer protein gene family. Curr. Genet. 57, 1–62. doi: 10.1007/s00294-010-0326-y
Burow, M., Zhang, Z.-Y., Ober, J. A., Lambrix, V. M., Wittstock, U., Gershenzon, J., et al. (2008). ESP and ESM1 mediate indol-3-acetonitrile production from indol-3-ylmethyl glucosinolate in Arabidopsis. Phytochemistry 69, 663–671. doi: 10.1016/j.phytochem.2007.08.027
Calderaro, F., Bevers, L. E., and van den Berg, M. A. (2021). Oxidative power: tools for assessing LPMO activity on cellulose. Biomolecules 11:1098. doi: 10.3390/biom11081098
Champigny, M. J., Isaacs, M., Carella, P., Faubert, J., Fobert, P. R., and Cameron, R. K. (2013). Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 4:230. doi: 10.3389/fpls.2013.00230
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Beltrán-Peña, E., Herrera-Estrella, A., and López-Bucio, J. (2011). Trichoderma-induced plant immunity likely involves both hormonal- and camalexin-dependent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungi Botrytis cinerea. Plant Signal. Behav. 6, 1554–1563. doi: 10.4161/psb.6.10.17443
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Cortés-Penagos, C., and López-Bucio, J. (2009). Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. doi: 10.1104/pp.108.130369
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Real-Santillán, R. O., López-Carmona, D., García-Gómez, G., Galicia-Gallardo, A. P., et al. (2021). In a belowground multitrophic interaction, Trichoderma harzianum induces maize root herbivore tolerance against Phyllophaga vetula. Pest Manag. Sci. 77, 3952–3963. doi: 10.1002/ps.6415
Crutcher, F. F., Moran-Diez, M. E., Ding, S., Liu, J., Horwitz, B. A., Mukherjee, P. K., et al. (2015). A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 119, 476–486. doi: 10.1016/j.funbio.2015.01.004
Djonovic, S., Vargas, W. A., Kolomiets, M. V., Horndeski, M., Wiest, A., and Kenerley, C. M. (2007). A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. doi: 10.1104/pp.107.103689
Druzhinina, I. S., Chenthamara, K., Zhang, J., Atanasova, L., Yang, D., Miao, Y., et al. (2018). Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PLoS Genet. 14:e1007322. doi: 10.1371/journal.pgen.1007322
Emanuelsson, O., Brunak, S., Heijne, G., and von Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Forsberg, Z., Sørlie, M., Petrović, D., Courtade, G., Aachmann, F. L., Vaaje-Kolstad, G., et al. (2019). Polysaccharide degradation by lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 59, 54–64.
Gage, M. J., Bruenn, J., Fischer, M., Sanders, D., and Smith, T. J. (2002). KP4 fungal toxin inhibits growth in Ustilago maydis by blocking calcium uptake. Mol. Microbiol. 41, 775–785. doi: 10.1046/j.1365-2958.2001.02554.x
Ghahremani, M., Tran, H., Biglou, S. G., O’Gallagher, B., She, Y. M., and Plaxton, W. C. (2019). A glycoform of the secreted purple acid phosphatase AtPAP26 co-purifies with a mannose-binding lectin (AtGAL1) upregulated by phosphate-starved Arabidopsis. Plant Cell Environ. 42, 1139–1157. doi: 10.1111/pce.13432
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
Gómez-Mendoza, D. P., Junqueira, M., do Vale, L. H., Domont, G. B., Ferreira Filho, E. X., Sousa, M. V., et al. (2014). Secretomic survey of Trichoderma harzianum grown on plant biomass substrates. J. Proteome Res. 13, 1810–1822. doi: 10.1021/pr400971e
González-López, M. D. C., Jijón-Moreno, S., Dautt-Castro, M., Ovando-Vázquez, C., Ziv, T., Horwitz, B. A., et al. (2021). Secretome analysis of Arabidopsis-Trichoderma atroviride interaction unveils new roles for the plant glutamate:glyoxylate aminotransferase GGAT1 in plant growth induced by the fungus and resistance against Botrytis cinerea. Int. J. Mol. Sci. 22:6804. doi: 10.3390/ijms22136804
Gu, F., Khimani, A., Rane, S. G., Flurkey, W. H., Bozarth, R. F., and Smith, T. J. (1995). Structure and function of a virally encoded fungal toxin from Ustilago maydis: a fungal and mammalian Ca2+ channel inhibitor. Structure 3, 805–814. doi: 10.1016/s0969-2126(01)00215-5
Guo, L., Yang, H., Zhang, X., and Yang, S. (2013). Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 64, 1755–1767. doi: 10.1093/jxb/ert040
Guzmán-Guzmán, P., Alemán-Duarte, M. I., Delaye, L., Herrera-Estrella, A., and Olmedo-Monfil, V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18:16. doi: 10.1186/s12863-017-0481-y
Hermosa, R., Rubio, M. B., Cardoza, R. E., Nicolás, C., Monte, E., and Gutiérrez, S. (2013). The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 16, 69–80. doi: 10.2436/20.1501.01.181
Hermosa, R., Viterbo, A., Chet, I., and Monte, E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25. doi: 10.1099/mic.0.052274-0
Imaizumi, C., Tomatsu, H., Kitazawa, K., Yoshimi, Y., Shibano, S., Kikuchi, K., et al. (2017). Heterologous expression and characterization of an Arabidopsis β-l-arabinopyranosidase and α-d-galactosidases acting on β-l-arabinopyranosyl residues. J. Exp. Bot. 68, 4651–4661. doi: 10.1093/jxb/erx279
Jagadeeswaran, G., Veale, L., and Mort, A. J. (2021). Do lytic polysaccharide monooxygenases aid in plant pathogenesis and herbivory? Trends Plant Sci. 26, 142–155. doi: 10.1016/j.tplants.2020.09.013
Kappel, L., and Gruber, S. (2020). “Chitin and chitosan—important structural components in Trichoderma cell wall remodeling,” in New and Future Developments in Microbial Biotechnology and Bioengineering. Recent Developments in Trichoderma Research, eds V. K. Gupta, S. Zeilinger, and H. B. Singh. (Amsterdam: Elsevier).
Kaushal, M. (2020). Insights into microbially induced Salt Tolerance and Endurance Mechanisms (STEM) in plants. Front. Microbiol. 11:1518. doi: 10.3389/fmicb.2020.01518
Kou, Y., Tan, Y. H., Ramanujam, R., and Naqvi, N. (2017). Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 214, 330–342. doi: 10.1111/nph.14347
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., Druzhinina, I. S., and Thon, M. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. doi: 10.1186/gb-2011-12-4-r40
Kulkarni, R. D., Kelkar, H. S., and Dean, R. A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121. doi: 10.1016/S0968-0004(03)00025-2
Lamdan, N. L., Shalaby, S., Ziv, T., Kenerley, C. M., and Horwitz, B. A. (2015). Secretome of Trichoderma interacting with maize roots: role in induced systemic resistance. Mol. Cell. Proteomics 14, 1054–1063. doi: 10.1074/mcp.M114.046607
Liu, Y., Wang, K., Cheng, Q., Kong, D., Zhang, X., Wang, Z., et al. (2020). Cysteine protease RD21A regulated by E3 ligase SINAT4 is required for drought-induced resistance to Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 71, 5562–5576. doi: 10.1093/jxb/eraa255
Lyou, S. H., Park, H. J., Jung, C., Sohn, H. B., Lee, G., Kim, C. H., et al. (2009). The Arabidopsis AtLEC gene encoding a lectin-like protein is up-regulated by multiple stimuli including developmental signal, wounding, jasmonate, ethylene, and chitin elicitor. Mol. Cells 27, 75–81. doi: 10.1007/s10059-009-0007-1
Manavalan, T., Stepnov, A. A., Hegnar, O. A., and Eijsink, V. G. H. (2021). Sugar oxidoreductases and LPMOs - two sides of the same polysaccharide degradation story? Carbohydr. Res. 505:108350. doi: 10.1016/j.carres.2021.108350
Mangiest, A. A. (2020). Endophytes: colonization, behaviour, and their role in defense mechanism. Int. J. Microbiol. 3:6927219. doi: 10.1155/2020/6927219
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol. 26, 553–602008. doi: 10.1038/nbt1403
McLaughlin, J. E., Bin-Umer, M. A., Widiez, T., Finn, D., McCormick, S., and Tumer, N. E. (2015). A lipid transfer protein increases the glutathione content and enhances arabidopsis resistance to a Trichothecene Mycotoxin. PLoS One 10:e0130204. doi: 10.1371/journal.pone.0130204
Mendoza-Mendoza, A., Zaid, R., Lawry, R., Hermosa, R., Monte, E., Horwitz, B. A., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85. doi: 10.1016/j.fbr.2017.12.001
Mengistu, A. A. (2020). Endophytes: colonization, behaviour, and their role in defense mechanism. Int. J. Microbiol. 2020:6927219.
Moneo-Sánchez, M., Alonso-Chico, A., Knox, J. P., Dopico, B., Labrador, E., and Martín, I. (2019). β-(1,4)-Galactan remodelling in Arabidopsis cell walls affects the xyloglucan structure during elongation. Planta 249, 351–362. doi: 10.1007/s00425-018-3008-5
Morán-Diez, M. E., Trushina, N., Lamdan, N. L., Rosenfelder, L., Mukherjee, P. K., Kenerley, C. M., et al. (2015). Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots. BMC Genomics 16:8. doi: 10.1186/s12864-014-1208-3
Mukherjee, P. K., Horwitz, B. A., and Kenerley, C. M. (2012). Secondary metabolism in Trichoderma - a genomic perspective. Microbiology 158, 35–45. doi: 10.1099/mic.0.053629-0
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nasser, L., Weissman, Z., Pinsky, M., Amartely, H., Dvir, H., and Kornitzer, D. (2016). Structural basis of haem-iron acquisition by fungal pathogens. Nat. Microbiol. 1:16156. doi: 10.1038/nmicrobiol.2016.156
Nieto-Jacobo, M. F., Steyaert, J. M., Salazar-Badillo, F. B., Nguyen, D. V., Rostás, M., Braithwaite, M., et al. (2017). Environmental growth conditions of trichoderma spp. affects indole acetic acid derivatives, volatile organic compounds, and plant growth promotion. Front. Plant Sci. 8:102. doi: 10.3389/fpls.2017.00102
Nogueira-Lopez, G., Greenwood, D. R., Middleditch, M., Winefield, C., Eaton, C., Steyaert, J. M., et al. (2018). The apoplastic secretome of Trichoderma virens during interaction with maize roots shows an inhibition of plant defence and scavenging oxidative stress secreted proteins. Front Plant Sci. 9:409. doi: 10.3389/fpls.2018.00409
Oide, S., Tanaka, Y., Watanabe, A., and Inui, M. (2019). Carbohydrate-binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae. Enzyme Microb. Technol. 125, 13–20. doi: 10.1016/j.enzmictec.2019.02.009
Ormancey, M., Thuleau, P., van der Hoorn, R. A. L., Grat, S., Testard, A., Kamal, K. Y., et al. (2019). Sphingolipid-induced cell death in Arabidopsis is negatively regulated by the papain-like cysteine protease RD21. Plant Sci. 280, 12–17. doi: 10.1016/j.plantsci.2018.10.028
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Peškan-Berghöfer, T., Shahollari, B., Giong, P. H., Hehl, S., Markert, C., Blanke, V., et al. (2004). Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant–microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol. Plant. 122, 465–477.
Pogorelko, G. V., Juvale, P. S., Rutter, W. B., Hütten, M., Maier, T. R., Hewezi, T., et al. (2019). Re-targeting of a plant defense protease by a cyst nematode effector. Plant J. 98, 1000–1014. doi: 10.1111/tpj.14295
Rani Singhania, R., Dixit, P., Kumar Patel, A., Shekher Giri, B., Kuo, C. H., Chen, C. W., et al. (2021). Role and significance of lytic polysaccharide monooxygenases (LPMOs) in lignocellulose deconstruction. Bioresour. Technol. 335:125261. doi: 10.1016/j.biortech.2021.125261
Rashid, A., and Deyholos, M. K. (2011). PELPK1 (At5g09530) contains a unique pentapeptide repeat and is a positive regulator of germination in Arabidopsis thaliana. Plant Cell Rep. 9, 1735–1745. doi: 10.1007/s00299-011-1081-3
Ruocco, M., Lanzuise, S., Lombardi, N., Woo, S. L., Vinale, F., Marra, R., et al. (2015). Multiple roles and effects of a novel Trichoderma Hydrophobin. Mol. Plant Microb. Interact. 28, 167–179. doi: 10.1094/MPMI-07-14-0194-R
Sabnam, N., and Barman, R. (2017). WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet. Biol. 105, 37–51. doi: 10.1016/j.fgb.2017.05.006
Sagarika, M. S., Parameswaran, C., Senapati, A., Barala, J., Mitra, D., Prabhukarthikeyan, S. R., et al. (2021). Lytic polysaccharide monooxygenases (LPMOs) producing microbes: a novel approach for rapid recycling of agricultural wastes. Sci. Total Environ. 806:150451. doi: 10.1016/j.scitotenv.2021.150451
Samolski, I., Rincón, A. M., Pinzón, L. M., Viterbo, A., and Monte, E. (2012). The qid74 gene from Trichoderma harzianum has a role in root architecture and plant biofertilization. Microbiology 158, 129–138. doi: 10.1099/mic.0.053140-0
Sarrocco, S., Matarese, F., Baroncelli, R., Vannacci, G., Seidl-Seiboth, V., Kubicek, C. P., et al. (2017). The constitutive endopolygalacturonase TvPG2 regulates the induction of plant systemic resistance by Trichoderma virens. Phytopathology 107, 537–544. doi: 10.1094/PHYTO-03-16-0139-R
Sato, Y., Tezuka, A., Kashima, M., Shimizu-Inatsugi, R., Yamazaki, M., et al. (2019). Transcriptional variation in glucosinolate biosynthetic genes and inducible responses to aphid herbivory on field-grown Arabidopsis thaliana. Front. Sci. 10:787. doi: 10.3389/fgene.2019.00787
Seidl, V., Marchetti, M., Schandl, R., Allmaier, G., and Kubicek, C. P. (2006). Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J. 273, 4346–4359. doi: 10.1111/j.1742-4658.2006.05435.x
Sharon, A., Fuchs, Y., and Anderson, J. D. (1993). The elicitation of ethylene biosynthesis by a Trichoderma xylanase is not related to the cell wall degradation activity of the enzyme. Plant Physiol. 102, 1325–1329. doi: 10.1104/pp.102.4.1325
Shoresh, M., Harman, G. E., and Mastouri, F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. doi: 10.1146/annurev-phyto-073009-114450
Silva, R. N., Monteiro, V. N., Steindorff, A. S., Gomes, E. V., Noronha, E. F., and Ulhoa, C. J. (2019). Trichoderma/pathogen/plant interaction in pre-harvest food security. Fungal Biol. 123, 565–583. doi: 10.1016/j.funbio.2019.06.010
Skamnioti, P., Furlong, R. F., and Gurr, S. J. (2008). The fate of gene duplicates in the genomes of fungal pathogens. Commun. Integr. Biol. 1, 196–198. doi: 10.4161/cib.1.2.7144
Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar. a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590. doi: 10.1002/pmic.200300776
Thürich, J., Meichsner, D., Furch, A. C. U., Pfalz, J., Krüger, T., Kniemeyer, O., et al. (2018). Arabidopsis thaliana responds to colonisation of Piriformospora indica by secretion of symbiosis-specific proteins. PLoS One 13:e0209658. doi: 10.1371/journal.pone.0209658
Tong, S. M., Chen, Y., Zhu, J., Ying, S. H., and Feng, M. G. (2016). Subcellular localization of five singular WSC domain-containing proteins and their roles in Beauveria bassiana responses to stress cues and metal ions. Environ. Microbiol. Rep. 8, 295–304. doi: 10.1111/1758-2229.12380
Tong, S.-M., Wang, D. Y., Gao, B. J., Ying, S. H., and Feng, M. G. (2019). The DUF1996 and WSC domain-containing protein Wsc1I acts as a novel sensor of multiple stress cues in Beauveria bassiana. Cell Microbiol. 21:e13100. doi: 10.1111/cmi.13100
Tsai, H.-J., Shao, K.-H., Chan, M. C., Cheng, C.-P., Yeh, K.-W., Oelmüller, R., et al. (2020). Piriformospora indica symbiosis improves water stress tolerance of rice through regulating stomata behavior and ROS scavenging systems. Plant Signal. Behav. 15:1722447. doi: 10.1080/15592324.2020.1722447
Tseng, Y. H., Rouina, H., Groten, K., Rajani, P., Furch, A. C. U., Reichelt, M., et al. (2020). An endophytic trichoderma strain promotes growth of its hosts and defends against pathogen attack. Front. Plant Sci. 11:573670. doi: 10.3389/fpls.2020.573670
Vaknin, Y., Shadkchan, Y., Levdansky, E., Morozov, M., Romano, J., and Osherov, N. (2014). The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence. Fungal Genet. Biol. 63, 55–64. doi: 10.1016/j.fgb.2013.12.005
van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433.
Viterbo, I., and Chet, T. (2006). Hyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol. Plant Pathol. 7, 249–258. doi: 10.1111/j.1364-3703.2006.00335.x
Vuong, T. V., and Master, E. R. (2021). Enzymatic upgrading of heteroxylans for added-value chemicals and polymers. Curr. Opin. Biotechnol. 73, 51–60. doi: 10.1016/j.copbio.2021.07.001
Wang, D., Li, Y., Zheng, Y., and Hsieh, Y. S. Y. (2021). Recent advances in screening methods for the functional investigation of lytic polysaccharide monooxygenases. Front. Chem. 9:653754. doi: 10.3389/fchem.2021.653754
Wawra, S., Fesel, P., Widmer, H., Neumann, U., Lahrmann, U., Becker, S., et al. (2019). FGB1 and WSC3 are in planta-induced β-glucan-binding fungal lectins with different functions. New Phytol. 222, 1493–1506. doi: 10.1111/nph.15711
Wu, L., and Bao, J. K. (2013). Anti-tumor and anti-viral activities of Galanthus nivalis agglutinin (GNA)-related lectins. Glycoconj. J. 30, 269–279. doi: 10.1007/s10719-012-9440-z
Yeats, T. H., and Rose, J. K. C. (2008). The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 17, 191–198. doi: 10.1110/ps.073300108
Zapparata, A., Baroncelli, R., Brandström Durling, M., Kubicek, C. P., Karlsson, M., Vannacci, G., et al. (2021). Fungal cross-talk: an integrated approach to study distance communication. Fungal Genet. Biol. 148:103518. doi: 10.1016/j.fgb.2021.103518
Zerva, A., Pentari, C., Ferousi, C., Nikolaivits, E., Karnaouri, A., and Topakas, E. (2021). Recent advances on key enzymatic activities for the utilisation of lignocellulosic biomass. Bioresour. Technol. 28:126058. doi: 10.1016/j.biortech.2021.126058
Zhang, Z.-N., Wu, Q.-Y., Zhang, G.-Z., Zhu, Y.-Y., Murphy, R. W., Liu, Z., et al. (2015). Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 5:3032. doi: 10.1038/srep13032
Zhao, Z., Cai, F., Gao, R., Ding, M., Jiang, S., Chen, P., et al. (2021). At least three families of hyphosphere small secreted cysteine-rich proteins can optimize surface properties to a moderately hydrophilic state suitable for fungal attachment. Environ. Microbiol. 23, 5750–5768. doi: 10.1111/1462-2920.15413
Keywords: Trichoderma, secretome, salt stress, endophyte, symbiosis, root colonization, lytic polysaccharide monooxygenases, PYK10
Citation: Rouina H, Tseng Y-H, Nataraja KN, Uma Shaanker R, Krüger T, Kniemeyer O, Brakhage A and Oelmüller R (2022) Comparative Secretome Analyses of Trichoderma/Arabidopsis Co-cultures Identify Proteins for Salt Stress, Plant Growth Promotion, and Root Colonization. Front. Ecol. Evol. 9:808430. doi: 10.3389/fevo.2021.808430
Received: 03 November 2021; Accepted: 26 November 2021;
Published: 04 January 2022.
Edited by:
Stephan Pollmann, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Paulina Guzmán-Guzmán, Universidad Michoacana de San Nicolás de Hidalgo, MexicoJoy Michal Johnson, Kerala Agricultural University, India
Copyright © 2022 Rouina, Tseng, Nataraja, Uma Shaanker, Krüger, Kniemeyer, Brakhage and Oelmüller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Oelmüller, YjdvZXJhQHVuaS1qZW5hLmRl
 Hamid Rouina
Hamid Rouina Yu-Heng Tseng
Yu-Heng Tseng Karaba N. Nataraja
Karaba N. Nataraja Ramanan Uma Shaanker
Ramanan Uma Shaanker Thomas Krüger4
Thomas Krüger4 Olaf Kniemeyer
Olaf Kniemeyer Axel Brakhage
Axel Brakhage Ralf Oelmüller
Ralf Oelmüller