- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Fujian Provincial Key Laboratory of Insect Ecology, Fujian Agriculture and Forestry University, Fuzhou, China
Adaptation to different host plants is considered to be an important driver of the divergence and speciation of herbivorous insects. The application of molecular data and integrated taxonomic practices in recent years may contribute to our understanding of population divergence and speciation, especially for herbivorous insects considered to be polyphagous. Aphis aurantii is an important agricultural and forestry pest with a broad range of host plants. In this study, samples of A. aurantii feeding on different host plants in the same geographical area were collected, and their population genetic divergence and morphological difference were analyzed. Phylogenetic analysis and haplotype network analysis based on five genes revealed that the population on Ficus exhibited significantly genetic divergence from populations on other host plants, which was also supported by the statistical analysis based on measurements of 38 morphological characters. Our results suggest that A. aurantii has undergone specialized evolution on Ficus, and the Ficus population may represent a lineage that is experiencing ongoing sympatric speciation.
Introduction
The mechanism of speciation has been a hot research topic in biology from Darwin’s time to present day (Darwin, 1859; Futuyma and Mayer, 1980; McKinnon et al., 2004; Li et al., 2015; Taylor and Friesen, 2017). Whether speciation can occur without geographical barriers, i.e., sympatric speciation, is one of the core points of the debate. During much of the twentieth century, sympatric speciation was considered to be more unreliable when compared with allopatric speciation (Futuyma and Mayer, 1980). However, due to the in-depth research on biogeography and phylogeny in recent years, the concept of sympatric speciation has been accepted gradually (Via, 2001; Berlocher and Feder, 2002; Drès and Mallet, 2002; Bolnick and Fitzpatrick, 2007; Li et al., 2015). The growing acceptance of sympatric divergence and speciation has crucial implications for the interpretation of high biodiversity on Earth and the optimization of systematic theory and practice (Berlocher and Feder, 2002). Compared with allopatric speciation, complete sympatric speciation events in nature may take a long time (Mallet, 2008), and much fewer empirical studies have been reported (Savolainen et al., 2006; Bolnick and Fitzpatrick, 2007). However, exploring divergence among sympatric populations, which may indicate ongoing sympatric speciation, can be helpful for understanding mechanisms of sympatric speciation (Drès and Mallet, 2002; Peccoud et al., 2009).
Phytophagous insects are considered as ideal candidates for the study of sympatric divergence due to intimate and specialized relationship with their host plants (Berlocher and Feder, 2002; Bolnick and Fitzpatrick, 2007; Peccoud et al., 2009; Lee et al., 2015). Differences in physical structure, nutritional composition, and chemical defense of different host plants may generate variant selection pressures on phytophagous insects that feeding on them (Egan and Ott, 2007). Moreover, the microenvironments provided by different host plants vary greatly, which may lead to different exposure probabilities to natural predators for phytophagous insects (Nosil, 2004; Nosil and Crespi, 2006; Rull et al., 2009). Therefore, for phytophagous insect populations in a sympatric area, long-term specialization on certain host plants may lead to adaptive evolution and reproductive isolation (Malausa et al., 2005; Xue et al., 2014; Lee et al., 2015). Host races of phytophagous insects are important evidence of sympatric genetic divergence driven by host plant (Peccoud et al., 2009).
Aphids exhibit varying degrees of host specialization. About half of all aphid species are specific to a single plant species, and at higher taxonomic levels, some aphid genera or families are strictly to a single plant genus or family (Eastop, 1973; Peccoud et al., 2010). There are also polyphagous aphid species in ecosystems, including many important agricultural pests such as Aphis gossypii, Myzus persicae and Acyrthosiphon pisum, having very high diversity of host plants (Blackman and Eastop, 2021). Host races or host-specialized populations with a relatively narrow host range are also frequently present in these polyphagous species (Via et al., 2000; Margaritopoulos et al., 2005; Carletto et al., 2009). This phenomenon indicates that these polyphagous species may have undergone population divergence or speciation events due to specialization on specific host plants (Peccoud et al., 2010).
Aphis aurantii (Hemiptera: Aphididae), known as the black citrus aphid or camellia aphid, is one of the most destructive pests of citrus and tea plants, mainly distributed in tropical and subtropical regions (Carver, 1978; Sevim et al., 2012; Blackman and Eastop, 2021). It is also a polyphagous species, which can feed on more than 120 plant species belonging to various families such as Rutaceae, Theaceae, Moraceae, Rosaceae, and Asteraceae (Blackman and Eastop, 2021). Although this aphid species can feed on phylogenetically and physiologically different host plants, at present there has been no report on host specificity or host races in it. However, previous studies discussed that the A. aurantii population on Ficus (Moraceae) exhibits some special features. Tao (1961) described Toxoptera schlingeri from Ficus, which was later considered as a synonym of Aphis (Toxoptera) aurantii by Raychaudhuri (1980) and Remaudière and Remaudiere (1997). Martin (1989) ever discussed that the validity of T. schlingeri might be supported by more in-depth study on the numbers and distribution of antennal rhinaria in more alatae samples. Qiao et al. (2008) found some morphological difference between T. schlingeri and A. aurantii specimens, but they suggested that morphological overlap would be found if more materials can be examined, and T. schlingeri was also regarded as a synonymy of A. aurantii in their paper. In subtropical and tropical areas of southern China, we observe that the morphology of A. aurantii varies across populations feeding on Ficus and other host plants, implying the Ficus population of A. aurantii may have undergone divergent evolution. Given the rapid advances in sequencing technology and new research methods such as DNA barcoding (Hebert et al., 2003; Foottit et al., 2008; Li et al., 2020) over the years, we think exploring the divergence of host-related populations of A. aurantii by integrating genetic evidence should be a worthwhile effort.
Considering A. aurantii and its host plants (especially Ficus) mainly distributed in subtropical and tropical areas (Volf et al., 2018; Blackman and Eastop, 2021), and with the aim to test sympatric population divergence of this species, our study took the subtropical Fujian province in southeastern China as target area. The specimens of A. aurantii were collected extensively to cover as many host plants as possible. Several molecular markers, including two mitochondrial genes (COI, cytochrome c oxidase subunit I; Cytb, cytochrome b), one nuclear gene (EF-1α, elongation factor-1α) and two genes (gnd, gluconate-6-phosphate dehydrogenase; 16S rDNA) of Buchnera, the primary endosymbiont of aphids, were analyzed to explore the genetic structure of A. aurantii sympatric host-populations. We also undertook morphometrics of A. aurantii samples feeding on Ficus and other host plants to test population divergence in morphology.
Materials and Methods
Specimen Sampling
A total of 48 A. aurantii specimens were collected from host plants of 11 families. The live morphology and habitats of A. aurantii in the field were photographed with digital cameras (Cannon EOS 7D plus Canon EF 100 mm f/2.8LMacro IS USM Lens). After recording the ecological information, aphid clones were stored in 95% ethanol and kept at −20°C for further morphological measurement and molecular experiments. All samples and voucher specimens were deposited in the Insect Systematics and Diversity Lab at Fujian Agriculture and Forestry University. Detailed information (host plant, voucher number, and GenBank accession number) of the specimens were listed in Supplementary Table 1.
DNA Extraction, PCR, and Sequencing
The genomic DNA of both aphids and Buchnera symbionts was extracted from each single specimen with the DNeasy Blood and Tissue Kit (QIAGEN, GERMANY). In order to obtain more accurate and comprehensive phylogenetic information, two mitochondrial genes (COI, Cytb), one nuclear gene (EF-1α) and two Buchnera genes (gnd and 16S rDNA) were amplified in this study. The primers for amplification of COI were LepF (5′-ATTCAACCAATCATAAAGATATTGG-3′) and LepR (5′-TAAACTTCTGGATGTCCAAAAAATCA-3′) (Foottit et al., 2008). Cytb sequences were amplified based on CP1 (5′-GATGATGAAATTGGATC-3′) and CP2 (5′-CTAATGCAATAACTCCTCC-3′) (Harry et al., 1998). The primers EF3 (5′-GAACGTGAACGTGGTATCAC-3′) and EF2 (5′-ATGTGAGCAGTGTGGCAATCCAA-3′) (von Dohlen et al., 2002) were used to amplify EF-1α sequences. gnd sequences were amplified based on BamHI (5′-CGCGGATCCGGWCCWWSWATWATGCCWGGWGG-3′) and ApaI (5′-CGCGGGCCCGTATGWGCWCCAAAATAATCW CKTTGWGCTTG-3′) (Clark et al., 1999). The primers 16SA1 (5’-AGAGTTTGATCMTGGCTCAG-3′) and 16SB1 (5′-TACGGYTACCTTGTTACGACTT-3′) were used to amplify 16S rDNA sequences (Weisburg et al., 1991).
PCR were performed in a final volume of 50 μl reaction mixture containing 28.5 μl dd H2O, 8 μl dNTPs, 5 μl 10Xbuffer, 4 μl of template DNA, 2 μl of both forward and reverse primers (10 μM) and 0.5 μl of Taq DNA polymerase (5 U/μl). An initial denaturation step (95°C, 5 min) and final extension step (72°C, 10 min) were included in all polymerase chain reactions. The cycling conditions for COI were 35 cycles of 20 s at 94°C, 30 s at 50°C and 2 min at 72°C. The thermal setup for Cytb was 35 cycles of 1 min at 92°C, 1.5 min at 48°C and 1 min at 72°C. The cycling conditions of EF-1α included 35 cycles of denaturation at 95°C for 30 s, annealing at 51°C for 30 s and extension at 72°C for 1 min. The conditions for 35 cycles of gnd were 95°C for 20 s, 53°C for 30 s, and 72°C for 2 min. The PCR conditions for 16S rDNA were according to the following procedure: 30 cycles at 94°C for 1 min; an annealing temperature of 50°C for 1 min; an extension at 72°C for 2 min. The PCR products were visualized by electrophoresis on a 1.5% agarose gel and then bidirectionally sequenced by Sangon Biotech (Shanghai). All sequences obtained in this study were uploaded to the GenBank, and the accession numbers were shown in Supplementary Table 1.
Based on the chromatograms, the raw sequences were corrected and assembled using BioEdit software (Hall, 1999). All sequences for each gene fragment were aligned using MAFFT (Kazutaka and Standley, 2013) and then verified manually. The introns of EF-1α sequences were removed based on GT-AG rule and the cDNA region of a reference sequence of Schizaphis graminum (GenBank accession number AF068479), and the coding regions of EF-1α were used in further phylogenetic analyses.
Genetic Distance and Phylogenetic Analysis
On the basis of current knowledge of the phylogenetic relationships within Aphididae, A. gossypii and A. spiraecola were chosen as outgroups for subsequent phylogenetic analyses. The COI, Cytb, EF-1α and gnd sequences of the outgroups were sequenced in our study, and the 16S rDNA for their Buchnera symbionts were downloaded from the GenBank (A. gossypii: KC897373, KC897372; A. spiraecola: KC897427, KT175934). The MEGA 7.0 software (Kumar et al., 2016) was used to calculate pairwise distance among nucleotide sequences based on the Kimura 2-parameter (K2P) model (Kimura, 1980). We also downloaded the COI sequences of Aphis species from the BOLD database,1 and then used SpeciesIdentifier software (Meier et al., 2006) to explore the distribution of intraspecific and interspecific genetic distances among Aphis species.
Phylogenetic analyses (Maximum likelihood, ML; Bayesian inference, BI) were performed based on three types of genes (mitochondrial: COI and Cytb; nuclear: EF-1α; Buchnera: gnd and 16S rDNA), respectively. The appropriate nucleotide substitution models were selected based on Akaike Information Criterion (AIC) by using PartitionFinder 2 (Lanfear et al., 2016) and ModelFinder (Kalyaanamoorthy et al., 2017). The best-fit model for COI was GTR + I, for Cytb was GTR + I, for EF-1α was GTR + F, for gnd was GTR + G, for 16S rDNA was GTR + I + G. RAxML (Stamatakis, 2014) was used to build the ML trees based on random starting trees with the GTRGAMMA substitution model and topological robustness was investigated using 1,000 non-parametric bootstrap replicates. Bayesian analyses were performed on all datasets using MrBayes 3.2.6 (Drummond et al., 2012). The combined dataset was divided into different gene partitions, and the best fitting models were assigned, respectively. For each dataset, two million generations MCMC (Markov Chain Monte Carlo) chains were run, with trees sampled every 100 generations. The first 5,000 trees (25%) for each dataset were discarded as burn-in to acquire posterior probability values (PP). The remaining trees were used to construct Bayesian consensus trees and viewed in iTOL (Letunic and Bork, 2016). In addition, the haplotypes based on the three gene datasets of different host-populations were analyzed using DnaSP 5.0 (Librado and Rozas, 2009). A median-joining network (MJ) was constructed using NETWORK 5.0.0.3 based on default setting (Bandelt et al., 1999).
Morphometry and Statistical Analysis
The samples used for morphometrics were collected from populations on eight genera of main host plants, including Ficus, Calliandra, Camellia, Citrus, Loropetalum, Michelia, Morinda, Xylosma. In principle, 3 clones of A. aurantii were selected from each genus of host plant, and then 10 adult apterous viviparous females were randomly selected from each aphid clone for morphological measurement. For some aphid clones with an insufficient number of adults, only those meeting above criteria were measured (Supplementary Table 2). All specimens were examined using Nikon SMZ18 stereomicroscope. A total of 20 morphological features were measured: body length (BL), body width (BW), length of 1st antennal segment (Ant1), length of 2nd antennal segment (Ant2), length of 3rd antennal segment (Ant3), hair length of 3rd antennal segment (Ant3_HL), width of 3rd antennal segment (Ant3_W), length of 4rth antennal segment (Ant4), length of 5th antennal segment (Ant5), base length of 6th antennal segment (Ant6_BL), processus terminalis of 6th antennal segment (Ant6_PT), whole antennal length (WA), hind femur (HF), siphunculi length (SIPH), basal width of siphunculi (SIPH_BW), distal width of siphunculi (SIPH_DW), cauda length (Cauda), basal width of cauda (Cauda_BW), length of dorsal hairs of tergite 1 (T1_DHL), hair length of tergite 8 (T8_HL).
We also calculated the ratios of WA/BL, Ant1/WA, Ant2/WA, Ant3/WA, Ant3_HL/WA, Ant3_W/WA, Ant4/WA, Ant5/WA, Ant6_BL/WA, Ant6_PT/WA, Ant3_HL/BL, Ant3_HL/Ant3, Ant3_HL/Ant3_W, Ant3_W/T1_DHL, Ant6_BL/Ant6_PT, T1_DHL/BL, SIPH/BL, SIPH/Cauda as supplementary morphological characters. The average as well as the minimum and maximum values of each morphological character for A. aurantii from different host plants were calculated separately (Supplementary Table 2). A one-way analysis of variance (ANOVA) was performed for 38 morphological characters to determine whether significant morphological difference among A. aurantii different host-populations can be found. In addition, post hoc multiple comparisons were performed based on LSD to detect the pairwise differences of each morphological feature between taxa. All statistical analyses were performed in SPSS ver. 24 (IBM, Chicago, IL, United States).
Results
Sequence Features and Genetic Variation
Five gene fragments of most samples were successfully amplified. The 610 bp long COI alignment with 48 sequences included 595 conserved sites, 15 variable sites, and 15 parsimony-informative sites. The 45 Cytb sequences were trimmed to a 732 bp long alignment with 681 conserved sites, 51 variable sites, and 22 parsimony-informative sites. The 44 exon sequences of EF-1α were aligned to a final length of 712 bp, which included 703 conserved sites, 9 variable sites, and 5 parsimony-informative sites. A total of 45 gnd sequences (821 bp, conserved sites: 765; variable sites: 56; parsimony-informative sites: 54) and 46 16S rDNA sequences (337 bp, conserved sites: 329; variable sites: 8; parsimony-informative sites: 4) were successfully generated. The nucleotide composition of mitochondrial gene (COI and Cytb) and gnd fragments showed a strong bias toward A + T content (76, 77.5, and 75.3%, respectively), while EF-1α and 16S have no similar bias (Supplementary Table 3).
The sequences of each gene were divided into two groups, group 1 feeding on nearly 20 other host plants and group 2 feeding on Ficus, then the genetic distances within and between groups of different genes were calculated, respectively (Supplementary Table 3). Using the COI gene as an example, we found that the genetic distance of samples between groups was much larger than that within groups. The genetic distance range of samples on Ficus was 0–0%, and that of samples feeding on other host plant was 0–0.8%. However, the genetic distances between samples from Ficus and other host plants could reach as high as 1.8%. Within group 1, the Theaceae population contributed the largest genetic distance (0.8%) with other host-populations (Supplementary Table 3).
The COI sequences of Aphis downloaded from BOLD database were collated and corrected, and 3,429 sequences of 99 species were obtained. The analysis of distribution of the intraspecific and interspecific genetic distances of Aphis species showed an obvious barcoding gap (Supplementary Figure 1). For the intraspecific distances, 97.56% of them were less than 1%, 98.04% less than 1.5, and 99.1% less than 2%. Besides, 99.18 and 98.68% of the interspecific distances were greater than 2 and 2.5% (Supplementary Table 4).
Phylogeny and Haplotype Network
The phylogenetic trees that inferred from mitochondrial (COI and Cytb) and Buchnera (gnd and 16S) genes showed that sympatric host-populations of A. aurantii were divided into two well-supported clades (Figures 1A,B), corresponding to the populations feeding on Ficus (G2) and the other plants (G1), respectively. All the samples feeding on Ficus were clustered into a separate clade at the base of the phylogenetic tree. In addition, some populations of A. aurantii feeding on Theaceae in the G1 clade also showed relatively obvious divergence (Figures 1A,B). The nuclear gene (EF-1α), which was most conserved among the five gene markers, however, showed a less unambiguous phylogenetic pattern (Figure 1C).
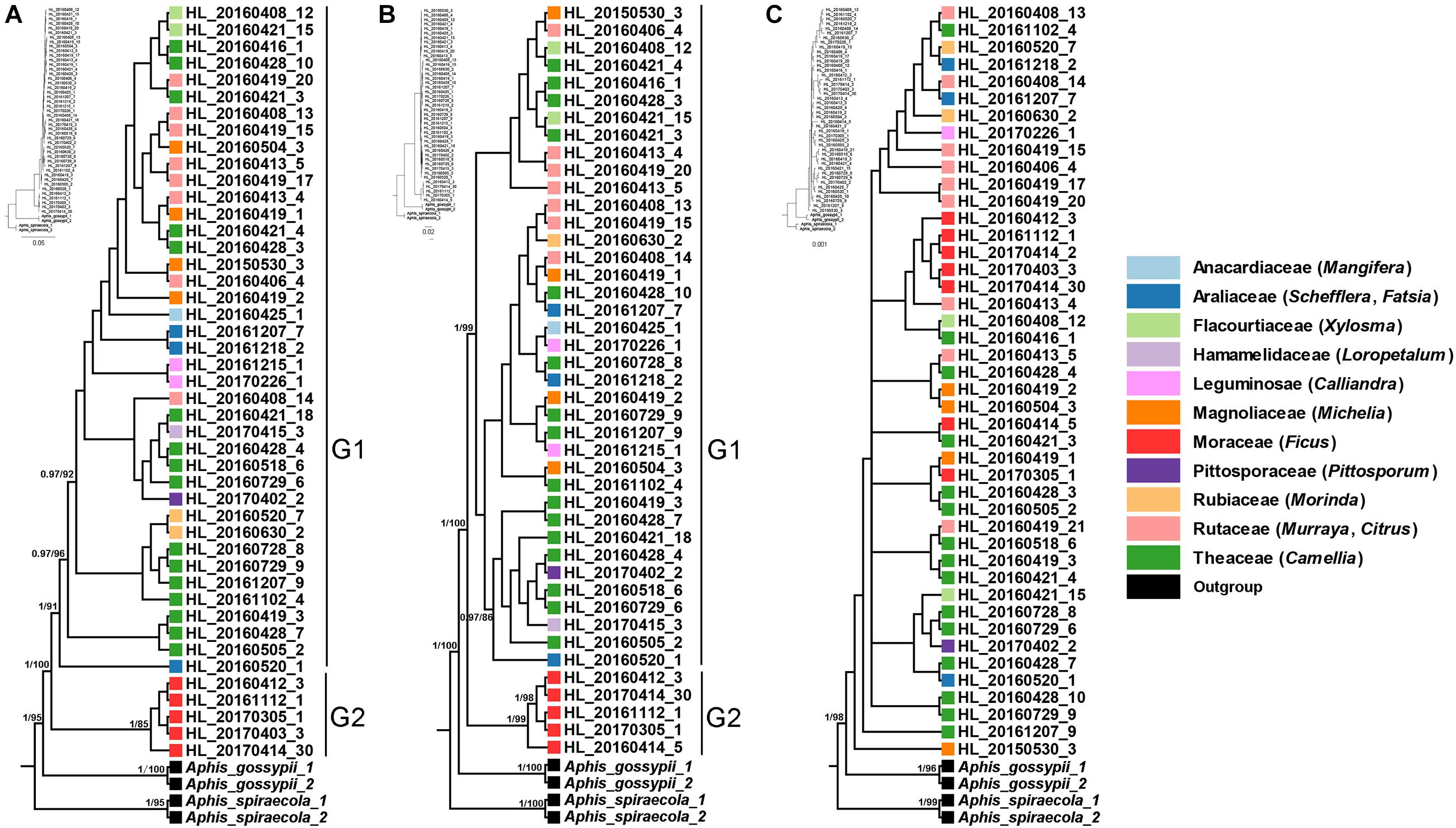
Figure 1. Phylogeny of Aphis aurantii populations inferred from Bayesian analyses (BI) based on COI + Cytb (A), gnd and 16S (B) and EF-1α (C). Different color represent samples from different host plant families. The bootstrap values (>80) and posterior probabilities (>0.9) are shown for main nodes.
A total of 16 haplotypes were identified among the 45 mitochondrial (COI and Cytb) sequences (Figure 2A). Haplotype H1 contains the most samples and host plant families. The most frequently observed host plant was Theaceae and appeared in several haplotypes (H1, H5, H6, H8, H12, H13, H14, H15). All the samples feeding on Ficus were assigned as haplotype H4, which showed greatest differentiation from other haplotypes. Similarly, in the haplotype network analyses of nuclear and Buchnera genes, there were significant genetic differences among the populations feeding on Ficus and other host plants (Figures 2B,C).
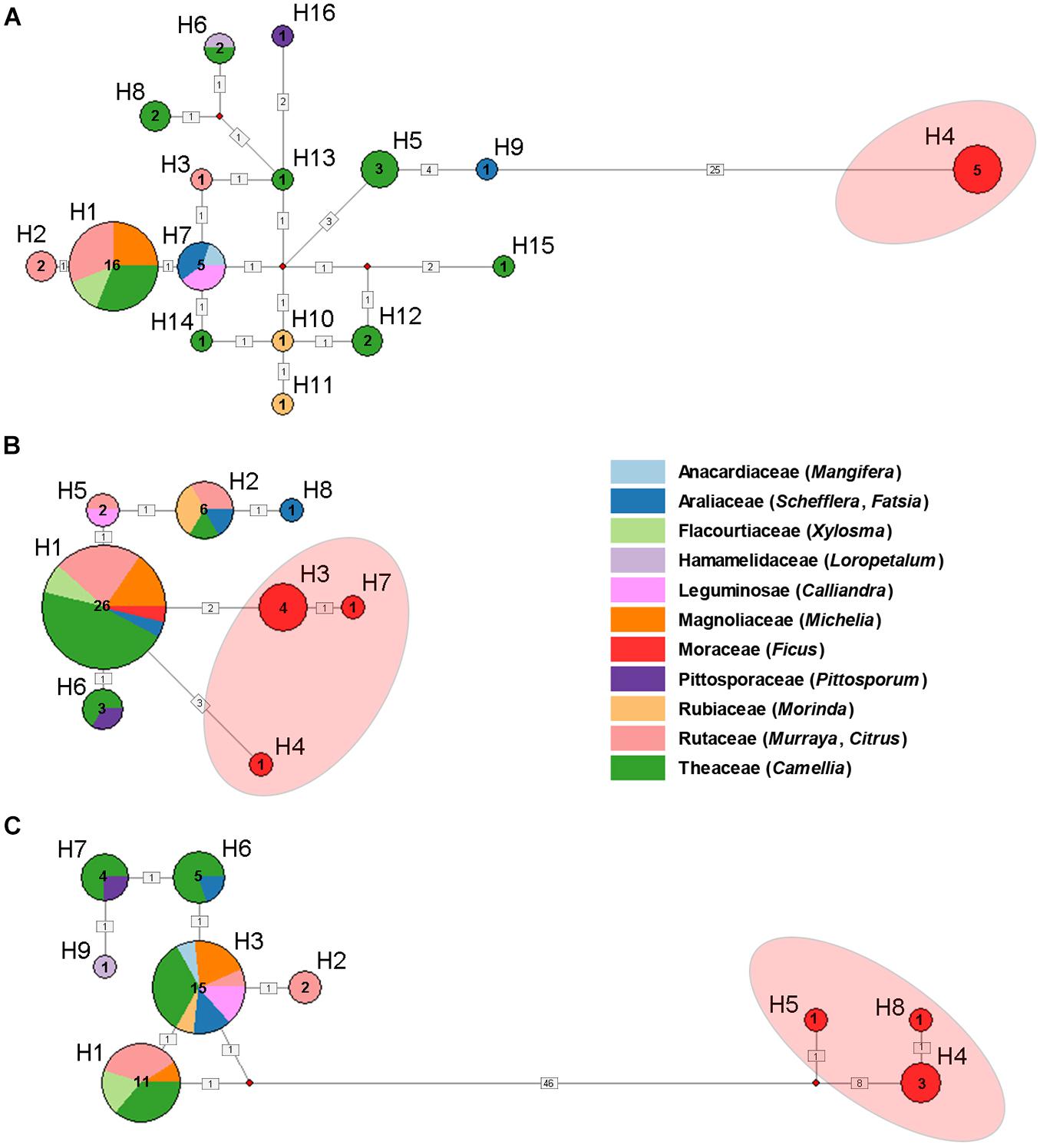
Figure 2. Haplotype network of Aphis aurantii populations based on COI + Cytb (A), EF-1α (B) and gnd and 16S (C). Numbers near the circle indicate haplotype numbers. Sample numbers and host plants (in different color) of haplotypes are annotated in the circles. The numbers in the black square indicate number of mutations. The red shading on the haplotype network represents Ficus samples.
Morphology and Statistics
The results of the ANOVA showed that most morphological characters exhibited significant difference between different host plant genera (P < 0.05), except for the length of Ant1, Ant2, Ant4, T8_HL and ratio of WA/BL, Ant6_BL/WA, Ant6_BL/Ant6_PT (Supplementary Table 5). With integration of the results of post hoc analysis by LSD, morphological characters having significant difference between populations on Ficus and other host plant genera were determined according to a rule that there should have samples from more than four different plant genera showing significant difference with the Ficus samples. Our results indicated that there were significant difference in eighteen morphological characters between A. aurantii populations on Ficus and other plant genera, including length of Ant3_HL, Ant5, Ant6_PT, WA, SIPH_DW, Cauda, T1_DHL; ratio of WA/BL, Ant3/WA, Ant3_HL/WA, Ant3_W/WA, Ant5/WA, Ant3_HL/Ant3, Ant3_HL/Ant3_W, Ant3_HL/BL, Ant3_W/T1_DHL, SIPH/Cauda, T1_DHL/BL (Figure 3 and Supplementary Table 5). Among these morphological characters, Ant3_HL, T1_DHL, Ant3_HL/WA, Ant3_HL/Ant3_W, Ant3_HL/BL, Ant3_W/T1_DHL, T1_DHL/BL of the Ficus population showed significant difference with samples from all other plant genera. That is to say, at least seven morphological characters can be used to distinguish the A. aurantii population feeding on Ficus from those feeding on other host plants.
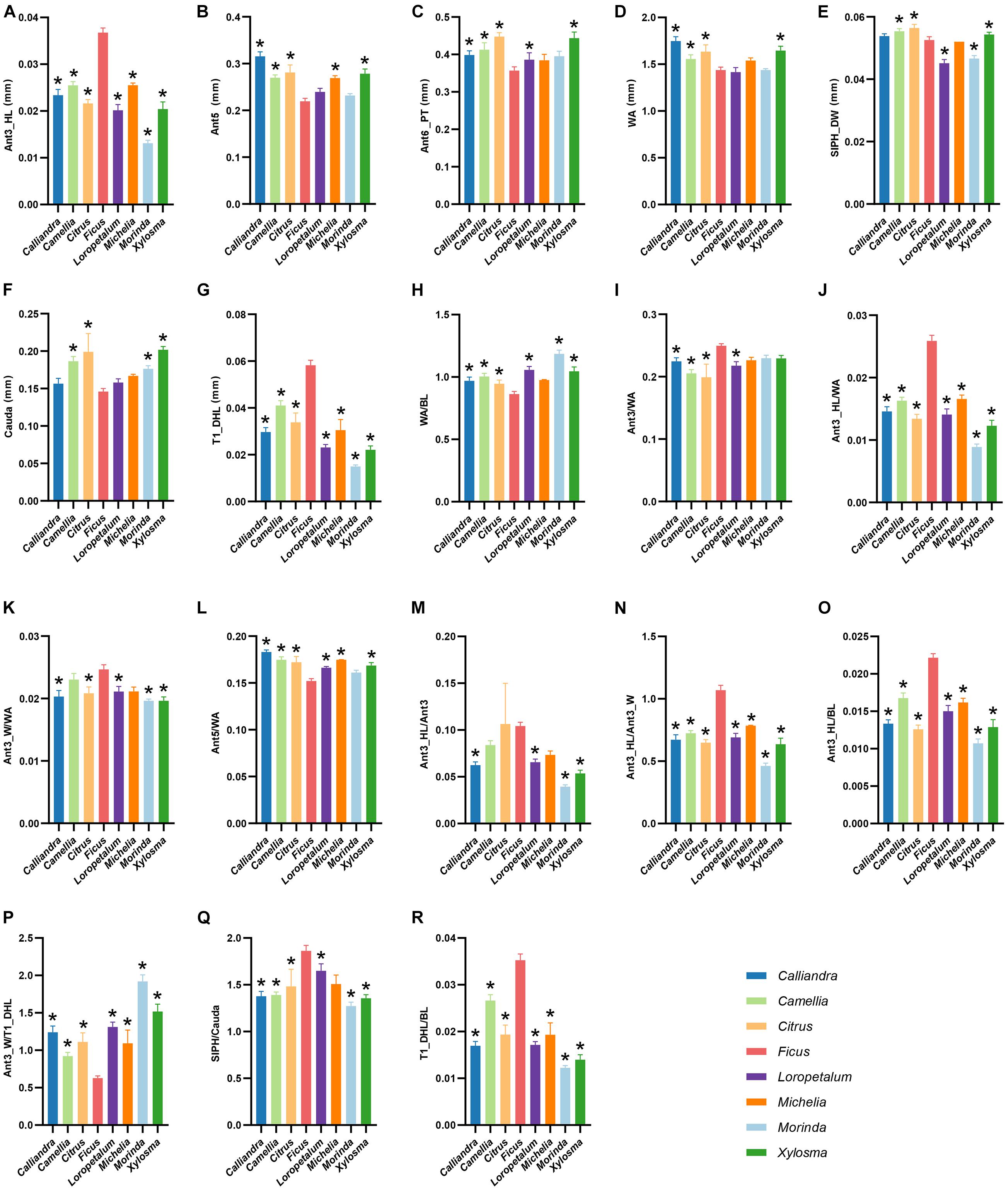
Figure 3. Eighteen morphological characters with significant difference between A. aurantii populations feeding on Ficus and other plant genera, including Ant3_HL (A), Ant5 (B), Ant6_PT (C), WA (D), SIPH_DW (E), Cauda (F), T1_DHL (G); ratios of WA/BL (H), Ant3/WA (I), Ant3_HL/WA (J), Ant3_W/WA (K), Ant5/WA (L), Ant3_HL/Ant3 (M), Ant3_HL/Ant3_W (N), Ant3_HL/BL (O), Ant3_W/T1_DHL (P), SIPH/Cauda (Q), T1_DHL/BL (R). Different color bars represent samples from different host plant genera. *Represents host-populations significantly different with the Ficus population.
Discussion
Host plants are considerable source of selective pressure for their associated aphids, constituting their only food resource, habitat, mating and oviposition sites (Peccoud and Simon, 2010; Peccoud et al., 2010; Blackman and Eastop, 2021). In the past three decades, the pea aphid A. pisum has been considered as a good model to investigate host specialization and sympatric speciation (Via, 1991a,b). In Western Europe, at least eight conspecific host races were found on different host plants, which showed that adaptation to different host plants could indeed play a very important role in sympatric speciation of insects (Peccoud et al., 2009). In the present study, by using the polyphagous A. aurantii as a model, we tested whether host-related divergence exist among sympatric populations. Our results based on mitochondrial (COI and Cytb) and Buchnera (gnd and 16S) genes clearly showed that all samples feeding on Ficus were clustered in a separate clade with relatively high divergence (Figure 1) and formed a unique haplotype (Figure 2), indicating that the Ficus population of A. aurantii have experienced a host specialization process. In addition, we also noted that the nuclear gene (EF-1α) did not show a clear evolutionary relationship. Considering EF-1α is a relatively conserved nuclear gene (Cho et al., 1995), we think the lack of accumulation of sufficient genetic variation at the early stage of species divergence may be an important factor leading to unclear phylogenetic pattern of this gene. The conflict of phylogenetic signals between different types of genes, in fact, indicates they these genes may undergo different histories of lineage sorting in the process of species divergence.
In previous DNA barcoding studies, COI genetic distance thresholds were used to determine different animal species (Hajibabaei et al., 2006; Zhou et al., 2010; Schmidt et al., 2015). Although the genetic distance thresholds vary slightly among different taxonomic groups, a threshold of 2–2.5% is generally accepted by aphid taxonomists (Liu et al., 2013; Zhu et al., 2017; Li et al., 2020). Our analysis of distribution of the intraspecific and interspecific genetic distances of Aphis species (Supplementary Figure 1 and Supplementary Table 4) also confirmed this threshold. For most Aphis species, 2–2.5% can be used as a reasonable genetic distance threshold to distinguish different species. In the present study, the maximum COI genetic distance between the G1 and G2 clades (Supplementary Table 3) reached 1.8%, which exceeds about 99% of the intraspecific genetic difference of the Aphis species dataset, indicating a relatively deep divergence of the Aphis aurantii lineage on Ficus.
The ecological adaptation of phytophagous insects may first occur at the physiological and genetic levels, and is sometimes relatively unobvious in phenotype (Doolittle and Sapienza, 1980; Kearney and Porter, 2009). Since identification of species in traditional taxonomy is mainly based on morphological characters, this phenomenon of insignificant phenotypic information might cause troubles for taxonomy, especially when specimens and characters are insufficiently sampled. Morphological comparison of different host-populations of A. aurantii showed that among the 38 characters, more characters were relatively similar. For instances, the body length, body width, hind femur and siphunculi did not show significant difference. Tao (1961) described T. schlingeri just based on a single sample collected from Ficus in Hong Kong. Qiao et al. (2008) compared the morphology of T. schlingeri and A. aurantii with more apterous viviparous female specimens, and found only the length of marginal hairs on abdominal tergite I and the widest diameter of antennal segment III were different. They suggested that further examination and thorough research were needed to determine the validity of T. schlingeri. Our measurement results of the length of Ant3_HL and T1_DHL were similar to those of Qiao et al. (2008), in addition, we also found that ratios of Ant3_HL/WA, Ant3_HL/Ant3_W, Ant3_HL/BL, Ant3_W/T1_DHL, T1_DHL/BL of the Ficus population of A. aurantii were significantly different from that of the populations on other host plants. By integrating the genetic divergence and morphological difference of A. aurantii on Ficus and other plants, and our field observations on this species, we think a reasonable explanation is that the specimen used to define T. schlingeri might be actually an individual from the population of A. aurantii specialized on Ficus. And the Ficus population of A. aurantii may represent a lineage that is experiencing ongoing sympatric speciation.
The physical and chemical characters of host plants are important factors that lead to the specialization of phytophagous insects. For example, the tobacco biotype of M. persicae can respond specifically to the volatile released by tobacco, whereas other biotypes do not, suggesting that difference in olfactory perception exists among different biotypes (Vargas et al., 2005). Furthermore, gustatory reception also affects the aphids’ selection of host plants, as aphids probe the phloem for nutritional composition and chemical information before deciding whether to feed (Caillaud and Via, 2000; Powell et al., 2006). Ficus is a special plant group. It can produce a broad range of chemical and physical defenses to against phytophagous insects (Cruaud et al., 2012). These include the production of latex, polyphenols and terpenoids. Ficus can produce some special chemical substances, including phenanthroindolizidine alkaloids (Damu et al., 2005) and cysteine proteases, which can interfere the digestive function of insects and increase the mortality of them (Konno et al., 2004). Considering A. aurantii shows obvious host specialization on Ficus, it can be a good model for investigating the mechanism of host specialization. Further studies on comparative transcriptomics among different host populations and detailed screening of symbiotic bacteria are extremely needed.
Data Availability Statement
The sequence data are publicly archived in the GenBank, and the accession numbers are provided in Supplementary Table 1.
Author Contributions
XH conceived and designed the study, reviewed and edited the manuscript, and contributed resources during the study. QLi, CC, JAS, YL, and QLiu performed the experiments. QLi, CC, YW, CL, and ZC analyzed the data. QLi, CC, CL, and ZC wrote the original draft. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 31772504) and the Fujian Provincial Department of Science and Technology (Grant No. 2015J06005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Xiaolan Lin and Lingda Zeng for help in sample collection and molecular experiment, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.786450/full#supplementary-material
Supplementary Figure 1 | Frequency histogram of intra- and inter-specific genetic distances of Aphis COI sequences downloaded from the BOLD database.
Supplementary Table 1 | Sample information including voucher number, host plant family and species, and GenBank accession number.
Supplementary Table 2 | Morphometrics of A. aurantii samples collected on different host plant genera.
Supplementary Table 3 | The nucleotide composition and genetic distances of A. aurantii feeding on different host plants.
Supplementary Table 4 | The distribution of intraspecific and interspecific distances among Aphis COI sequences.
Supplementary Table 5 | Result of one-way ANOVA and post hoc LSD for morphological characters of A. aurantii samples collected on different host plant genera. Calliandra (n = 15), Camellia (n = 30), Citrus (n = 28), Ficus (n = 30), Loropetalum (n = 20), Michelia (n = 20), Morinda (n = 10), Xylosma (n = 10).
Footnotes
References
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Berlocher, S. H., and Feder, J. L. (2002). Sympatric speciation in phytophagous insects: moving beyond controversy? Annu. Rev. Entomol. 47, 773–815. doi: 10.1146/annurev.ento.47.091201.145312
Blackman, R. L., and Eastop, V. F. (2021). Aphids on the World’s Plants: an online identification and information guide. Available Online at: http://www.aphidsonworldsplants.info/ (accessed June 23, 2021)
Bolnick, D. I., and Fitzpatrick, B. M. (2007). Sympatric speciation: models and empirical evidence. Annu. Rev. Ecol. Evol. Syst. 38, 459–487. doi: 10.1146/annurev.ecolsys.38.091206.095804
Caillaud, M. C., and Via, S. (2000). Specialized feeding behavior influences both ecological specialization and assortative mating in sympatric host races of pea aphids. Am. Nat. 156, 606–621. doi: 10.1086/316991
Carletto, J., Lombaert, E., Chavigny, P., Brévault, T., Lapchin, L., and Vanlerberghe Masutti, F. (2009). Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol. Ecol. 18, 2198–2212. doi: 10.1111/j.1365-294X.2009.04190.x
Carver, M. (1978). The black citrus aphids, Toxoptera citricidus (Kirkaldy) and T. aurantii (Boyer de Fonscolombe) (Homoptera: aphididae). Aust. J. Entomol. 17, 263–270. doi: 10.1111/j.1440-6055.1978.tb00156.x
Cho, S., Mitchell, A., Regier, J. C., Mitter, C., Poole, R. W., Friedlander, T. P., et al. (1995). A highly conserved nuclear gene for low-level phylogenetics: elongation factor-1 alpha recovers morphology-based tree for heliothine moths. Mol. Biol. Evol. 12, 650–656. doi: 10.1093/oxfordjournals.molbev.a040244
Clark, M. A., Moran, N. A., and Baumann, P. (1999). Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 16, 1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071
Cruaud, A., Rønsted, N., Chantarasuwan, B., Chou, L. S., Clement, W. L., Couloux, A., et al. (2012). An extreme case of plant-insect codiversification: figs and fig-pollinating wasps. Syst. Biol. 61, 1029–1047. doi: 10.1093/sysbio/sys068
Damu, A. G., Kuo, P. C., Shi, L. S., Li, C. Y., Kuoh, C. S., Wu, P. L., et al. (2005). Phenanthroindolizidine alkaloids from the stems of Ficus septica. J. Nat. Prod. 68, 1071–1075. doi: 10.1021/np050095o
Darwin, C. R. (1859). The origin of species by means of natural selection: or, the preservation of favored races in the struggle for life. London, UK: John Murray.
Doolittle, W. F., and Sapienza, C. (1980). Selfish genes, the phenotype paradigm and genome evolution. Nature 284, 601–603. doi: 10.1038/284601a0
Drès, M., and Mallet, J. (2002). Host races in plant-feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 357, 471–492. doi: 10.1098/rstb.2002.1059
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Eastop, V. (1973). “Deductions from the present day host plants of aphids and related insects,” in Insect/Plant relationships, ed. H. F. V. Emden (London: Symposia of the Royal Entomological Society of London), 157–178.
Egan, S. P., and Ott, J. R. (2007). Host plant quality and local adaptation determine the distribution of a gall-forming herbivore. Ecology 88, 2868–2879. doi: 10.1890/06-1303.1
Foottit, R. G., Maw, H. E., Von Dohlen, C. D., and Hebert, P. D. (2008). Species identification of aphids (Insecta: hemiptera: aphididae) through DNA barcodes. Mol. Ecol. Resour. 8, 1189–1201. doi: 10.1111/j.1755-0998.2008.02297.x
Futuyma, D. J., and Mayer, G. C. (1980). Non-allopatric speciation in animals. Syst. Biol. 29, 254–271. doi: 10.1093/sysbio/29.3.254
Hajibabaei, M., Janzen, D. H., Burns, J. M., Hallwachs, W., and Hebert, P. D. N. (2006). DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. U. S. A. 103, 968–971. doi: 10.1073/pnas.0510466103
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Harry, M., Solignac, M., and Lachaise, D. (1998). Molecular evidence for parallel evolution of adaptive syndromes in fig-breeding Lissocephala (Drosophilidae). Mol. Phylogenet. Evol. 9, 542–551. doi: 10.1006/mpev.1998.0508
Hebert, P. D., Ratnasingham, S., and De Waard, J. R. (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270, S96–S99. doi: 10.1098/rsbl.2003.0025
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kazutaka, K., and Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearney, M., and Porter, W. (2009). Mechanistic niche modelling: combining physiological and spatial data to predict species’ ranges. Ecol. Lett. 12, 334–350. doi: 10.1111/j.1461-0248.2008.01277.x
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Konno, K., Hirayama, C., Nakamura, M., Tateishi, K., Tamura, Y., Hattori, M., et al. (2004). Papain protects papaya trees from herbivorous insects: role of cysteine proteases in latex. Plant J. 37, 370–378. doi: 10.1046/j.1365-313X.2003.01968.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution formolecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lee, Y., Lee, W., Lee, S., and Kim, H. (2015). A cryptic species of Aphis gossypii (Hemiptera: aphididae) complex revealed by genetic divergence and different host plant association. Bull. Entomol. Res. 105, 40–51. doi: 10.1017/S0007485314000704
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, K., Hong, W., Jiao, H., Wang, G.-D., Rodriguez, K. A., Buffenstein, R., et al. (2015). Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proc. Natl. Acad. Sci. U. S. A. 112, 11905–11910. doi: 10.1073/pnas.1514896112
Li, Q., Deng, J., Chen, C., Zeng, L., Lin, X., Cheng, Z., et al. (2020). DNA Barcoding Subtropical Aphids and Implications for Population Differentiation. Insects 11:11. doi: 10.3390/insects11010011
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, Q. H., Jiang, L. Y., and Qiao, G. X. (2013). DNA barcoding of Greenideinae (Hemiptera: Aphididae) with resolving taxonomy problems. Invertebr. Syst. 27, 428–438. doi: 10.1071/IS13014
Malausa, T., Bethenod, M.-T., Bontemps, A., Bourguet, D., Cornuet, J.-M., and Ponsard, S. (2005). Assortative mating in sympatric host races of the European corn borer. Science 308, 258–260. doi: 10.1126/science.1107577
Mallet, J. (2008). Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2971–2986. doi: 10.1098/rstb.2008.0081
Margaritopoulos, J., Tsourapas, C., Tzortzi, M., Kanavaki, O., and Tsitsipis, J. (2005). Host selection by winged colonisers within the Myzus persicae group: a contribution towards understanding host specialisation. Ecol. Entomol. 30, 406–418. doi: 10.1111/j.0307-6946.2005.00700.x
Martin, J. (1989). Identification, occurrence and pest status of Toxoptera odinae (van der Goot) (Hemiptera: aphididae) in Africa. Bull. Entomol. Res. 79, 607–611. doi: 10.1017/S0007485300018757
McKinnon, J. S., Mori, S., Blackman, B. K., David, L., Kingsley, D. M., Jamieson, L., et al. (2004). Evidence for ecology’s role in speciation. Nature 429, 294–298. doi: 10.1038/nature02556
Meier, R., Shiyang, K., Vaidya, G., and Ng, P. K. (2006). DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst. Biol. 55, 715–728. doi: 10.1080/10635150600969864
Nosil, P. (2004). Reproductive isolation caused by visual predation on migrants between divergent environments. Proc. R. Soc. Lond. Ser. B Biol. Sci. 271, 1521–1528. doi: 10.1098/rspb.2004.2751
Nosil, P., and Crespi, B. J. (2006). Experimental evidence that predation promotes divergence in adaptive radiation. Proc. Natl. Acad. Sci. U. S. A. 103, 9090–9095. doi: 10.1073/pnas.0601575103
Peccoud, J., Ollivier, A., Plantegenest, M., and Simon, J. C. (2009). A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl. Acad. Sci. U. S. A. 106, 7495–7500. doi: 10.1073/pnas.0811117106
Peccoud, J., and Simon, J. C. (2010). The pea aphid complex as a model of ecological speciation. Ecol. Entomol. 35, 119–130. doi: 10.1111/j.1365-2311.2009.01147.x
Peccoud, J., Simon, J. C., Von Dohlen, C., Coeur D’acier, A., Plantegenest, M., Vanlerberghe Masutti, F., et al. (2010). Evolutionary history of aphid-plant associations and their role in aphid diversification. C. R. Biol. 333, 474–487. doi: 10.1016/j.crvi.2010.03.004
Powell, G., Tosh, C. R., and Hardie, J. (2006). Host plant selection by aphids: behavioral, evolutionary, and applied perspectives. Annu. Rev. Entomol. 51, 309–330. doi: 10.1146/annurev.ento.51.110104.151107
Qiao, G., Wang, J., and Zhang, G. (2008). Toxoptera Koch (Hemiptera: aphididae), a generic account, description of a new species from China, and keys to species. Zootaxa 1746, 1–14. doi: 10.11646/zootaxa.1746.1.1
Raychaudhuri, D. N. (1980). Aphids of North-East India and Bhutan. Calcutta: The Zoological Society.
Remaudière, G., and Remaudiere, M. (1997). Catalogue of the world’s Aphididae: homoptera Aphidoidea. Paris: Institut National de la Recherche Agronomique (INRA).
Rull, J., Wharton, R., Feder, J. L., Guillén, L., Sivinski, J., Forbes, A., et al. (2009). Latitudinal variation in parasitoid guild composition and parasitism rates of North American hawthorn infesting Rhagoletis. Environ. Entomol. 38, 588–599. doi: 10.1603/022.038.0310
Savolainen, V., Anstett, M. C., Lexer, C., Hutton, I., Clarkson, J. J., Norup, M. V., et al. (2006). Sympatric speciation in palms on an oceanic island. Nature 441, 210–213. doi: 10.1038/nature04566
Schmidt, S., Schmid-Egger, C., Morinière, J., Haszprunar, G., and Hebert, P. D. (2015). DNA barcoding largely supports 250 years of classical taxonomy: identifications for Central European bees (Hymenoptera, Apoidea partim). Mol. Ecol. Resour. 15, 985–1000. doi: 10.1111/1755-0998.12363
Sevim, E., Çelebi, Ö, and Sevim, A. (2012). Determination of the bacterial flora as a microbial control agent of Toxoptera aurantii (Homoptera: aphididae). Biologia 67, 397–404. doi: 10.2478/s11756-012-0022-0
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tao, C. C. (1961). Revision of the genus Toxoptera Koch, 1856 (Homoptera: aphididae). Q. J. Taiwan Mus. 14, 257–260.
Taylor, R. S., and Friesen, V. L. (2017). The role of allochrony in speciation. Mol. Ecol. 26, 3330–3342. doi: 10.1111/mec.14126
Vargas, R. R., Troncoso, A. J., Tapia, D. H., Olivares-Donoso, R., and Niemeyer, H. M. (2005). Behavioural differences during host selection between alate virginoparae of generalist and tobacco-specialist Myzus persicae. Entomol. Exp. Appl. 116, 43–53. doi: 10.1111/j.1570-7458.2005.00311.x
Via, S. (1991a). Specialized host plant performance of pea aphid clones is not altered by experience. Ecology 72, 1420–1427. doi: 10.2307/1941114
Via, S. (1991b). The genetic structure of host plant adaptation in a spatial patchwork: demographic variability among reciprocally transplanted pea aphid clones. Evolution 45, 827–852. doi: 10.1111/j.1558-5646.1991.tb04353.x
Via, S. (2001). Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 16, 381–390. doi: 10.1016/S0169-5347(01)02188-7
Via, S., Bouck, A. C., and Skillman, S. (2000). Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution 54, 1626–1637. doi: 10.1111/j.0014-3820.2000.tb00707.x
Volf, M., Segar, S. T., Miller, S. E., Isua, B., Sisol, M., Aubona, G., et al. (2018). Community structure of insect herbivores is driven by conservatism, escalation and divergence of defensive traits in Ficus. Ecol. Lett. 21, 83–92. doi: 10.1111/ele.12875
von Dohlen, C. D., Kurosu, U., and Aoki, S. (2002). Phylogenetics and evolution of the eastern Asian-eastern North American disjunct aphid tribe, Hormaphidini (Hemiptera: aphididae). Mol. Phylogenet. Evol. 23, 257–267. doi: 10.1016/S1055-7903(02)00025-8
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Xue, H.-J., Li, W.-Z., and Yang, X.-K. (2014). Assortative mating between two sympatric closely-related specialists: inferred from molecular phylogenetic analysis and behavioral data. Sci. Rep. 4:5436. doi: 10.1038/srep05436
Zhou, X., Jacobus, L. M., Dewalt, R. E., Adamowicz, S. J., and Hebert, P. D. (2010). Ephemeroptera, Plecoptera, and Trichoptera fauna of Churchill (Manitoba, Canada): insights into biodiversity patterns from DNA barcoding. J. N. Am. Benthol. Soc. 29, 814–837. doi: 10.1899/09-121.1
Keywords: adaptation, host plant, population divergence, phylogeny, speciation
Citation: Li Q, Chen C, Wu Y, Siddiqui JA, Lu C, Cheng Z, Li Y, Liu Q and Huang X (2021) Specialization on Ficus Supported by Genetic Divergence and Morphometrics in Sympatric Host-Populations of the Camellia Aphid, Aphis aurantii. Front. Ecol. Evol. 9:786450. doi: 10.3389/fevo.2021.786450
Received: 30 September 2021; Accepted: 21 October 2021;
Published: 18 November 2021.
Edited by:
Xin Zhou, China Agricultural University, ChinaReviewed by:
Cheng-Min Shi, Agricultural University of Hebei, ChinaQiang Xie, Sun Yat-sen University, China
Copyright © 2021 Li, Chen, Wu, Siddiqui, Lu, Cheng, Li, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolei Huang, aHVhbmd4bEBmYWZ1LmVkdS5jbg==
 Qiang Li1
Qiang Li1 Yangxue Wu
Yangxue Wu Junaid Ali Siddiqui
Junaid Ali Siddiqui Xiaolei Huang
Xiaolei Huang