
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Ecol. Evol. , 10 January 2022
Sec. Population, Community, and Ecosystem Dynamics
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.785019
This article is part of the Research Topic Kin Selection and Kin Cooperation in Plants View all 7 articles
Kin recognition, manifesting through various traits such as changes in root or shoot growth, has been documented in several species of plants. Identifying this phenomenon in plants has intrinsic value itself, understanding why plants recognize kin and how it might benefit them evolutionarily has been of recent interest. Here we explore studies regarding nutrient and resource allocation in regard to kin recognition as well as discuss how kin recognition is involved in multispecies interactions with an emphasis on how plant roots are involved in these processes. Future directions of this research are also discussed.
Throughout the biological world, we see diverse examples of animal populations interacting and often cooperating with tasks such as obtaining food, protecting young, building dens, etc. What many of these animals have in common is that they are often part of groups where many of the members share genetic relatedness and they have the ability to recognize the individuals in their group and identify them as kin (related) or non-kin (not related) (Hanggi and Schusterman, 1990; Mateo, 2003). Identification of other member’s relatedness may facilitate inbreeding avoidance or cooperative behaviors (Sharp et al., 2005; Gerlach and Lysiak, 2006; Carazo et al., 2014). Cooperative or altruistic behaviors that benefit the population can evolve if the benefit to a group of related individuals outweighs the direct costs to the individuals themselves (Hamilton, 1964). In general, cooperation between animals is more likely to occur between closely related rather than distantly or unrelated individuals (West et al., 2021). Cooperation between kin can be common in harsh environments, although not always as a result of the environment, but often because cooperation can be favored in these hard to survive locations (West et al., 2021). Additionally, the costs and benefits of cooperation may differ among animal species and be dependent upon unique situations such as competition with other species, food availability, climate and habitat availability (Lin et al., 2019; West et al., 2021).
Despite the fact that plants have evolved and thrived in the same harsh environments as animal species it was not until the last few decades that scientists have begun to understand the complexities of plant-plant interactions in their environment (Baldwin et al., 2006; Sanon et al., 2009; Kong et al., 2018). Unlike most animal species, plants have a more limited ability to relocate to a different site if a stressful situation such as disease, drought, or competition for nutrients or light arises. Plants have adapted to these biotic and abiotic challenges in a myriad of ways including immune responses, dynamic shoot growth, root plasticity, resource allocation, secretion of allelochemicals and shoot volatile emissions (Kessler and Baldwin, 2001; Jones and Dangl, 2006; Haig, 2008; Murphy and Dudley, 2009; Karban et al., 2013). We also know that plant responses in growth can be influenced by the genetic relatedness of their neighbors. Just as in certain animal species, several species of plants have been found to have the ability to distinguish individuals as related or unrelated plants (Dudley and File, 2007; Biedrzycki et al., 2010; Karban et al., 2013). Early kin studies by Dudley and File (2007) demonstrated that Cakile edentula plant root growth patterns differed when plants were grown with strangers (plants that were from the same species but a different mother) vs kin (plants that were siblings from the same mother). With the discovery that plants do indeed recognize kin and change their root growth based on the relatedness of the surrounding individuals, determining why and how plants were communicating during these behaviors were the next questions. The concept of kin recognition in plants is still considered controversial by some. In animal species “kin recognition” has been described as “the ability to recognize and cooperate with genetically close relatives” (Ho et al., 2013). However, here, this review will consider plant kin recognition, in a broad sense, as evidence of differences in plant growth patterns (roots, shoots, overall biomass, etc.) or physiological processes (nutrient acquisition, chlorophyll content, metabolite and root secretion profiles, etc.) between the plants when grown with related (kin) versus non-related (stranger or non-kin) plants that may contribute to direct or indirect benefits for the plants involved.
As a consequence of living a sessile lifestyle, many plant communities have adapted to resource partitioning in their environment, allowing for the coexistence of multiple species when nutrients are limited (McKane et al., 2002; Xu et al., 2011; Zhang et al., 2016; Yang et al., 2018). Although this adaptation can decrease competition between species, it may potentially increase competition within a species (Murphy and Dudley, 2009; Zhang et al., 2016). Zhang et al. (2016) examined whether increased competition or nutrient partitioning occurred when two species (Glycine max and Sorghum vulgare) were grown in the presence of kin vs strangers by investigating changes in biomass and nitrogen uptake. Researchers found that although both species showed evidence of kin recognition, that S. vulgare increased root allocation when growing with strangers as opposed to kin and did not have significant changes in the rate or quantity of nitrogen uptake whereas G. max did not have changes in biomass when grown with strangers vs kin but did have modifications to the rate and quantity of different forms of nitrogen when grown with strangers (Zhang et al., 2016). These results support previous studies that trait manifestation of kin vs stranger recognition may be species dependent and may present as physiological changes as well as morphological changes.
Further investigations continued to examine changes in biomass as well as nutrient niches in kin recognition in various species. Li et al. (2017) studied whether biomass and/or nutrient-uptake efficiency changed during kin vs stranger recognition in four species (perennials: Belamcanda chinensis, Caesalpinia pulcherrima, Populus tomentosa and annual: G. max). All four species exhibited varied responses to growth with kin plants vs with stranger plants. G. max plants grown with kin exhibited reduced nutrient-uptake efficiency in several nutrients and increased biomass indicating reduced competition between kin in these conditions. Although P. tomentosa reduced phosphorus uptake in the presence of kin vs strangers, they increased root allocation in the presence of kin as well, indicating that competition as well as kin recognition occurred simultaneously. The two other species, C. pulcherrima and B. chinesis, did not show any changes in biomass, however, saw inverse reactions to S uptake efficiency, where this efficiency decreased in C. pulcherrima and decreased in B. chinesis (Li et al., 2017). These results indicate that plants with different lifestyles may respond to kin recognition with increased or decreased competition. Perennial plants with a longer lifespan may not benefit from decreased competition among kin as much as an annual plant with shorter life cycle since nutrient storage maybe more important for a perennial species whereas an annual maybe more likely to utilize the nutrients directly (Zhu et al., 2011; Li et al., 2017).
These previous studies hint at the fact that plants may encounter situations where reduced competition with kin maybe more beneficial to growth of both plants. Along these lines, Pezzola et al. (2019) examined whether varying concentrations of nutrients can influence the degree of kin cooperation in Pisum sativum. The researchers grew plants with kin and strangers in low and high nitrogen conditions. Increased aboveground biomass and nutrient allocation was found in the plants grown with kin strangers after 30 days. However, at the end of the growth cycle, plants grown with strangers in low nutrient conditions showed decreased fruit production and biomass versus the plants grown with strangers in the high nutrient conditions. Therefore, the degree to which P. sativum plants cooperated was dependent upon the relatedness of individuals as well as the surrounding environment, indicating that there is a gradient to kin recognition and cooperation behaviors in some plants (Pezzola et al., 2019).
Li et al. (2018) took this line of research one step further when they examined how kin recognition responded to the effects of soil nutrient levels as well as heavy metal soil pollution. Working with S. vulgare, researchers compared leaf and root parameters as well as nutrient use efficiency and heavy metal concentrations in plants growth with kin or strangers. They found that when grown in high levels of nutrients, S. vulgare kin plants exhibited lower specific leaf area (SLA) and nutrient use efficiency (NUE) of nitrogen and sodium compared to S. vulgare grown with strangers. However, the plants grown with kin in low nutrient conditions showed higher root growth, SLA and NUE of phosphorus and sulfur. Addition of heavy metal cadmium resulted in a decrease in biomass and chlorophyll production in kin plants as compared to stranger plants. Conversely, addition of lead decreased production of physiological stress indicators in kin plants but also resulted in a decrease in chlorophyll content in kin plants as compared to strangers. These differing results between kin and strangers in regards to growth and stress responses under heavy metal stress indicate that kin recognition maybe a resource tradeoff and perhaps more beneficial under specific environmental stresses than others.
As nutrients are a limiting factor for plant growth, water acquisition is also essential for plant survival and Goddard et al. (2020) recently investigated whether kin recognition was influenced by water limitation. Using Glechoma hederacea, plants were grown alone, with kin and with strangers in water adequate and water limited conditions. G. hederacea plants exhibited root growth avoidance toward strangers but not toward roots of kin plants. Water limited conditions altered plant root growth patterns but was not influenced by relatedness of individuals. Therefore, in this study, presence of kin did not enhance growth in low water conditions, in contrast to many low nutrient conditions, where presence of kin aids in growth under stress. Taken together, these studies highlight that kin cooperation can be considered as a gradient of behaviors based upon environmental conditions and that manifestation of kin cooperation in plants may be species specific and is not simply an “all or nothing” behavior.
We see that kin recognition can play an important and varied role in plant competition in regard to nutrient partitioning and allocation but this leads to additional questions regarding plant competition in reference to other traits as well. In addition to nutrient uptake and root growth patterns, kin recognition has been identified to benefit several above ground traits as well. According to Torices et al. (2018) pollinator attraction can be considered a costly investment for plants pertaining to growth and resources. They investigated the role of kin recognition in an insect pollinated species, Moricandia moricandioides to determine whether plants growing with kin invest more resources into floral display versus plants grown with non-relatives. Their results showed that “advertising effort” (total mass allocated to petal mass) was significantly increased in plants grown with kin. These results point toward the possibility of higher pollination rate in kin plants as insects are more likely to be attracted to the larger floral displays (Torices et al., 2018). Thus, kin interactions may influence plant behaviors not only within the population but at different levels of an ecosystem if pollinator attraction behavior is also influenced by as well.
In addition to increasing floral display, researchers have examined the role of kin recognition on other above ground anatomical as well as chemical features. Yamawo (2015) investigated the role of neighbor relatedness on expression of defense traits that protect against herbivores. Indirect defense traits such as number of extrafloral nectaries, volume of extrafloral nectar, and number of food bodies as well as direct defense traits such as pellucid dot density, trichome density, and total phenolic content (Yamawo, 2015). Interestingly, kin plant growth was significantly decreased because of increased investment in direct defenses such as total phenolic compounds and pellucid dot production. However, plants grown with non-kin were found to invest more in trichome production, an indirect defense which may not be as costly to the plant to produce compared to the direct defenses (Yamawo, 2015). This experiment further supported earlier research by Karban et al. (2013) that Artemisia tridentata plants experienced less herbivory because of volatile cues emitted from wounded neighbors when the neighbors were more closely related versus distantly related. The results from these two studies support that plant allocation of resources for protection against herbivores may well be influenced by kin interactions.
Protection against herbivory is vital to the survival of many species, however, in addition to attacks from herbivores, many plants face chemical attacks from other plant species. Production of allelopathic compounds has been well documented in many plant species, including rice. Xu et al. (2021) recently investigated the influence of kin interactions on the allelopathic interactions between Oryza sativa and various species of paddy weeds. The study found that plants grown with close relatives shifted their resource allocation to increased growth of intrusive roots toward weeds and decreased their production of allelochemicals. Remarkably, the rice grown with closely related neighbors also had increased populations of root bacterial communities and the communities differed from those found on distantly related neighbors.
In addition to promoting bacterial community growth, kin recognition has been found to support mycorrhizal colonization. File et al. (2012) investigated the effect of kin recognition in Ambrosia artemisiifolia and mycorrhizal symbiosis with Glomus intraradices with the hypothesis that greater cooperation between kin plants would result in an increase in the common mycorrhizal network (CMN) as compared to plants grown with strangers and that this increase in the CMN would increase total nutrient acquisition and enhance overall fitness, including pathogen resistance, for the groups of kin (File et al., 2012). The results demonstrated that, indeed, A. artemisiifolia plants grown with kin had increased mycorrhizal growth, increased leaf nitrogen content and fewer lesions from bacterial infection as compared to the plants grown with strangers. Similar studies were conducted at higher and low soil phosphorus conditions. Plants grown with kin showed long soil hyphae growth in both high or low phosphorus levels whereas plants grown with strangers showed an increase in soil hyphae length under low phosphorus conditions. This highlights the previously mentioned research suggesting that altered nutrient conditions can result in tradeoffs between kin cooperation and other traits. Additionally, this research emphasizes the ecological significance of kin recognition and cooperation through influence on interspecies interactions.
As previously mentioned, Dudley and File (2007) first demonstrated clear evidence of kin versus stranger recognition in plants by growing C. edentula plants together in pots and it has been well established that plants roots secrete compounds into the surrounding soil or rhizosphere in response to biotic and abiotic conditions (Bais et al., 2006). Therefore, the next question to be explored was whether root secretions were involved in the kin recognition process. Biedrzycki et al. (2010) exposed Arabidopsis thaliana plants to root secretions rather than the roots themselves, of kin and stranger plants and determined that lateral root growth increased when exposed to stranger secretions as opposed to kin secretions. Additionally, when a root secretion inhibitor, sodium orthovanadate (NA3VO4), was applied to the plants, root growth differences between kin and stranger plants were eliminated further supporting the role of root exudates in kin recognition (Biedrzycki et al., 2010).
Further investigations regarding the role of root exudates in kin recognition were performed by Semchenko et al. (2014). Here, root exudates of Deschampsia cespitosa were collected and then exposed to siblings, strangers from the same species and plants of different species (Lychnis flos-cuculi) from both the same and different communities, in localized applications. Plants exposed to exudates from strangers from the same population had more branched roots and higher specific root length compared to plants that were exposed to sibling exudates. Conspecific plants exposed to root secretions from individuals from the same population had greater root density than when exposed to secretions from a different population and exudates from L. flos-cuculi did not show any significant differences. These results indicate a highly specific mechanism for recognition of rhizosphere neighbors facilitated by plant root exudates (Semchenko et al., 2014).
Root exudates have also been found to be involved in kin recognition in O. sativa cultivars. Yang et al. (2018) examined the presence of kin recognition in the crop species and found that two of the cultivars were able to recognize kin and altered root growth and biomass allocation in response. Additionally, exposure of seedlings to distantly related cultivars promoted growth of larger root systems as compared to when the seedlings were exposed to exudates of a closely related or their own cultivar. This growth response also ceased when the root secretion sodium orthovanadate was applied (Yang et al., 2018). The focal cultivars of O. sativa in this study were also found to significantly increase their secretion of allantoin (a nitrogen compound also known to protect plants from abiotic stress) in the present of distantly related cultivars versus when with the same or closely related cultivars. Furthermore, the O. sativa cultivars were capable of recognizing kin secreted allantoin, in the presence of non-kin plants, to stimulate growth of their root systems, suggesting that in some rice cultivars, allantoin is specifically involved in kin recognition (Yang et al., 2018). Identification of specific root exudates involved in kin recognition stresses the significance of this work.
Over the last decade, many studies, including those referenced here, have established that kin recognition does, indeed, occur in plants (Figure 1); but what is lacking from our greater understanding of these plant interactions are “big picture generalities” for plant kin recognition (West et al., 2021). As West et al. (2021) discussed, in regards to microorganisms and animal species, often details can be learned about kin recognition in a specific species but that we still have more to uncover about kin responses in organisms in general. Questions remain such as “Does cooperation actually occur in some species but not others?”, “When does kin recognition confer fitness benefits and do these benefits occur at the individual or population level?”, “What are the costs and benefits of kin recognition?” (Anten and Chen, 2021; West et al., 2021). Improved understanding of these plant kin interactions would also benefit from standardization of terminology such as what exactly constitutes as a “kin”, “stranger” or “non-kin” and what degree of recognition or change in growth or behavior is enough to qualify as true “kin recognition” as opposed to general cooperation or altruism (Penn and Frommen, 2010). Elucidating these fundamental ecological questions will aid in better understanding plant interactions.
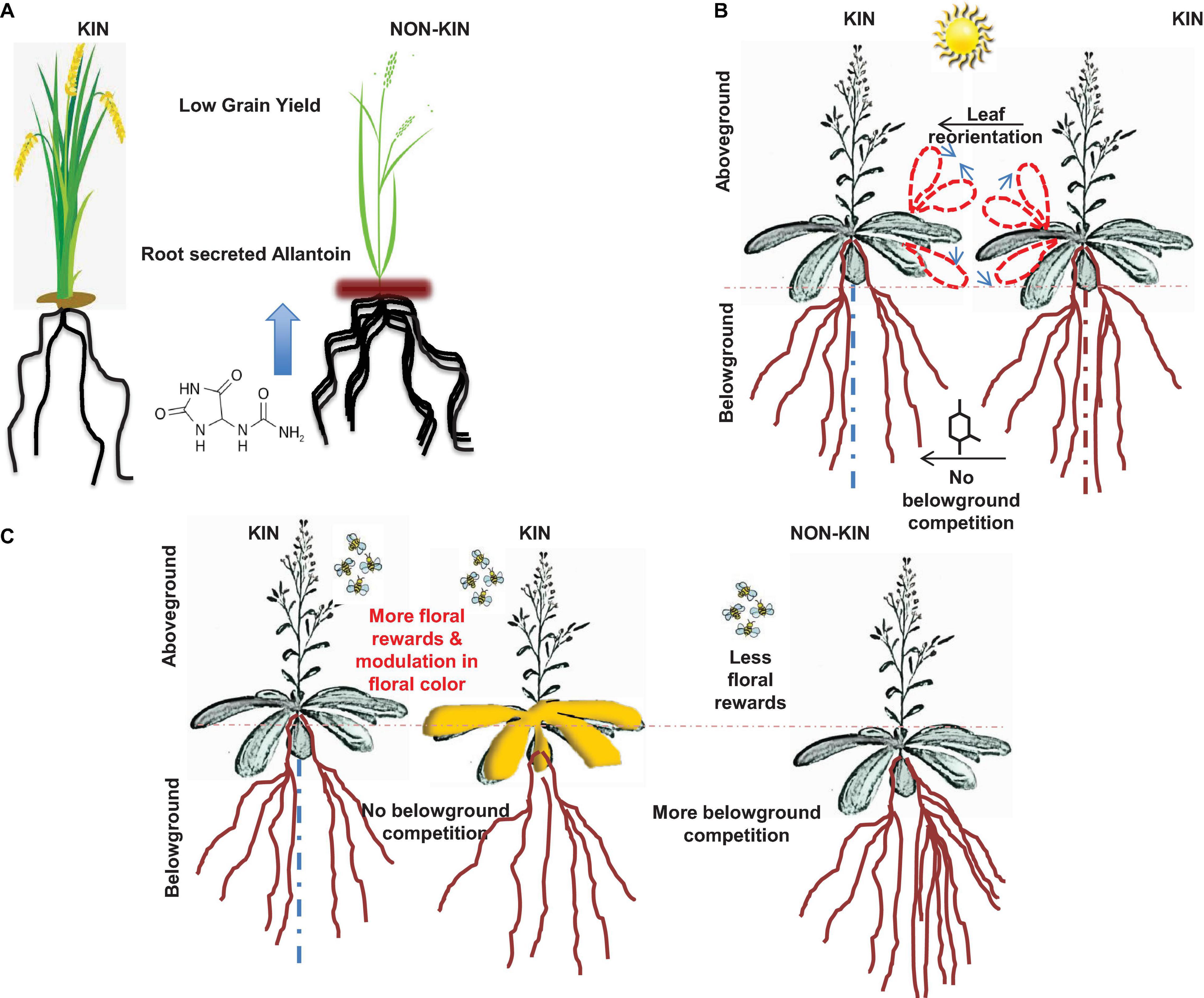
Figure 1. The schematic shown depicts variation in kin recognition and discrimination in plants with different trade-offs. (A) The image in panel A is adapted from Bais (2018), rice plants exposed to kin members secrete less allelochemical (allantoin) compared to interactions with non-kins (Yang et al., 2018). (B) Panel B shows aboveground trade-offs of kin recognition in Arabidopsis plants. The schematic is adapted from Bais (2015), wherein kin plants interactions with other kin showed reorientation of leaves (more light sharing) and less root allocation (Crepy and Casal, 2014). (C) Panel C is also adapted from Bais (2015), in here, plants reacting with kin showed more floral reward system compared to its interaction with non-kin members (Torices et al., 2018).
In addition to investigating broad kin recognition interactions, continued research to understand the tradeoffs between kin cooperation and macro and micronutrient acquisition in normal as well as in nutrient-depleted conditions would elucidate the process of nutrient allocation and plant growth plasticity. Along these lines, exploring the implications of kin interactions on competition and yield in crops species may prove for future crop breeding (Murphy et al., 2017; Anten and Chen, 2021). Additionally, more studies investigating how kin recognition influences plant interactions along multitrophic levels with beneficial insects and microbes as well as with herbivores and pathogenic microbes will help understand the greater ecological significance of plant kin recognition in plant survival (File et al., 2012; Torices et al., 2018). It would be valuable, not only to conduct these studies in lab settings but in field or natural conditions if possible, as well.
However, the area of plant kin recognition that would benefit the most from continued research would be biochemical studies of root exudates to elucidate the chemical signals involved. Furthermore, investigation of these chemical signals as well as genetic and metabolic studies to examine genes and proteins involved in root secretion need to resume (Badri et al., 2012). Growth plasticity reduced competition, improved associations with beneficial as well as decreased interactions with pathogens and herbivores have all been observed in plants during kin recognition, management of these traits of kin recognition mechanisms may lead to a deeper understanding of ecological interactions as well as the improvement of crop species.
HB conceived the idea of the review. MB wrote the review with the help of HB. All authors contributed to the article and approved the submitted version.
HB acknowledges the support from USDA NIFA 2016-67013-24846.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anten, N. P. R., and Chen, B. J. W. (2021). Detect thy family; Mechanisms, ecology, and agricultural aspects of kin recognition. Plant Cell Environ. 44, 1059–1071. doi: 10.1111/pce.14011
Badri, D. V., De-la-Pena, C., Lei, Z., Manter, D. K., Chaparro, J. M., Guimarães, R. L., et al. (2012). Root secreted metabolites and proteins are involved in the early events of plant-plant recognition prior to competition. PLoS One 7:e46640. doi: 10.1371/journal.pone.0046640
Bais, H. P. (2015). Shedding light on kin recognition response in plants. New Phytol. 205, 4–6. doi: 10.1111/nph.13155
Bais, H. P. (2018). We are family; kin recognition in crop plants. New Phytol. 220, 357–359. doi: 10.1111/nph.15399
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Baldwin, I. T., Halitschke, R., Paschold, A., von Dahl, C. C., and Preston, C. A. (2006). Volatile signaling in plant-plant interactions: “Talking Trees” in the Genomics Era. Science 311, 812–815. doi: 10.1126/science.1118446
Biedrzycki, M., Jilany, T., Dudley, S., and Bais, H. (2010). Root exudates mediate kin recognition in plants. Commun. Integr. Biol. 3, 1–8. doi: 10.4161/cib.3.1.10118
Carazo, P., Tan, C. K. W., Allen, F., Wigby, S., and Pizzari, T. (2014). Within-group male relatedness reduces harm to females in Drosophila. Nature 505, 672–675. doi: 10.1038/nature12949
Crepy, M. A., and Casal, J. J. (2014). Photoreceptor-mediated kin recognition in plants. New Phytol. 205, 329–338. doi: 10.1111/nph.13040
Dudley, S. A., and File, A. L. (2007). Kin recognition in an annual plant. Biol. Lett. 3, 435–438. doi: 10.1098/rsbl.2007.0232
File, A. L., Klironomos, J., Maherali, H., and Dudley, S. A. (2012). Plant kin recognition enhances abundance of symbiotic microbial partner. PLoS One 7:e45648. doi: 10.1371/journal.pone.0045648
Gerlach, G., and Lysiak, N. (2006). Kin recognition and inbreeding avoidance in zebrafish, Danio rerio, is based on phenotype matching. Anim. Behav. 76, 1371–1377. doi: 10.1016/j.anbehav.2005.10.010
Goddard, E. L., Varga, S., John, E. A., and Sousbury, C. D. (2020). Evidence for root kin recognition in the clonal plant species Glechoma hederacea. Front. Ecol. Evol. 8:578. doi: 10.2289/fevo.2020.578141
Haig, T. (2008). “Allelochemicals in Plants,” in Allelopathy in Sustainable Agriculture and Forestry, eds R. S. Zeng, A. U. Mallik, and S. M. Luo (New York: Springer), 63–104. doi: 10.1007/978-0-387-77337-7_4
Hamilton, W. D. (1964). The genetic evolution of social behaviour. I. J. Theor. Biol. 7, 1–16. doi: 10.1016/0022-5193(64)90038-4
Hanggi, E. B., and Schusterman, R. J. (1990). Kin recognition in captive California sea lions (Zalophus californianus). J. Comp. Psychol. 104, 368–372. doi: 10.1037/0735-7036.104.4.368
Ho, H. I., Hirose, S., Kuspa, A., and Shaulsky, G. (2013). Kin recognition protects cooperators against cheater. Curr. Biol. 23, 1590–1595. doi: 10.1016/j.cub.2013.06.049
Jones, J., and Dangl, J. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karban, R., Shiojiri, K., Ishizaki, S., Wetzel, W. C., and Evans, R. Y. (2013). Kin recognition affects plant communication and defence. Proc. R. Soc. B. 280:20123062.
Kessler, A., and Baldwin, I. T. (2001). Defensive function of herbivore-induced plant volatile emissions in nature. Science 291, 2141–2144. doi: 10.1126/science.291.5511.2141
Kong, C. H., Zhang, S. Z., Li, Y. H., Xia, Z. C., Yang, X. F., Meiners, S. J., et al. (2018). Plant neighbor detection and allelochemical response are driven by root-secreted signaling chemicals. Nat. Commun. 9:3867. doi: 10.1038/s41467-018-06429-1
Li, J., Xu, X., and Feng, R. (2018). Soil fertility and heavy metal pollution (Pb and Cd) alter kin interaction of Sorghum vulgare. Environ. Exp. Bot. 155, 368–377. doi: 10.1016/j.envexpbot.2018.05.009
Li, J., Xu, X., and Liu, Y. (2017). Kin recognition in plants with distinct lifestyles: implications of biomass and nutrient niches. Plant Grow. Reg. 84, 333–339. doi: 10.1007/s10725-017-0343-7
Lin, Y. H., Chan, S. F., Rubenstein, D. R., Liu, M., and Shen, S. F. (2019). Resolving the paradox of environmental quality and sociality; the ecological causes and consequences of cooperative breeding in two lineages of birds. Am. Nat. 194, 207–216. doi: 10.1086/704090
Mateo, J. M. (2003). Kin recognition in ground squirrels. J, Mammal. 84, 1163–1181. doi: 10.1644/BLe-011
McKane, R. B., Johnson, L. C., Shaver, G. R., Nadelhoffer, K. J., Rastetter, E. B., Fry, B., et al. (2002). Resource-based niches provide a basis for plant species diversity and dominance in arctic tundra. Nature 415, 68–71. doi: 10.1038/415068a
Murphy, G. P., and Dudley, S. A. (2009). Kin recognition: competition and cooperation in Impatiens (Balsaminaceae). Am. J. Bot. 96, 1990–1996. doi: 10.3732/ajb.0900006
Murphy, G. P., Swanton, C. J., Van Acker, R. C., and Dudley, S. A. (2017). Kin recognition, multilevel selection and altruism in crop sustainability. J. Ecol. 105, 930–934. doi: 10.1111/1365-2745.12787
Penn, D., and Frommen, J. (2010). “Kin recognition: an overview of conceptual issues, mechanisms and evolutionary theory,” in Animal Behaviour: Evolution and Mechanisms, ed. P. Kappeler (Berlin: Springer), 55–85. doi: 10.1007/978-3-642-02624-9_3
Pezzola, E., Pandolfi, C., and Mancuso, S. (2019). Resource availability affects kin selection in two cultivars of Pisum sativum. Plant Grow. Reg. 90, 321–329. doi: 10.1007/s10725-019-00562-7
Sanon, A., Andrianjaka, Z. N., Prin, Y., Bally, R., Thioulouse, J., Comte, G., et al. (2009). Rhizosphere microbiota interferes with plant-plant interactions. Plant Soil 321, 259–278. doi: 10.1007/s11104-009-0010-5
Semchenko, M., Saar, S., and Lepik, A. (2014). Plant root exudates mediate neighbor recognition and trigger complex behavioural changes. New Phytol. 204, 631–637. doi: 10.1111/nph.12930
Sharp, S., McGowan, A., Wood, M., and Hatchwell, B. J. (2005). Learned kin recognition cues in a social bird. Nature 434, 1127–1130. doi: 10.1038/nature03522
Torices, R., Gomez, J. M., and Pannell, J. R. (2018). Kin discrimination allows plants to modify investment towards pollinator attraction. Nat. Commun. 9:2018. doi: 10.1038/s41467-018-04378-3
West, S. A., Cooper, G. A., Ghoul, M. B., and Griffin, A. S. (2021). Ten recent insights for our understanding of cooperation. Nat. Ecol. Evol. 5, 419–430. doi: 10.1038/s41559-020-01384-x
Xu, X. L., Ouyang, H., Cao, G. M., Richter, A., Wanek, W., and Kuzyakov, Y. (2011). Dominant plant species shift their nitrogen uptake patterns in response to nutrient enrichment caused by a fungal fairy in an alpine meadow. Plant Soil 341, 495–504. doi: 10.1007/s11104-010-0662-1
Xu, Y., Cheng, H., Kong, C., and Meiners, S. (2021). Intraspecific kin recognition contributes to interspecific allelopathy: a case of allelopathic rice interference with paddy weeds. Plant Cell Environ. 44, 1–13. doi: 10.1111/pce.14083
Yamawo, A. (2015). Relatedness of neighboring plants alters the expression of indirect defense traits in an extrafloral nectary-bearing plant. Evol. Biol. 42, 12–19. doi: 10.1007/s11692-014-9295-2
Yang, X., Li, L., Xu, Y., and Kong, C. (2018). Kin recognition in rice (Orzyza sativa) lines. New Phytol. 220, 567–578. doi: 10.1111/nph.15296
Zhang, L., Liu, Q., Tian, Y., Xu, X., and Ouyang, H. (2016). Kin selection or resource partitioning for growing with siblings: implications from measurements of nitrogen uptake. Plant Soil 398, 79–86. doi: 10.1007/s11104-015-2641-z
Keywords: kin recognition, root allocation, kinship, root secretions, rhizosphere, kin recognition mechanisms
Citation: Biedrzycki ML and Bais HP (2022) Kin Recognition in Plants: Did We Learn Anything From Roots? Front. Ecol. Evol. 9:785019. doi: 10.3389/fevo.2021.785019
Received: 28 September 2021; Accepted: 06 December 2021;
Published: 10 January 2022.
Edited by:
Kamal Bawa, University of Massachusetts Boston, United StatesReviewed by:
Mitchel Daniel, Florida State University, United StatesCopyright © 2022 Biedrzycki and Bais. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harsh P. Bais, aGJhaXNAdWRlbC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.