- 1Department of Molecular Biology and Biotechnology, University of Dar es Salaam, Dar es Salaam, Tanzania
- 2Virus Vector Ecology Group, International Institute of Tropical Agriculture (IITA), Dar es Salaam, Tanzania
- 3Department of Biology, Lund University, Lund, Sweden
Cassava is a vital food-security crop in Sub-Saharan Africa. Cassava crops are, however, severely affected by viral diseases transmitted by members of the whitefly species complex Bemisia tabaci. We have here investigated the role of olfaction in host selection behavior of the cassava whitefly B. tabaci SSA-ESA biotype. Surprisingly, we find that the whiteflies appear to make little use of olfaction to find their favored host. The cassava whitely shows a highly reduced olfactory system, both at the morphological and molecular level. Whitefly antennae possess only 15 sensilla with possible olfactory function, and from the genome we identified just a handful of candidate chemoreceptors, including nine tuning odorant receptors, which would afford the whitefly with one of the smallest olfactomes identified from any insect to date. Behavioral experiments with host and non-host plants, as well as with identified specific volatiles from these sources, suggest that the few input channels present are primarily tuned toward the identification of unwanted features, rather than favored ones, a strategy quite unlike most other insects. The demonstrated repellence effect of specific volatile chemicals produced by certain plants unflavored by whiteflies suggests that intercropping with these plants could be a viable strategy to reduce whitefly infestations in cassava fields.
Introduction
Members of the cryptic whitefly species complex Bemisia tabaci (Figure 1A) are major agricultural pests and vectors of viruses causing diseases in wide range of crops (Martin et al., 2000; Navas-Castillo et al., 2011). Cassava (Manihot esculenta) is a key food-security staple in Sub-Saharan Africa and one of the crops severely affected by diseases caused by whitefly-transmitted viruses, particularly cassava mosaic disease (CMD) and cassava brown streak disease (CBSD) (Figures 1A,B) (Legg et al., 2015). CMD alone is estimated to result in more than US$1.25 billion in crop losses annually — reducing cassava yields by as much as 50% in affected regions (Thresh et al., 1998; Legg et al., 2006). Due to its hardiness, cassava is predicted to be resilient or even positively affected by upcoming climate changes, in contrast to other major African staples, such as maize, sorghum, and millets, all of which are projected to experience negative impacts from increased mean temperatures and/or reduced rainfall (Jarvis et al., 2012). Cassava’s importance for food security in many parts of Africa will hence likely increase greatly in the near future.
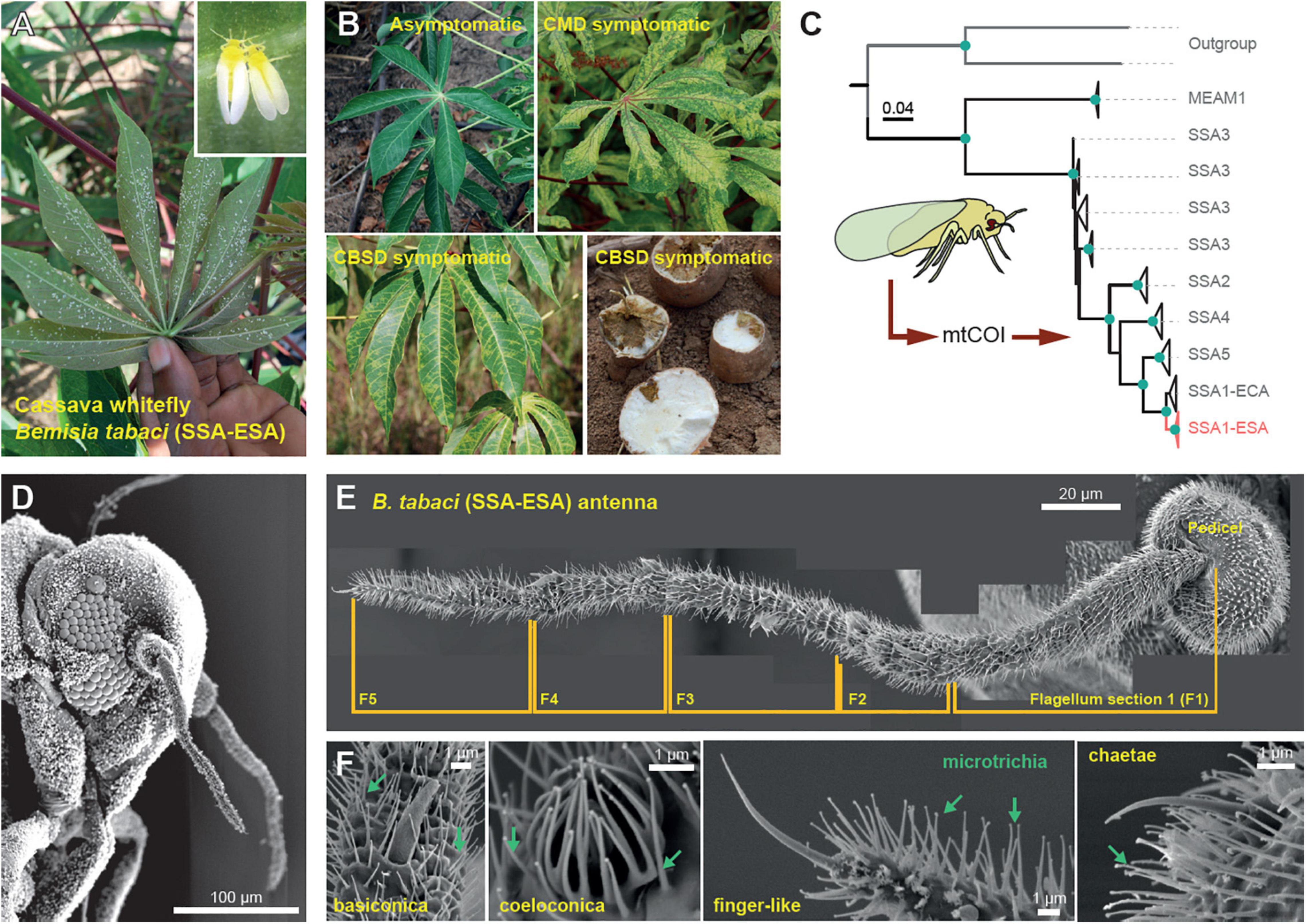
Figure 1. The cassava whitefly Bemisia tabaci. (A) The cassava whiteflies are usually found underneath plant leaves where they feed, mate and reproduce. (B) Symptoms of the viral diseases of cassava (cassava mosaic disease and cassava brown streak disease) transmitted by the cassava whitefly. (C) Maximum-likelihood phylogenetic tree showing the similarities between the 16S mtCOI sequences of the whiteflies from Sub-Saharan Africa (SSA), including the cassava whiteflies found in the Eastern and Southern Africa (SSA-ESA), the cassava whiteflies found in the Eastern and Central Africa (SSA-ECA) and the non-cassava whitefly Middle East Asia Minor (MEAM1). (D) Scanning electron microscopy (SEM) image of the head of the cassava whitefly (210x magnification) showing the position and relative size of the antenna. (E) External structure of the antenna of the cassava whitefly SSA-ESA showing different segments of the flagellum obtained from SEM. (F) Types of olfactory sensilla found on the antenna of the cassava whitefly SSA-ESA obtained from SEM.
The use of disease-free planting materials, of resistant or tolerant varieties, is presently the main method of controlling CMD and CBSD. The abundance of whiteflies, however, limits the effectiveness of this approach, since uninfected plants quickly become infected. Bemisia tabaci transmits cassava mosaic begomoviruses (CMBs) in a persistent manner (Dubern, 1994) and transmits cassava brown streak ipomoviruses (CBSIs) semi-persistently (Maruthi et al., 2005, 2017), although in both cases the viruses can be inoculated in less than 10 min. Development of practical and cost-effective methods for controlling whiteflies is therefore an urgent priority. This requires an understanding of how the cassava whiteflies interact with their host plants, including the mechanisms used for identification and selection of suitable hosts. Olfaction is the primary method used by many insects to discriminate between favorable and unfavorable hosts (Visser, 1988; Finch and Collier, 2003; Bernays and Chapman, 2007) and is suggested to be of importance for whiteflies (Mellor and Anderson, 1995; Togni et al., 2010). The present study investigated the role of olfaction-mediated host selection behavior of the whitefly genotype colonizing cassava in the coastal region of East Africa i.e., B. tabaci Sub-Saharan Africa, East Southern Africa Sub-group (SSA-ESA). We have revealed the role of olfaction-mediated host selection and identified some of the key plant-produced volatile chemicals that can influence host selection in the cassava whiteflies. Improved understanding of these critical interactions between whiteflies and their cassava hosts will contribute to the development of whitefly control measures that will in turn reduce virus incidence and enhance cassava productivity.
Materials and Methods
Insects
We investigated the cassava whiteflies found in the coastal zone of East Africa, which are known to be B. tabaci Sub-Saharan Africa Eastern and Southern African type (SSA-ESA). The whiteflies were collected from asymptomatic cassava plants grown in Chambezi plant fields (Bagamoyo, Tanzania) and reared on potted healthy cassava plants (of variety Albert) placed in 50 cm × 50 cm × 100 cm aluminum-framed cages covered with plastic nets of 15 cm × 15 cm hole size. The colonies were maintained in screen-houses at 25 ± 5°C and 65 ± 5% relative humidity (RH). We confirmed the genotype of the whiteflies captured from the field by amplifying and sequencing part of the mitochondrial COI gene (MtCOI) from the total nucleic acids extracted from some of the whiteflies.
Extraction of Total Nucleic Acids From the Whiteflies
Total nucleic acid was collected from individual whiteflies using the method described by Wosula et al. (2017). The whiteflies were anesthetized at −20°C for 1 min and then stored in 70% ethanol at −20°C prior to nucleic acid extraction. During the extraction individual whiteflies were placed in 1.5 mL eppendorf tubes and residual ethanol was removed by pipetting and air drying. Then 3 μL of ice-cold lysis buffer containing Tris–HCl (10 mM, pH 8.0), Proteinase K (60 μg/mL), MgCl2 (2.5 mM), KCl (50 mM), Tween-20 (0.45%) and Gelatine (0.01%) was added into the eppendorf tube and whiteflies were ground using sterile plastic pastels. Once ground, an additional lysis buffer (27 μL) was added and the solution was gently mixed, centrifuged and then incubated in a 55°C water bath for 30 min to allow for Proteinase K digestion. This was followed by incubation at 95°C for 10 min to inactivate the Proteinase K and then storage at −20°C.
PCR Amplification and Sequencing of the MtCOI Gene
We amplified a 859 bp fragment of the mtCO1 gene from the whiteflies using 2 μL of 10x diluted total nucleic acid extracted from cassava whitefly, 1x QuickLoad PCR Master mix (Cat# M0486S/L, New England Biolabs, United Kingdom) and 0.25 mM of each of the primers (BTSM-2-F 5′-TCTGGTTYTTTGGTCATTC-3′ and BTSM-3-R 5′-CACTTTCTGCCACATTAGA-3′) (Milenovic et al., 2019) made up to 25 μL reaction volume using sterile distilled water. The PCR amplification reactions were done using following thermocycling conditions: initial DNA denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s and extension at 68°C for 1 min followed by a final extension at 68°C for 10 min. The PCR products were analyzed by agarose gel electrophoresis using 1% Agarose gel made in 1x TAE buffer (Cat#15558042, Invitrogen, New York, NY, United States) and stained with GelRed™ (Cat#41003, Biotinum, Fremont, CA, United States). The electrophoresis was conducted in 1x TAE buffer at 70 eV for 30 min using 10 μL of the PCR products mixed with 2 μL of 6x Blue loading dye (Cat#: B7021S, New England Biolabs, United Kingdom) prior to loading onto the gel. Samples containing the bands of interest (∼860 bps) were sent to Macrogen Corp (MD, United States) for purification and sequencing the obtained sequences were compared to known sequences of B. tabaci SSA-ESA.
Comparison of Bemisia tabaci SS1-ESA Sequences
All the raw Sanger sequences of the mtCOI gene fragments of the cassava whiteflies collected in the coastal zone of Tanzania, were treated in MEGA X software (Sudhir et al., 2018). The sequences were first trimmed to remove low quality and ambiguous bases at each end and then the forward and reverse sequence pairs were aligned. The final genetic dataset was completed with the addition of mtCOI sequences of the B. tabaci SSA-ESA whiteflies downloaded from the NCBI Genbank (Supplementary Table 2). The dataset were used to generate a phylogenetic tree using a maximum likelihood approach using IQ-Tree2 (Minh et al., 2020). The best substitution model was obtained by ModelFinder (Kalyaanamoorthy et al., 2017) using “-m MFP+MERGE” option giving the codon position data of the sequence as maximum potential partitions. The topology of the tree was tested by aBayes (Anisimova et al., 2011) test and ultrafast bootstrap (Hoang et al., 2018), using “-abayes” and “-B 5000” options, to evaluate the nodal support of the obtained topology.
Plants
We used Cassava (Manihot esculenta) of variety Albert and Kiroba to represent host plants of different susceptibility to cassava diseases—Albert was highly susceptible to CBSD but resistant to CMD while Kiroba was highly susceptible to CMD but not CBSD. Tomato (Solanum lycopersicum), cowpea (Vigna unguiculata) and cotton (Gossypium hirsutum) were used to represent the alternative host plants while lemongrass (Cymbopogon citratus) represented the non-host plant. Tomato, cowpeas and cotton were grown from the certified seeds while lemongrass was grown from seedlings, all from local landraces in the coastal region of Tanzania. The cassava plants were grown from stem cuttings outsourced from asymptomatic cassava plants grown in the fields with low incidence for CMD and CBSD, these included in Mwele (Tanga) and Newala (Mtwara), both in Tanzania. The plants were grown in 2 L pots (filled with a mixture of soil and farmyard manure at 1:1 ratio) and maintained in the screen houses at 25 ± 5°C and 70 ± 10 % RH. We selected diseased cassava plants based on the presence of foliar symptoms for CMD or CBSD as well as though determining the presence of the disease-causing viruses (i.e., CMBs and CBSIs) by amplifying part of the viral genes from the nucleic acids extracted from asymptomatic leaf samples.
Extraction of Total Nucleic Acids From Cassava Mosaic Disease/Cassava Brown Streak Disease Symptomatic Cassava Leaves
We sampled two leaves, one from top and one from bottom of cassava plants, from 10 symptomatic and 10 asymptomatic cassava plants of variety Albert and Kiroba. The leaves were air dried and then ground using Geno-grinder (SPEX SamplePrepP 2010) at 1,500 rpm for 40 s. Total nucleic acid was extracted from the ground leaves using the cetyl trimethyl ammonium bromide (CTAB) extraction procedure adapted from Maruthi et al. (2002). This involved adding 1 mL of the CTAB buffer (containing 2.0%, w/v CTAB, 2.0% PVP, 25 mM EDTA, 2.0 M NaCl, 100 mM Tris HCL pH 8.0 and 0.2% β-mercaptoethanol, that was added immediately before extraction) into the ground cassava leaves and then mixing the solution by vortexing prior to incubation at 65°C for 15 min to lyse the cells. Afterward, an equal amount of chloroform:isoamyl alcohol (24:1) was added to the cell lysate and the solution was vortexed and the centrifuged at 13,800 × g for 10 min to separate the nucleic acids from the polysaccharides, polyphenols and the cellular material. The solution containing the nucleic acids (top layer) was removed by pipetting and placed into a new clean eppendorf tube prior to adding 0.6x volume of cold isopropanol. The nucleic acid containing isopropanol solution was gently mixed by tilting the Eppendorf tube prior to incubation at −20°C for 30 min and centrifugation at 13,000 × g and 4°C for 10 min to pellet the nucleic acids. The supernatant was discarded, and the nucleic acid pellet was washed twice by adding 700 μL of 70% ethanol, vortexing and incubating at −20°C prior to centrifugation at 13,000 × g for 5 min. Afterward, the ethanol solution was removed, the nucleic acid pellet was air-dried and then resuspended in 100 μL Tris-EDTA buffer (0.1x) on ice prior to storage at −20°C.
PCR Amplification of Viruses That Cause Cassava Mosaic Disease
We used conventional PCR to amplify a 500 bp fragment of DNA B component of EACMV and a 700 bp fragment from the coat protein of ACMV, since both viruses are the main causative agents of CMD in the coastal region of Tanzania (Morris et al., 1990; Bull et al., 2006). Amplification of the nucleic acid fragment from EACMV fragment was done using the EAB555 primer pair (forward primer sequence 5′-TACATCGGCCTTTGAGTCGCATGG-3′ and reverse primer sequence 5′- CTTATTAACGCCTATATAAACACC-3′ designed by Fondong et al. (2000) while amplification of the nucleic acid fragment from ACMV was done using the JSP primer pair (forward primer sequence 5′-ATGTCGAAGCGACCAGGAGAT-3′ and reverse primer sequence 5′-TGTTTATTAATTGCCAATACT-3′) designed by Pita et al. (2001). The PCR amplification reactions were performed using Veriti thermocycler (Applied Biosystems) and One Taq 2x Master mix with standard buffer (M0482S, New England Biolabs) at the following cycling conditions: initial DNA denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 68°C for 40 s followed by a final extension at 68°C for 10 min. The PCR products were analyzed by agarose gel electrophoresis using 1% (w/v) agarose gels and 1X TAE buffer as previously described. Samples containing DNA bands of about 550 bp were considered to be positive for EACMVs and samples containing DNA bands of about 700 bps were considered to be positive for ACMVs.
PCR Amplification of Viruses That Cause Cassava Brown Streak Disease
Detection of the virus that cause CBSD (cassava brown streak virus and Ugandan cassava brown streak virus), was done by real-time reverse transcription PCR using Taqman chemistry with primers and probes (Supplementary Table 3) developed by Adams et al. (2013). The PCR amplification reactions were performed in a reaction mixture containing 1x PCR buffer, 5.5 mM MgCl2, 0.5 mM dNTPs, 0.625 Units of Taq DNA polymerase and 0.4 Units of M-MLV- reverse transcriptase from Thermo Fisher Scientific as well as 300 nM primer, 100 nM probe, 30 nM Rox reference dye from Integrated DNA Technologies (IDT). For each 25 μL reaction, we used 4 μL of the RNA template obtained from the total nucleic acid extracted from cassava leaves of symptomatic and asymptomatic leaves. The total nucleic acid was pre-treated with DNAse 1 (Cat# AMPDI, Sigma Aldrich) to remove of DNA after nucleic acid extraction prior use. The RT-PCR amplification reactions were performed using Stratagene mX3000P (Agilent Technologies) with the following thermo-cycling conditions: 30 min incubation at 48° for reverse transcription, initial denaturation of the cDNA at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s and annealing and extension step at 60°C for 1 min. Fluorescent data were collected during the 60°C step using Stratagene MxPro Real-time QPCR software version 4 and samples which produced fluorescent before 36 cycles (Ct value) were considered to be positive for U/CBSIs.
External Structure of the Bemisia tabaci Antenna
Scanning electron microscopy (SEM) was used to identify the type and distribution of olfactory sensilla on the antennae of B. tabaci. Adult whiteflies, stored in 70% ethanol, were air dried and washed with acetone to remove the waxy coat from their surface. Afterward the whiteflies were air dried and then fixed in a solution containing 2% paraformaldehyde and 2.5% glutaraldehyde (in 0.1 M l–1 cacodylic buffer at pH 7.4) for 24 h at 4°C. After the 24 h the whiteflies were dehydrated in graded ethanol series by submerging the whiteflies in 25, 50, and 75% ethanol, each for 5 min, followed by 100% ethanol for 2 min. The dried whitefly samples were glued on SEM stubs and coated with gold (Cesington 108 auto, 45 s, 20 mA) prior to visualization by a scanning electron microscope (SEM; Hitachu SU3500) at 5 kV.
Molecular Composition of Bemisia tabaci’s Olfactory System
The draft genomes of the cassava whitefly B. tabaci SSA-ECA was used to identify the chemosensory proteins (odorant, gustatory and ionotropic receptors as well as the odorant binding proteins). This involves protein prediction from the genome using AUGUSTUS (v3.2.3) (Stanke and Morgenstern, 2005) and Drosophila melanogaster, Acyrthosiphon pisum, and Rhodnius prolixus as references. A consensus proteome was then generated by dereplication of the three predicted proteomes in a two-step process. First, output results from the three proteomes were merged in a single proteome. Next, proteins that shared an identity of at least 100% over their full length were clustered and duplicate sequences removed. Four Hidden Markov Models (HMMs) from were constructed from multiple sequence alignments from each chemosensory subfamily identified from A. pisum to identify chemoreceptors in the predicted proteome. The four HHMs models were validated by testing each HMM against the initial protein dataset for each subfamily prior to screening the predicted proteome for ORs, GRs, IRs, and OBPs. EggNOG mapper (v.2) (Huerta-Cepas et al., 2017) and InterPro (v.5) (Hunter et al., 2009) databases were used to functionally characterize the uncovered candidate proteins.
Transcriptome Analysis of Genes Expressed in the Heads of the Whitefly
We performed transcriptome analysis to determine the genes expressed in the heads of the whitefly using 150 heads of 3rd generation progeny of cassava whiteflies obtained from a single colony, to ensure that the whiteflies were of the same genetic make-up. The whitefly heads were isolated by immobilizing live whiteflies on ice plate and cutting their heads with tweezers, while observing through a light microscope. The heads were placed into RNAlater (Cat#AM7020, Ambion, United States) immediately after removal and then stored at −80°C. The head samples were shipped (in dry ice) to Beijing Genomics Institute (BGI Tech Solutions, Shenzhen, China) for RNA processing, sequencing and analysis to determine the genes expressed.
Validation of the or Sequences Found in the Cassava Whiteflies
We used primers generated from the Or gene sequences found in the whiteflies from the lake region (B. tabaci SSA-ECA) to screen and validate the presence of similar Or genes in the cassava whiteflies from the coastal region (B. tabaci SSA-ESA). These primers (Supplementary Table 4) were designed to amplify Or regions of about 800–900 bps from cDNA samples generated from RNA extracts of SSA-ESA whiteflies. The total RNA was extracted using Trizol reagent (Cat#15596026, Invitrogen Life Technologies, United States) and the Invitrogen Purelink RNA mini kit (Cat#121830, Thermofisher Scientific, United States) following the manufacturer’s instructions. The whiteflies, collected from cassava plants, were rinsed in 70% ethanol for 30 s prior to addition of 1 mL Trizol reagent and homogenization. The concentration and purity of the RNA in the extract was determined by measuring the absorbance of the solution at 230, 260 and 280 nm using Nanodrop 2000 (Thermofisher scientific, United States) prior to storage at −80°C. The M-MuLV Reverse Transcriptase (Cat#M0253S, New England Biolabs, United States) was used generate cDNA from the RNA extracts (using 5 μg RNA per 10 μL reaction volume) based on the manufacture’s instruction. The obtained cDNA solution was immediately used for PCR amplification or stored at −20°C.
The PCR amplification of the Or genes was performed using OneTaq® 2X Master Mix with Standard Buffer (Cat# M0482, New England Biolabs, United States) following manufacturer’s instructions and 10% v/v cDNA template. The PCR amplification reactions were performed using Veriti hot-lid thermocycler (Applied Biosystems, United States) based on thermocycling conditions recommended for OneTaq® 2X Master Mix with Standard Buffer and the primer-specific annealing temperatures (Supplementary Table 4). The PCR products were analyzed by agarose gel electrophoresis using 1% agarose gels in Tris-Acetate-EDTA (TAE) buffer, as previously described. The PCR products containing DNA fragments of interest, based on the size of expected DNA products were further analyzed by gel electrophoresis using 2% agarose gels to separate the DNA bands of interest, which were then extracted and purified from the gels by using the PureLink Quick gel purification kit (Cat#K220001, Invitrogen Life Technologies, United States) according to the manufacturer’s instruction.
The purified DNA fragments were cloned into Escherichia coli cells using the TOP10 competent E. coli cells and the pCR 2.1-TOPO vector provided with the TOPO TA cloning kit (Cat# K450002, Invitrogen Life Technologies, United States) following manufacturer’s instructions. The colonies containing the Or gene fragments of interest were identified by colony PCR using Or-specific gene-specific primers, 10-fold diluted E. coli colonies removed from agar plates and OneTaq 2X master mix with Standard Buffer (Cat# M0482, New England Biolabs, United States) based on the manufacturer’s instructions. Plasmids containing Or gene fragments of interest were purified from the colonies that contained the DNA fragments of interest, based on colony PCR results, by using the Invitrogen Purelink quick plasmid purification kit (Cat#k210010, Thermofisher Scientific, United States) as directed by the manufacturer. The Or gene fragments of interest were purified from the plasmids using M13 primer pairs (M13-forward 5′-GTAAAACGACGGCCAG-3′ and M13 reverse 5′- CAGGAAACAGCTATGAC-3′) and sequenced by Sanger sequencing (Macrogen, United States). The obtained sequences were inspected to identify poor quality reads, which were trimmed-off prior to comparison with the Or sequences from SSA-ECA through progressive pair-wise alignment method with 51% similarity threshold from Geneous Prime 2020.0.3(1).
Y-Tube Olfactometer
We conducted the odor-guided behavior assays with plant-produced volatile chemicals using a Y-tube olfactometer system consisting of a Y- tube connected to a stream of compressed air that delivers plant-produced volatile chemicals from head-space air of the tested plants. We used a glass Y-tube with internal diameter of 2.5 mm and 15 cm long arms at 60° from each other. The top ends of the Y-tubes were connected to the air inlets while the bottom end was used as an air outlet and inlet for the whiteflies. Plant-produced volatile chemicals were delivered to one top end of the Y-tube through Teflon tubes connected at the top end of an oven bag (Glad®2) enclosing the vegetative parts plants. The oven bags were tied at the bottom end of the stem to prevent entry and exit of air, except through a Teflon tubing that delivered compressed air (at 200 mL.min–1), which was passed through activated charcoal (for purification) and distilled water (for humidification) prior entry into the oven bag. The other end of the Y-tube was connected to a Teflon tubing delivering air from an empty oven bag, which acted as a control sample.
Two-Choice Behavior Assays With Plant-Produced Volatile Chemicals
We conducted two-choice behavior assays with head-space air from healthy cassava plant of variety Albert and Kiroba, CMD- and CBSD-infected cassava plants, tomato, cotton, cowpea, lemongrass, cassava whose leaves have been mechanical-damaged by crushing of the leaves, as well as mite-infested and whitefly-infested cassava plants. The two-choice behavior assays were performed indoors, between 14.00 and 17.00 pm in a room maintained at 25 ± 5°C and 65 ± 5% RH using adult whiteflies and Y-tube olfactometer system. Six repeats were done for each plant type and the behavior of 20 male and 20 female whiteflies was determined, individually, per each repeat. Two Y-tubes were used in each experiment, one for the male and the other for the female whiteflies to avoid the effect of con-specifics. Prior to the assay, the whiteflies were collected from colonies reared on cassava plants, anesthetized by placing on ice (4°C) for 5 min, separated based on their sex (using an aspirator) and then starved for 2 h. During the assays, the starved whiteflies were introduced into the Y-tube (one at a time) through the bottom end of the Y-tube using an aspirator. The path of the whiteflies within the Y-tube was observed for 5 min during which the whiteflies that moved to the end of either arms of the Y-tube were assumed to have made a choice and those that moved back and forth or remained at the center of the Y-tube after the 5 min were assumed to be indecisive. The number of whiteflies that made a decision was used to calculate the preference index of the whiteflies using Equation 1. After each assay the Y-tubes and Teflon tubings were cleaned by rinsing with 70% ethanol before and after soaking the tubes into acetone for at least 2 h and then dried at 40°C overnight.
Where: PI is Preference index, Wp is the number of whiteflies in the plants side and Wa is the number of whiteflies in the air side.
Two-Choice Behavior Assays With Individual Chemicals
We conducted two-choice behavior assays with pure volatile chemicals, individually or in combination, to determine the key chemicals responsible for odor-guided behavior of the whiteflies. The two-choice behavior assays were conducted using a glass Y-tube, similar to the one used in the two-choice behavior assays with plant-produced volatile chemicals, except that the ends of the Y-tubes were connected to clean sterile eppendorf tubes containing strips of filter membranes instead of air-inlets. The bottom end of the eppendorf tubes were cut such that they fit tightly around the top-ends of the Y-tubes and sterile filter papers strips (REF#31003, Sugi, Kettenbach Germany) were folded in half and inserted into the eppendorf tubes at a distance closer to the ends of the Y-tubes. All chemical compounds were supplied by Sigma Aldrich Inc., United States and of highest available purity. The chemicals were diluted to 1% in dimethyl sulfoxide (DMSO) (Cat#276855) prior the assays. The chemical solutions were added to one of the filter membranes that were inserted into one end of the Y-tubes and equal volume of DMSO was added to the other end of the Y-tube, this served as the control. Individual female whiteflies, that were starved for 2 h prior the assays, were inserted in the bottom end of the Y-tube and their movement within the Y-tube was observed for 5 min to determine their preference. We used 50 whiteflies (male and female) and five replicates (10 whiteflies per replicate) for each chemical tested, individual or in combination. One Y- tubes was used for each chemical and the preference index (PI) of the whiteflies was calculated based on the number of whiteflies that migrated to either end of the Y-tube using Equation 1. The behavior assays were performed indoors, between 11.00 am and 17.00 pm in a room maintained at 25 ± 5°C and 65 ± 5% RH.
Statistical Analysis
Statistical analyses were done in RStudio version 3.4.1 (RStudio Inc., Vienna, Austria) using Linear mixed-effects models (lmer4)—to determine variability in preference indices due to plant variety, conditions of the plants and sex of the insects—and Q-Q plots, the Shapiro–Wilk test and histogram plots were used to assess if the data points and their associated errors obtained from the different models were normally distributed. In the case where data points were not normally distributed, the data points were transformed using Box-Cox transformation. Once normal distribution was achieved, ANOVA and Tukey’s HSD test were used to compare the modals and identify the model that is significantly different from the others. T-test statistics (both one and two-way) were sued to determine the observed variability between and within groups and Boxplots were used to display the datasets and their statistics.
Collection of Plant-Produced Volatile Chemicals From Head-Space Air of Plants
We collected plant-produced volatile chemicals from the head-space air of the investigated plants using the “push-pull” dynamic head-space collection system and volatile collection traps (VCT) containing Tenax resins (Scientific Instruments, NJ, United States). We collected headspace air from six-eight-week-old plants (before the flowering stage) by covering the vegetative parts of the plants with oven bag and introduced compressed air (at 200 mL.min–1) into the plant using a Teflon tube inserted at the bottom (air inlet) oven bag as with the two-choice behavior assays. The top of the oven bag (air outlet) was connected to a VCT that was connected to a pump that pulled the air from the volatile collection trap. The VCTs were kept at −20°C when not in use and rinsed by with 100 μL of methanol, acetic acid and hexane (in the given order) prior to use. The volatile chemicals in the head-space air of the investigated plants were collected into the VCTs for 4 h, between 10.00 am and 14.00 pm in a room whose temperature and relative humidity was maintained at 25 ± 5°C and 65 ± 5% RH, respectively. Six samples were collected from each plant and three control samples (compressed air) for each plant type/condition. At the end of the collection period, the VCTs were removed from the system and immediately wrapped in aluminum foil and stored at −20°C.
Identification of Plant-Produced Volatile Chemicals From Head-Space Air of Plants
We performed gas-chromatography mass-spectrometry to identify the plant-produced volatile chemicals captured in the VCTs using the GCMS-QP2010 Ultra system with RTX-5MS capillary column (30 m long, 0.25 μm thickness, 0.25 mm diameter: Restek) and Helium gas, at a flow rate of 2.29 mL.min–1 as the carrier gas. The volatile chemicals trapped in the VCTs were eluted by passing 100 μL Hexane into the trap and the eluents were stored in ice prior analysis. During the analysis, an auto-injector was used to sample and introduce 1 μL of the eluents into the injector port that was maintained at 40°C. At the injector port, the samples were heated to 250°C at an increment rate of 8°C per minute and then held at 250°C for 8 min, after which they were carried into the capillary column for separation and then mass spectrometer for ionization using 70 eV electron charge. The spectra peaks obtained from the ionized volatile chemicals were manually annotated by comparing spectra profiles of the detected volatile chemicals to those of known chemicals found in the Flavor and Fragrance Natural and Synthetic Compounds (FFNSC3) library using the GCMS Solution software (Shimatzu, Kyoto, Japan). We performed automated peak integration to determine the area of each peak obtained from the spectra and we used the peak areas to calculate the relative abundance of the identified chemical compounds in each sample. We compared the distribution of the plant-produced volatile chemicals identified from the investigated plants using Non-metric Multidimensional Scaling (NMDS) ordination, as implemented in PAST3, and Heatmap analysis based on the “ComplexHeatMap” function on R studio.
Results and Discussion
The Cassava Whitefly Has a Reduced Peripheral Olfactory System
To establish a laboratory whitefly culture, we first sampled B. tabaci specimens from cassava plantations in Chambezi (Bagamayo, Tanzania). To ascertain the genotype, we sequenced mtCOI portions from 18 of the collected specimens, which we then compared against samples of the SSA-ESA B. tabaci genotype (Wosula et al., 2017) as well as suitable outgroups for which sequences were obtained from GenBank. A maximum-likelihood analysis of an alignment of this dataset revealed that our specimens grouped within the SSA clade, and more specifically together with samples from the SSA-ESA population, with strong bootstrap as well as approximate a-bayes test support (Figure 1C).
Having established a verified colony of B. tabaci SSA-ESA, we next examined the morphological configuration of their peripheral olfactory system. To this end, we used scanning electron microscopy to identify putative olfactory sensilla on the flagella and pedicels, the two parts of the antennae that are presumed to function as primary olfactory organs. We succeeded in examining two female and two male antennae from dorsal and ventral sides. An additional eight specimens examined were discarded from further analysis, due to difficulties in removing the thick wax coating covering these minute insects. Close examination of the antennae of the females revealed only 15 sensilla, in total, with a morphology suggesting olfactory function. The sensilla identified were 1 finger-like sensilla (situated at the tip of the flagellum), four basiconic and four coeloconic sensilla (also on the flagellum) as well as one short basiconic-like and five chaetae sensilla on the pedicel (Figures 1D–F). The same number of finger-like, basiconic, and coeloconic sensilla were also identified from the antennae of male whiteflies. The male whiteflies, however, had seven chaetae sensilla on the pedicel and lacked the short basiconic-like sensilla. The numbers and composition of sensilla noted here from B. tabaci SSA-ESA are in line with what has previously been reported from the non-cassava whitefly, the Middle East Asia Minor 1 (MEAM1) (Zhang et al., 2015). Although B. tabaci SSA-ESA (and MEAM1) seems to have the full complement of the typical sensilla types present in insects, the number of sensilla is remarkably reduced compared to most other insects.
The Cassava Whitefly Has a Greatly Reduced Olfactome
Having mapped the morphology, we next investigated the molecular composition of the olfactory system. Available draft genomes from B. tabaci SSA-ECA were mined for chemosensory proteins, specifically odorant receptors (OR) (Vosshall and Hansson, 2011), gustatory receptors (GR) (Montell, 2009), ionotropic receptors (IR) (Benton et al., 2009), and odorant binding proteins (OBP) (Pelosi, 1994). We first performed a protein prediction (proteome) analysis of the B. tabaci (SSA-ECA) genome using AUGUSTUS (v3.2.3) (Stanke and Morgenstern, 2005), with Drosophila melanogaster, Acyrthosiphon pisum, and Rhodnius prolixus as references. A consensus proteome was then generated by dereplication of the three predicted proteomes in a two-step process. First, output results from the three proteomes were merged in a single proteome. Next, proteins that shared an identity of at least 100% over their full length were clustered and duplicate sequences removed. To identify chemoreceptors in the predicted proteome, we constructed four Hidden Markov Models (HMM) from multiple sequence alignments from each chemosensory subfamily identified from A. pisum. To validate the final models, we tested each HMM against the initial protein dataset for each subfamily. We then used the Hidden Markov Models (HMMs) to screen the predicted proteome for ORs, GRs, IRs, and OBPs. Uncovered candidate proteins were compared against the eggNOG mapper (v.2) (Huerta-Cepas et al., 2017) and InterPro (v.5) (Mulder and Apweiler, 2007) databases for functional characterization. In the end, we identified 10 ORs, nine GRs, 11 IRs, and three OBPs, which would place B. tabaci among the insects with the smallest olfactome, a finding mirroring the reduced olfactory apparatus (Figure 2). The identified ORs included the odorant receptor co-receptor Orco, as well as eight tuning ORs.
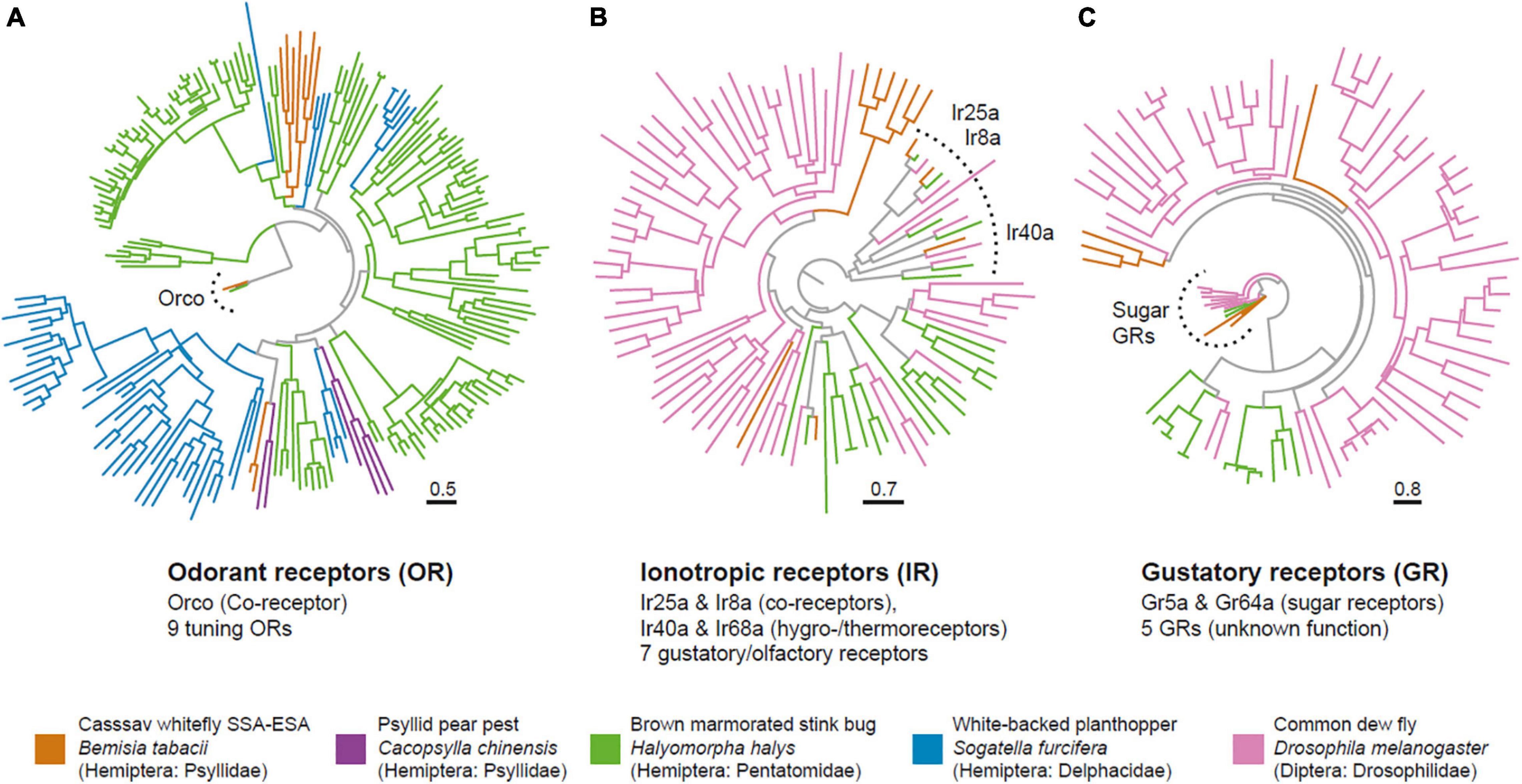
Figure 2. The cassava whitefly has a greatly reduced olfactome. Phylogenetic trees of chemosensory proteins identified from the cassava whitefly B. tabaci SSA-ESA and select other insects showing the odorant receptors (A), Ionotropic receptors (B) and Gustatory receptors (C). Trees were generated in IQ-Tree, and branch support tested with Shimodaira–Hasegawa approximate likelihood ratio test with 1,000 replicates.
A comparison of these ORs with other candidate ORs from Hemiptera reveal the typical pattern of this gene family with species specific expansions, with and very few direct orthologs present. We noted that two of the B. tabaci ORs clustered with Ors from Psyllid pear pest Cacopsylla chinensis, but whether the sequence homology also translates into functional similarity remains unclear. Of the IRs, we found orthologs of the co-receptors Ir25a and Ir8a, as well as copies of Ir40a, and Ir68a, which are involved in hygro- and thermo-sensation in Drosophila (Benton et al., 2009; Enjin et al., 2016; Frank et al., 2017). Apart from these, we noted seven additional genes with homology to known IRs. Of the nine identified GRs, two (Gr5a and Gr64a) fell together with known sugar receptors from Drosophila. Given the close relatedness between B. tabaci SSA-ECA and SSA-ESA we would expect both biotypes to carry similar numbers of chemosensory genes. Our approach here for identifying chemosensory genes was conservative. Hence, there might be additional members of the four chemosensory subfamilies among the predicted genes that did not pass the functional characterization scans.
For most of the identified chemosensory genes, it was not possible to determine full-length sequence and correct exon/intron structure. Thus, to get a better grasp of the gene structures, as well as to verify expression, we examined a transcriptome generated from 1,500 heads of B. tabaci SSA-ESA for transcripts corresponding to the putative chemosensory genes. The transcriptome, however, uncovered only the three OBPs and three GRs, as well as Orco but none of the tuning ORs, nor any IRs. The lack of transcripts corresponding to chemosensory genes is not wholly surprising. With the exception of OBPs, these genes are typically expressed at low levels, combined with the fact that B. tabaci possess few chemosensory neurons. We therefore used the ORs genes identified from the SSA-ECA genome to generate primers for amplification of these ORs in SSA-ESA whiteflies. We succeeded in partially amplifying six of the tuning ORs identified and the full-length sequence of Orco. The amino acid sequences of the amplified ORs were similar to those in silico determined from the SSA-ECA genome confirming that the ORs of whiteflies are conserved.
In summary, odor detection in the whiteflies appears to be highly reduced compared to other insects, including other Hemiptera, such as the white-backed planthopper Sogatella furcifera, with 63 ORs (He et al., 2015), the Asian citrus psyllid Diaphorina citri with 46 ORs (Wu et al., 2016) and the pear aphid A. gossypii with 45 ORs (Cao et al., 2014). Interestingly, however, similarly low numbers have also been reported from the Hemipteran relative—the pear pest Cacopsylla chinensis (Xu et al., 2019), which is another tiny and highly polyphagous insect pest.
Repulsion Is the Basis for Odor-Guided Responses in the Cassava Whitefly
Although reduced, cassava whiteflies evidently possess an olfactory system. To investigate this further, we examined whether olfactory cues are used for host-selection, more specifically to localize cassava plants. A two-choice behavioral assay (Figure 3A) was used to investigate the odor-guided behavior of the cassava whiteflies. Cassava whiteflies (both male and female SSA-ESA) displayed no preference for the host smell when provided a choice between headspace from cassava plants (Albert variety) and filtered air (Figure 3B). Volatiles from cowpea (Vigna unguiculata), also colonized by B. tabaci SSA-ESA, also failed to elicit attraction (Figure 3B). In short, olfaction does not appear to be the primary mode by which whiteflies are drawn to healthy host plants.
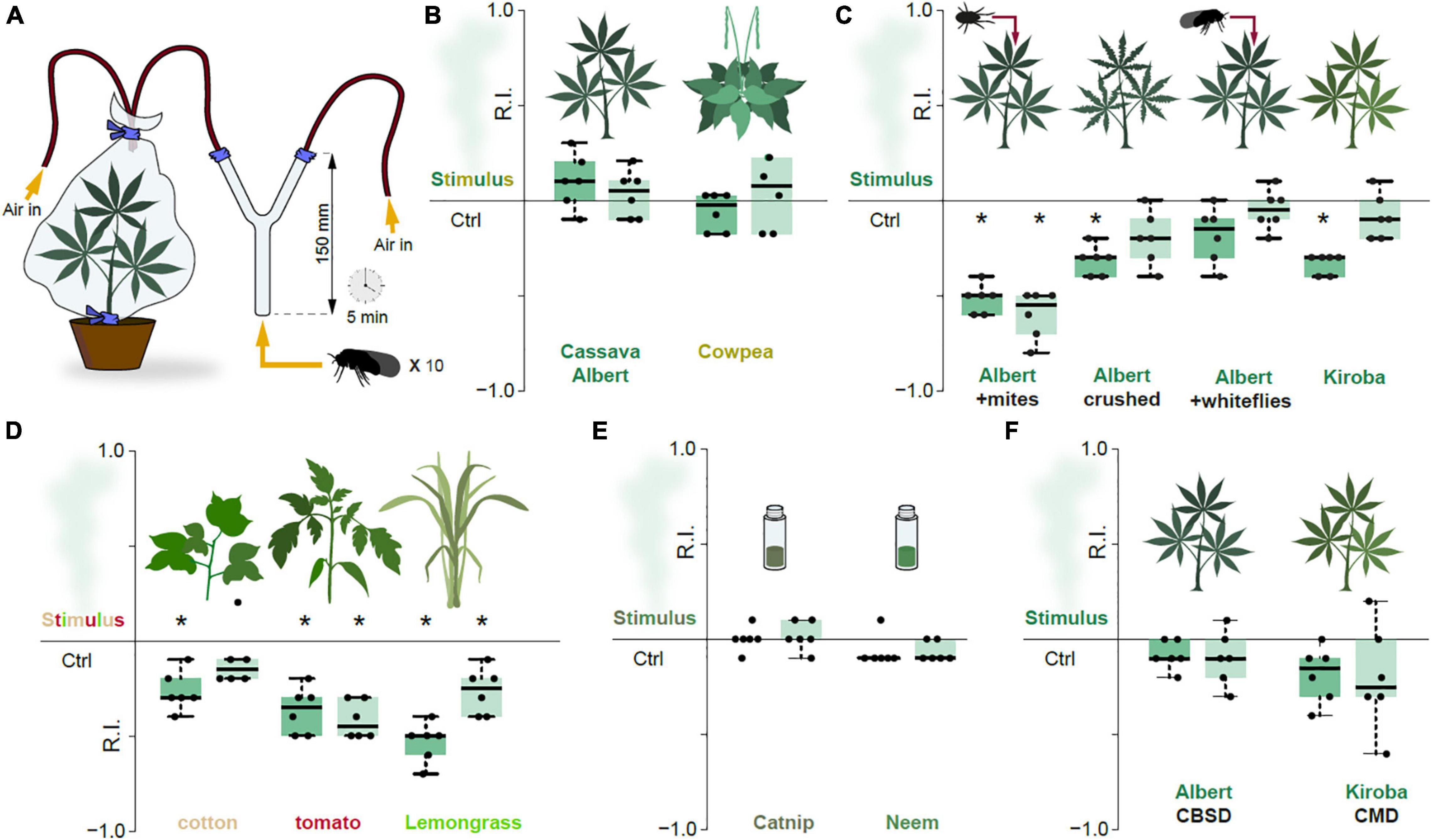
Figure 3. Repulsion is the basis for odor-guided response in the cassava whitefly. (A) Experimental set-up of the two-choice odor-guided behavior assays with 20 male and 20 female adult whiteflies with six repeats for each plant type. (B) Response indices (RI) obtained from the cassava whiteflies exposed to volatile chemicals from suitable host plants. The edges of the boxes are the first and third quartiles, thick lines mark the medians, and whiskers represent data range. (C) RI obtained from the cassava whiteflies exposed to volatile chemicals from suitable host plants mechanically damaged cassava plants. (D) RI obtained from the cassava whiteflies exposed to volatile chemicals from alternative host-pants. (E) RI obtained from the cassava whiteflies exposed to volatile chemicals from insect repellent essential oils, catnip and neem. (F) RI obtained from the cassava whiteflies exposed to volatile chemicals from virus infected cassava plants. For all Boxplots, the dark green boxes represented the preference indices of the male whiteflies while the light green boxes represented the preference indices of the female whiteflies. The preference was tested with one-sample Wilcoxon test, theoretical mean 0. *Denotes significantly different from 0; p < 0.05.
We accordingly wondered if the main function of the olfactory system in B. tabaci SSA-ESA might rather be to decipher cues produced by unsuitable hosts. Previous work has found that mite-infested cassava plants are repellent toward whiteflies, with presence of mites associated with a reduced population of adult whiteflies and number of eggs laid (Legg, 1994). We thus tested the response toward cassava plants (Albert) infested with cassava green spider mites (Mononychellus tanajoa) in our assay. Indeed, presence of mites resulted in significant repulsion (Figure 3C). Similarly, the whiteflies were also repelled by head-space air from cassava plants with crushed leaves (Albert), a process inducing plant defenses by mimicking mechanical damage caused by feeding of herbivores (Walling, 2000). Presence of conspecifics from whiteflies on the cassava leaves, however, had no effect on females, but was mildly repellent toward the males (Figure 3C). The aversion seen in the males was unlikely to be attributed to induced plant defenses, since whitefly feeding causes little plant damage (Walling, 2008) and given that the females were not repelled. Possibly, the males might react negatively to whitefly emitted semiochemicals, but whether such are indeed produced by members of the B. tabaci species complex remains to be determined.
The Kiroba variety of cassava, which according to reports from farmers and from unpublished field observations is less prone to B. tabaci SSA-ESA infestations than variety Albert, was also moderately but significantly repellent to the examined whiteflies (Figure 3C). Similar repulsion was recorded for headspace air from tomato and cotton, suboptimal host plants for B. tabaci SSA-ESA (Legg, 1996; Figure 3D). Lemongrass (Cymbopogon citratus), reportedly repellent to B. tabaci type Q—the Mediterranean (MED) genetic group (Deletre et al., 2015), was also repellent to B. tabaci SSA-ESA in our assay (Figure 3D). Neem and catnip, two efficient insect repelling plants were (Peterson and Coats, 2001; Brahmachari, 2004), however, not repellent to B. tabaci SSA-ESA (Figure 3E). In contrast to B. tabaci types B (Middle East–Minor Asia 1) and Q (Mediterranean genetic group) — vectors of the Tomato yellow leaf curl virus (TYLCV) — that prefers infected over non-infected tomato plants (Li et al., 2010), B. tabaci SSA-ESA showed no such preference, displaying no difference in behavior toward CMD- or CBSD- infected cassava plants compared to healthy controls (Figure 3F).
In summary, our results indicate that B. tabaci SSA-ESA appears to use olfactory cues primarily in the identification of suitable host plants, by discriminating volatiles indicative of what is not a suitable host. A strategy quite unlike most other insects, such as Drosophila melanogaster that has dedicated olfactory channels for favorable as well as unfavorable host features, with a clear majority tuned to the former (Semmelhack and Wang, 2009). For the extreme generalist members of the B. tabaci complex, devoting their reduced olfactory resources to detecting the seemingly few aspects they find aversive, rather than discriminating among the myriad of host plants they can thrive on would indeed seem like an efficient strategy.
Individual Volatiles Repel Cassava Whiteflies
To determine which volatiles mediate the observed repellence, we collected and analyzed head-space air via linked gas chromatography mass spectroscopy from host and non-host plants; more specifically from cassava (Albert and Kiroba varieties), mite-infested cassava (Albert), cowpea, cotton, tomato, as well as lemongrass. From these samples we identified a total of 172 volatiles. A non-metric multidimensional scaling (NMDS) analysis based on the volatile composition (Supplementary Table 1) revealed a separation between Albert and Kiroba, with the former grouping together with cowpea, and partly with cotton, whereas the latter grouped together with the mite-infested cassava (Figure 4A). Lemongrass and tomato were partly overlapping and clustered distinctly from the other groups. The NMDS hence indicated that the repellent Kiroba variety and the mite-infested cassava share certain volatile features, which are distinct from the preferred host plants (i.e., cassava of variety Albert and cowpea) host plants. Kiroba and the mite-infested cassava samples both consistently showed the presence of ß-ocimene, which was either absent or in trace amounts in the other examined samples (Figure 4B). Y-tube behavioral tests with this chemical indeed also elicited consistent aversion (Figure 4C). Methyl salicylate — a major volatile in the headspace samples from mite-infested cassava, which was either absent or found in traces in the other samples — likewise elicited aversion (Figure 4C). Geraniol and α-phellandrene — found in lemongrass and tomato, respectively, but not in the other samples- was also aversive (Figure 4C), as was α-caryophyllene, found in tomato and cotton (Figure 4C). 2-hexenal and 3-hexenol have previously been shown to be attractive cues to the sweet potato whitefly (Li et al., 2014). In B. tabaci SSA-ESA, and in the assays employed here, we failed to see any attraction. Whether or not this reflects a genuine difference in biotype behavior, or simply reflects bioassay design remains to be determined.
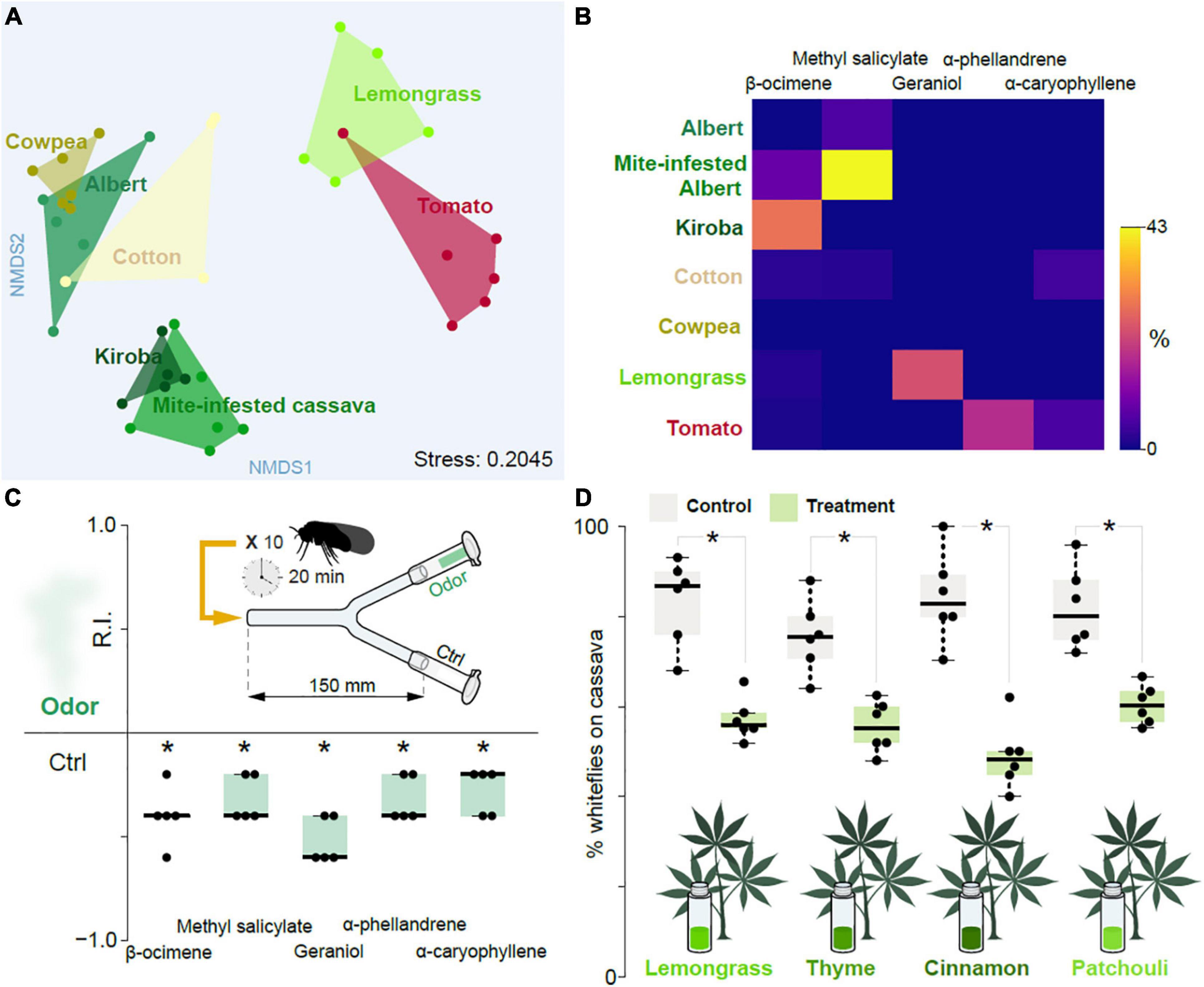
Figure 4. Influence of plant-produced volatile chemicals. (A) NMDS plot showing the inter- and intra- relationship between the plant-produced volatile chemicals obtained from host plants of the cassava whitefly (cassava of variety Albert and Kiroba), alternative host plants (cowpeas, cotton and tomato), unsuitable host plant (mite-infested cassava plants) and non-host plant (lemongrass). (B) Heat map of the volatile chemicals identified from the head-space air of cassava of variety Albert and Kiroba, cowpeas, cotton, tomato, mite-infested cassava plants and Lemongrass showing the relative abundance of the potentially key volatile chemicals as determined from GC-MS analysis. (C) The response indices obtained from two-choice behavior assays with some of the plant-produced volatile chemicals thought to be the key chemicals responsible for the repulsion of the cassava whiteflies. The assays were done using individual chemicals and five replicates for each chemical. DMSO (at 1% dilution) was used as the control. One-sample Wilcoxon test statistics, with theoretical mean 0, was used to test the preference displayed on the Boxplots where the * denotes significantly different from 0; p < 0.05. ANOSM and ANOVA were used to compare the results from the NMDS and Heatmap plots, respectively. (D) The response indices obtained from cassava leaves exposed to repulsive essential oils.
Essential Oils Cause an Effective but Unstable Repulsion to Whiteflies
Intercropping with aromatic plants can reduce populations of insect pests, either by repelling the pests, or by masking host plant odors (Song et al., 2010; Zhang et al., 2015; Hata et al., 2016). This approach has also been successful in reducing whitefly infestations. For example, intercropping cucumbers with celery and Malabar spinach (two non-preferred plants) reduced the numbers of whiteflies of B. tabaci biotypes B and Q (Zhao et al., 2014). Similarly, volatiles from coriander and celery also reduced numbers of whiteflies (biotype B) on tomato plants (Carvalho et al., 2017; Tu and Qin, 2017). To examine whether intercropping with aversive plants could also be used to combat whitefly infestations in cassava plants, we used essential oils from cinnamon, lemongrass, thyme, and patchouli to scent the air surrounding cassava plants leaves and monitored the cassava whiteflies’ behavior. Indeed, the headspace from these oils also reduced the number of whiteflies settling on the plants compared to the solvent control (Figure 4D). A viable approach for controlling, or at least reducing whitefly numbers, could accordingly be to intercrop cassava with unsuitable plants, although this would need to be verified with field experiments.
Conclusion
We demonstrate here that the cassava whitefly has a highly reduced olfactory apparatus compared to other insects, at the morphological as well as molecular level. Nevertheless, cassava whitefly do seem to rely on olfaction to identify suitable hosts, however, unlike most insects, the selection mechanism is not based on attraction but rather on repulsion. The repulsion-based host selection identified here concurs with previous findings for other members of the B. tabaci species complex such as Islam et al. (2017) who showed that more whiteflies settled on tomato plants that produced low amounts of terpenes indicating that negative selection, through repulsion, appears to be the basis of odor-guided behavior. A repulsion-based host selection mechanism could be more efficient for polyphagous insects that have a wide range of host plants as that would allow the cassava whiteflies to screen a broader number of plants and rule out unsuitable host plants prior to landing.
Several groups (Poveda and Kessler, 2012; Tang et al., 2013; Sulvai et al., 2016; Zhang et al., 2020) have demonstrated that intercropping with aromatic plants can reduce populations of insect pests either through repelling the pests, attracting their natural enemies or masking host-plant odors. Intercropping represents a lower cost and more practical potential control tactic compared to the direct use of volatile chemicals or essential oils for management of insect pests since it circumvents the challenges associated with the physiological properties of essential oils and volatile chemicals that would constrain their effectiveness (i.e., volatility, solubility and oxidation). Herbs like lemongrass and thyme could therefore be good candidates for intercropping with cassava as these plants will provide a continuous supply of volatile chemicals that can repel and reduce the population of whiteflies in the field. Hence further studies are recommended to determine the in-field effectiveness of locally grown herbs in minimizing and controlling the population of whiteflies as these might be more practical for smallholder farmers, who are the main cassava producers in Africa.
Data Availability Statement
All relevant data is contained within the article, further inquiries can be directed to the corresponding author/s.
Author Contributions
LM, JL, and MS were involved in the conceptualization, investigation, and writing of the original draft. JL, DM, and MS were involved in the reviewing of the article and supervision of the study. LM, ZO, HG, and MS performed formal analyses and prepared the graphics. JL and MS were also responsible for funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the U-Forsk program of the Swedish Research Council and supported by the International Institute of Tropical agriculture through its capacity building program. Contributions of JL were supported through the Roots, Tubers and Bananas Program of the CGIAR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Fortunatus Sunghwa, Everlyne Wosula, Helena Fritz, Dan-Dan Zhang, and Ola Gustafsson for assistance in experimental design and data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.775778/full#supplementary-material
Supplementary Table 1 | Variation in the type and amount of the plant produced volatile chemicals identified from the headspace air of the investigated plants, showing the relative abundance of the chemicals identified from each plant.
Supplementary Table 2 | The MtCO1 sequences of B. tabaci whiteflies used for phylogenetic comparison and confirmation of the type of B. tabaci used in the present study.
Supplementary Table 3 | Nucleotide sequences of the primers and probes used for amplification of part of the CBSI/UCBSI viral genomes from CBSD symptomatic cassava plants.
Supplementary Table 4 | Nucleotide sequence of the primers used to screen for the presence of Ors in B. tabaci SSA-ESA showing the forward (F) and reverse (R) primer sequence pairs and their melting temperatures, for each of the Or sequence.
Footnotes
References
Adams, I., Abidrabo, P., Miano, D., Alicai, T., Kinyua, Z., Clarke, J., et al. (2013). High throughput real-time RT-PCR assays for specific detection of cassava brown streak disease causal viruses, and their application to testing of planting material. Plant Pathol. 62, 233–242. doi: 10.1111/j.1365-3059.2012.02622.x
Anisimova, M., Gil, M., Dufayard, J. F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi: 10.1093/sysbio/syr041
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Bernays, E. A., and Chapman, R. F. (2007). Host-Plant Selection by Phytophagous Insects, Vol. 2. Switzerland: Springer Science & Business Media, 95–115.
Brahmachari, G. (2004). Neem—an omnipotent plant: a retrospection. Chembiochemistry 5, 408–421. doi: 10.1002/cbic.200300749
Bull, S. E., Briddon, R. W., Sserubombwe, W. S., Ngugi, K., Markham, P. G., and Stanley, J. (2006). Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 87, 3053–3065. doi: 10.1099/vir.0.82013-0
Cao, D., Liu, Y., Walker, W. B., Li, J., and Wang, G. (2014). Molecular characterization of the Aphis gossypii receptor gene families. PLoS One 9:e101187. doi: 10.1371/journal.pone.0101187
Carvalho, M. G., Bortolotto, O. C., and Ventura, M. U. (2017). Aromatic plants affect the selection of host tomato plants by Bemisia tabaci biotype B. Entomol. Exp. Appl. 162, 86–92.
Deletre, E., Mallent, M., Menut, C., Chandre, F., and Martin, T. (2015). Behavioral response of Bemisia tabaci (Hemiptera: Aleyrodidae) to 20 plant extracts. J. Econ. Entomol. 108, 1890–1901. doi: 10.1093/jee/tov118
Dubern, J. (1994). Transmission of African cassava mosaic geminivirus by the whitefly (Bemisia tabaci). Trop. Sci. 34, 82–91.
Enjin, A., Zaharieva, E. E., Frank, D. D., Mansourian, S., Suh, G. S., Gallio, M., et al. (2016). Humidity sensing in Drosophila. Curr. Biol. 26, 1352–1358.
Finch, S., and Collier, R. (2003). Host-plant selection by insects – a theory based on ‘appropriate/inappropriate landings’ by pest insects of cruciferous plants. Entomol. Exp. Appl. 96, 91–102. doi: 10.1046/j.1570-7458.2000.00684.x
Fondong, V. N., Pita, J. S., Rey, M. E. C., De Kochko, A., Beachy, R. N., and Fauquet, C. M. (2000). Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. Microbiology 81, 287–297. doi: 10.1099/0022-1317-81-1-287
Frank, D. D., Enjin, A., Jouandet, G. C., Zaharieva, E. E., Para, A., Stensmyr, M. C., et al. (2017). Early integration of temperature and humidity stimuli in the Drosophila brain. Curr. Biol. 27, 2381–2388. doi: 10.1016/j.cub.2017.06.077
Hata, F. T., Ventura, M. U., Carvalho, M. G., Miguel, A. L., Souza, M. S., Paula, M. T., et al. (2016). Intercropping garlic plants reduces Tetranychus urticae in strawberry crop. Exper. App. Acarol. 69, 311–321. doi: 10.1007/s10493-016-0044-3
He, M., Zhang, Y. N., and He, P. (2015). Molecular characterization and differential expression of an olfactory receptor gene family in the white-backed planthopper Sogatella furcifera based on transcriptome analysis. PLoS One 10:e0140605. doi: 10.1371/journal.pone.0140605
Hoang, T. D., Chernomor, O., Von Haeseler, A., Minh, Q. B., and Vinh, S. L. (2018). UFBoot2: improving the ultrafast Bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., Von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37(Suppl_1), D211–D215.
Islam, M., Hasanuzzaman, A. T. M., Zhang, Z., Zhang, Y., and Liu, T. (2017). High level of nitrogen makes tomato plants releasing less volatiles and attracting more Bemisia tabaci (Hemiptera: Aleyrodidae). Front. Plant Sci. 8:466. doi: 10.3389/fpls.2017.00466
Jarvis, A., Ramirez-Villegas, J., Campo, B. V. H., and Navarro-Racines, C. (2012). Is cassava the answer to African climate change adaptation? Trop. Plant Biol. 5, 9–29. doi: 10.1007/s12042-012-9096-7
Kalyaanamoorthy, S., Minh, Q. B., Wong, F. K. T., Von Haeseler, A., and Jermiin, S. L. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Legg, J. (1994). Bemisia tabaci: the whitefly vector of cassava mosaic geminiviruses in Africa: an ecological perspective. Afr. Crop Sci. J. 2, 437–448.
Legg, J. (1996). Host-associated strains within Ugandan populations of the whitefly Bemisia tabaci (Genn.), (Hom., Aleyrodidae). J. Appl. Entomol. 120, 523–527. doi: 10.1111/j.1439-0418.1996.tb01646.x
Legg, J. P., Kumar, P. L., Makeshkumar, T., Tripathi, L., Ferguson, M., Kanju, E., et al. (2015). Cassava virus diseases: biology, epidemiology, and management. Adv. Virus Res. 91, 85–142.
Legg, J., Owor, B., Sseruwagi, P., and Ndunguru, J. (2006). Cassava mosaic virus disease in East and Central Africa: epidemiology and management of a regional pandemic. Adv. Virus Res. 67, 355–418. doi: 10.1016/S0065-3527(06)67010-3
Li, M., Hu, J., Xu, F., and Liu, S. (2010). Transmission of tomato yellow leaf curl virus by two invasive biotypes and a Chinese indigenous biotype of the whitefly Bemisia tabaci. Int. J. Pest Manage. 56, 275–280. doi: 10.1080/09670871003743428
Li, Y., Zhong, S., Qin, Y., Zhang, S., Gao, Z., Dang, Z., et al. (2014). Identification of plant chemicals attracting and repelling whiteflies. Arthropod. Plant Interact. 8, 183–190. doi: 10.1007/s10886-010-9891-2
Martin, J. H., Mifsud, D., and Rapisarda, C. (2000). The whiteflies (Hemiptera: Aleyrodidae) of Europe and the Mediterranean Basin. Bull. Entomol. Res. 90, 407–448. doi: 10.1017/s0007485300000547
Maruthi, M. N., Jeremiah, S. C., Mohammed, I. U., and Legg, J. P. (2017). The role of the whitefly, Bemisia tabaci (Gennadius), and farmer practices in the spread of cassava brown streak ipomoviruses. J. Phytopathol. 165, 707–717. doi: 10.1111/jph.12609
Maruthi, M., Colvin, J., Seal, S., Gibson, G., and Cooper, J. (2002). Co-adaptation between cassava mosaic geminiviruses and their local vector populations. Virus Res. 86, 71–85. doi: 10.1016/s0168-1702(02)00051-5
Maruthi, M., Hillocks, R., Mtunda, K., Raya, M., Muhanna, M., Kiozia, H., et al. (2005). Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153, 307–312.
Mellor, H. E., and Anderson, M. (1995). Antennal sensilla of whiteflies: Trialeurodes vaporariorum (Westwood), the glasshouse whitefly, Aleyrodes proletella (Linnaeus), the cabbage whitefly, and Bemisia tabaci (Gennadius), the tobacco whitefly (Homoptera: Aleyrodidae). Part 1: external morphology. Int. J. Insect Morphol. Embryol. 24, 133–143. doi: 10.1016/0020-7322(94)00021-h
Minh, Q. B., Schmidt, A. H., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Milenovic, M., Wosula, E. N., Rapisarda, C., and Legg, J. P. (2019). Impact of host plant species and whitefly species on feeding behavior of Bemisia tabaci. Front. Plant Sci. 10:1. doi: 10.3389/fpls.2019.00001
Montell, C. (2009). A taste of the Drosophila gustatory receptors. Curr. Opin. Neurobiol. 19, 345–353.
Morris, B., Coates, L., Lowe, S., and Richardson, K. (1990). Nucleotide sequence of the infectious cloned DNA components of African cassava mosaic virus (Nigerian strain). Nucleic Acids Res. 18, 197–198. doi: 10.1093/nar/18.1.197
Mulder, N., and Apweiler, R. (2007). “Interpro and interproscan,” in Comparative Genomics, ed. N. Bergman (Totowa, NJ: Humana Press), 59–70.
Navas-Castillo, J., Fiallo-Olivé, E., and Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Peterson, C., and Coats, J. R. (2001). Insect repellents-past, present and future. Pestic. Outlook 12, 154–158.
Pita, J. S., Fondong, V. N., Sangare, A., Otim-Nape, G. W., Ogwal, S., and Fauquet, C. M. (2001). Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82, 655–665. doi: 10.1099/0022-1317-82-3-655
Poveda, K., and Kessler, A. (2012). New synthesis: plant volatiles as functional cues in intercropping systems. J. Chem. Ecol. 38, 1341–1341. doi: 10.1007/s10886-012-0203-x
Semmelhack, J. L., and Wang, J. W. (2009). Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature 459, 218–223.
Song, B. Z., Wu, H. Y., Kong, Y., Zhang, J., Du, Y. L., Hu, J. H., et al. (2010). Effects of intercropping with aromatic plants on the diversity and structure of an arthropod community in a pear orchard. Biocontrol 55, 741–751.
Stanke, M., and Morgenstern, B. (2005). AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Suppl_2), W465–W467. doi: 10.1093/nar/gki458
Sudhir, K., Glen, S., Michael, Li, Christina, K., and Koichiro, T. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Sulvai, F., Chaúque, B. J. M., and Macuvele, D. L. P. (2016). Intercropping of lettuce and onion controls caterpillar thread, Agrotis ípsilon major insect pest of lettuce. Chem. Biol. Technol. Agric. 3, 1–5.
Tang, G. B., Song, B. Z., Zhao, L. L., Sang, X. S., Wan, H. H., Zhang, J., et al. (2013). Repellent and attractive effects of herbs on insects in pear orchards intercropped with aromatic plants. Agrofor. Syst. 87, 273–285.
Thresh, J., Otim-Nape, G., and Fargette, D. (1998). “The control of African cassava mosaic virus disease: phytosanitation and/or resistance,” in Plant Virus Disease Control, eds A. Hadidi, R. K. Khetaspal, and H. Koganezawa (St Paul, MIN: American Phytopathological Society), 670–677.
Togni, P. H., Laumann, R. A., Medeiros, M. A., and Sujii, E. R. (2010). Odour masking of tomato volatiles by coriander volatiles in host plant selection of Bemisia tabaci biotype B. Entomol. Exp. Appl. 136, 164–173. doi: 10.1111/j.1570-7458.2010.01010.x
Tu, H., and Qin, Y. (2017). Repellent effects of different celery varieties in Bemisia tabaci (Hemiptera: Aleyrodidae) biotype Q. J. Econ. Entomol. 110, 1307–1316. doi: 10.1093/jee/tox110
Visser, J. (1988). Host-plant finding by insects: orientation, sensory input and search patterns. J. Insect Physiol. 34, 259–268. doi: 10.1007/s10886-010-9766-6
Vosshall, L. B., and Hansson, B. S. (2011). A unified nomenclature system for the insect olfactory coreceptor. Chem. Senses 36, 497–498. doi: 10.1093/chemse/bjr022
Walling, L. L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19, 195–216. doi: 10.1007/s003440000026
Walling, L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866. doi: 10.1104/pp.107.113142
Wosula, E. N., Chen, W., Fei, Z., and Legg, J. P. (2017). Unravelling the genetic diversity among cassava Bemisia tabaci whiteflies using NextRAD sequencing. Genome Biol. Evol. 9, 2958–2973. doi: 10.1093/gbe/evx219
Wu, Z., Zhang, H., Bin, S., Chen, L., Han, Q., and Lin, J. (2016). Antennal and abdominal transcriptomes reveal chemosensory genes in the Asian citrus psyllid, Diaphorina citri. PLoS One 11:e0159372. doi: 10.1371/journal.pone.0159372
Xu, J., Zhu, X., Chao, Q., Zhang, Y., Yang, Y., Wang, R., et al. (2019). Chemosensory gene families in the oligophagous pear pest Cacopsylla chinensis (Hemiptera: Psyllidae). Insects 10:175. doi: 10.3390/insects10060175
Zhang, X., Lövei, G. L., Ferrante, M., Yang, N., and Wan, F. (2020). The potential of trap and barrier cropping to decrease densities of the whitefly Bemisia tabaci MED on cotton in China. Pest Manag. Sci. 76, 366–374. doi: 10.1002/ps.5524
Zhang, X., Wang, S., Li, S., Luo, C., Li, Y., and Zhang, F. (2015). Comparison of the antennal sensilla ultrastructure of two cryptic species in Bemisia tabaci. PLoS One 10:e0121820. doi: 10.1371/journal.pone.0121820
Keywords: olfaction, whiteflies, cassava mosaic disease, cassava brown streak disease, Bemisia tabaci
Citation: Mrisho LM, Maeda DG, Ortiz ZM, Ghanavi HR, Legg JP and Stensmyr MC (2021) Influence of Olfaction in Host-Selection Behavior of the Cassava Whitefly Bemisia tabaci. Front. Ecol. Evol. 9:775778. doi: 10.3389/fevo.2021.775778
Received: 14 September 2021; Accepted: 21 October 2021;
Published: 18 November 2021.
Edited by:
Swayamjit Ray, Cornell University, United StatesReviewed by:
Hong Lei, Arizona State University, United StatesXin-Cheng Zhao, Henan Agricultural University, China
Copyright © 2021 Mrisho, Maeda, Ortiz, Ghanavi, Legg and Stensmyr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Latifa M. Mrisho, TC5tcmlzaG9AZ21haWwuY29t; James P. Legg, Si5sZWdnQGNnaWFyLm9yZw==; Marcus C. Stensmyr, TWFyY3VzLnN0ZW5zbXlyQGJpb2wubHUuc2U=
 Latifa M. Mrisho
Latifa M. Mrisho Daniel G. Maeda
Daniel G. Maeda Zaide M. Ortiz
Zaide M. Ortiz Hamid R. Ghanavi
Hamid R. Ghanavi James P. Legg
James P. Legg Marcus C. Stensmyr
Marcus C. Stensmyr