
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 20 December 2021
Sec. Environmental Informatics and Remote Sensing
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.762173
Citizen science is essential for nationwide ecological surveys of species distribution. While the accuracy of the information collected by beginner participants is not guaranteed, it is important to develop an automated system to assist species identification. Deep learning techniques for image recognition have been successfully applied in many fields and may contribute to species identification. However, deep learning techniques have not been utilized in ecological surveys of citizen science, because they require the collection of a large number of images, which is time-consuming and labor-intensive. To counter these issues, we propose a simple and effective strategy to construct species identification systems using fewer images. As an example, we collected 4,571 images of 204 species of Japanese dragonflies and damselflies from open-access websites (i.e., web scraping) and scanned 4,005 images from books and specimens for species identification. In addition, we obtained field occurrence records (i.e., range of distribution) of all species of dragonflies and damselflies from the National Biodiversity Center, Japan. Using the images and records, we developed a species identification system for Japanese dragonflies and damselflies. We validated that the accuracy of the species identification system was improved by combining web-scraped and scanned images; the top-1 accuracy of the system was 0.324 when trained using only web-scraped images, whereas it improved to 0.546 when trained using both web-scraped and scanned images. In addition, the combination of images and field occurrence records further improved the top-1 accuracy to 0.668. The values of top-3 accuracy under the three conditions were 0.565, 0.768, and 0.873, respectively. Thus, combining images with field occurrence records markedly improved the accuracy of the species identification system. The strategy of species identification proposed in this study can be applied to any group of organisms. Furthermore, it has the potential to strike a balance between continuously recruiting beginner participants and updating the data accuracy of citizen science.
Monitoring biodiversity is essential to evaluate the status of global ecosystems, as biodiversity is an indicator of climate change, environmental pollution, overexploitation of resources, invasive species, and natural disasters (van Klink et al., 2020; Hallmann et al., 2021). However, basic data to quantify recent degradation in biodiversity are insufficient. Surveillance of bioindicators, such as dragonflies, frogs, and birds, is the first step toward the methodical quantification of biodiversity because these species are well-known, and their conservation is a priority (Paoletti, 1999; Kadoya and Washitani, 2007; Kadoya et al., 2009; Parmar et al., 2016; Zaghloul et al., 2020).
A total of 204 species of dragonflies and damselflies (hereafter, odonates), belonging to the order Odonata, have been well-documented in Japan since ancient times (Sugimura et al., 1999; Futahashi et al., 2004, 2012; Naraoka, 2005; Kadoya et al., 2008; Ozono et al., 2021). The distribution surveys of well-known native species, such as odonates, are expensive and rely on amateur participants. In the 1990s, the Ministry of Environment of Japan conducted a comprehensive national survey of well-known animal groups, including odonates, in collaboration with more than 300 donate specialists throughout Japan to compile their distribution records (National Biodiversity Center of Japan, Ministry of the Environment, Japan, Tokyo, 2002). Although the survey was comprehensive, it could not collect adequate information on some species, especially a few abundant species. To quantify the dynamics of biodiversity loss, it is necessary to regularly update field occurrence records. Nevertheless, no similar biodiversity surveys have been conducted since the 1990s, possibly because of insufficient budget for nationwide surveys.
Citizen science, which includes scientific or ecological surveys conducted in collaboration with citizens, has been used to collect ecological data for scientific purposes in several countries and can improve the feasibility of sustainable nationwide surveys of biodiversity (Silvertown, 2009; Newman et al., 2012; Marzluff, 2013; Kobori et al., 2016). Furthermore, recent advances in information technology have dramatically reduced the barriers to public participation in ecological research. It is reported that more than 500 citizen-science projects have recruited more than 1.3 million participants for data collection, and the recruitment has particularly increased since 2010 for the projects wherein data is collected using smartphones (Theobald et al., 2015; Pocock et al., 2017). Using a smartphone, participants not only capture images using the equipped camera but also record additional information, such as date, time, and location. Thus, the advantages of smartphone technology are a boon for citizen-science projects.
Although citizen science can counter resource limitations, it has a major drawback, that is, data quality is not validated; compared to scientists and specialists, the low accuracy of species identification by amateur participants is an issue. Therefore, inaccurate data collected from citizen-science projects cannot be directly used to quantify biodiversity or its loss.
Recent advances in image recognition using artificial intelligence can improve the reliability of data collected from citizen-science projects (Langlotz et al., 2019). Image recognition, also called object recognition, with convolutional neural network (CNN) is a deep learning technique used to recognize the objects in an image or to classify the objects in an image (Emmert-Streib et al., 2020). This technique enables machines to self-learn from a large number of images and identify the key features for recognizing similar objects in different images. Since the development of image recognition model AlexNet in 2012, more than 20 different CNN architectures have been designed nowadays (Khan et al., 2020). For example, the visual geometry group network (VGGNet) architecture has been used since the beginning of the deep learning era (Simonyan and Zisserman, 2014), and the residual network (ResNet) architecture has deeper CNN layers and outperforms VGGNet under several conditions (Veit et al., 2016). VGGNet and ResNet architectures can have different number of CNN layers; there are 16 and 19 layers in VGGNet (VGGNet16 and VGGNet19, respectively) and 18 and 152 layers in ResNet (ResNet18 and ResNet152, respectively). Well-trained systems are equivalent to human experts for image recognition in several industrial fields (Goëau et al., 2018; Valan et al., 2019). Furthermore, deep learning-based image recognition systems have been reported to solve ecological problems, such as species identification (Kamilaris and Prenafeta-Boldú, 2018; Wäldchen and Mäder, 2018; Christin et al., 2019; Moen et al., 2019; Tabak et al., 2019; Hansen et al., 2020).
However, the accuracy of the species identification system is still an issue because collecting a large number of images to accurately identify species in the field is arduous (Perez and Wang, 2017; Shahinfar et al., 2020). To compensate for the low availability of images, researchers incorporate prior knowledges into image recognition systems (Berg et al., 2014; Ellen et al., 2019). Previous studies have reported that prior information of species distribution eliminates the probability of misidentification during the image recognition process and improves the accuracy of species identification (Berg et al., 2014; Aodha et al., 2019). Other studies have shown that combining image recognition systems with metadata, such as date, time, temperature, and location, can also effectively improve the accuracy of species identification systems (Ellen et al., 2019; Terry et al., 2020).
Along the line of these precedent studies, we propose an easy and simplified strategy to develop a species identification system with few images. In this study, we selected odonates as a representative species group, and developed three types of species identification systems using datasets of: (i) images collected from open-access websites (i.e., web scraping), (ii) combined web-scraped and scanned images obtained from books and specimens, and (iii) the combination of images and field occurrence records obtained from the National Biodiversity Center. We determined the accuracies of the three species identification systems to demonstrate how the accuracy of the species identification system can be improved.
Six image datasets, namely, F, W1, W2, W1F, W2F, and T, were used in this study to train the species identification system (Table 1). Images of the dataset F were obtained from open-access websites using the search engine Google (i.e., web scraping). We searched images using their scientific and Japanese names. Article 30 (iv) and 47 (v) of the Japanese Copyright Act allows the use of web-scraped images for implementation in machine learning and data analysis. By web scraping, 4,571 images captured in the field were collected. As the species were inaccurately identified in a few of these images, we directly verified the species in all the images and selected only reliable images. Then, rectangular sections of the original images, containing a single odonate, were trimmed using the image annotation software LabelImg (Lin, 2015) to obtain 4,589 images, as few original images contained multiple individuals. Most of the odonate images were captured from the lateral side of the individual, whereas few were captured from the dorsal and frontal sides. These images included 86 genera and 204 species of Japanese odonates (Supplementary Table 1). To improve the accuracy of the deep learning systems, we augmented the images using random affine transformation, adding noise, rotating, and applying flip-flop (Figure 1A). Image augmentation was repeated until 200 images of each species were obtained. All images were resized to a fixed size of 224 × 224 pixels. A total of 40,800 images of the dataset F were used for model training.
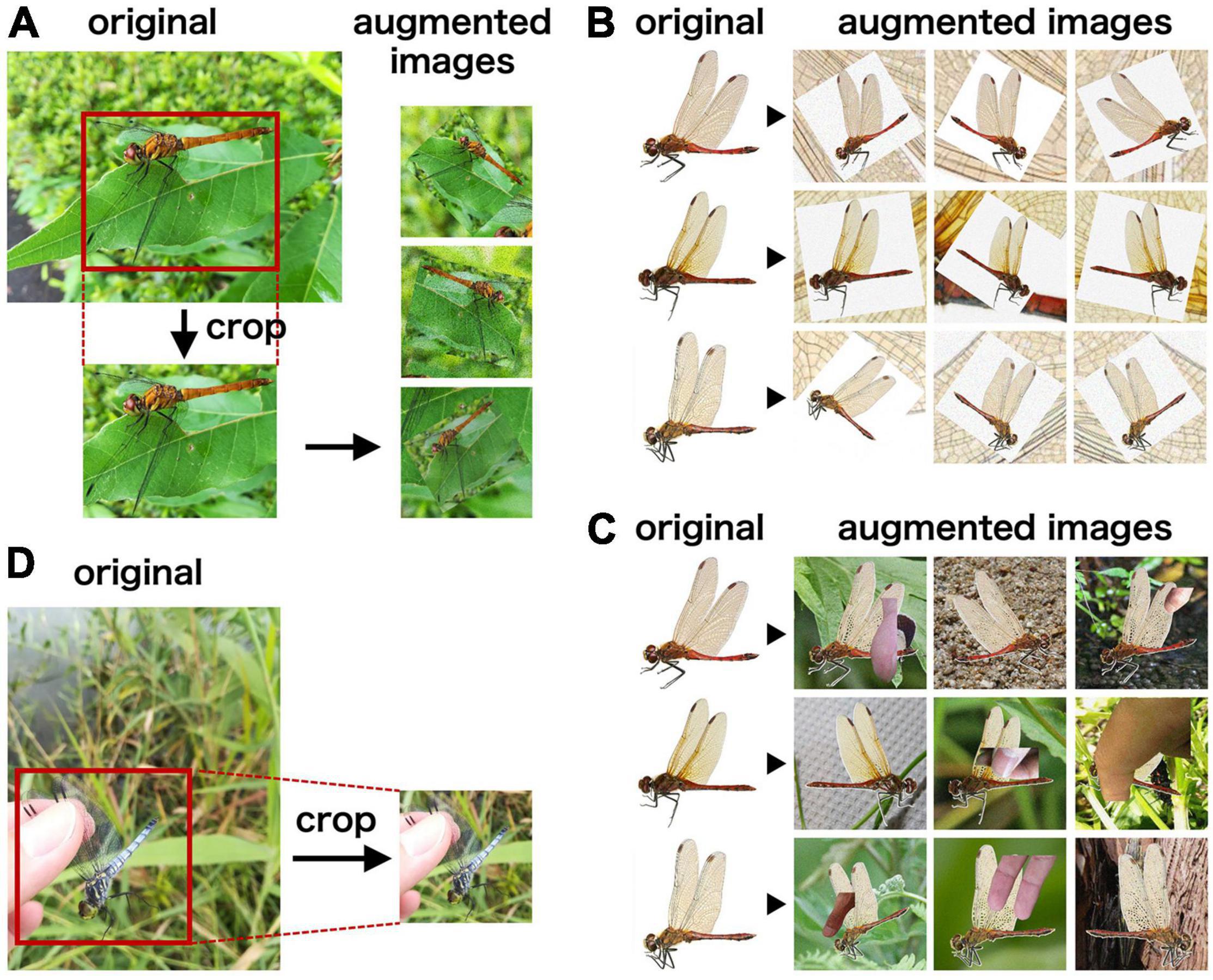
Figure 1. Examples of the images for model training and validation. (A–C) Examples of the original images and augmented images in dataset F (A), W1 (B), and W2 (C). (D) Examples of the original images from citizen surveys and the cropped images for model validation.
Images of the dataset W1 were obtained by scanning images from several books on Japanese odonates (Hamada and Inoue, 1985; Sugimura et al., 1999; Ozono et al., 2021) and specimens collected by the authors. We scanned 4,005 images in the RGB color mode and saved them in the PNG format. These images included 86 genera and 204 species of Japanese odonates (Supplementary Table 1). Each image contained a single odonate individual against a white background, and all images were captured from the lateral side of the individual. We then augmented the scanned images until 200 images of each species were obtained (Figure 1B), and resized them to a fixed size of 224 × 224 pixels.
Images of the dataset W2 were generated in the same way as for dataset W1. However, during image augmentation of dataset W2, the white background was replaced with images captured in the field, and images with human fingers were piled up (Figure 1C). This was because the images captured in the field contained natural background and photographers’ fingers. Like the dataset W1, the dataset W2 included 40,800 images, and all images were resized to a fixed size of 224 × 224 pixels.
The datasets W1F and W2F were generated by combining the images of the datasets W1 and F and W2 and F, respectively. To generate the dataset W1F, we randomly selected and merged 100 images of each species from the augmented images of both W1 and F, resulting in 200 images of each species and a total of 40,800 images in the dataset W1F. The dataset W2F was generated in a manner similar to that of W1F using the augmented images of W2 and F.
Images of the dataset T were obtained from citizen surveys, primarily using the data archive of Osawa et al., 2017. A total of 316 images captured using various types of smartphones were collected throughout Japan from August 2011 to December 2018 (Supplementary Table 2). The latitude and longitude of the location where the images were taken are stored in exchangeable image file format (EXIF) of the images. The sections of the original image, containing a single odonate, were manually trimmed and resized to a fixed size of 224 × 224 pixels (Figure 1D). Species identification for these images was performed by us. These images covered 41 genera and 62 species. The number of images in each genus and species can be found in Supplementary Table 1.
The field occurrence records of odonates were obtained from the National Biodiversity Center, the Ministry of Environment, Japan. The original dataset contained 1,07,717 occurrence records of 204 odonate species primarily recorded during the 1990s (National Biodiversity Center of Japan, Ministry of the Environment, Japan, Tokyo, 2002) in each secondary grid mesh of the Japanese national grid mesh system (Ministry of Internal Affairs and Communications, 1996). Each secondary mesh measured approximately 10 km × 10 km and corresponded to a square of 1:25,000 on the topographic map of Japan.
Four CNN architectures, namely, VGGNet16, VGGNet19, ResNet18, and ResNet152 (Simonyan and Zisserman, 2014; Veit et al., 2016), pre-trained using ImageNet datasets (Deng et al., 2009), were selected. This is because both VGGNet and ResNet architectures are implemented by different layers of neural networks, which can be used for evaluating the correlation between number of layers and accuracy. Each architecture was independently trained using the five datasets with 50 epochs and a batch size of 32. The learning rate was initially set at 0.001, which was automatically multiplied by 0.1 after every 10 epochs. The architecture optimisation was based on a categorical cross-entropy loss function using stochastic gradient descent. The output score of each trained architecture was transformed using the sigmoid function based on the one-versus-the-rest approach. The transformed scores represented probability-like scores (p) of the 204 species of odonates.
The trained architectures (i.e., models), were then validated using the dataset T, and the top-1 and top-3 accuracies were evaluated using the whole dataset, that is, the micro average of accuracy was evaluated. The training and validation processes were repeated ten times using identical codes and datasets but with different random seeds to ensure that the model accuracies were not affected by the initial-value problem.
A confusion matrix represents the number of true positives, true negatives, false positives, and false negatives among the data values of the validation results and is used to determine the class (i.e., species) in which the model prediction was inaccurate. To validate the prediction accuracy of the model with the highest top-1 accuracy, we computed a confusion matrix and analyzed the data.
To improve the accuracy of the species identification system, the output of the image-based model was combined with the field occurrence records (Figure 2). To evaluate the accuracy of the combined model, we used the trained image-based model (for example, ResNet152 trained using the dataset W2F) to determine the p of each image in the dataset T. p, which is a vector consisting of 204 elements, can be regarded as the probabilities of occurrence of 204 odonate species.
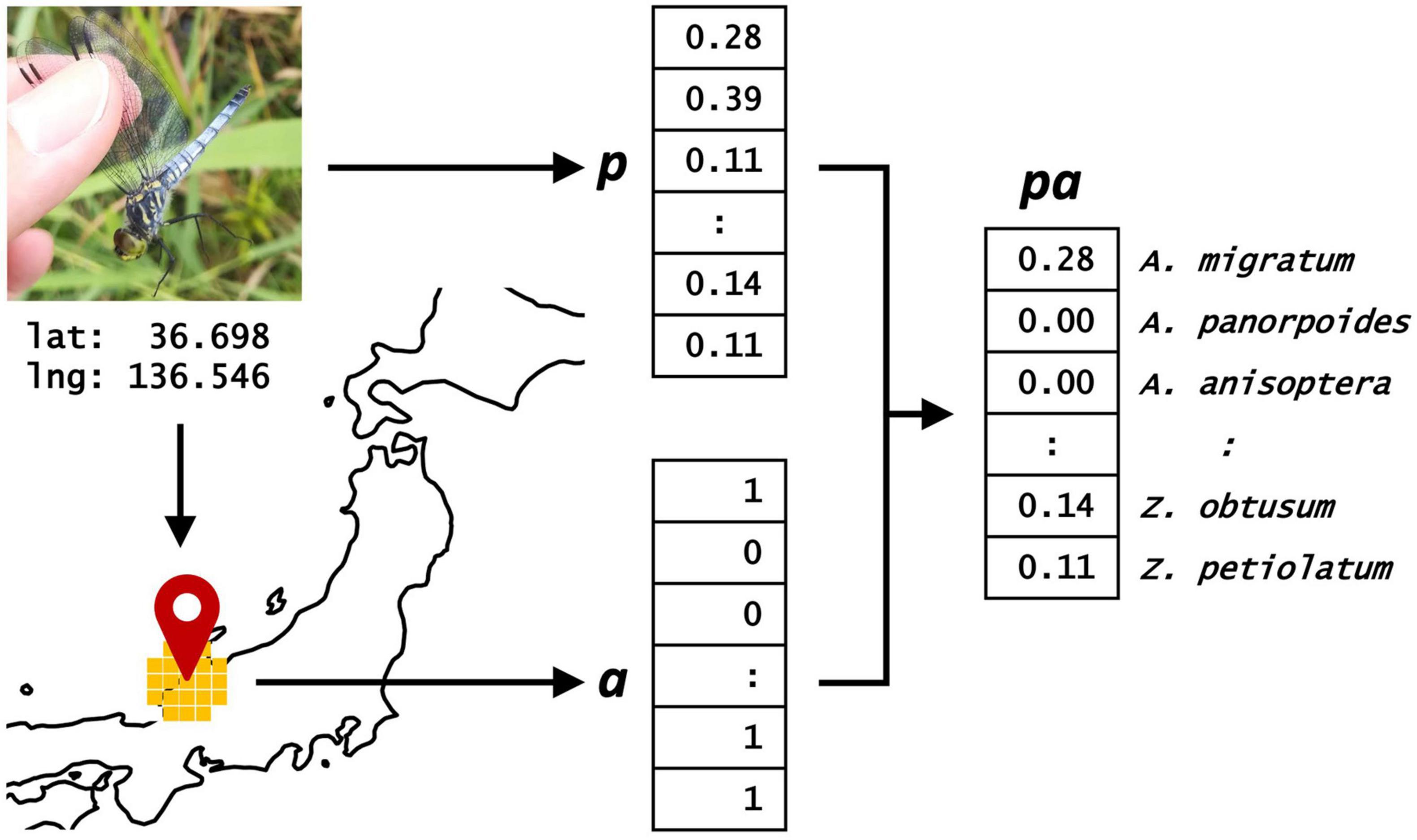
Figure 2. Combined model for species identification of odonates. Scores of 204 species (p) were calculated from the image-based model. Status of appearance of 204 species (a) were summarized from ecological survey data. Scores of the combined model were calculated by multiplying p and a.
Next, we obtained the geographic coordinates of each image using its EXIF. Based on the latitude and longitude, we determined the status of appearance of the 204 odonate species within a radius of 50 km from the location where the image was captured using the field occurrence records, as few migratory species (for example, Sympetrum frequens and Pantala flavescens) can fly more than 50 km during their lifetime (Sugimura et al., 1999). The status of appearance of the 204 species was then converted into a binary vector a, with 1 indicating presence, whereas 0 indicating the absence of the species within a radius of 50 km. However, the status of appearance was unavailable for the following 10 species: Anax junius, Agrionoptera sanguinolenta, Neurothemis fluctuans, N. ramburii, Rhyothemis phyllis, R. regia, Somatochlora exuberata, Sympetrum vulgatum, Tramea loewii, and T. basilaris. According to the field occurrence records, although these species were recently recorded, they were rarely observed in Japan. Therefore, we set their binary vectors at 0. In the combined model, the transformed scores for all odonates were calculated as pa, which were then used to evaluate the micro average of top-1 and top-3 accuracies for all images in the dataset T.
One point to note here is that the first step (i.e., species classification based on deep learning model) was trained with training image datasets but the latter step (i.e., get a from the occurrence records and calculate pa) was not trained. This is because the occurrence records were simply used as a filter against the outputs of the first step, thus it was unnecessary to be trained.
We evaluated the accuracy of genus identification by grouping the images of the datasets based on the genus rather than species. Then, we trained and validated the accuracy of the genus identification models using the same protocols that we used for species identification.
The experiments were performed on a Ubuntu 18.04 system equipped with an NVIDIA Tesla V100 SXM2 (Intel Xeon Gold 6254) and 128 GB memory. Python 3.6.8 (Python Software Foundation, 2018) coupled with PyTorch 1.5.1 (Paszke et al., 2019) was used to train and validate the CNN models. The source codes for model usages and image augmentation used in this study are downloadable at GitHub repository https://github.com/biunit/dragonfly under the MIT license.
The four CNN architectures (i.e., VGGNet16, VGGNet19, ResNet18, and ResNet152) were separately trained with the five types of datasets (i.e., W1, W2, F, W1F, and W2F). The execution time of the training process depended on the model architectures. The more layers in the architecture, the longer time was required (Supplementary Figure 1). The trained architectures, i.e., models, were then validated with dataset T, and the top-1 and top-3 accuracies were calculated.
The models trained using the datasets W1 and W2 exhibited lower accuracy, whereas that trained using the dataset F exhibited approximately two times higher accuracy than W1 and W2, although the number of original images was similar in the datasets W1 (4,005 images) and F (4,571 images). By contrast, the models trained using combined datasets (W1F and W2F) exhibited higher accuracy than those trained using independent datasets W1, W2, or F (Table 2). In summary, the training architectures using the integration of web-scraped images and scanned images showed higher accuracy than those trained individually.
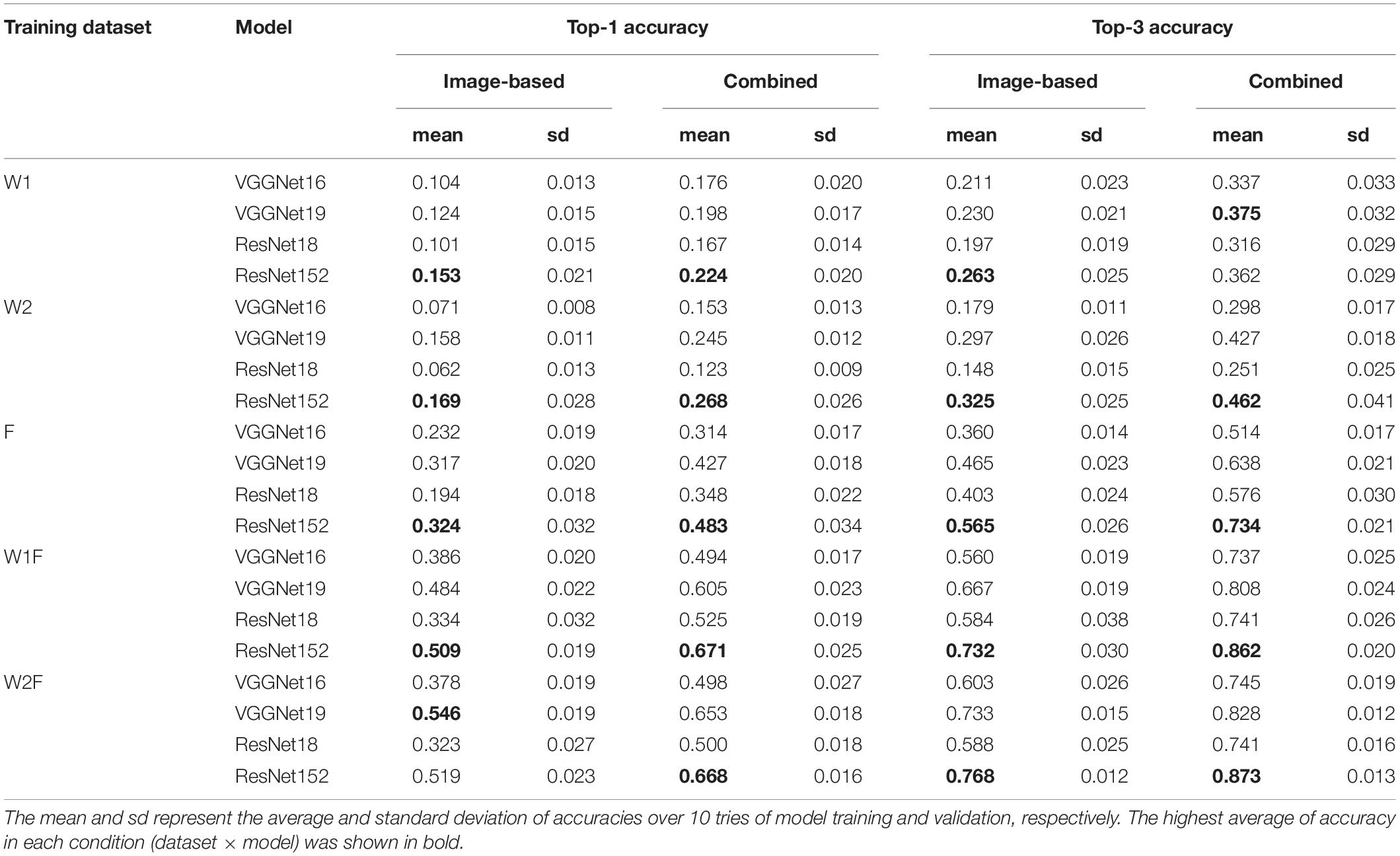
Table 2. The top-1 and top-3 accuracies of image-based and combined models for species identification.
VGGNet16 and VGGNet19, as well as ResNet18 and ResNet152, are technologically similar but vary in the number of CNN layers (Simonyan and Zisserman, 2014; Veit et al., 2016). The findings of this study indicated that although the training datasets were different, deeper architectures (i.e., VGGNet19 and ResNet152) exhibited higher accuracies than the shallow ones (i.e., VGGNet16 and ResNet18) (Table 2).
To further validate the prediction accuracy of the models, we computed a confusion matrix (Supplementary Figure 2A) and the identification accuracies of each species (Supplementary Figure 3) against the model ResNet152 trained using the dataset W2F, as this model exhibited the highest top-1 accuracy (Table 2). We found that species in the same genus tended to be confused, especially species that have highly similar morphological characteristics (Supplementary Figure 4). For example, images of Sympetrum frequens had tendency to be confused by the model with those of the same genus, S. striolatum, S. vulgatum, and S. depressiusculum. Similarly, Orthetrum albistylum was easily confused by the model with O. glaucum, O. poecilops and O. sabina.
The combination of image-based models with the field occurrence records resulted in improved accuracies for species identification under all conditions (Table 2). Compared to the highest top-1 (0.546) and top-3 accuracies (0.768) of the image-based models, the accuracies of the combined models were 0.668 and 0.873, respectively.
To further validate the prediction accuracy of the combined model, we computed a confusion matrix and identification accuracies of each species against the combined model ResNet152 trained using the dataset W2F (Supplementary Figures 2B, 3). We found that some images of S. frequens were correctly identified by the combining model although these images were incorrectly predicted as S. striolatum, S. vulgatum, and S. depressiusculum by the image-based model; also, some images of O. albistylum were correctly identified by the combining model although these images were incorrectly predicted as O. glaucum, O. poecilops, and O. sabina by the image-based model.
The trends of model accuracies for genus identification were similar to those of species identification (Table 3). Additionally, the top-1 and top-3 accuracies for genus identification were higher than those for species identification under all conditions. We found that the highest top-1 and top-3 accuracies for genus identification were 0.765 and 0.899, respectively, whereas 0.546 and 0.768 for species identification, respectively. The confusion matrix (Supplementary Figure 5) and identification accuracies of each genus (Supplementary Figure 6) of ResNet152 indicated that few classes (i.e., genera) were inaccurately identified. Moreover, similar to species identification, the combination of image-based model with field occurrence records for genus identification also improved the accuracy of the identification system (Table 3), with the highest top-1 and top-3 accuracies of 0.835 and 0.933, respectively, for the combined models.
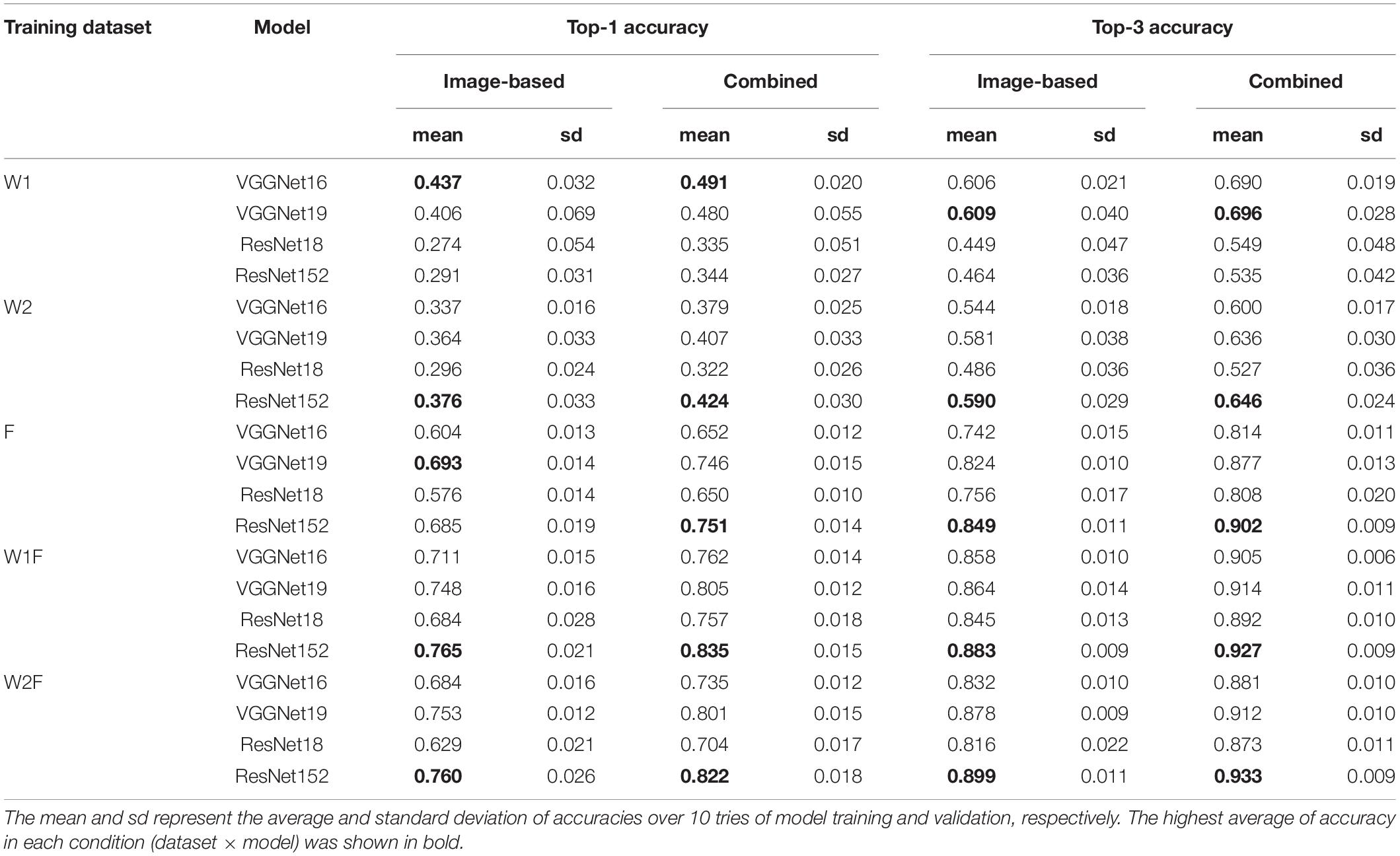
Table 3. The top-1 and top-3 accuracies of image-based and combined models for genus identification.
Image recognition using deep learning technologies exhibits great potential in many fields of biology and ecology (Kamilaris and Prenafeta-Boldú, 2018; Christin et al., 2019; Moen et al., 2019; Tabak et al., 2019; Hansen et al., 2020; Høye et al., 2021). However, a large number of images are required to calibrate a large number of parameters in a deep learning system (Perez and Wang, 2017; Shahinfar et al., 2020). Nonetheless, in field ecology, image collection is an issue because of resource limitations. Therefore, we proposed a strategy to develop a species identification model using approximately no image datasets.
To develop species identification models, we did not prepare any images by field work. Instead, we collected images by web scraping and scanning of books and specimens. In this case, it is essential to verify the species names are identical to the web-scraped images before machine learning. By using these images, we demonstrated that by using mixed image datasets (i.e., web-scraping images and scanning images) contributed to the accuracy for species identification (Table 2). We elucidated that this accuracy improvement is resulted in: (i) the increasing numbers of images; and (ii) compensating for features that are present in one (e.g., dataset F) and not in the other (e.g., dataset W1), that allowed the model to better capture the characteristics of odonates.
We also built identification models for the genus level. The number of odonate species was 204, while the number of genera was 86 (Supplementary Table 1). When images are grouped by genus, the number of images in each genus is increased; also, several species with the similar morphological characteristics were grouped into the same genus (Supplementary Figure 4). By grouping these similar odonates into a single class (i.e., genera), the deep learning models no longer need to distinguish trivial differences in the species level and can more readily capture the characteristics of the genus level. For example, similar species in the genus Sympetrum, e.g., S. frequens, S. striolatum, S. vulgatum, and S. depressiusculum, which are often confused by the species model, can be treated as one class in the genus model (Supplementary Figures 2, 5). Indeed, our result showed that the top-1 and top-3 accuracy of genus identification were 0.14–0.38 and 0.13–0.40 higher than that of species identification (Tables 2, 3), respectively, suggesting that mixing species with the similar morphological characteristics or targeting genus level have potential to improve the identification accuracy.
To compensate for the low availability of images, various strategies have been reported in image recognition studies (Berg et al., 2014; Aodha et al., 2019; Ellen et al., 2019; Terry et al., 2020). Several strategies, such as a linear combination of the image-based outputs and occurrence records with weights or a combination of the output values of the image-based model with the occurrence records, followed by using them as input for another neural network or support vector machine (SVM), are used for species identification (Terry et al., 2020). These approaches rely on a large number of images, along with their geographical coordinates. However, most images used in this study, which were web-scraped or scanned from books and specimens, did not include any geographic coordinate information, thereby the insufficiency of information prevented the execution of the training processes of a linear combination, neural networks, or SVM. Thus, we simply multiplied the image-based outputs and occurrence records to improve the accuracy of species and genus identification (Tables 2, 3). Despite this simple approach, the result suggests that occurrence records successfully distinguished morphologically similar but geographically isolated species.
Dataset T, which was used for accuracy validation in this study, was obtained from citizen surveys by amateur participants. Thus, the 62 species consisted in dataset T can be considered as common ondonate species in Japan, and reflects the actual frequency of odonate occurrence in nature. Therefore, it is natural to evaluate the accuracy of identification models using all images (i.e., micro average) rather than averaging the accuracy for identifying each species (i.e., macro average). We believe that this micro average reflects the accuracy of the identification system in the practical citizen science. However, it is important to note that the accuracy validated with dataset T only reflects the accuracy of these 62 species. The remained 142 species could not be validated due to lack of image data.
There were several studies of species classification of ondonate were performed recently (Ožana et al., 2019; Manoj et al., 2020). Ožana et al. (2019) developed a mathematical model for classification only based on occurrence records including date, location, surrounding environment type, suborder, and color. On the other hand, Manoj et al. (2020) developed a deep learning-based model based on images only. Compared to both previous studies, our study used both occurrence records and images. As we described above, using both information is expected to improve the accuracy of species classification rather than using one of them.
Citizen science has a great potential to determine the changes in biodiversity loss. However, citizen science has two major drawbacks: difficulty in ensuring data quality and the continuous recruitment of new participants (Dickinson et al., 2012). Veteran volunteers are required for accurate species identification, but it is very difficult to keep their willingness. Furthermore, recruiting new participants decreases the quality of species identification surveys. Thus, automated species identification systems provide an excellent solution for these problems in citizen science, that is, automation will validate their observations and contribute to the rationale of the participants. In addition, it would greatly enhance data quality in citizen science even when most participants would be amateurs.
The strategy proposed in this study is applicable to any organism because images obtained from open-access websites and books were utilized to develop the basic identification system with an appropriate accuracy. After deploying the basic identification system in surveys, it can collect large number of image data from across the nation using the identification system. By determining the species distribution using this system, researchers can assess the real-time status of biodiversity loss at low costs. Moreover, the image datasets can be used for further improving the system. Thus, nationwide backyard biodiversity surveys for all iconic species groups can be realized, which will address long-standing challenges in ecology and contribute to its advancement.
Image datasets used in this study are not publicly archived owing to image licensing and data protection constraints. However, most images of the dataset W1 can be found in the books of Japanese odonates (Hamada and Inoue, 1985; Sugimura et al., 1999; Ozono et al., 2021), and most images of the dataset T can be found in Osawa et al. (2017). Occurrence records of odonates are available from the National Biodiversity Centre of Japan, 2002.
JS and TY conceived the ideas and designed the methodology and led the writing of the manuscript. TY and RF collected and verified the datasets. JS developed the models and analyzed the data. All authors have read, revised, and approved the final manuscript.
This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 24651040 and 17K20068 to TY and 15K12154 to TY and RF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Computations were partially performed on the SHIHO supercomputer at the National Agriculture and Food Research Organization (NARO), Japan.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.762173/full#supplementary-material
Supplementary Figure 1 | The execution time of training for species identification model (A) and genus identification model (B).
Supplementary Figure 2 | Confusion matrix of ResNet152 trained with dataset W2F for species identification. (A) Confusion matrix of image-based model. (B) Confusion matrix of combined model.
Supplementary Figure 3 | Identification accuracy of each species of ResNet152 trained with dataset W2F. (A) Image-based model. (B) Combined model.
Supplementary Figure 4 | Specimen images of odonate species of Sympetrum (A) and Orthetrum (B) genera that were easily confused with each other by deep learning models.
Supplementary Figure 5 | Confusion matrix of ResNet152 trained with dataset W2F for genus identification. (A) Confusion matrix of image-based model. (B) Confusion matrix of combined model.
Supplementary Figure 6 | Identification accuracy of each genus of ResNet152 trained with dataset W2F. (A) Image-based model. (B) Combined model.
Supplementary Table 1 | Summary of image datasets saved with XLSX format. Sheet 1 contains the number of odonates images of dataset W1, F, and T, and the number of mesh codes that summarized according to species. Sheet 2 contains the number of odonates images of dataset W1, F, and T, and the number of mesh codes that summarized according to genus.
Supplementary Table 2 | Exchangeable image file format (EXIF) information of dataset T saved with XLSX format. Sheet 1 contains date, time, latitude, and longitude of location of photos taken. Sheet 2 contains a Japanese map which represents the location of photos taken.
Aodha, O. M., Cole, E., and Perona, P. (2019). Presence-only geographical priors for fine-grained image classification. arXiv [Preprint]. arXiv: 1906.05272.
Berg, T., Liu, J., Lee, S. W., Alexander, M. L., Jacobs, D. W., and Belhumeur, P. N. (2014). “Birdsnap: large-scale fine-grained visual categorization of birds,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH. doi: 10.1109/CVPR.2014.259
Christin, S., Hervet, É, and Lecomte, N. (2019). Applications for deep learning in ecology. Methods Ecol. Evol. 10, 1632–1644. doi: 10.1111/2041-210x.13256
Deng, J., Dong, W., Socher, R., Li, L. J., Li, K., and Li, F. (2009). “ImageNet: a large-scale hierarchical image database,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Miami, FL). doi: 10.1109/cvprw.2009.5206848
Dickinson, J. L., Shirk, J., Bonter, D., Bonney, R., Crain, R. L., Martin, J., et al. (2012). The current state of citizen science as a tool for ecological research and public engagement. Front. Ecol. Environ. 10:291–297. doi: 10.1890/110236
Ellen, J. S., Graff, C. A., and Ohman, M. D. (2019). Improving plankton image classification using context metadata. Limnol. Oceanogr. Methods 17, 439–461. doi: 10.1002/lom3.10324
Emmert-Streib, F., Yang, Z., Feng, H., Tripathi, S., and Dehmer, M. (2020). An introductory review of deep learning for prediction models with big data. Front. Artif. Intell. Appl. 3:4. doi: 10.3389/frai.2020.00004
Futahashi, R., Futahashi, H., Araki, K., and Negoro, H. (2004). The dragonflies and damselflies of Toyama prefecture, central Honshu, Japan. Bull. Toyama Sci. Museum 28, 97–107.
Futahashi, R., Yamanaka, T., Yoshinobu, U., and Hisamatsu, M. (2012). Collection and photographic data on dragonflies and damselflies from ibaraki prefecture. Nat. Museum 15, 13–38.
Goëau, H., Joly, A., Bonnet, P., Lasseck, M., Šulc, M., and Hang, S. T. (2018). Deep learning for plant identification: how the web can compete with human experts. Biodivers. Inform. Sci. Standards 2:e25637. doi: 10.3897/biss.2.25637
Hallmann, C. A., Ssymank, A., Sorg, M., de Kroon, H., and Jongejans, E. (2021). Insect biomass decline scaled to species diversity: general patterns derived from a hoverfly community. Proc. Natl. Acad. Sci. U.S.A. 118:e2002554117. doi: 10.1073/pnas.2002554117
Hansen, O. L. P., Svenning, J. C., Olsen, K., Dupont, S., Garner, B. H., Iosifidis, A., et al. (2020). Species-level image classification with convolutional neural network enables insect identification from habitus images. Ecol. Evol. 10, 737–747. doi: 10.1002/ece3.5921
Høye, T. T., Ärje, J., Bjerge, K., Hansen, O. L. P., Iosifidis, A., Leese, F., et al. (2021). Deep learning and computer vision will transform entomology. Proc. Natl. Acad. Sci. U.S.A. 118:e2002545117. doi: 10.1073/pnas.2002545117
Kadoya, T., and Washitani, I. (2007). An adaptive management scheme for wetland restoration incorporating participatory monitoring into scientific predictions using dragonflies as an indicator taxon. Glob. Environ. Res. 11, 179–185.
Kadoya, T., Suda, S. I., and Washitani, I. (2009). Dragonfly crisis in Japan: a likely consequence of recent agricultural habitat degradation. Biol. Conserv. 142, 1899–1905. doi: 10.1016/j.biocon.2009.02.033
Kadoya, T., Suda, S. I., Tsubaki, Y., and Washitani, I. (2008). The sensitivity of dragonflies to landscape structure differs between life-history groups. Landscape Ecol. 23, 149–158. doi: 10.1007/s10980-007-9151-1
Kamilaris, A., and Prenafeta-Boldú, F. X. (2018). Deep learning in agriculture: a survey. Comp. Electronics Agric. 147, 70–90. doi: 10.1016/j.compag.2018.02.016
Khan, A., Sohail, A., Zahoora, U., and Qureshi, A. S. (2020). A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 53, 5455–5516. doi: 10.1007/s10462-020-09825-6
Kobori, H., Dickinson, J. L., Washitani, I., Sakurai, R., Amano, T., Komatsu, N., et al. (2016). Citizen science: a new approach to advance ecology, education, and conservation. Ecol. Res. 31, 1–19. doi: 10.1007/s11284-015-1314-y
Langlotz, C. P., Allen, B., Erickson, B. J., Kalpathy-Cramer, J., Bigelow, K., Cook, T. S., et al. (2019). A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The academy workshop. Radiology 291, 781–791. doi: 10.1148/radiol.2019190613
Lin, T. (2015). LabelImg. GitHub. Available online at: https://github.com/tzutalin/labelImg (accessed September 10, 2020).
Manoj, B. J., Chinmaya, H. S., Ganesh, S. N., and Nithish, D. (2020). Dragonfly-net: dragonfly classification using convolution neural network. J. Appl. Inform. Sci. 10, 60–66.
Marzluff, J. M. (2013). Citizen science: public participation in environmental research. BioScience 63, 139–140. doi: 10.1525/bio.2013.63.2.10
Ministry of Internal Affairs and Communications (1996). Method of Demarcation for Grid Square. Available online at: https://www.stat.go.jp/english/data/mesh/05.html (accessed September 10, 2020).
Moen, E., Bannon, D., Kudo, T., Graf, W., Covert, M., and Van Valen, D. (2019). Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246. doi: 10.1038/s41592-019-0403-1
Naraoka, H. (2005). The life histories of dragonflies inhabit in irrigation pond, laied emphasis on the damselflies. Insects Nat. 40, 12–15.
National Biodiversity Center of Japan, Ministry of the Environment, Japan, Tokyo (2002). Data From: The National Survey on the Natural Environment Report of the Distributional Survey of Japanese Animals (Dragonflies). Available online at: http://www.biodic.go.jp/reports/4-09/h000.html (accessed January 21, 2021).
Newman, G., Wiggins, A., Crall, A., Graham, E., Newman, S., and Crowston, K. (2012). The future of citizen science: emerging technologies and shifting paradigms. Front. Ecol. Environ. 10:298–304. doi: 10.1890/110294
Osawa, T., Yamanaka, T., Nakatani, Y., Nishihiro, J., Takahashi, S., Mahoro, S., et al. (2017). A crowdsourcing approach to collecting photo-based insect and plant observation records. Biodivers. Data J. 5:e21271. doi: 10.3897/BDJ.5.e21271
Ožana, S., Burda, M., Hykel, M., Malina, M., Prášek, M., Bárta, D., et al. (2019). Dragonfly hunter CZ: mobile application for biological species recognition in citizen science. PLoS One 14:e0210370. doi: 10.1371/journal.pone.0210370
Ozono, A., Kawashima, I., and Futahashi, R. (2021). Dragonflies of Japan, Revised Edition. Tokyo: Bunichi Sogo Shuppan.
Paoletti, M. G. (1999). “Using bioindicators based on biodiversity to assess landscape sustainability,” in Invertebrate Biodiversity as Bioindicators of Sustainable Landscapes, ed. M. G. Paoletti (Amsterdam: Elsevier), 1–18. doi: 10.1016/B978-0-444-50019-9.50004-2
Parmar, T. K., Rawtani, D., and Agrawal, Y. K. (2016). Bioindicators: the natural indicator of environmental pollution. Front. Life Sci. 9:110–118. doi: 10.1080/21553769.2016.1162753
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., et al. (2019). PyTorch: an imperative style, high-performance deep learning library. arXiv [Preprint]. arXiv: 1912.01703.
Perez, L., and Wang, J. (2017). The effectiveness of data augmentation in image classification using deep learning. arXiv [Preprint]. arXiv: 1712.04621.
Pocock, M. J. O., Tweddle, J. C., Savage, J., Robinson, L. D., and Roy, H. E. (2017). The diversity and evolution of ecological and environmental citizen science. PLoS One 12:e0172579. doi: 10.1371/journal.pone.0172579
Python Software Foundation (2018). Python Language Reference (Version 3.6.8). Available online at: http://www.python.org (accessed January 6, 2021).
Shahinfar, S., Meek, P., and Falzon, G. (2020). How many images do I need?” Understanding how sample size per class affects deep learning model performance metrics for balanced designs in autonomous wildlife monitoring. Ecol. Inform. 57:101085. doi: 10.1016/j.ecoinf.2020.101085
Silvertown, J. (2009). A new dawn for citizen science. Trends Ecol. Evol. 24, 467–471. doi: 10.1016/j.tree.2009.03.017
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv [Preprint]. arXiv: 1409.1556.
Sugimura, M., Ishida, S., Kojima, K., Ishida, K., and Aoki, H. (1999). Dragonflies of the Japanese Archipelago in Color. Sapporo: Hokkaido University Press.
Tabak, M. A., Norouzzadeh, M. S., Wolfson, D. W., Sweeney, S. J., Vercauteren, K. C., Snow, N. P., et al. (2019). Machine learning to classify animal species in camera trap images: applications in ecology. Methods Ecol. Evol. 10, 585–590. doi: 10.1111/2041-210x.13120
Terry, J. C. D., Roy, H. E., and August, T. A. (2020). Thinking like a naturalist: enhancing computer vision of citizen science images by harnessing contextual data. Methods Ecol. Evol. 11, 303–315. doi: 10.1111/2041-210x.13335
Theobald, E. J., Ettinger, A. K., Burgess, H. K., DeBey, L. B., Schmidt, N. R., Froehlich, H. E., et al. (2015). Global change and local solutions: tapping the unrealized potential of citizen science for biodiversity research. Biol. Conserv. 181, 236–244. doi: 10.1016/j.biocon.2014.10.021
Valan, M., Makonyi, K., Maki, A., Vondráèek, D., and Ronquist, F. (2019). Automated taxonomic identification of insects with expert-level accuracy using effective feature transfer from convolutional networks. Syst. Biol. 68, 876–895. doi: 10.1093/sysbio/syz014
van Klink, R., Bowler, D. E., Gongalsky, K. B., Swengel, A. B., Gentile, A., and Chase, J. M. (2020). Meta-analysis reveals declines in terrestrial but increases in freshwater insect abundances. Science 368, 417–420. doi: 10.1126/science.aax9931
Veit, A., Wilber, M., and Belongie, S. (2016). Residual networks behave like ensembles of relatively shallow networks. arXiv [Preprint]. arXiv: 1605.06431.
Wäldchen, J., and Mäder, P. (2018). Machine learning for image based species identification. Methods Ecol. Evol. 9, 2216–2225. doi: 10.1111/2041-210x.13075
Keywords: citizen science, species identification, dragonfly, damselfly, deep learning, image recognition, web scraping
Citation: Sun J, Futahashi R and Yamanaka T (2021) Improving the Accuracy of Species Identification by Combining Deep Learning With Field Occurrence Records. Front. Ecol. Evol. 9:762173. doi: 10.3389/fevo.2021.762173
Received: 21 August 2021; Accepted: 23 November 2021;
Published: 20 December 2021.
Edited by:
Dmitry Efremenko, German Aerospace Center, Helmholtz Association of German Research Centers (HZ), GermanyReviewed by:
Lingxin Chen, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Sun, Futahashi and Yamanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianqiang Sun, ai5zdW5AYWZmcmMuZ28uanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.