
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 06 October 2021
Sec. Behavioral and Evolutionary Ecology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.736741
This article is part of the Research TopicLinking Primate Ecology and Conservation Science in a Changing EnvironmentView all 7 articles
 Elaine E. Guevara1,2*
Elaine E. Guevara1,2* Lydia K. Greene3,4,5
Lydia K. Greene3,4,5 Marina B. Blanco3,4
Marina B. Blanco3,4 Casey Farmer6
Casey Farmer6 Jeannin Ranaivonasy7
Jeannin Ranaivonasy7 Joelisoa Ratsirarson7
Joelisoa Ratsirarson7 Karine L. Mahefarisoa8,9
Karine L. Mahefarisoa8,9 Tsiky Rajaonarivelo8,10
Tsiky Rajaonarivelo8,10 Hajanirina H. Rakotondrainibe8
Hajanirina H. Rakotondrainibe8 Randall E. Junge8,11
Randall E. Junge8,11 Cathy V. Williams3,8
Cathy V. Williams3,8 Elodi Rambeloson12
Elodi Rambeloson12 Hoby A. Rasoanaivo12
Hoby A. Rasoanaivo12 Vololonirina Rahalinarivo13
Vololonirina Rahalinarivo13 Laza H. Andrianandrianina14
Laza H. Andrianandrianina14 Jonathan B. Clayton5,15,16,17
Jonathan B. Clayton5,15,16,17 Ryan S. Rothman18,19
Ryan S. Rothman18,19 Richard R. Lawler20
Richard R. Lawler20 Brenda J. Bradley2
Brenda J. Bradley2 Anne D. Yoder4
Anne D. Yoder4The lemurs of Madagascar include numerous species characterized by folivory across several families. Many extant lemuriform folivores exist in sympatry in Madagascar’s remaining forests. These species avoid feeding competition by adopting different dietary strategies within folivory, reflected in behavioral, morphological, and microbiota diversity across species. These conditions make lemurs an ideal study system for understanding adaptation to leaf-eating. Most folivorous lemurs are also highly endangered. The significance of folivory for conservation outlook is complex. Though generalist folivores may be relatively well equipped to survive habitat disturbance, specialist folivores occupying narrow dietary niches may be less resilient. Characterizing the genetic bases of adaptation to folivory across species and lineages can provide insights into their differential physiology and potential to resist habitat change. We recently reported accelerated genetic change in RNASE1, a gene encoding an enzyme (RNase 1) involved in molecular adaptation in mammalian folivores, including various monkeys and sifakas (genus Propithecus; family Indriidae). Here, we sought to assess whether other lemurs, including phylogenetically and ecologically diverse folivores, might show parallel adaptive change in RNASE1 that could underlie a capacity for efficient folivory. We characterized RNASE1 in 21 lemur species representing all five families and members of the three extant folivorous lineages: (1) bamboo lemurs (family Lemuridae), (2) sportive lemurs (family Lepilemuridae), and (3) indriids (family Indriidae). We found pervasive sequence change in RNASE1 across all indriids, a dN/dS value > 3 in this clade, and evidence for shared change in isoelectric point, indicating altered enzymatic function. Sportive and bamboo lemurs, in contrast, showed more modest sequence change. The greater change in indriids may reflect a shared strategy emphasizing complex gut morphology and microbiota to facilitate folivory. This case study illustrates how genetic analysis may reveal differences in functional traits that could influence species’ ecology and, in turn, their resilience to habitat change. Moreover, our results support the body of work demonstrating that not all primate folivores are built the same and reiterate the need to avoid generalizations about dietary guild in considering conservation outlook, particularly in lemurs where such diversity in folivory has probably led to extensive specialization via niche partitioning.
The primates of Madagascar include a remarkable number of folivores (Fleagle and Reed, 1996). The island is home to more than 50 extant species of folivorous lemur that are phylogenetically and ecologically diverse, as well as endangered (Fleagle and Reed, 1996; Mittermeier et al., 2010). Extant species include members of six genera in three families: Indriidae, Lepilemuridae, and Lemuridae (Figure 1A). Madagascar was also home to several larger bodied, extinct, “subfossil” lineages that were also likely characterized by folivory, including the “sloth lemurs” (family: Paleopropithecidae) and the koala lemurs (genus: Megaladapis) (Yoder, 1999; Fleagle, 2013; Kistler et al., 2015; Marciniak et al., 2021). It is difficult to estimate the number of times that folivory evolved independently in lemurs with confidence, especially given challenges to reconstructing the phylogenetic relationships among lemur families (Horvath et al., 2008; Perelman et al., 2011; McLain et al., 2012; Springer et al., 2012; Herrera and Dávalos, 2016), likely due to a series of ancient rapid divergences resulting in incomplete lineage sorting (Horvath et al., 2008; Marciniak et al., 2021). However, the ancestral lemurid is thought to be a generalist, with folivory in bamboo lemurs representing convergence with other lemur lineages (Ballhorn et al., 2016; Fulwood et al., 2021). Thus, folivory most likely evolved independently at several time-depths (dos Reis et al., 2018). Nutritional reliance on leaves, which contain plant structural carbohydrates and chemical defenses, is a challenging nutritional strategy (Feeny, 1969; Milton, 1999) and requires numerous anatomical, physiological, and behavioral adaptations (Hladik, 1978).
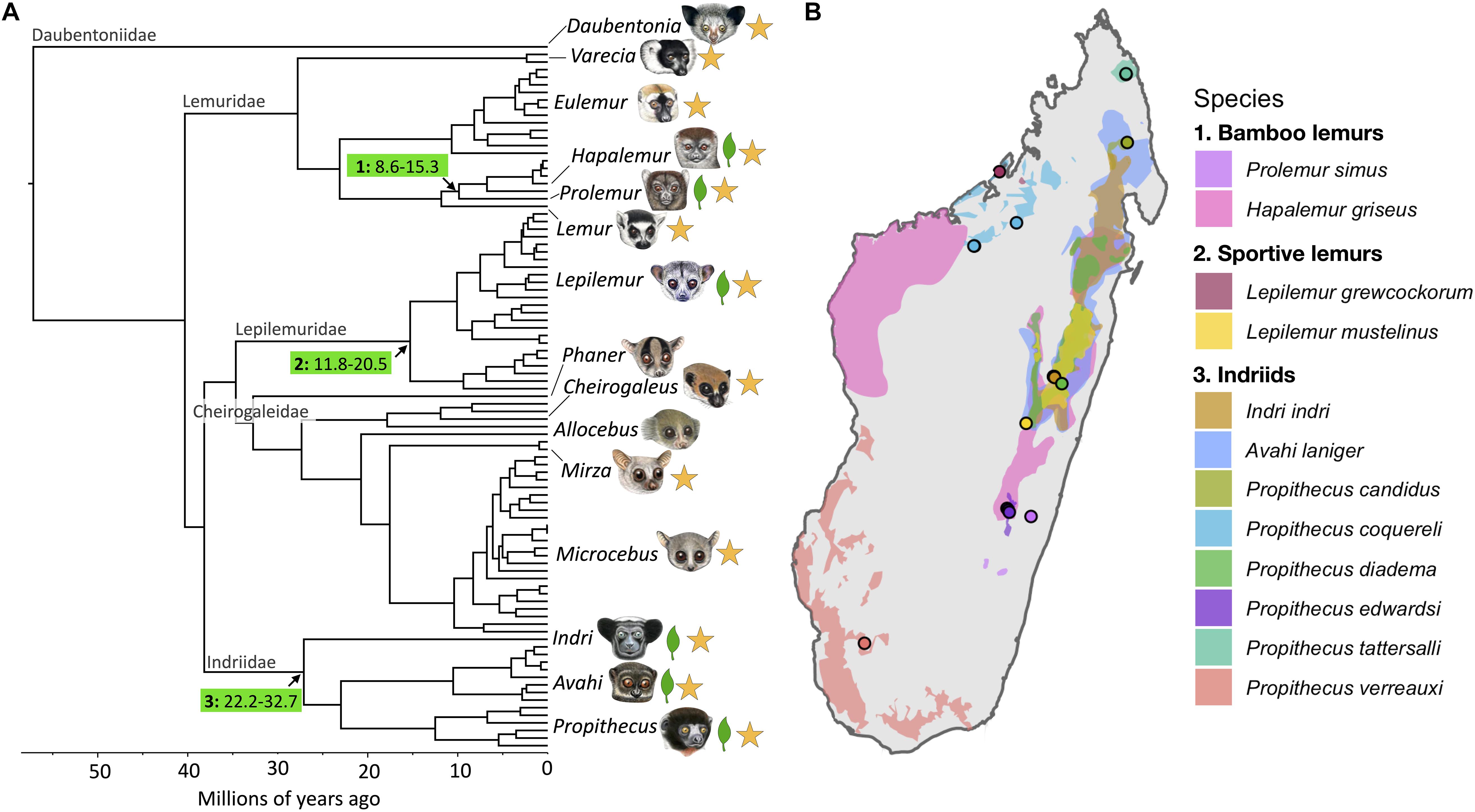
Figure 1. Species included in this study. (A) Lemur phylogeny adapted from dos Reis et al. (2018). Illustrations copyright 2020 Stephen D. Nash/IUCN SSC Primate Specialist Group. Used with permission. Arrows denote branches along which folivory emerged, with numbers in the green boxes reflecting the three folivorous lemur groups: 1: bamboo lemurs, 2: sportive lemurs and 3: indriids and number ranges indicating 95% confidence intervals for the time (in millions of years ago) of the basal split in that group from dos Reis et al. (2018). Leaf icons at tips denote folivorous genera and stars denote genera represented in this study. (B) Range maps of folivorous species included in this study from IUCN Red List of Threatened Species. Points indicate sampling localities for wild individuals sequenced.
Dietary strategy is of great importance for conservation outlook. Folivores are traditionally considered more resilient to habitat disturbance and change due to the relative abundance of leaves (Johns and Skorupa, 1987; Estrada and Coates-Estrada, 1996; Crockett, 1998). However, decades of research indicate that not all folivores are ecologically or physiologically equivalent (Glander, 1982; Chapman et al., 2012; Tombak et al., 2012). Phylogenetic constraints, for example on body size, the nature of available foliage in a given habitat, and niche partitioning influence which behavioral, anatomical, and physiological routes to folivory different taxa adopt (Ganzhorn, 1988; Cork, 1996).
For example, the many folivorous lemurs that live in sympatry (Figure 1B) across Madagascar’s remnant forested ecosystems avoid feeding competition through differentiation in activity patterns, microhabitat use, and selection of leaves with differing amounts of protein, structural carbohydrates, and plant secondary compounds, including tannins and alkaloids (Ganzhorn, 1988, 1989; Warren and Crompton, 1997; Powzyk and Mowry, 2003). Within the last several hundred years, many of these habitats were shared with even more folivores, which have since gone extinct (Fleagle, 2013). Niche partitioning likely involves specialization via varying underlying physiological adaptations to different aspects of leaf digestion, which may in turn influence species’ ability to cope with environmental limitations and change (Tombak et al., 2012; Jensen et al., 2014).
Genetic analysis offers an opportunity to get “under the hood” of different primate folivores to better understand their specific physiological adaptations that facilitate folivory. A prime example is the RNASE1 gene, which encodes the pancreatic ribonuclease enzyme (RNase1 or RNase A). RNASE1 is part of a ribonuclease gene superfamily that has expanded via gene duplication over mammalian evolution. Members of this family play diverse roles, including in pathogen defense, angiogenesis, and reproduction, in various mammalian lineages (Cho et al., 2005). In several folivorous mammalian lineages, RNase1 shows evidence of adaptive convergence at various evolutionary time depths. It has undergone independent gene duplications followed by rapid sequence evolution in bovine ruminants and colobine monkeys (Zhang et al., 2002; Schienman et al., 2006; Zhang, 2006), who share a common ancestor 85–105 million years ago (Ma) (Springer et al., 2012; dos Reis et al., 2018). Moreover, there have been independent duplications of RNASE1 followed by parallel amino acid substitutions in both the African and Asian colobine monkey clades, who share an ancestor ∼15 Ma (Schienman et al., 2006; Zhang, 2006; dos Reis et al., 2018). Functionally parallel amino acid substitutions in the duplicated RNase1 peptide in both ruminants and colobines result in a lowered optimal pH for enzymatic activity relative to the conserved peptide (Schienman et al., 2006; Zhang, 2006). Although RNASE1 is expressed across several tissues, it is much more highly expressed in the pancreas of ruminants than that of non-herbivorous mammals (Barnard, 1969; Beintema et al., 1988). The duplicated RNase1 is thought to play a key role in ruminant digestion by breaking down RNA produced by the large microbial communities that facilitate cellulose digestion to recover nitrogen for host absorption (Barnard, 1969; Zhang, 2006). The enzyme’s lowered optimal pH presumably boosts efficiency in the small intestine, which in ruminants has a lowered pH (Schienman et al., 2006; Zhang, 2006). Recent work in platyrrhine primates has uncovered evidence for an RNASE1 duplication in howler monkeys (Alouatta spp.) followed by amino acid substitutions and a lowered isoelectric point paralleling those in colobines (Janiak et al., 2019). Isoelectric point (pI) is the pH at which the peptide has no charge, and changes in pI reflect substitutions that alter charge and thus how the enzyme interacts with negatively charged RNA molecules. Howler monkeys are also highly folivorous, but do not have a ruminant digestive system, relying instead on hindgut fermentation (Milton and McBee, 1983). The finding of parallel evolution in howler monkeys and colobines thus raises the possibility of an alternative or additional role for RNase1 in hindgut-based foliage digestion.
Our previous work has uncovered positively selected sites in RNASE1 in sifaka lemurs (genus Propithecus) (Guevara et al., 2021). Sifakas, along with the indri (genus Indri) and woolly lemurs (genus Avahi), belong to the family Indriidae, all of whose members eat diets high in leafy material and show traits associated with obtaining nutrients from leaves. Such traits include shearing teeth and an enlarged caecum and complex colon to facilitate hindgut fermentation using commensurate specialized microbiota (Campbell et al., 2000; Yamashita, 2008; Fleagle, 2013; Greene et al., 2020).
In this study, we expanded our analysis to include additional folivorous lemurs to determine if they also show convergent changes in the RNASE1 gene. Specifically, we analyzed RNASE1 sequence data for 21 species of lemur, representing all five lemur families (Table 1), many of which are sympatric in eastern Madagascar (Figure 1B).
We include two representatives of the bamboo lemur clade, the greater bamboo lemur (Prolemur simus) and the eastern lesser bamboo lemur (Hapalemur griseus; Figure 1 and Table 1). Bamboo lemurs are members of the Lemuridae family, which shares a common ancestor with indriids at least 30 Ma (Figure 1A; Springer et al., 2012; dos Reis et al., 2018). Whereas most lemurids are largely omnivorous or frugivorous, bamboo lemurs are named for their dietary focus on bamboo (Fleagle, 2013). Bamboo lemurs have a relatively simple gastrointestinal tract compared with indriids and lack an enlarged cecum (Campbell et al., 2000). Eastern lesser bamboo lemurs nevertheless exhibit a very long gut passage time and fiber digestibility capacity greater than sifakas (Campbell et al., 2004), though greater bamboo lemurs may have a faster gut passage rate (Tan, 1999). Bamboo lemurs exhibit dental traits, including premolarized canines and molarized premolars, some of which are seen in other grass-eating mammals (Milton, 1978; Jernvall et al., 2008). These features likely allow them to efficiently process bamboo, which is highly fibrous and, like other grasses, gains structural integrity from silica bodies (Milton, 1978; Jernvall et al., 2008). Bamboo lemurs are also cathemeral, which has been proposed to be an adaptation to subsistence on a low quality diet, as it allows for round-the-clock intake (Eppley et al., 2011, 2015b).
The proportion of the diet comprised of bamboo varies by species, season, and geographic locale (Tan, 1999; Ballhorn et al., 2016). Members of the genus Hapalemur appear to exhibit a degree of dietary flexibility. For example, southern bamboo lemurs (H. meridionalis) and eastern lesser bamboo lemurs appear to successfully exploit several different habitat types and consume invasive species, including the broad-leaved paperbark tree (Melaleuca quinquenervia) in the case of southern bamboo lemurs (Eppley et al., 2015a) and guava in the case of eastern lesser bamboo lemurs (Grassi, 2006). Hapalemur spp. also seem to demonstrate considerable behavioral flexibility. For example, southern bamboo lemurs increase terrestrial foraging when preferred foods are not available (Eppley et al., 2016) and the Sambirano lesser bamboo lemur (H. occidentalis) was observed to exploit human-altered, agricultural landscapes (Martinez, 2008). Bamboo can be a pioneer species following anthropogenic disturbance and eastern lesser bamboo lemurs have been found to occupy such edge habitats (Arrigo-Nelson and Wright, 2004). It has been proposed that members of the genus Hapalemur are better considered “ubiquity specialists” or “facultative specialists” rather than bamboo specialists (Grassi, 2006; Ballhorn et al., 2016).
In contrast, the greater bamboo lemur is considered an “obligate specialist” (Ballhorn et al., 2016). Madagascar giant bamboo (Cathariostachys madagascariensis) comprises > 95% of its diet (Tan, 1999). Madagascar giant bamboo is the most cyanogenic species of bamboo in Madagascar and, given its specialization on this species, greater bamboo lemurs ingest high levels of cyanide, likely necessitating physiological specialization, though the precise mechanisms of detoxification remain unknown (Ballhorn et al., 2016). Greater bamboo lemurs focus on Madagascar giant bamboo shoots when available during the wet season and, during the dry season, are able to exploit the culm. Greater bamboo lemurs today have a very restricted and patchy geographic distribution, but the species used to be much more widespread. The lengthening of dry seasons resulting from climate change past the greater bamboo lemurs’ capacity to subsist on bamboo culm has been proposed to explain the species’ decline (Eronen et al., 2017).
We also include two sportive lemurs, the weasel sportive lemur of the eastern rainforests (Lepilemur mustelinus) and Anjiamangirana sportive lemur (L. grewcockorum) of the dry, deciduous forests of the northwest. Folivory evolved at least ∼12 Ma (Figure 1) in Lepilemuridae, a family of a single extant, specious genus (Lepilemur) (Mittermeier et al., 2010). The 26 currently-recognized sportive lemur species (Lepilemur spp.) occupy nearly all forest types in Madagascar (IUCN, 2020). They are of medium body size (600 g – 1 kg) (Fleagle, 2013) and are enigmatic folivores, as they push the lower boundary of expected body size for a mammal that subsists on leaves (Kay, 1984). Moreover, they have an energetically expensive, leaping locomotor style and are nocturnal (Warren and Crompton, 1997). Nocturnality is rare in folivores presumably due to lower sugar content in non-photosynthesizing leaves (Warren and Crompton, 1997) and possible increases in energetic demands of thermoregulation, especially during the cool season in Madagascar (Dröscher, 2015). Sportive lemurs further show little selectivity in leaf choice (Warren and Crompton, 1997; Dröscher, 2015) and appear to often eat leaves that are high in alkaloids, which may function as a form of niche partitioning with similarly-sized, nocturnal, folivorous woolly lemurs (Avahi spp.) (Ganzhorn, 1988, 1993). Sportive lemurs have relatively short small intestines relative to other folivorous lemurs, but greatly enlarged caeca, which may allow for the selective separation and retention of digestive material and represent convergence with arboreal marsupial consumers of mature leaves (Cork, 1996). Their relatively simple GI tract and fairly monotonous diet of mature leaves appears to be supported by a structurally simple gut microbiota largely comprised of taxa capable of cellulolysis (Greene et al., 2020). Sportive lemurs may partly meet their energetic needs from a low-quality diet by maintaining a markedly low resting metabolic rate, even among lemurs (Schmid and Ganzhorn, 1996). They further have a high dry matter intake for their body size (Dröscher, 2015), suggesting that they take a high-throughput strategy to nutrient acquisition, though, to our knowledge, gut passage time remains to be studied. Sportive lemurs also show potential behavioral adaptations to low nutrient intake, including very low activity levels (Hladik and Charles-Dominique, 1974), small home ranges sizes (Nash, 1998; Dröscher, 2015), short travel distances (Ganzhorn, 1993; Warren and Crompton, 1997), and infant parking (Dröscher, 2015). Loss of contiguous habitat may thus seriously compromise their survival (Ganzhorn, 1993). They also may practice caecotrophy, the reingestion of feces, as exhibited by other smaller-bodied folivores like rabbits (Hladik and Charles-Dominique, 1974; Andrews, 2019). Caecotrophy may lead to a ∼50% increase in digestive efficiency and could have co-evolved with the reduction in body size from a larger-bodied, folivorous ancestor (Andrews, 2019).
Finally, we also added representatives of the two other indriid genera, the indri (Indri indri) and the eastern woolly lemur (Avahi laniger), as well as two additional sifaka species, silky sifakas (P. candidus), and Milne-Edwards’ sifakas (P. edwardsi), thus covering the entire phylogenetic span of the indriid clade. Sifakas incorporate more fruit in their diet than do indris, which are considered flexible young leaf specialists, or woolly lemurs, which are dedicated young leaf specialists (Hladik, 1978; Ganzhorn et al., 1985; Britt et al., 2002; Powzyk and Mowry, 2003; Faulkner and Lehman, 2006; Irwin, 2006). Woolly lemurs, like sportive lemurs, are also small-bodied for folivores and nocturnal, but appear to have adopted a different folivory strategy, relying on selection of high quality leaves and exhibiting larger home ranges (Ganzhorn et al., 1985; Ganzhorn, 1988, 1993; Warren and Crompton, 1997). Given that enlarged caeca are found among all indriids, folivory with hindgut fermentation is likely the primitive condition (Fleagle, 2013). Indriids share a common ancestor ∼22–32 Ma, a time depth similar to that of the common ancestor of colobines (Springer et al., 2012; dos Reis et al., 2018). We thus sought to determine the pattern of amino acid substitutions over indriid evolution, including the degree to which substitutions observed in sifakas are primitive retentions from an indriid ancestor or whether species within Indriidae show parallel substitutions, as has been observed in Asian and African colobines (Zhang, 2006).
Our subjects were 21 species of lemur, representing five lemur families (Table 1). For several species, data came from previously released genome assemblies (Meyer et al., 2015; Larsen et al., 2017; Hawkins et al., 2018; Damas et al., 2020). We obtained the alignment of RNASE1 from the UCSC 30-way multi-Z alignment data and BLASTed the sequence of the closest relative against available primate genomes (e.g., the Eulemur macaco sequence for Prolemur simus and Lemur catta; the Microcebus murinus sequence for Cheirogaleus medius). In all cases, BLAST yielded a single best hit, which we retained. We generated new data for Avahi laniger, Indri indri, Propithecus candidus, Propithecus edwardsi, Lepilemur grewcockorum, Lepilemur mustelinus, Eulemur fulvus, Eulemur rufifrons, and Hapalemur griseus (Table 1). All new data were generated from wild individuals, with the exception of the H. griseus individual, who was born at the Duke Lemur Center (DLC). When possible (Table 1), multiple individuals per species were analyzed to assess whether substitutions were fixed or polymorphic in the species. P. candidus, P. edwardsi, E. fulvus, and E. rufifrons samples were collected and extracted as described in Greene et al. (2019) and A. laniger and I. indri samples as in Greene et al. (2020). H. griseus liver tissue was obtained from the DLC tissue biobank, whereas Lepilemur grewcockorum and Lepilemur mustelinus samples were field collected. DNA was extracted from samples using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). DNA concentrations were quantified on Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, United States). We designed primers—5′-GTCCTGTTCCCACTGCTAGTT-3′ (RN1-2-F, forward) and 5′-CGAAATGATGAGGTGGGGGTG-3′ (RN1-2-R, reverse)—to target a 489-bp portion of DNA including the coding portion of the RNASE1 gene. The 25 μL PCR reaction included 12.5 μL Qiagen HotStartTaq Master Mix, 2.0 μL Ambion Ultrapure non-acetylated Bovine Serum Albumin (20 mg/mL), 1.0 μL each of 10 μM forward and reverse primers and 4.0 μL of template DNA. Following an activation step at 95°C for 15 min, PCR cycling conditions (35 cycles) were: 94°C for 60 s, 60°C for 60 s, 72°C for 60 s. The final extension was at 72°C for 10 min. PCR products were visualized via agarose gel electrophoresis and then enzymatically purified and sequenced at the Duke DNA Analysis Facility on an Applied Biosystems 3730 Genetic Analyzer. Chromatograms were visually inspected using FinchTV v 1.5.0 (Geospiza).
We assessed the occurrence of a duplication of RNASE1 in bamboo lemurs and indriids by BLASTing (Altschul et al., 1990) the Microcebus murinus and Eulemur macaco RNASE1 sequences against the Prolemur simus (Prosim_1.0;1) and Propithecus coquereli (Pcoq_1.0;2) genome assemblies under both “highly similar” (megablast) and “more dissimilar” (discontiguous megablast) settings. We also designed primers based on conserved regions for amplification across species included in this analysis; as such we expected that if, in the case of a gene duplication followed by divergence, primers that successfully amplified diverged RNASE1 sequences in folivorous species would also amplify the conserved sequence, which could be evident as apparent heterozygosity in the Sanger sequencing results.
Ancestral sequences were reconstructed and amino acid substitutions mapped to a species tree using the codeml program in the PAML package (Yang, 2007) and visualized using the ggtree R package (Yu et al., 2017). We estimated a maximum likelihood gene tree under a general time reversible model of nucleotide substitution using RAxML v8.2.12 with the rapid bootstrap algorithm (n = 100). However, the resulting gene tree was not consistent with generally accepted genus-level relationships and seemed unlikely to reflect incomplete lineage sorting or introgression. This lack of resolution may be due to the short length of the alignment. There is not additional sequence data for this locus for some of the species included in our analyses; we were thus unable to estimate a gene tree using, for example, upstream or/and downstream DNA data. As such, we used a species tree (Figure 2A) consistent with well-supported taxonomic relationships and robustly estimated from a large genomic dataset (dos Reis et al., 2018) for analyses. We placed Propithecus verreauxi, whose taxonomic placement is uncertain, as sister to P. coquereli in this analysis, as their RNASE1 sequences are identical. The only species not included in this species tree was Propithecus candidus. There is little genomic data available for this taxon: we added this taxon to the tree (Figure 2A) in a way consistent with best available evidence (Mayor et al., 2004). Moreover, the Propithecus candidus sequences was identical to that of Propithecus diadema, which supports this placement in this analysis. We tested for positive selection on the bamboo lemur, sportive lemur, and indriid RNASE1 sequences using branch and branch-sites models in codeml (Yang, 2007). Many of the branches of interest, including the ancestral bamboo lemur branch, as well as the Propithecus tattersalli branch and the ancestral branch leading to A. laniger and the Propithecus spp., did not contain any synonymous substitutions, leading to dS = 0, but did contain 1–3 nonsynonymous substitutions, resulting in a nonzero dN. These conditions make dN/dS estimates unreliable, but likelihood ratio tests to compare models are nevertheless valid. We fit two positive selection branch models (Yang, 2002): H1PS (“ancestor model”) fit an elevated dN/dS on the ancestral bamboo lemur, sportive lemur, and indriid branches (Figure 2A) and H2PS (“clade model”) fit an elevated dN/dS on the all bamboo lemur, sportive lemur, and indriid branches. We compared these models by likelihood ratio test to a null model (M0) fitting a single dN/dS to the entire tree and neutral models (H1N and H2N) in which dN/dS was fixed to 1 on the foreground branches. We next used branch-sites models to test whether there might be a class of sites on the bamboo lemur, sportive lemur, and indriid branches (Figure 2A) that exhibit a dN/dS> 1 (Zhang, 2005). We compared this positive selection model (BSH1PS) to a neutral model (BSH1N) in which dN/dS for this additional class of sites on these branches is fixed to 1.
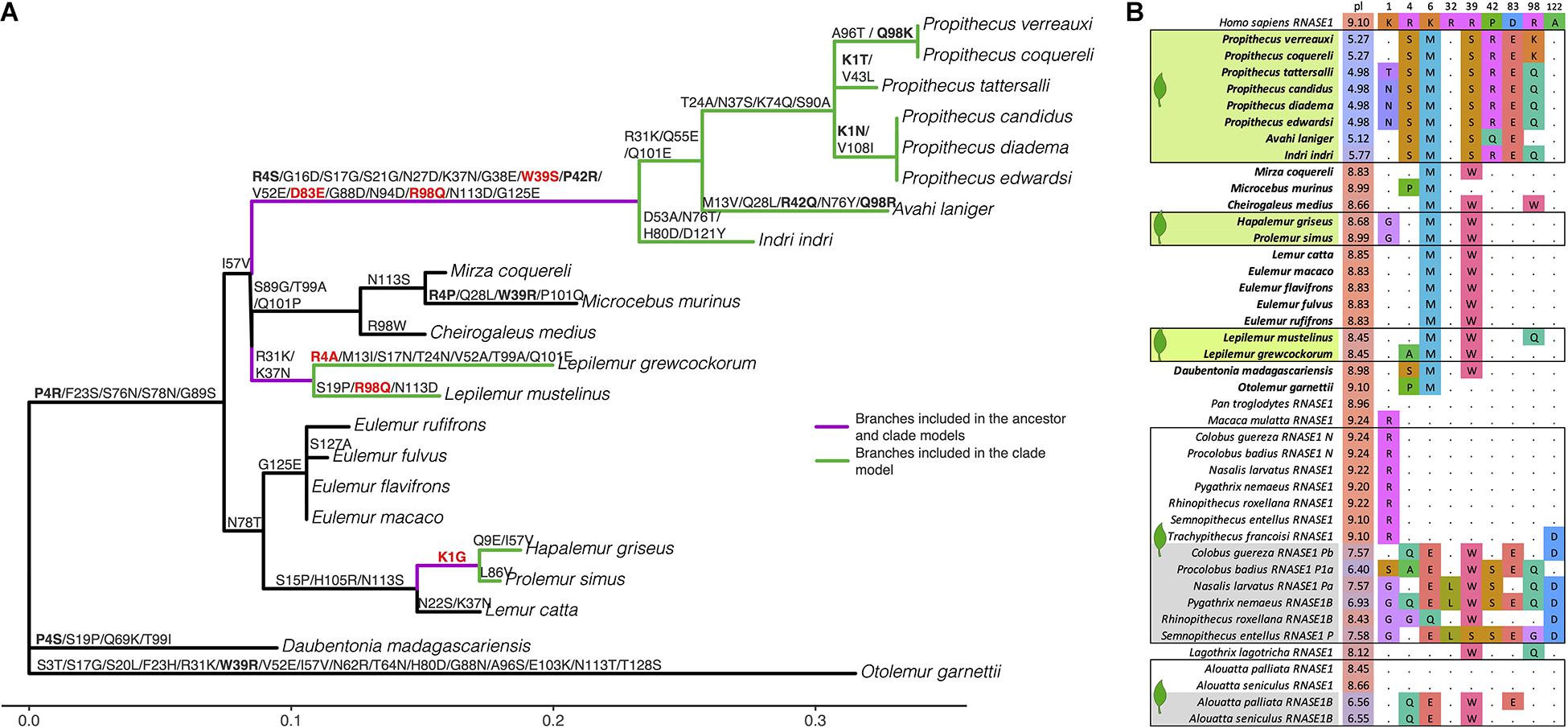
Figure 2. RNase1 substitutions. (A) unrooted phylogenetic tree used for analyses with branches scaled to the number of substitutions per codon. Nonsynonymous substitutions mapped to tree from ancestral sequences reconstructed using codeml. Bolded substitutions occur at sites predicted to impact enzymatic activity that also show substitutions in duplicated RNase1 sequences of colobine monkeys. Substitutions in red involve changes to the same amino acid as in duplicated colobine sequences. The purple branches were the foreground branches in the ancestor positive selection branch model (H1) and branches colored both purple and green were foreground branches in the clade positive selection branch model (H2) and positive selection branch-sites model. (B) Alignment of primate RNase1 sequences at sites predicted/known to influence enzymatic activity. Bolded taxon names are taxa included in the present study, taxon names highlighted in green are folivorous lemurs included in the present study, and taxon names highlighted in gray are the duplicated sequences of colobines and howler monkeys. All folivorous species are outlined and denoted with a leaf icon. pI = isoelectric point at pH = 7. Primate sequences in the alignment not in the present study are from the UCSC 30-way multi-Z alignment and (Zhang et al., 2002; Zhou et al., 2014; Janiak et al., 2019).
To assess the potential consequences of amino acid substitutions on enzymatic activity, we calculated the isoelectric point (pI) of RNase1 for each species using the ExPASy compute pI/Mw tool3.
We uncovered no evidence of a duplication of the RNASE1 gene in bamboo lemurs, sportive lemurs, or indriids. We did not observe any polymorphisms within species. We did, however, observe many amino acid substitutions along the indriid branches, including at sites that influence enzymatic activity in colobine monkeys (Figure 2). In particular, these included substitutions on the ancestral indriid branch at amino acid positions 39, 83, and 98 (Zhang et al., 2002; Zhou et al., 2014). The substitution occurring at residue 39 involves a change from a basic to non-basic amino acid, as in colobines and howler monkeys (R > S in indriids; R > W in colobines and howler monkeys; Figure 2; Janiak et al., 2019; Zhang et al., 2002), and is among the substitutions found to be most critical to altering enzymatic activity (Zhou et al., 2014). It is important, however, that many non-folivore lemurs are actually more similar to most colobines and howler monkeys at this position, exhibiting a R > W substitution. The indriid substitution at residue 83 involves the same exact change (D > E) as in colobines and howler monkeys (Zhang et al., 2002; Janiak et al., 2019) and the substitution at residue 98 involved the exact same change (R > Q) as observed in some colobines (Zhang et al., 2002). We moreover observed four losses of arginines across the indriid branches; the loss of these positively-charged amino acids is thought to be especially important for RNase1 function (Schienman et al., 2006).
We observed fewer amino acid substitutions on the bamboo lemur branches and no losses of arginines (Figure 2A). Further, bamboo lemurs’ RNase1 molecules do not demonstrate a lowered isoelectric point. Nevertheless, one substitution on the ancestral bamboo lemur branch occurring at residue 1 involves the same amino acid change (K > G) as observed in many duplicated colobine RNase1 sequences and that experimentally alters RNase1 enzymatic activity (Zhang et al., 2002; Figure 2).
Sportive lemurs display an intermediate scenario. They exhibit three losses of arginines, including at two sites that parallel substitutions that affect enzymatic activity in colobines (Figure 2A). One of these substitutions occurs in each of two included sportive lemur species (R4A in the Anjiamangirana sportive lemur and R98Q, also seen in indriids, in the weasel sportive lemur). An additional substitution is shared between the ancestral sportive lemur and ancestral indriid at residue 37 (K > N). The isoelectric points of sportive lemurs are slightly lower than other, non-indriid lemurs, but nevertheless much more similar to other lemurs than indriids.
The ancestor branch model (H1PS) in the codeml analysis revealed that a model including a significantly elevated dN/dS (dN/dS = 2.24) on the branches of the common ancestor of all bamboo lemurs, sportive lemurs, and indriids fit the data significantly better than did the null model (p = 0.0012; Table 2). However, this model was not a better fit to the data than the neutral model in which these branches have a dN/dS = 1 (p = 0.24). The clade branch model (H2PS; Methods) including an elevated dN/dS on all branches in the bamboo lemur, sportive lemur, and indriid clades was also better than the null model (p = 0.0005; Table 2); however, dN/dS on these branches was not greater than 1 (dN/dS = 0.69), indicating that most accelerated protein evolution occurred on the ancestral branches rather than in parallel within the folivore clades.
The positive selection branch-sites model identified one site (at position 1) showing a dN/dS> 1 (Table 2). Substitutions at this site are exhibited in the bamboo lemur ancestor, the ancestor to Propithecus candidus, P. diadema and P. edwardsi, and in P. tattersalli (Figure 2). However, the positive selection branch-sites model was not a better fit to the data than the neutral branch-sites model (p = 0.06).
This study reveals a probable difference in the physiological basis of folivory in the three folivorous lemur lineages, which may be relevant to better understanding the ability of these taxa to cope with habitat change. Our results indicate convergence of indriid RNase1 with duplicated RNase1s in folivorous monkeys. Specifically, as in colobines and howlers, we observe several losses of arginines, substitutions at sites predicted to influence enzymatic activity (D83E, R98Q/K) similar to those in some colobines, and a decreased peptide pI in indriid RNase1s (Figure 2). The elevated dN/dS (>3) on the ancestral indriid branch together with changes in amino acids and peptide pI paralleling those in colobines and howler monkeys indicate this is a likely example of molecular adaptation in indriids. Indeed, the probability of three charge-altering amino acid substitutions occurring at the same residues in this peptide in different lineages was calculated to be extremely low (<0.0026) (Zhang, 2006), thus emphasizing the probability of convergent adaptive function. Convergent change in RNASE1 in indriids primarily occurred in the indriid ancestor rather than in parallel along the various indriid branches.
Evidence for adaptive convergence in the RNASE1 gene is more limited in sportive lemurs and bamboo lemurs. Bamboo lemurs exhibit one substitution paralleling colobines and some indriids. Intriguingly, each sportive lemur lineage shows one substitution involving a loss of arginine and paralleling colobines and indriids, and the ancestral sportive lemur demonstrates additional substitutions involving losses of arginines or paralleling indriids. The results of the branch-sites tests indicate that a model including positive selection at some sites is significantly better than a null model, though not significantly better than a neutral model in which dN/dS = 1. A significantly better fit than a dN/dS of 1 at some sites, is a fairly conservative threshold; these results thus offer some provisional evidence for selection across all lineages.
However, neither bamboo lemurs nor sportive lemurs show a substantially lowered pI. The evolutionary shift to a foliage-based diet may be somewhat more recent in sportive lemurs and is certainly more recent in bamboo lemurs than in indriids (Figure 1A). Natural selection may thus have had more time to act on RNASE1 in indriids. RNASE1 evolution has been explored in other mammalian bamboo specialists, specifically giant and red pandas. Notably, red pandas (Ailurus fulgens) exhibit a duplication of this gene with one copy characterized by numerous amino acid substitutions and a slightly lowered isoelectric point. Giant pandas (Ailuropoda melanoleuca), however, show no evidence of gene duplication or change in isoelectric point (Liu et al., 2014).
Alternatively, the lack of convergence among lemur lineages may reflect differing pathways to folivory. It is also noteworthy that many indriids are arguably more ecologically similar to howler monkeys and colobines than the more specialized sportive and bamboo lemurs are. Like these folivorous monkeys, indris and sifakas are fairly large-bodied and, in sifakas, in particular, flexibly incorporate some fruit in the diet (Irwin, 2006). Molecular adaptation of RNASE1 may thus be part of a folivory strategy involving large body size, at least in the case of indriids and howlers, caeco-colonic fermentation, and relatively generalist feeding strategies. There is some evidence that sifakas, howler monkeys, and some colobines may be somewhat resilient to habitat disturbance due to their dietary flexibility and ability to rely on foliage as fallback foods (Estrada and Coates-Estrada, 1996; Bicca-Marques, 2003; Rimbach et al., 2013; Amato et al., 2020; Guevara et al., 2021).
Our results generally bolster the view that folivores do not represent an ecologically uniform guild. Folivores have generally been assumed to face less food limitations and competition (Wrangham, 1980; Isbell, 1991), which presumably make them more tolerant of habitat disturbance and climate change. This may be particularly true in the context of changing landscapes characterized by increasing anthropogenic disturbance (Ganzhorn, 1995). In support of this premise, extinction risk in primates has been positively associated with trophic level and negatively correlated with percent of mature leaves in the diet (Harcourt et al., 2002; Kamilar and Paciulli, 2008). Similarly, disturbance tolerance, is negatively correlated with degree of frugivory and body size in primates (Johns and Skorupa, 1987). Anthropogenic disturbance appears to restrict dispersal in more frugivorous macaques more so than in more folivorous langurs (Erinjery et al., 2019).
However, increasing evidence suggests that many folivores are often quite selective foragers (Garber, 1987; Hemingway, 1998) and are affected behaviorally and physiologically by nutritional limitations (Snaith and Chapman, 2005, 2007; Harris et al., 2010; Tombak et al., 2012; Gogarten et al., 2014). Rather than being evenly distributed, the biomass of several folivorous primates and marsupials is predicted by indicators of leaf quality, like protein to fiber ratio, secondary compound concentrations, and soil quality (Feeny, 1969; Ganzhorn, 1992; Oates, 1996; Peres, 1997; Chapman et al., 2004; Au et al., 2019). Nutritional studies suggest that diet quality cannot be straightforwardly predicted by dietary guild, undermining the predictive utility of broad dietary categories like folivore and frugivore (Barton et al., 1993; Rothman et al., 2007). Moreover, folivores generally are very specific in their choice of leaves based on species, season, tree, location within the tree, and leaf part (Glander, 1982; Chapman and Chapman, 2002; Chapman et al., 2012). It appears that many folivores adopt strategies of either specializing on a few species or being very selective about which specimens they consume of a range of species (Glander, 1982).
In particular, folivores with narrower dietary breadth may have limited resilience in the face of rapid change (Ganzhorn, 1993; Adams-Hosking et al., 2015). Specialization is associated with rarity in primates (Purvis et al., 2000). In Madagascar, the levels of extinction risk of the two bamboo lemur species included here correspond to degree of specialization, with highly specialized greater bamboo lemurs classified as Critically Endangered while the more generalized eastern lesser bamboo lemurs retain a fairly broad distribution and are classified as Vulnerable (Figure 1; Irwin et al., 2020; Ravaloharimanitra et al., 2020). Similarly, two of the more generalist folivores, sifakas and eastern lesser bamboo lemurs, show similar densities at two sites within Ranomafana National Park that differ in disturbance levels. More specialized woolly lemurs, however, show decreased densities at the more disturbed site (Herrera et al., 2011). Several fairly large-bodied primate folivores appear to have highly flexible diets and incorporate a high proportion of fruit when available (Rothman et al., 2007; Li et al., 2010; Owens et al., 2015). Notably, frugo-folivores show greater variation in habitat size and patchiness, indicating potential greater tolerance for habitat disturbance, than either frugivores or folivores (Eppley et al., 2020).
Understanding the many differing ways in which individual species select, process, and digest foliage is necessary to better anticipate and address resilience to habitat change. In some cases, genetic adaptation to aspects of folivory may enhance a species’ ability to cope with shifting abundances of food items (e.g., allowing a sifaka to switch between fruit and leaves), and in some cases, genetic adaptation may constrain a species to overly rely on a limited and diminishing set of food resources.
Demonstrated chemical separation of the foods selected by sympatric folivores indicates differential detoxification capacities, as well as sensory tuning to guide discrimination behavior (Garber, 1987; Ganzhorn, 1988). As such, additional genetic analyses of various genes with roles in detoxification capacity and taste and odor perception could provide insights into differences in functional capacity that could inform conservation assessments. For example, members of the cytochrome P450 monooxygenase gene superfamily are hypothesized to evolve in response to plant secondary compound composition (Gonzalez and Nebert, 1990), and have shown rapid evolution in folivorous mammals, including lemurs (Hu et al., 2017; Johnson et al., 2018; Guevara et al., 2021). Sifakas and extinct koala lemurs (Megaladapis edwardsi), the latter based on a recent innovative analysis of recovered ancient DNA, show evidence of molecular adaptation in genes involved in detoxification and nutrient absorption (Guevara et al., 2021; Marciniak et al., 2021). Bitter taste receptor evolution may also play a key role in fine-tuning primate plant food selection (Soranzo et al., 2005; Purba et al., 2017; Dong et al., 2021). Differences in morphology are also likely to reflect differing physiological strategies. For example, gut complexity and passage time likely coevolves with the gut microbiota. Indeed, the microbiota of the sympatric folivore lemurs studied to date are markedly distinct. Phylogenetic and functional microbiome analyses have revealed that different lemur species harbor microbial communities differentially enriched for functional pathways including cellulolysis, secondary compound degradation, and amino acid metabolism (Greene et al., 2020).
It is also possible that RNase1 is not involved in folivory in lemurs. Although RNase1 duplications have been implicated in herbivore digestion in ruminants and other primates, RNase1 duplication followed by rapid evolution of the duplicate gene has also been observed in other mammalian lineages, including bat, rodent, and mustelid lineages (Dubois et al., 2002; Yu and Zhang, 2006; Goo and Cho, 2013; Xu et al., 2013; Lang et al., 2017). These lineages are not characterized by herbivory and adaptation to viral pathogen pressure has been proposed as a more likely explanation in bats (Goo and Cho, 2013; Xu et al., 2013). Given the wide range of functions that members of the RNase superfamily perform, it is plausible that the changes to the indriid RNase1s reflect adaptation to something other than foliage digestion. Nevertheless, the lowered isoelectric point of indriid RNase1s suggests this function is unlikely to be antiviral, as duplicated RNase1s in bats, for example, exhibit increased pI, which boosts double-stranded RNA digestion efficiency (Xu et al., 2013). Ultimately, expression experiments and functional assays will be necessary to gain further insight into RNase1 expression patterns and function in indriids. Pending such validation, any interpretation drawn from our results can only be considered tentative.
Further, it is puzzling that we found amino acid changes in RNase1 in folivorous lemurs that parallel those in duplicated RNase1s of colobines and howler monkeys without parallel gene duplications, as seen in those other primate lineages and given this gene family’s predominant mode of evolution by expansion. Gene duplication can promote adaptive flexibility by allowing the original gene to retain its initial function, while duplicate copies are more likely to evolve new amino acid changes that result in novel genetic adaptation. The original function of RNase1 in vertebrates is thought to be in host defense via microbial double-stranded RNA degradation (Cho and Zhang, 2006) and, in colobines, the enzyme encoded by the original gene is thought to play a role in the immune system by digesting double-stranded viral RNA (Zhang, 2006). The enzymes encoded by duplicated genes in colobines have evolved amino acid changes that increase efficiency of single-stranded RNA digestion, characteristic of gut microbiota, and reduce the efficiency of double-stranded (viral) RNA digestion (Schienman et al., 2006; Janiak et al., 2019). The lack of duplication in lemurs suggests that RNase1 has not both maintained its original function and gained a new function in folivorous lemurs, as in colobines and howler monkeys. Rather, given the preponderance of amino acid substitutions in indriids paralleling those in colobines, the indriid enzyme is likely to efficiently degrade the gut microbiota’s single-stranded RNA but not retain much antiviral function. In the sportive and bamboo lemurs, the situation is less clear, but the enzyme might have intermediate efficiency in digesting both microbial single-stranded and viral double-stranded RNA. This raises the question of how the enzyme’s initial, antiviral function could be lost or reduced without a prohibitive fitness cost in indriids. It may be that the digestive benefit outweighs the cost or that other genes in the RNASE gene family have taken on compensatory roles. Gene duplication in this family is not limited to evolutionarily recent duplications of RNASE1; indeed, RNASE1 is but one member of this gene family, which contains 13 functional genes in humans (Cho and Zhang, 2006). The RNASE gene family’s evolution in mammals is characterized by expansion via successive duplications of an original gene and its duplicates (Cho and Zhang, 2006). Further study is warranted to better understand the RNASE gene family evolution and enzyme function in primates.
Moreover, our results, along with the findings in howler monkeys, suggest that RNase1 plays a role in types of folivory other than strictly ruminant digestion. The proposed role of RNase1 in foregut fermentation—the reabsorption of nitrogen derived from the degradation of microbial RNA—does not explain molecular adaptation of RNase1 in hindgut fermentation. Recent work suggests that RNase1 may regulate gut microbiota composition (Geng et al., 2021). The role of RNase1 in non-ruminant folivory requires further investigation.
Ultimately, relationships between dietary guild and conservation outlook are unlikely to be straightforward. In particular, folivory probably does not indiscriminately translate into resilience to climate and habitat change. Rather, degree and specific type of specialization may be more informative. It is important to note, however, that there are likely limitations to behavioral flexibility, even for generalists or species with functional traits that “match up” with directional ecological change. For example, increased folivory may have downstream consequences for plant communities, which may result in increased defense or population crashes stemming from disruptions to pollination (Kolb, 2008; Ballhorn et al., 2016). Nevertheless, host genetic and microbiome studies can offer a window into physiological functional capacity that, along with behavioral, anatomical, and life history information, is relevant to conservation. This study is an example of such an approach.
The datasets presented in this study can be found in NCBI at https://www.ncbi.nlm.nih.gov/genbank/ under accession numbers MZ129024, MZ129025, MZ129026, MZ129027, MZ129021, MZ129022, MZ129023, MZ129028, and MZ129029 (https://github.com/uncwmel/rnase1).
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Duke University (protocols: A028-14-02; A007-17-01), and Madagascar’s Ministère de l’Environnement, de l’Ecologie et des Forêts (MEEF/SG/DGF/DAPT/SCBT.Re permits numbers: 85/14; 68/15; 41/17; 83/17; 136/17).
EG and LG designed the study. EG, LG, MB, CF, and RL conducted the genetics lab work. EG conducted the data analysis. JeR, JoR, KM, TR, HHR, RJ, CW, ER, HAR, VR, RL, LA, JC, RR, and AY assisted with sample collection. BB and AY provided reagents and supplies. EG, LG, MB, RL, and AY wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by Duke University and grants from the Wenner–Gren Foundation, Yale Institute for Biospheric Studies, and the American Association of Physical Anthropologists (W. Montague Cobb Award) to EG, and an NSF Postdoctoral Fellowship in Biology to LG (DBI 1906416).
KM, TR, HHR, RJ, and CW were employed by Ambatovy Minerals S.A. ER and HAR were employed by Anjajavy le Lodge and Reserve.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Marojejy guides, Ambatovy Biocamp Agents, the Bezà Mahafaly Monitoring team, Josia Razafindramanana, Vanessa Mass, Alison Richard, Patricia Wright, Thomas Gillespie, and Madagascar National Parks for facilitating sample collection in Madagascar; the Duke Lemur Center Research Department for samples; Diane Genereux and Jeremy Johnson of the Broad Institute for allowing us to use lemur genomic data they generated; members of the Duke Tung lab and Yoder lab for helpful discussion; Ziheng Yang for helpful comments on the methods; and Scott Langdon of the Duke DNA Analysis Facility for expert technical assistance. We complied with the American Association of Physical Anthropologists Code of Ethics. Our procedures were approved by the Institutional Animal Care and Use Committee of Duke University (protocols: A028-14-02; A007-17-01), and by Madagascar’s Ministère de l’Environnement, de l’Ecologie et des Forêts (MEEF/SG/DGF/DAPT/SCBT.Re permits numbers: 85/14; 68/15; 41/17; 83/17; 136/17). This is Duke Lemur Center publication #1493.
Adams-Hosking, C., McAlpine, C. A., Rhodes, J. R., Moss, P. T., and Grantham, H. S. (2015). Prioritizing regions to conserve a specialist folivore: considering probability of occurrence, food resources, and climate change. Conserv. Lett. 8, 162–170. doi: 10.1111/conl.12125
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Amato, K. R., Kuthyar, S., Ekanayake-Weber, M., Salmi, R., Snyder-Mackler, N., Wijayathunga, L., et al. (2020). Gut microbiome, diet, and conservation of endangered langurs in Sri Lanka. Biotropica 52, 981–990. doi: 10.1111/btp.12805
Andrews, C. A. (2019). The Evolution Of The Lepilemuridae-Cheirogaleidae Clade. Ph.D. thesis. Gqeberha: Nelson Mandela University.
Arrigo-Nelson, S. J., and Wright, P. C. (2004). Survey results from Ranomafana National Park: new evidence for the effects of habitat preference and disturbance on the distribution of Hapalemur. Folia Primatol. (Basel). 75, 331–334. doi: 10.1159/000080210
Au, J., Clark, R. G., Allen, C., Marsh, K. J., Foley, W. J., and Youngentob, K. N. (2019). A nutritional mechanism underpinning folivore occurrence in disturbed forests. For. Ecol. Manag. 453, 117585. doi: 10.1016/j.foreco.2019.117585
Ballhorn, D. J., Rakotoarivelo, F. P., and Kautz, S. (2016). Coevolution of cyanogenic bamboos and bamboo lemurs on Madagascar. PLoS One 11:e0158935. doi: 10.1371/journal.pone.0158935
Barnard, E. A. (1969). Biological function of pancreatic ribonuclease. Nature 221, 340–344. doi: 10.1038/221340a0
Barton, R. A., Whiten, A., Byrne, R. W., and English, M. (1993). Chemical composition of baboon plant foods: implications for the interpretation of intra-and interspecific differences in diet. Folia Primatol. (Basel) 61, 1–20. doi: 10.1159/000156722
Beintema, J. J., Schüller, C., Irie, M., and Carsana, A. (1988). Molecular evolution of the ribonuclease superfamily. Prog. Biophys. Mol. Biol. 51, 165–192. doi: 10.1016/0079-6107(88)90001-6
Bicca-Marques, J. C. (2003). “How do howler monkeys cope with habitat fragmentation?,” in Primates in Fragments, ed. L. K. Marsh (Boston, MA: Springer), 283–303. doi: 10.1007/978-1-4757-3770-7_18
Britt, A., Randriamandratonirina, N. J., Glasscock, K. D., and Iambana, B. R. (2002). Diet and feeding behaviour of Indri indri in a low-altitude rain forest. Folia Primatol. (Basel) 73, 225–239. doi: 10.1159/000067455
Campbell, J. L., Eisemann, J. H., Williams, C. V., and Glenn, K. M. (2000). Description of the gastrointestinal tract of five lemur species: Propithecus tattersalli, Propithecus verreauxi coquereli, Varecia variegata, Hapalemur griseus, and Lemur catta. Am. J. Primatol. 52, 133–142. doi: 10.1002/1098-2345(200011)52:3<133::aid-ajp2>3.0.co;2-#
Campbell, J. L., Williams, C. V., and Eisemann, J. H. (2004). Use of total dietary fiber across four lemur species (Propithecus verreauxi coquereli, Hapalemur griseus griseus, Varecia variegata, and Eulemur fulvus): does fiber type affect digestive efficiency? Am. J. Primatol. 64, 323–335. doi: 10.1002/ajp.20081
Chapman, C. A., and Chapman, L. J. (2002). Foraging challenges of red colobus monkeys: influence of nutrients and secondary compounds. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133, 861–875.
Chapman, C. A., Chapman, L. J., Naughton-Treves, L., Lawes, M. J., and Mcdowell, L. R. (2004). Predicting folivorous primate abundance: validation of a nutritional model. Am. J. Primatol. 62, 55–69. doi: 10.1002/ajp.20006
Chapman, C. A., Rothman, J. M., and Lambert, J. E. (2012). “Food as a selective force in primates,” in The Evolution Of Primate Societies, eds J. B. Silk, J. C. Mitani, J. Call, P. M. Kappeler, and R. A. Palombit (Chicago, IL: University of Chicago Press), 145–168.
Cho, S., and Zhang, J. (2006). Ancient expansion of the ribonuclease A superfamily revealed by genomic analysis of placental and marsupial mammals. Gene 373, 116–125. doi: 10.1016/j.gene.2006.01.018
Cho, S., Beintema, J. J., and Zhang, J. (2005). The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics 85, 208–220. doi: 10.1016/j.ygeno.2004.10.008
Cork, S. J. (1996). Optimal digestive strategies for arboreal herbivorous mammals in contrasting forest types: why koalas and colobines are different. Aust. J. Ecol. 21, 10–20. doi: 10.1111/j.1442-9993.1996.tb00581.x
Damas, J., Hughes, G. M., Keough, K. C., Painter, C. A., Persky, N. S., Corbo, M., et al. (2020). Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. bioRxiv [Preprint] bioRxiv: 2020.04.16.045302, doi: 10.1073/pnas.2010146117
Dong, X., Liang, Q., Li, J., and Feng, P. (2021). Positive selection drives the evolution of a primate bitter taste receptor gene. Ecol. Evol. 11, 5459–5467.
dos Reis, M., Gunnell, G. F., Barba-Montoya, J., Wilkins, A., Yang, Z., and Yoder, A. D. (2018). Using phylogenomic data to explore the effects of relaxed clocks and calibration strategies on divergence time estimation: primates as a test case. Syst. Biol. 67, 594–615. doi: 10.1093/sysbio/syy001
Dröscher, I. (2015). Behavioral And Feeding Ecology Of A Small-Bodied Folivorous Primate (Lepilemur Leucopus) Ph.D. Thesis. Göttingen: Niedersächsische Staats-und Universitätsbibliothek Göttingen.
Dubois, J.-Y. F., Jekel, P. A., Mulder, P. P., Bussink, A. P., Catzeflis, F. M., Carsana, A., et al. (2002). Pancreatic-type ribonuclease 1 gene duplications in rat species. J. Mol. Evol. 55, 522–533. doi: 10.1007/s00239-002-2347-8
Eppley, T. M., Donati, G., and Ganzhorn, J. U. (2016). Determinants of terrestrial feeding in an arboreal primate: the case of the southern bamboo lemur (Hapalemur meridionalis). Am. J. Phys. Anthropol. 161, 328–342. doi: 10.1002/ajpa.23034
Eppley, T. M., Ganzhorn, J. U., and Donati, G. (2015b). Cathemerality in a small, folivorous primate: proximate control of diel activity in Hapalemur meridionalis. Behav. Ecol. Sociobiol. 69, 991–1002. doi: 10.1007/s00265-015-1911-3
Eppley, T. M., Donati, G., Ramanamanjato, J.-B., Randriatafika, F., Andriamandimbiarisoa, L. N., Rabehevitra, D., et al. (2015a). The use of an invasive species habitat by a small folivorous primate: implications for lemur conservation in Madagascar. PLoS One 10:e0140981. doi: 10.1371/journal.pone.0140981
Eppley, T. M., Santini, L., Tinsman, J. C., and Donati, G. (2020). Do functional traits offset the effects of fragmentation? The case of large-bodied diurnal lemur species. Am. J. Primatol. 82:e23104. doi: 10.1002/ajp.23104
Eppley, T. M., Verjans, E., and Donati, G. (2011). Coping with low-quality diets: a first account of the feeding ecology of the southern gentle lemur, Hapalemur meridionalis, in the Mandena littoral forest, southeast Madagascar. Primates 52, 7–13. doi: 10.1007/s10329-010-0225-3
Erinjery, J. J., Singh, M., and Kent, R. (2019). Diet-dependent habitat shifts at different life stages of two sympatric primate species. J. Biosci. 44, 1–12.
Eronen, J. T., Zohdy, S., Evans, A. R., Tecot, S. R., Wright, P. C., and Jernvall, J. (2017). Feeding ecology and morphology make a bamboo specialist vulnerable to climate change. Curr. Biol. 27, 3384–3389. doi: 10.1016/j.cub.2017.09.050
Estrada, A., and Coates-Estrada, R. (1996). Tropical rain forest fragmentation and wild populations of primates at Los Tuxtlas Mexico. Int. J Primatol. 17:759. doi: 10.1002/ajp.1350140402
Faulkner, A. L., and Lehman, S. M. (2006). Feeding patterns in a small-bodied nocturnal folivore (Avahi laniger) and the influence of leaf chemistry: a preliminary study. Folia Primatol. (Basel) 77, 218–227. doi: 10.1159/000091231
Feeny, P. P. (1969). Inhibitory effect of oak leaf tannins on the hydrolysis of proteins by trypsin. Phytochemistry 8, 2119–2126. doi: 10.1016/S0031-9422(00)88169-8
Fleagle, J. G., and Reed, K. E. (1996). Comparing primate communities: a multivariate approach. J. Hum. Evol. 30, 489–510.
Fulwood, E. L., Shan, S., Winchester, J. M., Kirveslahti, H., Ravier, R., Kovalsky, S., et al. (2021). Insights from macroevolutionary modelling and ancestral state reconstruction into the radiation and historical dietary ecology of Lemuriformes (Primates, Mammalia). BMC Ecol. Evol. 21:60. doi: 10.1186/s12862-021-01793-x
Ganzhorn, J. U. (1988). Food partitioning among Malagasy primates. Oecologia 75, 436–450. doi: 10.1007/BF00376949
Ganzhorn, J. U. (1989). Niche separation of seven lemur species in the eastern rainforest of Madagascar. Oecologia 79, 279–286. doi: 10.1007/BF00388489
Ganzhorn, J. U. (1992). Leaf chemistry and the biomass of folivorous primates in tropical forests. Oecologia 91, 540–547. doi: 10.1007/BF00650329
Ganzhorn, J. U. (1993). “Flexibility and constraints of Lepilemur ecology,” in Lemur Social Systems And Their Ecological Basis, eds P. M. Kappeler and J. U. Ganzhorn (Boston, MA: Springer), 153–165.
Ganzhorn, J. U. (1995). Low-level forest disturbance effects on primary production, leaf chemistry, and lemur populations. Ecology 76, 2084–2096.
Ganzhorn, J. U., Abraham, J. P., and Razanahoera-Rakotomalala, M. (1985). Some aspects of the natural history and food selection of Avahi laniger. Primates 26, 452–463.
Garber, P. A. (1987). Foraging strategies among living primates. Annu. Rev. Anthropol. 16, 339–364. doi: 10.1146/annurev.an.16.100187.002011
Geng, R., Liu, H., Tan, K., Wang, Z., and Wang, W. (2021). RNase1 can modulate gut microbiota and metabolome after Aeromonas hydrophila infection in blunt snout bream. Environ. Microbiol. 23, 5258–5272. doi: 10.1111/1462-2920.15564
Glander, K. E. (1982). The impact of plant secondary compounds on primate feeding behavior. Am. J. Phys. Anthropol. 25, 1–18. doi: 10.1002/ajpa.1330250503
Gogarten, J. F., Bonnell, T. R., Brown, L. M., Campenni, M., Wasserman, M. D., and Chapman, C. A. (2014). Increasing group size alters behavior of a folivorous primate. Int. J. Primatol. 35, 590–608.
Gonzalez, F. J., and Nebert, D. W. (1990). Evolution of the P450 gene superfamily: animal-plant ‘warfare’, molecular drive and human genetic differences in drug oxidation. Trends Genet. 6, 182–186. doi: 10.1016/0168-9525(90)90174-5
Goo, S. M., and Cho, S. (2013). The expansion and functional diversification of the mammalian ribonuclease a superfamily epitomizes the efficiency of multigene families at generating biological novelty. Genome Biol. Evol. 5, 2124–2140. doi: 10.1093/gbe/evt161
Grassi, C. (2006). Variability in habitat, diet, and social structure of Hapalemur griseus in Ranomafana National Park, Madagascar. Am. J. Phys. Anthropol. 131, 50–63. doi: 10.1002/ajpa.20423
Greene, L. K., Clayton, J. B., Rothman, R. S., Semel, B. P., Semel, M. A., Gillespie, T. R., et al. (2019). Local habitat, not phylogenetic relatedness, predicts gut microbiota better within folivorous than frugivorous lemur lineages. Biol. Lett. 15:20190028.
Greene, L. K., Williams, C. V., Junge, R. E., Mahefarisoa, K. L., Rajaonarivelo, T., Rakotondrainibe, H., et al. (2020). A role for gut microbiota in host niche differentiation. ISME J. 14, 1675–1687.
Guevara, E. E., Webster, T. H., Lawler, R. R., Bradley, B. J., Greene, L. K., Ranaivonasy, J., et al. (2021). Comparative genomic analysis of sifakas (Propithecus) reveals selection for folivory and high heterozygosity despite endangered status. Sci. Adv. 7:eabd2274. doi: 10.1126/sciadv.abd2274
Harcourt, A. H., Parks, S. A., and Coppeto, S. A. (2002). Rarity, specialization and extinction in primates. J. Biogeogr. 29, 445–456.
Harris, T. R., Chapman, C. A., and Monfort, S. L. (2010). Small folivorous primate groups exhibit behavioral and physiological effects of food scarcity. Behav. Ecol. 21, 46–56.
Hawkins, M. T. R., Culligan, R. R., Frasier, C. L., Dikow, R. B., Hagenson, R., Lei, R., et al. (2018). Genome sequence and population declines in the critically endangered greater bamboo lemur (Prolemur simus) and implications for conservation. BMC Genomics 19:445. doi: 10.1186/s12864-018-4841-4
Hemingway, C. A. (1998). Selectivity and variability in the diet of Milne-Edwards’ sifakas (Propithecus diadema edwardsi): implications for folivory and seed-eating. Int. J. Primatol. 19, 355–377.
Herrera, J. P., and Dávalos, L. M. (2016). Phylogeny and divergence times of lemurs inferred with recent and ancient fossils in the tree. Syst. Biol. 65, 772–791. doi: 10.1093/sysbio/syw035
Herrera, J. P., Wright, P. C., Lauterbur, E., Ratovonjanahary, L., and Taylor, L. L. (2011). The effects of habitat disturbance on lemurs at Ranomafana National Park, Madagascar. Int. J. Primatol. 32, 1091–1108.
Hladik, C. M. (1978). Adaptive Strategies Of Primates In Relation To Leaf Eating. Washington, DC: Smithsonian Institution Press.
Hladik, C. M., and Charles-Dominique, P. (1974). The Behaviour And Ecology Of The Sportive Lemur (Lepilemur mustelinus) In Relation To Its Dietary Peculiarities. In: Martin R, Doyle G, Walker A, editors. Prosimian Biology [Internet]. Duckworth; p. 25–37. Available online at: https://hal.archives-ouvertes.fr/hal-01237918 [Accessed Feb 5, 2021].
Horvath, J. E., Weisrock, D. W., Embry, S. L., Fiorentino, I., Balhoff, J. P., Kappeler, P., et al. (2008). Development and application of a phylogenomic toolkit: resolving the evolutionary history of Madagascar’s lemurs. Genome Res. 18, 489–499. doi: 10.1101/gr.7265208
Hu, Y., Wu, Q., Ma, S., Ma, T., Shan, L., Wang, X., et al. (2017). Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad, Sci. U.S.A. 114, 1081–1086. doi: 10.1073/pnas.1613870114
Irwin, M. T. (2006). “Ecologically enigmatic lemurs: the sifakas of the eastern forests (Propithecus candidus, P. diadema, P. edwardsi, P. perrieri, and P. tattersalli),” in Lemurs. Developments in Primatology: Progress and Prospect, eds L. Gould and M. L. Sauther (Boston, MA: Springer), 305–326. doi: 10.1007/978-0-387-34586-4_14
Irwin, M., Frasier, C. L., and Louis, E. E. (2020). Hapalemur griseus. The IUCN Red List of Threatened Species 2020: e.T9673A115564580. [Internet]. IUCN Red List of Threatened Species. Available from: https://dx.doi.org/10.2305/IUCN.UK.2020-2.RLTS.T9673A115564580.en [Accessed May 27, 2021].
Isbell, L. A. (1991). Contest and scramble competition: patterns of female aggression and ranging behavior among primates. Behav. Ecol. 2, 143–155. doi: 10.1002/ajp.22429
Janiak, M. C., Burrell, A. S., Orkin, J. D., and Disotell, T. R. (2019). Duplication and parallel evolution of the pancreatic ribonuclease gene (RNASE1) in folivorous non-colobine primates, the howler monkeys (Alouatta spp.). Sci. Rep. 9:20366. doi: 10.1038/s41598-019-56941-7
Jensen, L. M., Wallis, I. R., Marsh, K. J., Moore, B. D., Wiggins, N. L., and Foley, W. J. (2014). Four species of arboreal folivore show differential tolerance to a secondary metabolite. Oecologia 176, 251–258. doi: 10.1007/s00442-014-2997-4
Jernvall, J., Gilbert, C. C., and Wright, P. C. (2008). “Peculiar tooth homologies of the greater bamboo lemur (Prolemur = Hapalemur simus),” in Elwyn Simons: A Search For Origins, eds C. C. Gilbert and J. G. Fleagle (New York, NY: Springer), 335–342.
Johns, A. D., and Skorupa, J. P. (1987). Responses of rain-forest primates to habitat disturbance: a review. Int. J. Primatol. 8, 157–191. doi: 10.1126/sciadv.1701422
Johnson, R. N., O’Meally, D., Chen, Z., Etherington, G. J., Ho, S. Y., Nash, W. J., et al. (2018). Adaptation and conservation insights from the koala genome. Nat. Genet. 50, 1102–1111.
Kamilar, J. M., and Paciulli, L. M. (2008). Examining the extinction risk of specialized folivores: a comparative study of Colobine monkeys. Am. J. Primatol. 70, 816–827. doi: 10.1002/ajp.20553
Kay, R. F. (1984). “On the use of anatomical features to infer foraging behavior in extinct primates,” in Adapt Foraging Nonhum Primates, eds J. Cant and P. Rodman (New York, NY: Columbia University Press), 21–53.
Kistler, L., Ratan, A., Godfrey, L. R., Crowley, B. E., Hughes, C. E., Lei, R., et al. (2015). Comparative and population mitogenomic analyses of Madagascar’s extinct, giant ‘subfossil’ lemurs. J. Hum. Evol. 79, 45–54. doi: 10.1016/j.jhevol.2014.06.016
Kolb, A. (2008). Habitat fragmentation reduces plant fitness by disturbing pollination and modifying response to herbivory. Biol. Conserv. 141, 2540–2549. doi: 10.1016/j.biocon.2008.07.015
Lang, D.-T., Wang, X.-P., Wang, L., and Yu, L. (2017). Molecular evolution of pancreatic ribonuclease gene (RNase1) in Rodentia. J. Genet. Genomics. 44, 219–222. doi: 10.1016/j.jgg.2017.03.002
Larsen, P. A., Harris, R. A., Liu, Y., Murali, S. C., Campbell, C. R., Brown, A. D., et al. (2017). Hybrid de novo genome assembly and centromere characterization of the gray mouse lemur (Microcebus murinus). BMC Biol. 15:110. doi: 10.1186/s12915-017-0439-6
Li, Y., Jiang, Z., Li, C., and Grueter, C. C. (2010). Effects of seasonal folivory and frugivory on ranging patterns in Rhinopithecus roxellana. Int. J. Primatol. 31, 609–626.
Liu, J., Wang, X., Cho, S., Lim, B. K., Irwin, D. M., Ryder, O. A., et al. (2014). Evolutionary and functional novelty of pancreatic ribonuclease: a study of Musteloidea (order Carnivora). Sci. Rep. 4:5070. doi: 10.1038/srep05070
Marciniak, S., Mughal, M. R., Godfrey, L. R., Bankoff, R. J., Randrianatoandro, H., Crowley, B. E., et al. (2021). Evolutionary and phylogenetic insights from a nuclear genome sequence of the extinct, giant,“subfossil” koala lemur Megaladapis edwardsi. Proc. Natl. Acad. Sci. U.S.A. 118:e2022117118.
Martinez, B. (2008). Occurrence of bamboo lemurs, Hapalemur griseus occidentalis, in an agricultural landscape on the Masoala peninsula. Lemur. News. 13, 11–14.
Mayor, M. I., Sommer, J. A., Houck, M. L., Zaonarivelo, J. R., Wright, P. C., Ingram, C., et al. (2004). Specific status of Propithecus spp. Int. J. Primatol. 25, 875–900.
McLain, A. T., Meyer, T. J., Faulk, C., Herke, S. W., Oldenburg, J. M., Bourgeois, M. G., et al. (2012). An alu-based phylogeny of lemurs (infraorder: Lemuriformes). PLoS One 7:e44035. doi: 10.1371/journal.pone.0044035
Meyer, W. K., Venkat, A., Kermany, A. R., van de Geijn, B., Zhang, S., and Przeworski, M. (2015). Evolutionary history inferred from the de novo assembly of a nonmodel organism, the blue-eyed black lemur. Mol. Ecol. 24, 4392–4405. doi: 10.1111/mec.13327
Milton, K. (1978). Role of the upper canine and P 2 in increasing the harvesting efficiency of Hapalemur griseus Link, 1795. J. Mammal. 59, 188–190. doi: 10.2307/1379890
Milton, K. (1999). Nutritional characteristics of wild primate foods: do the diets of our closest living relatives have lessons for us? Nutrition 15, 488–498. doi: 10.1016/s0899-9007(99)00078-7
Milton, K., and McBee, R. H. (1983). Rates of fermentative digestion in the howler monkey, Alouatta palliata (Primates: Ceboidea). Comp. Biochem. Physiol. A Physiol. 74, 29–31. doi: 10.1016/0300-9629(83)90706-5
Mittermeier, R. A., Louis, E. E. Jr., Richardson, M. J., Schwitzer, C., Langrand, O., Rylands, A. B., et al. (2010). Lemurs of Madagascar. Arlington, TX: Conservation International.
Nash, L. T. (1998). Vertical clingers and sleepers1: seasonal influences on the activities and substrate use of Lepilemur leucopus at Beza Mahafaly Special Reserve, Madagascar. Folia Primatol. (Basel) 69(Suppl. 1) 204–217. doi: 10.1159/000052714
Oates, J. F. (1996). Habitat alteration, hunting and the conservation of folivorous primates in African forests. Aust. J. Ecol. 21, 1–9. doi: 10.1111/j.1442-9993.1996.tb00580.
Owens, J. R., Honarvar, S., Nessel, M., and Hearn, G. W. (2015). From frugivore to folivore: Altitudinal variations in the diet and feeding ecology of the Bioko Island drill (Mandrillus leucophaeus poensis). Am. J. Primatol. 77, 1263–1275. doi: 10.1002/ajp.22479
Perelman, P., Johnson, W. E., Roos, C., Seuánez, H. N., Horvath, J. E., Moreira, M. A., et al. (2011). A molecular phylogeny of living primates. PLoS Genet. 7:e1001342. doi: 10.1371/journal.pgen.1001342
Peres, C. A. (1997). Effects of habitat quality and hunting pressure on arboreal folivore densities in neotropical forests: a case study of howler monkeys (Alouatta spp.). Folia Primatol. (Basel) 68, 199–222. doi: 10.1159/000157247
Powzyk, J. A., and Mowry, C. B. (2003). Dietary and feeding differences between sympatric Propithecus diadema diadema and Indri indri. Int. J. Primatol. 24, 1143–1162. doi: 10.1023/B:IJOP.0000005984.36518.94
Purba, L. H. P. S., Widayati, K. A., Tsutsui, K., Suzuki-Hashido, N., Hayakawa, T., Nila, S., et al. (2017). Functional characterization of the TAS2R38 bitter taste receptor for phenylthiocarbamide in colobine monkeys. Biol. Lett. 13:20160834. doi: 10.1098/rsbl.2016.0834
Purvis, A., Gittleman, J. L., Cowlishaw, G., and Mace, G. M. (2000). Predicting extinction risk in declining species. Proc. R. Soc. Lond. B Biol. Sci. 267, 1947–1952.
Ravaloharimanitra, M., King, T., Wright, P. C., Raharivololona, B., Ramaherison, R. P., Louis, E. E., et al. (2020). Prolemur Simus. The IUCN Red List of Threatened Species 2020: e.T9674A115564770. [Internet]. IUCN Red List of Threatened Species. Available online at: https://dx.doi.org/10.2305/IUCN.UK.2020-2.RLTS.T9674A115564770.en (accessed May 27, 2021).
Rimbach, R., Link, A., Heistermann, M., Gómez-Posada, C., Galvis, N., and Heymann, E. W. (2013). Effects of logging, hunting, and forest fragment size on physiological stress levels of two sympatric ateline primates in Colombia. Conserv. Physiol. 1:cot031. doi: 10.1093/conphys/cot031
Rothman, J. M., Plumptre, A. J., Dierenfeld, E. S., and Pell, A. N. (2007). Nutritional composition of the diet of the gorilla (Gorilla beringei): a comparison between two montane habitats. J. Trop. Ecol. 23, 673–682.
Schienman, J. E., Holt, R. A., Auerbach, M. R., and Stewart, C.-B. (2006). Duplication and divergence of 2 distinct pancreatic ribonuclease genes in leaf-eating African and Asian colobine monkeys. Mol. Biol. Evol. 23, 1465–1479. doi: 10.1093/molbev/msl025
Schmid, J., and Ganzhorn, J. U. (1996). Resting metabolic rates of Lepilemur ruficaudatus. Am. J. Primatol. 38, 169–174. doi: 10.1002/(SICI)1098-2345(1996)38:2<169::AID-AJP5>3.0.CO;2-X
Snaith, T. V., and Chapman, C. A. (2005). Towards an ecological solution to the folivore paradox: patch depletion as an indicator of within-group scramble competition in red colobus monkeys (Piliocolobus tephrosceles). Behav. Ecol. Sociobiol. 59, 185–190. doi: 10.1007/s00265-005-0023-x
Snaith, T. V., and Chapman, C. A. (2007). Primate group size and interpreting socioecological models: do folivores really play by different rules? Evol. Anthropol. 16, 94–106. doi: 10.1002/evan.20132
Soranzo, N., Bufe, B., Sabeti, P. C., Wilson, J. F., Weale, M. E., Marguerie, R., et al. (2005). Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr. Biol. 15, 1257–1265.
Springer, M. S., Meredith, R. W., Gatesy, J., Emerling, C. A., Park, J., Rabosky, D. L., et al. (2012). Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS One 7:e49521. doi: 10.1371/journal.pone.0049521
Tan, C. L. (1999). Group composition, home range size, and diet of three sympatric bamboo lemur species (genus Hapalemur) in Ranomafana National Park Madagascar. Int. J. Primatol. 20, 547–566.
Tombak, K. J., Reid, A. J., Chapman, C. A., Rothman, J. M., Johnson, C. A., and Reyna-Hurtado, R. (2012). Patch depletion behavior differs between sympatric folivorous primates. Primates 53, 57–64. doi: 10.1007/s10329-011-0274-2
Warren, R. D., and Crompton, R. H. (1997). Locomotor ecology of Lepilemur edwardsi and Avahi occidentalis. Am. J. Phys. Anthropol. 104, 471–486. doi: 10.1002/(SICI)1096-8644(199712)104:4<471::AID-AJPA4>3.0.CO;2-V
Wrangham, R. W. (1980). An ecological model of female-bonded primate groups. Behaviour 75, 262–300. doi: 10.1163/156853980X00447
Xu, H., Liu, Y., Meng, F., He, B., Han, N., Li, G., et al. (2013). Multiple bursts of pancreatic ribonuclease gene duplication in insect-eating bats. Gene 526, 112–117. doi: 10.1016/j.gene.2013.04.035
Yamashita, N. (2008). Chemical properties of the diets of two lemur species in southwestern Madagascar. Int. J. Primatol. 29, 339–364. doi: 10.1007/s10764-008-9232-2
Yang, Z. (2002). Inference of selection from multiple species alignments. Curr. Opin. Genet. Dev. 12, 688–694. doi: 10.1016/S0959-437X(02)00348-9
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S. W., and Nielsen, R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118.
Yoder, A. D. (1999). “Ancient DNA in subfossil lemurs: methodological challenges and their solutions,” in New Directions in Lemur Studies, eds B. Rakotosamimanana, H. Rasamimanana, J. U. Ganzhorn, and S. M. Goodman (New York, NY: Plenum Press).
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T.-Y. (2017). ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36.
Yu, L., and Zhang, Y. (2006). The unusual adaptive expansion of pancreatic ribonuclease gene in carnivora. Mol. Biol. Evol. 23, 2326–2335. doi: 10.1093/molbev/msl101
Zhang, J. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. doi: 10.1093/molbev/msi237
Zhang, J. (2006). Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 38, 819–823. doi: 10.1038/ng1812
Zhang, J., Zhang, Y., and Rosenberg, H. F. (2002). Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415. doi: 10.1038/ng852
Keywords: Hapalemur, Prolemur, Indri, Avahi, Propithecus, Lepilemur, dietary ecology, herbivory
Citation: Guevara EE, Greene LK, Blanco MB, Farmer C, Ranaivonasy J, Ratsirarson J, Mahefarisoa KL, Rajaonarivelo T, Rakotondrainibe HH, Junge RE, Williams CV, Rambeloson E, Rasoanaivo HA, Rahalinarivo V, Andrianandrianina LH, Clayton JB, Rothman RS, Lawler RR, Bradley BJ and Yoder AD (2021) Molecular Adaptation to Folivory and the Conservation Implications for Madagascar’s Lemurs. Front. Ecol. Evol. 9:736741. doi: 10.3389/fevo.2021.736741
Received: 05 July 2021; Accepted: 08 September 2021;
Published: 06 October 2021.
Edited by:
Claudia Fichtel, Deutsches Primatenzentrum, GermanyReviewed by:
Mareike Janiak, University of Salford, United KingdomCopyright © 2021 Guevara, Greene, Blanco, Farmer, Ranaivonasy, Ratsirarson, Mahefarisoa, Rajaonarivelo, Rakotondrainibe, Junge, Williams, Rambeloson, Rasoanaivo, Rahalinarivo, Andrianandrianina, Clayton, Rothman, Lawler, Bradley and Yoder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elaine E. Guevara, Z29tZXpndWV2YXJhZUB1bmN3LmVkdQ==; ZWxhaW5lZWd1ZXZhcmFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.