- Department of Integrative Biology, University of Guelph, Guelph, ON, Canada
Programmed cell death (PCD) has been identified as a key process in the metamorphic transition of indirectly developing organisms such as frogs and insects. Many marine invertebrate species with indirect development and biphasic life cycles face the challenge of completing the metamorphic transition of the larval body into a juvenile when they settle into the benthic habitat. Some key characteristics stand out during this transition in comparison to frogs and insects: (1) the transition is often remarkably fast and (2) the larval body is largely abandoned and few structures transition into the juvenile stage. In sea urchins, a group with a drastic and fast metamorphosis, development and destruction of the larval body is regulated by endocrine signals. Here we provide a brief review of the basic regulatory mechanisms of PCD in animals. We then narrow our discussion to metamorphosis with a specific emphasis on sea urchins with indirect life histories and discuss the function of thyroid hormones and histamine in larval development, metamorphosis and settlement of the sea urchin Strongylocentrotus purpuratus. We were able to annotate the large majority of PCD related genes in the sea urchin S. purpuratus and ongoing studies on sea urchin metamorphosis will shed light on the regulatory architecture underlying this dramatic life history transition. While we find overwhelming evidence for hormonal regulation of PCD in animals, especially in the context of metamorphosis, the mechanisms in many marine invertebrate groups with indirect life histories requires more work. Hence, we propose that studies of PCD in animals requires functional studies in whole organisms rather than isolated cells. We predict that future work, targeting a broader array of organisms will not only help to reveal important new functions of PCD but provide a fundamentally new perspective on its use in a diversity of taxonomic, developmental, and ecological contexts.
Review of Cell Death Mechanisms
Cell death, whether active (apoptosis) or passive (necrosis), effectively results in the removal of cells from organisms and therefore stands in contrast to cell proliferation which results from cell division. Apoptosis and other forms of programmed cell death (PCD) occur in response to internal (cellular environment) signals and external environment cues, including stressors, and involve a broad array of now well established molecular and cellular responses, ultimately packaging the cellular material for removal (Kerr et al., 1972; Green and Fitzgerald, 2016). PCD plays an extremely important role in development and the molecular and genetic processes are tightly regulated. Still, many diseases have been shown to emerge due to dysregulation of cell suicide mechanisms and it is therefore assumed that over evolutionary time multiple complementary and redundant regulatory mechanisms have emerged in organisms. Furthermore, many of these processes and pathways also perform other functions in development and physiology (Amiesen, 2002).
Traditionally three upstream apoptotic pathways and one downstream apoptotic pathway are distinguished in the literature, however their functions are deeply intertwined (Figure 1). The extrinsic pathway involves death receptors of the tumor necrosis factor (TNF) receptor gene superfamily. Extracellular cell death signals trigger the binding of homologous trimeric ligands such as FasLG to death effector domain (DED)-containing receptors like Fas to activate the extrinsic pathway (Figure 1; Fulda and Debatin, 2006; Elmore, 2007). These complexes then bind to cytoplasmic adaptor proteins like FADD. After binding the ligands either FADD or TRADD with the assistance of RIPK1 and FADD associate with procaspase-8 to form the death-inducing signaling complex (DISC). DISC is responsible for the activation of CASP8 which then cleaves CASP3 to start the execution pathway (Elmore, 2007).
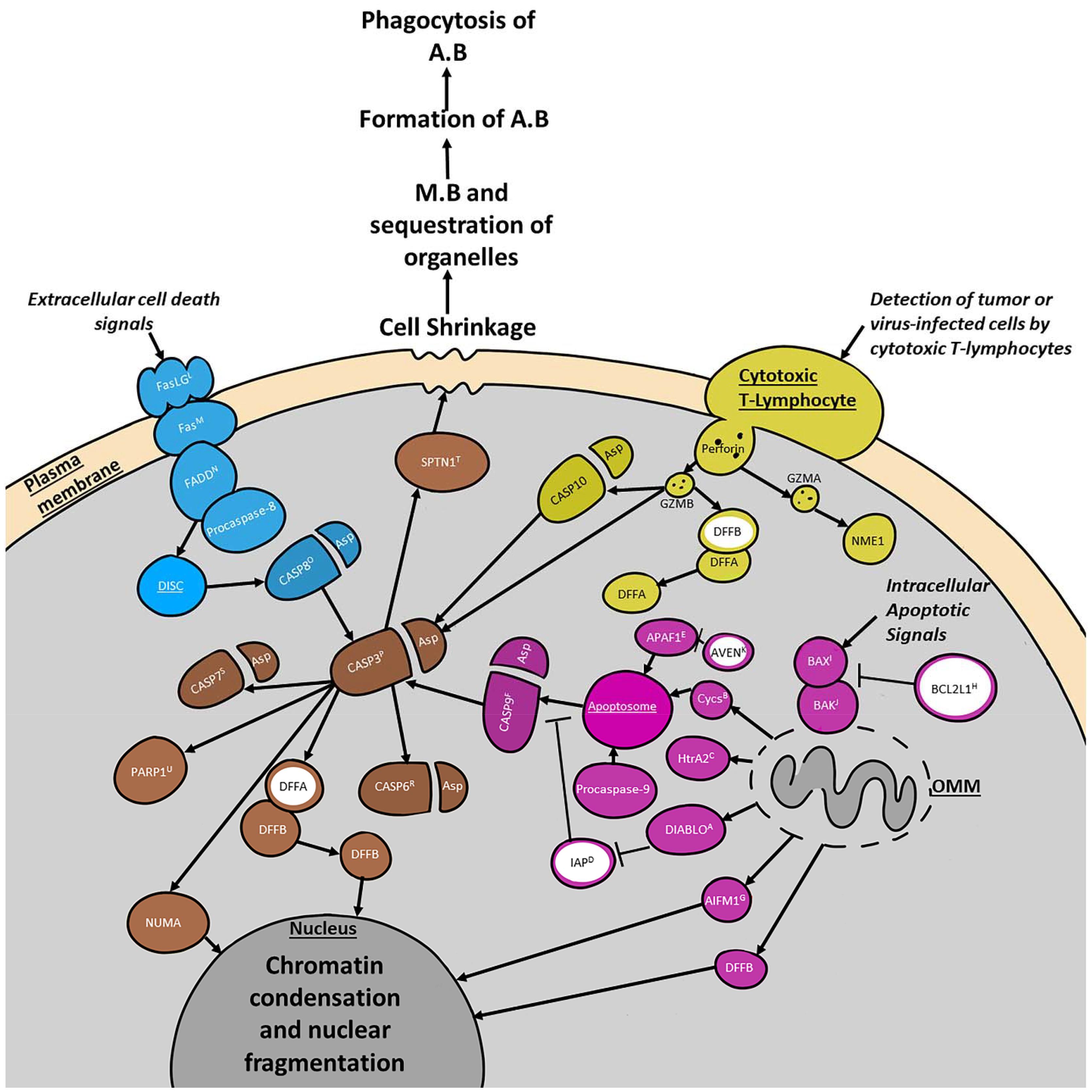
Figure 1. Summary of four major programmed cell death (PCD) pathways discussed in the text. Selected cell organelles are marked in dark gray. Proteins involved in the extrinsic pathway are blue. Proteins involved in the intrinsic pathway are purple. Proteins involved in the granzyme/perforin pathway are indicated in yellow. All three pathways converge at the execution pathway which is colored brown. Major inhibitory proteins are filled in white with a ring colored according to the pathway they are primarily involved in. Major complexes such as the Apoptosome and DISC are underlined. The intrinsic pathway is activated by intracellular apoptotic signals that either suppress anti-apoptotic proteins or increase pro-apoptotic proteins. This leads to the activation of pro-apoptotic BCL-2 family proteins such as BAK1 and BAX which triggers the mitochondrial aspect of the pathway by increasing the permeability of the outer mitochondrial membrane (OMM). The granzyme/perforin pathway is activated by the detection of tumor/virus-infected cells by cytotoxic t-cells. These cells secrete perforin which creates pores in the plasma membrane into which GZMA and GZMB can be secreted. The extrinsic pathway is activated by extracellular cell death signals that trigger the binding of homologous trimeric ligands like FasLG to DED-containing receptors such as Fas. Next Cytoplasmic adaptor proteins like FADD are recruited to the complex, along with procaspase-8. Together these proteins form the DISC. All three pathways meet at the execution pathway which is responsible for triggering chromatin condensation and nuclear fragmentation, as well as cell shrinkage. This shrinking leads to membrane blebbing (M.B) and sequestration of the organelles prior to the formation of the apoptotic bodies (A.B). The A.B are then phagocytosed and the chromatin fragments and organelles within them are degraded or incorporated into other cells. Protein superscripts indicate sea urchin (S. purpuratus) orthologous genes from echinobase. A, DIABLO (LOC574919); B, Cycs (LOC575421); C, HtrA2 (LOC115918650); D, IAP (LOC581540); E, APAF1 (LOC591503); F, CASP9 (LOC115924698); G, AIFM1 (LOC578253); H, BCL2L1 (LOC100890351); I, BAX (LOC586236); J, BAK1 (LOC115917937); K, AVEN (LOC762362); L, FasLG (LOC582167); M, Fas (LOC586563); N, FADD (LOC587131); O, CASP8 (LOC585496); P, CASP3 (LOC115918952); R, CASP6 (LOC584221); S, CASP7 (LOC580916); T, SPTAN1 (LOC580822); U, PARP1 (LOC752216).
The intrinsic signaling pathway initiates apoptosis through non-receptor mediated stimuli, acting either directly on pro-apoptotic targets upstream of the pro-apoptotic B-cell lymphoma 2 (Bcl-2) family, or within the cell by inhibiting anti-apoptosis targets (Figure 1). This signaling pathway is triggered by environmental stressors such as radiation, toxins, hypoxia, viral infections, and others. The intrinsic pathway is often also referred to as the mitochondrial pathway due to the role that the permeabilization of the mitochondrial outer membrane plays in the activation of CASP9. It responds to intracellular signals produced by non-receptor mediated stimuli. These lead to the activation of Bcl-2 family members such as the Bcl-2 effector proteins BAX and BAK1 (Figure 1), which in turn are responsible for mitochondrial outer membrane permeabilization (Degterev and Yuan, 2008; Kalkavan and Green, 2018). Mitochondrial outer membrane permeabilization can be prevented by anti-apoptotic members of the Bcl-2 family like Bcl-2 and BCL2L1 (Elmore, 2007; reviewed in Daniel et al., 2003). For apoptosis to proceed, anti-apoptotic members must be blocked by proteins such as BAG1 and Bik-like killer protein (BLK) (Figure 1 and Table 1; Hegde et al., 1998; Götz et al., 2005; Degterev and Yuan, 2008). Upon mitochondrial outer membrane permeabilization completion, several proteins are released from the outer mitochondrial membrane (i.e., cycs, HtrA2, DIABLO, AIFM1, and DFFB). One of the proteins released is cycs which forms the apoptosome with APAF1 and procaspase-9. Apoptosome formation is blocked by AVEN which binds BCL2L1 and APAF1 to prevent activation of procaspase-9 by the apoptosome. If the apoptosome can form, procaspase-9 is cleaved turning into activated CASP9, which can then trigger the execution pathway (Figure 1). Even once the apoptosome has cleaved CASP9 there are still a few ways the execution phase can be blocked. Inhibitor of apoptosis proteins (IAPs) can bind CASP9, CASP3, and CASP7 in order to prevent the start of the execution pathway (Obexer and Ausserlechner, 2014). Prior to this, DIABLO and HtrA2 can inhibit IAPs and encourage apoptosis (Figure 1; Du et al., 2000; Suzuki et al., 2001; Elmore, 2007). After IAPs have bound to caspases, BCL10 can cause dissociation of those caspases (Yui et al., 2001). The protein AIFM1, which is released from the outer mitochondrial membrane plays a role triggering DNA fragmentation (Elmore, 2007). Despite being independent processes, the extrinsic and intrinsic pathways share many of the same molecules and a molecule from one pathway has the potential to influence the other.
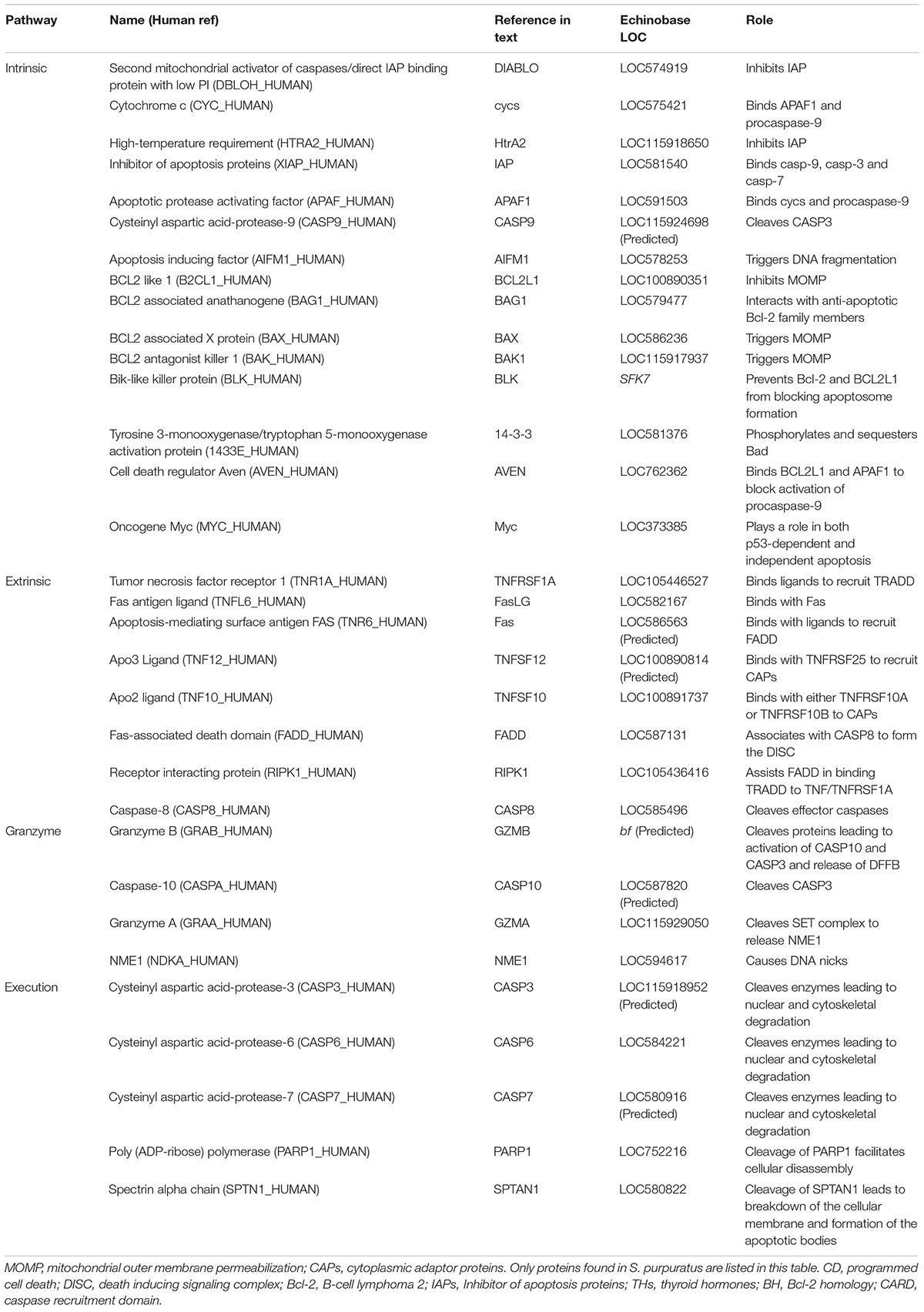
Table 1. Summary table listing proteins often involved in apoptosis and which pathway they are involved in, how they are referenced in the text, their LOC identification in echinobase, and their primary function in apoptosis.
The third, and least well-known pathway is the perforin/granzyme pathway which induces apoptosis through proteins secreted into the cell by Cytotoxic T-lymphocytes. This pathway is initiated when cytotoxic t-lymphocytes detect tumor or virus-infected cells and begin to secrete perforin in order to trigger pore-formation in the plasma membrane (Figure 1). Next, GZMA and GZMB are secreted into the cell where they begin to cleave other proteins. GZMA cleaves the 270–420 kDa endoplasmic reticulum-associated complex (SET complex) and GZMB begins cleaving CASP10 and DFFA. Once the SET complex is cleaved, NME1 is released and begins to cleave DNA (Elmore, 2007). CASP10 then triggers the execution pathway by cleaving CASP3 and DFFB is released from DFFA. There is also a significant overlap between the perforin/granzyme pathway and the intrinsic and extrinsic pathways due to the ability of GZMB to cleave proteins associated with mitochondrial permeability.
Finally, all three of these pathways meet up at the execution pathway which is activated with the cleavage of CASP3 and is responsible for the dismantling of the cell. When CASP3 is cleaved it begins to activate endonucleases and proteases in the cytoplasm that then degrade the nucleus and cytoskeleton (Coleman et al., 2001; Andrade et al., 2010). The other two effector caspases, CASP7, and CASP6 play a very similar role and can be activated by initiator caspases or CASP3. An important part of the execution pathway that requires more information is the role that cleavage of certain proteins such as PARP1 and SPTAN1 play int his process. Currently it is thought that cleavage of PARP1 and SPTAN1 may help facilitate cellular disassembly (Oliver et al., 1998; Williams et al., 2003).
Autophagic cell death (autophagic type II cell death) may serve as an alternate regulated mechanism of killing cells when apoptosis is blocked (Fuchs and Steller, 2011). It can be characterized by the cytoplasmic vacuolization, autophagosome formation and clearance of material via the lysosome. Autophagy (macroautophagy) fulfills an important function in cell metabolism as it blocks the buildup of toxic protein aggregates. It may also drive other forms of cell death, block anti-apoptotic factors, and play a role in ensuring proper engulfment of dead cells to minimize inflammation. Furthermore, it may also be involved in dying cells switching between different forms of cell death (Doherty and Baehrecke, 2018). Autophagic cell death and autophagy both serve as a good reminder that PCD is a highly regulated, conserved, and important process for development and homeostasis.
While apoptosis and autophagic cell death are the most well-known forms of programmed cell death, other types have recently been observed and characterized like pyroptosis, ferroptosis and oxeiptosis. Pyroptosis is a form of programmed necrosis in response to a strong inflammatory response caused by pathogenic infections (Hanson, 2016). Like apoptosis, pyroptosis requires caspase activation however, the caspases involved are completely different from those involved in apoptosis which cleave the inflammatory proteins pro-IL-1β and pro-IL-18 (Hanson, 2016). These pro-inflammatory cytokines then burst from the dying cell (Hanson, 2016). While little is known about the existence of pyroptosis outside of vertebrates, there is some evidence of a pyroptosis induction pathway in bivalves that is regulated by caspase-3 and gasdermin E-like proteins (Vogeler et al., 2021). Ferroptosis is another non-apoptotic form of programmed cell death which is executed by excessive production of reactive oxygen species (Hanson, 2016). Unlike pyroptosis which appears very apoptosis-like, ferroptosis appears very necrosis-like but is still the product of the activation of a unique pathway (Hanson, 2016). Ferroptosis is characterized by an overwhelming and iron-dependent accumulation of lipid lethal ROS (Dixon et al., 2012). Another type of non-apoptotic programmed cell death is oxeiptosis which is triggered by oxidative stress (Holze et al., 2018). Oxeiptosis is caspase-independent and, similar to ferroptosis, is activated by harmful amounts of ROS (Holze et al., 2018).
Still, not all cell death unfolds via a regulated mechanism. Necrosis is a passive, uncontrolled, toxic process that follows an energy-independent mode of death. Necrosis often also affects vast areas of cells where apoptosis usually affects individual cells or small clusters of cells (Elmore, 2007). It typically leads to organelle swelling, lysis, release of intracellular contents, and inflammation (Doherty and Baehrecke, 2018). Since necrosis can occur independently, sequentially, or simultaneously to PCD it can be difficult to distinguish these processes. Furthermore, there are forms of cell death that share morphological features of both necrosis and apoptosis called aponecrosis (Elmore, 2007). There is also necroptosis, which functions as a unique signaling pathway that is characterized by 2 major necroptotic death effector complexes, caspase inactivity and a series of morphological changes (Hanson, 2016). The necrosome and ripoptosome are the two major necroptotic death effector complexes and are induced by TNFR1 and toll like receptor 3 (TLR3) signaling (Hanson, 2016). During this process the cell undergoes several morphological and internal changes such as cellular rounding, an increase in cytosolic calcium ions and reactive oxygen species, depletion of ATP, intracellular acidification, cellular swelling and finally plasma membrane rupture, and the release of damage-associated molecular patterns (Hanson, 2016). Many of the proteins important for necroptosis are found across Deuterostomia however, it is unclear if the actual process of necroptosis occurs in some of the non-vertebrate deuterostomes (Dondelinger et al., 2016).
Traditionally, a large part of mechanistic understanding of cell death originates from cell culture experiments and less so from whole organism studies. Still, in recent years, new data on cell death regulation and function has emerged from a diversity of taxa and this data suggests that PCD mechanisms are used in a range of morphological, physiological, and developmental contexts. For example, animals with an indirect life-history, characterized by a drastic morphological and physiological transition (Tata, 1993; Balon, 1999; Bishop et al., 2006; Heyland and Moroz, 2006; Ishizuya-Oka et al., 2010; Heyland et al., 2018; Tettamanti and Casartelli, 2019) have been repeatedly shown to employ PCD for the removal of cells and the transformation of the larval body plan into a juvenile (Heyland and Moroz, 2006; Hodin et al., 2016; Lutek et al., 2018). Here we integrate available mechanistic information on PCD from whole organism studies with our understanding of the regulatory mechanisms of metamorphosis with an emphasis on endocrine regulation. We propose, that the metamorphic transition in animals provides an excellent model system to study the role of PCD in post-embryonic development and therefore provide critical functional background for PCD from an ecological, physiological, and developmental perspective.
PCD in Metamorphic Development and Its Regulation by Hormones
The importance of PCD in development is exemplified during the metamorphic transition of animals. This drastic and often rapid change in morphology, physiology and ecology of an organism is typically associated with substantial cellular remodeling, differentiation, and the removal of cells and tissues. Historically, frogs and insects have been extensively studied in this context and a significant amount of information on PCD regulation and functions exists in these groups (Tata, 1993; Bishop et al., 2006; Heyland and Moroz, 2006; Ishizuya-Oka et al., 2010; Heyland et al., 2018; Tettamanti and Casartelli, 2019). Even though metamorphosis evolved independently in insects and vertebrates (Hadfield, 2000; Heyland and Moroz, 2006; Hodin, 2006; Heyland et al., 2018), similar, convergent mechanisms are used in its regulation. Notably, hormones have emerged as a key regulator of cell death and these endocrine mechanisms continue to be investigated (Nakajima et al., 2005; Heyland and Moroz, 2006; Mané-Padrós et al., 2010; Heyland et al., 2018). In contrast, marine invertebrate species with indirect life histories undergo equally dramatic transitions but the cellular and molecular processes (including PCD) have received little attention, considering invertebrate diversity (but see Wray and Raff, 1990; Degnan, 2001; Davidson and Swalla, 2002; Bishop and Brandhorst, 2003; Bishop and Anderson, 2005; Heyland and Moroz, 2006; Hodin, 2006; Paris et al., 2008, 2010; Nakayama-Ishimura et al., 2009; Heyland et al., 2018; Wong et al., 2019). Here we briefly review the function of PCD in metamorphosis and settlement among animals to create a foundation for our discussion of PCD in sea urchin embryonic and post-embryonic development in the next section.
Frog tadpoles develop specific structures during the metamorphic transition that allow them to live a semi-aquatic life in the adult stage. This transformation impacts most physiological and morphological systems (Figure 2). Apoptosis has been studied in some detail during frog tail resorption, the rewiring of the nervous system, the digestive system, and gill remodeling (Ishizuya-Oka et al., 2010). As tadpoles transition to live on land, the aquatic tail of the tadpole is no longer required and rapidly disintegrates. The process of tail resorption is tightly regulated and can be roughly divided into three stages. First, the dorsal fins are resorbed, then the muscle fibers are fragmented and finally the notochord lamella is dissolved (Das et al., 2002; Brown et al., 2005). In addition to these morphological changes, there are several important cellular events that make up this process. In response to THs, tail muscle cells up-regulate and activate CASP3 and a high level of pro-apoptotic BAX expression in the tail during its resorption has been described in Xenopus laevis (Das et al., 2002, 2006). This is supported by the fact that overexpression of the anti-apoptotic Bcl-2 in tail muscle inhibits TH-induced cell death. Similarly, downstream of BAX, pro-apoptotic CASP9 is also upregulated in the brain, intestine and tail during metamorphic climax (Das et al., 2006). Still, the tadpole tail is not the only structure that is removed or modified by apoptosis. The gills also degenerate almost completely, and the intestine and pancreas are drastically remodeled to accommodate the change in diet from the herbivorous tadpole stage to the carnivorous adult stage (Sun et al., 2014). A similar remodeling must occur in the organs of the immune system like the spleen as the larval immune system is not equipped to defend against the same pathogens as an adult frog immune system (Ishizuya-Oka et al., 2010). Just like the tail resorption, regulation of these processes is under the control of thyroid hormones (THs) (Ishizuya-Oka et al., 2010). The thyroid hormones T3 and T4 are secreted by the developing tadpole’s thyroid gland and, in the case of T4, is converted to T3 by type I and II deiodinases in the target tissue. TH actions are mediated via thyroid hormone receptors, a type of nuclear hormone receptor that is necessary for mediating the metamorphic effects of TH in tadpoles. In frogs two thyroid hormone receptors have been identified and characterized, Thra and Thrb (Ishizuya-Oka et al., 2010; Faunes and Larraín, 2016). Apoptosis in larval tissue is primarily mediated by Thrb (Ishizuya-Oka et al., 2010). While it was previously thought that TH was both necessary and sufficient for metamorphosis, recent research has indicated that steroid hormones such as glucocorticoids may play an important role in the progression of tail resorption as well (Sachs and Buccholz, 2019).
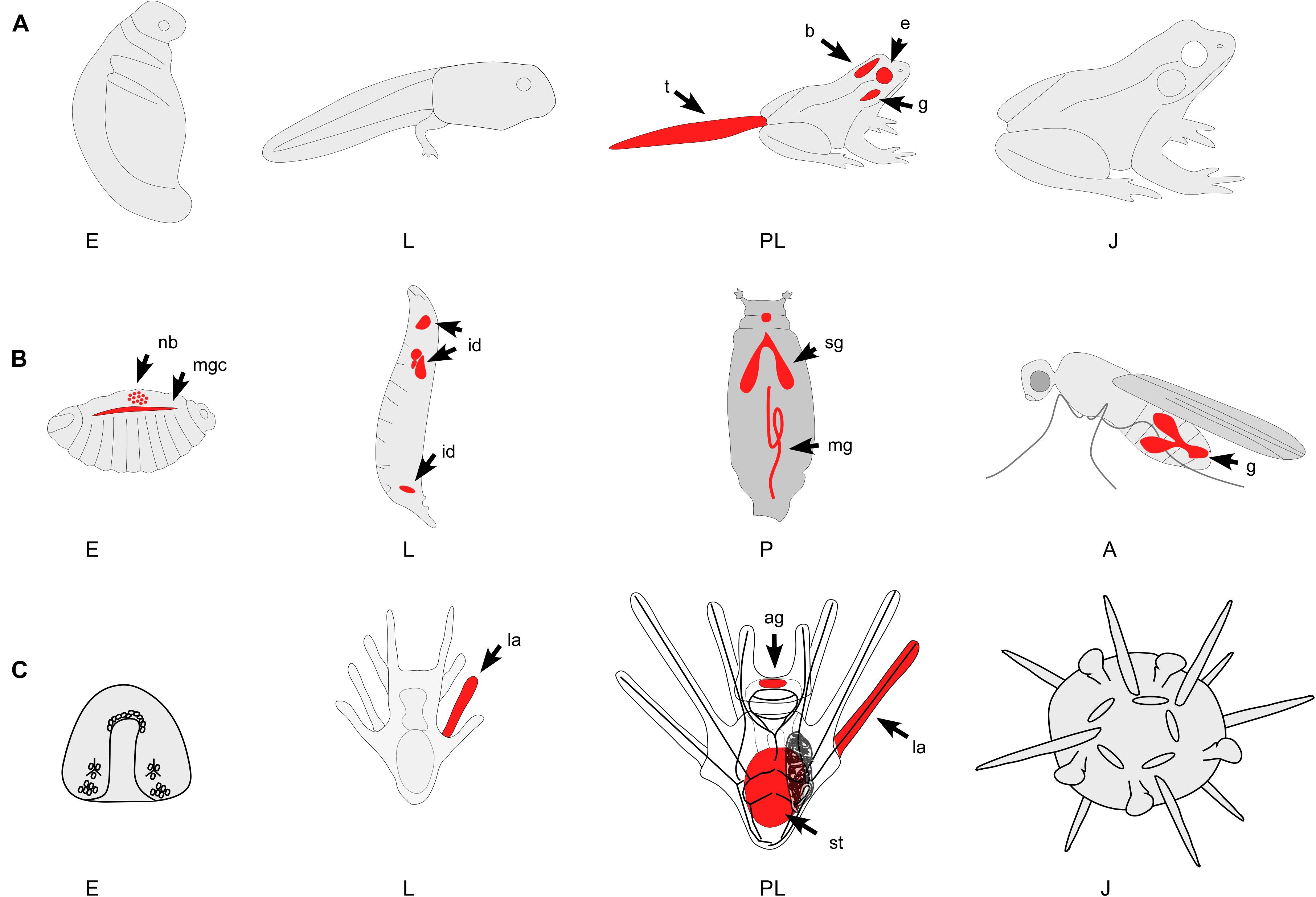
Figure 2. Three representative examples of animals with indirect life histories and metamorphic transitions. Red color indicates major areas of PCD during development. Note that only major areas of PCD are depicted in these diagrams and many more have been identified. (A) Indirectly developing frog. The metamorphic transition of frogs impacts most physiological and morphological systems. While PCD occurs during embryogenesis (E – gastrulation and early cleavage stages) most work has identified the mechanisms of PCD during tail (t) regression, gill (g) and brain (b) remodeling, and the restructuring of many other organ systems. (B) Holometabolous insect. PCD occurs throughout holometabolous development, including the larval epidermis, neuroblasts (nb), imaginal disks (id), salivary gland (sg), midgut (mg), mushroom bodies, and gonads (g). The pupal stage (P) is a time of active tissue remodeling and patterning that is strongly associated with PCD. (C) Indirectly developing sea urchin. PCD has been identified throughout sea urchin development, including the embryo (E) and the larval (L) tissues. During settlement, many larval structures such as the arms (la), the ciliated band and parts of the stomach (st) degenerate, a process that has been shown to involve PCD. A, adult; E, embryo; J, juvenile; L, larva; P, pupa; PL, pre-metamorphic larva; ag, apical ganglion; b, brain; e, eyes; g, gills; g, gonads; id, imaginal disks; la, larval arms; mg, midgut; mgc, midline glial cells; nb, neurblasts; sg, salivary gland; st, stomach; t, tail.
In insects, proliferation and apoptosis are tightly linked during the pupal stage, where many steps of differentiation occur (Eroglu and Derry, 2016). Apoptosis contributes to the patterning and development of almost all adult structures in the imaginal disks of the larva (Figure 2; Fuchs and Steller, 2011). Furthermore, epithelial cells in the larval skin and the abdominal region are removed through apoptosis and are replaced by proliferating and migrating neuroblasts – stem cell-like founder cells (Parasathy and Palli, 2008; Ninov and Martín-Blanco, 2009). In Drosophila, group apoptosis is observed in the posterior wing imaginal discs, haltere and leg discs (Eroglu and Derry, 2016). Group apoptosis occurs as dying cells emulate signals that are partially sufficient in stimulating apoptosis in their neighbors. Additionally, overexpression of both the anti-apoptotic Cdk5α and pro-apoptotic hid genes can trigger neighboring cells into undergoing apoptosis. This coordinated apoptosis likely plays an important role in regulating proper tissue growth and preventing group crowding during metamorphosis (Eroglu and Derry, 2016; Kawamoto et al., 2016). The insect steroid hormone ecdysone plays a fundamental role in the regulation of development, being responsible for cell proliferation during larval molts and the pupal transition. Specifically, ecdysone activates transcriptional cascades that lead to the expression of Reaper, Hid Grim (RHG) genes and active caspases in insects. Conversely, in adult neurons, ecdysone acts as a pro-survival factor (Fuchs and Steller, 2011).
In many marine invertebrate groups, the drastic transition from the planktonic larva a free living benthic juvenile body that occurs during settlement involves PCD (Leise et al., 2004; Gifondorwa and Leise, 2006; Hadfield, 2011; Heyland et al., 2011, 2018; Lutek et al., 2018; Hadfield et al., 2021). In indirectly developing mollusks, metamorphosis, and settlement is characterized by substantial tissue remodeling. For example, veliger larvae of bivalves, scaphopods and gastropods lose their velar lobes during this process and build the adult ganglionic nervous system (Vogeler et al., 2021). Meanwhile, the central component of the larval nervous system, the apical organ, degenerates likely through apoptosis (Kiss, 2010; Heyland et al., 2011). Cell death within this organ has been shown to be initiated by the neurotransmitter serotonin and inhibited by nitric oxide (NO) (Bishop and Brandhorst, 2001; Kiss, 2010). In fact, using nitric oxide to inhibit apoptosis in some mollusc species may inhibit metamorphosis itself in competent larvae (Gifondorwa and Leise, 2006). At the molecular level, similarities exist between the apoptotic networks of mollusks and vertebrates (Kiss, 2010; Heyland et al., 2011; Romero et al., 2011, 2015). Like vertebrates, mollusks use both intrinsic and extrinsic apoptotic mechanisms and the presence of perforin in several bivalves may suggest the existence of a perforin/granzyme apoptotic pathway as well (Romero et al., 2015). These pathways also include caspases that are homologous to vertebrate caspases such as, CASP1, CASP2, CASP3, CASP6, CASP8, and a combination caspase-3/7 (Vogeler et al., 2021). Another similarity between vertebrate and mollusk apoptotic networks is the common convergence of the execution pathway, as initiated by the cleavage of CASP3 (Romero et al., 2015). While apoptosis has not received much attention in invertebrates, there is evidence that it plays an important role in the development of many different species of marine invertebrates. In the hydrozoan Hydractinia echinate PCD activity in the larval nervous system has been described (Seipp et al., 2010). In ascidians, a marine basal chordate, apoptosis has been identified during the transition of the larval (tadpole) body to the juvenile (Karaiskou et al., 2015). Apoptosis acts as a primary driver of tissue remodeling in the larvae of the cnidarian Clytia hemisphaerica (Krasovec et al., 2021). Furthermore, an expansion of the caspase complement has been described in the ascidian Ciona intestinalis in comparison to closely related species and this expansion may be in part driven by tail regression in the tadpole (Weill et al., 2005).
PCD in Sea Urchin Development and Metamorphosis
Programmed cell death has also been studied in the development of several sea urchin species. As in other animal groups, PCD functions in oocyte development, embryonic development, and metamorphosis. Understanding the function of PCD in the developmental context of a whole animal will help to understand the relationships between PCD, hormones and metamorphosis. Here we briefly summarize this work to elucidate PCD function in the life cycle of sea urchins and then provide an up-to-date annotation of PCD related genes from the sea urchin S. purpuratus with insights into their potential functions in PCD regulation.
Sea urchins possess a functional apoptosis machinery that can act even during the development and maturation of oocytes and early embryos (Voronina and Wessel, 2001). Although PCD in actively dividing cells is widely described because of its role in tissue development, experimental evidence suggests that apoptosis occurs in cells that are not actively dividing but still metabolically active – often called quiescent cells (Voronina and Wessel, 2001). This means that not only is the apoptotic machinery present in germ cells, but it is functional as well (Voronina and Wessel, 2001). Still the function of apoptosis and other forms of programmed cell death in early embryonic development remain largely unclear. Similar to other, more studied organisms such as Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, cleavage stages are mostly characterized by necrotic and pathological cell death rather than regulated apoptosis and it is not until gastrulation that a marked increase of apoptosis has been observed (Thurber and Epel, 2007; Galasso et al., 2019). Abundant experimental evidence shows that cell death can also be induced in early sea urchin development via cytotoxic treatments, pollutants, and UV radiation (Lesser et al., 2003; Chiarelli et al., 2011). During sea urchin gastrulation apoptosis appears to be a necessary component of normal development, as pharmacological induction of apoptosis or the inhibition of caspases results in deformed larvae and reduced survival in Stronglyocentrotus purpuratus (Thurber and Epel, 2007; Heyland pers. obs.). Galasso et al. (2019) found that many death genes are differentially expressed during this important developmental process and that apoptosis and autophagy during gastrulation are likely involved in cavity formation and the removal of inner ectodermal cells. Intriguingly, induction of apoptosis in gastrulation can result in significant acceleration of larval development of the surviving embryos compared to controls (Thurber and Epel, 2007). In larval development, PCD, specifically apoptosis, play a prominent role in sculpting tissues in the arms and the ciliary band and a function of apoptosis has been proposed in the adaptive phenotypic plastic response of larval arm and ciliary band length to microalgae concentration in the environment (Thurber and Epel, 2007; Agnello and Roccheri, 2010).
Data from a broad range of animals suggests a prominent role of PCD in the process of metamorphosis as summarized above and links between PCD and sea urchin metamorphosis and settlement have been studied in several species [reviewed in Heyland and Moroz (2006)]. As in other marine invertebrate species, sea urchins with an indirect life history undergo a drastic ecological, developmental, and physiological transition between the larval and juvenile stage. Settlement, the transition from the planktonic to the benthic environment can be preceded by major morphological or developmental changes but does not have to. In fact, some species, such as cnidarian, ascidian, and echinoderm species complete a large portion of morphological changes during or after settlement (Rodriguez et al., 1993; Heyland and Hodin, 2014; Krasovec et al., 2021). In sea urchins, a hallmark of settlement is the larval arm retraction and subsequent transition from a bilateral to a pentaradial organism. During this process cell death, proliferation, migration, and differentiation occur in a relatively short period of time (Cameron et al., 1989; Heyland and Moroz, 2006; Hodin, 2006; Lutek et al., 2018). Other changes, such as the formation of the juvenile mouth and nervous system develop on a much longer time scale, taking typically days to week to fully form (Thet et al., 2004; Emlet, 2010; Fadl et al., 2017, 2019). Apoptotic cells have been identified in the arms and ciliary bands of metamorphically competent larvae (the developmental stage preceding settlement) as well as in juveniles (Roccheri et al., 2002; Lutek et al., 2018). Moreover, specific signaling molecules have been implicated in the induction of PCD in sea urchin metamorphosis and settlement. Glutamine and some of its derivatives, compounds that have been shown to accelerate metamorphic development, can induce apoptosis in the larval epithelium in the sea urchin Hemicentrotus pulcherrimus (Sato et al., 2006). Furthermore, recent research shows that histamine has a dual function in larval development of the sea urchin S. purpuratus. While it aids in the attainment of metamorphic competence, it also keeps larvae in that state by inhibiting the settlement process (Lutek et al., 2018). The latter process is, at least in part, the result of the inhibition of apoptosis in the larval arms by histamine signaling via the sea urchin histamine receptor (Lutek et al., 2018). Histamine may interact with TH signaling, a hormone that can be endogenously synthesized (Heyland et al., 2006b,a; Miller and Heyland, 2009, 2013) or derived from unicellular algae that echinoid larvae feed on (van Bergeijk et al., 2013) and that has been shown to regulate development, including metamorphic development in sea urchins and sand dollars (Heyland and Hodin, 2004; Heyland et al., 2004). Recent evidence from the sea urchin S. purpuratus also suggests that THs may signal via integrins in addition to their nuclear hormone receptor pathway (Taylor and Heyland, 2018). A distinct characteristic of the effect TH signaling in sea urchin and sand dollar larvae is the acceleration of juvenile structures while reducing the length of larval arms (Heyland and Hodin, 2004). It is the ladder effect of THs that is in part the result of PCD (Wynen and Heyland, 2021).
Apoptotic Genes in the Sea Urchin (S. purpuratus)
Due to the important role apoptosis plays in development and homeostasis, the apoptotic toolkit may become complex and contain many redundancies within specific lineages. As far as metazoans are concerned, there have been many documented cases of gene losses and expansion (Zmasek and Godzik, 2013; Romero et al., 2015; Green and Fitzgerald, 2016). This makes it difficult to provide a comprehensive overview of apoptosis in specific groups for which little functional data is available. Keeping this in mind, we used an earlier PCD annotation of the sea urchin genome as a starting point (Robertson et al., 2006) and further annotated the sea urchin apoptotic tool kit (sea urchin genome assembly and annotation v. 5.0).
As outlined above, the intrinsic pathway responds to intracellular signals produced by non-receptor mediated stimuli that leads to the activation of Bcl-2 family members. One type of non-receptor mediated stimuli that leads to intracellular signaling is exposure to ultraviolet radiation. As discussed earlier, DNA damage caused by radiation can lead to the activation of the gene p53 (Lesser et al., 2003). The protein p53 leads to a downregulation of anti-apoptotic Bcl-2 proteins which can lead to apoptosis (Bourgarel-Rey et al., 2009). The Bcl-2 family may also be responsible for the absence of early developmental apoptosis in sea urchins (Thurber and Epel, 2007). Bcl-2 members that were identified in the sea urchin genome included Bcl-2, BAX, BAK1, BAG1, BLK, and BCL10 (Table 1). In sea urchins, orthologs for both BAX and BAK1 have been identified (Table 1) and similarly to vertebrates, these proteins share a Bcl-2 homology 3 (BH3) motif at the N-terminus of the Bcl-2 domain (Robertson et al., 2006). While the function of a protein or domain can only be conclusively determined by experimental work, examining the presence or absence of specific domains or comparing their structure to known proteins can still provide a good foundation to generate hypotheses and predictions about their role. The Bcl-2 family of proteins has a few characteristic domains that, while difficult to presume their function, at least allow for possible identification. These include the BH1, BH2, BH3, and BH4 domains (Moroy et al., 2009). The BH3 domain is a particularly well-known part of the Bcl-2 protein family and has been identified as a mediator of cell death (Moroy et al., 2009). The BH3 domain appears in pro-apoptotic proteins such as BAX, BAD and BIK (Moroy et al., 2009). Another class of pro-apoptotic Bcl-2 family members called the BH3-only proteins bind with anti-apoptotic proteins like Bcl-2 and BCL2L1 through the BH3 domain in order to suppress their anti-apoptotic activity (Moroy et al., 2009). The BH domains are important for facilitating interactions between the Bcl-2 protein family members (Gurudutta et al., 2005; Aouacheria et al., 2013).
The proteins involved in the formation of the apoptosome, cycs, APAF1, and CASP9, are well-represented in sea urchins. While four predictions for APAF1-like proteins exist in the sea urchin genome, two are nearly identical and generally regarded as paralogs (Robertson et al., 2006) and one APAF1 -like protein lacks the N-terminal caspase recruitment domain (CARD) found in humans, flies and nematodes, and instead possessing a death domain (Robertson et al., 2006). The final APAF1 -like protein is very similar in architecture to human APAF1 (Table 1; Robertson et al., 2006). The CARD domain of APAF1 is important due to its interaction with the CARD domain of CASP9 (Wang et al., 2017). As mentioned previously, formation of the apoptosome can be blocked by AVEN which uses the BH1 domain of BCL2L1 to bind it and APAF1 to prevent activation of CASP9 by the apoptosome; all of which we found orthologs for (Table 1; Chau et al., 2000; Elmore, 2007). Should the apoptosome form, then CASP9 is activated (Elmore, 2007). The CASP9 family underwent an echinoderm-specific expansion which resulted in several CASP9-like proteins (Robertson et al., 2006; Agnello and Roccheri, 2010). When analyzed by Robertson et al. (2006) four CASP9-like proteins were reported and only one featured a recognizable CARD domain (LOC594735). In the updated genome, we were able to annotate three proteins that are similar to CASP9 that have a complete CARD domain (LOC586121, LOC115924698, and LOC584219) and one protein that is very similar in sequence to CASP9 and has an incomplete CARD domain (LOC586121). Furthermore, the protein that Robertson et al. (2006) originally identified, does not appear to have a CARD domain or show as much similarity to human CASP9 as the proteins reported above. The CARD domain is important for binding of procaspase-9 to APAF1 and may play a role in stabilizing interactions within the core of CASP9 (Huber et al., 2018). Since this domain is so important, it may help to identify the function of CASP9-like proteins. This function can be blocked however, when the baculoviral IAP repeat 3 (BIR3) domain of XIAP binds with CASP9 in order to prevent the start of the execution pathway (Shiozaki et al., 2003). This action can be blocked by DIABLO which competes with CASP9 for binding of the BIR3 domain (Gao et al., 2007). Additionally, binding of DIABLO with the BIR2 domain of XIAP can antagonize inhibition of CASP3 by XIAP (Gao et al., 2007). HtrA2 is able to inhibit XIAP in a similar way (Figure 1; Verhagen et al., 2002; Elmore, 2007; Obexer and Ausserlechner, 2014). The protein AIFM1, which is released from the outer mitochondrial membrane plays a role triggering DNA fragmentation and was also found upon analysis (Table 1; Elmore, 2007).
We found fewer orthologous proteins in the extrinsic complement than in the intrinsic pathway. Specifically, we were unable to find a clear ortholog of TNF, even though we found an ortholog for its receptor TNFRSF1A (Table 1). There are a few couplings that are well known in this pathway: TNF/TNFRSF1A, FasLG/Fas, TNFSF12/TNFRSF25, TNFSF10/TNFRSF10A, and TNFSF10/TNFRSF10B (Elmore, 2007). We were able to identify possible orthologs of FasLG, Fas, TNFSF12 and TNFSF10 however, we were unable to find clear orthologs for TNFRSF25, TNFRSF10A, and TNFRSF10B (Table 1). Interestingly, there seems to be similarity between TNFRSF10A, TNFRSF10B and S. purpuratus TNFR16 (LOC586563). While this similarity is relatively low, it may provide valuable insight into the extrinsic pathway of sea urchins and the absence of so many extrinsic receptors. Of the proteins that are involved in recruiting procaspase-8 to the DISC, possible orthologs for FADD and RIP were identified but not TRADD (Table 1). Like the CASP9-like protein family, there seems to have been some expansion of the CASP8 family in the sea urchin genome (Robertson et al., 2006; Agnello and Roccheri, 2010). Upon analysis, Robertson et al. (2006) found five caspase-like-proteins, two of which contained clear linkages to DEDs, similar to human CASP8. Of these two proteins, only one has a complete caspase domain and therefore is a strong candidate for a human CASP8 ortholog (Table 1; Robertson et al., 2006).
The granzyme/perforin pathway contains fewer proteins than the intrinsic and extrinsic pathway but still seems to be well represented in the sea urchin genome (Table 1). In the perforin/granzyme pathway, sea urchin orthologs for GZMA, GZMB, CASP10, and NME1 were identified (Table 1).
All three of these pathways converge at the execution pathway which is also well represented in the sea urchin genome, likely due to the important role caspases play in this part of apoptosis (Table 1). One of the biggest differences between mammals and sea urchins seems to be the lack of clear distinction between CASP3 and CASP7 (Robertson et al., 2006). Due to this fact, it is difficult to conclusively identify separate orthologs for these caspases and the LOCs represented in the table may indicate paralogs of one gene (Table 1). We have been able to identify orthologs for CASP6, PARP1 and SPTAN1 (Table 1).
Conclusion
As others have pointed out, endocrine mechanisms are intricately linked to the mechanisms underlying metamorphic transitions in animals and PCD is one important output of these mechanisms (Nakajima et al., 2005; Heyland and Moroz, 2006; Mané-Padrós et al., 2010; Heyland et al., 2018). In addition to frogs and flies, for which many of these mechanisms have been studied in detail, our analysis also revealed that PCD is an intricate part of metamorphosis and settlement of many marine invertebrate groups with indirect life histories. Specifically, the target of PCD in these groups is the larval nervous system and feeding and digestive structures such as the ciliated band and larval arms. Studies into the molecular and cellular mechanisms underlying these processes require further research. While the annotation of PCD related genes in target species can be difficult due to species specific expansions and diversification, it is a valuable exercise and can help shed light on the ecological and environmental factors driving these processes (Lespinet et al., 2002). For example, caspases in sea urchins have undergone such expansions and may play a role in important developmental processes such as metamorphosis and settlement by removing larval tissue. Although necessary to switch between environments, metamorphosis, and settlement can be rapid and resource intense processes and elucidating the putative function of caspases in these processes may help shed light onto the environmental conditions driving the expansion of the caspase family in sea urchins. Finally, the role of endocrine mechanisms in the regulation of metamorphosis and settlement as well as PCD may be significantly more wide-spread and future studies should explore this link in more details in marine invertebrate phyla.
Author Contributions
HW and AH contributed to the concept and writing of the article. Both authors contributed to the article and approved the submitted version.
Funding
Funding for this work was provided, in part by NSERC Discovery Grant to AH (400230).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank members of the Heylandlab for feedback on earlier versions of the manuscript and help with the annotation of apoptosis genes form the sea urchin.
References
Agnello, M., and Roccheri, M. C. (2010). Apoptosis: focus on sea urchin development. Apoptosis 15, 322–330. doi: 10.1007/s10495-009-0420-0
Amiesen, J. C. (2002). On the origin, evolution and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 9, 367–393. doi: 10.1038/sj.cdd.4400950
Andrade, R., Crisol, L., Prado, R., Boyano, M. D., Arluzea, J., and Aréchaga, J. (2010). Plasma membrane and nuclear envelope integrity during the blebbing stage of apoptosis: a time-lapse study. Biol. Cell 102, 25–35. doi: 10.1042/bc20090077
Aouacheria, A., Rech, de Laval, V., Combet, C., and Hardwick, M. J. (2013). Evolution of Bcl-2 homology motifs: homology versus homoplasy. Trends Cell Biol. 23, 103–111. doi: 10.1016/j.tcb.2012.10.010
Balon, E. (1999). Alternative ways to become a juvenile or a definitive phenotype (and on some persisting linguistic offenses). Environ. Biol. Fishes 56, 17–38. doi: 10.1007/978-94-017-3678-7_2
Bishop, A., and Anderson, J. E. (2005). NO signaling in the CNS: from the physiological to the pathological. Toxicology 208, 193–205. doi: 10.1016/j.tox.2004.11.034
Bishop, C. D., and Brandhorst, B. P. (2001). NO/cGMP signaling and HSP90 activity represses metamorphosis in the sea urchin Lytechinus pictus. Biol. Bull. 201, 394–404. doi: 10.2307/1543617
Bishop, C. D., and Brandhorst, B. P. (2003). On nitric oxide signaling, metamorphosis, and the evolution of biphasic life cycles. Evol. Dev. 5, 542–550. doi: 10.1046/j.1525-142x.2003.03059.x
Bishop, C. D., Huggett, M. J., Heyland, A., Hodin, J., and Brandhorst, B. P. (2006). Interspecific variation in metamorphic competence in marine invertebrates: the significance for comparative investigations into the timing of metamorphosis. Integr. Comp. Biol. 46, 662–682. doi: 10.1093/icb/icl043
Bourgarel-Rey, V., Savry, A., Hua, G., Carré, M., Bressin, C., Chacon, C., et al. (2009). Transcriptional down-regulation of Bcl-2 by vinorelbine: identification of a novel binding site of p53 on Bcl-2 promoter. Biochem. Pharmacol. 78, 1148–1156. doi: 10.1016/j.bcp.2009.06.025
Brown, D. D., Cai, L., Das, B., Marsh-Armstrong, N., Schreiber, A. M., and Juste, R. (2005). Thyroid hormone controls multiple independent programs required for limb development in Xenopus laevis metamorphosis. Proc. Natl. Acad. Sci. USA 35, 12455–12458. doi: 10.1073/pnas.0505989102
Cameron, A. R., Britten, R. J., and Davidson, E. H. (1989). Expression of two actin genes during larval development in the sea urchin Strongylocentrotus purpuratus. Mol. Reprod. Dev. 1, 149–155. doi: 10.1002/mrd.1080010302
Chau, B. N., Cheng, E. H., Kerr, D. A., and Hardwick, J. M. (2000). Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol. Cell 6, 31–40. doi: 10.1016/s1097-2765(05)00021-3
Chiarelli, R., Agnello, M., and Roccheri, M. C. (2011). Sea urchin embryos as a model system for studying autophagy induced by cadmium stress. Autophagy 7, 1028–1034. doi: 10.4161/auto.7.9.16450
Coleman, M. L., Sahai, E. A., Yeo, M., Bosch, M., Dewar, A., and Olson, M. F. (2001). Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3, 339–345. doi: 10.1038/35070009
Daniel, P. T., Schulze-Osthoff, K., Belka, C., and Güner, D. (2003). Guardians of cell death: the Bcl-2 family proteins. Essays Biochem. 39, 73–88. doi: 10.1042/bse0390073
Das, B., Cai, L., Carter, M. G., Piao, Y. L., Sharov, A. A., Ko, M. S. H., et al. (2006). Expression changes at metamorphosis induced by thyroid hormone in Xenopus laevis tadpoles. Dev. Biol. 291, 342–355. doi: 10.1016/j.ydbio.2005.12.032
Das, B., Schreiber, A. M., Huang, H., and Brown, D. D. (2002). Multiple thyroid hormone-induced muscle growth and death programs during metamorphosis in Xenopus laevis. Proc. Natl. Acad. Sci. USA. 99, 12230–12235. doi: 10.1073/pnas.182430599
Davidson, B., and Swalla, B. J. (2002). A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response. Development 129, 4739–4751. doi: 10.1242/dev.129.20.4739
Degnan, B. M. (2001). “Settlement and metamorphosis of the tropical ascidian Herdmania curvata,” in The Biology of Ascidians, eds H. Sawada, H. Yokosawa, and C. C. Lambert (Tokyo: Springer), 258–263. doi: 10.1007/978-4-431-66982-1_41
Degterev, A., and Yuan, J. (2008). Expansion and evolution of cell death programs. Nat. Rev. Mol. Cell Biol. 9, 378–290. doi: 10.1038/nrm2393
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. doi: 10.1016/j.cell.2012.03.042
Dondelinger, Y., Hulpiau, P., Saeys, Y., Bertrand, M. J. M., and Vandenabeele, P. (2016). An evolutionary perspective on the necroptotic pathway. Trends Cell Biol. 26, 721–732. doi: 10.1016/j.tcb.2016.06.004
Du, C., Fang, M., Li, Y., Li, L., and Wang, X. (2000). Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42. doi: 10.1016/s0092-8674(00)00008-8
Emlet, R. B. (2010). Morphological evolution of newly metamorphosed sea urchins—a phylogenetic and functional analysis. Integr. Comp. Biol. 50, 571–588. doi: 10.1093/icb/icq073
Eroglu, M., and Derry, W. B. (2016). Your neighbours matter – non-autonomous control of apoptosis in development and disease. Cell Death Differ. 23, 1110–1118. doi: 10.1038/cdd.2016.41
Fadl, A. E. A., Mahfouz, M. E., El-Gamal, M. M. T., and Heyland, A. (2017). New biomarkers of post-settlement growth in the sea urchin Strongylocentrotus purpuratus. Heliyon 3:e00412. doi: 10.1016/j.heliyon.2017.e00412
Fadl, A. E. A., Mahfouz, M. E., El-Gamal, M. M. T., and Heyland, A. (2019). Onset of feeding in juvenile sea urchins and its relation to nutrient signalling. Invertebr. Reprod. Dev. 63, 11–22. doi: 10.1080/07924259.2018.1513873
Faunes, F., and Larraín, J. (2016). Conservation in the involvement of heterochronic genes and hormones during developmental transitions. Dev. Biol. 416, 3–17. doi: 10.1016/j.ydbio.2016.06.013
Fuchs, Y., and Steller, H. (2011). Programmed cell death in animal development and disease. Cell 147, 742–758. doi: 10.1016/j.cell.2011.10.033
Fulda, S., and Debatin, K. M. (2006). Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 25, 4798–4811. doi: 10.1038/sj.onc.1209608
Galasso, C., D’Aniello, S., Sansone, A., and Romano, G. (2019). Identification of cell death genes in sea urchin Paracentrotus lividus and their expression patterns during embryonic development. Genome Biol. Evol. 11, 586–596. doi: 10.1093/gbe/evz020
Gao, Z., Tian, Y., Wang, J., Yin, Q., Wu, H., Li, Y. M., et al. (2007). A dimeric Smac/diablo peptide directly relieves caspase-3 inhibition by XIAP. dynamic and cooperative regulation of XIAP by Smac/Diablo. J. Biol. Chem. 282, 30718–30727. doi: 10.1074/jbc.m705258200
Gifondorwa, D. J., and Leise, E. M. (2006). Programmed cell death in the apical ganglion during larval metamorphosis of the marine mollusc Ilyanassa obsolete. Biol. Bull. 210, 109–120. doi: 10.2307/4134600
Green, D. R., and Fitzgerald, P. (2016). Just so stories about the evolution of apoptosis. Curr. Biol. 26, R620–R627.
Gurudutta, G. U., Verma, Y. K., Singh, V. K., Gupta, P., Raj, H. G., Sharma, R. K., et al. (2005). Structural conservation of residues in BH1 and BH2 domains of Bcl-2 family proteins. FEBS. Lett. 579, 3503–3507. doi: 10.1016/j.febslet.2005.05.015
Götz, R., Wiese, S., Takayama, S., Camarero, G., Rossol, W., Schweizer, U., et al. (2005). Bag1 is essential for differentiation and survival of hematopoietic and neuronal cells. Nat. Neurosci. 8, 1169–1178. doi: 10.1038/nn1524
Hadfield, M. G. (2000). Why and how marine-invertebrate larvae metamorphose so fast. Semin. Cell Dev. Biol. 11, 437–443. doi: 10.1006/scdb.2000.0197
Hadfield, M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Hadfield, M. G., Freckelton, M. L., and Nedved, B. T. (2021). The natural sequence of events in larval settlement and metamorphosis of Hydroides elegans (Polychaeta; Serpulidae). PLoS One 16:e0249692. doi: 10.1371/journal.pone.0249692
Hanson, B. (2016). Necroptosis: a new way of dying? Cancer Biol. Ther. 17, 899–910. doi: 10.1080/15384047.2016.1210732
Hegde, R., Srinivasula, S. M., Ahmad, M., Fernandes-Alnemri, T., and Alnemri, E. S. (1998). Blk, a BH3-containing mouse protein that interacts with Bcl-2 and Bcl-xL, is a potent death agonist. J. Biol. Chem. 273, 7783–7786. doi: 10.1074/jbc.273.14.7783
Heyland, A., and Hodin, J. (2004). Heterochronic developmental shift caused by thyroid hormone in larval sand dollars and its implications for phenotypic plasticity and the evolution of nonfeeding development. Evolution 58, 524–538. doi: 10.1554/03-243
Heyland, A., and Hodin, J. (2014). A detailed staging scheme for late larval development in Stronglyocentrotus purpuratus focused on readily-visible juvenile structures within the rudiment. BMC Dev. Biol. 14:22. doi: 10.1186/1471-213x-14-22
Heyland, A., and Moroz, L. L. (2006). Signaling mechanisms underlying metamorphic transitions in animals. Integr. Comp. Biol. 46, 743–759. doi: 10.1093/icb/icl023
Heyland, A., Price, D., Bodnarova-Buganova, M., and Moroz, L. (2006a). Thyroid hormone metabolism and peroxidase function in two non-chordate animals. J. Exp. Zool. 306B, 551–566. doi: 10.1002/jez.b.21113
Heyland, A., Reitzel, A. M., and Hodin, J. (2004). Thyroid hormones determine developmental mode in sand dollars (Echinodermata: Echinoidea). Evol. Dev. 6, 382–392. doi: 10.1111/j.1525-142x.2004.04047.x
Heyland, A., Reitzel, A. M., Price, D. A., and Moroz, L. L. (2006b). Endogenous thyroid hormone synthesis in facultative planktotrophic larvae of the sand dollar Clypeaster rosaceus: implications for the evolutionary loss of larval feeding. Evol. Dev. 8, 568–579. doi: 10.1111/j.1525-142x.2006.00128.x
Heyland, A., Schuh, N., and Rast, J. (2018). Sea urchin larvae as a model for postembryonic development. Mar. Organ. Model Syst. Biol. Med. 65, 137–161. doi: 10.1007/978-3-319-92486-1_8
Heyland, A., Vue, Z., Voolstra, C. R., Medina, M., and Moroz, L. L. (2011). Developmental transcriptome of Aplysia californica. J. Exp. Zool. B Mol. Dev. Evol. 316B, 113–134. doi: 10.1002/jez.b.21383
Hodin, J. (2006). Expanding networks: signaling components in and a hypothesis for the evolution of metamorphosis. Integr. Comp. Biol. 46, 719–742. doi: 10.1093/icb/icl038
Hodin, J., Lutek, K., and Heyland, A. (2016). A newly identified left-right asymmetry in larval sea urchins. R. Soc. Open. Sci. 3:160139. doi: 10.1098/rsos.160139
Holze, C., Michaudel, C., Mackowiak, C., Haas, D. A., Benda, C., Hubel, P., et al. (2018). Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 19, 130–140. doi: 10.1038/s41590-017-0013-y
Huber, K. L., Serrano, B. P., and Hardy, J. A. (2018). Caspase-9 CARD : core domain interactions require a properly formed active site. Biochem. J. 475, 1177–1196. doi: 10.1042/bcj20170913
Ishizuya-Oka, A., Hasebe, T., and Shi, Y. B. (2010). Apoptosis in amphibian organs during metamorphosis. Apoptosis 15, 350–364. doi: 10.1007/s10495-009-0422-y
Kalkavan, H., and Green, D. R. (2018). MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 25, 46–55. doi: 10.1038/cdd.2017.179
Karaiskou, A., Swalla, B. J., Sasakura, Y., and Chambon, J. P. (2015). Metamorphosis in solitary ascidians. Genesis 53, 34–47. doi: 10.1002/dvg.22824
Kawamoto, Y., Nakajima, Y. I., and Kuranaga, E. (2016). Apoptosis in cellular society: communication between apoptotic cells and their neighbors. Int. J. Mol. Sci. 17:2144. doi: 10.3390/ijms17122144
Kerr, J. F., Wyllie, A. H., and Currie, A. R. (1972). Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257. doi: 10.1038/bjc.1972.33
Kiss, T. (2010). Apoptosis and its functional significance in molluscs. Apoptosis 15, 313–321. doi: 10.1007/s10495-009-0446-3
Krasovec, G., Pottin, K., Rosello, M., Quéinnec, É, and Chambon, J. P. (2021). Apoptosis and cell proliferation during metamorphosis of the planula larva of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev. Dyn. doi: 10.1002/dvdy.376 Online ahead of print.
Leise, E. M., Kempf, S. C., Durham, N. R., and Gifondorwa, D. J. (2004). Induction of metamorphosis in the marine gastropod Ilyanassa Obsoleta: 5HT, NO and programmed cell death. Acta Biol. Hung. 55, 293–300. doi: 10.1556/abiol.55.2004.1-4.35
Lespinet, O., Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 12, 1048–1059. doi: 10.1101/gr.174302
Lesser, M. P., Kruse, V. A., and Barry, T. M. (2003). Exposure to ultraviolet radiation causes apoptosis in developing sea urchin embryos. J. Exp. Biol. 206, 4097–4103. doi: 10.1242/jeb.00621
Lutek, K., Dhaliwal, R. S., Van Raay, T. J., and Heyland, A. (2018). Sea urchin histamine 1 regulates programmed cell death in larval Stronglyocentrotus purpuratus. Sci. Rep. 8, 4002–4014.
Mané-Padrós, D., Cruz, J., Vilaplana, L., Nieva, C., Josefa Cruz, Ureña, E., et al. (2010). The hormonal pathway controlling cell death during metamorphosis in a hemimetabolous insect. Dev. Biol. 346, 150–160. doi: 10.1016/j.ydbio.2010.07.012
Miller, A. E. M., and Heyland, A. (2009). Endocrine interactions between plants and animals: implications of exogenous hormone sources for the evolution of hormone signaling. Gen. Comp. Endocrinol. 166, 455–461. doi: 10.1016/j.ygcen.2009.09.016
Miller, A. E. M., and Heyland, A. (2013). Iodine accumulation in sea urchin larvae is dependent on peroxide. J. Exp. Biol. 216, 915–926.
Moroy, G., Martin, E., Dejaegere, A., and Stote, R. H. (2009). Molecular basis for Bcl-2 homology 3 domain recognition in the Bcl-2 protein family: identification of conserved hot spot interactions. J. Biol. Chem. 284, 17499–17511. doi: 10.1074/jbc.m805542200
Nakajima, K., Fujimoto, K., and Yaoita, Y. (2005). Programmed cell death during amphibian metamorphosis. Semin. Cell Dev. Biol. 16, 271–280. doi: 10.1016/j.semcdb.2004.12.006
Nakayama-Ishimura, A., Chambon, J. P., Horie, T., Satoh, N., and Sasakura, Y. (2009). Delineating metamorphic pathways in the ascidian Ciona intestinalis. Dev. Biol. 326, 357–367. doi: 10.1016/j.ydbio.2008.11.026
Ninov, N., and Martín-Blanco, E. (2009). Changing gears in the cell cycle: histoblasts and beyond. Fly 3, 286–289. doi: 10.4161/fly.10443
Obexer, P., and Ausserlechner, M. J. (2014). X-linked inhibitor of apoptosis protein – a critical death resistance regulator and therapeutic target for personalized cancer therapy. Front. Oncol. 4:197.
Oliver, F. J., de la Rubia, G., Rolli, V., Ruiz-Ruiz, M. C., de Murcia, G., and Me’nissier-de Murcia, J. (1998). Importance of Poly(ADP-ribose) Polymerase and its cleavage in apoptosis: lesson from an uncleavable mutant. J. Biol. Chem. 50, 33533–33539.
Parasathy, R., and Palli, S. R. (2008). Proliferation and differentiation of intestinal stem cells during metamorphosis of the Red Flour Beetle, Tribolium castaneum. Dev. Dyn. 237, 893–908. doi: 10.1002/dvdy.21475
Paris, M., Escriva, H., Schubert, M., Brunet, F., Brtko, J., Ciesielski, F., et al. (2008). Amphioxus postembryonic development reveals the homology of chordate metamorphosis. Curr. Biol. 18, 825–830. doi: 10.1016/j.cub.2008.04.078
Paris, M., Hillenweck, A., Bertrand, S., Delous, G., Escriva, H., Zalko, D., et al. (2010). Active metabolism of thyroid hormone during metamorphosis of amphioxus. Integr. Comp. Biol. 50, 63–74. doi: 10.1093/icb/icq052
Robertson, A. J., Croce, J., Carbonneau, S., Voronina, E., Miranda, E., McClay, D. R., et al. (2006). The genomic underpinnings of apoptosis in Stronglyocentrotus purpuratus. Dev. Biol. 300, 321–334. doi: 10.1016/j.ydbio.2006.08.053
Roccheri, M. C., Tipa, C. T., Bonaventura, R., and Matranga, V. (2002). Physiological and induced apoptosis in sea urchin larvae undergoing metamorphosis. Int. J. Dev. Biol. 46, 801–806.
Rodriguez, S. R., Ojeda, F. P., and Inestrosa, N. C. (1993). Settlement of benthic marine invertebrates. Mar. Ecol. Prog. Ser. 97, 193–207. doi: 10.3354/meps097193
Romero, A., Estevez-Calvar, N., Dios, S., Figueras, A., Novoa, B., and Bergmann, A. (2011). New insights into the apoptotic process in mollusks: characterization of caspase genes in Mytilus galloprovincialis. PLoS. One 6:e17003. doi: 10.1371/journal.pone.0017003
Romero, A., Novoa, B., and Figueras, A. (2015). The complexity of apoptotic cell death in mollusks: an update. Fish Shellfish Immunol. 46, 79–87. doi: 10.1016/j.fsi.2015.03.038
Sachs, L. M., and Buccholz, D. R. (2019). Insufficiency of Thyroid Hormone in frog metamorphosis and the role of glucocorticoids. Front. Endocrinol. 10:287.
Sato, Y., Kaneko, H., Negishi, S., and Yazaki, I. (2006). Larval arm resorption proceeds concomitantly with programmed cell death during metamorphosis of the sea urchin Hemicentrotus pulcherrimus. Cell Tissue Res. 326, 851–860. doi: 10.1007/s00441-006-0212-6
Seipp, S., Schmich, J., Will, B., Schetter, E., Plickert, G., and Leitz, T. (2010). Neuronal cell death during metamorphosis of Hydractina echinata (Cnidaria, Hydrozoa). Invert. Neurosci. 10, 77–91. doi: 10.1007/s10158-010-0109-7
Shiozaki, E. N., Jijie, C., Rigotti, D. J., Riedl, S. J., Li, P., Srinivasula, S. M., et al. (2003). Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 11, 519–517. doi: 10.1016/s1097-2765(03)00054-6
Sun, G., Fu, L., and Shi, Y. B. (2014). Epigenetic regulation of thyroid hormone-induced adult intestinal stem cell development during anuran metamorphosis. Cell Biosci. 4:73. doi: 10.1186/2045-3701-4-73
Suzuki, Y., Imai, Y., Nakayama, H., Takahashi, K., Takio, K., and Takahashi, R. (2001). A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8, 613–621. doi: 10.1016/s1097-2765(01)00341-0
Tata, J. R. (1993). Gene expression during metamorphosis: an ideal model for post-embryonic development. Bioessays 15, 239–248. doi: 10.1002/bies.950150404
Taylor, E., and Heyland, A. (2018). Thyroid hormones accelerate initiation of skeletogenesis via MAPK (ERK1/2) in larval sea urchins (Stronglyocentrotus purpuratus). Front. Endocrinol. 9, 439–439. doi: 10.3389/fendo.2018.00439
Tettamanti, G., and Casartelli, M. (2019). Cell death during complete metamorphosis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 374:1783.
Thet, M. M., Noguchi, M., and Yazaki, I. (2004). Larval and juvenile development of the echinometrid sea urchin Colobocentrotus mertensii: emergence of the peculiar form of spines. Zool. Sci. 21, 265–274. doi: 10.2108/zsj.21.265
Thurber, R. V., and Epel, D. (2007). Apoptosis in early development of the sea urchin, Strongylocentrotus purpuratus. Dev. Biol. 303, 336–346. doi: 10.1016/j.ydbio.2006.11.018
van Bergeijk, S. A., Hernández Javier, L., Heyland, A., Manchado, N., and Pedro Cañavate, J. (2013). Uptake of iodide in the marine haptophyte Isochrysis sp. (T.ISO) driven by iodide oxidation. J. Phycol. 49, 640–647. doi: 10.1111/jpy.12073
Verhagen, A. M., Silke, J., Ekert, P. G., Moritz, R. L., Simpson, R. J., and Vaux, D. (2002). HtrA2 promotes cell death through Its Serine Protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 277, 445–454. doi: 10.1074/jbc.m109891200
Vogeler, S., Carboni, A., Li, X., and Joyce, A. (2021). Phylogenetic analysis of the caspase family in bivalves: implications for programmed cell death, immune response and development. BMC Genom. 22:80.
Voronina, E., and Wessel, G. M. (2001). Apoptosis in sea urchin oocytes, eggs, and early embryos. Mol. Reprod. Dev. 60, 553–561. doi: 10.1002/mrd.1120
Wang, L., Qiao, Q., and Wu, H. (2017). Understanding CARD tricks in apoptosomes. Structure 25, 575–577. doi: 10.1016/j.str.2017.03.013
Weill, M., Philips, A., Chourrout, D., and Fort, P. (2005). The caspase family in urochordates: distinct evolutionary fates in ascidians and larvaceans. Biol. Cell 97, 857–866. doi: 10.1042/bc20050018
Williams, S. T., Smith, A. N., Cianci, C. D., Morrow, J. S., and Brown, T. L. (2003). Identification of the primary caspase 3 cleavage site in alpha II-spectrin during apoptosis. Apoptosis 8, 353–361.
Wong, E., Mölter, J., Anggono, V., Degnan, S. M., and Degnan, B. M. (2019). Co-expression of synaptic genes in the sponge Amphimedon queenslandica uncovers ancient neural submodules. Sci. Rep. 10:15781.
Wray, G. A., and Raff, R. A. (1990). Origins of lineage founder cells in the direct-developing sea urchin Heliocidaris erythrogamma. Dev. Biol. 141, 41–54. doi: 10.1016/0012-1606(90)90100-w
Wynen, H., and Heyland, A. (2021). Thyroid hormones induce programmed cell death in sea urchin (Stronglyocentrotus purpuratus) larval arms prior to metamorphosis. (Manuscript in preparation).
Yui, D., Yoneda, T., Oono, K., Katayama, T., Imaizumi, K., and Tohyama, M. (2001). Interchangeable binding of Bcl10 to TRAF2 and cIAPs regulates apoptosis signaling. Oncogene 20, 4317–4323. doi: 10.1038/sj.onc.1204576
Keywords: apoptosis, metamorphosis, PCD, hormones, settlement, life history, evolution
Citation: Wynen H and Heyland A (2021) Hormonal Regulation of Programmed Cell Death in Sea Urchin Metamorphosis. Front. Ecol. Evol. 9:733787. doi: 10.3389/fevo.2021.733787
Received: 30 June 2021; Accepted: 06 August 2021;
Published: 08 September 2021.
Edited by:
Sarah Kelly McMenamin, Boston College, United StatesReviewed by:
Yun-Bo Shi, National Institutes of Health (NIH), United StatesMarkus Friedrich, Wayne State University, United States
Copyright © 2021 Wynen and Heyland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannah Wynen, aHd5bmVuQHVvZ3VlbHBoLmNh; Andreas Heyland, YWhleWxhbmRAdW9ndWVscGguY2E=
 Hannah Wynen
Hannah Wynen Andreas Heyland
Andreas Heyland