- 1Department of Biology, University of British Columbia, Kelowna, BC, Canada
- 2Ecosystem Conservation Team, Protected Areas Establishment and Conservation Directorate, Parks Canada Agency, Gatineau, QC, Canada
- 3Advanced Conservation Strategies, Córdoba, Spain
- 4Coastal Conservation, Tappen, BC, Canada
Invasive species are major contributors to global biodiversity decline. Invasive mammalian species (IMS), in particular, have profound negative effects in island systems that contain disproportionally high levels of species richness and endemism. The eradication and control of IMS have become important conservation tools for managing species invasions on islands, yet these management operations are often subject to failure due to knowledge gaps surrounding species- and system-specific characteristics, including invasion pathways and contemporary migration patterns. Here, we synthesize the literature on ways in which genetic and genomic tools have effectively informed IMS management on islands, specifically associated with the development and modification of biosecurity protocols, and the design and implementation of eradication and control programs. In spite of their demonstrated utility, we then explore the challenges that are preventing genetics and genomics from being implemented more frequently in IMS management operations from both academic and non-academic perspectives, and suggest possible solutions for breaking down these barriers. Finally, we discuss the potential application of genome editing to the future management of invasive species on islands, including the current state of the field and why islands may be effective targets for this emerging technology.
Introduction
When first discovered 378 years ago (c. 1502), St. Helena was densely covered with a luxuriant forest vegetation, the trees overhanging the seaward precipices and covering every part of the surface with an evergreen mantle… the general aspect of the island is now so barren and forbidding that some persons find it difficult to believe it was once green and fertile…This irreparable destruction was caused in the first place by goats, which were introduced by the Portuguese in 1513, and increased so rapidly that in 1588 they existed in thousands. These animals are the greatest of all foes to trees, because they eat off the young seedlings, and thus prevent the natural restoration of the forest.
Alfred Russel Wallace, Island Life (1880) pp. 284–285
The devastating impacts of invasive species on native biotas have long been recognized, as documented by 18th and 19th century naturalists-explorers (Simberloff, 2013), including Wallace (1880) in the passage above. Today, invasive species are considered among the most significant threats to global biodiversity, impacting native species, community structures, and entire ecosystems (Clavero and Garcia-Berthou, 2005; Bellard et al., 2016; Doherty et al., 2016; Spatz et al., 2017). The economic consequences of invasive species are just as severe, with costs associated with mitigating their impacts exceeding $220 billion annually in the United States alone (Pimentel, 2011). Invasive species can also negatively impact the physical and spiritual well-being of humans through disease transmission (Kosoy et al., 2015; Chinchio et al., 2020), decline in food security, and loss of connection to indigenous food systems (Huambachano, 2019).
Islands, in particular, have suffered an accelerated loss of species, mainly caused by the introduction of invasive species (Bellard et al., 2016). Hosting disproportionately high levels of biodiversity and endemicity relative to their area (∼15% of all terrestrial species on ∼5.5% of terrestrial surface area; Kier et al., 2009), islands account for 61% of all documented extinctions since 1500 CE (Tershy et al., 2015). In addition to biodiversity loss, invasive species contribute to the critically endangered conservation status of insular flora and fauna, and can directly and indirectly affect island economies, livelihoods, food security, human and non-human community health, infrastructure and culture (Tershy et al., 2015; Russell et al., 2017; de Wit et al., 2020).
One of the most important threats to remaining insular biodiversity and the primary driver of animal extinctions on islands are invasive mammalian species (IMS), including rodents, goats and cats among others (Doherty et al., 2016). The eradication of IMS by governmental agencies and non-governmental organizations have led to substantial conservation gains, with positive distributional and/or demographic responses documented for ∼6% of all highly threatened mammals, birds and reptiles on the IUCN Red List (Jones et al., 2016). Yet, despite the growing frequency of IMS eradications as a conservation strategy, >10% fail to meet their targeted management outcomes (Simberloff et al., 2018). Given the demonstrated benefits to island biodiversity of IMS eradication/reduction, there is a need to consider alternative approaches for informing best practices and improving management success; here we argue that genetic and genomic tools can help fill some of these knowledge gaps to increase the scale, scope and pace of IMS eradications from islands.
In this review, we first summarize general management strategies related to IMS on islands including trends in eradication performance and reasons such activities may fail. We next synthesize the literature on ways in which genetic and genomic tools have effectively informed the development and modification of biosecurity protocols, and the design and implementation of eradication and control programs. In spite of their demonstrated utility, we then discuss impediments that are preventing genetic and genomic tools from becoming more routinely implemented into IMS management strategies and suggest potential solutions for breaking down these barriers. Lastly, we discuss the promise of genome editing for transforming IMS management moving forward and why islands may offer significant advantages for continued research and potential deployment.
Current IMS Management on Islands
General Management Strategies
Management of IMS on islands can be divided into two general strategies: (1) eradication; and (2) control. Eradication refers to the complete (100%) removal of the introduced species, while control involves sustained harvesting of the introduced species to mitigate negative impacts. The choice between these two general strategies depends on the specific management goals developed within the context of broader ecological (Oppel et al., 2019), economic (Epanchin-Niell, 2017), socio-political (La Morgia et al., 2017; Roberts et al., 2018), and cultural (Bellis et al., 2019) considerations. For example, eradication of IMS is not always necessary for achieving conservation success, which is usually evaluated based on endemic species recovery and/or ecosystem restoration (Simberloff et al., 2011). Alternatively, managers may employ control focused on minimizing impacts of IMS through removal of individuals (Cromarty et al., 2002) when the anticipated conservation gains fail to justify the higher initial costs of eradications (Fraser et al., 2006; Helmstedt et al., 2016) or when eradication is not feasible (Campbell et al., 2015).
Eradication Performance Trends
The past 20 years have seen tremendous progress in our understanding of the many threats posed by IMS on islands, conditions for eradication feasibility, and necessary steps for increasing the scale, scope and pace of management operations. This work has been effectively summarized in the proceedings of three international conferences on island invasives over that period (Veitch and Clout, 2002; Veitch et al., 2011, 2019). It remains unclear, however, whether these knowledge gains have translated into an improved frequency of IMS eradication success. Simberloff et al. (2018) used the Database of Island Invasive Species Eradications (DIISE) to evaluate the frequency and success of island-wide eradication attempts for various IMS, with entries dating back over a century. For invasive mammals, the authors identified 1086 eradication attempts, 67% of which occurred in recent times (after 1995; Simberloff et al., 2018). Despite the increased volume of attempts post-1995, the overall frequency of successful eradications during this period (84%) did not significantly differ from the overall rate (85.1%; Simberloff et al., 2018).
The increase in recent eradication attempts can be largely attributed to our improved understanding concerning the extent and magnitude of threats posed by IMS on islands (McCreless et al., 2016). Awareness of IMS impacts is greater than ever before and the attitude of the scientific community is becoming more optimistic and ambitious regarding the scale, scope and complexity of eradications (Veitch et al., 2019). Managers in IMS eradications are guided by well-established principles of eradication (Cromarty et al., 2002) that have been rendered into best practice guidelines for various taxa that simplify IMS management strategies on islands, contextualized using real-world applications (Ramsey and Will, 2012; Keitt et al., 2015; Broome et al., 2017). Together, this awareness has led to more initiatives and funding available for invasion biology research and applied IMS management, enabling innovations in control methods designed to be more effective in removing the target species and less detrimental to non-target species and surrounding ecosystems (Donlan et al., 2003; Campbell K. J. et al., 2015; Carter et al., 2016; Campbell et al., 2019). For small mammals, successful use of rodenticides in particular has greatly benefited from aerial broadcasting techniques that allow managers to target much larger islands (Howald et al., 2007). Use of helicopters and semi-automatic firearms has yielded similar successes for large mammal eradications, from primates (Hanson et al., 2019) to ungulates (Macdonald et al., 2019).
While these advances have resulted in larger, more complex, and more frequent eradications performed with similar levels of success (Veitch et al., 2019), approximately one in ten eradication attempts still fails (Simberloff et al., 2018). The outcome of failed eradications can be significant; targeted populations typically return to pre-eradication densities or higher, thereby rendering any conservation gains temporary. At the same time, failures lead to additional financial costs to complete the eradication, while also incurring future opportunity costs that delay other interventions necessary to combat the massive scope of global biodiversity loss (Gerber, 2016). In addition, failures can diminish confidence in eradication as a viable conservation tool, threatening social license for any future attempts. Consequently, improving the frequency of eradication success on islands requires a thorough understanding of why eradications fail in the first place.
Why Do Eradications Fail?
The three main causes of eradication failure include: (1) incomplete operation that allowed individuals to survive; (2) completed operation, but individuals survived; or (3) completed operation that was initially successful, but the target area was reinvaded (summarized in Kappes et al., 2019). Although the above framework was developed for invasive rodents, it is relevant to the eradication of IMS from islands writ large. For category 1 failures, many of the causes are similar across species and they consist of logistical (e.g., inadequate funding), social (e.g., loss of social license), and/or operational issues (e.g., use of unskilled teams or removal tools ineffectively applied; Kappes et al., 2019).
The remaining two categories, however, are each unique such that they can be heavily dependent on species-specific life history and/or characteristics of the local ecosystem. For instance, common drivers of category 2 failures (i.e., survival) of rats have been due to unexpected levels of bait competition from non-target species, perennial breeding of the target species, and more preferential/abundant food resources versus bait (Keitt et al., 2015). Likewise, influences that lead to failure to eradicate large mammals from islands differ from rodents partially due to differences in eradication methods (e.g., delivery of rodenticide to small mammals versus active hunting of large mammals), but also because of species-specific characteristics. For example, forest-dwelling ungulates such as deer may inhabit densely forested environments that limit their detection, even at high-densities. In turn, reduced detection probabilities render common eradication methods that rely on visual sighting, such as ground or aerial hunting, less effective. Overall, category 2 failures are perhaps most hindered by an inability to detect and respond to species at low densities, as recently evidenced in red deer (Cervus elaphus; Macdonald et al., 2019), mink (Neovison vison; Macleod et al., 2019), rats, as well as feral sheep (Ovis aries) and cats (Felis catus; Ortiz-Alcaraz et al., 2019).
Category 3 failures (i.e., reinvasion) differ in that they result from non-existent or ineffective biosecurity protocols that ideally should precede attempted eradications, or from inappropriately scaled eradications that do not encompass the entire eradication unit (e.g., islands that require simultaneous IMS removal to prevent reinvasion; Abdelkrim et al., 2005). Such protocols greatly benefit from tools that can reconstruct the source population of reinvaders, as this information may further point to the underlying cause of eradication failure (Kappes et al., 2019). To be effective, biosecurity measures must account for species reinvasion that are human-mediated (e.g., rats transported on boats) or a natural consequence of high dispersal (e.g., deer swimming from nearby islands). For example, conservation efforts often prioritize single islands for eradication from within a cluster of invaded islands, due to biological or other considerations such as targeting Important Bird Areas or World Heritage Sites. Yet, if IMS are readily moving within an island cluster, reinvasion of a cleared island is likely, leading to eradication failure. Likewise, successful eradication of IMS from an entire island cluster may still fail to avert reinvasion if effective biosecurity measures are not in place to prevent, detect and respond to an introduction from external sources.
Genetic and genomic tools exist that can address several challenges surrounding eradication failures, including but not limited to: (1) defining eradication units; (2) determining the source(s) of (re)invasion; (3) identifying invasion pathways; and (4) detecting IMS at low-density (Table 1). Integration of genetic information into biosecurity and control protocols has been recommended (Abdelkrim et al., 2005; Kappes et al., 2019), but is far from seeing regular application, warranting greater discussion of their utility for insular IMS management.
The Promise of Genetics/Genomics to Inform IMS Management on Islands
Genetic and Genomic Tools for Informing the Study and Management of Invasive Species
Since publication of The Genetics of Colonizing Species (Baker and Stebbins, 1965), genetics has played a key role in the study of invasive species, both from the standpoint of using such systems for investigating ecological and evolutionary principles related to colonization of novel environments, as well as for informing applied management strategies. Investigating the population genetics of invasive lineages has been instrumental for testing evolutionary theory related to adaptation and population bottlenecks, reconstructing the complex history of biological invasions to provide insights on the invasion process, and, more recently, uncovering the genetic basis of features associated with invasion success (Barrett, 2015). The sheer impact of invasion genetics over this period was recently highlighted by a symposium marking the 50th anniversary of the seminal work that covered the current state of the field, which was subsequently communicated in a special issue of Molecular Ecology (2015) as well as an expanded edited volume (Barrett, 2015). Aided by our ever-improving ability to mine the genomes of living organisms, such work has been carried out over a wide taxonomic breadth, from zooplankton, terrestrial plants and fungi, to fish, amphibians, reptiles, birds and mammals.
The use of genetics for informing applied management of invasive species was initially centered around plant invasions to clarify taxonomic uncertainties for biosecurity, address questions underpinning the impacts of hybridization and introgression, and identify sources of invasion and potential invasion pathways (Le Roux and Wieczorek, 2009). Different classes of molecular markers have been employed in invasive species research, historically including protein-based allozymes, polymorphic nuclear and organellar DNA fragments [e.g., restriction/amplified fragment length polymorphisms (RFLPs/AFLPs), mitochondrial/chloroplast DNA (mtDNA/cpDNA), respectively], and hypervariable tandemly repeated DNA sequences (e.g., microsatellites and minisatellites; Le Roux and Wieczorek, 2009). Microsatellites and mtDNA, in particular, have seen frequent application in IMS research (Browett et al., 2020), although they may not be sensitive enough to reconstruct invasion histories and delineate eradication units in recently diverged populations or those exhibiting contemporary gene flow (Cowen et al., 2019; Yoshida et al., 2020).
More recently, the advent of massively parallel (MPS; also known as next- or second generation; Margulies et al., 2005; Metzker, 2010) and single-molecule, long-read (also known as third-generation; Branton et al., 2008; Eid et al., 2009) DNA sequencing has elevated our ability to generate orders of magnitude more data and to make use of more robust molecular markers in the form of single nucleotide polymorphisms (SNPs), extending the depth and breadth of questions that can be addressed (Rius et al., 2015), while providing unprecedented insight into the processes underpinning biological invasions (McCartney et al., 2019). In particular, these technologies have enabled low-cost SNP discovery and genotyping-by-sequencing (GBS) of thousands of genetic markers for any species using a range of approaches including restriction site-associated DNA (RAD) sequencing (Baird et al., 2008) and others (e.g., nextRAD; Russello et al., 2015). For example, Bay et al. (2019) used GBS to investigate interactions of introgression and selection between hybridizing invasive rainbow trout (Oncorhynchus mykiss) and threatened westslope cutthroat trout (Oncorhynchus clarkii lewisi). The authors found evidence of parallel adaptation by identifying several loci under selection in admixed populations across multiple river basins that represent important tools for informing management action to prevent the genomic extinction of westslope cutthroat trout. In addition to GBS applications, leading-edge DNA sequencing technologies have greatly facilitated whole genome sequencing of any target organism. As a case in point, full genomes have now been sequenced for 27 of the 100 worst invasive species listed by the IUCN (McCartney et al., 2019). This number promises to increase as sequencing costs decrease and the advantages of reference genomes become more apparent, including providing greater resolution for reconstructing invasion histories, resolving fine-scale structure among recently diverged populations, and identifying targets for genome editing.
Utility of Genetics/Genomics to Design and Evaluate Eradication Programs
When performed prior to an eradication attempt, genetic surveys of a target species can provide an evidence-based platform to direct effective management strategies (Abdelkrim et al., 2005; Kappes et al., 2019). Genetic information generated from these surveys can be used to inform design of management strategies, define eradication units, improve cost-effectiveness of implementation, and evaluate outcomes.
In multi-island systems, estimates of population genetic structure and individual relatedness of IMS may reveal signatures of historical and contemporary gene flow (Escoda et al., 2017; Pichlmueller et al., 2020). Managers can use these data to infer the extent and direction of IMS dispersal within island systems; such information can minimize eradication failures due to reinvasion by helping to define eradication units and frame biosecurity plans and practices (Abdelkrim et al., 2005). For example, Sjodin et al. (2019) used population assignment tests and Bayesian clustering of SNP genotypes to investigate patterns of population connectivity of black and brown rats in the Haida Gwaii archipelago (British Columbia, Canada), identifying six eradication units that constituted single or clusters of islands. On-going rat management efforts in Haida Gwaii should target these eradication units to minimize potential for reinvasion.
On large islands, such as New Zealand’s South Island, genetic data can inform regional IMS management, despite the differences in dispersal barriers. For example, Adams et al. (2014) used Bayesian clustering of microsatellite genotypes to reveal two distinct population clusters of invasive common brushtail possums (Trichosurus vulpecula) in and around Dunedin City and on the rural Otago Peninsula. As a result, a single reinvasion pathway onto the Otago Peninsula was inferred from urban areas to the south; the researchers recommended that eradication of possums on the Otago Peninsula should be paired with on-going monitoring of an urban buffer zone and highlighted the importance of pre-eradication genetic surveys for informing management planning and maximizing eradication success (Adams et al., 2014).
When eradications are not successful, managers who conducted pre-eradication genetic surveys may have the ability to identify the cause of failure by comparing the genetic material from individuals before and after the eradication operation. A recent example was demonstrated by Sjodin et al. (2020a), where the cause of a failed brown rat eradication on the Bischof Islands in Haida Gwaii was due to reinvasion from nearby Lyell Island, rather than from surviving rats in situ (Figure 1B). In New Zealand, pre-eradication genetic surveys of three rat species from Pearl Island allowed genetic assignment of individuals post-eradication that revealed the cause of failure likely being reinvasion from Stewart Island, directing management to reconsider the eradication unit and not the procedure itself (Russell et al., 2010).
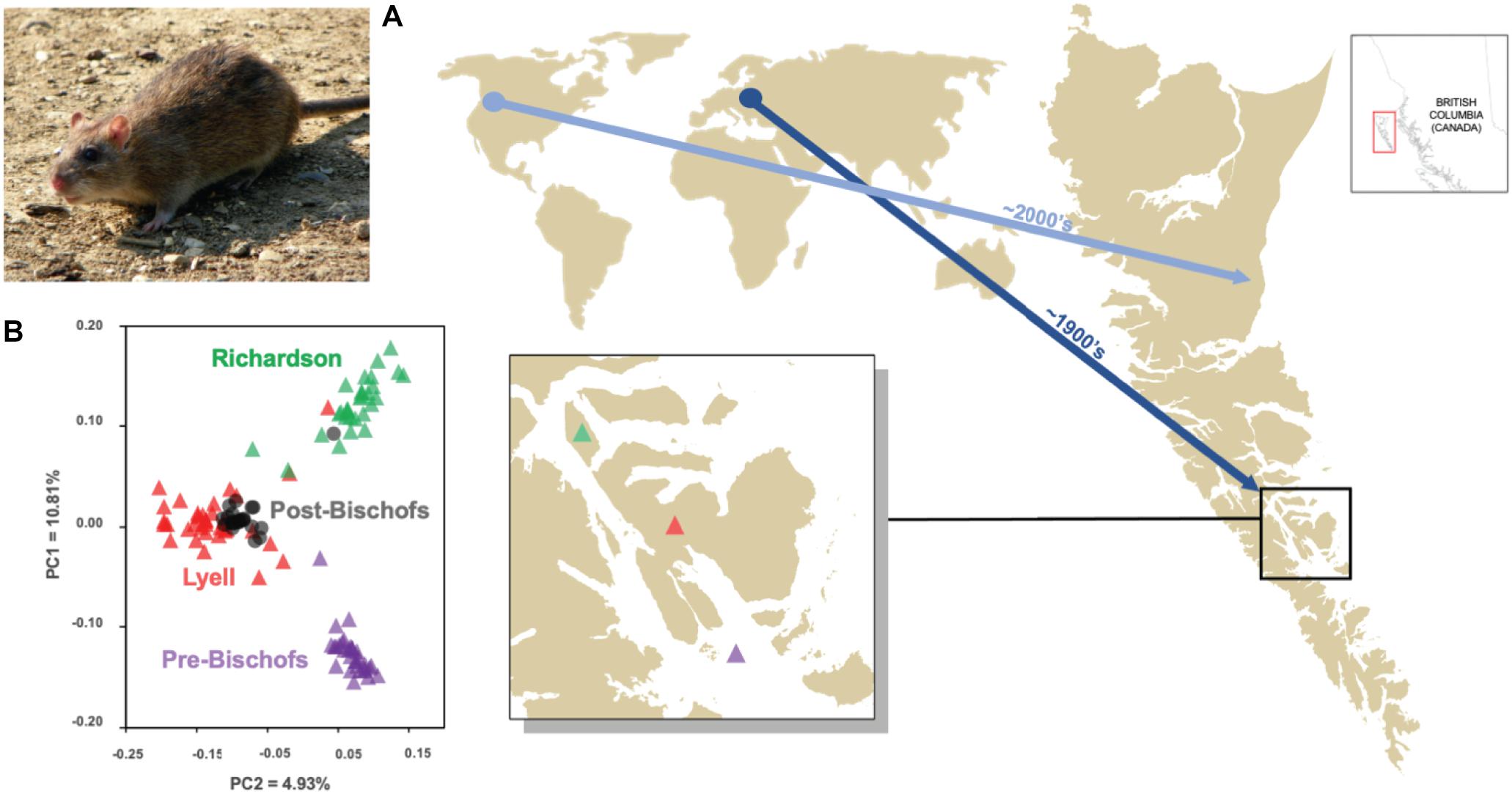
Figure 1. Examples of the use of genomic tools for informing biosecurity protocols and evaluating eradication operations, based on case studies involving invasive brown rats (Rattus norvegicus) in the Haida Gwaii archipelago off the coast of British Columbia, Canada. (A) Genome-wide single nucleotide polymorphism (SNP) data at 16,598 loci and Bayesian coalescent modeling were used to infer multiple invasions of brown rats in Haida Gwaii. The initial invasion into the southern portion of the archipelago was likely from Western Europe in the late 19th or early 20th century, with a second invasion into northern Haida Gwaii from mainland British Columbia occurring in the early 21st century, suggesting the need for continued vigilance and enhanced biosecurity protocols (Sjodin et al., 2020b). (B) Targeted SNP data using the RapidRat Genotyping-in-Thousands by sequencing (GT-seq) panel (see Figure 2) combined with principal components analysis was used to determine that the cause of a failed brown rat eradication in the Bischof Islands in Haida Gwaii was due to reinvasion from nearby Lyell Island, rather than from surviving rats in situ (Sjodin et al., 2019, 2020a).
A previous limitation of genetic tools for informing the design and evaluation of eradication programs was related to obtaining a sufficient number of DNA samples for conducting pre-eradication surveys or post-eradication monitoring, especially in elusive species and/or those found at low densities. This impediment has been overcome in the past two decades as genetic and genomic tools have been optimized for use with non-invasive DNA samples, such as feces, hair and feathers (Kohn et al., 1999). Although such sample types characteristically contain low levels of target DNA that is often degraded due to environmental exposure, use of targeted, species-specific markers such as mtDNA (Taberlet and Waits, 1999), microsatellites (Waits and Paetkau, 2005) and, more recently, SNPs (Russello et al., 2015; Andrews et al., 2018; Schmidt et al., 2020) has circumvented these issues. Non-invasive samples are especially useful for island eradication programs that target species such as deer or feral pigs, which can easily escape direct detection in densely forested environments, but leave readily identifiable sign (e.g., feces). For instance, Brinkman et al. (2011) demonstrated the ability of microsatellites to estimate population density of Sitka black-tailed deer (Odocoileus hemionus sitkensis) using fecal pellets collected on Prince of Wales Island, Alaska. Likewise, Ferreira et al. (2018) used a similar approach for the much smaller and elusive Cabrera voles (Microtus cabrerae), finding that non-invasive genetic sampling generated better estimates of population density at one third of the cost of live trapping. Island eradication programs can benefit greatly by using such estimates of population density to prioritize areas for action and guide efficient resource allocation, such as the number of hunters hired or the duration of a hunting event. This application was demonstrated during the eradication of red deer (Cervus elaphus) from Secretary Island in New Zealand (Macdonald et al., 2019); after several failed eradications attempts, management strategies employed DNA profiling of fecal pellets to create an inventory of individual red deer present that helped guide hunting teams by comparing DNA of deer killed to those identified using fecal pellet DNA (Macdonald et al., 2019). This incorporation of genetic tools proved effective for mitigating the high costs of detecting individuals at low density during the critical period following the initial population reduction. Moving forward, the development and deployment of targeted SNP panels that harness leading-edge DNA sequencing technologies, such as in Genotyping-in-Thousands by sequencing (GT-seq; Campbell N. R. et al., 2015), can provide scalable, rapid and cost-effective tools for informing the design, implementation and evaluation of eradication operations (Sjodin et al., 2020a), even when paired with non-invasive genetic sampling (Schmidt et al., 2020; see Figure 2 for a generalized workflow for GT-seq SNP panel design). Further advances in DNA sequencing technologies have enabled portable genotyping in the field (e.g., Oxford Nanopore MinION; Lamb et al., 2020); although not yet opertionalized within the context of IMS eradication or control programs, this technology holds great promise if paired with pre-eradication inventories (e.g., Macdonald et al., 2019) for informing management operations in real-time.
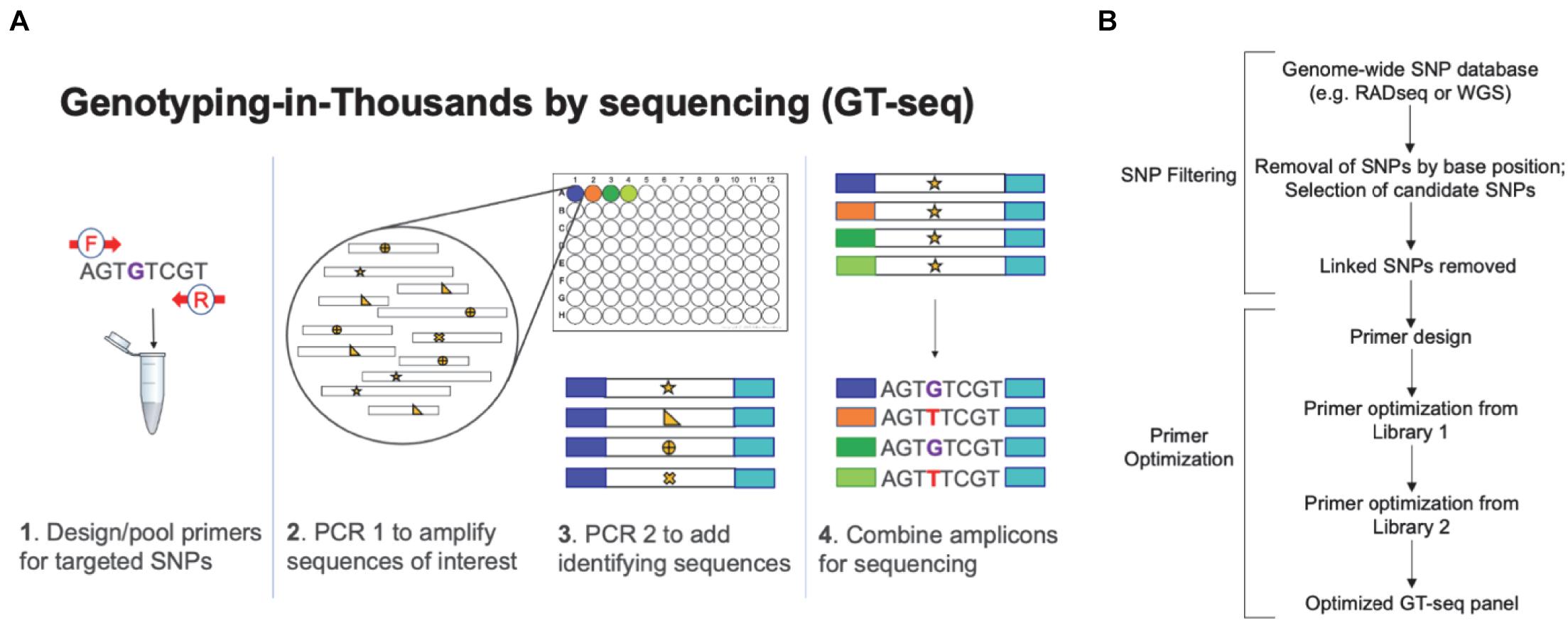
Figure 2. Schematics of Genotyping-in-Thousands by sequencing (GT-seq), a method that can simultaneously genotype few to thousands of individuals at hundreds of single nucleotide polymorphisms (SNPs). (A) Data collection workflow demonstrating multiplex amplification of loci containing SNPs of interest ( ,
,  ,
,  ,
,  ). The schematic is based on Figure 1 of Campbell N. R. et al. (2015). (B) Generalized workflow for developing a GT-seq panel, including initial source of data, filtering steps, and primer optimization. The flowchart is based on Figure 1 of Chang et al. (submitted).
). The schematic is based on Figure 1 of Campbell N. R. et al. (2015). (B) Generalized workflow for developing a GT-seq panel, including initial source of data, filtering steps, and primer optimization. The flowchart is based on Figure 1 of Chang et al. (submitted).
Utility of Genetics/Genomics to Inform and Modify Biosecurity Protocols
Biosecurity includes all efforts made to prevent the immigration, establishment, and spread of invasive species; thus, eradications that proceed without biosecurity protocols in place are often doomed to failure (Kennedy and Broome, 2019). The effectiveness of such protocols depends on evidence-based policy that directs each component of island biosecurity, including quarantines and inspections of invasion vectors (e.g., boat and air traffic), island surveillance for new invaders, and appropriate responses to new invasions (Reaser et al., 2020). Obstacles to effective biosecurity can include technical limitations that arise from inappropriate eradication design (see previous section) as well as insufficient practices for describing and monitoring invasive populations including: (1) incomplete knowledge of invasion pathway(s); (2) inaccurate low-density IMS detection; and/or (3) lack of continuous surveillance (Kennedy and Broome, 2019).
Genetic information can help overcome these obstacles to effective biosecurity. One increasingly common application has been to use genetic data to reconstruct important components of the invasion pathway, such as identifying the source(s) of invasion and point(s) of establishment, with the flexibility to do so at multiple scales. At a local scale, microsatellite analysis of invasive American mink (Neovison vison) in the Atlantic Islands National Park in Spain revealed high genetic affinity to individuals at a nearby fur-farm, suggesting the population was founded by escapees (Velando et al., 2017). At a broader scale, mtDNA-based analyses revealed the origins of invasive monk parakeets (Myiopsitta monachus) in the United States and Spain, and Siberian chipmunks (Eutamias sibiricus) in Italy were directly linked to the legal and illegal pet trades (Russello et al., 2008, 2021; Edelaar et al., 2015; Mori et al., 2018). Further, Puckett et al. (2016) compiled a global SNP database of brown rat (Rattus norvegicus) populations to reconstruct their world-wide invasion history, revealing natural and human-mediated dispersal pathways. This global brown rat SNP database now provides an important resource for identifying sources of invasions on islands around the world, which can be directly integrated into new or existing biosecurity protocols. For example, Sjodin et al. (2020b) used this database to identify multiple invasions of brown rats in the Haida Gwaii archipelago (Figure 1A). Genetic assignment and demographic modeling using several thousand SNP genotypes suggested an initial invasion into the southern portion of the archipelago likely originating from Western Europe in the late 19th or early 20th century, which coincided with active European whaling activities in the area (Figure 1A). This was followed by a second invasion into northern Haida Gwaii from mainland British Columbia in the early 21st century, suggesting the need for continued vigilance and enhanced biosecurity protocols (Figure 1A).
Effective biosecurity is often challenged by the difficulty of detecting IMS at low population densities, representing another area where genetic and genomic tools can assist. Upon establishment, many IMS will undergo a lag phase before exponential population growth and spread, depending on intrinsic genetic and extrinsic environmental conditions (Allendorf and Lundquist, 2003). Consequently, conventional methods such as direct observation, fecal counts, live-trapping, and camera-trapping may not provide sufficient sensitivity for IMS detection during the lag phase. Management may compensate within this critical period by increasing the amount of resources and effort applied, but this can dramatically increase costs (Macdonald et al., 2019). Non-invasive genetic sampling, as described above, can be beneficial for the detection of IMS at low densities because it avoids reliance on methods that involve direct sightings (Ferreira et al., 2018), and can take advantage of reliable and cost-effective methods for sample collection such as wildlife detection dogs (Orkin et al., 2016; Hollerbach et al., 2018; Roda et al., 2021). Additionally, non-invasive genetic sampling also provides opportunities for citizen science contributions that can greatly increase the area searched at much lower costs than field technicians (Biggs et al., 2015; Blåhed et al., 2019).
Another emerging area of research and development for low density species detection as well as surveillance is environmental DNA (eDNA) barcoding (Taberlet et al., 2012). Animals are constantly shedding DNA into their environment, leaving trace amounts of genetic material within environmental samples such as water, soil, snow, or air, with the majority of applications using water samples in aquatic systems (Ruppert, 2019). By optimizing species-specific genetic barcodes, researchers can use such environmental samples to determine the presence or absence of rare or elusive species indirectly. eDNA technology has been effectively applied to invasive species detection across a variety of taxa (Sarre et al., 2013; Hunter et al., 2015; Smart et al., 2015). For example, eDNA barcoding has been used to detect invasive American bullfrog (Rana catesbeiana) from pond water samples with greater sensitivity than traditional field surveys (Dejean et al., 2012), and has been adapted for surveillance of invasive cane toad (Rhinella marina) on islands off the coast of mainland Australia (Tingley et al., 2019). Applications of eDNA also show promise for terrestrial IMS management. Ushio et al. (2017) demonstrated the utility of MiMammal – a set of mammalian primers for eDNA applications – to detect and distinguish various mammalian taxa using water samples. Many of these taxa included widespread insular IMS such as sika deer (Cervus nippon), brown rats, and raccoons (Procyon lotor). Given that eDNA persistence has been estimated between 1 day to over 2 weeks depending on environmental conditions (Barnes et al., 2014), any detection of IMS would indicate that invasion is probable, supporting immediate management action. Nevertheless, avoiding false species detections is still a major hurdle for presence/absence eDNA applications, which must be carefully considered when designing an eDNA workflow for IMS detection (Ficetola et al., 2015, 2016; Roussel et al., 2015; Beng and Corlett, 2020).
What Is Keeping Promise From Practice?
Challenges
Conservation managers are regularly tasked with maximizing conservation returns using typically limited budgets (Gerber, 2016). Given that the collection and analysis of genetic samples has historically been relatively expensive, it is commonly easier to raise funds for eradication operations themselves rather than the necessary a priori science and biosecurity measures that could be enhanced by genetic information. In non-academic circles, genetics may still be perceived as too costly by some (Matzek et al., 2014; Taylor et al., 2017; Cook and Sgrò, 2018), despite the rapid development of laboratory infrastructures that increase the accessibility, applicability, and resolution of genetics for informing invasive species management at ever decreasing per sample costs (Rollins et al., 2006; Rius et al., 2015; McCartney et al., 2019; Browett et al., 2020; Martinez et al., 2020). In fact, the costs of generating genetic and genomic data are lower than ever before – data from the National Human Genome Research Institute suggest the cost of sequencing a human genome has dropped to $1000 in 2020, down from $100 million just 20 years ago (Wetterstrand, 2020). Therefore, the challenge here is not solely related to the costs of generating genetic data themselves, but to reducing the perception that genetics is inaccessible to support a range of evidence-based IMS management operations, regardless of the scale or scope.
In addition, knowledge gaps exist within and outside of academia that may inhibit more frequent implementation of genetics and genomics into IMS management operations. Within academia, geneticists may have an incomplete understanding of what information is actually needed and actionable to best guide IMS management operations. For example, land and conservation managers with decision-making authority in invasive species management programs in California (United States) reported a disparity between their need of scientific information and the research output being produced by scientists (Matzek et al., 2014). Furthermore, IMS management in practice combines several different value sets into decision-making, including social, cultural, economic, and ecological considerations that may not always receive attention in basic research. Outside of academia, conservation managers may lack the genetic expertise necessary to incorporate these approaches into the design and implementation of IMS management strategies. Survey results of conservation managers and scientists in New Zealand and Australia found that insufficient expertise was viewed as a major impediment to the inclusion of genetics within management operations (Taylor et al., 2017; Cook and Sgrò, 2018). Specifically related to the control of invasive species, Cook and Sgrò (2018) found that many conservation managers considered genetics to be irrelevant. This perception may again be attributed to a lack of genetic expertise and awareness, as genetic data have been a key part of informing invasive species management on islands for over 20 years (see section “The Promise of Genetics/Genomics to Inform IMS Management on Islands”). However, for IMS eradications to be more successful in preventing extinctions, we must increase the scale, scope and pace of eradications that is both informing and being responsive to ever-improving and diversifying genetic solutions (Le Roux and Wieczorek, 2009; Geller et al., 2010; Rius et al., 2015; Browett et al., 2020).
Contributing to the divide, conservation scientists and managers within academic and non-academic institutions have different roles and responsibilities; for example, universities and government agencies offer varying reward systems for job performance, require alternate timelines for deliverables, and contain different administrative hurdles that are necessary prior to undertaking research projects (Oliver and Cairney, 2019). Collectively, these disparities may hinder the integration of genetics and genomics into IMS management and contribute to what has been called the conservation genetics gap, a widely discussed division between academic researchers and conservation managers that prevents more frequent use of genetics into conservation practice (Hoban et al., 2013a; Shafer et al., 2015; Taylor et al., 2017; Britt et al., 2018; Holderegger et al., 2019; Sandström et al., 2019). A central argument underpinning many of these discussions is that academic researchers are not rewarded for active participation in conservation and/or engagement with managers because of the perceived reward system in academia (e.g., “publish or perish”). Some argue that the publication pressure for scientists in academia counteracts real world conservation applications because time and resources are consistently invested in projects that generate peer-reviewed publications rather than those that promise conservation success (Vernesi et al., 2008; Arlettaz et al., 2010). If publication output is indeed among the core measures of job performance in academia, contributions to insular IMS removal programs could be limited, as such projects may require great time and resource investment, can occur in remote locations, and always run the risk of failure or postponement due to loss of social license or other factors. Alternatively, government personnel can feel discouraged by working with academic researchers due to incongruence in timelines, perceived lack of interest in ‘practical or real world’ problems in the academy, and/or incomplete understanding of the applicability of the science to the conservation policy or management problem (Choi et al., 2005). Within the government agency or division, personnel may be discouraged by the administrative load of managing the contracting side of the relationship, prevented by senior managers who discourage deviation from conventional approaches, and/or by other cultural factors that are impediments to collaboration with academic scientists and institutions (Oliver and Cairney, 2019).
Solutions
We echo the call of many for greater collaboration between academic researchers and conservation managers to confront the most pressing future or latent conservation problems (Hoban et al., 2013b; Matzek et al., 2014; Shafer et al., 2015; Sutherland and Wordley, 2017; Taylor et al., 2017; Britt et al., 2018; Cook and Sgrò, 2019; Fabian et al., 2019; Holderegger et al., 2019; Browett et al., 2020); in this case, related to the increased scale, scope and pace of IMS eradications from islands and protecting these investments into the future. For example, collaborations provide the opportunity for academics and managers to pool their funding to maximize the effectiveness of removal programs while enhancing research studies. For academics, costs of obtaining field samples for research are substantially reduced, leaving greater funds available to expand data collection or pursue other lines of inquiry. Conversely, managers would no longer have to worry about purchasing research infrastructure, while also gaining access to graduate students for both field and lab work frequently at lower costs and higher skill levels than other personnel options. Thus, the funding pool mutualism may help to alleviate costs and this barrier to implementation frequently cited by conservation managers (Taylor et al., 2017; Cook and Sgrò, 2018).
Collaborations further enable academic researchers to bring the most advanced, cutting-edge science to address targeted questions raised by managers in order to help fill knowledge gaps that represent barriers to management success. IMS management on islands would benefit greatly from a culture of knowledge exchange that effectively communicates the advantages of genetics and genomics to decision-makers; managers learn more about the utility of genetic and genomic tools for conservation, and academics are incentivized to design innovative studies and solution-focused approaches that are relevant for successful management planning, implementation and monitoring. Academics can also benefit by undertaking research on islands that naturally provide excellent systems to study ecological and evolutionary patterns and processes. For example, by working with eradication or control programs, academics gain access to island systems for investigating microevolutionary questions underpinning local adaptation and consequences of low genetic diversity (e.g., founder effects and inbreeding), while further advancing the growing field of invasion biology. The prevention of extinctions on islands will become increasingly reliant on such research insights as global transportation networks continue to facilitate IMS introductions (Ricciardi et al., 2017). Thus, the solution space for greater collaboration then becomes framed by the question – how are fruitful collaborations developed?
Collaborations can generally be established by way of top-down or bottom-up approaches. The top-down approach is mostly driven by government policy and associated funding to promote university-government-industry relationships that are typically focused on innovation agendas (Etzkowitz and Leydesdorff, 2000). This framework has been successfully used in China for stimulating knowledge creation and commercialization through university-government collaborations (Abbas et al., 2019). As policies move toward addressing climate change and the drivers of biodiversity loss around the world, the possibility exists of shifting the goals of collaborations to conservation innovation and directing funding support to achieving that end. However, there is no need for academics and managers with shared interests in IMS to wait on policy to advance collaborations; a bottom-up approach to establishing collaborations can also be fruitful. Often government agencies hold long-term data that can yield ecological insights (Lindenmayer et al., 2012), but commonly lack time, the mandate or the capacity to apply current analytical tools to the data. Conversely, academic researchers often lack the understanding of the pressing and current issues in management. To provide such connections, there has been success with the use of databases to match interested parties. In a research context, the Federation of African Immunological Societies Legacy Project initiative has been established to match immunology graduate students in Africa with labs in Europe and North America with overlapping research interests (Gewin, 2019); the Canadian Parks Collective for Innovation and Leadership is attempting a similar program for conservation managers in Canada, but is in the very early stages of development. In such a way, government and academic colleagues can provide a mutually beneficial arrangement for data to be analyzed and understood. In such cases, the challenge may be to find a question that has management implications, while also providing the academic context to support a graduate student or allocate laboratory resources to the research. Co-generation of research questions gives ownership to all parties as does co-authorship of publications, patents or other outcomes; academics may not fully recognize the validation this may bring to managers and their programs outside of the academy.
Future Directions
The success of IMS eradications on islands is unequivocal, as evidenced by the recovery of threatened and endangered species, restoration of ecosystems, and demonstrated direct and indirect benefits to local communities; yet such management interventions have still only been conducted on ∼0.3% of invaded islands (DIISE, 2020). Conservation managers are hitting the upper limits of total islands that can be restored using current approaches and tools due to the size and complexity of operations. These impediments also limit the benefits of IMS eradications on islands to ∼15% of all threatened species (Campbell K. J. et al., 2015). Overcoming the barriers to implementation requires change to stem the extinction crisis. Genetic and genomic tools can inform, guide, and even catalyze the next generation of conservation efforts on islands.
There is considerable interest in emerging approaches in genome editing, including the use of synthetic gene drives to increase the scope and scale of restoration or vector control programs (Esvelt and Gemmell, 2017; Burt and Crisanti, 2018; Campbell et al., 2019). A gene drive is a naturally occurring phenomenon of biased inheritance that can be bioengineered into a target species to ensure that specific allele(s) are transmitted to the next generation at a much higher rate than the 50% expected under Mendelian inheritance (Burt and Trivers, 2006; Burt and Crisanti, 2018). The harnessing of gene drive technology is being explored as a means to potentially transform invasive species management by editing genomes to control populations in various ways, effectively eliminating the need for pesticides or other tools, while also increasing the efficiency and lessening the risks of IMS control or eradication on islands (Oh et al., 2021). The harnessing of CRISPR-Cas9 mediated genetic engineering has greatly enhanced the potential of gene drives by facilitating the spread of a targeted gene up to 100% in a population (National Academies of Sciences, Engineering, and Medicine (NASEM), 2016). Of the many research paths for invasive species, the most active programs to date have endeavored to reduce transmission of malaria (Kyrou et al., 2018) and to control the negative impacts of house mice on islands (Campbell et al., 2019). In both of these examples, the focus has been on biasing or skewing sex ratios toward all male or all female offspring, reducing the ability of the population to find mates and breed, and ultimately leading to population suppression through collapse.
Despite the increased attention directed toward adapting this technology for the control of invasive species, there are significant barriers to implementation. Given that gene drives are only transmitted each generation, the effects are more readily measured and seen in species that reproduce quickly (Burt and Crisanti, 2018; Campbell et al., 2019; Carballar-Lejarazú et al., 2020). The majority of IMS fail to meet this criterion, with some rodent species being notable exceptions. The time required for fixation of a drive allele has been estimated at 10–15 generations, depending on several factors (Unckless et al., 2015), suggesting species with low reproduction rates or longer development periods may not yet be suitable. Even within highly fecund species that reproduce prolifically, such as mice or rats, there are other challenges preventing the deployment of gene drives. One is the potential of species to evolve resistance through random mutation of the drive allele or other means (Gomulkiewicz et al., 2021), which could render the drive unsuccessful while permanently altering the invasive population (Rode et al., 2019). There is also significant concern about the potential risks, impacts, and consequences of introducing the drive allele into non-target species through hybridization (Webber et al., 2015). In light of these considerations and others (James et al., 2020), gene drives have yet to be developed for field use, and only the mosquito gene drive has been demonstrated in a laboratory setting (Kyrou et al., 2018), with limited success to date in mammals (Callaway, 2018).
Gene drive technology will undoubtably continue making advances toward deployment, and discussions are on-going involving the application of this technology to invasive species management. When the time comes, invaded islands will be at the forefront of these discussions, as they offer significant advantages over mainland systems for field trials (Godwin et al., 2019). Primarily, the inherent isolation of islands reduces some of the risks previously mentioned, such as transfer of the drive allele to non-target species through hybridization or non-target populations through natural dispersal (Leitschuh et al., 2018), assuming anthropogenic biosecurity risks can be addressed. Deploying a novel gene drive in mainland systems has greater risk of undesirable consequences as habitat is generally less fragmented and more conducive to gene flow. Additionally, island species contain relatively low levels of genetic diversity compared to mainland conspecifics due to historical founder effects, making insular populations more likely to carry locally fixed alleles that are excellent targets for gene drive technology (Sudweeks et al., 2019; Oh et al., 2021). Importantly, assessing the risks of synthetic biology remains a focus of discussion in global policy arenas such as the United Nations, where gene drives are currently regulated under the Cartagena Protocol on Biosafety to the Convention on Biological Diversity (Keiper and Atanassova, 2020). Considering the global ramifications and promise of this technology, decisions regarding the development and deployment of genome editing technology for IMS management (and other applications) must continue to be informed by way of open and transparent peer-reviewed research, comprehensive and case-by-case risk assessment, and inclusive discussion on an international scale.
Author Contributions
MR and BB conceived the project and prepared the first draft of the manuscript. BB, MR, RI, and GH performed revisions of the manuscript and completed the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Conservation and Restoration (CoRe) program at Parks Canada Agency for funding rat and deer genomic research to inform invasive species management in Gwaii Haanas National Park Reserve, National Marine Conservation Area, and Haida Heritage Site, which helped stimulate this collaboration. We are further grateful to Bryson Sjodin, Danielle Schmidt, and Sarah Chang for their contributions to Figures 1, 2. MR acknowledges the support of the NSERC Discovery Grant Programme (# RGPIN-2019-04621), which helps enable cross-disciplinary, cross-sector work as exemplified here.
References
Abbas, A., Avdic, A., Xiaobao, P., Hasan, M. M., and Ming, W. (2019). University-government collaboration for the generation and commercialization of new knowledge for use in industry. J. Innov. Knowl. 4, 23–31. doi: 10.1016/j.jik.2018.03.002
Abdelkrim, J., Pascal, M., Calmet, C., and Samadi, S. (2005). Importance of assessing population genetic structure before eradication of invasive species: examples from insular Norway Rat Populations. Conserv. Biol. 19, 1509–1518. doi: 10.1111/j.1523-1739.2005.00206.x
Adams, A. L., van Heezik, Y., Dickinson, K. J. M., and Robertson, B. C. (2014). Identifying eradication units in an invasive mammalian pest species. Biol. Invasions 16, 1481–1496. doi: 10.1007/s10530-013-0586-9
Allendorf, F. W., and Lundquist, L. L. (2003). Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30. doi: 10.1046/j.1523-1739.2003.02365.x
Andrews, K. R., De Barba, M., Russello, M. A., and Waits, L. P. (2018). “Advances in Using Non-invasive, Archival, and Environmental Samples for Population Genomic Studies,” in Population Genomics: Wildlife, eds P. A. Hohenlohe and O. P. Rajora (Cham: Springer International Publishing), 63–99.
Arlettaz, R., Schaub, M., Fournier, J., Reichlin, T. S., Sierro, A., Watson, J. E. M., et al. (2010). From publications to public actions: when conservation biologists bridge the gap between research and implementation. BioScience 60, 835–842. doi: 10.1525/bio.2010.60.10.10
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD Markers. PLoS One 3:e3376. doi: 10.1371/journal.pone.0003376
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., and Lodge, D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi: 10.1021/es404734p
Barrett, S. C. H. (2015). Foundations of invasion genetics: the Baker and Stebbins legacy. Mol. Ecol. 24, 1927–1941. doi: 10.1111/mec.13014
Bay, R. A., Taylor, E. B., and Schluter, D. (2019). Parallel introgression and selection on introduced alleles in a native species. Mol. Ecol. 28, 2802–2813. doi: 10.1111/mec.15097
Bellard, C., Cassey, P., and Blackburn, T. M. (2016). Alien species as a driver of recent extinctions. Biol. Lett. 12:20150623. doi: 10.1098/rsbl.2015.0623
Bellis, K. X. T., Peet, R. T., Irvine, R. L., Howald, G., and Alsop, G. J. (2019). “Beyond biodiversity: the cultural context of invasive species initiatives in Gwaii Haanas,” in Island Invasives: Scaling up to meet the challenge, eds C. R. Veitch, M. N. Clout, A. R. Martin, J. C. Russell, and C. J. West (Gland, Switzerland: IUCN), 494–496.
Beng, K. C., and Corlett, R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodivers. Conserv. 29, 2089–2121. doi: 10.1007/s10531-020-01980-0
Biggs, J., Ewald, N., Valentini, A., Gaboriaud, C., Dejean, T., Griffiths, R. A., et al. (2015). Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 183, 19–28. doi: 10.1016/j.biocon.2014.11.029
Blåhed, I.-M., Ericsson, G., and Spong, G. (2019). Noninvasive population assessment of moose (Alces alces) by SNP genotyping of fecal pellets. Eur. J. Wildl. Res. 65, 1–11. doi: 10.1007/s10344-019-1337-8
Branton, D., Deamer, D. W., Marziali, A., Bayley, H., Benner, S. A., Butler, T., et al. (2008). The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153. doi: 10.1038/nbt.1495
Brinkman, T. J., Person, D. K., Chapin, F. S., Smith, W., and Hundertmark, K. J. (2011). Estimating abundance of Sitka black-tailed deer using DNA from fecal pellets: estimating abundance of deer using DNA. J. Wildl. Manag. 75, 232–242. doi: 10.1002/jwmg.22
Brinkman, T. J., Person, D. K., Schwartz, M. K., Pilgrim, K. L., Colson, K. E., and Hundertmark, K. J. (2010). Individual identification of Sitka black-tailed deer (Odocoileus hemionus sitkensis) using DNA from fecal pellets. Conserv. Genet. Resour. 2, 115–118. doi: 10.1007/s12686-010-9176-7
Britt, M., Haworth, S. E., Johnson, J. B., Martchenko, D., and Shafer, A. B. A. (2018). The importance of non-academic coauthors in bridging the conservation genetics gap. Biol. Conserv. 218, 118–123. doi: 10.1016/j.biocon.2017.12.019
Broome, K., Golding, C., Brown, K., Corson, P., and Bell, P. (2017). Rat eradication using aerial baiting Current agreed best practice used in New Zealand. Wellington: New Zealand Department of Conservation.
Browett, S. S., O’Meara, D. B., and McDevitt, A. D. (2020). Genetic tools in the management of invasive mammals: recent trends and future perspectives. Mamm. Rev. 50, 200–210. doi: 10.1111/mam.12189
Burt, A., and Crisanti, A. (2018). Gene drive: evolved and synthetic. ACS Chem. Biol. 13, 343–346. doi: 10.1021/acschembio.7b01031
Burt, A., and Trivers, R. (2006). Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge: Belknap Press, Harvard University Press.
Callaway, E. (2018). Controversial CRISPR ‘gene drives’ tested in mammals for the first time. Nature 559:164. doi: 10.1038/d41586-018-05665-1
Campbell, K. J., Beek, J., Eason, C. T., Glen, A. S., Godwin, J., Gould, F., et al. (2015). The next generation of rodent eradications: Innovative technologies and tools to improve species specificity and increase their feasibility on islands. Biol. Conserv. 185, 47–58. doi: 10.1016/j.biocon.2014.10.016
Campbell, N. R., Harmon, S. A., and Narum, S. R. (2015). Genotyping-in-Thousands by sequencing (GT-seq): A cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Resour. 15, 855–867. doi: 10.1111/1755-0998.12357
Campbell, K. J., Saah, J. R., Brown, P. R., Godwin, J., Gould, F., Howald, G., et al. (2019). “A potential new tool for the toolbox: assessing gene drives for eradicating invasive rodent populations,” in Island Invasives: Scaling up to meet the challenge, eds C. R. Veitch, M. N. Clout, A. R. Martin, J. C. Russell, and C. J. West (Gland, Switzerland: IUCN), 6–14.
Canu, A., Apollonio, M., and Scandura, M. (2018). Unmasking the invader: Genetic identity of invasive wild boar from three minor islands off Sardinia (Italy). Mamm. Biol. 93, 29–37. doi: 10.1016/j.mambio.2018.07.008
Carballar-Lejarazú, R., Ogaugwu, C., Tushar, T., Kelsey, A., Pham, T. B., Murphy, J., et al. (2020). Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. 117, 22805–22814. doi: 10.1073/pnas.2010214117
Carter, A., Barr, S., Bond, C., Paske, G., Peters, D., and van Dam, R. (2016). Controlling sympatric pest mammal populations in New Zealand with self-resetting, toxicant-free traps: a promising tool for invasive species management. Biol. Invasions 18, 1723–1736. doi: 10.1007/s10530-016-1115-4
Chang, S., Ward, H., and Russello, M. (submitted). Genotyping-in-thousands by sequencing (GT-seq) panel development and application to inform kokanee salmon (Oncorhynchus nerka) fisheries management at multiple scales. PLoS ONE
Chinchio, E., Crotta, M., Romeo, C., Drewe, J. A., Guitian, J., and Ferrari, N. (2020). Invasive alien species and disease risk: An open challenge in public and animal health. PLoS Pathog. 16:e1008922. doi: 10.1371/journal.ppat.1008922
Choi, B. C. K., Pang, T., Lin, V., Puska, P., Sherman, G., Goddard, M., et al. (2005). Can scientists and policy makers work together? J. Epidemiol. Commun. Health 59, 632–637. doi: 10.1136/jech.2004.031765
Clavero, M., and Garcia-Berthou, E. (2005). Invasive species are a leading cause of animal extinctions. Trends Ecol. Evol. 20:110. doi: 10.1016/j.tree.2005.01.003
Cook, C. N., and Sgrò, C. M. (2018). Understanding managers’ and scientists’ perspectives on opportunities to achieve more evolutionarily enlightened management in conservation. Evol. Appl. 11, 1371–1388. doi: 10.1111/eva.12631
Cook, C. N., and Sgrò, C. M. (2019). Poor understanding of evolutionary theory is a barrier to effective conservation management. Conserv. Lett. 12:e12619. doi: 10.1111/conl.12619
Cowen, S., Clausen, L., Algar, D., and Comer, S. (2019). Using genetics to evaluate the success of a feral cat (Felis catus) control program in North-Western Australia. Animals 9, 1–15. doi: 10.3390/ani9121050
Cromarty, P. L., Broome, K. G., Cox, A., Empson, R. A., Hutchinson, W. M., and McFadden, I. (2002). “Eradication planning for invasive alien animal species on islands - the approach developed by the New Zealand Department of Conservation,” in Turning the Tide: The Eradication of Invasive Species, eds C. R. Veitch and M. N. Clout (Gland, Switzerland: IUCN SSC Invasive Species Specialists Group), 85–91.
de Wit, L. A., Zilliacus, K. M., Quadri, P., Will, D., Grima, N., Spatz, D., et al. (2020). Invasive vertebrate eradications on islands as a tool for implementing global sustainable development goals. Environ. Conserv. 47, 139–148. doi: 10.1017/S0376892920000211
Dejean, T., Valentini, A., Miquel, C., Taberlet, P., Bellemain, E., and Miaud, C. (2012). Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus: Alien invasive species detection using eDNA. J. Appl. Ecol. 49, 953–959. doi: 10.1111/j.1365-2664.2012.02171.x
DIISE. (2020). The Database of Island Invasive Species Eradications, developed by Island Conservation, Coastal Conservation Action Laboratory UCSC, IUCN SSC Invasive Species Specialist Group. Available Online at: http://diise.islandconservation.org (accessed March 13, 2021).
Doherty, T. S., Glen, A. S., Nimmo, D. G., Ritchie, E. G., and Dickman, C. R. (2016). Invasive predators and global biodiversity loss. Proc. Natl. Acad. Sci. 113, 11261–11265. doi: 10.1073/pnas.1602480113
Donlan, C. J., Howald, G. R., Tershy, B. R., and Croll, D. A. (2003). Evaluating alternative rodenticides for island conservation: roof rat eradication from the San Jorge Islands, Mexico. Biol. Conserv. 114, 29–34. doi: 10.1016/S0006-3207(02)00401-9
Edelaar, P., Roques, S., Hobson, E. A., Gonçalves da Silva, A., Avery, M. L., Russello, M. A., et al. (2015). Shared genetic diversity across the global invasive range of the monk parakeet suggests a common restricted geographic origin and the possibility of convergent selection. Mol. Ecol. 24, 2164–2176. doi: 10.1111/mec.13157
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-Time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Epanchin-Niell, R. S. (2017). Economics of invasive species policy and management. Biol. Invasions 19, 3333–3354. doi: 10.1007/s10530-017-1406-4
Escoda, L., González-Esteban, J., Gómez, A., and Castresana, J. (2017). Using relatedness networks to infer contemporary dispersal: Application to the endangered mammal Galemys pyrenaicus. Mol. Ecol. 26, 3343–3357. doi: 10.1111/mec.14133
Esvelt, K. M., and Gemmell, N. J. (2017). Conservation demands safe gene drive. PLoS Biol. 15:e2003850. doi: 10.1371/journal.pbio.2003850
Etzkowitz, H., and Leydesdorff, L. (2000). The dynamics of innovation: from National Systems and “Mode 2” to a Triple Helix of university–industry–government relations. Res. Policy 29, 109–123. doi: 10.1016/S0048-7333(99)00055-4
Fabian, Y., Bollmann, K., Brang, P., Heiri, C., Olschewski, R., Rigling, A., et al. (2019). How to close the science-practice gap in nature conservation? Information sources used by practitioners. Biol. Conserv. 235, 93–101. doi: 10.1016/j.biocon.2019.04.011
Ferreira, C. M., Sabino-Marques, H., Barbosa, S., Costa, P., Encarnação, C., Alpizar-Jara, R., et al. (2018). Genetic non-invasive sampling (gNIS) as a cost-effective tool for monitoring elusive small mammals. Eur. J. Wildl. Res. 64, 1–15. doi: 10.1007/s10344-018-1188-8
Ficetola, G. F., Pansu, J., Bonin, A., Coissac, E., Giguet-Covex, C., De Barba, M., et al. (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 15, 543–556. doi: 10.1111/1755-0998.12338
Ficetola, G. F., Taberlet, P., and Coissac, E. (2016). How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 16, 604–607. doi: 10.1111/1755-0998.12508
Fraser, R. W., Cook, D. C., Mumford, J. D., Wilby, A., and Waage, J. K. (2006). Managing outbreaks of invasive species: Eradication versus suppression. Int. J. Pest. Manag. 52, 261–268. doi: 10.1080/09670870600795930
Gatto-Almeida, F., Pichlmueller, F., Micheletti, T., Abrahão, C. R., Mangini, P. R., and Russell, J. C. (2020). Using genetics to plan black rat (Rattus rattus) management in Fernando de Noronha archipelago, Brazil. Perspect. Ecol. Conserv. 18, 44–50. doi: 10.1016/j.pecon.2020.01.001
Geller, J. B., Darling, J. A., and Carlton, J. T. (2010). Genetic perspectives on marine biological invasions. Annu. Rev. Mar. Sci. 2, 367–393. doi: 10.1146/annurev.marine.010908.163745
Gerber, L. R. (2016). Conservation triage or injurious neglect in endangered species recovery. Proc. Natl. Acad. Sci. 113, 3563–3566. doi: 10.1073/pnas.1525085113
Gewin, V. (2019). Meet the African researcher who is building a core of junior immunologists on the continent. Nature doi: 10.1038/d41586-019-02334-9
Godwin, J., Serr, M., Barnhill-Dilling, S. K., Blondel, D. V., Brown, P. R., Campbell, K., et al. (2019). Rodent gene drives for conservation: opportunities and data needs. Proc. R. Soc. B Biol. Sci. 286:20191606. doi: 10.1098/rspb.2019.1606
Gomulkiewicz, R., Thies, M. L., and Bull, J. J. (2021). Evading resistance to gene drives. Genetics 217:iyaa040. doi: 10.1093/genetics/iyaa040
Hanson, C. C., Hall, T. J., DeNicola, A. J., Silander, S., Keitt, B. S., and Campbell, K. J. (2019). Rhesus macaque eradication to restore the ecological integrity of Desecheo National Wildlife Refuge, Puerto Rico. In: Island Invasives: Scaling up to meet the challenge. Gland, Switzerland: International Union for Nature Conservation (IUCN), 249–255.
Helmstedt, K. J., Shaw, J. D., Bode, M., Terauds, A., Springer, K., Robinson, S. A., et al. (2016). Prioritizing eradication actions on islands: it’s not all or nothing. J. Appl. Ecol. 53, 733–741. doi: 10.1111/1365-2664.12599
Hoban, S., Arntzen, J. W., Bertorelle, G., Bryja, J., Fernandes, M., Frith, K., et al. (2013a). Conservation Genetic Resources for Effective Species Survival (ConGRESS): Bridging the divide between conservation research and practice. J. Nat. Conserv. 21, 433–437. doi: 10.1016/j.jnc.2013.07.005
Hoban, S. M., Hauffe, H. C., Pérez-Espona, S., Arntzen, J. W., Bertorelle, G., Bryja, J., et al. (2013b). Bringing genetic diversity to the forefront of conservation policy and management. Conserv. Genet. Resour. 5, 593–598. doi: 10.1007/s12686-013-9859-y
Holderegger, R., Balkenhol, N., Bolliger, J., Engler, J. O., Gugerli, F., Hochkirch, A., et al. (2019). Conservation genetics: linking science with practice. Mol. Ecol. 28, 3848–3856. doi: 10.1111/mec.15202
Hollerbach, L., Heurich, M., Reiners, T. E., and Nowak, C. (2018). Detection dogs allow for systematic non-invasive collection of DNA samples from Eurasian lynx. Mamm. Biol. 90, 42–46. doi: 10.1016/j.mambio.2018.02.003
Howald, G., Donlan, C. J., Galván, J. P., Russell, J. C., Parkes, J., Samaniego, A., et al. (2007). Invasive rodent eradication on islands. Conserv. Biol. 21, 1258–1268. doi: 10.1111/j.1523-1739.2007.00755.x
Huambachano, M. (2019). Indigenous food sovereignty: Reclaiming food as sacred medicine in Aotearoa New Zealand and Peru. New Zealand J. Ecol. 43, 1–6. doi: 10.20417/nzjecol.43.39
Hunter, M. E., Oyler-McCance, S. J., Dorazio, R. M., Fike, J. A., Smith, B. J., Hunter, C. T., et al. (2015). Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive burmese pythons. PLoS One 10:e0121655. doi: 10.1371/journal.pone.0121655
James, S. L., Marshall, J. M., Christophides, G. K., Okumu, F. O., and Nolan, T. (2020). Toward the definition of efficacy and safety criteria for advancing gene drive-modified mosquitoes to field testing. Vector-Borne Zoonotic Dis. 20, 237–251. doi: 10.1089/vbz.2019.2606
Jones, H. P., Holmes, N. D., Butchart, S. H. M., Tershy, B. R., Kappes, P. J., Corkery, I., et al. (2016). Invasive mammal eradication on islands results in substantial conservation gains. Proc. Natl. Acad. Sci. 113, 4033–4038. doi: 10.1073/pnas.1521179113
Kappes, P. J., Bond, A. L., Russell, J. C., and Wanless, R. M. (2019). Diagnosing and responding to causes of failure to eradicate invasive rodents. Biol. Invasions 21, 2247–2254. doi: 10.1007/s10530-019-01976-0
Keiper, F., and Atanassova, A. (2020). Regulation of synthetic biology: developments under the convention on biological diversity and its protocols. Front. Bioeng. Biotechnol. 8:310. doi: 10.3389/fbioe.2020.00310
Keitt, B., Griffiths, R., Boudjelas, S., Broome, K., Cranwell, S., Millett, J., et al. (2015). Best practice guidelines for rat eradication on tropical islands. Biol. Conserv. 185, 17–26. doi: 10.1016/j.biocon.2014.10.014
Kennedy, E. S., and Broome, K. G. (2019). “How do we prevent the obstacles to good island biosecurity from limiting our eradication ambitions?,” in Island Invasives: Scaling up to meet the challenge, eds C. R. Veitch, M. N. Clout, A. R. Martin, J. C. Russell, and C. J. West (Gland, Switzerland: IUCN), 478–483.
Kier, G., Kreft, H., Lee, T. M., Jetz, W., Ibisch, P. L., Nowicki, C., et al. (2009). A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. 106, 9322–9327. doi: 10.1073/pnas.0810306106
Kohn, M. H., York, E. C., Kamradt, D. A., Haught, G., Sauvajot, R. M., and Wayne, R. K. (1999). Estimating population size by genotyping faeces. Proc. R. Soc. Lond. B Biol. Sci. 266, 657–663. doi: 10.1098/rspb.1999.0686
Kosoy, M., Khlyap, L., Cosson, J.-F., and Morand, S. (2015). Aboriginal and invasive rats of genus rattus as hosts of infectious agents. Vector-Borne Zoonotic Dis. 15, 3–12. doi: 10.1089/vbz.2014.1629
Kyrou, K., Hammond, A. M., Galizi, R., Kranjc, N., Burt, A., Beaghton, A. K., et al. (2018). A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066. doi: 10.1038/nbt.4245
La Morgia, V., Paoloni, D., and Genovesi, P. (2017). Eradicating the grey squirrel Sciurus carolinensis from urban areas: an innovative decision-making approach based on lessons learnt in Italy: Innovative approach to grey squirrel eradication from urban areas. Pest. Manag. Sci. 73, 354–363. doi: 10.1002/ps.4352
Lamb, H. J., Hayes, B. J., Nguyen, L. T., and Ross, E. M. (2020). The future of livestock management: A review of real-time portable sequencing applied to livestock. Genes 11:1478. doi: 10.3390/genes11121478
Le Roux, J., and Wieczorek, A. M. (2009). Molecular systematics and population genetics of biological invasions: towards a better understanding of invasive species management: Systematics and population genetics of biological invasions. Ann. Appl. Biol. 154, 1–17. doi: 10.1111/j.1744-7348.2008.00280.x
Leitschuh, C. M., Kanavy, D., Backus, G. A., Valdez, R. X., Serr, M., Pitts, E. A., et al. (2018). Developing gene drive technologies to eradicate invasive rodents from islands. J. Responsible Innov. 5, S121–S138. doi: 10.1080/23299460.2017.1365232
Lindenmayer, D. B., Likens, G. E., Andersen, A., Bowman, D., Bull, C. M., Burns, E., et al. (2012). Value of long-term ecological studies. Austral Ecol. 37, 745–757. doi: 10.1111/j.1442-9993.2011.02351.x
Macdonald, N., Nugent, G., Edge, K.-A., and Parkes, J. P. (2019). Eradication of red deer from Secretary Island, New Zealand: changing tactics to achieve success. Lincoln: Landcare Research, 256–260.
Macleod, I. A., Maclennan, D., Raynor, R., Thompson, D. B. A., and Whitaker, S. (2019). Large scale eradication of non-native invasive American mink (Neovison vison) from the Outer Hebrides of Scotland. Inverness: Scottish Natural Heritage, 261–266.
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. doi: 10.1038/nature03959
Martinez, B., Reaser, J. K., Dehgan, A., Zamft, B., Baisch, D., McCormick, C., et al. (2020). Technology innovation: advancing capacities for the early detection of and rapid response to invasive species. Biol. Invasions 22, 75–100. doi: 10.1007/s10530-019-02146-y
Matzek, V., Covino, J., Funk, J. L., and Saunders, M. (2014). Closing the knowing–doing gap in invasive plant management: accessibility and interdisciplinarity of scientific research. Conserv. Lett. 7, 208–215. doi: 10.1111/conl.12042
McCartney, M. A., Mallez, S., and Gohl, D. M. (2019). Genome projects in invasion biology. Conserv. Genet. 20, 1201–1222. doi: 10.1007/s10592-019-01224-x
McCreless, E. E., Huff, D. D., Croll, D. A., Tershy, B. R., Spatz, D. R., Holmes, N. D., et al. (2016). Past and estimated future impact of invasive alien mammals on insular threatened vertebrate populations. Nat. Commun. 7, 1–11. doi: 10.1038/ncomms12488
Metzker, M. L. (2010). Sequencing technologies — the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/nrg2626
Mori, E., Milanesi, P., Menchetti, M., Zozzoli, R., Monaco, A., Capizzi, D., et al. (2018). Genetics reveals that free-ranging chipmunks introduced to Italy have multiple origins. Hystrix-Italian. J. Mamm. 29, 239–242. doi: 10.4404/hystrix-00116-2018
National Academies of Sciences, Engineering, and Medicine (NASEM). (2016). Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values. Washington, D.C: National Academies Press.
Oh, K., Shiels, A., Shiels, L., Blondel, D. V., Campbell, K. J., Royden Saah, J., et al. (2021). Population genomics of invasive rodents on islands: genetic consequences of colonization and prospects for localized synthetic gene drive. Evol. Appl. 14, 1421–1435. doi: 10.1111/eva.13210
Okuyama, M. W., Shimozuru, M., Nakai, M., Yamaguchi, E., Fujii, K., Shimada, K., et al. (2020). Genetic population structure of invasive raccoons (Procyon lotor) in Hokkaido, Japan: Unique phenomenon caused by pet escape or abandonment. Sci. Rep. 10:8108. doi: 10.1038/s41598-020-64526-y
Oliver, K., and Cairney, P. (2019). The dos and don’ts of influencing policy: a systematic review of advice to academics. Palgrave Commun. 5:21. doi: 10.1057/s41599-019-0232-y
Oppel, S., Havery, S. J., John, L., Bambini, L., Varnham, K., Dawson, J., et al. (2019). Maximising conservation impact by prioritising islands for biosecurity. Sandy, UK: Royal Society for the Protection of Birds, 659–665.
Orkin, J. D., Yang, Y., Yang, C., Yu, D. W., and Jiang, X. (2016). Cost-effective scat-detection dogs: unleashing a powerful new tool for international mammalian conservation biology. Sci. Rep. 6:34758. doi: 10.1038/srep34758
Ortiz-Alcaraz, A., Aguirre-Muñoz, A., Méndez-Sánchez, F., Rojas-Mayoral, E., Solís-Carlos, F., Rojas-Mayoral, B., et al. (2019). “Ecological restoration of Socorro Island, Revillagigedo Archipelago, Mexico: the eradication of feral sheep and cats,” in Island Invasives: Scaling up to meet the challenge, eds C. R. Veitch, M. N. Clout, and A. R. Martin (Gland, Switzerland: IUCN), 267–273.
Pichlmueller, F., Murphy, E. C., MacKay, J. W. B., Henderson, J., Fewster, R. M., and Russell, J. C. (2020). Island invasion and reinvasion: Informing invasive species management with genetic measures of connectivity. J. Appl. Ecol. 57, 2258–2270. doi: 10.1111/1365-2664.13727
Pimentel, D. (2011). Biological Invasions: Economic and Environmental Costs of Alien Plant, Animal, and Microbe Species, 2nd Edn. Boca Raton, FL: CRC Press.
Puckett, E. E., Magnussen, E., Khlyap, L. A., Strand, T. M., Lundkvist, Å, and Munshi-South, J. (2020). Genomic analyses reveal three independent introductions of the invasive brown rat (Rattus norvegicus) to the Faroe Islands. Heredity 124, 15–27. doi: 10.1038/s41437-019-0255-6
Puckett, E. E., Park, J., Combs, M., Blum, M. J., Bryant, E., Caccone, A., et al. (2016). Global population divergence and admixture of the brown rat (Rattus norvegicus). Proc. R. Soc. Lond. B Biol. Sci. 283:20161762. doi: 10.1098/rspb.2016.1762
Ramsey, D., and Will, D. (2012). Framework for undertaking eradication programs on insular populations of vertebrate pests. Unpublished Client Report for Island Conservation. Heidelberg, Victoria: Arthur Rylah Institute for Environmental Research, Department of Sustainability and Envionment.
Reaser, J. K., Burgiel, S. W., Kirkey, J., Brantley, K. A., Veatch, S. D., and Burgos-Rodríguez, J. (2020). The early detection of and rapid response (EDRR) to invasive species: a conceptual framework and federal capacities assessment. Biol. Invasions 22, 1–19. doi: 10.1007/s10530-019-02156-w
Ricciardi, A., Blackburn, T. M., Carlton, J. T., Dick, J. T. A., Hulme, P. E., Iacarella, J. C., et al. (2017). Invasion science: a horizon scan of emerging challenges and opportunities. Trends. Ecol. Evol. 32, 464–474. doi: 10.1016/j.tree.2017.03.007
Rius, M., Bourne, S., Hornsby, H. G., and Chapman, M. A. (2015). Applications of next-generation sequencing to the study of biological invasions. Curr. Zool. 61, 488–504. doi: 10.1093/czoolo/61.3.488
Roberts, M., Cresswell, W., and Hanley, N. (2018). Prioritising invasive species control actions: evaluating effectiveness, costs, willingness to pay and social acceptance. Ecol. Econ. 152, 1–8. doi: 10.1016/j.ecolecon.2018.05.027
Roda, F., Sentilles, J., Molins, C., Duchamp, C., and Hansen, É, and Jean, N. (2021). Wolf scat detection dog improves wolf genetic monitoring in new French colonized areas. J. Vertebr. Biol. 69:20102. doi: 10.25225/jvb.20102
Rode, N. O., Estoup, A., Bourguet, D., Courtier-Orgogozo, V., and Débarre, F. (2019). Population management using gene drive: molecular design, models of spread dynamics and assessment of ecological risks. Conserv. Genet. 20, 671–690. doi: 10.1007/s10592-019-01165-5
Rollins, L. A., Woolnough, A. P., and Sherwin, W. B. (2006). Population genetic tools for pest management: a review. Wildl. Res. 33, 251–261. doi: 10.1071/WR05106
Roussel, J.-M., Paillisson, J.-M., Tréguier, A., and Petit, E. (2015). The downside of eDNA as a survey tool in water bodies. J. Appl. Ecol. 52, 823–826. doi: 10.1111/1365-2664.12428
Ruppert, K. M. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17:e00547. doi: 10.1016/j.gecco.2019.e00547
Russell, J. C., Meyer, J.-Y., Holmes, N. D., and Pagad, S. (2017). Invasive alien species on islands: impacts, distribution, interactions and management. Environ. Conserv. 44, 359–370. doi: 10.1017/S0376892917000297
Russell, J. C., Miller, S. D., Harper, G. A., MacInnes, H. E., Wylie, M. J., and Fewster, R. M. (2010). Survivors or reinvaders? Using genetic assignment to identify invasive pests following eradication. Biol. Invasions 12, 1747–1757. doi: 10.1007/s10530-009-9586-1
Russello, M. A., Avery, M. L., and Wright, T. F. (2008). Genetic evidence links invasive monk parakeet populations in the United States to the international pet trade. BMC Evol. Biol. 8:217. doi: 10.1186/1471-2148-8-217
Russello, M. A., Smith-Vidaurre, G., and Wright, T. F. (2021). “Genetics of Invasive Parrot Populations,” in Naturalized Parrots of the World, ed. S. Pruett Jones (Princeton, NJ: Princeton University Press), 54–70.
Russello, M. A., Waterhouse, M. D., Etter, P. D., and Johnson, E. A. (2015). From promise to practice: pairing non-invasive sampling with genomics in conservation. Peer J. 3:e1106. doi: 10.7717/peerj.1106
Sandström, A., Lundmark, C., Andersson, K., Johannesson, K., and Laikre, L. (2019). Understanding and bridging the conservation-genetics gap in marine conservation. Conserv. Biol. 33, 725–728. doi: 10.1111/cobi.13272
Sarre, S. D., MacDonald, A. J., Barclay, C., Saunders, G. R., and Ramsey, D. S. L. (2013). Foxes are now widespread in Tasmania: DNA detection defines the distribution of this rare but invasive carnivore. J. Appl. Ecol. 50, 459–468. doi: 10.1111/1365-2664.12011
Savidge, J. A., Hopken, M. W., Witmer, G. W., Jojola, S. M., Pierce, J. J., Burke, P. W., et al. (2012). Genetic evaluation of an attempted Rattus rattus eradication on Congo Cay, U.S. Virgin Islands, identifies importance of eradication units. Biol. Invasions 14, 2343–2354. doi: 10.1007/s10530-012-0233-x
Schmidt, D. A., Campbell, N. R., Govindarajulu, P., Larsen, K. W., and Russello, M. A. (2020). Genotyping-in-Thousands by sequencing (GT-seq) panel development and application to minimally invasive DNA samples to support studies in molecular ecology. Mol. Ecol. Resour. 20, 114–124. doi: 10.1111/1755-0998.13090
Shafer, A. B. A., Wolf, J. B. W., Alves, P. C., Bergström, L., Bruford, M. W., Brännström, I., et al. (2015). Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 30, 78–87. doi: 10.1016/j.tree.2014.11.009
Simberloff, D. (2013). Invasive Species: What Everyone Needs to Know. New York: Oxford University Press.
Simberloff, D., Genovesi, P., Pysek, P., and Campbell, K. (2011). Recognizing conservation success. Science 332, 419–419. doi: 10.1126/science.332.6028.419-a
Simberloff, D., Keitt, B., Will, D., Holmes, N., Pickett, E., and Genovesi, P. (2018). Yes we can! exciting progress and prospects for controlling invasives on Islands and beyond. West North Am. Nat. 78:942. doi: 10.3398/064.078.0431
Sjodin, B. M. F., Irvine, R. L., Ford, A. T., Howald, G. R., and Russello, M. A. (2019). Rattus population genomics across the Haida Gwaii archipelago provides a framework for guiding invasive species management. Evol. Appl. 13, 889–904. doi: 10.1111/eva.12907
Sjodin, B. M. F., Irvine, R. L., and Russello, M. A. (2020a). RapidRat: Development, validation and application of a genotyping-by-sequencing panel for rapid biosecurity and invasive species management. PLoS One 15:e0234694. doi: 10.1371/journal.pone.0234694
Sjodin, B. M. F., Puckett, E. E., Irvine, R. L., Munshi-South, J., and Russello, M. A. (2020b). Global origins of invasive brown rats (Rattus norvegicus) in the Haida Gwaii archipelago. Biol. Invasions 23, 1–13. doi: 10.1007/s10530-020-02390-7
Smart, A. S., Tingley, R., Weeks, A. R., van Rooyen, A. R., and McCarthy, M. A. (2015). Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol. Appl. 25, 1944–1952. doi: 10.1890/14-1751.1
Spatz, D. R., Zilliacus, K. M., Holmes, N. D., Butchart, S. H. M., Genovesi, P., Ceballos, G., et al. (2017). Globally threatened vertebrates on islands with invasive species. Sci. Adv. 3:e1603080. doi: 10.1126/sciadv.1603080
Sudweeks, J., Hollingsworth, B., Blondel, D. V., Campbell, K. J., Dhole, S., Eisemann, J. D., et al. (2019). Locally fixed alleles: A method to localize gene drive to island populations. Sci. Rep. 9:15821. doi: 10.1038/s41598-019-51994-0
Sutherland, W. J., and Wordley, C. F. R. (2017). Evidence complacency hampers conservation. Nat. Ecol. Evol. 1, 1215–1216. doi: 10.1038/s41559-017-0244-1
Taberlet, P., Coissac, E., Hajibabaei, M., and Rieseberg, L. H. (2012). Environmental DNA. Mol. Ecol. 21, 1789–1793. doi: 10.1111/j.1365-294X.2012.05542.x
Taberlet, P., and Waits, L. P. (1999). Non-invasive genetic sampling. Trends Ecol. Evol. 13, 26–27. doi: 10.1016/S0169-5347(97)01276-7
Taylor, H., Dussex, N., and van Heezik, Y. (2017). Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Glob. Ecol. Conserv. 10, 231–242. doi: 10.1016/j.gecco.2017.04.001
Tershy, B. R., Shen, K.-W., Newton, K. M., Holmes, N. D., and Croll, D. A. (2015). The importance of islands for the protection of biological and linguistic diversity. BioScience 65, 592–597. doi: 10.1093/biosci/biv031
Tingley, R., Greenlees, M., Oertel, S., van Rooyen, A. R., and Weeks, A. R. (2019). Environmental DNA sampling as a surveillance tool for cane toad Rhinella marina introductions on offshore islands. Biol. Invasions 21, 1–6. doi: 10.1007/s10530-018-1810-4
Unckless, R. L., Messer, P. W., Connallon, T., and Clark, A. G. (2015). Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 201, 425–431. doi: 10.1534/genetics.115.177592
Ushio, M., Fukuda, H., Inoue, T., Makoto, K., Kishida, O., Sato, K., et al. (2017). Environmental DNA enables detection of terrestrial mammals from forest pond water. Mol. Ecol. Resour. 17, e63–e75. doi: 10.1111/1755-0998.12690
Veale, A. J., Clout, M. N., and Gleeson, D. M. (2012). Genetic population assignment reveals a long-distance incursion to an island by a stoat (Mustela erminea). Biol. Invasions 14, 735–742. doi: 10.1007/s10530-011-0113-9
Veitch, C. R., and Clout, M. N. (2002). Turning the Tide: The Eradication of Invasive Species. Proceedings of the International Conference on Eradication of Island Invasives. Gland, Switzerland and Cambridge, UK: International Union for Conservation of Nature.
Veitch, C. R., Clout, M. N., Martin, A. R., Russell, J. C., and West, C. J. (2019). Island Invasives: Scaling up to meet the challenge. Proceedings of the international conference on island invasives 2017. Gland, Switzerland: International Union for Conservation of Nature.
Veitch, C. R., Clout, M. N., and Towns, D. R. (2011). Island Invasives: Eradication and Management. Proceedings of the International Conference on Island Invasives. Gland, Switzerland: International Union for Conservation of Nature.
Velando, A., Morán, P., Romero, R., Fernández, J., and Piorno, V. (2017). Invasion and eradication of the American mink in the Atlantic Islands National Park (NW Spain): a retrospective analysis. Biol. Invasions 19, 1227–1241. doi: 10.1007/s10530-016-1326-8
Vernesi, C., Bruford, M. W., Bertorelle, G., Pecchioli, E., Rizzoli, A., and Hauffe, H. C. (2008). Where’s the conservation in conservation genetics? Conserv. Biol. 22, 802–804. doi: 10.1111/j.1523-1739.2008.00911.x
Waits, L. P., and Paetkau, D. (2005). Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate collection. J. Wildl. Manag. 69, 1419–1433.
Wallace, A. R. (1880). Island Life: Or, The Phenomena and Causes of Insular Faunas and Floras, Including a Revision and Attempted Solution of the Problem of Geological Eliminates. New York: Macmillan and Company.
Webber, B. L., Raghu, S., and Edwards, O. R. (2015). Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc. Natl. Acad. Sci. 112, 10565–10567. doi: 10.1073/pnas.1514258112
Wetterstrand, K. (2020). DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available Online at: www.genome.gov/sequencingcostsdata (accessed March 3, 2021).
Williams, K. E., Huyvaert, K. P., Vercauteren, K. C., Davis, A. J., and Piaggio, A. J. (2018). Detection and persistence of environmental DNA from an invasive, terrestrial mammal. Ecol. Evol. 8, 688–695. doi: 10.1002/ece3.3698
Wostenberg, D. J., Hopken, M. W., Shiels, A. B., and Piaggio, A. J. (2019). Using DNA to identify the source of invasive mongooses, herpestes auropunctatus (Carnivora: Herpestidae) Captured on Kaua‘i, Hawaiian Islands. Pac. Sci. 73:215. doi: 10.2984/73.2.3
Keywords: invasive alien species management, island biodiversity, species-at-risk, conservation genetics and genomics, gene drive, university-government-industry collaboration, biosecurity
Citation: Burgess BT, Irvine RL, Howald GR and Russello MA (2021) The Promise of Genetics and Genomics for Improving Invasive Mammal Management on Islands. Front. Ecol. Evol. 9:704809. doi: 10.3389/fevo.2021.704809
Received: 03 May 2021; Accepted: 18 June 2021;
Published: 03 August 2021.
Edited by:
Emiliano Mori, University of Siena, ItalyReviewed by:
Andrea Galimberti, University of Milano-Bicocca, ItalyDavide Sogliani, University of Pavia, Italy
Copyright © 2021 Burgess, Irvine, Howald and Russello. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael A. Russello, michael.russello@ubc.ca
 Brock T. Burgess
Brock T. Burgess