- Evolutionary Anthropology Department, Duke University, Durham, NC, United States
The overuse of man-made antibiotics has facilitated the global propagation of antibiotic resistance genes in animals, across natural and anthropogenically disturbed environments. Although antibiotic treatment is the most well-studied route by which resistance genes can develop and spread within host-associated microbiota, resistomes also can be acquired or enriched via more indirect routes, such as via transmission between hosts or via contact with antibiotic-contaminated matter within the environment. Relatively little is known about the impacts of anthropogenic disturbance on reservoirs of resistance genes in wildlife and their environments. We therefore tested for (a) antibiotic resistance genes in primate hosts experiencing different severities and types of anthropogenic disturbance (i.e., non-wildlife animal presence, human presence, direct human contact, and antibiotic treatment), and (b) covariation between host-associated and environmental resistomes. We used shotgun metagenomic sequencing of ring-tailed lemur (Lemur catta) gut resistomes and associated soil resistomes sampled from up to 10 sites: seven in the wilderness of Madagascar and three in captivity in Madagascar or the United States. We found that, compared to wild lemurs, captive lemurs harbored greater abundances of resistance genes, but not necessarily more diverse resistomes. Abundances of resistance genes were positively correlated with our assessments of anthropogenic disturbance, a pattern that was robust across all ten lemur populations. The composition of lemur resistomes was site-specific and the types of resistance genes reflected antibiotic usage in the country of origin, such as vancomycin use in Madagascar. We found support for multiple routes of ARG enrichment (e.g., via human contact, antibiotic treatment, and environmental acquisition) that differed across lemur populations, but could result in similar degrees of enrichment. Soil resistomes varied across natural habitats in Madagascar and, at sites with greater anthropogenic disturbance, lemurs and soil resistomes covaried. As one of the broadest, single-species investigations of wildlife resistomes to date, we show that the transmission and enrichment of antibiotic resistance genes varies across environments, thereby adding to the mounting evidence that the resistance crisis extends outside of traditional clinical settings.
Introduction
Antibiotic resistance genes (ARGs) occur naturally and are evolutionarily ancient (Aminov and Mackie, 2007; D’Costa et al., 2011), but the pervasive use of antibiotics has accelerated their global propagation and precipitated a resistance crisis (Ventola, 2015; Van Puyvelde et al., 2018). The ARGs within a microbial community, collectively known as the resistome, influence the function of native bacteria (Kim et al., 2020), increase pathogen morbidity and mortality rates (Howard et al., 2003; Lin et al., 2015), and diminish the efficacy of antibiotics (Rossolini et al., 2014; Martens and Demain, 2017). ARGs are thus well-studied in clinical populations or agricultural systems that receive antibiotic treatment (French, 2005; Ghosh and LaPara, 2007; Lerminiaux and Cameron, 2019). Although ARGs are also components of anthropogenic disturbance, such that contact with infected hosts or contaminated environments facilitates the transfer and incorporation of ARGs into natural microbial consortia (Hiltunen et al., 2017; Manaia, 2017), the presence and abundance of ARGs in wildlife and non-model animals remains relatively understudied (Dolejska and Literak, 2019; Ramey and Ahlstrom, 2020). Here, we use a site-comparative approach within a single host species, the endangered ring-tailed lemur (Lemur catta), to (a) characterize ARGs in the gut microbiota of multiple wild and captive populations, (b) determine if identified ARGs correlate with components of anthropogenic disturbance faced by these populations, and (c) test for covariation between ARGs in wild lemurs and their natural environments, as proxied by ARGs in soil microbes.
Antibiotic resistance genes evolved as defense mechanisms, so natural ARGs are diverse, present in low abundances, and persist in microbial communities with no known human contamination (Segawa et al., 2013; Jardine et al., 2019). Their propensity to be exchanged via horizontal gene transfer has allowed ARGs to pass between and persist within host-associated and environmental microbial communities (Baquero et al., 2009; Forsberg et al., 2012; Martínez, 2012). The associated arms race between the evolution of protective ARGs and antibiotic production has resulted in myriad, naturally occurring resistance mechanisms (Sengupta et al., 2013; Versluis et al., 2015; Van Goethem et al., 2018). The increased selective pressure from human antibiotic use, coupled with widespread human encroachment into natural environments, is thus expanding ARG reservoirs in wildlife and their environments (Gatica and Cytryn, 2013; Berglund, 2015; Anthony et al., 2018).
The most notable and well-studied route of ARG enrichment within a microbial community stems from treatment with man-made antibiotics (Alanis, 2005). This direct route of enrichment has greatly contributed to the spread of clinically relevant ARGs – those that confer resistance to multiple drug types or to last-resort antibiotics (Koch et al., 2014; Mühlberg et al., 2020). Once resistant microbes are present in a microbial community, they also can indirectly enrich the community resistome by persisting as community members and/or by transferring ARGs to native microbes (Chee-Sanford et al., 2009; Gao et al., 2018). Resistant microbes can be shared between humans and companion animals (Pomba et al., 2017), acquired through consumption of contaminated dietary items (Wang et al., 2006; Nawaz et al., 2011), or acquired via contact with human or agricultural waste that includes antibiotic residues or ARGs (Addison, 1984), the latter prompting investigation of soil microbiomes (Esiobu et al., 2002; D’Costa et al., 2007).
Antibiotic resistance genes are thus a newly recognized component of the Anthropocene, expanding the traditional definition of human disturbance (e.g., habitat degradation) to include perturbations of microbial communities (Tripathi and Cytryn, 2017; Zhu et al., 2017; Trevelline et al., 2019) that can influence host-associated microbiota, transfer ARGs to pathogens, and alter the host’s immune function (Bengtsson-Palme et al., 2018; Dafale et al., 2020). ARG spillover into wild animals also has the potential to subsequently re-infect exposed humans or other animals (Pomba et al., 2017). The risk of ARG enrichment in wildlife is thus linked to the potential for both direct (e.g., antibiotic administration) and indirect (e.g., human and agriculture presence) routes of transmission. As a first step to determining the respective roles of these enrichment routes, we will characterize ARGs in systems that portray varying types and severities of anthropogenic disturbance.
Madagascar is a prime study site. Its ongoing population and agricultural boom has increased the demand for antibiotics to treat human and agricultural diseases, thereby enriching ARGs in human and domestic animal populations (Gay et al., 2017; Padget et al., 2017; Randrianirina et al., 2010). Furthermore, antibiotics are broadly used to treat Madagascar’s semi-regular plague outbreaks (caused by the bacterium Yersinia pestis) (Boisier et al., 2002; Andrianaivoarimanana et al., 2013; Salam et al., 2020), which has fueled the demand for antibiotics (Rasamiravaka, 2020). Coupled with a lack of infrastructure to contain human and agricultural waste, ubiquitous antibiotic use in Madagascar poses a significant risk of widespread ARG propagation to its unique and endangered wildlife.
Unlike many ecologically specialized lemurs endemic to Madagascar, the ecologically flexible ring-tailed lemur is a semi-terrestrial omnivore that performs geophagy or earth-eating (Ganzhorn, 1987; Gould, 2006); it survives and reproduces successfully in greatly disturbed habitats and in a wide range of captivity settings (Jolly et al., 2006; Mason, 2010). Across their natural range, ring-tailed lemurs live along a gradient of disturbance from near-pristine forests, that have minimal human activity, to degraded habitats with heavy human encroachment, including logging, agricultural land use, and hunting. Similarly, captive ring-tailed lemurs live under conditions that range from naturalized settings (e.g., social housing in natural habitat enclosures) to artificial settings, some even living solitarily, as pets, in human dwellings, where they experience consistent human contact and substandard conditions (Reuter et al., 2015; Reuter and Schaefer, 2016; LaFleur et al., 2019).
Regarding direct routes of ARG enrichment, lemurs in natural and captivity settings have differential exposure to antibiotic treatment. Whereas most wild lemurs and lemurs kept as illegal pets in Madagascar will have never experienced antibiotic treatment (the latter owing to lack of veterinary care), populations at well-established field sites may have, on occasion, experienced antibiotic treatment during capture sessions or as part of a research protocol. At the other extreme, lemurs at research or rescue facilities will have experienced routine veterinary care, including potentially frequent antibiotic treatment. Regarding indirect routes of ARG enrichment, wild and captive lemurs differ in their exposure to humans and domestic animals, in their potential for social transmission between conspecifics, and in their potential to acquire ARGs from their environment.
We first characterize ARGs in the gut microbiota of wild and captive ring-tailed lemurs, by population. We predict that the abundance and diversity of ARGs across populations will positively correlate with the degree of overall anthropogenic disturbance, and that any clinically relevant ARGs in wild lemurs will reflect antibiotic use in human and agricultural settings in Madagascar. For captive lemurs, we expect overall ARG abundance to be greater than in wild conspecifics; however, the diversity and type of ARGs could reflect different routes of enrichment. In captive lemurs at facilities that provide naturalized settings and routine veterinary care, we expect ARGs to reflect treatment with common antibiotics. By contrast, in pet lemurs that are unlikely to receive antibiotic treatment, but have consistent and sustained direct contact with humans and domestic animals, we expect to observe ARGs that confer resistance to antibiotics commonly used in Madagascar (such as those used to treat the plague).
Similar to the patterns predicted for lemur ARGs, we expect ARGs in soil microbiota to also correlate with the degree of environmental disturbance and to also reflect the types of antibiotics used in human and agricultural settings in Madagascar. As regards ARG covariation between wild lemur resistomes and soil resistomes, we expect soil from the more disturbed habitats to have greater abundance and diversity of ARGs than would soil from the more pristine habitats. Consistent with evidence that hosts acquire microbes from their environments (Smith et al., 2015; Adair and Douglas, 2017; Selway et al., 2020), we expect wild lemur resistomes to be more similar to soil resistomes from their local habitat than to soil resistomes from outside their home ranges.
Materials and Methods
Study Sites and Subjects
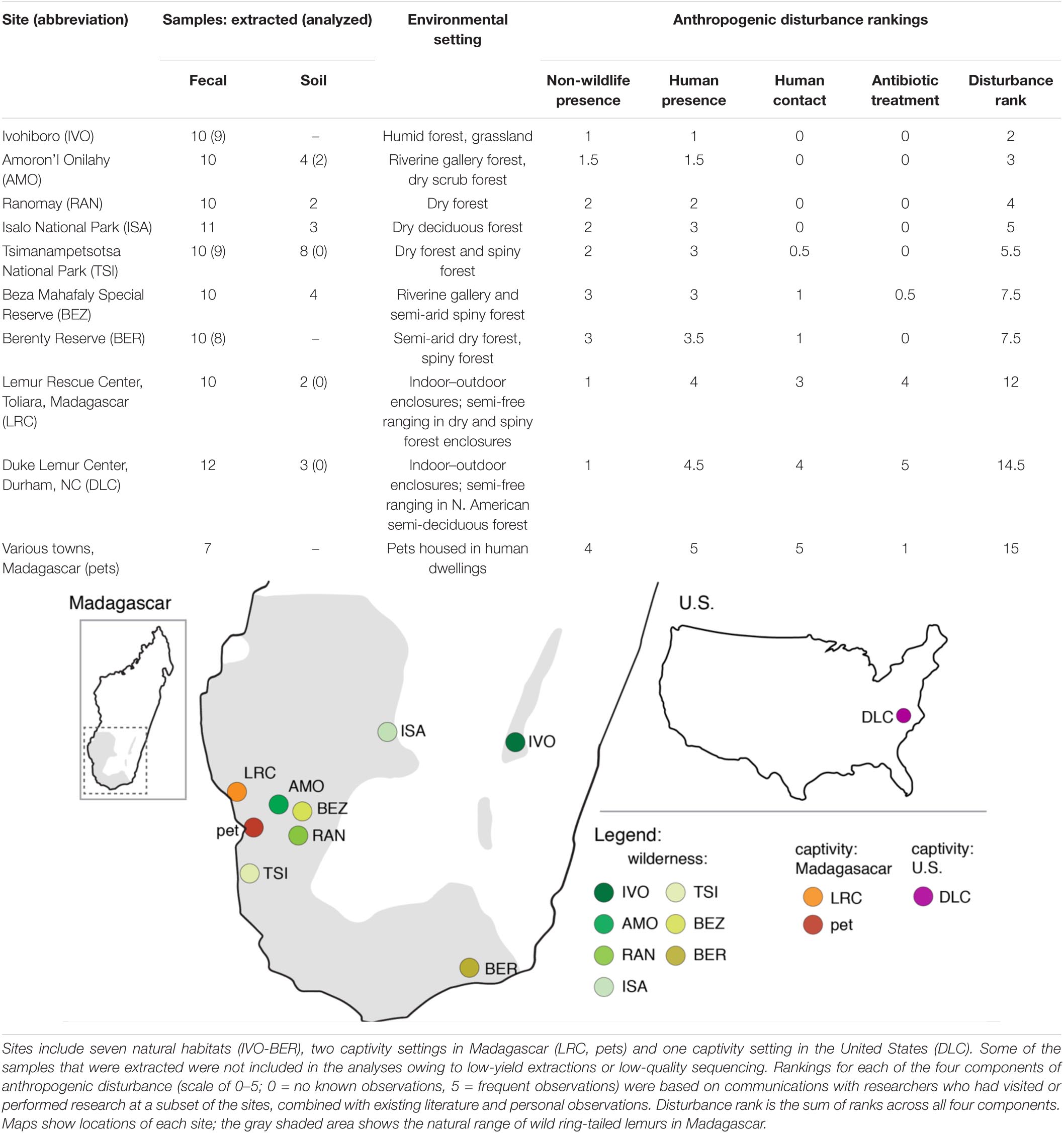
We collected lemur fecal samples and soil samples from the following 10 habitats or settings, hereafter called ‘sites’ (Table 1): seven natural sites (e.g., national parks, community-managed reserves across the lemurs’ natural range in Madagascar); two captivity sites in Madagascar (the Lemur Rescue Center or LRC and, collectively, households with pet lemurs), and; one captivity site in the United States (the Duke Lemur Center or DLC). Although the pet lemurs derived from different households in different townships, we categorized them as one population or site because of the specific, shared experience of pethood (see below and Supplementary Table 1 for more details). We selected these sites based on the presence of ring-tailed lemurs, feasibility of sample collection, and a priori predictions that these sites would span a gradient of anthropogenic disturbance.
The lemurs at the seven natural sites (n = 71) free-ranged in groups of 5–24 individuals. Those at the LRC (n = 10) and DLC (n = 12) were socially housed in indoor–outdoor enclosures, and some gained additional access to forested enclosures where they could semi-free range. The latter were provided facility-standardized diets (i.e., fresh produce and commercial chow: Mowry and Campbell, 2001). The pet lemurs (n = 7) were kept solitarily in human dwellings, were fed fruit, rice, and other foods marketed for human consumption, and had frequent contact with humans and domestic animals (personal observations and communications).
Assessing Anthropogenic Disturbance
We identified four components of potential anthropogenic disturbance across our 10 sites that we expected might influence ARG prevalence: (1) non-wildlife animal presence, (2) human presence, (3) direct human contact, and (4) antibiotic treatment. Examples of (1) exposure to non-wildlife animal presence in natural habitats include via grazing or the holding of agricultural animals, as well as via free-roaming domestic or feral dogs and cats. In more developed areas, non-wildlife animals are kept in proximity to human dwellings and/or can roam in areas shared with humans (e.g., companion animals, free-ranging poultry, and rats). As regards (2) human presence, lemurs are exposed to the local Malagasy and to foreigners (e.g., researchers and tourists), and to their waste (including feces). Most common examples of human activity within natural habitats are (a) shepherding agricultural animals, (b) harvesting natural resources (e.g., logging, hunting), (c) agriculture (e.g., tending rice paddies), and (d) foot traffic. In captivity settings, human presence encompasses animal caretakers, veterinarians, visitors, and/or pet owners. (3) Direct human contact with lemurs most commonly entails handling of the animals for research or veterinary purposes. Although it is rare for certain populations of wild lemurs to have human contact, some have been subjects of long-term research, involving annual captures. Pet lemurs have frequent contact with their owners and other humans (e.g., for promoting tourism). Lastly, (4) antibiotic treatment in wild lemurs is only applicable at certain established field sites (e.g., BEZ, BER) where antibiotics have been used during capture sessions for veterinary purposes. For captive animals, antibiotics are used for veterinary treatment and research purposes; for pet owners, antibiotics are cost prohibitive and seeking veterinary treatment may have legal consequences. Additional information (and citations) about each site is located in Supplementary Table 1.
For the purpose of ranking these threats, we weighted each component independently and equally (rankings in Table 1 are based on the criteria and information presented in Supplementary Table 1). Each of five researchers, who had visited or performed research at a subset of the sites, provided descriptions of the sites and information on the presence or frequency of the four disturbance components (Supplementary Table 1). We combined this information with any existing literature, and ranked each component on a scale of 0–5 (0 = no known observations, 5 = frequent observations) for each of the 10 sites. We then tallied the rankings across the four components to estimate overall disturbance at each site (Table 1 and Supplementary Table 1 list sites in ascending order of total estimated disturbance). Although differing in severity, agriculture and human presence were estimated to influence lemurs at nearly all sites. By contrast, direct human contact and antibiotic treatment were largely limited to impacting lemurs in captivity settings, with the exceptions being TSI, BEZ, and BER, where lemurs are infrequently handled for research purposes.
Sample Collection
The protocols associated with sample collection are detailed in Bornbusch et al., 2021. In brief, we opportunistically collected fresh fecal samples from lemurs at all 10 sites. We additionally collected soil samples from the lemurs’ core areas at four of the natural habitat sites (Table 1). Each soil sample was a composite of five aliquots of topsoil (2–3 cm deep) from a 2–3 m2 area of the resident lemurs’ core range. To avoid the confounding variable of seasonality, we collected all samples during the dry season in Madagascar (May–October, 2016–2020) and a single fall season in the United States (October 2017). Each sample was placed in an Omnigene tube containing a stabilizing buffer that preserves microbial communities at room temperature for 8 weeks (Omnigene.Gut tube, DNAgenotek, ON, Canada; Choo et al., 2015; Song et al., 2016) and, within that 8-week period, the samples were transported to the United States and stored at −80°C until analysis.
Microbial DNA Extraction, Sequencing, and Bioinformatics
Following the manufacturer’s protocols for the DNeasy Powersoil kit (QIAGAN, Frederick, MD, United States), we extracted bacterial genomic DNA from fecal and soil samples. We quantified DNA using a Fluorometer (broad-spectrum kit, Qubit 4, Thermo Fisher Scientific, Waltham, MA, United States). Extracted samples that did not yield sufficient, extracted DNA (<1000 ng/ml) were excluded from shotgun sequencing. Aliquots of extracted DNA were sent to CosmosID Inc. (Rockville, MD, United States) for shotgun metagenomic sequencing. DNA libraries were prepared using the Illumina Nextera XT library preparation kit, with a modified protocol (Hasan et al., 2014; Lax et al., 2014). Library quantity was assessed with Qubit (Thermo Fisher). Libraries were then sequenced on an Illumina HiSeq platform 2 × 150 bp. On average, samples yielded approximately 17 million total sequence reads per sample, with an average of ∼18 million and ∼10 million reads for fecal and soil samples, respectively. Sequences with fewer than five million reads were excluded from downstream analyses.
Unassembled sequencing reads were directly analyzed using CosmosID’s bioinformatics platform for identifying and profiling ARGs (described in Hasan et al., 2014; Lax et al., 2014; Ottesen et al., 2016). Briefly, the system uses curated genome databases and a high-performance, data-mining algorithm that rapidly disambiguates millions of metagenomic sequence reads. ARGs in the microbiome were identified by querying the unassembled sequence reads against the CosmosID curated antibiotic resistance and virulence-associated gene databases. Outputs include the identity and family, percent gene coverage, and frequency counts of ARGs within each sample. A total of ∼2.5 million ARGs were identified across all samples, with an average of ∼22,000 per sample, and were the only microbial genes analyzed in this study. We categorized clinically relevant ARGs (i.e., those that pose significant risk to human health) using existing literature on the global enrichment, mobility, and pathogenicity of ARGs (Martínez et al., 2015; Zhang et al., 2019).
Statistical Analyses
To calculate the relative abundance of ARGs within a fecal or soil sample, we divided the frequency count of all ARGs or specific gene families by the sample’s total read count. Because relative abundance is dependent on sequencing depth, whereas raw sequence counts are not, relative abundance can better account for sequencing-related biases. To determine the diversity of ARGs, we calculated Shannon–Weaver and Simpson diversity (R studio, ver. 4.0.2), two widely used diversity indices that reflect community richness and evenness, respectively (Legendre and Legendre, 2012). For fecal samples, we used linear mixed models (LMMs) of the relative abundance or diversity of ARGs to test for correlations with the disturbance ranks, while taking into account the research site as a random effect. To determine the composition of ARGs in lemur gut and soil microbiota, we calculated Bray–Curtis dissimilarities, a metric that takes into account the presence and abundance of features (Bray and Curtis, 1957). We used the resulting dissimilarity matrix to perform principal coordinate analyses, allowing us to visualize the clustering of ARG composition, and to compare ARG composition within and between sites (via Kruskal–Wallis tests with Benjamini–Hochberg adjustments). Because relative abundance and diversity metrics are influenced by sequencing depth, we ran the linear mixed models, detailed above, with sequencing depth as an additional fixed effect. As sequencing depth was non-significant, we removed it from our final models.
To test if soil-associated ARGs were present in lemur fecal samples, we used FEAST, a tool for microbial source tracking that relies on fast expectation-maximization (Shenhav et al., 2019). For this analysis, we used the four natural sites for which we had matched fecal (n = 41) and soil (n = 11) samples; the FEAST output includes an estimated proportion of soil-associated ARGs for each pairwise comparison (∼400) of the available samples. Because this analysis requires an assumption of directionality (i.e., from a source to a sink), we categorized soil samples as ‘sources’ of ARGs and lemur fecal samples as ‘sinks’ (Dias, 1996); however, we acknowledge the potential for bi-directional transmission of ARGs between lemurs and soils. For each fecal sample, we calculated the proportions of ARGs that were shared with each soil community or with a default, hypothetical “unknown source” that accounts for ARGs not found in the soil samples.
Results
Ring-Tailed Lemur Resistomes
We identified ARGs in the fecal samples of 89% (i.e., all but 11) of the ring-tailed lemurs in this study. Their relative abundances (i.e., proportion of all metagenomic sequence reads assigned to known ARGs) ranged from ∼0–1% (Figures 1A,B). Compared to wild lemurs, lemurs from the three captivity settings had significantly greater ARG relative abundances (Figure 1A and Supplementary Table 2); nevertheless, whether considering all of the environments surveyed (LMM; t = 7.081, p < 0.0001; Figure 1A) or only the wilderness sites (LMM; t = 2.248, p = 0.027; Figure 1B), disturbance rank significantly correlated with the relative abundance of ARGs in host guts. Qualitatively, among wild lemurs, those from the most disturbed wilderness sites (i.e., BEZ and BER) had the greatest ARG abundances. This trend was significant when comparing BER to four of the other wilderness sites (i.e., IVO, AMO, RAN, and TSI), but not when comparing to ISA (Supplementary Table 2). At most of the 10 sites, there were outlier individuals, all of which were included in the analyses; they appeared to have substantially greater ARG abundances than their site-mates (Figures 1A,B). For example, at IVO, ISA, BEZ, and BER, the relative abundance of ARGs in outlier individuals was 2–3 standard deviations greater than the average ARG abundance across the site-mates.
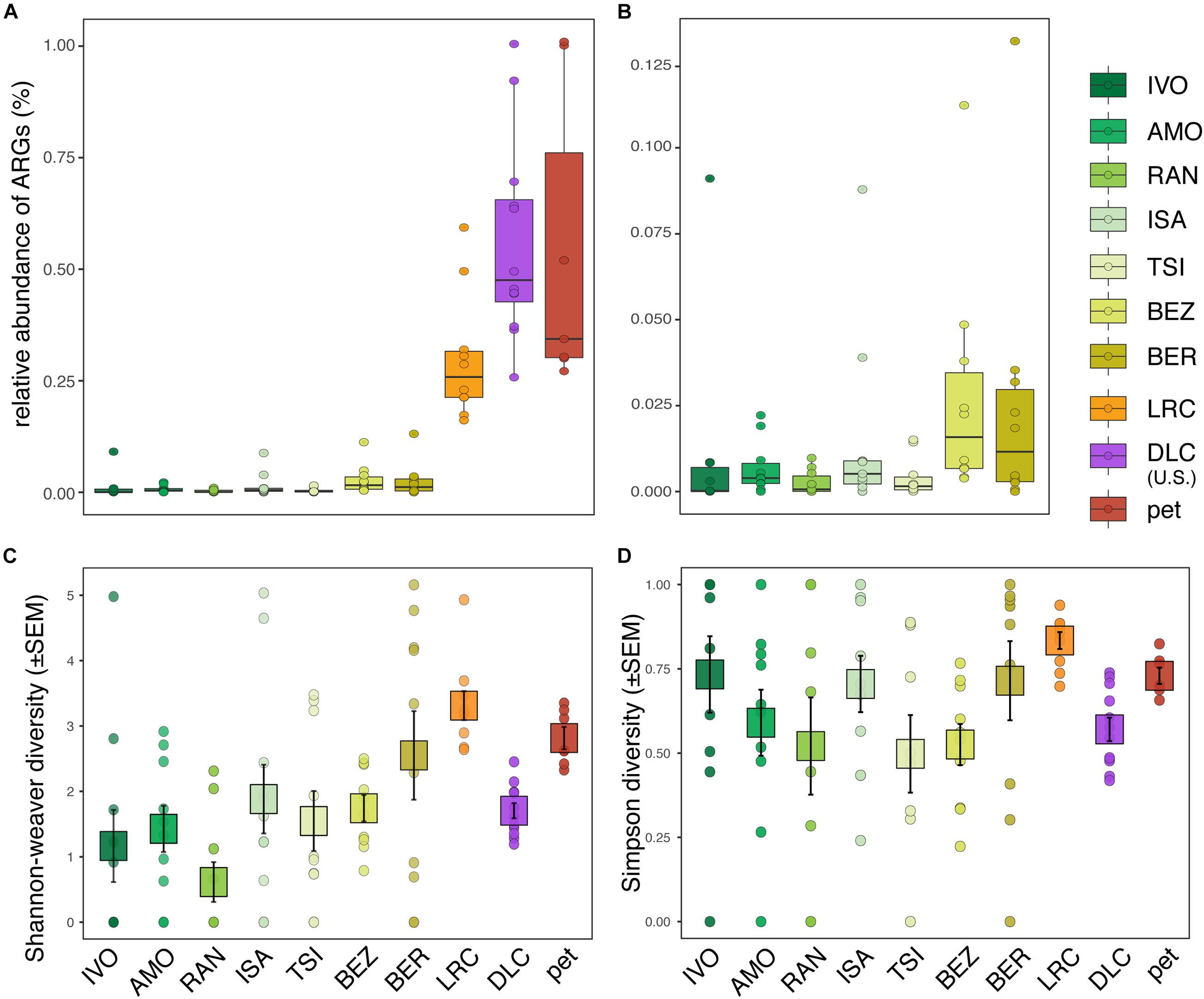
Figure 1. Relative abundance and diversity of antibiotic resistance genes (ARGs) in the gut microbiota of ring-tailed lemurs (Lemur catta) living at 10 sites: seven natural habitats, two captive settings in Madagascar, and one captive setting in the United States. Shown are the relative abundances of ARGs in (A) all lemurs and (B) wild lemurs only. Also shown are (C) Shannon–Weaver diversity and (D) Simpson diversity of ARGs across all lemurs. Sites are presented in ascending order according to their ranked level of anthropogenic disturbance (Table 1).
Compared to ARG relative abundance, the diversity metrics of ARGs in lemur fecal samples were more similar across sites, showing both high inter-individual variation within sites and substantial overlap between sites (Figures 1C,D). Disturbance rank was nonetheless a significant predictor of ARG Shannon diversity across all lemurs (LMM; all lemurs: t = 2.505, p = 0.034; Figure 1C), but not across wild lemurs (LMM; wild lemurs: t = 2.170, p = 0.081; Figure 1C). Disturbance rank did not significantly predict Simpson diversity at any scale (LMM; all lemurs: t = 0.679; p = 0.515, wild lemurs: t = −0.428, p = 0.685; Figure 1D).
Overall, lemur resistomes were largely clustered according to whether the lemur was wild or captive (Figure 2A). Within a site, wild lemurs had significantly greater interindividual variation than did captive lemurs (i.e., they clustered less tightly; Supplementary Table 3, highlighted in gray). Additionally, variation between lemur resistomes was significantly lower when comparing samples within sites versus between sites, a pattern that held true at all but one site (i.e., RAN; Supplementary Table 3, outlined in black). In other words, lemur resistomes were largely site-specific (Figures 2B–D and Supplementary Table 3). This pattern was most pronounced in the lemurs from all three captive settings, despite pet lemurs being housed at various locations, further indicating that the resistomes of captive lemurs differed dramatically from those of wild lemurs.
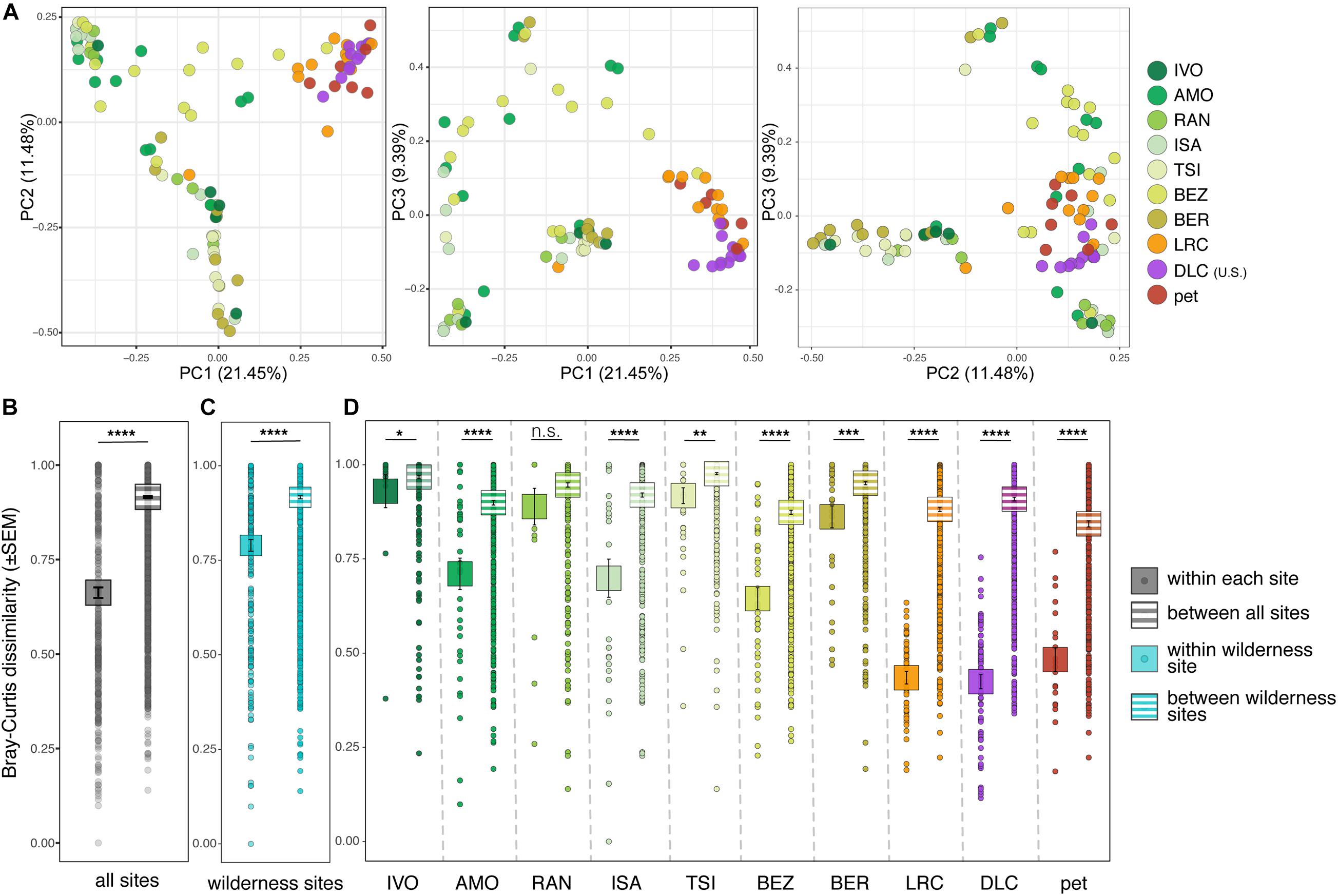
Figure 2. Beta diversity of ARGs in the gut microbiota of ring-tailed lemurs (Lemur catta) living at 10 sites (as identified in Table 1). (A) Principal coordinate analyses of Bray–Curtis dissimilarity including combinations of axes 1, 2, and 3. Analyses of dissimilarity within or between sites for (B) all lemurs, (C) only wild lemurs, and (D) by site. Sites are presented in ascending order according to the ranked level of anthropogenic disturbance (Table 1). Kruskal–Wallis test with pairwise comparisons and Benjamini–Hochberg correction; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s., nonsignificant.
Although the relative abundances of ARGs varied across lemur populations, lemur resistomes were largely dominated by ARGs in the tetracycline resistance gene family (μ = 53.47% ± σ = 36.12%), with additional notable contributions of ARGs from the vancomycin (8.45% ± 21.22%), multi-drug resistant (MDR; 7.96% ± 17.82%), beta-lactam (6.98% ± 16.51%), aminoglycoside (3.45% ± 11.29%), and macrolide (3.40% ± 8.64%) gene families (Figure 3). At the gene level, the five most prevalent ARGs included three tetracycline ARGs (tetW, tetQ, and tetO) and two vancomycin ARGs (vanH and vanS), which, when summed, accounted for 63.16% of ARGs across all of the lemurs. Certain ARGs were more prevalent in different lemur populations. Notably, lemurs from BER had marked proportions of ARGs (e.g., evgS, mdtF, and AcrF genes) that were not classified under a single gene family, but confer resistance to multiple antibiotic classes, including fluoroquinolones, penams, and macrolides (Alcock et al., 2020; Figure 3). In addition, lemurs from the LRC, predominantly, had notable proportions of phenicol resistance genes (6.88% ± 6.49%; Figure 3). Vancomycin ARGs were found almost exclusively in lemurs from Madagascar, notably in animals at the LRC, as well as in some pets (Figure 3). Although vancomycin ARGs dominated the resistomes of lemurs at IVO, it should be noted that their relative abundances were quite low.
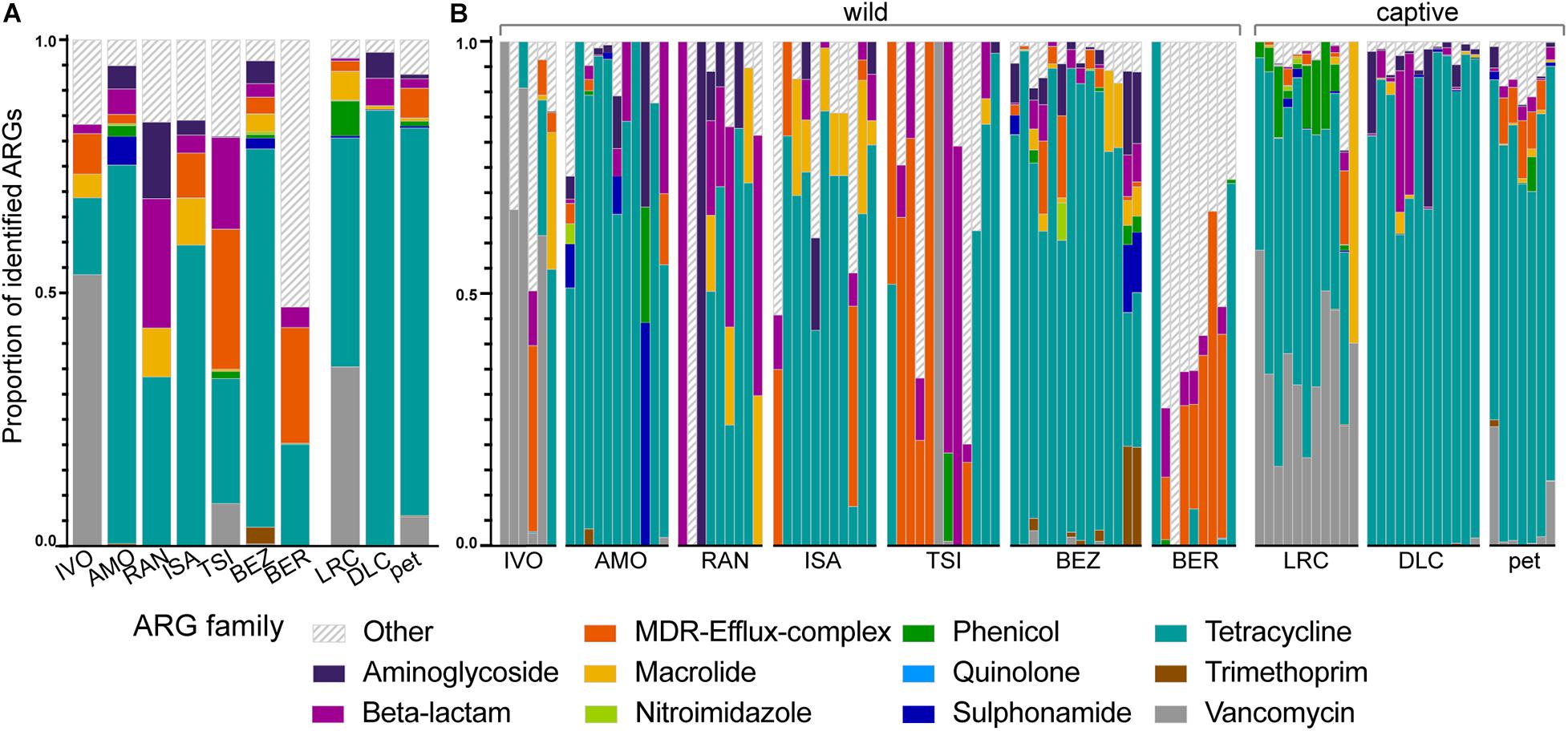
Figure 3. The proportions of identified ARGs, categorized by gene family, in the gut microbiota of ring-tailed lemurs (Lemur catta) at seven natural habitats, two captive settings in Madagascar, and one captive setting in the United States. Data are shown (A) averaged by ‘site’ and (B) by individual lemur within sites. Gene families were identified by color and those representing <1% of the ARGs were combined into the category “Other.” Sites are presented in ascending order according to the ranked level of anthropogenic disturbance (Table 1).
With respect to clinically relevant ARGs – those identified as posing significant risk to human health – relative abundance was significantly correlated with disturbance rank when comparing across all lemurs (LMM; t = 6.201, p = 0.0002; Figure 4A), but not when comparing across wild lemurs only (LMM; t = 1.459, p = 0.204; Figure 4B). This pattern was mirrored in the relative abundance of the tetW gene, the most prevalent of the clinically relevant ARGs (LMM; all lemurs: t = 5.724, p = 0.0003; wild lemurs: t = 1.281, p = 0.256; Figures 4C,D). Other clinically relevant ARGs, such as mdtE and vanY, also varied in relative abundance in lemurs across sites (Figures 4E,F).
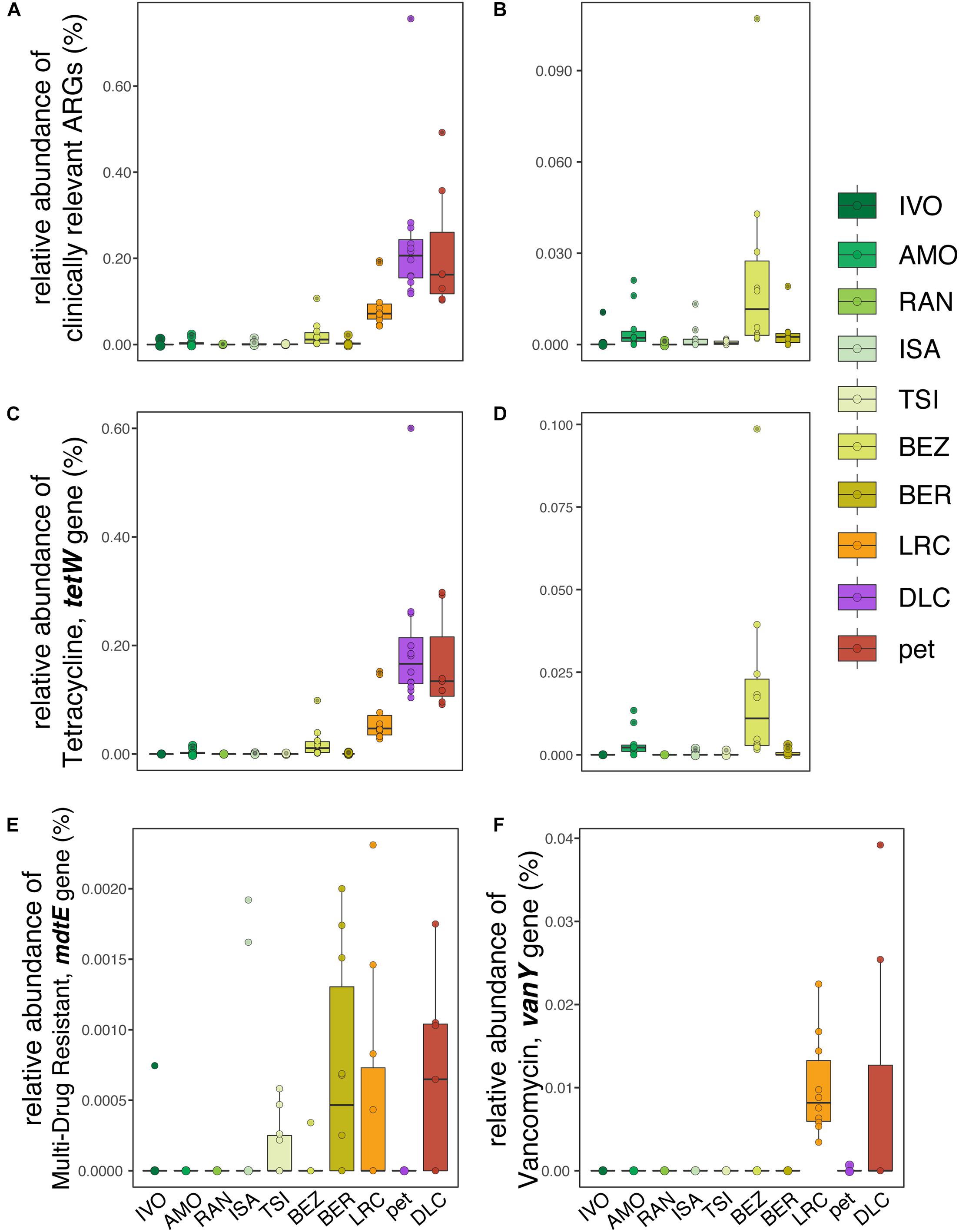
Figure 4. Relative abundances of (A,B) clinically relevant ARGs, including (C,D) tetracycline resistance gene, tetW in the gut microbiota of (A,C) all ring-tailed lemurs (Lemur catta) versus (B,D) only wild lemurs, across sites ranked by increasing level of anthropogenic disturbance (see Table 1). Also shown are the relative abundances of (E) multi-drug resistant gene mdtE and (F) vancomycin-resistant gene, vanY, for lemurs at all sites.
Soil Resistomes and Their Covariation With Lemur Resistomes
The relative abundance and diversity of ARGs in soil microbiota appeared to vary between the four sites analyzed, but the differences did not reach statistical significance (Figures 5A,B), likely owing to small sample sizes. One notable pattern consistent with predictions, however, was that the greatest relative abundance and diversity of ARGs occurred at BEZ, the most disturbed of these four sites (Figures 5A,B). Here, the resistomes of two soil samples clustered distinctly from all other samples (arrows in Figures 5C,D), and were the only ones to include notable proportions of phenicol, sulfonamide, and trimethoprim ARGs. Otherwise, similar to lemur resistomes, ARGs in soil microbiota were dominated by tetracycline resistance genes (μ = 61.044% ± σ = 30.091%), with additional substantial contributions of ARGs from macrolide (11.22% ± 11.06%) and beta-lactam (10.03% ± 16.39%) ARGs. Notably, soil from BEZ also included substantial proportions of aminoglycoside (3.59% ± 6.17%), trimethoprim (3.58% ± 7.96%), and sulfonamide (2.30% ± 5.13%) families (Figure 5D). Unlike lemur resistomes, however, the soil microbiota had low abundances of ARGs in the vancomycin and MDR gene families. The relative abundance of clinically relevant ARGs (Figure 5E), including the tetW gene (Figures 5E,F) reprised patterns observed in lemur fecal samples (Figures 4A–D).
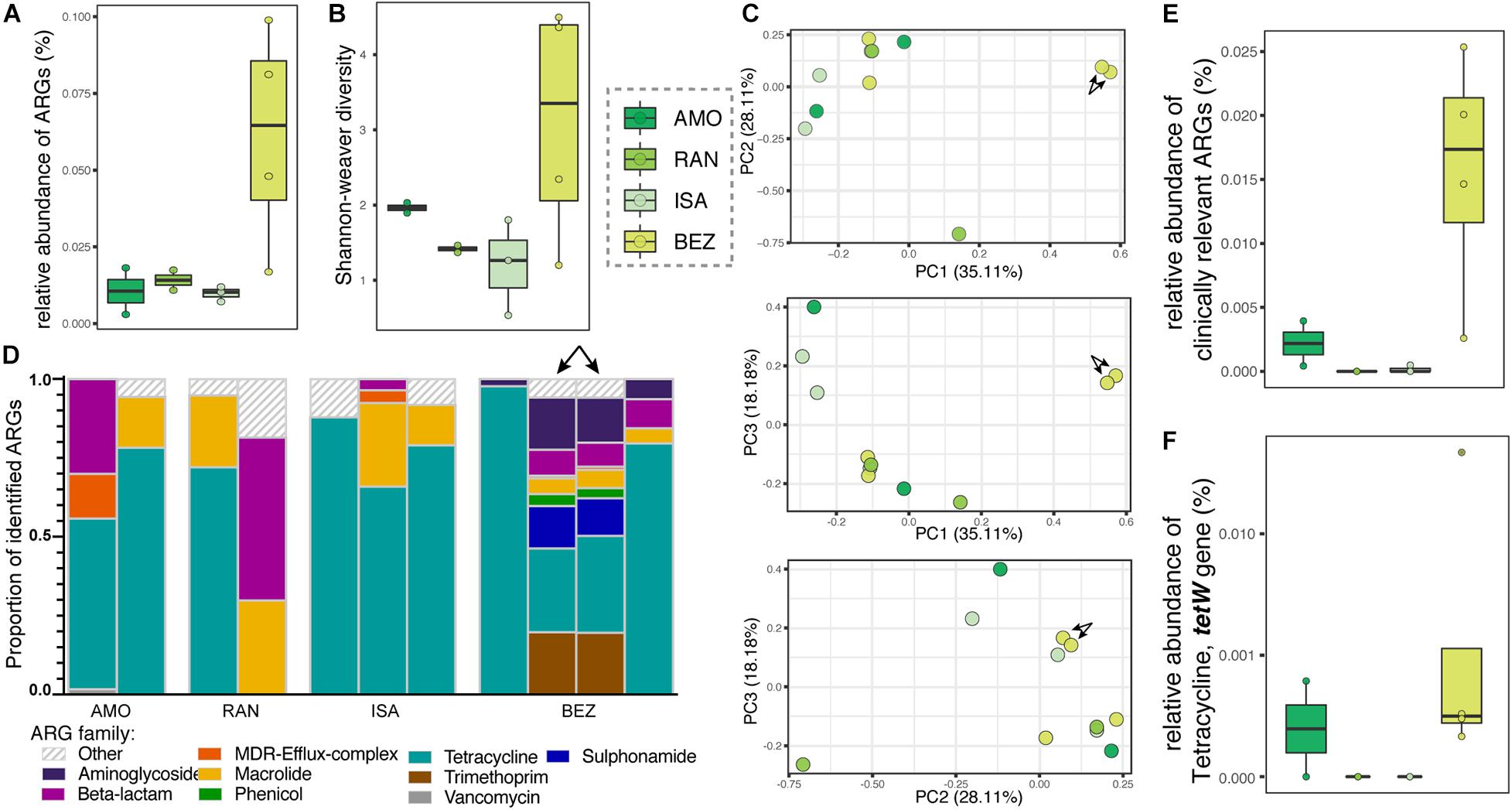
Figure 5. Antibiotic resistance genes in soil samples from four sites in Madagascar, representing different levels of anthropogenic disturbance. Shown are (A) relative abundance, (B) alpha diversity, (C) principal coordinate analyses of Bray–Curtis dissimilarity, including combinations of axes 1, 2, and 3, (D) proportions of identified ARGs by gene family, (E) relative abundances of all clinically relevant ARGs, and (F) relative abundances of the tetW gene. Gene families were identified by color and those representing <1% of the ARGs were combined into the category “Other.” Sites are presented in ascending order according to the ranked level of disturbance (Table 1). Arrows in panels (C,D) point to two soil samples from BEZ that cluster distinctly, reflecting their unique composition.
Regarding covariation between lemur and soil resistomes, there were no significant differences between the total proportion of soil-associated ARGs in lemur guts across the four sites from which soil samples were analyzed (Kruskal–Wallis test; χ2 = 2.781, p = 0.426). When using FEAST to compare all lemur and soil samples from the four sites, soil resistomes from within a population’s site accounted for, on average, significantly greater proportions of lemur ARGs than did soil resistomes from other sites (Kruskal–Wallis test; all sites: χ2 = 20.91, p < 0.0001). When comparing lemur and soil ARGs within and between specific sites, this pattern was not significant for all sites (Figure 6); whereas soil resistomes from ISA and BEZ had significantly greater representation in ISA and BEZ lemurs than did soil resistomes from other sites, the same pattern was nonsignificant from soil and lemurs from AMO and RAN (Pairwise Wilcoxon tests with Benjamini–Hochberg adjustment; ISA: p = 0.005; BEZ: p = 0.0001; AMO: p = 0.896; RAN: p = 0.896; Figure 6).
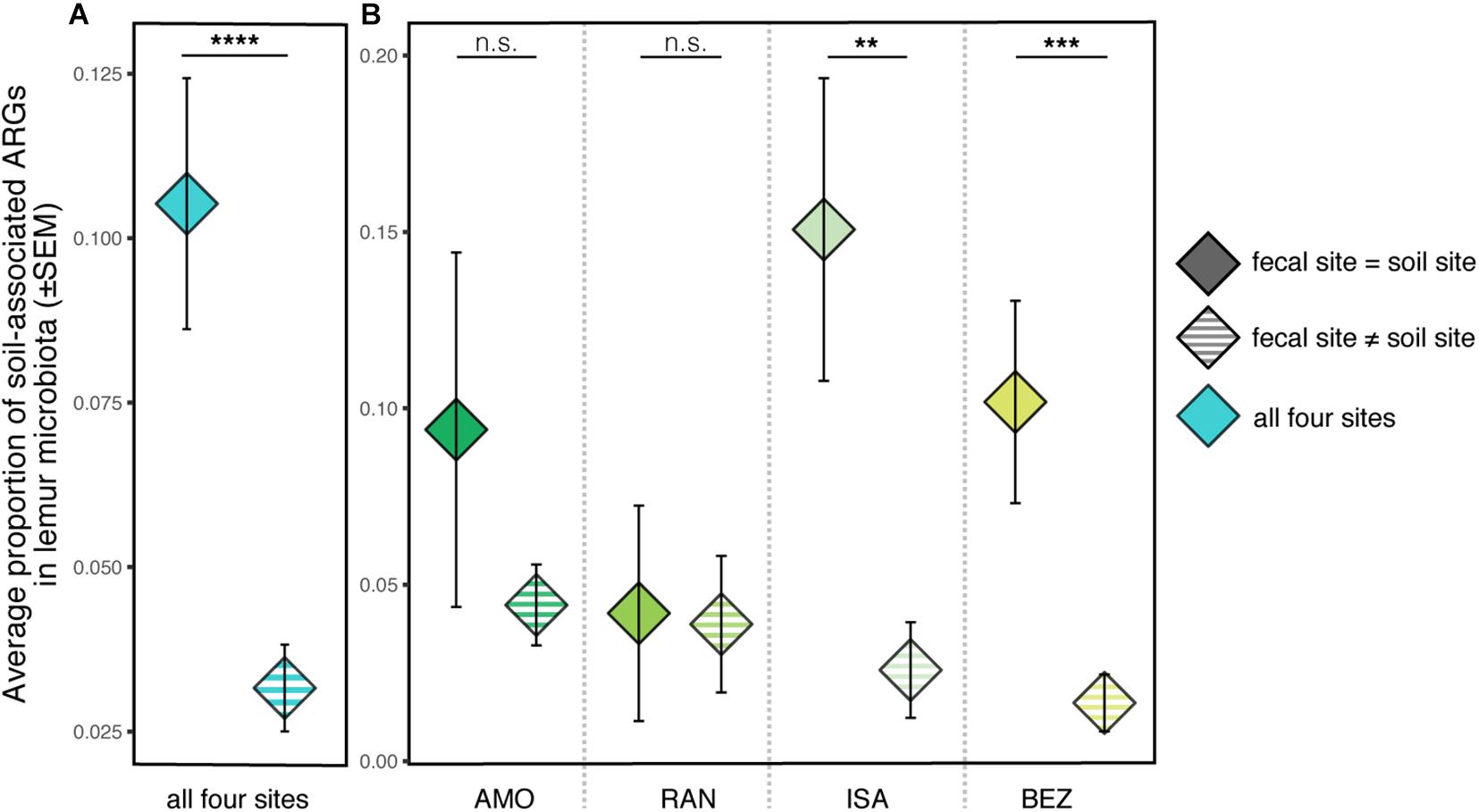
Figure 6. Proportions of soil-associated ARGs present in ring-tailed lemur (Lemur catta) gut microbiota depending on whether the soil was from the same site as the lemur (fecal site = soil site; solid color) or from a different site (fecal site ≠ soil site; striped). Data are shown (A) averaged across all four sites and (B) for each specific site. Kruskal–Wallis test with pairwise comparisons and Benjamini–Hochberg correction; **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s., nonsignificant.
Discussion
By assessing ARGs in multiple ring-tailed lemur populations living under variably disturbed conditions, we (a) shed light on the breadth of ARG presence and diversity outside of traditionally studied settings, (b) highlight the potentially different routes by which ARGs may be acquired by wild versus captive animals, and (c) demonstrate covariance between lemur and soil resistomes in a subset of wild populations. Consistent with previous findings on natural resistomes in wildlife microbiota (Vittecoq et al., 2016; Kipkorir et al., 2019; Marcelino et al., 2019) and environmental microbiota (Esiobu et al., 2002), ARGs were present in nearly all lemur and soil samples collected, including from animals that had potentially minimal exposure to introduced antibiotics (Pallecchi et al., 2008; McCann et al., 2019). As one of the broadest examinations of ARGs in an endangered species and its habitat, our study adds to the growing recognition that wildlife-associated and environmental consortia may act as inextricable reservoirs of ARGs, exemplifying both the ecological and conservation concerns associated with the resistance crisis.
At the lowest end of the anthropogenic disturbance gradient, relatively undisturbed, wild lemur populations (e.g., at IVO and AMO) nonetheless harbored diverse, low-abundance ARGs that likely reflect natural resistomes. Even in these low-risk populations, the composition of lemur resistomes was site-specific, indicating that naturally occurring ARGs are neither randomly present across individuals nor are they homogenized across populations. Thus, under conditions of minimal anthropogenic impact, selective pressures other than human influence may shape animal resistomes.
Although not directly addressed in the present study, one such pressure may involve host sociality. Notably, the site-specificity we observed in lemur resistomes mirrors the site- or group-specificity observed for microbiome composition in highly social species (Theis et al., 2012; Leclaire et al., 2014; Tung et al., 2015; Moeller et al., 2016), including the ring-tailed lemur (Bornbusch et al., 2021). The widely accepted mechanism underlying these patterns is the sharing of microbes between hosts, from many possible bodily sources, particularly between conspecific social partners (reviewed in Archie and Tung, 2015). If social vectors for microbial transmission have a homogenizing effect on the microbiomes of group members, the same pattern could be expected for specific microbial genes, such as ARGs. Our observation of site-specific resistomes in group-living lemurs is thus consistent with the potentially homogenizing impact of ARG social transmission.
Likewise, another pressure may involve components of the host’s diet. Omnivory and geophagy in the ring-tailed lemur (Canington, 2021) promotes ingestion of naturally occurring biomolecules and chemical elements, such as plant secondary compounds (e.g., tannins) and heavy metals, that can modify or co-occur with bacterial resistance mechanisms (Hatano et al., 2005; Compean and Ynalvez, 2014; Pal et al., 2015). The presence and abundance of these compounds could vary with food availability and dietary preferences across the wide range of habitats occupied by wild ring-tailed lemurs. The social transmission of microbes and the diets available within microhabitats are plausible mechanisms for shaping natural resistomes in wild populations and warrant further research to better characterize the potential for even ‘unperturbed’ wildlife to serve as reservoirs of ARGs.
Unexpectedly, we observed ‘outliers’ in most of our wild lemur populations, including those in the most undisturbed habitats, according to our criteria. These outliers appeared to be more ARG-enriched than were their site-mates. To the extent that they represent ‘sentinel’ animals – a concept widely used in epidemiology to describe individuals or species that manifest a specific risk and thus provide advance warning (Van der Schalie et al., 1999) – their resistomes may reflect increased potential of ARG exposure at given sites (Blanco and Bautista, 2020). Sentinel individuals or species may be differentially exposed to ARGs via their different behavioral patterns or biogeographical ranges (Rabinowitz et al., 2005; Vittecoq et al., 2016). Given the probability of host–host transmission of ARGs, sentinels may act as propagators of ARG within their population or community (Sacristán et al., 2020; Plaza-Rodríguez et al., 2021). Alternatively, whereas healthy hosts may be able to combat colonization by resistant bacteria, sentinels may have been those most susceptible to infection (Aguirre, 2009; Taur and Pamer, 2013). In this case, ARG enrichment might reflect vulnerability. Although we could not pinpoint the cause of this variation in our study populations, both scenarios are plausible. Longitudinal data on ARG prevalence in wildlife would help disentangle these two possibilities.
Although degree of anthropogenic disturbance has been suggested as a driver of ARG enrichment in other wildlife species (Tripathi and Cytryn, 2017; Sacristán et al., 2020), most ARG studies have been limited to single or few host populations. Here, across seven populations of wild ring-tailed lemurs, we show that ARG abundance in host gut microbiota positively correlated with our assessments of four anthropogenic components. Compared to subjects from other wilderness sites, those from BEZ and BER (i.e., the sites with the greatest disturbance rankings) had significantly greater abundances of overall and clinically relevant ARGs. Notably, because both of these sites have well-established, long-term research stations (Sauther et al., 1999; Jolly et al., 2006; Jolly, 2012), certain groups had been habituated, captured, and handled for research purposes, and, at BEZ, on rare occasions in the recent past, treated with antibiotics (personal communication with M. Sauther). Because routine captures ceased almost a decade ago, it is improbable that the lemurs sampled for this study received direct antibiotic treatment. Nonetheless, given the persistence and propagation of ARGs, earlier research practices may have left a signal in the lemur resistomes that could have been perpetuated, albeit dampened, over time. Lemurs at TSI have also been handled for research purposes, yet they showed no signal of ARG enrichment. Whereas researcher presence and animal handling at TSI began relatively recently (i.e., in the 10 years before sampling), researcher activity has been occurring for ∼47 and ∼60 years, respectively, at BEZ and BER. Perhaps our findings suggest that, when human contact is infrequent and sporadic, long-term exposure may be required for ARGs to accumulate and disseminate within wildlife populations.
Compared to the ARGs found in wild lemurs, those in captive lemurs were greatly enriched, in some cases by multiple orders of magnitude, likely owing to direct antibiotic treatment received by captive animals at the LRC and DLC. Moreover, the types of ARGs present in these two populations reflected the differences in antibiotic availability and use between the two countries. The resistomes of LRC lemurs, for example, included phenicol ARGs, which most commonly confer resistance to chloramphenicol, an antibiotic that is rarely used in developed countries (Wareham and Wilson, 2002; Fernández et al., 2012), but available in Madagascar for treating the plague (Chanteau et al., 2000; Godfred-Cato et al., 2020). In addition, LRC lemurs harbored notable proportions of ARGs conferring resistance to vancomycin, which, until recently, was considered a last-resort antibiotic for severe bacterial infections, including Staphylococcus aureus (i.e., MRSA: Gardete and Tomasz, 2014; Koch et al., 2014). A recent increase in vancomycin use in Madagascar has led to increased vancomycin resistance and decreased efficacy (Dhanda et al., 2018), including in isolates from humans and other animals (Gay et al., 2017).
By contrast, for DLC lemurs, tetracycline, aminoglycoside, and beta-lactam ARGs were the first, second, and third most prevalent, respectively, potentially reflecting the common use of all three antibiotic families in United States veterinary care (Schwarz and Chaslus-Dancla, 2001; Sarmah et al., 2006; personal communication with veterinarians at the DLC). Nonetheless, not all lemurs in these captivity settings had received equivalent antibiotic treatment. Moreover, DLC lemurs without any history of antibiotic treatment still harbored substantial ARGs that rivaled those seen in conspecifics with numerous previous antibiotics treatments (Bornbusch et al., 2020). In combination, these results indicate that antibiotic treatment produced predictable patterns in ARG enrichment, but those patterns are also present in untreated lemurs. The presence of specific types of ARG enrichment in treatment-naïve individuals perhaps again suggests an independent mechanism of social or environmental transmission.
Another line of evidence in favor of social or environmental transmission is the finding that, despite a lack of antibiotic treatment, pet lemurs harbored some of the greatest abundances of ARGs. Similar to the resistomes of wild and LRC lemurs, those of pet lemurs included ARGs that reflected human use of antibiotics in Madagascar, but at significantly higher abundances than that shown by their wild conspecifics. In a recent report, researchers indicated that ARGs associated with virtually every class of antibiotics have increased in Madagascar, both in humans and in domestic animals (Gay et al., 2017). Whereas for wild lemurs, intraspecific social contact likely facilitates the transmission of ARGs, for pet lemurs isolated from conspecifics, interspecific social contact with humans and domestic animals likely serves as a vector for ARG transmission. Pet lemurs live under unsuitable conditions (Reuter et al., 2015; Reuter and Schaefer, 2016; LaFleur et al., 2021); their poor health may present a particular vulnerability to ARG enrichment. Moreover, because the transmission of ARGs is not host-specific or even route (e.g., fecal-oral) specific, ARG enrichment in pet lemurs poses a significant risk of transmission to the humans who come into contact with these lemurs.
With growing recognition that animal hosts represent only one component of microbial landscapes, researchers are increasingly probing the relationships between host-associated and environmental microbiota. In a previous study on these same ring-tailed lemur populations, we showed that exposure to and acquisition of soil microbes likely contributed to interpopulation variation in gut microbiota (Bornbusch et al., 2021). Here, we expand on this finding, showing that the same pattern holds for ARGs: significant covariation between lemur gut and soil resistomes was found in two disturbed sites, ISA and BEZ. Although based on a directional, source-sink analysis, this covariation is likely to be bidirectional, such that lemurs can acquire ARGs from their environment and also shed them into the environment. Although ISA lemurs had relatively low-abundance resistomes and neither human contact nor antibiotic exposure, the significant covariation between lemur and soil ARGs indicates that the presence of non-wildlife animals and humans might contribute to ARG reservoirs both in lemurs and their environments.
Although antibiotic stewardship has rightly become a focus of western medicine, our study highlights that the resistance crisis extends outside of traditional clinical settings; antibiotic treatment should be considered as only one of many possible mechanisms that facilitate ARG enrichment. Indeed, unlike the curbing of antibiotic use, the exploitation and anthropogenic disturbance of natural ecosystems is accelerating. We show that an endangered primate harbors diverse and, in some cases, anthropogenically enriched resistomes. Our results reinforce the recent call for greater preservation of natural microbial communities (Trevelline et al., 2019; Banerjee et al., 2020) and highlight the need to incorporate antibiotic resistance into in vivo and ex vivo conservation strategies, particularly those that could facilitate the propagation of ARGs (e.g., animal reintroductions, translocations, and transfers). As a premier One Health issue, with relevance to the health of humans, other animals, and the environment, antibiotic resistance demands greater study in a wider range of systems, specifically in the context of wildlife and environmental reservoirs of ARGs.
Data Availability Statement
The data presented in the study are deposited in the Open Science Framework repository, link: https://osf.io/vkr2f/, doi: 10.17605/OSF.IO/VKR2F. The full metagenomic library is available upon reasonable request.
Ethics Statement
The animal study was reviewed and approved by Duke University Institutional Animal Care and Use Committee. Written informed consent for participation was not obtained from the pet owners because verbal consent was provided for collection of feces from pet lemurs.
Author Contributions
SB and CD conceived of this study. SB collected and analyzed samples, and drafted the original manuscript. Both authors contributed to the article and approved the submitted version.
Funding
Funding for this study came from an NSF Doctoral Dissertation Research Improvement Grant to SB and CD (award #1945776), an NSF Behavioral and Cognitive Sciences Grant to CD (award #1749465), a Graduate Student Research Grant to SB from the Triangle Center for Evolutionary Medicine, and a Research fellowship to SB from The Kenan Institute for Ethics at Duke University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
For their assistance in sample collection and transportation, and/or for contributing information on the disturbance components at each site, we are deeply grateful to Lydia Greene, Sylvia Rahobilalaina, Samantha Calkins, Ryan Rothman, Tara Clarke, Marnie LaFleur, and Michelle Sauther. We thank the current and past staff members of the LRC and DLC for their assistance with sample collection. We thank Patricia Wright and the Centre ValBio for providing access to the IVO collection site and assistance in sample transportation and storage. We are grateful to Karlis Graubics and Brian Fanelli at CosmosID for providing guidance and sequencing services. This is DLC publication number (#1484).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.704070/full#supplementary-material
References
Adair, K. L., and Douglas, A. E. (2017). Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 35, 23–29. doi: 10.1016/j.mib.2016.11.002
Addison, J. B. (1984). Antibiotics in sediments and run-off waters from feedlots. Residue Rev. 92, 1–28. doi: 10.1007/978-1-4612-5266-5_1
Aguirre, A. A. (2009). Wild canids as sentinels of ecological health: a conservation medicine perspective. Parasit. Vectors 2, 1–8.
Alanis, A. J. (2005). Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36, 697–705. doi: 10.1016/j.arcmed.2005.06.009
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525.
Aminov, R. I., and Mackie, R. I. (2007). Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161. doi: 10.1111/j.1574-6968.2007.00757.x
Andrianaivoarimanana, V., Kreppel, K., Elissa, N., Duplantier, J.-M., Carniel, E., Rajerison, M., et al. (2013). Understanding the persistence of plague foci in Madagascar. PLoS Negl. Trop. Dis. 7:e2382. doi: 10.1371/journal.pntd.0002382
Anthony, A., Adekunle, C. F., and Thor, A. S. (2018). Residual antibiotics, antibiotic resistant superbugs and antibiotic resistance genes in surface water catchments: public health impact. Phys. Chem. Earth 105, 177–183. doi: 10.1016/j.pce.2018.03.004
Archie, E. A., and Tung, J. (2015). Social behavior and the microbiome. Curr. Opin. Behav. Sci. 6, 28–34. doi: 10.1016/j.cobeha.2015.07.008
Banerjee, A., Cornejo, J., and Bandopadhyay, R. (2020). Emergent climate change impact throughout the world: call for “Microbiome conservation” before it’s too late. Biodivers. Conserv. 29, 345–348. doi: 10.1007/s10531-019-01886-6
Baquero, F., Alvarez-Ortega, C., and Martínez, J. L. (2009). Ecology and evolution of antibiotic resistance. Environ. Microbiol. Rep. 1, 469–476. doi: 10.1111/j.1758-2229.2009.00053.x
Bâtie, C., Kassie, D., Randravatsilavo, D. N. R. M., Baril, L., Waret Szkuta, A., and Goutard, F. L. (2020). Perception of drug vendors and pig and poultry farmers of imerintsiatosika, in Madagascar, toward risks related to antibiotic usage: a q-method approach. Front. Vet. Sci. 7:490. doi: 10.3389/fvets.2020.00490
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:fux053.
Berglund, B. (2015). Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 5:28564. doi: 10.3402/iee.v5.28564
Blanco, G., and Bautista, L. M. (2020). Avian scavengers as bioindicators of antibiotic resistance due to livestock farming intensification. Int. J. Environ. Res. Public Health 17:3620. doi: 10.3390/ijerph17103620
Boisier, P., Rahalison, L., Rasolomaharo, M., Ratsitorahina, M., Mahafaly, M., Razafimahefa, M., et al. (2002). Epidemiologic features of four successive annual outbreaks of bubonic plague in Mahajanga, Madagascar. Emerg. Infect. Dis. 8:311. doi: 10.3201/eid0803.010250
Booton, R. D., Meeyai, A., Alhusein, N., Buller, H., Feil, E., Lambert, H., et al. (2021). One Health drivers of antibacterial resistance: quantifying the relative impacts of human, animal and environmental use and transmission. One Health 12:100220. doi: 10.1016/j.onehlt.2021.100220
Bornbusch, S. L., Greene, L., Rahobilalaina, S., Calkins, S., Rothman, R., Clarke, T., et al. (2021). Gut microbiota of ring-tailed lemurs (Lemur catta) vary across natural and captive populations and correlate with environmental microbiota. bioRxiv [Preprint] doi: 10.1101/2021.06.27.450077
Bornbusch, S. L., Harris, R. L., Grebe, N. M., Dimac-Stohl, K., and Drea, C. M. (2020). Longitudinal effects of antibiotics and fecal transplant on lemur gut microbiota structure, associations, and resistomes. bioRxiv [Preprint] doi: 10.1101/2020.11.11.378349
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Campbell, T. P., Sun, X., Patel, V. H., Sanz, C., Morgan, D., and Dantas, G. (2020). The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 14, 1584–1599. doi: 10.1038/s41396-020-0634-2
Canington, S. L. (2021). Plant species fed on by wild ring-tailed lemurs (Lemur catta) at nine sites. Int. J. Primatol. 42, 16–25. doi: 10.1007/s10764-020-00184-1
Chanteau, S., Ratsitorahina, M., Rahalison, L., Rasoamanana, B., Chan, F., Boisier, P., et al. (2000). Current epidemiology of human plague in Madagascar. Microb. Infect. 2, 25–31. doi: 10.1016/s1286-4579(00)00289-6
Chee-Sanford, J. C., Mackie, R. I, Koike, S., Krapac, I. G., Lin, Y., Yannarell, A. C., et al. (2009). Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 38, 1086–1108. doi: 10.2134/jeq2008.0128
Choo, J. M., Leong, L. E., and Rogers, G. B. (2015). Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 5:16350.
Clarke, T. A., Reuter, K. E., LaFleur, M., and Schaefer, M. S. (2019). A viral video and pet lemurs on Twitter. PLoS One 14:e0208577. doi: 10.1371/journal.pone.0208577
Compean, K. L., and Ynalvez, R. A. (2014). Antimicrobial activity of plant secondary metabolites: a review. Res. J. Med. Plants 8, 204–213. doi: 10.3923/rjmp.2014.204.213
D’Costa, V. M., Griffiths, E., and Wright, G. D. (2007). Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10, 481–489. doi: 10.1016/j.mib.2007.08.009
D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W. L., Schwarz, C., et al. (2011). Antibiotic resistance is ancient. Nature 477, 457–461.
Dafale, N. A., Srivastava, S., and Purohit, H. J. (2020). Zoonosis: an emerging link to antibiotic resistance under “One health approach”. Indian J. Microbiol. 60, 139–152. doi: 10.1007/s12088-020-00860-z
De Souze Lopez, E., Maciel, W. C., Albuquerque, A. H., Machado, D. N., Bezerra, W. G. A., Vasconcelos, R. H., et al. (2015). Prevalence and antimicrobial resistance profile of enterobacteria isolated from psittaciformes of illegal wildlife trade. Acta Sci. Vet. 43:1313.
Dhanda, G., Sarkar, P., Samaddar, S., and Haldar, J. (2018). Battle against vancomycin-resistant bacteria: recent developments in chemical strategies. J. Med. Chem. 62, 3184–3205. doi: 10.1021/acs.jmedchem.8b01093
Dias, P. C. (1996). Sources and sinks in population biology. Trends Ecol. Evol. 11, 326–330. doi: 10.1016/0169-5347(96)10037-9
Dolejska, M., and Literak, I. (2019). Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 63:e01167-19.
Dulo, F., Feleke, A., Szonyi, B., Fries, R., Baumann, M. P. O., and Grace, D. (2015). Isolation of multidrug-resistant Escherichia coli O157 from goats in the Somali region of Ethiopia: a cross-sectional, abattoir-based study. PLoS One 10:e0142905. doi: 10.1371/journal.pone.0142905
Duquette, R. A., and Nuttall, T. J. (2004). Methicillin−resistant Staphylococcus aureus in dogs and cats: an emerging problem? J. Small Anim. Pract. 45, 591–597. doi: 10.1111/j.1748-5827.2004.tb00180.x
Epstein, C. R., Yam, W. C., Peiris, J. S. M., and Epstein, R. J. (2009). Methicillin-resistant commensal staphylococci in healthy dogs as a potential zoonotic reservoir for community-acquired antibiotic resistance. Infect. Genet. Evol. 9, 283–285. doi: 10.1016/j.meegid.2008.11.003
Esiobu, N., Armenta, L., and Ike, J. (2002). Antibiotic resistance in soil and water environments. Int. J. Environ. Health Res. 12, 133–144. doi: 10.1080/09603120220129292
Fernández, M., Conde, S., de la Torre, J., Molina-Santiago, C., Ramos, J.-L., and Duque, E. (2012). Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob. Agents Chemother. 56, 1001–1009. doi: 10.1128/aac.05398-11
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
French, G. L. (2005). Clinical impact and relevance of antibiotic resistance. Adv. Drug Deliv. Rev. 57, 1514–1527. doi: 10.1016/j.addr.2005.04.005
Ganzhorn, J. (1987). Soil consumption of two groups of semi-free-ranging lemurs (Lemur catta and Lemur fulvus). Ethology 74, 146–154. doi: 10.1111/j.1439-0310.1987.tb00927.x
Gao, H., Zhang, L., Lu, Z., He, C., Li, Q., and Na, G. (2018). Complex migration of antibiotic resistance in natural aquatic environments. Environ. Pollut. 232, 1–9. doi: 10.1016/j.envpol.2017.08.078
Gardete, S., and Tomasz, A. (2014). Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124, 2836–2840. doi: 10.1172/jci68834
Gatica, J., and Cytryn, E. (2013). Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome. Environ. Sci. Pollut. Res. 20, 3529–3538. doi: 10.1007/s11356-013-1505-4
Gay, N., Belmonte, O., Collard, J.-M., Halifa, M., Issack, M. I., Mindjae, S., et al. (2017). Review of antibiotic resistance in the Indian Ocean Commission: a human and animal health issue. Front. Public Health 5:162. doi: 10.3389/fpubh.2017.00162
Ghosh, S., and LaPara, T. M. (2007). The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1, 191–203. doi: 10.1038/ismej.2007.31
Gillings, M. R. (2013). Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front. Microbiol. 4:4. doi: 10.3389/fmicb.2013.00004
Godfred-Cato, S., Cooley, K. M., Fleck-Derderian, S., Becksted, H. A., Russell, Z., Meaney-Delman, D., et al. (2020). Treatment of human plague: a systematic review of published aggregate data on antimicrobial efficacy, 1939–2019. Clin. Infect. Dis. 70(Suppl. 1), S11–S19.
Gould, L. (2006). “Lemur catta ecology: what we know and what we need to know,” in Lemurs, eds L. Gould and M. L. Sauther (Boston, MA: Springer), 255–274. doi: 10.1007/978-0-387-34586-4_12
Hasan, N. A., Young, B. A., Minard-Smith, A. T., Saeed, K., Li, H., Heizer, E. M., et al. (2014). Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One 9:e97699. doi: 10.1371/journal.pone.0097699
Hatano, T., Kusuda, M., Inada, K., Ogawa, T., Shiota, S., Tsuchiya, T., et al. (2005). Effects of tannins and related polyphenols on methicillin-resistant Staphylococcus aureus. Phytochemistry 66, 2047–2055. doi: 10.1016/j.phytochem.2005.01.013
Higuera-Llantén, S., Vásquez-Ponce, F., Barrientos-Espinoza, B., Mardones, F. O., Marshall, S. H., and Olivares-Pacheco, J. (2018). Extended antibiotic treatment in salmon farms select multiresistant gut bacteria with a high prevalence of antibiotic resistance genes. PLoS One 13:e0203641. doi: 10.1371/journal.pone.0203641
Hiltunen, T., Virta, M., and Laine, A.-L. (2017). Antibiotic resistance in the wild: an eco-evolutionary perspective. Philos. Trans. R. Soc. B Biol. Sci. 372:20160039. doi: 10.1098/rstb.2016.0039
Howard, D. H., Scott, R. D., Packard, R., and Jones, D. (2003). The global impact of drug resistance. Clin. Infect. Dis. 36(Suppl. 1), S4–S10.
Iramiot, J. S., Kajumbula, H., Bazira, J., Kansiime, C., and Asiimwe, B. B. (2020). Antimicrobial resistance at the human–animal interface in the pastoralist communities of Kasese District, South Western Uganda. Sci. Rep. 10: 14737.
Jardine, J., Mavumengwana, V., and Ubomba-Jaswa, E. (2019). Antibiotic resistance and heavy metal tolerance in cultured bacteria from hot springs as indicators of environmental intrinsic resistance and tolerance levels. Environ. Pollut. 249, 696–702. doi: 10.1016/j.envpol.2019.03.059
Jolly, A. (2012). “Berenty reserve, Madagascar: a long time in a small space,” in Long-Term Field Studies of Primates, eds P. Kappeler and D. Watts (Berlin: Springer), 21–44. doi: 10.1007/978-3-642-22514-7_2
Jolly, A., Sussman, R. W., Koyama, N., and Rasamimanana, H. (2006). Ringtailed Lemur Biology: Lemur Catta in Madagascar. Berlin: Springer Science & Business Media.
Kang, K., Imamovic, L., Misiakou, M.-A., Bornakke Sørensen, M., Heshiki, Y., Ni, Y., et al. (2021). Expansion and persistence of antibiotic-specific resistance genes following antibiotic treatment. Gut Microb. 13, 1–19.
Kim, Y., Leung, M. H. Y., Kwok, W., Fournié, G., Li, J., Lee, P. K. H., et al. (2020). Antibiotic resistance gene sharing networks and the effect of dietary nutritional content on the canine and feline gut resistome. Anim. Microb. 2:4.
Kipkorir, K. C., Ang’ienda, P. O., Onyango, D. M., and Onyango, P. O. (2019). Antibiotic Resistance of Escherichia coli from Humans and Black Rhinoceroses in Kenya. EcoHealth 17, 41–51. doi: 10.1007/s10393-019-01461-z
Koch, G., Yepes, A., Förstner, K. U., Wermser, C., Stengel, S. T., Modamio, J., et al. (2014). Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell 158, 1060–1071. doi: 10.1016/j.cell.2014.06.046
LaFleur, M., Clarke, T. A., Reuter, K. E., and Schaefer, M. S. (2019). Illegal trade of wild-captured Lemur catta within Madagascar. Folia Primatol. 90, 199–214. doi: 10.1159/000496970
LaFleur, M., Reuter, K. E., Hall, M. B., Rasoanaivo, H. H., McKernan, S., Ranaivomanana, P., et al. (2021). Drug-rsistant tuberculosis in pet ring-tailed lemur, Madagascar. Emerg. Infect. Dis. 27:977. doi: 10.3201/eid2703.202924
Lax, S., Smith, D. P., Hampton-Marcell, J., Owens, S. M., Handley, K. M., Scott, N. M., et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052. doi: 10.1126/science.1254529
Leclaire, S., Nielsen, J. F., and Drea, C. M. (2014). Bacterial communities in meerkat anal scent secretions vary with host sex, age, and group membership. Behav. Ecol. 25, 996–1004. doi: 10.1093/beheco/aru074
Lemos, L. N., Pedrinho, A., de Vasconcelos, A. T. R., Tsai, S. M., and Mendes, L. W. (2021). Amazon deforestation enriches antibiotic resistance genes. Soil Biol. Biochem. 153:108110. doi: 10.1016/j.soilbio.2020.108110
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi: 10.1139/cjm-2018-0275
Li, J., Rettedal, E. A., Van Der Helm, E., Ellabaan, M., Panagiotou, G., and Sommer, M. O. A. (2019). Antibiotic treatment drives the diversification of the human gut resistome. Genomics Proteomic Bioinformatics 17, 39–51. doi: 10.1016/j.gpb.2018.12.003
Lin, J., Nishino, K., Roberts, M. C., Tolmasky, M., Aminov, R. I., and Zhang, L. (2015). Mechanisms of antibiotic resistance. Front. Microbiol. 6:34. doi: 10.3389/fmicb.2015.00034
Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 25, 173–181. doi: 10.1016/j.tim.2016.11.014
Marcelino, V. R., Wille, M., Hurt, A. C., González-Acuña, D., Klaassen, M., Schlub, T. E., et al. (2019). Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 17:31. doi: 10.1186/s12915-019-0649-1
Martens, E., and Demain, A. L. (2017). The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 70, 520–526. doi: 10.1038/ja.2017.30
Martínez, J. L. (2012). Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials. Front. Microbiol. 3:1. doi: 10.3389/fmicb.2012.00001
Martínez, J. L., Coque, T. M., and Baquero, F. (2015). Prioritizing risks of antibiotic resistance genes in all metagenomes. Nat. Rev. Microbiol. 13:396. doi: 10.1038/nrmicro3399-c2
Mason, G. J. (2010). Species differences in responses to captivity: stress, welfare and the comparative method. Trends Ecol. Evol. 25, 713–721. doi: 10.1016/j.tree.2010.08.011
McCann, C. M., Christgen, B., Roberts, J. A., Su, J.-Q., Arnold, K. E., Gray, N. D., et al. (2019). Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems. Environ. Int. 125, 497–504. doi: 10.1016/j.envint.2019.01.034
Moeller, A. H. A., Foerster, S., Wilson, M. L., Pusey, A. E., Hahn, B. H., and Ochman, H. (2016). Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2:e1500997. doi: 10.1126/sciadv.1500997
Mowry, C. B., and Campbell, J. L. (2001). AZA Nutrition Advisory Group TAG/SSP Husbandry Notebook Nutrition Section Lemur catta (Ring-tailed lemur). Silver Spring, MA: American Association of Zoos and Aquariums.
Mühlberg, E., Umstätter, F., Kleist, C., Domhan, C., Mier, W., and Uhl, P. (2020). Renaissance of vancomycin: approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 66, 11–16. doi: 10.1139/cjm-2019-0309
Nawaz, M., Wang, J., Zhou, A., Ma, C., Wu, X., Moore, J. E., et al. (2011). Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr. Microbiol. 62, 1081–1089. doi: 10.1007/s00284-010-9856-2
Ortega-Paredes, D., Haro, M., Leoro-Garzón, P., Barba, P., Loaiza, K., Mora, F., et al. (2019). Multidrug-resistant Escherichia coli isolated from canine faeces in a public park in Quito, Ecuador. J. Glob. Antimicrob. Resist. 18, 263–268. doi: 10.1016/j.jgar.2019.04.002
Ottesen, A., Ramachandran, P., Reed, E., White, J. R., Hasan, N., Subramanian, P., et al. (2016). Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 16:275. doi: 10.1186/s12866-016-0894-1
Padget, M., Tamarelle, J., Herindrainy, P., Ndir, A., Diene Sarr, F., Richard, V., et al. (2017). A community survey of antibiotic consumption among children in Madagascar and Senegal: the importance of healthcare access and care quality. J. Antimicrob. Chemother. 72, 564–573. doi: 10.1093/jac/dkw446
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864-015-2153-5
Pallecchi, L., Bartoloni, A., Paradisi, F., and Rossolini, G. M. (2008). Antibiotic resistance in the absence of antimicrobial use: mechanisms and implications. Expert Rev. Anti Infect. Ther. 6, 725–732. doi: 10.1586/14787210.6.5.725
Pehrsson, E. C., Tsukayama, P., Patel, S., Mejía-Bautista, M., Sosa-Soto, G., Navarrete, K. M., et al. (2016). Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216. doi: 10.1038/nature17672
Plaza-Rodríguez, C., Alt, K., Grobbel, M., Hammerl, J. A., Irrgang, A., Szabo, I., et al. (2021). Wildlife as sentinels of antimicrobial resistance in Germany? Front. Vet. Sci. 7:627821. doi: 10.3389/fvets.2020.627821
Pomba, C., Rantala, M., Greko, C., Baptiste, K. E., Catry, B., Van Duijkeren, E., et al. (2017). Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 72, 957–968.
Pruden, A., Pei, R., Storteboom, H., and Carlson, K. H. (2006). Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40, 7445–7450. doi: 10.1021/es060413l
Rabinowitz, P. M., Gordon, Z., Holmes, R., Taylor, B., Wilcox, M., Chudnov, D., et al. (2005). Animals as sentinels of human environmental health hazards: an evidence-based analysis. EcoHealth 2, 26–37. doi: 10.1007/s10393-004-0151-1
Rajgire, A. V. (2013). Open defecation: a prominent source of pollution in drinking water in villages. Int. J. Life Sci. Biotechnol. Pharma Res. 2, 238–246.
Rakotoharinome, M., Pognon, D., Randriamparany, T., Ming, J. C., Idoumbin, J.-P., Cardinale, E., et al. (2014). Prevalence of antimicrobial residues in pork meat in Madagascar. Trop. Anim. Health Prod. 46, 49–55. doi: 10.1007/s11250-013-0445-9
Ramey, A. M., and Ahlstrom, C. A. (2020). Antibiotic resistant bacteria in wildlife: perspectives on trends, acquisition and dissemination, data gaps, and future directions. J. Wildl. Dis. 56, 1–15. doi: 10.7589/2019-04-099
Randrianirina, F., Ratsima, E. H., Ramparany, L., Randremanana, R., Rakotonirina, H. C., Andriamanantena, T., et al. (2014). Antimicrobial resistance of bacterial enteropathogens isolated from stools in Madagascar. BMC Infect. Dis. 14:104. doi: 10.1186/1471-2334-14-104
Randrianirina, F., Vaillant, L., Ramarokoto, C. E., Rakotoarijaona, A., Andriamanarivo, M. L., Razafimahandry, H. C., et al. (2010). Antimicrobial resistance in pathogens causing nosocomial infections in surgery and intensive care units of two hospitals in Antananarivo, Madagascar. J. Infect. Dev. Ctries. 4, 74–82.
Rasamiravaka, T. (2020). Antimicrobial resistance in Madagascar: a review of the current situation and challenges. Afr. J. Clin. Exp. Microbiol. 21, 165–174.
Reuter, K. E., and Schaefer, M. S. (2016). Captive conditions of pet lemurs in Madagascar. Folia Primatol. 87, 48–63. doi: 10.1159/000444582
Reuter, K. E., Gilles, H., Wills, A. R., and Sewall, B. J. (2015). Live capture and ownership of lemurs in Madagascar: extent and conservation implications. Oryx 50, 344–354. doi: 10.1017/s003060531400074x
Richards, V. P., Velsko, I. M., Alam, M. T., Zadoks, R. N., Manning, S. D., Pavinski Bitar, P. D., et al. (2019). Population gene introgression and high genome plasticity for the zoonotic pathogen Streptococcus agalactiae. Mol. Biol. Evol. 36, 2572–2590. doi: 10.1093/molbev/msz169
Rossolini, G. M., Arena, F., Pecile, P., and Pollini, S. (2014). Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 18, 56–60. doi: 10.1016/j.coph.2014.09.006
Rousham, E. K., Unicomb, L., and Islam, M. A. (2018). Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. B Biol. Sci. 285:20180332. doi: 10.1098/rspb.2018.0332
Sacristán, I., Esperón, F., Acuña, F., Aguilar, E., García, S., López, M. J., et al. (2020). Antibiotic resistance genes as landscape anthropization indicators: using a wild felid as sentinel in Chile. Sci. Total Environ. 703:134900. doi: 10.1016/j.scitotenv.2019.134900
Salam, A. P., Raberahona, M., Andriantsalama, P., Read, L., Andrianarintsiferantsoa, F., Razafinambinintsoa, T., et al. (2020). Factors influencing atypical clinical presentations during the 2017 Madagascar pneumonic plague outbreak: a prospective cohort study. Am. J. Trop. Med. Hyg. 102, 1309–1315. doi: 10.4269/ajtmh.19-0576
Sarmah, A. K., Meyer, M. T., and Boxall, A. B. A. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. doi: 10.1016/j.chemosphere.2006.03.026
Sauther, M. L., Sussman, R. W., and Gould, L. (1999). The socioecology of the ring-tailed lemur: Thirty−five years of research. Evol. Anthropol. 8, 120–132. doi: 10.1002/(sici)1520-6505(1999)8:4<120::aid-evan3>3.0.co;2-o
Schwarz, S., and Chaslus-Dancla, E. (2001). Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32, 201–225. doi: 10.1051/vetres:2001120
Segawa, T., Takeuchi, N., Rivera, A., Yamada, A., Yoshimura, Y., Barcaza, G., et al. (2013). Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 5, 127–134. doi: 10.1111/1758-2229.12011
Selway, C. A., Mills, J. G., Weinstein, P., Skelly, C., Yadav, S., Lowe, A., et al. (2020). Transfer of environmental microbes to the skin and respiratory tract of humans after urban green space exposure. Environ. Int. 145:106084. doi: 10.1016/j.envint.2020.106084
Sengupta, S., Chattopadhyay, M. K., and Grossart, H.-P. (2013). The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4:47. doi: 10.3389/fmicb.2013.00047
Shenhav, L., Thompson, M., Joseph, T. A., Briscoe, L., Furman, O., Bogumil, D., et al. (2019). FEAST: fast expectation-maximization for microbial source tracking. Nat. Methods 16:627. doi: 10.1038/s41592-019-0431-x
Smith, C. C., Snowberg, L. K., Caporaso, J. G., Knight, R., and Bolnick, D. I. (2015). Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 9, 2515–2526. doi: 10.1038/ismej.2015.64
Song, S. J., Amir, A., Metcalf, J. L., Amato, K. R., Xu, Z. Z., Humphrey, G., et al. (2016). Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. MSystems 1:e00021-16.
Sussman, R. W., and Ratsirarson, J. (2006). “Beza Mahafaly special reserve: a research site in southwestern Madagascar,” in Ringtailed Lemur Biology: Lemur Catta in Madagascar, eds A. Jolly, R. W. Sussman, N. Koyama, and H. Rasamimanana (Boston, MA: Springer), 43–51. doi: 10.1007/978-0-387-34126-2_4
Taur, Y., and Pamer, E. G. (2013). The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr. Opin. Infect. Dis. 26:332. doi: 10.1097/qco.0b013e3283630dd3
Theis, K. R., Schmidt, T. M., and Holekamp, K. E. (2012). Evidence for a bacterial mechanism for group-specific social odors among hyenas. Sci. Rep. 2:615.
Trevelline, B. K., Fontaine, S. S., Hartup, B. K., and Kohl, K. D. (2019). Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B 286:20182448. doi: 10.1098/rspb.2018.2448
Tripathi, V., and Cytryn, E. (2017). Impact of anthropogenic activities on the dissemination of antibiotic resistance across ecological boundaries. Essays Biochem. 61, 11–21. doi: 10.1042/ebc20160054
Tung, J., Barreiro, L. B., Burns, M. B., Grenier, J. C., Lynch, J., Grieneisen, L. E., et al. (2015). Social networks predict gut microbiome composition in wild baboons. ELife 4:e05224.
Van den Honert, M. S., Gouws, P. A., and Hoffman, L. C. (2018). Importance and implications of antibiotic resistance development in livestock and wildlife farming in South Africa: a review. South Afr. J. Anim. Sci. 48, 401–412. doi: 10.4314/sajas.v48i3.1
Van der Schalie, W. H., Gardner, H. S. Jr., Bantle, J. A., De Rosa, C. T., Finch, R. A., Reif, J. S., et al. (1999). Animals as sentinels of human health hazards of environmental chemicals. Environ. Health Perspect. 107, 309–315. doi: 10.1289/ehp.99107309
Van Goethem, M. W., Pierneef, R., Bezuidt, O. K. I., Van De Peer, Y., Cowan, D. A., and Makhalanyane, T. P. (2018). A reservoir of ‘historical’antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6, 1–12.
Van Puyvelde, S., Deborggraeve, S., and Jacobs, J. (2018). Why the antibiotic resistance crisis requires a one health approach. Lancet Infect. Dis. 18, 132–134. doi: 10.1016/s1473-3099(17)30704-1
Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. Pharm. Ther. 40:277.
Versluis, D., D’Andrea, M. M., Garcia, J. R., Leimena, M. M., Hugenholtz, F., Zhang, J., et al. (2015). Mining microbial metatranscriptomes for expression of antibiotic resistance genes under natural conditions. Sci. Rep. 5:11981.
Vittecoq, M., Godreuil, S., Prugnolle, F., Durand, P., Brazier, L., Renaud, N., et al. (2016). Antimicrobial resistance in wildlife. J. Appl. Ecol. 53, 519–529.
Wang, F., Stedtfeld, R. D., Kim, O.-S., Chai, B., Yang, L., Stedtfeld, T. M., et al. (2016). Influence of soil characteristics and proximity to Antarctic research stations on abundance of antibiotic resistance genes in soils. Environ. Sci. Technol. 50, 12621–12629. doi: 10.1021/acs.est.6b02863
Wang, H. H., Manuzon, M., Lehman, M., Wan, K., Luo, H., Wittum, T. E., et al. (2006). Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 254, 226–231. doi: 10.1111/j.1574-6968.2005.00030.x
Wareham, D. W., and Wilson, P. (2002). Chloramphenicol in the 21st century. Hosp. Med. 63, 157–161. doi: 10.12968/hosp.2002.63.3.2061
Worsley−Tonks, K. E. L., Miller, E. A., Gehrt, S. D., McKenzie, S. C., Travis, D. A., Johnson, T. J., et al. (2020). Characterization of antimicrobial resistance genes in Enterobacteriaceae carried by suburban mesocarnivores and locally owned and stray dogs. Zoonoses Public Health 67, 460–466. doi: 10.1111/zph.12691
Xiong, W., Wang, Y., Sun, Y., Ma, L., Zeng, Q., Jiang, X., et al. (2018). Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 6, 1–11.
Zhang, A.-N., Li, L.-G., Yin, X., Dai, C. L., Groussin, M., Poyet, M., et al. (2019). Choosing your battles: which resistance genes warrant global action? bioRxiv [Preprint] doi: 10.1101/784322
Keywords: lemur, primate, antibiotic resistance, resistome, microbiome, environment, Anthropocene
Citation: Bornbusch SL and Drea CM (2021) Antibiotic Resistance Genes in Lemur Gut and Soil Microbiota Along a Gradient of Anthropogenic Disturbance. Front. Ecol. Evol. 9:704070. doi: 10.3389/fevo.2021.704070
Received: 01 May 2021; Accepted: 14 July 2021;
Published: 09 August 2021.
Edited by:
Ellen Decaestecker, KU Leuven, BelgiumReviewed by:
Devin Drown, University of Alaska System, United StatesAlison Gould, California Academy of Sciences, United States
Karine Mahefarisoa, Catholic University of Louvain, Belgium
Copyright © 2021 Bornbusch and Drea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sally L. Bornbusch, c2FsbHkuYm9ybmJ1c2NoQGdtYWlsLmNvbQ==
 Sally L. Bornbusch
Sally L. Bornbusch Christine M. Drea
Christine M. Drea