- 1Department of Public Health, Qingdao University, Qingdao, China
- 2Department of Nutrition and Food Hygiene, Qingdao Municipal Center for Disease Control and Prevention, Qingdao Institute of Preventive Medicine, Qingdao, China
- 3Department of Laboratory, Qingdao Central Hospital, Qingdao, China
Toxic plants have been a major threat to public health in China. However, identification and tracing of poisoned species with traditional methods are unreliable due to the destruction of plant morphology by cooking and chewing. DNA barcoding is independent of environmental factors and morphological limitations, making it a powerful tool to accurately identify species. In our study, a total of 83 materials from 26 genera and 31 species of 13 families were collected and 13 plant materials were subjected to simulated gastric fluid digestion. Four markers (rbcL, trnH-psbA, matK, and ITS) were amplified and sequenced for all untreated and mock-digested samples. The effectiveness of DNA barcoding for the identification of toxic plants was assessed using Basic Local Alignment Search Tool (BLAST) method, PWG-Distance method, and Tree-Building (NJ) method. Except for the matK region, the amplification success rate of the remaining three regions was high, but the sequencing of trnH-psbA and ITS was less satisfactory. Meanwhile, matK was prone to be more difficult to amplify and sequence because of simulated gastric fluid. Among the three methods applied, BLAST method showed lower recognition rates, while PWG-Distance and Tree-Building methods showed little difference in recognition rates. Overall, ITS had the highest recognition rate among individual loci. Among the combined loci, rbcL + ITS had the highest species recognition rate. However, the ITS region may not be suitable for DNA analysis of gastric contents and the combination of loci does not significantly improve species resolution. In addition, identification of species to the genus level is sufficient to aid in the clinical management of most poisoning events. Considering primer versatility, DNA sequence quality, species identification ability, experimental cost and speed of analysis, we recommend rbcL as the best single marker for clinical identification and also suggest the BLAST method for analysis. Our current results suggest that DNA barcoding can rapidly identify and trace toxic species and has great potential for clinical applications. In addition, we suggest the creation of a proprietary database containing morphological, toxicological and molecular information to better apply DNA barcoding technology in clinical diagnostics.
Introduction
As primary producers, plants have always been an important source of nutrition for humans. However, some herbs and ingredients are potentially toxic when mishandled, e.g., lectins contained in fresh Phaseolus vulgaris can cause neurological and digestive symptoms (De Mejia et al., 2005; He et al., 2018). As a result, toxic plants have become a major threat to public health in China. Hundreds of poison cases are reported by poison control centers every year (Fuchs et al., 2011; Krenzelok and Mrvos, 2011).
Several common toxins in plants may show overlapping symptoms (Petersen, 2011). An incorrect diagnosis may be obtained solely based on the patient’s clinical presentation (Li T. X. et al., 2019). Moreover, poisoning caused by different plants requires variable treatment. For example, the anti-digoxin Fab fragment, which is a safe and effective treatment for severe arrhythmias caused by yellow oleander (Eddleston et al., 2000), and physostigmine is the antidote of choice for severe poisoning by Datura (Doan et al., 2019). Therefore, rapid and effective determination of the etiology is important in clinical treatment (Shinozaki et al., 2018). However, species identification has been a difficult task in poison detection. 39.41% of the food poisoning cases reported by the Centers for Disease Control (CDC) in China from 2008 to 2010 were considered to be of unknown origin (Chu et al., 2012). Generally, clinicians collect food residues, vomit from patients, and make a diagnosis by morphological analysis of plant fragments (Müller and Desel, 2013). Chewing, however, can change the appearance of the plant, as can the human stomach environment (Romano et al., 2019). Even experienced botanists face the challenge of distinguishing species only by almost invisible features. Therefore, clinical identification based on traditional morphology is difficult. The recently proposed DNA barcoding technique is a possible solution to this problem.
DNA barcoding is a new molecular marker technique first proposed by Hebert et al. in 2003 for rapid and accurate species identification using a short DNA sequence or several DNA segments (Hebert et al., 2003). This technique can compensate for the shortcomings of traditional morphological identification because it distinguishes species by virtue of specific DNA segments that represent differences at the genomic level (Agarwal et al., 2008). DNA barcoding technology was first developed for the identification of animals, and the mitochondrial gene cytochrome c oxidase I (COI) is considered to be the core of the global animal biometric identification system (Hebert et al., 2003). Although recent studies have further shown that the COI gene can effectively distinguish various animal species (Hebert et al., 2016; Thu et al., 2019; Zangl et al., 2020), it is highly invariant in land plants and is not suitable as a universal DNA barcode for plants (Bruni et al., 2010). Therefore, the search for suitable candidate barcodes has focused on chloroplasts and ribosomes. Numerous studies have shown that chloroplast genomic matK, rbcL, trnH-psbA and internal transcribed spacer ITS are suitable barcode markers for molecular identification in land plants, and have been tested for recognition power in various families of plants (Group, 2009; China Plant et al., 2011; Li et al., 2015).
In this study, we collected common toxic plants with reported cases of poisoning in China and evaluated four candidate barcodes (trnH-psbA, rbcL, ITS, and matK). Our objectives were: (1) to examine the amplification and sequencing of DNA barcodes after simulated gastric fluid digestion. (2) to examine the resolution of these markers individually or in combination and to evaluate the most appropriate markers for clinical identification of phytotoxicosis, and (3) to establish a reference database to facilitate future application of DNA barcoding technology for clinical diagnosis.
Materials and Methods
Toxic Plant Materials
When poisonings occur, local hospitals often seek medical advice from poison control centers and report cases in relevant journals. Our sampling was based on plant poisoning incidents reported in the literature in China from 1994 to 2011, as compiled by Qian (2014). A total of 83 materials from 13 families, 26 genera and 31 species that are more easily accessible in daily life were finally collected. These samples were purchased at traditional markets in Qingdao, Shandong Province, or collected in the field in Zhejiang, Jiangsu and Guangxi provinces from June to December 2020. All samples were classified and identified by Professor Xin Hua of Qingdao Agricultural University. Fresh plant tissues were dried in a 40°C drying oven and stored in sealed bags with silica gel. Voucher specimens were stored in the laboratory of Qingdao CDC. In order to calculate genetic distances better, we downloaded 51 sequences from GenBank to ensure more than two individuals of each species. In addition, to assess the efficiency of identification by individual DNA barcodes in closely related species, we also downloaded 42, 30, and 33 sequences of ITS, trnH-psbA and rbcL fragments represented 16, 13, and 15 Subtrib. Phaseolinae species, respectively.
Simulated Gastric Fluid (SGF) Digestion
Thirteen plant materials were digested in SGF to simulate the stomach contents of a poisoned patient. According to the United States Pharmacopoeia (USP) (Pharmacopeia, 2020), 1000 mL of SGF consisted of 2.6 g of pepsin (806 U/mg; Sigma-Aldrich, Germany), 2.0 g of NaCl (Carl Roth, Germany), and distilled water adjusted to pH 1.2 with HCl (Carl Roth, Karlsruhe, Germany). Each crushed target plant weighing 200 mg was boiled in 100°C water for 15 min, 2 mL SGF was added and kept at 37°C for 120 min, 240 min and 360 min, respectively. Digestion was stopped with 0.7 mL of 0.2 M Na2CO3. DNA was stored at −20°C before extraction.
DNA Extraction, Amplification, and Sequencing
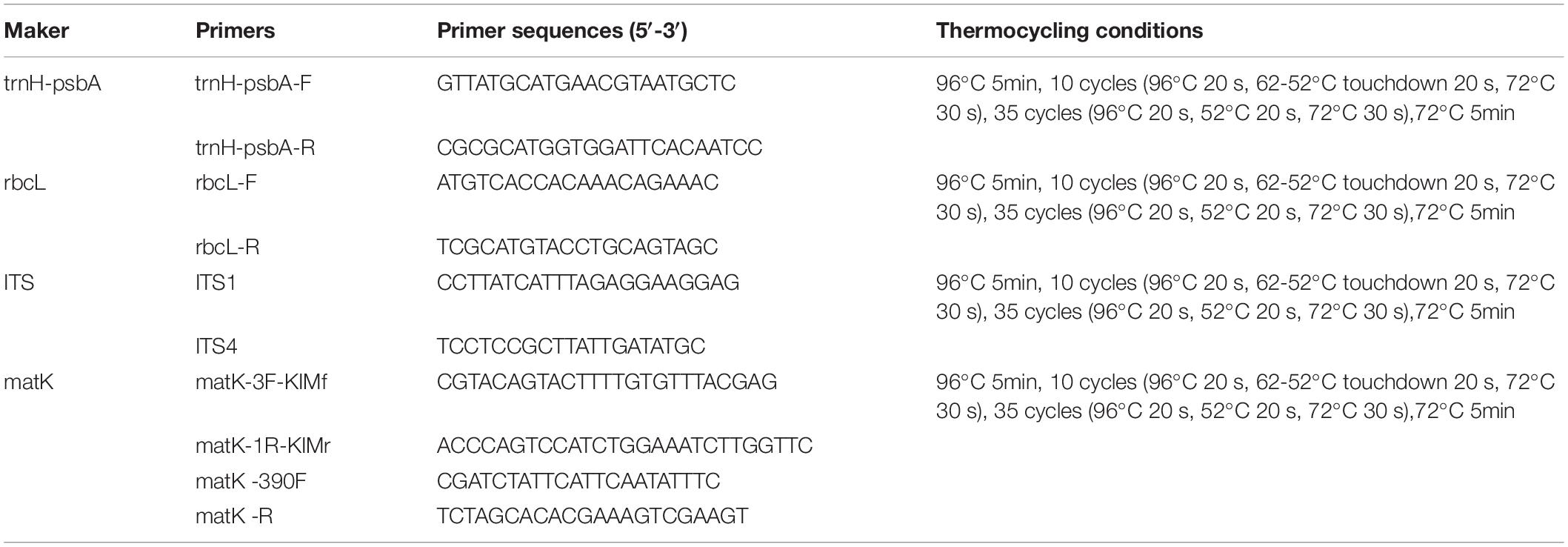
Total genomic DNA was extracted from all samples according to the instructions of the DNA extraction kit (Tiangen Biotech CO., LTD, Beijing, China). Amplification of DNA was performed by standard polymerase chain reaction (PCR). The PCR mixture (30 μL) contained template genomic DNA 2 μL, forward primer (10 μM) 1μL, reverse primer (10 μM)1 μL, Ex Taq (5U/μL) 0.2 μL, dNTP (2.5 mM each) 2 μL and H2O 20.8 μL. The primer sequences and thermocycling conditions for PCR amplification are listed in Table 1. Each PCR fragment was purified according to Millipore’s 96 purification plate procedure before sequenced by UW Genetics Technology Ltd (Beijing). High-quality PCR products are sequenced from both directions to reduce sequencing errors and improve accuracy.
Data Analysis
All sequences were spliced using CodonCode Aligner 6.0.2 to remove primer sequences and correct for ambiguous bases. All sequences were aligned using ClustalW. Insertion deletions (indels) were detected and counted for each DNA region using DNAsp5.0. To assess the species resolution of individual and combined barcodes, three different analytical methods were used: (1) sequence similarity-based methods (BLAST). (2) PWG-Distance, and (3) Tree-Building (NJ).
Sequence Similarity-Based Method
For the BLAST method, the sample sequence was used as the query sequence and the standard database in National Center for Biotechnology Information (NCBI) was used as the reference database. The BLAST program1 was used to perform base pairwise comparisons of the query sequences. Species discrimination was considered successful if a species had only one best hit for homozygous individuals.
PWG-Distance Method
The PWG distance method calculates distances to paired pairs by calculating explicit base substitutions. Interspecific distance and intraspecific distance were calculated in MEGA 7.0.26 using the Kimura two-parameter distance model (K2P). We considered that correct species differentiation was confirmed if the uncorrected minimum interspecific p distance of a species, involving at least more than one individual, was greater than its maximum intraspecific distance. In addition, we plotted the distribution of intra- and interspecific variation for each candidate barcode and its combinations to reveal barcode gaps. A barcode is considered suitable if there is a visible barcode gap.
Tree-Building Method
All regional sequences were tested for base substitution saturation using DAMBE. The evolutionary tree reflects the developmental relationships between species only if the sequences are proven to be unsaturated. Neighbor-Join (NJ) trees were constructed using the K2P model of MEGA 7.0.26. The node support is calculated based on 1000 bootstrap replicates. In general, a species can be considered successfully distinguished if all individuals of the species form independent clusters in the tree with bootstrap support greater than 70%.
Results
Amplification, Sequencing, and Sequence Analysis
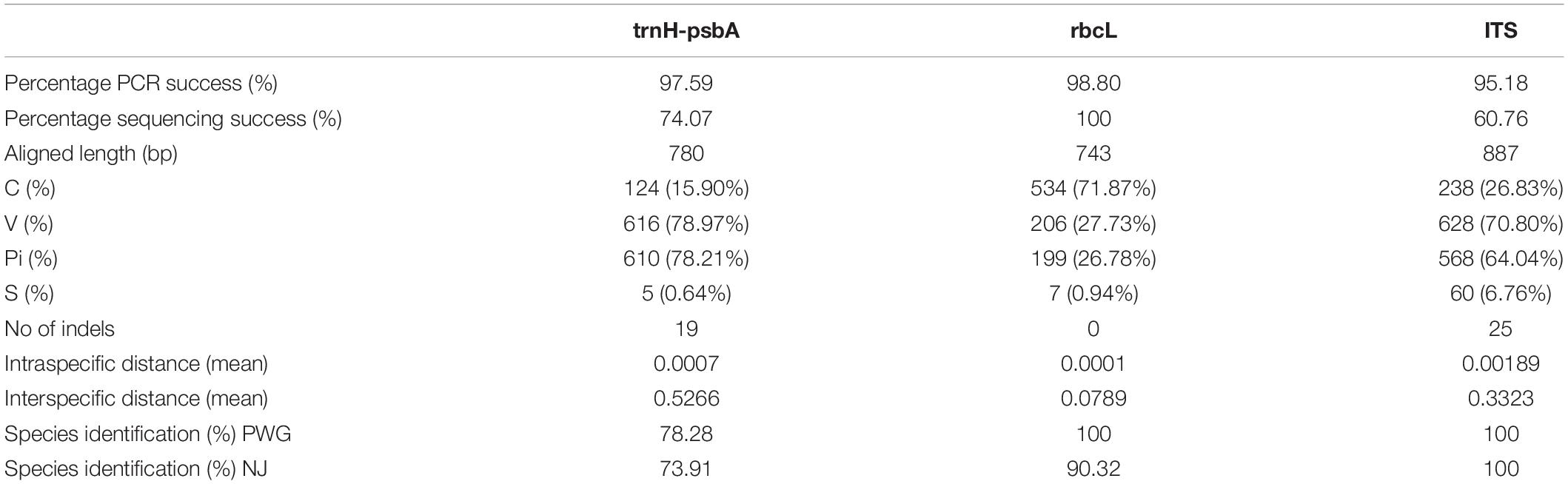
Polymerase chain reaction (PCR) amplification of the three DNA barcodes had high success rates, i.e., 97.59, 98.80, and 95.18% for trnH-psbA, rbcL, and ITS, respectively. The matK region was difficult to amplify and we could not obtain the target barcode for 54.22% of the samples even using two different primer pairs. Therefore, this barcode was not included in the subsequent barcode analysis. In addition, rbcL had the highest sequencing success rate (100%), followed by ITS (74.07%) and trnH-psbA (60.76%). In total, we obtained 220 new sequences from 83 materials, of which 60 were trnH-psbA, 82 were rbcL, 48 were ITS, and 30 were matK. All sequences were submitted to the NCBI accession number see Supplementary Table 1.
The aligned lengths of the trnH-psbA, rbcL, and ITS sequences are 744, 746, and 855, respectively. trnH-psbA (337–769) has the most significant length variation than the other two markers. Among the three markers, rbcL was the most conserved, with the highest percentage of conserved regions and the least indels. The sequence characteristics of all DNA barcodes are shown in Table 2.
Amplification and Sequencing Results After Simulated Gastric Fluid Digestion
Generally, nausea and vomiting occur within a few hours after ingestion of toxic plants. We digested the plant material with SGF for 120, 240, and 360 min to replace the stomach contents. The results showed that the amplification and sequencing of rbcL and trnH-psbA regions were not affected, but the matK region showed difficulties. Notably, the ITS region of two samples, P. vulgaris 08 and L. esculentum 01, showed better sequencing efficiency after SGF digestion. The amplification and sequencing are shown in Supplementary Table 3.
Intra-Specific and Inter-Specific Genetic Divergence Analyses
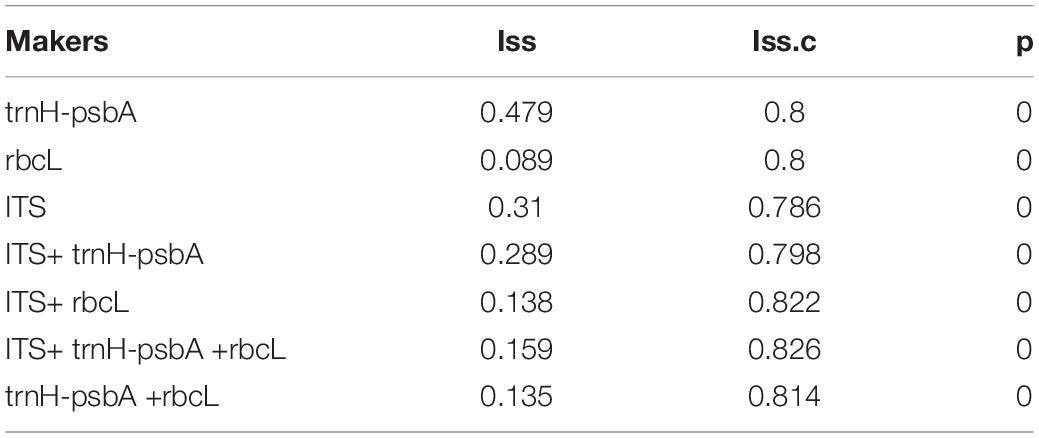
All three DNA regions exhibited higher genetic variability than within species (Table 2). The trnH-psbA region showed the greatest interspecific variation, followed by the ITS region, and rbcL the least. The barcoding gap was graphed based on the K2P model for each marker and their combinations. The results indicated that chloroplast barcode markers and their combinations appeared to be light overlapping without significant gaps. However, the combinations of ITS + single or composite chloroplast barcodes had obvious barcode gaps, with ITS + trnH-psbA showing the highest divergence (Figure 1).
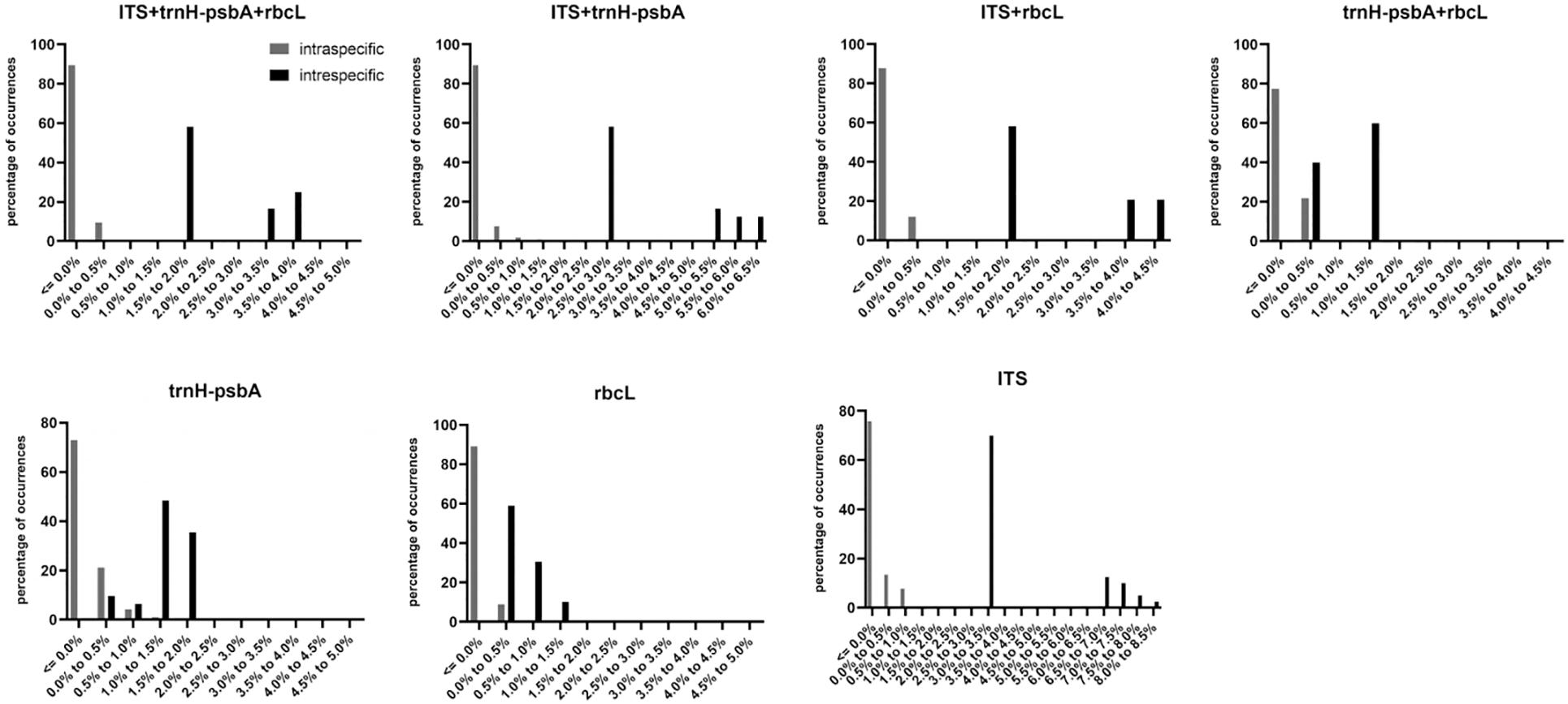
Figure 1. Histograms of the frequency (y-axis) of paired intraspecific and interspecific divergences based on the K2P distance (x-axis) of rbcL, trnH-psbA, ITS and their combined markers.
Species Discrimination
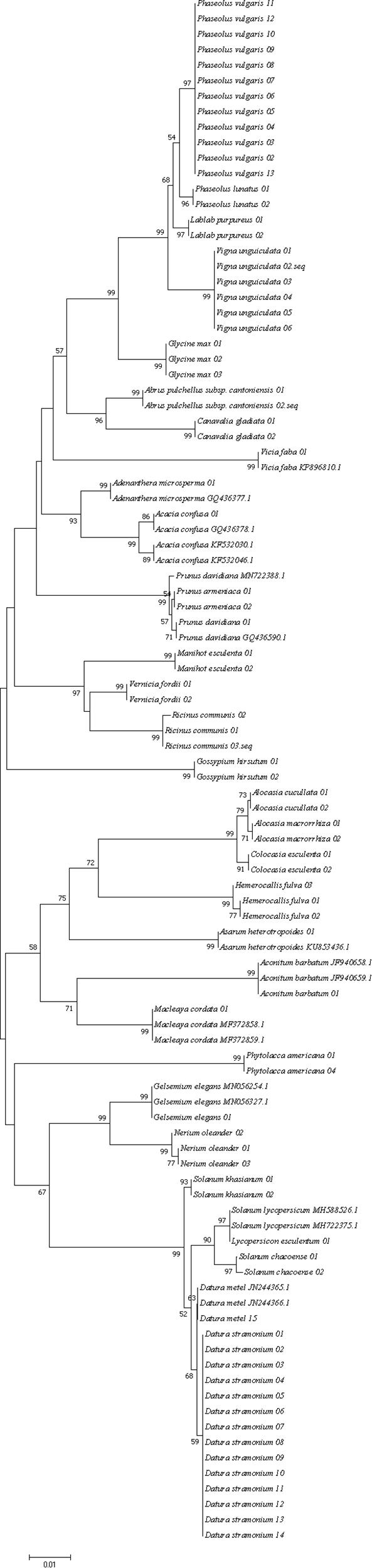
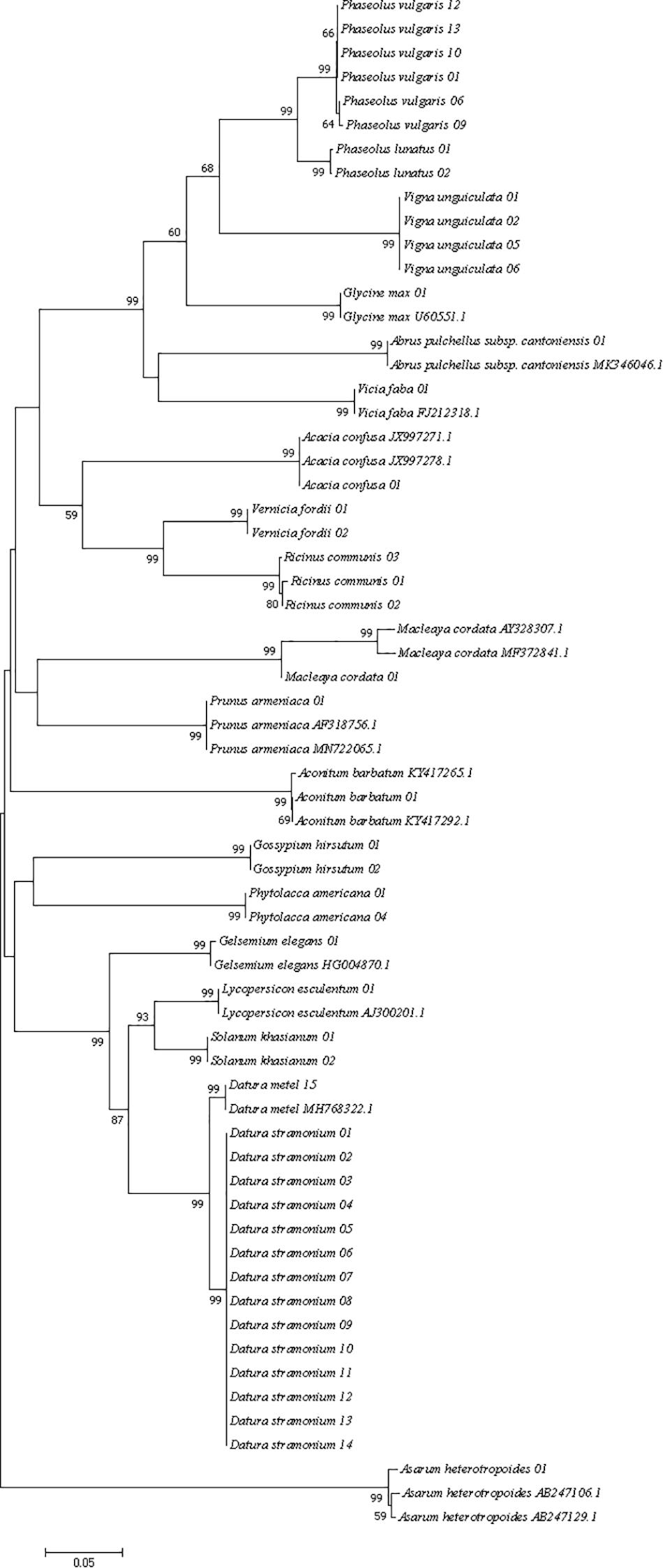
Three different analytical methods were used to assess the discriminatory ability of single and combined barcode markers for common toxic plants in China. In the BLAST method, species discrimination was high for all markers at the genus level, but at the species level, rbcL was not as good as trnH-psbA and ITS (Table 3). For the tree-building method (Figures 2–4), the Iss values for both single and combined markers were smaller than the Iss.c values (Min et al., 2020), indicating that the sequences were not saturated (Table 4). The species recognition power of all barcodes is shown in Figure 5. Among the single barcodes, ITS had the highest recognition rate (PWG:100%, NJ tree: 100%), followed by rbcL (PWG:100%, NJ tree: 90.32%), while trnH-psbA had a lower recognition rate (PWG:78.28%, NJ tree: 73.91%). When the barcode loci were combined, the resolution of any combination was higher than that of a single loci. The highest resolution was achieved by the combination of rbcL + ITS (PWG:100%, NJ tree:100%). Overall, in the Tree-Building and PWG-Distance analyses, there were similar results in the species discrimination power. The BLAST method has a lower species resolution but is faster and more intuitive.
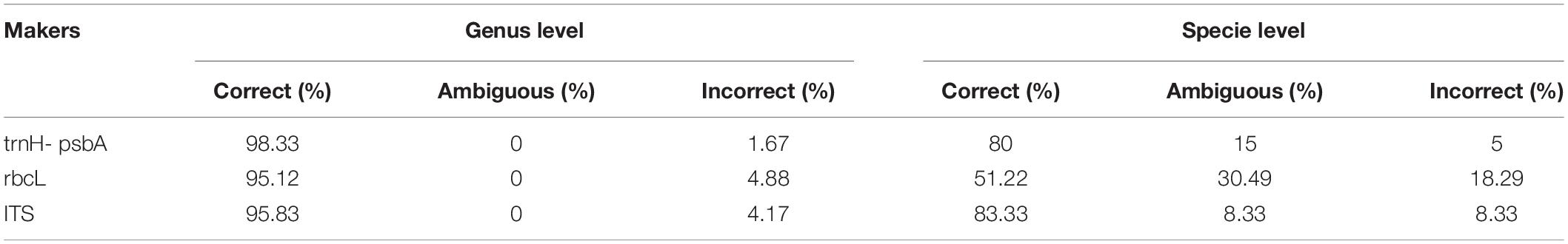
Table 3. Species resolution of three markers at the genus and species level based on the BLAST method.
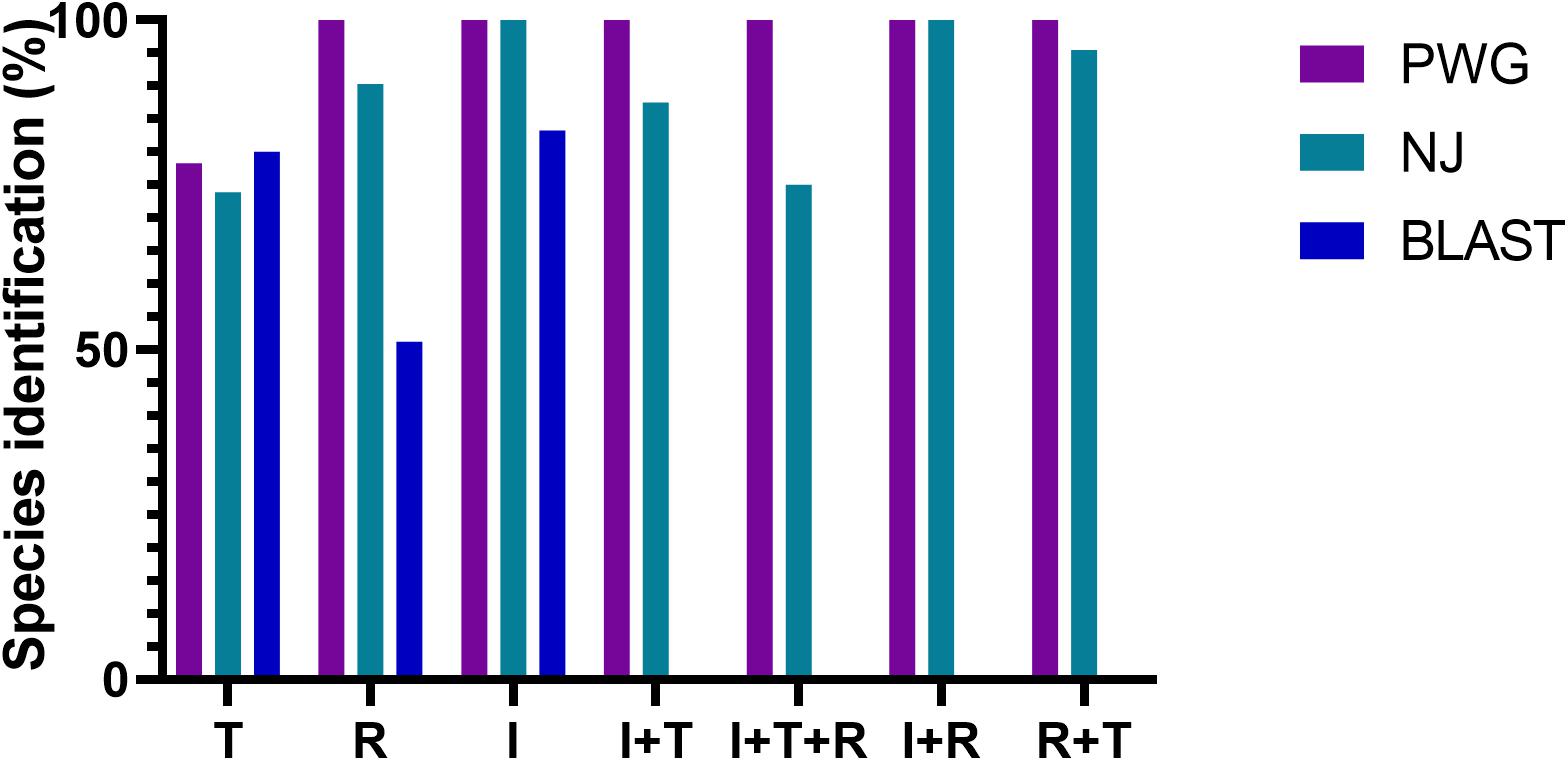
Figure 5. Species recognition rates for all tested single and combined markers, BLAST method was evaluated for single barcodes only rbcL(R), trnH-psbA(T), and ITS(I).
In addition, to assess the discriminatory ability of three single barcode markers in closely related species, we selected Subtrib. phaseolinae species as representative and used two analysis methods, NJ-tree (Supplementary Figures 1–3) and PWG. The results showed that ITS fragments had the highest resolution (PWG:100%, NJ-tree:100%), followed by psbA-trnH (PWG:80%, NJ-tree:80%) and rbcL (PWG:73.68%, NJ-tree:52.63%).
Discussion
Amplification and Sequencing Success Rate of Four Candidate Barcodes
The ideal DNA barcode should satisfy the following conditions (Group, 2009): (1) the presence of sufficient flanking sequences to enable the development of primers with high generality (2) relatively short nucleotide sequences for good amplification sequencing and little need for manual editing of sequence tracks. (3) Be able to provide a large degree of discrimination between species. In this study, PCR amplification using universal primer pairs had high success rates for each of the four DNA barcode regions, except for the matK region. In addition, rbcL had the best sequencing performance, while the difficulties in sequencing trnH-psbA and ITS were observed.
The chloroplast gene matK is the closest plant analog to the animal barcode (CO1) (Hollingsworth et al., 2011). However, the matK region requires more primers than other regions to be amplified due to the high variability of primer binding sites (Fazekas et al., 2008; Hollingsworth et al., 2011). In our study, only 36.14% of the samples were successfully amplified by using additional primers. Meanwhile, the amplification and sequencing of this region became harder after SGF treatment because the shorter length of the remaining fragment would hinder the PCR extension phase of the longer gene (Li Q. J. et al., 2019).
The trnH-psbA possesses a highly conserved flanking sequence and a non-coding region with a large number of base substitutions, making this region well suited as a plant DNA barcode (Li et al., 2015). However, the biggest problem of trnH-psbA marker is the variable-length distribution (337–769 bp) in various plant species (Pang et al., 2012). In addition, the single nucleotide repeat (PolyA, PolyT) fragment located in the middle of the marker leads to sequencing failure of the second half of the region in some individuals.
Successful sequencing of ITS region from plant samples can be difficult. In our study, only 60.76% of the samples were successfully sequenced. On the one hand, incomplete co-evolution of nuclear multicopy regions due to hybridization or other factors can affect amplification and sequencing efficiency (China Plant et al., 2011). On the other hand, fungal DNA is often amplified from samples inadvertently and eventually confused with plant sequences (Seifert, 2009; Li et al., 2015). The sequencing peak maps in our study overlapped from the middle, which may occur in impure samples. In addition, the ITS region of certain samples showed better sequencing efficiency after SGF digestion. The possible reason for this anomaly is that pepsin degrades the DNA of fungal contaminants.
In summary, we were unable to obtain the full sequences of all samples and do not recommend the use of the matK region for the identification of toxic plants considering the success of amplification and sequencing.
Resolution of the Three Single Barcodes and Their Combinations
The trnH-psbA region provided the highest inter-and intraspecific divergences (0.5266 and 0.0007, respectively), with similar species resolution in the three analysis methods (BLAST:80%, PWG:78.26% and NJ:73.91%). However, regions flanked by trnH-psbA with inverted repeats were frequently inverted (Whitlock et al., 2010) and insertions of pseudogenes (rps19) were very common (Pang et al., 2012), which may allow this region to overestimate differences between homologous sequences and misclassify relationships between closely related species when identifying species (Pang et al., 2012). Nevertheless, trnH-psbA is still considered as a promising DNA barcode in distinguishing plants of certain families, such as Fabaceae (Gao et al., 2013; Loera-Sanchez et al., 2020), Solanaceae (Feng et al., 2018), and Rhododendron (Liu et al., 2012). Therefore, the marker may be used for further studies of toxic plants.
The other chloroplast genome rbcL had the most conserved regions and the smallest interspecific genetic distance (0.0789). For the tree building method, the success rate of accurate identification to the genus level was high (95.12%), but only 51.22% at the species level. It is also less discriminatory between near-derived species than the other two regions (PWG:73.68%, NJ-tree:52.63%). This inadequate resolution may be due to the lack of variation in the rbcL region (Ng et al., 2016). However, species from the same genus usually have similar toxins (Xie et al., 2014), and identification of species to the genus level is helpful in the clinical management of most poisoning events. In our study, PWG and NJ trees had higher identification rates than BLAST, which is inconsistent with the findings of the Chinese plant Bol Group in angiosperms (China Plant et al., 2011). This contradiction may be related to the fact that our sampling considered only common toxic plants. Moreover, our reference database is the standard database from Genbank. The interference of redundant genes might have reduced the accuracy of BLAST.
In our study, ITS markers had the highest species resolution among all individual candidate DNA barcodes (BLAST:83.33%, PWG:100% and NJ:100%). The same result was demonstrated in a previous study (Zhang et al., 2016; Guo et al., 2017). However, ITS markers are considered to have drawbacks, including incomplete genealogical sequencing, homogeneous coevolution (Wirta et al., 2016) and fungal contamination (Liu et al., 2019). In addition, some studies suggest that ITS are not suitable for DNA analysis of gastric contents because their primers can bind to human ITS and 18S rRNA (Lee et al., 2009). Therefore, we considered that the ITS region is not a suitable barcode for clinical identification.
Combined barcodes can improve species resolution (Li Q. J. et al., 2019). As early as 2009, the Consortium for the Barcode of Life (CBOL) recommended the rbcL + matK combination as the core plant barcode and suggested supplementing with additional loci to distinguish closely related species (Group, 2009). In our study, the resolution of any combination of barcodes was higher than that of single markers (Figure 5). Similar results were confirmed in many studies (Gere et al., 2013; Han Y. W. et al., 2016). However, single barcodes may be more suitable for rapid identification and tracing of toxic plants in clinical settings. First, combining barcodes does not significantly improve the resolution of species, but increases the experimental cost and identification time significantly. Second, toxic plants from the one genus tend to have similar toxic chemicals. In our study, individual barcodes at the genus level all had sufficient resolution using the BLAST method (over 95%), which was sufficient to provide assistance in clinical treatment. Finally, the chloroplast DNA regions and nuclear ITS regions have different genetic patterns. The combination of DNA markers from different genomes may hinder our understanding of species delimitation (Li Q. J. et al., 2019).
Overall, the rbcL region is considered to be the most suitable candidate DNA barcode for clinical poisoning identification, and BLAST is recommended for analysis because it is faster and easier compared to other analysis methods.
Prospects and Problems of DNA Barcoding as a Useful Tool for Clinical Identification
Conventional identification methods based on external characteristics of plants (e.g., shape, size, color, flavor, composition, structure, texture, etc.) have many limitations (Yongfu et al., 2014). First, the method requires a high level of expertise and practical experience of the worker, so misidentification often occurs. Second, plants belonging to the same species may show considerable differences in morphology due to geographical location and some abiotic factors (Wäldchen et al., 2018). Both large intraspecific visual variation and smaller interspecific visual variation can confuse the identifier (Wäldchen et al., 2018). Finally, morphological identification is usually valid only for specific life stages or gender (Hebert et al., 2003), plants presented in propagule form are usually not identifiable.
DNA barcoding is independent of plant morphology and can help identify species in cases where complete biological evidence that can determine the cause of the disease is not available at the poisoning site. At the same time, the universality of universal primers makes DNA barcoding non-specific. It allows rapid targeting of species families when a hypothetical poison center needs to deal with an unknown poisoning event (Mezzasalma et al., 2017). In addition, the rapid development of “next-generation sequencing” (NGS) technologies is reducing the cost of sequencing (Sucher et al., 2012). Moreover, third-generation sequencing technology, an approach based on the detection of individual molecular signals without amplification of samples, is expected to solve the identification problems caused by low PCR efficiency. These indicate that DNA barcoding technology shows a strong potential for identifying toxic plants. However, there are still issues to be considered in using DNA barcoding as a clinical diagnostic tool for rapid identification of toxic plants. (1) A study has shown that the recovery of deoxyribonucleic acid from digested target plants is lower than that of untreated samples (Matsuyama and Nishi, 2011). In our study, some samples showed difficulties in amplifying the matK region after SGF digestion. However, Galimberti et al. successfully extracted plant DNA barcodes from the feces of herbivorous birds (Galimberti et al., 2016). Their study shows that it is possible to extract high-quality DNA from stomach contents by adjusting experimental conditions. (2) Some highly toxic species can be hazardous to health even in small amounts. For example, Abrin, a type II ribosomal inactivating protein from Abrus precatorius, is extremely toxic with an estimated lethal dose in humans of 0.1–1 μg/kg (Karthikeyan and Amalnath, 2017; Ninan and James, 2019). In fact, DNA barcoding could only amplify the major components of plant foods. False negatives may be observed in the management of such poisonings. Our experiments only simulated the digestion of individual plants and DNA barcoding should be further tested for its power to identify mixed plant material after different cooking methods.
At present, China has constructed a proprietary Traditional Chinese Medicine Database (TCMD) (Han J. et al., 2016; Gong et al., 2018) and the Chinese Rare and Endangered Plant Information System. It contains DNA sequences and its morphological information, which provides great help in identifying drug adulteration (Han J. et al., 2016) and protecting rare plants. Therefore, the establishment of a specialized toxic plant database containing primer information is considered necessary (Bruni et al., 2010). On the one hand, the sequences of the same gene currently registered in NCBI may have been read by different researchers from different primer pairs (Wei et al., 2019). Different PCR primer pairs binding to different loci of the same gene can make subtle differences in the reads. On the other hand, the identification of DNA barcoding relies on the availability of high-quality reference databases. Insufficient or missing sequence data can lead to ambiguous identification results (Tanaka and Ito, 2020). Misidentification due to missing data occupied a relatively high percentage (21.68%) in Lu Gong’s study (Gong et al., 2018).
The next steps in this study are to improve DNA extraction experiments to obtain high-quality DNA and to construct a proprietary database of toxic organisms containing species morphological information, molecular information, toxicological information and geographic location information.
Conclusion
In this study, four barcodes were selected to assess their applicability in the classification of several common toxic plants in China. We concluded that the single barcode rbcL was the most efficient and cost-effective marker for clinical identification after considering primer versatility, DNA sequence quality, species differentiation ability and experimental cost. In addition, the BLAST method is faster and more intuitive than other methods, and has a high recognition rate at the genus level, making it suitable as a clinical diagnostic analysis method. Future studies should aim to adopt a more comprehensive and balanced sampling scheme. Exploring and developing simple DNA barcode detection instruments and improving database information may be a future research trend.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WY conceived and designed the research. JW, JZ, SB, and SW performed the experiments. JW and JZ wrote the manuscript. WY, XZ, and XS revised the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by the Qingdao Minsheng Science and Technology Plan Project (18-6-1-67-nsh).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.698418/full#supplementary-material
Supplementary Figure 1 | Neighbor-joining tree (NJ) of Subtrib. Phaseolinae species based on trnH-psbA region.
Supplementary Figure 2 | Neighbor-joining tree (NJ) of Subtrib. Phaseolinae species based on rbcL region.
Supplementary Figure 3 | Neighbor-joining tree (NJ) of Subtrib. Phaseolinae species based on ITS region.
Supplementary Table 1 | Information and GenBank accession numbers of the samples in this study.
Supplementary Table 2 | GenBank accession numbers of samples from the NCBI.
Supplementary Table 3 | Amplification and sequencing of 13 species after simulated gastric fluid digestion.
Footnotes
References
Agarwal, M., Shrivastava, N., and Padh, H. (2008). Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 27, 617–631. doi: 10.1007/s00299-008-0507-z
Bruni, I., De Mattia, F., Galimberti, A., Galasso, G., Banfi, E., Casiraghi, M., et al. (2010). Identification of poisonous plants by DNA barcoding approach. Int. J. Legal Med. 124, 595–603. doi: 10.1007/s00414-010-0447-3
China Plant, B. O. L. G., Li, D. Z., Gao, L. M., Li, H. T., Wang, H., Ge, X. J., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U S A 108, 19641–19646. doi: 10.1073/pnas.1104551108
Chu, F. J., Ran, L., Ma, L., and Lin, X. H. (2012). Analysis on the reported food poisoning incidents in public health emergency events surveillance system in china,2008-2010. Chinese J. Food Hyg. 24, 387–390.
De Mejia, E. G., Carmen, M. Del, Valadez-Vega, R., and Reynoso-Camacho, G. (2005). Tannins, trypsin inhibitors and lectin cytotoxicity in tepary (Phaseolus acutifolius) and common (Phaseolus vulgaris) beans. Plant Foods Hum. Nutre 60, 137–145. doi: 10.1007/s11130-005-6842-0
Doan, U. V., Wu, M. L., Phua, D. H., Mendez Rojas, B., and Yang, C. C. (2019). Datura and Brugmansia plants related antimuscarinic toxicity: an analysis of poisoning cases reported to the Taiwan poison control center. Clin. Toxicol. 57, 246–253. doi: 10.1080/15563650.2018.1513527
Eddleston, M., Rajapakse, S., Jayalath, S., Sjöström, L., Santharaj, W., Thenabadu, P. N., et al. (2000). Anti-digoxin Fab fragments in cardiotoxicity induced by ingestion of yellow oleander: a randomised controlled trial. Lancet 355, 967–972. doi: 10.1016/s0140-6736(00)90014-x
Fazekas, A. J., Burgess, K. S., Kesanakurti, P. R., Graham, S. W., Newmaster, S. G., Husband, B. C., et al. (2008). Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One 3:e2802. doi: 10.1371/journal.pone.0002802
Feng, S., Jiao, K., Zhu, Y., Wang, H., Jiang, M., and Wang, H. (2018). Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: the chloroplast psbA-trnH intergenic region. Genome 61, 15–20. doi: 10.1139/gen-2017-0115
Fuchs, J., Rauber-Luthy, C., Kupferschmidt, H., Kupper, J., Kullak-Ublick, G. A., and Ceschi, A. (2011). Acute plant poisoning: analysis of clinical features and circumstances of exposure. Clin. Toxicol. 49, 671–680. doi: 10.3109/15563650.2011.597034
Galimberti, A., Spinelli, S., Bruno, A., Mezzasalma, V., De Mattia, F., Cortis, P., et al. (2016). Evaluating the efficacy of restoration plantings through DNA barcoding of frugivorous bird diets. Conserv. Biol. 30, 763–773. doi: 10.1111/cobi.12687
Gao, T., Ma, X., and Zhu, X. (2013). Use of the psbA-trnH region to authenticate medicinal species of Fabaceae. Biol. Pharm. Bull. 36, 1975–1979. doi: 10.1248/bpb.b13-00611
Gere, J., Yessoufou, K., Daru, B. H., Mankga, L. T., Maurin, O., and van der Bank, M. (2013). Incorporating trnH-psbA to the core DNA barcodes improves significantly species discrimination within Southern African Combretaceae. Zookeys 365, 129–147. doi: 10.3897/zookeys.365.5728
Gong, L., Qiu, X. H., Huang, J., Xu, W., Bai, J. Q., Zhang, J., et al. (2018). Constructing a DNA barcode reference library for southern herbs in China: A resource for authentication of southern Chinese medicine. PLoS One 13:e0201240. doi: 10.1371/journal.pone.0201240
Group, C. P. W. (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U. S. A. 106, 12794–12797.
Guo, M., Ren, L., and Pang, X. (2017). Inspecting the True Identity of Herbal Materials from Cynanchum Using ITS2 Barcode. Front. Plant Sci. 8:1945.
Han, J., Pang, X., Liao, B., Yao, H., Song, J., and Chen, S. (2016). An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep. 6:18723.
Han, Y. W., Duan, D., Ma, X. F., Jia, Y., Liu, Z. L., Zhao, G. F., et al. (2016). Efficient Identification of the Forest Tree Species in Aceraceae Using DNA Barcodes. Front Plant Sci 7:1707.
He, S., Simpson, B. K., Sun, H., Ngadi, M. O., Ma, Y., and Huang, T. (2018). Phaseolus vulgaris lectins: A systematic review of characteristics and health implications. Crit. Rev. Food Sci. Nutr. 58, 70–83. doi: 10.1080/10408398.2015.1096234
Hebert, P. D., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D., Ratnasingham, S., Zakharov, E. V., Telfer, A. C., Levesque-Beaudin, V., Milton, M. A., et al. (2016). Counting animal species with DNA barcodes: Canadian insects. Philos Trans. R. Soc. Lond. B. Biol. Sci. 371: 1702.
Hollingsworth, P. M., Graham, S. W., and Little, D. P. (2011). Choosing and using a plant DNA barcode. PLoS One 6:e19254. doi: 10.1371/journal.pone.0019254
Karthikeyan, A., and Amalnath, S. D. (2017). Abrus precatorius Poisoning: A Retrospective Study of 112 Patients. Indian J. Crit. Care Med. 21, 224–225. doi: 10.4103/ijccm.ijccm_320_16
Krenzelok, E. P., and Mrvos, R. (2011). Friends and foes in the plant world: a profile of plant ingestions and fatalities. Clin. Toxicol. 49, 142–149. doi: 10.3109/15563650.2011.568945
Lee, E. J., Kim, S. C. I, Hwang, K., Yang, H. J., Kim, Y. S., Han, M. S., et al. (2009). The identification of ingested dandelion juice in gastric contents of a deceased person by direct sequencing and GC-MS methods. J Forensic Sci 54, 721–727. doi: 10.1111/j.1556-4029.2009.01019.x
Li, Q. J., Wang, X., Wang, J. R., Su, N., Zhang, L., Ma, Y. P., et al. (2019). Efficient Identification of Pulsatilla (Ranunculaceae) Using DNA Barcodes and Micro-Morphological Characters. Front. Plant Sci. 10:1196.
Li, T. X., Cai, T. T., and Xue, Q. P. (2019). Misdiagnosis and Treatment of Solanine Poisoning. Clin. Misdiagn. Mistreat. 32, 8–10.
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. Camb. Philos. Soc. 90, 157–166. doi: 10.1111/brv.12104
Liu, M., Li, X. W., Liao, B. S., Luo, L., and Ren, Y. Y. (2019). Species identification of poisonous medicinal plant using DNA barcoding. Chin. J. Nat. Med. 17, 585–590. doi: 10.1016/s1875-5364(19)30060-3
Liu, Y., Zhang, L., Liu, Z., Luo, K., Chen, S., and Chen, K. (2012). Species identification of Rhododendron (Ericaceae) using the chloroplast deoxyribonucleic acid psbA-trnH genetic marker. Pharmacogn Mag 8, 29–36. doi: 10.4103/0973-1296.93311
Loera-Sanchez, M., Studer, B., and Kolliker, R. (2020). DNA barcode trnH-psbA is a promising candidate for efficient identification of forage legumes and grasses. BMC Res. Notes 13:35.
Matsuyama, S., and Nishi, K. (2011). Genus identification of toxic plant by real-time PCR. Int. J. Legal. Med. 125, 211–217. doi: 10.1007/s00414-010-0487-8
Mezzasalma, V., Ganopoulos, I., Galimberti, A., Cornara, L., Ferri, E., and Labra, M. (2017). Poisonous or non-poisonous plants? DNA-based tools and applications for accurate identification. Int. J. Legal. Med. 131, 1–19. doi: 10.1007/s00414-016-1460-y
Min, P., Yaoquan, H., Weijun, W., Yusen, L., Jun, S., Dapeng, W., et al. (2020). Genetic diversity analysis of the Leiocassis crassilabris population in the Xijiang River basin based on mitochondrial D-loop. Freshw. Fish. 50, 3–9.
Müller, D., and Desel, H. (2013). Common causes of poisoning: etiology, diagnosis and treatment. Dtsch. Arztebl. Int. 110, 690–699.
Ng, A. E., Sandoval, E., and Murphy, T. M. (2016). Identification and Individualization of Lophophora using DNA Analysis of the trnL/trnF Region and rbcL Gene. J. Forensic. Sci. 61(Suppl. 1), S226–S229.
Ninan, E. C., and James, E. (2019). Acute disseminated encephalomyelitis due to Abrus precatorius poisoning - A case report. Saudi Pharm. J. 27, 521–524. doi: 10.1016/j.jsps.2018.11.016
Pang, X., Liu, C., Shi, L., Liu, R., Liang, D., Li, H., et al. (2012). Utility of the trnH-psbA intergenic spacer region and its combinations as plant DNA barcodes: a meta-analysis. PLoS One 7:e48833. doi: 10.1371/journal.pone.0048833
Petersen, D. D. (2011). Common plant toxicology: a comparison of national and southwest Ohio data trends on plant poisonings in the 21st century. Toxicol. Appl. Pharmacol. 254, 148–153. doi: 10.1016/j.taap.2010.10.022
Qian, H. (2014). Literature Analysis of Poisoning Caused by Poisonous Organisms in China and Experimental Study on Factors Affecting Levels of Aconitine-type Alkaloids in Aconitum kusnezoffii. Master Master’s Thesis, Beijing: China Center for Disease Control.
Romano, M. C., Doan, H. K., Poppenga, R. H., Filigenzi, M. S., Bryant, U. K., and Gaskill, C. L. (2019). Fatal Amanita muscaria poisoning in a dog confirmed by PCR identification of mushrooms. J. Vet. Diagn. Invest. 31, 485–487. doi: 10.1177/1040638719842897
Seifert, K. A. (2009). Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. (9 Suppl.), 83–89. doi: 10.1111/j.1755-0998.2009.02635.x
Shinozaki, J., Kazuma, K., Satake, M., Kondo, K., and Konno, K. (2018). PCR-RFLP Method for Rapid Discrimination of Toxic Plants Involved in Food Poisoning. Shokuhin Eiseigaku Zasshi 59, 134–140. doi: 10.3358/shokueishi.59.134
Sucher, N. J., Hennell, J. R., and Carles, M. C. (2012). DNA fingerprinting, DNA barcoding, and next generation sequencing technology in plants. Methods Mol. Biol. 862, 13–22. doi: 10.1007/978-1-61779-609-8_2
Tanaka, S., and Ito, M. (2020). DNA barcoding for identification of agarwood source species using trnL-trnF and matK DNA sequences. J. Nat. Med. 74, 42–50. doi: 10.1007/s11418-019-01338-z
Thu, P. T., Huang, W. C., Chou, T. K., Van Quan, N., Van Chien, P., Li, F., et al. (2019). DNA barcoding of coastal ray-finned fishes in Vietnam. PLoS One 14:e0222631. doi: 10.1371/journal.pone.0222631
Wäldchen, J., Rzanny, M., Seeland, M., and Mäder, P. (2018). Automated plant species identification-Trends and future directions. PLoS Comput. Biol. 14:e1005993. doi: 10.1371/journal.pcbi.1005993
Wei, S., Luo, Z., Cui, S., Qiao, J., Zhang, Z., Zhang, L., et al. (2019). Molecular Identification and Targeted Quantitative Analysis of Medicinal Materials from Uncaria Species by DNA Barcoding and LC-MS/MS. Molecules 24:1.
Whitlock, B. A., Hale, A. M., and Groff, P. A. (2010). Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLoS One 5:e11533. doi: 10.1371/journal.pone.0011533
Wirta, H., Várkonyi, G., Rasmussen, C., Kaartinen, R., Schmidt, N. M., Hebert, P. D., et al. (2016). Establishing a community-wide DNA barcode library as a new tool for arctic research. Mol. Ecol. Resour. 16, 809–822. doi: 10.1111/1755-0998.12489
Xie, L., Wang, Y. W., Guan, S. Y., Xie, L. J., Long, X., and Sun, C. Y. (2014). Prospects and Problems for Identification of Poisonous Plants in China using DNA Barcodes. Biomed. Environ. Sci. 27, 794–806.
Yongfu, C., Hua, L., and Qia, C. (2014). Status and Prospect for the Study of Plants Identification Methods. Word Forest. Res. 27, 18–23.
Zangl, L., Daill, D., Schweiger, S., Gassner, G., and Koblmüller, S. (2020). A reference DNA barcode library for Austrian amphibians and reptiles. PLoS One 15:e0229353. doi: 10.1371/journal.pone.0229353
Keywords: DNA barcoding, phytotoxicity, rapid identification, simulated gastric fluid, rbcL
Citation: Wang J, Zhao J, Yu W, Wang S, Bu S, Shi X and Zhang X (2021) Rapid Identification of Common Poisonous Plants in China Using DNA Barcodes. Front. Ecol. Evol. 9:698418. doi: 10.3389/fevo.2021.698418
Received: 23 April 2021; Accepted: 30 July 2021;
Published: 23 August 2021.
Edited by:
Xiaofan Zhou, South China Agricultural University, ChinaReviewed by:
Jianping Han, Chinese Academy of Medical Sciences, ChinaJianqiang Zhang, Shaanxi Normal University, China
Copyright © 2021 Wang, Zhao, Yu, Wang, Bu, Shi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weisen Yu, eXV3ZWlzZW5AMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jie Wang
Jie Wang Jinquan Zhao2†
Jinquan Zhao2† Xuexiang Shi
Xuexiang Shi Xiuyun Zhang
Xiuyun Zhang