
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Ecol. Evol. , 22 July 2021
Sec. Chemical Ecology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.681913
 Divya Lakshmanan Mangalath1*
Divya Lakshmanan Mangalath1* Shabeer Ali Hassan Mohammed2,3
Shabeer Ali Hassan Mohammed2,3It remains a mystery why estrogen hormone receptors (ERs), which are highly specific toward its endogenous hormones, are responsive to chemically distinct exogenous agents. Does it indicate that ERs are environmentally regulated? Here, we speculate that ERs would have some common structural features with prokaryotic taxis receptor responsive toward environmental signals. This study addresses the low specificity and high responsiveness of ERs toward chemically distinct exogenous substances, from an evolutionary point of view. Here, we compared the ligand binding domain (LBD) of ER alpha (α) with the LBDs of prokaryotic taxis receptors to check if LBDs share any structural similarity. Interestingly, a high degree of similarity in the domain structural fold architecture of ERα and bacterial taxis receptors was observed. The pharmacophore modeling focused on ligand molecules of both receptors suggest that these ligands share common pharmacophore features. The molecular docking studies suggest that the natural ligands of bacterial chemotaxis receptors exhibit strong interaction with human ER as well. Although phylogenetic analysis proved that these proteins are unrelated, they would have evolved independently, suggesting a possibility of convergent molecular evolution. Nevertheless, a remarkable sequence divergence was seen between these proteins even when they shared common domain structural folds and common ligand-based pharmacophore features, suggesting that the protein architecture remains conserved within the structure for a specific function irrespective of sequence identity.
- ER-LBD shares structural folds with bacterial chemotaxis receptor LBD.
- Domain architecture is preserved by conserved structural folds, irrespective of sequence identity.
- Ligands for ER and bacterial chemotaxis receptors share common pharmacophore features.
- Ligands of bacterial chemotaxis receptors interacted with human ER and vice versa.
At present, the evolution of nuclear hormone receptors (NRs) is traced back up to protostomes such as plathelminths and mollusks (Kohler et al., 2007; Vogeler et al., 2014; Baker and Lathe, 2018; Wu and LoVerde, 2019). According to the present perspective, Amphioxus, a prochodate deuterostome, contains the most primitive estrogen receptor (ER) (Baker and Chandsawangbhuwana, 2008; Lecroisey et al., 2012). However, the full transcriptional function of the ER evolved hundreds of millions of years later after the evolution of early deuterostomes is reported (Kao et al., 2000; Baker and Chandsawangbhuwana, 2008; Callard et al., 2011). ERα is believed to be the most primitive among all the NRs reported so far. Sequence analysis indicates that the other NRs such as other ER subtypes, progesterone receptors, androgen receptors, glucocorticoid receptors, and minerocorticoid receptors evolved from ERα through gene duplication and sequence divergence (Elliston and Katzenellenbogen, 1988; Thornton, 2001; Guerriero, 2009; Baker et al., 2015; Baker, 2019).
All the known NRs discovered so far adopts common structural and functional characteristics and consists of evolutionarily conserved, structurally and functionally distinct three to six basic domains (Lakshmanan and Shaheer, 2020). ERs are made up of an N-terminal domain, central DNA binding domain, C-terminal LBD, and two distinct, conformationally active regions designated as activation function 1 (AF-1) and activation function 2 (AF-2) (Lakshmanan and Sadasivan, 2014). DNA-binding domain (DBD) is the most conserved among the domains, and the N-terminal domain is most variable in sequence and length. ER signaling depends on the ligand/hormone and begins with the binding of ligand to LBD (Lakshmanan and Shaheer, 2020).
LBDs of nuclear hormones are specific toward its endogenous ligand and normally do not cross-interact with other non-specific endogenous hormones (Sasson and Notides, 1983; Klinge, 2001; Razandi et al., 2004; Yang et al., 2004; Morissette et al., 2008). On the other hand, they are responsive to metabolites and precursors of steroid hormones and wide variety of chemically distinct exogenous substances grouped as xenohormones. Both ERα and ERβ exhibits similar affinities for the endogenous hormones estradiol, estriol, and estrone (Sasson and Notides, 1983; Morissette et al., 2008).
Estrogen hormone receptors exhibits least substrate specificity and strong binding affinities for most of the steroid/phenolic and few of the non-phenolic/non-steroid agents classified as xenoestrogens (Cauley, 2015; Gore et al., 2015; Shafei et al., 2018; Pacyga et al., 2019; Rosenfeld and Cooke, 2019; Basak et al., 2020; Lakshmanan and Shaheer, 2020; Park et al., 2020). It is interesting to explore why a receptor that shows high specificity to its endogenous ligand can be transactivated by other hormone metabolites or chemically distinct exogenous agents. The present study was initiated to address this question through bioinformatics tools.
Emergence of proteins with novel domain and/or domain combinations, generated by either homologous or non-homologous DNA repair or inserted into the genomes by transposition, is an important evolutionary mechanism, as it confers potentially diverse biological functions for the organisms (Itoh et al., 2007; Peisajovich et al., 2010; Forslund et al., 2019). Such domain accretion would have paved way in the emergence of novel proteins with specialized functions in the due course of eukaryotic evolution. While ERs are highly specific toward their endogenous hormone, they exhibit least substrate specificity and strong binding affinities toward array of exogenous ligand. This prompted us to speculate that LBD of ER alpha, the NR, would have some similarity with prokaryotic receptors responsive toward environmental signals (chemotaxis receptors). To prove this speculation, we downloaded the amino acid sequences and structures of LBDs of ERα and compared it with bacterial taxis receptors and sequences from other lower life forms such as protozoa and protostoma (Table 1) using Constraint-Based Multiple Alignment Tool (COBALT) and sequence-independent structural alignment (TM-align).
Analysis based on sequence similarity such as Basic Local Alignment Search Tool (BLAST), BLAST-Like Alignment Tool (BLAT), and CLUSTAL multiple sequence alignment tools are not reliable in detecting the homology of distantly related species if similarity of the protein sequence is <30% (Bhagwat et al., 2012; Moreno-Hagelsieb and Hudy-Yuffa, 2014; Ward and Moreno-Hagelsieb, 2014). Nevertheless, protein domain composition is anticipated to be conserved throughout the course of evolution due to functional constraints (Forslund et al., 2019; Yu et al., 2019). The function of a particular protein, to a great degree, is decided by the orderly arrangement of protein sequence to specific domains that constitutes domain architecture. Proteins with the same domain architecture may probably have similar structures and hence related cellular function (Koonin et al., 2002).
Constraint-based multiple alignment tool has advantage over other protein multiple sequence alignment tools in that it finds a set of pairwise constraints derived from conserved domain database and protein motif database and sequence similarity, using RPS-BLAST, BLASTP, and PHI-BLAST. Pair-wise constraints were then integrated into a progressive multiple alignment (Papadopoulos and Agarwala, 2007). All the protein sequences were downloaded from protein sequence database from NCBI. Protein structural files were downloaded from RCSB, protein databank, and COBALT and were used for multiple sequence alignment and prediction of common domain architecture. Molecular Evolutionary Genetics Analysis (MEGA) version X was used for constructing phylogenetic trees based on molecular evolution.
Interestingly, using COBALT, we found that LBDs of ER showed high conservation in domain architecture with all the selected sequences (Figure 1A and Supplementary Figure 1). Furthermore, we selected only the first four proteins shown in Table 1 and performed the COBALT alignment. All the four sequences, that is, LBDs of ER, nuclear receptor of Ciona sp., and two bacterial taxis receptors, exhibited high degree of conservation in domain architecture (Figure 1B and Supplementary Figure 2: first four sequence). As a validation to our hypothesis, we randomly selected some protein sequences (Supplementary Table 1) and repeated COBALT multiple sequence alignment. The result (Supplementary Figure 3) showed that the selected sequences have less conservation and more gaps.
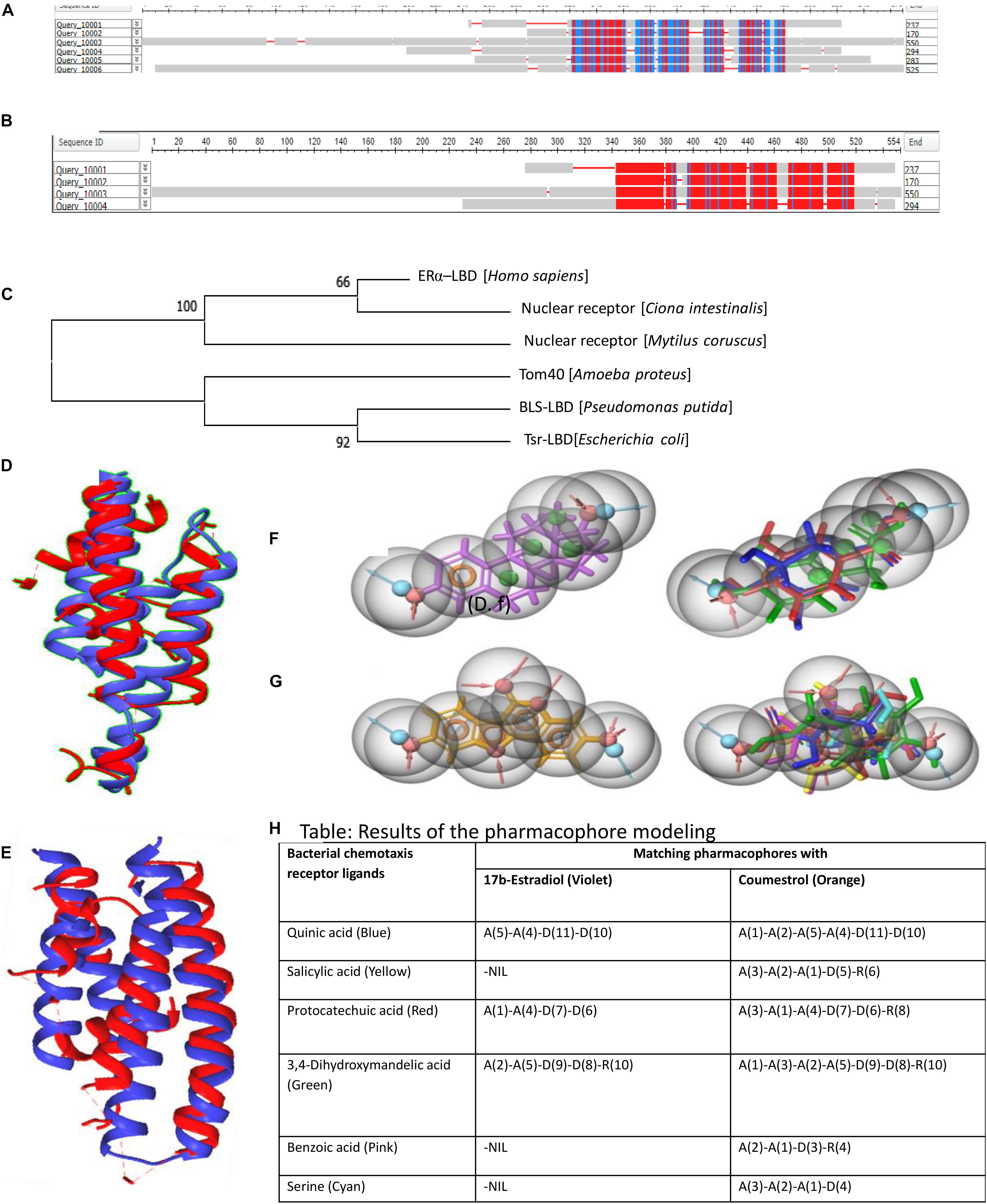
Figure 1. Constraint-Based Multiple Alignment Tool results for the selected sequences. (A) Six sequences. (B) Selected four sequences. Here, the highly conserved and least conserved regions are highlighted based on residues relative entropy threshold. Alignment columns with no gaps are colored in red or blue. Red indicates highly conserved regions, and blue indicates less conserved regions. (C) The gene tree constructed with MEGA version X is represented. (D,E) TM-align results: (D) structural alignment of human ER-α-LBD superposed on Tsr-LBD (E. coli); (E) structural alignment of human ER-α-LBD superposed on broad ligand chemoreceptor, LBD (P. putida). Blue chain represents bacterial taxis receptors, and red chain represents human ER-alpha. Superposed full-atom structure of the aligned region is represented as cartoon. (F,G) Pictorial representation of pharmacophore modeling results. (F) The pharmacophore of the reference ligand 17β-estradiol (left, violet) and the result of the ligand database screening (right) for the identification of matching pharmacophores. (G) The pharmacophore of the reference ligand coumestrol (left, orange) and the result of the ligand database screening (right) for the identification of matching pharmacophores. Detailed color codes are given in tabular form (H). (H) Tabular representation of pharmacophore modeling results.
The phylogenetic tree built for all the six sequences with mega gave an unrooted gene trees (Figure 1C) with clustering of genes into two separate groups. Human ER was clustered into a common clade with Ciona sp., and Mytilus sp. was an outgroup from this clade. Two bacterial chemoreceptors were clustered together. Tom40 of Amoeba was an outgroup from the clade of bacterial receptors. Phylogenetic analysis showed that ERs and bacterial chemotactic receptors are evolutionarily unrelated.
Irrespective of their sequence and structural and functional diversity, ERs and other bacterial chemoreceptors such as taxis to serine and repellents (Tsr) and broad ligand-specific (BLS) receptor of Pseudomonas putida used in this study have certain common features such as a ligand binding domain (LBD) and homodimerization of the LBDs to exert their downstream signaling.
TM-align has advantage over other pairwise sequence alignment tools, as it confers sequence-independent protein structure comparison (Zhang and Skolnick, 2005). For any given two protein structures of unidentified similarity, the program uses heuristic dynamic programming iterations and initially generates optimized amino acid-to-amino acid alignment based on structural similarity. The tool then returns an optimal superposition of the two structures with a TM-score that can be used to scale the similarity. We performed TM-align to find a sequence-independent alignment based on local backbone similarity using heuristic dynamic programming iterations. The structures of LBDs of the serine receptor (Tsr) (PDB ID: 3ATP) of Escherichia coli with the LBD of ER-alpha (PDB ID: 1ERE) and LBDs of the chemoreceptor (BLS) (PDB ID: 6S33) of P. putida with the LBD of ER-alpha (PDB ID: 1ERE) were selected for TM-align.
Structural alignments with TM-align confirmed similar structural folds between LBDs of ER and LBDs of bacterial chemoreceptors (Figures 1D,E). The superposition of two proteins using T-align showed a clear picture on the similarity of Domain Structural architecture between Tsr and ER and between BLS and ER, substantiating our hypothesis on preservation of common fold architecture between the LBDs between bacterial chemoreceptor and human ERα.
Pharmacophore models provide description of optimal supramolecular interactions that typically occur between small-molecule ligands and their respective protein receptor. The pharmacophores of ERs ligands and bacterial chemotaxis receptor ligands were compared to understand if these ligands shared any common pharmacophore features. Two pharmacophore hypotheses were developed in which the first hypothesis involves 17β-estradiol (human estrogen) as the reference ligand and the second hypothesis was focused on coumestrol (Phytoestrogen) as the reference ligand. The hypotheses were developed using the “Develop pharmacophore hypothesis” module of Maestro 11.0 Schrodinger suite. To develop the hypotheses using the reference ligands, hydrogen bond (HB) donors (D), HB acceptors (A), aromatic ring (R), hydrophobic/non-polar groups (H), and positive/negative ionizable groups were selected as the pharmacophore features. Furthermore, the ligands to be screened for matching pharmacophore features were subjected to “ligand-database pharmacophore screening” using the same software.
Interestingly, pharmacophore modeling uncovered a set of common features among the ligands specific for the ER and bacterial chemotaxis receptors (Figures 1F–H). All the bacterial chemotaxis receptor ligands possessed pharmacophore features that are common for 17β-estradiol and coumestrol (Figures 1F–H). The bacterial chemotaxis receptor ligands such as salicylic acid, benzoic acid, and serine exhibited matching pharmacophores with the phytoestrogen “coumestrol,” a known ER agonist.
Our finding that the structural architecture was conserved between the LBD of ERs and bacterial Taxis receptors along with the existence of common pharmacophore feature between the ligands of both receptors prompted us to check for the interactions of the ligands with both receptors. The molecular docking studies were performed using Schrodinger suite 11.0. The three dimensional structures of ER (PDB ID 1ERE), E. coli chemotaxis receptor Tsr (PDB ID 3ATP), and P. putida PcaY_PP LBD (PDB ID 6S33) were downloaded from Protein Data Bank. The downloaded protein structures were processed and energy minimized using the Protein Preparation Wizard. The protein preparation involved assigning bond orders, adding hydrogens, creating disulfide bonds, and creating zero-order bonds to metals. The het states of the proteins were generated using “Epik” at pH 7 ± 2.0, and restrained minimization was performed using OPLS3 forcefield. Furthermore, a receptor grid was generated from each structure by selecting the cocrystal ligand as the centroid of the receptor. The ligand molecules (Figure 2) were downloaded from NCBI-PubChem database and prepared for docking with the LigPrep module. The ligand molecules were structurally optimized at near neutral pH (7 ± 1) and subjected to energy minimization using OPLS3 force field.
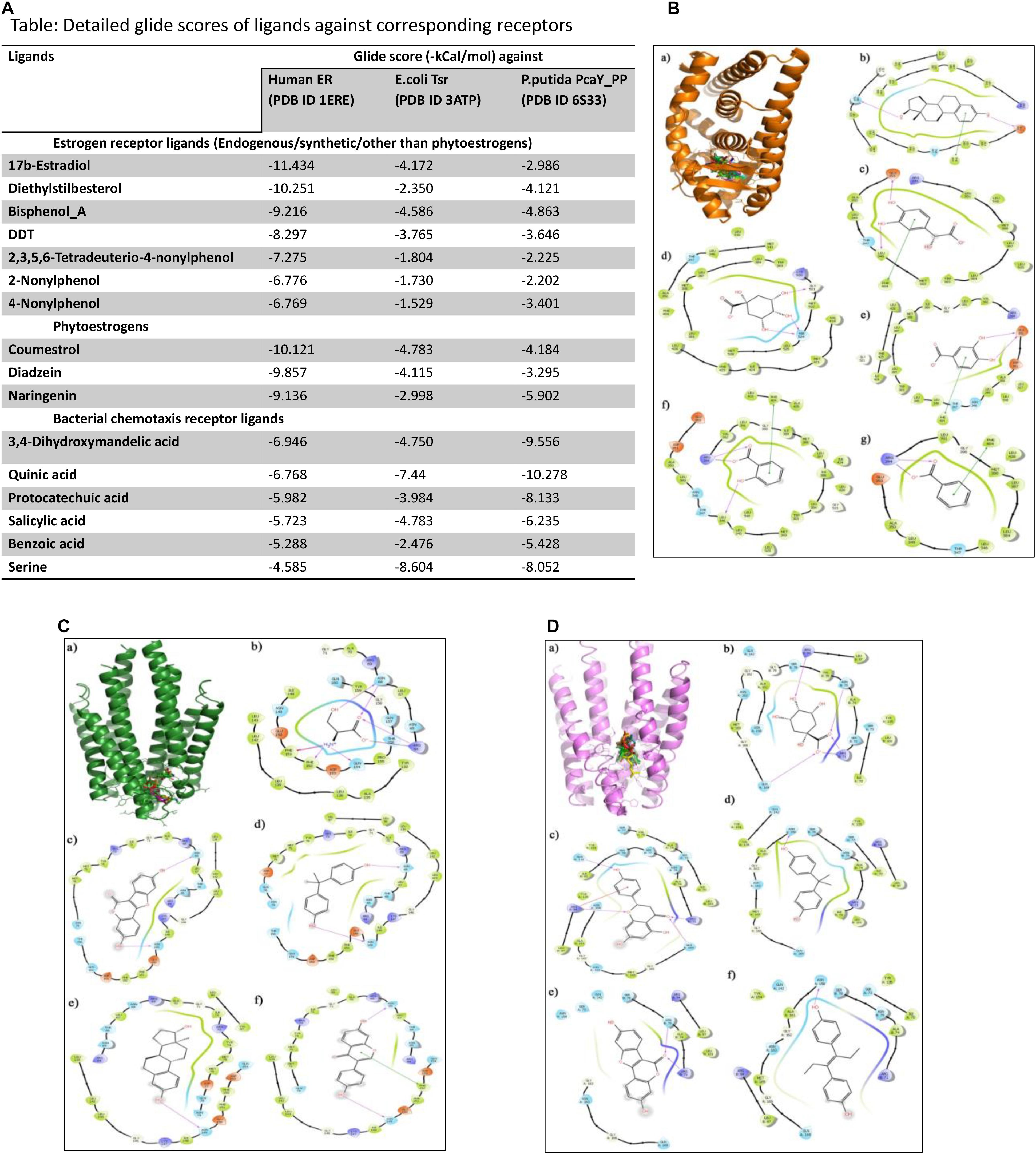
Figure 2. (A) Table representing detailed glide scores of ligands against corresponding receptors. (B) Docking of human ER (PDB ID: 1ERE) with ligands of bacterial chemotaxis receptors. (B.a) Binding of selected ligands in the receptor cavity of human ER (orange, PDB ID 1ERE). (B.b) Ligand interaction diagram of the natural ER ligand 17β-estradiol with binding site residues of ER. (B.c) Ligand interaction diagrams of the top scoring bacterial chemotaxis ligands 3,4-dihydroxy mandelic acid, (B.d) quinic acid, (B.e) protocatechuic acid, (B.f) salicylic acid, and (B.g) benzoic acid. (C) Docking of E. coli chemotaxis receptor Tsr (PDB ID: 3ATP) with ligands of human ER. (C.a) Binding cavity in the AB chain interface of E. coli chemotaxis receptor Tsr (green) occupied with the reference ligand serine and ER ligands. (C.b) The interaction of serine, (C.c) coumestrol, (C.d) bisphenol A, (C.e) estradiol, and (C.f) diadzeine. (D) Docking of bacterial chemotaxis receptor (PDB ID 6S33) with ligands of human ER. (D.a). Ligand binding domain in the AB chain interface of P. putida chemotaxis receptor PcaY_PP (violet, PDB ID 6S33) occupied with the reference ligand “quinic acid” and ER ligands. The interaction of (D.b) quinic acid, (D.c) naringenin, (D.d) bisphenol A, (D.e) coumestrol, and (D.f) diethylstilbestrol.
Furthermore, the prepared ligand sets were docked against the receptor grid of the above-mentioned target proteins. Extra precision (XP) flexible docking was performed using the GLIDE module, and the affinity of the ligands toward the target proteins were ascertained in terms of negative glide score (kcal/mol). Fifty docking poses were collected for each ligand. The binding poses were generated using Pymol software (free license). The glide score is an indirect measure of binding free energy of the ligand–receptor interaction. The more negative the glide score, the stronger the interaction.
For the molecular docking studies against human ER, 17β-estradiol was used as the reference ligand, and other ER ligands were used for validation of the experiment (Figure 2A). Similarly, quinic acid was used as the standard reference ligand for bacterial chemotaxis receptors, and other ligands specific for bacterial receptor was used for cross-validation of the experiment. The docking studies strongly support the findings of structural similarity between human ER and bacterial chemotaxis receptors that were presented in the structure alignment section. When discussing the target specificity of ER ligands against bacterial chemotaxis receptors, all the ligands exhibited thermodynamically favorable interaction (Figure 2B). Phytoestrogens such as coumestrol, diadzeine, and naringenin exhibited stronger interaction with high binding affinity toward bacterial chemotaxis receptors than the estradiol or synthetic ER ligands (Figures 2C,D). On the other hand, the natural ligands of bacterial chemotaxis receptors exhibited strong interaction with human ER (Figures 2A,B). The favorable interactions of all ER ligands toward bacterial receptors irrespective of their binding strength and vice versa were the key findings of this experiment.
We observed a remarkable similarity in the domain structural fold architecture of ERα and bacterial taxis receptors. The ligand molecules of both receptors also shared common pharmacophore features. The natural ligands of bacterial chemotaxis receptors exhibit favorable binding affinity with human ER. This holds true for ER–ligand interaction with bacterial receptors as well. However, phylogenetic analysis proves that these proteins are unrelated. Together, these results suggest that these receptors have evolved independently to respond against certain environmental agents, pointing toward a possibility of convergent molecular evolution. However, during the course of evolution, even when these sequences exhibited high divergence and acquired novel functions, the domain structural fold remained greatly preserved. Domain structural architecture for a particular acquired or specialized functions maybe conserved across species in due course of evolution even when the sequence composition varies. This hypothesis also explains the non-specific binding of the nuclear hormones toward an array of chemically distinct exogenous ligands and at the same time maintaining its high specificity toward the respective hormones.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.”
DL conceived the idea, developed the concept, designed the experiments, performed the experiments, and wrote the manuscript. SH contributed in doing revision experiments in the revised version of the manuscript. Both authors contributed to the article and approved the submitted version.
This research was funded with Seed grant (YU/Seed grant/065-2018), from Yenepoya (Deemed to be University).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge Ranajith Das, Yenepoya Research Centre, for his comments and suggestions on the phylogenetic tree construction and analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.681913/full#supplementary-material
Supplementary Figure 1 | Results for Constraint-based Multiple Alignment Tool. Sequences with no gaps are colored in red or blue. Red indicates highly conserved sequence and blue indicates less conserved sequence.
Supplementary Figure 2 | Results for Constraint-based Multiple Alignment Tool for random sequences. Sequences with no gaps are colored in red or blue. Red indicates highly conserved sequence and blue indicates less conserved sequence.
Supplementary Figure 3 | Graphical representation of the Constraint-based Multiple Alignment for random sequences. Sequences with no gaps are colored in red or blue. Red indicates highly conserved sequence and blue indicates less conserved sequence. Line indicates gaps. (A) Six random sequences. (B) Four selected random sequences.
Supplementary Table 1 | Sequence ID and their Function for the random sequences selected for COBALT analysis.
Baker, M. E. (2019). Steroid receptors and vertebrate evolution. Mol. Cell. Endocrinol. 496:110526. doi: 10.1016/j.mce.2019.110526
Baker, M. E., and Chandsawangbhuwana, C. (2008). Motif analysis of amphioxus, lamprey and invertebrate estrogen receptors: toward a better understanding of estrogen receptor evolution. Biochem. Biophys. Res. Commun. 371, 724–728. doi: 10.1016/j.bbrc.2008.04.152
Baker, M. E., and Lathe, R. (2018). The promiscuous estrogen receptor: evolution of physiological estrogens and response to phytochemicals and endocrine disruptors. J. Steroid Biochem. Mol. Biol. 184, 29–37. doi: 10.1016/j.jsbmb.2018.07.001
Baker, M. E., Nelson, D. R., and Studer, R. A. (2015). Origin of the response to adrenal and sex steroids: roles of promiscuity and co-evolution of enzymes and steroid receptors. J. Steroid Biochem. Mol. Biol. 151, 12–24. doi: 10.1016/j.jsbmb.2014.10.020
Basak, S., Das, M. K., and Duttaroy, A. K. (2020). Plastics derived endocrine-disrupting compounds and their effects on early development. Birth Defects Res. 112, 1308–1325. doi: 10.1002/bdr2.1741
Bhagwat, M., Young, L., and Robison, R. R. (2012). Using BLAT to find sequence similarity in closely related genomes. Curr. Protoc. Bioinformatics Chapter 10:Unit 10.8.
Callard, G. V., Tarrant, A. M., Novillo, A., Yacci, P., Ciaccia, L., Vajda, S., et al. (2011). Evolutionary origins of the estrogen signaling system: insights from amphioxus. J. Steroid Biochem. Mol. Biol. 127, 176–188. doi: 10.1016/j.jsbmb.2011.03.022
Cauley, J. A. (2015). Estrogen and bone health in men and women. Steroids 99(Pt A), 11–15. doi: 10.1016/j.steroids.2014.12.010
Elliston, J. F., and Katzenellenbogen, B. S. (1988). Comparative analysis of estrogen receptors covalently labeled with an estrogen and an antiestrogen in several estrogen target cells as studied by limited proteolysis. J. Steroid Biochem. 29, 559–569. doi: 10.1016/0022-4731(88)90152-5
Forslund, S. K., Kaduk, M., and Sonnhammer, E. L. L. (2019). Evolution of protein domain architectures. Methods Mol. Biol. 1910, 469–504. doi: 10.1007/978-1-4939-9074-0_15
Gore, A. C., Chappell, V. A., Fenton, S. E., Flaws, J. A., Nadal, A., Prins, G. S., et al. (2015). EDC-2: the endocrine society’s second scientific statement on endocrine-disrupting chemicals. Endocr. Rev. 36, E1–E150.
Guerriero, G. (2009). Vertebrate sex steroid receptors: evolution, ligands, and neurodistribution. Ann. N. Y. Acad. Sci. 1163, 154–168. doi: 10.1111/j.1749-6632.2009.04460.x
Itoh, M., Nacher, J. C., Kuma, K., Goto, S., and Kanehisa, M. (2007). Evolutionary history and functional implications of protein domains and their combinations in eukaryotes. Genome Biol. 8:R121.
Kao, Y. C., Higashiyama, T., Sun, X., Okubo, T., Yarborough, C., Choi, I., et al. (2000). Catalytic differences between porcine blastocyst and placental aromatase isozymes. Eur. J. Biochem. 267, 6134–6139. doi: 10.1046/j.1432-1327.2000.01705.x
Klinge, C. M. (2001). Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29, 2905–2919. doi: 10.1093/nar/29.14.2905
Kohler, H. R., Kloas, W., Schirling, M., Lutz, I., Reye, A. L., Langen, J. S., et al. (2007). Sex steroid receptor evolution and signalling in aquatic invertebrates. Ecotoxicology 16, 131–143. doi: 10.1007/s10646-006-0111-3
Koonin, E. V., Wolf, Y. I., and Karev, G. P. (2002). The structure of the protein universe and genome evolution. Nature 420, 218–223. doi: 10.1038/nature01256
Lakshmanan, M. D., and Sadasivan, C. (2014). “Selective estrogen receptor modulators (SERMs) from plants,” in Bioactive Natural Products: Chemistry and Biology, ed. G. Brahmachari (Weinheim: Wiley-VCH), 375–386. doi: 10.1002/9783527684403.ch13
Lakshmanan, M. D., and Shaheer, K. (2020). Endocrine disrupting chemicals may deregulate DNA repair through estrogen receptor mediated seizing of CBP/p300 acetylase. J. Endocrinol. Invest. 43, 1189–1196.
Lecroisey, C., Laudet, V., and Schubert, M. (2012). The cephalochordate amphioxus: a key to reveal the secrets of nuclear receptor evolution. Brief. Funct. Genomics 11, 156–166. doi: 10.1093/bfgp/els008
Moreno-Hagelsieb, G., and Hudy-Yuffa, B. (2014). Estimating overannotation across prokaryotic genomes using BLAST+, UBLAST, LAST and BLAT. BMC Res. Notes 7:651. doi: 10.1186/1756-0500-7-651
Morissette, M., Le Saux, M., D’Astous, M., Jourdain, S., Al Sweidi, S., Morin, N., et al. (2008). Contribution of estrogen receptors alpha and beta to the effects of estradiol in the brain. J. Steroid Biochem. Mol. Biol. 108, 327–338. doi: 10.1016/j.jsbmb.2007.09.011
Pacyga, D. C., Sathyanarayana, S., and Strakovsky, R. S. (2019). Dietary predictors of phthalate and bisphenol exposures in pregnant women. Adv. Nutr. 10, 803–815. doi: 10.1093/advances/nmz029
Papadopoulos, J. S., and Agarwala, R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. doi: 10.1093/bioinformatics/btm076
Park, C., Song, H., Choi, J., Sim, S., Kojima, H., Park, J., et al. (2020). The mixture effects of bisphenol derivatives on estrogen receptor and androgen receptor. Environ. Pollut. 260:114036. doi: 10.1016/j.envpol.2020.114036
Peisajovich, S. G., Garbarino, J. E., Wei, P., and Lim, W. A. (2010). Rapid diversification of cell signaling phenotypes by modular domain recombination. Science 328, 368–372. doi: 10.1126/science.1182376
Razandi, M., Pedram, A., Merchenthaler, I., Greene, G. L., and Levin, E. R. (2004). Plasma membrane estrogen receptors exist and functions as dimers. Mol. Endocrinol. 18, 2854–2865. doi: 10.1210/me.2004-0115
Rosenfeld, C. S., and Cooke, P. S. (2019). Endocrine disruption through membrane estrogen receptors and novel pathways leading to rapid toxicological and epigenetic effects. J. Steroid Biochem. Mol. Biol. 187, 106–117. doi: 10.1016/j.jsbmb.2018.11.007
Sasson, S., and Notides, A. C. (1983). Estriol and estrone interaction with the estrogen receptor. II. Estriol and estrone-induced inhibition of the cooperative binding of [3H]estradiol to the estrogen receptor. J. Biol. Chem. 258, 8118–8122. doi: 10.1016/s0021-9258(20)82036-5
Shafei, A., Ramzy, M. M., Hegazy, A. I., Husseny, A. K., El-Hadary, U. G., Taha, M. M., et al. (2018). The molecular mechanisms of action of the endocrine disrupting chemical bisphenol A in the development of cancer. Gene 647, 235–243. doi: 10.1016/j.gene.2018.01.016
Thornton, J. W. (2001). Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. U.S.A. 98, 5671–5676. doi: 10.1073/pnas.091553298
Vogeler, S., Galloway, T. S., Lyons, B. P., and Bean, T. P. (2014). The nuclear receptor gene family in the pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genomics 15:369. doi: 10.1186/1471-2164-15-369
Ward, N., and Moreno-Hagelsieb, G. (2014). Quickly finding orthologs as reciprocal best hits with BLAT, LAST, and UBLAST: how much do we miss? PLoS One 9:e101850. doi: 10.1371/journal.pone.0101850
Wu, W., and LoVerde, P. T. (2019). Nuclear hormone receptors in parasitic platyhelminths. Mol. Biochem. Parasitol. 233:111218. doi: 10.1016/j.molbiopara.2019.111218
Yang, S. H., Liu, R., Perez, E. J., Wen, Y., Stevens, S. M. Jr., Valencia, T., et al. (2004). Mitochondrial localization of estrogen receptor beta. Proc. Natl. Acad. Sci. U.S.A. 101, 4130–4135.
Yu, L., Tanwar, D. K., Penha, E. D. S., Wolf, Y. I., Koonin, E. V., and Basu, M. K. (2019). Grammar of protein domain architectures. Proc. Natl. Acad. Sci. U.S.A. 116, 3636–3645.
Keywords: estrogen receptor, ligand binding domain, taxis receptors, domain architecture, nuclear hormone receptor
Citation: Lakshmanan Mangalath D and Hassan Mohammed SA (2021) Ligand Binding Domain of Estrogen Receptor Alpha Preserve a Conserved Structural Architecture Similar to Bacterial Taxis Receptors. Front. Ecol. Evol. 9:681913. doi: 10.3389/fevo.2021.681913
Received: 18 March 2021; Accepted: 21 June 2021;
Published: 22 July 2021.
Edited by:
Ernesto Mollo, National Research Council (CNR), ItalyReviewed by:
Taisen Iguchi, Graduate University for Advanced Studies (Sokendai), JapanCopyright © 2021 Lakshmanan Mangalath and Hassan Mohammed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Divya Lakshmanan Mangalath, ZGl2eWFsbWFuZ2FsYXRoQGdtYWlsLmNvbQ==; ZGl2eWFAeWVuZXBveWEuZWR1Lmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.