- 1Department of Integrative Biology, Tampa, FL, United States
- 2H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, United States
- 3Plant Evolutionary Ecology Group, University of Tübingen, Tübingen, Germany
The concept of invasion is useful across a broad range of contexts, spanning from the fine scale landscape of cancer tumors up to the broader landscape of ecosystems. Invasion biology provides extraordinary opportunities for studying the mechanistic basis of contemporary evolution at the molecular level. Although the field of invasion genetics was established in ecology and evolution more than 50 years ago, there is still a limited understanding of how genomic level processes translate into invasive phenotypes across different taxa in response to complex environmental conditions. This is largely because the study of most invasive species is limited by information about complex genome level processes. We lack good reference genomes for most species. Rigorous studies to examine genomic processes are generally too costly. On the contrary, cancer studies are fortified with extensive resources for studying genome level dynamics and the interactions among genetic and non-genetic mechanisms. Extensive analysis of primary tumors and metastatic samples have revealed the importance of several genomic mechanisms including higher mutation rates, specific types of mutations, aneuploidy or whole genome doubling and non-genetic effects. Metastatic sites can be directly compared to primary tumor cell counterparts. At the same time, clonal dynamics shape the genomics and evolution of metastatic cancers. Clonal diversity varies by cancer type, and the tumors’ donor and recipient tissues. Still, the cancer research community has been unable to identify any common events that provide a universal predictor of “metastatic potential” which parallels findings in evolutionary ecology. Instead, invasion in cancer studies depends strongly on context, including order of events and clonal composition. The detailed studies of the behavior of a variety of human cancers promises to inform our understanding of genome level dynamics in the diversity of invasive species and provide novel insights for management.
Introduction
The concept of invasion is provocative across many levels of biology. In the context of biodiversity and ecology, microbial, plant, and animal species invade non-native ecosystems imposing ecological and economic problems and challenges on a global scale (Pimentel et al., 2000; Pyšek and Richardson, 2010; Simberloff et al., 2013; Strong and Ayres, 2013; van Kleunen et al., 2018; Bartz and Kowarik, 2019; Cuthbertab et al., 2021). In cancers, a primary tumor in one tissue can give rise to lineages that disperse to a wide variety of novel environments in other tissues of the host (Turajlic et al., 2018; Capp and Thomas, 2020), imposing a potentially lethal cost on the host (Pienta et al., 2020a; Dujon et al., 2021). Evolutionary processes are inherent to invasions as biological entities are exposed to environmental conditions that may vary from their original environments. The invasive species may experience population bottlenecks, and be subject to genetic drift (Bock et al., 2015; Sottoriva et al., 2017; Zahir et al., 2020). In studies that span the diversity of biological taxa and known human cancers, comparison of source and invasive populations of species and cancer cells has shed light on how rapid evolution can occur (Lee, 2002; Bossdorf et al., 2005; Prentis et al., 2008; Turajlic et al., 2018; Alexandrov et al., 2020; Gerstung et al., 2020).
The field of invasion genetics was established in ecology and evolution more than 50 years ago to understand the genetic mechanisms underlying invasion in natural systems (Baker and Stebbins, 1965). But even by 2002, evolutionary genetics was considered “relatively unexplored” in most invasive species (Lee, 2002). Despite some level of success in the last two decades, we have only a limited understanding of how genomic processes translate into phenotypic diversity across different taxa in response to complex environmental conditions (Bock et al., 2015; van Kleunen et al., 2018; Mounger et al., 2021a). Several studies have concluded that while population bottlenecks and genetic drift typically have a negative effect on invasion success, adaptive responses by invasive species are not limited by reduced genetic variation (Bock et al., 2015; Dlugosch et al., 2015; Estoup et al., 2016; Colautti et al., 2017). However, the study of most invasive species is constrained by a lack of information about complex genome level processes. We lack good reference genomes for most species. Genomic approaches are expensive, and typically studies have focused only on DNA sequences as the mechanism of inheritance (Bock et al., 2015; Richards and Pigliucci, 2020; Mounger et al., 2021a). Further, information about the genetic make-up of source populations is often limited, and what genetic changes occur during the “lag time” (see glossary in Table 1 for bolded terms) between introduction and invasion is virtually unexplored (Bock et al., 2015; van Kleunen et al., 2018).

Table 1. Glossary of relevant terms used to describe biological invasion in invasive species and cancers.
There are several ways to consider how cancer can be seen as a process of invasion. This could include the initiation of cancer as a cell lineage goes from being part of the whole organism program to becoming its own self-defined fitness function and unit of selection (Gatenby and Brown, 2017). Furthermore, once initiated, the expanding population of cancer cells evolves rapidly, and invades adjacent unoccupied tissue. Finally, some cells may metastasize to other regions of the same tissue or to novel organs other than that of the primary tumor. Here, we are interested in parallels between cancer and biological invasions with respect to an extant and expanding population of cancer cells as opposed to cancer initiation itself (Figure 1). In this context, metastases are described quite similarly to biological invasions in the cancer literature. There are also comparable outstanding questions in both fields of inquiry (Table 2). During the process of metastasis, cancer cells leave the primary tumor and establish new tumors either in the same or different tissue (Nowell, 1976). Understanding this process is critical considering that metastasis is linked to the majority of cancer-related deaths (Lambert et al., 2017; Birkbak and McGranahan, 2020). Besides creating lethal burdens or organ failures, the metastatic disease eventually evolves resistance to all known therapies (Pienta et al., 2020a).
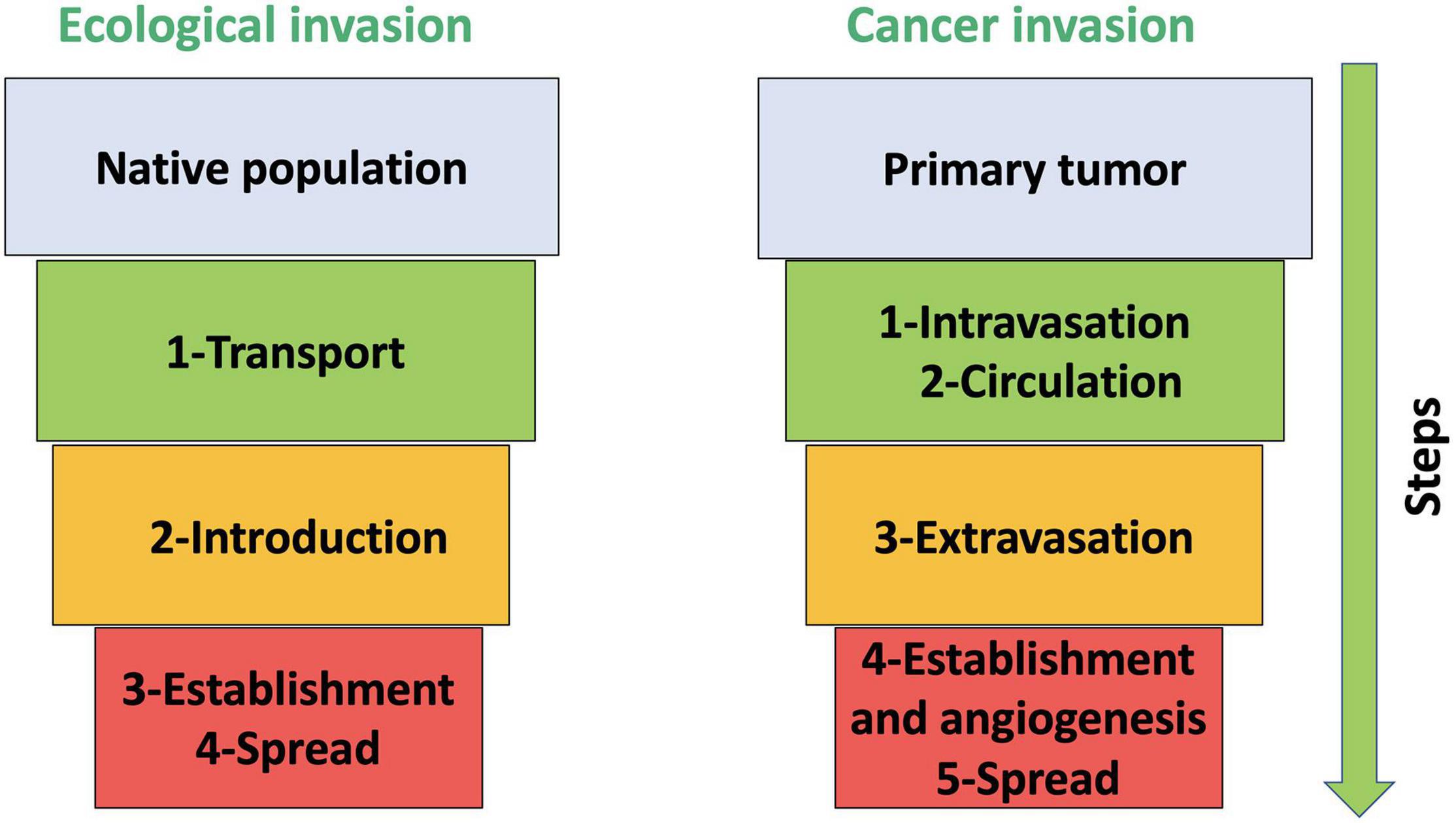
Figure 1. A schematic figure that compares the steps of biological invasion in invasive species (adopted from Theoharides and Dukes, 2007) and cancers (adopted from Gatenby et al., 2009). Native population and primary tumor (in light blue) refer to the source of ecological invasion and cancer invasion, respectively. The first step (green) for invasive species is transport in ecological invasion, which can be natural or by human-assisted dispersal. This step resembles two steps in cancer invasion: (1) intravasation and (2) circulation, where a non-random subset of cancer cells first enters the blood or lymphatic vessels and then travels to a distant organ(s). The following step in the ecological invasion (2) introduction (in orange) closely mimics step three in cancer invasion: extravasation. In invasive species and cancer cells, this step refers to the introduction to the new site. The final steps in the ecological invasion, (3) establishment and (4) spread, resembles in cancer (4) establishment and extension into the adjacent tissue, and (5) colony formation, angiogenesis, proliferation, and spread.
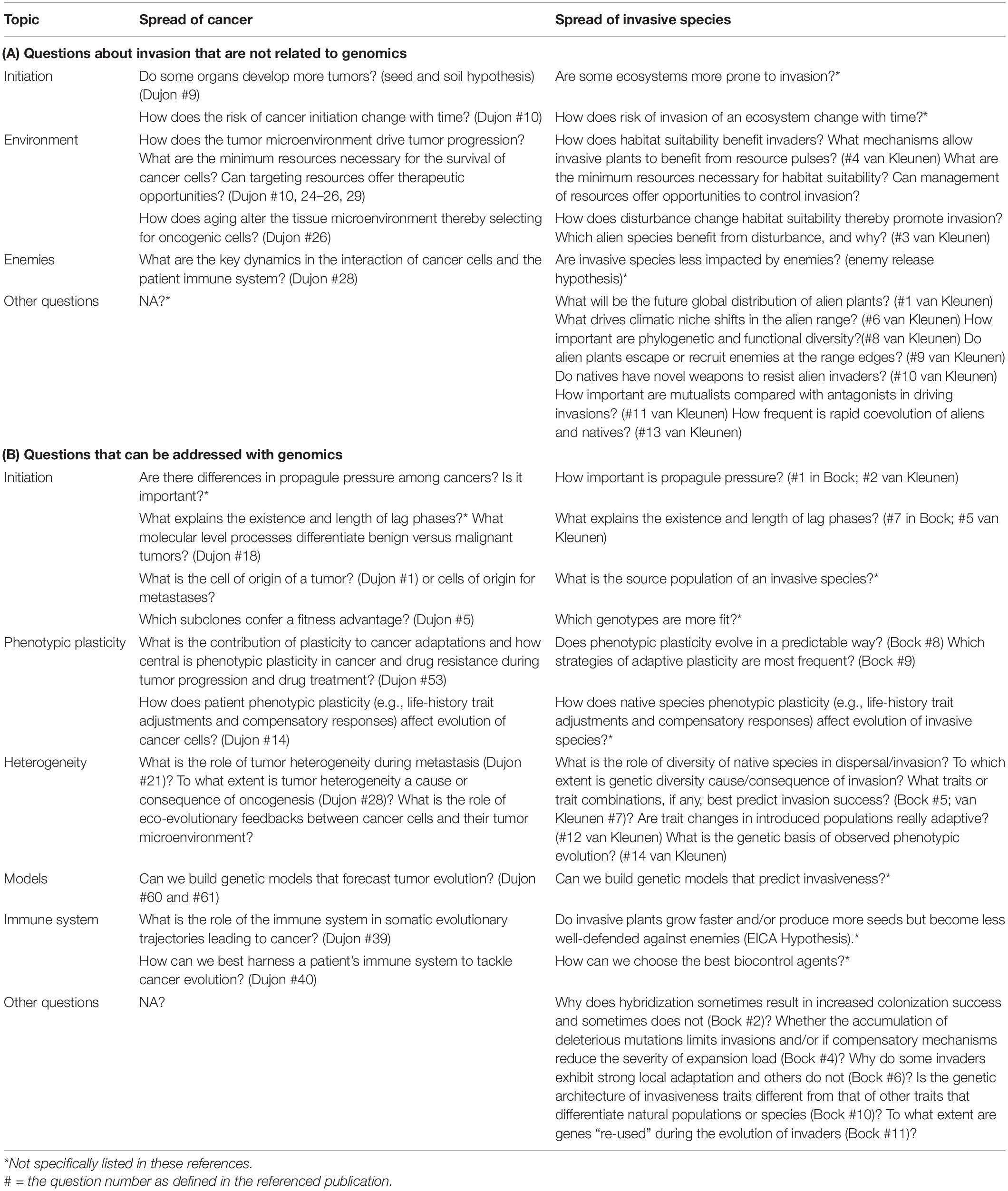
Table 2. Similarities in approaches to study invasions in cancer and ecology [based on selected questions identified in Bock et al. (2015), van Kleunen et al. (2018), and Dujon et al. (2021)].
The process of invasion in diverse cancers provides unique opportunities for studying contemporary evolution since cancer studies are fortified with extraordinary resources for studying genome level dynamics and the interactions among genetic and non-genetic mechanisms (Turajlic et al., 2018; Gerstung et al., 2020). Cancer studies have shown that compared to primary cancer cell counterparts, metastatic samples can have higher mutation rates, multiple types of mutations, and aneuploidy or whole genome doubling with attendant non-genetic effects (Patel and Vanharanta, 2017; Sansregret and Swanton, 2017; Turajlic et al., 2018; Pienta et al., 2020b, 2021; Patel et al., 2021). This suggests that these genome level processes can be important in the invasion process as has also been implicated in the evolutionary ecology of invasive species (Bock et al., 2015; Mounger et al., 2021a). At the same time, bottleneck events shape the genomics and evolution of metastatic cancers (Loeb et al., 2003; Szczurek et al., 2020; Patel et al., 2021), and clonal diversity varies by cancer type and the recipient tissues of the metastases (Turajlic et al., 2018). All the progress in cancer genomics notwithstanding, the cancer research community has been unable to identify any common events that provide a universal predictor of “metastatic potential,” but recent studies from the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium have found that very early events in cancer are associated with predictable sets of so called “drivers” (Gerstung et al., 2020; but see concerns raised in Plutynski, 2021). Moreover, invasion in cancer studies depends strongly on context, including order of events and clonal composition (Birkbak and McGranahan, 2020). These characteristics contribute to disease state, metastatic potential, location of metastasis, and even response to therapy. The detailed studies of the behavior of a variety of human cancers promises to inform our understanding of genome level dynamics in the diversity of invasive species and provide insight for management.
Here, we aim to review the concepts related to the process of invasion and how they can be applied in parallel to the study of a broad variety of invasive taxa as well as a broad variety of metastatic (invasive) cancers. We then briefly review the applications of genomics technologies in these different fields, highlighting similarities, and differences. We emphasize that many findings in cancer research have not yet been replicated or uncovered in invasive species due to various limitations. We also emphasize the opportunities and need for research into questions that have not been answered in either invasive species or cancer. In order to identify these questions, we explore parallels in the recent summaries of 11 (Bock et al., 2015) and 14 (van Kleunen et al., 2018) open questions in the ecology and evolution of invasive species and 84 outstanding questions in cancer research (Dujon et al., 2021; see Table 2 for summary).
The Concept of Invasion in Biology
Definitions for biological invasions vary with the diverse aims of ecological studies (van Kleunen et al., 2015, 2018), but similar language and concepts have also been applied to cancer (Amend et al., 2016; de Groot et al., 2017; Ibrahim-Hashim et al., 2017; Pienta et al., 2020a; Dujon et al., 2021). The idea that cancer progression is an eco-evolutionary process has been discussed for over 50 years (Cairns, 1975; Nowell, 1976). de Groot et al. (2017) recently suggested that “studying cancer as an invasive species provides insight into the necessary phenotypic characteristics of the metastatic ‘seed’ and how those traits are selected for.” They further describe similarities in migration to a “distant secondary habitat” through the use of “established dispersal corridors.” In the case of cancers, these are blood vessels, lymphatics, and nerves (de Groot et al., 2017). Many ecological studies argue that the invasion process depends on the status of communities which may not be at an Evolutionarily Stable Strategy (ESS) for several reasons [e.g., empty niches, or anthropogenic induced changes; (McGill and Brown, 2007; Maron and Marler, 2008; Thuiller et al., 2010; Pintor et al., 2011)], which also applies in cancer (de Groot et al., 2017). Other studies have focused on the mechanisms that allow for individual species to be invasive (Richardson and Pyšek, 2006). This approach could be compared to the study of different successful cancer metastases (e.g., Turajlic et al., 2018), which has even been referred to as a speciation event (Capp and Thomas, 2020; Pienta et al., 2020a; Dujon et al., 2021). Several authors in ecology have also emphasized that the process depends on propagule pressure and time since introduction (Simberloff, 2009; Bock et al., 2015), which is similar to the concept of “billions of failures” in circulating cancer cells that lead to very few successful metastases (de Groot et al., 2017; Tissot et al., 2019). Likewise, many studies of invasive species report that improved fecundity could contribute to their rapid spread and population growth in the invaded range (Bock et al., 2015), which has obvious parallels in metastatic cancers (Lloyd et al., 2016; Vittecoq et al., 2018). In both ecology and cancer, the goal is to understand the causal processes involved in the transport, establishment, spread, and eventual adaptations of invasive species. However, many experimental studies of species invasions are limited in scope, because they provide only a single snapshot of native and invasive populations. When genomics approaches have been used, only a very small representation of genome level mechanisms and dynamics were assayed (Bock et al., 2015; Richards et al., 2017; Paun et al., 2019; Richards and Pigliucci, 2020; Mounger et al., 2021a).
Although limited in genomics prowess, ecological studies across a tremendous diversity of species have developed an array of theoretical frameworks to understand the process of invasion and examine fundamental questions in ecology and evolution (Gurevitch et al., 2011; Bock et al., 2015; van Kleunen et al., 2018). There is intense pressure to understand the process of biological invasions due to their ecological and economic effects (Simberloff et al., 2013; Bellard et al., 2016b; Lodge et al., 2016; Diagne et al., 2020). These effects arise from three ecological characteristics of a successful species invasion: rapid increase in population, local dominance or monoculture, and rapid range expansion (Gurevitch et al., 2011). Research has resulted in many proposed causal explanations of these invasion dynamics, including propagule pressure (Simberloff, 2009; Britton and Gozlan, 2013), biotic resistance (Levine et al., 2004; Nunez-Mir et al., 2017), and evolution of increased competitive ability (Blossey and Notzold, 1995; Rotter and Holeski, 2018). The variety of support (and lack thereof) for these explanations makes it clear that no single factor can explain all biological invasions or contribute to all of them (Catford et al., 2009; Gurevitch et al., 2011; van Kleunen et al., 2018). However, efforts to synthesize these explanations have resulted in valuable conceptual frameworks that relate ecological and evolutionary processes to the steps and barriers of the invasion process.
These conceptual frameworks describe species invasion as a process with several steps, or filters, through which non-native species introduced to a new range must pass before exhibiting the characteristics of invasiveness (Lodge, 1993; Rejmánek, 2000; Richardson et al., 2000; Blackburn et al., 2011; van Kleunen et al., 2018). A species must be (1) transported and (2) introduced to the novel range via natural or human-assisted dispersal. Then, the species needs to (3) survive and become established in the novel range. Finally, an invasive species is (4) able to reproduce and spread (Figure 1; Theoharides and Dukes, 2007; Blackburn et al., 2011; Lloyd et al., 2017; van Kleunen et al., 2018).
The metastatic cascade follows closely the same steps of how invasive species disperse, colonize, and spread with the exception that the first step of “transport” is typically broken down into two parts (green boxes in Figure 1; Chen and Pienta, 2011; Lloyd et al., 2016). Metastasis includes: (1) intravasation (invasion into the bloodstream or lymphatics system), (2) circulation and evasion of the immune system, (3) extravasation (exiting the bloodstream), (4) establishment and angiogenesis, and (5) spread (accessing and establishing vasculature; Paterlini-Brechot and Benali, 2007; Gatenby et al., 2009; Hapach et al., 2019). Intravasation involves cancer cells from an established tumor entering or falling into the bloodstream. Such cells are not a random subset of the tumor’s cancer cells, and location within the tumor will likely matter (Lloyd et al., 2016, 2017; Ibrahim-Hashim et al., 2017). Those at the tumor’s edge will have access to larger, normal blood vessels while those in the interior will experience the disorganized and poorly perfused vasculature induced by angiogenesis. The cancer cell’s characteristics will also matter. For instance, the epithelial to mesenchymal transition in cancer cells generates a more motile phenotype capable of squeezing between cells including those forming the walls of blood vessels (Barriere et al., 2015). So, while intravasation involves accidental dispersers (Joosse et al., 2015), the cancer cells that enter the bloodstream as circulating tumor cells (CTCs) are a weighted average of cancer cell types within the tumor.
Ecological studies offer additional conceptual frameworks that could be explored more thoroughly with genomic approaches in both invasive species and cancers (see Table 2B). Forecasts of invasion risk have been made based on the relationship between phylogenetic distance between the invaders and members of the invaded community (van Kleunen et al., 2015, 2018). A related concept is that species may be “pre-adapted” if the recipient environment is a close match to the native environment and the breadth of the native range may be a good predictor of this possibility (Bossdorf et al., 2005; Bock et al., 2015). These predictions based on phylogenetic distance among species can be argued to support multiple outcomes leading to “Darwin’s naturalization conundrum” (Diez et al., 2008; Thuiller et al., 2010). For example, “Darwin’s naturalization hypothesis,” argues that invaders that are phylogenetically unrelated to local communities should be more successful because they can exploit unfilled ecological niches in native communities (Rejmánek, 1996; Thuiller et al., 2010). On the other hand, closely related species might share similar pre-adaptations to local environmental conditions or have similar biotic or abiotic requirements (Thuiller et al., 2010). Regardless, phylogeny does not always reflect trait differences or niche differentiation, and often the importance of these different components of invasion potential are unknown (Thuiller et al., 2010; van Kleunen et al., 2015, 2018). Recent application of genomic techniques to trace the “life history” of cancers have found parallels in a diversity of cancer types (Nik-Zainal et al., 2012; Turajlic et al., 2018; Gerstung et al., 2020), but this type of evolutionary ecology framework has not yet been fully explored in the study of cancers.
Many studies in both ecology and cancer have demonstrated that in addition to the importance of pre- or post-invasion sources of genetic differences, phenotypic plasticity can be important. A rich literature in evolutionary ecology has explored several different scenarios for how invasive species may benefit from phenotypic plasticity, some of which were outlined by Richards et al. (2006). Plasticity may allow for invasives to be a (1) “Jack-of-all trades” with positive fitness across many habitat types; (2) “Master-of-some” with highly positive fitness in the most favorable habitats and conditions, or (3) “Jack-and-Master” that combines both attributes. However, increased plasticity does not always translate into positive fitness outcomes (Levis and Pfennig, 2016; Matesanz et al., 2017; He et al., 2021). In fact, several meta-analyses (Van Buskirk and Steiner, 2009; Davidson et al., 2011; Palacio-López and Gianoli, 2011; Arnold et al., 2019) have not supported the hypothesis that plasticity contributes to the success of invasive species. In either case, the mechanisms underlying this plasticity will be mediated by genetic and non-genetic molecular level processes (Richards et al., 2010; Nicotra et al., 2010; Herman and Sultan, 2011, 2016; Banta and Richards, 2018). While the fine molecular details of plastic responses have not yet been assessed in any species (Richards et al., 2017; Bock et al., 2018; Laitinen and Nikoloski, 2019; Richards and Pigliucci, 2020; Sommer, 2020; Mounger et al., 2021a), the potential to do so is currently greatest in cancers.
Another important concept that has been explored in evolutionary ecology is the role of propagule pressure, or the rate of arrival of non-native individuals. Propagule pressure can be important not only to the initial step of introduction, but it can influence survival by overwhelming stochastic processes, and increasing survival, reproduction, and competitive dominance. High propagule pressure can enhance genetic diversity permitting the invasive species to persist and thrive in a new environment (Simberloff, 2009; Rius et al., 2015b). Invasive species often exhibit a lag time, where extended periods of time pass between initial establishment and later development of invasive characteristics (Simberloff, 2009; Aikio et al., 2010; Bock et al., 2015; van Kleunen et al., 2018). In ecology, the length of this lag time remains unpredictable and the mechanisms at work remain unknown, but rapid evolution has occurred on the scale of <50 years, and evidence shows that local adaptation occurs in invasive species (Bock et al., 2015). Similarly, disseminated cancer cells (DTC) that arrive at and survive in a novel organ may exhibit long lag periods before expanding from micrometastases into clinically detectable metastatic tumors (Birkbak and McGranahan, 2020).
Recent evidence from the Pan Cancer Atlas has traced the life-history of thousands of cancers and found the lag time from so called “driver events” to detection can be on the order of years to decades (Nik-Zainal et al., 2012; Alexandrov et al., 2013b; Gerstung et al., 2020). Extensive research has detailed several components of the metastatic process. The quantity of CTCs with potential to seed new tumors depends on survival while passing through the bloodstream (Box 1: Figure A). This involves tolerating potentially destructive shearing forces from moving swiftly through vasculature, as well as avoiding detection and mortality from the immune system in the blood (Lloyd et al., 2016). Additional characteristics such as immune evasion and the ability to be compressed may enhance the chances of a cancer cell surviving in the blood. Authors have suggested that several cancer cells traveling together as a raft (Aceto et al., 2014), or unusually large cancer cells such as those in a polyaneuploid state (polyaneuploid cancer cells, PACCs) may be able to circulate longer and more successfully (Pienta et al., 2021). However, regardless of characteristics, the half-life of a CTC remains unknown, yet is vitally important. Blood circulates throughout the human body on average every 45 seconds, and the number of times a CTC can circulate influences their number as well as their likelihood of entering other tissues. CTCs can reach 1–10 per ml of blood (Yu et al., 2011; Alix-Panabières and Pantel, 2021), and the number of CTCs in the blood correlates with progression free survival (5 or more CTCs per 7.5 ml of blood represents a poor prognosis for breast or prostate cancer patients; Rack et al., 2014; Pantel and Alix-Panabières, 2019).
BOX 1.
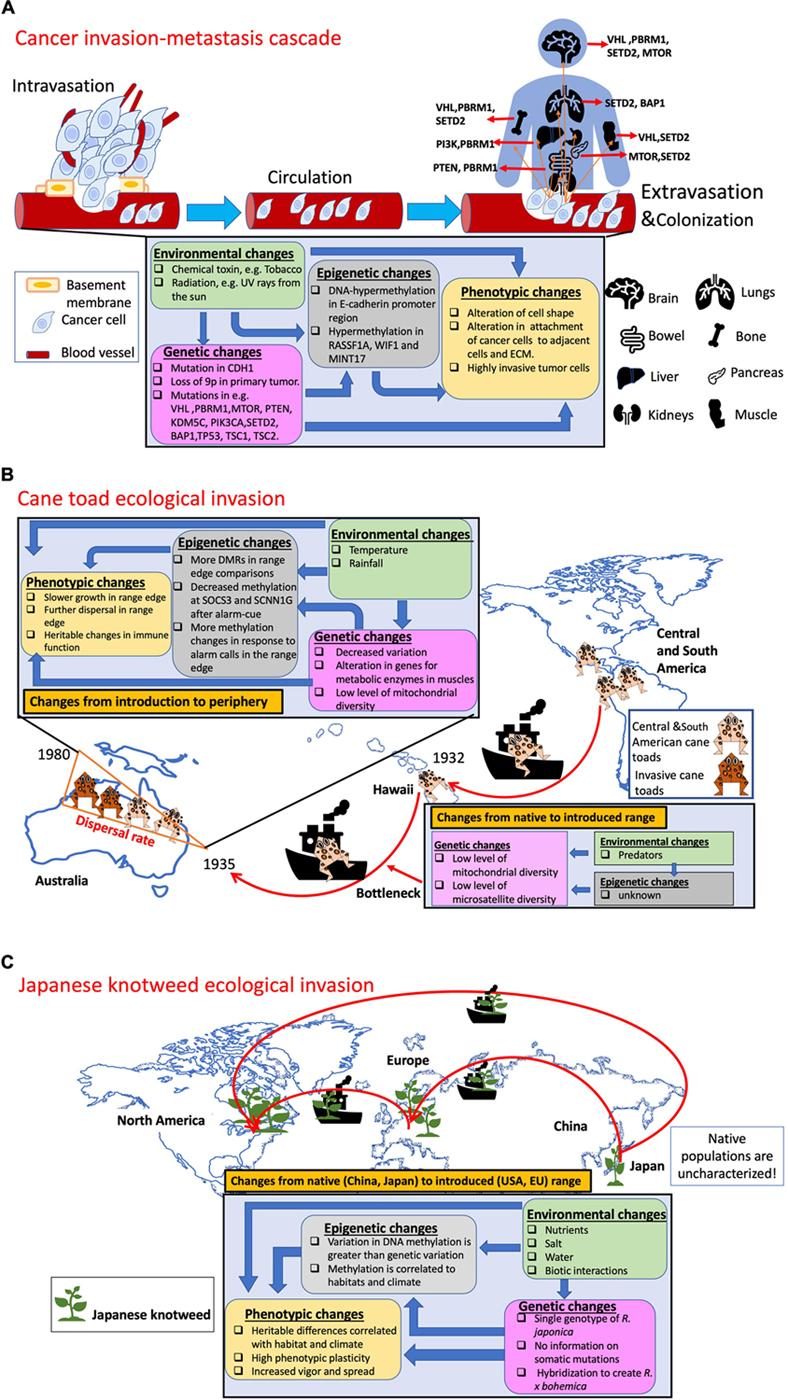
The invasion processes in cancer and ecology share commonalities but there are also important differences. The cancer metastasis and successful invasion process consists of a sequence of steps that are like the steps of ecological invasion (see Figure 1). Here, we highlight examples of genetic and epigenetic mechanisms involved in invasion in cancer and two different ecological systems. (A) Cancer invasion. Cancer cells invade the basement membrane and enter the blood vessels (intravasation), circulate in the blood and reach a distal organ (e.g., the liver). Genetic as well as epigenetic alterations govern cancer metastasis. One of the most studied alterations is the reduction of E-cadherin protein expression, which is responsible for the adhesions between cells. Mutations have been found in the CDH1 gene that codes for E-cadherin, as well as DNA hypermethylation. Specific site of metastasis has been associated with genomic driver mutations that occurred in the primary tumor: examples from Turajlic et al. (2018) are depicted where metastases from primary kidney tumors metastasized to lung, bone, liver, brain, pancreas, and muscle (adrenal, parotid, thyroid glands, skin, and soft tissue not shown). (B) Cane toad invasion. A total of 101 cane toads were introduced to Australia in 1935 from Central and North America. The invasion traveled from the northeast to southern and northwestern regions of Australia resulting in heritable differences in physiological, morphological, and behavioral traits. After already surviving a bottleneck from the initial invasion, genetic diversity declined between the initial site of establishment and the leading edge of the invasion (Selechnik et al., 2019). One potentially important alteration is that genes involved in metabolism and immune function were upregulated (Rollins et al., 2015). Experimental manipulations also support heritable epigenetic changes at the SCNN1G gene could be involved in the response to predators (Sarma et al., 2020, 2021). (C) Japanese knotweed invasion. This plant was first introduced from Japan to Europe in the 1840s and then to North America sometime before 1873 (Del Tredici, 2017). The invasive knotweed Reynoutria japonica has been reported to be a single widespread genotype, however, the molecular characterization of this species has been limited and cannot exclude potentially important single nucleotide polymorphisms (Mounger et al., 2021a). On the other hand, epigenetic markers have been associated with differences in habitat and climate (Richards et al., 2012; Zhang et al., 2016).
In addition to these characteristics that might contribute to the success of CTCs, only a small fraction of CTCs are able to exit a blood vessel and enter a novel tissue or novel location in the same tissue type. Actin dynamics within a cancer cell can permit pseudopodia facilitating motility and the ability to move through cell junctions (diapedesis) (Castro-Giner and Aceto, 2020). The establishment of a successful metastasis from one or several of such DTCs represents yet another hurdle. Several characteristics of the DTC may favor success albeit it is still a very small probability. Actin mechanics can be critical for degrading and constructing extra-cellular matrices (Kumar and Weaver, 2009). The establishing DTC must overcome additional threats from what can be a tissue specific immune response. To avoid failure, the DTC or emerging micrometastasis must overcome Allee effects (Johnson et al., 2019), and access and establish vasculature (angiogenesis) (Amend et al., 2016; Lloyd et al., 2016, 2017).
Despite these hurdles that are difficult to trace, the metastatic cascade is a simpler and perhaps more accessible microcosm for the evolutionary ecology of invasive species. The patient is the entirety of the system, i.e., the entire planet for the cancer (Pienta et al., 2020a). All the living cells within the patient, including the cancer cells and normal cells are relatively similar in size. The pathway to invasion in cancer is straightforward, occurring through the blood or lymphatics. The cancer cell will face roughly the same supply of resources, types of resources, and threats from the immune system. However, the different organs and tissues of the body differ structurally, functionally, and metabolically in ways that influence their susceptibility to metastases (Schild et al., 2018). Like in invasion ecology, the heritable characteristics of the metastasizing cancer cells matter (de Groot et al., 2017; Pienta et al., 2020a). They can carry adaptations for dispersal and pre-adaptations for surviving the rigors of dispersal and avoiding the immune system (Hanahan and Weinberg, 2011; de Groot et al., 2017). Similarly, ecological species invasions can be facilitated by dispersal adaptations such as burrs on seeds that stick to the fur of animals (or artificial surfaces) or dispersal events prompted by over-crowding. Also, there is often an association between the habitat characteristics of the donor and recipient locations (van Kleunen et al., 2018). This aligns with Paget’s (1889) seed-and-soil hypothesis for the relationship between the donor tumor’s tissue type and the recipient tissue where metastases are likely to occur. Some authors have even suggested that exosomes released by cancer cells in the primary tumor “prepare the soil” for the metastatic “seeds” (Rodrigues et al., 2019). A reasonable comparison in ecological invasions could be human habitat disturbances and constructs that favor the invasion of numerous species (van Kleunen et al., 2018).
Upon successful establishment, the cancer cells can evolve further adaptations to successfully exploit their novel environment. From the perspective of the emerging field of invasion genomics, principles from invasion ecology in natural systems offer a conceptual framework for metastases. In return, the extensive, replicated opportunities for measuring DNA sequence, epigenetic markers, gene expression, and heritable characteristics of cancer cells means that oncology offers unique opportunities for evaluating and testing the genomics of species invasions (e.g., Box 1: Figure A; Turajlic et al., 2018).
Genomics of Invasion
In both species level ecology and cancer, one major goal is to identify how genomic level processes translate into the ability of the organism to respond to complex environmental conditions. The genomic mechanisms that underlie invasions are particularly intriguing because of classic evolutionary theory that predicts how the ability to respond to environmental challenges rests on heritable phenotypic variation which is presumably genetically based. The fact that invasions by definition have been assumed to result from just a few individuals creates a so-called “genetic paradox” for understanding the success of invaders and their adaptations to new habitats (Allendorf and Lundquist, 2003; Estoup et al., 2016). This has been a mystery in both ecology and cancer research with several hypotheses about how the invaders overcome the negative consequences of limited genetic variation (Baker and Stebbins, 1965; Nowell, 1976; Vogelstein et al., 2013; Estoup et al., 2016).
In this context, both invasive species studies and cancer studies have largely focused on the importance of sequence based differences or mutations (Alexandrov et al., 2013b, 2020; Vogelstein et al., 2013; Bock et al., 2015). However, evolutionary responses to challenging environmental conditions rely on heritable phenotypes, regardless of the underlying mechanisms of inheritance (Jablonka and Lamb, 1998, 1999; West-Eberhard, 2005; Banta and Richards, 2018: Bonduriansky and Day, 2018). We now have evidence that the structural and functional dynamics of genomes along with a variety of epigenetic and other non-genetic effects can contribute to heritable variation and thus to adaptation and cancer metastasis (Feinberg et al., 2006; Jablonka and Raz, 2009; Johannes et al., 2009; Feinberg and Irizarry, 2010; Timp and Feinberg, 2013; Richards et al., 2017; Bonduriansky and Day, 2018; Kooke et al., 2019; Richards and Pigliucci, 2020).
Discoveries and Limitations of Genomic Studies of Diverse Invasive Species
Genetic markers have been used to identify the genetic make-up of invasive populations, and understand how genome level processes contribute to invasion of diverse natural ecosystems (Bock et al., 2015; Colautti and Lau, 2015; Dlugosch et al., 2015; Rius et al., 2015b; Mounger et al., 2021a). Studies in the native and introduced ranges have reported that many invasive populations undergo only modest reductions in genetic variation due to multiple introductions (Stepien et al., 2005; Dlugosch and Parker, 2008a; Snyder and Stepien, 2017; Flucher et al., 2021), hybridization (Fitzpatrick et al., 2009; Scascitelli et al., 2010; van Riemsdijk et al., 2018, 2020; Quilodrán et al., 2020), or Allee effects that result from reverse density dependence or cooperation (Kramer and Sarnelle, 2008; Aikio et al., 2010; Rius et al., 2015b). For example, sequences of mitochondrial DNA revealed multiple invasion sources for both dreissenid mussels and goby species of fish, and that this diversity was correlated to rapid spread and colonization success in a variety of habitats (Stepien et al., 2005). Further, invasive populations of zebra mussels, quagga mussels, round gobies, tubenose gobies, and Eurasian ruffe (another fish species) that have established in the Great Lakes had as much or greater genetic diversity as native populations (Stepien et al., 2005; Snyder and Stepien, 2017). Recently, reduced representation sequencing (Narum et al., 2013) has provided much more power to inform studies of invasion by demonstrating, for instance, the absence of strong population structure which could indicate repeated human-assisted dispersal across the invaded range such as in the pavement ant (Tetramorium immigrans) (Zhang et al., 2019).
On the other hand, lower diversity in invasive populations may reflect that there was a higher diversity of founding genotypes initially, and selection in the novel habitat filtered out unfit individuals (Dlugosch and Parker, 2008b; Vandepitte et al., 2014). Other studies argue that the process of genetic bottlenecks can purge deleterious alleles, reveal beneficial cryptic variation or create new beneficial interactions among genomic elements (Colautti and Lau, 2015; Dlugosch et al., 2015; Stapley et al., 2015; Estoup et al., 2016; van Kleunen et al., 2018). The invasive brown rat (Rattus norvegicus) is a globally successful invader, but recent studies in China discovered that local populations had experienced severe bottlenecks and then rapidly differentiated since establishment in the 1970s, including new alleles associated with lipid metabolism and immunity genes (Chen et al., 2021).
Despite such bottlenecks, loss of diversity measured by molecular markers does not necessarily reflect loss of quantitative trait variation (Dlugosch and Parker, 2008b; Estoup et al., 2016). A series of studies revealed that rapid phenotypic evolution facilitated the invasion of the widespread red macroalga Agarophyton vermiculophyllum despite the fact that the species experienced a severe genetic bottleneck and increased through clonal spreading (Krueger-Hadfield et al., 2016; Sotka et al., 2018; Flanagan et al., 2021). The European starling (Sturnus vulgaris) in North America was founded by only ∼180 individuals, but local populations seem to have evolved rapidly and now show the signature of only a moderate population bottleneck (Hofmeister et al., 2021). European starlings in North America show higher levels of genetic diversity than invasive populations in Australia or South Africa (Bodt et al., 2020). This is somewhat surprising since the invasion in Australia occurred across multiple introduction sites and the pattern of rapid differentiation appears to be explained by distance instead of response to environment (Rollins et al., 2016; Stuart et al., 2020).
Most genomics studies of invasive species lack the resources required for understanding the single base pair resolution of how specific genome level differences might translate into function and the species’ success. However, transcriptomic studies can now be conducted in almost any system, providing a measure of variation in gene expression at the level of mRNA, which contributes to the formation of proteins, cellular phenotypes and ultimately the organism’s phenotype (Alvarez et al., 2015). A few studies have identified candidate genes that were differentially expressed in invasive populations (Hodgins et al., 2013; Bock et al., 2015). Studies of populations of the Argentine ant (Linepithema humile) discovered consistent differences among invaded compared to native populations in expression of genes related to biogenic amines (which modulate behavioral traits like foraging and aggression) and immune function. Unfortunately, they could not associate these expression differences with behavioral differences in their study. Furthermore, interpretation of the functional relevance of some of the differentially expressed genes was limited by the need for better annotation (Felden et al., 2019).
Hodgins et al. (2015) compared transcriptomes across 35 species of plants in the Asteraceae, including six major invasive species. They found no support for the idea that there was consistent selection on genes that contributed to invasiveness [but see opposite results with a similar approach in the invasive green crab, Carcinus maenas (Tepolt and Palumbi, 2020)]. In a rare comparison of sequence variation and expression variation, a study of two independent invasions of the Pacific Oyster (Crassostrea gigas) into the North Sea found little overlap between differentially expressed genes and outlier loci. This suggested that differential gene expression did not necessarily correlate with changes in allele frequencies (Wegner et al., 2020). However, as is common in ecological genomics, this study suffered from limited annotation of the transcriptome and limited coverage of the genome (using reduced representation RADseq). In general, identifying underlying molecular level “drivers” is so far limited to very few studies across widely diverse taxa (Bock et al., 2015).
In addition to using transcriptomics approaches, several authors have argued that epigenetic mechanisms could be particularly important for invasive species (Ardura et al., 2017; Hawes et al., 2018; Marin et al., 2020; Mounger et al., 2021a; but see Eckert et al., 2021). The case has been made particularly for those invasive species that are clonal or have low genetic diversity. Epigenetic mechanisms could provide a non-genetic source of heritable variation (Verhoeven and Preite, 2014; Douhovnikoff and Dodd, 2015; Liebl et al., 2015; Rollins et al., 2015; Richards et al., 2017; Sarma et al., 2020, 2021; Mounger et al., 2021b). In the last 10 years, there has been an explosion of studies of natural populations and ecological experiments that provide some level of information about how epigenetic mechanisms (mainly DNA methylation) may contribute to organismal responses to environmental challenges (Jablonka, 2017; Richards et al., 2017; Richards and Pigliucci, 2020; Stajic and Jansen, 2021). But so far these ideas and approaches have rarely been applied to the understanding of invasive species (Hawes et al., 2018; Marin et al., 2020) and they are almost universally limited in scope (Paun et al., 2019; Richards and Pigliucci, 2020; Mounger et al., 2021a).
The recent work in cane toads (Rhinella marina) provides one excellent example of integrating genomic, transcriptomic, and epigenomic approaches to understand the process of invasion (Rollins et al., 2015; Selechnik et al., 2019; Sarma et al., 2020, 2021). A substantial bottleneck occurred during the introduction of only 101 cane toads into Queensland, Australia in the 1930s (Shine, 2018; Box 1: Figure B). At present, the toads at the edge of their range are less genetically diverse than those at the initial site of introduction, but show a wide variety of heritable differences in physiological, morphological, and behavioral traits (Phillips et al., 2008; Rollins et al., 2015; Selechnik et al., 2019). In addition, this team of researchers suspected that toads at the invasion front encounter more abundant predators than at the original site of introduction, which may contribute to higher mortality rates and select for larger toxin glands. By radio-tracking toads, researchers were able to see that some toads moved long distances almost every night in the first 2 years, and inheritance of rapid dispersal became spatially sorted (Shine et al., 2011). Rollins et al. (2015) argued that this resulted in assortative mating among individuals at the front of the invasion between fast-moving individuals which reinforced the evolution of this trait (Shine et al., 2011). By measuring genome wide levels of gene expression with RNAseq, they also showed that metabolic enzymes were upregulated at the invasion front and that many of the most highly differentially expressed genes involved energy production, immune function, and parasite resistance (e.g., PSME4 and RASGEF1B) (Rollins et al., 2015).
Remarkably, these researchers took advantage of this invasion gradient to experimentally examine the potential role of epigenetic mechanisms as well (Sarma et al., 2020, 2021). They exposed tadpoles to alarm cues and found elevated cortisone levels as well as decreased methylation at the suppressor of cytokine signaling 3 (SOC3) and the Sodium Channel Epithelial 1 Subunit Gamma gene (SCNN1G) genes. They further tested the idea that DNA methylation drives this pattern by manipulating methylation levels with the drug zebularine but could not associate changes in DNA methylation to the promoter region of the glucocorticoid (GC) receptor gene (NR3C1). However, they did find differences in single cytosine methylation in the promoter region of SOCS3, which may be involved in predator avoidance behavior (Sarma et al., 2020). This team then examined the inheritance of changes in methylation by running a breeding experiment. They showed that some shifts in DNA methylation in response to alarm cues were inherited by the next generation. In particular, they showed demethylation within SCNN1G, which regulates sodium in epithelial cells and may help to maintain the epidermis (Sarma et al., 2021). While this series of studies has not dissected every molecular detail of the invasion response, it is unparalleled in their exploration of various levels of response. This research team demonstrated the wide variety of questions that can be addressed with genomics approaches.
In the last few decades, we have gained increased data about the genetic structure of a variety of invasive species and how genetic variation is distributed on the landscape (Bock et al., 2015), but most of our information is limited to markers distributed across the genome and very few studies can evaluate the whole genome. Many studies have only described patterns of diversity and are limited in their ability to address underlying adaptive processes (Rius et al., 2015a,b). Rius et al. (2015a) report that the application of next generation sequencing (NGS) to invasive species started in 2008 and by 2015 resources had been developed for many species. NGS had even identified candidate genes like the detoxification gene cytochrome P450 and other stress related genes. However, how the translation of DNA sequence to phenotypes unfolds through the invasion process requires much more fine scale dissection of the entire genome and other molecular level processes over time (see e.g., Bock et al., 2018; reviewed in Pigliucci, 2010; Keller, 2014; Müller, 2017; Bonduriansky and Day, 2018; Richards and Pigliucci, 2020). The increasing application of sequencing and other “omics” technologies within appropriately designed experiments promises to provide more powerful insight into the molecular mechanisms underlying responses to selection and adaptation (Alvarez et al., 2015; Rius et al., 2015a,b; Mounger et al., 2021a), but it is yet unclear how far we can go with this approach and in how many unique species. In this context, cancer genomics studies provide some important insights.
On the other hand, the diversity represented among invasive species provides information about the potential for novel function, particularly in organisms with extreme phenotypes (Castoe et al., 2013; Bock et al., 2018). A study of invasive Burmese pythons before and after a major freeze event in Florida in 2010 found evidence for directional selection in genomic regions enriched for genes associated with thermosensation, behavior, and physiology. Several of these genes were linked to regenerative organ growth, which modulates feeding and fasting responses in pythons (Card et al., 2018). In addition, ecological experimental approaches can be quite creative in testing for the importance of some of these processes by combining genomics approaches with, for example, the creation of synthetic hybrids (Rosenthal et al., 2002; Rieseberg et al., 2003; Lai et al., 2006; Whitney et al., 2015; Nieto Feliner et al., 2020; Irimia et al., 2021), and synthetic or recent polyploids (Yoo et al., 2014; Nieto Feliner et al., 2020; Paape et al., 2020; Shan et al., 2020).
Are Clonal Plant Species a Particularly Useful Comparison to Cancer?
Some of the world’s most successful invasive plants are thought to spread by clonal reproduction (e.g., Japanese knotweed Box 1: Figure C), which at first brush might seem like a good analogy for invasion in cancer considering that cancers arise within a host who has the same genotype. Mounger et al. (2021a) recently reported that clonal plants are potentially over-represented among invasive plant species. This is surprising because asexual reproduction is predicted to result in slower rates of evolution. But clonality may also be adaptive under the circumstances faced by invasive species and serve as useful subjects to investigate how a single genome can respond to a myriad of environments (e.g., Geng et al., 2007; Verhoeven et al., 2010; Gao et al., 2010; Richards et al., 2012; Shi et al., 2018; Chen et al., 2020). A recent study in the perennial sunflower, Helianthus tuberosus, provided a powerful combination of approaches to demonstrate that there had been selection for the ability to increase clonality (Bock et al., 2018). The authors compared populations of ancestral lineages with invasive lineages and found support for increased ability to respond to well watered conditions by producing more clonal propagules. As such, this is one of the first studies to demonstrate the process of genetic accommodation during invasion (sensu West-Eberhard, 2005; see also Sultan, 2015; Levis and Pfennig, 2016). In this study, the researchers were able to link the genomic mechanisms of hybrid vigor and two specific QTL to the increased ability to respond to water content in the invasive habitat. We know of no study where the genomic level processes that underlie the success of entirely clonal lineages have been fully explored. Genetic variation that arises from somatic mutations in natural clonal lineages, albeit low, cannot be excluded since several studies have reported that high rates of somatic mutation may allow asexual species to maintain abundant genetic variation and adapt to changing environmental conditions (reviewed in Schoen and Schultz, 2019; see also discussions in Chen et al., 2020; Robertson et al., 2020).
Japanese knotweed (Reynoutria japonica aka Fallopia japonica) is one of the most well−known cases of an invasive clonal plant. A single octoploid clone of R. japonica has spread aggressively through a broad range of habitats in temperate Europe and North America (Box 1: Figure C; Beerling et al., 1994; Bailey and Conolly, 2000; Grimsby et al., 2007; Gerber et al., 2008; Bailey et al., 2009; Richards et al., 2012; Zhang et al., 2016). Unfortunately, not much is known about the levels of diversity in the native populations of China and Japan. In the United States, replicates of the same clone of R. japonica collected from different habitats had different DNA methylation patterns even after they were grown in a common garden in New York (Richards et al., 2012). Another study across central Europe showed that individuals from different populations of this same R. japonica clone harbored significant epigenetic and phenotypic variation which was associated with climate (Zhang et al., 2016). However, both studies were based on anonymous molecular markers (AFLP and methylation sensitive AFLP) which only survey a small portion of the genome. They cannot detect single DNA base changes even in the surveyed fragments (Schrey et al., 2013; Paun et al., 2019). A recent survey of the same samples in the United States populations suggested that within this R. japonica clone there were most likely some single nucleotide polymorphisms (Robertson et al., 2020; see also VanWallendael et al., 2020), but whether any of these polymorphisms are functional remains to be evaluated.
As in the knotweed studies, the low genomic resolution of studies of most organisms precludes pinpointing the actual accrual of sequence and methylation polymorphisms and therefore isolating the importance of genetic and epigenetic variation (Richards et al., 2017; Paun et al., 2019; Naciri and Linder, 2020). The whole genome sequencing studies that have been done (almost entirely in model species) reveal the importance of genomic redundancy, largely resulting from multiple episodes of whole genome duplication (polyploidy) followed by reduction processes (Doyle et al., 2008; Wendel et al., 2016), which play a major role in diversification and adaptation in plants and some animals (Van de Peer et al., 2017). Whole-genome studies in the model plant Arabidopsis thaliana (as in cancers) show that novel epigenetic variation can be dramatically shaped by de novo sequence mutation. For example, studies have found that single nucleotide polymorphisms can change the methylome by modifying a methyl transferase or a nucleotide context where methyltransferases act (Becker et al., 2011; Timp and Feinberg, 2013; Dubin et al., 2015; Feinberg et al., 2016; Sasaki et al., 2019).
The relevance of somatic mutations has been clearly documented in cancers (Nik-Zainal et al., 2012; Alexandrov et al., 2013b, 2020; Gerstung et al., 2020), and in cancer metastases (e.g., Turajlic et al., 2018). Mutation could contribute to the rapid generation of genetic or epigenetic variation in natural clonal lineages of plants, and in organisms more generally (Vonholdt et al., 2010; Exposito-Alonso et al., 2018; Hawkins et al., 2019; Schoen and Schultz, 2019; Yoder and Tiley, 2021). But it may be unclear how comparable mutation in cancers is to natural populations of plants and animals. Studies across plant species have reported a range of mutation rates from e.g., 7 × 10−9 per base per haploid genome per generation in Arabidopsis thaliana lines (Ossowski et al., 2010; Exposito-Alonso et al., 2018) and peach (Xie et al., 2016) to 4 × 10−8 in long lived poplar and oak species (Schmid-Siegert et al., 2017; Hofmeister et al., 2020). Mutation rates across a diversity of animal species ranged from 3.6 × 10−9 in bumblebee to 1.5 × 10−8 in chimpanzee (Yoder and Tiley, 2021). This is roughly comparable to the average generational mutation rate for single-base substitutions in humans 1–1.5 × 10−8 (Rahbari et al., 2016). Therefore, studies in human cancer could provide insight into the mechanisms that underlie rapid organismal response more generally.
Unlike in most species, cancer studies have unsurpassed power to document how mutation rate depends on location and nucleotide context in the genome as well as tissue types (Alexandrov et al., 2013a, 2020; Rahbari et al., 2016). Studies have also identified ‘mutational signatures’ that reflect age, mutagen exposures and DNA repair mechanisms (Alexandrov et al., 2013a, 2020; Rahbari et al., 2016). Although not as finely detailed in scale, one important study took advantage of herbarium samples to demonstrate that this type of de novo mutation occurred during colonization and expansion in the United States of a single lineage of A. thaliana (Exposito-Alonso et al., 2018). Arabidopsis thaliana is an annual selfing plant, which is therefore almost entirely homozygous across its diploid genome. Exposito-Alonso et al. (2018) discovered de novo mutations that were associated with genes related to adaptive traits that may have been selected during the establishment and expansion of this species.
Epimutations occur much more frequently than genetic mutations, they do not occur randomly across the genome, and they occur more often in genic regions than in transposable elements (reviewed in Richards et al., 2017). A recent whole genome survey of Populus trichocarpa showed epimutation rates that were very similar to A. thaliana on a per generation basis in the range of 10−3 to 10−4 (Hofmeister et al., 2020). Another study in maize showed that the forward epimutation rate was about 10 times larger than the backward epimutation rate, and two orders of magnitude larger than that of DNA mutation rate (Xu et al., 2020). In humans, the epimutation rate appears to be lower than in A. thaliana but was also estimated to be over two orders of magnitude greater than the germline genetic mutation rate (Carja et al., 2017). Unlike the extensive focus in cancer, how these mutation and epimutation rates translate into function has not been explored in invasive organisms, or in clonal plants more generally. Unfortunately, even with the most accurate sequencing platforms and assembly methods currently available, the technological challenges of accurately detecting mutation and epimutation indicate that this type of information is not yet within our reach for most non-model species (Yoder and Tiley, 2021).
What We Know About Genomics of Cancer Metastasis
Even before the era of cancer genomics, extensive studies had revealed that “genetic instability” was a hallmark of cancer (Coffey, 1998; Duesberg et al., 1998). Mutations rates are higher in cells with genetic instability (Weisenberger et al., 2006; Hanahan and Weinberg, 2011; Loeb, 2011). Such is the case for cancer cells. The increased genetic variation that results from this mutation can result in phenotypic variation that has different fitness benefits for cells based on their ability to divide, migrate, and survive environmental conditions (Amend et al., 2016; Lloyd et al., 2016; Ibrahim-Hashim et al., 2017; Somarelli, 2021). The microenvironment of the tumor selects on this variation in phenotype and determines which cell lines will die, proliferate, or metastasize. Cancer cells that metastasize start in the selective environment of the primary tumor, then travel through lymphatic tissue or blood vessels to a distant organ (Figure 1 and Box 1: Figure A). During this journey, metastatic cancer cells must evade immune cells and ultimately compete with healthy cells for resources when they reach a distal organ (Lloyd et al., 2016; Amend et al., 2016), all of which is mediated by genomic processes.
Cancer genomics was launched in 2006, but the sample information was limited in many cases (Ledford, 2010; Nature, 2020). Early analyses of cancer genomes showed that they carried thousands to tens of thousands of somatic mutations along with aneuploidies and genome doubling (∼30% of cases) (Stratton, 2011; Williams et al., 2019). While the vast majority of mutations were thought to have no biological function, they have been informative to understand the evolutionary history of cancers. Researchers have been able to develop algorithms that predict evolutionary fates of cell lineages based on population genetics concepts (Nik-Zainal et al., 2012; Williams et al., 2019). Nik-Zainal et al. (2012) identified a collection of somatic mutations shared by all cancer cells within a given breast cancer sample and used this concept to identify discrete clones and subclones. In order to do so, they examined the details of one patient (sequenced to 188× depth) and found that in the aneuploid tumor there were 70,690 somatic mutations genome-wide, many of which were in fewer than 5% of the reads for a given location in the genome. Their model predicted that 26,762 of these mutations (∼38%; including in genes TP53, PIK3CA, GATA3, MLL3, SMAD4, and NCOR1) along with trisomy 1q and several other rearrangements were found in every tumor cell indicating that some ancestral cell carried all of these somatic mutations. From this point, they could reconstruct the emergence of additional subclones as well [i.e., their subclone labeled cluster C represented 65% of the tumor cells, cluster B represented 18% of tumor cells, and cluster A accounted for 14% of tumor cells (Nik-Zainal et al., 2012)]. They concluded that large-scale chromosomal changes did not start to occur until after at least 15–20% of the point mutations had already occurred. Hence, instability at the chromosome level was not usually the earliest source of mutation in this breast cancer. Across the 20 breast cancer samples, they found a dominant subclonal lineage represented 50–95% of tumor cells, but a considerable proportion of somatic genetic variation was in only a fraction of tumor cells.
More recent studies of the PCAWG Consortium of the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas have provided further evidence of the power of cancer genomics to contribute to our understanding of shared and unique evolutionary genomic mechanisms (Alexandrov et al., 2020; Gerstung et al., 2020; ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium, 2020). Gerstung et al. (2020) used whole genome sequencing in cancer samples from 2,658 unique donors across 38 cancer types. They took advantage of the same approaches as Nik-Zainal et al. (2012) using sequence data to measure the number of copies of different alleles. They used this information to define categories of early and late clonal variants, the order in which variants arise, and the most recent common ancestor (MRCA) of all cancer cells in a tumor sample. Using phylogenetic reconstruction of mutations as a clock they mapped mutation timing estimates onto approximate real time to reconstruct the evolutionary trajectories of cancer to even before the point of diagnosis. They presented timing and typical sequences of mutations as well as how drivers and mutational signatures varied across each cancer type.
Among the many interesting findings, Gerstung et al. (2020) identified an increase in diversity of mutated driver genes at later stages of tumor development, and 50% of all early clonal driver mutations occured in just nine genes (although see concerns raised by Plutynski, 2021). In many cases, the earliest events included TP53 mutations, as well as losses of chromosome 17 and most other highly recurrent cancer genes, such as KRAS, TERT, and CDKN2A. Whole genome duplication events occurred after tumors had accumulated several driver mutations, and many chromosomal gains and losses typically occurred later. This confirmed a long-held prediction in colorectal cancer called the “APC-KRAS-TP53 progression model” of Fearon and Vogelstein (1990). This finding concurs with several previous studies that had reported that very early events in cancer evolution occur in a few common drivers, and a more diverse array of drivers is involved in late tumor development (Jamal-Hanjani et al., 2017; Hu et al., 2019). Overall, the study showed that the spectrum of mutations changed throughout tumor evolution in 40% of samples. There were some common trends among tumors as they evolved, but they all followed diverse paths. Other studies demonstrated large differences in the underlying mutation rate among individual tumors and tumor types and emphasized that only a handful of mutations occur at appreciable frequencies across all cancer types; for example, only mutations in TP53 and PIK3CA occurred at a frequency of greater than 10% across cancer types in one study (Kandoth et al., 2013). Further, Alexandrov et al. (2013b, 2020) recently evaluated 84,729,690 somatic mutations from 4,645 whole-genomes and 19,184 exomes across most cancer types and made associations of signatures to exogenous or endogenous exposures, as well as to defective DNA-maintenance processes. They identified positive correlations between the age at cancer diagnosis and the number of mutations attributable to a signature, and that the underlying mutational process was active throughout the entire evolution of the lineage from normal cells.
The application of genomics to the study of metastases is of particular interest for understanding the genomics of invasions. The genetic and non-genetic alterations underlying cancer metastasis vary depending on the type of tumor as well as the stage of metastasis (Nguyen and Massagué, 2007). Genome-wide analysis of gene-expression in tumors has been applied to hematological cancers (Golub et al., 1999; Alizadeh et al., 2000), followed by solid tumors (Ramaswamy et al., 2003) to find signatures for predicting metastasis. Genes involved in initiation of metastasis promote invasion of the basement membrane and entry into the circulatory system. For example, loss of CASp8 (caspase 8) activation can protect cancer cells from apoptosis during invasion (Stupack et al., 2006). Epigenetic modifications can also initiate the metastatic state, particularly since mutations in epigenetic machinery can reshape the epigenome (Feinberg et al., 2006; Timp and Feinberg, 2013). Chromatin regulators are often mutated in cancer [e.g., mutations in the SWI/SNF complex occur in over 20% of all cancers (Kadoch and Crabtree, 2015)]. Further, a study in patients of glioblastoma multiforme evaluating SNP-genotypes, methylation, copy number variants, and gene expression data found that whole genome DNA methylation was the most informative molecular level predictor of survival (Bernal Rubio et al., 2018).
Recent studies indicated that even when specific genetic mutations instigated the invasion process, completion of the process depended only on non-genetic changes, specifically epigenetic changes that complement the genetic mutations (Lambert et al., 2017). Metastatic progression depends on the expression of genes that have specific functions as the cancer cell first becomes a CTC, then a DTC and finally an expanding micrometastasis. The expression of such genes during the metastatic cascade may provide quite different functions than they do for cancer cells of the primary tumor. Such genes include EREG that encodes COX2 and MMP1 (Gupta et al., 2007; Kuramochi et al., 2012; Qu et al., 2016). These remodel the vasculature in sites of metastasis and simultaneously, facilitate intravasation and angiogenesis at the primary tumor site (Gupta et al., 2007). Genes that are not involved in the primary tumor but facilitate metastasis at distal sites are classified as “metastasis-virulent”. An example is CXCR4, a cytokine receptor that mediates cancer survival in a distant organ where its ligand CXCR12 is abundant in tissue microenvironments like bone marrow (Müller et al., 2001; Kang et al., 2003).
Metastatic samples can have higher mutation rates, specific types of mutations, aneuploidy or whole genome doubling and non-genetic effects compared to primary cancer cell counterparts (Alexandrov et al., 2013a,b, 2020; Martincorena et al., 2015). Chromosome instability also creates aneuploidy and promotes tumor evolution (Ben-David and Amon, 2020) and can result in dissimilarities between metastatic and primary tumors. However, a study of 118 biopsies of colorectal cancer with metastases to the liver or brain showed little divergence between the primary tumor and metastasis, and that “driver genes” were acquired early in the process of tumor progression. In fact, cells that “disseminated early” were more likely to seed metastases when the primary tumor was still clinically undetectable (Hu et al., 2019). Similar findings were reported in a study of two breast cancer patients: primary tumors and associated metastases were similar in gene expression and somatic mutation patterns (Hoadley et al., 2016). On the other hand, clones seeding metastasis in breast cancer in another study disseminated late from primary tumors and continued to acquire mutations. Further, distant metastases acquired driver mutations that were not seen in the primary tumor, including a wider repertoire of cancer genes than early drivers, e.g., inactivation of SWI-SNF and JAK2-STAT3 pathways (Yates et al., 2017).
For the purposes of understanding specific genomic response to invaded habitats, a particularly compelling study was recently completed on clear-cell renal cell carcinoma (ccRCC) tumors (Turajlic et al., 2018). Across 463 primary and 169 matched metastatic regions from 38 patients, Turajlic et al. (2018) found the number of driver events was lower in metastases (mean = 9), compared to primary tumors, and that metastases were significantly more homogeneous than primary tumors: 456 driver events were shared between primary tumors and metastases, 230 were only found in primary tumors, and 39 driver events were only found in metastases. They further determined behavior of clonal lineages within the primary tumor by dividing 253 clones in the 38 patients into: (1) clones that were not represented in the metastatic samples (n = 130 clones, defined as subclonal in the primary tumor and absent in metastasis), (2) clones that were maintained (n = 38 clones, defined as the MRCA clones, clonal in both primary tumor and metastasis), and (3) clones that were selected (n = 85 clones, defined as subclonal in the primary and clonal in metastasis; or absent in the primary and present in metastasis). Comparing selected versus unselected mutations, they found hallmark genomic alterations in ccRCC metastasis but also report the fascinating finding of specific and shared mutations associated with metastases across 18 different invaded tissues (Turajlic et al., 2018; Box 1: Figure A). This type of detailed information about genomic modifications associated with the invasion of “habitat types” is unprecedented in ecological studies and provides information not only about current status of populations but evolutionary history and therefore potential prevention. While each patient exhibited some unique features, this study also demonstrates the remarkable degree of parallel and convergent evolution in both the primary and metastatic tumors of different patients of this cancer type.
The Illusive Universal Predictor of Invasive Potential-Management Issues
There are often intensive management responses to both invasive species and cancer, although the stakes differ in notable ways. Untreated, metastatic cancer is inevitably deadly (Wells et al., 2013), while the impacts of invasive species generally include economic costs ranging from minor to immense (Bradshaw et al., 2016; Diagne et al., 2021), and disruption of ecological communities, in some cases including native species extinction (Bellard et al., 2016a). Another striking difference between cancer and invasive species is general agreement that cancer is bad whereas some or even all stakeholders may see an invasive species neutrally or positively (e.g., burros in the Grand Canyon, stocking non-native game species, and non-native biological control agents). Regardless, several of the similarities and differences in management approaches could be addressed by a better understanding of the genomic mechanisms in context.
It is widely argued that the best way to reduce ecological and economic costs of invasive species is to interfere with the transport and establishment steps of invasion (Keller et al., 2007; Bailey et al., 2011). A primary reason is that once species have become invasive in a new habitat, eradication is rarely feasible, except on some islands (Parkes and Panetta, 2009; Moon et al., 2015). Damage or costs related to damage reduction become recurring (Liebhold et al., 2016). Cancer treatment is similar with regards to the importance of cancer prevention and early detection. Ecologists use species traits or ecological niche models to identify species of concern and locations of high risk, whereas doctors can use individual traits, such as environmental exposures, lifestyle, age, or genetic predispositions to guide cancer surveillance (Dobson, 2013; Katzke et al., 2015; Bernal Rubio et al., 2018; Hu et al., 2021). But this often applies to preventing cancer initiation in the first place. With respect to preventing metastases, there is growing interest in therapies that target CTCs (Ortiz-Otero et al., 2020) as well as bolstering normal tissues and the immune system to prevent the establishment of DTCs (Risson et al., 2020).
Surveillance of invasive species aimed at preventing introduction of new propagules often lapses after a species is established. While understandable, this may be unwise. Further import of new individuals into an already invaded habitat likely provides additional heritable variation (e.g., Kolbe et al., 2004), potentially allowing for faster adaptation to new habitats or increased success in the invaded range. In cancer treatment, the focus on removing as many of the cancer cells as possible has the effect of reducing the variation present, potentially pushing cancer back toward earlier, less invasive stages, which seems to be effective for about 50% of the cases (Pienta et al., 2020a). However, this approach also selects for those few cells that are able to resist therapy and may require a different management strategy (Ibrahim-Hashim et al., 2017; Gatenby and Brown, 2020; Pressley et al., 2021). This line of reasoning suggests increased focus on preventing introductions may still be appropriate and cost-effective for existing invaders. Similarly identifying the molecular features associated with progression to invasions is a major objective in cancer research (Srivastava et al., 2018). The potential for this type of biomarker approach was recently highlighted in a study of colorectal cancer with metastases to the liver or brain. The early mutations in “driver genes” were associated with seeding metastases. These key mutations were in an independent cohort of 2,751 colorectal cancers (Hu et al., 2019) and could be the targets of therapy, enhancing a personalized medicine approach. Analysis of multi-omics data and development of new statistics approaches that can integrate these data will be an imperative to identify the relative contributions of different molecular level mechanisms that underlie cancer progression, invasion and response to environmental challenges more generally (Bernal Rubio et al., 2018; Hofmeister et al., 2020; Nam et al., 2021; Teschendorff and Feinberg, 2021; Yoder and Tiley, 2021).
Once invasion has taken place the management regime diverges between cancer treatment and invasive species management. Invasive species management tends to focus on slowing the spread (Sharov et al., 2002) or protecting specific habitats (e.g., Short et al., 1992), or even tolerance (Schlaepfer et al., 2011) and resignation (Regulations.gov, 2020). Cancer treatment initially tends to take a much more aggressive treatment approach, often combining several methods with the goal of eradication and long term suppression of tumors (Blagosklonny, 2004; Yap et al., 2013; Gatenby and Brown, 2020). These treatments are often expensive and have intense side effects, but genomic analysis of tumors may provide directly relevant information for selecting treatments most likely to be effective (Bozic et al., 2012).
When detected and treated early enough, most cancers are curable. Cure generally involves surgical resection of the tumor and/or radiation therapy. So long as the resected or irradiated tumor contains all of the cancer cells, then knowledge of the genomics becomes less relevant. However, identifying the whereabouts of all of the cancer cells involves some guesswork. Hence, neoadjuvant drug therapies prior to surgery help ensure a contained population of cancer cells, and adjuvant drug therapies after surgery aim to eliminate undetectable surviving fragments of the primary tumor or micrometastases elsewhere in the body. Both neoadjuvant and adjuvant therapies can be improved based on genetic and molecular markers of the cancer cells’ state and heterogeneity (Dressman et al., 2006; Duran et al., 2020; Oshi et al., 2020). Upon detection or at the start of a management program, complete elimination of an invasive species often succeeds or fails based on the ability to find and cull all individuals. For both cancer and invasive species, if the management regime does not kill them all, the cancer and pest species will evolve resistance (Pressley et al., 2021). In cancer, understanding the genomics of resistance promises insight to why initial therapies do not or cannot cure the patient.
Despite the limitations for understanding the translation of genome level processes into traits in most species, genomics is becoming part of the invasive species management toolbox because these approaches can provide accurate diagnostics of invasive species (Cristescu, 2015; Hamelin and Roe, 2020). We are unaware of any invasive species management regimes that have been truly shaped by genomic knowledge (see Stewart et al., 2009), but molecular analyses can provide taxonomic clarification, evidence of hybridization and cryptic species, population structure and origin of invasions for management purposes (Gaskin et al., 2011; Chown et al., 2015). Studies on knotweed for instance, have shown that closely related taxa had dramatically different responses to herbicide application and that the hybrid Reynoutria x. bohemica is particularly resilient (Bímová et al., 2001). The genomic mechanisms underlying these differences have not yet been examined. Genomics approaches are now being used to identify appropriate biocontrol agents (Sun et al., 2020; van Steenderen et al., 2021; Harms et al., 2021). For example, specific cochineal insects in the genus Dactylopius (Hemiptera: Dactylopiidae) are effective biocontrol agents of some invasive Opuntia cactus species. But the different Dactylopius species are so morphologically similar that numerous misidentifications have contributed to failed attempts at biological control. van Steenderen et al. (2021) report that nucleotide sequencing of three gene regions (12S rRNA, 18S rRNA, and COI) and two inter-simple sequence repeats (ISSR) were effective in identifying the target species Dactylopius opuntiae and Dactylopius tomentosus and even different lineages within D. tomentosus. A study of invasive Ambrosia artemisiifolia found that the genotype of the leaf beetle Ophraella communa determined potential success as a biocontrol agent, but the specific genomic mechanisms of that association were not investigated (Sun et al., 2020).
Conclusion
In the last 50 years, foundational concepts in ecological and evolutionary genetics have been applied to both the study of invasive species and the study of cancers. We have discussed many similarities in the application of genomics to cancer and invasive species (summarized in Table 2 with reference to questions outlined recently by Dujon et al., 2021 for cancer and Bock et al., 2015 and van Kleunen et al., 2018 for ecological invasions). Cancer cells and invasive species alter their environment and can cause extinctions of other cells or organisms because they alter the composition of their habitat or deplete resources. Ecological studies have a stronger history of describing this process across a diversity of species and habitat interactions, but the molecular mechanisms underlying this could be informed by genomics, as we have seen in cancer studies (e.g., Box 1: Figure A; Turajlic et al., 2018).
There are some important outstanding questions in invasive species ecology that do not easily find parallels in cancer studies. For example: van Kleunen et al. (2018) highlighted the importance of questions like “What will be the future global distribution of alien plants?,” “How important are mutualists compared with antagonists in driving invasions?” and “How frequent is rapid coevolution of aliens and natives?” which have some parallels in cancer but do not have obvious analogs in Dujon et al. (2021; Table 2A). These include, for instance, the cooperative interactions that might occur between DTCs, or how clusters of CTCs may increase likelihoods of metastases (Fabisiewicz and Grzybowska, 2017). This aligns with invasions in nature where success generally increases with the number of individuals introduced simultaneously to the novel location (Barney and Whitlow, 2008).
Bock et al. (2015) highlight important questions that can be specifically addressed with genomics approaches in invasive species studies like “why hybridization sometimes results in increased colonization success and sometimes does not,” “whether the accumulation of deleterious mutations limits invasions and/or if compensatory mechanisms reduce the severity of expansion load,” and “the extent of gene re-use during the evolution of invaders” which are not particularly relevant in cancers. However, both van Kleunen et al. (2018) and Bock et al. (2015) emphasize the importance of the outstanding question: “What explains the existence and length of lag phases?,” which is also unknown in cancers (e.g., “What molecular level processes differentiate benign versus malignant tumors?”; Table 2B) and has great potential for therapeutic targets. Other major common themes highlighted in the table address questions about the molecular level mechanisms involved in initiation and progression, the importance of plasticity at various stages, the importance of habitat suitability and our ability to use genomics in predictive modeling. Genomics approaches promise to inform our understanding about these outstanding questions in both cancer and ecological studies.
We have argued that the detailed studies of the behavior of a variety of human cancers can inform our understanding of genome level dynamics in the diversity of invasive species and provide predictive frameworks for management. However, despite the tremendous efforts of the last 15 years, the transitions from normal to cancerous conditions or from primary tumor to metastasis (Turajlic et al., 2018; Hu et al., 2019) are still not well understood. Even normal cells contain many mutations that accrue with age, and some genic regions have a higher mutation rate than others (Martincorena and Campbell, 2015; Martincorena et al., 2015, 2018; Gao et al., 2019; Goldmann et al., 2019; Zahir et al., 2020). It is particularly challenging that there is no discrete boundary between normal ageing processes and cancer evolution (Lee-Six et al., 2018, 2019; Moore et al., 2020). In addition, changes in methylation more strongly predicted survival in patients with glioblastoma multiforme than genetic polymorphisms and methylation was strongly associated with age (Bernal Rubio et al., 2018). Further, the microenvironment of the pre-tumor is also aging and this could contribute to tumorigenesis and subsequent progression (Zahir et al., 2020).
As with the pre-invasive stage of invasive species in ecology (Vandepitte et al., 2014), premalignancy in solid tumors has not been well studied, partly because of the challenge of early detection (Gerstung et al., 2020; Zahir et al., 2020). Several researchers have concluded that a comprehensive understanding of the progression of cancer requires understanding not only at the molecular level but also at the phenotypic and ecological level such as physiological, structural, and environmental information that occurs spatially and temporally (Ibrahim-Hashim et al., 2017, 2021; Zahir et al., 2020; Nam et al., 2021). While this is also potentially the holy grail for understanding the progression of invasive species, in the case of cancer, the stakes are higher and very immediate to the patient. Hence, Zahir et al. (2020) have reviewed how sophisticated techniques have been developed for multiplexing genomic, proteomic and transcriptomic analysis in situ, while preserving the spatial relationships between cells within their native tissue architecture and immune context. These include finely dissected spatial transcriptomic profiling and single-molecule fluorescence in situ hybridization (smFISH), where transcripts are directly labeled in tissue sections to image and visualize their subcellular locations. Cancer studies can also include manipulations to verify functional relationships thanks to the Cancer Cell Line Encyclopedia (CCLE), application of CRISPR loss-of-function methods, cell-viability data for thousands of compounds which define a “cancer dependency map,” and single cell sequencing technologies (Tsherniak et al., 2017; Williams et al., 2019; Teschendorff and Feinberg, 2021).
While this outstanding level of resources may never be available in any invasive species, the enormous amount of data that is already accumulated and will continue to accumulate could be combined with the nuanced, and crafty (out of necessity) approaches of evolutionary ecologists to provide a better understanding of the translation of genotype to phenotype. Doing so is likely to improve risk analysis and management interventions.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the Federal Ministry of Education and Research (BMBF; MOPGA Project ID 306055 to CR), NIH/NCI 1U54CA193489-01A1, Cancer as a Complex Adaptive System (to JB and AI-H), and NIH/NCI U54 Supplement, The tumor-host evolutionary arms race (to JB, AI-H, and CR).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past collaboration with one of the authors, JB.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the editors for the opportunity to pursue these ideas. We acknowledge support by the Open Access Publishing Fund of University of Tübingen.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.681100/full#supplementary-material
References
Aceto, N., Bardia, A., Miyamoto, D. T., Donaldson, M. C., Wittner, B. S., and Spencer, J. A. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. doi: 10.1016/j.cell.2014.07.013
Aikio, S., Duncan, R. P., and Hulme, P. E. (2010). Lag-phases in alien plant invasions: separating the facts from the artefacts. Oikos 119, 370–378. doi: 10.1111/j.1600-0706.2009.17963.x
Alexandrov, L. B., Australian Pancreatic, Cancer Genome, Initiative, Nik-Zainal, S., Wedge, D. C., et al. (2013a). Signatures of mutational processes in human cancer. Nature 500, 415–421.
Alexandrov, L. B., Kim, J., Haradhvala, N. J., Huang, M. N., Tian, Ng, A. W., et al. (2020). The repertoire of mutational signatures in human cancer. Nature 578, 94–101.
Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Campbell, P. J., and Stratton, M. R. (2013b). Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259. doi: 10.1016/j.celrep.2012.12.008
Alix-Panabières, C., and Pantel, K. (2021). Liquid biopsy: from discovery to clinical application. Cancer Discov. 11, 858–873. doi: 10.1158/2159-8290.cd-20-1311
Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., and Rosenwald, A. (2000). Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511.
Allendorf, F. W., and Lundquist, L. L. (2003). Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. J. Soc. Conserv. Biol. 17, 24–30. doi: 10.1046/j.1523-1739.2003.02365.x
Alvarez, M., Schrey, A. W., and Richards, C. L. (2015). Ten years of transcriptomics in wild populations: what have we learned about their ecology and evolution? Mol. Ecol. 24, 710–725. doi: 10.1111/mec.13055
Amend, S. R., Roy, S., Brown, J. S., and Pienta, K. J. (2016). Ecological paradigms to understand the dynamics of metastasis. Cancer Lett. 380, 237–242. doi: 10.1016/j.canlet.2015.10.005
Ardura, A., Zaiko, A., Morán, P., Planes, S., and Garcia-Vazquez, E. (2017). Epigenetic signatures of invasive status in populations of marine invertebrates. Sci. Rep. 7:42193.
Arnold, P. A., Nicotra, A. B., and Kruuk, L. E. B. (2019). Sparse evidence for selection on phenotypic plasticity in response to temperature. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374:20180185. doi: 10.1098/rstb.2018.0185
Bailey, J. P., and Conolly, A. P. (2000). Prize-winners to pariahs - a history of Japanese knotweed s.l. (Polygonaceae) in the British Isles. Watsonia 23, 93–110.
Bailey, J. P., Bímová, K., and Mandák, B. (2009). Asexual spread versus sexual reproduction and evolution in Japanese Knotweed s.l. sets the stage for the “Battle of the Clones. Biol. Invasions 11, 1189–1203. doi: 10.1007/s10530-008-9381-4
Bailey, S. A., Deneau, M. G., Jean, L., Wiley, C. J., Leung, B., and MacIsaac, H. J. (2011). Evaluating efficacy of an environmental policy to prevent biological invasions. Environ. Sci. Technol. 45, 2554–2561. doi: 10.1021/es102655j
Baker, H. G., and Stebbins, G. L. (1965). The Genetics of Colonizing Species. New York: Academic Press.
Banta, J. A., and Richards, C. L. (2018). Quantitative epigenetics and evolution. Heredity 121, 210–224 doi: 10.1038/s41437-018-0114-x
Barney, J. N., and Whitlow, T. H. (2008). A unifying framework for biological invasions: the state factor model. Biol. Invasions 10, 259–272. doi: 10.1007/s10530-007-9127-8
Barriere, G., Fici, P., Gallerani, G., Fabbri, F., and Rigaud, M. (2015). Epithelial mesenchymal transition: a double-edged sword. Clin. Transl. Med. 4:14.
Bartz, R., and Kowarik, I. (2019). Assessing the environmental impacts of invasive alien plants: a review of assessment approaches. NeoBiota 43, 69–99. doi: 10.3897/neobiota.43.30122
Becker, C., Hagmann, J., Müller, J., Koenig, D., Stegle, O., Borgwardt, K., et al. (2011). Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249. doi: 10.1038/nature10555
Beerling, D. J., Bailey, J. P., and Conolly, A. P. (1994). Fallopia Japonica (Houtt.) Ronse Decraene. J. Ecol. 82:959. doi: 10.2307/2261459
Bellard, C., Cassey, P., and Blackburn, T. M. (2016a). Alien species as a driver of recent extinctions. Biol. Lett. 12:20150623. doi: 10.1098/rsbl.2015.0623
Bellard, C., Genovesi, P., and Jeschke, J. M. (2016b). Global patterns in threats to vertebrates by biological invasions. Proc. Biol. Sci. 283:20152454. doi: 10.1098/rspb.2015.2454
Ben-David, U., and Amon, A. (2020). Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62. doi: 10.1038/s41576-019-0171-x
Bernal Rubio, Y. L., González-Reymúndez, A., Wu, K.-H. H., Griguer, C. E., Steibel, J. P., and de Los Campos, G. (2018). Whole-genome multi-omic study of survival in patients with glioblastoma multiforme. G3 8, 3627–3636. doi: 10.1534/g3.118.200391
Bímová, K., Mandák, B., and Pyšek, P. (2001). “Experimental control of Reynoutria congeners: a comparative study of a hybrid and its parents,” in Invasions: Species Ecology and Ecosystem Management, eds G. Brundu, J. Brock, I. Camarda, L. Child, Wade, and MPlant (Leiden: Backhuys Publishers).
Birkbak, N. J., and McGranahan, N. (2020). Cancer genome evolutionary trajectories in metastasis. Cancer Cell 37, 8–19. doi: 10.1016/j.ccell.2019.12.004
Blackburn, T. M., Pyšek, P., Bacher, S., Carlton, J. T., Duncan, R. P., Jarošík, V., et al. (2011). A proposed unified framework for biological invasions. Trends Ecol. Evol. 26, 333–339. doi: 10.1016/j.tree.2011.03.023
Blagosklonny, M. V. (2004). Analysis of FDA approved anticancer drugs reveals the future of cancer therapy. Cell Cycle 3, 1035–1042.
Blossey, B., and Notzold, R. (1995). Evolution of increased competitive ability in invasive nonindigenous plants: a hypothesis. J. Ecol. 83, 887–889. doi: 10.2307/2261425
Bock, D. G., Caseys, C., Cousens, R. D., Hahn, M. A., Heredia, S. M., Hübner, S., et al. (2015). What we still don’t know about invasion genetics. Mol. Ecol. 24, 2277–2297.
Bock, D. G., Kantar, M. B., Caseys, C., Matthey-Doret, R., and Rieseberg, L. H. (2018). Evolution of invasiveness by genetic accommodation. Nat. Ecol. Evol. 2, 991–999. doi: 10.1038/s41559-018-0553-z
Bodt, L. H., Rollins, L. A., and Zichello, J. M. (2020). Contrasting mitochondrial diversity of European starlings (Sturnus vulgaris) across three invasive continental distributions. Ecol. Evol. 10, 10186–10195. doi: 10.1002/ece3.6679
Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E., Siemann, E., and Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11. doi: 10.1007/s00442-005-0070-z
Bozic, I., Allen, B., and Nowak, M. A. (2012). Dynamics of targeted cancer therapy. Trends Mol. Med. 18, 311–316.
Bradshaw, C. J. A., Leroy, B., Bellard, C., Roiz, D., Albert, C., Fournier, A., et al. (2016). Massive yet grossly underestimated global costs of invasive insects. Nat. Commun. 7:12986.
Britton, J. R., and Gozlan, R. E. (2013). How many founders for a biological invasion? predicting introduction outcomes from propagule pressure. Ecology 94, 2558–2566. doi: 10.1890/13-0527.1
Cairns, J. (1975). Mutation selection and the natural history of cancer. Nature 255, 197–200. doi: 10.1038/255197a0
Capp, J.-P., and Thomas, F. (2020). A similar speciation process relying on cellular stochasticity in microbial and cancer cell populations. iScience 23:101531. doi: 10.1016/j.isci.2020.101531
Card, D. C., Perry, B. W., Adams, R. H., Schield, D. R., Young, A. S., and Andrew, A. (2018). Novel ecological and climatic conditions drive rapid adaptation in invasive Florida Burmese pythons. Mol. Ecol. 27, 4744–4757. doi: 10.1111/mec.14885
Carja, O., MacIsaac, J. L., Mah, S. M., Henn, B. M., Kobor, M. S., Feldman, M. W., et al. (2017). Worldwide patterns of human epigenetic variation. Nat. Ecol. Evol. 1, 1577–1583. doi: 10.1038/s41559-017-0299-z
Castoe, T. A., de Koning, A. P. J., Hall, K. T., Card, D. C., Schield, D. R., Fujita, M. K., et al. (2013). The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl. Acad. Sci. USA 110, 20645–20650. doi: 10.1073/pnas.1314475110
Castro-Giner, F., and Aceto, N. (2020). Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med. 12:31.
Catford, J. A., Jansson, R., and Nilsson, C. (2009). Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers. Distrib. 15, 22–40. doi: 10.1111/j.1472-4642.2008.00521.x
Chen, C., Zheng, Z., Bao, Y., Zhang, H., Richards, C. L., Li, J., et al. (2020). Comparisons of natural and cultivated populations of Corydalis yanhusuo indicate divergent patterns of genetic and epigenetic variation. Front. Plant Sci. 11:985.
Chen, K.-W., and Pienta, K. J. (2011). Modeling invasion of metastasizing cancer cells to bone marrow utilizing ecological principles. Theor. Biol. Med. Model. 8:36.
Chen, Y., Zhao, L., Teng, H., Shi, C., Liu, Q., Zhang, J., et al. (2021). Population genomics reveal rapid genetic differentiation in a recently invasive population of Rattus norvegicus. Front. Zool. 18:6.
Chown, S. L., Hodgins, K. A., Griffin, P. C., Oakeshott, J. G., Byrne, M., and Hoffmann, A. A. (2015). Biological invasions, climate change and genomics. Evol. Appl. 8, 23–46. doi: 10.1111/eva.12234
Coffey, D. S. (1998). Self-organization, complexity and chaos: the new biology for medicine. Nat. Med. 4, 882–885. doi: 10.1038/nm0898-882
Colautti, R. I., Alexander, J. M., Dlugosch, K. M., Keller, S. R., and Sultan, S. E. (2017). Invasions and extinctions through the looking glass of evolutionary ecology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:1712.
Colautti, R. I., and Lau, J. A. (2015). Contemporary evolution during invasion: evidence for differentiation, natural selection, and local adaptation. Mol. Ecol. 24, 1999–2017. doi: 10.1111/mec.13162
Cristescu, M. E. (2015). Genetic reconstructions of invasion history. Mol. Ecol. 24, 2212–2225. doi: 10.1111/mec.13117
Cuthbertab, R. N., Pattison, Z., Taylor, N. G., Verbrugge, L., Diagne, C., Ahmed, D. A., et al. (2021). Global economic costs of aquatic invasive alien species. Sci. Total Environ. 775:145238. doi: 10.1016/j.scitotenv.2021.145238
Davidson, A. M., Jennions, M., and Nicotra, A. B. (2011). Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis: Invasive species have higher phenotypic plasticity. Ecol. Lett. 14, 419–431. doi: 10.1111/j.1461-0248.2011.01596.x
de Groot, A. E., Roy, S., Brown, J. S., Pienta, K. J., and Amend, S. R. (2017). Revisiting seed and soil: examining the primary tumor and cancer cell foraging in metastasis. Mol. Cancer Res. 15, 361–370. doi: 10.1158/1541-7786.mcr-16-0436
Del Tredici, P. (2017). The introduction of Japanese knotweed, Reynoutria japonica, into North America. J. Torrey Bot. Soc. 144, 406–416. doi: 10.3159/torrey-d-17-00002.1
Diagne, C., Leroy, B., Gozlan, R. E., Vaissière, A.-C., Assailly, C., Nuninger, L., et al. (2020). InvaCost, a public database of the economic costs of biological invasions worldwide. Sci. Data 7:277.
Diagne, C., Leroy, B., Vaissière, A.-C., Gozlan, R. E., Roiz, D., Jarić, I., et al. (2021). High and rising economic costs of biological invasions worldwide. Nature 592, 571–576. doi: 10.1038/s41586-021-03405-6
Diez, J. M., Sullivan, J. J., Hulme, P. E., Edwards, G., and Duncan, R. P. (2008). Darwin’s naturalization conundrum: dissecting taxonomic patterns of species invasions. Ecol. Lett. 11, 674–681. doi: 10.1111/j.1461-0248.2008.01178.x
Dlugosch, K. M., and Parker, I. M. (2008a). Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449. doi: 10.1111/j.1365-294x.2007.03538.x
Dlugosch, K. M., and Parker, I. M. (2008b). Invading populations of an ornamental shrub show rapid life history evolution despite genetic bottlenecks. Ecol. Lett. 11, 701–709. doi: 10.1111/j.1461-0248.2008.01181.x
Dlugosch, K. M., Anderson, S. R., Braasch, J., Cang, F. A., and Gillette, H. D. (2015). The devil is in the details: genetic variation in introduced populations and its contributions to invasion. Mol. Ecol. 24, 2095–2111. doi: 10.1111/mec.13183
Douhovnikoff, V., and Dodd, R. S. (2015). Epigenetics: a potential mechanism for clonal plant success. Plant Ecol. 216, 227–233. doi: 10.1007/s11258-014-0430-z
Doyle, J. J., Flagel, L. E., Paterson, A. H., Rapp, R. A., Soltis, D. E., Soltis, P. S., et al. (2008). Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42, 443–461. doi: 10.1146/annurev.genet.42.110807.091524
Dressman, H. K., Hans, C., Bild, A., Olson, J. A., Rosen, E., Marcom, P. K., et al. (2006). Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin. Cancer Res. 12(3 Pt 1), 819–826. doi: 10.1158/1078-0432.ccr-05-1447
Dubin, M. J., Zhang, P., Meng, D., Remigereau, M.-S., Osborne, E. J., Paolo Casale, F., et al. (2015). DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4:e05255.
Duesberg, P., Rausch, C., Rasnick, D., and Hehlmann, R. (1998). Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl. Acad. Sci. USA 95, 13692–13697. doi: 10.1073/pnas.95.23.13692
Dujon, A. M., Aktipis, A., Alix-Panabières, C., Amend, S. R., Boddy, A. M., Brown, J. S., et al. (2021). Identifying key questions in the ecology and evolution of cancer. Evol. Appl. 14, 877–892.
Duran, G., Cruz, R., Simoes, A. R., Barros, F., Giráldez, J. M., Bernárdez, B., et al. (2020). Efficacy and toxicity of adjuvant chemotherapy on colorectal cancer patients: how much influence from the genetics? J. Chemother. 32, 310–322. doi: 10.1080/1120009x.2020.1764281
Eckert, S., Herden, J., Stift, M., Joshi, J., and van Kleunen, M. (2021). Manipulation of cytosine methylation does not remove latitudinal clines in two invasive goldenrod species in Central Europe. Mol. Ecol. 30, 222–236. doi: 10.1111/mec.15722
Estoup, A., Ravigné, V., Hufbauer, R., Vitalis, R., Gautier, M., and Facon, B. (2016). Is there a genetic paradox of biological invasion? Annu. Rev. Ecol. Evol. Syst. 47, 51–72.
Exposito-Alonso, M., Becker, C., Schuenemann, V. J., Reiter, E., Setzer, C., Slovak, R., et al. (2018). The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 14:e1007155. doi: 10.1371/journal.pgen.1007155
Fabisiewicz, A., and Grzybowska, E. (2017). CTC clusters in cancer progression and metastasis. Med. Oncol. 34:12.
Fearon, E. R., and Vogelstein, B. (1990). A genetic model for colorectal tumorigenesis. Cell 61, 759–767. doi: 10.1016/0092-8674(90)90186-i
Feinberg, A. P., and Irizarry, R. A. (2010). Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. USA 107(Suppl. 1), 1757–1764. doi: 10.1073/pnas.0906183107
Feinberg, A. P., Koldobskiy, M. A., and Göndör, A. (2016). Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299. doi: 10.1038/nrg.2016.13
Feinberg, A. P., Ohlsson, R., and Henikoff, S. (2006). The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 7, 21–33. doi: 10.1038/nrg1748
Felden, A., Paris, C., Chapple, D. G., Suarez, A. V., Tsutsui, N. D., Lester, P. J., et al. (2019). Native and introduced Argentine ant populations are characterised by distinct transcriptomic signatures associated with behaviour and immunity. Working Paper Series 49, 105–126. doi: 10.3897/neobiota.49.36086
Fitzpatrick, B. M., Johnson, J. R., Kump, D. K., Shaffer, H. B., Smith, J. J., and Voss, S. R. (2009). Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid tiger salamanders. BMC Evol. Biol. 9:176. doi: 10.1186/1471-2148-9-176
Flanagan, B. A., Krueger-Hadfield, S. A., Murren, C. J., Nice, C. C., Strand, A. E., and Sotka, E. E. (2021). Founder effects shape linkage disequilibrium and genomic diversity of a partially clonal invader. Mol. Ecol. 30, 1962–1978. doi: 10.1111/mec.15854
Flucher, S. M., Krapf, P., Arthofer, W., Suarez, A. V., Crozier, R. H., Steiner, F. M., et al. (2021). Effect of social structure and introduction history on genetic diversity and differentiation. Mol. Ecol. 30, 2511–2527. doi: 10.1111/mec.15911
Gao, L., Geng, Y., Li, B., Chen, J., and Yang, J. (2010). Genome-wide DNA methylation alterations of Alternanthera philoxeroides in natural and manipulated habitats: implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant Cell Environ. 33, 1820–1827. doi: 10.1111/j.1365-3040.2010.02186.x
Gao, Z., Moorjani, P., Sasani, T. A., Pedersen, B. S., Quinlan, A. R., Jorde, L. B., et al. (2019). Overlooked roles of DNA damage and maternal age in generating human germline mutations. Proc. Natl. Acad. Sci. USA 116, 9491–9500. doi: 10.1073/pnas.1901259116
Gaskin, J. F., Bon, M.-C., Cock, M. J. W., Cristofaro, M., Biase, A. D., De Clerck-Floate, R., et al. (2011). Applying molecular-based approaches to classical biological control of weeds. Biol. Control Theory Appl. Pest Manag. 58, 1–21. doi: 10.1016/j.biocontrol.2011.03.015
Gatenby, R. A., and Brown, J. (2017). Mutations, evolution and the central role of a self-defined fitness function in the initiation and progression of cancer. Biochim. Biophys. Acta Rev. Cancer 1867, 162–166. doi: 10.1016/j.bbcan.2017.03.005
Gatenby, R. A., and Brown, J. S. (2020). The evolution and ecology of resistance in cancer therapy. Cold Spring Harbor Perspect. Med. 8:a033415. doi: 10.1101/cshperspect.a033415
Gatenby, R. A., Brown, J., and Vincent, T. (2009). Lessons from applied ecology: cancer control using an evolutionary double bind. Cancer Res. 69, 7499–7502. doi: 10.1158/0008-5472.can-09-1354
Geng, Y.-P., Pan, X.-Y., Xu, C.-Y., Zhang, W.-J., Li, B., Chen, J.-K., et al. (2007). Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol. Invasions 9, 245–256. doi: 10.1007/s10530-006-9029-1
Gerber, E., Krebs, C., Murrell, C., Moretti, M., Rocklin, R., and Schaffner, U. (2008). Exotic invasive knotweeds (Fallopia spp.) negatively affect native plant and invertebrate assemblages in European riparian habitats. Biol. Conserv. 141, 646–654. doi: 10.1016/j.biocon.2007.12.009
Gerstung, M., Jolly, C., Leshchiner, I., Dentro, S. C., Gonzalez, S., Rosebrock, D., et al. (2020). The evolutionary history of 2,658 cancers. Nature 578, 122–128.
Goldmann, J. M., Veltman, J. A., and Gilissen, C. (2019). De novo mutations reflect development and aging of the human Germline. Trends Genet. 35, 828–839. doi: 10.1016/j.tig.2019.08.005
Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., et al. (1999). Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537. doi: 10.1126/science.286.5439.531
Grimsby, J. L., Tsirelson, D., Gammon, M. A., and Kesseli, R. (2007). Genetic diversity and clonal vs. sexual reproduction in Fallopia spp. (Polygonaceae). Am. J. Bot. 94, 957–964. doi: 10.3732/ajb.94.6.957
Gupta, G. P., Nguyen, D. X., Chiang, A. C., Bos, P. D., Kim, J. Y., Nadal, C., et al. (2007). Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 446, 765–770. doi: 10.1038/nature05760
Gurevitch, J., Fox, G. A., Wardle, G. M., Inderjit, K., and Taub, D. (2011). Emergent insights from the synthesis of conceptual frameworks for biological invasions. Ecol. Lett. 14, 407–418. doi: 10.1111/j.1461-0248.2011.01594.x
Hamelin, R. C., and Roe, A. D. (2020). Genomic biosurveillance of forest invasive alien enemies: a story written in code. Evol. Appl. 13, 95–115. doi: 10.1111/eva.12853
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hapach, L. A., Mosier, J. A., Wang, W., and Reinhart-King, C. A. (2019). Engineered models to parse apart the metastatic cascade. NPJ Precis. Oncol. 3:20.
Harms, N. E., Williams, D. A., and Purcell, M. F. (2021). The role of overseas genetic surveys to potentially accelerate biological control development for a new Hydrilla verticillata introduction in the USA. Biocontrol 66, 271–280. doi: 10.1007/s10526-020-10050-x
Hawes, N. A., Fidler, A. E., Tremblay, L. A., Pochon, X., Dunphy, B. J., and Smith, K. F. (2018). Understanding the role of DNA methylation in successful biological invasions: a review. Biol. Invasions 20, 2285–2300. doi: 10.1007/s10530-018-1703-6
Hawkins, N. J., Bass, C., Dixon, A., and Neve, P. (2019). The evolutionary origins of pesticide resistance. Biol. Rev. Camb. Philos. Soc. 94, 135–155. doi: 10.1111/brv.12440
He, F., Steige, K. A., Kovacova, V., Göbel, U., Bouzid, M., Keightley, P. D., et al. (2021). Cis-regulatory evolution spotlights species differences in the adaptive potential of gene expression plasticity. Nat. Commun. 12:3376.
Herman, J. J., and Sultan, S. E. (2011). Adaptive transgenerational plasticity in plants: case studies, mechanisms, and implications for natural populations. Front. Plant Sci. 2:102.
Herman, J. J., and Sultan, S. E. (2016). DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. R. Soc. B Biol. Sci. 283:20160988. doi: 10.1098/rspb.2016.0988
Hoadley, K. A., Siegel, M. B., Kanchi, K. L., Miller, C. A., Ding, L., Zhao, W., et al. (2016). Tumor evolution in two patients with basal-like breast cancer: a retrospective genomics study of multiple metastases. PLoS Med. 13:e1002174. doi: 10.1371/journal.pmed.1002174
Hodgins, K. A., Bock, D. G., Hahn, M. A., Heredia, S. M., Turner, K. G., and Rieseberg, L. H. (2015). Comparative genomics in the Asteraceae reveals little evidence for parallel evolutionary change in invasive taxa. Mol. Ecol. 24, 2226–2240. doi: 10.1111/mec.13026
Hodgins, K. A., Lai, Z., Nurkowski, K., Huang, J., and Rieseberg, L. H. (2013). The molecular basis of invasiveness: differences in gene expression of native and introduced common ragweed (Ambrosia artemisiifolia) in stressful and benign environments. Mol. Ecol. 22, 2496–2510. doi: 10.1111/mec.12179
Hofmeister, B. T., Denkena, J., Colomé-Tatché, M., Shahryary, Y., Hazarika, R., Grimwood, J., et al. (2020). A genome assembly and the somatic genetic and epigenetic mutation rate in a wild long-lived perennial Populus trichocarpa. Genome Biol. 21:259.
Hofmeister, N. R., Werner, S. J., and Lovette, I. J. (2021). Environmental correlates of genetic variation in the invasive european starling in North America. Mol. Ecol. 30, 1251–1263. doi: 10.1111/mec.15806
Hu, C., Hart, S. N., Gnanaolivu, R., Huang, H., Lee, K. Y., Na, J., et al. (2021). A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 384, 440–451.
Hu, Z., Ding, J., Ma, Z., Sun, R., Seoane, J. A., Scott Shaffer, J., et al. (2019). Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122. doi: 10.1038/s41588-019-0423-x
Ibrahim-Hashim, A., Luddy, K., Abrahams, D., Enriquez-Navas, P., Damgaci, S., Yao, J., et al. (2021). Artificial selection for host resistance to tumour growth and subsequent cancer cell adaptations: an evolutionary arms race. Br. J. Cancer 124, 455–465. doi: 10.1038/s41416-020-01110-1
Ibrahim-Hashim, A., Robertson-Tessi, M., Enriquez-Navas, P. M., Damaghi, M., Balagurunathan, Y., Wojtkowiak, J. W., et al. (2017). Defining cancer subpopulations by adaptive strategies rather than molecular properties provides novel insights into intratumoral evolution. Cancer Res. 77, 2242–2254. doi: 10.1158/0008-5472.can-16-2844
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (2020). Pan-cancer analysis of whole genomes. Nature 578, 82–93. doi: 10.1038/s41586-020-1969-6
Irimia, R. E., Hierro, J. L., Branco, S., Sotes, G., Cavieres, L. A., Eren, Ö, et al. (2021). Experimental admixture among geographically disjunct populations of an invasive plant yields a global mosaic of reproductive incompatibility and heterosis. J. Ecol. 109, 2152–2162. doi: 10.1111/1365-2745.13628
Jablonka, E. (2017). The evolutionary implications of epigenetic inheritance. Interface Focus 7:20160135. doi: 10.1098/rsfs.2016.0135
Jablonka, E., and Lamb, M. J. (1998). Epigenetic inheritance in evolution. J. Evol. Biol. 11, 159–183.
Jablonka, E., and Lamb, M. J. (1999). Epigenetic Inheritance and Evolution: the Lamarckian Dimension. Oxford: Oxford University Press.
Jablonka, E., and Raz, G. (2009). Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176. doi: 10.1086/598822
Jamal-Hanjani, M., Wilson, G. A., McGranahan, N., Birkbak, N. J., Watkins, T. B. K., Veeriah, S., et al. (2017). Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121.
Johannes, F., Porcher, E., Teixeira, F. K., Saliba-Colombani, V., Simon, M., Agier, N., et al. (2009). Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5:e1000530. doi: 10.1371/journal.pgen.1000530
Johnson, K. E., Howard, G., Mo, W., Strasser, M. K., Lima, E. A. B. F., Huang, S., et al. (2019). Cancer cell population growth kinetics at low densities deviate from the exponential growth model and suggest an Allee effect. PLoS Biol. 17:e3000399. doi: 10.1371/journal.pbio.3000399
Joosse, S. A., Gorges, T. M., and Pantel, K. (2015). Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol. Med. 7, 1–11. doi: 10.15252/emmm.201303698
Kadoch, C., and Crabtree, G. R. (2015). Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 1:e1500447. doi: 10.1126/sciadv.1500447
Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B., Lu, C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. doi: 10.1038/nature12634
Kang, Y., Siegel, P. M., Shu, W., Drobnjak, M., Kakonen, S. M., Cordón-Cardo, C., et al. (2003). A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–5496. doi: 10.1016/s1535-6108(03)00132-6
Keller, E. F. (2014). From gene action to reactive genomes. J. Physiol. 592, 2423–2429. doi: 10.1113/jphysiol.2014.270991
Keller, R. P., Lodge, D. M., and Finnoff, D. C. (2007). Risk assessment for invasive species produces net bioeconomic benefits. Proc. Natl. Acad. Sci. USA 104, 203–207. doi: 10.1073/pnas.0605787104
Kolbe, J. J., Glor, R. E., Rodríguez Schettino, L., Lara, A. C., Larson, A., and Losos, J. B. (2004). Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181. doi: 10.1038/nature02807
Kooke, R., Morgado, L., Becker, F., van Eekelen, H., Hazarika, R., Zheng, Q., et al. (2019). Epigenetic mapping of the Arabidopsis metabolome reveals mediators of the epigenotype-phenotype map. Genome Res. 29, 96–106. doi: 10.1101/gr.232371.117
Kramer, A., and Sarnelle, O. (2008). Limits to genetic bottlenecks and founder events imposed by the allee effect. Oecologia 157, 561–569. doi: 10.1007/s00442-008-1105-z
Krueger-Hadfield, S. A., Kollars, N. M., Byers, J. E., Greig, T. W., Hammann, M., Murray, D. C., et al. (2016). Invasion of novel habitats uncouples haplo-diplontic life cycles. Mol. Ecol. 25, 3801–3816. doi: 10.1111/mec.13718
Kumar, S., and Weaver, V. M. (2009). Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 28, 113–127. doi: 10.1007/s10555-008-9173-4
Kuramochi, H., Nakajima, G., Kaneko, Y., Nakamura, A., Inoue, Y., Yamamoto, M., et al. (2012). Amphiregulin and Epiregulin mRNA expression in primary colorectal cancer and corresponding liver metastases. BMC Cancer 12:88.
Lai, Z., Gross, B. L., Zou, Y., Andrews, J., and Rieseberg, L. H. (2006). Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 15, 1213–1227. doi: 10.1111/j.1365-294x.2006.02775.x
Laitinen, R. A. E., and Nikoloski, Z. (2019). Genetic basis of plasticity in plants. J. Exp. Bot. 70, 739–745. doi: 10.1093/jxb/ery404
Lambert, A. W., Pattabiraman, D. R., and Weinberg, R. A. (2017). Emerging biological principles of metastasis. Cell 168, 670–691. doi: 10.1016/j.cell.2016.11.037
Ledford, H. (2010). Big science: the cancer genome challenge. Nature 464, 972–974. doi: 10.1038/464972a
Lee, C. E. (2002). Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391. doi: 10.1016/s0169-5347(02)02554-5
Lee-Six, H., Øbro, N. F., Shepherd, M. S., Grossmann, S., Dawson, K., Belmonte, M., et al. (2018). Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478. doi: 10.1038/s41586-018-0497-0
Lee-Six, H., Olafsson, S., Ellis, P., Osborne, R. J., Sanders, M. A., Moore, L., et al. (2019). The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574, 532–537. doi: 10.1038/s41586-019-1672-7
Levine, J. M., Adler, P. B., and Yelenik, S. G. (2004). A meta-analysis of biotic resistance to exotic plant invasions: biotic resistance to plant invasion. Ecol. Lett. 7, 975–989. doi: 10.1111/j.1461-0248.2004.00657.x
Levis, N. A., and Pfennig, D. W. (2016). Evaluating “plasticity-first” evolution in nature: key criteria and empirical approaches. Trends Ecol. Evol. 31, 563–574. doi: 10.1016/j.tree.2016.03.012
Liebhold, A. M., Berec, L., Brockerhoff, E. G., Epanchin-Niell, R. S., Hastings, A., Herms, D. A., et al. (2016). Eradication of invading insect populations: from concepts to applications. Annu. Rev. Entomol. 61, 335–352. doi: 10.1146/annurev-ento-010715-023809
Liebl, A. L., Schrey, A. W., Andrew, S. C., Sheldon, E. L., and Griffith, S. C. (2015). Invasion genetics: lessons from a ubiquitous bird, the house sparrow Passer domesticus. Curr. Zool. 61, 465–476. doi: 10.1093/czoolo/61.3.465
Lloyd, M. C., Cunningham, J. J., Bui, M. M., Gillies, R. J., Brown, J. S., and Gatenby, R. A. (2016). Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Cancer Res. 76, 3136–3144. doi: 10.1158/0008-5472.can-15-2962
Lloyd, M. C., Gatenby, R. A., and Brown, J. S. (2017). “Ecology of the metastatic process,” in Ecology and Evolution of Cancer, eds B. Ujvari, B. Roche, and F. Thomas (Amsterdam: Elsevier).
Lodge, D. M. (1993). Biological invasions: lessons for ecology. Trends Ecol. Evol. 8, 133–137. doi: 10.1016/0169-5347(93)90025-k
Lodge, D. M., Simonin, P. W., Burgiel, S. W., Keller, R. P., Bossenbroek, J. M., Jerde, C. L., et al. (2016). Risk analysis and bioeconomics of invasive species to inform policy and management. Annu. Rev. Environ. Resour. 41, 453–488.
Loeb, L. A. (2011). Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer 11, 450–457. doi: 10.1038/nrc3063
Loeb, L. A., Loeb, K. R., and Anderson, J. P. (2003). Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA 100, 776–781.
Marin, P., Genitoni, J., Barloy, D., Maury, S., Gibert, P., Ghalambor, C. K., et al. (2020). Biological invasion: the influence of the hidden side of the (epi)genome. Funct. Ecol. 34, 385–400. doi: 10.1111/1365-2435.13317
Maron, J. L., and Marler, M. (2008). Effects of native species diversity and resource additions on invader impact. Am. Nat. 172(Suppl. 1), S18–S33.
Martincorena, I., and Campbell, P. J. (2015). Somatic mutation in cancer and normal cells. Science 349, 1483–1489. doi: 10.1126/science.aab4082
Martincorena, I., Fowler, J. C., Wabik, A., Lawson, A. R. J., Abascal, F., Hall, M. W. J., et al. (2018). Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917. doi: 10.1126/science.aau3879
Martincorena, I., Roshan, A., Gerstung, M., Ellis, P., Van Loo, P., McLaren, S., et al. (2015). Tumor evolution. high burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886. doi: 10.1126/science.aaa6806
Matesanz, S., Rubio Teso, M. L., García-Fernández, A., and Escudero, A. (2017). Habitat fragmentation differentially affects genetic variation, phenotypic plasticity and survival in populations of a gypsum endemic. Front. Plant Sci. 8:843.
McGill, B. J., and Brown, J. S. (2007). Evolutionary game theory and adaptive dynamics of continuous traits. Annu. Rev. Ecol. Evol. Syst. 38, 403–435. doi: 10.1146/annurev.ecolsys.36.091704.175517
Moon, K., Blackman, D. A., and Brewer, T. D. (2015). Understanding and integrating knowledge to improve invasive species management. Biol. Invasions 17, 2675–2689. doi: 10.1007/s10530-015-0904-5
Moore, L., Leongamornlert, D., Coorens, T. H. H., Sanders, M. A., Ellis, P., Dentro, S. C., et al. (2020). The mutational landscape of normal human endometrial epithelium. Nature 580, 640–646.
Mounger, J., Ainouche, M. L., Bossdorf, O., Cavé-Radet, A., Li, B., Parepa, M., et al. (2021a). Epigenetics and the success of invasive plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376:20200117. doi: 10.1098/rstb.2020.0117
Mounger, J., Teresa Boquete, M., Schmid, M. W., Granado, R., Robertson, M. H., Voors, S. A., et al. (2021b). Inheritance of DNA methylation differences in the mangrove Rhizophora mangle. Evol. Dev. 23, 351–374. doi: 10.1111/ede.12388
Müller, A., Homey, B., Soto, H., Ge, N., Catron, D., Buchanan, M. E., et al. (2001). Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56.
Müller, G. B. (2017). Why an extended evolutionary synthesis is necessary. Interface Focus 7:20170015. doi: 10.1098/rsfs.2017.0015
Naciri, Y., and Linder, H. P. (2020). The genetics of evolutionary radiations. Biol. Rev. Camb. Philos. Soc. 95, 1055–1072. doi: 10.1111/brv.12598
Nam, A. S., Chaligne, R., and Landau, D. A. (2021). Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 22, 3–18. doi: 10.1038/s41576-020-0265-5
Narum, S. R., Buerkle, C. A., Davey, J. W., Miller, M. R., and Hohenlohe, P. A. (2013). Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22, 2841–2847. doi: 10.1111/mec.12350
Nature. (2020). The era of massive cancer sequencing projects has reached a turning point [Review of the era of massive cancer sequencing projects has reached a turning point]. Nature 578, 7–8. doi: 10.1038/d41586-020-00308-w
Nguyen, D. X., and Massagué, J. (2007). Genetic determinants of cancer metastasis. Nat. Rev. Genet. 8, 341–352. doi: 10.1038/nrg2101
Nicotra, A. B., Atkin, O. K., Bonser, S. P., Davidson, A. M., Finnegan, E. J., Mathesius, U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. doi: 10.1016/j.tplants.2010.09.008
Nieto Feliner, G., Casacuberta, J., and Wendel, J. F. (2020). Genomics of evolutionary novelty in hybrids and polyploids. Front. Genet. 11:792.
Nik-Zainal, S., Van Loo, P., Wedge, D. C., Alexandrov, L. B., Greenman, C. D., Lau, K. W., et al. (2012). The life history of 21 breast cancers. Cell 149, 994–1007.
Nowell, P. C. (1976). The clonal evolution of tumor cell populations. Science 194, 23–28. doi: 10.1126/science.959840
Nunez-Mir, G. C., Liebhold, A. M., Guo, Q., Brockerhoff, E. G., Jo, I., Ordonez, K., et al. (2017). Biotic resistance to exotic invasions: its role in forest ecosystems, confounding artifacts, and future directions. Biol. Invasions 19, 3287–3299. doi: 10.1007/s10530-017-1413-5
Ortiz-Otero, N., Marshall, J. R., Lash, B. W., and King, M. R. (2020). Platelet mediated TRAIL delivery for efficiently targeting circulating tumor cells. Nanoscale Adv. 2, 3942–3953. doi: 10.1039/d0na00271b
Oshi, M., Katsuta, E., Yan, L., Ebos, J. M. L., Rashid, O. M., Matsuyama, R., et al. (2020). A Novel 4-Gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers 12:1148. doi: 10.3390/cancers12051148
Ossowski, S., Schneeberger, K., Lucas-Lledó, J. I., Warthmann, N., Clark, R. M., Shaw, R. G., et al. (2010). The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94. doi: 10.1126/science.1180677
Paape, T., Akiyama, R., Cereghetti, T., Onda, Y., Hirao, A. S., Kenta, T., et al. (2020). Experimental and field data support range expansion in an allopolyploid arabidopsis owing to parental legacy of heavy metal Hyperaccumulation. Front. Genet. 11:565854.
Paget, S. (1889). The distribution of secondary growths in cancer of the breast. Lancet 133, 571–573. doi: 10.1016/s0140-6736(00)49915-0
Palacio-López, K., and Gianoli, E. (2011). Invasive plants do not display greater phenotypic plasticity than their native or non-invasive counterparts: a meta-analysis. Oikos 120, 1393–1401. doi: 10.1111/j.1600-0706.2010.19114.x
Pantel, K., and Alix-Panabières, C. (2019). Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424. doi: 10.1038/s41571-019-0187-3
Parkes, J. P., and Panetta, F. D. (2009). “Eradication of invasive species: progress and emerging issues in the 21st century,” in Invasive Species Management: a Handbook of Principles and Techniques. Techniques in Ecology and Conservation Series. Chapter 4. Oxford: Oxford University Press. 47–60.
Patel, S. A., and Vanharanta, S. (2017). Epigenetic determinants of metastasis. Mol. Oncol. 11, 79–96. doi: 10.1016/j.molonc.2016.09.008
Patel, S. A., Rodrigues, P., Wesolowski, L., and Vanharanta, S. (2021). Genomic control of metastasis. Br. J. Cancer 124, 3–12.
Paterlini-Brechot, P., and Benali, N. L. (2007). Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 253, 180–204. doi: 10.1016/j.canlet.2006.12.014
Paun, O., Verhoeven, K. J. F., and Richards, C. L. (2019). Opportunities and limitations of reduced representation bisulfite sequencing in plant ecological epigenomics. New Phytol. 221, 738–742. doi: 10.1111/nph.15388
Phillips, B. L., Brown, G. P., Travis, J. M. J., and Shine, R. (2008). Reid’s paradox revisited: the evolution of dispersal kernels during range expansion. Am. Nat. 172, S34–S48.
Pienta, K. J., Hammarlund, E. U., Axelrod, R., Amend, S. R., and Brown, J. S. (2020a). Convergent evolution, evolving evolvability, and the origins of lethal cancer. Mol. Cancer Res. 18, 801–810. doi: 10.1158/1541-7786.mcr-19-1158
Pienta, K. J., Hammarlund, E. U., Axelrod, R., Brown, J. S., and Amend, S. R. (2020b). Poly-aneuploid cancer cells promote evolvability, generating lethal cancer. Evol. Appl. 13, 1626–1634. doi: 10.1111/eva.12929
Pienta, K. J., Hammarlund, E. U., Brown, J. S., Amend, S. R., and Axelrod, R. M. (2021). Cancer recurrence and lethality are enabled by enhanced survival and reversible cell cycle arrest of polyaneuploid cells. Proc. Natl. Acad. Sci. USA 118:e2020838118. doi: 10.1073/pnas.2020838118
Pigliucci, M. (2010). Genotype-phenotype mapping and the end of the “genes as blueprint” metaphor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 557–566. doi: 10.1098/rstb.2009.0241
Pimentel, D., Lach, L., Zuniga, R., and Morrison, D. (2000). Environmental and economic costs of nonindigenous species in the United States. BioScience 50, 53–65. doi: 10.1641/0006-3568(2000)050[0053:eaecon]2.3.co;2
Pintor, L. M., Brown, J. S., and Vincent, T. L. (2011). Evolutionary game theory as a framework for studying biological invasions. Am. Nat. 177, 410–423. doi: 10.1086/658149
Plutynski, A. (2021). Testing multi-task cancer evolution: How Do We test ecological hypotheses in cancer? Front. Ecol. Evol. 9:301.
Prentis, P. J., Wilson, J. R. U., Dormontt, E. E., Richardson, D. M., and Lowe, A. J. (2008). Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–294. doi: 10.1016/j.tplants.2008.03.004
Pressley, M., Salvioli, M., Lewis, D. B., Richards, C. L., Brown, J. S., and Stankova, K. (2021). Evolutionary dynamics of drug resistance in cancer informs understanding of rapid evolution in natural systems. Front. Ecol. Evol. doi: 10.3389/fevo.2021.681121
Pyšek, P., and Richardson, D. M. (2010). Invasive species, environmental change and management, and health. Annu. Rev. Environ. Resour. 35, 25–55. doi: 10.1146/annurev-environ-033009-095548
Qu, X., Sandmann, T., Frierson, H., Fu, L., Fuentes, E., Walter, K., et al. (2016). Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene 35, 6403–6415. doi: 10.1038/onc.2016.170
Quilodrán, C. S., Tsoupas, A., and Currat, M. (2020). The spatial signature of introgression after a biological invasion with hybridization. Front. Ecol. Evol. 8:32120.
Rack, B., Schindlbeck, C., Jückstock, J., Andergassen, U., Hepp, P., Zwingers, T., et al. (2014). Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl. Cancer Inst. 106:dju066.
Rahbari, R., Wuster, A., Lindsay, S. J., Hardwick, R. J., Alexandrov, L. B., Turki, S. A., et al. (2016). Timing, rates and spectra of human germline mutation. Nat. Genet. 48, 126–133. doi: 10.1038/ng.3469
Ramaswamy, S., Ross, K. N., Lander, E. S., and Golub, T. R. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54. doi: 10.1038/ng1060
Regulations.gov (2020). Available Online at: https://www.federalregister.gov/documents/2020/12/15/2020-26734/removal-of-emerald-ash-borer-domestic-quarantine-regulations (accessed January 20, 2021).
Rejmánek, M. (1996). A theory of seed plant invasiveness: the first sketch. Biol. Conserv. 78, 171–181. doi: 10.1016/0006-3207(96)00026-2
Rejmánek, M. (2000). Invasive plants: approaches and predictions: invasive plants. Austral Ecol. 25, 497–506. doi: 10.1046/j.1442-9993.2000.01080.x
Richards, C. L., Alonso, C., Becker, C., Bossdorf, O., Bucher, E., Colomé-Tatché, M., et al. (2017). Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecol. Lett. 20, 1576–1590. doi: 10.1111/ele.12858
Richards, C. L., and Pigliucci, M. (2020). Epigenetic inheritance. a decade into the extended evolutionary synthesis. Paradigmi 38, 463–494.
Richards, C. L., Bossdorf, O., and Pigliucci, M. (2010). What role does heritable epigenetic variation play in phenotypic evolution? Bioscience 60, 232–237. doi: 10.1525/bio.2010.60.3.9
Richards, C. L., Bossdorf, O., Muth, N. Z., Gurevitch, J., and Pigliucci, M. (2006). Jack of all trades, master of some? on the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9, 981–993. doi: 10.1111/j.1461-0248.2006.00950.x
Richards, C. L., Schrey, A. W., and Pigliucci, M. (2012). Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol. Lett. 15, 1016–1025. doi: 10.1111/j.1461-0248.2012.01824.x
Richardson, D. M., and Pyšek, P. (2006). Plant invasions: merging the concepts of species invasiveness and community invasibility. Prog. Phys. Geography Earth Environ. 30, 409–431. doi: 10.1191/0309133306pp490pr
Richardson, D. M., Pysek, P., Rejmanek, M., Barbour, M. G., Panetta, F. D., and West, C. J. (2000). Naturalization and invasion of alien plants: concepts and definitions. Divers. Distrib. 6, 93–107. doi: 10.1046/j.1472-4642.2000.00083.x
Rieseberg, L. H., Raymond, O., Rosenthal, D. M., Lai, Z., Livingstone, K., Nakazato, T., et al. (2003). Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216. doi: 10.1126/science.1086949
Risson, E., Nobre, A. R., Maguer-Satta, V., and Aguirre-Ghiso, J. A. (2020). The current paradigm and challenges ahead for the dormancy of disseminated tumor cells. Nat. Cancer 1, 672–680. doi: 10.1038/s43018-020-0088-5
Rius, M., Bourne, S., Hornsby, H. G., and Chapman, M. A. (2015a). Applications of next-generation sequencing to the study of biological invasions. Curr. Zool. 61, 488–504. doi: 10.1093/czoolo/61.3.488
Rius, M., Turon, X., Bernardi, G., Volckaert, F. A. M., and Viard, F. (2015b). Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biol. Invasions 17, 869–885. doi: 10.1007/s10530-014-0792-0
Robertson, M., Alvarez, M., van Gurp, T., Wagemaker, C. A. M., Yu, F., Amador, D. M., et al. (2020). Combining epiGBS markers with long read transcriptome sequencing to assess differentiation associated with habitat in Reynoutria (aka Fallopia). Biorxiv [preprint] doi: 10.1101/2020.09.30.317966
Rodrigues, G., Hoshino, A., Kenific, C. M., Matei, I. R., Steiner, L., Freitas, D., et al. (2019). Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 21, 1403–1412. doi: 10.1038/s41556-019-0404-4
Rollins, L. A., Richardson, M. F., and Shine, R. (2015). A genetic perspective on rapid evolution in cane toads (Rhinella marina). Mol. Ecol. 24, 2264–2276. doi: 10.1111/mec.13184
Rollins, L. A., Woolnough, A. P., Fanson, B. G., Cummins, M. L., Crowley, T. M., Wilton, A. N., et al. (2016). Selection on mitochondrial variants occurs between and within Individuals in an expanding invasion. Mol. Biol. Evol. 33, 995–1007. doi: 10.1093/molbev/msv343
Rosenthal, D. M., Schwarzbach, A. E., Donovan, L. A., Raymond, O., and Rieseberg, L. H. (2002). Phenotypic differentiation between three ancient hybrid taxa and their parental species. Int. J. Plant Sci. 163, 387–398. doi: 10.1086/339237
Rotter, M. C., and Holeski, L. M. (2018). A meta-analysis of the evolution of increased competitive ability hypothesis: genetic-based trait variation and herbivory resistance trade-offs. Biol. Invasions 20, 2647–2660. doi: 10.1007/s10530-018-1724-1
Sansregret, L., and Swanton, C. (2017). The role of aneuploidy in cancer evolution. Cold Spring Harbor Perspect. Med. 7:a028373. doi: 10.1101/cshperspect.a028373
Sarma, R. R., Crossland, M. R., Eyck, H. J. F., DeVore, J. L., Edwards, R. J., Cocomazzo, M., et al. (2021). Intergenerational effects of manipulating DNA methylation in the early life of an iconic invader. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 376:20200125. doi: 10.1098/rstb.2020.0125
Sarma, R. R., Edwards, R. J., Crino, O. L., Eyck, H. J. F., Waters, P. D., Crossland, M. R., et al. (2020). Do epigenetic changes drive corticosterone responses to alarm cues in larvae of an invasive amphibian? Integr. Compar. Biol. 60, 1481–1494. doi: 10.1093/icb/icaa082
Sasaki, E., Kawakatsu, T., Ecker, J. R., and Nordborg, M. (2019). Common alleles of CMT2 and NRPE1 are major determinants of CHH methylation variation in Arabidopsis thaliana. PLoS Genet. 15:e1008492. doi: 10.1371/journal.pgen.1008492
Scascitelli, M., Whitney, K. D., Randell, R. A., King, M., Buerkle, C. A., and Rieseberg, L. H. (2010). Genome scan of hybridizing sunflowers from Texas (Helianthus annuus and H. debilis) reveals asymmetric patterns of introgression and small islands of genomic differentiation. Mol. Ecol. 19, 521–541. doi: 10.1111/j.1365-294x.2009.04504.x
Schild, T., Low, V., Blenis, J., and Gomes, A. P. (2018). Unique metabolic adaptations dictate distal organ-specific metastatic colonization. Cancer Cell 33, 347–354. doi: 10.1016/j.ccell.2018.02.001
Schlaepfer, M. A., Sax, D. F., and Olden, J. D. (2011). The potential conservation value of non-native species. Conserv. Biol. J. Soc. Conserv. Biol. 25, 428–437. doi: 10.1111/j.1523-1739.2010.01646.x
Schmid-Siegert, E., Sarkar, N., Iseli, C., Calderon, S., Gouhier-Darimont, C., Chrast, J., et al. (2017). Low number of fixed somatic mutations in a long-lived oak tree. Nat. Plants 3, 926–929. doi: 10.1038/s41477-017-0066-9
Schoen, D. J., and Schultz, S. T. (2019). Somatic mutation and evolution in plants. Annu. Rev. Ecol. Evol. Syst. 50, 49–73. doi: 10.1146/annurev-ecolsys-110218-024955
Schrey, A. W., Alvarez, M., Foust, C. M., Kilvitis, H. J., Lee, J. D., Liebl, A. L., et al. (2013). Ecological epigenetics: beyond MS-AFLP. Integr. Compar. Biol. 53, 340–350. doi: 10.1093/icb/ict012
Selechnik, D., Richardson, M. F., Shine, R., DeVore, J. L., Ducatez, S., and Rollins, L. A. (2019). Increased adaptive variation despite reduced overall genetic diversity in a rapidly adapting invader. Front. Genet. 10:1221.
Shan, S., Boatwright, J. L., Liu, X., Chanderbali, A. S., Fu, C., Soltis, P. S., et al. (2020). Transcriptome dynamics of the inflorescence in reciprocally formed allopolyploid Tragopogon miscellus (Asteraceae). Front. Genet. 11:888.
Sharov, A. A., Leonard, D., Liebhold, A. M., Roberts, E. A., and Dickerson, W. (2002). “Slow The Spread”: a national program to contain the Gypsy moth. J. Forestry 100, 30–36.
Shi, W., Chen, X., Gao, L., Xu, C.-Y., Ou, X., Bossdorf, O., et al. (2018). Transient stability of epigenetic population differentiation in a clonal invader. Front. Plant Sci. 9:1851.
Shine, R., Brown, G. P., and Phillips, B. L. (2011). An evolutionary process that assembles phenotypes through space rather than through time. Proc. Natl. Acad. Sci. USA 108, 5708–5711. doi: 10.1073/pnas.1018989108
Short, J., Bradshaw, S. D., Giles, J., Prince, R. I. T., and Wilson, G. R. (1992). Reintroduction of macropods (Marsupialia: Macropodoidea) in Australia—a review. Biol. Conserv. 62, 189–204. doi: 10.1016/0006-3207(92)91047-v
Simberloff, D. (2009). The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 40, 81–102. doi: 10.1146/annurev.ecolsys.110308.120304
Simberloff, D., Martin, J.-L., Genovesi, P., Maris, V., Wardle, D. A., Aronson, J., et al. (2013). Impacts of biological invasions: what’s what and the way forward. Trends Ecol. Evol. 28, 58–66. doi: 10.1016/j.tree.2012.07.013
Snyder, M. R., and Stepien, C. A. (2017). Genetic patterns across an invasion’s history: a test of change versus stasis for the Eurasian round goby in North America. Mol. Ecol. 26, 1075–1090. doi: 10.1111/mec.13997
Somarelli, J. A. (2021). The hallmarks of cancer as ecologically driven phenotypes. Front. Ecol. Evol. 9:226.
Sommer, R. J. (2020). Phenotypic plasticity: from theory and genetics to current and future challenges. Genetics 215, 1–13. doi: 10.1534/genetics.120.303163
Sotka, E. E., Baumgardner, A. W., Bippus, P. M., Destombe, C., Duermit, E. A., Endo, H., et al. (2018). Combining niche shift and population genetic analyses predicts rapid phenotypic evolution during invasion. Evol. Appl. 11, 781–793. doi: 10.1111/eva.12592
Sottoriva, A., Barnes, C. P., and Graham, T. A. (2017). Catch my drift? Making sense of genomic intra-tumour heterogeneity. Biochim. Biophys. Acta Rev. Cancer 1867, 95–100. doi: 10.1016/j.bbcan.2016.12.003
Srivastava, S., Ghosh, S., Kagan, J., and Mazurchuk, R. (2018). The PreCancer Atlas (PCA). Trends Cancer 4, 513–514. doi: 10.1016/j.trecan.2018.06.003
Stajic, D., and Jansen, L. (2021). Empirical evidence for epigenetic inheritance driving evolutionary adaptation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. doi: 10.1098/rstb.2020.0121
Stapley, J., Santure, A. W., and Dennis, S. R. (2015). Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol. Ecol. 24, 2241–2252. doi: 10.1111/mec.13089
Stepien, C. A., Brown, J. E., Neilson, M. E., and Tumeo, M. A. (2005). Genetic diversity of invasive species in the Great lakes versus their Eurasian source populations: insights for risk analysis. Risk Anal. 25, 1043–1060. doi: 10.1111/j.1539-6924.2005.00655.x
Stewart, N. C., Tranel, P. J., Horvath, D. P., Anderson, J. V., Rieseberg, L. H., Westwood, J. H., et al. (2009). Evolution of weediness and invasiveness: charting the course for weed genomics. Weed Sci. 57, 451–462. doi: 10.1614/ws-09-011.1
Stratton, M. R. (2011). Exploring the genomes of cancer cells: progress and promise. Science 331, 1553–1558. doi: 10.1126/science.1204040
Strong, D. R., and Ayres, D. R. (2013). Ecological and evolutionary misadventures of spartina. Annu. Rev. Ecol. Evol. Syst. 44, 389–410. doi: 10.1146/annurev-ecolsys-110512-135803
Stuart, K. C., Cardilini, A. P. A., Cassey, P., Richardson, M. F., Sherwin, W. B., Rollins, L. A., et al. (2020). Signatures of selection in a recent invasion reveal adaptive divergence in a highly vagile invasive species. Mol. Ecol. 30, 1419–1434. doi: 10.1111/mec.15601
Stupack, D. G., Teitz, T., Potter, M. D., Mikolon, D., Houghton, P. J., Kidd, V. J., et al. (2006). Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature 439, 95–99. doi: 10.1038/nature04323
Sultan, S. E. (2015). Organism and Environment: Ecological Development, Niche Construction, and Adaptation. Oxford: Oxford University Press.
Sun, Y., Beuchat, C., and Müller-Schärer, H. (2020). Is biocontrol efficacy rather driven by the plant or the antagonist genotypes? a conceptual bioassay approach. Working Paper Series 63, 81–10062. doi: 10.3897/neobiota.63.54962
Szczurek, E., Krüger, T., Klink, B., and Beerenwinkel, N. (2020). A mathematical model of the metastatic bottleneck predicts patient outcome and response to cancer treatment. PLoS Comput. Biol. 16:e1008056. doi: 10.1371/journal.pcbi.1008056
Tepolt, C. K., and Palumbi, S. R. (2020). Rapid adaptation to temperature via a potential genomic island of divergence in the invasive green crab, Carcinus maenas. Front. Ecol. Evol. 8:411.
Teschendorff, A. E., and Feinberg, A. P. (2021). Statistical mechanics meets single-cell biology. Nat. Rev. Genet. 22, 459–476. doi: 10.1038/s41576-021-00341-z
Theoharides, K. A., and Dukes, J. S. (2007). Plant invasion across space and time: factors affecting nonindigenous species success during four stages of invasion. New Phytol. 176, 256–273. doi: 10.1111/j.1469-8137.2007.02207.x
Thuiller, W., Gallien, L., Boulangeat, I., De Bello, F., Münkemüller, T., Roquet, C., et al. (2010). Resolving Darwin’s naturalization conundrum: a quest for evidence: resolving Darwin’s naturalization conundrum. Divers. Distrib. 16, 461x–475x.
Timp, W., and Feinberg, A. P. (2013). Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510. doi: 10.1038/nrc3486
Tissot, T., Massol, F., Ujvari, B., Alix-Panabieres, C., Loeuille, N., and Thomas, F. (2019). Metastasis and the evolution of dispersal. Proc. R. Soc. B Biol. Sci. 286:20192186186.
Tsherniak, A., Vazquez, F., Montgomery, P. G., Weir, B. A., Kryukov, G., Cowley, G. S., et al. (2017). Defining a cancer dependency map. Cell 170, 564–576.e16.
Turajlic, S., Xu, H., Litchfield, K., Rowan, A., Chambers, T., Lopez, J. I., et al. (2018). Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e12.
Van Buskirk, J., and Steiner, U. K. (2009). The fitness costs of developmental canalization and plasticity. J. Evol. Biol. 22, 852–860. doi: 10.1111/j.1420-9101.2009.01685.x
Van de Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
van Kleunen, M., Bossdorf, O., and Dawson, W. (2018). The ecology and evolution of alien plants. Annu. Rev. Ecol. Evol. Syst. 49, 25–47. doi: 10.1146/annurev-ecolsys-110617-062654
van Kleunen, M., Dawson, W., and Maurel, N. (2015). Characteristics of successful alien plants. Mol. Ecol. 24, 1954–1968. doi: 10.1111/mec.13013
van Riemsdijk, I., Arntzen, J. W., Bucciarelli, G., McCartney-Melstad, E., Rafajlović, M., Scott, P. A., et al. (2020). Spatial variation in introgression along a toad hybrid zone in France. Biorxiv [preprint] doi: 10.1101/746073
van Riemsdijk, I., van Nieuwenhuize, L., Martínez-Solano, I., Arntzen, J. W., and Wielstra, B. (2018). Molecular data reveal the hybrid nature of an introduced population of banded newts (Ommatotriton) in Spain. Conserv. Genet. 19, 249–254. doi: 10.1007/s10592-017-1004-0
van Steenderen, C. J. M., Paterson, I. D., Edwards, S., and Day, M. D. (2021). Addressing the red flags in cochineal identification: the use of molecular techniques to identify cochineal insects that are used as biological control agents for invasive alien cacti. Biol. Control Theory Appl. Pest Manag. 152:104426. doi: 10.1016/j.biocontrol.2020.104426
Vandepitte, K., de Meyer, T., Helsen, K., van Acker, K., Roldán-Ruiz, I., Mergeay, J., et al. (2014). Rapid genetic adaptation precedes the spread of an exotic plant species. Mol. Ecol. 23, 2157–2164. doi: 10.1111/mec.12683
VanWallendael, A., Alvarez, M., and Franks, S. J. (2020). Patterns of population genomic diversity in the invasive Japanese knotweed species complex. Am. J. Bot. 108, 857–886. doi: 10.1002/ajb2.1653
Verhoeven, K. J. F., and Preite, V. (2014). Epigenetic variation in asexually reproducing organisms. Evolution 68, 644–655. doi: 10.1111/evo.12320
Verhoeven, K. J. F., Jansen, J. J., Van Dijk, P. J., and Biere, A. (2010). Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185, 1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x
Vittecoq, M., Giraudeau, M., Sepp, T., Marcogliese, D. J., Klaassen, M., Renaud, F., et al. (2018). Turning natural adaptations to oncogenic factors into an ally in the war against cancer. Evol. Appl. 11, 836–844. doi: 10.1111/eva.12608
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A., and Kinzler, K. W. (2013). Cancer genome landscapes. Science 339, 1546–1558.
Vonholdt, B. M., Pollinger, J. P., Lohmueller, K. E., Han, E., Parker, H. G., Quignon, P., et al. (2010). Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902. doi: 10.1038/nature08837
Wegner, K. M., Lokmer, A., and John, U. (2020). Genomic and transcriptomic differentiation of independent invasions of the pacific oyster Crassostrea gigas. Front. Ecol. Evol. 8:371.
Weisenberger, D. J., Siegmund, K. D., Campan, M., Young, J., Long, T. I., Faasse, M. A., et al. (2006). CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 38, 787–793. doi: 10.1038/ng1834
Wells, A., Grahovac, J., Wheeler, S., Ma, B., and Lauffenburger, D. (2013). Targeting tumor cell motility as a strategy against invasion and metastasis. Trends Pharmacol. Sci. 34, 283–289. doi: 10.1016/j.tips.2013.03.001
Wendel, J. F., Jackson, S. A., Meyers, B. C., and Wing, R. A. (2016). Evolution of plant genome architecture. Genome Biol. 17:37.
West-Eberhard, M. J. (2005). Developmental plasticity and the origin of species differences. Proc. Natl. Acad. Sci. USA 102(Suppl. 1), 6543–6549. doi: 10.1073/pnas.0501844102
Whitney, K. D., Broman, K. W., Kane, N. C., Hovick, S. M., Randell, R. A., and Rieseberg, L. H. (2015). Quantitative trait locus mapping identifies candidate alleles involved in adaptive introgression and range expansion in a wild sunflower. Mol. Ecol. 24, 2194–22114. doi: 10.1111/mec.13044
Williams, M. J., Sottoriva, A., and Graham, T. A. (2019). Measuring clonal evolution in cancer with genomics. Annu. Rev. Genomics Hum. Genet. 20, 309–329. doi: 10.1146/annurev-genom-083117-021712
Xie, Z., Wang, L., Wang, L., Wang, Z., Lu, Z., Tian, D., et al. (2016). Mutation rate analysis via parent-progeny sequencing of the perennial peach. I. a low rate in woody perennials and a higher mutagenicity in hybrids. Proc. Biol. Sci. 283:20161016. doi: 10.1098/rspb.2016.1016
Xu, G., Lyu, J., Li, Q., Liu, H., Wang, D., Zhang, M., et al. (2020). Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat. Commun. 11:5539. doi: 10.1038/s41467-020-19333-4
Yap, T. A., Omlin, A., and de Bono, J. S. (2013). Development of therapeutic combinations targeting major cancer signaling pathways. J. Clin. Oncol. 31, 1592–1605. doi: 10.1200/jco.2011.37.6418
Yates, L. R., Knappskog, S., Wedge, D., Farmery, J. H. R., Gonzalez, S., Martincorena, I., et al. (2017). Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32, 169–184.e705.
Yoder, A. D., and Tiley, G. P. (2021). The challenge and promise of estimating the de novo mutation rate from whole-genome comparisons among closely related individuals. Mol. Ecol. doi: 10.1111/mec.16007
Yoo, M.-J., Liu, X., Pires, J. C., Soltis, P. S., and Soltis, D. E. (2014). Nonadditive gene expression in polyploids. Annu. Rev. Genet. 48, 485–517. doi: 10.1146/annurev-genet-120213-092159
Yu, M., Stott, S., Toner, M., Maheswaran, S., and Haber, D. A. (2011). Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 192, 373–382. doi: 10.1083/jcb.201010021
Zahir, N., Sun, R., Gallahan, D., Gatenby, R. A., and Curtis, C. (2020). Characterizing the ecological and evolutionary dynamics of cancer. Nat. Genet. 52, 759–767. doi: 10.1038/s41588-020-0668-4
Zhang, Y. M., Vitone, T. R., Storer, C. G., Payton, A. C., Dunn, R. R., Hulcr, J., et al. (2019). From pavement to population genomics: characterizing a long-established non-native ant in north america through citizen science and ddRADseq. Front. Ecol. Evol. 7:453.
Zhang, Y.-Y., Parepa, M., Fischer, M., and Bossdorf, O. (2016). “Epigenetics of colonizing species? a study of japanese knotweed in central Europe,” in Invasion Genetics: The Baker and Stebbins Legacy, eds S. Barrett, R. I. Colautti, K. M. Dlugosch, and L. H. Rieseberg (Hoboken, NJ: Wiley-Blackwell), 328–340. doi: 10.1002/9781119072799.ch19
Keywords: clonal diversity, epigenomics and epigenetics, genomics, metastasis, non-genetic inheritance, invasion biology, invasive species
Citation: Neinavaie F, Ibrahim-Hashim A, Kramer AM, Brown JS and Richards CL (2021) The Genomic Processes of Biological Invasions: From Invasive Species to Cancer Metastases and Back Again. Front. Ecol. Evol. 9:681100. doi: 10.3389/fevo.2021.681100
Received: 15 March 2021; Accepted: 08 July 2021;
Published: 22 September 2021.
Edited by:
Sarah R. Amend, Johns Hopkins Medicine, United StatesReviewed by:
Aurora Mihaela Nedelcu, University of New Brunswick Fredericton, CanadaJavad Noorbakhsh, Broad Institute, United States
Copyright © 2021 Neinavaie, Ibrahim-Hashim, Kramer, Brown and Richards. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina L. Richards, Y2xyQHVzZi5lZHU=
 Fargam Neinavaie
Fargam Neinavaie Arig Ibrahim-Hashim
Arig Ibrahim-Hashim Andrew M. Kramer
Andrew M. Kramer Joel S. Brown
Joel S. Brown Christina L. Richards
Christina L. Richards