- Department of Animal Behaviour, Bielefeld University, Bielefeld, Germany
The sirtuin protein family plays a role in the lifespan of various species and is involved in numerous key metabolic processes. To understand the evolutionary role of sirtuins in marmots, a long-living rodent species group with remarkable metabolic shutdown during hibernation, we conducted a phylogeny-based substitution rate analysis of coding genes based on genetic information of seven marmot species. We show that sirtuin 1 (SIRT1) has evolved under positive selection in the marmot lineage. We pinpoint three amino acid changes in four different marmot species that underlie the signal of positive selection and that may favor increased longevity in marmots. Based on a computational structural analysis we can show that all three substitutions affect the secondary structure of the same region in human SIRT1. We propose that the identified region is close to the catalytic domain and that the potential structural changes may impact the catalytic activity of the enzyme and therefore might be playing a functional role in marmot's extended lifespan and metabolic shutdown.
1. Introduction
Maximum lifespans of mammals differ greatly between species (Kowalczyk et al., 2020). Although the rodent species evolved a tenfold variability in lifespan (Gorbunova et al., 2014), the underlying molecular and evolutionary mechanisms are still little understood. Studies on ageing processes have identified the highly conserved sirtuin family (Zhao et al., 2020), whose effect on lifespan was first demonstrated in yeasts and is suspected to have a similar influence in mammals (Haigis and Sinclair, 2010).
The mammalian sirtuins have seven members (SIRT1 to SIRT7) (Frye, 2000) and are a protein family of nicotinamide adenine dinucleotide (NAD+)-dependent deacylases (Haigis et al., 2006). While they have a conserved catalytic core domain in common (Frye, 2000), they differ by cellular localization, functions, and substrates (Maissan et al., 2021). The best studied protein of this family is SIRT1 which is predominately a nuclear protein and acts as a transcription regulator and deacetylates certain histones such as H3, H4, and H1 (Toiber et al., 2011), but also modifies more than 50 non-histone proteins (Nakagawa and Guarente, 2014), such as p53 (Luo et al., 2001). Generally, it influences energy metabolism, cell survival, DNA repair, and tissue regeneration as well as numerous other important cellular processes (Bonkowski and Sinclair, 2016). Like SIRT1, SIRT6, and SIRT7 are located in the nucleus and involved in histone modification. While SIRT6 controls the expression of multiple glycolytic genes and is a regulator of glucose homeostasis (Zhong et al., 2010), SIRT7 is a positive regulator of RNA polymerase transcription and is linked to cell proliferation (Ford et al., 2006). In contrast, SIRT2 is primarily located in the cytoplasm where it regulates various metabolic enzymes. However, under stress and during mitosis, it also migrates into the cell nucleus where it regulates the cell cycle and cell differentiation (Vaquero et al., 2006). The other mammalian sirtuins SIRT3, SIRT4, and SIRT5 are mitochondrially localized. There, SIRT3 and SIRT5 influence a variety of metabolic enzymes (e.g., in fatty acid oxidation) (Verdin et al., 2010), while SIRT4 is active in amino acid metabolism by inhibiting glutamate dehydrogenase (Haigis et al., 2006).
In addition to the functions already mentioned, sirtuins, especially SIRT1, have been investigated for their role in caloric restriction (Finkel et al., 2009; Derous et al., 2021). It has long been known that calorie restriction significantly delays the aging process and extends life span in a variety of animal species, including rodents, and has been shown to reduce the incidence of age-related disorders (e.g., diabetes, cancer, and cardiovascular disease) in mammals (Bordone and Guarente, 2005). In humans, the association between SIRT1 expression and caloric restriction (Civitarese et al., 2007) as well as an age-related increase in SIRT1 expression intensity (Kilic et al., 2015) has been demonstrated. Furthermore show transgenic mice with brain-specific SIRT1 overexpression a significant delay of the aging process and an extension of life span (Satoh et al., 2013).
Since sirtuins play an important role in aging processes it has been postulated that the evolution of sirtuins may be associated with the heterogeneity of maximum lifespan between species (Tian et al., 2017). Despite the numerous analyses of sirtuin functions, the insights of evolutionary processes of this important protein family is fairly limited. Currently existing studies mainly focus on model organisms, such as mice (Vanhooren and Libert, 2013). Exceptionally, a recent study pointed out amino acid substitutions in beaver SIRT6 relative to mouse SIRT6 leading to lifespan extension (Tian et al., 2019). Based on these promising results we were intrigued whether positive selection might have been played a role in the evolution of sirtuin protein members in other rodent species. A sequence-based analysis of positive selection might reveal particular amino acid with potential impact on sirtuin catalytic activity and may explain an association with longevity. As the sirtuins are associated with dietary restricted longevity here we focused on a species group that experiences long periods of dietary restriction in form of deep hibernation and conducted a phylogeny-based substitution rate analysis based on genetic information of seven marmot species (Cardini and O'Higgins, 2004). We find that sirtuin 1 (SIRT1) has evolved under positive selection in the marmot lineage.
2. Materials and Methods
2.1. Data Retrieval
For the analysis of the seven mammalian sirtuin protein family members SIRT1 to SIRT7 we obtained coding DNA sequences, including several isoforms or different prediction entries from NCBI Genbank (Clark et al., 2015). The sequences were retrieved for human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), American beaver (Castor canadensis), and guinea pig (Cavia porcellus). The coding DNA sequence for SIRT5 for rats could not be retrieved. Additionally, we obtained some sirtuin coding DNA sequences for marmots from Genbank, including sequences of SIRT1, SIRT3, SIRT5, and SIRT6 for yellow-bellied marmot (Marmota flaviventris) and the sequence of SIRT 5 for Alpine marmot (Marmota marmota marmota, Gossmann et al., 2019). A complete list of retrieved sirtuin sequences along with their respective identifiers can be seen in Supplementary Table 1 (see Supplementary Material). To conduct an analysis tailored to marmots, additional publically available coding sequences for SIRT1 to SIRT7 homologs were predicted. For this, coding sequences were predicted from genomic data based on sequence homology to the available Refseq sirtuin marmot sequences using Exonerate v2.2.0 (Slater and Birney, 2005). The genomes of the woodchuck (Marmota monax), the Himalayan marmot (Marmota himalayana) and the Vancouver Island marmot (Marmota vancouverensis) were retrieved from Genbank. Furthermore, coding sequences for the Mongolian marmot (Marmota mongolia), long-tailed marmot (Marmota caudata) and Altai marmot (Marmota baibacina) could be obtained by using re-sequencing information (Bai et al., 2019) (http://www.marmotdb.org/). Specifically, coding information were obtained by using the vcf's information obtained from mapping short read data onto the Himalayan reference genome by Bai et al. (2019). Each species for which sequence data was used was included in the phylogenetic tree that was provided obtained from Timetree (Kumar et al., 2017).
2.2. Sequence Preparation and Selection Analysis
To prepare the obtained sequences a customized python pipeline was designed based on available scripts from Afanasyeva et al. (2018). In brief, it contained general automated preparation steps, e.g., renaming data files, adjusting identifier information and storing the coding sequences for each sirtuin SIRT1 to SIRT7 from each species. Note, that this pipeline could be easily extended to handle large scale data, such as entire proteomes or across large trees (Afanasyeva et al., 2018; Yusuf et al., 2020). Then, each coding DNA sequence was translated into the corresponding protein sequence using the standard genetic code. All sequences containing a stop codon within the sequence were disregarded for further analysis. The translated sequences were then aligned using MUSCLE v3.8 using default parameter settings (Edgar, 2004). These alignments were used for a manual inspection and limited to the selection of one sequence per species and protein. This was necessary, because alignment artifacts may artificially create a signature of positive selection (Afanasyeva et al., 2018). Criterions for the exclusions of sequences included sequences with missing fragments in comparison to the human canonical sequence as well as sequences with potentially erroneous intron exon boundaries. The identifier of the selected sequences which were retrieved from Genbank are listed in Supplementary Table 2, while the selected sequences of the predicted sirtuin sequences for SIRT1 are shown in Supplementary Table 3 (Supplementary Material).
Alignments of the selected and translated sequences where further processed using PRANK v.170427 using default parameter settings (Loytynoja, 2013) and critical residues were masked with ZORRO (Wu et al., 2012) when the confidence score was smaller than nine. Combined with the previous coding DNA sequences the results were used as input for PAL2NAL v14 (Suyama et al., 2006) to create coding DNA sequence alignments. Thereby, output files with renamed identifier were created as input for PAML (Yang, 2007). Prepared codeml files and phylogenetic tree files pruned down to the number of available proteins were then used as input for codeml of the PAML package.
To conduct a selection analysis based on substitution rates, codeml from the PAML package version 4.9j was used. Two model types were considered. First, a branch model comparison was executed to determine heterogeneity in dN/dS between the marmot clade and the rest of the tree (two-ratio model) relative to a model where a single dN/dS ratio (one-ratio model) was assumed. Secondly, branch-site comparisons were used to test for the evidence of positive selection within the marmot clade. We compared the branch-site model A to a model where a potentially positively selected site category was restricted to drift (e.g., dN/dS = 1). Furthermore, the site models model 1 (M1a, NearlyNeutral, 2 categories) (Wong et al., 2004; Yang et al., 2005) and model 22 (M2a_rel, PositiveSelection, 3 categories) (Weadick and Chang, 2012), that allow the ω ratio to vary among sites (among codons or amino acids in the protein), were compared to a Clade model C (Bielawski and Yang, 2004).
2.3. Selection Model Comparison
The codeml-generated output files were further processed using a customized python script. For the branch test comparison dN/dS values were extracted from the one ratio and two ratio models and twice the log likelihood difference was assumed to be χ2 distributed with one degree of freedom. Same accounts for the three different branch-site model comparisons. Here, the dN/dS values were extracted for model A, model A fixed, model=1 (M1a) and model=22 (M2a_rel), as well as for Clade model C. Again, twice the log likelihood difference of model A against model A fixed (with ω = 1) was assumed to be χ2 distributed with one degree of freedom. For model M1a against Clade model C d.f.=3 was used while one degree of freedom was used for model=22 (M2a_rel) against Clade model C. Furthermore, the NEB (Naïve Empirical Bayes) and BEB (Bayes Empirical Bayes) values were extracted from rst-files from the codeml output for data plotting.
2.4. Structural Prediction
A 3D model was calculated using the SWISS-MODEL (Guex et al., 2009) (with access on SWISS-MODEL Interactive Workspace, n.d.) a template for human SIRT1 protein structure from Protein Data Bank (4I5I) (Zhao et al., 2013) and the coding sequence of human SIRT1. With FirstGlance (FirstGlance in Jmol, n.d.) the model was visually processed. The influence of the substitution of selected amino acids regarding the secondary structure of the protein was analyzed using Jpred4 (Drozdetskiy et al., 2015) as well as Porter 5.0 (Torrisi et al., 2019) submitting the protein sequence of human SIRT1 in comparison to the same sequence with single amino acid substitution [with substitution of lysin (K) to threonine (T) in amino acid sites 499, serine (S) to leucin (L) in amino acid sites 508 and substitution of arginine (R) to isoleucine (I) in amino acid sites 613] deducted from the marmot sequences of M. sibirica, M. himalayana and M. caudata, and M. flaviventris, respectively.
3. Results
3.1. Branch Test: Differences in Evolutionary Rates in Marmot Clade
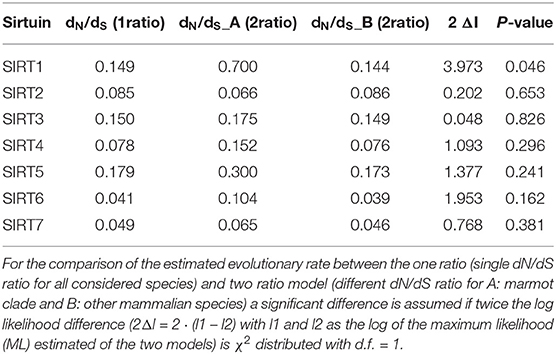
Differences in the evolutionary rates, recovered from between-species comparisons, may reveal molecular footprints of selective pressures. Here we investigated heterogeneity in the rate of molecular evolution of sirtuins between the marmots and other mammalian species (Figure 1).
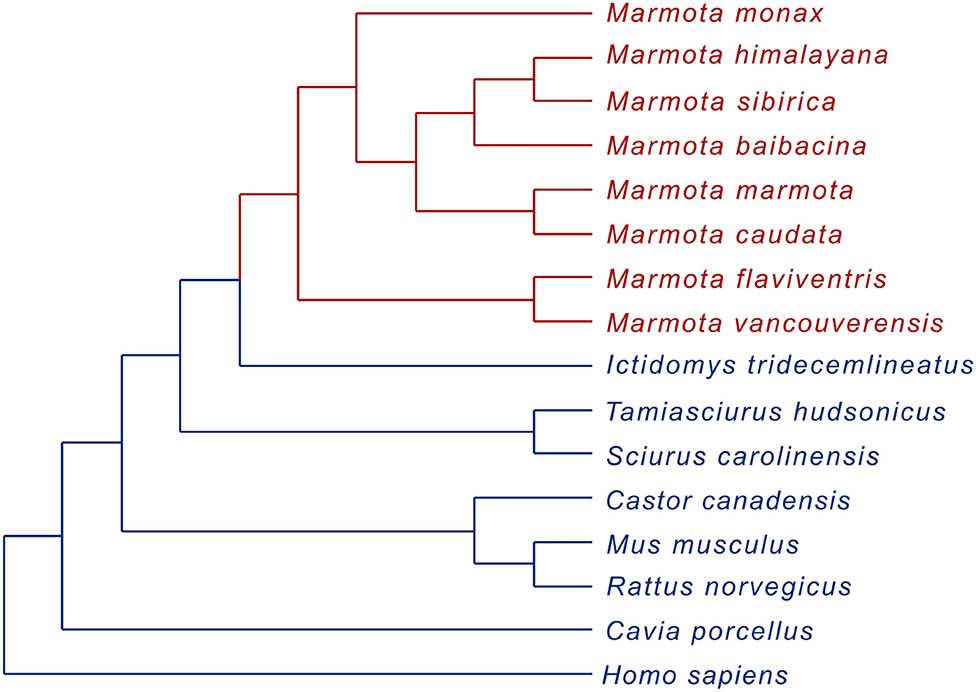
Figure 1. A cladogram of the phylogenetic relationship of the species used for substitution rate analyses, divided into A: marmot clade (red) and B: other mammalian species (blue).
The ratio of non-synonymous to synonymous substitutions ω = dN/dS was used as a measure for the rate of molecular evolution at the protein level. The dN/dS ratio indicates the driving selective forces in the evolutionary process, as elevated dN/dS ratios above one suggest the influence of positive selection (ω > 1) while a low ratio (ω <1) reveals a process driven mainly by negative selection (Gossmann and Schmid, 2011). The comparison of the estimated evolutionary rate between the one ratio (single dN/dS ratio for all considered species) and two ratio model (different dN/dS ratio for marmot clade) revealed higher dN/dS ratios in the marmot clade relative to the compared species for six of the seven sirtuin proteins (SIRT1, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7) (Table 1).
Indeed, we find a significant difference of the dN/dS ratios between the marmot clade and the remaining tree for SIRT1. This shows that the marmot clade has a different evolutionary rate regarding SIRT1 than the remaining branches of the phylogenetic tree. The explanation for the higher dN/dS ratios could be relaxed purifying selection, but it could also suggest a possible influence of positive selection specific to marmots.
3.2. Branch-Site Test: SIRT1 Repeatedly Evolved Under Positive Selection
Since the branch test revealed an increased rate of molecular evolution in the marmot clade relative to other mammalian species, a possible role of positive selection events was further analyzed. To investigate positive selection in the evolution of SIRT1 in the marmot clade three branch-site model comparisons were executed. The branch-site model A was compared to the model A fixed where a potentially positively selected site category was restricted to drift (with dN/dS = 1). Furthermore, Clade C was compared to model 1 (M1a, NearlyNeutral) and also to model 22 (M2a_rel, PositiveSelection). All three comparisons revealed a significant difference (Figure 2), indicating the occurrence of positive selection events within the marmot clade.
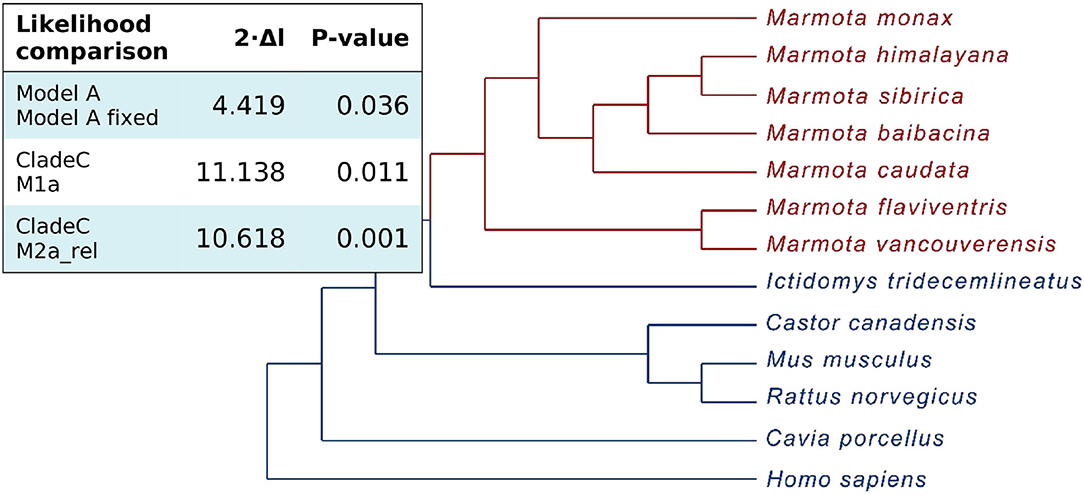
Figure 2. Results of branch-site model comparisons for SIRT1 protein coding genes with a cladogram of the phylogenetic relationship of the species used for analyses, divided into A: marmot clade (red) and B: other mammalian species (blue).
A significant difference for the three different likelihood comparisons is assumed if twice the log likelihood difference (2Δl = 2 · (l1 – l2) with l1 and l2 as the log of the maximum likelihood (ML) estimated of two models) is χ2 distributed. A likelihood comparisons between the branch-site model A and model A fixed, where a potentially positively selected site category was restricted to drift (e.g., dN/dS = 1) was performed to test for the evidence of positive selection within the marmot clade (χ2 distribution with d.f. = 1). Furthermore, results of a likelihood comparisons between Clade model C (Bielawski and Yang, 2004) and site models model 1 (M1a, NearlyNeutral, 2 categories) (Wong et al., 2004; Yang et al., 2005) (χ2 distribution with d.f. = 3) and a comparison between Clade model C and model 22 (M2a_rel, PositiveSelection (3 categories)) (Weadick and Chang, 2012) (χ2 distribution with d.f. = 1) differed significantly.
To identify individual amino acid substitutions that were likely promoted by positive selection, we determined every amino acid's probability to belong to a class with either low (dN/dS <1), neutral (dN/dS = 1) or a high dN/dS ratio (dN/dS > 1). As the different models revealed similar results, we focused on model A results. The gained NEB (Naive Emperical Bayes) probabilities show three amino acid sites with high posterior probabilities (> 75%) for site class 2 with ω = dN/dS = 1.68 (Figure 3), indicating that the footprints of positive selection can be pinpointed to particular amino-acid replacements. Similar results have been obtained for posterior probabilities based on BEB (Bayes empirical Bayes, Figure 3) probabilities. This approach accounts for sampling error (Yang et al., 2005) illustrating that the amino acids sites underlying the signal of positive selection can be robustly identified.
Figure 3. Quantitative representation of the Naive Empirical Bayesian (NEB) and Bayes Emperical Bayesian (BEB) class probabilities derived from the branch-site model comparison as a test for positive selection. The amino acid sites respective to human SIRT1 protein sequence fit into the classes with certain dN/dS ratios with the given posterior probabilities. Posterior probabilities for site class 2 indicate the probability for positive selection and are shown in red.
The three outlined amino acid sites presumably have undergone positive selection. The alignment of the analyzed sequences for SIRT1 shows the amino acid sites and the corresponding substitutions within the marmot clade (Figure 4). In respective to coding sequence of human SIRT1 the amino acid sites are 499 with substitution of lysin (K) to threonine (T) in M. sibirica, 508 with substitution of serine (S) to leucin (L) in M. himalayana and M. caudata and 613 with substitution of arginine (R) to isoleucine (I) in M. flaviventris.
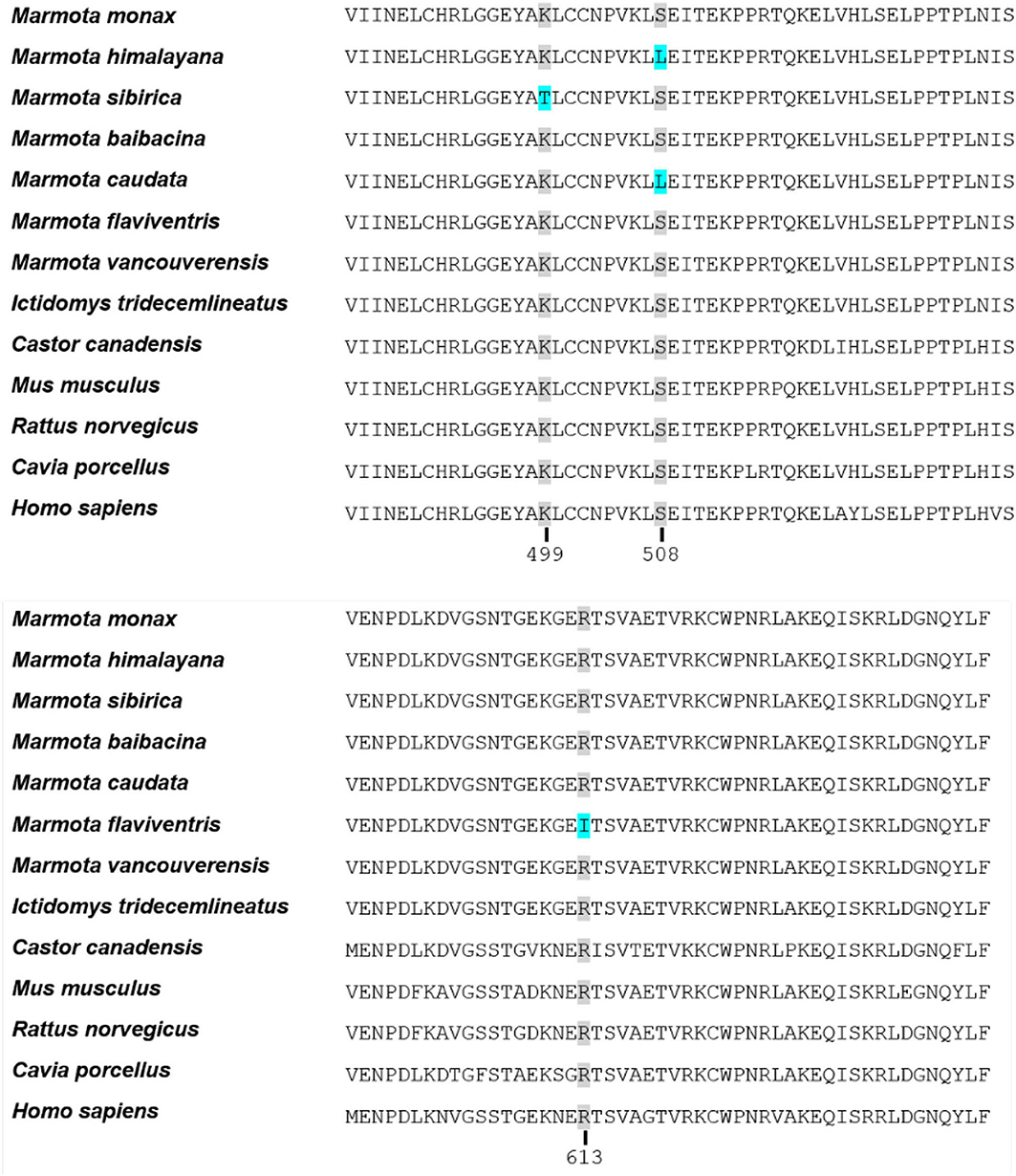
Figure 4. Relevant sections of the alignment of SIRT1 proteins from 13 mammals including seven different marmots. Substitutions with the signature of positive selection (analyzed by branch site model comparison with codeml of the PAML package) are highlighted in blue (amino acid sites 499, 508, and 613). The Alignment was performed with MUSCLE.
3.3. Secondary Structure Prediction
Since we identified substitutions that presumably evolved under positive selection, we were interested whether we could identify a possible impact of these residues on the structural level. To investigate this effect further, a secondary structure prediction of human SIRT1 protein was performed along with in-silico mutated variants of the human protein, substituting the three residues we identified from the marmot species. Intriguingly, we find that a differently predicted substructure within the amino acid sites 518 to 523 of the human SIRT1 sequence is observed for each of the 3 residues (Figure 5). Interestingly the secondary structure of this 6 amino acid long sequence stretch can be predicted only with low confidence, but it is embedded in a coiled coiled region that can be confidently predicted. The observed effect on the secondary structure is also remarkable, because one of the mutations is 270 bp (90 aa) away from the affected amino acid stretch. While the human sequence is predicted with a helix structure for the mentioned amino acid sites, the sequences with the single substitutions each are predicted with a predominantly coiled structure instead. We conducted control mutations where we randomly substituted amino acid residues nearby the identified positions and did not find such an effect on the structure prediction for serine and arginine (Supplementary Figures 3E–G). We find that changes of nearby lysin residues to the observed mutations had a similar impact that lead to a coiled-coiled prediction in the target region (Supplementary Figures 3H–J).
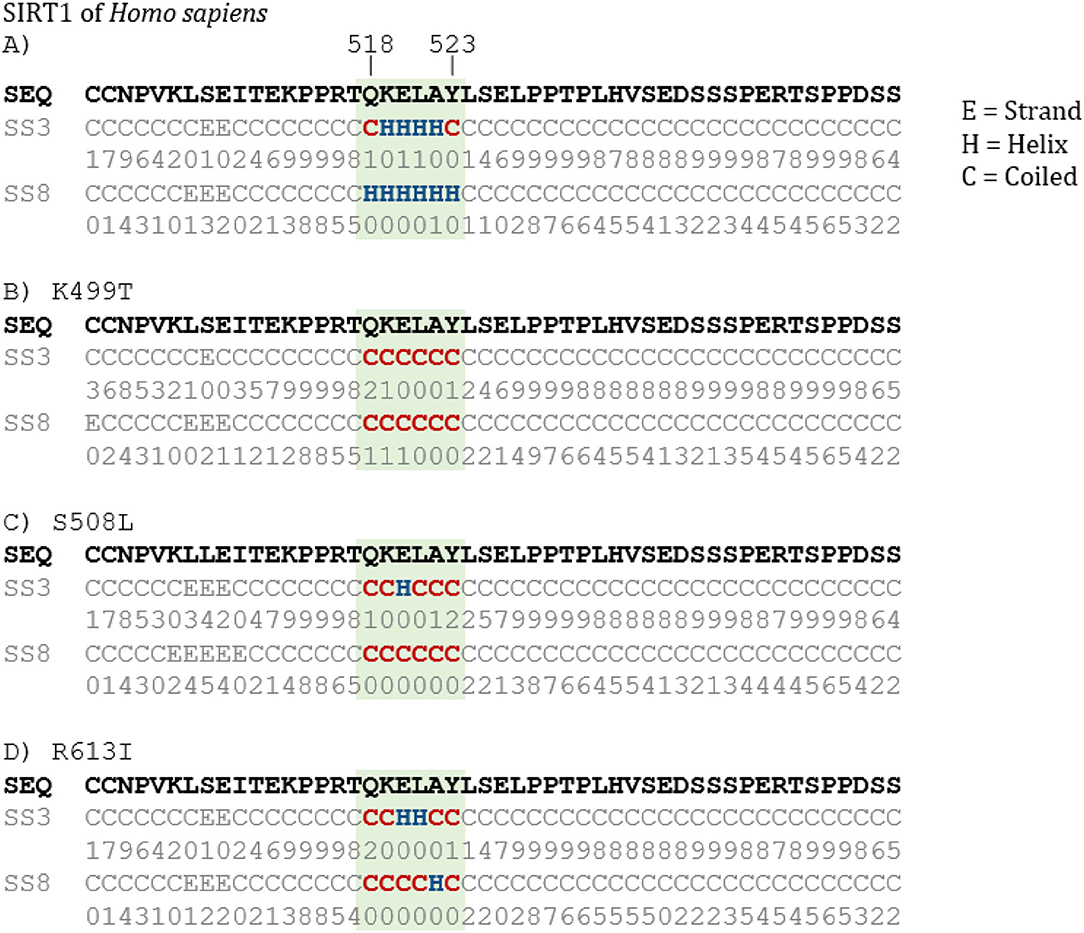
Figure 5. Relevant section of secondary structure prediction. Protein sequences for secondary structure prediction are (A) human SIRT1 protein sequence, and human SIRT1 protein sequences with substitution of (B) lysin (K) to threonine (T) in amino acid site 499, (C) serine (S) to leucin (L) in amino acid site 508, (D) arginine (R) to isoleucine (I) in amino acid site 613. Predictions were performed with Porter 5.0 (Torrisi et al., 2019). Secondary structures are divided into three classes: E, Strand; H, Helix; C, Coil; SS3, 3 class secondary structure; SS8, 8 class secondary structure, each followed by the line of confidence values for 3 and 8 class predictions, respectively (with 9, maximal confidence; 0, very little confidence).
This structural difference may alter the enzyme function. As Zhao et al. (2013) reported the catalytic domain of SIRT1 reaches up to the residue 516. Their estimated structure (shown in Supplementary Figure 1) which ends with the amino acid E 512 suggests a very close position of the subsequence QKELAY with amino acid sites 518 to 523 to the substrate bounded in the catalytic domain. Assuming a coiled structure for the subsequence (as predicted in Figure 5) instead of the shown strand in Supplementary Figure 1 it is conceivable that the subsequence QKELAY might have an influence on the substrate affinity.
4. Discussion
The association of sirtuins and dietary restricted longevity has been at the center of ageing research for more than a decade (Dali-Youcef et al., 2007; Guarente, 2007; Imai, 2007). Nevertheless, little is known about the role of molecular adaptations of sirtuins especially in non-model animals experiencing long periods of dietary restriction under natural conditions. These studies hardly deviate from the usual models, such as yeast, mouse and human, leading to a limited amount of insights (Vanhooren and Libert, 2013; Mitchell et al., 2015). Our analyses, as well as work of Tian et al. (2019) with highlighted results found in beaver SIRT6 protein, underline the importance of considering animals outside the common model organisms. To understand the influence of sirtuins on dietary restricted longevity, it is important to compare the different species with their different phenotypic characteristics regarding dietary restriction and longevity. Learning about the correlation of the genomic differences and phenotypic characteristics helps discovering the genetic basis for a long life span. In order to gain such important insights, species with extreme characteristics in the respective area, such as in this case animals with extreme dietary restriction, should be especially considered.
Here, we report differences in sirtuin evolution in the marmot clade relative to other mammalian species. We find a significantly increased rate of molecular evolution for one of the sirtuin family members, SIRT1. Increased rates of dN/dS might stem from relaxed purifying selection, or reflect an overall lower effective population size in the marmot lineage (Gossmann et al., 2019). We tested a third hyporthesis that could drive this pattern: positive selection. Indeed, We identify three amino acid substitutions occurring in four specific marmot species but not in humans or the common model organism mouse. Interestingly, for two species, M. himalayana and M. caudata the same amino acid substitution may have appeared independently or may stem from common ancient genetic variation, illustrating the possibility of parallel evolution. The fact that significant differences were observed for three branch-site model comparisons underlines the robustness of the analysis and demonstrates the independence of the result from the model assumptions.
With the huge difference in dietary restriction between these species the results outline the importance of an even broader analysis. Our phylogeny-based substitution rate analysis presented here can easily be extended to sequences of many other species to learn more about sirtuins and their evolution. The investigation of other deep hibernators such as dormice, chipmunks, hamsters, fat-tailed lemurs, bats and hedgehogs (Geiser, 2011) could provide further findings. For this gaining further insights depend on the genome sequences of additional species. Large scale genome projects such as the Bat1K (Teeling et al., 2018) will allow for more phylogenetic analyses in the future. Furthermore, population genetic analyses can be performed to better understand the recent evolutionary mechanisms underlying lineage specific mutations.
As experimentally shown in the work of Tian et al. (2019), the discovered amino acid substitutions have an influence on lifespan, demonstrated by swapping the critical amino acids of mouse and beaver SIRT6. Similar experiments using mouse models and inserting the amino acid substitutions highlighted in this work could verify the effect on molecular mechanisms and life span. The protein changes observed are in regions of human Sirt1 involved in NAD binding and a potential phosphorylation site at residue 530 (Shan et al., 2017; Ling et al., 2018). It would be intriguing to investigate further whether this could indicate the regulation of the Sirt1 interaction networks (Elibol and Kilic, 2018) being under selection. Due to lack of quality sequencing data of the annotated marmot genomes that were used in this study, we could not analyse the region that is homologous to the N-terminus of human/mouse Sirt1. Since the SIRT1 N-terminal region that has been shown to interact with histone proteins (Vaquero et al., 2004) future investigation could shed further light on the functional implications of evolutionary transitions in SIRT1.
In conclusion, this work demonstrates the ability of phylogenetic studies to determine the signature of molecular adaptation, especially when looking at a range of species with specific life-history features, which is enabled by large quantities of genome information and the use of computational approaches. However we note, that studies on a comparable scale involving other species clades will be crucial to shed light on the evolutionary signatures of extreme life styles to deduct the mechanistic underpinnings of metabolic adaptation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
NJ and TG contributed to conception and design of the study. NJ performed the analysis and wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank three reviewers for their helpful input that improved the quality of the manuscript. We acknowledge the financial support of the German Research Foundation (DFG) and the Open Access Publication Fund of Bielefeld University for the article processing charge.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.666564/full#supplementary-material
References
Afanasyeva, A., Bockwoldt, M., Cooney, C. R., Heiland, I., and Gossmann, T. I. (2018). Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. 28, 975–982. doi: 10.1101/gr.232645.117
Bai, L., Liu, B., Ji, C., Zhao, S., Liu, S., Wang, R., et al. (2019). Hypoxic and cold adaptation insights from the himalayan marmot genome. iScience 11, 519–530. doi: 10.1016/j.isci.2018.11.034
Bielawski, J. P., and Yang, Z. (2004). A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol, 59, 121–132. doi: 10.1007/s00239-004-2597-8
Bonkowski, M. S., and Sinclair, D. A. (2016). Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690. doi: 10.1038/nrm.2016.93
Bordone, L., and Guarente, L. (2005). Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 6, 298–305. doi: 10.1038/nrm1616
Cardini, A., and O'Higgins, P. (2004). Patterns of morphological evolution in marmota (Rodentia, Sciuridae): Geometric morphometrics of the cranium in the context of marmot phylogeny, ecology and conservation. Biol. J. Linnean Soc. 82, 385–407. doi: 10.1111/j.1095-8312.2004.00367.x
Civitarese, A. E., Carling, S., Heilbronn, L. K., Hulver, M. H., Ukropcova, B., Deutsch, W. A., et al. (2007). Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 4:e76. doi: 10.1371/journal.pmed.0040076
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Sayers, E. W. (2015). GenBank. Nucleic Acids Res. 44, D67–D72. doi: 10.1093/nar/gkv1276
Dali-Youcef, N., Lagouge, M., Froelich, S., Koehl, C., Schoonjans, K., and Auwerx, J. (2007). Sirtuins: The ‘magnificent seven,' function, metabolism and longevity. Ann. Med. 39, 335–345. doi: 10.1080/07853890701408194
Derous, D., Sahu, J., Douglas, A., Lusseau, D., and Wenzel, M. (2021). Comparative genomics of cetartiodactyla: energy metabolism underpins the transition to an aquatic lifestyle. Conserv. Physiol. 9:coaa136. doi: 10.1093/conphys/coaa136
Drozdetskiy, A., Cole, C., Procter, J., and Barton, G. J. (2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, 389–394. doi: 10.1093/nar/gkv332
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elibol, B., and Kilic, U. (2018). High levels of SIRT1 expression as a protective mechanism against disease-related conditions. Front. Endocrinol. 9:614. doi: 10.3389/fendo.2018.00614
Finkel, T., Deng, C. X., and Mostoslavsky, R. (2009). Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591. doi: 10.1038/nature08197
Ford, E., Voit, R., Liszt, G., Magin, C., Grummt, I., and Guarente, L. (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075–1080. doi: 10.1101/gad.1399706
Frye, R. A. (2000). Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798. doi: 10.1006/bbrc.2000.3000
Geiser, F. (2011). “Hibernation: endotherms,” in eLS (Chichester: John Wiley & Sons, Ltd). doi: 10.1002/9780470015902.a0003215.pub2
Gorbunova, V., Seluanov, A., Zhang, Z., Gladyshev, V. N., and Vijg, J. (2014). Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat. Rev. Genet. 15, 531–540. doi: 10.1038/nrg3728
Gossmann, T. I., and Schmid, K. J. (2011). Selection-driven divergence after gene duplication in arabidopsis thaliana. J. Mol. Evol. 73, 153–165. doi: 10.1007/s00239-011-9463-2
Gossmann, T. I., Shanmugasundram, A., Borno, S., Duvaux, L., Lemaire, C., Kuhl, H., et al. (2019). Ice-age climate adaptations trap the alpine marmot in a state of low genetic diversity. Curr. Biol. 29, 1712.e7–1720.e7. doi: 10.1016/j.cub.2019.04.020
Guarente, L. (2007). Sirtuins in aging and disease. Cold Spring Harbor Symp. Quant. Biol. 72, 483–488. doi: 10.1101/sqb.2007.72.024
Guex, N., Peitsch, M. C., and Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, S162–S173. doi: 10.1002/elps.200900140
Haigis, M. C., Mostoslavsky, R., Haigis, K. M., Fahie, K., Christodoulou, D. C., Murphy, A. J. J., et al. (2006). SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954. doi: 10.1016/j.cell.2006.06.057
Haigis, M. C., and Sinclair, D. A. (2010). Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295. doi: 10.1146/annurev.pathol.4.110807.092250
Imai, S. (2007). Is Sirt1 a miracle bullet for longevity? Aging Cell 6, 735–737. doi: 10.1111/j.1474-9726.2007.00344.x
Kilic, U., Gok, O., Erenberk, U., Dundaroz, M. R., Torun, E., Kucukardali, Y., et al. (2015). A remarkable age-related increase in SIRT1 protein expression against oxidative stress in elderly: SIRT1 gene variants and longevity in human. PLoS ONE 10:e0117954. doi: 10.1371/journal.pone.0117954
Kowalczyk, A., Partha, R., Clark, N. L., and Chikina, M. (2020). Pan-mammalian analysis of molecular constraints underlying extended lifespan. eLife 9:51089. doi: 10.7554/elife.51089
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Ling, H., Peng, L., Wang, J., Rahhal, R., and Seto, E. (2018). Histone deacetylase SIRT1 targets Plk2 to regulate centriole duplication. Cell Rep. 25, 2851.e3–2865.e3. doi: 10.1016/j.celrep.2018.11.025
Loytynoja, A. (2013). Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170. doi: 10.1007/978-1-62703-646-7_10
Luo, J., Nikolaev, A. Y., Imai, S., ichiro Chen, D., Su, F., Shiloh, A., et al. (2001). Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148. doi: 10.1016/S0092-8674(01)00524-4
Maissan, P., Mooij, E., and Barberis, M. (2021). Sirtuins-mediated system-level regulation of mammalian tissues at the interface between metabolism and cell cycle: a systematic review. Biology 10:194. doi: 10.3390/biology10030194
Mitchell, S. J., Scheibye-Knudsen, M., Longo, D. L., and Cabo, R. de (2015). Animal models of aging research: implications for human aging and age-related diseases. Annu. Rev. Anim. Biosci. 3, 283–303. doi: 10.1146/annurev-animal-022114-110829
Nakagawa, T., and Guarente, L. (2014). Targets of sirtuins NAD metabolism snapshot: sirtuins, NAD, and aging. Cell Metab. 20, 192.e1–192.e1. doi: 10.1016/j.cmet.2014.06.001
Satoh, A., Brace, C. S., Rensing, N., Cliften, P., Wozniak, D. F., Herzog, E. D., et al. (2013). Sirt1 extends life span and delays aging in mice through the regulation of Nk2 Homeobox 1 in the DMH and LH. Cell Metab. 18, 416–430. doi: 10.1016/j.cmet.2013.07.013
Shan, P., Fan, G., Sun, L., Liu, J., Wang, W., Hu, C., et al. (2017). SIRT1 functions as a negative regulator of eukaryotic poly(a)RNA transport. Curr. Biol. 27, 2271.e5–2284.e5. doi: 10.1016/j.cub.2017.06.040
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
Teeling, E. C., Vernes, S. C., Davalos, L. M., Ray, D. A., Gilbert, M. T. P., and Myers, E. (2018). Bat biology, genomes, and the Bat1K project: to generate chromosome-level genomes for all living bat species. Annu. Rev. Anim. Biosci. 6, 23–46. doi: 10.1146/annurev-animal-022516-022811
Tian, X., Firsanov, D., Zhang, Z., Cheng, Y., Luo, L., Tombline, G., et al. (2019). SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 177, 622.e22–638.e22. doi: 10.1016/j.cell.2019.03.043
Tian, X., Seluanov, A., and Gorbunova, V. (2017). Molecular mechanisms determining lifespan in short- and long-lived species. Trends Endocrinol. Metab. 28, 722–734. doi: 10.1016/j.tem.2017.07.004
Toiber, D., Sebastian, C., and Mostoslavsky, R. (2011). Characterization of nuclear sirtuins: Molecular mechanisms and physiological relevance. Handb. Exp. Pharmacol. 206, 189–224. doi: 10.1007/978-3-642-21631-2_9
Torrisi, M., Kaleel, M., and Pollastri, G. (2019). Deeper Profiles and Cascaded Recurrent and Convolutional Neural Networks for state-of-the-art Protein Secondary Structure Prediction. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-48786-x
Vanhooren, V., and Libert, C. (2013). The mouse as a model organism in aging research: usefulness, pitfalls and possibilities. Ageing Res. Rev. 12, 8–21. doi: 10.1016/j.arr.2012.03.010
Vaquero, A., Scher, M., Lee, D., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2004). Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16, 93–105. doi: 10.1016/j.molcel.2004.08.031
Vaquero, A., Scher, M. B., Dong, H. L., Sutton, A., Cheng, H. L., Alt, F. W., et al. (2006). SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20, 1256–1261. doi: 10.1101/gad.1412706
Verdin, E., Hirschey, M. D., Finley, L. W. S., and Haigis, M. C. (2010). Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669–675. doi: 10.1016/j.tibs.2010.07.003
Weadick, C. J., and Chang, B. S. W. (2012). An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol. Biol. Evol. 29, 1297–1300. doi: 10.1093/molbev/msr311
Wong, W. S. W., Yang, Z., Goldman, N., and Nielsen, R. (2004). Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168, 1041–1051. doi: 10.1534/genetics.104.031153
Wu, M., Chatterji, S., and Eisen, J. A. (2012). Accounting for alignment uncertainty in phylogenomics. PLoS ONE 7:e30288. doi: 10.1371/journal.pone.0030288
Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S. W., and Nielsen, R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yusuf, L., Heatley, M. C., Palmer, J. P. G., Barton, H. J., Cooney, C. R., and Gossmann, T. I. (2020). Noncoding regions underpin avian bill shape diversification at macroevolutionary scales. Genome Res. 30, 553–565. doi: 10.1101/gr.255752.119
Zhao, L., Cao, J., Hu, K., He, X., Yun, D., Tong, T., et al. (2020). Sirtuins and their biological relevance in aging and age-related diseases. Aging Dis. 11:927. doi: 10.14336/ad.2019.0820
Zhao, X., Allison, D., Condon, B., Zhang, F., Gheyi, T., Zhang, A., et al. (2013). The 2.5 Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD +) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J. Med. Chem. 56, 963–969. doi: 10.1021/jm301431y
Keywords: non-model species, dietery restriction, hibernation, positive selection, 3D structure
Citation: Junker N and Gossmann TI (2021) Adaptation-Driven Evolution of Sirtuin 1 (SIRT1), a Key Regulator of Metabolism and Aging, in Marmot Species. Front. Ecol. Evol. 9:666564. doi: 10.3389/fevo.2021.666564
Received: 10 February 2021; Accepted: 08 June 2021;
Published: 02 July 2021.
Edited by:
Patricia Izar, University of São Paulo, BrazilReviewed by:
Patrice Corneli, The University of Utah, United StatesJohannes Rack, University of Oxford, United Kingdom
Leon Hilgers, Centre for Molecular Biodiversity Research, Zoological Research Museum Alexander Koenig, Germany
Copyright © 2021 Junker and Gossmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toni I. Gossmann, dG9uaS5nb3NzbWFubkBnbWFpbC5jb20=
 Nora Junker
Nora Junker Toni I. Gossmann
Toni I. Gossmann