- 1Centre for Chemical Biology, Universiti Sains Malaysia, Bayan Lepas, Malaysia
- 2School of Biological Sciences, Universiti Sains Malaysia, Minden, Malaysia
- 3Department of Aquaculture, Faculty of Marine and Fisheries, Universitas Syiah Kuala, Banda Aceh, Indonesia
- 4Faculty of Mathematics and Natural Sciences, Universitas Mulawarman, Samarinda, Indonesia
Introduction
Miniaturization, or the evolution of small size, is widely seen in vertebrate species and best documented in amphibians and fishes (Weitzman and Vari, 1988; Hanken, 1993; Hanken and Wake, 1993). The highly acidic blackwater peat swamps in Southeast Asia are hotspots for an exceptionally large number of miniature fishes (Kottelat et al., 2006). Among these, members of the genus Paedocypris (Paedocypris progenetica, Paedocypris micromegethes, and Paedocypris carbunculus) display the most intriguing form of miniaturization with maximum adult sizes 10–12 mm in standard length (Kottelat et al., 2006; Britz and Kottelat, 2008). Miniaturization in Paedocypris stems from an event known as progenetic paedomorphosis or developmental truncation, which results in adult forms resembling the larval stage of its close relatives (Britz et al., 2014). The miniaturized phenotypes represent unique combinations of ancestral and derived traits such as reduction and structural simplification, morphological novelty, and increased morphological variability (Hanken, 1993; Hanken and Wake, 1993). With more than 3,000 species, Cyprinidae (Teleostei: Ostariophysi: Cypriniformes) is the most species-rich family of vertebrates, distributed in freshwaters throughout North America, Africa, and Eurasia (Nelson, 2006). Paedocypris fish are located within the subfamily Danioninae, which also includes two other paedomorphic groups, the Sundadanio and Danionella (Fang et al., 2009; Tang et al., 2010).
There has been considerable interest in resolving the phylogenetic position of Paedocypris, using morphology features (Britz and Conway, 2009; Britz et al., 2014), a combination of nuclear and or mitochondrial markers (Fang et al., 2009; Mayden and Chen, 2010; Tang et al., 2010, 2013; Hirt et al., 2017) or individual markers alone (Ruber et al., 2007). Consequently, Paedocypris was suggested to be within the Danioninae subfamily (Ruber et al., 2007; Fang et al., 2009; Tang et al., 2010, 2013), still within Cyprinidae but not within Danioninae (Yang et al., 2015), or as a lineage sister to Cypriniformes (Mayden and Chen, 2010; Stout et al., 2016; Hirt et al., 2017). These differences could be due to the simplified anatomical structure and some highly derived autamorphic characters of Paedocypris which complicates morphological classification (Britz and Conway, 2009). Base compositional heterogeneity is known to affect phylogenetic reconstruction. High molecular evolution rates may undermine the inference of the phylogenetic position of miniaturized taxa due to issues associated with long-branch attraction (Hirt et al., 2017).
The mitochondrial DNA (mtDNA) has been widely used for phylogenetic, molecular evolution and phylogeography studies because of its maternal inheritance, compact gene organization, low recombination frequency, and rapid evolutionary rate (Miya et al., 2003; Gissi et al., 2008). The circular mitogenomes of fish, having similar composition and structure to that of most vertebrates, are 15–20 Kb in size with 37 genes: 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), control region, and origin of L-strand replication (Boore, 1999; Satoh et al., 2016). The mitochondrial gene set, including 13 PCGs and two rRNAs, has been consistent used as markers to improve classification or to resolve higher-level relationships of fish species (Mullens et al., 2020; Sun et al., 2021). Mitogenome, with higher informative sites than shorter sequences is now a widely used tool to resolve deep branching lineages at the level of many taxa levels (Boore et al., 2005).
In view of the unresolved phylogenetic position of the Paeocypris genus and paucity of knowledge on the mitogenome content of miniaturized animals, the complete mitochondrial genomes of P. micromegethes and P. carbunculus were sequenced, assembled, and annotated. This enables the use of 13 mitochondrial PCGs and two rRNAs from complete mitochondrial genome to study the phylogenetic relationship of Paedocypris. In addition, a comparative mitogenome analysis with other miniature fish, Danioninae and Cyprinidae species was also carried out.
Materials and Methods
Sample Collection, DNA Extraction, and Mitogenome Sequencing
Specimens of P. micromegethes and P. carbunculus were collected from the peat swamp forests located at Sarawak, Malaysia (01° 34.78067′ N, 110° 16.04233′ E) and Kalimantan Tengah, Indonesia (2° 02.21977′ S, 113° 46.27098′ E), respectively. The specimens were deposited at the Zoological Reference Museum Collection, Universiti Sains Malaysia [Voucher number: USMFC(95)00001 (P. micromegethes) and USMFC(95)00002 (P. carbunculus)]. All experimental procedures complied with the current animal ethics guidelines and were approved by the USM Institutional Animal Care and Use Committee (USM IACUC). Total genomic DNA was extracted from the whole fish using the Genomic-tip 100/G kit (Qiagen, Germany). The DNA quality was assessed by gel electrophoresis and the DNA concentration was measured using the Qubit® 2.0 Fluorometer (Life Technologies, USA). Paired-end libraries were prepared for P. micromegethes and P. carbunculus, respectively using the TruSeq DNA PCR-free Library Prep kit (Illumina, USA). The constructed libraries were sequenced on a Hiseq platform (Illumina, USA) using 2 × 150 bp paired-end reads.
Mitochondrial Genome Assembly, Annotation, and Analyses
8.5 million reads totaling 1.3 Gb were used for the mitochondrial genome assemblies of P. micromegethes and P. carbunculus, respectively. The complete mitogenomes were assembled in MITObim v1.9 (Hahn et al., 2013) with the sequence of a closely-related species, P. progenetica, as reference (NC_020436). The assembled mitogenomes of P. micromegethes and P. carbunculus were annotated in the MitoAnnotator (Iwasaki et al., 2013) and MITOS (Bernt et al., 2013) web servers. The secondary structures of tRNAs were inferred using the mitochondrial tRNA finder as implemented in MITOS and visualized on the Forna webserver (Kerpedjiev et al., 2015). The secondary structures of the origin of L-strand replication and control region were analyzed using mfold software v3.6 (Zuker, 2003). The strand asymmetry was calculated according to the formulas: AT-skew = (A–T)/(A+T) and GC-skew = (G–C)/(G+C) (Perna and Kocher, 1995). The codon usage and relative synonymous codon usage for the PCGs were analyzed with MEGAX (Kumar et al., 2018) using the vertebrate mitochondrial genetic code. The ratios of non-synonymous (Ka) and synonymous substitution (Ks) of the PCGs were estimated by KaKs_Calculator v2.0 (Zhang et al., 2006) with genetic code 2 and γ-MYN model. The Ka and Ks values were based on pairwise comparisons between the four Paedocypris species and Sundadanio rubellus (AP011401, in GenBank listed under the trade name “Sundadanio axelrodi “red””), as outgroup. Sundadanio is the genus most closely related to Paedocypris.
Phylogenetic Analyses
The phylogenetic relationships were reconstructed based on the nucleotide sequences of 13 PCGs and two rRNA genes from 75 Cyprinidae. Along with P. micromegethes and P. carbunculus, sequences of selected Danioninae, Acheilognathinae, Culturinae, Xenocyprinae, Squaliobarbinae, Opsariichthys, Leuciscinae species were used for the phylogenetic analyses, and Gobioninae were used as the outgroups (Supplementary Table 1). The alignment was performed using MUSCLE, followed by alignment trimming using trimAl v1.2 and then concatenation of the trimmed sequences into a supermatrix file with FASconCAT v1.11. The best-fit model for the alignment was estimated with jModelTest v2.1.10 based on the Akaike Information Criterion. The concatenated nucleotide alignment was used to perform maximum likelihood (ML) phylogenetic analysis in RaxML software (Stamatakis, 2014) with the GTR+ G + I model and 1,000 bootstraps. For Bayesian inference (BI), the analysis was conducted with MrBayes v3.2.3 (Huelsenbeck and Ronquist, 2001) using the same supermatrix file in nexus format and running with the same model (GTR+ G + I). Four Markov chain Monte Carlo (MCMC) chains were run for 1 million generations with sampling every 100 generations, and the first 25% of the trees were discarded as burn-in. Tracer v1.7.1 was used to assess the chain convergence. FigTree v1.4.2 was used to visualize and edit the phylogenetic trees.
Results and Discussion
Genome Organization, Structure, and Composition
The mitochondrial genomes of P. micromegethes and P. carbunculus are circular DNA molecules of 17,208 bp and 17,280 bp in length, respectively (Figure 1A), well within the range of a typical vertebrate mitogenome size of 15–20 Kb. The Paedocypris mitogenomes also possess a complete gene set identical to that of the vertebrate mitogenomes (Boore, 1999), comprising 13 PCGs, 22 tRNA genes, two rRNA genes [rrnS (12S rRNA) and rrnL (16S rRNA)], control region and origin of L-strand replication (Supplementary Table 2). Our analysis of the mitochondrial genome size and content revealed that there are no significant changes in these two characteristics in the miniature fish mitogenomes compared to vertebrate mitogenomes. The complete mtDNA sequences of Paedocypris, on the other hand, are the largest in length of sequences of the mitogenomes in Danioninae examines (Supplementary Table 3). Since the length is conserved in PCGs, tRNAs, and rRNAs, the differences in mitogenome length are mainly attributed to variations in the control region. The mitochondrial control region has a higher evolutionary rate and tends to be under weaker purifying selection than protein-coding genes, allowing the region to accumulate length variations more readily (Tang et al., 2006; Resch et al., 2007). The nucleotide compositions of the complete mtDNA sequences of the Paedocypris are biased toward A and T, as has been noted in other Danioninae mitogenomes (Broughton et al., 2001; Chang et al., 2013; Kusuma and Kumazawa, 2016). In order to evaluate the base bias of the nucleotide composition, we measured skewness in the different gene regions of P. micromegethes and P. carbunculus mitogenomes and found that the AT-skew values were mostly positive, whereas the GC-skew values were mostly negative (Supplementary Table 4).
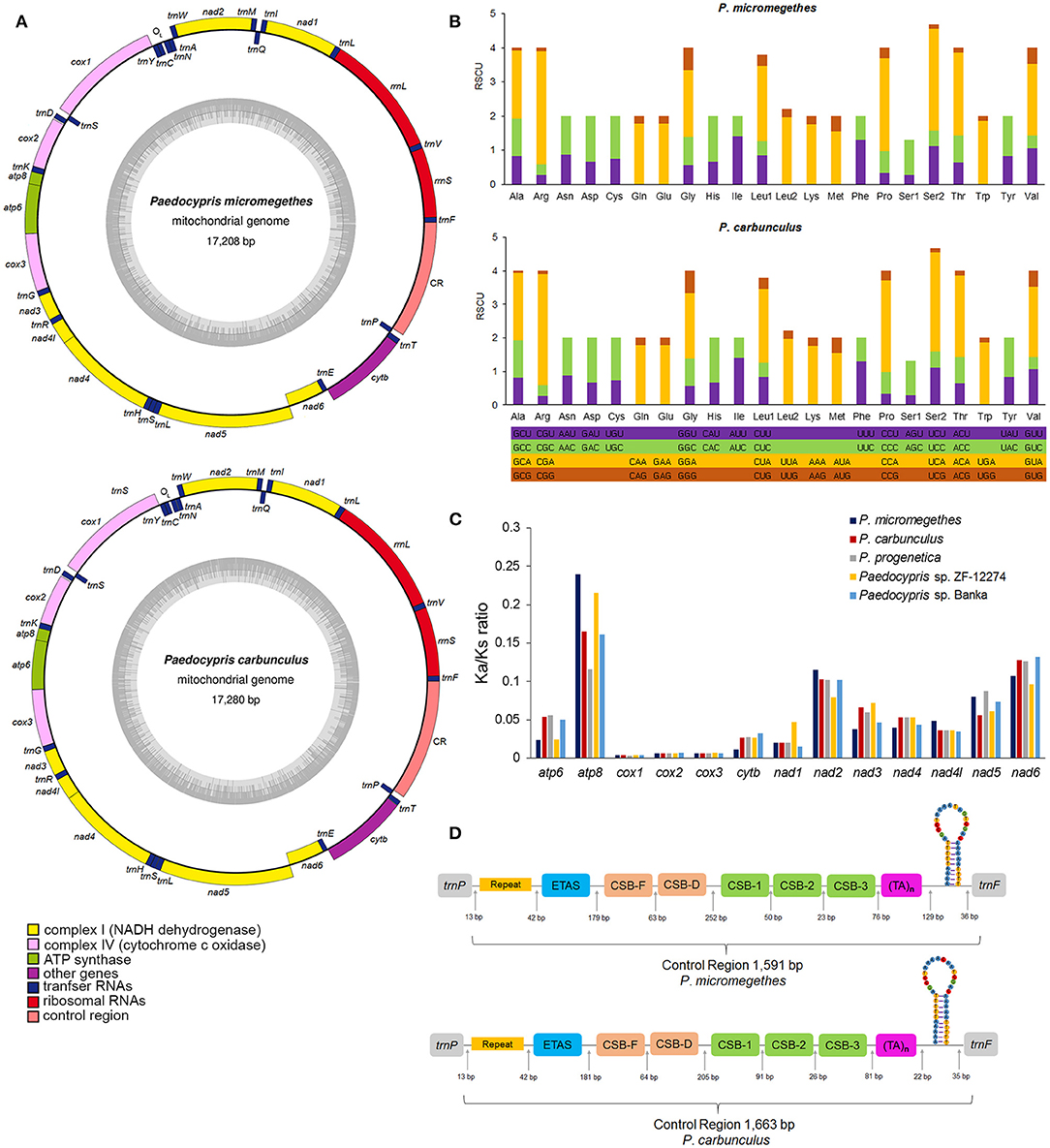
Figure 1. The complete mitochondrial genomes of P. micromegethes and P. carbunculus. (A) Circular map of the P. micromegethes and P. carbunculus mitogenomes. (B) Relative synonymous codon usage of protein-coding genes in P. micromegethes and P. carbunculus mitogenomes. (C) The ratio of non-synonymous and synonymous substitution (Ka/Ks) estimated in all 13 mitochondrial protein-coding genes of Paedocypris species. (D) Control region of P. micromegethes and P. carbunculus mitogenomes. The structural elements were displayed in different colors.
Protein-Coding Genes and Codon Usage
Most Paedocypris mitochondrial PCGs are transcribed from the positive strand of the molecules except for nad6 on the negative strand. All PCGs use start codon ATG except cox1 which uses start codon GTG. Seven PCGs (nad1, cox1, atp8, atp6, na4l, nad4, and nad5) ended with a complete and canonical stop codon (TAA or TAG). The genes cox2, cox3, nad2, and nad3 were found to have a truncated stop codon T. The presence of truncated stop codon is a common phenomenon among metazoan mitochondrial genes (Sheffield et al., 2010), and the truncated stop codon is hypothesized to be completed by post-transcriptional poly-adenylation (Ojala et al., 1981).
The relative synonymous codon usage values for the third position of the 13 PCGs are summarized in Figure 1B. The most frequently used codons found in the PCGs of Paedocypris were CGA (Arg), CCA (Pro), and UCA (Ser); ACG (Thr), CAG (Gln), and GCG (Ala) being rarely used. There was a bias in favor of A/T in the third position rather than G/C, as almost all frequently used codons ended with A/T. The A+T bias present in the third position of Paedocypris PCGs is consistent with the A+T bias in whole mitogenomes. To investigate the selective pressure across the Paedocypris species, the Ka/Ks ratios for the PCGs of each mitogenome were estimated (Figure 1C). The Ka/Ks values for all 13 protein-coding genes were <1, indicating the existence of purifying selection in these species.
Transfer RNAs, Ribosomal RNAs, and Control Region
The complete set of 22 tRNA genes typical of metazoan mitogenomes was identified in P. micromegethes and P. carbunculus. Most of the tRNAs could be folded into classic cloverleaf structures, except for trnC and trnS1 (Supplementary Figure 1). In trnC, the loop of the pseudoridine arm (T-arm) was missing, while trnS1 lacked the stem of the T-arm. The function of these aberrant tRNA genes might be complemented by coevolved interacting factors or post-transcriptional RNA-editing (Ohtsuki et al., 2002; Chimnaronk et al., 2005). The large ribosomal rrnL and small rrnS present in the mitogenomes of P. micromegethes and P. carbunculus were 1,672–1,673 and 947–949 nucleotides long, respectively, and were located close to each other between trnF and trnL1 but separated by trnV.
A single long intergenic space of 1,591–1,663 bp located between trnP and trnF in the mitogenomes of P. micromegethes and P. carbunculus is recognized as the control region (Figure 1D). Multiple copies of 34-bp long tandem repeats “TGGTATAGTGCATATTATGCTTAATACTACATAG” were detected at the 5′ end of this region (Supplementary Figure 2), contributing to a greater control region length in Paedocypris compared to other Danioninae. The secondary structure prediction analysis revealed possible folding configuration featuring a stem-loop structure near the 3′ end of this non-coding sequence. Paedocypris control region also contains conserved blocks including extended termination associated sequences (ETAS), central conserved domains (CSB-F and CSB-D), and conserved sequence block domains (CSB-1, CSB-2, and CSB-3). CSB-E reported in another teleost CSB domain (Lee et al., 1995) was not present in Paedocypris, indicating that the regulatory mechanism of transcription may be varied in the species. The origin of L-strand replication displaying a stem and loop structure was identified in a cluster of five tRNA genes (WANCY region) as in other cyprinid fishes (Broughton et al., 2001; Wei et al., 2016, Yu et al., 2019).
Gene Arrangements
The pattern of genome arrangement in miniature fish still remain elusive. Here, we analyzed the mitochondrial gene arrangement in representative miniature cyprinids (Paedocypris, Sundadanio, and Danionella), Danioninae (Danio, Horadandia, Boraras, Trigonostigma, and Rasbora), and Cyprinidae (Acheilognathus, Ischikauia, Xenocypris, Squaliobarbus, Opsariichthys, Leuciscus, and Gobio) species (Supplementary Figure 3). Comparative analysis revealed that the mitochondrial gene organization in these miniature taxa shows high stability, with the independent miniaturization in these lineages not influencing the mitochondrial gene order. The same mitochondrial gene order is found across the Danioninae subfamily and the Cyprinidae family. The shared mitochondrial gene order among miniature fish, Danioninae and Cyprinidae species is indicative of common ancestry.
Phylogenetic Analyses
The phylogenetic position of Paedocypris was analyzed based on the combined mitochondrial gene set of 13 PCGs and two rRNAs (Figure 2 and Supplementary Figure 4). The topologies of the ML and BI trees were mostly congruent, except for the position of Acheilognathinae. Based on the ML analysis, the 75 Cyprinidae species could be divided into five major clades corresponding to the subfamilies Danioninae, Acheilognathinae, [((Culturinae + Xenocyprinae) + Squaliobarbibinae) + Opsariichthys group], Leuciscinae, and Gobioninae. In the BI tree, subfamilies of Danioninae, [((Culturinae + Xenocyprinae) + Squaliobarbibinae) + Opsariichthys group], (Leuciscinae + Acheilognathinae), and Gobioninae were found, with Acheilognathinae clustering with Leuciscinae. The recognition of the Danioninae subfamily in its current usage follows Fang et al. consideration of Danioninae as the senior synonym of the cyprinid lineage that includes Danio and its closest relatives (Fang et al., 2009). Our mitogenome analyses confirm the previous exclusion of ex- Danioninae taxa (Candida, Nipponocypris, Opsariichthys, Parazacco, and Zacco) that are not part of Danioninae sensu stricto and have been reassigned to the Opsariichthys group (Liao et al., 2011; Huang et al., 2017). The 28 Danioninae species were paraphyletic, forming two clades in the phylogenetic trees. The first clade contains 21 species from the genera Rasbora, Trigonostigma, Boraras, Horadandia, Danio, and Danionella. The genus Paedocypris comprising five species, P. progenetica, P. carbunculus, Paedocypris sp. ZF-12274, P. micromegethes and Paedocypris sp. Banka was placed in the second clade sister to representatives from the genus Sundadanio. This finding shows that the genetically closest relationship exists between Paedocypris and Sundadanio. We found that the sequence of P. progenetica is almost identical to that of P. carbunculus, likely due to a misidentification in the GenBank specimen.
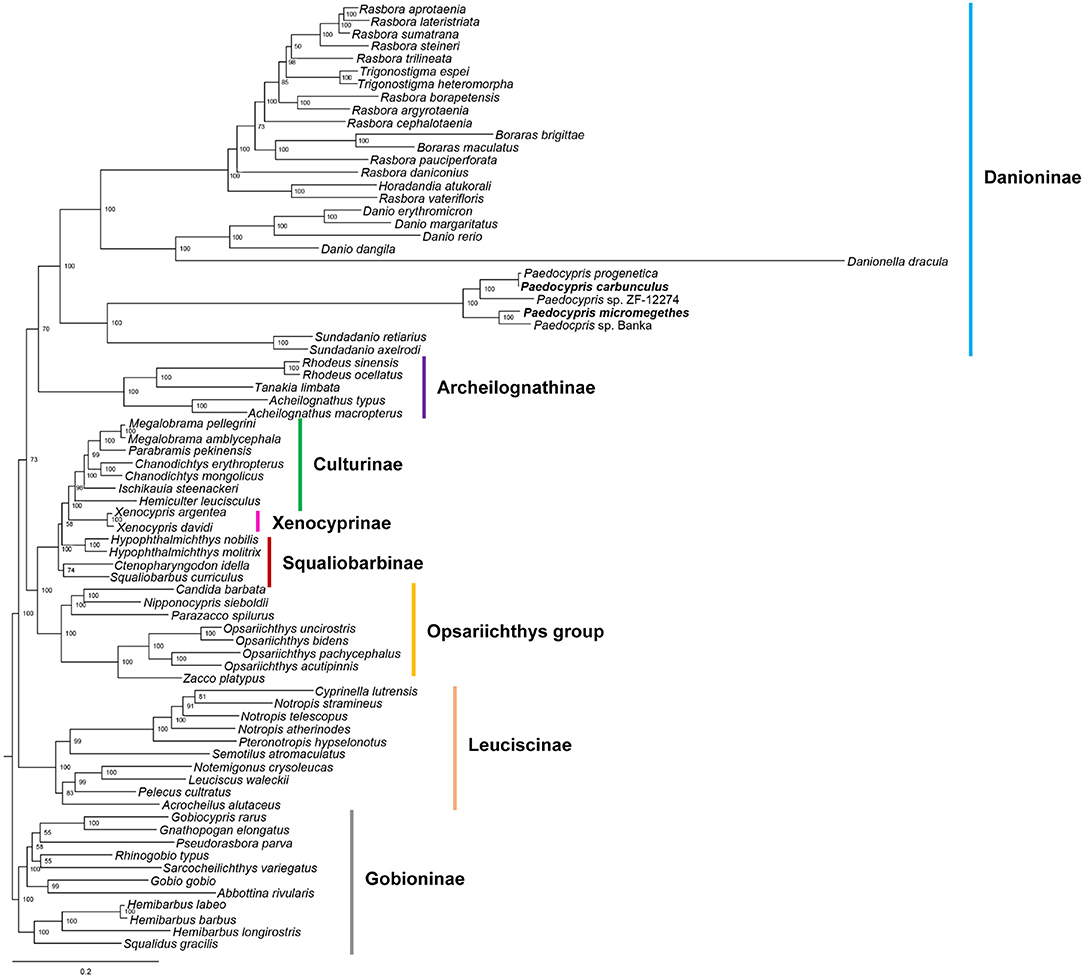
Figure 2. Phylogenetic tree inferred from nucleotide sequences of 13 protein-coding genes and two rRNAs of the mitogenomes using maximum likelihood analysis. The numbers along branches include bootstrap values. The sequences generated in this study are indicated in bold font.
Mitogenomic phylogeny has placed Paedocypris as a sister group of Sundadanio with high support (bootstrap 100% and posterior probability 1.00). This Paedocypris-Sundadanio clade was also resolved with high support (bootstrap 100% and posterior probability 1.00) as a sister group to the remaining Danioninae. This topology is consistent with the phylogenies constructed using cytochrome b, cytochrome c oxidase I, opsin, and recombination activating gene 1, respectively (Ruber et al., 2007; Fang et al., 2009; Tang et al., 2010, 2013). This topology is dissimilar to the findings of Yang et al. (2015) using nuclear and mitochondrial genes which place Paedocypris within Cyprinidae but not within Danioninae. Elsewhere, based on nuclear genes and multi-locus analysis Paedocypris was designated as lineage sister to Cypriniformes (Mayden and Chen, 2010; Stout et al., 2016; Hirt et al., 2017). Notably, the miniature taxa in our mitogenomic phylogenies are widely separated from each other. Danionella dracula is a sister to the Danio clade; Danio erythromicron is a member of the Danio clade; and Paedocypris + Sundadanio form the sister group to the remainder of the Danioninae. A clade of miniature taxa is refuted in our observations, with the phylogeny suggests morphological homoplasy. Convergent evolution of miniaturization has occurred recurrently in the Danioninae from a common ancestor leading to multiple distinct clades of miniature fish.
In this study, the complete mitochondrial genomes of the miniature fish genus Paedocypris (P. micromegethes and P. carbunculus) were analyzed and compared with other members of the Danioninae and Cyprinidae. Our results indicate that mitochondrial genome features, including genome size, gene content and gene arrangement in Paedocypris spp. were not influenced by miniaturization event. The placement of Paedocypris, Sundadanio, and Danionella in different positions in the phylogenetic tree provides clue to the convergent evolutionary trajectory of the miniature taxa. The complete mitogenome information including gene content, structure, gene arrangement, and phylogenetic presented here provide a basis for population genetic and evolutionary biology of the miniature fish and related groups.
Data Availability Statement
The complete mitogenome sequences of P. micromegethes and P. carbunculus have been deposited in the Genbank under the accession numbers MT909824 and MT909825, respectively.
Ethics Statement
All experiments procedures for this study complied with the current animal ethics guidelines and were approved by the USM Institutional Animal Care and Use Committee (USM IACUC).
Author Contributions
AS-C and N-SL conceived and designed the experiments. ZM and RN collected and identified the samples. K-KS and N-SL performed the experiments, analyzed the data, and wrote the manuscript. K-KS prepared the figures. All authors reviewed the manuscript.
Funding
This work was supported by the Universiti Sains Malaysia Research University Grant (1001/PCCB/8011126).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Fieldwork in Sarawak was conducted under permit issued by Forest Department Sarawak (NCCD/600-7/2/107). We are grateful to Michael L.S.H. for his valuable support in the fieldwork. K-KS acknowledges the financial assistance offered by Universiti Sains Malaysia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.662501/full#supplementary-material
References
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Boore, J. L., Macey, J. R., and Medina, M. (2005). Sequencing and comparing whole mitochondrial genomes of animals. Meth. Enzymol. 395, 311–348. doi: 10.1016/S0076-6879(05)95019-2
Britz, R., and Conway, K. W. (2009). Osteology of Paedocypris, a miniature and highly developmentally truncated fish (Teleostei: Ostariophysi: Cyprinidae). J. Morphol. 270, 389–412. doi: 10.1002/jmor.10698
Britz, R., Conway, K. W., and Rüber, L. (2014). Miniatures, morphology and molecules: Paedocypris and its phylogenetic position (Teleostei, Cypriniformes). Zool. J. Linnean Soc. 172, 556–615. doi: 10.1111/zoj12184
Britz, R., and Kottelat, M. (2008). Paedocypris carbunculus, a new species of miniature fish from Borneo (Teleostei: Cypriniformes: Cyprinidae). Raffles B. Zool. 56, 415–422. Available online at: https://lkcnhm.nus.edu.sg/wp-content/uploads/sites/10/app/uploads/2017/06/56rbz415-422.pdf
Broughton, R. E., Milam, J. E., and Roe, B. A. (2001). The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11, 1958–1967. doi: 10.1101/gr.156801
Chang, C. H., Tsai, C. L., and Jang-Liaw, N. H. (2013). Complete mitochondrial genome of the Chinese rasbora Rasbora steineri (Teleostei, Cyprinidae). Mitochondrial DNA 24, 183–185. doi: 10.3109/19401736.2012.744977
Chimnaronk, S., Gravers Jeppesen, M., Suzuki, T., Nyborg, J., and Watanabe, K. (2005). Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 24, 3369–3379. doi: 10.1038/sj.emboj.7600811
Fang, F., Norén, M., Liao, T. Y., Källersjö, M., and Kullander, S. O. (2009). Molecular phylogenetic interrelationships of the south Asian cyprinid genera Danio, Devario and Microrasbora (Teleostei, Cyprinidae, Danioninae). Zool. Scr. 38, 237–256. doi: 10.1111/j.1463-6409.2008.00373.x
Gissi, C., Iannelli, F., and Pesole, G. (2008). Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb) 101, 301–320. doi: 10.1038/hdy.2008.62
Hahn, C., Bachmann, L., and Chevreux, B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. doi: 10.1093/nar/gkt371
Hanken, J. (1993). “Adaptation of bone growth to miniaturization of body size,” in Bone Growth-B, ed B. Hall (Boca Raton: CRC Press), 79–104.
Hanken, J., and Wake, D. B. (1993). Miniaturization of body size: organismal consequences and evolutionary significance. Annu. Rev. Ecol. Syst. 24, 501–519. doi: 10.1146/annurev.es.24.110193.002441
Hirt, M. V., Arratia, G., Chen, W.-J., Mayden, R. L., Tang, K. L., Wood, R. M., et al. (2017). Effects of gene choice, base composition and rate heterogeneity on inference and estimates of divergence times in cypriniform fishes. Biol. J. Linn. Soc. 121, 319–339. doi: 10.1093/biolinnean/blw045
Huang, S. P., Wang, F. Y., and Wang, T. Y. (2017). Molecular phylogeny of the Opsariichthys group (Teleostei: Cypriniformes) based on complete mitochondrial genomes. Zool. Stud. 56:e40. doi: 10.6620/ZS.2017.56-40
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Iwasaki, W., Fukunaga, T., Isagozawa, R., Yamada, K., Maeda, Y., Satoh, T. P., et al. (2013). MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 30, 2531–2540. doi: 10.1093/molbev/mst141
Kerpedjiev, P., Hammer, S., and Hofacker, I. L. (2015). Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics 31, 3377–3379. doi: 10.1093/bioinformatics/btv372
Kottelat, M., Britz, R., Hui, T. H., and Witte, K.-E. (2006). Paedocypris, a new genus of Southeast Asian cyprinid fish with a remarkable sexual dimorphism, comprises the world's smallest vertebrate. P. Roy. Soc. B. Biol. Sci. 273, 895–899. doi: 10.1098/rspb.2005.3419
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kusuma, W. E., and Kumazawa, Y. (2016). Complete mitochondrial genome sequences of two Indonesian rasboras (Rasbora aprotaenia and Rasbora lateristriata). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 4222–4223. doi: 10.3109/19401736.2015.1022749
Lee, W. J., Conroy, J., Howell, W. H., and Kocher, T. D. (1995). Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 41, 54–66. doi: 10.1007/BF00174041
Liao, T.-Y., Kullander, S. O., and Fang, F. (2011). Phylogenetic position of rasborin cyprinids and monophyly of major lineages among the Danioninae, based on morphological characters (Cypriniformes: Cyprinidae). J. Zool. Syst. Evol. Res. 49, 224–232. doi: 10.1111/j.1439-0469.2011.00621.x
Mayden, R. L., and Chen, W.-J. (2010). The world's smallest vertebrate species of the genus Paedocypris: a new family of freshwater fishes and the sister group to the world's most diverse clade of freshwater fishes (Teleostei: Cypriniformes). Mol. Phylogenetics Evol. 57, 152–175. doi: 10.1016/j.ympev.2010.04.008
Miya, M., Takeshima, H., Endo, H., Ishiguro, N. B., Inoue, J. G., Mukai, T., et al. (2003). Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenet. Evol. 26, 121–138. doi: 10.1016/S1055-7903(02)00332-9
Mullens, N., Sonet, G., Decru, E., Virgilio, M., Snoeks, J., and Vreven, E. (2020). Mitogenomic characterization and systematic placement of the Congo blind barb Caecobarbus geertsii (Cypriniformes: Cyprinidae). Int. J. Biol. Macromol. 161, 292–298. doi: 10.1016/j.ijbiomac.2020.05.248
Ohtsuki, T., Kawai, G., and Watanabe, K. (2002). The minimal tRNA: unique structure of Ascaris suum mitochondrial tRNASerUCU having a short T arm and lacking the entire D arm. FEBS Lett. 514, 37–43. doi: 10.1016/S0014-5793(02)02328-1
Ojala, D., Montoya, J., and Attardi, G. (1981). tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474. doi: 10.1038/290470a0
Perna, N. T., and Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41, 353–358. doi: 10.1007/BF01215182
Resch, A. M., Carmel, L., Mariño-Ramírez, L., Ogurtsov, A. Y., Shabalina, S. A., Rogozin, I. B., et al. (2007). Widespread positive selection in synonymous sites of mammalian genes. Mol. Biol. Evol. 24, 1821–1831. doi: 10.1093/molbev/msm100
Ruber, L., Kottelat, M., Tan, H. H., Ng, P. K., and Britz, R. (2007). Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world's smallest vertebrate. BMC Evol. Biol. 7:38. doi: 10.1186/1471-2148-7-38
Satoh, T. P., Miya, M., Mabuchi, K., and Nishida, M. (2016). Structure and variation of the mitochondrial genome of fishes. BMC Genomics 17:719. doi: 10.1186/s12864-016-3054-y
Sheffield, N. C., Hiatt, K. D., Valentine, M. C., Song, H., and Whiting, M. F. (2010). Mitochondrial genomics in Orthoptera using MOSAS. Mitochondrial DNA 21, 87–104. doi: 10.3109/19401736.2010.500812
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stout, C. C., Tan, M., Lemmon, A. R., Lemmon, E. M., and Armbruster, J. W. (2016). Resolving Cypriniformes relationships using an anchored enrichment approach. BMC Evol. Biol. 16:244. doi: 10.1186/s12862-016-0819-5
Sun, C.-H., Liu, H.-Y., Xu, N., Zhang, X.-L., Zhang, Q., and Han, B.-P. (2021). Mitochondrial genome structures and phylogenetic analyses of two tropical characidae fishes. Front. Genet. 12:627402. doi: 10.3389/fgene.2021.627402
Tang, D., Di, B., Wei, G., Ni, I. H., Oh, I. S., and Wang, S. (2006). Spatial, seasonal and species variations of harmful algal blooms in the South Yellow Sea and East China Sea. Hydrobiologia 568, 245–253. doi: 10.1007/s10750-006-0108-1
Tang, K. L., Agnew, M. K., Hirt, M. V., Lumbantobing, D. N., Sado, T., Teoh, V. H., et al. (2013). Limits and phylogenetic relationships of East Asian fishes in the subfamily Oxygastrinae (Teleostei: Cypriniformes: Cyprinidae). Zootaxa 3681, 101–135. doi: 10.11646/zootaxa.3681.2.1
Tang, K. L., Agnew, M. K., Hirt, M. V., Sado, T., Schneider, L. M., Freyhof, J., et al. (2010). Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol. Phylogenet. Evol. 57, 189–214. doi: 10.1016/j.ympev.2010.05.021
Wei, M., Yu, P., Yang, Y., and Wan, Q. (2016). The complete mitochondrial genome of Parabotia fasciata (Cypriniformes: Cobitidae). Mitochondrial DNA A 27, 1831–1832. doi: 10.3109/19401736.2014.971246
Weitzman, S. H., and Vari, R. P. (1988). Miniaturization in South American freshwater fishes; an overview and discussion. Proc. Biol. Soc. Wash. 101, 444–465.
Yang, L., Sado, T., Vincent Hirt, M., Pasco-Viel, E., Arunachalam, M., Li, J., et al. (2015). Phylogeny and polyploidy: resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 85, 97–116. doi: 10.1016/j.ympev.2015.01.014
Yu, P., Zhou, L., Zhou, X. Y., Yang, W. T., Zhang, J., Zhang, X. J., et al. (2019). Unusual AT-skew of Sinorhodeus microlepis mitogenome provides new insights into mitogenome features and phylogenetic implications of bitterling fishes. Int. J. Biol. Macromol. 129, 339–350. doi: 10.1016/j.ijbiomac.2019.01.200
Zhang, Z., Li, J., Zhao, X.-Q., Wang, J., Wong, G. K.-S., and Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Keywords: mitogenome, miniature fish, Paedocypris, gene arrangement, phylogeny
Citation: Sam K-K, Lau N-S, Shu-Chien AC, Muchlisin ZA and Nugroho RA (2021) Complete Mitochondrial Genomes of Paedocypris micromegethes and Paedocypris carbunculus Reveal Conserved Gene Order and Phylogenetic Relationships of Miniaturized Cyprinids. Front. Ecol. Evol. 9:662501. doi: 10.3389/fevo.2021.662501
Received: 01 February 2021; Accepted: 29 March 2021;
Published: 23 April 2021.
Edited by:
Joong-Ki Park, Ewha Womans University, South KoreaReviewed by:
Wei-Kuang Wang, Feng Chia University, TaiwanElizabeth M. A. Kern, North Carolina State University, United States
Copyright © 2021 Sam, Lau, Shu-Chien, Muchlisin and Nugroho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander Chong Shu-Chien, YWxleEB1c20ubXk=
 Ka-Kei Sam
Ka-Kei Sam Nyok-Sean Lau
Nyok-Sean Lau Alexander Chong Shu-Chien
Alexander Chong Shu-Chien Zainal Abidin Muchlisin3
Zainal Abidin Muchlisin3