- 1Laboratory of Ecological and Evolutionary Genetics, Federal University of Viçosa, Campus Rio Paranaíba, Rio Paranaíba, Brazil
- 2Laboratory of Bioinformatics and Genomics, Federal University of Viçosa, Campus Rio Paranaíba, Rio Paranaíba, Brazil
- 3Integrated Nucleus of Biotechnology, University of Mogi das Cruzes, Mogi das Cruzes, Brazil
- 4Department of Genetics and Genome Biology, University of Leicester, Leicester, United Kingdom
Introduction
Stethaprioninae, a subfamily of characiform fish, comprises small fishes popularly known as tetras. Species in this subfamily were initially assigned to the genus Astyanax, which has a broad geographical distribution ranging from the southern United States to northern Argentina (Ornelas-Garcia et al., 2008). Some species of Astyanax, however, are hard to accurately identify based on the lack of diagnostic morphological characteristics (Weitzman and Malabarba, 1998). As noted by Weitzman and Malabarba (1998), the genus Astyanax is probably not monophyletic, and this thinking is reflected in the recent revision by Terán et al. (2020), who reassigned some species of Astyanax to six different genera. The species complexes A. fasciatus and A. scabripinnis, for instance, were placed in the genus Psalidodon, and Astyanax species from the coastal river basins of Brazil (e.g., A. giton) were placed in the genus Deuterodon. The only taxa remaining in the genus Astyanax were the species complex A. bimaculatus and North American species. The revision by Terán was predicted by previous studies on chromosomes and mitochondrial sequences. Molecular analysis of 16 species of Astyanax based on the mitochondrial gene ATPase 8 and chromosomal characteristics obtained four clades (Pazza et al., 2018), and based on a DNA barcoding approach, Rossini et al. (2016) identified five lineages of Astyanax separated by high levels of divergence.
Despite the great diversity and taxonomic issues, at least one species of the genus Astyanax, A. mexicanus (formerly cited as Astyanax fasciatus), has been used as a model to understand the development of its eyes and the evolution of complex traits (Borowsky, 2008; Jeffery, 2008). Astyanax mexicanus is represented by surface and troglobite populations that show similar traits for cave dwelling. According to Ornelas-Garcia et al. (2008), populations from Brazil are not the same species as those seen farther north in middle America and Mexico. Currently, the valid name for tetras from Brazil is Psalidodon fasciatus, a taxon probably containing cryptic species based on their chromosomal variation ranging from 2n = 45 to 49 plus B chromosomes and heterochromatin polymorphism (Pazza et al., 2006; Ferreira-Neto et al., 2012; Kavalco et al., 2013). These chromosomal variants display low molecular divergence (Kavalco et al., 2016). Morphologically, the populations of the São Francisco river basin are different from the specimens from Alto Paraná and Paraíba do Sul rivers (Melo, 2001) and the original morphological description is so broad that it certainly covers other species outside the complex (Melo and Buckup, 2006). In this case, P. fasciatus should be restricted to the specimens from the São Francisco river basin (original basin of the type species), while the others may be either considered cryptic species of the complex or even other species (Melo and Buckup, 2006). On the other hand, despite a clear genetic structure among populations from the Alto Paraná and São Francisco river basins, morphometric traits seem to be homoplasy (Pazza et al., 2017).
The A. bimaculatus complex is currently represented by Astyanax bimaculatus species along with others, such as A. altiparanae (considered as a junior synonym of A. lacustris by some authors), A. lacustris, A. assuncionensis, and A. abramis. With a preference for calm waters, these species inhabit mainly in the Alto Paraná, Paraguay, Iguassu, and São Francisco river basins (Domingues et al., 2007). Contrary to what has been observed in P. fasciatus and P. scabripinnis, A. bimaculatus shows a constant diploidy number in different populations, 2n = 50 chromosomes, which is considered a symplesiomorphic character in Gymnocharacini (Kavalco et al., 2011; Martinez et al., 2012; Fernandes et al., 2014). The diversity within the group refers to differences in its karyotypic formula, fundamental number, and general symmetry of the karyotypes (Kavalco et al., 2011; Fernandes et al., 2014). These cytogenetic data, associated with the molecular ones, suggest a relatively recent divergence, as well as the monophyletic status of this branch (Pazza et al., 2018).
Alongside with the P. fasciatus and A. bimaculatus, the third species complex explored in this work, P. scabripinnis, was proposed by Moreira-Filho and Bertollo (1991) based on morphological and chromosomal characteristics of specimens collected in the Paraná and São Francisco river basins. In a review of the P. scabripinnis group, Bertaco and Lucena (2006) pointed out the existence of 15 species, including P. paranae and P. rivularis. The species of this complex are known for their wide karyotypic diversity, with diploid numbers ranging from 2n = 46 to 50 chromosomes (Moreira-Filho and Bertollo, 1991; Fernandes and Martins-Santos, 2005). In recent studies using molecular phylogeography and geometric morphometry, Rocha et al. (2019) reinforced the validity of P. rivularis and P. paranae as sister species of the complex, inhabiting the São Francisco and Paraná river basins, respectively. However, among the populations from the Alto Paranaíba river, the existence of a new species of the complex was observed due to morphometric and mtDNA data (Alves R de et al., 2020). This new species, called Psalidodon rioparanaibanus, was collected only in a small tributary of the Paranaíba river, surrounded by populations of P. paranae. Moreover, within the P. paranae and P. rivularis groups, karyotypic diversity is also present (Maistro et al., 1998) indicating that, even though delimited by individual lineages, these groups still constitute compilations of cryptic species.
Hundreds of studies describing mitogenomes have been published in the last few years. Despite being the most sequenced genome nowadays (Smith, 2015), to this date, just the mitogenomes of A. mexicanus (Nakatani et al., 2011), P. paranae (Silva et al., 2016), Deuterodon giton (Barreto et al., 2017), P. fasciatus, and A. altiparanae (Calegari et al., 2019) are published. In an attempt to fill this gap, we focused on the three species complex of the group (A. bimaculatus, P. scabripinnis and P. fasciatus) to present here the complete sequences of the mitochondrial genome of 10 species/cytotypes of Astyanax/Psalidodon. Such data could be very useful in further phylogeny studies and to understand the diversity of the group.
Materials and Methods
Specimens from the Astyanax/Psalidodon genus were analyzed. We collected the samples in different locations throughout the major Brazilian rivers and their vouchers are deposited in the ichthyological collection of the Laboratory of Ecological and Evolutionary Genetics at Federal University of Viçosa, campus Rio Paranaíba, Brazil (Supplementary Table 1). After sampling, we brought the living specimens to the laboratory, euthanized them in accordance with the ethical standards of CONCEA, the Brazilian Council for the Control of Animal Experimentation and CEUA/UFV—Ethics Committee on Animal Use/Federal University of Viçosa (760/2018). We performed the sampling with licenses provided by SISBIO/ICMBIO—Biodiversity Authorization and Information System (1938128) and SISGEN—National System for the Management of Genetic Heritage and Associated Traditional Knowledge (A9FE946).
We extracted the total genomic DNA from the liver and heart tissues of six specimens according to the instructions of Invitrogen's PureLink DNA extraction and purification kit. After quality checking using fluorometer Qubit (Thermo Fisher Scientific), the Whole Genome Sequencing was performed using Novaseq 6000 (Illumina, San Diego, CA) at Novogene, UK.
For broader comparisons, we also assembled the mitogenome of two Astyanax/Psalidodon species with raw reads available on ENA (European Nucleotide Archive): P. fasciatus, from the Alto Paraná river basin (SRR8476332) and A. aeneus, from Mexico (SRR1927238). Aiming to validate our methodology, we reassembled the mitogenomes of P. paranae (SRR5461470) and A. mexicanus (SRR2040423). In the Bayesian analysis, we also included the mitochondrial complete sequence of Deuterodon giton (NC_044970.1) and added Brycon orbignianus (KY825192.1) as outgroup.
We assembled the mitogenome from raw reads on Novoplasty v3.7 (Dierckxsens et al., 2017) in a parallel cluster computer (64 Gb RAM) using the mitogenome of Psalidodon paranae available on GenBank (SRR5461470) as seed. We chose this approach because it is fast and assemble the mitogenomes “de novo” from raw data using a single mitochondrial sequence as seed, without the bias of a reference. We annotated the sequences obtained on MitoAnnotator (Iwasaki et al., 2013) at MitoFish (http://mitofish.aori.u-tokyo.ac.jp).
We performed comparative genomics analysis by BLAST comparison of all the Astyanax/Psalidodon mitochondrial genomes against a reference (Psalidodon paranae) generated by Blast Ring Image Generator (BRIG) (Alikhan et al., 2011). To assess the repetitive region, we analyzed the mitochondrial sequences with Tandem Repeats Finder (Benson, 1999), following we isolate and duplicate the repeats of D-loop, and we aligned at ClustalW (Thompson et al., 1994) to find the repeat motif.
To validate the mitogenome as a tool for understanding the phylogenetic relationships among the samples, we aligned Fasta sequences with ClustalW and calculated the p-distance with MEGA X software (Kumar et al., 2018). We used the 13 protein-coding genes (PCGs), extracted by hand from fasta file produced by Mitoannotator, in Bayesian phylogenetic inference with MrBayes 3.2.7 (Roquist et al., 2012) after calculating the best evolutionary models for each segment with Partition Finder 2.1.1 (Lanfear et al., 2016). All PCGs used on phylogeny was tested by saturation (Xia et al., 2003) on DAMBE v7. (Xia, 2018). Bayesian analyses were performed using four independent chains with 10-million-generations and the effective sample size (ESS) and strand convergence were, posteriorly, verified in the software Tracer 1.7 (Rambaut et al., 2018). The first 25% of the generations were discarded as burn-in. For the Maximum Likelihood analysis, we used the concatenated 13 protein-coding genes (PCGs) after testing for the best model in the software MEGA X (Kumar et al., 2018).
Data Description
Our results have shown that all mitogenomes content and gene order were identical (Figure 1), with 13 PCGs, 22 tRNA genes, and 2 rRNA genes, as already described for Characiformes mitogenomes. The same is true for other Astyanax/Psalidodon/Deuterodon species, such as A. mexicanus (Nakatani et al., 2011), P. paranae (Silva et al., 2016), and D. giton (Barreto et al., 2017). All PCGs, except the ND6 gene, are on the heavy chain. All but 8 tRNAs are on the heavy chain as well.
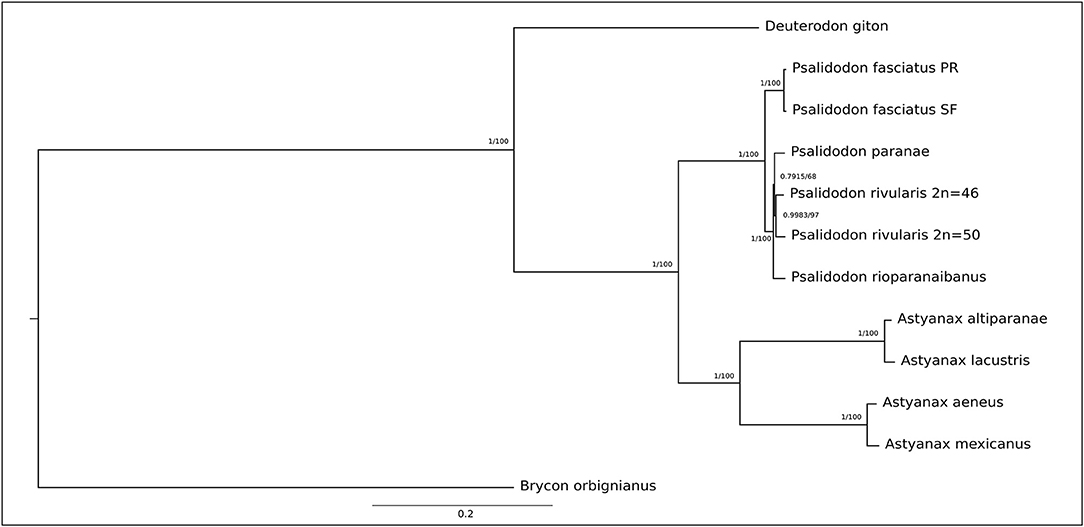
Figure 1. Phylogenetic tree based on 13 protein-coding genes (PCGs) showing the relationships among the Stethaprioninae fish using Brycon orbignianus as outgroup. The topology was the same in Maximum Likelihood (100 bootstrap replicates) after the test of the best model (General Time Reversible +G) and Bayesian Inference after calculation of best evolutionary models for each segment, using four independent chains with 10-million-generations (The first 25% of the generations were discarded as burn-in). The posterior probabilities/bootstrap are on the branches.
All the new sequences are deposited at GenBank (Supplementary Table 2). The length of mitochondrial sequences range from 16,626 bp in the Psalidodon fasciatus from the São Francisco river basin to 16,812 bp in Psalidodon rivularis 2n = 50. The average length of D-loop was 1,061 bp, ranging from 951 bp in Psalidodon fasciatus from São Francisco river basin to 1,136 bp in Psalidodon rivularis with 2n = 50 chromosomes. No differences between deposited mitogenome of P. paranae and our reassembling could be seen. On the other side, our pipeline could extend the D-loop region of A. mexicanus, a problematic region in the deposited sequence (named “almost complete” on the GenBank entry). The difference in the size of D-loop was due to a repeat of 35 bp in all D-loops, except in Deuterodon giton (Supplementary Table 3). For the alignment, we got a repeated motif (TATGTATTAGTACATATTATGCATAATTATACATA) slightly variable in some species.
Deepening the knowledge on the mitogenome control region, called D-loop, can play a fundamental role in understanding the evolutionary history in the Astyanax and Psalidodon genera. In this work, we observed that the size variation among different Astyanax/Psalidodon mitogenomes occurs mainly due to the extension of the D-loop. Neglecting this region in the reconstruction of mitogenomes can result in a valuable loss of information since, in addition to the variation in size, we found a repetitive sequence of 35 bp in nine of the 10 mitogenomes studied (Supplementary Table 3). On the other side, studies based on D-loop sequence must be aware of this kind of feature that can bias the analysis.
D. giton was the only species that did not present the repetitive sequence in the D-loop. In addition to occupying a sister group position in the recuperated phylogeny (Figure 2) when describing the mitogenome of the species, Barreto et al. (2017) observed in their phylogeny of Characidae that the species Grundulus bogotensis was closer to the Astyanax/Psalidodon group than to D. giton. Therefore, we not only confirm D. giton as a species outside the genus Astyanax as suggested by the taxonomic review (Terán et al., 2020) but also that the repetitive sequence found in the D-loop may correspond to a synapomorphy absent in the Deuterodon group. However, it is necessary to reassemble the genome of D. giton under our methodology and conduct complementary studies on species of the Deuterodon group to clarify this issue.
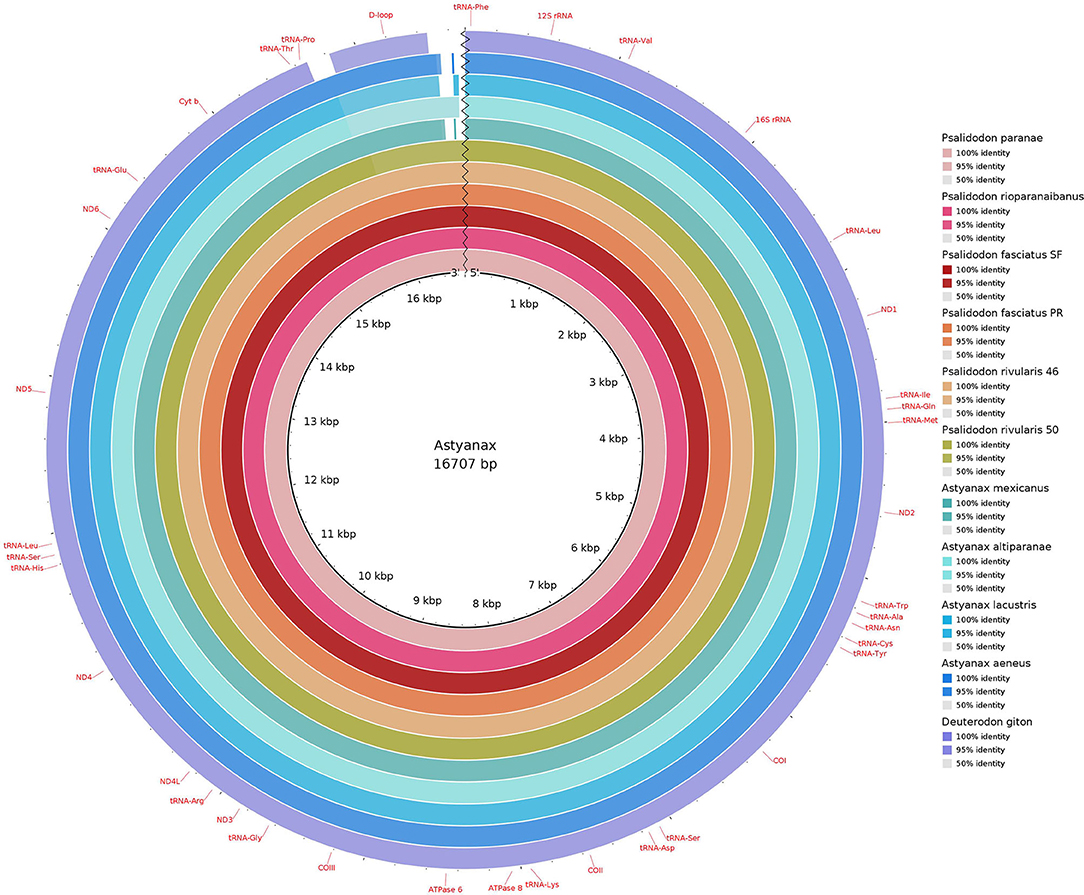
Figure 2. Comparative mitogenomics analysis of all the 10 Stethaprioninae fish against a reference (Psalidodon paranae), generated by Blast Ring Image Generator (BRIG). Gaps in rings correspond to regions with <50% identity to the reference sequence (BLAST comparison). Colors from the center: dd9998; cb3b54; a31418; e85e25; df9856; 95ac42; 38a67f; 79bdbe; 00a8c3; 016db8; 6c5ab1.
The genetic distance among species (Supplementary Table 4) is reflected in both the Maximum Likelihood and the Bayesian tree (Figure 2), which shows strong construction with high bootstrap value and posterior probabilities, respectively. Once no gene sequence show saturation, we did the phylogeny with all 13 PCGs. The Partition Finder analysis results in five subsets (Supplementary Table 5) used in bayesian phylogeny.
The topology of the tree is congruent with those inferred by Rossini et al. (2016) and Pazza et al. (2018), except for the North American clade, which appears as a sister group of A. altiparanae and A. lacustris here. Besides, our study reinforces the taxonomic review by Terán et al. (2020), meanwhile disagree with Lucena and Soares (2016) that describe A. altiparanae as a new junior synonyms of A. lacustris.
The methodology used to reconstruct the mitochondrial genome proved to be satisfactory and enabled the assessment of the length of this type of genome, plus the composition and nature of the D-loop, solving possible gaps in previous methodologies (Silva et al., 2016; Barreto et al., 2017; Calegari et al., 2019). Besides, the study of the complete mitochondrial genome proves to be a tool with the potential to solve taxonomic problems and to help understand the evolutionary relationships in species complexes, such as A. bimaculatus, P. fasciatus, and P. scabripinnis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, BK013055 MT428072 MT428067 BK013062 BK013061 MT428071 MT428068 MT428069 MT428070.
Ethics Statement
The animal study was reviewed and approved by Brazilian Council for the Control of Animal Experimentation and CEUA/UFV—Ethics Committee on Animal Use/Federal University of Viçosa (760/2018).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Henrique Peluzio for his support on the UFV cluster and to University of Leicester (UK) for the support and hospitality to RP and KK in their sabbatical.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.650783/full#supplementary-material
Abbreviations
ATP6/8, ATP Synthase Membrane Subunit 6/8; BRIG, Blast Ring Image Generator; CEUA/UFV, Comissão de Ética no Uso de Animais/Universidade Federal de Viçosa (Ethics Committee on Animal Use/Federal University of Viçosa); CONCEA, Conselho Nacional de Controle de Experimentação Animal (Brazilian Council for the Control of Animal Experimentation); COI, Cytochrome c oxidase subunit I; D-loop, Displacement Loop; ENA, Europe Nucleotide Archive; ESS, Effective Sample Size; ESU, Evolutionarily Significant Unit; mtDNA, mitochondrial DNA; ND6, NADH-ubiquinone oxidoreductase chain 6; PCG, Protein-coding genes; SISBIO/ICMBIO, Sistema de Autorização e Informação em Biodiversidade/Instituto Chico Mendes de Conservação da Biodiversidade (Biodiversity Authorization and Information System/Chico Mendes Institute for Biodiversity Conservation); SISGEN, Sistema Nacional de Gestão do Patrimônio Genético e do Conhecimento Tradicional Associado (Brazilian National System for the Management of Genetic Heritage and Associated Traditional Knowledge).
References
Alikhan, N. F., Petty, N. K., Zakour, N. L. B., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Alves R de, M., Oliveira, I. H. R., Pasa, R., and Kavalco, K. F. (2020). A new Psalidodon species related to the P. paranae complex (Characiformes, Characidae) from Upper Paranaíba river basin, Brazil, supported by genetic and morphometric data. Biodivers. J. 11, 807–814. doi: 10.31396/Biodiv.Jour.2020.11.4.807.814
Barreto, C. A. V., Granja, M. M. C., Vidigal, P. M. P., Carmo, A. O., and Dergam, J. A. (2017). Complete mitochondrial genome sequence of neotropical fish Astyanax giton Eigenmann 1908 (Ostariophysi; Characidae). Mitochondrial DNA B. 2, 839–840. doi: 10.1080/23802359.2017.1403869
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bertaco, V. A., and Lucena, C. A. (2006). Two new species of Astyanax (Ostariophysi: Characiformes: Characidae) from eastern Brazil, with a synopsis of the Astyanax scabripinnis species complex. Neotrop. Ichthyol. 4, 53–60. doi: 10.1590/S1679-62252006000100004
Borowsky, R. (2008). Astyanax mexicanus, the blind Mexican cave fish: a model for studies in development and morphology. Cold Spring Harb. Protoc. 11: pdb-emo107. doi: 10.1101/pdb.emo107
Calegari, R. M., Rodrigues, P. H. M., Santos, R. Z. D., Foresti, F., Utsunomia, R., and Porto-Foresti, F. (2019). The complete mitochondrial genome sequences of five Otophysi species (Vertebrata, Teleostei). Mitochondrial DNA B 4, 4198–4199. doi: 10.1080/23802359.2019.1693294
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. doi: 10.1093/nar/gkw955
Domingues, M. S., Vicari, M. R., Abilhoa, V., Wamser, J. P., Cestari, M. M., Bertollo, L. A. C., et al. (2007). Cytogenetic and comparative morphology of two allopatric populations of Astyanax altiparanae Garutti and Britski, 2000 (Teleostei: Characidae) from upper rio Paraná basin. Neotrop Ichthyol. 5, 37–44. doi: 10.1590/S1679-62252007000100005
Fernandes, C. A., da Rocha, R. H., Bailly, D., Guterres, Z. R., Soares, D., and Martins-Santos, I. C. (2014). Cytogenetic and Morphometric Analysis in the Species Astyanax altiparanae Garutti and Britski, 2000 (Teleostei, Characidae) from the Iguatemi River Basin, Brazil. Folia Biol. 62, 193–201. doi: 10.3409/fb62_3.193
Fernandes, C. A., and Martins-Santos, I. C. (2005). Sympatric occurrence of three cytotypes and four morphological types of B chromosomes of Astyanax scabripinnis (Pisces, Characiformes) in the River Ivaí Basin, state of Paraná, Brazil. Genetica 124, 301–306. doi: 10.1007/s10709-005-4751-7
Ferreira-Neto, M., Artoni, R. F., Vicari, M. R., Moreira-Filho, O., Camacho, J. P. M., Bakkali, M., et al. (2012). Three sympatric karyomorphs in the fish Astyanax fasciatus (Teleostei, Characidae) do not seem to hybridize in natural populations. Comp Cytogenet. 6, 29–40. doi: 10.3897/compcytogen.v6i1.2151
Iwasaki, W., Fukunaga, T., Isagozawa, R., Yamada, K., Maeda, Y., Satoh, T. P., et al. (2013). MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 30, 2531–2540. doi: 10.1093/molbev/mst141
Jeffery, W. R. (2008). Emerging model systems in evo-devo: cavefish and microevolution of development. Evol. Dev. 10, 265–272. doi: 10.1111/j.1525-142X.2008.00235.x
Kavalco, K. F., Pazza, R., Brandão, K. D. O., and de Almeida-Toledo, L. F. (2013). Biogeographic patterns in the chromosomal distribution of a satellite DNA in the banded tetra Astyanax fasciatus (Teleostei: Characiformes). Org. Divers. Evol. 13, 67–76. doi: 10.1007/s13127-012-0100-8
Kavalco, K. F., Pazza, R., Brandão, K. D. O., Garcia, C., and Almeida-Toledo, L. F. (2011). Comparative cytogenetics and molecular phylogeography in the group Astyanax altiparanae–Astyanax aff. bimaculatus (Teleostei, Characidae). Cytogenet Genome Res. 134, 108–119. doi: 10.1159/000325539
Kavalco, K. F., Pazza, R., Brandão, K. D. O., Garcia, C., Bertollo, L. A. C., and Almeida-Toledo, L. F. (2016). Chromosomal diversification higher than molecular variation in Astyanax aff. fasciatus (Teleostei, Characidae). Zebrafish 13, 345–353. doi: 10.1089/zeb.2016.1272
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lanfear, R., Frandsen, P. B., Wright, A. P., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lucena, C. A. S., and Soares, H. G. (2016). Review of species of the Astyanax bimaculatus “caudal peduncle spot” subgroup sensu Garutti and Langeani (Characiformes, Characidae) from the rio La Plata and rio São Francisco drainages and coastal systems of southern Brazil and Uruguay. Zootaxa 4072, 101–125. doi: 10.11646/zootaxa.4072.1.5
Maistro, E. L., Oliveira, C., and Foresti, F. (1998). Comparative cytogenetic and morphological analysis of Astyanax scabripinnis paranae (Pisces, Characidae, Tetragonopterinae). Genet Mol Biol. 21, 201–206. doi: 10.1590/S1415-47571998000200005
Martinez, E. R., Alves, A. L., Silveira, S. M., Foresti, F., and Oliveira, C. (2012). Cytogenetic analysis in the incertae sedis species Astyanax altiparanae Garutti and Britzki, 2000 and Hyphessobrycon eques Steindachner, 1882 (Characiformes, Characidae) from the upper Paraná river basin. Comp. Cytogenet. 6, 41–51. doi: 10.3897/compcytogen.v6i1.1873
Melo, F. A. G. (2001). Revisão taxonômica das espécies do gênero Astyanax Baird e Girard 1854 (Teleostei, Characiformes, Characidae) da região da Serra dos Órgãos. Arq Mus Nac Rio de Janeiro. 59, 1–46.
Melo, F. A. G., and Buckup, P. A. (2006). Astyanax henseli, a new name for Tetragonopterus aeneus Hensel (1870), from Southern Brazil (Teleostei, Characiformes). Neotrop. Ichthyol. 4, 45–52. doi: 10.1590/S1679-62252006000100003
Moreira-Filho, O., and Bertollo, L. A. C. (1991). Astyanax scabripinnis (Pisces, Characidae) a species complex. Rev. Brasil Gene. 142, 331–357.
Nakatani, M., Miya, M., Mabuchi, K., Saitoh, K., and Nishida, M. (2011). Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol. Biol. 11:177. doi: 10.1186/1471-2148-11-177
Ornelas-Garcia, C. P., Dominguez-Dominguez, O., and Doadrio, I. (2008). Evolutionary history of the fish genus Astyanax Baird and Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol. Biol. 8:340. doi: 10.1186/1471-2148-8-340
Pazza, R., Cruvinel, L. M., and Kavalco, K. F. (2017). Parallel evolution evidenced by molecular data in the banded-tetra (Astyanax fasciatus). Biochem. Syst. 70, 141-146. doi: 10.1016/j.bse.2016.10.024
Pazza, R., Dergam, J. A., and Kavalco, K. F. (2018). Trends in Karyotype Evolution in Astyanax (Teleostei, Characiformes, Characidae): Insights From Molecular Data. Front. Genet. 9:131. doi: 10.3389/fgene.2018.00131
Pazza, R., Kavalco, K. F., and Bertollo, L. A. C. (2006). Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). 1. Karyotype analysis, Ag-NORs and mapping of the 18S and 5S ribosomal genes in sympatric karyotypes and their possible hybrid forms. Cytogenet Genome Res. 112, 313–319. doi: 10.1159/000089886
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Rocha, R. R., Alves R de, M., Pasa, R., and Kavalco, K. F. (2019). Morphological and genetic structure of two equivalent Astyanax species (Characiformes: Characidae) in the region of Paranaíba Arc. Sci World J. 4:650795. doi: 10.1155/2019/6507954
Roquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rossini, B. C., Oliveira, C. A., Melo, F. A., Bertaco, V. A., Astarloa, J. M. D., Rosso, J. J., et al. (2016). Highlighting Astyanax Species Diversity through DNA Barcoding. PLoS ONE. 11:e0167203. doi: 10.1371/journal.pone.0167203
Silva, D. M. D. A., Utsunomia, R., Ruiz-Ruano, F. J., Oliveira, C., and Foresti, F. (2016). The complete mitochondrial genome of Astyanax paranae (Teleostei: characiformes). Mitochondrial DNA B 1, 586–587. doi: 10.1080/23802359.2016.1222251
Smith, D. R. (2015). The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief. Funct. Genom. 15, 47–54. doi: 10.1093/bfgp/elv027
Terán, G. E., Benitez, M. F., and Mirande, J. M. (2020). Opening the Trojan horse: phylogeny of Astyanax, two new genera and resurrection of Psalidodon (Teleostei: Characidae). Zool. J. Linnean Soc. 190, 1217–1234. doi: 10.1093/zoolinnean/zlaa019
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTALW: improving the sensitivity of progressive multiple sequence through weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Weitzman, S. H., and Malabarba, L. (1998). “Perspectives about the phylogeny and classification of the Characidae (Teleostei: Characiformes),” in Phylogeny and classification of Neotropical fishes edn, eds L.R. Malabarba, R. E. Reis, R. P. Vari, Z. M. S. Lucena, C. A. S. Lucena, (Edipucrs, Porto Alegre), 161–170.
Xia, X. (2018). DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Keywords: evolutionary genetics, criptic species, neotropical fish, molecular ecology, fish genomics
Citation: Pasa R, Menegídio FB, Rodrigues-Oliveira IH, da Silva IB, de Campos MLCB, Rocha-Reis DA, Heslop-Harrison JS, Schwarzacher T and Kavalco KF (2021) Ten Complete Mitochondrial Genomes of Gymnocharacini (Stethaprioninae, Characiformes). Insights Into Evolutionary Relationships and a Repetitive Element in the Control Region (D-loop). Front. Ecol. Evol. 9:650783. doi: 10.3389/fevo.2021.650783
Received: 08 January 2021; Accepted: 23 June 2021;
Published: 22 July 2021.
Edited by:
Rodney L. Honeycutt, Pepperdine University, United StatesReviewed by:
Bingjian Liu, Zhejiang Ocean University, ChinaEdna Judith Márquez, National University of Colombia, Medellin, Colombia
Copyright © 2021 Pasa, Menegídio, Rodrigues-Oliveira, da Silva, de Campos, Rocha-Reis, Heslop-Harrison, Schwarzacher and Kavalco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rubens Pasa, cnBhc2FAdWZ2LmJy
†These authors have contributed equally to this work
‡ORCID: Rubens Pasa orcid.org/0000-0002-3513-4071
Fabiano Bezerra Menegídio orcid.org/0000-0002-4705-8352
Igor Henrique Rodrigues-Oliveira orcid.org/0000-0003-1998-7768
Iuri Batista da Silva orcid.org/0000-0003-2788-5665
Matheus Lewi Cruz Bonaccorsi de Campos orcid.org/0000-0001-7176-9667
Dinaíza Abadia Rocha-Reis orcid.org/0000-0003-1762-8745
John Seymour Heslop-Harrison orcid.org/0000-0002-3105-2167
Trude Schwarzacher orcid.org/0000-0001-8310-5489
Karine Frehner Kavalco orcid.org/0000-0002-4955-2792
 Rubens Pasa
Rubens Pasa Fabiano Bezerra Menegídio
Fabiano Bezerra Menegídio Igor Henrique Rodrigues-Oliveira
Igor Henrique Rodrigues-Oliveira Iuri Batista da Silva
Iuri Batista da Silva Matheus Lewi Cruz Bonaccorsi de Campos
Matheus Lewi Cruz Bonaccorsi de Campos Dinaíza Abadia Rocha-Reis
Dinaíza Abadia Rocha-Reis John Seymour Heslop-Harrison
John Seymour Heslop-Harrison Trude Schwarzacher
Trude Schwarzacher Karine Frehner Kavalco
Karine Frehner Kavalco