
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 18 March 2021
Sec. Evolutionary Developmental Biology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.643661
This article is part of the Research Topic Evolution of Postembryonic Development View all 12 articles
 Amruta Tendolkar1†
Amruta Tendolkar1† Aaron F. Pomerantz2,3†
Aaron F. Pomerantz2,3† Christa Heryanto1†
Christa Heryanto1† Paul D. Shirk4
Paul D. Shirk4 Nipam H. Patel3†
Nipam H. Patel3† Arnaud Martin1*†
Arnaud Martin1*†The forewings and hindwings of butterflies and moths (Lepidoptera) are differentiated from each other, with segment-specific morphologies and color patterns that mediate a wide range of functions in flight, signaling, and protection. The Hox gene Ultrabithorax (Ubx) is a master selector gene that differentiates metathoracic from mesothoracic identities across winged insects, and previous work has shown this role extends to at least some of the color patterns from the butterfly hindwing. Here we used CRISPR targeted mutagenesis to generate Ubx loss-of-function somatic mutations in two nymphalid butterflies (Junonia coenia, Vanessa cardui) and a pyralid moth (Plodia interpunctella). The resulting mosaic clones yielded hindwing-to-forewing transformations, showing Ubx is necessary for specifying many aspects of hindwing-specific identities, including scale morphologies, color patterns, and wing venation and structure. These homeotic phenotypes showed cell-autonomous, sharp transitions between mutant and non-mutant scales, except for clones that encroached into the border ocelli (eyespots) and resulted in composite and non-autonomous effects on eyespot ring determination. In the pyralid moth, homeotic clones converted the folding and depigmented hindwing into rigid and pigmented composites, affected the wing-coupling frenulum, and induced ectopic scent-scales in male androconia. These data confirm Ubx is a master selector of lepidopteran hindwing identity and suggest it acts on many gene regulatory networks involved in wing development and patterning.
Hox genes are major players in the evolution of animal body plans, as they provide region-specific information to developing tissues that enables the specialization of body parts, including serial homologs that form the axial skeleton of vertebrates or the appendages of arthropods (Hughes and Kaufman, 2002; Pourquie, 2009). First identified and characterized in Drosophila, it has been shown that these genes are well conserved across metazoans, prompting a prolific effort to understand how Hox genes are embedded in the regulatory logic of development, and how they may mediate phenotypic change at evolutionary scales.
The diversity of insect wing configurations provides a good comparative system to tackle those questions. Wing appendage development occurs in the second and third thoracic segments (T2 and T3, syn. mesothorax and metathorax). The development of wing primordia includes segment-specific processes that give rise to marked functional differences, such as the T3 halteres of Diptera, the T2 protective shields of Coleoptera and Hemiptera (elytra, hemelytra), and the many examples of divergence in color pattern and flight functions between the T2/T3 wings of Lepidoptera (Williams and Carroll, 1993; Carroll, 1994; Jantzen and Eisner, 2008; Ohde et al., 2014; Tomoyasu et al., 2017). The homeotic selector gene Ultrabithorax (Ubx) is consistently expressed in the T3 but not in the T2 wing primordia of winged insects (Hughes and Kaufman, 2002; Tomoyasu, 2017), and instructs the differentiation of T3 relative to T2 wing derivatives. A complete loss of Ubx in the Drosophila melanogaster T3 imaginal disks replaces the halteres with an apparent second set of wings (Bender et al., 1983; Weatherbee et al., 1998; Hersh et al., 2007; Lipshitz, 2007; Pavlopoulos and Akam, 2011). This seminal discovery has led to detailed studies showing that Ubx represses forewing identity in the haltere disc, both through the transcriptional repression of wing genes and activation of haltere genes (Shashidhara et al., 1999; Crickmore and Mann, 2006; Mohit et al., 2006; Navas et al., 2006; Hersh et al., 2007; Makhijani et al., 2007). In conjunction with work in Drosophila legs and embryonic tissues, these data have established Ubx as a selector gene that specifies metathoracic structures from their Ubx-free homologs (Zandvakili et al., 2019).
This generic role of Ubx is supported in all insect wing systems for which functional data has been reported so far. RNAi knock-downs of Ubx in Tribolium castaneum (Tomoyasu et al., 2005, 2009), Oncopeltus fasciatus (Medved et al., 2015) and Nilaparvata lugens (Fu et al., 2020) all showed homeotic transformations of T3 wing tissue into a T2 identity. In Lepidoptera, two lines of evidence have confirmed the role of Ubx as a homeotic selector gene specifying T3/hindwing identity. A spontaneous mutation called Hindsight, isolated from a Junonia coenia laboratory colony, causes mosaic patches of forewing color pattern and scale identities on the ventral side of butterfly hindwings (Nijhout and Rountree, 1995; Weatherbee et al., 1999). It is unclear if Hindsight is caused by a mutation at the Ubx locus itself, but it induces dominant somatic clones that lack Ubx expression during development, and its restriction to the more posterior region of the T3 appendage is somewhat reminiscent of the Postbithorax and bithoraxoid alleles at the Drosophila Ubx locus, which result in clonal homeotic transformations of the posterior haltere (Adler, 1979, 1981). More recently, crispants (CRISPR/Cas9 somatic knock-outs) of Ubx in Bicyclus anynana revealed T3-to-T2 homeotic transformations on the hindwings (Matsuoka and Monteiro, 2020), where local homeotic shifts in scale types and pattern size mirrored the matching forewing condition. For instance, in B. anynana, silver scales are normally limited to the ventral posterior region of the forewings, and ectopic silver scales appeared in mutant hindwing clones that were restricted to the ventral posterior region. Likewise, eyespots that are present in the hindwing but are absent in forewings were reduced or missing upon mosaic KO, while conversely, eyespots that are prominent in the forewing were enlarged.
Overall, these butterfly wing data suggest that Ubx loss-of-function phenotypes vary across the wing (context-dependency), and that this Hox input is plugged into the differential control of a majority of hindwing-specific features. This includes homeotic shifts in both scale identities (e.g., a specific morphology and pigment) and in their pattern arrangement (e.g., eyespot size and color composition). Here we leveraged the diversity of lepidopteran wing traits to gain additional insights into the modalities of Ubx hindwing trait specification. Our study expanded the use of CRISPR Ubx mosaic knock-outs to two other nymphalid butterflies, J. coenia and Vanessa cardui, which display marked pattern organization and color differences between their two sets of wings, as well as in the pyralid moth Plodia interpunctella, a lepidopteran with folding and depigmented hindwings.
The nymphalid butterfly J. coenia shows marked forewing/hindwing (FW/HW) differences in the distribution and color of pattern elements (Figure 1A), including the Discalis stripes, the central symmetry system, eyespots (s.s. border ocelli), and the parafocal/marginal bands (Mazo-Vargas et al., 2017). Previously described homeotic clones from the Hindsight line affect those elements but are limited to the posterior compartment of the ventral hindwings. To formally test the role of Ubx in hindwing patterning in this species, we generated somatic loss-of-function mutants by injecting Cas9/sgRNA duplexes into syncytial embryos, targeting the Ubx coding sequence (Table 1, Supplementary Figure 1, and Supplementary Table 1). These experiments led to high-mortality rates at both embryonic and post-embryonic stages, but batches injected 3 h after egg-laying (AEL) showed enough mosaicism to allow survivors to reach adulthood. A total of 19 adult butterflies showed hindwing-to-forewing color pattern transformations, not only replicating the effects of Hindsight on the ventral posterior hindwings but also extending to the dorsal surface and to the anterior compartments (Supplementary Figures 2–4). Scales of the J. coenia ventral hindwing are normally serrated, contrasting with the rounded shape of their forewing homologs, which allows the delineation of homeotic clones, as in the Hindsight mutants (Nijhout and Rountree, 1995; Weatherbee et al., 1999). For all the pattern elements besides the eyespots, homeotic clones induced a color shift in a cell autonomous fashion, with mutant scale patches showing sharp demarcation from non-mutant scales (Figure 1). Those shifts in scale morphology were always accompanied by the acquisition of color states exactly matching the forewing state in the equivalent position. This is particularly evident in the induction of orange and black patterns known as the forewing Discalis elements in positions where in the wild type hindwing they are brown/beige (ventral) or absent (dorsal). Likewise, mutants show discrete clones transposing the white band from the forewing onto the corresponding hindwing territories. Similar homeoses are conspicuous in the stripe systems that border the margin of the wings, where in wild type they are continuous, but in the mutant clones they are interrupted and absent. The positioning and outlining of the aforementioned patterns all depend on positional information derived from Wnt ligands in Junonia (Martin and Reed, 2014; Mazo-Vargas et al., 2017). Overall, these cell-autonomous effects suggest that the two wing sets deploy equivalent spatial information and that differences in color patterns are mostly due to differential interpretation and color fate induction.
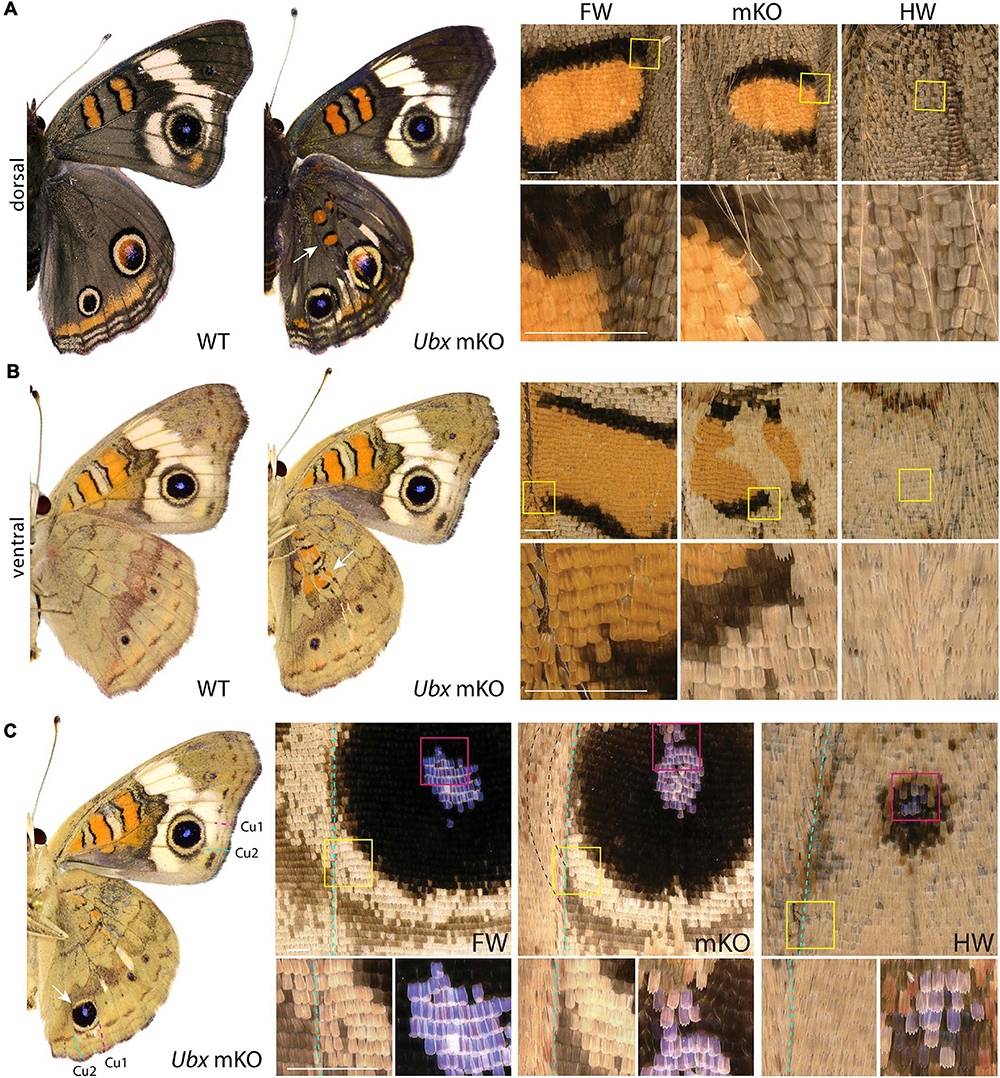
Figure 1. Ubx mosaic knock-outs show homeotic transformations of hindwing color patterns in J. coenia. Insets depict homeotic hindwing (HW)-to-forewing (FW) mutant patterns [mosaic knock-out (mKO), center column]. (A,B) Dorsal and ventral views of WT and an example Ubx mKO J. coenia, with insets showing the Discalis I pattern element. (C) Example of a ventral Cu1-Cu2 eyespot homeosis; mutant cells stopped at the Cu2 vein (dashed line), and non-mutant cells across that clone failed to induce outer rings. Scale bars = 500 μm.
Eyespots showed more complex and variegated effects than other pattern elements, depending on clone size, shape, and position. In J. coenia, the Cu1–Cu2 eyespot is particularly prominent in forewings (see Figures 2A,B for vein nomenclature). One Ubx mosaic knock-out induced the gain of a large eyespot in the hindwing Cu1–Cu2 region, almost reaching the size of its forewing homolog, but with its outer ring failing to cross the veins (Figure 1C). Scale shape shifts indicate that the mutant clone included the signaling focus and most of eyespot field, but did not cross the Cu1 and Cu2 veins, leading to an eyespot trimmed from the sections of its outer rings that normally extend into the neighboring M3vCu1 and Cu2–A1 regions. Thus, WT cells from those two regions were not responsive to the neighboring effect of the modified eyespot and mutant cells, indicating cell autonomy in the determination of the outer rings. We also obtained mosaic eyespot phenotypes, and speculate here on a few salient cases. A small satellite eyespot appeared in the R5–M1 hindwing region – normally devoid of eyespots – likely by transposing the morphogenetic activity of the small white R5–M1 forewing eyespot (Figure 2A). Remarkably, the induced eyespot was not transformed, but instead formed a complete black outer ring, suggesting this area retained the competency of WT dorsal hindwing tissue. M1–M2 eyespots are large in WT hindwings and reduced or cryptic in forewings. We did not obtain clones clearly overlapping with its induction focus, which may have reduced them in the mutant context; however, several individuals showed elongated clones streaking through this pattern, and resulting in its truncation (Figures 2B–D). The inner, orange-pink-blue-black sections from those eyespots were disorganized, while their outer buff and black rings were truncated, again pointing at a cell-autonomous mode of induction. A similar effect was observed in a partially transformed Cu1–Cu2 eyespot that showed discontinuity in its rings (Figure 2E). Overall, those composite eyespot phenotypes highlight the complex dynamics of eyespot development, and suggest that the determination of outer rings respond differently to eyespot morphogens between the two wing sets.
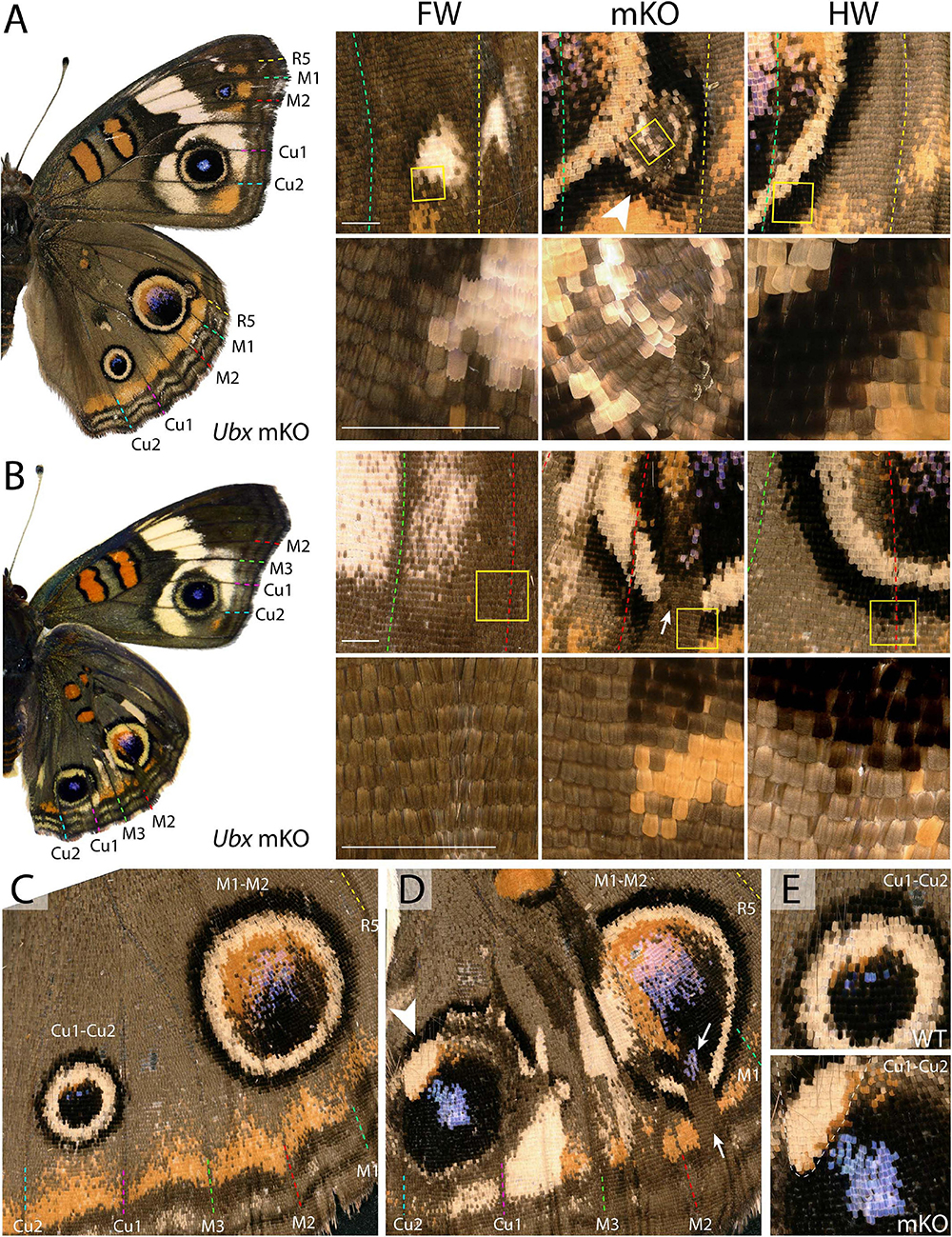
Figure 2. Variegated effect of partial mKO in the Junonia eyespot fields. (A) Ectopic R5-M1 eyespot appearing as a satellite to a WT M1–M2 eyespot, and forming rings not observed in the forewing (arrowhead). Lower panel depicts magnification of yellow insets in the upper panels. (B) Mosaic mutant with a complete Cu1–Cu2 eyespot homeosis, and partial mutant clones in the M1–M2 eyespot (insets). Arrow: missing outer rings and orange distal parafocal element in a mutant clone. (C) Magnified view of a WT hindwing (inset in E, upper panel: Cu1–Cu2 eyespot) (D) Equivalent view of a mosaic KO with partial Cu1–Cu2 and M1–M2 eyespot phenotype. Arrow: missing outer rings and orange distal parafocal element in a mutant clone; arrowhead: residual outer rings in a WT clone. Inset in (E, lower panel): magnified view of the Cu1–Cu2 eyespot, with residual outer rings in wild-type tissue (upper left, dashed line). Scale bars = 500 μm.
Next, we investigated the effects of Ubx mosaic knock-outs in V. cardui, another nymphalid butterfly. In V. cardui, injections earlier than 4 h AEL produced a low hatching rate at 250 ng/μL of sgRNA but a higher hatching rate at 125 ng/μL of sgRNA. The hatching rate was highest for injections done up to 6 h AEL at 250 ng/μL, however, penetrance was low (Table 1). A total of eight surviving adults showed hindwing-to-forewing homeosis, visible as shifts in scale morphology and color pattern organization (Figure 3 and Supplementary Figure 5). As in Junonia, transformed hindwings showed cell autonomous homeotic shifts equivalent to a visible transposition of the color patterns from the matching forewing position. For instance, we observed the transformation of the ventral hindwing central symmetry system into pink and black patterns from the forewing (Figures 3A–C). The distal parafocal elements of V. cardui (Otaki, 2012; Abbasi and Marcus, 2015) are chevrons that vary in color (black in the forewing, blue in the hindwing), and the same mutant showed a composite chevron with the forewing-like black section showing a thicker and pointier shape (Figure 3D). As the WT blue section was unaffected by the neighboring clone, we extrapolate that this pattern identity shift relied on a cell autonomous conversion of the cells to a forewing fate. Other cell autonomous effects were prominent on the dorsal surfaces, as seen in clones showing both the loss of hair-like scales, and the transposition of forewing color patterns such as the Discalis black patterns of the forewing and their surrounding orange-pink scales (Figures 3E–G).

Figure 3. Homeotic transformations of hindwing color patterns after Ubx mosaic knock-out in V. cardui. Panels (B–H) each show tripartite comparisons of positionally matched patterns from a forewing (left), a mutant hindwing (center), and a WT hindwing (right). Lower panel depicts magnification of yellow insets in the upper panel. (A) Ventral side of WT and an Ubx mKO individual in V. cardui, with magnified views in panels (B) (median Cu2), (C) (Cu1–Cu2 vein junction), and (D) (Cu1–Cu2 distal parafocal element). (E) Comparison of a dorsal forewing area, and the corresponding region of the mutant and WT hindwing, with an elongated mutant clone resulting in homeoses, as magnified in panels (F) (Cu1–M3 vein junction), (G) (Discalis II elements), and (H) (M2–M3 eyespot). The wildtype forewing in panel E was shrunk to 75% of its original length to adjust for allometric differences with the hindwings. Scale bars = 500 μm.
As in Junonia, mosaic knock-outs of Ubx varied in penetrance (percentage of mutant adults) and expressivity (clone size) depending on the injected stage interval and possibly the sgRNA dosage sequence. But more importantly they produced variegated effects in the eyespot field that depended on the affected vein compartment, on the nature of the WT pattern differences between the wing sets, and on clone size and position. The forewing eyespots are limited to the R5–M1, M1–M2, and M2–M3 intravenous compartments in Vanessa. These three white eyespots are peculiar as they are the only ones known to express WntA, suggesting a recent co-option in this lineage (Martin and Reed, 2014; Mazo-Vargas et al., 2017). Interestingly, we found several occurrences of mutant hindwing eyespot foci acquiring a white color in those compartments, including in a visibly elongated clone that encroached right through the focus on the mutant dorsal side (Figures 3E,H, 4A–D). On the ventral side, these three eyespots are bilaterally symmetric and white in the forewing, but radially symmetric with more color complexity in the hindwing, and mosaic knock-outs yielded predicted partial and complete transformations of the latter into the former (Figures 4A,C,D). Finally, we were surprised to observe a composite Cu1–Cu2 hindwing eyespot where most of the intravenous field took the orange/spotless identity of the forewing, as expected, but where a WT clone also left an eyespot remnant that included portions of the blue, black, and buff rings (Figures 4E,F’). This eyespot seems to have acted as its own signaling center, as suggested by a faded black ring in the mutant clone that radiates from its center (arrowhead in Figure 4F’). Thus, mosaic mutant eyespot field may displace the position of signaling centers and result in adjacent eyespots. The transformed tissue failed to recapitulate the ring organization observed across in the wild-type tissue, suggesting limited competency to respond to the satellite signaling activity. Together, these observations suggest that at least in the V. cardui Cu1–Cu2 space, Ubx is acting at two consecutive stages of eyespot development (Beldade and Monteiro, 2021): first it may be required of the correct positioning and induction of an eyespot signaling center, and later during the signaling phase, it may provide hindwing identity that is used for the integration of the signals into rings of the proper color and size.
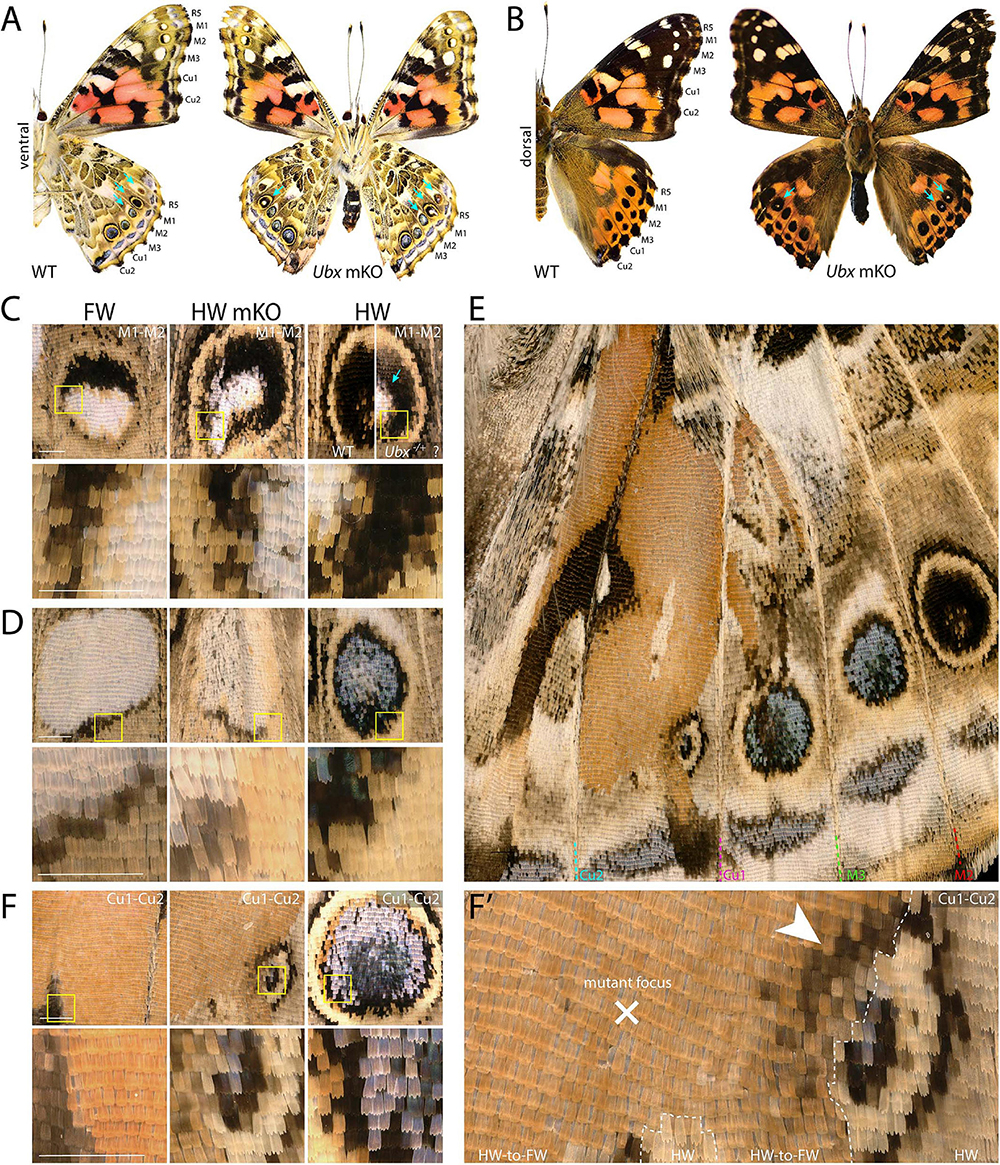
Figure 4. Dominant-white, partial and complete replacement, and large truncation effects on Vanessa eyespots. (A,B) Ubx mKO individual with multiple hindwing eyespots showing ectopic white. White scales are prominent in the three WT forewing eyespots between R5 and M3, but normally absent from hindwings (arrows). (C) Magnified views featuring two mutant M1–M2 eyespots from panel A; center: homeotic transformation of the left side, the rest of the eyespot may be heterozygous-mutant due to the forewing-like white center; right: comparison of a putative heterozygous mKO and WT (uninjected) eyespots. (D) Complete homeosis of an M2–M3 eyespot. (E–F’) Views at increasing magnification of truncated Cu1–Cu2. Most of the eyespot field is transformed, but unaffected tissue left an eyespot remnant that likely deployed its own signaling activity, as the partial induction of black rings in the mutant field implies (arrowhead). Panels (A,C–F’): ventral views. Scale bars = 500 μm.
Among the survivors that were derived from a batch of 212 V. cardui eggs injected between 2 and 4 h, five larvae showed ectopic limbs on the first abdominal segment (A1) at the fifth instar stage. Those rudimentary limbs showed all three podomers observed in a T3 leg, but were shorter and smaller (Figure 5). These results are consistent with RNAi and spontaneous mutations in the silkworm and tobacco hornworm, which established Ubx as a suppressor of larval leg development in the A1–A2 segments (Zheng et al., 1999; Masumoto et al., 2009), and with the known role of Ubx in repressing thoracic appendage development in the abdomen of other insects (Hughes and Kaufman, 2002; Angelini et al., 2005; Mahfooz et al., 2007). The effect of sgRNA dosage and time of injection on the penetrance and expressivity of this phenotype were untested, but we note a high embryonic lethality of CRISPR injections performed prior to 3 h AEL (Table 1), suggesting the level of mosaicism needed for generating larval phenotypes is optimal in injections performed at 3–4 h AEL.
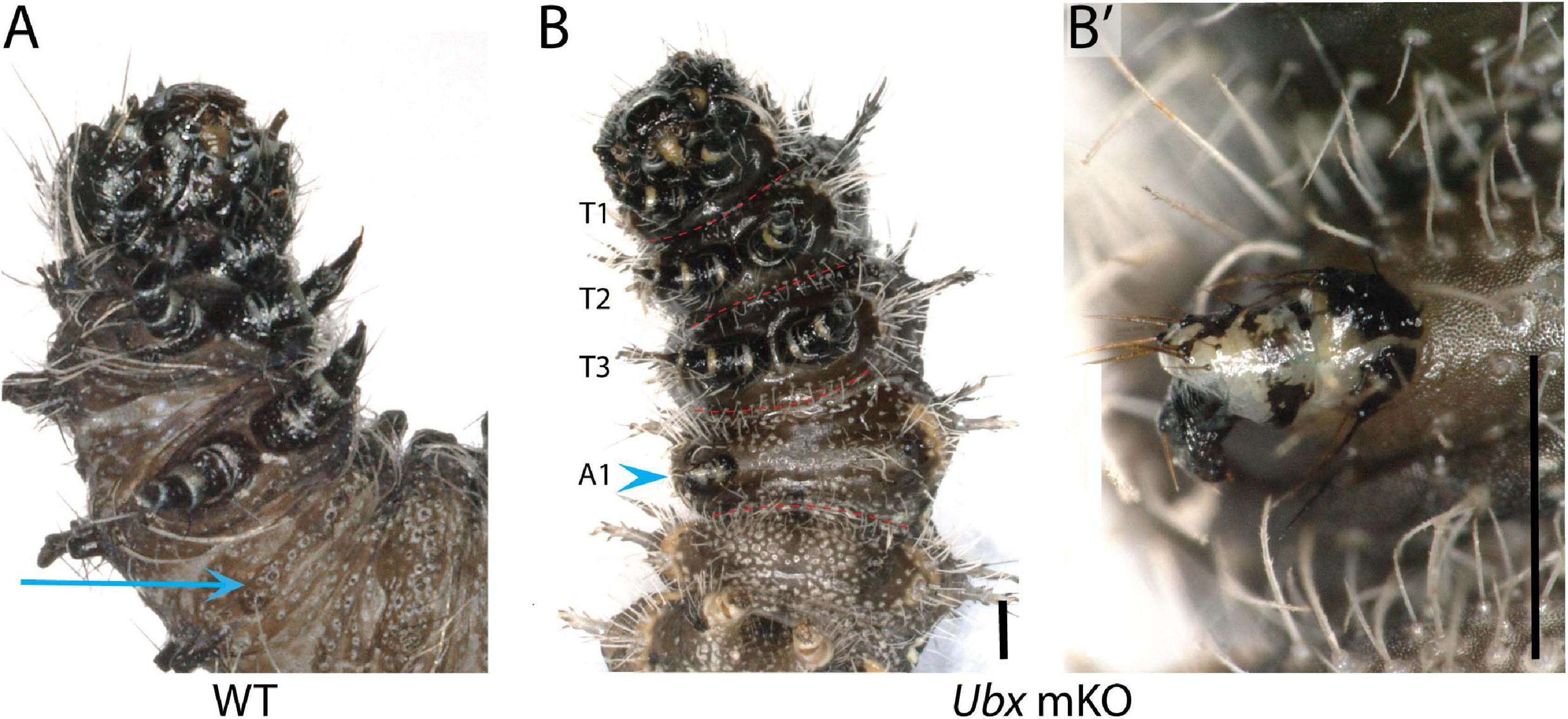
Figure 5. Loss of Ubx produces ectopic abdominal legs. (A) WT fifth instar larva, ventral view; arrow: A1 ventral epithelium. (B,B’) Partial limb (arrowhead, magnified in B’) on the A1 segment of a fifth instar Vanessa cardui Ubx mKO larva. Scale bars = 1000 μm.
Spontaneous color pattern homeoses similar to Hindsight have been described in museum specimens spanning many lineages of butterflies and moths (Sibatani, 1980, 1983a, b). However, it is unknown if homeotic transformations can also affect other aspects of wing morphology, as in the more radically T2/T3 specialized appendages of Drosophila (wing/haltere) and Tribolium (elytra/wing). Phycitine moths of the Pyralidae family offer a level of wing specialization that is somewhat analogous to elytral wing plans. The forewings of Phycitinae are thick, concave, and elongate beyond the caudal end of the insect, thus providing protection to the animal while at rest, while, akin to beetles, hindwings are thin and folded at rest. The hindwing has a hook-like frenulum that locks into the retinaculum, a tuft of bristles at the base of the ventral forewing (Braun, 1924; Zaspel, 2016). This frenulo-retinacular wing coupling mechanically pulls the folded hindwing open, and synchronizes the wings during flight (Wootton, 2002). The dorsal forewing is pigmented, as the only surface exposed at rest, while the hindwing is lightly pigmented. Here we used the phycitine Indian Mealmoth (P. interpunctella) as a model system to investigate additional aspects of lepidopteran wing specialization. First, we confirmed that the UbdA antigen (anti-Ubx/abd-A FP6.87 monoclonal antibody) is absent from the forewing imaginal disks, and expressed throughout the hindwing, thus corroborating the hindwing-specific expression of Ubx described in other lepidopterans (Weatherbee et al., 1998; Tong et al., 2014; Prasad et al., 2016). Then, we generated Ubx mosaic knock-outs and observed homeotic clones on the hindwing that were most conspicuous as transpositions toward black-and-brown pigmented states on the dorsal side (Figures 6A,B and Supplementary Figure 6). Importantly, the most transformed hindwings were imperfectly folded, and we often noticed a kink in the positioning of the overlying forewing in the live emerged adults. These hindwings also showed visible transformation in their overall architecture, with a thicker anterior edge, and clones in the anal region that prevented the fan-like folding along the abdomen (Figure 6C). The hindwing-specific frenulum hook was malformed in several individuals. We failed to detect ectopic retinaculum on mutant hindwings, possibly because it derives from a small patch of transformed scales that was not reached in the mosaics we screened. Finally, male forewings possess a scent-producing apparatus at the base of the ventral costa, consisting of a costal fold and external tuft of hairs, that protects androconial scales known as hairpencils, and exposes them by eversion during courtship (Richards and Thomson, 1932; Grant, 1978). In mosaic KOs, we recovered two males with a hindwing that acquired an ectopic, fully developed set of hairpencils at its base, as well as a reduced external tuft in what appears to be a incomplete costal fold (Figures 6C,D and Supplementary Figure 6D). Upon descaling, mosaic KO hindwings revealed additional homeotic transformations of their vein organization (Figure 6E). The costal area was transformed to recreate the thicker leading edge of forewing, visible by the ectopic gain of radial veins. In the anal region, the CuP and A3 veins are specific to the WT hindwing and involved in the fan-like folding of this wing, but those veins were lost in mutant hindwings, here again adopting a forewing morphology (Supplementary Figure 7). Overall the pyralid data show that Ubx is not only required for hindwing depigmentation but also for the repression of forewing states, including secondary sexual structures involved in male courtship behavior, and vein architectures that influence wing shape, rigidity, and foldability.
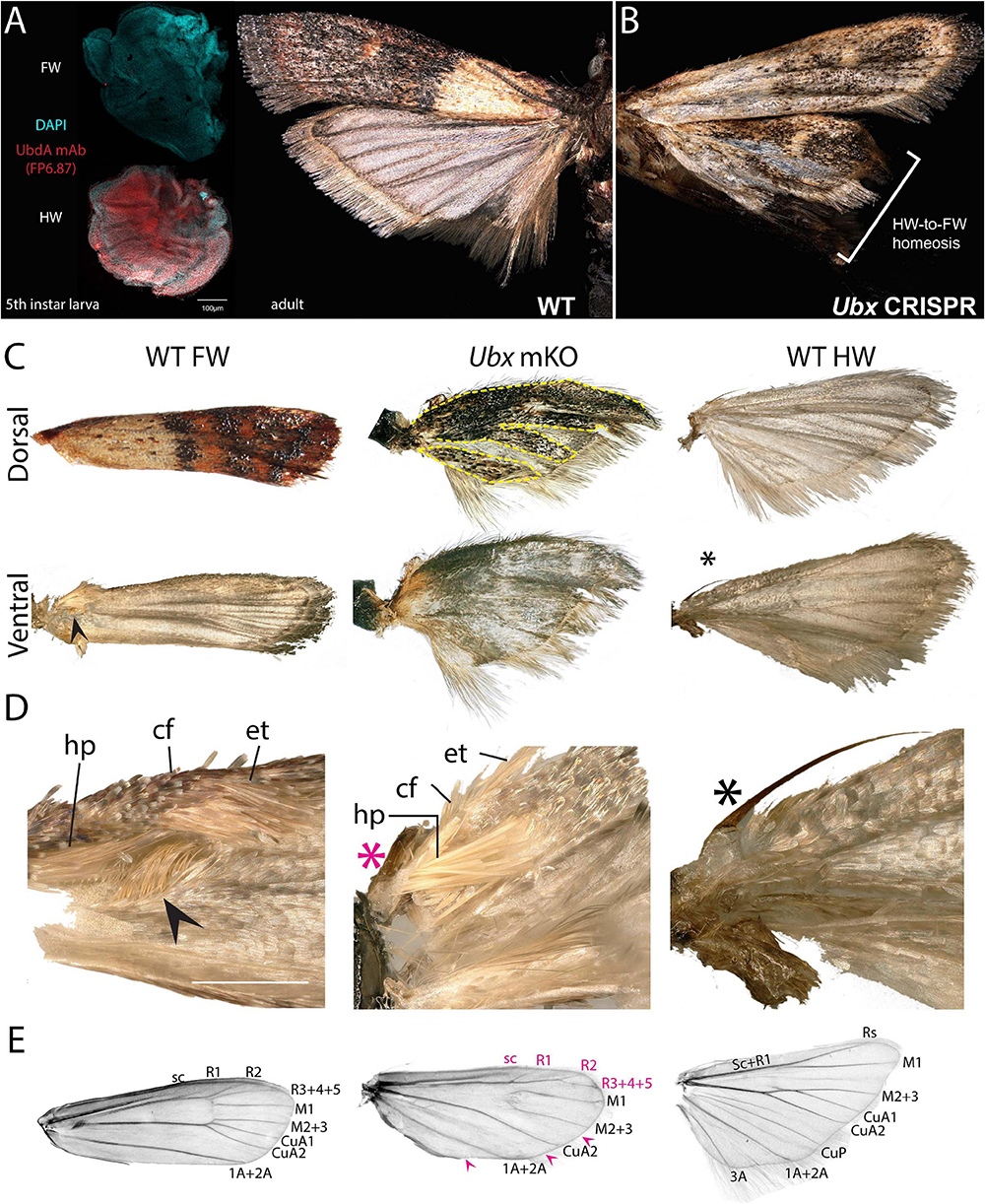
Figure 6. Ubx controls hindwing specification in Plodia. (A) Left: Immunodetection of UbdA in fifth instar larval wing disks (FP6.87 mAb, red; DAPI, cyan). Right: wild-type P. interpunctella (dorsal view). (B) Ubx mKO individual with transformation in pigmentation and wing rigidity. (C) Dorsal and ventral surfaces of a WT forewing, Ubx mKO and WT hindwing from male specimens. The mutant hindwing takes a concave appearance, loses its posterior folds and shows transformed dorsal black/brown patterns with a forewing identity (dashed lines). (D) Magnified ventral view of the wing base. Wing coupling uses a frenulum (black asterisk) engaging into a retinaculum (arrowhead). Male forewings also possess an extendable scent apparatus. The Ubx crispant hindwing shows a defective frenulum (magenta) and an ectopic scent apparatus with fully developed hairpencils (hp); cf: costal fold; et: external tuft of hair. (E) Wing venation of a de-scaled adult female forewing, Ubx mKO and hindwing. The Ubx mosaic KO shows homeoses of anterior veins toward the forewing venation (magenta letters) and loss of a few veins (magenta arrowheads). Vein nomenclature adapted from the phycitine moth wing plan (Solis and Neunzig, 2017). Scale bars: A = 100 μm; D = 500 μm.
Ubx loss-of-function was deleterious throughout development, with few individuals reaching the adult stage. Previous mosaic knock-outs of the WntA gene, which show no embryonic or larval defects, can generate crispants with a mutant phenotype that extends to all the wing surfaces (Mazo-Vargas et al., 2017; Concha et al., 2019). This contrasts with the bulk of our Ubx wing phenotypes, which showed small wing clones and no complete hindwing transformation. Mosaicism was thus needed to generate viable adults, and as a corollary, our failure to observe certain clone sizes or locations can be due to a form of “survivorship bias.” The observed lethality of those experiments is reminiscent of the low hatching rate of silkworm embryos injected with Ubx RNAi, perhaps due to the inferred role of Ubx in patterning the central nervous system (Warren et al., 1994; Zheng et al., 1999; Masumoto et al., 2009). As a visible manifestation of those functions in embryos and larvae, we observed Ubx crispant larvae exhibiting partially developed additional thoracic limbs on the A1 segment (Figure 5). These results are in line with a previous study that functionally characterized Ubx in Bombyx mori embryos through RNAi, suggesting it acts as a suppressor of leg development in the first abdominal segment (Masumoto et al., 2009). Homeotic mutants that map to the Ubx/abd-A region show deficient expressions of Ubx, and bear ectopic A1 (and occasionally A2) larval legs, have also been isolated in both B. mori and Manduca sexta (Ueno et al., 1995; Zheng et al., 1999; Tong et al., 2017). Previous data and our Ubx loss-of-function somatic mutations via CRISPR targeted mutagenesis thus indicate Ubx is a suppressor of A1 leg development. Importantly, the embryos of most insects including lepidopterans develop a transient A1 appendage, known as the pleuropod, which functions as a glandular organ involved in egg hatching (Konopová et al., 2020). Instead of blocking appendage development in A1, it is likely that Ubx is necessary for establishing the identity of the pleuropod appendage, and that ectopic A1 legs that have been observed in Hox perturbation experiments are pleuropod-to-leg homeoses (Lewis et al., 2000; Konopova and Akam, 2014).
Previous studies have highlighted that mosaic wing homeoses perturb the eyespot determination system, and have discussed how effects can vary based on the placement of the clone and whether or not it encompasses a focal signaling center (Sibatani, 1980; Nijhout and Rountree, 1995; Weatherbee et al., 1999). As previously reported, our Ubx mosaic knock-outs as well as the ones reported in Bicyclus (Matsuoka and Monteiro, 2020) are consistent with those previous findings: a large homeotic clone overlapping with the putative eyespot center will result in a gain or reduction of eyespot development depending on the corresponding forewing state in this vein compartment. In general, hindwings and forewings seem to respond to the same morphogenetic signal (Nijhout and Rountree, 1995). As a possible exception, Vanessa shows a recent, forewing specific evolutionary acquisition of focal WntA in the developing eyespots (Mazo-Vargas et al., 2017), and we speculate that this transposed signal causes the induction of white foci in the mutant hindwings. When they do not intersect with the signaling center, mutant clones can also reveal color pattern discontinuities that are informative about eyespot signaling and color ring induction. Our data suggest the hindwing cells require Ubx in a cell autonomous fashion to induce outer black and buff eyespot rings, in the dorsal hindwing eyespots of J. coenia, and in the ventral V. cardui Cu1–Cu2 eyespot. In contrast, the inner ring or “core” of those eyespots (Iwata and Otaki, 2016) were disrupted in a more unpredictable way in Junonia mosaics: in clones that streaked through the vicinity of the M1–M2 eyespot focus, the core eyespot showed a disorganization of the color field rather than a reversion to the eyespot-free state of the corresponding forewing field. The current model of eyespot formation involves the definition of Distal-less (Dll) positive foci in the midvein spaces from fifth instar imaginal disks, which becomes a signaling center during early pupal stages, and establishes morphogen gradients or signals that pattern boundaries between the surrounding rings, directly or sequentially (Monteiro, 2015; Connahs et al., 2019; Iwata and Otaki, 2019). Besides its role in focus activation, Distal-less is also involved in the determination of the core eyespot (Brunetti et al., 2001; Monteiro et al., 2013). Ubx represses Dll in insect abdomens (Vachon et al., 1992; Angelini et al., 2005; Uhl et al., 2016), and in Junonia pupal eyespot rings (Weatherbee et al., 1999), implying that the core eyespot disorganization may be linked to a depression of Dll. This phenomenon may not hold true in in satyrid butterflies like Bicyclus, which express Ubx and Antp in the eyespot foci, and where Ubx gain-of-function experiments suggest Ubx acts as an activator of Dll (Tong et al., 2014; Matsuoka and Monteiro, 2020). Moving forward, a better understanding of the eyespot determination networks and of the levels of Ubx integration within them may help to better understand the different Ubx mutant clones, as well as possible differences between different nymphalid lineages (Tong et al., 2014; Matsuoka and Monteiro, 2020; Monteiro, 2020).
Ubx somatic mutagenesis consistently yielded complete homeotic transformation of hindwing states into their forewing positional homolog. This included identity T3-to-T2 shifts in scale derivatives (color scales, wing-coupling scales, male-specific androconia), color pattern arrangements, eyespot size, and wing structure. Thus, consistently with other insect wing systems (Tomoyasu et al., 2009; Pavlopoulos and Akam, 2011; Kaschula et al., 2018), lepidopteran Ubx seems to act as a multitasking master gene or “micromanager” (Akam, 1998) that is embedded in a multitude of gene regulatory networks during wing development. As previously reviewed (Tomoyasu, 2017), similar roles of Ubx as a metathoracic specifier have been identified in a variety of insects, and probably indicate a conserved function across Holometabola. It would be challenging but providential to study Ubx expression and function in insect lineages with alternative wing sets such as Strepsiptera (T2 halteres, T3 wings), or in basal winged insects such as dragonflies and mayflies. At the molecular level, a characterization of the regulation, transcriptional targets, and chromatin-level interactions of Ubx is underway in a variety of systems (McKay and Lieb, 2013; Prasad et al., 2016; Sánchez-Higueras et al., 2019; Diaz-de-la-Loza et al., 2020), and we expect similar approaches in lepidopterans to yield valuable insights on the evolutionary specialization of serial homologs.
Insect stock origins, rearing conditions, larval diets and oviposition plants are summarized in Supplementary Table 2. Both butterfly species were reared in plastic cups following a previously described procedure (Martin et al., 2020). For J. coenia, the agar-based diet was prepared using water boiled with dry ground leaves of the host plant Plantago lanceolata. The modified larval diet for P. interpunctella (Silhacek and Murphy, 2008) was made by mixing (w/w) 50% coarse wheat bran, 22% glycerol, 13% dextrose, 8% water, 5% brewer’s yeast powder, and 2% canola oil.
Ubx sequences of J. coenia (AAL71873.1) and V. tameamea (XP_026485353.1) were obtained from NCBI and we used TBLASTN against the J. coenia genome (van der Burg et al., 2019), a V. cardui wing transcriptome (Connahs et al., 2016), and the P. interpunctella genome (Roberts et al., 2020) to obtain additional sequences. SgRNAs were designed against Ubx exon 1 in order to produce a frameshift mutation due to imperfect non-homologous end joining repair. The uniqueness of the guide was confirmed by performing a BLASTN against the available whole genome assemblies.
Cas9:sgRNA heteroduplexes were prepared as previously described (Martin et al., 2020). Before each experiment, frozen aliquots of 2.5 μL sgRNA (250 ng/μL or 333 ng/μL) and Cas9 (500 or 666 ng/μL) were mixed, incubated at room temperature for 10 min, and left on ice until injection.
Butterfly eggs laid on host plant leaves were collected at controlled time intervals before injection. To soften the chorion, J. coenia eggs were washed for 1 min in 5% benzalkonium chloride, rinsed with autoclaved water and dried using a gentle flow of compressed air. Eggs from both butterfly species were arranged with the micropyle side facing up on double-sided tape and injected on the side following a previously described procedure (Martin et al., 2020). Post injection, butterfly eggs were rested in a humidity chamber at their respective rearing temperatures until hatching and addition of artificial diet. P. interpunctella adults were anaesthetized with CO2 gas which triggers a release of freshly fertilized eggs. Eggs were collected and arranged horizontally on a parafilm strip for immediate injection in the posterior half, opposite to the micropyle. The egg injection wounds were sealed with cyanoacrylate glue and stored in a humidity chamber until hatching. All injections were performed using a compressed air micro-injection set-up and pulled borosilicate needles.
Adult butterflies were pinned with the mutant side up (ventral side up for adults with mutant clones on both surfaces). Full-mount photographs of butterflies were taken either on a Nikon D5300 digital camera mounted with an AF-S VR MicroNikkor 105 mm f/2.8G lens and an 80-LED ring light, or on a Keyence VHX-5000 digital microscope at 50X on a VH-Z00T lens. The high resolution images of butterflies and the wing mounts of moths were taken at 100X, 300X or 600X, and 50X, respectively, on the Keyence VHX-5000 fitted with a VH-Z100T lens.
Wing disks of P. interpunctella were dissected from fifth instar larvae that stopped feeding and had spun a silk chamber, fixed in 3.7% formaldehyde, and stained for immunolocalization of the UbdA epitope following a previously described procedure (Martin et al., 2020).
Wings were dissected and washed by repeating consecutive immersions in 90% ethanol and 10% freshly prepared bleach. Descaled wings were washed in 1X PBS to remove debris, mounted with coverslips in 100% ethanol, and immediately imaged for green autofluorescence on an Olympus SZX2 fluorescent stereomicroscope. Photographs were post-processed in Adobe Photoshop for contrast adjustment and the generation of inverted gray images.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AT, AFP, and CH conducted the research. PDS, NHP, and AM supervised the project. AT, AFP, and AM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the NSF awards IOS-1656553 and IOS-1923147 to AM, and a GWU Packer Graduate Fellowship to AT.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Chris Day, Nora Wolcott, Emily Earls, Damien Gailly, Ling Sheng Loh, Joseph Hanly, and Richard Furlong for their assistance with micro-injections and insect rearing throughout this project, Joseph Hanly for proof-reading the manuscript, and the Insect Genetic Transformation Research Coordination Network led by David O’Brochta and Rob Harrell for stimulating the initiation of this project via the Peer-to-Peer training program.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.643661/full#supplementary-material
Abbasi, R., and Marcus, J. M. (2015). Color pattern evolution in Vanessa butterflies (Nymphalidae: Nymphalini): non-eyespot characters. Evol. Dev. 17, 63–81. doi: 10.1111/ede.12109
Adler, P. N. (1979). Position-specific interaction between cells of the imaginal wing and haltere discs of Drosophila melanogaster. Dev. Biol. 70, 262–267. doi: 10.1016/0012-1606(79)90023-X
Adler, P. N. (1981). Bithoraxoid, postbithorax, and the determination system of the haltere disk. Dev. Biol. 86, 157–169. doi: 10.1016/0012-1606(81)90326-2
Akam, M. (1998). Hox genes: from master genes to micromanagers. Curr. Biol. 8, R676–R678. doi: 10.1016/S0960-9822(98)70433-6
Angelini, D. R., Liu, P. Z., Hughes, C. L., and Kaufman, T. C. (2005). Hox gene function and interaction in the milkweed bug Oncopeltus fasciatus (Hemiptera). Dev. Biol. 287, 440–455. doi: 10.1016/j.ydbio.2005.08.010
Beldade, P., and Monteiro, A. (2021). Eco-evo-devo advances with butterfly eyespots. Curr. Opin. Genet. Dev. 69, 6–13.
Bender, W., Akam, M., Karch, F., Beachy, P. A., Peifer, M., Spierer, P., et al. (1983). Molecular genetics of the bithorax complex in Drosophila melanogaster. Science 221, 23–29. doi: 10.1126/science.221.4605.23
Braun, A. F. (1924). The frenulum and its retinaculum in the Lepidoptera. Ann. Entomol. Soc. Am. 17, 234–257. doi: 10.1093/aesa/17.3.234
Brunetti, C. R., Selegue, J. E., Monteiro, A., French, V., Brakefield, P. M., and Carroll, S. B. (2001). The generation and diversification of butterfly eyespot color patterns. Curr. Biol. 11, 1578–1585. doi: 10.1016/S0960-9822(01)00502-4
Carroll, S. B. (1994). Developmental regulatory mechanisms in the evolution of insect diversity. Development 1994, 217–223.
Concha, C., Wallbank, R. W. R., Hanly, J. J., Fenner, J., Livraghi, L., Rivera, E. S., et al. (2019). Interplay between developmental flexibility and determinism in the evolution of mimetic heliconius wing patterns. Curr. Biol. 29, 3996.e–4009.e. doi: 10.1016/j.cub.2019.10.010
Connahs, H., Rhen, T., and Simmons, R. B. (2016). Transcriptome analysis of the painted lady butterfly, Vanessa cardui during wing color pattern development. BMC Genomics 17:270. doi: 10.1186/s12864-016-2586-5
Connahs, H., Tlili, S., Creij, J., van Loo, T. Y. J., Banerjee, T. D., Saunders, T. E., et al. (2019). Activation of butterfly eyespots by Distal-less is consistent with a reaction-diffusion process. Development 146:dev169367. doi: 10.1242/dev.169367
Crickmore, M. A., and Mann, R. S. (2006). Hox control of organ size by regulation of morphogen production and mobility. Science 313, 63–68. doi: 10.1126/science.1128650
Diaz-de-la-Loza, M.-C., Loker, R., Mann, R. S., and Thompson, B. J. (2020). Control of tissue morphogenesis by the HOX gene Ultrabithorax. Development 147:dev184564. doi: 10.1242/dev.184564
Fu, S.-J., Zhang, J.-L., Chen, S.-J., Chen, H.-H., Liu, Y.-L., and Xu, H.-J. (2020). Functional analysis of Ultrabithorax in the wing-dimorphic planthopper Nilaparvata lugens (Stål, 1854) (Hemiptera: Delphacidae). Gene 737:144446. doi: 10.1016/j.gene.2020.144446
Grant, G. G. (1978). Morphology of the presumed male pheromone glands on the forewings of tortricid and phycitid moths. Ann. Entomol. Soc. Am. 71, 423–431. doi: 10.1093/aesa/71.3.423
Hersh, B. M., Nelson, C. E., Stoll, S. J., Norton, J. E., Albert, T. J., and Carroll, S. B. (2007). The UBX-regulated network in the haltere imaginal disc of D. melanogaster. Dev. Biol. 302, 717–727. doi: 10.1016/j.ydbio.2006.11.011
Hughes, C. L., and Kaufman, T. C. (2002). Hox genes and the evolution of the arthropod body plan1. Evol. Dev. 4, 459–499. doi: 10.1046/j.1525-142X.2002.02034.x
Iwata, M., and Otaki, J. M. (2016). Spatial patterns of correlated scale size and scale color in relation to color pattern elements in butterfly wings. J. Insect Physiol. 85, 32–45. doi: 10.1016/j.jinsphys.2015.11.013
Iwata, M., and Otaki, J. M. (2019). Insights into eyespot color-pattern formation mechanisms from color gradients, boundary scales, and rudimentary eyespots in butterfly wings. J. Insect Physiol. 114, 68–82. doi: 10.1016/j.jinsphys.2019.02.009
Jantzen, B., and Eisner, T. (2008). Hindwings are unnecessary for flight but essential for execution of normal evasive flight in Lepidoptera. PNAS 105, 16636–16640. doi: 10.1073/pnas.0807223105
Kaschula, R., Pinho, S., and Alonso, C. R. (2018). MicroRNA-dependent regulation of hox gene expression sculpts fine-grain morphological patterns in a Drosophila appendage. Development 145:dev161133. doi: 10.1242/dev.161133
Konopova, B., and Akam, M. (2014). The hox genes Ultrabithorax and abdominal-A specify three different types of abdominal appendage in the springtail Orchesella cincta (Collembola). EvoDevo 5:2. doi: 10.1186/2041-9139-5-2
Konopová, B., Buchberger, E., and Crisp, A. (2020). Transcriptome of pleuropodia from locust embryos supports that these organs produce enzymes enabling the larva to hatch. Front. Zool. 17:4. doi: 10.1186/s12983-019-0349-2
Lewis, D. L., DeCamillis, M., and Bennett, R. L. (2000). Distinct roles of the homeotic genes Ubx and abd-A in beetle embryonic abdominal appendage development. PNAS 97, 4504–4509. doi: 10.1073/pnas.97.9.4504
Lipshitz, H. D. (2007). Genes, Development and Cancer: The Life and Work of E. B. Lewis. Dordrecht: Springer Netherlands.
Mahfooz, N., Turchyn, N., Mihajlovic, M., Hrycaj, S., and Popadić, A. (2007). Ubx regulates differential enlargement and diversification of insect hind legs. PLoS One 2:e866. doi: 10.1371/journal.pone.0000866
Makhijani, K., Kalyani, C., Srividya, T., and Shashidhara, L. S. (2007). Modulation of decapentaplegic gradient during haltere specification in Drosophila. Dev. Biol. 302, 243–255. doi: 10.1016/j.ydbio.2006.09.029
Martin, A., and Reed, R. D. (2014). Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev. Biol. 395, 367–378. doi: 10.1016/j.ydbio.2014.08.031
Martin, A., Wolcott, N. S., and O’Connell, L. A. (2020). Bringing immersive science to undergraduate laboratory courses using CRISPR gene knockouts in frogs and butterflies. J. Exp. Biol. 223:jeb208793. doi: 10.1242/jeb.208793
Masumoto, M., Yaginuma, T., and Niimi, T. (2009). Functional analysis of Ultrabithorax in the silkworm, Bombyx mori, using RNAi. Dev. Genes Evol. 219, 437–444. doi: 10.1007/s00427-009-0305-9
Matsuoka, Y., and Monteiro, A. (2020). Hox genes are essential for the development of eyespots in Bicyclus anynana butterflies. Genetics 217:iyaa005. doi: 10.1093/genetics/iyaa005
Mazo-Vargas, A., Concha, C., Livraghi, L., Massardo, D., Wallbank, R. W. R., Zhang, L., et al. (2017). Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc. Natl. Acad. Sci. U.S.A. 114:10701. doi: 10.1073/pnas.1708149114
McKay, D. J., and Lieb, J. D. (2013). A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27, 306–318. doi: 10.1016/j.devcel.2013.10.009
Medved, V., Marden, J. H., Fescemyer, H. W., Der, J. P., Liu, J., Mahfooz, N., et al. (2015). Origin and diversification of wings: insights from a neopteran insect. PNAS 112, 15946–15951. doi: 10.1073/pnas.1509517112
Mohit, P., Makhijani, K., Madhavi, M. B., Bharathi, V., Lal, A., Sirdesai, G., et al. (2006). Modulation of AP and DV signaling pathways by the homeotic gene Ultrabithorax during haltere development in Drosophila. Dev. Biol. 291, 356–367. doi: 10.1016/j.ydbio.2005.12.022
Monteiro, A. (2015). Origin, development, and evolution of butterfly eyespots. Annu. Rev. Entomol. 60, 253–271. doi: 10.1146/annurev-ento-010814-020942
Monteiro, A. (2020). Distinguishing serial homologs from novel traits: experimental limitations and ideas for improvements. BioEssays 43:e2000162. doi: 10.1002/bies.202000162
Monteiro, A., Chen, B., Ramos, D. M., Oliver, J. C., Tong, X., Guo, M., et al. (2013). Distal-less regulates eyespot patterns and melanization in Bicyclus butterflies. J. Exp. Zool. B Mol. Dev. Evol. 320, 321–331. doi: 10.1002/jez.b.22503
Navas, L. F., de Garaulet, D. L., and Sánchez-Herrero, E. (2006). The Ultrabithorax hox gene of Drosophila controls haltere size by regulating the Dpp pathway. Development 133, 4495–4506. doi: 10.1242/dev.02609
Nijhout, H. F., and Rountree, D. B. (1995). Pattern induction across a homeotic boundary in the wings of Precis coenia (Hbn.) (Lepidoptera: Nymphalidae). Int. J. Insect Morphol. Embryol. 24, 243–251. doi: 10.1016/0020-7322(95)00004-N
Ohde, T., Yaginuma, T., and Niimi, T. (2014). Wing serial homologs and the origin and evolution of the insect wing. Zoology 117, 93–94. doi: 10.1016/j.zool.2013.11.001
Otaki, J. M. (2012). Color pattern analysis of nymphalid butterfly wings: revision of the nymphalid groundplan. Zoolog. Sci. 29, 568–576. doi: 10.2108/zsj.29.568
Pavlopoulos, A., and Akam, M. (2011). Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. PNAS 108, 2855–2860. doi: 10.1073/pnas.1015077108
Pourquie, O. (2009). HOX Genes, 1st Edn. 88. Available online at: https://www.elsevier.com/books/hox-genes/pourquie/978-0-12-374529-3 [Accessed November 24, 2020]
Prasad, N., Tarikere, S., Khanale, D., Habib, F., and Shashidhara, L. S. (2016). A comparative genomic analysis of targets of Hox protein Ultrabithorax amongst distant insect species. Sci. Rep. 6:27885. doi: 10.1038/srep27885
Richards, O. W., and Thomson, W. S. (1932). A contribution to the study of the genera Ephestia, Gn. (including Strymax, Dyar), and Plodia, Gn. (Lepidoptera, Phycitidae), with notes on parasites of the larvae. Trans. R. Entomol. Soc. Lond. 80, 169–247. doi: 10.1111/j.1365-2311.1932.tb03306.x
Roberts, K. E., Meaden, S., Sharpe, S., Kay, S., Doyle, T., Wilson, D., et al. (2020). Resource quality determines the evolution of resistance and its genetic basis. Mol. Ecol. 29, 4128–4142. doi: 10.1111/mec.15621
Sánchez-Higueras, C., Rastogi, C., Voutev, R., Bussemaker, H. J., Mann, R. S., and Hombría, J. C.-G. (2019). In vivo hox binding specificity revealed by systematic changes to a single cis regulatory module. Nat. Commun. 10:3597. doi: 10.1038/s41467-019-11416-1
Shashidhara, L. S., Agrawal, N., Bajpai, R., Bharathi, V., and Sinha, P. (1999). Negative regulation of dorsoventral signaling by the homeotic gene Ultrabithorax during haltere development in Drosophila. Dev. Biol. 212, 491–502. doi: 10.1006/dbio.1999.9341
Sibatani, A. (1980). Wing homoeosis in Lepidoptera: a survey. Dev. Biol. 79, 1–18. doi: 10.1016/0012-1606(80)90069-X
Sibatani, A. (1983a). A compilation of data on wing homoeosis in Lepidoptera. J. Res. Lepid 22, 1–46.
Sibatani, A. (1983b). Compilation of data on wing homoeosis on Lepidoptera: supplement I. J. Res. Lepid 22, 118–125.
Silhacek, D., and Murphy, C. (2008). Moisture content in a wheat germ diet and its effect on the growth of Plodia interpunctella (Hübner). J. Stored Prod. Res. 44, 36–40. doi: 10.1016/j.jspr.2006.03.004
Solis, M. A., and Neunzig, H. H. (2017). A new phycitine genus and species of a pourouma-feeding moth (Lepidoptera: Pyralidae) from Panama. Proc. Entomol. Soc. Wash. 119, 464–470.
Tomoyasu, Y. (2017). Ultrabithorax and the evolution of insect forewing/hindwing differentiation. Curr. Opin. Insect Sci. 19, 8–15. doi: 10.1016/j.cois.2016.10.007
Tomoyasu, Y., Arakane, Y., Kramer, K. J., and Denell, R. E. (2009). Repeated co-options of exoskeleton formation during wing-to-elytron evolution in beetles. Curr. Biol. 19, 2057–2065. doi: 10.1016/j.cub.2009.11.014
Tomoyasu, Y., Ohde, T., and Clark-Hachtel, C. (2017). What serial homologs can tell us about the origin of insect wings. F1000Res 6:268. doi: 10.12688/f1000research.10285.1
Tomoyasu, Y., Wheeler, S. R., and Denell, R. E. (2005). Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature 433, 643–647. doi: 10.1038/nature03272
Tong, X., Fu, M. Y., Chen, P., Chen, L., Xiang, Z. H., Lu, C., et al. (2017). Ultrabithorax and abdominal-A specify the abdominal appendage in a dosage-dependent manner in silkworm, Bombyx mori. Heredity 118, 578–584. doi: 10.1038/hdy.2016.131
Tong, X., Hrycaj, S., Podlaha, O., Popadic, A., and Monteiro, A. (2014). Over-expression of Ultrabithorax alters embryonic body plan and wing patterns in the butterfly Bicyclus anynana. Dev. Biol. 394, 357–366. doi: 10.1016/j.ydbio.2014.08.020
Ueno, K., Toshifumi, N., and Suzuki, Y. (1995). “Roles of homeotic genes in the Bombyx body plan,” in Molecular Model Systems in the Lepidoptera, eds A. S. Wilkins and M. R. Goldsmith (Cambridge: Cambridge University Press), 165–180.
Uhl, J. D., Zandvakili, A., and Gebelein, B. (2016). A hox transcription factor collective binds a highly conserved Distal-less cis-Regulatory module to generate robust transcriptional outcomes. PLoS Genet. 12:e1005981. doi: 10.1371/journal.pgen.1005981
Vachon, G., Cohen, B., Pfeifle, C., McGuffin, M. E., Botas, J., and Cohen, S. M. (1992). Homeotic genes of the bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71, 437–450. doi: 10.1016/0092-8674(92)90513-C
van der Burg, K. R. L., Lewis, J. J., Martin, A., Nijhout, H. F., Danko, C. G., and Reed, R. D. (2019). Contrasting roles of transcription factors spineless and EcR in the highly dynamic chromatin landscape of butterfly wing metamorphosis. Cell Reports 27, 1027–1038.e3. doi: 10.1016/j.celrep.2019.03.092
Warren, R. W., Nagy, L., Selegue, J., Gates, J., and Carroll, S. (1994). Evolution of homeotic gene regulation and function in flies and butterflies. Nature 372, 458–461. doi: 10.1038/372458a0
Weatherbee, S. D., Halder, G., Kim, J., Hudson, A., and Carroll, S. (1998). Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 12, 1474–1482. doi: 10.1101/gad.12.10.1474
Weatherbee, S. D., Nijhout, H. F., Grunert, L. W., Halder, G., Galant, R., Selegue, J., et al. (1999). Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr. Biol. 9, 109–115. doi: 10.1016/S0960-9822(99)80064-5
Williams, J. A., and Carroll, S. B. (1993). The origin, patterning and evolution of insect appendages. BioEssays 15, 567–577. doi: 10.1002/bies.950150902
Wootton, R. J. (2002). Design, function and evolution in the wings of holometabolous insects. Zool. Scripta 31, 31–40. doi: 10.1046/j.0300-3256.2001.00076.x
Zandvakili, A., Uhl, J. D., Campbell, I., Salomone, J., Song, Y. C., and Gebelein, B. (2019). The cis-regulatory logic underlying abdominal Hox-mediated repression versus activation of regulatory elements in Drosophila. Dev. Biol. 445, 226–236. doi: 10.1016/j.ydbio.2018.11.006
Keywords: Hox genes, color patterns, evo-devo, CRISPR, wing evolution
Citation: Tendolkar A, Pomerantz AF, Heryanto C, Shirk PD, Patel NH and Martin A (2021) Ultrabithorax Is a Micromanager of Hindwing Identity in Butterflies and Moths. Front. Ecol. Evol. 9:643661. doi: 10.3389/fevo.2021.643661
Received: 18 December 2020; Accepted: 26 February 2021;
Published: 18 March 2021.
Edited by:
Patrícia Beldade, University of Lisbon, PortugalReviewed by:
Antonia Monteiro, National University of Singapore, SingaporeCopyright © 2021 Tendolkar, Pomerantz, Heryanto, Shirk, Patel and Martin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arnaud Martin, YXJuYXVkQGd3dS5lZHU=
†ORCID: Amruta Tendolkar, orcid.org/0000-0002-8496-3669; Aaron F. Pomerantz, orcid.org/0000-0002-6412-9001; Christa Heryanto, orcid.org/0000-0002-9917-5710; Nipam H. Patel, orcid.org/0000-0003-4328-654X; Arnaud Martin, orcid.org/0000-0002-5980-2249
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.