- 1Department of Ecology and Evolutionary Biology, The University of Tennessee, Knoxville, Knoxville, TN, United States
- 2Environmental Science Division and Climate Change Science Institute, Oak Ridge National Laboratory, Oak Ridge, TN, United States
Macroecological rules have been developed for plants and animals that describe large-scale distributional patterns and attempt to explain the underlying physiological and ecological processes behind them. Similarly, microorganisms exhibit patterns in relative abundance, distribution, diversity, and traits across space and time, yet it remains unclear the extent to which microorganisms follow macroecological rules initially developed for macroorganisms. Additionally, the usefulness of these rules as a null hypothesis when surveying microorganisms has yet to be fully evaluated. With rapid advancements in sequencing technology, we have seen a recent increase in microbial studies that utilize macroecological frameworks. Here, we review and synthesize these macroecological microbial studies with two main objectives: (1) to determine to what extent macroecological rules explain the distribution of host-associated and free-living microorganisms, and (2) to understand which environmental factors and stochastic processes may explain these patterns among microbial clades (archaea, bacteria, fungi, and protists) and habitats (host-associated and free living; terrestrial and aquatic). Overall, 78% of microbial macroecology studies focused on free living, aquatic organisms. In addition, most studies examined macroecological rules at the community level with only 35% of studies surveying organismal patterns across space. At the community level microorganisms often tracked patterns of macroorganisms for island biogeography (74% confirm) but rarely followed Latitudinal Diversity Gradients (LDGs) of macroorganisms (only 32% confirm). However, when microorganisms and macroorganisms shared the same macroecological patterns, underlying environmental drivers (e.g., temperature) were the same. Because we found a lack of studies for many microbial groups and habitats, we conclude our review by outlining several outstanding questions and creating recommendations for future studies in microbial ecology.
Introduction
Understanding the factors that determine the abundance, distribution, and diversity of organisms across spatial and temporal scales is a fundamental challenge in ecology. For plants and animals, many macroecological rules have been theorized to explain the physiological, ecological, and some evolutionary processes that underlie these patterns. Microorganisms also exhibit patterns in abundance, distribution, and diversity over space and time (Martiny et al., 2006, 2011; Hanson et al., 2012; Talbot et al., 2014; Kivlin, 2020). However, it is unclear if macroecological rules developed for plants and animals apply to microorganisms and if they could be used to improve predictions for abundance, distribution, and diversity of microorganisms (Prosser et al., 2007; Soininen, 2012; Shade et al., 2018; Kivlin et al., 2020). Recent review papers have highlighted how macroecological frameworks can be leveraged to investigate biogeographic patterns in microorganisms (Shoemaker et al., 2017; Xu et al., 2020). Yet, to our knowledge, no comprehensive literature search has defined when microbial biogeographic patterns conform to those expected by various macroecological rules or the mechanisms driving microbial biogeographic patterns. Defining which macroecological rules are universal across the tree of life will allow for ecosystem-wide development of theory of common mechanisms affecting community assembly, response to global change, and recovery from human-induced disturbance.
Some macroecological rules should apply across the spectrum of life. For example, those that describe patterns of abundance or biodiversity of plants and animals (e.g., latitudinal/elevational clines in biodiversity, species-area relationships, species-abundance distributions, and island biogeographic patterns) should easily translate to microorganisms since these rules share a common theoretical underpinning of applying the drivers of local, regional and continental community assembly to larger scales. Community assembly of all organisms, including microscopic ones, is driven by deterministic and stochastic processes (Vellend, 2010; Nemergut et al., 2013). Deterministic processes include environmental selection or filtering, and biotic interactions such as competition and facilitation (Funk et al., 2008; Goldford et al., 2018). Stochastic processes that affect community assembly include dispersal limitation, neutral processes of ecological and evolutionary drift (Hubbell, 2001; Martiny et al., 2011), priority effects of colonization (Fukami et al., 2010), legacy effects of previous environmental conditions (Hawkes et al., 2017), and historical vicariance of geographic position of land masses (Matheny et al., 2009) or suitable habitat (Takacs-Vesbach et al., 2008). Finally, selection regimes over long time periods can influence community assembly through evolution (Kraft et al., 2007; Starnawski et al., 2017). Because these explanatory drivers of community assembly apply regardless of the focal organism, community-level macroecological patterns driven by these processes should hold across the tree of life (Horner-Devine et al., 2004; Shade et al., 2018).
In contrast, other organismal and functional macroecological rules, such as the Metabolic Theory of Ecology (MTE; Brown et al., 2004), Species-Energy Theory (Wright, 1983), Bergmann’s Rule (Bergmann, 1847), Foster’s Rule (Foster, 1964), and Rapoport’s Rule (Stevens, 1989) may lack direct analogs to microbial consortia because the mechanisms posited to underlie these rules relate to traits (e.g., body size, thermoregulation, or homeostasis) or energy sources (e.g., thermodynamics) that may not exist or apply to microorganisms or that are difficult to measure. This is particularly true of macroecological rules that assume sexual dimorphism of two sexes (Rensch’s Rule; Rensch, 1950), which apply well to many animals but break down in organisms with simpler (e.g., asexual bacteria) or much more complex mating systems (e.g., fungal species with hundreds of mating types). Moreover, macroecological rules based on body size lack direct analogs for most microorganisms where body size could refer to cell size, colony size, or sporocarp size and therefore difficult to systematically investigate. Even if a consensus could be reached, DNA-based inference of microbial abundance is unlikely to represent organismal size when ribosomal copy number varies 100-fold among microbial taxa and lacks clear phylogenetic signal (Lofgren et al., 2019).
In addition to whether or not macroecological rules apply to microorganisms, microbial groups may also vary in their ability to conform to macroecological rules of plants and animals due to differences in habitat or due to difference in microbial groups (e.g., prokaryotes vs. multicellular eukaryotes). For example, dispersal limitation of soil-borne and plant-associated microorganisms, in general, should be much higher compared to air-borne, water-borne, or animal-associated microorganisms that are actively transported as animals explore surrounding landscapes. Dispersal limitation should also be greater for microbes with a multicellular life stage than those microbes that remain unicellular throughout their whole life. This could limit range sizes of terrestrial and plant-associated microorganisms relative to aquatic or animal-associated microorganisms, with the exception of isolated, island-like aquatic systems such as lakes or other small bodies of water. In addition, climate fluctuations of terrestrial ecosystems may be more pronounced than those in aquatic ecosystems, which could affect organism range sizes (Sorte et al., 2013). Finally, biotic interactions may be more coupled in confined terrestrial spaces (e.g., soil pores) compared to large or more well mixed aquatic and marine ecosystems.
Perhaps one of the most crucial distinctions among microbial lifestyles is that of free-living versus host-associated microorganisms (Figure 1). Some microorganisms are free-living in soil or aquatic habitats. Distributions of these taxa may be mostly affected by dispersal limitation and subsequent environmental filtering, where traits such as dormancy (Lennon and Jones, 2011) or reduced metabolic rates (Wisnoski et al., 2020) allow some microorganisms to cope with unfavorable environmental conditions. However, a large portion of microbial diversity lives in symbiosis inside (endosymbiotic) or on (episymbiotic) animal and plant tissues. For many vertically transmitted host-associated microorganisms in animals and some plants, dispersal of the microorganism often depends on dispersal of the host (Salerno et al., 2016; Shade et al., 2017), eventually leading to co-cladogenesis in each group (Takiya et al., 2006; Schardl et al., 2008) and a strong signal of biotic filtering on the microbiome among host taxa. This is especially true when host-associated microbes are more buffered from environmental conditions compared to free-living microorganisms. Multiple lines of evidence suggest that host-associated microorganisms (especially vertically transmitted microorganisms) often have narrower thermal tolerances (Dunbar et al., 2007; Kikuchi et al., 2016) and reduced genome sizes and functions (Moran et al., 2008). However, for other plant-associated microorganisms (e.g., mycorrhizal fungi), independent dispersal of plants and microorganisms allows dispersal limitation and environmental filtering (Hazard et al., 2013) to influence host-associated microbial communities.
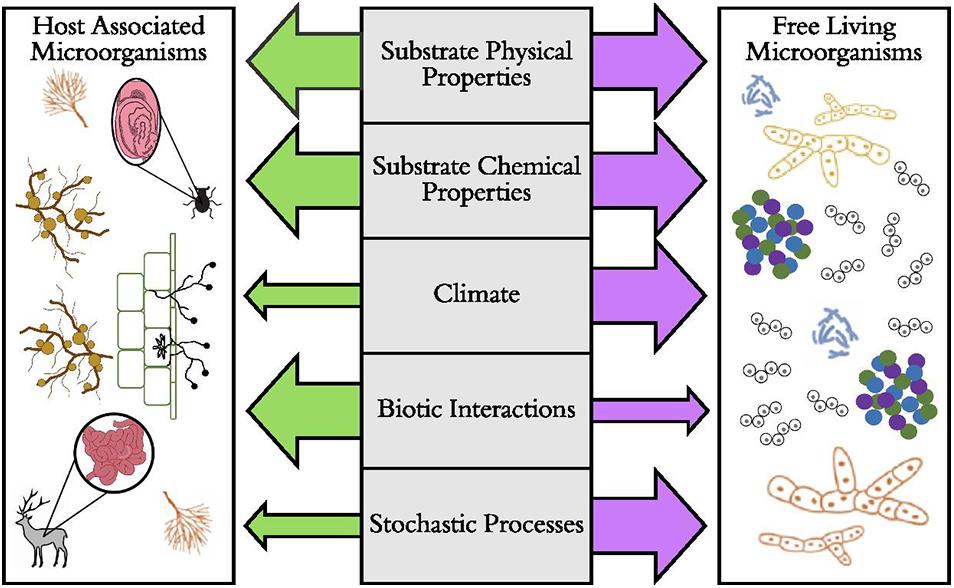
Figure 1. The predicted relative importance of ecological and evolutionary factors that drive the distributions of host-associated and free-living microorganisms. Here, the hypothesized strength of the factor is illustrated by the size of the arrow, with larger arrows representing stronger effects. Examples of host-associated microorganisms include bacterial microbiomes of beetle and deer digestive tracts, arbuscular mycorrhizal fungi, the absorptive hyphae of ectomycorrhizal fungi, and root nodule-forming bacteria (Rhizobium spp.). Examples of free-living microorganisms include soil and aquatic aggregates of bacteria, fungi, and archaea and protists such as diatoms.
In this review, we first introduce seven macroecological rules and discuss cases where they may apply to microorganisms. We then conduct a literature meta-analysis to synthesize available information on macroecological trends of free-living and host-associated microorganisms from terrestrial and aquatic ecosystems to understand the extent to which microorganisms follow the same large-scale macroecological patterns as animals and plants. The objectives of this review are to (1) determine to what extent macroecological rules explain the distribution of host-associated and free-living microorganisms, and (2) understand which environmental factors most affect the distributions of host-associated versus free-living microorganisms among microbial clades and habitats. We focus on four macroecological rules at the organismal level (Bergmann’s Rule, Rapoport’s Rule, Abundance/Occupancy Relationships, and Gloger’s Rule), three rules that describe/explain community assembly and biodiversity [Latitudinal Diversity Gradient (LDG), Theory of Island Biogeography, and Species-Area Relationships], and one functional rule (MTE).
For each rule we collected studies by querying Web of Science or Google Scholar with the search terms “rule name” AND “micro∗” or “fung∗” or “bacteri∗” or “protist∗” or “archaea∗” (query completed 06.15.2020). These queries returned studies on fungi, bacteria, archaea, and protists (including diatoms). Because many studies report trends consistent with these rules without studying them directly, we also surveyed all papers that were cited by or cited the papers in our query. We did not include the term “macroecology” because many papers including this term do not include primary data or did not survey a given macroecological rule of interest. We also did not include papers that collected appropriate data but did not conduct statistics to analyze a macroecological pattern as raw data availability varies and therefore was beyond the purview of this synthesis. We did not include human-associated microorganisms or green algae in our survey. Human travel, diet, and medicinal applications can affect microbiomes in complicated ways and therefore human microbiomes were not deemed as natural communities for the purposes of this review. Overall, this search resulted in 175 studies with 218 records (macroecological patterns for a given microbial group within a study) across all continents. For each study, we classified which macroecological rule(s) were surveyed and each study organism (archaea, bacteria, fungi, or protist), habitat (terrestrial or aquatic/marine), and niche (free-living or host-associated). We considered species-area relationships only for contiguous habitats and included species-area relationships among island-like habitats (e.g., lakes, tree holes, plant rhizosphere, and animals) with other island biogeography studies. When available, we classified which deterministic (e.g., resources and climate) and stochastic (e.g., dispersal limitation, priority effects) drivers were correlated with each observed macroecological pattern. Despite collecting 218 records for this synthesis, data limitation precluded a quantitative meta-analytical approach to addressing both adherence of microorganisms to macroecological rules and the environmental drivers influencing these distributions. Instead, we use vote counting (Hedges and Olkin, 1980) to define which macroecological rules were supported among microbial guilds and lifestyles and which environmental factors may have influenced these distributional patterns. Each time a study record confirmed a macroecological rule, it received one positive vote, and each time a study record provided contradictory evidence for a given macroecological rule, it received one negative vote. While vote counting techniques are sensitive to sample size within studies, this analysis clearly demonstrates both when microbial distributions conform to macroecological rules and more importantly when gaps in scientific inquiry do not allow us to make this conclusion.
Review of Macroecological Rules
Organismal Macroecological Rules
Bergmann’s Rule
Bergmann’s rule, which states that body size decreases among species with decreasing temperature, has primarily been studied in mammals and birds. Not all birds and mammals follow the rule, and even fewer smaller organisms, such as insects, conform to it (Blackburn et al., 1999a; Meiri and Dayan, 2003). A possible clue for ectotherms may be that Bergmann’s rule partially agrees with the temperature-size rule, which states that ectotherms develop more quickly with smaller body sizes under higher temperatures (Zuo et al., 2012); a species’ body size is a result of temperature dependent energy consumption and reallocation which can vary over elevation and across latitudes.
There is some evidence supporting Bergmann’s rule for microorganisms, such as bacterioplankton (Meador et al., 2009), with smaller cells at higher temperatures (Daufresne et al., 2009). The temperature-size relationship has also been examined as a possible mechanism behind a Bergmann’s rule pattern in microorganisms, stating that as temperature increases in aquatic systems and nucleic acid content is reduced, and thus microbes have smaller cells (Huete-Stauffer et al., 2016). Temperature-size relationships have also been studied with ubiquitous microbes, such as diatoms in geothermal streams, whereby there is no observed relationship of this relationship (Adams et al., 2013). While some evidence of Bergmann’s rule exists, significant challenges in defining the size of many microorganisms, such as multicellular fungal hyphae, must be standardized before rigorous comparisons can be drawn.
Rapoport’s Rule
Rapoport’s rule states that the latitudinal ranges of plants and animals are greater at higher compared to lower latitudes (Stevens, 1989). Stevens (1989) originally proposed a mechanism underlying this rule that relates to the climatic conditions under which different organisms have evolved. Specifically, species at higher latitudes must be able to cope with greater temporal variability in climate compared to species at lower latitudes. As a result, species that have evolved at higher latitudes should be able to occupy larger spatial extents than species at lower latitudes. The rule was later extended to elevational gradients with the expectation that at higher elevations, climate is more variable and thus high elevation species should have broader elevational ranges (Stevens, 1992). Other possible mechanisms for the rule have since been proposed, such as decreasing land area available to species at lower latitudes, reduced competition at higher latitudes, or extinction of species with narrow ranges at high latitudes due to glaciation (Gaston et al., 1998). Rapoport’s rule tends to be weaker in marine compared to terrestrial systems and in the Southern compared to Northern Hemisphere (Ruggiero and Werenkraut, 2007). The rule has received such mixed support that some researchers have questioned whether it is “time for an epitaph” for Rapoport’s rule (Gaston et al., 1998; Gaston and Chown, 1999). Despite this, ecologists are still intrigued by this macroecological rule and continue to examine whether it holds in a host of taxa, including microorganisms.
Thus far, support for Rapoport’s rule across latitude and elevation in microorganisms is equivocal at best. In terrestrial systems, the latitudinal ranges of fungal taxa (Tedersoo et al., 2014; Cox et al., 2016) and ciliates appear to increase toward the poles [W. Foissner, pers. comm. to Azovsky and Mazei (2013)]. By contrast, several studies have observed smaller range sizes of bacterial taxa at higher elevations and latitudes (Okie et al., 2015; Lear et al., 2017; Wu et al., 2018). In marine systems, a global study of bacteria found strong support for Rapoport’s rule, with bacteria in the tropics having smaller ranges than bacteria in temperate regions (Amend et al., 2013). The relationship was so strong that the most abundant bacteria in the tropics were completely absent from higher latitudes, but even narrowly distributed bacteria in temperate regions were found at lower latitudes (Amend et al., 2013). A second study of marine bacteria also found that bacteria showed narrower ranges at lower compared to higher latitudes, but this was only true for bacteria in the phyla Bacteroidetes and Cyanobacteria and the classes α-, β-, and γ- of the phylum Proteobacteria (Sul et al., 2013). Researchers have also found a reverse Rapoport’s rule in marine benthic ciliates. One study found that ciliates had narrower ranges at higher latitudes (Azovsky and Mazei, 2013). Support is also mixed for an elevational Rapoport’s rule across microbial taxa. In a study across fungal phyla (Basidiomycota, Ascomycota, Zygomycota, Chytridiomycota, and Glomeromycota), fungi at higher elevations had greater distributional ranges (Ogwu et al., 2019). In contrast, bacteria and diatoms in streams in Asia and Europe did not follow an elevational Rapoport’s rule (Teittinen et al., 2016; Wang and Soininen, 2017).
Researchers have identified several possible mechanisms leading to the distributions of microorganisms across latitudinal and elevational gradients. Range sizes of some taxa have been associated with variation in temperature and precipitation (Lear et al., 2017), suggesting that environmental selection determines range extents. However, there is some evidence that suggests dispersal limitation may be the primary driver of range sizes for some groups of microorganisms at larger spatial scales (Sul et al., 2013), while environmental selection, competition, and niche differentiation becoming more important factors at smaller spatial scales (Kivlin et al., 2014).
Abundance/Occupancy Relationships
The abundance–occupancy (A/O) relationship refers to the prevalent pattern among macroorganisms, whereby more abundant species occupy more locations within a given range (Gaston et al., 2000), both within and among species. Most A/O relationships report positive correlations but fail to investigate the potential reasons why this relationship forms. There are a handful of empirical studies that show weakly positive, or even negative trends amongst plants (Boeken and Shachak, 1998), fish (Marshall and Frank, 1994), and birds (Blackburn et al., 1999b). In addition, evidence of positive A/O relationships have been found for protists (Warren and Gaston, 1997), bacteria (Holt et al., 2004), and freshwater diatoms (Heino and Soininen, 2006; Spatharis et al., 2009; Soininen and Teittinen, 2019).
In general, the mechanisms resulting in the A/O pattern are not well understood. The A/O relationship can occur when extinction and colonization are density-dependent (Warren and Gaston, 1997). In experimental microcosms with constant resources, when dispersal was allowed, the relationship tracked the carrying capacity hypothesis (i.e., locally abundant species have lower extinction and higher colonization rates than species with low abundance; Holt et al., 2002). However, this study found that the relationship was not contingent on dispersal; without dispersal, the A/O relationship arose through extinction (Holt et al., 2002). Moreover, the A/O relationship does not always depend on environmental resource consistency. For example, the relationship held in heterogeneous environments, though to a lesser degree than in homogeneous environments (Holt et al., 2004). In field-based observations, the A/O relationship has been observed with soil fungal assemblages where dispersal had less of a role than environmental filtering on species distributions (Kivlin et al., 2014). Mechanisms behind this pattern were attributed to soil nutrients, though the relationship was weak (Kivlin et al., 2014).
Niche-based processes can also create A/O relationships among organisms. In microcosm experiments with protists, biotic interactions strengthened A/O relationships (Holt et al., 2002). Likewise, in natural systems Rocha et al. (2018) found that niche-based variables were the primary predictors for the positive A/O relationship observed in freshwater diatoms and that the effect of niche breadth on the relationship was less important than local environmental niche position. In animal host-associated intestinal bacteria, Green et al. (2016) found support for the A/O relationship that they attributed to multiple factors, including dispersal limitation, host selection, historical contingency, and stochastic processes.
Gloger’s Rule
Gloger (1833) observed a relationship between plumage coloration and climate variation in birds, predicting that darker coloration due to pigmentation occurred in warmer, more humid tropical regions while lighter colors occurred in cooler areas toward the poles. There is limited evidence for Gloger’s rule in organisms other than animals. However, pigments such as melanin are also an important protective mechanism for many microorganisms, since darker pigments can provide protection from ionizing radiation and desiccation (Dadachova and Casadevall, 2008; Fernandez and Koide, 2013). Pigmentation may also play a role in thermoregulation and energy exchange by increasing or decreasing light absorption or reflectance (Cordero et al., 2018). Consequently, pigmentation could potentially provide an important adaptive function for microbes.
However, rather than following Gloger’s rule, evidence from microbial distributions thus far supports the theory of thermal melanism, whereby greater melanism increases solar radiation absorption (e.g., Kettlewell, 1973; Brakefield, 1984). This has been shown both observationally and experimentally in yeast where pigmentation increases heat capture. For example, in contrast to Gloger’s rule, free-living, dark-pigmented yeasts were found more often outside of the tropics (Cordero et al., 2018). This suggests pigmentation provides an adaptive advantage for these microorganisms at higher latitudes by increasing heat capture. Similarly, a survey of ∼3,000 European macrofungal assemblages showed mushrooms were more darkly colored in colder climates (Krah et al., 2019). Mean temperature and in some cases, seasonality were identified as drivers of mushroom coloration, and increased reproductive success was suggested as a potential advantage of darker pigmentation in colder climates. While suggestive that Gloger’s rule may not apply to microorganisms, evidence thus far is mainly derived from free-living fungi and has not been examined in other microbial groups.
Community Level Macroecological Rules
Latitudinal Diversity Gradient
The LDG, whereby biodiversity increases from the poles to the equator, is a common large-scale pattern in ecology (Jablonski et al., 2006). Many invertebrate and vertebrate species exhibit this gradient, as do vascular plants and some fungi and marine bacteria (Hillebrand, 2004). However, many macroorganisms do not exhibit a LDG (Gillman et al., 2015), owing to legacies of environmental disturbance, dependence on environmental factors that do not trend with latitude, or dependence on biotic interactions with other organisms whose diversity also does not scale linearly with latitude. However there is more observed variation amongst studies that investigate species richness and latitude patterns for microorganisms than there is for macroorganisms (Thompson et al., 2017).
Early reports on microbial diversity suggested that some fungi and marine bacteria may follow the classic LDG pattern much as plants and animals do (Hillebrand, 2004). Further studies confirmed this pattern in stream diatoms, marine bacterioplankton, marine tintinnids, Streptomyces bacteria, freshwater fungi, and most fungal taxonomic groups (Fuhrman et al., 2008; Tedersoo et al., 2014; Andam et al., 2016; Hyde et al., 2016). The main drivers of this pattern were mean annual precipitation, temperature, carbon availability, and substrate pH, all of which can affect microbial activity, energy acquisition and longevity. However, microbes also tend to follow a hump-shaped diversity pattern with latitude within a hemisphere such that the greatest diversity is found at intermediate latitudes. This trend was demonstrated in planktonic bacteria (Milici et al., 2016), ectomycorrhizal fungi (Tedersoo et al., 2014), aquatic hyphomycete fungi (Jabiol et al., 2013; Duarte et al., 2016), saprotrophic stream fungi (Seena et al., 2019), and other fungal guilds (Větrovský et al., 2019). Potential drivers of this type of distribution include sea water temperature, thermoclines, day length, and phosphorus levels for aquatic species, dispersal limitation in terrestrial microbes, and plant host-related diversity and distribution in ectomycorrhizal fungi.
Other patterns of diversity have also been found across latitudes. A study of stream diatoms found that richness showed a U-shaped pattern. However, the study ranged only from subtropical to temperate latitudes in the northern hemisphere (Passy, 2010). Thus, a different pattern might have been observed had a larger latitudinal gradient been examined. In addition, for both the hump-shaped and U-shaped diversity distributions, sampling bias toward temperate areas may have influenced these results. Overall, a majority of studies found no pattern in microbial diversity across latitude. These include microbes in the Pacific Ocean (Baldwin et al., 2005), marine free-living ciliates (Azovsky and Mazei, 2013), freshwater diatoms (Soininen and Teittinen, 2019), heterotrophic bacterioplankton (Schiaffino et al., 2013), autotrophic eukaryotes (Schiaffino et al., 2016), soil bacteria (Fierer and Jackson, 2006; Hendershot et al., 2017), and soil fungi (Hendershot et al., 2017). Instead, soil pH was an important driver of distributions for terrestrial microorganisms in these studies, but localized climatic factors, nutrient availability, dispersal limitations, and scale of study may also play a role. Species richness of some ecto- and endoparasites also does not vary with latitude, since they are insulated from environmental conditions by their hosts (Rohde, 1978, 1999).
Theory of Island Biogeography
The dynamic equilibrium Theory of island Biogeography is an influential framework for understanding species diversity patterns. Its original proponents (MacArthur and Wilson, 1967) predicted two general patterns: (1) larger islands have more species at equilibrium, since extinction rates decline and colonization rates increase with increased area and (2) islands at a greater distance from the “mainland” or source population have fewer species at equilibrium, as colonization rates decrease with distance. Traditionally applied to groups of oceanic islands, the theory has recently been used to understand microbial biogeography across many scales on patchy, island-like systems. These include roots (Peay et al., 2007), leaves (Andrews et al., 1987; Kinkel et al., 1987), lakes or pools (Reche et al., 2005), and non-human animal organs (Loudon et al., 2016; Moeller et al., 2017). The theory has also been applied to other discrete habitats that are separated by sharp differences in temperature (Whitaker et al., 2003; Darcy et al., 2018), substrate textural or chemical qualities such as oxygen availability or host-secreted metabolites (Loudon et al., 2016), or other abiotic factors that have been shown to structure microbial distributions.
Results from several studies support the general relationship between microbial diversity and island size, although there is no consensus on how habitat size is measured amongst varying “island-like” systems (Itescu, 2019). For example, Darcy et al. (2018) observed that bacterial diversity within cryoconite holes in Antarctic glaciers is positively correlated with hole area, whereas Bell et al. (2005) found bacterial genetic diversity in water-filled tree holes increased with water volume. Studies have also found fungal species richness increases with tree host population size in arctic and alpine environments (Chlebicki and Olejniczak, 2007) and as a function of tree photosynthetic tissue volume for ectomycorrhizal fungi (Peay et al., 2007; Glassman et al., 2017a). Yet, some studies fail to detect a significant relationship between species diversity and island area, such as AM fungal communities on true islands, bacterial richness in oak litter patches and filamentous fungi on apple leaves (Kinkel et al., 1987; Davison et al., 2018; Spiesman et al., 2018). In some cases, the lack of an observed relationship is likely because environmental heterogeneity relevant for focal taxa does not scale with island size (Teittinen and Soininen, 2015). The application of the Theory of Island Biogeography across many scales (e.g., tree holes to oceanic islands) for microorganisms reflects the multitude of scales at which habitat heterogeneity and dispersal affect microbial distributions. Yet, how microbial distributions on island-like habitats differ across these scales warrants further attention.
The mechanisms that produce microbial diversity patterns across islands of various sizes are still debated. Recent studies have found these patterns hold even after explicitly accounting for environmental variables. For example, forest fragment size predicted diversity of root-associated fungi in a dominant tree even after accounting for the effects of soil carbon (Vannette et al., 2016). Dinnage et al. (2019) determined that tree size explained more variation in rhizobia diversity than other edaphic factors. Various host-associated microbial clades have differing responses to ecological drift, host size, and other environmental factors (Li et al., 2020). For example, bacterial alpha diversity increased with island area, which may be explained by differences in overall immigration and extinction rates (Dinnage et al., 2019). However, fungal beta diversity increased with area, which instead suggests an effect of greater microenvironmental variation in larger patch sizes (Chlebicki and Olejniczak, 2007). Further research into mechanisms underlying diversity-area patterns may clarify such differences across taxa, although studying processes such as local extinction in microbial communities may not be straightforward, particularly in natural systems (but see Andrews et al., 1987).
Empirical evidence that microbial diversity and community similarity decline as a function of distance from the source population is mixed across the literature, reflecting taxon-specific patterns as well as the need for more research. A significant challenge in applying island biogeographic theory to microbial communities in island-like systems is that the microorganism source, or “mainland” population, is often difficult to define, and thus most studies instead consider only distances among islands. Interestingly, most of the studies that assess the effects of distance from the mainland have used the roots of isolated or planted pine trees as “islands” and a denser, nearby pine forest as the “mainland” (Peay et al., 2010, 2012; Bahram et al., 2013; Glassman et al., 2017a, b). For example, Bahram et al. (2013) and Glassman et al. (2017a) both found that ectomycorrhizal fungal community similarity and diversity declined with increasing distance from the natural forest’s edge. The majority of studies examining how distance among “islands” shapes community similarity and diversity find spatial autocorrelation. For instance, communities of hot spring-dwelling archaea (Whitaker et al., 2003) and protists in lakes (Lepère et al., 2013) become more dissimilar as a function of distance, rather than variation in environmental factors. This suggests that for these organisms, the difficulties of dispersing among patches of suitable habitat are more important for community structure than environmental filtering.
Exceptions to the predictions of island biogeographic theory reveal that the relative importance of environmental filtering and dispersal limitation is taxon- and scale-specific. For example, Vannette et al. (2016) determined that the degree of forest fragment isolation was important in structuring the communities of some classes of root-associated fungi but not others. In other cases, lack of support may instead be due to the spatial scale considered. Darcy et al. (2018) observed positive spatial autocorrelation of cryoconite glacier hole bacterial communities among glaciers, but very little within glaciers, suggesting that had they sampled at a smaller scale, they may not have found evidence in line with island biogeographic theory. The importance of scale is also taxon-specific because taxa differ in their dispersing and colonizing abilities (Peay et al., 2012), likely explaining why Teittinen and Soininen (2015) did not find spatial structuring of diatom communities in springs. Overall, more manipulative experiments to assess why and how taxa differ in their dispersal capacities would help assess the relevance of an island-distance relationship for microorganisms.
Species-Area Relationships
The positive relationship between species richness (S) and the surveyed area (A) is one of the clearest, least disputed macroecological patterns and is fundamental to understanding the inherent heterogeneity of global biodiversity (Arrhenius, 1921). The power-law S = cAz has been used to compare the relationship between species number and area, where c is the intercept and the exponent z measures the rate of community turnover across a standardized spatial scale. This phenomenon has been explored across myriad taxa, including birds (Jetz and Rahbek, 2001), mammals (Lomolino, 1982), plants (Honnay et al., 1999), terrestrial microbial eukaryotes (Green et al., 2004), soil bacteria (Horner-Devine et al., 2004), aquatic bacteria (Reche et al., 2005; Martiny et al., 2011), ectomycorrhizal fungi (Peay et al., 2007), and vertebrate gut bacteria (Godon et al., 2016). Yet, the power-law often fails to predict species richness for many organisms (McCoy and Connor, 1976; Connor and McCoy, 1979) indicating that this relationship may only apply to a subset of spatial scales (i.e., local or regional), habitat types, or taxonomic groups.
Among microbial studies that investigate species-area or taxa-area relationships, microbial diversity often increases with spatial scale but to a much lower extent (slope z = 0.02–0.1) than observed for macroorganisms [slope z = 0.3–0.6, see Green and Bohannan (2006) and references therein]. The observed lower slopes in microorganisms in comparison to macroorganisms may be due to inconsistency in taxonomic comparisons made with direct observation versus DNA sequencing. For instance, 97% clustering is a fairly coarse clustering strategy for microorganisms that may obscure patterns of species turnover (Storch and Šizling, 2008). Spatial scaling of microbial diversity may vary from that of macroorganisms for many reasons including the higher diversity of microorganisms versus macroorganisms, loose species definitions for microorganisms, dormancy of microbial taxa (Jones and Lennon, 2010), or extracellular DNA confounding species richness estimates (Carini et al., 2016).
In addition to methodological concerns, many variables other than spatial extent affect the distribution of free-living and host-associated microorganisms. For example, microbes associated with marshland habitat respond to both geographic distance and sediment moisture (Horner-Devine et al., 2004). While species-area relationships are beginning to be explored across microbial systems, many confounding factors exist when comparing studies, such as differences in spatial extent or environmental heterogeneity. Illustrating this point is the finding that the rate of community turnover depends on whether microbial species richness is measured within or across habitats (Martiny et al., 2011). In addition, determining patterns of species richness over space is even more intractable for host-associated microbial taxa, such as ectomycorrhizal fungi, where host effects and dispersal limitation might conflate the relative importance of other environmental factors in prediction of species richness (Tanesaka, 2012). Nevertheless, despite inconsistencies in the strength of the relationship of species richness and area, taxa within most microbial guilds and lifestyles increase in diversity as more space is sampled.
Functional Macroecological Rules
Metabolic Theory of Ecology
The MTE predicts that the metabolism of individual organisms increases with environmental temperature, often with a 3/4th scaling to body size (Brown et al., 2004). Corollary predictions suggest that metabolic rates of all organisms are a combination of the allometric scaling of their body mass and biochemical kinetics (West et al., 1997) which leads to a negative association between population carrying capacity and temperature and therefore a positive association between temperature and species richness (Brown et al., 2004). While supported in some plant and animal systems, considerable controversy surrounds the universality of metabolic theory. For example, many underlying assumptions of metabolic theory, such as the actual scaling of surface area to volume ratio across organisms and enzymatic activation energies, remain untested or unproven across the tree of life (Duncan et al., 2007; Price et al., 2012).
In microbial systems, this theory has been extended to extracellular enzyme activities (Elias et al., 2014), which are assumed to reflect intracellular metabolic rates. Many studies across environmental temperature gradients find positive relationships between temperature and soil enzyme activities and respiration (Xu et al., 2017) as well as between temperature and microbial diversity (Okie et al., 2015; Zhou et al., 2016; Wu et al., 2018). Similarly, meta-analyses confirm that metabolic efficiency (i.e., the quantity of resources incorporated into biomass relative to uptake) scales with body size among microorganisms in terrestrial and aquatic ecosystems (Sinsabaugh et al., 2015). However, metabolic scaling in microorganisms rarely follows the 3/4th power to body size (DeLong et al., 2010) and studies are often confounded by other abiotic and biotic factors that also shift with temperature, such as variation in water, nutrient availability, plant diversity, or productivity (Zhou et al., 2016). Furthermore, thermal acclimation (Bradford et al., 2008) and adaptation (Alster et al., 2020) can alter microbial temperature responses in less than a decade. Thus, while evidence in support of metabolic scaling of microbial processes seems confirmatory, the rule should be examined more carefully across gradients that decouple temperature from other environmental drivers over evolutionary time. In addition, cell size is not linearly related to metabolic rates in marine phytoplankton communities (Marañón, 2015), suggesting that the relationship between organismal size and metabolism may apply to aggregates of microorganisms but break down at the smallest, cellular scales.
Literature Synthesis Results
Across all macroecological rules the majority of macroecological microbial studies (78%) have been conducted on free-living microorganisms, mainly in aquatic ecosystems (Table 1 and Figure 2). Citations corresponding with Table 1 can be found in Supplemental Material 1. In contrast, few studies have examined similar patterns in host-associated microorganisms with the notable exceptions of studies of latitudinal diversity and island biogeographic patterns focused on plant-associated fungi (e.g., mycorrhizal fungi and endophytes; Figure 2). Furthermore, the bulk of these studies examined community-level trends in microbial macroecology (71%), with less than 35% of studies occurring at the organismal level (Figure 2). Within community-based macroecological rules, LDGs were the more commonly studied, followed by the Theory of Island Biogeography and then species-area relationships.

Table 1. Evidence for each group of organisms is represented by kingdom for protists (P), fungi (F), bacteria (B), and archaea (A).
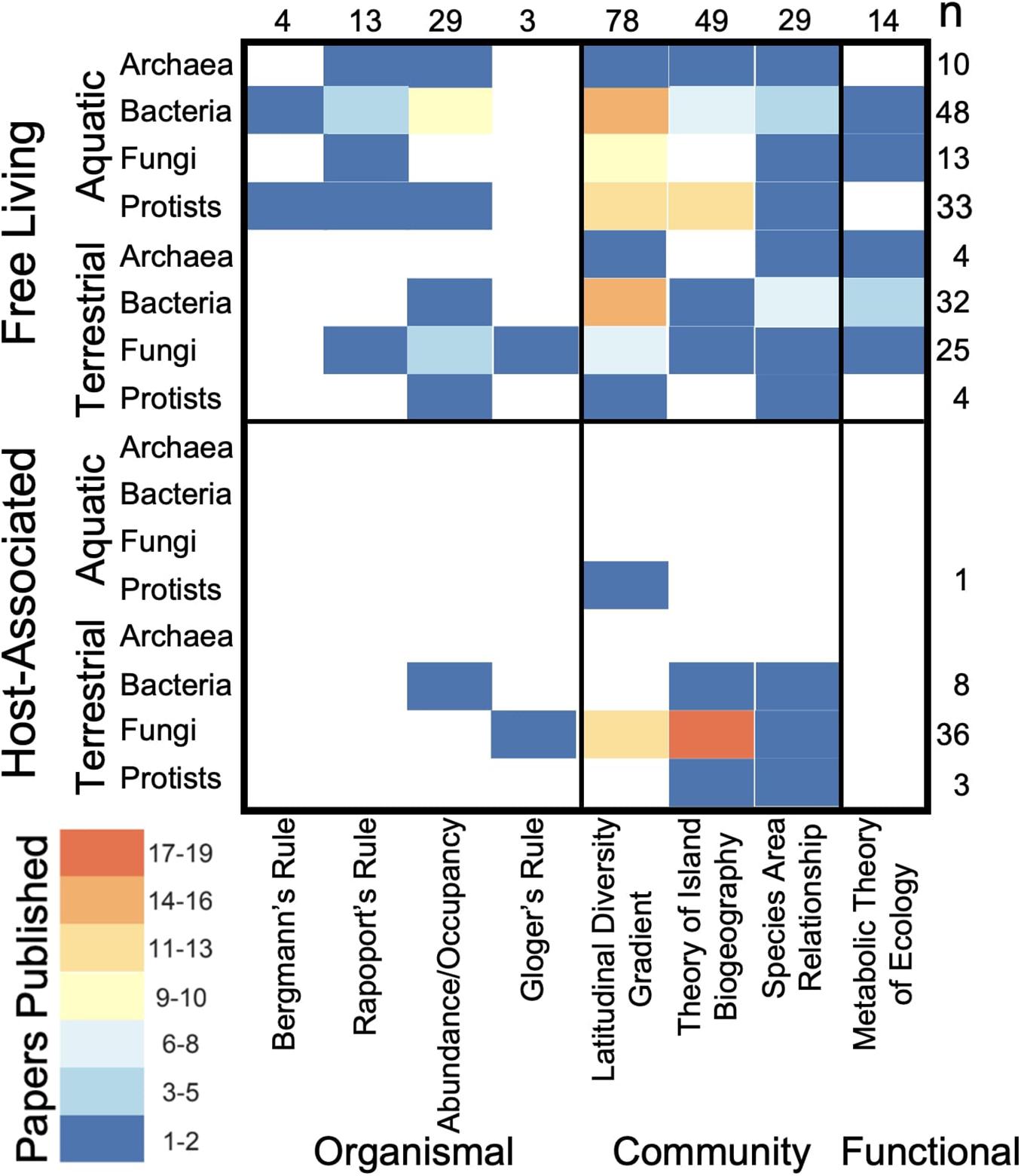
Figure 2. Numbers of publications for each queried macroecological rule across free-living and host-associated microorganisms in aquatic and terrestrial environments. Heat map indicates the number of supporting publications per grid square with total number of publications denoted for each row and column on the left and top sides of the chart.
Conformance to Macroecological Rules
Sixty-three percent of the studies we examined in this synthesis confirmed similar patterns of microbial and macro-organismal distributions. In particular, organismal-level microorganism patterns supported macroecological rules the most (Bergmann’s Rule – 75% confirm; Rapoport’s Rule – 85% confirm; and Area-Occupancy – 100% confirm). Notably, Gloger’s Rule of increasing pigmentation at lower latitudes was reversed for fungi in the only two published studies, which suggested that melanization was associated with thermal adaptation to colder, more polar latitudes. However, community-level patterns were not as consistent among taxa. For example, patterns of microbial diversity did not consistently decrease from the equator to the poles as expected from macroorganisms (32% confirm). Instead, microorganisms exhibited reverse, hump-shaped, and indistinct diversity patterns with latitude (Figure 3). In contrast, rules predicted by the dynamic equilibrium Theory of Island Biogeography, were largely upheld (74% confirm) across many systems (e.g., terrestrial host-associated mycorrhizal fungi, aquatic bacteria and protists) with increasing diversity on larger islands (but see notable exceptions, e.g., leaf endophytic fungi; Figure 3). Similarly, diversity was often negatively correlated with island distance from the source population, especially for plant-host associated microorganisms. All descriptions of microbial species-area relationships noted increased species diversity as more habitat area was sampled. Finally, for functional-based macroecological rules, evidence for metabolic scaling of microbial function confirmed expected positive temperature-dependence of microbial activity in 93% of studies but was examined in only 14 studies and never in host-associated microbial communities.
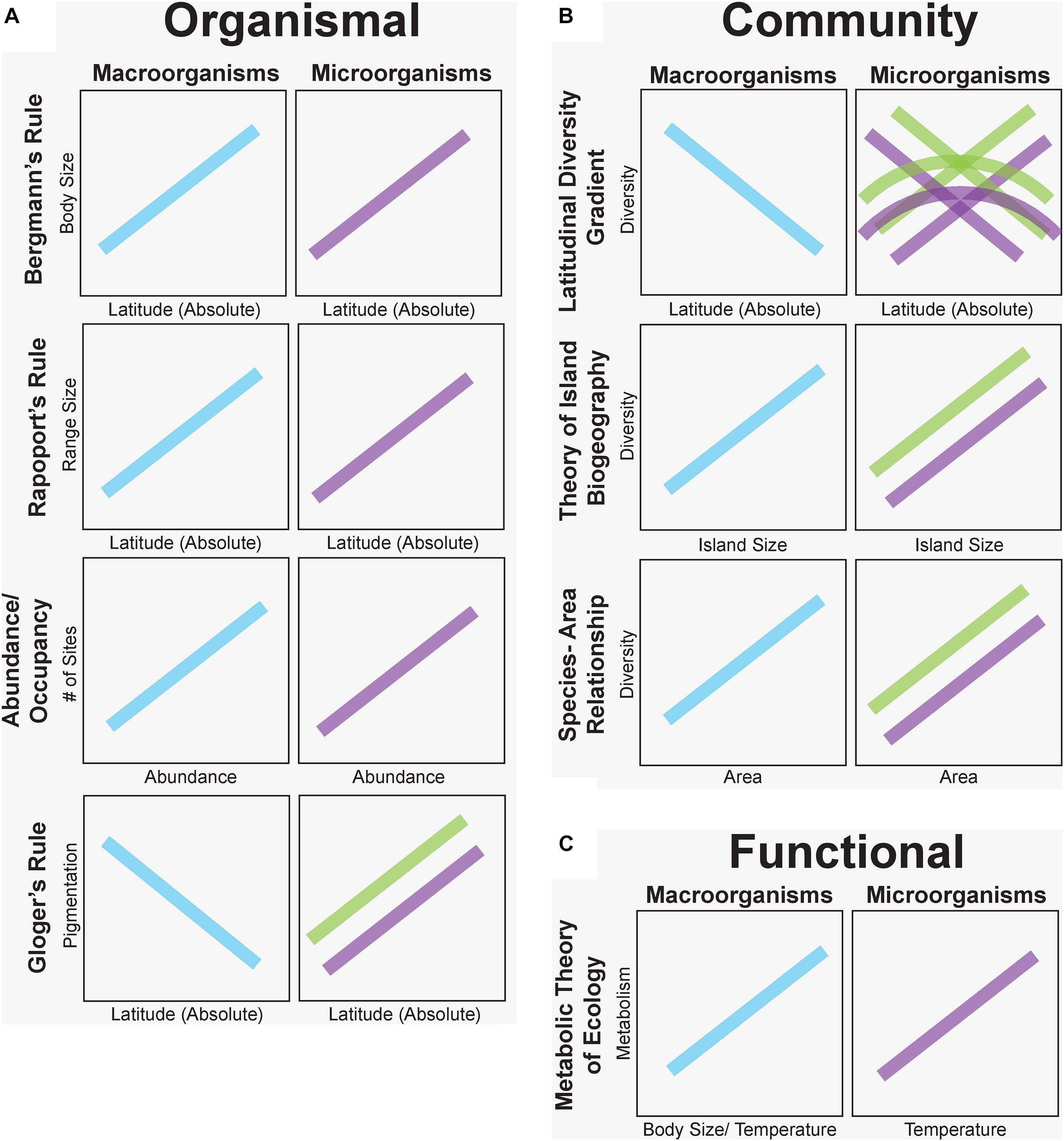
Figure 3. Predicted macroecological patterns based on proposed macroecological rules for plants and animals (blue) versus observed patterns for free-living (purple) and host-associated (green) microorganisms grouped at organismal (A), community (B), and functional (C) levels. Trends here are generated from the predominant pattern or all patterns from our synthesis when there was no consensus on vote counting. Microbial patterns match those of plants and animals for organismal (but see Gloger’s Rule) and functional levels, but community-level patterns are variable, reflecting different ecological drivers of macro- and microorganism distributions.
Drivers of Macroecological Rules
When macroecological rules were upheld in microorganisms, factors driving these patterns were mostly similar to those hypothesized for the macroorganisms studied to date (Table 1). For example, patterns consistent with the rules predicted by the dynamic equilibrium Theory of Island Biogeography were often correlated with island patch size and dispersal, whereas LDGs and metabolic scaling followed temperature gradients (but see Hawkins and Diniz-filho, 2004 for a discussion of confounding effects of temperature and latitude). However, there were notable exceptions. For example, we expected substrate pH to be a strong determining factor for microbial richness patterns due to its pivotal role in explaining beta diversity at large spatial scales. Instead, pH was only related to species-area relationships and island size in island biogeography (Fierer and Jackson, 2006). Other environmental factors related to climate and resources (carbon, nutrients) often were correlated with many microbial macroecological patterns. In addition, dispersal limitation and patch size were among the main correlates of microbial distributions, regardless of habitat or host-associated versus free-living status. While data were scarce, the distributions of host-associated microorganisms tended to be structured more heavily by biotic factors (e.g., host identity, microbial interactions) than were free-living taxa, as expected in Figure 1. Similarly, when comparisons were possible (e.g., LDG, Species Area Relationship, and Theory of Island Biogeography) there were no clear differences in drivers of terrestrial and aquatic microorganisms or among microbial kingdoms (Table 1). Clearly more investigation into several macroecological rules such as Gloger’s Rule and Bergmann’s Rule is warranted, as is more in-depth inquiry of the processes driving macroecological patterns, since over 30 studies (14%) found support for microbial congruence with macroecological rules but did not test or describe a process underlying these patterns.
Discussion
Many factors should be considered when one compares these microorganismal trends to those of macroorganisms. Many microbial studies did not explicitly test a macroecological rule and thus were not searchable in Web of Science or Google Scholar. In addition, null results are less likely to be published. These factors could have biased our findings toward rule confirmation rather than exception. Biologically, patterns of microbial distributions obtained from sequencing may sample a different subset of organisms than those derived from observational studies of plants and animals. DNA-based sampling encompasses both dormant and active states, which may increase estimates of microbial diversity as compared to plants and animals. An analog would be to sample all plant and animal gametes, which clearly is not feasible. Microorganisms that undergo horizontal gene transfer or have higher DNA substitution rates may also evolve faster than macroorganisms, allowing greater ability to respond to microclimates and shifts in environments than plants and animals. This could be why microbial distributions seem to track local nutrients or niches more than would be predicted by theories mentioned above for macroorganisms.
Understudied Macroecological Rules in Microorganisms
Many macroecological rules are still not addressable for microorganisms. For example, validation of the species-energy theory (Wright, 1983) is largely lacking for microorganisms that gain energy from multiple sources (e.g., photoautotrophic and chemolithotrophs). Examinations of center-periphery dynamics in which organisms are expected to have the highest abundance at the center of their range (Hengeveld and Haeck, 1982) are also lacking. Accurate range maps are not available for the majority of microorganisms (but see Kivlin, 2020), and when they are, do not contain information on microbial abundance. Other macroecological rules related to organismal size (e.g., Foster’s Rule; Foster, 1964) or sexual selection (e.g., Rensch’s Rule; Rensch, 1950) require detailed morphological measurements across space that are not generated from DNA-based microbial observations. As data on microbial distributions and microbial traits (e.g., Zanne et al., 2020) accumulates, many of these understudied rules may be addressable in the near future.
Outstanding Questions
Several outstanding questions and subsequent discussion points remain after our synthesis. First, if a given macroecological rule is supported, are there important environmental factors or interactions among environmental drivers specific to microbial taxa (e.g., soil pore size) that were not captured in previous studies? If so, can these explain some of the cases where microorganisms follow macroecological patterns but where the proposed causal mechanism for macroorganisms seems inapplicable? Second, how should we conceptualize traits of microorganisms, such as body size, to compare with macroorganisms? So far, microbial body size has been measured as cell size, but extensions to sporocarp or colony size could better reflect ideas of resource allocation and use underlying some theories of macroecology (Young, 2006). Third, how do the evolutionary rates of microbial speciation and extinction affect our interpretation of macroecological rules, such as those associated with the MTE or the dynamic equilibrium Theory of Island Biogeography? For example, are biologically diverse islands hotspots for speciation or cold spots for extinction, or are patterns relating to island biogeography theory solely driven by ecological dispersal trajectories? Finally, how does temporal turnover of microbial communities influence our conceptualization of macroecological patterns that are inferred from single time point studies? Are macroecological patterns of microorganisms consistent intra- and inter-annually? Data from multi-year studies may address most of these questions and can allow assessment of ecological and evolutionary turnover in microbial communities in a variety of habitats.
Best Practices in Microbial Macroecology
To spur future studies, we have developed a list of best practices for the future of microbial macroecology. First, we recommend hypothesis-driven inquiry where researchers use macroecological rules as null hypotheses instead of surveying DNA for de novo patterns. To illustrate this further, one could test the null hypothesis that rhizosphere microbes have no difference in mean range size at higher elevations than lower elevations (Rapoport’s Rule) given their association with their host. Second, macroecological rules should be used as a framework to connect multiple factors influencing biogeographic patterns (e.g., patch size and dispersal limitation among islands). Third, we should consider unique limitations and prospects of microbial ecology (microbial morphology and physiology, DNA-based studies, destructive sampling, horizontal gene transfer, the effect of spatial scale relative to the system, pooling across replicates within sites for DNA sequencing) and interpret findings in this light. For example, Dickey et al. (2020) shows that rhizosphere bacterial communities vary within the rhizosphere habitat and that DNA sequencing resolution affects estimates of microbial richness. When comparing macroecological rules across the tree of life, collection methods and statistics should be standardized as much as possible to avoid over-estimating microbial diversity. This could be accomplished by standardizing sampling effort among taxa through rarefaction or creating universal species concepts that encompass variable selection regimes instead of universal cutoff of 97% to define microbial taxa (e.g., Beaulieu et al., 2012). Furthermore, sampling efforts in tropical regions should be increased to counterbalance the sampling bias in temperate regions. Fourth, researchers should examine more than DNA (e.g., organismal traits, metabolomes) when surveying microorganisms to understand organismal trait-based and functional macroecological patterns. Finally, we should conduct longitudinal studies across years to understand microbial fluctuations due to ecological and evolutionary processes. The future is open for many lines of inquiry into the patterns and processes underlying microbial macroecology. The promising analogs to established macroecological rules for macroorganisms and notable exceptions we found here provide a roadmap to guide forecasts of microbial distributions under current future environmental conditions.
Author Contributions
All authors made substantial contributions to the concept and design of this study, contributed to the data acquisition for this study, and contributed in writing and revising the manuscript. JD and SK created figures and finalized revisions prior to submission.
Funding
CW acknowledges funding from the NSF GFRP 1650115. KS acknowledges funding from the NSF IOS 1930829 and The University of Tennessee, Knoxville. SK was funded by the NSF DEB 1936195, Texas Ecological Laboratory, and The University of Tennessee, Knoxville. RS acknowledges funding from the NSF GFRP 1452154.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the UTK Ecology and Evolutionary Biology and Microbiology departments for participating in the microbial macroecology seminars that inspired this synthesis. We also appreciate comments from Daniel Simberloff, Jennifer Schweitzer, and Colin Averill that substantially improved this work.
References
Aarnio, S., Teittinen, A., and Soininen, J. (2019). High diatom species turnover in a Baltic Sea rock pool metacommunity. Mar. Biodivers. 49, 2887–2899. doi: 10.1007/s12526-019-01016-z
Adams, G. L., Pichler, D. E., Cox, E. J., O’Gorman, E. J., Seeney, A., Woodward, G., et al. (2013). Diatoms can be an important exception to temperature-size rules at species and community levels of organization. Glob. Chang. Biol. 19, 3540–3552. doi: 10.1111/gcb.12285
Alster, C. J., von Fischer, J. C., Allison, S. D., and Treseder, K. K. (2020). Embracing a new paradigm for temperature sensitivity of soil microbes. Glob. Chang. Biol. 26, 3221–3229. doi: 10.1111/gcb.15053
Amend, A. S., Oliver, T. A., Amaral-Zettler, L. A., Boetius, A., Fuhrman, J. A., Horner-Devine, M. C., et al. (2013). Macroecological patterns of marine bacteria on a global scale. J. Biogeogr. 40, 800–811. doi: 10.1111/jbi.12034
Andam, C. P., Doroghazi, J. R., Campbell, A. N., Kelly, P. J., Choudoir, M. J., and Buckley, D. H. (2016). A latitudinal diversity gradient in terrestrial bacteria of the genus strepromyces. Am. Soc. Microbiol. 7, 1–9.
Andrews, J. H., Kinkel, L. L., Berbee, F. M., and Nordheim, E. V. (1987). Fungi, leaves, and the theory of island biogeography. Microb. Ecol. 14, 277–290. doi: 10.1007/bf02012947
Arandia-Gorostidi, N., Huete-Stauffer, T. M., Alonso-Sáez, L., and Xosé, X. A. (2017). Testing the metabolic theory of ecology with marine bacteria: different temperature sensitivity of major phylogenetic groups during the spring phytoplankton bloom. Environ. Microbiol. 19, 4493–4505. doi: 10.1111/1462-2920.13898
Arnold, A. E., and Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Azovsky, A. I. (2002). Size-dependent species-area relationships in benthos: is the world more diverse for microbes? Ecography 25, 273–282. doi: 10.1034/j.1600-0587.2002.250303.x
Azovsky, A., and Mazei, Y. (2013). Do microbes have macroecology? Large-scale patterns in the diversity and distribution of marine benthic ciliates. Glob. Ecol. Biogeogr. 22, 163–172. doi: 10.1111/j.1466-8238.2012.00776.x
Bahram, M., Kõljalg, U., Kohout, P., Mirshahvaladi, S., and Tedersoo, L. (2013). Ectomycorrhizal fungi of exotic pine plantations in relation to native host trees in Iran: evidence of host range expansion by local symbionts to distantly related host taxa. Mycorrhiza 23, 11–19. doi: 10.1007/s00572-012-0445-z
Baldwin, A. J., Moss, J. A., Pakulski, J. D., Catala, P., Joux, F., and Jeffrey, W. H. (2005). Microbial diversity in a Pacific Ocean transect from the Arctic to Antarctic circles. Aquat. Microb. Ecol. 41, 91–102. doi: 10.3354/ame041091
Barberán, A., and Casamayor, E. O. (2014). A phylogenetic perspective on species diversity, β-diversity and biogeography for the microbial world. Mol. Ecol. 23, 5868–5876. doi: 10.1111/mec.12971
Barreto, D. P., Conrad, R., Klose, M., Claus, P., and Enrich-Prast, A. (2014). Distance-decay and taxa-area relationships for bacteria, archaea and methanogenic archaea in a tropical lake sediment. PLoS One 9:e110128. doi: 10.1371/journal.pone.0110128
Beaulieu, J. M., Jhwueng, D. C., Boettiger, C., and O’Meara, B. C. (2012). Modeling stabilizing selection: expanding the ornstein-uhlenbeck model of adaptive evolution. Evolution 66, 2369–2383. doi: 10.1111/j.1558-5646.2012.01619.x
Belisle, M., Peay, K. G., and Fukami, T. (2012). Flowers as Islands: spatial distribution of nectar-inhabiting microfungi among plants of mimulus aurantiacus, a hummingbird-pollinated shrub. Microb. Ecol. 63, 711–718. doi: 10.1007/s00248-011-9975-8
Bell, T., Ager, D., Song, J. I., Newman, J. A., Thompson, I. P., Lilley, A. K., et al. (2005). Ecology: larger islands house more bacterial taxa. Science 308:1884. doi: 10.1126/science.1111318
Bergmann, C. (1847). “Über die verhältnisse der wärmeökonomie der tiere zu ihrer grösse,” in Göttinger Studien, Vol. 3, 595–708. (Charleston: Nabu Press).
Bickel, S., Chen, X., Papritz, A., and Or, D. (2019). A hierarchy of environmental covariates control the global biogeography of soil bacterial richness. Sci. Rep. 9:12129.
Bienhold, C., Zinger, L., Boetius, A., and Ramette, A. (2016). Diversity and biogeography of bathyal and abyssal seafloor bacteria. PLoS One 11:e0148016. doi: 10.1371/journal.pone.0148016
Blackburn, T. M., Gaston, K. J., and Loder, N. (1999a). Geographic gradients in body size-a clarification of Bergmann’s rule. Divers. Distrib. 5, 165–174. doi: 10.1046/j.1472-4642.1999.00046.x
Blackburn, T. M., Gaston, K. J., Quinn, R. M., and Gregory, R. D. (1999b). Do local abundances of British birds change with proximity to range edge? J. Biogeogr. 26, 493–505. doi: 10.1046/j.1365-2699.1999.00298.x
Boeken, B., and Shachak, M. (1998). The dynamics of abundance and incidence of annual plant species during colonization in a desert. Ecography 21, 63–73. doi: 10.1111/j.1600-0587.1998.tb00394.x
Boeraeve, M., Honnay, O., and Jacquemyn, H. (2019). Local abiotic conditions are more important than landscape context for structuring arbuscular mycorrhizal fungal communities in the roots of a forest herb. Oecologia 190, 149–157. doi: 10.1007/s00442-019-04406-z
Boeraeve, M., Honnay, O., Mullens, N., Vandekerkhove, K., De Keersmaeker, L., Thomaes, A., et al. (2018). The impact of spatial isolation and local habitat conditions on colonization of recent forest stands by ectomycorrhizal fungi. For. Ecol. Manage. 429, 84–92. doi: 10.1016/j.foreco.2018.06.043
Bolgovics, Á, Ács, É, Várbíró, G., Görgényi, J., and Borics, G. (2016). Species area relationship (SAR) for benthic diatoms: a study on aquatic islands. Hydrobiologia 764, 91–102. doi: 10.1007/s10750-015-2278-1
Bolgovics, Á, Viktória, B., Várbíró, G., Krasznai-K, E. Á, Ács, É, Kiss, K. T., et al. (2019). Groups of small lakes maintain larger microalgal diversity than large ones. Sci. Total Environ. 678, 162–172. doi: 10.1016/j.scitotenv.2019.04.309
Botnen, S. S., Davey, M. L., Aas, A. B., Carlsen, T., Thoen, E., Heegaard, E., et al. (2019). Biogeography of plant root-associated fungal communities in the North Atlantic region mirrors climatic variability. J. Biogeogr. 46, 1532–1546.
Bradford, M. A., Davies, C. A., Frey, S. D., Maddox, T. R., Melillo, J. M., Mohan, J. E., et al. (2008). Thermal adaptation of soil microbial respiration to elevated temperature. Ecol. Lett. 11, 1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x
Brakefield, P. M. (1984). Ecological studies on the polymorphic ladybird Adalia bipunctata in the Netherlands. II. Population dynamics, differential timing of reproduction and thermal melanism. J. Anim. Ecol. 53, 775–790. doi: 10.2307/4659
Brown, J. H., Gillooly, J. F., Allen, A. P., Savage, V. M., and West, G. B. (2004). Toward a metabolic theory of ecology. Ecology 85, 1771–1789. doi: 10.1890/03-9000
Carini, P., Marsden, P. J., Leff, J. W., Morgan, E. E., Strickland, M. S., and Fierer, N. (2016). Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 1–6.
Chlebicki, A., and Olejniczak, P. (2007). Symbiotic drift as a consequence of declining host plant populations. Acta Biol. Cracoviensia 49, 89–93.
Connor, E. F., and McCoy, E. D. (1979). The statistics and biology of the species-area relationship. Am. Nat. 113, 791–833. doi: 10.1086/283438
Cordero, R. J. B., Robert, V., Cardinali, G., Arinze, E. S., Thon, S. M., and Casadevall, A. (2018). Impact of yeast pigmentation on heat capture and latitudinal distribution. Curr. Biol. 28, 2657–2664. doi: 10.1016/j.cub.2018.06.034
Cox, F., Newsham, K. K., Bol, R., Dungait, J. A. J., and Robinson, C. H. (2016). Not poles apart: antarctic soil fungal communities show similarities to those of the distant Arctic. Ecol. Lett. 19, 528–536. doi: 10.1111/ele.12587
Da Silva, I. R., de Souza, F. A., da Silva, D. K. A., Oehl, F., and Maia, L. C. (2017). Patterns of arbuscular mycorrhizal fungal distribution on mainland and island sandy coastal plain ecosystems in Brazil. Microb. Ecol. 74, 654–669. doi: 10.1007/s00248-017-0979-x
Dadachova, E., and Casadevall, A. (2008). Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr. Opin. Microbiol. 11, 525–531. doi: 10.1016/j.mib.2008.09.013
Darcy, J. L., Gendron, E., Sommers, P., Porazinska, D. L., and Schmidt, S. K. (2018). Island biogeography of cryoconite hole bacteria in Antarctica’s Taylor Valley and around the world. Front. Ecol. Evol. 6:180. doi: 10.3389/fevo.2018.00180
Daufresne, M., Lengfellner, K., and Sommer, U. (2009). Global warming benefits the small in aquatic ecosystems. Proc. Natl. Acad. Sci. U.S.A. 106, 12788–12793. doi: 10.1073/pnas.0902080106
Davison, J., Moora, M., Öpik, M., Adholeya, A., Ainsaar, L., Bâ, A., et al. (2015). Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349, 970–973. doi: 10.1126/science.aab1161
Davison, J., Moora, M., Öpik, M., Ainsaar, L., Ducousso, M., Hiiesalu, I., et al. (2018). Microbial island biogeography: isolation shapes the life history characteristics but not diversity of root-symbiotic fungal communities. ISME J. 12, 2211–2224. doi: 10.1038/s41396-018-0196-8
DeLong, J. P., Okie, J. G., Moses, M. E., Sibly, R. M., and Brown, J. H. (2010). Shifts in metabolic scaling, production, and efficiency across major evolutionary transitions of life. Proc. Natl. Acad. Sci. 107, 12941–12945. doi: 10.1073/pnas.1007783107
de Menezes, A. A., da Silva Cáceres, M. E., Bastos, C. J. P., and Lücking, R. (2018). The latitudinal diversity gradient of epiphytic lichens in the Brazilian Atlantic Forest: does Rapoport’s rule apply? Bryologist 121, 480–497. doi: 10.1639/0007-2745-121.4.480
Delgado-Baquerizo, M., and Eldridge, D. J. (2019). Cross-biome drivers of soil bacterial alpha diversity on a worldwide scale. Ecosystems 22, 1220–1231. doi: 10.1007/s10021-018-0333-2
Delgado-Baquerizo, M., Eldridge, D. J., Hamonts, K., Reich, P. B., and Singh, B. K. (2018). Experimentally testing the species-habitat size relationship on soil bacteria: A proof of concept. Soil Biol. Biochem. 123, 200–206. doi: 10.1016/j.soilbio.2018.05.016
Deng, Y., Ning, D., Qin, Y., Xue, K., Wu, L., He, Z., et al. (2018). Spatial scaling of forest soil microbial communities across a temperature gradient. Environ. Microbiol. 20, 3504–3513. doi: 10.1111/1462-2920.14303
Dickey, J. R., Fordyce, J. A., and Lebeis, S. L. (2020). Bacterial communities of the Salvia lyrata rhizosphere explained by spatial structure and sampling grain. Microb. Ecol. 80, 846–858. doi: 10.1007/s00248-020-01594-7
Dickie, I. A., Wakelin, A., and Richardson, S. J. (2020). Rare species of wood inhabiting fungi are not local. Ecol. Appl. 30:e02156.
Dinnage, R., Simonsen, A. K., Barrett, L. G., Cardillo, M., Raisbeck-Brown, N., Thrall, P. H., et al. (2019). Larger plants promote a greater diversity of symbiotic nitrogen-fixing soil bacteria associated with an Australian endemic legume. J. Ecol. 107, 977–991. doi: 10.1111/1365-2745.13083
Dolan, J. R., Lemee, R., Gasparini, S., Mousseau, L., and Heyndrickx, C. (2006). Probing diversity in the plankton: using patterns in tintinnids (planktonic marine ciliates) to identify mechanisms. Hydrobiologia 555:143. doi: 10.1007/1-4020-4697-9_12
Dolan, J. R., Yang, E. J., Kang, S.-H., and Rhee, T. S. (2016). Declines in both redundant and trace species characterize the latitudinal diversity gradient in tintinnid ciliates. ISME J. 10, 2174–2183. doi: 10.1038/ismej.2016.19
Duarte, S., Bärlocher, F., Pascoal, C., and Cássio, F. (2016). Biogeography of aquatic hyphomycetes: current knowledge and future perspectives. Fungal Ecol. 19, 169–181. doi: 10.1016/j.funeco.2015.06.002
Duarte, S., Cássio, F., Pascoal, C., and Bärlocher, F. (2017). Taxa-area relationship of aquatic fungi on deciduous leaves. PLoS One 12:e0181545. doi: 10.1371/journal.pone.0181545
Dunbar, H. E., Wilson, A. C. C., Ferguson, N. R., and Moran, N. A. (2007). Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 5:e96. doi: 10.1371/journal.pbio.0050096
Duncan, R. P., Forsyth, D. M., and Hone, J. (2007). Testing the metabolic theory of ecology: allometric scaling exponents in mammals. Ecology 88, 324–333. doi: 10.1890/0012-9658(2007)88[324:ttmtoe]2.0.co;2
Elias, M., Wieczorek, G., Rosenne, S., and Tawfik, D. S. (2014). The universality of enzymatic rate–temperature dependency. Trends Biochem. Sci. 39, 1–7. doi: 10.1016/j.tibs.2013.11.001
Feinstein, L. M., and Blackwood, C. B. (2012). Taxa–area relationship and neutral dynamics influence the diversity of fungal communities on senesced tree leaves. Environ. Microbiol. 14, 1488–1499. doi: 10.1111/j.1462-2920.2012.02737.x
Fernandez, C. W., and Koide, R. T. (2013). The function of melanin in the ectomycorrhizal fungus Cenococcum geophilum under water stress. Fungal Ecol. 6, 479–486. doi: 10.1016/j.funeco.2013.08.004
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. 103, 626–631. doi: 10.1073/pnas.0507535103
Finlay, B. J., Esteban, G. F., and Fenchel, T. (1998). Protozoan diversity: converging estimates of the global number of free-living ciliate species. Protist 149, 29–37. doi: 10.1016/s1434-4610(98)70007-0
Fuhrman, J. A., Steele, J. A., Hewson, I., Schwalbach, M. S., Brown, M. V., Green, J. L., et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 7774–7778. doi: 10.1073/pnas.0803070105
Fukami, T., Dickie, I. A., Paula Wilkie, J., Paulus, B. C., Park, D., Roberts, A., et al. (2010). Assembly history dictates ecosystem functioning: evidence from wood decomposer communities. Ecol. Lett. 13, 675–684. doi: 10.1111/j.1461-0248.2010.01465.x
Funk, J. L., Cleland, E. E., Suding, K. N., and Zavaleta, E. S. (2008). Restoration through reassembly: plant traits and invasion resistance. Trends Ecol. Evol. 23, 695–703. doi: 10.1016/j.tree.2008.07.013
Gange, A. C., Allen, L. P., Nussbaumer, A., Gange, E. G., Andrew, C., Egli, S., et al. (2019). Multiscale patterns of rarity in fungi, inferred from fruiting records. Glob. Ecol. Biogeogr. 28, 1106–1117.
Gaston, K. J., and Chown, S. L. (1999). Why Rapoport’s rule does not generalise. Oikos 84, 309–312. doi: 10.2307/3546727
Gaston, K. J., Blackburn, T. I. M. M., Greenwood, J. D., Gregory, R. D., Quinn, M., and Lawton, J. H. (2000). Abundance-occupancy relationships. J. Appl. Ecol. 37, 39–59.
Gaston, K. J., Blackburn, T. M., and Spicer, J. I. (1998). Rapoport’s rule: time for an epitaph? Trends Ecol. Evol. 13, 70–74. doi: 10.1016/s0169-5347(97)01236-6
Ghiglione, J.-F., Galand, P. E., Pommier, T., Pedrós-Alió, C., Maas, E. W., Bakker, K., et al. (2012). Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 109, 17633–17638. doi: 10.1073/pnas.1208160109
Gillman, L. N., Wright, S. D., Cusens, J., McBride, P. D., Malhi, Y., and Whittaker, R. J. (2015). Latitude, productivity and species richness. Glob. Ecol. Biogeogr. 24, 107–117. doi: 10.1111/geb.12245
Glassman, S. I., Lubetkin, K. C., Chung, J. A., and Bruns, T. D. (2017a). The theory of island biogeography applies to ectomycorrhizal fungi in subalpine tree “islands” at a fine scale. Ecosphere 8:e01677. doi: 10.1002/ecs2.1677
Glassman, S. I., Wang, I. J., and Bruns, T. D. (2017b). Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol. Ecol. 26, 6960–6973. doi: 10.1111/mec.14414
Gloger, C. W. L. (1833). Das Abändern der Vögel durch Einfluss des Klima’s, etc. Charleston, CA: Nabu Press.
Glynou, K., Ali, T., Buch, A., Haghi Kia, S., Ploch, S., Xia, X., et al. (2016). The local environment determines the assembly of root endophytic fungi at a continental scale. Environ. Microbiol. 18, 2418–2434. doi: 10.1111/1462-2920.13112
Godon, J.-J., Arulazhagan, P., Steyer, J.-P., and Hamelin, J. (2016). Vertebrate bacterial gut diversity: size also matters. BMC Ecol. 16:12. doi: 10.1186/s12898-016-0071-2
Goldenberg Vilar, A., van Dam, H., van Loon, E. E., Vonk, J. A., van Der Geest, H. G., and Admiraal, W. (2014). Eutrophication decreases distance decay of similarity in diatom communities. Freshw. Biol. 59, 1522–1531. doi: 10.1111/fwb.12363
Goldford, J. E., Lu, N., Bajić, D., Estrela, S., Tikhonov, M., Sanchez-Gorostiaga, A., et al. (2018). Emergent simplicity in microbial community assembly. Science 361, 469–474. doi: 10.1126/science.aat1168
Green, H. C., Fisher, J. C., McLellan, S. L., Sogin, M. L., and Shanks, O. C. (2016). Identification of specialists and abundance-occupancy relationships among intestinal bacteria of Aves, Mammalia, and Actinopterygii. Appl. Environ. Microbiol. 82, 1496–1503. doi: 10.1128/aem.02456-15
Green, J., and Bohannan, B. J. M. (2006). Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 21, 501–507. doi: 10.1016/j.tree.2006.06.012
Green, J., Holmes, A., Westoby, M., Oliver, I., Briscoe, D., Dangerfield, M., et al. (2004). Spatial scaling of microbial eukaryote diversity. Nature 432, 747–750. doi: 10.1038/nature03034
Grilli, G., Urcelay, C., Galetto, L., Davison, J., Vasar, M., and Saks, Ü, et al. (2015). The composition of arbuscular mycorrhizal fungal communities in the roots of a ruderal forb is not related to the forest fragmentation process. Environ. Microbiol. 17, 2709–2720. doi: 10.1111/1462-2920.12623
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C., and Martiny, J. B. H. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506. doi: 10.1038/nrmicro2795
Hassett, B. T., Vonnahme, T. R., Peng, X., Jones, E. B. G., and Heuzé, C. (2020). Global diversity and geography of planktonic marine fungi. Bot. Mar. 63, 121–139. doi: 10.1515/bot-2018-0113
Hawkes, C. V., Waring, B. G., Rocca, J. D., and Kivlin, S. N. (2017). Historical climate controls soil respiration responses to current soil moisture. Proc. Natl. Acad. Sci. U.S.A. 114, 6322–6327. doi: 10.1073/pnas.1620811114
Hawkins, B. A., and Diniz-filho, J. A. F. (2004). “Latitude” and geographic patterns in species richness. Ecography 27, 268–272. doi: 10.1111/j.0906-7590.2004.03883.x
Hazard, C., Gosling, P., Van Der Gast, C. J., Mitchell, D. T., Doohan, F. M., and Bending, G. D. (2013). The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME J. 7, 498–508. doi: 10.1038/ismej.2012.127
Heděnec, P., Jílková, V., Lin, Q., Cajthaml, T., Filipová, A., Baldrian, P., et al. (2019). Microbial communities in local and transplanted soils along a latitudinal gradient. Catena 173, 456–464. doi: 10.1016/j.catena.2018.10.043
Hedges, L. V., and Olkin, I. (1980). Vote-counting methods in research synthesis. Psychol. Bull. 88, 359–369. doi: 10.1037/0033-2909.88.2.359
Heino, J., and Soininen, J. (2006). Regional occupancy in unicellular eukaryotes: a reflection of niche breadth, habitat availability or size-related dispersal capacity? Freshw. Biol. 51, 672–685. doi: 10.1111/j.1365-2427.2006.01520.x
Heino, J., and Tolonen, K. T. (2018). Ecological niche features override biological traits and taxonomic relatedness as predictors of occupancy and abundance in lake littoral macroinvertebrates. Ecography 41, 2092–2103. doi: 10.1111/ecog.03968
Helander, M., Ahlholm, J., Sieber, T. N., Hinneri, S., and Saikkonen, K. (2007). Fragmented environment affects birch leaf endophytes. New Phytol. 175, 547–553. doi: 10.1111/j.1469-8137.2007.02110.x
Hendershot, J. N., Read, Q. D., Henning, J. A., Sanders, N. J., and Classen, A. T. (2017). Consistently inconsistent drivers of microbial diversity and abundance at macroecological scales. Ecology 98, 1757–1763. doi: 10.1002/ecy.1829
Hengeveld, R., and Haeck, J. (1982). The distribution of abundance. I. Measurements. J. Biogeogr. 9, 303–316.
Hessen, D. O., Faafeng, B. A., Smith, V. H., Bakkestuen, V., and Walseng, B. (2006). Extrinsic and intrinsic controls of zooplankton diversity in lakes. Ecology 87, 433–443. doi: 10.1890/05-0352
Hillebrand, H. (2004). On the generality of the latitudinal diversity gradient. Am. Nat. 163, 192–211. doi: 10.1086/381004
Holt, A. R., Warren, P. H., and Gaston, K. J. (2002). The importance of biotic interactions in abundance-occupancy relationships. J. Anim. Ecol. 71, 846–854. doi: 10.1046/j.1365-2656.2002.00650.x
Holt, A. R., Warren, P. H., and Gaston, K. J. (2004). The importance of habitat heterogeneity, biotic interactions and dispersal in abundance-occupancy relationships. J. Anim. Ecol. 73, 841–851. doi: 10.1111/j.0021-8790.2004.00862.x
Holt, E. A., Bradford, R., and Garcia, I. (2015). Do lichens show latitudinal patterns of diversity? Fungal Ecol. 15, 63–72. doi: 10.1016/j.funeco.2015.03.004
Honnay, O., Hermy, M., and Coppin, P. (1999). Effects of area, age and diversity of forest patches in Belgium on plant species richness, and implications for conservation and reforestation. Biol. Conserv. 87, 73–84. doi: 10.1016/s0006-3207(98)00038-x
Horner-Devine, M. C., Lage, M., Hughes, J. B., and Bohannan, B. J. M. (2004). A taxa–area relationship for bacteria. Nature 432, 750–753. doi: 10.1038/nature03073
Hu, Y., Veresoglou, S. D., Tedersoo, L., Xu, T., Ge, T., Liu, L., et al. (2019). Contrasting latitudinal diversity and co-occurrence patterns of soil fungi and plants in forest ecosystems. Soil Biol. Biochem. 131, 100–110. doi: 10.1016/j.soilbio.2019.01.001
Hubbell, S. P. (2001). The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). Princeton, NJ: Princeton University Press.
Huete-Stauffer, T. M., Arandia-Gorostidi, N., Alonso-Sáez, L., and Morán, X. A. G. (2016). Experimental warming decreases the average size and nucleic acid content of marine bacterial communities. Front. Microbiol. 7:730. doi: 10.3389/fmicb.2016.00730
Humbert, J., Dorigo, U., Cecchi, P., Le Berre, B., Debroas, D., and Bouvy, M. (2009). Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ. Microbiol. 11, 2339–2350. doi: 10.1111/j.1462-2920.2009.01960.x
Hyde, K. D., Fryar, S., Tian, Q., Bahkali, A. H., and Xu, J. (2016). Lignicolous freshwater fungi along a north–south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. doi: 10.1016/j.funeco.2015.07.002
Ibarbalz, F. M., Henry, N., Brandão, M. C., Martini, S., Busseni, G., Byrne, H., et al. (2019). Global trends in marine plankton diversity across kingdoms of life. Cell 179, 1084–1097.
Ishtiaq, F., Clegg, S. M., Phillimore, A. B., Black, R. A., Owens, I. P. F., and Sheldon, B. C. (2010). Biogeographical patterns of blood parasite lineage diversity in avian hosts from southern Melanesian islands. J. Biogeogr. 37, 120–132. doi: 10.1111/j.1365-2699.2009.02189.x
Itescu, Y. (2019). Are island-like systems biologically similar to islands? A review of the evidence. Ecography 42, 1298–1314. doi: 10.1111/ecog.03951
Jabiol, J., Bruder, A., Gessner, M. O., Makkonen, M., Mckie, B. G., Peeters, E. T. H. M., et al. (2013). Diversity patterns of leaf-associated aquatic hyphomycetes along a broad latitudinal gradient. Fungal Ecol. 6, 439–448. doi: 10.1016/j.funeco.2013.04.002
Jablonski, D., Roy, K., and Valentine, J. W. (2006). Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314, 102–106. doi: 10.1126/science.1130880
Jetz, W., and Rahbek, C. (2001). Geometric constraints explain much of the species richness pattern in African birds. Proc. Natl. Acad. Sci. U.S.A. 98, 5661–5666. doi: 10.1073/pnas.091100998
Ji, M., Kong, W., Stegen, J., Yue, L., Wang, F., Dong, X., et al. (2020). Distinct assembly mechanisms underlie similar biogeographical patterns of rare and abundant bacteria in Tibetan Plateau grassland soils. Environ. Microbiol. 22, 2261–2272. doi: 10.1111/1462-2920.14993
Jiang, Y., Huang, H., Ma, T., Ru, J., Blank, S., Kurmayer, R., et al. (2019). Temperature response of planktonic microbiota in remote alpine lakes. Front. Microbiol. 10:1714.
Jiao, S., and Lu, Y. (2020). Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Chang. Biol. 26, 4506–4520. doi: 10.1111/gcb.15130
Jones, S. E., and Lennon, J. T. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U.S.A. 107, 5881–5886. doi: 10.1073/pnas.0912765107
Jüttner, I., Van de Vijver, B., Williams, D., Lange-Bertalot, H., and Ector, L. (2018). The genus Eunotia (Bacillariophyta) in the Falkland Islands and species-area relationships in sub-Antarctic islands. Diatom Res. 33, 413–452. doi: 10.1080/0269249x.2019.1570344
Kahlert, M., Rühland, K. M., Lavoie, I., Keck, F., Saulnier-Talbot, E., and Bogan, D. (2020). Biodiversity patterns of Arctic diatom assemblages in lakes and streams: current reference conditions and historical context for biomonitoring. Freshw. Biol. 1–25.
Kettlewell, B. (1973). The Evolution of Melanism: The Study of a Recurring necessity. Oxford: Clarendon Press Oxford.
Kikuchi, Y., Tada, A., Musolin, D. L., Hari, N., Hosokawa, T., Fujisaki, K., et al. (2016). Collapse of insect gut symbiosis under simulated climate change. mBio 7:e01578-16. doi: 10.1128/mBio.01578-16
Kinkel, L. L., Andrews, J. H., Berbee, F. M., and Nordheim, E. V. (1987). Leaves as islands for microbes. Oecologia 71, 405–408. doi: 10.1007/bf00378714
Kivlin, S. N. (2020). Global mycorrhizal fungal range sizes vary within and among mycorrhizal guilds but are not correlated with dispersal traits. J. Biogeogr. 47, 1994–2001. doi: 10.1111/jbi.13866
Kivlin, S. N., Fei, S., Kalisz, S., and Averill, C. (2020). Microbial ecology meets macroecology: developing a process-based understanding of the microbial role in global ecosystems. Bull. Ecol. Soc. Am. 101, 1–4. doi: 10.1007/978-1-4615-8219-9_1
Kivlin, S. N., Winston, G. C., Goulden, M. L., and Treseder, K. K. (2014). Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecol. 12, 14–25. doi: 10.1016/j.funeco.2014.04.004
Kraft, N. J. B., Cornwell, W. K., Webb, C. O., and Ackerly, D. D. (2007). Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am. Nat. 170, 271–283. doi: 10.1086/519400
Krah, F. S., Büntgen, U., Schaefer, H., Müller, J., Andrew, C., Boddy, L., et al. (2019). European mushroom assemblages are darker in cold climates. Nat. Commun. 10:2890.
Kranabetter, J. M., Berch, S. M., MacKinnon, J. A., Ceska, O., Dunn, D. E., and Ott, P. K. (2018). Species–area curve and distance–decay relationships indicate habitat thresholds of ectomycorrhizal fungi in an old-growth Pseudotsuga menziesii landscape. Divers. Distrib. 24, 755–764. doi: 10.1111/ddi.12720
Kruk, C., Segura, A. M., Peeters, E., Huszar, V. L. M., Costa, L. S., Kosten, S., et al. (2012). Phytoplankton species predictability increases towards warmer regions. Limnol. Oceanogr. 57, 1126–1135. doi: 10.4319/lo.2012.57.4.1126
Lara, E., Roussel-Delif, L., Fournier, B., Wilkinson, D. M., and Mitchell, E. A. D. (2016). Soil microorganisms behave like macroscopic organisms: patterns in the global distribution of soil euglyphid testate amoebae. J. Biogeogr. 43, 520–532. doi: 10.1111/jbi.12660
Lear, G., Lau, K., Perchec, A., Buckley, H. L., Case, B. S., Neale, M., et al. (2017). Following Rapoport’s Rule: the geographic range and genome size of bacterial taxa decline at warmer latitudes. Environ. Microbiol. 19, 3152–3162. doi: 10.1111/1462-2920.13797
Lennon, J. T., and Jones, S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130. doi: 10.1038/nrmicro2504
Lepère, C., Domaizon, I., Taïb, N., Mangot, J.-F., Bronner, G., Boucher, D., et al. (2013). Geographic distance and ecosystem size determine the distribution of smallest protists in lacustrine ecosystems. FEMS Microbiol. Ecol. 85, 85–94. doi: 10.1111/1574-6941.12100
Li, L., and Ma, Z. (2019). Global microbiome diversity scaling in hot springs with DAR (diversity-area relationship) profiles. Front. Microbiol. 10:118.
Li, S., Wang, P., Chen, Y., Wilson, M. C., Yang, X., Ma, C., et al. (2020). Island biogeography of soil bacteria and fungi: similar patterns, but different mechanisms. ISME J. 14, 1886–1896. doi: 10.1038/s41396-020-0657-8
Liao, J., Cao, X., Wang, J., Zhao, L., Sun, J., Jiang, D., et al. (2017). Similar community assembly mechanisms underlie similar biogeography of rare and abundant bacteria in lakes on Yungui Plateau, China. Limnol. Oceanogr. 62, 723–735. doi: 10.1002/lno.10455
Liu, J., Zhu, S., Liu, X., Yao, P., Ge, T., and Zhang, X.-H. (2020). Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship. ISME J. 14, 1463–1478. doi: 10.1038/s41396-020-0621-7
Liu, L., Yang, J., Yu, Z., and Wilkinson, D. M. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9, 2068–2077. doi: 10.1038/ismej.2015.29
Liu, S., Wang, H., Tian, P., Yao, X., Sun, H., Wang, Q., et al. (2020). Decoupled diversity patterns in bacteria and fungi across continental forest ecosystems. Soil Biol. Biochem. 144:107763. doi: 10.1016/j.soilbio.2020.107763
Lofgren, L. A., Uehling, J. K., Branco, S., Bruns, T. D., Martin, F., and Kennedy, P. G. (2019). Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 28, 721–730. doi: 10.1111/mec.14995
Logares, R., Lindström, E. S., Langenheder, S., Logue, J. B., Paterson, H., Laybourn-Parry, J., et al. (2013). Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J. 7, 937–948. doi: 10.1038/ismej.2012.168
Logue, J. B., Langenheder, S., Andersson, A. F., Bertilsson, S., Drakare, S., Lanzén, A., et al. (2012). Freshwater bacterioplankton richness in oligotrophic lakes depends on nutrient availability rather than on species–area relationships. ISME J. 6, 1127–1136. doi: 10.1038/ismej.2011.184
Lomolino, M. V. (1982). Species-area and species-distance relationships of terrestrial mammals in the Thousand Island Region. Oecologia 54, 72–75. doi: 10.1007/bf00541111
Loudon, A. H., Venkataraman, A., Van Treuren, W., Woodhams, D. C., Parfrey, L. W., McKenzie, V. J., et al. (2016). Vertebrate hosts as islands: dynamics of selection, immigration, loss, persistence, and potential function of bacteria on salamander skin. Front. Microbiol. 7:333. doi: 10.3389/fmicb.2016.00333
Lu, X., He, M., Ding, J., and Siemann, E. (2018). Latitudinal variation in soil biota: testing the biotic interaction hypothesis with an invasive plant and a native congener. ISME J. 12, 2811–2822. doi: 10.1038/s41396-018-0219-5
Ma, B., Dai, Z., Wang, H., Dsouza, M., Liu, X., He, Y., et al. (2017). Distinct biogeographic patterns for archaea, bacteria, and fungi along the vegetation gradient at the continental scale in Eastern China. Msystems 2:e174-16.
MacArthur, R. H., and Wilson, E. O. (1967). “The theory of island biogeography,” in Monographs in Population Biology I (Princeton, NJ: Princeton University Press).
Macingo, S. C., Kormas, K. A., Oikonomou, A., and Karayanni, H. (2019). Taxa–area and distance–decay relationships of unicellular eukaryotes along an elevation gradient of mountainous freshwater ecosystems. J. Plankton Res. 41, 821–834. doi: 10.1093/plankt/fbz066
Marañón, E. (2015). Cell size as a key determinant of phytoplankton metabolism and community structure. Annu. Rev. 7, 241–264. doi: 10.1146/annurev-marine-010814-015955
Marshall, C. T., and Frank, K. T. (1994). Geographic responses of groundfish to variation in abundance: methods of detection and their interpretation. Can. J. Fish. Aquat. Sci. 51, 808–816. doi: 10.1139/f94-079
Martiny, J. B. H., Bohannan, B. J. M., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
Martiny, J. B. H., Eisen, J. A., Penn, K., Allison, S. D., and Horner-Devine, M. C. (2011). Drivers of bacterial β-diversity depend on spatial scale. Proc. Natl. Acad. Sci. U.S.A. 108, 7850–7854. doi: 10.1073/pnas.1016308108
Matheny, P. B., Aime, M. C., Bougher, N. L., Buyck, B., Desjardin, D. E., Horak, E., et al. (2009). Out of the Palaeotropics? Historical biogeography and diversification of the cosmopolitan ectomycorrhizal mushroom family Inocybaceae. J. Biogeogr. 36, 577–592. doi: 10.1111/j.1365-2699.2008.02055.x
McCoy, E. D., and Connor, E. F. (1976). Environmental determinants of island species number in the British Isles. A reconsideration. J. Biogeogr. 3, 381–382. doi: 10.2307/3037980
Meador, J. A., Baldwin, A. J., Catala, P., Jeffrey, W. H., Joux, F., Moss, J. A., et al. (2009). Sunlight-induced DNA damage in marine micro-organisms collected along a latitudinal gradient from 70° N to 68° S. Photochem. Photobiol. 85, 412–420. doi: 10.1111/j.1751-1097.2008.00462.x
Meiri, S., and Dayan, T. (2003). On the validity of Bergmann’s rule. J. Biogeogr. 30, 331–351. doi: 10.1046/j.1365-2699.2003.00837.x
Meyer, K. M., Memiaghe, H., Korte, L., Kenfack, D., Alonso, A., and Bohannan, B. J. M. (2018). Why do microbes exhibit weak biogeographic patterns? ISME J. 12, 1404–1413. doi: 10.1038/s41396-018-0103-3
Milici, M., Tomasch, J., Wos-Oxley, M. L., Wang, H., Jáuregui, R., Camarinha-Silva, A., et al. (2016). Low diversity of planktonic bacteria in the tropical ocean. Sci. Rep. 6, 1–9.
Mo, Y., Zhang, W., Yang, J., Lin, Y., Yu, Z., and Lin, S. (2018). Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J. 12, 2198–2210. doi: 10.1038/s41396-018-0153-6
Moeller, A. H., Suzuki, T. A., Lin, D., Lacey, E. A., Wasser, S. K., and Nachman, M. W. (2017). Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 114, 13768–13773. doi: 10.1073/pnas.1700122114
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. doi: 10.1146/annurev.genet.41.110306.130119
Moss, J. A., Henriksson, N. L., Pakulski, J. D., Snyder, R. A., and Jeffrey, W. H. (2020). Oceanic microplankton do not adhere to the latitudinal diversity gradient. Microb. Ecol. 79, 511–515. doi: 10.1007/s00248-019-01413-8
Nemergut, D. R., Costello, E. K., Hamady, M., Lozupone, C., Jiang, L., Schmidt, S. K., et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ. Microbiol. 13, 135–144. doi: 10.1111/j.1462-2920.2010.02315.x
Nemergut, D. R., Schmidt, S. K., Fukami, T., O’Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356.
Noguez, A. M., Arita, H. T., Escalante, A. E., Forney, L. J., García-Oliva, F., and Souza, V. (2005). Microbial macroecology: highly structured prokaryotic soil assemblages in a tropical deciduous forest. Glob. Ecol. Biogeogr. 14, 241–248. doi: 10.1111/j.1466-822x.2005.00156.x
Ogwu, M. C., Takahashi, K., Dong, K., Song, H.-K., Moroenyane, I., Waldman, B., et al. (2019). Fungal elevational Rapoport pattern from a High Mountain in Japan. Sci. Rep. 9, 1–10.
Okie, J. G., Van Horn, D. J., Storch, D., Barrett, J. E., Gooseff, M. N., Kopsova, L., et al. (2015). Niche and metabolic principles explain patterns of diversity and distribution: theory and a case study with soil bacterial communities. Proc. R. Soc. B Biol. Sci. 282:20142630. doi: 10.1098/rspb.2014.2630
Olguin, H. F., Boltovskoy, D., Lange, C. B., and Brandini, F. (2006). Distribution of spring phytoplankton (mainly diatoms) in the upper 50 m of the Southwestern Atlantic Ocean (30–61 S). J. Plankton Res. 28, 1107–1128. doi: 10.1093/plankt/fbl045
Östman, Ö, Drakare, S., Kritzberg, E. S., Langenheder, S., Logue, J. B., and Lindström, E. S. (2010). Regional invariance among microbial communities. Ecol. Lett. 13, 118–127. doi: 10.1111/j.1461-0248.2009.01413.x
Pärtel, M., Öpik, M., Moora, M., Tedersoo, L., Szava-Kovats, R., Rosendahl, S., et al. (2017). Historical biome distribution and recent human disturbance shape the diversity of arbuscular mycorrhizal fungi. New Phytol. 216, 227–238. doi: 10.1111/nph.14695
Passy, S. I. (2010). A distinct latitudinal gradient of diatom diversity is linked to resource supply. Ecology 91, 36–41. doi: 10.1890/09-0545.1
Passy, S. I., Larson, C. A., Jamoneau, A., Budnick, W., Heino, J., and Leboucher, T. (2018). Biogeographical patterns of species richness and abundance distribution in stream diatoms are driven by climate and water chemistry. Am. Nat. 192, 605–617. doi: 10.1086/699830
Patrick, R. (1967). The effect of invasion rate, species pool, and size of area on the structure of the diatom community. Proc. Natl. Acad. Sci. U. S. A. 58:1335. doi: 10.1073/pnas.58.4.1335
Peay, K. G., Bruns, T. D., Kennedy, P. G., Bergemann, S. E., and Garbelotto, M. (2007). A strong species–area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecol. Lett. 10, 470–480. doi: 10.1111/j.1461-0248.2007.01035.x
Peay, K. G., Garbelotto, M., and Bruns, T. D. (2010). Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91, 3631–3640. doi: 10.1890/09-2237.1
Peay, K. G., Schubert, M. G., Nguyen, N. H., and Bruns, T. D. (2012). Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol. Ecol. 21, 4122–4136. doi: 10.1111/j.1365-294x.2012.05666.x
Pino, V., McBratney, A., Fajardo, M., Wilson, N., and Deaker, R. (2019). Understanding soil biodiversity using two orthogonal 1000km transects across New South Wales, Australia. Geoderma 354:113860. doi: 10.1016/j.geoderma.2019.07.018
Pommier, T., Canbäck, B., Riemann, L., Boström, K. H., Simu, K., Lundberg, P., et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16, 867–880. doi: 10.1111/j.1365-294x.2006.03189.x
Price, C. A., Weitz, J. S., Savage, V. M., Stegen, J., Clarke, A., Coomes, D. A., et al. (2012). Testing the metabolic theory of ecology. Ecol. Lett. 15, 1465–1474.
Prosser, J. I., Bohannan, B. J. M., Curtis, T. P., Ellis, R. J., Firestone, M. K., Freckleton, R. P., et al. (2007). The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392.
Raes, E. J., Bodrossy, L., Van De Kamp, J., Bissett, A., Ostrowski, M., Brown, M. V., et al. (2018a). Oceanographic boundaries constrain microbial diversity gradients in the South Pacific Ocean. Proc. Natl. Acad. Sci. U.S.A. 115, E8266–E8275.
Raes, E. J., Bodrossy, L., Van De Kamp, J., Bissett, A., and Waite, A. M. (2018b). Marine bacterial richness increases towards higher latitudes in the eastern Indian Ocean. Limnol. Oceanogr. Lett. 3, 10–19. doi: 10.1002/lol2.10058
Reche, I., Pulido-Villena, E., Morales-Baquero, R., and Casamayor, E. O. (2005). Does ecosystem size determine aquatic bacterial richness? Ecology 86, 1715–1722. doi: 10.1890/04-1587
Rensch, B. (1950). Die Abhängigkeit der relativen Sexualdifferenz von der Körpergrösse. Bonner Zool. Beiträge 1, 58–69.
Rocha, M. P., Bini, L. M., Siqueira, T., Hjort, J., Grönroos, M., Lindholm, M., et al. (2018). Predicting occupancy and abundance by niche position, niche breadth and body size in stream organisms. Oecologia 186, 205–216. doi: 10.1007/s00442-017-3988-z
Rohde, K. (1978). Latitudinal differences in host-specificity of marine Monogenea and Digenea. Mar. Biol. 47, 125–134. doi: 10.1007/bf00395633
Rohde, K. (1999). Latitudinal gradients in species diversity and Rapoport’s rule revisited: a review of recent work and what can parasites teach us about the causes of the gradients? Ecography 22, 593–613. doi: 10.1111/j.1600-0587.1999.tb00509.x
Ruggiero, A., and Werenkraut, V. (2007). One-dimensional analyses of Rapoport’s rule reviewed through meta-analysis. Glob. Ecol. Biogeogr. 16, 401–414. doi: 10.1111/j.1466-8238.2006.00303.x
Salerno, J. L., Bowen, B. W., and Rappé, M. S. (2016). Biogeography of planktonic and coral-associated microorganisms across the Hawaiian Archipelago. FEMS Microbiol. Ecol. 92:fiw109. doi: 10.1093/femsec/fiw109
Santos, J. B. O., Silva, L. H. S., Branco, C. W. C., and Huszar, V. L. M. (2016). The roles of environmental conditions and geographical distances on the species turnover of the whole phytoplankton and zooplankton communities and their subsets in tropical reservoirs. Hydrobiologia 764, 171–186. doi: 10.1007/s10750-015-2296-z
Schardl, C. L., Craven, K. D., Speakman, S., Stromberg, A., Lindstrom, A., and Yoshida, R. (2008). A novel test for host-symbiont codivergence indicates ancient origin of fungal endophytes in grasses. Syst. Biol. 57, 483–498. doi: 10.1080/10635150802172184
Schiaffino, M. R., Gasol, J. M., Izaguirre, I., and Unrein, F. (2013). Picoplankton abundance and cytometric group diversity along a trophic and latitudinal lake gradient. Aquat. Microb. Ecol. 68, 231–250. doi: 10.3354/ame01612
Schiaffino, M. R., Lara, E., Fernández, L. D., Balagué, V., Singer, D., Seppey, C. C. W., et al. (2016). Microbial eukaryote communities exhibit robust biogeographical patterns along a gradient of Patagonian and Antarctic lakes. Environ. Microbiol. 18, 5249–5264. doi: 10.1111/1462-2920.13566
Seena, S., Bärlocher, F., Sobral, O., Gessner, M. O., Dudgeon, D., McKie, B. G., et al. (2019). Biodiversity of leaf litter fungi in streams along a latitudinal gradient. Sci. Total Environ. 661, 306–315.
Shade, A., Dunn, R. R., Blowes, S. A., Keil, P., Bohannan, B. J. M., Herrmann, M., et al. (2018). Macroecology to unite all life, large and small. Trends Ecol. Evol. 33, 731–744. doi: 10.1016/j.tree.2018.08.005
Shade, A., Jacques, M.-A., and Barret, M. (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22. doi: 10.1016/j.mib.2017.03.010
Shi, L.-L., Mortimer, P. E., Slik, J. W. F., Zou, X.-M., Xu, J., and Feng, W.-T. (2014). Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers. 64, 305–315. doi: 10.1007/s13225-013-0270-5
Shoemaker, W. R., Locey, K. J., and Lennon, J. T. (2017). A macroecological theory of microbial biodiversity. Nat. Ecol. Evol. 1, 1–6. doi: 10.1038/s41559-017-0107
Shu, Q., and Jiao, N. (2008). Different Planctomycetes diversity patterns in latitudinal surface seawater of the open sea and in sediment. J. Microbiol. 46, 154–159. doi: 10.1007/s12275-008-0002-9
Sinsabaugh, R. L., Shah, J. J. F., Findlay, S. G., Kuehn, K. A., and Moorhead, D. L. (2015). Scaling microbial biomass, metabolism and resource supply. Biogeochemistry 122, 175–190. doi: 10.1007/s10533-014-0058-z
Sintes, E., De Corte, D., Ouillon, N., and Herndl, G. J. (2015). Macroecological patterns of archaeal ammonia oxidizers in the Atlantic Ocean. Mol. Ecol. 24, 4931–4942. doi: 10.1111/mec.13365
Smith, V. H., Foster, B. L., Grover, J. P., Holt, R. D., Leibold, M. A., and deNoyelles, F. (2005). Phytoplankton species richness scales consistently from laboratory microcosms to the world’s oceans. Proc. Natl. Acad. Sci. U.S.A. 102, 4393–4396. doi: 10.1073/pnas.0500094102
Soininen, J. (2012). Macroecology of unicellular organisms–patterns and processes. Environ. Microbiol. Rep. 4, 10–22. doi: 10.1111/j.1758-2229.2011.00308.x
Soininen, J., and Kokocinski, M. (2006). Regional diatom body size distributions in streams: Does size vary along environmental, spatial and diversity gradients? Ecoscience 13, 271–274. doi: 10.2980/i1195-6860-13-2-271.1
Soininen, J., and Teittinen, A. (2019). Fifteen important questions in the spatial ecology of diatoms. Freshw. Biol. 64, 2071–2083. doi: 10.1111/fwb.13384
Soininen, J., Jamoneau, A., Rosebery, J., and Passy, S. I. (2016). Global patterns and drivers of species and trait composition in diatoms. Glob. Ecol. Biogeogr. 25, 940–950. doi: 10.1111/geb.12452
Soininen, J., Kokocinski, M., Estlander, S., Kotanen, J., and Heino, J. (2007). Neutrality, niches, and determinants of plankton metacommunity structure across boreal wetland ponds. Ecoscience 14, 146–154. doi: 10.2980/1195-6860(2007)14[146:nnadop]2.0.co;2
Sommaruga, R., and Casamayor, E. O. (2009). Bacterial ‘cosmopolitanism’and importance of local environmental factors for community composition in remote high-altitude lakes. Freshw. Biol. 54, 994–1005. doi: 10.1111/j.1365-2427.2008.02146.x
Sorte, C. J. B., Ibáñez, I., Blumenthal, D. M., Molinari, N. A., Miller, L. P., Grosholz, E. D., et al. (2013). Poised to prosper? A cross-system comparison of climate change effects on native and non-native species performance. Ecol. Lett. 16, 261–270. doi: 10.1111/ele.12017
Spatharis, S., Dolapsakis, N. P., Economou-Amilli, A., Tsirtsis, G., and Danielidis, D. B. (2009). Dynamics of potentially harmful microalgae in a confined Mediterranean Gulf—Assessing the risk of bloom formation. Harmful Algae 8, 736–743. doi: 10.1016/j.hal.2009.03.002
Spiesman, B. J., Stapper, A. P., and Inouye, B. D. (2018). Patch size, isolation, and matrix effects on biodiversity and ecosystem functioning in a landscape microcosm. Ecosphere 9:e02173. doi: 10.1002/ecs2.2173
Spurgin, L. G., Illera, J. C., Padilla, D. P., and Richardson, D. S. (2012). Biogeographical patterns and co-occurrence of pathogenic infection across island populations of Berthelot’s pipit (Anthus berthelotii). Oecologia 168, 691–701. doi: 10.1007/s00442-011-2149-z
Starnawski, P., Bataillon, T., Ettema, T. J. G., Jochum, L. M., Schreiber, L., Chen, X., et al. (2017). Microbial community assembly and evolution in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 114, 2940–2945. doi: 10.1073/pnas.1614190114
Stevens, G. C. (1989). The latitudinal gradient in geographical range: how so many species coexist in the tropics. Am. Nat. 133, 240–256. doi: 10.1086/284913
Stevens, G. C. (1992). The elevational gradient in altitudinal range: an extension of Rapoport’s latitudinal rule to altitude. Am. Nat. 140, 893–911. doi: 10.1086/285447
Stomp, M., Huisman, J., Mittelbach, G. G., Litchman, E., and Klausmeier, C. A. (2011). Large-scale biodiversity patterns in freshwater phytoplankton. Ecology 92, 2096–2107. doi: 10.1890/10-1023.1
Stopnisek, N., and Shade, A. (2020). Prioritizing persistent microbiome members in the common bean rhizosphere: an integrated analysis of space, time, and plant genotype. bioRxiv [Preprint] doi: 10.1101/727461
Storch, D., and Šizling, A. L. (2008). The concept of taxon invariance in ecology: do diversity patterns vary with changes in taxonomic resolution? Folia Geobot. 43, 329–344. doi: 10.1007/s12224-008-9015-8
Sul, W. J., Oliver, T. A., Ducklow, H. W., Amaral-Zettler, L. A., and Sogin, M. L. (2013). Marine bacteria exhibit a bipolar distribution. Proc. Natl. Acad. Sci. U.S.A. 110, 2342–2347. doi: 10.1073/pnas.1212424110
Svensson, F., Norberg, J., and Snoeijs, P. (2014). Diatom cell size, coloniality and motility: trade-offs between temperature, salinity and nutrient supply with climate change. PLoS One 9:e109993. doi: 10.1371/journal.pone.0109993
Takacs-Vesbach, C., Mitchell, K., Jackson-Weaver, O., and Reysenbach, A. (2008). Volcanic calderas delineate biogeographic provinces among Yellowstone thermophiles. Environ. Microbiol. 10, 1681–1689. doi: 10.1111/j.1462-2920.2008.01584.x
Takiya, D. M., Tran, P. L., Dietrich, C. H., and Moran, N. A. (2006). Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15, 4175–4191. doi: 10.1111/j.1365-294x.2006.03071.x
Talbot, J. M., Bruns, T. D., Taylor, J. W., Smith, D. P., Branco, S., Glassman, S. I., et al. (2014). Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. U.S.A. 111, 6341–6346. doi: 10.1073/pnas.1402584111
Tanesaka, E. (2012). Colonizing success of saprotrophic and ectomycorrhizal basidiomycetes on islands. Mycologia 104, 345–352. doi: 10.3852/11-009
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688.
Tedersoo, L., Bahram, M., Toots, M., Diedhiou, A. G., Henkel, T. W., Kjøller, R., et al. (2012). Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol. Ecol. 21, 4160–4170. doi: 10.1111/j.1365-294x.2012.05602.x
Tedersoo, L., May, T. W., and Smith, M. E. (2010). Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20, 217–263. doi: 10.1007/s00572-009-0274-x
Teittinen, A., and Soininen, J. (2015). Testing the theory of island biogeography for microorganisms patterns for spring diatoms. Aquat. Microb. Ecol. 75, 239–250. doi: 10.3354/ame01759
Teittinen, A., Kallajoki, L., Meier, S., Stigzelius, T., and Soininen, J. (2016). The roles of elevation and local environmental factors as drivers of diatom diversity in subarctic streams. Freshw. Biol. 61, 1509–1521. doi: 10.1111/fwb.12791
Terrat, S., Dequiedt, S., Horrigue, W., Lelievre, M., Cruaud, C., Saby, N. P. A., et al. (2015). Improving soil bacterial taxa–area relationships assessment using DNA meta-barcoding. Heredity 114, 468–475. doi: 10.1038/hdy.2014.91
Terrat, S., Horrigue, W., Dequietd, S., Saby, N. P. A., Lelièvre, M., Nowak, V., et al. (2017). Mapping and predictive variations of soil bacterial richness across France. PLoS One 12:e0186766. doi: 10.1371/journal.pone.0186766
Thompson, F. L., Bruce, T., Gonzalez, A., Cardoso, A., Clementino, M., Costagliola, M., et al. (2011). Coastal bacterioplankton community diversity along a latitudinal gradient in Latin America by means of V6 tag pyrosequencing. Arch. Microbiol. 193, 105–114. doi: 10.1007/s00203-010-0644-y
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463.
Tian, J., He, N., Hale, L., Niu, S., Yu, G., Liu, Y., et al. (2018). Soil organic matter availability and climate drive latitudinal patterns in bacterial diversity from tropical to cold temperate forests. Funct. Ecol. 32, 61–70. doi: 10.1111/1365-2435.12952
Timling, I., Dahlberg, A., Walker, D. A., Gardes, M., Charcosset, J.-Y., Welker, J. M., et al. (2012). Distribution and drivers of ectomycorrhizal fungal communities across the North American Arctic. Ecosphere 3, 1–25.
Tu, Q., Deng, Y., Yan, Q., Shen, L., Lin, L., He, Z., et al. (2016). Biogeographic patterns of soil diazotrophic communities across six forests in the North America. Mol. Ecol. 25, 2937–2948. doi: 10.1111/mec.13651
Valverde, A., Makhalanyane, T. P., and Cowan, D. A. (2014). Contrasting assembly processes in a bacterial metacommunity along a desiccation gradient. Front. Microbiol. 5:668.
van der Gast, C. J., Gosling, P., Tiwari, B., and Bending, G. D. (2011). Spatial scaling of arbuscular mycorrhizal fungal diversity is affected by farming practice. Environ. Microbiol. 13, 241–249. doi: 10.1111/j.1462-2920.2010.02326.x
Van Der Linde, S., Suz, L. M., Orme, C. D. L., Cox, F., Andreae, H., Asi, E., et al. (2018). Environment and host as large-scale controls of ectomycorrhizal fungi. Nature 558, 243–248.
Vannette, R. L., Leopold, D. R., and Fukami, T. (2016). Forest area and connectivity influence root-associated fungal communities in a fragmented landscape. Ecology 97, 2374–2383. doi: 10.1002/ecy.1472
Várbíró, G., Görgényi, J., Tóthmérész, B., Padisák, J., and Hajnal, É, and Borics, G. (2017). Functional redundancy modifies species–area relationship for freshwater phytoplankton. Ecol. Evol. 7, 9905–9913. doi: 10.1002/ece3.3512
Vellend, M. (2010). Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206. doi: 10.1086/652373
Větrovský, T., Kohout, P., Kopecký, M., Machac, A., Man, M., Bahnmann, B. D., et al. (2019). A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat. Commun. 10, 1–9. doi: 10.1038/s41467-019-13164-8
Vilmi, A., Tolonen, K. T., Karjalainen, S. M., and Heino, J. (2019). Niche position drives interspecific variation in occupancy and abundance in a highly-connected lake system. Ecol. Indic. 99, 159–166. doi: 10.1016/j.ecolind.2018.12.029
Vogt, J. C., Abed, R. M. M., Albach, D. C., and Palinska, K. A. (2019). Latitudinal gradient of cyanobacterial diversity in tidal flats. PLoS One 14:e0224444. doi: 10.1371/journal.pone.0224444
Vyverman, W., Verleyen, E., Sabbe, K., Vanhoutte, K., Sterken, M., and Hodgson, D. A. (2007). Historical processes constrain patterns in global diatom diversity. Ecology 88, 1924–1931. doi: 10.1890/06-1564.1
Wang, J., and Soininen, J. (2017). Thermal barriers constrain microbial elevational range size via climate variability. Environ. Microbiol. 19, 3283–3296. doi: 10.1111/1462-2920.13823
Warren, P. H., and Gaston, K. J. (1997). Interspecific abundance-occupancy relationships: a test of mechanisms using microcosms. J. Anim. Ecol. 112, 730–742. doi: 10.2307/5924
Weiser, M. D., Michaletz, S. T., Buzzard, V., Deng, Y., He, Z., Shen, L., et al. (2018). Toward a theory for diversity gradients: the abundance–adaptation hypothesis. Ecography 41, 255–264. doi: 10.1111/ecog.02314
West, G. B., Brown, J. H., and Enquist, B. J. (1997). A general model for the origin of allometric scaling laws in biology. Science 276, 122–126. doi: 10.1126/science.276.5309.122
Wetzel, C. E., Bicudo, D., de, C., Ector, L., Lobo, E. A., Soininen, J., et al. (2012). Distance decay of similarity in neotropical diatom communities. PLoS One 7:e45071. doi: 10.1371/journal.pone.0045071
Whitaker, R. J., Grogan, D. W., and Taylor, J. W. (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301, 976–978. doi: 10.1126/science.1086909
Wisnoski, N. I., Muscarella, M. E., Larsen, M. L., Peralta, A. L., and Lennon, J. T. (2020). Metabolic insight into bacterial community assembly across ecosystem boundaries. Ecology 101:e02968.
Wright, D. H. (1983). Species-energy theory: an extension of species-area theory. Oikos 41, 496–506. doi: 10.2307/3544109
Wu, B., Liu, F., Weiser, M. D., Ning, D., Okie, J. G., Shen, L., et al. (2018). Temperature determines the diversity and structure of N2O-reducing microbial assemblages. Funct. Ecol. 32, 1867–1878. doi: 10.1111/1365-2435.13091
Xia, Z., Bai, E., Wang, Q., Gao, D., Zhou, J., Jiang, P., et al. (2016). Biogeographic distribution patterns of bacteria in typical Chinese forest soils. Front. Microbiol. 7:1106.
Xu, X., Schimel, J. P., Janssens, I. A., Song, X., Song, C., Yu, G., et al. (2017). Global pattern and controls of soil microbial metabolic quotient. Ecol. Monogr. 87, 429–441. doi: 10.1002/ecm.1258
Xu, X., Wang, N., Lipson, D., Sinsabaugh, R., Schimel, J., He, L., et al. (2020). Microbial macroecology: In search of mechanisms governing microbial biogeographic patterns. Glob. Ecol. Biogeogr. 29, 1870–1886. doi: 10.1111/geb.13162
Young, K. D. (2006). The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703. doi: 10.1128/mmbr.00001-06
Zanne, A. E., Abarenkov, K., Afkhami, M. E., Aguilar-Trigueros, C. A., Bates, S., Bhatnagar, J. M., et al. (2020). Fungal functional ecology: bringing a trait-based approach to plant-associated fungi. Biol. Rev. 95, 409–433. doi: 10.1111/brv.12570
Zhang, J., Zhang, B., Liu, Y., Guo, Y., Shi, P., and Wei, G. (2018). Distinct large-scale biogeographic patterns of fungal communities in bulk soil and soybean rhizosphere in China. Sci. Total Environ. 644, 791–800. doi: 10.1016/j.scitotenv.2018.07.016
Zhou, J., Deng, Y., Shen, L., Wen, C., Yan, Q., Ning, D., et al. (2016). Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun 7:12083.
Zhou, J., Kang, S., Schadt, C. W., and Garten, C. T. (2008). Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. U.S.A. 105, 7768–7773. doi: 10.1073/pnas.0709016105
Zhou, Z., Liu, Y., Lloyd, K. G., Pan, J., Yang, Y., Gu, J.-D., et al. (2019). Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). ISME J. 13, 885–901. doi: 10.1038/s41396-018-0321-8
Zinger, L., Boetius, A., and Ramette, A. (2014). Bacterial taxa–area and distance–decay relationships in marine environments. Mol. Ecol. 23, 954–964. doi: 10.1111/mec.12640
Zuo, W., Moses, M. E., West, G. B., Hou, C., and Brown, J. H. (2012). A general model for effects of temperature on ectotherm ontogenetic growth and development. Proc. R. Soc. B Biol. Sci. 279, 1840–1846. doi: 10.1098/rspb.2011.2000
Glossary
Endosymbiotic: Microorganisms that colonize inter- or intracellular space of plants, animals, and fungi.
Episymbiotic: Microorganisms that live on the surface of plants, animals, and fungi.
Historical vicariance: Contemporaneously observed geographic disjunction in species ranges caused by historical severance of a contiguous species range due to separation of land masses or geological events (e.g., river gorges).
Island: An island is both a true island of land surrounded by water and island-like habitats that are spatially and temporally isolated from habitats of the same type (sensu Itescu, 2019).
Legacy effects: Effects of prior environmental conditions on subsequent organismal physiology, diversity, composition, and distributions after environmental conditions have changed.
Macroecological rules: Generalizable patterns of organismal distributions, abundances, and diversity supported by known processes of organismal evolution, physiology, community assembly, and/or dispersal.
Microorganisms: Here, we define microorganisms as microscopic fungi, bacteria, archaea, and protists.
Priority effects: When the order of arrival at a given habitat gives primary colonizers an advantage in using resources to be able to outcompete subsequent colonizers.
Keywords: microorganisms, Rapoport’s rule, Abundance/Occupancy, Bergmann’s rule, Theory of Island Biogeography, Species-Area Relationship, Latitudinal Diversity Gradient, macroecology
Citation: Dickey JR, Swenie RA, Turner SC, Winfrey CC, Yaffar D, Padukone A, Beals KK, Sheldon KS and Kivlin SN (2021) The Utility of Macroecological Rules for Microbial Biogeography. Front. Ecol. Evol. 9:633155. doi: 10.3389/fevo.2021.633155
Received: 24 November 2020; Accepted: 12 March 2021;
Published: 01 April 2021.
Edited by:
Sidney F. Gouveia, Federal University of Sergipe, BrazilReviewed by:
Fabricio Villalobos, Instituto de Ecología, MexicoPeter Zee, University of Mississippi, United States
Catalina Cuellar-Gempeler, Humboldt State University, United States
Copyright © 2021 Dickey, Swenie, Turner, Winfrey, Yaffar, Padukone, Beals, Sheldon and Kivlin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan R. Dickey, amRpY2tleTlAdm9scy51dGsuZWR1; Stephanie N. Kivlin, c2tpdmxpbkB1dGsuZWR1
†Present address: Claire C. Winfrey, Department of Ecology and Evolutionary Biology, University of Colorado, Boulder, Boulder, CO, United States
 Jonathan R. Dickey
Jonathan R. Dickey Rachel A. Swenie
Rachel A. Swenie Sophia C. Turner
Sophia C. Turner Claire C. Winfrey
Claire C. Winfrey Daniela Yaffar
Daniela Yaffar Anchal Padukone
Anchal Padukone Kendall K. Beals
Kendall K. Beals Kimberly S. Sheldon1
Kimberly S. Sheldon1 Stephanie N. Kivlin
Stephanie N. Kivlin