- Parental Investment and Immune Dynamics, Marine Evolutionary Ecology, GEOMAR Helmholtz Centre for Ocean Research Kiel, Kiel, Germany
Genetic adaptation and phenotypic plasticity facilitate the migration into new habitats and enable organisms to cope with a rapidly changing environment. In contrast to genetic adaptation that spans multiple generations as an evolutionary process, phenotypic plasticity allows acclimation within the life-time of an organism. Genetic adaptation and phenotypic plasticity are usually studied in isolation, however, only by including their interactive impact, we can understand acclimation and adaptation in nature. We aimed to explore the contribution of adaptation and plasticity in coping with an abiotic (salinity) and a biotic (Vibrio bacteria) stressor using six different populations of the broad-nosed pipefish Syngnathus typhle that originated from either high [14–17 Practical Salinity Unit (PSU)] or low (7–11 PSU) saline environments along the German coastline of the Baltic Sea. We exposed wild caught animals, to either high (15 PSU) or low (7 PSU) salinity, representing native and novel salinity conditions and allowed animals to mate. After male pregnancy, offspring was split and each half was exposed to one of the two salinities and infected with Vibrio alginolyticus bacteria that were evolved at either of the two salinities in a fully reciprocal design. We investigated life-history traits of fathers and expression of 47 target genes in mothers and offspring. Pregnant males originating from high salinity exposed to low salinity were highly susceptible to opportunistic fungi infections resulting in decreased offspring size and number. In contrast, no signs of fungal infection were identified in fathers originating from low saline conditions suggesting that genetic adaptation has the potential to overcome the challenges encountered at low salinity. Offspring from parents with low saline origin survived better at low salinity suggesting genetic adaptation to low salinity. In addition, gene expression analyses of juveniles indicated patterns of local adaptation, trans-generational plasticity and developmental plasticity. In conclusion, our study suggests that pipefish are locally adapted to the low salinity in their environment, however, they are retaining phenotypic plasticity, which allows them to also cope with ancestral salinity levels and prevailing pathogens.
Introduction
Genetic adaptation and phenotypic plasticity (Chevin et al., 2010) facilitate the migration of organisms into new habitats and permit coping with changing environmental conditions (Brierley and Kingsford, 2009; Poloczanska et al., 2013; Urban, 2015). Genetic adaptation is a multigenerational evolutionary process spreading in a population over the rise and fixation of novel mutations (Chatterjee et al., 2014), or over selection on standing genetic variation and shifts in allele frequency (Barrett and Schluter, 2008; Eizaguirre et al., 2012; Torda et al., 2017). In contrast, phenotypic plasticity is an individual trait that enables organisms of one genotype to show multiple, alternative phenotypes in response to biotic or abiotic conditions (West-Eberhard, 1989). The environment influences the phenotype (Chevin et al., 2010) and elicits changes in gene expression, which have the ability to impact individual development, morphology, physiology, and behavior (Angers et al., 2010). Phenotypic responses occur within the life-time of an organism (reversible and developmental plasticity) and can persist across one or several generations (trans-generational plasticity) (Sunday et al., 2014).
Trans-generational plasticity (TGP) is the non-genetic inheritance of an alternative phenotype by transferring nutrients, hormones, proteins, or epigenetic marks from the parent to the offspring generation (Sunday et al., 2014). The impact of TGP may differ among species, life stages and abiotic conditions (Uller et al., 2013; Laland et al., 2014) as well as the biotic interaction partners (e.g., parasite type or strain) (Beemelmanns and Roth, 2016a,b, 2017; Roth et al., 2018). TGP can be adaptive and result in increased offspring performance when environmental conditions of parental and offspring generations match (Sunday et al., 2014). This has been shown for instance in wild Atlantic silversides exposed to ocean acidification (Murray et al., 2014) or in three-spined sticklebacks exposed to heat stress (Shama and Wegner, 2014). However, TGP can also induce negative carry-over effects, i.e., if parents are exposed to challenging conditions, juveniles may be negatively affected by the transfer of stress hormones from the parents to the eggs or adult allocate resources in their own survival rather than in the fitness of their offspring (Eriksen et al., 2006; Marshall, 2008). For example, mortality increased in the early life stages of sticklebacks upon changes in salinity levels of the parental generation (Heckwolf et al., 2018).
On an evolutionary time scale, phenotypic plasticity was suggested to slow down genetic adaptation by buffering the effects of natural selection (Kelly, 2019). However, the specific impact of phenotypic plasticity on genetic adaptation is driven by various factors including the species, the traits assessed and the level of current environmental variability and predictability (Reed et al., 2010). The outcome of the interacting genetic adaptation and phenotypic plasticity on the adaptation toward environmental change is thus still debated, as it has been rarely addressed. Instead, the two processes have been mainly studied in isolation (Gienapp et al., 2008; Lind et al., 2020). To depict and understand biological responses to environmental change, we need models (Donelson et al., 2019) and experiments (Kelly, 2019) addressing these processes simultaneously. An approach to study the interaction between genetic adaptations and phenotypic plasticity are space-for-time experiments (Blois et al., 2013; Kelly, 2019), where organisms living along a natural environmental gradient can serve as a prediction for how organisms can cope with future environmental conditions (Reusch et al., 2018).
The main focus of climate change research relies on warming and ocean acidification (but see DeFaveri and Merila, 2014; Hasan et al., 2017; Heckwolf et al., 2018). It is often neglected that changing ocean salinities (Meier et al., 2006; Andersson et al., 2015; Kniebusch et al., 2019) will have strong impacts on coastal and polar ecosystem (Gibson and Najjar, 2000; Loder et al., 2015) by impacting the physiology, metabolism, growth development, immunity and reproduction of marine organisms (Morgan and Iwama, 1991; Haddy and Pankhurst, 2000; Boeuf and Payan, 2001; Velasco et al., 2019), among them teleost fishes like the common goby (Mück and Heubel, 2018), turbot (Nissling et al., 2006), cod (Nissling and Westin, 1997) and flounder (Ustups et al., 2013; Nissling et al., 2017). Teleosts can cope with distinct salinity levels over a costly adjustment of their osmoregulation, which consumes up to 50% of the total energy budget (Boeuf and Payan, 2001) and results in metabolic trade-offs (DeWitt et al., 1998). Strong selection is thus expected to drive the adaptation toward novel salinities.
The Baltic Sea is particularly prone to future reductions in salinity due to little water exchange with the North Sea and river runoffs from the surrounding countries. Increased precipitation in the northern part may cause a decrease by up to 30% in surface salinity by the end of the century (Meier et al., 2006; Andersson et al., 2015). Already today, the Baltic Sea is characterized by a strong salinity gradient ranging from 30 PSU in the transition to the North Sea to an almost freshwater environment in the north-eastern parts making it an ideal setting for space for time experiments (Blanquart and Gandon, 2013; Heckwolf et al., 2018). The stability of the salinity gradient (Janssen et al., 1999; Hinrichs et al., 2019), the energetic cost of both, osmoregulation (Boeuf and Payan, 2001), and phenotypic plasticity (DeWitt et al., 1998), promote genetic adaptation in teleost fishes toward different salinity levels in the Baltic Sea (DeFaveri and Merila, 2014; Berg et al., 2015; Guo et al., 2015, 2016). In relatively stable new salinity environments, genetic assimilation was suggested to result in reduced plasticity and more adaptive genotypes (Angers et al., 2010). Adaptation to the low salinity conditions of the Baltic Sea and the isolation from the Atlantic source population is also accompanied by a loss of genetic diversity (Johannesson and Andre, 2006; Holmborn et al., 2011). Therefore, adaptation to low salinity can result in reduced osmoregulatory plasticity, such as changes in kidney morphology and gene expression (Hasan et al., 2017), and thus hamper the ability to cope with further salinity fluctuations. TGP was predicted to not be sufficient to buffer the negative impacts of salinity change (Heckwolf et al., 2018), in particular if salinity is subject to strong fluctuations and if populations are locally adapted. In contrast, increased selection due to negative carry-over effects may facilitate rapid adaptation (Heckwolf et al., 2018).
A suitable organism to study the interactive contribution of genetic adaptation and phenotypic plasticity is the broad-nosed pipefish Syngnathus typhle (Syngnathidae, Teleostei) (Wilson et al., 2020). S. typhle inhabits a wide range of waters with different salinity levels along the European coastline from the Black Sea in Eastern Europe to the Mediterranean Sea and the Eastern Atlantic (Wilson and Veraguth, 2010). After the last glacial maximum and thus in less than 8000 years, the pipefish migrated from the full marine salinity environment of the North Sea and Atlantic (33–35 PSU) into the brackish Baltic Sea with relatively low salinity levels (Björck, 1995; Wilson and Veraguth, 2010). TGP in response to immune and temperature challenges has been demonstrated in broad-nosed pipefish in numerous studies (Beemelmanns and Roth, 2016b, 2017; Roth and Landis, 2017) as well as the impairing effect of low salinity on the immune system (Birrer et al., 2012). Beyond the direct impact of salinity changes on organisms and populations (genotype × environment interaction, G×E), salinity shifts may increase or decrease the virulence of parasites and pathogens (genotype × genotype × environment interaction, G×G×E) (Stockwell et al., 2011; Hall et al., 2013; Poirier et al., 2017) and alter co-evolutionary dynamics between host and pathogens (Mostowy and Engelstadter, 2011; Molnar et al., 2013; Brunner and Eizaguirre, 2016; Kutzer and Armitage, 2016).
The abundance and virulence of opportunistic and omnipresent marine pathogens, such as several strains of the Vibrio bacteria clade (Baker-Austin et al., 2017) are modulated by salinity and temperature (Chen et al., 2011; Oberbeckmann et al., 2011; Baker-Austin et al., 2017). Vibrio alginolyticus frequently infects pipefish in the Baltic Sea (Roth et al., 2012a) and is known to cause higher mortality in artemia and herring at low salinity (Dayma et al., 2015; Poirier et al., 2017). Increases in bacterial virulence are evoked due to a combination of phenotypic changes, including bacterial biofilm formation (Dayma et al., 2015; Kim and Chong, 2017) and the expression of bacterial motility and virulence factors (Hase and Barquera, 2001; Wendling et al., 2017). We hypothesized that genetic adaptation of the pipefish to local salinity and the prevailing pathogens may compensate for the previously observed drop of immunological activity in case of exposure to decreasing salinities (Birrer et al., 2012; Poirier et al., 2017) and, hence, has the potential to reduce the negative impact of pathogens like Vibrio bacteria (Roth et al., 2012a).
To explore how the pipefish S. typhle has genetically adapted to long-term salinity changes and how this adaptation influences its phenotypic plasticity in coping with short-term shifts in salinity, we compared the potential of pipefish originating from relatively high and relatively low salinity environments in the Baltic Sea to react toward salinity shifts (Meier et al., 2006; Kniebusch et al., 2019) with developmental and trans-generational plasticity. Furthermore, we investigated how adaptation and acclimation of the pipefish host and the bacterial Vibrio pathogen to high and low salinity changes the host-pathogen interaction. We tested the following hypotheses: (1) S. typhle populations are genetically adapted to the salinity in their local habitat, (2) adaptive trans-generational plasticity in matching parental and offspring salinity results in enhanced juvenile survival and matching gene expression pattern in the parental and offspring generation, (3) S. typhle populations locally adapted to low salinity have reduced phenotypic plasticity and are not able to cope with ancestral salinity levels, and (4) bacterial virulence is higher at low salinity.
To investigate how S. typhle have adapted toward their local salinity and local pathogens in the past (genetic adaptation) and to assess their consecutive acclimation potential (phenotypic plasticity) toward salinity shifts and their immune response toward a bacterial infection, we collected six S. typhle populations in the Baltic Sea. Fish were collected at three sampling sites with high saline conditions and at three sampling sites with low saline conditions. In a laboratory aquaria experiment, animals were exposed to either their native salinity (high or low, respectively) or the salinity of the other three populations (novel salinity). Upon successful male pregnancy, offspring were exposed to either native or novel salinity conditions, in a fully reciprocal design. Subsequently, juvenile fish were injected with a V. alginolyticus strain that evolved for 90 days either at low or high salinity in the laboratory. In addition to life-history traits and mortality, we investigated the expression of 47 target genes involved in (i) general metabolism, (ii) immune response, (iii) gene regulation (DNA and histone modification), and (iv) osmoregulation.
Materials and Methods
Sampling of Adult Pipefish Populations
The parental Syngnathus typhle generation (females and non-pregnant males) was caught in seagrass meadows of six sampling sites along the German coastline of the Baltic Sea in spring 2017 before the onset of the reproductive season (Figure 1 and Table 1). Three sampling sites are characterized by relatively high salinity conditions (14–17 PSU; high origin salinity; H) and three sampling sites by relatively low salinity conditions (7–11 PSU; low origin salinity, L; Table 1). Salzhaff was assigned to the category low because salinity drops are common after rainfall accompanied with freshwater discharge due to enclosed morphology of the inlet. Therefore, pipefish in Salzhaff are often exposed to salinity levels below 10 PSU. Pipefish collected in high origin salinity are predicted to experience salinity levels of pipefish collected in low origin salinity considering the predicted decrease of Baltic Sea surface salinity (Meier et al., 2006; Kniebusch et al., 2019) and short term salinity fluctuations of about 5 PSU below and above the average (Bock and Lieberum, 2017). For the experiment, the six sample sites were assigned to two replicated salinity categories with one above and one below the isosmotic point of approximately 12 PSU. A minimum of 30 non-pregnant males and 30 females were caught snorkeling with hand nets at each sampling site at depths ranging between 0.5 and 2.5 m. At each sampling site water temperature and salinity were measured from water collected about 1 m below the surface using a salinometer (WTW Cond 330i). Pipefish (females and non-pregnant males) were transported in large aerated coolers from the sampling site to the aquaria facilities of the GEOMAR (Westshore) in Kiel (Germany). Females and non-pregnant males were kept separately to avoid mating prior to the experiment. Females of each sampling site were split into two groups and placed into twelve 80-liter tanks with 14 to 16 individuals per aquarium. Males of each sampling site were split into four and placed in total into 24 80-liter with seven to eight individuals per aquarium. These 36 tanks were connected to two independent circulating water systems containing either high saline (15 PSU; n = 18) or low saline water (7 PSU, n = 18) and equipped with artificial seagrass. Pipefish from high origins salinity were kept at 15 PSU (Baltic Sea water) and those from low origins salinity at 7 PSU (Baltic Sea water, diluted with deionized water and tap water (ratio 2:1:1) to keep the water alkalinity constant). The water temperature throughout the experiment was 18°C and illumination was set to a 16:8 h day and night cycle. Pipefish adults were fed twice a day with frozen and occasionally with live mysids.
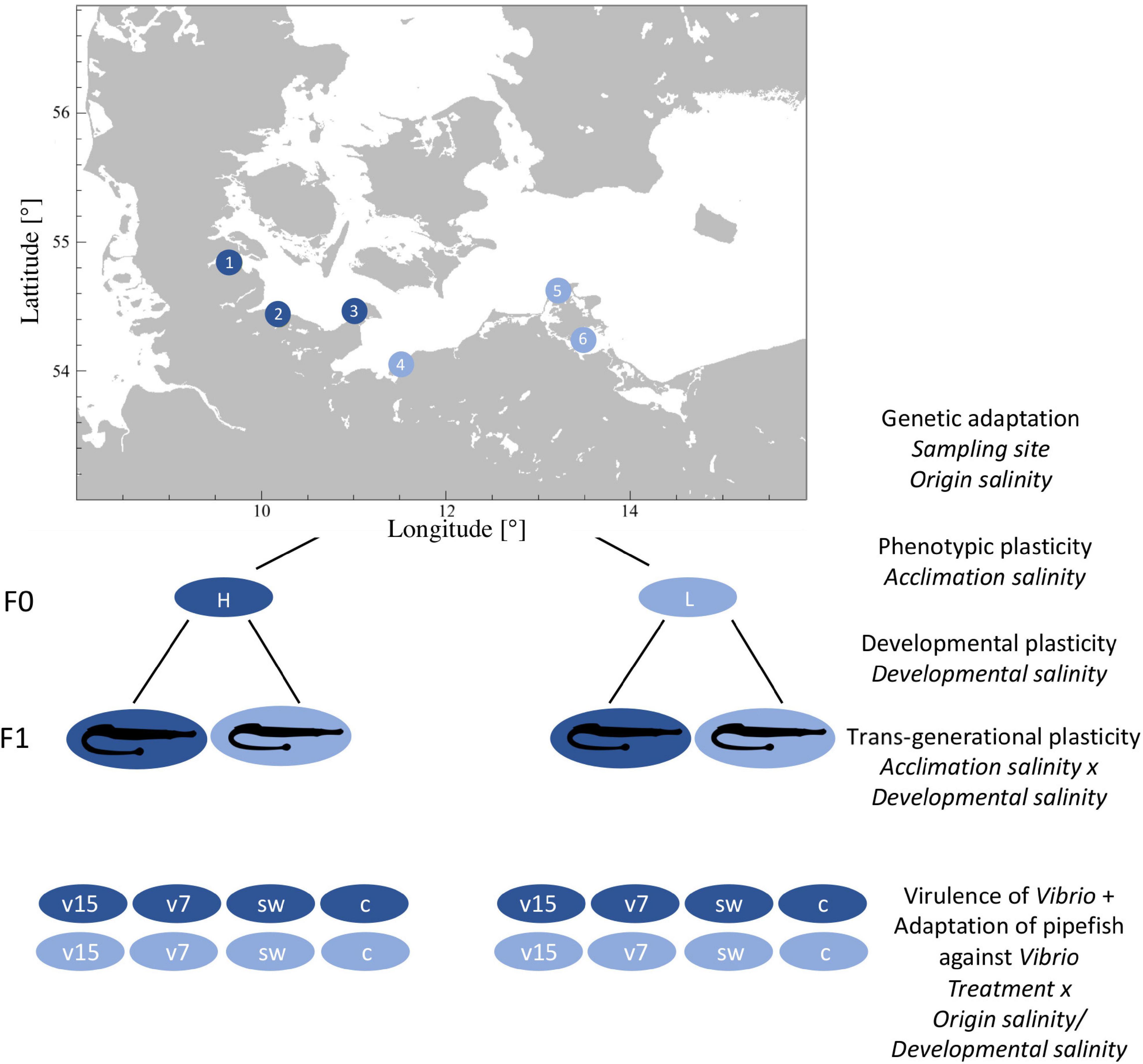
Figure 1. Fully reciprocal experimental design. We sampled pipefish along the Baltic Sea coast, at three sampling sites from a relatively high saline environment (high origin salinity: 14–17 PSU; dark blue circles; subsequently labeled as italic H): (1) Flensburg Fjord, (2) Falckenstein Strand and (3) Fehmarn and three sampling sites with a relatively low salinity level (low origin salinity: 7–11 PSU; light blue circles; subsequently labeled as italic L): (4) the Salzhaff and (5) Ruegen North and (6) Ruegen South. In the laboratory males and females were kept separately and acclimated to the opposing salinity [acclimation salinity: 15 PSU (H, dark blue), 7 PSU (L, light blue)] or their respective native salinity. Subsequently, males and females were allowed to mate and pregnant males were kept at constant conditions. Half of the F1 generation was either exposed to high (h) or low (l) salinity within 24 h after birth (developmental salinity). Ten days post hatch juveniles were injected with Vibrio alginolyticus evolved at 15 PSU (v15) or at 7 PSU (v7), sham injected with sterile seawater (sw) or left naive (c) (treatment). Label in italic on the right side correspond to the factors that were considered in the statistical models.
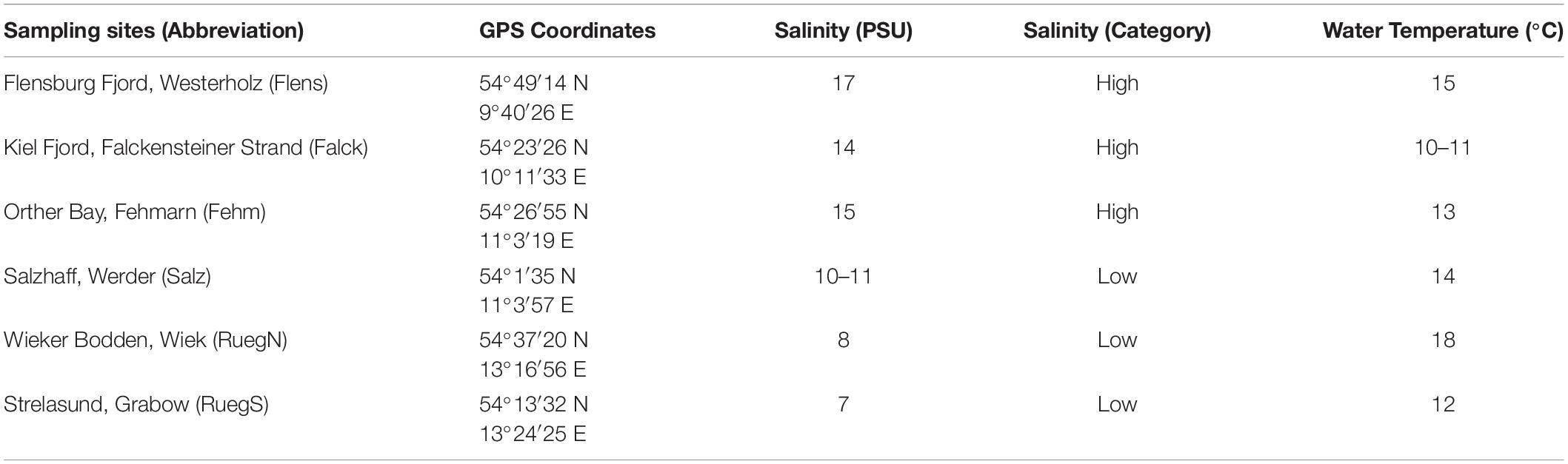
Table 1. Pipefish sampling sites with coordinates, sampling date and ambient salinity and water temperature.
After pipefish were acclimated to laboratory conditions for at least 2 days, half of the individuals from each sampling site were gradually acclimated to the novel salinity over 4 days. Each day, tanks were briefly connected to the 15 PSU or the 7 PSU circulating system to either increase or decrease the salinity by 1.5 to 2 PSU. The other half of the fish remained in their native salinity. Apart from the salinity adjustment, all 36 tanks remained disconnected from the circulation system during the time of salinity acclimation.
One day after the final salinity acclimation, four to six randomly chosen males and four to six females originating from the same sampling site and acclimated to the same salinity, were placed together in one of the 36 tanks connected to circulating water systems of either high or low acclimation salinity (Figure 1). During mating and male pregnancy, fish maintenance and aquaria set-up remained as described.
One week after mating, some pipefish males started to show signs of infection with a fungus growing inside and on the brood pouch. The causative agent could be the oomycete Saprolegnia, given prevalent Saprolegnia water molds on fish eggs in low saline waters (Lehtonen and Kvarnemo, 2015). Three weeks after mating, we visually assessed and photographically documented the prevalence of the fungus.
Sampling of Adult Pipefish for Targeted Gene Expression, Population Genetics, and Life History Traits
Four days after mating, females were removed from the tanks and immediately euthanized using anesthetic tricaine methane sulfonate (MS-222, 500 mg/L). We measured standard body length and total weight and removed the gills to store them in RNAlater at 4°C overnight and subsequently at −80°C. Fin clips were taken and placed in 96% Ethanol for population genetic analysis. Standard body length and total weight of males were measured one to three days after males gave birth.
Population Genetics Using Microsatellites
DNA Isolation and Preparation
Genomic DNA was isolated from fin clips of F0 female pipefish using the DNeasy 96 Blood and Tissue Kit (QIAGEN, Venlo, Netherlands) following the manufacturer’s protocol. All samples were incubated and eluted twice to obtain a higher extraction yield. A subset of the isolated genomic DNA was quantified using NanoDrop (Spectrometer; Peqlab, Erlangen, Germany) and visually evaluated by gel electrophoresis on a 1.2% agarose gel [GelRed nucleic acid stain, Lambda DNA/HindIII Marker and 1 kb DNA marker (Invitrogen; Thermo Fisher Scientific, Germany)].
All 144 female S. typhle samples were genotyped for 11 microsatellite loci, with a minimum of 20 individuals per sampling site. Genotyping was performed in three pooled reactions, each containing 3–4 primer pairs that were designed on an expressed sequence tag (EST) library of S. typhle [Pool A: Sy_ty_1, Sy_ty_4, Sy_ty_6, Sy_ty_7; Pool B: Sy_ty_11, Sy_ty_22, Sy_ty_23; Pool C: Sy_ty_16, Sy_ty_17, Sy_ty_21, Sy_ty_24 (Jones et al., 1999; Roth et al., 2012a)]. Microsatellites, the associated primer pairs and the Multiplex PCR protocol can be found in GenBank under accession numbers JQ598279–JQ598290 and in the Supplementary Material 1. Primers had an initial concentration of 5 pmol and were color labeled with either Hex green or Fam blue to allow differentiation during fragment analysis. In a 10 μl reaction, several loci were amplified simultaneously from 1 μl of extracted DNA using 5 μl of the Multiplex PCR Master Mix (QIAGEN) and varying amounts of the pooled primer mixes (Pool A: 1.75 μl, Pool B: 0.75 μl, Pool C: 1.5 μl). Three negative controls (ddH2O) were added onto each 96-well plate.
Capillary electrophoresis and fragment analysis were performed using the 3130xl Genetic Analyzer (Applied Biosystems/Thermo Fisher Scientific). A loading mix containing 8.75 μl HiDi Formamide and 0.25 μl GeneScan 350 ROX dye Size Standard (Applied Biosystems/Thermo Fisher Scientific) was added to 1 μl of each PCR product. Prior to the fragment run, samples were denatured in a thermo cycler for 2 min at 90°C.
Microsatellite Analysis
Raw fragment data were scored using the GeneMarker Genotyping Software (Hulce et al., 2011). The software displays allele frequency panels that identify the alleles for each locus in each sample, thus provides an overview of whether individuals are homozygous or heterozygous for certain alleles at a locus. Additionally, the raw data were screened using the Microsatellite Data Checking Software Micro-Checker (Oosterhout et al., 2004). Micro-Checker identifies genotyping errors caused by non-amplified null-alleles that either appear due to mutations in the primer binding regions or generally occur in fragment analysis because PCR shows greater efficiency in longer sequences. GENETIX (Belkhir, 2004) was used to describe the level to which the genotype frequency differed from the expected Hardy-Weinberg equilibrium (HWE) frequency by calculating a global FST value as a correlation of inbreeding in the substructure vs. in the entire population. For completeness, pairwise FST values were calculated to display distances between pairs of haplotypes and a FIS value was calculated as a correlation of inbreeding vs. random mating within the population. Although GENETIX has a greater statistical power, the population structure within the multi-locus genotype data was further investigated by the STRUCTURE Software for Population Genetics Inference (Pritchard et al., 2000). Based on the Bayesian clustering method, STRUCTURE creates an admixture model, which provides likelihood scores for each individual of belonging to a certain population. The model was tested with varying numbers of expected populations ranging from a minimum of two (high salinity vs. low salinity) to a maximum of six (number of sampling stations). Visualization of the population clustering was performed using the PHYLogeny Inference Package PHYLIP (Felsenstein, 1989). PHYLIP provides a pipeline of programs to randomize comparisons, create randomized trees, which are then assembled to a final phylogeographic tree that is based on the most frequent combinations found within the randomized trees. As the retrieved fragment data did not provide any lineage data that allows to draw conclusions with regard to a common ancestor, we created an unrooted phylogeographic tree.
Candidate Gene Expression of Females
To assess local adaptation to salinity and the potential of S. typhle to cope with novel salinity conditions, we selected candidate genes from three different functional categories, i.e., (i) immune response, (ii) metabolism, and (iii) gene regulation (DNA and histone modification) (Supplementary Table 2). Immune genes were further subdivided into innate, adaptive and complement system genes and gene regulation genes into activating and silencing genes.
RNA Extraction and Reverse Transcription
RNA was extracted from gill tissue of adult female pipefish that was stabilized in RNAlater using the RNeasy® Universal Tissue kit (QIAGEN, Venlo, Netherlands). Tissue samples were homogenized by adding a 5 mm stainless steel bead into each collection tube and placing them into a homogenizer shaking for two times 30 s at 25 Hertz. Thereafter, we followed the manufacturer’s protocol “Purification of Total RNA from Animal Tissues Using Spin Technology.” RNA concentration (extraction yield) and purity of the samples were checked by spectrophotometry (NanoDrop ND-1000 Spectrometer; Peqlab, Erlangen, Germany). Protein contamination was quantified using the absorption ratio of 260/280 nm (target > 2.0) and the ratio 260/230 nm (target > 1.8) was used to detect organic contamination. A fixed amount of RNA (300 ng/sample = 50 ng/μl) was then reverse transcribed into cDNA using the QuantiTect Reverse Transcriptase kit (QIAGEN, Venlo, Netherlands).
Preamplification of cDNA and Candidate Gene Expression
For each sample, 1.4 μl target cDNA was pre-amplified with 0.5 μl primer pool mix of all 48 genes (500 nM), 2.5 μl TaqMan PreAmp Master Mix (Applied Biosystems, Waltham, MA, United States) and 0.7 μl H2O (10 min at 95°C, 14 cycles: 15 s at 95°C followed by 4 min at 60°C). Afterward, the PCR product was diluted 1:10 with low TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8). The sample mix for the 96.96 Dynamic ArrayTM IFCs chips contained 3.1 μl pre-amplified and diluted PCR product, 3.55 μl SsoFast-EvaGreen Supermix with Low ROX (Bio-Rad Laboratories, Hercules, CA, United States) and 0.37 μl 20 x DNA Binding Dye Sample Loading Reagent (Fluidigm) per sample. The assay mix for the chip contained 0.7 μl primer pair mix (50 μM), 3.5 μl Assay Loading Reagent (Fluidigm) and 2.8 μl low TE buffer per primer pair. Chips were loaded with 5 μl sample mix and 5 μl assay mix. To measure gene expression, the chips were placed into the BioMark system (Fluidigm, South San Francisco, CA, United States) applying the “GE fast 96 × 96 PCR+Melt v2.pcl” protocol (Fluidigm). Each of the chips contained two technical replicates per sample and gene, two no-template controls (H2O), one control for gDNA contamination (−RT) and one between plate control.
Juvenile Infection Experiment
Experimental Design and Treatment Groups
Within the first 24 h after birth, juveniles were removed from the adult tanks. Half of the brood was transferred to native salinity conditions and the other half to novel salinity conditions in a fully reciprocal design. Juveniles were fed twice a day with freshly hatched, nutrient enriched (Aqua Biotica orange+TM) Artemia salina nauplii. Siblings were kept together in one non-aerated 1.5 l tank, of which one third of the water was exchanged daily. Once a day, left-over food was removed using single-use pipettes and mortality was documented.
Ten days post-hatch, juveniles received one of the four following infection treatments: (i) no injection – control (c), (ii) sham injection of autoclaved seawater (sw) with the equivalent salinity, i.e., 15 or 7 PSU, (iii) injection of Vibrio alginolyticus strain K01M1, which evolved for 90 days under laboratory condition at 15 PSU (v15) or (iv) Vibrio alginolyticus strain K01M1 that had evolved for 90 days under laboratory condition at 7 PSU (v7) (Goehlich, Roth et al., unpublished data). Pipefish were briefly taken out of the water and 2 μl of sterile seawater with or without bacteria was injected in the ventral part of the juveniles, using a MonojectTM insulin syringe (Coviden) with a sterile 30 Gauge needle. Subsequently, all juvenile siblings with the same treatment were placed in one 500 ml Kautex bottle containing seawater with the respective salinity of the 1.5 l tanks. Survival of juveniles was documented for 6 days and fish maintenance was according to the procedure described for 1.5 l tanks. One day post infection, one juvenile from each treatment (Kautex bottle) was euthanized and decapitated to assess expression of candidate genes. Standard body length was measured and whole-body samples were stored in RNAlater overnight at 4°C and subsequently at −80°C.
Characterization and Evolution of Vibrio alginolyticus Strain Used for Injection
The Vibrio alginolyticus strain K01M1 used for injection of pipefish juveniles was isolated from a healthy pipefish caught in the Kiel Fjord (Roth et al., 2012a) and fully sequenced (Chibani et al., 2020b). The strain was evolved for 90 days either at 15 or 7 PSU [medium 101: 0.5% (w/v) peptone, 0.3% (w/v) meat extract, 1.5% (w/v) or 0.7% (w/v) NaCl in Milli-Q deionized water] (Goehlich, Roth et al., unpublished data). We used the same strain and evolved it at two different salinities to ensure that salinity is the only driver for potential differences in bacterial virulence, which could also be influenced by the presence of filamentous phages (Waldor and Mekalanos, 1996; Ilyina, 2015; Chibani et al., 2020a).
After 90 days the bacterial populations were diluted and plated onto Vibrio selective Thiosulfate-Citrate-Bile-Saccharose (TCBS) agar plates (Fluka AnalyticalTM). The next day, single colonies from each plate were picked and grown overnight in medium 101 with the respective salinity. Subsequently, cultured bacteria were stored at −80°C as 33% glycerol stocks. For the infection experiment, part of the glycerol stocks were plated onto TCBS agar and one clone was grown in a 50 ml Falcon tube containing 30 ml medium 101 in the respective salinity for 24 h, at 25°C with shaking at 230 rpm. Overnight cultures were centrifuged for 20 min at 2,000 rpm. The supernatant was discarded and the cell pellet was resuspended in 3 ml sterile seawater (7 or 15 PSU, respectively) to achieve similar bacterial densities of 5 × 1010 ml–1.
Juvenile Gene Expression
We measured gene expression of juveniles to assess the effect of (a) genetic adaptation (i.e., origin salinity) on gene expression, (b) trans-generational effects driven by an interaction between acclimation salinity and developmental salinity and (c) developmental plasticity induced by developmental salinity. Furthermore, we investigated (d) whether virulence differed in V. alginolyticus evolved at 15 or 7 PSU and whether juveniles from parents originating from a matching salinity were better adapted to Vibrio strains evolved at the respective salinity. Therefore, we selected genes from three functional categories, namely (i) immune response (ii), general metabolism, and (iii) gene regulation (DNA and histone modification) as described above for female pipefish S. typhle (section “Candidate Gene Expression of Females”). Compared to female gene expression, eleven genes from the categories (i–iii) were replaced by osmoregulation genes (iv). We selected osmoregulatory genes from teleost studies (Supplementary Material 3) and designed specific primers with Primer3Web (Koressaar and Remm, 2007; Untergasser et al., 2012; Supplementary Material 4 and Supplementary Table 4). RNA extraction and quantification of gene expression were conducted as described in section “RNA Extraction and Reverse Transcription” with the following modifications due to a higher RNA yield: the fixed amount of RNA that was reverse transcribed into cDNA was 400 ng/sample (67 ng/μl) instead of 300 ng/sample (50 ng/μl) and pre-amplified cDNA was diluted 1:10 and instead of 1:20.
Statistics
All statistical analyses and visualizations were performed in the R 3.6.1 environment (RCoreTeam, 2020).
Life-History Traits
We used two-way ANOVAs to assess size and weight differences between males and females from different origin salinity as well as differences in brood size and in total length between juveniles at 10 days post-hatch. Fixed factors included origin salinity (salinity at samling sites of origin (two levels): High or Low), acclimation salinity (High or Low), sex of the pipefish (male or female) and the sampling site (Flens, Fehm, Falck, Salz, RuegN, or RuegS) nested in origin salinity. ANOVA of brood size additionally included the average body length of males exposed to a given treatment. Homogeneity of variances was tested by Fligner test and normal distribution of data by using the Cramer-von Mises normality test. The brood size was square root transformed to achieve normal distribution of residuals.
We performed two spearman-rank correlations using the function “ggscatter” (package: “ggpubr”) to test for (1) correlation between the total length of adult pipefish and the salinity measured at the sampling site on the day of capture as well as (2) between brood size and average male size of sampling site. The brood size of males originating from high salinity and acclimated to low salinity conditions was removed from the correlation due to fungus infection. Post hoc tests were carried out using Tukey’s “honest significant difference” (Tukey’s HSD, package: “multcomp”) (Hothorn et al., 2020).
Gene Expression of Parental Generation and Juveniles
From the Fluidigm output data, mean cycle time (Ct) and standard deviation (SD) for each of the two technical replicates were calculated. Expression measurements with a coefficient of variation (CV; CV = SD/Ct) larger than 4% were excluded from the study (Bookout and Mangelsdorf, 2003). For females, the combination of HDAC1 and HDAC3 were identified as the optimal reference genes [geNorm V < 0.15 (Vandesompele et al., 2002)] with high reference target stability (geNorm M ≤ 0.5), based on 155 samples (Supplementary Table 5) and 34 target genes (Supplementary Table 2) in Qbase+3.0 (Hellemans et al., 2007).
In the analyses of juvenile gene expression one osmoregulation gene (15% NAs) and 36 samples were excluded from the study due to failed reactions on the Fluidigm chip in at least one of the duplicates. Samples with more than 10% excluded genes were omitted from the analysis, as many missing Ct values are indicative for insufficient sample quality. Remaining missing Ct values were substituted by the mean Ct for the given gene calculated from all other samples, as subsequent analyses are sensitive to missing data. Based on 559 samples and 47 target genes, the reference genes ASH and HDAC1 were selected using the same criteria as for pipefish females. From the geometrical mean of the two reference genes –ΔCt values were calculated to quantify relative gene expression.
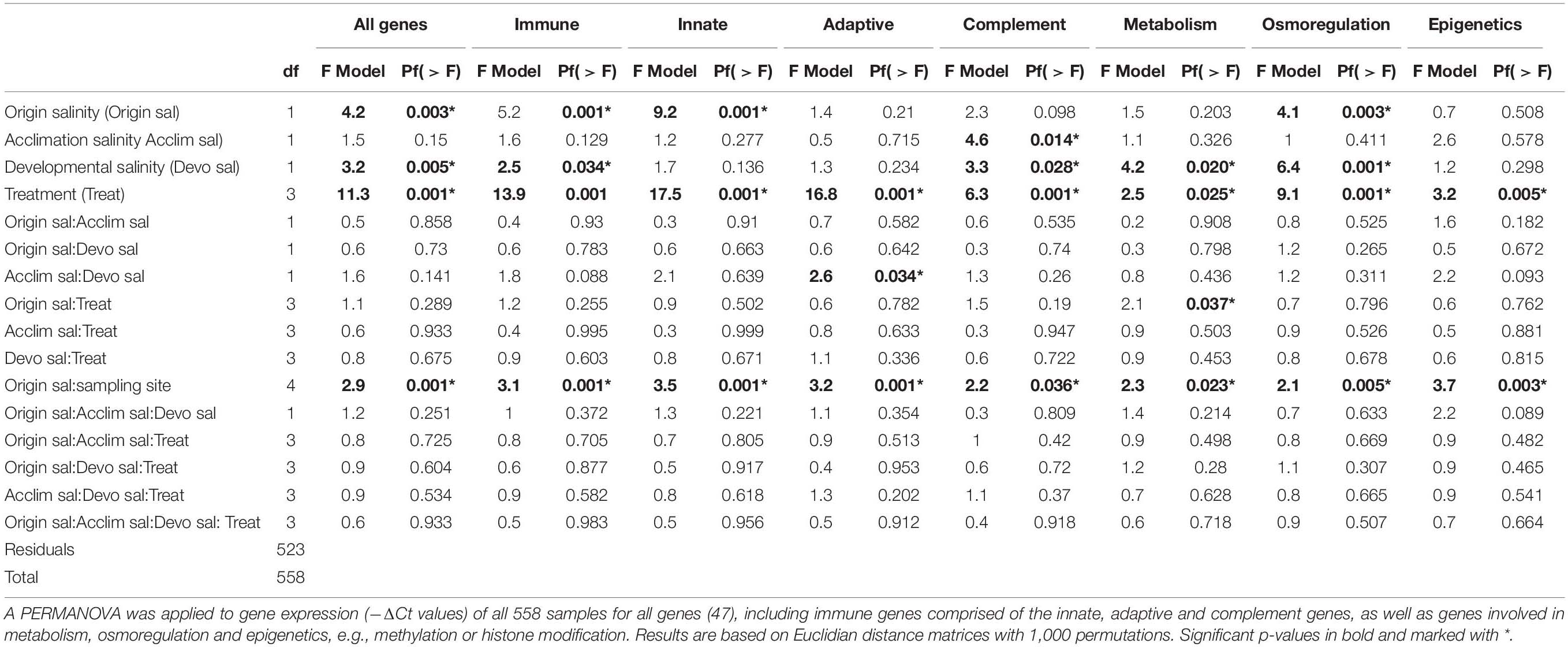
Origin salinity (High or Low), acclimation salinity (High or Low) and developmental salinity (high or low) were defined as fixed factors, whereas sampling site (Flens, Fehm, Falck, Salz, RuegN, or RuegS) was nested within origin salinity. A Permutational Multivariate Analysis of Variance (PERMANOVA) was applied to gene expression (–ΔCt values) of all samples and target genes for each factor and every interaction of the fixed factors. The PERMANOVA [package: “vegan,” function “adonis” in R (Oksanen et al., 2019)] was based on Euclidean distance matrixes with 1000 permutations (Beemelmanns and Roth, 2016b).
A post-hoc analysis of variance (ANOVA) for every gene was applied; though, to account for multiple testing, only factors and factor interactions identified as significant by the PERMANOVA were considered.
To visualize similarity/dissimilarity in gene expression among treatment groups, we performed PCAs [package: “ade4,” function: “dudi.pca” and “s.class” (Dray and Dufour, 2007)]. To visualize significant differential gene expression among groups in heatmaps (package: “NMF,” function: “aheatmap”), –ΔΔCt values for each gene were calculated as follows (Yuan et al., 2006):
Mortality of Juveniles Within the First 10 Days and Post-infection
Ten days post-hatch endpoint mortality of juveniles was analyzed as a ratio of “alive” vs. “dead” fish using a generalized linear model (package: “lme4”, function: “glm”) with binomial error and the following fixed factors: Origin salinity (High or Low), acclimation salinity (High or Low) and developmental salinity (high or low) and the sampling site nested in origin salinity. Significance was tested using ANOVA type two partial sums of squares, and models were simplified using Akaike information criterion (AIC) (Akaike, 1976). Post hoc tests were carried out using Tukey’s honest significant difference (Tukey’s HSD, package: “multcomp”, function: “glht” (Hothorn et al., 2020)). Endpoint mortality of juveniles used in the infection experiment was analyzed as described above including infection treatment [control (c), sea water injection (sw), Vibrio 7 PSU (v7), and Vibrio 15 PSU (v15) injection] as an additional factor.
Results
Pipefish Population Structure
Allele frequencies obtained at 11 microsatellite loci of 144 female pipefish sampled at six sampling sites along the German Baltic Sea coastline indicated gene flow or recently isolated populations with no or very little divergence on neutral genetic markers. The findings are based on a Bayesian clustering method using the software STRUCTURE (Figure 2A), global fixation index (FST) of 0.002 and pairwise FST (Figure 2B and Supplementary Table 6). Overall, the pairwise FST were low for all comparisons and with the exception of Falckenstein-Fehmarn (FST = 0.024) and Falckenstein-Flensburg (FST = 0.016) pairwise comparisons had a FST ≤ 0.01 (Supplementary Table 5). Phylogenetic analyses suggest three branches in an unrooted tree with clusters comprised of pipefish from i) Ruegen North and Ruegen South, ii) Salzhaff and Flackenstein as well as iii) Flensburg and Fehmarn (Figure 2C).
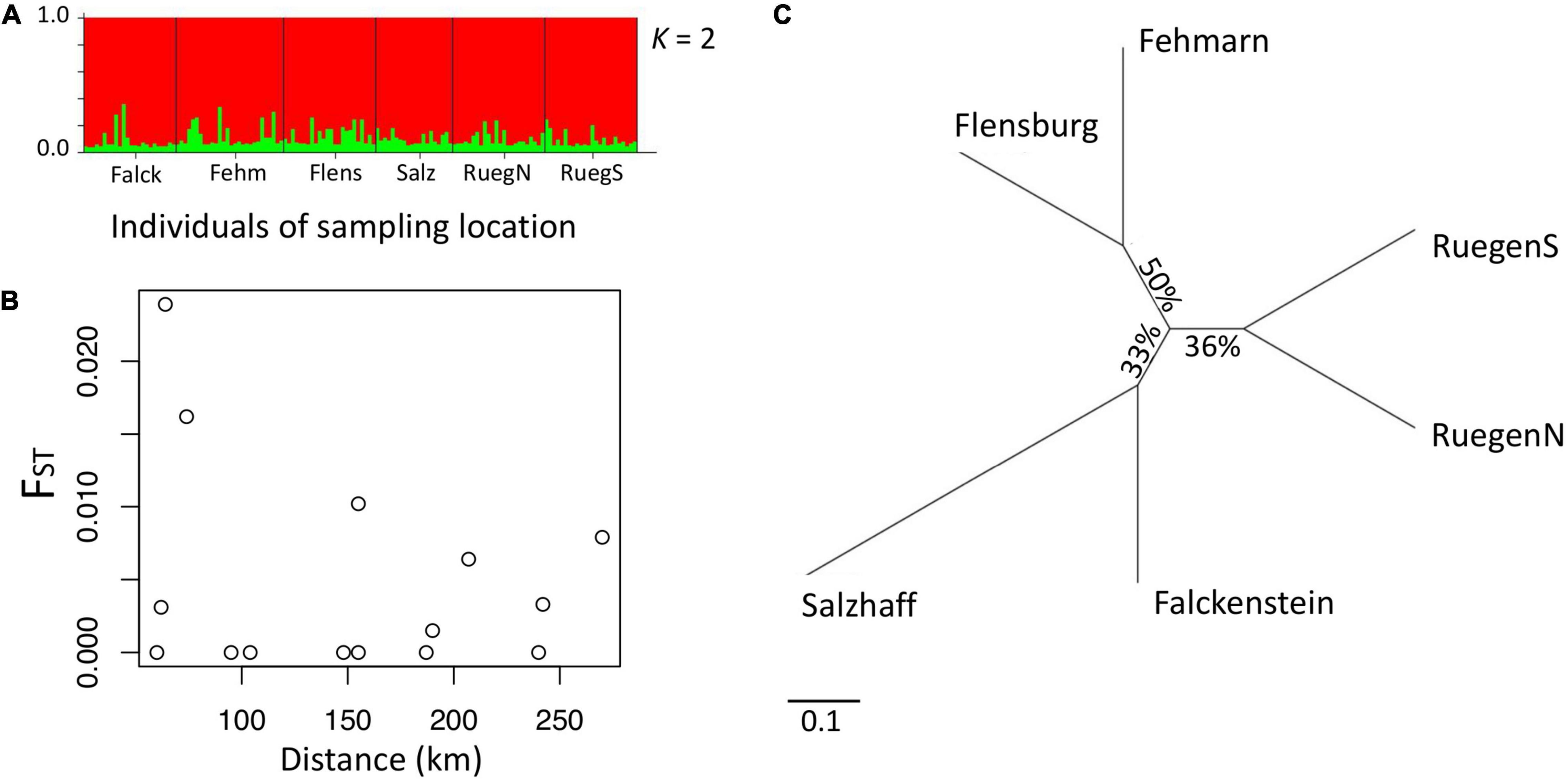
Figure 2. Population structure of pipefish in the southern Baltic Sea. (A) STRUCTURE software results based on 11 microsatellite loci. Each individual is represented by a vertical line, which is colored according to the assigned groups (K = 2). (B) Plotting pairwise FST (y-axis) against distance between sampling sites (x-axis) does not reveal isolation by distance (waterway). (C) Unrooted phylogeographic PHYLIP tree. Distances indicate the relative divergence of microsatellite loci in pipefish between sampling sites. Scale bar represents nucleotide substitutions per site.
Life-History Traits and Fungus Infection of Parental Generation
Pipefish Adults From Low Saline Environment Have a Smaller Body Size
Pipefish males and females caught in high origin salinity of the Baltic Sea were on average larger [mean ± SD, all high salinity sampling sites: 14.2 ± 2.1 cm; Flens: 14.4 ± 2.1 cm (n = 61), Falck: 14.5 ± 2.0 cm (n = 59), Fehm: 13.8 ± 2.1 cm (n = 59)] than those from the low origin salinity [all low salinity sampling sites: 12.8 ± 2.0 cm; Salz: 14.0 ± 2.1 cm (n = 53), RuegN: 11.7 ± 1.6 cm (n = 48), RuegS: 12.4 ± 1.5 cm (n = 52): Figure 3A]. We found an interaction in the total length of adult pipefish between origin salinity and acclimation salinity [ANOVA F(1,320) = 7.4, p < 0.01; Supplementary Table 7A] indicating that parental acclimation salinity negatively affected growth of adult pipefish depending on the origin salinity. Adults from high origin salinity tended to grow slower at low acclimation salinity compared to high acclimation salinity (Tukey’s HSD, HH – HL: p = 0.085; Supplementary Table 7B), whereas acclimation salinity did not affect size of pipefish from low origin salinity (Tukey’s HSD, LL – LH: p = 0.535: Supplementary Table 7B). Furthermore, all pairwise comparisons suggested that pipefish from high origin salinity were in general larger than pipefish from low origin salinity (Tukey’s HSD, LL – HH: p < 0.001, LL – HH: p < 0.001, LH – HL: p < 0.001; Supplementary Table 7B). The significant factor sampling site, which was nested in origin salinity [ANOVA F(4,320) = 11.2, p < 0.01; Supplementary Table 7A] indicates that individuals from Salzhaff were larger compared to individuals from Ruegen North and Ruegen South but did not differ from pipefish caught at high origin salinity (Tukey’s HSD, Salz – RuegN: p < 0.001; Salz – RuegS: p < 0.001; Supplementary Table 7C).
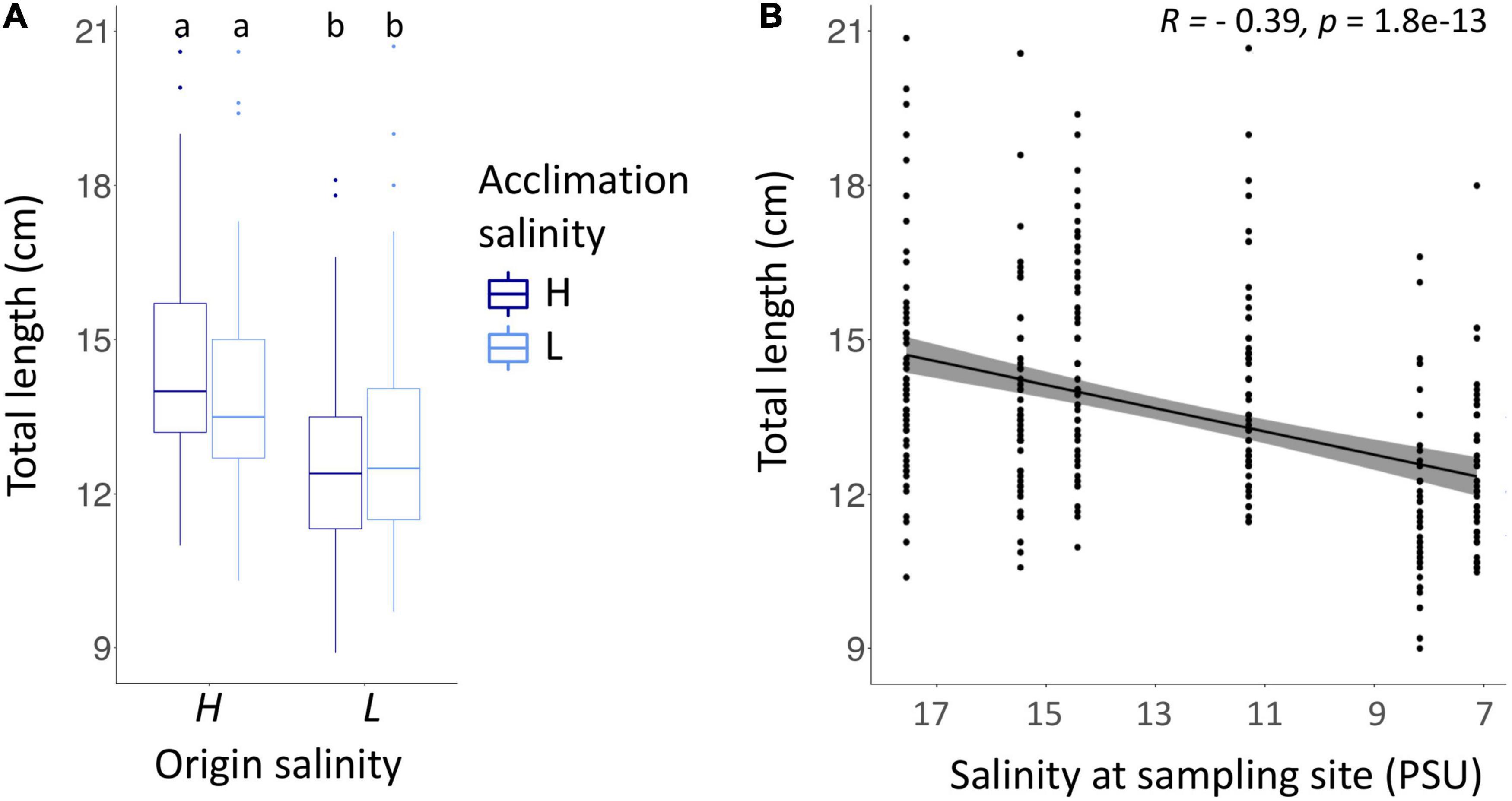
Figure 3. Body length of adult pipefish (A) correlated with salinity at the sampling site (B). (A) The total length of pipefish is shown for both origin salinities (high (H) – 15 PSU, low (L) – 7 PSU). Colors indicate whether the parental generation was exposed to a high (H, dark blue) or to low acclimation salinity (L, light blue). (B) The total length of pipefish after acclimation correlated with the salinity measured at the sampling site using spearman rank correlation. Gray bar indicates 95% confidence interval.
Pipefish from low origin salinity were smaller compared to pipefish from low origin salinity (correlation between salinity at sampling site and adult size, i.e., length (Figure 3B) and weight (Supplementary Figure 7B). Length and weight of animals were not corrected for age. When caught in spring, pipefish are usually all in the same age, as they were born the previous summer and reach sexual maturity around the catching date.
Pipefish From High Saline Environments Were More Susceptible to Fungus Infection When Exposed to Low Saline Conditions
Visible fungus infections of the brood pouch occurred in almost half of the pipefish males (47%) caught at high origin salinity and kept at low acclimation salinity (Figure 4). Fungus infections ranged from mild infections in the brood pouch not affecting brood size to a complete loss of the brood. Males from a high origin salinity that remained at high acclimation salinity as well as males from the low origin salinity had no symptoms of fungus infection regardless of the acclimation salinity.

Figure 4. Brood pouch fungus infections were present in 47% of fathers from high origin salinity kept at low acclimation salinity. Pipefish from low origin salinity independent of acclimation salinity and pipefish from high origin salinity kept at high acclimation salinity did not show any signs of fungus infection (A). In males caught at high origin salinity and kept at low acclimation salinity fungi infections ranged from mild (B) to extreme (C) resulting in the complete loss of eggs and offspring.
Brood Size
Males from a high origin salinity kept at high acclimation salinity had the largest brood size (HH, mean ± SD., 41.8 ± 23.4, n = 68, Tukey’s TSD; Supplementary Table 8B) followed by males from low origin salinity kept at high salinity (LH, 27.8 ± 13.2, n = 58) or low salinity (LL, 25.2 ± 16.9, n = 58), which corresponds to the lower body size at low salinity (Figure 3). Pipefish from high origin salinity exposed to low acclimation salinity were frequently infected by a brood pouch fungus which reduced the brood size (HL 19.4 ± 15.2, n = 54; Tukey’s TSD, HL-HH, t = −4.8, p < 0.001; Supplementary Table 8B). In contrast, acclimation salinity did not affect brood size of parents from low origin salinity (Tukey’s TSD, LL-LH, t = −0.4, p = 0.976; Supplementary Table 8B). This divergent patterns caused an origin salinity:acclimation salinity interaction [ANOVA F(1,109) = 9.0, p = 0.003; Supplementary Table 8A]. Larger brood size was in general driven by a larger total body length of male pipefish (Spearman rank correlation, R = 0.43, p < 0.001, n = 234; Figure 5).
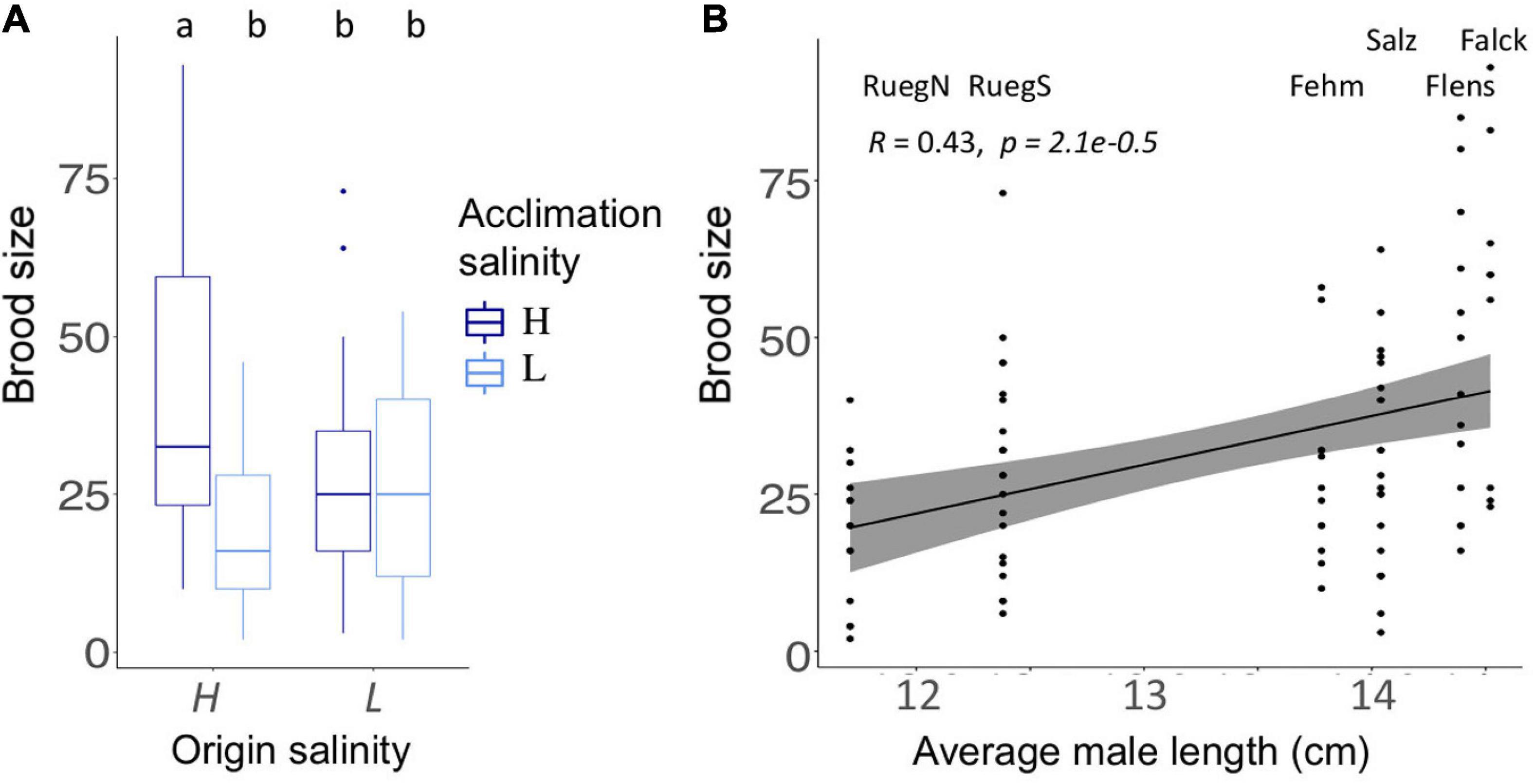
Figure 5. Parental life-history traits. (A) Number of juveniles (brood size) is shown for pipefish with high (H) or low (L) origin salinity. Color indicates acclimation salinity in the lab (H: 15 PSU – dark blue and L: 7 PSU – light blue). Different letters indicate significant differences (Tukey’s HSD, p < 0.05). (B) Brood size correlated positively with the average size of male pipefish from each sampling site. The sampling sites are presented above the data points. The gray bar along the regression line indicates the 95% confidence interval. A more detailed figure visualizing the differences between the single origins can be found in the Supplementary Table 7C.
Two Immune Genes Are Upregulated in Females From a High Origin Salinity
Origin salinity had an impact on the expression of immune genes in female pipefish (PERMANOVA, immune F(1,146) = 3.1, p = 0.010, n = 155, Supplementary Table 9, Supplementary Figure 9A), in particular of the innate immune system [PERMANOVA, innate F(1,146) = 4.0, p = 0.002; Supplementary Table 9]. Lectin protein type I (Lecpt1), a pathogen recognition receptor, and chemokine 7 (ck7), a gene encoding a protein responsible for chemotaxis in blood cells, were upregulated in pipefish from low origin salinity in contrast to high origin salinity females. Low acclimation salinity caused a slight upregulation in the expression of histone modification gene histone deacetylase 6-like (hdac6) [PERMANOVA, silencing F(1,146) = 2.9, p = 0.044; Supplementary Table 9, Supplementary Figure 9A].
Life-History Traits, Survival and Gene Expression of Offspring Generation
Juveniles From Low Origin Salinity Parents Have Higher Survival Rates and Are Smaller
In the first 10 days after hatching, juvenile survival was impacted by origin salinity and acclimation salinity of the parental generation origin salinity:acclimation salinity interaction, GLM, χ21 = 6.1 (p = 0.013, n = 235; Supplementary Table 10A; Figure 6). There was no difference in survival of juveniles from parents that were continuously exposed to the same salinity (i.e., matching origin and acclimation salinity) (Tukey’s HSD, LL – HH: z = −3.4, p = 0.783; Supplementary Table 10B). Similarly, juvenile survival did not differ for non-matching origin salinity and acclimation salinity LL – LH: z = −4.7, p = 0.842; HH – HL: z = −2.1, p = 0.148; Supplementary Table 10B). However, juveniles from high origin salinity parents exposed to high acclimation salinity in the lab (LL) had higher survival rates compared to juveniles from high origin salinity exposed to low acclimation salinity (Tukey’s HSD, LL – HL: z = −3.4, p = 0.043; Supplementary Table 10B). The origin salinity:sampling site effect suggest that patterns at single sampling sites differ. In particular, Flensburg (origin salinity: 17 PSU “high”) offspring exposed to high developmental salinity had reduced survival rates, when parents were acclimated to low instead of high salinity (Tukey’s HSD; Hh – Lh: z = −4.4, p = 0.046; Supplementary Table 10B). Following the same pattern of non-matching acclimation salinity, Ruegen South (origin salinity: 7 PSU “low”) offspring exposed to low developmental salinity had reduced survival, when parents were kept at high acclimation salinity (Tukey’s HSD; Hl – Ll: z = 5.8, p < 0.001; Supplementary Table 10B). This suggests that exposure of parents to novel salinities can negatively impact juvenile survival when juveniles experience salinity conditions, which did not match parental acclimation salinity.
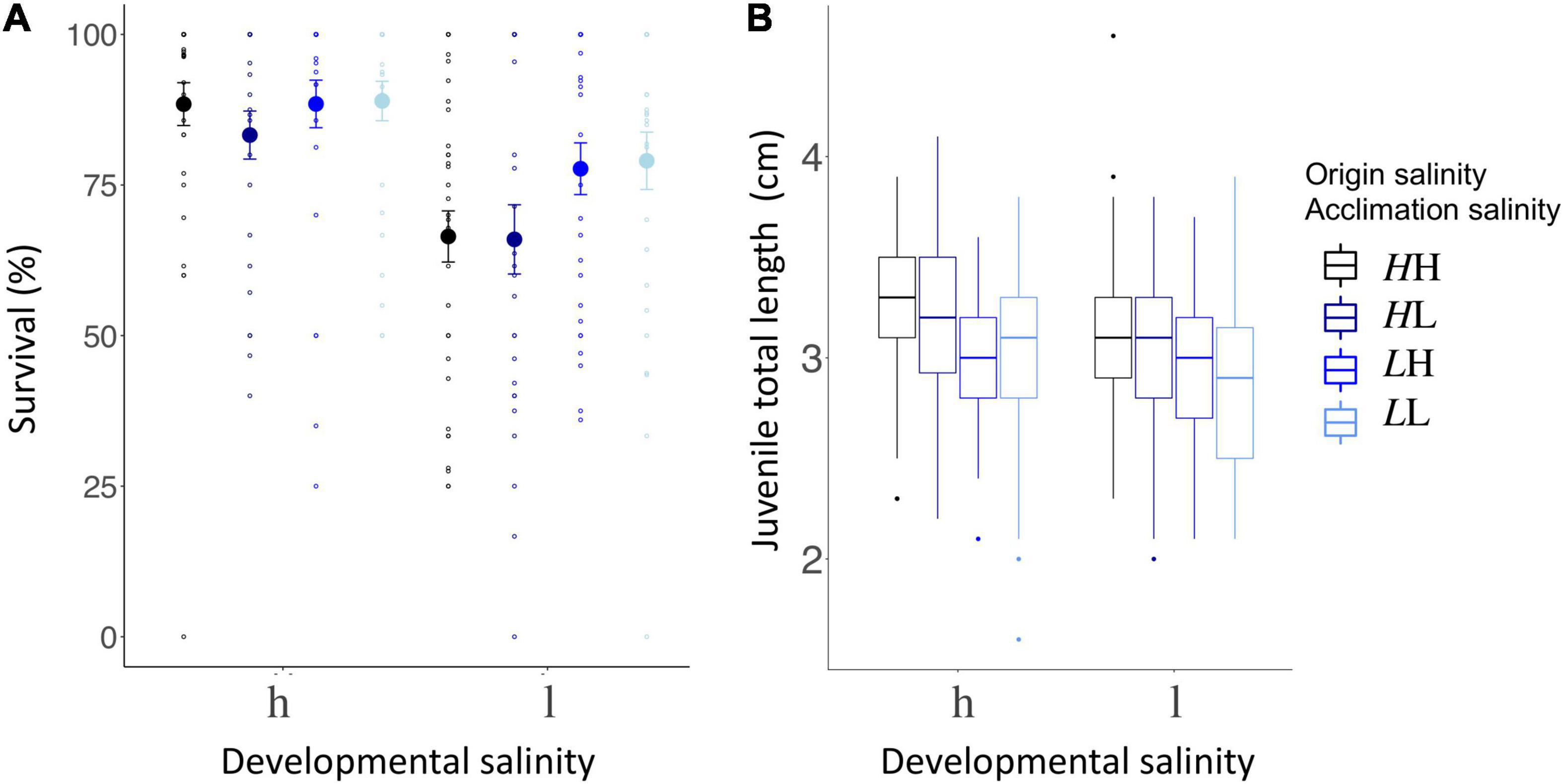
Figure 6. Juvenile life history parameters (A) survival rate (%), (B) body length (cm). (A) The percentage of juveniles surviving the first 10 days post-hatch (%) are plotted on the y-axis. The x-axis indicates the developmental salinity after hatching (h – high/15 PSU, l – low/7 PSU). Italic letters and colors specify the origin salinity of the parental generation (H: 15 PSU, black and dark blue; L: 7 PSU blue and light blue). The 2nd letter indicates the acclimation salinity in the lab (H: 15 PSU, black and blue; L: 7 PSU, dark and light blue). (B) Juvenile size (cm) 10 days post hatch is plotted on the y-axis. Labeling and color code correspond to panel (A).
Overall higher survival at high developmental salinity compared to low developmental salinity (developmental salinity, GLM, χ21 = 192.8, p = 0.031; Figure 6; Supplementary Table 10A) indicates that low salinity imposes stress on pipefish juveniles regardless of the origin salinity. An exception are juveniles from Ruegen North (sampling site, GLM, χ24 = 24.1, p < 0.001; Supplementary Figure 9A) where juvenile survival was not affected by developmental salinity (GLM, χ21 = 0.1, p = 0.766).
Ten days after hatching, juveniles from high origin salinity were larger (3.18 cm ± 0.37, n = 408; Figure 6) than juveniles from low origin salinity sampling sites (2.95 cm ± 0.37, n = 405) [origin salinity, ANOVA F(1,782) = 86.2, p < 0.001; Supplementary Table 11; Figure 6]. While acclimation salinity, i.e., mating and male pregnancy, had no effect on size of juveniles [acclimation salinity, ANOVA F(1,782) = 2.2, p < 0.136; Supplementary Table 11], low developmental salinity reduced offspring size [developmental salinity, ANOVA F(1,782) = 17.4, p < 0.001; Supplementary Table 11] suggesting that low salinity levels are stressful for pipefish offspring and reduce their growth rates.
Juvenile Survival Is Reduced After Injections and at Low Salinity
Ten days post hatch, juvenile pipefish were challenged either with Vibrio alginolyticus bacteria evolved at 15 PSU, 7 PSU, autoclaved seawater (sham injection) or not treated at all (control) and survival was measured 6 days post infection, i.e., approximately 16 days post hatch. Non-challenged control groups had the highest survival rates (Mean ± SD.; 83.0% ± 32.2, n = 237; Figure 7). The injection itself decreased survival of juveniles by at least 10% in all salinity treatments combined, regardless whether seawater (66.9% ± 38.2, n = 192), Vibrio evolved at 15 PSU (73.0% ± 36.8, n = 190) or 7 PSU (66.7% ± 38.6, n = 192; Figure 7) was administered. Vibrio strains evolved at 15 PSU caused a higher mortality in juveniles from high origin salinity regardless of acclimation salinity compared to juveniles from low origin salinity with low parental acclimation salinity (origin salinity × acclimation salinity × treatment, GLM, χ21 = 13.0, p = 0.005; Supplementary Table 12A; Tukey’s HSD; LLv15 – HHv15: z = −3.4, p = 0.046; LLv15 – HLv15: z = −3.5, p = 0.038; Supplementary Table 12B). When fathers from low origin salinity were exposed to high acclimation salinity these positive effects on offspring survival were lost (Tukey’s HSD, LHv15 – HHv15 z = −1.5, p = 0.971; Supplementary Table 12B). This suggests that mis-matching salinity levels between the parental and juvenile generation can lead to reduced survival rates.
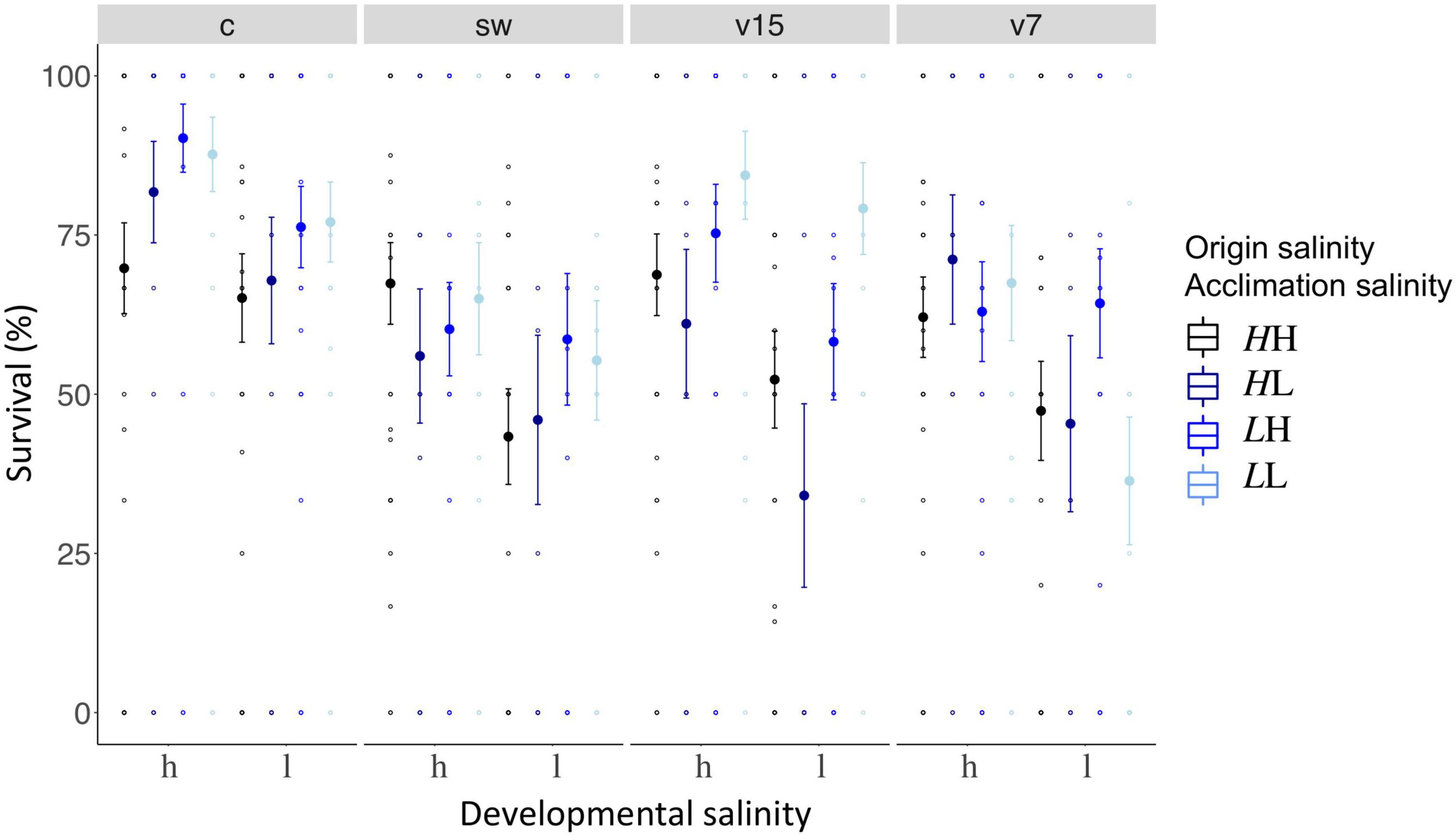
Figure 7. Juvenile survival 6 days post infection. Juvenile survival 6 days post infection is plotted on the y-axis. The x-axis indicates the developmental salinity (h – high/15 PSU, l – low/7PSU). Italic letters and colors specify the origin salinity of the parental generation (H: 15 PSU, black and dark blue; L: 7 PSU, blue and light blue). The 2nd letter and colors indicate the acclimation salinity in the lab (H: 15 PSU, black and blue; L: 7 PSU, dark and light blue). Each treatment is represented by one panel, i.e., control (c) or injection with seawater (sw), Vibrio strain evolved at 15 PSU (v15), or at 7 PSU (v7).
Juvenile survival was in general higher in high developmental salinity conditions (71.1% ± 36.7, n = 421) compared to low developmental salinity conditions (58.6% ± 36.7, n = 392) (GLM, developmental salinity, χ21 = 40.2, p = 0.001; Supplementary Table 12A) suggesting that low salinity levels are a stressful environment for pipefish development. An adaptation to low salinity may result in an increased fitness as juveniles from low origin salinity fathers (69.8% ± 40.1, n = 405) had in general a higher survival rate compared to juveniles from high origin salinity fathers (60.6% ± 37.7, n = 408; GLM, origin salinity, χ21 = 9.1, p = 0.003, Supplementary Table 12A).
An effect of sampling site nested in origin salinity (GLM, χ24 = 39.5, p < 0.001; Supplementary Table 12A) indicates that survival patterns for each sampling site within the origin salinity categories are diverse. The statistical diversity may be a result of the high variation in survival rates within a single treatment, which sometimes ranged from 0 to 100%. Combining the survival rates of all three sample sites of one origin salinity resulted in more robust and conclusive results as described in the first paragraph of this section.
Matching Parental Acclimation and Juvenile Developmental Salinity Results in Similar Juvenile Gene Expression Patterns of Adaptive Immune Genes
An origin salinity effect indicates that gene expression of juveniles differs depending on the salinity they originated from [PERMANOVA, all genes; origin salinity F(1,523) = 4.2, p = 0.003, n = 559; Figure 8, Table 2]. Such signs of genetic adaptation were found in genes associated with the innate immune system [PERMANOVA, innate, F(1,523) = 9.2, p = 0.001] and with osmoregulation [PERMANOVA, osmo, F(1,523) = 4.1, p = 0.003; Figure 8, Table 2]. Single ANOVAs suggested that this effect was driven by five genes. Whereas the expression of the pathogen reception recognition gene lectin protein type II (lectpt2) was induced in juveniles from low origin salinity parents, the expression of the following genes was upregulated in juveniles from high origin salinity parents: immunoglobulin light chain (IgM; pathogen recognition), heat shock protein 70 kDa (hsp70; osmotic stress response); voltage gated potassium channel (kcnh8; cell volume regulation), prolactin (prl; ion uptake promotion and ion secretion inhibition). The genetically induced upregulation of osmoregulatory genes suggests an adaptation to low salinity levels.
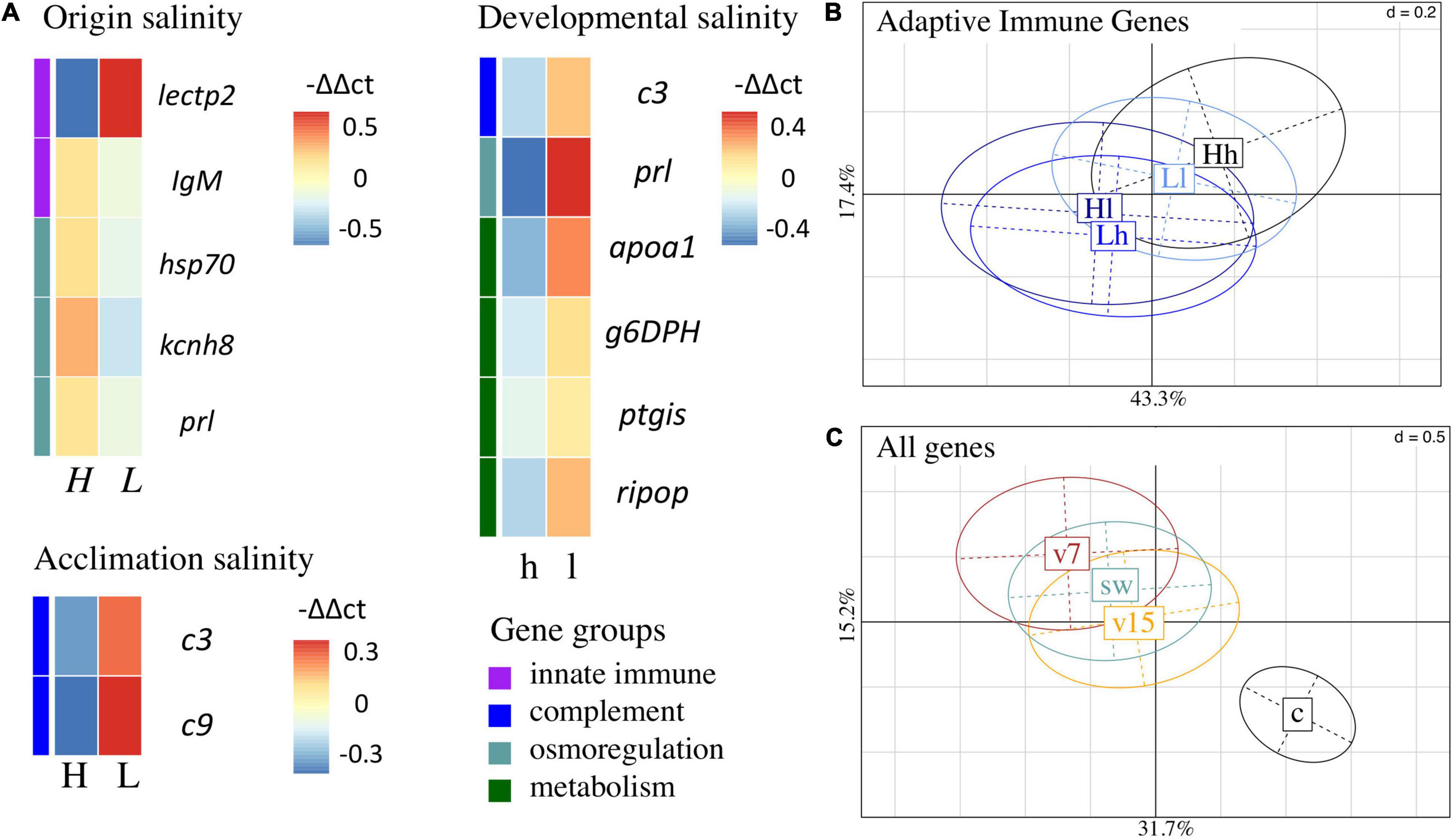
Figure 8. Gene expression patterns of juveniles. (A) Non-hierarchical gene expression heatmap for genes showing differential expression (-ΔΔCt) in response to origin salinity, acclimation salinity or developmental salinity. Genes are sorted by gene groups which are assigned with different colors (purple: innate immune system, blue: complement system, turquoise: osmoregulation, green: metabolism). (B) Principal component analysis (PC1: 43.3%; PC2: 17.4%) of adaptive immune genes for significant interaction between parental acclimation salinity [H – high (15 PSU), L – low (7 PSU)] and developmental salinity (h, l). Four out of seven adaptive immune gene were upregulated (Supplementary Figure 13), when parental acclimation salinity and juvenile developmental salinity did not match, i.e., Hl and Lh, compared to a matching acclimation and developmental salinity, i.e., Hh and Ll [PERMANOVA, adaptive F(1,523) = 2.6, p = 0.034]. The confidence ellipse explains 20% of the variability. (C) Principal component analysis (PC1: 31.7%; PC2: 15.2%) of gene expression patterns caused by juvenile treatment, i.e., injection with Vibrio alginolyticus evolved at 15 PSU (v15, orange), V. alginolyticus evolved at 7 PSU (v7, brown), sham injection with seawater (sw, blue, green) or untreated control (c, black). The confidence ellipse explains 20% of the variability.
Juvenile gene expression did not provide further evidence for genetic adaptation, tested as an interaction between origin salinity and acclimation salinity [PERMANOVA, all genes F(1,523) = 0.5, P = 0.858]. Trans-generational plasticity could also not be detected in the interaction between acclimation salinity and developmental salinity [PERMANOVA, all genes F(1,523) = 0.6, P = 0.730].
Exposing parents to low acclimation salinity led to an expression induction of two genes in juveniles [PERMANOVA, complement; acclimation salinity F(1,523) = 4.6, p = 0.014]. Both genes are associated with the complement system: Complement component 3 (c3, complement system activation) and Complement component 9 (c9, membrane attack complex). In addition to this parental effect a trans-generational effect was observed as a parental acclimation salinity: developmental salinity interaction effect on adaptive immune gene expression [PERMANOVA, adaptive F(1,523) = 2.6, p = 0.034; Figure 8B, Table 2]. Gene expression was lower in four out of seven adaptive immune genes when acclimation salinity and developmental salinity were matching compared to non-matching conditions (Supplementary Figure 13): Human immunodeficiency virus type l enhancer 2 (hivep2, transcription factor, MHC enhancer binding) and 3 (hivep3; transcription factor, MHC enhancer binding), B-cell receptor-associated protein (becell.rap31, T- and B-cell regulation activity) and immunoglobulin light chain (igM; antigen/pathogen recognition). The reduction in gene expression of immune genes can hint at a reduced stress level in offspring when parents were acclimated to the same salinity as their offspring.
Developmental plasticity allows juveniles to quickly respond to present salinity levels. Low developmental salinity resulted in higher expression of six genes. Complement component 3 (c3) is involved in the complement system [PERMANOVA, comple; developmental salinity F(1,523) = 4.6, p = 0.014; Figure 8, Table 2], prolactin (prl, ion uptake promotion and ion secretion inhibition) is associated with osmoregulation [PERMANOVA, osmo; developmental salinity F(1,523) = 3.3, p = 0.028] and apolipoprotein A1 (apoa1, antimicrobial activity), glucose 6 phosphate dehydrogenase (g6DPH, pentose phosphate pathway), prostaglandin I2 Synthase (ptgis, Lipid and fatty acid metabolism), ribosomal protein (ripop, Translation process) are related to the metabolism [PERMANOVA, meta; developmental salinity F(1,523) = 4.6, p = 0.014].
Finally, we wanted to test whether genetic background, i.e., origin salinity, acclimation salinity of the parents and the developmental salinity influenced the ability of juveniles to cope with infections of the opportunistic pathogen V. alginolyticus, which evolved in the lab at either 15 or 7 PSU. However, we found no interaction between any salinity regime of parents or juveniles interacting with gene expression after juvenile infection. An injection, regardless of the component, i.e., autoclaved seawater, V. alginolyticus evolved at 15 PSU or 7 PSU caused similar changes in gene expression patterns that could only be differentiated from the untreated control group. In 24 genes injections induced gene expression [PERMANOVA, all genes; F(3,523) = 11.3, p = 0.001; Supplementary Figure 14], including genes from all groups. In five genes, injections reduced gene expression compare to the control group. Using post hoc tests on ANOVAs of single genes, we found no differences in gene expression between the three injection treatments.
Discussion
We have investigated the role of genetic adaptation and phenotypic plasticity as well as their interaction on the ability of the broad-nosed pipefish Syngnathus typhle to cope with changes in salinity levels. S. typhle is a marine teleost, which originally invaded from the North Sea into the Baltic Sea after its formation about 8000 years ago (Wilson and Veraguth, 2010). The brackish salinity environment in the Baltic Sea imposes osmoregulatory stress on marine animals and is thus assumed to be an important driver for genetic divergence and adaptation to local condition (Berg et al., 2015; Guo et al., 2015; Johannesson et al., 2020). Strong genetic clines over the Baltic Sea salinity gradient were shown for a diversity of species (Johannesson et al., 2020) with strong patterns of local salinity adaptation (Leder et al., 2021). Here, we focused on six pipefish populations from the German coastline of the Baltic Sea, out of which three originated from a relatively high saline environment (14–17 PSU), and three from a relatively low saline environment (7–11 PSU). By taking the two salinity regimes into account, our experiment permitted to test both for local adaptation and for phenotypic plastic acclimation to different salinities. Pipefish from all sampling sites displayed low levels of neutral differentiation. This provides good opportunities for studying local adaptation of phenotypically differentiated populations because observed phenotypic differences are likely to reflect genes under selection rather than genetic differences resulting from stochastic processes such as drift (Sunde et al., 2020).
Syngnathus typhle caught in the Baltic Sea high salinity environments (14–17 PSU) were smaller (mean size in this study: 14.2 ± 2.1 cm) than those populating the marine realm with more than 28 PSU (mean size animals caught between 28 and 36 PSU: 18.7 cm (Rispoli and Wilson, 2008) or 15.5 cm (Gurkan and Taskavak, 2007), but larger than those sampled in Baltic Sea low salinity environments, i.e., mean size in this study: 12.8 cm ± 2.0 cm, pipefish sampled at 5.5 PSU around Askö (Sweden): 14.5 cm (Rispoli and Wilson, 2008) or at 7 PSU around Gotland: 14.0 ± 1.1 cm (females), 13.4 ± 1.1 cm (males) (Berglund et al., 2017). This suggests that osmoregulation is costly (Rolfe and Brown, 1997; Boeuf and Payan, 2001) and that the negative impact of low salinity can potentially not be fully compensated through local adaptation. As such, trade-offs for osmoregulation may reduce growth rates, which ultimately result in a decreased fitness. Studies of other marine teleosts that originated from fully marine environments, e.g., sticklebacks and cod, suggested that high growth rates at intermediate salinity levels (10–20 PSU) are possible, especially when the salinity levels reclose or slightly above isosmotic levels (Dutil et al., 1997; Imsland et al., 2001; Heckwolf et al., 2018).
In this previous study (Nygård et al., 2019), the body weight or length of the offspring correlated with the parental size. Larger females produced larger eggs, and fathers with larger body size gave birth to larger juveniles (Nygård et al., 2019). To this end, both the parental body size and a resource-allocation trade off imposed by an increased energy demand for osmoregulation can explain the reduced embryonal growth in the low saline environment (Boeuf and Payan, 2001). In the here presented survival experiment, juveniles from low origin salinity parents, i.e., RuegS, RuegN and Salzhaff, survived better compared to high origin salinity parents, i.e., Flensburg, Falckenstein, Fehmarn. Their induced survival was neither influenced by the parental acclimation salinity nor by the developmental salinity or the exposure to Vibrio bacteria. We thus suggest two alternative parental care strategies: (i) large broad-nosed pipefish parents can invest in larger brood and offspring size as suggested for the larger animals originating from high saline populations that produced more offspring of a larger size, while (ii) small parents may rather invest in offspring survival (Nygård et al., 2019) via genetically determined gene expression patterns as suggested by the induced survival of offspring from the generally smaller parents originating from low saline environments.
Such genetically determined gene expression patterns that are inherited from generation to generation can be indicative signs for local adaptation (Larsen et al., 2011; Fraser, 2013; Heckwolf et al., 2020). In this study, juveniles from animals caught in high saline environments generally had an induced expression of osmoregulation genes. We suggest that the here observed genetically determined level of osmoregulation gene expression in pipefish is an adaptation to local salinity levels. The upregulation of the immune gene lecpt2 in the juveniles from low salinity parents may be an indicator for an adaptation towards the specific local pathogen community. Adaptive gene expression levels in the immune system can result in a faster and stronger and eventually more effective immune responses (Swaggerty et al., 2008). In contrast, pipefish from low origin salinity may rather suffer stress induced by the above stated resource allocation trade-off, which decreases the resources available for the innate immune system.
Under stress, animals are more susceptible to infections with pathogens, which may turn opportunistic pathogens into causative agents of deadly diseases (Boyett et al., 2007; Poirier et al., 2017; Sullivan and Neigel, 2018). Furthermore, low saline environments have been suggested to select for increased pathogenic virulence, e.g., due to changes in gene expression (Hase and Barquera, 2001) and biofilm formation (Dayma et al., 2015). This is in line with the observed brood pouch infections during pregnancy that massively impacted fathers adapted to a high origin salinity but exposed to low acclimation salinity. The number of offspring was reduced and the offspring had a smaller body size. Fathers caught at low origin salinity (both LL: low origin salinity, low acclimation salinity and LH: low origin salinity, high acclimation salinity) did not show signs of brood pouch infection, which gives support for our hypothesis that these animals were locally adapt to low saline environments and the associated pathogens. Juveniles exposed to Vibrio bacteria survived better when their parents originated from low saline waters. This suggests that in low saline waters selection induced by the potentially more virulent and more prevalent microbial infectious agents must have resulted in pipefish that are less susceptible (i.e., either more resistant or more tolerant) to infections with opportunistic Vibrio bacteria.
Juveniles are expected to have advantages when exposed to the same environment as their parents (Sunday et al., 2014; Roth et al., 2018). An interaction of the parental acclimation salinity and the juvenile developmental salinity is generally interpreted as an indicator for trans-generational plasticity (Uller et al., 2013; Heckwolf et al., 2018). In contrast to previous experiments focusing on trans-generational plasticity and immune priming in pipefish (Beemelmanns and Roth, 2016b; Beemelmanns and Roth, 2017; Roth and Landis, 2017), the adaptive trans-generational plastic effects identified in this study were limited. Even though survival of juveniles was higher in matching parental acclimation and developmental salinity, the effect was driven by the genetic adaptation and not the parental acclimation, as indicated by the strong origin salinity effect. The same applied to juvenile growth, which was imposed both by origin salinity and by the developmental salinity, but not by acclimation salinity. However, parental acclimation shifted expression of genes involved in complement and adaptive immune systems. As such, parental acclimation to low salinity (main effect) induced the expression of genes of the complement system. Non-matching parental acclimation and developmental salinity (interaction) upregulated genes of the adaptive immune system compared to matching parental acclimation and developmental salinity. In contrast to the above discussed upregulation of innate immune genes, an upregulation of the complement and adaptive immune system is indicative for a clear response toward prevailing parasites and pathogens, due to the specificity of the adaptive immune system (Janeway, 2005). The complement system links the innate to the specific adaptive immune system. Their joint induction could give evidence for a shift in the microbial pathogen community in non-matching environments to which the specific arm of the immune system has to react. However, final support would enquire the genotyping of the microbial pipefish gut community.
The limited presence of trans-generational plasticity gives only marginal support for our hypotheses and is in strong contrast to previous experiments performed with the same model system, where the genetic background was mostly ignored and experiments focused only on one population [Roth et al., 2012b; Beemelmanns and Roth, 2016b (Gotland, Sweden); Beemelmanns and Roth, 2017; Wackerballig (Gelting, Germany), Roth and Landis, 2017; Wackerballig (Gelting, Germany)]. The here performed experiment allows us to at least partially disentangle genetic adaptation and trans-generational plasticity and suggests that selection and the resulting genetic adaptation is a lot stronger than the impact of trans-generational plasticity. To this end, the unexpected limited identification of trans-generational plastic effects could indicate that we are generally overestimating trans-generational plasticity in experiments that ignore genetic background, as genetic adaptation is intermingled with the phenotypic plastic components. Alternatively, we have potentially not identified all present signs of trans-generational plasticity in this experiment as the populations are too distinct due to their history of genetic adaptation hindering the identification of trans-generational plastic effects. By taking the genetic adaptation into account, we suggest that the probability to identify existing phenotypic plastic effects is lower, as the impact of phenotypic plastic effects is weaker than the impact of genetic differences among populations.
Populations that migrate into a new habitat are under strong selection for genetic adaptation toward the novel environmental condition. They go through a bottleneck, which results in populations that are diverged from their ancestral populations (Johannesson et al., 2020) and are characterized by a reduced genetic diversity (Johannesson and Andre, 2006). In another study this reduced genetic diversity as a consequence of genetic adaptation negatively impacted the individual phenotypic plasticity of sticklebacks populating low salinity regions of the Baltic Sea (DeFaveri and Merila, 2014; Hasan et al., 2017). In a stable salinity environment, we would thus expect that genetic adaptation had resulted in reduced phenotypic plasticity and lower performance in the ancestral environment (DeWitt et al., 1998; Schneider and Meyer, 2017). In contrast to our expectation, juvenile survival of parents from low salinity origins was not reduced at high developmental salinity suggesting that genetic adaptation toward low salinity conditions did not result in a reduction of phenotypic plasticity. Along the same line, the smaller size of juveniles from parents originating from low salinity environments is no indicator for reduced plasticity either. The smaller phenotype (at the same age) was more likely a result of the reduced parental size (Nygård et al., 2019) which can be an adaptation to low salinities (McGuigan et al., 2011). The strong horizontal and vertical salinity gradients, the variations in precipitation and erratic saltwater inflow from the North Sea are causing strong salinity fluctuations in the coastal environments across the Baltic Sea (Bock and Lieberum, 2017) most likely selecting against the loss of phenotypic plasticity.
The isosmotic level of many marine fish is equivalent to around 12 PSU (Schaarschmidt et al., 1999) or a couple of units higher, depending on the ambient salinity conditions (Quast and Howe, 1980; Partridge et al., 2007). This suggests that the here applied high salinity treatment is rather hyper- to isosmotic, whereas the low salinity treatment is hypoosmotic. The hormone prolactin is involved in many metabolic pathways in vertebrates and highly relevant for fish in hypoosmotic conditions as it prevents the loss of ions and the uptake of water. Both mechanisms are crucial in hypoosmotic conditions to maintain homeostasis (McCormick, 2001; Manzon, 2002; Breves et al., 2014). In our study, prolactin (prl) was the gene with the strongest upregulation in juveniles at low developmental salinity conditions underlining the ability of pipefish to quickly respond to prevailing salinity conditions. Similar patterns in the upregulation of prolactin in marine fish have been identified in black porgy Acanthopagrus schlegelii (Tomy et al., 2009) and rainbow trout Oncorhynchus mykiss (Prunet et al., 1990). This implies that higher prl expression under low salinity conditions could be indicative for adaptive developmental plasticity and suggest that juvenile fish are able to cope with short term salinity changes.
Some strains of the species Vibrio alginolyticus have been shown to become more virulent under low saline conditions (Dayma et al., 2015; Poirier et al., 2017). Drivers for this increased virulence can be trade-offs in the host (Birrer et al., 2012; Poirier et al., 2017), a phenotypic response of the bacteria (Hase and Barquera, 2001; Dayma et al., 2015) or a genetic adaptation of the bacteria to low salinity (Brown et al., 2012). Under low saline condition, we thus expected strong selection for immunological adaptation toward the prevailing pathogens that potentially resulted in a higher tolerance or a more effective immune defense against Vibrio bacteria. In line with this expectation, we found that pipefish offspring from parents caught at low origin salinity survived better when exposed to Vibrio bacteria than offspring from parents caught at high saline origins. This suggests that local adaptation to low saline conditions allows pipefish to allocate sufficient resources toward their immune system for fighting Vibrio infections. To this end, we found support for our hypothesis that increased Vibrio virulence in marine host organism can result from resource allocation tradeoffs toward osmoregulation, impairing the host’s immune system (Birrer et al., 2012).
The bacteria used in this experiment were previously evolved at the respective high (v15: 15 PSU) or low (v7: 7 PSU) Baltic Sea salinity condition. If genetic adaptation of bacteria to low salinity induces their virulence, we would have expected that the bacteria evolved at 7 PSU (v7) are more virulent, in particular for the pipefish offspring from parents originating from high saline locations. In contrast to our expectation, we have identified that v15 caused a higher mortality in juveniles originating from a high saline environment than in juveniles coming from low origin salinity and low parental salinity acclimation, while the impact of v7 was not differentiable across all groups. Gene expression measurements were not appropriate to answer the question of induced virulence and a corresponding stronger host immune response against bacteria evolved at low or high salinity depending on pipefish local salinity adaptation. Instead, the injury imposed by the injection had the strongest impact on the gene expression pattern. This implies that we did not identify a difference in gene expression of sham-injected animals and those animals injected either with v7 or v15. This is an unexpected limitation of our study. However, given that Vibrio bacteria had an impact on pipefish mortality, we assume that we have simply not chosen the time point when immune reactions against the Vibrio infection would have been best mirrored in the gene expression patterns but rather the timepoint when inflammation or stress responses induced by the injection were strongest. Bearing these limitations in mind, in combination with the higher survival of juveniles originating from low salinity parents, we suggest that the increased virulence of the V. alginolyticus strain is mainly driven by trade-offs impairing the pipefish’s immune system. A deficiency that can potentially be overcome by local adaptation.
The patterns identified here have to be interpreted with care. Due to the unintended brood pouch infection that negatively affected 47% of the pregnant males originating from high saline conditions and parentally acclimated at low saline conditions, we are dealing with distinct selection intensities on the different treatment groups (Roth et al., 2018). In the treatment affected by the brood pouch fungus (Origin Salinity: H, Acclimation salinity: L) multiple clutches have at least been partially lost and potentially all infected fathers were suffering stress levels that can seriously confound the results from this study. The brood pouch fungus has severely impacted offspring development such that only the strongest will have survived. Addressing life history traits in the offspring and their gene expression will thus only be done in the strongest animals within the HL group, which does not resemble the original cohort, and makes interpretation of the data in the offspring generation difficult. We are aware of this limitation and have been taking this into account when interpreting our data.
Conclusion
After the last glacial maximum, broad-nosed pipefish have successfully populated the low salinity areas of the Baltic Sea. The results of our study suggest that the components of this success story are a mixture of genetic adaptation and the maintenance of a high degree of phenotypic plasticity of locally adapted pipefish enabling them to deal with present and ancestral salinity levels and re-occurring salinity fluctuations. Future genome scans will give closer insights into local adaptation of the pipefish to the salinity gradient of the Baltic Sea. Evidence for adaptive trans-generational effects were limited to expression changes in adaptive immune genes. Pipefish individuals with suitable alleles for low salinity conditions can inhabit low saline environments. The adaptation and adjustment of life-history strategies to lower salinity also enable pipefish to cope with prevailing pathogens such as Vibrio bacteria or aquatic fungi. Pipefish of the species S. typhle inhabiting the Baltic Sea are thus expected to genetically adapt to predicted further drops in salinity and retain phenotypic plasticity to cope with temporarily varying salinity levels. The findings are underlined by the fact that S. typhle also inhabit areas in the northern part of the Baltic Sea with salinity levels below 5 PSU.
Data Availability Statement
The datasets presented in this study can be found in the online repository PANGAEA: https://doi.pangaea.de/10.1594/PANGAEA.926923.
Ethics Statement
The animal study was reviewed and approved by the Experimental work was conducted in agreement with the German animal welfare law and approved by the Ministerium für Energiewende, Landwirtschaft, Umwelt, Natur und Digitalisierung under permission MELUR V 312–7224.121-19 (67–5/13), “komparative Vergleichsstudie von Immunantwort-Transfer von Eltern zu Nachkommen in Fischarten mit extremer Brutpflege”). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
OR and HG designed the study with input from CW. HG, K-SW, and OR collected fish in the field, conducted the experiment, and sampled the fish. LS designed the primers. LS, HG, and K-SW did the molecular lab work. HG, LS, OR, and K-SW analyzed the data. HG, LS, and OR wrote the manuscript with input from all authors. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by a DFG grant (WE 5822/1-1) within the priority program SPP1819 given to CW and OR, and a DFG grant (RO4628/3-1) to OR. Furthermore, this study was supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (Grant agreement No: 755659 – acronym: MALEPREG). HG received career and financial support from the International Max Planck Research School for Evolutionary Biology (IMPRS EvolBio).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the help of Kristina Dauven, Andreas Ebner, Janina Röckner, and Paulina Urban for fish collection in the field and fish maintenance. Furthermore, we thank Fabian Wendt for setting up the aquaria system and Tatjana Liese, Paulina Urban, Jakob Gismann, and Thorsten Reusch for support with DNA extraction and analysis of pipefish population structure. The authors acknowledge support of Isabel Tanger, Agnes Piecyk, Jonas Müller, Grace Walls, Sebastian Albrecht, Julia Böge, and Julia Stefanschitz for their support in preparing cDNA and running of Fluidigm chips. A special thank goes to Diana Gill for general lab support, ordering materials and just being the good spirit of our molecular lab, to Till Bayer for bioinformatics support and to Melanie Heckwolf for fruitful discussion and feedback on the manuscript. HG is very grateful for inspirational office space with ocean view provided by Lisa Hentschel and family. This manuscript has been released as a pre-print at BIORXIV https://biorxiv.org/cgi/content/short/2020.11.12.379305v1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.626442/full#supplementary-material
References
Andersson, A., Meier, H. E. M., Ripszam, M., Rowe, O., Wikner, J., Haglund, P., et al. (2015). Projected future climate change and Baltic Sea ecosystem management. Ambio 44, 345–356. doi: 10.1007/s13280-015-0654-8
Angers, B., Castonguay, E., and Massicotte, R. (2010). Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 19, 1283–1295. doi: 10.1111/j.1365-294x.2010.04580.x
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N., and Martinez-Urtaza, J. (2017). Non-cholera vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84. doi: 10.1016/j.tim.2016.09.008
Barrett, R. D. H., and Schluter, D. (2008). Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44. doi: 10.1016/j.tree.2007.09.008
Beemelmanns, A., and Roth, O. (2016a). Bacteria-type-specific biparental immune priming in the pipefish Syngnathus typhle. Ecol. Evol. 6, 6735–6757. doi: 10.1002/ece3.2391
Beemelmanns, A., and Roth, O. (2016b). Biparental immune priming in the pipefish Syngnathus typhle. Zoology 119, 262–272. doi: 10.1016/j.zool.2016.06.002
Beemelmanns, A., and Roth, O. (2017). Grandparental immune priming in the pipefish Syngnathus typhle. BMC Evol. Biol. 17:44. doi: 10.1186/s12862-017-0885-3
Belkhir, K. (2004). Genetix 4.05, logiciel sous Windows TM pour la Genetique des Populations. Available online at: http://www.genetix.univ-montp2.fr/genetix/genetix.htm (accessed June 22, 2018).
Berg, P. R., Jentoft, S., Star, B., Ring, K. H., Knutsen, H., Lien, S., et al. (2015). Adaptation to low salinity promotes genomic divergence in atlantic cod (Gadus morhua L.). Genome Biol. Evol. 7, 1644–1663. doi: 10.1093/gbe/evv093
Berglund, A., Sundin, J., and Rosenqvist, G. (2017). Baltic pipefish females need twice as many males as they get. Behav. Ecol. 28, 827–832. doi: 10.1093/beheco/arx046
Birrer, S. C., Reusch, T. B. H., and Roth, O. (2012). Salinity change impairs pipefish immune defence. Fish Shellfish Immunol. 33, 1238–1248. doi: 10.1016/j.fsi.2012.08.028
Björck, S. (1995). A review of the history of the Baltic Sea, 13.0-8.0 ka BP. Quat. Int. 27, 19–40. doi: 10.1016/1040-6182(94)00057-c
Blanquart, F., and Gandon, S. (2013). Time-shift experiments and patterns of adaptation across time and space. Ecol. Lett. 16, 31–38. doi: 10.1111/ele.12007
Blois, J. L., Williams, J. W., Fitzpatrick, M. C., Jackson, S. T., and Ferrier, S. (2013). Space can substitute for time in predicting climate-change effects on biodiversity. Proc. Natl. Acad. Sci. U.S.A. 110, 9374–9379. doi: 10.1073/pnas.1220228110
Bock, G., and Lieberum, C. (2017). Neobiota in Ausgewählten Häfen der Schleswig-Holsteinischen Ostsee, Kiel: Landesamtes für Landwirtschaft, Umwelt und ländliche Räume des Landes Schleswig-Holstein. doi: 10.1073/pnas.1220228110
Boeuf, G., and Payan, P. (2001). How should salinity influence fish growth? Comp. Biochem. Physiol. C Toxicol. Pharmacol. 130, 411–423. doi: 10.1016/s1532-0456(01)00268-x
Bookout, A. L., and Mangelsdorf, D. J. (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nuclear Recept. Signal. 1:e012.
Boyett, H. V., Bourne, D. G., and Willis, B. L. (2007). Elevated temperature and light enhance progression and spread of black band disease on staghorn corals of the Great Barrier Reef. Mar. Biol. 151, 1711–1720. doi: 10.1007/s00227-006-0603-y
Breves, J. P., McCormick, S. D., and Karlstrom, R. O. (2014). Prolactin and teleost ionocytes: new insights into cellular and molecular targets of prolactin in vertebrate epithelia. Gen. Comp. Endocrinol. 203, 21–28. doi: 10.1016/j.ygcen.2013.12.014
Brierley, A. S., and Kingsford, M. J. (2009). Impacts of climate change on marine organisms and ecosystems. Curr. Biol. 19, 602–614.
Brown, S. P., Cornforth, D. M., and Mideo, N. (2012). Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 20, 336–342. doi: 10.1016/j.tim.2012.04.005
Brunner, F. S., and Eizaguirre, C. (2016). Can environmental change affect host/parasite-mediated speciation? Zoology 119, 384–394. doi: 10.1016/j.zool.2016.04.001
Chatterjee, K., Pavlogiannis, A., Adlam, B., and Nowak, M. A. (2014). The time scale of evolutionary innovation. PLoS Comput. Biol. 10:e1003818. doi: 10.1371/journal.pcbi.1003818
Chen, M. X., Li, H. Y., Li, G., and Zheng, T. L. (2011). Distribution of Vibrio alginolyticus-like species in shenzhen coastal waters, China. Braz. J. Microbiol. 42, 884–896. doi: 10.1590/s1517-83822011000300007
Chevin, L. M., Lande, R., and Mace, G. M. (2010). Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biology 8: e1000357. doi: 10.1371/journal.pbio.1000357
Chibani, C. M., Hertel, R., Hoppert, M., Liesegang, H., and Wendling, C. C. (2020a). Closely related vibrio alginolyticus strains encode an identical repertoire of caudovirales-like regions and filamentous phages. Viruses 12:1359. doi: 10.3390/v12121359
Chibani, C. M., Roth, O., Liesegang, H., and Wendling, C. C. (2020b). Genomic variation among closely related Vibrio alginolyticus strains is located on mobile genetic elements. BMC Genomics 21:354. doi: 10.1186/s12864-020-6735-5
Dayma, P. I., Raval, H., Joshi, N., Patel, N. P., Haldar, S., and Mody, K. H. (2015). Influence of low salinity stress on virulence and biofilm formation potential in Vibrio alginolyticus, isolated from the Gulf of Khambhat, Gujarat India. Aquat. Living Resour. 28, 99–109. doi: 10.1051/alr/2016004
DeFaveri, J., and Merila, J. (2014). Local adaptation to salinity in the three-spined stickleback? J. Evol. Biol. 27, 290–302. doi: 10.1111/jeb.12289
DeWitt, T. J., Sih, A., and Wilson, D. S. (1998). Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 13, 77–81. doi: 10.1016/s0169-5347(97)01274-3
Donelson, J. M., Sunday, J. M., Figueira, W. F., Gaitan-Espitia, J. D., Hobday, A. J., Johnson, C. R., et al. (2019). Understanding interactions between plasticity, adaptation and range shifts in response to marine environmental change. Philos. Trans. R. Soc. B Biol. Sci. 374, 14.
Dray, S., and Dufour, A.-B. (2007). The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22:15378.
Dutil, J. D., Lambert, Y., and Boucher, E. (1997). Does higher growth rate in Atlantic cod (Gadus morhua) at low salinity result from lower standard metabolic rate or increased protein digestibility? Can. J. Fish. Aquat. Sci. 54, 99–103. doi: 10.1139/f96-148
Eizaguirre, C., Lenz, T. L., Kalbe, M., and Milinski, M. (2012). Rapid and adaptive evolution of MHC genes under parasite selection in experimental vertebrate populations. Nat. Commun. 3:621.
Eriksen, M. S., Bakken, M., Espmark, A., Braastad, B. O., and Salte, R. (2006). Prespawning stress in farmed Atlantic salmon Salmo salar: maternal cortisol exposure and hyperthermia during embryonic development affect offspring survival, growth and incidence of malformations. J. Fish Biol. 69, 114–129. doi: 10.1111/j.1095-8649.2006.01071.x
Fraser, H. B. (2013). Gene expression drives local adaptation in humans. Genome Res. 23, 1089–1096. doi: 10.1101/gr.152710.112
Gibson, J. R., and Najjar, R. G. (2000). The response of Chesapeake Bay salinity to climate-induced changes in streamflow. Limnol. Oceanogr. 45, 1764–1772. doi: 10.4319/lo.2000.45.8.1764
Gienapp, P., Teplitsky, C., Alho, J. S., Mills, J. A., and Merila, J. (2008). Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 17, 167–178. doi: 10.1111/j.1365-294x.2007.03413.x
Guo, B. C., DeFaveri, J., Sotelo, G., Nair, A., and Merila, J. (2015). Population genomic evidence for adaptive differentiation in Baltic Sea three-spined sticklebacks. BMC Biol. 13:19. doi: 10.1186/s12915-015-0130-8
Guo, B. C., Li, Z. T., and Merila, J. (2016). Population genomic evidence for adaptive differentiation in the Baltic Sea herring. Mol. Ecol. 25, 2833–2852. doi: 10.1111/mec.13657
Gurkan, S., and Taskavak, E. (2007). Length-weigth relationships for syngnathid fishes of the Aegean Sea, Turkey. Belgian J. Zool. 137, 219–222.
Haddy, J. A., and Pankhurst, N. W. (2000). The effects of salinity on reproductive development, plasma steroid levels, fertilisation and egg survival in black bream Acanthopagrus butcheri. Aquaculture 188, 115–131. doi: 10.1016/s0044-8486(00)00326-4
Hall, M. D., Vettiger, A., and Ebert, D. (2013). Interactions between environmental stressors: the influence of salinity on host-parasite interactions between Daphnia magna and Pasteuria ramosa. Oecologia 171, 789–796. doi: 10.1007/s00442-012-2452-3
Hasan, M. M., DeFaveri, J., Kuure, S., Dash, S. N., Lehtonen, S., Merila, J., et al. (2017). Sticklebacks adapted to divergent osmotic environments show differences in plasticity for kidney morphology and candidate gene expression. J. Exp. Biol. 220, 2175–2186. doi: 10.1242/jeb.146027
Hase, C. C., and Barquera, B. (2001). Role of sodium bioenergetics in Vibrio cholerae. Biochim. Biophys. Acta Bioenerg. 1505, 169–178. doi: 10.1016/s0005-2728(00)00286-3
Heckwolf, M. J., Meyer, B. S., Doring, T., Eizaguirre, C., and Reusch, T. B. H. (2018). Transgenerational plasticity and selection shape the adaptive potential of sticklebacks to salinity change. Evol. Appl. 11, 1873–1885. doi: 10.1111/eva.12688
Heckwolf, M. J., Meyer, B. S., Hasler, R., Hoppner, M. P., Eizaguirre, C., and Reusch, T. B. H. (2020). Two different epigenetic information channels in wild three-spined sticklebacks are involved in salinity adaptation. Sci. Adv. 6:13.
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:19.
Hinrichs, I., Jahnke-Bornemann, A., Andersson, A., Ganske, A., Gouretski, V., Jensen, C., et al. (2019). The Baltic and North Seas Climatology (BNSC) – a comprehensive, observation-based data product of atmospheric and hydrographic parameters. Front. Earth Sci. 7:158. doi: 10.3389/feart.2019.00158
Holmborn, T., Goetze, E., Pollupuu, M., and Pollumae, A. (2011). Genetic species identification and low genetic diversity in Pseudocalanus acuspes of the Baltic Sea. J. Plankton Res. 33, 507–515. doi: 10.1093/plankt/fbq113
Hothorn, T., Bretz, F., Westfall, P., Heiberger, R. M., Schetzenmeister, A., and Scheibe, S. (2020). multcomp: Simultaneous Inference in General Parametric Models. Available online at: https://cran.r-project.org/web/packages/multcomp/index.html (accessed June 29, 2020).
Hulce, D., Li, X., Snyder-Leiby, T., and Johathan Liu, C. S. (2011). GeneMarker® genotyping software: tools to increase the statistical power of DNA fragment analysis. J. Biomol. Tech. 22, S35–S36.
Ilyina, T. S. (2015). Filamentous bacteriophages and their role in the virulence and evolution of pathogenic bacteria. Mol. Genet. Microbiol. Virol. 30, 1–9. doi: 10.3103/s0891416815010036
Imsland, A. K., Foss, A., Gunnarsson, S., Berntssen, M. H. G., FitzGerald, R., Bonga, S. W., et al. (2001). The interaction of temperature and salinity on growth and food conversion in juvenile turbot (Scophthalmus maximus). Aquaculture 198, 353–367. doi: 10.1016/s0044-8486(01)00507-5
Janeway, C. (2005). Immunobiology: The Immune System in Health And Disease. New York, NY: Garland Science.
Janssen, F., Schrum, C., and Backhaus, J. O. (1999). A climatological data set of temperature and salinity for the Baltic Sea and the North Sea. Deutsche Jydrpgrafische Zeitschrift 51:5. doi: 10.1007/bf02933676
Johannesson, K., and Andre, C. (2006). Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol. Ecol. 15, 2013–2029. doi: 10.1111/j.1365-294x.2006.02919.x
Johannesson, K., Le Moan, A., Perini, S., and André, C. (2020). A Darwinian laboratory of multiple contact zones. Trends Ecol. Evol. 35, 1021–1036. doi: 10.1016/j.tree.2020.07.015
Jones, A. G., Rosenqvist, G., Berglund, A., and Avise, J. C. (1999). The genetic mating system of a sex-role-reversed pipefish (Syngnathus typhle): a molecular inquiry. Behav. Ecol. Sociobiol. 46, 357–365. doi: 10.1007/s002650050630
Kelly, M. (2019). Adaptation to climate change through genetic accommodation and assimilation of plastic phenotypes. Philos. Trans. R. Soc. B Biol. Sci. 374:10.
Kim, L. H., and Chong, T. H. (2017). Physiological responses of salinity-stressed Vibrio sp and the effect on the biofilm formation on a nanofiltration membrane. Environ. Sci. Technol. 51, 1249–1258. doi: 10.1021/acs.est.6b02904
Kniebusch, M., Meier, H. E. M., and Radtke, H. (2019). Changing salinity gradients in the Baltic Sea as a consequence of altered freshwater budgets. Geophys. Res. Lett. 46, 9739–9747. doi: 10.1029/2019gl083902
Koressaar, T., and Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kutzer, M. A. M., and Armitage, S. A. O. (2016). Maximising fitness in the face of parasites: a review of host tolerance. Zoology 119, 281–289. doi: 10.1016/j.zool.2016.05.011
Laland, K., Uller, T., Feldman, M., Sterelny, K., Muller, G. B., Moczek, A., et al. (2014). Does evolutionary theory need a rethink? - POINT Yes, urgently. Nature 514, 161–164. doi: 10.1038/514161a
Larsen, P. F., Schulte, P. M., and Nielsen, E. E. (2011). Gene expression analysis for the identification of selection and local adaptation in fishes. J. Fish Biol. 78, 1–22. doi: 10.1111/j.1095-8649.2010.02834.x
Leder, E. H., Andr, C., Le Moan, A., Topel, M., Blomberg, A., Havenhand, J. N., et al. (2021). Post-glacial establishment of locally adapted fish populations over a steep salinity gradient. J. Evol. Biol. 34, 138–156.
Lehtonen, T. K., and Kvarnemo, C. (2015). Infections may select for filial cannibalism by impacting egg survival in interactions with water salinity and egg density. Oecologia 178, 673–683. doi: 10.1007/s00442-015-3246-1
Lind, M. I., Zwoinska, M. K., Andersson, J., Carlsson, H., Krieg, T., Larva, T., et al. (2020). Environmental variation mediates the evolution of anticipatory parental effects. Evol. Lett. 4, 371–381. doi: 10.1002/evl3.177
Loder, J. W., van der Baaren, A., and Yashayaev, I. (2015). Climate comparisons and change projections for the Northwest Atlantic from six CMIP5 models. Atmosphere Ocean 53, 529–555. doi: 10.1080/07055900.2015.1087836
Manzon, L. A. (2002). The role of prolactin in fish osmoregulation: a review. Gen. Comp. Endocrinol. 125, 291–310. doi: 10.1006/gcen.2001.7746
Marshall, D. J. (2008). Transgenerational plasticity in the sea: context-dependent maternal effects across the life history. Ecology 89, 418–427. doi: 10.1890/07-0449.1
McCormick, S. D. (2001). Endocrine control of osmoregulation in teleost fish. Am. Zool. 41, 781–794. doi: 10.1093/icb/41.4.781
McGuigan, K., Nishimura, N., Currey, M., Hurwit, D., and Cresko, W. A. (2011). Cryptic genetic variation and body size evolution in threespine stickleback. Evolution 65, 1203–1211. doi: 10.1111/j.1558-5646.2010.01195.x
Meier, H. E. M., Kjellstrom, E., and Graham, L. P. (2006). Estimating uncertainties of projected Baltic Sea salinity in the late 21st century. Geophys. Res. Lett. 33:4.
Molnar, P. K., Kutz, S. J., Hoar, B. M., and Dobson, A. P. (2013). Metabolic approaches to understanding climate change impacts on seasonal host-macroparasite dynamics. Ecol. Lett. 16, 9–21. doi: 10.1111/ele.12022
Morgan, J. D., and Iwama, G. K. (1991). Effects of salinity on growth, metabolism, and ion regulation in juvenile rainbow and steelhead trout (Oncorhynchus mykiss) and fall chinook salmon (Oncorhynchus tshawytscha). Can. J. Fish. Aquat. Sci. 48, 2083–2094. doi: 10.1139/f91-247
Mostowy, R., and Engelstadter, J. (2011). The impact of environmental change on host-parasite coevolutionary dynamics. Proc. R. Soc. B Biol. Sci. 278, 2283–2292. doi: 10.1098/rspb.2010.2359
Mück, I., and Heubel, K. U. (2018). Ecological variation along the salinity gradient in the Baltic Sea Area and its consequences for reproduction in the common goby. Curr. Zool. 64, 259–270. doi: 10.1093/cz/zoy006
Murray, C. S., Malvezzi, A., Gobler, C. J., and Baumann, H. (2014). Offspring sensitivity to ocean acidification changes seasonally in a coastal marine fish. Mar. Ecol. Prog. Series 504, 1–11. doi: 10.3354/meps10791
Nissling, A., Johansson, U., and Jacobsson, M. (2006). Effects of salinity and temperature conditions on the reproductive success of turbot (Scophthalmus maximus) in the Baltic Sea. Fish. Res. 80, 230–238. doi: 10.1016/j.fishres.2006.04.005
Nissling, A., Nyberg, S., and Petereit, C. (2017). Egg buoyancy of flounder, Platichthys flesus, in the Baltic Sea–adaptation to salinity and implications for egg survival. Fish. Res. 191, 179–189. doi: 10.1016/j.fishres.2017.02.020
Nissling, A., and Westin, L. (1997). Salinity requirements for successful spawning of Baltic and Belt Sea cod and the potential for cod stock interactions in the Baltic Sea. Mar. Ecol. Prog. Ser. 152, 261–271. doi: 10.3354/meps152261
Nygård, M., Kvarnemo, C., Ahnesjö, I., and Braga Goncalves, I. (2019). Pipefish embryo oxygenation, survival, and development: egg size, male size, and temperature effects. Behav. Ecol. 30, 1451–1460. doi: 10.1093/beheco/arz101
Oberbeckmann, S., Wichels, A., Wiltshire, K. H., and Gerdts, G. (2011). Occurrence of Vibrio parahaemolyticus and Vibrio alginolyticus in the German Bight over a seasonal cycle. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 100, 291–307. doi: 10.1007/s10482-011-9586-x
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2019). vegan: Community Ecology Package. R packae version 2.5-4.
Oosterhout, C. V., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Partridge, C., Shardo, J., and Boettcher, A. (2007). Osmoregulatory role of the brood pouch in the euryhaline Gulf pipefish, Syngnathus scovelli. Comp. Biochem. Physiol. 147, 556–561. doi: 10.1016/j.cbpa.2007.02.007
Poirier, M., Listmann, L., and Roth, O. (2017). Selection by higher-order effects of salinity and bacteria on early life-stages of Western Baltic spring-spawning herring. Evol. Appl. 10, 603–615. doi: 10.1111/eva.12477
Poloczanska, E. S., Brown, C. J., Sydeman, W. J., Kiessling, W., Schoeman, D. S., Moore, P. J., et al. (2013). Global imprint of climate change on marine life. Nat. Clim. Change 3, 919–925.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using Multilocus genotype data. Genetics 155, 945.
Prunet, P., Avelia, M., Föstier, A., Björnsson, B. T., Boeuf, G., and Haux, C. (1990). Roles of prolactin in salmonids. Prog. Comp. Endocrinol. 342, 547–552.
Quast, W. D., and Howe, N. R. (1980). The osmotic role of the brood pouch in the pipefish Syngnathus scovelli. Comp. Biochem. Physiol. Physiol. 67, 675–678. doi: 10.1016/0300-9629(80)90259-5
RCoreTeam (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reed, T. E., Waples, R. S., Schindler, D. E., Hard, J. J., and Kinnison, M. T. (2010). Phenotypic plasticity and population viability: the importance of environmental predictability. Proc. R. Soc. B Biol. Sci. 277, 3391–3400. doi: 10.1098/rspb.2010.0771
Reusch, T. B. H., Dierking, J., Andersson, H. C., Bonsdorff, E., Carstensen, J., Casini, M., et al. (2018). The Baltic Sea as a time machine for the future coastal ocean. Sci. Adv. 4:eaar8195.
Rispoli, V. F., and Wilson, A. B. (2008). Sexual size dimorphism predicts the frequency of multiple mating in the sex-role reversed pipefish Syngnathus typhle. J. Evol. Biol. 21, 30–38. doi: 10.1111/j.1420-9101.2007.01470.x
Rolfe, D. F. S., and Brown, G. C. (1997). Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 77, 731–758. doi: 10.1152/physrev.1997.77.3.731
Roth, O., Beemelmanns, A., Barribeau, S. M., and Sadd, B. M. (2018). Recent advances in vertebrate and invertebrate transgenerational immunity in the light of ecology and evolution. Heredity 121, 225–238. doi: 10.1038/s41437-018-0101-2
Roth, O., Keller, I., Landis, S. H., Salzburger, W., and Reusch, T. B. H. (2012a). Host are ahead in a marine host-parasite coevolutionary arms race: innate ummune system adapatation in pipefish Syngnathus typhle agains Vibrio phylotypes. Evolution 66, 2528–2539. doi: 10.1111/j.1558-5646.2012.01614.x
Roth, O., Klein, V., Beemelmanns, A., Scharsack, J. P., and Reusch, T. B. H. (2012b). Male pregnancy and biparental immune priming. Am. Nat. 180, 802–814. doi: 10.1086/668081
Roth, O., and Landis, S. H. (2017). Trans-generational plasticity in response to immune challenge is constrained by heat stress. Evol. Appl. 10, 514–528. doi: 10.1111/eva.12473
Schaarschmidt, T., Meyer, E., and Jurss, K. (1999). A comparison of transport-related gill enzyme activities and tissue-specific free amino acid concentrates of Baltic Sea (brackish water) and freshwater threespine sticklebacks, Gasterosteus aculeatus, after salinity and temperature acclimation. Mar. Biol. 135, 689–697. doi: 10.1007/s002270050670
Schneider, R. F., and Meyer, A. (2017). How plasticity, genetic assimilation and cryptic genetic variation may contribute to adaptive radiations. Mol. Ecol. 26, 330–350. doi: 10.1111/mec.13880
Shama, L. N. S., and Wegner, K. M. (2014). Grandparental effects in marine sticklebacks: transgenerational plasticity across multiple generations. J. Evol. Biol. 27, 2297–2307. doi: 10.1111/jeb.12490
Stockwell, C. A., Purcell, K. M., Collyer, M. L., and Janovy, J. (2011). Effects of salinity on Physa acuta, the intermediate host for the parasite Posthodiplostomum minimum: implications for the translocation of the protected white sands pupfish. Trans. Am. Fish. Soc. 140, 1370–1374. doi: 10.1080/00028487.2011.620499
Sullivan, T. J., and Neigel, J. E. (2018). Effects of temperature and salinity on prevalence and intensity of infection of blue crabs, Callinectes sapidus, by Vibrio cholerae, V. parahaemolyticus, and V. vulnificus in Louisiana. J. Invertebr. Pathol. 151, 82–90. doi: 10.1016/j.jip.2017.11.004
Sunde, J., Yildirim, Y., Tibblin, P., and Forsman, A. (2020). Comparing the performance of microsatellites and RADseq in population genetic studies: analysis of data for pike (Esox lucius) and a synthesis of previous studies. Front. Genet. 11:218. doi: 10.3389/fgene.2020.00218
Sunday, J. M., Calosi, P., Dupont, S., Munday, P. L., Stillman, J. H., and Reusch, T. B. H. (2014). Evolution in an acidifying ocean. Trends Ecol. Evol. 29, 117–125. doi: 10.1016/j.tree.2013.11.001
Swaggerty, C. L. I, Pevzner, Y., Kaiser, P., and Kogut, M. H. (2008). Profiling pro-inflammatory cytokine and chemokine mRNA expression levels as a novel method for selection of increased innate immune responsiveness. Vet. Immunol. Immunopathol. 126, 35–42. doi: 10.1016/j.vetimm.2008.06.005
Tomy, S., Chang, Y. M., Chen, Y. H., Cao, J. C., Wang, T. P., and Chang, C. F. (2009). Salinity effects on the expression of osmoregulatory genes in the euryhaline black porgy Acanthopagrus schlegeli. Gen. Comp. Endocrinol. 161, 123–132. doi: 10.1016/j.ygcen.2008.12.003
Torda, G., Donelson, J. M., Aranda, M., Barshis, D. J., Bay, L., Berumen, M. L., et al. (2017). Rapid adaptive responses to climate change in corals. Nat. Clim. Change 7, 627–636. doi: 10.1038/nclimate3374
Uller, T., Nakagawa, S., and English, S. (2013). Weak evidence for anticipatory parental effects in plants and animals. J. Evol. Biol. 26, 2161–2170. doi: 10.1111/jeb.12212
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Urban, M. C. (2015). Accelerating extinction risk from climate change. Science 348, 571–573. doi: 10.1126/science.aaa4984
Ustups, D., Müller-Karulis, B., Bergstrom, U., Makarchouk, A., and Sics, I. (2013). The influence of environmental conditions on early life stages of flounder (Platichthys flesus) in the central Baltic Sea. J. Sea Res. 75, 77–84. doi: 10.1016/j.seares.2012.05.001
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:12.
Velasco, J., Gutierrez-Canovas, C., Botella-Cruz, M., Sanchez-Fernandez, D., Arribas, P., Carbonell, J. A., et al. (2019). Effects of salinity changes on aquatic organisms in a multiple stressor context. Philos. Trans. R. Soc. B Biol. Sci. 374:9.
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Wendling, C. C., Piecyk, A., Refardt, D., Chibani, C., Hertel, R., Liesegang, H., et al. (2017). Tripartite species interaction: eukaryotic hosts suffer more from phage susceptible than from phage resistant bacteria. BMC Evolutionary Biology 17:98. doi: 10.1186/s12862-017-0930-2
West-Eberhard, M. J. (1989). Phenotypic plasticity and the origins of diversity. Annu. Rev. Ecol. Syst. 20, 249–278.
Wilson, A. B., and Veraguth, I. E. (2010). The impact of Pleistocene glaciation across the range of a widespread European coastal species. Mol. Ecol. 19, 4535–4553. doi: 10.1111/j.1365-294x.2010.04811.x
Wilson, A. B., Wegmann, A., Ahnesjo, I., and Goncalves, J. M. S. (2020). The evolution of ecological specialization across the range of a broadly distributed marine species. Evolution 74, 629–643. doi: 10.1111/evo.13930
Keywords: trans-generational plasticity, genetic adaptation, local adaptation, phenotypic plasticity, Baltic Sea, climate change, salinity, Syngnathids
Citation: Goehlich H, Sartoris L, Wagner K-S, Wendling CC and Roth O (2021) Pipefish Locally Adapted to Low Salinity in the Baltic Sea Retain Phenotypic Plasticity to Cope With Ancestral Salinity Levels. Front. Ecol. Evol. 9:626442. doi: 10.3389/fevo.2021.626442
Received: 05 November 2020; Accepted: 05 February 2021;
Published: 25 March 2021.
Edited by:
Hope Klug, University of Tennessee at Chattanooga, United StatesCopyright © 2021 Goehlich, Sartoris, Wagner, Wendling and Roth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henry Goehlich, aGdvZWhsaWNoQGdlb21hci5kZQ==; aGVucnlnb2VobGljaEBnbXguZGU=
†Present address: Linda Sartoris, Social Immunity, Institute of Science and Technology Austria (IST Austria), Klosterneuburg, Austria; Carolin C. Wendling, Institute of Integrative Biology, ETH Zürich, Zurich, Switzerland
 Henry Goehlich
Henry Goehlich Linda Sartoris
Linda Sartoris Kim-Sara Wagner
Kim-Sara Wagner Carolin C. Wendling
Carolin C. Wendling Olivia Roth
Olivia Roth