
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 29 March 2021
Sec. Coevolution
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.618331
This article is part of the Research TopicSymbiotic Relationships as Shapers of BiodiversityView all 14 articles
In the last decade, the influence of microbial symbionts on ecological and physiological traits of their hosts has been increasingly recognized. However, most of these effects have been revealed under laboratory conditions, which oversimplifies the complexity of the factors involved in the dynamics of symbiotic associations in nature. The pea aphid, Acyrthosiphon pisum, forms a complex of plant-adapted biotypes, which strongly differ in the prevalence of their facultative endosymbionts. Some of the facultative endosymbionts of A. pisum have been shown to confer protection against natural enemies, among which Hamiltonella defensa is known to protect its host from parasitoid wasps. Here, we tested under natural conditions whether the endosymbiont communities of different A. pisum biotypes had a protective effect on their hosts and whether endosymbiotic associations and parasitoid communities associated with the pea aphid complex were linked. A space-time monitoring of symbiotic associations, parasitoid pressure and parasitoid communities was carried out in three A. pisum biotypes respectively specialized on Medicago sativa (alfalfa), Pisum sativum (pea), and Trifolium sp. (clover) throughout the whole cropping season. While symbiotic associations, and to a lesser extent, parasitoid communities were stable over time and structured mainly by the A. pisum biotypes, the parasitoid pressure strongly varied during the season and differed among the three biotypes. This suggests a limited influence of parasitoid pressure on the dynamics of facultative endosymbionts at a seasonal scale. However, we found a positive correlation between the α and β diversities of the endosymbiont and parasitoid communities, indicating interactions between these two guilds. Also, we revealed a negative correlation between the prevalence of H. defensa and Fukatsuia symbiotica in co-infection and the intensity of parasitoid pressure in the alfalfa biotype, confirming in field conditions the protective effect of this symbiotic combination.
Antagonistic interactions such as host–parasite or prey–predator are major drivers of ecological and evolutionary processes and affect more globally biodiversity patterns and ecosystem functioning. Antagonistic interactions usually involve arms race between protagonists whereby the enemy evolves new weapons to counteract victim’s defensive strategies (Abrams, 2000). Selection of these adaptive responses acts on variation encoded by enemy or victim’s genomes and relying on different mechanisms such as toxins released by pathogens into the infected host or immunity host defenses triggered upon parasite attacks. More recently, evidence accumulated that some microbial symbionts hosted by eukaryotes can also actively participate in antagonistic interactions by extending the arsenal of enemies or the defensive strategies of victims (Flórez et al., 2015).
Protective microbial symbionts have been particularly well-studied in insects with the best examples being the Spiroplasma bacterial endosymbiont which protects a mushroom-feeding fly Drosophila neotestacea from parasitic nematodes (Jaenike et al., 2010) or the Hamiltonella defensa bacterial endosymbiont which confers protection to aphids against hymenopteran parasitoids (Oliver et al., 2003). In both cases, the mechanisms of the symbiont-mediated protection involve the production of microbial toxins active against the parasite (Oliver and Perlman, 2020). The infection with these protective symbionts confers an immediate fitness advantage to the host when enemy pressure exists. However, hosting protective symbionts may come with some fitness costs (Oliver et al., 2008; Simon et al., 2011; Vorburger and Perlman, 2018). This cost/benefit balance has been repeatedly invoked to explain intermediate frequencies of protective symbionts in natural populations of their hosts (Oliver et al., 2014). In addition, these bacterial symbionts being transmitted occasionally through horizontal transfer events, they may rapidly spread across both host populations and species, leading to rapid adaptations and invasion processes (Jaenike et al., 2010; Himler et al., 2011). Finally, these protective symbionts may modify the diversity, structure and function of ecological networks that link their hosts to lower or higher trophic levels, through bottom-up and top-down effects (Mclean and Godfray, 2015; Rothacher et al., 2016; Ye et al., 2018; Mclean, 2019). Most studies on protective symbionts in insects have been carried out in laboratory conditions or in simplified field experiments. Although these studies have been crucial in investigating the costs and benefits in hosting protective symbionts in presence or absence of parasitism, only few assessed the response of host–symbiont associations to varying pressure intensities of natural enemies and by extent, the link between symbiont communities and parasitoid communities in real and complex natural environments (Smith et al., 2015; Ye et al., 2018).
The pea aphid Acyrthosiphon pisum is a good model to study how symbiotic associations are influenced by the parasitism exerted on their hosts and in return, how symbionts affect diversity and community structure of their host’s natural enemies. This sap-feeding insect feeds on legumes and forms at least 15 biotypes differentiated by host plant utilization (Peccoud et al., 2009, 2015; Ferrari et al., 2012). A large variation in symbiotic associations is found in natural populations of pea aphids with seven heritable facultative endosymbionts being hosted alone or in co-infections in addition to the primary symbiont Buchnera aphidicola (Gauthier et al., 2015; Guyomar et al., 2018). These endosymbionts can have different phenotypic effects on their hosts, including protection from natural enemies. For example, H. defensa, Fukatsuia symbiotica (also named PAXS for Pea Aphid X-type Symbiont), and Serratia symbiotica have been reported as protective against parasitoids (Oliver et al., 2003; Ferrari et al., 2004; Guay et al., 2009; Leclair et al., 2016), while Regiella insecticola, Rickettsiella viridis, Rickettsia sp., Spiroplasma sp., and F. symbiotica confer protection against fungal pathogens (Scarborough et al., 2005; Łukasik et al., 2013; Heyworth and Ferrari, 2015) and R. viridis against predators (Polin et al., 2015). Beyond the phenotype induced by one endosymbiont, other phenotypes are observed when multiple symbionts coexist in a single host (Leclair et al., 2017; Mclean et al., 2018). For example, multiple infections with H. defensa and F. symbiotica can give stronger protection against parasitoids than each endosymbiont alone (Guay et al., 2009; Heyworth and Ferrari, 2015; Leclair et al., 2016), although this may not be always the case (Doremus and Oliver, 2017). This variation in phenotypic effects induced by the different endosymbionts could result from strain differences as showed in earlier works (Oliver et al., 2003; Cayetano et al., 2015; Leclair et al., 2016).
Population cage experiments showed that pea aphids infected with H. defensa increased rapidly in frequency when exposed to Aphidius ervi, the main parasitoid of A. pisum, but declined when parasitoid pressure was removed (Oliver et al., 2008). Recent independent studies exposed laboratory clones carrying or not H. defensa to field conditions in order to analyze the temporal dynamics of protected and unprotected aphid clones when facing a wider range of natural enemies (Hrcek et al., 2016; Rothacher et al., 2016). While parasitism rates were lower in protected clones, this advantage did not translate into higher population growth rate. Overall, these results indicate that although symbiont protection is expressed in field conditions, it is mitigated by multiple factors that may include the rate of parasitism, the range of natural enemies and the costs entailed by harboring the protective symbionts. The work of Smith et al. (2015) was the first to analyze the dynamics of protective endosymbionts in relation with parasitism pressures in natural populations of the pea aphid. Important seasonal shifts in symbiont composition were found, possibly driven by changes in parasitism rates. In particular, the frequency of H. defensa was related with parasitoid pressure while the frequency of R. insecticola was linked with fungal-induced mortality. Additionally, the aforementioned studies highlighted the influence of protective endosymbionts on diversity and structure of parasitoid communities, with potential consequences on food web functioning (Mclean et al., 2016; Mclean, 2019). More recently, in an elegant experimental evolution study (Hafer-Hahmann and Vorburger, 2020), it was discovered that symbiont diversity could be driven by parasitoid diversity, suggesting potential reciprocal influence of these two communities through their shared aphid hosts.
In the present study, we tested under natural conditions whether the endosymbiont communities of different A. pisum biotypes had an actual protective effect on their hosts and whether endosymbiont communities and parasitoid communities associated with the pea aphid complex were linked. To this end, we measured parasitoid pressure on various pea aphid populations, examined whether facultative symbiotic associations responded to parasitoid pressures of varying intensity, and analyzed the relationships between symbiotic and parasitoid communities in terms of both α-diversity (i.e., the species diversity in a given sample) and β-diversity (i.e., the change in diversity of species from one sample to another). In essence, our study is similar to that of Smith et al. (2015) but differs in that we performed field surveys in the native range of A. pisum and its natural enemies [and not in the introduced range as in Smith et al. (2015)]. We therefore expect a wider diversity in aphid-endosymbiont–enemy interactions that may drive different patterns of protective symbiont dynamics. To address these issues, we carried out a field survey, spanning twelve dates across the growing season, and encompassing three biotypes of A. pisum, each specialized to distinct legume crops and harboring or not protective facultative endosymbionts. For each date and crop, we estimated parasitoid pressure exerted on pea aphids using two proxies, determined the structure of the parasitoid community, recorded the frequencies of each facultative endosymbiont in the pea aphid populations and their distribution among host individuals (i.e., symbiont communities), analyzed how the prevalence of symbiont communities influenced parasitism rate within aphid populations, and measured how diversities of both parasitoid and facultative symbiont communities were related. Finally, because parasitoids may also drive symbiont diversity at the population level (Hafer and Vorburger, 2019), we examined in a subsample of our aphid collections whether variation in the strain of the protective endosymbiont H. defensa was related to the intensity of parasitism pressures.
Pea aphid individuals were collected in crop fields of alfalfa (Medicago sativa), clover (Trifolium sp.), and pea (Pisum sativum) in Western France (25 km around Rennes, Brittany) between May and October 2014 during the parthenogenetic phase of the aphid life cycle. Locations of the sampled fields and their coding are given in Supplementary Figure 1 and Supplementary Table 1. When possible, three distant fields per legume species were sampled every two weeks starting on May 7 and ending on October 8, 2014, resulting in 12 sampling dates (Supplementary Table 2). As in Brittany pea is harvested by the end of July, no sample was available for this legume in August. By chance, some pea regrowth colonized by aphids were found at beginning of September and were sampled until October 8. While we aimed to monitor the same crop fields during our survey, some fields were sometimes not available because of either the absence of insects or the crop harvesting (see Supplementary Table 1 for details). From here, one sampling refers to an insects’ collection made at one date in one field of a given legume species. Overall, we made a total of 34, 31, and 19 samplings for alfalfa, clover, and pea, respectively. At each sampling, alive pea aphid adults and nymphs were collected using beat sampling from plants separated by approximately 5 m to cover a larger spatial/genetic diversity at the field scale. Aphid mummies (i.e., a dead aphid containing an immature parasitoid) were also collected by view in order to extract parasitoid specimens from field.
To estimate the parasitism pressure exerted on aphid populations, we used two different proxies. For the first one, we collected on average 80 adult aphids at each field sampling (see Supplementary Table 2 for a precise number of sampled adults per field) that were then installed by batch of ten on plants of broad bean (Vicia faba), which is used as a host plant by the different pea aphid biotypes (Peccoud and Simon, 2010), under laboratory conditions (20°C and 16hD:8hN regime). Ten days after the sampling, the rate of parasitism was assessed by calculating the ratio between the number of aphid mummies obtained and the total number of installed aphids for each field per date sample. Overall, 3,074 aphid adults from alfalfa, 1,830 from clover and 1,637 from pea were followed for their parasitism status. A second proxy of parasitism pressure was considered in order to reduce the potential estimate bias caused by aphid resistance against parasitoids. For this second proxy, we sampled the field canopy by using a leaf vacuum cleaner during five seconds at three different sites randomly chosen in a given field. The three canopy samples were kept in a plastic bag separately and stored at −20°C. In the laboratory, pea aphids (all instars combined) were counted and parasitoids were numbered and preserved in 70% ethanol for species identification. The second proxy of parasitism pressure consisted on calculating the ratio between the number of parasitoids belonging to the guild using pea aphids as hosts (Starý, 2006) and the number of pea aphids.
To estimate the diversity and structure of parasitoid communities, we considered parasitoid specimens emerging from pea aphid mummies collected from the fields or obtained in laboratory. Those parasitoid specimens were determined at species level (when possible) based on morphological criteria (Müller et al., 1999; Starý, 2006). The present study excluded all hyperparasitoids from the analyses (258 specimens were found). Overall, we determined 744 primary parasitoids from alfalfa, 794 from clover, and 373 from pea, respectively. From all these specimens, six parasitoid taxa were identified: A. ervi, Aphidius eadyi, Aphidius avenae, Praon volucre, Praon barbatum, and Aphelinus sp.
For each sampling, 60 pea aphid nymphs were collected in order to determine the symbiotic composition in the sampled aphid population. Young aphid nymphs (second-third instars) were chosen instead of older stages to reduce the bias induced by differential mortality caused by parasitoids. After sampling, the aphid nymphs were preserved in 95% ethanol. A random subsample of about 30 aphids per sampling was then inspected for their bacterial symbiont composition (see Supplementary Table 2 for precise number of aphids inspected per field). A total of 993 of aphids from alfalfa, 831 from clover and 509 from pea were screened for the presence of the seven facultative endosymbionts reported in the pea aphid: S. symbiotica, H. defensa, R. insecticola, F. symbiotica (or “Pea Aphid X-type Symbiont”), R. viridis, Rickettsia sp., and Spiroplasma sp. Their presence was detected using species-specific PCR primers according to Peccoud et al. (2014) and the detection of the obligatory bacterial endosymbiont B. aphidicola was used as a control for DNA extraction. Note that we also searched for Wolbachia because of some reports for the pea aphid in the literature (Russell et al., 2013; Wang et al., 2014; Gauthier et al., 2015) but we did not detect any positive aphids. As in the past, we have conducted extensive surveys of the microbial communities associated with the pea aphid, using either targeted or whole-genome metagenomics approaches (Gauthier et al., 2015; Guyomar et al., 2018), which have confirmed the presence of seven facultative endosymbionts considered in our paper, we are confident that we have surveyed the main fraction of the pea aphid symbionts. For each sampling, we thus estimated the prevalence of these seven facultative heritable endosymbionts and their distribution among aphid individuals. The endosymbiont infection status of each aphid individual (i.e., identity and number of endosymbionts) was then determined.
In order to monitor possible changes in H. defensa strain diversity throughout the cropping season that could result from a variation in the intensity of parasitoid pressure, a subsample of 72 pea aphid nymphs from the alfalfa biotype and infected with H. defensa singly (30 aphids) or in coinfection with F. symbiotica (42 aphids) was selected among the samplings done before (36 aphids) and after the peak (36 aphids) of parasitoids’ activity. All aphid individuals were genotyped at seven microsatellite markers following (Peccoud et al., 2008) in order to distinguish aphid clones based on their multilocus genotype. As the parasitism protection due to H. defensa infection is associated with the presence of a bacteriophage (APSE), which encodes toxins potentially responsible for parasitoid development arrest (Degnan and Moran, 2008), phage presence was checked by PCR using the primer pair P3 (forward) 5′-TCGGGCGTAGTGTTAATGAC-3′ (reverse) 5′-TTCCATAGCGGAATCAAAGG-3′ and P51 (forward) 5′-AG GTGCGATTACCCTGTTTG-3′ (reverse) 5′-GATAAAACATCG CCGTTTGC-3′ (Mclean and Godfray, 2015). A multilocus sequence-typing (MLST) was then performed for the characterization of H. defensa strains with partial DNA sequences of housekeeping genes accD and murE (Henry et al., 2013). Fragments were amplified by PCR using H. defensa-specific primers and cycling conditions described in Henry et al. (2013). Amplicons were sent to Genoscreen for Sanger sequencing. Sequences obtained were cleaned and aligned using Geneious® v.7.1.5 (Kearse et al., 2012). For each sample and each gene (accD and murE) sequences were used to build a phylogenetic tree using the Neighbor Joining method (Tamura-Nei distance). Bootstrap values were computed for each branch node (N = 1,000).
The first analysis consisted in assessing the quality of parasitism pressure estimates by calculating the Spearman correlation coefficient between the two proxies. Then, we analyzed the temporal dynamics of parasitoid activity in the three pea aphid biotypes. From the latter analysis, we found three distinct parasitism periods arbitrarily defined as “pre-parasitism,” “maximal parasitism,” and “post-parasitism.” To test whether the overall parasitism rate differed between the three biotypes, we used Generalized Linear Mixed Model (GLMM) (glmer function of the lme4 package (Bates et al., 2015) with a binomial error distribution (logit link function). Both the field ID and session number were fit as random factor to include data substructure.
For each combination of parasitism period and pea aphid biotype, the occurrence and co-occurrence of parasitoid species and facultative endosymbionts were visualized using the R package Mondrian (Siberchicot et al., 2016). For each combination biotype/session (i.e., we pooled the data obtained in the fields of a given legume crop during one sampling session), the α-diversity of parasitoid communities was estimated using the Shannon Index that accounts for both abundance and evenness of the species. Also, we calculated the α-diversity of the aphid endosymbiont infection statuses using the Shannon Index by considering the relative abundance of each infection status found in a given biotype/session combination. By using a General Linear Models, we tested whether the parasitism period (i.e., three levels factor: “pre-parasitism,” “maximal parasitism,” and “post-parasitism”) or the pea aphid biotype (i.e., three levels factor: “alfalfa,” “clover,” and “pea” biotypes) affected the Shannon index of parasitoid communities or endosymbiont infection statuses of aphids. Finally, to test whether α-diversities of both parasitoid communities and aphid endosymbiont infection statuses were linked, a Pearson correlation coefficient between their Shannon indexes was calculated.
We quantified the dissimilarity between the parasitoid communities or the endosymbiont infection statuses of aphids (β-diversity) between all pea aphid samples using the Bray–Curtis distance. The Bray–Curtis dissimilarities between all pairwise combinations of pea aphid samples were calculated, ordinated following a non-metric multidimensional scaling (nMDS) and represented on a scatter graph where the position of an endosymbiont status or a parasitoid community depended on its distance from all other points in the analysis. Then, the effects of the pea aphid biotype (i.e., three levels factor: “alfalfa”, “clover,” and “pea” biotypes) and the parasitism period (i.e., three levels factor: “pre-parasitism,” “maximal parasitism,” and “post-parasitism”) on endosymbiont infection statuses of aphids or parasitoid community dissimilarity were tested by performing a permutational multivariate analysis of variance (PERMANOVA) on the Bray–Curtis dissimilarity matrix. Pairwise comparisons between levels of factors were performed using pairwise Adonis tests with Bonferroni corrections (Martinez-Arbizu, 2017). Finally, the correlation between the two dissimilarity matrices (β-diversity of endosymbiont infection statuses of aphids vs β-diversity of parasitoid communities) was calculated by using a Mantel test. The Mantel test, nMDS, and PERMANOVA were implemented using the R package vegan (Oksanen et al., 2019), and the graphical representation of the nMDS was generated using the R package ggplot2 (Wickham, 2016).
To search for a link between symbiont associations and parasitoid pressures, the parasitism rate due to a dominant parasitoid species or all parasitoid species was tested against the frequency of the symbiotic associations in each biotype using General Linear Mixed Models (LMM) (lme function of the nlme package; Pinheiro et al., 2020). Note that we did not test all the correlations as some infection states had too few observations. As some fields were considered several times during our survey, the field ID was fit as a random factor to include data dependency. Before each statistical modeling, we checked linearity assumption between the response and the explanatory covariate. In case of linearity departure, we used Generalized Additive Mixed Models (GAMM) (gamm function of the mgcv package; Wood, 2017). Model assumptions were verified by plotting residuals versus fitted values for each model.
To analyze the effect of parasitism pressure on the H. defensa strain diversity, we compared the frequencies of each H. defensa strain found before and after the parasitism peak in alfalfa fields using a χ2 test.
The two proxies of parasitism pressure were positively correlated (Spearman correlation: rho = 0.452, p < 0.001) and showed similar dynamics during the survey (Figure 1): a low parasitism (<0.3) early in the season, then a maximal parasitism in June and July (up to 0.5) and a decrease below 0.2 from late July to October. The same temporal dynamics of parasitism activity was found in the three legume crops and from these results, three distinct periods were arbitrarily defined as “pre-parasitism,” “maximal parasitism” (0.45–0.65 parasitism rate), and “post-parasitism” (Figure 1). Overall, the rates of parasitism were the highest in clover fields (30.3%), intermediate in pea (22.4%), and the lowest in alfalfa (15.9%) (GLMM: χ2 = 10.26, df = 2, p < 0.005).
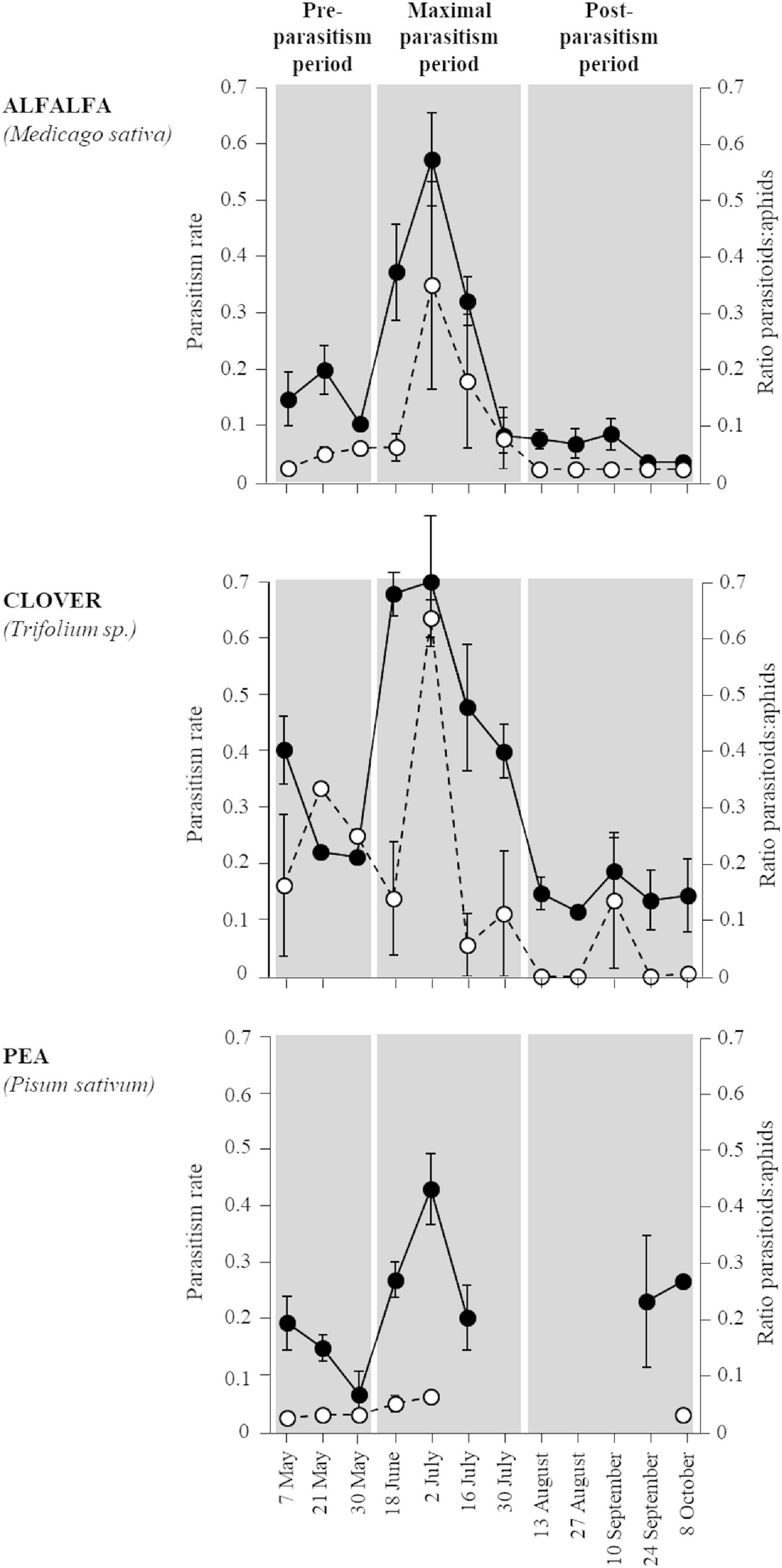
Figure 1. Temporal variation in parasitism pressure in alfalfa, clover, and pea crops estimated using two proxies: parasitism rate (black dots, —) and the ratio of the number of parasitoids/pea aphid individuals (white dots, –). The three shaded areas within each graph correspond to the three parasitism periods arbitrarily defined: “pre-parasitism,” “maximum parasitism,” and “post-parasitism” periods. Error bars represent standard errors.
Overall, A. ervi was the dominant parasitoid species in both alfalfa and clover crops (respectively 79.23 and 94.29% of emerging parasitoids over all the season) and the second most abundant species in pea crops (36.44%), after A. eadyi (45.48%) and before A. avenae (15.16%) (Figure 2). Both P. barbatum and Aphelinus sp. were exclusive to the clover and alfalfa biotypes, with P. barbatum accounting for 7.34% of parasitoids in alfalfa crops. The Shannon index in the parasitoid communities varied according to the pea aphid biotype and the parasitism period (LM: interaction term, F = 4.00, df = 4, p = 0.013). Overall, the α-diversity in parasitoid communities was the lowest in the clover fields, intermediate in alfalfa and the highest in pea. Although the dominant parasitoid species remained the same during the season in alfalfa and clover biotypes, species richness of their parasitoid communities increased throughout the season (Figure 3). In pea crops, A. eadyi increased in frequency, reaching 82% of parasitoids in post-parasitism period, replacing A. ervi as dominant species in the guild and leading to a decline in the Shannon index at this period.
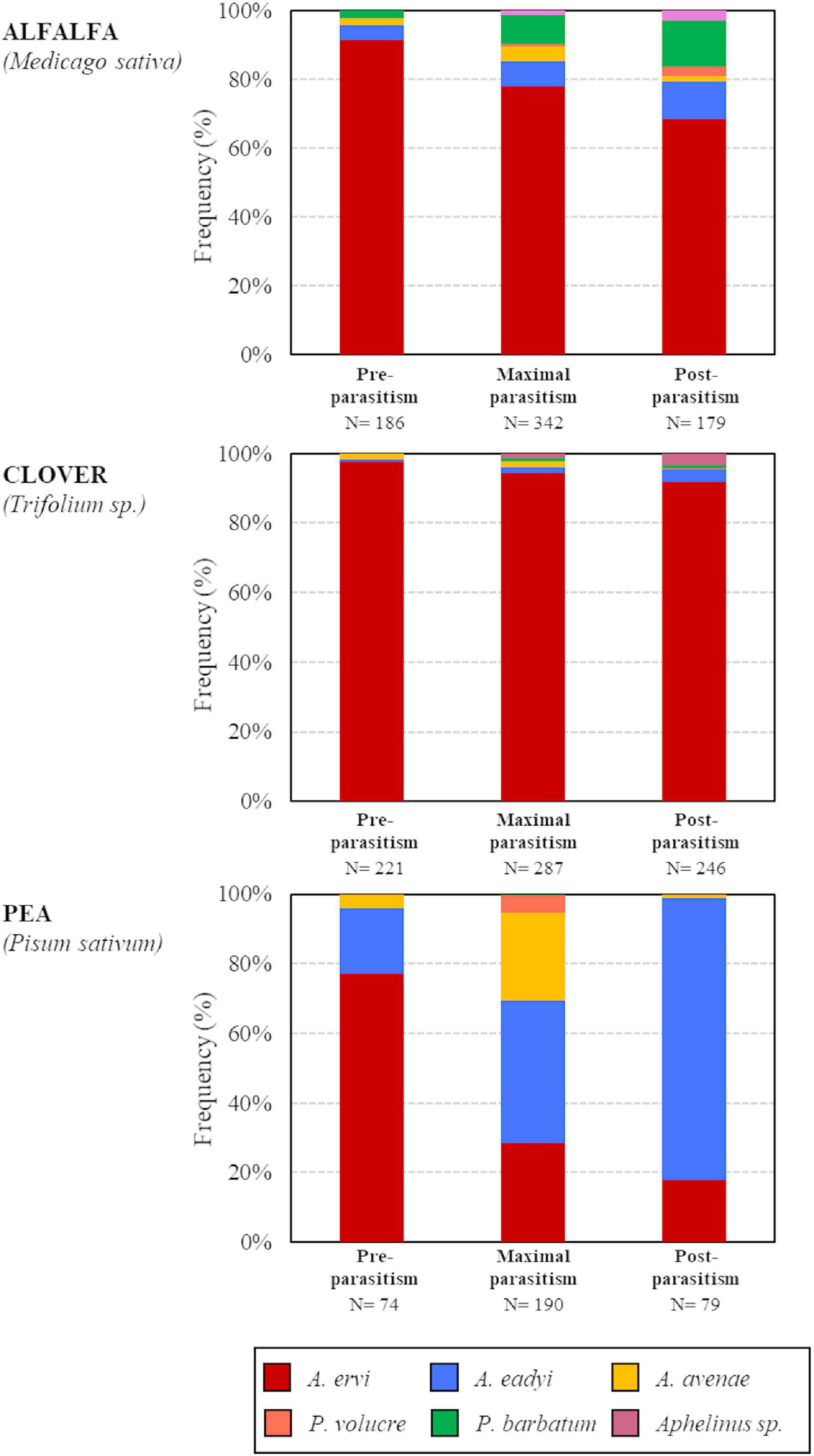
Figure 2. Frequencies of the parasitoid species in the different pea aphid biotypes (“alfalfa,” “clover,” and “pea”) at different parasitism period (“pre-parasitism,” “maximal parasitism,” and “post-parasitism”).
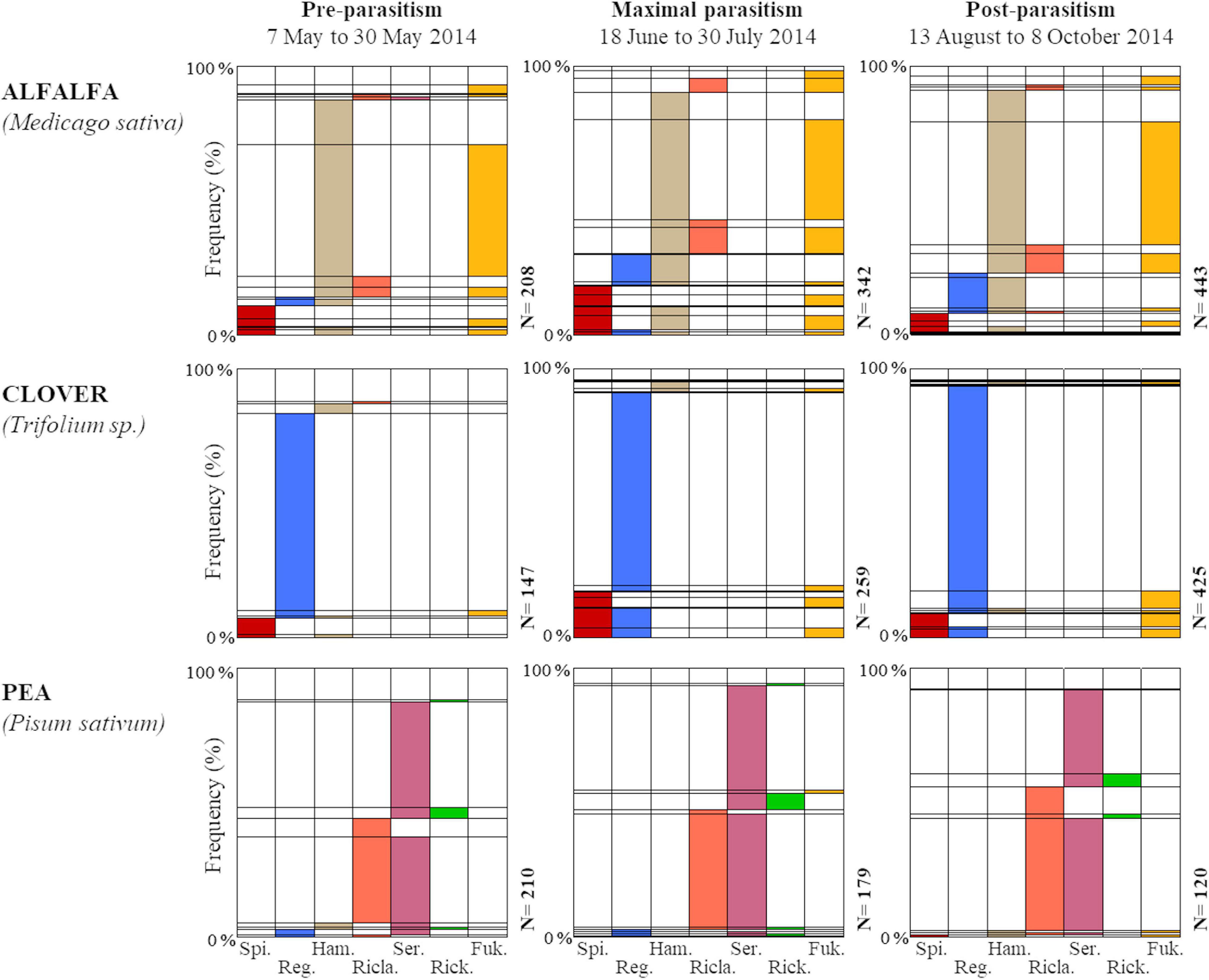
Figure 3. Shannon Index (H) of parasitoid communities and endosymbiont infection statuses, and covariation between both Shannon Indices, considering both the pea aphid biotypes and the parasitism periods.
Overall, 94.7% of pea aphids (96.5% for aphids from alfalfa, 94.7% from clover and 91.3% from pea) were infected by at least one facultative bacterial endosymbiont (Figure 4). Seven facultative endosymbiont species were detected with prevalence varying strongly between A. pisum biotypes. On average aphid individuals harbored 1.8, 1.1, and 1.4 facultative endosymbionts in alfalfa, clover and pea crops, respectively. In alfalfa, coinfection of H. defensa with F. symbiotica was the most frequent symbiotic association across the field survey (accounting for 43.70% of aphids) followed by H. defensa in monoinfection (12.28%). Two other associations involving H. defensa were also noted in alfalfa fields: coinfection with R. insecticola (8.96%) and triple infection with R. viridis and F. symbiotica (7.25%). Aphids free of secondary endosymbionts in alfalfa represented only 3.5%. In clover fields, R. insecticola in monoinfection represented 74.48% of surveyed aphids while individuals free of any facultative endosymbiont represented only 5.29%. In pea fields, infection with S. symbiotica singly (37.25%) or in coinfection with R. viridis (37.72%) predominated. Also, 5.89% were singly infected with R. viridis and 8.64% were deprived of facultative endosymbiont. The α-diversity in aphid endosymbiont infection statuses varied according to the pea aphid biotype only (LM: F = 26.13, df = 2, p < 0.001): the Shannon index was the highest in the alfalfa fields, intermediate in pea and the lowest in clover (Figure 3).
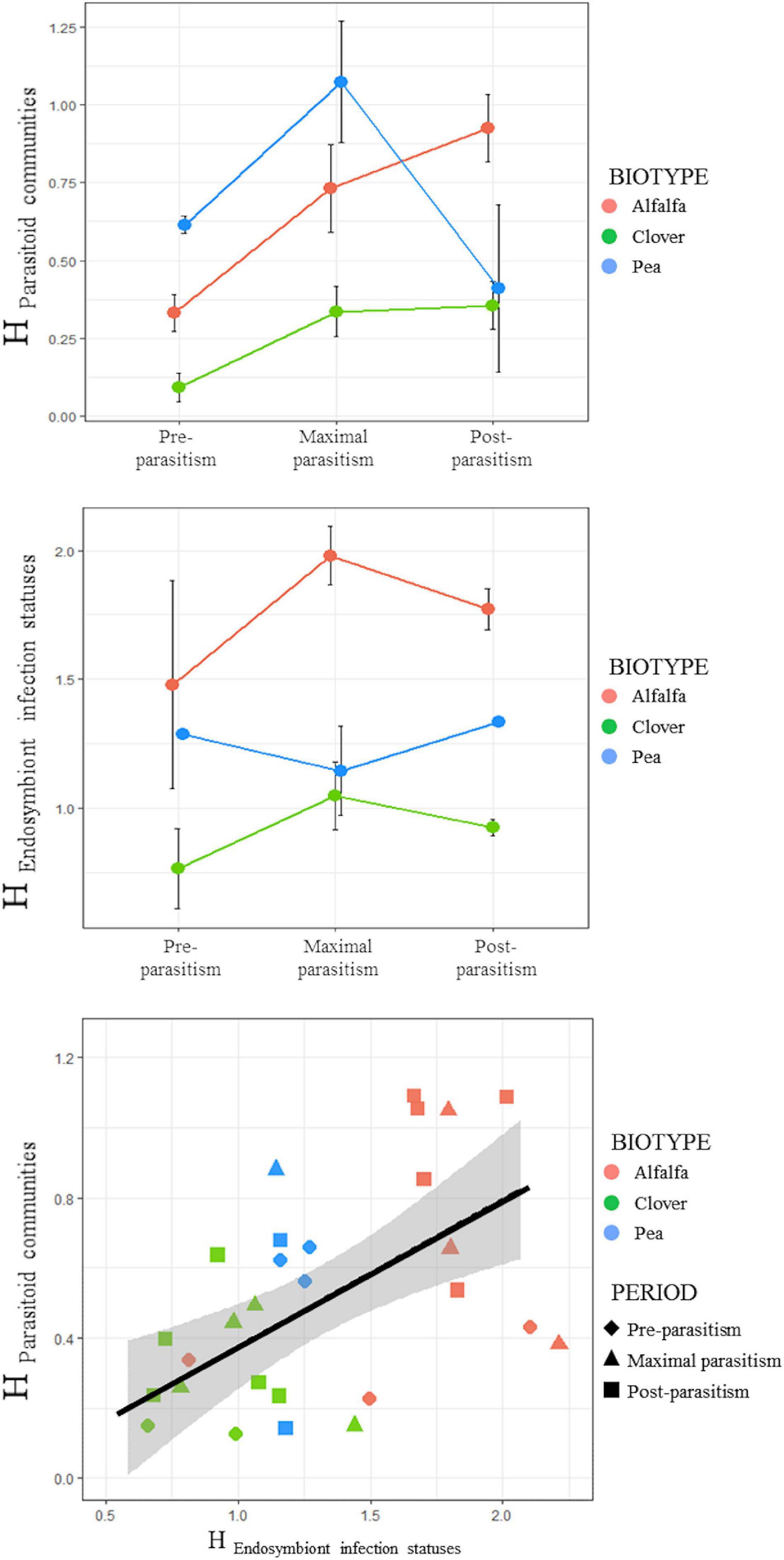
Figure 4. The diversity and relative abundance of endosymbiont infection statuses of pea aphids in relation with aphid biotype (“alfalfa,” “clover,” and “pea”) and parasitism period (“pre-parasitism,” “maximal parasitism,” and “post-parasitism”). For each column, the colored area corresponds to the percentage of aphid individuals harboring the corresponding symbiont species. When colored areas are present in several columns, it indicates a co-infection with multiple symbiont species. (Spi: Spiroplasma sp.; Reg: Regiella insecticola; Ham: Hamiltonella defensa; Rica: Rickettsiella viridis; Ser: Serratia symbiotica; Rick: Rickettsia sp.; Fuk: Fukatsuia symbiotica).
When we analyzed the relationship between the Shannon Index (H) of parasitoid communities and the Shannon Index (H) of endosymbiont infection statuses of aphids, we found a significantly positive covariation (Pearson correlation, r = 0.499, p = 0.004) in α-diversities (Figure 3).
When the Bray–Curtis distances were calculated between all pairwise combinations of parasitoid communities, we found that they varied significantly in composition between A. pisum biotypes (Figure 5). PERMANOVA detected a significant effect of aphid biotype on the parasitoid community assemblages (F = 3.84, df = 2, p = 0.003) and this factor accounted for 21% of the variance in the data. Pairwise comparisons between biotypes showed that the structure of parasitoid communities emerging from pea crops differed from the two other crops. No temporal dynamics in β-diversity in parasitoid communities was found (PERMANOVA, period effect: F = 1.565, df = 2, p = 0.150).
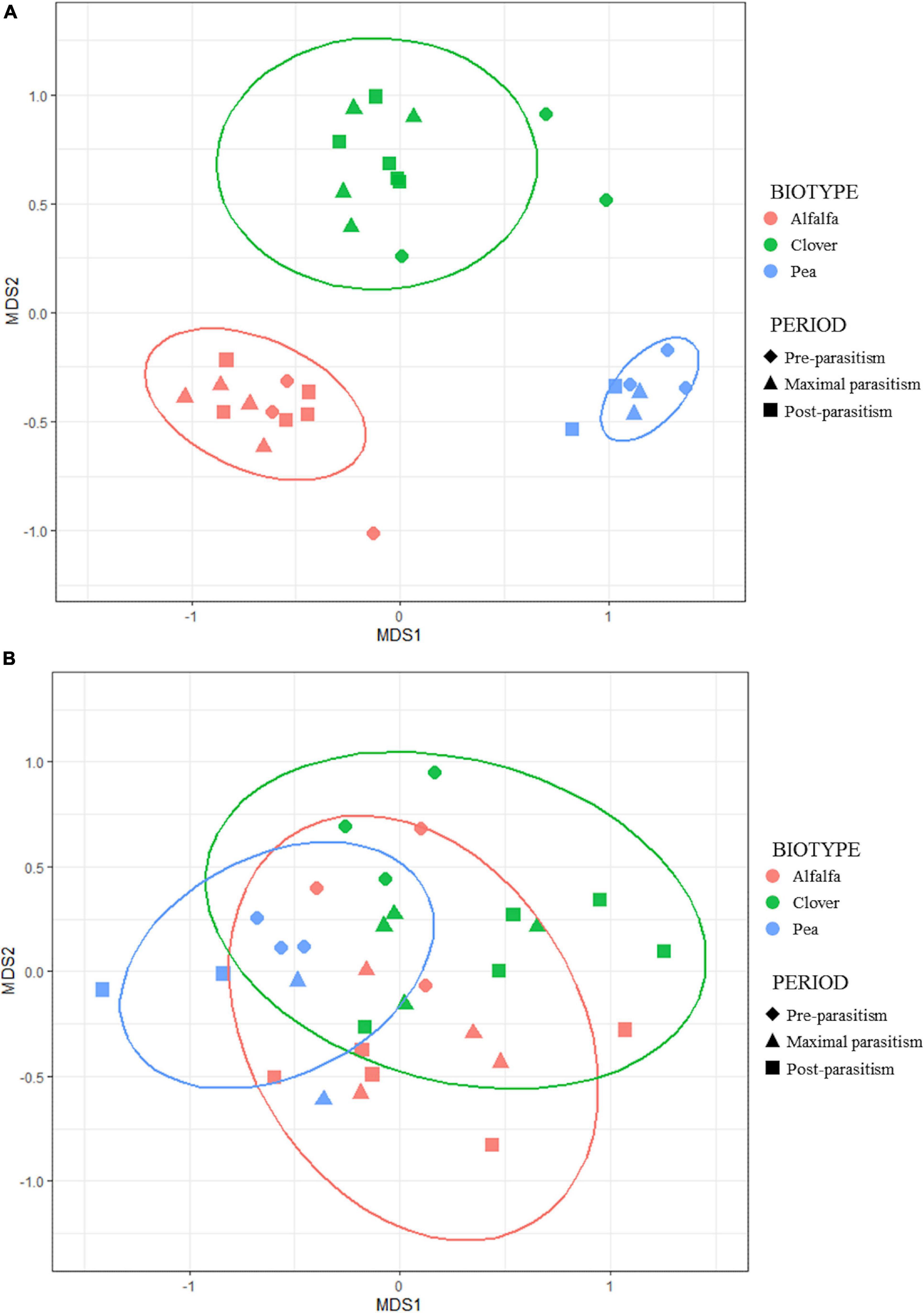
Figure 5. Non-metric MDS ordination plot comparing (A) endosymbiont infection statuses and (B) parasitoid communities from different pea aphid samples. Each data point represents the (A) symbiont/(B) parasitoid community identified from aphid samples collected at one date for a given biotype. The shape of the dots refers to the parasitism periods. The Bray-Curtis dissimilarity index was used to rank distances calculated using the abundance community data. Stress of the nMDS: (A) symbiont infection status = 0.083; (B) parasitoid community = 0.164.
The Bray–Curtis dissimilarities between endosymbiont infection statuses of aphids presented highly contrasted values and showed that they differed strongly between biotypes while being stable during the season within each biotype (Figure 5). PERMANOVA confirmed this pattern since the biotype effect on infection statuses dissimilarity was highly significant (F = 79.08, df = 2, p = 0.001). This factor accounted for 83% of the variance in the data. No temporal dynamics in β-diversity of endosymbiont infection statuses of aphids was found (PERMANOVA, period effect: F = 740, df = 2, p = 0.596), confirming their stability throughout the cropping season.
A positive correlation between the two dissimilarity matrices was found (Mantel test: zM = 0.186, p = 0.002, see Supplementary Figure 2). The dissimilarity between parasitoid communities was therefore correlated with the dissimilarity between endosymbiont infection statuses of aphids.
In both clover and pea crops, the parasitism rate of all parasitoid species or of the dominant ones did not vary according to the prevalence of the most frequent symbiotic associations (Table 1 and Supplementary Figure 3). For the alfalfa biotype, while the prevalence of H. defensa in monoinfection did not influence the parasitism rates (Table 1 and Supplementary Figure 3), the coinfection with H. defensa and F. symbiotica had a significantly negative effect on parasitism rates: when pea aphids from alfalfa fields presented high prevalence of coinfection with both protective endosymbionts, the parasitism rate of A. ervi and the overall parasitism rate declined although non-linearly (Table 1 and Figure 6).
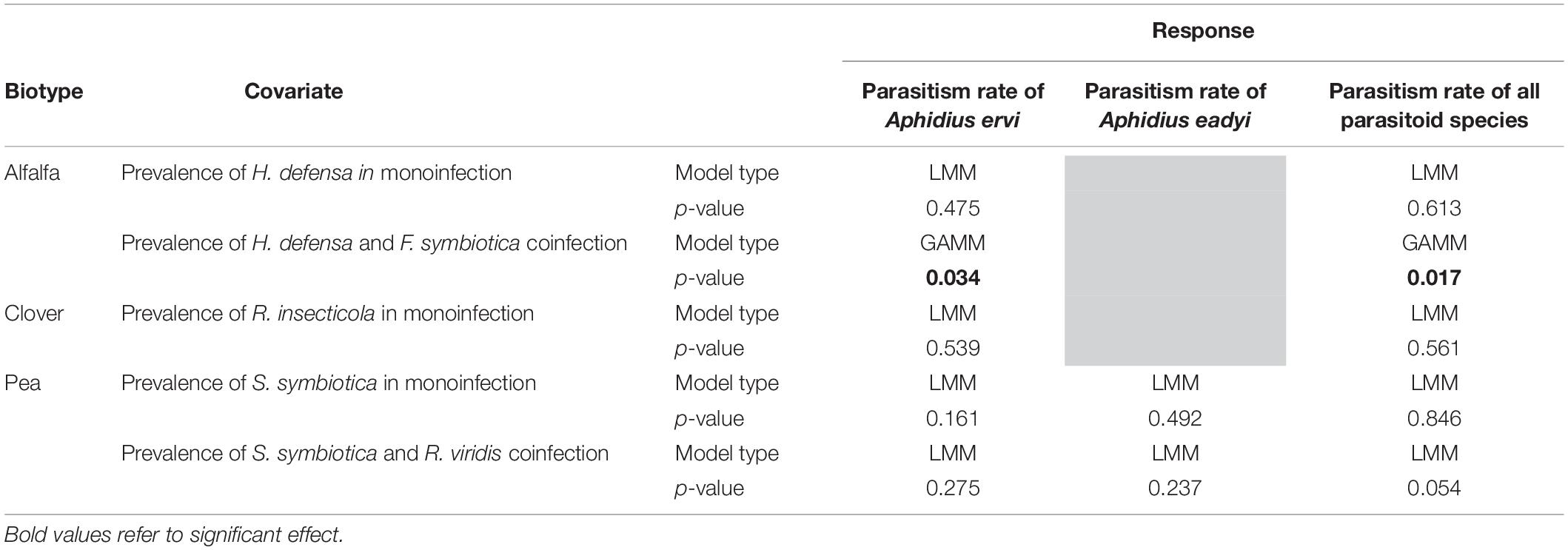
Table 1. Results of statistical models (LMM: General Linear Mixed Models; GAMM: Generalized Additive Mixed Models) testing the effect of the most prevalent symbiotic associations on parasitism rate (for the dominant parasitoid species and all parasitoids).
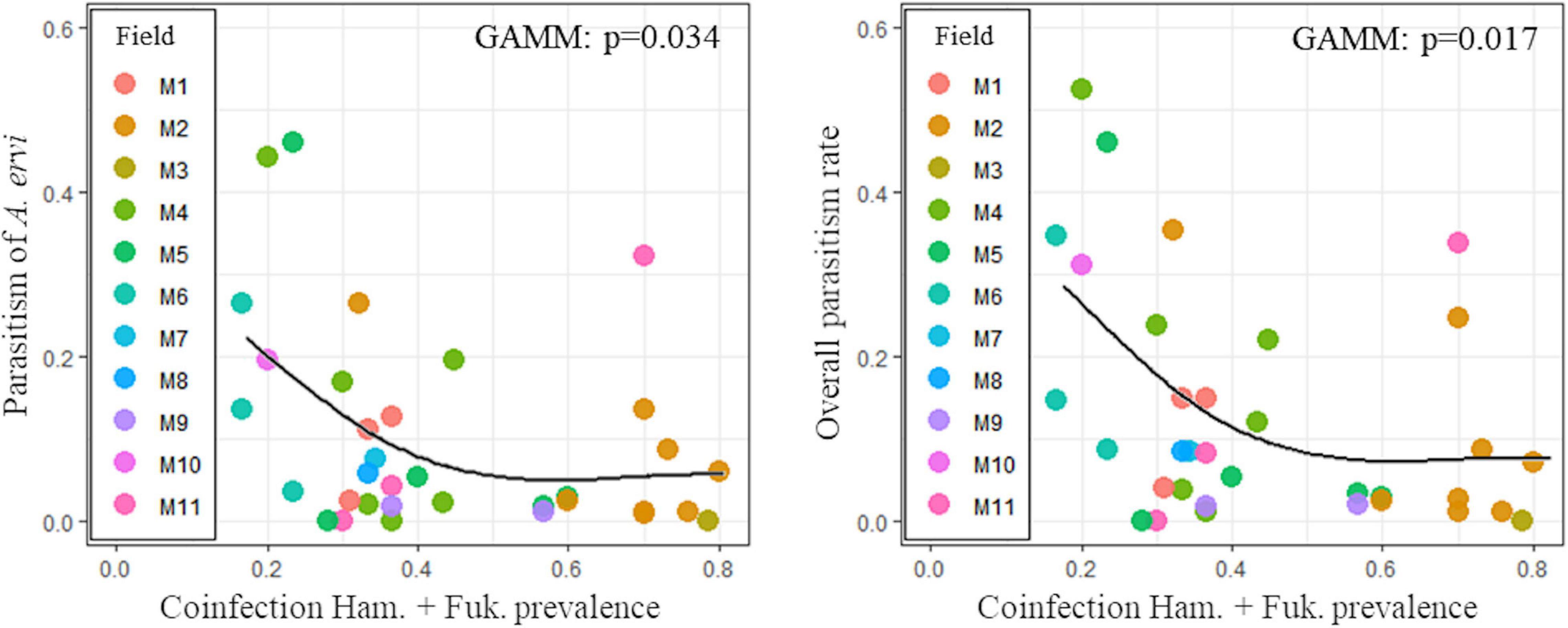
Figure 6. Relationship between Hamiltonella defensa/Fukatsuia symbiotica coinfection and parasitism rate (assessed for the dominant parasitoid species, Aphidius ervi, and for all parasitoids) in pea aphids from alfalfa fields. ID refers to the alfalfa field. Solid line represents the predicted values from the fitted generalized additive mixed models.
A subsample of 72 A. pisum from different alfalfa fields and infected with H. defensa singly or co-infected with H. defensa and F. symbiotica was analyzed to characterize the strain variation of H. defensa. Among the pea aphid individuals, 36 aphids were selected before the peak of parasitism and 36 aphids after the peak. The APSE phage associated with H. defensa was consistently detected in all aphids. Seven genetically different strains of H. defensa were characterized (Table 2). Two main haplotypes, representing 73% of the H. defensa strains, dominated aphid populations before and after parasitism peak. A greater diversity of strains was observed after the peak of parasitism (0.11 before and 0.19 after) and a similar pattern was observed for the aphid clonal diversity (0.58 before and 0.64 after). However, the frequencies of H. defensa strains did not differ significantly before and after the parasitism peak in alfalfa fields (χ2 test: χ2 = 6.65, df = 6, p = 0.645).
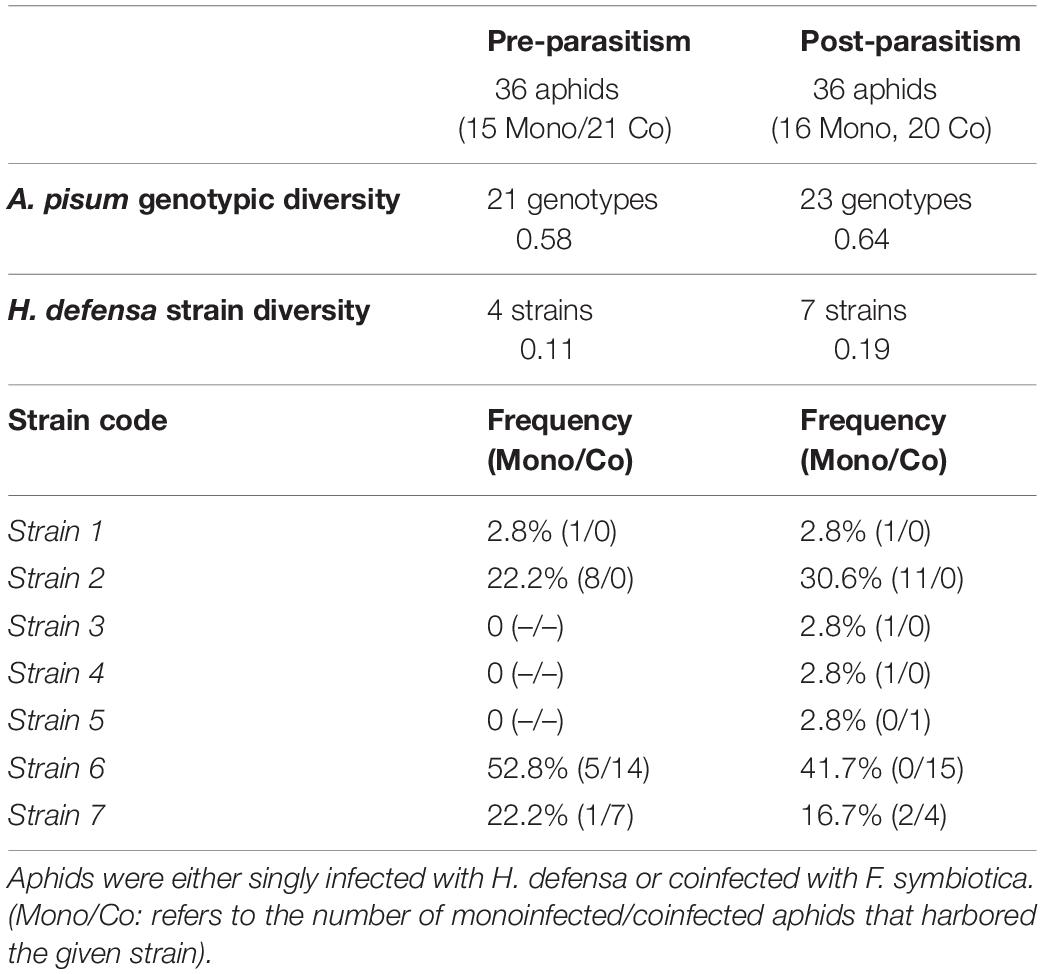
Table 2. Aphid clonal diversity, H. defensa strain diversity and haplotypes in the “pre-parasitism” and “post-parasitism” periods for pea aphids collected in alfalfa fields.
Since some microbial symbionts confer a protection against natural enemies to their hosts that can potentially alter food web interactions (Hafer and Vorburger, 2019; Mclean, 2019), our objectives were to test under natural conditions whether the parasitism rate of different A. pisum biotypes depended on their endosymbiont communities and whether symbiont communities and parasitoid communities associated with pea aphids were related. We found that facultative endosymbiont communities were highly structured by biotype and stable in time while the parasitoid communities showed moderate differences between pea aphid biotypes and some change in structure over time. At the level of the pea aphid complex, we revealed a correlation between diversities (i.e., α- and β-diversities) of endosymbiont infection statuses of aphids and parasitoid communities. Interestingly, we found a negative correlation between the prevalence of H. defensa and F. symbiotica in co-infection and the intensity of parasitoid pressure in the alfalfa biotype, confirming in field conditions the protective effect of this symbiotic combination.
The strong associations between endosymbiont communities and pea aphid biotypes are not novel and have been recurrently reported in various studies (Simon et al., 2003; Ferrari et al., 2004; Russell et al., 2013; Henry et al., 2015; Smith et al., 2015). Beyond the absence or the presence of each facultative bacterial endosymbiont in natural populations, each pea aphid biotype presented dominant symbiotic associations during the season: more than 80% of the aphids feeding on clover were singly infected with R. insecticola; in alfalfa, the infection with H. defensa alone or in coinfection with another facultative symbionts predominated the natural populations; and almost all aphids specialized on pea harbored S. symbiotica singly or in coinfection with R. viridis. Several hypotheses have been put forward for such differences in symbiotic associations and involve either ecological filters exerting selective pressure on aphid symbioses, symbiont-symbiont interactions or the effect of drift on symbiont associations (Mathé-Hubert et al., 2019).
An essential step for linking facultative endosymbionts with parasitoid communities was to reliably assess the parasitism pressure exerted on aphid populations. Here, we used two proxies to reduce the bias of underestimating the parasitoid pressure by measuring only the mummification rate (Oliver et al., 2003). For the three biotypes, both parasitoid pressure proxies were highly positively correlated and showed the same temporal dynamics, indicating that the actual parasitoid pressure exerted on field populations of the pea aphid in different ecological situations was reliably assessed. Overall, the parasitism pressure over time presented a single peak of high parasitoids’ activity in early July. This temporal dynamic was similar between alfalfa, clover and pea biotypes but varied quantitatively: the pressure exerted by parasitoids was the highest in clover, intermediate in pea and the lowest in alfalfa crops. One explanation for these quantitative differences could be that clover is typically grown in a more complex agricultural mosaic than the other crops, which would favor biological regulations. An alternative could be that clover is more attractive to parasitoids.
Despite these temporal dynamics and inter-biotype variations in parasitism pressure, the relative abundance of the endosymbiont infection statuses of aphids changed very little over the season. These results contradict those obtained in a previous study done on both alfalfa and clover biotypes in the United States (Smith et al., 2015). In this earlier work, considerable seasonal shifts were indeed observed in the frequencies of endosymbionts, especially in Pennsylvania fields. This difference may result from the fact that this previous work considered each symbiont species individually while we analyzed them as communities. It could also be due to differences in population and community composition of endosymbionts and parasitoids, which exist between the native and introduced range of A. pisum. For example, we found six species of parasitoids in our survey while Smith et al. found only two. In addition, while there is evidence of some protection against parasitoids conferred by F. symbiotica in western Europe (Heyworth and Ferrari, 2015; Leclair et al., 2016), this does not appear to be the case in the United States (Doremus and Oliver, 2017), suggesting strain differences between countries. The temporal stability of symbiotic associations we observed suggests a good fidelity in vertical transmission of symbiont combinations in natural conditions, confirming previous results on field estimates of maternal transmission rate of pea aphid endosymbionts (Rock et al., 2018). This seasonal stability was also observed at the symbiont population level as we did not detect differences in the frequencies of H. defensa strains throughout the alfalfa growing season. Temporal fluctuations in symbiotic associations may, however, occur on a longer time scale (between years or beyond), although the same symbiotic associations have been found repeatedly in pea aphid populations in independent studies and in different years (Henry et al., 2013; Rock et al., 2018; Mathé-Hubert et al., 2019).
Interestingly, we found a negative non-linear relationship between the frequency of co-infections with H. defensa and F. symbiotica and the parasitism rate (estimated for all parasitoid species or for A. ervi alone). Surprisingly, the single infection with H. defensa alone did not appear related to the rate of parasitism. However, such results are consistent with observations made in controlled conditions. Indeed, it has been shown that the protection conferred by H. defensa alone varies greatly according to the host and genotypes of H. defensa (Oliver and Higashi, 2019), whereas co-infections with F. symbiotica give the aphids a high or even total protection against parasitism (Guay et al., 2009; Leclair et al., 2016), but see Doremus and Oliver (2017). Overall, this endosymbiont protection may be responsible for the lower parasitism rate (between 30 and 50% reduction) observed in the alfalfa biotype compared to the two other biotypes. Negative correlations between protective endosymbionts in pea aphids and parasitoid rates have been reported in earlier field studies (Smith et al., 2015; Hrcek et al., 2016; Rothacher et al., 2016). Our study and the previous ones thus confirm the effectiveness of symbiont-mediated protection in natural environments exploited by complex and diverse parasitoid communities.
One explanation given for the absence of H. defensa in clover and pea biotypes is because of a lower parasitoid pressure in natural populations of these biotypes (Oliver et al., 2008). However, our results showed that parasitoid wasps severely attacked these two pea aphid biotypes, with parasitism rates up to 70%. Given this parasitism pressure, an alternative protection against parasitoids may exist in these aphid populations; for instance, another facultative endosymbiont could confer an alternative protection to H. defensa. However, we did not observe a relationship between endosymbiont communities in the pea and clover biotypes and the rate of parasitism, although S. symbiotica (predominant in the pea biotype) and a strain of R. insecticola (the dominant symbiont in the clover biotype) have been reported to confer some parasitoid resistance in several laboratory studies (Oliver et al., 2003; Vorburger et al., 2010; Heyworth and Ferrari, 2015). Also, we showed in controlled conditions that the endosymbiont communities that dominated clover and pea biotypes in our field survey conferred very limited protection to A. ervi (Leclair et al., pers. obs.). Other ecological and non-ecological factors would better explain the prevalent symbiotic associations observed in these biotypes as discussed earlier (Mathé-Hubert et al., 2019). For example (Smith et al., 2015) suggested that the high prevalence of R. insecticola in the clover biotype was due to the strong incidence of fungal pathogen-induced mortality in clover fields, which would select this symbiont because of the fungal protection it confers (Scarborough et al., 2005). More field works are needed to assess the influence of environmental factors on symbiont composition of the various pea aphid biotypes.
Two earlier field studies showed that protective symbionts could influence the third trophic level by shaping the structure of parasitoid community attacking aphid populations (Hrcek et al., 2016; Rothacher et al., 2016). In addition, an experimental evolution study showed that parasitoid diversity could maintain diversity in protective symbionts (Hafer-Hahmann and Vorburger, 2020). Our study is in line with this body of work linking symbiont and parasitoid diversities. Indeed, we found that both α- and β-diversities of symbionts and parasitoids were correlated, suggesting some interactions between these two communities through their aphid hosts or other environmental factors (i.e., local habitats).
Given the high prevalence of H. defensa in the alfalfa biotype and the negative correlation we found only between H. defensa-F. symbiotica co-infection and parasitoid pressure, one might have expected to find a strong effect of the symbiotic protection found in the alfalfa biotype on the community structure of parasitoids attacking the pea aphid. However, we showed that parasitoid communities varied little between the clover and alfalfa biotypes but more between these two and the pea biotype. A. ervi is often cited as the parasitoid species exerting the highest pressure on pea aphid populations (Kavallieratos et al., 2004; Smith et al., 2015). The alfalfa biotype was predominantly attacked by this parasitoid species despite the high prevalence of protective symbioses in this biotype. Previous works showed an evolutionary potential of parasitoids to counteract the symbiont protection conferred by H. defensa (Dion et al., 2011; Rouchet and Vorburger, 2014); some genotypes of A. ervi may thus have evolved virulence factors against H. defensa mediated protection in the wild (Dennis et al., 2020). Our results showed that the parasitoid community associated with the pea biotype strongly differed from the two others, in particular by the abundance of A. eadyi and A. avenae. Ferrari et al. (2004) have showed that H. defensa could provide resistance to A. eadyi in A. pisum populations. However, this endosymbiont was not present in the pea biotype in our survey and it is not known whether S. symbiotica or R. viridis, which, on the other hand, were both well represented in A. pisum from pea, confer resistance to A. eadyi. More work is needed to test whether the most prevalent symbiotic combination in each biotype confers an optimal protection to the corresponding parasitoid communities.
In conclusion, this study showed a temporal stability in symbiont populations and communities, in sharp contrast with a strong seasonality in parasitoid activities. This weak response of symbiont communities to parasitoid pressures could be explained by the limited costs of carrying protective symbionts on this timescale. However, further field surveys are needed to determine whether the composition in protective symbionts is maintained over longer period or is rather driven by an ecological-evolutionary dynamics resulting from selection for or against resistance, as suggested earlier (Smith et al., 2015) and recently demonstrated in a manipulated agricultural system (Ives et al., 2020). We also showed that the three pea aphid biotypes, despite their distinct endosymbiont composition, were exposed to similar range of parasitism pressures, suggesting that other protective alternatives than hosting H. defensa alone or with F. symbiotica, and involving symbiont or host mechanisms, may be used by the various biotypes of the pea aphid complex, which merits further work. Finally, we detected a link between communities of parasitoids and symbionts, suggesting interactions through shared resources or other environmental filters. The study of other communities of natural enemies of aphids (e.g., predators, pathogens) could reveal more such links and allow to better measure the importance of symbionts in food webs.
The datasets generated and analyzed in this article are available in Zenodo at https://doi.org/10.5281/zenodo.4548282.
ML, J-CS, and YO conceived and designed the field works and experiments and managed the field samplings. FM and ML performed both molecular studies and symbiont detection. CB performed species identification. YO analyzed the data. YO and J-CS contributed to the supervision of this study. All authors contributed critically to the drafts and gave final approval for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are very grateful to all people who helped in aphid sampling, to Anne-Lise Boixel and Antoine Briand for their strong investment in field surveys and laboratory works during summer 2014, and to Bernard Chaubet for his help in parasitoid species determination. This work was supported by the French “Ministère de l’Enseignement Supérieur et de la Recherche,” the Plant Health and Environment division of INRAE and a grant from the French Research Agency (HMICMAC project 16-CE02-0014).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.618331/full#supplementary-material
Supplementary Figure 1 | Location of the sampled fields. The color and the shape of the dots represent the legume species (“Alfalfa,” “Clover,” “Pea”) on which pea aphids and parasitoids were collected.
Supplementary Figure 2 | Bray-Curtis dissimilarity matrix of (a) endosymbiont infection statuses and (b) parasitoid communities in the different pea aphid biotypes (“Alfalfa,” “Clover,” “Pea”) at different parasitism period (P1: “pre-parasitism,” P2: “maximum parasitism,” and P3: “post-parasitism”).
Supplementary Figure 3 | Effect of the most prevalent symbiotic associations on the parasitism rate (dominant parasitoid species and all parasitoid together) in pea aphid individuals from the alfalfa, clover and pea biotypes. ID refers to the field crop sampled. For all these panels, no significant relationship was found.
Supplementary Table 1 | Sampling information: code, location and sampling frequency of the fields considered in our study. For a given legume crop, the field code assignment depends on the geographical location: we numbered from the westernmost to the easternmost field.
Supplementary Table 2 | Sampling information: dates, field identity (i.e. field), number of aphid nymphs used to assess symbiotic composition (i.e. nymph), number of adult aphids kept to measure parasitism rate (i.e. adult) and its value for each sampling (i.e. para) (when “-”, no sampling).
Abrams, P. A. (2000). The evolution of predator-prey interactions: theory and evidence. Ann. Rev. Ecol. Syst. 31, 79–105. doi: 10.1146/annurev.ecolsys.31.1.79
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Software 67:48.
Cayetano, L., Rothacher, L., Simon, J.-C., and Vorburger, C. (2015). Cheaper is not always worse: strongly protective isolates of a defensive symbiont are less costly to the aphid host. Proc. Biol. Sci. 282:20142333. doi: 10.1098/rspb.2014.2333
Degnan, P. H., and Moran, N. A. (2008). Diverse phage-encoded toxins in a protective insect endosymbiont. Appl. Environ. Microbiol. 74, 6782–6791. doi: 10.1128/aem.01285-08
Dennis, A. B., Ballesteros, G. I., Robin, S., Schrader, L., Bast, J., Berghöfer, J., et al. (2020). Functional insights from the GC-poor genomes of two aphid parasitoids, aphidius ervi and Lysiphlebus fabarum. BMC Genom. 21:376. doi: 10.1186/s12864-020-6764-0
Dion, E., Polin, S. E., Simon, J.-C., and Outreman, Y. (2011). Symbiont infection affects aphid defensive behaviours. Biol. Lett. 7, 743–746. doi: 10.1098/rsbl.2011.0249
Doremus, M. R., and Oliver, K. M. (2017). Aphid heritable symbiont exploits defensive mutualism. Appl. Environ. Microbiol. 83, e3216–e3276.
Ferrari, J., Darby, A. C., Daniell, T. J., Godfray, H. C. J., and Douglas, A. E. (2004). Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29, 60–65. doi: 10.1111/j.1365-2311.2004.00574.x
Ferrari, J., West, J. A., Via, S., and Godfray, H. C. J. (2012). Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66, 375–390. doi: 10.1111/j.1558-5646.2011.01436.x
Flórez, L. V., Biedermann, P. H. W., Engl, T., and Kaltenpoth, M. (2015). Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936. doi: 10.1039/c5np00010f
Gauthier, J.-P., Outreman, Y., Mieuzet, L., and Simon, J.-C.(2015). Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS One 10:e0120664. doi: 10.1371/journal.pone.0120664
Guay, J. F., Boudreault, S., Michaud, D., and Cloutier, C. (2009). Impact of environmental stress on aphid clonal resistance to parasitoids: Role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect Physiol. 55, 919–926. doi: 10.1016/j.jinsphys.2009.06.006
Guyomar, C., Legeai, F., Jousselin, E., Mougel, C., Lemaitre, C., and Simon, J.-C. (2018). Multi-scale characterization of symbiont diversity in the pea aphid complex through metagenomic approaches. Microbiome 6:181.
Hafer, N., and Vorburger, C. (2019). Diversity begets diversity: do parasites promote variation in protective symbionts? Curr. Opin. Insect Sci. 32, 8–14. doi: 10.1016/j.cois.2018.08.008
Hafer-Hahmann, N., and Vorburger, C. (2020). Parasitoids as drivers of symbiont diversity in an insect host. Ecol. Lett. 23, 1232–1241. doi: 10.1111/ele.13526
Henry, L. M., Maiden, M. C. J., Ferrari, J., and Godfray, H. C. J. (2015). Insect life history and the evolution of bacterial mutualism. Ecol. Lett. 18, 516–525. doi: 10.1111/ele.12425
Henry, L. M., Peccoud, J., Simon, J. C., Hadfield, J. D., Maiden, M. J. C., Ferrari, J., et al. (2013). Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 23, 1713–1717. doi: 10.1016/j.cub.2013.07.029
Heyworth, E. R., and Ferrari, J. (2015). A facultative endosymbiont in aphids can provide diverse ecological benefits. J. Evol. Biol. 28, 1753–1760. doi: 10.1111/jeb.12705
Himler, A. G., Adachi-Hagimori, T., Bergen, J. E., Kozuch, A., Kelly, S. E., Tabashnik, B. E., et al. (2011). Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332, 254–256. doi: 10.1126/science.1199410
Hrcek, J., Mclean, A. H. C., and Godfray, H. C. J. (2016). Symbionts modify interactions between insects and natural enemies in the field. J. Anim. Ecol. 85, 1605–1612. doi: 10.1111/1365-2656.12586
Ives, A. R., Barton, B. T., Penczykowski, R. M., Harmon, J. P., Kim, K. L., Oliver, K., et al. (2020). Self-perpetuating ecological-evolutionary dynamics in an agricultural host-parasite system. Nat. Ecol. Evol. 4, 702–711. doi: 10.1038/s41559-020-1155-0
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M., and Perlman, S. J. (2010). Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. doi: 10.1126/science.1188235
Kavallieratos, N. G., Tomanovi, Ž, Stary, P., Athanassiou, C. G., Sarlis, G. P., Petroviæ, O., et al. (2004). A survey of aphid parasitoids (hymenoptera: braconidae: aphidiinae) of southeastern europe and their aphid-plant associations. Appl. Entomol. Zool. 39, 527–563. doi: 10.1303/aez.2004.527
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Leclair, M., Polin, S., Jousseaume, T., Simon, J.-C., Sugio, A., Morlière, S., et al. (2017). Consequences of coinfection with protective symbionts on the host phenotype and symbiont titres in the pea aphid system. Insect Sci. 24, 798–808. doi: 10.1111/1744-7917.12380
Leclair, M., Pons, I., Mahéo, F., Morlière, S., Simon, J.-C., and Outreman, Y. (2016). Diversity in symbiont consortia in the pea aphid complex is associated with large phenotypic variation in the insect host. Evol. Ecol. 30, 925–941. doi: 10.1007/s10682-016-9856-1
Łukasik, P., Van Asch, M., Guo, H., Ferrari, J., Charles, J., and Godfray, H. (2013). Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 16, 214–218. doi: 10.1111/ele.12031
Martinez-Arbizu, P. (2017). pairwise Adonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.3. [Online]. Available Online at: https://github.com/pmartinezarbizu/pairwiseAdonis [Accessed].
Mathé-Hubert, H., Kaech, H., Hertaeg, C., Jaenike, J., and Vorburger, C. (2019). Nonrandom associations of maternally transmitted symbionts in insects: the roles of drift versus biased cotransmission and selection. Mol. Ecol. 28, 5330–5346. doi: 10.1111/mec.15206
Mclean, A. H. C. (2019). Cascading effects of defensive endosymbionts. Curr. Opin. Insect Sci. 32, 42–46. doi: 10.1016/j.cois.2018.10.003
Mclean, A. H. C., and Godfray, H. C. J. (2015). Evidence for specificity in symbiont-conferred protection against parasitoids. Proc. Biol. Sci. 282:20150977. doi: 10.1098/rspb.2015.0977
Mclean, A. H. C., Parker, B. J., Hrček, J., Henry, L. M., and Godfray, H. C. J. (2016). Insect symbionts in food webs. Philos. Trans. R. Soc. Lond B Biol. Sci. 371:20150325.
Mclean, A. H. C., Parker, B. J., Hrček, J., Kavanagh, J. C., Wellham, P. A. D., and Godfray, H. C. J. (2018). Consequences of symbiont co-infections for insect host phenotypes. J. Anim. Ecol. 87, 478–488. doi: 10.1111/1365-2656.12705
Müller, C. B., Adriaanse, I. C. T., Belshaw, R., and Godfray, H. C. J. (1999). The structure of an aphid–parasitoid community. J. Anim. Ecol. 68, 346–370. doi: 10.1046/j.1365-2656.1999.00288.x
Oksanen, J. F., Blanchet, G. F., Kindt, R., Legendre, P., Mcglinn, D., Minchin, P. R., et al. (2019). Vegan: Community Ecology Package [Online]. Available Online at: https://github.com/vegandevs/vegan [Accessed].
Oliver, K. M., Campos, J., Moran, N. A., and Hunter, M. S. (2008). Population dynamics of defensive symbionts in aphids. Proc. Biol. Sci. 275, 293–299. doi: 10.1098/rspb.2007.1192
Oliver, K. M., and Higashi, C. H. V. (2019). Variations on a protective theme: hamiltonella defensa infections in aphids variably impact parasitoid success. Curr. Opin. Insect Sci. 32, 1–7. doi: 10.1016/j.cois.2018.08.009
Oliver, K. M., and Perlman, S. J. (2020). “Chapter Eight - Toxin-mediated protection against natural enemies by insect defensive symbionts,” in Advances in Insect Physiology, eds K. M. Oliver and J. A. Russell (Cambridge: Academic Press), 277–316. doi: 10.1016/bs.aiip.2020.03.005
Oliver, K. M., Russell, J. A., Moran, N. A., and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. 100, 1803–1807. doi: 10.1073/pnas.0335320100
Oliver, K. M., Smith, A. H., and Russell, J. A. (2014). Defensive symbiosis in the real world – advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 28, 341–355. doi: 10.1111/1365-2435.12133
Peccoud, J., Bonhomme, J., Mahéo, F., De La Huerta, M., Cosson, O., and Simon, J. C. (2014). Inheritance patterns of secondary symbionts during sexual reproduction of pea aphid biotypes. Insect Sci. 21, 291–300. doi: 10.1111/1744-7917.12083
Peccoud, J., Mahéo, F., de la Huerta, M., Laurence, C., and Simon, J. C. (2015). Genetic characterisation of new host-specialised biotypes and novel associations with bacterial symbionts in the pea aphid complex. Insect Cons. Div. 8, 484–492. doi: 10.1111/icad.12131
Peccoud, J., Figueroa, C. C., Silva, A. X., Ramirez, C. C., Mieuzet, L., Bonhomme, J., et al. (2008). Host range expansion of an introduced insect pest through multiple colonizations of specialized clones. Mol. Ecol. 17, 4608–4618. doi: 10.1111/j.1365-294x.2008.03949.x
Peccoud, J., Ollivier, A., Plantegenest, M., and Simon, J. C. (2009). A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl. Acad. Sci. U S A. 106, 7495–7500. doi: 10.1073/pnas.0811117106
Peccoud, J., and Simon, J. C. (2010). The pea aphid complex as a model of ecological speciation. Ecol. Entomol. 35, 119–130. doi: 10.1111/j.1365-2311.2009.01147.x
Pinheiro, J., Bates, D., Debroy, S., Sarkar, D., and Team, R. C. (2020). nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-149 [Online]. Available Online at: https://CRAN.R-project.org/package=nlme [Accessed].
Polin, S., Le Gallic, J.-F., Simon, J.-C., Tsuchida, T., and Outreman, Y. (2015). Conditional reduction of predation risk associated with a facultative symbiont in an insect. PLoS One 10:e0143728. doi: 10.1371/journal.pone.0143728
Rock, D. I., Smith, A. H., Joffe, J., Albertus, A., Wong, N., O’connor, M., et al. (2018). Context-dependent vertical transmission shapes strong endosymbiont community structure in the pea aphid, acyrthosiphon pisum. Mol. Ecol. 27, 2039–2056. doi: 10.1111/mec.14449
Rothacher, L., Ferrer-Suay, M., and Vorburger, C. (2016). Bacterial endosymbionts protect aphids in the field and alter parasitoid community composition. Ecology 97, 1712–1723. doi: 10.1890/15-2022.1
Rouchet, R., and Vorburger, C. (2014). Experimental evolution of parasitoid infectivity on symbiont-protected hosts leads to the emergence of genotype specificity. Evolution 68, 1607–1616. doi: 10.1111/evo.12377
Russell, J. A., Weldon, S., Smith, A. H., Kim, K. L., Hu, Y., Łukasik, P., et al. (2013). Uncovering symbiont-driven genetic diversity across north american pea aphids. Mol. Ecol. 22, 2045–2059. doi: 10.1111/mec.12211
Scarborough, C. L., Ferrari, J., and Godfray, H. C. J. (2005). Aphid protected from pathogen by endosymbiont. Science 310:1781. doi: 10.1126/science.1120180
Siberchicot, A., Charif, D., Terraz, G., and Vavre, F. (2016). Mondrian: a Simple graphical representationof the relative occurrence and co-occurrence of events [Online]. Available Online at: https://CRAN.R-project.org/package=Mondrian [Accessed].
Simon, J.-C., Boutin, S., Tsuchida, T., Koga, R., Le Gallic, J.-F., Frantz, A., et al. (2011). Facultative symbiont infections affect aphid reproduction. PloS One 6:e21831. doi: 10.1371/journal.pone.0021831
Simon, J.-C., Carré, S., Boutin, M., Prunier–Leterme, N., Sabater–Muñoz, B., Latorre, A., et al. (2003). Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc. Biol. Sci. 270, 1703–1712. doi: 10.1098/rspb.2003.2430
Smith, A. H., Łukasik, P., O’connor, M. P., Lee, A., Mayo, G., Drott, M. T., et al. (2015). Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol. Ecol. 24, 1135–1149. doi: 10.1111/mec.13095
Starý, P. (2006). Aphid Parasitoids of the Czech Republic: (Hymenoptera: Braconidae, Aphidiinae). Czech: Akademie věd České republiky.
Vorburger, C., Gehrer, L., and Rodriguez, P. (2010). A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol. Lett. 6, 109–111. doi: 10.1098/rsbl.2009.0642
Vorburger, C., and Perlman, S. J. (2018). The role of defensive symbionts in host-parasite coevolution. Biol. Rev. Camb. Philos. Soc. 93, 1747–1764. doi: 10.1111/brv.12417
Wang, Z., Su, X. M., Wen, J., Jiang, L. Y., and Qiao, G. X. (2014). Widespread infection and diverse infection patterns of wolbachia in chinese aphids. Insect. Sci. 21, 313–325. doi: 10.1111/1744-7917.12102
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. [Online]. New York: Springer-Verlag.
Wood, S. (2017). Mixed GAM Computation Vehicle with Automatic Smoothness Estimation [Online]. Available Online at: https://www.maths.ed.ac.uk/swood34/ [Accessed]
Keywords: protective symbioses, insect parasitoids, parasitism pressure, symbiont communities, field approach, Acyrthosiphon pisum, Hamiltonella defensa
Citation: Leclair M, Buchard C, Mahéo F, Simon J-C and Outreman Y (2021) A Link Between Communities of Protective Endosymbionts and Parasitoids of the Pea Aphid Revealed in Unmanipulated Agricultural Systems. Front. Ecol. Evol. 9:618331. doi: 10.3389/fevo.2021.618331
Received: 16 October 2020; Accepted: 10 March 2021;
Published: 29 March 2021.
Edited by:
Roxanne Beinart, University of Rhode Island, United StatesReviewed by:
Jorge Doña, Prairie Research Institute, University of Illinois at Urbana–Champaign, United StatesCopyright © 2021 Leclair, Buchard, Mahéo, Simon and Outreman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yannick Outreman, eWFubmljay5vdXRyZW1hbkBhZ3JvY2FtcHVzLW91ZXN0LmZy
†These authors have contributed equally to this work
‡ORCID: Jean-Christophe Simon, orcid.org/0000-0003-0620-5835; Yannick Outreman, orcid.org/0000-0002-7337-3049
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.