- 1Department of Biology, Integrated Science Center, College of William and Mary, Williamsburg, VA, United States
- 2Department of Molecular and Cell Biology, University of California, Merced, Merced, CA, United States
Symbiotic bacteria in the Vibrionaceae are a dynamic group of γ-Proteobacteria that are commonly found throughout the world. Although they primarily are free-living in the environment, they can be commonly found associated with various Eukarya, either as beneficial or pathogenic symbionts. Interestingly, this dual lifestyle (free-living or in symbiosis) enables the bacteria to have enormous ecological breadth, where they can accommodate a variety of stresses in both stages. Here, we discuss some of the most common stressors that Vibrio bacteria encounter when in their free-living state or associated with an animal host, and how some of the mechanisms that are used to cope with these stressors can be used as an evolutionary advantage that increases their diversity both in the environment and within their specific hosts.
Introduction
The Vibrionaceae are a cosmopolitan family of gram-negative bacteria that form special associations with metazoan organisms (Thompson et al., 2006). These associations can be beneficial, commensal, and even pathogenic (Gomez-Gil et al., 2014). Close study of associations between the Vibrionaceae and their metazoan hosts have led to many important discoveries about the evolution of such unique partnerships, the molecular cross-talk that is established between different organisms, and how the partnership is more resilient to changes in the surrounding environment (Bourne et al., 2009). Examining how the entire “holobiome” (host + symbionts) responds to changes in the surrounding milieu can provide a barometer not only for the maintenance and selection of host-microbe relationships, but communities of other nested organisms that rely on each other for ecosystem health (McFall-Ngai, 2007; McFall-Ngai et al., 2013). This in turn produces a wide variety of host-symbiont assemblages, that would not exist without both partners being in association with one another. The holobiome thus is an independent “super organism” that can survive under conditions where neither symbiont or host cannot- thereby increasing the diversity and breadth of their habitat.
When not associated with a host, free-living, planktonic vibrios are subjected to multiple abiotic and biotic factors that select against various traits involved in colonization, infection, and eventually persistence (Figure 1; Piculell et al., 2008; Bright and Bulgheresi, 2010; Medina and Sachs, 2010). The distribution of vibrios that associate with animal hosts has been shown to be independent of host availability, while ecological factors such as temperature and salinity have been shown to impact Vibrio proliferation in the water column (Takemura et al., 2014). Recent reviews of the impact that environmental factors have on Vibrio biodiversity have focused primarily on human disease, aquaculture, or the population dynamics between the benign and pathogenic states of environmental vibrios (Takemura et al., 2014; Vezzulli et al., 2015). For example, a recent, long term evaluation of pathogenic Vibrio populations demonstrated that rising sea surface temperatures (SST) correlate with increased numbers of human infections (Vezzulli et al., 2012). Thus, temperature is just one abiotic factor that has tremendous effects on ecosystem health (Pantos et al., 2015). How Vibrio bacteria respond to such changes, and whether stress increases bacterial fitness and their ability to diversify among different associations with various eukaryotes, has yet to be determined. Most recently, the definition of “stress” in Vibrio was previously defined in an earlier review (Soto et al., 2010), focusing on environmental vibrios when not associated with a host.
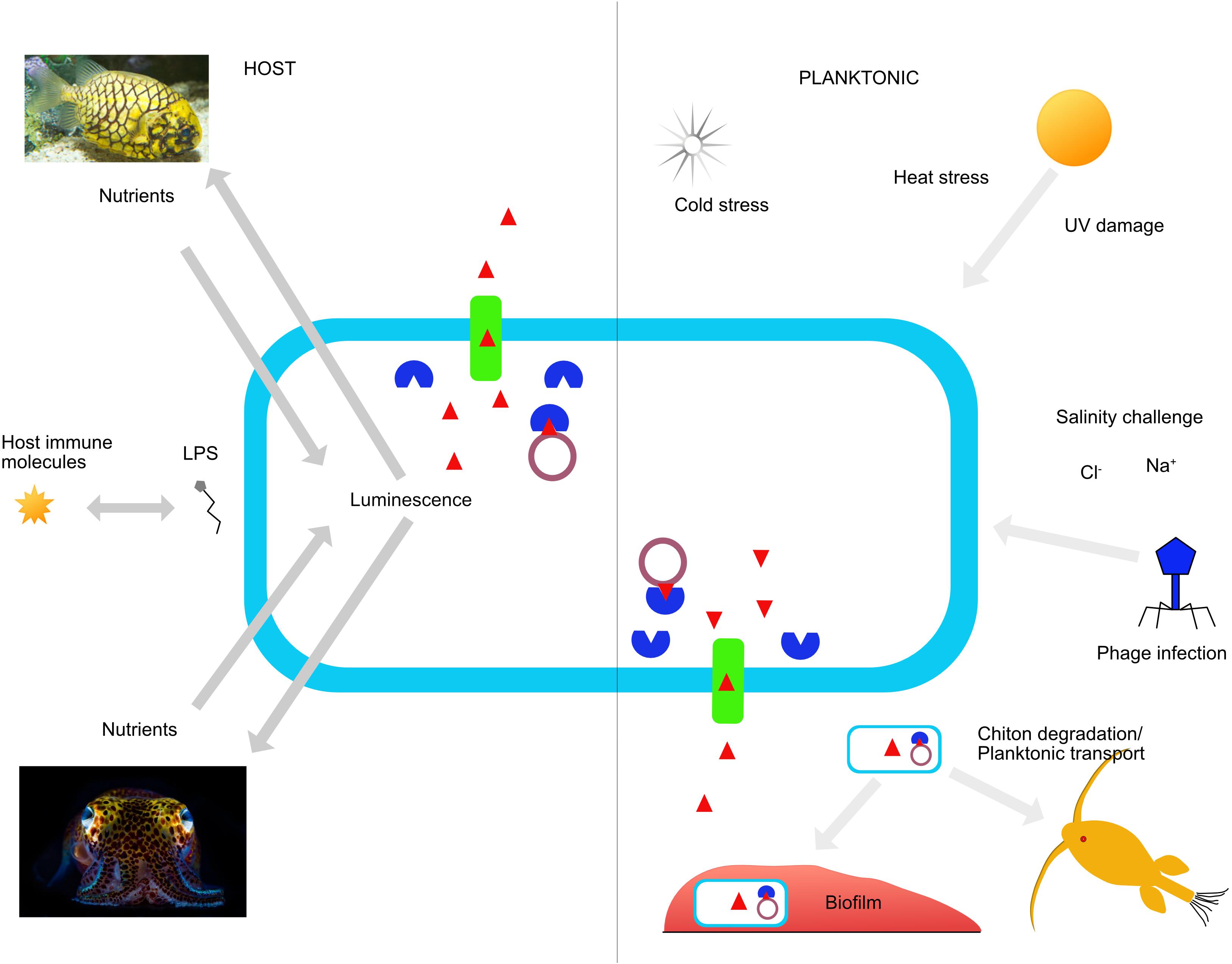
Figure 1. Response to environmental cues inside and outside the host by V. fischeri. A diagrammatic representation of the various impacts and responses that V. fischeri endures as a result of planktonic or host associated factors. Some mechanisms are universal, but induce different responses according to the environmental context. One example is quorum sensing, used for light generation and host immune response suppression inside a host, while these same signaling and response pathways are employed planktonically to ensure survival and persistence when nutrients are limiting. Host factors drive specificity among certain ecotypes of V. fischeri, thus creating a hierarchy of diverse strains that range in host compatibility. See Figure 2 for key.
The importance of studying the dynamics of these Vibrio-animal associations cannot be overstated. Impacting human health and economy, Vibrio bacteria are being probed for answers to some of the most pressing questions about how environmental factors influence bacterial fitness and biodiversity. Studies using models where both Vibrio bacteria and animal host can be studied independently or in symbioses, researchers have examined the effects of various abiotic and biotic factors that eventually shape these relationships (Chavez-Dozal and Nishiguchi, 2011; Chavez-Dozal et al., 2015). Evaluating not only both abiotic and biotic factors that facilitate Vibrio-animal relationships, but how various stressors influence genotype, physiology, environment, and geography, may help determine the basis for diversity among these unique symbiotic relationships (Piculell et al., 2008; Bright and Bulgheresi, 2010; Medina and Sachs, 2010; Pantos et al., 2015). Herein, the most common stressors that Vibrio bacteria encounter is discussed, including the free-living environment and animal hosts. Moreover, how these stressors can drive increased bacterial diversity in the free-living state and hosts is addressed. Three interesting empirical case studies have been completed with temperature and pH quite recently, which readers are encouraged to read (Cohen et al., 2019, 2020; Nourabadi and Nishiguchi, 2021).
Temperature Drives Bacterial Speciation
Vibrio bacteria have been observed in a wide range of habitats that vary in temperature, creating conditions that select for specific phenotypes that can survive and proliferate, both in their free-living mode as well as in their symbiotic lifestyle (Kimbell et al., 2002; Soto et al., 2009). Most notably, populations can vary, particularly when there are specific temperature gradients commonly found in nature (Jones et al., 2006; Coryell et al., 2018). For example, when sampling populations of Euprymna tasmanica, the sepiolid host squid for Vibrio fischeri in Australia, a distinct population break occurs within the symbionts where the habitat changes, particularly due to currents from the south (colder) and those from the north (warmer; Jones et al., 2006). This can also be observed in environments that have seasonal changes in water temperature, or those driven by currents (Coryell et al., 2018). Interestingly, when vibrio bacteria are “forced” into specific temperature regimes, they can rapidly adapt to survival outside the host, which oftentimes promotes the coevolution between hosts and their symbionts (Cohen et al., 2019). This temperature switch can subsequently facilitate gene expression of a wide variety of mechanisms that allow the bacterium to become more virulent to hosts (Kimes et al., 2002) as well as invade new host populations by relocating and surviving at a new environmental temperature (González-Escalona, 2015). Gene expression of such proteins such as Hsps (heat shock proteins), which are molecular chaperones that are upregulated in bacteria during heat stress (Madigan et al., 2018) are found to have an important role in the symbioses between nitrogen-fixing rhizobia (e.g., Rhizobium and Bradyrhizobium) and leguminous plants (Alexandre and Oliveira, 2016), but are not well studied in the Vibrionaceae. Albeit most stress comes in the form of higher temperatures (inciting stress in the host), drops in temperature can also elicit a change in gene expression (Liu et al., 2016), creating new ecological niches for those populations of Vibrio bacteria (Figure 2). This creates a situation where Vibrio bacteria must either adapt to these conditions or migrate to better environments (Richardson et al., 2012; Sunday et al., 2012).
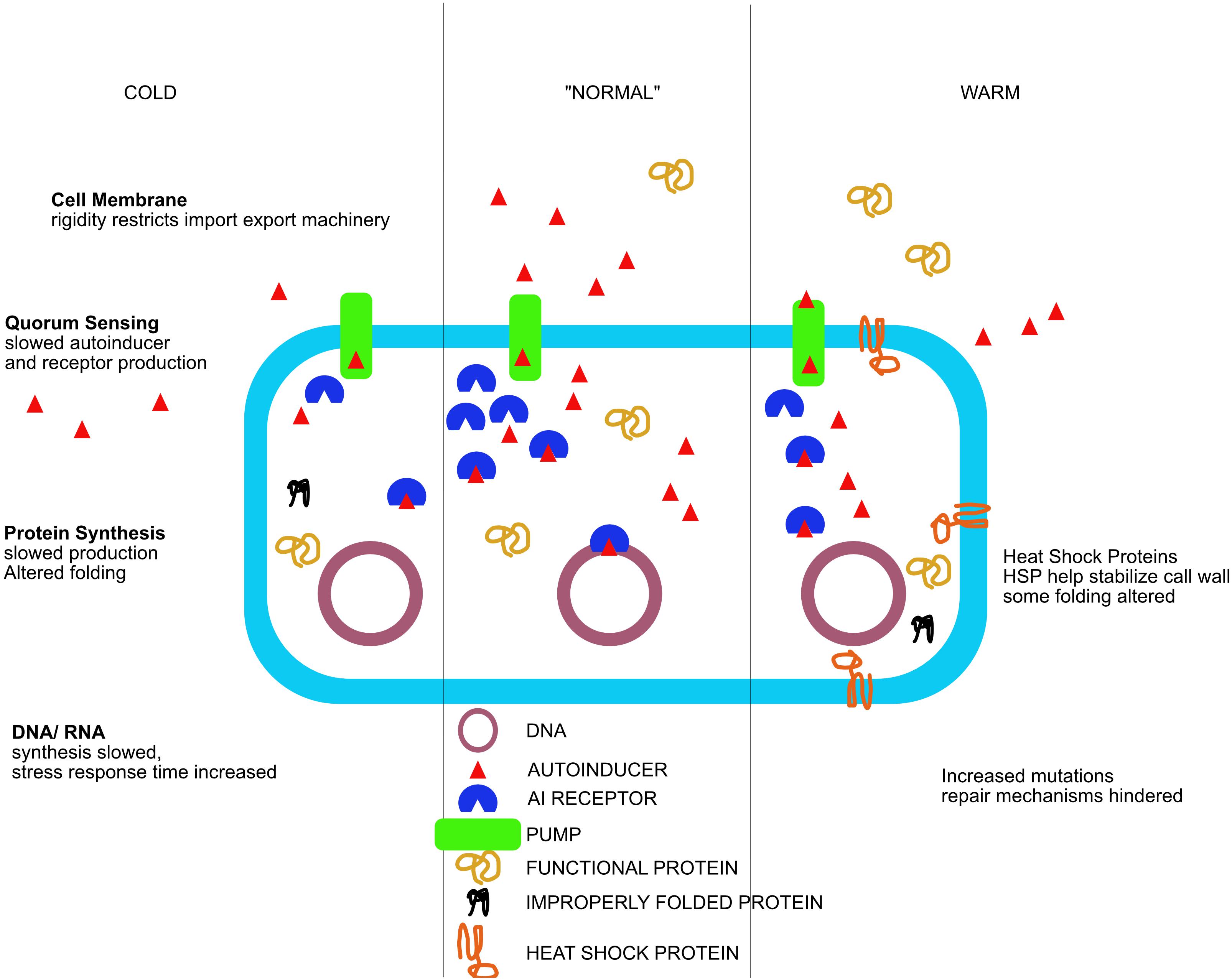
Figure 2. The response of environmentally transmitted Vibrio fischeri to temperature stress as a driver for diversification. Cell features and processes are affected differently when presented with a cold (left side of figure), or hot (right side of figure) environmental challenge. Metabolic and signaling processes are altered in response to these temperature changes, effecting both genotype and phenotype. Subsequently, these adapted strains diversify and become a novel ecotype, possibly leading to speciation.
Osmotic Stress Changes Phenotypic Variation
The Vibrionaceae have evolved mechanisms to cope with hypertonic and hypotonic stress. During hypotonic stress, cells must regulate to maintain the proper cytoplasmic concentrations of metabolites and ions, prevent lysis, and preserve ionic strength and pH (Bartlett, 2006; Nourabadi and Nishiguchi, 2021). Under hypotonicity, some vibrios may increase putrescine content to compensate for decreased K+ that are necessary to stabilize the phosphate backbones of nucleic acids (Soto et al., 2010). Hypertonicity, however, promotes dehydration and shriveling of cells. Microorganisms must be able to import or synthesize counterbalancing solutes that are compatible with metabolic and physiological functions. K+ uptake is a common mechanism to compensate for increased external osmolarity. Additionally, negative counter-ions (e.g., glutamate) must also be concurrently imported into the cell or synthesized de novo to sustain the same intracellular net charge (Sleator and Hill, 2001). Alternatively, cells can forgo K+ uptake and import or synthesize neutral compatible solutes, as they carry no charge. Ectoine is such an example and its biosynthesis may be unique to the genus Vibrio (Bartlett, 2006). Interestingly, Vibrio fischeri can synthesize disaccharide trehalose, which is also a neutral compatible solute. Vibrios can also incorporate polyunsaturated fatty acids in the cell membrane. This allows excess Na+ to exit the cell during hypertonic stress (Valentine and Valentine, 2004). Unsurprisingly, more avenues are available to bacteria to neutralize high osmolarity than hypotonicity. As a result, there are few vibrios that tolerate freshwater, Vibrio cholerae and Vibrio mimicus are two exceptions (Soto and Nishiguchi, 2014). Evidence exists that osmolar stress can affect host-microbe interactions—including pathogenesis, mutualisms, and commensalisms.
Generally, microbial diversity peaks at optimal environments and decreases at extreme ones (Ruhl et al., 2018). However, this conclusion has not always held true for environments where microbial diversity was evaluated with respect to osmotic conditions. For example, habitat salinity has been found to be linearly and inversely related to diversity for all three domains of life (Ruhl et al., 2018). In fact, salinity explained 44% of the variation in numerous diversity metrics (OTUs, Shannon index, and Phylogenetic Diversity) within the domain Bacteria. In the domain Archaea, diversity has even been found to increase at higher salinities (Ruhl et al., 2018). Since a diversity of compatible solutes can be used as osmolytes (e.g., ions, sugars, polyols, amino acids, and their respective derivatives such as ectoines and betaines) within the domains Bacteria and Archaea, there are myriad ways microbial diversification can proceed at extreme salinities (Da Costa et al., 2006; Empadinhas and Da Costa, 2006).
Ultraviolet Light and DNA Damage Induce Phenotypic Change
In bacteria, the SOS response is the change in gene expression that occurs when there is extensive DNA damage (Fry et al., 2005). Consequently, many genes involved in DNA repair are upregulated, including DNA polymerases IV (dinB) and V (umuCD). In the Vibrionaceae, the SOS response involves the expression of at least twenty genes (Sanchez-Alberola et al., 2012). For microorganisms that routinely engage in host-microbe interactions, the microbial chromosome can be a consistent target for inflicting substantial DNA damage by host immune cells via a respiratory burst (see oxidative stress) (O’Rourke et al., 2003). In the Vibrionaceae, induction of the SOS response is frequently associated with increased virulence and pathogenicity. However, the SOS response is not necessary for toxin production and intestinal colonization in suckling mice (Quinones et al., 2006), which is perplexing given the multitude of genes governed by the SOS system in Vibrio cholerae (Krin et al., 2018). Perhaps Vibrio cholerae utilizes other chromosomal repair systems such as base excision repair and mismatch repair, which can function independently of SOS (Davies et al., 2011). There is also the photolyase reaction, which is capable of DNA repair and requires light for activation (Thiagarajan et al., 2011; Yamamoto et al., 2017). Vibrio cholerae strains exist that are bioluminescent (Zo et al., 2009), and the visible light produced from bioluminescence can conceivably stimulate photolyase DNA repair. Photolyase DNA repair has also been proposed as an additional mechanism that is available to bioluminescent bacteria at night (Czyz et al., 2000). Thus, bioluminescent bacteria can continue to ameliorate DNA damage in the dark. Although light can be used as an activator for DNA repair, the roles of the SOS response and most bacterial DNA repair systems in the squid-Vibrio mutualism have not yet been rigorously investigated. Nonetheless, the photolyase reaction does not appear to have a major contribution to for combating stress in vibrio bacteria (Walker et al., 2006).
DNA damage caused by ultraviolet light can also lead to mutations that generate transient diversity via various mechanisms, which ultimately enhances a microbial population’s chance of survival (Aertsen and Michiels, 2005). These mechanisms include transient and constitutive mutators and the activation of contingency loci. These processes can facilitate the targeting of mutations in the genome that are customized to the specific needs of a certain time and particular environment (Massey and Buckling, 2002). Additionally, mutations induced by ultraviolet light can also reveal genetic variation within a microbial population that was previously latent due to “genetic buffering” mechanisms (Aertsen and Michiels, 2005). Consequently, the revelation of cryptic polygenic variation becomes possible in a microbial population as a result of ultraviolet light as an environmental stressor. Thus, even within a clonal population, intrapopulation diversity can be manufactured to ensure survival after prolonged exposure to ultraviolet light (Booth, 2002).
Oxidative Stress
Reactive oxygen species (ROS) include superoxide anion, peroxides, hydroxyl radical, and other toxic chemical agents containing oxygen (Imlay, 2019). ROS are highly unstable and cause deleterious effects by oxidizing cellular components. Bacteria are constantly exposed to ROS due to endogenous biochemical reactions (oxidative phosphorylation) and extrinsic processes external to the cell. Hydrogen peroxide produced from the photooxidation of water, via UV light from the sun, is a common cause of exogenous ROS (Imlay, 2019). Bacteria have evolved a diverse array of tools to neutralize ROS. Two regulons that combat oxidative stress include oxyR and soxRS, which have been well characterized is Escherichia coli (Chiang and Schellhorn, 2012). The regulons oxyR and soxRS control at least 30 and 100 genes, respectively. OxyR is mainly activated by the presence of hydrogen peroxide, while soxRS is stimulated by multiple ROS and other oxidizing agents. Catalase gene expression is also governed by oxyR. SoxRS is hypothesized to combat superoxide anion, including the upregulation of superoxide dismutase (Schellhorn et al., 2016). Both oxyR and soxRS are present within the Vibrionaceae (Vattanaviboon et al., 2003; Chiang and Schellhorn, 2012). Since superoxide anion and hydrogen peroxide are two ROS that regularly arise in living cells, bacteria have evolved regulons that are highly sensitive, including oxyR and soxRS. In Vibrio vulnificus, the soxRS regulon encompasses cadBA, a locus that encodes for a lysine-cadaverine antiporter and lysine decarboxylase, respectively (Kim et al., 2006). cadBA is involved in superoxide detoxification. The physiological response to oxidative stress at least partially overlaps with other bacterial stress responses, including heat shock (Storz et al., 1990). Several genes involved in the oxidative stress response are also controlled by rpoS, which is a stationary phase and general stress response regulator (Chiang and Schellhorn, 2012). Neutralizing the ROS produced by the respiratory burst of host immune cells is necessary for microorganisms involved in host-microbe interactions, including Vibrio fischeri during sepiolid squid colonization (Visick and Ruby, 1998). Additionally, bioluminescence, a key symbiotic phenotype in the squid-Vibrio mutualism is controlled by the redox-responsive regulator ArcA (Bose et al., 2007), where the ArcAB system represses the lux operon by ArcA binding and regulating the transcription of the luxCDABEG promoter. Although the deletion of arcA in V. fischeri increases luminescence in vitro, it does not repress symbiotic luminescence in vivo but rather causes a decrease in colonization competitiveness (Bose et al., 2007). Given that there may be alternative metabolic pathways that ArcAB regulates, as well as other genes responsible for reducing stress to oxygen and nitric oxide exposure (Aox; Dunn, 2018), the mechanisms of combating oxidative stress are complex. Oxidative stress also causes DNA damage that produces diverse subpopulations that can extend the range of conditions in which communities can thrive (Boles and Singh, 2008). These genetic variants are created due to double-stranded DNA breaks which are repaired by recombinatorial genes. In fact, this process is known to occur in biofilms, where endogenous oxidative stress is responsible for increasing the mutation rate as much as 10-fold (Boles and Singh, 2008; Steenackers et al., 2016). Little is known about the role of oxidative stress in Vibrio biofilms, but given the number of genetic regulators that modulate how the cell responds to oxygen and other respiratory stressors, they are more likely to have a major role in modulating the viability of biofilms in the environment and in possibly the development of the host light organs during symbiosis (Patelunas and Nishiguchi, 2018).
Grazing and Predation Select for More Diverse Phenotypes
Bacteria are prone to grazing and predation from numerous organisms. “Grazing” generally refers to consuming numerous prey individuals at once, whereas “predation” is used more commonly when the prey item is one individual (Atlas and Bartha, 1998). Vibrio bacteria have evolved numerous strategies to hinder being the prey of other organisms, especially against eukaryotic microbes such as ciliates and amoebas (Matz and Kjelleberg, 2005). After being ingested by protists, some vibrio species produce toxins which can kill grazers or predators. Other strains can block digestion and grow within the grazer or predator trying to feed on them. The grazer or predator is then killed or made ill by a bacterial infection (Matz and Kjelleberg, 2005). Additionally, oversizing and microcolony formation are other phenotypes that are selected to resist being preyed upon by microbial eukaryotes (Matz and Kjelleberg, 2005). Vibrios can also utilize their ability to develop complex biofilms that enables bacteria to escape grazing or predation, especially in aquatic habitats (Matz et al., 2005; Chavez-Dozal et al., 2013). Although vibrio bacterial biofilms can still be devoured, biofilms are generally less susceptible to bacterivory than their planktonic counterparts (cells in the water column) and the capability to produce strong biofilm varies from strain to strain. Recent data supports that predation selects for the hearty biofilm phenotype, and vibrio bacteria can use this strategy to their advantage to settle on various substrates or within their hosts (Chavez-Dozal et al., 2013; Chavez-Dozal et al., 2021).
Biofilm Regulation and Variability In Vibrio
Vibrio biofilms may be regulated by various quorum sensing mechanisms. This entails the use of a secreted autoinducer (AI), namely a homoserine lactone (HSL) derivative that is produced by each individual cell and is secreted to the exterior environment. Small concentrations of autoinducer do not have any type of effect on the Vibrio bacteria themselves, but once they are in high enough concentration (e.g., a quorum), the HSLs are able to induce or inhibit gene regulatory pathways by their interaction with the regulatory gene for a specific operon(s). Thus, this chemical communication amongst same or similar species of Vibrio bacteria allow them to regulate specific genetic mechanisms that are important for the entire population of Vibrio bacteria to thrive and outcompete other bacteria for resources. Biofilms are particularly important for bacterial survival in the environment, since they provide a refuge for exposure to antibiotics, phage predation, and protozoan grazing than their planktonic counterparts. Many of the components that define Vibrio biofilms such as their extracellular matrix (Faruque et al., 2006; Smith et al., 2015), polysaccharides, and attachment pili are regulated by AIs, as well as their dispersal once the biofilm community is no longer viable. Since biofilms are especially important for Vibrio bacteria to colonize and persist in a specific host species or for their subsequent dispersal, the molecular signals which induce the production and continuity of these biofilms are tightly regulated and specific. Biofilm communities are also varied, thus contributing to the diversity of species and species complexes that are represented in both mono and multi-type biofilms.
The most well studied type of regulation that is linked to biofilm production is that found in Vibrio cholerae. Determining how cholerae is able to transition between the environment (where no virulence factors are expressed) to the human gut (where virulence is expressed) has been the forefront of most cholera research. Studies focusing on temperature acclimation as a cue as well as other abiotic changes have shown that the molecular mechanisms that transition the bacterium to a free-living, non-pathogenic state to its ability to inflict damage in the host gastrointestinal tract are intricately related to complex signals that determine the behavior of the bacterium. For example, the temperature “sensor” that is capable of regulating pili and other virulence related loci is important in how V. cholerae can be innocuous in the water column, yet benign until it is ingested by the proper host where conditions are ripe for growth and exploitation of host resources (Townsley et al., 2016). V. cholerae bacteria are also able to detect important sources of energy, such as chitin in the surrounding environment (Meibom et al., 2004; Markov et al., 2015). V. cholerae can metabolize chitin as an carbon source, and at the same time, produce high levels of ammonia and antiprotozoan compounds that reduce grazing pressure on biofilm structures (Sun et al., 2015). Moreover, recent studies investigating filamentous V. cholerae strains have found that these morphotypes are capable of producing biofilms without the presence of matrix proteins, and instead use filaments to attach to chitin when in their free-living (environmental) state (Wucher et al., 2019). Additionally, changes in osmotic pressure between the external environment and the V. cholerae biofilm matrix promotes expansion on certain surfaces by increasing the colony size and subsequently enhances nutrient uptake and expanding on areas where other bacteria may be growing (Yan et al., 2017). This change in osmotic pressure in the matrix allows the cells that produce the matrix proteins to outcompete the non-matrix competing strains (cheaters) through physical exclusion (Yan et al., 2017). Thus, V. cholerae biofilms are capable of adapting to various abiotic or biotic factors to establish and maintain biofilm structure and maintain a colony that is more resistant to planktonic cells trying to invade the biofilm. Given that not all V. cholerae strains are capable of expressing these behaviors, selection of these given morphotypes demonstrates the breadth and diversity that Vibrio bacteria and in particular, V. cholerae has obtained for its bimodal life-style.
Interestingly, marine mutualistic bacteria such as V. fischeri, form biofilms inside their host squids during colonization, and are maintained until the squid “vents” the bacteria from the light organ with the cue of dawn. The bioluminescent bacterium V. fischeri, which is in association with sepiolid squids (Cephalopoda: Sepiolidae) is beneficial to the host by providing light for a behavior termed counterillumination (Jones and Nishiguchi, 2004). Sepiolid squids use the bioluminescence to match downwelling moonlight at night when they are out hunting or finding mates. In return, the squid host provides nutrients and increases the growth rate by as much as four times compared to when the bacteria are in the environment (Soto et al., 2009). This mutualism has been a model to study beneficial symbiosis for over 30 years, and the mechanisms of biofilm formation and light production have been well studied (Visick, 2009). Biofilm formation in V. fischeri has been closely tied to the success of colonization and proliferation in squid light organs; bacteria first form a biofilm on the outside of the pores that eventually lead into the crypt spaces that lie within the light organ of the squid (Nyholm and Nishiguchi, 2008). Differences exist between symbiotic V. fischeri, and those strains that are free-living and cannot colonize host squids (Nishiguchi et al., 1998; Nishiguchi, 2002; Chavez-Dozal and Nishiguchi, 2011), which may be due to the fact that these free-living strains lack specific regulators for biofilm production that are responsible for colonization and persistence (Chavez-Dozal et al., 2021; Mandel et al., 2009; Thompson et al., 2018). Additionally, biotic factors outside of the squid, such as protozoan grazing, have impacted how V. fischeri (both symbiotic and non-symbiotic) is susceptible to selective pressures that influence biofilm production and are thought to have driven the diversity of various biofilm phenotypes observed in nature (Chavez-Dozal et al., 2013).
Bacteriophages
Phages definitely impose a tremendous burden on bacterial populations as ecological parasites or even predators (Atlas and Bartha, 1998). Phages can impose negative frequency-dependent selection on their hosts, which can in turn maintain high levels of diversity in bacterial communities within various ecosystems. However, phages can also benefit bacterial populations in numerous ways (Taylor et al., 2018). Some virologists even hypothesize that phage can even serve as mutualists for their bacterial hosts under certain circumstances. As prophages inserted into bacterial chromosomes, host cells can gain antibiotic resistance, immunity to other phages, beneficial alterations to motility and chemotaxis, and enhanced quorum sensing properties (Taylor et al., 2018). Moreover, prophages might enable lysogenized bacteria to utilize metabolites or nutrients that were previously inaccessible. Prophages can even permit lysogens to colonize novel eukaryotic hosts or to acquire new virulence factors (e.g., CTXΦ phage for V. cholerae). Prophages as lysogen symbionts might be especially relevant for prokaryotes colonizing eukaryotic hosts, including animals (Taylor et al., 2018). For instance, prophages sometimes enable bacteria and archaea to evade or suppress the immune systems of multicellular hosts (Mirzaei and Maurice, 2017). The role of phages in the mutualism between sepiolid squid and Vibrio fischeri is largely unexplored. Since seawater is continuous with the fluid in the squid light organ, phages in the ocean could conceivably enter the light organ crypts to influence the squid-Vibrio symbiosis (Soto and Nishiguchi, 2014).
Phage introduce genetic diversity in bacteria by promoting horizontal gene transfer via transduction (Jiang and Paul, 1998). Transduction can facilitate the dissemination of genes for antibiotic resistance, the catabolism of novel carbon substrates, detoxification of deleterious substances, and virulence factors (Qiu et al., 2009; Von Wintersdorff et al., 2016; Bhandari and Karn, 2019). Conceivably, the spread of loci involved in stress response and elevated tolerance to drastic environmental fluctuations can also occur through transduction. However, the exact role of phage transduction in the lateral transfer of stress response genes is unclear. Since stress response in bacteria may involve operons if not regulons, generalized transduction would perhaps be a better candidate than specialized transduction, since more genetic material can be passed onto new host cells by phage (Popa et al., 2017). If transduction is able to promote the horizontal transfer of stress genetic networks, phage and environmental stressors will have an even larger role in impacting prokaryotic diversity than previously considered. Interestingly, one mechanism involved in managing cell membrane stress is the phage shock response (Joly et al., 2010). Consequently, stress regulons might even be able to suppress their own transduction by phage to some extent. For instance, phage infection in Vibrio alginolyticus leads to upregulation of molecular chaperones that bind to nucleic acid (RNA and DNA), which could inhibit phage genome replication (Luo et al., 2018).
Conclusion
There is a need to ascertain how quickly organisms can adapt to different environments; with the noticeable change in global climate, it is to our benefit to determine if these changes will increase or decrease biodiversity (Duarte, 2014). Using model symbiotic associations to examine how microbial diversity responds to environmental change can potentially offer insights into the organisms, communities, and ecosystems that these microbes interact with. Understanding the interactions between genes, function, and the factors which select for specific Vibrio genotypes will help determine whether microbial associations with other organisms will survive in response to stress from the environment.
Author Contributions
WS and MN designed and wrote the manuscript equally. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by NASA EXO-80NSSC18K1053 to MN and the School of Natural Sciences at UC Merced. WS was supported by startup funds from the College of William and Mary.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank R. Coryell with help for the figures.
References
Aertsen, A., and Michiels, C. W. (2005). Diversify or die: generation of diversity in response to stress. Crit. Rev. Microbiol. 31, 69–78. doi: 10.1080/10408410590921718
Alexandre, A., and Oliveira, S. (2016). “Heat shock response in bacteria with large genomes: lessons from rhizobia,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, ed. F. J. de Bruijn (Hoboken, NJ: JohnWiley & Sons, Inc), 737–746. doi: 10.1002/9781119004813.ch70
Atlas, R. M., and Bartha, R. (1998). Microbial Ecology: Fundamentals and Applications, 4th Edn. California: Benjamin Cummings, Menlo Park.
Bartlett, D. H. (2006). “Extremophilic vibrionaceae,” in The Biology of the Vibrios, eds F. L. Thompson, B. Austin, and J. Swings (Washington, DC: ASM Press).
Bhandari, G., and Karn, S. K. (2019). “Evaluation of horizontal gene transfer of catabolic genes and its application in bioremediation,” in Smart Bioremediation Technologies: Microbial Enzymes, ed. P. Bhatt (Cambridge: Academic Press), 359–372. doi: 10.1016/b978-0-12-818307-6.00019-6
Boles, B. R., and Singh, P. K. (2008). Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 105, 12503–12508. doi: 10.1073/pnas.0801499105
Booth, I. R. (2002). Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78, 19–30. doi: 10.1016/s0168-1605(02)00239-8
Bose, J. L., Kim, U., Bartkowski, W., Gunsalus, R. P., Overley, A. M., Lyell, N. L., et al. (2007). Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65, 538–553. doi: 10.1111/j.1365-2958.2007.05809.x
Bourne, D. G., Garren, M., Work, T. M., Rosenberg, E., Smith, G. W., and Harvell, C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Bright, M., and Bulgheresi, S. (2010). A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. doi: 10.1038/nrmicro2262
Chavez-Dozal, A., Gorman, C., Erken, M., Steinberg, P. D., McDougald, D., and Nishiguch, M. K. (2013). Predation response of Vibrio fischeri biofilms to bacterivorus protists/phagotrophic protozoa. Appl. Environ. Microbiol. 79, 553–558. doi: 10.1128/aem.02710-12
Chavez-Dozal, A., Soto, W., and Nishiguchi, M. K. (2021). Identification of a transcriptomic network underlying the wrinkly and smooth phenotypes of Vibrio fischeri. J. Bacteriol. 203:e00259-20. doi: 10.1128/JB.00259-20
Chavez-Dozal, A. A., Gorman, C., and Nishiguchi, M. K. (2015). Proteomic and metabolomic profiles demonstrate variation among free-living and symbiotic Vibrio fischeri biofilms. BMC Microbiol. 15:226.
Chavez-Dozal, A. A., and Nishiguchi, M. K. (2011). Variation in biofilm formation among symbiotic and free−living strains of Vibrio fischeri. J. Basic Microbiol. 51, 452–458. doi: 10.1002/jobm.201000426
Chiang, S. M., and Schellhorn, H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169. doi: 10.1016/j.abb.2012.02.007
Cohen, M. L., Mashanova, E. V., Jagannathan, S. V., and Soto, W. (2020). Adaptation to pH stress by Vibrio fischeri can affect its symbiosis with the Hawaiian bobtail squid (Euprymna scolopes). Microbiology 166, 262–277. doi: 10.1099/mic.0.00088
Cohen, M. L., Mashanova, E. V., Rosen, N. M., and Soto, W. (2019). Adaptation to temperature stress by Vibrio fischeri facilitates this microbe’s symbiosis with the Hawaiian bobtail squid (Euprymna scolopes). Evolution 73, 1885–1897. doi: 10.1111/evo.13819
Coryell, R. L., Turnham, K. E., de Jesus Ayson, E. G., Lavilla-Pitogo, C., Alcala, A. C., Sotto, F., et al. (2018). Phylogeographic patterns in the Philippine Archipelago influence symbiont diversity in the bobtail Squid-Vibrio mutualism. Ecol. Evol. 8, 7421–7435. doi: 10.1002/ece3.4266
Czyz, A., Wróbel, B., and Węgrzyn, G. (2000). Vibrio harveyi bioluminescence plays a role in stimulation of dna repair. Microbiology 146, 283–288. doi: 10.1099/00221287-146-2-283
Da Costa, M. S., Santos, H., and Galinski, E. A. (2006). An overview of the role and diversity of compatible solutes in bacteria and archaea. Adv. Biochem. Eng. Biotechnol. 61, 117–153. doi: 10.1007/bfb0102291
Davies, B. W., Bogard, R. W., Dupes, N. M., Gerstenfeld, T. A. I., Simmons, L. A., and Mekalanos, J. J. (2011). Dna damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 7:e1001295. doi: 10.1371/journal.ppat.1001295
Duarte, C. M. (2014). Global change and the future of the ocean: a grand challenge for marine sciences. Front. Mar. Sci. 1, 1–16.
Dunn, A. K. (2018). Alternative oxidase activity reduces stress in Vibrio fischeri cells exposed to nitric oxide. J. Bacteriol. 200:e00797-17. doi: 10.1128/JB.00797-17
Empadinhas, N., and Da Costa, M. S. (2006). Diversity and biosynthesis of compatible solutes in hyper/thermophiles. Int. Microbiol. 9, 199–206.
Faruque, S. M., Biswas, K., Nashir Udden, S. M., Ahmad, Q. S., Sack, D. A., Nair, G. B., et al. (2006). Transmissibility of cholera: In vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U.S.A. 103, 6350–6355. doi: 10.1073/pnas.0601277103
Fry, R. C., Begley, T. J., and Samson, L. D. (2005). Genome-wide responses to dna-damaging agents. Annu. Rev. Microbiol. 59, 357–377. doi: 10.1146/annurev.micro.59.031805.133658
Gomez-Gil, B., Thompson, C. C., Matsumura, Y., Sawabe, T., Iida, T., Christen, R., et al. (2014). “The famlily vibrionaceae,” in The Prokaryotes: Gammaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 659–747. doi: 10.1007/978-3-642-38922-1_225
González-Escalona, N., Gavilan, R. G., Brown, E. W., and Martinez-Urtaza, J. (2015). Transoceanic spreading of pathogenic strains of Vibrio parahaemolyticus with distinctive genetic signatures in the recA gene. PLoS One 10:e0117485. doi: 10.1371/journal.pone.0117485.g004
Imlay, J. A. (2019). Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 21, 521–530. doi: 10.1111/1462-2920.14445
Jiang, S. C., and Paul, J. H. (1998). Gene Transfer by transduction in the marine environment. Appl. Environ. Microbiol. 64, 2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998
Joly, N., Engl, C., Jovanovic, G., Huvet, M., Toni, T., Sheng, X., et al. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827. doi: 10.1111/j.1574-6976.2010.00240.x
Jones, B. W., and Nishiguchi, M. K. (2004). Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes (Mollusca: Cephalopoda). Mar. Biol. 144, 1151–1155. doi: 10.1007/s00227-003-1285-3
Jones, B. W., Lopez, J. E., Huttenberg, J., and Nishiguchi, M. K. (2006). Population structure between environmentally transmitted Vibrios and bobtail squids using nested clade analysis. Mol. Ecol. 15, 4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x
Kim, J. S., Choi, S. H., and Lee, J. K. (2006). Lysine decarboxylase expression by Vibrio vulnificus is induced by soxR in response to superoxide stress. J. Bacteriol. 188, 8586–8592. doi: 10.1128/jb.01084-06
Kimes, N. E., Grim, C. J., Johnson, W. R., Hasan, N. A., Tall, B. D., Kothary, M. H., et al. (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6, 835–846. doi: 10.1038/ismej.2011.154
Kimbell, J. R., McFall-Ngai, M. J., and Roderick, G. K. (2002). Two genetically distinct populations of bobtail squid. Euprymna scolopes, exist on the island of O’ahu. Pac. Sci. 56, 347–355. doi: 10.1353/psc.2002.0024
Krin, E., Pierlé, S. A., Sismeiro, O., Jagla, B., Dillies, M.-A., Varet, H., et al. (2018). Expansion of the sos regulon of Vibrio cholerae through extensive transcriptome analysis and experimental validation. BMC Genomics 19:373.
Liu, R., Chen, H., Zhang, R., Zhou, Z., Hou, Z., Gao, D., et al. (2016). Comparative transcriptome analysis of Vibrio splendidus JZ6 reveals the mechanism of its pathogenicity at low temperatures. Appl. Environ. Microbiol. 82, 2050–2061. doi: 10.1128/aem.03486-15
Luo, P., Liu, Q., Deng, Y., Tian, Y., Yun, L., and Hu, C. (2018). Strand-specific RNA-Seq analysis provides first insight into transcriptome response of Vibrio alginolyticus to phage infection. Mar. Genomics 38, 5–8. doi: 10.1016/j.margen.2017.05.011
Madigan, M. T., Bender, K. S., Buckley, D. H., Sattley, W. M., and Stahl, D. A. (2018). Brock Biology of Microorganisms, 15th Edn. New York, NY: Pearson.
Mandel, M. J., Wollenberg, M. S., Stabb, E. V., Visick, K. L., and Ruby, E. G. (2009). A single regulatory gene is sufficient to alter bacterial host range. Nature 458, 215–218. doi: 10.1038/nature07660
Markov, E. Y., Kulikalova, E. S., Urbanovich, L. Y., Vishnyakov, V. S., and Balakhonov, S. V. (2015). Chitin and products of its hydrolysis in Vibrio cholerae ecology. Biochem (Moscow) 80, 1109–1116. doi: 10.1134/s0006297915090023
Massey, R. C., and Buckling, A. (2002). Environmental regulation of mutation rates at specific sites. Trends Microbiol. 10, 580–584. doi: 10.1016/s0966-842x(02)02475-7
Matz, C., and Kjelleberg, S. (2005). Off the hook – how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. doi: 10.1016/j.tim.2005.05.009
Matz, C., McDougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., and Kjelleberg, S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 102, 16819–16824. doi: 10.1073/pnas.0505350102
McFall-Ngai, M. J., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Loso, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236.
Medina, M., and Sachs, J. L. (2010). Symbiont genomics, our new tangled bank. Genomics 95, 129–137. doi: 10.1016/j.ygeno.2009.12.004
Meibom, K. L., Li, X. B., Nielsen, A. T., Wu, C.-Y., Roseman, S., and Schoolnik, G. K. (2004). The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529. doi: 10.1073/pnas.0308707101
Mirzaei, M. K., and Maurice, C. F. (2017). Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15, 397–408. doi: 10.1038/nrmicro.2017.30
Nishiguchi, M. K. (2002). Host-symbiont recognition in the environmentally transmitted sepiolid squid-Vibrio mutualism. Microb. Ecol. 44, 10–18. doi: 10.1007/bf03036870
Nishiguchi, M. K., Ruby, E. G., and McFall-Ngai, M. J. (1998). Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in Sepiolid squid-Vibrio symbioses. Appl. Environ. Microbiol. 64, 3209–3213. doi: 10.1128/aem.64.9.3209-3213.1998
Nourabadi, N., and Nishiguchi, M. K. (2021). pH adaptation drives diverse phenotypes in a beneficial bacterium-host mutualism. Front. Ecol. Evol. 9:611411. doi: 10.3389/fevo.2021.611411
Nyholm, S. V., and Nishiguchi, M. K. (2008). The evolutionary ecology of a sepiolid squid-Vibrio association: From cell to environment. Vie et Milieu 58, 175–184.
O’Rourke, E. J., Chevalier, C., Pinto, A. V., Thiberge, J. M., Ielpi, L., Labigne, A., et al. (2003). Pathogen dna as target for host-generated oxidative stress: role for repair of bacterial dna damage in Helicobacter pylori colonization. Proc. Natl. Acad. Sci. U.S.A. 100, 2789–2794. doi: 10.1073/pnas.0337641100
Pantos, O., Bongaerts, P., Dennis, P. G., Tyson, G. W., and Hoegh-Guldberg, O. (2015). Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. Int. Soc. Microb. Ecol. J. 9, 1916–1927. doi: 10.1038/ismej.2015.3
Patelunas, A. J., and Nishiguchi, M. K. (2018). Vascular architecture in the bacteriogenic light organ of Euprymna tasmanica (Cephalopoda: Sepiolidae). Inverteb. Biol. 137, 240–249. doi: 10.1111/ivb.12223
Piculell, B. J., Hoeksema, J. D., and Thompson, J. N. (2008). Interactions of biotic and abiotic environmental factors in an ectomycorrhizal symbiosis, and the potential for selection mosaics. BMC Biol. 6:23. doi: 10.1186/1741-7007-6-23
Popa, O., Landan, G., and Dagan, T. (2017). Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 11, 543–554. doi: 10.1038/ismej.2016.116
Qiu, X., Kulasekara, B. R., and Lory, S. (2009). Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Microb. Pathog. 6, 126–139. doi: 10.1159/000235767
Quinones, M., Davis, B. M., and Waldor, M. K. (2006). Activation of the Vibrio cholerae sos response is not required for intestinal cholera toxin production or colonization. Infect. Immunity 74, 927–930. doi: 10.1128/iai.74.2.927-930.2006
Richardson, A. J., Brown, C. J., Brander, K., Bruno, J. F., Buckley, L., Burrows, M. T., et al. (2012). Climate change and marine life. Biol. Lett. 8, 907–909.
Ruhl, I. A., Grasby, S. E., Haupt, E. S., and Dunfield, P. F. (2018). Analysis of microbial communities in natural halite springs reveals a domain−dependent relationship of species diversity to osmotic stress. Environ. Microbiol. Rep. 10, 695–703. doi: 10.1111/1758-2229.12695
Sanchez-Alberola, N., Campoy, S., Barbé, J., and Erill, I. (2012). Analysis of the sos response of Vibrio and other bacteria with multiple chromosomes. BMC Genomics 13:58. doi: 10.1186/1471-2164-13-58
Schellhorn, H. E., Mohiuddin, M., Hammond, S. M., and Steven Botts, S. (2016). “Regulators of oxidative stress response genes in Escherichia coli and their conservation in bacteria,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, ed. F. J. de Bruijn (Hoboken, NJ: JohnWiley & Sons, Inc), 632–637. doi: 10.1002/9781119004813.ch60
Sleator, R. D., and Hill, C. (2001). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
Smith, D. R., Maestre-Reyna, M., Lee, G., Gerard, H., Wang, A., and Watnick, P. I. (2015). In situ proteolysis of the Vibrio cholerae matrix protein RbmA promotes biofilm recruitment. Proc. Natl. Acad. Sci. U.S.A. 112, 10491–10496. doi: 10.1073/pnas.1512424112
Soto, W., Gutierrez, J., Remmenga, M. D., and Nishiguchi, M. K. (2009). Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb. Ecol. 57, 140–150. doi: 10.1007/s00248-008-9412-9
Soto, W., Lostroh, C. P., and Nishiguchi, M. K. (2010). “Physiological responsesto stress in the vibrionaceae,” in Cooperation and Stress in Biology, eds J. Seckback and M. Grube (New York, NY: Springer).
Soto, W., and Nishiguchi, M. K. (2014). Microbial experimental evolution as a novel researchapproach in the vibrionaceae and squid-Vibrio symbiosis. Front. Microbiol. 5:593.
Steenackers, H. P., Parijs, I., Foster, K. R., and Vanderleyden, J. (2016). Experimental evolution in biofilm populations. FEMS Microbiol. Rev. 40, 373–397. doi: 10.1093/femsre/fuw002
Storz, G., Tartaglia, L. A., Farr, S. B., and Ames, B. N. (1990). Bacterial defenses against oxidative stress. Trends Genet. 6, 363–368. doi: 10.1016/0168-9525(90)90278-e
Sun, S., Tay, Q., Kjelleberg, S., Rice, S. A., and McDougald, D. (2015). Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J. 9, 1812–1820. doi: 10.1038/ismej.2014.265
Sunday, J. M., Bates, A. E., and Dulvy, N. K. (2012). Thermal tolerance and the global redistribution of animals. Nat. Clim. Change 2:686. doi: 10.1038/nclimate1539
Takemura, A. F., Chien, D. M., and Polz, M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38.
Taylor, V. L., Fitzpatrick, A. D., Islam, Z., and Maxwell, K. L. (2018). The diverse impacts of phage morons on bacterial fitness and virulence. Adv. Virus Res. 103, 1–31. doi: 10.1016/bs.aivir.2018.08.001
Thiagarajan, V., Byrdin, M., Eker, A. P., Müller, P., and Brette, K. (2011). Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV. Proc. Natl. Acad. Sci. U.S.A. 108, 9402–9407. doi: 10.1073/pnas.1101026108
Thompson, C. M., Marsden, A. E., Tischler, A. H., Koo, J., and Visick, K. L. (2018). Vibrio fischeri biofilm formation prevented by a trio of regulators. Appl. Environ. Microbiol. 84, e1257–e1218. doi: 10.1128/AEM.01257-18
Thompson, F. L., Austin, B., and Swings, J. (2006). The Biology of Vibrios. Washington, DC: ASM Press.
Townsley, L., Mangus, M. P., Mehic, S., and Yildiz, F. H. (2016). Response of Vibrio cholerae to low-temperature shifts: CspV regulation of type VI secretion, biofilm formation, and association with zooplankton. Appl. Environ. Microbiol. 82, 4441–4452. doi: 10.1128/aem.00807-16
Valentine, R. C., and Valentine, D. L. (2004). Omega-3 fatty acids in cellular membranes: a unified concept. Prog. Lipid Res. 43, 383–402. doi: 10.1016/j.plipres.2004.05.004
Vattanaviboon, P., Panmanee, W., and Mongkolsuk, S. (2003). Induction of peroxide and superoxide protective enzymes and physiological crossprotection against peroxide killing by a superoxide generator in Vibrio harveyi. FEMS Microbiol. Lett. 221, 89–95. doi: 10.1016/s0378-1097(03)00172-1
Vezzulli, L., Brettar, I., Pezzati, E., Reid, P. C., Colwell, R. R., Höfle, M. G., et al. (2012). Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J. 6:21. doi: 10.1038/ismej.2011.89
Vezzulli, L., Höfle, M., Pruzzo, C., Pezzati, E., and Brettar, I. (2015). Effects of global warming on vibrio ecology. Microbiol. Spect. 3.
Visick, K. L. (2009). An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol. Microbiol. 74, 782–789. doi: 10.1111/j.1365-2958.2009.06899.x
Visick, K. L., and Ruby, E. G. (1998). The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180, 2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998
Von Wintersdorff, C. J. H., Penders, J., Van Niekerk, J. M., Mills, N. D., Majumder, S., VanAlphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173.
Walker, E. L., Bose, J. L., and Stabb, E. V. (2006). Photolyase confers resistance to UV Light but does not contribute to the symbiotic benefit of bioluminescence in Vibrio fischeri es114. Appl. Environ. Microbiol. 72, 6600–6606. doi: 10.1128/aem.01272-06
Wucher, B. R., Bartlett, T. M., Hoyos, M., Papenfort, K., Persat, A., and Nadell, C. D. (2019). Vibrio cholerae filamentation promotes chitin surface attachment at the expense of competition in biofilms. Proc. Natl. Acad. Sci. U.S.A. 116, 14216–14221. doi: 10.1073/pnas.1819016116
Yamamoto, J., Shimizu, K., Kanda, T., Hosokawa, Y., Iwai, S., Plaza, P., et al. (2017). Loss of fourth electron-transferring tryptophan in animal (6-4) photolyase impairs dna repair activity in bacterial cells. Biochemistry 56, 5356–5364. doi: 10.1021/acs.biochem.7b00366
Yan, J., Nadell, C. D., Stone, H. A., Wingreen, N. S., and Bassler, B. L. (2017). Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat. Commun. 8:327.
Keywords: symbiosis, stress, Vibrio, temperature, selection
Citation: Soto W and Nishiguchi MK (2021) Environmental Stress Selects for Innovations That Drive Vibrio Symbiont Diversity. Front. Ecol. Evol. 9:616973. doi: 10.3389/fevo.2021.616973
Received: 13 October 2020; Accepted: 21 May 2021;
Published: 17 June 2021.
Edited by:
Kimberly B. Ritchie, University of South Carolina Beaufort, United StatesReviewed by:
David D. Dunigan, University of Nebraska-Lincoln, United StatesAlison Gould, California Academy of Sciences, United States
Copyright © 2021 Soto and Nishiguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele K. Nishiguchi, bmlzaEB1Y21lcmNlZC5lZHU=
 William Soto
William Soto Michele K. Nishiguchi
Michele K. Nishiguchi