
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Ecol. Evol. , 13 May 2021
Sec. Conservation and Restoration Ecology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.613200
This article is part of the Research Topic Environmental DNA Innovations for Conservation View all 19 articles
 Taylor M. Wilcox1*
Taylor M. Wilcox1* Anthony Caragiulo2
Anthony Caragiulo2 Joseph C. Dysthe1
Joseph C. Dysthe1 Thomas W. Franklin1
Thomas W. Franklin1 Daniel H. Mason1
Daniel H. Mason1 Kevin S. McKelvey1
Kevin S. McKelvey1 Katherine E. Zarn1
Katherine E. Zarn1 Michael K. Schwartz1
Michael K. Schwartz1Jaguar (Panthera onca) are of conservation concern and occur at very low densities in the northern portion of their range in northern Mexico and the southwestern United States. Environmental DNA sampling to detect genetic material from drinking water may be an effective approach for jaguar detection in these arid landscapes. Here we develop a qPCR assay for the detection of jaguar mitochondrial DNA, show that large quantities of DNA (mean 66,820 copies/L) can be found in the drinking water of captive animals, and observe detectable levels of DNA (80 copies/L) in a wild habitat with known jaguar populations. We suggest that environmental DNA sampling may represent a useful, complementary sampling tool for detection of rare jaguars, although effective application would require careful consideration of DNA persistence time in the environment.
Jaguar (Panthera onca) are of conservation concern, particularly in the northern portion of their range in northern Mexico and the southwestern United States (Brown and González, 2000). Within the United States, the species is Federally protected (U.S. Federal Register 37 FR 6476) and occurs at extremely low abundances, with individuals rarely being detected. Current jaguar monitoring in the United States primarily uses camera traps and genetic testing of scat samples which may be located with the use of detection dogs (Culver, 2016). These approaches can be labor intensive, involving hundreds of cameras and tens of thousands of images (Culver, 2016). Additional sampling tools could help build a better understanding of jaguar distributions at the northern margin of their range and enable more effective protection of rare individuals and management of their habitat.
Environmental DNA (eDNA) sampling—the inference of species presence from genetic material in the environment—has been rapidly adopted for rare aquatic species sampling (e.g., Cristescu and Hebert, 2018; Sepulveda et al., 2020). Recent studies suggest that under the right circumstances, eDNA sampling of water may also be an effective approach for the detection of terrestrial species (e.g., Williams et al., 2018; Franklin et al., 2019). Harper et al. (2019) and Sales et al. (2020) documented the ability to detect terrestrial species from water samples, in some cases with detection rates comparable to camera trapping. Although there are likely multiple routes of transmission, DNA deposited when drinking is likely a major contributor of terrestrial species eDNA in aquatic habitats (Rodgers and Mock, 2015). In arid landscapes, drinking water may be scarce and represent relatively concentrated sources of jaguar eDNA.
Here we describe a hydrolysis assay developed for detection of jaguar mitochondrial DNA in environmental samples, then demonstrate this tool on water samples taken from known and suspected jaguar drinking water sources.
Using sequence data from the NCBI GenBank database and the R (R Core Development Team, 2020) package DECIPHER (Wright, 2016), we designed and tested candidate quantitative PCR (qPCR) primers in silico, targeting jaguar mitochondrial DNA to the exclusion of 14 other felid species that either potentially co-occur with jaguar in North America or are closely related to jaguar (Table 1). Although only four jaguar mitochondrial genome sequences were included in this initial screen, there are low levels of genetic diversity and structure within the northern range of this species (e.g., Wultsch et al., 2016). We then selected one of these primer sets within the mitochondrial gene ATP6 for hydrolysis probe development using PrimerExpress software (Thermo Fisher Scientific). These primers were selected based on a scan of the entire mitochondrial genome, but this same locus has also been found to have particularly good species discriminatory power across Carnivora (Chaves et al., 2012). We also conducted an in silico analysis of primer specificity through a BLAST (Atschul et al., 1990) search against GenBank to identify any unexpected cross-amplification with non-felids, but we did not attempt to use an in silico approach to evaluate potential cross-amplification of other rare felids which are not expected to be found in North America. We sourced the primers from Integrated DNA Technologies and obtained a FAM-labeled minor groove-binding (MGB) non-fluorescent quencher (NFQ) probe from Thermo Fisher Scientific (Table 2).
We validated assay specificity in vitro by testing tissue-extracted DNA from felids which may be found in the southwestern U.S. (Felis catus, Puma concolor, Lynx rufus, Leopardus pardalis, Puma yagouaroundi) or are closely related to jaguar [Acinonyx jubatus (n = 2), Leptailurus serval (n = 2), Panthera leo (n = 3), Panthera tigris (n = 3)]. These samples were sourced from the collections of partners for other projects and were collected in accordance with any relevant animal care guidelines. We extracted DNA using the DNeasy Blood and Tissue Kit (QIAGEN), quantified DNA using a Qubit fluorometer (Invitrogen), and diluted extracts to approximately 0.1 ng genomic DNA per microliter (0.4 ng gDNA per reaction), then analyzed with qPCR as described below. There was low-level amplification in one African lion sample. Low-level contamination of tissue-derived DNA samples that is only detected when used for eDNA-type applications is common (Rodgers, 2017) and was suspected in this case because there are many basepair mismatches between African lion and the jaguar assay (Table 1). To confirm this, we ensured that the assay did not amplify a synthetic gene fragment with the same sequence as African lion (gBlock; Integrated DNA Technologies), diluted to 6,250 and 1,250 copies per reaction.
We tested the ability of the assay to amplify jaguar DNA by analyzing DNA extracted from 10 jaguar scats that were collected in Belize and included in Menchaca et al. (2019; AC; Sackler Institute for Comparative Genomics). We tested the ability to detect jaguar DNA from drinking water by analyzing DNA extracted from six water samples provided to captive animals at the Phoenix Zoo in Phoenix, Arizona (n = 5; Figure 1) and Banana Bank Lodge in Belize (n = 1, Table 3). This sampling required minimal animal disturbance and sampling at the Phoenix Zoo was approved by the Arizona Center for Nature Conservation Research Committee. Finally, we analyzed five water samples from ponds in Belize where wild jaguar are known to occur regionally, but whose recent use of these habitats is unknown. Water samples were collected as described in Carim et al. (2016b). Briefly, for each sample, 5 L of water was drawn through a 47 mm diameter, 1.5 micron pore size glass microfiber filter paper using an electric peristaltic pump at the sampling site. The filter paper was then stored in silica desiccant until received at the lab (<2 weeks) when they were archived at −20°C until extraction. Filters were handled with sterile forceps. Filter cups, forceps, and all other sampling supplies were prepared in a dedicated, restricted-access room at the National Genomics Center for Wildlife and Fish Conservation and were sterilized using a 50% household bleach solution and ultrapure water (4% hypochlorite solution).

Figure 1. Image of a jaguar drinking at the Phoenix Zoo, AZ, United States. Photo credit: David Bissegger.
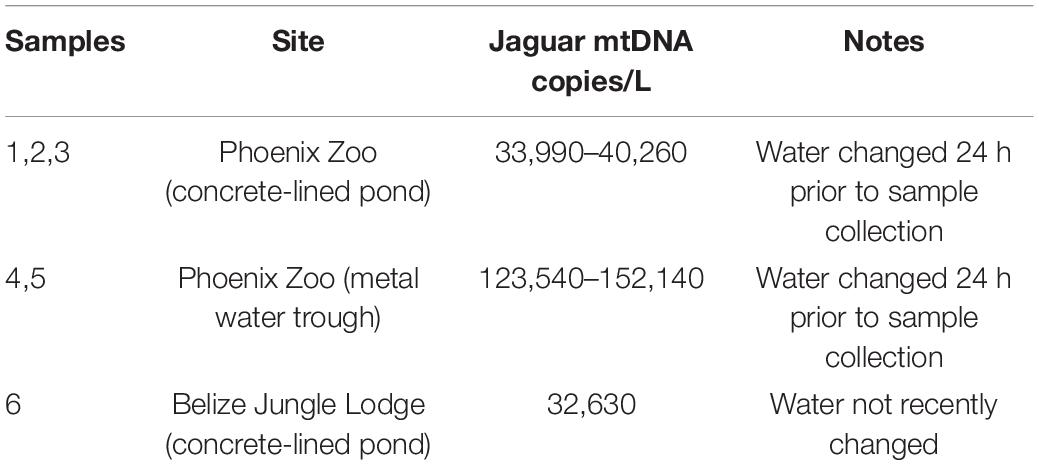
Table 3. Jaguar mtDNA concentration for five samples collected from drinking water sources provided to captive animals.
We then extracted DNA from half filters using DNeasy Blood and Tissue Kit (QIAGEN) with modifications as described in Carim et al. (2016a). In steps 1 and 2, we doubled the amount of ATL and proteinase K, and incubated samples for 48 h. We doubled the amount of AL buffer in step 3 and added 400 μl of ethanol simultaneously. We repeated step 4 loading using a single spin column for each sample until all elution for a given sample had been processed through the spin column. Additionally, we loaded each filter using sterile forceps into a QIAshredder spin column and centrifuged for 2 min a 20,000 × g. The elution from the QIAshredder was also loaded and processed through the corresponding spin column for that sample. Between steps 5 and 6, we added an additional wash of 500 μl ethanol and centrifuged for 2 min at 20,000 × g. In step 6, we increased spin time to 4 min. In step 7, we eluted DNA in 100 μl of 70°C TE and allowed to incubate at room temperature for 10 min before the final centrifuge step. All environmental samples were extracted in a dedicated space where no high concentration sources of DNA are handled.
Quantitative PCRs contained 7.5 μl TaqMan Environmental Master Mix 2.0 (Thermo Fisher Scientific), 900 nM each primer, 250 nM hydrolysis probe (also known as “taqman” probe), 4 μl template DNA, and molecular grade water to a total volume of 15 μl. Environmental and scat samples (described below) also contained an internal positive control template and assay to test for the presence of PCR inhibitors (indicated by a > 1 CT shift in amplification relative to the control samples; TaqMan Exogenous Internal Positive Control Kit from Thermo Fisher Scientific). On the PCR plate with environmental samples, a triplicate no-template control was included to test for contamination. All reactions were run in triplicate. We optimized primer concentrations by testing all possible combinations of forward and reverse primers at 100, 300, 600, and 900 nM concentration (n = 16 combinations) and selecting the combination with the lowest CT value and highest end-point fluorescence (900:900 nM combination).
We also quantified jaguar mitochondrial DNA based on comparison with a standard curve. The standard curve was a dilution series (2, 10, 50, 250, 1,250, 6,250, and 31,250 copies per reaction) of a synthetic jaguar gene covering the target amplicon (gBlock; Integrated DNA Technologies) which was quantified on a Qubit 2.0 fluorometer (dsDNA Broad Range Kit; Invitrogen). Each dilution level was run in six replicates.
In silico and in vitro testing showed our assay to be highly specific against potential non-target taxa in North America. Additional specificity testing of the assay may be necessary prior to application in the southern portion of the species’ range. Although there was low-level amplification in one African lion tissue sample, we were able to verify that this was due to sample contamination using a synthetic gene covering the target amplicon. In our in silico assessment, the closest non-felid present in North America was Equus caballus (GenBank accession# AY584828) with a total of 12 bp mismatches (8 primer mismatches, including the 3′ end of the forward primer, and 4 probe mismatches). Thus, non-felids are unlikely to cross-amplify. The assay also amplified all ten jaguar scat samples from Belize and had 100% amplification of the standard curve down to 10 copies/reaction (2/6 amplifications at a concentration of two copies/reaction). The standard curve slope implied an amplification efficiency of 85.3% (r2 = 0.996). Based on this information, we estimated the Limit of Detection (LOD) to be 10 copies/reaction (minimum concentration with a 95% amplification rate) and the Limit of Quantification (LOQ) to be 50 copies/reaction (minimum concentration with a coefficient of variation < 35%) as described in Klymus et al. (2020).
All drinking water samples were strongly positive with a mean of 6,682 copies/reaction (range 3,251–14,254), or 66,820 copies/L of water. One of the five unknown Belize pond water samples was also positive, but at much lower concentration (8 copies/reaction or 80 copies/L of water), which is in line with concentrations of other terrestrial species detections from eDNA (e.g., Williams et al., 2018). Negative control reactions all showed no amplification.
The high concentrations of jaguar DNA in captive animal drinking water sources (over 30,000 copies/L) suggest that eDNA sampling may have reasonable detection probabilities in more natural settings, as long as sampling has occurred soon after the site was visited by an animal. Harper et al. (2019) and Sales et al. (2020) used eDNA sampling for detection of terrestrial mammals using a community-wide, metabarcoding approach. They found that low-density, wide-ranging taxa like jaguar tend to have lower detection rates than common and evenly distributed taxa. Aquatic organisms, even at low abundances, provide a constant input source of eDNA. In contrast, terrestrial species deposit their DNA at a drinking site over a very brief period of time. Thus, detection probably relies on sampling within hours or days of visitation—before the DNA has degraded beyond detectable levels.
In the studies described by Harper et al. (2019) and Sales et al. (2020), drinking water sources were abundant on the landscape. For sampling low-density jaguars, a key landscape characteristic influencing detection might be the number of drinking water sources within a single home-range. When there are only several water sources, the mean frequency with which each water source is visited is relatively high. When there are many, it could be days or weeks between visits to any one site. Hence, we see eDNA sampling for rare terrestrial species as being particularly promising in arid landscapes such as the southwestern United States and northern Mexico.
Camera trapping, in contrast to eDNA sampling, provides a more continuous view of habitat use. However, the initial equipment investment and ongoing maintenance costs may limit sampling effort over large scales. Other authors have suggested that eDNA sampling might be particularly useful in lower probability areas, such as at the presumed range periphery of a taxon, where investment in camera traps is impractical (Harper et al., 2019). Environmental DNA sampling might also become more useful as recent advances in continuous or autonomous eDNA sampling are refined and provide a more continuous view of DNA inputs (e.g., Yamahara et al., 2019; Kirtane et al., 2020).
We imagine eDNA sampling being a useful complement to existing sampling approaches and conveniently added to other monitoring efforts on the landscape. For example, technicians performing vegetation surveys in a remote area could also opportunistically collect water samples without much additional effort (e.g., <20 min per water sample in this study). Much of the cost of eDNA sampling, particularly in remote areas, comes from travel time (Smart et al., 2016). In the arid southwestern United States the relative cost for sample collection versus analysis might be even greater. Often in these remote and arid landscapes, the hydroperiod of ephemeral springs and tanks are unknown. There is a high level of risk during dry seasons of hiking into a remote habitat only to find that there is no water. This risk is mitigated when technicians are performing other fieldwork simultaneously.
Further savings might be gained when the genetic analyses for jaguar is conducted on eDNA samples initially collected to survey for the presence of other species. In this case, the cost of sample collection and DNA extraction is already covered. For example, the jaguar qPCR assay described here might complement a more community-wide survey effort with metabarcoding. Metabarcoding tends to be less sensitive than single-species qPCR and even reasonably abundant taxa might be missed (Harper et al., 2019; Sales et al., 2020). Targeted analysis used in conjunction with metabarcoding has been found to be a useful approach for picking up rare-but-important taxa (Simmons et al., 2016). Alternatively, an analysis for jaguar eDNA could even be included as part of a larger high throughput qPCR (HT-qPCR) panel which includes assays for detection of various aquatic and terrestrial species (Wilcox et al., 2020). In this case, the expense invested in analyzing samples for the presence of jaguar is likely <10% of the overall laboratory cost. Generally, our hope is that this new molecular tool can be leveraged in creative ways to build cost-effective study designs composed of multiple, complementary sampling approaches.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was reviewed and approved by Arizona Center for Nature Conservation Research Committee.
TW, AC, KM, KZ, and MS conceived the study design. TW, JD, TF, DM, and KZ conducted sampling and analyses. All authors contributed to writing of the manuscript.
This work was funded by the SERDP (RC18-1348).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Phoenix Zoo for assistance with sampling and the Banana Bank Lodge for allowing sample collection on their property. Samples collected in central Belize as part of the Valley of Peace Archaeology Project directed by Dr. Lisa J. Lucero with permission of the Institute of Archaeology, National Institute of Culture and History, and the Government of Belize. We thank the Mills Lab at the University of Montana and Bill Murphy at Texas A&M for sharing DNA samples from various non-target felids. This manuscript was improved by suggestions made by John Kronenberger and Torrey Rodgers.
Atschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Brown, D. E., and González, C. A. L. (2000). Notes on the occurrences of jaguars in Arizona and New Mexico. Southwestern Nat. 45, 537–542. doi: 10.2307/3672607
Carim, K. J., Dysthe, J. C. S., Young, M. K., McKelvey, K. S., and Schwartz, M. K. (2016a). An environmental DNA assay for detecting Arctic grayling in the upper Missouri River basin, North America. Conserv. Genet. Resour. 8, 197–199. doi: 10.1007/s12686-016-0531-1
Carim, K. J., McKelvey, K. S., Young, M. K., Wilcox, T. M., and Schwartz, M. K. (2016b). A Protocol for Collecting Environmental DNA Samples from Streams. Gen. Tech. Rep. RMRS-GTR-355. Fort Collins, CO: US Department of Agriculture, Forest Service, Rocky Mountain Research Station, 18.
Chaves, P. B., Graeff, V. G., Lion, M. B., Oliveira, L. R., and Eizirik, E. (2012). DNA barcoding meets molecular scatology: short mtDNA sequences for standardized species assignment of carnivore noninvasive samples. Mol. Ecol. Resour. 12, 18–35. doi: 10.1111/j.1755-0998.2011.03056.x
Cristescu, M. E., and Hebert, P. D. N. (2018). Uses and misuses of environmental DNA in biodiversity science and conservation. Annu. Rev. Ecol. Evol. Syst. 49, 209–230. doi: 10.1146/annurev-ecolsys-110617-062306
Culver, M. (2016). Jaguar Surveying and Monitoring in the United States (No. 2016-1095). Reston, VA: US Geological Survey. doi: 10.3133/ofr20161095
Franklin, T. W., McKelvey, K. S., Golding, J. D., Mason, D. H., Dysthe, J. C., Pilgrim, K. L., et al. (2019). Using environmental DNA methods to improve winter surveys for rare carnivores: DNA from snow and improved noninvasive techniques. Biol. Conserv. 229, 50–58. doi: 10.1016/j.biocon.2018.11.006
Harper, L. R., Handley, L. L., Carpenter, A. I., Ghazali, M., DiMuri, C., Mcgregor, C. J., et al. (2019). Environmental DNA (eDNA) metabarcoding of pond water as a tool to survey conservation and management priority mammals in the UK. Biol. Conserv. 238:108225. doi: 10.1016/j.biocon.2019.108225
Kirtane, A., Atkinson, J. D., and Sassoubre, L. (2020). Design and validation of passive environmental DNA samplers using granular activated carbon and Montmorillonite Clay. Environ. Sci. Technol. 54, 11961–11970. doi: 10.1021/acs.est.0c01863
Klymus, K. E., Merkes, C. M., Allison, M. J., Goldberg, C. S., Helbing, C. C., Hunter, M. E., et al. (2020). Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2, 271–282. doi: 10.1002/edn3.29
Menchaca, A., Rossi, N. A., Froidevaux, J., Dias-Freedman, I., Caragiulo, A., Wultsch, C., et al. (2019). Population genetic structure and habitat connectivity for jaguar (Panthera onca) conservation in Central Belize. BMC Genet. 20:100. doi: 10.1186/s12863-019-0801-5
R Core Development Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rodgers, T. (2017). Proper fin-clip sample collection for molecular analyses in the age of eDNA. J. Fish Biol. 91, 1265–1267. doi: 10.1111/jfb.13485
Rodgers, T., and Mock, K. E. (2015). Drinking water as a source of environmental DNA for the detection of terrestrial wildlife species. Conserv. Genet. Resour. 7, 693–696. doi: 10.1007/s12686-015-0478-7
Sales, N. G., McKenzie, M. B., Drake, J., Harper, L. R., Browett, S. S., Coscia, I., et al. (2020). Fishing for mammals: landscape-level monitoring of terrestrial and semi-aquatic communities using eDNA from riverine systems. J. Appl. Ecol. 57, 707–716. doi: 10.1111/1365-2664.13592
Sepulveda, A. J., Nelson, N. M., Jerde, C. L., and Luikart, G. (2020). Are environmental DNA methods ready for aquatic invasive species management? Trends Ecol. Evol. 35, 668–678. doi: 10.1016/j.tree.2020.03.011
Simmons, M., Tucker, A., Chadderton, W. L., Jerde, C. L., and Mahon, A. R. (2016). Active and passive environmental DNA surveillance of aquatic invasive species. Can. J. Fish. Aquat. Sci. 73, 76–83. doi: 10.1139/cjfas-2015-0262
Smart, A. S., Weeks, A. R., van Rooyen, A. R., Moore, A., McCarthy, M. A., and Tingley, R. (2016). Assessing the cost-efficiency of environmental DNA sampling. Methods Ecol. Evol. 7, 1291–1298. doi: 10.1111/2041-210x.12598
Wilcox, T. M., McKelvey, K. S., Young, M. K., Engkjer, C., Lance, R. F., Lahr, A., et al. (2020). Parallel, targeted analysis of environmental samples via high-throughput quantitative PCR. Environ. DNA 2, 544–553. doi: 10.1002/edn3.80
Williams, K. E., Huyvaert, K. P., Vercauteren, K. C., Davis, A. J., and Piaggio, A. J. (2018). Detection and persistence of environmental DNA from an invasive, terrestrial mammal. Ecol. Evol. 08, 688–695. doi: 10.1002/ece3.3698
Wright, E. S. (2016). Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 8, 352–359. doi: 10.32614/rj-2016-025
Wultsch, C., Caragiulo, A., Dias-Freedman, I., Quigley, H., Rabinowitz, S., and Amato, G. (2016). Genetic diversity and population structure of Mesoamerican jaguars (Panthera onca): implications for conservation and management. PLoS One 11:e0162377. doi: 10.1371/journal.pone.0162377
Keywords: environmental DNA, eDNA, non-invasive genetics, wildlife, monitoring
Citation: Wilcox TM, Caragiulo A, Dysthe JC, Franklin TW, Mason DH, McKelvey KS, Zarn KE and Schwartz MK (2021) Detection of Jaguar (Panthera onca) From Genetic Material in Drinking Water. Front. Ecol. Evol. 9:613200. doi: 10.3389/fevo.2021.613200
Received: 01 October 2020; Accepted: 21 April 2021;
Published: 13 May 2021.
Edited by:
Matthew A. Barnes, Texas Tech University, United StatesReviewed by:
Antoinette J. Piaggio, National Wildlife Research Center, Animal and Plant Health Inspection Service (USDA), United StatesCopyright © 2021 Wilcox, Caragiulo, Dysthe, Franklin, Mason, McKelvey, Zarn and Schwartz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taylor M. Wilcox, taylor.wilcox@usda.gov
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.