
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 09 February 2021
Sec. Evolutionary Developmental Biology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.583136
This article is part of the Research TopicEvo-Devo of Color Pattern FormationView all 20 articles
 Asier Ullate-Agote1,2,3
Asier Ullate-Agote1,2,3 Athanasia C. Tzika1,2,3*
Athanasia C. Tzika1,2,3*Albinism and leucism are phenotypes resulting from impaired melanin pigmentation in the skin and skin appendages. However, melanin pigmentation of eyes remains unaffected in leucism. Here, using transmission electron microscopy, we show that the leucistic morph of the Texas rat snake (Pantherophis obsoletus lindheimeri) lacks both melanophores and xanthophores in its skin and exhibits a uniform ivory white color generated by iridophores and collagen fibers. In addition, we sequenced the full genome of a leucistic individual and obtained a highly-contiguous near-chromosome quality assembly of 1.69 Gb with an N50 of 14.5 Mb and an L50 of 29 sequences. Using a candidate-gene approach, we then identify in the leucistic genome a single-nucleotide deletion that generates a frameshift and a premature termination codon in the melanocyte inducing transcription factor (MITF) gene. This mutation shortens the translated protein from 574 to 286 amino acids, removing the helix-loop-helix DNA-binding domain that is highly conserved among vertebrates. Genotyping leucistic animals of independent lineages showed that not all leucistic individuals carry this single-nucleotide deletion. Subsequent gene expression analyses reveal that all leucistic individuals that we analyzed exhibit a significantly decreased expression of MITF. We thus suggest that mutations affecting the regulation and, in some cases, the coding sequence of MITF, the former probably predating the latter, could be associated with the leucistic phenotype in Texas rat snakes. MITF is involved in the development and survival of melanophores in vertebrates. In zebrafish, a classical model species for pigmentation that undergoes metamorphosis, larvae and adults of homozygous mitfa mutants lack melanophores, show an excess of iridophores and exhibit reduced yellow pigmentation. On the contrary, in the leucistic Texas rat snake, a non-metamorphic species, only iridophores persist. Our results suggest that fate determination of neural-crest derived melanophores and xanthophores, but not of iridophores, could require the expression of MITF during snake embryonic development.
Vertebrates display a wide variety of adaptive color patterns on their skin and skin appendages. Coloration is involved in the thermoregulation of ectotherms, ultraviolet radiation protection and in intra- and inter-specific interactions, including sexual selection and mating, camouflage for predator evasion and as a warning of dangerous traits. Melanophores are found in all vertebrate lineages and they are the only chromatophores in birds and mammals. Melanophores synthesize melanin from tyrosine and store the resulting black/brown pigment in melanosomes, a type of lysosomal-related organelles (LROs) (Wasmeier et al., 2008). Albinism and leucism, two distinct phenotypes, result from impaired melanin pigmentation. Albinism is characterized by the absence (amelanism) or reduction (hypomelanism) of melanin pigment in the skin, skin appendages and eyes. Mutations causing this condition can affect either genes involved in melanin production per se or the biogenesis and maturation of melanosomes. Known variants in genes such as tyrosinase (TYR), oculocutaneous albinism II (OCA2), solute carrier family 45 member 2 (SLC45A2), and tyrosinase related protein 1 (TYRP1) are known to affect melanin production and cause oculocutaneous albinism in humans (Montoliu et al., 2014). On the other hand, mutations affecting melanosome biogenesis/maturation can impact on other LROs, hence, affecting different cell types and impacting other organs and functions. For example, in humans, mutations in subunits of the lysosomal-trafficking protein complexes cause the Hermansky-Pudlak syndrome (Wei and Li, 2013) and a non-functional lysosomal trafficking regulator (LYST) results in the Chédiak-Higashi syndrome (Barbosa et al., 1996). In both syndromes, decreased or absent pigmentation is associated with other symptoms, such as platelet abnormality and peripheral neuropathy. Recently, we have shown that the Lavender phenotype in corn snakes is likely caused by a mutation in LYST and impacts the formation of (i) pterinosomes in xanthophores, (ii) reflecting platelets of guanine crystals in iridophores, and (iii) melanosomes, resulting in lighter overall coloration without affecting the geometrical elements of the color (Ullate-Agote et al., 2020). Spontaneously-occurring albino phenotypes are widespread among vertebrates, from fish to snakes and mammals (e.g., Chang et al., 2006; Anistoroaei et al., 2013; Ullate-Agote et al., 2014; Saenko et al., 2015; Hart and Miller, 2017; Nakayama et al., 2017; Woodcock et al., 2017; Iwanishi et al., 2018).
In leucistic mammals, the eye pigmentation is preserved, while skin coloration is absent or reduced, resulting in a fully white or piebald phenotype (Reissmann and Ludwig, 2013). Apart from the retinal pigment epithelium (RPE), a one-cell layer derived from the optic neuroepithelium (Fuhrmann et al., 2014), all chromatophores originate from the neural crest. Note that the iris coloration can be affected in leucism because, while the back layer of the iris is an extension of the RPE and thus maintains its pigmentation, the front of the iris is populated by neural crest-derived chromatophores which are impacted in leucistic individuals (Bharti et al., 2006). Therefore, leucism is caused by mutations in genes involved in the migration, differentiation or survival of the neural crest-derived chromatophores during development, rather than melanin production. Examples of such genes are the melanocyte inducing transcription factor (MITF), paired box 3 (PAX3), Sry-Box transcription factor 10 (SOX10), endothelin 3 (EDN3), and KIT proto-oncogene receptor tyrosine kinase (KIT). Mutations in all but KIT cause the Waardenburg syndrome in humans, in which pigmentation abnormalities are associated with hearing loss (Pingault et al., 2010).
The plethora of color skin patterns in squamates, i.e., snakes and lizards, originates from different combinations of chromatophore cells (Kuriyama et al., 2006, 2013; Saenko et al., 2013; Teyssier et al., 2015; Manukyan et al., 2017; Andrade et al., 2019; Ullate-Agote et al., 2020). In addition to melanophores, squamates exhibit xanthophores and erythrophores, jointly referred to as xanthophores. These are pigment-containing cells producing yellow and red coloration, respectively. The pigments are either pteridines, synthesized from purines and stored in pterinosomes (Ziegler, 2003), or carotenoids absorbed from the diet and stored in carotenoid vesicles. Color differences may also arise from variations in the pH of the organelle or redox state of the pigments within (Saenko et al., 2013). The third chromatophore type is the iridophore, a cell type that contains stacks of crystallized guanine forming reflective platelets, and which produce structural colors through light interference.
Albino snakes lack melanin in their skin and eyes, while preserving their yellow and red coloration and overall pattern (Ullate-Agote et al., 2014; Saenko et al., 2015; Iwanishi et al., 2018). However, it has been suggested that leucistic snakes are missing both xanthophores and melanophores, resulting in a uniform ivory white coloration generated by iridophores (Bechtel, 1991). On the other hand, leucistic axolotl (Ambystoma mexicanum) individuals, named “white,” have defects in the morphogenesis of both melanophores and xanthophores, whereas iridophores are absent (Woodcock et al., 2017). The endothelin 3 (edn3) transcripts, encoding a peptide that functions as a ligand for the Endothelin receptor type B, consistently lack exon 2 in “white” axolot. The interaction of these two proteins is essential for the migration of the neural crest-derived cells (Baynash et al., 1994), and it is probably for this reason that all types of chromatophores are lacking in “white” axolotl.
Here, we focus on the Texas rat snake (Pantherophis obsoletus lindheimeri), a non-venomous colubrid found in central North America, characterized by black blotches on an orange background. Similarly to the closely related corn snake (Pantherophis guttatus), spontaneously-occurring color morphs, including xanthic, albino, hypomelanistic, and leucistic, have been recognized in the Texas rat snake (Bechtel and Bechtel, 1985). First, we use transmission electron microscopy (TEM) to show that the leucistic phenotype (Figure 1), an autosomal recessive trait (Bechtel and Bechtel, 1985) in Texas rat snakes, is associated with the lack of both melanophores and xanthophores in the skin. Second, we suggest that the uniform ivory white color in these animals is generated by iridophores and collagen fibers. Third, we perform genomic analyses to identify the causative variant responsible for the leucistic phenotype. Mapping by sequencing is an outstanding approach to determine the genomic interval where a mutation of interest resides (Schneeberger, 2014; Ullate-Agote et al., 2014, 2020; Saenko et al., 2015). However, it is a time-consuming protocol that can span over several years for species with seasonal breeding and long generation time. The advent of deep-sequencing technologies has greatly facilitated the genome assembly of non-classical model species, in a fast and affordable manner and with sufficient quality and completeness to annotate complete gene structures. Here, we assembled and annotated a high-quality genome of a leucistic Texas rat snake individual using 10x Genomics linked-reads (Zheng et al., 2016). Then, using a candidate gene approach based on the extensive literature on mutations responsible for leucism, mainly in mammals and birds (Reissmann and Ludwig, 2013; Kinoshita et al., 2014; Woodcock et al., 2017), we identified a single nucleotide deletion in MITF that introduces a frameshift and a premature stop codon in the leucistic genome. As this deletion is not present in all the genotyped leucistic animals of independent lineages, we additionally carried out gene expression analyses that revealed a significantly-decreased expression of MITF in all leucistic individuals, even in those lacking the above-mentioned single-nucleotide deletion in the coding sequence. We thus suggest that the leucistic phenotype in Texas rat snakes might be associated to mutations affecting the regulation of MITF, and the single-nucleotide deletion might have appeared subsequently as a secondary consequence of an under- or mis-expressed MITF. MITF regulates the expression of genes involved in melanocyte migration, survival and differentiation during development (Fock et al., 2019). The absence of xanthophores and melanophores in the leucistic Texas rat snake suggests that in snakes MITF might be necessary for the differentiation of neural-crest derived melanophores and xanthophores, but not of iridophores.

Figure 1. Coloration of wild type (A) and leucistic (B) Texas rat snakes. A dorsal overview (left), close ups of lateral and ventral views (center), and a head dorsal view are provided. The inset shows the iris coloration.
Texas rat snakes are housed and bred at the LANE animal facility running under the veterinary cantonal permit no. 1008. The individuals were sampled and imaged following Swiss law regulations and under the experimentation permit GE/169/17.
Eyes from wild type and leucistic individuals were fixed in PFA 4% and washed in MetOH. Paraffin sections of 7 μm were either stained with hematoxylin/eosin or dewaxed with xylene and mounted directly to visualize the pigmentation. Skin pieces of 1 mm2 were fixed and sectioned as described in Ullate-Agote et al. (2020). Micrographs were taken with a transmission electron microscope Philips CM100 (Thermo Fisher Scientific) at an acceleration voltage of 80 kV with a TVIPS TemCam-F416 digital camera (TVIPS GmbH). Large montage alignments were performed using Blendmont command-line from the IMOD software (Kremer et al., 1996).
High molecular weight (HMW; >50 Kb) genomic DNA (gDNA) was extracted with the QIAGEN Genomic-tip 20/G kit (10223, QIAGEN) from nucleated erythrocytes of an adult female leucistic individual. The resulting gDNA was used for a 10x Chromium library preparation (Novogene Co., Ltd.) with the Chromium Genome HT Library Kit and Gel Bead Kit v2 (Zheng et al., 2016). We sequenced the resulting barcoded fragments with a HiSeq X instrument, obtaining 522 million 150 bp paired-end reads.
We de novo assembled all the reads of the 10x Chromium library with Supernova v2.1.1 (Weisenfeld et al., 2017). The total sequencing data corresponded to a 77x raw coverage, above the recommended 56x (Weisenfeld et al., 2017). However, the length-weighted mean of HMW DNA molecule lengths estimated by Supernova is 35.8 Kb, below the ideal 50–100 Kb range. Downstream analyses were based on the pseudo-haplotype Supernova output. We removed highly similar and duplicated scaffolds by running CD-HIT-EST (Li and Godzik, 2006) to cluster sequences with 99% of sequence identity, keeping the longest one in the final assembly (parameters: -g 1 -c 0.99 -s 0.99 -aS 0.99). Finally, we removed mitochondrial DNA scaffolds and looked for contaminants running blastn searches (Camacho et al., 2009) against the closely-related corn snake mtDNA (GenBank accession number: AM236349.1), as well as the “UniVec Build 10,” Saccharomyces cerevisiae, Escherichia coli, and reptilian ferlaviruses databases with the NCBI VecScreen parameters.
We assessed the gene completeness of the assembly with BUSCO v3.0.2 (Waterhouse et al., 2018) focusing on the 3,950 single-copy orthologous genes of the Tetrapoda set from OrthoDB v9. We also modeled repeat families for the Texas rat snake using RepeatModeler v1.0.11 (Smit, 2008–2019)1. We masked the assembly with RepeatMasker v4.0.7 (Smit, 2013–2019)2 in sensitive search mode with rmblastn v2.2.27+ in three sequential steps, taking the masked output genome of each run as an input for the following one. First, we considered all the Tetrapoda repeats from the RepeatMasker database (Repbase release 20170127). Second, we looked for classified Texas rat snake repetitive elements coming from RepeatModeler and, finally, we retrieved the repeats with unknown classification.
We predicted and annotated protein-coding genes in the Texas rat snake genome assembly using the BRAKER v2.1.4 fully automated pipeline (Hoff et al., 2019). Due to the lack of RNA-Seq data, we built a dataset including the non-mitochondrial proteins of five snakes in the NCBI RefSeq database (accessed in November 2019): Pseudonaja textilis, Notechis scutatus, Protobothrops mucrosquamatus, Python bivittatus, and Thamnophis sirtalis. After removing identical sequences, we obtained 123,537 proteins. They were aligned to the soft masked Texas rat snake genome using GenomeThreader v1.7.1 (Gremme et al., 2005) with compatible parameters for BRAKER (-gff3out -skipalignmentout). Then, we input the soft-masked genome assembly (adding the–softmasking flag) and the alignments generated with GenomeThreader (using the–prot_aln option) into BRAKER, with the–prg=gth and–trainFromGth parameters to indicate that the input is GenomeThreader alignments used to train the Augustus gene predictor. This corresponds to the BRAKER annotation pipeline for proteins of short evolutionary distance (Hoff et al., 2019), in which Augustus v3.3.3 (Stanke et al., 2006) is the only gene predictor and it is automatically trained with the protein alignments.
We used the MAKER v3 (Holt and Yandell, 2011) annotation pipeline to calculate the Annotation Edit Distance (AED) quality-control metric (Yandell and Ence, 2012) of the genes predicted by BRAKER as explained in (Campbell et al., 2014). AED measures the concordance between the annotated gene models and the supporting evidence data, with a value of one corresponding to lack of support. We input the BRAKER gene annotation into the MAKER pipeline in GFF format, together with the RefSeq snake protein database as evidence and the complex repetitive elements information from RepeatMasker, masking simple repeats during the MAKER run. Then, we ran InterProScan-5.39-77.0 (Jones et al., 2014) to look for Pfam protein domains. The final gene set includes the BRAKER gene models that are supported by evidence (AED < 1) or have a Pfam protein domain identified. Finally, we performed blastp searches against UniProtKB/SwissProt and Squamata proteins in RefSeq to annotate the genes (both accessed on 26.11.2019). A protein was considered complete if it covered at least 95% of its best blastp hit.
We built a list of nine candidate genes responsible for leucistic phenotypes in other species which are mainly involved in the migration, survival, and differentiation of neural crest-derived melanocytes during development (Table 1). We obtained from the NCBI RefSeq database the protein sequences of these genes for the corn snake Pantherophis guttatus and the eastern brown snake Pseudonaja textilis (accessed in July 2020). We aligned them to the leucistic Texas rat snake genome assembly using exonerate v2.2.0 (Slater and Birney, 2005) in protein2genome mode to retrieve any disruptive modifications in their coding sequence, i.e., frameshifts, premature stop codons, or modification of intron-exon boundaries.
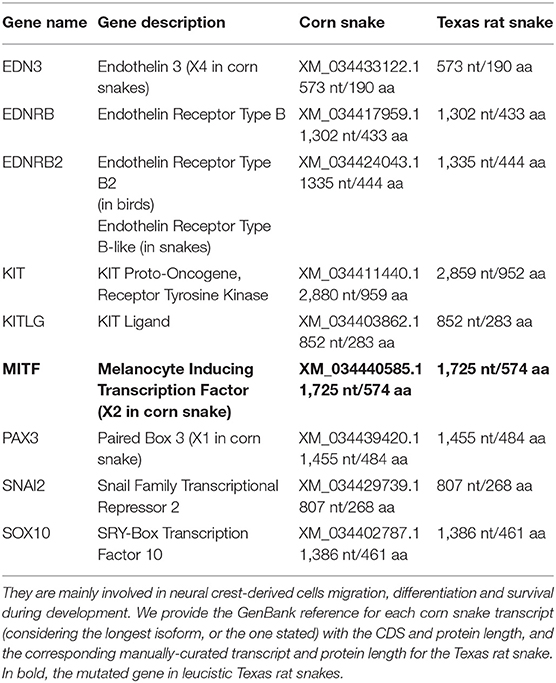
Table 1. List of the nine candidate genes known to be responsible for leucistic phenotypes in vertebrates.
We extracted genomic DNA from animal sheds using the DNeasy Blood & Tissue Kit (Qiagen 69504) and genotyped them for the point mutation identified by deep-sequencing. We extracted mRNA with Direct-zol RNA Miniprep kit (Zymo Research R2050) from dorsal skin and prepared complementary DNA with PrimeScript™ Reverse Transcriptase (Takara Bio 2680A). We then amplified and sequenced the MITF transcript in leucistic and wild type individuals. All primers are provided in Supplementary Table 1.
We obtained 50 μl of whole blood and mixed it with 1 ml DNA/RNA Shield (Zymo Research) before storing at −80°C. We extracted RNA with the Quick-RNA™Whole Blood kit (Zymo Research). Reverse transcription starting with 500 ng of RNA was performed with the PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara Bio) which includes a step for the elimination of any gDNA contamination. We used the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) for the quantitative RT-PCR run on a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). All samples were run in triplicates. We verified the PCR efficiency for the amplification of MITF and TUBB, the reference gene, as described in Pfaffl (2001). Biogazelle qbasePLUS 3.2 software was used to analyze the results and perform statistical analyses (Biogazelle, Zwijnaarde, Belgium - www.qbaseplus.com).
Wild type Texas rat snakes have black blotches on a red to orange background and a white belly with some traces of yellow (Figure 1A). On the other hand, the dorsal and ventral skin of leucistic individuals is ivory white, their iris coloration is light gray (instead of red as in the wild-type) and the pupil remains black (Figure 1B). TEM analyses indicate that the dorsal skin of wild type individuals contains epidermal melanocytes, dermal xanthophores close to the epidermal basal membrane, as well as iridophores and melanocytes localized deeper in the dermis (Figure 2A and Supplementary Figure 1). No dermal melanophores could be found in the background sections, but we cannot exclude their presence in scarce numbers. The disposition and density of iridophores looks similar in the background and blotch dorsal skin. In comparison to the closely-related corn snake (Ullate-Agote et al., 2020), Texas rat snakes exhibit the following differences in chromatophore composition and distribution: (i) xanthophores are more scarce and can be found deeper in the dermis, instead of forming a continuous layer just under the basal membrane, (ii) iridophores and melanocytes co-exist, instead of melanocytes being restricted at the bottom of the loose dermis, and (iii) the concentric lamellae of the pterinosomes are less evident and we cannot exclude the presence of carotenoid vesicles in some xanthophores. Indeed, the background coloration of the Texas rat snakes is darker compared to the corn snake, most likely because melanophores are situated higher in the dermis. On the other hand, Texas rat snake leucistic individuals only exhibit iridophores and collagen fibers (Figure 2B) that together produce the ivory white coloration of the animal. Sagittal sections of the Texas rat snake wild type and leucistic eye (Figure 3) both revealed the presence of melanophores in the RPE and the posterior part of the iris, consistent with an optic neuroepithelium origin of these melanophores. In the wild type, melanophores are also present in the sclera and the choroid and we observe red pigmentation in the ciliary body. Iridophores are present in the anterior part of the iris for both wild type and leucistic animals. Therefore, both melanophores and xanthophores of neural crest origin are impacted in the leucistic phenotype, but not the iridophores.
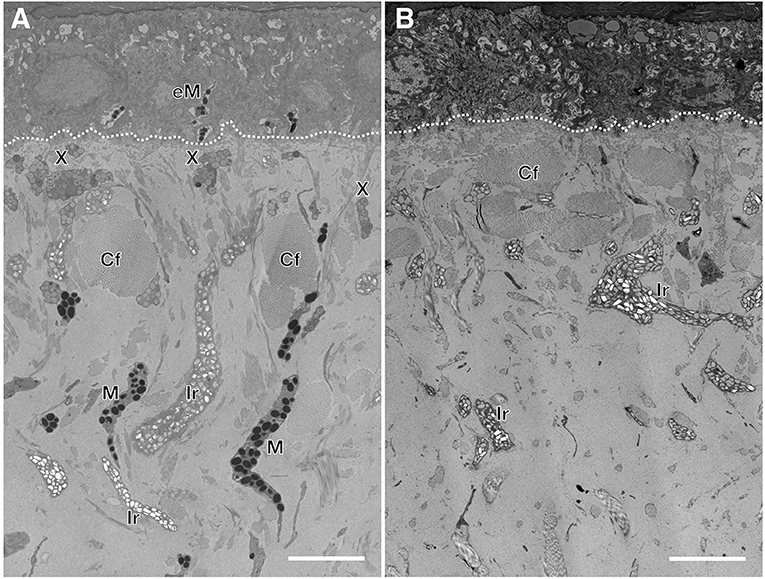
Figure 2. Semi-thin sections of wild type (A) and leucistic (B) Texas rat snake dorsal skin. In the wild type, xanthophores (X) are situated closer to the basal membrane (dashed line) whereas the iridophores (Ir) and the melanophores (M) are spread in the loose dermis among collagen fibers (Cf). In the leucistic skin, only iridophores and collagen fibers are observed. Epidermal melanophores (eM) are observed only in the wild type. Scale bar: 5 μm.
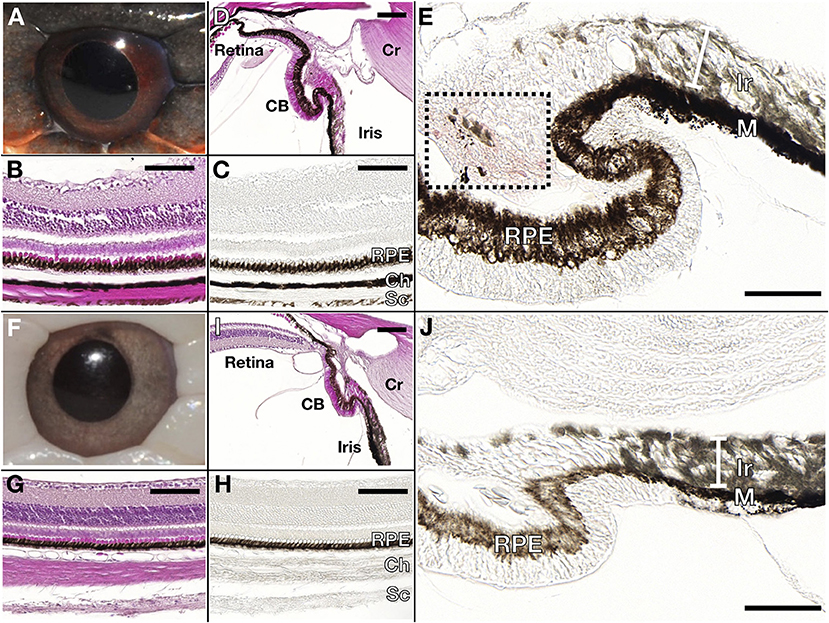
Figure 3. Wild type (A–E) and leucistic (F–J) eye anatomy. Hematoxylin/Eosin stained (B,G) and unstained (C,H) paraffin sections through the retina show that pigmentation is present in the Retinal Pigmented Epithelium (RPE) of both wild type and leucistic snakes, whereas only the wild type choroid (Ch) and sclera (Sc) contain melanophores. (D,I) The RPE of the retina extends in the ciliary body (CB) and in the posterior part of the iris. Red pigmentation (area in black box) is visible in the CB of the wild type (E) but not in the leucistic snake (J), whereas iridophores (Ir) are found in the anterior part of the iris in both phenotypes (E,J). M: melanophores, Cr: cornea. Scale bar: 100 μm.
The karyotype of the Texas rat snake consists of 18 pairs of chromosomes, of which 10 are micro-chromosomes (Baker et al., 1972). We sequenced a 10x Chromium library and obtained 522 million paired-end reads from an adult leucistic female individual, female being the heterogametic sex, and we assembled them using the Supernova software (Weisenfeld et al., 2017). After removing redundant and contaminant sequences, the final rat snake genome assembly (GenBank accession number GCA_012654085.1) is 1.69 Gb long, consists of 36,795 scaffolds >1 Kb and presents only 3.6% of gaps. The N50 is 14.5 Mb with an L50 of 29 sequences. We thus obtained a highly contiguous genome in a single step, presenting similar statistics to other high-quality snake genomes, such as Pantherophis guttatus, Notechis scutatus, and Pseudonaja textilis (Table 2).
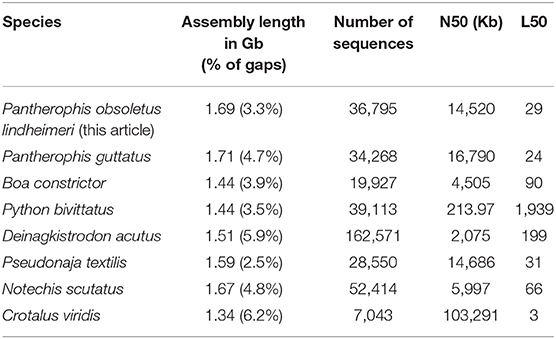
Table 2. Statistics of the final Texas rat snake genome assembly compared to other published snake genomes.
We assessed gene completeness using the 3,950 Tetrapoda single-copy gene set of BUSCO v3.0.2: we retrieved 3,481 full genes (88.1%), 270 partial genes (6.9%), and only 199 (5.04%) were missing. These values indicate high levels of completeness similar to other Squamata genomes (Supplementary Figure 2). The RepeatModeler library for the Texas rat snake consists of 1,496 repeat families, of which 687 are classified. Repetitive elements cover 45.18% of the genome and their diversity resembles that of the closely-related corn snake [(Ullate-Agote et al., 2020); Supplementary Figure 3 and Supplementary Table 2]. The two species have an estimated evolutionary divergence time of 13 Mya (Kumar et al., 2017); hence, it is not surprising that the repeat content is highly conserved.
Using the BRAKER pipeline and snake proteins from RefSeq, we predicted 24,107 genes with a total of 39,255 transcript isoforms. We only kept the gene models that were either supported by protein evidence data [i.e., with an Annotation Edit Distance lower than one or that had a Pfam domain (Supplementary Figure 4A)]. The annotation of 38,323 proteins is supported by other Squamata or UniProtKB/SwissProt proteins, with 21,435 being complete (Supplementary Figure 4B). Thus, most annotated genes are complete, which is crucial for the retrieval of variants affecting protein-coding sequences, as we attempt in this study. We provide a genome browser of the Texas rat snake genome to visualize the assembly and annotation (https://www.reptilomics.org/jbrowse/Pantherophis_obsoletus).
The leucistic phenotype of the Texas rat snake is caused by a recessive allele, so the assembled genome of a leucistic individual should harbor the causative variant in a homozygous state. The persistent coloration of the retinal pigment epithelium in leucistic animals (Figure 1B) rules out genes related to pigment synthesis or the biogenesis of color-producing organelles. Thus, we focused on genes involved in the migration, survival and differentiation of neural crest-derived chromatophores during development, i.e., genes whose variants can cause similar pigmentation phenotypes, like white skin and colored eyes, in other vertebrate species (Table 1). In mice, mutations in Edn3 and its receptor Endothelin receptor type B (Ednrb) affect the migration of neural crest-derived melanocytes and enteric ganglion neurons precursors from E10.5 to E12.5, resulting in both leucism and an aganglionic megacolon (Lee et al., 2003). The Endothelin receptor type B2 gene (EDNRB2), absent in mammals, is a paralog of EDNRB. In birds, it is known to interact with EDN3 and mutations in EDNRB2 have been associated with leucistic plumage in chicken and the Japanese quail (Miwa et al., 2007; Kinoshita et al., 2014). MITF is the key transcription factor for melanocyte differentiation and survival and regulates the transcription of melanocyte-specific genes, such as TYR and TYRP1 (Steingrimsson et al., 2004). PAX3 and SOX10 transcription factors are activators of MITF transcription, binding a proximal region of its promoter (Bondurand et al., 2000). Moreover, SOX10 is expressed early in pre-migratory neural crest cells and then during migration, influencing the fate specification of certain neural crest derivatives, such as melanocytes (Kelsh, 2006). The Snail Family Transcriptional Repressor 2 (SNAI2) gene is involved in the epithelial-to-mesenchymal transition and it is expressed in migrating neural crest cells, impairing melanoblast migration and survival when absent (Cobaleda et al., 2007). Mutations in any of the genes listed above, besides EDNRB2 which is absent in mammals, can cause different types of Waardenburg syndrome in humans (Pingault et al., 2010). Finally, the KIT receptor and its ligand (KITLG) are involved in melanogenesis, gametogenesis and hematopoiesis (Besmer et al., 1993; Fleischman, 1993): in mice, mutations in these genes cause leucism and sterility and, in some cases, are lethal.
We aligned the closely-related corn snake and the eastern brown snake proteins of the nine candidate genes to the leucistic genome and discovered a single nucleotide deletion in the coding sequence (CDS) of MITF. Genotyping of wild type and leucistic Texas rat snakes in our colony (Figure 4A and Supplementary Table 3) confirmed that indeed a cytosine is missing in exon 6 at position 825 of the coding sequence in all leucistic individuals, introducing a frameshift and a premature stop codon downstream (Figure 4B). As a result, the leucistic MITF protein is only 286 amino acids long, instead of 574, with the last 12 aa being different from the ones in the wild type (Figure 4C). The sequences of the other candidate genes do not present any obvious disruptions of the gene structure (Supplementary Text).
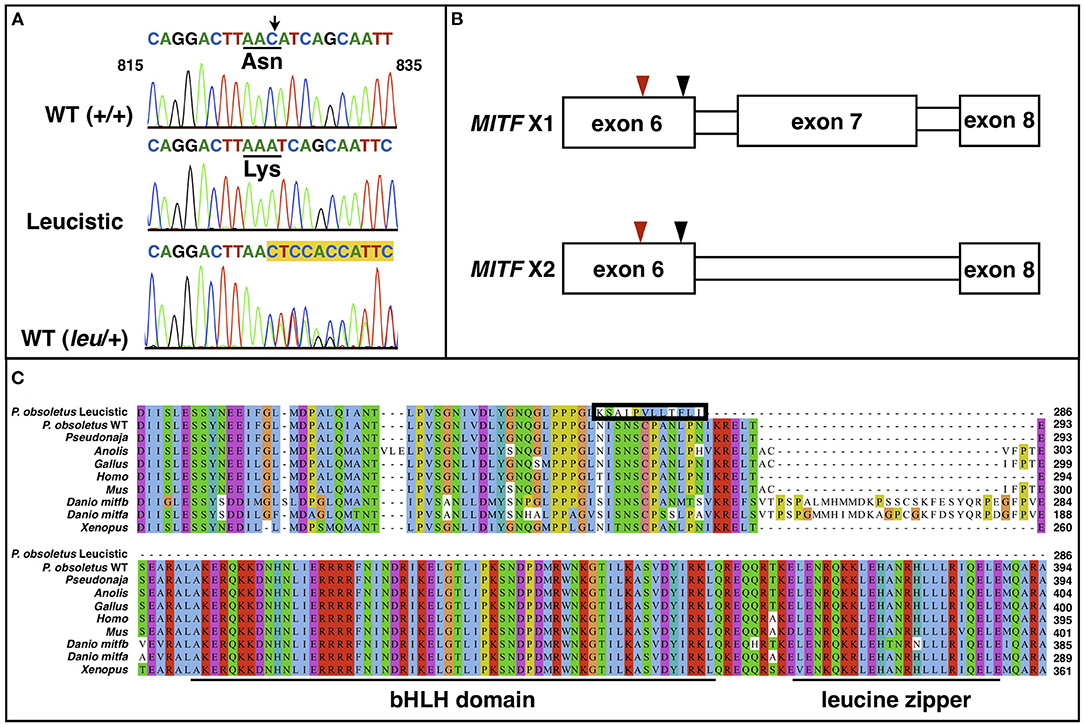
Figure 4. (A) Chromatogram sequences of MITF between positions 815 and 835 of a WT (+/+), a leucistic and an heterozygous WT snake. The black arrow points to the deleted cytosine. The amino acid change is shown. Highlighted in yellow are the double peaks in the heterozygous individual. (B) Representation of the two MITF isoforms, which only differ by the presence/absence of exon 7. The red arrowhead points to the location of the cytosine deletion, while the black one indicates the location of the premature stop codon. (C) Multiple sequence alignment of MITF proteins from different vertebrate lineages along with the wild type (isoform X2 in 4B) and leucistic Pantherophis obsoletus MITF. The 12 amino acids that are modified in the leucistic MITF after the frameshift and before the premature stop codon are included in the black box. The basic helix-loop-helix and the leucine zipper domain are underlined.
There are six annotated MITF isoforms for the corn snake. We confirmed the presence of two of them—isoform 2 (XM_034440585.1), which is one of the longest, and isoform 5 (XM_034440588.1), where exon 7 is missing—in the wild-type Texas rat snake by sequencing complementary DNA of multiple animals. As the premature stop codon in MITF of leucistic snakes is located in exon 6, we expect only one truncated protein to be produced in these animals (Figure 4B). The truncated protein misses the bHLH-Zip domain, which includes a DNA binding region, the helix-loop-helix domain and the leucine-zipper domain, with the latter two being involved in homo- and heterodimerization (Tachibana, 2000; Steingrimsson et al., 2004). These domains are highly conserved among all vertebrates (Figure 4C), suggesting that the mutated MITF protein is probably non-functional. A similar mutation has been identified in mice [in two different genetic backgrounds, midi and mice (West et al., 1985; Zimring et al., 1996)]: a C to T transition at position 916 bp that introduces a stop codon after amino acid 262. These mutants feature a white coat and reduced iris pigmentation. Depending on the genetic background, it has been reported that females have reduced reproduction.
As all the leucistic individuals of our colony originate from the same four related founders (Supplementary Table 3; individuals OBS01, 02, 05, and 06), we investigated if the mutation of the MITF CDS described above is present in other lineages of leucistic Texas rat snakes. Indeed, as discussed in (Bechtel and Bechtel, 1985), leucistic individuals exist in the wild and they have been independently introduced to the pet trade in different occasions. We thus purchased four additional leucistic snakes from three different sources and lineages (Supplementary Table 3; individuals OBS42-45). One of the animals had red pupils (OBS42; Supplementary Figure 5A) and we thus assume that it is a double mutant: amelanistic and leucistic. When we genotyped these new individuals for the single-nucleotide deletion in MITF, we found that only one of them was homozygous for it (OBS42). The other three were either heterozygous (siblings OBS44 and 45) or homozygous for the wild type allele (OBS43). Both MITF isoforms were present in these four individuals without any other modification of the transcript, besides the presence in some cases of the single-nucleotide deletion as seen in their gDNA. Based on this genotypic heterogeneity, one could argue that the single-nucleotide deletion introducing a stop codon in the MITF protein cannot be solely responsible for the leucistic phenotype, as it is absent in some leucistic individuals (OBS43), or it is present on only one allele in others (OBS44 and 45).
To assess whether MITF is involved in the determination of the leucistic phenotype, we performed gene expression analyses. Red blood cells in reptiles are nucleated and on average 25,000 proteins are transcribed in the blood, making it a valid starting material for gene expression analyses when minimally invasive sampling is required (Waits et al., 2020). We thus performed our analyses with this type of samples. It was not possible to collect more adequate target tissues (namely, embryonic skin) because the leucistic individuals in our colony produce a very small number of eggs, compared to their heterozygous and wild-type counterparts.
We extracted RNA from the blood of all living individuals in our colony, as well as from the four new animals, and performed quantitative RT-PCR on all samples (Figure 5 and Supplementary Figures 6, 7). The results clearly show that MITF expression is significantly reduced in all leucistic individuals (p = 3.9e−4; non-paired Mann-Whitney test with two-sided significance), independently of the presence or absence of the single-nucleotide deletion in the coding sequence. We thus hypothesize that a regulatory mutation could down-regulate MITF expression in all leucistic snakes and a more recent single-nucleotide deletion might have appeared in another lineage. Indeed, a down-regulated expression or mis-expression of MITF could facilitate the accumulation of mutations in the coding part of the gene. The advantage of using blood as starting material for these analyses is that the expression levels detected are independent of the number of chromatophores present in the skin. Indeed, in leucistic individuals, a reduced MITF expression in the skin could also be due to the absence of melanophores and xanthophores.
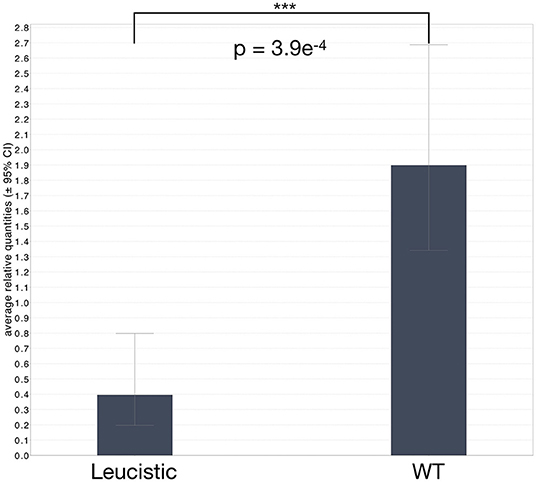
Figure 5. Average relative levels of MITF expression in leucistic and non-leucistic individuals. Whiskers correspond to the ± 95% confidence interval. Statistical significance was assessed with a non-paired Mann-Whitney test and a two-sided significance; ***corresponds to P ≤ 0.001.
Vertebrate pigmentation has been extensively studied, mainly focusing on human related syndromes and mouse mutants in mammals, the zebrafish in teleosts and Xenopus in amphibians [for example Jackson (1997), Baxter and Pavan (2013), Mitros et al. (2019), Patterson and Parichy (2019)]. Mammals have limited interest for studying the development of chromatophores as they exhibit only melanophores. Fishes and amphibians go through metamorphosis and the origin of the embryonic and adult chromatophores is not the same for the different types. Squamates are important new models for understanding chromatophore development because they display remarkable coloration patterns that are produced by different combinations of the three chromatophore types: melanophores, xanthophores, and iridophores. Although color patterns can evolve over time, as is the case of the ocellated lizard [Timon lepidus; (Manukyan et al., 2017)], squamates do not undergo metamorphosis such that the time evolution of color patterns must be associated to spatial reorganization of existing cell lineages or the re-distribution of their pigmented organelles (Szydłowski et al., 2016). Here, we focus on the Texas rat snake whose pigmentation pattern, produced by the three types of chromatophores, does not change after hatching. Whole-genome sequencing and assembly, in combination with a candidate-gene approach, suggest that reduced MITF expression could be associated with leucism in this species. Additional expression analyses focusing on the developmental stage at which the cell fate of the neural-crest cells is determined would be necessary to provide additional support to our observations. Obviously, one cannot exclude the possibility that additional and independent mutations in MITF exist in other leucistic animals of the species, as is the case of the numerous MITF mutations that have been identified in humans and which all cause the Waardenburg syndrome type 2 and the Tietz syndrome (Grill et al., 2013). Both syndromes are characterized by generalized hypopigmentation and hearing loss.
In humans, mutations in MITF cause the Waardenburg syndrome type 2A (Pingault et al., 2010), an autosomal-dominant condition where depigmented skin patches are associated with sensorineural deafness and sometimes Heterochromia Iridis (the two eyes presenting different colors). Some Mitf variants in mice cause, in addition to the Waardenburg syndrome traits, abnormalities in eye development, such as microphthalmia. Zebrafish is a model species for coloration with similar chromatophore types as in snakes. The zebrafish mitfa mutants lack melanophores, while they exhibit an excess of iridophores and a reduction in xanthophores (Lister et al., 1999). Despite being expressed in the RPE of wild type individuals, mitfa mutations appear to have no effect on the RPE pigmentation or development (Lister et al., 1999). Indeed, following the whole genome duplication at the origin of the teleost lineage, zebrafish have acquired a functional paralog gene, named mitfb. The latter is not expressed in neural crest cells but only in the RPE, compensating for the lack of mitfa expression in this structure in mitfa mutants (Lister et al., 2001).
One major developmental difference between snakes and zebrafish is that the latter undergo metamorphosis. As a result, the early larval and adult patterns in zebrafish are distinct; the former is generated directly from the neural crest cells, whereas the latter mainly originates from multipotent pigment progenitors located in the dorsal root ganglia (Singh et al., 2016). However, extensive support exists on the dual origin of adult xanthophores. Most larval xanthophores lose their pigmentation during metamorphosis and become cryptic, but they re-acquire pigmentation later in development and participate to the adult pattern (McMenamin et al., 2014; Saunders et al., 2019). Some of the adult xanthophores also originate from the multipotent pigment progenitors, as do most melanophores and iridophores.
Single-cell RNA sequencing analyses performed during the post-embryonic zebrafish development show that mitfa is expressed in pigment cell progenitors, melanophores and xanthophores, but not in iridophores (Saunders et al., 2019). Among xanthophores, the expression is higher in the actively differentiating population [xanthophores 2 in Saunders et al. (2019)] rather than in the cryptic one [xanthophores 1 in Saunders et al. (2019)]. This observation coupled with the reduced yellow coloration observed in mitfa zebrafish mutants suggests that the differentiation of xanthophores originating from the multipotent pigment cell progenitors is more impacted by the lack of mitfa than the state of the cryptic xanthophores, whose fate has already been defined during early larval development. Thus, the xanthophores that persist in mitfa mutants might be the early-larval xanthophores, containing pteridines. Although, the carotenoid content of mitfa mutants was not quantified, it was shown that their xanthophores maintain their pteridine pigmentation as adults (Lister et al., 1999), consistent with the presence of early-larva xanthophores.
The common origin of chromatophores from a multipotent precursor has been suggested long ago (Bagnara et al., 1979). Detailed lineage marker experiments propose the presence of a melanophore/iridophore bi-potent precursor in zebrafish (Curran et al., 2010; Petratou et al., 2018), where the expression of mitfa inhibits iridophore fate and promotes the melanophore differentiation. The co-expression of melanoblast and xanthoblasts markers, as well as the increase of melanophores in the absence of xanthophores (Minchin and Hughes, 2008) suggests that there might exist a melanophore/xanthophore bi-potent precursor as well. A common melanophore/xanthophore lineage has also been suggested with clonal experiments during the fin regeneration of the zebrafish (Tu and Johnson, 2011). Our observations in the leucistic Texas rat snake substantiate the latter suggestion. Reduced expression of MITF, coupled in some individuals with a truncated MITF protein production, could result in the absence of melanophores and xanthophores, whereas the iridophore lineage remains intact. Indeed, the density of iridophores seems to be the same between wild type and leucistic individuals, whereas zebrafish mitfa mutants lacking melanophores have an increased number of iridophores (Lister et al., 1999). Our results corroborate the possible involvement of MITF in the differentiation of melanophores and xanthophores in a non-metamorphic species, that does not bear a paralogue of this gene. We did not observe an increase of iridophores in the leucistic samples, so we cannot deduce how MITF expression could impact iridophore fate. Melanophores persist in the RPE of leucistic snakes (Figure 3), so we can assume that their fate determination is probably independent of MITF, as is the case in other leucistic species (Bharti et al., 2006). On the other hand, in leucistic animals, both xanthophores and melanophores are absent from the anterior part of the iris whereas iridophores persist (Figure 3), as is the case in the skin. In conclusion, neural-crest derived melanophores and xanthophores are absent and only iridophores remain.
Knowledge is building up on the development of Squamate pigmentation (McLean et al., 2017; Andrade et al., 2019; Kuriyama et al., 2020; Ullate-Agote et al., 2020), clearly showing that there are similarities with the zebrafish in terms of gene regulation and expression. Thus, it is now important to understand how different chromatophores are specified and migrate in these species. Using known neural crest and chromatophore markers established in the zebrafish, it is conceivable to study the spatial and temporal distribution of these cells during development. These analyses would improve our understanding of the role of MITF in chromatophore differentiation in squamates. It would also allow to visualize the routes along which chromatophores migrate to populate the skin and generate the remarkable diversity of colors and color patterns in snakes and lizards.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/assembly/, GCA_012654085.1.
The animal study was reviewed and approved by the Geneva Cantonal Veterinary Authorities.
AT conceived and directed the study and organized all wet lab experiments. AT and AU-A designed the study, analyzed all data, and wrote the manuscript. All authors read and approved the final manuscript.
Funding was provided by the University of Geneva and the Georges et Antoine Claraz Foundation. AU-A received an iGE3 Ph.D. salary award.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Ingrid Burgelin, Carine Langrez, and Valerie Haechler for technical assistance, as well as Adrien Debry and Florent Montange for maintaining and imaging the animals. Jean Deraspe and Rodrigue Peraldi participated in the TEM sample processing and imaging. The TEM sample processing and imaging were performed at the Electron Microscopy Facility, University of Lausanne (Switzerland). The computations were run on the Baobab cluster (https://baobabmaster.unige.ch/iface/) of the University of Geneva (Switzerland). Michel Milinkovitch commented on an earlier version of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.583136/full#supplementary-material
1. ^RepeatModeler Open-1.0.11. Available online at: http://www.repeatmasker.org/RepeatModeler/.
2. ^RepeatMasker Open-4.0.7. Available online at: http://www.repeatmasker.org.
Andrade, P., Pinho, C., Perez, Id. L. G., Afonso, S., Brejcha, J., Rubin, C. J., et al. (2019). Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. U. S. A. 116, 5633–5642. doi: 10.1073/pnas.1820320116
Anistoroaei, R., Krogh, A. K., and Christensen, K. (2013). A frameshift mutation in the LYST gene is responsible for the Aleutian color and the associated Chediak-Higashi syndrome in American mink. Anim. Genet. 44, 178–183. doi: 10.1111/j.1365-2052.2012.02391.x
Bagnara, J. T., Matsumoto, J., Ferris, W., Frost, S. K., Turner, W. A. Jr, Tchen, T. T., et al. (1979). Common origin of pigment cells. Science 203, 410–415. doi: 10.1126/science.760198
Baker, R. J., Bull, J. J., and Mengden, G. A. (1972). Karyotypic studies of 38 species of North-American snakes. Copeia 257:1442486. doi: 10.2307/1442486
Barbosa, M. D., Nguyen, Q. A., Tchernev, V. T., Ashley, J. A., Detter, J. C., Blaydes, S. M., et al. (1996). Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 382, 262–265. doi: 10.1038/382262a0
Baxter, L. L., and Pavan, W. J. (2013). The etiology and molecular genetics of human pigmentation disorders. Wiley Interdiscip. Rev. Dev. Biol. 2, 379–392. doi: 10.1002/wdev.72
Baynash, A. G., Hosoda, K., Giaid, A., Richardson, J. A., Emoto, N., Hammer, R. E., et al. (1994). Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell 79, 1277–1285. doi: 10.1016/0092-8674(94)90018-3
Bechtel, H. B. (1991). Inherited color defects. Comparison between humans and snakes. Int. J. Dermatol. 30, 243–246. doi: 10.1111/j.1365-4362.1991.tb04628.x
Bechtel, H. B., and Bechtel, E. (1985). Genetics of color mutations in the snake, Elaphe obsoleta. J. Heredity 76, 7–11. doi: 10.1093/oxfordjournals.jhered.a110026
Besmer, P., Manova, K., Duttlinger, R., Huang, E. J., Packer, A., Gyssler, C., et al. (1993). The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Development. Supplement, 125–137. doi: 10.1538/expanim1992.9.1
Bharti, K., Nguyen, M. T., Skuntz, S., Bertuzzi, S., and Arnheiter, H. (2006). The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment. Cell Res. 19, 380–394. doi: 10.1111/j.1600-0749.2006.00318.x
Bondurand, N., Pingault, V., Goerich, D. E., Lemort, N., Sock, E., Le Caignec, C., et al. (2000). Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum. Mol. Genet. 9, 1907–1917. doi: 10.1093/hmg/9.13.1907
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Campbell, M. S., Holt, C., Moore, B., and Yandell, M. (2014). Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinform. 48, 4–39. doi: 10.1002/0471250953.bi0411s48
Chang, C. M., Coville, J. L., Coquerelle, G., Gourichon, D., Oulmouden, A., and Tixier-Boichard, M. (2006). Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genom. 7:19. doi: 10.1186/1471-2164-7-19
Cobaleda, C., Perez-Caro, M., Vicente-Duenas, C., and Sanchez-Garcia, I. (2007). Function of the zinc-finger transcription factor SNAI2 in cancer and development. Ann. Rev. Genet. 41, 41–61. doi: 10.1146/annurev.genet.41.110306.130146
Curran, K., Lister, J. A., Kunkel, G. R., Prendergast, A., Parichy, D. M., and Raible, D. W. (2010). Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 344, 107–118. doi: 10.1016/j.ydbio.2010.04.023
Fleischman, R. A. (1993). From white spots to stem cells: the role of the Kit receptor in mammalian development. Trends Genet. 9, 285–290. doi: 10.1016/0168-9525(93)90015-A
Fock, V., Gudmundsson, S. R., Gunnlaugsson, H. O., Stefansson, J. A., Ionasz, V., Schepsky, A., et al. (2019). Subcellular localization and stability of MITF are modulated by the bHLH-Zip domain. Pigment. Cell Melanoma Res. 32, 41–54. doi: 10.1111/pcmr.12721
Fuhrmann, S., Zou, C., and Levine, E. M. (2014). Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 123, 141–150. doi: 10.1016/j.exer.2013.09.003
Gremme, G., Brendel, V., Sparks, M. E., and Kurtz, S. (2005). Engineering a software tool for gene structure prediction in higher organisms. Inform. Softw. Tech. 47, 965–978. doi: 10.1016/j.infsof.2005.09.005
Grill, C., Bergsteinsdottir, K., Ogmundsdottir, M. H., Pogenberg, V., Schepsky, A., Wilmanns, M., et al. (2013). MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Hum. Mol. Genet. 22, 4357–4367. doi: 10.1093/hmg/ddt285
Hart, J. C., and Miller, C. T. (2017). Sequence-based mapping and genome editing reveal mutations in stickleback Hps5 cause oculocutaneous albinism and the casper phenotype. G3 7, 3123–3131. doi: 10.1534/g3.117.1125
Hoff, K. J., Lomsadze, A., Borodovsky, M., and Stanke, M. (2019). Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962, 65–95. doi: 10.1007/978-1-4939-9173-0_5
Holt, C., and Yandell, M. (2011). MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 12:491. doi: 10.1186/1471-2105-12-491
Iwanishi, S., Zaitsu, S., Shibata, H., and Nitasaka, E. (2018). An albino mutant of the Japanese rat snake (Elaphe climacophora) carries a nonsense mutation in the tyrosinase gene. Genes Genet. Syst. 93, 163–167. doi: 10.1266/ggs.18-00021
Jackson, I. J. (1997). Homologous pigmentation mutations in human, mouse and other model organisms. Hum. Mol. Genet. 6, 1613–1624. doi: 10.1093/hmg/6.10.1613
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W. Z., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kelsh, R. N. (2006). Sorting out Sox10 functions in neural crest development. Bioessays 28, 788–798. doi: 10.1002/bies.20445
Kinoshita, K., Akiyama, T., Mizutani, M., Shinomiya, A., Ishikawa, A., Younis, H. H., et al. (2014). Endothelin receptor B2 (EDNRB2) is responsible for the tyrosinase-independent recessive white [mo(w) ] and mottled (mo) plumage phenotypes in the chicken. PLoS ONE 9:e86361. doi: 10.1371/journal.pone.0086361
Kremer, J. R., Mastronarde, D. N., and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. doi: 10.1006/jsbi.1996.0013
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Kuriyama, T., Misawa, H., Miyaji, K., Sugimoto, M., and Hasegawa, M. (2013). Pigment cell mechanisms underlying dorsal color-pattern polymorphism in the Japanese four-lined snake. J. Morphol. 274, 1353–1364. doi: 10.1002/jmor.20182
Kuriyama, T., Miyaji, K., Sugimoto, M., and Hasegawa, M. (2006). Ultrastructure of the dermal chromatophores in a lizard (Scincidae: Plestiodon latiscutatus) with conspicuous body and tail coloration. Zool. Sci. 23, 793–799. doi: 10.2108/zsj.23.793
Kuriyama, T., Murakami, A., Brandley, M., and Hasegawa, M. (2020). Blue, black, and stripes: evolution and development of color production and pattern formation in lizards and snakes. Front Ecol Evol. 8:232. doi: 10.3389/fevo.2020.00232
Lee, H. O., Levorse, J. M., and Shin, M. K. (2003). The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev. Biol. 259, 162–175. doi: 10.1016/S0012-1606(03)00160-X
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lister, J. A., Close, J., and Raible, D. W. (2001). Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev. Biol. 237, 333–344. doi: 10.1006/dbio.2001.0379
Lister, J. A., Robertson, C. P., Lepage, T., Johnson, S. L., and Raible, D. W. (1999). nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757–3767.
Manukyan, L., Montandon, S. A., Fofonjka, A., Smirnov, S., and Milinkovitch, M. C. (2017). A living mesoscopic cellular automaton made of skin scales. Nature 544, 173–179. doi: 10.1038/nature22031
McLean, C. A., Lutz, A., Rankin, K. J., Stuart-Fox, D., and Moussalli, A. (2017). Revealing the biochemical and genetic basis of color variation in a polymorphic lizard. Mol. Biol. Evol. 34, 1924–1935. doi: 10.1093/molbev/msx136
McMenamin, S. K., Bain, E. J., McCann, A. E., Patterson, L. B., Eom, D. S., Waller, Z. P., et al. (2014). Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science 345, 1358–1361. doi: 10.1126/science.1256251
Minchin, J. E., and Hughes, S. M. (2008). Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 317, 508–522. doi: 10.1016/j.ydbio.2008.02.058
Mitros, T., Lyons, J. B., Session, A. M., Jenkins, J., Shu, S., Kwon, T., et al. (2019). A chromosome-scale genome assembly and dense genetic map for Xenopus tropicalis. Dev. Biol. 452, 8–20. doi: 10.1016/j.ydbio.2019.03.015
Miwa, M., Inoue-Murayama, M., Aoki, H., Kunisada, T., Hiragaki, T., Mizutani, M., et al. (2007). Endothelin receptor B2 (EDNRB2) is associated with the panda plumage colour mutation in Japanese quail. Anim. Genet. 38, 103–108. doi: 10.1111/j.1365-2052.2007.01568.x
Montoliu, L., Gronskov, K., Wei, A. H., Martinez-Garcia, M., Fernandez, A., Arveiler, B., et al. (2014). Increasing the complexity: new genes and new types of albinism. Pigm. Cell Melanoma R. 27:12167. doi: 10.1111/pcmr.12167
Nakayama, T., Nakajima, K., Cox, A., Fisher, M., Howell, M., Fish, M. B., et al. (2017). No privacy, a Xenopus tropicalis mutant, is a model of human Hermansky-Pudlak Syndrome and allows visualization of internal organogenesis during tadpole development. Dev. Biol. 426, 472–486. doi: 10.1016/j.ydbio.2016.08.020
Patterson, L. B., and Parichy, D. M. (2019). Zebrafish pigment pattern formation: insights into the development and evolution of adult form. Ann. Rev. Genet. 53, 505–530. doi: 10.1146/annurev-genet-112618-043741
Petratou, K., Subkhankulova, T., Lister, J. A., Rocco, A., Schwetlick, H., and Kelsh, R. N. (2018). A systems biology approach uncovers the core gene regulatory network governing iridophore fate choice from the neural crest. PLoS Genet. 14:e1007402. doi: 10.1371/journal.pgen.1007402
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pingault, V., Ente, D., Dastot-Le Moal, F., Goossens, M., Marlin, S., and Bondurand, N. (2010). Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 31, 391–406. doi: 10.1002/humu.21211
Reissmann, M., and Ludwig, A. (2013). Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Semin. Cell Dev. Biol. 24, 576–586. doi: 10.1016/j.semcdb.2013.03.014
Saenko, S. V., Lamichhaney, S., Martinez Barrio, A., Rafati, N., Andersson, L., and Milinkovitch, M. C. (2015). Amelanism in the corn snake is associated with the insertion of an LTR-retrotransposon in the OCA2 gene. Sci. Rep. 5:17118. doi: 10.1038/srep17118
Saenko, S. V., Teyssier, J., van der Marel, D., and Milinkovitch, M. C. (2013). Precise colocalization of interacting structural and pigmentary elements generates extensive color pattern variation in Phelsuma lizards. BMC Biol. 11:105. doi: 10.1186/1741-7007-11-105
Saunders, L. M., Mishra, A. K., Aman, A. J., Lewis, V. M., Toomey, M. B., Packer, J. S., et al. (2019). Thyroid hormone regulates distinct paths to maturation in pigment cell lineages. Elife 8:40. doi: 10.7554/eLife.45181.040
Schneeberger, K. (2014). Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15, 662–676. doi: 10.1038/nrg3745
Singh, A. P., Dinwiddie, A., Mahalwar, P., Schach, U., Linker, C., Irion, U., et al. (2016). Pigment cell progenitors in zebrafish remain multipotent through metamorphosis. Dev. Cell. 38, 316–330. doi: 10.1016/j.devcel.2016.06.020
Slater, G. S., and Birney, E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6:31. doi: 10.1186/1471-2105-6-31
Stanke, M., Schoffmann, O., Morgenstern, B., and Waack, S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 7:62. doi: 10.1186/1471-2105-7-62
Steingrimsson, E., Copeland, N. G., and Jenkins, N. A. (2004). Melanocytes and the microphthalmia transcription factor network. Ann. Rev. Genet. 38, 365–411. doi: 10.1146/annurev.genet.38.072902.092717
Szydłowski, P., Madej, J. P., and Mazurkiewicz-Kania, M. (2016). Ultrastructure and distribution of chromatophores in the skin of the leopard gecko (Eublepharis macularius). Acta Zool. 97, 370–375. doi: 10.1111/azo.12132
Tachibana, M. (2000). MITF: a stream flowing for pigment cells. Pigm. Cell Res. 13, 230–240. doi: 10.1034/j.1600-0749.2000.130404.x
Teyssier, J., Saenko, S. V., van der Marel, D., and Milinkovitch, M. C. (2015). Photonic crystals cause active colour change in chameleons. Nat. Commun. 6:6368. doi: 10.1038/ncomms7368
Tu, S., and Johnson, S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725–732. doi: 10.1016/j.devcel.2011.04.013
Ullate-Agote, A., Burgelin, I., Debry, A., Langrez, C., Montange, F., Peraldi, R., et al. (2020). Genome mapping of a LYST mutation in corn snakes indicates that vertebrate chromatophore vesicles are lysosome-related organelles. Proc. Natl. Acad. Sci. U. S. A. 2020:117. doi: 10.1073/pnas.2003724117
Ullate-Agote, A., Milinkovitch, M. C., and Tzika, A. C. (2014). The genome sequence of the corn snake (Pantherophis guttatus), a valuable resource for EvoDevo studies in squamates. Int. J. Dev. Biol. 58, 881–888. doi: 10.1387/ijdb.150060at
Waits, D. S., Simpson, D. Y., Sparkman, A. M., Bronikowski, A. M., and Schwartz, T. S. (2020). The utility of reptile blood transcriptomes in molecular ecology. Mol. Ecol. Resour. 20, 308–317. doi: 10.1111/1755-0998.13110
Wasmeier, C., Hume, A. N., Bolasco, G., and Seabra, M. C. (2008). Melanosomes at a glance. J. Cell Sci. 121, 3995–3999. doi: 10.1242/jcs.040667
Waterhouse, R. M., Seppey, M., Simao, F. A., Manni, M., Ioannidis, P., Klioutchnikov, G., et al. (2018). BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. doi: 10.1093/molbev/msx319
Wei, A. H., and Li, W. (2013). HermanskyPudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigm. Cell Melanoma R. 26:12051. doi: 10.1111/pcmr.12051
Weisenfeld, N. I., Kumar, V., Shah, P., Church, D. M., and Jaffe, D. B. (2017). Direct determination of diploid genome sequences. Genome Res. 27, 757–767. doi: 10.1101/gr.214874.116
West, J. D., Fisher, G., Loutit, J. F., Marshall, M. J., Nisbet, N. W., and Perry, V. H. (1985). A new allele of microphthalmia induced in the mouse: microphthalmia–defective iris (midi). Genet Res. 46, 309–324. doi: 10.1017/S0016672300022801
Woodcock, M. R., Vaughn-Wolfe, J., Elias, A., Kump, D. K., Kendall, K. D., Timoshevskaya, N., et al. (2017). Identification of mutant genes and introgressed tiger salamander DNA in the laboratory Axolotl, Ambystoma mexicanum. Sci. Rep. 7:6. doi: 10.1038/s41598-017-00059-1
Yandell, M., and Ence, D. (2012). A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 13, 329–342. doi: 10.1038/nrg3174
Zheng, G. X., Lau, B. T., Schnall-Levin, M., Jarosz, M., Bell, J. M., Hindson, C. M., et al. (2016). Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 34, 303–311. doi: 10.1038/nbt.3432
Ziegler, I. (2003). The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigm. Cell Res. 16, 172–182. doi: 10.1034/j.1600-0749.2003.00044.x
Keywords: MITF, leucism, chromatophores, reptiles, melanophores, xanthophores, genome, pigmentation
Citation: Ullate-Agote A and Tzika AC (2021) Characterization of the Leucistic Texas Rat Snake Pantherophis obsoletus. Front. Ecol. Evol. 9:583136. doi: 10.3389/fevo.2021.583136
Received: 14 July 2020; Accepted: 15 January 2021;
Published: 09 February 2021.
Edited by:
Marcus Kronforst, University of Chicago, United StatesReviewed by:
Florian Raible, University of Vienna, AustriaCopyright © 2021 Ullate-Agote and Tzika. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Athanasia C. Tzika, YXRoYW5hc2lhLnR6aWthQHVuaWdlLmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.