
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 08 March 2021
Sec. Conservation and Restoration Ecology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.570454
This article is part of the Research Topic Trait-Based Plant Community Assembly, Ecological Restoration, and the Biocontrol of Invasive Exotic Plant Species View all 12 articles
Plant functional traits are fundamental to the understanding of plant adaptations and distributions. Recently, scientists proposed a trait-based species selection theory to support the selection of suitable plant species to restore the degraded ecosystems, to prevent the invasive exotic species and to manage the sustainable ecosystems. Based on this theory, in a previous study, we developed a species screening model and successfully applied it to a project where plant species were selected for restoring a tropical coral island. However, during this process we learned that a software platform is necessary to automate the selection process because it can flexible to assist users. Here, we developed a generalized software platform called the “Restoration Plant Species Selection (RPSS) Platform.” This flexible software is designed to assist users in selecting plant species for particular purposes (e.g., restore the degraded ecosystems and others). It is written in R language and integrated with external R packages, including the packages that computing similarity indexes, providing graphic outputs, and offering web functions. The software has a web-based graphical user interface that allows users to execute required functions via checkboxes and buttons. The platform has cross-platform functionality, which means that it can run on all common operating systems (e.g., Windows, Linux, macOS, and others). We also illustrate a successful case study in which the software platform was used to select suitable plant species for restoration purpose. The objective of this paper is to introduce the newly developed software platform RPSS and to provide useful guidances on using it for various applications. At this step, we also realized that the software platform should be constantly updated (e.g., add new features) in the future. Based on the existing successful application and the possible updates, we believe that our RPSS software platform will have broader applications in the future.
Climate change and human activities (e.g., ore mining and agricultural use) have caused the land ecosystem degraded worldwide (Chapin et al., 2000). These degraded ecosystems are known to seriously affect human economic and social life (Laughlin and Laughlin, 2013; Laughlin, 2014). As such, ecological restoration has rapidly developed to solve the urgent and complex degraded problems (Brown and Amacher, 1999; Cardinale et al., 2012). Among all, identifying the suitable species that can successfully restore the degraded ecosystems is the primary challenge in restoration science (Fry et al., 2013). However, this step requires a comprehensive understanding of the ecological restoration theory, including information on species interactions, successional processes, and resource-use patterns. Because these processes differ greatly across different ecosystems, it remains an enormous challenge. As this type of information is typically lacking, the selection of candidate species for restoration purposes are typically chosen using traditional trial-and-error method. Nevertheless, the trial-and-error method based largely on expert knowledge of the ecosystem (Rosenthal, 2003), and such information requires the involvement of a restoration practitioner who has a wealth of practical experience which is based on years of training (Padilla et al., 2009; Ostertag et al., 2015).
Plant functional traits (including morphological, physiological and phenological characteristics that are linked to plant life history strategies) are fundamental to understanding plant adaptations and distributions. In theory, plant functional traits can be useful for selecting suitable species to restore degraded ecosystems by computing the similarity index between the target species (e.g., species have been proved to have high survival rates in the degraded ecosystems) and the potential species (e.g., all possible suitable species) (Zhang et al., 2018; Wang et al., 2020). Several studies have successfully used functional traits to select suitable plant species for restoring degraded ecosystems (Bochet and García-Fayos, 2015; Ostertag et al., 2015; Guimarães et al., 2018; Werden et al., 2018; Rayome et al., 2019; Wang et al., 2020). However, each of these studies used different screening methods or different sets of functional traits to select species. A generalized framework and a software platform which can assist this selection process is necessary to people which are new to this field.
Recently, Laughlin (2014) proposed a quantitative trait-based species selection process for selecting suitable species to restore degraded ecosystems. In a previous study (Wang et al., 2020), we developed a species screening model based on this quantitative trait-based theory (Shipley et al., 2006). Also, we successfully applied this model to select best fit plant species for restoring a tropical coral island which is part of Hainan Island, China (Wang et al., 2020). However, along this way we realized that a software platform which can help automate this modeling process is necessary as it will save time for beginners.
In this study, we introduced a newly developed, web-based species selection software platform and named it as “Restoration Plant Species Selection (RPSS) Platform.” The platform aims to select suitable plant species for restoration purpose based on plant functional traits. It was developed using the high-level R programming language1 which is popular for statistical computing and graphics. It is a web-based application that can run on a wide variety of operating systems, like Windows, Linux, and macOS and it can work based on various general browsers (e.g., IE, Chrome, Firefox, and others). The software platform makes use of several external R packages that perform various functions, including computing similarity rankings and drawing multiple graphic functions. For users, there are only two sets of information needed to run the software: (1) the trait values of the target species (species which proved to meet the specific goals) and (2) the trait values of the potential species (species to be chosen from). In addition, the RPSS platform also has a graphic user interface that helps users execute each of the functions.
In this section, we will introduce the abiotic filtering theory which is the key selection theory for our software platform, and the trait-based plant species selection process in details.
Abiotic filtering theory claims that for adapting well to the limiting abiotic environment (e.g., light and soil nutrient), all species have to develop similar traits to optimize fitness and performance (Laughlin, 2014). If we have found at least one or two species that can adapt well to the specific environment in the degraded ecosystem, many species that are appropriate for restoring the degraded ecosystem can easily be selected, as long as we compare the similarities in functional traits among them.
The target species are needed as a prerequisite in this process. Generally, the target species are species that have met certain standards of durability in ecosystems. For restoration purposes, the target species are which we already know have high survival rates in the degraded ecosystems. However, in some cases, the restoration is initiated from bare soil conditions which means that the target species are not easily found based on poor background knowledge. We therefore use the traditional trial-and-error method to determine them (Wang et al., 2020).
Potential species pool consists all the possible plant species which may be suitable for our specific purposes. For restoration purposes, the potential species pool should include both historical native species and non-native species that have the potential to restore the degraded ecosystems. The non-native species are which found in regions that have similar environments with the study site (Ostertag et al., 2015). However, in some cases, native species or non-native species are not easily found. For example, the study site may be located on a highly degraded island on which almost no native plants can be found. In such situations, only non-native species will be considered for the potential species pool.
The selected functional traits should capture the key characteristics of the degraded ecosystems’ environmental conditions. For example, leaf turgor loss point should be measured to reflect plant resistance to the drought stress, if drought stress is a key characteristic for the degraded ecosystem. Additionally, the appropriate traits should be well measurable for laboratories. If there is not any information on the characteristics already, the users may try to measure all possible functional traits (e.g., commonly measured morphological and physiological traits). We focused on different species that expressed similar trait values that were relevant to our objectives.
All trait information was obtained from field observations and laboratory measurements. Many previous studies have addressed how to collect and measure the functional traits. For the detailed information of how to prepare the functional trait dataset, please refer to some previous studies (Zhang et al., 2019; Wang et al., 2020) and Supplementary Tables 2,3 in the Supplementary Materials.
In our software platform, the maximum entropy (Maxent) model (Shipley et al., 2006) was used to screen plant species that are most functionally similar to the target species. The model outputs relative abundances to screen suitable species from the potential species pool. We defined relative abundance as the index of similarity between the target species and the potential species. The larger the value of similarity index, the more ecologically similar the two species are, and vice versa. Using this index, we assumed that the most similar species to the target species was also the most suitable species for restoring the degraded ecosystems.
At the final step, the survival rates of the selected species should be monitored to check (1) if they have the high survival rates than the unselected species; and (2) if they have the comparable survival rates to the target species. If both requirements are met, that means our species screening process is succeed.
In this study, we developed a web-based software platform named “Restoration Plant Species Selection (RPSS) Platform” to aid in the trait-based plant species selection process. The RPSS platform is written in R language, and the main page of the platform is shown in Figure 1. The user only needs to prepare trait values for the target and potential species in text format (see data examples in Supplementary Tables 2,3) when using the software platform. The software outputs both text and figure results of similarity rankings for each potential plant species, ranked from highest to lowest. The returned similarity index can indicate the similarity rank of each candidate plant species to target species. To establish the whole functions of the platform, we make use of numerous external R packages to perform various functions. For example, the FactoMineR package2 is used to perform the PCA analysis, the FD package3 (Laliberté and Legendre, 2010; Laliberté and Shipley, 2010; Legendre, 2010) is used to compute the similarity rankings based on the Maxent model (Shipley et al., 2006), the ggplot2 package4 is used for graphic outputs, and the shiny package5 helps accomplish the web functions. The software is open source. Anyone who needs the software can contact the corresponding author to acquire the whole software platform.
In this section, we illustrate how to use our RPSS software platform successfully selects suitable plant species to restore a degraded ecosystem. Our study site is located in a tropical coral island which is part of Hainan Island, China. Because of multiple harsh environmental characteristics (e.g., high temperatures, strong light, drought, no soil, and other harsh conditions) (Zhang et al., 2019; Wang et al., 2020), it is difficult for plants to establish and grow in this area. For human habitation and economic development, restoration of the vegetation on this island is important and urgent. Plantations of some species are considered a quick way to vegetate the island and make the island suitable for the development of plant communities native to the region (Wolfe et al., 2015; Wang et al., 2020). The objective of this section is to use our newly developed software platform to select suitable plant species that could be used to restore the degraded ecosystem of this area. Specifically, this island can be comparable to primary succession, as it is only made of rock and sand and does not have any soil for plants to colonize. If finally, the species selected by our software platform can indeed be suitable for restoring this extremely degraded island, we believe that our software can be widely used for restoring multiple different types of degraded ecosystems.
Our study site is located in a tropical coral island which lays to the southern of Hainan Island (lying between 108°37′—111°03′ E and 18°10′—20°10′ N), China. The study site has an area of approximately 1 km2 and the mean altitude is about 5 meters (Wang et al., 2020).
The coral island our study site located has a tropical monsoon oceanic climate with a mean annual temperature of 28°C. The average annual precipitation on the island is about 2,800 mm, and most precipitation occurs between April and September. The adverse environments in this study site are characterized by high temperatures, intense light, drought and high salinity and alkalinity soil, in which it is difficult for plants to colonize and grow (Zhang et al., 2019). For human habitation and economic development, developing successful restoration management strategies for this area is exceedingly important.
As described earlier, the trait-based species screening process requires target species as a baseline. However, our restoration is initiated from bare soil condition which no native plants can be found. Only the traditional trial-and-error method can be used to acquire the target species. So, we have identified 20 species on a nearby island with similar environmental conditions to the study site. These species were cultivated (watering and fertilizing) in Wenchang City, Hainan Island for 1–3 months and then transplanted to the study site. Three years later (from the year 2014 to 2017), target species were defined as those with survival rates >90% based on trial-and-error method. Ultimately, three species Scaevola sericea, Ipomoea pes-caprae, and Cynodon dactylon‘Yangjiang’ were selected as target species for trees, vines, and herbs (Li et al., 2016; Luo et al., 2018; Wang et al., 2020).
In order to prepare the potential species pool for restoring the degraded coral island, we reviewed literatures and studied surveys of plants in four tropical regions which the climatic and environmental conditions are similar to our study site in global. The four regions are the South China Sea Islands, the South Pacific Islands/Hawaii, the Indian Ocean Islands, and the Caribbean Sea Islands. At last, 66 species were selected to form the potential species pool. These species included a wide range of plant groups containing trees, shrubs, herbs, vines, legumes, semi-mangrove plants, and some medicinal and edible plants. The list of the 66 candidate species is shown in Supplementary Table 1 (Wang et al., 2020). The seedlings of these potential species were cultivated mainly including watering and fertilizing in Wenchang City, Hainan Island.
We want to find out key traits that can assist plant species adapt well to the specific environments of the degraded ecosystem. Thus, the traits will help to select suitable species to restore this place well. Based on our previous studies (Luo et al., 2018; Wang et al., 2020), we identified 28 traits that associated with the harsh environmental conditions on the tropical coral island. The selected functional traits are relate to drought resistance, resource allocation, antioxidant and photosynthetic capacity: specific leaf area (g/cm2), leaf water conductance (mmol m-2 s-1 MPa-1), leaf dry matter content (%), stomatal conductance (mmol m–2 s–1), stomatal density (numbers mm–2), upper epidermic thickness (μm), palisade tissue (μm); spongy tissue (μm), lower epidermic thickness (μm), maximum photosynthetic rate (mol m–2 s–1), stomatal conductance (mmol m–2 s–1), instantaneous water use efficiency (μmol mol–1), transpiration rate (mol m–2 s–1) and others. The 28 selected key traits are listed in Table 1. Because we don’t have enough fresh herb leaf samples, some leaf related traits were not able to measure for herb species. As a result, we only measured 19 traits for herbs, but 28 traits for wood species. The omitted nine leaf structure-related traits are leaf/palisade/spongy tissue thickness, palisade tissue width, upper epidermis thickness, guard cell length, stomatal density, and stomatal area index. For three target species, only Cynodon dactylon‘Yangjiang’ had measured 19 traits while others all had 28 traits. Among all 66 potential species, 6 species had only 19 traits, others had 28 traits. For the selected target and potential species, traits on mature and healthy leaves of ten individuals for each species in a growing season were measured. The measurement methods and trait dataset can be available in the supporting files.
We input the trait values of target and potential species into the RPSS platform, the input file format can be found in Supplementary Tables 2,3. The software returned the final screening results in both text and graphic formats. In this case study, we used the Windows operating system and Google Chrome browser to accomplish the whole analysis process. The example graphic results are shown in Figures 2–4. For the target species Scaevola sericea, four plant species (Artocarpus heterophyllus, Pluchea indica, Hibiscus tiliaceus, and Ficus microcarpa) were selected as the potential species. For target species Ipomoea pes-caprae, six plant species (Ipomoea tuba, Pluchea indica, Pandanus tectorius, Hibiscus tiliaceus, Medicago sativa, and Sesuvium portulacastrum) were screened to be more similar to the target species. For target species Cynodon dactylon‘Yangjiang,’ four plant species (Spinifex littoreus, Lepturus repens, Miscanthus sinensis, and Cerbera manghas) were outperformed than others. After removing duplicate species, we selected a total of 12 species for vegetation restoration in this study.
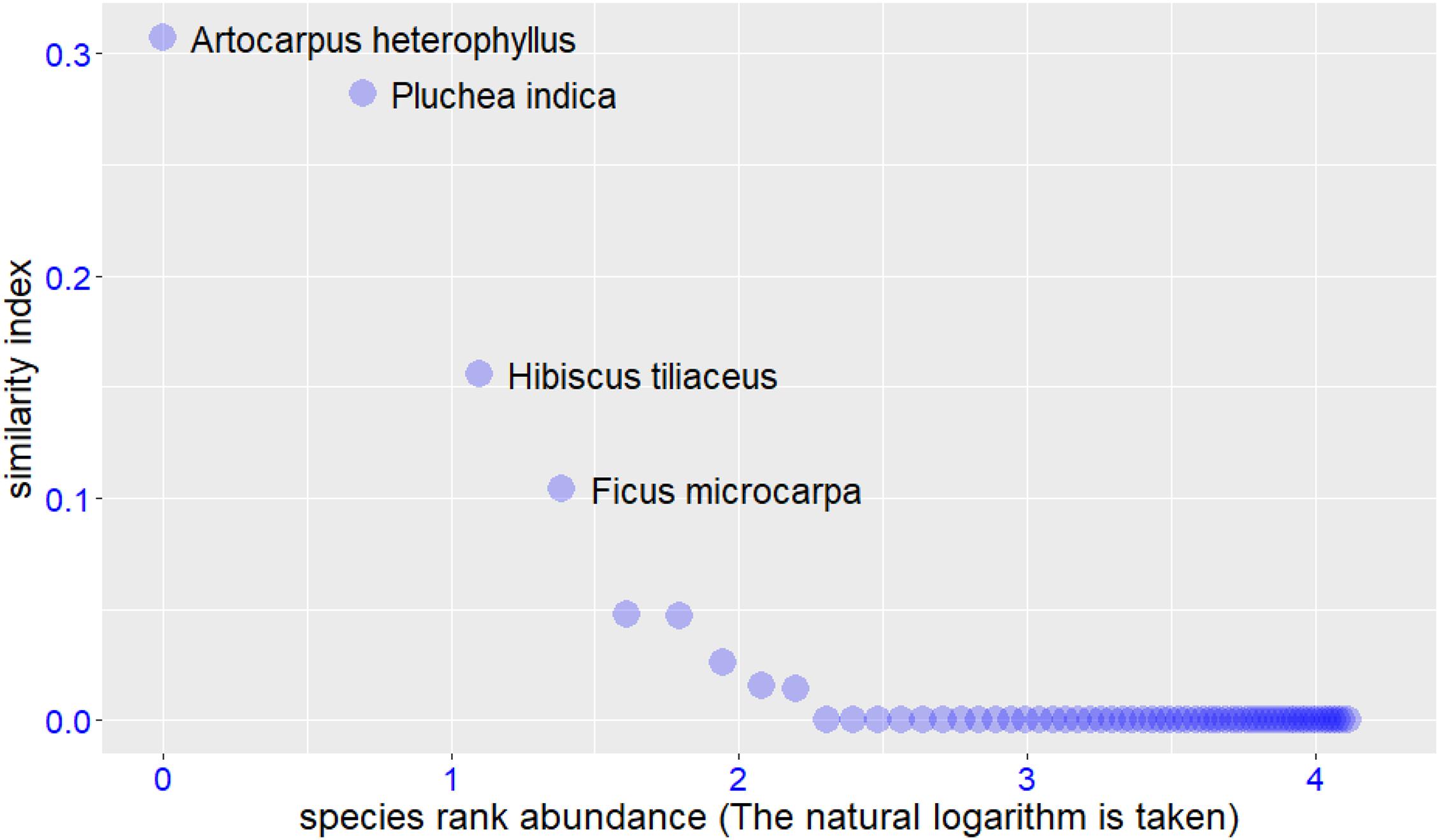
Figure 2. Species selection results for the target species Scaevola sericea. The similarity index used in this study are relative abundances which output from the Maxent model.
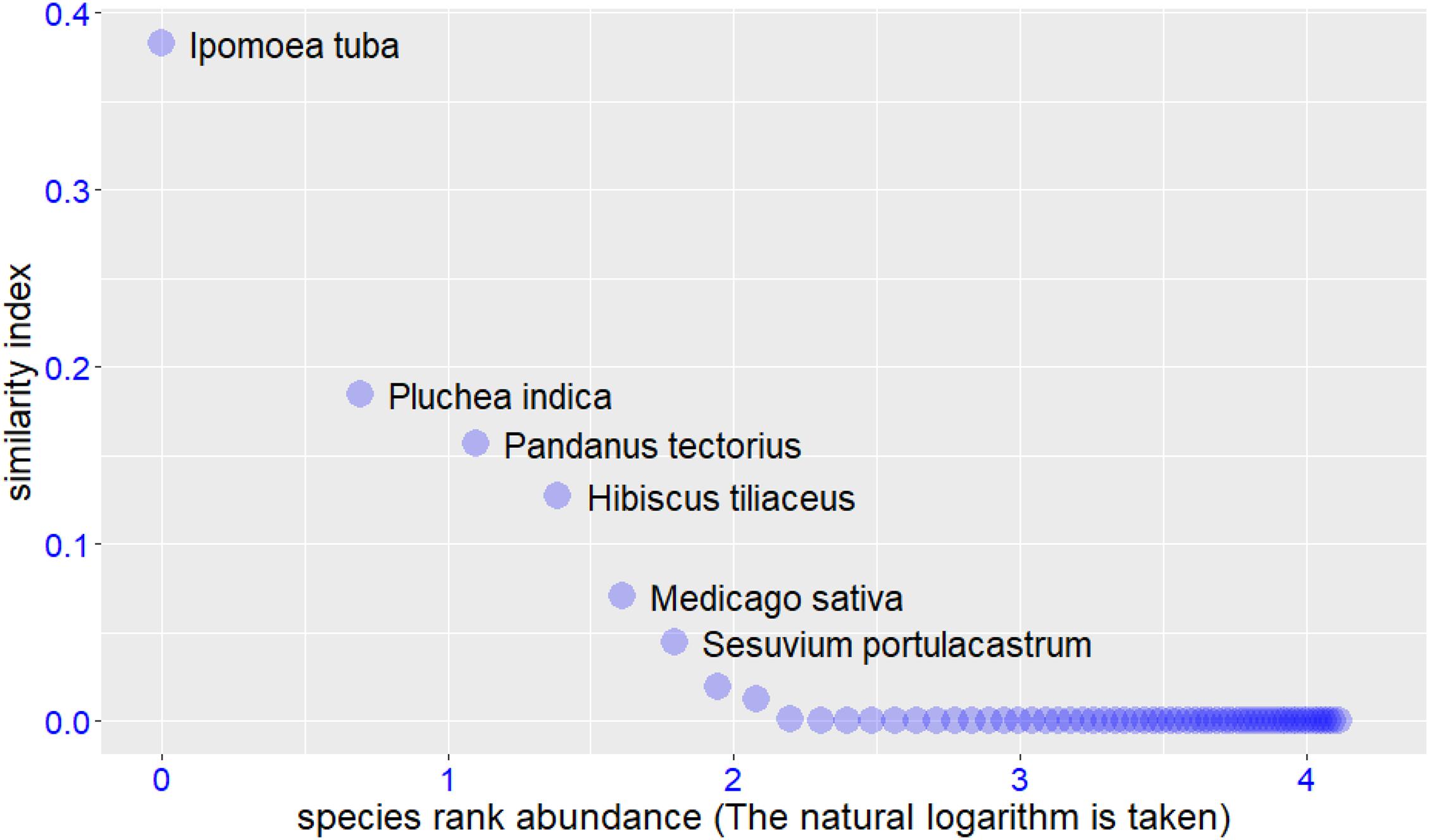
Figure 3. Similar to figure 2, but the species selection results for the target species Ipomoea pes-caprae.
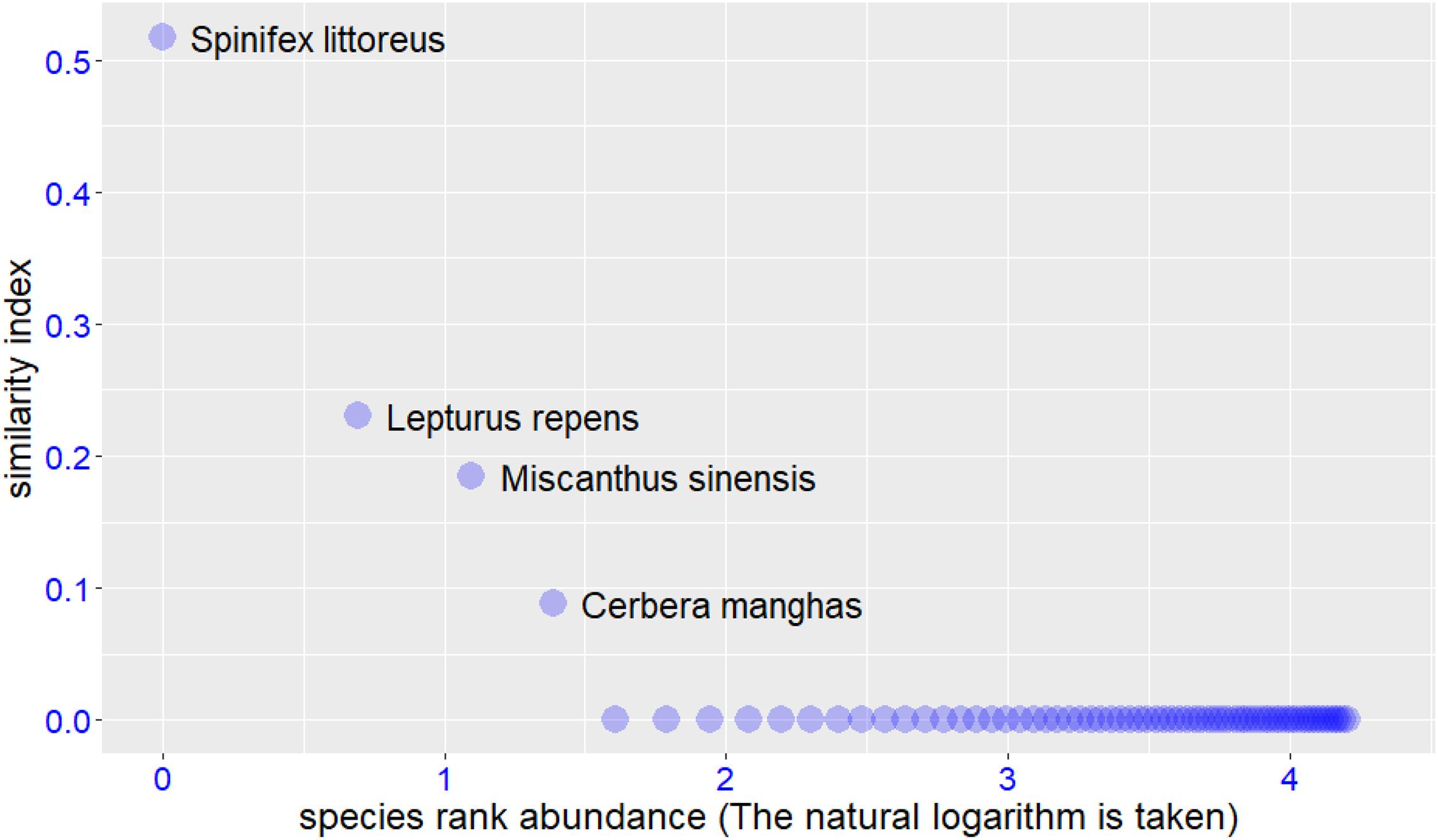
Figure 4. Similar to figure 2, but the species selection results for the target species Cynodon dactylon‘Yangjiang.’
Finally, we monitored seedling survival for all species in order to test (1) whether the selected species had the comparable survival rates with the target species; and (2) whether the selected species had higher survival rates than the unselected species. We cultivated 1,000 seedlings for each species in a nursery in Wenchang city, Hainan Island, China. In July 2017, we transplanted seedlings from our nursery to the tropical island. Each plant was planted on the same substrate type, and each row was divided into three repeat planting areas. The planting density was maintained at 80–100 plants per hectare. We recorded each plant’s annual growth rates. After 2 years’ follow-up, we recorded the final survival rate for each species using the following formula:
In final results, comparably high survival rates were found among the three target species and the twelve selected species (about 86 to 94%). However, the survival rates of the non-selected species were significantly lower than the target species in all cases (see Supplementary Figure 1). The results showed that the species selected by our software platform had significantly better survival rates than the non-selected species. This means that the software platform successfully performed the species selection for the purpose of restoration in our study site.
In this study, we introduce a newly developed web-based species screening software platform – the RPSS platform. It uses a trait-based framework to select suitable plant species for particular purposes (e.g., restore the degraded ecosystem and others). The software is implemented in the high-level R language and uses some external R packages (e.g., FD package, ggplot2 package, and shiny package) to realize the model calculations, the picture productions, and the web functions. The GUI equipped in the software allows an easy execution of purposed analyses. Because it is a web-based platform, it can run across different operating systems including Windows, Linux, and macOS. Compare with other similar work (e.g., Restoring Ecosystem Services Tool, REST, Rayome et al., 2019), our software is a web-based platform which don’t need to install any programs on the local computer. In addition, our RPSS platform not only equipped with a web-based GUI to facilitate execution of various functions, it can also run as a script program in R language batch mode. Thus, users can clearly see which commands are executed, so as to better understand the science nature behind the plant species screening process.
We demonstrated a successful application of this software platform for selecting suitable species to restore a highly degraded coral island in Hainan Island, China. The example shown in the paper is designed to demonstrate the capabilities of the software platform. We have tested our software platform in different operating systems (e.g., Windows, Linux, and macOS) and various browsers (e.g., IE, Firefox, Google Chrome), all the test samples ran normally and the results were the same which proved that our software platform was stable across different systems. The RPSS platform is an evolving program with many directions for future development (e.g., add new features, output multiple types of figures). We believe that our software platform will have broad applications in the future, especially for selecting many appropriate plant species to restore degraded ecosystems.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
CW conceived the ideas and designed the software platform, analyzed the data, and wrote the manuscript. NL collected the data. All authors contributed critically to the drafts and have final approval for publication.
This study was supported by the National Natural Science Foundation of China (Grant No. 41905094), a start-up fund from Hainan University, China [Grant No. KYQD (ZR) 1876], NSFC-Guangdong Joint Fund, China (Grant No. U1701246), and Youth Innovation Promotion Association, Chinese Academy of Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.570454/full#supplementary-material
Bochet, E., and García-Fayos, P. (2015). Identifying plant traits: A key aspect for species selection in restoration of eroded roadsides in semiarid environments. Ecol. Eng. 83, 444–451. doi: 10.1016/j.ecoleng.2015.06.019
Brown, R. W., and Amacher, M. C. (1999). “Selecting plant species for ecological restoration: a perspective for land managers,” in Revegetation with native species: Proceedings, 1997 Society for Ecological Restoration annual meeting; 1997 November 12- 15; Ft Lauderdale, FL. Ogden (UT): USDA Forest Service, Rocky Mountain Research Station. Proceedings RMRS-P-8, eds L. K. Holzworth, R. Brown, and compilers (Washington, D.C: USDA), 1–16.
Cardinale, B. J., Duffy, J. E., Gonzalez, A., Hooper, D. U., Perrings, C., Venail, P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486, 59–67. doi: 10.1038/nature11148
Chapin, F. S., Zavaleta, E. S., Eviner, V. T., Naylor, R. L., Vitousek, P. M., Reynolds, H. L., et al. (2000). Consequences of changing biodiversity. Nature 405, 234–242.
Fry, E. L., Power, S. A., and Manning, P. (2013). Trait-based classification and manipulation of plant functional groups for biodiversity–ecosystem function experiments. J. Veg. Sci. 25, 248–261. doi: 10.1111/jvs.12068
Guimarães, Z. T. M., dos Santos, V. A. H. F., Nogueira, W. L. P., and Martins, N. O. de A., and Ferreira, M. J. (2018). Leaf traits explaining the growth of tree species planted in a Central Amazonian disturbed area. For. Ecol. Manage. 430, 618–628. doi: 10.1016/j.foreco.2018.08.048
Laliberté, E., and Legendre, P. (2010). A distance-based framework for measuring functional diversity from multiple traits. Ecology 91, 299–305.
Laliberté, E., and Shipley, B. (2010). FD: Measuring Functional Diversity From Multiple Traits, and Other Tools for Functional Ecology. Vienna: R Foundation for Statistical Computing.
Laughlin, D. C. (2014). Applying trait-based models to achieve functional targets for theory-driven ecological restoration. Ecol. Lett. 17, 771–784. doi: 10.1111/ele.12288
Laughlin, D. C., and Laughlin, D. E. (2013). Advances in modelling trait-based plant community assembly. Trends Plant Sci. 18, 584–593. doi: 10.1016/j.tplants.2013.04.012
Legendre, L. P. (2010). A distance-based framework for measuring functional diversity from multiple traits. Ecology 91, 299–305. doi: 10.1890/08-2244.1
Li, J., Liu, N., Ren, H., Shen, W. J., and Jian, S. G. (2016). Ecological adaptability of seven plant species to tropical coral island habitat. Ecol. Environ. Sci. 25, 790–794.
Luo, Q., Liu, H., Wu, G. L., He, P. C., Hua, L., Zhu, L. W., et al. (2018). Using functional traits to evaluate the adaptability of five plant species on tropical coral islands. Acta Ecol. Sin. 38, 1256–1263.
Ostertag, R., Warman, L., Cordell, S., and Vitousek, P. M. (2015). Using plant functional traits to restore Hawaiian rainforest. J. Appl. Ecol. 52, 805–809. doi: 10.1111/1365-2664.12413
Padilla, F. M., Ortega, R., Sánchez, J., and Pugnaire, F. I. (2009). Rethinking species selection for restoration of arid shrublands. Basic Appl. Ecol. 10, 640–647. doi: 10.1016/j.baae.2009.03.003
Rayome, D., Dimanno, N., Ostertag, R., Cordell, S., Fung, B., Vizzone, A., et al. (2019). Restoring Ecosystem Services Tool (Rest): A Program For Selecting Species For Restoration Projects Using A Functional-Trait Approach. Gen. Tech. Rep. PSW-GTR-262. Albany, CA: U.S. Department of Agriculture, Forest Service, Pacific Southwest Research Station, 47.
Rosenthal, G. (2003). Selecting target species to evaluate the success of wet grassland restoration. Agric. Ecosyst. Environ. 98, 227–246. doi: 10.1016/s0167-8809(03)00083-5
Shipley, B., Vile, D., and Garnier, É (2006). From plant traits to plant communities: a statistical mechanistic approach to biodiversity. Science 314, 812–814. doi: 10.1126/science.1131344
Wang, C., Zhang, H., Liu, H., Jian, S., Yan, J., and Liu, N. (2020). Application of a trait-based species screening framework for vegetation restoration in a tropical coral island of China. Funct. Ecol. 6, 1193–1204. doi: 10.1111/1365-2435.13553
Werden, L. K., Alvarado, J. P., Zarges, S., Calderón, M. E., Schilling, E M., Gutiérrez, L. M., et al. (2018). Using soil amendments and plant functional traits to select native tropical dry forest species for the restoration of degraded vertisols. J. Appl. Ecol. 55, 1019–1028. doi: 10.1111/1365-2664.12998
Wolfe, B. T., Dent, D. H., Deago, J., and Wishnie, M. H. (2015). Forest regeneration under Tectona grandis and Terminalia Amazonia plantation stands managed for biodiversity conservation in western panama. New Forests 46, 157–165. doi: 10.1007/s11056-014-9448-2
Zhang, H., Chen, H., Lian, J. Y., Robert, J., Li, R. H., Liu, H., et al. (2018). Using functional trait diversity patterns to disentangle the scale-dependent ecological processes in a subtropical forest. Funct. Ecol. 32, 1379–1389. doi: 10.1111/1365-2435.13079
Keywords: vegetation restoration, plant functional traits, maximum entropy model, restoration plant species selection platform, R language
Citation: Wang C, Jian S, Ren H, Yan J and Liu N (2021) A Web-Based Software Platform for Restoration-Oriented Species Selection Based on Plant Functional Traits. Front. Ecol. Evol. 9:570454. doi: 10.3389/fevo.2021.570454
Received: 08 June 2020; Accepted: 15 February 2021;
Published: 08 March 2021.
Edited by:
Wenxing Long, Hainan University, ChinaReviewed by:
Tao Su, Nanjing Forestry University, ChinaCopyright © 2021 Wang, Jian, Ren, Yan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Liu, bGl1bmFuQHNjYmcuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.