
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 15 February 2021
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.537398
Sugar transporters, an important class of transporters for sugar function, regulate many processes associated with growth, maturation, and senescence processes in plants. In this study, a total of 35 NuMSTs were identified in the Nelumbo nucifera genome and grouped by conserved domains and phylogenetic analysis. Additionally, we identified 316 MST genes in 10 other representative plants and performed a comparative analysis with Nelumbo nucifera genes, including evolutionary trajectory, gene duplication, and expression pattern. A large number of analyses across plants and algae indicated that the MST family could have originated from STP and Glct, expanding to form STP and SFP by dispersed duplication. Finally, a quantitative real-time polymerase chain reaction and cis-element analysis showed that some of them may be regulated by plant hormones (e.g., abscisic acid), biotic stress factors, and abiotic factors (e.g., drought, excessive cold, and light). We found that under the four abiotic stress conditions, only NuSTP5 expression was upregulated, generating a stress response, and ARBE and LTR were present in NuSTP5. In summary, our findings are significant for understanding and exploring the molecular evolution and mechanisms of NuMSTs in plants.
Sugars (sucrose, monosaccharides, and polyols), as the main product of photosynthesis, occupy an important place in plant growth and development. Sugars constitute not only osmotic and signaling molecules but also metabolites and nutrients. The movement of sugars on the whole plant occurs in two different ways (Aje, 2003). One way is the loading and unloading of transport tissues (Aje, 2003). In long-distance carbon partitioning, sugar alcohols can be transported on sucrose (Zimmermann and Ziegler, 1975). The other form of movement is the sugar transporter, which controls the distribution of sugars in the sink and source, such as mediating the transport of sucrose (Kühn, 2003, 2010), monosaccharides (Büttner, 2007) or polyols (Noiraud et al., 2001; Juchauxcachau et al., 2007). Sugars exist in different organelles of source and sink cells or more biochemically for the transport of hexoses into the vacuoles (Martinoia et al., 2000), chloroplasts (Weber and Flügge, 2000), and Golgi apparatus (Wang et al., 2006). Thus, they have constructed sink and source organs (Chaffey, 2015). At present, the most studied sugar transporter belongs to the main promoter superfamily (MFS), which has a classic structure with 12 putative transmembrane helices and 11 loops (Chaudhry et al., 2007). Sugar transporters (STs) are responsible for the transmembrane transport of sucrose and play a key role in the sour-sink transport mediated by phloem and the sucrose supply of the sink tissues (Lemoine, 2000). In a number of fauna and flora studies, some ST genes have been identified, for instance, hexose transporters in Juglans regia (Decourteix et al., 2008), Vitis (Fillion et al., 1999; Hayes and Dry, 2007) and some polyol transporters in Prunus cerasus (Maurousset et al., 2003), Malus domestica (Watari et al., 2004), and Olea europea (Conde et al., 2007). In many plants, the sugar transporters, principally monosaccharide transporters (MSTs) and sucrose transporters (SUTs), have been identified. These species with sugar transporters include rice (Oryza sativa) (Aoki et al., 2003), Arabidopsis thaliana (Wormit et al., 2006), wheat (Triticum aestivum) (Aoki et al., 2002), populus (Payyavula et al., 2011), sorghum (Sorghum bicolor) (Milne et al., 2013), Medicago truncatula (Doidy et al., 2012), tomato (Reuscher et al., 2014a), and Solanum lycopersicum (Moore and Purugganan, 2003; Hackel et al., 2006), as well as woody plants such as Rosa hybrida (Henry et al., 2011). In recent years, SWEETs (Sugars Will Eventually Be Exported Transporters) have been identified as newly discovered sugar transporter proteins from Arabidopsis and Oryza sativa (Chen et al., 2010).
The MST gene family has important functions. The monosaccharide transporter encoded by the MST gene mediates the transport of multiple monosaccharides (such as glucose, fructose, mannose, galactose, and xylose). According to the location of monosaccharide transporters and the transport substances, MSTs contain seven distinct subfamilies (STP, VGT, PLT, INT, SFP, TMT, and Glct) (Büttner, 2007). In previous studies, 53 and 69 MST members were identified in Arabidopsis and pear, respectively (Li J.M. et al., 2015). Afterward, 58, 52, 64, and 76 monosaccharide transporters were identified from sweet orange (Citrus sinensis), tomato (Solanum lycopersicum), rice, and peanut (Arachis hypogaea L.) sugar transporter genes, respectively (Reuscher et al., 2014a; Zheng et al., 2014; Deng et al., 2019; Wan et al., 2020). In plants, STPs that play important roles in sugar accumulation have been proven by related studies. The STP protein belongs to the main facilitator superfamily, usually has 12 transmembrane domains and is considered a H+/sugar transporter (Ming et al., 2013). For example, the three hexose transporters were RNAi-mediated knockdown in the tomato, resulting in a significant decrease in hexose content in fruits, which proved that they could control hexoses in tomato fruits (Mccurdy et al., 2010). MdSTP13a takes up both hexose and sucrose for sorbitol-modulated pollen tube growth in apple, revealing a situation where the acquisition of sugars for pollen tube growth is regulated by a sugar alcohol (Li et al., 2020). TaSTP6, a sugar transporter protein in wheat (Triticum aestivum), was previously shown to exhibit enhanced expression in leaves upon infection by Puccinia striiformis f. sp. tritici (Pst), the causal agent of wheat stripe rust (Huai et al., 2019). Overexpression of TaSTP13 promoted Arabidopsis susceptibility to powdery mildew and led to increased glucose accumulation in the leaves (Huai et al., 2020). Furthermore, other MST subfamilies have been identified as located in the tonoplast. In Arabidopsis and rice, AtTMTs (tonoplast monosaccharide transporter), AtVGTs (vacuolar glucose transporter) and OsTMTs, which play key roles in vacuole sugar partitioning, were proven to import hexoses into the vacuole (Michael, 2007). Studies have also shown that BvTST2.1 is a sucrose-specific transporter. Current evidence shows that it also functions as a proton reversal transporter, combining sucrose input into the vacuole with proton output. Because BvTST2.1 has a high degree of amino acid sequence similarity with members of the Arabidopsis tonoplast monosaccharide transporter family, this group of proteins has been renamed vacuole sugar transporters (TSTs) (Jung et al., 2015). PpTST1 out of the TST gene family in the peachis likely responsible for sucrose accumulation in fruit. It is worth noting that silencing of PpTST1 in peach fruit also inhibited hexose accumulation suggesting that it may also have influence on hexose accumulation (Peng et al., 2020). Previous studies have indicated that AtERDL6 is a tonoplast-localized glucose exporter that releases glucose from vacuoles into the cytosol (Poschet and Büttner, 2011). In addition, sugar transporters respond to abiotic stress in plants. After cold stress treatment, the expression of AtTMT1 was upregulated, and more glucose and fructose accumulated in the vacuole membrane of Arabidopsis leaf cells to increase the stress resistance of the plant (Wormit et al., 2006; Wingenter et al., 2010). In rice, the expression of the Golgi monosaccharide transporter OsGMST1 is positively correlated to salt treatment (Cao et al., 2011). The overexpression of OsMST6 gene can confer greater tolerance to drought and salinity stress in A. thaliana (Monfared et al., 2020). In cucumber, CsTST2 is responsive to various abiotic stresses, inducing salt, drought and cold (Hu et al., 2019). Although the expression patterns and functional analysis of the sugar transporter gene family have been previously studied in a variety of plants, knowledge of the sugar transporter gene family is lacking in lotus roots.
Lotus root (Nelumbo nucifera Gaertn) is an ancient perennial aquatic herb plant that originated from India and China (Xue et al., 2012). Archeological research has estimated that the history of lotus cultivation in China is more than 2,000 years old (Shen-Miller, 2002; Na et al., 2009). In the long-term evolution of lotus, various types of mutations have appeared. After long-term artificial selection, the plants were grouped into three major types: ornament, seed, and rhizome lotus. Lotus roots, powder and seeds are recognized as nourishing foods that are rich in starch, protein, vitamins and mineral substances, have high edible value and medicinal value and are popular among many people (Pagels, 2005; Liu et al., 2010). Starch is the main storage material of lotus roots, accounting for more than 70% of the dry matter weight. Starch is the main factor that affects the quality of lotus root products (Libao et al., 2013).
In plants, sugar transporters in sugar transport, absorption and utilization affect the growth and development of plants (Wu and Zhu, 2004). Sucrose synthesized in the source organ enters the phloem under the action of sugar transporters, and after long-distance transportation, it enters the sink organ through the apoplast or extracellular pathway (Carpaneto et al., 2005). In the storage organ of lotus roots, synthetic starch from sucrose is produced by the synthesis or degradation of starch in photosynthetic tissue. It enters the storage organ via sugar transporters and synthesizes starch with the action of a series of enzymes.
However, little information is currently available on the expression of the sugar transporter gene family in different tissues and developmental stages of lotus roots. Due to the significant functions of sugar transporters in lotus roots, it is of great significance to study the gene families of sugar transporters in lotus roots. In this study, we comprehensively analyzed the influences of drought, exogenous hormones, extreme temperatures, and salinity on NuMST. In addition, based on cis-regulatory elements and qRT-PCR, we further analyzed the roles of NuMST in N. nucifera. Finally, we constructed NuMST gene interaction networks and analyzed NuMST gene expression patterns through comparative genomics. This is the first study to report on MST genes in N. nucifera, extending our comprehension of the MST gene family in evolution stress responses and providing basic information for improving the edible quality of lotus.
The N. nucifera and 10 representative genome sequences Were used for comparative analyses in this study. the genome sequences of N. nucifera Were downloaded From the NCBI genome database Under the link https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=4432. the Arabidopsis sequences Were downloaded From the TAIR database Under the link http://www.Arabidopsis.org/. the sequences of the other 9 species Were downloaded From Phytozome (Goodstein et al., 2012).
The Pfam database Was used to identify MST genes From all protein sequences of the examined species, With a threshold of e < 1e–5. ST genes Have the typical MFS domain (PF07690). Using SMART to further verify the retrieved ST candidates (Song et al., 2018). MEME Was used to search for conserved motifs (Bailey et al., 2009).
Sequence alignment and phylogenetic analyses uses MUSCLE with default parameters for Multiple sequence alignment (Tong et al., 2013). Based on alignment, we generated phylogeny using a previously reported method (Li Q. et al., 2015). Phylogenetic analyses were conducted using Maximum Likelihood (Kühn, 2010).
The NuMST genes were mapped to N. nucifera chromosomes using Circos. The Multiple Collinearity Scan toolkit (MCScanX) exhibited the collinear relationship of the NuMST genes and other selected species, and uses the Dual Systeny Plotter software1 was used to construct the syntenic analysis maps (Chen et al., 2020). Non-synonymous and synonymous (ks) were calculated using KaKs_Calculator 2.0 (Reuscher et al., 2014a).
Experimental samples were used cultivar N. nucifera seedlings “MeiRenHong,” which grew in plastic containers containing water in a controlled-environment growth chamber. Under the same condition, seedlings at the leaf stage were transferred to the following treatment. Including (1) control; (2) ABA 18.9 mM; (3) polyethylene glycol (PEG) 300 mM; (4) NaCl 102.7 mM; (5) 4°C. The samples were collected seedling leaves at the leaf stage at 0, 8, 16, 24 h after treatment and frozen in liquid nitrogen, and stored at −70°C and used for bioassays. The material sample is repeated three times.
Total RNA was isolated from treated leaves using TaKaRa MiniBEST Plant RNA Extraction Kit (Takara, Dalian, China). Total RNA was reverse transcribed into cDNA for RT-PCR (Takara, Dalian, China). To verify the primer specificity, we used the program BLAST against the Euryale ferox Salisb genome. qRT-PCR was performed in 20 μl reactions consisting of 10 μl 2 × ChamQ SYBR qPCR Master Mix (Vazyme, NanJin, China), 0.8 μl of a mixture of the forward and reverse primers, 1.0 μl cDNA template, and 8.2 μl ddH2O. The reactions were performed on a CFX-96 real-time PCR system (Bio-Rad). According to the instructions of ChamQ SYBR qPCR Master Mix (there is no need to add 50 × ROX Reference Dye when using Bio-Rad CFX96) (Liu et al., 2018; Hong et al., 2019). The qRT-PCR assays were carried out with three biological and technical replicates. According to the 2–Δ Δ CT method, gene expression levels were calculated (Pfaffl, 2001).
To identify all putative MST proteins in the N. nucifera genome, we used the HMM profile of the MFS domain (PF07690) and Blastp to search against the database. A total of 35 MSTs were identified and subjected to Pfam and SMART analyses, which are shown in Supplementary Table S1. The MST genes were clustered into seven groups: STP, VGT, PLT, INT, SFP, TMT, and Glct. The SUT subfamily was not found in N. nucifera. To study the classification and phylogenetic relationship of the MST gene family in N. nucifera, we constructed a phylogenetic tree using the MST protein. According to the phylogenetic tree (Figure 1), all the MST proteins were consistent with Arabidopsis groups.
To explore the diversity in each group, we used the MEME program to identify motifs. As shown in Figure 1, the MST proteins that share common motifs 1–10 were conversed in MST. Motifs 1, 4, 7, 8, and 10 were representative of MFS domains. Each subgroup that shared similar motifs and motif compositions was clustered into the same group. The results showed that the MST gene family has highly conserved domains and motifs (Figure 1 and Supplementary Table S1).
To study the evolution of the MST gene family in the kingdom of plants, we chose the 9 most representative Angiospermaes from low to high (6 eudicots and 1 basal angiosperm, 1 Pteridophyta and 1 Bryophyta species) for comparative analysis (Figure 2). According to the whole-genome level, the number of MSTs in each species was counted (Figure 2A). Land plants have a relatively large number of MST genes. Moreover, species that have a larger genome seem to contain more MST genes except for Am. Trichopoda and P. patens. In all species, the densities of MST proteins in A. thaliana (0.3926 number/Mb) were the highest, followed by B. rapa (0.2403 number/Mb) and C. papaya (0.1704 number/Mb), which were higher than those in lower plants. The reason is that C. papaya, Vitis vinifera, and P. trichocarpa do not have α and β duplications; Am. trichopoda, a basal angiosperm, is missing the γ duplication event. Furthermore, some species have more members of the MST gene family due to specific WGT events (Tuskan et al., 2006; Wang et al., 2011). We found that no MSTs were detected in V. carteri and C. reinhardtii. Then, a phylogenetic tree of the MST genes was constructed to analyze the evolutionary relationships of these species (Figure 2B). The phylogenetic tree indicated that the MST gene family can be divided into 7 distinct groups (STP, VGT, PLT, INT, SFP, TMT, and Glct), which is consistent with the result for A. thaliana (Figure 2C). The expansion occurred in the process of evolution from lower to higher plants, and the density of MST proteins increased as the plants evolved. From algae to angiosperms, we found that the MST gene family has highly conserved domains and motifs (Figure 2D). According to these findings, the evolutionary history of MST in the plant kingdom was constructed.
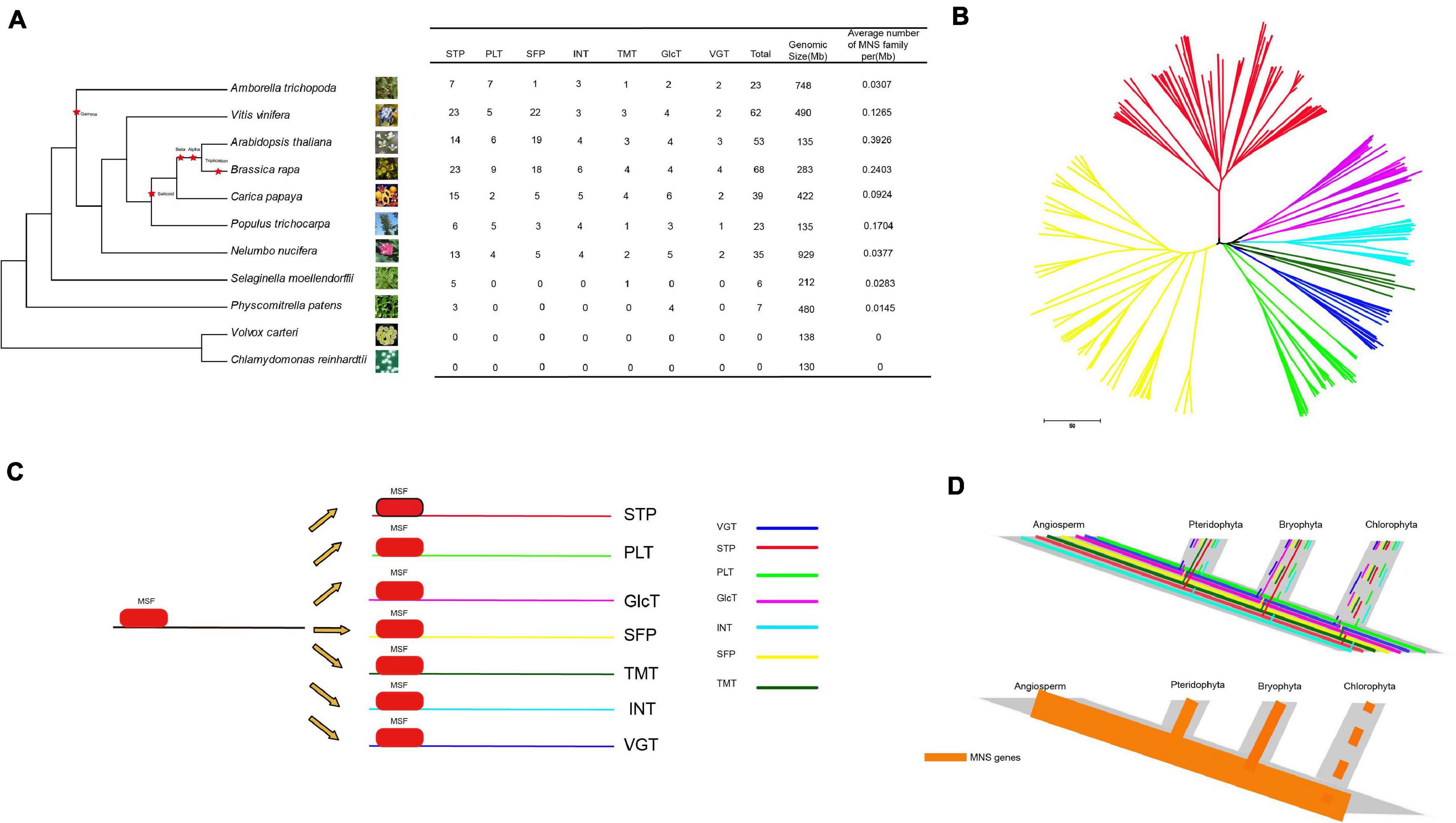
Figure 2. The analysis of NNUST genes evolution (A) comparison of the percentage of ST gene in representative species. (B) Phylogenetic relationships among MST genes. (C,D) The evolutionary pattern of STs.
The NuMST genes were unevenly mapped on the 11 scaffolds (Figure 3). Some scaffolds have more genes, whereas others have few. The scaffold contained the largest number of NuMST genes (11). In other scaffolds, the highest numbers of MST genes were found in scaffolds6 (4), followed by scaffolds2 and scaffolds3 (3), and scaffolds10, scaffolds14, scaffolds5, and scaffolds8 had only one gene. The NuMST duplicate genes were identified with PlantDGD. The duplicate genes were derived from four modes of gene duplication, including 7 whole-genome duplications (WGD), 2 tandem duplications (TD), 7 transposed duplications (TRD), and 17 dispersed duplications (DSD). These results indicate that dispersed duplication may be a major driving force for NuMST gene evolution.
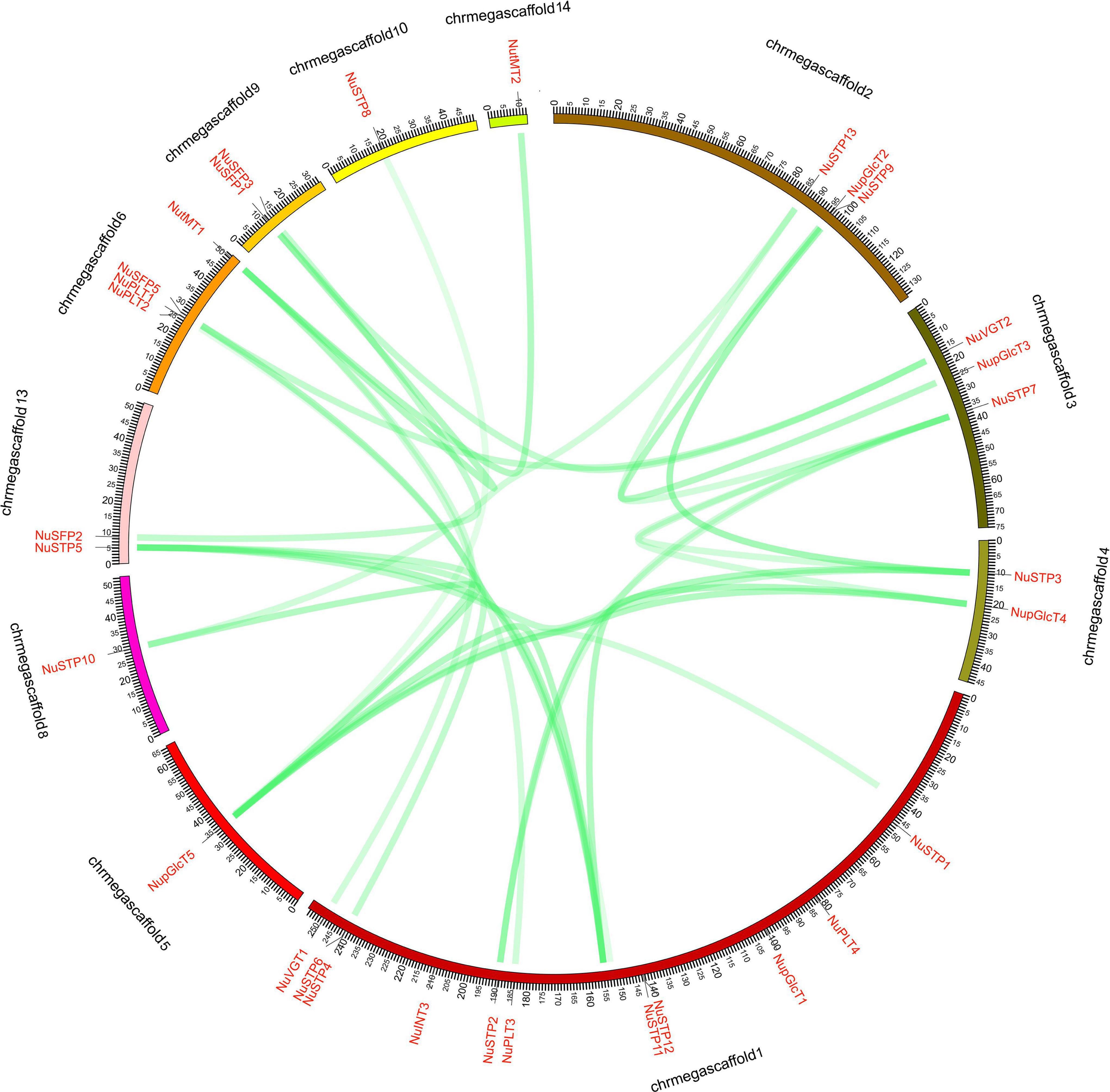
Figure 3. NNUST syntenic gene pairs of N. nucifera. The collinear pairs of gene (green lines) are shown between the 11 scaffolds.
To investigate the evolution of the NuMST family, three dicots (Arabidopsis, S. tuberosum and grape) and one monocot (maize) were constructed from four comparative microsynteny maps with N. nucifera (Figure 4). The collinear gene pairs showed syntenic relationships in maize (135), followed by grape (107), Arabidopsis (89), S. tuberosum (22) (Supplementary Table S2). In these syntenic gene pairs, we found that some genes correspond to at least 4 collinear genes, especially in maize and grape, such as NuSTP7 and NuSFP5. Some NuMST collinear gene pairs (corresponding to at least 4 collinear genes) were identified in N. nucifera/Arabidopsis, N. nucifera/grape and N. nucifera/maize, indicating that these genes may have already existed before ancestral divergence and played a vital role in the NuMST gene family during evolution. In contrast, subgroups of INT collinear gene pairs were not identified between N. nucifera and any of the other four species, indicating that they may have occurred after the divergence of dicotyledonous and monocotyledonous plants.
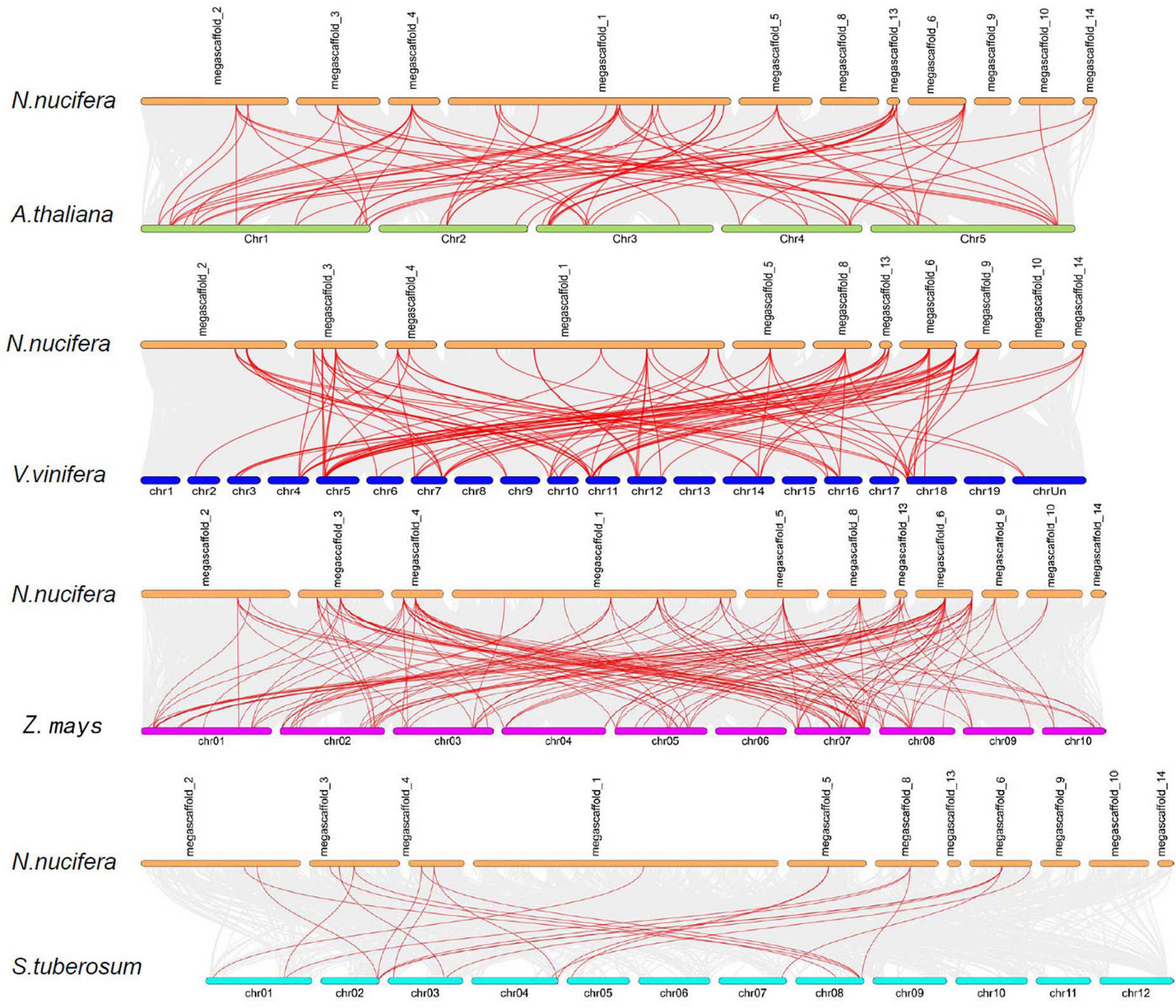
Figure 4. Synteny of NNUSTs in maize, grape, S. tuberosum and Arabidopsis. Gray lines in the background indicate the collinear blocks within N. nucifera and other plant genomes, while the red lines highlight the syntenic NNUST gene pairs.
To further investigate the evolution footprint of the NuMST family, the Ka/Ks ratios of the NuMST gene pairs were calculated between N. nucifera and Arabidopsis (Supplementary Table S3). All collinear gene pairs NuMST gene pairs had Ka/Ks < 1. Thus, these findings suggest that the NuMST gene family might have had a purifying selective pressure in the process of evolution (Supplementary Figure S1).
Comparing the expression of different sugar transporters in different tissues, we investigated divergence expression patterns (Figure 5 and Supplementary Tables S4, S6). We detected different MST gene expression levels, including the leaves, petioles, flowers, and rhizome. In the SFP subfamily, all genes were downregulated in leaves and petioles, of which the expression of the NuSFP3 gene was upregulated. The expression of the NuSFP3 gene was upregulated in the rhizome, followed by NuSFP4, NuSFP5, and NuSFP2, which remained almost unchanged. In the pGlcT subfamily, all genes were upregulated in flowers, of which the upregulated expression of NupGlcT2 and NupGlcT4 was most significant. NupGlcT4 was the predominantly expressed member in this gene family and was expressed in all four tissues, followed by NupGlcT5. In the STP subfamily, the NuSTP6 gene is a member with low activity because it is not detectable. NuSTP3 and NuSTP9 were the main members of the subfamily, showing relative levels in all four tissues, of which the upregulated expression of the STP9 gene was most significant, especially in flowers. NuSTP1, NuSTP4, NuSTP5, NuSTP7, and NuSTP8 expression was detected in leaves, but these genes were not expressed or expressed at lower levels in other tissues. In the INT and PLT subfamilies, some genes (including NuINT2, NuINT3, NuPLT1, and NuPLT2) were detected in only leaves. Both the NuINT4 and NuPLT4 genes were upregulated in only flowers. The NuINT1 and NuPLT3 genes were not expressed or downregulated in any of the four tissues. In the tMT subfamily, the expression of all genes was detected in all four tissues, and NutMT2 was the most active. In the VGT subfamily, both NuVGT1 and NuVGT2 were upregulated in leaves and flowers but expressed at lower levels in other tissues.
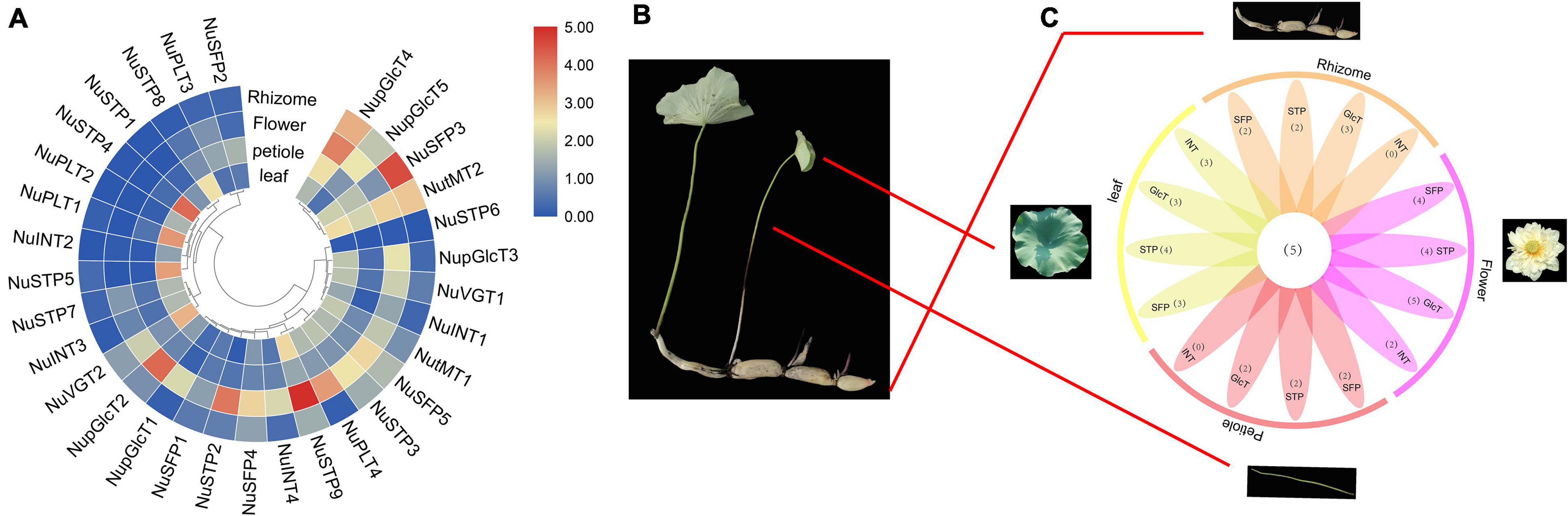
Figure 5. Phylogenetic relationships, expression among four N. nucifera tissues of ST proteins. (A) ST genes among four N. nucifera tissues. (B) N. nucifera tissues. (C) Venn diagram depicting the distribution of shared expression of the ST.
The MST gene of cis-elements was identified at the 1.5 kb promoter region. Then, we analyzed the cis-elements by the Plant Cis-acting Regulatory DNA Elements (PLACE) website. We identified the 10 most common cis-elements in MST genes in N. nucifera (Supplementary Figure S2).
In the promoter regions of the MST gene, we found a total of 10 common cis-regulatory elements, which were highly conserved among all the studied MST genes in N. nucifera (Supplementary Figure S2). We found that ARBE, TGACG, and GARE motifs responded to plant hormones, such as GA, ABA, and JA. A number of common cis-regulatory elements were responsive to both abiotic and biotic stresses, including a drought-responsive element (MBS), defense and stress responsiveness (TC-rich repeats), a light-responsive element pathogen (G-Box), low-temperature responsiveness (LTR), and one fungal elicitor-responsive element (W-box), indicating the importance of MST genes in stress tolerance. The MST genes were responsive to drought, cold, salinity and other stresses, which may be due to upstream gene specificity and the combination of corresponding cis-elements regulating the expression of MST genes. This speculation needs further verification in future work.
In plants, the MST gene family plays particularly important roles in development and stress responses. Abiotic stress conditions such as drought, extreme temperatures, and salinity, adversely affects plant growth and crop productivity. Therefore, we chose NaCl, PEG, ABA, and cold treatments to identify the stress-responsive MST genes (Figure 6 and Supplementary Tables S5, S6). With ABA treatment, we found that 23 MST genes were upregulated, and two MST genes were downregulated. Among the 31 MST genes, NuSFP5, NuSTP2, NuSTP4, and NuSTP5, their expression was over 6 times that of the control at 8 h. With NaCl treatment, 25 MST genes were upregulated. We found that NuSFP2, NuSFP3, NuSTP3, NuSTP4, NuSTP6, NuSTP7, and NuVGT2 expression was upregulated at 8 h or 16 h but downregulated at 24 h. By contrast, when PEG treatment was applied, we found that upregulated NuSTP5 and NuSTP8 expression reached the highest levels at 24 h and was more than 6 times that of the control. Including the 12 MST genes that were upregulated at 8 h. Under Cold treatment, 7 MST genes were upregulated at 8 h and reached their highest levels. Among the 31 MST genes, especially NuINT3, NuSTP2, and NuSTP5, their upregulated expression was over 5 times that of the control at 8 h; the upregulated expression of NuINT1, NuINT2, NuSTP3, NuSTP5, and NuSTP8 was over 4 times that of the control at 24 h. Specifically, we observed that NuVGTs did not respond or responded less to the four stress treatments. Under the four abiotic stress conditions, only NuSTP5 expression was upregulated at 8 h and 24 h, generating a stress response. Combined with cis-element analysis, ARBE and LTR were present in NuSTP5.
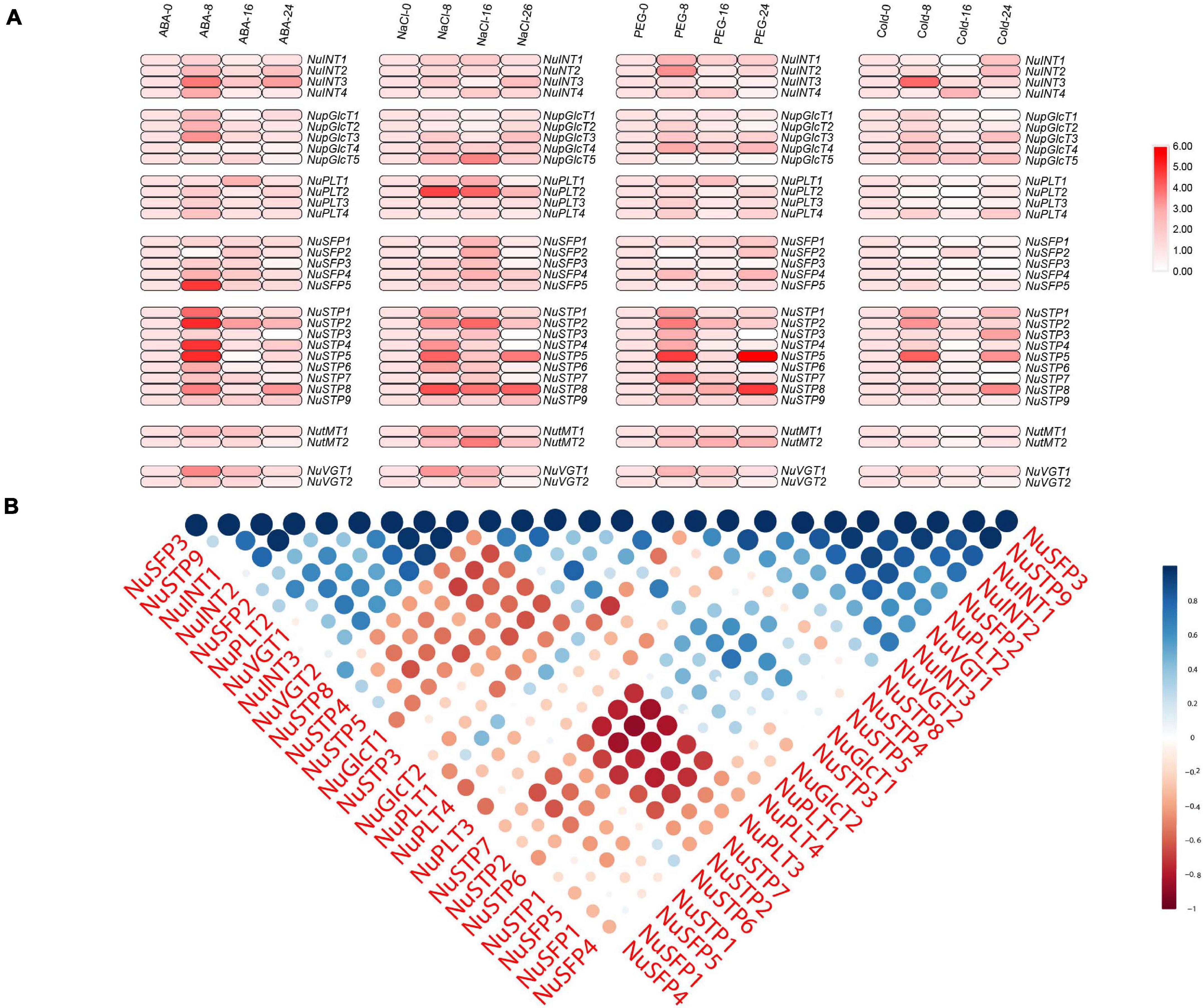
Figure 6. Expression analysis of NNUSTs under abiotic stresses. (A) Heat map representation and hierarchical clustering of NNUSTs during a ABA stress, Nacl stress, PEG stress, cold stress. (B) Correlation analysis by using the R package program.
To further investigate the link between these MST genes, PCCs based on the relative expression of these genes established a related and co-regulatory network (Figure 6B). We collected NuMST gene pairs evaluated at the p ≥ 0.05 significance level and visualized them to build a hormone and abiotic stress co-regulatory network (Supplementary Figure S3). In the co-regulatory network, a total of 35 nodes were positively significantly correlated with the gene pairs. The NuMST gene interaction network shows a very complex correlation with other genes of N. nucifera, which may reveal that NuMST genes take part in a number of basic mechanisms and are regulated by many upstream genes and/or regulate many downstream factors. In this described network, increasing cooperation and obtaining new functions help plants adapt to abiotic stresses and changing environments.
The production of sugar by photosynthesis in plants is a very important process. Sugar transporters are distributed in various tissues and organs and expressed in different tissues. Sugar transporters play different roles in the growth and development of plants. In the current study, we found that the MST gene family was likely to be involved in growth, development, and responses to stress.
If gene products are involved in complex regulatory networks, these genes may be retained (Birchler and Veitia, 2007; Aad et al., 2012). Previous research has shown that gene duplicates generate new functions or expression through the processes of neo- and subfunctionalization, and they are retained more frequently than the classical model permits. In our study, we found 35 NuMST genes encoding putative sugar transporters in the N. nucifera genome. The NuSTP (13) genes were preferably retained in N. nucifera. The duplication genes went through three fates: subfunctionalization, neofunctionalization, or nonfunctionalization (deletion or pseudogenization). In subfunctionalization, the expression or function of the progenitor gene is divided into daughter genes by complementary mutations in regulatory or coding regions (Hughes and Friedman, 2005). In neofunctionalization, through preliminary relaxation of purification choices and accumulation of mutations, under either neutral or positive selection, related or new functions may appear in one of the duplicates, endowing it with a new function (Gu et al., 2002). The ST genes provided opportunities to gain functional diversification.
Expression analysis of the MST gene family of N. nucifera in different tissues was performed to identify the most important MSTs. In our study, NuINT2 and NuINT3 were expressed at relatively low levels in all tested tissues but responded to abiotic stresses and sugar treatments very rapidly, which indicated that NuINT2 and NuINT3 responded to environmental changes. The NuINT4 gene was expressed only in flowers of N. nucifera. In the tomato, SlINT4 was expressed in all tested tissues, apart from fruits (days after flowering) (Reuscher et al., 2014b). pGlcT has a significant contribution to the output of chloroplast starch degradation products in Arabidopsis leaves and the starch-mediated export of pGlcT photoassimilates (Cho et al., 2010). A pGlcT protein was first discovered in olive trees during fruit development (Rafal et al., 2003). CspGlcT2 and CspGlcT4 were upregulated as fruit ripened in the orange (Citrus sinensis Osbeck) (Zheng et al., 2014). In our study, all pGlcT genes were expressed in flowers, of which the expression of NupGlcT2 and NupGlcT4 was most significant, where NupGlcT4 was a ubiquitously expressed gene with high expression, especially at ripening rhizomes, consistent with the results in oranges. NupGlcT5 showed a strong response to salt treatment, which supported its role in tolerance to salt stress (Cao et al., 2011). NuPLT1, NuPLT2, and NuPLT4 were expressed in leaves, among which NuPLT4 was the most active. However, NuPLTs were not detected in rhizomes, indicating that these genes may not play an important role in the accumulation of sugar during the development of rhizomes, which is consistent with the results of pear research (Li J.M. et al., 2015). NuPLT2 was strongly induced by NaCl treatment, which suggests an important role of NuPLT2 in tolerance to salt stress. We found that NuSTP3 and NuSTP9 were ubiquitously expressed genes with high expression in flowers. In particular, NuSTP9 was not sensitive to the abiotic stress response, which is consistent with the research results of sweet orange (Zheng et al., 2014). NuSTP1 and NuSTP6 were expressed at relatively low levels in all tested tissues but responded to ABA treatment. This has the same result as the wheat study. In wheat research, it was found that Pst infection caused increased accumulation of abscisic acid (ABA) and that application of exogenous ABA significantly enhanced TaSTP6 expression (Huai et al., 2019). NuSTP4, NuSTP5, and NuSTP8 were only active in leaves, but they were all induced by ABA, NaCl, and PEG, suggesting that they might be important emergency response genes (Figure 7). NuSTP8 is an important abiotic stress gene, which is consistent with the results of rice (Mamun et al., 2006). In previous studies, TMTs of Arabidopsis and rice have been identified. In Arabidopsis, AtTMT1 and AtTMT2 are located in the VM and considered to be antiporters of glucose or fructose/H+, which introduce sugar into vacuoles (Wormit et al., 2006; Wingenter et al., 2010). In sweet orange, CsTMT2 showed relatively high expression levels in fruits (Zheng et al., 2014). In our study, NutMT1 and NutMT2 were ubiquitously expressed, but NutMT2 showed more redundancy in the petiole and rhizome sheath than NutMT1. NutMT2 plays a role in vacuolar sugar accumulation during fruit ripening, which is consistent with the results of sweet orange studies. Moreover, NutMT2 was induced by salt treatment, which suggests an important role of NutMT2 in tolerance to salt stress. Both NuVGT1 and NuVGT2 are expressed in leaves and flowers, with NuVGT1 responding to ABA and salt. There are few studies on sugar transporters in lotus roots, which provides a good reference for future research on lotus root sugar transporters.
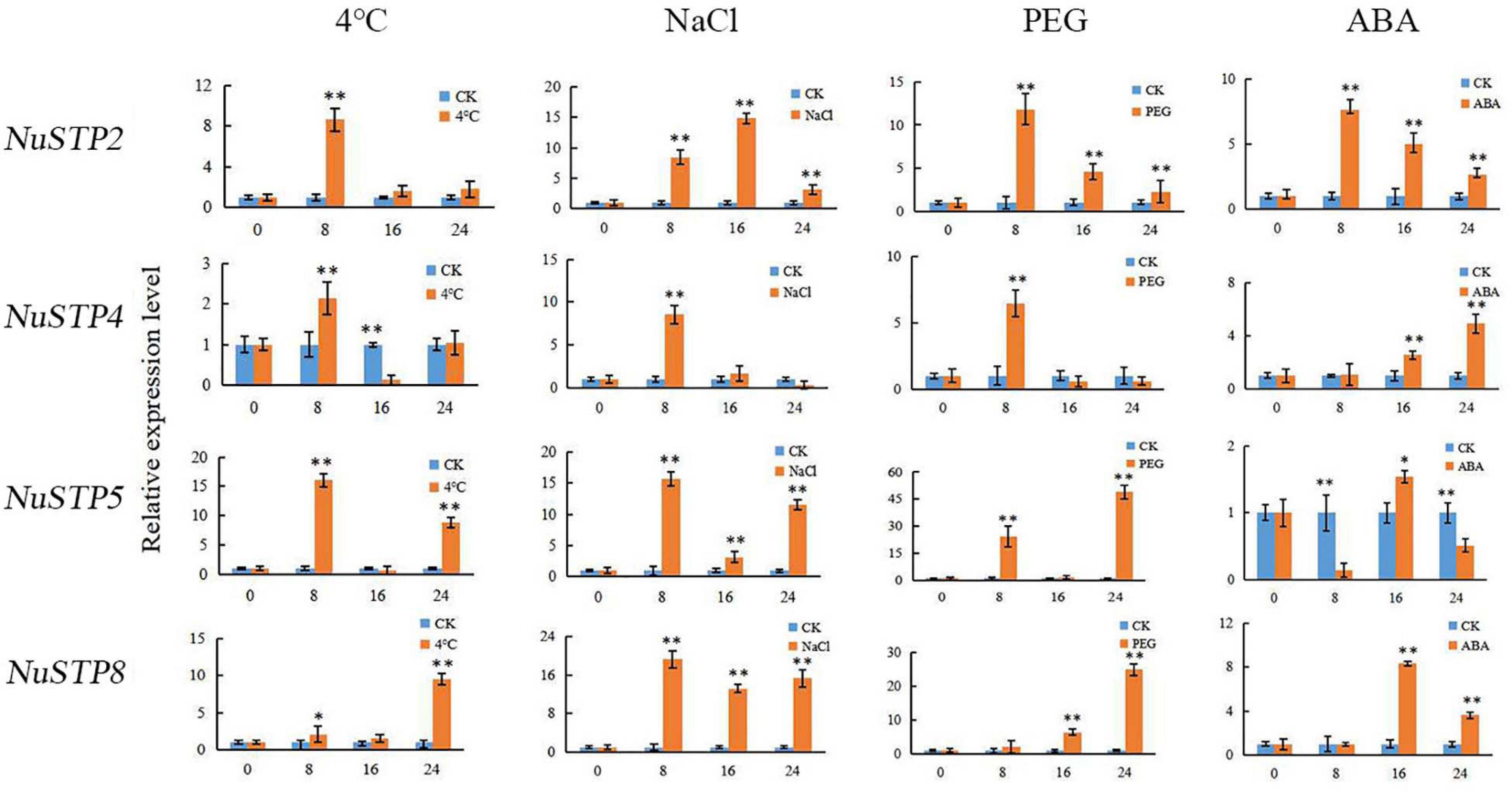
Figure 7. Relative expression levels of NNUSTP2, NNUSTP4, NNUSTP5 and NNUSTP8 under four abiotic stresses. ** indicate a significant difference at P < 0.01.
According to the sequences of cis-elements and motifs analyzed, MST gene promoters included common cis-regulatory elements and several common motifs and domains. The MFS domain was originally thought to play a major role in sugar absorption. These observations have led to MFS being much broader in nature and more diverse in function than previously thought. The common motifs of STs, such as DOFCOREZM, suggest that DNA binding may play a significant role in regulating gene expression at the level of ST gene activity. For instance, the expression of AtSUT2 is regulated by a putative HD-Zip transcription factor and a DOFCOREZM protein binding site in close cooperation (Schneidereit and Imlau, 2008). In previous studies, the concentration of sugar-responsive elements was regulated by some transporter genes. Transcriptional regulation via sugars, such as the transcriptional regulation of VvHT1 by glucose, was demonstrated (Atanassova and Delrot, 2003; Conde et al., 2006). Furthermore, the MYBCOREATCYCB1 promoter was found in ST members of pear, which showed that different species might have different transcriptional regulation mechanisms in the ST gene family.
Today, the great diversity of terrestrial plants on Earth provides an opportunity to study the evolution of gene families in major lineages that vary in functional complexity and life histories (Kenrick and Crane, 1997; Karol et al., 2001). In plants, gene families of evolution which prevalently combined of tandem, segmental and whole genome duplication (polyploidy) events. N. nucifera experienced whole genome triplication (WGT) events (Wang et al., 2011; Cheng et al., 2013). The ancient ST gene family was discovered in the moss lineage and isolated from vascular plants > 410 million years ago (Johnson et al., 2006). In our study, the MST gene family was detected in P. patens, which was consistent with ancient MST from the moss lineage. At the same time, the MST of the gene family shared similar evolution with MST. The expansion of gene families in plants is often due to multiple gene duplications caused by ancestral genes. In Arabidopsis, the large expansion of the MST gene family is attributed to tandem duplications. However, MST gene duplications in N. nucifera belong to dispersed duplications (Figure 4). Therefore, it can be concluded that the MST family experienced large expansions caused by WGDs and that most of the MST family began to expand after the differentiation of bryophytes and chlorophyta.
In summary, we comprehensive analyzed the evolutionary pattern, gene synteny, gene duplication or losses, stress treatments, and interaction network of MST genes involved in the sugar transport pathway. A total of 316 MST genes were identified among 10 representative species. The expansion of MST occurs during the evolution from low plants to high plants. The analysis of promoter sequences showed that the MST gene families of different species have different transcriptional regulatory effects. The expression analysis indicated that most MST genes respond to different stress treatments. Among these, NuSTP2, NuSTP4, NuSTP5, and NuSTP8 could be used as excellent candidate genes to improve various stress tolerances of Nelumbo nucifera. This speculation needs further verification in future work. Our study can provide comprehensive functional characterization of MST genes genes through molecular genetics studies and reverse genetic methods.
The N. nucifera and 10 representative genome sequences were used for comparative analyses in this study. The genome sequences of N. nucifera were downloaded from NCBI genome database under the link https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=4432. The Arabidopsis sequences were downloaded from TAIR database under the link http://www.Arabidopsis.org/. The sequences of the other 9 species were downloaded from phytozome (Goodstein et al., 2012).
PW conceived and designed the experimental design. LL, SZ, and YZ contributed to the experimental work. All authors read and approved the final manuscript.
This work was supported by the China Agriculture Research System (CARS-24). The funding bodies had no role in the design, collection and analysis, interpretation of data, or in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.537398/full#supplementary-material
Supplementary Figure 1 | The Ka/Ks ratios of the NNUST gene pairs between N. nucifera and Arabidopsis.
Supplementary Figure 2 | The ST genes of cis-elements.
Supplementary Figure 3 | The phylogeny and motif of 6 species.
DAF, days after flowering; WGT, whole genome triplication; MST, monosaccharide transporters; ST, sugar transporter; SUT, sucrose transporter; FPKM, Fragments per kilobase of exon per million fragments mapped; PCC, Pearson Correlation Coefficient.
Aad, G., Abajyan, T., Abbott, B., Abdallah, J., Khalek, S. A., Abdelalim, A., et al. (2012). Observation of a new particle in the search for the Standard Model Higgs boson with the ATLAS detector at the LHC. Phys. Lett. B 716, 1–29.
Aje, V. B. (2003). The phloem, a miracle of ingenuity. Plant Cell Environ. 26, 125–149. doi: 10.1046/j.1365-3040.2003.00963.x
Aoki, N., Hirose, T., Scofield, G. N., Whitfeld, P. R., and Furbank, R. T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44, 223–232. doi: 10.1093/pcp/pcg030
Aoki, N., Whitfeld, P., Hoeren, F., Scofield, G., Newell, K., Patrick, J., et al. (2002). Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol. Biol. 50, 453–462.
Atanassova, R., and Delrot, S. (2003). Sugar-regulated expression of a putative hexose transport gene in grape. Plant Physiol. 131, 326–334. doi: 10.1104/pp.009522
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208.
Birchler, J. A., and Veitia, R. A. (2007). The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19, 395–402. doi: 10.1105/tpc.106.049338
Büttner, M. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581, 2318–2324. doi: 10.1016/j.febslet.2007.03.016
Cao, H., Guo, S., Xu, Y., Jiang, K., Jones, A. M., and Chong, K. (2011). Reduced expression of a gene encoding a Golgi localized monosaccharide transporter (OsGMST1) confers hypersensitivity to salt in rice (Oryza sativa). J. Exp. Bot. 62, 4595–4604. doi: 10.1093/jxb/err178
Carpaneto, A., Geiger, D., Bamberg, E., Sauer, N., Fromm, J., and Hedrich, R. (2005). Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. J. Biol. Chem. 280, 21437–21443. doi: 10.1074/jbc.m501785200
Chaffey, N. (2015). Strasburger’s Plant Sciences: Including Prokaryotes and Fungi [Book Review], Vol. 2, Berlin: Springer, 3–6.
Chaudhry, M. T., Huang, Y., Shen, X. H., Poetsch, A., Jiang, C. Y., and Liu, S. J. (2007). Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology 153, 857–865. doi: 10.1099/mic.0.2006/002501-0
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532.
Cheng, F., Mandáková, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Cho, M. H., Lim, H., Shin, D. H., Jeon, J. S., Bhoo, S. H., Park, Y. I., et al. (2010). Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol. 190, 101–112. doi: 10.1111/j.1469-8137.2010.03580.x
Conde, C., Agasse, A., Glissant, D., Tavares, R., Gerós, H., and Delrot, S. (2006). Pathways of glucose regulation of monosaccharide transport in grape cells. Plant Physiol. 141, 1563–1577. doi: 10.1104/pp.106.080804
Conde, C., Silva, P., Agasse, A., Lemoine, R., Delrot, S., Tavares, R., et al. (2007). Utilization and transport of mannitol in Olea europaea and implications for salt stress tolerance. Plant Cell Physiol. 48, 42–53. doi: 10.1093/pcp/pcl035
Decourteix, M., Alves, G., Bonhomme, M., Peuch, M., Ben, B. K., Brunel, N., et al. (2008). Sucrose (JrSUT1) and hexose (JrHT1 and JrHT2) transporters in walnut xylem parenchyma cells: their potential role in early events of growth resumption. Tree Physiol. 2008, 215–224. doi: 10.1093/treephys/28.2.215
Deng, X., An, B., Zhong, H., Yang, J., and Li, Y. (2019). A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 10:239. doi: 10.3390/genes10030239
Doidy, J., Grace, E., Kühn, C., Simon-Plas, F., Casieri, L., and Wipf, D. (2012). Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 17, 413–422. doi: 10.1016/j.tplants.2012.03.009
Fillion, L., Picaud, S., Coutos-Thevenot, P., Romieu, C., Delrot, S., Ageorges, A., et al. (1999). Cloning and expression of a hexose transporter gene expressed during the ripening of grape berry. Plant Physiol. 120, 1083–1093. doi: 10.1104/pp.120.4.1083
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186.
Gu, Z., Nicolae, D., Lu, H. H., and Li, W. H. (2002). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18, 609–613. doi: 10.1016/s0168-9525(02)02837-8
Hackel, A., Schauer, N., Carrari, F., Fernie, A. R., Grimm, B., and Kühn, C. (2006). Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. Cell Mol. Biol. 45, 180–192. doi: 10.1111/j.1365-313x.2005.02572.x
Hayes, M. A., and Dry, I. B. (2007). Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. J. Exp. Bot. 58, 1985–1997. doi: 10.1093/jxb/erm061
Henry, C., Rabot, A., Laloi, M., Mortreau, E., Sigogne, M., Leduc, N., et al. (2011). Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud burst in Rosa sp. Plant Cell Environ. 34, 1776–1789. doi: 10.1111/j.1365-3040.2011.02374.x
Hong, Y., Wang, Z., Liu, X., Yao, J., and Zhu, J. K. (2019). Two chloroplast proteins negatively regulate plant drought resistance through separate pathways. Plant Physiol. 182, 1007–1021. doi: 10.1104/pp.19.01106
Hu, B., Huang, W., Dong, L., Liu, S., and Zhou, Y. (2019). Molecular cloning and functional analysis of a sugar transporter gene (CsTST2) from cucumber (Cucumis sativus L.). Biotechnol. Biotechnol. Equip. 33, 118–127. doi: 10.1080/13102818.2018.1555011
Huai, B., Yang, Q., Qian, Y., Qian, W., and Liu, J. (2019). ABA-induced sugar transporter TaSTP6 promotes wheat susceptibility to stripe rust. Plant Physiol. 181, 1328–1343. doi: 10.1104/pp.19.00632
Huai, B., Yang, Q., Wei, X., Pan, Q., Kang, Z., and Liu, J. (2020). TaSTP13 contributes to wheat susceptibility to stripe rust possibly by increasing cytoplasmic hexose concentration. BMC Plant Biol. 20:49. doi: 10.1186/s12870-020-2248-2
Hughes, A. L., and Friedman, R. (2005). Expression patterns of duplicate genes in the developing root in Arabidopsis thaliana. J. Mol. Evol. 60, 247–256. doi: 10.1007/s00239-004-0171-z
Johnson, D. A., Hill, J. P., and Thomas, M. A. (2006). The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evol. Biol. 6:64. doi: 10.1186/1471-2148-6-64
Juchauxcachau, M., Landouararsivaud, L., Pichaut, J. P., Campion, C., Porcheron, B., Jeauffre, J., et al. (2007). Characterization of AgMaT2, a plasma membrane mannitol transporter from celery, expressed in phloem cells, including phloem parenchyma cells. Plant Physiol. 145, 62–74. doi: 10.1104/pp.107.103143
Jung, B., Ludewig, F., Schulz, A., Meißner, G., Wöstefeld, N., Flügge, U. I., et al. (2015). Identification of the transporter responsible for sucrose accumulation in sugar beet taproots. Nat. Plants 1:14001.
Karol, K. G., Mccourt, R. M., Cimino, M. T., and Delwiche, C. F. (2001). The closest living relatives of land plants. Science 294, 2351–2353. doi: 10.1126/science.1065156
Kenrick, P., and Crane, P. R. (1997). The origin and early diversification of land plants: a cladistic study. Int. J. Plant Sci. 392:393.
Kühn, C. (2003). A comparison of the sucrose transporter systems of different plant Species – Kühn – 2008 – plant biology – wiley online library. Plant Biol. 5, 215–232. doi: 10.1055/s-2003-40798
Kühn, C. (2010). A comparison of the sucrose transporter systems of different plant species. Plant Biol. 5, 215–232. doi: 10.1055/s-2003-40798
Lemoine, R. (2000). Sucrose transporters in plants: update on function and structure. Biochim. Biophys. Acta 1465, 246–262. doi: 10.1016/s0005-2736(00)00142-5
Li, C., Meng, D., Piñeros, M. A., Mao, Y., Dandekar, A. M., and Cheng, L. (2020). A sugar transporter takes up both hexose and sucrose for sorbitol-modulated in vitro pollen tube growth in apple. Plant Cell 32, 449–469. doi: 10.1105/tpc.19.00638
Li, J. M., Zheng, D. M., Li, L. T., Qiao, X., Wei, S. W., Bai, B., et al. (2015). Genome-wide function, evolutionary characterization and expression analysis of sugar transporter family genes in pear (Pyrus bretschneideri Rehd). Plant Cell Physiol. 56, 1721–1737. doi: 10.1093/pcp/pcv090
Li, Q., Zhang, N., Zhang, L., and Ma, H. (2015). Differential evolution of members of the rhomboid gene family with conservative and divergent patterns. New Phytol. 206, 368–380. doi: 10.1111/nph.13174
Libao, C., Shuyan, L., Jingjing, Y., Li, L., and Chen, X. (2013). Genome-wide analysis of differentially expressed genes relevant to rhizome formation in lotus root (Nelumbo nucifera Gaertn). PLoS One 8:e67116. doi: 10.1371/journal.pone.0067116
Liu, J., Zhang, M., and Wang, S. (2010). Processing characteristics and flavour of full lotus root powder beverage. J. Sci. Food Agric. 90, 2482–2489. doi: 10.1002/jsfa.4110
Liu, X., He, Z., Yin, Y., Xu, X., Wu, W., and Li, L. (2018). Transcriptome sequencing and analysis during seed growth and development in Euryale ferox Salisb. BMC Genomics 19:343. doi: 10.1186/s12864-018-4707-9
Mamun, E. A., Alfred, S., Cantrill, L. C., Overall, R. L., and Sutton, B. G. (2006). Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 30, 583–591. doi: 10.1016/j.cellbi.2006.03.004
Martinoia, E., Massonneau, A., and Frangne, N. (2000). Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol. 41, 1175–1186. doi: 10.1093/pcp/pcd059
Maurousset, L., Lemoine, R., Yoo, S. D., and Nocker, S. V. (2003). Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol. 131, 1566–1575. doi: 10.1104/pp.102.016725
Mccurdy, D. W., Dibley, S., Cahyanegara, R., Martin, A., and Patrick, J. W. (2010). Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Mol. Plant 3, 1049–1063. doi: 10.1093/mp/ssq050
Michael, B. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581, 2318–2324. doi: 10.1016/j.febslet.2007.03.016
Milne, R. J., Byrt, C. S., Patrick, J. W., and Grof, C. P. L. (2013). Are sucrose transporter expression profiles linked with patterns of biomass partitioning in Sorghum phenotypes? Front. Plant Sci. 4:223. doi: 10.3389/fpls.2013.00223
Ming, R., VanBuren, R., Liu, Y., Yang, M., Han, Y., Li, L. T., et al. (2013). Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14:R41.
Monfared, H. H., Chew, J. K., Azizi, P., Xue, G. P., Ee, S. F., Kadkhodaei, S., et al. (2020). Overexpression of a rice monosaccharide transporter gene (OsMST6) confers enhanced tolerance to drought and salinity stress in Arabidopsis thaliana. Plant Mol. Biol. Rep. 38, 151–164. doi: 10.1007/s11105-019-01186-x
Moore, R. C., and Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Na, A. N., Hong-Bo, G., and Wei-Dong, K. E. (2009). Genetic variation in rhizome lotus (Nelumbo nucifera Gaertn. ssp. nucifera) germplasms from China assessed by RAPD markers. Agric. Sci. China 8, 31–39. doi: 10.1016/s1671-2927(09)60006-7
Noiraud, N., Maurousset, L., and Lemoine, R. (2001). Transport of polyols in higher plants. Plant Physiol. Biochem. 39, 717–728. doi: 10.1016/s0981-9428(01)01292-x
Payyavula, R. S., Tay, K. H. C., Tsai, C. J., and Harding, S. A. (2011). The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 65, 757–770. doi: 10.1111/j.1365-313x.2010.04463.x
Peng, Q., Wang, L., Ogutu, C., Liu, J., Liu, L., Mollah, M. D. A., et al. (2020). Functional analysis reveals the regulatory role of PpTST1 encoding tonoplast sugar transporter in sugar accumulation of peach fruit. Int. J. Mol. Sci. 21:1112. doi: 10.3390/ijms21031112
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45.
Poschet, G., and Büttner, M. (2011). A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol. 157, 1664–1676. doi: 10.1104/pp.111.186825
Rafal, B., David, G., and Rodríguez-García María, I. (2003). A putative plastidic glucose translocator is expressed in heterotrophic tissues that do not contain starch, during olive (Olea europea L.) fruit ripening. Plant Cell Physiol. 44, 1152–1161. doi: 10.1093/pcp/pcg149
Reuscher, S., Akiyama, M., Yasuda, T., Makino, H., Aoki, K., Shibata, D., et al. (2014a). The sugar transporter inventory of tomato: genome-wide identification and expression analysis. Plant Cell Physiol. 55, 1123–1141. doi: 10.1093/pcp/pcu052
Reuscher, S., Akiyama, M., Yasuda, T., Makino, H., Koh, A., Daisuke, S., et al. (2014b). The sugar transporter inventory of tomato: genome-wide identification and expression analysis. Plant Cell Physiol. 55, 1123–1141. doi: 10.1093/pcp/pcu052
Schneidereit, A., and Imlau, A. N. (2008). Conserved cis-regulatory elements for DNA-binding-with-one-finger and homeo-domain-leucine-zipper transcription factors regulate companion cell-specific expression of the Arabidopsis thaliana sucrose transporter 2 gene. Planta 228, 651–662. doi: 10.1007/s00425-008-0767-4
Shen-Miller, J. (2002). Sacred lotus, the long-living fruits of China antique. Seed Sci. Res. 12, 131–143. doi: 10.1079/ssr2002112
Song, X., Ma, X., Li, C., Hu, J., Yang, Q., Wang, T., et al. (2018). Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genomics 19:346. doi: 10.1186/s12864-018-4744-4
Tong, C., Wang, X., Yu, J., Wu, J., Li, W., Huang, J., et al. (2013). Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 14:689. doi: 10.1186/1471-2164-14-689
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Wan, L., Ren, W., Miao, H., Zhang, J., and Fang, J. (2020). Genome-wide identification, expression, and association analysis of the monosaccharide transporter (MST) gene family in peanut (Arachis hypogaea L.). 3 Biotech 10, 1–16. doi: 10.1016/j.plgene.2017.11.001
Wang, H. X., Weerasinghe, R. R., Perdue, T. D., Cakmakci, N. G., Taylor, J. P., Marzluff, W. F., et al. (2006). A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol. Biol. Cell 17, 4157–4591.
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 43, 1035–1039.
Watari, J., Kobae, Y., Yamaki, S., Yamada, K., Toyofuku, K., Tabuchi, T., et al. (2004). Identification of sorbitol transporters expressed in the phloem of apple source leaves. Plant Cell Physiol. 45, 1032–1041. doi: 10.1093/pcp/pch121
Weber, A., and Flügge, U. I. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12, 787–801. doi: 10.2307/3871001
Wingenter, K., Schulz, A., Wormit, A., Wic, S., Trentmann, O., Hoermiller, I. I., et al. (2010). Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol. 154, 665–677. doi: 10.1104/pp.110.162040
Wormit, A., Trentmann, O., Feifer, I., Lohr, C., Tjaden, J., Meyer, S., et al. (2006). Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18, 3476–3490. doi: 10.1105/tpc.106.047290
Wu, C., and Zhu, N. (2004). Advances in research on sugar transport pathway, sugar transporters and their physiological functions in plants. J. Hubei Agric. Coll. 24, 294–301.
Xue, J., Dong, W. A., Cheng, T., and Zhou, S. I. (2012). Nelumbonaceae: systematic position and species diversification revealed by the complete chloroplast genome. J. Syst. Evol. 50, 477–487. doi: 10.1111/j.1759-6831.2012.00224.x
Zheng, Q. M., Tang, Z., Xu, Q., and Deng, X. X. J. (2014). Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis). Plant Cell Tissue Organ Cult. 119, 609–624. doi: 10.1007/s11240-014-0560-y
Keywords: gene rentain, conserved synteny, evolutionary model, gene expression pattern analysis, abiotic stresses
Citation: Wu P, Zhang Y, Zhao S and Li L (2021) Comprehensive Analysis of Evolutionary Characterization and Expression for Monosaccharide Transporter Family Genes in Nelumbo nucifera. Front. Ecol. Evol. 9:537398. doi: 10.3389/fevo.2021.537398
Received: 02 November 2020; Accepted: 18 January 2021;
Published: 15 February 2021.
Edited by:
Liangsheng Zhang, Zhejiang University, ChinaReviewed by:
Ai-Sheng Xiong, Nanjing Agricultural University, ChinaCopyright © 2021 Wu, Zhang, Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangjun Li, bGpsaUB5enUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.