
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 22 December 2020
Sec. Coevolution
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.605304
This article is part of the Research Topic Symbiotic Relationships as Shapers of Biodiversity View all 14 articles
 Rhea Sanders-Smith1,2†
Rhea Sanders-Smith1,2† Bianca Trevizan Segovia1,2*†
Bianca Trevizan Segovia1,2*† Coreen Forbes1,3
Coreen Forbes1,3 Margot Hessing-Lewis1,2
Margot Hessing-Lewis1,2 Evan Morien1,2
Evan Morien1,2 Matthew A. Lemay1,2
Matthew A. Lemay1,2 Mary I. O'Connor1,3
Mary I. O'Connor1,3 Laura Wegener Parfrey1,2,3*
Laura Wegener Parfrey1,2,3*The seagrass Zostera marina is a widespread foundational species in temperate coastal ecosystems that supports diverse communities of epiphytes and grazers. Bacteria link the production of seagrass to higher trophic levels and are thought to influence seagrass biology and health. Yet, we lack a clear understanding of the factors that structure the seagrass microbiome, or whether there is a consistent microbial community associated with seagrass that underpins functional roles. We sampled surface microbiome (epibiota) from new and old growth seagrass leaves and the surrounding seawater in eight meadows among four regions along the Central Coast of British Columbia, Canada to assess microbiome variability across space and as leaves age. We found that the seagrass leaf microbiome differs strongly from seawater. Microbial communities in new and old growth leaves are different from each other and from artificial seagrass leaves we deployed in one meadow. The microbiome on new leaves is less diverse and there is a small suite of core OTUs (operational taxonomic units) consistently present across regions. The overall microbial community for new leaves is more dispersed but with little regional differentiation, while the epiphytes on old leaves are regionally distinct. Many core OTUs on old leaves are commonly associated with marine biofilms. Together these observations suggest a stronger role for host filtering in new compared to old leaves, and a stronger influence of the environment and environmental colonization in old leaves. We found 11 core microbial taxa consistently present on old and new leaves and at very low relative abundance on artificial leaves and in the water column. These 11 taxa appear to be strongly associated with Z. marina. These core taxa may perform key functions important for the host such as detoxifying seagrass waste products, enhancing plant growth, and controlling epiphyte cover.
Seagrass meadows are highly productive coastal ecosystems that provide numerous ecosystem functions and services (Duarte and Chiscano, 1999; Fourqurean et al., 2012). Bacteria link seagrass productivity to higher trophic levels by metabolizing organic carbon and nitrogen released by the host, suggesting a symbiotic relationship between the seagrass and associated microbes (McRoy and Goering, 1974; Kirchman et al., 1984). The seagrass microbiome is diverse, with 10 to 100 s of taxa identified per leaf sample (Ugarelli et al., 2017; Crump et al., 2018) and shaped by seagrass species identity (Crump et al., 2018), space (Ugarelli et al., 2017) and local environment (Bengtsson et al., 2017). A better understanding of the factors that structure the seagrass microbiome and a catalog of symbionts tightly associated with seagrass is needed to improve our knowledge of the complex interactions between seagrass and their symbionts in a changing world.
Microbial symbionts influence the growth and health of a wide range of hosts (Fitzpatrick et al., 2018; Longford et al., 2019). For example, isolated beneficial bacteria have been shown to improve resistance of corals to bleaching under high temperatures (Rosado et al., 2019). Similarly, seagrass symbionts are thought to influence host health by consuming toxic by-products (i.e., methanol and ethanol) and by producing agarases which limit epiphyte overgrowth (Crump et al., 2018). We use the broad definition of symbiont, microbes that live in association with a host, as the fitness consequence of most microbes is unknown and often variable in different contexts (Leung and Poulin, 2008; Wilkins et al., 2019). On the other side of the symbiotic spectrum, the pathogenic protist Labyrinthula zosterae is the best studied symbiont in seagrass ecosystems, putatively causing the seagrass wasting disease responsible for extensive declines in Zostera marina meadows in the Atlantic Ocean in the past (Short et al., 1987) and still of concern today.
One path toward making sense of the vast diversity in the seagrass microbiome and identifying candidate symbionts that may be essential to host functioning is investigating the core microbes that are tightly associated with the host (Shade and Handelsman, 2012; Wilkins et al., 2019; Risely, 2020). The definition of core taxa varies (Risely, 2020), but typically only a few microbial symbionts are consistently found at high prevalence on host populations across regions (Hernandez-Agreda et al., 2018).
At the level of the whole community, better understanding of how symbiont communities are assembled can provide foundational insights such as whether the microbiome is likely to shift or remain stable in changing environments. In marine ecosystems, hosts are submerged in a sea of microbes and horizontal transmission (acquisition of microbes from the environment) is likely the main route (Turon et al., 2018; Russell, 2019). Environmental acquisition might lead to microbial communities on marine hosts that mirror those from the surrounding environment, as is reported in one global study of seagrass leaves (Fahimipour et al., 2017). But, much more often selective processes lead to host microbial communities that are distinct from their surroundings and specific to host species (Wilkins et al., 2019). These patterns are observed for diverse seaweeds (Roth-Schulze et al., 2016; Lemay et al., 2018a), kelps (Lemay et al., 2018b; Weigel and Pfister, 2019), seagrass (Crump et al., 2018), sponges (Thomas et al., 2016), and others. In fact, seaweeds can select or deter microorganisms that colonize from the environment via their exudates (Lam et al., 2008; Salaün et al., 2012). Seagrass leaves also release exudates that attract microbes as food sources, such as methanol and cellulose, and others that deter colonization. Seagrass leaves are highly defended by phenols (Harrison, 1982) and other compounds that have been shown to modulate settlement and growth of microbes in lab settings (Papazian et al., 2019).
We sampled microbial communities from new and old seagrass leaves and the surrounding seawater in eight meadows across four regions along the Central Coast of British Columbia, Canada, to test host specificity and identify core seagrass taxa that are consistently present. If host selectivity is strong, we expect: (1) seagrass leaf microbiome to differ from the microbiome of the surrounding environment, (2) seagrass leaf microbiome to be similar across regions with different source pools, and (3) a consistent core assemblage found across tissue ages and regions. We further compare the role of selective processes influencing microbial biofilm communities using data from artificial seagrass leaves made of plastic and shaped like natural seagrass that were deployed in one of the regions. We used these artificial leaves to assess the similarity between the biofilm communities on this passive surface and natural seagrass shoots.
The Hakai Lúxvbálís conservancy encompasses 120,000 acres of land and sea and is the largest provincial marine protection area on the Coast of British Columbia. The conservancy hosts relatively low human impacted Zostera marina meadows. We collected 54 Zostera marina shoots and 35 seawater samples by scuba diving among eight paired meadows nested within four geographical regions (Figure 1), within two weeks during the summer of 2015.
Figure 1. Map showing the sampled seagrass meadows across four regions along the Central Coast of BC. A total of 54 Zostera marina shoots and 35 seawater samples were collected.
For seagrass leaf samples, we collected one shoot just outside each quadrat and placed it inside a sterile Ziploc bag underwater at the location of collection. New leaves of Zostera marina shoots are encompassed within a protective leaf sheath as they develop from a basal meristem. The sheath bundles all the leaves of an individual shoot by surrounding the blades at the shoot base and minimizes exposure to the environmental pool of microbes for new leaves. We accessed the new leaf growth by carefully opening the sheath from its upward edge and locating the youngest leaf tip within the sheath bundle. The old growth (emerging beyond the sheath) was standardized across replicates by sampling a 10 cm length of the mid-section of the third oldest leaf close to the sheath (frequently the longest leaf), in an area with minimal macro-epibiota such as bryozoan species and the red algal epiphyte, Smithora naiadum. Both new and old growth leaves, from the same shoot, were rinsed with 0.22 μm filtered seawater for 10 seconds to remove passive microbes and then sampled for another 10 seconds with a Puritan® sterile swab and transferred to a sterile cryovial (VWR). Seawater samples (500 mL; 3-5 reps) were collected by scuba diving in Nalgene water bottles from each meadow at seagrass collection depth (3–5 meters) within ~ 1 meter of the eelgrass canopy before the seagrass sampling. All samples were stored on ice in the field and transferred to a −80°C freezer upon return. Water samples were filtered using 0.22 μm membrane filters using a peristaltic pump the same day of collection.
In order to further explore biological and physical environmental filtering processes in seagrass ecosystems we sampled microbial communities from artificial seagrass units deployed in the Choked region. A single unit consisted of a 15 cm2 plastic grid for securing the artificial plants on which were attached 4 seagrass “shoots” (1 centimeter wide strips of high density polyethylene) each consisting of two 50 centimeter long “blades.” This provided a blade width, length and shoot density within the range observed in the same location in July 2014 (unpublished data, Hakai Institute). Artificial seagrass units were anchored to the substrate within the eelgrass meadow along a transect such that they were 40 meters apart and 10 m away from the meadow edge. When retrieved, a single blade on one shoot of each artificial seagrass unit was sampled 1 week after deployment and processed the same way as seagrass samples.
DNA was extracted from swabs and water filters using the MoBio PowerSoilVR –htp 96 well DNA extraction kit (Carlsbad, CA) following the manufacturers recommended protocol. The V4 region of 16S rRNA gene in Bacteria and Archaea was targeted for amplification using redesigned versions of the primers 515f/806r (Caporaso et al., 2012): 515f: 5'–GTGYCAGCMGCCGCGGTAA−3', 806r: 5'–GGACTACNVGGGTWTCTAAT−3' as previously described by Lemay et al. (2018a). Equal amounts (25 ng) of each sample were pooled and then purified using the MoBio UltaCleanVR PCR clean-up kit. Pooled library quantitation and paired-end Illumina MiSeq sequencing (2 × 300 bp) was carried out at the Integrated Microbiome Resource facility in the Center for Genomics and Evolutionary Bioinformatics at Dalhousie University (Halifax, Canada).
Raw sequencing reads were demultiplexed using the Split Libraries function from the Quantitative Insights into Microbial Ecology (QIIME v.1.9) analysis pipeline (Caporaso et al., 2010b). Demultiplexed reads were quality trimmed, then truncated to a uniform length of 250 bp with a quality score of 20 using FastX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), and processed into taxonomic units using the Minimum Entropy Decomposition (MED) method (Eren et al., 2015). MED does not rely on sequence similarity thresholds such as a standard 97% OTU, but instead uses Shannon entropy to separate out meaningful patterns of nucleotide diversity from sequencing noise and partition the data into MED nodes, which in practice are roughly analogous to ASVs or ≥99% OTUs. This analysis was carried out with the minimum substantive abundance parameter (–M) set at 500 reads while other parameters were run with default settings. Taxonomy was assigned to the resulting MED-nodes as implemented in the Assign Taxonomy function of QIIME v.1.9 using SILVA 128 database (Quast et al., 2013) clustered at 99% similarity. OTUs annotated to either chloroplast or mitochondrial sequences were removed as host contamination. Additionally, we removed OTUs with fewer than three reads per sample and fewer than 250 total reads, and samples with low read count (fewer than 1000 reads). After filtering, the final dataset consisted of 1206 OTUs with sequences/sample counts ranging from 3,130 to 103,313. Representative sequences for the remaining OTUs were aligned with PyNAST v.1.2.2 (Caporaso et al., 2010a), and a tree was constructed using FastTree (Price et al., 2010) within Qiime v 1.9. Alpha and beta diversity analyses were performed after rarefying the data to 3,000 reads/sample.
We calculated OTU richness using the non-parametric Chao1 index (Chao, 1984) and Pielou's evenness (Pielou, 1966). We used Analysis of Variance (ANOVA) to test for differences in alpha diversity metrics among sample types (new and old seagrass leaf growth, seawater) with geographical regions as a random effect, using function “lm” in the stats R package (R Core Team, 2020). Additionally, we ran an ANOVA with seagrass individual as a random effect to confirm differences between new and old seagrass leaves after accounting for sampling from the same plant. Because artificial seagrass samples were only taken in the Choked region, comparisons with real seagrass and seawater were restricted to samples from this region only. We performed pairwise comparisons using Tukey HSD correction for multiple comparisons. In cases where we found heterogeneity in the data, we randomly subsampled the sample types to get equal sample sizes, so that F-tests are robust (Keppel, 1991). The rarefaction curves for new and old seagrass leaves and artificial seagrass are all saturated. Several seawater samples do not appear to have reached saturation; therefore, richness comparisons with seawater samples were not discussed in depth.
To further compare microbial community composition, we visualized the similarity of sample types across regions with a non-metric multidimensional scaling (NMDS) using weighted UniFrac distance (phylogenetic relatedness of the species) (Lozupone and Knight, 2005) and used Permutational Multivariate Analysis of Variance (PERMANOVA) to test for differences in community structure between sample types for all regions, using both the weighted UniFrac distance and the Bray-Curtis dissimilarity index (abundance data). We should find similar patterns in both metrics when communities are composed of distantly related species, whereas when communities have a large share of closely related species, we should find a higher overlap in the UniFrac distances compared to Bray Curtis dissimilarities (Ortmann and Ortell, 2014). We further used Jaccard dissimilarity (presence-absence data) on PERMANOVA to test if differences were also related to community composition. PERMANOVA was performed using function “adonis2” in the vegan R package (Oksanen et al., 2018) to account for marginal effects of regions in each sample type (new and old seagrass leaf growth, seawater), and for Choked region including comparisons with artificial seagrass. We further compared new and old growth seagrass leaves to account for the effect of the individuals, since they were sampled from the same plant. Results of PERMANOVA analyses are potentially sensitive to heterogeneity in dispersions among treatments when there is an unbalanced design (Anderson and Walsh, 2013). Therefore, when our data was heterogeneous, we ran PERMANOVA tests by randomly subsampling the sample types to get a balanced design. We also tested for differences in the microbial community of each sample type among regions. Pairwise comparisons with Benjamini and Hochberg correction were conducted using function “adonis.pair” in the EcolUtils package (Salazar, 2019). Test statistics were generated using 10,000 permutations. Moreover, we calculated the percentage of taxa shared between sample types and regions, based on OTUs found in at least two samples of each sample type or region to avoid accounting for noise.
To determine the core microbes consistently associated with both new and old growth leaves of Zostera marina, we used Indicator Species Analysis (IndVal) (Dufrêne and Legendre, 1997), with the function “multipatt” in the Indicspecies R package (De Caceres and Legendre, 2009). IndVal combines both specificity (i.e., if a species is found in higher relative abundances in a particular habitat) and fidelity (i.e., if a species is prevalent across samples from that habitat) to determine indicator species (Legendre, 2013). This approach has been used to identify habitat specialists of coral compartments (Li et al., 2014) and of the mucus and surrounding habitats of corals (Glasl et al., 2016). We considered OTUs as core taxa if they were significant according to the permutation tests showing high association values (IndVal statistics > 0.7) (Demircan et al., 2018; Kruger, 2020), present in over 50% of the samples from that sample type, and present in all regions. This means that for an OTU to be considered core of Zostera marina, for example, it would have to be found in relatively higher abundance in both new and old leaves when compared to seawater and artificial seagrass, be present in all regions and across most of the seagrass samples. We ran this analysis including seawater and artificial seagrass samples to determine the core seagrass microbes that were not generally found in either seawater or other marine surfaces. Significance (P < 0.05) was tested by 10,000 random permutations.
All data statistical analyses were performed in R version 3.6.3 (R Core Team, 2020). Graphs were constructed in R using package ggplot2 (Wickham, 2016).
Microbial communities associated with seagrass differ in diversity from the surrounding seawater. Both richness and evenness differed between new and old growth leaves and seawater [ANOVA Richness F(2,99) = 31.51, p < 0.0001; Evenness F(2,99) = 30.34, p < 0.0001] (Figure 2A; Supplementary Table 1). New and old leaves were sampled from each plant, but differences in richness and evenness remain significant even after controlling for individual (Supplementary Table 1). Microbial communities in new growth leaves showed the lowest OTU richness and highest dominance (a few species were highly abundant, and most were rare). Richness in old growth leaves was the highest, and the abundance was more evenly distributed between species than in new growth leaves. Artificial seagrass was similar to old growth leaves in terms of richness and evenness (Figure 2B; Supplementary Table 1).
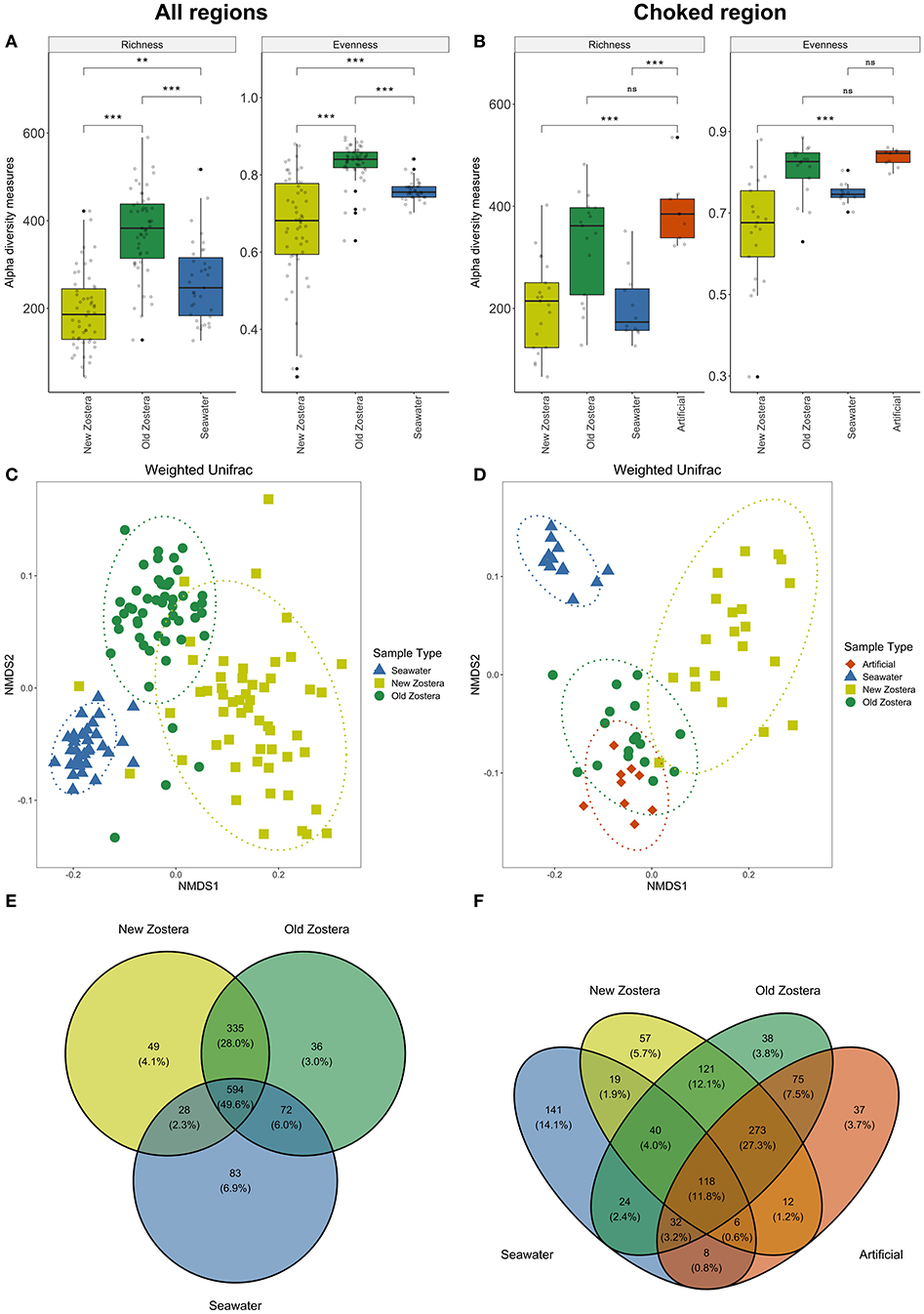
Figure 2. Alpha diversity metrics, NMDS plots based on weighted UniFrac distances, and Venn diagrams of shared taxa of microbial communities on Z. marina new growth leaves, old growth leaves, and seawater for all regions (A,C,E), and for Choked region including artificial seagrass (B,D,F). Asterisks in boxplots represent significant difference, where *indicates p > 0.05, **indicates p > 0.01, and ***indicates p > 0.001; ns, not significant. Ellipses in NMDS plots represent ordination confidence intervals (95%).
PERMANOVA analyses showed a clear distinction in microbial community structure between seawater, new and old growth seagrass leaves [weighted UniFrac distance; F(2,99) = 70.75, P < 0.0001, R2 = 0.55] (Figure 2C, Supplementary Table 2). These community distinctions are underlain by differences in relative abundance as well as composition as the differences are also significant when using non-phylogenetic Bray-Curtis and Jaccard dissimilarity indices (Supplementary Table 2). Differences in microbial community structure between new and old growth leaves persist after accounting for individuals as a random effect [PERMANOVA weighted UniFrac distance; F(1, 43) = 49.82, P < 0.0001, R2 = 0.29; Supplementary Table 2]. Almost 50% of all taxa were shared among Zostera marina and seawater samples across all regions, though shared taxa where generally much more abundant in one sample type, and new and old growth leaves shared around 28% of taxa (Figure 2E). We used data for the Choked region to compare microbiome on natural (new and old growth) and artificial seagrass leaves. We found that all sample types were distinct from each other [Supplementary Table 2: PERMANOVA weighted UniFrac F(3,32) = 18.98, P <0.0001, R2 = 0.64], but artificial seagrass appear more similar to old growth (Figure 2D). Artificial seagrass shared the vast majority of OTUs with seagrass, with only 3.7% unique OTUs and <1% shared only with water (Figure 2F). Still, old and new leaves share more OTUs than do old and artificial (Figure 2F).
Microbial community structure varied among regions on seagrass but not in the water column, with the most pronounced differences observed in old seagrass leaves (Figure 3). Microbiome significantly differ by region using both taxon and phylogenetic-based metrics for old growth [PERMANOVA Bray Curtis F(3,40) = 5.87, P < 0.0001, R2 = 0.30; Weighted UniFrac F(3,40) = 8.41, P < 0.0001, R2 = 0.38] and new growth [PERMANOVA Bray Curtis F(3,50) = 2.78, P < 0.0001, R2 = 0.14; Weighted UniFrac F(3,50) = 2.35, P = 0.0018, R2 = 0.12] leaves. However, pairwise comparisons using weighted UniFrac distances indicated greater regional overlap in microbial communities for new growth leaves (Figures 3A,D; Supplementary Table 3); this contrast between Bray Curtis and Weighted Unifrac results indicate that new leaves have a large share of closely related species across regions (Ortmann and Ortell, 2014). Old growth leaves remained quite distinct (Figures 3B,E; Supplementary Table 3). There was also a lower degree of taxa sharing between regions, with 19.7 and 31.7% of taxa shared among regions for new and old growth, respectively (Figure 3). Microbial community structure in seawater samples was somewhat distinct across regions when we considered both Bray-Curtis and UniFrac distance (Figures 3C,F; Supplementary Table 3), and around 32.1% of taxa were shared among all regions (Figure 3I).
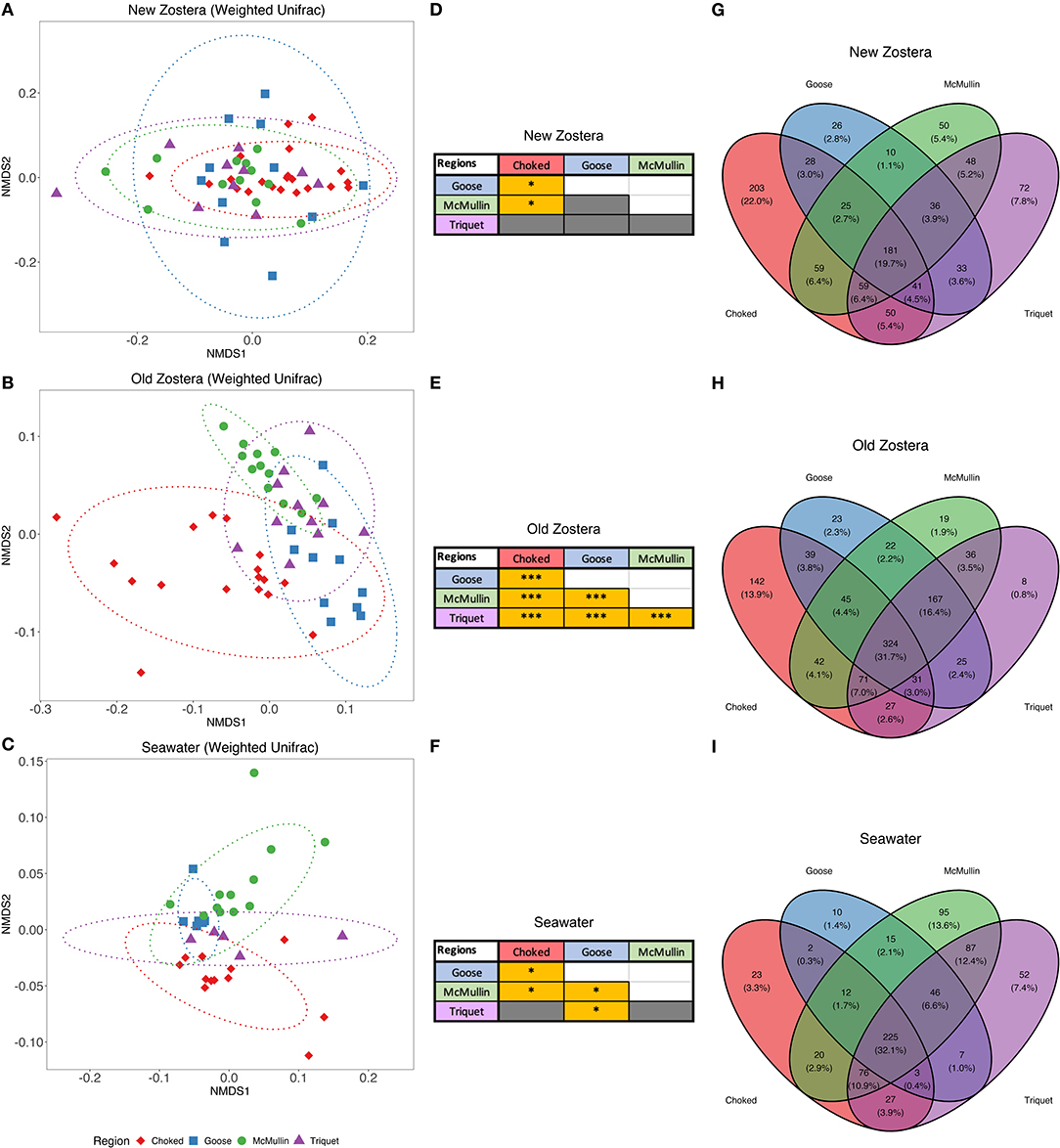
Figure 3. NMDS plots based on weighted UniFrac distances, pairwise PERMANOVA results table and Venn diagrams of shared taxa of microbial communities of Z. marina new growth leaves (A,D,G), old growth leaves (B,E,H), and seawater (C,F,I) samples among regions. Pairwise PERMANOVA grids colored in gray are not significant, whereas grids colored in yellow are significant, where * indicates p > 0.05 and *** indicates p > 0.001. Ellipses represent ordination confidence intervals (95%).
We used indicator species analysis (Dufrêne and Legendre, 1997) to define core bacteria, with the criteria that the core are significantly enriched on seagrass compared to seawater and artificial seagrass, at 50% or greater prevalence, and present across all regions. We identified core taxa across Zostera marina in general and separately on new and old growth leaves to better understand persistent taxa and changes with tissue age.
Zostera marina core: We found 11 core OTUs belonging to six families on Zostera marina leaves (enriched and prevalent in both new and old growth leaves; Figure 4). Six core OTUs come from the Methylophilaceae family, genus Methylotenera. One core OTU (genus Marinomonas) was undetected in seawater samples (Figure 4). Alteromonas and Paraglaciecola (Alteromonadaceae), Rubidimonas (Saprospiraceae), and Rhodobacteraceae round out the core (Figure 4).
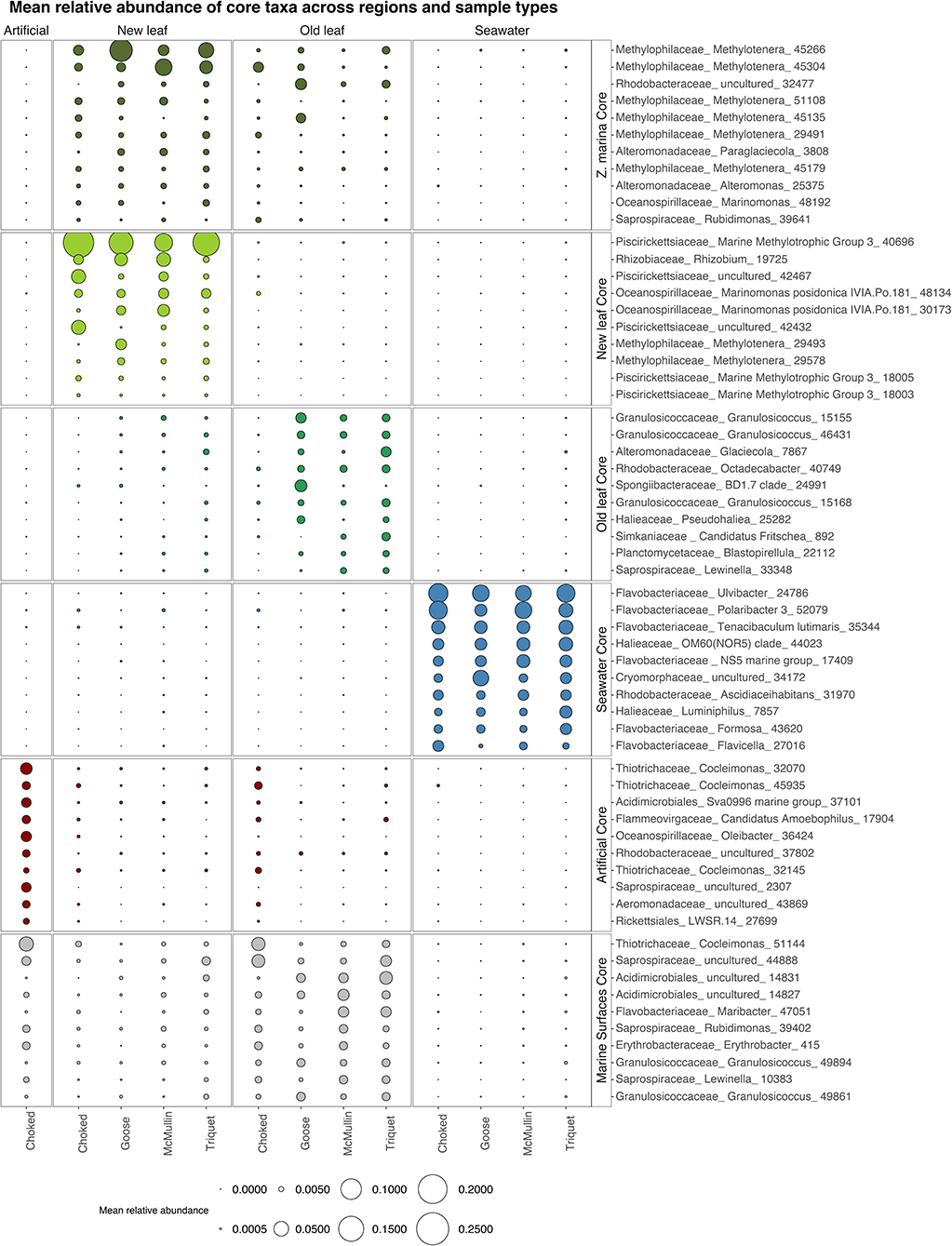
Figure 4. Bubble plot showing the mean relative abundance of the 10 most abundant core taxa across regions and sample types: New growth leaves, Old growth leaves, Zostera marina (core in both new and old growth, here all 11 core taxa are shown), seawater, artificial and marine surfaces (core in both old leaves and artificial seagrass). We considered core taxa the ones significant in Indval analysis, present in at least 50% of the samples and present in all regions. Taxa are labeled by Family names along with the lowest taxonomy level identified.
New growth core: For new growth seagrass leaves, we found a small and taxonomically consistent set of 16 core OTUs. The new growth core includes several representatives from Methylotenera (Methylophylaceae) and Marinomonas posidonica (Oceanospirillaceae), the Marine Methylotrophic group 3 in the Piscirickettsiaceae family, and Rhizobium (Figure 4; the complete figure showing all core taxa can be found in the Supplementary Figure 1).
Old growth core: For old growth seagrass leaves we found 57 core OTUs. Core taxa for old growth leaves come from many different clades common on marine surfaces, including Granulosicoccaceae, Rhodobacteraceae, and Planctomycetaceae (Figure 4; Supplemenatary Figure 1).
Artificial seagrass core: The core genera for artificial seagrass include taxa typical of marine biofilms such as Saprospiraceae, as well as Cocleimonas (Thiotrichaceae) and taxa that are likely degrading these strips of plastic bags Oleibacter (Oceanospiralles).
Detailed information on indicator species analysis statistics, taxonomy, and sequences of the core taxa can be found in the Supplementary Table 4.
Seagrass meadows are highly productive ecosystems. The microbes associated with seagrass likely influence seagrass health and cycling of carbon and other nutrients through the ecosystem. We investigated the specificity of seagrass microbiome compared to the water column and artificial seagrass, and the consistency of the microbiome across plant tissues of different age and across regions with different environmental conditions. We identified a core community associated with Z. marina. We hypothesized that if host selectivity is strong, we would find: (1) seagrass microbiome that differ from the microbiome of the surrounding environment, (2) seagrass microbiome that are similar across regions with different source pools, and (3) a consistent core assemblage across tissue ages and regions. We found good evidence for predictions 1 and 3: the seagrass leaf microbiome is distinct from their surrounding environment (Figure 2) and is colonized by a consistent suite of core taxa (Figure 4). Evidence for prediction 2 is mixed: we detected differences across regions in old leaves but little differentiation across regions for new leaves (Figure 3), that had a larger share of closely related species across regions. This points to seagrass leaves being more selective in newer tissues that are more highly defended. As seagrass blades age the species pool of colonizers available in each region may more strongly influence the leaf microbiome.
Seagrass leaves harbor microbial communities that are distinct from seawater. The strong differences in microbiome composition hold using taxonomic and phylogenetic dissimilarity measures (Figure 2; Supplementary Table 2), and different bacterial lineages are dominant on seagrass compared to seawater. This result is consistent with previous studies reporting distinct microbial communities on a range of marine hosts compared to the surrounding seawater communities, including seaweed (Michelou et al., 2013; Lemay et al., 2018a), sponges (Thomas et al., 2016), and seagrass (Bengtsson et al., 2017; Crump et al., 2018; Ugarelli et al., 2019). Our results contrast with a global study of seagrass that found similar microbiome on seagrass leaves using different methods (Fahimipour et al., 2017), as discussed in (Crump et al., 2018).
Differences between seagrass and seawater microbiome point to the importance of selective mechanisms in microbial community assembly. The physicochemical properties of seagrass leaves and seagrass exudates enrich for or select against particular taxa from the surrounding environment. Seagrass surfaces are chemically defended by compounds such as flavonoids, phenolic compounds, and carboxylic acids (Papazian et al., 2019). Seagrass also exudes nutrients and dissolved organic carbon, including methanol, that support abundant bacterial production (Penhale and Thayer, 1980; Kirchman et al., 1984). Experimental work in seagrass shows that different cultured isolates grow on seagrass blades compared to abiotic surfaces (Kurilenko et al., 2007; Papazian et al., 2019), supporting the notion that seagrass leaves are selective surfaces. Metatranscriptomic analysis highlights matches between seagrass leaf bacterial metabolism and the plant chemistry/exudates (Crump et al., 2018).
Almost half of the OTUs in our study were shared between seagrass and seawater (Figure 2E) and most of the seagrass core taxa are found at low relative abundance, but still present, in the water column (Figure 4). Thus, seagrass leaf microbiome seems to be primarily acquired horizontally via transmission through the water column, as is also the case for sponges (Turon et al., 2018), seaweeds, and other marine surfaces (Russell, 2019). Studies of the microbiome in seeds and/or shoots grown from seed in sterile conditions are needed to investigate vertical transmission. Transmission of microbes is bidirectional; some of the microbes detected in seawater, which was sampled just above the seagrass meadow canopy, surely originated on seagrass. An additional layer of complication arises because seagrass and seaweed exudates can influence the microbial composition of the water column (Lam et al., 2008; Lamb et al., 2017; Chen and Parfrey, 2018).
Our data is consistent with initial colonization of a handful of seagrass specialists that feed on the metabolic by-products of seagrass—particularly methanol and 1-carbon compounds such as Methylotenera (Kalyuhznaya et al., 2009) and Piscirickettsiaceae Methylotrophic group 3 (Krolicka et al., 2017), as well as a potential seagrass symbiont Marinomonas posidonica (Lucas-Elió et al., 2011) and predicted nitrogen fixing Rhizobium (Avis et al., 2008). We expected new leaves to represent a more selective microbial environment because they are more chemically defended and have lower microbial biomass (Harrison, 1982; Kurilenko et al., 2001). Further, new leaves were fully within the protective sheath at the time of sampling, and thus in much reduced contact with the water column microbial source pool. Consistent with expectations, new leaves of Zostera marina harbor reduced microbial diversity and their microbiome composition is distinct from old leaves (Figure 2).
As seagrass blades age, they accumulate higher bacterial loads (Harrison, 1982) and are colonized by diverse micro- and macroalgae. These biofilms support a diverse array of bacteria and heterotrophic grazers such as ciliates, copepods, and rotifers (Segovia et al., 2020) that feed on metabolic by-products or directly on microbial or algal biomass. Although only 11 core taxa are shared between new and old growth leaves, many more are shared between old growth leaves and artificial seagrass (Supplementary Figure 1). Our data suggest that as Zostera marina leaves age, seagrass microbiome composition shifts to include more generalist biofilm taxa, along with seagrass specialists. In contrast with Ettinger et al. (2017), we found similarities between seagrass leaf- and seaweed microbiome even at lower taxonomic ranks, since the genera Blastopirellula and Granulosicoccus, both core to old growth leaves (Figure 4), are also commonly found on seaweeds (Bondoso et al., 2017; Lemay et al., 2018a; Weigel and Pfister, 2019). Research on influence of plant age in terrestrial plants also show that leaves harbor distinct communities over time, but contrary to our findings, they seem to resemble the surrounding (i.e., air and sediment) communities during the first stages while converging to more unique communities as they age (Maignien et al., 2014; Copeland et al., 2015). This difference could be due to new leaves in seagrass being protected within the sheath and/or more chemically defended, as mentioned above.
Many taxa overlap between old growth and artificial leaves (Figure 4 Marine surfaces core), including representatives of Saprospiraceae, Flavobacteriaceae and additional Granulosicoccus OTUs, all of which are widespread on marine surfaces (Burke et al., 2011; Florez et al., 2017; Van De Water et al., 2018; James et al., 2020). The increase in generalists is likely due to decreased selectivity of the leaves with age (Harrison, 1982), coupled with the formation of a biofilm that enables generalists to colonize (Datta et al., 2016). It is likely that other marine surfaces are an important source pool for seagrass leaves as they age, as many core genera present in old growth leaves are shared with other hosts.
Additional research, particularly experimental manipulations, are necessary to uncover the relative importance of host selectivity and the environment in the microbiome assembly of seagrass leaves.
We identified core bacteria as those found in more than 50% of samples for a given sample type, found across regions and enriched in relation to other sample types. Only a few taxa associated with seagrass met these criteria and represent the Zostera core microbiome (Figure 4). Corals also have a small core microbiome (Hernandez-Agreda et al., 2018) and this is likely a common phenomenon. Below we discuss potential functions and distribution of select members of the core.
Only 11 taxa are consistently part of the Zostera core for both new and old leaves. Most of these core taxa belong to methylotrophic clades within the Methylophilaceae (genus Methylotenera) and Piscirickettsiaceae, while Rhizobium, is usually involved in nitrogen fixation and is known for promoting plant growth (Avis et al., 2008). Methylophilaceae are non-methane oxidizing methylotrophs—which use single-carbon compounds such as methanol and methylamine as carbon and energy sources. They have been suggested to use the methanol released by the methane oxidizing methanotrophs (Chistoserdova and Kalyuzhnaya, 2018) and by the seagrass itself—which is a toxic by-product for the plant—and were found to be an important component of Z. marina leaf microbiome (Crump et al., 2018). Other methylotrophic bacteria—Gammaproteobacteria, family Piscirickettsiaceae—encompassed half of all core taxa found in new leaves. Pseudoalteromonas, a common marine genus that is part of the core of new leaves (Supplementary Figure 1), shows antibacterial and antialgal activity, and thus may modulate the establishment of other species (Lage and Bondoso, 2014). The microbes present in new leaves could influence the assembly of communities in old leaves.
Marinomonas appears to be a true associate of seagrass, and we find Marinomonas in the new growth and overall Zostera core (Figure 4). This genus has been recently isolated from Zostera marina leaf tissues from Bodega Bay, California (Ettinger and Eisen, 2020). Several species of Marinomonas have also been isolated from the seagrass Posidonia oceanica on the coast of Spain (Espinosa et al., 2010; Lucas-Elió et al., 2011), including M. posidonica, which is in the core of new leaves here (Figure 4). Marinomonas posidonica was shown to enhance leaf growth in P. oceanica likely due to the release of metabolites and nutrients (Celdrán et al., 2012). It has also been registered in leaves of the seagrass Enhalus acoroides in Papua New Guinea (Hassenrück et al., 2015). This indicates that this taxon may have an affinity for seagrass leaf microbiomes.
Several taxa we identified in the core for old growth and Zostera overall are common colonizers of marine plants and algae (Figure 4). The genus Granulosicoccus (Gammaproteobacteria) is part of the Z. marina core, as well as the marine surfaces core, encompassing most of the old growth leaves core taxa. Granulosicoccus was also pervasive across leaf microbiomes of two Zostera species on the coast of Oregon, US (Crump et al., 2018), and has been commonly found in association with seaweeds in the Calvert Island region where we sampled (Lemay et al., 2018a). Rhodobacteraceae are frequently the first colonizers of marine surfaces (Dang et al., 2008) and are found here on both new and old leaves. Several taxa from this family are in seagrass leaf microbiomes across different regions [German Baltic Sea coast: (Bengtsson et al., 2017); Bodega Bay, CA: (Ettinger et al., 2017); coast of Oregon, US: (Crump et al., 2018)]. Similarly, Blastopirellula (Planctomycetes) is commonly found on macroalgae across the globe (Bondoso et al., 2017) and in the core for old leaves here.
Interestingly, some core OTUs found in old growth leaves and artificial seagrass are endosymbionts of protists and invertebrates. This suggests their eukaryotic hosts must also be common in the seagrass microbiome, demonstrating that exchange with the microbiome of other organisms also contributes to the overall diversity of seagrass microbial communities. For instance, OTUs assigned to “Candidatus Fritschea” in the order Chlamydiales (family Simkaniaceae) are among the core of old leaves. Candidatus Fristchea are obligatory intracellular endosymbionts first found living in the gut of terrestrial insects (Thao et al., 2003), while two closely related Chlamydiales symbionts have been identified from marine environments living in association with the marine worm Xenoturbella westbladi and the gills of Atlantic salmon (Israelsson, 2007; Everett, 2014). Caedibacter caryophilus, core to artificial seagrass, are endosymbionts of the protist Acanthamoeba (Horn et al., 1999).
Further comparative studies including samples from seagrass roots, nearby sediments and other marine surfaces are needed to identify the true habitat range of these core taxa and their influence on host health.
Differences between the seagrass leaf microbiome and the surrounding seawater and artificial seagrass, together with the consistent core assemblage across tissue age and regions, all point to host selectivity being an important process in the microbiome assembly in seagrass leaves. Microbiome community structure on new and old growth leaves is distinct; new leaves are also less diverse with a smaller suite of core, and show little regional differentiation, in contrast with old leaves that are spatially more distinct. This points to seagrass leaves being more selective in newer tissues that are more highly defended. As seagrass blades age the species pool of colonizers available in each region more strongly influences the leaf microbiome. Despite those differences, we found 11 core microbial taxa that persisted throughout development, showing a strong association with Z. marina. These core taxa may perform key functions important for the host such as detoxifying seagrass waste products, enhancing plant growth, and controlling epiphyte cover. The functional role of these core taxa should be further investigated in follow up studies. These findings provide baseline knowledge of the seagrass microbiome that will inform our understanding of marine microbiome in this foundational species.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: https://www.ebi.ac.uk/ena/browser, PRJEB25267. More details about the code and figure construction can be found in GitHub: https://github.com/biatsegovia/Core_Z.marina_leaf_microbiome.git; a copy of this repository is archived at Zenodo: https://doi.org/10.5281/zenodo.4287628.
RS-S analyzed the data and wrote the first draft of the manuscript. BS analyzed the data, interpreted the results, and led the writing. EM performed bioinformatic analysis. CF, MO'C, and MH-L collected the host data. RS-S, ML, and LP collected and generated the microbial data. LP, MO'C, and MH-L designed and conceived the study. All authors contributed to manuscript corrections and improved the final version.
This work was supported by the Hakai Institute (Microbes to Macrophytes grant to LP and MO'C). BS was supported by a Mitacs Accelerate Postdoctoral Fellowship, in partnership with the Tula Foundation (IT09704). Further support provided by NSERC Discovery Grants to LP (RGPIN-2014-05461) and MO'C (#402006-13).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the Hakai Institute Calvert marine station field teams and staff for their support. We acknowledge Cassandra Jensen and Marcus Campbell for their contributions to lab work. We thank reviewers for their helpful comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.605304/full#supplementary-material
Anderson, M. J., and Walsh, D. C. I. (2013). Permanova, anosim, mantel test face heterogeneous dispersions : what null hypothesis are you testing? Ecol. Monogr. 83, 557–574. doi: 10.1890/12-2010.1
Avis, T. J., Gravel, V., Antoun, H., and Tweddell, R. J. (2008). Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 40, 1733–1740. doi: 10.1016/j.soilbio.2008.02.013
Bengtsson, M. M., Bühler, A., Brauer, A., Dahlke, S., Schubert, H., and Blindow, I. (2017). Eelgrass leaf surface microbiomes are locally variable and highly correlated with epibiotic eukaryotes. Front. Microbiol. 8:1312. doi: 10.3389/fmicb.2017.01312
Bondoso, J., Godoy-Vitorino, F., Balagué, V., Gasol, J. M., Harder, J., and Lage, O. M. (2017). Epiphytic planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol. Ecol. 93:fiw255. doi: 10.1093/femsec/fiw255
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Caporaso, J. G., Bittinger, K., Bushman, F. D., Desantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high- throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth0510-335
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Celdrán, D., Espinosa, E., Sánchez-Amat, A., and Marín, A. (2012). Effects of epibiotic bacteria on leaf growth and epiphytes of the seagrass Posidonia oceanica. Mar. Ecol. Prog. Ser. 456, 21–27. doi: 10.3354/meps09672
Chao, A. (1984). Non-parametric estimation of the classes in a population. Scand. J. Stat. 11, 265–270. doi: 10.2307/4615964
Chen, M. Y., and Parfrey, L. W. (2018). Incubation with macroalgae induces large shifts in water column microbiota, but minor changes to the epibiota of co-occurring macroalgae. Mol. Ecol. 27, 1966–1979. doi: 10.1111/mec.14548
Chistoserdova, L., and Kalyuzhnaya, M. G. (2018). Current trends in methylotrophy. Trends Microbiol. 26, 703–714. doi: 10.1016/j.tim.2018.01.011
Copeland, J. K., Yuan, L., Layeghifard, M., Wang, P. W., and Guttman, D. S. (2015). Seasonal community succession of the phyllosphere microbiome. Mol. Plant-Microbe Interact. 28, 274–285. doi: 10.1094/MPMI-10-14-0331-FI
Crump, B. C., Wojahn, J. M., Tomas, F., and Mueller, R. S. (2018). Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 9:388. doi: 10.3389/fmicb.2018.00388
Dang, H., Li, T., Chen, M., and Huang, G. (2008). Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/AEM.01400-07
Datta, M. S., Sliwerska, E., Gore, J., Polz, M. F., and Cordero, O. X. (2016). Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7:11965. doi: 10.1038/ncomms11965
De Caceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Demircan, T., Ovezmyradov, G., Yildirim, B., Keskin, I., and Ilhan, A. E., Fesçioglu, E. C., et al. (2018). Experimentally induced metamorphosis in highly regenerative axolotl (Ambystoma mexicanum) under constant diet restructures microbiota. Sci. Rep. 8:10974. doi: 10.1038/s41598-018-29373-y
Duarte, C. M., and Chiscano, C. L. (1999). Seagrass biomass and production: a reassessment. Aquat. Bot. 65, 159–174. doi: 10.1016/S0304-3770(99)00038-8
Dufrêne, M., and Legendre, P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366.
Eren, A. M., Morrison, H. G., Lescault, P. J., Reveillaud, J., Vineis, J. H., and Sogin, M. L. (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 9, 968–979. doi: 10.1038/ismej.2014.195
Espinosa, E., Marco-Noales, E., Gómez, D., Lucas-Elío, P., Ordax, M., Garcías-Bonet, N., et al. (2010). Taxonomic study of marinomonas strains isolated from the seagrass Posidonia oceanica, with descriptions of Marinomonas balearica sp. nov. and Marinomonas pollencensis sp. nov. Int. J. Syst. Evol. Microbiol. 60, 93–98. doi: 10.1099/ijs.0.008607-0
Ettinger, C. L., and Eisen, J. A. (2020). Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PLoS ONE 15:e0236135. doi: 10.1371/journal.pone.0236135
Ettinger, C. L., Voerman, S. E., Lang, J. M., Stachowicz, J. J., and Eisen, J. A. (2017). Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 5:e3246. doi: 10.7717/peerj.3246
Everett, K. D. (2014). “The family simkaniaceae,” in The Prokaryotes: Other Major Lineages of Bacteria and The Archaea, eds. E. Rosenberg, E. F. De Long, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer), 891–906.
Fahimipour, A. K., Kardish, M. R., Lang, J. M., Green, J. L., Eisen, J. A., and Stachowicz, J. J. (2017). Global-scale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83:e03391–16. doi: 10.1128/aem.03391-16
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U. S. A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Florez, J. Z., Camus, C., Hengst, M. B., and Buschmann, A. H. (2017). A functional perspective analysis of macroalgae and epiphytic bacterial community interaction. Front. Microbiol. 8:2651. doi: 10.3389/fmicb.2017.02561
Fourqurean, J. W., Duarte, C. M., Kennedy, H., Marbà, N., Holmer, M., Mateo, M. A., et al. (2012). Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 5, 505–509. doi: 10.1038/ngeo1477
Glasl, B., Herndl, G. J., and Frade, P. R. (2016). The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292. doi: 10.1038/ismej.2016.9
Harrison, P. G. (1982). Control of microbial growth and of amphipod grazing by water-soluble compounds from leaves of Zostera marina. Mar. Biol. 67, 225–230. doi: 10.1007/BF00401288
Hassenrück, C., Hofmann, L. C., Bischof, K., and Ramette, A. (2015). Seagrass biofilm communities at a naturally CO <inf>2</inf>-rich vent. Environ. Microbiol. Rep. 7, 516–525. doi: 10.1111/1758-2229.12282
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., Herrera, C., and Ainsworth, T. D. (2018). Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. MBio 9:e00812-18. doi: 10.1128/mbio.00812-18
Horn, M., Fritsche, T. R., Gautom, R. K., Schleifer, K. H., and Wagner, M. (1999). Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ. Microbiol. 1, 357–367. doi: 10.1046/j.1462-2920.1999.00045.x
Israelsson, O. (2007). Chlamydial symbionts in the enigmatic Xenoturbella (Deuterostomia). J. Invertebr. Pathol. 96, 213–220. doi: 10.1016/j.jip.2007.05.002
James, A. K., English, C. J., Nidzieko, N. J., Carlson, C. A., and Wilbanks, E. G. (2020). Giant kelp microbiome altered in the presence of epiphytes. Limnol. Oceanogr. Lett. 5, 354–362. doi: 10.1002/lol2.10157
Kalyuhznaya, M. G., Martens-Habbena, W., Wang, T., Hackett, M., Stolyar, S. M., Stahl, D. A., et al. (2009). Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ. Microbiol. Rep. 1, 385–392. doi: 10.1111/j.1758-2229.2009.00046.x
Keppel, G. (1991). Design and Analysis: A Researcher's Handbook. Englewood Cliffs, NJ: Prentice-Hall, Inc.
Kirchman, D., Mazzella, L., Alberte, R., and Mitchell, R. (1984). Epiphytic bacterial production on Zostera marina. Mar. Ecol. Prog. Ser. 15, 117–123. doi: 10.3354/meps015117
Krolicka, A., Boccadoro, C., Nilsen, M. M., and Baussant, T. (2017). Capturing early changes in the marine bacterial community as a result of crude oil pollution in a mesocosm experiment. Microbes Environ. 32, 358–366. doi: 10.1264/jsme2.ME17082
Kruger, A. (2020). Frog skin microbiota vary with host species and environment but not chytrid infection. Front. Microbiol. 11:1330. doi: 10.3389/fmicb.2020.01330
Kurilenko, V. V., Ivanova, E. P., and Mikhailov, V. V. (2001). Zonal distribution of epiphytic microorganisms on the eelgrass Zostera marina. Microbiology 70, 372–373. doi: 10.1023/A:1010475916224
Kurilenko, V. V., Ivanova, E. P., and Mikhailov, V. V. (2007). Characteristics of adhesion of epiphytic bacteria on leaves of the seagrass Zostera marina and on abiotic surfaces. Mikrobiologiia 76, 502–506. doi: 10.1134/S0026261707040091
Lage, O. M., and Bondoso, J. (2014). Planctomycetes and macroalgae, a striking association. Front. Microbiol. 5:267. doi: 10.3389/fmicb.2014.00267
Lam, C., Stang, A., and Harder, T. (2008). Planktonic bacteria and fungi are selectively eliminated by exposure to marine macroalgae in close proximity. FEMS Microbiol. Ecol. 63, 283–291. doi: 10.1111/j.1574-6941.2007.00426.x
Lamb, J. B., van de Water, J. A. J. M., Bourne, D. G., Altier, C., Hein, M. Y., Fiorenza, E. A., et al. (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733. doi: 10.1126/science.aal1956
Legendre, P. (2013). “Indicator species: computation,” in Encyclopedia of Biodiversity, Vol. 4, 2nd Edn, ed Levin S (San Diego, CA: Academic Press), 264-268.
Lemay, M. A., Martone, P. T., Hind, K. R., Lindstrom, S. C., and Wegener Parfrey, L. (2018a). Alternate life history phases of a common seaweed have distinct microbial surface communities. Mol. Ecol. 27, 3555–3568. doi: 10.1111/mec.14815
Lemay, M. A., Martone, P. T., Keeling, P. J., Burt, J. M., Krumhansl, K. A., Sanders, R. D., et al. (2018b). Sympatric kelp species share a large portion of their surface bacterial communities. Environ. Microbiol. 20, 658–670. doi: 10.1111/1462-2920.13993
Leung, T. L. F., and Poulin, R. (2008). Parasitism, commensalism, and mutualism: exploring the many shades of symbioses. Vie Milieu 58, 107–115.
Li, J., Chen, Q., Long, L. J., Dong, J., De, Y. J., and Zhang, S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 4:7320. doi: 10.1038/srep07320
Longford, S. R., Campbell, A. H., Nielsen, S., Case, R. J., Kjelleberg, S., and Steinberg, P. D. (2019). Interactions within the microbiome alter microbial interactions with host chemical defences and affect disease in a marine holobiont. Sci. Rep. 9:1363. doi: 10.1038/s41598-018-37062-z
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lucas-Elió, P., Marco-Noales, E., Espinosa, E., Ordax, M., López, M. M., Garcías-Bonet, N., et al. (2011). Marinomonas alcarazii sp. nov., M. rhizomae sp. nov., M. foliarum sp. nov., M. posidonica sp. nov. and M. aquiplantarum sp. nov., isolated from the microbiota of the seagrass Posidonia oceanica. Int. J. Syst. Evol. Microbiol. 61, 2191–2196. doi: 10.1099/ijs.0.027227-0
Maignien, L., DeForce, E. A., Chafee, M. E., Murat Eren, A., and Simmons, S. L. (2014). Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 5:e00682-13. doi: 10.1128/mBio.00682-13
McRoy, C. P., and Goering, J. J. (1974). Nutrient transfer between the seagrass Zostera marina and its epiphytes. Nature 248, 173–174. doi: 10.1038/248173a0
Michelou, V. K., Caporaso, J. G., Knight, R., and Palumbi, S. R. (2013). The ecology of microbial communities associated with Macrocystis pyrifera. PLoS ONE 8:67480. doi: 10.1371/journal.pone.0067480
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2018). vegan: Community Ecology Package. R package version 2.5-6. Available online at: https://CRAN.R-project.org/package=vegan (accessed November 25, 2020).
Ortmann, A. C., and Ortell, N. (2014). Changes in free-living bacterial community diversity reflect the magnitude of environmental variability. FEMS Microbiol. Ecol. 87, 291–301. doi: 10.1111/1574-6941.12225
Papazian, S., Parrot, D., Burýšková, B., Weinberger, F., and Tasdemir, D. (2019). Surface chemical defence of the eelgrass Zostera marina against microbial foulers. Sci. Rep. 9:3323. doi: 10.1038/s41598-019-39212-3
Penhale, P. A., and Thayer, G. W. (1980). Uptake and transfer of carbon and phosphorus by eelgrass (Zostera marina L.) and its epiphytes. J. Exp. Mar. Bio. Ecol. 42, 113–123. doi: 10.1016/0022-0981(80)90170-7
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE 5:9490. doi: 10.1371/journal.pone.0009490
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
R Core Team (2020). R: A Language and Environment for Statistical Computing. Available online at: https://www.r-project.org/ (accessed November 25, 2020).
Risely, A. (2020). Applying the core microbiome to understand host–microbe systems. J. Anim. Ecol. 89, 1549–1558. doi: 10.1111/1365-2656.13229
Rosado, P. M., Leite, D. C. A., Duarte, G. A. S., Chaloub, R. M., Jospin, G., Nunes da Rocha, U., et al. (2019). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Roth-Schulze, A. J., Zozaya-Valdés, E., Steinberg, P. D., and Thomas, T. (2016). Partitioning of functional and taxonomic diversity in surface-associated microbial communities. Environ. Microbiol. 18, 4391–4402. doi: 10.1111/1462-2920.13325
Russell, S. L. (2019). Transmission mode is associated with environment type and taxa across bacteria-eukaryote symbioses: a systematic review and meta-analysis. FEMS Microbiol. Lett. 366:efnz013. doi: 10.1093/femsle/fnz013
Salaün, S., la Barre, S., Santos-Goncalvez, M., Dos, P. P., Haras, D., and Bazire, A. (2012). Influence of exudates of the kelp laminaria digitata on biofilm formation of associated and exogenous bacterial epiphytes. Microb. Ecol. 64, 359–369. doi: 10.1007/s00248-012-0048-4
Salazar, G. (2019). EcolUtils: Utilities for Community Ecology Analysis. R package version 0.1. Available online at: https://github.com/GuillemSalazar/EcolUtils (accessed November 25, 2020).
Segovia, B. T., Sanders-Smith, R., Adamczyk, E. M., Forbes, C., Hessing-Lewis, M., O'Connor, M. I., et al. (2020). Microeukaryotic communities associated with the seagrass Zostera marina are spatially structured. J. Eukaryot. Microbiol. doi: 10.1111/jeu.12827. [Epub ahead of print].
Shade, A., and Handelsman, J. (2012). Beyond the Venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Short, F. T., Muehlstein, L. K., and Porter, D. (1987). Eelgrass wasting disease: cause and recurrence of a marine epidemic. Biol. Bull. 173, 557–562. doi: 10.2307/1541701
Thao, M. L. L., Baumann, L., Hess, J. M., Falk, B. W., Ng, J. C. K., Gullan, P. J., et al. (2003). Phylogenetic evidence for two new insect-associated chlamydia of the family Simkaniaceae. Curr. Microbiol. 47, 46–50. doi: 10.1007/s00284-002-3953-9
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870. doi: 10.1038/ncomms11870
Turon, M., Cáliz, J., Garate, L., Casamayor, E. O., and Uriz, M. J. (2018). Showcasing the role of seawater in bacteria recruitment and microbiome stability in sponges. Sci. Rep. 8:15201. doi: 10.1038/s41598-018-33545-1
Ugarelli, K., Chakrabarti, S., Laas, P., and Stingl, U. (2017). The seagrass holobiont and its microbiome. Microorganisms 5:81. doi: 10.3390/microorganisms5040081
Ugarelli, K., Laas, P., and Stingl, U. (2019). The microbial communities of leaves and roots associated with turtle grass (Thalassia testudinum) and manatee grass (Syringodium filliforme) are distinct from seawater and sediment communities, but are similar between species and sampling sites. Microorganisms 7:4. doi: 10.3390/microorganisms7010004
Van De Water, J. A. J. M., Allemand, D., and Ferrier-Pagès, C. (2018). Host-microbe interactions in octocoral holobionts - recent advances and perspectives. Microbiome 6:64. doi: 10.1186/s40168-018-0431-6
Weigel, B. L., and Pfister, C. A. (2019). Successional dynamics and seascape-level patterns of microbial communities on the canopy-forming kelps Nereocystis luetkeana and Macrocystis pyrifera. Front. Microbiol. 10:346. doi: 10.3389/fmicb.2019.00346
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag. Available online at: https://ggplot2.tidyverse.org (accessed November 25, 2020).
Keywords: core microbiome, 16S rRNA gene, eelgrass (Zostera marina), biodiversity, symbiosis
Citation: Sanders-Smith R, Segovia BT, Forbes C, Hessing-Lewis M, Morien E, Lemay MA, O'Connor MI and Parfrey LW (2020) Host-Specificity and Core Taxa of Seagrass Leaf Microbiome Identified Across Tissue Age and Geographical Regions. Front. Ecol. Evol. 8:605304. doi: 10.3389/fevo.2020.605304
Received: 11 September 2020; Accepted: 25 November 2020;
Published: 22 December 2020.
Edited by:
Carlos Prada, University of Rhode Island, United StatesReviewed by:
Cassie L. Ettinger, University of California, Riverside, United StatesCopyright © 2020 Sanders-Smith, Segovia, Forbes, Hessing-Lewis, Morien, Lemay, O'Connor and Parfrey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bianca Trevizan Segovia, YmlhdHNlZ292aWFAZ21haWwuY29t; Laura Wegener Parfrey, bHdwYXJmcmV5QGJvdGFueS51YmMuY2E=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.