- Institute of Plant Sciences, University of Regensburg, Regensburg, Germany
Global changes in land use are threatening the diversity of many ecosystems on both the intra- and interspecific levels. Among these ecosystems are the species-rich hay meadows, which have drastically declined in quality and quantity, due to land use intensification or abandonment in recent decades. The remaining genetic resources of their plant species must therefore be protected. To determine the driving forces impacting genetic variation in common hay meadow species (Dactylis glomerata, Heracleum sphondylium, and Trifolium pratense), we used data on the land use history, historic and present landscape structure and habitat quality. Our results showed average genetic diversity within the study sites, with low differentiation levels and a high gene flow among grasslands. Land use history, landscape structure and habitat quality were found to be related to the distribution of genetic diversity in the studied species, highlighting the complex forces acting in these ecosystems and showing the specific impact of litter accumulation on genetic diversity. Both historic and current environmental variables influence genetic diversity, demonstrating the importance of the land use history of a habitat. The most important group of variables impacting genetic variation in all three species was the landscape structure (e.g., distance to the nearest-located urban area or grassland). Also important was the influence of litter cover on genetic diversity in D. glomerata, which provides an interesting starting point for further research.
Introduction
Current global developments in land use are having detrimental effects on many ecosystems and are therefore also a threat to genetic variation. Especially species-rich and extensively managed European grasslands have declined drastically in recent decades, despite their relevance to species diversity and ecosystem services. Through land use intensification and abandonment these habitats are facing an ongoing decline in quality and quantity on the species as well as the genetic level (Poschlod et al., 2005; Hejcman et al., 2013). In the protection of these species-rich habitats, information on the distribution of genetic variation and its driving forces can contribute significantly to the success of nature conservation measures (Vellend et al., 2014).
Among these species-rich habitats are the oat-grass meadows, a type of lowland hay meadow of anthropogenic origin. The use of meadows for hay making was practiced on a larger scale since the Medieval Ages. It has been assumed that the first meadows were established in river floodplains, giving rise to the formation of different meadow types through the ages. With the introduction of the name-giving oat-grass Arrhenatherum elatius at the end of the Seventeenth century, oat-grass meadows became the dominant meadow type until the second half of the Twentieth century (Poschlod et al., 2009). The species diversity of these meadows depends on a traditional management scheme, consisting of one or two cuttings per year and low fertilization rates (Oberdorfer and Müller, 1983). However, after the invention and application of mineral fertilizers in the Twentieth century, these meadows were often transformed into intensively managed grasslands with up to seven cuttings per year (Poschlod et al., 2009; Kapfer, 2010), which has drastically reduced the species diversity of these meadows (Gaujour et al., 2012) and potentially decreased genetic diversity in the species present (Kölliker et al., 1998).
Traditionally managed hay meadows, in addition to hosting rare species, include “wild” populations of agriculturally relevant species such as Dactylis glomerata and Trifolium pratense, which occur in a broad range of habitat and soil conditions. Populations in these grasslands harbor genotypes adapted to their local, diverse habitat conditions. These genotypes provide resources useful to plant breeding efforts, as future climatic conditions may make specific genotypes better suited than others for agricultural use. Locally adapted populations are also important for the production of autochthonous seed material for conservation and restoration purposes, which require locally produced seed material to avoid outbreeding effects (McKay et al., 2005; Durka et al., 2017, 2019; Wang et al., 2020).
As it is time and cost intensive to study a wide range of populations in order to find those of highest interest, it is of considerable value to study the factors having general impacts on genetic variation in grasslands. Therefore, we investigated factors possibly impacting genetic variation in hay meadow species: (1) land use history, (2) historic and present landscape structure, and (3) current habitat quality and population size. To investigate the importance of these three factors we analyzed populations of three different grassland species (D. glomerata, H. sphondylium, and T. pratense) from traditionally managed hay meadows on the Swabian Alb. All of these factors have been previously reported to impact genetic variation in grassland species.
The land use history, often referred to as habitat age, is one factor frequently studied in grassland ecosystems (Rosengren et al., 2013). As nearly all grassland ecosystems in Central Europe are of anthropogenic origin, the management history of these systems has an impact on genetic variation. Grasslands with a long history of traditional management practices (“ancient” sites), like grazing or hay making and transfer, show higher species and genetic diversity, due to the effects of gene flow between sites and the accumulation of different genotypes over time (Jacquemyn et al., 2004; Cousins et al., 2009). The species and genetic diversity in more recently established grasslands (“recent” sites) is expected to be lower, due to the shorter time available for the accumulation of diversity. Based on these previous studies, we expected to find differences in genetic diversity among ancient and recent sites due to their land use history.
Another frequently studied variable is the historic and present landscape structure (Jacquemyn et al., 2004; Helm et al., 2009). The area of surrounding grasslands or human settlements may have an impact on genetic diversity of typical grassland species, especially in fragmented landscapes. Because of habitat fragmentation and thus reduced connectivity among populations, gene flow is reduced, and genetic drift or bottleneck events are possible. These processes lead to a decrease of genetic diversity within grassland patches and genetic differentiation among them. Besides connectivity, anthropogenic influence, and disturbance, as measured by the distance to and the area of settlements in the landscape, can impact genetic diversity. Historic management intensity, inferred from historic population density, was found to be correlated to genetic diversity in the grass species Briza media (Helm et al., 2009). Therefore, grasslands surrounded by many other grassland patches may show higher genetic diversity, due to their higher connectivity, than more isolated grasslands. Historic settlement area and distances are thought to have a positive effect on genetic diversity, while current anthropogenic disturbance can be expected to have negative impacts.
The third aspect often focused on in grassland studies is habitat quality and population size, which is frequently observed to be correlated with genetic variation in plant species (Schleuning et al., 2009; Vellend et al., 2014). Grasslands with a high species diversity, and therefore high habitat quality, may also show high genetic diversity, as a result of processes working on both diversity levels (Vellend, 2005). Also, the cover of vascular plants and litter may impact the germination of seeds. It has been found that litter acts as a seed trap, thereby reducing species diversity (Kahmen et al., 2002; Ruprecht and Szabó, 2012) by diminishing successful seedling establishment. Thus, the establishment of new genotypes is also reduced. Additionally, plant population size is a parameter frequently associated with genetic diversity. Larger populations tend to contain higher genetic diversity, due to the decreased risk of inbreeding and genetic drift (Vergeer et al., 2003b). However the effect of population size can be overshadowed by stronger effects, e.g., habitat quality (Vergeer et al., 2003a). Overall, therefore, we expected to find effects of both habitat quality and population structure on genetic diversity for the studied species.
Several studies have focused on one or two of these groups of explanatory variables, often with different conclusions (Helm et al., 2009; Reitalu et al., 2010; Münzbergová et al., 2013). Additionally, most studies were restricted to single species and focused on small geographical regions. Many previous studies in this context have been conducted on grazed grasslands such as calcareous grasslands, but studies in mown grasslands such as oat-grass meadows are rare. As previously observed effects are habitat- and species-specific, it is valuable to include several species in a study design, so as to uncover more general effects. Based on previous studies we expected to find differences among species based on their life history traits, with D. glomerata as a wind pollinated species, likely showing the least correlation with land use history and landscape structure, while the other two studies were more likely to show the impacts of these variables.
Methods
Study Sites
In the present study, we focused on hay meadows, e.g., oat-grass meadows, a comparably young grassland habitat, established for hay production at the beginning of the Eighteen century with the introduction of oat-grass (Arrhenatherum elatius) into central Europe (Poschlod et al., 2009). With the invention of mineral fertilizers, these habitats were transformed either into arable land or more intensively used grasslands, with up to seven cuttings per year (Kapfer, 2010), or on less profitable sites afforested or abandoned.
The studied hay meadows are located on the Swabian Alb in Southwestern Germany. The Swabian Alb belongs to the low mountain range in Southern Germany, with heights up to more than 1,000 m above sea level.
Twenty traditionally managed hay meadows were chosen for this study (Figures 1, 2b, Supplementary Table 3).
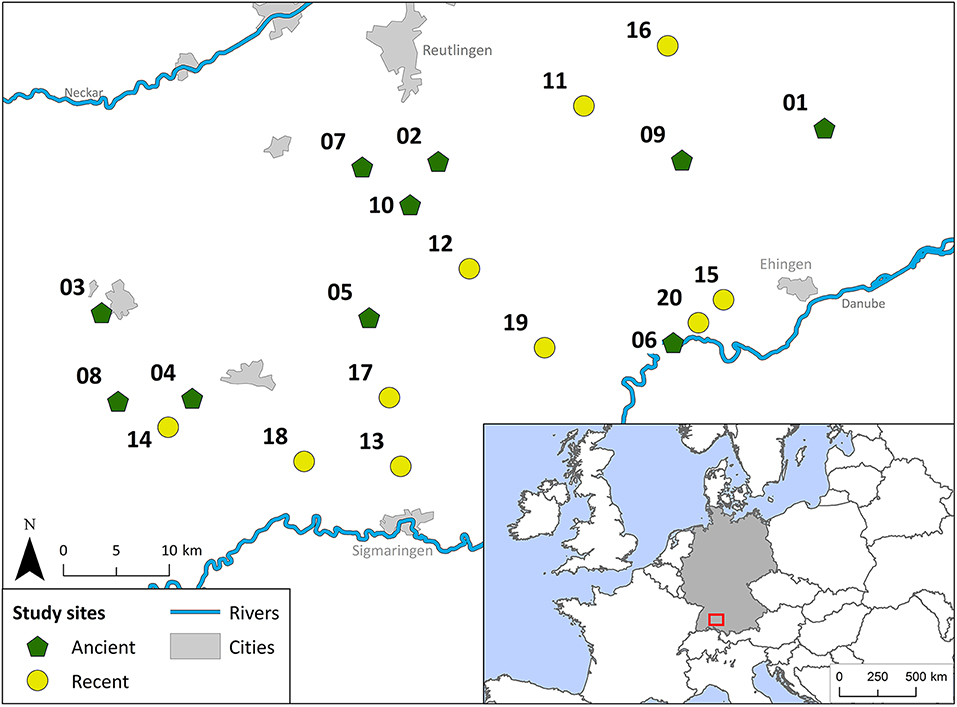
Figure 1. Geographic location of all 20 study sites on the Swabian Alb within the administrative district of Tübingen in Southwest Germany. Ancient grasslands are shown with a green pentagon (1–10) and recent grasslands with a yellow circle (11–20).

Figure 2. Pictures of the studied species [D. glomerata (a), H. sphondylium (c), and T. pratense (d)] and study site 01, as an example of a typical study site. Pictures were taken by EP (b) and TL (a,c,d) during sampling.
Analysis of Land Use History, Historic and Current Landscape Structure, and Present Vegetation Structure
In a first step the land use history of the studied meadows over the last ~200 years was accessed. Meadows with a long history of continuous use as managed grassland, dating back at least to the beginning of the Nineteen century, were classified as “ancient” grasslands (Karlík and Poschlod, 2009) and were compared with meadows located on sites used as arable land until after WWII (1950ies), here called “recent” grasslands. This classification was based on several topographic maps of the area between 1820 and 2018 (Supplementary Table 1). By using maps from different time points, we controlled for a continuous historic land use as meadow or arable field for the ancient and recent grasslands, respectively. In total ten ancient and ten recent grasslands were included in the study design.
In a second step, we collected data on the historic and current landscape structure. The historic landscape structure was analyzed based on maps from circa 1820, while the current landscape structure was inferred from current (2018) topographic maps. Within a three-kilometer radius around each studied grassland, the area of managed grassland (Area.G), as well as urban areas (Area.S) were digitized, using ArcGIS (Version 10.4.1). From these areas the distance of the study sites to the nearest-located grassland and the nearest-located human settlement was calculated (Dist.G & Dist.S).
Finally, to investigate the present habitat quality at the sites, vegetation surveys were conducted, using the extended Braun-Blanquet scale in six plots (2 ×2 m, 24 m2 in total) per study site (Reichelt and Wilmanns, 1973). The Braun-Blanquet scale is a method commonly used for vegetation surveys in Central Europe that uses the abundance of each occurring species to study vegetation assemblages. The extended Braun-Blanquet scale allows for a more detailed categorization of species with low cover. Plots were placed evenly across the meadow on homogenous vegetation representing the habitat, so as to represent the diversity within the whole meadow as best as possible. For the six vegetation survey plots per study site the coverage of total vegetation and of litter were estimated as percentages. The mean cover of vegetation and plant litter per study site was used in the later data analysis to test for correlations between vegetation characteristics and genetic variation.
Study Species and Sampling
In our study we analyzed three common grassland species, frequently occurring in hay meadows: Dactylis glomerata L. s. str., Heracleum sphondylium L., and Trifolium pratense L (Figures 2a,c,d). All three species were chosen due to their frequent occurrence in hay meadows. They also represent species with different life history traits and D. glomerata and T. pratense are also important from an agricultural perspective.
D. glomerata, commonly known as cock's-foot or orchard grass, is a perennial Poaceae, growing on e.g., fresh meadows and semidry grasslands. This species is a widespread grass with a high fodder value (Sebald et al., 1998). D. glomerata prefers good nutrient availability and depends on light for seed germination (Oberdorfer et al., 2001; Rothmaler, 2017).
The Apiaceae H. sphondylium (hogweed) grows up to 1.50 m tall, with white to yellow-greenish flowers. It occurs in fresh meadows and along ditches and roads (Rothmaler, 2017). Pollination mainly occurs by flies and bees. H. sphondylium thrives in nutrient-rich meadows (Sebald et al., 1992), but its fodder value and grazing tolerance are low (Oberdorfer et al., 2001; Rothmaler, 2017).
The Fabaceae T. pratense (red clover), along with its symbiotic bacteria, acts as a soil improver. Like D. glomerata this species is a valuable fodder plant (Sebald et al., 1992). T. pratense is mainly pollinated by bumblebees and is self-sterile (Oberdorfer et al., 2001; Rothmaler, 2017).
At each study site, plant leaf material of up to 16 healthy individuals of each species was collected for genetic analysis in order to cover more than 90% of the total genetic diversity present (Leipold et al., 2020). In total, we sampled 953 individuals (Supplementary Table 3). To prevent sampling clones or closely related individuals, leaf samples were taken at least five meters apart (20 m on average), and sampling was distributed across the whole study area so as to represent the present genetic variation in the best way possible. Samples were dried and stored on silica gel at room temperature until DNA extraction.
Additionally, the population size per species was estimated at each site. Within 10 to 15 randomly placed 1 m2-plots (depending on grassland size) the occurrence of each species was counted. Plots were at least 10 m apart and spread across the whole study site. The present population size of each species was calculated by multiplying the mean number of individuals over all plots by the present habitat size (Reisch et al., 2018) (Supplementary Table 3). At two study sites, individuals of H. sphondylium did not occur within the plots, although the species had been known to occur at the site. Therefore, as an approximation for the calculation of the population size, we assumed one individual occurring in one plot.
Molecular Analyses
From the dried leaf material, DNA was extracted following the CTAB protocol by Rogers and Bendich (1994) with modifications by Reisch (2007). Extracts were diluted with water to 7.8 ng/μL and used for AFLP analysis, carried out in accordance with the protocol provided by Beckman Coulter as previously described by Vogler and Reisch (2011). After screening 36 primer combinations for eight randomly chosen individuals per species, three primer combinations were chosen for each study species (Supplementary Table 2) to be used in the selective PCR step. Samples were analyzed using automated capillary electrophoresis. Results were surveyed using the GeXP software (Beckman Coulter), exported as synthetic gel files (.crv), and analyzed using the software Bionumerics 4.6 (Applied Maths, Kortrijk, Belgium). Samples yielding unclear or weak band patterns, or obviously representing PCR artifacts, were repeated once. In total 318, 315 and 320 samples of each respective species were used for subsequent statistical analyses (Supplementary Table 3). Band scoring resulted in 185 fragments for D. glomerata, 184 for H. sphondylium and 163 for T. pratense (see Supplementary Informations 2–4).
The reproducibility of the AFLP analysis was tested by calculating the genotyping error rate, by reanalyzing 10% of all studied samples (Bonin et al., 2004). Following this analysis, a genotyping error rate of 2.96% for D. glomerata, 3.65% for H. sphondylium and 5.23% for T. pratense was determined.
Data Analysis
To evaluate the differences between historic and current landscape structure variables (Dist.S, Dist.G, Area.S, Area.G) and habitat quality indicators (Cov_Veg, Cov_Litter, Popsize), Dunn's test was performed in R using the “PMCMR” package (Pohlert, 2014), which checked for significant decreases or increases in the observed variables, as well as differences between ancient and recent grasslands.
Using the AFLP fragment data, a binary matrix was created, representing the presence/absence of the respective fragments for each studied individual. Based on this 0/1 matrix, genetic variation within populations of each species was calculated using the software PopGene 32 (Yeh et al., 1997). This program, among other functions, allows for the calculation of Nei's Gene Diversity (H = 1–Σ (pi)2).
Additionally, to analyses the genetic variation within and among all populations and among populations from ancient and recent grasslands, a hierarchical Analysis of Molecular Variance (AMOVA) (Excoffier et al., 1992), based on pairwise Euclidian distances between samples, was calculated using GenAlEx 6.5 (Peakall and Smouse, 2012). Based on the genetic distance values (?PT values) produced by the AMOVA and the geographic distance among populations, a Mantel test was performed with 999 permutations (Mantel, 1967) in the GenAlEx software, to test for correlations of genetic and geographic distances and thus to test for isolation by distance.
Finally, to investigate the effects of various environmental factors (Table 1) multivariate linear models were run in RStudio 1.1.423 (RStudio Team, 2016) for each studied species using the “l m” function. Before constructing the full model, Pearson correlation coefficients were calculated for all explanatory variables (Supplementary Table 5) to check for intercorrelations between variables; however, we did not exclude variables prior to model construction. From the full model, containing (1) historic and current total grassland and settlement area, (2) historic and current distances to nearest-located grassland and settlement, (3) land use history, (4) population size, as well as (5) vegetation and litter cover, an AIC based backward selection procedure was used to identify the model best fitting the data, using the “AICc” method (Akaike Information Criterion corrected for small sample sizes) from the “MuMIn” R-package (Burnham and Anderson, 2002; Kamil, 2018). To account for the difference in scale of the predictor variables used in the models, the function “scale()” was used on all variables except land use history. This function scales and centers each variable to allow for a better comparison among predictor variables.
Additionally we calculated a Redundancy analysis (RDA) for all three species and all explanatory variables used in the linear models using the “rda” function implemented in the “vegan” package (Oksanen et al., 2019).
Results
Historic and Current Landscape Structure
Around the study sites settlement area increased to six times its previous extent from 1820 to the present day. With this expansion, the distance of the study sites to the nearest-located urban area decreased (Supplementary Table 4). Thus, recent grasslands were located significantly closer to current settlement areas (mean distance = 343.8 m), than ancient grasslands (mean distance = 746.6 m, Figure 3A, p = 0.028).
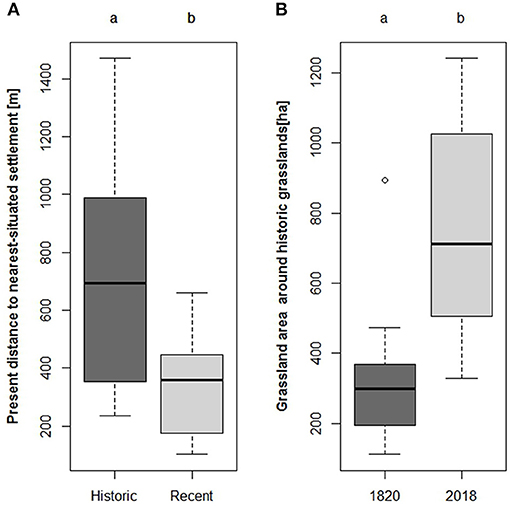
Figure 3. (A) Present distance to the nearest-located settlement or city separated for the two age categories of the investigated grasslands. A Dunne test revealed ancient grasslands to be located further away from settlements than recent grasslands (p = 0.028). (B) Grassland area [ha] around ancient grasslands in 1,820 and at present. The amount of grassland around ancient grasslands increased significantly (p <0.01).
Total area of grassland increased around all study sites between 1820 and 2018, however the increase was only significant for ancient grasslands (Supplementary Table 4, Figure 3B, p <0.01). As the total grassland area increased, the distance between the study sites and the nearest-located grassland decreased between 1820 and 2018 (Supplementary Table 4).
Current Habitat Quality and Population Size
The vegetation surveys showed a vegetation cover of ~90% for both ancient and recent grasslands and did no show significant differences. Litter cover showed an overall mean of 2.2%, which did not vary significantly among ancient and recent grasslands (Supplementary Table 4). The vegetation cover and litter cover were negatively correlated (Supplementary Table 5).
The size of the studied populations varied greatly among sites, with species population sizes ranging from a few hundred individuals up to several hundred thousand. We found no difference in population size between ancient and recent grasslands in any of the studied species (Supplementary Table 3).
Genetic Diversity and Differentiation
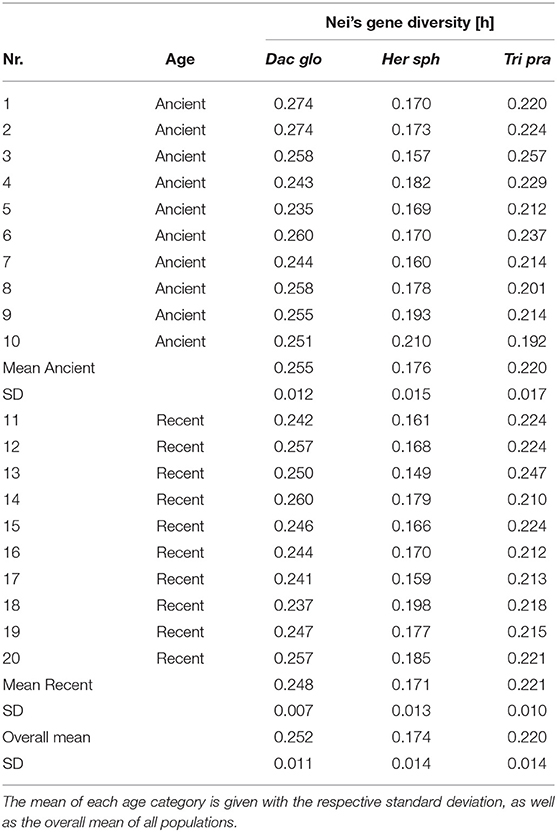
The Poaceae D. glomerata showed a mean genetic diversity of 0.252. The herbaceous species H. sphondylium showed a mean Nei's gene diversity of 0.174, while the legume T. pratense showed a mean diversity of 0.220 (Table 2). Nei's gene diversity did not differ significantly among ancient and recent populations. The AMOVA showed low levels of differentiation (2–5%) among populations and no differentiation between populations on ancient and recent grassland; most variation was found within populations (Table 3). The Mantel-test revealed no isolation by distance for either plant species (Dac glo: R2 = 0.0051, p = 0.240; Her sph: R2 = 0.0051, p = 0.223; Tri pra: R2 = 0.0006, p = 0.387).
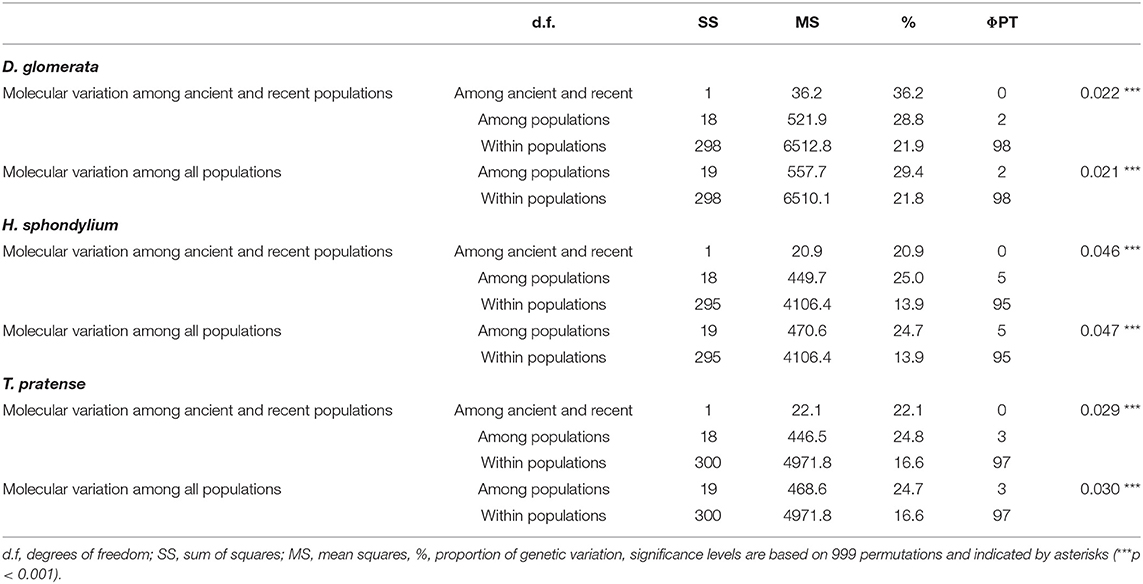
Table 3. Results of the Three-Level AMOVA given as the genetic variation among ancient and recent populations, as well as among all and within the respective populations of the studied species.
Multivariate Analysis
A significant linear model could be found for each species, but the final models, as calculated by the AICc method, differed among species (Table 4). Genetic diversity in D. glomerata was influenced by litter cover (Cov_Litter) on the grassland, with increasing litter cover leading to lower levels of genetic diversity. Additionally, in this model, land use history (Age) was significantly correlated with genetic diversity. In the model populations of D. glomerata from ancient grasslands showed higher genetic variation than populations sampled from recent grasslands. This model explained 29.4% of the observed variation. The model for H. sphondylium included only one significant variable. Genetic diversity of this species was positively related to the current distance to the nearest-located settlement (Dist.S_2018). This association explained 18.8% of the observed variation.
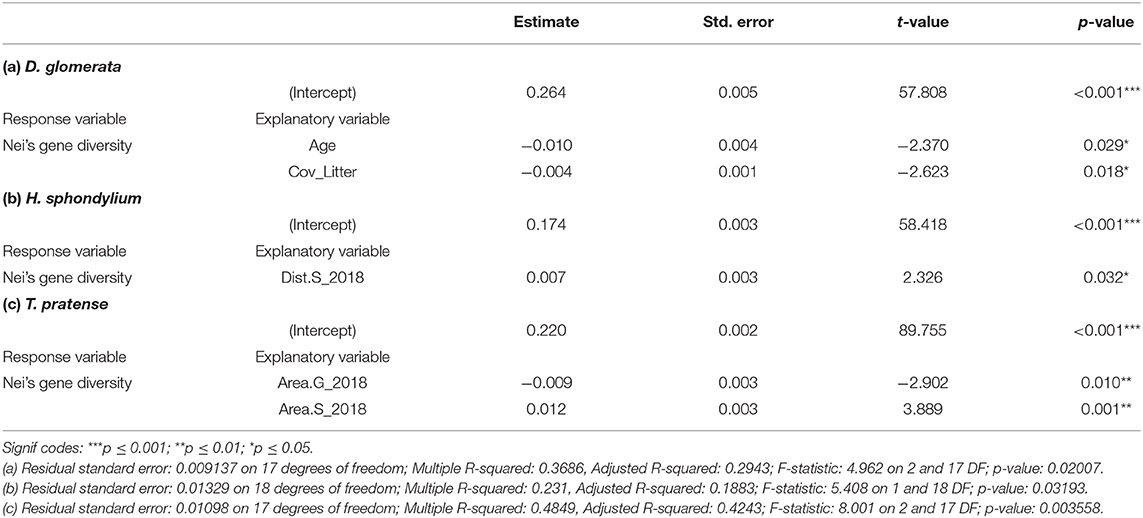
Table 4. Linear models explaining genetic diversity of D. glomerata (a), H. sphondylium (b), and T. pratense (c).
The model for T. pratense explained 42.4% of the observed variation with a positive association between genetic diversity and present settlement area (Area.S_2018), as well as a negative association with current grassland area (Area.G_2018).
The RDA showed correlations of AFLP fragment frequencies with landscape structure variables (Figure 4). Fragment frequencies in D. glomerata populations were correlated with Dist.S_2018 (r = 0.031, p = 0.041) and Area.S_2018 (r = 0.35, p = 0.037), while Dist.S_1820 was only marginally significant (r = 0.28, p = 0.064). For H. sphondylium and T. pratense only a correlation with Dist.G_1820 was observed (Hs: r = 0.505, p = 0.003; Tp: r = 0.505, p = 0.007).
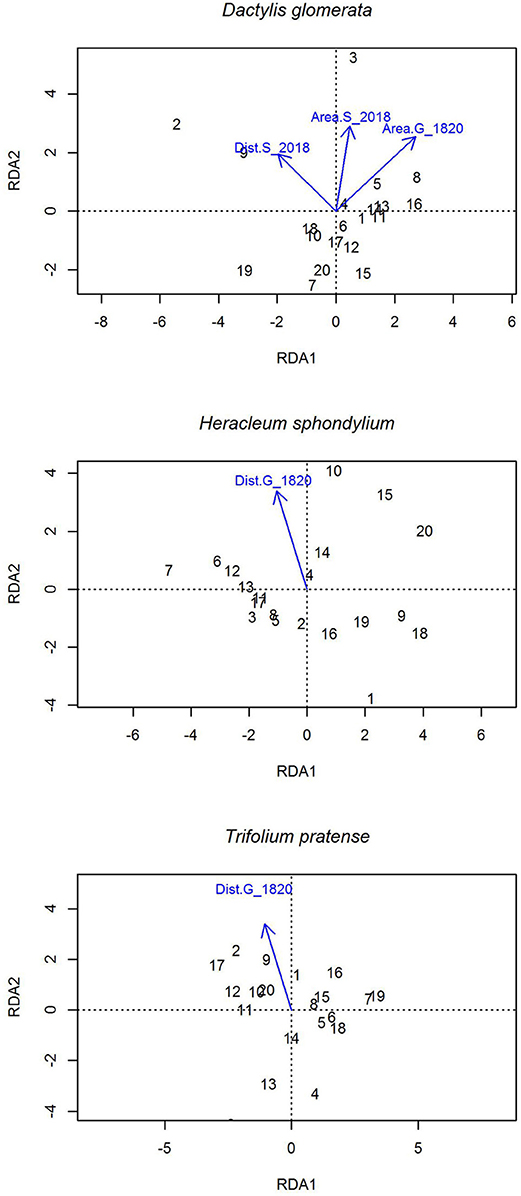
Figure 4. Redundancy analysis (RDA) of AFLP fragment abundance with the explanatory variables used in the linear models for each species (Table 1). Fragment frequencies in D. glomerata populations were correlated with Dist.S_2018 (r = 0.031, p = 0.041) and Area.S_2018 (r = 0.35, p = 0.037) and Dist.S_1820 was marginally significant (r = 0.28, p = 0.064). For H. sphondylium and T. pratense only a correlation with Dist.G_1820 was observed (Hs: r = 0.505, p = 0.003; Tp: r = 0.505, p = 0.007).
Discussion
The observed genetic diversity among and within populations varied only slightly in the three studied species. Overall, D. glomerata, H. sphondylium, and T. pratense showed moderate levels of genetic diversity within the range observed for species with similar life history traits (Reisch and Bernhardt-Römermann, 2014) comparable to those reported by other studies (Kölliker et al., 2003; Last et al., 2014). As expected due to its wind pollination, D. glomerata showed the highest genetic diversity, while the insect pollinated H. sphondylium and T. pratense showed lower values. For all three species, the highest genetic diversity was found in populations from ancient grasslands.
Genetic differentiation among populations was low, and we did not detect isolation by distance within the study area for any of the three species. As the distribution of the investigated species is not limited to hay meadows, or more specifically oat-grass meadows, but extends to road margins, hedges, and other grassland types, there are many possibilities for gene flow among populations. Pollen of D. glomerata has been shown to be transported by wind as far as 10 km (Willerding and Poschlod, 2002), explaining the low genetic differentiation observed here. Additionally, honey bees have been found to forage over long distances (up to 9.5 km), providing long-distance pollen dispersal for insect-pollinated species like T. pratense (Beekman and Ratnieks, 2000). Moreover, oat-grass meadows are a comparably young habitat, with a well-documented history of seed transfer events, seed trade across Europe, and frequent re-sowing as a means of increasing productivity (Kauter, 2001; Hejcman et al., 2013), all of which may contribute to explaining the comparably low genetic differentiation among populations observed here.
However, genetic diversity values did differ among populations, and the linear models revealed several associations of land use history, landscape structure, and habitat quality with genetic diversity.
We found land use history to be associated with genetic diversity in D. glomerata. Plant populations of ancient meadows, continually used as grassland for over 200 years, showed overall higher genetic diversity, than populations in meadows established on arable fields after WWII. An impact of land use history has previously been shown on the level of plant species diversity in European grasslands (Cousins et al., 2009; Reitalu et al., 2010), as well as on the genetic level for typical grassland species (Jacquemyn et al., 2004; Rosengren et al., 2013). For example Rosengren et al. (2013) found a positive relationship between habitat age and genetic diversity in the moss Homalothecium lutescens explained by the effects of grazing continuity on the ancient grassland patches. Similarly, our findings for typical hay meadow species, can be explained by the long tradition of sowing and re-sowing practices in historic meadows. Due to the repeated introduction of new genotypes, genetic diversity accumulated in ancient grasslands, giving them a higher genetic diversity. Recent meadows were established on arable fields only within the last 60 years, and have thus experienced fewer introductions, resulting in lower accumulated genetic diversity. Additionally, the management practices changed simultaneously with the establishment of the recent meadows in the middle of the Twentieth century, with modern agricultural machines and more intensive fertilization, all of which led to a more homogenous land use (Poschlod, 2017). These modern land use practices at the beginning of the meadow establishment decreased small-scale variations in the disturbance regime, thus providing fewer opportunities for the establishment of new genotypes.
The landscape structure of the Swabian Alb has changed dramatically during the last 200 years. Urban areas have increased, leading to a decrease in the distance of the studied grasslands to the nearest-located settlements, suggesting a larger human impact on these ecosystems (Frey et al., 2016). Additionally, the total grassland cover in the study area has increased, and with it, the connectivity of grassland patches. As the distribution of our studied plant species is not limited to extensively used grasslands, we assume them to be potentially present in most of the grasslands in this area. We found significant associations between present landscape configuration and genetic diversity in H. sphondylium and T. pratense in the linear models and for D. glomerata in the redundancy analysis. Similar associations were also found in other studies focusing on the influence of historic and current landscape configuration on genetic diversity (Münzbergová et al., 2013; Reisch et al., 2017). Helm et al. (2009) found that current landscape structure was an important predictor for genetic variation in Briza media, where grasslands with a high connectivity index also exhibited high genetic diversity. Additionally, this study found a negative correlation between genetic diversity with current human population density. Similarly, we found a positive association between genetic diversity in H. sphondylium and present distance to the nearest-located human settlement, meaning that populations that are less influenced by human disturbance exhibit higher genetic diversity. This relationship might best be explained by the mowing sensitivity of hogweed (Dierschke and Briemle, 2002), as mowing frequency, at least in traditionally managed grasslands, likely decreases with increased distance to the nearest-located settlement area.
Contrastingly, genetic diversity in T. pratense was negatively associated with present grassland area and positively with present settlement area. The negative relationship with grassland area can be explained by considering that the topographic maps do not include information on the management intensity of the investigated grasslands. The areas surrounding our studied grasslands include intensively managed grasslands, sown with industrial seed mixtures, which likely do not include different genotypes. Thus, grasslands that are surrounded by a larger proportion of intensively managed grasslands will decrease in genetic diversity over time through gene flow.
Reisch et al. (2017) found historic landscape configuration to be more important for genetic variation in typical calcareous grassland species than present habitat conditions.
Similarly, in our study, historic landscape structure (Dist.G_1820 & Area.S_1820) was correlated with genetic variation of D. glomerata, H. sphondylium, and T. pratense in the RDA analysis, supporting the idea that historic human impact (e.g., mowing and sowing events), and not only current landscape structure, contributed to the build-up of current genetic diversity.
On the level of habitat quality the linear models revealed a negative association of litter cover with genetic diversity in D. glomerata. To our knowledge, a relationship between plant litter cover and genetic variation has not yet been reported in any published studies. Several studies have found negative effects of plant litter accumulation on plant species variation, seed germination, and seedling establishment (Schleuning et al., 2009; Ruprecht et al., 2010; Ruprecht and Szabó, 2012). As litter prevents the establishment of new individuals genetic variation will be lost in the long term due to genetic drift. These findings have important implications for the conservation and restoration of grasslands. Proper management practices that reduce litter accumulation, might not only improve species richness, as shown by Ruprecht et al. (2010) and others, but potentially also prevent the loss of genetic variation within individual species.
Conclusion
The results of our study highlight the possibilities afforded by a multi-species approach. By including several species and a range of different explanatory variables, we were able to show that, while individual species showed unique associations with all three groups of explanatory variables, the main factors overall were linked to historical and current landscape structure. While the level of genetic diversity seemed to be mainly associated with current landscape and habitat quality parameters, fragment frequencies were mainly associated with historic landscape structure variables. These results show that conservation and restoration of grassland ecosystems should also take historic developments into account, so as to better understand the patterns observed today. Additionally, the association of genetic diversity with litter accumulation merits further research.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author's Note
Maps throughout this article were created using ArcGIS® software by Esri. ArcGIS® and ArcMap™ are the intellectual property of Esri and are used herein under license. Copyright © Esri. All rights reserved. For more information about Esri® software, please visit www.esri.com.
Author Contributions
CR and PP designed the study. EP and TL collected the data and performed the genetic analysis. EP performed the data analysis. EP and CR led the writing of the manuscript. All authors contributed to manuscript writing and approved it for publication.
Funding
The study presented in this manuscript was funded by the Bundesanstalt für Landwirtschaft und Ernährung.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Lina Begemann, Franziska Parzefall, Laila Pfaettisch, and Petra Schitko for their support in the field and the lab. Many thanks go also to Lukas Feichtmeier and Sabine Fischer for their support with ArcGIS. Cornelia Straubinger and Eva Rosa Wagner we thank for the vegetation surveys and the cooperative efforts in this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.593302/full#supplementary-material
References
Beekman, M., and Ratnieks, F. L. W. (2000). Long-range foraging by the honey-bee, Apis mellifera L. Funct. Ecol. 14, 490–496. doi: 10.1046/j.1365-2435.2000.00443.x
Bonin, A., Bellemain, E., Eidesen, P. B., Pompanon, F., Brochmann, C., and Taberlet, P. (2004). How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 13, 3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x
Burnham, K. P., and Anderson, D. R. (2002). Model Selection and Multimodel Inference : A Practical Information-Theoretic Approach. 2nd Edn. New York, NY: Springer.
Cousins, S. A. O., Lindborg, R., and Mattsson, S. (2009). Land use history and site location are more important for grassland species richness than local soil properties. Nord. J. Bot. 27, 483–489. doi: 10.1111/j.1756-1051.2009.00472.x
Dierschke, H., and Briemle, G. (2002). Kulturgrasland–Wiesen, Weiden und Verwandte Staudenfluren. Stuttgart: Ökosysteme Mitteleuropas aus geobotanischer Sicht.
Durka, W., Michalski, S. G., Berendzen, K. W., Bossdorf, O., Bucharova, A., Hermann, J. M., et al. (2017). Genetic differentiation within multiple common grassland plants supports seed transfer zones for ecological restoration. J. Appl. Ecol. 54, 116–126. doi: 10.1111/1365-2664.12636
Durka, W., Bossdorf, O., Bucharova, A., Frenzel, M., Hermann, J.-M., Hölzel, N., et al. (2019). Regionales Saatgut von Wiesenpflanzen: genetische Unterschiede, regionale Anpassung und Interaktion mit Insekten. Nat. Landsc. 94, 146–153. doi: 10.17433/4.2019.50153679
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Frey, D., Arrigo, N., Granereau, G., Sarr, A., Felber, F., and Kozlowski, G. (2016). Parallel declines in species and genetic diversity driven by anthropogenic disturbance: a multispecies approach in a French Atlantic dune system. Evol. Appl. 9, 479–488. doi: 10.1111/eva.12351
Gaujour, E., Amiaud, B., Mignolet, C., and Plantureux, S. (2012). Factors and processes affecting plant biodiversity in permanent grasslands. A review. Agron. Sustain. Dev. 32, 133–160. doi: 10.1007/s13593-011-0015-3
Hejcman, M., Hejcmanová, P., Pavlu, V., and Beneš, J. (2013). Origin and history of grasslands in Central Europe - a review. Grass Forage Sci. 68, 345–363. doi: 10.1111/gfs.12066
Helm, A., Oja, T., Saar, L., Takkis, K., Talve, T., and Pärtel, M. (2009). Human influence lowers plant genetic diversity in communities with extinction debt. J. Ecol. 97, 1329–1336. doi: 10.1111/j.1365-2745.2009.01572.x
Jacquemyn, H., Honnay, O., Galbusera, P., and Roldán-Ruiz, I. (2004). Genetic structure of the forest herb Primula elatior in a changing landscape. Mol. Ecol. 13, 211–219. doi: 10.1046/j.1365-294X.2003.02033.x
Kahmen, S., Poschlod, P., and Schreiber, K. F. (2002). Conservation management of calcareous grasslands. Changes in plant species composition and response of functional traits during 25 years. Biol. Conserv. 104, 319–328. doi: 10.1016/S0006-3207(01)00197-5
Kamil, B. (2018). MuMIn: Multi-Model Inference. R package version 1.42.1. Available online at: https://cran.r-project.org/package=MuMIn.
Karlík, P., and Poschlod, P. (2009). History or abiotic filter: which is more important in determining the species composition of calcareous grasslands? Preslia 81, 321–340.
Kauter, D. (2001). “Sauergras” und “Wegbreit”: die Entwicklung der Wiesen in Mitteleuropa zwischen 1500 und 1900. Berichte des Instituts für Lanschafts- und Pflanzenökologie der Universität Hohenheim, Beiheft 14, Stuttgart.
Kölliker, R., Herrmann, D., Boller, B., and Widmer, F. (2003). Swiss Mattenklee landraces, a distinct and diverse genetic resource of red clover (Trifolium pratense L.). Theor. Appl. Genet. 107, 306–315. doi: 10.1007/s00122-003-1248-6
Kölliker, R., Stadelmann, F. J., Reidy, B., and Nösberger, J. (1998). Fertilization and defoliation frequency affect genetic diversity of Festuca pratensis Huds. In permanent grasslands. Mol. Ecol. 7, 1557–1567. doi: 10.1046/j.1365-294x.1998.00486.x
Last, L., Lüscher, G., Widmer, F., Boller, B., and Kölliker, R. (2014). Indicators for genetic and phenotypic diversity of Dactylis glomerata in Swiss permanent grassland. Ecol. Indic. 38, 181–191. doi: 10.1016/j.ecolind.2013.11.004
Leipold, M., Tausch, S., Hirtreiter, M., Poschlod, P., and Reisch, C. (2020). Sampling for conservation genetics: how many loci and individuals are needed to determine the genetic diversity of plant populations using AFLP? Conserv. Genet. Resour. 12, 99–108. doi: 10.1007/s12686-018-1069-1
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220.
McKay, J. K., Christian, C. E., Harrison, S., and Rice, K. J. (2005). “How local is local?”A review of practical and conceptual issues in the genetics of restoration. Restor. Ecol. 13, 432–440. doi: 10.1111/j.1526-100X.2005.00058.x
Münzbergová, Z., Cousins, S. A. O., Herben, T., Plačková, I., Mildén, M., and Ehrlén, J. (2013). Historical habitat connectivity affects current genetic structure in a grassland species. Plant Biol. 15, 195–202. doi: 10.1111/j.1438-8677.2012.00601.x
Oberdorfer, E., and Müller, T. (1983). Süddeutsche Pflanzengesellschaften/ 3 Wirtschaftswiesen und Unkrautgesellschaften. 2nd Edn. Stuttgart: Fischer.
Oberdorfer, E., Schwabe, A., and Müller, T. (2001). Pflanzensoziologische Exkursionsflora - Für Deutschland und angrenzende Gebiete. 8th Edn. Ulmer, Stuttgart.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). Vegan: Community Ecology Package. Available online at: https://cran.r-project.org/package=vegan
Peakall, R., and Smouse, P. E. (2012). GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pohlert, T. (2014). The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). Available online at: https://cran.r-project.org/package=PMCMR
Poschlod, P., Bakker, J. P., and Kahmen, S. (2005). Changing land use and its impact on biodiversity. Basic Appl. Ecol. 6, 93–98. doi: 10.1016/j.baae.2004.12.001
Poschlod, P., Baumann, A., and Karlik, P. (2009). “Origin and development of grasslands in Central Europe,” in Grasslands in Europe of High Nature Value (Leiden: KNNV Publishing), 15–25. doi: 10.1163/9789004278103_003
Reisch, C. (2007). Genetic structure of Saxifraga tridactylites (Saxifragaceae) from natural and man-made habitats. Conserv. Genet. 8, 893–902. doi: 10.1007/s10592-006-9244-4
Reisch, C., and Bernhardt-Römermann, M. (2014). The impact of study design and life history traits on genetic variation of plants determined with AFLPs. Plant Ecol. 215, 1493–1511. doi: 10.1007/s11258-014-0409-9
Reisch, C., Schmid, C., and Hartig, F. (2018). A comparison of methods for estimating plant population size. Biodivers. Conserv. 27, 2021–2028. doi: 10.1007/s10531-018-1522-1
Reisch, C., Schmidkonz, S., Meier, K., Schöpplein, Q., Meyer, C., Hums, C., et al. (2017). Genetic diversity of calcareous grassland plant species depends on historical landscape configuration. BMC Ecol. 17:19. doi: 10.1186/s12898-017-0129-9
Reitalu, T., Johansson, L. J., Sykes, M. T., Hall, K., and Prentice, H. C. (2010). History matters: village distances, grazing and grassland species diversity. J. Appl. Ecol. 47, 1216–1224. doi: 10.1111/j.1365-2664.2010.01875.x
Rogers, S. O., and Bendich, A. J. (1994). “Extraction of total cellular DNA from plants, algae and fungi,” in Plant Molecular Biology Manual, eds S. B. Gelvin, and R. A. Schilperoort (Dordrecht: Springer Netherlands), 183–190. doi: 10.1007/978-94-011-0511-8_12
Rosengren, F., Cronberg, N., Reitalu, T., and Prentice, H. C. (2013). Genetic variation in the moss Homalothecium lutescens in relation to habitat age and structure. Botany 91, 431–441. doi: 10.1139/cjb-2012-0258
Rothmaler, W. (2017). Rothmaler - Exkursionsflora von Deutschland. Gefäßpflanzen: Grundband. 21st Edn, ed E. J. Jäger (Berlin: Springer Berlin Heidelberg).
RStudio Team. (2016). RStudio: Integrated Development for R. Available at: http://www.rstudio.com
Ruprecht, E., Enyedi, M. Z., Eckstein, R. L., and Donath, T. W. (2010). Restorative removal of plant litter and vegetation 40 years after abandonment enhances re-emergence of steppe grassland vegetation. Biol. Conserv. 143, 449–456. doi: 10.1016/j.biocon.2009.11.012
Ruprecht, E., and Szabó, A. (2012). Grass litter is a natural seed trap in long-term undisturbed grassland. J. Veg. Sci. 23, 495–504. doi: 10.1111/j.1654-1103.2011.01376.x
Schleuning, M., Niggemann, M., Becker, U., and Matthies, D. (2009). Negative effects of habitat degradation and fragmentation on the declining grassland plant Trifolium montanum. Basic Appl. Ecol. 10, 61–69. doi: 10.1016/j.baae.2007.12.002
Sebald, O., Seybold, S., and Philippi, G. (1992). Die Farn- und Blütenpflanzen Baden-Württembergs 4: Spezieller Teil (Spermatophyta, Unterklasse Rosidae), Haloragaceae bis Apiaceae. Stuttgart: Ulmer.
Sebald, O., Seybold, S., Philippi, G., and Wörz, A. (1998). Die Farn- und Blütenpflanzen Baden-Württembergs. 7: Spezieller Teil (Spermatophyta, Unterklassen Alismatidae, Liliidae Teil 1, Commelinidae Teil 1), Butomaceae bis Poaceae. Stuttgart: Ulmer.
Vellend, M. (2005). Species diversity and genetic diversity: parallel processes and correlated patterns. Am. Nat. 166, 199–215. doi: 10.1086/431318
Vellend, M., Lajoie, G., Bourret, A., Múrria, C., Kembel, S. W., and Garant, D. (2014). Drawing ecological inferences from coincident patterns of population- and community-level biodiversity. Mol. Ecol. 23, 2890–2901. doi: 10.1111/mec.12756
Vergeer, P., Rengelink, R., Copal, A., and Ouborg, N. J. (2003a). The interacting effects of genetic variation, habitat quality and population size on performance of Succisa pratensis. J. Ecol. 91, 18–26. doi: 10.1046/j.1365-2745.2003.00736.x
Vergeer, P., Rengelink, R., Ouborg, N. J., and Roelofs, J. G. M. (2003b). Effects of population size and genetic variation on the response of Succisa pratensis to eutrophication and acidification. J. Ecol. 91, 600–609. doi: 10.1046/j.1365-2745.2003.00785.x
Vogler, F., and Reisch, C. (2011). Genetic variation on the rocks - the impact of climbing on the population ecology of a typical cliff plant. J. Appl. Ecol. 48, 899–905. doi: 10.1111/j.1365-2664.2011.01992.x
Wang, J., Shi, J., Liu, S., Sun, X., Huang, J., Qiao, W., et al. (2020). Conservation recommendations for oryza rufipogon griff. in China based on genetic diversity analysis. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-70989-w
Willerding, C., and Poschlod, P. (2002). Does seed dispersal by sheep affect the population genetic structure of the calcareous grassland species Bromus erectus? Biol. Conserv. 104, 329–337. doi: 10.1016/S0006-3207(01)00198-7
Keywords: European grasslands, conservation genetics, habitat quality, land use history, AFLP (amplified fragment length polymorphism), landscape structure, hay meadows
Citation: Pagel E, Lehmair TA, Poschlod P and Reisch C (2020) Genetic Variation of Typical Plant Species in Hay Meadows: The Effect of Land Use History, Landscape Structure, and Habitat Quality. Front. Ecol. Evol. 8:593302. doi: 10.3389/fevo.2020.593302
Received: 10 August 2020; Accepted: 02 December 2020;
Published: 21 December 2020.
Edited by:
Rosane Garcia Collevatti, Universidade Federal de Goiás, BrazilReviewed by:
Carolina Da Silva Carvalho, Vale Technological Institute (ITV), BrazilBruno Baur, University of Basel, Switzerland
Copyright © 2020 Pagel, Lehmair, Poschlod and Reisch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ellen Pagel, ZWxsZW4ucGFnZWxAdXIuZGU=
†ORCID: Ellen Pagel orcid.org/0000-0002-4257-2897
Theresa A. Lehmair orcid.org/0000-0003-4595-867X
Christoph Reisch orcid.org/0000-0003-4098-6516
Peter Poschlod orcid.org/0000-0003-4473-7656
 Ellen Pagel
Ellen Pagel Theresa A. Lehmair†
Theresa A. Lehmair† Peter Poschlod
Peter Poschlod