- 1Institute of Fisheries Science, National Taiwan University, Taipei, Taiwan
- 2Germark Biotechnology Co., Ltd., Taichung, Taiwan
- 3Council of Agriculture, Executive Yuan, Taiwan Agricultural Chemicals and Toxic Substances Research Institute, Taichung, Taiwan
- 4Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 5WellGenetics Inc., Taipei, Taiwan
- 6Bioinformatics Program, Institute of Information Science, Taiwan International Graduate Program, Academia Sinica, Taipei, Taiwan
- 7Institute of Molecular and Cellular Biology, National Tsing Hua University, Hsinchu, Taiwan
- 8Department of Bioresources Engineering, National Institute of Technology, Okinawa College, Okinawa, Japan
- 9Kuroshio Biological Research Foundation, Kochi, Japan
- 10Okinawa Churashima Foundation Research Center, Okinawa, Japan
The Kuroshio Current (KC) is one of the fastest water currents in the world, running through the western boundary of the North Pacific Ocean. The KC strongly influences the regional hydroclimate by creating temperature, salinity, and pH gradients from tropical to subtropical and temperate zones, including regions with rich coral reef habitats. Microbial community composition of corals is influenced by various environmental factors, including salinity, pH, and geographical location. However, to date, it is unclear how coral-associated microbial communities would respond to the same water current running through different locations with a time lag. Therefore, we hypothesized that the locations subjected to the KC at higher latitudes experience similar but sequential lag in environmental variability compared to those at lower latitudes, and thus the coral communities of both will respond similarly, but at different times. In this year-long study, we compared the bacterial communities of Acropora muricata at Taiwan, Okinawa, and Kochi subjected to the KC. We found that site-specific conditions and site latitude may have stronger effects on bacterial composition and dynamics than a water current. Consequently, we suggest that latitude largely determines the temperature tolerance range of coral microbiota. Additionally, among the dominant coral associated bacteria, Endozoicomonas from A. muricata and Stylophora pistillata forms distinct phylogroups, while Acinetobacter is more likely a host generalist.
Introduction
Coral associated microbes, together with their coral host, form an integrated holobiont, and have diverse interactions with their host and maintain coral holobiont function, such as nutrient acquisition and coral’s health (Rosenberg et al., 2007). With respect to the coral-associated microbial community, Gammaproteobacteria is dominant (Bourne et al., 2013), and the putative symbiontEndozoicomonas-like group in particular has received notable attention for its potential role in coral fitness (Meyer et al., 2014; Morrow et al., 2015). Based on genomics evidence, Endozoicomonas is proposed being beneficial to coral host by modulating coral metabolisms to prevent mitochondrial dysfunction (Ding et al., 2016), and furthermore by participating in coral-associated carbohydrate cycling and protein provision (Neave et al., 2017). The relative abundance of coral microbiome varies depending on different factors, such as temperature (Shiu et al., 2017) and coral species (Neave et al., 2016). Our recent report investigating the same coral species from different locations around Taiwan further demonstrated that the coral-associated microbiome was dominated by different bacterial taxa (Yang et al., 2017), indicating local environmental factors playing an important role affecting microbial composition in corals.
Coral-associated microbial communities were reportedly influenced by environmental factors such as seawater temperature, pH, salinity, and nutrients (Guppy and Bythell, 2006; Klaus et al., 2007; Hong et al., 2009; Littman et al., 2009), all of which are largely affected by ocean currents. In fact, ocean currents transport a massive amount of waters (of distinct properties) and nutrients across a long distance and even different climate zones, resulting in latitudinal gradients of water parameters (Qu et al., 2018). Furthermore, the change in current speed and current structure also contributed to the variation of oceanic nutrient flux (Guo et al., 2012). Along the Western Australian coast, the Leeuwin Current has been proposed to explain the conserved microbial community in corals at geographically distant locations (Ceh et al., 2011). This report thus aroused the curiosity about to what extent the coral microbial communities in different climate zones are affected by the same ocean current that results in differential temperature, pH, and salinity gradients.
The Kuroshio Current (KC) is a strong northward current that runs through the western boundary of the North Pacific Ocean, transporting heat, salt, and water from low to high latitude regions. The KC is warm, low in nutrients, has a small plankton population and a high flow speed (roughly two to four knots). It passes through three climate zones, from tropical (the Philippines and southern Taiwan) to subtropical (northern Taiwan and Okinawa, Japan), and finally to the temperate zone (mainland Japan), resulting in temperatures of 15–29°C (Abbot et al., 2003) and a pH of 7.8–8.5 (Dai, 1991; Chen et al., 1995) at shallow depth above 300 m. The KC is analogous to a high-speed “freeway” that moves marine organisms from low to high latitudes in the western Pacific Ocean. Warming temperatures are associated with a poleward coral expansion (Yamano et al., 2011), and the coral reefs around Taiwan and Japan are therefore implicitly linked by the warm KC water (Fujiwara et al., 2000; Chen and Shashank, 2009).
Given its large latitudinal distance and wide temperature and pH gradients, the KC provides a unique opportunity to investigate the relationship between environmental factors driven by the current and their impacts on coral-associated microbial communities. We hypothesized that the locations subjected to the KC at higher latitudes experience similar but delayed environmental conditions compared to those at lower latitudes, and thus the coral communities of both respond similarly, but at different times.
To test this hypothesis, simultaneous observation and sampling (of the same coral species) were performed at different locations. Although the locational variations in soft coral-associated microbial composition among six sites subjected to the KC (Woo et al., 2017) were reported, there is still a knowledge gap that can be fulfilled by repetitive sampling for investigating microbial dynamics in corals under the same seasonal conditions. In addition, coral-associated bacteria can be coral species- or colony-specific (Hernandez-Agreda et al., 2018), making it difficult to compare observations obtained from different coral species. Therefore, in this study, we chose Acropora muricata (Linné, 1789) as our monitoring coral species to investigate the impacts of KC temperature and pH on coral-associated microbial communities across different climate zones. The sampling was performed bimonthly throughout 1 year in coastal areas of southern Taiwan and Okinawa and Kochi, Japan, which are all subjected to the KC, and their reefs are all dominated by A. muricata. A. muricata-associated microbiota is reportedly influenced by the environment (Casey et al., 2015). Therefore, we further explored the abundance variation and dynamics of Endozoicomonas and other genera in A. muricata in different climate zones but subjected to the same water current.
Materials and Methods
Sample Collection and Measurement of Environmental Parameters
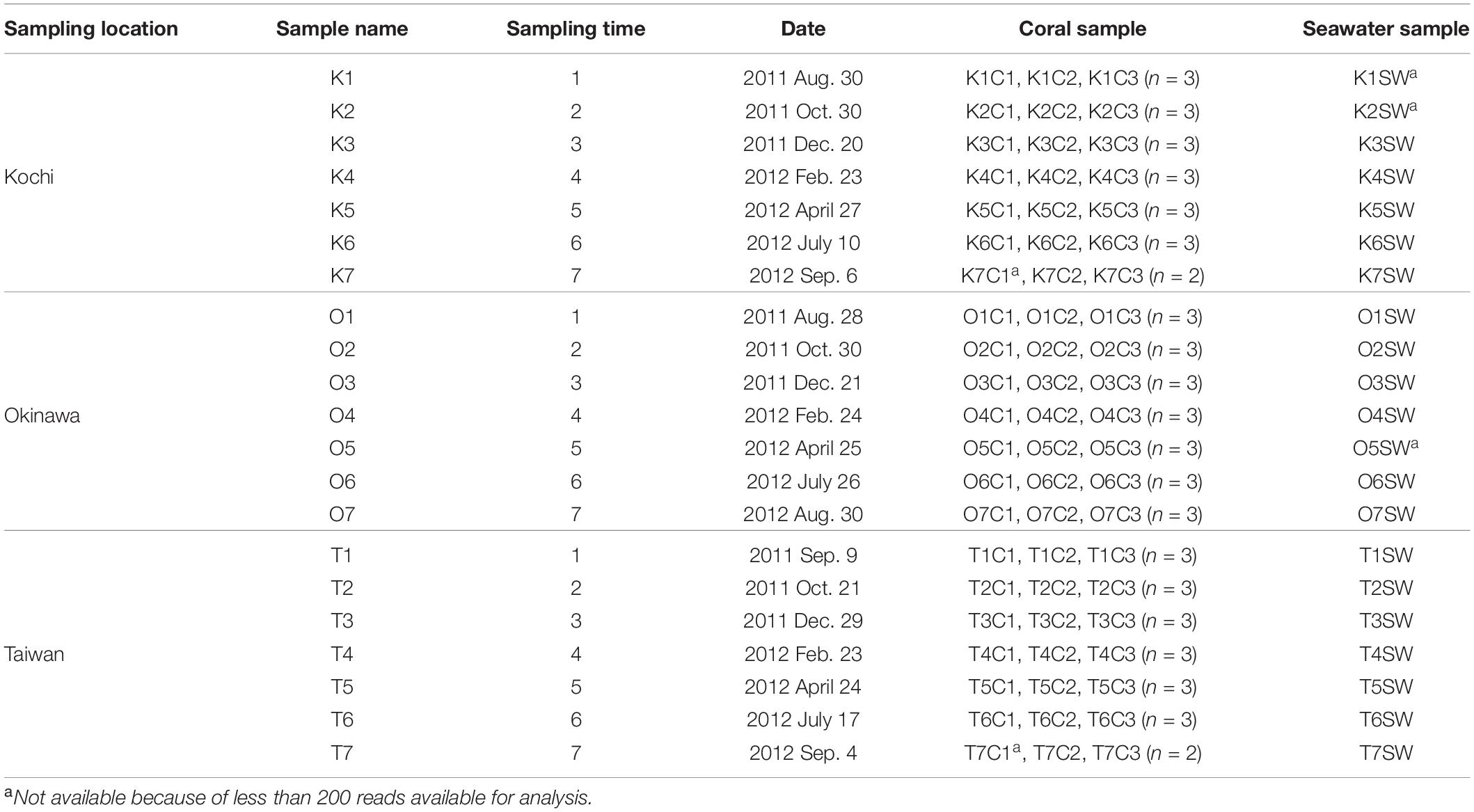
A total of 189 A. muricata coral and 21 seawater samples were collected in Kochi (32°46′42.95″N, 132°43′56.06″E), Okinawa (26°41′39.1″N, 127°52′31.8″E), and Taiwan (21°56′58.3″N, 120°45′11.9″E) from August 2011 to September 2012 (Table 1). During the sampling period, a typhoon occurred 1 month earlier than sampling O6, and shortly before O7, T1, and T7. At each site, three branches from three distinct colonies were randomly selected to represent biological replicates (n = 9) for the local ecology of A. muricata. The coral branches were fixed with 95% ethanol after sampling. Seawater (1 L) was collected and its parameters (including pH, salinity, and temperature) were measured at the time of sampling. Seawater was filtered with a 0.2 μm membrane (0.2 μm pore size; Adventec, Japan) to collect microbial particulate and stored at 4°C until DNA extraction.
DNA Extraction, Sequencing, and Processing
Coral and seawater samples were sent to the laboratory at Academia Sinica, Taiwan, for DNA extraction within 1 week of collection. Microbial DNA of samples was extracted via the cetyltrimethylammonium bromide (CTAB) method (Wilson, 2001). The hypervariable V1–V2 regions of bacterial 16S rRNA genes were amplified by polymerase chain reaction (PCR) using universal primers 27F (F: forward) and 341R (R: reverse), both of which were tagged with 4-base barcodes at their 5’-end as described previously (Chen et al., 2011). Amplified DNA was quantified by Qubit Fluorometer (Thermo Fisher Scientific, United States) and pooled in equal amounts. Multiplexed sequencing was performed using the Genome Sequencer FLX Titanium System (Roche 454 Life Sciences, Branford, CT, United States) at Mission Biotech (Taipei, Taiwan).
The raw sequencing reads were sorted into different samples according to the barcodes using an in-house script, which also removed the barcode and primer sequences. Reads (1) of lengths shorter than 280 bp or longer than 350 bp, (2) with an average quality score less than 20, (3) with a homopolymer longer than 8 bp, or (4) containing ambiguous bases were quality-filtered using Mothur (Schloss et al., 2009). Chimeric reads predicted by UCHIME (Edgar et al., 2011) implemented in USEARCH (v8.0.1623; reference mode, rdp_gold database, mindiv of 3) (Edgar, 2010) were also eliminated. Non-chimeric reads (including borderline cases by UCHIME) were retained for further analysis. Reads of coral branches from the same colony were pooled as one sample, resulting in a total of 84 samples. The read processing code is available in Supplementary Data.
For operational taxonomic unit (OTU) analysis, quality-filtered and non-chimeric reads were analyzed on a per-sample basis with UPARSE (Edgar, 2013); the chimera removal step was excluded, as it had been performed in previous quality filtering steps. The OTU approach has been shown to have comparable validity of results to the more stringent amplicon sequence variants (i.e., ASV) methods (Glassman and Martiny, 2018). OTUs were generated at a 97% identity threshold. Each OTU was searched (with global alignment) using USEARCH against the Ribosomal Database Project’s database (training set 16 and release 11.5) to find the corresponding taxonomy of the best hit. OTUs with no hit or weak hits (i.e., the average of % identity and % query coverage <93) were excluded. After filtering OTUs of the chloroplast, which occupied a mean (± SD) of 1.35% (± 1.65%) in coral and 27.67% (± 25.41%) in seawater samples, samples containing ≥ 200 reads were used for downstream analysis.
Statistical Analysis
All the statistical analyses were performed in R software1 unless stated otherwise. Community data were handled with the R package phyloseq (McMurdie and Holmes, 2013), by which alpha diversity indices were estimated. Non-parametric tests were performed to compare alpha diversities between groups; Kruskal-Wallis test was used for three-group and Wilcoxon test for two-group comparison. Hierarchical clustering with average linkage was performed and visualized using the R package pheatmap (Kolde, 2012). At the genus level, samples were clustered based on the Euclidean distance of a base-10 logarithmic transformation of percent relative abundance, while taxa were based on the Bray–Curtis distance of non-transformed percent relative abundance. During the distance calculation, a pseudo-count of 1 × 10–8 was added to each taxon (Costea et al., 2014), and taxa with unknown taxonomy or <0.1% average relative abundance were classified as “Others.” The distance matrix was calculated using the R package vegan (Dixon, 2003). The analysis of similarities (ANOSIM) was performed to statistically test whether there is a significant difference in community composition between two or more groups.
OTU Analysis of Acinetobacter and Endozoicomonas
Endozoicomonas and Acinetobacter OTUs were originally defined on a per-sample basis and were difficult to pool directly by taxonomy because most species of these two genera are unclassified. Therefore, the cross-sample OTUs were analyzed. After extracting amplicon reads of Endozoicomonas and Acinetobacter OTUs, reads of each genus were clustered at 97% identity by USEARCH (with –cluster_smallmem), and the abundance profiles were constructed according to the sampling source information retained on each read.
Correlation (Pearson) heatmaps were generated in R with the corrplot package (Wei and Simko, 2017) using the relative abundance and presence-absence profiles of Endozoicomonas and Acinetobacter OTUs across all samples. Correlation significance was tested at 95% confidence intervals.
Beta diversity was assessed and visualized using non-metric multidimensional scaling (nMDS) on Primer 6 (PRIMER-E, Lutton, Lvybridge, United Kingdom) and fitted with environmental factors through constrained correspondence analysis (CCA) on the R package vegan. The top-20 Endozoicomonas and Acinetobacter OTUs (in abundance) were used for phylogenetic analysis, using the maximum likelihood method based on the Jukes-Cantor model (Jukes and Cantor, 1969) to obtain the tree. Phylogenetic analyses were conducted in MEGA7 (Kumar et al., 2016).
Data Availability
All sequence data generated in this study have been deposited in NCBI under the Bioproject accession PRJNA636267.
Results
Comparison of Environmental Parameters
Salinity, pH, and temperature were measured from concurrently sampled seawater as environmental parameters. Kochi, Okinawa, and Taiwan were the sampling sites from north to south. The seawater salinity in Kochi exhibited large seasonal variation (showing a statistical variance of 42.2, defined as the average of the squares of difference between the data points and the mean) and remained relatively stable throughout the year in Okinawa (showing a variance of 2.2) and Taiwan (showing a variance of 6.4) (Supplementary Figure S1A). The pH values varied widely during summertime in all three sites, with particularly high fluctuations in Okinawa (from sampling time 6, 7, and 1; Supplementary Figure S1B). A distinct time-lag was observed for temperature; in Taiwan, the temperature began to rise at the third sampling, which was, however, observed in Okinawa and Kochi at the fourth sampling (Supplementary Figure S1C). In general, Okinawa and Taiwan shared similar salinity, pH, and temperature values. Kochi showed large fluctuations in salinity and pH, and consistently had lower temperatures than the other two sites.
Bacterial Diversity and Composition
Hierarchical clustering revealed that bacterial composition (at the genus level) was distinct in coral and seawater samples (ANOSIM R = 0.41, p = 0.01) (Figure 1). The bacterial Shannon diversities in coral were significantly higher in Taiwan than the other two sites (p = 0.0012 by Kruskal-Wallis); seawater samples were also higher in Taiwan but not significant (p = 0.84 by Kruskal-Wallis). Regarding each location, although with only a marginal significance, the bacterial Shannon diversities were higher in seawater than coral samples in Kochi (p = 0.17 by Wilcoxon) and Okinawa (p = 0.049 by Wilcoxon) (Supplementary Figure S2), while higher in coral than seawater samples in Taiwan (p = 0.57 by Wilcoxon) (Supplementary Figure S2), suggesting that bacteria at the low latitude site thrived better in corals than seawater. Results of the Inverse Simpson, richness, and Shannon analyses suggested that the coral bacterial diversities did not show any time-lag trend among the three sampling sites (Supplementary Figure S3). The highest diversity for each site was found during the fifth sampling in Kochi and Taiwan, and the seventh sampling in Okinawa (Supplementary Figure S3).
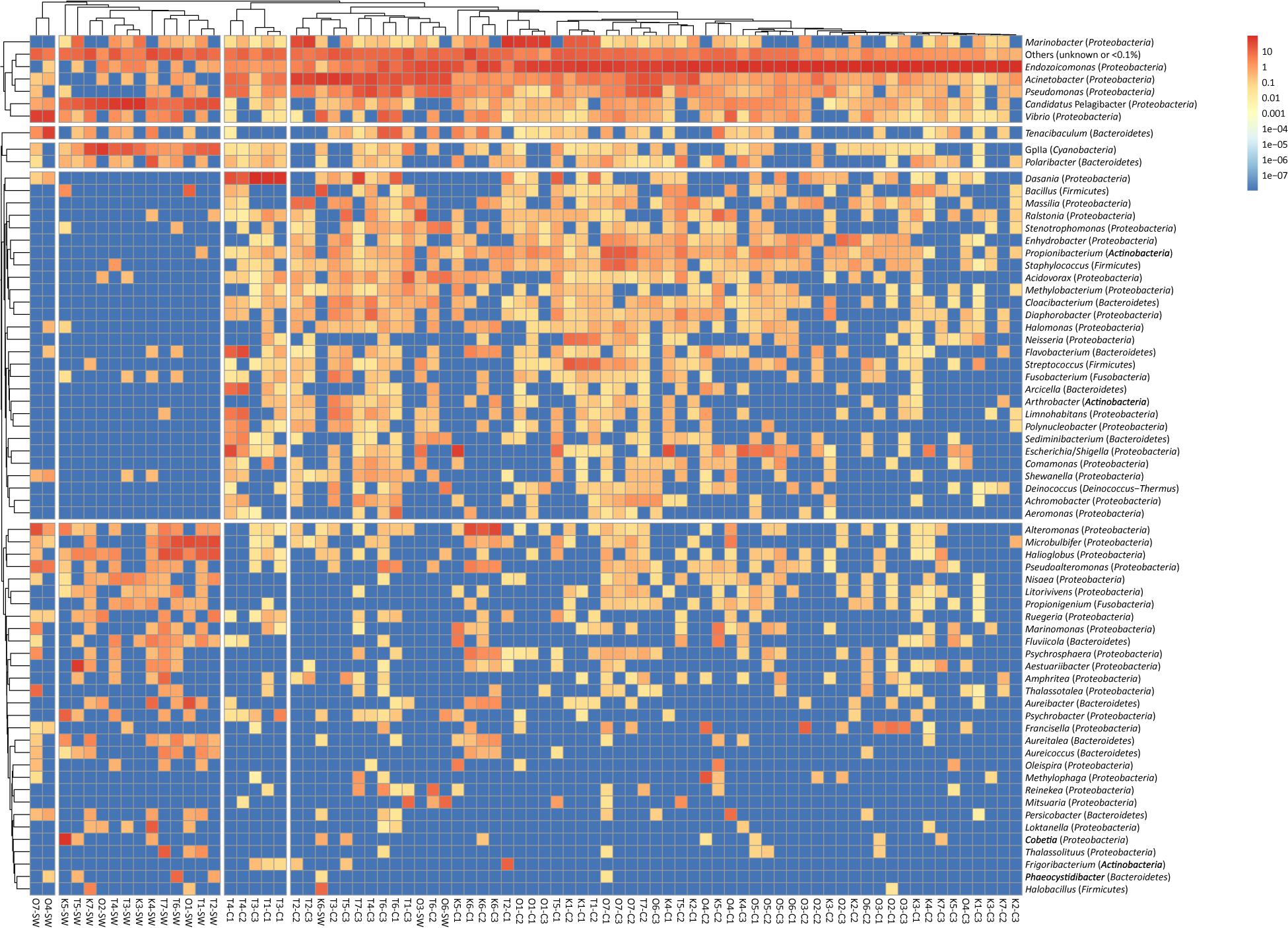
Figure 1. Clustering analysis of bacterial genera in Acropora muricata and seawater from Kochi, Okinawa, and Taiwan. A total of 68 genera remained in the analysis after excluding genera of unknown taxonomy or < 0.1% relative abundance. Trees were cut at four and five groups in columns and rows, respectively. The color scale indicates the relative abundance (percentage). Columns are clustered by Bray–Curtis distance and rows by Euclidean distance of their abundance profile.
The nMDS analysis showed that the coral bacterial compositions in Kochi and Okinawa were relatively stable compared to that of Taiwan (Figure 2). High variation in bacterial composition was found in coral samples in Taiwan during autumn and winter (Figure 2A), while in Kochi similar results were identified during spring and summer (Figure 2B). In seawater samples, the bacterial compositions in Taiwan were similar to those in Kochi (ANOSIM R = 0.22, p = 0.059), but significantly different from those in Okinawa (ANOSIM R = 0.26, p < 0.01) (Supplementary Figure S4). No significant difference was found between the compositions of Kochi and Okinawa (ANOSIM R = 0.04, p = 0.33).
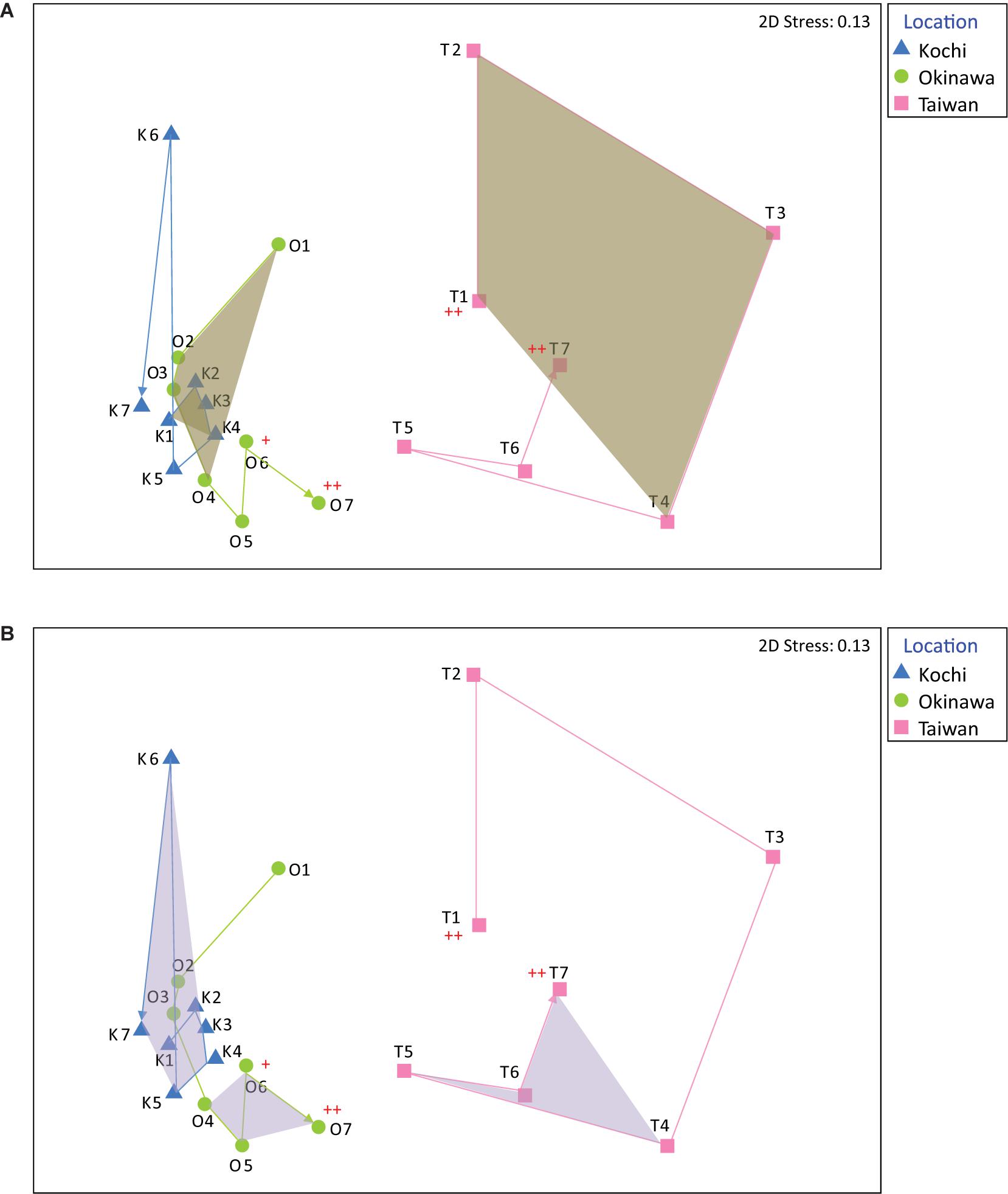
Figure 2. Non-metric multidimensional scaling (nMDS) analysis of bacterial genera in Acropora muricata from Kochi, Okinawa, and Taiwan during (A) autumn and winter (samples K1-K4, O1-O4, and T1-T4) and during (B) spring and summer (samples K4-K7, O4-O7, and T4-T7). A cross indicates sampling after typhoon about 1 month and two crosses indicate sampling just after the typhoon.
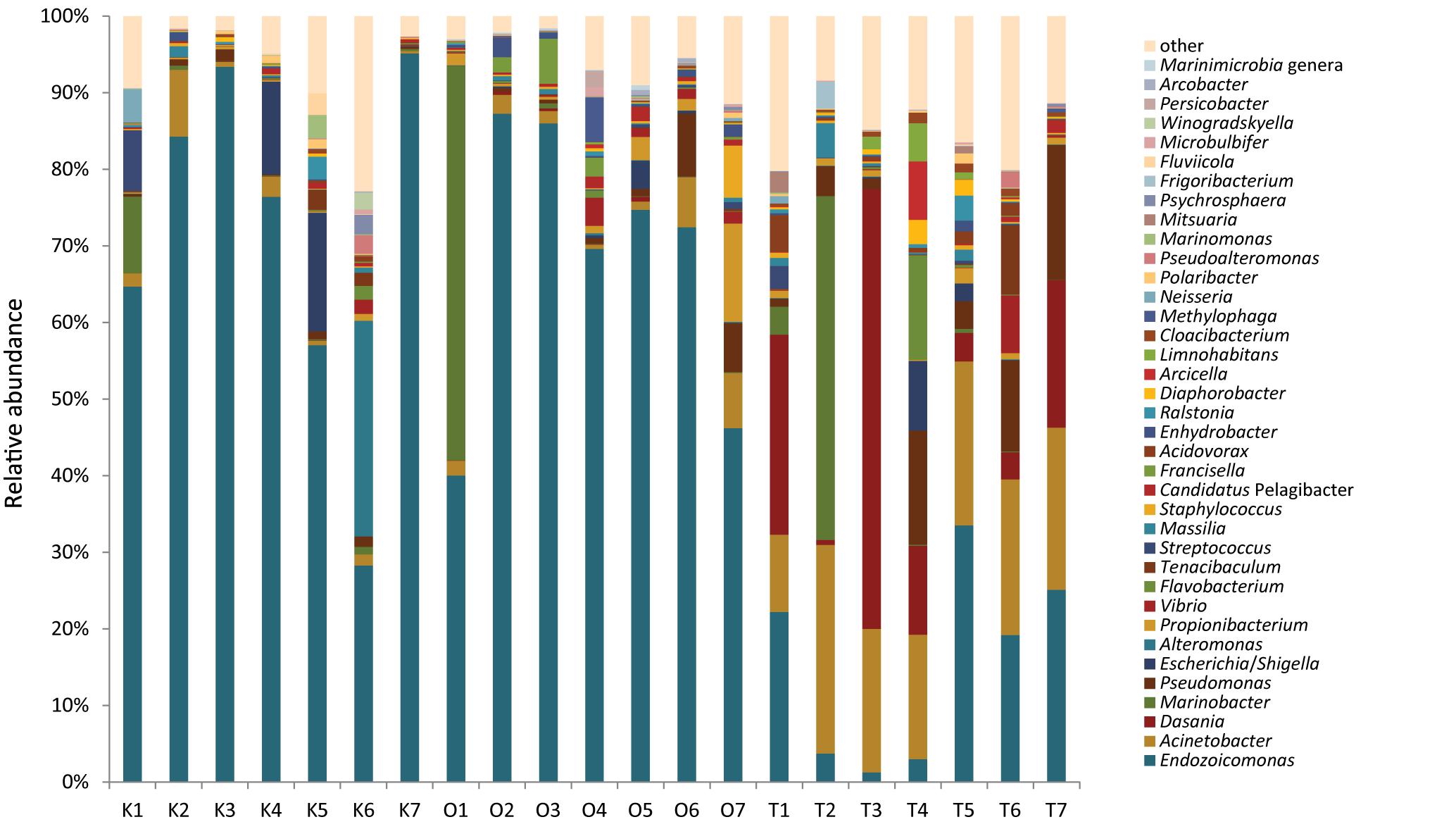
Among the 20 most abundant bacterial genera, corals in Taiwan harbored a greater diversity than the other two sites (Figure 3). Endozoicomonas was consistently dominant in corals from Kochi and Okinawa but not Taiwan, where it was particularly low in abundance during autumn and winter months (T1–T4 in Figure 3). Inspired by our recent study which reported that Endozoicomonas acroporae is an Acropora-associated dimethylsulfoniopropionate (DMSP) degrader (Tandon et al., 2020), we further investigated how many of our Endozoiocmonas amplicons were associated with E. acroporae. By analyzing the best BLASTn match, we observed 6.35 and 4.36% relative abundance of E. acroporae in Okinawa and Taiwan, respectively, but only 0.3% in Kochi (Supplementary Figure S5).
The CCA results showed that temperature and salinity had larger influences on coral bacterial composition than pH when considering all samples (Supplementary Figure S6A). In the site-specific analysis, salinity and temperature had the largest effect on Kochi (Supplementary Figures S1A,C) and were the main factors contributing to its coral bacterial community composition (Supplementary Figure S6B). In Kochi, the abundance of many genera was negatively correlated with temperature, indicating that they acclimated to the lower water temperature (<20°C). In Okinawa, temperature and pH were the major factors influencing the coral bacterial community (Supplementary Figures S1, S6C). In Taiwan, most genera were positively correlated with temperature, indicating an acclimation to the warmer environment (Supplementary Figure S6D). As a result, temperature, pH, and salinity had site-specific and differential effects, suggesting that ambient conditions have an important role in the coral-associated bacterial composition.
Endozoicomonas and Acinetobacter
Endozoicomonas and Acinetobacter were the most dominant genera in corals (Figure 3), so we further analyzed them to resolve their relationships to sampling times from all three sites in greater detail (see section “Materials and Methods”). Both Endozoicomonas and Acinetobacter abundance (log-transformed) demonstrated higher correlation values within sites than between sites, indicating that the compositional differences were site-specific (Figure 4A; ANOSIM R = 0.695, p = 0.0001 for Endozoicomonas; R = 0.416, p = 0.0001 for Acinetobacter), but the correlations diminished in Boolean transformation (i.e., presence and absence) (Figure 4B). Neither Endozoicomonas nor Acinetobacter demonstrated any time-lag phenomenon in OTU compositions across locations, indicating that the time-lag succession did not apply to dominant bacteria in corals along the KC. The site-specific CCA analysis revealed that temperature and salinity were the main factors affecting genera composition in both Kochi (Supplementary Figure S7A,D) and Okinawa (Supplementary Figures S7B,E), while temperature and pH were the major factors in Taiwan (Supplementary Figures S7C,F).
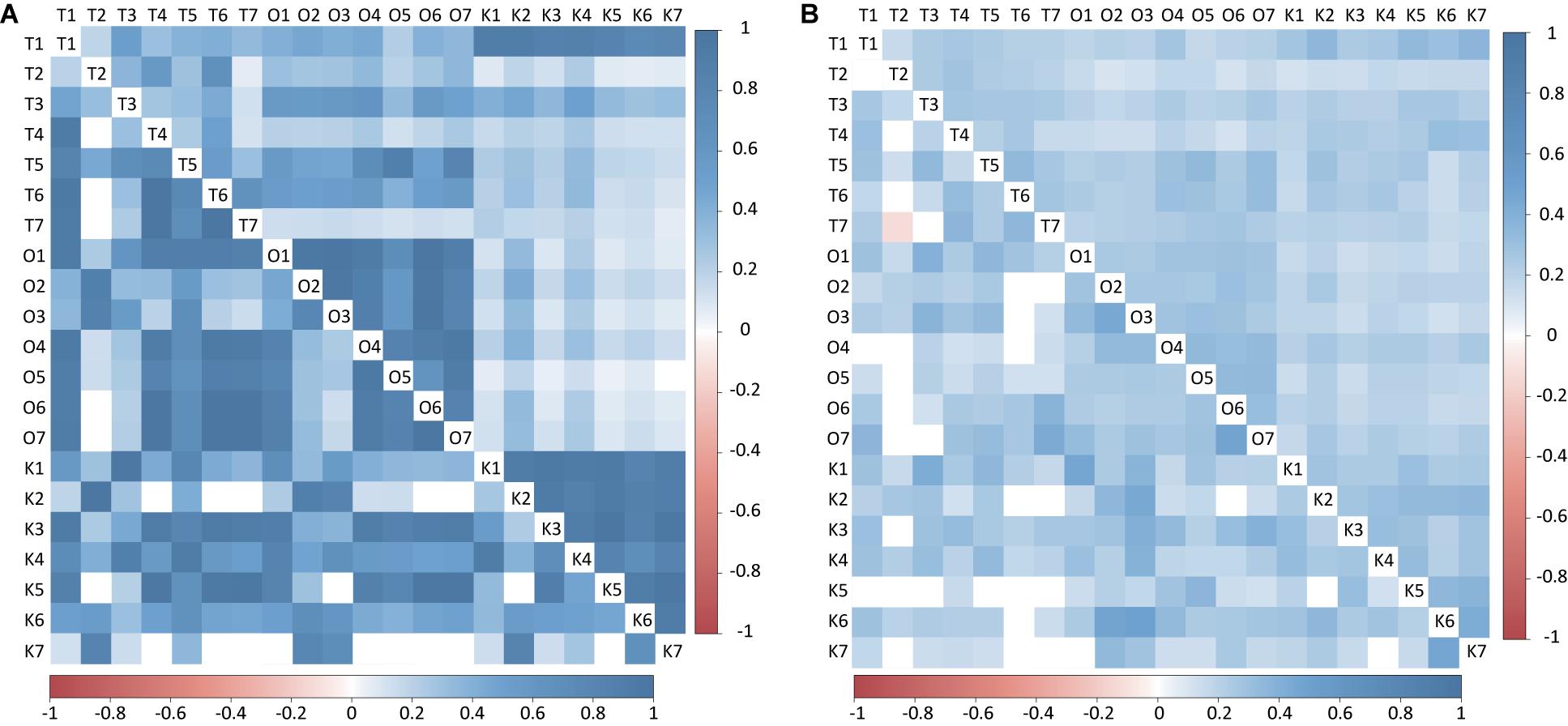
Figure 4. Heatmap of Pearson’s correlation between Endozoicomonas and Acinetobacter among different sampling time from Kochi, Okinawa, and Taiwan. (A) matrix with log transformation. (B) matrix with present/absent transformation. The values above the diagonal represented Endoizoicomonas abundance correlation values and below the diagonal represented Acinetobacter abundance correlation.
Lastly, the 20 most abundant OTUs of Endozoicomonas and Acinetobacter were phylogenetically analyzed with their counterparts from a different coral species, Stylophora pistillata, obtained from our previous study (Yang et al., 2017), to verify the existence of host specificity. Results showed that Endozoicomonas from S. pistillata and A. muricata formed distinct phylogroups (Figure 5A), but Acinetobacter OTUs did not (Figure 5B), suggesting that Endozoicomonas has a higher host specificity than Acinetobacter.
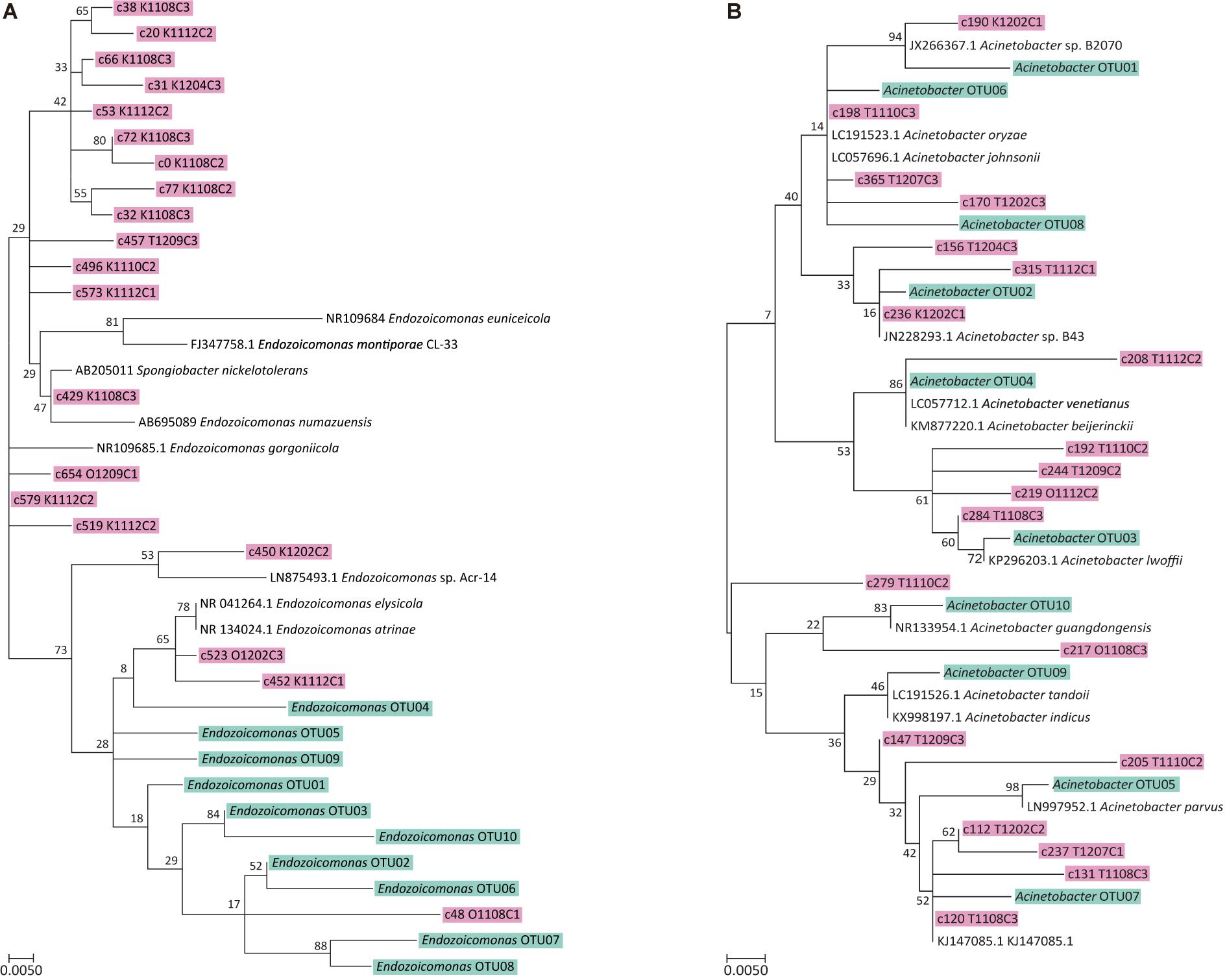
Figure 5. Maximum likelihood trees of top 20 OTUs Endozoicomonas and Acinetobacter. (A) ML tree of top 20 Endozoicomonas OTUs from this study (shown in pink) and top 10 Endozoicomonas OTUs from coral Stylophora pistillata in Yang et al. (2017) (shown in blue). (B) ML tree of top 20 Acinetobacter OTUs from this study (shown in pink) and top 10 Acinetobacter OTUs from coral S. pistillata in Yang et al. (2017) (shown in blue). The trees with the highest log likelihood (−1227.6999) and 1,000 bootstraps. The analysis involved 42 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 295 positions in the final dataset.
Discussion
Influences of Current and Location
Current is driven by gradients of temperature, pH, and salinity and was expected to result in time-lagged shifts in microbial composition. However, the present study did not find enough evidence to support the hypothesis that the microbiota of coral and seawater samples can be influenced by water current from low to high latitude. Along the path of the KC, the sampling sites were more or less influenced by its water (Chu, 1974; Andres et al., 2008; Kuroda et al., 2008), but the coral-associated bacterial composition exhibited significant site-specific differences and no obvious sign of time-lag succession. In addition, further comparisons between the two most dominant genera, Endozoicomonas and Acinetobacter, repeatedly found site-specific and season-independent correlations in abundance profiles without time-lag succession. These results collectively suggest that local conditions may have stronger effects on bacterial compositions and dynamics than does the KC.
The coral-associated bacterial community in Taiwan demonstrated higher seasonal variation than those of Okinawa and Kochi, plausibly attributable to the water flow velocity. Advection in currents has been suggested to influence microbial community assemblies (Doblin and Van Sebille, 2016), and increased water flow velocity has also been shown to modify the bacterial composition in the coral surface mucus and tissue layers (Lee et al., 2016). Compared to Kochi, the sampling sites in Taiwan and Okinawa had faster current flow because of their physical proximity to the KC mainstream (according to the Ocean Data Bank, Ministry of Science and Technology, Taiwan)2. Moreover, the site in Taiwan is affected not only by the KC but also waters from the South China Sea (Hu et al., 2000), which in combination may impact coral-associated bacterial compositions. Last but not least, extreme hydrodynamics (e.g., typhoons) can cause a shift in the coral ecosystem (White et al., 2017) and phytoplankton composition (Blanco et al., 2008), and lead to disease in corals (Bright et al., 2016) by disrupting local environments for coral holobionts. In Taiwan and Kochi, the first and last sampling (T1 and T7, K1 and K7) were similar in community composition (Figure 2), which was not observed in Okinawa (i.e., O1 and O7 were separated from each other). As O7 sampling was performed shortly after a typhoon, it was very likely that the observed microbial community has been substantially disrupted.
The coral microbiota is also associated with site latitude. Our previous study found that Stylophora pistillata-associated bacterial compositions differed between tropical and subtropical zones (Yang et al., 2017). A one-degree latitudinal difference was proposed to be the distribution boundary for similar bacterial compositions in the same coral (i.e., two corals of the same species separated by more than one degree would have different bacterial compositions) (Woo et al., 2017). Pollock et al. (2018) found that the coral microbiota has lower richness at higher latitudes, corresponding to the lower coral bacterial diversity in Okinawa and Kochi than Taiwan. The coral microbiota in Kochi and Okinawa shared similar compositions, but largely differed from those in Taiwan, suggesting that coral bacterial dynamics correspond with the climate zone. However, the geographic location is still attributable to this observation as they may differentially be influenced by the water currents from the South China Sea.
Several variation patterns associated with seasons in coral microbiota were discovered. Being a middle-latitude site, the coral microbiota in Okinawa highly overlapped with those in Kochi (the high-latitude site) during autumn and winter but became closer to those of Taiwan (the low-latitude site) during spring and summer. Also, compared to autumn and winter, the coral microbiota in Kochi largely fluctuated during spring and summer, while those in Taiwan demonstrated the opposite trend. The coral microbiota has been shown to tolerate a temperature range similar to ambient thermal history (Shiu et al., 2017). In this study, the Acropora-associated microbial composition in the low-latitude site (i.e., Taiwan) demonstrated a relatively small fluctuation during spring and summer; the counterparts in the high-latitude site (i.e., Kochi) demonstrated a relatively small fluctuation during autumn and winter. Therefore, the latitude largely determines the temperature tolerance range of coral microbiota. Although coral microbiota is slightly fluctuating from time to time, this phenomenon is escalated while the ambient temperature varies beyond the tolerance range.
Site and Host Specificities of Endozoicomonas and Acinetobacter
Endozoicomonas is a ubiquitous taxon in coral (Neave et al., 2016). Its high abundance has been suggested as an indicator of good coral health (Bourne et al., 2008). However, in the present study performed on healthy A. muricata, the abundance of Endozoicomonas varied across sampling sites. As our sampling was performed at inshore sites, the impact of terrestrial nutrient influx (e.g., P and N) on coral microbiota cannot be ignored (Vega Thurber et al., 2014).
It has been suggested that ubiquitously dominant taxa may be eurythermic environmental generalists or composed of multiple subtypes selected through different environmental factors (Yung et al., 2015). Although Endozoicomonas is common in corals, at low latitude the A. muricata-associated microbiota was simultaneously dominated by other genera, such as Acinetobacter, Dasania, and Pseudomonas. Acinetobacter was found particularly more in S. pistillata around Taiwan than was Endozoicomonas (Yang et al., 2017).
Among the Endozoicomonas species, Endozoicomonas amplicons associated with E. acroporae were detected at all three sites, and its abundance in Okinawa and Taiwan increased during the warm seasons (spring and summer). E. acroporae is a DMSP degrader (Tandon et al., 2018, 2020), and DMSP and its breakdown products can quickly scavenge hydroxyl radicals and mitigate oxidative stress to protect corals from bleaching (Sunda et al., 2002; Lesser, 2006). However, the coral microbiota in Taiwan was not overwhelmed by Endozoicomonas, and the second-most abundant genus, Acinetobacter, is proposed to be a DMSP degrader performing a similar protective function for A. muricata (Raina et al., 2010). Acinetobacter was abundant in different stony corals in various regions, including the tropics (Littman et al., 2009; McKew et al., 2012; Li et al., 2014; Leite et al., 2018), and its role is still elusive as being reportedly beneficial (Shnit-Orland and Kushmaro, 2009) and detrimental (Sweet et al., 2013) to corals. Given the observation that Acinetobacter abundance was higher and more stable in low-latitude (Taiwan) than high-latitude (Okinawa and Kochi) sites, the question of whether Acinetobacter could edge Endozoicomonas out of the microbial assemblage dominance in Acropora warrants further study, especially with the rising seawater temperature in the age of global warming.
Conclusion
This year-long study compared the bacterial communities of A. muricata at three sites subjected to the KC at different climate zones (Taiwan, Okinawa, and Kochi), and found an inverse correlation between coral bacterial diversity and site latitude. The bacterial composition in coral has no obvious time-lag succession along with the KC, but a strong correlation with ambient conditions, indicating that environmental parameters are more influential to the indigenous bacterial community. Endozoicomonas were substantially detected in temperate and subtropical sites but outnumbered by Acinetobacter in tropics, indicating Acinetobacter having a lower host specificity and better adaptability to the warmer environment than Endozoicomonas.
Manipulation of the coral microbiome was recently proposed as a promising means to improve coral health by strengthening coral resistance and resilience to environmental stress (Damjanovic et al., 2017; Sweet et al., 2017), and Endozoicomonas has been proposed as an environmental probiotic for this purpose (Peixoto et al., 2017). According to the site-specific distribution of Endozoicomonas, the microbial manipulation in coral would be more effective if we factor in local environmental parameters and host species. Finally, the dominance of Acinetobacter in tropical corals suggests its potential role of being another environmental probiotic candidate.
Data Availability Statement
The datasets presented in this study can be found in online repositories, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA636267.
Author Contributions
S-LT designed the study and edited the draft manuscript. S-HY, C-HT, and KT performed data analysis and manuscript writing. H-PL, P-WC, H-JC, J-HS, H-CL, NI, TM, and HY performed sampling. All authors read and approved the final version of the manuscript.
Funding
We greatly thank Biodiversity Research Center, Academia Sinica for its funding from Thematic Grant AS-100-TP2-A02-3. S-HY is partially funded by the Ministry of Education, Taiwan, R.O.C., under Grant number MOST 109-2621-B-002-005-MY3. C-HT is partially supported by the Economic Development Bureau, Taichung City Government, Taiwan, R.O.C., under Grant number 107SBIR-23.
Conflict of Interest
C-HT and H-CL were employed by the companies Germark Biotechnology Co. Ltd. and WellGenetics Inc., respectively.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly thank Okinawa Churaumi Aquarium for coral sampling in Okinawa. We would also like to thank Noah Last of Third Draft Editing for his English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.569107/full#supplementary-material
Footnotes
References
Abbot, P., Celuzza, S., Dyer, I., Gomes, B., Fulford, J., and Lynch, J. (2003). Effects of east china sea shallow-water environment on acoustic propagation. IEEE J. Ocean. Eng. 28, 192–211. doi: 10.1109/JOE.2003.811901
Andres, M., Park, J. H., Wimbush, M., Zhu, X. H., Chang, K. I., and Ichikawa, H. (2008). Study of the kuroshio/ryukyu current system based on satellite-altimeter and in situ measurements. J. Oceanography 64, 937–950. doi: 10.1007/s10872-008-0077-2
Blanco, A. C., Nadaoka, K., and Yamamoto, T. (2008). Planktonic and benthic microalgal community composition as indicators of terrestrial influence on a fringing reef in Ishigaki Island, Southwest Japan. Mar. Environ. Res. 66, 520–535. doi: 10.1016/j.marenvres.2008.08.005
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D. G., Dennis, P. G., Uthicke, S., Soo, R. M., Tyson, G. W., and Webster, N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. IEEE J. Ocean. Eng. 7, 1452–1458. doi: 10.1038/ismej.2012.172
Bright, A. J., Rogers, C. S., Brandt, M. E., Muller, E., and Smith, T. B. (2016). Disease prevalence and snail predation associated with swell-generated damage on the threatened coral, Acropora palmata (Lamarck). Front. Mar. Sci. 3:77. doi: 10.3389/fmars.2016.00077
Casey, J. M., Connolly, S. R., and Ainsworth, T. D. (2015). Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci. Rep. 5:11903. doi: 10.1038/srep11903
Ceh, J., Van Keulen, M., and Bourne, D. G. (2011). Coral-associated bacterial communities on ningaloo reef, Western Australia. FEMS Microbiol. Ecol. 75, 134–144. doi: 10.1111/j.1574-6941.2010.00986.x
Chen, C. A., and Shashank, K. (2009). Taiwan as a connective stepping-stone in the kuroshio traiangle and the conservation of coral ecosystems under the impacts of climate change. Kuroshio Sci. 3, 15–22.
Chen, C. P., Tseng, C. H., Chen, C. A., and Tang, S. L. (2011). The dynamics of microbial partnerships in the coral isopora palifera. ISME J. 5, 728–740. doi: 10.1038/ismej.2010.151
Chen, C. T. A., Ruo, R., Paid, S. C., Liu, C. T., and Wong, G. T. F. (1995). Exchange of water masses between the east china sea and the kuroshio off Northeastern Taiwan. Continent. Shelf Res. 15, 19–39. doi: 10.1016/0278-4343(93)E0001-O
Chu, T. Y. (1974). The fluctuations of the kuroshio current in the eastern sea area of Taiwan. Acta Oceanogr. Taiwan. 4, 1–12.
Costea, P., Zeller, G., Sunagawa, S., and Bork, P. (2014). A fair comparison. Nat. Methods 11:359. doi: 10.1038/nmeth.2897
Dai, C. F. (1991). Reef environment and coral fauna of Southern Taiwan. Atoll Res. Bull. 345, 1–24. doi: 10.5479/si.00775630.354.1
Damjanovic, K., Blackall, L. L., Webster, N. S., and van Oppen, M. J. H. (2017). The contribution of microbial biotechnology to mitigating coral reef degradation. Microb. Biotechnol. 10, 1236–1243. doi: 10.1111/1751-7915.12769
Ding, J. Y., Shiu, J. H., Chen, W. M., Chiang, Y. R., and Tang, S. L. (2016). Genomic insight into the host–endosymbiont relationship of Endozoicomonas montiporae CL-33(T) with its coral host. Front. Microbiol. 7:251. doi: 10.3389/fmicb.2016.00251
Dixon, P. (2003). Vegan, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Doblin, M. A., and Van Sebille, E. (2016). Drift in ocean currents impacts intergenerational microbial exposure to temperature. Proc. Natl. Acad. Sci. U.S.A. 113, 5700–5705. doi: 10.1073/pnas.1521093113
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than blast. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). Uparse: highly accurate otu sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). Uchime improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fujiwara, S., Shibuno, T., Mito, K., Nakai, T., Sasaki, Y., Dai, C. F., et al. (2000). Status of coral reefs of East and North asia: China, Japan and Taiwan. Status Coral Reefs World 2000, 131–140.
Glassman, S. I., and Martiny, J. B. (2018). Broadscale ecological patterns are robust to use of exact sequence variants versus operational taxonomic units. mSphere 3:e00148-18. doi: 10.1128/mSphere.00148-18
Guo, X., Zhu, X. H., Wu, Q. S., and Huang, D. (2012). The kuroshio nutrient stream and its temporal variation in the East China Sea. J. Geophys. Res. 117:C01026. doi: 10.1029/2011JC007292
Guppy, R., and Bythell, J. C. (2006). Environmental effects on bacterial diversity in the surface mucus layer of the reef coral montastraea faveolata. Mar. Ecol. Prog. Ser. 328, 133–142. doi: 10.3354/meps328133
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., Herrera, C., and Ainsworth, T. D. (2018). Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9:e00812-18. doi: 10.1128/mBio.00812-18
Hong, M. J., Yu, Y. T., Chen, C. A., Chiang, P. W., and Tang, S. L. (2009). Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl. Environ. Microbiol. 75, 7797–7806. doi: 10.1128/AEM.01418-09
Hu, J., Kawamura, H., Hong, H., and Qi, Y. (2000). A review on the currents in the south china sea: seasonal circulation, south China sea warm current and kuroshio intrusion. J. Oceanography 56, 607–624. doi: 10.1023/A:1011117531252
Jukes, T., and Cantor, C. (1969). Evolution of Protein Molecules. New York, NY: Academic Press. doi: 10.1016/B978-1-4832-3211-9.50009-7
Klaus, J. S., Janse, I., Heikoop, J. M., Sanford, R. A., and Fouke, B. W. (2007). Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9, 1291–1305. doi: 10.1111/j.1462-2920.2007.01249.x
Kumar, S., Stecher, G., and Tamura, K. (2016). Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuroda, H., Shimizu, M., Hirota, Y., Ambe, D., and Akiyama, H. (2008). Surface current and vertical thermal structure on the continental slope in Tosa Bay. J. Oceanogr. 64, 81–91. doi: 10.1007/s10872-008-0006-4
Lee, S., Davy, S. K., Tang, S. L., and Kench, P. S. (2016). Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front. Microbiol. 7:371. doi: 10.3389/fmicb.2016.00371
Leite, D. C., Salles, J. F., Calderon, E. N., Castro, C. B., Bianchini, A., Marques, J. A., et al. (2018). Coral bacterial-core abundance and network complexity as proxies for anthropogenic pollution. Front. Microbiol. 9:833. doi: 10.3389/fmicb.2018.00833
Lesser, M. P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Li, J., Chen, Q., Long, L. J., Dong, J. D., Yang, J., and Zhang, S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral porites lutea during different seasons. Sci. Rep. 4, 1–8. doi: 10.1038/srep07320
Linné, C. V. (1789). Systema Naturae Per Regna Tria Naturae: Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis. Lugduni: Apud JB Delamolliere. doi: 10.5962/bhl.title.36932
Littman, R. A., Willis, B. L., Pfeffer, C., and Bourne, D. G. (2009). Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the great barrier reef. FEMS Microbiol. Ecol. 68, 152–163. doi: 10.1111/j.1574-6941.2009.00666.x
McKew, B., Dumbrell, A., Daud, S., Hepburn, L., Thorpe, E., Mogensen, L., et al. (2012). Characterization of geographically distinct bacterial communities associated with coral mucus produced by Acropora spp. and Porites spp. Appl. Environ. Microbiol. 78, 5229–5237. doi: 10.1128/AEM.07764-11
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meyer, J. L., Paul, V. J., and Teplitski, M. (2014). Community shifts in the surface microbiomes of the coral porites astreoides with unusual lesions. PLoS One 9:e100316. doi: 10.1371/journal.pone.0100316
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural Volcanic Co 2 seeps reveal future trajectories for host–microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Neave, M. J., Michell, C. T., Apprill, A., and Voolstra, C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7:40579. doi: 10.1038/srep40579
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2016). Differential specificity between closely related corals and abundant endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Peixoto, R. S., Rosado, P. M., Leite, D. C., Rosado, A. S., and Bourne, D. G. (2017). Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 7:341. doi: 10.3389/fmicb.2017.00341
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 1–13. doi: 10.1038/s41467-018-07275-x
Qu, B., Song, J., Yuan, H., Li, X., and Li, N. (2018). Carbon chemistry in the mainstream of kuroshio current in eastern taiwan and its transport of carbon into the East China Sea Shelf. Sustainability 10:791. doi: 10.3390/su10030791
Raina, J. B., Dinsdale, E. A., Willis, B. L., and Bourne, D. G. (2010). Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18, 101–108. doi: 10.1016/j.tim.2009.12.002
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shiu, J. H., Keshavmurthy, S., Chiang, P. W., Chen, H. J., Lou, S. P., Tseng, C. H., et al. (2017). Dynamics of coral-associated bacterial communities acclimated to temperature stress based on recent thermal history. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-14927-3
Shnit-Orland, M., and Kushmaro, A. (2009). Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol. Ecol. 67, 371–380. doi: 10.1111/j.1574-6941.2008.00644.x
Sunda, W., Kieber, D., Kiene, R., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320. doi: 10.1038/nature00851
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral stephanocoenia intersepta. PLoS One 8:e62580. doi: 10.1371/journal.pone.0062580
Sweet, M. J., Ramsey, A., and Bulling, M. T. (2017). Designer reefs and coral probiotics: great concepts but are they good practice? Biodiversity 18, 19–22.
Tandon, K., Chiang, P. W., Chen, W. M., and Tang, S. L. (2018). Draft genome sequence of Endozoicomonas acroporae strain Acr-14T, isolated from acropora coral. Genome Announc. 6:e001576-17. doi: 10.1128/genomeA.01576-17
Tandon, K., Lu, C. Y., Chiang, P. W., Wada, N., Yang, S. H., Chan, Y. F., et al. (2020). Comparative genomics: dominant coral-bacterium endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303. doi: 10.1038/s41396-020-0610-x
Vega Thurber, R. L., Burkepile, D. E., Fuchs, C., Shantz, A. A., McMinds, R., and Zaneveld, J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Change Biol. 20, 544–554. doi: 10.1111/gcb.12450
Wei, T., and Simko, V. (2017). R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84).
White, K. N., Weinstein, D. K., Ohara, T., Denis, V., Montenegro, J., and Reimer, J. D. (2017). Shifting communities after typhoon damage on an upper mesophotic reef in Okinawa, Japan. PeerJ 5:e3573. doi: 10.7717/peerj.3573
Wilson, K. (2001). Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 1, 2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56
Woo, S., Yang, S. H., Chen, H. J., Tseng, Y. F., Hwang, S. J., De Palmas, S., et al. (2017). Geographical variations in bacterial communities associated with soft coral Scleronephthya gracillimum. PLoS One 12:e0183663. doi: 10.1371/journal.pone.0183663
Yamano, H., Sugihara, K., and Nomura, K. (2011). Rapid poleward range expansion of tropical reef corals in response to rising sea surface temperatures. Geophys. Res. Lett. 38:L04601. doi: 10.1029/2010GL046474
Yang, S. H., Tseng, C. H., Huang, C. R., Chen, C. P., Tandon, K., Lee, S., et al. (2017). Long-term survey is necessary to reveal various shifts of microbial composition in corals. Front. Microbiol. 8:1094. doi: 10.3389/fmicb.2017.01094
Keywords: Kuroshio Current, coral-associated bacteria, Endozoicomonas, Acinetobacter, generalist and specialist, Acropora muricata
Citation: Yang S-H, Tseng C-H, Lo H-P, Chiang P-W, Chen H-J, Shiu J-H, Lai H-C, Tandon K, Isomura N, Mezaki T, Yamamoto H and Tang S-L (2020) Locality Effect of Coral-Associated Bacterial Community in the Kuroshio Current From Taiwan to Japan. Front. Ecol. Evol. 8:569107. doi: 10.3389/fevo.2020.569107
Received: 16 June 2020; Accepted: 17 September 2020;
Published: 28 October 2020.
Edited by:
David Michael Baker, The University of Hong Kong, Hong KongReviewed by:
Matthew R. Nitschke, University of Aveiro, PortugalAngela Poole, Berry College, United States
Copyright © 2020 Yang, Tseng, Lo, Chiang, Chen, Shiu, Lai, Tandon, Isomura, Mezaki, Yamamoto and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sen-Lin Tang, c2x0YW5nQGdhdGUuc2luaWNhLmVkdS50dw==
†These authors have contributed equally to this work and share first authorship
 Shan-Hua Yang
Shan-Hua Yang Ching-Hung Tseng
Ching-Hung Tseng Hsueh-Ping Lo3
Hsueh-Ping Lo3 Pei-Wen Chiang
Pei-Wen Chiang Jia-Ho Shiu
Jia-Ho Shiu Kshitij Tandon
Kshitij Tandon Naoko Isomura
Naoko Isomura Takuma Mezaki
Takuma Mezaki Sen-Lin Tang
Sen-Lin Tang