
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Ecol. Evol., 14 August 2020
Sec. Evolutionary Developmental Biology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00261
This article is part of the Research TopicEvo-Devo of Color Pattern FormationView all 20 articles
Developmental modularity has long been viewed as a hierarchical organization that facilitates evolution through modification or reuse of preexisting modules. More recently, developmental modularity has been proposed as a mechanism capable of driving rapid evolution of novel color pattern phenotypes between closely related taxa. In this scenario, recombination between modular cis-regulatory elements (CREs) generates novel phenotypes by shuffling genetic variation at preexisting color pattern modules into new arrangements. Recent functional evidence from Drosophila flies and Heliconius butterflies, however, provides a series of examples in which CREs function in multiple developmental contexts and are thus highly pleiotropic. The potential prevalence of pleiotropy in CRE function could be a barrier to the proposed importance of CRE modules as a mechanism for rapid evolutionary change. Here we review the concept of developmental modularity, some examples that suggest developmental modularity underlies pattern evolution, and recent evidence that indicates modular CREs may be less common than previously expected. This leads us to suggest that alternative, non-modular hypotheses should be considered alongside proposals of modular CREs. We then propose the concept of evolutionary modularity as a specific alternative to developmental modularity when discrete, seemingly modular, phenotypes occur in hybridizing taxa. We suggest that evolutionary modularity provides a potentially important pathway for exchange of phenotypic elements between hybridizing taxa independent of the underlying developmental architecture.
Diversification of animal coloration has been often used as a model for the genetics and ecology of adaptive evolution. In numerous taxa, adaptive color pattern diversity has repeatedly mapped to relatively few genomic loci. Bird plumage coloration (Toews et al., 2016; Campagna et al., 2017), cryptic hair pigmentation in mice (Steiner et al., 2007; Manceau et al., 2011), and aposematic color patterns in Heliconius butterflies (Reed et al., 2011; Martin et al., 2012; Nadeau et al., 2016; Westerman et al., 2018) are just a few key examples where the loci that differentiate color morphs include only a handful of developmental genes. Consistent with the well-known trend that gene regulatory mechanisms evolve faster than coding sequence change, many of the loci driving differentiation of adaptive coloration show the strongest signal of divergence between morphs at non-coding loci presumed to capture cis-regulatory variants. “Combinatorial” adaptation – the restructuring of existing genetic variation in cases of rapid diversification – has become a common observation in studies of diversification and adaptive radiation (Marques et al., 2019). That is, genomic comparisons often show signals suggesting that combinations of extant alternative alleles at closely linked loci underlie the diversity seen in these color patterns. In some cases of combinatorial evolution, the architecture of cis-regulatory adaptation has been coined as modular and modularity of cis-regulatory elements (CREs) has been suggested as a potent genetic architecture to explain the rapidly evolving diversity found in these systems (Wallbank et al., 2016; Campagna et al., 2017; Van Belleghem et al., 2017).
Recent studies on the genetic basis of Drosophila morphology and Heliconius butterfly wing color pattern evolution provide the first lines of evidence that putative cis-regulatory modules may, instead, be pleiotropic and non-modular. Here we review the case for modular elements inferred from genomic comparisons, then consider how recent counterexamples question the ubiquity of CRE modularity. We propose that, while exchange of modular elements may indeed underlie the transfer of adaptive phenotypes, recombination of developmental modules should not be the only hypothesis considered. Instead, a genetic model combining hybrid zone homogenization of trans acting factors with evolutionary modules can reconcile the apparent modular exchange of alternate alleles with developmental pleiotropy and non-modular genetics.
Modularity has long been a mainstay of developmental biology and evo-devo. Examples of modular genetic mechanisms that underlie trait development, such as melanin patterning across Drosophila species (Rebeiz et al., 2009) and Hox gene expression domains (Kuratani, 2009), have demonstrated the importance of modular architectures in determining organismal form. More recently, comparative and evolutionary genomics have begun to propose modularity as a mechanism capable of facilitating phenotypic diversification (e.g., Hughes and Leips, 2017; Van Belleghem et al., 2017; Mason et al., 2020). Modularity as a combinatorial mechanism of diversification has been associated with exchange of modular CREs, such as enhancer, promoter, insulator, and silencer elements. In this model of diversification, transfer of autonomous CREs via hybridization and recombination of specific genomic loci from one population to another allows for swapping of discrete phenotypic elements to generate new phenotypes from ancestral genetic components (Wallbank et al., 2016; Van Belleghem et al., 2017).
The concept of a “module” or “modular element” is critical to grasping the evidence for modularity as a mechanism of evolution. Despite our focus here on modular CREs, modularity is not specific to non-coding regulatory elements, and thus we aim for a more general definition that fits both non-coding and coding loci alike. While the abstract concept “modularity” is a difficult term to define, autonomy of function (i.e., module components mostly function independently; von Dassow et al., 2000; Schlosser, 2004; Wagner et al., 2007; Monteiro and Podlaha, 2009; Espinosa-Soto and Wagner, 2010; Lacquaniti et al., 2013; Melo et al., 2016) and sufficiency (i.e., modules include all novel elements necessary to induce a phenotype; Monteiro and Podlaha, 2009; Arnoult et al., 2013; Henry et al., 2015; Koshikawa, 2015; Merrill et al., 2019) are common requirements of developmental modules and we continue this practice here. For this perspective, we adopt the definition of a developmental module as: A genomic locus or set of loci sufficient to semi-autonomously induce a phenotype when activated in any common genetic background within a species. Developmental modularity then refers to a developmental system consisting of one or more distinct modules [e.g., induced eyeless expression is sufficient to produce ectopic eyes in non-retinal tissues (Weasner et al., 2009)]. This can be contrasted against non-modular architectures, such as seen in many polygenic traits (e.g., Gudbjartsson et al., 2008) and additive cis-regulatory control of non-specific gene expression (e.g., Fulco et al., 2016).
When considered as a mechanism for producing novel adaptive phenotypes, modular loci seem most likely to underlie variation in phenotypes with discrete pattern elements, such as bird plumage patches, fish scale pigmentation, or butterfly color pattern elements. Specific examples indicative of recombination of modular elements include capuchino seedeaters in Argentina (Campagna et al., 2017) and North American warblers (Toews et al., 2016; Figure 1A). In both species groups, discrete plumage color pattern elements appear to be exchanged via hybridization and the strongest signal of genomic differentiation almost exclusively maps to intergenic, putatively cis-regulatory, loci. A modular mechanism of pattern diversification has been proposed in cichlids (Maan and Sefc, 2013), and stripe variation among clownfishes (Litsios and Salamin, 2014; Salis et al., 2018) appears modular as well (Figure 1A). Consistent with modular stripe evolution in fish, CRE differences at the gene csf1 can explain the transitions between striped and non-striped zebrafish species (Patterson et al., 2014). Direct evidence for the mechanism of phenotypic exchange in fish and birds is sparse, however, and studies have limited speculation on the role of modular loci in generating novel color pattern phenotypes. Nonetheless, closely related, hybridizing taxa that appear to exchange color pattern elements to produce novel phenotypes provide an ideal scenario for evolution via modular genomic loci.
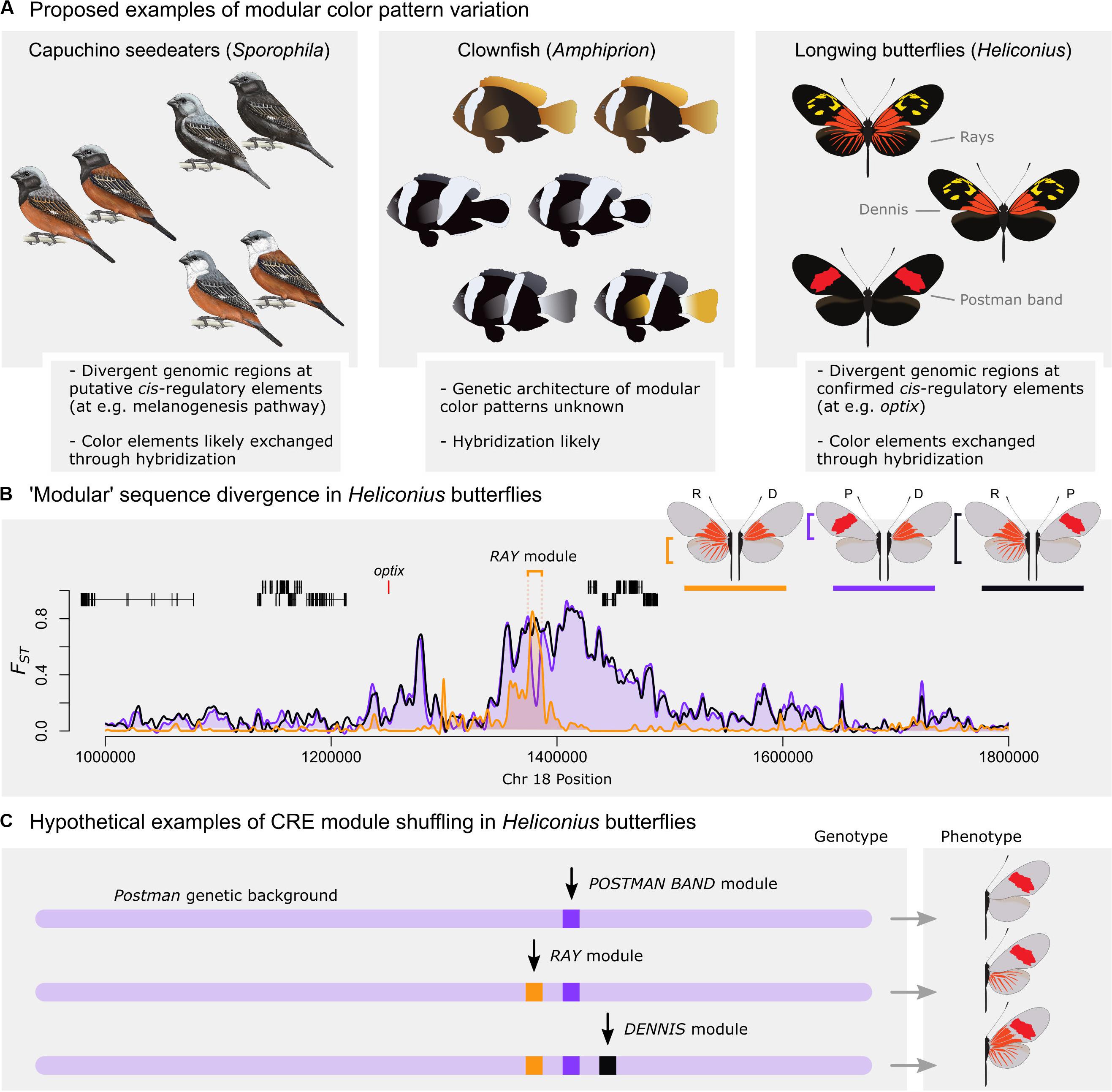
Figure 1. Examples of putative modular color pattern diversity. (A) Examples of potential exchange of modular phenotypes via recombination include plumage patterns in capuchino seedeaters (left, reprinted from Campagna et al., 2017), white stripes in clownfish (middle, modified from Salis et al., 2018), and red wing color patterns in longwing butterflies (right). (B) Genomic differentiation (Fst) between red wing pattern morphs in Heliconius erato at the optix locus (data from hybrid zones in French Guiana; Van Belleghem et al., 2017). Orange: Differentiation between radiate (R) and dennis (D) morphs; Purple: Differentiation between postman (P) and dennis (D) morphs; and Black: Differentiation between radiate (R) and postman (P) morphs. A single locus, called Ray module, shows a putative cis-regulatory module controlling differentiation between the radiate and dennis morphs. (C) Model of CRE module shuffling assuming these divergent loci function as developmental modules. In this case, different combinations of CRE modules would generate modular alternative phenotypes. Note however that the proposed phenotypes with both a red forewing band and dennis/rays on the right have not been found in a homozygous state in nature and suggest that recombination has not occurred (F1 hybrids can have these phenotypes due to dominance effects of optix).
Perhaps the best argument for adaptive evolution via transfer of modular CREs comes from Heliconius hybrid zones in two co-mimetic species. In both Heliconius erato and Heliconius melpomene, regional butterfly populations converge on the same mimicry-related phenotypes to form local morphs with discrete aposematic color pattern elements. In the H. melpomene clade, some subspecies have evolved to contain partial phenotypes completely present in neighboring subspecies. This includes, for example, the sole presence of rayed hindwing or red forewing triangle patterns in the absence of the other element (Wallbank et al., 2016). This scenario repeats in H. erato, where butterfly morphs again form narrow hybrid zones in which discrete red wing pattern phenotypes appear to be exchanged between morphs (Van Belleghem et al., 2017; Figure 1A). Importantly, in both species, the exchange of red pattern elements: (A) maps back to a cis-regulatory region distal to the “switch” gene optix, and (B) associates with recombination of specific genomic haplotypes at these loci that transfer via recombination between neighboring populations with shared wing color pattern elements (Figure 1B). In cases similar to Heliconius wing patterns, where hybridization and recombination of extant alleles can produce phenotype diversity, exchange of modular elements is an attractive hypothesis that posits a simple mechanistic process by which new adaptive phenotypes can be rapidly gained or lost.
While indirect evidence for recombination of modular CREs between geographically close and genetically related taxa is abundant, recent studies in Drosophila, cell lines, and Heliconius butterflies suggest pervasive CRE pleiotropy and interdependence. These results present several counterexamples to key principles that support phenotype modularity derived from modular CREs. We consider evidence against the following two principles: (A) CREs are independent and highly tissue-specific and that (B) recombination of existing CREs is more important for rapid evolutionary change compared to de novo mutations. We discuss these findings and their consequences for CRE and phenotypic modularity in more detail below.
The first principle favoring exchange of modular CREs can be stated as: genes are frequently found to be highly pleiotropic, where a single gene often affects multiple characteristics, while cis-regulatory loci are often assumed to be highly tissue-specific (Prud’homme et al., 2007; Carroll, 2008). This principle provides a foundation for the exchange of phenotype-specific CREs without any corresponding alteration of fitness from undesirable pleiotropic effects. In support of this principle, many early studies of trait evolution found that evolution of one or two enhancer elements underlies variation in phenotypes. Yet recent evidence suggests that cis-regulatory loci may often be substantially more pleiotropic than initially expected. The overall prevalence of enhancer pleiotropy has been well covered by Sabarís et al. (2019) and the potential interpretations of “pleiotropy” in Paaby and Rockman (2013). For this perspective, we adopt the standard genetic definition of pleiotropy, where a locus is pleiotropic if it affects multiple characteristics. Some aspects of CRE biology that suggest pleiotropic elements may frequently play an important role in generating novel traits are worth considering further.
For example, pioneering work by the ENCODE project found that CRE availability, an accessible chromatin state important for CRE activity, tends to be maintained through cell lineages (Stergachis et al., 2013). Thus, enhancers activated in earlier cell types are often available for use through much of the remainder of development. It is likely, then, that these elements could be reutilized or co-opted by evolutionary processes to drive new expression patterns instead of generating a suite of novel CREs de novo (Monteiro and Podlaha, 2009). Important for evolution, CREs active in multiple tissues or during extended periods of development show increased conservation between taxa and provide an evolutionarily stable set of pre-wired regulatory loci (Lewis et al., 2016; Fish et al., 2017). The availability of accessible CREs in many tissues – a likely precursor for the evolution of enhancer pleiotropy – thus appears frequent enough to be found in meta-analyses of CREs. But does this context result in actual enhancer pleiotropy? The recent discovery of pleiotropic enhancers associated with development of leg bristles and trichome patterning explicitly demonstrates that pleiotropic CREs targeting developmental genes can and do underlie more than one important traits in Drosophila (Nagy et al., 2018; Preger-Ben Noon et al., 2018).
The second principle guiding the prediction of modular CRE transfer is that recombination of extant CRE modules is a logical mechanism for the rapid introduction of new alleles into a population while mitigating deleterious effects likely to occur with coding sequence variation (Prud’homme et al., 2007; Wallbank et al., 2016). While this is undoubtedly true, past studies suggest that transfer of modular elements should not necessarily be the default assumption. In many cases, such as loss of stickleback spines (Chan et al., 2010) and horizontal stripes in cichlids (Kratochwil et al., 2018), adaptive trait evolution is driven by loss of function mutations when an organism is exposed to a novel environment. Adaptive loss of phenotype requires no assumptions regarding the modularity of a trait, as a simple deletion can be sufficient to break the regulatory architecture that underlays trait development in both modular and non-modular scenarios (Prud’homme et al., 2007). In the gain of function case, numerous studies have highlighted the relatively rapid rate of cis-regulatory evolution (e.g., Villar et al., 2015; Lewis et al., 2016). Multiple studies have shown that mutations within enhancer elements drive variation in complex phenotypes (e.g., Gompel et al., 2005; Nagy et al., 2018). Similarly, convergent evolution can occur from independent mutations at the same loci, such as the repeated evolution of warning coloration in bumblebees (Tian et al., 2019). Thus, recombination of existing CREs is not required for rapid evolutionary change. Conditional on the number of CREs, distance between loci, and strength of selection against partial recombinants, evolution via directional selection on novel variants or some alternate process may potentially be faster than precise exchange of multiple CRE modules.
Evidence of pleiotropic enhancers and non-modular evolution of novel phenotypes does not reject the transfer of modular CREs. Instead, these counterexamples and arguments suggest we need additional studies to explicitly test for adaptation via exchange of putatively modular elements. Fortunately, recent work on the evolution of mimicry phenotypes in Heliconius provides the perfect case study for how evolution of non-modular genetic architectures may drive variation in apparently modular traits.
Heliconius wing patterns have been proposed as a key example of how modular cis-regulatory alleles can generate novel color pattern adaptations (Mallet and Clarke, 1989; Wallbank et al., 2016; Van Belleghem et al., 2017). In Heliconius erato, red wing pattern phenotypes appear to be shuffled between hybridizing populations to produce morphs with various combinations of these pattern components. The entire list of hybrid zones described in Van Belleghem et al. (2017) is extensive, so we focus here on a particularly clear example of apparent modularity.
In French Guiana and Suriname, three red pattern morphs – named radiate, dennis, and postman (Figure 1A) – form a complex hybrid zone. Dennis, which has the forewing pattern of the radiate morph and the hindwing pattern of the postman, appears by eye to be a simple shuffling of these two other phenotypes via recombination of forewing and hindwing pattern-associated alleles. Consistent with this, DNA sequence analysis of these morphs has shown that a single locus downstream of the red switch gene optix is the only site that differentiates red wing phenotypes between the radiate and dennis morphs (Van Belleghem et al., 2017; Figure 1B). When adding the postman phenotype to this comparison, we see that the locus differentiating dennis from radiate morphs contains the postman allele, rather than the radiate allele. This suggests a simple evolutionary mechanism where a presumptive postman hindwing allele (lacking the Rays module) has been recombined into the radiate haplotype, resulting in the loss of the hindwing rays to produce the dennis phenotype. Thus, the evolution of the dennis morph appears to be a clear example of modular CRE transfer to create a novel phenotype. CRE modules have been similarly suggested for the other red color pattern elements, including a module for the red Dennis and Band color pattern elements (Figure 1C).
Recent findings, however, suggest that the origin and subsequent evolution of the radiate morph is much more complicated than expected from a few simple modular CREs (Lewis et al., 2019). These results show that at least five distinct cis-regulatory loci drive adaptive evolution of the radiate mimicry phenotype. Consistent with the maintenance of CRE availability through cell-lineages, wing CREs rarely differ between forewings and hindwings (Lewis and Reed, 2018; van der Burg et al., 2019). Thus, the same CRE landscape is shared near optix by both forewing and hindwing color patterns (Lewis et al., 2019). Inconsistent with modularity, pattern associated CREs are all interdependent and pleiotropic – CRE mutants alter both the rays and the dennis components of the radiate morph (Figure 2A). While hybridization seems to have spread the radiate phenotype between multiple geographic populations, recombination of a single CRE is insufficient to produce a partial or complete radiate phenotype in a postman-like morph. Perhaps most surprisingly, deletion of one CRE showed the effects of both enhancer (phenotype suppression in the mutant) and silencer (phenotype activation in the mutant) activity in different wing sections. This underscores the long-standing view that epistatic effects, and thus the genetic background, can play an important part in how traits evolve (Phillips, 2008). The dual-modality of individual CREs in Heliconius is consistent with evidence from Drosophila that activating or repressing behavior from CREs can be context dependent (Gisselbrecht et al., 2019). Similarly, epistatic control of wing-pattern CREs is supported by recent observations in various Heliconius species that a single gene can modulate phenotype expression associated with other unlinked color pattern loci (Concha et al., 2019).
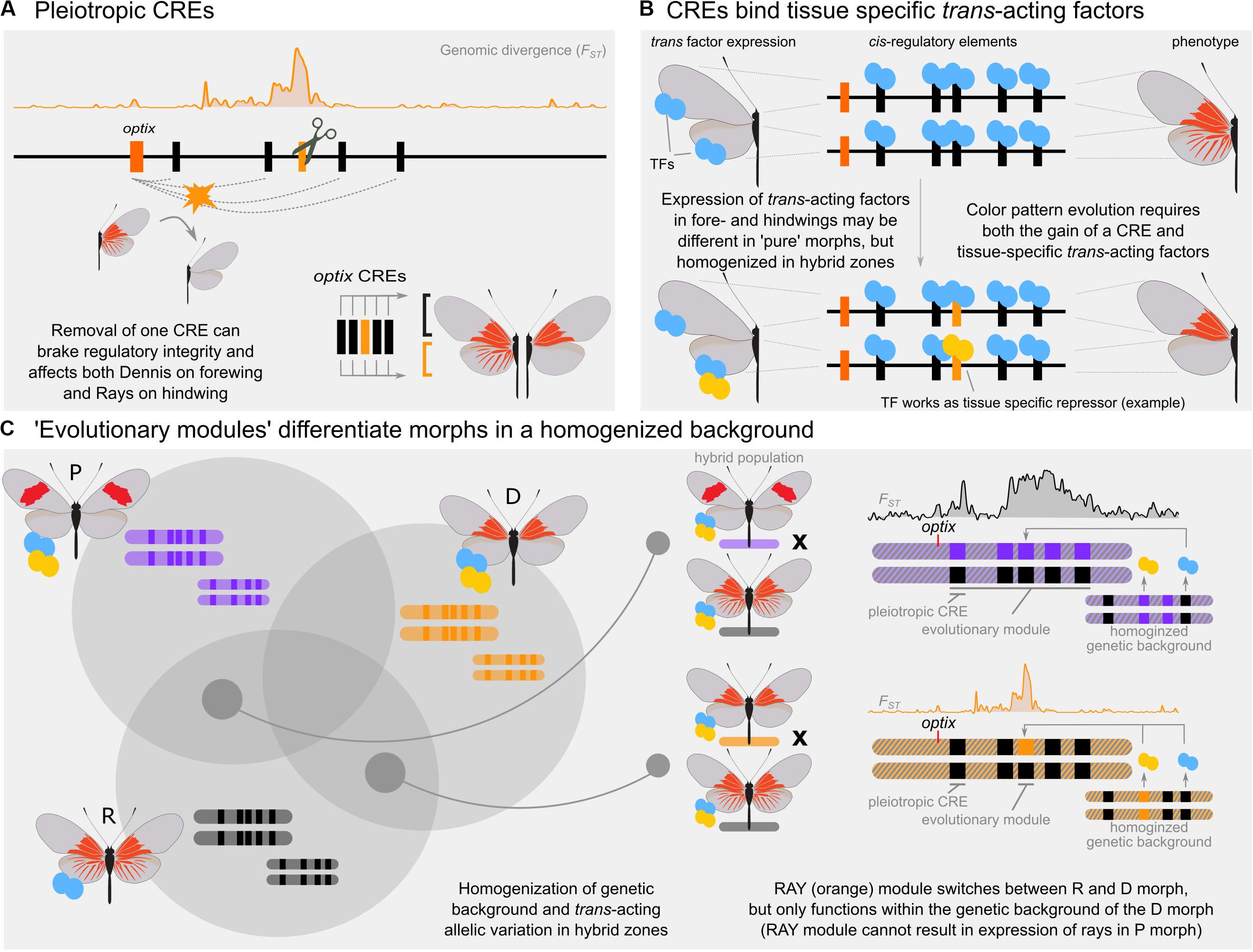
Figure 2. Conceptual overview of evolutionary modularity. (A) CRISPR/Cas9 experiments of five putative CREs demonstrate that red wing patterns in Heliconius erato are controlled by an interdependent set of enhancers. Loss of any enhancer causes loss of red pattern phenotype in both fore- and hindwings and are pleiotropic (Lewis et al., 2019). (B) Hypothetical mechanism by which a novel, tissue specific phenotype (dennis) might evolve through the acquisition of both a CRE and a tissue-specific transcription factor. Blue and yellow symbols indicate tissue specific transcription factors (TFs) which may be unique to a “pure” population but homogenized in hybrid zones. The evolutionary module in this example includes a CRE that is accessible to TFs in both the fore- and hindwing, but may bind a TF unique to the hindwing in the dennis morph only. (C) Depiction of hypothesized evolutionary modularity in the East Amazon H. erato hybrid zone. Multiple unlinked alleles control divergence between all three morphs (Left). These loci become homogenized in narrow regions of hybridization, allowing a single locus to modulate wing pattern phenotype (Right). These homogenized loci can include trans-acting factors, which may not show any signal of differentiation in genomic hybrid zone comparisons.
Taken together, pleiotropic CRE activity and indication of epistasis between color pattern loci runs counter to the developmental modularity of red color pattern CREs in Heliconius erato. These experiments can, specifically, reject the hypothesis that a single locus is sufficient to induce wing pattern components in the absence of additional CRE alleles and the necessary genetic background. This work does, however, raise the question: Why do specific loci appear to control wing phenotypes in a modular fashion in genome sequence comparisons?
It is important to reconcile the apparent conflict between the genomic sequence comparisons and experimental data in our case study. It is our view that comparative genomic analyses indicating modular transfer of phenotype components capture an important aspect of adaptive evolution and phenotype stability in the face of gene flow. While this approach does not demonstrate developmental modularity, it can provide strong evidence for a similar concept – evolutionary modularity. By evolutionary modules, we mean: Any locus sufficient to modulate the gain or loss of phenotype components in the local genetic context of two or more hybridizing populations. Evolutionary modularity differs from developmental modularity by making no requirements for the autonomy or sufficiency of a module, and may be specific only to a single geographic region or pair of hybridizing taxa. That is, an evolutionary module may require a specific genetic background found only within specific geographic regions. This concept does not make any assumptions about the true developmental genetic architecture of a trait, but instead suggests that many architectures can be utilized in a modular fashion by evolutionary processes.
To parse out how evolutionary modularity would work, we return to our example of the East Amazon Heliconius hybrid zone. In the admixed genetic background of the hybrid zone, many combinations of pleiotropic and epistatic loci are likely to occur due to hybridization of “pure” parental phenotypes and recombination in hybrid and backcrossed offspring. When most genetic elements for a trait are homogenous among all three morphs, a single, variable locus may be sufficient to create a novel phenotype and modulate between wing pattern morphs. The apparent gain of a modular phenotype in the dennis morph can be explained as the product of this scenario: In the admixed genetic background of the hybrid zone, trans-acting factors are shared by both morphs and a single, cis-regulatory domain provides a module-like switch for swapping between phenotypes (Figures 2B,C). This single locus may be sufficient to maintain differentiation between the derived dennis morph and the radiate population, while more complicated differentiation patterns would separate radiate from postman. Thus, a single locus, insufficient for producing a phenotype in the absence of a specific genetic context, may act modular in localized population structures. This is different from the scenario of modular CRE shuffling presented in Figure 1C, as evolutionary modules require the interdependent regulatory genetic architecture to be present and may require additional trans factors.
The concept of evolutionary modularity points to an important feature of adaptive evolution: Evolutionary novelty arises at a specific time and place. The process of refining or separating phenotype components in derived taxa can be distinct from the processes that generate the ancestral form. This, in turn, suggests that individual hybrid zones – where modular phenotypes appear most likely – can be a breeding ground for phenotype diversity via evolutionary modularity from either modular or non-modular developmental landscapes.
Here we consider a single case study of whether modular CREs drive adaptation of novel phenotypes. Many more studies will be necessary before we can parse the relative significance of modular and non-modular genetic architectures for phenotypic novelty and diversification. Importantly, we are not suggesting that modular genetic elements cannot or do not underlie novel phenotypes. Our perspective simply suggests that developmental modularity should not be the default assumption, even in cases where discrete phenotypes are swapped between hybridizing populations. The larger evolutionary implications of developmental CRE modularity, or the lack thereof, are substantial: modular CREs would favor adaptation via exchange of a few large-effect loci and the potential for simple adaptive introgression of genetic elements. If, however, our concept of evolutionary modularity accurately captures a common evolutionary scenario, we might expect to see an increasing number of studies mapping traits to oligogenic and polygenic adaptive architectures. Similarly, combinatorial adaptation would thus require multiple genetically distinct recombination events.
It will be important that future cases of putative developmental modularity be demonstrated with empirical assays, rather than assumed from sequence comparisons. While consideration of genetic background is not new, and is even quite common in many studies of developmental genetics, we predict that analysis of genomic interactions and pleiotropic effects will be increasingly important during future studies of phenotypic adaptation. We also suspect, though only time will tell, that evolutionary modularity will be an important process in the production of novel phenotypes. As a deeper understanding of the genetic basis of adaptive evolution emerges, we anticipate that complex developmental architectures will repeatedly be processed in fairly simple evolutionary scenarios via hybridization and recombination to produce ecologically significant phenotypes.
JL and SV contributed equally to the presented ideas, writing of the manuscript, and making the figures. Both authors contributed to the article and approved the submitted version.
JL was supported by the NASA 17-EXO-17-2-0112 and NSF DEB-1546049. SV was supported by the NSF EPSCoR RII Track-2 FEC (OIA 1736026) and in part by National Institutes of Health-NIGMS COBRE Phase 2 Award – Center for Neuroplasticity at the University of Puerto Rico (Grant No. 1P20GM103642). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Leo Campagna and Vincent Laudet for providing us with images for capuchino seedeaters and clownfish, and Karin van der Burg, Arnaud Martin, and Joe Hanly for helpful comments.
Arnoult, L., Su, K. F. Y., Manoel, D., Minervino, C., Magriña, J., Gompel, N., et al. (2013). Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339, 1423–1426. doi: 10.1126/science.1233749
Campagna, L., Repenning, M., Silveira, L. F., Fontana, C. S., Tubaro, P. L., and Lovette, I. J. (2017). Repeated divergent selection on pigmentation genes in a rapid finch radiation. Sci. Adv. 3:e1602404. doi: 10.1126/sciadv.1602404
Carroll, S. B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36. doi: 10.1016/j.cell.2008.06.030
Chan, Y. F., Marks, M. E., Jones, F. C., Villarreal, G., Shapiro, M. D., Brady, S. D., et al. (2010). Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305. doi: 10.1126/science.1182213
Concha, C., Wallbank, R. W. R., Hanly, J. J., Fenner, J., Livraghi, L., Rivera, E. S., et al. (2019). Interplay between developmental flexibility and determinism in the evolution of mimetic Heliconius wing patterns. Curr. Biol. 29, 3996.e4–4009.e4. doi: 10.1016/j.cub.2019.10.010
Espinosa-Soto, C., and Wagner, A. (2010). Specialization can drive the evolution of modularity. PLoS Comput. Biol. 6:e1000719. doi: 10.1371/journal.pcbi.1000719
Fish, A., Chen, L., and Capra, J. A. (2017). Gene regulatory enhancers with evolutionarily conserved activity are more pleiotropic than those with species-specific activity. Genome Biol. Evol. 9, 2615–2625. doi: 10.1093/gbe/evx194
Fulco, C. P., Munschauer, M., Anyoha, R., Munson, G., Grossman, S. R., Perez, E. M., et al. (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 354, 769–773. doi: 10.1126/science.aag2445
Gisselbrecht, S. S., Palagi, A., Kurland, J. V., Rogers, J. M., Ozadam, H., Zhan, Y., et al. (2019). Transcriptional silencers in Drosophila serve a dual role as transcriptional enhancers in alternate cellular contexts. Mol. Cell 77, 324.e8–337.e8. doi: 10.1016/j.molcel.2019.10.004
Gompel, N., Prud’homme, B., Wittkopp, P. J., Kassner, V. A., and Carroll, S. B. (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487. doi: 10.1038/nature03235
Gudbjartsson, D. F., Walters, G. B., Thorleifsson, G., Stefansson, H., Halldorsson, B. V., Zusmanovich, P., et al. (2008). Many sequence variants affecting diversity of adult human height. Nat. Genet. 40, 609–615. doi: 10.1038/ng.122
Henry, K. F., Kawashima, T., and Goldberg, R. B. (2015). A cis-regulatory module activating transcription in the suspensor contains five cis-regulatory elements. Plant Mol Biol. 88, 207–217. doi: 10.1007/s11103-015-0308-z
Hughes, K. A., and Leips, J. (2017). Pleiotropy, constraint, and modularity in the evolution of life histories: insights from genomic analyses. Ann. N.Y. Acad. Sci. 1389, 76–91. doi: 10.1111/nyas.13256
Koshikawa, S. (2015). Enhancer modularity and the evolution of new traits. Fly 9, 155–159. doi: 10.1080/19336934.2016.1151129
Kratochwil, C. F., Liang, Y., Gerwin, J., Woltering, J. M., Urban, S., Henning, F., et al. (2018). Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362, 457–460. doi: 10.1126/science.aao6809
Kuratani, S. (2009). Modularity, comparative embryology and evo-devo: Developmental dissection of evolving body plans. Dev. Biol. 332, 61–69. doi: 10.1016/j.ydbio.2009.05.564
Lacquaniti, F., Ivanenko, Y. P., d’Avella, A., Zelik, K. E., and Zago, M. (2013). Evolutionary and developmental modules. Front. Comput. Neurosci. 7:61. doi: 10.3389/fncom.2013.00061
Lewis, J. J., Geltman, R. C., Pollak, P. C., Rondem, K. E., Van Belleghem, S. M., Hubisz, M. J., et al. (2019). Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc. Natl. Acad. Sci. U.S.A 116, 24174–24183. doi: 10.1073/pnas.1907068116
Lewis, J. J., and Reed, R. D. (2018). Genome-wide regulatory adaptation shapes population-level genomic landscapes in Heliconius. Mol. Biol. Evol. 36, 159–173. doi: 10.1093/molbev/msy209/
Lewis, J. J., van der Burg, K. R. L., Mazo-Vargas, A., and Reed, R. D. (2016). ChIP-seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Rep. 16, 2855–2863. doi: 10.1016/j.celrep.2016.08.042
Litsios, G., and Salamin, N. (2014). Hybridisation and diversification in the adaptive radiation of clownfishes. BMC Evol. Biol. 14:245. doi: 10.1186/s12862-014-0245-5
Maan, M. E., and Sefc, K. M. (2013). Colour variation in cichlid fish: developmental mechanisms, selective pressures and evolutionary consequences. Semin. Cell Dev. Biol. 24, 516–528. doi: 10.1016/j.semcdb.2013.05.003
Mallet, J., and Clarke, C. (1989). The genetics of warning colour in Peruvian hybrid zones of Heliconius erato and H. melpomene. Proc. R. Soc. London B Biol. Sci. 236, 163–185. doi: 10.1098/rspb.1989.0019
Manceau, M., Domingues, V. S., Mallarino, R., and Hoekstra, H. E. (2011). The developmental role of Agouti in color pattern evolution. Science 331, 1062–1065. doi: 10.1126/science.1200684
Marques, D. A., Meier, J. I., and Seehausen, O. (2019). A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34, 531–544. doi: 10.1016/j.tree.2019.02.008
Martin, A., Papa, R., Nadeau, N. J., Hill, R. I., Counterman, B. A., Halder, G., et al. (2012). Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. U.S.A. 109, 12632–12637. doi: 10.1073/pnas.1204800109
Mason, A. J., Margres, M. J., Strickland, J. L., Rokyta, D. R., Sasa, M., and Parkinson, C. L. (2020). Trait differentiation and modular toxin expression in palm-pitvipers. BMC Genomics 21:147. doi: 10.1186/s12864-020-6545-9
Melo, D., Porto, A., Cheverud, J. M., and Marroig, G. (2016). Modularity: genes, development and evolution. Annu. Rev. Ecol. Evol. Syst. 47, 463–486. doi: 10.1146/annurev-ecolsys-121415-032409
Merrill, R. M., Rastas, P., Martin, S. H., Melo, M. C., Barker, S., Davey, J., et al. (2019). Genetic dissection of assortative mating behavior. PLoS Biol. 17:e2005902. doi: 10.1371/journal.pbio.2005902
Monteiro, A., and Podlaha, O. (2009). Wings, horns, and butterfly eyespots: how do complex traits evolve? PLoS Biol. 7:e1000037. doi: 10.1371/journal.pbio.1000037
Nadeau, N. J., Pardo-Diaz, C., Whibley, A., Supple, M. A., Saenko, S. V., Wallbank, R. W. R., et al. (2016). The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534, 106–110. doi: 10.1038/nature17961
Nagy, O., Nuez, I., Savisaar, R., Peluffo, A. E., Yassin, A., Lang, M., et al. (2018). Correlated evolution of two copulatory organs via a single cis-regulatory nucleotide change. Curr. Biol. 28, 3450.e13–3457.e13. doi: 10.1016/j.cub.2018.08.047
Paaby, A. B., and Rockman, M. V. (2013). The many faces of pleiotropy. Trends Genet. 29, 66–73. doi: 10.1016/j.tig.2012.10.010
Patterson, L. B., Bain, E. J., and Parichy, D. M. (2014). Pigment cell interactions and differential xanthophore recruitment underlying zebrafish stripe reiteration and Danio pattern evolution. Nat. Commun. 5:5299. doi: 10.1038/ncomms6299
Phillips, P. C. (2008). Epistasis–the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867. doi: 10.1038/nrg2452
Preger-Ben Noon, E., Sabarís, G., Ortiz, D. M., Sager, J., Liebowitz, A., Stern, D. L., et al. (2018). Comprehensive analysis of a cis-regulatory region reveals pleiotropy in enhancer function. Cell Rep. 22, 3021–3031. doi: 10.1016/j.celrep.2018.02.073
Prud’homme, B., Gompel, N., and Carroll, S. B. (2007). Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. U.S.A. 104(Suppl. 1), 8605–8612. doi: 10.1073/pnas.0700488104
Rebeiz, M., Pool, J. E., Kassner, V. A., Aquadro, C. F., and Carroll, S. B. (2009). Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663–1667. doi: 10.1126/science.1178357
Reed, R. D., Papa, R., Martin, A., Hines, H. M., Counterman, B. A., Pardo-Diaz, C., et al. (2011). optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141. doi: 10.1126/science.1208227
Sabarís, G., Laiker, I., Preger-Ben Noon, E., and Frankel, N. (2019). Actors with multiple roles: pleiotropic enhancers and the paradigm of enhancer modularity. Trends Genet. 35, 423–433. doi: 10.1016/j.tig.2019.03.006
Salis, P., Roux, N., Soulat, O., Lecchini, D., Laudet, V., and Frédérich, B. (2018). Ontogenetic and phylogenetic simplification during white stripe evolution in clownfishes. BMC Biol. 16:90. doi: 10.1186/s12915-018-0559-7
Schlosser, G. (2004). The role of modules in development and evolution. Modul. Dev. Evol. 1, 519–582.
Steiner, C. C., Weber, J. N., and Hoekstra, H. E. (2007). Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5:e219. doi: 10.1371/journal.pbio.0050219
Stergachis, A. B., Neph, S., Reynolds, A., Humbert, R., Miller, B., Paige, S. L., et al. (2013). Developmental fate and cellular maturity encoded in human regulatory dna landscapes. Cell 154, 888–903. doi: 10.1016/j.cell.2013.07.020
Tian, L., Rahman, S. R., Ezray, B. D., Franzini, L., Strange, J. P., Lhomme, P., et al. (2019). A homeotic shift late in development drives mimetic color variation in a bumble bee. Proc. Natl. Acad. Sci. U.S.A. 116, 11857–11865. doi: 10.1073/pnas.1900365116
Toews, D. P. L., Taylor, S. A., Vallender, R., Brelsford, A., Butcher, B. G., Messer, P. W., et al. (2016). Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr. Biol. 26, 2313–2318. doi: 10.1016/j.cub.2016.06.034
Van Belleghem, S. M., Rastas, P., Papanicolaou, A., Martin, S. H., Arias, C. F., Supple, M. A., et al. (2017). Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1:0052. doi: 10.1038/s41559-016-0052
van der Burg, K. R. L., Lewis, J. J., Martin, A., Nijhout, H. F., Danko, C. G., and Reed, R. D. (2019). Contrasting roles of transcription factors Spineless and EcR in the highly dynamic chromatin landscape of butterfly wing metamorphosis. Cell Rep. 27, 1027.e3–1038.e3. doi: 10.1016/j.celrep.2019.03.092
Villar, D., Berthelot, C., Aldridge, S., Rayner, T. F., Lukk, M., Pignatelli, M., et al. (2015). Enhancer evolution across 20 mammalian species. Cell 160, 554–566. doi: 10.1016/j.cell.2015.01.006
von Dassow, G., Meir, E., Munro, E. M., and Odell, G. M. (2000). The segment polarity network is a robust developmental module. Nature 406, 188–192. doi: 10.1038/35018085
Wagner, G. P., Pavlicev, M., and Cheverud, J. M. (2007). The road to modularity. Nat. Rev. Genet. 8, 921–931. doi: 10.1038/nrg2267
Wallbank, R. W. R., Baxter, S. W., Pardo-Diaz, C., Hanly, J. J., Martin, S. H., Mallet, J., et al. (2016). Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 14:e1002353. doi: 10.1371/journal.pbio.1002353
Weasner, B. M., Weasner, B., Deyoung, S. M., Michaels, S. D., and Kumar, J. P. (2009). Transcriptional activities of the Pax6 gene eyeless regulate tissue specificity of ectopic eye formation in Drosophila. Dev. Biol. 334, 492–502. doi: 10.1016/j.ydbio.2009.04.027
Keywords: modularity, pleiotropy, cis-regulatory element, Heliconius, color pattern, hybridization, evolutionary modularity
Citation: Lewis JJ and Van Belleghem SM (2020) Mechanisms of Change: A Population-Based Perspective on the Roles of Modularity and Pleiotropy in Diversification. Front. Ecol. Evol. 8:261. doi: 10.3389/fevo.2020.00261
Received: 23 December 2019; Accepted: 24 July 2020;
Published: 14 August 2020.
Edited by:
Marie Manceau, Collège de France, FranceReviewed by:
Ariel D. Chipman, The Hebrew University of Jerusalem, IsraelCopyright © 2020 Lewis and Van Belleghem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James J. Lewis, ampsMzM2QGNvcm5lbGwuZWR1; Steven M. Van Belleghem, VmFuQmVsbGVnaGVtU3RldmVuQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.