
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Ecol. Evol. , 04 August 2020
Sec. Evolutionary Developmental Biology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00215
This article is part of the Research Topic Evo Devo Cell by Cell View all 8 articles
Evolutionary developmental biology focuses on understanding the origin and evolution of extant biological variation, and the genetic basis for this variation. The genetic toolkit appears largely finite across animals, such that a combination of regulatory evolution, gene recruitment (co-option) and genetic modularity often allow morphological and developmental diversity to arise. Here we summarize a number of observations from across animals, which together suggest that many genes and gene product interaction “modules” originally characterized for their role in the germ line also have neural roles. We explore potential explanations for this observation, noting that in the context of the germ line, these genes appear to have molecular and biochemical properties that make them well-suited to breaking symmetry within cells. The resulting asymmetry is often caused by gene products co-localizing asymmetrically to sub-cellular, non-membrane bound, electron dense compartments known as ribonucleoprotein (RNP) granules. RNP granules contain high concentrations of translationally quiescent messenger RNAs and proteins and are thought to act as hubs of localized translational control. We propose that the use of strict translational control, which may be achieved via molecular processes important for RNP granule formation and/or small RNA-related processes, is an important property of and a commonality between the germ line and nervous tissues, and helps explain, at least in part, the close relationship between these two tissue types.
Understanding the genomic basis of extant biological variation over evolutionary time scales has been the main focus of modern evolutionary developmental biology (evo-devo) research. In the pre-genomic era, it was unclear to what extent genes unique to an organism were the basis of their morphological, cellular and biological diversity (King and Wilson, 1975). Over the years, however, a large body of evo-devo work has led to the realization that much of the biological variation in extant animals has evolved based on an ancestral genetic toolkit (Peterson and Davidson, 2000). Genes in such shared ancestral toolkits are often conserved both in sequence and developmental function across animals (e.g., Hox genes) (Hrycaj and Wellik, 2016). In other cases, conserved genes have been co-opted for additional, distinct biological roles, leading to pleiotropic gene functions (e.g., distal-less, yellow) (Panganiban et al., 1997; Gompel et al., 2005; Moczek and Rose, 2009; Khila et al., 2012). Both scenarios ultimately contribute to morphological diversity between species, within species, and between cell types within an organism, underpinned by a combination of differences in developmental gene regulation and modularity. Pleiotropy is widespread in genomes, can contribute to phenotypic variation, and may occur through a variety of molecular mechanisms (Guillaume and Otto, 2012), including alternative splicing, different substrate or binding partner affinities, localization to different cellular compartments or tissues, or the same gene product having more than one distinct biochemical property. Barring the extreme cases of “housekeeping” genes (usually ubiquitously expressed) and so-called “luxury” genes (expressed in only one tissue type) (King et al., 2013), most animal genes likely exhibit some degree of pleiotropy (Hodgkin, 1998).
Over the past two decades, multiple primary data observations and some synthetic overviews of the literature (see for example Broadus et al., 1998; Roegiers and Jan, 2000) have hinted at a potentially underappreciated example of pleiotropy that we wish to draw further attention to with this review: namely, that a number of genes sometimes dubbed “germ line genes” based on the initial primary characterization of their roles in the germ lines of animals, also have roles in the development and/or functioning of the nervous system. For example, on a genome-wide scale, tissue-specific transcriptome studies in both humans and mice have shown that the testes and nervous system are two tissues that share a larger overlap in their gene expression profiles and proteomes than they do with any other tissue types within the animal (Guo et al., 2005). Here we gather evidence for this dual tissue expression pattern across multiple metazoans. In cases where “germ line genes” are documented as playing a neural role in one species, we ask whether there is evidence that these genes play any neural role in additional species, and whether they share the same set of interactors. We provide possible molecular mechanistic explanations and suggest that these observations may be explained by co-option of pre-existing molecular interactions to new developmental contexts. Both germ cells and neurons use subcellular compartmentalization of gene products as a mechanism for proper cellular functioning. Neurons are highly compartmentalized cells, and localized translational control within and between synapses is an important mechanism regulating neuronal function (Holt et al., 2019). Likewise, germ cells often require subcellular localization of specific gene products for normal functioning of germ cells or patterning of early embryos, which can be achieved by localized translational control (Pushpa et al., 2017). In addition, both these tissues rely on small RNAs for proper functioning. Small RNAs are important for maintaining the genomic integrity of the germ line and also have key roles in memory and synaptic plasticity in the nervous system of animals (Saxe and Lin, 2011; Posner et al., 2019). Thus, we aim to summarize and synthesize data that may be relevant to understanding both the molecular and cellular basis of pleiotropy in this specific context. More generally, this approach may help shed light on the origins of cell type diversity and evolutionary novelty.
In the following sections, we present evidence from primary literature for genes best known for their role in the germ line, that are also expressed in the nervous system, either singly or in groups of gene products with conserved molecular interaction. Wherever possible, we present currently available data for the functions of these genes in both these tissues. For each example, we briefly summarize their roles in the germ line and in the nervous system (Table 1), providing a list of reported molecular interaction partners in both tissues (Table 2). In cases where, to our knowledge, no molecular interaction data are available, we point out evidence of their co-expression, acknowledging that co-expression may not reflect conserved molecular interactions. This gene list is not exhaustive, as it is necessarily limited to those that, to our knowledge, have been specifically examined in the context of both tissue types across animals. For each gene discussed, we note whether it predates animals or not, based on OrthoMCL-DB predictions (Chen et al., 2006). Because we aim to point out conserved molecular interactions reported in both cell types, we discuss those genes with more abundant co-IP and other interaction data in both germ line and nervous system first, and end with genes for which data are available primarily for only one tissue type. We discuss genes in groups, to indicate reported conserved molecular interactions between their gene products.
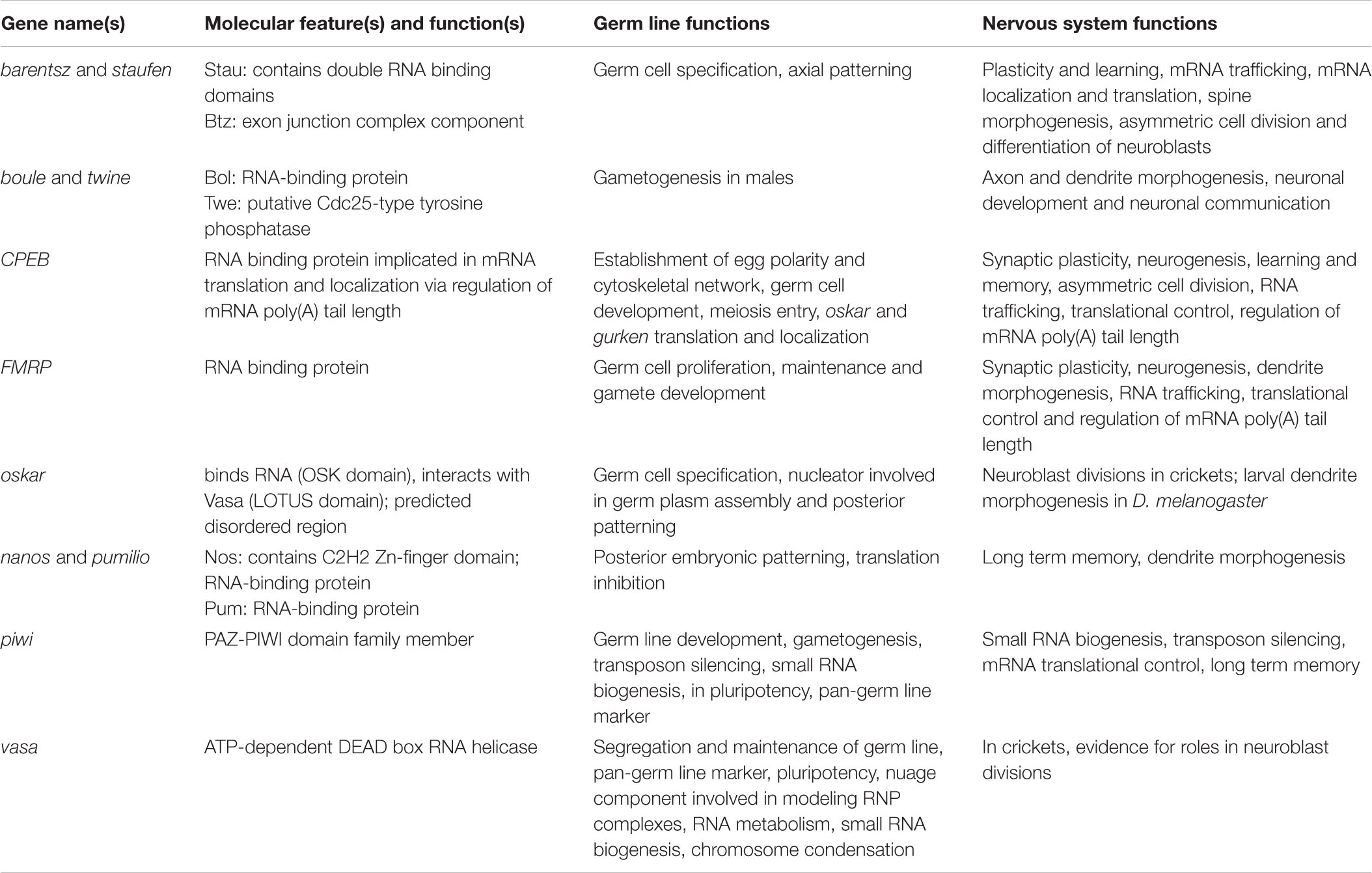
Table 1. Summary of germ line and nervous system roles of genes discussed in the text, listed here in alphabetical order.
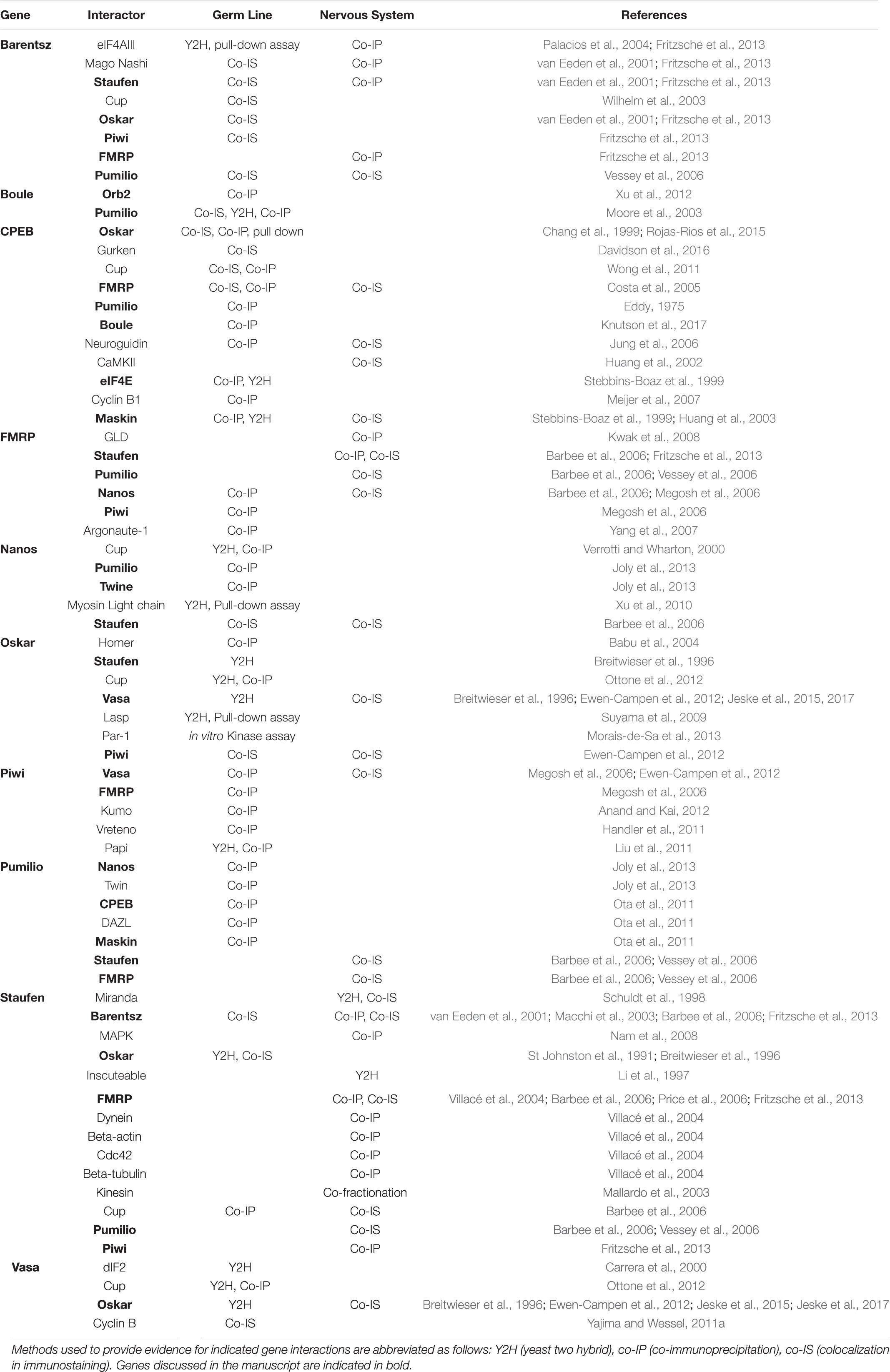
Table 2. Summary of selected gene products and their reported physical interactors in the germ line and in nervous system tissues, listed here in alphabetical order.
staufen (stau) was first identified in a maternal effect genetic screen for Drosophila melanogaster mutants with anterior-posterior body polarity defects, while barentsz (btz) was identified in a female sterile chromosome screen for mutants with defects in localization of Stau protein (Schüpbach and Wieschaus, 1986; van Eeden et al., 2001). Stau belongs to a conserved family of animal proteins (Chen et al., 2006) that contain multiple double-stranded RNA binding domains, and stau orthologs are present in bilaterian outgroups, protostomes and deuterostomes (Heraud-Farlow and Kiebler, 2014). btz genes also appear to be animal-specific (Chen et al., 2006), and Btz protein is a component of the exon junction complex (Ariz et al., 2009), which regulates spliced mRNAs (Bono et al., 2006).
During D. melanogaster oogenesis, Stau and Btz localize to the cytoplasm at the posterior of the oocyte, where they both have a role in primordial germ cell (PGC) specification and establishment of the anterior-posterior axis of the embryo (St Johnston et al., 1991; van Eeden et al., 2001). Stau is required for the posterior localization and translation of the mRNAs of oskar (osk), another gene whose products likely have evolved similar molecular interactions in both germ line and nervous systems [see “oskar (osk), nanos (nos), piwi and vasa (vas)” below] mRNA, and Stau and Btz form a complex and move together during this posterior localization event (van Eeden et al., 2001). Additionally, btz null mutants show defects in Stau protein and osk mRNA localization to the posterior of the oocyte (van Eeden et al., 2001).
Evidence from multiple animals suggests that Stau and Btz function together in neuronal cells via mechanisms similar to those observed in the germ line. Stau is concentrated in ribonucleoprotein (RNP) granules within D. melanogaster neurites in the larval nervous system, where it co-localizes with Btz and dFMR1 (Barbee et al., 2006). Such Stau-Btz-containing neuronal granules also contain molecules that are found in yeast and mammalian somatic P-bodies (e.g., Dcp1p, Xrn1p), suggesting that neuronal and germ line Stau-containing granules may be similar to somatic P-bodies in molecular composition (Barbee et al., 2006). Stau is also present in the D. melanogaster neuromuscular junction (NMJ). At the NMJ, it is localized to the post-synaptic compartment, where it regulates localization and translation of coracle (cora) mRNA (Gardiol and St Johnston, 2014). cora in turn promotes synaptic bouton formation, and accordingly loss of stau leads to a reduction in synaptic bouton number (Macchi et al., 2003). The same Stau domain that is required in oocytes for the translation and localization of the mRNA of osk [see oskar (osk), nanos (nos), piwi and vasa (vas) below] (Micklem et al., 2000), called “dsRNA binding domain 5,” is also required for local cora translation at the NMJ, and Cora protein fails to localize to the NMJ in stau mutants lacking this domain (Gardiol and St Johnston, 2014). Furthermore, Tropomyosin II, which, like Stau, localizes osk to the oocyte posterior (Erdelyi et al., 1995), is also required for cora’s NMJ localization (Gardiol and St Johnston, 2014). Stau plays a critical role in asymmetric neuroblast divisions (Jia et al., 2015) and long-term memory formation in D. melanogaster (Dubnau et al., 2003), a role that appears conserved in the mollusk Aplysia californica (Liu et al., 2006).
In mouse and rat neurons, Stau is contained within RNP particles distributed along the somatodendrites of hippocampal neurons (Tang et al., 2001; Macchi et al., 2003). Btz also co-localizes with Stau in these granular RNPs in hippocampal neurons, and these two proteins co-immunoprecipitate from doubly transfected Baby Hamster Kidney fibroblasts (Macchi et al., 2003). Fritzsche et al. (2013) have recently reported a protein interactome for Stau- and Btz-RNPs in the rat brain, which includes some proteins also found in germ cells, such as Pum [see “nanos (nos) and pumilio (pum)” below] and FMRP [see “Fragile Mental Retardation Protein (FMRP), argonatue (AGO) piwi and staufen (stau)” below]. In mice and rats, Stau is implicated in spatial learning, novelty preference and explorative behavior (Berger et al., 2017; Popper et al., 2018), and in humans Stau is required for normal dendritic arborization during neuroblastoma cell differentiation in vitro (Peredo et al., 2014). Interestingly, when expressed in vivo in D. melanogaster, GFP-tagged mouse Btz localizes to the oocyte posterior, suggesting that it can interact with D. melanogaster Stau (Macchi et al., 2003). However, despite this colocalization with D. melanogaster Stau, mouse Btz is unable to perform the function of D. melanogaster Btz in localizing osk mRNA to the posterior of the oocyte cytoplasm (Macchi et al., 2003), suggesting that not all Btz/Stau functional molecular interactions are conserved across species.
stau is expressed or required in the germ line outside of fruit flies as well. In zebrafish, morpholino-mediated knockdowns of the stau paralogs stau1/2 abrogate the formation of Vasa-positive PGCs (Ramasamy et al., 2006). In mice, stau mRNA is expressed in oocytes and during meiosis in males (Saunders et al., 2000). In human oocytes, immunofluorescence studies show that STAU protein is present throughout all stages of oocyte maturation, and that its subcellular localization changes throughout oogenesis, initially dispersed throughout the cytoplasm and later localized into large discrete granules at the cortex (De Santis et al., 2015). As in D. melanogaster oocytes (St Johnston et al., 1991; van Eeden et al., 2001), Stau localization to a specific region of the Xenopus laevis oocyte cytoplasm is required to specify PGCs (Yoon and Mowry, 2004). Human Staufen (STAU1/2) and Barentsz (CASC3) are both expressed in multiple tissues outside of the germ line and nervous system (Uhlen et al., 2015).
Fragile Mental Retardation Proteins (FMRP) are conserved RNA binding proteins that may have origins predating animals, based on the prediction of a putative ortholog in the green alga Micromonas (Chen et al., 2006). FMRPs underlie human Fragile X syndrome, which is an X-linked dominant disorder causing mental retardation and cognitive impairment (Ashley et al., 1993; Inoue et al., 2000). This defect is caused by an expansion of a CGG trinucleotide repeat in the FMR1 gene, correlated with transcriptional silencing and loss of the gene product FMRP (Verkerk et al., 1991; Verheij et al., 1993). Mammalian FMRP is a member of a small protein family consisting of members FMRP, FXR1 and FXR2, all of which are RNA binding proteins containing two K homology (KH) domains and one RGG box (Siomi et al., 1995; Zhang et al., 1995). FMRP/FXR proteins also contain protein-protein interaction and 60S ribosomal subunit interaction domains (Ashley et al., 1993; Siomi et al., 1996; Wan et al., 2000). FMRP is predominantly detected in the cytoplasm of cells in multiple human tissues (Uhlen et al., 2015), including neurons, glial cells, and spermatogonia, but can also be detected in the nucleus (Devys et al., 1993; Verheij et al., 1993). The presence of nuclear localization (NLS) and export (NES) signals (Eberhart D. E. et al., 1996), suggest that it may function as a nucleo-cytoplasmic shuttle protein for RNA. In vitro experiments suggest that FMRP binds a selective but abundant fraction of brain RNA, but little is currently known about the identity of these targets (Ashley et al., 1993; Brown et al., 1998). FMRP associates with polyribosomes (Khandjian et al., 1996; Tamanini et al., 1996; Feng et al., 1997) and negatively regulates translation (Laggerbauer et al., 2001; Li et al., 2001; Zhang et al., 2001). While all three orthologs of the FMRP/FXR family are found in multiple vertebrates, only one homolog, called dfmr1, has been reported in D. melanogaster (Wan et al., 2000).
FMRP plays roles in germ line development in D. melanogaster and mammals, in both cases via interactions with Piwi or Piwi-related proteins of the Argonaute family (AGO). In D. melanogaster Dfmr1 protein co-immunoprecipitates with Ago1 in ovaries and in adult testes (Yang et al., 2007; Bozzetti et al., 2015). Similarly, in embryos Dfmr1 forms a complex with Piwi during the formation of the specialized cytoplasm, called germ plasm, that ensures PGC specification in D. melanogaster (Megosh et al., 2006). In dfmr1 homozygous null mutants, the ovaries contain fewer germ line stem cells (GSC) than controls, suggesting that dfmr1 is required for GSC maintenance (Yang et al., 2007). dfmr1 and piwi mutants show similar phenotypes of defective pole plasm and reduced PGC number (Megosh et al., 2006). In mice, FMR1 knockout mice display macroorchidism, a disorder in which males have abnormally large testes, in this case caused by an increased postnatal proliferation of Sertoli cells (Slegtenhorst-Eegdeman et al., 1998), which are associated with and required for correct development of male gametes. In mouse testes and in human embryonal carcinoma cell lines derived from testes, FMR1 and AGO1 regulate miRNA-383, implicating FMRP in small RNA-mediated gene regulation in the mammalian germ line (Tian et al., 2013).
FMRP is implicated in multiple neuronal functions in fruit flies and mice, including synaptic plasticity (Padmashri et al., 2013; Feuge et al., 2019), dendritic morphogenesis (Feuge et al., 2019), and olfactory learning and memory (Nimchinsky et al., 2001; Bolduc et al., 2008; Sears et al., 2019). Some studies report that homozygous FMR1 knockout mice display defects in dendritic spine morphology (e.g., Nimchinsky et al., 2001; Bolduc et al., 2008) [but see Feuge et al. (2019) for a report of no abnormal dendritic spine morphology in FMRP knockout mice]. FMRP also appears important for adult mouse neurogenesis: FMR1 knockout mice show misregulation of multiple genes expressed in adult neural progenitor cells (Liu et al., 2018), increased neural progenitor cell proliferation and incorrect neuronal fate specification (Luo et al., 2010), significant reduction in hippocampal neurogenesis (Guo et al., 2011), and reduced hippocampal-dependent learning (Guo et al., 2011). Furthermore, in D. melanogaster, Dfmr1 and Ago1 are required for the regulation of synaptic plasticity (McBride et al., 2005; Bolduc et al., 2008; Sudhakaran et al., 2014). Dfrm1 loss of function mutants show ectopic axon growth (Tessier and Broadie, 2008), and trans-heterozygotes for dfmr1 and Ago1 have overgrown synapses and abnormally elaborate synaptic terminals compared to wild type flies and single heterozygotes (Jin et al., 2004). This phenotype is reminiscent of that of homozygous FMR1 knockout mice, which some researchers report have dendritic spines that are longer than controls (Comery et al., 1997; Nimchinsky et al., 2001) [but see Feuge et al. (2019)]. The molecular functions of FMRP in neurons include trafficking RNA in both fruit fly (Estes et al., 2008) and mouse (Antar et al., 2005; Dictenberg et al., 2008) neurons, regulating length of the mRNA poly(A) tail (Bienkowski et al., 2017), and local translational regulation in both dendrites and cell bodies of neurons (Darnell et al., 2011; Darnell and Klann, 2013). FMRP also co-immunoprecipitates with Stau in rat neurons (Price et al., 2006), and complexes with Stau in transfected human cells and differentiated human neuroblasts (Villacé et al., 2004). An ortholog of FMRP has been identified in the cnidarian Hydractinia echinata (HyFMR1), where it is expressed in neural precursors and nerve cells in the mature polyp (Guduric-Fuchs et al., 2004).
nanos (nos) and pumilio (pum) were first identified in genetic screens for D. melanogaster embryos with posterior and abdominal specification defects (Lehmann and Nüsslein-Volhard, 1987; Nüsslein-Volhard et al., 1987; Lehmann and Nusslein-Volhard, 1991). Pum belongs to a conserved RNA-binding protein family that is found across eukaryotes (Zamore et al., 1997; Zhang et al., 1997; Gamberi et al., 2002; Chen et al., 2006). Its signature PUF domain is named after D. melanogaster Pumilio and the Caenorhabditis elegans translational regulator FBF (fem-3-binding factor) (Zhang et al., 1997). PUF proteins are implicated in post-transcriptional gene regulation (Wang et al., 2018), stem cell maintenance (Lin and Spradling, 1997; Forbes and Lehmann, 1998; Crittenden et al., 2002; Ariz et al., 2009), axial patterning (Lehmann and Nüsslein-Volhard, 1987; Nüsslein-Volhard et al., 1987; Lehmann and Nusslein-Volhard, 1991), and learning and memory (Dubnau et al., 2003). nos is an animal-specific gene (Chen et al., 2006) maternally required for the development and maintenance of the D. melanogaster germ line, and zygotically for embryonic patterning and PGC migration in the developing embryo (Wang and Lehmann, 1991; Wang et al., 1994; Kobayashi et al., 1996). Pum proteins often function together with Nos proteins during development (Sonoda and Wharton, 1999; Parisi and Lin, 2000; Jaruzelska et al., 2003), including in the germ line and nervous system, as detailed below.
In D. melanogaster, nos and pum act together as inhibitors to repress hunchback and bicoid translation in the posterior of the embryo (Wharton and Struhl, 1991; Zamore et al., 1999). Pum is thought to directly bind hunchback and bicoid mRNAs, and to bring Nos to the repression complex (Murata and Wharton, 1995; Sonoda and Wharton, 1999). nos and pum are required in the germ line for continued egg chamber production during oogenesis, by regulating the germ line stem cell to cystoblast fate transition via translational repression of oocyte differentiation genes (Wang et al., 1994; Lin and Spradling, 1997; Forbes and Lehmann, 1998; Szakmary et al., 2005; Joly et al., 2013). nos is required in embryonic development for PGC survival and migration (Kobayashi et al., 1996; Sano et al., 2001; Hayashi et al., 2004; Sato et al., 2007), as well as for patterning the abdomen and embryo posterior (Wang and Lehmann, 1991; Wang et al., 1994).
Requirements for, and genetic and physical interactions between, Nos and Pum in the germ line are conserved in many animals. In C. elegans, nos-1, nos-2 and a subset of pumilio-related genes (fbf-1/fbf-2, puf-6/puf-7 and puf-8) are required for various aspects of PGC development, including PGC migration, cell death and proliferation (Subramaniam and Seydoux, 1999). In X. laevis oocytes, Pum protein co-immunoprecipitates with a X. laevis ortholog of nos (Nanos1; also called Xcat2) (Lai et al., 2011), and binds cyclin B transcripts (Nakahata et al., 2001). In addition to their conserved physical interaction, at least some targets of Nos/Pum may also be conserved: in D. melanogaster, these proteins also bind to and repress translation of cyclin B1 (Kadyrova et al., 2007). In zebrafish, Pum2 is expressed in male and female gonads, and is important for germ cell and nervous tissue development (Wang et al., 2012). Furthermore, a zebrafish homolog of nos is involved in PGC maintenance and migration into the future gonad (Koprunner et al., 2001).
nos and pum also play roles in the development and function of the nervous system of multiple taxa. For example, in D. melanogaster, Nos colocalizes with RNA granules in dendrites, and both nos and pum are needed for appropriate dendrite morphogenesis, suggesting that they may repress mRNA translation in the nervous system as they do in the germ line (Ye et al., 2004). In larval class IV neurons, nos mRNA requires osk for appropriate localization, as described below (Xu et al., 2013). In addition, long-term memory in D. melanogaster requires pum (Dubnau et al., 2003; Chen et al., 2008).
In mice, Pum2 is localized with RNP particles in the somatodendritic region of hippocampal neurons (Vessey et al., 2006), and Pum1 and Pum2 are required for hippocampal neurogenesis and proper functioning (Zhang et al., 2017). Furthermore, mouse Pum2 is implicated in forming stress granules under metabolic stress in neurons, in dendritic morphogenesis, and in regulating the synaptic function along dendritic shafts (Vessey et al., 2006, 2010). Interestingly, nos1 knockdown mice show no detectable neural defects in terms of behavior or fertility (Haraguchi et al., 2003). In the C. elegans genome, there are three nos-related genes and at least ten PUF-domain proteins (Lynch et al., 2011), and PUF-domain proteins have been shown to play memory-related important roles in axonal and presynaptic regions (Lee and Schedl, 2006; Arey et al., 2019). One of these Pum-like proteins, FBF-1, is needed for the change in C. elegans odor sensitivity that comes with prolonged exposure, known as odor adaptation (Kaye et al., 2009). Pum also binds to the 3′UTR of the cGMP-dependent kinase EGL-4 and promotes its translation (Kaye et al., 2009). Of the three nos-related genes, NOS-1 is required for odor adaptation (Kaye et al., 2009).
Outside of bilaterians, there is also evidence for expression and function of nos and pum orthologs in the germ line. In the sexual polyp of the hydroid H. echinata, a pum ortholog and the nanos ortholog nos2 are both expressed in oocytes (Kanska and Frank, 2013), as are nos orthologs in the jellyfish Podocoryne carnea (Torras et al., 2004) and Clytia hemisphaerica (Leclère et al., 2012). In H. magnipapillata, nos orthologs Cnnos1 and Cnnos2, are both expressed in the germ line (Mochizuki et al., 2000). In the anthozoan Nematostella vectensis, the nos ortholog Nvnos2 is expressed in putative germ cells during embryogenesis and in developing oocytes (Extavour et al., 2005; Torras and Gonzalez-Crespo, 2005). nos orthologs are also expressed in developing gametes in the sponges Sycon ciliatum (Leininger et al., 2014) and Oscarella lobularis (Fierro-Constain et al., 2017). In zebrafish, Pum2 is expressed in the brain (Wang et al., 2012). In H. echinata, reduction of Nos2 causes a reduction in nematogenesis (production of stinging cells called nematocytes, considered a type of neural cell) and an increase in neurogenesis (Kanska and Frank, 2013). In sponges, while putative neural tissues remain difficult to identify based on bilaterian-centric cell type criteria (Dunn et al., 2015), expression of nos has been reported in globular cells and cross cells, two candidate sensory cell types unique to sponges (Mah and Leys, 2017).
The insect-specific gene oskar (osk) was first identified in the fruit fly D. melanogaster as a maternal-effect gene that is necessary and sufficient for specifying both the germ line and the posterior abdomen of the embryo (Lehmann and Nüsslein-Volhard, 1986; Ephrussi et al., 1991; Chen et al., 2006). Osk proteins have two conserved, well-folded domains on either side of a region of predicted high disorder (Jeske et al., 2015; Yang et al., 2015). The N terminal domain is a LOTUS domain (also called an OST-HTH domain) (Anantharaman et al., 2010) similar to that of TUDOR5 and TUDOR7 proteins (Ewen-Campen et al., 2012), and is predicted to dimerize (Jeske et al., 2015; Yang et al., 2015) and bind Vasa protein (Markussen et al., 1995; Breitwieser et al., 1996; Vanzo and Ephrussi, 2002; Jeske et al., 2017). The C terminal domain is known as the OSK domain and is implicated in binding nanos (see below), oskar, germ cell less and polar granule component mRNAs (Jeske et al., 2015; Yang et al., 2015).
In D. melanogaster, osk is expressed from the maternal genome during oogenesis, and osk mRNA is deposited into the developing oocyte in a process dependent on Splicing oskar Location Elements (SOLE) in its 3′UTR (Ghosh et al., 2012). SOLE recruitment of Exon Junction Complex components, including barentsz, mago nashi, and tsunagi, is required for proper osk ribonucleoprotein (RNP) granule motility into the oocyte, and for posterior localization of osk within the oocyte (Ghosh et al., 2012). Posterior localization of osk also requires interactions with Staufen (St Johnston et al., 1991; see below) and Kinesin proteins (Brendza et al., 2002). Posteriorly localized osk mRNA is translated into two protein isoforms, Short Osk and Long Osk (Markussen et al., 1995). Short and Long Osk differ by an N terminal 138 amino acid (aa) addition (Markussen et al., 1995). The current model of the distinct functions of these isoforms is as follows: Long Osk localizes to endocytic membranes at the oocyte posterior (Vanzo et al., 2007; Tanaka and Nakamura, 2008), anchors both osk mRNA and Short Osk (Vanzo and Ephrussi, 2002; Tanaka et al., 2011), and stabilizes mitochondrial accumulation (Hurd et al., 2016). Short Osk localizes to electron-dense organelles called polar granules and recruits products of genes required for germ cell and posterior identity specification including vasa, nanos, and piwi (see below) (Markussen et al., 1995; Breitwieser et al., 1996; Vanzo et al., 2007). Although osk likely evolved in a last common insect ancestor (Lynch et al., 2011; Ewen-Campen et al., 2012; Blondel et al., 2020), the Long Osk domain and isoform appear to have evolved only within the Diptera (Blondel et al., 2020).
Evidence for a role for osk in the nervous system comes from studies of two insects, D. melanogaster and the cricket Gryllus bimaculatus. In the cricket, Gb-osk mRNA and protein are enriched in neuroblasts in the embryonic nervous system (Ewen-Campen et al., 2012) and in the adult brain (Ewen-Campen and Extavour, unpublished). First identified in a grasshopper (Wheeler, 1891), neuroblasts are neural stem cells found in all pancrustaceans (insects and crustaceans) (Lear, 2001; Richter et al., 2010). Neuroblasts arise from the ventral ectoderm during embryogenesis and divide asymmetrically to produce all of the neurons of the nervous system. Gb-osk RNAi in cricket embryos results in broken or reduced lateral axon tracts, a phenotype that is consistent with neuronal division defects (Ewen-Campen et al., 2012). Neuroblasts of G. bimaculatus also express Vasa and Piwi proteins (Ewen-Campen et al., 2012), raising the possibility that Osk may interact with these proteins in neuroblasts, as it does in the germ line in other contexts (see section on Vasa below). In D. melanogaster, osk co-localizes with nanos (nos) mRNA in larval class IV neurons, and is required for correct localization of nos mRNA within these neurons (Xu et al., 2013).
Gene expression data suggest that osk also specifies germ cells in the ant Messor pergandei, and osk knockdown experiments in the wasp Nasonia vitripennis show that the germ cell and posterior identity specification roles of osk are conserved in this insect as well (Lynch et al., 2011). However, osk is not required for germ line establishment, maintenance or function in the cricket G. bimaculatus (Orthoptera) (Ewen-Campen et al., 2012).
PIWI proteins are evolutionarily conserved RNA binding proteins (e.g., Bohmert et al., 1998; Moussian et al., 1998; reviewed in Thomson and Lin, 2009; Ku and Lin, 2014) found across metazoan and plant genomes (Chen et al., 2006). The founder ortholog of this group was first identified in a D. melanogaster screen for genes that abolish asymmetrical divisions in germ line stem cells (GSCs) (Lin and Spradling, 1997), and named after the male sterility phenotype caused by loss of function mutations (PIWI: P-element induced wimpy testis). The PIWI clade of proteins belongs to the Argonaut/PIWI protein family (AGO/PIWI, also known as the PAZ-PIWI domain or PPD family of proteins) (Thomson and Lin, 2009; Ku and Lin, 2014).
PIWI proteins are expressed in germ cells or their progenitors in many animals, and their functions in the germ line have been extensively studied in a wide range of animals (Juliano et al., 2011). PIWI germ line functions include germ line determination, germ line stem cell (GSC maintenance), spermiogenesis, and silencing transposon expression in the germ line genome both at the epigenetic and post-transcriptional levels (Thomson and Lin, 2009; Ku and Lin, 2014). The latter role is performed via interaction with small RNAs, including but not limited to PIWI-associated small RNAs (piRNAs) (Iwasaki et al., 2015; Furrer et al., 2017; Rojas-Rios and Simonelig, 2018). Like vasa, piwi is also expressed in multiple somatic stem cell types outside of bilaterians.
PIWI-related proteins play critical functions in the soma as well as the germ line (Ross et al., 2014). This includes roles in the central nervous system of all major groups of animals, including deuterostomes, protostomes, and bilaterian outgroups (Juliano et al., 2011), as illustrated by the following examples: In the sea slug A. californica, Piwi protein interacts with a DNA methyltransferase to control the expression of CREB2, a long-term memory repressor, during long-term memory formation (Rajasethupathy et al., 2012). The zebrafish piwi ortholog ziwi is expressed in the eye, the forebrain, and the midbrain during organogenesis (Tan et al., 2002). In the nematode C. elegans, the PIWI protein PRG-1 represses axonal regeneration in adult mechanosensory neurons (Kim et al., 2018). Mouse piwi orthologs (miwi genes) are expressed in the adult brain (Leighton et al., 2019), and miwi colocalizes with piRNAs to form RNP puncta in the dendrites of cultured hippocampal neurons (Lee et al., 2011). LNA-based antisense inhibition of one of these piRNAs results in a significant decrease in dendrite spine area (Lee et al., 2011). Further, knockdown of piwi-like genes in the mouse hippocampus affects adult behavior, as assayed in an experimental fear-conditioning paradigm (Leighton et al., 2019). In D. melanogaster, PIWI-related proteins Argonaute (Ago3) and Aubergine (Aub) are expressed at different levels in distinct subsets of neurons in the mushroom body (Perrat et al., 2013), the substrate for learning and memory within the insect brain (Heisenberg, 2003). Lower expression levels of Ago3 and Aub correlate with increased expression of transposable elements in the adult fly brain (Perrat et al., 2013), consistent with the proposed role of Piwi-related proteins in suppressing transposable element mobility (Thomson and Lin, 2009; Ku and Lin, 2014). This heterogeneity of Aub and Ago expression levels is speculated to contribute to behavioral variability (Perrat et al., 2013).
Expression of piwi orthologs during gametogenesis has been documented in multiple cnidarians (Seipel et al., 2004; Leclère et al., 2012; Plickert et al., 2012). The homoscleromorph sponge O. lobularis expresses a piwi ortholog in germ cells during spermatogenesis and oogenesis (Fierro-Constain et al., 2017). In another sponge, the demosponge Ephydatia fluviatilis, a piwi homolog is expressed in choanocytes and archeocytes (Funayama, 2010; Funayama et al., 2010; Alié et al., 2015), which is relevant to the sponge germ line because gametogenic cells are thought to be derived from one or both of these cell types in these animals (Funayama, 2010). In the ctenophore Pleurobrachia pileus, piwi is expressed in the adult male and female germ line (Alié et al., 2010). Neural cell type expression of piwi orthologs is also present in non-bilaterians. In the cnidarian Clytia hemisphaerica, piwi is expressed in nematogenic and neural stem cells (Denker et al., 2008), and in the ctenophore P. pileus, piwi is expressed in the apical organ, which is an aboral sensory organ (Alié et al., 2010).
vasa encodes a highly conserved DEAD box-containing ATP-dependent RNA helicase (Hay et al., 1988; Lasko and Ashburner, 1988) that is expressed in the germ line of every animal studied to date (Ewen-Campen et al., 2010; Gustafson and Wessel, 2010; Yajima and Wessel, 2011b). DEAD box helicases predate animals (Chen et al., 2006) and are implicated in a broad range of biological functions including transcription, translation, splicing, ribosome biogenesis, nuclear export, and mRNA degradation (Linder, 2006; Lasko, 2013). vasa expression is also a hallmark of many types of stem cells, where it is proposed to interact with the products of the piwi, bruno, and PL10 genes in a conserved gene network to help maintain pluripotency (Alié et al., 2010; Juliano et al., 2010; Fierro-Constain et al., 2017).
First discovered for its role in abdomen formation during embryonic development in D. melanogaster (Schüpbach and Wieschaus, 1986), vasa encodes a protein found in the cytoplasm of animal germ cells and required for one or both of germ cell specification and germ line development in multiple animals (reviewed in Yajima and Wessel, 2011b). Vasa protein is a component of germ line RNP granules, and has predicted roles in regulating mRNA translation, including that of nanos (Gavis et al., 1996; see below) and gurken (Tomancak et al., 1998), potentially by interacting with initiation factor dIF2 (Carrera et al., 2000). During the cell cycle, vasa may be regulated by the meiotic checkpoint pathway (Ghabrial and Schupbach, 1999), can associate with the spindle (Carré et al., 2002; Oyama and Shimizu, 2007), and is implicated in regulation of mitotic chromosome condensation (Pek and Kai, 2011; Yajima and Wessel, 2011a, b; Schwager et al., 2015). Vasa protein interacts physically with Piwi protein in the germ line of mice (Kirino et al., 2010) and D. melanogaster (Megosh et al., 2006), and in cultured ovarian cells of the silkworm Bombyx mori (Xiol et al., 2014). Vasa, like Piwi, is involved in the small RNA biogenesis pathway in many animals (Vagin et al., 2004; Shirayama et al., 2014; Xiol et al., 2014; Dehghani and Lasko, 2016; Spracklin et al., 2017). Vasa and Osk proteins also physically interact in the germ line, where the LOTUS domain of Osk binds Vasa and facilitates its helicase activity (Jeske et al., 2015; Yang et al., 2015; Jeske et al., 2017).
To our knowledge, the only reported examples of a role for vasa in the nervous system come from (1) the cricket G. bimaculatus, where it is found co-expressed along with piwi and osk in neuroblasts (Ewen-Campen et al., 2012), and (2) cells of the apical sensory organ in the ctenophore Pleurobrachia pileus (Alié et al., 2010). Its function in these invertebrate nervous systems remains to be elucidated.
We note that in multiple animals, vasa expression is also a hallmark of pluripotent and somatic stem cell lineages, which can give rise to both germ line and neural cells. These include the archaeocytes of the sponge E. fluviatilis (Alié et al., 2015), the interstitial cells of the cnidarians H. magnipapillata and H. echinata (Mochizuki et al., 2001; Rebscher et al., 2008), the presumptive founder cells of the larval posterior growth zone of the annelid Platynereis dumerilii (Rebscher et al., 2007), the stem cells of the colonial tunicate Botryllus schlosseri (Sunanaga et al., 2006; Rosner et al., 2009; Kawamura and Sunanaga, 2011), and the neoblasts of the platyhelminths Macrostomum lignano (Pfister et al., 2008), Dugesia japonica (Shibata et al., 1999), Schmidtea mediterranea (Wagner et al., 2012), and Schistosoma mansoni (Wang et al., 2013).
boule (bol) is a member of the Deleted in Azoospermia (DAZ) RNA-binding protein family, which contains the autosomal dazl and bol genes, and the human Y-linked DAZ gene (Shah et al., 2010). Although not reported in plant or fungal genomes to date, bol may predate animals based on identification of a putative ortholog in the slime mold Dictyostelium discoideum (Chen et al., 2006). In the bony fish lineage, a bol duplication likely gave rise to the Daz-like gene (Dazl), which then underwent a transposition to the Y chromosome in primates to give rise to DAZ (Shah et al., 2010). DAZ family members display predominant male germ line expression patterns, and DAZ family genes are crucial for germ cell development and meiotic progression across animals (summarized in Kim and Rhee, 2016).
bol was first identified in a mutagenesis screen for D. melanogaster male-sterile mutants (Castrillon et al., 1993). twe was identified by multiple independent studies (Jimenez et al., 1990; Alphey et al., 1992; Courtot et al., 1992) that searched for orthologous or functionally analogous genes to the cdc25 phosphatase that regulates mitotic entry in Schizosaccharomyces pombe (Russell and Nurse, 1986). bol mutants fail to undergo male meiosis, but homozygous female bol mutants are fertile (Eberhart C. G. et al., 1996). Bol controls the translation of twe, allowing meiotic entry in males (Courtot et al., 1992; Maines and Wasserman, 1999). The D. melanogaster meiotic entry defect can be rescued by the X. laevis bol ortholog Xdazl (Houston et al., 1998), and human and mouse DAZ can also partially rescue D. melanogaster bol loss of function (Houston et al., 1998; Xu et al., 2003). Orthologs of bol and twe also play a role in sperm maturation in haploid males in the sawfly Athalia rosae (Hymenoptera), which normally abort meiosis I but maintain meiosis II to produce haploid sperm (Sekine et al., 2015). As in D. melanogaster, bol knockdowns in A. rosae show no apparent defects in females (Sekine et al., 2015). Bol is also expressed in the testis in male mammals. In mice and humans, Bol protein is present in the cytoplasm of developing spermatocytes and can be detected through meiosis (Xu et al., 2001). Loss of dazl function in mice leads to defects in gametogenesis in both sexes (Ruggiu et al., 1997). As in D. melanogaster, bol homozygous mutant male mice are infertile, but females are viable and fertile (Shah et al., 2010). Bol also co-localizes to RNPs that form under stress (called stress granules) in mouse male germ cells (Kim and Rhee, 2016). In X. laevis, knockdown of the maternally expressed ortholog Xdazl reduces the number of PGCs and perturbs PGC migration during embryogenesis (Houston and King, 2000). In contrast to the fly, mouse, human and frog bol genes, the C. elegans bol ortholog daz-1 plays a role in oocyte determination rather than in spermatogenesis (Karashima et al., 2000). In wild type hermaphroditic worms, germ cells undergo two developmental decisions, the first from mitotic proliferation to meiosis in the L4 larval stage, and the second from sperm to oocyte production in young adults (Karashima et al., 2000). RNAi against daz-1 in C. briggsae leads to continuous sperm production, indicating a disruption in the spermatogenesis/oogenesis switch (Karashima et al., 2000).
While bol expression is not detected in the human brain (Uhlen et al., 2015), bol and twe also function in the nervous system in adult D. melanogaster (Joiner and Wu, 2004), where an isoform of bol that is not found in the testis is expressed in the cytoplasm and extending neurites of most cells throughout the adult brain (Joiner and Wu, 2004). Bol negatively regulates developmental axon pruning in D. melanogaster (Hoopfer et al., 2008). Over-expression of bol throughout the nervous system leads to defects in neuronal communication between the retina and the lamina, abnormal locomotory behavior in wandering larvae, and lethality before the third larval stage (Joiner and Wu, 2004). The neuronal bol isoform interacts genetically with twe in the nervous system, just as bol does in the germ line (Joiner and Wu, 2004).
Cytoplasmic polyadenylation element binding protein (CPEB) is a member of an animal protein family implicated in binding the 3’UTRs of mRNAs at cytoplasmic polyadenylation element (CPE) sites, and in controlling their translation and cytoplasmic localization via regulation of their poly(A) tail lengths (Hake and Richter, 1994; Wells et al., 2000). Some animals have multiple paralogs of CPEB genes in their genomes: D. melanogaster has two CPEB genes, whereas X. laevis, mice, humans and C. elegans have four (Chen et al., 2006). The C-terminal half of the CPEB protein contains RNA binding domains (RBDs), including two RNA-recognition motifs (RRM domains) and a zinc finger domain (ZZ domain), which are used to establish CPEB gene relationships (Hake et al., 1998; Mendez and Richter, 2001; Fernandez-Miranda and Mendez, 2012). Pairwise sequence alignments of the RBDs of different CPEB genes show that CPEB genes form two subgroups (Hake et al., 1998; Mendez and Richter, 2001; Fernandez-Miranda and Mendez, 2012). One subgroup, which includes the D. melanogaster oo18 RNA binding protein (orb) (Christerson and McKearin, 1994; Lantz et al., 1994), mouse CPEB1 (Tay and Richter, 2001) and X. laevis CPEB1 (Hake and Richter, 1994), are expressed and required in the germ line for initiation of translation of CPE-containing mRNAs. CPEB genes in the second group are more broadly expressed in several somatic tissues, including the nervous system, in addition to the germ line. Their examples include D. melanogaster orb2 (Hafer et al., 2011), mouse CPEB2-4 (Kurihara et al., 2003; Theis et al., 2003), and human CPEB3 and CPEB4 (Kikuno et al., 2004). Given that CPEB genes control mRNA expression across tissues, developmental stages and species, some have speculated that they do so via a mechanism of local translational control that is evolutionarily conserved, involving the cytoskeleton, eukaryotic initiation factor (eIF4E) and the eIF4E binding protein Maskin (Stepien et al., 2016).
D. melanogaster orb was the first identified member of the CPEB family of translational regulators and is required to establish polarity in developing eggs and early embryos (Lantz et al., 1992, 1994; Christerson and McKearin, 1994). orb controls translation and polyadenylation of mRNAs including oskar and gurken (Chang et al., 1999, 2001; Tan et al., 2001; Castagnetti and Ephrussi, 2003; Norvell et al., 2015; Davidson et al., 2016), and organizes and repolarizes the microtubule cytoskeleton during D. melanogaster oogenesis by interacting with Actin, Dynein and Kinesin (Barr et al., 2019a, b). In C. elegans, CPEB homologs (called CPB-1,2,3 and FOG-1) are required for the switch from sperm to egg production during germ cell development, and control germ cell fate by regulating the translation of specific mRNAs (Luitjens et al., 2000; Jin et al., 2001). CPEB interactions are also well studied in X. laevis oocytes, where CPEB homologs are required for normal oocyte maturation, and also regulate the cell cycle in early embryos (Stebbins-Boaz et al., 1996; Groisman et al., 2000; Groisman et al., 2002; Igea and Mendez, 2010). Co-immunoprecipitation, protein pull downs and yeast two-hybrid assays have shown that in X. laevis, CPEB1 directly binds both the eukaryotic translation initiation factor eIF4E, and the 4E-binding protein Maskin (Stebbins-Boaz et al., 1999; Cao and Richter, 2002; Meijer et al., 2007). It has been suggested that such a CPEB-Maskin-eIF4E interaction may serve as a typical example for 3′UTR-mediated translational repression across metazoans (Stebbins-Boaz et al., 1999). Indeed, D. melanogaster Orb from ovary extracts has also been shown to immuno precipitate with eIF4E (Wong et al., 2011).
D. melanogaster orb2 is expressed in several somatic tissues, including the nervous system at all stages of development (Hafer et al., 2011). orb2 mRNA and protein expression are detectable in the central and peripheral embryonic nervous systems (Hafer et al., 2011). In the central nervous system of embryos and larvae, Orb2 protein expression is largely limited to cell bodies, and functions in asymmetrical cell division (Hafer et al., 2011). In adult neurons, orb2 is localized at the synaptic terminals, and is required for learning and memory (Keleman et al., 2007; Kruttner et al., 2012; Majumdar et al., 2012). In the sea slug A. californica, CPEB forms prion-like multimers in neurons. D. melanogaster Orb2 injected into A. californica neurons also forms such aggregates (Si et al., 2003, 2010), suggesting that these aggregates may be relevant to learning and memory in these animals, as they may contribute to synapse-specific differences (Fiumara et al., 2015). In sensory neurons, ApCPEB co-localizes in RNA granules that also contain eIF4E, FMRP, and Stau (Barbee et al., 2006; Chae et al., 2010). A second A. californica CPEB homolog, ApCPEB4, has a role in long-term facilitation, although it lacks a prion-like domain (Lee et al., 2016). In both mammalian and A. californica neurons, CPEB is required for mRNA shuttling, and it co-localizes with and polyadenylates multiple mRNAs (Huang et al., 2002, 2003; Chae et al., 2010). In X. laevis and mouse neurons, CPEB colocalizes with Maskin in a complex containing Kinesin and Dynein, suggesting that it may regulate mRNA transport and translation in dendrites (Huang et al., 2003) similar to its role in the germ line. In mice, CPEB3 interacts with the Actin cytoskeleton and has been shown to act as a functional prion as well (Stephan et al., 2015), with CPEB expression at synapses in rodent brains being required for synaptic plasticity (Wu et al., 1998), the cellular basis for memory and learning.
Here we have highlighted many genes that, following their initial characterization in the germ line, were discovered to also have neural roles. For many such genes with a neural role in one species, there is evidence for a neural role in other species as well, often with the same set of core molecular interaction partners (Table 2). We consider that the data currently available are too limited for us to propose whether the germ line roles or the neural roles of these genes represent their putative ancestral functions in a last common ancestor of animals [but see Ewen-Campen et al. (2012) for a proposal that oskar’s role in the insect germ line is derived, resulting from co-option from a putative neural role]. It is clear that relying on single gene expression patterns alone to identify homologies can be misleading (Wagner et al., 2012; Wang et al., 2013), and we are not proposing to use such data as the sole criteria for this purpose (Tautz, 1998; Nielsen and Martinez, 2003). Instead, our aim here is to suggest possible explanations for the molecular and cellular basis for this pleiotropy by looking at the properties of the molecular mechanisms of these shared genes, which may be linked to the evolution of cell-type specific functions.
We begin by highlighting some independent yet interesting similarities between the germ line and the nervous system. First, germ cells, pluripotent stem cells, and undifferentiated or abnormally organized embryonic cells have been reported to differentiate towards neural cell fate under a number of circumstances. For example, in C. elegans, germ cells that lose P-granules can ectopically express somatic markers, including neuronal markers (Knutson et al., 2017). In induced human PGC-like cells generated from pluripotent stem cells, BLIMP1 is actively required to promote PGC fate and to repress neuronal differentiation (Sasaki et al., 2015). Dissociated X. laevis embryonic animal cap cells are able to upregulate the neural marker N-CAM despite the absence of normal spatial organization (Sato and Sargent, 1989) Embryonic stem (ES) cells spontaneously and readily exhibit aspects of neural identity under specific culture conditions (Tropepe et al., 2001). When plated at low densities in phosphate buffered saline, mouse ESCs can express nestin and Sox1, which is suggestive of neural stem cell differentiation (Smukler et al., 2006). It has therefore been suggested that neuronal fate is a preferred differentiation program for cells that lose their germ line identity or pluripotency (Knutson et al., 2017).
Second, the gene expression profiles of human and mouse testes and brain are highly similar to each other (Guo et al., 2003, 2005). Whether or how the two tissues communicate to regulate this similar gene expression is unclear, although Guo and colleagues (Guo et al., 2005) speculate that the hypothalamus-pituitary-gonadal axis (Plant, 2015; Kaprara and Huhtaniemi, 2018) may play a role. Finally, such observations may also help explain the link between disruption of genes with known roles in the germ line, and neural disease phenotypes. For example, the D. melanogaster tumor suppressor gene brain tumor (brat), together with nos and pum, represses translation in female germ line stem cells (Sonoda and Wharton, 2001), and brat loss of function mutations also cause tumors in the brain (Arama et al., 2000). Additionally, ectopic expression of at least 26 genes normally expressed in the germ line, may be linked to malignant brain tumor growth in D. melanogaster (Janic et al., 2010). Thus, it is possible that some shared or similar biological processes link these genes to both germ line and neural tissue types outside of mammals as well.
In this review we have summarized some of the evidence for the expression and functional requirements for a number of genes in the above mentioned two cell types. However, in most cases the molecular mechanisms linking the function of these genes to the cellular execution of neural or germ line fate remain unclear. It is therefore difficult to determine whether this pleiotropy is a result of the same molecular function in apparently unrelated biological processes, or because some or all of these genes have multiple molecular functions per gene. In principle, it could be the case that these genes have the same immediate downstream partners in both tissue contexts, but their subsequent interactors or secondary targets are different, leading to differences in cellular responses to the activities of these genes within each tissue. Nevertheless, in the following section, we propose some possible explanations, based on shared molecular functions of these genes, for the potentially close or labile relationship between germ line and neural cell fates.
One way to understand the repeated conservation of expression, molecular function and interactions of these genes in neural tissues and germ lines, is by considering whether the products of these genes have functional or biochemical properties that could make them particularly suited for use by these cell types. We note that products of most of the genes discussed here share three notable properties. First, they are RNA binding proteins (e.g., Osk, Piwi, Vasa, Stau, Nos, FMRP), and play multiple roles in RNA biology including localization (e.g., Stau, Osk), translational activation (e.g., Vas, and Stau), and translational repression (e.g., Nos, Pum, Stau). Second, many of them break cellular symmetry by becoming asymmetrically localized within the cytoplasm or facilitating the asymmetrical localization of other molecules (e.g., Osk, Stau/Btz, Nos). Third, the majority catalyze the formation of and/or localize to RNP granule complexes, which are in turn sometimes asymmetrically distributed within the cell (e.g., germ granules in D. melanogaster). RNP granules are electron dense, non-membrane bound cytoplasmic aggregates of RNAs and proteins (Eddy, 1975; Ikenishi, 1998). The assembly of proteins within RNPs is often transient or reversible, and RNPs are important for the localization, stability and translational control of their RNA (and protein) cargo (Arkov and Ramos, 2010; Voronina et al., 2011; Schisa, 2012; Gao and Arkov, 2013). Moreover, in addition to giving RNP granules their functionality in translational control, RNA Binding Proteins (RBPs) have been noted to commonly have regions of low sequence complexity and prion-like domains, both of which can mediate RNP granule assembly and disassembly (Brangwynne et al., 2009; Han et al., 2012; Kato et al., 2012; Molliex et al., 2015; Sudhakaran and Ramaswami, 2017).
RNP granules are found in both germ line and somatic cells. Depending on the tissue they are found in, RNP granules are referred to in the literature by various names, including polar or germinal granules in germ cells, stress granules and processing bodies in somatic cells, and neuronal granules in neurons (reviewed in Voronina et al., 2011). All described classes of RNP granules share multiple components with each other (reviewed in Kulkarni and Extavour, 2017). Functional amyloid-like assemblies like RNP granules can govern cellular processes both in the germ line, including PGC specification and spermatogenesis (reviewed in Voronina et al., 2011), and in the soma, including in the consolidation of memory in the nervous system (Si et al., 2003, 2010; Si and Kandel, 2016). In the latter context, proteins with prion-like domains, which may facilitate amyloid-like assemblies, localize at neuronal synapses and form active, stable complexes with self-perpetuating properties central to memory storage (Si et al., 2003, 2010; Sudhakaran and Ramaswami, 2017). We note that Oskar and FMRP have predicted prion-like domains (McBride et al., 2012; Boke et al., 2016). Germ line and neural cells also share the commonality of regulating translation at specific sites within the cell, e.g., the oocyte posterior in the case of germ plasm formation (Lehmann, 2016), or at select neuronal synapses in the case of neurons, leading to synaptic plasticity (Kang and Schuman, 1996; Si et al., 2003).
Piwi, its related protein Aubergine, and Vasa are among the many RNA binding proteins that are associated with and indispensable for small RNA biogenesis in the germ line (Ku and Lin, 2014). piRNAs are endogenous small non-coding RNAs that are proposed to maintain the genomic integrity of germ cells by limiting transposon mobility (Aravin et al., 2001, 2006; Girard et al., 2006; Grimson et al., 2008). piRNAs associate with the Argonaute family member Piwi (e.g., Mochizuki et al., 2002), and other members of this family (e.g., Ago3) interact with other small RNAs, including miRNAs and siRNAs (Girard et al., 2006; Vagin et al., 2006; Brennecke et al., 2007; Houwing et al., 2007; Kim et al., 2009). Small RNA-mediated gene silencing occurs at both transcriptional and post-transcriptional levels, and is an important mechanism controlling gene expression (Holoch and Moazed, 2015). piRNAs were first characterized in the germ line, but recent reports support their existence in somatic tissues as well, including neural tissues (Lee et al., 2011; Rajasethupathy et al., 2012; Ross et al., 2014). Indeed, in A. californica, after the germ line, the nervous system is amongst the tissue types that show relatively high selective enrichment for piRNAs (Rajasethupathy et al., 2012). There is evidence for primary piRNA biogenesis in the germ line and neurons (Rajasethupathy et al., 2012; Mani and Juliano, 2013; Kim et al., 2018) consistent, with a functional role for piRNAs in both cell types. For example, Piwi and piRNAs regulate Myosin-Va in the central nervous system of mammals (Naisbitt et al., 2000; Lee et al., 2011), control local translation in mouse neuronal dendrites (Lee et al., 2011), mouse dendritic spine development (Lee et al., 2011), neuronal migration (Viljetic et al., 2017), and may be linked to growth of malignant brain tumors (Janic et al., 2010). Finally, piRNAs regulate transposon activity both in the brain and in the germ line (reviewed in Mani and Juliano, 2013). Retrotransposons are highly active in neural tissues and contribute to proper neuronal differentiation and generation of somatic mosaicism in the brain (Muotri et al., 2005; Coufal et al., 2009). Thus, piRNAs are crucial both for the germ line, and for normal development and function of the nervous system, which may help explain why we observe that genes important for their biogenesis are expressed in both tissue types.
Co-option of partial or complete gene networks in different biological contexts is common (Jacob, 1977). Novel traits may evolve either by the co-option of pre-existing gene networks that operate in functional modules, or by building a new gene network for each new developmental context (Sanetra et al., 2005; Monteiro and Podlaha, 2009). Based on the observations summarized herein, we propose that the germ line and nervous tissues of animals contain examples of gene network co-option, given that the genes involved are pleiotropic, and that we do not think it likely that the germ line and nervous system are homologous organ systems. In principle, one way of co-opting a gene network could be by recruiting an upstream regulator of an existing network into a new developmental context. This is what we previously proposed may have happened in the case of oskar in germ plasm (Ewen-Campen et al., 2012). In both cricket (Ewen-Campen et al., 2012) and fly (Xu et al., 2013) nervous systems, oskar is co-expressed with vasa, piwi and/or nanos, genes whose products function together in multiple other cellular contexts as discussed above. Given that germ plasm in insects is likely a derived mechanism of PGC specification (Extavour and Akam, 2003; Lynch et al., 2011; Ewen-Campen et al., 2013), we propose that the functional links among these genes are likely to predate the evolution of insect germ plasm, suggesting that they were co-opted to the germ line context from a preexisting somatic role (Ewen-Campen et al., 2012).
When moving beyond insects to consider all animals, because there have been fewer reported instances to date of the expression or function of these genes in the nervous system outside of bilaterians, one might wish to hypothesize that the germ line functions of these gene evolved first, and then were co-opted to the nervous system in Bilateria. However, the functions of these genes have been explored primarily in a small number of study systems, heavily biased toward the Bilateria. Moreover, the diversity of cell types, including neural cell types, outside of Bilateria are not as well studied at the molecular level as are those of bilaterians. The evidence that the earliest metazoans were highly complex animals is mounting (Halanych, 2015; Whelan et al., 2017; Paps, 2018; Laumer et al., 2019), and may well displace the traditional view that early animals were “simple” with few differentiated cell types, lacking complex reproductive or sensory systems. We therefore consider it premature to speculate on whether the ancestral function of these genes in animals, was in the germ line or in the nervous system. Rather than thinking about the patterns in their putative ancestral functions in establishing a particular cell type, we could consider the hypothesis that the cellular function of translational control in RNP granules is the relevant conserved ancestral role of this machinery in eukaryotes. This could explain why striking phenotypes are particularly or easily observed in neurons and germ cells, because these cell types rely heavily on translational regulation for their biological functions. The advent of animal-specific genes like nos and osk may have permitted the emergence of tissue-specific versions of this machinery, deployed specifically in germ lines and nervous systems to refine or augment their regulation of translation.
We note that an association between many of the genes discussed herein and “stemness” or cellular multipotency, has already been pointed out by several researchers: the general proposal is that these genes may have been components of an ancestral animal toolkit in stem cells, regardless of the fate of their differentiated progeny (e.g., Alié et al., 2010, 2015; Juliano et al., 2010; Fierro-Constain et al., 2017). Here we speculate that if, as in many extant animals, ancient metazoans generated gametes from germ line stem cells, and/or neurons from neuroblasts, the observed association of these genes with pluripotency may also help explain the gene expression overlap in germ line and nervous tissues. Going forward, technical advances including single-cell RNA sequencing, chromatin architecture analysis and proteomics, and improved microscopy and computational methodologies including machine learning, might make it possible to test such hypotheses experimentally (e.g., Siebert et al., 2019). The case we have discussed here, of the germ line and the nervous system, is an example of the broader, fundamental question of how the same molecular mechanisms can underlie different cell identities. Once putative ancient cell type inventories are reconstructed for important evolutionary nodes, we can perhaps begin to unravel how ancient cell types, in some cases expressing highly similar machinery, diversified into extant cell types that make up the tissues and organ systems of living animals (Kin, 2015; Arendt et al., 2016), helping answer some of the questions that we have raised here.
CE conceived of the project. DL, AK, and CE compiled the evidence from primary literature. AK and CE wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Harvard University. DL was supported by the Harvard GSAS Research Scholar Initiative.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Samuel Church and Seth Donoughe from the Extavour lab for helpful comments, critical reading, and discussions on earlier versions of this manuscript. We also thank the reviewers for their comments and suggestions which helped improve the manuscript.
Alié, A., Hayashi, T., Sugimura, I., Manuel, M., Sugano, W., Mano, A., et al. (2015). The ancestral gene repertoire of animal stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E7093–E7100.
Alié, A., Leclère, L., Jager, M., Dayraud, C., Chang, P., Le Guyader, H., et al. (2010). Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev. Biol. 350, 183–197. doi: 10.1016/j.ydbio.2010.10.019
Alphey, L., Jimenez, J., White-Cooper, H., Dawson, I., Nurse, P., and Glover, D. M. (1992). twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69, 977–988. doi: 10.1016/0092-8674(92)90616-k
Anand, A., and Kai, T. (2012). The tudor domain protein Kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 31, 870–882. doi: 10.1038/emboj.2011.449
Anantharaman, V., Zhang, D., and Aravind, L. (2010). OST-HTH: a novel predicted RNA-binding domain. Biol. Direct 5:13. doi: 10.1186/1745-6150-5-13
Antar, L. N., Dictenberg, J. B., Plociniak, M., Afroz, R., and Bassell, G. J. (2005). Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 4, 350–359. doi: 10.1111/j.1601-183x.2005.00128.x
Arama, E., Dickman, D., Kimchie, Z., Shearn, A., and Lev, Z. (2000). Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 19, 3706–3716. doi: 10.1038/sj.onc.1203706
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. doi: 10.1038/nature04916
Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M., and Gvozdev, V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. doi: 10.1016/s0960-9822(01)00299-8
Arendt, D., Musser, J. M., Baker, C. V. H., Bergman, A., Cepko, C., Erwin, D. H., et al. (2016). The origin and evolution of cell types. Nat. Rev. 17, 744–757.
Arey, R. N., Kaletsky, R., and Murphy, C. T. (2019). Nervous system-wide profiling of presynaptic mRNAs reveals regulators of associative memory. Sci. Rep. 9:20314.
Ariz, M., Mainpal, R., and Subramaniam, K. (2009). C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 326, 295–304. doi: 10.1016/j.ydbio.2008.11.024
Arkov, A. L., and Ramos, A. (2010). Building RNA-protein granules: insight from the germline. Trends Cell Biol. 20, 482–490. doi: 10.1016/j.tcb.2010.05.004
Ashley, C. T. Jr., Wilkinson, K. D., Reines, D., and Warren, S. T. (1993). FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262, 563–566. doi: 10.1126/science.7692601
Babu, K., Cai, Y., Bahri, S., Yang, X., and Chia, W. (2004). Roles of bifocal, homer, and f-actin in anchoring oskar to the posterior cortex of Drosophila oocytes. Genes Dev. 18, 138–143. doi: 10.1101/gad.282604
Barbee, S. A., Estes, P. S., Cziko, A. M., Hillebrand, J., Luedeman, R. A., Coller, J. M., et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009. doi: 10.1016/j.neuron.2006.10.028
Barr, J., Charania, S., Gilmutdinov, R., Yakovlev, K., Shidlovskii, Y., and Schedl, P. (2019a). The CPEB translational regulator, orb, functions together with Par proteins to polarize the Drosophila oocyte. PLoS Genet. 15:e1008012. doi: 10.1371/journal.pgen.1008012
Barr, J., Gilmutdinov, R., Wang, L., Shidlovskii, Y., and Schedl, P. (2019b). The Drosophila CPEB protein orb specifies oocyte fate by a 3’UTR-dependent autoregulatory loop. Genetics 213, 1431–1446. doi: 10.1534/genetics.119.302687
Berger, S. M., Fernandez-Lamo, I., Schonig, K., Fernandez Moya, S. M., Ehses, J., Schieweck, R., et al. (2017). Forebrain-specific, conditional silencing of Staufen2 alters synaptic plasticity, learning, and memory in rats. Genome Biol. 18:222.
Bienkowski, R. S., Banerjee, A., Rounds, J. C., Rha, J., Omotade, O. F., Gross, C., et al. (2017). The conserved, disease-associated RNA binding protein dNab2 interacts with the fragile X protein ortholog in drosophila neurons. Cell Rep. 20, 1372–1384. doi: 10.1016/j.celrep.2017.07.038
Blondel, L., Jones, T. E. M., and Extavour, C. G. (2020). Bacterial contribution to genesis of the novel germ line determinant oskar. eLife 9:e45539.
Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. doi: 10.1093/emboj/17.1.170
Boke, E., Ruer, M., Wuhr, M., Coughlin, M., Lemaitre, R., Gygi, S. P., et al. (2016). Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650. doi: 10.1016/j.cell.2016.06.051
Bolduc, F. V., Bell, K., Cox, H., Broadie, K. S., and Tully, T. (2008). Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat. Neurosci. 11, 1143–1145. doi: 10.1038/nn.2175
Bono, F., Ebert, J., Lorentzen, E., and Conti, E. (2006). The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725. doi: 10.1016/j.cell.2006.08.006
Bozzetti, M. P., Specchia, V., Cattenoz, P. B., Laneve, P., Geusa, A., Sahin, H. B., et al. (2015). The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J. Cell Sci. 128, 2070–2084. doi: 10.1242/jcs.161810
Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. doi: 10.1126/science.1172046
Breitwieser, W., Markussen, F.-H., Horstmann, H., and Ephrussi, A. (1996). Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 10, 2179–2188. doi: 10.1101/gad.10.17.2179
Brendza, R., Serbus, L., Saxton, W., and Duffy, J. (2002). Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 12, 1541–1545. doi: 10.1016/s0960-9822(02)01108-9
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi: 10.1016/j.cell.2007.01.043
Broadus, J., Fuerstenberg, S., and Doe, C. Q. (1998). Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 391, 792–795. doi: 10.1038/35861
Brown, V., Small, K., Lakkis, L., Feng, Y., Gunter, C., Wilkinson, K. D., et al. (1998). Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem. 273, 15521–15527. doi: 10.1074/jbc.273.25.15521
Cao, Q., and Richter, J. D. (2002). Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 21, 3852–3862. doi: 10.1093/emboj/cdf353
Carré, D., Djediat, C., and Sardet, C. (2002). Formation of a large vasa-positive granule and its inheritance by germ cells in the enigmatic chaetognaths. Development 129, 661–670.
Carrera, P., Johnstone, O., Nakamura, A., Casanova, J., Jackle, H., and Lasko, P. (2000). VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell. 5, 181–187. doi: 10.1016/s1097-2765(00)80414-1
Castagnetti, S., and Ephrussi, A. (2003). Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development 130, 835–843. doi: 10.1242/dev.00309
Castrillon, D. H., Gönczy, P., Alexander, S., Rawson, R., Eberhat, C. G., Viswanathan, S., et al. (1993). Toward a molecular genetics analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single p element mutagenesis. Genetics 135, 489–505.
Chae, Y. S., Lee, S. H., Cheang, Y. H., Lee, N., Rim, Y. S., Jang, D. J., et al. (2010). Neuronal RNA granule contains ApCPEB1, a novel cytoplasmic polyadenylation element binding protein, in Aplysia sensory neuron. Exp. Mol. Med. 42, 30–37.
Chang, J. S., Tan, L., and Schedl, P. (1999). The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev. Biol. 215, 91–106. doi: 10.1006/dbio.1999.9444
Chang, J. S., Tan, L., Wolf, M. R., and Schedl, P. (2001). Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 128, 3169–3177.
Chen, F., Mackey, A. J., Stoeckert, C. J. Jr., and Roos, D. S. (2006). OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–D368.
Chen, G., Li, W., Zhang, Q. S., Regulski, M., Sinha, N., Barditch, J., et al. (2008). Identification of synaptic targets of Drosophila pumilio. PLoS Computat. Biol. 4:e1000026. doi: 10.1371/journal.pcbi.1000026
Christerson, L. B., and McKearin, D. M. (1994). orb is required for anteroposterior and dorsoventral patterning during Drosophila oogenesis. Genes Dev. 8, 614–628. doi: 10.1101/gad.8.5.614
Comery, T. A., Harris, J. B., Willems, P. J., Oostra, B. A., Irwin, S. A., Weiler, I. J., et al. (1997). Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl. Acad. Sci. U.S.A. 94, 5401–5404. doi: 10.1073/pnas.94.10.5401
Costa, A., Wang, Y., Dockendorff, T. C., Erdjument-Bromage, H., Tempst, P., Schedl, P., et al. (2005). The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev. Cell 8, 331–342. doi: 10.1016/j.devcel.2005.01.011
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Yeo, G. W., Mu, Y., Lovci, M. T., et al. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131. doi: 10.1038/nature08248
Courtot, C., Fankhauser, C., Simanis, V., and Lehner, C. F. (1992). The Drosophila cdc25 homolog twine is required for meiosis. Development 116, 405–416.
Crittenden, S. L., Bernstein, D. S., Bachorik, J. L., Thompson, B. E., Gallegos, M., Petcherski, A. G., et al. (2002). A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660–663. doi: 10.1038/nature754
Darnell, J. C., and Klann, E. (2013). The translation of translational control by FMRP: therapeutic targets for FXS. Nat. Neurosci. 16, 1530–1536. doi: 10.1038/nn.3379
Darnell, J. C., Van Driesche, S. J., Zhang, C., Hung, K. Y., Mele, A., Fraser, C. E., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi: 10.1016/j.cell.2011.06.013
Davidson, A., Parton, R. M., Rabouille, C., Weil, T. T., and Davis, I. (2016). Localized translation of gurken/TGF-alpha mRNA during axis specification is controlled by access to Orb/CPEB on processing bodies. Cell Rep 14, 2451–2462. doi: 10.1016/j.celrep.2016.02.038
De Santis, L., Gandolfi, F., Pennarossa, G., Maffei, S., Gismano, E., Intra, G., et al. (2015). Expression and intracytoplasmic distribution of staufen and calreticulin in maturing human oocytes. J. Assist. Reprod. Genet. 32, 645–652. doi: 10.1007/s10815-015-0437-y
Dehghani, M., and Lasko, P. (2016). C-terminal residues specific to Vasa among DEAD-box helicases are required for its functions in piRNA biogenesis and embryonic patterning. Dev. Genes Evol. 226, 401–412. doi: 10.1007/s00427-016-0560-5
Denker, E., Manuel, M., Leclère, L., Le Guyader, H., and Rabet, N. (2008). Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa. Cnidaria). Dev. Biol. 315, 99–113. doi: 10.1016/j.ydbio.2007.12.023
Devys, D., Lutz, Y., Rouyer, N., Bellocq, J. P., and Mandel, J. L. (1993). The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 4, 335–340. doi: 10.1038/ng0893-335
Dictenberg, J. B., Swanger, S. A., Antar, L. N., Singer, R. H., and Bassell, G. J. (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939. doi: 10.1016/j.devcel.2008.04.003
Dubnau, J., Chiang, A.-S., Grady, L., Barditch, J., Gossweiler, S., McNeil, J., et al. (2003). The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13, 286–296. doi: 10.1016/s0960-9822(03)00064-2
Dunn, C. W., Leys, S. P., and Haddock, S. H. (2015). The hidden biology of sponges and ctenophores. Trends Ecol. Evol. 30, 282–291. doi: 10.1016/j.tree.2015.03.003
Eberhart, C. G., Maines, J. Z., and Wasserman, S. A. (1996). Meiotic cell cycle requirement for a fly homologue of human deleted in azoospermia. Nature 381, 783–785. doi: 10.1038/381783a0
Eberhart, D. E., Malter, H. E., Feng, Y., and Warren, S. T. (1996). The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 5, 1083–1091. doi: 10.1093/hmg/5.8.1083
Eddy, E. M. (1975). Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 43, 229–280. doi: 10.1016/s0074-7696(08)60070-4
Ephrussi, A., Dickinson, L. K., and Lehmann, R. (1991). Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66, 37–50. doi: 10.1016/0092-8674(91)90137-n
Erdelyi, M., Michon, A. M., Guichet, A., Glotzer, J. B., and Ephrussi, A. (1995). Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature 377, 524–527. doi: 10.1038/377524a0
Estes, P. S., O’Shea, M., Clasen, S., and Zarnescu, D. C. (2008). Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol. Cell. Neurosci. 39, 170–179. doi: 10.1016/j.mcn.2008.06.012
Ewen-Campen, B., Donoughe, S., Clarke, D. N., and Extavour, C. G. (2013). Germ cell specification requires zygotic mechanisms rather than germ plasm in a basally branching insect. Curr. Biol. 23, 835–842. doi: 10.1016/j.cub.2013.03.063
Ewen-Campen, B., Schwager, E. E., and Extavour, C. G. (2010). The molecular machinery of germ line specification. Mol. Reprod. Dev. 77, 3–18. doi: 10.1002/mrd.21091
Ewen-Campen, B., Srouji, J. R., Schwager, E. E., and Extavour, C. G. (2012). oskar Predates the evolution of germ plasm in insects. Curr. Biol. 22, 2278–2283. doi: 10.1016/j.cub.2012.10.019
Extavour, C. G., and Akam, M. E. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884. doi: 10.1242/dev.00804
Extavour, C. G., Pang, K., Matus, D. Q., and Martindale, M. Q. (2005). Vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol. Dev. 7, 201–215. doi: 10.1111/j.1525-142x.2005.05023.x
Feng, Y., Absher, D., Eberhart, D. E., Brown, V., Malter, H. E., and Warren, S. T. (1997). FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1, 109–118. doi: 10.1016/s1097-2765(00)80012-x
Fernandez-Miranda, G., and Mendez, R. (2012). The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res. Rev. 11, 460–472. doi: 10.1016/j.arr.2012.03.004
Feuge, J., Scharkowski, F., Michaelsen-Preusse, K., and Korte, M. (2019). FMRP modulates activity-dependent spine plasticity by binding Cofilin1 mRNA and regulating localization and local translation. Cereb. Cortex 29, 5204–5216. doi: 10.1093/cercor/bhz059
Fierro-Constain, L., Schenkelaars, Q., Gazave, E., Haguenauer, A., Rocher, C., Ereskovsky, A., et al. (2017). The conservation of the germline multipotency program, from sponges to vertebrates: a stepping stone to understanding the somatic and germline origins. Genome Biol. Evol. 9, 474–488.
Fiumara, F., Rajasethupathy, P., Antonov, I., Kosmidis, S., Sossin, W. S., and Kandel, E. R. (2015). MicroRNA-22 gates long-term heterosynaptic plasticity in aplysia through presynaptic regulation of CPEB and downstream targets. Cell Rep. 11, 1866–1875. doi: 10.1016/j.celrep.2015.05.034
Forbes, A., and Lehmann, R. (1998). Nanos and pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690.
Fritzsche, R., Karra, D., Bennett, K. L., Ang, F. Y., Heraud-Farlow, J. E., Tolino, M., et al. (2013). Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 5, 1749–1762. doi: 10.1016/j.celrep.2013.11.023
Funayama, N. (2010). The stem cell system in demosponges: insights into the origin of somatic stem cells. Dev. Growth. Differ 52, 1–14. doi: 10.1111/j.1440-169x.2009.01162.x
Funayama, N., Nakatsukasa, M., Mohri, K., Masuda, Y., and Agata, K. (2010). Piwi expression in archeocytes and choanocytes in demosponges: insights into the stem cell system in demosponges. Evol. Dev. 12, 275–287. doi: 10.1111/j.1525-142x.2010.00413.x
Furrer, D. I., Swart, E. C., Kraft, M. F., Sandoval, P. Y., and Nowacki, M. (2017). Two sets of piwi proteins are involved in distinct sRNA pathways leading to elimination of germline-specific DNA. Cell Rep. 20, 505–520. doi: 10.1016/j.celrep.2017.06.050
Gamberi, C., Peterson, D. S., He, L., and Gottlieb, E. (2002). An anterior function for the Drosophila posterior determinant pumilio. Development 129, 2699–2710.
Gao, M., and Arkov, A. L. (2013). Next generation organelles: structure and role of germ granules in the germline. Mol. Reprod. Dev. 80, 610–623. doi: 10.1002/mrd.22115
Gardiol, A., and St Johnston, D. (2014). Staufen targets coracle mRNA to Drosophila neuromuscular junctions and regulates GluRIIA synaptic accumulation and bouton number. Dev. Biol. 392, 153–167. doi: 10.1016/j.ydbio.2014.06.007
Gavis, E. R., Lunsford, L., Bergsten, S. E., and Lehmann, R. (1996). A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122, 2791–2800.
Ghabrial, A., and Schupbach, T. (1999). Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nature Cell Biol. 1, 354–357. doi: 10.1038/14046
Ghosh, S., Marchand, V., Gaspar, I., and Ephrussi, A. (2012). Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat. Struct. Mol. Biol. 19, 441–449. doi: 10.1038/nsmb.2257
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202. doi: 10.1038/nature04917
Gompel, N., Prud’homme, B., Wittkopp, P. J., Kassner, V. A., and Carroll, S. B. (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487. doi: 10.1038/nature03235
Grimson, A., Srivastava, M., Fahey, B., Woodcroft, B. J. H., Chiang, R., King, N., et al. (2008). Early origins and evolution of microRNAs and piwi-interacting RNAs in animals. Nature 455, 1193–1197. doi: 10.1038/nature07415
Groisman, I., Huang, Y. S., Mendez, R., Cao, Q., Theurkauf, W., and Richter, J. D. (2000). CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103, 435–447. doi: 10.1016/s0092-8674(00)00135-5
Groisman, I., Jung, M. Y., Sarkissian, M., Cao, Q., and Richter, J. D. (2002). Translational control of the embryonic cell cycle. Cell 109, 473–483. doi: 10.1016/s0092-8674(02)00733-x
Guduric-Fuchs, J., Mohrlen, F., Frohme, M., and Frank, U. (2004). A fragile X mental retardation-like gene in a cnidarian. Gene 343, 231–238. doi: 10.1016/j.gene.2004.10.007
Guillaume, F., and Otto, S. P. (2012). Gene functional trade-offs and the evolution of pleiotropy. Genetics 192, 1389–1409. doi: 10.1534/genetics.112.143214
Guo, J., Zhu, P., Wu, C., Yu, L., Zhao, S., and Gu, X. (2003). In silico analysis indicates a similar gene expression pattern between human brain and testis. Cytogenet. Genome Res. 103, 58–62. doi: 10.1159/000076290
Guo, J. H., Huang, Q., Studholme, D. J., Wu, C. Q., and Zhao, Z. (2005). Transcriptomic analyses support the similarity of gene expression between brain and testis in human as well as mouse. Cytogenet. Genome. Res. 111, 107–109. doi: 10.1159/000086378
Guo, W., Allan, A. M., Zong, R., Zhang, L., Johnson, E. B., Schaller, E. G., et al. (2011). Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat. Med. 17, 559–565. doi: 10.1038/nm.2336
Gustafson, E. A., and Wessel, G. M. (2010). Vasa genes: emerging roles in the germ line and in multipotent cells. BioEssays 32, 626–637. doi: 10.1002/bies.201000001
Hafer, N., Xu, S., Bhat, K. M., and Schedl, P. (2011). The Drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function. Genetics 189, 907–921. doi: 10.1534/genetics.110.123646
Hake, L. E., Mendez, R., and Richter, J. D. (1998). Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell. Biol. 18, 685–693. doi: 10.1128/mcb.18.2.685
Hake, L. E., and Richter, J. D. (1994). CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79, 617–627. doi: 10.1016/0092-8674(94)90547-9
Halanych, K. M. (2015). The ctenophore lineage is older than sponges? That cannot be right! Or can it? J. Exp. Biol. 218, 592–597. doi: 10.1242/jeb.111872
Han, T. W., Kato, M., Xie, S., Wu, L. C., Mirzaei, H., Pei, J., et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. doi: 10.1016/j.cell.2012.04.016
Handler, D., Olivieri, D., Novatchkova, M., Gruber, F. S., Meixner, K., Mechtler, K., et al. (2011). A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 30, 3977–3993. doi: 10.1038/emboj.2011.308
Haraguchi, S., Tsuda, M., Kitajima, S., Sasaoka, Y., Nomura-Kitabayashid, A., Kurokawa, K., et al. (2003). nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech. Dev. 120, 721–731. doi: 10.1016/s0925-4773(03)00043-1
Hay, B., Jan, L. Y., and Jan, Y. N. (1988). A Protein Component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55, 577–587. doi: 10.1016/0092-8674(88)90216-4
Hayashi, Y., Hayashi, M., and Kobayashi, S. (2004). Nanos suppresses somatic cell fate in Drosophila germ line. Proc. Natl. Acad. Sci. U.S.A. 101, 10338–10342. doi: 10.1073/pnas.0401647101
Heisenberg, M. (2003). Mushroom body memoir: from maps to models. Nat. Rev. Neurosci. 4, 266–275. doi: 10.1038/nrn1074
Heraud-Farlow, J. E., and Kiebler, M. A. (2014). The multifunctional staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 37, 470–479. doi: 10.1016/j.tins.2014.05.009
Holoch, D., and Moazed, D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84. doi: 10.1038/nrg3863
Holt, C. E., Martin, K. C., and Schuman, E. M. (2019). Local translation in neurons: visualization and function. Nat. Struct. Mol. Biol. 26, 557–566. doi: 10.1038/s41594-019-0263-5
Hoopfer, E. D., Penton, A., Watts, R. J., and Luo, L. (2008). Genomic analysis of Drosophila neuronal remodeling: a role for the RNA-binding protein Boule as a negative regulator of axon pruning. J. Neurosci. 28, 6092–6103. doi: 10.1523/jneurosci.0677-08.2008
Houston, D. W., and King, M. L. (2000). A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development 127, 447–456.
Houston, D. W., Zhang, J., Maines, J. Z., Wasserman, S. A., and King, M. L. (1998). A xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development 125, 171–180.
Houwing, S., Kamminga, L. M., Berezikov, E., Cronembold, D., Girard, A., van den Elst, H., et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129, 69–82. doi: 10.1016/j.cell.2007.03.026
Huang, Y. S., Carson, J. H., Barbarese, E., and Richter, J. D. (2003). Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 17, 638–653. doi: 10.1101/gad.1053003
Huang, Y. S., Jung, M. Y., Sarkissian, M., and Richter, J. D. (2002). N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 21, 2139–2148. doi: 10.1093/emboj/21.9.2139
Hurd, T. R., Herrmann, B., Sauerwald, J., Sanny, J., Grosch, M., and Lehmann, R. (2016). Long oskar controls mitochondrial inheritance in Drosophila melanogaster. Dev. Cell 39, 560–571. doi: 10.1016/j.devcel.2016.11.004
Igea, A., and Mendez, R. (2010). Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 29, 2182–2193. doi: 10.1038/emboj.2010.111
Ikenishi, K. (1998). Germ plasm in Caenorhabditis elegans. Drosophila and Xenopus. Dev. Growth. Differ. 40, 1–10. doi: 10.1046/j.1440-169x.1998.t01-4-00001.x
Inoue, S. B., Siomi, M. C., and Siomi, H. (2000). Molecular mechanisms of fragile X syndrome. J. Med. Invest. 47, 101–107.
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-Interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433. doi: 10.1146/annurev-biochem-060614-034258
Janic, A., Mendizabal, L., Llamazares, S., Rossell, D., and Gonzalez, C. (2010). Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 330, 1824–1827. doi: 10.1126/science.1195481
Jaruzelska, J., Kotecki, M., Kusz, K., Spik, A., Firpo, M., and Reijo, R. A. (2003). Conservation of a pumilio-nanos complex from Drosophila germ plasm to human germ cells. Dev. Genes Evol. 213, 120–126. doi: 10.1007/s00427-003-0303-2
Jeske, M., Bordi, M., Glatt, S., Muller, S., Rybin, V., Muller, C. W., et al. (2015). The crystal structure of the Drosophila germline inducer oskar identifies two domains with distinct vasa helicase- and RNA-binding activities. Cell Rep. 12, 587–598. doi: 10.1016/j.celrep.2015.06.055
Jeske, M., Muller, C. W., and Ephrussi, A. (2017). The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 31, 939–952. doi: 10.1101/gad.297051.117
Jia, M., Shan, Z., Yang, Y., Liu, C., Li, J., Luo, Z. G., et al. (2015). The structural basis of miranda-mediated staufen localization during Drosophila neuroblast asymmetric division. Nat. Comm. 6:8381.
Jimenez, J., Alphey, L., Nurse, P., and Glover, D. M. (1990). Complementation of fission yeast cdc2ts and cdc25ts mutants identifies two cell cycle genes from Drosophila: a cdc2 homologue and string. EMBO J. 9, 3565–3571. doi: 10.1002/j.1460-2075.1990.tb07567.x
Jin, P., Zarnescu, D. C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T. A., et al. (2004). Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117. doi: 10.1038/nn1174
Jin, S. W., Arno, N., Cohen, A., Shah, A., Xu, Q., Chen, N., et al. (2001). In Caenorhabditis elegans, the RNA-binding domains of the cytoplasmic polyadenylation element binding protein FOG-1 are needed to regulate germ cell fates. Genetics 159, 1617–1630.
Joiner, M. L., and Wu, C. F. (2004). Nervous system function for the testis RNA-binding protein Boule in Drosophila. J. Neurogenetics 18, 341–363. doi: 10.1080/01677060490477435
Joly, W., Chartier, A., Rojas-Rios, P., Busseau, I., and Simonelig, M. (2013). The CCR4 deadenylase acts with nanos and pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 1, 411–424. doi: 10.1016/j.stemcr.2013.09.007
Juliano, C., Wang, J., and Lin, H. (2011). Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 45, 447–469. doi: 10.1146/annurev-genet-110410-132541
Juliano, C. E., Swartz, S. Z., and Wessel, G. M. (2010). A conserved germline multipotency program. Development 137, 4113–4126. doi: 10.1242/dev.047969
Jung, M. Y., Lorenz, L., and Richter, J. D. (2006). Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol. Cell. Biol. 26, 4277–4287. doi: 10.1128/mcb.02470-05
Kadyrova, L. Y., Habara, Y., Lee, T. H., and Wharton, R. P. (2007). Translational control of maternal cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519–1527. doi: 10.1242/dev.002212
Kang, H., and Schuman, E. M. (1996). A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273, 1402–1406. doi: 10.1126/science.273.5280.1402
Kanska, J., and Frank, U. (2013). New roles for Nanos in neural cell fate determination revealed by studies in a cnidarian. J. Cell Sci. 126, 3192–3203. doi: 10.1242/jcs.127233
Kaprara, A., and Huhtaniemi, I. T. (2018). The hypothalamus-pituitary-gonad axis: tales of mice and men. Metabolism 86, 3–17. doi: 10.1016/j.metabol.2017.11.018
Karashima, T., Sugimoto, A., and Yamamoto, M. (2000). Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 127, 1069–1079.
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. doi: 10.1016/j.cell.2012.04.017
Kawamura, K., and Sunanaga, T. (2011). Role of Vasa, Piwi, and Myc-expressing coelomic cells in gonad regeneration of the colonial tunicate, Botryllus primigenus. Mech. Dev. 128, 457–470. doi: 10.1016/j.mod.2011.09.001
Kaye, J. A., Rose, N. C., Goldsworthy, B., Goga, A., and L’Etoile, N. D. (2009). A 3’UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61, 57–70. doi: 10.1016/j.neuron.2008.11.012
Keleman, K., Kruttner, S., Alenius, M., and Dickson, B. J. (2007). Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 10, 1587–1593. doi: 10.1038/nn1996
Khandjian, E. W., Corbin, F., Woerly, S., and Rousseau, F. (1996). The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 12, 91–93. doi: 10.1038/ng0196-91
Khila, A., Abouheif, E., and Rowe, L. (2012). Function, developmental genetics, and fitness consequences of a sexually antagonistic trait. Science 336, 585–589. doi: 10.1126/science.1217258
Kikuno, R., Nagase, T., Nakayama, M., Koga, H., Okazaki, N., Nakajima, D., et al. (2004). HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucleic Acids Res. 32, D502–D504.
Kim, B., and Rhee, K. (2016). BOULE, a deleted in azoospermia homolog, is recruited to stress granules in the mouse male germ cells. PLoS ONE 11:e0163015. doi: 10.1371/journal.pone.0163015
Kim, K. W., Tang, N. H., Andrusiak, M. G., Wu, Z., Chisholm, A. D., and Jin, Y. (2018). A neuronal piRNA pathway inhibits axon regeneration in C. elegans. Neuron 97:e6.
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 10, 126–139. doi: 10.1038/nrm2632
Kin, K. (2015). Inferring cell type innovations by phylogenetic methods-concepts, methods, and limitations. J. Exp. Zool. B Mol. Dev. Evol. 324, 653–661. doi: 10.1002/jez.b.22657
King, M. C., and Wilson, A. C. (1975). Evolution at two levels in humans and chimpanzees. Science 188, 107–116. doi: 10.1126/science.1090005
King, R. C., Mulligan, P. K., and Stansfield, W. D. (2013). A Dictionary of Genetics. New York, NY: Oxford University Press.
Kirino, Y., Vourekas, A., Kim, N., de Lima Alves, F., Rappsilber, J., Klein, P. S., et al. (2010). Arginine methylation of Vasa protein is conserved across phyla. J. Biol. Chem. 285, 8148–8154. doi: 10.1074/jbc.m109.089821
Knutson, A. K., Egelhofer, T., Rechtsteiner, A., and Strome, S. (2017). Germ granules prevent accumulation of somatic transcripts in the adult Caenorhabditis elegans germline. Genetics 206, 163–178. doi: 10.1534/genetics.116.198549
Kobayashi, S., Yamada, M., Asaoka, M., and Kitamura, T. (1996). Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380, 708–711. doi: 10.1038/380708a0
Koprunner, M., Thisse, C., Thisse, B., and Raz, E. (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877–2885.
Kruttner, S., Stepien, B., Noordermeer, J. N., Mommaas, M. A., Mechtler, K., Dickson, B. J., et al. (2012). Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron 76, 383–395. doi: 10.1016/j.neuron.2012.08.028
Ku, H. Y., and Lin, H. (2014). PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl. Sci. Rev. 1, 205–218. doi: 10.1093/nsr/nwu014
Kulkarni, A., and Extavour, C. G. (2017). Convergent evolution of germ granule nucleators: a hypothesis. Stem Cell Res. 24, 188–194. doi: 10.1016/j.scr.2017.07.018
Kurihara, Y., Tokuriki, M., Myojin, R., Hori, T., Kuroiwa, A., Matsuda, Y., et al. (2003). CPEB2, a novel putative translational regulator in mouse haploid germ cells. Biol. Reprod. 69, 261–268. doi: 10.1095/biolreprod.103.015677
Kwak, J. E., Drier, E., Barbee, S. A., Ramaswami, M., Yin, J. C., and Wickens, M. (2008). GLD2 poly(A) polymerase is required for long-term memory. Proc. Natl. Acad. Sci. U.S.A. 105, 14644–14649. doi: 10.1073/pnas.0803185105
Laggerbauer, B., Ostareck, D., Keidel, E. M., Ostareck-Lederer, A., and Fischer, U. (2001). Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10, 329–338. doi: 10.1093/hmg/10.4.329
Lai, F., Zhou, Y., Luo, X., Fox, J., and King, M. L. (2011). Nanos1 functions as a translational repressor in the Xenopus germline. Mech. Dev. 128, 153–163. doi: 10.1016/j.mod.2010.12.001
Lantz, V., Ambrosio, L., and Schedl, P. (1992). The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding protiens and has localized transcripts in ovaries and early embryos. Development 115, 75–88.
Lantz, V., Chang, J. S., Horabin, J. I., Bopp, D., and Schedl, P. (1994). The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8, 598–613. doi: 10.1101/gad.8.5.598
Lasko, P. (2013). The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta 1829, 810–816. doi: 10.1016/j.bbagrm.2013.04.005
Lasko, P. F., and Ashburner, M. (1988). The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 335, 611–617. doi: 10.1038/335611a0
Laumer, C. E., Fernandez, R., Lemer, S., Combosch, D., Kocot, K. M., Riesgo, A., et al. (2019). Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. Biol. Sci. 286:20190831. doi: 10.1098/rspb.2019.0831
Lear, B. (2001). Roles of Intrinsic Factors During Cell Fate Decisions in the Insect Central Nervous System, Department of Molecular Genetics and Cell Biology. Chicago: University of Chicago, 150.
Leclère, L., Jager, M., Barreau, C., Chang, P., Le Guyader, H., Manuel, M., et al. (2012). Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian clytia. Dev. Biol. 364, 236–248. doi: 10.1016/j.ydbio.2012.01.018
Lee, E. J., Banerjee, S., Zhou, H., Jammalamadaka, A., Arcila, M., Manjunath, B. S., et al. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099. doi: 10.1261/rna.2565011
Lee, M.-H., and Schedl, T. (2006). RNA-binding Proteins. WormBook: The Online Review of C. elegans Biology. Pasadena, CA: WormBook, 1–13.
Lee, S. H., Shim, J., Cheong, Y. H., Choi, S. L., Jun, Y. W., Lee, S. H., et al. (2016). ApCPEB4, a non-prion domain containing homolog of ApCPEB, is involved in the initiation of long-term facilitation. Mol. Brain 9:91.
Lehmann, R. (2016). Germ plasm biogenesis–an oskar-centric perspective. Curr. Top. Dev. Biol. 116, 679–707. doi: 10.1016/bs.ctdb.2015.11.024
Lehmann, R., and Nüsslein-Volhard, C. (1986). Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell 47, 144–152.
Lehmann, R., and Nüsslein-Volhard, C. (1987). Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature 329, 167–170. doi: 10.1038/329167a0
Lehmann, R., and Nusslein-Volhard, C. (1991). The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112, 679–691.
Leighton, L. J., Wei, W., Marshall, P. R., Ratnu, V. S., Li, X., Zajaczkowski, E. L., et al. (2019). Disrupting the hippocampal Piwi pathway enhances contextual fear memory in mice. Neurobiol. Learn. Mem. 161, 202–209. doi: 10.1016/j.nlm.2019.04.002
Leininger, S., Adamski, M., Bergum, B., Guder, C., Liu, J., Laplante, M., et al. (2014). Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat. Comm. 5:3905.
Li, P., Yang, X., Wasser, M., Cai, Y., and Chia, W. (1997). Inscuteable and staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 90, 437–447. doi: 10.1016/s0092-8674(00)80504-8
Li, Z., Zhang, Y., Ku, L., Wilkinson, K. D., Warren, S. T., and Feng, Y. (2001). The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 29, 2276–2283. doi: 10.1093/nar/29.11.2276
Lin, H., and Spradling, A. C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476.
Linder, P. (2006). Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 34, 4168–4180. doi: 10.1093/nar/gkl468
Liu, B., Li, Y., Stackpole, E. E., Novak, A., Gao, Y., Zhao, Y., et al. (2018). Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E11397–E11405.
Liu, J., Hu, J. Y., Wu, F., Schwartz, J. H., and Schacher, S. (2006). Two mRNA-binding proteins regulate the distribution of syntaxin mRNA in Aplysia sensory neurons. J. Neurosci. 26, 5204–5214. doi: 10.1523/jneurosci.4917-05.2006
Liu, L., Qi, H., Wang, J., and Lin, H. (2011). PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138, 1863–1873. doi: 10.1242/dev.059287
Luitjens, C., Gallegos, M., Kraemer, B., Kimble, J., and Wickens, M. (2000). CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14, 2596–2609. doi: 10.1101/gad.831700
Luo, Y., Shan, G., Guo, W., Smrt, R. D., Johnson, E. B., Li, X., et al. (2010). Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 6:e1000898. doi: 10.1371/journal.pgen.1000898
Lynch, J. A., Özüak, O., Khila, A., Abouheif, E., Desplan, C., and Roth, S. (2011). The phylogenetic origin of oskar Coincided with the origin of maternally provisioned germ plasm and pole cells at the base of the holometabola. PLoS Genet. 7:e1002029. doi: 10.1371/journal.pgen.1002029
Macchi, P., Kroening, S., Palacios, I. M., Baldassa, S., Grunewald, B., Ambrosino, C., et al. (2003). Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J. Neurosci. 23, 5778–5788. doi: 10.1523/jneurosci.23-13-05778.2003
Mah, J. L., and Leys, S. P. (2017). Think like a sponge: the genetic signal of sensory cells in sponges. Dev. Biol. 431, 93–100. doi: 10.1016/j.ydbio.2017.06.012
Maines, J. Z., and Wasserman, S. A. (1999). Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nature Cell Biol. 1, 171–174. doi: 10.1038/11091
Majumdar, A., Cesario, W. C., White-Grindley, E., Jiang, H., Ren, F., Khan, M. R., et al. (2012). Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148, 515–529. doi: 10.1016/j.cell.2012.01.004
Mallardo, M., Deitinghoff, A., Muller, J., Goetze, B., Macchi, P., Peters, C., et al. (2003). Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. U.S.A. 100, 2100–2105. doi: 10.1073/pnas.0334355100
Mani, S. R., and Juliano, C. E. (2013). Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 80, 632–664. doi: 10.1002/mrd.22195
Markussen, F. H., Michon, A. M., Breitwieser, W., and Ephrussi, A. (1995). Translational control of oskar generates short OSK, the isoform that induces pole plasm assembly. Development 121, 3723–3732.
McBride, S. M., Bell, A. J., and Jongens, T. A. (2012). Behavior in a Drosophila model of fragile X. Results Probl. Cell Differ. 54, 83–117. doi: 10.1007/978-3-642-21649-7_6
McBride, S. M., Choi, C. H., Wang, Y., Liebelt, D., Braunstein, E., Ferreiro, D., et al. (2005). Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron 45, 753–764. doi: 10.1016/j.neuron.2005.01.038
Megosh, H. B., Cox, D. N., Campbell, C., and Lin, H. (2006). Campbell, and Lin, H., the role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 16, 1884–1894. doi: 10.1016/j.cub.2006.08.051
Meijer, H. A., Radford, H. E., Wilson, L. S., Lissenden, S., de, C. H., and Moor (2007). Translational control of maskin mRNA by its 3’ untranslated region. Biol. Cell 99, 239–250. doi: 10.1042/bc20060112
Mendez, R., and Richter, J. D. (2001). Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529. doi: 10.1038/35080081
Micklem, D. R., Adams, J., Grunert, S., and St Johnston, D. (2000). Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 19, 1366–1377. doi: 10.1093/emboj/19.6.1366
Mochizuki, K., Fine, N. A., Fujisawa, T., and Gorovsky, M. A. (2002). Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110, 689–699. doi: 10.1016/s0092-8674(02)00909-1
Mochizuki, K., Nishimiya-Fujisawa, C., and Fujisawa, T. (2001). Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev. Genes Evol. 211, 299–308. doi: 10.1007/s004270100156
Mochizuki, K., Sano, H., Kobayashi, S., Nishimiya-Fujisawa, C., and Fujisawa, T. (2000). Expression and evolutionary conservation of nanos-related genes in Hydra. Dev. Genes Evol. 210, 591–602. doi: 10.1007/s004270000105
Moczek, A. P., and Rose, D. J. (2009). Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc. Natl. Acad. Sci. U.S.A. 106, 8992–8997. doi: 10.1073/pnas.0809668106
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. doi: 10.1016/j.cell.2015.09.015
Monteiro, A., and Podlaha, O. (2009). Wings, horns, and butterfly eyespots: how do complex traits evolve? PLoS Biol. 7:e1000037. doi: 10.1371/journal.pbio.1000037
Moore, F. L., Jaruzelska, J., Fox, M. S., Urano, J., Firpo, M. T., Turek, P. J., et al. (2003). Human pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 538–543. doi: 10.1073/pnas.0234478100
Morais-de-Sa, E., Vega-Rioja, A., Trovisco, V., and St Johnston, D. (2013). Oskar is targeted for degradation by the sequential action of Par-1, GSK-3, and the SCF(-)Slimb ubiquitin ligase. Dev. Cell. 26, 303–314. doi: 10.1016/j.devcel.2013.06.011
Moussian, B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799–1809. doi: 10.1093/emboj/17.6.1799
Muotri, A. R., Chu, V. T., Marchetto, M. C., Deng, W., Moran, J. V., and Gage, F. H. (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910. doi: 10.1038/nature03663
Murata, Y., and Wharton, R. P. (1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756. doi: 10.1016/0092-8674(95)90353-4
Naisbitt, S., Valtschanoff, J., Allison, D. W., Sala, C., Kim, E., Craig, A. M., et al. (2000). Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J. Neurosci. 20, 4524–4534. doi: 10.1523/jneurosci.20-12-04524.2000
Nakahata, S., Katsu, Y., Mita, K., Inoue, K., Nagahama, Y., and Yamashita, M. (2001). Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276, 20945–20953. doi: 10.1074/jbc.m010528200
Nam, Y. J., Cheon, H. S., Choi, Y. K., Kim, S. Y., Shin, E. Y., Kim, E. G., et al. (2008). Role of mitogen-activated protein kinase (MAPK) docking sites on Staufen2 protein in dendritic mRNA transport. Biochem. Biophys. Res. Commun. 372, 525–529. doi: 10.1016/j.bbrc.2008.05.047
Nielsen, C., and Martinez, P. (2003). Patterns of gene expression: homology or homocracy? Dev. Genes Evol. 213, 149–154. doi: 10.1007/s00427-003-0301-4
Nimchinsky, E. A., Oberlander, A. M., and Svoboda, K. (2001). Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci. 21, 5139–5146. doi: 10.1523/jneurosci.21-14-05139.2001
Norvell, A., Wong, J., Randolph, K., and Thompson, L. (2015). Wispy and Orb cooperate in the cytoplasmic polyadenylation of localized gurken mRNA. Dev. Dyn. 244, 1276–1285. doi: 10.1002/dvdy.24311
Nüsslein-Volhard, C., Frohnhöfer, H. G., and Lehmann, R. (1987). Determination of anteroposterior polarity in Drosophila. Science 238, 1675–1681. doi: 10.1126/science.3686007
Ota, R., Kotani, T., and Yamashita, M. (2011). Biochemical characterization of pumilio1 and pumilio2 in Xenopus oocytes. J. Biol. Chem. 286, 2853–2863. doi: 10.1074/jbc.m110.155523
Ottone, C., Gigliotti, S., Giangrande, A., Graziani, F., Verrotti, and di Pianella, A. (2012). The translational repressor Cup is required for germ cell development in Drosophila. J. Cell Sci. 125, 3114–3123. doi: 10.1242/jcs.095208
Oyama, A., and Shimizu, T. (2007). Transient occurrence of vasa-expressing cells in nongenital segments during embryonic development in the oligochaete annelid Tubifex tubifex. Dev. Genes Evol. 217, 675–690. doi: 10.1007/s00427-007-0180-1
Padmashri, R., Reiner, B. C., Suresh, A., Spartz, E., and Dunaevsky, A. (2013). Altered structural and functional synaptic plasticity with motor skill learning in a mouse model of fragile X syndrome. J. Neurosci. 33, 19715–19723. doi: 10.1523/jneurosci.2514-13.2013
Palacios, I. M., Gatfield, D., St Johnston, D., and Izaurralde, E. (2004). An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427, 753–757. doi: 10.1038/nature02351
Panganiban, G., Irvine, S. M., Lowe, C., Roehl, H., Corley, L. S., Sherbon, B., et al. (1997). The origin and evolution of animal appendages. Proc. Natl. Acad. Sci. U.S.A. 47, 5162–5166.
Paps, J. (2018). What makes an animal? The molecular quest for the origin of the animal kingdom. Integr. Comp. Biol. 58, 654–665. doi: 10.1093/icb/icy036
Parisi, M., and Lin, H. (2000). Translational repression: a duet of Nanos and Pumilio. Curr. Biol. 10, R81–R83.
Pek, J. W., and Kai, T. (2011). A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr. Biol. 21, 39–44. doi: 10.1016/j.cub.2010.11.051
Peredo, J., Villacé, P., Ortin, J., and de Lucas, S. (2014). Human Staufen1 associates to miRNAs involved in neuronal cell differentiation and is required for correct dendritic formation. PLoS ONE 9:e113704. doi: 10.1371/journal.pone.0113704
Perrat, P. N., DasGupta, S., Wang, J., Theurkauf, W., Weng, Z., Rosbash, M., et al. (2013). Transposition-driven genomic heterogeneity in the Drosophila brain. Science 340, 91–95. doi: 10.1126/science.1231965
Peterson, K. J., and Davidson, E. H. (2000). Regulatory evolution and the origin of the bilaterians. Proc. Natl. Acad. Sci. U.S.A. 97, 4430–4433. doi: 10.1073/pnas.97.9.4430
Pfister, D., De Mulder, K., Hartenstein, V., Kuales, G., Borgonie, G., Marx, F., et al. (2008). Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev. Biol. 319, 146–159. doi: 10.1016/j.ydbio.2008.02.045
Plickert, G., Frank, U., and Muller, W. A. (2012). Hydractinia, a pioneering model for stem cell biology and reprogramming somatic cells to pluripotency. Int. J. Dev. Biol. 56, 519–534. doi: 10.1387/ijdb.123502gp
Popper, B., Demleitner, A., Bolivar, V. J., Kusek, G., Snyder-Keller, A., Schieweck, R., et al. (2018). Staufen2 deficiency leads to impaired response to novelty in mice. Neurobiol. Learn. Mem. 150, 107–115. doi: 10.1016/j.nlm.2018.02.027
Posner, R., Toker, I. A., Antonova, O., Star, E., Anava, S., Azmon, E., et al. (2019). Neuronal small RNAs control behavior transgenerationally. Cell 177:e15.
Price, T. J., Flores, C. M., Cervero, F., and Hargreaves, K. M. (2006). The RNA binding and transport proteins staufen and fragile X mental retardation protein are expressed by rat primary afferent neurons and localize to peripheral and central axons. Neuroscience 141, 2107–2116. doi: 10.1016/j.neuroscience.2006.05.047
Pushpa, K., Kumar, G. A., and Subramaniam, K. (2017). Translational control of germ cell decisions. Results Probl. Cell Differ. 59, 175–200. doi: 10.1007/978-3-319-44820-6_6
Rajasethupathy, P., Antonov, I., Sheridan, R., Frey, S., Sander, C., Tuschl, T., et al. (2012). A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149, 693–707. doi: 10.1016/j.cell.2012.02.057
Ramasamy, S., Wang, H., Quach, H. N., and Sampath, K. (2006). Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev. Biol. 292, 393–406. doi: 10.1016/j.ydbio.2006.01.014
Rebscher, N., Volk, C., Teo, R., and Plickert, G. (2008). The germ plasm component Vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia echinata. Dev. Dyn. 237, 1736–1745. doi: 10.1002/dvdy.21562
Rebscher, N., Zelada-Gonzalez, F., Banisch, T. U., Raible, F., and Arendt, D. (2007). Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev. Biol. 306, 599–611. doi: 10.1016/j.ydbio.2007.03.521
Richter, S., Loesel, R., Purschke, G., Schmidt-Rhaesa, A., Scholtz, G., Stach, T., et al. (2010). Invertebrate neurophylogeny: suggested terms and definitions for a neuroanatomical glossary. Front. Zool. 7:29. doi: 10.1186/1742-9994-7-29
Roegiers, F., and Jan, Y. N. (2000). Staufen: a common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 10, 220–224. doi: 10.1016/s0962-8924(00)01767-0
Rojas-Rios, P., Chartier, A., Pierson, S., Severac, D., Dantec, C., Busseau, I., et al. (2015). Translational control of autophagy by orb in the Drosophila germline. Dev. Cell 35, 622–631. doi: 10.1016/j.devcel.2015.11.003
Rojas-Rios, P., and Simonelig, M. (2018). piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145:dev161786. doi: 10.1242/dev.161786
Rosner, A., Moiseeva, E., Rinkevich, Y., Lapidot, Z., and Rinkevich, B. (2009). Vasa and the germ line lineage in a colonial urochordate. Dev. Biol. 331, 113–128. doi: 10.1016/j.ydbio.2009.04.025
Ross, R. J., Weiner, M. M., and Lin, H. (2014). PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359. doi: 10.1038/nature12987
Ruggiu, M., Speed, R., Taggart, M., McKay, S. J., Kilanowski, F., Saunders, P., et al. (1997). The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389, 73–77. doi: 10.1038/37987
Russell, P., and Nurse, P. (1986). cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45, 145–153. doi: 10.1016/0092-8674(86)90546-5
Sanetra, M., Begemann, G., Becker, M. B., and Meyer, A. (2005). Conservation and co-option in developmental programmes: the importance of homology relationships. Front. Zool. 2:15. doi: 10.1186/1742-9994-2-15
Sano, H., Mukai, M., and Kobayashi, S. (2001). Maternal nanos and pumilio regulate zygotic vasa expression autonomously in the germ-line progenitors of Drosophila melanogaster embryos. Dev. Growth Diff. 43, 545–552. doi: 10.1046/j.1440-169x.2001.00593.x
Sasaki, K., Yokobayashi, S., Nakamura, T., Okamoto, I., Yabuta, Y., Kurimoto, K., et al. (2015). Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 17, 178–194. doi: 10.1016/j.stem.2015.06.014
Sato, K., Hayashi, Y., Ninomiya, Y., Shigenobu, S., Arita, K., Mukai, M., et al. (2007). Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 104, 7455–7460. doi: 10.1073/pnas.0610052104
Sato, S. M., and Sargent, T. D. (1989). Development of neural inducing capacity in dissociated Xenopus embryos. Dev. Biol. 134, 263–266. doi: 10.1016/0012-1606(89)90096-1
Saunders, P. T., Pathirana, S., Maguire, S. M., Doyle, M., Wood, T., and Bownes, M. (2000). Mouse staufen genes are expressed in germ cells during oogenesis and spermatogenesis. Mol. Hum. Reprod. 6, 983–991. doi: 10.1093/molehr/6.11.983
Saxe, J. P., and Lin, H. (2011). Small noncoding RNAs in the germline. Cold Spring Harb. Perspect. Biol. 3:a002717. doi: 10.1101/cshperspect.a002717
Schisa, J. A. (2012). New insights into the regulation of RNP granule assembly in oocytes. Int. Rev. Cell Mol. Biol. 295, 233–289. doi: 10.1016/b978-0-12-394306-4.00013-7
Schuldt, A. J., Adams, J. H., Davidson, C. M., Micklem, D. R., Haseloff, J., St Johnston, D., et al. (1998). Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 12, 1847–1857. doi: 10.1101/gad.12.12.1847
Schüpbach, T., and Wieschaus, E. (1986). Maternal-effect mutations altering the anterior-posterior patterns of the Drosophila embryo. Roux’s Arch. Dev. Biol. 195, 302–317. doi: 10.1007/bf00376063
Schwager, E. E., Meng, Y., and Extavour, C. G. (2015). vasa and piwi are required for mitotic integrity in early embryogenesis in the spider Parasteatoda tepidariorum. Dev. Biol. 402, 276–290. doi: 10.1016/j.ydbio.2014.08.032
Sears, J. C., Choi, W. J., and Broadie, K. (2019). Fragile X mental retardation protein positively regulates PKA anchor rugose and PKA activity to control actin assembly in learning/memory circuitry. Neurobiol. Dis. 127, 53–64. doi: 10.1016/j.nbd.2019.02.004
Seipel, K., Yanze, N., and Schmid, V. (2004). The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int. J. Dev. Biol. 48, 1–7. doi: 10.1387/ijdb.15005568
Sekine, K., Furusawa, T., and Hatakeyama, M. (2015). The boule gene is essential for spermatogenesis of haploid insect male. Dev. Biol. 399, 154–163. doi: 10.1016/j.ydbio.2014.12.027
Shah, C., Vangompel, M., Naeem, J. W. V., Chen, Y., Lee, T., Angeloni, N., et al. (2010). Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function. PLoS Genet. 6:e1001022. doi: 10.1371/journal.pgen.1001022
Shibata, N., Umesono, Y., Orii, H., Sakurai, T., Watanabe, K., and Agata, K. (1999). Expression of vasa (vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 206, 73–87. doi: 10.1006/dbio.1998.9130
Shirayama, M., Stanney, W. III, Gu, W., Seth, M., and Mello, C. C. (2014). The vasa homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr. Biol. 24, 845–851. doi: 10.1016/j.cub.2014.03.008
Si, K., Choi, Y. B., White-Grindley, E., Majumdar, A., and Kandel, E. R. (2010). Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140, 421–435. doi: 10.1016/j.cell.2010.01.008
Si, K., and Kandel, E. R. (2016). The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb. Perspect. Biol. 8:a021774. doi: 10.1101/cshperspect.a021774
Si, K., Lindquist, S., and Kandel, E. R. (2003). A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 115, 879–891. doi: 10.1016/s0092-8674(03)01020-1
Siebert, S., Farrell, J. A., Cazet, J. F., Abeykoon, Y. L., Primack, A. S., Schnitzler, C. E., et al. (2019). Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365:eaav9314. doi: 10.1126/science.aav9314
Siomi, M. C., Siomi, H., Sauer, W. H., Srinivasan, S., Nussbaum, R. L., and Dreyfuss, G. (1995). FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 14, 2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x
Siomi, M. C., Zhang, Y., Siomi, H., and Dreyfuss, G. (1996). Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol. 16, 3825–3832. doi: 10.1128/mcb.16.7.3825
Slegtenhorst-Eegdeman, K. E., de Rooij, D. G., Verhoef-Post, M., van de Kant, H. J., Bakker, C. E., Oostra, B. A., et al. (1998). Macroorchidism in FMR1 knockout mice is caused by increased Sertoli cell proliferation during testicular development. Endocrinology 139, 156–162. doi: 10.1210/endo.139.1.5706
Smukler, S. R., Runciman, S. B., Xu, S., and van der Kooy, D. (2006). Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J. Cell Biol. 172, 79–90. doi: 10.1083/jcb.200508085
Sonoda, J., and Wharton, R. P. (1999). Recruitment of nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712. doi: 10.1101/gad.13.20.2704
Sonoda, J., and Wharton, R. P. (2001). Drosophila brain tumor is a translational repressor. Genes Dev. 15, 762–773. doi: 10.1101/gad.870801
Spracklin, G., Fields, B., Wan, G., Becker, D., Wallig, A., Shukla, A., et al. (2017). The RNAi inheritance machinery of Caenorhabditis elegans. Genetics 206, 1403–1416. doi: 10.1534/genetics.116.198812
St Johnston, D., Beuchle, D., and Nusslein-Volhard, C. (1991). Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66, 51–63. doi: 10.1016/0092-8674(91)90138-o
Stebbins-Boaz, B., Cao, Q., de Moor, C. H., Mendez, R., and Richter, J. D. (1999). Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell. 4, 1017–1027. doi: 10.1016/s1097-2765(00)80230-0
Stebbins-Boaz, B., Hake, L. E., and Richter, J. D. (1996). CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 15, 2582–2592. doi: 10.1002/j.1460-2075.1996.tb00616.x
Stephan, J. S., Fioriti, L., Lamba, N., Colnaghi, L., Karl, K., Derkatch, I. L., et al. (2015). The CPEB3 protein is a functional prion that interacts with the actin cytoskeleton. Cell Rep. 11, 1772–1785. doi: 10.1016/j.celrep.2015.04.060
Stepien, B. K., Oppitz, C., Gerlach, D., Dag, U., Novatchkova, M., Kruttner, S., et al. (2016). RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc. Natl. Acad. Sci. U.S.A. 113, E7030–E7038.
Subramaniam, K., and Seydoux, G. (1999). nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorgabditis elegans. Development 126, 4861–1871.
Sudhakaran, I. P., Hillebrand, J., Dervan, A., Das, S., Holohan, E. E., Hulsmeier, J., et al. (2014). FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl. Acad. Sci. U.S.A. 111, E99–E108.
Sudhakaran, I. P., and Ramaswami, M. (2017). Long-term memory consolidation: the role of RNA-binding proteins with prion-like domains. RNA Biol. 14, 568–586. doi: 10.1080/15476286.2016.1244588
Sunanaga, T., Saito, Y., and Kawamura, K. (2006). Postembryonic epigenesis of vasa-positive germ cells from aggregated hemoblasts in the colonial ascidian, Botryllus primigenus. Dev. Growth Diff. 48, 87–100. doi: 10.1111/j.1440-169x.2006.00849.x
Suyama, R., Jenny, A., Curado, S., Pellis-van Berkel, W., and Ephrussi, A. (2009). The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development 136, 95–105. doi: 10.1242/dev.027698
Szakmary, A., Cox, D. N., Wang, Z., and Lin, H. (2005). Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 15, 171–178. doi: 10.1016/j.cub.2005.01.005
Tamanini, F., Meijer, N., Verheij, C., Willems, P. J., Galjaard, H., Oostra, B. A., et al. (1996). FMRP is associated to the ribosomes via RNA. Hum. Mol. Genet. 5, 809–813. doi: 10.1093/hmg/5.6.809
Tan, C.-H., Lee, T.-C., Weeraratne, S. D., Korzh, V., Lim, T.-M., and Gong, Z. (2002). Ziwi, the zebrafish homologue of the Drosophila piwi: co-localization with vasa at the embryonic genital ridge and gonad-specific expression in the adults. Gene Exp. Patterns 2, 257–260. doi: 10.1016/s1567-133x(02)00052-2
Tan, L., Chang, J. S., Costa, A., and Schedl, P. (2001). An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development 128, 1159–1169.
Tanaka, T., Kato, Y., Matsuda, K., Hanyu-Nakamura, K., and Nakamura, A. (2011). Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development 138, 2523–2532. doi: 10.1242/dev.062208
Tanaka, T., and Nakamura, A. (2008). The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development 135, 1107–1117. doi: 10.1242/dev.017293
Tang, S. J., Meulemans, D., Vazquez, L., Colaco, N., and Schuman, E. (2001). A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron 32, 463–475. doi: 10.1016/s0896-6273(01)00493-7
Tay, J., and Richter, J. D. (2001). Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell 1, 201–213. doi: 10.1016/s1534-5807(01)00025-9
Tessier, C. R., and Broadie, K. (2008). Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development 135, 1547–1557. doi: 10.1242/dev.015867
Theis, M., Si, K., and Kandel, E. R. (2003). Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 100, 9602–9607. doi: 10.1073/pnas.1133424100
Thomson, T., and Lin, H. (2009). The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Ann. Rev. Cell Dev. Biol. 25, 355–376. doi: 10.1146/annurev.cellbio.24.110707.175327
Tian, H., Cao, Y. X., Zhang, X. S., Liao, W. P., Yi, Y. H., Lian, J., et al. (2013). The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis. 4:e617. doi: 10.1038/cddis.2013.138
Tomancak, P., Guichet, A., Zavorsky, P., and Ephrussi, A. (1998). Oocyte polarity depends on regulation of gurken by Vasa. Development 125, 1723–1732.
Torras, R., and Gonzalez-Crespo, S. (2005). Posterior expression of nanos orthologs during embryonic and larval development of the anthozoan Nematostella vectensis. Int. J. Dev. Biol. 49, 895–899. doi: 10.1387/ijdb.051980rt
Torras, R., Yanze, N., Schmid, V., and Gonzalez-Crespo, S. (2004). Nanos expression at the embryonic posterior pole and the medusa phase in the hydrozoan Podocoryne carnea. Evol. Dev. 6, 362–371. doi: 10.1111/j.1525-142x.2004.04044.x
Tropepe, V., Hitoshi, S., Sirard, C., Mak, T. W., Rossant, J., and van der Kooy, D. (2001). Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron 30, 65–78. doi: 10.1016/s0896-6273(01)00263-x
Uhlen, M., Fagerberg, L., Hallstrom, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics, tissue-based map of the human proteome. Science 347:1260419.
Vagin, V. V., Klenov, M. S., Kalmykova, A. I., Stolyarenko, A. D., Kotelnikov, R. N., and Gvozdev, V. A. (2004). The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 1, 54–58.
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. doi: 10.1126/science.1129333
van Eeden, F. J., Palacios, I. M., Petronczki, M., Weston, M. J., and St Johnston, D. (2001). Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J. Cell Biol. 154, 511–523.
Vanzo, N., Oprins, A., Xanthakis, D., Ephrussi, A., and Rabouille, C. (2007). Stimulation of endocytosis and actin dynamics by Oskar polarizes the Drosophila oocyte. Dev. Cell. 12, 543–555. doi: 10.1016/j.devcel.2007.03.002
Vanzo, N. F., and Ephrussi, A. (2002). Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development 129, 3705–3714.
Verheij, C., Bakker, C. E., de Graaff, E., Keulemans, J., Willemsen, R., Verkerk, A. J., et al. (1993). Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature 363, 722–724. doi: 10.1038/363722a0
Verkerk, A. J., Pieretti, M., Sutcliffe, J. S., Fu, Y. H., Kuhl, D. P., Pizzuti, A., et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. doi: 10.1016/0092-8674(91)90397-h
Verrotti, A. C., and Wharton, R. P. (2000). Nanos interacts with cup in the female germline of Drosophila. Development 127, 5225–5232.
Vessey, J. P., Schoderboeck, L., Gingl, E., Luzi, E., Riefler, J., Di Leva, F., et al. (2010). Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc. Natl. Acad. Sci. U.S.A. 107, 3222–3227. doi: 10.1073/pnas.0907128107
Vessey, J. P., Vaccani, A., Xie, Y., Dahm, R., Karra, D., Kiebler, M. A., et al. (2006). Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci. 26, 6496–6508. doi: 10.1523/jneurosci.0649-06.2006
Viljetic, B., Diao, L., Liu, J., Krsnik, Z., Wijeratne, S. H., Kristopovich, R., et al. (2017). Multiple roles of PIWIL1 in mouse neocorticogenesis. bioRXiv [Preprint] doi: 10.1101/106070
Villacé, P., Marión, R. M., and Ortin, J. (2004). The composition of staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32, 2411–2420. doi: 10.1093/nar/gkh552
Voronina, E., Seydoux, G., Sassone-Corsi, P., and Nagamori, I. (2011). RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3:a002774. doi: 10.1101/cshperspect.a002774
Wagner, D. E., Ho, J. J., and Reddien, P. W. (2012). Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299–311. doi: 10.1016/j.stem.2012.01.016
Wan, L., Dockendorff, T. C., Jongens, T. A., and Dreyfuss, G. (2000). Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol. 20, 8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000
Wang, B., Collins, J. J. III, and Newmark, P. A. (2013). Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. eLife 2:e00768.
Wang, C., Dickinson, L. K., and Lehmann, R. (1994). Genetics of nanos localization in Drosophila. Dev. Dyn. 199, 103–115.
Wang, C., and Lehmann, R. (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66, 637–647. doi: 10.1016/0092-8674(91)90110-k
Wang, H. N., Xu, Y., Tao, L. J., Zhou, J., Qiu, M. X., Teng, Y. H., et al. (2012). Identification and characterization of the pumilio-2 expressed in zebrafish embryos and adult tissues. Mol. Biol. Rep. 39, 2811–2819. doi: 10.1007/s11033-011-1040-7
Wang, M., Oge, L., Perez-Garcia, M. D., Hamama, L., and Sakr, S. (2018). The PUF protein family: overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 19:410. doi: 10.3390/ijms19020410
Wells, D. G., Richter, J. D., and Fallon, J. R. (2000). Molecular mechanisms for activity-regulated protein synthesis in the synapto-dendritic compartment. Curr. Opin. Neurobiol. 10, 132–137. doi: 10.1016/s0959-4388(99)00050-1
Wharton, R. P., and Struhl, G. (1991). RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67, 955–967. doi: 10.1016/0092-8674(91)90368-9
Wheeler, W. M. (1891). Neuroblasts in the arthropod embryo. J. Morphol. 4, 337–343. doi: 10.1002/jmor.1050040305
Whelan, N. V., Kocot, K. M., Moroz, T. P., Mukherjee, K., Williams, P., Paulay, G., et al. (2017). Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746. doi: 10.1038/s41559-017-0331-3
Wilhelm, J. E., Hilton, M., Amos, Q., and Henzel, W. J. (2003). Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 163, 1197–1204. doi: 10.1083/jcb.200309088
Wong, L. C., Costa, A., McLeod, I., Sarkeshik, A., Yates, J. III, Kyin, S., et al. (2011). The functioning of the Drosophila CPEB protein Orb is regulated by phosphorylation and requires casein kinase 2 activity. PLoS ONE 6:e24355. doi: 10.1371/journal.pone.0024355
Wu, L., Wells, D., Tay, J., Mendis, D., Abbott, M. A., Barnitt, A., et al. (1998). CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron 21, 1129–1139. doi: 10.1016/s0896-6273(00)80630-3
Xiol, J., Spinelli, P., Laussmann, M. A., Homolka, D., Yang, Z., Cora, E., et al. (2014). RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157, 1698–1711. doi: 10.1016/j.cell.2014.05.018
Xu, E. Y., Lee, D. F., Klebes, A., Turek, P. J., Kornberg, T. B., Reijo, R. A., et al. (2003). Human BOULE gene rescues meiotic defects in infertile flies. Hum. Mol. Genet. 12, 169–175. doi: 10.1093/hmg/ddg017
Xu, E. Y., Moore, F. L., and Pera, R. A. (2001). A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc. Natl. Acad. Sci. U.S.A. 98, 7414–7419. doi: 10.1073/pnas.131090498
Xu, S., Hafer, N., Agunwamba, B., and Schedl, P. (2012). The CPEB protein Orb2 has multiple functions during spermatogenesis in Drosophila melanogaster. PLoS Genet. 8:e1003079. doi: 10.1371/journal.pgen.1003079
Xu, X., Brechbiel, J. L., and Gavis, E. R. (2013). Dynein-dependent transport of nanos RNA in Drosophila sensory neurons requires rumpelstiltskin and the germ plasm organizer oskar. J. Neurosci. 33, 14791–14800. doi: 10.1523/jneurosci.5864-12.2013
Xu, Y., Wang, H., Zhou, J., Lei, Y., Zhou, Y., Yang, Q., et al. (2010). Zebrafish nanos interacts with and regulates the phosphorylation of Mylz2. Biochimie 92, 1812–1817. doi: 10.1016/j.biochi.2010.07.010
Yajima, M., and Wessel, G. M. (2011a). The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development 138, 2217–2222. doi: 10.1242/dev.065052
Yajima, M., and Wessel, G. M. (2011b). The multiple hats of Vasa: its functions in the germline and in cell cycle progression. Mol. Reprod. Dev. 78, 861–867. doi: 10.1002/mrd.21363
Yang, L., Duan, R., Chen, D., Wang, J., Chen, D., and Jin, P. (2007). Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Hum. Mol. Genet. 16, 1814–1820. doi: 10.1093/hmg/ddm129
Yang, N., Yu, Z., Hu, M., Wang, M., Lehmann, R., and Xu, R. M. (2015). Structure of Drosophila Oskar reveals a novel RNA binding protein. Proc. Natl. Acad. Sci. U.S.A. 112, 11541–11546. doi: 10.1073/pnas.1515568112
Ye, B., Petritsch, C., Clark, I. E., Gavis, E. R., Jan, L. Y., and Jan, Y. N. (2004). nanos and pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14, 314–321. doi: 10.1016/j.cub.2004.01.052
Yoon, Y. J., and Mowry, K. L. (2004). Xenopus staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131, 3035–3045. doi: 10.1242/dev.01170
Zamore, P. D., Bartel, D. P., Lehmann, R., and Williamson, J. R. (1999). The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38, 596–604. doi: 10.1021/bi982264s
Zamore, P. D., Williamson, J. R., and Lehmann, R. (1997). The pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3, 1421–1433.
Zhang, B., Gallegos, M., Puoti, A., Durkin, E., Fields, S., Kimble, J., et al. (1997). A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390, 477–484. doi: 10.1038/37297
Zhang, M., Chen, D., Xia, J., Han, W., Cui, X., Neuenkirchen, N., et al. (2017). Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev. 31, 1354–1369. doi: 10.1101/gad.298752.117
Zhang, Y., O’Connor, J. P., Siomi, M. C., Srinivasan, S., Dutra, A., Nussbaum, R. L., et al. (1995). The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14, 5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x
Keywords: genetic toolkit, co-option, modularity, developmental function evolution, germ line, nervous system, RNP granules, pleiotropy
Citation: Kulkarni A, Lopez DH and Extavour CG (2020) Shared Cell Biological Functions May Underlie Pleiotropy of Molecular Interactions in the Germ Lines and Nervous Systems of Animals. Front. Ecol. Evol. 8:215. doi: 10.3389/fevo.2020.00215
Received: 10 December 2019; Accepted: 09 June 2020;
Published: 04 August 2020.
Edited by:
Jordi Solana, Oxford Brookes University, United KingdomReviewed by:
Paul Lasko, McGill University, CanadaCopyright © 2020 Kulkarni, Lopez and Extavour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arpita Kulkarni, YWt1bGthcm5pQGZhcy5oYXJ2YXJkLmVkdQ==; Cassandra G. Extavour, ZXh0YXZvdXJAb2ViLmhhcnZhcmQuZWR1
†Present address: Davys H. Lopez, Department of Genetics and Development, Columbia University, New York, NY, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.