
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 26 June 2020
Sec. Biogeography and Macroecology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00208
This article is part of the Research Topic Ecological, Behavioral and Genomic Consequences in the Rodent Family Sciuridae: Why Are Squirrels So Diverse? View all 14 articles
A surprising amount of hidden phylogenetic diversity exists in the small to medium size, drab colored squirrels of the genus Sundasciurus. This genus is endemic to Sundaland and the Philippines, where it is widespread. An earlier revision of this genus found that the high elevation ‘populations’ of the widespread, lowland slender squirrel (S. tenuis) were different species. Previous phylogenies based on mitochondrial cytochrome b sequences also suggested that the widespread, lowland Low’s squirrel (S. lowii) and the narrow endemic Fraternal squirrel (S. fraterculus) are not reciprocally monophyletic. Additionally, deep divergences have been identified between lineages within Low’s squirrel that date to the early Pliocene. Here we focus on evaluating the relationships and differences within and between populations of these two nominal species using whole mitochondrial genome sequences, nuclear intron sequences, and morphology. We reassess the taxonomy of this group, revalidate the species status of Robinson’s squirrel (Sundasciurus robinsoni Bonhote, 1903) support the species level recognition of the Natuna squirrel (Sundasciurus natunensis Thomas, 1895) and identify three other lineages that require further study. We estimate times of divergence and integrate geologic history to find that most of the divergences are pre-Pleistocene, and thus predate the Pleistocene flooding of Sundaland. Biogeographic, and ecological factors may have played a more important role than climatic factors in generating these patterns. While divergence in allopatry seems to be the main process driving speciation in lowland Sundaland squirrels (Sundasciurus), ecomorphological and behavioral adaptations in this clade suggest an important role of niche divergence.
The small brown tree squirrels of the genus Sundasciurus are endemic to Sundaland and the Philippines. This genus was described by Moore (1958) based on the presence of a Y-shaped trans-bullar bony septum and includes species previously assigned to Sciurus and Tomeutes (Thomas, 1915; Chasen, 1940). Most species were widespread across multiple landmasses in Sundaland. This genus has received more attention than other genera in the subfamily (Callosciurinae/Nannosciurinae). Deep genetic divergences associated with habitat (such as highland or lowland forest) or geography (such as land mass) were obscured by conservative morphology and have resulted in multiple revisions of the genus in the last few years, increasing the number of species from 11 to 17 (Heaney, 1979; Den Tex et al., 2010; Thorington et al., 2012; Hawkins et al., 2016a). These latest systematic revisions have largely focused on high elevation ‘populations’ of widespread taxa which in all cases turned out to be very divergent and merited specific status. The recognition of these species was supported by both genetic and morphological lines of evidence. The genus was divided into two subgenera by Moore (1958). These subgenera, Aletesciurus and Sundasciurus, were defined based on morphological criteria (presence/absence of a sagittal crest, an inconspicuous/inflated antero-mesial lobe, and skull size), but these were not found to be reciprocally monophyletic with molecular data (Den Tex et al., 2010; Hawkins et al., 2016a). At that time, taxonomic rearrangements were not performed because of limited sampling and low support of key nodes in the tree, suggesting that additional data were necessary. The subgenus Aletesciurus contains one widely distributed species (Sundasciurus hippurus of the Malay Peninsula, Sumatra, and Borneo), a narrowly distributed Bornean mountain endemic (S. everetti), and nine species distributed in the Philippines (S. samarensis, S. philippinensis, S. mindanensis, S. rabori, S. steerii, S. moellendorffi, S. davensis, S. juvencus, and S. hoogstraali) (Chasen, 1937; Den Tex et al., 2010; Hawkins et al., 2016a). The subgenus Sundasciurus includes seven species restricted to Sundaland in two clades: a mainly highland clade on a long branch (S. altitudinis, S. brookei, S. jentinki, S. tahan, and S. tenuis) and a widespread lowland clade sister to the other two (S. fraterculus and S. lowii) (Thorington and Hoffmann, 2005; Den Tex et al., 2010; Thorington et al., 2012; Hawkins et al., 2016a). Many of these widespread species contain multiple subspecies described from a limited number of specimens that were generally identified by differences in pelage coloration (Corbet and Hill, 1992; Thorington et al., 2012). In order to evaluate the diversity in the genus and uncover other unidentified lineages, we used populations as the unit of analysis.
Despite recent research on the phylogenetics of some of the members of the genus Sundasciurus, less attention has been paid to lowland Sunda squirrels. Widespread across Sundaland, lowland species of rodents have been shown to have more and stronger genetic structure associated with land masses, as compared to larger mammals but these divergences have not yet led to taxonomic revisions. Given the forest-dependent nature of these squirrels, an improved knowledge of their evolutionary history is key for our understanding of the dynamics of Sundaland rainforests (Den Tex et al., 2010). Deep divergences and speciation events may be the consequence of past isolation of landmasses or forest pockets while lack of genetic structure could shed light on recent episodes of land or forest connection. Conflicting patterns have been shown across different phylogeographic studies on mammals distributed across Sundaland (Mason et al., 2011; Leonard et al., 2015). Some species of murids, carnivores, suids, greater mouse deer and pangolins have weak or no genetic structure across Sundaland and/or within Borneo (Gorog et al., 2004; Achmadi et al., 2013; Patel et al., 2017; Mason et al., 2019; Veron et al., 2020) while others, such as Sunda rats, viverrids, colugos or the lesser mouse deer, show much deeper divergences some of which predate to the Pleistocene (Den Tex et al., 2010; Mason et al., 2011; Camacho-Sánchez, 2017; Veron et al., 2019). Recently, Husson et al. (2019) provided substantial evidence for a new paleogeographic scenario in which prior to 400 thousand years ago (ka) the Sunda shelf would have been continuously exposed. According to this study, dispersal across this exposed shelf would have been possible through this period for terrestrial species. Nevertheless, geological connectivity does not necessarily imply population connectivity, different dates of divergences between populations on different land masses might reflect different dispersal capabilities across non-forested ecological barriers such as the possible “savanna corridor” in interior Sundaland (Heaney, 1991; Sheldon et al., 2015).
Within the genus Sundasciurus, previous phylogenies based on mitochondrial cytochrome b sequences have identified deep divergences between lineages within Low’s squirrel (S. lowii), and a lack of reciprocal monophyly between Low’s squirrel and the Fraternal squirrel (S. fraterculus) from the Mentawai islands (Den Tex et al., 2010; Hawkins et al., 2016a). Low’s squirrel is a good example of a widespread, lowland forest dwelling tree squirrel. It is currently listed as a Species of Least Concern by the IUCN, largely based on its wide historic distribution (Meijaard, 2016). However, the lowland tropical forest is rapidly disappearing in this region due to human induced habitat modifications and its current status and distribution need to be re-evaluated. Furthermore, it is possible that the widely distributed Low’s squirrel is actually a complex of multiple species each with a more restricted range. Low’s squirrel contains seven nominal subspecies distributed across Borneo (S. l. lowii), northern Borneo’s offshore islets Banguey and Balambangan (S. l. bangueyae), the Malay Peninsula (S. l. robinsoni), southern Malay Peninsula Riau offshore islands (S. l. seimundi), Sumatra (S. l. humilis), west Sumatra Batu offshore islands (S. l. balae), and the Natuna islands (S. l. natunensis) (Thorington and Hoffmann, 2005; Thorington et al., 2012) all based on morphology (Figure 1).
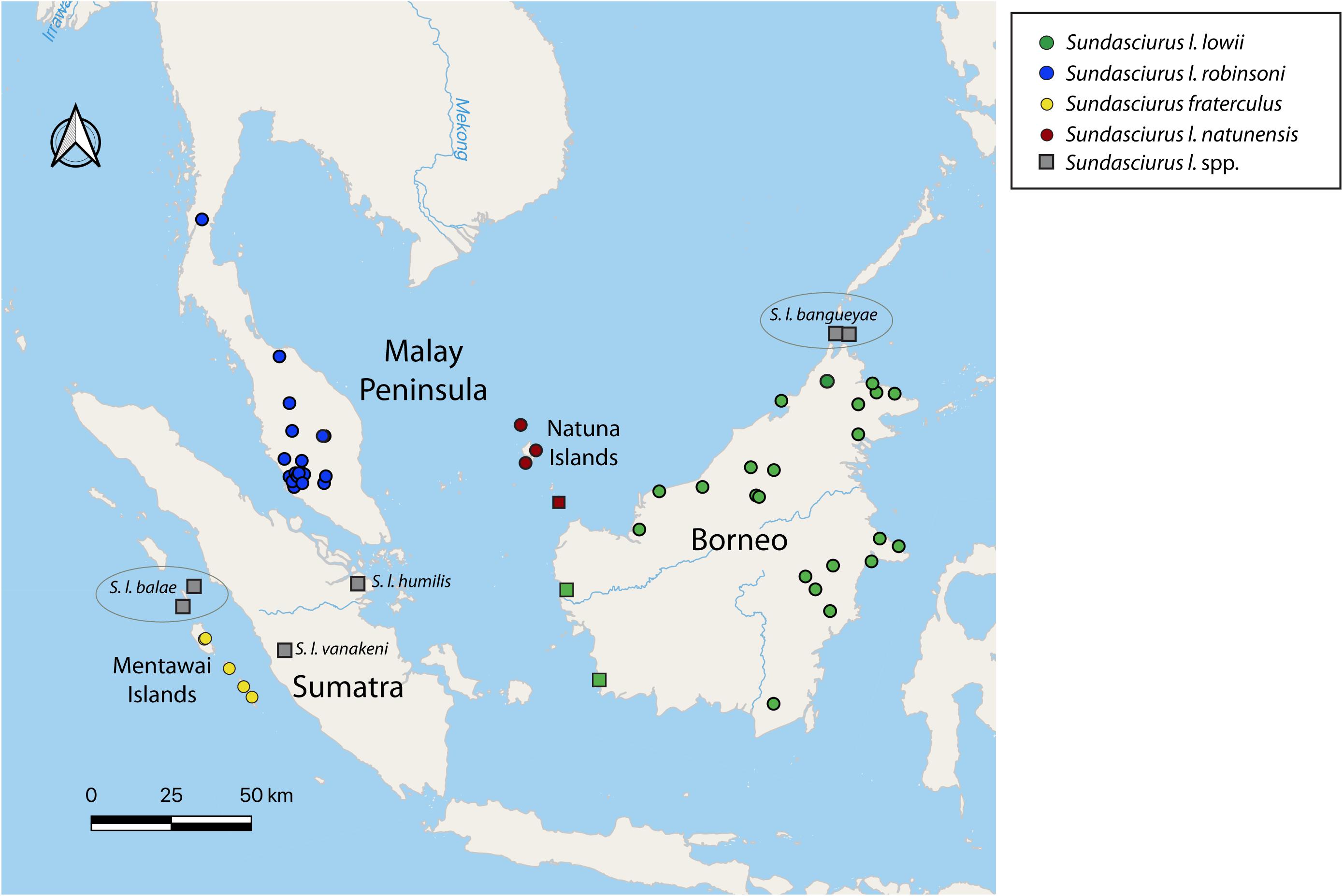
Figure 1. Map of Sundaland showing the distribution of the different subspecies of the Low-Fraternal squirrel species complex, with dots indicating sample collection locations of specimens included in the morphometric analyses. Samples are colored according to the four highly supported major clades recovered in the mitogenome-based phylogenies of Figures 2, 3. Populations indicated by a red circle have been considered S. l. natunensis but the morphological data in this study suggests they are more similar to S. l. lowii. Populations indicated with a green square have been considered S. l. lowii but the morphological data in this study suggests these they are more similar to S. l. natunensis. Molecular evidence from these populations is needed to clarify their taxonomic status. Populations included in the morphological analyses but not the genetic analyses are indicated in gray.
In this study, we use genetic data including complete mitochondrial genomes and twelve nuclear loci from across the Sundasciurus lowii and S. fraterculus distribution to construct a well-resolved phylogeny of the group, and to date divergences. These results are placed in the context of the morphological variation and distribution of the different taxa in order to re-evaluate and resolve the number of species in this group. Additionally, a comprehensive taxonomic evaluation of this species complex will have important conservation implications that could lead to the re-assessment of the IUCN Red List category for some of these squirrels based on their updated distributional ranges. It will also provide a better understanding of the ecological and evolutionary processes that led to the high diversification rates of these forest dependent squirrels in this region.
A total of 11 samples of Sundasciurus lowii and S. fraterculus from the major landmasses and isolated archipelago populations (Malay Peninsula, Borneo, Mentawai Islands and Natuna Islands) and 29 outgroups were included in the molecular part of the study (Supplementary Table S1). Populations from Sumatra and satellite islands (Batu and Riau) were not sampled, because only type specimens are available (S. l. humilis, S. l. balae, S. l. seimundi). In any case, different studies have suggested recent connections among Sumatra and Peninsular Malaysia (Ruedi and Fumagalli, 1996; Mason et al., 2011; Camacho-Sánchez et al., 2017) and among these satellite islands and the nearest landmasses (Mason et al., 2019) so our geographic coverage could potentially represent most of the genetic structure of this complex. Twelve samples were derived from tissue collected from historical specimens housed in three museums, 10 samples were modern tissue coming from museum loans and 14 sample sequences were downloaded from Genbank. Four samples were obtained from specimens collected during a previous expedition to Borneo (Camacho-Sánchez et al., 2019) and these were all taken according to the guidelines of the American Society of Mammalogists (Sikes, 2016) and as approved by Institutional Animal Care and Use Committees (Smithsonian Institution, National Museum of Natural History, proposal number 2012-04 and Estación Biológica de Doñana proposal number CGL2010-21424). Field research was approved by Sabah Parks [TS/PTD/5/4 Jld. 45 (33) and TS/PTD/5/4 Jld. 47 (25)], the Economic Planning Unit (100-24/1/299) and the Sabah Biodiversity Council (Ref:TK/PP:8/8Jld.2). All recognized species and subspecies of this complex were included in the morphological analyses (Figure 1 and Supplementary Table S2).
DNA was extracted using phenol-chloroform with ethanol precipitation. Amicon Ultra 0.5 mL filters were used for the final clean instead of ethanol precipitation for historic samples. All museum samples were processed in an isolated ancient DNA laboratory following strict protocols to control for contamination.
Twelve nuclear loci (introns) previously found to be present in single copies and informative in closely related mammalian species (Igea et al., 2010) were targeted. Primer sets for four of those introns developed for Tamiasciurus squirrels by Chavez et al. (2014) worked in our taxa as a multiplex. For the other eight loci, we designed primers following the steps of Forcina et al. (2020). We developed 15 primer sets to amplify smaller fragments (ranging from 88-233 base pairs) of these same loci for the low quality specimens (Supplementary Table S1). PCR conditions for the two modern DNA and four historic DNA multiplexes are in Supplementary Table S3. PCR products were purified using streptavidin magnetic ‘SPRI’ beads (Rohland and Reich, 2012) and visualized on agarose gels. Given the degraded nature of historic DNA, three replicate PCRs were run per sample. Amplicon libraries were cleaned and checked on a gel with reference standards for quantification (Image Lab 5.2.1 software), and were pooled at equimolar concentrations. Illumina TruSeq adapters with an individual index were added to the purified PCR products in a second PCR as performed in Camacho-Sánchez et al. (2017).
We used target capture to recover complete mitochondrial genomes. We used an Illumina modification to the protocol based on Maricic et al. (2010) to enrich complete mitogenomes from historic samples. Enrichment conditions followed kit protocols as described in Hawkins et al. (2016b). Briefly, samples were denatured, incubated for ∼24 h, recovered the baits, and amplified from the p5/p7 priming site on the Illumina adapters for a limited number of PCR cycles to generate concentrations of mitochondrial DNA which could be sequenced. PCR products were purified as above.
Dual-indexed shotgun libraries were sequenced on an Illumina MiSeq using 2 × 300 (600 cycle) PE at the Center for Conservation Genomics, Smithsonian Conservation Biology Institute.
Adaptor removal and quality trimming was performed with Trimmomatic (Bolger et al., 2014) with the sliding window parameter set to 5:20, read minimum length parameter to 50 bp and leading and trailing to 5. Single quality scans were run on the raw fastq files before and after trimming with FastQC (Andrews, 2010) and MultiQC (Ewels et al., 2016) was run to create a single report visualizing the output from all the samples of each run, enabling global trends and biases to be quickly identified. Mitochondrial genome reads were preprocessed following the protocol described in Hawkins et al. (2016b).
Quality trimmed reads were mapped to a reference mitochondrial genome with BWA-MEM algorithm or with Geneious Mapper iterative algorithm (up to 10 iterations and medium-sensitivity) for some divergent samples. The output of BWA was converted to BAM files with SAMtools (Li et al., 2009). These were later sorted, merged and PCR duplicates removed. Finally, BAM files were imported to Geneious where consensus sequences were called (minimum 5x and 75% threshold). Samples mapped in Geneious were inspected for duplicate removal with Dedupe plugin of the BBTools package v. 35.82 (Bushnell, 2015). We aligned our sequences with Callosciurinae whole mitochondrial sequences available in Genbank (Supplementary Table S1) with MAFFT v7.450 Geneious plugin automatic algorithm (Katoh and Standley, 2013) under default parameters (scoring matrix:200PAM/K = 2, gap open penalty = 1.53, offset value = 0.123). The control region was removed from the mitogenome assemblies because it was poorly assembled in many historic samples and has been shown to provide low phylogenetic resolution and overestimation of divergence times (Duchêne et al., 2011). ND6 is present in the light strand and was thereby reverse complemented. Protein-coding regions were translated and inspected for frameshift mutations and for the presence of unexpected stop codons to prevent inclusion of NUMTs. We computed summary statistics of the alignment with AMAS (Borowiec, 2016).
For nuclear sequences, we imported the trimmed reads to Geneious and mapped these to the closest homologous sequences found in Genbank (Marmota marmota (NW_015351277.1, NW_015351233, XM_027934759, NW_015351257, NW_015351271, NW_01535121) and Tamasciurus hudsonicus (KF883544, KF883925, KF885067) with Geneious Mapper (low-medium sensitivity and up to 5 iterations). We called consensus sequences in Geneious with a minimum 5× coverage and 75% consensus threshold. Each locus was aligned independently with MAFFT v7.450 Geneious plugin automatic algorithm (Katoh and Standley, 2013) under default parameters (scoring matrix:200PAM/K = 2, gap open penalty = 1.53, offset value = 0.123). We translated the alignments to amino acids and inspected them for insertions, deletions and premature stop codons to prevent inclusion of paralogous sequences. Amplicon sizes exceeded the length generated by the Illumina sequencing for two loci (GDAP1, 795 bp and P4HA2, 758 bp). This generated missing data in the center of those alignments (203 and 295 bp), which were trimmed.
All nuclear sequences were resolved into statistically probable haplotypes using PHASE 2.1.1 (Stephens, 2005) with an acceptance threshold of 0.90. The online application SeqPHASE (Flot, 2010) was used to convert FASTA files to PHASE input files, as well as to convert PHASE output back to FASTA format. Due to the different size of modern and historic sample sequences, the initial gene alignments contained sections of missing data for the shorter historic samples. Since PHASE can not deal with missing data in heterozygous sites, two alignments (historical and modern samples) were generated for each gene to remove most missing data and overcome this problem. Genotypes that were still unresolved following PHASE were phased manually based on the original BAM alignment (Camacho-Sánchez, 2017).
We performed a likelihood ratio test (LRT) with MEGA 7 to test the clock-like behavior of the mitochondrial sequences. A strict molecular clock was rejected. The optimal partitioning scheme and substitution model was selected by PartitionFinder 2.1.1.2 (Lanfear et al., 2016). We followed Mackiewicz et al. (2019) considering 3 codon positions for each individual protein coding gene and separate partitions for each of the RNA genes, but retained contiguous t-RNAs because individual t-RNA alignments would be too short to be informative (Nicolas et al., 2019). This reduced the number of initial partitions from 67 to 53. We used the “greedy” algorithm with branch length estimated as “unlinked”, AICc criteria and “–raxml” command line option to search for the best-fit partitioning scheme (11 partitions) and substitution model (GTR + GAMMA). We tested for saturation plotting transitions and transversions of each alignment partition against each other and against raw/uncorrected pairwise genetic distances with the APE R package and “dist.dna” command. We also performed Xia substitution saturation tests in Dambe 7 (Xia, 2018) for the 3rd codon of some fast-evolving genes (e.g., Cytochrome b) as a complementary line of evidence. We did not find saturation so all partitions were kept in downstream analyses. We performed phylogenetic inference through a maximum likelihood framework with RAxML (Stamatakis, 2014) on the mitochondrial genome dataset. We specified the output of PartitionFinder2 as input for RAxML (Stamatakis, 2014). We selected the rapid bootstrapping algorithm, GTR GAMMA substitution model and followed the extended majority rule stopping criterion (-autoMRE). The maximum likelihood phylogeny was used for the BEAST analyses.
Divergence times were estimated with our mitochondrial dataset in BEAST2 (Bouckaert et al., 2014). The mitochondrial genome alignment was split into the best partition schemes as identified by Partitionfinder2 with AMAS. We performed site modeling through a Bayesian approach with bmodeltest, selecting mutation rate estimation and empirical frequencies priors (Bouckaert and Drummond, 2017). Following MEGA7 tests, a lognormal (uncorrelated) relaxed clock was used under a Yule speciation tree, with linked partitions and default operators. Only one sequence per putative species was kept to comply with Yule speciation prior assumptions. Three independent runs of Markov chains (MCMC) for Monte Carlo simulations were run for 50,000,000 generations, with parameters and trees sampled every 5000 generations. Convergence was checked using Tracer 1.7.1 (Rambaut et al., 2018). For each run, the first 10% of sampled trees were discarded as burn-in. Two fossil constraints were selected to calibrate the phylogeny. A fossil of Callosciurus sp. (specimen number: YGSP 21682; left upper third molar that conforms to the proportions of living Callosciurus) from locality Y589 (Chinji formation, Siwaliks, Pakistan) dated by paleomagnetic correlation to 14 Ma (Flynn, 2003) for the Callosciurus and Sundasciurus clade; and a stem form of Tamiops (specimen numbers: Z2438, Z2439, Z2440, Z2441) from Z122 (Vihowa Formation) dated by paleomagnetic correlation to 18.7 Ma (L. Flynn, pers. comm.) as the oldest known fossil for the Callosciurus, Sundasciurus, Tamiops, and Dremomys clade. All constraints were set using hard minimum bounds and soft upper bounds using a lognormal prior, as suggested by Parham et al. (2012).
We performed TCS haplotype networks with popART (Leigh and Bryant, 2015) separately, for the modern and historic alignments of the mitochondrial genomes and nuclear intron sequences. We computed uncorrected pairwise genetic distances on the Cytochrome b gene with the APE R package and dist. dna command (Paradis et al., 2004). Results were visualized with ade4:table.paint. Finally, a multidimensional scaling analysis was performed for both datasets with stats:cmdscale and ggplot2. We defined the a priori groups for the species tree analysis within our ingroup following four criteria: Cytochrome b uncorrected genetic distances > 5% (Baker and Bradley, 2006) reciprocal monophyly in mitochondrial genome tree, clusters in MDS, presence of private alleles in the intron haplotype networks and phenotype differentiation. Based on these criteria we considered six groups within our ingroup as input for the ∗BEAST species tree: Sundasciurus fraterculus, S. lowii robinsoni, S. l. natunensis, S. l. lowii from Sabah, S. l. lowii from Sarawak, and S. l. lowii from East Kalimantan.
We included all intron phased alleles available per group in the ∗BEAST analysis, and unlinked partitions, selected an uncorrected lognormal clock, bmodeltest with empirical frequencies and mutation rate estimation, analytical population size integration model and Yule speciation prior. Three independent runs were conducted for 50000000 generations Markov chains (MCMC) for Monte Carlo simulations, with parameters and trees sampled every 5000 generations.
Species delimitation was conducted using the program BPP X1.2.2 (Yang, 2015) details regarding this analysis are specified in the Supplementary Material File 1.
The specimens examined here are housed in the following natural history repositories: American Museum of Natural History, New York, United States (AMNH); Natural History Museum, London, United Kingdom (BMNH); Estación Biológica de Doñana, Seville, Spain (EBD); Field Museum of Natural History, Chicago, Illinois (FMNH); Museum of Comparative Zoology, Harvard University, Cambridge, Massachusetts (MCZ); and National Museum of Natural History, Smithsonian Institution, Washington, DC, United States (USNM) Supplementary Table S2. Skins were carefully examined to avoid any misidentifications and a geographically balanced subset was photographed for further evaluation. During our initial surveys of the squirrel collections, we found that a high proportion of the skulls were broken and therefore decided to undertake traditional morphometric analyses instead of geometric morphometrics. Dental eruption and wear patterns were checked to age each specimen. Fusion of the presphenoid-basisphenoid and basisphenoid-basioccipital sutures were not informative to age individuals in this taxon since some large-sized adults with very worn molars had non-fused sutures. Only adults were included in the morphometric analyses. We combined data from males and females in the morphometric analyses because sexual dimorphism is uncommon in tree squirrels (Moore and Tate, 1965; Tenzin et al., 2013; C̀anády et al., 2015). However, if certain characters were sexually dimorphic, results would be interpreted in the context of phenotypic variation among putative species inclusive of potential patterns of sexual dimorphism, thereby providing a more conservative assessment of divergence (Meik et al., 2018). Seventeen cranial measurements were taken with a Fowler High Precision electronic digital caliper to the nearest 0.01 mm by AH (Supplementary Table S2). These were occipitonasal length (ONL), zygomatic breadth (ZB), interorbital breadth (IB), length of nasals (LN), anterior nasal breadth (ANB), length of rostrum (LR), height of rostrum (HR), mastoid breadth (MB), height of braincase (HBC), length of orbit (LO), length of diastema (LD), length of bony palate (LBP), breadth of bony palate at fourth premolar (BBP), length of auditory bulla (LB), crown length of maxillary cheek teeth (CLPMM), median palatal length (MPL) and least breadth of caudal point of zygomatic process of frontal bone (LBC). All measurements are as defined in Musser et al. (2010) except median palatal length (MPL) and least breadth of caudal point of zygomatic process of frontal bone (LBC), which are defined in Hayashida et al. (2007). In addition, given the curved rostrum of our target taxa, the height of rostrum (HR) was modified after conducting preliminary error measuring tests. We define it as the distance from dorsal surface of the nasals to ventral surface of the premaxillary at the premaxillary-maxillary suture and taken perpendicularly to the molar row axis. External measurements, head-body length (HB), tail (T), hindfoot (HF), ear (E) and weight (W), were taken directly from specimen labels (Supplementary Table S2). We examined, measured and photographed all of the types of our target taxa.
We performed standard univariate descriptive statistics (mean, standard deviation and observed range) for each of the subspecies (Thorington and Hoffmann, 2005) and also for each of the S. lowii Borneo populations in a separate analysis (Supplementary Table S4). We log-transformed each measurement to its standard deviation prior to computing the principal component analyses (PCA) so that the data was analyzed on the basis of correlations instead of covariances. PCA was implemented by the prcomp command in R studio (Allaire, 2012; R Core Team, 2018). Results were extracted and visualized with the following functions of the factoextra package (Kassambara and Mundt, 2017): fviz_pca_ind, plots PCA results; fviz_pca_biplot, biplot of individuals and variables; get_eigenvalue, extracts eigenvalues, variance percentage and cumulative variance percentage; get_pca_var, outputs each variable’s contribution to variance.
We conducted a discriminant analysis of the principal components (DAPC) with the dapc function of adegenet (Jombart and Ahmed, 2011). Identification of the clusters was made with find.clusters. This function first transforms the data using PCA, asking the user to specify the number of retained principal components (PCs) interactively. Then, it runs k-means algorithm with increasing values of k and computes associated summary statistics (by default, BIC). We performed a cross-validation with the xvalDapc() function with default parameters to decide the number of PCs to retain. This function runs 30 replicates of cross-validation for a number of PCs less than the total number of variables. After having an approximate idea of the PCA axes range to be retained, we re-ran the cross-validation with 1000 replicates. Different number of clusters had similar BIC values so a DAPC was run for each scenario (3-10 clusters). Different DAPCs (retaining 13-3 PCs and 6-3 discriminant functions) with a priori grouping for each of the subspecies (Wiley, 1978; Thorington and Hoffmann, 2005) and for each of the S. lowii Borneo populations (in a separate analysis) were also performed. We interpreted group memberships with the functions summary, compoplot and assignplot to test how clear-cut clusters were. These functions indicate the proportions of successful reassignment (based on the discriminant functions) of individuals to their original clusters. To increase the number of samples (from 55 to 152) and include the types, we made bivariate plots of craniodental variables based on the output of the DAPC and standard external measurements.
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser at https://www.urn:lsid:zoobank.org:pub:727A0DA1-2B92-4400-A3C2-CE816BE00B1F
Whole mitochondrial genomes were constructed from 24 individuals, and their alignment (without control region) was 15735 bp, with 5557 parsimony informative sites and 8.97% missing data. Missing data was mainly in the sequences from the eleven historic samples (15–47%). Mitochondrial genomes of all ingroup taxa were successfully assembled, although only 53% of one Sundasciurus lowii natunensis from two attempted samples could be reconstructed. Similarly, Sundasciurus lowii natunensis was the only taxon from which we failed to amplify any introns. Three intron loci were not informative or did not amplify correctly (GAD2, SLC17A9, Ulk1b) and were excluded from downstream analyses.
All nodes were strongly supported (Bootstrap support (BP) > 84/Posterior probability (PP) > 0.98) in the mitochondrial genome maximum likelihood tree (ML) and Bayesian maximum clade credibility tree (BMCC) and the topology was consistent between approaches (Figures 2, 3). Only the relationships among S. l. lowii had weak support in the ML tree (BP = 0.66) but strong support in the BMCC (PP = 0.99). These tree topologies were consistent with previous studies (Den Tex et al., 2010; Hawkins et al., 2016a). The Low’s squirrel complex contained two divergent clades from east and west Sundaland. The former included S. l. lowii from Borneo and S. l. natunensis from the Natunas, while the latter had S. l. robinsoni from peninsular Malaysia and S. fraterculus from Mentawai Islands. The sister relationship of S. fraterculus with S. l. robinsoni makes S. lowii sensu lato paraphyletic as in Den Tex et al. (2010). Within S. l. lowii, the population from Sarawak was sister to the clade containing the ones from Sabah and East Kalimantan. All genera (Tamiops, Dremomys, Callosciurus and Sundasciurus) were recovered as monophyletic. Previously suggested subgenera within Sundasciurus (Aletesciurus and Sundasciurus; Moore, 1958) were not found to be reciprocally monophyletic. Previous studies either could not resolve the lower nodes or found them not to be monophyletic (Den Tex et al., 2010; Hawkins et al., 2016a) and thus had suggested suspending that nomenclature (Hawkins et al., 2016a).
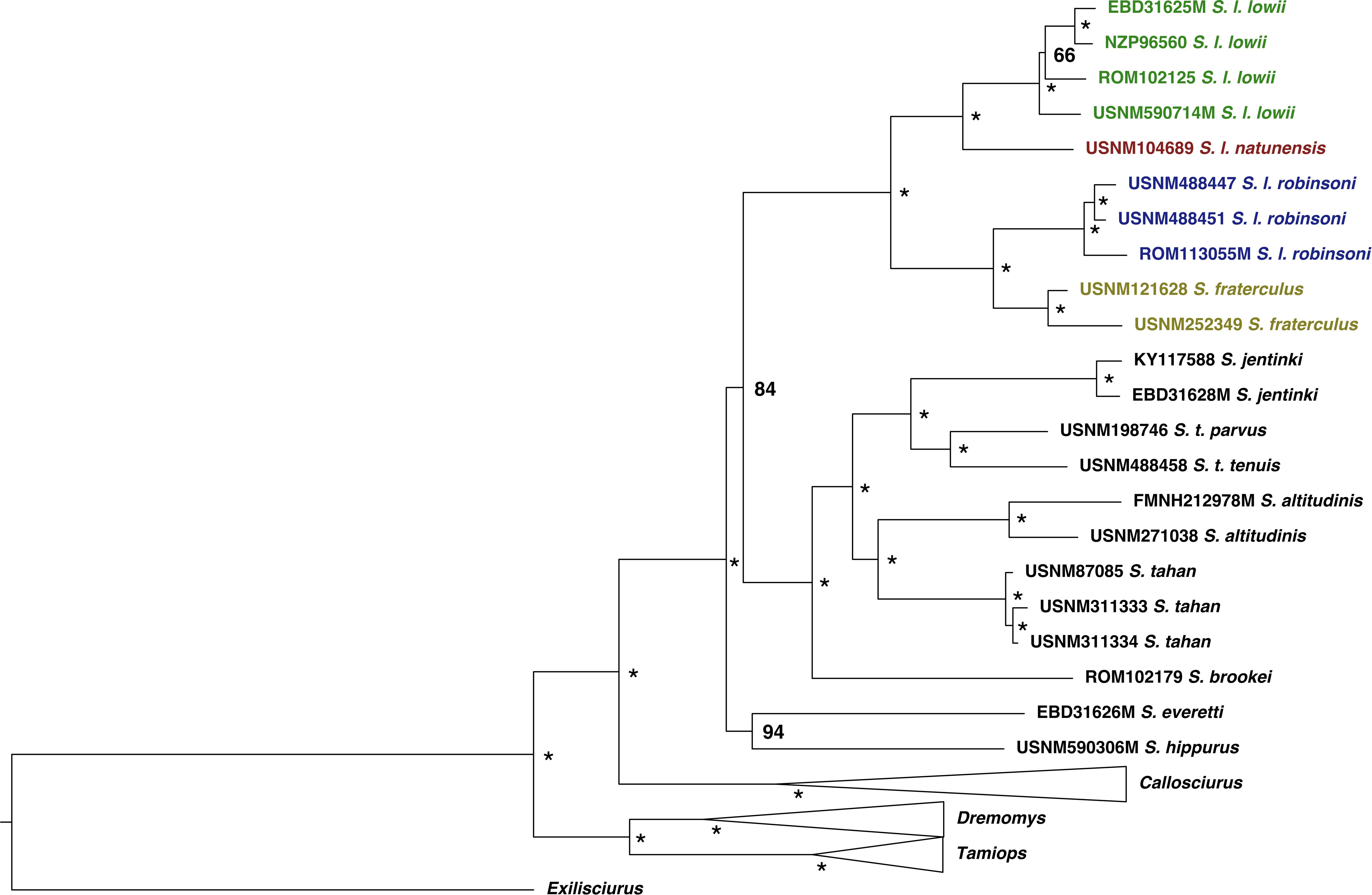
Figure 2. Mitogenome-based maximum likelihood phylogeny for Sunda squirrels. Highly supported nodes (PP > 0.95) are shown with an asterisk. Posterior probabilities are specified in less supported nodes. Ingroup samples of the four main clades are colored to match map in Figure 1. Outgroup samples are shown in black. Outgroup genera clades are collapsed for clarity.
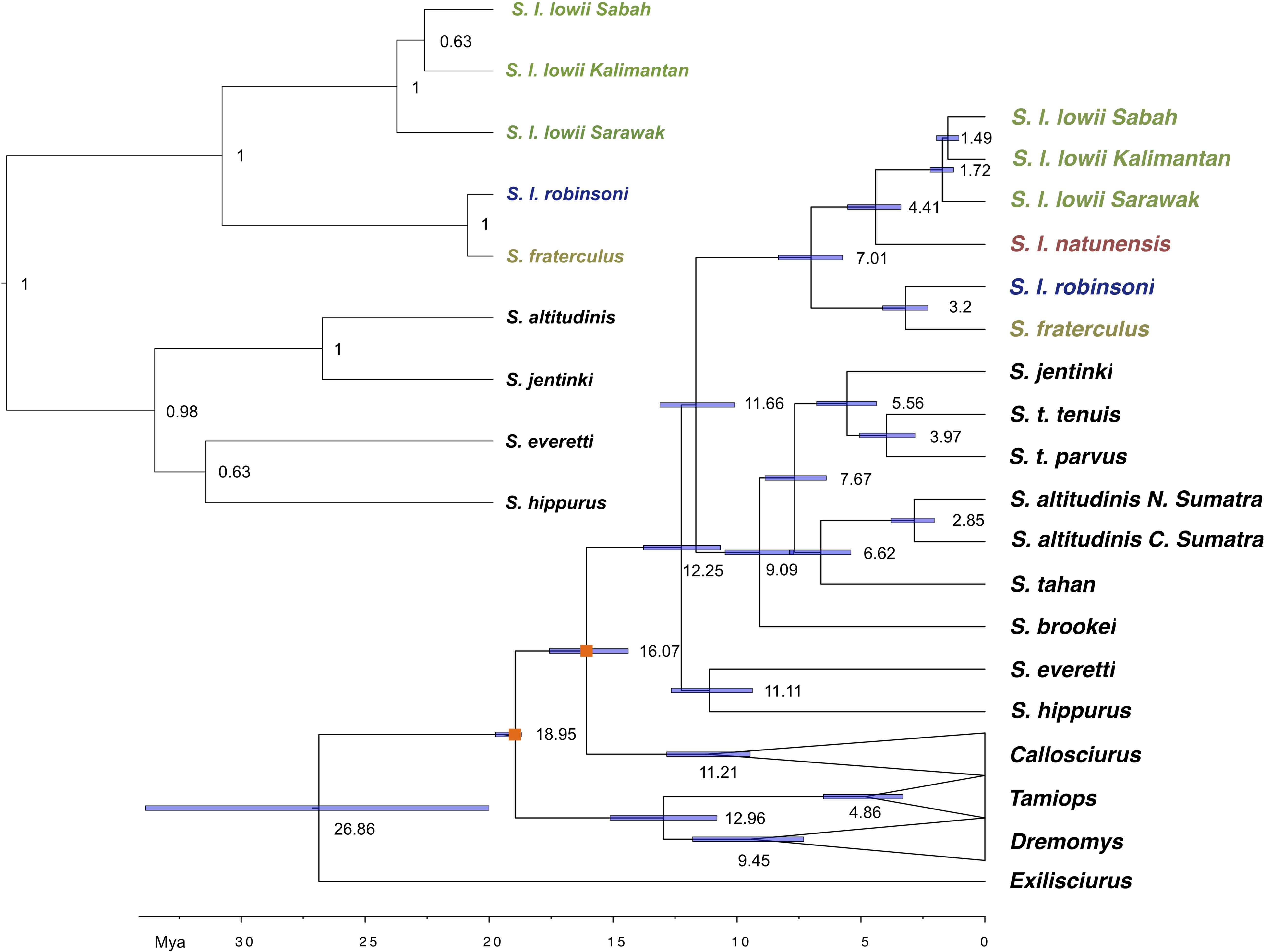
Figure 3. Mitochondrial genome Bayesian maximum clade credibility tree including all Sundasciurus species from Sundaland. Dates are indicated on the bar at the bottom of the figure in millions of years before present (Mya) and on nodes. Nodes used to calibrate the dating are marked with an orange box. All nodes are fully supported (PP = 1). Inset shows a species tree of the Low’s and Fraternal squirrel species complex, as estimated from nine nuclear loci using *BEAST. Ingroup samples of the four main clades are colored to match Figure 1 map. Outgroup samples are shown in black. Outgroup genera clades are collapsed for clarity.
The time to the most recent common ancestor (TMRCA) of Callosciurinae and Sundasciurus was estimated at 26.86 (95% HPD: 20.00–33.85) and 12.25 (95% HPD: 10.67–13.67) million years ago (MYA). These dates are older than those previously estimated by Mercer and Roth (2003) who associated the diversification of Sundaland genera with the lowest pre-Pleistocene sea stand in the Cenozoic about 11 MYA. TRMCA for the subgenus Sundasciurus and Sundasciurus lowii complex were estimated at 11.65 (95% HPD: 10.09–13.11) and 7.01 (95% HPD: 5.74–8.33) MYA. At 4.41 (95% HPD: 3.39–5.53) MYA one of Sundasciurus lowii complex lineages split into the ancestors of S. l. lowii clade and S. l. natunensis. At 3.20 (95% HPD: 2.30–4.12) MYA the other Sundasciurus lowii complex lineage split into the ancestors of S. fraterculus and S. robinsoni. The lowland Sundasciurus tenuis and highland S. altitudinis exhibited remarkable deep levels of intraspecific divergence dated at 3.96 (95% HPD: 2.82–5.05) and 2.85 (95% HPD: 2.04–3.79) MYA. These dates are older to what was previously estimated from Cytochrome b by Den Tex et al. (2010).
Many subspecies had private alleles (Figure 4 and Supplementary Figure S1). The taxa S. l. robinsoni with S. fraterculus and S. l. lowii had private alleles for all introns. Within Borneo S. l. lowii of Sabah, S. l. lowii of Kalimantan and S. l. lowii of Sarawak had private alleles for seven out of nine introns. The narrow endemic S. fraterculus had private alleles for three of nine introns.
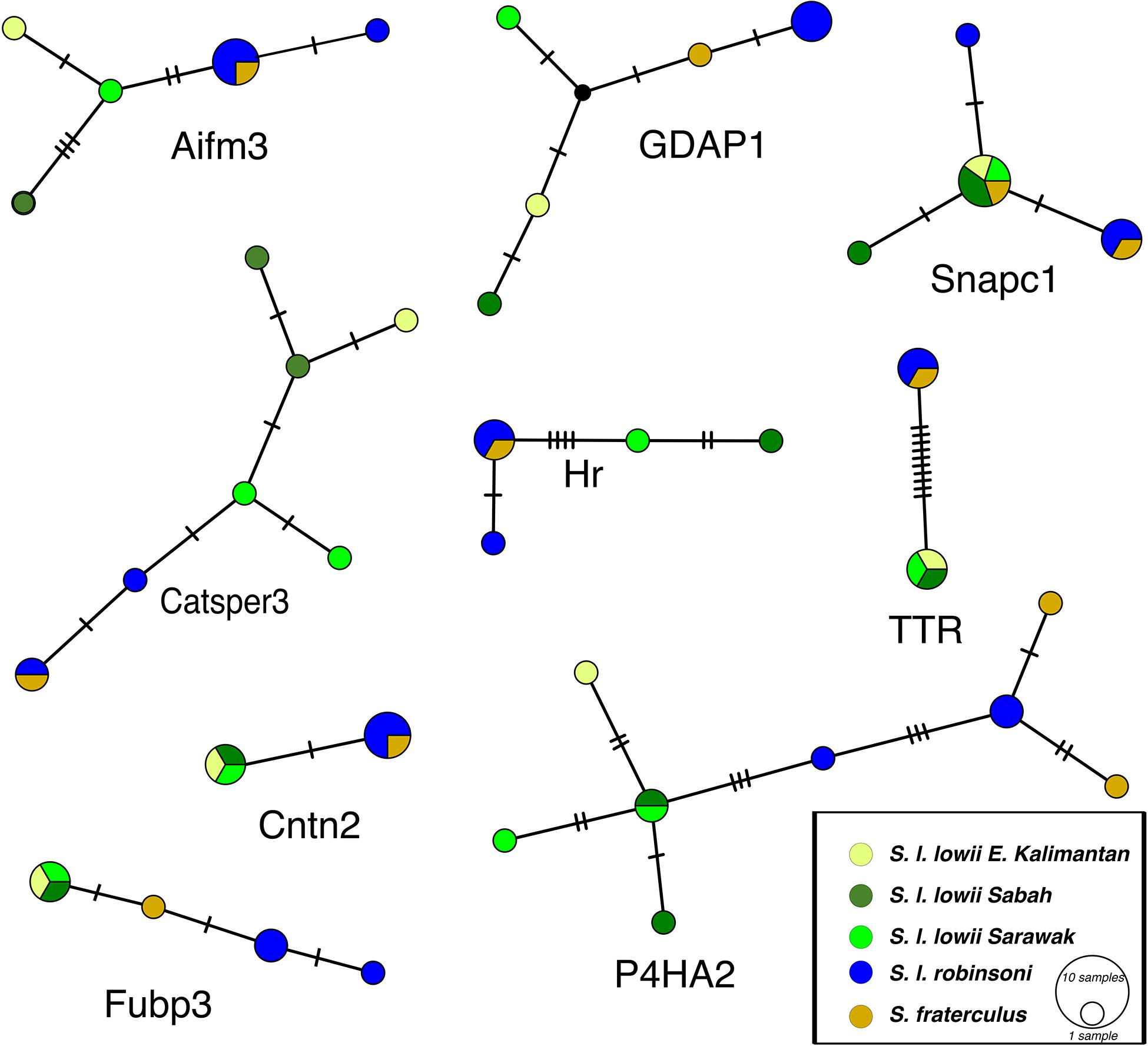
Figure 4. TCS haplotype networks for each of the nine polymorphic introns studied. The size of the pie charts corresponds to the frequency of the haplotype; transversal lines represent single base pair differences; black dots represent missing haplotypes.
Cytochrome b uncorrected genetic distances among some of the taxa (S. l. robinsoni, S. l. natunensis, S. l. lowii of Sabah and S. l. lowii of Sarawak) were above 5% (Supplementary Figure S2), a scale of divergence which may suggest a revision of the status of those taxa could be of interest (Baker and Bradley, 2006). Genetic distances among these taxa were higher than that observed among other recognized species (Supplementary Figure S2). These results further highlight that while Cytochrome b distances might indicate potential unrecognized genetic species, it should never be used alone to define species. We note that high percent sequence divergence levels were also observed between S. t. tenuis and S. t. parvus, and among populations within S. hippurus, S. mindanensis, and S. altitudinis (Supplementary Figure S2), suggesting that additional work on these groups in the future could shed more light on the evolution of this genus.
Phylogenetic relationships inferred by the intron species tree constructed in ∗BEAST were concordant with the mitochondrial genome tree topology. Only paraphyly of the subgenus Sundasciurus in the nuclear species tree was inconsistent with the mitochondrial topology, but concordant with Hawkins et al. (2016a)Figure 3. However, deep nodes such as the MRCA of S. everetti and S. hippurus or shallow ones such as the relationships among Bornean S. lowii were weakly supported (PP = 0.62-0.63) while the other nodes were strongly supported (PP > 0.99).
Our analyses of nuclear data were consistent across the independent BPP runs and among the five prior combinations. We inferred a posterior probability of 1.0 for all species postulated by the ∗BEAST guide tree except S. fraterculus and S. l. lowii from E. Kalimantan. The former was not supported (PP = 0.07–0.26) and the latter went from weakly to moderately supported (PP = 0.51–0.95). The most likely scenario in every run supported all species except S. fraterculus (PP = 0.41–0.88).
A high degree of morphometric differentiation among the different populations was observed (Figures 5, 6) with a large part (82%) of the variance explained by the first principal component. PC1 exhibited different loadings for most variables, with crown length of maxillary cheek teeth (17%), length of nasals (14%) and breadth of bony palate (10%) driving most of the variation. PC1 discriminated the larger-sized subspecies S. l. lowii and S. l. bangueyae from the smaller-sized subspecies S. l. robinsoni, S. l. humilis, S. l. balae and S. fraterculus. PC2 explained 5.7% of the variance and was mainly correlated with shape robustness/broadness, bulla and molar row length. The variables with a higher contribution were crown length of maxillary cheek teeth (26%), interorbital breadth (21%), anterior nasal breadth (20%), length of bulla (11%), and least breadth of caudal point of zygomatic process of frontal bone (7%). It discriminated the relatively gracile and large molar S. l. robinsoni, southern Natuna S. l. natunensis, east and west Kalimantan S. l. lowii and S. l. bangueyae, from the relatively robust/broad shaped and small molar S. fraterculus, S. l. balae, S. humilis, Sarawak and most Sabah samples of S. l. lowii, and northern Natuna S. l. natunensis. PC3 explained 3.3% of the variance and was correlated with anterior nasal breadth (35%), crown length of maxillary cheek teeth (14%), length of bulla (10%) and mastoid breadth (9%). It discriminated the narrow-nasal shaped S. l. robinsoni, S. l. balae, S. humilis, northern Natuna S. l. natunensis and east and west Kalimantan S. l. lowii from broad-nasal shaped S. fraterculus, S. l. bangueyae, southern Natuna S. l. natunensis and Sarawak and east-central Kalimantan S. l. lowii. Sabah S. l. lowii fell in between.
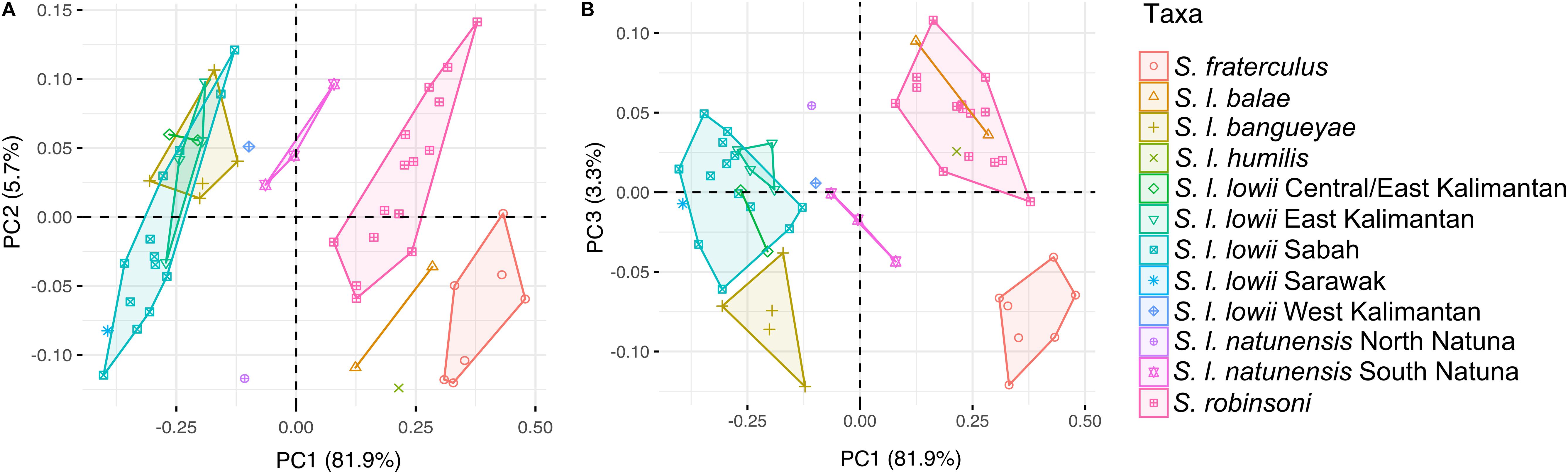
Figure 5. Inter- and intraspecific morphometric variation among species of the Low’s and Fraternal squirrel complex. (A) shows first and second principal components from an analysis of 17 cranial dimensions, while (B) shows first and third principal components of that same analysis. Samples are labeled and colored according to the different subspecies but subspecies with high genetic/phenotypic variation (S. l. lowii and S. l. natunensis) have been split into geography-based categories.
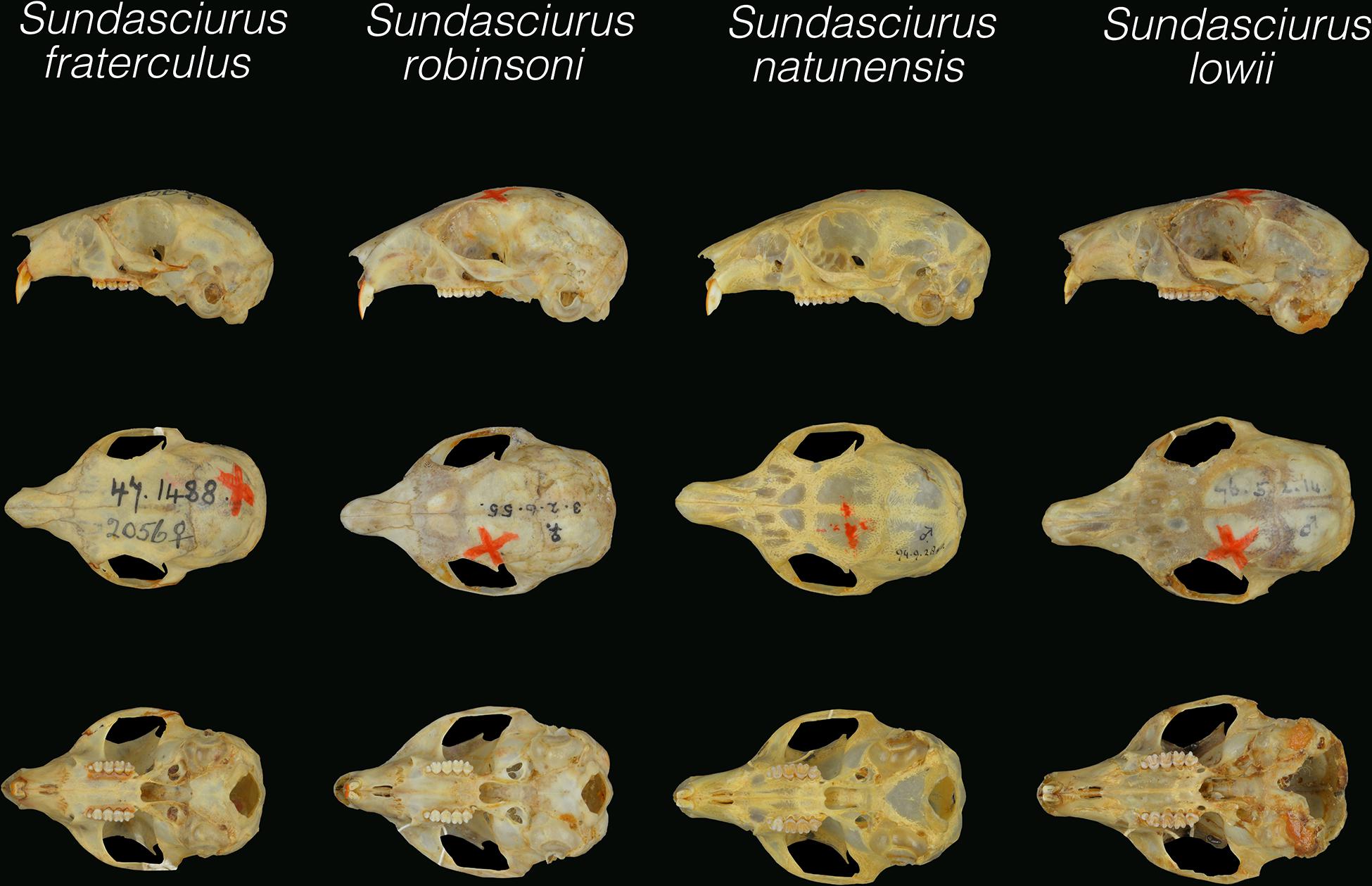
Figure 6. Dorsal, palatal, and lateral views of the holotype skulls of Sundasciurus lowii (NHM76.5.2.14), Sundasciurus natunensis (NHM94.9.28.40) and Sundasciurus robinsoni (NHM3.2.6.55). Given the highly damaged state of the holotype skull of Sundasciurus fraterculus (NHM95.1.9.12), the type of Sundasciurus fraterculus siberu (NHM47.1488) is shown instead for the sake of comparison. All pictures are at the same scale.
The DAPC discriminated most subspecies (Supplementary Figure S3). Some overlap was observed between S. l. balae and S. l. robinsoni, and between S. l. lowii and S. l. bangueyae. The Mount Kerinci sample of S. l. humilis (belonging to the junior synonym S. l. vanakeni) had an intermediate position between S. fraterculus and S. l. robinsoni, while the only west Bornean sample of S. l. lowii was clustered between other Borneo S. l. lowii and S. l. natunensis. Only samples from East Kalimantan could be discriminated from the remaining Bornean populations. This analysis identified crown length of maxillary cheek teeth, bony palate breadth, orbit length, rostrum length, interorbital breadth, and bulla length, as the measurements most useful for distinguishing among all taxa (Supplementary File S1).
Sundasciurus robinsoni (Bonhote, 1903) – Robinson’s squirrel
Sciurus robinsoni Bonhote (1903):24.—Type locality. Bukit Besar, Nawngchik, Patani, Peninsular Thailand, 2500 ft.
Holotype. BMNH 3.2.6.55, skull, skin, adult female, collected 30 August 1901 by C. H. Robinson and N. Annandale
Sciurus balae Miller, 1903:14.—Type locality. Tana Bala, Batu Islands, Sumatra
Sciurus piniensis Miller, 1903:14.—Type locality. Pulo Pinie, Batu Islands, Sumatra
Sciurus seimundi Thomas and Wroughton, 1909:440.—Type locality. Kundur Island, Riau Islands
Sciurus humilis Miller, 1913:24.—Type locality. Kateman river, East Sumatra
Sciurus vanakeni Robinson and Kloss, 1916:270.—Type locality. Barong Bharu, Korinchi, Sumatra, 4000 ft.
Sundasciurus robinsoni (Bonhote, 1903)
Sciurus robinsoni alacris Thomas, 1908e: 306.—Type locality. Selangor-Pahang border, Malaya, 3000ft.
Sciurus seimundi Thomas and Wroughton, 1909:440.—Type locality. Kundur Island, Riau Islands
Sundasciurus robinsoni balae (Miller, 1903)
Sciurus balae Miller, 1903:14.—Type locality. Tana Bala, Batu Islands, Sumatra
Sciurus piniensis Miller, 1903:14.—Type locality. Pulo Pinie, Batu Islands, Sumatra
Sciurus humilis Miller, 1913:24.—Type locality. Kateman river, East Sumatra
Sundasciurus robinsoni vanakeni (Robinson and Kloss, 1916)
Sciurus vanakeni Robinson and Kloss, 1916:270.—Type locality. Barong Bharu, Korinchi, Sumatra, 4000 ft.
The species Sundasciurus robinsoni has a dorsum that ranges from medium brown with orange agouti to dark brown (S. r. vanakeni), and its venter ranges from white to pale yellow/buff white, with a reduction in the extent of this pale coloration and a lack of distinct margins in the case of S. r. vanakeni. Some populations (S. r. balae and S. r. vanakeni) have a grayish ventral coloration in limbs while others do not (S. r. robinsoni). It can be easily distinguished from other medium-sized western Sundaland Sundasciurus based on its ventral coloration and tail. All populations of S. fraterculus except Siberut, S. tahan, and S. altitudinis have a venter fur coloration homogeneously admixed with gray. The only other medium sized squirrel found in syntopy, S. tenuis, is also usually ventrally darker (admixed with gray) and dorsally lighter, with reddish-brown coloration on the shoulders and hips, white/pallid yellow hair tips present on tail, and a relatively thinner and longer tail (85–95 % of head-body length; Corbet and Hill, 1992) than S. robinsoni (56–84% of head-body length). Males of S. fraterculus, S. tahan and S. tenuis have a darker orange wash in the scrotal area than S. robinsoni, which is peach colored. Bonhote (1903) pointed out S. robinsoni has a “paler tinge” than S. lowii and S. natunensis, who have a more reddish brown agouti fur. The taxa also differ in variation of fur length on the tail, and body size and proportions. Moderate variation of hair length along the tail is observed in S. robinsoni, S. fraterculus, S. natunensis (and S. lowii Northwest Borneo populations) and S. tenuis. Extensive variation with short hair close to the base and longer hair at the tips giving a tapered appearance is observed in S. lowii and S. tahan. A tapered tail, but thickest at the base is found in S. fraterculus (Supplementary Figure S4). S. robinsoni is smaller (head-body range: 112–140 mm; body mass range: 49–77 g) than S. lowii (head-body range: 127–169 mm; body mass range: 60–125 g), somewhat smaller but overlapping in size with S. natunensis (head-body range: 133–152 mm) and slightly larger but quite overlapping in size with S. fraterculus (head-body range: 109–125 mm). Although the absolute tail length of S. robinsoni is about the same as S. lowii, S. fraterculus, and S. natunensis, it is relatively longer (56–85% of head-body length as opposed to 53–76%, 62 – 72%, and 57 – 67%, respectively). Within S. robinsoni, S. r. balae has a relatively shorter tail than the other two subspecies. S. robinsoni also has a relatively larger ear length (9–15% of head-body length) than S. lowii (7–13% of head-body length) and absolutely and relatively larger ear length than S. fraterculus (9% of head-body length) and S. natunensis (8% of head-body length) (Supplementary Table S4).
As with overall body size, the skull of S. robinsoni is smaller (33.60–36.43 mm) than S. lowii (36.93–41.03 mm); somewhat smaller but overlapping in size with S. natunensis (35.35–36.55 mm), and somewhat larger but overlapping in size with S. fraterculus (32.46–34.51 mm) (Supplementary Table S4). The skull of all of the small/medium-sized Sundasciurus are quite similar and conserved in proportion. Not many discrete characters have been found to distinguish the different species. Bonhote’s statement “the bony palate extends well back the last molar which is not the case with S. lowii” can be considered accurate for S. robinsoni, and be extended to S. fraterculus and S. natunensis as well. However, the bony palate does not always end in line with the last molars in S. lowii, and does sometimes end after, as in the other species, but quite closer. Another feature described by Bonhote, “the bullae are more flattened and rounded in S. robinsoni than S. lowii and do not project so far downwards” does not apply when additional specimens are examined. Incisor size and projection, however, do seem to be quite diagnostic. Bonhote (1903) pointed out “the molar series of S. robinsoni is much shorter and smaller than S. lowii, but incisors are about the same size.” In line with this statement, the height of the molars of S. robinsoni is clearly shorter than that of S. lowii and S. natunensis. While the incisors of S. robinsoni are less orthodont than S. tenuis, they are more orthodont than S. lowii, S. fraterculus, S. natunensis and S. tahan, which are conspicuously proodont. Finally, S. robinsoni has chisel-shaped incisors with sharp-angled beveled edges as in S. fraterculus, S. natunensis, S. lowii lingungensis and some specimens from west Borneo, but in contrast to S. lowii lowii from the remainder of Borneo (Figure 6). S. robinsoni was characterized by the PCA analysis as having the most gracile skull, with a relatively short interorbital breadth, anterior nasal breadth and least frontal bone breadth (Figure 5). It also has a shorter molar row (5.57–6.16 mm), palatal length (16.03–17.83 mm) and breadth (6.51–7.5 mm) than S. lowii (6.63–7.81 mm, 18.15–20.65 mm, 7.99–8.89 mm), but a relatively larger braincase.
Sundasciurus robinsoni has a shorter muzzle than S. natunensis, so it can be differentiated based on a shorter rostrum length (13.13–14.86/14.84–15.36 mm), palatal length (16.03–17.83/17.48–18.44 mm), and relative and absolute narrower bony palate (6.51–7.5/7.81–8.07 mm). S. robinsoni has a relative and absolute shorter molar row (5.57–6.16 mm) and smaller bullae (6.17–7.34 mm) than S. natunensis (6.72–7.15 mm; 7.26–7.65 mm) but a relatively larger proportion of bony palate of the total palatal length, relatively larger height of rostrum and relatively larger braincase (Supplementary Figure S5). Postorbital processes are relatively shorter and less pointed in S. robinsoni than S. natunensis.
There are major distinctions between almost all populations of S. robinsoni and S. fraterculus analyzed here. However, the intermediate position of the incertae sedis Kerinci population, that we have tentatively considered a subspecies (Sundasciurus robinsoni vanakeni) until molecular lines of evidence are available, generates an overlap among S. robinsoni and S. fraterculus for most diagnostic characters. The nominal S. r. robinsoni and S. r. balae have larger tympanic bullae (6.46–7.34 mm) than S. fraterculus (5.88–6.43 mm) but there is overlap with the inclusion of S. r. vanakeni (6.17–7.34 mm) in the comparison. The larger maxillary cheek teeth of S. robinsoni (5.83–6.16 mm) than S. fraterculus (5.32–5.79 mm) becomes another invalid character once S. r. vanakeni (5.6–6.16 mm) is included in the comparison. Finally, the shorter anterior nasal breadth (4.27–4.88 mm) of S. robinsoni as compared to S. fraterculus (4.79–5.31 mm) also loses its diagnostic power with the inclusion of S. r. vanakeni (4.27–5.06 mm).
Although there are clear distinctions among these two species there are no discrete craniodental measurements to distinguish them. However, the first and third components of the PCA effectively separated these, especially the latter. PC3 loadings confirm that despite the previously mentioned overlapping measurements (anterior nasal breadth, maxillary cheek teeth and tympanic bullae length), their combination can still efficiently differentiate these taxa. Mastoid breadth, diastema length and interorbital breadth also seem to be diagnostic to a lesser degree with moderate contribution in the PC3 loadings (Supplementary File S1). S. robinsoni seems to have a somewhat broader mastoid breadth and narrower interorbital breadth than S. fraterculus. S. robinsoni also exhibits a relatively narrower least frontal bone breadth than S. fraterculus. Finally, S. robinsoni has a relatively shorter muzzle than S. fraterculus, which is reflected in a relatively shorter length and height of rostrum (Supplementary Figure S5).
Distribution: Malay Peninsula, Sumatra, and Batu and Rhio Archipelagos. The subspecies S. r. robinsoni and S. r. balae have been recorded from 0–610 m, on the Malay Peninsula and Rhio and Sumatra and Batu, respectively, while S. r. vanakeni has been recorded between 900–1372 m on Mount Kerinci.
Sundasciurus natunensis (Thomas, 1895)- Natuna squirrel.
Sciurus lowii natunensis Thomas, 1895:26.—Type locality. “Sirhassen Island” (Serasan), Natuna Islands.
Holotype. BMNH 94.9.28.40, skull, skin, adult male, collected 23 September 1893 by A. H. Everett.
Emended comparison with other Sundasciurus species.
Comparisons of S. natunensis with S. robinsoni are detailed in the previous section.
The slight brown pinkish flush described by Thomas (1895) was only found to be present in the type series and is likely an effect of the preservation of these specimens. As Thomas reasoned, these were “skinned out of spirit” and certain yellows turned into red in other specimens of that same expedition. Therefore, this species exhibits the same coloration as S. lowii, and can be distinguished from S. fraterculus based on paler ventral coloration. A major external distinction is that S. natunensis (together with west Borneo S. l. lowii) lack that tapered appearance in its tail (described in the previous section) that characterizes S. lowii (from remaining Borneo and northern Natunas), with a thinner tail base, and S. fraterculus, with a thicker tail base (Supplementary Figure 4). Thomas (1895) described S. natunensis, in relation to S. lowii, as having “smaller size, shorter feet and longer ears, which in the typical variety is a mere low rim, while the Natuna subspecies has a distinct upstanding conch”. The data presented here show smaller but largely overlapping head-body size (133.3–152. mm) and shorter but overlapping feet (33–35 mm with claw) than S. lowii (127–169 mm; 32–40 mm with claw). However, the only ear measurement available for S. natunensis (provided in the original description) is smaller (12 mm) than S. lowii (12–17 mm), not larger. Nevertheless, in relation to this, perhaps the most distinctive characteristic of S. natunensis is its relatively larger bullae than S. lowii. Besides this, the skulls of S. natunensis and S. lowii are quite similar and conserved in proportion, and only differ on the relatively thinner and less deep rostrum and narrower palate of S. natunensis (Figure 6missing and Supplementary Figure S5). The skull of S. natunensis (33.60–36.43 mm) is smaller than that of S. lowii (36.93–41.03 mm), as Thomas (1895) pointed out. The other non-overlapping characters are braincase height (12.34–13.01 mm) and orbit length (8.33–9.05 mm), which are also larger in S. lowii (13.07–14.30 mm; 9.03 – 10.06 mm). Our expanded dataset also supports Thomas’ statement that S. natunensis “postorbital processes are longer and slenderer” than those of S. lowii (except West Borneo populations), but also than S. fraterculus and S. robinsoni. The presence of chisel-shaped incisors with sharp-angled beveled edges are an additional diagnostic feature to differentiate S. natunensis from S. lowii.
The species S. natunensis has a larger skull (35.35–36.55 mm), rostrum (14.84–15.36 mm), mastoid breadth (15.66 – 16.56 mm), diastema length (8.91–9.68 mm), bony palate breadth (7.81–8.03 mm), upper molar row length (6.72–7.15 mm) and bulla length (7.26–7.65) than S. fraterculus (32.46–34.51 mm; 13.550 – 14.83 mm; 14.48–15.43 mm; 7.84–8.67 mm; 6.40–6.99 mm; 5.32–5.79 mm; 5.88–6.43 mm). In addition, these taxa also differ in skull proportions, with S. natunensis having a relatively more gracile skull than S. fraterculus, which exhibits a broader skull (IB and LBC) and thicker rostrum (HR) relative to skull length.
Distribution: Southern Natuna islands (only recorded in Sirhassen Island), and possibly in west Borneo. Despite discrete characters such as postorbital processes and tail shape of the latter populations closely resemble S. natunensis, we currently consider the taxonomic status of west Borneo populations as incertae sedis given the intermediate phenotypic position of these among S. natunensis and S. lowii shown in the PCA and DAPC and the lack of genetic evidence. Genetic data from these, southern Sumatra and northern Natuna populations is needed to clarify the taxonomy and distribution of this group.
Sundasciurus lowii (Thomas, 1895) -Low’s squirrel.
Sciurus lowii Thomas, 1895: 253.—Type locality. “Lumbidan, on the mainland opposite Labuan”, Sarawak, Borneo.
Holotype. BMNH 76. 5. 2. 14., skull, skin, adult male, collected by H. Low (unknown date).
Sciurus lingungensis Miller, 1901:123.—Type locality. Lingung Island, Near Bunguran Island, Natuna Islands.
Sundasciurus lowii lowii (Thomas, 1895)
Sundasciurus lowii bangueyae (Thomas, 1908)
Sciurus lowii bangueyae Thomas, 1908:387.— Type locality. “Banguey Island” (Banggi)
Sundasciurus lowii lingungensis (Miller, 1901)
Sciurus lingungensis Miller, 1901:123.—Type locality. Lingung Island, Near Bunguran Island, Natuna Islands
Distribution: S. l. lowii is present in Borneo, S. l. bangueyae in Banggi and Balambangan islands, (and possibly Malawali island as well) and S. l. lingungensis in the Northern Natunas (recorded in Bunguran, Lingung and Laut islands).
Corbet and Hill (1992) pointed out that geographical variation in Sundasciurus lowii sensu lato is slight. Many of the currently recognized subspecies are not morphologically distinct from others. We consider Sundasciurus robinsoni balae and Sundasciurus robinsoni vanakeni valid subspecies based on their ventral coloration and craniodental differentiation (Figure 5 and Supplementary Figure S3). We synonymize former Sundasciurus lowii humilis with Sundasciurus robinsoni balae due to external resemblance (ventral grayish coloration in limbs). We also synonymize based on external resemblance (lack of ventral grayish coloration in limbs) Sundasciurus lowii seimundi with Sundasciurus robinsoni robinsoni. Finally, we consider Sundasciurus lowii lingungensis and Sundasciurus lowii bangueyae valid subspecies given their craniodental differentiation (Figure 5 and Supplementary Figure S3).
Molecular species delimitation analyses suggest the presence of three species in Borneo within S. lowii (in Sabah, Sarawak and East Kalimantan). However, phenotypic divergence seems to be slight and the fine scale distribution of the morphs is unknown. The only Sarawak sample included in the PCA was clustered separately but close to Sabah + east Kalimantan, that largely overlapped. Regarding the DAPC, only east Kalimantan samples were differentiated from the remaining overlapping populations. Relative size of interorbital breadth seems to differentiate a small number of Sarawak specimens from the remaining S. lowii, but these samples are clustered very close to the other S. lowii. Sarawak specimens also seem to have darker fur than Sabah and East Kalimantan S. lowii and a lack of blond tips on the tail, which are only observed in Sabah populations. We consider these populations unconfirmed candidate species (Padial et al., 2010) until a better molecular and morphological sampling is performed in terms of geographic coverage and number of specimens included.
As pointed out by Hawkins et al. (2016a) the two subgenera, Aletesciurus and Sundasciurus, proposed by Moore (1958) are not supported as reciprocally monophyletic groups. The discrete characters (presence of sagittal crest, skull size, and shape of anterior-mesial lobe of auditory bullae) described as distinguishing subgenera were not valid for all species. For instance, the Palawan mountain squirrel (Sundasciurus rabori) belongs to Aletesciurus but lacks a sagittal crest and has an intermediate antero-mesial lobe and skull size (40.9-43.6 mm) among both subgenera (Heaney, 1979; Hawkins et al., 2016a). The northern Palawan tree squirrel (Sundasciurus juvencus) shows an antero-mesial lobe that resembles that of the subgenus Sundasciurus despite being assigned to Aletesciurus. Finally, different species of the subgenus Sundasciurus such as S. lowii, S. tahan or S. altitudinis have skulls that reach sizes (41–43 mm) that overlap with those of S. rabori (authors unpublished data). Although these discrete characters do not support the subgenera Aletesciurus and Sundasciurus, Heaney (1979) found support for them in his morphometric analyses. Molecular phylogenies suggest that there are 5–6 major phylogenetic lineages within Sundasciurus, which correspond to monophyly in the subgenus Aletesciurus (which contains 3 divergent clades), and paraphyly in the subgenus Sundasciurus (which also contains 2–3 divergent clades) (Den Tex et al., 2010; Hawkins et al., 2016a). The lack of universally valid diagnostic features and the phylogenetic evidence from this and previous studies demonstrate that current Sundasciurus subgeneric classification is invalid, so we synonymize Aletesciurus with Sundasciurus.
Deep genetic and moderate phenotypic divergences between the lowland Sunda tree squirrels formerly recognized under the species Sundasciurus lowii were confirmed. The consistency between these two lines of evidence along with geography required the revalidation of Sundasciurus robinsoni and species level recognition of S. natunensis.
The patterns of divergence suggest ancient diversification events between these taxa that predate the Pleistocene. This does not support the Pleistocene speciation-pump hypothesis as the main driver behind the high levels of diversity found in Sundaland (Brown et al., 2013; Husson et al., 2019). The ancient patterns of divergence that we found in Sunda squirrels are consistent with those found in other small vertebrates and invertebrates (Klaus et al., 2013; Grismer et al., 2017; Karin et al., 2017; Manawatthana et al., 2017). This is particularly interesting in light of recent geological work suggesting that the Sunda shelf was continuously exposed until 400 ka (Husson et al., 2019). This suggests that dispersal should have been fluid across the entire shelf until this point. The much older times of divergence suggest that there was substantial structure, and likely speciation prior to the inundation of the Sunda shelf in the Middle Pleistocene.
The dated phylogeny also suggests that Sunda squirrels experienced a rapid radiation, given the almost simultaneous diversification event during the middle-late Miocene transition among major lineages of Sundasciurus. The mito-nuclear discordant relationship among these major lineages supports this hypothesis. There are two potential processes that might have driven this gene discordance, incomplete lineage sorting and hybridization, both of which have been viewed as byproducts of rapid radiations (Degnan and Rosenberg, 2006; Wiens et al., 2006; McLean et al., 2016). The latter has proved to be a widespread process across Sciuridae (Garroway et al., 2010; Chang et al., 2011; Chavez et al., 2014; McLean et al., 2016; Hawkins et al., 2016b), and may be the result of speciation outpacing the evolution of reproductive incompatibilities (Wiens et al., 2006). Interestingly, similar diversification patterns have also been shown during this narrow timeframe in stream toads, halfbeaks and colugos (De Bruyn et al., 2013; Grismer et al., 2017; Mason et al., 2019).
Land bridges that were formed after the inundation of the Sunda shelf may have allowed disparate dispersal rates between the different land masses. Whether or not a species successfully dispersed during these periods was likely due to a number of factors including the available habitat on the exposed shelf, distance between land masses, and presence or absence of close relatives/competitors for the same resources. Previous studies have suggested that during this time, despite shelf exposure, many rainforest taxa remained isolated (Den Tex et al., 2010; Leonard et al., 2015; Mason et al., 2019).
The Mentawai Islands are geographically very close to Sumatra, but were not connected by land bridges to Sumatra when other land masses were connected due to a deep-sea trench between them (Voris, 2000; Roberts et al., 2011). The endemic fraternal squirrel became isolated from Robinson’s squirrel, which was distributed across Sumatra and the Malay Peninsula in the Pliocene. This level of deep divergence between taxa on Mentawai and sister taxa elsewhere in Sundaland has also been identified in other taxa such as gibbons, civets and skinks (Chan et al., 2010; Patou et al., 2010; Karin et al., 2017) and even deeper divergence was found in macaques (Evans et al., 2017). This sister relationship between Mentawai and Sumatra-Malay Peninsula populations (Meijaard and Groves, 2004; Chan et al., 2010; Patou et al., 2010; Arifin et al., 2018) however, is not the most common in mammals (Wilting et al., 2012). For example, the Mentawai treeshrew (Tupaia chrysogaster) is sister to the endemic Bornean Long-footed treeshrew (T. longipes, Roberts et al., 2011) and the Mentawai endemic Koopman’s pencil-tailed tree mouse (Chiropodomys karlkoopmani) is sister to the Bornean endemic greater pencil-tailed tree mouse (C. major/C. calamianensis) (Musser and Carleton, 2005).
Although bathymetry suggests that the Mentawai islands have not been directly connected to Sumatra, it is possible that they have been connected to the Batu Islands which have been connected to Sumatra, creating a peninsula parallel to the latter (Voris, 2000; Roberts et al., 2011). Potential episodes of gene flow across this past connection in both directions could explain the lack of differentiation in nuclear introns but high mitochondrial divergence and important phenotypic differentiation among populations. The intermediate phenotype of the Siberut population, the closest Mentawai island to the Batu archipelago, which externally resembles Robinson’s squirrel but is clustered within the craniodental variation of the Fraternal squirrel would also support this. In accordance with this peninsular dispersal corridor, Karin et al. (2017) suggest a divergence of ∼2.60 Mya among skink populations of Nias (Batu Islands) and Siberut (Mentawai Islands) which would be slightly after to the divergence among Mentawai and Malay Peninsula squirrels. Given that western Sumatra and Mentawai have been suggested to be a forest refuge (Morley, 2000; Lim et al., 2010) it is not straight-forward to assess the direction of the potential dispersal events. Intron haplotype networks suggest that S. fraterculus is within the diversity of S. robinsoni, which could suggest a Malay Peninsula to Sumatra to Batu to Mentawai colonization. However, within Mentawai, just South Pagi was sampled, so data from other populations from Mentawai, Sumatra and Batu islands are needed to confirm this.
There is a deep, Late Miocene divergence between the western Robinson’s squirrel and Fraternal squirrel and the eastern Low’s squirrel and Natuna squirrel. At about the same time the western high elevation Sunda squirrels (S. tahan and S. altitudinis) diverged from the western slender and Jentink’s squirrels. This period was also characterized by diversification across Sundaland in semiarboreal rough-skinned skinks, bulbuls, barbets and Sundaic freshwater crabs (Den Tex et al., 2010; Klaus et al., 2013; Karin et al., 2017; Manawatthana et al., 2017). These common patterns could be due to dispersal barriers following the development of the Bornean highlands and river systems into their more modern representation by ∼8–10 Mya (Hall, 2013; Mason et al., 2011; Yang and Rannala, 2010). Alternatively, vicariance during fragmentation of the Sunda shelf/oceanic dispersal between islands during periods of high sea-levels by ∼5–6 Mya (Karin et al., 2017) if there were early inundations, could also explain this pattern.
The Natuna Islands are located in the South China Sea between Borneo and mainland Asia, a key location for understanding the biogeography of the region. Few studies to date (Den Tex et al., 2010; Mason et al., 2011, 2019) have been able to gather any information from historic specimen samples from this archipelago possibly due to poor specimen and DNA preservation. A deep divergence between populations of colugos from the northern and southern Natuna islands has been identified, with the former populations closely related to northeastern Borneo populations and the latter populations to the rest of Borneo (Mason et al., 2019). Here we show that the Natuna squirrels from the southern islands are deeply divergent from Low’s squirrels of central and east Borneo (Figure 3). These squirrels from the southern Natuna islands are also morphologically distinct from the northern Natuna squirrels, which are more similar to central and east Borneo specimens (Figure 5). Finally, populations from west Borneo have an intermediate phenotype (Supplementary Figure S3). These squirrels might have a similar pattern to the colugos, with one species on the southern Natuna islands and possibly extreme west Borneo (the Natuna tree squirrels, S. natunensis) and the other on the northern islands and rest of Borneo (S. lowii). However, additional genetic and morphologic data from the Natuna islands and Borneo is needed in order to confirm this hypothesis and the taxonomic status of west Borneo and north Natuna populations.
While divergence in allopatry seems to be the main process driving the speciation of Low’s squirrels, ecomorphological and behavioral adaptations in this clade suggest an important role of post-vicariance/colonization niche divergence, and a lack of niche conservatism. Previous studies have shown a significant correlation between locomotion modes and morphological traits in squirrels and other rodents. Tail length is thought to be associated with arboreality in squirrels (Hayssen, 2008) and other small mammals such as deer mice (Horner, 1954; Kingsley et al., 2017) and treeshrews (Martin, 1968). Nations et al. (2019) found a positive relationship of tail length with climbing in Philippine murines, but also a large variance and low predictive power for this trait across all locomotor categories. Thorington and Heaney (1981) provided a comparative table with different ratios that also shows this trend (e.g., the most terrestrial Dremomys rufigenis and S. lowii exhibit the relative shortest tail among Callosciurinae). Arboreal Sigmondontines and tree squirrels are known to have a more rounded and inflated vault and shorter nasals than terrestrial species (Lu et al., 2014; Camargo et al., 2019). Arboreal squirrels and Procyonids also seem to have shorter nasals or slightly shorter olfactory bulbs compared to terrestrial ones (Lu et al., 2014; Ahrens, 2014; Bertrand et al., 2019). A longer rostrum could imply a better sense of smell, which might be useful when living on the ground (Lu et al., 2014) while brain enlargement is an adaptation in arboreal species that use the space in three dimensions and need to integrate information of complex environments (Camargo et al., 2019). The relatively long tail, larger braincase and shorter rostrum of S. robinsoni as compared to the terrestrial S. lowii and S. natunensis might suggest arboreality. Ecological field studies (Kemper and Bell, 1985; Sharma, 1992; Abdullah et al., 2001; Yasuda et al., 2003; Saiful and Nordin, 2004; Jayaraj et al., 2013) and diet data finding bark, fruit and insects (Harrison, 1962; Abdullah et al., 2001) are also consistent with an arboreal life style for S. robinsoni. On the other hand, S. lowii on Borneo seems to be much more terrestrial (Davis, 1962; Wells et al., 2004; Camacho-Sánchez et al., 2019) with a diet including small arthropods, fruits and fungus (Davis, 1962). The Fraternal squirrel seems to have intermediate habits, since it uses the ground extensively, but also climbs showing a preference for heights up to five meters, but avoids the upper canopy (Whitten, 1981). It has only been observed eating bark, moss and lichen, but stomach content analyses also found annelid remains, in accordance with more terrestrial behavior.
The relatively longer rostrum of S. lowii, S. fraterculus and S. natunensis with respect to S. robinsoni might be due to its terrestrial foraging behavior, reflecting a more developed olfactory bulb as an adaptation to a more insectivorous diet. In addition, the more robust skull of S. fraterculus with regard to S. robinsoni might reflect a possible adaptation to the harder dietary elements such as an increased consumption of chitin from insects, lichens and bark, and an absence of soft food items such as fruit (Whitten, 1981). These findings suggest that not only has there been a high degree of genetic and phenotypic divergence, but also that morphological changes have possibly been the result of natural selection driven by adaptation to different niches, and are not only due to drift caused by geographic isolation (Mayr, 1963). It is difficult to interpret if the different degrees of scansoriality found among these species are due to different levels of interspecific competition, predator avoidance, food limitation or a combination of some of these. In any case, differences in the squirrel and treeshrew communities among islands are notable. On the Mentawai islands, S. fraterculus has to share its habitat with just one diurnal treeshrew (Tupaia chrysogaster), one terrestrial squirrel (Lariscus obscurus) and one arboreal diurnal squirrel (Callosciurus melanogaster), allowing it a greater range of vertical habitat than its sister species (Whitten, 1981). On Borneo, S. lowii has to share its habitat with up to six diurnal treeshrews and seven species of diurnal squirrels. Two of the treeshrews (Tupaia minor and Tupaia gracilis) and most of the squirrels are arboreal, and three of the latter are bark gouging specialists (Sundasciurus tenuis, Nannosciurus melanotis and Exilisciurus exilis) (Emmons, 2000; Zelditch et al., 2017). However, the only two terrestrial squirrels (Lariscus insignis or L. hosei and Rhinosciurus laticaudatus) are in very low densities or possibly absent where Low’s squirrel is present (Wells et al., 2007; Yasuda and Tsuyuki, 2012; Fitzmaurice, 2014; Chapman et al., 2018; Camacho-Sánchez et al., 2019; authors unpublished data) and are specialized on fruits and insects. On Sumatra and the Malay peninsula S. robinsoni is co-distributed with diurnal arboreal and terrestrial treeshrews (Tupaia minor and T. glis/T. ferruginea), and up to ten diurnal squirrels, three of which are terrestrial (Lariscus insignis, Menetes berdmorei and Rhinosciurus laticaudatus) (Lekagul and McNeely, 1977). These terrestrial squirrels seem to be more common in mainland field-studies than in Borneo (Kemper and Bell, 1985; Yasuda et al., 2000; Syakirah et al., 2001; Yasuda et al., 2003; Jayaraj et al., 2013). On the other hand, the bark gouging niche seems to be less exploited in Peninsular Malaysia, with just S. tenuis (Zelditch et al., 2017). This could have potentially added evolutionary selective pressures on the ancestor of S. robinsoni to alter its diet to avoid competition with other species. The relatively larger and more orthodont incisors of this species with regard to its closest relatives are also in line with this hypothesis. Convergence towards a bark gouging diet seems to have arisen at least seven times in Sciuridae and it exhibits higher rates of convergence relative to the number of living species (10 species) within that dietary niche (Zelditch et al., 2017).
Within Borneo, deep genetic divergences were detected across Sundasciurus lowii. Bornean populations exhibit a high level of genetic and some morphological diversity, but the fine scale distribution of this diversity is not yet characterized. Molecular species delimitation analyses suggest candidate species found in Sabah, Central Sarawak and East Kalimantan. However, much more sampling will be required to clarify their taxonomy. As in previous studies on Sunda squirrels, our results also point towards a possible underestimation of highland species diversity given the high genetic divergence observed among populations of S. altitudinis and high phenotypic divergence found between Mount Kerinci mid-elevation (900–1400 m) populations (S. r. vanakeni) and the remaining Sumatra and Malay Peninsula lowland populations (S. r. robinsoni and S. r. balae). The lack of molecular evidence for this pelage and craniodentally distinct population suggests to consider S. r. vanakeni as an unconfirmed candidate species (Padial et al., 2010). Given that a previous attempt failed to amplify DNA from the only known series of specimens of this taxon (Den Tex et al., 2010) future collection of new specimens along an elevational gradient in Kerinci will be needed to confirm the specific status of this population.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Smithsonian Institution, National Museum of Natural History, proposal number 2012-04 and Estación Biológica de Doñana proposal number CGL2010-21424.
AH, MH, JM, and JL designed the study. AH, MH, and AA performed field and museum data collection. AH and MH performed laboratory work and statistical analyses. AH and JL wrote the manuscript with input from MH, AA, and JM. All authors approved the submitted version.
AH was supported by an Ernst Mayr Travel grant and AMNH Collection Study Grant during museum data collection. This research received support from the SYNTHESYS Project financed by European Community Research Infrastructure Action under the FP7 “Capacities” Program. AH was also supported by a Spanish Ministry of Economy contract CGL2014-58793-P. The Spanish Ministry of Science and Innovation grant CGL2017-86068-P to JL supported this study. MH was supported by discretionary research funding from Humboldt State University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
L. Heaney provided important advice and guidance during specimen examination and morphometric data collection. G. Forcina provided valuable insights during marker development. L. Flynn and J. Barry provided expertise and guidance regarding fossil calibrations and A. Jaramillo and A. Sánchez regarding taxonomical nomenclature. M. Camacho, P. Carro, M. Lopez and different field assistants participated in fieldwork. Sabah Parks, Sabah Forestry Department, and Sabah Biodiversity Council provided research permits and logistic support, especially FTY Yuh. R. Portela, L. Heaney, D. Lunde, R. Voss, C. Urdiales, and M. Omura provided assistance at NHMUK, FMNH, USNM, AMNH, EBD-CSIC, and Harvard MCZ collections. L. Heaney, J. Esselstyn, A. Ferguson, L. Smith, B. Lim, and D. Lunde granted access to several tissue samples hosted at FMNH, ROM, and USNM. Nancy McInerney for help with MiSeq runs at the CCG. B. Lim, O. Johnson, M. La, R. J. Backhouse, and C. Lee shared pictures. Logistical support was provided by Laboratorio de Ecología Molecular, Estación Biológica de Doñana, CSIC (LEM-EBD) and Doñana ICTS-RBD.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00208/full#supplementary-material
Abdullah, S. A., Yusoff-Rashid, N., and Idris, A. H. (2001). Niche segregation among three sympatric species of squirrels inhabiting a lowland dipterocarp forest, Peninsular Malaysia. Mamm. Study 26, 133–144. doi: 10.3106/mammalstudy.26.133
Achmadi, A. S., Esselstyn, J. A., Rowe, K. C., Maryanto, I., and Abdullah, M. T. (2013). Phylogeny, diversity, and biogeography of Southeast Asian spiny rats (Maxomys). J. Mammal. 94, 1412–1423. doi: 10.1644/13-mamm-a-092.1
Ahrens, H. E. (2014). Morphometric study of phylogenetic and ecologic signals in procyonid (Mammalia: Carnivora) endocasts. Anat. Rec. 297, 2318–2330. doi: 10.1002/ar.22996
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed January, 2020).
Arifin, U., Cahyadi, G., Smart, U., Jankowski, A., and Haas, A. (2018). A new species of the genus Pulchrana Dubois, 1992 (Amphibia: Ranidae) from Sumatra, Indonesia. Raffles Bull. Zool. 66, 277–299.
Baker, R. J., and Bradley, R. D. (2006). Speciation in mammals and the genetic species concept. J. Mammal. 87, 643–662. doi: 10.1644/06-mamm-f-038r2.1
Bertrand, O. C., San Martin-Flores, G., and Silcox, M. T. (2019). Endocranial shape variation in the squirrel-related clade and their fossil relatives using 3D geometric morphometrics: contributions of locomotion and phylogeny to brain shape. J. Zool. 308, 197–211. doi: 10.1111/jzo.12665
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borowiec, M. L. (2016). AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4:e1660. doi: 10.7717/peerj.1660
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Bouckaert, R. R., and Drummond, A. J. (2017). bModelTest: bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 17:42. doi: 10.1186/s12862-017-0890-6
Brown, R. M., Siler, C. D., Oliveros, C. H., Esselstyn, J. A., Diesmos, A. C., Hosner, P. A., et al. (2013). Evolutionary processes of diversification in a model island archipelago. Annu. Rev. Ecol. Evol. Syst. 44, 411–435. doi: 10.1146/annurev-ecolsys-110411-160323
Bushnell, B. (2015). BBTools Package. Available online at: http://www.sourceforge.net/projects/bbmap/ (accessed January, 2020).
Camacho-Sánchez, M. (2017). Evolution in Sundaland: Insights From Comparative Phylogeography of Rattus and Sundamys Rats.
Camacho-Sánchez, M., Hawkins, M. T., Tuh, F. Y., Maldonado, J. E., and Leonard, J. A. (2019). Endemism and diversity of small mammals along two neighboring Bornean mountains. PeerJ 7:e7858. doi: 10.7717/peerj.7858
Camacho-Sánchez, M., Leonard, J. A., Fitriana, Y., Tilak, M. K., and Fabre, P. H. (2017). The generic status of Rattus annandalei (Rodentia, Murinae) and its evolutionary implications. J. Mammal. 98, 1340–1355.
Camargo, N. F., Machado, L. F., Mendonça, A. F., and Vieira, E. M. (2019). Cranial shape predicts arboreal activity of Sigmodontinae rodents. J. Zool. 308, 128–138. doi: 10.1111/jzo.12659
C̀anády, A., Mošanský, L., and Krišovský, P. (2015). Cranial dimorphism in Eurasian red squirrels, Sciurus vulgaris from Slovakia. Zool. Anzeiger J. Compar. Zool. 257, 96–102. doi: 10.1016/j.jcz.2015.05.004
Chan, Y. C., Roos, C., Inoue-Murayama, M., Inoue, E., Shih, C. C., Pei, K. J. C., et al. (2010). Mitochondrial genome sequences effectively reveal the phylogeny of Hylobates gibbons. PLoS One 5:e14419. doi: 10.1371/journal.pcbi.14419
Chang, S. W., Oshida, T., Endo, H., Nguyen, S. T., Dang, C. N., Nguyen, D. X., et al. (2011). Ancient hybridization and underestimated species diversity in Asian striped squirrels (genus Tamiops): inference from paternal, maternal and biparental markers. J. Zool. 285, 128–138. doi: 10.1111/j.1469-7998.2011.00822.x
Chapman, P. M., Wearn, O. R., Riutta, T., Carbone, C., Rowcliffe, J. M., Bernard, H., et al. (2018). Inter-annual dynamics and persistence of small mammal communities in a selectively logged tropical forest in Borneo. Biodiver. Conserv. 27, 3155–3169. doi: 10.1007/s10531-018-1594-y
Chavez, A. S., Maher, S. P., Arbogast, B. S., and Kenagy, G. J. (2014). Diversification and gene flow in nascent lineages of island and mainland North American tree squirrels (Tamiasciurus). Evolution 68, 1094–1109. doi: 10.1111/evo.12336
Corbet, G. B., and Hill, J. E. (1992). The Mammals Of The Indomalayan Region: A Systematic Review, Vol. 488. Oxford: Oxford University Press.
Davis, D. D. (1962). Mammals of the lowland rainforests of North Borneo. Bull. Nat. Mus. Singapore 31, 1–129.
De Bruyn, M., Rüber, L., Nylinder, S., Stelbrink, B., Lovejoy, N. R., Lavoué, S., et al. (2013). Paleo-drainage basin connectivity predicts evolutionary relationships across three Southeast Asian biodiversity hotspots. Syst. Biol. 62, 398–410. doi: 10.1093/sysbio/syt007
Degnan, J. H., and Rosenberg, N. A. (2006). Discordance of species trees with their most likely gene trees. PLoS Genet. 2:e68. doi: 10.1371/journal.pgen.0020068
Den Tex, R. J., Thorington, R. W., Maldonado, J. E., and Leonard, J. A. (2010). Speciation dynamics in the SE Asian tropics: putting a time perspective on the phylogeny and biogeography of Sundaland tree squirrels. Sundasciurus. Mol. Phylogenet. Evol. 55, 711–720. doi: 10.1016/j.ympev.2009.12.023
Duchêne, S., Archer, F. I., Vilstrup, J., Caballero, S., and Morin, P. A. (2011). Mitogenome phylogenetics: the impact of using single regions and partitioning schemes on topology, substitution rate and divergence time estimation. PLoS One 6:e27138. doi: 10.1371/journal.pone.0027138
Emmons, L. H. (2000). Tupai: A Field Study Of Bornean Treeshrews. California: University of California Press.
Evans, B. J., Tosi, A. J., Zeng, K., Dushoff, J., Corvelo, A., and Melnick, D. J. (2017). Speciation over the edge: gene flow among non-human primate species across a formidable biogeographic barrier. R. Soc. Open Sci. 4, 170351. doi: 10.1098/rsos.170351
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fitzmaurice, A. (2014). The Direct And Indirect Impacts Of Logging On Mammals In Sabah, Borneo. Dissertation, Imperial College London, London.
Flot, J. F. (2010). SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol. Ecol. Resour. 10, 162–166. doi: 10.1111/j.1755-0998.2009.02732.x
Flynn, L. J. (2003). “Small mammal indicators of forest paleoenvironment in the siwalik deposits of the potwar plateau, Pakistan,” in Distribution and Migration Of Tertiary Mammals In Eurasia. A Volume In Honour Of Hans De Bruijn, eds J. W. F. Reumer and W. Wessels (Harvard, MA: DEINSEA), 183–196.
Forcina, G., Camacho-Sanchez, M., Tuh, F. Y. Y., Moreno, S., and Leonard, J. A. (2020). Markers For Genetic Change. Submitted.
Garroway, C. J., Bowman, J., Cascaden, T. J., Holloway, G. L., Mahan, C. G., Malcolm, J. R., et al. (2010). Climate change induced hybridization in flying squirrels. Glob. Chang. Biol. 16, 113–121. doi: 10.1111/j.1365-2486.2009.01948.x
Gorog, A. J., Sinaga, M. H., and Engstrom, M. D. (2004). Vicariance or dispersal? Historical biogeography of three shelf murine rodents (Maxomys surifer, Leopoldamys sabanus and Maxomys whiteheadi). Biol. J. Linn. Soc. 81, 91–109. doi: 10.1111/j.1095-8312.2004.00281.x
Grismer, L. L., Wood, P. L. Jr., Aowphol, A., Cota, M., Grismer, M. S., Murdoch, M. L., et al. (2017). Out of Borneo, again and again: biogeography of the Stream Toad genus Ansonia Stoliczka (Anura: Bufonidae) and the discovery of the first limestone cave-dwelling species. Biol. J. Linnean Soc. 120, 371–395.
Hall, R. (2013). The palaeogeography of sundaland and wallacea since the late jurassic. J. Limnol. 72:e1.
Hawkins, M. T., Helgen, K. M., Maldonado, J. E., Rockwood, L. L., Tsuchiya, M. T., and Leonard, J. A. (2016a). Phylogeny, biogeography and systematic revision of plain long-nosed squirrels (genus Dremomys, Nannosciurinae). Mol. Phylogenet. Evol. 94, 752–764. doi: 10.1016/j.ympev.2015.10.023
Hawkins, M. T., Leonard, J. A., Helgen, K. M., McDonough, M. M., Rockwood, L. L., and Maldonado, J. E. (2016b). Evolutionary history of endemic Sulawesi squirrels constructed from UCEs and mitogenomes sequenced from museum specimens. BMC Evol. Biol. 16:80. doi: 10.1186/s12862-017-0890-80
Hayashida, A., Endo, H., Sasaki, M., Oshida, T., Kimura, J., Waengsothorn, S., et al. (2007). Geographical variation in skull morphology of gray-bellied squirrel Callosciurus caniceps. J. Vet. Med. Sci. 69, 149–157. doi: 10.1292/jvms.69.149
Hayssen, V. (2008). Patterns of body and tail length and body mass in Sciuridae. J. Mammal. 89, 852–873. doi: 10.1644/07-mamm-a-217.1
Heaney, L. R. (1979). A new species of tree squirrel (Sundasciurus) from Palawan island, Philippines (Mammalia: Sciuridae). Proc. Biol. Soc. Wash. 92, 280–286.
Heaney, L. R. (1991). A synopsis of climatic and vegetational change in Southeast Asia. Trop. Forest. Clim. 19, 53–61. doi: 10.1007/bf00142213
Horner, E. B. (1954). Arboreal adaptations of peromyscus with special reference to use of the tail. Contribut. Lab. Verteb. Biol. Univ. Mich. 61, 1–84.
Husson, L., Boucher, F. C., Sarr, A. C., Sepulchre, P., and Cahyarini, S. Y. (2019). Evidence of Sundaland’s subsidence requires revisiting its biogeography. J. Biogeogr. 47:13762.
Igea, J., Juste, J., and Castresana, J. (2010). Novel intron markers to study the phylogeny of closely related mammalian species. BMC Evol. Biol. 10:369. doi: 10.1186/s12862-017-0890-369
Jayaraj, V. K., Daud, S. H. M., Azhar, M. I., Sah, S. A. M., Mokhtar, S. I., and Abdullah, M. T. (2013). Diversity and conservation status of mammals in Wang Kelian State Park, Perlis, Malaysia. Check List 9, 1439–1448.
Jombart, T., and Ahmed, I. (2011). adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. doi: 10.1093/bioinformatics/btr521
Karin, B. R., Das, I., Jackman, T. R., and Bauer, A. M. (2017). Ancient divergence time estimates in Eutropis rugifera support the existence of Pleistocene barriers on the exposed Sunda Shelf. PeerJ 5:e3762. doi: 10.7717/peerj.3762
Kassambara, A., and Mundt, F. (2017). Package ‘Factoextra’. Extract and Visualize the Results of Multivariate Data Analyses, 76. Available online at: https://cran.r-project.org/web/packages/factoextra/factoextra.pdf (accessed January, 2020).
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kemper, C., and Bell, D. T. (1985). Small mammals and habitat structure in lowland rain forest of Peninsular Malaysia. J. Trop. Ecol. 1, 5–22. doi: 10.1017/s0266467400000043
Kingsley, E. P., Kozak, K. M., Pfeifer, S. P., Yang, D. S., and Hoekstra, H. E. (2017). The ultimate and proximate mechanisms driving the evolution of long tails in forest deer mice. Evolution 71, 261–273. doi: 10.1111/evo.13150
Klaus, S., Selvandran, S., Goh, J. W., Wowor, D., Brandis, D., Koller, P., et al. (2013). Out of borneo: neogene diversification of sundaic freshwater crabs (Crustacea: Brachyura: Gecarcinucidae: Parathelphusa). J. Biogeogr. 40, 63–74. doi: 10.1111/j.1365-2699.2012.02771.x
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773.
Leigh, J. W., and Bryant, D. (2015). popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210x.12410
Leonard, J. A., den Tex, R.-J., Hawkins, M. T. R., Muñoz-Fuentes, V., Thorington, R., and Maldonado, J. E. (2015). Phylogeography of vertebrates on the Sunda Shelf: a multi-species comparison. J. Biogeogr. 42, 871–879. doi: 10.1111/jbi.12465
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lim, H. C., Zou, F., Taylor, S. S., Marks, B. D., Moyle, R. G., Voelker, G., et al. (2010). Phylogeny of magpie-robins and shamas (Aves: Turdidae: Copsychus and Trichixos): implications for island biogeography in Southeast Asia. J. Biogeogr. 37, 1894–1906.
Lu, X., Ge, D., Xia, L., Huang, C., and Yang, Q. (2014). Geometric morphometric study of the skull shape diversification in Sciuridae (Mammalia, Rodentia). Integr. Zool. 9, 231–245. doi: 10.1111/1749-4877.12035
Mackiewicz, P., Urantówka, A. D., Kroczak, A., and Mackiewicz, D. (2019). Resolving phylogenetic relationships within passeriformes based on mitochondrial genes and inferring the evolution of their mitogenomes in terms of duplications. Genome Biol. Evol. 11, 2824–2849. doi: 10.1093/gbe/evz209
Manawatthana, S., Laosinchai, P., Onparn, N., Brockelman, W. Y., and Round, P. D. (2017). Phylogeography of bulbuls in the genus Iole (Aves: Pycnonotidae). Biol. J. Linnean Soc. 120, 931–944. doi: 10.1093/biolinnean/blw013
Maricic, T., Whitten, M., and Pääbo, S. (2010). Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5:e14004. doi: 10.1371/journal.pcbi.14004
Martin, R. D. (1968). Reproduction and Ontogeny in treeshrews (Tupaia belangeri), with reference to their general behaviour and taxonomic relationships 1. Zeitschrift Tierpsychol. 25, 409–495. doi: 10.1111/j.1439-0310.1968.tb00023.x
Mason, V. C., Helgen, K. M., and Murphy, W. J. (2019). Comparative phylogeography of forest-dependent mammals reveals Paleo-forest corridors throughout Sundaland. J. Hered. 110, 158–172. doi: 10.1093/jhered/esy046
Mason, V. C., Li, G., Helgen, K. M., and Murphy, W. J. (2011). Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Res. 21, 1695–1704. doi: 10.1101/gr.120196.111
McLean, B. S., Jackson, D. J., and Cook, J. A. (2016). Rapid divergence and gene flow at high latitudes shape the history of Holarctic ground squirrels (Urocitellus). Mol. Phylogenet. Evol. 102, 174–188. doi: 10.1016/j.ympev.2016.05.040
Meijaard, E., and Groves, C. P. (2004). The biogeographical evolution and phylogeny of the genus Presbytis. Prim. Rep. 68, 71–90.
Meik, J. M., Schaack, S., Flores-Villela, O., and Streicher, J. W. (2018). Integrative taxonomy at the nexus of population divergence and speciation in insular speckled rattlesnakes. J. Nat. Hist. 52, 989–1016. doi: 10.1080/00222933.2018.1429689
Mercer, J. M., and Roth, V. L. (2003). The effects of cenozoic global change of squirrel phylogeny. Science 299, 1568–1572. doi: 10.1126/science.1079705
Miller, G. S. (1901). Mammals Collected by Dr. W.L. Abbott on the Natuna Islands. Cambridge, MA: Academy Press.
Moore, J. C., and Tate, G. H. H. (1965). A study of the diurnal squirrels, Sciurinae, of the Indian and indochinese subregions. Fieldiana Zool 48, 37, 139.
Morley, R. J. (2000). Origin and Evolution Of Tropical Rain Forests. New York, NY: John Wiley & Sons.
Musser, G. G., Durden, L. A., Holden, M. E., and Light, J. E. (2010). Systematic review of endemic Sulawesi squirrels (Rodentia, Sciuridae), with descriptions of new species of associated sucking lice (Insecta, Anoplura), and phylogenetic and zoogeographic assessments of sciurid lice. Bull. Am. Museum Nat. Hist. 2010, 1–261.
Musser, G.G., and Carleton, M.D. (2005). “Superfamily muroidea,” in Mammal Species Of The World: A Geographic And Taxonomic Reference,” eds D.E. Wilson and D.A. Reeder (Baltimore, MD: The John Hopkins University Press), 894–1531.
Nations, J. A., Heaney, L. R., Demos, T. C., Achmadi, A. S., Rowe, K. C., and Esselstyn, J. A. (2019). A simple skeletal measurement effectively predicts climbing behaviour in a diverse clade of small mammals. Biol. J. Linnean Soc. 128, 323–336.
Nicolas, V., Fabre, P. H., Bryja, J., Denys, C., Verheyen, E., Missoup, L., et al. (2019). The phylogeny of the African wood mice (Muridae, Hylomyscus) based on complete mitochondrial genomes and five nuclear genes reveals their evolutionary history and undescribed diversity. Mol. Phylogenet. Evol. 144:106703. doi: 10.1016/j.ympev.2019.106703
Padial, J. M., Miralles, A., De la Riva, I., and Vences, M. (2010). The integrative future of taxonomy. Front. Zool. 7:16. doi: 10.1186/1742-9994-7-16
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Parham, J. F., Donoghue, P. C., Bell, C. J., Calway, T. D., Head, J. J., Holroyd, P. A., et al. (2012). Best practices for justifying fossil calibrations. Syst. Biol. 61, 346–359.
Patel, R. P., Lenz, D., Kitchener, A. C., Fickel, J., Förster, D. W., and Wilting, A. (2017). Threatened but understudied: supporting conservation by understanding the genetic structure of the flat-headed cat. Conserv. Genet. 18, 1423–1433. doi: 10.1007/s10592-017-0990-2
Patou, M. L., Wilting, A., Gaubert, P., Esselstyn, J. A., Cruaud, C., Jennings, A. P., et al. (2010). Evolutionary history of the Paradoxurus palm civets – a new model for Asian biogeography. J. Biogeogr. 37, 2077–2097. doi: 10.1111/j.1365-2699.2010.02364.x
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Roberts, T. E., Lanier, H. C., Sargis, E. J., and Olson, L. E. (2011). Molecular phylogeny of treeshrews (Mammalia: Scandentia) and the timescale of diversification in Southeast Asia. Mol. Phylogenet. Evol. 60, 358–372. doi: 10.1016/j.ympev.2011.04.021
Robinson, H. C., and Kloss, C. B. (1916). Preliminary diagnoses of some new species and subspecies of mammals and birds obtained in korinchi, west sumatra, Feb.-June 1914. J. Straits Branch Asiat. Soc. 73, 269–278.
Rohland, N., and Reich, D. (2012). Cost-effective, high-throughput DNA sequencing. Genome Res. 22, 939–946. doi: 10.1101/gr.128124.111
Ruedi, M., and Fumagalli, L. (1996). Genetic structure of Gymnures (genus Hylomys; Erinaceidae) on continental islands of Southeast Asia: historical effects of fragmentation. J. Zool. Syst. Evol. Res. 34, 153–162. doi: 10.1111/j.1439-0469.1996.tb00821.x
Saiful, A. A., and Nordin, M. (2004). Diversity and density of diurnal squirrels in a primary hill dipterocarp forest, Malaysia. J. Trop. Ecol. 20, 45–49. doi: 10.1017/s0266467404006169
Sharma, D. (1992). Survey of Mata Ayer Forest Reserve and the proposed Bukit Wang Mu Forest Reserve, Perlis. Kuala Lumpur: WWF Malaysia.
Sheldon, F. H., Lim, H. C., and Moyle, R. G. (2015). Return to the malay archipelago: the biogeography of Sundaic rainforest birds. J. Ornithol. 156, 91–113. doi: 10.1007/s10336-015-1188-3
Sikes, R. S. (2016). Guidelines of the american society of mammalogists for the use of wild mammals in research and education. J. Mammal. 97, 663–688. doi: 10.1093/jmammal/gyw078
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stephens, M. (2005). PHASE: Software for Haplotype Reconstruction, and Recombination Rate Estimation From Population Data. Available online at: http://www.stat.washington.edu/stephens/software.html (accessed January, 2020).
Syakirah, S., Zubaid, A., Lopez, A., Prentice, C., Azmin, M. R., and Mohd Yusof, A. (2001). A small-mammal survey at Tasek Bera, Pahang, Malaysia’s first Ramsar site. Malay. Nat. J. 54, 31–41.
Tenzin, N., Satasook, C., Bumrungsri, S., and Bates, P. J. J. (2013). Does sexual dimorphism exist in squirrels? J. Wildl. Thail. 20, 15–37.
Thomas, O. (1908). XXIII.–On mammals form the Malay Peninsula and Islands. J. Nat. Hist. 2, 301–306. doi: 10.1080/00222930808692485
Thomas, O. (1915). The penis-bone, or “baculum,” as a guide to the classification of certain squirrels. Ibid. 8, 383–387. doi: 10.1080/00222931508693653
Thomas, O., and Wroughton, R. C. (1909). Diagnoses of new mammals collected by Mr. HC Robinson in the Malay Peninsula and Rhio Archipelago. Journal of Natural History 3, 440–441.
Thorington, R. W., and Heaney, L. R. (1981). Body proportions and gliding adaptations of flying squirrels (Petauristinae). J. Mammal. 62, 101–114. doi: 10.2307/1380481
Thorington, R. W., and Hoffmann, R. S. (eds) (2005). “Family sciuridae,” in Mammal Species of the World, A Taxonomic And Geographic Reference, 3rd Edn, (Baltimore, MA: Johns Hopkins University Press).
Thorington, R. W., Koprowski, J. L., Steele, M. A., and Whatton, J. F. (2012). Squirrels of the world. Baltimore. Maryland: Johns Hopkins University Press.
Veron, G., Debruille, A., Kayser, P., Fernandez, D. A. P., and Bourgeois, A. (2019). Genetic diversity and structure of the binturong Arctictis binturong (Carnivora: Viverridae)–status of the elusive Palawan binturong and implications for conservation. Zool. J. Linnean Soc. 188, 302–318.
Veron, G., Debruille, A., Kayser, P., Fernandez, D. A. P., and Bourgeois, A. (2020). Genetic diversity and structure of the binturong Arctictis binturong (Carnivora: Viverridae)–status of the elusive Palawan binturong and implications for conservation. Zool. J. Linnean Soc. 188, 302–318.
Voris, H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27, 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x
Wells, K., Kalko, E. K., Lakim, M. B., and Pfeiffer, M. (2007). Effects of rain forest logging on species richness and assemblage composition of small mammals in Southeast Asia. J. Biogeogr. 34, 1087–1099. doi: 10.1111/j.1365-2699.2006.01677.x
Wells, K., Pfeiffer, M., Lakim, M. B., and Linsenmair, K. E. (2004). Use of arboreal and terrestrial space by a small mammal community in a tropical rain forest in Borneo, Malaysia. J. Biogeogr. 31, 641–652. doi: 10.1046/j.1365-2699.2003.01032.x
Whitten, J. E. (1981). Ecological separation of three diurnal squirrels in tropical rainforest on Siberut Island, Indonesia. J. Zool. 193, 405–420. doi: 10.1111/j.1469-7998.1981.tb03453.x
Wiens, J. J., Engstrom, T. N., and Chippindale, P. T. (2006). Rapid diversification, incomplete isolation, and the “speciation clock” in North American salamanders (genus Plethodon): testing the hybrid swarm hypothesis of rapid radiation. Evolution 60, 2585–2603. doi: 10.1111/j.0014-3820.2006.tb01892.x
Wilting, A., Sollmann, R., Meijaard, E., Helgen, K. M., and Fickel, J. (2012). Mentawai’s endemic, relictual fauna: is it evidence for Pleistocene extinctions on Sumatra? J. Biogeogr. 39, 1608–1620. doi: 10.1111/j.1365-2699.2012.02717.x
Xia, X. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Yang, Z. (2015). The BPP program for species tree estimation and species delimitation. Curr. Zool. 61, 854–865. doi: 10.1093/czoolo/61.5.854
Yang, Z., and Rannala, B. (2010). Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. U.S.A. 107, 9264–9269. doi: 10.1073/pnas.0913022107
Yasuda, M., Ishii, N., Okuda, T., and Hussein, N. A. (2003). “Small mammal community: habitat preference and effects after selective logging,” in Pasoh, eds T. Okuda, N. Manokaran, Y. Matsumoto, K. Niiyama, S. C. Thomas, and P. S. Ashton (Tokyo: Springer), 533–546. doi: 10.1007/978-4-431-67008-7_37
Yasuda, M., Miura, S., and Hussein, N. A. (2000). Evidence for food hoarding behaviour in terrestrial rodents in Pasoh Forest Reserve, a Malaysian lowland rain forest. J. Trop. Forest Sci. 12, 164–173.
Yasuda, M., and Tsuyuki, S. (2012). Comparison of mammalian communities in a human-disturbed tropical landscape in East Kalimantan, Indonesia. Mamm. Study 37, 299–311. doi: 10.3106/041.037.0404
Keywords: Sundasciurus, mammal, Borneo, biogeography, speciation, systematics, rodent, Sciuridae
Citation: Hinckley A, Hawkins MTR, Achmadi AS, Maldonado JE and Leonard JA (2020) Ancient Divergence Driven by Geographic Isolation and Ecological Adaptation in Forest Dependent Sundaland Tree Squirrels. Front. Ecol. Evol. 8:208. doi: 10.3389/fevo.2020.00208
Received: 28 February 2020; Accepted: 05 June 2020;
Published: 26 June 2020.
Edited by:
Toni Gossmann, Bielefeld University, GermanyReviewed by:
David Lee James Vendrami, Bielefeld University, GermanyCopyright © 2020 At least a portion of this work is authored by Melissa Hawkins and Jesús E. Maldonado on behalf of the U.S. Government and, as regards Dr. Hawkins and Dr. Maldonado and the U.S. Government, is not subject to copyright protection in the United States. Foreign and other copyrights may apply. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer A. Leonard, Skxlb25hcmRAZWJkLmNzaWMuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.