
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 14 July 2020
Sec. Conservation and Restoration Ecology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00188
This article is part of the Research Topic Understanding the Impact and Invasion Success of Aquatic Non-native Species: How they Interact with Novel Environments and Native Biota View all 11 articles
 Dubravka Škraba Jurlina1
Dubravka Škraba Jurlina1 Ana Marić1
Ana Marić1 Danilo Mrdak2
Danilo Mrdak2 Tamara Kanjuh1
Tamara Kanjuh1 Ivan Špelić3
Ivan Špelić3 Vera Nikolić1
Vera Nikolić1 Marina Piria3
Marina Piria3 Predrag Simonović1,4*
Predrag Simonović1,4*The diversity of native trout fish Salmo spp. comprises a variety of nominal taxa in Serbia, Montenegro, and Bosnia and Herzegovina. Recent mapping of the resident trout populations detected the presence of brown trout Salmo trutta (sensu stricto) of the Atlantic (AT) mtDNA lineage introduced into populations of both tentative Danubian trout Salmo labrax and of tentative Adriatic trout Salmo farioides belonging to the Danubian (DA) and Adriatic (AD) mtDNA lineages, respectively. Introduction of the tentative Macedonian trout Salmo macedonicus of the AD lineage was also detected in a native population of the tentative S. labrax. In almost all recipient nonmigratory trout populations, a cross-breeding between native and introduced trout was detected by heterozygosity in either only the LDH-C nuclear locus or the LDH-C and specific microsatellite loci. The only exception was a population where both resident and migratory, lake-dwelling individuals of the tentative Adriatic trout spawned in a downstream section of a stream in Montenegro, as no microsatellite alleles of Atlantic brown trout that had been introduced upstream were detected. The occurrence of cross-breeding between Adriatic and brown trout was evident in the isolated, upstream section. It appears that migrating, lake-dwelling Adriatic trout in combination with their resident, stream-dwelling conspecifics suppress the introgression of genes from those situated upstream. In this regard, consideration should be given to the occurrence of the migratory brown trout in the Danube River at the broader Iron Gate Gorge area. They migrate in late summer and early fall from the Iron Gate One reservoir to the lower sections of tributaries devoid of any trout fish. However, some of these streams house very special native trout of the DA lineage in their short-extending upper sections. These native trout populations are, so far, still out of contact with the reservoir-dwelling trout. However, given the resilience of trout and their migratory life history, the outcome of this introduction could be deleterious for those native fish that are very precious in the conservation sense.
Some 25 nominal trout Salmo spp. taxa have been described throughout the dispersal area of this complex species (Kottelat, 1997). This illustrates the great variety in their traits, but also introduces confusion in their taxonomy from the conceptual point of view. There are taxonomic studies and reviews (Bernatchez, 2001; Simonović et al., 2007; LoBrutto et al., 2010; Vera et al., 2011; Meraner et al., 2013) that consider Salmo trutta L., 1758, to be a species complex comprising phylogenetically closely related species, while others (Abolhasan et al., 2017; Kalayci et al., 2018; Sanz, 2018; Rezaei et al., 2019; Whiteley et al., 2019) consider this to be a single highly polymorphic species. Whiteley et al. (2019) stated that the current taxonomic richness within the S. trutta was promoted by Kottelat (1997), who followed the evolutionary species concept of Simpson (1961) to assign local brown trout subspecies originally described on the basis of slight differences in a limited number of traits between local populations as nominal species (Kottelat and Freyhof, 2007).
The Western Balkans is a region known for its prominent trout diversity, as seen in the many trout taxa described in all three main drainage areas found here. Marble trout Salmo marmoratus (Cuvier, 1829), soft-muzzled trout Salmo obtusirostris (Heckel, 1851), Adriatic trout Salmo farioides (Karaman, 1938), and Salmo montenegrinus (Karaman, 1933) are species endemic to the eastern Adriatic Basin in Montenegro, Dalmatia, and Herzegovina, whereas the Lake Ohrid trout Salmo letnica (Karaman, 1924) (together with the tentative Salmo lumi (Poljakov et al., 1958), and Salmo aphelios (Kottelat, 1997), belvica trout Salmo ohridanus (Steindachner, 1892), and the Lake Prespa trout Salmo peristericus (Karaman, 1938), are endemic to the Adriatic Basin in Northern Macedonia. Taler's trout Salmo taleri (Karaman, 1933) and Danubian salmon Salmo labrax (Pallas, 1814), tentatively occur in the Black Sea Basin, while the Macedonian trout Salmo macedonicus (Karaman, 1924) and Pelagonian trout Salmo pelagonicus (Karaman, 1938), are found in the Vardar River (Axios) and Struma River (Strymon) in the Aegean Sea Basin.
The nominal taxonomy of Salmo spp. presented here was recently supplemented with a prominent insight into their diversity, assessed using molecular markers, both mitochondrial, [e.g., cytochrome b and control region (CR), i.e., the D-loop (Bernatchez et al., 1992; Bernatchez, 2001), and nuclear, e.g., LDH-C* and several microsatellite loci (Hamilton et al., 1989; Richard and Thorpe, 2001)]. The basic mapping of indigenous populations of nominal trout taxa accomplished using CR as a marker in the Adriatic Sea Basin assigned them to the Adriatic (AD), marmoratus (MA), and Danubian (DA) mtDNA lineages (Marić et al., 2006; Mrdak, 2011; Mrdak et al., 2012; Simonović et al., 2017b; Škraba Jurlina et al., 2018); those native to the Black Sea Basin belonged to the DA lineage (Marić et al., 2006; Mrdak, 2011; Tošić et al., 2014, 2016; Simonović et al., 2017b; Škraba et al., 2017), whereas in the Aegean Sea Basin, they belonged to the AD lineage (Marić et al., 2006, 2017; LoBrutto et al., 2010).
Every introduction poses a risk to the loss of original genetic structure in the recipient local brown trout population (Templeton, 1986; Laikre and Ryman, 1996; Laikre et al., 2010; Hansen, 2002; Ferguson, 2006a). The effect of introductions of domesticated, hatchery-reared brown trout of the DA haplogroup is difficult to detect using diploid genetic markers. This is an especially sensitive matter, given the fact that very few stocks at brown trout hatcheries in the Western Balkans have been genotyped. Though some countries have prescribed mandatory marking and registration of brood fish, this is poorly enforced. It is more feasible to detect the effects of introduction of brown trout of the AT haplogroup into the dispersal range of trout of either DA or AD haplogroups by seeking out the LDH-C*90 allele they transfer to their offspring by admixture with trout of DA and AD haplogroups. Subsequent analysis of microsatellite alleles can provide insight into the changes in the genetic structure of the recipient brown trout population by detecting private alleles specific to the introduced brown trout strain.
Since the introduction of brown trout worldwide (MacCrimmon and Marshall, 1968), it has become one of the most widely introduced fish species (Welcome, 1992; Fausch, 2007). Its invasiveness has been noticed everywhere, resulting in its proclamation as one of the World's 100 worst invasive alien species (Lowe et al., 2000). The most invasive impact of brown trout on the recipient fish fauna was recorded in New Zealand (Townsend, 1996; McIntosh et al., 2010; Jones and Closs, 2018), where the local native galaxiids approached extirpation after the introduction of the brown trout into their native streams. Behnke (2007) reported a similar effect in the Yellowstone National Park ecosystem, where introduction of the brown trout strongly suppressed the native West slope cutthroat trout Oncorhynchus clarkii (Richardson, 1836).
The economic impact of their invasive character is closely tied to conservational issues and trout fisheries, where fishery management is the main driver of the increased risk of introduction of non-native trout strains. Recently, activities to build small hydropower plants as renewable energy sources have been focused primarily on trout streams, which threatens very fragile mountain stream ecosystems and their native trout (Simonović, 2019). The adverse effects of small hydropower plants onto native trout populations in the sections of streams along the derivations are compensated through planned stocking, which perpetuates and augments the risk of introducing alien brown trout. This widely adopted stocking practice in trout stream management accompanied by the flow of money among fishery managers, trout fish farmers, and trout fishermen is the primary obstacle to effective conservation of indigenous and often unique native brown trout stocks (Simonović et al., 2017b), both in common fisheries (Simonović, 2019) and even in those situated in protected areas (Simonović et al., 2014).
The life history plasticity of brown trout makes the species highly resilient and able to adapt to abrupt alterations in habitat and to long-term harsh conditions when resources are scarce, which sustains adverse effects and maintains the genetic variability inherent to local populations (Ferguson et al., 2019). Three life forms of trout known as morphae (i.e., sea trout, m. marinus; lake trout, m. lacustris; and river trout, m. fario) generally depict that plasticity, which extends also to the life history in sea and lake morphs featuring migratory behavior-anadromy and lymnodromy, respectively. In the Balkans, migratory behavior in native trout is found in only a few populations. In the Eastern Balkans, [e.g., sea trout S. labrax migrate to spawn in the small tributaries on the Bulgarian coast of the Black Sea (Kohout et al., 2013)]. Migratory trout in the Western Balkans include lake-dwelling forms belonging to the Adriatic trout Salmo farioides (Karaman, 1938) in Montenegro, [e.g., the Lake Skadar “strun” (Mrdak et al., 2006) and to the marble trout S. marmoratus in Herzegovina, e.g., the tentative “zubatak” nominally described as Salmo dentex Heckel and Kner (1958)]. Snoj et al. (2002) reported that sea trout recorded in the Northern Adriatic Sea are brown trout of the AT lineage, as escapees from trout farms in streams draining to the sea. They considered them distinct from the sea trout, reported by Chiereghini (1818), which Kolombatović (1890) described as the tentative Trutta adriatica. Contrary to the mapping of brown trout of the AT lineage introduced into streams of the Danube River catchment (Simonović et al., 2017a), there are no reliable records on the introduction of the hatchery-reared brown trout of the Atlantic origin into streams at the Adriatic Sea watershed in Montenegro in the last 50 years at least. In the inland waters of the Danube River catchment, reservoir-dwelling migrating trout were recorded soon after the construction of dams, though it is not yet known whether those individuals belong to native trout of the DA lineage or to introduced trout of the AT lineage or whether they are an admixture of both of them. The discovery of brown trout individuals in the Djerdap (Iron Gate) One reservoir and in the lowermost sections of streams draining into the Danube River there (Figure 1, localities 1–7) confirmed they belong to the AT lineage or are admixed with it (Marić et al., 2012; Simonović et al., 2015, 2017a; Tošić et al., 2016). There are no records of these trout in the upstream, headwater sections, where pure brown trout of the very specific Da23c haplotype still occur (Tošić et al., 2016).
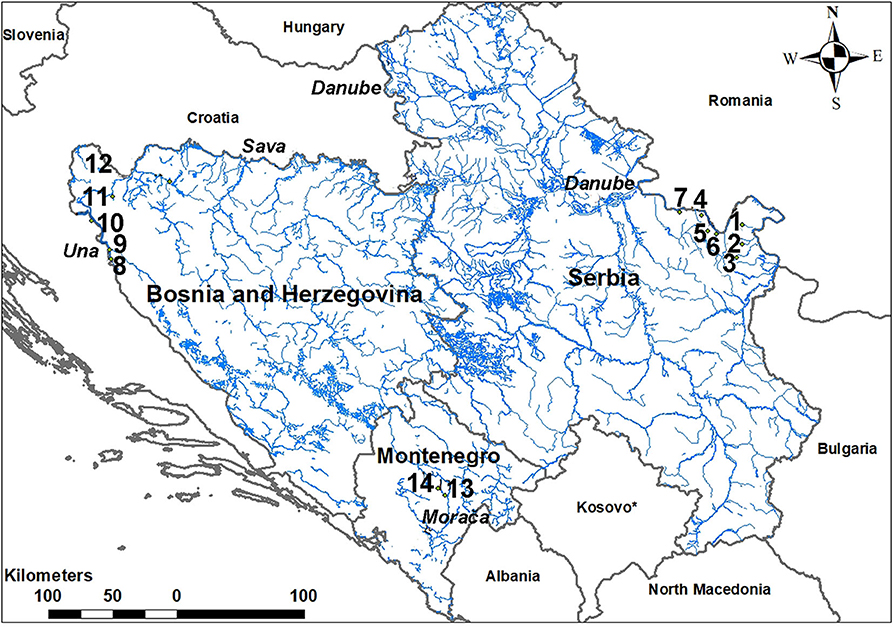
Figure 1. Map of the study area in the Western Balkans where brown trout populations were sampled (1. Zamna River; 2. Rečka River; 3. Vratna River; 4. KoŽica River; 5. Mala Boljetinska River; 6. Zlatica River; 7. Brnjica River; 8. Krka River; 9. Upper Una River at Martinbrod; 10. Upper Una River at Loskun; 11. Svetinja River; 12. Krušnica River; 13. lower Mrtvica River; 14. upper Mrtvica River).
The aims of this study were: (1) to consider how alternative life histories and occurrence of migratory behavior in native brown trout of the AD lineage responded to the introduction of an alien strain of brown trout of the AT lineage into their home streams; (2) how introduced alien brown trout of the AT lineage affected the genetic structure of the stream-dwelling native individuals by introgressing into their gene pools; and (3) how migratory behavior in alien brown trout of the AT lineage acted in their favor when invading the recipient streams with resident, native, stream-dwelling brown trout of the DA lineage in the Western Balkans, regarding either the loss of original genetic diversity, or increased migratory behavior in the native populations. The results should allow scientists to anticipate the risks faced by the native brown trout population and the expected effects in real circumstances occurring in the area.
Trout sampling was conducted by fly fishing and by electrofishing using either an engine-powered electrofishing gear Suzuki-Bosch™ (220 V DC, Imax = 6 A) or battery-powered portable electrofishing gear AquaTech™ IG200/1® (380/600 V DC, Imax = 15A).
Sampling was conducted in the Svetinja and Krušnica Rivers, two tributaries of the middle reach of the Una River in the Black Sea Basin (Figure 1, localities 11 and 12), in autumn of 2012 and 2013. All individuals were small [of the standard length (SL) <25 cm] resident, stream-dwelling, with coloration common for brown trout in the Black Sea Basin: dark brown back and golden-yellow flanks, with the moderately large black spots scattered along the back and upper, dark flanks, and with uniformly sized red spots aligned in two almost regular rows along the golden mid-flanks of the body. Sampling in the lower Mrtvica River in Montenegro in the Adriatic Sea Basin (Figure 1, locality 13) was conducted by angling, from 2004 to 2007, in the peak of the spawning season in late autumn, when both life history forms co-occur in the spawning grounds. The sample consisted of both small, stream-dwelling fish of both sexes, but predominantly males, up to 20 cm in SL, with dark blue back and light bluish flanks scattered with tiny black and reddish spots, and large (some individuals exceeded 40 cm SL), smoltified (i.e., silvery in color, with only tiny, black spots on the back and flanks), lake-dwelling, exclusively female individuals, as seen by freely leaking roe. Samples from the upper Mrtvica River (Figure 1, locality 14) were collected in the spring of 2014 by electrofishing and consisted of only small, stream-dwelling fish, not exceeding 15 cm in SL. Samplings in the streams draining into the Danube River at the Djerdap Gorge (Iron Gate) area (Figure 1, localities 1–7) were conducted during the late summer–early autumn period from 2011 to 2015, and most samples in all streams consisted of typical stream-dwelling, small (of SL ≤ 15 cm) brown trout that resembled those from Svetinja and Krušnica Rivers. Only a couple of brown trout from the Brnjica River were slightly larger (of SL >25 cm SL) and silvery in color, with numerous, tiny black spots on the back and flanks, resembling the smoltified brown trout. After sampling, all fish were released into their home streams. Therefore, the determination of sex (except for individuals from the lower Mrtvica River) was not possible.
From each fish, an anal fin clip was taken and stored in microtubes filled with 96% ethanol. Fin clips were collected from 10 individuals from the Svetinja River, from 11 caught in the Krušnica River, from 11 individuals caught in the lower reach, and 12 from the upper reach of the Mrtvica River, where the size of samples was limited most likely by extensive poaching occurring there (Škraba Jurlina et al., 2018). As for the Danube River tributaries in the Iron Gate Gorge area, fin clips were collected from each of a very small number of individuals due to their small size and the scarcity of water and brown trout in those streams (Tošić et al., 2016), [e.g., two samples from the Brnjica River, 18 from the KoŽica River, six from the Mala Boljetinska River, 12 from the Zlatica River, 11 from the north fork of the Rečka River, 10 from the Vratna River, and seven from the Zamna River (Figure 1, localities 7, 4, 5, 6, 3, 2, and 1, respectively) (Supplementary Table)].
DNA was extracted using either the High-Salt Extraction technique of Miller et al. (1988) or Quick-gDNA™ MiniPrep extraction kit following the manufacturer's instructions (Zymo Research Corporation, Irvine, CA).
To identify introductions of non-native brown trout stocks, we amplified and analyzed the CR or D-loop that has been proven to be a good phylogeographic and taxonomic marker (Bernatchez et al., 1992; Cortey and García-Marín, 2002; Tougard et al., 2018). It is used for a range of purposes in combination with other molecular markers, including determining implication for conservation and management (Vera et al., 2011), migrating strains (Habibi et al., 2013), and analyzing the consequences of stocking (Škraba et al., 2017).
Amplification of the CR of mtDNA was carried out using the forward primers 28Riba (Snoj et al., 2000) and Trutta_mt_F (5′-TGAATGAACCTGCCCTAGTAGC-3′, designed by M. Brkušanin), and the reverse primer HN20 (Bernatchez and Danzmann, 1993), following the protocol from Tošić et al. (2014). PCR products were purified and sequenced at Macrogen Europe. Sequencing reactions were performed in a DNA Engine Tetrad 2 Peltier Thermal Cycler (BIO-RAD) using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), following the protocols supplied by the manufacturer by single-pass sequencing on each template using the forward (Trutta_mt_F) primer. Sequences were aligned with those from the GenBank using program Mega 7.0.21 (Larkin et al., 2007).
In addition to mtDNA showing maternal inheritance, other nuclear markers that complement with paternal inheritance in brown trout were used to infer the relationships between their populations. The lactate dehydrogenase (LDH) gene (Hamilton et al., 1989), specifically its eye-specific locus (LDH-C1*), was mostly used for this purpose. It is found only within the genus Salmo with two codominant alleles: LDH-C*100 and LDH-C*90. The first has an ancestral character and features a DA haplogroup, while the second features exclusively in northwestern European populations of the AT mtDNA haplogroup (McMeel et al., 2001). This was used to analyze introgression of alien individuals in the native populations.
Part of the LDH gene was amplified using Ldhxon3F and Ldhxon4R primers (McMeel et al., 2001). The final concentrations of PCR components were: 1 × PCR buffer (Invitrogen™, SAD), 1.5 mM MgCl2, 0.2 mM of each dNTPs, 0.5 μM for both primers (Metabion, DE), 1.5 U of Taq polymerase (Invitrogen™, USA), and ~100 ng of DNA. PCR amplification started with initial denaturation (95°C, 5 min), continued with 30 cycles of denaturation (94°C, 1 min), followed by primer annealing (62°C, 1 min) and DNA extension (72°C, 1 min), and final elongation (72°C, 10 min) in the ProFlex™ PCR System (Applied Biosystems®, USA). A 2.5% gel was used for DNA electrophoresis. Restriction fragment length polymorphism (RFLP) was analyzed using BselI endonuclease that cuts DNA at the CCCNNNNN/NNGGG position. PCR products were incubated for 16 h with BselI endonuclease (Thermo Fisher Scientific, USA) at 55°C according to instructions. The mixture contained ~100 ng of DNA, 10 U of enzyme, 10 × Tango Buffer and molecular water. A 2.5% gel was used for DNA electrophoresis with ® SYBR Green™ for visualization.
Microsatellite markers are often used for conservational and population genetic purposes, specifically to estimate population genetic diversity, influences of stocking on native populations (Hansen et al., 2000, 2001), effective population size (Serbezov et al., 2012), and broodstock formation (Hansen et al., 2000).
Analysis of the samples from both the Svetinja and Krušnica Rivers and from streams in the broader Iron Gate Gorge area was based on eight microsatellite loci—SsaD190, SsaD71 (King et al., 2005), Str73INRA (Estoup et al., 1998), Ssa410Uos (Cairney et al., 2000), Ssa85 (O'Reilly et al., 1996), SSsp2216 (Paterson et al., 2004), OMM1064 (Rexroad et al., 2002), and SsoSL438 (Slettan et al., 1995). All were amplified in four duplex reactions. The last microsatellite locus, SsoSL438, was excluded from the analysis of specimens from the Mrtvica River, while the remaining seven loci were combined in one single and three duplex reactions, with the forward primer labeled with a fluorescent dye (FAM or NED) (Škraba Jurlina et al., 2018). Fragment analysis was performed using GeneScan 500 LIZ Size Standard (Applied Biosystems, USA) on an ABI-3130 Genetic Analyzer (Applied Biosystems, USA). Analysis was performed using GeneMapper ID v3.2.1 (Applied Biosystems, USA).
Statistical analysis was performed on the microsatellite data to determine the differences in genetic structure between populations whose members revealed an alternative life history by partial migration and populations consisting exclusively of resident, stream-dwelling brown trout. Genetic structure from microsatellite data was determined as reported in Škraba Jurlina et al. (2018). Factorial correspondence analysis (FCA) was carried out using the program GENETIX 4.05 (Belkhir et al., 2004). Program FSTAT 2.9.3.2 (Goudet, 2002) was used to calculate allelic richness and values for Fisher's F statistics per locus (FIS, FIT, FST) and to test Hardy–Weinberg equilibrium within populations. Population structure was analyzed using the STRUCTURE 2.3.4 (Pritchard et al., 2000), with the proposed number of clusters K = 10. The length of burn in periods was set to 20,000, with the number of Markov Chain Monte Carlo (MCMC) repeats of 10 for each K depending on convergence after burning was set to 500,000. Structure Harvester software (Earl and VonHoldt, 2012) was used to estimate the most probable K according to Evanno et al. (2005).
Sequencing of the mtDNA CR revealed the introduction of brown trout belonging to the AT haplogroup in all three areas. In the streams feeding the middle course of the Una River, where the native brown trout belongs to the Da22 haplotype (GenBank accession number AF321993; Duftner et al., 2003), three out of 10 brown trout from the Krušnica River (33%) and three out of 10 from the Svetinja River (33%) belonged to the Atcs1 haplotype (#AF321990; Weiss et al., 2001; Škraba et al., 2017).
In the upper course of the Mrtvica River, where Adriatic trout is the native brown trout taxon bearing the Adcs11 haplotype (#AY836340; Cortey et al., 2004), three of 12 (25%) fish were of the A17 haplotype (#HQ848368; Kohout et al., 2012), and all of 11 fish from the lower course were native Adriatic trout (Škraba Jurlina et al., 2018).
In all streams in the Iron Gate Gorge area, brown trout of the AT haplogroup were recorded in the Brnjica River, Dobra River, and Porečka River and in the Danube River near the confluence with the Dobra River, while in the Vratna River, they were found in sympatry with the brown trout of the Da23c haplotype (#KC630984; Tošić et al., 2014), [i.e., three of 10 (33%) brown trout in total were of this haplotype (Tošić et al., 2016)].
RFLP analysis of the partial LDH-C gene in brown trout samples from the middle course of the Una River showed that two fishes of the AT haplogroup were homozygotes for the slower allele (LDH-C*90/90), and the remaining brown trout of the AT haplogroup were heterozygotes LDH-C*100/90. The slower allele was also dominant in the DA haplogroup, where only four brown trout from the Krušnica River were homozygotes for the faster allele (LDH-C*100/100). That high proportion of slow allele occurrence in 75% of brown trout implied strong introgression of brown trout of the AT haplogroup into the gene pool of the native DA haplogroup.
In the upper course of the Mrtvica River, two of three fishes (66%) with the A17 haplotype were heterozygous (LDH-C*90/100), as were two of the remaining of nine fishes (22%) with the Adcs11 haplotype, indicating their paternal origin from the AT lineage. Other fishes in the upper course and all fishes in the lower course were homozygous for the LDH-C*100 allele. The remaining individuals belonging either to the ADcs11 or to the A17 haplotypes were homozygotic for the faster allele (LDH-C*100/100).
Of the three brown trout of the AT haplogroup from the Vratna River, only one (33%) was heterozygous (LDH-C*90/100), while two were homozygous for the faster allele (LDH-C*100/100) characteristic to brown trout of the DA haplogroup. This implies a paternal DA origin for those fishes and a long-lasting introgression with the recurrent cross-breeding between brown trout of the DA and AT lineages.
In the (sub)populations from the Svetinja and Krušnica Rivers in the middle course of the Una River catchment, genetic diversity in brown trout individuals was greater, but with the lower genetic differentitation, as revealed by their fixation indices FIS = 0.1250 (p < 0.02) and 0.1926 (p < 0.001), respectively (Table 1), in comparison with FIS values ranging between−0.01695 and 0.04296 (p > 0.1) in brown trout from the (sub)populations in the upper Una River, consisting exclusively of native fishes. AMOVA results showed the greatest proportion of genetic variation to be in individuals (71.0%, FST = 0.705, p < 0.01), followed by the component of variability inherent to populations (22.5%, FIT = 0.290). Several private alleles were found in fish belonging to the AT haplogroup. Private alleles found in samples from the Svetinja River were mostly heterozygous for two loci, Ssa410UoS and OMM1064, and one sample was in a homozygous state for the locus Ssa85 (Table 2). All private alleles from the Krušnica River were heterozygous. Pairwise distance FST values between the populations gave clear depiction of their position in the Una River catchment; the only deviation was for the sample from the Una River at Martinbrod (Table 3A). In the FCA, the first four correspondent factors explained 18.35% of the total genetic variability. Correspondent factors 1 and 2 explained 6.22 and 4.36% of the variability, respectively, and clustered the brown trout from the Svetinja and Krušnica Rivers separately from all others (Figure 2, top). STRUCTURE analysis revealed two (with greatest value of Delta K = 31.759468) distinct subpopulations (Figure 3, top): the first one consisting of brown trout from the Svetinja and Krušnica Rivers, and the second one of brown trout from the Krka River and upper Una River (localities Martinbrod and Loskun).
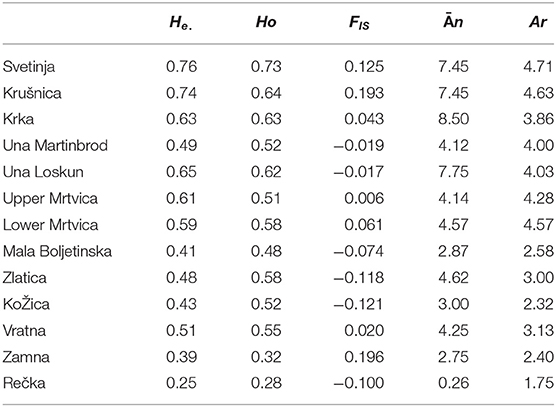
Table 1. Population genetics of brown trout populations from streams situated in the Una River catchment (Svetinja, Krušnica, and Krka Rivers, upper Una River at the sites Martinbrod and Loskun sites), Mrtvica River (upper and lower), and in the Iron Gate Gorge area (Mala Boljetinska, Zlatica, KoŽica, Vratna, and Zamna Rivers), as assessed from microsatellite loci (He., expected heterozygozity; Ho., observed heterozygosity; FIS, intrapopulation fixation index; n, mean allelic diversity per population; Ar, allelic richness; H-W, deviation from the Hardy–Weinberg equilibrium).
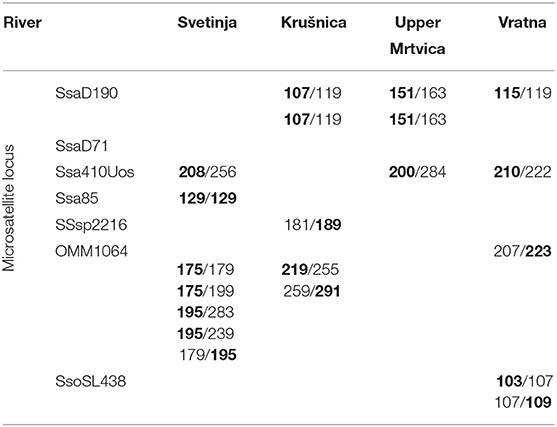
Table 2. Microsatellite loci revealing private alleles (in bold letters) for the AT haplogroup in brown trout from four populations: in the middle course of the Una River (Svetinja and Krušnica Rivers), Mrtvica River (upper Mrtvica River), and middle Danube River at the Iron Gate Gorge area (Vratna River).
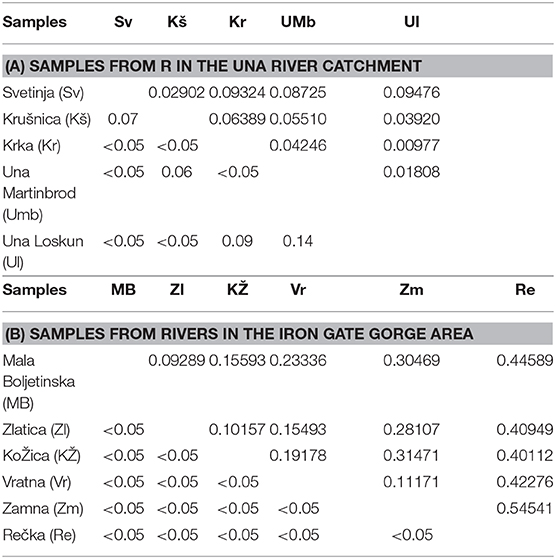
Table 3. Pairwise FST distance values (above and right of the diagonal) and their significance (p-values) levels between samples from (A) Una River catchment and (B) Iron Gate Gorge area (the pairwise FST distance value between the upper and lower Mrtvica River populations is 0.22060, p < 0.05).
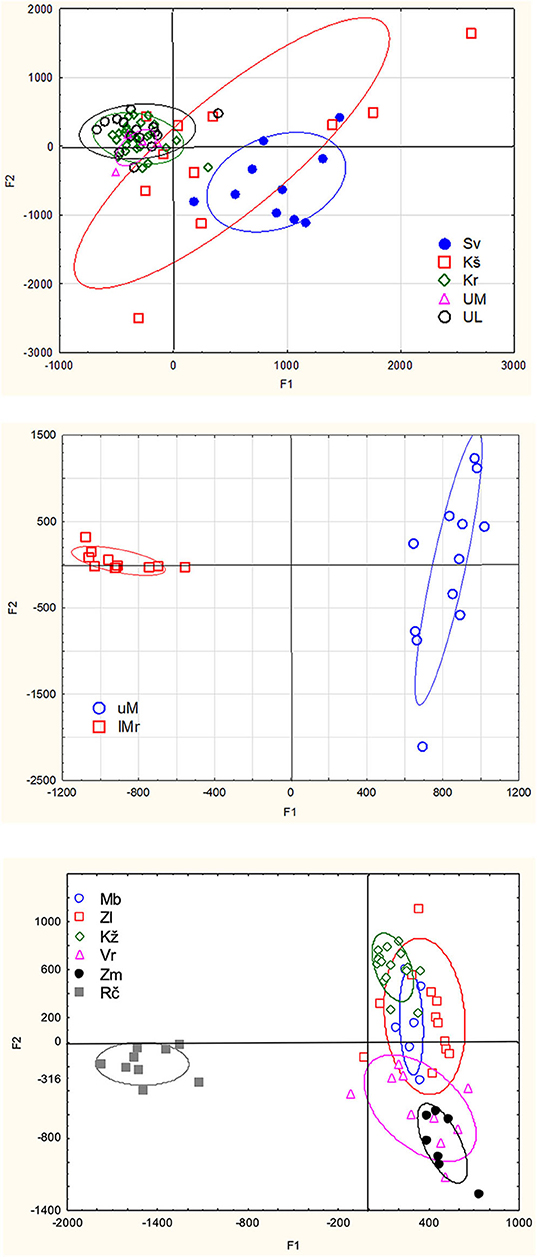
Figure 2. Correspondence analysis of brown trout populations from the Una River catchment (top), Mrtvica River (in the middle), and streams in the Iron Gate Gorge area (bottom) (Sv, Svetinja River; Kš, Krušnica River; Kr, Krka River; UM, Una River at Martinbrod; UL, Una River at Loskun; uM, upper Mrtvica River; lM, lower Mrtvica River; MB, Mala Boljetinska River; Zl, Zlatica River; KŽ, KoŽica River; Vr, Vratna River; Zm, Zamna River; Rč, Rečka River).
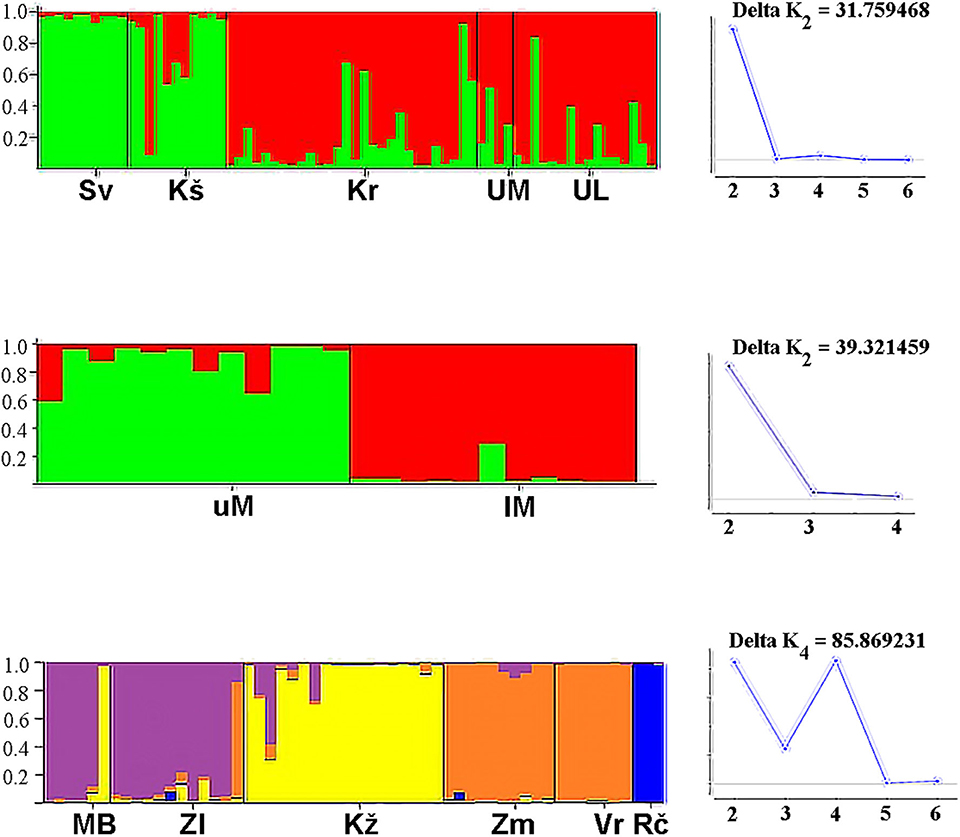
Figure 3. STRUCTURE analysis of brown trout populations from the Una River catchment (top), Mrtvica River (in the middle), and streams in the Iron Gate Gorge area (bottom) (Sv, Svetinja River; Kš, Krušnica River; Kr, Krka River; UM, Una River at Martinbrod; UL, Una River at Loskun; uM, upper Mrtvica River; lM, lower Mrtvica River; MB, Mala Boljetinska River; Zl, Zlatica River; KŽ, KoŽica River; Vr, Vratna River; Zm, Zamna River; Rč, Rečka River).
In the upper Mrtvica River where brown trout of the AT haplogroup were admixed with the native AD trout, the observed heterozygosity Ho = 0.16667 was significantly lower than the expected He = 0.72826 for the locus SsaD71, demonstrating a significant (p < 0.001) deviation from the Hardy–Weinberg equilibrium, with the greater genetic diversity, [i.e., a weak differentiation within that population (FIS = 0.779), compared with the lower Mrtvica River population (FIS = 0.135) (Table 1)]. Pairwise distance between the upper and lower Mrtvica populations (FST = 0.221, p < 0.05) showed their significant differentiation. AMOVA showed that the majority of genetic variability to be that of individuals in the two populations (77.9 %, FST = 0.221, p < 0.01), whereas the within-population component was lower (22.1%, FIT = 0.247, p < 0.01). Private alleles specific for the AT haplogroup were found in brown trout of the AT haplogroup at two microsatellite loci and were heterozygous for the loci SsaD190 (151) and Ssa410Uos (200), whereas a heterozygous private allele for the locus OMM1064 (215) was recorded in one fish of the AD haplogroup, also in the heterozygous state. None of these private alleles for the AT haplogroup were recorded in any of the fish of the AD haplogroup in the lower Mrtvica River population (Table 2). In the FCA, the first four correspondent factors explained 36.02% of the total genetic variability. The first and second correspondent factors explained 14.40 and 8.10% of the variability, respectively, and clearly separated the two populations (Figure 2, middle). STRUCTURE analysis demonstrated that brown trout in the upper and lower Mrtvica River are two distinct subpopulations (with greatest value of Delta K = 39.321459) (Figure 3, middle).
In the brown trout population from the Vratna River in the Iron Gate Gorge area, the expected average heterozygosity for all microsatellites' loci was Hexp = 0.51, the highest among all populations in the area (Table 1). Several private alleles were found in individuals belonging both to the AT and DA haplogroups. AMOVA revealed that the majority of genetic variability in samples from the Iron Gate Gorge area was inherent to individuals (62.1%, FIT = 0.621, p < 0.01), and then to samples, [i.e., the populations (37.9%, FST = 0.379, p < 0.05)]. The allelic, [i.e., genetic diversity was highest in the brown trout population from the Vratna River, e.g., that obtained for the loci Ssa410Uos (AR = 5.348, FIS = 0.875), SsaD190 (AR = 2.860, FIS = −0.213), SSsp2216 (AR = 4.528, FIS = 0.034), and SsoSL438 (AR = 2.698, FIS = 0.191)]. One brown trout from the Vratna River was of the AT haplogroup and was homozygous for the LDH-C* allele characteristic for the DA haplogroup (100/100) and had private microsatellite alleles of the AT haplogroup at the loci SsaD190 (115) and SsoSL438 (109), which were likely of maternal origin (Table 2). In two brown trout of the DA haplogroup, several other private alleles were found to occur exclusively in these fish and no others of that haplogroup from any population in the area. These alleles were recorded at three loci (Table 2) and were probably of paternal origin. Private alleles found at all microsatellite loci were in heterozygous state. In the FCA, the first four correspondent factors explained 34.51% of the total genetic variability of brown trout in the Iron Gate Gorge area. Correspondent factor 1 explained 12.55% of the total variability and clearly separated the Rečka River population from all others, while correspondent factor 2 explained 8.14% of the variability and separated populations from the Mala Boljetinska, Zlatica, and KoŽica Rivers from populations in the Vratna and Zamna Rivers (Figure 2, bottom), which was supported by pairwise FST distances between populations (Table 3B). STRUCTURE analysis revealed four (with greatest value of Delta K = 85.869231) distinct brown trout subpopulations (Figure 3, bottom): one in the Mala Boljetinska and Zlatica Rivers, the second in the KoŽica River, the third in the Zamna and Vratna Rivers, and the fourth in the Rečka River.
The relatively small sample sizes from certain localities due either to extensive poaching (e.g., in the Mrtvica River) or very small size of streams and extreme scarcity of water during sampling (e.g., the Svetinja River and all streams in the Iron Gate Gorge area) can in general weaken the conclusions of their population-genetic features. Interpretation of results for population genetics features (Table 1) should include caution, as a stronger effect of admixing with brown trout of the AT haplogroup is expected on the Ho. The high inbreeding coefficient FIS (Table 1) in line with the reduction of heterozygosity that depicts their fixation for particular alleles, e.g., in the populations from the Svetinja, Krušnica. and Zamna Rivers could be a consequence of the small sample size. However, the use of three molecular markers and occurrence of heterozygous alleles specific for brown trout of the AT haplogroup (Table 2) unequivocally demonstrated admixing with the resident, native brown trout. This showed the invasive risk that non-native, migrating brown trout of the AT haplogroup pose on the resident brown trout populations in their native dispersal areas and demonstrated the resilience in native Adriatic trout population which have partial migratory behavior.
In contrast to the positive effects that sustainable management in trout fisheries have [e.g., the conditional and unconditional catch-and-release (Simonović et al., 2018)], both fishery managers and trout farmers fostered stocking of farmed brown trout fry and even spent brood fish (Simonović et al., 2014) to quickly enhance brown trout fisheries and accelerate return of investments (Simonović et al., 2017b). Based on recent knowledge of the native diversity of brown trout in the Western Balkans (Marić et al., 2006; Simonović et al., 2017a) and the risks posed to them by the introduction of non-native strains of brown trout and related species might pose to them (Simonović et al., 2013, 2015), recent surveys have revealed the occurrence of permanent, long-lasting introduction caused by stocking streams attractive for trout fisheries (Simonović et al., 2014; Škraba Jurlina et al., 2018), streams originally devoid of trout prior to stocking (Jadan et al., 2007; Simonović et al., 2018), and even streams that are not at all suitable for brown trout fisheries (Tošić et al., 2016; Škraba et al., 2017). Introduction of trout fish of the non-native haplogroups and of the non-native haplotypes in the dispersal area of brown trout of the DA haplogroup in the Western Balkans has been a long-standing practice since the mid-nineteenth century at least, when Slovenia, Croatia, and Bosnia and Herzegovina were parts of the Austro–Hungarian Empire (Razpet et al., 2007; Simonović et al., 2017b). The nominal genetic diversity assessed from microsatellite loci and the FIS and AR values in the populations of stream-dwelling, resident trout where brown trout of the AT lineage introgressed suggests that alien brown trout of the AT lineage have impacted brown trout populations, which could lead to a loss of native haplotypes and co-adapted gene complexes created through the long-term adaptation of aboriginal brown trout populations in their native habitats (Templeton, 1986). In that way, non-native brown trout of the AT haplogroup unequivocally reveal the invasive effect as defined by Kolar and Lodge (2002) and Copp et al. (2005), justifying their assessment as having high sensu stricto invasive potential (Simonović et al., 2015).
Salmonids commonly possess a migratory instinct. In many species, populations partition between resident, stream-dwelling, predominantly male individuals, and migrating, lake- or sea-dwelling, predominantly female individuals (Fleming and Gross, 1990; Ferguson, 2006a) that admix (Hansen, 2002). This is known for many brown trout populations in the Atlantic Ocean basin. Unlike the differences in morphological, demographical, and ecological characteristics (Klemetsen et al., 2003) between resident and migratory fish, no genetic differences have been found between them (Hindar et al., 1991; Hansen, 2002; Charles et al., 2005), demonstrating their common population status. The common genetic structure and gender proportion in the stream- and lake-dwelling Adriatic trout in the lower Mrtvica River are in agreement with population features reported for other migratory salmonid stocks. The low value of gene flow between two (sub-)populations supports the view that brown trout of the AT haplogroup from the upper Mrtvica River failed to admix with native Adriatic trout (Škraba Jurlina et al., 2018), despite a lack of any physical or reproductive obstacles. The stocked fish of the AT haplogroup have certainly not bred and have hence vanished from the lower Mrtvica River, accessible to the spawning migratory, lake-dwelling Adriatic trout (“strun”). In contrast, brown trout of the AT haplogroup still remain in the upper Mrtvica River and spawn with the native, stream-dwelling, resident Adriatic trout, producing hybrids of reduced heterozygosity (Table 1), a feature also reported by Ryman et al. (1995), Laikre and Ryman (1996), and Ferguson (2006b). Snoj et al. (2010) reported an occurrence of brown trout of the Atlantic haplogroup (At-s1, #M97969, Bernatchez et al., 1992) in the lower Neretva River, where the lake-dwelling form of marble trout S. marmoratus (locally known as “dentex”) occurs together with the resident marble trout and Adriatic trout. Regardless of the similarity of Adriatic trout and the tentative “dentex” for the commonly shared AdN haplotype (#DQ297172), the genetic structure of “dentex” was more similar to marble than to Adriatic trout, suggesting that “dentex” is the lake-dwelling, migratory form of marble trout, and not a form of Adriatic trout. As the AT haplogroup was not detected in the phenotypic, resident marble trout (Snoj et al., 2010), it seems that their life history plasticity helped to preserve their genetic structure, whereas the lack of a lake-dwelling form of Adriatic trout in the lower Neretva River made them susceptible to hybridization with the stocked brown trout of the AT haplogroup.
In the native, stream-dwelling brown trout of the two haplotypes belonging to the DA haplogroup (Da23c in the Vratna River in Eastern Serbia, as well as Da22 in the Svetinja and Krušnica Rivers in Bosnia and Herzegovina), all diploid genetic markers revealed an undisturbed admixture with stocked brown trout of the AT haplogroup and their persistence, as in the upper Mrtvica River. Moreover, it seems that brown trout of the narrowly dispersed and very specific modern haplotype Da23c found in several streams in the Iron Gate Gorge area are strongly threatened by the occurrence of the lake- (actually, a reservoir-) dwelling brown trout of the AT haplogroup (Marić et al., 2012) that were detected in the lowermost section of those streams in the late summer and early autumn periods (Tošić et al., 2016). Their silver body coloration and numerous black dots on the back and flanks suggest they smoltified prior to descending downstream to the reservoir. If they have already started returning to the closest available spawning grounds upon achieving maturity, there is a high risk of admixing with the native brown trout of the DA haplogroup and introgression into their gene pools in those Iron Gate streams where there are no physical obstacles for upstream migration, e.g., in the Brnjica, KoŽica, Mala Boljetinska, and Zlatica Rivers. In time, they might produce hybrid offspring which could begin to alter their life history and produce both stream- and reservoir-dwelling individuals and to strongly impact the original brown trout of the native DA haplogroup. Makhov et al. (2018) reported the strong capability of the recently resident, stream-dwelling Black Sea trout in the Mzymta River population, which they assigned S. labrax, to retain migratory behavior, despite being cut off from the sea by damming. The lack of reproductive isolation increases the invasive risk from brown trout strains that have retained a capacity to alter their life history. In addition to the restoration measures outlined in Simonović et al. (2015) that are applicable for already affected native trout stocks, constructing physical barriers (insurmountable cascades) may be a reasonable precautionary measure to preserve the highly valuable stocks in the conservation sense from the upstream migrating, reservoir-dwelling brown trout of the AT haplogroup.
In conclusion, it seems that migratory behavior fortunately provides native trout stocks a mechanism to cope with the alien strains and/or species introduced into their home streams, but also enables non-native brown trout to intrude into the recipient streams and introgress into their resident trout stocks successfully. This feature supports the evaluation of brown trout of the AT haplogroup as of the high risk sensu stricto in the FISK evaluation performed by Simonović et al. (2015) for non-native trout fish species and strains in the rivers of Serbia as a recipient area. The migratory instinct and life history plasticity inherent to various nominal trout taxa facilitate them in overcoming the scarcity of resources in their native, home streams (Gross et al., 1988), thereby preserving the original genetic structure of locally adapted populations. Current knowledge about brown trout stocks in the Western Balkans suggests that these features act as a stabilizing population mechanism that can also facilitate them to deal with alien trout strains that intrude into streams in their native area of dispersal either naturally or through human intervention.
The datasets generated for this study can be found in the GenBank: KC630984.1, https://www.ncbi.nlm.nih.gov/nuccore/506485009.
Ethical review and approval was not required for the animal study because According to Serbian legal acts, such approval isn't needed. Each year, the sampling license for scientific purposes was issued by Ministry for Environment Protection that is in charge for Fisheries. All fish were returned alive to their home streams after taking the fin clip for DNA analyses.
The authors participated in fieldwork at the following localities: DŠ, AM, VN, MP, and PS at the Una River catchment. DŠ, DM, and PS at the Morača River catchment. AM, TK, VN, and PS at streams in the Iron Gate Gorge broader area. VN organized the database. DNA extraction and amplification of particular loci were accomplished by DŠ, AM, DM, TK, and PS. Statistical analyses of laboratory results were performed by DŠ, AM, TK, and PS, and their preparation for publication by VN. Manuscript design and its first draft were by PS, while DŠ and AM contributed suggestions during drafting. DM, IŠ, and MP reviewed the first draft. All authors read and approved the submitted version.
This study was supported by grant #173025 Evolution in Heterogeneous Environments: Adaptation Mechanisms, Monitoring, and Biodiversity Conservation, grants #451-03-02263/2018-09/19 Sustainable utilization of water courses in Montenegro and Serbia and conservation of genetic diversity of their fish fauna and #337-00-205/2019-09/04 Status of Diversity in Brown Trout in the Danube River Basin and Implications for Fisheries and Conservation, of the Ministry of Education, Science and Technological Development of Serbia in cooperation with the Ministry of Science of Montenegro and Ministry of Science and Education of Croatia, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Željko Mirković and Ãuro Mirković provided complete logistic support for the fieldwork in the Una River catchment. Vukoica Despotović assisted in the field in the Morača River catchment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00188/full#supplementary-material
Abolhasan, R., Sheyda, A., and Reza, J. H. (2017). Studies on the mitochondrial genomics in Salmo trutta caspius population in three rivers of South Caspian Sea. Res. J. Biotechnology 12, 49–61.
Behnke, R. J. (2007). About Trout: The Best of Robert Behnke from Trout Magazine. Guilford, CT: Globe Pequot.
Belkhir, K. P., Borsa, P., Chikhi, L., Raufaste, N., Bonhomme, F., and Belkhirr, K. (2004). GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions. Montpellier: Université de Montpellier II.
Bernatchez, L. (2001). The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55, 351–379. doi: 10.1111/j.0014-3820.2001.tb01300.x
Bernatchez, L., and Danzmann, R. G. (1993). Congruence in control-region sequence and restriction–site variation in mitochondrial DNA of brook charr (Salvelinus fontinalis Mitchill). Mol. Biol. Evol. 10, 1002–1014. doi: 10.1093/oxfordjournals.molbev.a040062
Bernatchez, L., Guyomard, R., and Bonhomme, F. (1992). DNA sequence variation of the mitochondrial control region among geographically and morphologically remote European brown trout Salmo trutta populations. Mol. Ecol. 1, 161–173. doi: 10.1111/j.1365-294X.1992.tb00172.x
Cairney, M., Taggart, J. B., and Høyheim, B. (2000). Characterization of microsatellite and minisatellite loci in Atlantic salmon (Salmo salar L.) and cross-species amplification in other salmonids. Mol. Ecol. 9, 2155–2234. doi: 10.1046/j.1365-294X.2000.105312.x
Charles, K., Guyomard, R., Hoyheim, B., Ombredane, D., and Baglinie‘re, J. L. (2005). Lack of genetic differentiation between anadromous and resident sympatric brown trout (Salmo trutta) in a Normandy population. Aquat. Living Resour. 18, 65–69. doi: 10.1051/alr:2005006
Chiereghini, A.b S. (1818). Descrizione de'crostacei, de'testacei e de'pesci che abitano le lagune e golfo Veneto. Op. ms. 128.
Copp, G. H., Garthwaite, R., and Gozlan, R. E. (2005). Risk Identification and Assessment of Non-Native Freshwater Fishes: Concepts and Perspectives on Protocols for the UK. Cefas Science Technical Report. Lowestoft, UK, 36. Available online at: http://www.cefas.co.uk/publications/techrep/tech129.pdf (accessed 13 May, 2013).
Cortey, M., and García-Marín, J. L. (2002). Evidence for phylogeographically informative sequence variation in the mitochondrial control region of Atlantic brown trout. J. Fish Biol. 60, 1058–1063. doi: 10.1111/j.1095-8649.2002.tb02429.x
Cortey, M., Pla, C., and García-Marín, J. L. (2004). Historical biogeography of Mediterranean trout. Mol. Phylogenet. Evol. 33, 831–844. doi: 10.1016/j.ympev.2004.08.012
Duftner, N., Weiss, S., Medgyesy, N., and Sturmbauer, C. (2003). Enhanced phylogeographic information about Austrian brown trout populations derived from complete mitochondrial control region sequences. J. Fish Biol. 62, 427–435. doi: 10.1046/j.1095-8649.2003.00038.x
Earl, D. A., and VonHoldt, B. M. (2012). Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359-361. doi: 10.1007/s12686-011-9548-7
Estoup, A., Rousset, F., Michalakis, Y., Cornuet, J.-M., Adriamanga, M., and Guyomard, R. (1998). Comparative analysis of microsatellite and allozyme markers: a case study investigating microgeographic differentiation in brown trout (Salmo trutta). Mol. Ecol. 7, 339-353. doi: 10.1046/j.1365-294x.1998.00362.x
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611-2620. doi: 10.1111/j.1365-294X.2005.02553.x
Fausch, K. D. (2007). Introduction, establishment and effects of non-native salmonids: considering the risk of rainbow trout invasion in the United Kingdom. J. Fish Biol. 71, 1–32. doi: 10.1111/j.1095-8649.2007.01682.x
Ferguson, A. (2006a). “Genetics of sea trout, with particular reference to Britain and Ireland,” in Sea Trout. Biology, Conservation and Management. eds H. Harris and Miller N (Oxford: Blackwell Publishing), 155–182.
Ferguson, A. (2006b). Genetic Impacts of Stocking on Indigenous Brown Trout Populations. Science Report. Bristol: UK Environmental Agency.
Ferguson, A., Reed, T. E., Cross, T. F., McGinnity, P., and Prodöhl, P. A. (2019). Anadromy, potamodromy and residency in brown trout Salmo trutta: the role of genes and the environment. J. Fish Biol. 95, 692–718. doi: 10.1111/jfb.14005
Fleming, I. A., and Gross, M. R. (1990). Latitudinal clines: a trade-off between egg number and size in Pacific salmon. Ecology 71, 1–11. doi: 10.2307/1940241
Goudet, J. (2002). FSTAT version 2.9. 3.2, a Program to Estimate and Test Gene Diversities and Fixation Indices. Lausanne: Institute of Ecology. Available online at: http://www2.unil.ch/popgen/softwares/fstat. htm (accessed August 20, 2012).
Gross, M. R., Coleman, R. M., and McDowall, R. M. (1988). Aquatic productivity and the evolution of diadromous fish migration. Science 239, 1291–1293. doi: 10.1126/science.239.4845.1291
Habibi, E., Kalbassi, M. R., Hosseini, S. J., and Qasemi, S. A. (2013). Feasibility of identification of fall and spring migrating Caspian trout (Salmo trutta caspius) by using AFLP molecular marker. Turk. J. Fish. Aquat. Sci. 13, 241–258. doi: 10.4194/1303-2712-v13_2_06
Hamilton, K. E., Fergusson, A., Taggart, J. B., Tómasson, T., Walker, A., and Fahy, E. (1989). Post-glacial colonization of brown trout, Salmo trutta L.: Ldh-5 as a phylogenetic marker locus. J. Fish Biol. 35, 651–664. doi: 10.1111/j.1095-8649.1989.tb03017.x
Hansen, M. M. (2002). Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Mol. Ecol. 11, 1003–1015. doi: 10.1046/j.1365-294X.2002.01495.x
Hansen, M. M., Ruzzante, D. E., Nielsen, E. E., and Mensberg, K.-L. D. (2000). Microsatellite and mitochondrial DNA polymorphism reveals life-history dependent interbreeding between hatchery trout and wild brown trout (Salmo trutta L.). Mol. Ecol. 9, 583–594. doi: 10.1046/j.1365-294x.2000.00898.x
Hansen, M. M., Ruzzante, D. E., Nielsen, E. E., and Mensberg, K.-L. D. (2001). Brown trout (Salmo trutta) stocking impact assessment using microsatellite DNA markers. Ecol. Appl. 11, 148–160. doi: 10.1890/1051-0761(2001)011[0148:BTSTSI]2.0.CO;2
Hindar, K., Jonsson, B., Ryman, N., and Stähl, G. (1991). Genetic relationships among landlocked, resident, and anadromous Brown Trout, Salmo trutta L. Heredity 66, 83–91. doi: 10.1038/hdy.1991.11
Jadan, M., CoŽ-Rakovac, R., Topić Popović, N., and Strunjak-Perović, I. (2007). Presence of unsexpected phylogenetic lineages of brown trout Salmo trutta L. in Gacka River, Croatia. Aquac. Res. 38, 1682-1685. doi: 10.1111/j.1365-2109.2007.01832x
Jones, P., and Closs, G. (2018). “The introduction of brown trout to New Zealand and their impact on native fish communities,” in Brown Trout: Biology, Ecology and Management, eds H. Lobon-Cervia and N. Sanz (London: John Wiley and Sons Ltd), 545–567. doi: 10.1002/9781119268352.ch21
Kalayci, G., Ozturk, R. C., Capkin, E., and Altinok, I. (2018). Genetic and molecular evidence that brown trout Salmo trutta belonging to the Danubian lineage are a single biological species. J. Fish Biol. 93, 792–804. doi: 10.1111/jfb.13777
King, T. L., Eackles, M. S., and Letcher, B. H. (2005). Microsatellite DNA markers for the study of Atlantic salmon (Salmo salar) kinship, population structure, and mixed-fishery analyses. Mol. Ecol. Notes 5, 130-132. doi: 10.1111/j.1471-8286.2005.00860.x
Klemetsen, A., Amundsen, P. A., Dempson, J. B., Jonsson, B., Jonsson, N., O'Connell, M. F., et al. (2003). Atlantic salmon Salmo salar L., brown trout Salmo trutta L. and Arctic charr Salvelinus alpinus (L.): a review of aspects of their life histories. Ecol. Freshw. Fish 12, 1–59. doi: 10.1034/j.1600-0633.2003.00010.x
Kohout, J., Jašková, I, Papoušek, I., Šedivá, A, and Šlechta, V. (2012). Effects of stocking on the genetic structure of brown trout, Salmo trutta, in Central Europe inferred from mitochondrial and nuclear DNA markers. Fish. Manag. Ecol. 19, 252–263. doi: 10.2478/s11756-013-0271-6
Kohout, J., Šedivá, A., Apostolou, A., Stefanov, T., Marić, S., Gaffarolu, M., et al. (2013). Genetic diversity and phylogenetic origin of brown trout Salmo trutta populations in Eastern Balkans. Biologia 68, 1229–1237. doi 10.2478/s11756-013-0271-6
Kolar, C. S., and Lodge, D. M. (2002). Ecological predictions and risk assessment for alien fishes in North America. Science 298, 1233–1236. doi: 10.1126/science.1075753
Kolombatović, J. (1890). Notizie ittiologiche. I. Seconda trota marina pescata nel mare di Vranjic, Trutta adriatica n. sp. II. Cattura di Lophotes cepedianus (Giorna), nelle acque di Trappano (Dalmazia). III. Sui Mullus dell'Adriatico. Glasnik Hrvatskoga Naravoslovnoga Društva 5, 165–174.
Kottelat, M. (1997). European freshwater fishes. An heuristic checklist of the freshwater fishes of Europe (exclusive of former USSR), with an introduction for non-systematists and comments on nomenclature and conservation. Biologia (Suppl.) 5, 1–271.
Kottelat, M., and Freyhof, J. (2007). Handbook of European Freshwater Fishes. Cornol: Kottelat; Berlin: Freyhof.
Laikre, L., Antunes, A., Apostolidis, A., Berrebi, P., Duguid, A., Ferguson, A., et al. (2010). Conservation genetic management of brown trout (Salmo trutta) in Europe. report by the concerted action on identification, management and exploitation of genetic resources in the brown trout (Salmo trutta). (TROUTCONCERT; EU FAIR CT97–3882).
Laikre, L., and Ryman, N. (1996). Effects on intraspecific biodiversity from harvesting and enhancing natural populations. Ambio 25, 504–509.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and CLustal X version 2.0. Bioinformatics 23, 2947-2948. doi: 10.1093/bioinformatics/btm404
LoBrutto, S., Hristovski, N., and Arculeo, M. (2010). Genetic difference between morphological forms of brown trout Salmo trutta L. in the Balkan region of Macedonia. J. Fish Biol. 76, 1220–1227. doi: 10.1111/j.1095-8649.2010.02595.x
Lowe, S., Browne, M., Boudjelas, S., and Poorter, M. D. (2000). “100 of the world's worst invasive alien species: a selection from the Global invasive species database,” in Invasive Species Specialist Group (ISSG), a specialist group of the Species Survival Commission (SSC) of the World Conservation Union (IUCN) (Auckland: World Conservation Union), 12.
MacCrimmon, H. R., and Marshall, T. L. (1968). Distribution of brown trout, Salmo trutta. J. Fish. Res. Board Can. 25, 2527–2548. doi: 10.1139/f68-225
Makhov, A. A., Artamonova, V. S., Murza, I. G., Pashkov, A. N., Ponomareva, V. N., Reshetnikov, S. I., et al. (2018). Ecological forms of Black Sea brown trout (Salmo trutta labrax) in the Mzymta River as manifestation of ontogenetic plasticity. Russ. J. Dev. Biol. 49, 117–127. doi: 10.1134/S1062360418020054
Marić, S., Nikolić, V., Tošić, A., and Simonović, P. (2012). Record of the brown trout Salmo trutta L., 1758 in the main riverbed of the Serbian part of the Danube River. J. Appl. Ichthyol. 28, 135–137. doi: 10.1111/j.1439-0426.2011.01881.x
Marić, S., Sušnik Bajec, S., Schöffmann, J., Kostov, V., and Snoj, A. (2017). Phylogeography of stream-dwelling trout in the Republic of Macedonia and a molecular genetic basis for revision of the taxonomy proposed by S. Karaman. Hydrobiologia 785, 249–260. doi: 10.1007/s10750-016-2930-4
Marić, S., Sušnik, S., Simonović, P., and Snoj, A. (2006). Phylogeographic study of brown trout from Serbia, based on mitochondrial DNA control region analysis. Genet. Select. Evol. 38, 411–430. doi: 10.1186/1297-9686-38-4-411
McIntosh, A. R., McHugh, P. A., Dunn, N. R., Goodman, J. R., Howard, S. W., Jellyman, P. G., et al. (2010). The impact of trout on galaxiid fishes in New Zealand. N Z J. Ecol. 34, 195–206.
McMeel, O., Hoey, E. M., and Marshall, T. L. (2001). Partial nucleotide sequences, and routine typing by polymerase chain reaction–restriction fragment length polymorphism, of the brown trout (Salmo trutta) lactatedehydrogenase, LDH-C1*90 and *100 alleles. Mol. Ecol. 10, 29–34. doi: 10.1046/j.1365-294X.2001.01166.x
Meraner, A., Gratton, P., Baraldi, F., and Gandolfi, A. (2013). Nothing but a trace left? Autochthony and conservation status of Northern Adriatic Salmo trutta inferred from PCR multiplexing, mtDNA control region sequencing and microsatellite analysis. Hydrobiologia 702, 201–213. doi: 10.1007/s10750-012-1321-8
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. doi: 10.1093/nar/16.3.1215
Mrdak, D. (2011). Trout (Salmo L., 1758) of rivers in Montengro-diversity, taxonomic status and phylogenetic relationships. (dissertation), University of Belgrade, Belgrade, Serbia.
Mrdak, D., Nikolić, V., Tošić, A., and Simonović, P. (2012). Molecular and ecological features of the soft-muzzled trout Salmo obtusirostris (Heckel, 1852) in the Zeta River, Montenegro. Biologia 67, 222–233. doi: 10.2478/s11756-011-0150-y
Mrdak, D., Simonović, P., Sušnik, S., and Snoj, A. (2006). “The existence of ≪strun≫ - Salmo dentex (Heckel, 1851) as distinct species from Salmo trutta fario (Linnaeus, 1758), in Adriatic rivers of Montenegro,” in II International Symposium of Ecologists of Montenegro – The Book of Abstracts and Programme. (Žabljak), 48.
O'Reilly, P. T., Hamilton, L. C., McConnell, S. K., and Wright, J. M. (1996). Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can. J. Fish. Aquat. Sci. 53, 2292–2298. doi: 10.1139/f96-192
Paterson, S., Piertney, SAMOVA. B., Knox, D., Gilbey, J., and Verspoor, E. (2004). Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol. Ecol. Notes 4, 160-162. doi: 10.1111/j.1471-8286.2004.00598.x
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945-959.
Razpet, A., Marić, S., Parapot, T., Nikolić, V., and Simonović, P. (2007). Re-evaluation of Salmo data by Gridelli (1936) – description of stocking, hybridization and repopulation in the River Sočabasin. Ital. J. Zool. 74, 63-70. doi: 10.1080/11250000601090081.
Rexroad, C. E., Coleman, R. L., Hershberger, W. K., and Killefer, J. (2002). Rapid communication: thirty-eight polymorphic microsatellite markers for mapping in rainbow trout. J. Anim. Sci. 80, 541-542. doi: 10.2527/2002.802541x
Rezaei, A., Akhshabi, S., and Jamalzadeh, H. R. (2019). Studies on the mitochondrial genomics in Salmo trutta caspius population in three rivers of Caspia Sea. J. Fish. Livestock Prod. 5:213. doi: 10.4172/2332-2608.1000213
Richard, M., and Thorpe, R. S. (2001). Can microsatellites be used to infer phylogenies? Evidence from population affinities of the Western Canary Island lizard (Gallotia galloti). Mol. Phyl. Evol. 20, 351–360. doi: 10.1006/mpev.2001.0981
Ryman, N., Jorde, P. E., and Laikre, L. (1995). Supportive breeding and variance effective population size. Conserv. Biol. 9, 1619-1628.
Sanz, N. (2018). “Phylogeographic structure of brown trout: a review,” in Brown Trout: Biology, Ecology and Management. eds H. Lobón-Cerviá, and N. Sanz (London: John Wiley and Sons Ltd), 17–63. doi: 10.1002/9781119268352.ch2
Serbezov, D., Jorde, P. E., Bernatchez, L., Olsen, E. M., and Vøllestad, L. A. (2012). Short-term genetic changes: evaluating effective population size estimates in a comprehensively described brown trout (Salmo trutta) population. Genetics 191, 579-592. doi: 10.1534/genetics.111.136580
Simonović, P. (2019). “Uticaj derivacionih malih hidroelektrana na zajednice riba i drugih akvatičnih organizama ekosistema planinskih reka Republike Srbije [Impact of derivative small hydropower plants on communities of fish and other aquatic organisms in the mountain stream ecosystems of the Republic of Serbia],” in Proceedings of the Symposium “Uticaj malih hidroelektrana na Životnu sredinu [Effects of Small Hydropower Plants on Environment],” Belgrade – Serbian Academy of Sciences and Arts in press (in Serbian with English Summary).
Simonović, P., Grubić, G., Tošić, A., Canak Atlagić, J., Škraba Jurlina, D., and Nikolić, V. (2018). Justification for retention of the Catch-and-Release in the wild brown trout Salmo cf. trutta fishery. Fish. Res. 200, 17-24. doi: 10.1016/j.fishres.2017.12.010
Simonović, P., Marić, S., and Nikolić, V. (2007). Trout Salmo spp. complex in Serbia and adjacent regions of western Balkans: reconstruction of evolutionary history from external morphology. J. Fish Biol. 70(Suppl.C): 359–380. doi: 10.1111/j.1095-8649.2007.01516.x
Simonović, P., Mrdak, D., Tošić, A., Škraba, D., Grujić, S., and Nikolić, V. (2014). Effects of stocking with brood fish to manage resident stream dwelling brown trout Salmo cf. trutta stock. J. Fish. Sci. 8, 139–152. doi: 10.3153/jfscom.201418
Simonović, P., Toši,ć, A., Vassilev, M., Apostolou, A., Mrdak, D., Ristovska, M., et al. (2013). Risk assessment of non-native fishes in the Balkans region using FISK, the invasiveness screening tool for non-native freshwater fishes. Med. Mar. Sci. 14, 369–376. doi: 10.12681/mms.337
Simonović, P., Tošić, A., Škraba Jurlina, D., Canak Atlagić, J., and Nikolić, V. (2017a). “Stakeholders participation in conservation of brown trout stocks in Serbia,” in Proceedings of the Wild Trout XII Symposium Science, Politics, and Wild Trout Management: Who's Driving and Where are we Going? (MT: West Yellowstone), 55–62.
Simonović, P., Tošić, A., Škraba Jurlina, D., Nikolić, V., Piria, M., Tomljanović, T., et al. (2017b). Diversity of brown trout Salmo cf. trutta (L.) in the Danube River basin of Western Balkans as assessed from the structure of their mitochondrial control region haplotypes. J. Ichthyol. 57, 603–616. doi: 10.1134/S0032945217040154
Simonović, P., Vidović, Z., Tošić, A., Škraba, D., Canak Atlagić, J., and Nikolić, V. (2015). Risks to stocks of native trout of the genus Salmo (Actinopterygii: Salmoniformes: Salmonidae) of Serbia and management for their recovery. Acta Ichthyol. Pisc. 45, 161–173. doi: 10.3750/AIP2015.45.2.06
Simpson, G. G. (1961). Principles of Animal Taxonomy. New York, NY: Columbia University Press. doi: 10.7312/simp92414
Škraba Jurlina, D., Marić, A., Karanović, J., Nikolić, V., Brkušanin, M., Kanjuh, T., et al. (2018). Effect of the introgression of Atlantic brown trout, Salmo trutta, into Adriatic trout, Salmo farioides in a stream at the drainage area of the Adriatic Sea basin of Montenegro. Acta Ichthyol. Pisc. 48, 363–372. doi: 10.3750/AIEP/02491
Škraba, D., Bećiraj, A., Šarić, I., Ićanović, I., DŽaferović, A., Piria, M., et al. (2017). Genotypization of brown trout (Salmo trutta L.) populations from River Una drainage area in Bosnia and Herzegovina and implications for conservation and fishery management. Acta zool. bulg. 69, 25–30.
Slettan, A., Olsaker, I., and Lie, Ø. (1995). Atlantic salmon, Salmo salar, microsatellites at the SSOSL25, SSOSL85, SSOSL311, SSOSL417 loci. Anim. Genet. 26, 281–282. doi: 10.1111/j.1365-2052.1995.tb03262.x
Snoj, A., Glamuzina, B., Razpet, A., Zablocki, J., Bogut, I., Lerceteau-Köhler, E., et al. (2010). Resolving taxonomic uncertainties using molecular systematics: Salmo dentex and the Balkan trout community. Hydrobiologia 651, 199–212. doi: 10.1007/s10750-010-0297-5
Snoj, A., Jug, T., Melkič, E, Sušnik, S., Pohar, J., Dovč, P., et al. (2000). Mitochondrial and microsatellite DNA analysis of marble trout in Slovenia. J. Fish Biol. (Quaderni ETP) 29, 5-11.
Snoj, A., Marčeta, B., Sušnik, S., Melkič, E., and Dovč, P. (2002). The taxonomic status of the ‘sea trout' from the north Adriatic Sea, as revealed by mitochondrial and nuclear DNA analysis. J. Biogeogr. 29, 1179–1185. doi: 10.1046/j.1365-2699.2002.00735.x
Templeton, A. R. (1986). “Coadaptation and outbreeding depression,” in Conservation Biology. The Science of Scarcity and Diversity. ed M. E. Soulé (Sunderland, MA: Sinauer Associates), 105–116.
Tošić, A., Škraba, D., Nikolić, V., Canak Atlagić, J., Mrdak, D., and Simonović, P. (2016). Haplotype diversity of brown trout in the broader Iron Gate area. Turk. J. Zool. 40, 655–662. doi: 10.3906/zoo-1510-54
Tošić, A., Škraba, D., Nikolić, V., Mrdak, D., and Simonović, P. (2014). New mitochondrial DNA haplotype of brown trout Salmo trutta L. from Crni Timok drainage area in Serbia. Turk. J. Fish. Aquat. Sci.14, 37–42. doi: 10.4194/1303-2712-v14_1_05
Tougard, C., Justy, F., Guinard, B., Douzery, J. P., and Berreby, P. (2018). Salmo macrostigma (Teleostei, Salmonidae): nothing more than a brown trout (S. trutta) lineage? J. Fish Biol. 93, 302–310. doi: 10.1111/jfb.13751/
Townsend, C. R. (1996). Invasion biology and ecological impact of brown trout Salmo trutta in New Zealand. Biol.Cons. 78, 13–22. doi: 10.1016/0006-3207(96)00014-6
Vera, M., Sourinejad, I., Bouza, C., Vilas, R., Pino-Querido, A., Kalbassi, M. R., et al. (2011). Phylogeography, genetic structure, and conservation of the endangered Caspian brown trout, Salmo trutta caspius (Kessler, 1877), from Iran. Hydrobiologia 664, 51–67. doi: 10.1007/s10750-010-0581-4
Weiss, S., Schlotterer, C., Waidbacher, H., and Jungwirth, M. (2001). Haplotype (mtDNA) diversity of brown trout Salmo trutta in tributaries of the Austrian Danube: massive introgression of Atlantic basin fish–by man or nature? Mol. Ecol. 10, 1241–1246. doi: 10.1046/j.1365-294X.2001.01261.x
Welcome, R. L. (1992). A history of international introductions of inland aquatic species. ICES Marine Sci. Symp. 194, 3–14.
Whiteley, A. R., Penaluna, B. E., Taylor, E. B., Weiss, S., Abadia-Cardoso, A., Gomez-Uchida, D., et al. (2019). “Trout and char: taxonomy, systematics and biogeography,” in Trout and Charr of the World. eds J. L. Kershner, N. E. Williams, R. E.Gresswell, and H. Lobón-Cerviá (Bethesda, MD: American Fisheries Society), 95–140.
Keywords: migratory trout, resident trout, non-indigenous strains, invasiveness, threats, conservation
Citation: Škraba Jurlina D, Marić A, Mrdak D, Kanjuh T, Špelić I, Nikolić V, Piria M and Simonović P (2020) Alternative Life-History in Native Trout (Salmo spp.) Suppresses the Invasive Effect of Alien Trout Strains Introduced Into Streams in the Western Part of the Balkans. Front. Ecol. Evol. 8:188. doi: 10.3389/fevo.2020.00188
Received: 09 October 2019; Accepted: 26 May 2020;
Published: 14 July 2020.
Edited by:
Eleni Kalogianni, Hellenic Centre for Marine Research (HCMR), GreeceReviewed by:
L. Asbjørn Vøllestad, University of Oslo, NorwayCopyright © 2020 Škraba Jurlina, Marić, Mrdak, Kanjuh, Špelić, Nikolić, Piria and Simonović. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Predrag Simonović, cGVkamFAYmlvLmJnLmFjLnJz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.