- 1Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden
- 2Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX, United States
- 3Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences, Uppsala, Sweden
The rich phenotypic diversity in coat and plumage color in domestic animals is primarily caused by direct selection on pigmentation phenotypes. Characteristic features are selection for viable alleles with no or only minor negative pleiotropic effects on other traits, and that alleles often evolve by accumulating several consecutive mutations in the same gene. This review provides examples of mutations that disrupt or create pigmentation patterns. White spotting patterns in domestic animals are often caused by mutations in KIT, microphthalmia transcription factor (MITF), or endothelin receptor B (EDNRB), impairing migration or survival of melanoblasts. Wild boar piglets are camouflage-colored and show a characteristic pattern of dark and light longitudinal stripes. This pattern is disrupted by mutations in Melanocortin 1 receptor (MC1R), implying that a functional MC1R receptor is required for wild-type camouflage color in pigs. The great majority of pig breeds carry MC1R mutations disrupting wild-type color and different mutations causing dominant black color were independently selected in European and Asian domestic pigs. The European allele evolved into a new allele creating a pigmentation pattern, black spotting, after acquiring a second mutation. This second mutation, an insertion of two C nucleotides in a stretch of 6 Cs, is somatically unstable and creates black spots after the open reading frame has been restored by somatic mutations. In the horse, mutations located in an enhancer downstream of TBX3 disrupt the Dun pigmentation pattern present in wild equids, a camouflage color where pigmentation on the flanks is diluted. A fascinating example of the creation of a pigmentation pattern is Sex-linked barring in chicken which is caused by the combined effect of both regulatory and coding mutations affecting the function of CDKN2A, a tumor suppressor gene associated with familial forms of melanoma in human. These examples illustrate how evolution of pigmentation patterns in domestic animals constitutes a model for evolutionary change in natural populations.
Introduction
Pigmentation phenotypes have been under strong selection in domestic animals throughout their evolutionary history, and references to variation in pigmentation are indicated already in ancient literature and illustrations. For instance, the Greek historian Herodotus described that the Persian emperor Xerxes (in reign 485 to 465 BC) kept sacred white horses, most likely white horses caused by the Graying with age mutation (Rosengren Pielberg et al., 2008). Coat color variation is also described in old Roman literature (Forster and Heffner, 1968). Pigmentation must have been one of the first traits that were altered after domestication was initiated and extensive color diversity is a hallmark for domestic animals. The molecular characterization of mutations underlying these changes has given insight about mechanisms underlying pigmentation patterns. The evolution of pigmentation patterns in domestic animals constitutes a model for evolutionary change in natural populations. This review provides examples of mutations that disrupt pigmentation patterns and others that create pigmentation patterns. In addition to the patterns described here, it is worth noticing the Himalayan pattern that occurs in several species like cat, rabbit, mouse, and gerbil (Lyons et al., 2005), and is caused by temperature-sensitive mutations of tyrosinase producing white color but with dark pigmentation in cooler areas of the body, like the tips of the ears.
White Spotting Patterns in Domestic Animals
White spotting patterns occur frequently in domestic animals. The most common causes are mutations in KIT encoding the KIT tyrosine kinase receptor, microphthalmia transcription factor (MITF), or endothelin receptor B (EDNRB), all with a crucial role for melanoblast migration and survival. Thus, a common reason for white spotting is lack of pigment cells in skin and/or in the hair/feather follicles.
The majority of KIT mutations causing pigment patterns in domestic animals are structural rearrangements. There are two reasons why these are common in domestic animals. One is that structural rearrangements that do not touch the coding sequence may give a spectacular pigmentation pattern without causing negative pleiotropic effects, because KIT function is also essential for development of hematopoietic cells and for germ cells. The second reason is because some regulatory elements affecting KIT expression are located hundreds of kb both upstream and downstream of the coding sequence, disruption of these often gives spectacular spotting patterns. This is well illustrated by the domestic pig where a 450 kb duplication encompassing the entire coding sequence and more than 100 kb upstream and downstream of the coding sequence is causing the Patch phenotype characterized by large areas of the coat lacking pigmentation (Johansson Moller et al., 1996; Giuffra et al., 2002). Further, the Belt phenotype, characterized by a white belt across the foreleg, is associated with several duplications in non-coding regions of KIT (Rubin et al., 2012). The top dominant KIT allele, present in billions of pigs used for meat production world-wide, is Dominant white causing complete or near complete absence of skin and hair pigmentation. The Dominant white allele carries multiple causal mutations, including the different duplications associated with the Patch and Belt phenotypes, and in addition a splice mutation in one of the copies that leads to skipping of exon 17 encoding the tyrosine kinase domain. Thus, this results in a dominant negative receptor with normal ligand binding but inactivated tyrosine kinase signaling (Marklund et al., 1998; Rubin et al., 2012). The Dominant white allele is affecting pigmentation based on the combined effect of regulatory mutations (the duplications) and a coding change (splice mutation) in one of the copies. Due to this combination, it is the most dominant KIT allele as regards its effect on pigmentation in any mammal and with no or only very mild pleiotropic effects on hematopoiesis and fertility. Other examples of KIT structural rearrangements causing striking pigmentation patterns in domestic animals are Tobiano white spotting in horses caused by a 40 Mb inversion where one of the inversion breakpoints is located about 100 kb downstream of KIT (Brooks et al., 2008), and color sidedness in cattle caused by two serial translocations affecting KIT expression (Durkin et al., 2012).
In contrast to pigs where there is an allelic series at the KIT locus, white spotting in dogs is largely determined by an allelic series at the MITF locus (Karlsson et al., 2007). This Spotting (S) locus was first described by Little (1957) and is composed of four alleles Solid (S, wild-type), Irish spotting (Si), Piebald (Sp), and Extreme white (Sw). Irish spotting occurs in breeds like Bernese mountain dogs, Collie and Basenji, and is characterized by limited white spotting on the chest and often with a white ring around the neck. The Piebald phenotype occurs in for instance Beagles and Fox terriers and is characterized by more extensive white spotting across the body. Finally, Extreme white occurs in Dalmatians, white Boxers, and white Bull terriers and presents as a near total absence of pigmentation but remaining spots of pigmentation show normal pigmentation implying no defect in pigment production per se. In contrast to the situation in mice where the majority of described alleles affect the coding sequence and is associated with severe negative pleiotropic effects in other tissues where MITF function is essential, none of the MITF alleles in dogs affect the coding sequence and they have no or only mild negative effects (Karlsson et al., 2007); a fraction of the Extreme white dogs show deafness. Furthermore, an interesting aspect of the dog MITF alleles is that the three mutant alleles do not represent three independent mutations but show haplotype sharing strongly suggesting that the three alleles have evolved by consecutive accumulation of several causal mutations in the non-coding part of MITF. Functional characterization indicated that a simple repeat polymorphism in the MITF promoter is likely one of the causal variants affecting white spotting patterns (Baranowska Körberg et al., 2014). A non-coding variant in the 5’ region of MITF is also associated with a white spotting pattern in cattle (Hofstetter et al., 2019). In horses, mutations in both MITF and PAX3 are associated with the Splashed white pigmentation pattern (Hauswirth et al., 2012).
A missense mutation in EDNRB Ile118Lys is causing the Overo white spotting pattern in horses and in the homozygous condition the Overo lethal white syndrome, where lethality is caused by intestinal aganglionosis (Metallinos et al., 1998; Santschi et al., 1998; Yang et al., 1998). This horse syndrome corresponds to the form of Hirschsprung disease in humans caused by mutations in the same gene. A missense mutation in EDNRB2 is also associated with a feather pigmentation pattern in chicken (see below).
MC1R Mutations in Pigs Both Disrupt and Create Pigmentation Patterning
Melanocortin 1 receptor (MC1R) is one of the major coat color loci in the domestic pig. The wild boar piglets show a striking camouflage color composed of longitudinal dark- and light-colored stripes (Figure 1). In the great majority of pig breeds of the world, this camouflage color is disrupted by MC1R mutations, either dominant black or recessive red mutations (Kijas et al., 1998, 2001). Fang et al. (2009) tested pigs from 68 different breeds from Europe and China and found that pigs from only one, the Hungarian Mangalica, were homozygous for the wild-type allele. Domestication of pigs occurred in parallel in Europe and Asia from two different subspecies of the wild boar, the European wild boar and the Asian wild boar that separated from each other about one million years ago (Giuffra et al., 2000; Kijas and Andersson, 2001; Groenen et al., 2012). Two different missense mutations in MC1R causing dominant black color were selected in European and Asian domestic pigs, D124N and L102P, respectively. A comprehensive screen of the MC1R coding sequences in wild boars and domestic pigs from both Europe and Asia led to the conclusion that there is purifying selection to maintain camouflage in wild boars and selection to disrupt camouflage in domestic pigs (Fang et al., 2009). A conclusion based on the observation that seven out of seven nucleotide substitutions among European and Asian wild boars were all synonymous, whereas nine out of 10 nucleotide substitutions among domestic pigs were non-synonymous changes.

Figure 1. Wild boar sow with piglets. The piglets show the characteristic camouflage pattern which requires the presence of a wild-type MC1R allele. Photo: Annelie Andersson.
The disruption of camouflage pattern in pigs carrying dominant black or recessive red alleles at MC1R suggests strongly that a wild-type MC1R receptor, whose signaling activity is controlled by the relative abundance of melanocyte-stimulating hormone (MSH) and agouti (ASIP), is required for the development of this pattern. The most likely explanation is that differential expression of agouti is causing patterning as recently reported for periodic feather patterning in juvenile galliform birds (Haupaix et al., 2018). Furthermore, differential expression of the transcription factor ALX3 is associated with the development of periodic dorsal stripes in the African striped mouse, which resemble the camouflage pattern in piglets (Mallarino et al., 2016).
One of the MC1R alleles in pigs is also creating a stochastic pigment pattern. That is the black-spotting EP allele that evolved from the European dominant black allele (ED1), carrying the D124N missense mutation, by the insertion of two C nucleotides at codon 22 creating a mononucleotide repeat of 8 C (Kijas et al., 2001). A frameshift mutation at codon 22 is expected to result in a complete loss-of-function and lack of black eumelanin in the coat. But that is not the case, the most common phenotype is red with a more or less random distribution of black spots across the body or white coat with larger black spots; whether the black spots occur on a white or red background is determined by one or more other genetic factors that have not yet been identified. The black-spotting phenotype associated with this allele ranges from almost no spots at all, in particular on the red background as in Tamworth pigs, to an entire black coat with six white points (tail, nose, and four white feet) in Berkshire. So, how is this possible? The explanation is that the 8 C mononucleotide repeat is somatically unstable and may lose two nucleotides or gain one nucleotide and thereby restore the open reading frame. When that happens, constitutive MC1R signaling is reactivated due to the presence of the D124N missense mutation. This somatic instability of the mononucleotide repeat was confirmed by RT-PCR analysis (Kijas et al., 2001). The phenotypic range associated with the black-spotting allele is most likely explained by sequence variants affecting the probability for somatic reversion to occur as well as loci affecting the proliferation of melanocytes after reversion has occurred.
A similar stochastic pattern of pigmented spots occurs in white horses carrying the dominant graying with age mutation. These horses are born normally colored but start to gray already during the first year of life and they are usually completely white before they are 10 years of age. Graying with age is caused by a 4.6 kb tandem duplication in an intron of syntaxin 17 (Rosengren Pielberg et al., 2008). Many horses that are heterozygous for this mutation show large number of small pigmented spots and are called flea-bitten gray. It appears plausible that this phenotype is caused by somatic loss of one of the duplicated copies or that it is inactivated by an epigenetic mechanism. A third example of a stochastic generation of a pigmentation pattern in domestic animals is the merle patterning in dogs in which pigmentation is diluted by a retrotransposon insertion in PMEL (previously denoted SILV), somatic deletion of the retrotransposon restores normal pigmentation (Clark et al., 2006).
Regulatory Mutations in TBX3 Disrupt Camouflage Color in Horses
The majority of domestic horses have a non-dun phenotype characterized by intense pigmentation and caused by homozygosity for a recessive allele at the Dun locus. The dominant Dun phenotype occurs in some horses like Icelandic horses and the Norwegian Fjord horse, but this is in fact a wild-type color present also in the Przewalski’s horse, a close relative to the ancestor of domestic horses. Dun is causing a pattern of dilution on the flanks but leaves a dark dorsal stripe and may be associated with other dark patterns which may include facial mask, shoulder cross, and zebra-like stripes on the legs (Imsland et al., 2016). There are many mutations described in the pigmentation literature causing pigment dilution caused by various defects in the pigment machinery. It is worth noticing that Dun in horses is in fact a wild-type phenotype contributing to camouflage by reducing the intensity of pigmentation.
Histological studies revealed that the difference between hairs from Dun and non-dun horses is that the latter have a symmetric deposition of pigment whereas hair from Dun horses have an asymmetric deposition of pigments on the outward-facing side of the hair (Imsland et al., 2016). Thus, this is a pigmentation pattern affecting the individual hair. High-resolution genetic mapping combined with whole genome sequencing data from Dun and non-dun horses revealed that the non-dun phenotype is caused by cis-acting regulatory mutations affecting tissue-specific expression of the TBX3 transcription factor gene (Imsland et al., 2016). TBX3 had never before been associated with pigmentation, but it is important during development and loss-of-function mutations cause the ulnar-mammary syndrome in humans that involves defects in limb, apocrine gland, tooth, and genital development (Bamshad et al., 1997). Imsland et al. (2016) first showed that the majority of non-dun horses, including the reference horse used for the horse genome assembly, were homozygous for an 1609 bp deletion located about 5 kb downstream of TBX3, in a region showing high sequence conservation among mammals. The fact that not all non-dun horses were homozygous required further analysis which revealed the presence of two different alleles non-dun1 (lacking the deletion) and non-dun2 (with the deletion). The causal mutation for non-dun1 is a single nucleotide substitution within the region deleted in non-dun2! Furthermore, genotyping more than 1000 horses for these two causal variants explained a phenotypic heterogeneity among non-dun horses where horses homozygous for non-dun2 have the most intense pigmentation and non-dun1 horses show an intermediate phenotype often with a weak dorsal stripe (Imsland et al., 2016). Interestingly, non-dun1 is in fact also a wild-type allele since it was found in two ancient horses (4400 and 42,700 years old). This implies that there existed two different color morphs of the ancestor of domestic horses, Dun and non-dun1, possibly adapted to different environmental conditions.
Imsland et al. (2016) also established a plausible molecular mechanism underlying camouflage color in Dun horses, which is disrupted in non-dun horses. In Dun horses, TBX3 has an asymmetric expression in the hair follicle that matches the asymmetric deposition of pigment. KIT ligand (KITL) shows downregulation in the area where TBX3 is expressed, which in turn means that pigment cells are not attracted to this part of the hair follicle explaining the lack of pigment deposition. In contrast, TBX3 is not expressed in the hair follicle in non-dun horses explaining the symmetric deposition of pigment. Thus, the results suggest that the deleted region contains an enhancer required for TBX3 expression in the hair follicle.
The work on the Dun horse coat color revealed a previously unknown function for TBX3 and a previously unknown mechanism for generation of camouflage color in mammals. This mechanism is present at least in all equids including zebras. For instance, the Somali wild ass, the wild ancestor of the donkey, shows a very clear Dun phenotype with diluted pigmentation on the flanks, a dorsal black stripe and zebra-like leg stripes. It is possible that this mechanism for camouflage pattern is also active in other mammals including different species of deer and antelopes.
Mutations in the CDKN2A Tumor Suppressor Gene Create Pigmentation Pattern in Chicken
Sex-linked barring is an iconic plumage phenotype present in breeds like Barred Plymouth Rock and Coucou de Rennes (Figure 2A). This phenotype shows sex-linked dominant inheritance and is characterized by feathers with periodic black and white bars. In the initial identification of the causal gene for this phenotype, Hellström et al. (2010) mapped this locus to a 12 kb region containing only the CDKN2A tumor suppressor gene encoding two transcripts INK4b and ARF. CDKN2A had never before been associated with a pigmentation phenotype but has a well-established link to pigment cell biology because heterozygosity for loss-of-function mutations in this gene is a major risk factor for familiar forms of malignant melanoma in humans (Hussussian et al., 1994). Sequence analysis of the 12 kb region across many breeds of chicken revealed three CDKN2A alleles: wild-type (N), Sex-linked barring (B1), and Sex-linked dilution (B2); the latter is a variant of Sex-linked barring but causing a more diluted pigmentation and not as sharp contrast between the black and white bars as in Sex-linked barring. Four sequence variants, all in or near the ARF transcript, were unique to the B1 and B2 alleles and not found in any of the sequenced wild-type chromosomes (Figure 2B). B1 carried a V9D missense mutation while B2 was associated with an R10C missense mutation, and both carried two SNPs in non-coding sequences. The two missense mutations were very strong candidates for being causal because they were both non-conservative and affected the MDM2-binding domain of the ARF protein. However, it was a mystery why both mutations occurred on the same very rare haplotype characterized by two non-coding changes not found among wild-type chromosomes.
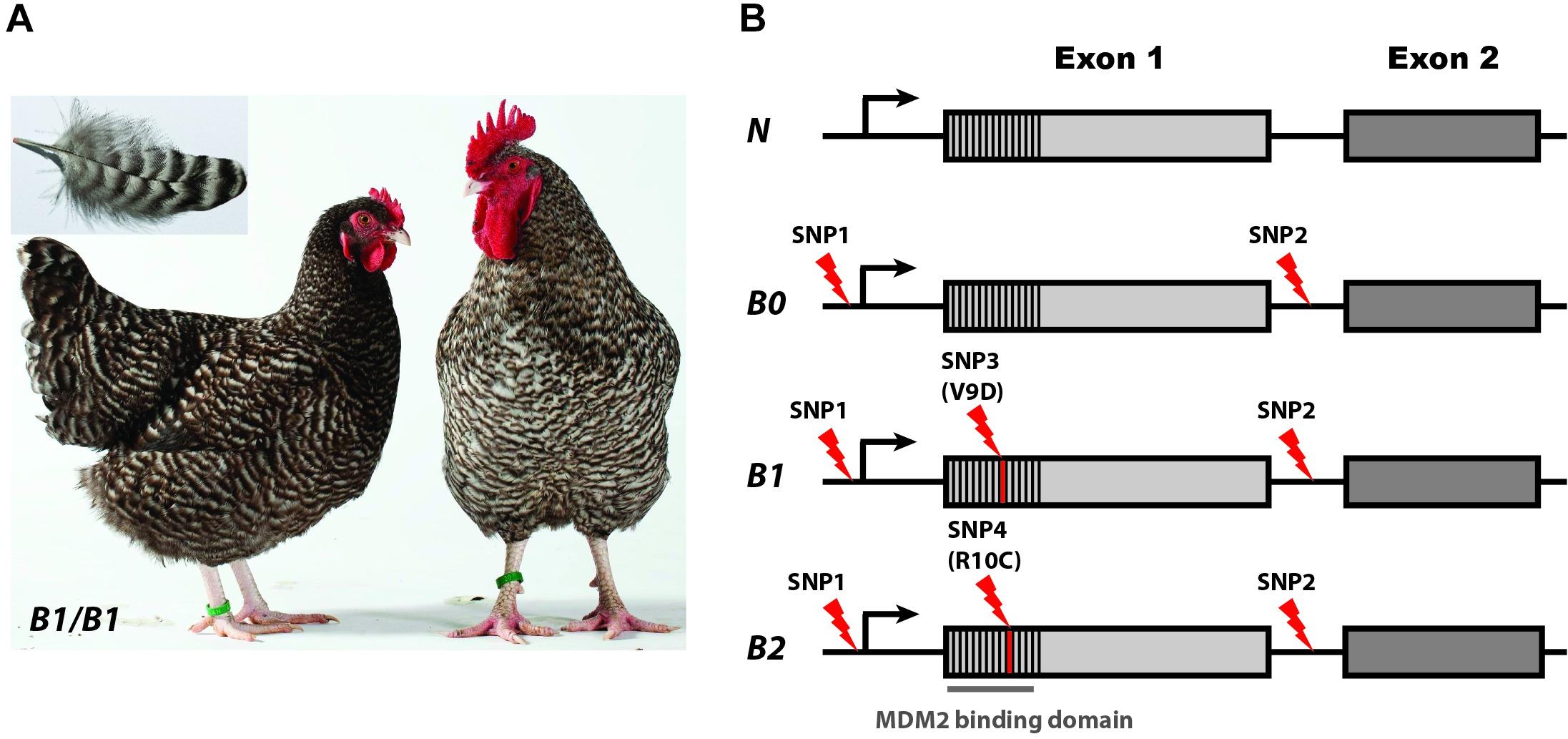
Figure 2. Phenotypes and alleles at the Sex-linked barring (CDKN2A) locus in chicken. (A) Female and male Coucou de Rennes chicken illustrating the Sex-linked barring phenotype caused by the B1 allele. (B) Sex-linked barring alleles and associated sequence variants. SNP1 and SNP2 are non-coding while SNP3 and SNP4 constitute non-synonymous changes in the region encoding the MDM2 binding domain. Photo credit: (A) Hervé Ronné, Ecomusée du pays de Rennes. From Schwochow-Thalmann et al. (2017).
A second study (Schwochow-Thalmann et al., 2017) characterized a fourth allele, B0, that carried only the two non-coding changes (Figure 2B). The B0 allele was associated with the most extreme suppression of pigmentation with a weak barring pattern and was therefore named Sex-linked extreme dilution. Functional analysis revealed allelic imbalance associated with the B0, B1, and B2 alleles, with higher expression of the mutant allele compared with the wild-type allele in feather follicles but not in skin or in liver. The results imply that one of the two non-coding changes or the combined effect of the two constitutes a cis-acting regulatory mutation. Furthermore, functional characterization using far-UV circular dichroism and isothermal titration calorimetry (ITC) of the different protein variants (N, B1, and B2) showed that the missense mutations impair the interaction between ARF and MDM2 (Schwochow-Thalmann et al., 2017) and thus counteract the effect of the upregulated expression of ARF caused by the regulatory mutation. The results are consistent with an evolutionary scenario where the regulatory mutation occurs first resulting in the B0 allele followed by two independent missense mutations causing the B1 and B2 alleles.
Schwochow-Thalmann et al. (2017) proposed a plausible mechanism of action of the mutant alleles (Figure 3), partially based on the finding by Lin et al. (2013) that Sex-linked barring is associated with premature differentiation of pigment cells. First, upregulation of ARF expression blocks MDM2 and leads to upregulation of the p53 tumor suppressor and shorter life-span of pigment cells. Thus, this is the opposite effect compared with the consequence of CDKN2A loss-of-function mutations predisposing for human melanoma. Second, the missense mutations in the B1 and B2 alleles impair the interaction between ARF and MDM2 (Figure 3B). The Sex-linked barring phenotype is most likely caused by a repeated process where melanocyte progenitor cells are recruited, differentiate and produce pigment resulting in a black bar, followed by exhaustion of pigment cells due to premature differentiation resulting in a white bar, and then the process start again with the recruitment of new progenitor cells (Figure 3C).
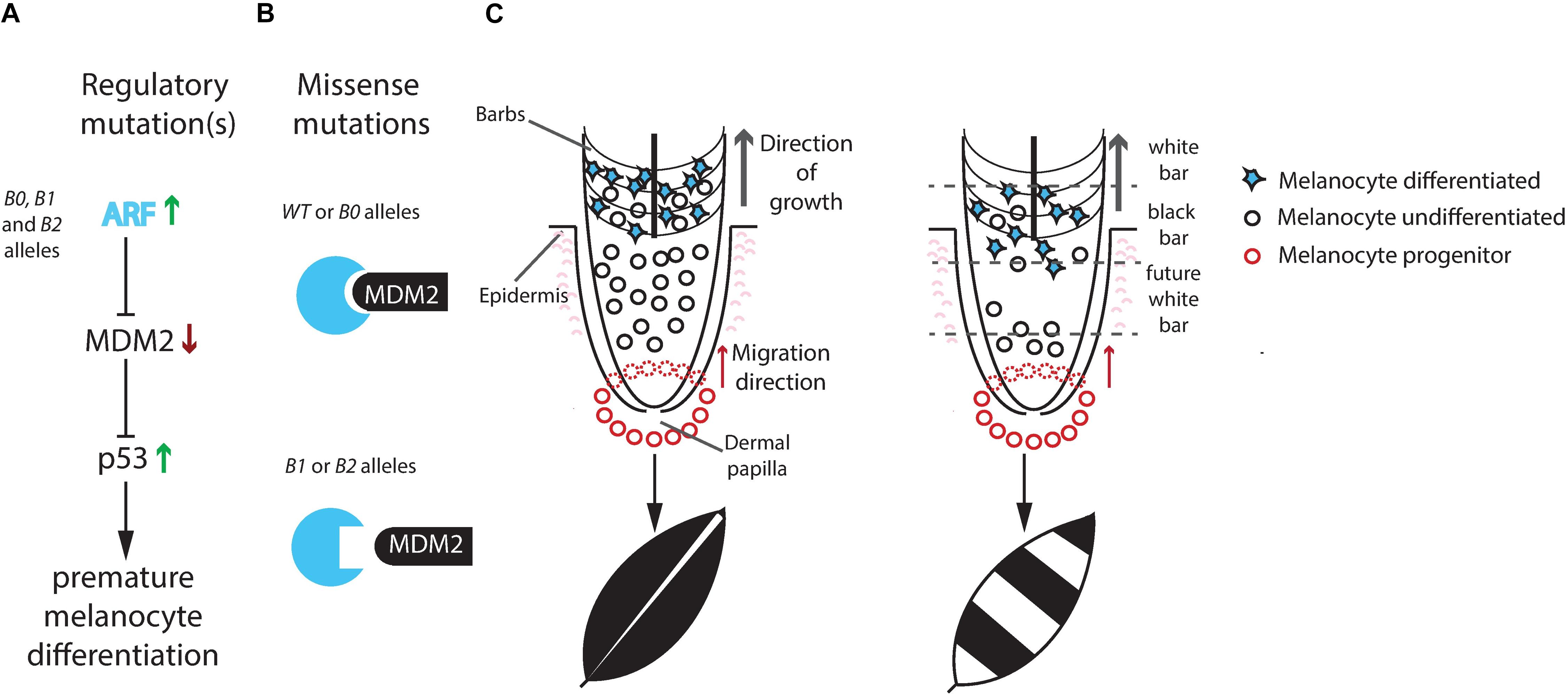
Figure 3. Proposed mechanism for development of the Sex-linked barring phenotype. (A) The non-coding mutation(s) present in the B0, B1, and B2 alleles cause a tissue-specific upregulation of CDKN2A encoding the ARF protein. ARF inhibits MDM2-mediated degradation of p53. p53 will activate downstream targets possibly initiating premature melanocyte differentiation resulting in a loss of mature pigment cells. (B) The amino acid substitutions associated with the B1 and B2 alleles impair the ARF/MDM2 interaction, which counteracts the consequences of upregulated ARF expression. (C) In wild-type feathers, melanocyte progenitor cells migrate up from the feather base and start expressing ARF in the barb region leading to differentiation of melanocytes and pigment production without exhausting the pool of undifferentiated melanocytes. In Sex-linked barred feathers, upregulated ARF expression may lead to premature differentiation of pigment cells and a lack of undifferentiated melanocytes that can replenish the ones producing pigment. As the feather keeps on growing, no more melanocytes are available to produce pigment resulting in the white bar. A plausible explanation for the periodic appearance of white and black bars is that new recruitment of melanocyte progenitor cells takes place after the undifferentiated melanocytes have been depleted. From Schwochow-Thalmann et al. (2017).
In addition to Sex-linked barring, there is an extensive diversity in intra-feather pigmentation patterns in the domestic chicken. Smyth (1990) described the following main types: stippling (wild-type), penciling, autosomal barring, single lacing, double lacing, spangling, and mottling. The difference between stippling and the other variants is caused by mutations at distinct loci or by the combined effect of multiple loci, some of these genes have been identified at the molecular level. Autosomal barring resembles Sex-linked barring but the difference is that the white bars in Sex-linked barring lack pigment whereas the light-colored bars in Autosomal barring usually have pigment deposition. According to Smyth (1990), autosomal barring is caused by the combined effect of mutations at the Extension, Dark brown, Patterning, and Columbian loci. Extension corresponds to MC1R in which an allelic series has been identified at the molecular level (Takeuchi et al., 1996; Kerje et al., 2003; Dávila et al., 2014). Dark brown is caused by an 8.3 kb deletion upstream of SOX10 (Gunnarsson et al., 2011). The SOX10 transcription factor plays an important role during melanocyte development, and SOX10 mutations is causing some forms of Waardenburg syndrome, involving pigmentary disturbances of hair, skin, and eyes (Pingault et al., 1998). The Patterning and Columbian loci have not yet been identified at the molecular level. Mottling (white tip of the feather) has been associated with a missense mutation in EDNRB2 encoding endothelin receptor B2 (Kinoshita et al., 2014).
Discussion
An extensive phenotypic diversity of pigmentation patterns is a characteristic feature in domestic animals. It has been speculated that this, in particular white spotting patterns, is a by-product of selection for tameness (Wilkins et al., 2014). The argument is based on proposed pleiotropic effects of mutations in genes expressed in neural-crest derived cells affecting both brain function and pigmentation. However, to the best of my knowledge, there are no convincing empirical data supporting this hypothesis as a major explanation for coat color diversity in domestic animals. For instance, the white spotting locus in dogs (S/MITF) contradicts this hypothesis, because mutations affecting melanocyte migration with no or minor pleiotropic effects have been selected. Furthermore, some of the friendliest dogs, like Labradors and Golden Retrievers, tend to be homozygous for the Solid (wild-type) allele at this locus. In fact, a recent study confirms the degree of white pigmentation do not covary with differences in behavior across dog breeds (Wheat et al., 2019). The most important reason for the extensive variation in pigmentation pattern in domestic animals is positive selection for variation (Fang et al., 2009). Selection against camouflage like the striping patterns in piglets may have facilitated animal husbandry when domestic piglets were roaming freely around early settlements. Furthermore, Columella, the Roman authority on agriculture, wrote almost 2000 years ago that shepherds prefer dogs with white color because it helps them to distinguish dogs from wolves at low light conditions (Forster and Heffner, 1968). There has also been a selection for pure beauty, exampled by the phenotype of the white horses (Rosengren Pielberg et al., 2008) and Sex-linked barring in the domestic chicken (Schwochow-Thalmann et al., 2017). Finally, relaxed purifying selection, related to the importance of pigmentation for camouflage and mate choice in natural populations, has most certainly contributed to the observed diversity in domestic animals.
A characteristic feature of alleles affecting pigmentation in domestic animals is that alleles with strong effects on pigmentation but no or only mild negative pleiotropic effects have been preferred. This is well illustrated by the MITF white spotting alleles in dogs, and by KIT alleles in pigs where a drastic effect on pigmentation without strong negative effect on hematopoiesis and fertility became possible after the gene was duplicated. The relatively long evolutionary history of domestic animals, compared with experimental organisms, has resulted in another characteristic feature of alleles affecting pigmentation in domestic animals, namely, the common occurrence of alleles differing by more than one sequence change from the wild-type allele. This is illustrated by four examples given in this review, including KIT alleles in pigs and cattle, MC1R alleles in pigs, and CDKN2A alleles in chicken. In this aspect, domestic animals are better models than experimental organisms for pigmentation phenotypes in natural populations. For traits that have been under selection for many generations, the presence of multiple causal variant is expected to be the rule rather than the exception. A possible example of this is the MC1R allele associated with white ornamental feathers in Satellite males in the ruff that differs by four derived missense mutations compared with the wild-type allele (Lamichhaney et al., 2016).
Author Contributions
LA wrote the manuscript.
Funding
LA was supported by The Swedish Research Council and The Knut and Alice Wallenberg Foundation.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bamshad, M., Lin, R. C., Law, D. J., Watkins, W. S., Krakowiak, P. A., Moore, M. E., et al. (1997). Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 16, 311–315. doi: 10.1038/ng0797-311
Baranowska Körberg, I., Sundström, E., Meadows, J. R. S., Rosengren, P. G., Gustafson, U., and Hedhammar, Å (2014). A simple repeat polymorphism in the MITF-M promoter Is a key regulator of white spotting in dogs. PLoS One 9:e104363. doi: 10.1371/journal.pone.0104363
Brooks, S. A., Lear, T. L., Adelson, D. L., and Bailey, E. (2008). A chromosome inversion near the KIT gene and the Tobiano spotting pattern in horses. Cytogenet. Genome Res. 119, 225–230. doi: 10.1159/000112065
Clark, L., Wahl, J., Rees, C., and Murphy, K. (2006). Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc. Natl. Acad. Sci. U.S.A. 103, 1376–1381. doi: 10.1073/pnas.0506940103
Dávila, S. G., Gil, M. G., Resino-Talaván, P., and Campo, J. L. (2014). Association between polymorphism in the melanocortin 1 receptor gene and E locus plumage color phenotype. Poult. Sci. 93, 1089–1096. doi: 10.3382/ps.2013-03611
Durkin, K., Coppieters, W., Drögemüller, C., Ahariz, N., Cambisano, N., Druet, T., et al. (2012). Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature 482, 81–84. doi: 10.1038/nature10757
Fang, M., Larson, G., Soares Ribeiro, H., Li, N., and Andersson, L. (2009). Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5:e1000341. doi: 10.1371/journal.pgen.1000341
Forster, E. S., and Heffner, E. H. (1968). Lucius Junius Moderatus(Columella)on Agriculture. London: Heinemann.
Giuffra, E., Kijas, J. M. H., Amarger, V., Carlborg, O., Jeon, J.-T., and Andersson, L. (2000). The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154, 1785–1791.
Giuffra, E., Törnsten, A., Marklund, S., Bongcam-Rudloff, E., Chardon, P., Kijas, J. M. H., et al. (2002). A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm. Genome 13, 569–577. doi: 10.1007/s00335-002-2184-5
Groenen, M. A., Archibald, A. L., Uenishi, H., Tuggle, C. K., Takeuchi, Y., Rothschild, M. F., et al. (2012). Pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398. doi: 10.1038/nature11622
Gunnarsson, U., Kerje, S., Bed’hom, B., Sahlqvist, A. S., Ekwall, O., Tixier-Boichard, M., et al. (2011). The Dark brown plumage color in chickens is caused by an 8.3 kb deletion upstream of SOX10. Pigment Cell Melanoma Res. 24, 268–274. doi: 10.1111/j.1755-148X.2011.00825.x
Haupaix, N., Curantz, C., Bailleul, R., Beck, S., Robic, A., and Manceau, M. (2018). The periodic coloration in birds forms through a prepattern of somite origin. Science 361:eaar4777. doi: 10.1126/science.aar4777
Hauswirth, R., Haase, B., Blatter, M., Brooks, S. A., Burger, D., Drögemüller, C., et al. (2012). Mutations in MITF and PAX3 cause “Splashed White” and other white spotting phenotypes in horses. PLoS Genet. 8:e1002653. doi: 10.1371/journal.pgen.1002653
Hellström, A. R., Sundström, E., Gunnarsson, U., Bed’Hom, B., Tixier-Boichard, M., Honaker, C. F., et al. (2010). Sex-linked barring in chickens is controlled by the CDKN2A/B tumour suppressor locus. Pigment Cell Melanoma Res. 23, 521–530. doi: 10.1111/j.1755-148X.2010.00700.x
Hofstetter, S., Seefried, F., Häfliger, I. M., Jagannathan, V., Leeb, T., and Drögemüller, C. (2019). A non-coding regulatory variant in the 5’-region of the MITF gene is associated with white-spotted coat in Brown Swiss cattle. Anim. Genet. 50, 27–32. doi: 10.1111/age.12751
Hussussian, C. J., Struewing, J. P., Goldstein, A. M., Higgins, P. A. T., Ally, D. S., Sheahan, M. D., et al. (1994). Germline p16 mutations in familial melanoma. Nat. Genet. 8, 15–21.
Imsland, F., McGowan, K., Rubin, C.-J., Henegar, C., Sundstrom, E., Berglund, J., et al. (2016). Regulatory mutations in TBX3 disrupt asymmetric hair pigmentation that underlies Dun camouflage color in horses. Nat. Genet. 48, 152–158. doi: 10.1038/ng.3475
Johansson Moller, M., Chaudhary, R., Hellmen, E., Hoyheim, B., Chowdhary, B., and Andersson, L. (1996). Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 7, 822–830. doi: 10.1007/s003359900244
Karlsson, E. K., Baranowska, I., Wade, C. M., Salmon Hillbertz, N. H. C., Zody, M. C., Andersson, N., et al. (2007). Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 39, 1321–1328. doi: 10.1038/ng.2007.10
Kerje, S., Lind, J., Schütz, K., Jensen, P., and Andersson, L. (2003). Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim. Genet. 34, 241–248. doi: 10.1046/j.1365-2052.2003.00991.x
Kijas, J. M. H., and Andersson, L. (2001). A phylogenetic study of the origin of the domestic pig estimated from the near complete mtDNA genome. J. Mol. Evol. 52, 302–308. doi: 10.1007/s002390010158
Kijas, J. M. H., Moller, M., Plastow, G., and Andersson, L. (2001). A frameshift mutation in MC1R and a high frequency of somatic reversions cause black spotting in pigs. Genetics 158, 779–785.
Kijas, J. M. H., Wales, R., Törnsten, A., Chardon, P., Moller, M., and Andersson, L. (1998). Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 150, 1177–1185.
Kinoshita, K., Akiyama, T., Mizutani, M., Shinomiya, A., Ishikawa, A., Younis, H. H., et al. (2014). Endothelin receptor B2 (EDNRB2) is responsible for the tyrosinase-independent recessive white (mow) and mottled (mo) plumage phenotypes in the chicken. PLoS One 9:e86361. doi: 10.1371/journal.pone.0086361
Lamichhaney, S., Fan, G., Widemo, F., Gunnarsson, U., and Schwochow Thalmann, D. (2016). Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Gen. 48, 84–88. doi: 10.1038/ng.3430
Lin, S. J., Foley, J., Jiang, T. X., Yeh, C. Y., Wu, P., Foley, A., et al. (2013). Topology of feather melanocyte progenitor niche allows complex pigment patterns to emerge. Science 340, 1442–1445. doi: 10.1126/science.1230374
Lyons, L. A., Imes, D. L., Rah, H. C., and Grahn, R. A. (2005). Tyrosinase mutations associated with siamese and burmese patterns in the domestic cat (Felis catus). Anim. Genet. 36, 119–126. doi: 10.1111/j.1365-2052.2005.01253.x
Mallarino, R., Henegar, C., Mirasierra, M., Manceau, M., Schradin, C., Vallejo, M., et al. (2016). Developmental mechanisms of stripe patterns in rodents. Nature 539, 518–523. doi: 10.1038/nature20109
Marklund, S., Kijas, J., Rodriguez-Martinez, H., Rönnstrand, L., Funa, K., Moller, M., et al. (1998). Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 8, 826–833. doi: 10.1101/gr.8.8.826
Metallinos, D. L., Bowling, A. T., and Rine, J. (1998). A missense mutation in the endothelin-B receptor gene is associated with lethal white foal syndrome: an equine version of hirschsprung disease. Mamm. Genome 9, 426–431. doi: 10.1007/s003359900790
Pingault, V., Bondurand, N., Kuhlbrodt, K., Goerich, D. E., Préhu, M.-O., Puliti, A., et al. (1998). SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 18, 171–173. doi: 10.1038/ng0298-171
Rosengren Pielberg, G., Golovko, A., Sundström, E., Curik, I., Lennartsson, J., Seltenhammer, M. H., et al. (2008). A cis-acting regulatory mutation causes premature hair greying and susceptibility to melanoma in the horse. Nat. Genet. 40, 1004–1009. doi: 10.1038/ng.185
Rubin, C.-J., Megens, H., Martinez Barrio, A., Maqbool, K., Sayyab, S., Schwochow, D., et al. (2012). Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. U.S.A. 109, 19529–19536. doi: 10.1073/pnas.1217149109
Santschi, E. M., Purdy, A. K., Valberg, S. J., Vrotsos, P. D., Kaese, H., and Mickelson, J. R. (1998). Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mamm. Genome 9, 306–309. doi: 10.1007/s003359900754
Schwochow-Thalmann, D., Ring, H., Sundström, E., Cao, X., Larsson, M., Kerje, S., et al. (2017). The evolution of Sex-linked barring alleles in chickens involves both regulatory and coding changes in CDKN2A. PLoS Genet. 13:e1006665. doi: 10.1371/journal.pgen.1006665
Smyth, J. R. Jr. (1990). “Genetics of plumage, skin and pigmentation in chickens,” in Poultry Breeding and Genetics, ed. R. D. Crawford (New York, NY: Elsevier Science), 109–167.
Takeuchi, S., Suzuki, H., Yabuuchi, M., and Takahashi, S. (1996). A possible involvement of melanocortin 1-receptor in regulating feather color pigmentation in the chicken. Biochim. Biophys. Acta 1308, 164–168. doi: 10.1016/0167-4781(96)00100-5
Wheat, C. H., van der Bijl, W., and Wheat, C. W. (2019). Morphology does not covary with predicted behavioural correlations of the domestication syndrome in dogs. bioRxiv [Preprint]. doi: 10.1101/660829
Wilkins, A. S., Wrangham, R. W., and Fitch, W. T. (2014). The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197, 795–808. doi: 10.1534/genetics.114.165423
Keywords: pigmentation, domestic animals, patterning, domestication, selection
Citation: Andersson L (2020) Mutations in Domestic Animals Disrupting or Creating Pigmentation Patterns. Front. Ecol. Evol. 8:116. doi: 10.3389/fevo.2020.00116
Received: 08 December 2019; Accepted: 09 April 2020;
Published: 13 May 2020.
Edited by:
Ricardo Mallarino, Princeton University, United StatesReviewed by:
Salvatore D’Aniello, Stazione Zoologica Anton Dohrn, ItalyClaudius Frank Kratochwil, University of Konstanz, Germany
Copyright © 2020 Andersson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leif Andersson, TGVpZi5BbmRlcnNzb25AaW1iaW0udXUuc2U=
 Leif Andersson
Leif Andersson