
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 26 March 2020
Sec. Evolutionary Developmental Biology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00080
This article is part of the Research TopicEvo-Devo of Color Pattern FormationView all 20 articles
 Jesse T. Hughes1
Jesse T. Hughes1 Melissa E. Williams1
Melissa E. Williams1 Rachel Johnson1
Rachel Johnson1 Sumant Grover1
Sumant Grover1 Mark Rebeiz2*
Mark Rebeiz2* Thomas M. Williams1,3*
Thomas M. Williams1,3*Traits that appear discontinuously along phylogenies may be explained by independent origins (homoplasy) or repeated loss (homology). While discriminating between these models is difficult, the dissection of gene regulatory networks (GRNs) which drive the development of such repeatedly occurring traits can offer a mechanistic window on this fundamental problem. The GRN responsible for the male-specific pattern of Drosophila (D.) melanogaster melanic tergite pigmentation has received considerable attention. In this system, a metabolic pathway of pigmentation enzyme genes is expressed in spatial and sex-specific (i.e., dimorphic) patterns. The dimorphic expression of several genes is regulated by the Bab transcription factors, which suppress pigmentation enzyme expression in females, by virtue of their high expression in this sex. Here, we analyzed the phylogenetic distribution of species with male-specific pigmentation and show that this dimorphism is phylogenetically widespread among fruit flies. The analysis of pigmentation enzyme gene expression in distantly related dimorphic and monomorphic species shows that dimorphism is driven by the similar deployment of a conserved metabolic pathway. However, sexually dimorphic Bab expression was found only in D. melanogaster and its close relatives. These results suggest that dimorphism evolved by parallel deployment of differentiation genes, but was derived through distinct architectures at the level of regulatory genes. This work demonstrates the interplay of constraint and flexibility within evolving GRNs, findings that may foretell the mechanisms of homoplasy more broadly.
Recurring traits are widespread in nature, suggesting that evolution has predictable solutions to certain ecological challenges (Conway Morris, 2003; Losos, 2017; Blount et al., 2018). This discontinuous presence of similar phenotypes on phylogenies can result from different historical processes. Notably, the trait in question could be ancestral, and repeated loss events could explain the recurrent absence of the trait (Wiens, 2001). Alternatively, the trait could be derived and evolved independently in multiple lineages, a phenomenon known as homoplasy (Wake et al., 2011). Although methods for ancestral character reconstruction can help discriminate losses from gains, these methods are often inconclusive and are sensitive to estimated differences in the relative rates of trait gain and loss (Cunningham et al., 1998; Joy et al., 2016). Distinguishing between these two scenarios ultimately comes down to whether the genetic processes that build the trait are homologous. Thus, dissecting the individual genetic components underlying trait formation represents the most granular way to determine the elusive historical nature of recurring traits.
For morphological traits, their construction during development is recognized to depend upon precise spatial and temporal patterns of gene expression among the genes within gene regulatory networks (GRNs) (Peter and Davidson, 2011; Rebeiz et al., 2015). Each GRN utilizes numerous regulatory genes, many encoding transcription factors that govern the expression of the differentiation genes that produce the morphological feature. The patterns of expression for genes within any GRN depends upon combinations of transcription factors binding to cis-regulatory elements (CREs) that control the activation of target genes. For well-studied traits, dozens to more than hundreds of genes are known to comprise the GRNs (Bonn and Furlong, 2008). In addition to explaining how phenotypes develop, GRNs are the lens through which we can observe important aspects of a phenotype’s evolution. Important here, GRNs provide a critical context in which we might be able to detect the presence (common GRNs) or absence (distinct GRNs) of homology relationships (Wagner, 2016). Moreover, the examination of GRNs that govern recurring traits could reveal whether and how similar traits converge at the molecular level.
One of the premiere morphological traits for the study of GRN evolution is the rapidly diverging patterns of body pigmentation in Drosophila. Melanic pigmentation is widespread in nature, playing important ecological roles. Pigmentation has many uses for insects, including wound healing, desiccation resistance, thermal regulation, and sexual selection (Majerus, 1998). In fruit flies of the Drosophila (D.) genus, pigmentation traits are quite diverse (Wittkopp et al., 2003; Werner et al., 2010; Arnoult et al., 2013; Pham et al., 2017). Pigmentation patterns exist on the wings, legs, thorax, and the cuticle plates (known as tergites) that cover the dorsal abdomen surface. Work on Drosophila pigmentation traits has advanced rapidly by virtue of the ability to study the GRNs for pigmentation in the highly genetically tractable Drosophila melanogaster model system.
D. melanogaster belongs to the melanogaster species group within the subgenus Sophophora (Markow and O’Grady, 2006). Tergite pigmentation in this species is sexually dimorphic (Kopp et al., 2000), with males possessing fully melanic A5 and A6 tergite pigmentation, whereas female A5 and A6 tergites are only partially pigmented (Figure 1A). Species with similar dimorphic patterns of pigmentation are common among all three clades of the melanogaster species group, while members of more distantly related Sophophora groups such as D. pseudoobscura and D. willistoni bear monomorphic tergite pigmentation patterns (Figure 1A). Bolstered by an ancestral character reconstruction analysis (Jeong et al., 2006), it was inferred that dimorphic pigmentation is a derived trait that evolved in the lineage of the melanogaster species group (Rebeiz and Williams, 2017). Additionally, species outside of Sophophora are known to have dimorphic patterns of tergite pigmentation that resemble D. melanogaster (Gompel and Carroll, 2003). Hence, this system represents a tractable system to study homoplasy at the level of participating GRNs.
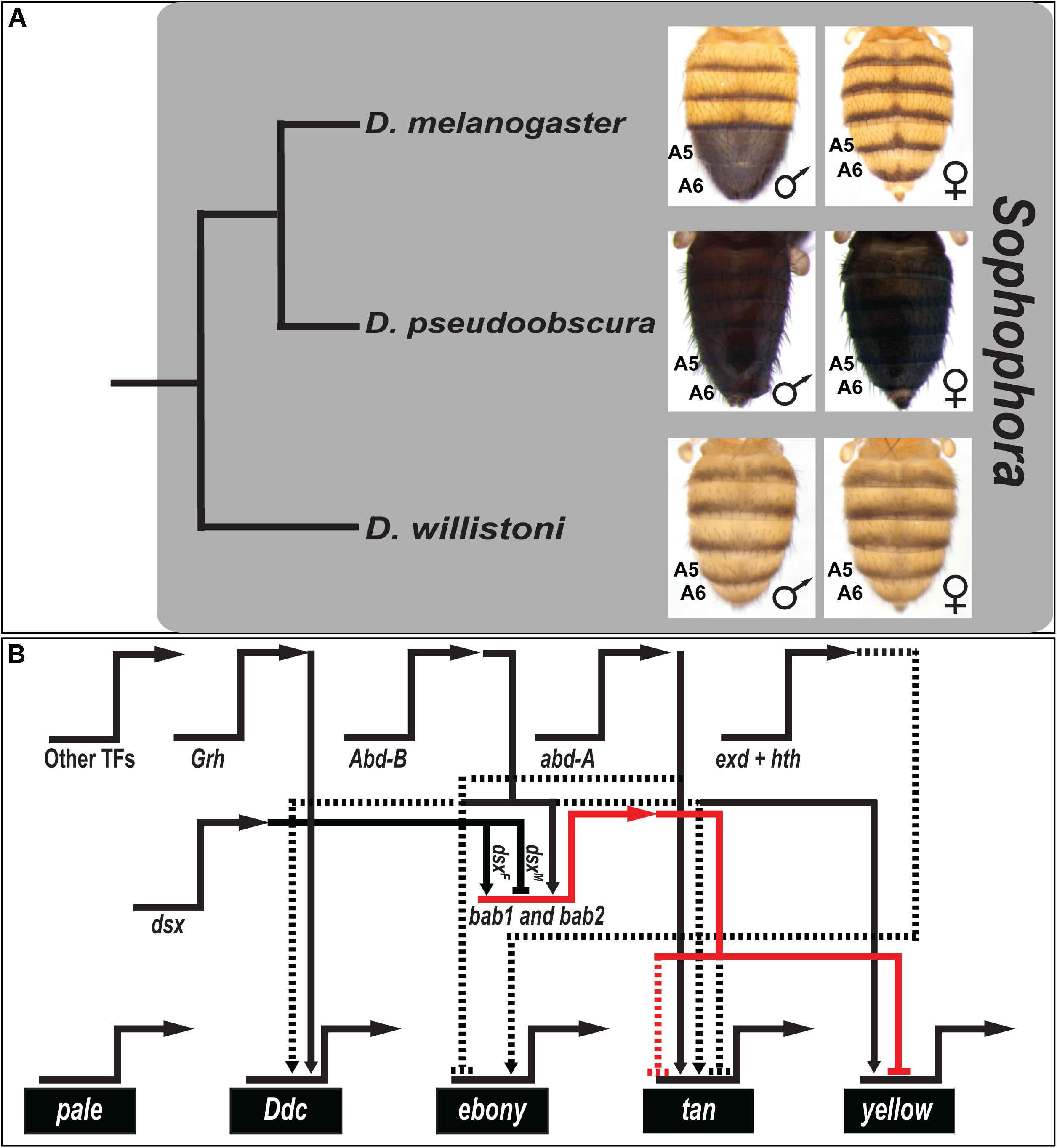
Figure 1. The canonical model for the origin of sexually dimorphic abdomen pigmentation of D. melanogaster, and its underlying gene regulatory network. (A) Phylogenetic relationship of model species of the Sophophora subgenus. Here, monomorphism is presumed to be ancestral, and dimorphism derived. (B) The current understanding of the gene regulatory network for the D. melanogaster phenotype has key transcription factor genes (regulatory genes) driving the expression of genes that encode proteins that participate in a pigment metabolic pathway (differentiation genes). The horizontal-pointing arrows represent expression occurring from the named loci. Key nodes within this network drive the male-limited expression of the tan and yellow genes, whose expression in females is repressed by the activity of the bab1 and bab2 transcription factor genes. Solid connectors between genes represent known direct interactions between the encoded transcription factor and a binding site(s) in the target gene’s cis-regulatory element. Dashed lines represent regulation not confirmed as direct. Activation is indicated by connectors terminating with an arrowhead, and repression as a nail head shape.
The D. melanogaster abdominal pigmentation GRN has received considerable attention, which includes a metabolic pathway of differentiation genes whose expression correlates or anti-correlates with the dimorphic phenotype. Among the first acting genes in this pathway are pale and Ddc, which are expressed monomorphically in the abdominal epidermis (Grover et al., 2018), and their enzyme activities catalyze the production of Dopamine. ebony is expressed in a dimorphic pattern, with prominent expression in the female A5 and A6 segments (Rebeiz et al., 2009), and its encoded enzyme converts Dopamine to NBAD that is used to make yellow-colored sclerotin. NBAD can be converted back into Dopamine by the enzyme encoded by tan, and converting Dopamine into black Dopamine-melanin requires the activity of the protein encoded by yellow (Grover et al., 2018). Both tan and yellow expression is upregulated in the male A5 and A6 segments (Camino et al., 2015).
The temporal, spatial, and sex-specific expression patterns of the pigmentation pathway genes are achieved by the regulatory genes of the GRN (Figure 1B). Two key regulators are bab1 and bab2, collectively referred to as bab, which encode the Bab1 and Bab2 transcription factor proteins, which function as dominant repressors of black pigmentation (Kopp et al., 2000; Couderc et al., 2002; Roeske et al., 2018). In D. melanogaster and species of all three clades of the melanogaster species group, the Bab1 and Bab2 proteins are expressed in a dimorphic pattern. Expression in the abdominal epidermis of males is downregulated, while expression can be observed throughout the A2–A6 segments of females (Salomone et al., 2013). One key function of Bab is to directly bind to the body element CRE of the yellow gene and thereby repress yellow expression in the female A5 and A6 segments (Roeske et al., 2018). Bab additionally represses tan expression in females, though the mechanism of action remains unknown. A previous study showed that Bab2 expression is broadly downregulated in the abdomen epidermis underlying melanic tergite regions during early pupal development, including distantly related species with dimorphic pigmentation (Gompel and Carroll, 2003). These results were interpreted to indicate that Bab has a long history as a suppressor of melanic pigmentation, and thus its expression might generally evolve to shape diverse patterns of tergite coloration.
Here, we performed an expanded survey of tergite pigmentation phenotypes in the Drosophila genus (Figure 2). Our results draw attention to how dimorphic pigmentation exists in most Sophophora species groups, but is also common outside this subgenus. To determine whether processes proximal to pigmentation phenotypes are generated by shared or divergent enzyme pathway uses, we characterized the expression patterns of known D. melanogaster pigmentation pathway genes in phylogenetically disparate cases of dimorphism. This revealed how dimorphic pigmentation evolved through the formation of GRNs that similarly deploy a conserved pathway of pigmentation genes. In contrast, analysis of Bab1 expression revealed that dimorphism in these similarly implemented pathways is mediated by different regulatory genes. We suggest that dimorphic pigmentation is indeed a product of parallel evolution in Drosophila, and provide an example where constraint exists at the level of the differentiation genes in contrast to the regulatory tier of this GRN.
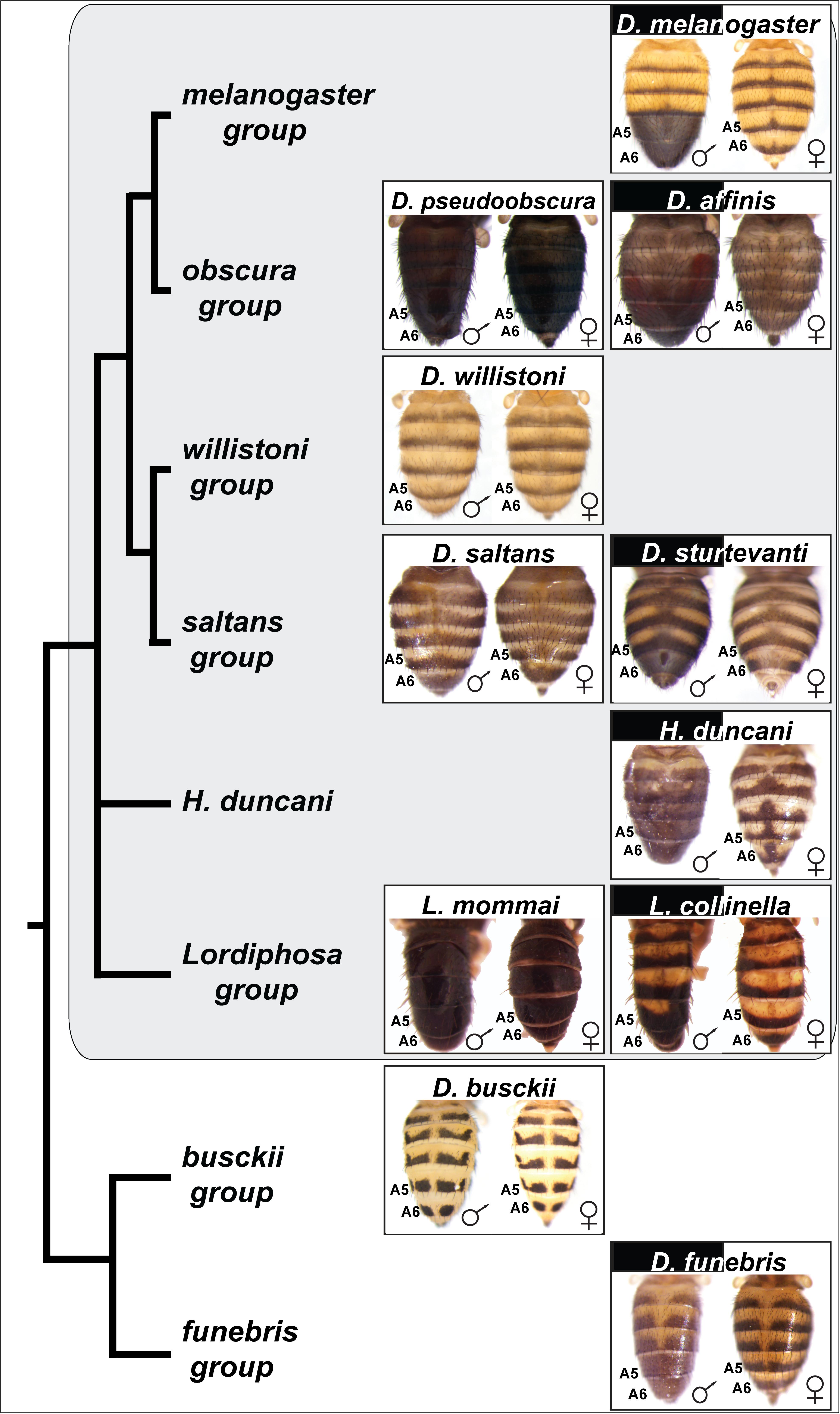
Figure 2. Species with dimorphic tergite pigmentation are widespread throughout the Drosophila genus. Sophophora subgenus species groups and species are indicated by the gray background. D. busckii and D. funebris are included as non-Sophophora species from the Drosophila genus that respectively exhibit monomorphic and dimorphic patterns of tergite pigmentation. The homologous A5 and A6 segment tergites are indicated for each species, the segments bearing the dimorphic pigmentation in D. melanogaster. While the obscura, saltans, and Lordiphosa groups are predominately populated by monomorphic species (e.g., D. pseudoobscura, D. saltans, and L. mommai), they possess a few dimorphic species, including D. affinis, D. sturtevanti, and L. collinella.
Fly stocks were maintained at 25°C on a sugar food medium (Salomone et al., 2013). Species stocks used in this study were D. melanogaster (14021-0231.04), D. willistoni (14030-08 11.24), D. affinis (14012-0141.09), D. algonquin (14012- 0161.04a), D. azteca (14012.0171.08), D. persimilis (14011-0111.00), D. miranda (14011-0101.08), D. ambigua (14013-1011.00), D. bifasciata (14012-0181.02), D. guanche (14011-0095.01), D. sturtevanti (14043-0871.07), D. nebulosa (14030-0761.03), D. milleri (14043-0861.00), D. saltans (14043-0871.01), D. lusaltans (14045-0891.00), D. prosaltans (14045-0901.02), D. emarginata (14042-0841.09), D. neocordata (14041-0831.00), D. tropicalis (14030-0801.00), D. paulistorum (14030-0771.06), D. equinoxialis (14030-0741.00), D. sucinea (14030-0791.00), and H. duncani (92000-0075.00) were obtained from the National Drosophila Species Stock Center. D. capricorni, D. fumipenis, D. obscura, D. pseudoobscura, D. funebris, and D. busckii were obtained from the lab of Dr. Sean B. Carroll. L. collinella and L. mommai specimens were obtained from Dr. Masanori J. Toda.
In situ hybridization was performed as described previously in greater detail (Jeong et al., 2008). In brief, Digoxigenin labeled RNA probes for pale, Ddc, ebony, tan, and yellow were prepared through in vitro transcription of species-specific PCR templates amplified from genomic DNA (PCR primers listed in Supplementary Table S1). Dorsal abdomens were dissected at various pupal developmental stages between P10 and P15ii (P15ii being newly eclosed adults). Stages were identified by the presence of various morphological markers (Ashburner et al., 2005; Grover et al., 2018) (Supplementary Figure S1). Male and female samples were pooled, and females were distinguished by the removal of their wings. All following steps were done with the male and female samples of the same stages together in the same tubes or plate and thereby experiencing identical conditions. Probe hybridizations were detected using an anti-digoxigenin antibody (Roche Diagnostics) and visualized by alkaline phosphatase color reaction using BCIP/NBT (Promega). Samples were allowed to stain in the dark, and once the staining reactions were stopped, the specimens were transferred to glycerol mountant (80% glycerol and 100 mM Tris pH 8) before being placed between a slide and coverslip for imaging.
Dorsal abdomens were dissected from pupae at the P10 and P14-15i developmental stages (Ashburner et al., 2005; Grover et al., 2018) (Supplementary Figure S1). Male and female samples were pooled, and females were distinguished by the removal of their wings. All following steps were done with the male and female samples of the same stages together in the same tubes or plate and thereby experiencing identical conditions. Samples were fixed for 35 min in PBST solution (phosphate buffered saline with 0.3% Triton X-100) that additionally contained 4% paraformaldehyde (Electron Microscopy Services). Following fixation, samples were washed twice with PBST and then blocked for 1 h at room temperature in blocking solution (PBST and 1% Bovine Serum Albumin). The abdomen specimens were then incubated overnight at 4°C with rabbit anti-Bab1 primary antibody (Williams et al., 2008) at a 1:200 dilution in PBST. Following four washes with PBST and then 1 h in blocking solution, specimens were incubated with goat anti-rabbit Alexa Fluor 647 (Invitrogen) secondary antibody at a dilution of 1:500 in PBST. After four washes with PBST, samples were incubated for ten minutes at room temperature in Glycerol Mount:PBST (50:50) solution. Samples were then transferred to glycerol mount before finally being placed between a glass cover slip and slide for imaging with a confocal microscope. Although we were unable to acquire a Bab2 antibody, we found in a previous study that Bab1 and Bab2 expression are indistinguishable in the abdominal epidermis in a variety of species (Salomone et al., 2013), and these paralogs are suspected to be under the regulatory control of the same CREs (Williams et al., 2008). Therefore, the Bab1 expression shown here is anticipated to reflect Bab2 expression as well.
Images for the specimens taken through the immunohistochemistry protocol, to visualize Bab1 expression in pupal dorsal abdominal tissue, were obtained using an Olympus FV3000 confocal microscope and FV31S-SW imaging software. Samples were imaged with microscope settings as follows: 10% laser power (647 laser), HV between 650 and 700, offset equal to 1, gain equal to 1, aperture set at 180 microns, Z-series step size of 5 microns, and Kalman line averaging set to 2.
Images of adult fruit fly abdomen pigmentation patterns (between two and four days old) and in situ hybridization specimens, were obtained using an Olympus SZX16 zoom stereoscope, running the Olympus cellSens Standard 2.2 software package, with a mounted DP72 digital camera. All samples were imaged at 63X magnification with a 1X objective lens.
Confocal projection images and stereoscope images were exported in TIFF, and processed with the consistent processing steps in Adobe Photoshop CS3. Figures were assembled by the use of Adobe Illustrator CS3. The Image J program (Abràmoff et al., 2004) was used to measure pixel intensity from Bab1 immunohistochemistry images. Similar epidermal regions of the A5 segment were selected that lack confounding expression from muscle and oenocyte cells. For each species, the pixel intensity values were measured for three separate male and female specimens. Mean pixel intensity values were calculated, and differences between the male and female means were evaluated by a two-sided t-test.
Previous phylogenetic analyses of the origin of D. melanogaster male-specific A5 and A6 tergite pigmentation suggested that monomorphism is the ancestral character state in the Sophophora subgenus, and dimorphism was derived. The origin of this trait was suspected to have occurred in the lineage of D. melanogaster at some point following its split from the lineage of D. pseudoobscura of the obscura species group (Figure 1; Jeong et al., 2006; Rebeiz and Williams, 2017). This conclusion was derived from a limited sampling of Sophophora species diversity (Figure 2A), including only a single taxon from the saltans and willistoni groups.
The melanogaster species group has been well characterized and includes a preponderance of species that possess a male-specific pattern of tergite pigmentation (Kopp et al., 2000; Jeong et al., 2006). This suggests that the common ancestor for this group possessed the dimorphic trait. The obscura group is most closely related to the melanogaster group. Males and females of D. pseudoobscura have a similar monomorphic pattern of melanic abdominal tergites. To see whether such monomorphism is typical of this species group, we inspected the coloration phenotypes of another nine of its member species (Supplementary Figure S2). Although monomorphic melanic tergite color was the most common phenotype, we observed male-limited pigmentation phenotypes in D. affinis (Figure 2) and D. algonquin (Supplementary Figure S2).
More distantly related to the melanogaster species group are both the saltans and willistoni groups (Figure 2) (Markow and O’Grady, 2006). Their charter species, D. saltans and D. willistoni, are characterized by monomorphic patterns of tergite pigmentation (Supplementary Figures S3, S4). To see whether such monomorphism is typical of these groups, we inspected the coloration phenotypes of another six species from the saltans group (Supplementary Figure S3) and seven species from the willistoni group (Supplementary Figure S4). Although monomorphic non-melanic tergite color was the most common phenotype among saltans species (Supplementary Figure S3) (De Magalhaes, 1956), we observed male-limited pigmentation phenotypes for D. sturtevanti (Figure 2) and D. emarginata (Supplementary Figure S3). Within the willistoni species group, all species analyzed (Supplementary Figure S4) or reported in the literature (Zanini et al., 2015) exhibit a monomorphic non-melanic tergite color.
Phylogenetic studies have expanded the number of species and lineages within Sophophora. One study supported a topology that places Hirtodrosophila (H.) duncani as an outgroup to the clade containing melanogaster, obscura, saltans, and willistoni groups (van der Linde et al., 2010). This inclusion in the Sophophora subgenus is consistent with findings that H. duncani genital morphology is most similar to species of the obscura group (Nater, 1950, 1953), and has been considered close to or within Sophophora in other analyses (Throckmorton, 1962). H. duncani is a species with an extensive male-limited pattern of melanic tergite pigmentation (Gompel and Carroll, 2003), including the A5 and A6 tergites (Figure 2). Phylogenetic studies have supported a branching structure that places species of the Lordiphosa genus within Sophophora (Figure 2; Hu and Toda, 2001; Gao et al., 2011). We inspected the coloration phenotypes of seven species from the Lordiphosa group phylogeny (Supplementary Figure S5) (personal communication from Dr. Masanori J. Toda) (Gao et al., 2011; Katoh et al., 2018). Although monomorphic non-melanic tergite color was the prevalent common phenotype, we observed a modest male-limited pigmentation phenotype for L. collinella on the A6 tergite (Figure 2 and Supplementary Figure S5). Elsewhere, four additional species have been recently described as having monomorphic patterns of tergite coloration (Fartyal et al., 2017).
Outside of the Sophophora subgenus exist a wealth of species with monomorphic tergite phenotypes, such as D. busckii (Figure 2). However, dimorphic species can be found among the diverse lineages, including D. funebris of the funebris group (Figure 2). This species possesses male-limited patterns of tergite coloration that include the A5 and A6 tergites. Thus, from this exploration of species representing diverse branches of the Sophophora subgenus and Drosophila genus, it is apparent that male-limited tergite pigmentation is widespread. This raises the question regarding whether the dimorphic trait was ancestral, or evolved on independent occasions.
In order to discern how D. melanogaster develops the robust male-specific melanic pigmentation of the A5 and A6 segment tergites (Figure 2), we contrasted the expression of its core pigment metabolic pathway genes to the orthologs in the monomorphic D. willistoni (Figure 3). To form black melanin or yellow-colored sclerotin from this pathway, Tyrosine is first converted to Dopa by the activity of the enzyme Pale, then Dopa is converted to Dopamine by the activity of Ddc (Wright, 1987). In D. melanogaster, pale and Ddc are expressed robustly during pupal development, and in patterns that appear monomorphic (Figures 3A,A’,B,B’). In D. willistoni, pale expression appears to be similarly robust, though Ddc expression was less pronounced as revealed by in situ hybridization (Figures 3H,H’,I,I’).
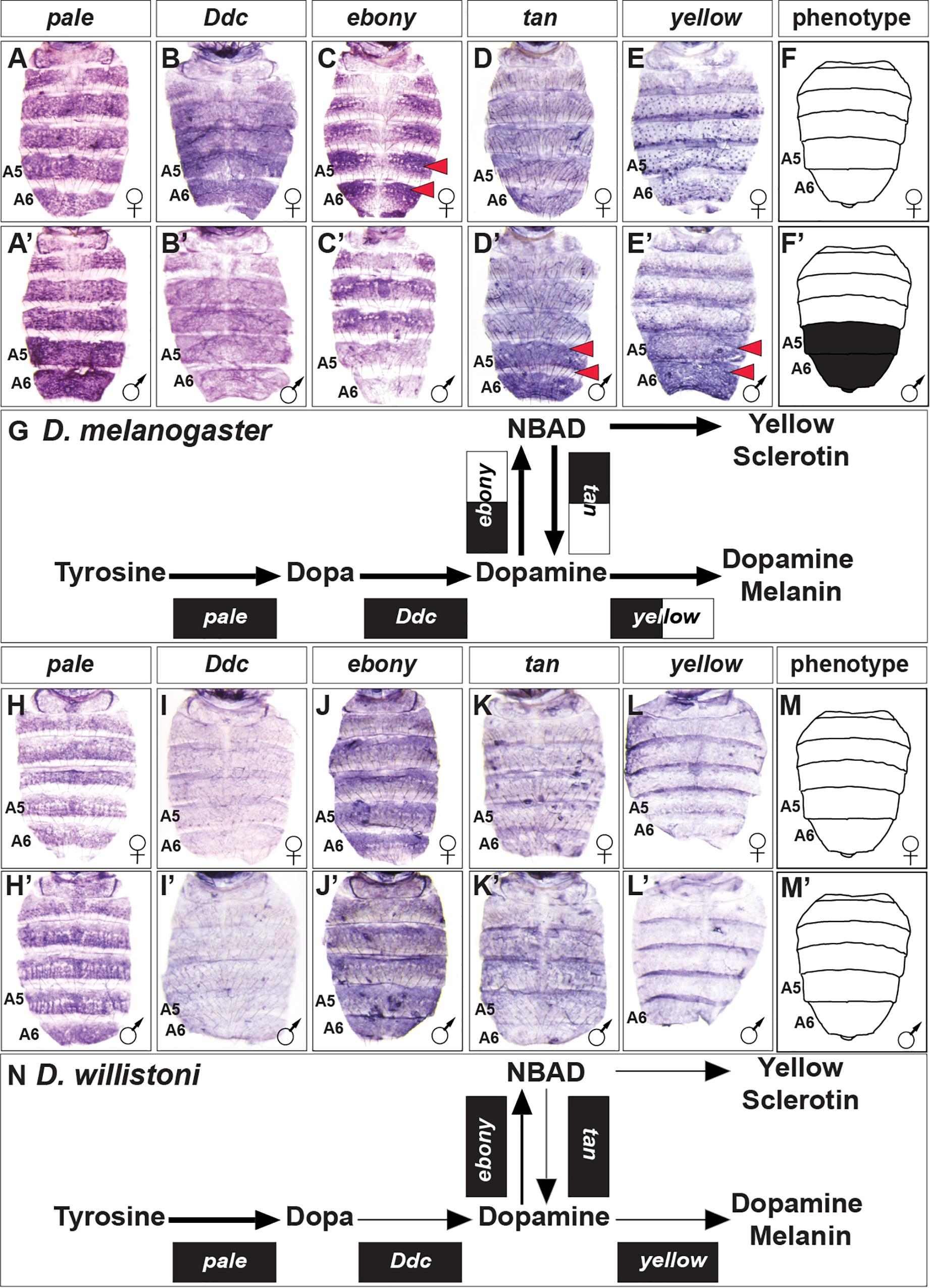
Figure 3. Contrast in the pigment metabolic pathway utilization between the dimorphic D. melanogaster and the monomorphic D. willistoni. (A–E) Female and (A’–E’) male expressions of D. melanogaster pigment metabolic pathway genes, and (F,F’) cartoon representation of the pigmentation phenotype. (G) Summary of the D. melanogaster pathway use includes robust expression of all genes, with dimorphic expressions of ebony, tan, and yellow. (H–L) Female and (H’–L’) male expressions of D. willistoni pigment metabolic pathway genes, and (M,M’) cartoon of the pigmentation phenotype. (N) Summary of the D. willistoni pathway use includes modest and monomorphic expression of most genes. (A,A’,H,H’) pale, (B,B’,I,I’) Ddc, (C,C’,J,J’) ebony, (D,D’,K,K’) tan, and (E,E’,L,L’) yellow. Red arrowheads indicate robust patterns of dimorphic expression in the A5 and A6 segment epidermis. (G,N) Black arrow thickness represents the level of expression for the underlying gene. Monomorphic gene expression is represented by the gene names being placed upon a black rectangle. Dimorphic gene expression is represented by the gene names being placed upon a black and white colored rectangle.
Dopamine can be converted to NBAD by the activity of Ebony, which provides the substrate to make the more yellow-colored sclerotin (Hovemann et al., 1998). In D. melanogaster, ebony expression occurs in a pattern that demarcates where the yellow-colored regions of the tergites will form. ebony expression is dimorphic, as it is absent from the male A5 and A6 segments (Figures 3C,C’). In D. willistoni, ebony expression is similarly robust, though monomorphic in a pattern that mirrors its tergite color phenotype (Figures 3J,J’). To facilitate the production of black melanin, NBAD can be converted back into Dopamine by the enzyme Tan (True et al., 2005), and then converted into Dopamine melanin through the involvement of Yellow. In D. melanogaster, both tan and yellow expression is upregulated in the male A5 and A6 segment epidermis to promote the final development of black tergites (Figures 3D,D’,E,E’). In contrast, tan and yellow expression is modest and monomorphic in D. willistoni (Figures 3K,K’,L,L’).
Our gene expression comparisons of the pigmentation pathway genes of D. melanogaster and D. willistoni revealed apparent expression differences across this conserved pathway that we would have reasonably predicted based upon phenotype alone (Figures 3G,N). We were curious whether other distantly related species with dimorphic or monomorphic tergite color phenotypes would show predictable patterns of pathway deployment.
The obscura group is the species group most closely related to that of the melanogaster group (Figure 2). This group is predominately populated by species with broadly melanic and monomorphic pigmentation, albeit with two species for which pigmentation is sexually dimorphic (Supplementary Figure S2). We investigated the expression of the pigmentation pathway genes for the dimorphic species D. affinis and the monomorphic melanic species D. pseudoobscura (Figure 4). The dimorphic pigmentation patterns differ somewhat between D. melanogaster and D. affinis, the latter displaying broad tergite pigmentation that extends to the male A4 and A3 segment tergites (Figure 2). Among the pigmentation pathway genes of D. affinis, pale, and Ddc are expressed monomorphically in males and females (Figures 4A,A’,B,B’). Similar to D. melanogaster, D. affinis expresses ebony and yellow in sex-specific patterns. Here, ebony is upregulated in the female abdomen consistent with their yellow-colored anterior tergite regions (Figures 4C,C’), and yellow is upregulated in males, prefiguring their melanic color (Figures 4E,E’). In contrast to D. melanogaster, D. affinis tan expression appears modest in level and monomorphic (Figures 4D,D’).
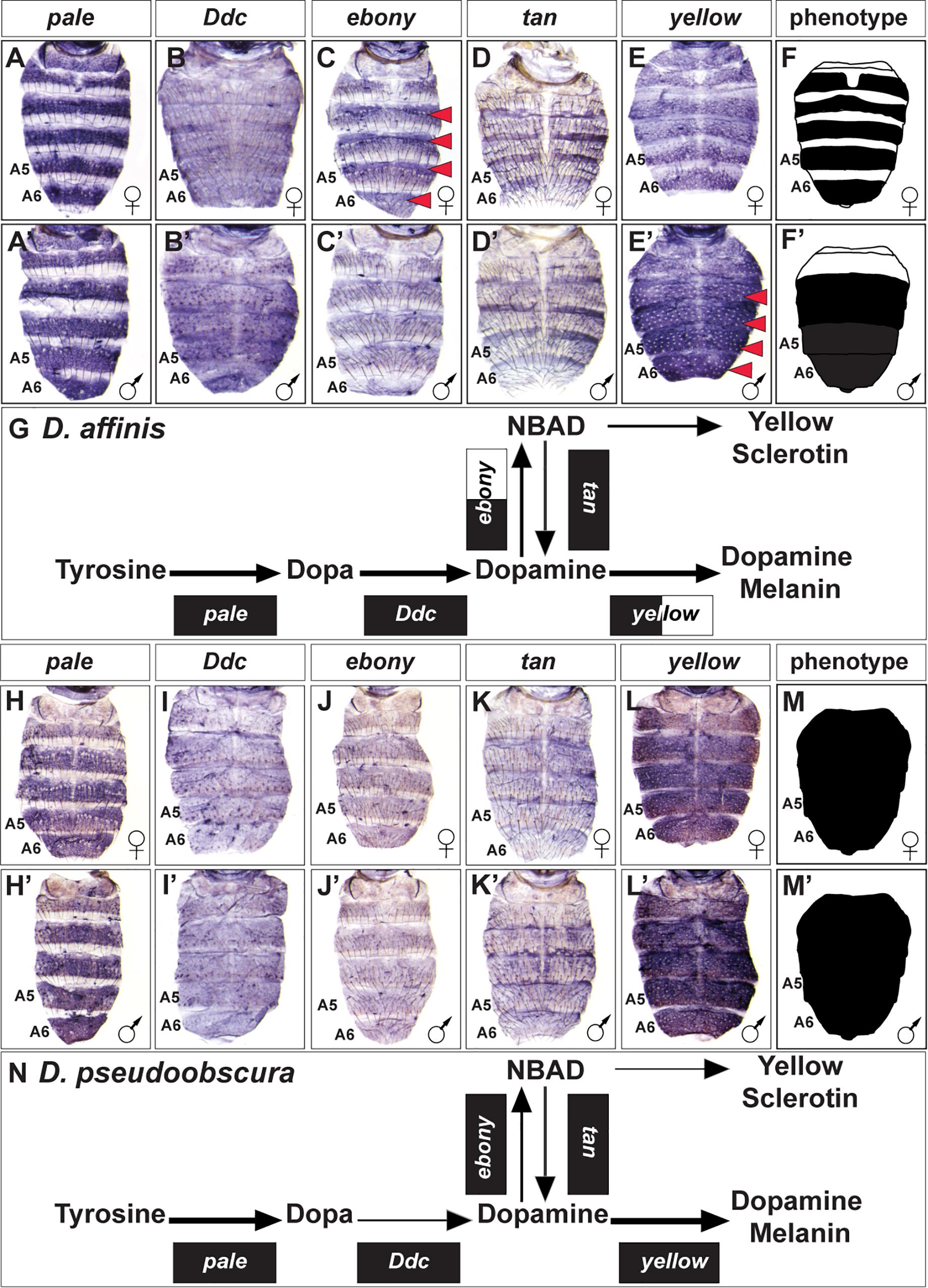
Figure 4. Contrast in the pigment metabolic pathway utilization between the dimorphic D. affinis and the monomorphic D. pseudoobscura. (A–E) Female and (A’–E’) male expressions of D. affinis pigment metabolic pathway genes, and (F,F’) cartoon representation of the pigmentation phenotype. (G) Summary of the D. affinis pathway use includes robust expression of all genes, with dimorphic expressions of ebony, and yellow. (H–L) Female and (H’–L’) male expressions of D. pseudoobscura pigment metabolic pathway genes, and (M,M’) cartoon representation of the pigmentation phenotype. (N) Summary of the D. pseudoobscura pathway use includes monomorphic expression of all genes. (A,A’,H,H’) pale, (B,B’,I,I’) Ddc, (C,C’,J,J’) ebony, (D,D’,K,K’) tan, and (E,E’,L,L’) yellow. Red arrowheads indicate robust patterns of dimorphic expression in the A3-A6 segment epidermis. (G,N) Black arrow thickness represents the level of expression for the underlying gene. Monomorphic gene expression is represented by the gene names being placed upon a black rectangle. Dimorphic gene expression is represented by the gene names being placed upon a black and white colored rectangle.
While D. pseudoobscura can be considered a monomorphic species with regards to its pigmentation, this species’ tergites are melanic rather than the light yellow-brown color of D. willistoni (Figure 1). This broadly melanic phenotype is associated with monomorphic adjustments to the expression of pigmentation pathway genes. We found all genes to be similarly expressed between males and females (Figures 4H–L,H’–L’). The melanic coloration appears to be shaped by reduced levels of ebony and tan expression (Figures 4J,J’,K,K’) and elevated expression of yellow (Figures 4L,L’).
H. duncani is a distant relative of D. melanogaster within Sophophora (van der Linde et al., 2010), which exhibits a striking male-specific pattern of melanic pigmentation on the A5 and A6 tergites, and this dimorphism extends to a lesser extent to the A4 and A3 tergites (Figure 2). Similar to D. melanogaster and D. affinis, pale expression is robust and monomorphic (Figures 5A,A’), ebony is upregulated in the female abdomen in the epidermis regions underlying where the yellow cuticle forms (Figures 5C,C’), and tan (Figures 5D,D’) and yellow (Figures 5E,E’) are upregulated in the male abdominal epidermis of segments A3–A6. One conspicuous difference with the H. duncani pigment metabolic pathway is the apparent upregulation of Ddc in the male A5 and A6 segments (Figures 5B,B’).
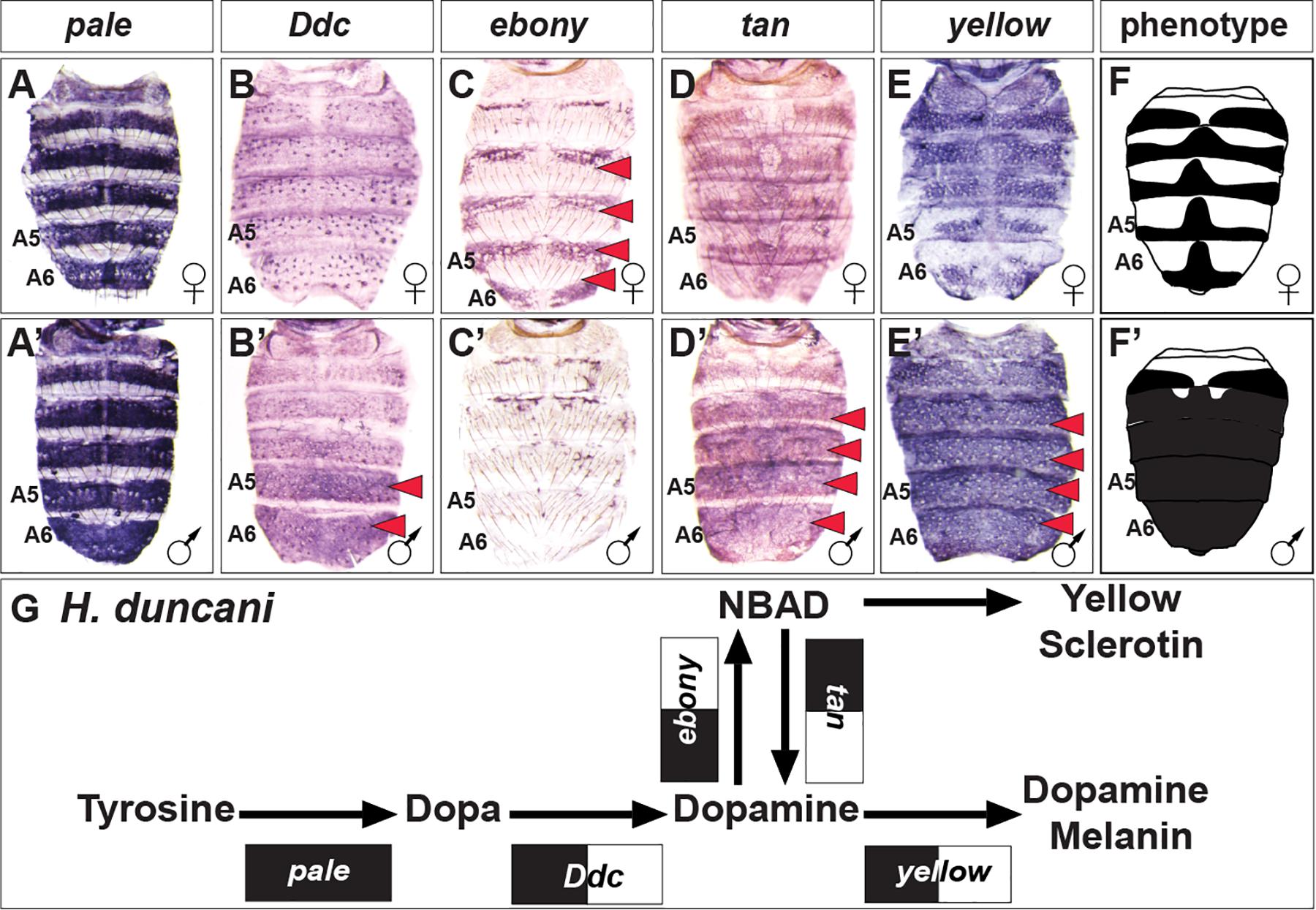
Figure 5. The pigment metabolic pathway utilization for the dimorphic H. duncani. (A–E) Female and (A’–E’) male expressions of H. duncani pigment metabolic pathway genes, and (F,F’) cartoon representation of the pigmentation phenotype. (G) Summary of the H. duncani pathway use includes robust expression of all genes, with dimorphic expressions of Ddc, ebony, tan, and yellow. (A,A’) pale, (B,B’) Ddc, (C,C’) ebony, (D,D’) tan, and (E,E’) yellow. Red arrowheads indicate robust patterns of dimorphic expression in the dorsal abdominal epidermis. (G) Black arrow thickness represents the level of expression for the underlying gene. Monomorphic gene expression is represented by the gene names being placed upon a black rectangle. Dimorphic gene expression is represented by the gene names being placed upon a black and white colored rectangle.
Overall, this comparison reveals how the dimorphism of D. affinis and H. duncani involves a similar deployment of the pigmentation pathway genes (Figures 4G, 5G) compared to D. melanogaster (Figure 3G), and how a related species develops broadly melanic and monomorphic tergite coloration through the modified use of this same metabolic pathway (Figure 4N). These trends in expression raise the suspicion that we can generally predict the patterns of expression for the pigmentation pathway more broadly among Sophophora. However, whether this predictability extends to more distantly related non-Sophophora species was the next question we sought to address.
To gain a perspective of pigment metabolic pathway gene expression outside of Sophophora, we focused our attention on the dimorphic species D. funebris, and the monomorphic species D. busckii (Figure 2). D. funebris exhibits a conspicuous male-specific pattern of melanic pigmentation on the A5 and A6 tergites, and this dimorphism extends to a lesser extent to the A4 and A3 tergites (Figure 2). Similar to D. melanogaster and D. affinis, pale and Ddc expression are monomorphic (Figures 6A,A’,B,B’), and ebony is upregulated in the female abdomen in the epidermis regions underlying where the non-melanic cuticle forms (Figures 6C,C’). tan (Figures 6D,D’) and more prominently yellow (Figures 6E,E’) are upregulated in the male abdominal epidermis of segments A3–A6. The monomorphic D. busckii pigment metabolic pathway genes are expressed in patterns similar to orthologs from the monomorphic D. willistoni (Compare Figure 6 to Figure 3). D. busckii possesses melanic interrupted stripes along the posterior region of the tergites (Figure 2). Interestingly, each of the five pigmentation genes were expressed in patterns that correlate (or anti-correlate in the case of ebony) with these stripes (Figures 6H–L,H’–L’).
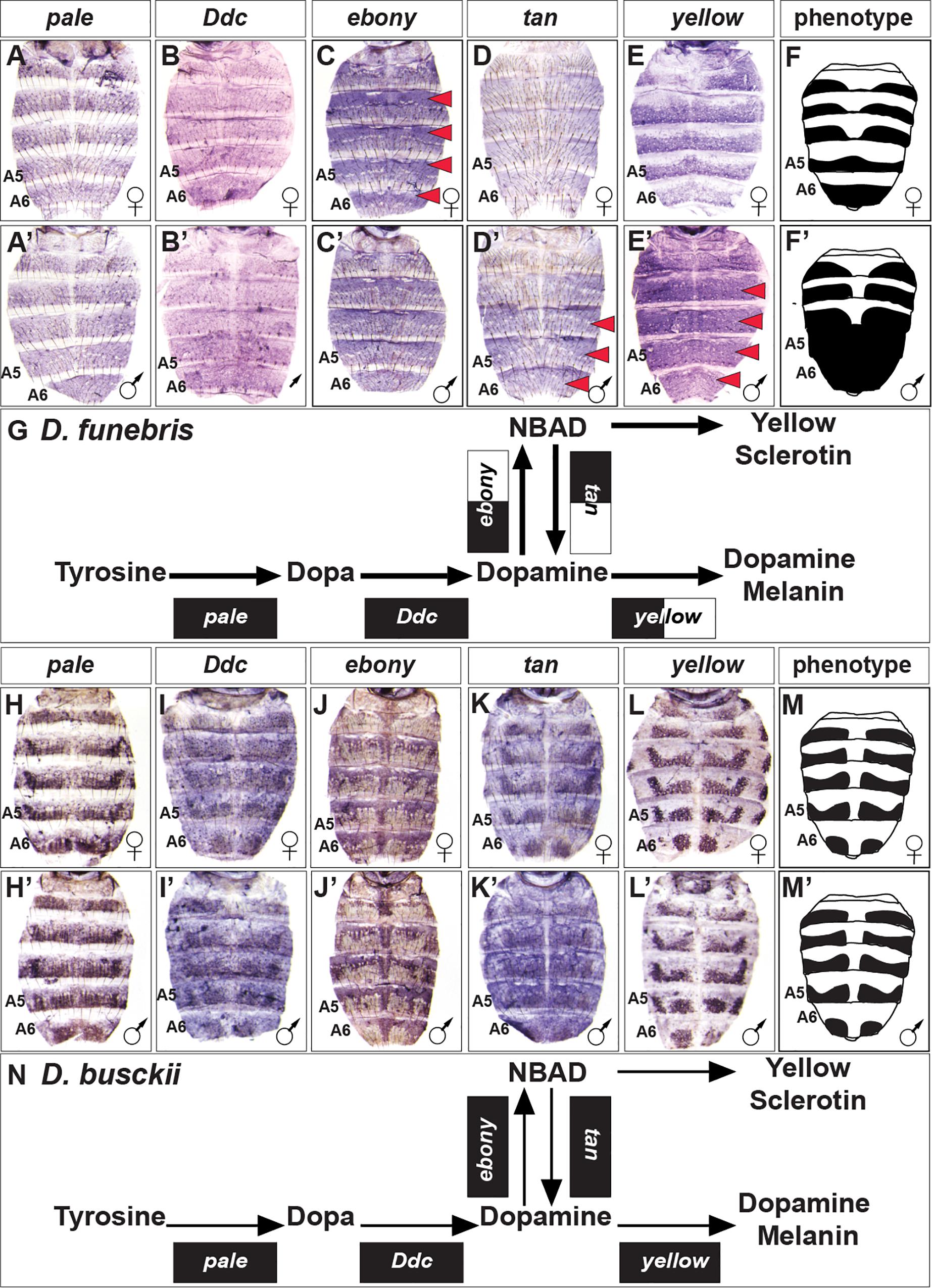
Figure 6. Contrast in the pigment metabolic pathway utilization between the dimorphic D. funebris and the monomorphic D. busckii. (A–E) Female and (A’–E’) male expressions of D. funebris pigment metabolic pathway genes, and (F,F’) cartoon representation of the pigmentation phenotype. (G) Summary of the D. funebris pathway use includes robust expression of all genes, with dimorphic expressions of ebony, tan, and yellow. (H–L) Female and (H’–L’) male expressions of D. busckii pigment metabolic pathway genes, and (M,M’) cartoon representation of the pigmentation phenotype. (N) Summary of the D. busckii pathway use includes robust and monomorphic expression of all genes. (A,A’,H,H’) pale, (B,B’,I,I’) Ddc, (C,C’,J,J’) ebony, (D,D’,K,K’) tan, and (E,E’,L,L’) yellow. Red arrowheads indicate robust patterns of dimorphic expression in the dorsal abdominal epidermis. (G,N) Black arrow thickness represents the level of expression for the underlying gene. Monomorphic gene expression is represented by the gene names being placed upon a black rectangle. Dimorphic gene expression is represented by the gene names being placed upon a black and white colored rectangle.
The patterns of pigmentation pathway deployment in the abdomen epidermis of these outgroup Drosophila species (Figures 6F,L) reinforce the impression that stereotypic patterns of gene expression evolved to mediate monomorphic and dimorphic tergite color patterns. We were curious whether such similarities extend to the level of transcription factors within this gene regulatory network. Thus, we examined the Bab1 transcription factor that plays an essential role in shaping dimorphism in D. melanogaster.
The sexually dimorphic expression of the Bab1 and Bab2 transcription factors is an essential feature of the GRN shaping the male-limited tergite pigmentation of D. melanogaster (Roeske et al., 2018). This dimorphism extends broadly among species of the melanogaster group, stimulating the interpretation that dimorphic Bab expression existed in the most recent common ancestor of this group (Kopp et al., 2000; Salomone et al., 2013). Here, we explored the expression of Bab1 in more distantly related species, including several with dimorphic tergite phenotypes, to investigate whether this is an ancestral feature of dimorphic pigmentation GRNs (Figures 7, 8, and Supplementary Figures S6–S17). To be consistent with a previous study, we first assessed Bab1 expression at the P14-P15i stage of pupal development (∼85–88 h after puparium formation or hAPF) (Salomone et al., 2013). During this stage, Bab1 expression is highly reduced in the dorsal epidermis of D. melanogaster males compared to females (Figures 7A’,B’ and Supplementary Figure S6). This time point is concurrent with the regulation of yellow, which is a direct Bab target (Roeske et al., 2018), and just after tan expression initiated. In contrast, Bab1 expression is monomorphic in D. affinis (Figures 7C’,D’ and Supplementary Figure S7), and D. pseudoobscura (Figures 7E’,F’ and Supplementary Figure S8). This suggested that despite the dimorphic pigmentation of D. affinis, dimorphism in pigmentation genes is achieved through a different regulatory mechanism.
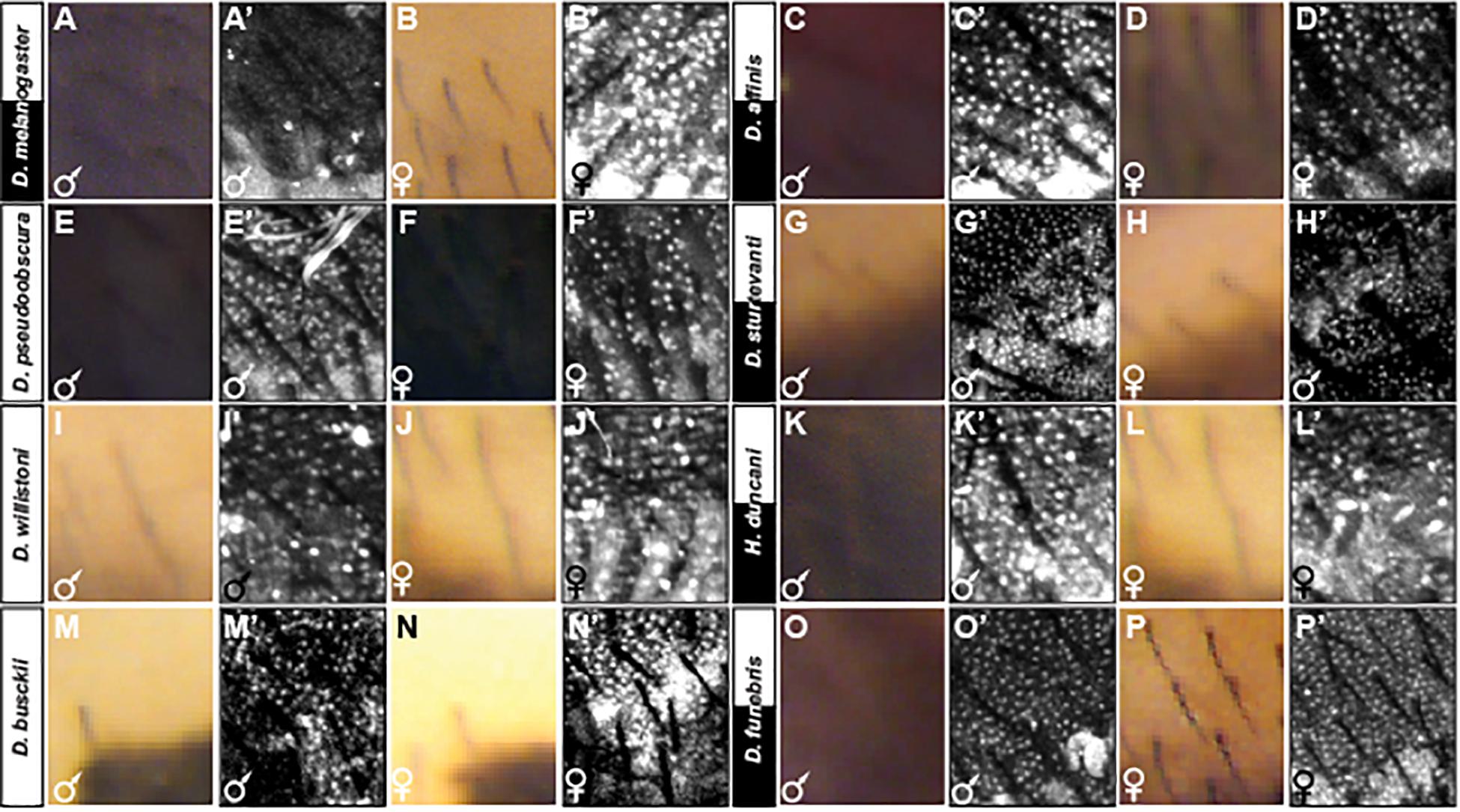
Figure 7. Adult pigmentation phenotypes and P14-P15i stage pupal Bab1 expression in a representative region of the A5 segment. (A) Male and (B) female tergite pigmentation, and (A’) male (B’) female Bab1 expression for D. melanogaster. (C) Male and (D) female tergite pigmentation, and (C’) male (D’) female Bab1 expression for D. affinis. (E) Male and (F) female tergite pigmentation, and (E’) male (F’) female Bab1 expression for D. pseudoobscura. (G) Male and (H) female tergite pigmentation, and (G’) male (H’) female Bab1 expression for D. sturtevanti. (I) Male and (J) female tergite pigmentation, and (I’) male (J’) female Bab1 expression for D. willistoni. (K) Male and (L) female tergite pigmentation, and (K’) male (L’) female Bab1 expression for H. duncani. (M) Male and (N) female tergite pigmentation, and (M’) male (N’) female Bab1 expression for D. busckii. (O) Male and (P) female tergite pigmentation, and (O’) male (P’) female Bab1 expression for D. funebris. Tergite pigmentation is dimorphic for D. melanogaster, D. affinis, D. sturtevanti, H. duncani, and D. funebris. Bab1 expression was dimorphic for D. melanogaster (little to no expression in males), while monomorphic for all other species examined here, including those that exhibit dimorphic pigmentation.
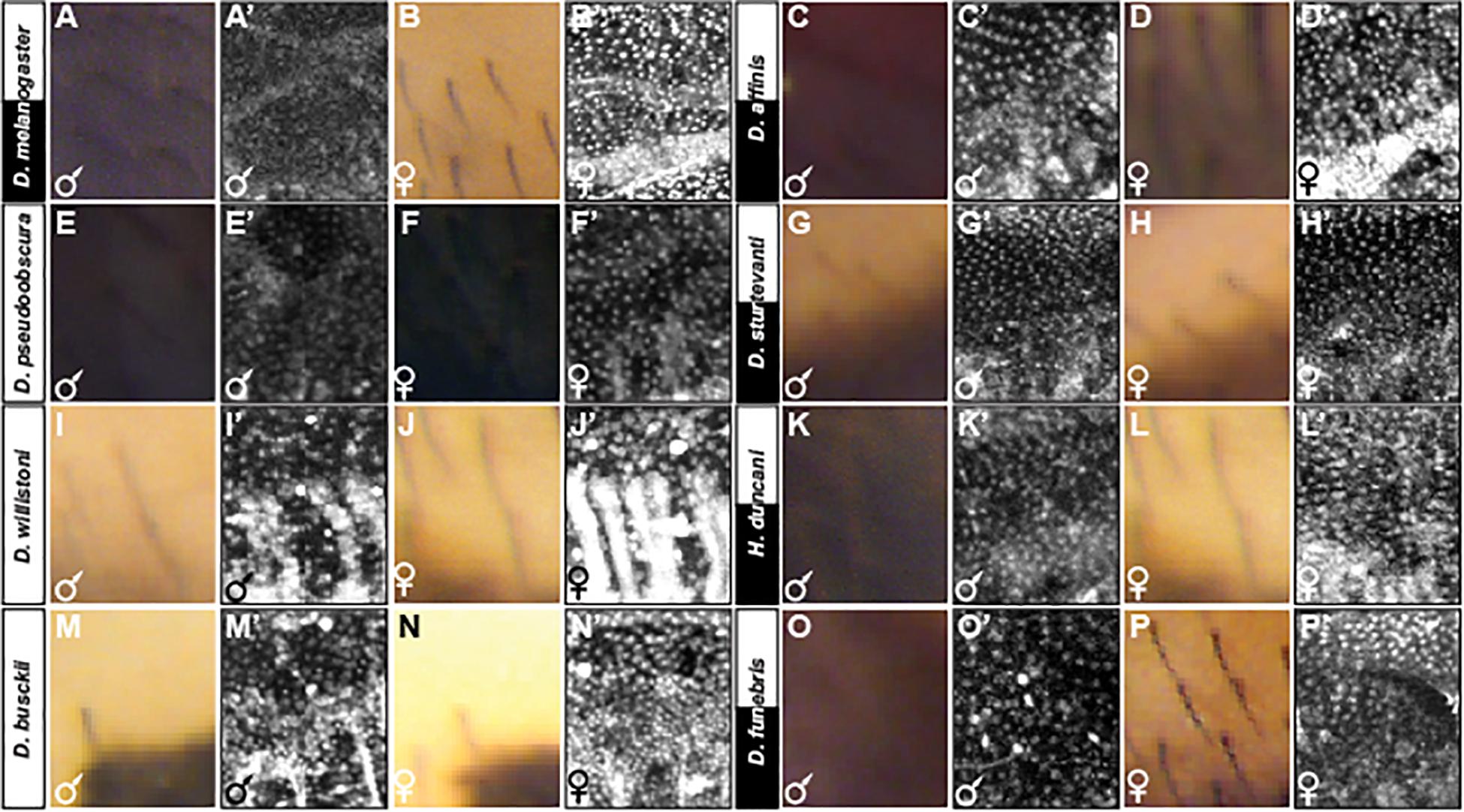
Figure 8. Adult pigmentation phenotypes and P10 stage pupal Bab1 expression in a representative region of the A5 segment. (A) Male and (B) female tergite pigmentation, and (A’) male (B’) female Bab1 expression for D. melanogaster. (C) Male and (D) female tergite pigmentation, and (C’) male (D’) female Bab1 expression for D. affinis. (E) Male and (F) female tergite pigmentation, and (E’) male (F’) female Bab1 expression for D. pseudoobscura. (G) Male and (H) female tergite pigmentation, and (G’) male (H’) female Bab1 expression for D. sturtevanti. (I) Male and (J) female tergite pigmentation, and (I’) male (J’) female Bab1 expression for D. willistoni. (K) Male and (L) female tergite pigmentation, and (K’) male (L’) female Bab1 expression for H. duncani. (M) Male and (N) female tergite pigmentation, and (M’) male (N’) female Bab1 expression for D. busckii. (O) Male and (P) female tergite pigmentation, and (O’) male (P’) female Bab1 expression for D. funebris. Tergite pigmentation is dimorphic for D. melanogaster, D. affinis, D. sturtevanti, H. duncani, and D. funebris. Bab1 expression was dimorphic for D. melanogaster (little to no expression in males), while monomorphic for all other species examined here, including those that exhibit dimorphic pigmentation.
We next analyzed Bab1 expression in species with either dimorphic or monomorphic tergite pigmentation that are more distantly related to D. melanogaster. D. sturtevanti of the saltans group is one such dimorphic species; however, Bab1 expression was found to be monomorphic (Figures 7G’,H’ and Supplementary Figure S9). This monomorphic expression is comparable to that observed for the monomorphically pigmented D. willistoni of the willistoni group (Figures 7I’,J’ and Supplementary Figure S10). While H. duncani is distantly related to D. melanogaster within Sophophora, this species possesses a comparable male-specific pattern of tergite pigmentation (Figure 2). For this species, Bab1 expression is monomorphic at the P14-P15i stage (Figures 7K’,L’ and Supplementary Figure S11).
To see whether dimorphic Bab expression might occur outside the Sophophora subgenus, we investigated Bab1 expression in the monomorphically pigmented D. busckii, and the dimorphically pigmented D. funebris (Figure 2). At the P14-15i developmental stage, monomorphic expression was observed in both species (Figures 7M’,N’,O’,P’ and Supplementary Figures S12, S13). These results suggest that dimorphic Bab expression is limited to the melanogaster species group at this stage which has been shown to be critical for pigment formation (Salomone et al., 2013).
We were concerned that the widespread observation of monomorphic Bab1 expression outside of the melanogaster group was due to the late developmental stage that we assessed. Thus, we investigated Bab1 expression at the P10 stage (Figure 8 and Supplementary Figures S6–S17), which corresponds to ∼65 hAPF in D. melanogaster. This is the stage that coincides with the initiation of male-specific yellow in D. melanogaster through the activity of a CRE that is directly repressed by Bab in females (Roeske et al., 2018), and when Bab expression has been shown to be relevant to the phenotype (Salomone et al., 2013). With the exception of D. melanogaster (Figures 8A’,B’ and Supplementary Figure S6) and perhaps D. pseudoobscura to a lesser extent (Figures 8E’,F’ and Supplementary Figure S8), Bab1 expression was found to be monomorphic in the other species studied here that possess dimorphic patterns of tergite pigmentation (Figures 8C’,D’,G’,H’,K’,L’,O’,P’, and Supplementary Figures S9, S11, and S13), as well for those with monomorphic patterns of tergite pigmentation (Figures 8I’,J’,M’,N’, and Supplementary Figures S10, S12). These dimorphic and monomorphic patterns of Bab1 expression were replicated in independent specimens (Supplementary Figures S14–S17). These results indicate that robust sexually dimorphic Bab expression is limited to the melanogaster species group and that a mild dimorphism may extend to some species of the most closely-related obscura species group (Figures 2, 8). However, monomorphism is broadly found across the Sophophora subgenus and Drosophila genus, indicating monomorphism as the ancestral state for Bab expression in the abdominal epidermis.
The existence of constantly recurring morphological characters within animal phylogenies raises a rarely mentioned, but important concern about our ability to infer whether such traits arose by independent gains or rampant patterns of loss from a common ancestral state. GRNs offer a unique perspective to distinguish these possibilities at multiple levels of organization and granularity: are the same genes expressed to produce the trait? If so, are they activated by the same CREs? And are homologous binding sites used to generate similar expression patterns? Here, we explored the gene expression patterns underlying sexually monomorphic and dimorphic patterns of abdominal tergite pigmentation across a phylogeny in an extensively studied trait that has repeatedly changed states. This revealed common changes associated with dimorphism, namely dimorphic patterns of ebony, tan, and yellow expression. In contrast, a critical sex-specific regulator of this trait in the D. melanogaster species group is notably absent in other observed instances of this trait. Combining these results with previous studies in this system, we discuss how the GRN perspective reveals the developmental basis for a trait to repeatedly flicker in and out of existence.
For traits in which patterned gene expression is an important feature of their development, analysis of these patterns can provide critical information concerning trait gain or loss. If completely different genes were deployed to generate dimorphic pigmentation patterns, this would support the independent convergence of these traits through separate genetic mechanisms (Stern, 2013). On the other hand, if the same genes are deployed, this could indicate ancestral homology coupled to loss or perhaps parallelism in which the same developmental mechanisms have been independently assembled multiple times. Our expression analyses of enzymes and their regulators in this system reveals a combination of these two outcomes.
Among the five enzyme-encoding genes we analyzed, we observed common themes in the deployment of this battery among dimorphic species. yellow and tan expression were upregulated in males, while ebony expression was reciprocally upregulated in females. pale and Ddc expression were generally monomorphic, presumably since Dopamine is a precursor for both yellow and black cuticle in males and females. The only exceptions were the dimorphic expression of Ddc in H. duncani and the underwhelming expression of tan in D. affinis. For H. duncani, this may reflect subtle differences in how the throughput of the pathway was arranged. In the case of D. affinis, this species has a dull color reminiscent of tan mutants (True et al., 2005), and may reflect differences relevant to generating its precise phenotype.
The patterning of the same enzymes in apparently separate instances of dimorphism is perhaps to be expected. These enzymes are certainly older than the genus Drosophila and encode proteins that perform the same enzymatic function in distantly related insects (Wright, 1987), including butterflies (Zhang et al., 2017) and the hemimetabolous milkweed bug Oncopeltus fasciatus (Liu et al., 2014, 2016). Thus, there is likely only a small number of potential paths by which a melanic trait could evolve at the enzymatic level. However, analysis of a key regulator in this system reveals a stark contrast.
While dozens of transcription factor genes have been implicated as being a part of the D. melanogaster GRN (Rogers et al., 2014), two factors, Abd-B and Bab are highly patterned and play particularly important and well-understood roles. The Bab proteins play a key role in regulating the dimorphic output of the D. melanogaster GRN (Figure 1B). Previously, we have shown Bab1 and Bab2 expression to be indistinguishable for D. melanogaster and related melanogaster group species in the abdominal epidermis, with expression virtually absent from the male epidermis during the latter half of pupal development (Salomone et al., 2013). The reduction of either protein results in masculinized pigmentation in females (Couderc et al., 2002), while ectopic expression of either protein feminizes the pigmentation of males (Kopp et al., 2000; Roeske et al., 2018). Previous work had suggested correlations of Bab expression with dimorphic pigmentation in instances outside of Sophophora (Gompel and Carroll, 2003). However, those studies were focused on very early stages that do not coincide with the physiologically relevant expression of Bab (Salomone et al., 2013). Thus, Bab was an excellent candidate regulator with which to evaluate homology in the production of this trait.
However, our results provide no such evidence for this repeated inclusion of Bab in other dimorphic GRNs. Rather our work in combination with a previous study indicates that dimorphic Bab expression evolved in the ancestry of the melanogaster species group, perhaps originating as early as the lineage in common between D. melanogaster and D. pseudoobscura. Thus, the dimorphic pigmentation for H. duncani and D. funebris, amongst other dimorphic species, were shaped by the origin of GRNs with another regulatory gene or genes shaping the sex-specific expression of the same pigmentation genes (Figure 9).
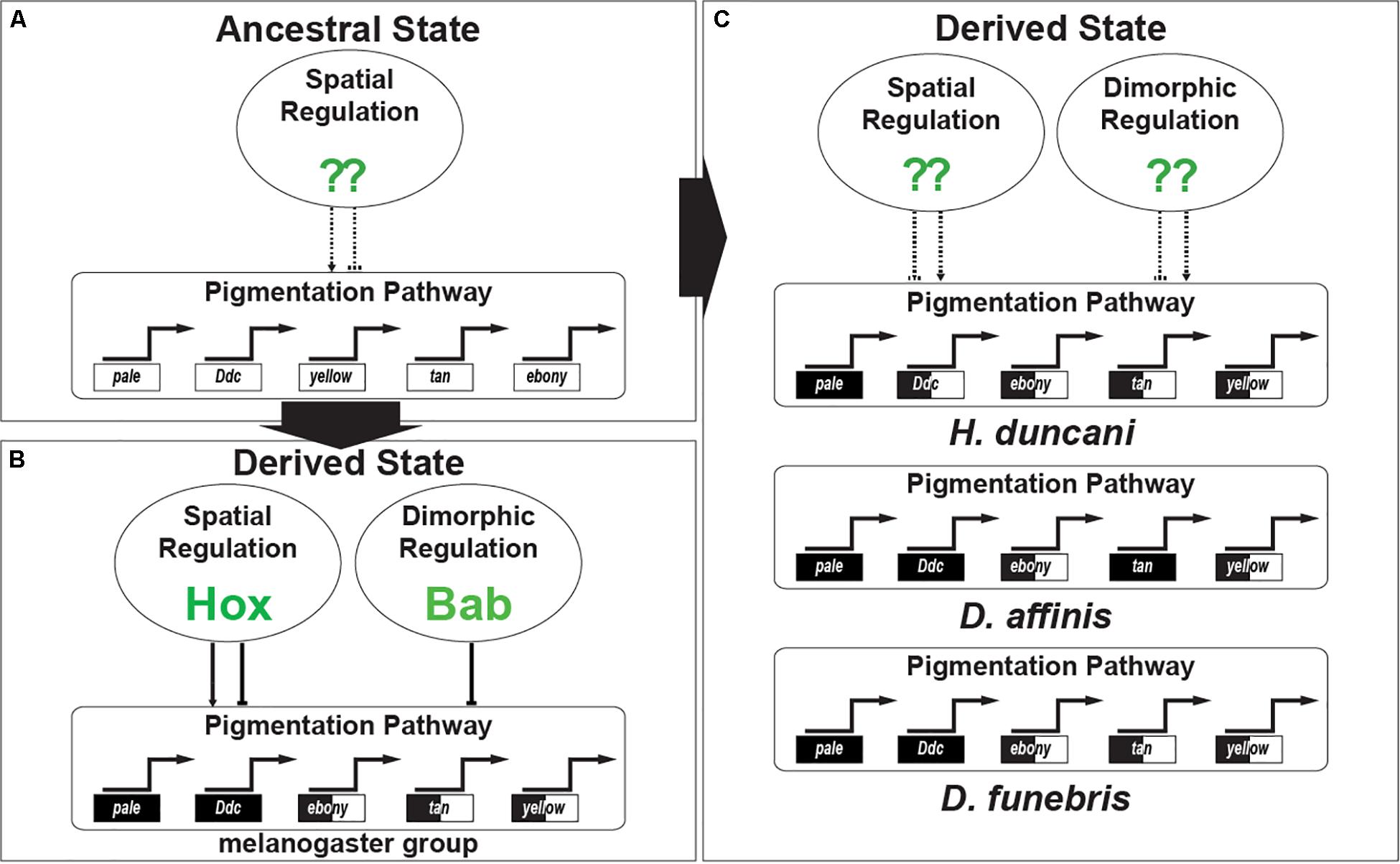
Figure 9. The convergent evolution of Gene Regulatory Networks responsible for sexually dimorphic tergite pigmentation. (A) Evidence supports an interpretation where monomorphic pigmentation was ancestral among fruit flies. In such an ancestor, the GRN’s pigmentation genes were under the regulatory control of transcription factors driving spatial patterns of expression. (B) The origin of dimorphic pigmentation for the melanogaster species group involved select pigmentation genes of the GRN adopting spatial regulation by the Hox proteins Abd-A and Abd-B, and sexually dimorphic regulation imparted by the Bab proteins. (C) Convergent gains of dimorphic pigmentation involved similar patterns of pigmentation gene expression being shaped by distinct transcription factors whose identity await identification.
While comparisons of gene expression can indicate whether the same genes underlie a recurring trait, analysis of the CREs which activate these gene expression patterns can provide much higher resolution concerning homology or homoplasy. If the same CRE drives the expression in two potentially parallel instances of a trait, it would strongly support loss from an ancestor that possessed the trait. On the other hand, if distinct CREs positioned in different parts of the gene are responsible, such data would support the hypothesis of parallelism. Combining the gene expression analysis presented here with previous work suggests that the CREs underlying recurring similar patterns of enzyme expression are unique and rapidly arise, while the apparatus that could mediate dimorphic Bab expression is quite old.
CREs for the patterned pigmentation genes yellow and tan appear to have evolved uniquely with this trait in the melanogaster subgroup (Camino et al., 2015). This included the integration of Hox genes as spatial patterning factors, such as direct activation of yellow through the gain of Abd-B binding sites in the yellow body element CRE (Jeong et al., 2006), and Abd-A and Abd-B regulating dimorphic tan expression through its CRE with male-specific activity (Camino et al., 2015). Previous work has shown that other abdominal CREs for yellow can rapidly arise within the phylogeny (Kalay and Wittkopp, 2010) and that fragments of its regulatory regions may contain “pre-enhancers” that are sufficient to drive patterns when tested in isolated reporter constructs (Kalay et al., 2019). Thus, the repeated GRN inclusion of yellow, tan, and ebony may indeed be due to convergence at the level of its CREs, a hypothesis that can now be tested more rigorously.
The CREs underlying the dimorphic expression of Bab are quite old, and very well could have been easily recruited to dimorphic pigmentation, but apparently were not. Dimorphic expression of Bab is mediated by the joint action of two CREs, one which drives a monomorphic pattern in anterior body segments, while a dimorphic element drives expression in female A5 and A6 segments (Williams et al., 2008). The inferred ancestral function of this element is to drive expression in the female genitalia, an activity that expanded the domain of its dimorphic activity to include the A5 and A6 segments in the melanogaster subgroup (Williams et al., 2008). The dimorphic element CRE and its Abd-B and DSX binding sites are conserved throughout the genus (Williams et al., 2008), and mutations in this CRE have impacts upon female pigmentation (Rogers et al., 2013). Thus, while Bab could easily have been recruited to mediate dimorphic pigmentation, its absence in these other networks is all the more surprising and suggests the existence of alternative ways to evolve the dimorphic regulation of pigmentation enzymes.
Ultimately, the question of trait gain or loss could be resolved at the level of individual transcription factor binding sites within CREs mediating recurrent traits. Our previous work on this topic, however, reveals how this question may nevertheless only weakly support homoplasy. We traced the binding of the Bab transcription factors to the yellow upstream regulatory region of D. melanogaster and found that only species closely related to D. melanogaster contain this binding site (Roeske et al., 2018). Our experiments supported an evolutionary scenario in which Bab binding evolved contemporaneously with the inferred origin of dimorphism in the lineage leading to the melanogaster species group. However, at the sequence level, the Bab-binding regions were not alignable outside of this group, and functional transgenic reporter assays confirmed that these distant relatives do not respond to Bab. For now, we believe that this is the best one can do to infer the origins of a transcription factor linkage within a GRN. This is because sequence divergence can erase traces of homology at the binding site level, while binding site turnover maintains ancestral functions without any trace of homology (Ludwig et al., 2000; Swanson et al., 2011). Thus, the absence of evidence supporting binding site conservation offers a poor readout for homoplasy. For this reason, we propose that analyses of expression patterns, coupled to functional assays of CRE activity or genetic tests of necessity represent the most fruitful ways to assess trait gain and loss within GRNs (Rebeiz and Williams, 2011).
It has been suggested that homoplasy and homology (loss) are two extremes on a continuum (Hall, 2007). In between these two extremes lies parallelism in which the similar traits flicker on and off through the deployment of the same developmental programs. This process has been posited to occur most often for mesoevolutionary comparisons that represent differences between closely related species (Abouheif, 2008). The work we describe here shows how such flickering may developmentally manifest, with rapid evolution at the extremities of networks, and dramatic differences in the internal architecture of the regulators deployed. Future investigations into these parallel pigmentation patterns and their GRNs should be prioritized to identify the regulatory gene or genes driving dimorphism. The outcomes of such investigations will inform whether there are any common features to the genes that were recruited to play a pivotal role in sexually dimorphic patterning, or whether any transcription factor will suffice.
The results here may also bear upon the repeated origins of other morphological traits. Specifically, the occurrence of hotspot genes that are predictable evolutionary targets of phenotype-modifying mutations are likely to be features of GRNs for the loss or diversification of homologous traits. This was shown for the repeated loss of trichomes on Drosophila larvae, and flowering time for Arabidopsis plants, where modifications occurred to the shavenbaby (Sucena et al., 2003; McGregor et al., 2007; Frankel et al., 2012) and frigida transcription factor genes repeatedly (Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Shindo et al., 2005; Stern, 2010). As for morphological novelties, the origin of their GRNs are likely to involve changes in the expression and function of transcription factors that are far less predictable.
The datasets generated for this study are available on request to the corresponding author.
JH and TW designed the project. JH, MW, RJ, and SG performed the experiments characterized the gene expression. JH performed all further experiments and analysis. RJ and MW took care of the fruit fly stocks. JH, MR, and TW analyzed and interpreted the data, and wrote the manuscript. All authors read and approved the submitted version of the manuscript.
RJ received support from a Dean’s Summer Fellowship from the University of Dayton, and SG and JH were supported by fellowships from the University of Dayton Graduate School. JH received support from a Graduate Research Fellowship from the National Science Foundation (DGE-1439647). MR and TW received support from a National Science Foundation grant (IOS-1555906). The funders had no role in data collection and analysis, study design, decision to publish, or manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Species stocks were purchased from the San Diego Drosophila Stock Center or provided by S.B. Carroll. Specimens and information on pigmentation phenotypes for Lordiphosa species were provided by Dr. Masanori J. Toda.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00080/full#supplementary-material
Abouheif, E. (2008). Parallelism as the pattern and process of mesoevolution. Evol. Dev. 10, 3–5. doi: 10.1111/j.1525-142X.2007.00208.x
Abràmoff, M. D., Hospitals, I., Magalhães, P. J., and Abràmoff, M. (2004). Image Processing with ImageJ. Biophotonics Int. 11, 36–42.
Arnoult, L., Su, K., Manoel, D., Minervino, C., Magrina, J., Gompel, N., et al. (2013). Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339, 1423–1426. doi: 10.1126/science.1233749
Ashburner, M., Golic, K. G., and Hawley, R. S. (2005). Drosophila: A Laboratory Handbook, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Blount, Z. D., Lenski, R. E., and Losos, J. B. (2018). Contingency and determinism in evolution: replaying life’s tape. Science 362:eaam5979. doi: 10.1126/science.aam5979
Bonn, S., and Furlong, E. E. M. (2008). cis-Regulatory networks during development: a view of Drosophila. Curr. Opin. Genet. Dev. 18, 513–520. doi: 10.1016/j.gde.2008.09.005
Camino, E. M., Butts, J. C., Ordway, A., Vellky, J. E., Rebeiz, M., and Williams, T. M. (2015). The evolutionary origination and diversification of a dimorphic gene regulatory network through parallel innovations in cis and trans. PLoS Genet. 11:e1005136. doi: 10.1371/journal.pgen.1005136
Conway Morris, S. (2003). Life’s Solution. Inevitable Humans in a Lonely Universe, 1st Edn. Cambridge: Cambridge University Press.
Couderc, J.-L., Godt, D., Zollman, S., Chen, J., Li, M., Tiong, S., et al. (2002). The bric à brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development 129, 2419–2433.
Cunningham, C. W., Omland, K. E., and Oakley, T. H. (1998). Reconstructing ancestral character states: a critical reappraisal. Trends Ecol. Evol. 13, 361–366. doi: 10.1016/s0169-5347(98)01382-2
De Magalhaes, L. E. (1956). Description of four new species of the “Saltans” Group Of “Drosophila” (Diptera). Rev. Bras. Bio. 16, 273–280.
Fartyal, R. S., Sati, P. C., Pradhan, S., and Kandpal, M. C. (2017). A review of the genus Lordiphosa Basden in India, with descriptions of four new species from the Himalayan region (Diptera. Drosophilidae). Zookeys 79, 49–79. doi: 10.3897/zookeys.688.12590
Frankel, N., Wang, S., and Stern, D. L. (2012). Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proc. Natl. Acad. Sci. US.A. 109, 20975–20979. doi: 10.1073/pnas.1207715109
Gao, J., Hu, Y., Toda, M. J., Katoh, T., and Tamura, K. (2011). Phylogenetic relationships between Sophophora and Lordiphosa, with proposition of a hypothesis on the vicariant divergences of tropical lineages between the Old and New Worlds in the family Drosophilidae. Mol. Phylogenet. Evol. 60, 98–107. doi: 10.1016/j.ympev.2011.04.012
Gazzani, S., Gendall, A. R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions 1 [w]. Plant Physiol. 132, 1107–1114. doi: 10.1104/pp.103.021212.Allelic
Gompel, N., and Carroll, S. B. (2003). Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424, 931–935. doi: 10.1038/nature01863.1
Grover, S., Williams, M. E., Kaiser, R., Hughes, J. T., Gresham, L., Rebeiz, M., et al. (2018). Augmentation of a wound response element accompanies the origin of a Hox-regulated Drosophila abdominal pigmentation trait. Dev. Biol. 441, 159–175. doi: 10.1016/j.ydbio.2018.07.001
Hall, B. K. (2007). Homoplasy and homology: dichotomy or continuum? J. Hum. Evol. 52, 473–479. doi: 10.1016/j.jhevol.2006.11.010
Hovemann, B. T., Ryseck, R. P., Walldorf, U., Stortkuhl, K. F., Dietzel, I. D., and Dessen, E. (1998). The Drosophila ebony gene is closely related to microbial peptide synthetases and shows specific cuticle and nervous system expression. Gene 221, 1–9. doi: 10.1016/S0378-1119(98)00440-5
Hu, Y., and Toda, M. J. (2001). Polyphyly of Lordiphosa and its relationships in Drosophilinae (Diptera: Drosophilidae). Syst. Entomol. 26, 15–31. doi: 10.1046/j.1365-3113.2001.00135.x
Jeong, S., Rebeiz, M., Andolfatto, P., Werner, T., True, J., and Carroll, S. B. (2008). The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132, 783–793. doi: 10.1016/j.cell.2008.01.014
Jeong, S., Rokas, A., and Carroll, S. B. (2006). Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387–1399. doi: 10.1016/j.cell.2006.04.043
Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of frigida, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–348.
Joy, J. B., Liang, R. H., Mccloskey, R. M., Nguyen, T., and Art, F. (2016). Ancestral reconstruction. PLoS Comput. Biol. 12:e1004763. doi: 10.1371/journal.pcbi.1004763
Kalay, G., Lachowiec, J., Rosas, U., Dome, M. R., and Wittkopp, P. J. (2019). Redundant and cryptic enhancer activities of the drosophila yellow gene. Genetics 212, 343–360. doi: 10.1534/genetics.119.301985
Kalay, G., and Wittkopp, P. J. (2010). Nomadic enhancers: tissue-specific cis-regulatory elements of yellow have divergent genomic positions among Drosophila species. PLoS Genet. 6:e1001222. doi: 10.1371/journal.pgen.1001222
Katoh, T. K., Zhang, G., Toda, M. J., Zhang, W.-X., and Gau, J.-J. (2018). The Lordiphosa denticeps species group (Diptera: Drosophilidae) in China, with redescriptions of four known species and descriptions of nine new species. Zootaxa 4471, 37–75. doi: 10.11646/zootaxa.4471.1.2
Kopp, A., Duncan, I., and Carroll, S. B. (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559. doi: 10.1038/35046017
Le Corre, V., Roux, F., and Reboud, X. (2002). DNA Polymorphism at the FRIGIDA Gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol. Biol. Evol. 19, 1261–1271. doi: 10.1093/oxfordjournals.molbev.a004187
Liu, J., Lemonds, T. R., Marden, J. H., and Popadić, A. (2016). A pathway analysis of melanin patterning in a hemimetabolous insect. Genetics 203, 403–413. doi: 10.1534/genetics.115.186684
Liu, J., Lemonds, T. R., and Popadić, A. (2014). The genetic control of aposematic black pigmentation in hemimetabolous insects: insights from Oncopeltus fasciatus. Evol. Dev. 277, 270–277. doi: 10.1111/ede.12090
Losos, J. B. (2017). Improbable Destinies: Fate, Chance, and the Future of Evolution, 1st Edn. New York, NY: Riverhead Books.
Ludwig, M. Z., Bergman, C., Patel, N. H., and Kreitman, M. (2000). Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564–567. doi: 10.1038/35000615
Markow, T. A., and O’Grady, P. M. (2006). Drosophila: A Guide to Species Identification and Use. Cambridge, MA: Academic Press.
McGregor, A. P., Orgogozo, V., Delon, I., Zanet, J., Srinivasan, D. G., Payre, F., et al. (2007). Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448, 587–590. doi: 10.1038/nature05988
Nater, H. (1953). Vergleichend-morphologische Unter- suchung des ausseren Geschlechtsapparates innerhalb der Gattung Drosophila. Zool. Jahrbucher 81, 437–486.
Nater, H. (1950). Der Samenpumpen-Sklerit von Drosophila als taxonomisches Merkmal. Arch. der Jul. Klaus-Stiftung fur̈ Verebungsforschung. Sozialanthro- pologie und Rassenhygiene 25, 623–625.
Peter, I. S., and Davidson, E. H. (2011). Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985. doi: 10.1016/j.cell.2011.02.017
Pham, T., Day, S. M., Glassford, W. J., Williams, T. M., and Rebeiz, M. (2017). The evolutionary origination of a novel expression pattern through an extreme heterochronic shift. Evol. Dev. 19, 43–55. doi: 10.1111/ede.12215
Rebeiz, M., Patel, N. H., and Hinman, V. F. (2015). Unraveling the tangled skein: the evolution of transcriptional regulatory networks in development. Annu. Rev. Genomics Hum. Genet. 16, 103–131. doi: 10.1146/annurev-genom-091212-153423
Rebeiz, M., Pool, J. E., Kassner, V. A., Aquadro, C. F., and Carroll, S. B. (2009). Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663–1667. doi: 10.1126/science.1178357
Rebeiz, M., and Williams, T. M. (2011). “experimental approaches to evaluate the contributions of candidate Cis- regulatory mutations to phenotypic evolution,” in Methods in Molecular Biology, eds V. Orgogozo and M. V. Rockman (Totowa, NJ: Humana Press), 351–375. doi: 10.1007/978-1-61779-228-1_21
Rebeiz, M., and Williams, T. M. (2017). Using Drosophila pigmentation traits to study the mechanisms of cis-regulatory evolution. Curr. Opin. Insect Sci. 19, 1–7. doi: 10.1016/j.cois.2016.10.002
Roeske, M. J., Camino, E. M., Grover, S., Rebeiz, M., and Williams, T. M. (2018). Cis-regulatory evolution integrated the bric-a-brac transcription factors into a novel fruit fly gene regulatory network. Elife 7, 1–28. doi: 10.7554/eLife.32273
Rogers, W. A., Grover, S., Stringer, S. J., Parks, J., Rebeiz, M., and Williams, T. M. (2014). A survey of the trans-regulatory landscape for Drosophila melanogaster abdominal pigmentation. Dev. Biol. 385, 417–432. doi: 10.1016/j.ydbio.2013.11.013
Rogers, W. A., Salomone, J. R., Tacy, D. J., Camino, E. M., Davis, K. A., Rebeiz, M., et al. (2013). Recurrent modification of a conserved Cis-Regulatory element underlies fruit fly pigmentation diversity. PLoS Genet. 9:e1003740. doi: 10.1371/journal.pgen.1003740
Salomone, J. R., Rogers, W. A., Rebeiz, M., and Williams, T. M. (2013). The evolution of Bab paralog expression and abdominal pigmentation among Sophophora fruit fly species. Evol. Dev. 15, 442–457. doi: 10.1111/ede.12053
Shindo, C., Aranzana, M. J., Lister, C., Baxter, C., Nicholls, C., Nordborg, M., et al. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138, 1163–1173. doi: 10.1104/pp.105.061309.1
Stern, D. L. (2010). Evolution, Development, & the Predictable Genome, 1st Edn. Greenwood Village: Roberts & Company Publishers.
Stern, D. L. (2013). The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764. doi: 10.1038/nrg3483
Sucena, E., Delon, I., Jones, I., Payre, F., and Stern, D. L. (2003). Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424, 935–938. doi: 10.1038/nature01768
Swanson, C. I., Schwimmer, D. B., and Barolo, S. (2011). Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr. Biol. 21, 1186–1196. doi: 10.1016/j.cub.2011.05.056
Throckmorton, L. H. (1962). The problem of phylogeny in the genus Drosophila. Univ. Texas Publ. 6205, 207–343. doi: 10.1016/j.ympev.2011.04.012
True, J. R., Yeh, S.-D., Hovemann, B. T., Kemme, T., Meinertzhagen, I. A., Edwards, T. N., et al. (2005). Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 1:e63. doi: 10.1371/journal.pgen.0010063
van der Linde, K., Houle, D., Spicer, G. S., and Steppan, S. J. (2010). A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet. Res. 92, 25–38. doi: 10.1017/S001667231000008X
Wagner, G. P. (2016). What Is “Homology Thinking” and what Is it for? J. Exp. Zool. 326B, 3–8. doi: 10.1002/jez.b.22656
Wake, D. B., Wake, M. H., and Specht, C. D. (2011). Homoplasy: from detecting pattern and mechanism of evolution. Science 331, 1032–1035. doi: 10.1126/science.1188545
Werner, T., Koshikawa, S., Williams, T. M., and Carroll, S. B. (2010). Generation of a novel wing colour pattern by the Wingless morphogen. Nature 464, 1143–1148. doi: 10.1038/nature08896
Wiens, J. J. (2001). Widespread loss of sexually selected traits: how the peacock lost its spots. Trends Ecol. Evol. 16, 517–523. doi: 10.1016/s0169-5347(01)02217-0
Williams, T. M., Selegue, J. E., Werner, T., Gompel, N., Kopp, A., and Carroll, S. B. (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623. doi: 10.1016/j.cell.2008.06.052
Wittkopp, P. J., Carroll, S. B., and Kopp, A. (2003). Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 19, 495–504. doi: 10.1016/S0168-9525(03)00194-X
Wright, T. R. F. (1987). “The Genetics of Biogenic Amine Metabolism, Sclerotization, and Melanization in Drosophila melanogaster,” in Advances in Genetics, eds J. G. Scandalios and E. W. Caspari (San Diego, CA: Harcourt Brace Jovanovich), 127–222. doi: 10.1016/s0065-2660(08)60008-5
Zanini, R., Deprá, M., and Lúcia, V. (2015). Can sibling species of the Drosophila willistoni subgroup be recognized through combined microscopy techniques? Rev. Bras. Entomol. 59, 323–331. doi: 10.1016/j.rbe.2015.09.006
Keywords: Drosophila, pigmentation, gene regulatory network (GRN), evo-devo (evolution and development), morphological evolution, gene expression, homoplasy, homology
Citation: Hughes JT, Williams ME, Johnson R, Grover S, Rebeiz M and Williams TM (2020) Gene Regulatory Network Homoplasy Underlies Recurrent Sexually Dimorphic Fruit Fly Pigmentation. Front. Ecol. Evol. 8:80. doi: 10.3389/fevo.2020.00080
Received: 13 December 2019; Accepted: 12 March 2020;
Published: 26 March 2020.
Edited by:
Ricardo Mallarino, Princeton University, United StatesReviewed by:
Élio Sucena, Universidade de Lisboa, PortugalCopyright © 2020 Hughes, Williams, Johnson, Grover, Rebeiz and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark Rebeiz, cmViZWl6QHBpdHQuZWR1; Thomas M. Williams, dHdpbGxpYW1zMkB1ZGF5dG9uLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.