
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 18 March 2020
Sec. Chemical Ecology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00063
Insect antennae are important olfactory organs that house high concentrations of odorant-binding proteins (OBPs) in the sensillum lymph. Previous studies in other insects have shown that OBPs play important roles in transporting odorants and enhancing the sensitivity of the olfactory system. However, the functions of OBPs in the oriental fruit fly, Bactrocera dorsalis, especially those specifically expressed in antennae, have not been fully elucidated. In this study, cDNA libraries were constructed from both the male and female antennal transcriptome, and twenty OBPs were identified in total. The expression profiles of these OBPs were examined in the adult antenna, head, thorax, leg, and abdomen of both sexes. Seven of the identified OBP genes had significantly higher expression in both the male and female antennae than in other tissues, while the transcript levels of the remaining OBPs varied across different tissues. Regarding the function of antenna-specific OBPs, we targeted Bdorsobp2 as a representative for further RNA interference (RNAi) and identified via electrophysiology a decrease in detection of a potential species-specific a potent attractant, methyl eugenol. Moreover, subsequent behavioral assay data showed that the behavioral response of B. dorsalis toward this odorant decreased when Bdorobp2 was silenced with injection of double-stranded RNA (dsRNA). Combined, these results support our initial hypothesis that antennae-specific OBPs are of critical importance for insect odorant detection, sensitivity, and behavior.
Antennae are the most important olfactory organs for insects to detect chemosensory compounds from the environment and to regulate their behaviors, including sexual attraction, mating or courtship, aggregation, host plant orientation, and the selection of oviposition sites. These semiochemicals enter the antennae via pores on the olfactory sensilla, which contain high concentrations of odorant-binding proteins (OBPs) in the sensillum lymph. It has been hypothesized that these OBPs in turn bind odorants from the environment and efficiently shuttle specific semiochemicals to the olfactory receptors (ORs), perhaps by changing their three-dimensional structure. In addition, it has been shown that OBPs can enhance the sensitivity of insects toward particular odorants (Leal, 2013). Because of the important roles they play in insect olfaction, OBPs have been thoroughly studied in various insect orders, including Lepidoptera, Coleoptera, and Diptera (especially the genus Drosophila) (Leal, 2005; Pelosi et al., 2006). Moreover, both physiological and behavioral evidence has shown that insect OBPs were important for odorant recognition and sensitivity. These characteristics make OBPs potential candidates for insect pest management (IPM), and provide potential applications that offer more environmentally friendly solutions than insecticides.
Antennae-specific OBPs are intimately involved in the olfactory system, and are responsible for the detection of pheromones and/or host plant volatiles (Maida et al., 1993; Große-Wilde et al., 2006; Sun et al., 2013). It has also been found that OBPs expressed in organs other than the antenna are involved in gustatory perception (Jeong et al., 2013). Male insects are able to find and copulate with potential females with the aid of pheromone-binding proteins (PBPs; OBPs specifically expressed in the male antennae that detect pheromones) (Chen et al., 2018), while the OBPs that are expressed in both sexes or expressed predominantly in female antennae are general OBPs that bind with more diverse odorants, which function in other behaviors such as oviposition and host plant selection (Leal, 2005; Choo et al., 2018; Ju et al., 2018; Li et al., 2018). In Drosophila melanogaster, the antennae-specific OBP, LUSH (DmelOBP69), is required for the pheromone detection of cis-vaccenyl acetate (cVA) (Kim et al., 1998). DsechOBP57d and DsechOBP57e, which are expressed in the tarsi, have been reported to be able to affect the taste perception and preference of D. sechellia (Matsuo et al., 2007; Harada et al., 2008). Thus, the investigation of tissue-specific expression profiles of OBPs has been successful in regards to building a link between these proteins and their potential chemosensory functions for the insect.
The oriental fruit fly, Bactrocera dorsalis, is a polyphagous pest in several countries, including China (Hsu et al., 2004; Siderhurst and Jang, 2006; Jin et al., 2011). This pest can inflict severe damage across a range of economically important tropical and subtropical fruit plants, including guava (Psidium guajava), mango (Mangifera indica), and citrus (Citrus grandis) (Steiner et al., 1965; Jin et al., 2011; Jayanthi et al., 2012). Unfortunately, highly insecticide-resistant populations have emerged in both China and Pakistan (Jin et al., 2011; Xie et al., 2013; Khan and Akram, 2018), which reduces pesticide efficiency in controlling this agricultural pest. As such, there is an urgent need to find an alternative, and more environmentally friendly method for control, perhaps including mass trapping using semiochemicals. The commercially available traps with the species-specific odorant, methyl eugenol, have been used to monitor and eradicate this pest for many years (Metcalf et al., 1975; Vargas et al., 2000). However, there has been no prior research on the OBPs' expression profiles that are necessary to clarify their function in olfaction, especially in regards to the detection of this important compound. Previously, several OBPs were identified in B. dorsalis, but none of them proved to be involved directly in olfaction (Zheng et al., 2013; Wu et al., 2015; Chen et al., 2019).
Here we hypothesize that B. dorsalis antennae-specific OBPs are likely involved in the perception of methyl eugenol and other semiochemicals. To build a classification system based on the expression profile of the OBPs in B. dorsalis, and to deduce their associated function, we first identified the full repertoire of OBPs in the adult antennae, and then investigated their expression profiles across various tissues within several different developmental stages. With the hypothesis that the antennae-specific OBPs are involved in olfaction, one classical OBP, Bdorsobp2, which is specifically expressed in the antennae, was selected to study its function using a combination of RNA interference (RNAi) and electrophysiological experiments. Here we confirm that this OBP is necessary for the detection and high sensitivity toward this species-specific odorant (i.e., methyl eugenol). Moreover, this study improves our understanding of B. dorsalis OBPs and their potential ecological functions, which will continue to help researchers to develop more efficient and environmentally friendly strategies for the control of this destructive insect species.
The B. dorsalis used in our experiments were collected from the Fujian Province, China and were maintained with artificial diet in a climate-controlled chamber at 28 ± 1°C under a constant photoperiod (L:D = 14:10 h) and relative humidity (75%). The 5-day old male and female adults were separately collected in order to extract the total RNA from both male and female antenna, head (without antennae), thorax, abdomen, legs, as well as different developmental stages, including the egg, larva, and pupa. Extractions were conducted with Trizol (Invitrogen, CA, USA) following the manufacturer's instructions. The extracted RNA samples were examined using 2% agarose electrophoresis and spectrophotometry (NanoVue, GE Healthcare Bio-Sciences, Uppsala, Sweden). The single-strand cDNA templates were synthesized with the extracted RNA using the First Strand cDNA Synthesis Kit (Takara Biotechnology (Dalian) Co., Ltd., Dalian, China), and the synthesized cDNA templates were used as a template in quantitative polymerase chain reaction (qPCR) experiments.
Twenty OBPs were again identified as in our previous research (Liu et al., 2016). The phylogenetic tree of these OBPs was constructed using neighbor joining and Poisson correction of distance, with the aid of MEGA 5.0 software (default settings and 1,000 bootstrap replicates), based on its amino acid sequence aligned by Clustal W.
To determine the OBP expression profile in different tissues, the RNA of both male and female antennae, heads, thoraxes, legs, abdomens, and different life stages, including the egg (E), larva (L), pupae (P), and adults (female:male = 1:1), were extracted to synthesize the cDNA. These procedures were followed by qPCR, which was performed in a Stratagene Mx3000P thermocycler (Agilent Technologies, Inc., Wilmington, NC, USA). Primer pairs were designed from the nucleotide sequences with Primer3 web software (Version 4.0.0, http://primer3.ut.ee), and the sequence of primers is listed in Supplementary Material. The reaction mixture included 10 μl iQTM SYBR Green Supermix (BIO-RAD, Hercules, CA, USA), 1 μl cDNA templates, 1μl of each primer, and 7 μl of double distilled water. The amplification reaction program was as follows: 2 min of initial denaturation at 95°C, 40 cycles for 5 s at 95°C, for 30 s at 60°C, and a melting cycle (from 60 to 95°C) was included at the end. The reactions were performed with three technical repeats and three biological repeats for each OBP gene. The expression level of each OBP gene was calculated according to the 2–ΔΔCT method, with α-tubulin as the control gene. The expression differences between tissues were examined with Tukey's honest significant difference test using SPSS software (Version 16).
The synthesized cDNA was used as the template for dsRNA synthesis. A fragment of Bdorobp2 from B. dorsalis was amplified by PCR using cDNA templates. The dsBdorobp2 was generated with an Ambion® MEGAscript® RNAi Kit (Ambion, Carlsbad, CA, USA) using specific primers containing a T7 promotor sequence (Bdorobp2 Forward: 5′- CGACTCACTATAGGGTTCACGAAGACGAGCTGTTG-3′ Bdorobp2 Reverse: 5′-CGACTCACTATAGGGTAATGCTTTGGATCCGCTTC-3′), according to the manufacturer's instructions. The dsBdorobp2 was cleaned and dissolved in RNase-free water. Lastly, the concentration was determined using NanoDrop One Microvolume UV-Vis spectrophotometers (Thermo Fisher Scientific, Madison, WI, USA).
To examine the gene silence efficacy, the dsRNA (4.2 μg/μl × 350 nl) was injected into the 3-day-old B. dorsalis adult via the abdomen with a glass capillary mounted on a Nanoject II Auto-Nanoliter Injector (Drummond Scientific, Broomall, PA, USA). Ten heads were dissected from the dsRNA-treated adult to extract the RNA 72 h after the injection. The control RNA was extracted from the green fluorescent protein (GFP)-injected B. dorsalis. The generated cDNA (synthesized as described previously) was used as the template for qPCR with Bdorobp2-specific primer pairs. The mRNA abundance was determined, averaged, and normalized with α-tubulin as the control gene. Six replicates were examined, and the expression level changes were determined using a t-test (SPSS, Version 16).
To explore the Bdorobp2 function in olfaction, the dsRNA-injected male and female adults (which were 3 days old; n = 6) were separately subjected to electroantennography (EAG), and recorded 3 days after the dsRNA injection. The reported B. dorslias attractant, methyl eugenol, was diluted using hexane to produce solutions of 0.01, 0.1, and 1% (v:v) concentrations. The EAG setup was similar to previously described procedures, with a few adjustments (Liu et al., 2010). The antennae were excised from the most distal segment of the antennae and connected to the recording electrode, which was filled with 0.1 M KCl and 0.5% polyvinylpyrrolidone solution; the reference electrode, filled with the same saline solution as the recording glass capillary, was inserted into the head of the insect from the occipital opening. A stimulus controller, CS-55 (Syntech, Kirchzarten, Germany), was used to produce a 2.2 mL/s airflow containing the attractant (a constant charcoal-filtered humidified airflow at 2.0mL/s and the stimulus pulse of 0.2 mL/s) was exhausted from the 5 ml polypropylene syringe. A time gap of 1 min was allowed between each response stimulation. The EAG signals were recorded and analyzed with EAG 2000 software (Syntech). The control insects were injected with the same volume of GFP solution, and tested accordingly. Six total replicates were tested, and the responses were analyzed with the Student's t-test using SPSS (Version 16).
In total, 20 previously identified OBP candidate sequences were obtained from the B. dorsalis antennae transcriptome, including 16 sequences that had the full length of open reading frames (ORFs) and the hallmarks of the classic OBP subfamily: a signal peptide and six conserve cysteine residues (Figure 1). Based on the phylogenetic tree analyses, the identified OBP genes were classified into two subgroups. Subgroup I included Bdorobp2, Bdorobp11, Bdorobp12, Bdorobp13, Bdorobp14, Bdorobp18, Bdorobp19, Bdorobp21, Bdorobp22, Bdorobp23, Bdorobp24, Bdorobp25, Bdorobp26, and Bdorpbprp, while subgroup II included Bdorobp1, Bdorobp7, Bdorobp10, Bdorobp17, Bdorobp20, and Bdorobp27 (Figure 2).

Figure 1. Alignment of deduced amino acid sequences of B. dorsalis OBP members. The alignment was performed with the Clustal X 1.80 tool, and the homologous areas were marked with the Boxshade 3.21 software.
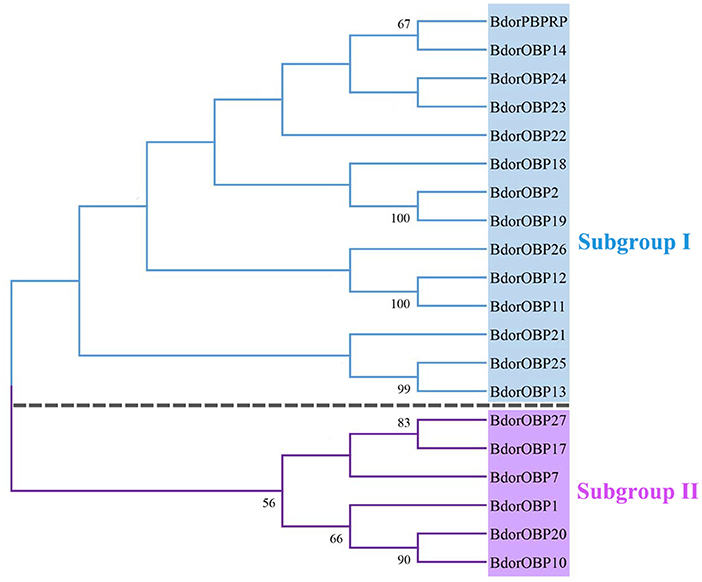
Figure 2. Phylogenetic relationships of the OBP proteins. Unrooted phylogenetic tree of OBP protein sequences from B. dorsalis constructed with the MEGA 5.0 tool. The identified OBPs are classified into subgroup I and subgroup II based on the sequences. The bootstrap values above 50 are shown.
Starting with the hypothesis that OBPs are vital for insect olfaction, and should be abundantly or exclusively expressed in olfactory tissues, the transcript levels of these candidate OBP genes were examined in different tissues, including the antennae, heads, thoraxes, legs, and abdomens of both male and female adults by qPCR experiments. The results showed that there were two expression patterns of these OBPs (Figure 3). The subgroup I includes seven antenna-specific genes, Bdorobp2, Bdorobp11, Bdorobp12, Bdorobp13, Bdorobp20, Bdorobp25, and Bdorobp26. Among these genes, the Bdorobp20 gene was more highly expressed in female antennae than in male antennae, in contrast with Bdorobp25, which was more highly expressed in male antennae than in female antennae; the remaining five OBP genes were highly expressed in both male and female antennae (Figure 3).
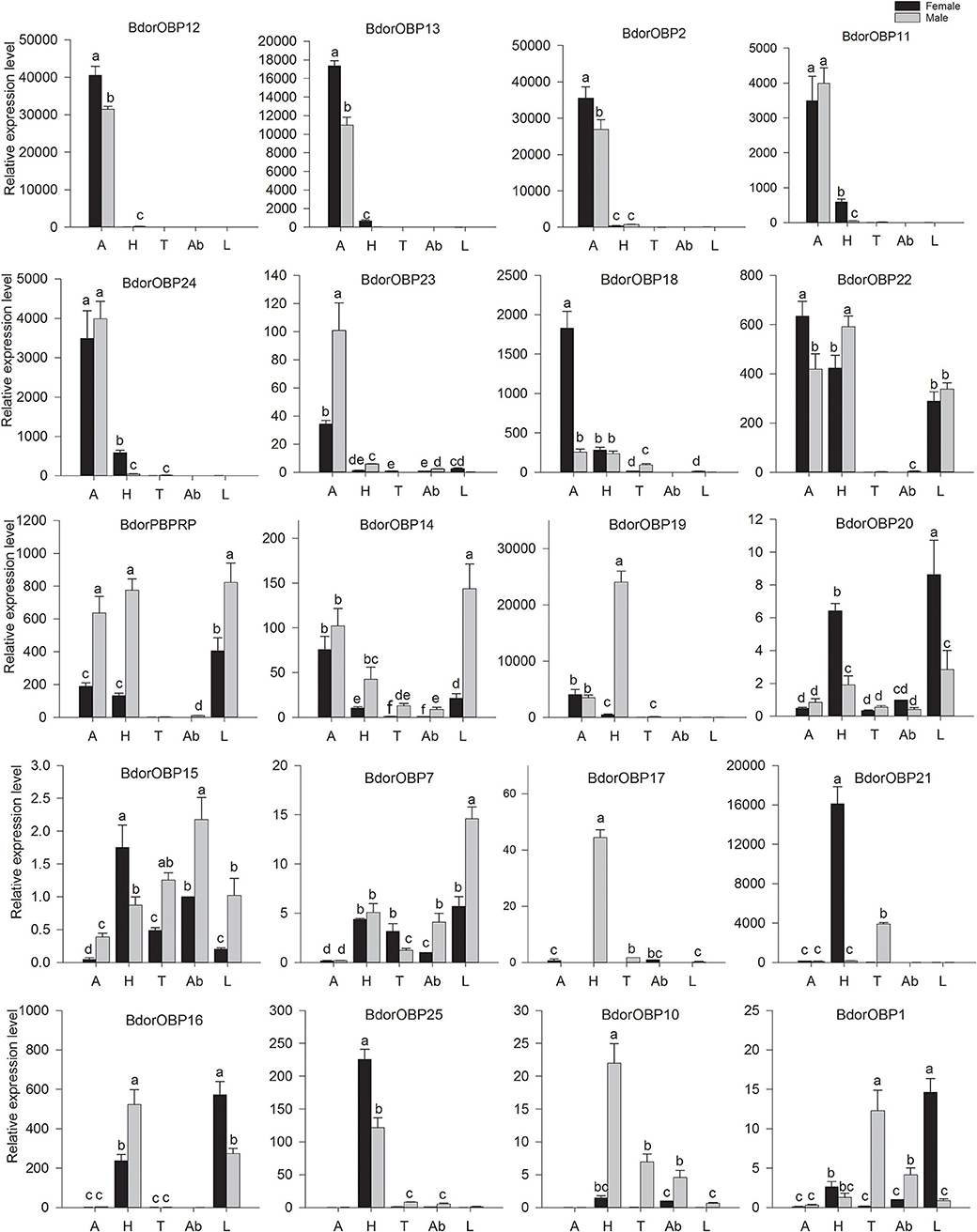
Figure 3. Expression profile of B. dorsalis OBPs in different tissues. Total RNA was separately extracted from both male and female antennae (A), heads (H), thoraxes (T), abdomens (Ab), and legs (L). The black rectangles show the expression level of the OBPs in male tissues, and the gray rectangles show the expression level of the OBPs in female tissues. The bars represent mean ± standard deviation (SD), and the letters indicate statistically significant differences between the groups (Tukey's honest significant difference test).
The subgroup II includes Bdorobp1, Bdorobp7, Bdorobp10, Bdorobp14, Bdorobp17, Bdorobp18, Bdorobp19, Bdorobp21, Bdorobp22, Bdorobp23, Bdorobp24, Bdorobp27, and Bdorpbprp were expressed mainly (if not exclusively) in non-olfactory tissues (i.e., heads without antennae, thoraxes, legs, and abdomens) rather than in olfactory tissues (e.g., the antennae). Among these genes, three (Bdorobp24, Bdorpbprp, and Bdorobp14) were more highly expressed in the heads, thoraxes, and legs than in other tissues. Three additional OBP genes (Bdorobp17, Bdorobp21, and Bdorobp22) had lower transcript levels in the antennae than in other tissues. Bdorobp27 was only expressed in the heads, and was more highly expressed in the male head than in the female. Bdorobp19 had an exclusively higher transcript level in male heads than in any of the other tissues examined (Figure 3).
The expression levels of the candidate OBP genes were also examined in different developmental stages, including eggs, larvae, pupae, and adults. The results showed that the expression levels of the tested candidate OBP genes varied in different developmental stages (Figure 4). These 17 candidate OBP genes (including Bdorobp1, Bdorobp2, Bdorobp7, Bdorobp10, Bdorobp12, Bdorobp13, Bdorobp14, Bdorobp24, Bdorobp18, Bdorobp20, and Bdorpbprp) were expressed mainly in the adult stage. However, some of these adult abundant genes (Bdorobp1, Bdorobp7, Bdorobp20, and Bdorobp25) were also weakly detected in other developmental stages, including eggs, larvae, and pupae. For example, the Bdorobp23 and Bdorobp27 genes were detected in all developmental stages, including with high transcript levels in the adult stage. In addition, the Bdorobp21 gene was highly expressed in both the larval and adult stages (Figure 4).
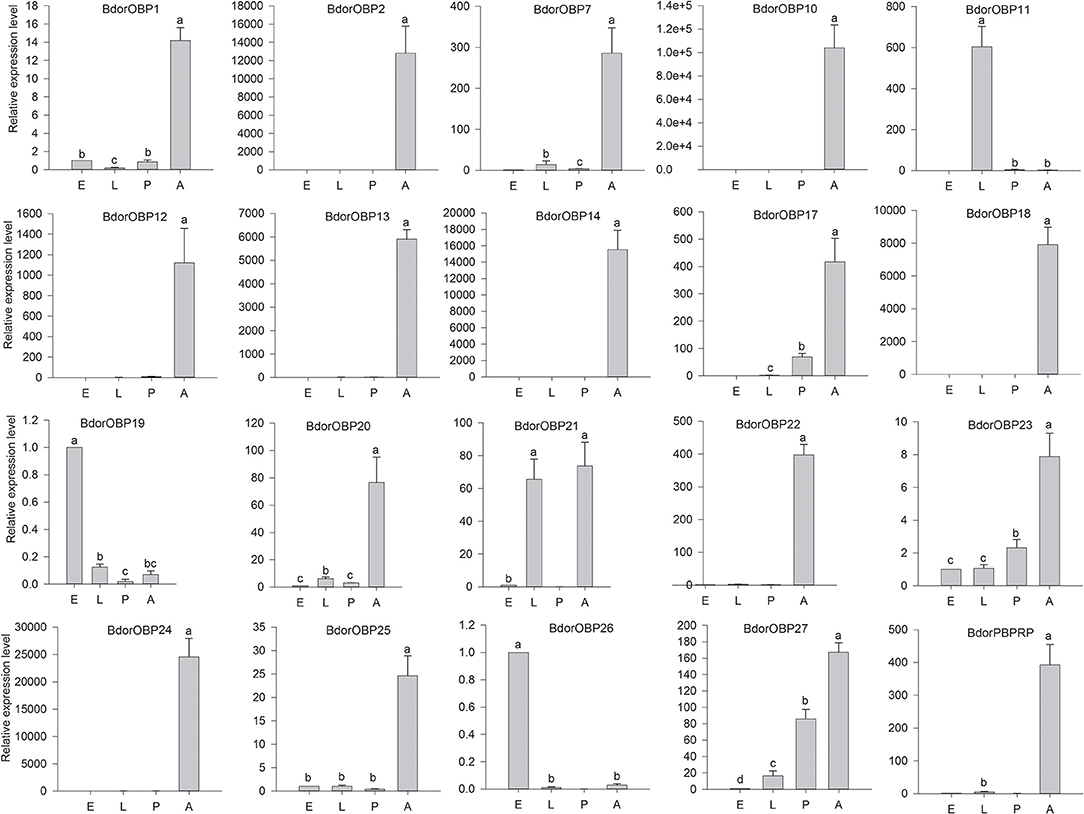
Figure 4. Expression profile of B. dorsalis OBPs in different developmental stages. Total RNA was extracted from eggs (E), larvae (L), pupae (P), and adults (A). The black rectangles show the expression level of the OBPs in different developmental stages. The bars represent mean ± SD, and the letters indicate statistically significant differences between the groups (Tukey's honest significant difference test).
Except for the genes with high transcript levels in the adult stage, some genes were highly expressed in other developmental stages. Here, for example, the Bdorobp11 gene was highly expressed in the larval stage and had a low expression in the adult stage. Two genes, Bdorobp19 and Bdorobp26, were highly expressed in the egg stage and had a lower expression in other tested developmental stages.
To examine the silencing of genes with dsRNA injection, the transcript level of Bdorobp2 was investigated in dsRNA-injected heads and control heads of B. dorsalis. Using qPCR experiments, it was confirmed that Bdorobp2 expression was significantly silenced with dsRNA injection (Figure 5). Compared with the control heads, the expression of Bdorobp2 in dsRNA-injected heads was reduced to 26.16 ± 7.65% in males and to 25.89 ± 4.24% in females. Thus, nearly three quarters of the total expression of these OBPs was successfully reduced in our experiments.
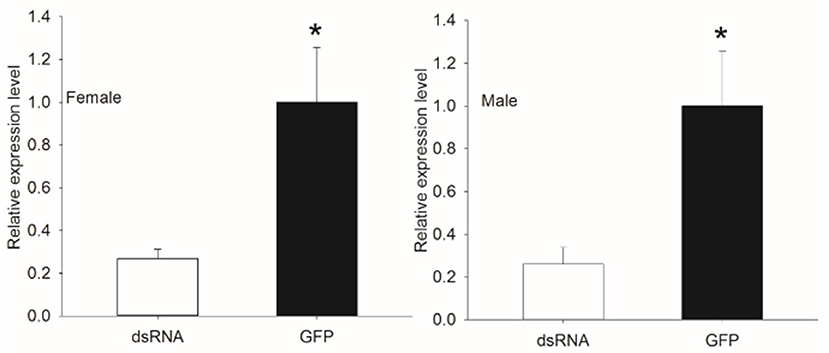
Figure 5. Decrease in Bdorobp2 gene expression after dsRNA injection. The dsRNA of Bdorobp2 was injected into the male and female abdomens. The change in the transcript level was examined by qPCR with three replicates. The expression of Bdorobp2 decreased significantly in the dsRNA-injected heads of adults (shown by the black rectangle), compared with the gene expression in the heads injected with green fluorescent protein (GFP) solution (shown by the blank rectangle). Vertical bars indicate the SD of the means (n = 6), and the asterisks (*) indicate the significant difference at the level of p < 0.05 based on the Student's t-test.
To compare the olfactory sensitivity of dsRNA-injected adults with the control flies, the electrophysiological responses of B. dorsalis toward methyl eugenol were examined using EAG. The results show that the antennal responses to the methyl eugenol were significantly reduced 3 days after dsRNA injection, which corresponds with the reduction of BdorOBP2 expression. When stimulated with 1% methyl eugenol, the control male adults showed an average response of 5.57 ± 0.43 mV, while the response decreased to 1.75 ± 0.15 mV when the Bdorobp2 gene was silenced with dsRNA injection (Figure 6). The responses of control females and dsRNA-injected females showed similar decreases in detection of this odorant. Moreover, the response to 1% methyl eugenol decreased from 4.64 ± 0.80 mV (control female adults) to 1.30 ± 0.29 mV (dsRNA-injected female adults) when treated with methyl eugenol.
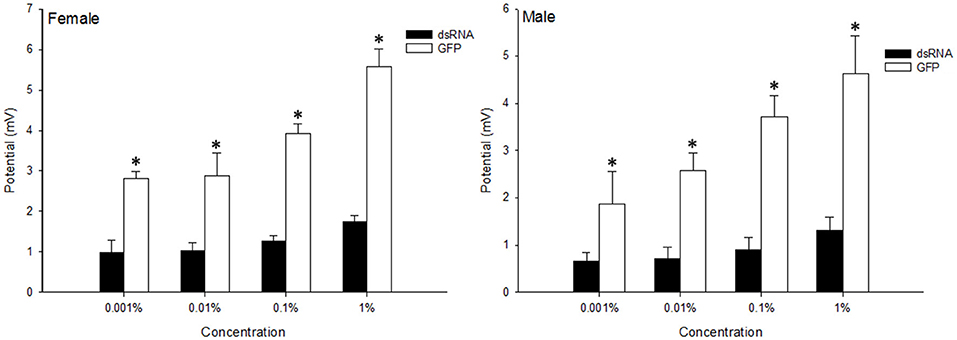
Figure 6. Electroantennographic (EAG) response of B. dorsalis to methyl eugenol attractant following dsRNA injection, compared with that of the insect injected with GFP solution as control. The x-bar represents the concentration of the attractant. Vertical bars indicate the SD of the means (n = 6), and the asterisks (*) indicate the significant difference at the level of p < 0.05 based on the Student's t-test.
OBPs are hypothesized to bind and more efficiently shuttle specific semiochemicals to the olfactory receptors in insect antennae. Extensive research has shown that OBPs play important roles in odorant perception and sensitivity (Kim et al., 1998; Zhou et al., 2008; Pelletier and Leal, 2009; Pelletier et al., 2010; Swarup et al., 2011; Sun et al., 2014). More recently, with the development of the genomic studies and transcriptome analyses, increasing numbers of OBPs have been identified from various insect species, and their functions have been proposed based on their molecular sequences. In our previous research, we identified 20 OBPs from B. dorsalis antennae (Liu et al., 2016), and an even larger number of OBPs (approximately 49) have been identified in other body tissues of this insect (Jin et al., 2011; Chen et al., 2019). However, no classification systems have been established to analyze the functions of antennae-specific OBPs and non-antennae expressed OBPs. In previous research, it was shown that many OBPs that were specifically expressed in the antennae were in fact associated with olfactory functions, and this has also been shown in many other insect species (Biessmann et al., 2010; Pelletier et al., 2010). To explore the functions of these identified OBPs, we built a OBPs classification system, and classified the OBPs identified in the antennae into two subgroups, I and II, based on the results of qPCR analysis and phylogenetic relationships.
The OBPs in subgroup I are hypothesized to be involved in olfaction because they are highly expressed in the classical olfactory organs, the antennae. The importance of tissue-specific OBP expression has been shown previously in the research related to oviposition and pheromone detection in the mosquito (Pelletier et al., 2010) as well as by pheromone detection in Drosophila (Xu et al., 2005). Thus, the location of highest expression has often been associated with the function of those OBPs. Moreover, we proposed that OBPs that are highly expressed in the antenna (i.e., the main olfactory organ) would be most likely associated with olfaction. To examine the hypothesis that the OBPs that are highly expressed in antennae are involved in B. dorsalis olfaction, we setup a system combining RNAi and EAG to examine the functions of a gene that is highly expressed in the antennae, namely, Bdorobp2. The species-specific chemical, methyl eugenol, which attracts the B. dorsalis adults at low concentrations (Metcalf et al., 1975), was used to compare the behavioral differences between the control and dsRNA treated B. dorsalis adults. The associated EAG response of the antennae toward the tested attractant (methyl eugenol) decreased significantly when Bdorobp2 was silenced in both male and female insects (Figure 6). Our observation is consistent with the previous reports that reduction of vital, antennae-specific OBP transcript levels can result in a decrease in EAG response for our insect, B. dorsalis (Wu et al., 2016). In our experiments, both the male and female B. dorsalis adults show a significantly decreased electrophysiological response to the olfactory stimulation with the attractant odor, methyl eugenol, after dsRNA injection. Based on these results, we expect that the OBPs in subgroup I are most likely involved in odorant detection and are true OBPs that function directly in odorant perception and sensitivity. Here, for example, our data suggest that the Bdorobp20 is most enriched in the female antennae, and is perhaps involved with the detection of oviposition signals or reception of other attractants, which would be consistent with the similar OBPs that have been reported in other species (Biessmann et al., 2010; Mao et al., 2010; Pelletier et al., 2010; Xu et al., 2010; Yin et al., 2015). However, the specific role of Bdorobp20 in B. dorsalis olfaction needs to be continued exploring in future research. However, future research needs to continue exploring whether Bdorobp20 in B. dorsalis can be directly linked to olfactory function and odor-dependent behavior.
At present, it is difficult to determine the function of other OBPs that are highly expressed in non-olfactory tissues without further experimental evidence. Some researchers have suggested that these OBPs were not “true” olfactory binding proteins because they have no obvious or direct function in olfaction. It has been shown, for example, that the DsechOBP57d and DsechOBP57e proteins, which are highly expressed in the non-olfactory tissues of D. sechellia and D. melanogaster have a function in gustatory perception (Galindo and Smith, 2001; Matsuo et al., 2007; Harada et al., 2008). Based on these previous researches, it is possible to conclude that the OBPs we identify within our current study (i.e., those that are expressed in non-olfactory tissues) may have other physiological or chemosensory functions. Here we propose that these proteins may participate in other chemosensory functions, such as the modulation of gustation, as opposed to (or in conjunction with) any role in olfaction (Blomquist and Vogt, 2003; Leal, 2005; Ling et al., 2014; Gonzalez-Gonzalez et al., 2016; Zhu et al., 2016). The additional OBPs we identify that are abundantly or specifically expressed in the B. dorsalis head may also function in olfaction because the maxillary palps were not explicitly removed in our experiments. As such, future work should also examine OBP expression across the labial or maxillary palps of this insect, especially in comparison to those found within the antenna in our current study. Thus, in summary, while we specifically identify the role of Bdorobp2 in the detection of and olfactory sensitivity toward the species-specific attractant (i.e., methyl eugenol), additional work is still needed to pair other highly expressed genes with their function across additional proteins from our subset of 20 OBPs identified within B. dorsalis. Moreover, while the function of these other OBPs is difficult to determine without further experiments, this study paves the way forward for our continued advancement of ecologically relevant odorants and chemosensory channels that in turn may prove vital in the control of this and other agricultural pest species.
The datasets generated for this study can be found in the https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0147783.s003&type=supplementary.
ZL, J-JW, and Z-RL conceived and designed the experiments. ZL, X-FL, and LX carried out the experiments. ZL, IK, J-JW, Z-RL, and GS analyzed the data and wrote the manuscript or contributed as native English speakers. All of the authors agreed to publish this paper.
This research is supported by the National Key Research and Development Program of China (2017YFD0200907) and the Natural Science Foundation of Chongqing, China (cstc2018jcyjAX0141).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00063/full#supplementary-material
Figure S1. Primers used for qPCR.
Biessmann, H., Andronopoulou, E., Biessmann, M. R., Douris, V., Dimitratos, S. D., Eliopoulos, E., et al. (2010). The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS ONE 5:e9471. doi: 10.1371/journal.pone.0009471
Blomquist, G. J., and Vogt, R. G. (2003). “The biosynthesis and detection of pheromones and plant volatiles,” in Insect Pheromone Biochemistry and Molecular Biology, eds G. J. Blomquist, R. G. Vogt (London: Elsevier Academic Press), 3–18. doi: 10.1016/B978-012107151-6/50003-7
Chen, X. F., Xu, L., Zhang, Y. X., Wei, D., Wang, J. J., and Jiang, H. B. (2019). Genome-wide identification and expression profiling of odorant-binding proteins in the oriental fruit fly, Bactrocera dorsalis. Com. Biochem. Phys. D 31:100605. doi: 10.1016/j.cbd.2019.100605
Chen, X. L., Li, G. W., Xu, X. L., and Wu, J. X. (2018). Molecular and functional characterization of odorant binding protein 7 from the oriental fruit moth Grapholita molesta (Busck) (Lepidoptera: Tortricidae). Front. Physiol. 9:1762. doi: 10.3389/fphys.2018.01762
Choo, Y. M., Xu, P., Hwang, J. K., Zeng, F., Tan, K., Bhagavathy, G., et al. (2018). Reverse chemical ecology approach for the identification of an oviposition attractant for Culex quinquefasciatus. Proc. Natl. Acad. Sci. U.S.A. 115, 714–719. doi: 10.1073/pnas.1718284115
Galindo, K., and Smith, D. P. (2001). A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159, 1059–1072.
Gonzalez-Gonzalez, A., Palma-Millanao, R., Yanez, O., Rojas, M., Mutis, A., Venthur, H., et al. (2016). Virtual screening of plant volatile compounds reveals a high affinity of Hylamorpha elegans (Coleoptera: Scarabaeidae) odorant-binding proteins for sesquiterpenes from its native host. J. Insect. Sci. 16, 1–15. doi: 10.1093/jisesa/iew008
Große-Wilde, E., Svatoš, A., and Krieger, J. (2006). A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses 31, 547–555. doi: 10.1093/chemse/bjj059
Harada, E., Haba, D., Aigaki, T., and Matsuo, T. (2008). Behavioral analyses of mutants for two odorant-binding protein genes, OBP57d and OBP57e, in Drosophila melanogaster. Genes Genet. Syst. 83, 257–264. doi: 10.1266/ggs.83.257
Hsu, J. C., Feng, H. T., and Wu, W. J. (2004). Resistance and synergistic effects of insecticides in Bactrocera dorsalis (Diptera: Tephritidae) in Taiwan. J. Econ. Entomol. 97, 1682–1688. doi: 10.1603/0022-0493-97.5.1682
Jayanthi, P. D. K., Woodcock, C. M., Caulfield, J., Birkett, M. A., and Bruce, T. J. (2012). Isolation and identification of host cues from mango, Mangifera indica, that attract gravid female oriental fruit fly, Bactrocera dorsalis. J. Chem. Ecol. 38, 361–369. doi: 10.1007/s10886-012-0093-y
Jeong, Y. T., Shim, J., Oh, S. R., Yoon, H. I., Kim, C. H., Moon, S. J., et al. (2013). An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron 79, 725–737. doi: 10.1016/j.neuron.2013.06.025
Jin, T., Zeng, L., Lin, Y., Lu, Y., and Liang, G. (2011). Insecticide resistance of the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), in mainland China. Pest Manag. Sci. 67, 370–376. doi: 10.1002/ps.2076
Ju, Q., Li, X., Guo, X. Q., Du, L., Shi, C. R., and Qu, M. J. (2018). Two odorant-inding proteins of the dark black chafer (Holotrichia parallela) display preferential binding to biologically active host plant volatiles. Front. Physiol. 9:769. doi: 10.3389/fphys.2018.00769
Khan, H. A. A., and Akram, W. (2018). Trichlorfon and spinosad resistance survey and preliminary determination of the resistance mechanism in Pakistani field strains of Bactrocera dorsalis. Sci. Rep. 8:11223. doi: 10.1038/s41598-018-29622-0
Kim, M. S., Repp, A., and Smith, D. P. (1998). LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 150, 711–721. doi: 10.1534/genetics.107.081331
Leal, W. S. (2005). “Pheromone reception,” in The Chemistry of Pheromones and Other Semiochemicals II, ed S. Schulz (Berlin: Springer) Vol. 240, 1–36. doi: 10.1007/b98314
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li, Z. Q., Zhang, S., Cai, X. M., Luo, J. Y., Dong, S. L., Cui, J. J., et al. (2018). Distinct binding affinities of odorant-binding proteins from the natural predator Chrysoperla sinica suggest different strategies to hunt prey. J. Insect Physiol. 111, 25–31. doi: 10.1016/j.jinsphys.2018.10.004
Ling, F., Dahanukar, A., Weiss, L. A., Kwon, J. Y., and Carlson, J. R. (2014). The molecular and cellular basis of taste coding in the legs of Drosophila. J. Neurosci. 34, 7148–7164. doi: 10.1523/JNEUROSCI.0649-14.2014
Liu, Z., Smagghe, G., Lei, Z. R., and Wang, J. J. (2016). Identification of male- and female-specific olfaction genes in antennae of the oriental fruit fly (Bactrocera dorsalis). PLoS ONE 11:e0147783. doi: 10.1371/journal.pone.0147783
Liu, Z., Vidal, D., Syed, Z., Ishida, Y., and Leal, W. (2010). Pheromone binding to general odorant-binding proteins from the navel orangeworm. J. Chem. Ecol. 36, 787–794. doi: 10.1007/s10886-010-9811-5
Maida, R., Steinbrecht, A., Ziegelberger, G., and Pelosi, P. (1993). The pheromone binding protein of Bombyx mori: Purification, characterization and immunocytochemical localization. Insect Biochem. Mol. Biol. 23, 243–253. doi: 10.1016/0965-1748(93)90005-D
Mao, Y., Xu, X. Z., Xu, W., Ishida, Y., Leal, W. S., Ames, J. B., et al. (2010). Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc. Natl. Acad. Sci.U.S.A. 107, 19102–19107. doi: 10.1073/pnas.1012274107
Matsuo, T., Sugaya, S., Yasukawa, J., Aigaki, T., and Fuyama, Y. (2007). Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5:e118. doi: 10.1371/journal.pbio.0050118
Metcalf, R. L., Mitchell, W. C., Fukuto, T. R., and Metcalf, E. R. (1975). Attraction of the oriental fruit fly, Dacus dorsalis, to methyl eugenol and related olfactory stimulants. Proc. Natl. Acad. Sci. U.S.A. 72, 2501–2505. doi: 10.1073/pnas.72.7.2501
Pelletier, J., Guidolin, A., Syed, Z., Cornel, A. J., and Leal, W. S. (2010). Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J. Chem. Ecol. 36, 245–248. doi: 10.1007/s10886-010-9762-x
Pelletier, J., and Leal, W. S. (2009). Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLos ONE 4:e6237. doi: 10.1371/journal.pone.0006237
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Siderhurst, M. S., and Jang, E. B. (2006). Female-biased attraction of oriental fruit fly, bactrocera dorsalis (Hendel), to a blend of host fruit volatiles from Terminalia catappa L. J. Chem. Ecol. 32, 2513–2524. doi: 10.1007/s10886-006-9160-6
Steiner, L. F., Mitchell, W. C., Harris, E. J., Kozuma, T. T., and Fujimoto, M. S. (1965). Oriental fruit fly eradication by male annihilation. J. Econ. Entomol. 58, 961–964. doi: 10.1093/jee/58.5.961
Sun, L., Gu, S. H., Xiao, H. J., Zhou, J. J., Guo, Y. Y., Liu, Z. W., et al. (2013). The preferential binding of a sensory organ specific odorant binding protein of the alfalfa plant bug adelphocoris lineolatus alinobp10 to biologically active host plant volatiles. J. Chem. Ecol. 39, 1221–1231. doi: 10.1007/s10886-013-0333-9
Sun, L., Xiao, H. J., Gu, S. H., Zhou, J. J., Guo, Y. Y., Liu, Z. W., et al. (2014). The antenna-specific odorant-binding protein AlinOBP13 of the alfalfa plant bug Adelphocoris lineolatus is expressed specifically in basiconic sensilla and has high binding affinity to terpenoids. Insect Mol. Biol. 23, 417–434. doi: 10.1111/imb.12089
Swarup, S., Williams, T. I., and Anholt, R. R. H. (2011). Functional dissection of odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 10, 648–657. doi: 10.1111/j.1601-183X.2011.00704.x
Vargas, R. I., Stark, J. D., Kido, M. H., Ketter, H. M., and Whitehand, L. C. (2000). Methyl eugenol and cue-lure traps for suppression of male oriental fruit flies and melon flies (Diptera: Tephritidae) in Hawaii: effects of lure mixtures and weathering. J. Econ. Entomol. 93, 81–87. doi: 10.1603/0022-0493-93.1.81
Wu, Z., Lin, J., Zhang, H., and Zeng, X. (2016). BdorOBP83a-2 mediates responses of the oriental fruit fly to semiochemicals. Front. Physiol. 7:452. doi: 10.3389/fphys.2016.00452
Wu, Z. Z., Zhang, H., Wang, Z. B., Bin, S. Y., He, H. L., and Lin, J. T. (2015). Discovery of chemosensory genes in the oriental fruit fly, Bactrocera dorsalis. PLoS ONE 10:e0129794. doi: 10.1371/journal.pone.0129794
Xie, Y. F., Yang, W. J., Dou, W., and Wang, J. J. (2013). Characterization of the cDNA encoding membrane-bound trehalase, its expression and enzyme activity in Bactrocera dorsalis (Diptera: Tephritidae). Florida Entomol. 96, 1233–1242. doi: 10.1653/024.096.0401
Xu, P., Atkinson, R., Jones, D. N., and Smith, D. P. (2005). Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200. doi: 10.1016/j.neuron.2004.12.031
Xu, W., Cornel, A. J., and Leal, W. S. (2010). Odorant-binding proteins of the malaria mosquito Anopheles funestus sensu stricto. PLoS ONE 5:e15403. doi: 10.1371/journal.pone.0015403
Yin, J., Choo, Y.-M., Duan, H., and Leal, W. S. (2015). Selectivity of odorant-binding proteins from the southern house mosquito tested against physiologically relevant ligands. Front. Physiol. 6:56. doi: 10.3389/fphys.2015.00056
Zheng, W., Peng, W., Zhu, C., Zhang, Q., Saccone, G., and Zhang, H. (2013). Identification and expression profile analysis of odorant binding proteins in the oriental fruit fly Bactrocera dorsalis. Int. J. Mol. Sci. 14, 14936–14949. doi: 10.3390/ijms140714936
Zhou, J. J., He, X. L., Pickett, J. A., and Field, L. M. (2008). Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol. Biol. 17, 147–163. doi: 10.1111/j.1365-2583.2007.00789.x
Keywords: oriental fruit fly, odorant-binding proteins, olfaction, antennae, dsRNA
Citation: Liu Z, Liang X-F, Xu L, Keesey IW, Lei Z-R, Smagghe G and Wang J-J (2020) An Antennae-Specific Odorant-Binding Protein Is Involved in Bactrocera dorsalis Olfaction. Front. Ecol. Evol. 8:63. doi: 10.3389/fevo.2020.00063
Received: 10 October 2019; Accepted: 26 February 2020;
Published: 18 March 2020.
Edited by:
Li Chen, Institute of Zoology (CAS), ChinaReviewed by:
Zhongjun Gong, Henan Academy of Agricultural Science, ChinaCopyright © 2020 Liu, Liang, Xu, Keesey, Lei, Smagghe and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong-Ren Lei, bGVpemhyQHNpbmEuY29t; Jin-Jun Wang, d2FuZ2ppbmp1bkBzd3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.