- 1Department of Life and Consumer Sciences, University of South Africa, Pretoria, South Africa
- 2Department of Geography, Environmental Management and Energy Studies, University of Johannesburg, Johannesburg, South Africa
- 3Department of Environmental Sciences, University of South Africa, Pretoria, South Africa
Biogeographically, cycads were once widely distributed but the extant cycads are restricted to tropical and subtropical regions. They originated ∼ 300 Ma and re-diversified recently around 12 Ma, with the genus Cycas being the most rapidly diversified and largely distributed lineage. However, the forces that shaped the diversification and biogeography of the genus remain to be fully understood. Here, we first retrieved and used DNA sequences from GenBank (nuclear: PHYP, RPB1, HZP, AC3, F3H, SAMS, and GTP; chloroplasts: plant barcode trnH-psbA, trnL-trnF, trnS-trnG, and psbM-trnD) to assemble a complete dated phylogeny of Cycas. Then, we employed the Bayesian Binary Method to reconstruct the historical biogeography of the extant Cycas and finally, using the Bayesian approach for diversification analysis, we explored the evolutionary events that might have shaped the rapid diversification and large distribution of Cycas across the Pacific Islands. Our analysis pointed to Indo-China as the origin of the genus, which may have dispersed firstly across the Pacific Islands during the late Miocene aided by multiple excursions of sea levels and the development of a key innovation, i.e., a spongy endocarp particularly in the seeds of the subsection Rumphiae. The colonization of South China, which was thought to be the origin of the genus, may have occurred more recently aided by both dispersal and vicariance events. However, no significant shifts in the evolutionary events (speciation, extinction, mass extinction) that shaped the diversity of the genus were observed. Overall, our study re-clarifies the historical biogeography and the evolutionary forces that shaped the current diversity of the genus Cycas.
Introduction
Cycads are dioecious and entomophilous plants that developed palm-like habit with stout trunks and large evergreen pinnate leaves (Jones, 2002). They share some characteristics with the ferns (e.g., spermatozoa with flagella) and angiosperms (e.g., seed productions; Guan, 1996; Norstog and Nicholls, 1997). The dispersal of cycad seeds is limited to 2–7 km, and is mostly mediated by rodents, small fruit-eating bats (Yang and Meerow, 1996) and long dispersal via the sea (Keppel et al., 2009). Cycads represent the oldest lineage of plants, originating ∼ 300 million years ago (Ma) in the mid-Permian (Hendricks, 1987; Gao and Thomas, 1989; Calonje et al., 2017) and reaching their greatest diversity in the Jurassic era (Jones, 2002; Nagalingum et al., 2011). Geographically, cycads are restricted to tropical and subtropical or warm temperate regions with predominantly summer rainfalls (Jones, 2002). In total, 10 genera diversified within the cycad group, with the genus Cycas being the largest of all (Osborne et al., 2012; Calonje et al., 2017).
Specifically, Cycas is the only genus in the family Cycadaceae, an early diverging lineage to the cycad phylogenetic tree (Stevenson, 1992; Nagalingum et al., 2011). This genus is comprised of six Sections, including Asiorientales, Panzhihuaenses, Wadeanae, Strongyloides, Indosinenses, and Cycas (Hill, 2004). The genus Cycas is the most rapidly diversified clade in the cycad group with ∼ 112 species (Yessoufou et al., 2017). Fossil evidence points to Asia as the origin of the genus (Hill, 1995; see also Xiao and Möller, 2015). From Asia, the genus Cycas is further distributed southward to Australia, eastern Africa and the Pacific Islands (Hill, 2004).
In Asia, the genus is distributed across the Red River Fault between South China and the Indochina block, with Red River potentially constituting a geographical barrier for gene flow (Xiao and Möller, 2015). If this barrier was effective, we would expect to detect the signature of vicariance events in the evolutionary history of the genus Cycas (Keppel et al., 2008; Xiao and Möller, 2015). Then, the widespread distribution of Cycas from Asia to Africa, Australia and across the Pacific regions might have been mediated through long distance dispersal events across the ocean. However, the sample analyzed in a recent study that tested this hypothesis (Xiao and Möller, 2015) was taxonomically limited (only 31 species out of 112), although they included representatives of all six Sections of the genus in their analysis. Even in Keppel et al.’s (2008) study, only the Subsection Rumphiae of the Section Cycas was analyzed. As such, inferences on the evolutionary and ecological processes that shaped the biogeography of Cycas may require further investigations. In addition, in their recent analysis of the diversification rate comparison across the cycad tree of life, Yessoufou et al. (2017) revealed a diversification rate heterogeneity across the tree with the genus Cycas identified as the most rapidly diversifying clade, and they suggested that this rapid diversification might have mediated their widest geographic distribution. Unfortunately, they did not go further to elucidate the patterns of diversification events within this clade.
In the present study, our aim is to provide a refined understanding of the evolutionary and ecological processes that shaped the biogeography of the genus Cycas. Specifically, we assembled the most comprehensive dated phylogeny of the genus, which was then used to elucidate its historical biogeography as well as the ecological forces that mediated the observed diversity patterns.
Materials and Methods
A Complete List of Cycas Species Used to Reconstruct a Dated Cycas Phylogeny
The full list of Cycas species is still a matter of debate. However, a recent study analyzed a large dataset of informative markers (DNA data) to estimate the total cycad diversity to 116 (100 accepted, 7 subspecies and 9 controversial species; Liu et al., 2018). To assemble a complete phylogeny for the 116 Cycas species, we retrieved from GenBank/EBI (accessed October 2018; Liu et al., 2018) DNA sequences of seven nuclear regions (PHYP, RPB1, HZP, AC3, F3H, SAMS, and GTP) and four plastid regions (including a complementary plant DNA barcode trnH-psbA as well as psbM-trnD, trnL-trnF and trnS-trnG) of Cycas species. The molecular matrix is available as Supplementary Material (DNA matrix; available at https://doi.org/10.5061/dryad.1g1jwstrn, Mankga et al., 2020b); accession numbers as well as the species names are presented in Supplementary Table S1. The dated phylogeny was assembled for 135 species including outgroups (Bowenia Hook.ex Hook.f., Ceratozamia Brongn, Dioon Lindl., Encephalartos Lehm., Lepidozamia Regel, Macrozamia Miq., Microcycas calocoma (Miq.) A. DC., Stangeria eriopus (Kunze) Baill., Zamia L., Ginkgo biloba L.) following the traditional Bayesian approach implemented in the BEAST program (Rambaut and Drummond, 2007).
The following steps were followed for the BEAST analysis. Firstly, an XML file using BEAUti (Drummond and Rambaut, 2007) was generated. Secondly, the best model GTR + I + Γ (based on Akaike information criterion evaluated using MODELTEST; Nylander, 2004) was selected as well as the birth-death process prior with uncorrelated relaxed lognormal model for rate variation among branches, following Condamine et al.’s (2015) recommendations. To calibrate the Cycas tree, uniform priors with minimum and maximum age estimates for nodes calibration were selected as the normal priors bias the node age estimates (Schenk, 2016). The following uniform calibration points were used following Condamine et al. (2015) for cycad group: Cycas SG (15.8–257.2 Ma), Cycads SG (273.9–364.9 Ma), Dioon SG (107–207.9 Ma), Bowenia SG (88.7–174.3 Ma), Lepidozamia SG (33.9–55 Ma), Ceratozamia SG (19.2–84.9 Ma), Zamia SG (14.6–57 Ma), Encephalartos SG (97.7–192.5 Ma). Lastly, MCMC was run for 100 million with trees sampled every 10,000 generations. At the end of the process of dated tree reconstruction, the ESS values ranged from 200 to 901 for the age estimates; the first 2,000 trees were burnt and the remaining 8,000 trees were combined using TREEANNOTATOR (Rambaut and Drummond, 2007) to generate a maximum clade credibility (MCC) tree. The node support on this MCC tree is interpreted as follows: not supported (PP < 0.50), supported (PP = 0.60) and strongly supported (PP > 0.60). In addition, the bootstrap node supports on the phylogeny were assessed using PAUP v40b10 (Swofford, 2002) approach. These node supports were assessed by reconstructing the Maximum Parsimony (MP) tree based on the heuristic search with 1000 random sequences additions keeping 10 trees. The bootstrap values were interpreted as follows: BS > 70% indicates strong support and BS < 70% indicate weak support (Hillis and Bull, 1993; Wilcox et al., 2002).
Ancestral Area Reconstruction States: Historical Biogeography of Cycas
To reconstruct the historical biogeography of the genus Cycas, we grouped all species into three categories based on their current geographic distribution (Osborne et al., 2012) and following Xiao and Möller (2015). The category (A) includes species from South China, Taiwan-Ryukyu Archipelago, and Palawan islands (we refer henceforth to category A simply as South China). The category (B) includes species from Indochina, and (C) include Islands of Southeast Asia plus the Malay Peninsula, the Indian subcontinent, East Africa and North Australia.
We used Bayesian Binary Model (BBM) analysis implemented in RASP to reconstruct the possible ancestral ranges of the genus Cycas on the phylogenetic trees. In this analysis, the frequencies of an ancestral range at a node in ancestral reconstructions are averaged over all trees generated by RASP in Bayesian analysis (Yan et al., 2010). To account for uncertainties in phylogeny, we used 20,000 trees from MCMC output generated with BBM model. The MCMC chains were run simultaneously for 5,000,000 generations. The state was sampled every 1000 generations. Fixed JC + G (Jukes-Cantor + Gamma) were used with null root distribution and the maximum number of areas for this analysis was kept as 3.
Diversification Analysis
All the diversification analyses were run using R library TESS (Höhna et al., 2015). Firstly, we identified the branching model that fits the diversification of the genus Cycas and then compared the number of taxa of the Cycas tree to the posterior-predictive distribution of 1000 simulated trees under a constant-rate birth-death model. The constant-rate birth-death model was parameterized by drawing rate parameters from the joint posterior densities inferred from the phylogenetic tree. This parameterized model was used to simulate 1000 phylogenies, which were then used to calculate the expected number of taxa. If the actual number of taxa falls near the center of the posterior-prediction distribution, then the model can be used to simulate the Cycas trees, indicating that it provides a good absolute fit and the diversification rates of Cycas are constant over time. Conversely, if the summary statistics fell outside the 95% credible interval of the posterior-predictive distributions, then the constant rate birth-death model is not suitable to predict the simulated trees and the diversification has significantly changed over time (Höhna et al., 2015).
In addition, we plotted the posterior-predictive distribution of the lineage accumulation curves (LTT plots for simulated trees) and compared the predictive distribution to the LTT plot of the observed tree. If the observed LTT plot falls within the simulated LTT plots, then the diversification rate of the genus Cycas has been constant over time and if not, this means that the diversification has experienced some evolutionary shifts.
Finally, the evolutionary models that explain the diversification patterns depicted by the observed LTT plot were identified. The models tested include a constant-rate birth-death model and three rate-variation models. The rate-variation models include a birth-death model with an exponentially decreasing speciation rate, a birth-death model with piecewise-constant rates (i.e., rates of speciation and extinction change over time but the diversification rate remains constant; Höhna et al., 2015) and a birth-death model of evolution punctuated by a mass-extinction event. Using Bayes Factors (BF; Baele et al., 2013), a pairwise comparison of these models was done to select the best model. For two models M0 and M1, BF values were interpreted following Jeffreys (1961). Specifically, BF(M0,M1) < 1 means the model M1 is supported; 1 < BF(M0,M1) < 3.2 suggests that M0 is barely worth-mentioning; 3.2 < BF(M0,M1) < 10 indicates a substantial support for M0, 10 < BF(M0,M1) < 100 is indicative of a strong support for M0, and BF(M0,M1) > 100 is interpreted as decisive support for M0 (Jeffreys, 1961).
The Compound Poisson Process Mass-Extinction Times (CoMET) Analysis
To investigate whether the genus Cycas has experienced some mass extinctions events (if so, when?), the CoMET [Compound Poisson Process (CPP) on Mass Extinction Time)] approach was employed (May et al., 2016). This approach has the advantage to fit not only all possible birth-death models to the data at hand but also to specifically model mass extinction events. The CoMET approach treats the number of speciation-rate shifts, extinction-rate shifts, mass-extinction events as well as the parameters associated with these events as random variables, and then estimates their joint posterior distribution. For this analysis, hyperpriors were set both empirically and a priori.
Results
Phylogenetic Tree of Cycas
The combined DNA data consisting of seven nuclear genes (PHYP, RPB1, HZP, AC3, F3H, SAMS, and GTP) and four chloroplasts (trnH-psbA, trnL-trnF, trnS-trnG, and psbM-trnD) includes 10788 characters, 3947 potential parsimony informative sites and 3584 constant characters (Supplementary Table S2). The missing data is less than 5% (Supplementary Table S1).
The phylogenetic tree reconstructed is, in general, well supported. Among all the nodes whose support values are reported on Figure 1, 77% of them have PP ≥ 0.80, whereas 59% of these nodes have BP > 70% (Figure 1). Further, the ESS values ranged from 200 to 901 for the age estimates, suggesting convergence between posterior distributions and the MCMC estimates. The dated tree suggests that the genus Cycas may have diverged around 12 Ma (95% HPD, 10.4 – 14.7; Figure 1). Even though the origin of the genus dates back to 12 Ma, most Cycas diversification was initiated in the Pleistocene and reached its peak in the Holocene (Figure 1).
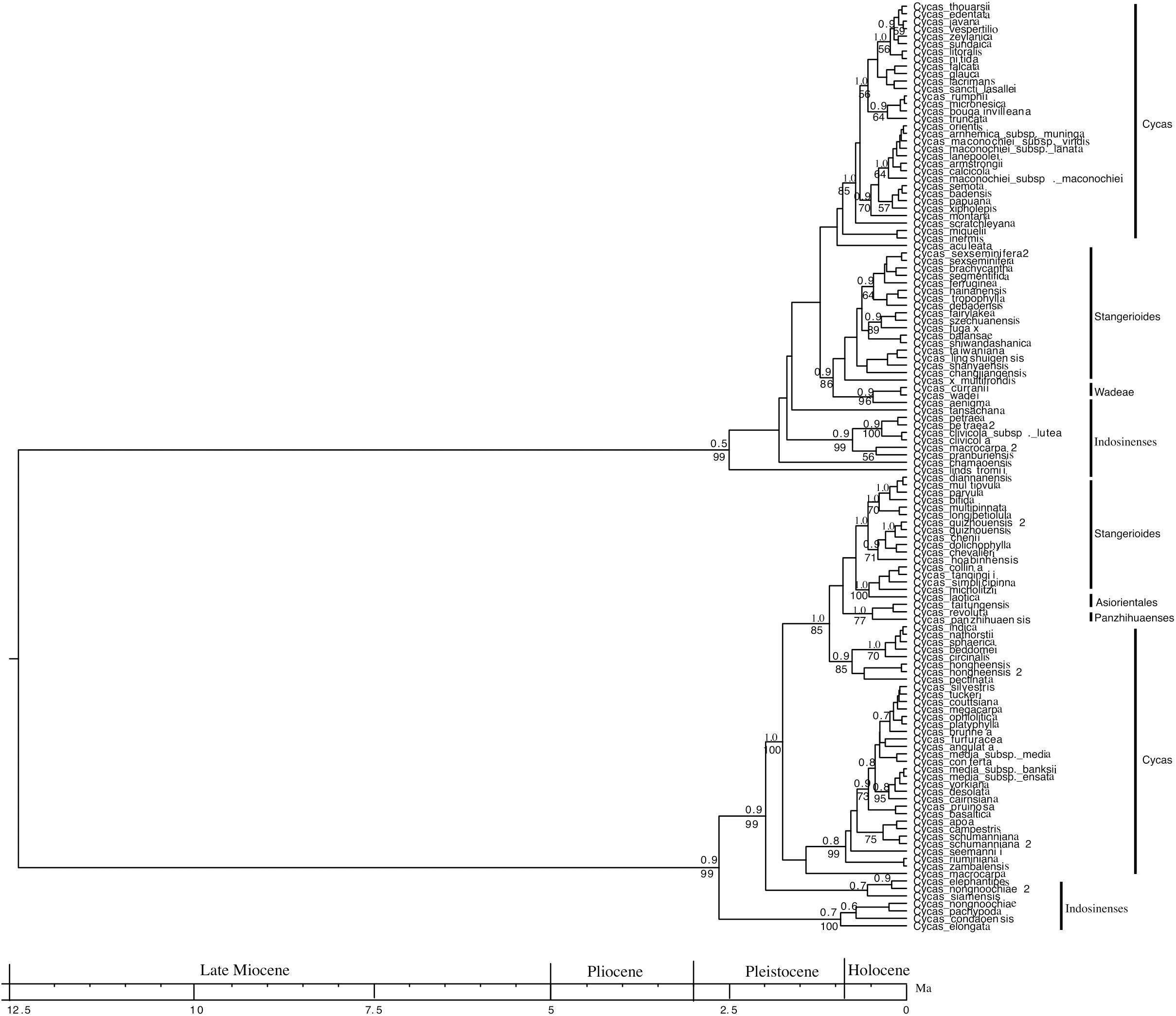
Figure 1. A complete dated phylogenetic tree of the genus Cycas from combined seven nuclear genes and three chloroplasts based on Bayesian Inference. The numbers above the branches represent Bayesian Posterior Probability (PP) and numbers below the branches represent the bootstrap value (BP).
In addition, of all the six sections of the genus (Cycas, Wadeae, Asiorientales, Stangerioides, Panzhihuaenses and Indosinenses), the section cycas is the largest (67 species out of 116 species), is polyphyletic and radiated ∼2 Ma (95% HPD, 1.09 – 2.6; Figure 1). The sections Stangerioides and Indosinenses are not monophyletic and most species in these sections radiated ∼1 Ma (95% HPD, 0.61–1.90; 0.17–2.4 and 1.09–3.03, respectively). However, Panzhihuaenses and Asiorientales sections are monophyletic with a strong support (PP/BP = 1.0/77). Finally, the section Wadeae, consisting of two species that are monophyletic with a strong support (PP/BP = 0.9/96), is the most recently radiated section (95% HPD, 0.01–0.37; Figure 1). All the six sections form two clearly defined major clades which both diverged in the Pleistocene (Figure 1). Each of these two clades is subtended by a long stem branch (phylogenetic fuse) connecting each clade to the origin of the genus. The early diverging clade (age = 2.75 Ma) is made up of the sections Indosinenses, Cycas, Panzhihuaenses, Asiorientales and Stangerioides. The sections Indosinenses, Wadeae, Stangerioides and Cycas (age = 2.5 Ma) form the second major clade (Figure 1).
Historical Biogeography
Our analysis points to Indochina (∼99%) as the origin of the genus Cycas, which dated back to around 12 Ma (node I, Figure 2). Around 2 Ma, the genus diverged from Indochina to the Islands of Southeast Asia, including the Malay Peninsula, the Indian subcontinent, East Africa and North Australia where the diversification was mostly mediated through vicariance (Figures 2, 3), but the origin is uncertain (node II; probability <50%). Around the same time period, the genus further diversified within Indochina (nodes III, probability 99%), and colonized South China around ∼ 1.5 Ma (node IV, probability 90%) aided by vicariance (Figures 2, 3).
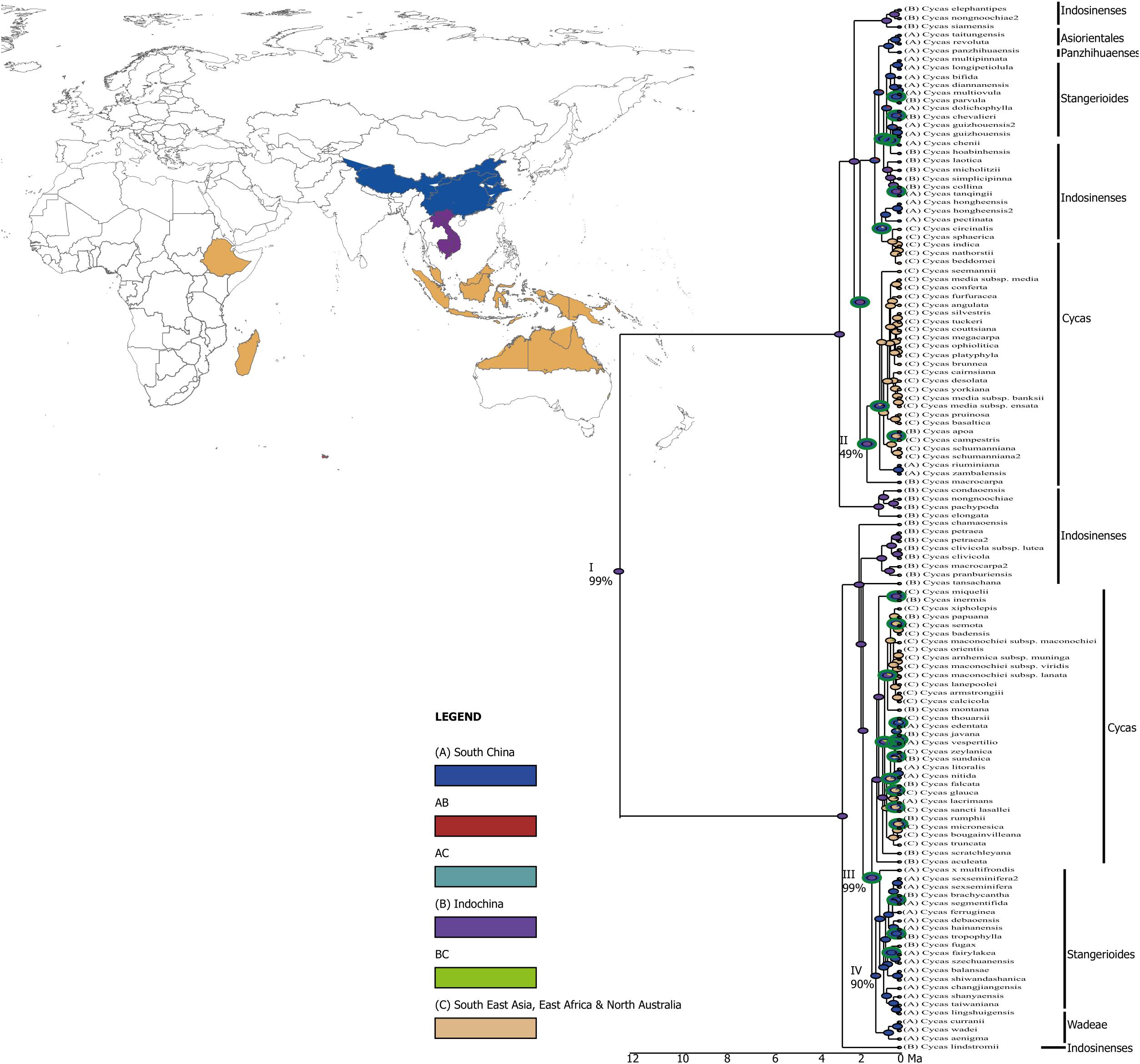
Figure 2. A graphical output from RASP showing the results of ancestral reconstruction area from BBM (Bayesian Binary Method) analysis. Pie charts at each node show probabilities of alternative ancestral ranges. The green circles around the node represent vicariance events and the blue circles represent dispersal events. Key major nodes representing historical origin of different taxonomic sections are noted I-IV and the probability of the origin at these nodes are also indicated (%).
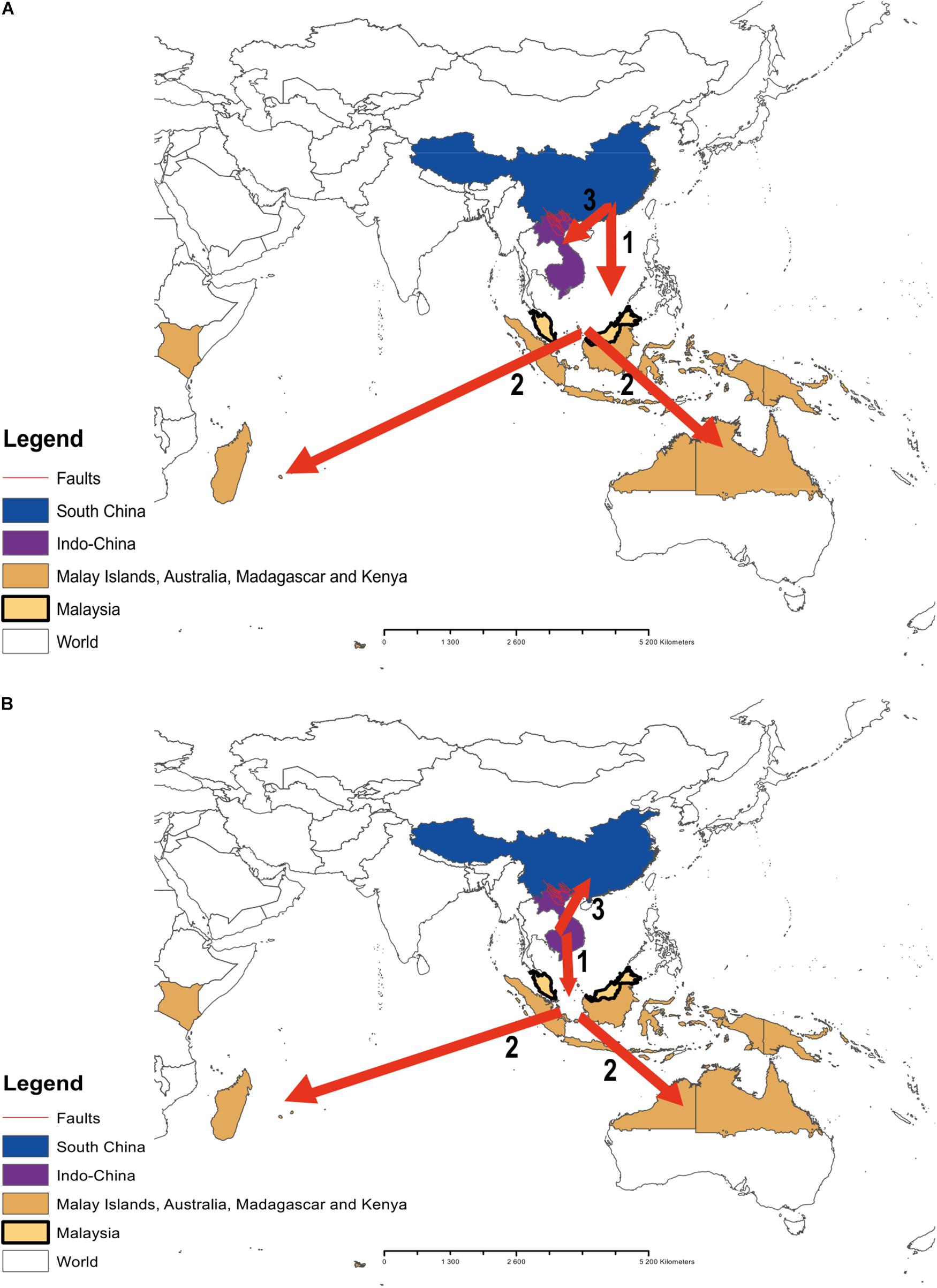
Figure 3. A representation of the potential colonization routes of the genus Cycas across the Pacific regions. Numbers 1, 2, and 3 indicate the probable sequences of colonization routes. (A) route reconstruction by Xiao and Möller (2015); (B) our route reconstruction.
Diversification Analysis
Most of the diversification events occurred in the last two million years (Pleistocene; Figures 1, 4A,B). These diversification events may have followed a constant diversification model as revealed in the following findings. The actual number of taxa (116) falls within the 95% credible interval of its posterior predictive distributions (Figure 5; left panel). This means that the constant-rate birth-death model used to reconstruct the posterior predictive distributions provides a good absolute fit to the evolutionary diversification of the genus Cycas. In addition, our LTT-plot does not depart significantly from those of the simulated trees under a constant-rate birth-death model (Figure 5; right panel). This is an additional support for the constant diversification over time. Finally, when testing alternative models using Bayes Factors to select the best diversification model, we found that a constant birth-death model is once more strongly supported (BF = 72.40; Supplementary Table S3).
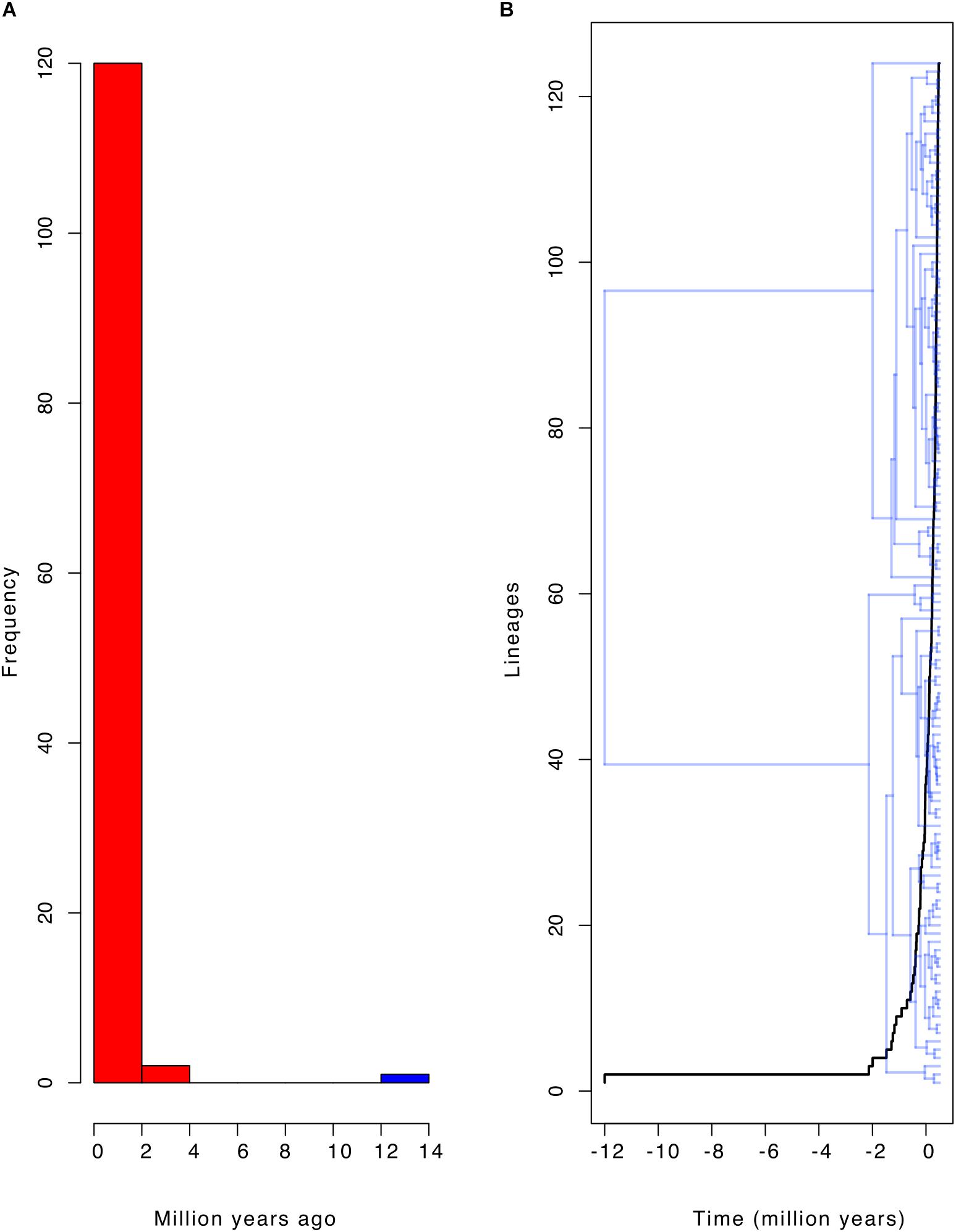
Figure 4. The diversification frequency of the genus Cycas. (A) Histogram showing the frequency of branching time of the phylogeny of Cycas; red color indicates earlier branching events, which shows that most branching events occurred in the last two million years; (B) Lineage-through-time plot matching the branching time on the phylogeny of Cycas.
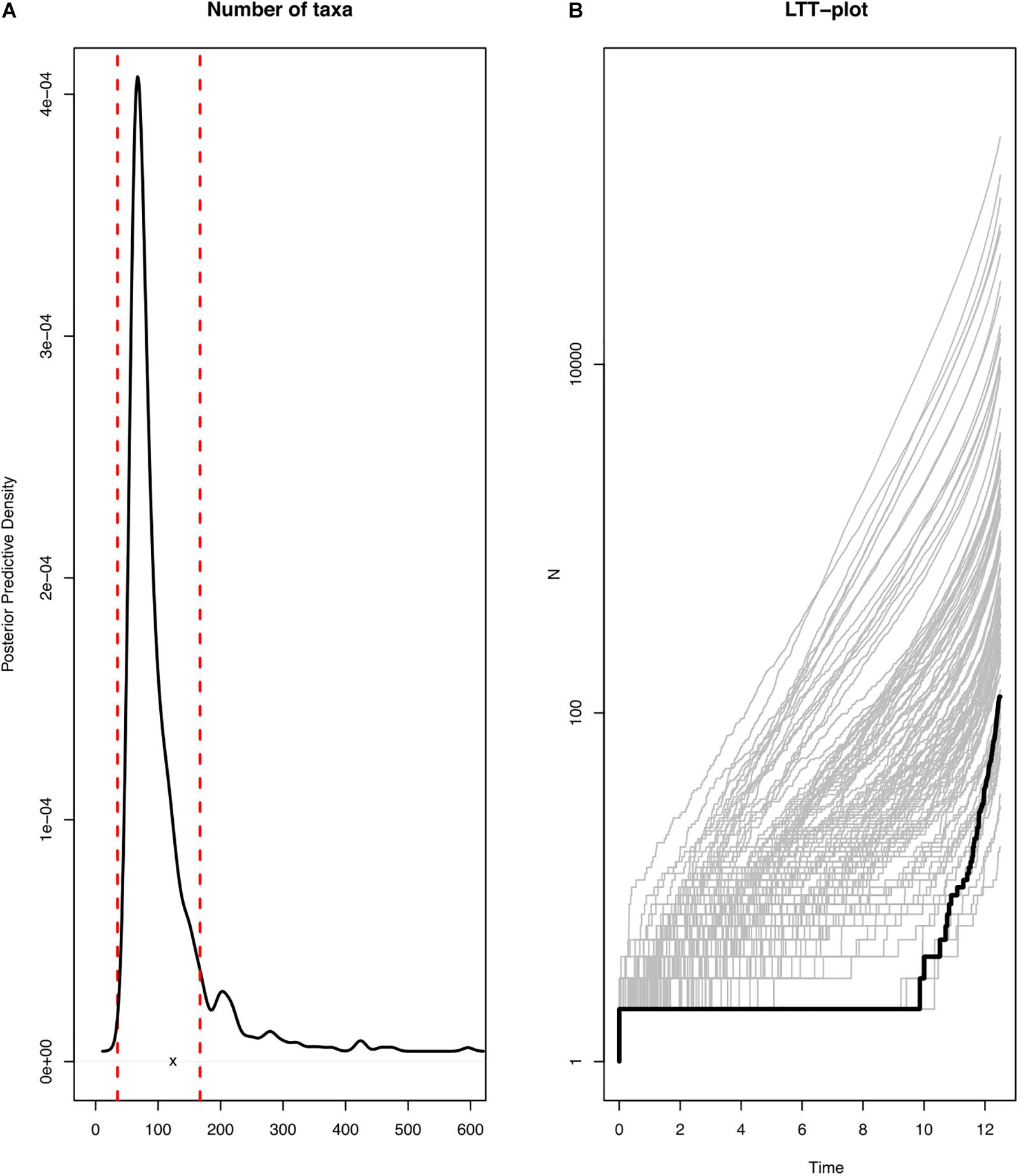
Figure 5. Evidence of constant-rate diversification of the genus Cycas. (A) actual number of species in the phylogeny (x) in comparison to the simulated number of species (posterior predictive distribution); (B) actual Lineage Through Time (LTT) plot in comparison to the simulated LLT plots.
The COMET Results
We tested several diversification events that might shape the biogeographical patterns. The diversification hyperpriors were specified a priori and empirically. Only the results of a priori hyperpriors are reported below as these are similar to those of the empirically set priors. The analysis indicates a general trend of increased speciation rate within the window of 2 to 4 species per million years (Myr; Figure 6A). These multiple speciation events did not correspond to any significant or dramatic shift (Figure 6B). Although the extinction rate remains roughly constant at 4 species Myr–1 from 12 to ∼4 Ma, it decreases gradually during the last period of diversification (4-0 Ma) to 3 species Myr–1 (Figure 6C). Again, none of these extinction events was significant, and there was no evidence of any significant shift in mass extinction events (2lnBF < 6; Figures 6D–F).
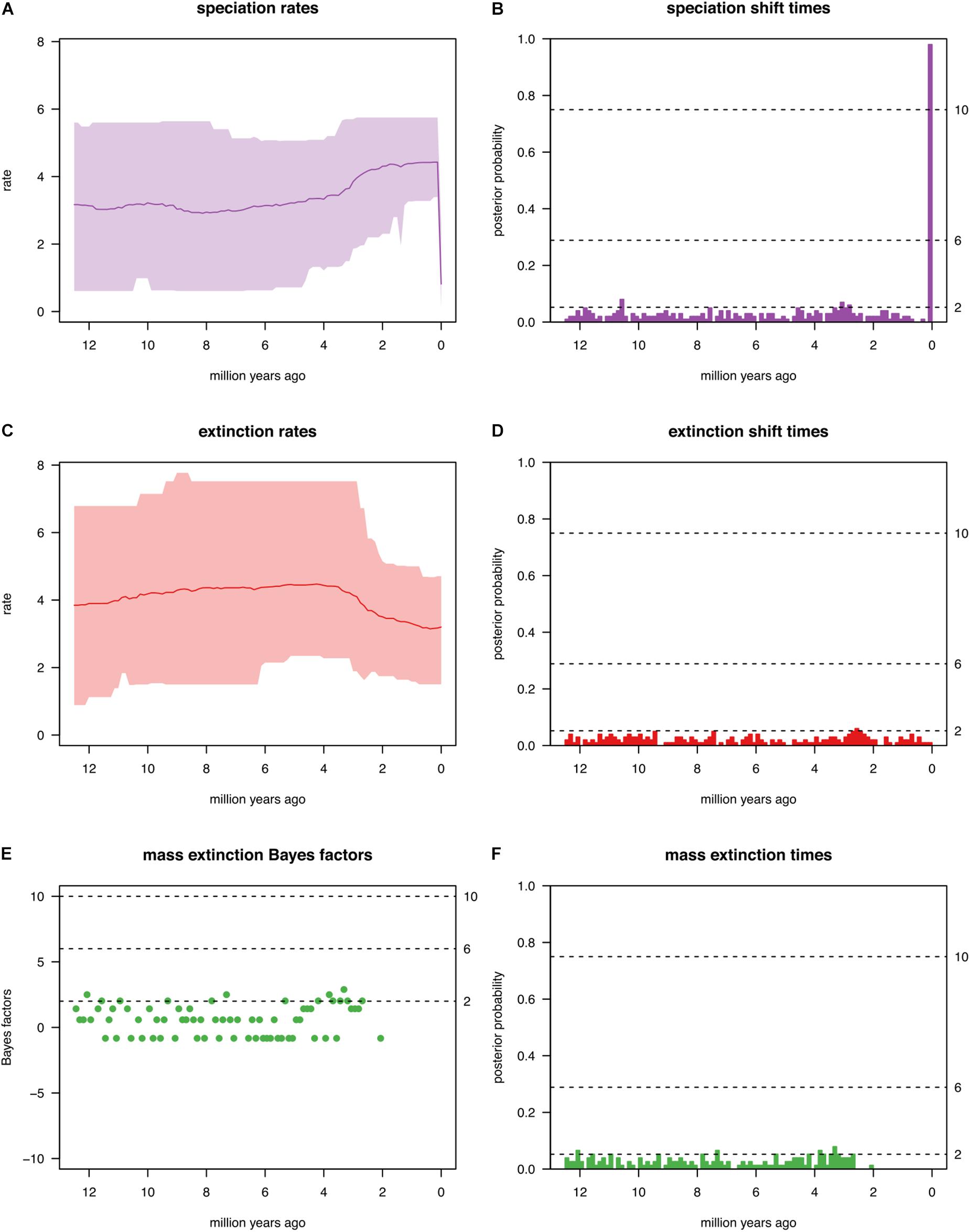
Figure 6. Summary of all evolutionary events reported in this study by fitting the CoMET model. Results reported are for the diversification hyperpriors specified a priori. (A) Speciation rate; (B) Speciation shift times; (C) Extinction rates; (D) Extinction shift times; (E) Mass extinction Bayes factors; (F) Mass extinction times.
Discussion
Phylogenetic Tree of Cycas
In comparison with the phylogeny reported in Liu et al. (2018), our phylogeny is similar in terms of the topology and the node support. This is not surprising because we used their DNA sequences. Three of the six Sections of the genus are polyphyletic (Cycas, Stangerioides, Indosinense) and the remaining sections are monophyletic as previously reported (Xiao and Möller, 2015; Liu et al., 2018). There are a few points worth highlighting. In our phylogeny, the species Cycas macrocarpa and Cycas pranburiensis are nested within the section Indosinenses, but they were included in the section Cycas in previous studies (Hill and Yang, 1999; Liu et al., 2018). Our finding is likely due to the following reason. The sections Cycas and Indosinenses have overlapping distribution pattern in Southeast China and India that might have caused a gene flow within the two sections (Yang and Meerow, 1996), making it difficult to distinguish species of these two sections on a phylogeny.
Diversification and Historical Biogeography
The Dispersal-Extinction-Cladogenesis (DEC) model provides alternative option to BBM for historical biogeographic analysis as it takes into account, unlike the BBM, the adjacent configuration of the areas through time (Ree and Smith, 2008; Beeravolu and Condamine, 2016). However, we reported only the results of BBM based on the following reasons. First, Xiao and Möller (2015) conducted a similar study on the same genus but with limited sampling size; in their study they used BBM, and for us to be able to compare our findings with theirs, we used the same BBM model. Our findings are indeed different from theirs. Second, we did run the DEC analysis, but the results point to an uncertain origin for the genus (60% of uncertainty) as opposed to the finding of the BBM (10% of uncertainty). In addition to the differences in sampling size between both studies (ours and that of Xiao and Möller, 2015), it is important to highlight the influence of priors (e.g., Yule vs. birth-death) on age estimates or divergence times. This has been showed in recent studies. For example, Condamine et al. (2015) demonstrated striking differences in node age estimates between the Yule versus the birth-death priors employed on the same dataset to assemble a phylogenetic tree of cycads (see also Couvreur et al., 2010). This is an additional but essential justification for the present study to re-investigate the biogeography of the genus Cycas.
Indeed, the biogeography of the genus Cycas has been investigated in recent studies (e.g., Keppel et al., 2008; Xiao and Möller, 2015). In their study, Xiao and Möller (2015) indicated, with high level of confidence (∼94%), that South China is the origin of the genus. Our analysis, instead, pointed to Indo-China as the origin of the genus, which dating back to ∼ 12 Ma (evolutionary age of the genus). They also indicated that Indo-China was the first geographic region to be colonized by the genus through vicariance and dispersal from South China (with a relatively low level of confidence of 46%) with a series of late dispersal events across the Malay archipelagos through to Australia and East Africa. In our study, the colonization routes are different. Specifically, we found that the colonization route might actually have started first from Indo-China (ancestral area B; Figure 2) to the ancestral area (C) (Malay islands southward to Australia and westward to Madagascar, East-Africa) and last from Indo-China (B) to South China (A).
Indeed, the historical biogeography of the Pacific Island’s flora has always been a matter of debate (e.g., see Keppel et al., 2009). Our study adds to this debate specifically with regard to the origin and the ecological forces that might have driven the distribution of the genus Cycas across the island. The differences between our findings and those of Xiao and Möller (2015) could be linked to the differences in the sampling size between both studies. Although they included representatives of the major sections of the genus into their analysis, only 31 species were analyzed whilst ours includes the complete sample (116 species) of the genus. In addition, our analysis further contradicts theirs in term of the sequences of the colonization events. As opposed to Xiao and Möller (2015), we found that the colonization of South China occurred actually not at an early stage but at the last, after the rest of the genus distribution ranges across the Pacific Islands has been colonized. However, our study agrees with Xiao and Möller (2015) concerning the ecological processes (dispersal and vicariance) that might have mediated the colonization. On this aspect, the Red River Fault may have played an important role, which may include the role of a geographic barrier between Indo-China and South China (Xiao and Möller, 2015; Zheng et al., 2016). This barrier may account for the delay of the colonization of South China in comparison to the early colonization of the Malay archipelagos and the distribution ranges of the genus previously reported (Xiao and Möller, 2015).
In this early colonization of the Malay archipelagos, Malaysia might have played the role of a source area from which the genus might have dispersed westward to East Africa and eastward into the Pacific Islands (centre-periphery hypothesis, Brown, 1984; Hampe and Petit, 2005; Kawecki, 2008; Gaston, 2009). The centre-periphery hypothesis provides an explanation to the biogeographical distribution of species from their centre of origin to their peripheral ranges. The hypothesis predicts that populations are more isolated and less abundant toward the periphery of their distribution (Sexton et al., 2009). Although we did not explicitly test this hypothesis in this study, early studies reported an overall decrease in taxonomic diversity of various plant groups from Malesia eastward in the Pacific region (Corner, 1963; van Balgooy, 1969; Woodroffe, 1987). Even this report holds for the genus Cycas as, for example, most Cycas species in the subsection Rumphiae are centred in or around Malesia (Hill, 1996a, b; Keppel et al., 2008).
The debate on the colonization process of the Pacific Islands (Keast and Miller, 1996; Ebach and Tangey, 2006) revolves around vicariance and long-distance dispersal events (see Keppel et al., 2009). The vicariance biogeography (Nelson and Platnick, 1981) was initially believed to be the major force structuring the flora of the Pacific regions (Whitmore, 1973; Ladiges et al., 2003; Heads, 2006, 2008; Ladiges and Cantrill, 2007). However, the long distance dispersal process has also been central in the early debate (Darwin, 1859; Guppy, 1906; Ridley, 1930; Mayr, 1954; Carlquist, 1967). Interestingly, mounting evidence, including molecular data, supports the long distance dispersal scenario (Turner et al., 2001; Price and Clague, 2002; Winkworth et al., 2002, 2005; Perrie and Brownsey, 2007). For the genus Cycas, the long distance dispersal is more likely the main event through which the entire geographic ranges of Cycas has been colonized (Keppel et al., 2008; Xiao and Möller, 2015). There are various scenarios for this mode of dispersal event, including the hitch-hiking, stepping-stones, and long distance dispersal scenarios (Keppel et al., 2009) mediated through a floatation-facilitating layer in the seeds of Cycas (Xiao and Möller, 2015; Zheng et al., 2016).
To further elucidate this historical biogeographic process, we first tested for the diversification model explaining the diversification history of the genus. All the tests point to an overall constant diversification history over time. Such constant diversification has recently been reported for another cycad genus, the African-endemic genus Encephalartos (Yessoufou et al., 2014; Mankga et al., 2020b). This suggests that cycads in general, not only diverged globally at the similar period (Nagalingum et al., 2011), but their diversification may have, perhaps, followed a similar overall pattern of constant-rate diversification. We explored several evolutionary events that might shape the diversification of Cycas, including speciation, extinction, and mass extinctions. Around 12 Ma, we found an initial speciation rate that is very similar to the overall speciation rate reported for gymnosperm in general (Crisp and Cook, 2011). However, the overall speciation corresponds to the late Miocene (Tortonian-Messinian), a period characterized in the Pacific regions by frequent sea level excursions (e.g., eight sea level excursions; Aharon et al., 1993). These multiple frequent rises and falls of sea level would likely promoted long-distance dispersal of Cycas seeds across the Pacific Islands through to Australia, Madagascar and East Africa. For example, species in the subsection Rumphiae developed seeds with spongy layer inside the sclerotesta (de Laubenfels and Adema, 1998); the “spongy” characteristic of the seeds facilitates the floatation of the seeds, potentially promoting a long trans-oceanic dispersal across the Pacific Islands (de Laubenfels and Adema, 1998; Xiao and Möller, 2015; Zheng et al., 2017).
Cycads have a fascinating evolutionary history (Mankga et al., 2020b) starting around 300 Ma (Hendricks, 1987), and the extant cycads re-diversified around 12-2 Ma (Nagalingum et al., 2011). They share morphological characteristics of ferns and angiosperms (Norstog and Nicholls, 1997; Brenner et al., 2003), and these characteristics make them a unique taxonomic and evolutionary group. In this group, the genus Cycas has recently been identified as the most rapidly diversified and widely distributed clade (Yessoufou et al., 2017). Here we build upon this knowledge to reconstruct the historical biogeography and the evolutionary events that might shape the rapid diversification and wide distribution across the Pacific Islands. Our analysis indicated that Indo-China may have been the origin of the genus (but see Xiao and Möller, 2015), and that the pacific island may have been first colonized through dispersal before the genus reaches South China. This dispersal may have been facilitated by multiple excursions of sea level and the development of a key innovation, a spongy endocarp. Our study therefore clarifies the historical biogeography and the evolutionary events that shaped the current diversity of the genus.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found in the Supplementary Material.
Author Contributions
KY designed the project and wrote the manuscript with editorial works from all other authors. LM collected the data. LM and KY analyzed the data. MC did the editorial works. TM did the GIS mapping.
Funding
We acknowledge the South Africa’s National Research Foundation (NRF) for funding (Research Development Grants for Y-Rated Researchers Grant No: 112113) to Dr. KY. Ms. LM acknowledges the staff grants support from the University of South Africa.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the two reviewers including the editor whose comments improve greatly an initial draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00044/full#supplementary-material
TABLE S1 | The species names and GenBank accession numbers of sequences used in the data analyses. All the sequences trnHpsbA, trnLF, trnS-trnG,psbM-trnD, RPB1, HZP, AC3, F3H, SAMS, PHYP and GTP were retrieved from GenBank. “—” indicates DNA sequences that are not available.
TABLE S2 | Summary statistics of the aligned DNA matrix.
TABLE S3 | The Bayes factor (BF) values calculated for each birth-death model tested for the phylogeny of the genus Cycas. ConstBD, constant-rate birth-death model; DecrBD, continuously variable-rate birth-death model; EpisodicBD, episodically variable-rate birth-death model; MassExtinctionBD, explicit mass-extinction birth-death model.
References
Aharon, P., Goldstein, S. L., Wheeler, C. W., and Jacobson, G. (1993). Sea level events in the South Pacific linked with the messinian salinity crisis. Geology 21, 771–775.
Baele, G., Lemey, P., and Vansteelandt, S. (2013). Make the most of your samples: bayes factor estimators for high-dimensional models of sequence evolution. BMC Bioinform. 14:85. doi: 10.1186/1471-2105-14-85
Beeravolu, C. R., and Condamine, F. L. (2016). An extended maximum likelihood inference of geographic range evolution by dispersal, local extinction and cladogenesis. BioRxiv [Preprint]. doi: 10.1101/038695
Brenner, E. D., Stevenson, D. W., and Twigg, R. W. (2003). Cycads: evolutionary innovations and the role of plant-derived neurotoxins. Trends Plant Sci. 8, 446–452. doi: 10.1016/s1360-1385(03)00190-0
Brown, J. H. (1984). On the relationship between abundance and distribution of species. Am. Natl. 124, 255–279. doi: 10.1086/284267
Calonje, M., Stevenson, D. W., and Stanberg, L. (2017). The World List of Cycads, Online edition [Internet]. 2013-2017. Available at: http://www.cycadlist.org (accessed January, 2017).
Carlquist, S. (1967). The biota of long-distance dispersal V. plant dispersal to Pacific Islands. Bull. Torrey Bot. Club 94, 129–162.
Condamine, F. L., Nagalingum, N. S., Marshall, C. R., and Morlon, H. (2015). Origin and diversification of living cycads: a cautionary tale on the impact of the branching prior in Bayesian molecular dating. BMC Evol. Biol. 15:65. doi: 10.1186/s12862-015-0347-8
Corner, E. J. H. (1963). “Ficus in the pacific region,” in Pacific Basin Biogeography – A Symposium, ed. J. L. Gressitcpesnm, (Hawaii: Bishop Museum Press), 233–245.
Couvreur, T. L. P., Franzke, A., Al-Shehbaz, I. A., Bakker, F., Koch, M., and Mummenhoff, K. (2010). Molecular phylogenetics, temporal diversification and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27, 55–71. doi: 10.1093/molbev/msp202
Crisp, M. D., and Cook, L. G. (2011). Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperm. New Phytol. 192, 997–1009. doi: 10.1111/j.1469-8137.2011.03862.x
Darwin, C. (1859). On the Origin of Species by Means of Natural Selection or the Preservation of Favored Races in the Struggle for Life. London: John Murray. doi: 10.1111/j.1469-8137.2011.03862.x
de Laubenfels, D. J., and Adema, F. A. (1998). Taxonomic revision of the genera Cycas and Epicycas gen. nov. (Cycadaceae). Blumea 43, 351–400.
Drummond, A. J., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Ebach, M. C., and Tangey, R. S. (2006). Biogeography in a Changing World. Boca Raton, FL: CRC Press. doi: 10.1186/1471-2148-7-214
Gao, Z. F., and Thomas, B. A. (1989). A review of fossil cycad megasporophylls, with new evidence of crossozamia pommel and its associated leaves from the lower Permian of Taiyuan, China. Rev. Palaeobot. Palynol. 60, 205–223. doi: 10.1016/0034-6667(89)90044-4
Gaston, K. J. (2009). Geographic range limits: achieving synthesis. Proc. R. Soc. B Biol. Sci. 276, 1395–1406. doi: 10.1098/rspb.2008.1480
Guan, Z. T. (1996). Cycads of China. Chengdu: Sichuan Science and Technology Press. doi: 10.1098/rspb.2008.1480
Guppy, H. B. (1906). Observations of a Naturalist in the Pacific between 1896 and 1899. Volume II: Plant Dispersal. London: Macmillan and Co. doi: 10.1098/rspb.2008.1480
Hampe, A., and Petit, R. J. (2005). Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 8, 461–467. doi: 10.1111/j.1461-0248.2005.00739.x
Heads, M. (2006). Seed plants of Fiji: an ecological analysis. Biol. J. Linnean Soc. 89, 407–431. doi: 10.1111/j.1095-8312.2006.00682.x
Heads, M. (2008). Panbiogeography of New Caledonia, southwest Pacific: basal angiosperms on basement terranes, ultramafic endemics inherited from volcanic island arcs and old taxa endemic to young islands. J. Biogeogr. 35, 2153–2175. doi: 10.1111/j.1365-2699.2008.01977.x
Hill, K. D. (1995). “Infrageneric relationships, phylogeny and biogeography of the genus Cycas (Cycadaceae),” in CYCAD 93, The 3rd International Conference on Cycad Biology, Proceedings, ed. P. Vorstercpesnm, (Stellenbosch: Cycad Society), 139–162.
Hill, K. D. (1996a). “Cycas, an evolutionary perspective. Biology and conservation of cycads,” in Proceedings of the Fourth International Conference on Cycad Biology, Panzhihua.
Hill, K. D. (1996b). “Cycads in the Pacific. The origin and evolution of Pacific island biotas,” in New Guinea to Eastern Polynesia: Patterns and Processes, eds A. Keastcpesnm, and S. E. Millercpesnm, (Amsterdam: SPB Academic Publishing), 267–274.
Hill, K. D. (2004). “Character evolution, species recognition and classification concepts in the Cycadaceae,” in Cycad Classification, Concepts and Recommendations, eds T. Walterscpesnm, and R. Osbornecpesnm, (Wallingford: CABI Publishing), 23–44. doi: 10.1079/9780851997414.0023
Hill, K. D., and Yang, S. L. (1999). The genus cycas (Cycadaceae) in Thailand. Brittonia 51, 48–73. doi: 10.1016/j.ympev.2018.05.019
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Höhna, S., May, M. R., and Moore, B. R. (2015). TESS: an R package for efficiently simulating phylogenetic trees and performing Bayesian inference of lineage diversification rates. Bioinform 32, 789–791. doi: 10.1093/bioinformatics/btv651
Jeffreys, H. (1961). The Theory of Probability. Oxford: Oxford University Press. doi: 10.1093/bioinformatics/btv651
Jones, D. L. (2002). Cycads of the World-Ancient Plant in Today’s Landscape. Washington, DC: Smithsonian Institution Press. doi: 10.1093/bioinformatics/btv651
Kawecki, T. J. (2008). Adaptation to marginal habitats. Ann. Rev. Ecol. Evol. Syst. 39, 321–342. doi: 10.1146/annurev.ecolsys.38.091206.095622
Keast, A., and Miller, S. E. (1996). The Origin and Evolution of Pacific Island Biotas, New Guinea to Eastern Polynesia: Patterns and Processes. Amsterdam: SPB Academic Publishing. doi: 10.1146/annurev.ecolsys.38.091206.095622
Keppel, G., Hodgskiss, P. D., and Plunkett, G. M. (2008). Cycads in the insular Southwest Pacific: dispersal or vicariance? J. Biogeogr. 35, 1004–1015. doi: 10.1111/j.1365-2699.2007.01869.x
Keppel, G., Lowe, A. J., and Possingham, H. P. (2009). Changing perspective on the biogeographical of the tropical South Pacific: influences of dispersal, vicariance and extinction. J. Biogeogr. 36, 1035–1054. doi: 10.1111/j.1365-2699.2009.02095.x
Ladiges, P. Y., and Cantrill, D. (2007). New Caledonia–Australia connections: biogeographic patterns and geology. Aust. Syst. Bot. 20, 383–389.
Ladiges, P. Y., Udovicic, F., and Nelson, G. (2003). Australian biogeographical connections and the phylogeny of large genera in the plant family Myrtaceae. J. Biogeogr. 30, 989–998. doi: 10.1046/j.1365-2699.2003.00881.x
Liu, J., Zhang, S., Nagalingum, N. S., Chiang, Y., Lindstrom, A., and Gong, X. (2018). Phylogeny of the gymnosperms genus Cycas L. (Cycadaceae) as inferred from plastid and nuclear loci based on a large-scale sampling: evolutionary relationships and taxonomical implications. Mol. Phylogenet. Evol. 127, 87–97. doi: 10.1016/j.ympev.2018.05.019
Mankga, L. T., Yessoufou, K., and Chitakira, M. (2020a). On the origin and diversification history of the African genus Encephalartos. South Afr. J. Bot. 130, 231–239. doi: 10.1016/j.sajb.2019.12.007
Mankga, L. T., Yessoufou, K., Mugwena, T., and Chitakira, M. (2020b). The genus cycas may have diversified from Indochina and occupied its current ranges through vicariance and dispersal events. Dryad, Dataset. https://doi.org/10.5061/dryad.1g1jwstrn
May, M. R., Höhna, S., and Moore, B. R. (2016). A Bayesian approach for detecting mass-extinction events when rates of lineage diversification vary. Syst. Biol. 7, 947–959. doi: 10.1111/2041-210x.12563
Mayr, E. (1954). “Change of genetic environment and evolution,” in Evolution as a Process, eds J. Huxley, A. C. Hardy, and E. B. Fordcpesnm, (London: Allen and Unwin), 157–180.
Nagalingum, N. S., Marshall, C. R., Quental, T. B., Rai, H. S., Little, D. P., and Mathews, S. (2011). Recent synchronous radiation of a living fossil. Science 334, 796–799. doi: 10.1126/science.1209926
Nelson, G., and Platnick, N. I. (1981). Recent synchronous radiation of a living fossil. Systematics and Biogeography: Cladistics and Vicariance. New York, NY: Columbia University Press. doi: 10.1126/science.1209926
Norstog, K. J., and Nicholls, T. J. (1997). “The fossil Cycadophytes,” in The Biology of the Cycads, eds K. J. Norstogcpesnm and T. J. Nichollscpesnm, (Ithaca, NY: Cornell University Press), 169–201.
Nylander, J. A. A. (2004). Modeltest v2. Program Distributed by the Author. Sweden: Evolutionary Biology Centre.
Osborne, R., Calonje, M. A., Hill, K. D., Stanberg, L., and Stevenson, D. W. (2012). The world list of Cycads. Mem. N.Y. Bot. Gard. 106, 480–510.
Perrie, L. R., and Brownsey, P. (2007). Molecular evidence for long-distance dispersal in the New Zealand pteridophyte flora. J. Biogeogr. 34, 2028–2038. doi: 10.1111/j.1365-2699.2007.01748.x
Price, J. P., and Clague, D. A. (2002). How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc. R. Soc. B Biol. Sci. 269, 2429–2435. doi: 10.1098/rspb.2002.2175
Rambaut, A., and Drummond, A. J. (2007). Tree Annotator (Version 1.5.4). Available at http://beast.bio.ed.ac.uk (accessed January, 2017).
Ree, R. H., and Smith, S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. doi: 10.1080/10635150701883881
Ridley, N. H. (1930). Dispersal of Plants Throughout the World. Ashford: L. Reeve and Co. doi: 10.1080/10635150701883881
Schenk, J. J. (2016). Consequences of secondary calibrations on divergence time estimates. PLoS One 11:e0148228. doi: 10.1371/journal.pone.0148228
Sexton, J. P., McIntyre, P. J., Angert, A. L., and Rice, K. J. (2009). Evolution and ecology of species range limits. Ann. Rev. Ecol. Evol. Syst. 4, 415–436. doi: 10.1146/annurev.ecolsys.110308.120317
Swofford, D. L. (2002). PAUP: Phylogenetic Analysis Using Parsimony (and Other methods), Version 4.0b10. Sunderland, MA: Sinauer Associates.
Turner, H., Hovenkamp, P. H., and van Welzen, P. C. (2001). Biogeography of Southeast Asia and the West Pacific. J. Biogeogr. 28, 217–230. doi: 10.1046/j.1365-2699.2001.00526.x
van Balgooy, M. M. J. (1969). A study on the diversity of island floras. Biodiv. Evol. Biogeogr. Plants 17, 139–178.
Whitmore, T. C. (1973). Plate tectonics and some aspects of Pacific plant geography. New Phytol. 72, 1185–1190. doi: 10.1111/j.1469-8137.1973.tb02095.x
Wilcox, T. P., Zwicki, D. J., Heath, T. A., and Hillis, D. M. (2002). Phylogenetic relationships of the dwarf boas and a comparison of Bayesian and bootstrap measures of phylogenetic support. Mol. Phylogenet. Evol. 25, 361–371. doi: 10.1016/s1055-7903(02)00244-0
Winkworth, R. C., Wagstaff, S. J., Glenny, D., and Lockhart, P. J. (2002). Plant dispersal N.E.W.S. from New Zealand. Trends Ecol. Evol. 17, 514–520.
Winkworth, R. C., Wagstaff, S. J., Glenny, D., and Lockhart, P. J. (2005). Evolution of the New Zealand mountain flora: origins diversification and dispersal. Org. Diver. Evol. 5, 237–247. doi: 10.1016/j.ode.2004.12.001
Woodroffe, C. D. (1987). Pacific Island mangroves: distribution and environmental settings. Pac. Sci. 41, 166–185.
Xiao, L. Q., and Möller, M. (2015). Nuclear ribosomal ITS functional paralogs resolve the phylogenetic relationships of a late-Miocene radiation cycad Cycas (Cycadaceae). PLoS One 10:e0117971. doi: 10.1371/journal.pone.0117971
Yan, Y., Harris, A. J., and Xingjin, H. (2010). S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850. doi: 10.1016/j.ympev.2010.04.011
Yang, S. L., and Meerow, A. W. (1996). The Cycas pectinata (Cycadaceae) complex: genetic structure and gene flow. Int. J. Plant Sci. 157, 468–483. doi: 10.1086/297364
Yessoufou, K., Bamigboye, S. O., Daru, B. H., and Van der Bank, M. (2014). Evidence of constant diversification punctuated by a mass extinction in the African cycads. Ecol. Evol. 4, 50–58. doi: 10.1002/ece3.880
Yessoufou, K., Daru, B. H., Tafirei, R., Elansary, H. O., and Rampedi, I. (2017). Integrating biogeography, threat and evolutionary data to explore extinction crisis in the taxonomic group of cycads. Ecol. Evol. 7, 2735–2746. doi: 10.1002/ece3.2660
Zheng, Y., Liu, J., Feng, X., and Gong, X. (2017). The distribution, diversity, and conservation status of Cycas in China. Ecol. Evol. 7, 3212–3224. doi: 10.1002/ece3.2910
Keywords: cycads, DNA barcode, evolutionary diversification, historical biogeography, late Miocene, sea-level excursions
Citation: Mankga LT, Yessoufou K, Mugwena T and Chitakira M (2020) The Cycad Genus Cycas May Have Diversified From Indochina and Occupied Its Current Ranges Through Vicariance and Dispersal Events. Front. Ecol. Evol. 8:44. doi: 10.3389/fevo.2020.00044
Received: 04 November 2019; Accepted: 11 February 2020;
Published: 28 February 2020.
Edited by:
Rodney L. Honeycutt, Pepperdine University, United StatesReviewed by:
Michael Wink, Heidelberg University, GermanyFabien L. Condamine, University of Gothenburg, Sweden
Copyright © 2020 Mankga, Yessoufou, Mugwena and Chitakira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kowiyou Yessoufou, a293aXlvdXlAdWouYWMuemE=
 Ledile T. Mankga
Ledile T. Mankga Kowiyou Yessoufou2*
Kowiyou Yessoufou2* Thendo Mugwena
Thendo Mugwena Munyaradzi Chitakira
Munyaradzi Chitakira