- 1Korean Entomological Institute, Korea University, Seoul, South Korea
- 2Molecular Ecology and Evolution Laboratory, Department of Biological Science, Sangji University, Wonju, South Korea
- 3GenCube, Gimpo, South Korea
- 4Division of Ocean Climate and Ecology Research, National Institute of Fisheries Science, Busan, South Korea
- 5School of Marine Biomedical Science (BK21 PLUS), Jeju National University, Jeju City, South Korea
- 6Biodiversity Research Center, Academia Sinica (BRCAS), Nankang, Taipei, Taiwan
- 7Shimoda Marine Research Centre, University of Tsukuba, Shizuoka, Japan
- 8Molecular Invertebrate Systematics and Ecology Laboratory, Graduate School of Engineering and Science, University of the Ryukyus, Nishihara, Japan
- 9Tropical Biosphere Research Center, University of the Ryukyus, Nishihara, Japan
- 10Laboratory of Estuarine and Coastal Ecology, Department of Marine Life Sciences, Jeju National University, Jeju City, South Korea
Marine ecosystems in temperate regions have been significantly affected by rising seawater temperatures due to climate change. Alveopora japonica, a small zooxanthellate scleractinian coral, occurs in the northwestern Pacific including Taiwan, Japan, and Jeju Island in Korea. The northern populations around Jeju Island have recently undergone rapid growth in numbers, with negative impacts on local biodiversity. However, it is unclear whether these Korean populations occurred historically and where they originate from. In this study, we investigated the phylogenetic relationships of A. japonica along with its endosymbiont Symbiodiniaceae across five geographic regions, including Jeju Island in Korea, Taiwan, and Japan. Nuclear internal transcribed spacer (ITS) sequences revealed unexpected species-level divergence among three distinct phylogenetic clusters (Korea, Taiwan, and Japan) with no sharing of haplotypes among lineages, suggesting these may each represent cryptic species in Alveopora. 23S ribosomal DNA of Symbiodiniaceae showed two well-separated phylogenetic clusters, in which Korean and Japanese symbionts shared the same clade and Taiwanese ones formed a distinct clade. Given the deep phylogenetic divergences among the lineages for both corals and Symbiodiniaceae, the Korean populations appear to have existed for a long evolutionary time period rather than representing a poleward migration from subtropical environments following recent climate change. Our study highlights cryptic species diversity in Alveopora at high-latitude environments.
Introduction
Over the past century, ocean warming driven by global climate change has led to shifts in geographic ranges toward higher latitude environments for a number of marine species (O’Connor et al., 2015; Poloczanska et al., 2016). Northward range shifts, i.e., range expansions in response to rising sea surface temperatures, have also been frequently reported for tropical reef scleractinian corals (Precht and Aronson, 2004; Greenstein and Pandolfi, 2008; Yamano et al., 2011). In addition to poleward migration, climate changes have strong negative impacts on the scleractinian corals even in temperate areas, as they may suffer from ocean acidification in the future (Mollica et al., 2018). However, climate changes can also have positive effects on population growth (i.e., population expansion) of corals via modulating growth, reproduction and recruitment (Denis et al., 2014; Vieira et al., 2016) or by creating novel coral assemblages sometimes resistant to disease (Yakob and Mumby, 2011), particularly in temperate regions. It has been suggested that high-latitude environments in the northern hemisphere might serve as climate change “refugia” for tropical coral reef species threatened by the negative effects of climate change (Yamano et al., 2011; Beger et al., 2014).
“Sub-tropicalization” of high-latitude (or temperate) marine ecosystems accompanied by ocean warming has driven dramatic alterations in community structures and functions, and has been reported from around the world including in the Pacific, the Atlantic and the Mediterranean sea (Vergés et al., 2014). In the northwestern Pacific (NWP), the sub-tropicalization of pelagic and benthic communities in Korean waters, especially around Jeju Island located approximately 155–200 km off the southern tip of the Korean Peninsula, is believed to be currently under way (National Institute of Fisheries Science, 2012). This region has been reported as one of the fastest warming regions in the world, and the sea surface temperature around Jeju Island has risen by 1.6 to 2.1°C over the last 100 years (Takatsuki et al., 2007). Recently, incidences of sub-tropical fish and scleractinian coral species in temperate Korean waters have been frequently reported, even further north from Jeju Island (Kim et al., 2015).
The subtropical scleractinian coral A. japonica Eguchi, 1968 (Scleractinia; Acroporidae) (Eguchi, 1968) is a hermaphroditic brooding coral known to be the northernmost ranged species within the genus Alveopora (Harii et al., 2001). This species occurs in subtidal benthic habitats in NWP ranging from southern Taiwan through Jeju Island, Korea and to mainland Japan (Figure 1; Veron, 2000). A. japonica has benefited from recent environmental changes such as the increase in seawater temperatures in high-latitude habitats (Denis et al., 2013; Denis et al., 2014; Vieira et al., 2016). The Jeju Island populations have rapidly repopulated recently due to the accelerated recruitment rate along with a superior competitive ability for space (Denis et al., 2013; Denis et al., 2014). A recent ecological survey revealed that Jeju Island populations in Korea are now propagating rapidly, which has resulted in strong competition for space with the regionally dominant and ecologically and economically important kelp species Ecklonia cava (Denis et al., 2013). This may be due to parallel occurrences of kelp decreasing and scleractinian coral increasing due to the rising seawater temperature. Nonetheless, A. japonica appears to be competitively superior to E. cava, which has led to a dramatic decline in kelp populations, causing a significant loss in regional biodiversity, particularly for economically important fisheries (Denis et al., 2013). Hence, public concern over the potential adverse effects of A. japonica populations on fisheries production has increased in the local community of Jeju Island. However, whether A. japonica previously occurred around Jeju Island and where these populations have originated from remain unclear. Two alternative, mutually exclusive, scenarios can be postulated: either “poleward migration” in the NWP from subtropical habitats (e.g., Taiwan) by either historical or contemporary gene flow following climate change; or recent expansion in numbers from a small population previously unnoticed by researchers.
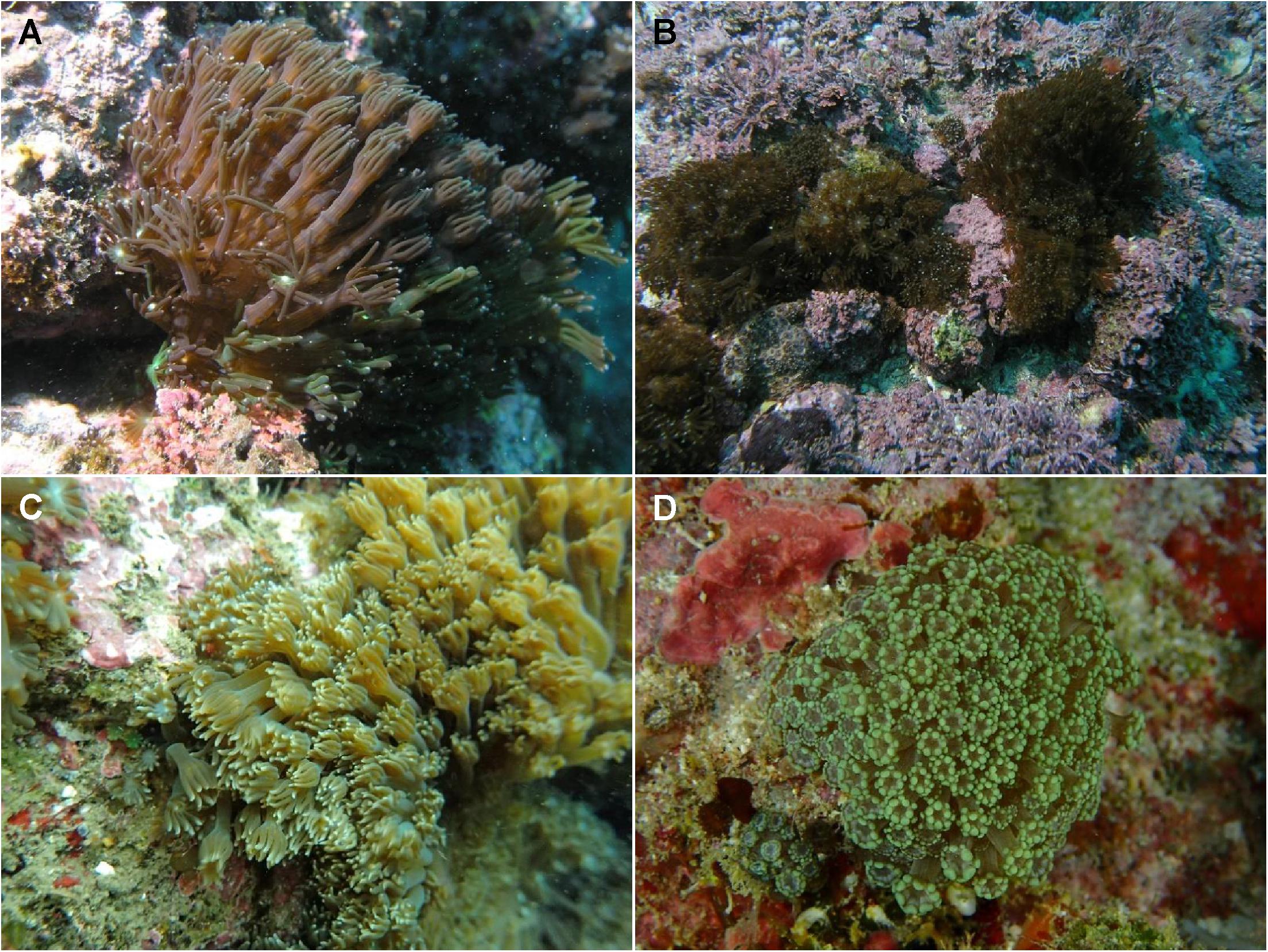
Figure 1. Photos of Alveopora japonica. Bomok (A) and Geumneung (B) populations around Jeju Island, off the southern coast of Korea; Shimoda (C) on the central southern coast of Japan; Green Island (D) in Taiwan. Skeletal features of these samples are shown in Supplementary Figure S1.
Molecular marker-based phylogeographic and phylogenetic analyses offer a powerful approach for tracking population or species histories (i.e., historical demography), and for inferring colonization patterns and evolutionary origins of populations and species (Avise, 2004). Phylogeographic and phylogenetic studies have been performed to understand the evolutionary history or phylogenetic patterns as well as phylogeographic structure for several marine invertebrate species, such as gastropods Littorina spp. (Lee and Boulding, 2007, 2009) and Thais clavigera (Guo et al., 2015), shrimps Palaemon spp. (Wood et al., 2017), the sponge Crambe crambe (Duran et al., 2004), the ctenophore Mnemiopsis leidyi (Bayha et al., 2015), and scleractinian corals such as Pocillopora damicornis (Schmidt-Roach et al., 2013) and Seriatopora hystrix (Warner et al., 2015). In addition, genetic surveys can allow for the identification of previously unrecognized, cryptic species diversity, particularly in morphologically variable species such as scleractinian corals (Schmidt-Roach et al., 2013; Boulay et al., 2014; Warner et al., 2015; Sheets et al., 2018).
Alveopora japonica has an obligate mutualistic relationship with dinoflagellate algae of the family Symbiodiniaceae (LaJeunesse et al., 2018). This association is a critical component in coral responses to environmental changes such as increasing seawater temperature as different genetic strains of Symbiodiniaceae display distinct eco-physiological characteristics (Cooper et al., 2011). Studies have revealed that the family Symbiodiniaceae contains several distinct genera (formerly “Clades” A–I) (Pochon and Gates, 2010; LaJeunesse et al., 2018). Subclade F2, which is exclusively associated with A. japonica (De Palmas et al., 2015), is a member of an undescribed genus (LaJeunesse et al., 2018). Although many studies have been conducted to examine the specificities and associations of different coral species and their Symbiodiniaceae (Chang et al., 2011; De Palmas et al., 2015), within-genus phylogeographic and phylogenetic patterns of Symbiodiniaceae remain poorly understood for many host taxa, including Alveopora.
In this study, we determined the phylogeographic and phylogenetic relationships of A. japonica and its endosymbiont Symbiodiniaceae by utilizing specimens from Jeju Island, off the southernmost region of Korea, as well as from the more subtropical/tropical environments of Taiwan and Japan. We tested the two alternative hypotheses of poleward migration or population expansion of A. japonica around Jeju Island. We further examined whether phylogenetic patterns of host A. japonica were congruent with those of their Symbiodiniaceae (i.e., phylogenetic concordance). We would predict that based on the poleward migration hypothesis, some A. japonica haplotypes should at least be shared among Taiwan, Korea and Japan sites due to historical or contemporary gene flow, with high haplotype diversity in ancestral populations (e.g., Taiwan) and lower diversity in derived populations (e.g., Korea, Japan) due to founder effects. Alternately, based the population expansion hypothesis, we would predict relatively deep and distinct genealogical structure to be present within each A. japonica regional group.
Materials and Methods
Coral Sampling
We collected a total of 121 individuals of A. japonica from five different localities in the NWP by scuba diving between May and July 2016 (Figures 1, 2). Those samples were collected in water depth between two and 15 meters. Specimens were collected from Bomok (BM) and Geumneung (GN) on the southern island of Korea (Jeju Island, located off the southern tip of Korea), Green Island (GI) and Kenting (KT) in Taiwan and Shimoda (SH) on the central southern coast of Japan (Figure 2). All collected colonies were identified as A. japonica following Veron (2000) and Kitano et al. (2014), based on colony size and shape, and skeletal features. They were always less than 4 cm in diameter and hemispherical or encrusting, and their septa had long and short fine spines which seldom connect. The skeletal structures of these A. japonica samples in SEM (Scanning Electron Microscope) images are shown in Supplementary Figure S1. No permits were required for the collection of Korean samples since this species is not listed as an endangered species in Korea. Taiwanese sampling was conducted in a non-marine protected area under the permission of a research project by Dr. Chaolun Allen Chen at the Biodiversity Research Centre, Academia Sinica (BRCAS). Japanese sampling was performed in the framework of a research project by Dr. Sylvain Agostini with a sampling permit granted by the prefectural government of Shizuoka, Japan. For each specimen, a small piece of coral tissue was collected by forceps from each specimen, preserved immediately in 99% ethanol and stored at 4°C until genetic analyses.
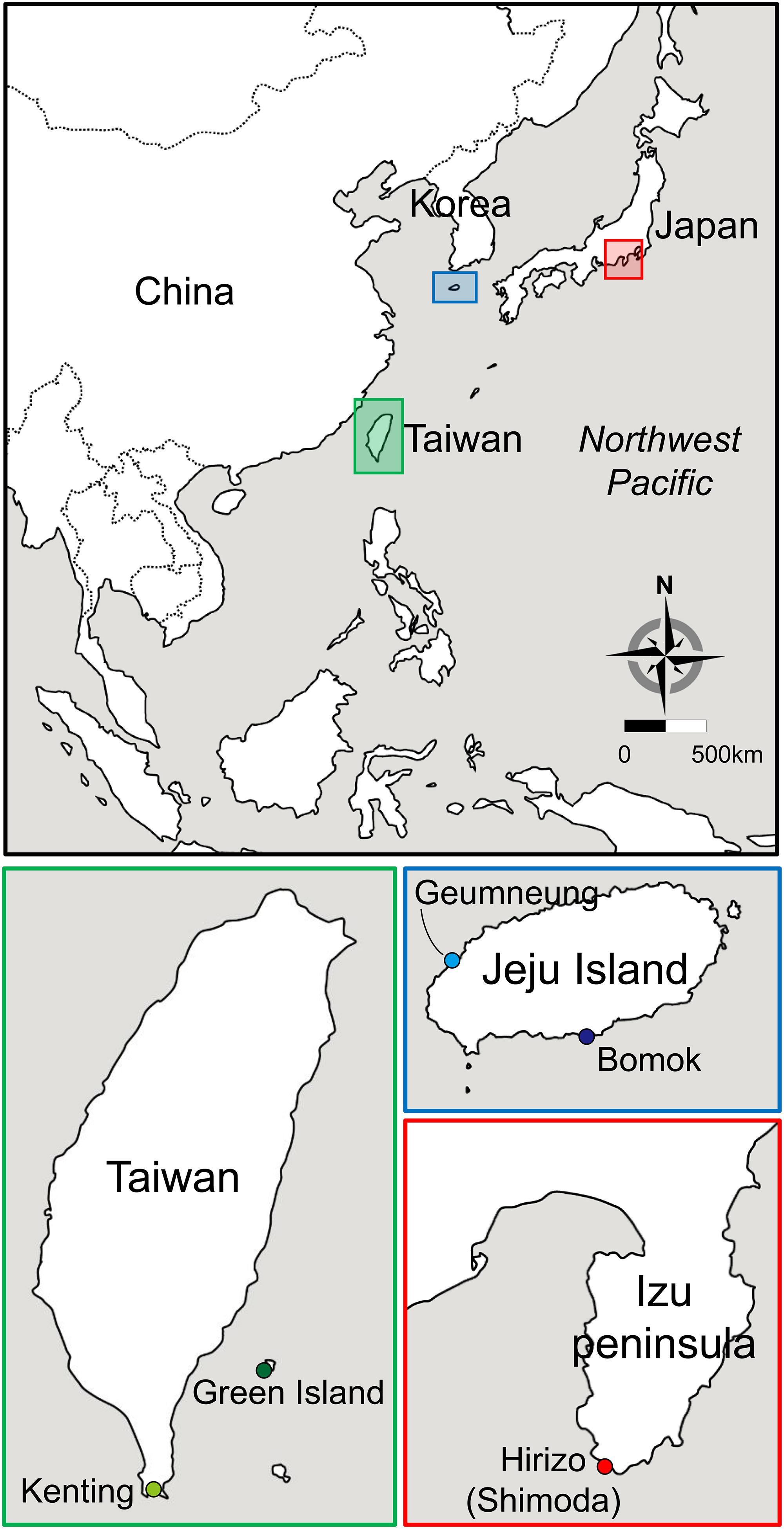
Figure 2. Map of five sampling sites of A. japonica and its symbiont Symbiodiniaceae in the northwestern Pacific Ocean [Jeju Island: Bomok (BM, 33°14′22.69′′N; 126°35′25.40′′E) and Geumneung (GN, 33°23′51.20′′N; 126°13′19.30′′E); Japan: Shimoda (SH, 34°36′55.27′′N; 138°48′42.87′′E); Taiwan: Green Island (GI, 22°39′14.00′′N; 121°28′23.63′′E); and Kenting (KT, 21°56′53.87′′N; 120°46′41.61′′E)]. Different colors denote the geographic regions where the corals and symbionts were sampled (Korea in blue, Japan in red, and Taiwan in green).
DNA Extraction, PCR, and Sequencing
Ethanol preserved tissues were used for genomic DNA extraction for both A. japonica (N = 121) and its endosymbiont Symbiodiniaceae (N = 110) using the DNeasy Blood and Tissue Kit (Qiagen, United States) following the manufacturer’s directions. DNA concentrations were determined using a Qubit® 2.0 Fluorometer (Invitrogen, United States). For A. japonica, mitochondrial (mt) DNA cytochrome oxidase I (COI) (Forsman et al., 2009) and nuclear DNA ribosomal internal transcribed spacer (ITS) (Chen et al., 1995; Wei et al., 2006) regions were amplified using published primers (COI, forward ZCOI: 5′-TCA ACT AAT CAT AAA GAT ATT GGT ACG-3′ and reverse ZCO1R: 5′-TAA ACC TCT GGA TGC CCA AA-3′; ITS, forward 1S: 5′-GGT ACC CTT TGT ACA CAC CGC CCG TCG CT-3′ and reverse 2SS: 5′-GCT TTG GGC GGC AGT CCC AAG CAA CCC GAC TC-3′). For Symbiodiniaceae, the chloroplast (cp) DNA 23S rDNA region was amplified using the following primers (Santos et al., 2002): forward 23S1M13: 5′-CAC GAC GTT GTA AAA CGA CGG CTG TAA CTA TAA CGG TCC-3′ and reverse 23S2M13: 5′-GGA TAA CAA TTT CAC ACA GGC CAT CGT ATT GAA CCC AGC-3′. All PCR amplifications were performed in a reaction volume of 15 μl containing 25 μM of each dNTP (Bio Basic, Canada), 0.6 μM each of the forward and reverse primers, 0.2 units of Taq DNA polymerase (Thermo Fisher Scientific, United States), 1 × PCR buffer, and approximately 5-10 ng of template DNA. PCR cycling conditions comprised an initial denaturation phase at 94°C for 5 min, followed by 37 cycles of 94°C for 20 sec (denaturation), 54–57°C for 30 sec (annealing), and 72°C for 30 sec (extension), followed by a terminal extension phase at 72°C for 10 min in a 2720 thermal cycler (Applied Biosystems, United States). Each PCR product was evaluated on a 2% agarose gel stained with RedsafeTM (iNtRON Biotechnology, Korea). The amplified PCR products were purified enzymatically with exonuclease I (New England BioLabs, United States) and shrimp alkaline phosphatase (New England BioLabs). The purified mtDNA, nuclear DNA and cpDNA fragments were then sequenced in both directions using an ABI 3730xl automated DNA sequencer (Applied Biosystems, United States) at the Humanizing Genomics Center of Macrogen in Seoul, South Korea.
Genetic Data Analyses
The obtained A. japonica COI (619 bp) and ITS (668 bp), and Symbiodiniaceae 23S rDNA (661 bp) sequences were aligned using Clustal W implemented with BioEdit v.7.0.1 (Hall, 1999) and manually edited. Due to no variation observed in COI sequences, this marker was not used for downstream analyses. Ambiguous regions in the alignment of ITS due to high levels of genetic variation were cut from the phylogenetic and phylogeographic analyses. Phylogenetic alignments were conducted separately using 123 coral ITS and 112 23S rDNA symbiont sequences. Goniopora albiconus (GenBank accession number AB906945) and G. djiboutiensis (AB748700) were used as outgroup species for the ITS of A. japonica, while sequences from Protodinium simplex (JN558033) and Polarella glacialis (JN558036) were used as outgroups for the 23S rDNA of Symbiodiniaceae.
The best nucleotide substitution model was tested using initial searches according to the AICc criterion with jModelTest v2.1.7 (Darriba et al., 2012), which selected the K80 + I + G and F81 + G model for host ITS and symbiont 23S rDNA, respectively. Maximum likelihood (ML) and neighbor joining (NJ) trees were inferred for all obtained sequences with PhyML v.3.0 (Guindon et al., 2010) and MEGA 7.0 (Kumar et al., 2016), respectively. Statistical supporting values were obtained with 1000 bootstrap pseudoreplicates.
Further phylogenetic analyses (NJ and ML) was conducted to determine the clades of endosymbiotic Symbiodiniaceae using five haplotypes determined from the five different sampling sites, along with 39 23S rDNA sequences representing eight different genera/clades [i.e., Symbiodinium (former Clade A), Breviolum (Clade B), Cladocopium (Clade C), Durusdinium (Clade D), Effrenium (Clade E), Clade F2-4, Fugacium (Clade F5), Gerakladium (Clade G), and Clade H] from the GenBank database (Supplementary Figure S2). Symbiodiniaceae Clade I was excluded for this phylogenetic analysis due to an ambiguous phylogenetic position with a long branch. Alignment and phylogenetic reconstruction with HKY + G + I as the best fit model were performed in the same manner as other phylogenetic analyses in this study.
The gametic phase of the nuclear ITS haplotypes was determined with the Excoffier-Laval-Balding (ELB) algorithm (Excoffier et al., 2003) from 121 individuals of A. japonica from all sampling localities using DnaSP v6 (Rozas and Rozas, 1999). The haplotype network was inferred using HAPSTAR v0.7 (Teacher and Griffiths, 2011) to identify the phylogenetic relationships among haplotypes. A further analysis was conducted using Haplowebs (Flot et al., 2010; Etoundi et al., 2019) with the online tool HaplowebMaker1 to show haplotype networks with additional connections between haplotypes found co-occurring in heterozygous individuals (Supplementary Figure S3). The genetic diversity indices of three separate clades (Korea, Taiwan, and Japan) were estimated using ARLEQUIN v3.5 (Excoffier and Lischer, 2010) including number of haplotypes (NH), haplotype diversity (h), nucleotide diversity (π), and number of polymorphic sites (NP). Haplotype richness (HR) was calculated by applying a rarefaction method using CONTRIB v1.02 (Petit et al., 1998) to correct for unequal sample sizes among the three regional groups. However, genetic diversity indices were not analyzed for Symbiodiniaceae since all the populations except for the KT population in Taiwan consisted of only a single haplotype.
To determine the ITS sequence divergence between the Korean, Taiwanese and Japanese clades in A. japonica, pairwise distances were estimated from Kimura-2-Parameter (K2P) distance model (Kimura, 1980), with 1000 bootstrap replications, using MEGA 7.0 (Kumar et al., 2016). Genetic differentiation among the three regional groups of A. japonica was estimated by pairwise FST statistics in ARLEQUIN v3.5 (Excoffier and Lischer, 2010). These sequence divergence and FST estimates were also performed for two sites within regions. The 95% significance levels for the pairwise population comparisons were adjusted using a Bonferroni correction. We also estimated the 23S rDNA divergence between the two clades of Taiwan and Korea/Japan for Symbiodiniaceae using Jukes-Cantor (JC) distance model (Jukes and Cantor, 1969) in MEGA.
Results
Phylogenetic Relationships and Cryptic Species Diversity in Alveopora
Obtained COI sequences (N = 50) were not polymorphic. Therefore, this marker was not used for downstream analyses. However, acquired ITS sequences (669 bp) of A. japonica (N = 121) showed 229 parsimonious informative sites and the overall mean diversity was 0.056. A total of 32 nuclear ITS haplotypes (=alleles), determined with the ELB algorithm (Excoffier et al., 2003), were identified from the five sampling localities.
The phylogenetic reconstruction of the ITS alignment of A. japonica uncovered three distinct phylogenetic clades, which corresponded to the respective geographic locations; Jeju Island in Korea (BM and GN), Taiwan (GI and KT), and Japan (SH) (Figures 2, 3). These three well-separated clades of A. japonica had high supporting values (91–100%) in both NJ and ML trees (Figure 3). In regions with two sites (BM and GN in Korea, GI and KT in Taiwan), sites did not form separate subclades. A. japonica lineages from Korea and Japan had a closer phylogenetic relationship to each other than to the lineage from Taiwan (Figure 3). Genetic diversity indices (NH, h, π, HR, and NP) of the ITS region within the three phylogenetic groups of A. japonica are summarized in Table 1.
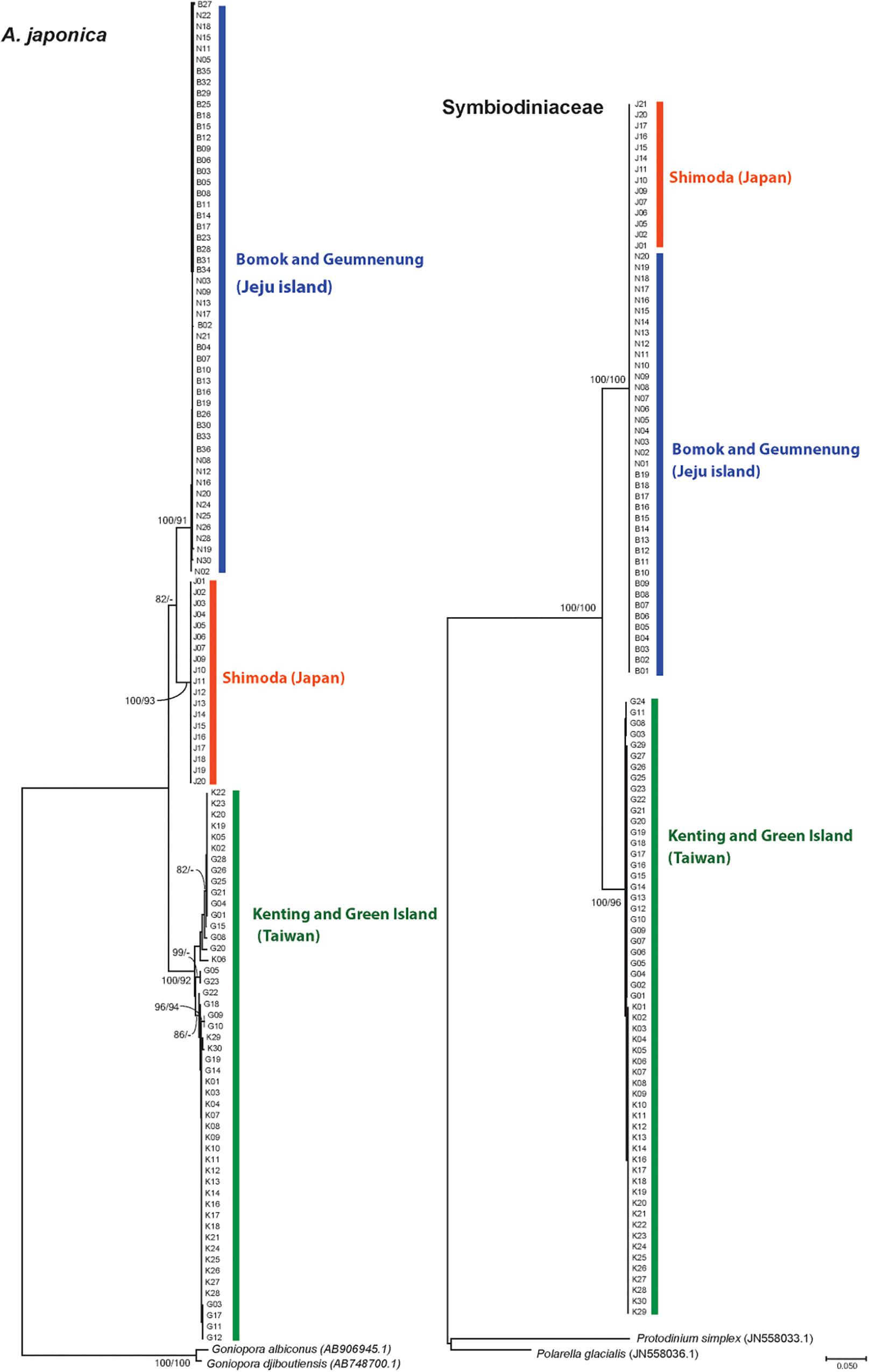
Figure 3. Phylogenetic trees of A. japonica (left, nuclear; ITS, internal transcribed spacer) and Symbiodiniaceae (right, chloroplast 23S rDNA) based on neighbor joining (NJ) and maximum likelihood (ML) methods. The values above the branches represent NJ and ML bootstrap supporting values. The nodes with values <50% are shown as “−.”
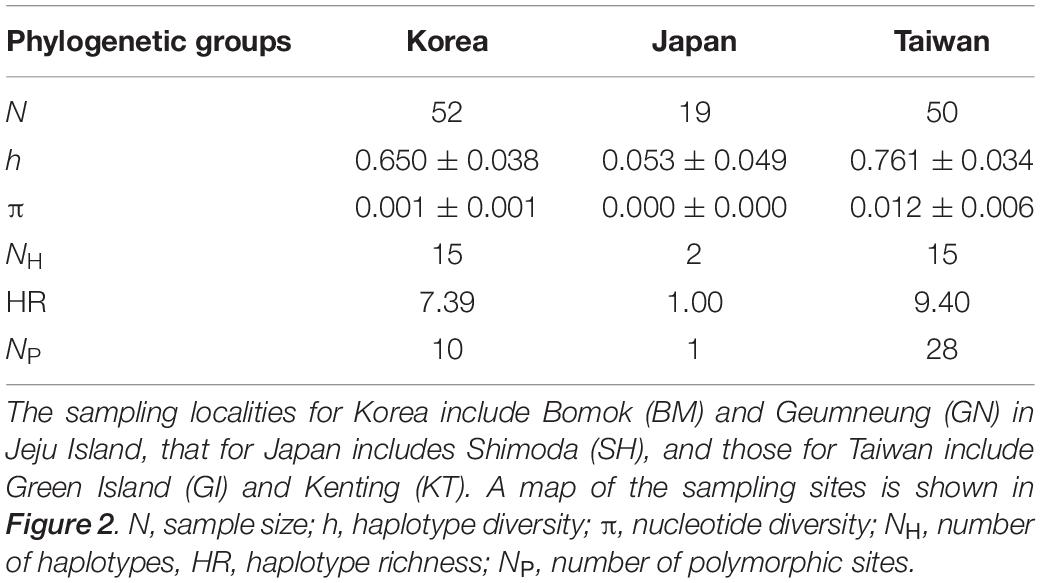
Table 1. Summary of the levels of genetic diversity in three phylogenetic groups of Alveopora japonica from Korea, Japan, and Taiwan at the nuclear ribosomal internal transcribed spacer (ITS) region (668 bp).
The haplotype network based on the ITS dataset of A. japonica also identified three deep phylogenetic groups comprising Jeju Island in Korea (BM and GN), Taiwan (GI and KT), and Japan (SH) (Figure 4; Supplementary Figure S3). The ITS sequence divergence levels between Taiwanese and Korean (42–75 mutational steps), and Korean and Japanese (26–31 steps) clades were large enough to be considered as separate species (“cryptic species complex”) based on the suggested species boundaries of >2–3% divergences in most animal taxa (Hebert et al., 2003). The average K2P distances between lineages from Korea and Taiwan, Korea and Japan, and Taiwan and Japan were 7.41% (6.59–8.43%), 4.21% (4.00–4.48%), and 7.62% (7.24–8.26%), respectively (Table 2). Those values between sites from Korea (BM vs. GN) and Taiwan (GI vs. KT) were 0.12% (0.00–0.45%), 1.35% (0.00–3.06%), respectively, which was much lower than between regional lineages.
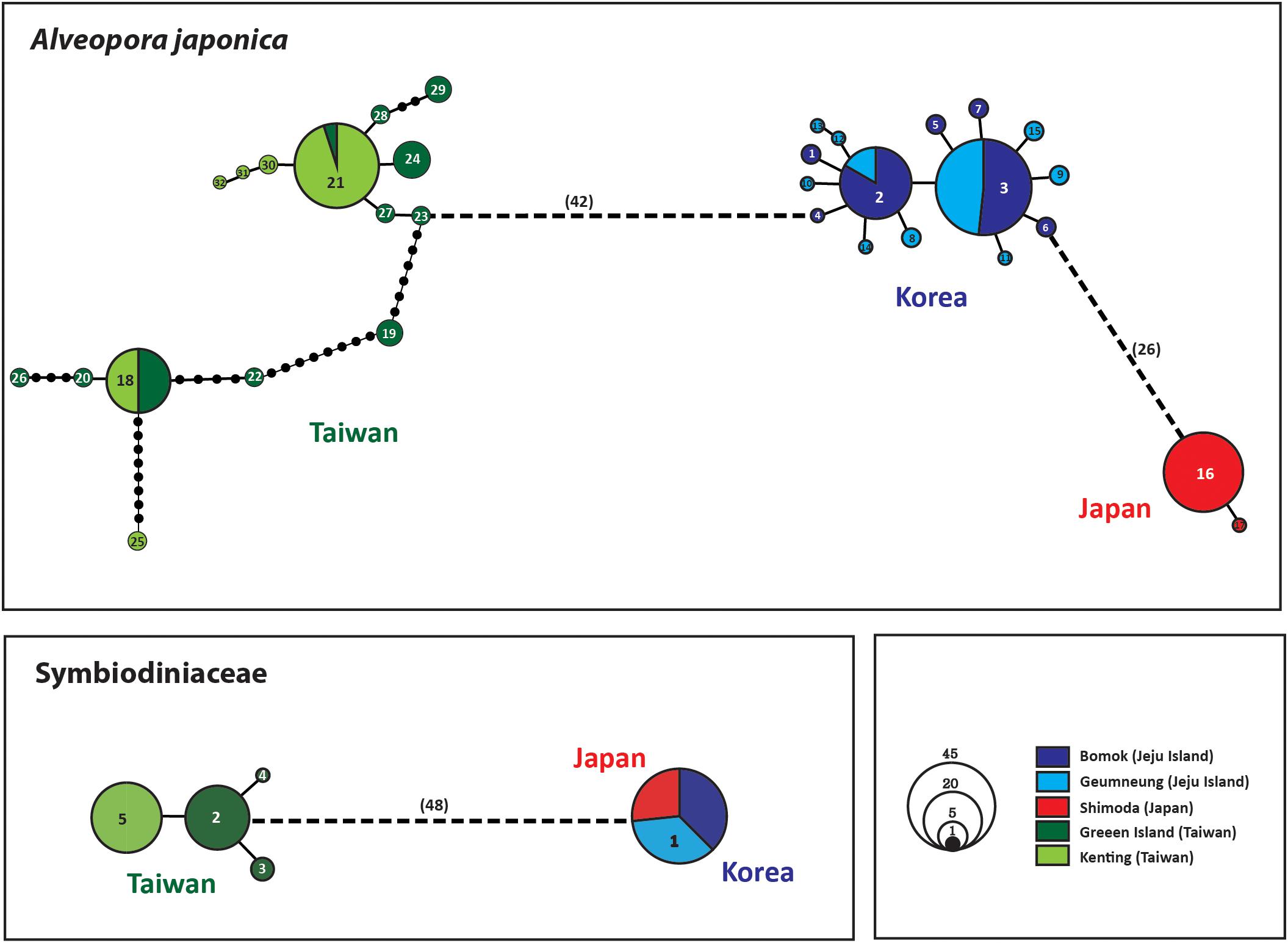
Figure 4. Statistical parsimony haplotype network of ITS haplotypes of A. japonica and 23S rDNA of Symbiodiniaceae. The area of the circle is proportional to the individual numbers found for the respective haplotype; numbers shown inside a circle represent haplotype numbers; each line in the network represents a single mutational step between haplotypes irrespective of its length (unless indicated otherwise). Different colors denote the geographic regions.

Table 2. Pairwise genetic differentiation (FST) based on haplotypes (above diagonal) and Kimura-2-parameter (K2P) distances (%) among the three phylogenetic groups (below diagonal) in Alveopora japonica using the ITS sequences.
Analyses of pairwise genetic differentiation (FST) based on ITS haplotypes showed strong divergence among the groups with a range of FST = 0.88–0.98, supporting the hypothesis that A. japonica from the NWP constitutes “three” cryptic genetic species (Table 2). However, much weaker differentiation was detected between two populations within regions (BM vs. GN in Korea: FST = 0.042; GI vs. KT in Taiwan: FST = 0.103), albeit a statistical significance. No haplotype was shared among the three different lineages, suggesting a complete lack of gene flow among these geographic regions. There were more mutational steps between Korea and Taiwan (42: H4–H23) than between Korea and Japan (26: H6–H16) (Figure 4). While 15 haplotypes were detected in each regional group from Korea and Taiwan, only 2 haplotypes were found in Japan, but the sample sizes analyzed were unequal among the regions (Korea: N = 52; Taiwan: N = 50; Japan: N = 19) (Table 1). More mutational steps among the haplotypes within Taiwan [1 to 36 mutational steps (H25–H29)] than among those within Korea and Japan [1 to 4 mutational steps (H9–H13)] indicated more genetic divergence and diversity present within the Taiwanese clade. The one or two most widely shared haplotypes were observed in each region. H21 (42%) for Taiwanese and H3 (51.9%) for Korean lineages were the most widely shared haplotypes for each regional group. The majority of individuals from Japan had the H16 haplotype (97.6%).
Phylogenetic Relationships in the Endosymbiotic Symbiodiniaceae
Acquired 23S rDNA sequences (661 bp) of Symbiodiniaceae (N = 110) showed 87 parsimonious informative sites and the overall mean diversity was 0.045. The phylogenetic relationships of Symbiodiniaceae showed the formation of two distinct clades from Taiwan and Korea/Japan, respectively, with high supporting values (ML, NJ, 96–100%) (Figure 3). Symbiodiniaceae from Korean and Japanese A. japonica shared the same clade, which is distinct from the one hosted by corals in Taiwan (Figure 3). When two sites from the same regions were examined (Korea and Taiwan), they did not form separate clades (Figure 3).
Only a few haplotypes (N = 5) were identified in the Symbiodiniaceae 23S rDNA dataset. Two deep phylogenetic groups, Taiwan and Korea/Japan, were identified with 48–49 mutational steps between haplotypes (Figure 4). The average JC distance between the Symbiodiniaceae lineages from Taiwan and Korea/Japan was 8.67% (8.56–8.78%). Four haplotypes were found in Taiwanese Symbiodiniaceae, while only a single identical haplotype (H1) was shared between Korean and Japanese specimens (Figure 4).
Additionally, phylogenetic analyses of the endosymbiotic Symbiodiniaceae revealed that populations from Korea (BM and GN) and Japan (SH) belong to the Clade F2 with relatively high supporting values (71%). However, Symbiodiniaceae from Taiwan (GI and KT) clustered within Cladocopium (former Clade C) with high supporting values (100%) (Supplementary Figure S2).
Discussion
Recently, much attention has been paid to range shifts or population expansions of organisms in a diverse set of marine ecosystems across the globe, as the observed shift in marine taxa distribution is faster than of terrestrial taxa (72 km per decade vs. 17 km per decade) (Pecl et al., 2017). For scleractinian corals, focus has been made on the negative effects of increased seawater temperature through mass coral bleaching and mortality, although moderate warming can also benefit some coral populations (Hoey et al., 2016). A recent investigation suggests that the populations at Jeju Island, Korea of the subtropical coral, A. japonica are substantially increasing in numbers, with damaging effects on local biodiversity (Denis et al., 2013). Based on the molecular-based phylogenetic analyses, we here tested two probable scenarios for the origins of A. japonica in Korean waters: (1) poleward migration from subtropical habitats, or (2) a recent expansion of a previously unnoticed A. japonica population of small numbers.
Multiple lines of evidence – distinct phylogenetic clusters between host and symbiont clades, no gene flow among the three regional populations in Korea, Taiwan and Japan, and relatively high genetic diversity within the Korean population – strongly suggest that the northern A. japonica population at Jeju Island has been present for a relatively longer evolutionary time period. These findings provide evidence supporting the scenario of recent population expansion of Korean A. japonica. Both analyses of ITS phylogeny and haplotype network clearly indicate that the Korean lineages (BM and GN on Jeju Island) of A. japonica are substantially evolutionarily divergent from those from Taiwan (GI and KT) and Japan (SH), and also show closer phylogenetic relationships with Japanese lineage relative to Taiwanese one (Figures 3, 4). The most plausible explanation is that each genetic cluster of the three geographic regions represents phylogenetic lineages that evolutionarily substantially diverged from one another. The ITS haplotype network shows that Taiwanese haplotypes are connected with Korean haplotypes through a large number of mutational steps (Figure 4; Supplementary Figure S3). Moreover, Korean clades are connected with Japanese ones through 26–31 mutational steps, suggesting that a rather long period of time has elapsed since the beginning of each clade’s divergence. Given the relatively higher levels of genetic diversity and their basal position of Taiwanese clades in the phylogeny, these populations might represent the more ancient lineages while the Korean and Japanese lineages are more relatively recently evolved.
The observed levels of divergence among the three lineages reveal that the NWP A. japonica constitutes three cryptic species rather than a single species. Average ITS divergence between the lineages ranged from 4.21% to 7.62%. A previous study of ITS2 of scleractinian corals found that interspecific genetic distances varied significantly across different genera (Chen et al., 2004). The smallest interspecific average distance of 1.34% was observed for the genus Madracis, whereas the distances of 23.53% were observed for Acropora. However, in general, >2–3% divergences have been suggested to be appropriate for identifying species boundaries in most animals (Hebert et al., 2003). The higher FST values among the lineages further support the hypothesis that they indeed represent cryptic Alveopora species. A recent report on genetic distances between five cryptic Acropora hyacinthus-related species showed FST levels of 0.18 to 0.55 (Sheets et al., 2018). Our results of FST = 0.88–0.98 are well beyond those levels, confirming A. japonica in the NWP comprises separate species.
Yet, analysis of skeletal morphology using SEM photos (Supplementary Figure S1) shows variation in calice diameter between localities and even between samples from the same locations. A. japonica can be distinguished from other Alveopora species in having calices 2.5–3.5 mm in diameter (Kitano et al., 2014). This is true for most of the samples we analyzed, but some show less than 2.5 mm. This is not surprising given environmental effects may contribute to variation in skeletal features (Barnes, 1973). The 2.5 mm cut off for the calice diameter as a diagnostic key to distinguish A. japonica from other species (e.g., A. excelsa or A. tizardi) therefore needs to be updated. Also, Taiwanese corals do not appear to have white tentacle tips, although A. japonica in Taiwan are usually dark green, with white stripes on the top of tentacle tips (Veron, 2000; Dai and Horng, 2009). Due to the unresolved morphological taxonomy in Alveopora species, we cannot rule out the possibility of different Alveopora species present in Taiwan and further studies need to clarify this issue.
No sharing of ITS haplotypes among the three lineages allows us to rule out the possibility of current poleward migration or range expansion of A. japonica from subtropical regions of southern Taiwan to Korea and Japan. Genetically distinct phylogenetic groups with no shared haplotypes among the three geographic regions suggest a complete lack of historical or ongoing gene flow among these regional groups, despite the possibility of gene flow by means of larval dispersal via oceanic currents, given a reported maximum of a 3-month planktonic larval phase for A. japonica (Harii et al., 2001). Alternatively, if current poleward migration takes place, no gene flow we identified may provide evidence for reproductive isolation among the three regional populations with cryptic species relationships.
The relatively high genetic diversity within the Korean population further suggests that it has been established around Jeju Island for a relatively long evolutionary time, supporting the recent population expansion hypothesis. The Taiwanese lineage harbored the highest levels of haplotype and nucleotide diversity, whereas Korean lineage showed intermediate levels and Japanese one the lowest (Table 1). If the Korean lineage was established by poleward migration along the NWP via historical or contemporary gene flow, its diversity level is predicted to be considerably lower due to founder effects, but this was not observed in this study.
The star-like tree topology of the Korean clade in the haplotype network of A. japonica supports the scenario of population expansion from a small number of individuals. The Korean lineage has two common haplotypes (i.e., H2 and H3) with 13 more recent haplotypes connected with one or two mutational steps (Figure 4). This type of haplotype network can be considered a “star-shape” network, which is interpreted as a phylogenetic signal of a population that has recently expanded in size from one or a small number of individuals following a population bottleneck (Slatkin and Hudson, 1991). Several genetic surveys of marine species in the northern hemisphere have often found this a star-shaped phylogenetic pattern, which is suggested to be the outcome of the demographic effects of glaciations, particularly the most recent last glacial maximum (LGM), approximately 20,000 years ago (Lee and Boulding, 2007; Guo et al., 2015). Our results also support a demographic scenario of sudden population expansion in Korean populations of A. japonica (Rogers and Harpending, 1992).
The molecular phylogenetic signal of Symbiodiniaceae with its host A. japonica found in this study may suggest localized adaptation to the host coral’s respective environments. The Symbiodiniaceae phylogeny based on 23S rDNA sequences shows two distinct clades, in which Korean and Japanese symbionts share one clade and Taiwanese ones form the other clade (Figures 3, 4). Clade F2 has been reported from A. japonica in Korea, and both Clades F2 and Cladocopium were from Japan (Rodriguez-Lanetty et al., 2003; Lien et al., 2012). In the current study, only Clade F2 was found in both Korean and Japanese lineages, whereas Cladocopium was found only in Taiwan. Symbiodiniaceae 23S rDNA haplotypes between lineages of Taiwan (e.g., H2) and Korea/Japan (e.g., H1) are connected with 48 mutational changes, which indicates that they have diverged over a rather long evolutionary history, as seen in A. japonica (Figures 3, 4). This concordant phylogenetic pattern between the host scleractinian coral and endosymbiont Symbiodiniaceae highlights the co-evolutionary relationships of the hosts and endosymbionts during the last 140–200 mya (LaJeunesse et al., 2018).
A further explanation is that the phylogeographic concordance of Symbiodiniaceae with A. japonica would be most likely caused by the geographic distribution patterns of Symbiodiniaceae genera/clades in the NWP. Corals associated with Symbiodiniaceae of genera Symbiodinium, Breviolum, Durusdinium, Fugacium, Gerakladium and Clade F comprising F2, F3 and F4 are known to be evolutionarily more recent and therefore genetically more similar to one another than those of Cladocopium, which is basal to all other clades (a dominant coral symbiont lineage) (Stat et al., 2008). A single shared and recent lineage between the Symbiodiniaceae populations of Korea and Japan, which in this study is Clade F2 (Supplementary Figure S2) and the most ancestral lineage within Taiwan might represent the evolutionary outcomes of local adaptation to their regional environments. The similarity between host coral and symbiont phylogenetic patterns is suggestive of evolutionary outcomes of natural selection, i.e., host corals with the local adaptation of specific symbiont lineages to heterogeneous environmental conditions of their habitats. Given cryptic Alveopora species found in this study, the differences could reflect different host coral species with local adaptations of different symbionts to their regional environments.
In summary, we show that each of the NWP regional populations of A. japonica forms their own genetic clades with deep evolutionary divergence among them, pinpointing previously unrecognized A. japonica cryptic species diversity in this region. The results also support the scenario of a recent expansion in numbers around Jeju Island from a small previously existing population. Populations of A. japonica around Jeju Island show a shallow phylogeographic pattern with a star-shape topology, indicative of rapid recent increases in population size. Our study also highlights the similarity of phylogenetic patterns between the host corals and their endosymbiont Symbiodiniaceae. These findings might suggest that the host was selected for the most adapted symbiont lineages to their local environments.
Data Availability Statement
One hundred twenty one nuclear DNA ITS sequences of Alveopora japonica and five chloroplast 23S rDNA sequences of Symbiodiniaceae can be accessed via GenBank with accession numbers (ITS: MN610250–MN610370; 23S rDNA: MN626570–MN626574) (https://ncbi.nlm.nih.gov/nuccore).
Author Contributions
JKa, K-SC, SP, and HL conceived and designed the study. SKe, SKi, SA, JR, CC, K-SC, and SP supported the field expedition for coral sampling. JKa, JJ, JKi, SP, and HL performed the experiments and analyzed the data. JKa, JJ, SKe, SA, JR, CC, and HL drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Ministry of Oceans and Fisheries of Korea (Project title: Long-Term Changes in Structure and Function in the Marine Ecosystems of Korea) to K-SC, SP, and HL. This research was also a part of the project titled “Improvement of Management Strategies on Marine Ecosystem Disturbing and Harmful Organisms” funded by the Ministry of Oceans and Fisheries, Korea. This study was also supported by a National Research Foundation of Korea (NRF) grant (NRF-2017R1C1B1010741) from the Basic Science Research Program funded by the Ministry of Science, ICT and Future Planning to JKa. SKe was funded by the Japanese Association for Marine Biology research grant for sampling in Shimoda.
Conflict of Interest
JKi was employed by the company GenCube.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Molecular Ecology and Evolution Laboratory of Sangji University, the Estuarine and Coastal Ecology Laboratory, and the Laboratory of K-SC (especially Hee-Jung Lee) at Jeju National University for helping to collect samples in the field.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00012/full#supplementary-material
Footnotes
References
Avise, J. (2004). Molecular Markers, Natural History, and Evolution. New York, NY: Chapman and Hall.
Bayha, K., Chang, M., Mariani, C., Richardson, J., Edwards, D., Deboer, T., et al. (2015). Worldwide phylogeography of the invasive ctenophore Mnemiopsis leidyi (Ctenophora) based on nuclear and mitochondrial DNA data. Biol. Invasions. 17, 827–850. doi: 10.1007/s10530-014-0770-6
Beger, M., Sommer, B., Harrison, P. L., Smith, S. D., and Pandolfi, J. M. (2014). Conserving potential coral reef refuges at high latitudes. Divers. Distrib. 20, 245–257. doi: 10.1111/ddi.12140
Boulay, J. N., Hellberg, M. E., Cortés, J., and Baums, L. B. (2014). Unrecognized coral species diversity masks differences in functional ecology. Proc. Biol. Sci. 281:20131580. doi: 10.1098/rspb.2013.1580
Chang, S.-J., Rodriguez-Lanetty, M., Yanagi, K., Nojima, S., and Song, J.-I. (2011). Two anthozoans, Entacmaea quadricolor (order Actiniaria) and Alveopora japonica (order Scleractinia), host consistent genotypes of Symbiodinium spp. across geographic ranges in the northwestern Pacific Ocean. Anim. Cells Syst. 15, 315–324. doi: 10.1080/19768354.2011.611174
Chen, C., Odorico, D., Tenlohuis, M., Veron, J., and Miller, D. (1995). Systematic relationships within the Anthozoa (Cnidaria: Anthozoa) using the 5’-end of the 28S rDNA. Mol. Phylogenet. Evol. 4, 175–183. doi: 10.1006/mpev.1995.1017
Chen, C. A., Chang, C.-C., Wei, N. V., Chen, C.-H., Lein, Y.-T., Lin, H.-E., et al. (2004). Secondary structure and phylogenetic utility of the ribosomal internal transcribed spacer 2 (ITS2) in scleractinian corals. Zool. Stud. 43, 759–771.
Cooper, T. F., Ulstrup, K. E., Dandan, S. S., Heyward, A. J., Kühl, M., Muirhead, A., et al. (2011). Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc. Biol. Sci. 278, 1840–1850. doi: 10.1098/rspb.2010.2321
Dai, C., and Horng, S. (2009). Scleractinia Fauna of Taiwan. I. The Complex Group. Taipei: National Taiwan University.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9:772. doi: 10.1038/nmeth.2109
De Palmas, S., Denis, V., Ribas-Deulofeu, L., Loubeyres, M., Woo, S., Hwang, S., et al. (2015). Symbiodinium spp. associated with high-latitude scleractinian corals from Jeju Island, South Korea. Coral Reefs 34, 919–925. doi: 10.1007/s00338-015-1286-y
Denis, V., Chen, C., Song, J., and Woo, S. (2013). Alveopora japonica beds thriving under kelp. Coral Reefs 32:503. doi: 10.1007/s00338-013-1019-z
Denis, V., Ribas-Deulofeu, L., Loubeyres, M., De Palmas, S., Hwang, S.-J., Woo, S., et al. (2014). Recruitment of the subtropical coral Alveopora japonica in the temperate waters of Jeju Island. South Korea. Bull. Mar. Sci. 91, 85–96. doi: 10.5343/bms.2014.1032
Duran, S., Giribet, G., and Turon, X. (2004). Phylogeographical history of the sponge Crambe crambe (Porifera, Poecilosclerida): range expansion and recent invasion of the Macaronesian islands from the Mediterranean Sea. Mol. Ecol. 13, 109–122. doi: 10.1046/j.1365-294x.2003.02022.x
Eguchi, M. (1968). The Hydrocorals and Scleractinian Corals of Sagami Bay Collected by HM the Emperor of Japan. Tokyo: Maruzen-Yushodo Company, Limited
Etoundi, E., Marescaux, J., Vastrade, M., Debortoli, N., Hedtke, S. M., and Pigneur, J. (2019). Distinct biogeographic origins of androgenetic Corbicula lineages followed by genetic captures. bioRxiv 590836.
Excoffier, L., Laval, G., and Balding, D. (2003). Gametic phase estimation over large genomic regions using an adaptive window approach. Hum. Genomics. 1, 7–19.
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Flot, J.-F., Couloux, A., and Tillier, S. (2010). Haplowebs as a graphical tool for delimiting species: a revival of Doyle’s “field for recombination” approach and its application to the coral genus Pocillopora in Clipperton. BMC Evol. Biol. 10:372. doi: 10.1186/1471-2148-10-372
Forsman, Z. H., Barshis, D. J., Hunter, C. L., and Toonen, R. J. (2009). Shape-shifting corals: molecular markers show morphology is evolutionarily plastic in Porites. BMC Evol. Biol. 9:45. doi: 10.1186/1471-2148-9-45
Greenstein, B. J., and Pandolfi, J. M. (2008). Escaping the heat: range shifts of reef coral taxa in coastal Western Australia. Glob. Change. Biol. 14, 513–528. doi: 10.1111/j.1365-2486.2007.01506.x
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, X., Zhao, D., Jung, D., Li, Q., Kong, L.-F., Ni, G., et al. (2015). Phylogeography of the rock shell Thais clavigera (Mollusca): evidence for long-distance dispersal in the northwestern Pacific. PLoS One 10:e0129715. doi: 10.1371/journal.pone.0129715
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT Nucleic. Nucl. Acids. Symp. Ser. 41, 95–98.
Harii, S., Omori, M., Yamakawa, H., and Koike, Y. (2001). Sexual reproduction and larval settlement of the zooxanthellate coral Alveopora japonica Eguchi at high latitudes. Coral Reefs 20, 19–23. doi: 10.1007/s003380000134
Hebert, P. D., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hoey, A., Howells, E., Johansen, J., Hobbs, J. P., Messmer, V., Mccowan, D., et al. (2016). Recent advances in understanding the effects of climate change on coral reefs. Diversity 8:12. doi: 10.3390/d8020012
Kim, S. W., Chung, M., and Park, H.-S. (2015). Tropical fish species thriving in temperate Korean waters. Mar. Biodivers. 2, 147–148. doi: 10.1007/s12526-014-0247-y
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/bf01731581
Kitano, Y. F., Benzoni, F., Arrigoni, R., Shirayama, Y., Wallace, C. C., and Fukami, H. (2014). A phylogeny of the family Poritidae (Cnidaria, Scleractinia) based on molecular and morphological analyses. PLoS One 9:e98406. doi: 10.1371/journal.pone.0098406
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., and Voolstra, C. R. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570.e6–2580.e6 doi: 10.1016/j.cub.2018.07.008
Lee, H. J., and Boulding, E. G. (2007). Mitochondrial DNA variation in space and time in the northeastern Pacific gastropod, Littorina keenae. Mol. Ecol. 16, 3084–3103. doi: 10.1111/j.1365-294x.2007.03364.x
Lee, H. J., and Boulding, E. G. (2009). Spatial and temporal population genetic structure of four northeastern Pacific littorinid gastropods: the effect of mode of larval development on variation at one mitochondrial and two nuclear DNA markers. Mol. Ecol. 18, 2165–2184. doi: 10.1111/j.1365-294X.2009.04169.x
Lien, Y.-T., Fukami, H., and Yamashita, Y. (2012). Symbiodinium clade C dominates zooxanthellate corals (Scleractinia) in the temperate region of Japan. Zool. Sci. 29, 173–180. doi: 10.2108/zsj.29.173
Mollica, N. R., Guo, W., Cohen, A. L., Huang, K. F., Foster, G. L., Donald, H. K., et al. (2018). Ocean acidification affects coral growth by reducing skeletal density. Proc. Natl. Acad. Sci. U.S.A. 115, 1754–1759. doi: 10.1073/pnas.1712806115
National Institute of Fisheries Science (2012). National Investigation of Marine Ecosystem. Seoul: Ministry of Oceans and Fisheries of Korea.
O’Connor, M. I., Holding, J. M., Kappel, C. V., Duarte, C. M., Brander, K., Brown, C. J., et al. (2015). Strengthening confidence in climate change impact science. Glob. Ecol. Biogeogr. 24, 64–76. doi: 10.1111/geb.12218
Pecl, G. T., Araujo, M. B., Bell, J. D., Blanchard, J., Bonebrake, T. C., Chen, I. C., et al. (2017). Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 355:eaai9214. doi: 10.1126/science.aai9214
Petit, R. J., Mousadik, A. E., and Pons, O. (1998). Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855. doi: 10.1111/j.1523-1739.1998.96489.x
Pochon, X., and Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol. Phylogenet. Evol. 56, 492–497. doi: 10.1016/j.ympev.2010.03.040
Poloczanska, E. S., Burrows, M. T., Brown, C. J., García Molinos, J., Halpern, B. S., Hoegh-Guldberg, O., et al. (2016). Responses of marine organisms to climate change across oceans. Front. Mar. Sci. 3:62. doi: 10.3389/fmars.2016.00062
Precht, W. F., and Aronson, R. B. (2004). Climate flickers and range shifts of reef corals. Front. Ecol. Environ. 2, 307–314. doi: 10.1890/1540-92952004002[0307:CFARSO]2.0.CO;2
Rodriguez-Lanetty, M., Chang, S.-J., and Song, J.-I. (2003). Specificity of two temperate dinoflagellate–anthozoan associations from the north-western Pacific Ocean. Mar. Biol. 143, 1193–1199. doi: 10.1007/s00227-003-1165-x
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569.
Rozas, J., and Rozas, R. (1999). DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175. doi: 10.1093/bioinformatics/15.2.174
Santos, S. R., Taylor, D. J., Kinzie Iii, R. A., Sakaj, K., and Coffroth, M. A. (2002). Evolution of length variation and heteroplasmy in the chloroplast rDNA of symbiotic dinoflagellates (Symbiodinium, Dinophyta) and a novel insertion in the universal core region of the large subunit rDNA. Phycologia 41, 311–318. doi: 10.2216/i0031-8884-41-4-311.1
Schmidt-Roach, S., Lundgren, P., Miller, K. J., Gerlach, G., Noreen, A. M., and Andreakis, N. (2013). Assessing hidden species diversity in the coral Pocillopora damicornis from Eastern Australia. Coral Reefs 32, 161–172. doi: 10.1007/s00338-012-0959-z
Sheets, E. A., Warner, P. A., and Palumbi, S. R. (2018). Accurate population genetic measurements require cryptic species identification in corals. Coral Reefs 37, 549–563. doi: 10.1007/s00338-018-1679-9
Slatkin, M., and Hudson, R. R. (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129, 555–562.
Stat, M., Morris, E., and Gates, R. D. (2008). Functional diversity in coral-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. U.S.A. 105, 9256–9261. doi: 10.1073/pnas.0801328105
Takatsuki, Y., Kuragano, T., Shiga, T., Bungi, Y., Inoue, H., Fujiwara, H., et al. (2007). Long-term trends in sea surface temperature adjacent to Japan. Sokko Jiho 74, S33–S87.
Teacher, A. G. F., and Griffiths, D. J. (2011). HapStar: automated haplotype network layout and visualization. Mol. Ecol. Res. 11, 151–153. doi: 10.1111/j.1755-0998.2010.02890.x
Vergés, A., Steinberg, P. D., Hay, M. E., Poore, A. G., Campbell, A. H., Ballesteros, E., et al. (2014). The tropicalization of temperate marine ecosystems: climate-mediated changes in herbivory and community phase shifts. Proc. Biol. Sci. 281:20140846. doi: 10.1098/rspb.2014.0846
Veron, J. E. (2000). Corals of the World, Vol. 1–3. Townsville: Australian Institute of Marine Science.
Vieira, C., Keshavmurthy, S., Ju, S.-J., Hyeong, K., Seo, I., Kang, C.-K., et al. (2016). Population dynamics of a high-latitude coral Alveopora japonica Eguchi from Jeju Island, off the southern coast of Korea. Mar. Freshw. Res. 67, 594–604.
Warner, P. A., Van Oppen, M. J., and Willis, B. L. (2015). Unexpected cryptic species diversity in the widespread coral Seriatopora hystrix masks spatial−genetic patterns of connectivity. Mol. Ecol. 24, 2993–3008. doi: 10.1111/mec.13225
Wei, N.-W. V., Wallace, C. C., Dai, C.-F., Pillay, K. R. M., and Chen, C. A. (2006). Analyses of the ribosomal internal transcribed spacers (ITS) and the 5.8 S gene indicate that extremely high rDNA heterogeneity is a unique feature in the scleractinian coral genus Acropora (Scleractinia; Acroporidae). Zool. Stud. 45, 404–418.
Wood, L. E., De Grave, S., and Daniels, S. R. (2017). Phylogeographic patterning among two codistributed shrimp species (Crustacea: Decapoda: Palaemonidae) reveals high levels of connectivity across biogeographic regions along the South African coast. PLoS One 12:e0173356. doi: 10.1371/journal.pone.0173356
Yakob, L., and Mumby, P. J. (2011). Climate change induces demographic resistance to disease in novel coral assemblages. Proc. Natl. Acad. Sci. U.S.A. 108, 1967–1969. doi: 10.1073/pnas.1015443108
Keywords: climate change, hidden diversity, Jeju Island, northward migration, phylogenetic concordance, subtropical corals, symbiont Symbiodiniaceae
Citation: Kang JH, Jang JE, Kim JH, Kim S, Keshavmurthy S, Agostini S, Reimer JD, Chen CA, Choi K-S, Park SR and Lee HJ (2020) The Origin of the Subtropical Coral Alveopora japonica (Scleractinia: Acroporidae) in High-Latitude Environments. Front. Ecol. Evol. 8:12. doi: 10.3389/fevo.2020.00012
Received: 09 August 2019; Accepted: 16 January 2020;
Published: 04 February 2020.
Edited by:
Annie Machordom, National Museum of Natural Sciences (MNCN), SpainReviewed by:
Roberto Arrigoni, Joint Research Centre, ItalyHironobu Fukami, University of Miyazaki, Japan
Copyright © 2020 Kang, Jang, Kim, Kim, Keshavmurthy, Agostini, Reimer, Chen, Choi, Park and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyuk Je Lee, bGhqazYyMkBnbWFpbC5jb20=
 Ji Hyoun Kang
Ji Hyoun Kang Ji Eun Jang
Ji Eun Jang Jae Hwan Kim2,3
Jae Hwan Kim2,3 Sangil Kim
Sangil Kim Shashank Keshavmurthy
Shashank Keshavmurthy Sylvain Agostini
Sylvain Agostini James D. Reimer
James D. Reimer Chaolun Allen Chen
Chaolun Allen Chen Sang Rul Park
Sang Rul Park Hyuk Je Lee
Hyuk Je Lee