- 1Department of Entomology, University of Georgia, Athens, GA, United States
- 2Department of Genetics, University of Georgia, Athens, GA, United States
DNA methylation is an important chromatin modification that can stably alter gene expression in cells and maintain genome integrity in plants and vertebrates. The function of DNA methylation outside of these well-studied systems, however, is unclear. Insects, in particular, represent an understudied group. Variation in the level of DNA methylation and gains and losses in the maintenance methyltransferase, DNMT1, across the insect tree of life suggests that there is much we don't understand about DMNT1 function and evolution. One constant across the studies examining patterns of Dnmt1 expression in insects is that expression is consistently high in reproductive tissues compared to somatic tissue. The explanation for this has been that DNMT1 is required in tissues that have high levels of cell division. Our previous study found that downregulation of Dnmt1 expression in the milkweed bug Oncopeltus fasciatus results in the expected reduction of DNA methylation, no global changes in gene expression reflecting changes in DNA methylation, and the loss of the ability to produce viable oocytes. Here, we show that females treated with ds-Dnmt1 RNA during larval development have a more extreme phenotype; they lack oocytes entirely but develop a normal somatic ovary. Our results indicate a specific role for DNMT1 in the formation of gametes and are consistent with data from other systems, including Tribolium castaneum, a species does not have DNA methylation. We propose that DNMT1 has multiple functional roles in addition to methylating DNA, which explains its complex patterns of evolution.
Introduction
DNA methylation is an important chromatin modification that can stably alter gene expression in cells and it functions in some species in the maintenance of genome integrity (Law and Jacobsen, 2010; Schmitz et al., 2019). Although diverse roles for DNA methylation have been identified in some plant, animal, and fungal species (Schmitz et al., 2019), the function of DNA methylation in animals outside of well-studied vertebrate systems is unclear. In particular, DNA methylation in the insects has been understudied. This is unfortunate, as there is extensive variation in levels of DNA methylation, from undetectable levels of methylation to 5 mC detection at around 15% of CpGs, across the insect tree of life (Bewick et al., 2016; Glastad et al., 2019). Variation exists in the amount of methylated DNA as well as the enzymatic toolkit required to establish and maintain it (Bewick et al., 2016). DNA methylation is associated with a suite of DNA methyltransferases (DNMT; Lyko, 2018), which is generally found across eukaryotic genomes (Schmitz et al., 2019). The canonical DNMT enzymes include DNMT1, which maintains existing patterns of symmetrical DNA methylation by restoring the opposing methyl group to a cytosine residue in a hemimethylated CpG following DNA replication and DNMT3, which can methylate cytosines de novo. Insects even vary in these typically conserved genes. The phylogenetic distribution of DNMT1 in insects is more widespread than DNMT3 and it is duplicated in some groups. Curiously, DNMT1 is found in insect taxa with low or no DNA methylation, including the red flour beetle Tribolium castaneum (Schulz et al., 2018). This variation raises questions both about the function of DNA methylation in insects and the selection leading to differences among taxa.
In this study we have investigated the function of DNMT1 in the large milkweed bug, Oncopeltus fasciatus, an evolution and development model system with molecular genetic resources, a methylated genome and the full complement of DNA methyltransferases identified within its genome (Bewick et al., 2016). Using parental RNAi, it has previously shown that knockdown of Dnmt1 in sexually mature females results in developmental arrest in embryos arising from eggs laid by Dnmt1 knockdown females (Bewick et al., 2019). Eventually, females injected with ds-Dnmt1 stopped laying eggs and their ovarian structure was significantly disrupted. Using RNA-seq we further found that reduction of DNA methylation was not directly associated with changes to global gene expression patterns within the ovary and that DNA methylation patterns in the somatic tissues, the gut and muscles, were unaffected by the knockdown. Our parental RNAi experiments raise the question as to why the morphological defects of Dnmt1 knockdown are confined to the ovary if the role of Dnmt1 is to maintain a methylated genome in all cells.
The most parsimonious hypothesis for why only the ovary displays a phenotype following Dnmt1 knockdown is that there are not enough rounds of DNA replication in the somatic cells between injection of the ds-Dnmt1 into the adult and sampling of the tissues for any phenotype to be realized. An alternative hypothesis is that there is an additional role for Dnmt1 in reproductive tissue. This latter hypothesis is partially supported by expression patterns of Dnmt1 in other insects. A common feature of Dnmt1 and Dnmt3 expression patterns in several species of insects is that it is often correlated with reproductive tissues (Kay et al., 2018), leading to the hypothesis that these enzymes may play a role in gametogenesis, gamete quality, or early embryogenesis. While only a few studies have examined the functional role of Dnmt1 knockdown on DNA methylation levels in insects, similar morphological phenotypes have been observed in other insects following Dnmt1 knockdown. In the jewel wasp Nasonia vitripennis, there are three distinct Dnmt1 genes, all of which are expressed and maternally provided to the embryo. One of these, Dnmt1a is required for early embryogenesis (Zwier et al., 2012). Tribolium castaneum, which lacks a methylated genome but retains Dnmt1, also has increased expression of Dnmt1 in female reproductive tissue. While Dnmt1 is expressed at all life stages, it is upregulated in ovaries, suggesting a role in reproduction. Finally, parental RNAi of Dnmt1 demonstrated that T. castaneum requires maternal expression of Dnmt1 for early embryogenesis (Schulz et al., 2018). Together, these studies suggest a germ cell specific function for Dnmt1 in insects.
In this paper, we examine whether there is a specific role for Dnmt1 in the development of germ cells outside of its function in maintaining DNA methylation. To differentiate among our two alternative hypotheses, we injected individuals during larval development to allow for sufficient time and rounds of cell division for DNA methylation to be lost in somatic cell genomes and for any somatic morphological phenotype to be manifest. We could reject the insufficient rounds of cell division explanation, so we then tested if Dnmt1 is required for oogenesis, but not required for somatic cell function.
In the first set of experiments, we injected fourth instar nymphs to provide adequate rounds of cell division between downregulation of Dnmt1 expression and adult stages. O. fasciatus is a hemimetabolous insect, undergoing incomplete metamorphosis. It develops through five immature instar stages; at each stage there is further development of color, wings and genitalia. O. fasciatus have telotrophic, meroistic ovaries with seven ovarioles per ovary. Female first instar nymphs hatch with primordial ovaries in which the female germ cells, the oogonia, have been set aside during embryogenesis (Wick and Bonhag, 1955). During the second and third instar stages, the ovaries increase in size and there is an increase in the numbers of oogonia by mitotic cell division. During the fourth instar stage, some oogonia divide mitotically to form trophocytes while others divide meiotically to form gametes, the primary oocytes. The trophic tissues (equivalent to nurse cells) differentiate during the fifth and final instar stage and at adult emergence, the ovary consists of nutritive trophic tissue, oogonia, primary oocytes, small growing oocytes connected to the nutritive tissue by trophic cords, and prefollicular cells that will envelop the developing oocytes. As the female undergoes sexual maturation, the oocytes move into the ovariole and complete maturation.
Our results established that knockdown of Dnmt1 earlier in development did not result in somatic morphological phenotypes, but rather exacerbated the impact on germ cell development. Given these results, we focused our experiments on testing the hypothesis that Dnmt1 is required for oogenesis, but is not required for somatic cell function. As a control, we included Boule, a gene known to be required for oogenesis in O. fasciatus (Ewen-Campen et al., 2013) for comparison. Downregulation of Boule expression in adult females using RNAi results in an almost identical phenotype to that of downregulating Dnmt1 (Ewen-Campen et al., 2013); initially, sexually mature females injected with ds-Boule lay eggs that don't develop, but after a few clutches, females stop producing eggs. Boule is a widely conserved gene required for reproduction across the bilateral animals (Shah et al., 2010). Mutations in Boule cause arrest in meiosis prior to metaphase in Drosophila melanogaster males and Caenorhabditis elegans females (Karashima et al., 2000). Thus, Boule provided an excellent comparison for evaluating the hypothesis that Dnmt1 is required for oogenesis, and perhaps the transition from oogonia to oocytes in O. fasciatus. We examined this hypothesis by testing for the following predictions arising from our overall hypothesis. First, Dnmt1 should be most highly expressed when and where oogenesis is occurring. Second, downregulating Dnmt1 expression during a critical developmental stage for oogenesis would result in the loss of oocyte production. Third, downregulating Dnmt1 expression will affect reproductive function without affecting somatic function and lifespan.
Methods
Animal Husbandry
To produce nymphs of known age and social history, O. fasciatus adults from our mass colony were randomly mated, and resulting eggs were removed and stored in individual plastic containers. Upon hatching, nymphs were transferred into 2-cup Rubbermaid containers supplied with organic, raw sunflower seeds and deionized water. Nymph colonies were housed under 12:12 h light/dark conditions at 27°C. Nymphs were staged utilizing a visual development chart from Chesebro et al. (2009).
Nymph RNAi Treatment
DNA templates of Dnmt1 and Boule was prepared using a PCR reaction with gene-specific primers (Table 1). Double stranded RNA was synthesized with an Ambion MEGAscript kit (ThermoFisher Sci, Waltham, MA) to generate sense and anti-sense RNA which were allowed to anneal. The ds-Dnmt1 RNA was 404 bp in length and the ds-Boule RNA was 716 bp in length. The concentration of ds-RNA was adjusted to 4 μg/μL in injection buffer (5 mM KCl, 0.1 mM NaH2PO4) for both Dnmt1 and Boule. Female fourth instar nymphs, hatched within 2–3 days of each other, were selected for injection. Treatments were ds-Dnmt1, ds-Boule, and control injections with injection buffer alone. Nymphs were injected with 2 μL using a syringe and a pulled glass capillary needle (Sutter Instrument Company micropipette puller model P-97, Novato, CA) between the third and fourth abdominal segments (Chesebro et al., 2009). Injected nymphs were housed in petri dishes with ad-libitum sunflower seeds and water under standard rearing conditions until they emerged as adults.
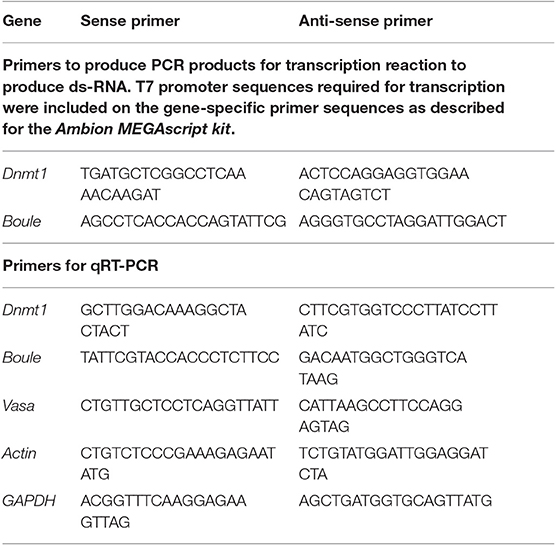
Table 1. Primer sequences used to generate RNAi for injection and to quantify expression levels using quantitative Real Time PCR.
Previous work from our group has shown that there is no difference between controls using a ds-RNA with no specificity to any O. fasciatus sequence (ds-RED) and buffer injections (Bewick et al., 2019), therefore we did not include this control in our current study. Further, we have previously used two different regions of Dnmt1 as targets for our ds-RNA. RNAi targeted against either the cytosine-specific DNA methyltransferase replication foci domain (RFD) or the DNA methylase domain (AdoMet) of Dnmt1 gave rise to identical phenotypes, arguing against any off-target effects (Bewick et al., 2019). Thus, for this study, we only used the RNAi against the RFD consensus domain. We are convinced that the ovary phenotype is specific to the ds-Dnmt1 and ds-Boule and not a generic RNAi response because in our laboratory we have used other ds-RNAs, including against Dnmt3 and the pigmentation gene ebony (Liu et al., 2014). In both of these cases females that emerged from nymph injections are fully fertile.
qRT-PCR, Development Series, and Tissue Specificity
To confirm that our RNAi treatment during nymphal development effectively knocked down expression of Dnmt1, total RNA was extracted from whole bodies of 10 flash frozen adults from either control or ds-Dnmt1 treated individuals using a Qiagen RNA Easy kit with Qiazol (Qiagen, Venlo, The Netherlands) as per manufacturer's instructions. Adults were dissected at 21 days post-adult emergence (35 to 31 days post-injection). Complementary DNA (cDNA) was synthesized from 500 ng RNA with qScript cDNA Super-Mix (Quanta Biosciences, Gaithersburg, MD). Expression levels of Dnmt1 were quantified by quantitative real-time PCR (qRT-PCR). Primers were designed using the O. fasciatus genome as a reference (Panfilio et al., 2019) and actin and GAPDH were used as reference genes (Table 1). We have validated these reference genes for our experimental samples and they are accepted as robust reference genes in O. fasciatus (Meinzer et al., 2019). We used a Roche LightCycler 480 with the SYBR Green Master Mix (Roche Applied Science Indianapolis, IN). All samples were run with 3 technical replicates using 10 μL reactions. Primer efficiency calculations, genomic contamination testing, and endogenous control gene selection were performed as described in Cunningham et al. (2015). We used the ΔΔCT method to compare levels of gene expression across the samples (Livak and Schmittgen, 2001).
To examine expression levels of Dnmt1 across development, groups of nymphs were staged and flash frozen in liquid nitrogen, and stored at −80°C. The earliest stages of nymphs are difficult to sex accurately, so the 2nd and 3rd instar nymphs were not separated by sex. By the 4th instar stage of development, females can be identified, so only female 4th and 5th instar nymphs were collected. To ensure that there was a sufficient amount of tissue for RNA isolation, smaller nymphs were pooled for each sample. A sample of 2nd, 3rd, 4th, and 5th instar nymphs contained 10, 5, 4, and 3 individuals, respectively. The other two developmental stages we collected were newly emerged females, collected within 24 h of adult emergence, and sexually mature females, collected 7–10 days post-adult emergence. All adult females were housed in a container without males to ensure that they had not mated prior to RNA isolation. A sample of adults consisted of a single individual. Total RNA was extracted and cDNA was synthesized as described above. Ten samples were analyzed for each developmental stage.
Expression levels of Dnmt1, Boule, and Vasa were quantified by quantitative real-time PCR (qRT-PCR). Vasa was chosen as a conserved marker of the germ line that is expressed at high levels in oocytes in O. fasciatus (Ewen-Campen et al., 2013). Boule and Vasa primers were designed using the O. fasciatus genome as a reference (Panfilio et al., 2019) and actin and GAPDH were used as endogenous reference genes (Table 1). Expression levels were quantified by qRT-PCR as described above. We used ANOVA followed by Tukey-Kramer HSD to compare all pairs in JMP Pro 14.1 to determine significant differences in levels of gene expression among the different stages. Data met the assumptions for ANOVA.
To examine for tissue specific expression of Dnmt1, we dissected adult, virgin females 7–10 days post adult emergence. We flash froze ovaries, gut, and thorax (muscle) from individual females. Total RNA was extracted and complementary DNA (cDNA) was synthesized as described above. RNA from eight females was analyzed for each tissue. Expression of Dnmt1 was quantified and analyzed as described for the development series.
Quantification of DNA Methylation
To determine the effect of downregulation of Dnmt1 expression from the fourth instar stage, total DNA was extracted from gut, muscle, and ovarian tissue samples from adult females (N = 3) that had been treated with ds-Dnmt1 or control injections at the fourth instar using a Qiagen Allprep DNA/RNA Mini Kit (Qiagen, Venlo, The Netherlands). MethylC-seq libraries were prepared from the DNA isolated from the gut, muscle, and ovary tissue from females as described (Ulrich et al., 2015). Briefly, genomic DNA was sonicated to 200 bp using a Covaris S-series focused ultrasonicator and end-repaired using an End-It DNA end-repair kit (Epicentre, Madison, WI). The end-repaired DNA was subjected to A-tailing using Klenow 3′-5′ exo–(New England Biolabs, Ipswich, MA) and ligated to methylated adapters using T4 DNA ligase (New England Biolabs, Ipswich, MA). The ligated DNA was subsequently bisulfite converted using an EZ DNA Methylation-Gold (Zymo Research, Irvine, CA) kit as per the manufacturer's instructions and amplified using KAPA HiFi Uracil + Readymix Polymerase (ThermoFisher Sci, Waltham, MA). Samples were sequenced on a NextSeq500 and qualified reads were aligned to the O. fasciatus genome assembly 1.0 as described (Schmitz et al., 2013). Chloroplast DNA (which is fully unmethylated) was used as a control to calculate the sodium bisulfite reaction non-conversion rate of unmodified cytosines (Table S1). The percent DNA methylation was calculated by dividing the number of methylated CpG sites by the total CpG sites. Further information on numbers of reads and alignment rates are provided in Table S1.
Fertility and Fecundity Assays
After the injected nymphs enclosed, the females were placed in individual petri dishes with ad-libitum food and water and provided with loose cotton wool as an oviposition substrate. Injected females were paired with stock males from the mass colony for mating. Eggs were collected three times a week over a period of 2 weeks. Eggs were incubated in solo plastic cups at 27°C for 1 week to allow for embryonic development and hatching. The total numbers of eggs and the number of eggs that were fertilized and developed were counted. Oncopeltus fasciatus eggs are pale yellow in color when they are first laid. Fertilized eggs turn a deep orange as development proceeds, while unfertilized eggs remain pale yellow.
Ovarian Structure Analysis
A separate set of female nymphs injected with either control, Dnmt1, or Boule ds-RNA was used to analyze the effect of knockdown on ovarian structure. Virgin females were dissected 7–10 days post-adult emergence. At this age, females are sexually receptive and have maturing oocytes in their ovaries. Females were dissected and the ovaries removed. For some females, whole mounts of ovaries were imaged using Leica M60 stereomicroscope and LAS v4 imaging software. All ovaries were processed for microscopy using the technique described in Duxbury et al. (2018). Briefly, ovaries were fixed for 30 min in 4% formaldehyde in Phosphate Buffered Saline plus 0.1% Triton-X100 (PBT) and stained for evidence of cells in the M phase of the cell cycle using an α-pHH3 primary antibody (Millipore antibody 06-570, Sigma-Aldrich, St. Louis, MO). The secondary antibody was an Alexa Fluor goat-anti-rabbit 647 (ThermoFisher Scientific, Waltham, MA). Following antibody staining the ovaries were stained with Invitrogen fluorescein labeled phalloidin (0.2 units/mL PBT; ThermoFisher Scientific, Waltham, MA) to visualize actin and DAPI (0.5 μg/mL PBT) to visualize nucleic acids. Stained ovaries were mounted in Mowiol 4-88 mounting medium (Sigma-Aldrich, St. Louis, MO) and imaged with a Zeiss LSM 710 Confocal Microscope (Zeiss) at the UGA Biomedical Microscopy core or an EVOS FI Cell Imaging system (ThermoFisher Scientific, Waltham, MA).
Lifespan Analysis
Control and experimental females from the fertility and fecundity assays were checked every 24 h for viability. The date of adult emergence and the date of death were recorded for each female and lifespan was calculated as the number of days between adult emergence and death. We tested for differences in female longevity relative to treatment using a Wilcoxon Rank Sum test with JMP Pro V14.1.
Results
Dnmt1 expression is significantly knocked down in adults injected with ds-Dnmt1 as fourth instar nymphs (ANOVA; F = 19.038, d.f. = 1, 18, p < 0.001). Fourth instar nymphs injected with ds-Dnmt1 are equally likely to survive to adulthood as control injected fourth instar nymphs. At least 80% of nymphs injected at the fourth instar stage emerged as adults in both control and ds-Dnmt1 injection treatments. There was no difference in the likelihood of adult emergence among control and Dnmt1 knockdown treatments (Contingency analysis: N = 80, d.f. = 1, Pearson χ2 = 0.082, p = 0.775). Furthermore, there was no obvious difference in morphology amongst the control and Dnmt1 knockdown emerging adults. Adults from all treatments appeared normal, including apparently normal wing development. There were no visible phenotypic effects of reduced methylation in somatic tissue.
As predicted, injecting earlier in development resulted in reduced DNA methylation of all tissues due to an increased number of mitotic division cycles required within the somatic tissues for growth during development between treatment at the nymphal stage and sampling at the adult stage. In both somatic tissues we tested, gut and muscle, and in the ovary, methylated CpG levels went from around 12.5% in controls to around 5% in Dnmt1 knockdowns (Figure 1).
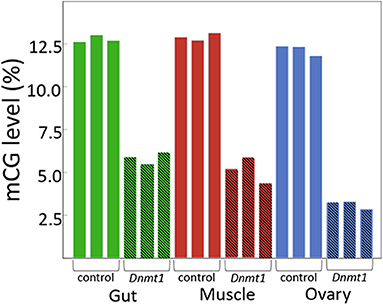
Figure 1. Dnmt1 is required for the maintenance of mCpG in somatic and reproductive tissue in O. fasciatus. Across all tissues the percent of mCpG is consistently reduced in the genomes of individuals in which Dnmt1 has been knocked down at the 4th instar stage of development (hatched bars) compared to control individuals (solid bars).
Dnmt1 Knockdown During Ovarian Development Exacerbated Impact on Oogenesis
Examination of the ovaries from females emerging from fourth instar injected nymphs demonstrated a significant impact on the ovary: although the somatic ovary appeared to have developed normally, there were no eggs present (Figures 2A,B). Dnmt1 knockdown clearly impacted female fertility. Only one out of 11 ds-Dnmt1 treated female ever laid eggs across their entire lifespan, whereas all control females (N = 15) laid eggs. The number of eggs laid by this single ds-Dnmt1 treated female was 86, whereas the control females laid a mean of 200 ± 42 eggs.
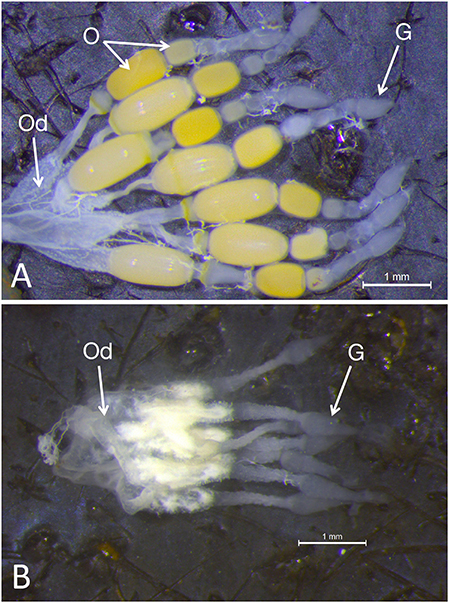
Figure 2. Whole mounts of ovaries from control (A) and Dnmt1 knockdown (B) females demonstrated a significant impact on the ovary. The somatic ovary appeared to have developed normally in the ds-Dnmt1 females. The ovaries from both the control and ds-Dnmt1 females contained seven ovarioles within each ovary, each connecting on to a common oviduct (Od). Further, at the anterior end of the ovarioles from females of both treatments, there was a teardrop shaped germarium (G) where the trophocytes, or nurse cells, would reside. However, there were no oocytes (O) present in the ovaries from adult females injected with ds-Dnmt1 at the 4th instar stage and these females never produced eggs over their lifespan. Scale bars equal to 1 mm.
Dnmt1 Is Expressed When and Where Oogenesis Is Occurring
Dnmt1 expression went up in sexually mature females and expression pattern mirrored that of the two other oocyte specific genes we tested, Vasa and Boule (Figure 3). For all three genes, expression was statistically significantly affected by developmental stage, with expression being highest in the sexually mature females [Dnmt1, F(5,53) = 89.749, p < 0.001; Vasa, F(5,53) = 61.289, p < 0.001; Boule, F(5,53) = 99.598, p < 0.001], with similar patterns in all three genes. Dnmt1 is expressed in all tissues. However, expression levels significantly depend on tissue type (Figure 4; F = 51.661, d.f. = 2, 21, p < 0.001). Tukey-HSD pairwise comparison of the different tissues showed that expression is highest in the ovary, and did not differ between the two somatic tissues.
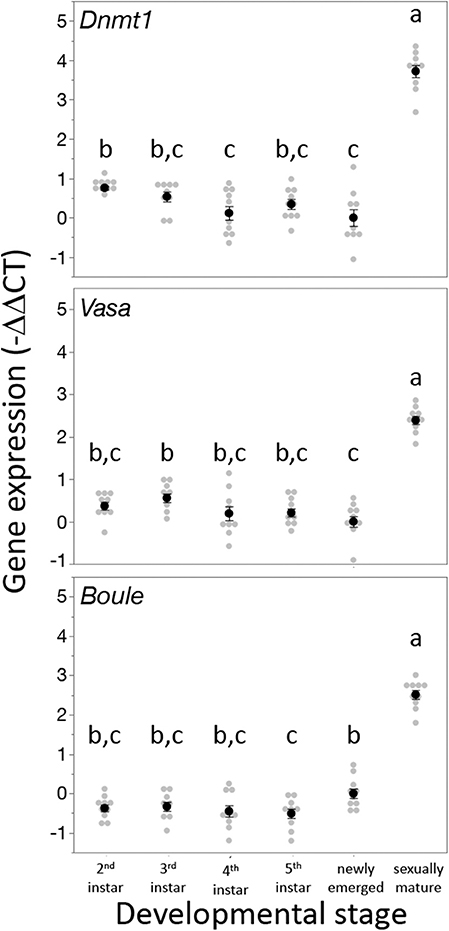
Figure 3. Dnmt1 expression is highest in sexually mature females and the expression pattern of Dnmt1 across development is the same as the germ cell specific genes Vasa and Boule. RNA was isolated from whole bodies of individuals at each instar stage of development from the 2nd instar to the 5th (final) instar. 2nd and 3rd instar nymphs cannot be sexed, but RNA was collected only from female 4th and 5th instar nymphs. Females were also sampled on the day of adult emergence and 10 days after adult emergence when they are sexually mature. Black dots and bars represent mean and SE. Gray dots represent data points for each individual female or nymph tested. Letters designate significant differences (p < 0.05) among pairwise comparisons using post-hoc Tukey-Kramer HSD test.
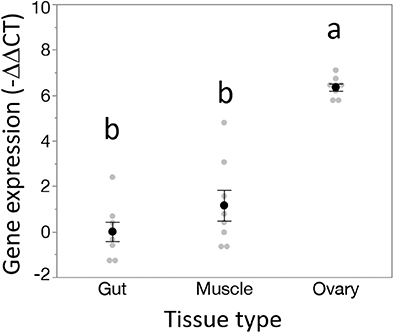
Figure 4. Dnmt1 shows tissue specific expression. While Dnmt1 is expressed in all tissues, it is more highly expressed in the ovary than in the two somatic tissues tested, gut and muscle. Black dots and bars represent mean and SE. Gray dots represent data points for each individual sample tested. Letters designate significant differences (p < 0.05) among pairwise comparisons using post-hoc Tukey-Kramer HSD test.
Dnmt1 Knockdown Leads to Loss of Developing Oocytes, but Not the Supporting Somatic Cells
Closer examination of the structure of the ovaries from control, Dnmt1 knockdown, and Boule knockdown females revealed that the somatic tissues within the ovary were all present in the ds-Dnmt1 and ds-Boule treated females. The O. fasciatus ovary can be divided into a terminal filament, a germarium containing trophocytes, oogonia, primary oocytes, and prefollicular tissue, the vitellarium where oocytes mature, and the pedicel that connects the germarium to the oviduct and in which the developing oocytes mature (Figure 5A; see also Bonhag and Wick, 1953). Low magnification images revealed that the somatic cells of the ovary, including the terminal filament, the trophic tissue, and the sheath cells that surround the developing ooctyes and extend to form the pedicel, were present in the ovaries of control females as well as Dnmt1 females (Figures 5B,C). Trophic cells were easily recognized by their distinctive large nuclei in aggregates around the trophic core. Dnmt1 knockdown females had trophic tissue with recognizable nuclear structure within the germarium region of the ovary and normal looking pedicels (Figure 5C). We also examined the structure of ovarioles from females in which Boule was knocked down from the fourth instar stage to compare the results from the Dnmt1 knockdown females to those with a knockdown of a gene with a known function in gametogenesis (Ewen-Campen et al., 2013). As with the ds-Dnmt1 treated females, females treated with ds-Boule as fourth instar nymphs never formed oocytes, yet had recognizable trophic tissue in the germarium and normal pedicel structure (Figure 5D).
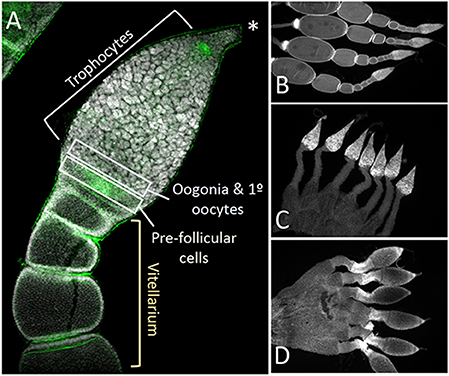
Figure 5. The structure of the somatic ovary, including the trophocytes, which develop from oogonia via mitotic division, was relatively normal in both Dnmt1 and Boule knockdown females. (A) Overview of the tip of an ovariole from a control female with the significant regions marked. The terminal tip (asterix) anchors the ovariole to the body wall. The majority of the germarium contains trophocytes that surround a trophic core. At the base of the germarium there is a group of cells with smaller nuclei. These cells are oogonia and primary ooctyes and pre-follicular cells that will envelope the developing oocyte as it moves into the vitellarium. During early stages of oocyte development, ooctyes remain connected to the trophocytes through trophic cords (arrowheads). Low magnification images (2X) of ovaries from control (B), Dnmt1 knockdown (C), and Boule knockdown (D) females stained with DAPI show that the germarium and the pedicel cells were present in the two knockdowns. However, no oocytes or their associated follicle cells ever entered the vitellarium.
Higher magnification of the germarium region of the ovarioles of control, ds-Dnmt1, and ds-Boule treated females (Figure 6) show that the large, aggregated nuclei of the trophocytes were present around a trophic core lined with actin in all three treatments. However, there are notable differences among the ds-Dnmt1 treated females and the ds-Boule treated females in the region containing the oogonia and primary ooctyes. The germarium tissue in the Boule knockdown females remained organized and the different regions of cell types were recognizable. The small nuclei of the oogonia are apparent at the base of the germarium (Figure 6G). The germarium tissue of the Dnmt1 knockdown females, however, was clearly disorganized and there was clear evidence of tissue disruption and cell death in the region where the oogonia and primary ooctyes would have been found (Figure 6D). Although the appearance of the control and ds-Boule germarium tissue was highly repeatable, there was significant variation in the amount of both cell death, evidenced by highly condensed and fragmented nuclei, and tissue disruption in the germarium of ds-Dnmt1 treated females (Figure S1). Despite the specific expression of the phenotype, all Dnmt1 knockdown females exhibited defects in the same region of the germarium between the trophocytes and ovariole sheath cells where the oogonia and primary ooctyes would be located.
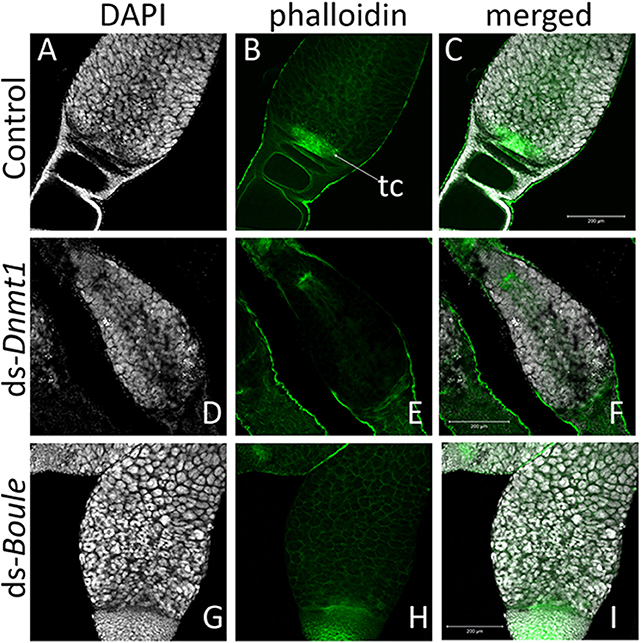
Figure 6. The structure of the germarium depends on the gene knockdown. In the control ovary (A–C), the large, aggregate nuclei of the trophocytes (A) were readily apparent, as are the smaller nuclei at the base of the germarium, consisting of the oogonia, primary ooctyes, and prefollicular cells. Actin staining (B) reveals both the early stages of oocyte development as the young oocytes become enveloped by follicle cells and lined by actin (B). The other structures revealed by actin staining are the developing trophic cords at the base of the germarium (tc), and the increased actin concentration within the trophic core. In ovaries from Dnmt1 knockdown females (D–F), the trophic cells were apparent (D), as was the trophic core (E). However, there were few small nuclei at the base of the germarium, and there often showed evidence of cell death. Ovaries from Boule knockdown females (G–I) contained both the large trophic nuclei (G) and smaller nuclei associated with the oogonia and pre-follicular cells. However, there was no evidence of developing oocytes or trophic cords forming in either the Dnmt1 females or Boule females. All images taken at 20X with 0.7 optical zoom. Scale bars are all 200 μm in length.
The other cell type located in this region is the pre-follicular cells. To determine if these cells were also affected by Dnmt1 knockdown, we examined cell division patterns by labeling the ovarioles with anti-phosphohistone H3 antibody, which labels dividing cells. The pre-follicular cells divide to provide follicle cells that will envelope the developing oocyte as it moved down through the ovariole. In our control females, division of the pre-follicle cells was apparent at the junction between the trophocytes and developing ovaries (Figure 7A). In the Boule knockdown females, the pre-follicular cells were particularly clear as they were not obscured by the developing ooctyes (Figure 7C). Although the germarium tissue disruption in the Dnmt1 knockdown made it difficult to identify the pre-follicular cells structurally, there was clear evidence that they were present based on the presence of dividing cells within this region of the germarium (Figure 7B).
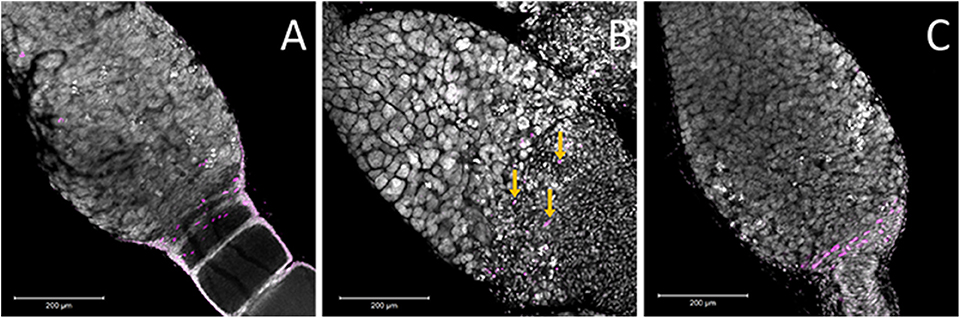
Figure 7. Ovaries stained for the M-phase of the cell cycle suggested that there are pre-follicular cells within all three treatments. (A) Within the control ovary, the pre-follicular cells are present in the tissues that surrounded the ooctyes at the early stages of development and the majority of the nuclei that stained positive for pHH3 were present within this region of the germarium. In the Boule knockdown ovaries (C), the prefollicular cells were very apparent as they were not obscured by any developing ooctyes. While the structure of the germarium in the Dnmt1 knockdown females (B) made it difficult to identify particular cell types, there was evidence that prefollicular cells remained in these germarium, as there are nuclei of the right size at the base end of the germarium and these were frequently stained for cell division (yellow arrows), although they were somewhat obscured by the condensed nuclei within this region of the germarium. All images were stained with DAPI and anti-phosphohistone H3 antibody and imaged at 20X with 0.7 optical zoom. Scale bars are all 200 μm in length.
Dnmt1 Knockdown Does Not Significantly Affect Lifespan
We predicted that if Dnmt1 plays a role in somatic function, we would see reduction in lifespan in Dnmt1 downregulated females due to the reduction of Dnmt1 and DNA methylation observed in the somatic tissues as indicated by our results with gut and muscle tissue. To control for any impact on lifespan due to reduced reproductive effort, we included ds-Boule treated females in our lifespan analysis. We found no statistically significant differences in lifespan among the three treatments (Figure 8; Wilcoxon χ2 = 0.882, d.f. = 2, p = 0.643), indicating that somatic tissues were not negatively impacted by reduction of DNA methylation or Dnmt1 expression.
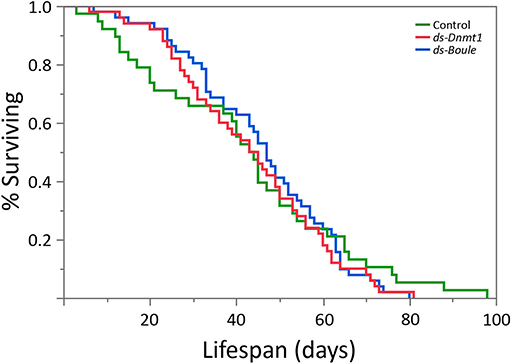
Figure 8. Neither the loss of egg production via Boule knockdown nor loss of egg production combined with loss of DNA methylation by Dnmt1 knockdown affected female lifespan. All females were treated during the 4th instar larval stage and housed individually with a male from the day of adult emergence. Lifespan was measured as the number of days between adult emergence and death.
Discussion
There has been much discussion about the role of DNA methylation and the function of the DNA methyltransferases in insects (Lyko and Maleszka, 2011; Yan et al., 2015; Lo et al., 2018; Glastad et al., 2019), given the diversity in both methylation state and patterns of evolution of the DNA methyltransferases across the insect tree of life (Bewick et al., 2016). There have been fewer experimental studies. In this study, we asked whether the phenotype observed following adult injection with ds-Dnmt1 was restricted to the ovary because this was the only tissue that had sufficient rounds of DNA replication to reduce DNA methylation in the adult tissues or if Dnmt1 actually has an additional function in the ovaries. We found that the germ cell specific phenotype produced by Dnmt1 knockdown is exacerbated by knocking down Dnmt1 function at the developmental stage at which oocytes are being born (Wick and Bonhag, 1955). Thus, our results support a pleiotropic function for DNMT1, functioning as an ovary-specific gene regulating development of eggs through either the production or maintenance of oogonia as well as in the maintenance of methylation.
As evidence from more insect systems emerge, it is clear that DNMT1 is required to restore methylation following semi-conservative replication in the insects. Knocking down Dnmt1 expression results in a reduction of DNA methylation (Bewick et al., 2019). Further, DNMT1 is required for embryo development in some insect species. Expression of Dnmt1 is high in eggs and embryos relative to other tissues in several insect species (Yan et al., 2015; Schulz et al., 2018, this study). Knocking down Dnmt1 expression in eggs and embryos results in embryo lethality (Zwier et al., 2012; Schulz et al., 2018; Bewick et al., 2019). Third, there is a suggestion that DNA methylation may be important in stability of germ cells (Takashima et al., 2009; Bewick et al., 2019). In honey bees, DNA methylation is more efficiently maintained in the germline than in the soma (Harris et al., 2019). This could indicate that preservation of germline DNA methylation patterns may be more critical than those of somatic methylation patterns, although it could also reflect other mechanisms that stabilize the genome of the germline (Maklakov and Immler, 2016). Knockdown of the DNMT1 associated protein 1 is also associated with a breakdown in ovaries and a loss of viable oocytes in Harmonia axyridis (Gegner et al., 2019). And loss of Dnmt1 function leads to loss of the ability to produce ooctyes (Bewick et al., 2019).
Much of what we know about DNA methylation in insects comes from correlational studies, based on expression patterns of the DNA methyltransferases. The few studies that explicitly disrupt expression of Dnmt1 assume that the functions revealed by this manipulation are mediated by the resulting change in DNA methylation patterns. Is DNMT1 function always related to DNA methylation? In Bewick et al. (2019), no phenotype is apparent within the somatic tissues such as the gut and muscle. However, rates of cell division in adult insects tend to be low and so the somatic tissues assayed did not lose DNA methylation. Thus, the lack of phenotype could have been due to the experimental design in which the timing of knockdown relative to assay was insufficient to allow any effect to be manifest. This study was specifically designed to address this issue; we treated nymphs to allow sufficient rounds of cell division during growth and development into adults to enable reduction of DNA methylation in both reproductive tissues and somatic tissues. We did indeed observe that the levels of DNA methylation were reduced over two-fold in all the tissues tested. However, despite this over two-fold reduction in DNA methylation in somatic tissues, we saw no disruption in the function of the somatic tissues. An altered function should have resulted in an impact on development and/or lifespan. Thus, DNA methylation and phenotype were uncoupled in this experiment. This is consistent with similar results in other insects. Tribolium castaneum is an insect with no detectable DNA methylation (Zemach et al., 2010; Cunningham et al., 2015; Bewick et al., 2016; Schulz et al., 2018), but which has maintained the Dnmt1 gene in its genome (Bewick et al., 2016). When Dnmt1 is knocked down during pupal development, it results in embryo lethality (Schulz et al., 2018). The data showing that DNA methylation can be lost without a phenotype and that DNMT1 plays a functional role in an insect that lacks DNA methylation make it likely that the functional role of DNMT1 is not always mediated through DNA methylation.
Our results establish that Dnmt1 has a specific function in germ cells. The comparison with the Boule knockdown females indicates that Dnmt1 is required for maintenance of oogonia or primary oocytes. In the Boule knockdown females, the overall structure of the germarium is preserved, although no ooctyes ever form. Given that Boule is required for entry into meiosis (Eberhart et al., 1996), one likely interpretation of our data is that healthy oogonia are waiting for the signal that it is time to divide meiotically to form primary ooctyes, but that signal never comes. The tissue disruption and evidence of cell death in the Dnmt1 knockdown females, however, indicate that Dnmt1 is required to maintain oogonia and/or primary ooctyes and that if Dnmt1 function is missing, these cells are not viable. While we are not able to differentiate among oogonia and primary ooctyes in our samples, the fact that the trophic tissue develops normally provides a clue as to where Dnmt1 is required. During the first, second, and third instar stages of development, the germ cells within the somatic ovary increase in number through mitosis (Wick and Bonhag, 1955). In the fourth instar stage, oogonia either divide mitotically to form trophocytes or divide meiotically to form ooctyes. The presence of a normal appearing population of trophocytes suggest that mitotic divisions are not affected by the lack of Dnmt1 expression, as these cells increase significantly in number through mitosis during the fifth instar (Wick and Bonhag, 1955) following Dnmt1 knockdown.
We suggest that Dnmt1 may be required for proper progression through meiosis or for stability of the germ cells. In Dnmt1 knockdown females, oogonia that have not been able to advance properly through meiosis will die or be targeted for destruction. Alternatively, oogonia that divide to form primary ooctyes may require Dnmt1 to maintain genome integrity. In the absence of Dnmt1 newly born oocytes may degenerate. We currently are not able to determine if this is due to the loss of methylation or a pleiotropic function of DNMT1. While it is clear that DNA methyltransferases are associated with the reproductive cells in insects, the role of DNA methylation in germ cell development in male mice has been more closely examined. DNA methyltransferase 3-like (Dnmt3L) is required for meiosis in male mice (Bourc'his and Bestor, 2004). Dnmt3L is required in cells prior to meiosis, perhaps involving a premeiotic genome scan that occurs in prospermatagonial cells, and the authors conclude that the defect that arises through the knockdown of expression of Dnmt3L is likely to arise because normal methylation patterns on dispersed repeated sequences are not properly established (Bourc'his and Bestor, 2004). Knockdown of Dnmt1 expression in male mice also impacts male fertility. Loss of Dnmt1 expression induces apoptosis in germline stem cells, without evidence of aberrant gene expression (Takashima et al., 2009). It is possible that this is due to hypomethylation, but the authors point out that the few rounds of cell division between knockdown and sampling did not provide an opportunity for significant loss of CpG methylation. They thus conclude that mouse germline stem cells are either extremely sensitive to slight demethylation or require DNMT1 activity outside of its function in maintaining methylation state.
Given the extreme impact on fitness of Dnmt1 in O. fasciatus, N. vitripennis, and T. castaneum, it is curious that this gene shows such variation across the insect tree of life where it is duplicated or lost across multiple taxa (Bewick et al., 2016). We expect genes with strong fitness effects, including genes essential in development, to experience strong stabilizing selection (Charlesworth, 1991; Ellegren and Parsch, 2007). For example, Boule, which is required for entry into meiosis, is a highly conserved gene across the bilaterian and experiences strong purifying selection (Shah et al., 2010). If Dnmt1 is required for oogenesis and early development in phylogenetically disparate species, how is so much variation in this gene tolerated? Variable DNA methylation within somatic cells is typical within insects, suggesting this is not an essential function. In honey bees, levels of methylation within somatic cells vary and only have to be maintained above a threshold level within exons (Harris et al., 2019). We found a similar tolerance for changes in somatic cell levels of DNA methylation here, where an over two-fold change in CpG methylation had no discernable effect on somatic function. It is likely, however, that there is less flexibility in the pathways required for maintaining a stable germline (Maklakov and Immler, 2016), which is expected to be strongly related to fitness and therefore under strong stabilizing selection. If Dnmt1 has a pleiotropic function in germline stability, independent of DNA methylation, it suggests that alternative or multiple pathways for stabilization of the germline genome must exist. Those pathways are yet to be discovered but examining insects that lack Dnmt1 (Bewick et al., 2019) provides a potential model.
Conclusion
We have demonstrated that Dnmt1 functions to maintain a healthy germline in female O. fasciatus. Dnmt1 knockdown, while resulting in a reduction in DNA methylation across both the germ and soma, only has a morphological phenotype in reproductive cells. We propose that Dnmt1 has a pleiotropic function independent of DNA methylation in the germ cells and could be required to maintain the genome integrity of germ cells or be required to progress through meiosis. These results raise a number of questions that need to be addressed in future experiments.
Data Availability Statement
The datasets analyzed for this study will be made available by the authors through the publicly available Dryad Digital Repository (https://datadryad.org/stash/); https://doi.org/10.5061/dryad.kwh70rz0m.
Author Contributions
PM, AM, and RS contributed conception and design of the study. AA, JW, EM, and PM collected the data. AA wrote the first draft of the manuscript. All authors contributed to the analysis of the data, manuscript revision, and read and approved the submitted version.
Funding
AA and ZS received funding for undergraduate research from the College of Agriculture and Environmental Sciences and from the University of Georgia Honors College to carry out this research. RS was a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trust.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Aleksandar Popadić and Lisa Hanna for helpful conversations about the injection protocol for larval stages of O. fasciatus and Christina Ethridge for preparing and analyzing low throughput whole genome bisulfite sequencing data. This manuscript has been released as a Pre-Print at https://doi.org/10.1101/824052.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00004/full#supplementary-material
References
Bewick, A. J., Sanchez, Z., McKinney, E. C., Moore, A. J., Moore, P. J., and Schmitz, R. J. (2019). Dnmt1 is essential for egg production and embryo viability in the large milkweed bug, Oncopeltus fasciatus. Epigenetics Chromatin 12:6. doi: 10.1186/s13072-018-0246-5
Bewick, A. J., Vogel, K. J., Moore, A. J., and Schmitz, R. J. (2016). Evolution of DNA methylation across insects. Mol. Biol. Evol. 34, 654–665. doi: 10.1093/molbev/msw264
Bonhag, P. F., and Wick, J. R. (1953). The functional anatomy of the male and female reproductive systems of the milkweed bug, Oncopeltus fasciatus (Dallas) (Heteroptera: Lygaeidae). J. Morphol. 93, 177–283. doi: 10.1002/jmor.1050930202
Bourc'his, D., and Bestor, T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99. doi: 10.1038/nature02886
Charlesworth, B. (1991). The evolution of sex chromosomes. Science 251, 1030–1033. doi: 10.1126/science.1998119
Chesebro, J., Hrycaj, S., Mahfooz, N., and Popadić, A. (2009). Diverging functions of Scr between embryonic and post-embryonic development in a hemimetabolous insect, Oncopeltus fasciatus. Dev. Biol. 329, 142–151. doi: 10.1016/j.ydbio.2009.01.032
Cunningham, C. B., Ji, L., Wiberg, R. A. W., Shelton, J., McKinney, E. C., Parker, D. J., et al. (2015). The genome and methylome of a beetle with complex social behavior, Nicrophorus vespilloides (Coleoptera: Silphidae). Genome Biol. Evol. 7, 3383–3396. doi: 10.1093/gbe/evv194
Duxbury, A. E., Weathersby, B., Sanchez, Z., and Moore, P. J. (2018). A study of the transit amplification divisions during spermatogenesis in Oncopeltus fasciatus to assess plasticity in sperm numbers or sperm viability under different diets. Ecology and Evol. 8, 10460–10469. doi: 10.1002/ece3.4511
Eberhart, C. G., Maines, J. Z., and Wasserman, S. A. (1996). Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 381, 783–785. doi: 10.1038/381783a0
Ellegren, H., and Parsch, J. (2007). The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8, 689–698. doi: 10.1038/nrg2167
Ewen-Campen, B., Jones, T. E., and Extavour, C. G. (2013). Evidence against a germ plasm in the milkweed bug Oncopeltus fasciatus, a hemimetabolous insect. Biol. Open 2, 556–568. doi: 10.1242/bio.20134390
Gegner, J., Gegner, T., Vogel, H., and Vilcinskas, A. (2019). Silencing of the DNA methyltransferase 1 associated protein 1 (DMAP1) gene in the invasive ladybird Harmonia axyridis implies a role of the DNA methyltransferase 1-DMAP1 complex in female fecundity. Insect Mol. Biol. doi: 10.1111/imb.12616. [Epub ahead of print].
Glastad, K. M., Hunt, B. G., and Goodisman, M. A. D. (2019). Epigenetics in insects: genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 64, 185–203. doi: 10.1146/annurev-ento-011118-111914
Harris, K. D., Lloyd, J. P., Domb, K., Zilberman, D., and Zemach, A. (2019). DNA methylation is maintained with high fidelity in the honey bee germline and exhibits global non-functional fluctuations during somatic development. Epigenetics Chromatin 12:62. doi: 10.1186/s13072-019-0307-4
Karashima, T., Sugimoto, A., and Yamamoto, M. (2000). Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 127, 1069–1079.
Kay, S., Skowronski, D., and Hunt, B. G. (2018). Developmental DNA methyltransferase expression in the fire ant Solenopsis invicta. Insect Sci. 25, 57–65. doi: 10.1111/1744-7917.12413
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Liu, J., Lemonds, T. R., and Popadić, A. (2014). The genetic control of aposematic black pigmentation in hemimetabolous insects: insights from Oncopeltus fasciatus. Evol. Dev. 16, 270–277. doi: 10.1111/ede.12090
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, N., Simpson, S. J., and Sword, G. A. (2018). Epigenetics and developmental plasticity in orthopteroid insects. Curr. Opin. Insect Sci. 25, 25–34. doi: 10.1016/j.cois.2017.11.003
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 doi: 10.1038/nrg.2017.80
Lyko, F., and Maleszka, R. (2011). Insects as innovative models for functional studies of DNA methylation. Trends Genet. 27, 127–131. doi: 10.1016/j.tig.2011.01.003
Maklakov, A. A., and Immler, S. (2016). The expensive germline and the evolution of ageing. Curr. Biol. 26, R577–R586. doi: 10.1016/j.cub.2016.04.012
Meinzer, F., Dobler, S., Donath, A., and Lohr, J. N. (2019). Robust reference gene design and validation for expression studies in the large milkweed bug, Oncopeltus fasciatus, upon cardiac glycoside stress. Gene 710, 66–75. doi: 10.1016/j.gene.2019.05.032
Panfilio, K. A., Jentzsch, I. M. V., Benoit, J. B., Erezyilmaz, D., Suzuki, Y., Colella, S., et al. (2019). Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Biol. 20:64. doi: 10.1186/s13059-019-1660-0
Schmitz, R. J., He, Y., Valdés-López, O., Khan, S. M., Joshi, T., Urich, M. A., et al. (2013). Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23, 1663–1674. doi: 10.1101/gr.152538.112
Schmitz, R. J., Lewis, Z. A., and Goll, M. G. (2019). DNA methylation: shared and divergent features across eukaryotes. Trends Genet. 35, 818–827. doi: 10.1016/j.tig.2019.07.007
Schulz, N. K., Wagner, C. I., Ebeling, J., Raddatz, G., Diddens-de Buhr, M. F., Lyko, F., et al. (2018). Dnmt1 has an essential function despite the absence of CpG DNA methylation in the red flour beetle Tribolium castaneum. Sci. Rep. 8:16462. doi: 10.1038/s41598-018-34701-3
Shah, C., VanGompel, M. J., Naeem, V., Chen, Y., Lee, T., Angeloni, N., et al. (2010). Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function. PLoS Genet. 6:e1001022. doi: 10.1371/journal.pgen.1001022
Takashima, S., Takehashi, M., Lee, J., Chuma, S., Okano, M., Hata, K., et al. (2009). Abnormal DNA methyltransferase expression in mouse germline stem cells results in spermatogenic defects. Biol. Reprod. 81, 155–164. doi: 10.1095/biolreprod.108.074708
Ulrich, M. S., Nery, J. R., Lister, R., Schmitz, R. J., and Ecker, J. R. (2015). MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat. Protocols 10, 475–483. doi: 10.1038/nprot.2014.114
Wick, J. R., and Bonhag, P. F. (1955). Postembryonic development of the ovaries of Oncopeltus fasciatus (Dallas). J. Morphol. 96, 31–59 doi: 10.1002/jmor.1050960103
Yan, H., Bonasio, R., Simola, D. F., Liebig, J., Berger, S. L., and Reinberg, D. (2015). DNA methylation in social insects: how epigenetics can control behavior and longevity. Annu. Rev. Entomol. 60, 435–452. doi: 10.1146/annurev-ento-010814-020803
Zemach, A., McDaniel, I. E., Silva, P., and Zilberman, D. (2010). Genome-wide evolutionary analysis of DNA methylation. Science 328, 916–919. doi: 10.1126/science.1186366
Keywords: DNA methylation, epigenetics, Dnmt1, oogenesis, germ cells, Oncopeltus fasciatus
Citation: Amukamara AU, Washington JT, Sanchez Z, McKinney EC, Moore AJ, Schmitz RJ and Moore PJ (2020) More Than DNA Methylation: Does Pleiotropy Drive the Complex Pattern of Evolution of Dnmt1? Front. Ecol. Evol. 8:4. doi: 10.3389/fevo.2020.00004
Received: 18 November 2019; Accepted: 07 January 2020;
Published: 28 January 2020.
Edited by:
David Ellard Keith Ferrier, University of St Andrews, United KingdomReviewed by:
Elizabeth Jenness Duncan, University of Leeds, United KingdomNora Kristin Elisa Schulz, Department of Biological Sciences, Vanderbilt University, United States
Copyright © 2020 Amukamara, Washington, Sanchez, McKinney, Moore, Schmitz and Moore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia J. Moore, cGptb29yZUB1Z2EuZWR1
 Ashley U. Amukamara
Ashley U. Amukamara Joshua T. Washington
Joshua T. Washington Zachary Sanchez
Zachary Sanchez Elizabeth C. McKinney
Elizabeth C. McKinney Allen J. Moore
Allen J. Moore Robert J. Schmitz
Robert J. Schmitz Patricia J. Moore
Patricia J. Moore