
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 06 December 2019
Sec. Evolutionary Developmental Biology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00468
This article is part of the Research Topic Evo Devo Cell by Cell View all 8 articles
Wnt signaling pathways are involved in many important cellular processes including proliferation and differentiation. Wnt ligands are released by source cells and signal to target cells by binding to the Frizzled receptor family and triggering changes in downstream target gene expression. Wnt signaling appeared at the base of metazoans and there was an early expansion in the repertoire of Wnt ligands to the 13 known subfamilies. However, little is known about functionality of these ligands in many animal lineages. Understanding the roles of these important signaling molecules in a wider range of animals is crucial to understand the regulation and evolution of cell fate during development and how this can lead to diversification. Here, we analyzed the Wnt repertoire among lepidopterans, where the embryological functionality of these ligands is understudied compared to other insect orders. To be able to explore Wnt gene roles during butterfly embryogenesis we first established a staging system for the butterfly model, Bicyclus anynana, and assayed the expression pattern of all eight lepidopteran Wnt genes during early butterfly development. We detected expression of Wnt1, Wnt10, and WntA in several expression domains, such as segmental stripes as well as expression of Wnt7 in the nervous system and Wnt11 in several head structures. Overall, our study provides, a basis for future research into butterfly embryogenesis and much needed new insights into the potential roles of Wnt genes in specifying cell fate in these animals as well as how this compares to other animals.
The regulation of cell fate is crucial for development and modifications of this regulation underlie various aspects of the evolution of animal development and phenotypic diversification. Wnt signaling (both canonical and non-canonical) plays key roles in the regulation of cell fate in most aspects of animal development including cell proliferation, cell differentiation, planar cell polarity and cell death (Logan and Nusse, 2004; Clevers, 2006; Wiese et al., 2018). Wnt ligands trigger downstream signaling cascades by binding to receptors such as those of the Frizzled (Fz) family (Bhanot et al., 1996; Kikuchi et al., 2007; Janda et al., 2012). There are 13 subfamilies of Wnt ligands in animals, namely Wnt1-11, 16, and A (Kusserow et al., 2005; Stefanik et al., 2014). No Wnt genes have been found in any non-metazoans suggesting they evolved and diversified early in metazoan evolution (Holstein, 2012). It is thought that this large repertoire of ligands confers specificity and robustness to cell fate decisions during development, but conversely changes in their copy number and expression can also facilitate the evolution of developmental processes (Logan and Nusse, 2004; Holstein, 2012). Therefore, understanding the repertoire and roles of Wnt ligands in a wide range of species can inform the evolution and development of animals more broadly.
In insects, the repertoire and expression patterns of Wnt genes has been described in detail in the fruit fly Drosophila melanogaster [e.g., Swarup and Verheyen (2012) and Murat et al. (2010)] and the red flour beetle Tribolium castaneum (Bolognesi et al., 2008a,b; Janssen et al., 2010). However, Wnts have hardly been studied in other insect orders such as the Lepidoptera (i.e., butterflies and moths). Differences between moths and butterflies are based on morphological characteristics although they do not form monophyletic groups (Kawahara and Breinholt, 2014; Mitter et al., 2017). While the embryology of a variety of lepidopteran species has been described (Krause and Krause, 1964; Ando and Tanaka, 1980; Kobayashi and Ando, 1990; Broadie et al., 1991; Miya, 2003; Masci and Monteiro, 2005), we know very little about gene expression patterns in early butterfly embryogenesis in particular, and curiously there are indications that it may be rather divergent with respect to other insects such as D. melanogaster and T. castaneum (Carter et al., 2013).
Previously, seven Wnt subfamilies were described in the butterfly Heliconius melpomene (Martin et al., 2012) and their expression was analyzed in larval wing discs (Martin and Reed, 2014). More recently, a study found representatives of eight subfamilies in several ditrysian butterfly and moth species (Ding et al., 2019). All used species in this study are part of the polyphyletic taxonomic suborder of the Ditrysia. However, embryonic Wnt gene expression has only been studied in the moths Bombyx mori (Ding et al., 2019) and Manduca sexta (Dow et al., 1988; Broadie et al., 1991; Miya, 2003).
The squinting bush brown butterfly, Bicyclus anynana (Butler, 1879 Lepidoptera, Ditrysia, Nymphalidae) (Figure 1), has been a very successful butterfly model species for the elucidation of wing pattern development and evolution (Monteiro, 2017; Özsu and Monteiro, 2017; Matsuoka and Monteiro, 2018; Prakash and Monteiro, 2018). Given the genomic resources for this species (Nowell et al., 2017), as well as its amenability for gene functional assays (Banerjee and Monteiro, 2018; Matsuoka and Monteiro, 2018; Prakash and Monteiro, 2018), it is an ideal model system to elucidate the functionality of butterfly Wnt genes during early embryogenesis. Therefore, in this study, we studied the Wnt repertoire of lepidopterans and compared the expression of these genes in B. anynana with what is known for the model insect species D. melanogaster and T. castaneum. These species were chosen for comparison because the repertoire, expression and function of Wnt genes is best understood in these two insects and they represent the two most closely related orders to lepidopterans.
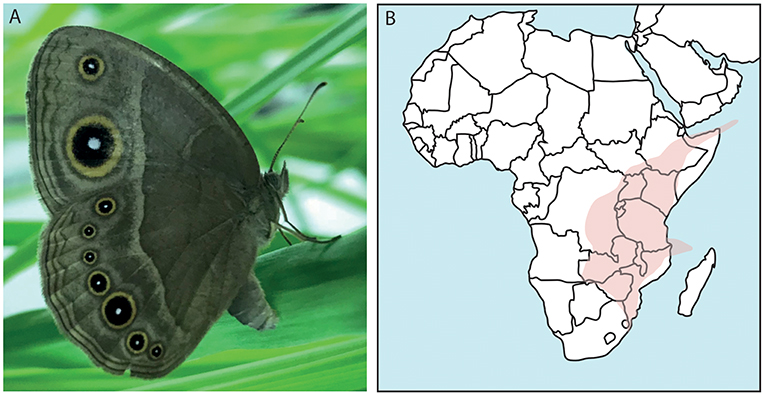
Figure 1. The African butterfly Bicyclus anynana and its distribution. (A) Shown is a female specimen of B. anynana with the characteristic eyespots on its wings. (B) Confirmed findings of B. anynana in Southeast Africa in the area indicated. This species can be found in: Democratic Republic of Congo, Kenya, Rwanda, Burundi, Tanzania, Uganda, Zambia, Malawi, Mozambique, Socotra islands, Comoro islands, and South Africa (Otto, 2015; Brattström et al., 2016).
Experimental animals were derived from an outbred B. anynana culture at Oxford Brookes University (established from an outbred stock at the University of Cambridge). Stocks were kept on the grass species Brachypodium sylvaticum in netted cages under controlled temperature, light and humidity (26°C; RH 70%; LD: 12:12 with dawn and dusk transition periods). A fresh potted host plant was provided for egg deposition. Eggs for the embryonic staging were collected every hour from 1 h AEL to 51 h AEL. The determination of the completion rate of development of the staging series is based on Dorn et al. (1987). Embryos for the analysis of the developmental stages were stained with 4′,6-diamidino-2-phenylindole (DAPI) and imaged using an Axio Zoom.V16 microscope with an Axiocam 506 color camera (Zeiss, Germany). Embryos from these collections were also used for in situ hybridisations and RNA extractions. Embryos and eggs were fixed according to Brakefield et al. (2009) and stored in 100% methanol at −20°C.
All butterfly and moth sequences used were extracted from Lepbase (Release 4) using a tBLASTn search with previously annotated lepidopteran Wnt gene sequences as queries (Challi et al., 2016). The final lepidopteran sequence set analyzed included only species of the largest lepidopteran clade, the Ditrysia: Plutella xylostella, Papilio glaucus, Phoebis sennae, Pieris napi, Lerema accuis, B. anynana, Heliconius melpomene, Danaus plexippus, Junonia coenia, Melitaea cinxia, Calycopis cercrops, Amyelois transitella, Plodia interpunctella, Operohtera brumata, B. mori and M. sexta. Anopheles gambiae, T. castaneum, Apis mellifera, and D. melanogaster were included as outgroups. In addition, sequences for B. mori were obtained from NCBI tBLASTn searches and transcriptomic data for M. sexta (Cao and Jiang, 2017) was mined using a local tBLASTn search. All nucleotide sequences were translated via EMBOSS Transeq (http://www.ebi.ac.uk/Tools/st/emboss_transeq/) and aligned using local ClustalX 2.1 (Larkin et al., 2007). All gene numbers of sequences used can be found in Supplement Table S1. The alignment was edited manually using SeaView version 4.6.1 (Gouy et al., 2010) and the full alignment file can be found in the Supplement Data Sheet 1. Phylogenetic analysis was performed using RAxML version 8.0.0 (released Dec 2013) including rapid bootstrapping with 1000 replicates (Stamatakis et al., 2008; Stamatakis, 2014). The gap proportion and completely undetermined characters in the lepidopteran dataset was 46.77%. The best protein substitution model was detected using local ProtTest 2.4.3 (Abascal et al., 2005) and the best fitting model for estimating the amino acid replacement frequencies during molecular evolution was VT+G (Muller and Vingron, 2000). This model is an extension of the Markov model previously estimated by Dayhoff et al. (1978) and allows prediction of the amino acid substitution for more divergent alignments. The +G (Gamma) added a category of change for each amino acid site and allowed rating into low, medium and high rate of change (Yang, 1993), here used with an accuracy of 0.1 log likelihood units. The maximum likelihood tree was calculated using the default alpha parameters of the RAxML analysis.
For whole mount in situ hybridization, the published protocol for P. aegeria (Ferguson et al., 2014) was used with minor modifications. Ribonucleotide probes were generated from cDNA derived from RNA of mixed embryonic stages (0–72 h) (see primer list and size of probes in Supplement Table S2). RNA was extracted using QIAzol reagent (Qiagen) and reverse transcribed using Quantitect Reverse transcription kit (Qiagen) into cDNA. Embryos were rehydrated from 100% methanol in to 100% PBS-T (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 2H2O, and 1.47 mM KH2PO4 plus 0.1% Tween20), digested with Proteinase K (1:1,000) for 12 min on ice which was stopped by replacing the solution with PBS+Glycine (2 mg/ml). Embryos were fixed for 20 min in 4% formaldehyde. Subsequently, embryos were hybridized at 56°C overnight with the probe (hybridization buffer with 50% formamide, 5x SSC pH 7.0, 10 mg/ml salmon sperm, 1 mg/ml tRNA, 100 μg/ml heparin, and 0.1% Tween 20, adjust pH 6.5 with 1M HCl). The next day embryos were brought back to PBS-T, blocked with 1× blocking reagent (Roche) and incubated with the Anti-Fab AP Fragment antibody (Roche, 1:2,000) in 1× blocking reagent (Roche). After washing in PBS-T overnight, embryos were stained using NBT/BCIP (Roche) in the AP staining buffer (100 mM NaCl, 50 mM MgCl2, 100 mM Tris HCl pH 9.5, and 0.1% Tween 20). Staining was stopped by several washes with PBS-T and the nuclei were additionally stained with DAPI (Roche) for 30 min. Embryos were kept in PBS or mounted in 80% glycerol. All images were taken using an Axio Zoom.V16 microscope with an Axiocam 506 color camera (Zeiss, Germany). For all Wnt genes, anti-sense and sense probes were used but a specific signal was only ever detected at any embryonic stage with the anti-sense probes.
To further explore the repertoire of Wnt genes among Lepidoptera, we mined 16 different ditrysian genomes for Wnt genes and performed a maximum likelihood analysis with several insects as outgroups (Figures 2, 3). The bootstrap values were all above 80 for the nine Wnt subfamilies in the tree indicating good phylogenetic support for the placement of the lepidopteran Wnt genes within eight of these subfamilies (Figure 2). The deeper branches of the tree topology were not very highly supported by the bootstrap values (ranging from 40 to 80), however, which makes it difficult to pinpoint an evolutionary relationship between Wnt subfamilies compared to previous studies that suggested close relationships between, for example, Wnt1 and Wnt6 (Figure 2) (Janssen et al., 2010).
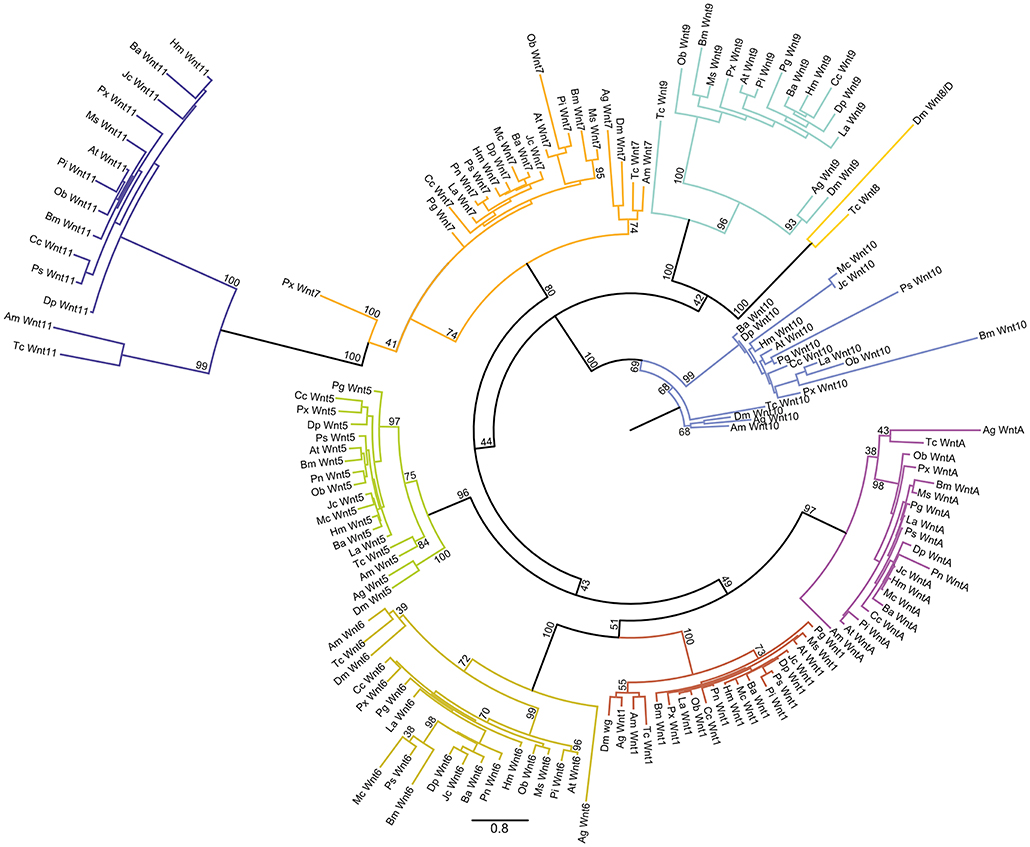
Figure 2. Maximum likelihood analysis of Lepidopteran Wnt genes. Circular maximum likelihood tree of all Wnt sequences used in our study from the 16 lepidopteran species as well as the four other insects. The bootstrap support for branches is indicated. Scale bar showing the number of substitutions compared to the length of branches between the sequences.

Figure 3. Summary of the Wnt repertoire in all examined insect species. Simplified cladogram based on previously published phylogenetic data (Kawahara and Breinholt, 2014; Mitter et al., 2017) with the presence of Wnt genes indicated by colored boxes. Framed boxes indicate the potential presence of a Wnt ligand, but absence from the used dataset. Gray boxes indicate a loss of a Wnt subfamily. Ag, A. gambiae; Tc, T. castaneum; Am, A. mellifera; Dm, D. melanogaster; Px, P. xylostella; Pg, P. glaucus; Ps, P. sennae; Pn, P. napi; La, L. accius; Ba, B. anynana; Hm, H. melpomene; Dp, D. plexippus; Jc, J. coenia; Mc, M. cinxia; Cc, C. cecrops; At, A. transitella; Pi, P. interpunctella; Ob, O. brumata; Bm, B. mori; Ms, M. sexta.
For eight of the lepidopteran species, we confirmed the presence of genes of the eight Wnt subfamilies: Wnt1, 5–7, 9–11, and A (Figure 3). For the other eight butterflies and one moth, not all subfamilies were found, with different subfamilies missing in different species. Wnt5 and Wnt10 are missing in P. interpunctella and M. sexta, Wnt11 is missing in P. glaucus, L. accius and M. cinxia, and Wnt9 is missing from P. sennae and J. coenia while Wnt9, 10 and 11 are missing from P. napi (Figure 3). These sequences could be missing because they could not reliably be identified in the genomic data or they could represent true species-specific losses.
Despite missing sequences for eight of our focal species, it seems very likely that eight Wnt genes is the ancestral ditrysian Wnt repertoire. These results are consistent with the previously published Wnt repertoire for B. mori (Ding et al., 2019). This could indicate that eight Wnt genes were also present in the common ancestor of all lepidopterans including non- ditrysian species, but further analysis of additional species is needed to verify this.
In flies and beetles Wnt expression and function during embryogenesis have been widely studied (Nusslein-Volhard et al., 1984; Bolognesi et al., 2008a,b; Murat et al., 2010; Swarup and Verheyen, 2012). To compare these data with Wnt expression in butterflies and compare the tissues and cell types involved, we first described the developmental stages of B. anynana embryos in detail.
The early development of B. anynana is similar to the previously described developmental course in B. mori (Krause and Krause, 1964; Kobayashi and Ando, 1990; Miya, 2003). As soon as a fertilized egg is laid, embryogenesis begins and the egg is initially syncytial with the dividing energids distributed throughout (Carter et al., 2013) around 4–5 h AEL (Figure 4A). Subsequently, the energids move to the egg surface where they cellularise and form the blastoderm. The blastoderm differentiates into presumptive embryonic and extra-embryonic regions. As the presumptive embryonic region sinks into the yolk, it becomes enveloped by a rapidly expanding extra-embryonic tissue constituting the serosa (5–7 h AEL; Figures 4B–D″).
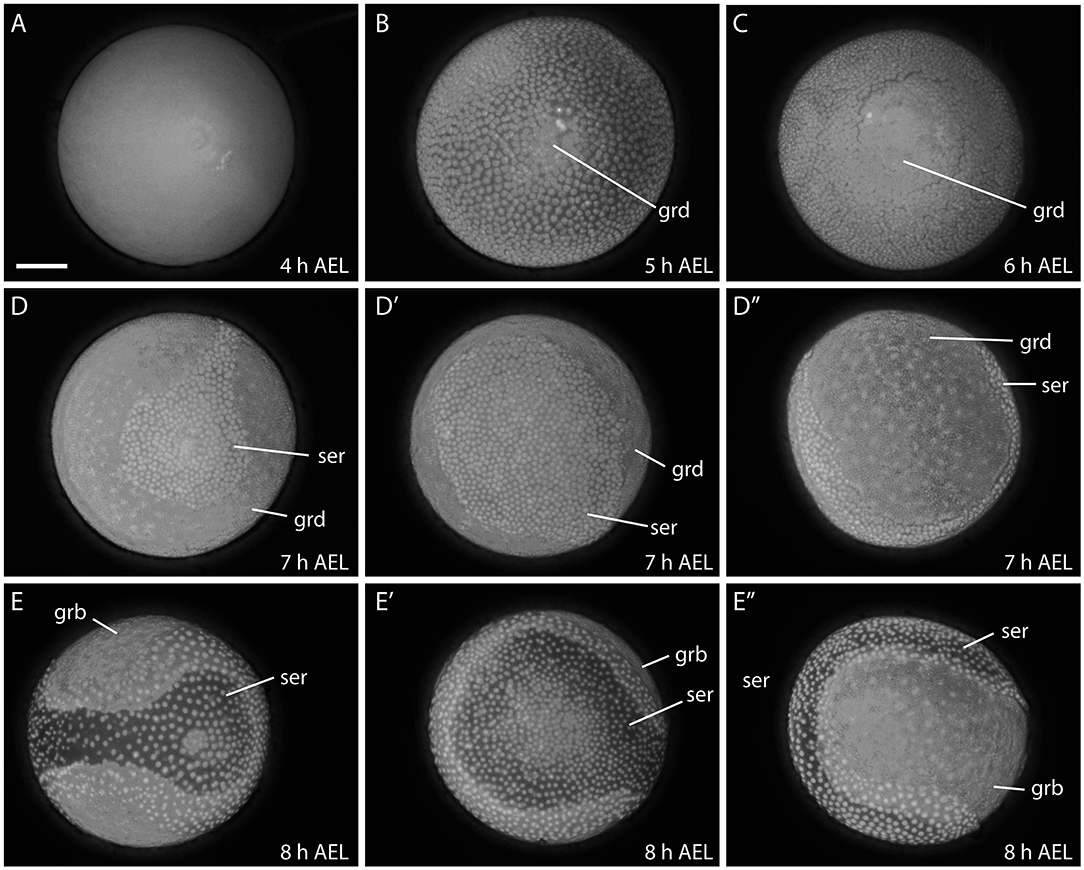
Figure 4. Early developmental stages of B. anynana from 4 to 8 h. The germ disc starts to form around 5 h AEL (A,B), expands at 6 h AEL (C) and forms the germband at around 8 h AEL (D-E″). See detailed descriptions in the main text. All embryos are stained with DAPI. AEL, after egg laying; grd, germ disc; grb, germ band; ser, serosa. Scale bar: 400 μm; all panels are at the same magnification.
The germ disc becomes enlarged at around 6–7 h AEL and covers more than two thirds of the egg surface (Figures 4C–D″). Subsequently, the germ disc elongates, becomes covered by yolk and enveloped by the serosa and is now referred to as the germ band (8 h AEL; Figures 4E–E″). Eventually, the serosa envelopes the whole egg, forming one of the protective egg layers (i.e., embryo and yolk; data not shown; Ferguson et al., 2014).
At 9–10 h AEL, the margins of the germ band begin to curl inwards (Figures 5A–B″). This process indicates the formation of the germ layers and the initiation of ectodermal and mesodermal differentiation (Krause and Krause, 1964). The patterning of the germ band begins at the anterior where at first, the protocephalon is visible. The primitive groove invaginates along the midline from anterior to posterior - the groove is deeper at the anterior than at the posterior end. Around this time, the first gnathal segments are visible (11 h AEL; Figures 5C–C″) (Krause and Krause, 1964; Kobayashi and Ando, 1990; Miya, 2003). The stomodeum, an invagination between the brain and the following gnathal segments, then became visible, and will form the future mouth region (Figure 5C″) (Krause and Krause, 1964).
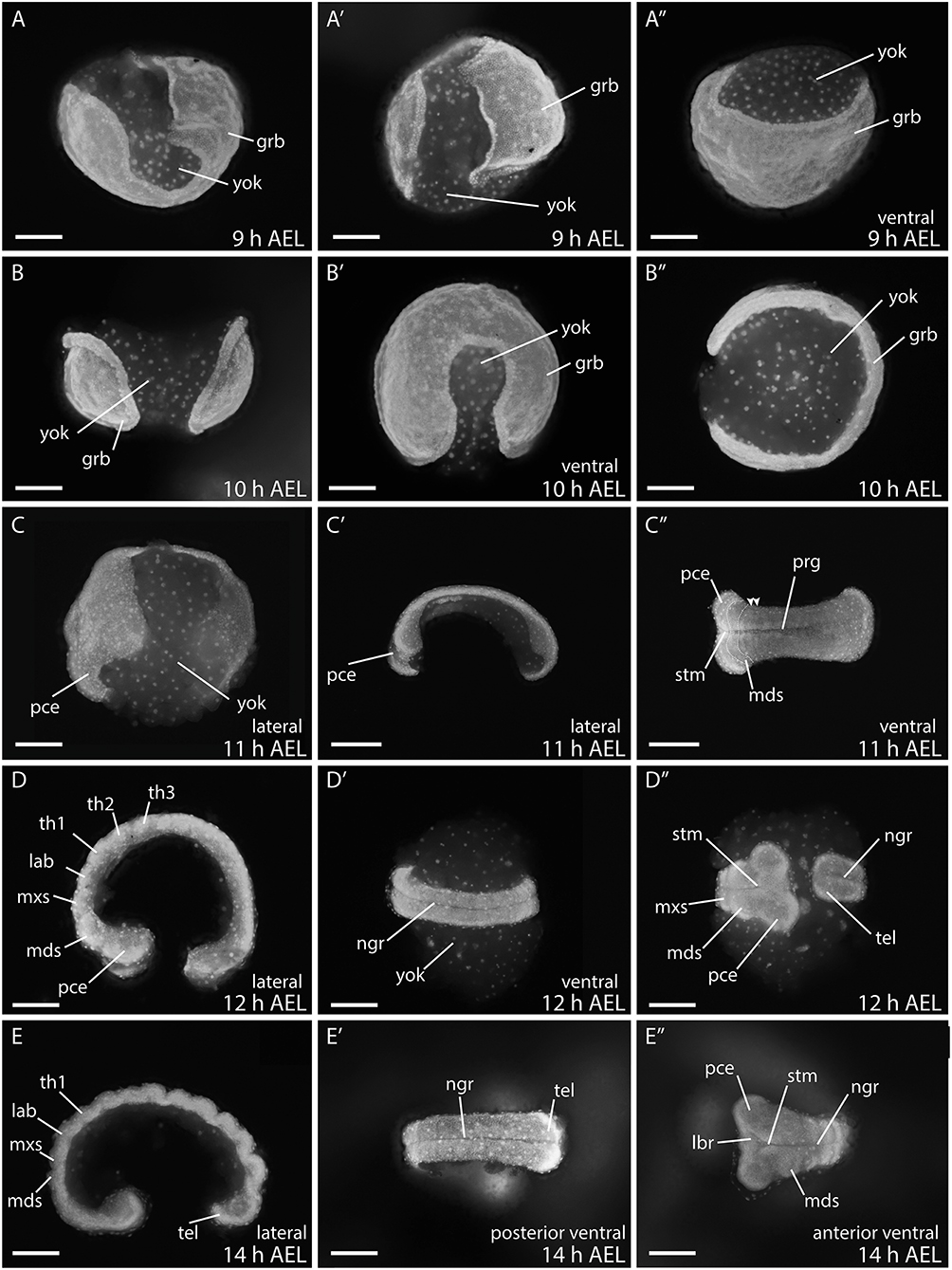
Figure 5. Embryonic stages of B. anynana from 9 to 14 h AEL. During this time the germ band develops into a basic patterned embryo with all gnathal, thoracic, and abdominal segments as well as the telson. (A–E″) See text for detailed description. AEL, after egg laying; grb, germ band; lab, labial segment; lbr, labrum; mds, mandibular segment; mxs, maxillary segment; ngr, neurogenic furrow; pce, protocephalon; pco, protocorm; prg, primitive grove; stm, stomodeum; tel, telson; th1-3, thoracic segment 1-3; yok, yolk. Orientation of embryos is indicated with anterior always to the left. Scale bar: 400 μm.
During the next three hours all gnathal and thoracic segments are determined (12–14 h AEL; Figures 5D–E″) and thus the protocephalon is followed by the mandibular, maxillary, and labial segments. The neurogenic furrow has formed by around 12 h AEL (12 h AEL; Figures 5D–D″), while the terminal region, the telson, differentiates and submerges into the yolk (14 h AEL; Figures 5E–E″) (Krause and Krause, 1964; Kobayashi and Ando, 1990; Miya, 2003). At 14 h AEL, the abdominal segments start to form as well as a terminal segment, the telson.
The abdominal segments becomes visible between 14 h AEL (Figures 5E–E″) to 18 h AEL (Figures 6A–B″), and by 20 h AEL all 10 abdominal segments have formed (Figure 6C). Further differentiation and outgrowth of the gnathal segments commences around 18 h AEL when the mandibular and maxillary segments become pronounced, the head lobes become enlarged, the labrum, and antennal rudiments become visible (Figures 6A–C″).
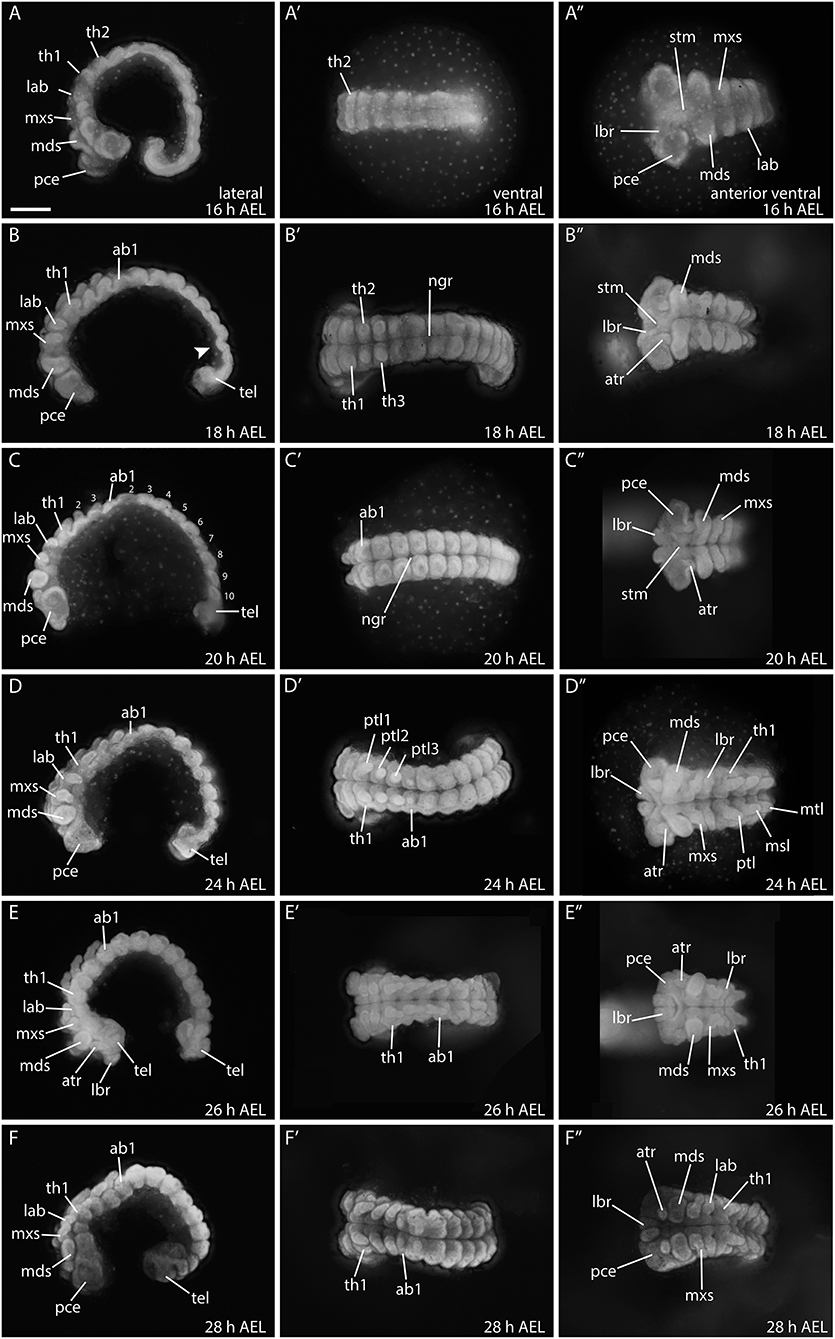
Figure 6. Embryonic development from 16 to 28 hours in B. anynana. (A–F″) See detailed description in text. ab1, abdominal segment 1; AEL, after egg laying; atr, antennal rudiment; lab, labial segment; lbr, labrum; mds, mandibular segment; mxs, maxillary segment; pce, protocephalon; pco, protocorm; ptl, prothoracic leg 1-3; stm, stomodeum; tel, telson; th1, thoracic segment. All columns are orientated in the same way as indicated in the top row. Anterior is always to the left and the ventral side is to the top. All pictures are to the same scale. Scale bar: 400 μm.
Between 24–28 h AEL, head structures continue to differentiate, and the prothoracic appendages elongate while the abdominal proleg buds become visible (Figures 6D–F″). Development of the prothoracic legs is complete by around 28 h AEL (Figures 6E–F″). At this stage the telson is still buried in the yolk and not fully extended (Figures 6F–F″).
Between 30 and 36 h AEL the embryo becomes compressed and thickens. The head structures continue to differentiate and become integrated in the protocephalon (Figures 7A–B″). Around 42 h AEL, embryonic reversion (blastokinesis) begins from the posterior end of the embryo. During this process the whole embryo goes through a dorsal extension while it rotates on its transversal layer along the anterior posterior axis. The embryo ends up with the previous ventral side facing inwards and the dorsal side facing outwards by around 51 h AEL (Figures 7C–F″) (Krause and Krause, 1964; Kobayashi and Ando, 1990; Miya, 2003). Figure 7D shows an embryo with the characteristic S-shape during this movement. After full rotation of the embryo, the dorsal side faces outwards. However, the dorsal opening is not yet closed, and the embryo still incorporates the yolk via this opening (around 44 h AEL; arrowheads) (Krause, 1939). At 48 h AEL, the dorsal opening becomes smaller (Figure 7E) and only a small amount of yolk remains attached to the embryo (Figure 7E′; asterisks). Full dorsal closure occurs at around 51 h AEL (Figures 7F–F″).
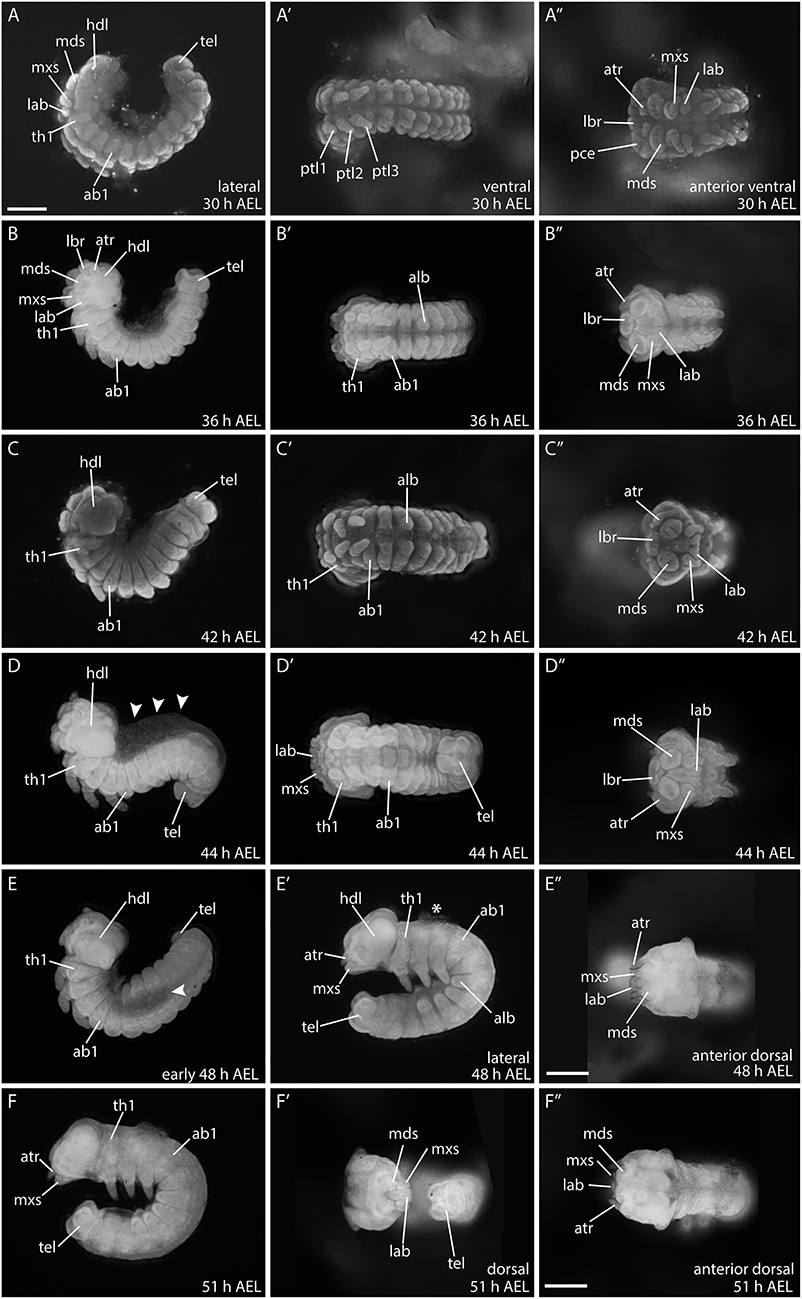
Figure 7. Developmental stages of B. anynana from 30 to 51 h AEL. (A–F″) See detailed description in text. ab1, abdominal segment 1; AEL, after egg laying; alb, abdominal limb bud; atr, antennal rudiment; hdl, head lobe; lab, labial segment; lbr, labrum; mds, mandibular segment; mxs, maxillary segment; pce, protocephalon; pco, protocorm; prg, primitive grove; ptl, protoracic leg 1-3; stm, stomodeum; tel, telson; th1, thoracic segment. All pictures are orientated anterior to the left and the dorsal side to the top as indicated in top row for each column if not otherwise stated. All pictures are to the same scale indicated in (A) (scale bar: 400 μm), except (E″) and (F″), which have individual scale bars (scale bar: 400 μm).
At around 48 h AEL, and after blastokinesis, the morphology of the embryonic head seems to be completed, including all gnathal appendages. Since this staging was performed to analyse Wnt gene expression during early embryogenesis, we did not extend staging beyond 50% of developmental time (51 h AEL) (Dorn et al., 1987; Broadie et al., 1991).
Overall, in terms of the order of development, the morphogenesis of the butterfly B. anynana embryo appears to proceed similarly to that of moth species, such as B. mori and M. sexta, described previously (Krause, 1939; Krause and Krause, 1964; Kobayashi and Ando, 1990; Broadie et al., 1991). To study the expression of Wnt genes in B. anynana we focused on developmental stages between 9 and 28 h AEL because this is the period during which the segments are formed and patterning occurs along the axes in general—thus allowing comparison to previously characterized Wnt gene expression in D. melanogaster and T. castaneum (e.g., Janson et al., 2001; Bolognesi et al., 2008a,b; Janssen et al., 2010; Swarup and Verheyen, 2012).
In situ hybridisations in B. anynana embryos showed expression of wg in a very thin and faint domain lining the anterior margin of the germ disc at 9 h AEL (Figures 8A,A′). This was the earliest expression observed for any Wnt gene in butterfly embryos. As soon as the germ band was formed and the head lobes, gnathal and thoracic segments were visible, expression in segmental stripes and in the head lobes was observed at 10 h AEL (Figure 8B). The previously described expression patterns in moths are similar to the segmental stripes detected in B. anynana (Kraft and Jäckle, 1994; Dhawan and Gopinathan, 2003; Nakao, 2010; Zhang et al., 2015). Two domains were observed in the labrum, which is again very similar to expression in the moth at this stage of embryogenesis (Figures 8C,D) (Nakao, 2010). At 16 h AEL, expression was observed in stripes in all segments, which was consistent with the segmental expression of wg in B. mori, M. sexta, D. melanogaster, T. castaneum, and other arthropods at equivalent developmental stages (Figure 8C) (Nusslein-Volhard et al., 1984; Kraft and Jäckle, 1994; Bolognesi et al., 2008a; Janssen et al., 2010; Nakao, 2010).
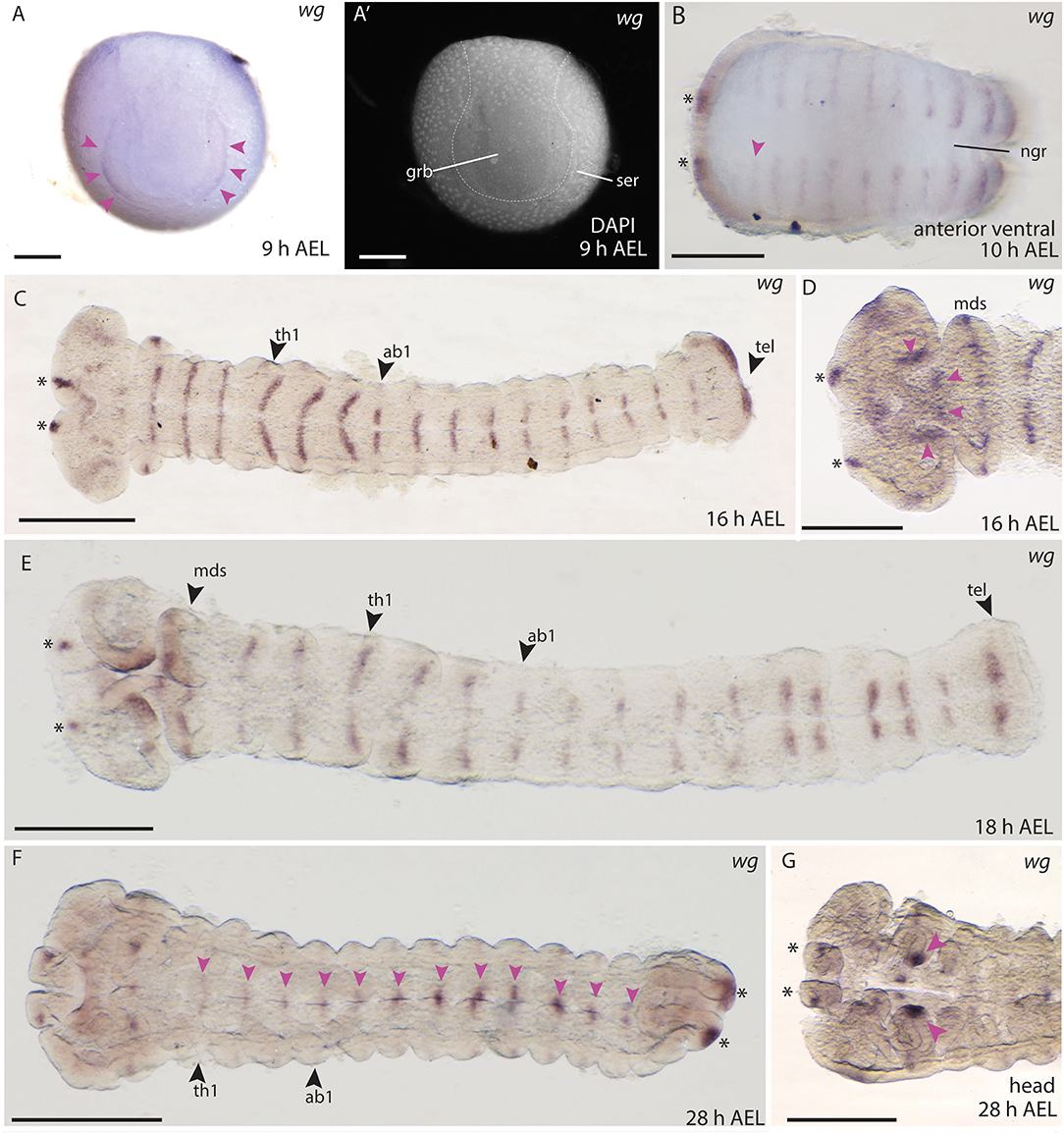
Figure 8. Wnt1/wg expression during embryogenesis in B. anynana. (A,A′) Early expression of wg at the anterior rim of the germ disc. (A′) Indicating morphology with DAPI. (B) Expression in segmental stripes at 10 h AEL. (C) Segmental expression of wg at 16 h AEL. (D) Higher magnification of head staining at 16 h AEL. (E) Expression at 18 h AEL showing segmental stripes. (F) wg expression at 28 h AEL along the A-P axis (pink arrowheads) and in the telson (asterisks). (G) Magnification of the head at 28 h AEL indicating strong expression in mandibular segments (pink arrowheads). All pictures are orientated with anterior to the left if not otherwise stated. ab1-3, abdominal segments 1-3; AEL, after egg laying; grb, germ band; h, hour; hdl, head lobe; lbs, labial segments; mds, mandibular segments; mxs, maxillary segments; ngr, neurogenic furrow; ser, serosa; th1-3, thoracic segments 1-3. Scale bar: 400 μm.
Expression in the developing head was observed at 16–18 h AEL in the most anterior part of the head lobes (Figures 8C,D), and around the stomodeum (Figures 8D,E) as well as two distinct domains in the telson (Figures 8C,E). During subsequent development the expression of wg in the segments was reduced and became concentrated at the neuronal furrow along the A-P axis (28 h AEL, Figure 8F). Strong domains of expression were seen at the tips of the mandibles (Figure 8G) as well as in the proximal parts of the labrum (Figure 8G). Although embryonic stages older than 30 h AEL were excluded from the expression domain analyses here (due to possible staining artifacts present in the embryonic cuticle in older embryos), we include observed wg expression at 42 to 48 h AEL in the thoracic legs and abdominal limb buds in (Supplement Figure S1). Overall, the expression of wg described during B. anynana embryogenesis was consistent with wg expression at similar stages in moths and other arthropods [e.g., Janssen et al. (2010)], where wg appears to have an early and conserved role in segmentation.
No embryonic expression was observed for Wnt5 in B. anynana embryos using several different in situ hybridization probes (see Supplement Figure S2). While Wnt5 is expressed at several embryonic stages in the nervous system in D. melanogaster (Fradkin et al., 1995, 2004) and in e.g., limb buds, head lobes, and segmental stripes T. castaneum (Bolognesi et al., 2008a) we did not detect any expression for this Wnt gene in B. anynana. In a recent study of B. mori, Wnt5 transcripts were also only observed in very late embryonic stages (6 days AEL) as well as in larval tissues such as head, ovaries and the wing discs (Ding et al., 2019). Therefore, we propose, that Wnt5, contrary to Drosophila and Tribolium, is not expressed in the embryonic stages of Bicyclus included in this study.
We did not detect any Wnt6 expression during embryogenesis in B. anynana using probes of different lengths and sequence (Supplement Figure S2). In T. castaneum embryos and D. melanogaster larvae, Wnt6 is expressed in a pattern similar to wg (Janson et al., 2001; Bolognesi et al., 2008a,b), which is also the case in wing discs of the butterfly H. melpomene (Martin and Reed, 2014). Embryonic Wnt6 expression is detected in the foregut and hindgut in D. melanogaster (Janson et al., 2001), which does not overlap with wg expression, but again we did not observe any such expression for Wnt6 in B. anynana at the stages assayed.
In B. anynana embryos, Wnt7 expression was first detected from around 16 h AEL in each of the thoracic and abdominal segments in lateral domains (Figures 9A,B). In D. melanogaster and T. castaneum, Wnt7 is expressed in mesodermal cells during development (Kozopas et al., 1998; Bolognesi et al., 2008a), and in T. castaneum Wnt7 is also observed in the developing central nervous system (CNS) (Bolognesi et al., 2008a). It is possible that the lateral domains of Wnt7 expression we observed in B. anynana correspond to the cells of the nervous system like in T. castaneum.
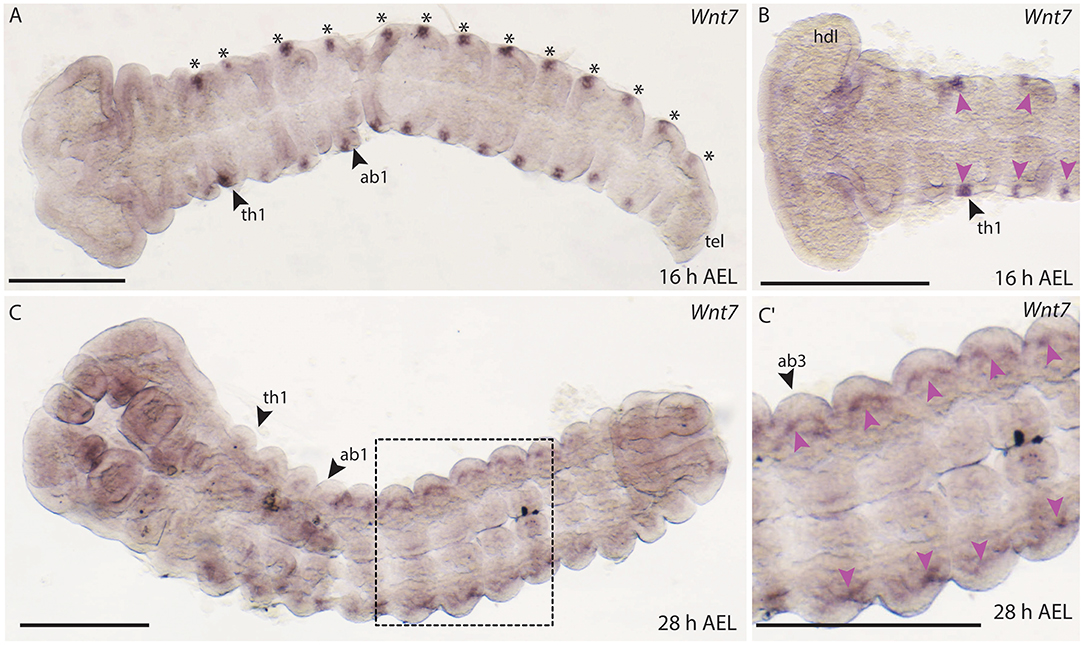
Figure 9. Expression of Wnt7 during development in B. anynana. (A) The first expression of Wnt7 occurs around 16 h AEL (asterisks). (B) Close up of the anterior head region indicating small lateral domains in each segment (pink arrowheads). (C) Around 28 h AEL faint expression can still be detected lateral in each segment. (C′) Close up of the ventral abdominal part of the embryo around 28 h AEL. Expression indicated by pink arrowheads. All pictures are orientated with anterior to the left if not otherwise stated. ab1, abdominal segment 1; AEL, after egg laying; atr, antennal rudiment; h, hours; hdl, head lobe; tel, telson; th1-3, thoracic segment 1-3. All pictures are orientated with anterior left. Scale bar: 400 μm.
Faint expression of Wnt7 was also observed at 28 h AEL in lateral regions of thoracic and abdominal segments (Figures 9C,C′). In later embryonic stages, expression of Wnt7 was observed in the abdominal limb buds of B. anynana (Supplement Figure S1).
No embryonic expression of Wnt9 was detected in B. anynana despite the application of different probes (Supplement Figure S2). In T. castaneum, expression was detected in the larval gut (Bolognesi et al., 2008a) while, in D. melanogaster embryos Wnt9 is expressed in a segment polarity pattern similar to Wnt1 as well as in in several other embryonic tissues such as the labrum, foregut and the ocular segment primordial, and subsequently in the CNS and in the gut of larval stages (Graba et al., 1995). In B. mori, Wnt9 transcripts were only detected in the testis and malphigian tubules at larval stages (Ding et al., 2019) which could potentially support our findings.
Expression of B. anynana Wnt10 was first detected at around 16 h AEL in the head lobes, the labrum and in stripes in the mandibular and maxillary segments (Figures 10A,A′). Stripes were also observed in the thoracic and abdominal segments, but this staining was very faint (data not shown). Later, at 26 h AEL, small domains of faint expression were detectable along the A-P axis in abdominal segments 2–6 (Figures 10B,B′). Expression was also observed in the mandibular, maxillary, and labial lobes at 28 h AEL (Figure 10C). Small domains of expression were also detected along the neurogenic furrow as well as two discrete domains in the telson (Figure 10C).
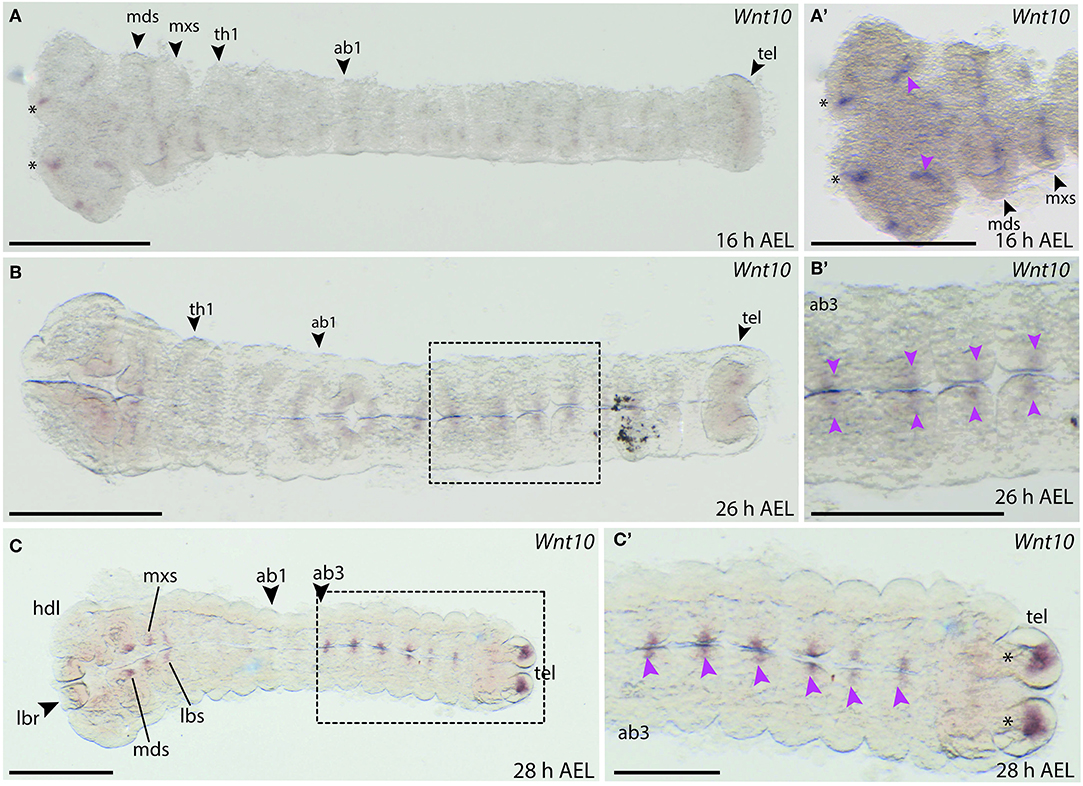
Figure 10. Wnt10 expression during B. anynana embryogenesis. (A) Expression around 16 h AEL is only detectable in the head structures of the embryo. (A′) Higher magnification of the head region of this embryo at 16 h AEL, indicating expression in the labrum (asterisks), the head lobes (pink arrowheads) and in the segments. (B) Expression at 26 h AEL becomes more restricted on either side of the neurogenic furrow, along the neurogenic furrow. (B′) Magnification of Wnt10 expression at 28 h AEL (pink arrowheads). (C,C′) Expression around 28 h AEL only in the abdominal segments, indicated by pink arrowheads. All images are orientated with anterior to the left if not otherwise stated. H, hour; AEL, after egg laying; th1, thoracic segment 1; ab1, abdominal segment 1; hdl, head lobe; mds, mandibular segment; mxs, maxillary segment; lbr, labrum; lbs, labial segment; tel, telson. Scale bar: 400 μm.
The expression in the neurogenic furrow and the head region could indicate that Wnt10 plays a role in the developing nervous system. Indeed, expression of Wnt10 in the developing CNS is seen in D. melanogaster and T. castaneum at similar developmental stages (Janson et al., 2001; Bolognesi et al., 2008a).
Expression of B. anynana Wnt11 was only observed during very late stages of embryogenesis (Supplement Figures S1D–F). Around 48 h AEL, expression could be seen in the proximal region of the mandibular segments as well as in the antennal regions (Supplement Figure S1E). Expression was also observed at the distal tips of the thoracic limb buds at this stage (Supplement Figure S1F), similar to that observed in T. castaneum (Bolognesi et al., 2008a).
The embryonic expression of WntA in B. anynana is similar to wg/Wnt1. WntA expression was observed as early as 10 h AEL and appears in segmental stripes in the gnathal and thoracic segments (Figure 11A). In addition, two distinct anterior domains were detectable in the forming head lobes, similar to Wnt1 expression (Figures 11A,A′). A faint expression domain was also observed within the head lobes (Figure 11A). Expression was absent from the primitive groove and did not transverse the entire segments at 10 h AEL (Figure 11A). Around 16 h AEL when all segments were formed, stripe-like WntA expression could be seen in all gnathal, thoracic and abdominal segments (Figures 11B,B′). Interestingly the length of the stripes in each segment was different between thoracic and abdominal segments (Figure 11B′).
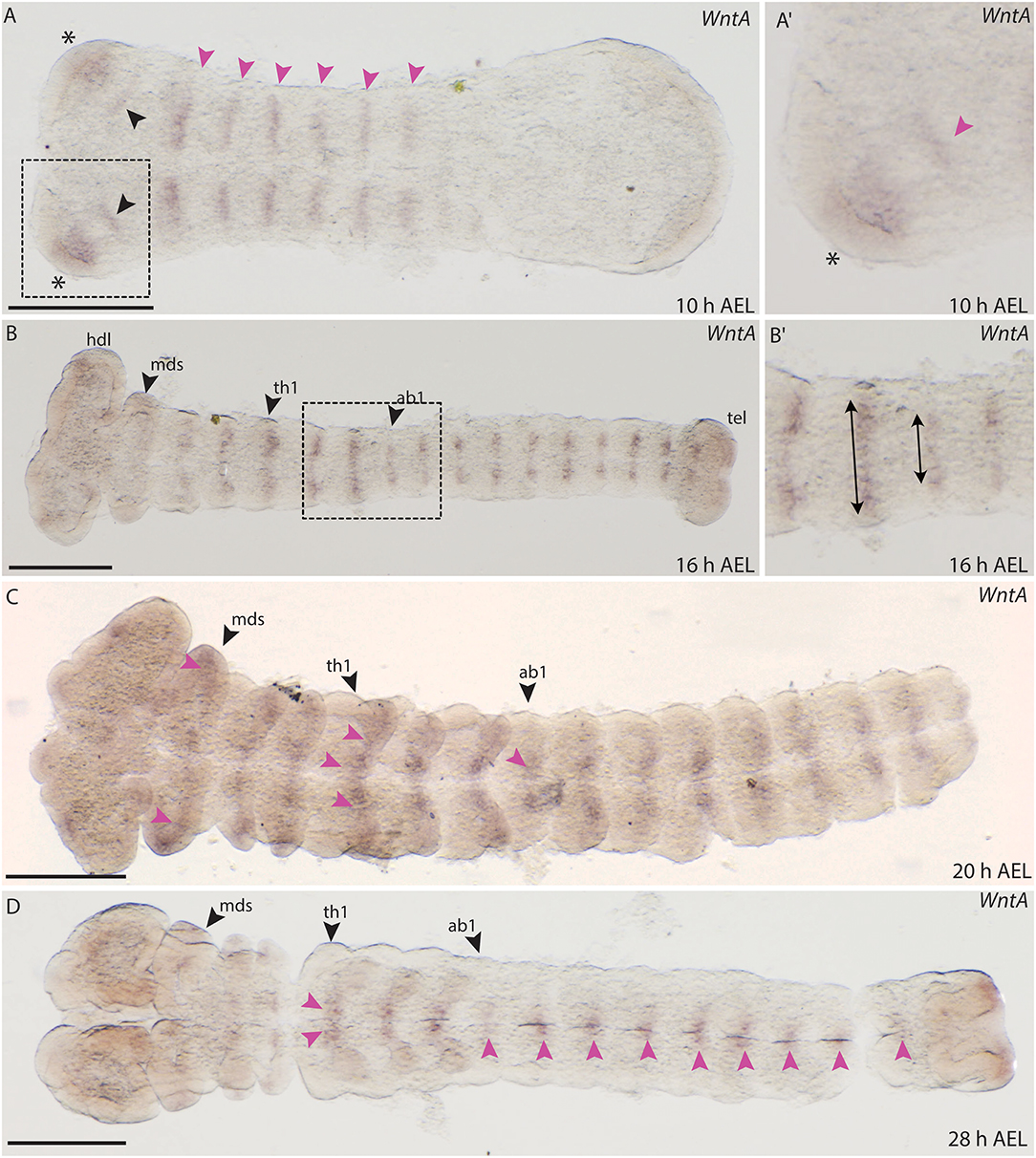
Figure 11. Expression of WntA during the development of B. anynana. (A) Expression in segmental stripes at 10 h AEL (pink arrowheads). (A′) Magnification of the head at 10 h AEL, expression can be detected in the head lobe (asterisks) and proximal to this domain (pink arrowhead). (B) Expression around 16 h AEL, segmental expression labeled by pink arrowheads. (B′) Magnification of segments at 16 h AEL indicating the different height of the stripes in the abdominal segments compared to the thoracic segments (arrows). (C) At 20 h AEL expression in all segments is still detectable (pink arrowheads). (D) At 28 h AEL, small expression domains along the A-P axis at the neurogenic furrow are seen (pink arrowheads). All pictures are orientated with anterior to the left if not otherwise stated. h, hours; AEL, after egg laying; th1, thoracic segment 1; ab1, abdominal segment 1; mds, mandibular segment. Scale bar: 400 μm.
Around 20 AEL, WntA expression retracts toward the neurogenic furrow and was detected in small domains in each segment (Figure 11C). At 28 h AEL, expression decreases but was concentrated in the thoracic limb buds as well as in very small domains along the A-P axis (Figure 11D). The expression observed for B. anynana WntA in segmental stripes that later concentrate along the midline is similar to the previously described expression of T. castaneum WntA (Bolognesi et al., 2008a).
The early embryonic development of the butterfly B. anynana is very similar to that described for the ditrysian moths M. sexta and B. mori (Krause and Krause, 1964; Kobayashi and Ando, 1990; Broadie et al., 1991; Miya, 2003), and therefore, early development is very likely conserved in Ditrysia. The staging system will be useful for further comparative studies and could be supplemented in the future by analyses of later developmental stages including larvae—especially with respect to the development of the gut or CNS. Here this staging system allowed us to begin to describe the embryonic expression of the repertoire of eight Wnt genes we have confirmed for butterflies and moths.
Overall, expression of five Wnt genes was observed during embryogenesis leaving three Wnt genes, which are probably expressed at later stages as shown by RT-qPCR analysis for B. mori (Ding et al., 2019). All the embryonically expressed butterfly Wnts (Wnt1, Wnt7, Wnt10, Wnt11, and WntA) have similar patterns to their homologs in T. castaneum during comparable developmental stages (Bolognesi et al., 2008a). Only expression of Wnt1 and potentially Wnt7 have similar embryonic expression to their homologs in D. melanogaster. We did not observe expression for Wnt5, Wnt6, and Wnt9 in the embryonic stages we observed in this study. Still, we were able to extract all of these genes form a cDNA pool of mixed embryonic stages—indicating that expression of these genes might be observed in different tissues and stages than the ones we have examined. Expression has been shown in several tissues of these genes in Drosophila and Tribolium which is why we would expect their involvement in embryogenesis. Therefore, we conclude that further analysis of these genes is warranted to characterize their possible divergent functional role in butterfly embryogenesis in more detail. These results make a further comparison of the Wnt gene function between Coleoptera and Lepidoptera very interesting, whereas the expression of some Wnts may be more evolutionary derived in Diptera.
Describing Wnt gene expression patterns during early embryogenesis and other aspects of development in Lepidoptera is the first step to understand the roles of these genes in these animals and their potential functional conservation and divergence compared to other arthropods. Given the important roles of these genes in cell-to-cell communication this also provides an excellent opportunity to generate much needed new insights into cell behavior during lepidopteran development. However, to achieve this our study needs to be followed up by functional analysis of these genes. Therefore, it would be interesting to knock out Wnt genes using CRISPR/Cas9 in B. anynana; a technique which was recently optimized for several butterfly and moth species (Livraghi et al., 2018; Matsuoka and Monteiro, 2018; Prakash and Monteiro, 2018).
The datasets generated for this study can be found in the a supplementary table file (all accession numbers).
MH and CB designed the project. MH and NB performed the experiment for the embryonic staging. MH performed all further experiments and analysis. OB and CB provided and took care of the animal culture. MH, CB, and AM analyzed and interpreted the data, and wrote the manuscript. All authors read and approved the submitted version of the manuscript.
MH and NB were kindly funded by the Nigel Groome Ph.D. scholarship of Oxford Brookes University, Oxford, UK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00468/full#supplementary-material
Supplement Data Sheet 1. Alignment file of all used predicted Wnt sequences from 16 lepidopteran species. The dataset also includes the insect outgroups D. melanogaster, T. castaneum, A. mellifera and A. gambiae. Sequence code can be found in Supplement Table S1.
Supplement Figure S1. Expression pattern of Wnt genes in older embryonic stages. (A) wg, (B) Wnt7, (C) WntA and (D-F) Wnt11 gene expression in late embryos (48–51 h AEL). H, hours; AEL, after egg lay. Scale bar: 400 μm.
Supplement Figure S2. Embryos stained with Wnt5, Wnt6 and Wnt9 probes. No expression was observed in the stages assayed. (A-C) Embryos stained for Wnt5. (D-F) Embryos stained for Wnt6. (G-I) Embryos stained for Wnt9. H, hours; AEL, after egg lay. Scale bar: 400 μm.
Supplement Table S1. Protein accession numbers for all used sequences in this study.
Supplement Table S2. Primer sequences for all Wnt probes used in this study. Capital letters show the gene specific sequence, small letters are T7 overhangs used for the cloning method of the probes.
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Ando, H., and Tanaka, M. (1980). Early embryonic development of the primitive moths, Endoclyta signifer walker and E. excrescens butler (Lepidoptera: Hepialidae). Int. J. Insect Morphol. Embryol. 9, 67–77. doi: 10.1016/0020-7322(80)90037-9
Banerjee, T. D., and Monteiro, A. (2018). CRISPR-Cas9 mediated genome editing in Bicyclus anynana butterflies. Methods Protoc. 1:16. doi: 10.3390/mps1020016
Bhanot, P., Brink, M., Samos, C. H., Hsieh, J. C., Wang, Y., Macke, J. P., et al. (1996). A new member of the frizzled family from Drosophila functions as a wingless receptor. Nature 382, 225–230. doi: 10.1038/382225a0
Bolognesi, R., Beermann, A., Farzana, L., Wittkopp, N., Lutz, R., Balavoine, G., et al. (2008a). Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev. Genes. Evol. 218, 193–202. doi: 10.1007/s00427-007-0170-3
Bolognesi, R., Farzana, L., Fischer, T. D., and Brown, S. J. (2008b). Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr. Biol. 18, 1624–1629. doi: 10.1016/j.cub.2008.09.057
Brakefield, P. M., Beldade, P., and Zwaan, B. J. (2009). In situ hybridization of embryos and larval and pupal wings from the African butterfly Bicyclus anynana. Cold Spring Harb Protoc. 2009:pdb.prot5208. doi: 10.1101/pdb.prot5208
Brattström, O., Aduse-Poku, K., Collins, S. C., Di Micco De Santo, T., and Brakefield, P. M. (2016). Revision of the Bicyclus sciathis species group (Lepidoptera: Nymphalidae) with descriptions of four new species and corrected distributional records. Syst. Entomol. 41, 207–228. doi: 10.1111/syen.12150
Broadie, K. S., Bate, M., and Tublitz, N. J. (1991). Quantitative staging of embryonic development of the tobacco hawkmoth, Manduca sexta. Roux's Arch. Dev. Biol. 199, 327–334. doi: 10.1007/BF01705925
Butler, A. G. (1879). On a Collection of Lepidoptera from the Island of Johanna. Ann. Mag. nat. Hist. 3, 186–192.
Cao, X., and Jiang, H. (2017). An analysis of 67 RNA-seq datasets from various tissues at different stages of a model insect, Manduca sexta. BMCGenomics 18:796. doi: 10.1186/s12864-017-4147-y
Carter, J.-M., Baker, S. C., Pink, R., Carter, D. R. F., Collins, A., Tomlin, J., et al. (2013). Unscrambling butterfly oogenesis. BMC Genomics 14, 283–283. doi: 10.1186/1471-2164-14-283
Challi, R. J., Kumar, S., Dasmahapatra, K. K., Jiggins, C. D., and Blaxter, M. (2016). Lepbase: the Lepidopteran genome database. bioRxiv [Preprint]. doi: 10.1101/056994
Clevers, H. (2006). Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480. doi: 10.1016/j.cell.2006.10.018
Dayhoff, M., Schwartz, R., and Orcutt, B. (1978). A Model of Evolutionary Change in Proteins. Washington, DC: Atlas Protein Sequence Structure, 345–352.
Dhawan, S., and Gopinathan, K. P. (2003). Expression profiling of homeobox genes in silk gland development in the mulberry silkworm Bombyx mori. Dev. Genes Evol. 213, 523–533. doi: 10.1007/s00427-003-0357-1
Ding, X., Liu, J., Zheng, L., Song, J., Li, N., Hu, H., et al. (2019). Genome-wide identification and expression profiling of Wnt family genes in the silkworm, Bombyx mori. Int. J. Mol. Sci. 20:1221. doi: 10.3390/ijms20051221
Dorn, A., Bishoff, S. T., and Gilbert, L. I. (1987). An incremental analysis of the Embryonic development of the tobacco hornworm, Manduca sexta. Int. J. Invertebrate Reprod. Dev. 11, 137–157. doi: 10.1080/01688170.1987.10510274
Dow, R. C., Carlson, S. D., and Goodman, W. G. (1988). A scanning electron microscope study of the developing embryo of Manduca sexta (L.) (Lepidoptera: Sphingidae). Int. J. Insect Morphol. Embryol. 17, 231–242. doi: 10.1016/0020-7322(88)90040-2
Ferguson, L., Marlétaz, F., Carter, J.-M., Taylor, W. R., Gibbs, M., Breuker, C. J., et al. (2014). Ancient expansion of the Hox cluster in Lepidoptera generated four homeobox genes implicated in extra-embryonic tissue formation. PLoS Genet 10:e1004698. doi: 10.1371/journal.pgen.1004698
Fradkin, L. G., Noordermeer, J. N., and Nusse, R. (1995). The drosophila Wnt protein DWnt-3 is a secreted glycoprotein localized on the axon tracts of the Embryonic CNS. Dev. Biol. 168, 202–213. doi: 10.1006/dbio.1995.1072
Fradkin, L. G., van Schie, M., Wouda, R. R., de Jong, A., Kamphorst, J. T., Radjkoemar-Bansraj, M., et al. (2004). The Drosophila Wnt5 protein mediates selective axon fasciculation in the embryonic central nervous system. Dev. Biol. 272, 362–375. doi: 10.1016/j.ydbio.2004.04.034
Gouy, M., Guindon, S., and Gascuel, O. (2010). Seaview version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Graba, Y., Gieseler, K., Aragnol, D., Laurenti, P., Mariol, M. C., Berenger, H., et al. (1995). DWnt-4, a novel Drosophila Wnt gene acts downstream of homeotic complex genes in the visceral mesoderm. Dev. 121, 209–218.
Holstein, T. W. (2012). The evolution of the Wnt pathway. Cold Spring Harb. Perspect. Biol. 4:a007922. doi: 10.1101/cshperspect.a007922
Janda, C. Y., Waghray, D., Levin, A. M., Thomas, C., and Garcia, K. C. (2012). Structural basis of Wnt recognition by Frizzled. Science 337, 59–64. doi: 10.1126/science.1222879
Janson, K., Cohen, E. D., and Wilder, E. L. (2001). Expression of DWnt6, DWnt10, and DFz4 during drosophila development. Mech. Dev. 103, 117–120. doi: 10.1016/S0925-4773(01)00323-9
Janssen, R., Le Gouar, M., Pechmann, M., Poulin, F., Bolognesi, R., Schwager, E. E., et al. (2010). Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol. Biol. 10:374. doi: 10.1186/1471-2148-10-374
Kawahara, A. Y., and Breinholt, J. W. (2014). Phylogenomics provides strong evidence for relationships of butterflies and moths. Proc. R. Soc. B Biol. Sci. 281:20140970. doi: 10.1098/rspb.2014.0970
Kikuchi, A., Yamamoto, H., and Kishida, S. (2007). Multiplicity of the interactions of Wnt proteins and their receptors. Cell Signal 19, 659–671. doi: 10.1016/j.cellsig.2006.11.001
Kobayashi, Y., and Ando, H. (1990). Early embryonic development and external features of developing embryos of the caddisfly, Nemotaulius admorsus (trichoptera: Limnephilidae). J. Morphol. 203, 69–85. doi: 10.1002/jmor.1052030108
Kozopas, K. M., Samos, C. H., and Nusse, R. (1998). DWnt-2, a Drosophila Wnt gene required for the development of the male reproductive tract, specifies a sexually dimorphic cell fate. Genes Dev. 12, 1155–1165. doi: 10.1101/gad.12.8.1155
Kraft, R., and Jäckle, H. (1994). Drosophila mode of metamerization in the embryogenesis of the lepidopteran insect manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 91, 6634–6638. doi: 10.1073/pnas.91.14.6634
Krause, G., and Krause, J. (1964). Schichtenbau und Segmentierung junger Keimanlagen vonBombyx mori L. (Lepidoptera) in vitro ohne dottersystem. Wilhelm Roux' Arch. Entwickl. Org. 155, 451–510. doi: 10.1007/BF00577653
Kusserow, A., Pang, K., Sturm, C., Hrouda, M., Lentfer, J., Schmidt, H. A., et al. (2005). Unexpected complexity of the WNT gene family in a sea anemone. Nature 433, 156–160. doi: 10.1038/nature03158
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Livraghi, L., Vodă, R., Evans, L. C., Gibbs, M., Dincă, V., Holland, P. W. H., et al. (2018). Historical and current patterns of gene flow in the butterfly pararge aegeria. J. Biogeogr. 45, 1628–1639. doi: 10.1111/jbi.13354
Logan, C. Y., and Nusse, R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810. doi: 10.1146/annurev.cellbio.20.010403.113126
Martin, A., Papa, R., Nadeau, N. J., Hill, R. I., Counterman, B. A., Halder, G., et al. (2012). Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. U.S.A. 109, 12632–12637. doi: 10.1073/pnas.1204800109
Martin, A., and Reed, R. D. (2014). Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev. Biol. 395, 367–378. doi: 10.1016/j.ydbio.2014.08.031
Masci, J., and Monteiro, A. (2005). Visualization of early embryos of the butterfly Bicyclus anynana. Zygote 13, 139–144. doi: 10.1017/S0967199405003163
Matsuoka, Y., and Monteiro, A. (2018). Melanin pathway genes regulate color and morphology of butterfly wing scales. Cell Rep. 24, 56–65. doi: 10.1016/j.celrep.2018.05.092
Mitter, C., Davis, D. R., and Cummings, M. P. (2017). Phylogeny and evolution of lepidoptera. Annu. Rev. Entomol. 62, 265–283. doi: 10.1146/annurev-ento-031616-035125
Miya, K. (2003). The Early Embryonic Development of Bombyx morii-An Ultrastructual Point of View. Sagamihara: Gendaitosho.
Monteiro, A. (2017). “Physiology and evolution of wing pattern plasticity in bicyclus butterflies: a critical review of the literature,” in Diversity and Evolution of Butterfly Wing Patterns: An Integrative Approach, eds T. Sekimura and H. F. Nijhout (Singapore: Springer, 91–105.
Muller, T., and Vingron, M. (2000). Modeling amino acid replacement. J. Comput. Biol. 7, 761–776. doi: 10.1089/10665270050514918
Murat, S., Hopfen, C., and McGregor, A. P. (2010). The function and evolution of Wnt genes in arthropods. Arthropod Struct. Dev. 39, 446–452. doi: 10.1016/j.asd.2010.05.007
Nakao, H. (2010). Characterization of Bombyx embryo segmentation process: expression profiles of engrailed, even-skipped, caudal, and Wnt1/wingless homologues. J. Exp. Zool. B Mol. Dev. Evol. 314B, 224–231. doi: 10.1002/jez.b.21328
Nowell, R. W., Elsworth, B., Oostra, V., Zwaan, B. J., Wheat, C. W., Saastamoinen, M., et al. (2017). A high-coverage draft genome of the mycalesine butterfly Bicyclus anynana. Gigascience 6, 1–7. doi: 10.1093/gigascience/gix035
Nusslein-Volhard, C., Wieschaus, E., and Kluding, H. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: I. Zygotic loci on the second chromosome. Wilehm Roux. Arch. Dev. Biol. 193, 267–282. doi: 10.1007/BF00848156
Otto, H. (2015). Butterflies of the Kruger National Park and Surrounds. Cape Town: Penguin Random House South Africa.
Özsu, N., and Monteiro, A. (2017). Wound healing, calcium signaling, and other novel pathways are associated with the formation of butterfly eyespots. BMC Genomics 18:788. doi: 10.1186/s12864-017-4175-7
Prakash, A., and Monteiro, A. (2018). apterous A specifies dorsal wing patterns and sexual traits in butterflies. Proc. Biol. Sci. 285:20172685. doi: 10.1098/rspb.2017.2685
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Stefanik, D. J., Lubinski, T. J., Granger, B. R., Byrd, A. L., Reitzel, A. M., DeFilippo, L., et al. (2014). Production of a reference transcriptome and transcriptomic database (EdwardsiellaBase) for the lined sea anemone, Edwardsiella lineata, a parasitic cnidarian. BMC Genomics 15:71. doi: 10.1186/1471-2164-15-71
Swarup, S., and Verheyen, E. M. (2012). Wnt/Wingless signaling in Drosophila. Cold Spring Harb Perspect Biol. 4:a007930. doi: 10.1101/cshperspect.a007930
Wiese, K. E., Nusse, R., and van Amerongen, R. (2018). Wnt signalling: conquering complexity. Development 145:dev165902. doi: 10.1242/dev.165902
Yang, Z. (1993). Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10, 1396–1401.
Keywords: embryogenesis, Wnt ligands, lepidoptera, Bicyclus anynana, evolution and development, butterfly development
Citation: Holzem M, Braak N, Brattström O, McGregor AP and Breuker CJ (2019) Wnt Gene Expression During Early Embryogenesis in the Nymphalid Butterfly Bicyclus anynana. Front. Ecol. Evol. 7:468. doi: 10.3389/fevo.2019.00468
Received: 31 July 2019; Accepted: 20 November 2019;
Published: 06 December 2019.
Edited by:
Aziz Aboobaker, University of Oxford, United KingdomReviewed by:
Kristen A. Panfilio, University of Warwick, United KingdomCopyright © 2019 Holzem, Braak, Brattström, McGregor and Breuker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alistair P. McGregor, YW1jZ3JlZ29yQGJyb29rZXMuYWMudWs=; Casper J. Breuker, Y2JyZXVrZXJAYnJvb2tlcy5hYy51aw==
†Present address: Michaela Holzem, Division of Signaling and Functional Genomics, German Cancer Research Centre (DKFZ), Heidelberg, Germany; Department of Cell and Molecular Biology, Medical Faculty Mannheim, Heidelberg University, Heidelberg, Germany
Oskar Brattström, The School of Life Sciences, University of Glasgow, Glasgow, Scotland
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.