- 1Disease Vector Group, Chemical Ecology, Department of Plant Protection Biology, Swedish University of Agricultural Sciences, Alnarp, Sweden
- 2School of Life Sciences, Arizona State University, Tempe, AZ, United States
- 3Division of Entomology, Department of Plant Protection, Gorgan University of Agricultural Sciences and Natural Resources, Gorgan, Iran
Background: Aedes aegypti is a prominent and highly competent vector of several arboviral diseases, including dengue, yellow fever, and Zika. Behaviors associated with reproductive feeding, both pre- and post-blood meal, directly influence disease transmission capacity. Odors mediate host seeking pre-blood meal, while post-blood meal females are refractory to host odors for at least 24 h. During this time, flight activity is substantially reduced. Two key host odors, carbon dioxide and (R)-1-octen-3-ol, are detected by the maxillary palps in mosquitoes. In the search for future vector control tools, the identification of genes that are regulated in the maxillary palps between host seeking and 24 h post-blood meal may provide an informative pool of targets.
Results: The blood meal-induced regulation of chemosensory, neuromodulatory, and other signal transduction genes was investigated in the maxillary palps of 24 h post-blood fed Ae. aegypti females, 6 days after emergence, and compared to host-seeking females of the same age using a transcriptomic approach. Genes-of-interest implicated in the behavioral switch from host seeking to post-blood meal quiescence were identified from multiple gene families investigated: odorant receptors, ionotropic receptors, pickpocket receptors, transient receptor potential receptors, odorant binding proteins, chemosensory proteins, neuromodulators, and their receptors, as well as constituents of second messenger signaling pathways. Reflecting the change in transcript abundance of families involved in CO2 signaling, the neural sensitivity to this key kairomone compound was found to decrease in blood fed mosquitoes compare with their on-blood fed counter parts.
Conclusions: Sensory-associated gene expression is regulated in the maxillary palps of Ae. aegypti females in response to blood feeding. The concerted regulation of multiple genes within the sensory pathways of the maxillary palps likely play a key role in modulating the behavioral changes observed post-blood meal. Future functional characterization of the proteins generated by the genes-of-interest identified in this study may provide both a better understanding of the regulation of gonotrophic feeding and a pool of potential targets for vector control strategies.
Introduction
The host-seeking behavior of mosquitoes depends heavily on the olfactory system, and is intimately linked to disease transmission (Mescher and De Moraes, 2017). Mosquito activation and medium-to-long range attraction to, and recognition of, hosts is partly mediated by two odors, carbon dioxide (CO2) (Dekker and Cardé, 2011; Majeed et al., 2017) and (R)-1-octen-3-ol (Majeed et al., 2016). These volatiles are detected by a series of chemosensory proteins expressed in an olfactory organ of relatively low complexity, the maxillary palps (Lu et al., 2007; Bohbot and Dickens, 2009; Grant and Dickens, 2011; Erdelyan et al., 2012; Manoharan et al., 2013; McMeniman et al., 2014; Hill et al., 2015). Short range attraction and the ultimate acceptance of a host also rely on other sensory modalities, including thermal and mechanosensory detection, and gustation (Montell and Zwiebel, 2016). In this study, we identify potential candidate genes important for generating the behavioral switch between host seeking and resting through their differential abundance in the maxillary palps of host-seeking and 24 h post-blood fed female Ae. aegypti mosquitoes.
Capitate peg sensilla, the only chemosensory sensillum type found on the maxillary palps, house three olfactory sensory neurons (OSNs) (Grant et al., 1995), and cover the fourth of the five segments comprising the maxillary palp in female Ae. aegypti (McIver and Charlton, 1970). Fourteen chemosensory gene families are expressed by the OSNs and accessory cells of these sensilla (Bohbot et al., 2014; Matthews et al., 2016). Of the chemoreceptors, the odorant receptors (Ors), the ionotropic receptors (Irs), and the gustatory receptors (Grs) are well-characterized (Hallem and Carlson, 2004; Hallem et al., 2004; Kent et al., 2008; Benton et al., 2009; Carey et al., 2010; Croset et al., 2010; Wang et al., 2010; Pitts et al., 2017). The Ors constitute a family of seven transmembrane domain proteins that function as heteromeric ionotropic and/or metabotropic receptors (for review see Stengl and Funk, 2013) composed of the highly conserved Or co-receptor (Orco) and unique Or-subunits, tuned to a defined set of odorants (Hallem and Carlson, 2004; Hallem et al., 2004; Ditzen et al., 2008; Carey et al., 2010; Wang et al., 2010; Bohbot et al., 2011a,b; Xu et al., 2013; McBride et al., 2014). Within this rapidly evolving gene family (Benton, 2015), only three are expressed in the maxillary palps of Ae. aegypti; Orco, the (R)-1-octen-3-ol-sensitive Or8 (Cook et al., 2011; Grant and Dickens, 2011) and the as yet orphan Or49 (Bohbot et al., 2014). While originally described as “antennal Irs” (Croset et al., 2010), a subset of Irs, including two (Ir25a, Ir76b) of the three Ir co-receptors are expressed in the maxillary palps (Matthews et al., 2016; Pitts et al., 2017). Although the primary role described for Grs is the detection of tastants, two of the three Grs expressed in the maxillary palp are involved in transducing CO2 reception, Gr1 and Gr3, but not Gr2 (Erdelyan et al., 2012). Carbon dioxide elicits activation of host-seeking behavior and long-range attraction in mosquitoes (Grant et al., 1995; Dekker and Cardé, 2011; Majeed et al., 2014, 2017).
Besides Ors, Irs, and Grs, two additional receptor protein families, the pickpocket and the transient receptor potential receptors (Pkps and Trps, respectively), have recently received attention for their involvement in chemosensation. The Ppks robustly expressed in the maxillary palps of Ae. aegypti (AAEL008053, AAEL000926) (Bohbot et al., 2014) are from subfamily V, and are likely to be involved in mechanosensation (Walker et al., 2000; Tracey et al., 2003; Johnson and Carder, 2012; Kim et al., 2012; Zelle et al., 2013; Guo et al., 2014). The Trps are involved in sensing auditory, proprioreceptive, geotactic, thermal and chemical cues (Fowler and Montell, 2013), and references therein). The TrpA, Trpc, and TrpL subfamilies are involved in chemosensation, and appear to be sensitive to ligands that can be modulated by Or and/or Gr activated G-protein Gq activating phospholipase C (PLC) signaling cascades (Kang et al., 2010; Kim et al., 2010; Kwon et al., 2010; Badsha et al., 2012; Fowler and Montell, 2013). In addition, painless is required for the avoidance of isothiocyanate (Al-Anzi et al., 2006). Of these, Trp, painless, and TrpA1 are expressed in the maxillary palps of Ae. aegypti (Bohbot et al., 2014).
While not recognized as receptors, sensory neuron membrane proteins (SNMPs), are expressed in the OSN membranes (Vogt, 2003; Nichols and Vogt, 2008). Sensory neuron membrane proteins are involved in modulating the interaction between odorants, particularly pheromones, and Ors (Benton et al., 2007; Jin et al., 2008). Orthologs of both Snmp1 and Snmp2 have been identified in mosquitoes (Nichols and Vogt, 2008), and shown to be expressed in the maxillary palps of host-seeking female Ae. aegypti (Bohbot et al., 2014; Matthews et al., 2016).
Signaling cascades amplify and modulate chemical signals, producing the final electrical output of the OSN. All current models indicate that the Or/Orco complex comprises a ligand binding receptor, a cation permeable ion channel, and connection to metabotropic pathways (Sato et al., 2008; Nakagawa and Vosshall, 2009; Sargsyan et al., 2011; Getahun et al., 2013). Of the two main metabotropic cascades, the first involves the G-protein Gq activating phospholipase c-ß (Plc-ß) to increase intracellular calcium. This follows the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to inositol trisphosphate (IP3) and diacylglycerol (DAG), leading to IP3-dependant calcium channel activation, increasing intracellular Ca2+, and activating protein kinase C (Pkc) phosphorylating cation channels, including Orco. The second cascade involves the G-protein Gs, which activates adenylate cyclase and triggers the cAMP signaling cascade, activating ion channels, including Orco, through Plc and Pkc pathways (Getahun et al., 2013). The activation of protein kinase A (Pka) by these pathways can lead to long-term effects through the activation of transcription factors that regulate gene expression.
Members of three chemosensory protein families have been shown to be secreted by the accessory cells into the sensillum lymph surrounding the OSNs: the odorant binding proteins (Obps), the chemosensory proteins (Csps) and the odorant degrading enzymes (Odes) (Pelosi et al., 2005; Leal, 2013). The Obps and Csps are small soluble proteins that play a role in the selective recognition, gain control and transport of odorants from the cuticular pores to the membrane receptors on the OSNs (Vogt et al., 1999; Vogt, 2003; Pelosi et al., 2005; Larter et al., 2016). There are three subfamilies of Obps, the classical, the Plus-C and the two-domain Obps (Zhou et al., 2008; Manoharan et al., 2013). The most robustly expressed Obps in the Ae. aegypti maxillary palps are from the classical and Plus-C subfamilies (Bohbot et al., 2014). Half of the identified Csps are expressed in the maxillary palps of host-seeking Ae. aegypti females (Matthews et al., 2016). Following the dissociation of the odorants from the receptors, the Odes act to rapidly clear the lymph of odorants (Younus et al., 2014). The Odes are not well-defined and are generally described by those members of the cytochrome P450 and carboxylic esterases expressed in olfactory tissues.
The sensory tuning of the peripheral olfactory system in mosquitoes is likely generated by the sensitivity and specificity of these proteins, acting in concert (Carey et al., 2010; Wang et al., 2010; Rinker et al., 2013; Bohbot et al., 2014; McBride et al., 2014; Omondi et al., 2015). In this study, the chemosensory genes that are modulated in the maxillary palps of host seeking (nbf) and 24 h post-blood meal (bf) female Ae. aegypti of the same age, 6 days-post-emergence (dpe), are identified as genes-of-interest for further functional investigation. The function and role in signal transduction are discussed for genes subject to modulation, and put in the context of the neuronal activity of the maxillary palp capitate peg neurons. In particular, the differential expression of the chemosensory-related genes is discussed in the context of the ligand sensitivity of their concordant OSNs, in both host-seeking and 24 h pbm states.
Materials and Methods
Animal Rearing and Tissue Collection
Aedes aegypti (Rockefeller strain) were reared at 27 ± 1°C, 65 ± 5% relative humidity under a 12 h:12 h light:dark period, as previously described (Majeed et al., 2017). To control for potential age- and circadian-related differences in all experiments, 6-day post-emergence (dpe) adult female mosquitoes with ad libitum access to sucrose (10%) were used between ZT 5 and 8. The mosquitoes were anesthetized on ice for at least 1.5 min prior to dissection. Maxillary palps were dissected from 6-dpe females under two conditions: those blood fed 24 h prior to dissection (bf) and those which were not (nbf). All mosquitoes had ad libitum access to 10% sucrose throughout. Tissues (500 pairs per condition) were stored at −20°C in RNAlater (Life Technologies, Sweden). Blood meals were provided to the mosquitoes using a Hemotek membrane feeder. The blood used was from donor sheep and was commercially purchased from a fully certified facility (Håtunalab, Bro, SE). No additional ethical approval from the university was required.
RNA Extraction and Illumina Sequencing
Tissues were sonicated (Vibra-Cell sonicator, Sonics and Materials, VCX-130; 10 cycles; 70% amplitude; 1 s pulses), incubated (30 s) on ice, and then the whole cycle was repeated twice. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Sweden), including the RNase-free DNase I on-column treatment, according to the manufacturer's protocol, and then stored at −80°C. Total RNA was quantified using fluorometric analysis (Qubit, Invitrogen, Sweden), and the quality assessed by NanoDrop and standard gel electrophoresis, prior to sending to Eurofins (Germany) for 3′ fragment cDNA library generation and sequencing on an Illumina HiSeq 2500 (single read, 1 × 100 nt). Bar coded fragment libraries were generated following the standard Illumina protocol. Four libraries were constructed, two replicates per condition, which were run in separate channels. Each channel contained a single replicate of the maxillary palp from female host-seeking (nbf), and 24 h post-blood meal (24 h pbm) bar coded libraries (channel 003: nbf 15505650 and bf 11263612 reads; channel 004: nbf 15927640 and bf 11518245 reads).
Read Mapping
Prior to mapping, the raw reads were analyzed and quality filtered. The adapter sequences and low-quality bases (Phred score < 20) were removed from both ends of each single read. Reads that did not fulfill an average quality threshold were clipped (sliding window, window size 20 nt, minimum quality 4) using Trimmomatic 0.20 (http://www.usadellab.org, u.d.), and reads shorter than 40 nt were removed. Reads were mapped using CLC Genomics Workbench 10 (http://www.clcbio.com, Qiagen), and the new Ae. aegypti Liverpool AGWG annotation, genome assembly AaegL5 and gene set AaegL5.1 (https://www.vectorbase.org/organisms/aedes-aegypti/lvp_agwg/aaegl5). While this genome database represents the current overall annotation, there has been a considerable upheaval in the chemosensory gene annotations, which has removed several long-standing, and empirically supported genes. Of note for the current study is the lack of annotation of the conserved culicine odorant receptor, Or49 (AAEL001303-RA in AaegL3.5), previously demonstrated to be expressed in the adult maxillary palps (Bohbot et al., 2007, 2014) and labrum (Jung et al., 2015), as well as in the larval antennae (Mysore et al., 2014). Indeed, RNAi knockdown of Or49 results in delayed blood feeding behavior (Jung et al., 2015). To account for this lack of annotation, reads mapping to the AaegL5 genome region, where the three-prime sequence of Or49 is located (2:448700040-448700246;448699296-448699784), were counted manually. Equally, the 3′ untranslated regions (UTRs) of multiple genes are truncated. Of note for this study, the 3′ UTR of Ir76b (AAEL006360), Ir93a (AAEL021659), Ir41a (AAEL000041), Csp05687 (AAEL005687), Scrb9 (AAEL000256), Scrb3 (AAEL005979), and Obp60 (AAEL008012) are truncated in the L5 annotation, and as we did quantitative sequencing of 3′ transcript ends, an incorrect 3′ annotation can result in false negatives (i.e., Irs), and underrepresented read abundances (i.e., Csp, Scrbs, and Obps). The 3′ UTRs are extended by 4850 bp (3: 396663291-396668151), 1356 bp (3:325201503-325202859), 1101 bp (3:103203960-103205061), 1159 bp (3:115197984-115199143), 3210 bp (2:227337205-227340415), 233 bp (2:8159347-8159580), and 1367 bp (1:164107819-164109186) from the L5 annotation, respectively. Additionally, the truncation of the 3′ UTR annotation of AAEL002924-RC, may have resulted in false positives attributed to Ir8a, as the 3′ UTRs of Ir8a and AAEL002924-RC overlap (1:85989081-85989566), albeit on complementary strands. In this study, all several hundred reads in this region are attributed to AAEL002924-RC, according to the strand onto which the reads mapped.
Abundance and Differential Analyses
Single-end RNA-Seq libraries produced reads from the 3′ end of each transcript present. To quantify transcript abundance in each library, the mean read counts were normalized using an upper-quantile-based normalization (Bolstad et al., 2003) prior to between-library comparisons (see below). Transcripts with a mean abundance of more than 20 normalized read counts were considered detectible and thus included in further analysis. This threshold is similar to the <1 RPKM filter that is commonly employed to reduce noise (Rinker et al., 2013). The upper-quantile-based normalization was performed on each library to increase the detection sensitivity of differential expression for low abundance transcripts (Bolstad et al., 2003). Kal's Z-type test, which assumes a ß-binomial distribution on the normalized read counts, generated fold changes (FC) and probability estimates that were used to detect differential expression (Kal et al., 1999). Comparisons were made between nbf vs. bf maxillary palp libraries. Strict criteria were used to determine significant differential abundance. Genes that exhibited a FC>2 and a P-value corrected for false discovery rate (FDR) <0.05 were considered to be significantly differentially abundant. Genes with 1.5 > FC < 2 and an FDR P-value <0.05, were considered potential genes-of-interest. The means of normalized read counts were transformed with the addition of a constant (1.1) to prevent values of infinitive fold change when one of the two libraries being compared exhibited no detectible reads.
Quantitative Real Time PCR
The MIQE guidelines were adhered to in this study (Bustin et al., 2009). Selected genes that demonstrated differential abundance according to the transcriptome analyses were assessed and verified using quantitative real time polymerase chain reaction (qPCR). Primer pairs for qPCR were designed using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (20-22 nt, Tm ~60°C, GC 50% ± 10%, GC-clamp 2 nt, amplicon 80–150 nt spanning exon-exon regions) and supplied by Eurofins Genomics (Munich, Germany) (Supplementary File 1). Six new cDNA libraries (nbf: 3; bf: 3) were constructed by Oligo(dT) priming from the total RNA extracted from 6 × 300 pairs of maxillary palps (extraction protocol described above in section RNA Extraction and Illumina Sequencing), using SuperScript III First-Strand Synthesis System for RT-PCR (Thermo Fischer Scientific, CA, USA), according to the manufacturer's protocol, and stored at−20°C. This resulted in three biological replicates for each of the nbf and bf conditions. Quantitative PCR was performed with minor modifications, according to manufacturers' protocol (Platinum SYBR Green qPCR SuperMix-UDG w/ROX, Bio-Rad Laboratories, CA, USA), in a total volume of 25 μl (12.5 μl Supermix, 0.5 μl each primer, 1 μl of cDNA, 11 μl DNAse-RNAse free water). Amplification was performed on a BioRad CFX 96 (Bio-Rad Laboratories, CA, United States), using the following parameters: 2 min at 50°C, then 2 min at 95°C, followed by 40 cycles (30 s at 95, 58, and 72°C, respectively). During elongation at 72°C, fluorescence was measured for each cycle. After the final cycle, specificity of the product amplification was assessed using melting curve analyses (65–94°C in 0.5°C increments). Controls (water alone and samples containing all reactants except cDNA) were included for each plate. For each of the three biological replicates for both nbf and bf, every primer pair was assessed three times (technical replicates), resulting in a total of nine replicates for each cohort. Levels of gene expression were determined using the ΔΔCq method (Livak and Schmittgen, 2001), and normalized to a reference factor. The geometric mean of three verified stable reference genes, Elfa1 (AAEL017096), RpL8 (AAEL000987), and RpS7 (AAEL009496) (Christ et al., 2017) comprised the reference factor for each replicate. The level of differential gene abundance was presented relative to that of the nbf female maxillary palps, verified for normality and homogeneity of residuals using D'Agostino-Pearson test, and then compared using a two-tailed paired Student's t-test with significance that was adjusted for multiple comparisons.
Electrophysiology
Each capitate peg sensillum contains three OSNs (McIver and Charlton, 1970). The highest and intermediate spiking amplitude neurons, by convention referred to as the A and B cells, respond to CO2 (Grant et al., 1995) and (R)-1-octen-3-ol (Lu et al., 2007; Cook et al., 2011), respectively. Electrophysiological recordings from these neurons were made using single sensillum recordings, as described previously (Majeed et al., 2014, 2017). The OSNs were stimulated by introducing various concentrations of either CO2, from gas cylinders containing metered amounts of CO2 and oxygen (20%) balanced by nitrogen (Strandmöllen, AB, Ljungby, Sweden) (Majeed et al., 2017), or racemic 1-octen-3-ol (Sigma Aldrich, Sweden) in a background of 150 ppm CO2. This background of 150 ppm was used to clarify the visualization of the B cell response by reducing the response in the A cell. The stimuli were introduced into the humid airstream passing over the preparation 11 cm upstream of the maxillary palps. The resulting dose response curves were generated by non-linear regression, with a variable slope for nbf and bf mosquitoes using maximum likelihood, and compared using the extra sum-of-squares F-test (GraphPad Prism v. 5.0a, GraphPad Software, San Diego, US).
Results and Discussion
Transcript Abundance in Varies With Blood Meal Status
Transcriptome Quality
The CEGMA gene set is a set of core eukaryotic genes (CEGs) identified as expressed in all eukaryotic animals and likely involved in essential cell processes (Parra et al., 2007). These genes were assessed to determine whether the depth of sequencing was sufficient to reliably identify expressed transcripts and to assess the stability of transcript abundance across libraries (Taparia et al., 2017). Ninety-nine percent of the CEGs were found at detectable levels in the maxillary palps, indicating that the sequencing depth was sufficient to reliably identify gene expression (Figure 1; Supplementary File 2). The fold change in abundance for 99% and >92% of the CEGs transcripts, between physiological states and between replicates, was <2.0 and <1.5, respectively, as would be expected for housekeeping genes (Figure 1) (Dzaki et al., 2017; Shakeel et al., 2017).
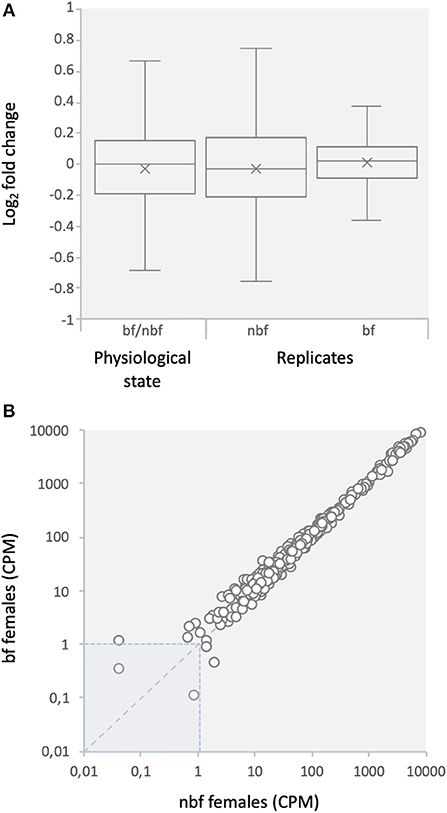
Figure 1. Expression of core eukaryotic genes (CEGs). (A) Comparison of fold changes between blood fed (bf) and non-blood fed (nbf) female mosquitoes, and between replicates of the maxillary palp transcript libraries of Aedes aegypti. (B) Comparison of counts per million (CPM) between bf and nbf females.
Transcript Abundance
A total of 17,330 genes are annotated in the Ae. aegypti genome database (AaegL5.1), of which 9,252 were reliably detected in the maxillary palps of the 6-dpe females. Previous studies report between 10,796 (Bohbot et al., 2014) and 11,052 (Matthews et al., 2016) transcripts, from earlier annotations, to be expressed in the maxillary palps of host-seeking females. The observed discrepancy in expression found between our study and those previous is likely due to methodological differences, including the wider age range assayed, earlier reference gene sets, and the paired-end libraries generated in previous studies. A mapping to the AaegRU database used in Matthews et al. (2016) resulted in the reliable detection of 363 more genes. While paired-end libraries are likely to identify more transcripts, since the sequencing covers the entire length of the transcript, the accuracy of the abundance estimates is reduced compared to the 3′ end quantitative sequencing used in this study (Moll et al., 2014).
The number of genes with detectable transcript abundance in the maxillary palps of bf females (8,899) was similar to that found in nbf females (8,834). An analysis of the transcripts with annotated gene ontologies (GOs) (nbf: 8,879, bf: 8,803) revealed that the most represented molecular functions in these olfactory organs, both by number and by read abundance were genes encoding for proteins with ligand binding capabilities (small molecule-, protein-, DNA-, and cofactor binding), enzyme (hydrolases, transferases, and oxidoreductases) and enzyme inhibitor activities, as well as transmembrane transporter activity (Figures 2A,B). The only GO class demonstrating differential overall read abundance was odorant binding (GO:0005549; Figure 2B). As maxillary palps are involved in olfaction, it is not surprising that this organ has a relatively high number of odorant binding genes expressed (2–3%). The proportion of odorant binding genes demonstrated significantly higher overall normalized read abundance [t-ratio(2) = 151.6; q = 6.590 × 10−4; Figure 2B], and significantly more genes with >2-fold higher abundance, in the maxillary palps of bf than in nbf females [χ2 = 3.89; P < 0.0486; Figure 2C]. The only other GO class showing differential numbers of genes with >2-fold difference in normalized read abundance was the structural constituent of cuticle (GO:0042302), which was more abundant in the maxillary palps of nbf than bf females [χ2 = 9.50; P < 0.0021; Figure 2C]. This is likely a reflection of an overall down-regulation of cuticular proteins in post-blood fed mosquitoes (Dana et al., 2005).
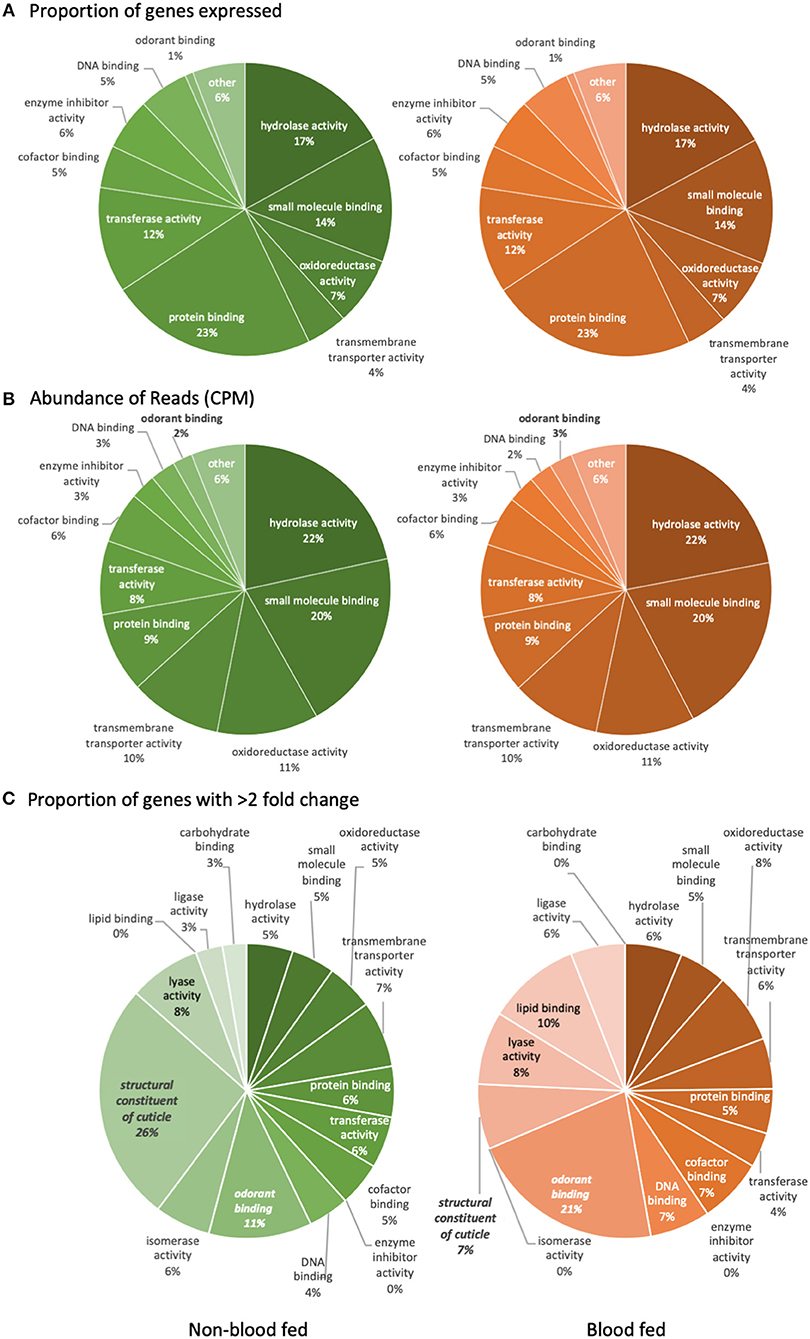
Figure 2. Level three gene ontology analysis of molecular functions. The analysis represents (A) the proportion of reliably expressed genes, (B) the abundance of reads (CPM), and (C) the proportion of genes with >2-fold change in the maxillary palps transcriptomes of non-blood fed (left) and blood fed (right) female Aedes aegypti. Significant difference (q < 0.05; multiple t-tests) is indicated in bold/italics. Gene ontology classes that were represented by <1% of the total were not named.
The 20 genes with the highest transcript abundance in the maxillary palps include the large ribosomal RNA subunit (lsu rRNA; AAEL018689), the most highly represented gene in both libraries, and 11 other ribosomal proteins present in both physiological states. As none of these ribosomal transcripts differed in abundance between nbf and bf mosquitoes, this suggests that protein translation is a major and ongoing process in both physiological states, indicating that the observed differences expression of other genes may be a result of targeted regulation and not an overall regulation of gene expression. The other top abundance transcripts included an Obp (Obp63), a haemolymph juvenile hormone binding protein (AAEL001306) and an ADP/ATP carrier protein (AAEL004855), similar to that previously described (Bohbot et al., 2014; Matthews et al., 2016).
Chemosensation
Host preference, driven by the necessity to detect and locate hosts to obtain blood meals, has exerted a strong selection pressure on the olfactory system of vector mosquitoes (Takken and Knols, 1999). The olfactory system of female mosquitoes is highly tuned to salient host volatiles (Takken and Knols, 1999; McBride et al., 2014), together with generic cues, such as CO2 and (R)-1-octen-3-ol, which are detected by two of the three maxillary palp OSNs, providing sufficient information to enable identification of, and discrimination among, potential host species (Majeed et al., 2016, 2017). The third OSN is not yet functionally characterized.
Modulation of Carbon Dioxide Signaling
The Grs involved in transducing CO2 reception in female Ae. aegypti mosquitoes are Gr1 (AAEL002380) and Gr3 (AAEL010058) (Erdelyan et al., 2012; McMeniman et al., 2014). Both of these CO2-sensitive receptors are up-regulated during adult maturation, indicating a role for the peripheral regulation of host seeking in both Ae. aegypti (Bohbot et al., 2013) and An. gambiae (Omondi et al., 2015). Interestingly, the transcript abundance of both of these receptor genes was significantly reduced in the maxillary palps of bf compared with that of nbf females of the same age (1.2 and 1.7-fold; FDR P < 0.0001, respectively; Figure 3A). This change in transcript abundance was confirmed by qPCR (Supplementary Data Sheet 1abc); in fact, there was an overall congruence between the qPCR and RNA-seq datasets amongst all genes tested. To assess the biological relevance of this <2-fold reduction in Gr expression, the response of the CO2-sensitive OSN, the A-cell, was analyzed in both nbf and bf females using single sensillum recordings. This showed that the sensitivity of the A-cell was reduced in bf compared to nbf females [F(4, 77) = 5.958, FDR P = 0.0003; Figure 3B], reflecting the reduction in Gr1 (150.80 to 125.66 CPM) and Gr3 (106.79 to 60.64 CPM) abundance (Supplementary File 2). Alternatively, the observed changes in the neuronal sensitivity may have been generated by post-transcriptional regulation, such as regulation of translation and other cellular regulation processes such as phosphorylation.
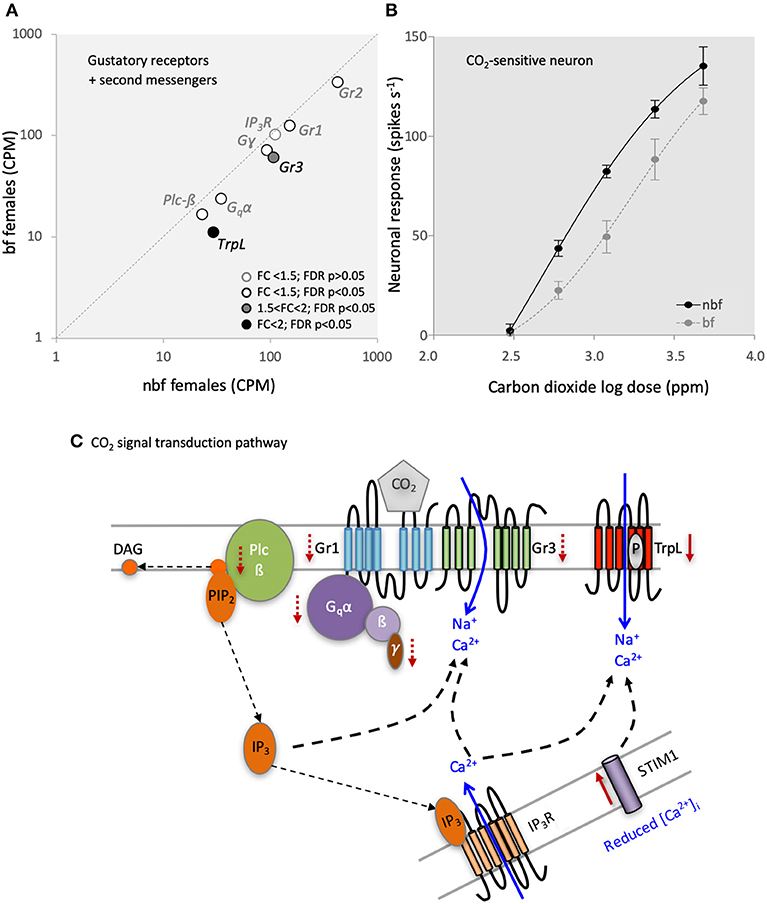
Figure 3. Carbon dioxide (CO2) signaling in the maxillary palps. (A) Differential transcript regulation of CO2-sensitive gustatory receptors (Gr1 and Gr3), Gr2 and the second messenger genes, phospholipase c beta (Plc-ß), Gq-protein alpha subunit (Gqα), G-protein gamma subunit (Gγ), and inositol trisphosphate receptor (IP3R) in the maxillary palps of blood fed (bf) and non-blood fed (nbf) Aedes aegypti females is shown. Transcripts that exhibit significant differences in abundance (Kal's test; FDR P < 0.05), are denoted according to their fold change (FC); FC > 2 in black-filled circles; 1.5 < FC < 2 in gray-filled circles; and FC < 1.5 in unfilled circles. (B) Differential response of the CO2-sensitive neuron to various concentrations of CO2 is shown. (C) Model of the putative CO2 signaling pathway. Solid red arrows indicate significant (FC > 2; FDR P > 0.05) and dashed red arrows indicate potential (FC < 2; FDR P > 0.05) changes in regulation. Diacylglycerol (DAG); phosphatidylinositol 4,5-bisphosphate (PIP2); G-protein beta subunit (ß); G-protein gamma subunit (γ); Stromal interaction molecule 1 (STIM1); transient receptor potential receptor L (TrpL).
The third Gr to be detected in the maxillary palps of both nbf and bf females was Gr2 (Figure 3A), a paralogue of Gr1, which also was less abundant in the maxillary palps of bf females (1.3-fold; FDR P = 0.0257; confirmed by qPCR, Supplementary Data Sheet 1b). The Gr2 appears not to be required for CO2 reception in Ae. aegypti (Erdelyan et al., 2012), and the function of this receptor has yet to be determined.
Since the abundance of the CO2-sensitive Gr transcripts was significantly lower in bf females, yet the fold change was <2-fold, the observed differential response in the CO2-sensitive OSNs 24 h post-blood meal may be a result of regulatory changes in both the translation and translocation of the receptors, and/or changes in the internal amplification of the signal via the second messenger cascades. The signal transduction pathway for CO2 detection has been partially described in D. melanogaster, implicating G-protein signaling via a Gαq (DmGα49B) (Yao and Carlson, 2010), which activates the phospholipase c-ß (Plc-ß) pathway via DmPlc21c to modulate the downstream Trps, DmTRP, and DmTRPL. The knockdown/knockout of any of the genes in this pathway results in a reduction/loss of the CO2 behavioral phenotype in D. melanogaster. Recently, Bohbot et al. (2014) proposed that a similar transduction pathway underlies CO2 detection in female Ae. aegypti mosquitoes, citing similar transcript abundances of the homologous genes: Gr1, Gr3, Gα49B (AAEL010506), Plc-ß (AAEL009380), and TrpL (AAEL005575). The comparison here between 6 dpe nbf and bf female maxillary palp transcriptomes lends support to this hypothesis (Figure 3A; Supplementary Data Sheet 1). The transcript abundance of the G-proteins Gα49B (AAEL010506; 1.4-fold; FDR P = 0.0002) and Gɤ (AAEL006685; 1.3-fold; FDR P = 0.0024) was significantly lower in bf females (confirmed by qPCR; Supplementary Data Sheet 1de), while G (AAEL019779) maintained a consistent abundance (FDR P = 0.1906). In the phospholipase c pathway, Plc-ß-A (AAEL009380) transcript abundance was significantly lower in bf females (1.3-fold change; FDR P = 0.0060), confirmed in subsequent qPCR analyses (Supplementary Data Sheet 1f), while the inositol 1,4,5-triphosphate receptor (AAEL02744) was not regulated in either the transcriptomic (1.1-fold, FDR P = 0.6572; Figure 3A) or qPCR analysis (Supplementary Data Sheet 1g). Of the downstream Trps, TrpL (AAEL005575) displayed a reduced abundance in the maxillary palps of bf females (2.6-fold; FDR P < 0.0001; confirmed by qPCR, Supplementary Data Sheet 1f), while trp (AAEL005437) was not expressed in the maxillary palps of 6-dpe females. In 6-dpe Orlando strain females, trp was reported to be robustly expressed, however it is not known whether the abundance is regulated post-blood meal (Bohbot et al., 2014). The other homologous gene implicated in CO2 signal transduction in D. melanogaster is Stim1 (AAEL018261), which displayed a higher abundance in bf compared to nbf (4-fold; FDR P = 0.0226). Combined, our results demonstrate that most of the transcripts in the putative CO2 signal transduction pathway are less abundant 24 h post-blood meal (Figures 3A,C), corresponding with the physiological reduction in sensitivity to CO2 (Figure 3B), when females no longer seek for hosts (Klowden and Blackmer, 1987; Christ et al., 2017). As genetic manipulation of non-model organisms becomes more accessible, it is becoming possible to directly test the link between the CO2 physiology/behavior and the CO2-related signal transduction pathway gene expression (McMeniman et al., 2014).
Differential Modulation of Or-Mediated Signaling
Ligand transduction in the B- and C-cells of Ae. aegypti is mediated by Or8, and tentatively by Or49, together with Orco, respectively (Bohbot and Dickens, 2009; Bohbot et al., 2013). Similar to the CO2-sensitive Grs, the gene expression of these Ors is up-regulated during adult maturation (Bohbot et al., 2013), emphasizing the importance of Or8 in host recognition (Majeed et al., 2016) and reflecting a likely role for Or49 in the acquisition of host seeking. The modulation in expression of these Ors in bf maxillary palps, however, differs between Or8 (1.2-fold change; FDR P = 0.1511; Figure 4A) and Or49 (1.2-fold change; FDR P = 0.0007; Figure 4A), as also confirmed by qPCR (Supplementary Data Sheet 1ik). This demonstrates that these Ors are differentially regulated in bf maxillary palps. Both Or8 and its cognate OSN, the B-cell, selectively detect (R)-1-octen-3-ol (Lu et al., 2007; Bohbot and Dickens, 2009; Hill et al., 2015), one of the most extensively studied mosquito host cues (e.g., Bohbot and Dickens, 2009; Cook et al., 2011; Bohbot et al., 2013; Majeed et al., 2016). Reflecting the consistent levels of transcript expression for Or8, as well as Orco (1.1-fold change; FDR P = 0.6517; Figure 4A; confirmed by qPCR, Supplementary Data Sheet 1j), the (R)-1-octen-3-ol-sensitive OSN, the B-cell, was not differentially sensitive to racemic 1-octen-3-ol in nbf and bf females (Figure 4B). Despite efforts to identify the ecologically relevant ligand for Or49, the function of Or49 and the C-cell is not yet known.
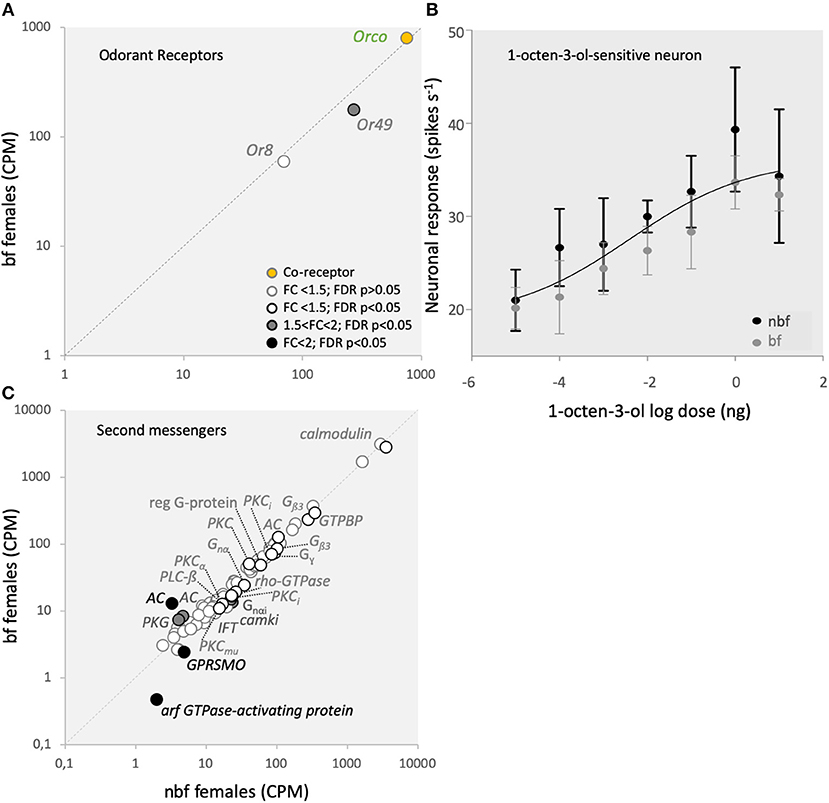
Figure 4. Odorant receptor (Or) mediated signaling. (A) Transcript regulation in the maxillary palps of blood fed (bf) and non-blood fed (nbf) Aedes aegypti females of the co-receptor, Orco (orange-filled circle), the (R)-1-octen-3-ol-sensitive Or8, and the orphan receptor Or49. (B) Response of the (R)-1-octen-3-ol-sensitive neuron to various concentrations of (R)-1-octen-3-ol. The trendline represents the non-linear regression among all responses and concentrations. (C) Differential regulation of second messenger gene expression in the maxillary palps. Transcripts that exhibit significant differences in abundance (Kal's test; FDR P < 0.05) are denoted according to their fold change (FC); FC > 2 in black-filled circles; 1.5 < FC < 2 in gray-filled circles; and FC < 1.5 in unfilled circles. The gray dotted line indicates a hypothetical equal transcript abundance between the states. Chimerin (rho-GTPase-activating protein; rho-GTPase); guanylate cyclase (GC); nucleolar GTP-binding protein (GTPBP); cGMP-dependent protein kinase (PKG); cAMP-dependent protein kinase type ii regulatory subunit (PKA); smoothened G-protein coupled receptor (GPRSMO); intraflagellar transport protein (IFT); G-protein, gamma-subunit (Gγ); GTP-binding protein alpha subunit (Gnα); GTP-binding protein (i) alpha subunit (Gnαi); protein kinase C inhibitor (PKCi); phospholipase c beta (PLC-ß); G-protein β3 (Gβ3); calcium/calmodulin-dependent protein kinase type 1 (camki); regulator of g protein signaling (reg G-protein); protein kinase C (PKC; PKCmu; PKCα); adenylate cyclase (AC); calmodulin.
Whilst the second messenger signaling pathway has not been determined for either Or8 or Or49, several second messengers have been implicated in Or signaling cascades, particularly in D. melanogaster and the hawkmoth, Manduca sexta (Sato et al., 2008; Nakagawa and Vosshall, 2009; Sargsyan et al., 2011; Getahun et al., 2013; Stengl and Funk, 2013). In addition to the pathway described for CO2 signal transduction above, the G-protein coupled cyclic AMP-dependent pathway (Wicher et al., 2008; Getahun et al., 2013), as well as feedback onto the Or complex from phosphokinase c (Pkc) (Sargsyan et al., 2011; Getahun et al., 2013), have also been proposed for the regulation of Ors (for review see Stengl, 2010; Stengl and Funk, 2013). In the adenylate cyclase cascade, the only regulated member in the maxillary palps was rho-GTPase-activating protein (AAEL011253), which demonstrated a lower abundance in bf than nbf (1.4-fold; FDR P = 0.0030; Figure 4C). This suggests that this pathway may not be a major pathway in regulating Ors in the maxillary palps. On the other hand, two (AAEL001549, AAEL002892) of the six Pkcs expressed in the maxillary palps exhibited changes in abundance between nbf and bf, albeit not above the 1.5-fold cutoff (1.2-fold higher in bf, FDR P = 0.0206; 1.4-fold high in nbf, FDR P = 0.0425; respectively; Figure 4C).
Other Receptor Families
Five Ir transcripts were detected in the maxillary palps of females (Figure 5A). The co-receptor Ir25a in Ae. aegypti was found to be the most abundant Ir in the maxillary palps (Figure 5A) (Bohbot et al., 2014; Matthews et al., 2016), suggesting the pre-dominance of this co-receptor in Ir complexes in this olfactory organ. While Ir8a was not expressed above threshold levels in the maxillary palps, the third Irco, Ir76b, was the fourth highest Ir in abundance, one tenth of that of Ir25a, similar to what was shown previously (Matthews et al., 2016) (Figure 5A). Neither co-receptor was shown to be differentially abundant (Figure 5A). Five other Irs were consistently expressed in the maxillary palps of nbf and bf females, of which Ir100e.1 demonstrated a >2-fold change in abundance in bf compared with nbf females (32.9-fold; FDR P < 0.0001), and Ir75k.2 was shown to be abundant only in nbf females (3.85 ± 0.76 CPM; FDR P = 0.0472; Figure 5A).
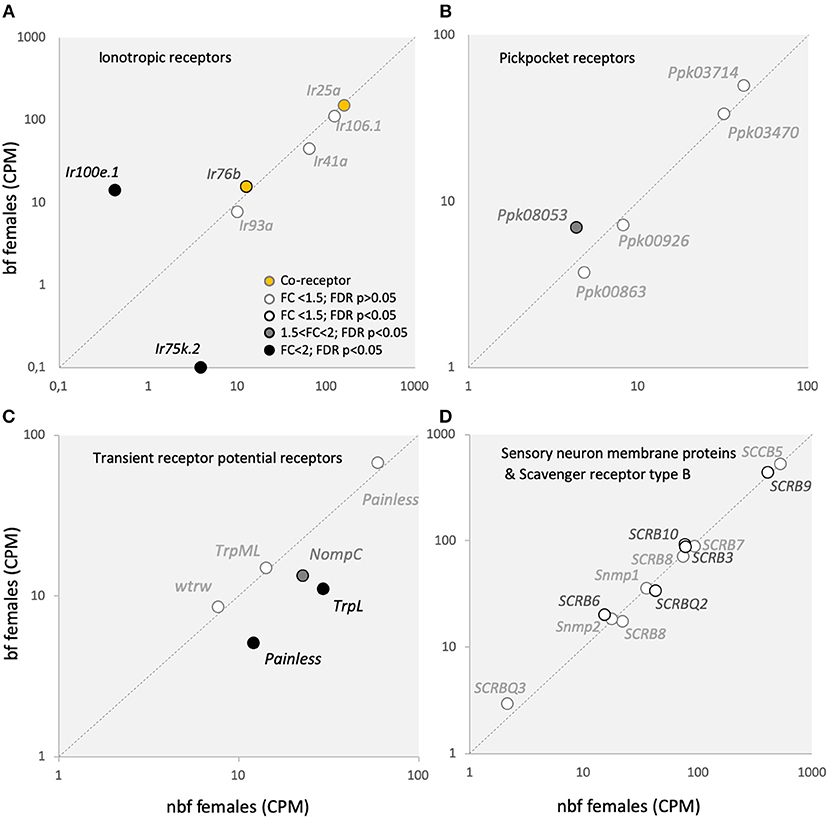
Figure 5. Sensory receptor transcript abundance. Transcript regulation of (A) ionotropic receptor (Ir) co-receptors, Ir25a and Ir76b (orange-filled circles), as well as other Irs; (B) pickpocket receptors (Ppks); (C) transient receptor potential receptors (Trps); and (D) sensory neuron membrane proteins (Snmps) and scavenger receptors type B (SCRBs) is presented as the normalized number of reads in the maxillary palps of blood fed (bf) and non-blood fed (nbf) Aedes aegypti females. Transcripts that exhibit significant differences in abundance (Kal's test; FDR P < 0.05) are denoted according to their fold change (FC); FC > 2 in black-filled circles; 1.5 < FC < 2 in gray-filled circles; and FC < 1.5 in unfilled circles. The gray dotted line indicates a hypothetical equal transcript abundance between the states.
In Ae. aegypti, members of the expanded Ir75 subfamily are expressed in the maxillary palp (Figure 5A) (Bohbot et al., 2014; Matthews et al., 2016), antenna (Matthews et al., 2016), as well as in the labella and tarsi (Sparks et al., 2014), however the function of these receptors in Ae. aegypti is not known. In An. gambiae and D. melanogaster, the Ir75 subfamily responds to short chain carboxylic acids (Yao et al., 2005; Abuin et al., 2011; Pitts et al., 2017). The subfamily expansion in mosquitoes emphasizes the ecological significance of carboxylic acids in driving host attraction (Davis, 1984; Bosch et al., 2000; Smallegange et al., 2005). The lack of expression of Ir75k.2 in the maxillary palps of bf females, and its presence in nbf females, suggests a role for this receptor in host detection. The Ir100s subfamily is expressed in the larval and adult internal gustatory organs of D. melanogaster (Croset et al., 2010), and may therefore play some role in the detection of taste compounds. In Ae. aegypti, Ir100e has been found to be expressed in the maxillary palps (Figure 5A) (Bohbot et al., 2014; Matthews et al., 2016) and in the antennae (Matthews et al., 2016), but not in the labella or tarsi (Sparks et al., 2014), suggesting that while the role in taste detection may be conserved in the pharyngeal organ, it is likely that the role for Ir100e has expanded. Since Ir100e.1 was more abundant in the maxillary palps of bf females (Figure 5A), it will be of future interest to determine the ligand sensitivity of this Ir, together with its co-receptor Ir76b, and its role in the maxillary palps of Ae. aegypti following a blood meal. While the results reported here are from the Rockefeller strain, those from the Liverpool strain (LVPIB12) (Matthews et al., 2016), and from those reported from the Orlando strain (Bohbot et al., 2014) each differ from one another. The sole unique Ir detected in the maxillary palps of all three strains was Ir100e. Of the other five Irs reported expressed in the Orlando strain (Ir92a, Ir41a.1, Ir75l, Ir7y.2, and Ir7h.2), but not in the Rockefeller strain, only Ir92a and Ir41a.1 were found to be consistently detected above threshold levels in the Liverpool strain. Whether these differences are strain-related, or whether they are a result of the ongoing re-annotation of the Ae. aegypti genome, will need to be further addressed as time and study concretise the annotations. For example, Ir106.1 represents the collapsing of 7 putative genes (AAEL005231; AAEL008226; AAEL008231; AAEL017163; AAEL018066; AAEL018068; AAEL018069) from previous annotations (Matthews et al., 2016) into a single gene (AAEL019784) in the current L5 annotation. Two of these putative genes from the earlier annotation were shown to have abundances above threshold in the maxillary palps of nbf females (Matthews et al., 2016). Additionally, the Ir75 subfamily has been extensively re-annotated, particularly the Ir75k paralogues, so that specific, direct comparisons between the transcriptomes are not feasible. In addition, those genes previously identified as Ir41a.1 (L3 AAEL000007) and Ir41a.2 (L3 AAEL000031) are no longer supported in the L5 annotation, however, AAEL000041 (currently annotated as a forkhead protein) shares strong similarity with the other mosquito Ir41as and with Ir41a in Drosophila, and is reported here as Ir41a.
The Ppks and Trps have recently been added to the list of chemosensory receptors (Al-Anzi et al., 2006; Kang et al., 2010; Badsha et al., 2012; Fowler and Montell, 2013). None of the five Ppk transcripts expressed in the maxillary palps (Figure 5B) are homologous to those shown to be chemosensitive in D. melanogaster (Joseph and Carlson, 2015). However, of the six Trps expressed in the maxillary palps of Ae. aegypti (Figure 5C), only painless (AAEL006835; AAEL004397), and TRPL (AAEL005575) have been implicated in chemosensation (Al-Anzi et al., 2006; Kang et al., 2010; Badsha et al., 2012; Fowler and Montell, 2013). In Ae. aegypti maxillary palps, of the two painless transcripts expressed, one had higher in abundance in nbf females (AAEL004397: 2.4-fold; FDR P < 0.0001). In D. melanogaster, painless is involved in noxious chemical sensing, particularly of isothiocyanate (Al-Anzi et al., 2006; Kang et al., 2010). The transcript abundance of TrpL, which likely plays a role as Gq/Plc activated ion channels downstream of Ors or Grs (Badsha et al., 2012; Fowler and Montell, 2013), was reduced in bf females (see above). The other Trp that exhibited a lower abundance in the maxillary palps of nbf females, NompC (1.7-fold; FDR P < 0.0001), likely plays a role in mechanosensation, together with painless and Ppk0863 (Joseph and Carlson, 2015). Taken together, this expression pattern of the Trps and Ppks suggests a decrease in sensitivity to chemical and mechanical stimuli in the maxillary palp in bf females, which reflects the ecology of the mosquito during this period in the gonotrophic cycle.
None of the 13 ligand-binding scavenger receptor, type B, genes (SCRB/CD36) (Vogt and Riddiford, 1981; Vogt et al., 2009), including the two chemosensory-related members of the gene family, the Snmps (Snmp1 and Snmp2) were regulated in the maxillary palps of nbf and bf females (Figure 5D). The SNMPs have been implicated in mediating pheromone detection, acting as molecular bridges (SNMP1) (Bohbot et al., 2010), and in the clearance of lipophilic components from the sensillum lymph (SNMP2) (Forstner et al., 2008; Jiang et al., 2016). The expression of the Snmps in the maxillary palps of the Rockefeller strain (Figure 5D) was found to be in line with that reported in the Liverpool strain (Matthews et al., 2016), however Snmp1 transcripts were reported as absent from the maxillary palps in the host-seeking females of the Orlando strain (Bohbot et al., 2014). The presence of Snmp transcripts in the maxillary palps suggests that this chemosensory organ may play a role in the proposed aggregation and oviposition pheromone detection by Ae. aegypti females (Cabrera and Jaffe, 2007; Seenivasagan et al., 2009).
Soluble Odor Binding Proteins
Chemosensory proteins belong to a conserved arthropod protein family that is poorly understood, but may play a role in mediating the movement of lipophilic volatiles through the aqueous environment of the sensillum lymph (Vogt, 2003). Of the 18 identified Csps in Ae. aegypti, nine are expressed in the maxillary palps (Figure 6A) (Matthews et al., 2016). Three of the Csps were significantly more abundant in maxillary palps of bf females (AAEL002007: 8.2-fold; FDR P < 0.0001; AAEL002003: 6.2-fold; FDR P = 0.0288; AAEL001991: 20.9-fold; FDR P = 0.0105), and two more Csps were identified as genes-of-interest as being significantly more abundant 24 h pbm (AAEL005710: 1.6-fold; FDR P < 0.0001; AAEL012383: 1.5-fold; FDR P = 0.0008). The functional significance of the abundance of Csps is unknown.
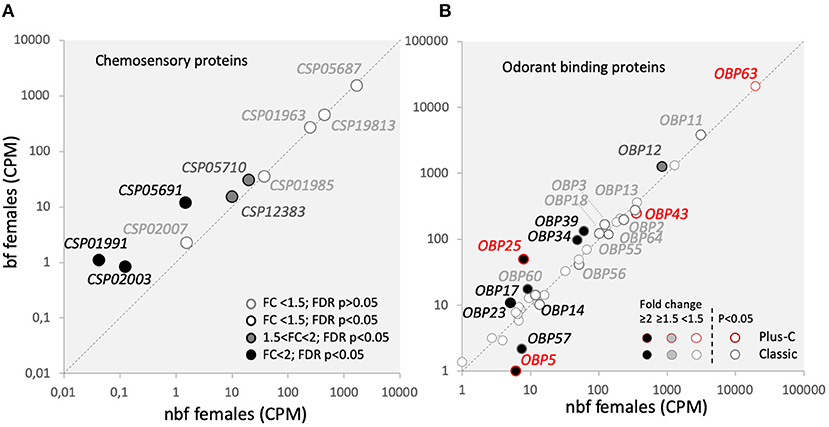
Figure 6. Olfactory binding proteins transcript abundance. Transcript regulation in the maxillary palps of blood fed (bf) and non-blood fed (nbf) Aedes aegypti females is presented as the normalized number of reads of (A) chemosensory proteins (Csps); and (B) odorant binding proteins (Obps) of two classes, outlined in greyscale and red for classic and plus-C Obps, respectively. Transcripts that exhibit significant differences in abundance (Kal's test; FDR P < 0.05), are denoted according to their fold change (FC); FC > 2 in black-filled circles; 1.5 < FC < 2 in gray-filled circles; and FC < 1.5 in unfilled circles. The gray dotted line indicates a hypothetical equal transcript abundance between the states. So that (B) is legible, the cutoff for including Obps FC < 1.5 is 25 reads.
Odorant binding proteins are the largest family of chaperone proteins, which may solubilise odorants, interact with chemoreceptors (Vogt, 2003), and play a role in gain control (Larter et al., 2016). In Ae. aegypti, there are 111 Obps annotated among the three classes of Obps: the Classic, the Plus-C and the Two-Domain Obps (Zhou et al., 2008; Manoharan et al., 2013). Classic and Plus-C Obp transcripts were expressed in the maxillary palps of both nbf and bf female, almost half of which were differentially abundant (Figure 6B). No Two-Domain Obp transcripts were above threshold levels in the maxillary palps (Figure 6B) (Bohbot et al., 2014; Matthews et al., 2016). While actual functional data for these regulated Obps is not yet available in Ae. aegypti, homology modeling for the classic Obps has suggested that these regulated Obps are predominantly binding amine-containing compounds and permethrin (Manoharan et al., 2013).
Modulators and Their Receptors
Modulation of host-seeking behavior following blood feeding has been shown to be regulated by various neuromodulators, predominantly neuropeptides (Brown et al., 1994; Liesch et al., 2013; Christ et al., 2017). Of the 13 neuropeptide transcripts that were detected in the maxillary palps, four were significantly more abundant in the maxillary palps of bf females (ITG-like, AAEL010262, 1.6-fold, FDR P < 0.0001; PK1, AAEL012060, 4.7-fold, FDR P < 0.0001; RYa, AAEL011702, 8.2-fold, FDR P = 0.0055; AKH, AAEL011996, 47.5-fold, FDR P = 0.0004) and four in nbf females (sulfakinin, AAEL006451, 2.4-fold, FDR P = 0.0004; ACP, AAEL010950, 7.8-fold, FDR P < 0.0001; PK2, AAEL005444, 2.6-fold, FDR P = 0.004; Agatoxin-like, AAEL022190, 8.0-fold, FDR P = 0.0089), while two neuropetides were only found to be expressed in the maxillary palps of nbf females (CCHa2, AAEL026488; PTTH, AAEL026383). Four neuropeptides were found to be expressed together with their cognate receptors (ASTC:GPRSMS, RYa:GPRNPY6, AKH:AKHR, PK2:GPRHP3). The ASTC transcripts were not regulated in the maxillary palps post-blood meal (Figure 7A), which is in line with that described in the brain (Mayoral et al., 2010). However, one of the two mosquito ASTC cognate receptors, GPRSMS, was more abundant in bf females (2.3-fold, FDR P = 0.03; Figure 7B). While ASTC signaling is known to be involved in inhibiting juvenile hormone synthesis in the corpora allata, its role in the maxillary palps of Ae. aegypti females is unknown. Recently identified, RYamide and its cognate receptor, GPRGNPY6 (12.2-fold, FDR P = 0.0012), were more abundant in the maxillary palps of bf females. RYamide signaling has been suggested to be involved in regulating response to food (Ohno et al., 2017), which may indicate a role for its expression and regulation post-blood meal in the maxillary palps of Ae. aegypti females. Similarly, both of the regulated neuropeptides Akh and Pyrokinin 2 were expressed along with their cognate receptors AKHR (AAEL011325) and GPRGHP3 (AAEL012796), respectively, however, neither receptor exhibited differential abundance (Figure 7B). The abundance of transcripts of both peptide precursor and receptor in the maxillary palps suggests that the signaling of these peptide systems is located, at least in part, within the maxillary palps, and/or that individual cells in the maxillary palps express both the peptide and the receptor, creating the possibility for autoregulation, similar to that which is observed in the autoregulation of antennal OSNs in D. melanogaster (Ko et al., 2015). The cognate receptors of the remaining neuropeptide transcripts abundant in the maxillary palps (Figure 7A) were not detected (Figure 7B). The presence of the neuropeptide precursor transcripts, and the absence of the receptor transcripts, in the maxillary palps suggests that neuropeptide-expressing cells are afferent neurons which will release the neuropeptides within the CNS. The most likely cells that fit this category in the maxillary palps are the OSNs. Additionally, there are 18 neuropeptide/GPCR receptor transcripts present in the maxillary palps, without the detection of their cognate neuropeptide transcript (Figure 7B). Of these, four (diuretic hormone CRF-like receptor, GPRDIH1, 1.8-fold, FDR P = 0.0430; vasopressin receptor, GPRVPR2, 1.8-fold, FDR P = 0.0129; Sex peptide receptor, Sex peptide R, 2.7-fold, FDR P = 0.00104; diuretic hormone CRF-like receptor, AAEL019757, 15.9-fold, FDR P = 0.04711) were more abundant in nbf, and two (diuretic hormone calcitonin-like receptor, GPRCAL1, 1.7-fold, FDR P = 0.0331; orphan GPCR class D, GPRNND3, 2.6-fold, FDR P = 0.0070) were more abundant in bf females (Figure 7B). Detection of these receptors suggests that the maxillary palps are sensitive to modulation from neural and/or humoral neuropeptides.
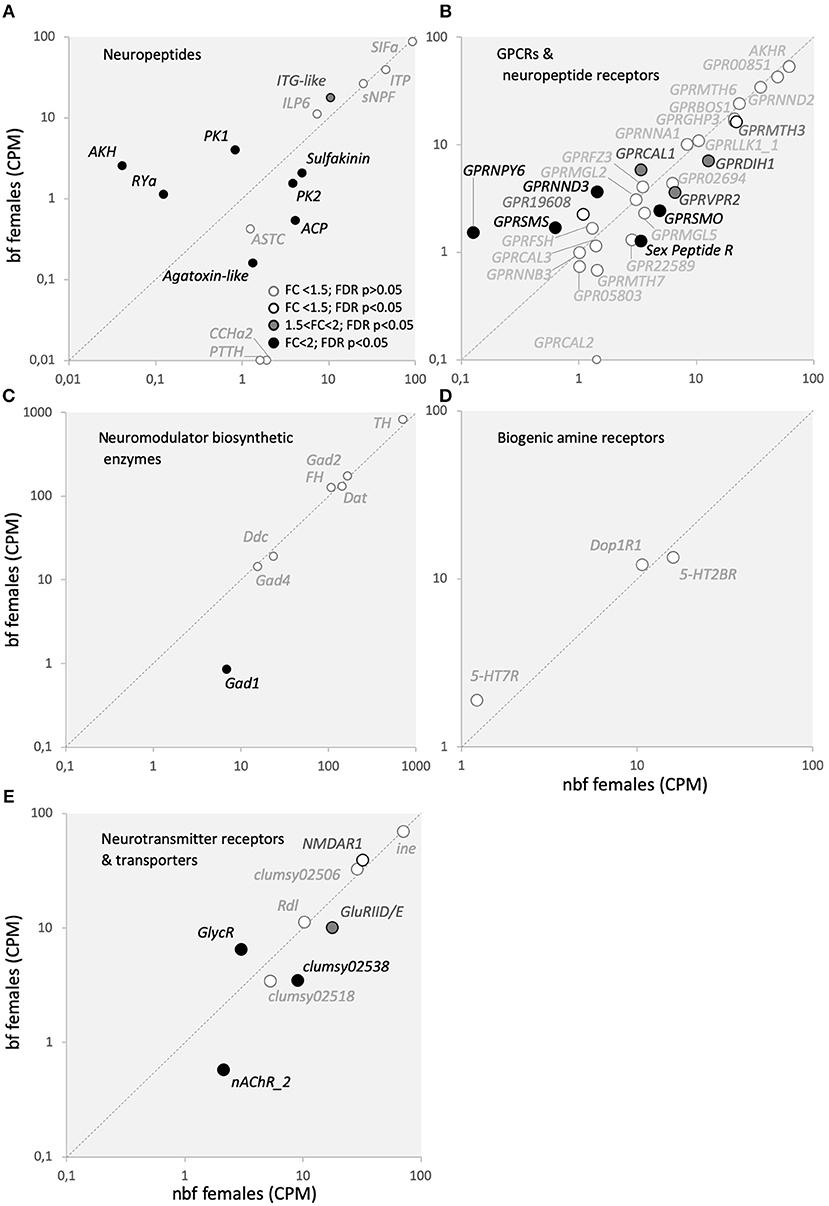
Figure 7. Neurotransmitters and modulators with their cognate receptors, transporters, and biosynthetic enzymes transcript abundance. Transcript regulation in the maxillary palps of blood fed (bf) and non-blood fed (nbf) Aedes aegypti females is presented as the normalized number of reads of (A) neuropeptides, (B) GPCR and neuropeptide receptors, (C) neuromodulator biosynthetic enzymes, (D) biogenic amine receptors, (E) neurotransmitter receptors and transporters. Transcripts that exhibit significant differences in abundance (Kal's test; FDR P < 0.05), are denoted according to their fold change (FC); FC > 2 in black-filled circles; 1.5 < FC < 2 in gray-filled circles; and FC < 1.5 in unfilled circles. The gray dotted line indicates a hypothetical equal transcript abundance between the states. (A) SIFamide (SIFa; AAEL009858); ion-transport peptide (IPT; AAEL019725); short neuropeptide F (sNPF; AAEL012542); ITG-like peptide (ITG-like; AAEL010262); insulin-like peptide 6 (ILP6; AAEL004547); sulfakinin (sulfakinin; AAEL006451); adipokinetic/corazonin peptide (ACP; AAEL010950); pyrokinin 2 (PK2; AAEL005444); CCHamide 2 (CCHa2; AAEL026488); prothoracicotropic hormone (PTTH; AAEL026383); Agatoxin-like neuropeptide (Agatoxin-like; AAEL022190); allatostatin C (ASTC; AAEL005747); pheromone/pyrokinin biosynthesis-activating neuropeptide (PK1; AAEL012060); RYamide (RYa; AAEL011702); adipokinetic hormone (AKH; AAEL011996); (B) gonadotropin-releasing hormone receptor (AKHR, AAEL011325); predicted G-protein coupled receptor (GPR00851, AAEL000851; GPR05803, AAEL005803; GPR22589, AAEL022589); GPCR Orphan/Putative Class A Family (GPRNNA1, AAEL003378; GPR19608, AAEL019608); GPCR Orphan/Putative Class B Family (GPRNNB3, AAEL001724); GPCR Orphan/Putative Class D Family (GPRNND2, AAEL001782; GPRNND3, AAEL06232); GPCR Growth Hormone Releasing Hormone Family (GPRGHP3, AAEL012796); GPCR Diuretic Insect Hormone (GPRDIH1, AAEL008292; GPRCAL1, AAEL010043; GPRCAL2, AAEL006490; GPRCAL3, AAEL009024); GPCR Glycoprotein Hormone Family (GPRFSH, AAEL004399); GPCR Somatostatin Family (GPRSMS, AAEL012356); GPCR Neuropeptide Y Family (GPRNPY6, AAEL017005); GPCR Leukokinin Family (GPRLLK1_1, AAEL006636); GPCR Vasopressin Family (GRPVPR2, AAEL008655); Sex Peptide Receptor (Sex Peptide R, AAEL019881); GPCR Metabotropic glutamate Family (GPRMGL2, AAEL004533; GPRMGL5, AAEL009822); GPCR Methuselah Family (GPRMTH3, AAEL017374; GPRMTH6, AAEL011521; GPRMTH7, AAEL000811); GPCR Bride of Sevenless Family (GPRBOS1, AAEL007004); GPCR Frizzled/Smoothened Family (GPRSMO, AAEL006669; GPRFZ4, AAEL008322); (C) tyrosine 3-monooxygenase (TH, AAEL017098); phenylalanine-4-hydroxylase (FH, AAEL017029); dopamine N-acetyltransferase (Dat, AAEL011088); aromatic L-amino acid decarboxylase (Ddc, AAEL014238); glutamic acid decarboxylase (Gad1, AAEL011981; Gad2, AAEL007542; Gad4, AAEL010951); (D) GPCR Serotonin Family (5-HT2BR, AAEL019805; 5-HT7R, AAEL027242); GPCR Dopamine Family (Dop1R1, AAEL019437); and (E) sodium- and chloride-dependent neurotransmitter transporter (ine, AAEL006412); glutamate receptors (NMDAR1, AAEL; clumsy02506, AAEL02506; clumsy02538; AAEL02538; clumsy02518, AAEL02518; GluRIID/E, AAEL022196); nicotinic acetylcholine receptor (nAChR_2, AAEL018352); glycine receptor beta precursor (GlyR, AAEL001568); GABA receptor (Rdl, AAEL008354).
Another class of neuromodulators implicated in regulating mosquito behavior is the biogenic amines. Unlike neuropeptides, which are encoded in the genome, biogenic amines are synthesized from an array of enzymes. It is the abundance of these enzyme transcripts that are reported to indirectly indicate biogenic amine level in transcriptome analyses. Four of the seven detected biosynthetic enzymes are from the families responsible the generation of biogenic amines, dopamine N-acetyltransferase (Dat), tyrosine hydroxylase (TH), phenylalanine-4-hydroxylase (FH), and dopa decarboxylase (Ddc) (Figure 7C) (Matthews et al., 2016), none of which were differentially abundant (Figure 7C). Biogenic amine receptor transcripts were detected for serotonin (5-HT7R; 5-HT2BR) and dopamine (Dop1R1), which also demonstrated consistent abundance pre- and post-blood meal (Figure 7D). These findings are supported by prior immunohistological investigations demonstrating that serotonergic neurons are present in the maxillary palps of female Ae. aegypti mosquitoes (Siju et al., 2008).
Neurotransmitters and their receptors are the primary signaling molecules at synapses, responsible for normal synaptic activity. Similar to biogenic amines, neurotransmitters are synthesized via enzymatic pathways, rather than being encoded in the genome. Three of these biosynthetic enzymes from the glutamate decarboxylase family (Gad1, 2, and 4), were detected in the maxillary palps (Figure 7C) (Matthews et al., 2016). Of these, only Gad1 was found to be more abundant in the maxillary palps of nbf females (8.2-fold, FDR P < 0.0001; Figure 7C), which is indicative of the biogenesis of the neurotransmitter GABA. It is interesting to note that while the biosynthetic enzymes for GABA were present, none of those for acetylcholine (choline O-acetyltransferase, ChAT, AAEL010471) were detected, indicating that the primary neurotransmitter produced in the maxillary palps of 6 dpe female Ae. aegypti is GABA, which primarily is involved in inhibitory signaling pathways. While GABA receptors have been described in OSNs to be involved in gain control (Olsen and Wilson, 2008; Root et al., 2008) and the biosynthetic enzymes that produce GABA can be found in olfactory tissues (Matthews et al., 2016), but GABA signaling from OSNs has not been described. A GABA-gated chloride receptor (Rdl, AAEL008354) was detected at similar levels in the maxillary palps of nbf and bf females (Figure 7E). Ten neurotransmitter receptor genes were expressed in the maxillary palps (Figure 7E). The predominant type of receptor detected had glutamate as its cognate ligand, the transcripts of which either did not differ in abundance (NMDAR1, clumsy02506, clumsy02518) or were more abundant in the maxillary palps of nbf females (GluRIID/E, 1.8-fold, FDR P < 0.0001; clumsy02538, 2.6-fold, FDR P < 0.0001). Two other neurotransmitter receptors were detected in the maxillary palps, with one acetylcholine receptor being more abundant in nbf females (nAChR_2; AAEL018308, 3.6-fold, FDR P = 0.0064), and one glycine receptor being more abundant in bf females (GlycR; AAEL001568, 2.1-fold, FDR P = 0.0012), and commonly mediating synaptic excitation and inhibition, respectively. The one neuromodulator transporter that was detected in the maxillary palps, a sodium- and chloride-dependent GABA transporter (ine, AAEL006412), was not differentially regulated (Figure 7E), and lends support to the importance of GABA signaling in this chemosensory tissue.
Conclusions
In this study, chemosensory, neuromodulatory, and signal transduction-related genes in the maxillary palps of Ae. aegypti females have been shown to be regulated post-blood meal. Differentially regulated genes in the maxillary palps between nbf and bf females are highlighted. Additional screens for gene expression in the future may consider adding a non-blood membrane feeding condition to clarify the role of the experience of other sensory modalities in producing differential gene expression. The future functional characterization of the proteins generated by the genes-of-interest identified in this study may offer important insights in the regulation of vector-related odor coding in female Ae. aegypti, and its impact on gonotrophic behaviors that may in turn lead to new innovations in vector control.
Data Availability
The datasets generated for this study can be found in the NCBI project database, BioProject, with the ID PRJNA545481.
Author Contributions
SH and RI conceived. SH designed the gene expression study and conducted the analyses and drafted the manuscript. RI conceived and designed the electrophysiological study. SH conducted all the experiments, except the physiological single sensillum recordings, which were conducted, and analyzed by MG. All authors provided constructive input toward the manuscript and participated in revising it critically for important intellectual content.
Funding
This work was funded by The Swedish Research Council Formas and supported by the Linnaeus initiative Insect Chemical Ecology, Ethology and Evolution IC-E3 (Formas and Swedish University of Agricultural Sciences). The funding body has had no role in the design of the study and collection, analysis, and interpretation of data, or in writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Thomas Svensson for his help in tissue collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00336/full#supplementary-material
References
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi: 10.1016/j.neuron.2010.11.042
Al-Anzi, B., Tracey, W. D., and Benzer, S. (2006). Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr. Biol. 16, 1034–1040. doi: 10.1016/j.cub.2006.04.002
Badsha, F., Kain, P., Prabhakar, S., Sundaram, S., Padinjat, R., Rodrigues, V., et al. (2012). Mutants in Drosophila TRPC channels reduce olfactory sensitivity to carbon dioxide. PLoS ONE 7:e49848. doi: 10.1371/journal.pone.0049848
Benton, R. (2015). Multigene family evolution: perspectives from insect chemoreceptors. Trends Ecol. Evol. 30, 590–600. doi: 10.1016/j.tree.2015.07.009
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Benton, R., Vannice, K. S., and Vosshall, L. B. (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293. doi: 10.1038/nature06328
Bohbot, J., Pitts, R. J., Kwon, H. W., Rützler, M., Robertson, H. M., and Zwiebel, L. J. (2007). Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol. Biol. 16, 525–537. doi: 10.1111/j.1365-2583.2007.00748.x
Bohbot, J. D., and Dickens, J. C. (2009). Characterization of an enantioselective odorant receptor in the yellow fever mosquito Aedes aegypti. PLoS ONE 4:e7032. doi: 10.1371/journal.pone.0007032
Bohbot, J. D., Durand, N. F., Vinyard, B. T., and Dickens, J. C. (2013). Functional development of the octenol response in Aedes aegypti. Front. Physiol. 4:39. doi: 10.3389/fphys.2013.00039
Bohbot, J. D., Fu, L., LE, T. C., Chauhan, K. R., Cantrell, C. L., and Dickens, J. C. (2011a). Multiple activities of insect repellents on odorant receptors in mosquitoes. Med. Veter. Entomol. 25, 436–444. doi: 10.1111/j.1365-2915.2011.00949.x
Bohbot, J. D., Jones, P. L., Wang, G., Pitts, R. J., Pask, G. M., and Zwiebel, L. J. (2011b). Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem. Senses 36, 149–160. doi: 10.1093/chemse/bjq105
Bohbot, J. D., Lu, T., and Zwiebel, L. J. (2010). “Molecular regulation of olfaction in mosquitoes,” in Olfaction in Vector-Host Interactions, eds W. Takken and B. G. J. Knols (Wageningen: Wageningen Academic Publishers, 17–38.
Bohbot, J. D., Sparks, J. T., and Dickens, J. C. (2014). The maxillary palp of Aedes aegypti, a model of multisensory integration. Insect Biochem. Mol. Biol. 48, 29–39. doi: 10.1016/j.ibmb.2014.02.007
Bolstad, B. M., Irizarry, R. A., Astrand, M., and Speed, T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. doi: 10.1093/bioinformatics/19.2.185
Bosch, O. J., Geier, M., and Boeckh, J. (2000). Contribution of fatty acids to olfactory host finding of female Aedes aegypti. Chem. Senses 25, 323–330. doi: 10.1093/oxfordjournals.chemse.a014042
Brown, M. R., Klowden, M. j., Crim, J. W., Young, L., Shrouder, L. A., and Lea, A. O. (1994). Endogenous regulation of mosquito host-seeking behavior by a neuropeptide. J. Insect Physiol. 40, 399–406. doi: 10.1016/0022-1910(94)90158-9
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cabrera, M., and Jaffe, K. (2007). An aggregation pheromone modulates lekking behavior in the vector mosquito Aedes aegypti (Diptera: Culicidae). J. Am. Mosquito Control Assoc. 23, 1–10. doi: 10.2987/8756-971X(2007)23[1:AAPMLB]2.0.CO;2
Carey, A. F., Wang, G., Su, C. Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1038/nature08834
Christ, P., Reifenrath, A., Kahnt, J., Hauser, F., Hill, S. R., Schachtner, J., et al. (2017). Feeding-induced changes in allatostatin-A and short neuropeptide F in the antennal lobes affect odor-mediated host seeking in the yellow fever mosquito, Aedes aegypti. PLoS ONE 12:e0188243. doi: 10.1371/journal.pone.0188243
Cook, J. I., Majeed, S., Ignell, R., Pickett, J. A., Birkett, M. A., and Logan, J. G. (2011). Enantiomeric selectivity in behavioural and electrophysiological responses of Aedes aegypti and Culex quinquefasciatus mosquitoes. Bull. Entomol. Res. 101, 541–550. doi: 10.1017/S0007485311000162
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
Dana, A. N., Hong, Y. S., Kern, M. K., Hillenmeyer, M. E., Harker, B. W., Lobo, N. F., et al. (2005). Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genom. 6:5. doi: 10.1186/1471-2164-6-5
Davis, E. E. (1984). Development of lactic acid-receptor sensitivity and host-seeking behaviour in newly emerged female Aedes aegypti mosquitoes. J. Insect Physiol. 30, 211–215. doi: 10.1016/0022-1910(84)90005-2
Dekker, T., and Cardé, R. T. (2011). Moment-to-moment flight manoeuvres of the female yellow fever mosquito (Aedes aegypti L.) in response to plumes of carbon dioxide and human skin odour. J. Exp. Biol. 214, 3480–3494. doi: 10.1242/jeb.055186
Ditzen, M., Pellegrino, M., and Vosshall, L. B. (2008). Insect odorant receptors are molecular targets of the insect repellent DEET. Science 319, 1838–1842. doi: 10.1126/science.1153121
Dzaki, N., Ramli, K. N., Azlan, A., Ishak, I. H., and Azzam, G. (2017). Evaluation of reference genes at different developmental stages for quantitative real-time PCR in Aedes aegypti. Sci. Rep. 7:43618. doi: 10.1038/srep43618
Erdelyan, C. N., Mahood, T. H., Bader, T. S., and Whyard, S. (2012). Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 21, 119–127. doi: 10.1111/j.1365-2583.2011.01120.x
Forstner, M., Gohl, T., Gondesen, I., Raming, K., Breer, H., and Krieger, J. (2008). Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem. Senses 33, 291–299. doi: 10.1093/chemse/bjm087
Fowler, M. A., and Montell, C. (2013). Drosophila TRP channels and animal behavior. Life Sci. 92, 392–403. doi: 10.1016/j.lfs.2012.07.029
Getahun, M. N., Olsson, S. B., Lavista-Llanos, S., Hansson, B. S., and Wicher, D. (2013). Insect odorant response sensitivity is tuned by metabotropically autoregulated olfactory receptors. PLoS ONE 8:e58889. doi: 10.1371/journal.pone.0058889
Grant, A. J., and Dickens, J. C. (2011). Functional characterization of the octenol receptor neuron on the maxillary palps of the yellow fever mosquito, Aedes aegypti. PLoS ONE 6:e21785. doi: 10.1371/journal.pone.0021785
Grant, A. J., Wigton, B. E., Aghajanian, J. G., and O'Connell, R. J. (1995). Electrophysiological responses of receptor neurons in mosquito maxillary palp sensilla to carbon dioxide. J. Comp. Physiol. 177, 389–396. doi: 10.1007/BF00187475
Guo, Y., Wang, Y., Wang, Q., and Wang, Z. (2014). The role of PPK26 in Drosophila larval mechanical nociception. Cell Rep. 9, 1183–1190. doi: 10.1016/j.celrep.2014.10.020
Hallem, E. A., and Carlson, J. R. (2004). The odor coding system of Drosophila. Trends Genet. 20, 453–459. doi: 10.1016/j.tig.2004.06.015
Hallem, E. A., Nicole Fox, A., Zwiebel, L. J., and Carlson, J. R. (2004). Olfaction: mosquito receptor for human-sweat odorant. Nature 427, 212–213. doi: 10.1038/427212a
Hill, S. R., Majeed, S., and Ignell, R. (2015). Molecular basis for odorant receptor tuning: a short C-terminal sequence is necessary and sufficient for selectivity of mosquito Or8. Insect Mol. Biol. 24, 491–501. doi: 10.1111/imb.12176
Jiang, X., Pregitzer, P., Grosse-Wilde, E., Breer, H., and Krieger, J. (2016). Identification and characterization of two “sensory neuron membrane proteins” (SNMPs) of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae). J. Insect Sci. 16:33. doi: 10.1093/jisesa/iew015
Jin, X., Ha, T. S., and Smith, D. P. (2008). SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. 105, 10996–11001. doi: 10.1073/pnas.0803309105
Johnson, W. A., and Carder, J. W. (2012). Drosophila nociceptors mediate larval aversion to dry surface environments utilizing both the painless TRP channel and the DEG/ENaC subunit, PPK1. PLoS ONE 7:e32878. doi: 10.1371/journal.pone.0032878
Joseph, R. M., and Carlson, J. R. (2015). Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31, 683–695. doi: 10.1016/j.tig.2015.09.005
Jung, J. W., Baeck, S. J., Perumalsamy, H., Hansson, B. S., Ahn, Y. J., and Kwon, H. W. (2015). A novel olfactory pathway is essential for fast and efficient blood-feeding in mosquitoes. Sci. Rep. 5:13444. doi: 10.1038/srep13444
Kal, A. J., van Zonneveld, A. J., Benes, V., van den Berg, M., Koerkamp, M. G., Albermann, K., et al. (1999). Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol. Cell Biol. 10, 1859–1872. doi: 10.1091/mbc.10.6.1859
Kang, K., Pulver, S. R., Panzano, V. C., Chang, E. C., Griffith, L. C., Theobald, D. L., et al. (2010). Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464, 597–600. doi: 10.1038/nature08848
Kent, L. B., Walden, K. K., and Robertson, H. M. (2008). The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem. Senses 33, 79–93. doi: 10.1093/chemse/bjm067
Kim, S. E., Coste, B., Chadha, A., Cook, B., and Patapoutian, A. (2012). The role of Drosophila Piezo in mechanical nociception. Nature 483, 209–212. doi: 10.1038/nature10801
Kim, S. H., Lee, Y., Akitake, B., Woodward, O. M., Guggino, W. B., and Montell, C. (2010). Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc. Natl. Acad. Sci. 107, 8440–8445. doi: 10.1073/pnas.1001425107
Klowden, M. J., and Blackmer, J. L. (1987). Humoral control of pre-oviposition behaviour in the mosquito, Aedes aegypti. J. Insect Physiol. 33, 689–692. doi: 10.1016/0022-1910(87)90052-7
Ko, K. I., Root, C. M., Lindsay, S. A., Zaninovich, O. A., Shepherd, A. K., Wasserman, S. A., et al. (2015). Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. eLife 4:e08298. doi: 10.7554/eLife.08298
Kwon, Y., Kim, S. H., Ronderos, D. S., Lee, Y., Akitake, B., Woodward, O. M., et al. (2010). Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Curr. Biol. 20, 1672–1678. doi: 10.1016/j.cub.2010.08.016
Larter, N. K., Sun, J. S., and Carlson, J. R. (2016). Organization and function of Drosophila odorant binding proteins. Elife 5:e20242. doi: 10.7554/eLife.20242
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Ann. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Liesch, J., Bellani, L. L., and Vosshall, L. B. (2013). Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Neglect. Tropic. Dis. 7:e2486. doi: 10.1371/journal.pntd.0002486
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2 - ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, T., Qiu, Y. T., Wang, G., Kwon, J. Y., Rutzler, M., Kwon, H. W., et al. (2007). Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544. doi: 10.1016/j.cub.2007.07.062
Majeed, S., Hill, S. R., Birgersson, G., and Ignell, R. (2016). Detection and perception of generic host volatiles by mosquitoes modulate host preference: context dependence of (R)-1-octen-3-ol. R. Soc. Open Sci. 3:160467. doi: 10.1098/rsos.160467
Majeed, S., Hill, S. R., Dekker, T., and Ignell, R. (2017). Detection and perception of generic host volatiles by mosquitoes: responses to CO 2 constrains host-seeking behaviour. R. Soc. Open Sci. 4, 170–189. doi: 10.1098/rsos.170189
Majeed, S., Hill, S. R., and Ignell, R. (2014). Impact of elevated CO2 background levels on the host-seeking behaviour of Aedes aegypti. J. Exp. Biol. 217, 598–604. doi: 10.1242/jeb.092718
Manoharan, M., Ng Fuk Chong, M., Vaïtinadapoulé, A., Frumence, E., Sowdhamini, R., and Offmann, B. (2013). Comparative genomics of odorant binding proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. Genome Biol. Evol. 5, 163–180. doi: 10.1093/gbe/evs131
Matthews, B. J., McBride, C. S., DeGennaro, M., Despo, O., and Vosshall, L. B. (2016). The neurotranscriptome of the Aedes aegypti mosquito. BMC Genom. 6:32. doi: 10.1186/s12864-015-2239-0
Mayoral, J. G., Nouzova, M., Brockhoff, A., Goodwin, M., Hernandez-Martinez, S., Richter, D., et al. (2010). Allatostatin-C receptors in mosquitoes. Peptides 31, 442–450. doi: 10.1016/j.peptides.2009.04.013
McBride, C. S., Baier, F., Omondi, A. B., Spitzer, S. A., Lutomiah, J., Sang, R., et al. (2014). Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515, 222–227. doi: 10.1038/nature13964
McIver, S., and Charlton, C. (1970). Studies on the sense organs on the palps of selected culicine mosquitoes. Canad. J. Zool. 48, 293–295. doi: 10.1139/z70-048
McMeniman, C. J., Corfas, R. A., Matthews, B. J., Ritchie, S. A., and Vosshall, L. B. (2014). Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 156, 1060–1071. doi: 10.1016/j.cell.2013.12.044
Mescher, M. C., and De Moraes, C. M. (2017). Editorial overview: ecology: the chemical ecology of human disease transmission by mosquito vectors. Curr. Opin. Insect Sci. 20, v–vi. doi: 10.1016/j.cois.2017.04.003
Moll, P., Ante, M., Seltze, A., and Reda, T. (2014). QuantSeq 3 [prime] mRNA sequencing for RNA quantification. Nat. Methods 11, i–iii. doi: 10.1038/nmeth.f.376
Montell, C., and Zwiebel, L. J. (2016). Mosquito sensory systems. Adv. Insect Physiol. 51, 293–328. doi: 10.1016/bs.aiip.2016.04.007
Mysore, K., Andrews, E., Li, P., and Duman-Scheel, M. (2014). Chitosan/siRNA nanoparticle targeting demonstrates a requirement for single-minded during larval and pupal olfactory system development of the vector mosquito Aedes aegypti. BMC Dev. Biol. 14:9. doi: 10.1186/1471-213X-14-9
Nakagawa, T., and Vosshall, L. B. (2009). Controversy and consensus: noncanonical signaling mechanisms in the insect olfactory system. Curr. Opin. Neurobiol. 19, 284–292. doi: 10.1016/j.conb.2009.07.015
Nichols, Z., and Vogt, R. G. (2008). The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 398–415. doi: 10.1016/j.ibmb.2007.11.003
Ohno, H., Yoshida, M., Sato, T., Kato, J., Miyazato, M., Kojima, M., et al. (2017). Luqin-like RYamide peptides regulate food-evoked responses in C. elegans. Elife 296:e28877. doi: 10.7554/eLife.28877
Olsen, S. R., and Wilson, R. I. (2008). Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Neuron 452, 956–962. doi: 10.1038/nature06864
Omondi, B. A., Majeed, S., and Ignell, R. (2015). Functional development of carbon dioxide detection in the maxillary palp of Anopheles gambiae. J. Exp. Biol. 218, 2482–2488. doi: 10.1242/jeb.116798
Parra, G., Bradnam, K., and Korf, I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067. doi: 10.1093/bioinformatics/btm071
Pelosi, P., Calvello, M., and Ban, L. (2005). Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 30:i291. doi: 10.1093/chemse/bjh229
Pitts, R. J., Derryberry, S. L., Zhang, Z., and Zwiebel, L. J. (2017). Variant ionotropic receptors in the malaria vector mosquito Anopheles gambiae tuned to amines and carboxylic acids. Sci. Rep. 7:40297. doi: 10.1038/srep40297
Rinker, D. C., Pitts, R. J., Zhou, X., Suh, E., Rokas, A., and Zwiebel, L. J. (2013). Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc. Natl. Acad. Sci. 110, 8260–8265. doi: 10.1073/pnas.1302562110
Root, C. M., Masuyama, K., Green, D. S., Enell, L. E., Nässel, D. R., Lee, C. H., et al. (2008). A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron 59, 311–321. doi: 10.1016/j.neuron.2008.07.003
Sargsyan, V., Getahun, M. N., Llanos, S. L., Olsson, S. B., Hansson, B. S., and Wicher, D. (2011). Phosphorylation via PKC regulates the function of the Drosophila odorant co-receptor. Front. Cell. Neurosci. 5:5. doi: 10.3389/fncel.2011.00005
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Seenivasagan, T., Sharma, K. R., Sekhar, K., Ganesan, K., Prakash, S., and Vijayaraghavan, R. (2009). Electroantennogram, flight orientation, and oviposition responses of Aedes aegypti to the oviposition pheromone n-heneicosane. Parasitol. Res. 104, 827–833. doi: 10.1007/s00436-008-1263-2
Shakeel, M., Rodriguez, A., Tahir, U. B., and Jin, F. (2017). Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 9, 1–10. doi: 10.1007/s10529-017-2465-4
Siju, K. P., Hansson, B. S., and Ignell, R. (2008). Immunocytochemical localization of serotonin in the central and peripheral chemosensory system of mosquitoes. Arthropod Struct. Dev. 37, 248–259. doi: 10.1016/j.asd.2007.12.001
Smallegange, R. C., Qiu, Y. T., van Loon, J. J., and Takken, W. (2005). Synergism between ammonia, lactic acid and carboxylic acids as kairomones in the host-seeking behaviour of the malaria mosquito Anopheles gambiae sensu stricto (Diptera: Culicidae). Chem. Senses 30, 145–152. doi: 10.1093/chemse/bji010
Sparks, J. T., Bohbot, J. D., and Dickens, J. C. (2014). The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem. Mol. Biol. 48, 8–16. doi: 10.1016/j.ibmb.2014.02.004
Stengl, M. (2010). Pheromone transduction in moths. Front. Cell. Neurosci. 4:133. doi: 10.3389/fncel.2010.00133
Stengl, M., and Funk, N. W. (2013). The role of the coreceptor Orco in insect olfactory transduction. J. Compar. Physiol. A 199, 897–909. doi: 10.1007/s00359-013-0837-3
Takken, W., and Knols, B. G. (1999). Odor-mediated behavior of Afrotropical malaria mosquitoes. Ann. Rev. Entomol. 44, 131–157. doi: 10.1146/annurev.ento.44.1.131
Taparia, T., Ignell, R., and Hill, S. R. (2017). Blood meal induced regulation of the chemosensory gene repertoire in the southern house mosquito. BMC Genom. 18:393. doi: 10.1186/s12864-017-3779-2
Tracey, W. D., Wilson, R. I., Laurent, G., and Benzer, S. (2003). painless, a Drosophila gene essential for nociception. Cell 113, 261–273. doi: 10.1016/S0092-8674(03)00272-1
Vogt, R. G. (2003). “Biochemical diversity of odor detection: OBPs, ODEs and SNMPs,” in Insect Pheromone Biochemistry and Molecular Biology, eds G. J. Blomquist and R. G. Vogt (Amsterdam: Elsevier), 391–445. doi: 10.1016/B978-012107151-6/50016-5
Vogt, R. G., Callahan, F. E., Rogers, M. E., Dickens, J. C., Vogt, R. G., Callahan, F. E., et al. (1999). Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris (Hemiptera, Heteroptera). Chem. Senses 24, 481–495. doi: 10.1093/chemse/24.5.481
Vogt, R. G., Miller, N. E., Litvack, R., Fandino, R. A., Sparks, J., Staples, J., et al. (2009). The insect SNMP gene family. Insect Biochem. Mol. Biol. 39, 448–456. doi: 10.1016/j.ibmb.2009.03.007
Vogt, R. G., and Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Walker, R. G., Willingham, A. T., and Zuker, C. S. (2000). A Drosophila mechanosensory transduction channel. Science 287, 2229–2234. doi: 10.1126/science.287.5461.2229
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. 107, 4418–4423. doi: 10.1073/pnas.0913392107
Wicher, D., Schäfer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Xu, P., Choo, Y. M., Pelletier, J., Sujimoto, F. R., Hughes, D. T., Zhu, F., et al. (2013). Silent, generic and plant kairomone sensitive odorant receptors from the Southern house mosquito. J. Insect Physiol. 59, 961–966. doi: 10.1016/j.jinsphys.2013.07.004
Yao, C. A., and Carlson, J. R. (2010). Role of G-proteins in odor-sensing and CO2-sensing neurons in Drosophila. J. Neurosci. 30, 4562–4572. doi: 10.1523/JNEUROSCI.6357-09.2010
Yao, C. A., Ignell, R., and Carlson, J. R. (2005). Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J. Neurosci. 25, 8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005
Younus, F., Chertemps, T., Pearce, S. L., Pandey, G., Bozzolan, F., Coppin, C. W., et al. (2014). Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 53, 30–43. doi: 10.1016/j.ibmb.2014.07.003
Zelle, K. M., Lu, B., Pyfrom, S. C., and Ben-Shahar, Y. (2013). The genetic architecture of degener in epithelial sodium channels in Drosophila. Genes Genomes Genet. 3, 441–450. doi: 10.1534/g3.112.005272
Keywords: chemosensory, mosquito, transcriptome, host seeking, blood feeding, resting
Citation: Hill SR, Ghaninia M and Ignell R (2019) Blood Meal Induced Regulation of Gene Expression in the Maxillary Palps, a Chemosensory Organ of the Mosquito Aedes aegypti. Front. Ecol. Evol. 7:336. doi: 10.3389/fevo.2019.00336
Received: 30 May 2019; Accepted: 23 August 2019;
Published: 10 September 2019.
Edited by:
Aijun Zhang, United States Department of Agriculture (USDA), United StatesReviewed by:
Jonathan Daniel Bohbot, Hebrew University of Jerusalem, IsraelJackson Sparks, High Point University, United States
Copyright © 2019 Hill, Ghaninia and Ignell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharon R. Hill, c2hhcm9uLmhpbGxAc2x1LnNl
 Sharon R. Hill
Sharon R. Hill Majid Ghaninia
Majid Ghaninia Rickard Ignell
Rickard Ignell