- Centre for Molecular Therapeutics, Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD, Australia
The phylum Cnidaria contains a wide variety of unique organisms that possess interesting adaptations evolved over many years to help them survive in a competitive environment. One of these adaptations is the presence of venom, which has been of particular interest for studies aimed at identifying novel drug leads and for understanding the mechanisms involved in envenomation. The potency of the venom varies significantly amongst cnidarians, and although corals are often overshadowed by the jellyfish and sea anemone toxins, they also possess a range of interesting bioactive compounds. In this mini-review, we provide an overview of the toxins present in corals, highlighting the diverse structures and bioactivities.
Introduction
The organisms in the phylum Cnidaria represent some of the oldest living venomous creatures on the planet, including jellyfish, hydroids, sea anemones, and corals (Rachamim et al., 2015). The phylum is primarily defined by the presence of nematocytes, or stinging cells, in the tissue of these organisms (Ozbek, 2011). Nematocyte cells contain a tubule with a capsule of venom that, when stimulated, is everted, striking the prey/predator, penetrating the outer membrane and delivering the venom (Ozbek, 2011). These stinging cells have a variety of purposes but are mainly used for prey capture and defense (Greenwood, 2009).
Venoms often contain a complex mixture of compounds, including small molecules, peptides and proteins. These compounds can be highly potent and specific for biological targets, and the peptides in venoms are generally stable because of the presence of disulfide bonds making them of interest in drug design (Vetter et al., 2011; Utkin, 2015). Venomous creatures, such as spiders, scorpions, and cone snails have been well-studied and several databases have been established to collate the data on the toxins present (Kaas et al., 2008, 2012; Kuzmenkov et al., 2016; Pineda et al., 2018). Although cnidarians are generally less well-studied (Macrander et al., 2018), information about the toxins present is currently expanding.
Perhaps the most well-known of the cnidarian organisms are jellyfish. Their venom can be extremely potent and act, not only on small marine prey organisms, but can also have severe physiological effects on humans (Tibballs, 2006; Tibballs et al., 2011; Remigante et al., 2018). Although not as harmful as some jellyfish, other cnidarians, such as select sea anemones can elicit a stinging sensation in humans when the nematocytes in the tentacles are stimulated (Lubbock et al., 1981; Garcia-Arredondo et al., 2016). Several sea anemone toxins have been well-characterized, including an analog of a ShK toxin from Stichodactyla helianthus, which has entered Phase 2 trials for autoimmune diseases (Pennington et al., 2009; Chi et al., 2012; Prentis et al., 2018). There have been several recent reviews of sea anemone toxins regarding their bioactivity as well as their potential uses in the field of pharmaceutical development (Prentis et al., 2018; Thangaraj et al., 2018; Madio et al., 2019; Utkin et al., 2019). Corals are often overshadowed by the highly potent and potentially life-threatening toxins from jellyfish and clinically relevant sea anemones, but they too possess toxins of interest.
Much of the research on corals has focused on climate change impacts and secondary metabolites. In the past year alone there have been more than 3,750 journal articles published on “corals and climate change” (GoogleScholar) while there are around 388 articles published on “corals and toxins” (2 August 2019). The majority of research on corals and their secondary metabolites is not necessarily venom related. For example, metabolites from the gorgonian coral Erythropodium caribaeorum have been shown to act as a deterrent against reef fishes predating on the coral (Fenical and Pawlik, 1991). However, more recent studies have characterized venom derived coral toxins, with potential in the field of drug development (Radwan et al., 2002; Frazao et al., 2012; Rodriguez et al., 2012; Garcia-Arredondo et al., 2016). There is significant scope for future studies aimed at characterizing coral toxins, and in this mini review we highlight some of the structural and functional diversity that has already been uncovered.
Coral Toxins
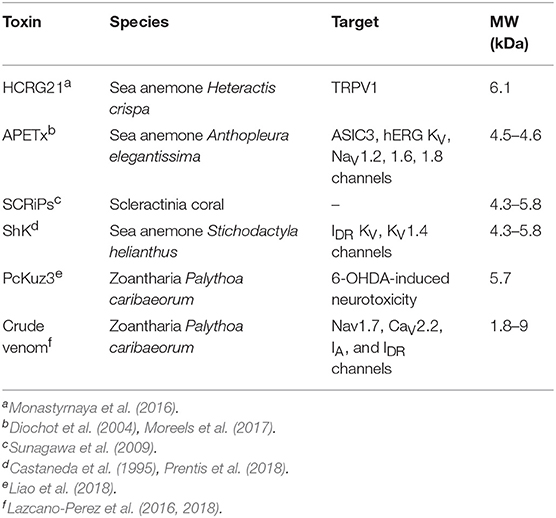
Corals, primarily grouped into stony corals and soft corals, are members of the Anthozoa class of the phylum Cnidaria as shown in the phylogenetic tree in Figure 1. Toxins have been characterized from four of the Anthozoa order and examples of these toxins are given in Table 1 to highlight the structural diversity, range of bioactivities and potential applications. The majority of research into toxins from organisms in the Anthozoa class has focused on sea anemones, because of the exciting therapeutic potential of some of the toxins, and several recent reviews have been published in this area (Prentis et al., 2018; Liao et al., 2019; Madio et al., 2019). Although there are distinct differences between sea anemones and corals, with stony corals having calcium carbonate skeletons in contrast to sea anemones (Shick, 1991; Stanley, 2003), the two organisms are in the same class and might have similar compounds to each other. We are only just beginning to appreciate the diversity of compounds present in the nematocysts of corals and potentially toxins present elsewhere in the tissue of these organisms. There is evidence that toxins can be delivered from anatomical structures other than nematocytes in sea anemones, with differences in localization between species (Moran et al., 2012; Bastos et al., 2016). It is possible a similar phenomenon also occurs in corals. While sea anemone toxins, such as the ShK toxins have not been found in corals to date, there are examples of other toxins and toxin families as outlined below for stony corals and soft corals and highlights the importance of characterizing coral toxins.
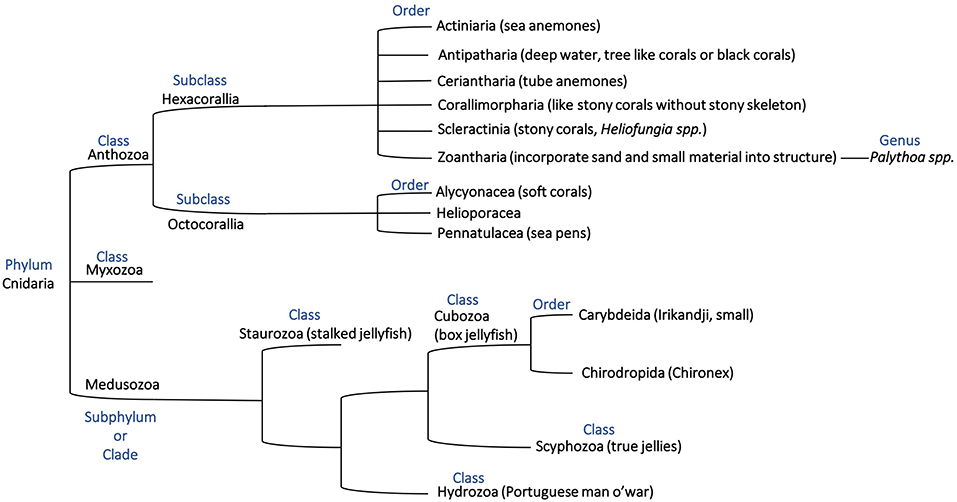
Figure 1. Phylogenetic tree representing the phylum Cnidaria based on previous descriptions (Bridge et al., 1992, 1995; Berntson et al., 1999; Marques and Collins, 2004; Collins et al., 2006; Daly et al., 2007; Jimenez-Guri et al., 2007; Feng et al., 2014; Zapata et al., 2015) and showing the relationship of organisms discussed in this mini-review.
Scleractinia (Stony Coral)
Stony corals (Scleractinia) are reef building corals that absorb calcium carbonate from the water to form a hard skeleton, and occur in colonial or solitary aggregates (Stanley, 2003). The most well-characterized toxins from stony corals are a family of peptides termed small cysteine-rich peptides (SCRiPs) found in the ectoderm of a common stony coral, Acropora millepora (Sunagawa et al., 2009). SCRiPs contain a conserved eight cysteine framework, similar to the rattlesnake myotoxin domain in crotamine toxins (Jouiaei et al., 2015a) but there is limited sequence conservation beyond the cysteine residues. SCRiPs were originally thought to be found strictly in Scleractinia corals but recent studies have shown that homologs of SCRiPs are found in the sea anemones Anemonia viridis and Metridium senile (Jouiaei et al., 2015a). SCRiPS were also originally thought to be involved with biomineralization in reef building corals by playing a role in calcification of the skeleton of the coral but there is now evidence that they are a family of toxins rather than calcifying proteins (Jouiaei et al., 2015a). Two SCRiPs originally found in the ectoderm of the coral A. millepora were recombinantly expressed and incubated with zebrafish larvae. The larvae became paralyzed and insensitive to touch, consistent with neurotoxic action (Jouiaei et al., 2015a). The presence of SCRiPS in the ectoderm of A. millepora is also consistent with a role in prey envenomation as the ectoderm is lined by nematocytes (Grasso et al., 2011; Jouiaei et al., 2015a). Overall, SCRiPS remain of interest for further research because of their interesting framework and their presence in a variety of Anthozoa organisms.
It appears likely that SCRiPs are not the only toxins present in stony corals. Analysis of extracts from 11 different Scleractinian coral families collected from Heron Island on the Great Barrier Reef showed variable levels of toxicity in several assays including mice toxicity, haemolytic activity, and antimicrobial activity (Gunthorpe and Cameron, 1990). While the toxicity was variable, the majority of the species tested displayed some level of toxicity. Although the compounds responsible for the bioactivity were not characterized in this study, a recent study on the analysis of extracts from the nematocysts of three stony corals (Scleractinia corals), Pseudodiploria strigosa, Porites astreoides, and Siderastrea sidereal indicated the presence of a range of toxins and provided insight into the chemical composition. Extracts from all three corals were lethal to crickets, had haemolytic and nociceptive activity to varying extents, and exhibited PLA2 and serine protease activities (Garcia-Arredondo et al., 2016). Interestingly, although these corals are not considered harmful to humans, the activity of these extracts is consistent with the physiological effects caused in humans by some hydroids, such as Millepora alcicornis and Millepora complanate, where the toxins work as lysins on erythrocytes (Garcia-Arredondo et al., 2016). Analysis of the extracts with SDS-PAGE indicated the presence of a broad range of proteins that differ under reducing conditions, and mass spectrometry analysis of a low molecular weight fraction indicated the presence of peptides with molecular weights in the range 3,000–6,000 Da. These peptide fractions were subsequently shown to be lethal to crickets and cause vasoconstriction. Further study is required to characterize these toxins, but it could be possible that some will have similarity to the SCRiPs family or the proteins with protease activity might have similarity to proteases present in other venoms, such as snake venom.
Toxins have also been predicted from the proteomics analysis of proteins discharged from nematocysts the stony coral Acropora digitifera, and the genome of this organism (Gacesa et al., 2015). A total of 55 potential toxins were predicted based on the genome but only 12 were found based on the proteomic analysis of the nematocyst extracts (Gacesa et al., 2015). These toxins are suggested to be phospholipases and toxic peptidases based on their similarity to other known toxins found on Tox-Prot (Gacesa et al., 2015). Furthermore, an haemolytic toxin from the actinoporin family has recently been characterized in Stylophora pistilata and was suggested to be a non-nematocyst protein (Ben-Ari et al., 2018).
Alcyonacea (Soft Coral)
Soft corals (Alcyonacea), formerly known as gorgonian corals, contrast stony corals in that they do no create a calcium carbonate skeleton (Alarif et al., 2019). In a similar study to the Scleractinia coral extract analyses, Radwan et. al demonstrated the effects of venom from three different soft corals on mice. The corals, Nephthea sp., Dendronephthya sp., and Heteroxenia fuscescens, were collected from the Red Sea and are known to cause a stinging effect in humans (Radwan et al., 2002). The data showed that extracts from nematocysts of all three corals resulted in fractions with bioactive effects including lethality to mice, haemolysis, vasopermeability, or dermonecrosis, with toxins from H. fuscescens the most lethal to mice (Radwan et al., 2002). Similar to the studies on stony corals, SDS-PAGE analysis of the venom extracts indicates the presence of a wide range of proteins ranging from ~200 kDa to <6,000 Da. The bioactivity was not restricted to one class of protein, with two fractions from the H. fuscescens extract showing potent haemolytic activity with one fraction containing a protein of 116 kDa and the other containing a peptide of <6 kDa. The addition of a variety of lipid membrane components to the venom followed by addition of this mixture to human red blood cells instigated a protective response of the cells against the crude venom toxins (Radwan et al., 2002). The inhibition of a physiological response in the cells suggests that the binding site is occupied, with the most effective inhibition occurring by the addition of dihydrocholesterol (Radwan et al., 2002). Occupation of the binding site prevents the toxin binding and therefore eliciting a physiological response on the cells. Furthermore, this research also tested the mice for antibody production. Mice injected with a dose of venom, and provided with boosters throughout the study, produced an immune response in 15 days with high levels of antibodies present in the blood (Radwan et al., 2002). Further studies are required to fully characterize the bioactive peptides and proteins present in the extracts.
Non-proteinaceous toxins are also present in soft corals. For example, the small molecule toxin sarcophine was isolated from the soft coral Sarcophyton glaucum and is toxic to fish as well as mice, rats and guinea pigs (Ne'eman et al., 1974). Ingestion by the animal led to a decrease in cardiac and pulmonary function as well as motor function and body temperature of the animals (Ne'eman et al., 1974). Using guinea pig ileum, it was shown that sarcophine acts as a competitive inhibitor of cholinesterase (Ne'eman et al., 1974). This coral was originally studied for its ecological characteristics when Ne'eman et al. observed that the fish in the area were not predating on this specific coral (Ne'eman et al., 1974) and subsequent studies lead to the characterization of sarcophine.
There have also been large scale studies on toxins of soft corals found on the Great Barrier Reef in which 136 different specimens from 15 different genera were analyzed (Coll et al., 1982). In this study two genera were found to be the most toxic and lethal to the fish species tested: Lemnalia and Sarcophyton (Coll et al., 1982). The different genera of coral tested exhibited a large range of affects, from no noticeable effect on the fish to causing death. This study and the studies on sarcophine involved extraction of coral tissue rather than nematocysts extracts, which makes it difficult to identify where the compound comes from in the coral, but these studies demonstrate the diversity of the compounds from corals and the potent activity they can possess.
Evolution of Coral Toxins
Insight into the evolution of coral toxins is primarily based on the SCRiP family of peptides, as these are the most well-characterized to date. As mentioned above, in contrast to the original suggestion, SCRiPS are not only found in corals but have been found in the sea anemones Anemonia viridis and Metridium senile (Jouiaei et al., 2015b). The toxin τ-AnmTx Ueq 12-1 isolated from the sea anemone Urticina eques also shows similarity to SCRiPs, in particular one cDNA matched the SCRiP Anthopleura elegantissima comp63456_c0_seq1 (Logashina et al., 2017). The presence of SCRiPs in corals and sea anemones suggests that these proteins evolved more than 500 million years ago, the estimated time when coral diverged from sea anemones (Shinzato et al., 2011). Furthermore, molecular evolutionary assessments indicate that coral SCRiPs have evolved under negative selection as no sites were found that were positively selected based on the Bayes Empirical Bayes approach (Jouiaei et al., 2015a). The role of negative selection in coral toxin evolution is supported by studies on the toxins of Acropora digitifera and Stylophora pistilata (Gacesa et al., 2015; Ben-Ari et al., 2018). Interestingly, this appears to be a general phenomenon for venoms of ancient lineages, whereas toxins from lineages that have evolved more recently appear to evolve under positive selection (Lynch, 2007; Casewell et al., 2011, 2012; Sunagar et al., 2012, 2013, 2014; Brust et al., 2013; Dutertre et al., 2014; Jouiaei et al., 2015a; Sunagar and Moran, 2015).
Given the large number of toxins that appear to be present in coral venom, further molecular characterization is likely to provide further insight into the evolution of this ancient lineage. In particular, characterization of some of the larger toxins might provide insight into origins of a range of toxins, as analysis of the venom from the sea anemone Stichodactyla haddoni showed that some venom peptides have similar sequences to housekeeping proteins involved in regulatory biological functions (Madio et al., 2017). This is a common trend across many venomous taxa because it is suggested that the main ways that toxins are recruited is via gene modification of regulatory proteins, such as sequence duplications (Fry et al., 2009). It has been shown that cnidarian organisms rely on similar structural frameworks of their toxins and then modify these toxins for activity on specific targets (Honma and Shiomi, 2006; Prentis et al., 2018). Because of this evolution from non-toxin related proteins, we see variability in the size of peptides found in nematocyte venom as the evolution of each peptide differs greatly (Table 1).
Despite the similarities between cnidarian organisms, such as coral and sea anemones, it is also likely that significant differences in the evolution of toxins will be found based on analysis of proteins found in the nematocysts of organisms from three different classes of Cnidaria, namely Anthozoa, Scyphozoa, and Hydrozoa (Rachamim et al., 2015). The organisms analyzed from these three classes were the sea anemone Anemonia viridis (Anthozoa), the jellyfish Aurelia aurita (Scyphozoa) and the hydrozoan Hydra magnipapillata (Hydrozoa). Although this analysis led to the identification of hundreds of proteins, only six proteins were common in all three species (Rachamim et al., 2015). Of these, most were structural proteins and only one of the six proteins common to all three species, the dickkopf protein, is predicted to function as a toxin (Rachamim et al., 2015). The A. aurita and H. magnipapillata venom showed the most similarities, mainly composed of cytotoxins and enzymes, while the A. viridis venom proteome composition was predominantly related to peptide neurotoxins (Rachamim et al., 2015). The general lack of conservation across these cnidarians might point to significant evolutionary differences and further cnidarian toxins promises to provide interesting insights into toxin evolution in general.
Challenges in Cnidarian Toxin Analyses
In the study of venomous creatures, such as spiders and cone snails it can often be quite straightforward to isolate the venom with limited contamination from the environment or other tissues. Indeed, Australian funnel-web spiders (Atracidae) can release microlitres of venom onto their fangs that can be “easily” recovered (Wilson and Alewood, 2004). However, for corals and cnidarian organisms in general this is not always the case, and can complicate the toxin extraction process (Garcia-Arredondo et al., 2016). A range of extraction methods have been used in the analysis of corals and sea anemones, including extraction of the toxins from the nematocyte in the tissue of the coral (Garcia-Arredondo et al., 2016) and homogenization of the whole tentacle of the sea anemone (Prentis et al., 2018). Homogenization of the whole tentacle will clearly yield more than just venom toxins, but even extraction of the toxins from the nematocyst can be complicated, given the small size of the nematocysts. Furthermore, it can be difficult to separate out the calcareous skeleton of the coral from the tissue itself (Garcia-Arredondo et al., 2016). The cellular contents can have implications for interpretation of the results because some components, such as minicollagens share characteristics similar to the cysteine-rich toxins of interest (Madio et al., 2017).
The difficulties in defining the origin of compounds, either from the nematocyst or other tissue, has significant implications for elucidating evolutionary relationships for these toxins. In particular, it is difficult to determine a common ancestor (Kayal et al., 2018). Genome and transcriptomic analyses are likely to provide further insight into the evolution of cnidarian toxins. Using an integrative approach of both genomic and transcriptomic analyses allows for a better understanding of the active toxins found in organisms compared to the potential toxins seen in the genome and will also allow comparison with other toxins from a range of venomous creatures.
Conclusion
Coral venoms are a source of interesting novel bioactive molecules with significant scope for further characterization of novel toxins. Further application of analysis technologies (e.g., genomics, transcriptomics, and proteomics) is likely to significantly enhance the knowledge in this field and identify novel classes of peptides/proteins. The well-characterized coral toxins, SCRiPs, have now been identified as likely neurotoxins, but given the highly microbial environment in which corals exist, further analysis of the nematocyst components of corals is likely to provide a new and unique source of antimicrobial molecules. Studies on coral extracts have indicated that such compounds exist. In addition, determining the composition of peptides that make up the venom from corals may provide insight into the overlap and differences between cnidarian groups.
Author Contributions
CS, ND, and DW wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alarif, W. M., Abdel-Lateff, A., Alorfi, H. S., and Alburae, N. A. (2019). Alcyonacea: a potential source for production of nitrogen-containing metabolites. Molecules 24:286. doi: 10.3390/molecules24020286
Bastos, C. L. Q., Varela, A. S., Ferreira, S. P., Nornberg, B. F., and Boyle, R. T. (2016). Who knows not where an anemone does wear his sting? Could polypeptides released from the columnar vesicles of Bunodosoma cangicum induce apoptosis in the ZF-L cell line? Toxicon 124, 73–82. doi: 10.1016/j.toxicon.2016.10.014
Ben-Ari, H., Paz, M., and Sher, D. (2018). The chemical armament of reef-building corals: inter- and intra-specific variation and the identification of an unusual actinoporin in Stylophora pistilata. Sci. Rep. 8:251. doi: 10.1038/s41598-017-18355-1
Berntson, E. A., France, S. C., and Mullineaux, L. S. (1999). Phylogenetic relationships within the class Anthozoa (phylum Cnidaria) based on nuclear 18S rDNA sequences. Mol. Phylogenet. Evol. 13, 417–433. doi: 10.1006/mpev.1999.0649
Bridge, D., Cunningham, C. W., DeSalle, R., and Buss, L. W. (1995). Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol. Biol. Evol. 12, 679–689. doi: 10.1093/oxfordjournals.molbev.a040246
Bridge, D., Cunningham, C. W., Schierwater, B., DeSalle, R., and Buss, L. W. (1992). Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc. Natl. Acad. Sci. U.S.A. 89, 8750–8753. doi: 10.1073/pnas.89.18.8750
Brust, A., Sunagar, K., Undheim, E. A., Vetter, I., Yang, D. C., Casewell, N. R., et al. (2013). Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol. Cell. Proteomics 12, 651–663. doi: 10.1074/mcp.M112.023135
Casewell, N. R., Huttley, G. A., and Wuster, W. (2012). Dynamic evolution of venom proteins in squamate reptiles. Nat. Commun. 3:1066. doi: 10.1038/ncomms2065
Casewell, N. R., Wagstaff, S. C., Harrison, R. A., Renjifo, C., and Wuster, W. (2011). Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 28, 2637–2649. doi: 10.1093/molbev/msr091
Castaneda, O., Sotolongo, V., Amor, A. M., Stocklin, R., Anderson, A. J., Harvey, A. L., et al. (1995). Characterization of a potassium channel toxin from the Caribbean Sea anemone Stichodactyla helianthus. Toxicon 33, 603–613. doi: 10.1016/0041-0101(95)00013-C
Chi, V., Pennington, M. W., Norton, R. S., Tarcha, E. J., Londono, L. M., Sims-Fahey, B., et al. (2012). Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon 59, 529–546. doi: 10.1016/j.toxicon.2011.07.016
Coll, J. C., Labarre, S., Sammarco, P. W., Williams, W. T., and Bakus, G. J. (1982). Chemical defenses in soft corals (Coelenterata, Octocorallia) of the great barrier-reef–a study of comparative toxicities. Mar. Ecol. Prog. Ser. 8, 271–278. doi: 10.3354/meps008271
Collins, A. G., Schuchert, P., Marques, A. C., Jankowski, T., Medina, M., and Schierwater, B. (2006). Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst. Biol. 55, 97–115. doi: 10.1080/10635150500433615
Daly, M., Brugler, M. R., Cartwright, P., Collins, A. G., Dawson, M. N., Fautin, D. G., et al. (2007). The phylum Cnidaria: a review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 1668, 127–182. doi: 10.11646/zootaxa.1668.1.11
Diochot, S., Baron, A., Rash, L. D., Deval, E., Escoubas, P., Scarzello, S., et al. (2004). A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 23, 1516–1525. doi: 10.1038/sj.emboj.7600177
Dutertre, S., Jin, A. H., Vetter, I., Hamilton, B., Sunagar, K., Lavergne, V., et al. (2014). Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 5:3521. doi: 10.1038/ncomms4521
Feng, J. M., Xiong, J., Zhang, J. Y., Yang, Y. L., Yao, B., Zhou, Z. G., et al. (2014). New phylogenomic and comparative analyses provide corroborating evidence that Myxozoa is Cnidaria. Mol. Phylogenet. Evol. 81, 10–18. doi: 10.1016/j.ympev.2014.08.016
Fenical, W., and Pawlik, J. R. (1991). Defensive properties of secondary metabolites from the Caribbean gorgonian coral Erythropodium caribaeorum. Mar. Ecol. Prog. Ser. 75, 1–8. doi: 10.3354/meps075001
Frazao, B., Vasconcelos, V., and Antunes, A. (2012). Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an overview. Mar. Drugs 10, 1812–1851. doi: 10.3390/md10081812
Fry, B. G., Roelants, K., Champagne, D. E., Scheib, H., Tyndall, J. D. A., King, G. F., et al. (2009). The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511. doi: 10.1146/annurev.genom.9.081307.164356
Gacesa, R., Chung, R., Dunn, S. R., Weston, A. J., Jaimes-Becerra, A., Marques, A. C., et al. (2015). Gene duplications are extensive and contribute significantly to the toxic proteome of nematocysts isolated from Acropora digitifera (Cnidaria: Anthozoa: Scleractinia). BMC Genomics 16:774. doi: 10.1186/s12864-015-1976-4
Garcia-Arredondo, A., Rojas-Molina, A., Ibarra-Alvarado, C., Lazcano-Perez, F., Arreguin-Espinosa, R., and Sanchez-Rodriguez, J. (2016). Composition and biological activities of the aqueous extracts of three scleractinian corals from the Mexican Caribbean: Pseudodiploria strigosa, Porites astreoides and Siderastrea siderea. J. Venom. Anim. Toxins Incl. Trop. Dis. 22:32. doi: 10.1186/s40409-016-0087-2
Grasso, L. C., Negri, A. P., Foret, S., Saint, R., Hayward, D. C., Miller, D. J., et al. (2011). The biology of coral metamorphosis: molecular responses of larvae to inducers of settlement and metamorphosis. Dev. Biol. 353, 411–419. doi: 10.1016/j.ydbio.2011.02.010
Greenwood, P. G. (2009). Acquisition and use of nematocysts by cnidarian predators. Toxicon 54, 1065–1070. doi: 10.1016/j.toxicon.2009.02.029
Gunthorpe, L., and Cameron, A. M. (1990). Widespread but variable toxicity in scleractinian corals. Toxicon 28, 1199–1219. doi: 10.1016/0041-0101(90)90120-V
Honma, T., and Shiomi, K. (2006). Peptide toxins in sea anemones: structural and functional aspects. Mar. Biotechnol. 8, 1–10. doi: 10.1007/s10126-005-5093-2
Jimenez-Guri, E., Philippe, H., Okamura, B., and Holland, P. W. H. (2007). Buddenbrockia is a cnidarian worm. Science 317, 116–118. doi: 10.1126/science.1142024
Jouiaei, M., Sunagar, K., Federman Gross, A., Scheib, H., Alewood, P. F., Moran, Y., et al. (2015a). Evolution of an ancient venom: recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol. Biol. Evol. 32, 1598–1610. doi: 10.1093/molbev/msv050
Jouiaei, M., Yanagihara, A. A., Madio, B., Nevalainen, T. J., Alewood, P. F., and Fry, B. G. (2015b). Ancient venom systems: a review on Cnidaria toxins. Toxins (Basel) 7, 2251–2271. doi: 10.3390/toxins7062251
Kaas, Q., Westermann, J. C., Halai, R., Wang, C. K., and Craik, D. J. (2008). ConoServer, a database for conopeptide sequences and structures. Bioinformatics 24, 445–446. doi: 10.1093/bioinformatics/btm596
Kaas, Q., Yu, R., Jin, A. H., Dutertre, S., and Craik, D. J. (2012). ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 40, D325–D330. doi: 10.1093/nar/gkr886
Kayal, E., Bentlage, B., Pankey, M. S., Ohdera, A. H., Medina, M., Plachetzki, D. C., et al. (2018). Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 18:68. doi: 10.1186/s12862-018-1142-0
Kuzmenkov, A. I., Krylov, N. A., Chugunov, A. O., Grishin, E. V., and Vassilevski, A. A. (2016). Kalium: a database of potassium channel toxins from scorpion venom. Database (Oxford) 2016:baw056. doi: 10.1093/database/baw056
Lazcano-Perez, F., Castro, H., Arenas, I., Garcia, D. E., Gonzalez-Munoz, R., and Arreguin-Espinosa, R. (2016). Activity of Palythoa caribaeorum venom on voltage-gated ion channels in mammalian superior cervical ganglion neurons. Toxins (Basel) 8:135. doi: 10.3390/toxins8050135
Lazcano-Perez, F., Zavala-Moreno, A., Rufino-Gonzalez, Y., Ponce-Macotela, M., Garcia-Arredondo, A., Cuevas-Cruz, M., et al. (2018). Hemolytic, anticancer and antigiardial activity of Palythoa caribaeorum venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 24:12. doi: 10.1186/s40409-018-0149-8
Liao, Q., Feng, Y., Yang, B., and Lee, S. M. (2019). Cnidarian peptide neurotoxins: a new source of various ion channel modulators or blockers against central nervous systems disease. Drug Discov. Today 24, 189–197. doi: 10.1016/j.drudis.2018.08.011
Liao, Q., Li, S., Siu, S. W. I., Yang, B., Huang, C., Chan, J. Y., et al. (2018). Novel kunitz-like peptides discovered in the Zoanthid Palythoa caribaeorum through transcriptome sequencing. J. Proteome Res. 17, 891–902. doi: 10.1021/acs.jproteome.7b00686
Logashina, Y. A., Solstad, R. G., Mineev, K. S., Korolkova, Y. V., Mosharova, I. V., Dyachenko, I. A., et al. (2017). New disulfide-stabilized fold provides sea anemone peptide to exhibit both antimicrobial and TRPA1 potentiating properties. Toxins (Basel) 9:154. doi: 10.3390/toxins9050154
Lubbock, R., Gupta, B. L., and Hall, T. A. (1981). Novel role of calcium in exocytosis: mechanism of nematocyst discharge as shown by X-ray microanalysis. Proc. Natl. Acad. Sci. U.S.A. 78, 3624–3628. doi: 10.1073/pnas.78.6.3624
Lynch, V. J. (2007). Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol. Biol. 7:2. doi: 10.1186/1471-2148-7-2
Macrander, J., Panda, J., Janies, D., Daly, M., and Reitzel, A. M. (2018). Venomix: a simple bioinformatic pipeline for identifying and characterizing toxin gene candidates from transcriptomic data. PeerJ 6:e5361. doi: 10.7717/peerj.5361
Madio, B., King, G. F., and Undheim, E. A. B. (2019). Sea anemone toxins: a structural overview. Mar. Drugs 17:E325. doi: 10.3390/md17060325
Madio, B., Undheim, E. A. B., and King, G. F. (2017). Revisiting venom of the sea anemone Stichodactyla haddoni: omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteomics 166, 83–92. doi: 10.1016/j.jprot.2017.07.007
Marques, A. C., and Collins, A. G. (2004). Cladistic analysis of Medusozoa and cnidarian evolution. Invertebr. Biol. 123, 23–42. doi: 10.1111/j.1744-7410.2004.tb00139.x
Monastyrnaya, M., Peigneur, S., Zelepuga, E., Sintsova, O., Gladkikh, I., Leychenko, E., et al. (2016). Kunitz-type peptide HCRG21 from the Sea anemone Heteractis crispa is a full antagonist of the TRPV1 receptor. Mar. Drugs 14:E229. doi: 10.3390/md14120229
Moran, Y., Genikhovich, G., Gordon, D., Wienkoop, S., Zenkert, C., Ozbek, S., et al. (2012). Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc. Biol. Sci. 279, 1351–1358. doi: 10.1098/rspb.2011.1731
Moreels, L., Peigneur, S., Galan, D. T., De Pauw, E., Beress, L., Waelkens, E., et al. (2017). APETx4, a novel sea anemone toxin and a modulator of the cancer-relevant potassium channel KV10.1. Mar. Drugs 15:E287. doi: 10.3390/md15090287
Ne'eman, I., Fishelson, L., and Kashman, Y. (1974). Sarcophine–a new toxin from the soft coral Sarcophyton glaucum (Alcyonaria). Toxicon 12, 593–598. doi: 10.1016/0041-0101(74)90192-5
Ozbek, S. (2011). The cnidarian nematocyst: a miniature extracellular matrix within a secretory vesicle. Protoplasma 248, 635–640. doi: 10.1007/s00709-010-0219-4
Pennington, M. W., Beeton, C., Galea, C. A., Smith, B. J., Chi, V., Monaghan, K. P., et al. (2009). Engineering a stable and selective peptide blocker of the Kv1.3 channel in T lymphocytes. Mol. Pharmacol. 75, 762–773. doi: 10.1124/mol.108.052704
Pineda, S. S., Chaumeil, P. A., Kunert, A., Kaas, Q., Thang, M. W. C., Le, L., et al. (2018). ArachnoServer 3.0: an online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 34, 1074–1076. doi: 10.1093/bioinformatics/btx661
Prentis, P. J., Pavasovic, A., and Norton, R. S. (2018). Sea anemones: quiet achievers in the field of peptide toxins. Toxins (Basel) 10:E36. doi: 10.3390/toxins10010036
Rachamim, T., Morgenstern, D., Aharonovich, D., Brekhman, V., Lotan, T., and Sher, D. (2015). The dynamically evolving nematocyst content of an anthozoan, a scyphozoan, and a hydrozoan. Mol. Biol. Evol. 32, 740–753. doi: 10.1093/molbev/msu335
Radwan, F. E., Aboul-Dahab, H. M., and Bumett, J. W. (2002). Some toxicological characteristics of three venomous soft corals from the Red Sea. Comp. Biochem. Phys. C 132, 25–35. doi: 10.1016/S1532-0456(02)00045-5
Remigante, A., Costa, R., Morabito, R., La Spada, G., Marino, A., and Dossena, S. (2018). Impact of scyphozoan venoms on human health and current first aid options for stings. Toxins (Basel) 10:E133. doi: 10.3390/toxins10040133
Rodriguez, A. A., Cassoli, J. S., Sa, F., Dong, Z. Q., de Freitas, J. C., Pimenta, A. M. C., et al. (2012). Peptide fingerprinting of the neurotoxic fractions isolated from the secretions of sea anemones Stichodactyla helianthus and Bunodosoma granulifera. New members of the APETx-like family identified by a 454 pyrosequencing approach. Peptides 34, 26–38. doi: 10.1016/j.peptides.2011.10.011
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Stanley, G. D. (2003). The evolution of modern corals and their early history. Earth Sci. Rev. 60, 195–225. doi: 10.1016/S0012-8252(02)00104-6
Sunagar, K., Fry, B. G., Jackson, T. N., Casewell, N. R., Undheim, E. A., Vidal, N., et al. (2013). Molecular evolution of vertebrate neurotrophins: co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenalf. PLoS ONE 8:e81827. doi: 10.1371/annotation/accecc73-91b2-45d4-bb33-774b1f394ca1
Sunagar, K., Johnson, W. E., O'Brien, S. J., Vasconcelos, V., and Antunes, A. (2012). Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol. Biol. Evol. 29, 1807–1822. doi: 10.1093/molbev/mss058
Sunagar, K., and Moran, Y. (2015). The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 11:e1005596. doi: 10.1371/journal.pgen.1005596
Sunagar, K., Undheim, E. A., Scheib, H., Gren, E. C., Cochran, C., Person, C. E., et al. (2014). Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteomics 99, 68–83. doi: 10.1016/j.jprot.2014.01.013
Sunagawa, S., DeSalvo, M. K., Voolstra, C. R., Reyes-Bermudez, A., and Medina, M. (2009). Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS ONE 4:e4865. doi: 10.1371/journal.pone.0004865
Thangaraj, S., Bragadeeswaran, S., and Gokula, V. (2018). Bioactive compounds of sea anemones: a review. Int. J. Pept. Res. Ther. 1–12. doi: 10.1007/s10989-018-9786-6
Tibballs, J. (2006). Australian venomous jellyfish, envenomation syndromes, toxins and therapy. Toxicon 48, 830–859. doi: 10.1016/j.toxicon.2006.07.020
Tibballs, J., Yanagihara, A. A., Turner, H. C., and Winkel, K. (2011). Immunological and toxinological responses to jellyfish stings. Inflamm. Allergy Drug Targets 10, 438–446. doi: 10.2174/187152811797200650
Utkin, Y., Vassilevski, A., Kudryavtsev, D., and Undheim, E. A. B. (2019). Editorial: animal toxins as comprehensive pharmacological tools to identify diverse ion channels. Front. Pharmacol. 10:423. doi: 10.3389/fphar.2019.00423
Utkin, Y. N. (2015). Animal venom studies: current benefits and future developments. World J. Biol. Chem. 6, 28–33. doi: 10.4331/wjbc.v6.i2.28
Vetter, I., Davis, J. L., Rash, L. D., Anangi, R., Mobli, M., Alewood, P. F., et al. (2011). Venomics: a new paradigm for natural products-based drug discovery. Amino Acids 40, 15–28. doi: 10.1007/s00726-010-0516-4
Wilson, D., and Alewood, P. (2004). Australian funnel-web spider venom analyzed with on-line RP-HPLC techniques. Methods Mol. Biol. 251, 307–322. doi: 10.1385/1-59259-742-4:307
Keywords: coral, sea anemone, toxin, nematocyst, venom
Citation: Schmidt CA, Daly NL and Wilson DT (2019) Coral Venom Toxins. Front. Ecol. Evol. 7:320. doi: 10.3389/fevo.2019.00320
Received: 31 March 2019; Accepted: 08 August 2019;
Published: 27 August 2019.
Edited by:
Maria Vittoria Modica, Stazione Zoologica Anton Dohrn, ItalyReviewed by:
Adam Michael Reitzel, University of North Carolina at Charlotte, United StatesYehu Moran, Hebrew University of Jerusalem, Israel
Copyright © 2019 Schmidt, Daly and Wilson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norelle L. Daly, bm9yZWxsZS5kYWx5QGpjdS5lZHUuYXU=; David T. Wilson, ZGF2aWQud2lsc29uNEBqY3UuZWR1LmF1
 Casey A. Schmidt
Casey A. Schmidt Norelle L. Daly
Norelle L. Daly David T. Wilson
David T. Wilson