
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 06 August 2019
Sec. Urban Ecology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00288
The impacts of extended flooding on microbial communities and their activities in natural and agricultural wetlands have been well-documented, but there is little basis for predicting the responses of urban soil microbial communities to infrequent, short-term flooding. To assess these responses, surface soil samples (0–1 cm) and intact soil cores (10 cm depth) were collected from an urban meadow in Baton Rouge, LA subsequent to an unprecedented flood during August 2016. During the flood, a topographically low region of the meadow (LM) was inundated for at least several days, while an elevated area (upper meadow or UM) was not flooded. Microbial community composition and diversity at each site were assessed for soils collected from cores at various depths over the upper 10 cm before and after 3 days of experimental flooding ex situ. Cores from LM and UM were also used to assess methane fluxes before and after the experimental flooding. The results indicated that methane fluxes differed between LM and UM sites, and that they were affected by flooding. LM cores emitted methane prior to flooding, and rates increased substantially post-flooding; UM cores consumed methane to levels below ambient atmospheric concentrations prior to flooding, but emitted methane post-flooding. In contrast both LM and UM microbial communities were resistant to short-term flooding, with no significant changes observed at either site, or at any depth interval from the surface to 10 cm. However, LM and UM soil communities differed significantly, with distinct distributions of Acidobacteria, Nitrospirae, and Thaumarchaeota among others. Based on responses of soil cores to experimental flooding, the differences between sites in microbial communities did not appear to be residual effects of the August, 2016 flood, but rather appeared to arise from physical, chemical, and biological variables that change along a 4-m elevation gradient. Collectively, the results suggest that the composition and diversity for some urban soils might be insensitive to short-term flooding, but that important biogeochemical processes, e.g., methane fluxes, might respond rapidly.
Due to long-term shifts from rural areas, more than 80% of America's population lives in cities, of which many are vulnerable to climate change (e.g., McMichael et al., 2006; Myers and Patz, 2009; Lau et al., 2010; Depietri et al., 2011; Douglas et al., 2012). The impacts of climate change, including temperature and precipitation extremes, more energetic storms, water insecurity, changing patterns of infectious diseases, and increased flooding risks, pose major challenges for urban design and engineering, energy and resource use, and healthcare (e.g., Lin N. et al., 2012).
With respect to flooding, research has focused primarily on mitigation, or on health impacts due to toxic chemical or pathogen exposures in floodwaters (e.g., Phanuwan et al., 2006; Bhardwaj et al., 2008; Harmon and Wyatt, 2008; El-Dahr et al., 2009; Socolovschi et al., 2011; Taylor et al., 2011; de Man et al., 2014; He et al., 2014; Emerson et al., 2015; Hammond et al., 2015). Little attention has been given to the response of urban soil microbes, even though they play significant roles in regional scale greenhouse gas fluxes, which are sensitive to soil water contents (Kaye et al., 2004, 2005; Hall et al., 2008; Livesley et al., 2010; Townsend-Small and Czimczik, 2010; Chen et al., 2013; Zhang et al., 2013).
Although the impacts of flooding on natural and agricultural wetlands, especially rice paddy soils, have been studied extensively using cultivation-dependent, cultivation-independent, and biogeochemical and functional analyses (e.g., Bronson et al., 1997; Hengstmann et al., 1999; Noll et al., 2005; Hansel et al., 2008; Unger et al., 2009), the results offer limited predictive insights for urban soils. In contrast to paddy soils, which are flooded regularly on a seasonal basis, and for extended periods allowing for microbial community adaptation, urban soils are flooded irregularly for relatively short periods and likely are not flood adapted. Floodplain soils might represent a more suitable model, since they too are flooded intermittently for short periods, but relatively little is known about the microbial communities in them (e.g., Katayama and Kuwatsuka, 1991; Baldwin and MItchell, 2000; Brettar et al., 2002; Rinklebe and Langer, 2006; Langer and Rinklebe, 2009; Parsons et al., 2013), which limits extrapolations to urban systems.
Additional constraints arise from the fact that urban soils are comprised of a mosaic of soil types with markedly different physical and chemical properties, histories, and a wide range of uses and management (Byrne, 2007). Furthermore, phenomena such as compaction can limit floodwater infiltration and subsequent perturbations in urban soils relative to non-urban soils, while inorganic and organic pollutants in urban floodwater can inhibit microbial activity (Bayer and Schaumann, 2007; Schaumann et al., 2007; Pitt et al., 2008). Thus, understanding the impacts and significance of flooding on urban soils requires analyses conducted within urban systems.
In order to better understand flooding impacts on urban soils, we conducted an analysis of microbial community responses to experimental flooding ex situ using intact soil cores from an urban meadow. Cores were collected from two sites; one site had been inundated about 3 weeks prior to core collection during a major flood (beginning 12 August 2016), while the second site was unflooded. We assayed microbial community composition and diversity along with methane fluxes prior to and after flooding cores with surface water from an adjacent creek. Methane fluxes were chosen for analysis, since they are sensitive to changes in methane oxidation, which depends on gas exchange and oxygen availability, as well as methanogenesis, which requires prolonged anoxia (e.g., King, 1990a,b, 1992; King et al., 1990; Adamsen and King, 1993; Angel et al., 2012). In addition, we analyzed microbial community composition and diversity for surface soils collected approximately 3 and 6 weeks after the natural flood event in an effort to determine whether distinct temporal shifts could be detected in the flooded site (LM) relative the unflooded site (UM).
Surface soil and soil cores were obtained from two sites (upper meadow, UM; lower meadow, LM) in the Pennington meadow, Baton Rouge, LA. The region experiences an average of 153.9 cm of precipitation annually (relatively evenly distributed) and an annual average temperature of 20.250C. The Pennington meadow is about 8 hectares, and is bounded on the north and east by multi-lane city roads; a woodlot bounds the west, while Dawson Creek bounds the south (see Supplemental Figure 1). The site is not used actively, but is mowed for hay annually.
The UM site (30.400, −91.120) occurs at a topographic high along the upper margin of the watershed for Dawson Creek. This site rarely experiences flooding (none recorded), and precipitation percolates through the soils and flows down a shallow gradient toward the LM site (30.382, −91.128), which is adjacent to Dawson Creek. Because of its proximity to the creek, LM appears subject to fluctuations in groundwater levels as well as intermittent flooding. The LM site was approximately 30 m from Dawson Creek, while the UM site was approximately 0.7 km distant.
Two soil types occur in the meadow. The UM site is characterized by Oprairie series soils. These soils are classified as fine-silty, mixed, semiactive, thermic Fragiaquic Glossudalfs that are somewhat poorly drained and form in loess deposits on Pleistocene age terraces. The LM site is characterized by Deerfield-Verdun series soils. These soils are classified as fine-silty, mixed, superactive, thermic Albic Glossic Natraqualfs (Deerfield) or Glossic Natraqualfs (Verdun), which also form in Pleistocene age deposits.
During the August 2016 Baton Rouge Flood (see https://en.wikipedia.org/wiki/2016_Louisiana_floods for a general description), a large portion of the meadow was inundated by floodwater from Dawson Creek as indicated by flood maps and photographic evidence (Supplemental Figures 1, 2). However, due to its elevation UM was not flooded. The period of inundation for LM is unknown, but lasted for at least several days to 1 week based on anecdotal reports.
To assess the potential impacts of flooding, six surface soil samples (0–1 cm depth) were obtained from the LM site on 2 and 25 September 2016, and six were obtained from the UM site on 3 and 25 September 2016. Surface soil samples (0–1 cm) from both sites were collected at arbitrary points within an area of about 100 m2. Soil was transferred to 100-ml Whirlpak sample bags that were stored at −80°C after collection.
In addition, six replicate intact cores (tubes 7.5 cm diameter x 30 cm length, soil about 10 cm depth) were collected from each site on 3 September 2016; the core bottoms were capped for transport to a laboratory at LSU where they maintained at ambient laboratory temperature (about 25°C) with an air headspace for the duration of the experimental manipulations.
Prior to experimental flooding, soil-atmosphere methane exchanges were assayed by sealing the cores to produce headspaces (about 700–800 cm3) that were sub-sampled (0.5 cm3) at intervals for immediate analysis with an SRI model 8610 gas chromatograph fitted with a flame ionization detector and standardized with a 10-ppm methane standard in hydrocarbon-free air prepared by diluting a SCOTTY 100-ppm methane in nitrogen certified standard (Sigma-Aldrich, St. Louis, MO). After terminating the gas exchange assays, three cores from each site were “flooded” with 120 ml of surface water obtained from Dawson Creek resulting in a water layer about 3 cm thick; the remaining cores were used as untreated controls. Flooded cores were not stirred during an incubation of 72 h.
Dawson Creek water for the flooding treatment was obtained on 3 September by filling a sterile 2-l bottle with water from the middle of the creek. The water was held at 5°C prior to use, and stirred with a magnetic stirrer. Coincident with addition to the soil cores, triplicate 20-ml sub-samples of Creek water were filtered through 0.2-μm pore size filters. The filters were stored at −80°C for subsequent extraction of genomic DNA (see below). As necessary, Dawson Creek water was added to the flooded cores (20–30 ml total) to maintain a 3-cm depth.
At the termination of the incubation, floodwater was removed from the cores using a pipet. A 20-ml sub-sample from each core was filtered as above. Cores were then sealed for assays of soil-atmosphere methane exchange as described above. Gas exchange incubations continued for approximately 40 h.
At the termination of the gas exchange assays, cores were extruded and sectioned into depth intervals of 0–1, 1–2, 2–4, 4–6, and 8–10 cm depths. Soil sub-samples from the interior of these depth intervals were transferred to Whirlpak bags, which were stored at −80°C until they were processed for genomic extractions. The remaining soil was sub-sampled for gravimetric water content, and stored at 4°C prior to drying at 80°C for 24 h. Additional sub-samples were used to measure pH. Briefly, soil samples were mixed with deionized water in a 1:2 ratio (mass of fresh soil:volume of water); pH of the mixture was assayed with a combination electrode and a Beckman Φ71 meter. Organic matter contents were assayed using a “loss on ignition” method (Davies, 1974). Briefly, dried soil samples from each of the core sections were weighed into porcelain crucibles and combusted at 550°C for a minimum of 8 h. The mass loss during combustion was considered an estimate or organic matter. Two cores (one each from flooded and non-flooded LM) were lost and not extracted.
Differences between sites, sampling dates, and flooding treatments, and among depths for pH, organic matter, and water content were assessed using the ANOVA module of XLSTAT (Addinsoft, Inc.). A Bonferroni correction was applied to the p-values for post-hoc comparisons. The independent variables (e.g., site, date, flooding treatment, and depth) were analyzed separately, and for interactions (e.g., site and depth) using a fixed effects model.
Genomic DNA was extracted from all soil samples (24 soil surface samples and 5 depths for each of 10 cores) using a PowerSoil kit (Qiagen Inc.) following the manufacturer's instructions. Genomic DNA was extracted from the 12 filters used for water sampling following the same procedure. After purification, DNA extracts were visualized by gel electrophoresis and then shipped overnight on dry ice to the Research Technology Support Facility of Michigan State University for 16S rRNA amplification (using primers 515f and 806r), barcoding, library preparation and sequencing on a Miseq platform (Illumina, Inc.) with 2 × 250 bp paired-end chemistry following the Kozich et al. (2013) protocol.
Sequences were analyzed using mothur (1.36) following the Miseq protocol (Kozich et al., 2013) to make contigs, trim, filter, align, remove chimeras, classify and cluster OTUs (d = 0.03). The SILVA v 128 database was used for both alignment and classification (Quast et al., 2013). Alpha diversity metrics for each sample group, including Chao index, ACE index, Shannon index, inverse Simpson's index and coverage, were estimated after removing singletons and normalizing the data set. Beta diversity metrics indicating shared diversity were based on Yue-Clayton theta (Yue and Clayton, 2005) and Jaccard (1912) dissimilarities. Pairwise distances were used to generate a dendrogram representing relationships among samples. Guided by these outcomes, additional analyses were conducted using the Microbiomeanalyst platform (Dhariwal et al., 2017). The most informative OTUs were selected using a filtering protocol based on mean abundances for OTU counts, and the inter-quantile range function for OTU variance. Data were transformed using the centered log ratio, but samples were not rarefied because the variation in library sizes was small. Analyses included standard estimates of alpha diversity (Chao1 [richness] and Shannon indices [abundance and evenness]) and beta diversity (Bray-Curtis plus unweighted and weighted UniFrac distance metrics) with ordination by principle coordinates analyses (PCoA) with permutational analysis of variance (Permanova) and permuted homogeneity of multivariate dispersions (Permdisp), plus Linear Discriminant Analysis Effect Size (LEfSe) to assess features (OTUs) that served as biomarkers for the various sample groups. Sequences have been deposited in the NCBI SRA as SRP176467.
Soil pH varied between 4.73 and 5.69 for upper meadow cores, and between 5.28 and 5.89 for lower meadow cores (Supplemental Figure 3); the pH of Dawson Creek water samples was somewhat higher, 6.60–6.72. Analysis of variance (ANOVA) of soil pH values with a Bonferroni post hoc test (critical value = 0.008) (Supplemental Table 2) revealed no statistically significant impact of experimental flooding (p > 0.215), although flooded soils tended to have lower pH values. Experimental flooding in LM cores was associated with higher pH values than in the unflooded cores, but the differences were only marginally significant (p = 0.09). However, the pH of the experimentally flooded LM soils was significantly higher than both sets of UM cores (p < 0.0001).
Organic contents were greatest in the surface interval, and decreased rapidly with increasing depth (Supplemental Figure 4). For the upper 1 cm, values varied between 15.4 and 20.1% for UM cores, and 29.1 and 21.4% for LM cores. Values at 9 cm were <4.3% for both sites and treatments. Organic matter contents were otherwise similar among sites and treatments (p > 0.210) (Supplemental Table 2).
Water contents varied significantly with increasing depth for soil cores (p < 0.0001, Figure 1), but did not vary between LM and UM sites when experimentally flooded or unflooded cores were compared separately. However, flooding for 3 days at both sites significantly increased water contents over the 0–4 cm depth interval relative to unflooded soils (p < 0.0001, Figure 1), while lower depths were unaffected (Supplemental Table 2).
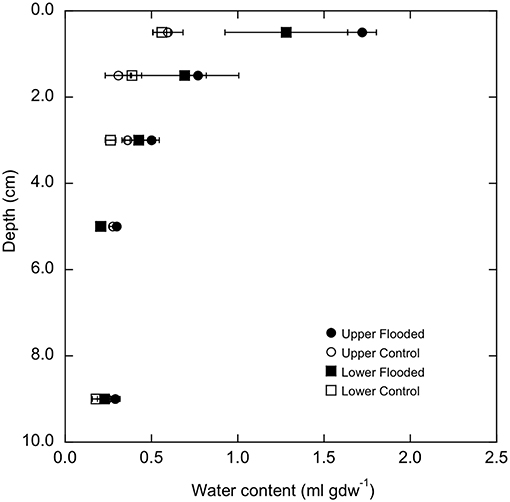
Figure 1. Water content (ml gdw−1) as a function of depth in flooded and control cores from lower and upper meadow sites (square and square symbols, respectively). Data represent means ± 1 standard error for triplicate determinations.
Soil-atmosphere methane fluxes differed distinctly between LM and UM, and responded differentially to experimental flooding at the two sites. In particular, LM cores prior to flooding consistently emitted methane (Figure 2; 1.3 ± 0.8 mg m−2 d−1 [mean ± 1 standard error of the mean], n = 3), while UM cores consumed it to values less than ambient atmospheric concentrations (Figure 2; −0.3 ± 0.2 mg m−2 d−1, n = 3). Subsequent to flooding, methane fluxes in LM cores increased relative to pre-flooding values (Figure 2; 6.2 ± 0.6 mg m−2 d−1, n = 3), while unflooded cores continued to emit methane, but at rates lower than for flooded cores (Figure 2; 1.5 ± 1.2 mg m−2 d−1, n = 3). UM cores that previously consumed atmospheric methane emitted it after flooding (Figure 2; 1.6 ± 0.7 mg m−2 d−1, n = 3), while consumption continued for unflooded cores (Figure 2; −0.8 ± 0.5 mg m−2 d−1, n = 3).
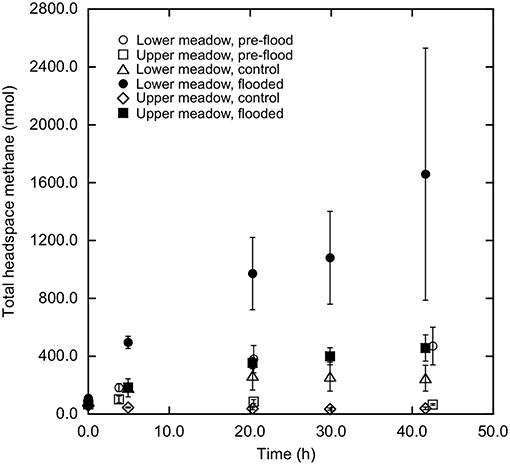
Figure 2. Methane accumulation (nmol total) vs. time (h) in headspaces of cores from lower and upper meadow sites. Lower meadow cores pre-flood and after flooding are represented by open and closed circles, respectively. Upper meadow cores pre-flood and after flooding are represented by open and closed squares, respectively. Control cores are represented by TRI and DIA for lower and upper meadow, respectively. Data represent means ± 1 standard error for triplicate determinations.
Genomic DNA was successfully extracted from 74 soil samples. Fifty of these were from 10 cores, each sampled at 5 depths; two of the initial 12 cores were lost during processing, one each from the LM flooded and unflooded sets. An additional 24 surface samples were collected from the LM and UM sites on 2 and 25 September, 2016. Of the 74 soil extracts, sequence libraries for two were excluded from the analysis pipeline due to low read yields. In addition, genomic DNA was extracted from 12 Dawson Creek water samples, including 3 that were filtered immediately after water collection, 3 that were stored at 15°C during the experimental period, and 3 each from the LM and UM cores that were flooded. A total of 7,140,700 reads were obtained from the Miseq run. Of these, 3,765,465 passed quality and filtering screens, and were assembled into contigs (mean library size = 45,367 reads, standard deviation = 9,546). After eliminating singletons, 64,525 OTUs defined at an evolutionary distance of 0.03 remained from mothur's cluster split algorithm.
Irrespective of sample location, date or treatment, four phyla dominated all communities. Acidobacteria and Proteobacteria were most abundant, followed by Verrucomicrobia and Bacteroidetes. These phyla collectively accounted for 74.7%-89.9% of all OTUs. Relative abundances varied somewhat among soils as a function of location, sampling date and depth, with ranges of 21.0–28.7%, 24.1–32.0%, 14.1–20.1%, and 4.1–11.5% for Acidobacteria, Proteobacteria, Verrucomicrobia, and Bacteroidetes, respectively (Supplemental Table 1). For Dawson Creek and the experimental floodwater samples, Proteobacteria and Bacteroidetes were generally more abundant than in soils, while Acidobacteria and Verrucomicrobia were lower (Supplemental Table 1).
Numerous phyla occurred at abundances <1%, including Actinobacteria, Armatamonadetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Planctomycetes, and Spirochaetes (Supplemental Table 1). Various candidate divisions and phyla were also present at low abundances, e.g., BRC1, Microgenomates, Latescibacteria, Parcubacteria, Saccharibacteria (formerly TM7), and WPS-1, WPS-2 (Supplemental Table 1). No distinct patterns for distribution of these phyla were evident.
The Acidobacteria were notable for the numerous phylogenetic groups that were represented in all samples (Figure 3). Groups 1, 3, 4, and 6 were particularly abundant, but 2, 5, 7, and 10 were also common, while Groups 11, 13, 15–18, 22, and 25 were present at low abundances (<0.1% overall). Alphaproteobacteria were generally more abundant in soil samples than Delta- and Betaproteobacteria, which were comparable in abundance, and substantially greater than Gammaproteobacteria; Epsiloproteobacteria were rare (Figure 3). In Dawson Creek and experimental floodwater samples, Betaproteobacteria were typically more abundant than other Proteobacteria classes (Figure 3). Among the Verrucomicrobia, the genus Opitutus was relatively abundant (0.6% overall), but unclassified OTUs belonging to Spartobacteria and Subdivision 3 represented large fractions of all libraries (7.3 and 5.3% overall, respectively), and dominated the Verrucomicrobia (Figure 3).
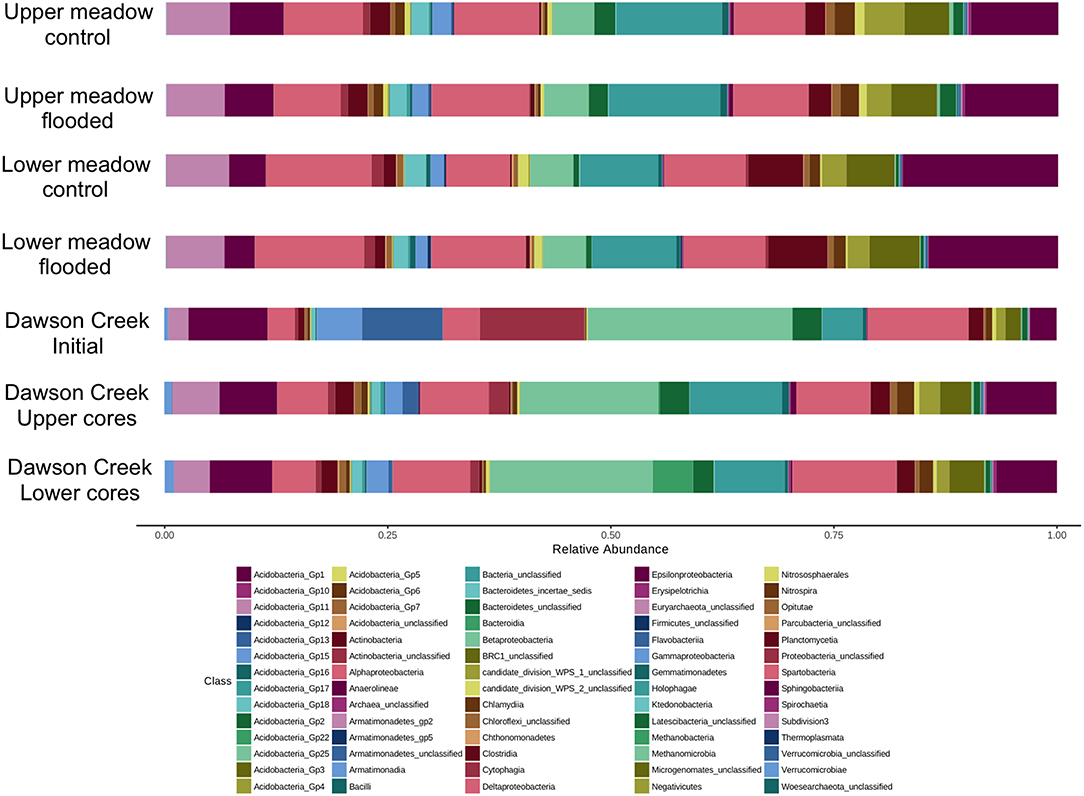
Figure 3. Taxonomic compositions (classes) for control and flooded upper and lower meadow soil cores integrated over 10 cm depth; also shown is the composition of surface water from Dawson Creek, and of the standing water on flooded cores at the termination of the flooding incubation.
Numerous OTUs were classified in functionally distinct families or genera. Methanotrophs were represented in the Alpha- and Gammaproteobacteria, but at a low incidence (0.04% each overall) with no distinct patterns among samples. OTUs classified as methanogenic Euryarchaeota were also present at a low abundance in soil (0.04% overall) but not in water samples. OTUs associated with nitrification were dominated by Nitrospirae (0.44% overall) and Nitrososphaera (Thaumarchaeota, 0.25% overall); proteobacterial nitrifiers (e.g., Nitrobacter, Nitrosomonas) were rare. In contrast, proteobacterial nitrogen fixers classified as members of the Bradyrhizobiaceae were relatively abundant (1.09% overall), though nitrogen fixers classified as Rhizobiaceae were considerably less common (0.06% overall).
The 20 most abundant OTUs in the dataset collectively accounted for 21.9% of all of the reads (Table 1). They included representatives from Acidobacteria, Actinobacteria, Bacteroidetes, Proteobacteria, Verrucomicrobia, and unclassified Bacteria. Two unclassified Spartobacteria (Verrucomicrobia) were the most abundant OTUs overall. Together with a representative of Verrucomicrobia subdivision 3, they accounted for 7.26% of total abundance. Acidobacteria (8 OTUs in the top 20) were represented by OTUs classified in Groups 1, 3, 4, and 6, collectively accounting for 7.47% of the total abundance (Table 1). Proteobacteria, the third most abundant phylum among the top 20 OTUs, was represented by 6 OTUs from the Alpha-, Beta-, and Deltaproteobacteria that collectively accounted for 3.39% of the total abundance. While many of the OTUs in this study could not be classified below a family level, four genera were found among the top 20 OTUs. They included Terrimonas (Bacteroidetes), Bradyrhizobium (Alphaproteobacteria), and Actinoallomurus (Actinobacteria), which collectively accounted for 2.78% of all reads, while 2 Geobacter OTUs (Deltaproteobacteria) accounted for an additional 1.56%.
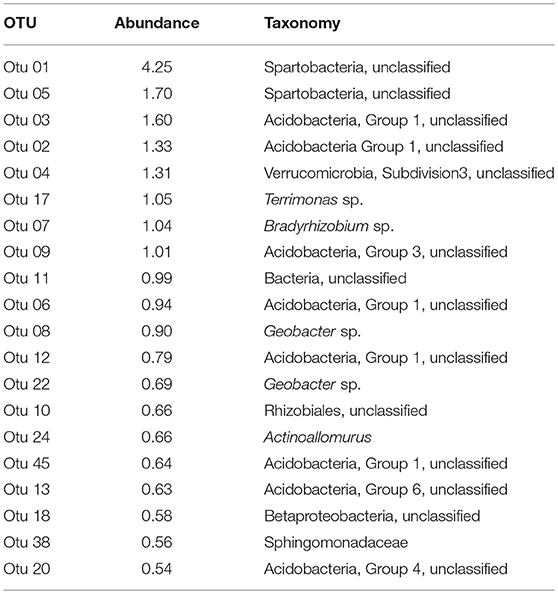
Table 1. Taxonomic identification and relative abundances of the 20 most abundant OTUs among all sequences, ranked by relative abundance.
Alpha and beta diversity indices were generated for the various samples using mothur after removing singletons and normalizing library abundances. Chao1 estimates of richness ranged from a minimum of 5,213 ± 1,556 for stored Dawson Creek samples to a maximum of 10,526 ± 747 for values integrated over the UM experimentally flooded cores (Table 2). However, there were no statistically significant differences in Chao1 among sample sources, dates or treatments, even though water samples consistently harbored lower richness, and UM soils were richer than LM soils. Similarly, no statistically significant trends were observed for Chao1 as a function of core depth.
Inverse Simpsons and Shannon diversity indices also did not vary significantly among sample sources, dates or treatments. The former varied from a minimum of 96.8 ± 24.7 to a maximum of 235.2 ± 23.2 in LM experimentally flooded cores and 2 September 2016 LM surface soils, respectively (Table 2). The distribution of Shannon diversity values largely paralleled that for the inverse Simpsons index, but with the highest value, 6.945 ± 0.044, observed for 2 September 2016 UM surface soils. Both indices tended to decrease with increasing soil depth, and both were higher in UM than LM soils. However, neither trend was statistically significant.
Relationships among soil microbial communities were initially explored with the Yue-Clayton theta similarity index (Yue and Clayton, 2005) as implemented in mothur with sample groups clustered using a UPGMA algorithm (Supplemental Figure 5). The initial results indicated that LM and UM soils were distinct, but that the flooding treatment had no notable impact on microbial community composition at either site. The results also indicated that communities in surface soils collected on 2 September 2016 differed from those collected on 25 September 2016, especially for LM. In addition, the results revealed that the composition of Dawson Creek samples differed from all of the soil samples. However, two of three water samples collected from each set of the flooded cores clustered with soil samples rather than Dawson Creek samples (Supplemental Figure 5).
Based on the initial findings, principal coordinates analyses (PCoA) were used to explore relationships among communities based on sample date (2 and 25 September 2016 surface samples) and location (LM vs. UM) using Bray-Curtis, unweighted UniFrac, and weighted UniFrac beta diversity indices based on filtered data sets with the most informative OTUs transformed using a centered log ratio algorithm as implemented by Microbiomeanalyst. The results were largely comparable to those obtained using mothur's tree.shared command and the thetaYC index (Supplemental Figure 5). PCoA analyses for LM and UM surface soils collected on 2 and 25 September 2016 were based on 1,827 significant features. Results showed that surface soils from the two sites were distinct, and that communities from 2 and 25 September differed for LM, but not UM (Figure 4; unweighted unifrac, Permanova: R2 = 0.452; p < 0.001; Permdisp: p = 0.467). PCoA results also showed that LM and UM communities as represented by experimentally flooded and unflooded cores at all depths (1928 significant features) were distinct, e.g., Figure 5 (Permanova: R2 = 0.187, p < 0.001; Permdisp: p = 0.002), with the first two axes accounting for 34.3% of community variability. However, experimental flooding did not alter communities in either location (Supplemental Figure 5, Figure 6).
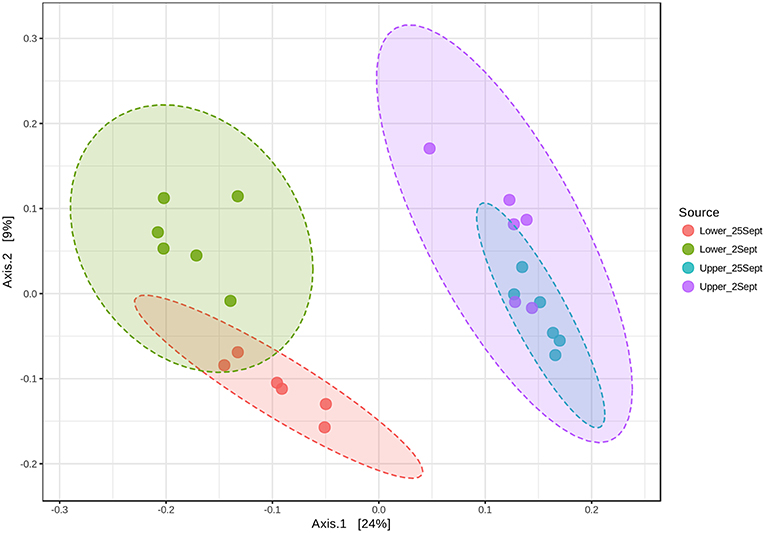
Figure 4. Principal Coordinates Analysis of surface soil samples from lower and upper meadow sites grouped by location (site) and sampling date using the weighted UniFrac distance metric. Permanova: r2 = 0.452; p < 0.001; Permdisp: p = 0.467.
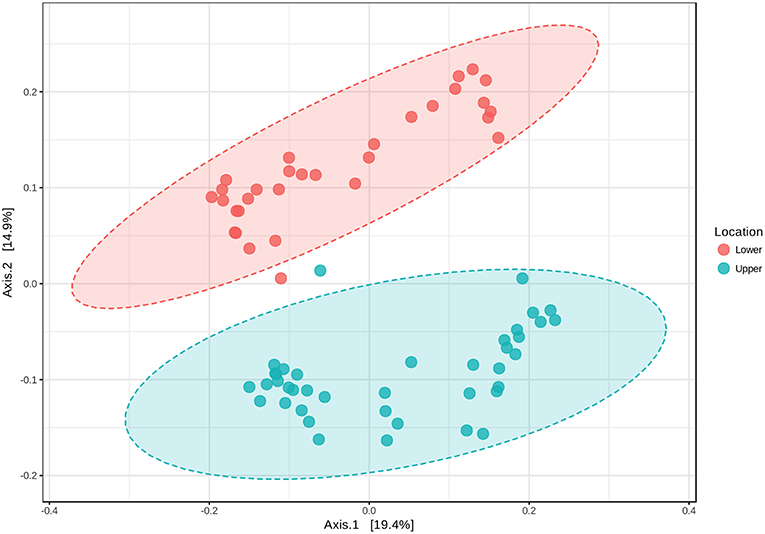
Figure 5. Principal Coordinates Analysis of all surface and core soil samples from lower and upper meadow sites grouped by location (site) using the weighted UniFrac distance metric. Permanova: r2 = 0.187, p < 0.001; Permdisp: p = 0.002.
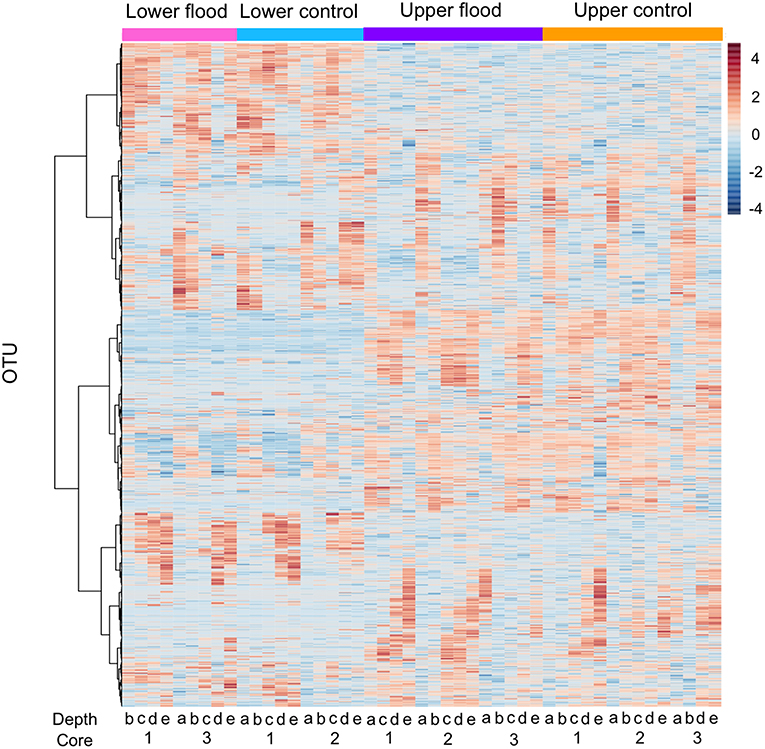
Figure 6. Heatmap of OTU abundance for upper and lower meadow flooded and control cores based on Pearson's r and Ward clustering. Core numbers (replicates) and depth intervals for each treatment (flooded and controls) are indicated below the figure. Depth intervals are designated: a, 0–1 cm; b, 1–2 cm; c, 2–4 cm; d, 4–6 cm; e, 8–10 cm.
The structure, composition, and biogeochemical activities of microbial communities in regularly flooded agroecosystems (e.g., rice paddy soils) have been documented extensively (e.g., Wang et al., 1993; Peters and Conrad, 1996; Hengstmann et al., 1999; Noll et al., 2005; Shrestha et al., 2008). Flooding history and duration, organic matter availability, fertilization, and management during unflooded and flooded periods all affect communities and their functions (e.g., Schütz et al., 1989; Rui et al., 2009; Watanabe et al., 2009). In contrast, relatively little is known about urban soil responses to episodic flooding that occurs with limited duration in landscapes that are considerably more heterogeneous and managed very differently than agroecosystems. However, an increasingly active hydrologic cycle in response to a warming climate and rising sea levels could alter both flooding frequency and duration in urban systems (Douglas et al., 2008; Wheater and Evans, 2009; Lau et al., 2010; Miller and Hutchins, 2017). Since urban soils play important roles in regional scale greenhouse gas fluxes (N2O and methane), understanding responses to flooding is essential for the design of both experimental and modeling studies of gas fluxes (Kaye et al., 2004, 2005; Hall et al., 2008; Townsend-Small and Czimczik, 2010).
Results from this study showed that soil-atmosphere methane fluxes were sensitive to flooding, but that outcomes were site dependent. All UM cores prior to experimental flooding, and unflooded UM control cores consumed atmospheric methane. The observed rates were relatively low compared to forest and other unmanaged soils, but similar to values reported for soils affected by agricultural use (e.g., King, 1992). In contrast, experimentally flooded UM cores emitted methane (Figure 2). The transition from uptake to emission likely reflects a decrease in methanotrophy coupled with a low rate of methanogenesis. Flooding increased water contents in the upper 5 cm of the cores (Figure 1), which would limit oxygen availability, thereby reducing methane oxidation. Although decreased oxygen availability could also stimulate methanogenesis, results from other studies have indicated that the response time of methanogens is typically much longer than the 72 h of flooding in this study (Peters and Conrad, 1996; Noll et al., 2005; Angel et al., 2012).
For the LM site, methane was emitted by all cores prior to experimental flooding, and also by the unflooded control cores (Figure 2); methane emission was greatly enhanced for experimentally flooded cores (Figure 2). These results indicate that methane production was more active in the LM than the UM. Net methane emission from LM cores might reflect the flooding history of the site and its proximity to Dawson Creek, which might promote methanogenesis relative to the better drained UM site that is rarely inundated, if ever. In addition other as yet unexplored factors, including hydrology, could promote methanogenesis in the LM relative to the UM site.
In spite of differences in methane fluxes between the sites before and after experimental flooding, there were no distinct differences in relative abundances or the depth distribution of OTUs identified as methanogens or methanotrophs. Both groups were represented by the same OTUs in lower and upper meadow soils, and their relative abundances did not change significantly with flooding. This suggests that responses to short-term flooding were due to changes in the activities of specific populations, which can occur rapidly relative to changes in abundance (e.g., Shrestha et al., 2008; Angel et al., 2012).
The relative abundances of other taxonomic groups were also largely unaffected by experimental flooding, which resulted in no differentiation by treatment (e.g., Figure 6, Supplemental Figure 5). The fact that water contents did not change for soil depths >5 cm likely accounts for the lack of treatment effects on communities from 4 to 6 cm and 8 to 10 cm depth intervals. Although soil at these depths might have experienced short-term hypoxia or anoxia, the duration of experimental flooding appears to have been insufficient to generate shifts in community composition. Similarly, flooding duration appears to have been insufficient to generate changes in communities for soil depths <5 cm, even though water contents in the upper sections of the cores were significantly greater after flooding (Figure 1), and direct air-soil gas exchange was precluded.
In contrast, during intervals of hours to days after rice paddy soils have been flooded, the relative abundances of numerous populations change, with differences based on 16S rRNA more pronounced than those based on 16S rRNA genes (Noll et al., 2005). In addition, molecular oxygen declines rapidly, fermentation end products accumulate, and methane fluxes increase (Noll et al., 2005). These and other differences between rice paddy and urban meadow soil responses to short-term flooding likely reflect adaptations by the former to seasonal inundation for several months. In addition, management practices for rice cultivation, including fertilization and organic amendments, might potentiate responses to flooding that occur more slowly in unmanaged soils.
Although experimental flooding did not affect community composition (e.g., Figures 3, 6), LM and UM communities differed distinctly based on comparisons of the surface soils and cores collected at each site (Figures 4, 5). Differentiation was apparent in analyses of the communities as a whole, as well as in analyses of specific phyla. For example, the relative abundances of two of the dominant phyla, e.g., Acidobacteria and Proteobacteria, did not differ significantly between LM and UM samples (ANOVA, p = 0.262 and 0.275, respectively), but each was comprised of distinct assemblages at the sub-phylum level as indicated by PCoA (Figure 7). For example, Acidobacteria group 1 was significantly more abundant in LM (p < 0.0001), while groups 2, 5, 6, 7, and 10 were significantly more abundant in the UM (p ≤ 0.003 for each). ANOVA also revealed that Verrucomicrobia and Bacteroidetes were more abundant in LM (p = 0.001 and 0.002, respectively), while Planctomycetes and Actinobacteria were more abundant in the UM (p < 0.0001 for both).
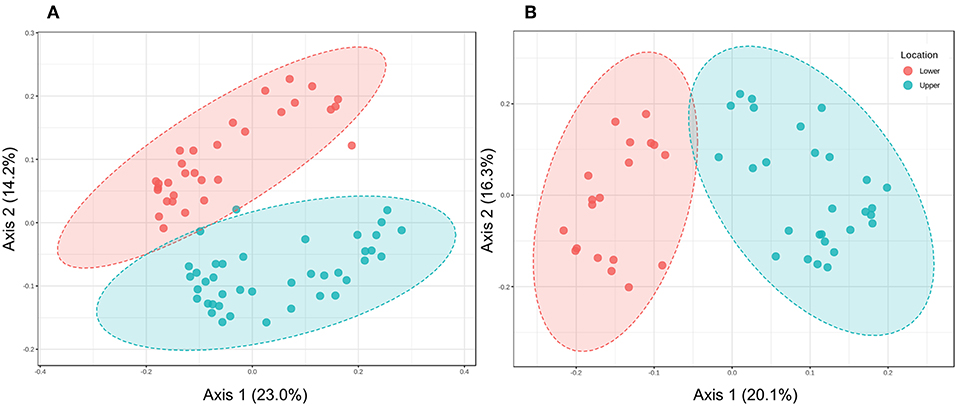
Figure 7. (A) Principal Coordinates Analysis of Acidobacteria in soil samples from lower and upper meadow cores grouped by location using the weighted UniFrac distance metric. Permanova: R-squared: 0.15; p < 0.001; Permdisp: p-value: 0.066. (B) Principal Coordinates Analysis of Acidobacteria in soil samples from lower and upper meadow cores grouped by location using the weighted UniFrac distance metric. Permanova: R-squared: 0.247; p < 0.001; Permdisp: p < 0.0001.
These differences in community membership and relative abundances could reflect residual effects of flooding prior to sample collection, but that is inconsistent with the lack of impacts observed for the experimental flooding. A more likely explanation is that different populations in LM and UM develop in response to habitat variations that result from the 4-m elevation gradient between sites. Although soil pH and organic matter have been documented as important variables for community composition and diversity in other studies (e.g., for Acidobacteria, Jones et al., 2009), neither varied consistently for LM and UM, which raises questions about their significance in the Pennington meadow.
The distributions of specific Acidobacteria groups also raise questions about the role of pH. Acidobacteria groups 5, 6, 7, and 10, which are typically found in greatest abundance above pH 6 (Jones et al., 2009), were relatively more abundant in UM, where pH tended to be lower than in LM (Supplemental Figure 3). In addition, Acidobacteria Groups 1 and 2 respond similarly to pH, but were distributed differently between LM and UM as noted above. Thus, the correlates with elevation that account for community differences are unknown at present. Nonetheless, previous studies have established that microbial communities and their activities vary with topography and elevation in unmanaged grasslands (Smith et al., 2002; Florinsky et al., 2004; Yang et al., 2014), and similar phenomena appear to occur in urban ecosystems as well, even across moderate distances.
The functional or biogeochemical impacts of different communities in LM and UM sites are uncertain. Differences in the relative abundances and composition of Acidobacteria, Actinobacteria, Chloroflexi, and Proteobacteria might affect organic matter transformations and other processes, since functional redundancy cannot be assumed across communities (Strickland et al., 2009), but the impacts might be subtle and difficult to observe experimentally. In contrast, impacts of differences in the distribution of Nitrospirae and Thaumarchaeota should be more readily observable.
Nitrospirae, which oxidize nitrite to nitrate or carry out comammox (Daims and Wagner, 2018) were undetectable or present at very low relative abundances in LM soils, but accounted for 0.4–2.2% of the reads in the UM. Similarly, relative abundances of Thaumarchaeota, which contribute to ammonia oxidation and co-occur with Nitrospirae in some soils (Attard et al., 2010; Daebeler et al., 2014), were significantly greater in UM (0.15–3.2%%) than LM (0–0.1%). This suggests that canonical ammonia and nitrite oxidizers (e.g., Proteobacteria) might be responsible for LM nitrification in spite of their low abundances, while Nitrospirae and Thaumarchaea might be more important in the UM. Assays of bacterial and archaeal amoA, and Nitrobacter and Nitrospirae nitrite oxidoreductases (nxrA and nxrB, respectively) would help confirm these differences and their significance (Poly et al., 2008; Pester et al., 2014).
Results presented here indicate that the composition of nitrifying communities, and perhaps their activities, can vary substantially over moderate spatial scales in an urban meadow. Similar variability might occur more generally across urban landscapes, resulting in a mosaic of communities and potentials corresponding to varied soil characteristics and management. Nitrospirae and Thaumarchaeota have both been reported at low relatively abundances in soil from Central Park, New York (Ramirez et al., 2014) and Beijing (Lin W. et al., 2012; Yan et al., 2016), but correspondence with canonical nitrifiers and nitrogen cycling rates were not addressed. Since urban nitrification can contribute to locally significant fluxes of NO and N2O (Kaye et al., 2004, 2005; Livesley et al., 2010; Zhang et al., 2013), and since changes in nitrifier composition affect nitrification potential (Attard et al., 2010), a greater understanding of the composition and activities of urban nitrifiers is warranted.
In summary, microbial community composition in urban meadow soils proved resistant to short-term experimental flooding. However, flooding rapidly altered methane fluxes resulting in a shift from net atmospheric uptake to net emission for soils from an elevated site (UM), and increased emission for soils from a lower site near a creek (LM). Although flooding had no impact on community composition, and although pH and organic matter concentrations were similar for the two sites, their communities were significantly different, with differences in some populations (e.g., Nitrospirae and Thaumarchaeota) likely to be associated with differences in nitrogen transformations. The results overall emphasize the need for additional studies in urban systems that include a wide range of soils, flooding frequencies and durations, and processes likely to respond to changes in water saturation.
GK collected samples, designed the research, collected and analyzed data, and wrote the manuscript. KH processed samples, collected and analyzed data and contributed to manuscript preparation.
GK acknowledges partial support from National Science Foundation EAR-1565499.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Pennington Biomedical Research Center for access to the Pennington Meadow.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00288/full#supplementary-material
Adamsen, A. P. S., and King, G. M. (1993). Methane consumption in temperate and sub-arctic forest soils: rates, vertical zonation and response to water and nitrogen. Appl. Environ. Microbiol. 59, 485–490.
Angel, R., Claus, P., and Conrad, R. (2012). Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 6, 847–862. doi: 10.1038/ismej.2011.141
Attard, E., Poly, F., Commeaux, C., Laurent, F., Terada, A., Smets, B. F., et al. (2010). Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12, 315–326. doi: 10.1111/j.1462-2920.2009.02070.x
Baldwin, D. S., and MItchell, A. M. (2000). The effects of drying and reflooding on the sediment and soil nutrient dynamics of lowland river - floodplain systems: a synthsis. Regul. Rivers: Res. Mgmt. 16, 457–467. doi: 10.1002/1099-1646(200009/10)16:5<457::AID-RRR597>3.0.CO;2-B
Bayer, J. V., and Schaumann, G. E. (2007). Development of soil water repellency in the course of isothermal drying and upon pH changes in two urban soils. Hydrol. Process. 21, 2266–2275. doi: 10.1002/hyp.6759
Bhardwaj, P., Kosambiya, J. K., and Desai, V. (2008). A case control study to explore the risk factors for acquisition of leptospirosis in Surat city, after flood. Indian J. Med. Sci. 62:431. doi: 10.4103/0019-5359.48454
Brettar, I., Sanchez-Perez, J.-M., and Trémolières, M. (2002). Nitrate elimination by denitrification in hardwood forest soils of the Upper Rhine floodplain - correlation with redox potential and organic matter. Hydrobiol 469, 11–21. doi: 10.1023/A:1015527611350
Bronson, K. F., Neue, H. U., Abao, E. B., and Singh, U. (1997). Automated chamber measurements of methane and nitrous oxide flux in a flooded rice soil: I. residue, nitrogen, and water management. Soil Sci. Soc. Am. J. 61, 981–987. doi: 10.2136/sssaj1997.03615995006100030038x
Byrne, L. B. (2007). Habitat structure: a fundamental concept and framework for urban soil ecology. Urban Ecosys 10, 255–274. doi: 10.1007/s11252-007-0027-6
Chen, W., Jia, X., Zha, T., Wu, B., Zhang, Y., Li, C., et al. (2013). Soil respiration in a mixed urban forest in China in relation to soil temperature and water content. Eur. J. Soil Biol. 54, 63–68. doi: 10.1016/j.ejsobi.2012.10.001
Daebeler, A., Bodelier, P. L., Yan, Z., Hefting, M. M., Jia, Z., and Laanbroek, H. J. (2014). Interactions between Thaumarchaea, Nitrospira and methanotrophs modulate autotrophic nitrification in volcanic grassland soil. ISME J. 8, 2397–2410. doi: 10.1038/ismej.2014.81
Daims, H., and Wagner, M. (2018). Nitrospira. Trends Microbiol. 26, 462–463. doi: 10.1016/j.tim.2018.02.001
Davies, B. E. (1974). Loss on ignition as an estimate of soil organic matter. Soil Sci. Soc. Am. Proc. 38, 150–151. doi: 10.2136/sssaj1974.03615995003800010046x
de Man, H., van den Berg, H. H., Leenen, E. J., Schijven, J. F., Schets, F. M., van der Vliet, J. C., et al. (2014). Quantitative assessment of infection risk from exposure to waterborne pathogens in urban floodwater. Water Res. 48, 90–99. doi: 10.1016/j.watres.2013.09.022
Depietri, Y., Renaud, F. G., and Kallis, G. (2011). Heat waves and floods in urban areas: a policy-oriented review of ecosystem services. Sustain Sci. 7, 95–107. doi: 10.1007/s11625-011-0142-4
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., and Xia, J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual, and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188. doi: 10.1093/nar/gkx295
Douglas, E. M., Kirshen, P. H., Paolisso, M., Watson, C., Wiggin, J., Enrici, A., et al. (2012). Coastal flooding, climate change and environmental justice: identifying obstacles and incentives for adaptation in two metropolitan Boston, Massachusetts communities. Mitig. Adapt. Strateg. Glob. Change 17, 537–562. doi: 10.1007/s11027-011-9340-8
Douglas, I., Alam, K., Maghenda, M., McDonnell, Y., McLean, L., and Campbell, J. (2008). Unjust waters: climate change, flooding and the urban poor in Africa. Environ. Urban. 20, 187–205. doi: 10.1177/0956247808089156
El-Dahr, J. M., Paris, K., Sikora, M. M., Lichtveld, M. Y., and Mitchell, H. E. (2009). Sensitivity to mold and environmental allergens in post-Katrina New Orleans. J. Allergy Clin. Immunol. 123, S18–S18. doi: 10.1016/j.jaci.2008.12.081
Emerson, J. B., Keady, P. B., Brewer, T. E., Clements, N., Morgan, E. E., Awerbuch, J., et al. (2015). Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 colorado front range flood. Environ. Sci. Technol. 49, 2675–2684. doi: 10.1021/es503845j
Florinsky, I. V., McMahon, S., and Burton, D. L. (2004). Topographic control of soil microbial activity: a case study of denitrifiers. Geoderma 119, 33–53. doi: 10.1016/S0016-7061(03)00224-6
Hall, S. J., Huber, D., and Grimm, N. B. (2008). Soil N2O and NO emissions from an arid, urban ecosystem. J Geophys Res. 113:11. doi: 10.1029/2007JG000523
Hammond, M. J., Chen, A. S., Djordevic, S., Butler, D., and Mark, O. (2015). Urban flood impact assessment: a state-of-the-art review. Urban Water J. 12. doi: 10.1080/1573062X.2013.857421
Hansel, C. M., Fendorf, S., Jardine, P. M., and Francis, C. A. (2008). Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74, 1620–1633. doi: 10.1128/AEM.01787-07
Harmon, S. M., and Wyatt, D. E. (2008). Evaluation of post-Katrina flooded soils for contaminants and toxicity to the soil invertebrates Eisenia fetida and Caenorhabditis elegans. Chemosphere 70, 1857–1864. doi: 10.1016/j.chemosphere.2007.08.007
He, C., Salonen, H., Ling, X., Crilley, L., Jayasundara, N., Cheung, H. C., et al. (2014). The impact of flood and post-flood cleaning on airborne microbiological and particle contamination in residential houses. Environ. Int. 69, 9–17. doi: 10.1016/j.envint.2014.04.001
Hengstmann, U., Chin, K.-J., Janssen, P. H., and Liesack, W. (1999). Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65, 5050–5058.
Jaccard, P. (1912). The distribution of the flora in the Alpine zone. New Phytol. 11, 37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x
Jones, R. T., Robeson, M. S., Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453. doi: 10.1038/ismej.2008.127
Katayama, A., and Kuwatsuka, S. (1991). Effect of pesticides on cellulose degradation in soil under upland and flooded conditions. Soil Sci. Plant Nutr. 37, 1–6. doi: 10.1080/00380768.1991.10415003
Kaye, J. P., Burke, I. C., Mosier, A. R., and Guerschman, J. P. (2004). Methane and nitrous oxide fluxes from urban soils to the atmosphere. Ecol. App. 14, 975–981. doi: 10.1890/03-5115
Kaye, J. P., McCulley, R. L., and Burke, I. C. (2005). Carbon fluxes, nitrogen cycling, and soil microbial communities in adjacent urban, native and agricultural ecosystems. Global Change Biol. 11, 575–587. doi: 10.1111/j.1365-2486.2005.00921.x
King, G. M. (1990a). Regulation by light of methane emission in a wetland. Nature 345, 513–515. doi: 10.1038/345513a0
King, G. M. (1990b). Dynamics and controls of methane oxidation in a Danish wetland sediment. FEMS Microbiol. Ecol. 74, 309–323 doi: 10.1016/0378-1097(90)90684-I
King, G. M. (1992). Ecological aspects if methane oxidation, a key determinant of global methane dynamics. Adv. Microb. Ecol. 12, 431–468. doi: 10.1007/978-1-4684-7609-5_9
King, G. M., Roslev, P., and Skovgaard, H. (1990). The distribution and rate of methane oxidation in sediments of the Florida Everglades. Appl. Environ. Microbiol. 56, 2902–2911.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Langer, U., and Rinklebe, J. (2009). Lipid biomarkers for assessment of microbial communities in floodplain soils of the Elbe River (Germany). Wetlands 29, 353–362. doi: 10.1672/08-114.1
Lau, C. L., Smythe, L. D., Craig, S. B., and Weinstein, P. (2010). Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 104, 631–638. doi: 10.1016/j.trstmh.2010.07.002
Lin, N., Emanuel, K., Oppenheimer, M., and Vanmarcke, E. (2012). Physically based assessment of hurricane surge threat under climate change. Nat. Clim. Chang. 2, 462–467. doi: 10.1038/nclimate1389
Lin, W., Li, J., and Pan, Y. (2012). Newly isolated but uncultivated magnetotactic bacterium of the phylum Nitrospirae from Beijing, China. Appl. Environ. Microbiol. 78, 668–675. doi: 10.1128/AEM.06764-11
Livesley, S. J., Dougherty, B. J., Smith, A. J., Navaud, D., Wylie, L. J., and Arndt, S. K. (2010). Soil-atmosphere exchange of carbon dioxide, methane and nitrous oxide in urban garden systems: impact of irrigation, fertiliser and mulch. Urban Ecosys 13, 273–293. doi: 10.1007/s11252-009-0119-6
McMichael, A. J., Woodruff, R. E., and Hales, S. (2006). Climate change and human health: present and future risks. Lancet 367, 859–869. doi: 10.1016/S0140-6736(06)68079-3
Miller, J. D., and Hutchins, M. (2017). The impacts of urbanisation and climate change on urban flooding and urban water quality: a review of the evidence concerning the United Kingdom. J. Hydrol. 12, 345–362. doi: 10.1016/j.ejrh.2017.06.006
Myers, S. S., and Patz, J. A. (2009). Emerging threats to human health from global environmental change. Annu. Rev. Environ. Resour. 34, 223–254. doi: 10.1146/annurev.environ.033108.102650
Noll, M., Matthies, D., Frenzel, P., Derakshani, M., and Liesack, W. (2005). Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 7, 382–395. doi: 10.1111/j.1462-2920.2005.00700.x
Parsons, C. T., Couture, R. M., Omoregie, E. O., Bardelli, F., Greneche, J. M., Roman-Ross, G., et al. (2013). The impact of oscillating redox conditions: arsenic immobilisation in contaminated calcareous floodplain soils. Environ. Pollut. 178, 254–263. doi: 10.1016/j.envpol.2013.02.028
Pester, M., Maixner, F., Berry, D., Rattei, T., Koch, H., Lucker, S., et al. (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 16, 3055–3071. doi: 10.1111/1462-2920.12300
Peters, V., and Conrad, R. (1996). Sequential reduction processes and initiation of CH4 production up flooding of oxic upland soils. Soil Biol. Biochem. 28, 371–382. doi: 10.1016/0038-0717(95)00146-8
Phanuwan, C., Takizawa, S., Oguma, K., Katayama, H., Yunika, A., and Ohgaki, S. (2006). Monitoring of human enteric viruses and coliform bacteria in waters after urban flood in Jakarta, Indonesia. Water Sci. Technol. 54, 203–10. doi: 10.2166/wst.2006.470
Pitt, R., Chen, S.-E., Clark, S. E., Swenson, J., and Ong, C. K. (2008). Compaction's impacts on urban storm-water infiltration. J. Irrig. Drain Eng. 134, 652–658. doi: 10.1061/(ASCE)0733-9437(2008)134:5(652)
Poly, F., Wertz, S., Brothier, E., and Degrange, V. (2008). First exploration of Nitrobacter diversity in soils by a PCR cloning-sequencing approach targeting functional gene nxrA. FEMS Microbiol. Ecol. 63, 132–140. doi: 10.1111/j.1574-6941.2007.00404.x
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramirez, K. S., Leff, J. W., Barberán, A., Bates, S. T., Betley, J., Crowther, T. W., et al. (2014). Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc. R. Soc. B 281:20141988. doi: 10.1098/rspb.2014.1988
Rinklebe, J., and Langer, U. (2006). Microbial diversity in three floodplain soils at the Elbe River (Germany). Soil Biol. Biochem. 38, 2144–2151. doi: 10.1016/j.soilbio.2006.01.018
Rui, J., Peng, J., and Lu, Y. (2009). Succession of bacterial populations during plant residue decomposition in rice field soil. Appl. Environ. Microbiol. 75, 4879–4886. doi: 10.1128/AEM.00702-09
Schaumann, G. E., Braun, B., Kirchner, D., Rotard, W., Szewzyk, U., and Grohmann, E. (2007). Influence of biofilms on the water repellency of urban soil samples. Hydrol. Process. 21, 2276–2284. doi: 10.1002/hyp.6746
Schütz, H., Holzapfel-Pschorn, A., Conrad, R., Rennenberg, H., and Seiler, W. (1989). A 3-year continuous record on the influence of daytime, season, and fertilizer treatment on methane emission rates from an Italian rice paddy. J. Geophys. Res. Atm. 94, 16405–16416. doi: 10.1029/JD094iD13p16405
Shrestha, M., Abraham, W. R., Shrestha, P. M., Noll, M., and Conrad, R. (2008). Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ. Microbiol. 10, 400–412. doi: 10.1111/j.1462-2920.2007.01462.x
Smith, J. L., Halvorson, J. J., and Bolton, H. Jr. (2002). Soil properties and microbial activity across a 500 m elevation gradient in a semi-arid environment. Soil Biol. Biochem. 34, 1749–1757. doi: 10.1016/S0038-0717(02)00162-1
Socolovschi, C., Angelakis, E., Renvoise, A., Fournier, P. E., Marie, J. L., Davoust, B., et al. (2011). Strikes, flooding, rats, and leptospirosis in Marseille, France. Int. J. Infect. Dis. 15, e710–e715. doi: 10.1016/j.ijid.2011.05.017
Strickland, M. S., Lauber, C. L., Fierer, N., and Bradford, M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90, 441–451. doi: 10.1890/08-0296.1
Taylor, J., Lai, K. M., Davies, M., Clifton, D., Ridley, I., and Biddulph, P. (2011). Flood management: prediction of microbial contamination in large-scale floods in urban environments. Environ. Int. 37, 1019–1029. doi: 10.1016/j.envint.2011.03.015
Townsend-Small, A., and Czimczik, C. I. (2010). Carbon sequestration and greenhouse gas emissions in urban turf. Geophys. Res. Lett. 37:5. doi: 10.1029/2009GL041675
Unger, I. M., Kennedy, A. C., and Muzika, R.-M. (2009). Flooding effects on soil microbial communities. Appl. Soil Ecol. 42, 1–8. doi: 10.1016/j.apsoil.2009.01.007
Wang, Z. P., DeLaune, R. D., Patrick, W. H., and Masscheleyn, P. H. (1993). Soil redox and pH effects on methane production in a flooded rice soil. Soil Sci. Soc. Am. J. 57:382. doi: 10.2136/sssaj1993.03615995005700020016x
Watanabe, T., Kimura, M., and Asakawa, S. (2009). Distinct members of a stable methanogenic archaeal community transcribe mcrA genes under flooded and drained conditions in Japanese paddy field soil. Soil Biol. Biochem. 41, 276–285. doi: 10.1016/j.soilbio.2008.10.025
Wheater, H., and Evans, E. (2009). Land use, water management and future flood risk. Land Use Policy 26, S251–S264. doi: 10.1016/j.landusepol.2009.08.019
Yan, B., Li, J., Xiao, N., Qi, Y., Fu, G., Liu, G., et al. (2016). Urban-development-induced changes in the diversity and composition of the soil bacterial community in Beijing. Sci. Rep. 6:38811. doi: 10.1038/srep38811
Yang, Y., Gao, Y., Wang, S., Xu, D., Yu, H., Wu, L., et al. (2014). The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 8, 430–440. doi: 10.1038/ismej.2013.146
Yue, J. C., and Clayton, M. K. (2005). A similarity measure based on species proportions. Commun. Stat. Theor. Methods 34, 2123–2131. doi: 10.1080/STA-200066418
Keywords: urban microbiome, urban meadow, flooding, methane, soil microbial community
Citation: King GM and Henry K (2019) Impacts of Experimental Flooding on Microbial Communities and Methane Fluxes in an Urban Meadow, Baton Rouge, Louisiana. Front. Ecol. Evol. 7:288. doi: 10.3389/fevo.2019.00288
Received: 04 February 2019; Accepted: 16 July 2019;
Published: 06 August 2019.
Edited by:
Stephanie Ann Yarwood, University of Maryland, College Park, United StatesReviewed by:
Ariane L. Peralta, East Carolina University, United StatesCopyright © 2019 King and Henry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gary M. King, Z2tpbmdtZUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.