
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 27 June 2019
Sec. Chemical Ecology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00238
This article is part of the Research TopicThe Role of Plant Secondary Metabolites in Plant Cross-Kingdom Interactions View all 5 articles
Benzoxazinoids, secondary metabolites of several Poaceae, and some benzoxazinoid downstream metabolites are bioactive compounds that act as allelochemicals and natural pesticides. Since a short lifetime of the substances is crucial to avoid long-term environmental effects, total degradation by microorganisms is of exceptional importance. We performed a screening with cultivable microorganisms (Species names and strain numbers: Mycobacterium fortuitum, 7; Bacillus aryabhattai, 34; Bacillus cereus, 59; Bacillus megaterium, 21, 48; Bacillus methylotrophicus, 58; Lysinibacillus xylanilyticus, 56; Paenibacillus polymyxa, 51; Aminobacter aminovorans, 49; the fungi Papulaspora sepedonioides, 12 and Trichoderma viride, 47) isolated from soil previously used for wheat and Persian clover mixed-culture systems to assess their behavior in the presence of the compounds. The microorganisms were exposed to glucosylated benzoxazinones, the benzoxazinones HBOA, DIBOA, and DIMBOA, the benzoxazolinones BOA, BOA-6-OH, and MBOA, and to several downstream products (AP, AAP, oHPMA, glucoside carbamate) in liquid culture to avoid interferences with soil minerals and other organisms. The microorganisms differed strongly in their metabolic activities in terms of growth, compound modification, and degradation. We observed degradation with DIBOA and GDIMBOA but rarely with DIMBOA, whereas BOA and MBOA showed almost no degradation when directly applied. Hydroxylation of BOA and demethylation of MBOA by the plant, resulting in BOA-6-OH, activated the benzoxazolinones for bacterial nitration. The resulting NBOA-6-OH was short-lived but could function temporarily as an allelochemical by inhibiting photosynthesis, e.g., in young seedlings of cress and kohlrabi. The BOA downstream products AP and oHPMA were converted to AAP, which can be nitrated to N-(2-OH-5-nitrophenyl)-acetamide and then degraded by A. aminovorans (49) and P. polymyxa (51). Only P. sepedonioides (12) and P. polymyxa (51) failed in the conversion of HBOA into AAP. While DIBOA, DIMBOA, MBOA, NBOA-6-OH, AP, AAP, and oHPMA reduced the growth of most microorganisms, glucoside carbamate promoted their growth. GDIMBOA had a stimulatory effect toward the fungi and three bacterial species. These findings lead to the hypothesis that in a natural habitat, such as the root surface, microorganisms may cooperate, perhaps by involving the plant, for the successful elimination of benzoxazinoids and their downstream metabolites.
Benzoxazinones are bioactive secondary metabolites of several Poaceae, including the crops maize, rye, and wheat, as well as some dicotyledonous species. The compounds are produced in young tissues of roots and shoots, and they are glucosylated and then stored in vacuoles or exuded by roots. Benzoxazinones, for instance GDIBOA (2-(2,4-dihydroxy-1,4-benzoxazin-3-one)-β-D-glucopyranose, 1) (Table S1) and GDIMBOA [(2R)-2-β-D-glucopyranosyloxy-4-hydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, 2], are unstable when deglucosylated. Their aglycones DIBOA (3) and DIMBOA (4) undergo heterocyclic ring contraction, releasing the benzoxazolinones BOA (5) and MBOA (6), which are more stable and, therefore, detectable for longer durations in the soil (Sicker and Schulz, 2002; Fomsgaard et al., 2004). Degradation of benzoxazinoids in soil involves most probably microorganisms. Since the degradation of phenolic compounds is also influenced by soil minerals (Nannipieri et al., 2002), benzoxazinoids may be degraded or polymerized, perhaps partly, by abiotic processes or by extracellular enzymes adsorbed at soil minerals. Thus, in soil, biotic and abiotic degradation steps may be miscellaneous.
The effects and functions of benzoxazinones have been investigated for decades, and they remain a focus as possible natural herbicides, lead structures for the development of new herbicides, and anti-herbivory compounds (Fomsgaard et al., 2004; Macias et al., 2005; Macías et al., 2009; Niemeyer, 2009; Niculaes et al., 2018). The allelopathic properties of benzoxazinoides have been intensively studied (for review Schulz et al., 2013). Weeds and crops differ in their sensitivity to the compounds, depending on the seedling age, dosage, and species. The ability to detoxify the compounds successfully is highly important for plants and microorganisms to meliorate and to survive injuries caused by allelopathic benzoxazinoids.
Figure 1 illustrates an overview of the presently known, major detoxification and degradation products derived from benzoxazinoids (see also list S1). HBOA (2-hydroxy-2H-1,4-benzoxazin-3(4H)-one) is not shown because its origin is controversially presented in the literature (see below). The benzoxazinones DIBOA and DIMBOA are precursors of a variety of natural derivatives. Some of them can be spontaneously formed, such as the BOA and MBOA. The instable, highly reactive intermediate formed upon ring opening of DIBOA leads to AP (13), which undergoes oxidative dimerization to yield APO (16). APO can be modified by fungi and bacteria by hydroxylation, acetylation, nitration, and other reactions. Plants, fungi, and bacteria convert BOA, MBOA, and AP to numerous derivatives by hydroxylation and subsequent O-glucosylation (plants), by N-glucosylation followed by rearrangement to carbamates (plants, e.g., maize: 9, not shown: carbamates 10, 12), by acetylation of AP to AAP (fungi and bacteria), and by conjugation to malonic acid after opening of the BOA heterocycle to N-(2-hydroxyphenyl)malonamic acid (oHPMA, 15) (a few fungi). The plant and microbial products are subjected to further alterations and pathways, named A, B, C, and D. Pathways A, C, and D can finally result in the complete degradation or polymerization of the compounds. Pathway B was not considered in this study. The following descriptions will give more detailed insights into plant and microbial reactions resulting in the detoxification and degradation products.
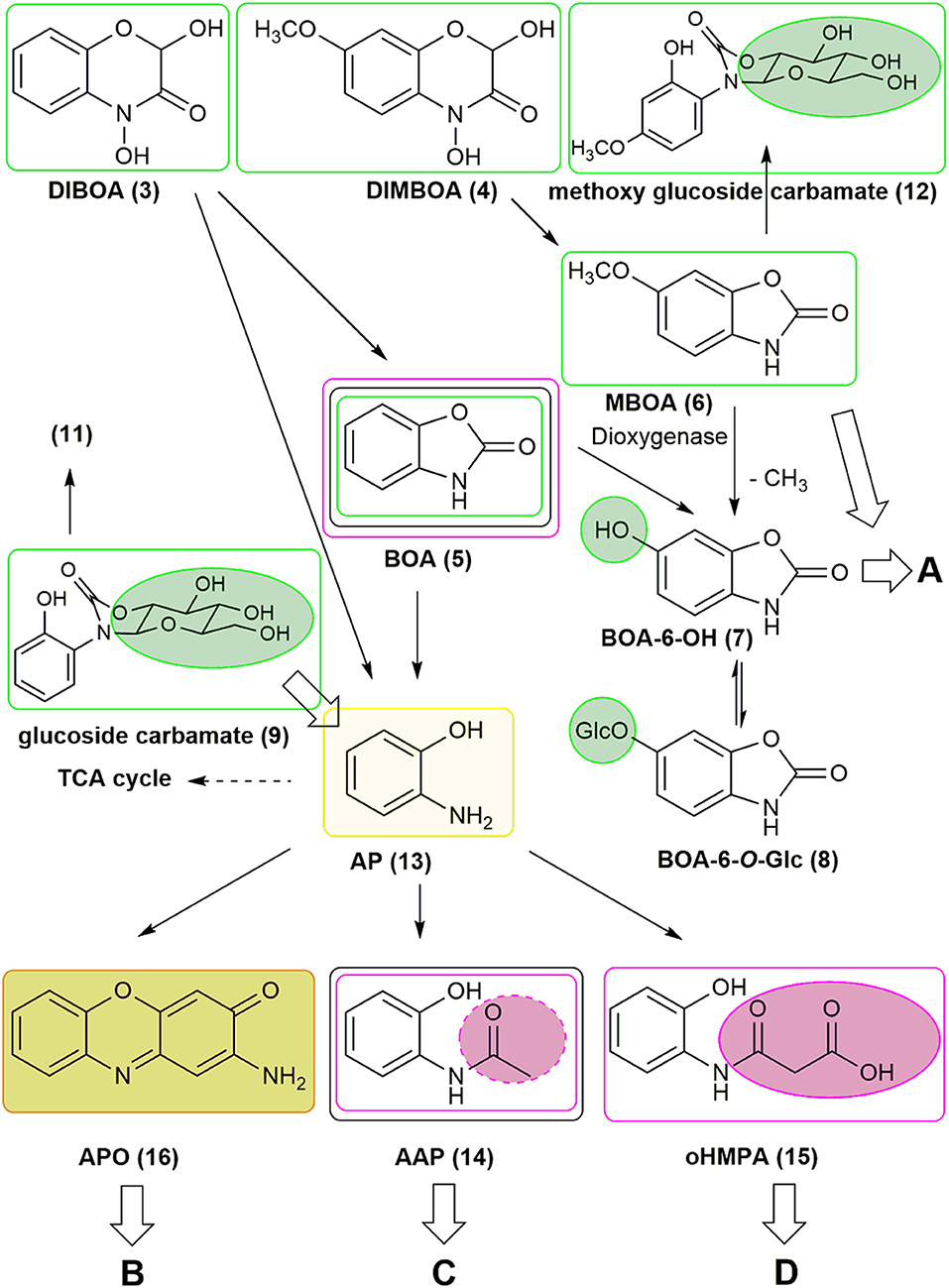
Figure 1. Metabolism of benzoxazinoids by plants and microorganisms. The pathways A, C, and D are further explored in Figures 7, 11. Green circles and lines: plant compounds and derivatives produced by plants. Purple circles and lines: fungal compound conversions. Gray lines: bacterial compound conversions. The light green line marks AP as a pivotal molecule in degradation processes and as a precursor for subsequent conversions. The olive green background depictures APO as a precursor for subsequent conversions (pathway B, not considered in this study).
Plants absorb benzoxazolinones, and many species are able to inactivate the compounds by hydroxylation and subsequent glucosylation. MBOA, as found in maize, can be demethylated to yield BOA-6-OH (7), the intermediate of BOA detoxification by O-glucosylation. Maize exudes GDIMBOA, which is rapidly deglucosylated to DIMBOA by GDIMBOA-β-glucosidase in the apoplast of the young maize root, but the abundance of the enzyme in this compartment is reduced in BOA-incubated roots (Schulz et al., 2016). BOA-6-O-glucoside (8) was first found in Avena sativa roots and later in many other plants after BOA exposure (Wieland et al., 1998; Schulz and Wieland, 1999). In most dicotyledonous species, BOA-6-O-glucoside is the major detoxification product. In many Poaceae, particularly in maize, the synthesis of BOA-6-O-glucoside presents a ready-to-go mechanism that is quickly surrogated by another detoxification pathway, namely by switching to BOA-N-glucosylation. The final detoxification product, glucoside carbamate (9), results from the molecular rearrangement of BOA-N-glucoside (Sicker et al., 2001; Schulz et al., 2016). In maize, three other carbamates can be found, gentiobioside carbamate (10), 1-(2-hydroxyphenylamino)-6-O-malonyl-1-deoxy-β-glucoside 1,2-carbamate (malonyl glucoside carbamate, 11), and as a minor detoxification product of MBOA, methoxy glucoside carbamate (12) (Hofmann et al., 2006). In dicotyledonous species, glucoside carbamates are not found at all or often exhibit subordinate accumulation, but they are rarely present as a major compound. Soluble detoxification products of benzoxazolinones accumulate in plants only transiently, and they are exuded or bound to cell wall constituents. Considered a consequence of microbial activities, immobilized benzoxazolinone can be released from dead plant material (Ozaki and Kato-Noguchi, 2016; Tabaglio et al., unpublished).
The further fate of benzoxazolinones is partly known. One pathway begins with opening of the heterocycle. This enzymatic reaction is performed by a γ-lactamase that is present in Fusarium species and possibly in Gaeumannomyces graminis (Friebe et al., 1998; Kettle et al., 2015; Glenn et al., 2016). Currently, only one bacterium, Acinetobacter calcoaceticus, is known to perform BOA heterocycle opening. Heterocycle cleavage leads to the generation of AP, which can be acetylated by fungi and bacteria to AAP. Fusarium species conjugate AP enzymatically via arylamine N-acetyltransferase to yield oHPMA. Several bacteria degrade AP into fragments that are able to enter the TCA cycle. Thus, the compound serves as an energy source. Picolinic acid, a bioactive compound with neuroprotective and immuno-modulating properties in vertebrates, is a spontaneous by-product of this pathway. Picolinic acid inhibits the sporulation of Bacillus cereus, indicating that its bioactive properties are not restricted to animals. The compound is degraded by numerous microorganisms, including the yeast species Rhodotorula sp. (Gollakota and Halvorson, 1960; Dagley and Johnson, 1963; Grant et al., 2009; Chirino et al., 2013; Qiu et al., 2017). Another very common reaction of AP is the oxidative dimerization to the lipophilic compound APO. Despite its stability, APO can be destroyed quickly by H2O2/Fenton chemistry or slowly by photo-oxidation, and is also converted to several derivatives by fungi (Zikmundová et al., 2002). Nevertheless, APO can be detected in agricultural soils for several months after wheat culture. The compound possesses phytotoxic properties; however, regarding the molecular targets, the mode of action is not well understood. The compound was recently described as a histone deacetylase inhibitor (Venturelli et al., 2015, 2016).
The plant-produced detoxification intermediate BOA-6-OH has been found to be a substrate for nitration to NBOA-6-OH (17) by Pantoea ananatis. P. ananatis is a member of a microbial consortium with the zygomycete Actinomucor elegans as the dominant species that colonizes the roots of Abutilon theophrasti, a weed in maize and beet fields (Haghikia et al., 2014; Schulz et al., 2017a,b). MBOA is another substrate for bacterial nitration, which lead to 6-methoxy-4-nitro-benzoxazolin-2(3H)-one (NMBOA, 18). Nitration of MBOA also occurs spontaneously, as found for the generation of NBOA-6-OH, but at a much slower rate (Schulz et al., 2018). Therefore, the nitration of MBOA is a rate limiting step. The reduction of nitrate presents a key step for NBOA-6-OH formation, whereas the subsequent nitration of BOA-6-OH occurs spontaneously but is dramatically accelerated by peroxidase (Schulz et al., 2017b). Hydroxylation of BOA by plants is a prerequisite for nitration by bacteria. The introduction of the nitro group destabilizes the BOA-6-OH molecule and allows subsequent degradation. NMBOA was identified in the culture media of the A. elegans consortium when incubated with MBOA. The compound can be attached to cellular surfaces, as found with the yeast Papiliotrema baii. NBOA-6-OH and NMBOA are degraded by P. ananatis and the A. elegans consortium (Schulz et al., 2017b, 2018). Hydroxylation of BOA prevents its degradation to AP and subsequent formation of APO or AAPO (19). NBOA-6-OH was detected in wash liquids obtained from Abutilon root surfaces after incubation of the seedlings with BOA, also glucosylated BOA-6-OH was found. A UDPG-glucosyltransferase activity able to catalyze the glucosylation of BOA-6-OH could be measured in root protein preparations (Schulz et al., 2017a). These results led to the assumption that Abutilon cooperate with root colonizing microorganisms and the rhizobiome for compound conversions.
Nitrated detoxification products are also known from fungi, for instance N-(2-hydroxy-5-nitrophenyl) acetamide (20) is synthesized from HBOA (21, 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one) by several endophytic fungi isolated from Aphelandra tetragona (Zikmundová et al., 2002). HBOA is the direct precursor of DIBOA in the benzoxazinone biosynthetic pathway (Frey et al., 1997; Nomura et al., 2002). It accumulates in maize in low amounts (Woodward et al., 1978) and was detected in soil after incorporation of wheat and rye residues in several studies. HBOA was assumed to be a bacterial degradation product of DIBOA, (Fomsgaard et al., 2004). The lactam HBOA is stable and no conversion to benzoxazolinone takes place. Regarding the mentioned fungi, HBOA serves as a source for nitrated acetamidophenols. In contrast, Fusarium sambucinum produces oHPMA from HBOA. HBOA degradation by bacteria was not yet investigated.
Based on previous studies, it is rather evident that microorganisms are of exceptional importance in plant secondary metabolite conversion and degradation processes. In this study, we focus mainly on cultivable microorganisms isolated from soil of an organic farming system (Siebers et al., 2018). Excluding B. megaterium (21), all microorganisms are known to possess plant growth-promoting properties. The microorganisms were chosen to acquire clues concerning their capacity to cope with benzoxazolinones, downstream metabolites, and derived compounds, particularly as they were isolated from soil that had previously been used to culture benzoxazinone-producing wheat and Persian clover as a catch crop (Siebers et al., 2018). We included additional microorganisms (Fusarium verticillioides, Pseudomonas putida, and Variovorax paradoxus) to assess their abilities to cope with several compounds. The aim of this study was to elucidate the role of microorganisms in eliminating benzoxazinoids and derived compounds that protect plant partners from the long-term accumulation of phytotoxins. The screening emphasizes also on plants for compound activation, preparing them for further metabolism, or, conversely, for their temporary inactivation. We investigated single microorganisms cultured in liquid medium to avoid interferences with strains of other species and with reactions triggered by soil minerals.
Compounds used for incubations or as references were either purchased (AP, AAP, BOA, MBOA, BOA-6-OH from Sigma-Merck, Munich, Germany) or synthesized (NBOA-6-OH, APO, according to the methods described in Schulz et al., 2017b). GDIBOA, DIBOA, DIMBOA glucoside carbamate, oHPMA and HBOA were either isolated from plants or synthesized (Sicker et al., 1989; Hartenstein et al., 1993; Friebe et al., 1998; Wieland et al., 1998). All compounds were tested for their stability in Czapek medium under the culture conditions. The amounts of relevant compounds generated by spontaneous formation were subtracted from the amounts synthesized by microbial activities. Calculations were based on external standard curves established with the above-mentioned compounds. External standards were preferred to avoid possible co-elution with unknown products. Furthermore, the maize endophyte Fusarium verticillioides (Knop et al., 2007) was used for additional oHPMA production by addition of BOA to the culture. The aqueous phase of the culture medium was concentrated, and was purified by TLC. GDIMBOA was isolated according to the method described by Hartenstein et al. (1993). For MS/MS experiments, the commercially available N-(2-hydroxy-5-nitrophenyl)-acetamide was obtained from Fluorochem (Hadfield, UK), and the internal standard AAP was from Sigma-Merck (Munich, Germany).
The cultivable species used in this study were the numbered strains of the bacteria Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), and the fungi Papulaspora sepedonioides (12) and Trichoderma viride (47). All of them were isolated and identified in a previous study (Siebers et al., 2018). The bacteria were precultured overnight in LB medium; the fungi were cultured in YEP medium. Five hundred microliter of the cultures were transferred to 15 mL Czapek medium supplemented with 0.5 mM DIBOA, DIMBOA, BOA, MBOA, AP, AAP, oHPMA, BOA-6-OH, glucoside carbamate, NBOA-6-OH, and HBOA. Other applied concentrations of the nitro aromatic compound are mentioned in the text. For some experiments with GDIBOA and GDIMBOA the Czapek medium contained gluconate instead of sucrose. The controls contained none of the compounds. The growth of the microorganisms was monitored by OD600 determination over a period of 72 h if not otherwise noted. Next, 200μL aliquots were obtained at t0 and every 24 h for HPLC/DAD (Shimadzu) analyses using the method described by Schulz et al. (2018). At the end of the incubation period, the cultures including the microorganisms were extracted with ethyl acetate and the organic and aqueous phases evaporated. The residues were dissolved in methanol or water and used as total extracts after centrifugation for 3 min at 15,000 rpm for HPLC analysis.
In the case of P. polymyxa (51) and A. aminovorans (49), the bacteria were collected by centrifugation, the supernatants extracted with ethyl acetate, the organic phases evaporated, and the residues dissolved in methanol. The solutions were transferred to glass tubes, freeze-dried, and analyzed for structural elucidation of the major compound. The identification of known compounds was performed using reference substances for co-chromatography. Other microorganisms subjected to incubations were Pseudomonas putida accession no. HM488364 and Variovorax paradoxus DSM-30034, which was purchased (DSMZ, Braunschweig). Compounds used for incubation of these microorganisms are indicated in the text. Previously isolated endophytic F. verticillioides from maize roots (Knop et al., 2007) was cultured on Czapek agar plates. Agar plugs of 1 cm2 were placed in a flask containing 20 mL liquid Czapek medium and cultured for 1 day in the dark at 25°C. F. verticillioides was incubated with NBOA6-OH, BOA-6-OH, and BOA.
Germinating seeds of zucchini were incubated with BOA-6-OH, (0.5 mM), and the incubation medium was analyzed after 24 h for the presence of NBOA-6-OH. Open seed coats of zucchini were placed with the inside down on Czapek agar plates, and well-developed plaques of microorganisms were further cultivated on Czapek agar at 22°C. Single colonies served as template for touchdown-PCR using bacterial 16S primers (27f AGAGTTTGATCMTGGCTCAG, 1492r(s) CGGTTGTTACGACTT). The PCR products were purified with the NucleoSpin Gel and PCR Clean-up Kit (Macherey & Nagel, Düren, Germany) and sequenced. Identification was conducted by searching against the nucleotide database at the NCBI Nucleotide database using BLASTN. The most abundant cultivable species was identified as Bacillus licheniformis (accession No: MK290600), which was incubated with BOA-6-OH as described above.
For MBOA modifications, three- and four-day-old seedlings of Brassica oleracea var. gongylodes L. (kohlrabi), Lepidium sativum (cress), and five-day-old seedlings of Cucumis sativus (cucumber) were used. Seeds of kohlrabi, cress, and cucumber were purchased from local garden centers. Seedlings were grown on watered filter paper placed in Petri dishes.
Incubation of cress, kohlrabi, and cucumber was performed with 0.5 and 2 mM MBOA. After incubation, the roots and shoots were separated, weighed, and homogenized with 70% methanol (weight/volume: 1:3). The homogenates were centrifuged at 20.000 rpm for 5 min, and the supernatants were transferred into caps and either directly used for analysis or frozen at −20°C until used for HPLC analysis. MBOA-derived products were identified by co-chromatography with the reference compounds (BOA-6-OH and its O-glucoside).
Three-day-old cress and kohlrabi seedlings and five-day-old cucumber seedlings were incubated with 2 mM NBOA-6-OH for 24 and 48 h. From each incubation, 30 cress or kohlrabi seedlings were separated in roots and shoots and extracted with 70% methanol as described above. Cucumber seedlings were separated in roots, hypocotyl and shoots including the cotyledons and leaves and extracted. The extracts were analyzed by HPLC for NBOA-6-OH contents (Figure S8). Ten-day-old A. theophrasti seedlings were incubated with 2 mM NBOA-6-OH for 24 h and the roots extracted and analyzed the same way.
Seedlings of cress, kohlrabi, and cucumber were incubated with 2 mM NBOA-6-OH, BOA-6-OH and BOA for evaluation of the compound's effects on photosynthesis. The quantum yields were estimated with a pulse-amplitude modulated fluorometer (Walz). Measurements were performed after a 30 min dark-adaptation period, and the maximum quantum yield of Photosystem II was determined.
Chromatographic separation was performed on an UltiMate3000 UHPLC system from ThermoFischer Scientific (Waltham, MA, USA) equipped with an Acquity UPLC BEH C18 column (2.1 × 100 mm/1.7 μm, Waters) at a flow rate of 450 μL/min at 30°C. The mobile phase consisted of H2O + 0.1% formic acid (A) and acetonitrile + 0.1% formic acid (B). A gradient elution was performed from 5 to 30% B in 8 min, increase to 99.5% B in 1 min and flushing at 99.5% B during 2 min. The UHPLC system was connected to a high-resolution QExactive orbitrap mass spectrometer (ThermoFischer Scientific) equipped with a heated electrospray ion source. Masses were calibrated below <1 ppm accuracy using ThermoFischer Pierce calibration solution. Data were acquired in (+)- and (–)-ESI mode, with a mass range from m/z 100 to 800 and 35,000 resolution. MS/MS experiments were performed with a normalized collision energy of 30 eV.
Each data point represents at least three biological replicates from three independent experiments, unless otherwise noted. Error bars are based on the SD. For statistical analysis of all the data, the t-test was used. Data in the figures are presented as the mean ± standard deviation. Significant differences compared with the control samples are indicated (t-test; *p < 0.05; **p < 0.005; ***p < 0.0005).
The growth behavior of the microorganisms exposed to benzoxazinoids (DIBOA, DIMBOA, BOA, and MBOA) and derived compounds (AP, AAP, oHPMA) in Czapek medium with sucrose was first monitored (Figure 2). The growth of M. fortuitum (7) was most reduced by the benzoxazinones and benzoxazolinones DIBOA mainly affected M. fortuitum (7), B. megaterium (21), B. megaterium (48), B. methylotrophicus (58) and B. cereus (59), and BOA had almost no effect. The compounds that resulted in the greatest growth inhibition were DIMBOA [in particular P. polymyxa (51), L. xylanilyticus (56), B. cereus (59) and T. viride (47)], MBOA, AP, and AAP, especially for B. aryabhattai (34) and the two B. megaterium strains (21, 48). oHPMA was inhibitory, except for P polymyxa (51). T. viride (47) was mainly affected by AP and MBOA was fatal to the fungus P. sepedonioides (12). In summary, the growth behavior revealed remarkable differences in sensitivity to the compounds, whereby A. aminovorans (49) and P. polymyxa (51) showed the lowest sensitivity.
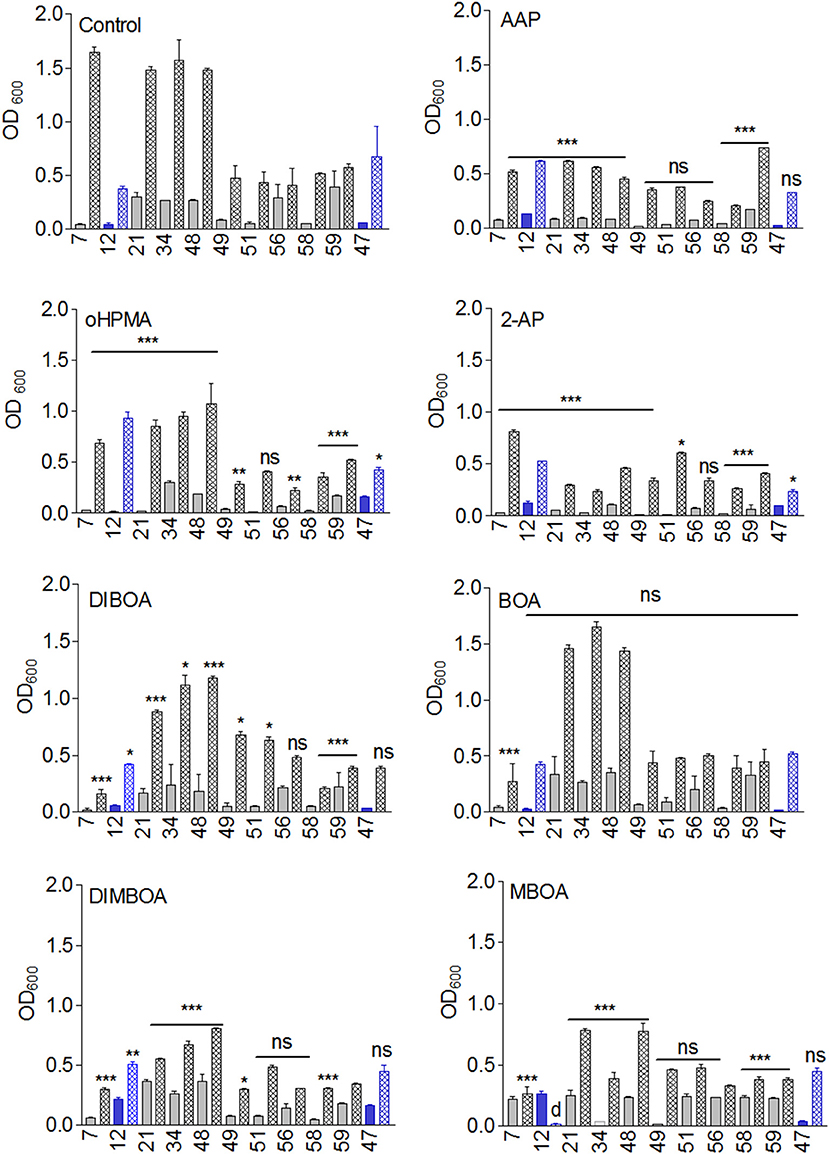
Figure 2. Growth of the microorganisms in the presence of 7.5 μmol DIBOA, DIMBOA, BOA, MBOA, AP, AAP, and oHPMA and control (n = 5). Ns, not significant; *p < 0.05; **p < 0.005; ***p < 0.0005). Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria; OD 600 culture start,  ; OD600 after 72h,
; OD600 after 72h,  .
.
The microorganisms were unable to hydroxylate BOA under the used culture conditions. Most of the applied BOA at t0 could be extracted from the incubation medium after 72 h of exposure (Figure 3A). Although some microorganisms might cleave the BOA heterocycle with low efficiency, no AAP, one of the AP detoxification products, was detected in the culture medium. AP might be metabolized immediately by some strains, similar as observed for Burkholderia xenovorans LB400 (Chirino et al., 2013). Pseudomonas putida HM488364 degraded AP rather rapidly, avoiding the accumulation of higher amounts of APO (data not shown). The pathway has been described for P. putida by Zhao et al. (2000). Since the microorganisms failed to degrade BOA with good efficiency, we started experiments with the precursor compound DIBOA.
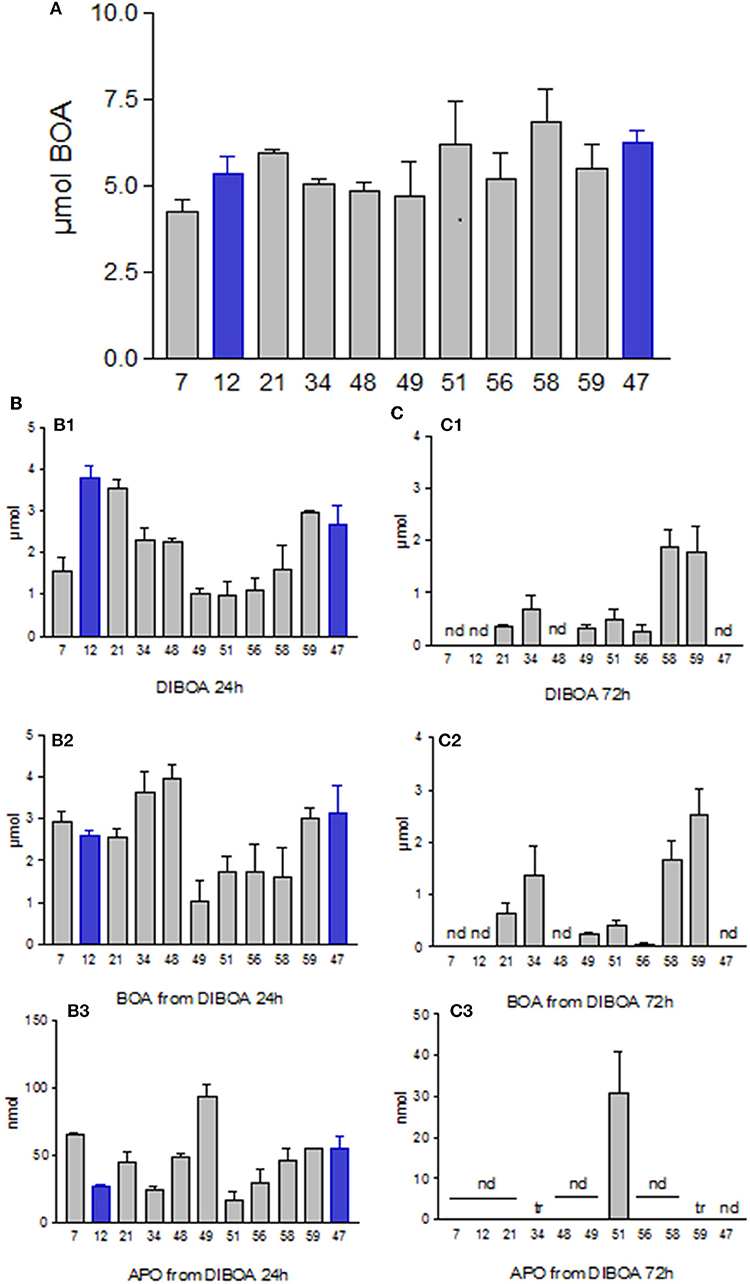
Figure 3. (A) Amounts of BOA recovered from the culture media after 72 h of incubation with 7.5 μmol. (B,C) Incubations with DIBOA. (B1) DIBOA recovered from media after 24 h. (C1) after 72 h. (B2) amounts of BOA produced from DIBOA after 24 h. (C2) after 72 h. (B3) amounts of APO produced from DIBOA after 24 h. (C3) after 72 h. Nd, not detectable; tr, traces. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria.
When 7.5 μmol of DIBOA was applied for 24 h, all cultures contained BOA in the μmol range and nmol amounts of APO, (Figures 3B,C). Again, none of the microorganisms produced AAP. BOA was much faster degraded when present as an intermediate. Prolonged exposure to DIBOA resulted in almost complete degradation after 72 h, except for L. xylanilyticus (56) and B. cereus (59). Additionally, APO disappeared over time, with the exception of P. polymyxa (51). With this strain and A. aminovorans (49), traces of another, not further identified, phenoxazinone were found. The total extracts of the latter strain contained traces of picolinic acid, and those of L. xylanilyticus (56), B. aryabhattai (34), and B. cereus (59) contained some AAP. These products indicate that the bacteria are able to open the BOA heterocycle over the course of DIBOA degradation. BOA degradation presented an apparent bottleneck for the tested microorganisms during direct exposure under the tested culture conditions. The slow conversion did not seem to be concentration-dependent (at least up to 0.5 mM), as the direct application of 0.1 mM BOA led to the same slow conversion.
For comparison, we tested some species for their ability to bio-transform GDIBOA, but only P. polymyxa (51) transformed GDIMBOA via an unknown, hydrophilic intermediate, which is presently under investigation. With Czapek medium containing gluconate, P. polymyxa (51) and B. megaterium (48) degraded all the GDIMBOA with low accumulation of BOA within 72 h. Some degradation was observed with strain B. aryabhattai (34) but not with B. megaterium (21). The growth behavior of the strains was not in accordance with GDIBOA degradation (Figure S4). Since DIBOA is degradable within three days, we did not focus further on GDIBOA degradation.
MBOA remained almost undegraded over a period of 3 days by most microorganisms (Figure 4B1). The original amount was reduced in cultures containing B. megaterium (48), L. xylanilyticus (56), and B. methylotrophicus (58), without an accumulation of degradation products. P. polymyxa (51) produced traces of NMBOA (see below).
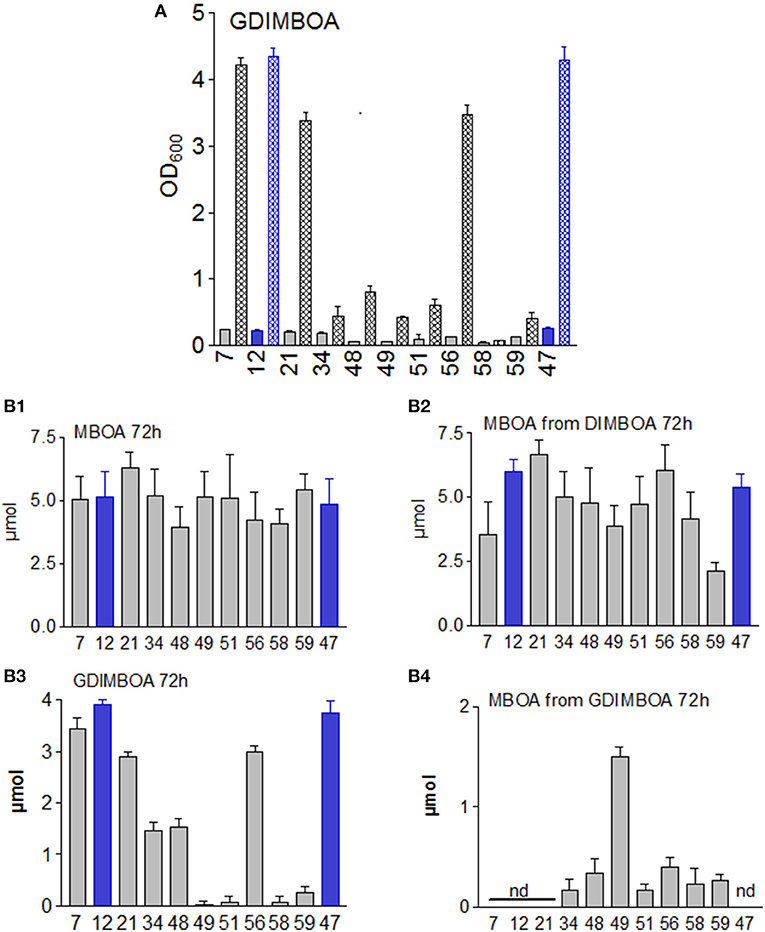
Figure 4. (A) Growth of the microorganisms in presence of 5 μmol GDIMBOA. (B1) Amounts of MBOA in culture media after 72 h of incubation with 7.5 μmol 6. (B2) Amounts of MBOA in the culture media after 72 h of incubation with 7.5 μmol DIMBOA. (B3) Amounts of DIMBOA after 72 h of incubation with 5μmol 2 in media containing gluconate. (B4) Amounts of MBOA in gluconate containing media after 72 h of incubation with 2. n = 4. Nd, not detectable. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria. OD 600 culture start,  ; OD600 after 72 h,
; OD600 after 72 h,  .
.
Analogous to DIBOA, we assumed that the microorganisms might degrade MBOA more rapidly upon exposure to the precursor DIMBOA. In fact, DIMBOA was completely converted to MBOA. However, in contrast to DIBOA, no complete degradation occurred (Figure 4). No other compounds accumulated in amounts that allowed proper identification, even when the concentration of MBOA decreased during the incubation [in particular for M. fortuitum (7), B. aryabhattai (34), A. aminovorans (49), and B. cereus (59)]. The presence of the O-methyl group heavily influenced the degradation by the microbial species. In contrast to DIBOA, several microorganisms developed colored cultures upon exposure to DIMBOA, indicating that colored derivatives were bound to the surfaces of the bacteria (Figure S1).
Regarding the possibility that the glucosylated precursor might be necessary to initiate a complete degradation, we incubated the strains with 5 μmol GDIMBOA. We used Czapek medium, replacing the sucrose with gluconate (10 g/L), (Figure 4). No colored cultures developed with GDIMBOA (Figure S1). The addition of GDIMBOA strongly stimulated the growth of M. fortuitum (7), B. megaterium (21), L. xylanilyticus (56) and the two fungi P. sepedonioides (12), and T. viride (47), which were unable or almost unable [L. xylanilyticus (56)] to degrade GDIMBOA (Figure 4; Figures S1, S3). B. aryabhattai (34) and B. megaterium (48) were the only ones that were inhibited without degrading the compound. In contrast, A. aminovorans (49), P. polymyxa (51), B. methylotrophicus (58), and B. cereus (59) converted all the GDIMBOA, while MBOA accumulated in well detectable amounts with B. aryabhattai (34), B. methylotrophicus (58), and B. cereus (59). A mechanism for complete degradation of GDIMBOA therefore seems plausible with B. aryabhattai (34), B. megaterium (48), P. polymyxa (51), B. methylotrophicus (58), and B. cereus (59), and likely with A. aminovorans (49). These strains showed no growth stimulation with GDIMBOA, and B. methylotrophicus (58) did not grow at all. The other strains seemed to require support from additional species to open the pathways for MBOA elimination. Thus, glucosylation of DIBOA and DIMBOA does not necessarily protect the compounds against degradation by special microorganisms, but the degradation is apparently influenced by ambient carbohydrates.
Hydroxylation of BOA at position 6, a modification performed by plants allowing subsequent O-glucosylation, is sufficient to prevail over the low reactivity of the BOA molecule. In most bacterial cultures supplemented with BOA-6-OH, NBOA-6-OH was identified when nitrate was present in the medium (Figure 5). The fungi P. sepedonioides (12) and T. viride (47) did not produce the compound, nor did the maize endophyte F. verticillioides, and thus they did not release nitrite into the medium. Incubation of the microorganisms with 170 μM NBOA-6-OH revealed the ability of most of the microorganisms, including the two ascomycetes, to degrade the compound (Figure 5), with the most rapid degradation by B. megaterium (48) and by the fungus P. sepedonioides (12). Some of the bacteria seemed to start the degradation early, during ongoing synthesis [M. fortuitum (7), B. megaterium (21), B. aryabhattai (34), and B. megaterium (48)]. Nitration of BOA-6-OH was not restricted to the used cultivable microorganisms isolated from soil. For instance, incubation of germinating seeds of Cucurbita pepo subsp. pepo convar. giromontiina (zucchini) with BOA-6-OH led to new compounds in the incubation medium, including NBOA-6-OH), (Figure S2). Here, production of the nitro aromatic compound is regarded to be due to seed-born microorganisms. However, in cultures with the most abundant bacterium isolated from zucchini seed coats (B. licheniformis), NBOA-6-OH did not accumulate. Additionally, in the presence of V. paradoxus, NBOA-6-OH was not found. The amount of added BOA-6-OH stayed unchanged during the entire incubation time of three days. With B. methylotrophicus (58), only traces of NBOA-6-OH were detectable (Figure 5A), and when the compound was directly applied, it was not degraded but de-nitrated back to BOA-6-OH. It is presently unclear whether B. licheniformis and V. paradoxus reconvert NBOA-6-OH immediately to BOA. There are apparently alternative strategies to cope with BOA and NBOA-6-OH.
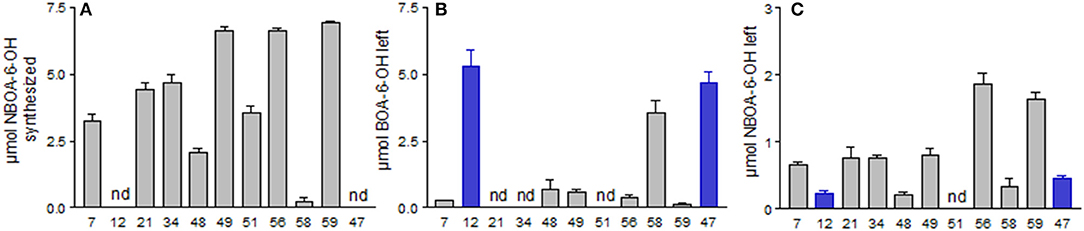
Figure 5. Nitration of BOA-6-OH and degradation of NBOA-6-OH by the microorganisms. (A) Formation of NBOA-6-OH. (B) Residual 7 after 72 h of incubation in the medium. (C) Residual NBOA-6-OH after 72 h of incubation with the compound. Nd, not detectable. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria.
NMBOA was not detected in the media of the microorganisms, except for P. polymyxa (51) where traces of the compound were found. The question arises whether plants can support microorganisms in MBOA elimination by modifying the molecule. For instance, demethylation would activate the molecule for subsequent nitration. Incubation of cress, kohlrabi, and cucumber revealed that MBOA was indeed demethylated, yielding BOA-6-OH. Thus, the low reactivity of MBOA can be bypassed by demethylation. In roots, the resulting BOA-6-OH was glucosylated (Figure 6; Table 1).
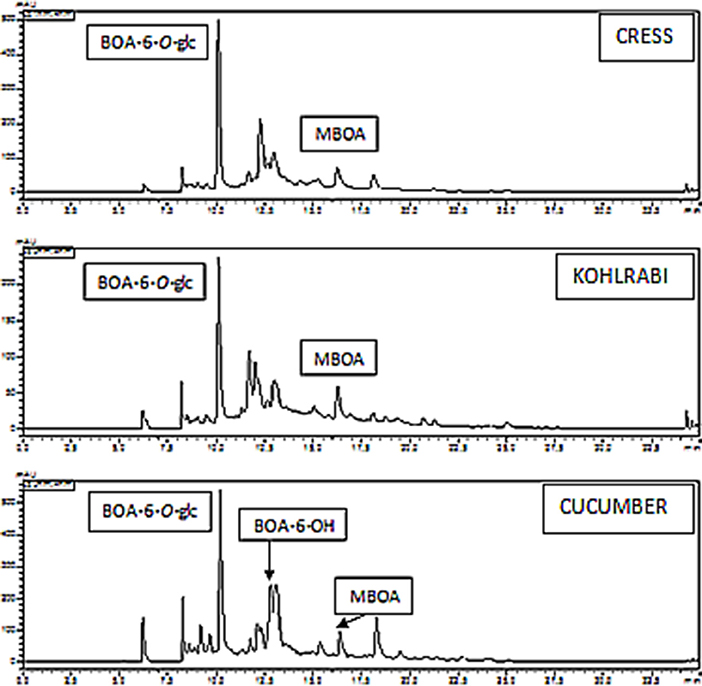
Figure 6. Cucumber, kohlrabi, and cress demethylate MBOA; the demethylation product BOA-6-OH is glucosylated, and the glucoside accumulates in the roots prior to exudation.
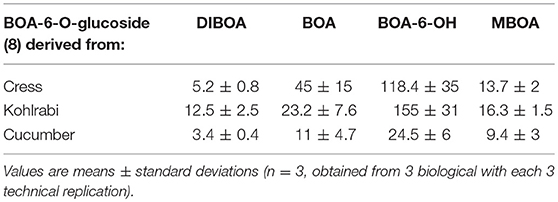
Table 1. HPLC analyses were performed to determine the contents of BOA-6-O-glucoside, (μmol/gFW) in the roots of cress, kohlrabi and cucumber seedlings when exposed for 48 h to 0.5 mM DIBOA, BOA, BOA-6-OH, and MBOA.
As mentioned, demethylation of MBOA, resulting in BOA-6-O-glucoside, has been described previously in maize (Schulz et al., 2016).
Nevertheless, the resulting BOA-6-OH is a substrate for subsequent nitration and can be eliminated via pathway A (Figure 7), regardless of whether it originated from BOA or MBOA. Pathway A is not the only possible elimination route of the compound. A. theophrasti, for instance, polymerizes NBOA-6-OH completely at the root surface (Figure S7), where the compound can be produced. Presently there are hints that some plants absorb and modify NBOA-6-OH.
For some microorganisms, NBOA-6-OH was harmful, as growth inhibition in the incubation medium supplemented with the compound was observed (Figure 8). M. fortuitum (7), B. megaterium (21, 48), B. aryabhatti (34), and T. viride (47) were significantly inhibited, in contrast to the other species. B. cereus (59) was weakly stimulated, whereas the others, including the fungus P. sepedonioides (12), showed no significant effect or even a tendency toward stimulation (Figure 8). Bacterial species that were significantly inhibited showed the highest rates of NBOA-6-OH synthesis, concomitant with the fastest degradation (Figure 5). This high metabolic activity has apparently negative influences on the growth of some species.
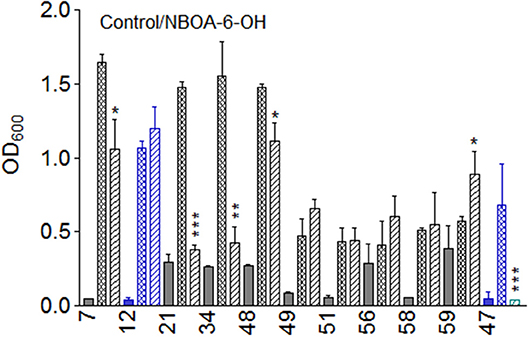
Figure 8. Growth of the microorganisms in the presence of NBOA-6-OH. n = 5; *p < 0.05; **p < 0.005; ***p < 0.0005). Nd, not detectable. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 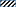 48); Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria. OD 600 culture start,
48); Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria. OD 600 culture start,  ; OD600 after 72 h: control,
; OD600 after 72 h: control,  ; OD600 in presence of 17.
; OD600 in presence of 17.
NBOA-6-OH can be harmful to plants as shown in a previous study, Schulz et al. (2017b). Growth reactions were strongly dependent on the concentration. Since the influence of nitro aromatic compounds on photosynthesis is known (Brack and Frank, 1998), we examined the effects of NBOA-6-OH on photosystem II (PSII) of cress, kohlrabi and cucumber using pulse-amplitude-modulated (PAM) fluorometry. Three-day-old cress seedlings showed a significant reduction of the quantum yield in the presence of 0.5 mM NBOA-6-OH (Figure 9). The quantum yield was lowest with 2 mM. The inhibitory effect was attributed to the nitro group, since the same amounts of BOA and BOA-6-OH had no or very weak inhibitory effects on the photosynthesis of the three plant species. Only low amounts of NBOA-6-OH were identified in extracts of kohlrabi and cress roots and shoots. In large seedlings of cucumber, NBOA-6-OH did not reach the upper parts of the stem and the leaves in sufficient amounts, explaining why no inhibition of the quantum yield occurred (Figure S8). Due to the longer stems of four-day-old compared with three-day-old cress and kohlrabi seedlings, the effect of NBOA-6-OH on quantum yields was also no longer measurable in these plants. Regarding the amounts of NBOA-6-OH found in the seedlings, the effective dose for quantum yield reduction in chloroplasts was estimated to be in the low nmol range and, thus, far below the applied concentration (2 mM). These results are in agreement with the known effects of nitro aromatic compounds on photosynthesis, such as blockade of the electron-transport chain and fluorescence quenching (Brack and Frank, 1998). The results support a high but temporary allelopathic potential of the nitro compound for the youngest and small seedlings. The negative effects should be further reduced by plant-associated microorganisms that are able to degrade the compound.
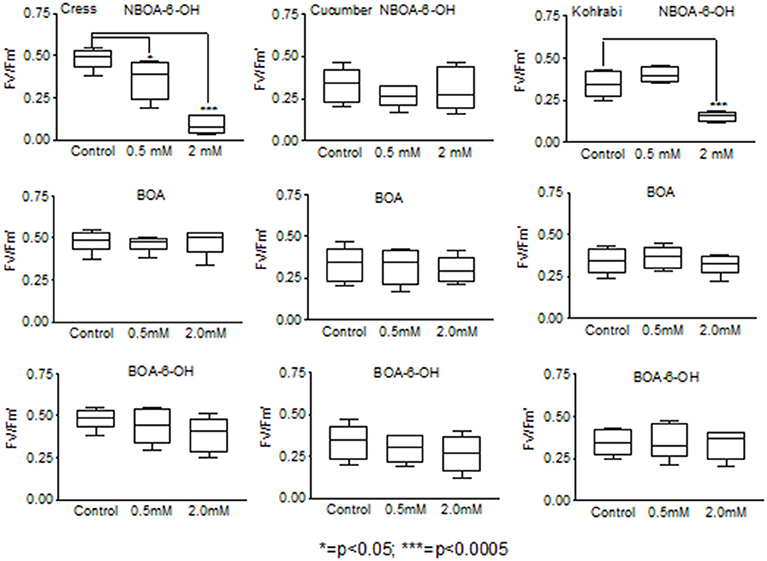
Figure 9. Reduction of the quantum yield by NBOA-6-OH is most pronounced in 3-day-old cress seedlings. p < 0.05; **, p < 0.005; ***, p < 0.0005.
As mentioned, the degradation of DIBOA led to the formation of APO as a by-product (Figure 3), an indication that AP) was generated during degradation. We incubated the microorganisms with AP and also with the microbial derivatives AAP and oHPMA to elucidate the further fate of the products.
When AP was applied, the compound was converted to AAP. The highest amounts were found with P. polymyxa (51) and A. aminovorans (49), but AAP was not present in the culture media of L. xylanilyticus (56) and B. cereus (59) and was present in low amounts in the cultures of P. sepedonioides (12) and B. megaterium (21) (Figure 10). APO was a major product in the cultures of M. fortuitum (7), P. sepedonioides (12), A. aminovorans (49), P. polymyxa (51), and L. xylanilyticus (56), and the only compound accumulating in the cultures of B. cereus (59). A portion of it was converted to AAPO by M. fortuitum (7), B. megaterium (21), T. viride (47), B. megaterium (48), B. methylotrophicus (58) and B. cereus (59). A still unidentified, hydrophilic product accumulated in the cultures of strains B. megaterium (21), B. aryabhattai (34), A. aminovorans (49), P. polymyxa (51), and B. methylotrophicus (58).
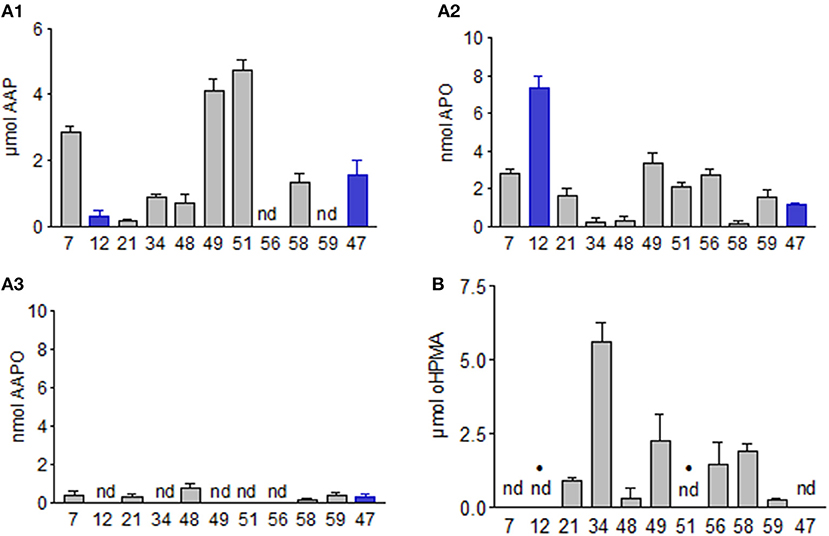
Figure 10. (A1) AAP, (A2) APO, and (A3) AAPO production of the microorganisms when incubated for 72 h with 7.5 μmol AP. (B) Residual oHPMA after incubation with 7.5 μmol, with and without accumulation of AAP (dots). Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria.
When AAP was directly applied, it was not converted by most of the microorganisms. A. aminovorans (49) and P. polymyxa (51) developed pinkish-purple colored cultures (Figure 11). These microorganisms were the most successful in reducing AAP in the culture medium, accompanied by the accumulation of new compounds (see below and Figure 12). We assumed that cell surface-bound conversion products were responsible for the colored bacteria.
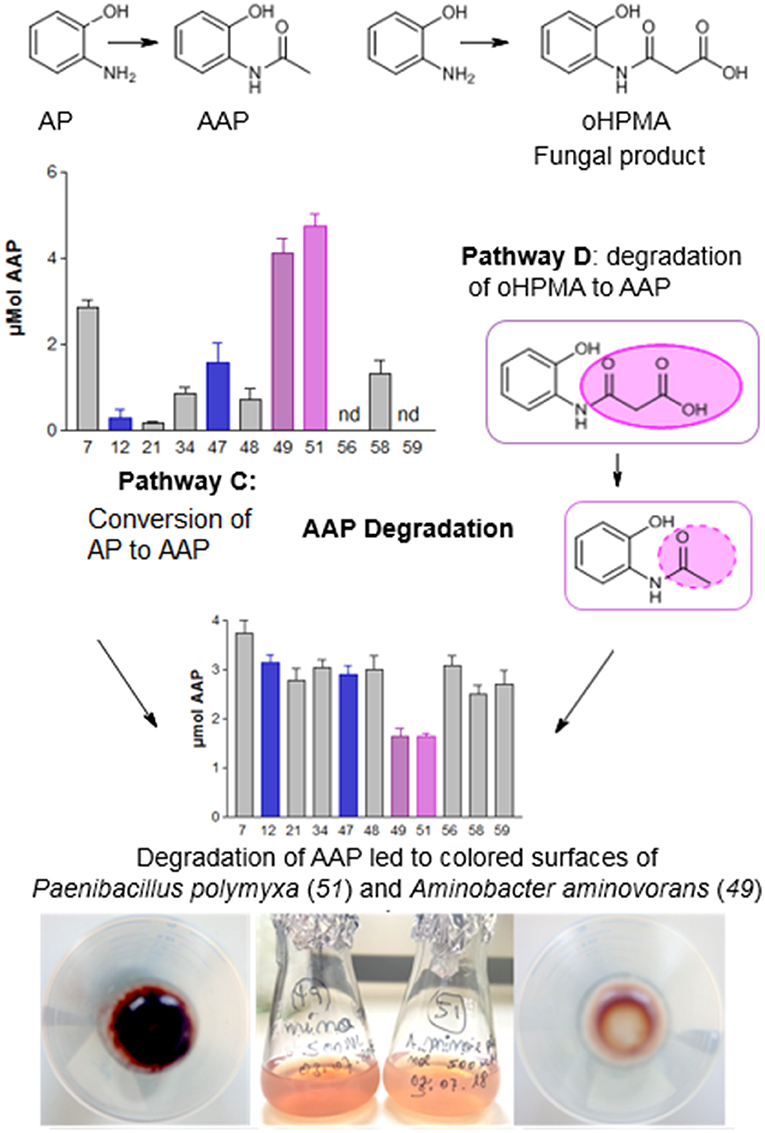
Figure 11. Pathways C and D. The scheme depicts AAP formation from AP and decrease of the compound in the media by Paenibacills polymyxa (51) and Aminobacter aminovorans (49). The rose color results from purple compounds on the bacterial surface.
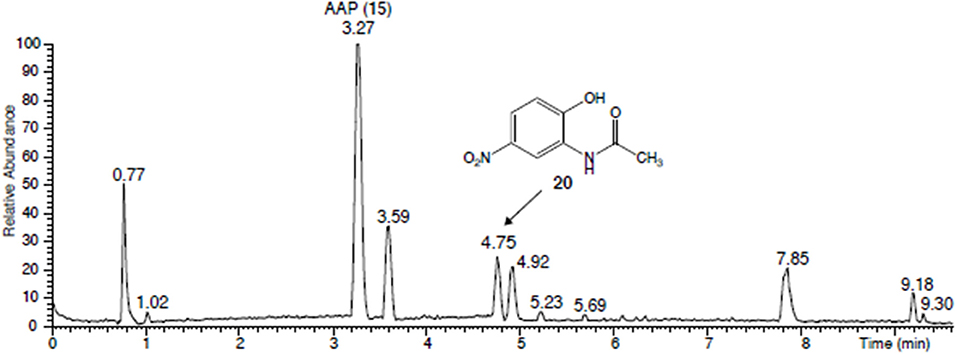
Figure 12. UHPLC chromatogram of the brown-yellowish supernatant of the centrifuged cultures of Paenibacillus polymyxa (51) and Aminobacter aminovorans (49) after incubation with AAP. Compound eluting at 4.75 min: N-(2-hydroxy-5-nitrophenyl)-acetamide.
The fungal BOA detoxification product oHPMA was completely converted by the fungus P. sepedonioides (12) and the bacterium P. polymyxa (51), (Figures 10, 11). The conversion of oHPMA resulted in the production of AAP. Rather low amounts of AAP were detected in the media of A. aminovorans (49), B. megaterium (48), and M. fortuitum (7). T. viride (47) converted all the oHPMA to AAP, which was not further degraded. A similar behavior was observed for B. megaterium (21), B. methylotrophicus (58), and B. cereus (59). In the culture medium of A. aminovorans (49) the same conversion products were found as with AAP (see below). Consequently, A. aminovorans (49) and P. polymyxa (51) developed pinkish-purple cultures with oHPMA as observed with AAP when directly applied. L. xylanilyticus (56) diminished oHPMA, but no degradation product was detected. B. aryabhattai (34) converted the compound, but only in low amounts. In the culture medium of M. fortuitum (7), B. megaterium (48), A. aminovorans (49), and P. polymyxa (51) conversion products were found, which have not yet been further characterized. The microorganisms show remarkable differences in coping with oHPMA, (Figure 10). AAP, as the first intermediate, is converted more easily by several microorganisms than when directly applied. Thus, malonylation opens a possibility for pathway D, leading to the degradation and modification of the original benzoxazolinone derivatives.
The cultures of A. aminovorans (49) and P. polymyxa (51) were centrifuged, and the obtained purple pellets of the microorganisms indicated that the bacteria were responsible for the color of the cultures (Figure 11). The supernatants had a brownish-yellowish color and contained, as mentioned, several compounds (Figure 12). The collected and freeze-dried culture supernatants of A. aminovorans (49) and P. polymyxa (51) were reconstituted in MeOH/acetonitrile 25:75 and analyzed by UHPLC-HR-MS. Four main peaks were detected at a tR of 3.27 (AAP, ISTD), 3.59, 4.75, and 4.92 min (Figure 12). A m/z signal of 197.0556 was detected in the peak at tR = 4.75 min, allowing deduction of the chemical formula of C8H9O4N2 (Δ = − 0.64 ppm) and identification of the protonated N-(2-OH-5-nitrophenyl)-acetamide (20) according to the previous work of Zikmundová et al. (2002). The identity of this structure was confirmed by analysis of the corresponding commercially available reference material and included the match of the chromatographic retention time and the MS/MS fragmentation pattern (Figure S5). Two additional compounds were detected, C8H9O3N2 at tR of 3.59 and C8H9O4N2 at tR 4.9. The HR-MS data revealed two protonated metabolites with the chemical formula of C8H9O3N2. Additional NMR and MS/MS experiments are in progress to elucidate their structure. Since nitration of the compounds may again initiate further degradation, A. aminovorans (49) and P. polymyxa (51) were incubated with the concentrated culture supernatant containing all the compounds. After 72 h, none of the compounds was left. Thus, nitration allows the rapid degradation of 20 by A. aminovorans (49) and P. polymyxa (51).
In cultures supplemented with HBOA (21), only M. fortuitum (7) was severely inhibited (Figure 13). Growth of B. megaterium (21), B. aryabhattai (34) and B. megaterium (48) was not significantly reduced. All others, except for T. viride (47), were significantly stimulated. All culture media contained still HBOA after 72 h of incubation. P. sepedonioides (12) and P. polymyxa (51) did not convert HBOA. In contrast, the compound was diminished in the media of all other microorganisms. Only about a third of the original amount was found with M. fortuitum (7), B. aryabhattai (34) and B. cereus (59). AAP was the only degradation product which accumulated in the media except for T. viride (47), A. aminovorans (49) and P. polymyxa (51) (Figure 13; Figure S6) where no product was found. The medium of P. sepedonioides (12) contained traces of AAP. According to these results, P. polymyxa (51) is the only bacterium which is unable to convert HBOA to AAP. The strain is however able to nitrate AAP and to degrade the resulting nitroaromatic compound, thus able to remove AAP produced by other species perhaps under natural conditions. The results demonstrate that HBOA is degraded to AAP not only by fungi but also by bacteria.
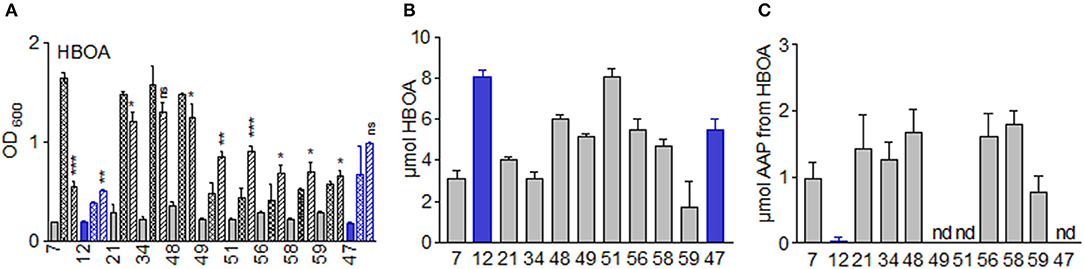
Figure 13. Incubations of the microorganisms with HBOA. (A) Growth of the microorganisms in presence of 7.5 μmol HBOA; OD 600 culture start,  ; OD600 after 72 h: control,
; OD600 after 72 h: control,  ; OD600 in presence of HBOA,
; OD600 in presence of HBOA, 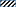 . (B) residual amounts of HBOA after 72 h of incubation with 7.5 μmol HBOA. (C) accumulation of the HBOA degradation product AAP after 72 h of incubation with HBOA. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria. *p < 0.05; **p < 0.005; ***p < 0.0005.
. (B) residual amounts of HBOA after 72 h of incubation with 7.5 μmol HBOA. (C) accumulation of the HBOA degradation product AAP after 72 h of incubation with HBOA. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars, fungi; gray bars, bacteria. *p < 0.05; **p < 0.005; ***p < 0.0005.
We tested the plant BOA detoxification product glucoside carbamate for its influence on microbial growth (Figure 14). With the exception of M. fortuitum (7) the growth of the microorganisms was stimulated. In the culture media of several microorganisms, glucoside carbamate was re-converted to the precursor BOA-N-glucoside, but the rate of re-conversion differed between repetitions. The rearrangement rates were higher for the two fungi and M. fortuitum (7), A. aminovorans (49), P. polymyxa (51), and B. methylotrophicus (58). With M. fortuitum (7), traces of glucosylated carbamic acid were detected (Schulz et al., 2013), resulting from lactone ring opening without rearrangement. Regarding the applied amount of glucoside carbamate (10 μmol), only M. fortuiutum (7), P. polymyxa (51), and B. methylorophicus (58) showed a considerably lower content of the compounds (glucoside carbamate and BOA-N-glucoside), indicating complete degradation without accumulation of BOA.
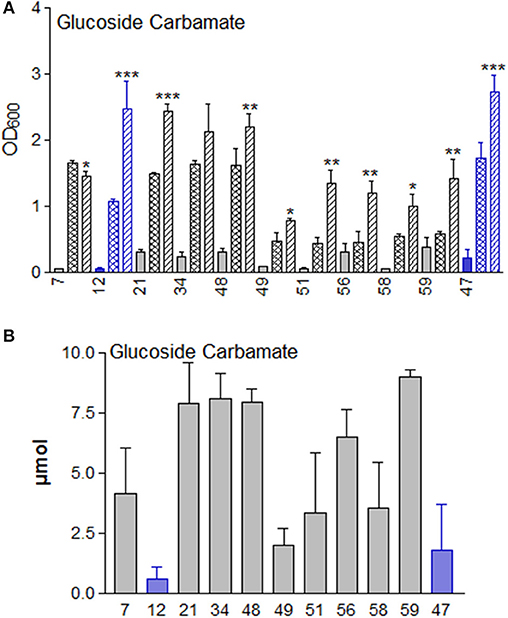
Figure 14. (A) Glucoside carbamate supplementation significantly stimulates the growth of the microorganisms in Czapek medium, except for strain 7, in comparison to the controls without glucoside carbamate. OD 600 culture start,  ; OD600 after 72 h: control,
; OD600 after 72 h: control,  ; OD600 in presence of glucoside carbamate,
; OD600 in presence of glucoside carbamate,  . Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars: fungi; gray bars: bacteria. (B) Glucoside carbamate can be converted back to BOA-N-glucoside by M. fortuitum (7), P. sepedonioides (12), A. aminovorans (49), P. polymyxa (51) and T. viride (47). M. fortuitum (7), P. polymyxa (51) and B. methylotrophicus (58) reduced the content of the compounds without BOA accumulation. *p < 0.05; **p < 0.005; ***p < 0.0005.
. Microorganisms: Mycobacterium fortuitum (7), Bacillus aryabhattai (34), Bacillus cereus (59), Bacillus megaterium (21, 48), Bacillus methylotrophicus (58), Lysinibacillus xylanilyticus (56), Paenibacillus polymyxa (51), Aminobacter aminovorans (49), the fungi Papulaspora sepedonioides (12), and Trichoderma viride (47). Blue bars: fungi; gray bars: bacteria. (B) Glucoside carbamate can be converted back to BOA-N-glucoside by M. fortuitum (7), P. sepedonioides (12), A. aminovorans (49), P. polymyxa (51) and T. viride (47). M. fortuitum (7), P. polymyxa (51) and B. methylotrophicus (58) reduced the content of the compounds without BOA accumulation. *p < 0.05; **p < 0.005; ***p < 0.0005.
This study illustrates the high input of metabolic activities, and thus energy, necessary to eliminate naturally occurring benzoxazinoids and their downstream metabolites from the environment, often at the cost of growth. Microorganisms differed markedly in their compound-modifying and degradation abilities, with P. polymyxa (51) and A. aminovorans (49) being the most active representatives. Generation of APO from DIBOA is most likely attributed to the degradation of α-oxo-aldehyde, the intermediate formed upon ring opening of DIBOA (Sicker and Schulz, 2002; Zikmundová et al., 2002; Niemeyer, 2009; Wouters et al., 2016). On root surfaces, consortia of microorganisms may act in concert to achieve more rapid compound elimination, for instance for HBOA degradation via AAP and nitro-AAP. This assumption is supported by previous studies (Haghikia et al., 2014; Schulz et al., 2017a,b, 2018). We did not include studies of the compositions of microorganisms due to our understanding that natural consortia are hierarchically structured and organized, perhaps by fungi and fungus/plant associations.
The microorganisms studied herein demonstrated difficulties degrading BOA and MBOA upon direct exposure, despite, according to the soil history, the prior exposure of the microorganisms to benzoxazinoids. As mentioned above, benzoxazolinones and phenoxazinones have been found in agricultural soils months after wheat culture, suggesting that elimination of the compounds occurs rather slowly. Thus, our results are in line with the known stability of the compounds in soil.
This study gives rise to the hypothesis that the conversion of BOA and MBOA by microorganisms can be facilitated by plant activities, in particular by the hydroxylation of BOA and de-methylation of MBOA. Both mechanisms require compound exchanges by still unknown mechanisms between plants and microbial organisms (Figure S9). The removal of O-linked methyl groups by plant 2-oxoglutarate/Fe (II)-dependent dioxygenase (ODD) has been recently described by Farrow and Facchini (2013). Peroxidase is another enzyme that is able to perform O-demethylation. Enzymes that catalyze O- and N-demethylation are common in bacteria. The demethylases responsible for caffeine demethylation by P. putida provide a well-known example (Summers et al., 2012). Presently, it is not clear why the microorganisms used in this study did not perform demethylation of MBOA. In cases in which the environmental conditions of the natural habitat are not suitable to activate microbial activities for MBOA demethylation, the deficit can be bypassed by plants. Another explanation would be that microorganisms perform the MBOA demethylation step only when associated with plants. Glucosylation and de-methylation of the downstream products by plant enzymes may maintain consistent steady state levels of the reactive intermediates. Further studies will substantiate the posited cooperation.
When nitrate is available, bacterial nitrite production leads to nitration of the plant product BOA-6-OH, which is an introductory step leading to complete degradation by many bacteria and fungi. Here, several pathways, which were not directed in this study, are possible, depending on the oxygen availability (Ju and Parales, 2010). For some bacteria, we found no or extremely low nitration [for instance B. methylotrophicus (58)], although strains of these species have been described to reduce nitrate (B. licheniformis: Konohama et al., 1993; V. paradoxus: Han et al., 2013; B. methylotrophicus: Madhaiyan et al., 2010). We assume that the oxygen content in the cultures may be crucial for nitration, with species-dependent thresholds. Strongly anaerobic conditions consume nitrate as an alternative electron acceptor, similar to the phenomenon described for the denitrifying Paracoccus denitrificans (Lycus et al., 2018). In this case, nitration of BOA-6-OH may not occur. This assumption remains to be challenged by subsequent studies.
Among the applied detoxification products, only glucoside carbamate had a stimulatory influence on the growth of almost all the microorganisms, GDIMBOA was stimulatory for five of them and GDIBOA was stimulatory for the B. megaterium strains. The underlying mechanism is presently elusive, since it does not seem to be directly related to the catabolism of glucoside carbamate, GDIBOA or GDIMBOA. Regarding GDIMBOA, interference with gluconic acid pathways is possible for several microorganisms. For a proper explanation, further investigations are necessary. Glucoside carbamate, which possesses a lactone bond, does not require β-glucosidases specific for the hydrolysis of O-bound glucose. Opening of the lactone ring results in glucosylated carbamic acid as a degradation intermediate. This compound, which is sufficiently stable, in contrast to the free acid, was formerly identified as a degradation intermediate in fungal cultures supplemented with glucoside carbamate (Schulz et al., 2013). However, we found only traces of this compound with M. fortuitum (7), instead, the two fungi and several bacterial strains reversed glucose carbamate to BOA-N-glucoside. We assume however, that opening of the lactone bond is a prerequisite for further degradation.
GDIMBOA was transformed without DIMBOA but with MBOA accumulation by four of the bacteria in a gluconate-rich environment. Microorganisms release organic acids for solubilization of phosphate from minerals (Siebers et al., 2018), of which gluconic acid is often a dominant acid. It is therefore realistic to assume that gluconic acid is an available metabolite in the natural environment. The microorganisms, which cannot degrade GDIMBOA, may participate in compound elimination by metabolizing degradation intermediates produced by other organisms, when they are members of plant-associated microbial consortia. Thus, even when they are unable to initiate degradation pathways, they can contribute to or take over subsequent degradation steps. The growth-promoting effect of GDIMBOA seems to be more species-specific than that of glucoside carbamate. The results led to the question whether plants can modulate the growth of associated microorganisms by glucosylation/ deglucosylation of key secondary metabolites, when moonlighting glucosyltransferases/ glucosidases are shifted to the sites where cross-kingdom communications take place. Moonlighting enzymes catalyze different reactions and accept other substrates when shifted to other compartments (Jeffery, 1999; Hernández et al., 2012). Maize, for instance, can prime DIBOA degradation of GDIBOA (1) by deglucosylation. In this context, the chloroplast-located glucosidases ZmGLU1 and ZmGLU2 (Nestler et al., 2011; Hannemann et al., 2018), may have moonlighting functions when shifted to the apoplast as well as glucosyltransferase BX9 at positions within the cell wall (Schulz et al., 2015, 2016).
Plant glucosides were found to promote growth of the gut bacterium Lactobacillus acidophilus and other Lactobacilli; the release of mono- and disaccharides requires phosphorylation of the sugar moiety prior to hydrolysis by a special phospho-β-glucosidase (Michlmayr and Kneifel, 2014; Theilmann et al., 2017). The released aglycones also affects other gut bacteria, modulates the species composition, and can undergo further conversions. Phosphorylation of the secondary glucoside products seems to more common than thought in animals. Salicin phosphate and catechol glucoside phosphate were recently identified in the feces of gypsy moths (Boeckler et al., 2016). In contrast to animals such as vertebrates, which avoid the sacrifice of valuable glucose and prefer glucuronic acid or sulfate for detoxification steps (Hanhineva et al., 2014; Hodges and Minich, 2015), plants often repeatedly glucosylate modified compounds. The glucosides present in this case molecules without bioactivity, but they can be re-activated by deglucosylation. After exudation, plant-modified compounds were again transformed and finally degraded by the microorganisms. Whether bacteria, fungi, and plants cooperate to perform the biochemical reaction sequences using strategies that prepare the compounds for degradation is still hypothetical.
The BOA breakdown product AP was acetylated to AAP by almost all the microorganisms, but with different rates of success. AAP was not produced by strains L. xylanilyticus (56) and B. cereus (59) and only in low amounts by P. sepedonioides (12) and B. aryabhattai (34). A.aminovorans (49), and P. polymyxa (51) showed outstanding AAP production. These strains initiated nitrite production for nitration of AAP. We conclude that nitration of naturally occurring aromatic compounds is a more common reaction than thought, especially in agricultural systems with high nitrate input.
AAP is found as an intermediate of oHPMA degradation. Only a few fungi perform malonyl conjugation to yield oHPMA. When AP is conjugated to malonic acid, most microorganisms can degrade the compound rapidly. Thus, the conjugation to malonic acid bypasses the low biological degradability observed with directly applied AAP, resembling the degradation mechanism of DIBOA. It has been suggested that malonate is converted to acetyl-CoA (Lee et al., 2000; Suvorova et al., 2012). For oHPMA degradation, a similar mechanism may be used, explaining why AAP was an intermediate in oHPMA degradation by several strains. Malonate conjugation is a common detoxification reaction for xenobiotics in plants (Taguchi et al., 2010). Interestingly, soluble glucoside carbamate is malonylated in maize at late stages of the BOA detoxification process (Hofmann et al., 2006), but this compound accumulates only in low amounts and was therefore not available for degradation studies. We assume that malonylation may promote the degradation activities of microorganisms for secondary metabolites.
The results of this study provide insights in the complex benzoxazinoid degradation processes. The degradation capacity for benzoxazinoids may present a selective driver for the microbial species composition and diversity in habitats with benzoxazinone-exuding plant species. Benzoxazinones are therefore thought to act as modifiers of the variable part of the microbiomes belonging to benzoxazinone-containing plant species (Hu et al., 2018). For instance, DIMBOA attracts Pseudomonas putida to the maize rhizosphere (Neal et al., 2012). The metabolic capacities of these species may also influence allelopathic interactions by the generation of short-lived toxic intermediates, including radicals, during compound degradation demanding antecedent molecule modification. Radicals occur, for instance, during polymerization reactions of BOA-6-OH (Schulz et al., 2017a). Microbial degradation and compound modification activities may also be important in the recently more intensively investigated phenomenon of Horizontal Natural Product Transfer (Selmar et al., 2018), which resembles in part the behavior of some Leptidoptera when storing toxic secondary metabolites absorbed from plants for defense. Plant-microbe secondary compound networks are certainly much more complex regarding the huge amount of microbial secondary compounds, which could be also involved and thus far have been less investigated in context with plant secondary metabolites. Under natural conditions, processes are most likely supported by spontaneous, abiotic chemical reactions and by extracellular enzymes adsorbed on soil minerals (Datta et al., 2017).
Although the present results may not mirror the complex processes in the soil and rhizosphere, they give rise to assume interactions for the successful elimination of bioactive natural compounds such as benzoxazinone. A high biodiversity of microorganisms is of extraordinary importance because it guarantees flexibility for the elimination of allelochemicals belonging to different chemical classes. Our findings also show that the elimination of benzoxazinoids is a time-consuming process demanding a high energy input and which may vary depending on oxygen availability. In this context, it is noteworthy that the plant detoxification product glucoside carbamate functions as a microbial growth stimulator. Thus, the abundance of some microorganisms could be increased, resulting in a faster degradation of compounds.The availability of nitrate is another factor in the modulation of degradation pathways. High inputs of nitrate may favor the synthesis of nitroaromatic compounds in agricultural systems. Nitroaromatic compounds can be extremely toxic to animals and to certain microorganisms, and thus rapid degradation will reduce negative effects on these organisms. The different metabolic abilities of a microbial community with a high species diversity also alleviate variations in environmental conditions, such as variable oxygen contents in the soil. We postulate that modulation of the microbiome species composition is a primary function of this class of secondary plant metabolites, enriching the soil with microorganisms that are compatible with benzoxazinone-containing plants and with accompanying plants belonging to the same plant community. Species of the accompanying flora can absorb the compounds (Schulz and Wieland, 1999). These interactions are also thought to drive co-evolutionary processes between plants and microoganisms. Functions in microbiome modulation do not discount the insecticidal and allelopathic/herbicidal properties of benzoxazinoids.
MS designed the project, synthesized NBOA-6-OH, conducted the experiments with plants, isolated the AAP conversion products, and did HPLC analyses. VS conducted and supervised culture incubations and PAM measurements. LL cultivated V. paradoxus. VS and LL performed the experiments for bacterial identification. LB and SG conducted the experiments for nitro AAP identification and structure analysis. DS provided compounds for the screening and helps with figures. MS and LB wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Scarlett Geunes-Boyer for language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00238/full#supplementary-material
Boeckler, G. A., Paetz, C., Feibicke, P., Gershenzon, J., and Unsicker, S. B. (2016). Metabolism of poplar salicinoids by the generalist herbivore Lymantria dispar (Lepidoptera). Insect Biochem. Mol. Biol. 78, 39–49. doi: 10.1016/j.ibmb.2016.08.001
Brack, W., and Frank, H. (1998). Chlorophyll a fluorescence: a tool for the investigation of toxic effects in the photosynthetic apparatus. Ecotoxicol. Environ. Saf. 40, 34–41. doi: 10.1006/eesa.1997.1639
Chirino, B., Strahsburger, E., Agulló, L., González, M., and Seeger, M. (2013). Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS ONE 8:e75746. doi: 10.1371/journal.pone.0075746
Dagley, S., and Johnson, P. A. (1963). Microbial oxidation of kynurenic, xanthurenic and picolinic acids. Biochim. Biophys. Acta 78, 577–587. doi: 10.1016/0006-3002(63)91023-0
Datta, R., Anand, S., Moulick, A., Baraniya, D., Pathan, S. I., Rejsek, K., et al. (2017). How enzymes are adsorbed on soil solid phase and factors limiting its activity: a review. Int. Agrophys. 31, 287–302. doi: 10.1515/intag-2016-0049
Farrow, S. C., and Facchini, P. J. (2013). Dioxygenases catalyze O-demethylation and O,O-demethylenation with widespread roles in benzylisoquinoline alkaloid metabolism in opium poppy. J. Bio.l Chem. 288, 28997–29012. doi: 10.1074/jbc.M113.488585
Fomsgaard, I. S., Mortensen, A. G., and Carlsen, S. C. (2004). Microbial transformation products of benzoxazolinone and benzoxazinone allelochemicals, a review. Chemosphere 54, 1025–1038. doi: 10.1016/j.chemosphere.2003.09.044
Frey, M., Chomet, P., Glawischnig, E., Stettner, G., Gruen, S., Winklmair, A., et al. (1997). Analyses of a chemical plant defense mechanism in grasses. Science 277, 696–699. doi: 10.1126/science.277.5326.696
Friebe, A., Vilich, V., Hennig, L., Kluge, M., and Sicker, D. (1998). Detoxification of benzoxazolinone allelochemicals from wheat by Gaeumannomyces graminis var. tritici, G. graminis var. graminis, G. graminis var. avenae, and Fusarium culmorum. Appl. Environ. Microbiol. 64, 2386–2391.
Glenn, A. E., Davis, C. B., Gao, M., Gold, S. E., Mitchell, T. R., Proctor, R. H., et al. (2016). Two horizontally transferred xenobiotic resistance gene clusters associated with detoxification of benzoxazolinones by Fusarium species. PLoS ONE 11:e0147486. doi: 10.1371/journal.pone.0147486
Gollakota, K. G., and Halvorson, H. O. (1960). Biochemical changes occurring during sporulation of Bacillus cereus. Inhibition of sporulation by alpha-picolinic acid. J. Bacteriol. 79, 1–8.
Grant, R. S., Coggan, S. E., and Smythe, G. (2009). The physiological action of picolinic acid in the human brain. Int. J. Tryptophan Res. 2, 71–79. doi: 10.4137/IJTR.S2469
Haghikia, S., Schulz, M., Ayah, E., Schouten, A., Müllenborn, C., Paetz, C., et al. (2014). Abutilon theophrasti's defense against the allelochemical benzoxazolin-2(3H)-one: Support by Actinomucor elegans. J. Chem. Ecol. 40, 1286–1298. doi: 10.1007/s10886-014-0529-7
Han, J. I., Spain, J. C., Leadbetter, J. R., Ovchinnikova, G., Goodwin, L. A., Han, C. S., et al. (2013). Genome of the root-associated plant growth-promoting bacterium Variovorax paradoxus Strain EPS. Genome Announ. 24;1:e00843–13. doi: 10.1128/genomeA.00843-13
Hanhineva, K., Keski-Rahkonen, P., Lappi, J., Katina, K., Pekkinen, J., Savolainen, O., et al. (2014). The postprandial plasma rye fingerprint includes benzoxazinoid-derived phenylacetamide sulfates. J. Nutr. 144, 1016–1022. doi: 10.3945/jn.113.187237
Hannemann, L., Lucaciu, C. R., Sharma, S., Rattei, T., Mayer, K. F. X., Gierl, A., et al. (2018). A promiscuous beta-glucosidase is involved in benzoxazinoid deglycosylation in Lamium galeobdolon. Phytochemistry 156, 224–233. doi: 10.1016/j.phytochem.2018.10.012
Hartenstein, H., Klein, J., and Sicker, D. (1993). Efficient isolation procedure for (2R)-2-β-D-glucopyranosyloxy-4-hydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one from maize. Ind. J. Heterocyc. Chem. 2, 151–153. doi: 10.1016/S0031-9422(00)90618-6
Hernández, S., Amela, I., Cedano, J., Piñol, J., Perez-Pons, J. A., Mozo-Villarias, A., et al. (2012). Do moonlighting proteins belong to the intrinsic disordered proteins class? J. Proteom. Bioinform. 5, 262–264. doi: 10.4172/jpb.1000247
Hodges, R. E., and Minich, D. M. (2015). Modulation of metabolic detoxification pathways using foods and food-derived components: a scientific review with clinical application. J. Nutr. Metab. 2015:760689. doi: 10.1155/2015/760689
Hofmann, D., Knop, M., Hao, H., Hennig, L., Sicker, D., and Schulz, M. (2006). Glucosides from MBOA and BOA detoxification by Zea mays and Portulaca oleracea. J. Nat. Prod. 69, 34–37. doi: 10.1021/np0580762
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Comm. 9:2738. doi: 10.1038/s41467-018-05122-7
Jeffery, C. J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24, 8–11. doi: 10.1016/S0968-0004(98)01335-8
Ju, K. S., and Parales, R. E. (2010). Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Biol. Res. 74, 250–272. doi: 10.1128/MMBR.00006-10
Kettle, A. J., Batley, J, Benfield, A. H., Manners, J. M., Kazan, K, and Gardinera, D. M. (2015). Degradation of the benzoxazolinone class of phytoalexins is important for virulence of Fusarium pseudograminearum towards wheat. Mol. Plant Pathol. 16, 946–962. doi: 10.1111/mpp.12250
Knop, M., Pacyna, S., Voloshchuk, N., Kant, S., Müllenborn, C., Steiner, U., et al. (2007). Zea mays: benzoxazolinone detoxification under sulfur deficiency conditions - a complex allelopathic alliance including endophytic Fusarium verticillioides. J. Chem. Ecol. 33, 225–237. doi: 10.1007/s10886-006-9226-5
Konohama, T., Murakami, S., Nanmori, T., Aoki, K., and Shinke, R. (1993). Nitrate reduction in Bacillus licheniformis in the presence of ammonium salt by shaking culture. Biosci. Biotech. Biochem. 57, 1082–1086. doi: 10.1271/bbb.57.1082
Lee, H. Y., An, J. H., and Kim, Y. S. (2000). Identification and characterization of a novel transcriptional regulator, MatR, for malonate metabolism in Rhizobium leguminosarum bv. Trifolii. Eur. J. Biochem. 267, 7224–7229. doi: 10.1046/j.1432-1327.2000.01834.x
Lycus, P., Soriano-Laguna, M. J, Kjos, M., Richardson, D. J., Gates, A. J., Milligan, D. A., et al. (2018). A bet-hedging strategy for denitrifying bacteria curtails their release of N2O. Proc Nat Acad Sci. U.S.A. 115, 11820–11825. doi: 10.1073/pnas.1805000115
Macías, F. A., Marín, D., Oliveros-Bastidas, A., and Molinillo, J. M. (2009). Rediscovering the bioactivity and ecological role of 1,4-benzoxazinones. Nat. Prod. Rep. 26, 478–89. doi: 10.1039/b700682a
Macias, F. A., Oliveros-Bastidas, A., Marin, D., Castellano, D., Simonet, A. M., and Molinillo, J. M. G. (2005). Degradation studies on benzoxazinoids. Soil degradation dynamics of (2R)-2-ß-D-glucopyranosyl-4-hydroxy(2H)-1,4-phenoxazin-3-(4H)-one (DIBOA-Glc) and its degradation products, phytotoxic allelochemicals from Gramineae. J. Agric. Food Chem. 53, 538–548. doi: 10.1021/jf048702l
Madhaiyan, M., Poonguzhali, S, Kwon, S. W., and Sa, T. M. (2010). Bacillus methylotrophicus sp. nov., a methanol-utilizing, plant-growth-promoting bacterium isolated from rice rhizosphere soil. Int. J. Sys. Evol. Microbiol. 60, 2490–2495. doi: 10.1099/ijs.0.015487-0
Michlmayr, H., and Kneifel, W. (2014). β-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS Microbiol. Lett. 352, 1–10. doi: 10.1111/1574-6968.12348
Nannipieri, P., Kandeler, E., and Ruggiero, P. (2002). “Enzyme activities and microbiological and biochemical processes in soil,” in Enzymes in the environment, eds R. G. Burns and R. P. Dick (New York, NY: Marcel Dekker), 1–33.
Neal, A. L., Ahmad, S., Gordon-Weeks, R., and Ton, J. (2012). Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 7:e35498. doi: 10.1371/journal.pone.0035498
Nestler, J., Schütz, W., and Hochholdinger, F. (2011). Conserved and unique features of the maize (Zea mays L.) root hair proteome. J. Proteome Res. 10, 2525–2537. doi: 10.1021/pr200003k
Niculaes, C., Abramov, A., Hannemann, L., and Frey, M. (2018). Plant protection by benzoxazinoids - Recent insights into biosynthesis and function. Agronomy 8:143. doi: 10.3390/agronomy8080143
Niemeyer, H. M. (2009). Hydroxamic acids derived from 2-hydroxy-2h-1,4-benzoxazin-3(4H) one: Key defense chemicals of cereals. J. Agric. Food. Chem. 57, 1677–1696. doi: 10.1021/jf8034034
Nomura, T., Ishihara, A., Imaishi, H., Endo, T., OhKawa, H., and Iwamura, H. (2002). Molecular characterization and chromosomal localization of cytochrome P450 genes involved in the biosynthesis of cyclic hydroxamic acids in hexaploid wheat. Molec. Genet. Genom. 267, 210–217. doi: 10.1007/s00438-002-0653-x
Ozaki, Y., and Kato-Noguchi, H. (2016). Effects of benzoxazinoids in wheat residues may inhibit the germination, growth and gibberellin-induced α-amylase activity in rice. Acta Physiol. Plant. 38:24. doi: 10.1007/s11738-015-2041-2
Qiu, J., Zhang, J., Zhang, Y., Wang, Y., Tong, L., Hong, Q., et al. (2017). Biodegradation of picolinic acid by a newly isolated bacterium Alcaligenes faecalis Strain JQ135. Curr. Microbiol. 74, 508–514. doi: 10.1007/s00284-017-1205-2
Schulz, M., Filary, B., Kühn, S., Colby, T., Harzen, A., Schmidt, J., et al. (2015). Benzoxazolinone detoxification by N-Glucosylation: the multi-compartment- network of Zea mays L. Plant Signal. Behav. 11:e1119962. doi: 10.1080/15592324.2015.1119962
Schulz, M., Hofmann, D., Sicker, D., Hennig, L., Schütz, V., Preusche, M., et al. (2018). Pantoea ananatis converts MBOA to 6-methoxy-4-nitro-benzoxazolin-2(3H)-one (NMBOA) for cooperative degradation with its native root colonizing microbial consortium. Nat. Prod. Com. 13, 1271–1275. doi: 10.1177/1934578X1801301010
Schulz, M., Kant, S., Colby, T., Harzen, A., Schmidt, J., Sicker, D., et al. (2016). Zea mays glucosyltransferase BX9 - An essential enzyme for benzoxazolinone detoxification. J. Allelochem. Interact. 2, 25–38.
Schulz, M., Sicker, D., Schackow, O., Hennig, L., Hofmann, D., Disko, U., et al. (2017b). 6-Hydroxy-5-nitrobenzo[d]oxazol-2(3H)-one - A degradable derivative of natural 6-hydroxybenzoxazolin-2(3H)-one produced by Pantoea ananatis. Commun. Int. Biol. 10:e1302633. doi: 10.1080/19420889.2017.1302633
Schulz, M., Sicker, D., Schackow, O., Hennig, O., Yurkov, A., Siebers, M., et al. (2017a). Cross-cooperations of Abutilon theophrasti Medik. and root surface colonizing microorganisms disarm phytotoxic hydroxy-benzoxazolin-2(3H)-ones. Plant Signal. Behav. 12:e1358843. doi: 10.1080/15592324.2017.1358843
Schulz, M., Tabaglio, V., Marocco, A., Macias, F. A., and Molinillo, J. M. (2013). Benzoxazinoids in rye allelopathy – from discovery to application in sustainable weed control and organic farming. J. Chem. Ecol. 39, 154–174. doi: 10.1007/s10886-013-0235-x
Schulz, M., and Wieland, I. (1999). Variations in metabolism of BOA among species in various field communities - biochemical evidence for co-evolutionary processes in plant communities? Chemoecology 9, 133–141. doi: 10.1007/s000490050044
Selmar, D., Abouzeid, S., Radwan, A., Hijazin, T, Yahyazadeh, M., Lewerenz, L., et al. (2018). “Horizontal natural product transfer: a novel attribution in allelopathy,” in Ref. Series. Phytochem Co-Evolution of Secondary Metabolites eds J. M. Mérillon and K. G. Ramawat (Heidelberg: Springer Nature).
Sicker, D., Prätorius, B., Mann, G., and Meyer, L. (1989). Convenient synthesis of 2,4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one. Synthesis 1989, 211–212. doi: 10.1055/s-00000084
Sicker, D., Schneider, B., Hennig, L., Knop, M., and Schulz, M. (2001). Glycoside carbamates from benzoxazolin-2(3H)-one detoxification in extracts and exudates of corn roots. Phytochemistry 58, 819–825. doi: 10.16/S0031-94220(01)00299-0
Sicker, D., and Schulz, M. (2002). “Benzoxazinones in plants: occurrence, synthetic access and biological activity,” in Studies in Natural Products Chemistry 27 Bioactive Natural Products Part H, ed Atta-ur-Rahman (Amsterdam: Elsevier, 185–232.
Siebers, M., Rohr, T., Ventura, M., Schütz, V., Thies, S., Kovacic, F., et al. (2018). Disruption of microbial community composition and identification of plant growth promoting microorganisms after exposure of soil to rapeseed-derived glucosinolates. PLoS ONE 13:e0200160. doi: 10.1371/journal.pone.0200160
Summers, R. M., Louie, T. M., Yu, C. L., Gakhar, L., Louie, K. C., and Subramanian, M. (2012). Novel, highly specific N-demethylases enable bacteria to live on caffeine and related purine alkaloids. J. Bacteriol. 194, 2041–2049. doi: 10.1128/JB.06637-11
Suvorova, I. A., Ravcheev, D. A., and Gelfand, M. S. (2012). Regulation and evolution of malonate and propionate catabolism in proteobacteria. J. Bacteriol. 194, 3234–3240. doi: 10.1128/JB.00163-12
Taguchi, G., Ubukata, T., Nozue, H., Kobayashi, Y., Takahi, M., Yamamoto, H., et al. (2010). Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 63, 1031–1041. doi: 10.1111/j.1365-313X.2010.04298.x
Theilmann, M. C., Goh, Y. J., Nielsen, K. F., Klaenhammer, T. R., Barrangou, R., and Abou Hachem, M. (2017). Lactobacillus acidophilus metabolizes dietary plant glucosides and externalizes their bioactive phytochemicals. mBio 8, e01421–e01417. doi: 10.1128/mBio.01421-17
Venturelli, S., Belz, R. G., Kämper, A., Berger, A., von Horn, K., Wegner, A., et al. (2015). Plants release precursors of histone deacetylase inhibitors to suppress growth of competitors. Plant Cell 27, 3175–3189. doi: 10.1105/tpc.15.00585
Venturelli, S., Petersen, S., Langenecker, T., Weigel, D., Lauer, U. M., and Becker, C. (2016). Allelochemicals of the phenoxazinone class act at physiologically relevant concentrations. Plant Signal. Behav. 11:e1176818. doi: 10.1080/15592324.2016.1176818
Wieland, I., Kluge, M., Schneider, B., Schmidt, J., Sicker, D., and Schulz, M. (1998). 3-ß-D-Glucopyranosyl-benzoxazolin-2(3H)-one - A detoxification product of benzoxazolin2(3H) -one in oat roots. Phytochemistry 48, 719–722. doi: 10.1016/S0031-9422(97)91012-8
Woodward, M. D., Corcuera, L. J., Helgeson, J. P., and Upper, C. D. (1978). Decomposition of 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one in aqueous solution. Plant Physiol. 61, 796–802. doi: 10.1104/pp.61.5.796
Wouters, F. C., Gershenzon, J., and Vassao, D. G. (2016). Benzoxazinoids: Reactivity and modes of action of a versatile class of plant chemical defenses. J. Braz. Chem. Soc. 27, 1379–1397. doi: 10.5935/0103-5053.20160177
Zhao, J. S., Singh, A., Huang, X. D., and Ward, O. P. (2000). Biotransformation of hydroxylaminobenzene and aminophenol by Pseudomonas putida 2NP8 cells grown in the presence of 3-nitrophenol. Appl. Environ. Microbiol. 66, 2336–2342. doi: 10.1128/AEM.66.6.2336-2342.2000
Keywords: microorganisms, benzoxazinoids, nitration, BOA-6-OH, degradation
Citation: Schütz V, Bigler L, Girel S, Laschke L, Sicker D and Schulz M (2019) Conversions of Benzoxazinoids and Downstream Metabolites by Soil Microorganisms. Front. Ecol. Evol. 7:238. doi: 10.3389/fevo.2019.00238
Received: 22 January 2019; Accepted: 07 June 2019;
Published: 27 June 2019.
Edited by:
Panagiotis Milonas, Benaki Phytopathological Institute, GreeceReviewed by:
Stine Krogh Steffensen, Aarhus University, DenmarkCopyright © 2019 Schütz, Bigler, Girel, Laschke, Sicker and Schulz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margot Schulz, dWxwNTA5QHVuaS1ib25uLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.