- 1Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland
- 2Musée Cantonal de Zoologie, Palais de Rumine, Lausanne, Switzerland
- 3Zoologische Staatssammlung München, Munich, Germany
- 4Department of Biological Sciences, Boise State University, Boise, ID, United States
- 5Department of Computational Biology, University of Lausanne, Lausanne, Switzerland
- 6Geneva Museum of Natural History, Geneva, Switzerland
Understanding processes that have driven the extraordinary high level of biodiversity in the tropics is a long-standing question in biology. Here we try to assess whether the large lineage richness found in a New Guinean clade of mayflies (Ephemeroptera), namely the Thraulus group (Leptophlebiidae) could be associated with the recent orogenic processes, by applying a combination of phylogenetic, biogeographic and ecological shift analyses. New Guinean representatives of the Thraulus group appear monophyletic, with the possible exception of a weakly-supported early-diverging clade from the Sunda Islands. Dating analyses suggest an Eocene origin of the Thraulus group, predating by several million years current knowledge on the origin of other New Guinean aquatic organisms. Biogeographic inferences indicate that 27 of the 28 inferred dispersals (96.4%) occurred during the Eocene, Oligocene and Miocene, while only one dispersal (3.6%) took place during the Pliocene-Pleistocene. This result contrasts with the higher number of altitudinal shifts (15 of 22; 68.2%) inferred during the Pliocene-Pleistocene. Our study illustrates the role played by—potentially ecological—diversification along the elevation gradient in a time period concomitant with the establishment of high-altitude ecological niches, i.e., during orogenesis of the central New Guinean mountain range. This process might have taken over the previous main mode of diversification at work, characterized by dispersal and vicariance, by driving lineage divergence of New Guinean Leptophlebiidae across a wide array of habitats along the elevation gradient. Additional studies on organisms spanning the same elevation range as Thraulus mayflies in the tropics are needed to evaluate the potential role of the ecological opportunity or taxon cycles hypotheses in partly explaining the latitudinal diversity gradient.
Introduction
Tropical biomes are the most diverse among all ecosystems (Myers et al., 2000; Dirzo and Raven, 2003). Understanding the underlying mechanisms shaping such extraordinary diversity and more generally the processes associated with the latitudinal diversity gradient (Gaston, 2000) is an exciting challenge for evolutionary ecologists. A potential explanation may reside in the diversity of habitats found along tropical elevation gradients (Kricher, 2011), quoted as the “ecological opportunity hypothesis” (Schluter, 2000). On the other hand, mountainous regions and more specifically sky islands can be considered as natural laboratories to test the taxon cycle hypothesis (Wilson, 1961). They promote speciation and species dispersal linked to the varying habitats that organisms encounter as their populations experience sequential phases of range expansion and contraction, generally associated with shifts in ecological distribution (Ricklefs and Bermingham, 2002; Matos-Maraví et al., 2018a).
The formation of mountain ranges is thus a potentially important driver of speciation as it produces the ecological heterogeneity to study diversification processes (Luebert and Muller, 2015), creating, along new dispersal routes and vicariance events, a vast variety of new ecological opportunities to adapt to Gueuning et al. (2017). This is especially true in the tropics where the number of stratified ecosystems found along the elevation gradient is larger than anywhere else (Körner, 2003). One such tropical and mountainous region is the island of New Guinea (NG), which is long known to be exceedingly diverse in terms of species numbers and other aspects of biological diversity (Gressitt, 1982).
The current morphology of NG is the result of a complex geotectonic history and the Cenozoic tectonic evolution of New Guinea is still subject to debate (Baldwin et al., 2012). Two scenarios have been suggested to explain the formation of the current landmass (reviewed in Toussaint et al., 2014). In the first scenario, a primary orogeny of the eastern New Guinea was caused by underthrusting of the Australian continent beneath the Inner Melanesian around 35–30 million years ago (Mya). Then the Central Range reached its present elevation around 5 Mya after a secondary orogeny that lasted from 15 to 5 Mya (van Ufford and Cloos, 2005). The second scenario suggests a more rapid and recent formation of the current island, explained by the convergence between the Pacific and the Australian plates around 5 Mya (Hall, 2002; Hill and Hall, 2003). This convergence would have created a fold-and-thrust belt, which corresponds to the Central Range. Toussaint et al. (2014) highlighted that these two principal biological scenarios have different biogeographic implications, in terms of timing, directionality, and location of lineage diversification. In the first scenario, the Papuan Peninsula in eastern New Guinea is recognized as the first colonized area (Kalkman et al., 2017), with successive lineages arising along the central range out of the Papuan Peninsula. In the second scenario, the biogeographic model would rather suggest colonization of the fold and thrust belts corresponding to present-day central mountain range, followed by colonization of surrounding areas. As an alternative to these two scenarios, Hall's paleogeographic models suggest the emergence of small islands or archipelagos along current northern New Guinea prior to the formation of the island as of today (Hall, 2013), which would make possible colonization of the proto-Papuan archipelagos as early as the Oligocene and serve as drivers of diversification for a large array of lineages (Jønsson et al., 2011; Matos-Maraví et al., 2018b). To provide a first larger scale empirical framework to test these, Toussaint et al. (2014) conducted biogeographic and diversification rate analyses using a phylogeny of diving beetles (Exocelina). They suggest that this radiation was mainly driven by processes around the central New Guinea orogeny around 10 Mya. There was no evidence for an ancient geological origin of the Papuan Pensinsula that could be derived from the diving beetle phylogeny as all the species there arrived through recent dispersal. On the other hand, Kalkman et al. (2017) studied damselflies and suggested that the East Papuan Composite Terrane played an important role in the early diversification of the New Guinea taxa, in line with predictions derived from the geological model proposed by van Ufford and Cloos (2005). Similarly, corvoid birds were found to have originated in the proto-Papuan archipelago during the Eocene, from an Australian ancestor (Jønsson et al., 2011). In contrast, using a fossil-calibrated phylogeny, Matos-Maraví et al. (2018b) inferred ancestral geographical ranges that suggested that the first colonization of the proto-Papuan archipelago by Prenolepis ants occurred ca. 20 Mya from continental Asia.
Here, we test whether and how far these findings can be detected and then generalized in a mayfly clade. We investigated the biogeographic history of a group of paleotropical Leptophlebiidae mayflies, well-diversified in Southeast Asia and the Sahul shelf, namely the Thraulus clade, which is represented in New Guinea and in Sunda Islands with a monophyletic radiation (see Appendix S1). Because of a lack of consensus in the taxonomy of genera and species in the Thraulus clade (M. Sartori, unpublished data), we adopt, here, a clade-oriented rather than a genus-oriented approach.
Mayflies (ca. 3,500 species worldwide) are among the most ancient insect lineages, as they most likely appeared during the Triassic (Grimaldi et al., 2005). Specifically, we study lotic (running water) Leptophlebiidae of New Guinea, which all belong to the Thraulus clade (M. Sartori, unpublished data). To date, seven species belonging to the Thraulus clade were known from New Guinea (Kluge, 2013), but additional cryptic or yet non-identified diversity might remain to be described (M. Balke and M. Sartori, unpublished data). Our working hypothesis was that the Thraulus group should be associated with a broad lineage diversity. We identified the extent of such diversity by using a molecular phylogeny of the New Guinea Thraulus clade, dated according to the concept of the molecular clock. Putative species, which we hereafter refer to as operational taxonomic units (OTUs), were then identified by applying two different Poisson tree processes algorithms. The resulting framework was eventually used to perform a biogeographical reconstruction for the Thraulus clade using a Bayesian binary MCMC approach (Yu et al., 2015). Finally, shifts of regimes in abiotic ecological factors along the phylogeny—using altitude as a proxy — were inferred to examine if adaptation to different elevations was a driver of diversification and if it is the case, when did these shifts appear.
Materials and Methods
Sampling
Larvae were manually picked or washed from stones and logs or swept from sandy/rocky substrate and then preserved in 96% Ethanol. Larvae were sorted between the two known genera occurring in NG, namely Nonnullidens Grant and Peters, 1993 and Thraulus Eaton, 1881, based on the following morphological characteristics (Grant and Peters, 1993; Kluge, 2013): Thraulus: first pair of gills different in shape from the following (II to VII); Nonnullidens: seven pairs of identical gills. The specimens were then identified to 18 morphospecies (M. Sartori). The specimens (N = 92) used here are deposited at the Museum of Zoology in Lausanne (Table 1). Eighty-four larvae belong to the Thraulus ingroup while 8 larvae in the genera Choroterpes Eaton, 1881, Euthraulus Barnard, 1932 and Isca Gillies, 1951 (Leptophlebiidae) were used as outgroup taxa. Our data are deposited at ZENODO: https://doi.org/10.5281/zenodo.1123279.
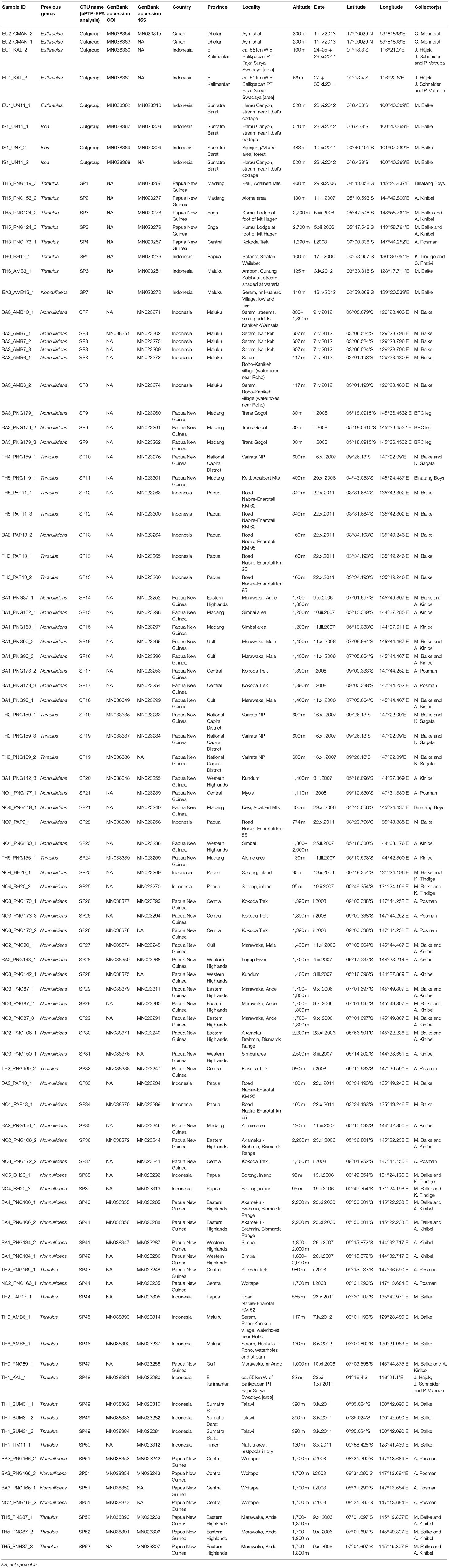
Table 1. Sampling information on specimens used in this study, with lineages putatively associated with species to be described (M. Sartori, unpublished data).
DNA Extraction and Sequencing
Whole larvae were digested overnight in 200 μL of a digestion solution (180 μL Qiagen ATL Buffer + 20 μL Proteinase K). DNA was then purified using a DNA extraction robot (Biosprint 96, Qiagen, Hombrechtikon, Switzerland), according to the manufacturer's instructions. We amplified a fragment of cytochrome c oxidase I (COI) using Jerry (5′-CAACATTTATTTTGATTTTTTGG-3′) and Pat (5′-TCCAATGCACTAATCTGC CATATT-3′) primers (Simon et al., 1994) and a fragment of 16S ribosomal DNA using 16Sar (5′-GCCTGTTTAACAAAAACAT-3′; (Simon et al., 1994)) and 16S2 primers (5′-GGAGCTCCGGTTTGAACTCAGATC-3′; Giessler et al., 1999) in separate reactions. Reactions were run in a volume of 10 μL including 200 μM dNTPs, 500 μM primers, 2 μL of 5X Q5® Reaction Buffer, 2 μL of 5X Q5® High GC Enhancer, 0.2U of Q5® High Fidelity DNA Polymerase (New England BioLabs) and 1 μL of DNA at the following conditions: 98°C for 30 s followed by 30 cycles of 98°C for 10 s, 50°C (COI) or 52.5°C (16S) for 30 s and 72°C for 20 s, with a final extension at 72°C for 2 min. Amplification products were purified using the Zymo purification Kit 96-well format, according to manufacturer's instructions (Zymo Research, Irvine, California, US).
To prepare the samples for Illumina sequencing, we used an adapted version of a protocol by Meyer and Kircher (2010) for Illumina library preparation, modified by C. Pitteloud and A. Mastretta-Yanes (see Appendix S2). After PCR amplification of the markers and purification of PCR products, this protocol consists in the following main steps: phosphorylation, ligation of the adapters containing the 6-mer tagging barcodes (known sequence of 6 nucleotides), pooling of the products, adapter fill-in and indexing. This method allows a high number of fragments to be sequenced at the same time, since every individual is tagged. The library was then sequenced on the Illumina MiSeq (Illumina, San Diego, California, USA) platform at the Lausanne Genomic Technologies Facility (Center for Integrative Genomics, CIG).
Demultiplexing and Sequence Calling
The sequencing output was processed using a custom bioinformatics pipeline (see Appendix S3). In brief, the reads were first sorted according to their combination of barcodes/index, filtered according to PHRED scores and cleaned (clipping out of primer and the barcode sequences). Clustering of reads with 99% similarity was performed using usearch (Edgar, 2010). Then, these sequences were blasted in GenBank. This “sequence calling” step allows to compare our sequences with archived sequences and returns information on the taxa (at the order level) and on the marker of the best hit. This provides a supplementary control of the quality of the sequences. This information was then associated with the reads and the hit scores. Then, the reads were processed according to the following steps: merging of the forward and reverse reads, conversion into Fasta files, control of the read orientation, dereplication of full length and prefix modes and sorting by cluster size. Finally, a double-filter was imposed on these sequences to keep the best candidates: the sequences had to be covered by a minimum of three reads, and only the sequences with a coverage of 2/3 of the coverage of the most abundant sequence were kept. We eventually produced one file for each gene prior to the alignment procedure.
Phylogenetic Inferences and Estimation of Divergence Times
Sequence alignment was performed using the online server MAFFT Version 7 (Katoh and Standley, 2013). The sequences were concatenated in a total evidence approach (Sanderson et al., 1998). In order to verify the monophyly of the Thraulus clade, we included all samples of the ingroup and outgroup listed above, as well as additional taxa from the following closely-related genera: Isca, Choroterpes, Euthraulus, Habrophlebiodes, Caenis, in a broader phylogenetic analysis. We applied RAxML (Stamatakis, 2014) using the GTR + G substitution model as the best model estimated by MrModeltest v. 2.3 (Nylander, 2004) and the Akaike Information criterion. We then applied Bayesian inference on the ingroup and the strict outgroup using BEAST v.1.8.3 (Drummond et al., 2012) with the same substitution model. Divergence time estimates were obtained using an uncorrelated relaxed molecular clock with a lognormal distribution of rates and a Yule speciation model. Applied substitution rates were 2.3% My−1 for COI (Brower, 1994) and 0.61% My−1 for 16S% My−1 (Borer et al., 2010). The analysis was run twice on the CIPRES portal (Miller et al., 2010) for 100 million generations, sampling one tree every 1,000th generation. The MCMC sampling was considered sufficient when the effective sampling size (ESS) was higher than 200, as verified in Tracer v. 1.4 (Rambaut and Drummond, 2007). Following a burn-in period of 20 million generations, a maximum clade credibility tree with median branch lengths and 95% highest posterior density (HPD) interval on nodes was reconstructed using TreeAnnotator v. 1.8.0 (Drummond and Rambaut, 2007). HPD was only inferred for nodes with Bayesian Posterior Probability (BPP) ≥ 0.5.
Species Delimitation and Biogeographical Inferences
Putative new species in the Thraulus clade remain difficult to be defined morphologically. Thus, we applied the Bayesian Poisson tree process (bPTP) model on our data to aid OTUs delineation (Zhang et al., 2013), in addition to morphology. The bPTP model was applied on the dated maximum credibility clade tree. We followed the recommendations by Zhang et al. (2013) and integrated the bPTP model with their evolutionary placement algorithm (EPA) to count the number of OTUs on the phylogeny (bPTP-EPA). This method was previously successfully applied to several organismal groups (e.g., Kenyon et al., 2015; Morard et al., 2016; Nieto-Montes de Oca et al., 2017). The analysis was conducted using the portal provided by the authors (http://species.h-its.org/ptp/) based on the default parameters. As recommended by the program's authors, a list of outgroup taxa was provided and those were automatically discarded to optimize species delimitations. As a complementary analysis, we applied the newly developed multi-rate Poisson tree process (Kapli et al., 2017) implemented in mPTP (https://github.com/Pas-Kapli/mptp/). In the mPTP model, each species has its own lambda, which means that the model assumes a different rate of evolution for each species and aims at maximizing the likelihood. Since the mPTP compares models with a different number of parameters (i.e., a separate lambda is set for each species), the Akaike Information Criterion has to be applied to identify which number of species best fits the phylogenetic tree. Such an approach tends to avoid over-splitting of the tips into too many species compared to the single-rate model implemented in the original PTP model (Zhang et al., 2013). Here we have taken advantage of the Markov Chain Monte Carlo (MCMC) sampling method implemented into mPTP to assess the confidence of the Maximum Likelihood delimitation scheme. Two independent 5-million steps MCMC analyses were ran with a sampling frequency of 100 and each run was started with the delimitation obtained by the Maximum Likelihood heuristic.
Since very little is known on the relationships and connectivity between biogeographical areas in New Guinea through time, we favored a neutral biogeographic model, not considering connectivity probabilities among geographical ranges. We used the Bayesian Binary MCMC (BBM) analysis as implemented in RASP (Yu et al., 2015) to investigate the biogeographical history of the Thraulus clade in New Guinea and considered geographical regions following the freshwater ecoregions described by Abell et al. (2008), with one slight modification, regarding a sample collected in Timor, which was considered belonging to the neighboring Maluku ecoregion for balanced sampling purpose. To further test our contrasting biogeographical hypotheses, dispersals, extinctions and vicariance events (including peripheral isolation) were recorded in three time slices, roughly corresponding to the main geological eras dividing the time since the origin of the Thraulus clade until today (40–23 Mya [Bartonian + Priabonian Eocene and Oligocene], 23–5 Mya [Miocene] and 5–0 Mya [Pliocene and Pleistocene]).
Altitudinal Shifts
To represent a crucial aspect of niche differentiation (Gueuning et al., 2017), we retraced shifts of altitudinal optima by analyzing our phylogeny with the R package “surface” (Ingram and Mahler, 2013; R Core Team, 2014). This method uses the Ornstein-Uhlenbeck stabilizing selection model to identify cases of convergent evolution, fitted with the stepwise Akaike Information Criterion, here, using elevation as a quantitative variable.
Results
The final alignments for COI and 16S were 474 and 554 bp, respectively. Both markers yielded a relatively large number of variable sites, respectively 216 and 389. Total number of reads was 4 458 392 for the two genes. Only amplicons for which a minimum coverage of 3 was obtained were kept in the analyses. These data were stored in Genbank (accession numbers: 16S: MN038347 to MN038393; COI: MN023233 to MN023316; see Table 1).
Phylogenetic analyses supported monophyly of the ingroup clade (Appendix S1). Our rate-based nodal age inference suggests an origin of the New Guinea radiation in the Eocene (ca. 41.1 Mya, 95% HPD range: 25.3–64.1 Mya).
We define here three major clades, two of which are strongly (clade α) and moderately (clade β) supported, whereas the third clade (clade γ, which includes two species from the Sunda Islands) is not well-supported. The bPTP analysis suggested presence of 52 OTUs. The 52 segregated lineages are distributed as follows: 13 in clade α, 34 in clade β and five in clade γ (Figure 1). None of the clades corresponded to genera as previously defined and our phylogenetic analyses support the polyphyly of the two genera (Nonnullidens and Thraulus). Regarding mPTP results, inspection of the combined likelihood log file summarizing the two independent MCMC analyses confirmed convergence of the runs. When both runs are accounted for, 29 species were recognized by the mPTP model (see brackets in Figure 1). However, before suggesting putative changes in the systematics of the Thraulus group, lineage definition should be examined in light of morphological traits. For instance, we defined one morphospecies as “BA1” based on gill shape, size and abdominal coloration; our results and a posteriori microscopic analyses suggest that this morphospecies would actually be composed of 7 (mPTP) to 8 (PTP) OTUs (Table 1; Figure 1).
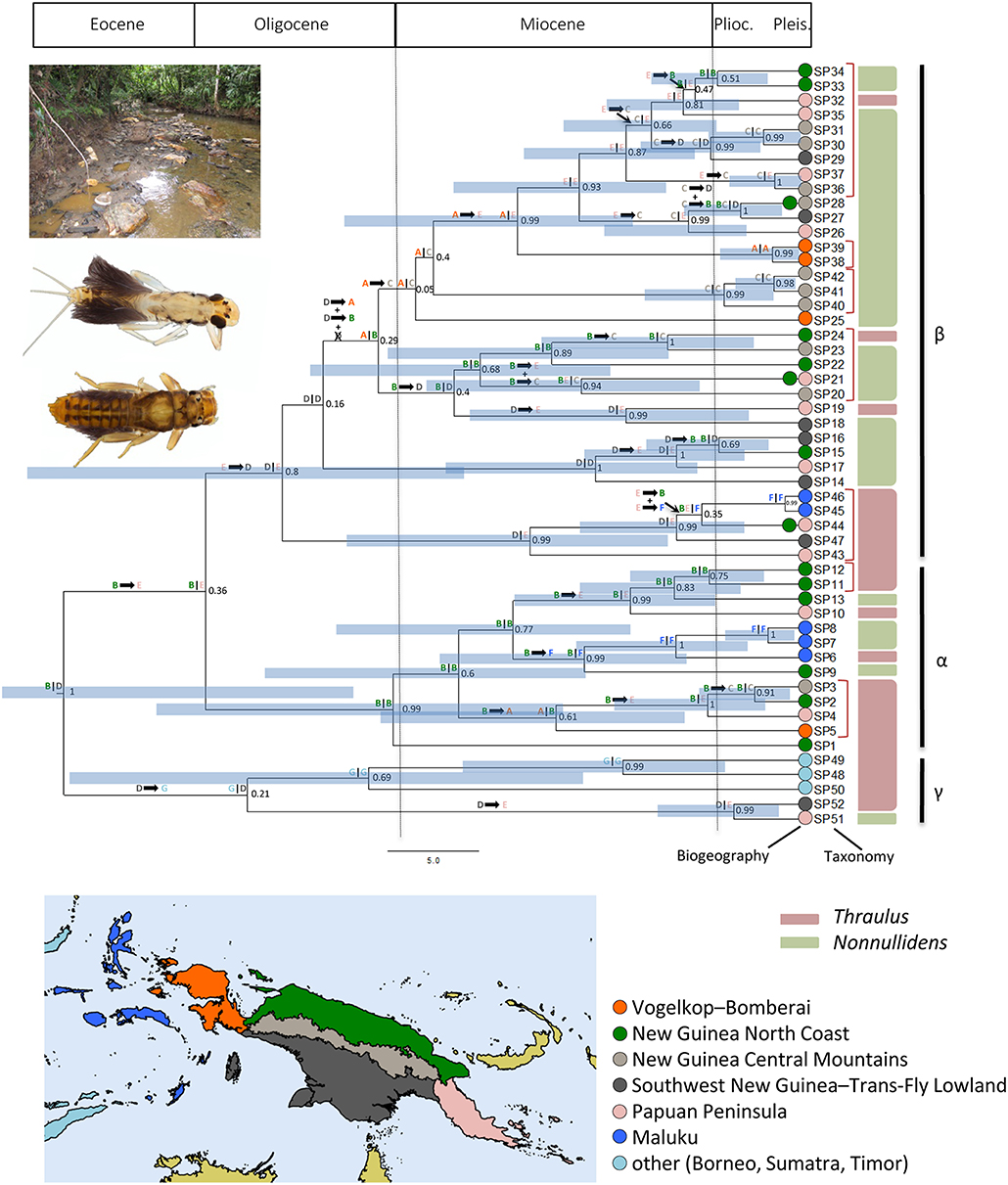
Figure 1. Chronogram of the Thraulus clade as inferred from BEAST v.1.8.3 (Drummond et al., 2012), after pruning the outgroups, with Bayesian Posterior Probabilities (BPPs) indicated at each node. Tips show lineages associated with OTUs as inferred from bPTP analyses (Zhang et al., 2013). Tips clustered by red brackets are lineages that were grouped into OTUs by the mPTP analyses (Kapli et al., 2017). Current taxonomic assignment to genera is indicated using a color frame on the right of the tree. Time is shown along the X-axis (with an indication of geological eras at the top of the figure). On each branch are shown dispersal events among biogeographic areas as inferred from the Bayesian Binary MCMC analysis (Yu et al., 2015). Above each node is shown the geographical context of lineage divergence (either in vicariance or within the same area). Blue rectangles over nodes represent 95% HPD ranges (values given only for nodes with BPP > 0.5). Colors shown at the tips of the tree represent the geographic area(s) corresponding to freshwater ecoregions of New Guinea following Abell et al. (2008), where the lineages are found. Areas are illustrated below on a map, created using the software QGIS (QGIS Development Team, 2009). Major clades are shown with vertical bars on the right of the tree.
A new classification based on a suite of morphological characters delimiting generic boundaries is in preparation and will be the subject of a forthcoming paper (M. Sartori, unpublished data). In the meantime, we will be referring to the clades and OTUs provided in Figure 1. With the exception of three OTUs occurring in two areas (i.e., 21, 28 and 44 all in clade β), all taxa were confined to one area.
For the following analyses, in order to favor an upper-bound approach of the OTUs delineation, we used the bPTP output as it produced the largest number of OTUs. The biogeographical analysis inferred 28 dispersal events distributed as follows: six in time slice 40-23 Mya (spanning part of middle and upper Eocene, and Oligocene), 21 in time slice 23-5 Mya (Miocene) and one in time slice 5-0 Mya (Pliocene and Quaternary; Figures 1, 2)—the 95% HPD ranges of each node with BPP > 0.5 are shown on Figure 1. For the sake of clarity, the next paragraph considers the nodes' mean dates, but one should keep in mind that these are subject to uncertainties. Clade α possibly originated in the New Guinean North Coast (area B) sometime at the boundary between the Oligocene and the Miocene and subsequently dispersed eastwards and westwards in Vogelkop-Bomberoi (area A) and Maluku (area F), respectively, as well as to the Papuan Peninsula (area E). Only one OTU, SP3, successfully colonized the New Guinean Central Mountain Range (area C) from the New Guinean North Coast (area B) during the Pliocene. A widespread ancestral area in the Papuan Peninsula and the Southwest New Guinea—Trans-Fly lowland (areas ED) is inferred at the most recent common ancestor of clade β sometime during the Oligocene. The subclade including SP14-17 (Figure 1) shows evidence of a dispersal route between the New Guinean North Coast (area B) and the Southwest New Guinea—Trans-Fly lowland (area D) prior to the establishment of the New Guinean Central Mountain Range (area C) during the Miocene. OTUs currently restricted to the Central Mountain Range (area C) do not form a monophyletic subclade within clade β, suggesting multiple colonization events to this area from the New Guinean North Coast (area B), the Southwest New Guinea—Trans-Fly lowland (area D) and the Papuan Peninsula (area E), from the Middle Miocene onwards. Sympatric divergence (hereafter used sensu lato; i.e., within a single biogeographical area) within the New Guinean Central Mountain Range (area C) has also been inferred, especially in the subclade including SP40-42 (Figure 1). Finally, clade γ includes all the OTUs recorded in the Sunda Islands (area G), which originated sometime during the Oligocene and likely dispersed from the Southwest New Guinea—Trans-Fly lowland (area D).
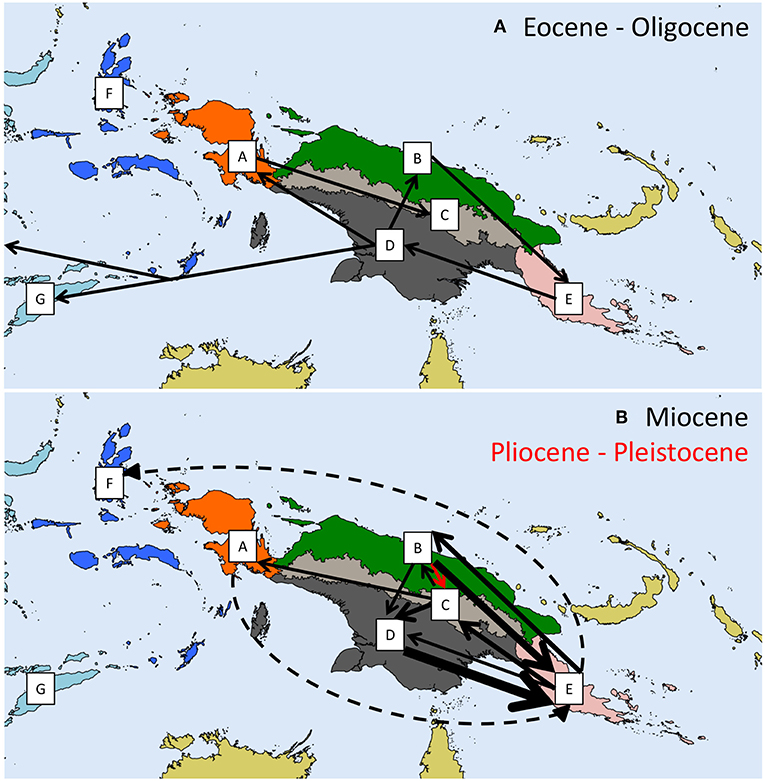
Figure 2. Biogeographic dispersal events inferred from the Bayesian Binary MCMC analysis Yu et al. (2015) for each of the three considered time slices: (A) Eocene-Oligocene; (B) Miocene and Pliocene-Pleistocene. Biogeographic areas correspond to freshwater ecoregions of New Guinea (Abell et al., 2008) as in Figure 1. Dashed arrows represent dispersal from geographically distant areas, which may involve stepping stone dispersal through in-between areas. The width of an arrow is proportional to the number of dispersal events (ranging between 1 and 8 events).
We found 11 altitudinal optima, with 23 shifts of altitudinal optimum. Fifteen of these shifts occurred during the last 5 My, i.e., during the Pliocene and Pleistocene time eras (see Figure 3).
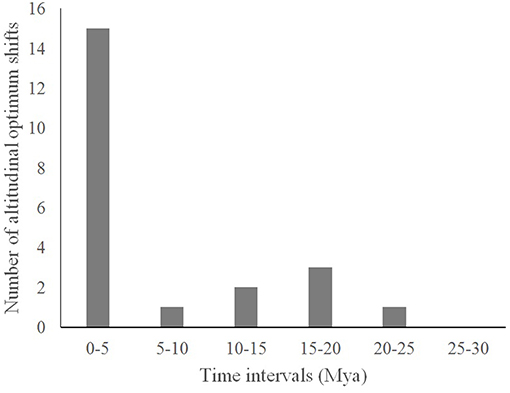
Figure 3. Number of altitudinal optimum shifts per 5 million years intervals, as identified using the “surface” R package.
Discussion
In this study, we made a first attempt to infer the biogeography of leptophlebiid mayflies from New Guinea by studying the patterns of dispersal and altitudinal shifts using a molecular clock-based dated phylogeny. This phylogeny revealed that the previous definition of genera (based on gill morphology) does not stand firm in light of the phylogenetic relationships.
We suggest that the colonization of New Guinea by the Thraulus clade might have occurred as early as the Eocene. This colonization timing is earlier than previous hypotheses proposed for Prenolepis ants and Ptilinopus birds, for which a colonization of the proto-Papuan archipelago was found to take place during the Miocene (Matos-Maraví et al., 2018b) and early-Oligocene (Cibois et al., 2014), respectively, but consistent with Jønsson et al. (2011) who identified an arrival of corvoid birds during the late Eocene / early Oligocene. Our reconstruction of the initial diversification phase remains ambiguous. It might have started either from the NG North Coast (area B) or from the SW New Guinea-Trans-Fly lowland (area D); we are therefore unable to favor one or the other scenario presented in the introduction. The major clades α, β, and γ are likely to have originated during the Eocene-Oligocene, respectively, in the New Guinean North Coast (area B), in the Papuan Peninsula and the SW New Guinea-Trans-Fly lowland (areas ED), and in the SW New Guinea-Trans-Fly lowland (area D). Three quarters (21 of 28) of the inferred dispersal events occurred during the Miocene, with most connections occurring between adjacent areas. The Papuan Peninsula was mainly colonized during the Miocene, with one third (7 of 21) of all dispersal events recorded during the Miocene occurring from the New Guinean North Coast (area B) and the SW New Guinea-Trans-Fly lowland (area D) to the Papuan Peninsula (area A). Such a scenario for the colonization of the island is not consistent with results by Toussaint et al. (2014) who found that the radiation of Exocelina diving beetles was likely to have originated in the Central Range. The importance of our results is however mitigated by the low support of several basal nodes across the phylogeny.
Our results show that for leptophlebiid mayflies, the Papuan Peninsula (area E) is only secondarily colonized from already existing terranes, nowadays likely corresponding to the New Guinean North Coast (area B) and the Southwest New Guinea – Trans-Fly lowland (area D). This is in agreement with Toussaint et al. (2014), who also found relatively late colonization of the Papuan Peninsula. While the node age estimations might prove inaccurate, both the latter and our work do however provide no evidence that the oldest lineages occurred on the Papuan Peninsula.
Respectively, 19 vicariant and 14 sympatric divergence events took place during the Miocene, a ratio similar to the scenario retrieved for the Eocene-Oligocene, with five vicariant and three sympatric divergence events. Such a proportion contrasts with the scenario highlighted during the Pliocene-Pleistocene, with five vicariant and seven sympatric divergence events. In addition, only one dispersal event was recorded during the Pliocene-Pleistocene.
Whereas most speciation events occurred through dispersal or vicariance during the Eocene-Oligocene and the Miocene, the majority of divergence events occurred in sympatry during the Pliocene-Pleistocene. Interestingly, more than two thirds (15 of 22) of all recorded altitudinal shifts occurred during the Pliocene-Pleistocene. Most differentiation therefore occurred within geographical areas, in the last 5 My, at the moment the main orogenesis took place in the island. Contrasting with previous evidence showing that NG OTUs arose since the Eocene, most of the diversification in the Thraulus group seems to be restricted to the last 5 My. Considering altitudinal shifts as potential indicators of niche differentiation (Pitteloud et al., 2017), this scenario suggests that ecological opportunity could be a driver of diversification in New Guinean Thraulus clade, as it was also found for diving beetles by Toussaint et al. (2014, 2015). Thus, the massive New Guinea orogeny with the associated formation of foothills and formation of one large landmass with the docking of oceanic north coast terranes might have created a natural laboratory for extensive and recent radiations.
As an alternative to the classical ecological opportunity hypothesis, we could invoke the taxon cycle as a driver of the diversification pattern observed across the Pliocene and Pleistocene. The pattern found here is indeed comparable to findings by Economo et al. (2015) and Jønsson et al. (2014), who showed that Pheidole ant lineages and Pachycephala passerine birds, respectively, have expanded their ranges across biogeographic regions, followed by ecological filtering and a cascade pattern of dispersal from higher to lower diversity areas during these range expansions. Although ecological opportunity and taxon cycle hypotheses seem plausible explanation for the observed pattern of recent diversification associated with altitudinal shifts, we cannot exclude that it reflects the results of neutral (non-adaptive) processes (Czekanski-Moir and Rundell, 2019): spatial isolation following dispersal can also drive divergence, especially in species with low dispersal ability such as mayflies (Rutschmann et al., 2014) and in strongly structured habitat as New Guinean mountains (Janda et al., 2016; Lam et al., 2018).
Overall, our results suggest that the Thraulus clade ancestor colonized the proto-Papuan archipelago during the Eocene, diversified during the colonization of emerging regions as well as by vicariance, and more recently by ecological shifts driven by the emergence of new habitats in association with mountain uplift. Despite conclusions could be biased given the relatively poor node supports retrieved and the fact that the two sequenced genes are associated with slightly different evolutionary rates, the dating of these events suggests a more ancient colonization of NG than often considered, followed by a more recent, potentially ecological, diversification. In the future, increasing the support of several nodes in our phylogeny, for instance through the incorporation of additional nuclear markers, might help reinforcing our biogeographic scenario. Integrating variation in the molecular clock (Papadopoulou et al., 2010) for divergence time estimate analyses should also inform on the validity of our finding of an ancient (Eocene) origin of the Thraulus clade. Finally, using comparative methods to investigate patterns of lineage accumulation and trait evolution in the Thraulus group as well as in other tropical organisms would help assessing the validity of the ecological opportunity hypothesis as a driver of diversification in the tropics.
Author Contributions
MB sampled the mayflies. MS, NaA, and MB conceived the ideas. CP designed the wetlab protocol. MG and C-SC did the labwork. NiA designed the script used for data treatments. NaA, SB, NS, and C-SC analyzed the data and led the writing.
Funding
This work was funded by a Swiss National Science Foundation grant (PP00P3_172899) awarded to NaA. MB fieldwork was supported by a UK Darwin Initiative grant Training the next generation of PNG conservation biologists as well as DFG BA2152/3-1, 6-1, 11-1.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all collectors listed in Table 1 for their precious contribution. We thank the Lausanne Genomic Technologies Facility for providing the Illumina sequencing service and N. Kluge, M. Gamboa, and P. Matos-Maraví, as well as five reviewers for insightful comments on earlier versions of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00233/full#supplementary-material
References
Abell, R., Thieme, M. L., Revenga, C., Bryer, M., Kottelat, M., Bogutskaya, N., et al. (2008). Freshwater ecoregions of the world: a new map of biogeographic units for freshwater biodiversity conservation. Bioscience 58, 403–414. doi: 10.1641/B580507
Baldwin, S. L., Fitzgerald, P. G., and Webb, L. E. (2012). Tectonics of the new guinea region. Annu. Rev. Earth Planet. Sci. 40, 495–520. doi: 10.1146/annurev-earth-040809-152540
Barnard, K. H. (1932). South African may-flies (Ephemeroptera). Trans. R. Soc. South Afr. 20, 201–259. doi: 10.1080/00359193209518858
Borer, M., Alvarez, N., Buerki, S., Margraf, N., Rahier, M., and Naisbit, R. E. (2010). The phylogeography of an alpine leaf beetle: divergence within Oreina elongata spans several ice ages. Mol. Phylogenet. Evol. 57, 703–709. doi: 10.1016/j.ympev.2010.08.017
Brower, A. V. (1994). Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. U.S.A. 91, 6491–6495. doi: 10.1073/pnas.91.14.6491
Cibois, A., Thibault, J. C., Bonillo, C., Filardi, C. E., Watling, D., and Pasquet, E. (2014). Phylogeny and biogeography of the fruit doves (Aves: Columbidae). Mol. Phylogenet. Evol. 70, 442–453. doi: 10.1016/j.ympev.2013.08.019
Czekanski-Moir, J. E., and Rundell, R. J. (2019). The ecology of nonecological speciation and nonadaptive radiations. Trends Ecol. Evol. 34, 400–415. doi: 10.1016/j.tree.2019.01.012
Dirzo, R., and Raven, P. H. (2003). Global state of biodiversity and loss. Annu. Rev. Environ. Resour. 28, 137–167. doi: 10.1146/annurev.energy.28.050302.105532
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Economo, E. P., Sarnat, E. M., Janda, M., Clouse, R., Klimov, P. B., Fischer, G., et al. (2015). Breaking out of biogeographical modules: range expansion and taxon cycles in the hyperdiverse ant genus Pheidole. J. Biogeogr. 42, 2289–2301. doi: 10.1111/jbi.12592
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Giessler, S., Mader, E., and Schwenk, K. (1999). Morphological evolution and genetic differentiation in Daphnia species complexes. J. Evol. Biol. 12, 710–723. doi: 10.1046/j.1420-9101.1999.00065.x
Gillies, M. T. (1951). Further notes on Ephemeroptera from India and South East Asia. Proc. R. Ent. Soc. Lond. B 20, 121–130. doi: 10.1111/j.1365-3113.1951.tb01009.x
Grant, P. M., and Peters, W. L. (1993). Description of four new genera of Thraulus group mayflies from the Eastern Hemisphere and redescription of Simothraulus and Chiusanophlebia (Ephemeroptera: Leptophlebiidae: Atalophlebiinae). Am. Entomol. Soc. 119, 131–168.
Gressitt, J. L. (1982). “Zoogeographical summary” in Biogeography and Ecology of New Guinea: Part One −7th Edn. ed J. L. Gressitt (Dordrecht: Springer Netherlands, 897–918.
Grimaldi, D., Engel, M. S., and Engel, M. S. (2005). Evolution of the Insects. Cambridge: Cambridge University Press.
Gueuning, M., Suchan, T., Rutschmann, S., Gattolliat, J. L., Jamsari, J., Kamil, A. I., et al. (2017). Elevation in tropical sky islands as the common driver in structuring genes and communities of freshwater organisms. Sci. Rep. 7:16089. doi: 10.1038/s41598-017-16069-y
Hall, R. (2002). Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer-based reconstructions, model and animations. J. Asian Earth Sci. 20, 353–431. doi: 10.1016/S1367-9120(01)00069-4
Hall, R. (2013). The palaeogeography of Sundaland and Wallacea since the Late Jurassic. J. Limnol. 72, 1–17. doi: 10.4081/jlimnol.2013.s2.e1
Hill, K. C., and Hall, R. (2003). “Mesozoic-Cenozoic evolution of Australia's New Guinea margin in a west Pacific context”, in Evolution and Dynamics of the Australian Plate, eds R. Hillis and R. D. Muller (Sydney, NSW; Boulder, NV: Geological Society of Australia Special Publication 22 and Geological Society of America Special Paper, 265–289.
Ingram, T., and Mahler, D. L. (2013). SURFACE: Detecting convergent evolution from comparative data by fitting Ornstein-Uhlenbeck models with stepwise Akaike Information Criterion. Methods Ecol. Evol. 4, 416–425. doi: 10.1111/2041-210X.12034
Janda, M., Matos-Marav,í, P., Borovanska, M., Zima, J., Youngerman, E., and Pierce, N. E. (2016). Phylogeny and population genetic structure of the ant genus Acropyga (Hymenoptera: Formicidae) in Papua New Guinea. Invertebr. Syst. 30, 28–40. doi: 10.1071/IS14050
Jønsson, K. A., Fabre, P. H., Ricklefs, R. E., and Fjeldså, J. (2011). Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc. Natl. Acad. Sci. U.S.A. 108, 2328–2333. doi: 10.1073/pnas.1018956108
Jønsson, K. A., Irestedt, M., Christidis, L., Clegg, S. M., Holt, B. G., and Fjeldså, J. (2014). Evidence of taxon cycles in an Indo-Pacific passerine bird radiation (Aves: Pachycephala). Proc. R. Soc. B. Biol. Sci. 281:20131727. doi: 10.1098/rspb.2013.1727
Kalkman, V. J., Dijkstra, K. B., Dow, R. A., Stokvis, F. R., and van Tol, J. (2017). Out of Australia: the Argiolestidae reveal the Melanesian Arc System and East Papua Composite Terrane as possible ancient dispersal routes to the Indo-Australian Archipelago (Odonata: Argiolestidae). Int. J. Odonatol. 21, 1–14. doi: 10.1080/13887890.2017.1402825
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A., et al. (2017). Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics. 33, 1630–1638. doi: 10.1093/bioinformatics/btx025
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kenyon, S. G., Buerki, S., Hansson, C., Alvarez, N., and Benrey, B. (2015). Uncovering cryptic parasitoid diversity in Horismenus (Chalcidoidea, Eulophidae). PLoS ONE 10: e0136063. doi: 10.1371/journal.pone.0136063
Kluge, N. (2013). Mayflies of the genus Nonnullidens Grant and Peters 1993 (Ephemeroptera: Leptophlebiidae) from New Guinea. Zootaxa 3722:33–49. doi: 10.11646/zootaxa.3722.1.3
Lam, A. W., Gueuning, M., Kindler, C., Van Dam, M., Alvarez, N., Panjaitan, R., et al. (2018). Phylogeography and population genomics of a lotic water beetle across a complex tropical landscape. Mol. Ecol. 27, 3346–3356. doi: 10.1111/mec.14796
Luebert, F., and Muller, L. A. (2015). Effects of mountain formation and uplift on biological diversity. Front. Genet. 6:54. doi: 10.3389/fgene.2015.00054
Matos-Maraví, P., Clouse, R. M., Sarnat, E. M., Economo, E. P., LaPolla, J. S., Borovanska, M., et al. (2018b). An ant genus-group (Prenolepis) illuminates the biogeography and drivers of insect diversification in the Indo-Pacific. Mol. Phylogenet. Evol. 123, 16–25. doi: 10.1016/j.ympev.2018.02.007
Matos-Maraví, P., Matzke, N. J., Larabee, F. J., Clouse, R. M., Wheeler, W. C., Sorger, D. M., et al. (2018a). Taxon cycle predictions supported by model-based inference in Indo-Pacific trap-jaw ants (Hymenoptera: Formicidae: Odontomachus). Mol. Ecol. 27, 4090–4107. doi: 10.1111/mec.14835
Meyer, M., and Kircher, M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010:prot5448. doi: 10.1101/pdb.prot5448
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Cipres science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA.
Morard, R., Escarguel, G., Weiner, A. K., Andr,é, A., Douady, C. J., Wade, C. M., et al. (2016). Nomenclature for the nameless: a proposal for an integrative molecular taxonomy of cryptic diversity exemplified by planktonic foraminifera. Syst. Biol. 65, 925–940. doi: 10.1093/sysbio/syw031
Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Nieto-Montes de Oca, A., Barley, A. J., Meza-Lázaro, R. N., García-Vázquez, U. O., Zamora-Abrego, J. G., Thomson, R. C., et al. (2017). Phylogenomics and species delimitation in the knob-scaled lizards of the genus Xenosaurus (Squamata: Xenosauridae) using ddRADseq data reveal a substantial underestimation of diversity. Mol. Phylogenet. Evol. 106, 241–253. doi: 10.1016/j.ympev.2016.09.001
Nylander, J. A. A. (2004). MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala: Uppsala University.
Papadopoulou, A., Anastasiou, I., and Vogler, A. P. (2010). Revisiting the insect mitochondrial molecular clock: the mid-aegean trench calibration. Mol. Biol. Evol. 27, 1659–1672. doi: 10.1093/molbev/msq051
Pitteloud, C., Arrigo, N., Suchan, T., Mastretta-Yanes, A., Vila, R., Dincă, V., et al. (2017). Climatic niche evolution is faster in sympatric than allopatric lineages of the butterfly genus Pyrgus. Proc. Biol. Sci. 284:20170208. doi: 10.1098/rspb.2017.0208
QGIS Development Team (2009). QGIS Geographic Information System. Open Source Geospatial Foundation. Available online at: http://qgis.osgeo.org
R Core Team (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. Available online at: http://www.R-project.org
Rambaut, A., and Drummond, A. J. (2007). Tracer v1. 4: MCMC trace analyses tool. Available online at: http://tree.bio.ed.ac.uk/software/tracer/
Ricklefs, R. E., and Bermingham, E. (2002). The concept of the taxon cycle in biogeography. Glob. Ecol. Biogeogr. 11, 353–361. doi: 10.1046/j.1466-822x.2002.00300.x
Rutschmann, S., Gattolliat, J. L., Hughes, S. J., Báez, M., Sartori, M., and Monaghan, M. T. (2014). Evolution and island endemism of morphologically cryptic Baetis and Cloeon species (Ephemeroptera, Baetidae) on the Canary Islands and Madeira. Freshw. Biol. 59, 2516–2527. doi: 10.1111/fwb.12450
Sanderson, M. J., Purvi, A., and Henze, C. (1998). Phylogenetic supertrees: assembling the trees of life. Trends Ecol. Evol. 13, 105–109. doi: 10.1016/S0169-5347(97)01242-1
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651–701. doi: 10.1093/aesa/87.6.651
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Toussaint, E. F., Hall, R., Monaghan, M. T., Sagata, K., Ibalim, S., Shaverdo, H. V., et al. (2014). The towering orogeny of New Guinea as a trigger for arthropod megadiversity. Nat. Commun. 5, 1–10. doi: 10.1038/ncomms5001
Toussaint, E. F. A., Hendrich, L., Shaverdo, H., and Balke, M. (2015). Mosaic patterns of diversification dynamics following the colonization of Melanesian islands. Sci. Rep. 5:16016. doi: 10.1038/srep16016
van Ufford, A. Q., and Cloos, M. (2005). Cenozoic tectonics of New Guinea. AAPG Bull. 89, 119–140. doi: 10.1306/08300403073
Wilson, E. O. (1961). The Nature of the Taxon Cycle in the Melanesian Ant Fauna. Am. Nat. 95, 169–193. doi: 10.1086/282174
Yu, Y., Harris, A. J., Blair, C., and He, X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Keywords: amplicon-sequencing, altitudinal shifts, freshwater insects, tropical diversity, Thraulus
Citation: Cozzarolo C-S, Balke M, Buerki S, Arrigo N, Pitteloud C, Gueuning M, Salamin N, Sartori M and Alvarez N (2019) Biogeography and Ecological Diversification of a Mayfly Clade in New Guinea. Front. Ecol. Evol. 7:233. doi: 10.3389/fevo.2019.00233
Received: 18 July 2018; Accepted: 05 June 2019;
Published: 03 July 2019.
Edited by:
Oana Moldovan, Emil Racovita Institute of Speleology, RomaniaReviewed by:
Maribet Gamboa, Ehime University, JapanPável Matos-Maraví, Institute of Entomology (ASCR), Czechia
Copyright © 2019 Cozzarolo, Balke, Buerki, Arrigo, Pitteloud, Gueuning, Salamin, Sartori and Alvarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Camille-Sophie Cozzarolo, Y2FtaWxsZS1zb3BoaWUuY296emFyb2xvQHVuaWwuY2g=
†These authors share last authorship
 Camille-Sophie Cozzarolo
Camille-Sophie Cozzarolo Michael Balke3
Michael Balke3 Sven Buerki
Sven Buerki Morgan Gueuning
Morgan Gueuning Nicolas Salamin
Nicolas Salamin Michel Sartori
Michel Sartori Nadir Alvarez
Nadir Alvarez