- 1Department of Botany, University of Hawaii, Manoa, HI, United States
- 2Department of Ecology and Evolutionary Biology, University of Colorado, Boulder, CO, United States
- 3Department of Entomology and Nematology, University of Florida, Gainesville, FL, United States
Cryoconite holes are holes in a glacier's surface caused by sediment melting into the glacier. These holes are self-contained ecosystems that include abundant bacterial life within their sediment and liquid water, and have recently gained the attention of microbial ecologists looking to use cryoconite holes as “natural microcosms” to study microbial community assembly. Here, we explore the idea that cryoconite holes can be viewed as “islands,” in the same sense that an island in the ocean is an area of habitat surrounded by a barrier to entry. In the case of a classic oceanic island, the ocean is a barrier between islands, but in the case of cryoconite holes, the ocean is comprised of impermeable solid ice. We test two hypotheses, born out of island biogeographic theory, that can be readily applied to cryoconite hole bacteria. First, we ask to what extent the size of a cryoconite hole is related to the amount of bacterial diversity found within it. Second, we ask to what extent cryoconite holes exhibit distance decay of similarity, meaning that geographically close holes are expected to harbor similar bacterial communities, and distant holes are expected to harbor more different bacterial communities. To test the island size hypothesis, we measured the sizes of cryoconite holes on three glaciers in Antarctica's Taylor Valley and used DNA sequencing to measure diversity of bacterial communities within them. We found that for two of these glaciers, there is a strong relationship between hole size and bacterial phylogenetic diversity, supporting the idea that cryoconite holes on those glaciers are “islands.” The high biomass dispersing to the third glacier we measured could explain the lack of size-diversity relationship, remaining consistent with island biogeography. To test the distance decay of similarity hypothesis, we used DNA sequence data from several previous studies of cryoconite hole bacteria from across the world. Combined with our Taylor Valley data, those data showed that cryoconite holes have strong spatial structuring at scales of one to several hundred kilometers, also supporting the idea that these dirty holes on glaciers are really islands in the cryosphere.
Introduction
Cryoconite holes are microbial oases within the extreme environment of a glacier's surface ice. These holes form when sediment is blown onto the ice and is heated by solar energy, causing it to melt into the glacier's surface. Because cryoconite holes can be easily visually identified on a glacier's surface, and because they are discrete, self-contained, isolated pockets of life occurring in an otherwise inhospitable environment, cryoconite holes have attracted the interest of ecologists studying microbial community assembly who wish to use them as natural microcosm experiments (Ambrosini et al., 2016; Sommers et al., 2018). Much like an island within a sea of glacial ice, each cryoconite hole harbors its own microbial community (Stanish et al., 2013), which is active at least during the summer months when they contain liquid water (Telling et al., 2014). Cryoconite holes on the glaciers of Antarctica's Taylor Valley are even more akin to islands than those on arctic or alpine glaciers in that significant dispersal barriers exist between them, since older holes are “lidded” by ice year-round (Fountain et al., 2004), meaning that their captive microbiota cannot easily escape, and new species cannot easily immigrate. Understanding the extent to which these “natural microcosms” are indeed islands within glaciers is important to the study of cryoconite holes and to their use as model systems for studying microbial community assembly. This is because biogeographic structure of microbial comminutes is a result of community assembly processes (Nemergut et al., 2013; Darcy et al., 2017), and theory for community assembly in island systems is well developed (Rominger et al., 2016). Here, we investigate the extent to which bacterial communities living within cryoconite holes match two predictions made by island biogeographic theory (MacArthur and Wilson, 1967; Nekola and White, 1999).
The first prediction is that larger islands are more species-rich than smaller islands (MacArthur and Wilson, 1967; Rosenzweig, 1995). This pattern has been shown to be true for bacteria several times, most notably in two model systems with “island sizes” comparable to those of cryoconite holes. Bell et al. (2005) surveyed bacterial diversity as a function of “island” size in tree-holes, which are permanent or semi-permanent pools of water that form at the base of European beech trees due to the buttressing of the tree's roots. They found that bacterial genetic diversity was strongly associated with the volume of water in tree holes, obeying the species-area power law, S = cAZ, where S is species richness or species diversity (alpha-diversity), c is a constant that is specific to the location or taxon in question, A is island area (or volume, in the case of tree wells), and Z is the slope of the line relating species to area. Species-area relationships are usually visualized and tested as linear fits of log-transformed S vs. log-transformed A, which is why Z, an exponent, is the slope. The other model system in which island size was a strong predictor of bacterial genetic diversity was in the oil reservoirs of machines that are used to cut metal, such as lathes and mills (Van Der Gast et al., 2005). This study used the total volume of the oil sump tank as area, and also found a highly significant relationship between island size and bacterial genetic diversity, even when temporal variation in diversity was accounted for. These patterns have been shown for other microbes as well, including a significant relationship between the size of trees and the diversity of fungi that live among the tree's roots (Glassman et al., 2017). Given previous successful demonstrations of bacteria and other microbes following the island biogeographic species-area relationship, if cryoconite holes are indeed island-like, larger cryoconite holes should have more species-rich bacterial communities than smaller holes.
However, relationships between island size and diversity are not comparable between tree holes and machine oil reservoirs, because those two very different microbiomes harbor different sorts of organisms with different baseline diversities for islands of the same size. Similarly, we do not expect bacterial communities from different glaciers to necessarily fall on the same species-area curve. Here, we intentionally sampled three glaciers whose characteristics differ, and used each glacier separately to test our hypothesis that larger cryoconite holes have more diverse bacterial communities than small cryoconite holes. The glaciers we analyze here are all located in Antarctica's Taylor Valley, and have each been the subject of previous microbiological study (Foreman et al., 2007; Stanish et al., 2013; Sommers et al., 2018). These previous studies have found that the Commonwealth, Canada, and Taylor glaciers represent a biomass and diversity gradient across the Taylor Valley (Sommers et al., 2018), making them ideal to study as separate island biogeographic experiments.
The second island biogeographic prediction we test here is that of distance decay of similarity. Simply stated, islands that are geographically close together should have similar communities living on them, and islands that are far apart should have communities that are less similar (more different) from each other (Nekola and White, 1999; Soininen et al., 2007). Microbial communities often follow this pattern, as they are spatially autocorrelated even when sample locations are not “islands” in a strict sense (Mackas, 1984; Franklin and Mills, 2003; Robeson et al., 2011; Wang et al., 2013). There can be many factors that confound this pattern, but it is commonly understood that the main drivers of community-level distance decay of similarity patterns (sometimes called “isolation by distance”) (Green et al., 2004; Martiny et al., 2011) are dispersal barriers (the inability of species to travel between distant islands) and environmental dissimilarity (Nekola and White, 1999). In the latter case, distant islands may be less likely to have similar environments than islands that are close together, and species distributions are often functions of their environment, even for bacteria (Fierer and Jackson, 2006; Nemergut et al., 2013). Although this type of spatial pattern is not diagnostic for islands since many non-island study systems exhibit strong spatial autocorrelation (Robeson et al., 2011; Darcy et al., 2017), it is expected that islands communities will be strongly spatially autocorrelated.
Unlike our first hypothesis, which we test here using three glaciers within the same valley, for the distance decay of similarity hypothesis we compiled a global dataset of cryoconite hole bacterial DNA sequence data from past studies. To test for patterns of distance decay of similarity, which are expected if cryoconite holes are truly island-like, we obtained geographic coordinates obtained for each sample from the cryoconite holes we sampled in Taylor Valley and from the other studies we used to assemble the global data set. Using these data, we tested the distance decay of similarity hypothesis at the global scale, but also at the regional scale for Antarctica's McMurdo Dry Valleys.
Methods
Sample Collection
Cryoconite hole sediment samples were collected frozen in November 2016, from the Taylor, Canada, and Commonwealth glaciers in Taylor Valley, Antarctica (Figure 1). More detailed information about these glaciers can be found elsewhere (Sommers et al., 2018). On each of the three glaciers, 30 sample sites were selected in a spatially nested sampling scheme (modified from King et al., 2010), containing geographic scales of roughly 2, 15, and 100 m (Figure 1). These 30 samples consisted of 5 triangles with side length 15 m, spaced 100 m from each other, and each of these 5 triangles contained 6 samples, two at each vertex, roughly 2 m apart. For each ice core taken, first the dimensions of the cryoconite hole were measured using a measuring tape. Since most holes were slightly ellipsoid, diameters were measured along each hole's north-south and east-west axes. Geographic locations of cryoconite holes were recorded using precision GPS measurements.
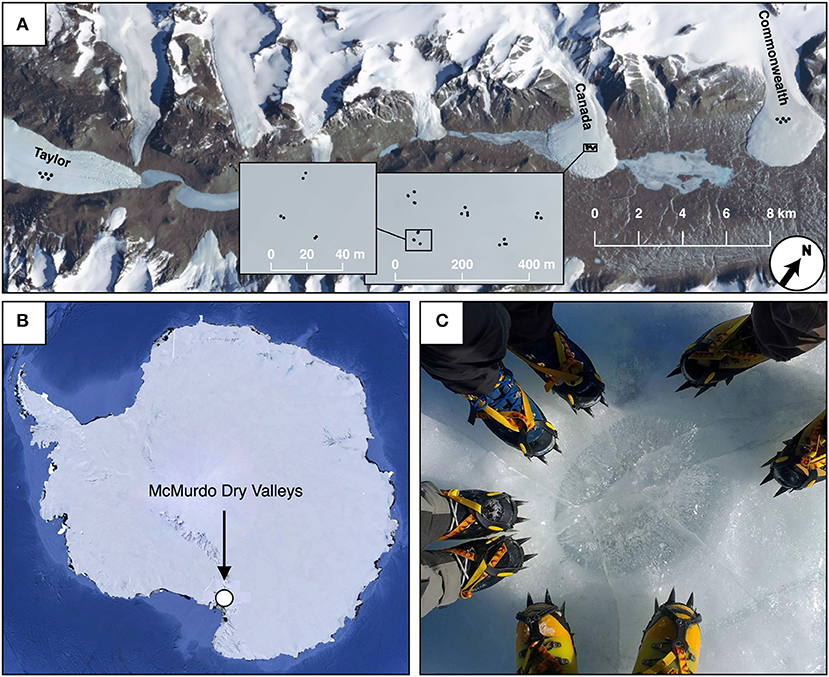
Figure 1. Map and sampling design for Taylor Valley cryoconite holes. Samples on Taylor, Canada, and Commonwealth glacier (A) used spatially nested sampling designs, consisting of pairs of samples roughly 5 meters apart (varying depending on cryoconite availability), in triangles with sides measuring 25 meters (A, inset). Five of these triangles were nested within a triangular grid, with 150 m between each triangle (A, inset). The position of the McMurdo Dry valleys (where Taylor Valley is) within Antarctica is shown (B), as well as a photograph of a cryoconite hole on Canada Glacier with the authors' boots for scale (C).
Next, a 10 cm SIPRE corer attached to a handheld gasoline-powered motor was used to extract the cryoconite from the glacier. The corer was centered over the cryoconite hole, and the motor was run until the swarf (debris displaced by the corer and ejected through its flutes) turned brown to indicate it passed through the sediment, and then white again, to indicate it drilled past the sediment and into the ice below. Cores were placed in Whirl-Pak bags (Nasco) and kept in a freezer at −20°C until further processing. Some ice cores broke apart immediately during sampling, but in all cases the sediment “puck” was entirely intact.
Sample Processing and DNA Sequencing
Each cryoconite hole core was processed in the Crary Laboratory at McMurdo Station to remove excess ice from the sediment, homogenize the sediment, and allocate a subset of the sediment for DNA sequencing. Excess ice was removed from each core using a chisel, then sterile water was used to melt away ~2 mm of the core's surface to avoid potential contaminants. Cores were then placed into acid-washed plastic beakers and thawed overnight at 4°C, with additional thawing taking place at room temperature where necessary. Thawed cores were then homogenized vigorously using a sterilized spatula, and ~0.3–0.5 g of homogenized sediment from each core was placed in PowerSoil DNA extraction kit bead-beating tubes (MoBio, USA), and genomic DNA was extracted following the manufacturer's instructions. Extracted DNA was kept at −80°C until it was shipped frozen from McMurdo Station to Boulder, Colorado, USA with a temperature data logger included. The temperature of DNA samples never exceeded −10°C during transit. The 16S ribosomal RNA gene was PCR amplified in triplicate from each sample using the bacterial universal primers 515f and 806r with attached Illumina barcodes unique to each sample (Caporaso et al., 2012). Amplified DNA was pooled and normalized to equimolar concentrations using SequalPrep normalization Plate Kits (Invitrogen, Carlsbad CA, USA), and was sequenced on the Illumina MiSeq platform using 2 × 250 bp chemistry at the BioFrontiers Sequencing Core Facility at the University of Colorado, Boulder.
Bioinformatics and DNA Sequence Processing
Raw data from the Taylor Valley cryoconite holes (above) were de-multiplexed and quality filtered, and paired-end reads were joined together using QIIME v.1.9.1 (Caporaso et al., 2010). DNA sequencing data from other studies of cryoconite holes on different glaciers across the Earth (Table 1) were downloaded from their respective repositories, and a metadata table was compiled to keep track of each sample and its geographic origins. Spatial locations for each individual sample were either taken directly from manuscripts, supplementary materials, or published datasets, but when locations were not specific (e.g., one geographic coordinate given for several samples), only the stated value was used, as such many pairs of samples had distances of 0 between them even though this was not true at the time of sampling. These 0-distance pairs were excluded from downstream analyses, but pairs containing the same samples at larger distances (>0 m) were retained. Although all studies aggregated here used the same 16S ribosomal RNA gene in their analyses, not all studies used the same regions within that gene. Therefore, in order to compare those different studies, we used QIIME to perform reference-based OTU (operational taxonomic unit, used instead of “species”) clustering, which enables analysis of DNA sequencing data generated using different regions of the 16S gene. We used the GreenGenes (DeSantis et al., 2006) database as a reference, because it contains a phylogeny for the sequences within it, and each sequence in the database covers the full length of the 16S gene. This resulted in an OTU table (species × site table), which was then rarefied (randomly downsampled) to 2,000 sequences per sample, so that samples that were sequenced more deeply (e.g., Ambrosini et al., 2016) would not have artificially higher phylogenetic diversity values than samples that were sequenced at a lower depth. Samples with < 2000 sequences were discarded.
Island Size and Phylogenetic Diversity Analysis
The “island size” of each cryoconite hole was calculated as A = π*r1*r2, where r1 and r2 are the two radii of the cryoconite hole (half of the measured diameters). Diversity of each hole was calculated with QIIME using Faith's (Faith, 1992) phylogenetic diversity metric, which uses the sum of branch-lengths in a phylogenetic tree as a diversity index. The phylogenetic tree in this analysis was provided by the GreenGenes database (DeSantis et al., 2006). Faith's (Faith, 1992) phylogenetic diversity was used because it is less sensitive to clustering artifacts (i.e. spurious OTUs) than OTU richness, although this analysis was done with OTU richness as well. Because the three glaciers we studied have significantly different geography and biogeochemistry (Sommers et al., 2018), each glacier was analyzed separately using linear regression. For each of the three glaciers, a linear model was fit to the log10-transformed phylogenetic diversity values as modeled by log10-transformed cryoconite hole areas, and statistically tested using R 3.33 (R Core Team, 2016). This analysis was not performed on data from glaciers outside the Taylor Valley because measurements of cryoconite hole area were not available for the other samples.
Spatial Autocorrelation Analysis
Compositional dissimilarity was calculated for each pair-wise comparison between samples for the entire data set using unweighted UniFrac distance (Lozupone and Knight, 2005) in QIIME 1.9 (Caporaso et al., 2010). UniFrac distance is a distance metric that is similar to Faith's (Faith, 1992) phylogenetic diversity (used above) because UniFrac distance takes the phylogeny of microbial communities into account. UniFrac distance is the fraction of branch lengths in a phylogenetic tree containing all OTUs from two communities that are not shared by those two communities. The GreenGenes tree (DeSantis et al., 2006) was used for this calculation. The resulting dissimilarity matrix was used in the analysis of spatial autocorrelation of bacterial community structure for each of the three glaciers we sampled in this study (Figure 1). We used Mantel tests to test for statistical significance in R 3.33 with the vegan package (Oksanen et al., 2016). Mantel tests were also run for all samples from the McMurdo Dry valleys (all samples from Antarctic glaciers except Ecology, Table 1) and for all samples worldwide (all samples from all glaciers in Table 1). Because the global pattern appeared to plateau at small (< 103 m) and large (>106 m) geographic scales while being driven by turnover at intermediate distances, we fit a logistic model to the data using:
where a is the upper asymptote, b is the lower asymptote, r is the exponential rate of change, and i is the x-coordinate of the curve's inflection point. This model was fit to the data using log-transformed geographic distances in R 3.33 (R Core Team, 2016) using downhill simplex optimization as implemented in R's optim function. A linear model was fit as well.
Results and Discussion
Island Size and Bacterial Diversity
Previous studies have shown that cryoconite hole microbial communities in the Taylor Valley follow a biodiversity gradient related to both spatial and environmental variables (Sommers et al., 2018), and our results here agree with those previous findings (Figure 1A). Taylor, Canada, and Commonwealth glaciers each had a significantly different mean bacterial phylogenetic diversity within its cryoconite holes, even though the sizes of the cryoconite holes selected for sampling were fairly uniform across glaciers. Thus, similar to other island biogeographic studies, the baseline diversity that an environment can support is a more important determinant of diversity than island size is. For example, one would not expect tree hole bacterial diversity (Bell et al., 2005) to have the same relationship to island size as machine oil reservoir bacteria (Van Der Gast et al., 2005), because other factors of those environments such as nutrient availability, temperature, and immigration may be very different and may influence diversity more strongly than island size. While different glaciers within the same valley are clearly not as different of ecosystems as the aforementioned examples, there is still a large effect of the glacier itself. This effect of local geography or biogeochemistry has been previously documented for cryoconite holes both within the Dry Valleys (Webster-Brown et al., 2015; Sommers et al., 2018) and elsewhere (Liu et al., 2015; Ambrosini et al., 2016).
When we analyzed the glaciers separately, both Canada Glacier and Taylor Glacier exhibited statistically significant relationships between cryoconite hole (island) size and bacterial phylogenetic diversity (Figure 2), and a significant relationship with species (OTU) richness as well (Supplementary Figure 1). In both of these cases, cryoconite hole surface area explained roughly 30% of the variance in bacterial phylogenetic diversity, indicating that although geography is likely the largest determinant of diversity [through climate and biogeochemical processes; (Bagshaw et al., 2013; Sommers et al., 2018)], island size is an important part of microbial community assembly within cryoconite holes. Island size can change over time as a cryoconite hole ages, since the growth of phototrophs can add to the volume of sediment and new sediments can be added during the rare times that the holes are open to the atmosphere. Diversity in these holes likely fluctuates as a function of hole age as well, so temporal variation in size and diversity may both be limitations of our analysis. Nonetheless, our analysis of Canada and Taylor Glacier diversity data supports our hypothesis that cryoconite hole bacterial communities exhibit a relationship between island size and diversity.
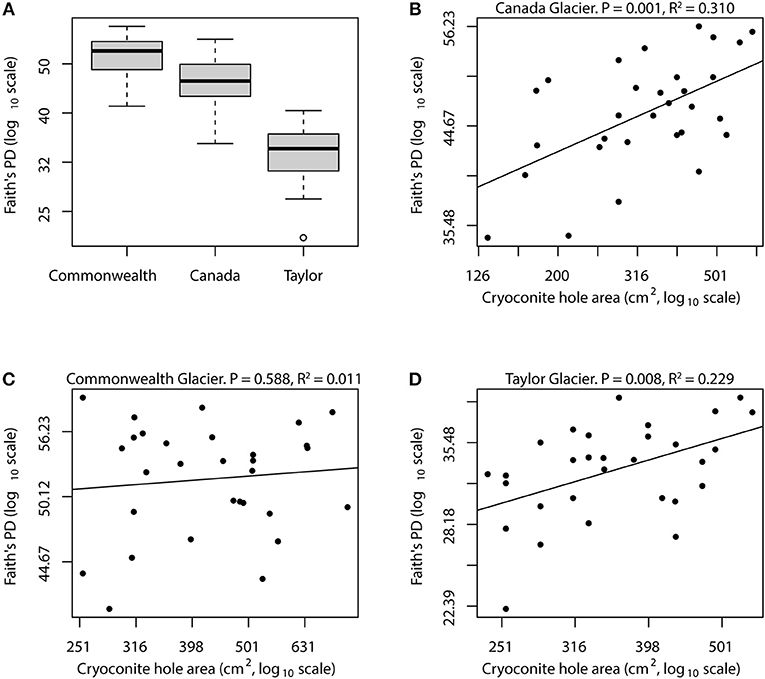
Figure 2. Relationships between island size and bacterial diversity for Taylor Valley cryoconite holes. Mean bacterial phylogenetic diversity is highest on Commonwealth glacier, followed by Canada and Taylor glaciers (A). Each of the within-glacier mean diversities was significantly different than the other two (Tukey's HSD, all P < 0.02). Canada and Taylor glaciers showed strong relationships between island size and bacterial diversity, with linear regression models explaining roughly 23 and 30% of diversity variance on log-log scales (B,D). However, the relationship on Commonwealth Glacier was not statistically significant (C), possibly because that glacier receives more biological input from nearby streams, causing the island size relationship to be confounded by high immigration rate.
This was not the case for Commonwealth glacier, however, where cryoconite hole size had no detectable qualitative or quantitative relationship with diversity (Figure 2). During our sample collection atop Commonwealth Glacier, we frequently observed pieces (up to 1 cm) of cyanobacterial mat littering the glacier's surface. These same cyanobacteria were present in the cryoconite sediment from Commonwealth glacier as well, and these conspicuous glacial denizens provided a stark contrast between Commonwealth glacier and the two more inland glaciers we sampled. Other researchers have noticed these cyanobacterial mats as well, and have jokingly named Commonwealth Glacier “The Jungle of the Dry Valleys.” These cyanobacterial mats are blown onto Commonwealth Glacier from nearby streams (McKnight et al., 1999; Taton et al., 2003), but they are not so common atop the glacier that they are abundant in every cryoconite hole. As we melted out cryoconite sediment from commonwealth glacier during sample processing, we noticed that not all samples contained visually conspicuous cyanobacterial mat. This wealth of wind-dispersed microbial biomass and diversity may explain why we detected no significant relationship between island size and bacterial diversity on Commonwealth Glacier: each cryoconite hole may encounter so much immigration that the slope of the relationship is significantly lowered, making it more difficult to detect.
Islands that are close to large sources of biodiversity like large islands or continents have increased immigration, meaning that they are at less risk of extinction (loss of diversity) and have a greater chance of recruiting new diversity. In this way, cryoconite holes on Commonwealth Glacier are close to the streams where cyanobacterial mats and other microbial life are abundant, so they essentially have a large nearby continent providing a source of new microbial diversity, and the other two glaciers do not have this feature. So it is our hypothesis that the lack of a relationship between island size and bacterial diversity in Commonwealth Glacier cryoconite holes is the result of the immigration rate being so high for Commonwealth Glacier that island size is not an important factor. The higher average bacterial phylogenetic diversity we and others observed on Commonwealth Glacier supports this hypothesis qualitatively. While cryoconite holes on Canada and Taylor glaciers appear very island-like, cryoconite holes on Commonwealth Glacier may be islands too, albeit islands close to continental shore.
Spatial Autocorrelation of Bacterial Communities
Within the McMurdo Dry Valleys, cryoconite hole bacterial communities that were more geographically close together had similar bacterial communities, while cryoconite holes that were more geographically distant had more different bacterial communities (Figure 3), supporting the distance decay of similarity hypothesis. This pattern was statistically significant (Mantel test; P = 0.001), and also the correlation coefficient was fairly large (rM = 0.718), indicating that the distance decay of similarity relationship is fairly robust even at the regional scale within the Dry Valleys.
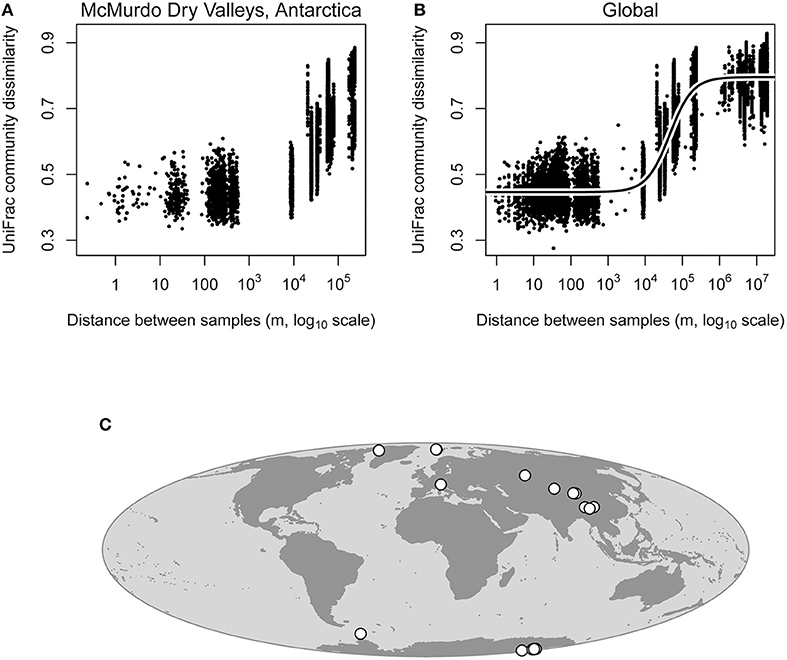
Figure 3. Distance decay of similarity analysis for cryoconite hole bacterial communities within the McMurdo Dry Valleys and globally. Note that UniFrac distance (Y-axes) is a distance metric rather than similarity, thus it increases as a function of geographic distance. Distance decay of similarity within the Dry Valleys was highly statistically significant (Mantel test; P = 0.001, rM = 0.718), meaning that geographically close cryoconite holes are likely to harbor similar bacterial communities, and geographically distant cryoconite holes are more likely to harbor more different bacterial communities (A). Distance decay of similarity in the global data set was also statistically significant (Mantel test; P = 0.001, rM = 0.888), meaning that the pattern observed in Antarctic Dry Valleys glaciers still holds when glaciers from around the Earth are included in the analysis (B). These data fit a logistic model much better than it fit a linear model (Supplementary Figure 3, R2 = 0.879), owing to the fact that the pattern observed is driven predominantly by spatial turnover at the regional scale (103-106 m, B). Locations of glaciers used in our analysis are shown on the map (C), and coordinates are given in Table 1.
While this relationship was statistically significant at smaller, within-glacier scales, its effect size was extremely low (Supplemental Figure 2) compared to analyses including multiple glaciers. We sampled at multiple nested spatial scales within each of the three Taylor Valley glaciers (Figure 1), and we expected the spatial structure of cryoconite hole bacterial communities to be fine-scale, occurring at spatial scales on the order of tens or hundreds of meters. Spatial structuring at small spatial scales (< 1 km) is often found in biogeographic analyses of supraglacial microbial communities, like in debris atop debris-covered glaciers (Franzetti et al., 2013; Darcy et al., 2017). But in the present study, this was not the case, as very little spatial structuring was observed for each of those three glaciers when analyzed separately (all rM values < 0.09). This contrasts with other supraglacial microbial biogeography, such as that of debris-covered glaciers, where strong spatial structuring of microbial communities is observed at scales < 1 km (Franzetti et al., 2013; Darcy et al., 2017). However, is assumed that spatial structuring on debris-covered glaciers is caused by biogeochemical gradients over space, rather than dispersal limitation. Indeed, for many organisms including microbes, patterns of distance decay of similarity are likely caused more by spatially autocorrelated environmental gradients than by dispersal limitation (Soininen et al., 2007; Astorga et al., 2012). While the physical isolation of Antarctic cryoconite holes made strong spatial structuring seem likely at small spatial scales, it may be that biogeochemical conditions within cryoconite holes on a given glacier are too uniform to see that spatial structuring.
At the global scale, significant spatial structuring of cryoconite hole bacterial communities is strong (Mantel test; P = 0.001, rM = 0.888). All data points from smaller spatial scales that we analyzed for the Dry Valleys data are still present in the global analysis, even though in this analysis comparisons between close-together pairs of cryoconite holes from different regions across the globe are overlaid at short distances in the plot (Figure 3). In other words, comparisons at 100 meters come from Antarctic, Arctic, and other glaciers worldwide. Therefore, across the world, cryoconite holes that are close together harbor similar bacterial communities, and perhaps more importantly, cryoconite holes that are farther apart do not harbor similar bacterial communities. Without a global-scale analysis of cryoconite hole biogeochemical conditions (which would require uniform global-scale data), it is not feasible to disentangle the effects of dispersal limitation and selection by environmental variables in driving this pattern.
Above distances of 1,000 kilometers, there is no longer any effect of geographic distance on cryoconite hole bacterial community composition (Figure 3, Supplemental Figure 4), suggesting that even though spatial structuring at geographic distances < 1,000 kilometers is a global phenomenon, at larger scales geographic distance no longer plays a role in shaping bacterial community composition of cryoconite holes. This is supported by our logistic model fit (Figure 3), which fit our data much better than a linear model considering the linear model's biased residuals (Supplementary Figure 3, R2 = 0.879). The logistic model shows how there is very little distance-decay pattern at small spatial scales (< 1 km), a strong pattern at the regional spatial scale (1 to 1,000 km), and very little pattern at larger spatial scales (>1,000 km). This lack of spatial structuring at larger scales is not surprising, since for some microbes, a similar dropoff has been observed at distances of several hundred meters (Robeson et al., 2011). This pattern is not diagnostic of the underlying process creating spatial structuring in cryoconite hole bacterial communities, but may inform future efforts to use cryoconite holes as model systems. Studies wishing to capture the spatial structuring of these bacterial communities (or the potential underlying biogeochemical features of the holes) should sample at scales from one to several hundred kilometers, and not to expect strong spatial structuring at smaller or much larger scales.
Conclusions
Cryoconite hole bacterial communities appear “island like,” because they exhibit a significant relationship between island size (hole area) and bacterial phylogenetic diversity (Figure 2), and show significant distance decay of similarity patterns both at the regional (McMurdo Dry Valleys) scale and at the global scale (Figure 3). However, these patterns are not entirely perfect or clear-cut, since not all glaciers analyzed exhibited a relationship between island size and diversity, and distance decay of similarity patterns for cryoconite hole microbial communities were weak (but significant) within glaciers yet very strong at larger spatial scales. We conclude that cryoconite hole bacterial communities do have island biogeography, but the strength of this pattern depends on spatial scale.
Author Contributions
JD, PS, DP, and SS designed experiments, and JD, PS, and DP collected and processed samples. JD, EG, and PS collected and analyzed data. JD primarily wrote the manuscript, and all authors contributed text and revisions.
Funding
This work was funded by NSF Polar Programs Award 1443578.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Diana Nemergut, who began this project. She will be missed. The authors also thank F. Zamora, A. Fountain, and UNAVCO for help in the field and help designing experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00180/full#supplementary-material
References
Ambrosini, R., Musitelli, F., Navarra, F., Tagliaferri, I., Gandolfi, I., Bestetti, G., et al. (2016). Diversity and assembling processes of bacterial communities in cryoconite holes of a Karakoram glacier. Microb. Ecol. 73, 827–837. doi: 10.1007/s00248-016-0914-6
Astorga, A., Oksanen, J., Luoto, M., Soininen, J., Virtanen, R., and Muotka, T. (2012). Distance decay of similarity in freshwater communities: do macro- and microorganisms follow the same rules? Glob. Ecol. Biogeogr. 21, 365–375. doi: 10.1111/j.1466-8238.2011.00681.x
Bagshaw, E. A., Tranter, M., Fountain, A. G., Welch, K., Basagic, H. J., and Lyons, W. B. (2013). Do cryoconite holes have the potential to be significant sources of C, N, and P to downstream depauperate ecosystems of Taylor Valley, Antarctica? Arctic, Antarct. Alp. Res. 45, 440–454. doi: 10.1657/1938-4246-45.4.440
Bell, T., Ager, D., Song, J. I., Newman, J. A., Thompson, I. P., Lilley, A. K., et al. (2005). Ecology: larger islands house more bacterial taxa. Science 308:1884. doi: 10.1126/science.1111318
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W., a, Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Darcy, J. L., King, A. J., Gendron, E. M. S., and Schmidt, S. K. (2017). Spatial autocorrelation of microbial communities atop a debris-covered glacier is evidence of a supraglacial chronosequence. FEMS Microbiol. Ecol. 93, 7287–7290. doi: 10.1093/femsec/fix095
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631.doi: 10.1073/pnas.0507535103
Foreman, C. M., Sattler, B., Mikucki, J. A., Porazinska, D. L., and Priscu, J. C. (2007). Metabolic activity and diversity of cryoconites in the Taylor Valley, Antarctica. J. Geophys. Res. Biogeosciences 112:G04S32. doi: 10.1029/2006JG000358
Fountain, A. G., Tranter, M., Nylen, T. H., Lewis, K. J., and Mueller, D. R. (2004). Evolution of cryoconite holes and their contribution to meltwater runoff from glaciers in the McMurdo Dry Valleys, Antarctica. J. Glaciol. 50, 35–45. doi: 10.3189/172756504781830312
Franklin, R. B., and Mills, A. L. (2003). Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol. Ecol. 44, 335–346. doi: 10.1016/S0168-6496(03)00074-6
Franzetti, A., Navarra, F., Tagliaferri, I., Gandolfi, I., Bestetti, G., Minora, U., et al. (2017). Potential sources of bacteria colonizing the cryoconite of an Alpine glacier. PLoS ONE 12:e0174786. doi: 10.1371/journal.pone.0174786
Franzetti, A., Tatangelo, V., Gandolfi, I., Bertolini, V., Bestetti, G., Diolaiuti, G., et al. (2013). Bacterial community structure on two alpine debris-covered glaciers and biogeography of Polaromonas phylotypes. ISME J. 7, 1483–1492. doi: 10.1038/ismej.2013.48
Glassman, S. I., Lubetkin, K. C., Chung, J. A., and Bruns, T. D. (2017). The theory of island biogeography applies to ectomycorrhizal fungi in subalpine tree “islands” at a fine scale. Ecosphere 8:e01677. doi: 10.1002/ecs2.1677
Green, J. L., Holmes, A. J., Westoby, M., Oliver, I., Briscoe, D., Dangerfield, M., et al. (2004). Spatial scaling of microbial eukaryote diversity. Nature 432, 747–750. doi: 10.1038/nature03034
King, A. J., Freeman, K. R., McCormick, K. F., Lynch, R. C., Lozupone, C., Knight, R., et al. (2010). Biogeography and habitat modelling of high-alpine bacteria. Nat. Commun. 1:53. doi: 10.1038/ncomms1055
Liu, Q., Zhou, Y.-G., and Xin, Y.-H. (2015). High diversity and distinctive community structure of bacteria on glaciers in China revealed by 454 pyrosequencing. Syst. Appl. Microbiol. 38, 578–585. doi: 10.1016/J.SYAPM.2015.09.005
Lozupone, C., and Knight, R. (2005). UniFrac : a new phylogenetic method for comparing microbial communities UniFrac : a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228
MacArthur, R. H., and Wilson, E. O. (1967). Theory of Island Biogeography. Princeton University Press. Available online at: https://books.google.com/books?hl=en&lr=&id=YBDWCgAAQBAJ&pgis=1 (Accessed April 12, 2016).
Mackas, D. L. (1984). Spatial autocorrelation of plankton community composition in a continental shelf ecosystem. Limnol. Oceanogr. 29, 451–471. doi: 10.4319/lo.1984.29.3.0451
Martiny, J. B., Eisen, J. A., Penn, K., Allison, S. D., and Horner-Devine, M. C. (2011). Drivers of bacterial beta-diversity depend on spatial scale. Proc. Natl. Acad. Sci. U.S.A. 108, 7850–7854. doi: 10.1073/pnas.1016308108
McKnight, D. M., Niyogi, D. K., Alger, A. S., Bomblies, A., Conovitz, P. A., and Tate, C. M. (1999). Dry valley streams in antarctica: ecosystems waiting for water. Bioscience 49, 985–995. doi: 10.1525/bisi.1999.49.12.985
Nekola, J. C., and White, P. S. (1999). The distance decay of similarity in biogeography and ecology. J. Biogeogr. 26, 867–878. doi: 10.1046/j.1365-2699.1999.00305.x
Nemergut, D. R., Schmidt, S. K., Fukami, T., O'Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGinn, D., et al. (2016). vegan: Community Ecology Package. Available online at: https://github.com/vegandevs/vegan
R Core Team (2016). R. A. language and environment for statistical computing. R Found. Stat. Comput. 1:409. doi: 10.1007/978-3-540-74686-7
Robeson, M. S., King, A. J., Freeman, K. R., Birky, C. W., Martin, A. P., and Schmidt, S. K. (2011). Soil rotifer communities are extremely diverse globally but spatially autocorrelated locally. Proc. Natl. Acad. Sci. U.S.A. 108, 4406–4410. doi: 10.1073/pnas.1012678108
Rominger, A. J., Goodman, K. R., Lim, J. Y., Armstrong, E. E., Becking, L. E., Bennett, G. M., et al. (2016). Community assembly on isolated islands: macroecology meets evolution. Glob. Ecol. Biogeogr. 25, 769–780. doi: 10.1111/geb.12341
Rosenzweig, M. L. (1995). Species Diversity in Space and Time. Cambridge, UK: Cambridge University Press.
Soininen, J., McDonald, R., and Hillebrand, H. (2007). The distance decay of similarity in ecological communities. Ecography 30, 3–12. doi: 10.1111/j.0906-7590.2007.04817.x
Sommers, P., Darcy, J. L., Gendron, E. M. S., Stanish, L. F., Bagshaw, E. A., Porazinska, D. L., et al. (2018). Diversity patterns of microbial eukaryotes mirror those of bacteria in Antarctic cryoconite holes. FEMS Microbiol. Ecol. 94:fix167. doi: 10.1093/femsec/fix167
Stanish, L. F., Bagshaw, E. A., McKnight, D. M., Fountain, A. G., and Tranter, M. (2013). Environmental factors influencing diatom communities in Antarctic cryoconite holes. Environ. Res. Lett. 8:045006. doi: 10.1088/1748-9326/8/4/045006
Taton, A., Grubisic, S., Brambilla, E., De Wit, R., and Wilmotte, A. (2003). Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 69, 5157–5169. doi: 10.1128/AEM.69.9.5157-5169.2003
Telling, J., Anesio, A. M., Tranter, M., Fountain, A. G., Nylen, T., Hawkings, J., et al. (2014). Spring thaw ionic pulses boost nutrient availability and microbial growth in entombed Antarctic Dry Valley cryoconite holes. Front. Microbiol. 5:694. doi: 10.3389/fmicb.2014.00694
Uetake, J., Tanaka, S., Segawa, T., Takeuchi, N., Nagatsuka, N., Motoyama, H., et al. (2016). Microbial community variation in cryoconite granules on Qaanaaq Glacier, NW Greenland. FEMS Microbiol. Ecol. 92:fiw127. doi: 10.1093/femsec/fiw127
Van Der Gast, C. J., Lilley, A. K., Ager, D., and Thompson, I. P. (2005). Island size and bacterial diversity in an archipelago of engineering machines. Environ. Microbiol. 7, 1220–1226. doi: 10.1111/j.1462-2920.2005.00802.x
Vonnahme, T. R., Devetter, M., Žárský, J. D., Šabacká, M., and Elster, J. (2016). Controls on microalgal community structures in cryoconite holes upon high-Arctic glaciers, Svalbard. Biogeosciences 13, 659–674. doi: 10.5194/bg-13-659-2016
Wang, J., Shen, J., Wu, Y., Tu, C., Soininen, J., Stegen, J. C., et al. (2013). Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J. 7, 1310–1321. doi: 10.1038/ismej.2013.30
Webster-Brown, J. G., Hawes, I., Jungblut, A. D., Wood, S. A., and Christenson, H. K. (2015). The effects of entombment on water chemistry and bacterial assemblages in closed cryoconite holes on Antarctic glaciers. FEMS Microbiol. Ecol. 91:fiv144. doi: 10.1093/femsec/fiv144
Keywords: Antarctica, cryoconite, biogeography, islands, microbial ecology
Citation: Darcy JL, Gendron EMS, Sommers P, Porazinska DL and Schmidt SK (2018) Island Biogeography of Cryoconite Hole Bacteria in Antarctica's Taylor Valley and Around the World. Front. Ecol. Evol. 6:180. doi: 10.3389/fevo.2018.00180
Received: 14 July 2018; Accepted: 22 October 2018;
Published: 20 November 2018.
Edited by:
Anton Pieter Van de Putte, Royal Belgian Institute of Natural Sciences, BelgiumReviewed by:
Jianjun Wang, Nanjing Institute of Geography and Limnology (CAS), ChinaAna M. C. Santos, University of Alcalá, Spain
Copyright © 2018 Darcy, Gendron, Sommers, Porazinska and Schmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John L. Darcy, amRhcmN5QGhhd2FpaS5lZHU=
 John L. Darcy
John L. Darcy Eli M. S. Gendron2
Eli M. S. Gendron2 Pacifica Sommers
Pacifica Sommers Steven K. Schmidt
Steven K. Schmidt