- 1Biodiversity and Conservation Science, Department of Biodiversity, Conservation & Attractions, Kensington, WA, Australia
- 2Division of Ecology & Evolution, Research School of Biology, Australian National University, Acton, ACT, Australia
- 3Centre for Biodiversity Analysis, Canberra, ACT, Australia
Current approaches to biodiversity conservation are largely based on geographic areas, ecosystems, ecological communities, and species, with less attention on genetic diversity and the evolutionary continuum from populations to species. Conservation management generally rests on discrete categories, such as identified species, and, for threated taxa, intraspecific units. Species, in particular, provide a common measure of biodiversity yet in both theory and nature, speciation is typically a protracted process progressing from connected populations to unambiguous species with variable rates of phenotypic, ecological and genetic divergence. Thus, most recognized species are not genetically uniform and are sometimes highly structured into historically isolated populations worthy of consideration as intraspecific units that represent unique genetic diversity for conservation. Genome screens offer unprecedented resolution of structure across taxonomic boundaries in species complexes, and have the potential to oversplit species if not interpreted conservatively. This highlights the blurred line between populations and species, and can confound simple dichotomies of “species” vs. “not species.” At the same time, like plants, there is increasing evidence that even distantly related animal species can hybridize and exchange genes. A review of conservation legislation reveals that legal definitions of “species” are quite flexible and can accommodate a range of infra-specifictaxa and divergent populations, as well as taxonomically recognized species. For example, the legislative definition of a species around the world can include: species, subspecies, varieties, and geographically and/or genetically distinct populations. In principle, this flexibility allows for protection of genetic diversity and maintenance of evolutionary processes at a broad range of infra-specific levels. However, evolutionary biologists often fail to adequately justify and then translate their evidence for genetically defined units into categories suited to assessment under local legislation. We recommend that (i) genomic data should be interpreted conservatively when formally naming species, (ii) concomitantly, there should be stronger impetus and a more uniform approach to identifying clearly justified intraspecific units, (iii) guidelines be developed for recognizing and labeling intraspecific data that align with best scientific practice, and (iv) that the more nuanced view of species and speciation emerging from genomic analyses is communicated more effectively by scientists to decision makers.
Introduction
International conservation policy recognizes biodiversity at three levels, ecosystem, species and genetic, and that management should aim to retain all three (Convention on Biological Diversity, 2007). This is clearly reflected in the Convention on Biological Diversity, Aichi Biodiversity Targets, agreed in 2010 (https://www.cbd.int/sp/targets/), where there is specific reference in goals and targets, not only to ecosystems and species, but also to genetic diversity. Yet current approaches to biodiversity conservation are largely based on geographic areas, ecosystems, ecological communities, and species, with less attention on genetic diversity and the species-population continuum. Indeed the majority of conservation practitioners focus on species as the metric of biodiversity, with some consideration of intraspecific units within intensively managed threatened species. In this context, there is a natural desire to employ simple categorizations—named species, and other named taxa (i.e., subspecies), and sometimes conservation units within species. Species in particular provide a common measure of biodiversity that underpins much scientific and management endeavor (Mace, 2004; Wilson, 2017). As a fundamental unit of conservation, species are often used to quantify biodiversity value through lists of species within protected areas, identification of threatened species within jurisdictions, and as a basis for biosecurity procedures. In addition, species are typically the entities with which we communicate conservation issues to the public.
This impetus to simply identify discrete units in nature runs counter to several, related realities. First, speciation is not instantaneous. Rather it is typically a protracted process (Dynesius and Jansson, 2014; Roux et al., 2016), progressing from isolated populations, perhaps subject to different selection pressures, to unambiguous species over long time scales, with frequent reversals caused by environmental change or simple genetic merging as ranges reconnect (Figure 1; Seehausen et al., 2008; Rosenblum et al., 2012). Second, most recognized species are not genetically uniform. Divergence (drift and protracted isolation) and reticulation (geneflow, hybridization, introgression, and recombination) effectively shape geographic and genomic variation within species (Cutter and Payseur, 2013; Edwards et al., 2016) and some species can be structured into long isolated populations, some of which could warrant recognition as full but perhaps morphologically cryptic species (Bickford et al., 2007; Jorger and Schrödl, 2013). Third, as long recognized, named species are not equivalent in terms of how much evolutionary legacy they represent—put simply, a platypus (as representative of an anciently diverged lineage) may be considered to be worth more than a recently diverged rodent (Isaac et al., 2007). Fourth, species designations are not cast in stone, rather they are hypotheses to be tested as new evidence becomes available (Hey et al., 2003). Each of these issues creates complexity and ambiguity in delimiting species in general, and assigning conservation attention to them or their component populations. In addition, the delimitation of species is also subject to different species concepts and ways of diagnosing them, which has caused great consternation among conservation biologists (Mace, 2004; Frankham et al., 2012; Garnett and Christidis, 2017).
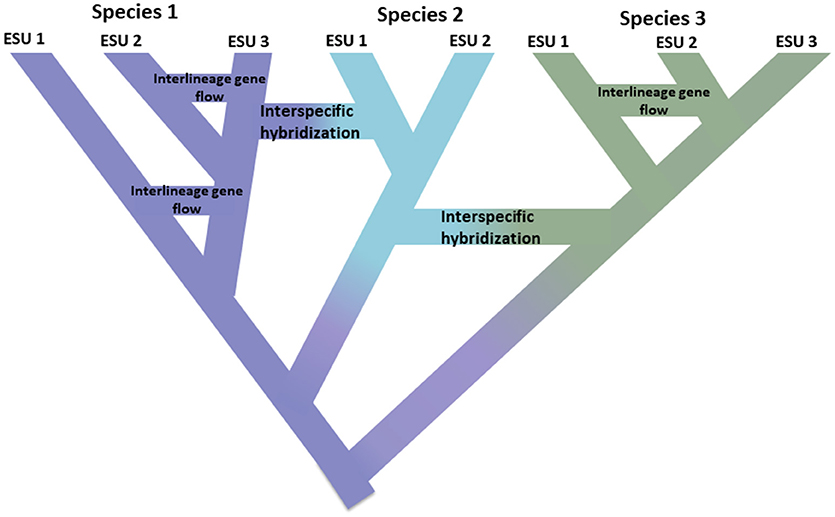
Figure 1. Speciation as a continuum progressing from isolated populations to unambiguous species shown by the shift in color with the colors representing different species. Some lineages that arise due to population isolation develop into true species with the internal nodes of the tree representing initiation of the speciation process. Other lineages do not develop into true species but diverge over time indicating that species are not genetically uniform. These lineages are shown as Evolutionarily Significant Units (ESUs). Inter-lineage gene flow is indicated for Species 1 and Species 3. In this case, evolutionary independence may be transient because two lineages belonging to the same species are not reproductively isolated. While they remain as distinct lineages (species), interspecific hybridization may occur with limited gene exchange (shift in color) between species.
Implementation of effective conservation strategies that aim to protect both species and genetic diversity would benefit from both a more consistent view of how we delimit species and other units across the population-species continuum, and how these units are interpreted in different legislation globally. New capabilities from genomics provide both opportunities and challenges for achieving these goals. In the following we consider: (i) how genome-scale analyses can be reconciled with a conservative approach to the delimitation and naming of species, while also identifying major components of genetic diversity within species, (ii) how existing conservation legislation across several continents, that defines “species” to include a broad range of infra-specific categories, can be used given the high resolution offered by genome screens, and (iii) how the insights into speciation processes and structuring of variation from genomics align with legislation and then into management. We then suggest some ways forward in aligning conservation units with legislative approaches in order to protect the full spectrum of diversity from populations through species.
Genomics, Speciation, and Taxonomy
Like many fields of biological science, systematics, the practice of naming taxa, and understanding their relationships and how they form, is being significantly advanced by genomics and statistical models. Whether through whole genome sequencing (e.g., Nater et al., 2017) or genome sampling (SNPs, target capture-sequencing; Jones and Good, 2016; Leache and Oaks, 2017), we now have unprecedented resolution of patterns of genetic diversity (Funk et al., 2012). Coupled with increasingly powerful statistical models for inferring histories of genomic and species divergence, these data are providing new insights into the evolutionary processes that generate species and genetic variation.
Application of these new tools generates two opposing insights. On one hand, many studies are revealing highly divergent genetic populations within named species, so called cryptic species (Bickford et al., 2007; Jorger and Schrödl, 2013; Struck et al., 2017). This is nothing new, as it builds on a long history of spatial population genetics and molecular phylogeography that has been a productive endeavor for three decades (Avise et al., 1987). What genomics brings to this endeavor is both increased resolution of population structure and data that are sufficient to statistically infer population histories and test alternative models of divergence (Degnan and Rosenberg, 2009; Sukumaran and Knowles, 2017). On the other hand, genome-scale analyses are also revealing that (as long been recognized for plants), genetic exchange (introgression) among animal species is more common than previously thought, both during and after speciation, and can even drive new adaptive radiations (Mallet, 2007; Rieseberg and Willis, 2007; Arnold and Kunte, 2017) Figure 1. Put together, these insights further emphasize that speciation is protracted (Roux et al., 2016) and reveal the opposing forces operating through the phylogeography-phylogenetics continuum (Edwards et al., 2016).
How does all this connect with taxonomy and conservation objectives? There is strong and justifiable concern that the increased resolution afforded by genome screens could lead to rampant over-splitting of species (Isaac et al., 2004), potentially restricting management flexibility and consigning small genetically divergent populations to inbreeding and eventual extinction (Frankham et al., 2012). Conversely, Gippoliti et al. (2018) argue that taxonomic inertia, resulting in previous failure to recognize many species of African ungulates, had resulted in outbreeding depression following mixing and the failure to protect geographically restricted taxa. Their view (and see also Morrison et al., 2009) is that there is no evidence that taxonomic inflation is having negative effects on ungulate conservation (Gippoliti et al., 2018). A related concern is that unstable taxonomy retards both the practice of conservation and public perceptions (Garnett and Christidis, 2017). These concerns are in part driven by the long running debate around species concepts and diagnostic methods in systematic biology, in particular application of the Phylogenetic Species Concept that holds that species are minimally diagnosable units (Cracraft, 1983). However, the majority of taxonomists are inherently conservative, only naming species when there is concordance across multiple lines of evidence (Dayrat, 2005; Padial et al., 2010; Schlick-Steiner et al., 2010: Yeates et al., 2011). Our view is that species should represent robust independently evolving lineages that remain largely intact when in contact with close relatives—in accord with the intent of the Evolutionary Species Concept (Wiley, 1978) and the Generalized Lineage Species Concept (de Queiroz, 1998, 2007). In practice, this requires direct evidence of reproductive isolation or reasonable surrogates of the same, while allowing for limited genetic exchange (Singhal et al., 2018).
Genomic data provides greater power than before to identify differentiation and divergence within and among species, requiring greater consideration of how this affects delineation of taxonomic units. Where populations are both strongly genetically and phenotypically divergent relative to already named species, there is a strong case for naming these as separate species. If populations are strongly divergent phenotypically with little genomic separation, then there is a judgement call on whether those phenotypic differences are (i) heritable, and (ii) likely to cause substantive reproductive isolation. Genome scans can resolve such situations by testing for restricted gene flow. The more challenging issue is where morphologically similar, but genetically divergent populations are detected. Such populations can readily be diagnosed as separate evolutionary lineages by applying statistical delimitation methods to multilocus data (Carstens et al., 2013; Rannala, 2015), but are they ephemeral populations or durable species (Sukumaran and Knowles, 2017)? It certainly is possible, within the rules of nomenclature, to recognize species from DNA sequences alone, providing sequence-based diagnostics (Tautz et al., 2003; Cook et al., 2010). For example, Murphy et al. (2015) delimit and describe six Australian desert spring amphipod species based on DNA sequence data despite no clear morphological differences. However, this is a general area for improvement where systematists need to apply the term “cryptic species” with greater consistency and rigor (Struck et al., 2017).
In situations where DNA sequence data provides a potential key defining feature for recognition of a species, we suggest that it is especially important to apply other lines of evidence to avoid oversplitting (Oliver et al., 2015). One approach is to test for substantial reproductive isolation where such populations form natural contact zones. For example, in a comparative genomic analysis of contact zones among lineage-pairs of Carlia lizards that are phenotypically indistinguishable, indices of genetic isolation were strongly correlated with divergence time (Singhal and Moritz, 2013; Singhal and Bi, 2017) (Figure 2). The more divergent of these lineage-pairs have now been formally recognized as separate species, whereas more closely related (yet statistically diagnosable) lineages with evidence of genomically extensive genetic exchange across contact zones were not (Singhal et al., 2018). Given the strong empirical relationship between divergence and reproductive isolation in these taxa, these authors further separated two allopatric and cryptic isolates (of a closely related species) that had even higher genomic divergence than those delimited above. Extending this reproductive isolation informed metric to other congeneric species with deeply divergent but morphologically similar phylogeographic lineages (Potter et al., 2016, 2018; Afonso Silva et al., 2017a) suggests that more species remain to be described, especially on islands, (Figure 2, Carlai amax and C. rufilatus), and indeed, some such species have been described following complementary phenotypic analysis (Afonso Silva et al., 2017b). Other lineages with more recent divergences, but still statistically delimited as separate lineages using a large set of nuclear genes, can be represented as Evolutionarily Significant Units (ESUs) within species.
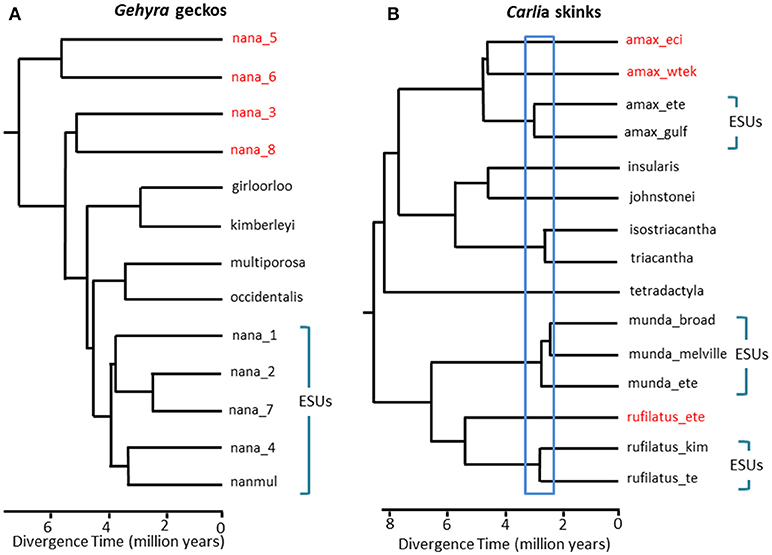
Figure 2. Phylogenomic resolution of species boundaries in lizards from the Australian Monsoonal Tropics (AMT). In each case relationships and relative divergence times are estimated from 100 exons using the multispecies coalescent implemented in StarBEAST2, but the time scales between the two examples are not necessarily the same. (A) Results for the nana group of Gehyra geckos (Moritz et al., 2018). The phylogenetic diversity of lineages within G. nana, as previously recognized, included four other already described and morphologically distinctive species. Subsequent analyses of range overlaps and morphology has led to recognition of each of the most phylogenetically divergent lineages (red taxon labels) as separate species (Doughty et al., 2018). The (morphologically inseparable) lineages remaining in G. nana as now recognized can be considered as ESUs. (B) Ongoing resolution of species boundaries in Carlia skinks from the AMT. Some lineages have already been recognized as species based on a combination of genetic species delimitation and post hoc identification of diagnostic morphological differences (C. insularis and C. isostriacantha; Afonso Silva et al., 2017b). For other complexes (Potter et al., 2016, 2018), taxonomic revisions are in progress. The blue rectangle indicates the range of divergence times where substantial reproductive isolation, correlated with divergence, has been observed in contact zones between morphologically cryptic lineages of related species (Singhal and Bi, 2017). Conservatively, and following (Singhal et al., 2018), we recognize as candidate species three genetic lineages with divergences greater than this empirical threshold. This would leave genetically distinct ESUs within each of Carlia amax, Carlia munda, and Carlia rufilatus (blue brackets). The scale bars represent estimates of divergence time in millions of years.
In extreme cases, genomic methods reveal single taxonomic species to not only be genetically heterogenous, but, in a phylogenetic sense, to also contain other named species. Such “paraphyly” can arise though recent speciation from a single, genetically subdivided species (Patton and Smith, 1994), but it also generates the hypothesis that the genetically heterogeneous taxon may represent multiple species. Again, more evidence is required. As a case in point, phylogenomic analysis of a complex of Gehyra gecko lizards from northern Australia delimited nine evolutionary lineages, with four already named species phylogenetically embedded among them (Moritz et al., 2018; Figure 2). Subsequent evidence of geographic overlap (while retaining phylogenetic separation) and subtle morphological differences supported naming of the four most phylogenetically divergent lineages (Doughty et al., 2018). Conversely, the more closely related (yet statistically separable) lineages had adjacent rather than overlapping distributions, but were not morphologically diagnosable, and accordingly, were retained as a single species (G. nana sensu stricto), albeit with multiple ESUs, pending further investigation (Figure 2). Subsequent genome scans of samples across the boundaries of these ESUs revealed that at least some do have substantial genetic exchange, validating the original, conservative approach taken for recognizing species.
In other cases, plant genomic studies, even through simple ordination of SNPs, can reveal stark, and unanticipated divergences among population samples, suggesting the presence of cryptic taxa that can then be analyzed for diagnostic phenotypic characters. For example, SNP-based evidence for genomic divergence among populations and ecotypic differences in Eucalyptus salubris revealed two distinct molecular lineages that maintained their distinctiveness in an area of geographic overlap, with ecotypic adaptation considered to be an important factor in minimizing gene flow between the lineages. (Steane et al., 2015; Figure 3). In another case, in Australian Pelargonium, genomic analysis, combined with morphological and reproductive studies, has led to significant changes in taxonomy, both splitting and lumping of taxa, especially where previously used morphological traits are found to be labile and discordant with phylogeny (Nicotra et al., 2016).

Figure 3. Genomic analysis using SNP data identified cryptic lineages in the widespread Eucalyptus salubris. The location of samples populations of each lineage are shown across an aridity index gradient in south west Western Australia. The PCoA based on binary genetic distances derived from 16,122 DArTseq markers, shows two distinct lineages. Adapted from Steane et al. (2015).
The above cases illustrate the progressive hypothesis-testing approach that has been used in integrative taxonomy (Padial et al., 2010; Yeates et al., 2011). Further, these cases use some form of genome reduction sequencing to obtain high quality data from 1000's of independent genes, giving much greater confidence in the outcomes than is the case using just plastid loci or a few nuclear genes. An example of how applying integrative taxonomy can dramatically alter perceptions of diversity comes from a combined genetic, acoustic and morphological survey of frogs from Madagascar that nearly doubled the known species diversity (Vieites et al., 2009). These authors proposed a series of terms reflecting the state of taxonomic resolution and congruency: Confirmed Candidate Species, for genetically and phenotypically divergent, but not yet named taxa; Unconfirmed Candidate Species for genetically divergent taxa where morphology and acoustics had not yet been examined; and Deeply Conspecific Lineages for entities known to be genetically divergent yet phenotypically cryptic. Genomic analyses are yet to be applied to these taxa, but would surely inform decisions about conspecific lineages vs. candidate species.
Genomic approaches, even when applied conservatively, will no doubt lead to further splitting of morphologically conservative taxa. But, conversely, there are clear examples where the same approaches reveal over-split or incorrectly diagnosed species (Nicotra et al., 2016). Of course there are already many molecular studies on plant and animal species complexes based on smaller-scale datasets (e.g., microsatellite, mtDNA, and cpDNA) that have identified where species have been misdiagnosed, or where multiple species have been circumscribed taxonomically but are now considered to be a single species. For example, Rabosky et al. (2014) used mitochondrial and nuclear gene phylogenies to revise the scincid Ctenotus inornatus complex, resulting in a net reduction of species and realignment of species boundaries; the morphological traits applied previously, while convenient for field biologists, were highly labile and largely failed to match species boundaries as diagnosed genetically. In plants some of the most well-known examples are found in the orchids, such as the European orchid genus Ophrys and the Australian orchid genus Caladenia, where molecular studies indicate significantly fewer entities that would be recognized as species, than those based on morphological traits and pseudocopulatory pollination syndromes (see Devey et al., 2008; Swarts et al., 2014).
Genome-scale data, when interpreted conservatively and ideally with parallel analyses of phenotypes, will continue to lead to taxonomic changes, especially in morphologically conservative taxa (Fišer et al., 2018). But, in contrast to concerns expressed by some (Garnett and Christidis, 2017), we consider that this is the proper pursuit of evolutionary systematics, even if outcomes are temporarily inconvenient.
Genomics and Intraspecific Conservation Units
Although less contentious than taxonomic delimitation and the species level debate, but in many ways just as complex, has been the significant discussion over the definition and delimitation of conservation units as important elements of intraspecific diversity that need consideration in conservation actions (Ryder, 1986; Waples, 1991; Dizon et al., 1992; Moritz, 1994; Vogler and de Salle, 1994; Crandall et al., 2000; Fraser and Bernatchez, 2001; Avise, 2004; Funk et al., 2012) (Figure 4). Given the cautious approach to recognizing species from genomic evidence that we advocate above (and see also Oliver et al., 2015; Singhal et al., 2018), it is inevitable that some taxonomic species will still contain long isolated populations within them. Concepts such as Evolutionarily Significant Units (ESUs) or Designatable Units (DUs) have been proposed to complement existing taxonomy (Ryder, 1986; Waples, 1991; Moritz, 1994; Mee et al., 2015). As with species, interpretations and criteria vary, but the common intent is to recognize major elements of intraspecific diversity for conservation actions (Funk et al., 2012). Moritz (1994, 2002) advocated a simplistic but practical molecular approach to identifying independently evolving segments within species, the “historical” axis of diversity; whereas others put more emphasis on the “adaptive” axis to include functional diversity (Ryder, 1986; Crandall et al., 2000; Fraser and Bernatchez, 2001; Waples et al., 2001; Mee et al., 2015) (Figure 4). Whatever the relative merits of these alternatives, it is clear that the technical and analytical tools of population genomics can now robustly identify both components of intraspecific diversity (Funk et al., 2012; Hoban et al., 2013; Whitlock, 2014; Yang and Rannala, 2014).
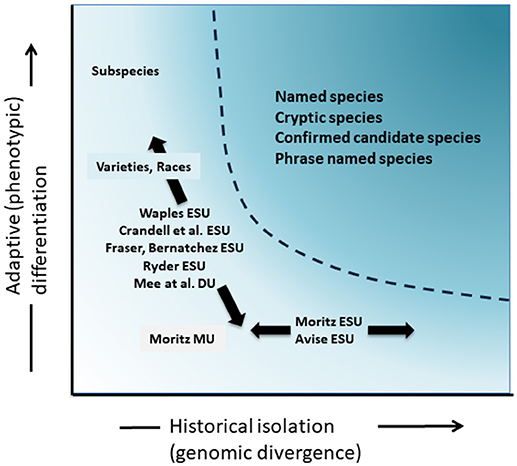
Figure 4. Separation of genetic diversity into two components: adaptive variation that arises through natural selection, and assayed though analysis of phenotypes and historical isolation or neutral divergence assessed through genomic divergence. The darker shaded area above the curve indicates conditions where populations are likely to be considered as separate species under most concepts. Various conservation unit concepts and infra-specific taxonomic categories are shown relative to the two axes of genetic diversity (after Moritz, 2002).
This leads to the question: why not just name ESUs as subspecies? Subspecies have traditionally been defined and viewed by both botanists and zoologists as phenotypically distinct, allopatric sets of populations that may intergrade into each other at geographic boundaries (Mayr, 1942; Grant, 1981). As such they potentially reflect adaptive divergence without necessarily having been historically isolated. Subspecies designations are widely used in plants, with geographical, and ecological differences the primary means by which they are distinguished (Hamilton and Reichard, 1992). In so far as subspecies manifest local adaptation, with or without ongoing gene flow, we suggest that phenotypically defined subspecies should continue to be recognized in conservation, and not conflated with ESUs, at least not ESUs as defined to represent historically isolated populations.
In the same way as population genomics provides far greater precision in the identification of ESUs by utilizing both neutral and adaptive loci it also provides a means for utilizing large numbers of neutral loci to more readily delimit Management Units (MUs) as demographically independent units characterized by restricted gene flow (Funk et al., 2012). While most of our focus here is on ESU's and analogous units such as DUs that can be recognized as the major intra-specific units for conservation and have been accepted for protection under some jurisdictions, MUs have also been identified representing demographically independent sets of populations (see Palsbøll et al., 2007) that can be managed to retain the larger ESU (Moritz, 1994; Funk et al., 2012). Management Units can be important in monitoring and managing populations of species and have also been referred to as “stocks,” particularly relevant to fisheries management (Avise, 2004; Laikre et al., 2005). The definition of MUs by Moritz (1994) as “populations with significant divergence of allele frequencies at nuclear or mitochondrial loci, regardless of the phylogenetic distinctiveness of the alleles” is generally widely accepted, although, as pointed out by Palsbøll et al. (2007), demographic connectivity is the key factor not the level of historical gene flow.
Delimiting conservation units as important elements of intraspecific diversity also has significant implications for genetic rescue of small inbred populations through restoration of gene flow or by augmentation and it is becoming increasingly clear that genetic rescue needs to be considered more broadly if increased population extinction is to be averted (Love Stowell et al., 2017; Ralls et al., 2018). It was proposed by Moritz (1999) that mixing MUs but not ESUs could be considered an appropriate strategy for genetic rescue. As emphasized by Frankham et al. (2012) mixing of taxonomically recognized species would generally not be considered acceptable by managers, in which case taxonomic oversplitting could restrict opportunities to rescue small inbred populations. We suggest that while a more rigorous approach to delimiting intraspecific units such as MUs could help alleviate this problem, it is also clear that genomic estimation of migration rates and of adaptive divergence has the potential to provide much more robust guidance in not only defining species boundaries and intraspecific variation, but in designing genetic rescue strategies.
Phylogenomics and Conservation
Genomics combined with phylogenetic methods can readily resolve relationships and evolutionary distance among taxa, as well as delimiting species. This is best achieved using species tree methods, rather than concatenation, as the former account for gene tree heterogeneity and avoid over-inflation of tip lengths due to ancestral polymorphism (Edwards et al., 2016; Ogilvie et al., 2016). While massive numbers of loci are often not essential, including more loci often improves resolution of both relationships and branch lengths (e.g., Blom et al., 2016). The importance to conservation is twofold. First, phylogenetic assessments enable consideration of the distinctiveness of species or ESUs, not just whether they are described. Thus, the EDGE program (Isaac et al., 2007) advocates weighting of species for conservation based on how much unique evolutionary history they represent, in addition to usual IUCN criteria. Second, branch lengths themselves can represent biodiversity values, independent of taxonomy (Faith, 1992), which has demonstrated value in prioritizing areas for conservation (Laity et al., 2015; Rosauer et al., 2016, 2018). In this sense, adopting phylogenetic metrics using measures such as phylogenetic diversity and phylogenetic endemism make it possible to highlight areas of high evolutionary diversity that can be targeted for protection, and be given high priority in any planning process for improved biodiversity conservation. This approach can enhance the protection of genetic diversity and evolutionary processes for diverse groups of organisms across a range of geographical scales (Laity et al., 2015) making decisions about areas to prioritize more robust to the vagaries of how genomic evidence is translated to recognition of species or intra specific units.
The Real World—Legislative Definitions
Policy and legislative frameworks for biodiversity conservation range from international conventions and strategies, through national strategies and legislation, and state or regional legislation and strategies within countries. The major international instruments on biological conservation include the Convention on Biological Diversity implemented through the Convention of the Parties (COP), the World Heritage Convention, the Global Biodiversity Strategy, the Montreal Process for Forest Conservation, the Ramsar Convention on Wetlands, the Convention on Migratory Species, the Convention on International Trade in Endangered Species (CITES), the International Union for Conservation of Nature (IUCN) red list on threatened species, and the United Nations Framework convention on climate change. All these charters embody the principal of conserving species and genetic diversity as a basic goal of biodiversity conservation, with species being the unit broadly defined for protection. However, the definitions of species under legislation and international directives are extremely variable. They incorporate taxonomic recognition of species and intraspecific taxa, but may also incorporate genetic/evolutionary concepts that are often covered under the legal definition of a “species” (Table 1).
Given the broad range of infra-specific levels that can be protected (Table 1), the legislation and associated listing processes of countries should facilitate the conservation of genetic diversity. For example, the definition of a “species,” when one considers legislation around the world (Table 1), can include: species, subspecies, varieties, and geographically and/or genetically distinct populations, while under South African legislation it may also include cultivars, geographic races, strains and hybrids. Thus, under most legislation globally a “species” includes entities that reflect various infra-specific levels that are all part of a broader species distribution and reflect to varying degrees the population-species continuum. This parallels the inevitable difficulty that taxonomists, evolutionary biologists and conservation geneticists face in finding a more consistent and uniform way to specify units within this continuum.
Some argue that concerns regarding the policy implications of species and infra-specific uncertainty are misplaced (Hey et al., 2003), but we are still of the view that while legislation can provide a mechanism for protecting genetic diversity at a range of levels, the onus is on biologists to provide more clarity and consistency on how those levels should be delimited from species through to individual populations. At present managers and legislators are left with a bewildering array of species concepts and infra-specific units provided by scientists when delimiting units across the population-species continuum. For example, all jurisdictions consider species recognized by taxonomists, as key units for conservation (Table 1), yet there is still broad debate among taxonomists around species concepts, and how species are diagnosed in practice varies within and across groups. This has led to a recent call for a more regulated approach to taxonomy (Garnett and Christidis, 2017), to which others have countered that regulation of taxonomy is not the answer (Thomson et al., 2018). Although we concur with some of the points made by Thomson et al. (2018) we suggest it is rather naive to propose that legislation itself needs to change to explicitly reference the specific taxon concept implied by a name. These and previous such debates highlight the need for evolutionary biologists and taxonomists to take the lead in ensuring clarity (transparency) in how they translate genomic and other evidence to delimitation of species or intraspecific units, and how these taxa relate to relevant legislation for conservation and protection.
Translation to Management
As discussed above, all legislation and directives recognize the species level as the fundamental unit of conservation, but also provide for the recognition of various infra-specific units. Many also have an implied requirement for the formal naming and taxonomic description of species when considered for conservation prioritization and formal protection. For example, under IUCN Red Listing Guidelines listing of unnamed species is discouraged and there is a requirement that the new species description will be published within four years of the species being included on the IUCN Red List or it would be removed. This highlights the importance of taxonomic naming and description of entities, at both species and infra-specific levels, the limited taxonomic information for many plant and animal groups (Mace, 2004; Oliver and Lee, 2010; Wilson, 2017), and the need to accelerate taxonomic effort, particularly in biodiversity rich areas (Riedel et al., 2013; Wege et al., 2015; Wilson, 2017).
Taxonomically named species and intraspecific taxonomic categories, such as subspecies and varieties, generally provides the basic units for formal listing, conservation prioritization and protection, yet in some jurisdictions other categories of intraspecific conservation units are formally recognized. These are generally identified using combined genomic and phenotypic data. For example, the USA Endangered Species Act recognizes Distinct Population Segments (DPSs) that are broadly based on ESUs (Waples, 1991), while the Canadian Species at Risk Act recognizes DUs. An operational interpretation of “distinct populations” under the Canadian Species at Risk Act, DUs aim to identify discrete sets of populations that represent important components of the evolutionary legacy of species and that are unlikely to be replaced through natural dispersion (Mee et al., 2015).
Both DUs and DPSs overlap conceptually with ESUs (Waples et al., 2013) and have been applied most extensively to north American fish, including north-west USA Salmonids (Waples, 1995) and Canadian coregonids (Mee et al., 2015) Interestingly DPSs in the USA only apply to vertebrates while in Canada DUs can be applied to any organism that can be listed under its Act (see Waples et al., 2013), with DUs currently listed for arthropods, molluscs, and plants. In Australia, like Canada, distinct populations can be listed for any organism recognized under the Environment Protection and Biodiversity Conservation Act 1999 (EPBC Act) although they are not a specifically named category, such as DUSs and DPSs. However, to date they have only been recognized for vertebrates. In the USA there are 127 vertebrate DPSs listed for conservation (https://ecos.fws.gov/ecp/); in Canada 70 DUs have been listed covering all organisms (http://www.registrelep-sararegistry.gc.ca/species/schedules_e.cfm?id=1); and in Australia populations of 7 vertebrate species covering mammals, fish and birds have been recognized as having specific conservation status under the EPBC Act (http://www.environment.gov.au/cgi-bin/sprat/public/publicthreatenedlist.pl). While assessments of these populations in US and Canada generally use genetic information (Waples et al., 2013), in Australia they are usually based on geographic and jurisdictional identification, although in some of these cases they have been supported by analysis of genetic differentiation for mammals (Firestone et al., 1999; Armstrong, 2006) and fish (Colgan and Paxton, 1997; Ahonen et al., 2009).
The listing of infra-specific conservation units for plants and invertebrates in Canada (DUs) but not in the USA or Australia highlights another issue not specifically related to the delimitation of conservation units but more broadly to significant inconsistencies in biodiversity conservation prioritization; iconic vertebrates such as birds, mammals, and fish are a focus in many jurisdictions while other organisms are not. This preferential listing of populations of certain organisms reflects a range of issues that are less to do with the science of delimiting conservation units and more to do with the listing of populations based on public awareness, political expediency and the charismatic nature of the organism to be listed (see Waples et al., 2013).
While in Australia some populations of well-known mammals, birds and fish are protected under the EPBC Act, there is now a significant body of information based on molecular studies in reptiles, invertebrates and plants where populations can be recognized as ESUs or MUs, and many of these are of conservation significance given their narrow geographic range, small population size, rate of decline, and susceptibility to threatening processes. For example, striking examples of highly structured populations and deeply divergent lineages within species that would readily fit the requirement of an ESU can be found in a range of low dispersal vertebrates from the tropics (Figure 2) and elsewhere. Similarly in Australian invertebrates, such as amphipods and millipedes, significant genetic structure, and phylogeographic patterns support the delimitation of discrete population clusters as ESUs that are geographically disparate, with narrow geographic ranges, restricted to specific habitats and represented by only a few populations (Finston et al., 2007; Murphy et al., 2013; Nistelberger et al., 2014a). Often, these short-range endemic are taxa of conservation concern (Harvey et al., 2011) and are co-located in endemism hotspots (e.g., Oliver et al., 2017), which are themselves high priorities for multi-taxon conservation attention (Rosauer et al., 2018), such as the Kimberley Region, and the Stirling Range in south west Western Australia.
In Australian plants there are also a significant number of population genetic and phylogeographic studies identifying distinct population groups within a range of species from different genera reflecting disjunct and historically isolated population systems, geological and edaphic complexities, and occupying contrasting habitats in terms of vegetation and climate (see Broadhurst et al., 2017). Genetically distinct populations are particularly evident in species from south west Western Australia (Coates, 2000; Byrne et al., 2014) with a number of studies highlighting significant genetic structure and highly divergent lineages in a range of plant genera and families including: Acacia (Mimosaceae; Coates, 1988), Atriplex (Amaranthaceae; Shepherd et al., 2015), Banksia (Proteaceae; Coates et al., 2015), Lambertia (Proteaceae; Byrne et al., 2002), Hakea (Proteaceae; Sampson et al., 2014), Eremophila (Scrophulariaceae; Llorens et al., 2015), Pultanaea (Fabaceae; Millar and Byrne, 2013), and Calothamnus (Myrtaceae; Nistelberger et al., 2014b). In many instances these populations are recognized in conservation planning by the State government agencies and given consideration in environmental impact assessments (see Shepherd et al., 2015) even though they are not listed for protection under legislation. However, we acknowledge that formal recognition of these units would improve their conservation in the long term.
These Australian examples highlight the increasing need to recognize this layer of genetic diversity below the species level. The challenge is to ensure that highly structured populations and deeply divergent lineages that are of conservation concern are protected in the face of competing demands for conservation attention. Beyond iconic species, this challenge is also evident globally with the exception of Canada. Failure to protect clearly delineated conservation units will likely increase the risk of cryptic extinction and loss of significant genetic diversity, which in many cases may involve lineages and historically isolated populations that have persisted within species for millions of years (Moritz, 2002).
Conclusions and Recommendations
The listing of species and various infra-specific categories under threatened species legislation or under international organizations such as IUCN, indicates a commitment to protecting genetic diversity across the species population continuum. Yet current approaches to biodiversity conservation largely focus on the species end of this continuum with less attention given to genetic diversity and infra-specific variation. This more limited focus on intra-species level genetic variation suggests that while conservation managers increasingly recognize the value of explicitly considering genetic information in management actions and prioritization, they often lack actionable information from biodiversity scientists (Waples et al., 2008; Laikre et al., 2010; Cook and Sgrò, 2017; Taylor et al., 2017). This gap between science and conservation practice impedes a more effective global approach to conserving and managing genetic diversity.
We suggest that the lack of clarity from biodiversity scientists over units of conservation is largely based on inconsistencies at two levels. The first concerns the multiple species concepts and the various diagnostic methods used to delimit species that creates inconsistency across taxonomic groups. The second relates to a lack of consistency and agreement by taxonomists, evolutionary biologists, and conservation biologists on how to treat infra-specific taxa and other entities such as ESUs, DUs, and MUs.
While we consider that legislation is often already sufficiently flexible in terms of how taxa (or “species”) are defined, we argue that there is a need to systematically address the population species continuum, and more clearly define units for conservation with greater recognition given to concepts such as ESU's and how these relate to relevant legislation. We emphasize that this issue will no doubt become more critical given the power of genomics to detect differentiation at progressively finer scales and its increasing use to delimit species and infra-specific entities.
From the preceding analysis, we offer the following recommendations:
(i) Genomic data should be interpreted conservatively when delimiting species, using an integrative taxonomic approach that typically includes evidence beyond the genetic data alone.
(ii) Where geneticists identify multiple lineages or distinct populations within a species, and these do not warrant elevation to species level and are not taxonomically recognized, that there be stronger impetus and a more uniform approach to identifying clearly justified intraspecific units for practical assessment of conservation status or impact assessments, and that these be explicitly recognized as units for conservation in the context of legislation relevant to that jurisdiction.
(iii) Consideration be given by the IUCN, through its Conservation Genetics Specialist Group and its regional sub-groups, to developing guidelines for recognizing and labeling intraspecific units that align with best scientific practice.
(iv) The more nuanced view emerging from evolutionary genomics of species and the continuum from populations to species should be communicated more effectively by scientists to conservation managers and policy makers.
More broadly we echo recommendations made recently in a number reviews (Frankham, 2010; Santamaría and Méndez, 2012; Hoban et al., 2013; Moritz and Potter, 2013; Laity et al., 2015; Pierson et al., 2016; Cook and Sgrò, 2017; Taylor et al., 2017) that highlight the need for scientists to improve communication and raise awareness with policy makers, government agencies, non-government organizations, and practitioners that outcomes from molecular systematics, molecular taxonomy, phylogeography, and population genetic research can provide critical information about conserving genetic diversity and managing evolutionary processes.
Author Contributions
DC, MB, and CM all contributed to the preparation and revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was supported by the Australian Research Council (ARC Linkage Grant No. LP120200063), the Australian National University and the Western Australian Department of Biodiversity, Conservation, and Attractions.
References
Afonso Silva, A. C., Bragg, J. G., Potter, S., Fernandes, C., Coelho, M. M., and Moritz, C. (2017a). Tropical specialist versus climate generalist: diversification and demographic history of sister species of Carlia skinks from northwestern Australia. Mol. Ecol. 26:4045–4058. doi: 10.1111/mec.14185
Afonso Silva, A. C., Santos, N., Ogilvie, H. A., and Moritz, C. (2017b). Validation and description of two new north-western Australian Rainbow skinks with multispecies coalescent methods and morphology. PeerJ 5:e3724. doi: 10.7717/peerj.3724
Ahonen, H., Harcourt, R. G., and Stow, A. J. (2009). Nuclear and mitochondrial DNA reveals isolation of imperilled grey nurse shark populations (Carcharias taurus). Mol. Ecol. 18, 4409–4421. doi: 10.1111/j.1365-294X.2009.04377.x
Armstrong, K. N. (2006). Phylogeographic structure in Rhinonicteris aurantia (Chiroptera: Hipposideridae): implications for conservation. Acta Chiropterologica 8, 63–81. doi: 10.3161/1733-5329(2006)8[63:PSIRAC]2.0.CO
Arnold, M. L., and Kunte, K. (2017). Adaptive genetic exchange: a tangled history of admixture and evolutionary innovation. Trends Ecol. Evol. 32, 601–611. doi: 10.1016/j.tree.2017.05.007
Avise, J. C. (2004). Molecular Markers, Natural History, and Evolution. 2nd Edn. Sunderland, MA; Oxford: Oxford University Press.
Avise, J. C., Arnold, J., Ball, R. M., Bermingham, E., Lamb, T., Neigel, J. E., et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Ann. Rev. Ecol. Sys. 18, 489–522. doi: 10.1146/annurev.es.18.110187.002421
Bickford, D., Lohman, D. J., Sodhi, N. S, Ng, P. K., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. doi: 10.1016/j.tree.2006.11.004
Blom, M. P. K., Bragg, J. G., Potter, S., and Moritz, C. (2016). Accounting for uncertainty in gene tree estimation: summary-coalescent species tree inference in a challenging radiation of Australian lizards. Syst. Biol. 66, 352–366. doi: 10.1093/sysbio/syw089
Broadhurst, L., Breed, M., Lowe, A., Bragg, J., Catullo, R., Coates, D. J., et al. (2017). Genetic diversity and structure of the Australian flora. Divers. Distrib. 23, 41–52. doi: 10.1111/ddi.12505
Byrne, M., Coates, D. J., Forest, F., Hopper, S. D., Krauss, S. L., Sniderman, J. M. K., et al. (2014). “A diverse flora - species and genetic relationships,” in Plant Life on the Sandplains in Southwest Australia, a Global Biodiversity Hotspot, editors H. Lambers (Crawley, WA: University of Western Australia Publishing), 81–99.
Byrne, M., Macdonald, B., and Coates, D. J. (2002). Phylogeographic patterns and high cpDNA diversity within the Acacia acuminata (Leguminosae:Mimosoideae). complex in Western Australia. J. Evol. Biol. 15, 576–587. doi: 10.1046/j.1420-9101.2002.00429.x
Carstens, B. C., Pelletier, T. A., Reid, N. M., and Satler, J. D. (2013). How to fail at species delimitation. Mol. Ecol. 22, 4369–4383. doi: 10.1111/mec.12413
Coates, D. J. (1988). Genetic diversity and population gene structure in the rare Chittering grass wattle, Acacia anomala Court. Aust. J. Bot. 36:273–286. doi: 10.1071/BT9880273
Coates, D. J. (2000). Defining conservation units in a rich and fragmented flora: implications for management of genetic resources and evolutionary processes in south-west Australia plants. Aust. J. Bot. 48, 329–339. doi: 10.1071/BT99018
Coates, D. J., McArthur, S. L., and Byrne, M. (2015). Significant genetic diversity loss following pathogen driven population extinction in the rare endemic Banksia brownii (Proteaceae). Biol. Conserv. 192, 353–360. doi: 10.1016/j.biocon.2015.10.013
Colgan, D. J., and Paxton, J. R. (1997). Biochemical genetics and recognition of a western stock of the common gemfish, Rexea solandri (Scombroidea: Gempylidae)., in Australia. Mar. Freshw. Res. 48, 103–118. doi: 10.1071/MF96048
Convention on Biological Diversity (2007). Available online at: https://www.cbd.int/doc/meetings/cop-bureau/cop-bur-2007/cop-bur-2007-10-14-en.pdf
Cook, C. N., and Sgrò, C. M. (2017). Aligning science and policy to achieve evolutionarily enlightened conservation. Conserv. Biol. 31, 501–512. doi: 10.1111/cobi.12863
Cook, L. G., Edwards, R. D., Crisp, M. D., and Hardy, N. B. (2010). Need morphology always be required for new species descriptions? Invert. Sys. 24, 322–332. doi: 10.1071/IS10011
Cracraft, J. (1983). Species concepts and speciation analysis. Curr. Ornithol. 1, 159–187. doi: 10.1007/978-1-4615-6781-3_6
Crandall, K. A., Bininda-Emonds, O. R., Mace, G. M., and Wayne, R. K. (2000). Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 15, 290–295. doi: 10.1016/S0169-5347(00)01876-0
Cutter, A. D., and Payseur, B. A. (2013). Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14, 262–274. doi: 10.1038/nrg3425
Dayrat, B. (2005). Towards integrative taxonomy. Biol. J. Linn. Soc. 85, 407–415. doi: 10.1111/j.1095-8312.2005.00503.x5
de Queiroz, K. (1998). “The general lineage concept of species, species criteria, and the process of speciation,” in Forms: Species and Speciation, D. J. Howard,r S. H. Berlocher (New York, NY: Oxford University Press), 57–75.
de Queiroz, K. (2007). Species concepts and species delimitation. Syst. Biol. 56, 879–886. doi: 10.1080/10635150701701083
Degnan, J. H., and Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340. doi: 10.1016/j.tree.2009.01.009
Devey, D. S., Bateman, R. M., Fay, M. F., and Hawkins, J. A. (2008). Friends or relatives? Phylogenetics and species delimitation in the controversial European orchid genus Ophrys. Ann. Bot. 101, 385–402. doi: 10.1093/aob/mcm299
Dizon, A. E., Lockyer, C, Perrin, W. F., Demaster, D. P., and Sisson, J. (1992). Rethinking the stock concept – a phylogeographic approach. Conserv. Biol. 6, 24–36. doi: 10.1046/j.1523-1739.1992.610024.x
Doughty, P., Bourke, G., Tedeschi, L., Pratt, R., Oliver, P., Palmer, R., et al. (2018). Species delimitation in the Gehyra nana (Squamata: Gekkonidae). complex: cryptic and divergent morphological evolution in the Australian Monsoonal Tropics, with the description of four new species. Zootaxa, 4403:201–244. doi: 10.11646/zootaxa.4403.2.1
Dynesius, M., and Jansson, R. (2014). Persistence of Within-species lineages: a neglected control of speciation rates. Evolution 68, 923–934. doi: 10.1111/evo.12316
Edwards, S. V., Potter, S., Schmitt, C. J., Bragg, J. G., and Moritz, C. (2016). Reticulation, divergence, and the phylogeography–phylogenetics continuumc. Proc. Natl. Acad. Sci. U.S.A. 113, 8025–8032. doi: 10.1073/pnas.1601066113
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Cons. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Finston, T. L., Johnson, M. S., Humphreys, W. F., Eberhard, S. M., and Halse, S. A. (2007). Cryptic speciation in two widespread subterranean amphipod genera reflects historical drainage patterns in an ancient landscape. Mol. Ecol. 16, 355–365. doi: 10.1111/j.1365-294X.2006.03123.x
Firestone, K. B., Elphinstone, M. S., Sherwin, W. B., and Houlden, B. A. (1999). Phylogeographical population structure of tiger quolls Dasyurus maculatus (Dasyuridae: Marsupalia), an endangered carnivorous marsupial. Mol. Ecol. 8, 1613–1625. doi: 10.1046/j.1365-294x.1999.00745.x
Fišer, C., Robinson, C. T., and Malard, F. (2018). Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 27, 613–635. doi: 10.1111/mec.14486
Frankham, R. (2010). Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143,1919–1927. doi: 10.1016/j.biocon.2010.05.011
Frankham, R., Ballou, J. D., Dudash, M. R., Eldridge, M. D., Fenster, C. B., Lacy, R. C., et al. (2012). Implications of different species concepts for conserving biodiversity. Biol. Conserv. 153, 25–31. doi: 10.1016/j.biocon.2012.04.034
Fraser, D. J., and Bernatchez, L. (2001). Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol. Ecol. 10, 2741–2752. doi: 10.1046/j.1365-294X.2001.t01-1-01411.x
Funk, W. C., Mckay, J. K., Hohenlohe, P. A., and Allendorf, F. W. (2012). Harnessing genomics for delineating conservation units. Trends Ecol. Evol. 27, 489–496. doi: 10.1016/j.tree.2012.05.012
Garnett, S. T., and Christidis, L. (2017). Taxonomy anarchy hampers conservation. Nature 546, 25–27. doi: 10.1038/546025a
Gippoliti, S., Cotterill, F. P. D., Zinner, D., and Groves, C. P. (2018). Impacts of taxonomic inertia for the conservation of African ungulate diversity: an overview. Biol. Rev. 93, 115–130. doi: 10.1111/brv.12335
Hamilton, C. W., and Reichard, S. H. (1992). Current practice in the use of subspecies, varieties and forma in the classification of wild plants. Taxon 41, 485–498. doi: 10.2307/1222819
Harvey, M. S., Rix, M. G., Framenau, V. W., Hamilton, Z. R., Johnson, M. S., Teale, R. J., et al. (2011). Protecting the innocent: studying short-range endemic taxa enhances conservation outcomes. Invert. Syst. 25, 1–10. doi: 10.1071/IS11011
Hey, J., Waples, R. S., Arnold, M. L., Butlin, R. K., and Harrison, R. G. (2003). Understanding and confronting species uncertainty in biology and conservation. Trends Ecol. Evol. 18, 597–603. doi: 10.1016/j.tree.2003.08.014
Hoban, S. M., Hauffe, H. C., Perez-Espona, S., Arntzen, J. W., Bertorelle, G., Bryja, J., et al. (2013). Bringing genetic diversity to the forefront of conservation policy and management. Conserv. Genet. Resour. 5, 593–598. doi: 10.1007/s12686-013-9859-y
Isaac, N. J., Mallet, J., and Mace, G. M. (2004). Taxonomic inflation: its influence on macroecology and conservation. Trends Ecol. Evol. 19, 464–469. doi: 10.1016/j.tree.2004.06.004
Isaac, N. J., Turvey, S. T., Collen, B., Waterman, C., and Baillie, J. E. (2007). Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2:e296. doi: 10.1371/journal.pone.0000296
Jones, M. R., and Good, J. M. (2016). Targeted capture in evolutionary and ecological genomics. Mol. Ecol. 25, 185–202. doi: 10.1111/mec.13304
Jorger, K. M., and Schrödl, M. (2013). How to describe a cryptic species? Practical challenges of molecular taxonomy. Front. Zool. 10:59. doi: 10.1186/1742-9994-10-59
Laikre, L., Allendorf, F., Aroner, L. C., Baker, S., Gregovich, D. P., Hansen, M. M., et al. (2010). Neglect of genetic diversity in implementation of the convention on biological diversity. Conserv. Biol. 24, 86–88. doi: 10.1111/j.1523-1739.2009.01425.x
Laikre, L., Palm, S., and Ryman, N. (2005). Genetic population structure of fishes: implications for coastal zone management. Ambio 34,111–119. doi: 10.1579/0044-7447-34.2.111
Laity, T., Laffan, S. W., González-Orozco, C. E., Faith, D. P., Rosauer, D. F., Byrne, M., et al. (2015). Phylodiversity to inform conservation policy: An Australian example. Sci. Total Environ. 534, 131–143. doi: 10.1016/j.scitotenv.2015.04.113
Leache, A. D., and Oaks, J. R. (2017). The utility of single nucleotide Polymorphism (SNP). data in phylogenetics. Ann. Rev. Ecol. Syst. 48, 69–84. doi: 10.1146/annurev-ecolsys-110316-022645
Llorens, T. M., Macdonald, B., McArthur, S., Coates, D. J., and Byrne, M. (2015). Disjunct, highly divergent genetic lineages within two rare Eremophila (Scrophulariaceae: Myoporeae). species in a biodiversity hotspot: implications for taxonomy and conservation. Biol. J. Linn. Soc. 177, 96–111. doi: 10.1111/boj.12228
Love Stowell, S. M., Pinzone, C. A., and Martin, A. P. (2017). Overcoming barriers to active interventions for genetic diversity. Biodivers. Conserv. 26, 1753–1765. doi: 10.1007/s10531-017-1330-z
Mace, G. M. (2004). The role of taxonomy in species conservation. Philos. Trans. R. Soc. B: Biol. Sci. 359, 711–719. doi: 10.1098/rstb.2003.1454
Mee, J. A., Bernatchez, L., Reist, J. D., Rogers, S. M., and Taylor, E. B. (2015). Identifying designatable units for intraspecific conservation prioritization: a hierarchical approach applied to the lake whitefish species complex (Coregonus spp.). Evol. Appl. 8, 423–441. doi: 10.1111/eva.12247
Millar, M. A., and Byrne, M. (2013). Cryptic divergent lineages of Pultenaea pauciflora M.B. Scott (Fabaceae: Mirbelieae) exhibit different evolutionary history. Biol. J. Linn. Soc. 108, 871–881. doi: 10.1111/bij.12002
Moritz, C. (1994). Defining evolutionarily significant units for conservation. Trends Ecol. Evol. 9, 373–375. doi: 10.1016/0169-5347(94)90057-4
Moritz, C. (1999). Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas 130, 217–228. doi: 10.1111/j.1601-5223.1999.00217.x
Moritz, C. (2002). Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 51, 238–254. doi: 10.1080/10635150252899752
Moritz, C., and Potter, S. (2013). The importance of an evolutionary perspective in conservation policy planning. Mol. Ecol. 22, 5969–5971. doi: 10.1111/mec.12565
Moritz, C., Pratt, R. C., Bank, S., Bourke, G., Bragg, J. G., Doughty, P., et al. (2018). Cryptic lineage diversity, body size divergence, and sympatry in a species complex of Australian lizards (Gehyra). Evolution 27, 54–66. doi: 10.1111/evo.13380
Morrison, W. R., Lohr, J. L., Duchen, P., Wilches, R., Trujillo, D., Mair, M., et al. (2009). The impact of taxonomic change on conservation: Does it kill, can it save, or is it just irrelevant? Biol. Cons. 142, 3201–3206. doi: 10.1016/j.biocon.2009.07.019
Murphy, N. P., Adams, M., Guzik, M. T., and Austin, A. D. (2013). Extraordinary micro-endemism in Australian desert spring amphipods. Mol. Phylogenet. Evol. 66, 645–653. doi: 10.1016/j.ympev.2012.10.013
Murphy, N. P., King, R. A., and Delean, S. (2015). Species, ESUs or populations? Delimiting and describing morphologically cryptic diversity in Australian desert spring amphipods. Invert. Syst. 29, 457–467. doi: 10.1071/IS14036
Nater, A., Mattle-Greminger, M. P., Nurcahyo, A., Nowak, M. G., de Manual, M., Desai, T., et al. (2017). Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr. Cell Biol. 27, 3487–3498. doi: 10.1016/j.cub.2017.09.047
Nicotra, A. B., Chong, C., Bragg, J. G., Ong, C. R., Aitken, N. C., Chuah, A., et al. (2016). Population and phylogenomic decomposition via genotyping-by-sequencing in Australian Pelargonium. Mol. Ecol. 25, 2000–2014. doi: 10.1111/mec.13584
Nistelberger, H., Byrne, M., Coates, D., and Roberts, J. D. (2014a). Strong phylogeographic structure in a millipede indicates Pleistocene vicariance between populations on banded iron formations in semi-arid Australia. PLoS ONE 9:e0093038. doi: 10.1371/journal.pone.0093038
Nistelberger, H., Gibson, N., Macdonald, B., Tapper, S.-L., and Byrne, M. (2014b). Phylogeographic evidence for two mesic refugia in a biodiversity hotspot. Heredity 113, 454–463. doi: 10.1038/hdy.2014.46
Ogilvie, H. A., Heled, J., Xie, D., and Drummond, A. J. (2016). Computational performance and statistical accuracy of beast and comparisons with other methods. Syst. Biol. 65, 381–396. doi: 10.1093/sysbio/syv118
Oliver, P. M., Keogh, J. S., and Moritx, C. (2015). New approaches to cataloguing and understanding evolutionary diversity: a perspective from Australian herpetology. Aust. J. Zool. 62, 417–430. doi: 10.1071/ZO14091
Oliver, P. M., Laver, R. J., Demello Martins, F., Pratt, R. C., Hunjan, S., and Moritz, C. (2017). A novel hotspot of vertebrate endemism and an evolutionary refugium in tropical Australia. Diver. Distrib. 23, 53–66. doi: 10.1111/ddi.12506
Oliver, P. M., and Lee, M. S. Y. (2010). The botanical and zoological codes impede biodiversity research by discouraging publication of unnamed new species. Taxon 59, 1201–1205. doi: 10.2307/20773990
Padial, J., Miralles, A., De la Riva, I., and Vences, M. (2010). The integrative future of taxonomy. Front. Zool. 7:16. doi: 10.1186/1742-9994-7-16
Palsbøll, P. J., Bérubé, M., and Allendorf, F. W. (2007). Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16. doi: 10.1016/j.tree.2006.09.003
Patton, J. L., and Smith, M. F. (1994). Paraphyly, polyphyly, and the nature of species boundaries in Pocket Gophers (Genus Thomomys). Syst. Biol. 43, 11–26. doi: 10.1093/sysbio/43.1.11
Pierson, J. C., Coates, D. J., Oostermeijer, J. G. B., Beissinger, S. R., Bragg, J. G., Sunnucks, P., et al. (2016). Genetic factors in threatened species recovery plans on three continents. Front. Ecol. Environ. 14, 433–440. doi: 10.1002/fee.1323
Potter, S., Bragg, J. G., Peter, B. M., Bi, K., and Moritz, C. (2016). Phylogenomics at the tips: inferring lineages and their demographic history in a tropical lizard, Carlia amax. Mol. Ecol. 25, 1367–1380. doi: 10.1111/mec.13546
Potter, S., Xue, A. T., Bragg, J. G., Rosauer, D. F., Roycroft, E. J., and Moritz, C. (2018). Pleistocene climatic changes drive diversification across a tropical savanna. Mol. Ecol. 27, 520–532. doi: 10.1111/mec.14441/full
Rabosky, D. L., Hutchinson, M. N., Donnellan, S. C., Talaba, A. L., and Lovette, I. J. (2014). Phylogenetic disassembly of species boundaries in a widespread group of Australian skinks (Scincidae: Ctenotus). Mol. Phylogenet. Evol. 77, 71–82. doi: 10.1016/j.ympev.2014.03.026
Ralls, K., Ballou, J. D., Dudash, M. R., Eldridge, M. D. B., Fenster, C. B., Lacy, R. C., et al. (2018). Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 11, 1–6. doi: 10.1111/conl.124126
Rannala, B. (2015). The art and science of species delimitation. Curr. Zool. 61, 846–853. doi: 10.1093/czoolo/61.5.846
Riedel, A., Sagata, K., Suhardjono, Y. R., Tänzler, R., and Balke, M. (2013). Integrative taxonomy on the fast track – towards more sustainability in biodiversity research. Front. Zool. 10:15. doi: 10.1186/1742-9994-10-15
Rieseberg, L. H., and Willis, J. H. (2007). Plant speciation. Science 317, 910–914. doi: 10.1126/science.1137729
Rosauer, D. F., Blom, M. P. K., Bourke, G., Catalano, S., Donnellan, S., Mulder, E., et al. (2016). Phylogeography, hotspots and conservation priorities: an example from the Top End of Australia. Biol. Cons. 204, 83–93. doi: 10.1016/j.biocon.2016.002
Rosauer, D. F., Byrne, M., Blom, M. P. K., Coates, D. J., Donnellan, S., Doughty, P., et al. (2018). Real-world conservation planning for evolutionary diversity in the Kimberley, Australia, sidesteps uncertain taxonomy Conserv. Lett. 11:e12438. doi: 10.1111/conl.12438
Rosenblum, E. B., Sarver, B. A. J., Brown, J. W., Des Roches, S., Hardwick, K. M., Hether, T. D., et al. (2012). Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol. 39, 255–261. doi: 10.1007/s11692-012-9171-x
Roux, C., Fraïsse, C., Romiguier, J., Anciaux, Y., Galtier, N., and Bierne, N. (2016). Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 14:e2000234. doi: 10.1371/journal.pbio.2000234
Ryder, O. A. (1986). Species conservation and systematics – the dilemma of subspecies. Trends Ecol. Evol. 1, 9–10. doi: 10.1016/0169-5347(86)90059-5
Sampson, J. F., Byrne, M., Yates, C. J., Gibson, N., Thavornkanlapachai, R., Stankowski, S., et al. (2014). Contemporary pollen-mediated gene immigration reflects the historical isolation of a rare shrub in a fragmented landscape. Heredity 112, 172–181. doi: 10.1038/hdy.2013.89
Santamaría, L., and Méndez, P. F. (2012). Evolution in biodiversity policy: current gaps and future needs. Evol. Appl. 5, 202–218. doi: 10.1111/j.1752-4571.2011.00229.x
Schlick-Steiner, B. C., Steiner, F. M., Seifert, B., Stauffer, C., Christian, E., and Crozier, R. H. (2010). Integrative taxonomy: a multisource approach to exploring biodiversity. Annu. Rev. Entomol. 55, 421–438. doi: 10.1146/annurev-ento-112408-085432
Seehausen, O, Takimoto, G., Roy, D., and Jokela, J. (2008). Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol. 17, 30–44. doi: 10.1111/j.1365-294X.2007.03529.x
Shepherd, K. A., Thiele, K. R., Sampson, J., Coates, D. J., and Byrne, M. (2015). A rare, new species of Atriplex (Chenopodiaceae) comprising two genetically distinct but morphologically cryptic populations in arid Western Australia: implications for taxonomy and conservation. Aust. Syst. Bot. 28, 234–245. doi: 10.1071/SB15029
Singhal, S., and Bi, K. (2017). History cleans up messes: The impact of time in driving divergence and introgression in a tropical suture zone. Evolution 71, 1888–1899. doi: 10.1111/evo.13278
Singhal, S., Hoskin, C., Couper, P., Potter, S., and Moritz, C. (2018). Framework for resolving cryptic species: a case study from the lizards of the Australian wet tropics. Syst. Biol. [Epub ahead of print]. doi: 10.1093/sysbio/syy
Singhal, S., and Moritz, C. (2013). Reproductive isolation between phylogeographic lineages scales with divergence. Proc. Roy. Soc B 280:20132246. doi: 10.1098/rspb.2013.2246026
Steane, D. A., Potts, B. M., McLean, E., Collins, L., Prober, S. M., Stock, W. D., et al. (2015). Genome-wide scans reveal cryptic population structure in a dry-adapted eucalypt. Tree Genet. Genomes 11:33. doi: 10.1007/s11295-015-0864-z
Struck, T. H., Feder, J. L., Bendiksby, M., Birkeland, S., Cerca, J., Gusarov, V. I., et al. (2017). Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 33, 153–163. doi: 10.1016/j.tree.2017.11.007
Sukumaran, J., and Knowles, L. L. (2017). The multispecies coalescent delimits structure not species. Proc. Nat. Aca. Sci. U.S.A. 114, 1607–1612. doi: 10.1073/pnas.1607921114
Swarts, N. D., Clements, M. A., Bower, C. C., and Miller, J. T. (2014). Defining conservation units in a complex of morphologically similar, sexually deceptive, highly endangered orchids. Biol. Cons. 174, 55–64. doi: 10.1016/j.biocon.2014.03.017
Tautz, D., Arctander, P., Minelli, A., Thomas, R. H., and Vogler, A. P. (2003). A plea for DNA taxonomy. Trends Ecol. Evol. 18, 70–74. doi: 10.1016/S0169-5347(02)00041-1
Taylor, H. R., Dussex, N., and van Heezik, Y. (2017). Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Glob. Ecol. Conserv. 10, 231–242. doi: 10.1016/j.gecco.2017.04.001
Thomson, S. A., Pyle, R. L., Ahyong, S. T., Alonso-Zarazaga, M., Ammirati, J., Araya, J. F., et al. (2018). Taxonomy based on science is necessary for global conservation. PLoS Biol. 16:e2005075. doi: 10.1371/journal.pbio.2005075
Vieites, D. R., Wollenberg, K. C., Andreonec, F., Köhler, J., Glaw, F., and Vences, M. (2009). Vast underestimation of Madagascar's biodiversity evidenced by an integrative amphibian inventory. Proc. Natl. Acad. Sci. U.S.A. 106, 8267–8272. doi: 10.1073/pnas.0810821106
Vogler, A. P., and de Salle, R. (1994). Diagnosing Units of Conservation Management. Cons. Biol. 8, 354–363. doi: 10.1046/j.1523-1739.1994.08020354.x
Waples, R. S. (1991). Pacific salmon, Oncorhynchus spp., and the definition of “species” under the Endangered Species Act. Mar. Fish. Rev. 53, 11–22.
Waples, R. S. (1995). “Evolutionarily significant units and the conservation of biological diversity under the Endangered Species Act,” in Evolution and the Aquatic Ecosystem: Defining Unique Units in Population Conservation, eds J. L. Nielsen (Bethesda, MD:American Fisheries Society), 8–27.
Waples, R. S., Gustafson, R. G., Weitkamp, L. A., Myers, J. M., Jjonson, O. W., Busby, P. J., et al. (2001). Characterizing diversity in salmon from the Pacific Northwest. J. Fish Biol. 59, 1–41. doi: 10.1111/j.1095-8649.2001.tb01376.x
Waples, R. S., Nammack, M., Cochrane, J. F., and Hutchings, J. A. (2013). A tale of two acts: endangered species listing practices in Canada and the United States. BioScience 63, 723–734. doi: 10.1093/bioscience/63.9.723
Waples, R. S., Punt, A. E., and Cope, J. M. (2008). Integrating genetic data into management of marine resources: how can we do it better? Fish 9, 423–449. doi: 10.1111/j.1467-2979.2008.00303.x
Wege, J. A., Thiele, K. R., Shepherd, K. A., Butcher, R., Macfarlane, T. D., and Coates, D. J. (2015). Strategic taxonomy in a biodiverse landscape: a novel approach to maximizing conservation outcomes for rare and poorly known flora. Biodivers. Conserv. 24, 17–32. doi: 10.1007/s10531-014-0785-4
Whitlock, R. (2014). Relationships between adaptive and neutral genetic diversity and ecological structure and functioning: a meta-analysis. J. Ecol. 102, 857–872. doi: 10.1111/1365-2745.12240
Wiley, E. O. (1978). The evolutionary species concept reconsidered. Syst. Biol. 27, 17–26. doi: 10.2307/2412809
Wilson, E. O. (2017). Biodiversity research requires more boots on the ground. Nat. Ecol. Evol. 1, 1590–1591. doi: 10.1038/s41559-017-0360-y
Yang, Z. H., and Rannala, B. (2014). Unguided species delimitation using DNA sequence data from multiple loci. Mol. Biol. Evol. 31, 3125–3135. doi: 10.1093/molbev/msu279
Keywords: conservation units, genetic diversity, genomics, speciation, legislation, infra-specific variation
Citation: Coates DJ, Byrne M and Moritz C (2018) Genetic Diversity and Conservation Units: Dealing With the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 6:165. doi: 10.3389/fevo.2018.00165
Received: 02 August 2018; Accepted: 01 October 2018;
Published: 23 October 2018.
Edited by:
Umesh Srinivasan, Princeton University, United StatesReviewed by:
William Ernest Magnusson, National Institute of Amazonian Research, BrazilSpartaco Gippoliti, Storia della Fauna, Italy
Copyright © 2018 Coates, Byrne and Moritz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David J. Coates, RGF2ZS5Db2F0ZXNAZGJjYS53YS5nb3YuYXU=
 David J. Coates
David J. Coates Margaret Byrne
Margaret Byrne Craig Moritz2,3
Craig Moritz2,3