
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Ecol. Evol. , 20 August 2018
Sec. Evolutionary and Population Genetics
Volume 6 - 2018 | https://doi.org/10.3389/fevo.2018.00117
This article is part of the Research Topic Polyploid Population Genetics and Evolution - From Theory to Practice View all 13 articles
The duplication of an entire genome is no small affair. Whole genome duplication (WGD) is a dramatic mutation with long-lasting effects, yet it occurs repeatedly in all eukaryotic kingdoms. Plants are particularly rich in documented WGDs, with recent and ancient polyploidization events in all major extant lineages. However, challenges immediately following WGD, such as the maintenance of stable chromosome segregation or detrimental ecological interactions with diploid progenitors, commonly do not permit establishment of nascent polyploids. Despite these immediate issues some lineages nevertheless persist and thrive. In fact, ecological modeling commonly supports patterns of adaptive niche differentiation in polyploids, with young polyploids often invading new niches and leaving their diploid progenitors behind. In line with these observations of polyploid evolutionary success, recent work documents instant physiological consequences of WGD associated with increased dehydration stress tolerance in first-generation autotetraploids. Furthermore, population genetic theory predicts both short- and long-term benefits of polyploidy and new empirical data suggests that established polyploids may act as “sponges” accumulating adaptive allelic diversity. In addition to their increased genetic variability, introgression with other tetraploid lineages, diploid progenitors, or even other species, further increases the available pool of genetic variants to polyploids. Despite this, the evolutionary advantages of polyploidy are still questioned, and the debate over the idea of polyploidy as an evolutionary dead-end carries on. Here we broadly synthesize the newest empirical data moving this debate forward. Altogether, evidence suggests that if early barriers are overcome, WGD can offer instantaneous fitness advantages opening the way to a transformed fitness landscape by sampling a higher diversity of alleles, including some already preadapted to their local environment. This occurs in the context of intragenomic, population genomic, and physiological modifications that can, on occasion, offer an evolutionary edge. Yet in the long run, early advantages can turn into long-term hindrances, and without ecological drivers such as novel ecological niche availability or agricultural propagation, a restabilization of the genome via diploidization will begin the cycle anew.
Whole genome duplication (WGD) is a pervasive event in the evolution of eukaryotes, with an especially strong representation throughout the plant kingdom. Despite this prevalence, however, WGD is no easy victory: the presence of an extra set of chromosomes creates numerous biological challenges ranging from chromosome mis-segregation to altered gene expression, changes in cell size or in intracellular physiology (Ramsey and Schemske, 2002; Osborn et al., 2003; Adams and Wendel, 2005; Comai, 2005; Chen and Ni, 2006; Chen, 2007; Otto, 2007; Parisod et al., 2010; Brownfield and Köhler, 2011; Hollister, 2015). Numerous studies have shown evidence of dysfunction in newly formed polyploids across kingdoms: in plants, fungi and animals, including notably cancer cells (Ramsey and Schemske, 2002; Storchova and Pellman, 2004; Comai, 2005; Yant and Bomblies, 2017). Even more, polyploid establishment is substantially constrained by overall low chances of polyploid mutants persisting among their diploid progenitors (Levin, 2002; Ramsey and Schemske, 2002). This stems from both direct competition between the two cytotypes (Yamauchi et al., 2004) and frequency dependent selection (Levin, 1975), which suggests most autopolyploids are likely to go extinct before establishment (Levin, 1975; Husband, 2000).
Despite these challenges, WGD events have occurred repeatedly throughout the evolution of eukaryotes (Gregory and Mable, 2005; Wood et al., 2009; Wendel, 2015), leading to an abundance of established polyploid species in the wild. There is clear evidence of WGD in the ancestry of most plant lineages (Vision et al., 2000; Bowers et al., 2003; Paterson et al., 2004; Schlueter et al., 2004; Pfeil et al., 2005; Barker et al., 2008; Burleigh et al., 2008; Shi et al., 2010). Among angiosperms, it is estimated that 30–70% have undergone additional WGD events (Stebbins, 1938; Grant, 1963; Goldblatt, 1980; Masterson, 1994; Wood et al., 2009; Mayrose et al., 2011; Ruprecht et al., 2017). Polyploidy is also common among important crops (e.g., wheat, maize, sugar cane, coffee, cotton, potato, and tobacco), suggesting that WGD is often a key factor in successful crop domestication (Salman-Minkov et al., 2016). Obtaining precise estimates of the extent of polyploidy can be complicated, in part due to difficulties obtaining direct empirical evidence. For example, autopolyploids, resulting from within-species genome duplication, are often not considered a separate species from their diploid progenitors. As a result, their overall abundance compared to allopolyploids (where two distinct genomes are combined) have historically been underestimated (Soltis et al., 2007; Barker et al., 2016).
Frequency estimates of WGD increase in habitats affected by environmental disturbances (Favarger, 1984; Brochmann et al., 2004; Parisod et al., 2010). Concordant with this observation, in diploid-polyploid systems overlapping former glaciation limits, polyploids are found more frequently in the previously glaciated areas while their diploid progenitors commonly remain or retreat within former refugia (Ehrendorfer, 1980; Kadereit, 2015). For example, in Arabidopsis, both auto and allopolyploidization events were estimated to coincide with glacial maxima (Novikova et al., 2018). Beyond the fact that environmental stochasticity both increases the rate of WGD and provides new space for colonization (Baack, 2005; Fawcett et al., 2009; Oswald and Nuismer, 2011), such observations implicate WGD in speciation and adaptive radiation (Wood et al., 2009) and support the long-standing hypothesis that WGD per se can potentiate evolutionary adaptation, although evidence for this is somewhat mixed. Clear empirical evidence from in vitro evolution experiments in yeast demonstrated that tetraploids adapted faster than lower ploidies (Selmecki et al., 2015) and has bolstered this hypothesis. However, complementary approaches such as ecological niche modeling do not always support niche innovation in polyploids (Glennon et al., 2014). For example in primroses, the niches occupied by the three polyploid species (tetraploid, hexaploidy and octoploid) were distinct relative to the diploid progenitor but they were also narrower (Theodoridis et al., 2013).
Here we synthesize recent advances in polyploid research from new genomic, ecological, and cytological analyses with older observations and theoretical arguments into two primary dimensions (Figure 1): consequences (challenges vs. gains) of WGD and their time-span (short-term vs. long-term). To address specifically the effects of WGD, we focus on autopolyploids, which arise from within-species WGD events and thus carry four or more homologous copies of each chromosome (for a clear depiction, see Bomblies and Madlung, 2014). We thus largely set aside allopolyploids (polyploid hybrids), which strongly confound the effects of WGD with hybridization. On many fronts, recent results from autopolyploid systems have confirmed earlier theoretical predictions, but some have unveiled surprising new results in the context of a wide range of biological processes. Most strikingly, the population genetic consequences of WGD have been the subject of ample theoretical arguments despite thin experimental support to date, but this is changing. Our synthesis paints an overall picture of autopolyploidy as a high-risk high-gain path, where long-term complications often outweigh initial benefits while paving the way for re-diploidization. This depiction strengthens the idea of polyploidy as a transitionary state, a “hop,” which has seen growing support from the polyploid community (Escudero et al., 2014; Wendel, 2015).
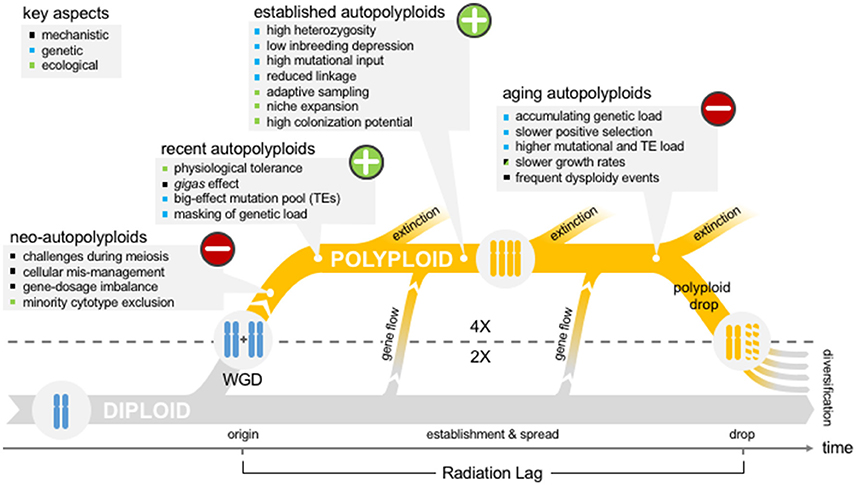
Figure 1. The polyploid hop. Schematic representation of the temporal evolution of a diploid-autopolyploid system accompanied with the shifting benefits and pitfalls linked with WGD and that can contribute to the different stages of polyploid evolution.
Perhaps the most stringent challenge faced by a nascent autopolyploid is directly tied to the very process of reproduction. A sudden doubling of homologous chromosome number disrupts regular meiotic pairing and segregation: instead of each chromosome having only one homolog with which to pair, in autotetraploids there are suddenly three.
The situation in most diploids is relatively straightforward, with proper chromosome pairing during synapsis typically relying on programed double-strand DNA breaks and a sequence-based homology search for the homolog using these broken fragments (Grelon et al., 2001; Page and Hawley, 2003; Stacey et al., 2006; Hartung et al., 2007). Once homologous chromosomes have aligned, a small fraction of these breaks mature into crossovers (COs) between homologes, thus creating bivalent chromosome pairs physically linked by the CO. These bivalents then align parallel to the poles at which point the COs are essential for the creation of mechanical tension between the assembling spindles via connections to each centromere. This tension transmitted through the obligate COs ensures the correct orderly segregation of chromosomes and further acts as an essential cell cycle checkpoint allowing progression to anaphase (Lampson and Cheeseman, 2011; Campbell and Desai, 2013).
In newly formed polyploids however, the presence of multiple equivalent partners for each chromosome leads to more complex arrays of CO formations. If there is more than one crossover per bivalent, a single homolog can have two separate partners, resulting in a multivalent. Most multivalent configurations are not conducive to the formation of regular tension (Bomblies et al., 2016) and some entirely fail to involve one homolog, leading to mis-segregation and aneuploidy. Compounding this, in nascent autopolyploids entanglements and interlocks can occur between non-homologs. If left unresolved these can result in catastrophic chromosome damage, losses and rearrangements. Such entanglements and interlocks do occur in diploids but are much more commonly resolved (Storlazzi et al., 2010). Thus, meiotic stability is a key hurdle that must be overcome following WGD and is one of the hallmarks of an adapted polyploid. Indeed, loci that encode genes controlling meiotic recombination and crossovers are strongly implicated in adaptation to WGD (Hollister et al., 2012; Yant et al., 2013).
Of course, one way to bypass unstable meiosis is to simply not use it. It has long been recognized that asexual reproduction (vegetative propagation and agamospermy) and WGD are correlated, with polyploids displaying elevated rates of asexual propagation compared to diploid relatives (Manning and Dickson, 1986; Schinkel et al., 2016; Herben et al., 2017). Such a reproductive strategy may even confer short-term benefits. Asexual reproduction is considered advantageous during range expansion (Baker, 1967), and this may go some way in explaining the invasive nature of many polyploids. Likewise, vegetative propagation could be a short-term fix; buying time for stable meiosis to evolve (Vallejo-Marín and Hiscock, 2016) and reducing the frequency of mating with the diploid progenitors (see section Cytotype and Competitive Exclusion below). Despite potential short-term gains, however, asexuality may not be a viable long-term strategy, and many polyploids are sexual.
In established sexual autopolyploids meiotic instability is often resolved (Yant et al., 2013; Bomblies et al., 2015), with chromosomes forming bivalents or multivalents that segregate regularly. Indeed, there are multiple conceivable ways to modify meiosis and escape genomic instability, but there is considerable empirical work yet to do to learn how many of these exist in nature, much less the mechanistic basis of different solutions. An elegant theory suggests that simply increasing the degree of CO interference could solve this problem (Bomblies et al., 2016). Under this theory if the range of CO interference is greater than the distance to the end of the chromosome, the number of COs would be reduced to one, and if the range of CO interference is only slightly smaller than the chromosome length the COs will be terminalized. This would favor conformations that produce appropriate tension leading to orderly anaphase (Bomblies et al., 2016). Whatever the mechanism, stabilizing meiosis would seem the best solution given the advantages of sex in the long run.
It was historically expected that an increase in gene number would result in a uniform increase in gene expression (Comai, 2005). This would correspond to a 1:1 dosage effect where 1x diploid expression per genome results in double the total gene expression per cell in an autotetraploid. However, other dosage responses are also possible (Coate and Doyle, 2010): for example, dosage compensation could reduce the per genome expression by half to match overall diploid expression levels per cell (0.5:1 in autotetraploids). This compensation could be partial (ratio between 0.5:1 and 1:1) or even negative (ratios below 0.5:1). In the other direction, dosage effects could also amplify the expression level increase resulting from polyploidy (ratios above 1:1). The impact of these effects could vary across the transcriptome with some gene categories more likely to follow a 1:1 response while others respond differently. The evidence for an overall 1:1 dosage response to WGD in autopolyploids stems primarily from one study of a synthetic polyploid series in maize (Guo et al., 1996). Among the 18 genes followed, most exhibited a 1:1 dosage response, but there were several exceptions and the dosage compensation response of some genes varied over different ploidy levels. Compared to the extensive literature on gene expression changes in allopolyploids (e.g., expression level dominance, genome-wide transcriptomic rewiring, biased fractionation) this represents a major gap in our understanding in autopolyploids. This gap should be closed, as recent empirical evidence points clearly to selective sweeps in transcription-related loci. This suggests that adaptation of the transcriptional machinery to cope with gene dosage effects may be important in neo-autopolyploids. Indeed, one of the most dramatic genome-wide selective sweeps in response to adaptation to WGD in A. arenosa is in the locus encoding the Transcription initiation factor IIF (TFIIF) beta subunit, a key member of the complex that drives RNA synthesis during the transcription (Hollister et al., 2012; Yant et al., 2013).
This gap in our understanding of the mechanistic basis behind dosage compensation is partly the result of technical difficulties. Methods commonly used to evaluate genome-wide expression patterns (microarrays and RNAseq) rely on extensive normalization of the RNA input and therefore are perfectly appropriate to detect relative changes in gene expression but not at all to measure variations in absolute transcriptome size (Lovén et al., 2012; Coate and Doyle, 2015). Only recently has a directed investigation of expression differences between diploids and autotetraploids been reported, using three normalization procedures to take into account transcriptome size, biomass, and cell density (Visger et al., 2017). This allows a clearer discrimination of expression differences than was previously possible, as concentration-based normalizations can mask up to 50% of expression differences. Indeed, in previous relative transcriptome comparisons (Stupar et al., 2007; del Pozo and Ramirez-Parra, 2014; Zhang et al., 2014) < 10% of the transcriptome undergoes expression changes in response to WGD. However, when absolute differences were measured by Visger et al. approximately 1.5 times more genes showed expression differences; 80% of which had a dosage sensitive response with a ratio >0.5:1 (overexpressed in autotetraploid cells compared to diploids) thus compensating for the lower cell density of tetraploids (similar expression per biomass). As the first global analysis of gene dosage response to WGD properly taking into account potential changes in overall transcriptome sizes, this study by Visger et al. effectively demonstrates that 1) most genes are compensating for gene dosage (83% with no differences between diploid and tetraploid cells) and 2) that the genes which do not (17%) mostly present increased expression per cell (ratio >0.5:1) somehow compensating for gene dosage per biomass. This data thus does not support a general trend for 1:1 dosage effects, and also shows other responses are possible (underexpression per cell in tetraploids). More empirical work in this area is required (Figure 2) as antagonistic dosage effects in particular may open possibilities for WGD-induced transcriptomic innovations. In particular, some functional categories were enriched for dosage-sensitivity, mostly in relation to photosynthesis or the chloroplast.
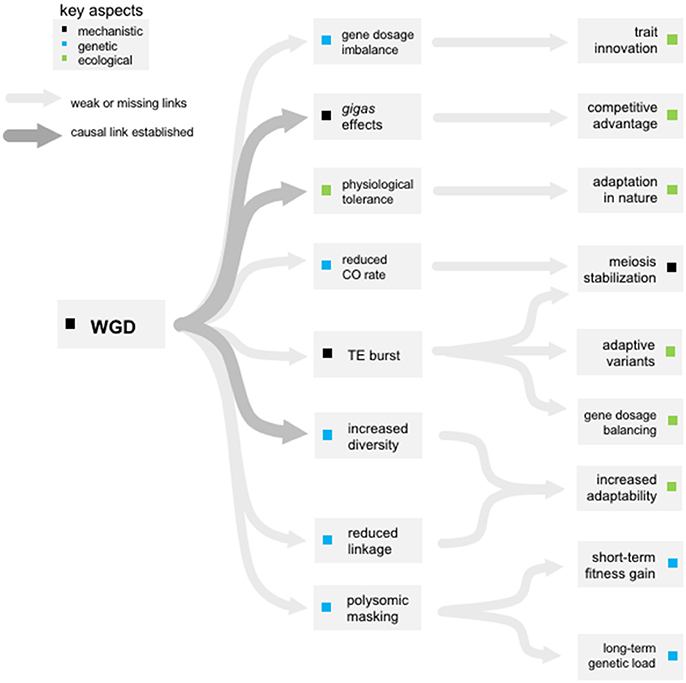
Figure 2. The missing links. The major direct and indirect consequences of WGD either firmly established (dark gray) or still in need of experimental confirmation (light gray lines).
Indeed, parallel evidence from the study of patterns of gene retention following allo-polyploidization (Maere et al., 2005; Thomas et al., 2006; Coate et al., 2011) or from more manageable experimental systems like the X chromosome (Birchler, 2014) support the idea that dosage responses are selectively constrained by genetic pathways. This idea was formalized as the dosage balance hypothesis by Papp et al. (2003), who argued that greater fitness loss would result from perturbing the relative abundance of components of a signaling cascade or of a multi-subunit protein complex than from absolute but concerted concentration changes that would maintain overall stoichiometry. For genes under this selection for gene dosage balance, WGD itself may not greatly disrupt relative abundances, as all genes would see their dosage increased more or less proportionally to one another. If that is the case, however, gene dosage compensation mechanisms (from reduction of gene expression to loss of gene duplicates) would be strongly constrained following WGD, as these would need to be concerted across all interacting subunits to maintain stoichiometry. Hence, the gene duplicates of interacting proteins have been well-preserved even over long time-spans following polyploidization (Papp et al., 2003; Thomas et al., 2006; Birchler and Veitia, 2010). This could be a major hindrance for polyploids as this selection would limit an ability to rectify deleterious gene dosage effects. One way to circumvent this selection would be a uniform reduction of gene expressions across the genome, which, Freeling et al. (2015) argue, is precisely one of the effects predicted for transposition bursts (see section Transposition Burst and the Generation of a High-Effect Mutation Pool below). Supporting this potential importance of TEs in dosage response, dosage-dependent genes in A. thaliana × A. arenosa polyploid series containing from 4 to 0 copies of the A. thaliana genome were depleted of TEs while dosage-independent genes were enriched in TEs (Shi et al., 2015). Under the assumption that TEs are equally likely to insert near both categories of genes, this data suggests that TE insertions near dosage-independent genes are selected against. This is consistent with the gene dosage balance hypothesis, as genes within a network would be unlikely affected synchronously, except in the case of a transposition burst where the effect of TEs would be evenly scattered across the genome. This hypothesis remains to be explored further (Figure 2), as there could also be reverse effects, where presence of TEs in the vicinity of a gene might itself influences the gene-dosage response of that gene. Such effects are yet to be tested but it could help explain the diversity of transcriptomic responses to WGD observed even between accessions of the same species. In A. thaliana, Yu et al. (2010) detected ~500 genes differentially expressed in the Col-0 genotype after WGD but only nine in Ler. Notably, the three genes Yu et al. identify as highly but differentially ploidy-responsive across Col-0, Ler, and a panel of seven other accessions (AT1G53480, AT4G32280, AT5G18030) are all located within 1 kb of a TE insertion (anno-j.org).
Newly formed polyploids are expected to suffer a mating disadvantage when they are relatively rare in the population (Husband, 2000). This type of frequency-dependent disadvantage is known as minority cytotype exclusion (Levin, 1975). The minority cytotype principle is based on the idea that, under random mating, rare cytotypes are expected to be involved in interploidy matings more often than common cytotypes. Assuming that interploidy matings are more likely to produce inviable or sterile offspring, rare cytotypes should have reduced relative fitness. Such a frequency-dependent mating disadvantage was described from experimental and natural mixed-ploidy populations (Hagberg and Elleström, 1959; Maceira et al., 1993; Husband, 2000; Baack and Stanton, 2005; Mráz et al., 2012), but only a few studies further evaluated its significance for polyploid establishment. Interestingly, studies of mixed-ploidy populations of Chamerion angustifolium indicate a surprising asymmetry in this relationship between ploidies. In experimental arrays in field conditions, diploid fitness was frequency-dependent, while fitness in tetraploids was unaffected by their relative frequency. This was likely a result of pollinators preferentially visiting flowers of tetraploid individuals, particularly when rare, and also due to skewed pollen competition favoring tetraploids (Husband, 2000; Husband et al., 2002). Interestingly the effect was mirrored in natural mixed-ploidy populations, where tetraploid mothers produced fewer triploid hybrids than diploid mothers (Husband and Sabara, 2003; Sabara et al., 2013). These studies thus provide a demonstration of minority cytotype exclusion in action and a novel mechanism by which polyploids may avoid its consequences through assortative mating. Indeed, given that WGD can yield larger flowers through the gigas effect (Ramsey and Schemske, 2002; Simon-Porcar et al., 2017), and pollinators often show a preference for visiting larger flowers, non-random mating in mixed-ploidy populations may be important for alleviating the costs of rarity. Additional mechanisms to reduce the costs of inter-cytotype mating are to shift toward self-pollination (Barringer, 2007), or bypass sex altogether either through agamospermy (Thompson and Whitton, 2006; Kao, 2007), or increased vegetative reproduction, as shown both by association studies, (Herben et al., 2017), and in synthetic polyploids (Drunen and Husband, 2018).
In addition, direct competition between the parental diploid and its derivative autopolyploid can hinder the establishment of a nascent polyploid as predicted by theory (Rodríguez, 1996; Yamauchi et al., 2004). This of course depends on niche overlap between the diploid and polyploid. Unlike allopolyploids, where hybridization is expected to create novel genetic combinations unique to the hybrids, autopolyploids may not immediately possess such dramatic genetic differentiation from their progenitors. On the other hand, ploidy-altered traits may translate to better polyploid performance in competition either with its diploid progenitor or with other species. Studies experimentally addressing competition between diploids and their naturally-occurring, recently arisen autopolyploid derivatives are, however, very rare and either support this view (Maceira et al., 1993) or show no difference (Thompson et al., 2015). Alternatively, ploidy-altered traits may also help to cope with competition with other species and may broaden niches, opening the possibility to escape from minority cytotype exclusion. This notion is supported by theoretical models (Rodríguez, 1996) and the observation that polyploids are more frequent in competitive, demanding, and human-disrupted habitats than their diploid relatives (Ehrendorfer, 1980). However, despite the frequent invocation of superior competitive ability to explain polyploid success, this has only rarely been addressed experimentally and available results speak against this trend in autopolyploids (Münzbergová, 2007; Fialová and Duchoslav, 2014), in contrast to allopolyploids (Rey et al., 2017).
Haldane pointed out in 1933 that in the short-term polyploidy should greatly reduce the effect of genetic load by masking recessive or partially-recessive deleterious mutations behind an increased allelic multiplicity (Haldane, 1933). Indeed, at a given allele frequency q, the proportion of homozygotes in a diploid population will be q2 but this drops exponentially to q4 in an autotetraploid population. Thus, for deleterious recessive alleles, the frequency of autotetraploids expressing the associated phenotype will be an order of magnitude lower (or two if q is already small). This means that deleterious recessive alleles can reach much higher frequencies in autotetraploid populations before being exposed to strong selection and equilibrium frequencies are higher in autotetraploids vs. diploids. As newly formed polyploids initially inherit genetic load from a diploid genomic background where the equilibrium frequency is much lower, genetic load will be relieved in young polyploids, providing an early benefit (Figure 1). As a result, as long as most deleterious alleles are at least partially recessive (which is the case in both Mimulus and yeast; Willis, 1999; Agrawal and Whitlock, 2011), WGD is predicted to lead to temporary fitness increases (Korona, 1999; Otto and Whitton, 2000; Mable and Otto, 2001), although empirical evidence for this is still missing (Figure 2).
Both population genetic theory and emerging empirical evidence suggest that a broad set of factors interact to alter the genomic landscape of autopolyploids. However, understanding the effect of WGD in isolation from separate yet correlated effects has only recently made major progress. While it was suggested 35 years ago that biochemical and physiological changes resulting from WGD might underlie polyploid adaptability (Levin, 1983), the best evidence of a direct link took three decades to emerge, when Chao et al., 2013) elegantly demonstrated that A. thaliana first generation autotetraploids have instantaneously enhanced salt tolerance compared to isogenic diploids. Neo-autotetraploid A. thaliana lines were shown to experience a tradeoff, being less fit compared to diploid progenitors under non-saline conditions, but more fit in response to saline challenge (Chao et al., 2013). The authors proposed that in conditions of salinity stress the autopolyploid lineages would benefit from a fitness advantage that could contribute to their establishment and persistence, thanks to an improved ability to accumulate potassium and exclude sodium. Indeed, the following year it was shown that autotetraploid A. thaliana are additionally more drought tolerant (del Pozo and Ramirez-Parra, 2014). A major challenge now is to determine the molecular events that bind WGD to this enhanced stress tolerance. It appears that the key tissue to investigate is the root, where salinity and drought tolerance meet potassium homeostasis and ABA signaling (Saleh et al., 2008; Meng et al., 2011; Allario et al., 2013; Chao et al., 2013; Wang et al., 2013; del Pozo and Ramirez-Parra, 2014). Work there promises to reveal mechanistically how WGD has an immediate effect on cellular physiology that is independent of increased genetic diversity.
There is also good evidence that somatic WGD may enhance stress resilience. For example, in Medicago and sorghum root endopolyploidy correlates with salt tolerance (Ceccarelli et al., 2006; Elmaghrabi et al., 2013) and can be induced by salt in tolerant, but not sensitive, strains (Ceccarelli et al., 2006). Thus, the ability to induce endopolyploidy may be responsible for salinity tolerance, perhaps due to cell size changes in the roots that alter ion uptake. Higher proportions of endopolyploid cells also correlate with greater drought tolerance (Cookson et al., 2006; Saleh et al., 2008; Meng et al., 2011; Chao et al., 2013).
Equally importantly, some effects have been disentangled from polyploidy and shown to be unrelated. A recent study by Solhaug et al. (2016) demonstrated that the allopolyploid Arabidopsis suecica had enhanced carbon assimilation via photosynthesis and elevated respiration rates relative to its progenitors A. arenosa and A. thaliana. This enhanced photosynthetic capacity was environment specific (dependent on high light levels) suggesting a potential mechanism for range expansion by the allopolyploid into novel niches. This advantage was not the direct result of polyploidization, as shown by comparing 12 accessions of isogenic diploid A. thaliana to colchicine-generated neo-polyploids. These autopolyploids showed no difference in carbon assimilation by photosynthesis compared to their diploid progenitors, suggesting that the photosynthetic vigor of A. suecica is a result of hybridization and not WGD.
Despite this progress, the majority of functional studies do not capture the final link: proof that the observed WGD-associated change is adaptive in the natural environment (Figure 2). An exception was provided by Ramsey, who used reciprocal transplants involving tetraploids, hexaploids, and neo-hexaploids (produced from the tetraploids) of Achillea borealis to show a link between WGD and increased fitness in the native environment (Ramsey, 2011). There, WGD itself accounted for 70% of the fitness difference, while the remaining variation (i.e., difference between neo-hexaploids and native hexaploids) could be ascribed to subsequent evolution of the native polyploid. However, the physiological mechanism and its genetic basis in this case remains unknown, which highlights the difficulty of comprehensive inter-disciplinary studies combining genetics, physiology, and ecology.
The hypothesis that WGD presents a genomic shock that activates transposable elements (TEs) across the genome was first proposed by Barbara McClintock (1984) to explain the association between polyploidy and increased TE content. Resident in virtually all genomes, TEs are highly mobile, making them powerful endogenous mutagens. To repress their activity, organisms target TEs with epigenetic silencing mechanisms such as DNA methylation (Bennetzen and Wang, 2014; Ito and Kakutani, 2014). However, the efficiency of TE silencing can be influenced by a number of factors, including environmental or cellular stressors. In some cases, the reactivation of TEs can be explained by the presence of stress-associated transcription factor binding sites in TE promoters (reviewed by Horváth et al., 2017). However, a more global impact of stress on the efficiency of TE-silencing mechanisms has also been suggested (Tittel-Elmer et al., 2010). In particular, genomic stress brought on by hybridization or polyploidization has global effects on epigenetic regulation and may thereby lead to TE reactivation (Kashkush et al., 2003; Madlung et al., 2005; Lopes et al., 2013; Springer et al., 2016; Edger et al., 2017). However, genome shock in polyploids has been studied primarily in allopolyploid contexts, where hybridization is the major contributor, as observed in Senecio cambrensis (Hegarty et al., 2006) and Spartina anglica (Parisod et al., 2009). To date, very few studies addressed the effect of WGD per se on TE transpositions apart from Bardil et al. (2015) who demonstrated an activation of LTR-retroelements following WGD, along with a contribution of gene-flow at the origin of polyploids.
Although most of the mutations TEs generate are deleterious, there is some evidence that TE insertions can be beneficial. The best example of this adaptive potential can be found in the classic case of industrial melanism in the peppered moth, where a young TE insertion that appeared and rapidly rose to fixation during the industrial revolution (~200 years ago) has been proposed as responsible for the dark morph providing camouflage from predators (Van't Hof et al., 2016). The variation produced by TE activity can thus become a fruitful target of natural selection, providing adaptive solutions to the very stresses that initiated their reactivation (Ito et al., 2016). Thus, a global transposition burst triggered by genomic shock could immediately provide nascent polyploids with a pool of high-effect mutations to test against new challenges. In addition, the reactivation of TEs in young polyploids may also contribute to the stabilization of the neo-polyploid genome. First, TE insertions close to genes are known to have an impact, mostly negative (Hollister and Gaut, 2009) but sometimes positive (Quadrana et al., 2016), on the expression of nearby genes. Therefore, the global array of new insertions resulting from a transposition burst might result in broad re-wiring of gene expression and thereby contribute to the rebalancing of gene-dosages (Kashkush et al., 2003; Freeling et al., 2015), as was suggested by recent observations in rice neopolyploids (Zhang et al., 2015; see section Gene Dosage above). Second, TE content in centromeric regions contributes to the bulk of centric heterochromatin that is essential for the separation of sister chromatids during meiosis. Heterochromatin resists the pull exerted by microtubules and the resultant tension silences the spindle checkpoint, allowing meiosis to proceed (Stephens et al., 2013). Increased TE content generated from a transposition burst in neo-polyploids and distributed across chromosomes may thus lead to an overall strengthening of the meiotic spindles and contribute to stabilizing chaotic meiosis following WGD (see Meiosis section above).
Polyploids are over-represented among invasive plants. While in many cases diploids and tetraploids co-exist in the native range, the tetraploids are more often found alone in the invaded range than the contrary (e.g., Hollingsworth and Bailey, 2000). Consistent with this, polyploidy is associated with a potential for habitat colonization and transitions to weediness (Brown and Marshall, 1981; Soltis and Soltis, 2000; Pandit et al., 2006, 2011; Prentis et al., 2008). Common physiological factors contributing to invasiveness are associated with the necessary tolerance to environmental variation including stress resilience, phenotypic plasticity, or rapid cycling (early and prolific flowering aids in coping with or escaping from unpredictable environmental conditions; Baker, 1965; Grotkopp et al., 2002; Blair and Wolfe, 2004; Burns, 2004; Hall and Willis, 2006; Sherrard and Maherali, 2006; Franks et al., 2007). Such life history adaptations can help mediate trade-offs between resource accumulation and stress-avoidance and are important for wild species as well as for crops (Jung and Müller, 2009).
Are polyploids pre-adapted or innately more capable of acquiring such traits? This is an open question, but the many cases where both cytotypes occur in their native range but only polyploids do in the invasive ranges (Lafuma et al., 2003; Mandák et al., 2005; Kubátová et al., 2007; Schlaepfer et al., 2008; Treier et al., 2009) suggest a potential pre-adaptation of polyploids for invasiveness (te Beest et al., 2012). However, environmental stresses also increase the rate of unreduced gamete formation and thus of polyploidization events (Bretagnolle and Thompson, 1995; Ramsey and Schemske, 1998). Therefore, polyploidization has also been viewed as a post-colonization process (Mandák et al., 2003) even if through hybridization (e.g., Hahn et al., 2012). Here we focus on the genetic factors implicated in invasiveness that are likely impacted by WGD (Figure 1). In particular, because the invasion of novel habitats typically proceeds from a small number of founders, some genetic properties of autopolyploids can enhance their chances of successful colonization. These include larger effective population sizes, a greater tolerance for selfing (and inbreeding depression), the ability to recover from the genetic bottlenecks, potentially enhanced sampling from existing standing variation, as well as expected lower levels of linked selection (below and te Beest et al., 2012).
In allopolyploids, where two distinct genomes are united, fitness advantages have often been attributed to interspecific hybridization rather than WGD (Barker et al., 2016). A conservative back-of-the-envelope calculation by Barker et al. (2016) estimated that the rate of production of autopolyploid cytotypes could be 40–80 times greater than that of allopolyploids. Given the approximate parity of allo- and auto-polyploids in nature, this suggests a large advantage to hybridization over the benefits directly attributable to WGD.
Because the two sub-genomes typically do not recombine, allopolyploids can continue to enjoy the advantage of heterosis and a stable multi-allelic state over many generations. Autopolyploids on the contrary, do not benefit from fixed heterozygosity. Nevertheless, it has been proposed that polysomic inheritance alone leads to higher genetic diversity (Haldane, 1932), and experimental comparisons between autotetraploids with tetrasomic inheritance and their diploid parents validate this theoretical expectation (Soltis and Soltis, 2000). This increased diversity has been linked to both an immediate increase in the number of mutational targets (doubled number of chromosomes in the case of autotetraploids), that in the long-run provide increased effective population sizes, and an expected reduced efficiency of purifying selection (Ronfort, 1999). This rise in genetic diversity in autopolyploids is proposed as a cause of the observed successes of tetraploids compared to their diploid sister lineages (Roose and Gottlieb, 1976; Soltis and Soltis, 1989, 2000; Soltis et al., 1993; Brochmann et al., 2004). The positive relationship between phenotypic plasticity and invasiveness introduced first by Baker (1965) is now well-documented through numerous physiological and morphological comparisons of invasive and native species (reviewed in Richards et al., 2006). Increased diversity in polyploids is often invoked to explain an increased plasticity and ability of polyploids to sustain range expansions into disturbed habitats. This is a tempting speculation, but a causal link demonstrating that the increased diversity in tetraploids confers an adaptive advantage is lacking (Figure 2).
In addition, reduced homozygosity in autopolyploids is expected to reduce the potential inbreeding depression associated with genetic load (Charlesworth and Charlesworth, 1987). This is because at any locus the increase in copy number in autopolyploids increases the probability of heterozygous offspring, even during selfing (Moody et al., 1993). As a result, the fitness cost associated with selfing (inbreeding depression) may be ameliorated (Lande and Schemske, 1985; Schemske and Lande, 1985) or at worst unchanged (Ronfort, 1999) depending on the range of dominance effects impacting fitness. This prediction has been confirmed in ferns, where the self-fertilization of the gametophyte makes it possible to directly measure the impact of selfing on survival rates in the resulting sporophytes. In two different diploid-tetraploid fern pairs, selfing survival rates were nearly 100% in the tetraploid races, while it ranged from 5 to 60% in the diploids (Masuyama and Watano, 1990). Similarly, a reduction of inbreeding depression is observed in other polyploid-diploid comparisons (Husband and Schemske, 1997; Galloway et al., 2003; Husband et al., 2008), even though there are cases where the opposite is observed (Johnston and Schoen, 1996).
Tolerance to selfing is of major importance in the ability of a population to colonize new habitats, a consideration known as “Baker's rule” (Baker, 1967). Indeed, during the colonization process, early invaders are likely to be isolated with little opportunity for outcrossing. Selfing therefore provides reproductive assurance for the dispersed invaders (Barrett et al., 2008), and this translates to a high rate of co-occurrence between selfing or asexual propagation and low-density conditions or frequent colonization bouts (Baker, 1967; Price and Jain, 1981; Pannell and Barrett, 1998). For example, Daehler found that low inbreeding depression in hexaploid smooth cordgrass populations invading the San Francisco Bay area in California was associated with higher self-fertility and a higher fitness advantage for founding populations in the field (Daehler, 1998; Renny-Byfield et al., 2010).
As a mirror image to the reduced efficiency of selection against deleterious mutations, the increase in frequencies of beneficial alleles, even when dominant, will be slower in polyploids under tetrasomic inheritance than in diploids (Hill, 1971). Therefore, the time to fixation for an allele during a selective sweep can be greatly increased in autopolyploids. This prolonged rise in allele frequency might lead to more opportunities for mutation and recombination with other haplotypes, which are even further enhanced by the increased mutation and recombination rates resulting from greater ploidy levels. Weaker linkage thus may promote adaptation through reduced interference between alleles, allowing greater opportunity for a beneficial allele to recombine onto haplotypes with fewer deleterious mutations (Figure 1). However, increased recombination can lead to lower fitness in constant environments by breaking down beneficial associations (Lewontin, 1971; Feldman et al., 1980). Therefore, increased recombination may only be selected for in environments with fluctuating conditions (Charlesworth, 1976; Otto and Michalakis, 1998; Lenormand and Otto, 2000), which also happen to be environments with higher incidences of polyploids (Favarger, 1984; Brochmann et al., 2004; Parisod et al., 2010). This association between increased recombination and adaptation to environmental variation would strongly favor long-term evolution of autopolyploids, but remains to be experimentally tested (Figure 2).
As an autopolyploid lineage expands its range, it may encounter populations of its diploid progenitor or other species with which hybridization is possible. Provided that such populations are locally adapted, introgression may then supply genetic variants that facilitate persistence. Although polyploidization is traditionally viewed as a means of instant speciation (Coyne and Orr, 2004), the ploidy barrier is permeable (reviewed in Ramsey and Schemske, 1998; Kolár et al., 2017). While adaptive introgression is increasingly recognized as an important force in the evolution of haploid and diploid organisms by genomic studies (reviewed by Arnold and Kunte, 2017; Schmickl et al., 2017), empirical genomic evidence for gene flow among a diploid and its autopolyploid derivative is lacking. The ability to accept genetic variation from alternative cytotype might be beneficial, as it could provide preadapted local alleles upon which selection may act and/or may alleviate inbreeding associated with founding events during range expansions (Parisod et al., 2010). We however lack well-documented examples of traits and underlying loci that may explain evolutionary significance of gene flow for establishment and further spread of a polyploid. The only case to our knowledge, although confined to allopolyploids, is across-ploidy transfer of potentially adaptive floral genes, RAY1 and RAY2, from diploid Senecio squalidus into the allotetraploid Senecio vulgaris that has given rise to a novel variety of S. vulgaris with ray florets (Chapman and Abbott, 2010). An additional hint, coming from an autopolyploid system, is the likely uptake of a diploid-like CONSTANS allele during the colonization of railways by a distinct lineage of autotetraploid A. arenosa (Baduel et al., 2018). This allele may allow the railway ecotype to escape the repression exacted on flowering by FLOWERING LOCUS C and underlie the observed rapid and repeated flowering. These two examples indicate that this may be a fruitful area for future research. An alternative benefit of interploidy hybrids for polyploid establishment may result from their contribution to recurrent formation of polyploids. Triploid hybrids, if fertile, often produce unreduced (2n = 2x) gametes (Ramsey and Schemske, 1998; Chrtek et al., 2017) that can merge with reduced (n = 2x) gametes of a tetraploid leading to formation of novel tetraploids (Husband, 2004). We note, however, that much gene flow may be neutral or even maladaptive. For example, if a tetraploid has adapted to problems associated with meiotic segregation during its establishment (Yant et al., 2013), later diluting of such co-adapted gene networks by introgression of diploid-like alleles would lead to reductions in fitness.
Even when assuming beneficial consequences, interploidy gene flow would provide relative advantages to the polyploid only in cases when (potentially adaptive) alleles flow more often into the polyploid than into progenitor diploids (Figure 1). Indeed, this seems to be the case and it was recognized as early as 1971 by Stebbins that gene flow among cytotypes is asymmetrical (Stebbins, 1971). A mechanistic explanation for this asymmetry is that while there are direct pathways for gene flow in the 2x –> 4x direction, the reverse is more convoluted. The unreduced 2n = 2x gamete of a diploid and the reduced n = 2x gamete of a tetraploid can combine leading to one-step formation of a tetraploid interploidy hybrid (2x + 2x = 4x; Koutecký et al., 2011; Chrtek et al., 2017; Sutherland and Galloway, 2017). However, a triploid hybrid, capable of forming reduced n = x gametes, is an essential stepping-stone for the creation of a diploid hybrid (Kolár et al., 2017). Thus, gene flow in the 4x –> 2x direction is less likely as it involves two separate crossing steps (4x –> 3x and 3x –> 2x). Moreover, the triploid hybrid is often either non-viable (triploid block) or unfit (Ramsey and Schemske, 1998; Köhler et al., 2010). Indeed, the few available empirical genetic studies document either much stronger (in autopolyploid systems: Ståhlberg, 2009; Jørgensen et al., 2011; Arnold et al., 2015) or exclusively unidirectional gene flow from the diploid into the polyploid (in allopolyploid systems: Slotte et al., 2008; Chapman and Abbott, 2010; Zohren et al., 2016). In a longer evolutionary timespan recurrent origins of autopolyploid lineages from different diploid sources followed by hybridization among these polyploids (Soltis and Soltis, 2009) would also lead to enrichment of the tetraploid gene pool by alleles from distinct diploid lineages, similar to direct unidirectional gene flow from diploid to polyploid.
If higher polyploids are formed (i.e., hexaploids, octoploid, etc.) they may hybridize with the tetraploid or among one another and further enhance variation of the polyploid lineages. The few empirical studies available show that the postzygotic barrier, both in terms of rate of hybrid formation and its fitness, is lower among the various polyploid cytotypes than it is between diploids and their polyploid derivatives (Greiner and Oberprieler, 2012; Sonnleitner et al., 2013; Hülber et al., 2015; Sutherland and Galloway, 2017). This corresponds well with the explanation of maternal: paternal genome imbalance in the endosperm as a primary cause of the postzygotic barrier (Köhler et al., 2010; Greiner and Oberprieler, 2012). Because the magnitude of endosperm imbalance in tetraploid–hexaploid hybrids is approximately one third lower than in diploid–tetraploid hybrids (Sonnleitner et al., 2013) these higher-ploidy hybrids may be more fit than diploid–tetraploid hybrids.
Aside from intraspecific gene flow, polyploidy may also break down systems of reproductive isolation present in diploid progenitors and thus increase interspecific gene flow. For example, although the reproductive isolation in diploid lineages of Arabidopsis arenosa and Arabidopsis lyrata is near complete, tetraploid A. lyrata can form viable hybrids with both diploid and tetraploid A. arenosa, likely due to the disruption of an endosperm-based barrier (Lafon-Placette et al., 2017). Interestingly, hybridization between those species appears to have donated beneficial alleles contributing to local adaptation to harsh serpentine soils in the tetraploid A. arenosa (Arnold et al., 2016). In this study Arnold et al. (2016) found that several genes exhibiting signatures of selection for adaptation to serpentine soils also appeared to have been introgressed from A. lyrata. Finally, the tendency of polyploids to expand into novel niches may further increase chances of encountering foreign lineages with which hybridization may occur. Although the cause of this is unclear, the heightened adaptability of many polyploids fueled by introgression may provide positive feedback, allowing further spread and hybridization. Altogether, these examples illustrate a tendency for polyploids to act as evolutionary “sponges,” accumulating variation through introgression across both ploidy and species barriers. This supports the view of polyploids as diverse evolutionary amalgamates from multiple distinct ancestral lineages—a property advantageous for further expansions.
This begs the question: if WGD events are common, and polyploids display advantageous traits, why are established autopolyploids relatively uncommon and paleo-polyploids so frequent? Transitions to polyploidy tend to be observed at the tips of phylogenies (Escudero et al., 2014), suggesting that polyploid lineages typically do not survive as such over longer evolutionary timescales. Consequently, the growing consensus is that polyploidy is an ephemeral but repeatedly appearing state (Wendel, 2015). This could be the result of both pervasive polyploid extinction, as there is some suggestion that recently arisen polyploids experience lower diversification rates and higher extinction rates relative to congeneric diploids (Mayrose et al., 2011, 2015; Arrigo and Barker, 2012), as well as repeated returns to diploidy and disomic inheritance after transitionary polyploid phases (e.g., Haufler, 1987; Wendel, 2015; Soltis et al., 2016). To date, such a transition has only been mathematically modeled in autopolyploids (Le Comber et al., 2010); empirical evidence is lacking. Thus, in addition to the short term biological challenges faced by newly-arisen polyploids, longer-term challenges may help explain the transience of the polyploid state, even reviving the idea of polyploidy as an evolutionary “dead-end” (Wagner, 1970; Stebbins, 1971; Mayrose et al., 2011, 2015). Ironically, many of these postulated longer-term negative effects result from the continuation of earlier beneficial population genetic mechanisms.
If in the short-term polysomic masking results in a fitness increase, a reduced strength of purifying selection (Ronfort, 1999) would eventually lead to the slow rise of recessive deleterious mutations until mutational load reaches a new, higher equilibrium (Otto and Whitton, 2000). This new polyploid equilibrium may take hundreds of thousands of generations to establish (Otto and Whitton, 2000), but would ultimately produce a genetic load proportional to ploidy level and the mutation rate μ per haploid genome (Haldane, 1937). A particularly strong effect of this would be on TEs, as their distribution of fitness effects is much more heavily skewed toward highly-deleterious mutations compared to single-nucleotide polymorphisms and thus are strongly affected by purifying selection. This has been demonstrated recently in A. thaliana (Quadrana et al., 2016), where it was shown that TEs insert throughout the genome, but are rapidly purged from genic rich regions and chromosome arms, most likely due to the deleterious consequences of insertions near or within genes (Quadrana et al., 2016). Even if evidence of this mostly comes from allopolyploid systems (wheat, Brassica, etc.), these long-term effects are likely to be similar in autopolyploids and in the long run we can expect the initial differences in transposition burst triggered by the two modes of polyploidization to be less important compared to the relaxation of purifying selection shared by both systems. Indeed, in the allotetraploid Capsella bursa-pastoris, an increase in TE content was observed around genes compared to its two parental diploid species, C. grandiflora and C. orientalis, which was attributed primarily to a relaxation of purifying selection and not to any change in TE activity (Ågren et al., 2016), and there is accumulating evidence of TE proliferation over long timespans following polyploidization (Sarilar et al., 2011; Yaakov and Kashkush, 2012; Piednoël et al., 2015). However, this doesn't seem to apply to all TE families equally. For example some gypsy-like retro-elements proliferated in Aegilops tetraploids while others remained quiescent (Senerchia et al., 2014). This could be due to differences in insertion preferences between TE families or more simply to the fact that many TEs are actually defective. Indeed, most of the TEs carried by a genome have lost their transpositional capacities and are fossilized: in the human genome < 0.05% of TEs remain active (Mills et al., 2007). Between two active families, differences in their regulation, copy number, chromosome localization, etc. may also explain different responses to relaxed purifying selection. For example, a family inserting more commonly into genes will be more strongly purified and therefore more strongly affected by a relaxation of purifying selection than in TE families that inherently avoid inserting into gene-coding loci. Such differences in insertion preferences have been observed in one LTR retrotransposon family between A. thaliana where genic insertions are strongly selected against, and A. lyrata, where gene-poor centromeric regions are preferentially targeted, reaching much higher copy numbers (Tsukahara et al., 2012).
Eventually, it is thought that the reduced strength of purifying selection from polysomic masking may overshadow the early advantages of low mutational load, which begins at the lower diploid equilibrium levels immediately following WGD (Otto, 2007; Gerstein and Otto, 2009). At equilibrium (Figure 1), polyploids are predicted to suffer from the increased frequency of deleterious mutations, which are introduced in higher numbers (doubled in the case of autotetraploids). However, given the difficulties of finding an ancient enough system where autopolyploids have reached such an equilibrium but are still ecologically comparable to their diploid progenitors, empirical support for this remains sparse (Figure 2).
In addition to hampering selection against deleterious mutations, polysomic masking can also prevent recessive beneficial mutations from reaching fixation. This may even effectively counter the increased input of beneficial mutations arising from the increased number of haploid genomes (Haldane, 1932; Gerstein and Otto, 2009). Whereas in haploids the rate of fitness increase only depends on the rate of appearance of beneficial mutations and their fitness effect (Haldane et al., 1927), in diploids it also depends on the dominance level of mutations. This is further intensified in polyploids (Gerstein and Otto, 2009). For example, in autotetraploids with tetrasomic inheritance, the rate of fitness increase (w) can be written as a function of the rate of appearance, the fixation probability, and the fitness effect (s) as follows:
Where N is the population size, ν is the beneficial mutation rate, and h1 is the dominance of the new allele in simplex (for example Aaaa for tetraploids). Therefore, polyploids would adapt faster only when mutations are at least partially dominant (h1 > 0.5) and thus not hindered by polysomic masking. In an attempt to test this prediction experimentally, Schoustra et al. (2007) observed the fastest rates of loss of a costly resistance allele in diploid strains of the fungus Aspergillus nidulans that periodically reverted to haploidy. These strains accumulated multiple recessive beneficial mutations in the diploid state that were exposed to positive selection in the haploid state. This pattern is reminiscent of the transitionary polyploid phases postulated to have occurred throughout the evolution of plants (Haufler, 1987; Wendel, 2015; Soltis et al., 2016).
Further amplifying this effect of reduced positive selection, lower linkage in autopolyploids (see Reductions in Linked Selection: An Advantage in Changing Environmental Conditions?) increases the likelihood of recombination breaking down favorable haplotypes as they slowly rise in frequency. As a consequence, beneficial mutations in close vicinity and positively selected in autotetraploids are unlikely to remain linked to each other for long. This may be beneficial in the early stages of invasion (directional selection) or under fluctuating environments, but in the long run it is predicted to be unfavorable. Indeed, once established in their new range and closer to a new fitness optimum, selection is predicted to favor increased linkage and reduced recombination (Feldman et al., 1980, 1996).
With their doubled genomes, autopolyploids are likely to face the general rule in animals and plants dictating that increased genomic content results in decreased growth and division rates (Gregory and Mable, 2005; Otto, 2007). If the impact of genome size is clear at the cellular level it is less evident at the organism level (Knight and Beaulieu, 2008) and exceptions to this rule can easily be found: first in the growth form (e.g., trees have small genomes, Beaulieu et al., 2008) and in the environment (Zörgö et al., 2013). This led some to suggest the overall negative relationship between genome size and metabolic rate across gymnosperms and angiosperms may be the result of a rather indirect effect through other traits such as growth form (Beaulieu et al., 2007a). It should be noted, however, that these rules are based on the observation of established polyploids and therefore the impact of genome size itself remains to be directly assessed independently of the potentially confounding effects of subsequent evolution. On the cellular level, it seems clear that increased nuclear content leads to increased cell volume (Beaulieu et al., 2008; Knight and Beaulieu, 2008) and slower growth rates (Cavalier-Smith, 1978; Gregory, 2001), which have long been observed in polyploids as well (Müntzing, 1936; Stebbins, 1971; Garbutt and Bazzaz, 1983). At the organismal level, older observations have illustrated that polyploids often flower later (Smith, 1946) and are more frequently perennial (Hagerup, 1932; Müntzing, 1936; Sano et al., 1980), but the role of WGD itself has been rarely experimentally evaluated since then. For example, in synthetic A. thaliana tetraploids, there was no consistent trend in flowering time over 12 ecotypes investigated in two common gardens (Solhaug et al., 2016) and similarly no differences in this trait were found between diploid and synthetic polyploids of Chamerion angustifolium (Husband et al., 2016). On the other hand, a recent study leveraged parallel altitudinal clines and intraspecific genome size variation in maize landraces to show repeated reductions in genome size in high-altitude populations most likely via selection on flowering time (Bilinski et al., 2018). Furthermore, in growth chamber experiments Bilinski et al. were able to confirm an association, even if modest, between genome size, cell production, and cell sizes. Therefore, such constraints may turn out to be particularly costly for polyploids that successfully switched to invasiveness thanks to early advantages (see section Enhanced Invasiveness and Colonization Potential above). Indeed, invasive species commonly exhibit early flowering (Pyšek et al., 2009), lower seed sizes with higher dispersal abilities and annual life cycles which are also the prerogative of small-genome species (Grotkopp et al., 2002; Knight et al., 2005; Beaulieu et al., 2007b). Even if more research is needed to clarify the direct impact of polyploidy, evidence so far suggests that the potential slowing of growth rates may impact negatively long-term fitness. Thus, selection would likely push for a reduction of genome size, especially in transitions to invasiveness. This process may be very long and stochastic, however, as evidence in the Nicotiana genus shows genome downsizing is minimal in young polyploids (~200,000 years old) only appearing in polyploids approximately 4.5 million years old, at which point genome size increases are also observed (Leitch et al., 2008).
It is now clear that nearly all plant lineages are paleo-polyploids, with their evolutionary histories including at least one round of WGD (Wendel, 2015). However, numbers of past WGD events do not correlate with chromosome numbers nor genome sizes. For example, given the three rounds of genomic multiplications that have occurred in Brassica genomes (α, β, and γ, Franzke et al., 2011; Jiao et al., 2011), and assuming ancestral angiosperms had between 5 and 7 chromosomes (Stebbins, 1971; Raven, 1975), we would expect, without reductions in chromosome numbers along the way, resultant species to carry between 40 and 56 chromosomes, when some carry as few as six (Anderson and Warwick, 1999). A similar reasoning holds with genome sizes (Wendel, 2015): thus it is apparent that past polyploidization events were followed by massive genome downsizing, both in chromosome numbers and in absolute size (Leitch and Bennett, 2004; Leitch and Leitch, 2008). This genome downsizing ultimately leads to the diploidization of descendants (Soltis et al., 2015). These paleo-polyploids then commonly undergo further rounds of polyploidization, generating a cyclical process described as the “wondrous cycles of polyploidy” and occurring repeatedly over long evolutionary timescales (Wendel, 2015).
Several mechanisms have been proposed to underlie the diploidization process, all of them relying on non-homologous translocations (Mandáková and Lysak, 2018). One contributor to these illegitimate recombination events are TEs, since homology between TE copies can lead to spurious recombination events between non-homologous chromosomes (Vicient and Casacuberta, 2017). Some of the rearrangements resulting from these non-homologous recombinations (inversions, reciprocal translocations, deletions, and duplications) do not affect chromosome numbers, but others (end-to-end translocations, EETs, nested chromosome insertions, NCIs, and Robertsonian translocations) are seen as the mechanistic basis of “polyploid drop” (Mandáková and Lysak, 2018). Indeed, all three processes result in the merger of two chromosomes into one via non-homologous recombinations between two distal regions (EETs), two distal regions with a pericentromeric region (NCIs), or between a distal and a pericentromeric region (Robertsonian translocations). Distal and pericentromeric regions are particularly prone to ectopic homologies due to their enrichment in repetitive elements, in particular TEs (Quadrana et al., 2016; Vicient and Casacuberta, 2017). Therefore, even though most recombination events between TEs will lead to small indels, the possibility of large-scale chromosomal rearrangements may represent a major driver of genome restructuring during diploidization (Vicient et al., 1999). In fact, evidence supporting a role for TEs during diploidization has been observed in Nicotiana (Lim et al., 2007) and maize (Bruggmann et al., 2006). However, these dysploidy events have an immediate fitness cost, as the merging of two chromosomes leads to obvious chromosome segregation issues. In outcrossers in particular, the probability of forming non-aneuploid offspring is very low, and newly formed dysploids are likely to suffer from woes similar to newly-formed autopolyploids (Mandáková and Lysak, 2018). This is why it was theorized that the establishment of dysploids would be relatively favored in selfers (Charlesworth, 1992). By increasing homozygosity of the offspring, selfing indeed reduces the fitness cost of dysploidy by increasing the probability of producing offspring homozygous for the merged chromosome. Extending this reasoning, we can expect higher rates of dysploidy among weedy invasives, due to both their propensity for selfing and their often faster cycling (e.g., Grant, 1981), which increases the probability of spurious recombinations (Mandáková and Lysak, 2018). This relationship between life-history and dysploidy rate has been confirmed (e.g., Luo et al., 2015) even though some examples show this is not always straightforward (slow polyploid drop rate in rice despite being annual, Murat et al., 2010). Furthermore, the advantage of a reduced number of chromosomes may be particularly valuable for colonizers (see section Bigger Genomes, Slower Growth Rates above).
These considerations become particularly relevant for aging polyploids, which both carry an increased TE content (see sections Transposition Burst and the Generation of a High-Effect Mutation Pool and Increased Genetic Load above) and are more likely to tolerate selfing (section Increased Diversity and Tolerance for Selfing above). Thus, factors that initially represented an advantage for the establishment of recent autopolyploids may transform into the very drivers of polyploid drop and return to diploidy (Figure 1).
Compared to WGD, which leads to an exact doubling of chromosome numbers, polyploid drop is more erratic and can produce broad variation in chromosome number. In Brassica for example, the variation in base chromosome numbers is the result of multiple and independent diploidizations from the mesohexaploid ancestor (Lysak et al., 2007; Mandáková et al., 2017). Indeed, the stochasticity of polyploid drop, not WGD, is thought to be a major contributor to speciations and radiations (Figure 1). However, polyploidy drop is of course not possible without an earlier WGD. Accordingly, recently revised phylogenetic evidence convincingly supports the occurrence of WGDs significantly before large angiosperm radiations, sometimes by millions of years (Tank et al., 2015; Clark and Donoghue, 2017). These reports strengthen the WGD Radiation Lag-Time Model formalized by Schranz et al. (2012), who found that in six examples of angiosperm radiations a species-poor sister-group shared a WGD event with the species-rich crown group, directly contradicting the notion that WGD was the sole immediate cause of these radiations. The lag between WGD and subsequent radiations thus has been proposed as evidence that the long and stochastic process of polyploid drop is the proximal engine of speciation and cladogenesis (Dodsworth et al., 2016; Clark and Donoghue, 2017; Mandáková and Lysak, 2018).
As best expressed by Johnathan Wendel, the “wondrous cycles” of polyploidy have gained increasing attention and support, both theoretical and empirical, over the earlier ideas that polyploids were evolutionary dead-ends. Excellent recent reviews have discussed the complex mixture of advantages and disadvantages of polyploidy (see especially Spoelhof et al., 2017), and here we aimed to extend this with the most recent evidence considered explicitly in the scope of the dynamic temporal nature of shifting costs and benefits. In doing so, we hope to bring to light the importance of the timescales at which evolutionary dynamics are at play over the lifespan, from dawn till dusk, of any given genome duplication, thus creating the conditions for these wondrous cycles to emerge. We see a picture of each cycle of WGD-diploidization as a temporary but powerful engine of evolutionary diversification. Eventually, without specific selective pressures maintaining a strong advantage for polyploids, each hop to polyploidy is restabilized in a drop to diploid form, but there are plenty of evolutionary opportunities for speciation and radiation along the way.
PB drafted the manuscript and realized the figures with help from all other authors. SB drafted the manuscript with help from all other authors. MV-M drafted the manuscript with help from all other authors. FK drafted the manuscript with help from all other authors. LY drafted the manuscript with help from all other authors.
LY acknowledges funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement 679056) and the UK Biological and Biotechnology Research Council (BBSRC) via grant BB/P013511/1 to the John Innes Centre. Additional support was provided by Czech Science Foundation (project 17-20357Y to FK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ågren, J. A., Huang, H.-R., and Wright, S. I. (2016). Transposable element evolution in the allotetraploid Capsella bursa-pastoris. Am. J. Bot. 103, 1197–1202. doi: 10.3732/ajb.1600103
Adams, K. L., and Wendel, J. F. (2005). Novel patterns of gene expression in polyploid plants. Trends Genet. 21, 539–543. doi: 10.1016/j.tig.2005.07.009
Agrawal, A. F., and Whitlock, M. C. (2011). Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187, 553–566. doi: 10.1534/genetics.110.124560
Allario, T., Brumos, J., Colmenero-Flores, J. M., Iglesias, D. J., Pina, J. A., Navarro, L., et al. (2013). Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 36, 856–868. doi: 10.1111/pce.12021
Anderson, J. K., and Warwick, S. I. (1999). Chromosome number evolution in the tribeBrassiceae (Brassicaceae): evidence from isozyme number. Plant Syst. Evol. 215, 255–285. doi: 10.1007/BF00984659
Arnold, B., Kim, S.-T., and Bomblies, K. (2015). Single geographic origin of a widespread autotetraploid Arabidopsis arenosa lineage followed by interploidy admixture. Mol. Biol. Evol. 32, 1382–1395. doi: 10.1093/molbev/msv089
Arnold, B. J., Lahner, B., DaCosta, J. M., Weisman, C. M., Hollister, J. D., Salt, D. E., et al. (2016). Borrowed alleles and convergence in serpentine adaptation. Proc. Natl. Acad. Sci. 113, 8320–8325. doi: 10.1073/pnas.1600405113
Arnold, M. L., and Kunte, K. (2017). Adaptive genetic exchange: a tangled history of admixture and evolutionary innovation. Trends Ecol. Evol. 32, 601–611. doi: 10.1016/J.TREE.2017.05.007
Arrigo, N., and Barker, M. S. (2012). Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15, 140–146. doi: 10.1016/j.pbi.2012.03.010
Baack, E. J. (2005). To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity 94, 538–546. doi: 10.1038/sj.hdy.6800656
Baack, E. J., and Stanton, M. L. (2005). Ecological factors influencing tetraploid speciation in snow buttercups (ranunculus adoneus): niche differentiation and tetraploid establishment. Evolution 59, 1936. doi: 10.1554/05-168.1
Baduel, P., Hunter, B., Yeola, S., and Bomblies, K. (2018). Genetic basis and evolution of rapid cycling in railway populations of tetraploid Arabidopsis arenosa. PLoS Genet. 14:e1007510. doi: 10.1371/journal.pgen.1007510
Baker, H. G. (1965). “Characteristics and modes of origin of weeds,” in The Genetics of Colonizing Species (Asilomar, CA: Academic Press), 147–168. Available online at: http://www.cabdirect.org.ezp-prod1.hul.harvard.edu/abstracts/19661605504.html;jsessionid=0903F3BC5F41C10D0D2169FF9710AE2D?freeview=true (Accessed October 6, 2015).
Bardil, A., Tayalé, A., and Parisod, C. (2015). Evolutionary dynamics of retrotransposons following autopolyploidy in the Buckler Mustard species complex. Plant J. 82, 621–631. doi: 10.1111/tpj.12837
Barker, M. S., Arrigo, N., Baniaga, A. E., Li, Z., and Levin, D. A. (2016). On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210, 391–398. doi: 10.1111/nph.13698
Barker, M. S., Kane, N. C., Matvienko, M., Kozik, A., Michelmore, R. W., Knapp, S. J., et al. (2008). Multiple paleopolyploidizations during the evolution of the compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 25, 2445–2455. doi: 10.1093/molbev/msn187
Barrett, S. C. H., Colautti, R. I., and Eckert, C. G. (2008). Plant reproductive systems and evolution during biological invasion. Mol. Ecol. 17, 373–383. doi: 10.1111/j.1365-294X.2007.03503.x
Barringer, B. C. (2007). Polyploidy and self-fertilization in flowering plants. Am. J. Bot. 94, 1527–1533. doi: 10.3732/ajb.94.9.1527
Beaulieu, J. M., Leitch, I. J., and Knight, C. A. (2007a). Genome size evolution in relation to leaf strategy and metabolic rates revisited. Ann Bot. 99, 495–505. doi: 10.1093/aob/mcl271
Beaulieu, J. M., Leitch, I. J., Patel, S., Pendharkar, A., and Knight, C. A. (2008). Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179, 975–986. doi: 10.1111/j.1469-8137.2008.02528.x
Beaulieu, J. M., Moles, A. T., Leitch, I. J., Bennett, M. D., Dickie, J. B., and Knight, C. A. (2007b). Correlated evolution of genome size and seed mass. New Phytol. 173, 422–437. doi: 10.1111/j.1469-8137.2006.01919.x
Bennetzen, J. L., and Wang, H. (2014). The contributions of transposable elements to the structure, function, and evolution of plant genomes. Ann. Rev. Plant Biol. 65, 505–530. doi: 10.1146/annurev-arplant-050213-035811
Bilinski, P., Albert, P. S., Berg, J. J., Birchler, J. A., Grote, M. N., Lorant, A., et al. (2018). Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLOS Genet. 14:e1007162. doi: 10.1371/journal.pgen.1007162
Birchler, J. A. (2014). Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma 123:459–469. doi: 10.1007/s00412-014-0478-5
Birchler, J. A., and Veitia, R. A. (2010). The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186, 54–62. doi: 10.1111/j.1469-8137.2009.03087.x
Blair, A. C., and Wolfe, L. M. (2004). The evolution of an invasive plant: an experimental study with Silene latifolia. Ecology 85, 3035–3042. doi: 10.1890/04-0341
Bomblies, K., Higgins, J. D., and Yant, L. (2015). Tansley review Meiosis evolves: adaptation to external and internal environments. New Phytol. 208, 306–323. doi: 10.1111/nph.13499
Bomblies, K., Jones, G., Franklin, C., Zickler, D., and Kleckner, N. (2016). The challenge of evolving stable polyploidy: could an increase in “crossover interference distance” play a central role? Chromosoma 125, 287–300. doi: 10.1007/s00412-015-0571-4
Bomblies, K., and Madlung, A. (2014). Polyploidy in the arabidopsis genus. Chromosome Res. 22, 117–134. doi: 10.1007/s10577-014-9416-x
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Bretagnolle, F., and Thompson, J. D. (1995). Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 129, 1–22. doi: 10.1111/j.1469-8137.1995.tb03005.x
Brochmann, C., Brysting, A. K., Alsos, I. G., Borgen, L., Grundt, H. H., Scheen, A.-C., et al. (2004). Polyploidy in arctic plants. Biol. J. Linn. Soc. 82, 521–536. doi: 10.1111/j.1095-8312.2004.00337.x
Brown, A. H. D., and Marshall, D. S. (1981). “Evolutionary changes accompanying colonization in plants,” in Evolution Today: Proceedings of 2nd International Congress of Systematics and Evolutionary Biology, 351–363.
Brownfield, L., and Köhler, C. (2011). Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62, 1659–1668. doi: 10.1093/jxb/erq371
Bruggmann, R., Bharti, A. K., Gundlach, H., Lai, J., Young, S., Pontaroli, A. C., et al. (2006). Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16, 1241–51. doi: 10.1101/gr.5338906
Burleigh, J. G., Bansal, M. S., Wehe, A., and Eulenstein, O. (2008). “Locating multiple gene duplications through reconciled trees,” in Research in Computational Molecular Biology (Berlin; Heidelberg: Springer), 273–284.
Burns, J. H. (2004). A comparison of invasive and non-invasive dayflowers (Commelinaceae) across experimental nutrient and water gradients. Divers. Distributions 10, 387–397. doi: 10.1111/j.1366-9516.2004.00105.x
Campbell, C. S., and Desai, A. (2013). What the hec is up with mouse oocyte meiosis? Dev. Cell 25, 3–4. doi: 10.1016/j.devcel.2013.03.018
Cavalier-Smith, T. (1978). Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J. Cell Sci. 34, 247–278.
Ceccarelli, M., Santantonio, E., Marmottini, F., Amzallag, G. N., and Cionini, P. G. (2006). Chromosome endoreduplication as a factor of salt adaptation in Sorghum bicolor. Protoplasma 227, 113–118. doi: 10.1007/s00709-005-0144-0
Chao, D., Dilkes, B., Luo, H., Douglas, A., Yakubova, E., Lahner, B., et al. (2013). Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341, 658–659. doi: 10.1126/science.1240561
Chapman, M. A., and Abbott, R. J. (2010). Introgression of fitness genes across a ploidy barrier. New Phytol. 186, 63–71. doi: 10.1111/j.1469-8137.2009.03091.x
Charlesworth, B. (1976). Recombination modification in a fluctuating environment. Genetics 83, 181–195.
Charlesworth, B. (1992). Evolutionary rates in partially self-fertilizing species. Am. Nat. 140, 126–48. doi: 10.1086/285406
Charlesworth, D., and Charlesworth, B. (1987). Inbreeding depression and its evolutionary consequences. Ann. Rev. Ecol. Syst. 18, 237–268. doi: 10.1146/annurev.es.18.110187.001321
Chen, Z. J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann. Rev. Plant Biol. 58, 377–406. doi: 10.1146/annurev.arplant.58.032806.103835
Chen, Z. J., and Ni, Z. (2006). Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28, 240–252. doi: 10.1002/bies.20374
Chrtek, J., Herben, T., Rosenbaumová, R., Münzbergová, Z., Dočkalová, Z., Zahradníček, J., et al. (2017). Cytotype coexistence in the field cannot be explained by inter-cytotype hybridization alone: linking experiments and computer simulations in the sexual species Pilosella echioides (Asteraceae). BMC Evol. Biol. 17:87. doi: 10.1186/s12862-017-0934-y
Clark, J. W., and Donoghue, P. C. J. (2017). Constraining the timing of whole genome duplication in plant evolutionary history. Proc. Biol. Sci. 284, 20170912. doi: 10.1098/rspb.2017.0912
Coate, J. E., and Doyle, J. J. (2010). Quantifying whole transcriptome size, a prerequisite for understanding transcriptome evolution across species: an example from a plant allopolyploid. Genome Biol. Evol. 2, 534–546. doi: 10.1093/gbe/evq038
Coate, J. E., and Doyle, J. J. (2015). Variation in transcriptome size: are we getting the message? Chromosoma 124, 27–43. doi: 10.1007/s00412-014-0496-3
Coate, J. E., Schlueter, J. A., Whaley, A. M., and Doyle, J. J. (2011). Comparative evolution of photosynthetic genes in response to polyploid and nonpolyploid duplication. Plant Physiol. 155, 2081–2095. doi: 10.1104/pp.105.073304
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Cookson, S. J., Radziejwoski, A., and Granier, C. (2006). Cell and leaf size plasticity in arabidopsis: what is the role of endoreduplication? Plant Cell Environ. 29, 1273–1283. doi: 10.1111/j.1365-3040.2006.01506.x
Daehler, C. C. (1998). Variation in self-fertility and the reproductive advantage of self-fertility for an invading plant (Spartina alterniflora). Evol. Ecol. 12, 553–568. doi: 10.1023/A:1006556709662
del Pozo, J. C., and Ramirez-Parra, E. (2014). Deciphering the molecular bases for drought tolerance in A rabidopsis autotetraploids. Plant Cell Environ. 37, 2722–2737. doi: 10.1111/pce.12344
Dodsworth, S., Chase, M. W., and Leitch, A. R. (2016). Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Bot. J. Linn. Soc. 180, 1–5. doi: 10.1111/boj.12357
Drunen, W. E. Van, and Husband, B. C. (2018). Immediate vs. evolutionary consequences of polyploidy on clonal reproduction in an autopolyploid plant. Ann. Bot. 122, 195–205. doi: 10.1093/aob/mcy071
Edger, P. P., Smith, R., McKain, M. R., Cooley, A. M., Vallejo-Marin, M., Yuan, Y., et al. (2017) Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell 29, 2150–2167. doi: 10.1105/tpc.17.00010
Ehrendorfer, F. (1980). “Polyploidy and distribution,” in Polyploidy (Boston, MA: Springer US), 45–60.
Elmaghrabi, A. M., Ochatt, S., Rogers, H. J., and Francis, D. (2013). Enhanced tolerance to salinity following cellular acclimation to increasing NaCl levels in Medicago truncatula. Plant Cell Tissue Organ Cult. 114, 61–70. doi: 10.1007/s11240-013-0306-2
Escudero, M., Martín-Bravo, S., Mayrose, I., Fernández-Mazuecos, M., Fiz-Palacios, O., Hipp, A. L., et al. (2014) Karyotypic changes through dysploidy persist longer over evolutionary time than polyploid changes. PLoS ONE 9:e85266. doi: 10.1371/journal.pone.0085266
Favarger, C. (1984). Cytogeography and biosystematics. Plant Biosyst. 453–476. doi: 10.1016/B978-0-12-295680-5.50033-0
Fawcett, J. A., Maere, S., and Van de Peer, Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. U.S.A. 106, 5737–5742. doi: 10.1073/pnas.0900906106
Feldman, M. W., Christiansen, F. B., and Brooks, L. D. (1980). Evolution of recombination in a constant environment. Proc. Natl. Acad. Sci. U.S.A. 77, 4838–4841. doi: 10.1073/PNAS.77.8.4838
Feldman, M. W., Otto, S. P., and Christiansen, F. B. (1996). Population genetic perspective on the evolution of recombination. Ann. Rev. Genet. 30, 261–295. doi: 10.1146/annurev.genet.30.1.261
Fialová, M., and Duchoslav, M. (2014). Response to competition of bulbous geophyte Allium oleraceum differing in ploidy level. Plant Biol. 16, 186–196. doi: 10.1111/plb.12042
Franks, S. J., Sim, S., and Weis, A. E. (2007). Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. U.S.A. 104, 1278–1282. doi: 10.1073/pnas.0608379104
Franzke, A., Lysak, M. A., Al-Shehbaz, I. A., Koch, M. A., and Mummenhoff, K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16, 108–116. doi: 10.1016/J.TPLANTS.2010.11.005
Freeling, M., Xu, J., Woodhouse, M., and Lisch, D. (2015). A solution to the c-value paradox and the function of junk DNA: The genome balance hypothesis. Mol. Plant 8, 899–910. doi: 10.1016/j.molp.2015.02.009
Galloway, L. F., Etterson, J. R., and Hamrick, J. L. (2003). Outcrossing rate and inbreeding depression in the herbaceous autotetraploid, Campanula americana. Heredity 90, 308–315. doi: 10.1038/sj.hdy.6800242
Garbutt, K., and Bazzaz, F. A. (1983). Leaf demography, flower production and biomass of diploid and tetraploid populations of phlox drummondii hook. on a soil moisture gradient. New Phytol. 93, 129–141. doi: 10.1111/j.1469-8137.1983.tb02698.x
Gerstein, A. C., and Otto, S. P. (2009). Ploidy and the causes of genomic evolution. J. Heredity 100, 571–581. doi: 10.1093/jhered/esp057
Glennon, K. L., Ritchie, M. E., and Segraves, K. A. (2014). Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecol. Lett. 17, 574–582. doi: 10.1111/ele.12259
Goldblatt, P. (1980). “Polyploidy in angiosperms: monocotyledons,” in Polyploidy (Boston, MA: Springer US), 219–239.
Grant, V. (1981). Plant Speciation. New York, NY: Columbia University Press. xii, 563p.-illus., maps, chrom. nos. En 2nd Edn. Maps, Chromosome numbers. General (KR, 198300748).
Gregory, T. R. (2001). The bigger the C-value, the larger the cell: Genome size and red blood cell size in vertebrates. Blood Cells Mol. Dis. 27, 830–843. doi: 10.1006/bcmd.2001.0457
Gregory, T. R., and Mable, B. K. (2005). Chapter 8—polyploidy in animals. Evol. Genome 427–517. doi: 10.1016/B978-012301463-4/50010-3
Greiner, R., and Oberprieler, C. (2012). The role of inter-ploidy block for reproductive isolation of the diploid Leucanthemum pluriflorum Pau (Compositae, Anthemideae) and its tetra- and hexaploid relatives. Flora Morphol. Distrib. Funct. Ecol. Plants 207, 629–635. doi: 10.1016/J.FLORA.2012.07.001
Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. doi: 10.1093/emboj/20.3.589
Grotkopp, E., Rejmánek, M., and Rost, T. L. (2002). Toward a causal explanation of plant invasiveness: seedling growth and life-history strategies of 29 pine (Pinus) species. Am. Nat. 159, 396–419. doi: 10.1086/338995
Guo, M., Davis, D., and Birchler, J. A. (1996). Dosage effects on gene expression in a maize ploidy series. Genetics 142, 1349–1355.
Hagberg, A., and Elleström, S. (1959). The competition between diploid, tetraploid and aneuploid rye: theoretical and practical aspects. Hereditas 45, 369–416. doi: 10.1111/j.1601-5223.1959.tb03058.x
Hagerup, O. (1932). Über polyploidie in Beziehung zu Klima, Ökologie und phylogenie. Hereditas 16, 19–40. doi: 10.1111/j.1601-5223.1932.tb02560.x
Hahn, M. A., Buckley, Y. M., and Müller-Schärer, H. (2012). Increased population growth rate in invasive polyploid Centaurea stoebe in a common garden. Ecol. Lett. 15, 947–954. doi: 10.1111/j.1461-0248.2012.01813.x
Haldane, J. B. S. (1933). The part played by recurrent mutation in evolution. Am. Nat. 67, 5–19. doi: 10.1086/280465
Haldane, J. B. S. (1937). The effect of variation of fitness. Am. Nat. 71, 337–349. doi: 10.1086/280722
Haldane, J. B. S., Harrison, J. W. H., Garrett, F. C., Gager, C. S., Blakeslee, A. F., and Metz, C. W. (1927). A mathematical theory of natural and artificial selection, part V: selection and mutation. Math. Proc. Camb. Philos. Soc. 23, 838. doi: 10.1017/S0305004100015644
Hall, M. C., and Willis, J. H. (2006). Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution 60, 2466–2477. doi: 10.1111/j.0014-3820.2006.tb01882.x
Hartung, F., Wurz-Wildersinn, R., Fuchs, J., Schubert, I., Suer, S., and Puchta, H. (2007). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19, 3090–3099. doi: 10.1105/tpc.107.054817
Haufler, C. H. (1987). Electrophoresis is modifying our concepts of evolution in homosporous pteridophytes. Am. J. Bot. 74, 953. doi: 10.2307/2443877
Hegarty, M. J., Barker, G. L., Wilson, I. D., Abbott, R. J., Edwards, K. J., and Hiscock, S. J. (2006). Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr. Biol. 16, 1652–1659. doi: 10.1016/J.CUB.2006.06.071
Herben, T., Suda, J., and Klimešová, J. (2017). Polyploid species rely on vegetative reproduction more than diploids: a re-examination of the old hypothesis. Ann. Bot. 120, 341–349. doi: 10.1093/aob/mcx009
Hill, R. R. (1971). Selection in autotetraploids. Theoret. Appl. Genet. 41, 181–186. doi: 10.1007/bf00277621
Hollingsworth, M. L., and Bailey, J. P. (2000). Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed). Bot. J. Linn. Soc. 133, 463–472. doi: 10.1006/bojl.2000.0359
Hollister, J. D. (2015). Polyploidy: adaptation to the genomic environment. New Phytol. 205, 1034–1039. doi: 10.1111/nph.12939
Hollister, J. D., Arnold, B. J., Svedin, E., Xue, K. S., Dilkes, B. P., and Bomblies, K. (2012). Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet. 8:e1003093. doi: 10.1371/journal.pgen.1003093
Hollister, J. D., and Gaut, B. S. (2009). Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19, 1419–1428. doi: 10.1101/gr.091678.109
Horváth, V., Merenciano, M., and González, J. (2017). Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends Genet. 33, 832–841. doi: 10.1016/j.tig.2017.08.007
Hülber, K., Sonnleitner, M., Suda, J., Krejčíková, J., Schönswetter, P., Schneeweiss, G. M., et al. (2015). Ecological differentiation, lack of hybrids involving diploids, and asymmetric gene flow between polyploids in narrow contact zones of Senecio carniolicus (syn. Jacobaea carniolica, Asteraceae). Ecol. Evol. 5, 1224–1234. doi: 10.1002/ece3.1430
Husband, B. C. (2000). Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proc. R. Soc. B Biol. Sci. 267, 217–223. doi: 10.1098/rspb.2000.0990
Husband, B. C. (2004). The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biol. J. Linn. Soc. 82, 537–546. doi: 10.1111/j.1095-8312.2004.00339.x
Husband, B. C., Baldwin, S. J., and Sabara, H. A. (2016). Direct vs. indirect effects of whole-genome duplication on prezygotic isolation in Chamerion angustifolium: implications for rapid speciation. Am. J. Bot. 103, 1259–1271. doi: 10.3732/ajb.1600097
Husband, B. C., Ozimec, B., Martin, S. L., and Pollock, L. (2008). Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. Int. J. Plant Sci. 169, 195–206. doi: 10.1086/523367
Husband, B. C., and Sabara, H. A. (2003). Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytol. 161, 703–713. doi: 10.1046/j.1469-8137.2003.00998.x
Husband, B. C., and Schemske, D. W. (1997). The effect of inbreeding in diploid and tetraploid populations of Epilobium angustifolium (onagraceae): implications for the genetic basis of inbreeding depression. Evolution 51, 737–746. doi: 10.1111/j.1558-5646.1997.tb03657.x
Husband, B. C., Schemske, D. W., Burton, T. L., and Goodwillie, C. (2002). Pollen competition as a unilateral reproductive barrier between sympatric diploid and tetraploid Chamerion angustifolium. Proc. R. Soc. B Biol. Sci. 269, 2565–2571. doi: 10.1098/rspb.2002.2196
Ito, H., and Kakutani, T. (2014). Control of transposable elements in Arabidopsis thaliana. Chromosome Res. 22, 217–223. doi: 10.1007/s10577-014-9417-9
Ito, H., Kim, J. M., Matsunaga, W., Saze, H., Matsui, A., Endo, T. A., et al. (2016). A stress-activated transposon in arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 6, 1–12. doi: 10.1038/srep23181
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Johnston, M. O., and Schoen, D. J. (1996). Correlated evolution of self-fertilization and inbreeding depression: an experimental study of nine populations of Amsinckia (Boragainaceae). Evolution 50, 1478–1491.
Jørgensen, M. H., Ehrich, D., Schmickl, R., Koch, M. A., and Brysting, A. K. (2011). Interspecific and interploidal gene flow in Central European Arabidopsis (Brassicaceae). BMC Evol. Biol. 11:346. doi: 10.1186/1471-2148-11-346
Jung, C., and Müller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–73. doi: 10.1016/j.tplants.2009.07.005
Kadereit, J. W. (2015). The geography of hybrid speciation in plants. Taxon 64, 673–687. doi: 10.12705/644.1
Kao, R. H. (2007). Asexuality and the coexistence of cytotypes. New Phytol. 175, 764–772. doi: 10.1111/j.1469-8137.2007.02145.x
Kashkush, K., Feldman, M., and Levy, A. A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106. doi: 10.1038/ng1063
Knight, C. A., and Beaulieu, J. M. (2008). Genome size scaling through phenotype space. Ann. Bot. 101, 759–766. doi: 10.1093/aob/mcm321
Knight, C. A., Molinari, N. A., and Petrov, D. A. (2005). The large genome constraint hypothesis: evolution, ecology and phenotype. Ann. Bot. 95, 177–190. doi: 10.1093/aob/mci011
Köhler, C., Mittelsten Scheid, O., and Erilova, A. (2010). The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 26, 142–148. doi: 10.1016/J.TIG.2009.12.006
Kolár, F., Certner, M., Suda, J., Schönswetter, P., and Husband, B. C. (2017). Mixed-ploidy species: progress and opportunities in polyploid research. Trends Plant Sci. 22, 1041–1055. doi: 10.1016/J.TPLANTS.2017.09.011
Korona, R. (1999). Unpredictable fitness transitions between haploid and diploid strains of the genetically loaded yeast Saccharomyces cerevisiae. Genetics 151, 77–85.
Koutecký, P., Badurová, T., Štech, M., Košnar, J., and Karásek, J. (2011). Hybridization between diploid Centaurea pseudophrygia and tetraploid C. jacea (Asteraceae): the role of mixed pollination, unreduced gametes, and mentor effects. Biol. J. Linn. Soc. 104, 93–106. doi: 10.1111/j.1095-8312.2011.01707.x
Kubátová, B., Trávníček, P., Bastlová, D., Curn, V., Jarolímová, V., and Suda, J. (2007). DNA ploidy-level variation in native and invasive populations of Lythrum salicaria at a large geographical scale. J. Biogeogr. 35, 167–176. doi: 10.1111/j.1365-2699.2007.01781.x
Lafon-Placette, C., Johannessen, I. M., Hornslien, K. S., Ali, M. F., Bjerkan, K. N., Bramsiepe, J., et al. (2017). Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrata and Arabidopsis arenosa in Central Europe. Proc. Natl. Acad. Sci. U.S.A. 114, E1027–E1035. doi: 10.1073/pnas.1615123114
Lafuma, L., Balkwill, K., Imbert, E., Verlaque, R., and Maurice, S. (2003). Ploidy level and origin of the European invasive weed Senecio inaequidens (Asteraceae). Plant Syst. Evol. 243, 59–72. doi: 10.1007/s00606-003-0075-0
Lampson, M. A., and Cheeseman, I. M. (2011). Sensing centromere tension: aurora B and the regulation of kinetochore function. Trends Cell Biol. 21, 133–140. doi: 10.1016/J.TCB.2010.10.007
Lande, R., and Schemske, D. W. (1985). The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39, 24–40. doi: 10.2307/2408514
Le Comber, S. C., Ainouche, M. L., Kovarik, A., and Leitch, A. R. (2010). Making a functional diploid: from polysomic to disomic inheritance. New Phytol. 186, 113–122. doi: 10.1111/j.1469-8137.2009.03117.x
Leitch, A. R., and Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320, 481–3. doi: 10.1126/science.1153585
Leitch, I. J., and Bennett, M. D. (2004). Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 82, 651–663. doi: 10.1111/j.1095-8312.2004.00349.x
Leitch, I. J., Hanson, L., Lim, K. Y., Kovarik, A., Chase, M. W., Clarkson, J. J., et al. (2008). The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann. Bot. 101, 805–814. doi: 10.1093/aob/mcm326
Lenormand, T., and Otto, S. P. (2000). The evolution of recombination in a heterogeneous environment. Genetics 156, 423–438.
Levin, D. A. (1975). Minority cytotype exclusion in local plant populations. Taxon 24, 35. doi: 10.2307/1218997
Levin, D. A. (2002). The Role of Chromosomal Change in Plant Evolution. New York, NY: Oxford University Press. Available online at: https://books.google.fr/books?hl=en&lr=&id=XinRCwAAQBAJ&oi=fnd&pg=PR9&dq=levin+2002+polyploidy&ots=yKoYFhdKcV&sig=nvaDxt6y7DE9mD6QXttyO6Ermao&redir_esc=y#v=onepage&q&f=false (Accessed April 14, 2018).
Lewontin, R. C. (1971). The effect of genetic linkage on the mean fitness of a population. Proc. Natl. Acad. Sci. U.S.A. 68, 984–986. doi: 10.1073/PNAS.68.5.984
Lim, K. Y., Kovarik, A., Matyasek, R., Chase, M. W., Clarkson, J. J., Grandbastien, M. A., et al. (2007). Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 175, 756–763. doi: 10.1111/j.1469-8137.2007.02121.x
Lopes, F. R., Jjingo, D., Da Silva, C. R. M., Andrade, A. C., Marraccini, P., Teixeira, J. B., et al. (2013). Transcriptional activity, chromosomal distribution and expression effects of transposable elements in Coffea genomes. PLoS ONE 8:e78931. doi: 10.1371/journal.pone.0078931
Lovén, J., Orlando, D. A., Sigova, A. A., Lin, C. Y., Rahl, P. B., Burge, C. B., et al. (2012). Revisiting global gene expression analysis. Cell 151, 476–482. doi: 10.1016/J.CELL.2012.10.012
Luo, M. C., You, F. M., Li, P., Wang, J. R., Zhu, T., Dandekar, A. M., et al. (2015). Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials. BMC Genomics 16:707. doi: 10.1186/s12864-015-1906-5
Lysak, M. A., Cheung, K., Kitschke, M., and Bures, P. (2007). Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 145, 402–410. doi: 10.1104/pp.107.104380
Mable, B. K., and Otto, S. P. (2001). Masking and purging mutations following EMS treatment in haploid, diploid and tetraploid yeast (Saccharomyces cerevisiae). Genet. Res. 77, 9–26.
Maceira, N. O., Jacquard, P., and Lumaret, R. (1993). Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytol. 124, 321–328.
Madlung, A., Tyagi, A. P., Watson, B., Jiang, H., Kagochi, T., Doerge, R. W., et al. (2005). Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41, 221–230. doi: 10.1111/j.1365-313X.2004.02297.x
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Mandák, B., Bímová, K., Pyšek, P., Štěpánek, J., and Plačková, I. (2005). Isoenzyme diversity in Reynoutria (Polygonaceae) taxa: escape from sterility by hybridization. Plant Syst. Evol. 253, 219–230. doi: 10.1007/s00606-005-0316-6
Mandák, B., Pyšek, P., Lysák, M., Suda, J., Krahulcová, A., and Bímová, K. (2003). Variation in DNA-ploidy levels of reynoutria taxa in the Czech Republic. Ann. Bot. 92, 265–272. doi: 10.1093/aob/mcg141
Mandáková, T., Li, Z., Barker, M. S., and Lysak, M. A. (2017). Diverse genome organization following 13 independent mesopolyploid events in Brassicaceae contrasts with convergent patterns of gene retention. Plant J. 91, 3–21. doi: 10.1111/tpj.13553
Mandáková, T., and Lysak, M. A. (2018). Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 42, 55–65. doi: 10.1016/j.pbi.2018.03.001
Manning, J. T., and Dickson, D. P. E. (1986). Asexual reproduction, polyploidy and optimal mutation rates. J. Theoret. Biol. 118, 485–489. doi: 10.1016/S0022-5193(86)80166-7
Masterson, J. (1994). Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264, 421–424. doi: 10.1126/science.264.5157.421
Masuyama, S., and Watano, Y. (1990). Trends for inbreeding in polyploid pteridophytes. Plant Species Biol. 5, 13–17.
Mayrose, I., Zhan, S. H., Rothfels, C. J., Arrigo, N., Barker, M. S., Rieseberg, L. H., et al. (2015). Methods for studying polyploid diversification and the dead end hypothesis: a reply to Soltis et al. (2014). New Phytol. 206, 27–35. doi: 10.1111/nph.13192
Mayrose, I., Zhan, S. H., Rothfels, C. J., Magnuson-Ford, K., Barker, M. S., Rieseberg, L. H., et al. (2011). Recently formed polyploid plants diversify at lower rates. Science 333, 1257. doi: 10.1126/science.1207205
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. doi: 10.1126/science.15739260
Meng, H. B., Jiang, S. S., Hua, S. J., Lin, X. Y., Li, Y. L., Guo, W. L., et al. (2011). Comparison between a tetraploid turnip and its diploid progenitor (Brassica rapa L.): the adaptation to salinity stress. Agric. Sci. China 10, 363–375. doi: 10.1016/S1671-2927(11)60015-1
Mills, R. E., Bennett, E. A., Iskow, R. C., and Devine, S. E. (2007). Which transposable elements are active in the human genome? Trends Genet. 23, 183–191. doi: 10.1016/j.tig.2007.02.006
Moody, M. E., Mueller, L. D., and Soltis, D. E. (1993). Genetic variation and random drift in autotetraploid populations. Genetics 134, 649–657.
Mráz, P., Garcia-Jacas, N., Gex-Fabry, E., Susanna, A., Barres, L., and Müller-Schärer, H. (2012). Allopolyploid origin of highly invasive Centaurea stoebe s.l. (Asteraceae). Mol. Phylogenet. Evol. 62, 612–623. doi: 10.1016/J.YMPEV.2011.11.006
Müntzing, A. (1936). The evolutionary significance of autopolyploidy. Hereditas 21, 363–378. doi: 10.1111/j.1601-5223.1936.tb03204.x
Münzbergová, Z. (2007). Population dynamics of diploid and hexaploid populations of a perennial herb. Ann. Bot. 100, 1259–1270. doi: 10.1093/aob/mcm204
Murat, F., Xu, J.-H., Tannier, E., Abrouk, M., Guilhot, N., Pont, C., et al. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20, 1545–57. doi: 10.1101/gr.109744.110
Novikova, P. Y., Hohmann, N., and Van de Peer, Y. (2018). Polyploid Arabidopsis species originated around recent glaciation maxima. Curr. Opin. Plant Biol. 42, 8–15. doi: 10.1016/j.pbi.2018.01.005
Osborn, T. C., Chris Pires, J., Birchler, J. A., Auger, D. L., Jeffery Chen, Z., Lee, H.-S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. doi: 10.1016/S0168-9525(03)00015-5
Oswald, B. P., and Nuismer, S. L. (2011). A unified model of autopolyploid establishment and evolution. Am. Nat. 178, 687–700. doi: 10.1086/662673
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. doi: 10.1016/j.cell.2007.10.022
Otto, S. P., and Michalakis, Y. (1998). The evolution of recombination in changing environments. Trends Ecol. Evol. 13, 145–151. doi: 10.1016/S0169-5347(97)01260-3
Otto, S. P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34:401–437. doi: 10.1146/annurev.genet.34.1.40
Page, S. L., and Hawley, R. S. (2003). Chromosome chreography: the meiotic ballet. Science 301, 785–789. doi: 10.1126/science.1086605
Pandit, M. K., Pocock, M. J. O., and Kunin, W. E. (2011). Ploidy influences rarity and invasiveness in plants. J. Ecol. 99, 1108–1115. doi: 10.1111/j.1365-2745.2011.01838.x
Pandit, M. K., Tan, H. T. W., and Bisht, M. S. (2006). Polyploidy in invasive plant species of Singapore. Bot. J. Linn. Soc. 151, 395–403. doi: 10.1111/j.1095-8339.2006.00515.x
Pannell, J. R., and Barrett, S. C. H. (1998). Baker's law revisited: reproductive assurance in a metapopulation. Evolution 52, 657–668. doi: 10.1111/j.1558-5646.1998.tb03691.x
Papp, B., Pál, C., and Hurst, L. D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. doi: 10.1038/nature01771
Parisod, C., Holderegger, R., and Brochmann, C. (2010). Evolutionary consequences of autopolyploidy. New Phytol. 186, 5–17. doi: 10.1111/j.1469-8137.2009.03142.x
Parisod, C., Salmon, A., Zerjal, T., Tenaillon, M., Grandbastien, M.-A., and Ainouche, M. (2009). Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 184, 1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x
Paterson, A. H., Bowers, J. E., and Chapman, B. A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U.S.A. 101, 9903–9908. doi: 10.1073/pnas.0307901101
Pfeil, B. E., Schlueter, J. A., Shoemaker, R. C., and Doyle, J. J. (2005). Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst. Biol. 54, 441–454. doi: 10.1080/10635150590945359
Piednoël, M., Sousa, A., and Renner, S. S. (2015). Transposable elements in a clade of three tetraploids and a diploid relative, focusing on Gypsy amplification. Mob. DNA 6, 5. doi: 10.1186/s13100-015-0034-8
Prentis, P. J., Wilson, J. R. U., Dormontt, E. E., Richardson, D. M., and Lowe, A. J. (2008). Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–94. doi: 10.1016/j.tplants.2008.03.004
Price, S. C., and Jain, S. K. (1981). Are inbreeders better colonizers? Oecologia 49, 283–286. doi: 10.1007/BF00349202
Pyšek, P., Jarošík, V., Pergl, J., Randall, R., Chytrý, M., Kühn, I., et al. (2009). The global invasion success of Central European plants is related to distribution characteristics in their native range and species traits. Divers. Distributions 15, 891–903. doi: 10.1111/j.1472-4642.2009.00602.x
Quadrana, L., Silveira, A. B., Mayhew, G. F., LeBlanc, C., Martienssen, R. A., Jeddeloh, J. A., et al. (2016). The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5, 1–25. doi: 10.7554/eLife.15716
Ramsey, J. (2011). Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. 108, 7096–7101. doi: 10.1073/pnas.1016631108
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Ramsey, J., and Schemske, D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639. doi: 10.1146/annurev.ecolsys.33.010802.150437
Raven, P. H. (1975). The bases of angiosperm phylogeny: cytology. Ann. Missouri Bot. Garden 62, 724. doi: 10.2307/2395272
Renny-Byfield, S., Ainouche, M., Leitch, I. J., Lim, K. Y., Le Comber, S. C., and Leitch, A. R. (2010). Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of S. alterniflora and S. maritima origin. Ann. Bot. 105, 527–533. doi: 10.1093/aob/mcq008
Rey, P. J., Manzaneda, A. J., and Alcántara, J. M. (2017). The interplay between aridity and competition determines colonization ability, exclusion and ecological segregation in the heteroploid Brachypodium distachyon species complex. New Phytol. 215, 85–96. doi: 10.1111/nph.14574
Richards, C. L., Bossdorf, O., Muth, N. Z., Gurevitch, J., and Pigliucci, M. (2006). Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9, 981–993. doi: 10.1111/j.1461-0248.2006.00950.x
Rodríguez, D. J. (1996). A model for the establishment of polyploidy in plants. Am. Nat. 147, 33–46. doi: 10.2307/2463222
Ronfort, J. (1999). The mutation load under tetrasomic inheritance and its consequences for the evolution of the selfing rate in autotetraploid species. Genet. Res. 74, 31–42. doi: 10.1017/S0016672399003845
Roose, M. L., and Gottlieb, L. D. (1976). Genetic and biochemical consequences of polyploidy in tragopogon. Evolution 30, 818. doi: 10.2307/2407821
Ruprecht, C., Lohaus, R., Vanneste, K., Mutwil, M., Nikoloski, Z., Van De Peer, Y., and Persson, S. (2017). Revisiting ancestral polyploidy in plants. Science Adv. 3, 1–7. doi: 10.1126/sciadv.1603195
Sabara, H. A., Kron, P., and Husband, B. C. (2013). Cytotype coexistence leads to triploid hybrid production in a diploid-tetraploid contact zone of Chamerion angustifolium (Onagraceae). Am. J. Bot. 100, 962–970. doi: 10.3732/ajb.1200583
Saleh, B., Allario, T., Dambier, D., Ollitrault, P., and Morillon, R. (2008). Tetraploid citrus rootstocks are more tolerant to salt stress than diploid. C. R. Biol. 331, 703–710. doi: 10.1016/j.crvi.2008.06.007
Salman-Minkov, A., Sabath, N., and Mayrose, I. (2016). Whole-genome duplication as a key factor in crop domestication. Nat. Plants 2, 16115. doi: 10.1038/nplants.2016.115
Sano, Y., Morishima, H., and Oka, H.-I. (1980). Intermediate perennial-annual populations ofOryza perennis found in Thailand and their evolutionary significance. Bot. Mag. (Tokyo) 93, 291–305. doi: 10.1007/BF02488735
Sarilar, V., Marmagne, A., Brabant, P., Joets, J., and Alix, K. (2011). BraSto, a Stowaway MITE from Brassica: recently active copies preferentially accumulate in the gene space. Plant Mol. Biol. 77, 59–75. doi: 10.1007/s11103-011-9794-9
Schemske, D. W., and Lande, R. (1985). The evolution of self-fertilization and inbreeding depression in plants. II. Empirical observations. Evolution 39, 41. doi: 10.2307/2408515
Schinkel, C. C. F., Kirchheimer, B., Dellinger, A. S., Klatt, S., Winkler, M., Dullinger, S., et al. (2016). Correlations of polyploidy and apomixis with elevation and associated environmental gradients in an alpine plant. AoB Plants 8, plw064. doi: 10.1093/aobpla/plw064
Schlaepfer, D. R., Edwards, P. J., Semple, J. C., and Billeter, R. (2008). Cytogeography of Solidago gigantea (Asteraceae) and its invasive ploidy level. J. Biogeogr. 35, 2119–2127. doi: 10.1111/j.1365-2699.2008.01937.x
Schlueter, J. A., Dixon, P., Granger, C., Grant, D., Clark, L., Doyle, J. J., et al. (2004). Mining EST databases to resolve evolutionary events in major crop species. Genome 47, 868–876. doi: 10.1139/g04-047
Schmickl, R., Marburger, S., Bray, S., and Yant, L. (2017). Hybrids and horizontal transfer: introgression allows adaptive allele discovery. J. Exp. Bot. 68, 5453–5470. doi: 10.1093/jxb/erx297
Schoustra, S. E., Debets, A. J. M., Slakhorst, M., and Hoekstra, R. F. (2007). Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet. 3:e68. doi: 10.1371/journal.pgen.0030068
Schranz, M. E., Mohammadin, S., and Edger, P. P. (2012). Ancient whole genome duplications, novelty and diversification: the WGD radiation lag-time model. Curr. Opin. Plant Biol. 15, 147–153. doi: 10.1016/J.PBI.2012.03.011
Selmecki, A. M., Maruvka, Y. E., Richmond, P. A., Guillet, M., Shoresh, N., Sorenson, A. L., et al. (2015) Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352. doi: 10.1038/nature14187.
Senerchia, N., Felber, F., and Parisod, C. (2014). Contrasting evolutionary trajectories of multiple retrotransposons following independent allopolyploidy in wild wheats. New Phytol. 202, 975–985. doi: 10.1111/nph.12731
Sherrard, M. E., and Maherali, H. (2006). The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution 60, 2478–2489. doi: 10.1111/j.0014-3820.2006.tb01883.x
Shi, T., Huang, H., and Barker, M. S. (2010). Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Ann. Bot. 106, 497–504. doi: 10.1093/aob/mcq129
Shi, X., Zhang, C., Ko, D. K., and Chen, Z. J. (2015). Genome-wide dosage-dependent and -independent regulation contributes to gene expression and evolutionary novelty in plant polyploids. Mol. Biol. Evol. 32, 2351–2366. doi: 10.1093/molbev/msv116
Simon-Porcar, V. I., Silva, J. L., Higgins, J. D., and Vallejo-Marin, M. (2017). Recent autopolyploidisation in a wild population of Mimulus guttatus (Phrymaceae). Bot. J. Linn. Soc. 185, 189–207. doi: 10.1093/botlinnean/box052
Slotte, T., Huang, H., Lascoux, M., and Ceplitis, A. (2008). Polyploid speciation did not confer instant reproductive isolation in Capsella (Brassicaceae). Mol. Biol. Evol. 25, 1472–1481. doi: 10.1093/molbev/msn092
Smith, H. E. (1946). Sedum pulchellum: a physiological and morphological comparison of diploid, tetraploid, and hexaploid races. Bull. Torrey Bot. Club 73, 495. doi: 10.2307/2481337
Solhaug, E. M., Ihinger, J., Jost, M., Gamboa, V., Marchant, B., Bradford, D., et al. (2016). Environmental regulation of heterosis in the allopolyploid Arabidopsis suecica. Plant Physiol. 170, 2251–2263. doi: 10.1104/pp.16.00052
Soltis, D. E., and Soltis, P. S. (1989). Genetic consequences of autopolyploidy in Tolmiea (Saxifragaceae). Evolution 43, 586. doi: 10.2307/2409061
Soltis, D. E., Soltis, P. S., and Rieseberg, L. H. (1993). Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci. 12, 243–273. doi: 10.1080/07352689309701903
Soltis, D. E., Soltis, P. S., Schemske, D. W., Hancock, J. F., Thompson, J. N., Husband, B. C., et al. (2007). Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56, 13–30. doi: 10.2307/25065732
Soltis, D. E., Visger, C. J., Marchant, D. B., and Soltis, P. S. (2016). Polyploidy: pitfalls and paths to a paradigm. Am. J. Bot. 103, 1146–66. doi: 10.3732/ajb.1500501
Soltis, P. S., Marchant, D. B., Van de Peer, Y., and Soltis, D. E. (2015). Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125. doi: 10.1016/J.GDE.2015.11.003
Soltis, P. S., and Soltis, D. E. (2000). The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. 97, 7051–7057. doi: 10.1073/pnas.97.13.7051
Soltis, P. S., and Soltis, D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588. doi: 10.1146/annurev.arplant.043008.092039
Sonnleitner, M., Weis, B., Flatscher, R., García, P. E., Suda, J., Krejčíková, J., et al. (2013) Parental ploidy strongly affects offspring fitness in heteroploid crosses among three cytotypes of autopolyploid Jacobaea carniolica (Asteraceae). PLoS ONE 8:e78959. doi: 10.1371/journal.pone.0078959
Spoelhof, J. P., Soltis, P. S., and Soltis, D. E. (2017). Pure polyploidy: closing the gaps in autopolyploid research. J. Syst. Evol. 55, 340–352. doi: 10.1111/jse.12253
Springer, N. M., Lisch, D., and Li, Q. (2016). Creating order from Chaos: epigenome dynamics in plants with complex genomes. Plant Cell 28, 314–325. doi: 10.1105/tpc.15.00911
Ståhlberg, D. (2009). Habitat differentiation, hybridization and gene flow patterns in mixed populations of diploid and autotetraploid Dactylorhiza maculata s.l. (Orchidaceae). Evol. Ecol. 23, 295–328. doi: 10.1007/s10682-007-9228-y
Stacey, N. J., Kuromori, T., Azumi, Y., Roberts, G., Breuer, C., Wada, T., et al. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48, 206–216. doi: 10.1111/j.1365-313X.2006.02867.x
Stebbins, G. L. (1938). Cytological characteristics associated with the different growth habits in the dicotyledons. Am. J. Bot. 25, 189–198.
Stephens, A. D., Snider, C. E., Haase, J., Haggerty, R. A., Vasquez, P. A., Gregory Forest, M., et al. (2013). Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J. Cell Biol. 203, 407–416. doi: 10.1083/jcb.201307104
Storchova, Z., and Pellman, D. (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45–54. doi: 10.1038/nrm1276
Storlazzi, A., Gargano, S., Ruprich-Robert, G., Falque, M., David, M., Kleckner, N., et al. (2010). Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141, 94–106. doi: 10.1016/j.cell.2010.02.041
Stupar, R. M., Bhaskar, P. B., Yandell, B. S., Rensink, W. A., Hart, A. L., Ouyang, S., et al. (2007). Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 176, 2055–67. doi: 10.1534/genetics.107.074286
Sutherland, B. L., and Galloway, L. F. (2017). Postzygotic isolation varies by ploidy level within a polyploid complex. New Phytol. 213, 404–412. doi: 10.1111/nph.14116
Tank, D. C., Eastman, J. M., Pennell, M. W., Soltis, P. S., Soltis, D. E., Hinchliff, C. E., et al. (2015). Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol. 207, 454–467. doi: 10.1111/nph.13491
te Beest, M., Le Roux, J. J., Richardson, D. M., Brysting, A. K., Suda, J., Kubesova, M., et al. (2012). The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 109, 19–45. doi: 10.1093/aob/mcr277
Theodoridis, S., Randin, C., Broennimann, O., Patsiou, T., and Conti, E. (2013). Divergent and narrower climatic niches characterize polyploid species of European primroses in Primula sect. Aleuritia. J. Biogeogr. 40, 1278–1289. doi: 10.1111/jbi.12085
Thomas, B. C., Pedersen, B., and Freeling, M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16, 934–46. doi: 10.1101/gr.4708406
Thompson, K. A., Husband, B. C., and Maherali, H. (2015). No influence of water limitation on the outcome of competition between diploid and tetraploid Chamerion angustifolium (Onagraceae). J. Ecol. 103, 733–741. doi: 10.1111/1365-2745.12384
Thompson, S. L., and Whitton, J. (2006). Patterns of recurrent evolution and geographic parthenogenesis within apomictic polyploid Easter daises (Townsendia hookeri). Mol. Ecol. 15, 3389–3400. doi: 10.1111/j.1365-294X.2006.03020.x
Tittel-Elmer, M., Bucher, E., Broger, L., Mathieu, O., Paszkowski, J., and Vaillant, I. (2010). Stress-induced activation of heterochromatic transcription. PLoS Genet. 6:e1001175. doi: 10.1371/journal.pgen.1001175
Treier, U. A., Broennimann, O., Normand, S., Guisan, A., Schaffner, U., Steinger, T., et al. (2009). Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology 90, 1366–1377. doi: 10.1890/08-0420.1
Tsukahara, S., Kawabe, A., Kobayashi, A., Ito, T., Aizu, T., Shin-i, T., et al. (2012). Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes Dev. 26, 705–13. doi: 10.1101/gad.183871.111
Vallejo-Marín, M., and Hiscock, S. J. (2016). Hybridization and hybrid speciation under global change. New Phytol. 211, 1170–1187. doi: 10.1111/nph.14004
Van't Hof, A. E., Campagne, P., Rigden, D. J., Yung, C. J., Lingley, J., Quail, M. A., et al. (2016). The industrial melanism mutation in British peppered moths is a transposable element. Nature 534, 102–105. doi: 10.1038/nature17951
Vicient, C. M., and Casacuberta, J. M. (2017). Impact of transposable elements on polyploid plant genomes. Ann. Bot. 120, 195–207. doi: 10.1093/aob/mcx078
Vicient, C. M., Suoniemi, A., Anamthawat-Jónsson, K., Tanskanen, J., Beharav, A., Nevo, E., et al. (1999). Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11, 1769–1784. doi: 10.1105/TPC.11.9.1769
Visger, C. J., Wong, G. K., Zhang, Y., Soltis, P. S., Soltis, D. E., Affiliation, A., et al. (2017). Divergent gene expression levels between diploid and autotetraploid. bioRxiv. doi: 10.1101/169367
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Wang, Z., Wang, M., Liu, L., and Meng, F. (2013). Physiological and proteomic responses of diploid and tetraploid black locust (Robinia pseudoacacia L.) subjected to salt stress. Int. J. Mol. Sci. 14, 20299–20325. doi: 10.3390/ijms141020299
Wendel, J. F. (2015). The wondrous cycles of polyploidy in plants. Am. J. Bot. 102, 1753–6. doi: 10.3732/ajb.1500320
Willis, J. H. (1999). Inbreeding load, average dominance and the mutation rate for mildly deleterious alleles in Mimulus guttatus. Genetics 153, 1885–1898.
Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., and Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. U.S.A. 106, 13875–9. doi: 10.1073/pnas.0811575106
Yaakov, B., and Kashkush, K. (2012). Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol. Biol. 80, 419–427. doi: 10.1007/s11103-012-9957-3
Yamauchi, A., Hosokawa, A., Nagata, H., and Shimoda, M. (2004). Triploid bridge and role of parthenogenesis in the evolution of autopolyploidy. Am. Nat. 164, 101–112. doi: 10.1086/421356
Yant, L., and Bomblies, K. (2017). Genomic studies of adaptive evolution in outcrossing Arabidopsis species. Curr. Opin. Plant Biol. 36, 9–14. doi: 10.1016/j.pbi.2016.11.018
Yant, L., Hollister, J. D., Wright, K. M., Arnold, B. J., Higgins, J. D., Franklin, F. C. H., et al. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23, 2151–2156. doi: 10.1016/j.cub.2013.08.059
Yu, Z., Haberer, G., Matthes, M., Rattei, T., Mayer, K. F. X., Gierl, A., et al. (2010). Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 17809–17814. doi: 10.1073/pnas.1000852107
Zhang, J., Liu, Y., Xia, E.-H., Yao, Q.-Y., Liu, X.-D., and Gao, L.-Z. (2015). Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. U.S.A. 112, E7022–E7029. doi: 10.1073/pnas.1515170112
Zhang, X., Deng, M., and Fan, G. (2014). Differential transcriptome analysis between Paulownia fortunei and its synthesized autopolyploid. Int. J. Mol. Sci. 15, 5079–5093. doi: 10.3390/ijms15035079
Zohren, J., Wang, N., Kardailsky, I., Borrell, J. S., Joecker, A., Nichols, R. A., et al. (2016). Unidirectional diploid-tetraploid introgression among British birch trees with shifting ranges shown by restriction site-associated markers. Mol. Ecol. 25, 2413–2426. doi: 10.1111/mec.13644
Keywords: polyploidy, selection, population genetics, evolution, autopolyploidy, genome duplication
Citation: Baduel P, Bray S, Vallejo-Marin M, Kolář F and Yant L (2018) The “Polyploid Hop”: Shifting Challenges and Opportunities Over the Evolutionary Lifespan of Genome Duplications. Front. Ecol. Evol. 6:117. doi: 10.3389/fevo.2018.00117
Received: 25 April 2018; Accepted: 23 July 2018;
Published: 20 August 2018.
Edited by:
Richard John Abbott, University of St Andrews, United KingdomReviewed by:
Barbara K. Mable, University of Glasgow, United KingdomCopyright © 2018 Baduel, Bray, Vallejo-Marin, Kolář and Yant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Levi Yant, bGV2aXlhbnRAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.