
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Ecol. Evol. , 18 July 2018
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 6 - 2018 | https://doi.org/10.3389/fevo.2018.00100
This article is part of the Research Topic Evolution, Phylogeny and Systematics of Euglenozoa View all 4 articles
Several lineages of euglenozoans are enveloped with epibiotic bacteria and live in low oxygen and anoxic marine sediments, such as Bihospites bacati and Calkinsia aureus. A combination of shared ultrastructural traits and molecular phylogenetic inferences demonstrate that these lineages belong to a clade called the “Symbiontida.” Bihospites and Calkinsia possess all of the synapomorphies for the Euglenozoa plus several novel traits. Bihospites has a distinctive cell surface organization reminiscent of the pellicle strips in euglenids, a robust C-shaped feeding apparatus that encircles the nucleus, and a diverse community of epibiotic bacteria. Calkinsia has a novel “extrusomal pocket” and a thick (orange) extracellular matrix beneath a uniform layer of epibiotic bacteria. Despite the absence of molecular phylogenetic data, similar ultrastructural traits in Postgaardi mariagerensis and its epibiotic bacteria strongly suggest that this species is also a member of the Symbiontida. Molecular phylogenetic trees inferred from small subunit (SSU) ribosomal DNA sequences have shown that Bihospites and Calkinsia group strongly with a diverse set of environmental DNA sequences (eDNA) generated from low-oxygen marine samples collected at different depths from different locations around the world. These data demonstrate a diverse array of symbiontids that have yet to be characterized at the genomic, cellular, and behavior levels, which underscores how poorly we currently understand the biology and ecology of the group. Moreover, current data suggest that the communities of epibiotic bacteria associated with Bihospites, Calkinsia, and Postgaardi co-evolved with their hosts and are metabolically integrated with modified mitochondria positioned immediately beneath the host's plasma membrane. No symbiontid species has ever been cultivated, so improved knowledge about these eukaryotic organisms and their intimate relationships with bacteria in low oxygen environments will likely be achieved using culture-independent approaches, such as isolated-cell metagenomics.
Advances in molecular phylogenetic analyses and next generation sequence data from diverse environmental samples have revealed that a large number of uncharacterized lineages of microbial eukaryotes—protists—exist in aquatic environments around the world. Although environmental DNA (eDNA) surveys help demonstrate the extent of protist diversity, the cellular and behavioral traits of these organisms remain invisible from DNA sequence data alone. The diversity of several different groups of relatively tiny (<20 microns) heterotrophic protists is known primarily from eDNA data, such as so-called “MAST” (marine stramenopiles) clades, “MALV” (marine alveolate) clades and diplonemid clades (Massana et al., 2004, 2014; Guillou et al., 2008; Flegontova et al., 2016). The cellular identities of eDNA clades are (1) ultimately established through cultivation and culture-independent approaches to the ultrastructural characterization of individually isolated cells and (2) fundamental to understanding evolutionary history, ecological/symbiotic interactions, and morphological and behavioral innovations. Environmental DNA sequences have also demonstrated that a recently recognized group of euglenozoan protists, called the Symbiontida, is much more diverse than previously recognized. Our goal is to provide an overview of known symbiontid diversity from both eDNA data and ultrastructural data in order to highlight a relatively underappreciated component of marine biodiversity and avenues for future research.
The Euglenozoa is a group of flagellates with diverse modes of feeding and locomotion associated with different habitats within marine and freshwater environments. Different clades of euglenozoans reflect either a phagotrophic, photosynthetic or osmotrophic mode of nutrition and use either a gliding or a swimming mode of motility (Leander, 2004). The Euglenozoa as a whole is distinguished by several ultrastructural traits, such as tubular extrusomes, a configuration of three microtubular roots stemming from two basal bodies, and two flagella reinforced with heteromorphic paraxonemal rods, and consists of four major subgroups: the Euglenida, Diplonemida, Kinetoplastida and Symbiontida (Simpson, 1997; Leander, 2004; Adl et al., 2012; Yubuki and Leander, 2012; Yubuki et al., 2013).
The Euglenida is monophyletic and morphologically diverse, consisting mainly of a paraphyletic stem group of phagotrophic lineages and a monophyletic group of photosynthetic lineages. The group is distinguished from other euglenozoans by a system of proteinaceous pellicle strips that subtend the plasma membrane (Leander et al., 2007). Most phagotrophic euglenids inhabit marine environments, and most photosynthetic lineages inhabit freshwater environments; however, the three earliest branching lineages of photosynthetic euglenids, Rapaza, Eutreptia, and Eutreptiella, also inhabit marine environments (Yamaguchi et al., 2012). The largest (>20 microns) phagotrophic euglenids tend to be eukaryovorous and possesses a flexible pellicle and a robust feeding apparatus. Eukaryovorous modes of feeding facilitated the secondary endosymbiotic origin of euglenid chroloplasts from a close relative of Pyramimonas, a marine lineage of green algae (Leander, 2004; Yamaguchi et al., 2012; Jackson et al., 2018). Eukaryovore euglenids are the closest relatives to the clade of photosynthetic euglenids, and the earliest branching lineage within photosynthetic euglenids, namely Rapaza viridis, is a mixotroph capable of eukaryovory (Yamaguchi et al., 2012).
The Kinetoplastida is a diverse group of euglenozoans with a distinctive mitochondrial DNA inclusion, called a “kinetoplast.” This monophyletic group contains bacterivorous lineages (e.g., Bodo) and endoparasitic lineages (e.g., Trypanosoma and Lieshmania) that inhabit a wide range of marine, freshwater and terrestrial environments. The most recent ancestor of kinetoplastids was a marine, free-living bacterivore, and parasitic lineages of kinetoplastids evolved from free-living relatives several times independently (Lukeš et al., 2014). The Diplonemida is a small monophyletic group of relatively streamlined euglenozoans with only three described genera: Diplonema, Rhynchopus, and Hemistasia. Members of the group contain a complex feeding apparatus involved in eukaryovory but lack paraxonemal rods, pellicle strips, and kinetoplasts. Environmental DNA sequences from marine water samples collected at different depths suggest that diplonemids are among the most abundant eukaryotes in the oceans and contain several clades that currently lack cellular identities (Flegontova et al., 2016).
The Symbiontida is a monophyletic group consisting of several environmental DNA sequences and two species that have been characterized at the molecular phylogenetic level: Calkinsia aureus and Bihospites baccati. Although a third species, Postgaardi mariagerensis has yet to be characterized at the molecular phylogenetic level, it has the ultrastructural traits that define the Symbiontida (Yubuki et al., 2013). The group is distinguished from other euglenozoans by novel cytoskeletal features associated with the feeding apparatus and symbiotic relationships with epibiotic bacteria (Figures 1C,F,G). Symbiontids are found in low oxygen marine environments; C. aureus and B. baccati live in sediments, and P. mariagerensi lives in the water column. The molecular phylogenetic position of the Symbiontida within the Euglenozoa remains unclear (Figure 2). Although one author has proposed multiple taxonomic schemes involving Postgaardi, Calkinsia, and Bihospites (Cavalier-Smith, 1998, 2016), we use the clade name “Symbiontida” because (1) it is clearly defined at both the molecular and morphological levels (Yubuki et al., 2009, 2013), (2) it is not linked to phylogenetic uncertainty (e.g., the unknown molecular phylogenetic position of Postgaardi within the Euglenozoa), and (3) it is the name most widely used by the protistological community for this clade. We view the originally proposed (typified) taxon Postgaardea Calvalier-Smith as a synonym for P. mariagerensis.
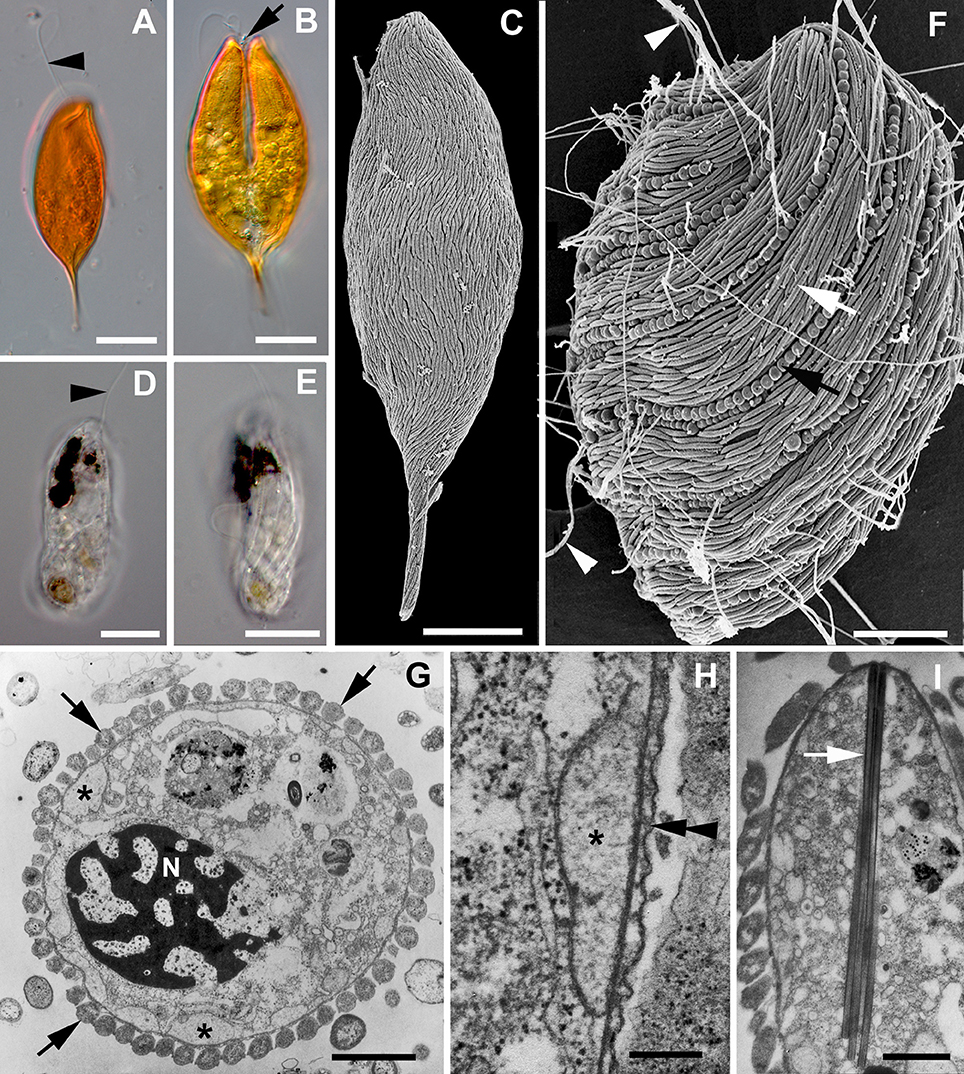
Figure 1. Images of symbiontids and their epibionts. (A,B) Differential Interference Contrast (DIC) light micrographs of Calkinsia aureus showing the orange extracellular matrix, the anterior flagellum (arrowhead) and the longitudinal cleavage furrow (arrow) of a dividing cell (scale bars = 10 microns). (C) Scanning electron micrograph (SEM) of Calkinsia aureus showing the cell surface enveloped with rod-shaped proteobacteria. (D,E) DIC light micrographs of Bihospites bacati showing the anterior flagellum (arrowhead) and longitudinally arranged ridges indicative of pellicle strips (scale bar = 10 microns). (F) SEM of Bihospites bacati showing two different morphotypes of epibiotic bacteria on the cell surface: rod-shaped proteobacteria (white arrow) and longitudinally arranged spherical verrucomicrobia (black arrow) capable of discharging threads of DNA (arrowheads) through an apical pore (scale bar = 15 microns). (G–I) Transmission electron micrographs (TEMs) of Postgaardi mariagerensis. (G) Low magnification transverse TEM showing the nucleus (N) with permanently condensed chromosomes, a uniform layer of epibiotic bacteria in cross section (arrows), and a superficial layer of mitochondrion-derived organelles (asterisks) positioned underneath the plasma membrane (scale bar = 3 microns). (H) High magnification TEM showing the mitochondrion-derived organelles (asterisk) positioned underneath the plasma membrane (double arrowhead) (scale bar = 0.25 microns). (I) Longitudunal TEM showing a bundle of tubular extrusomes (arrow) positioned near the anterior end of the cell (scale bar = 2 microns).
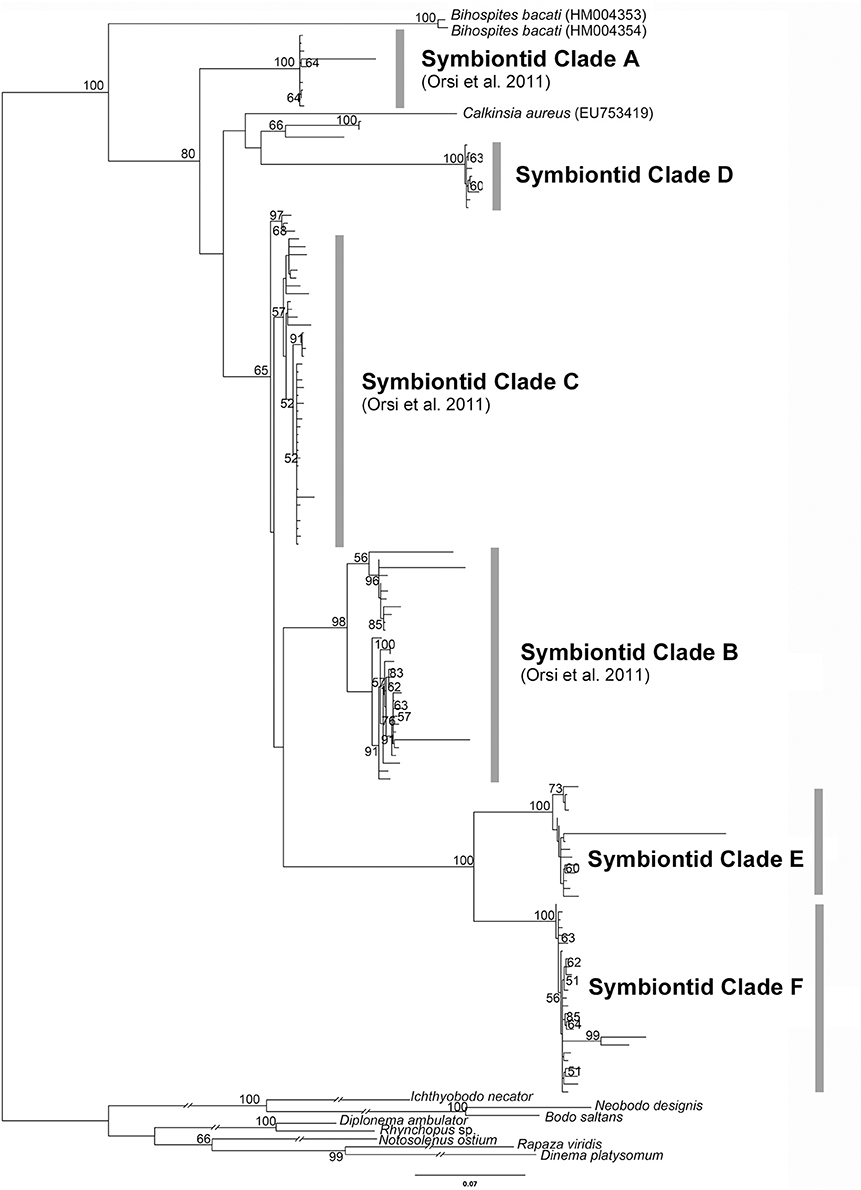
Figure 2. Phylogenetic tree inferred from a 153-taxon alignment of SSU rRNA gene sequences. The tree was inferred with Maximum Likelihood analysis using GTR+G+I model. ML boostrap values less than 50% are not shown. Symbiontid Clades A-C and E were demonstrated in Orsi et al. (2011). Outgroup branch lengths were shortened to half-length for illustrative purposes.
Calkinsia aureus, B. Baccati, and P. mariagerensis are marine heterotrophs, presumably phagotrophs based on the presence of a robust feeding apparatus consisting of microtubules and fibrous structures. They have two flagella inserted within a flagellar pocket located in a subapical region of the cell. A uniform layer of hydrogenosome-like, mitochondrion-derived organelles with reduced cristae is located beneath the cell membrane (Figures 1G,H). Elongated, rod-shaped epibiotic bacteria envelop the entire surface of the cell and likely exchange metabolites with the underlying layer of mitochondrion-derived organelles (Figures 1C,F–H). It is also possible that the epibiotic bacteria provide a food source for the symbiontids as reported in some ciliates (Fenchel and Finlay, 1989; Bauer-Nebelsick et al., 1996; Breglia et al., 2010).
As shown in Figure 2, our molecular phylogenetic analyses of eDNA sequences recovered from all over the world have established a robust and diverse symbiontid clade, where most eDNA sequences currently lack a cellular identity. The Symbiontida often branches within the Euglenida (Lax and Simpson, 2013; Lee and Simpson, 2014a,b) but, in some cases, it forms an independent clade from euglenids, diplonemids and kinetoplastids (Yubuki et al., 2009; Breglia et al., 2010). Ultrastructural evidence, such as a cell surface organized with S-shaped folds (B. baccati) and relatively long basal bodies, suggest a close relationship between symbiontids and euglenids (Breglia et al., 2010; Yubuki et al., 2013). Bihospites and Calkinsia have been characterized with light microscopy (LM), scanning and transmission electron microscopy (SEM and TEM, respectively) and molecular phylogenetic data (Yubuki et al., 2009; Breglia et al., 2010); Postgaardi has only been described at the morphological level using SEM and TEM (Fenchel et al., 1995; Simpson et al., 1996; Yubuki et al., 2013) (Figures 1G–I).
Calkinsia aureus is the type species of the Symbiontida (Figures 1A–C). The original description of C. aureus came from isolates near Woods Hole (MA, USA), and at that time, this species was classified as a member of the Petalomonidae within the Euglenida (Lackey, 1960) based on the rigidity of the cells and longitudinal striations on the cell surface, which were reminiscent of pellicle strips. However, subsequent SEM data from isolates collected from the Santa Barbara Basin (CA, USA) demonstrated that the striations reported by Lackey (1960) were actually a uniform layer of longitudinally arranged and elongated epibiotic bacteria on the cell surface (Bernhard et al., 2000; Yubuki et al., 2009). Cells of C. aureus are spindle-shaped with a pointed posterior end, 42–71 μm long and 15–23 μm wide. This species is easily recognized under light microscopy because it is bright orange, which reflects a thick extracellular matrix (Yubuki et al., 2009; Figures 1A,B). The extracellular matrix contains an arrangement of distinct conduits that link the epibiotic bacteria to the plasma membrane of the host cell, which is inferred to facilitate metabolic exchanges between the epibiotic bacteria and the underlying layer of modified mitochondria. The cells also contain a robust cluster of tubular extrusomes that is organized within an “extrusomal pocket” near the nucleus; the cluster of extrusomes is aligned along the longitudinal axis of the cell and is discharged from the anterior end of the cell when disturbed.
Bihospites baccati was collected from low oxygen sediments in the tidal flats of Cenntenial beach, British Columbia (Canada) and the Wadden Sea, Germany (Breglia et al., 2010). The cells are fragile, capable of metabolic euglenoid movement, 40–120 μm long, and 15–30 μm wide (Figures 1D–F). A robust feeding apparatus consisting of microtubules and fibers encircles a nucleus with permanently condensed chromosomes. Bihospites baccati is colorless, except for several black inclusions (Figures 1D,E) and glides along surfaces using a long anterior flagellum that pulls the cell forward. The cell surface is organized with folds that are S-shaped in cross-section and reminiscent of pellicle strips. The surface is also enveloped by two morphotypes of epibiotic bacteria: elongated rod-shaped bacteria and distinct longitudinal rows of spherical bacteria (Figure 1F).
Postgaardi mariagerensis was originally described from low oxygen water collected in a Danish Fjord (Fenchel et al., 1995) and classified as a close relative of Anisonema (Euglenids) based on the presence of longitudinal surface ridges, a colorless appearance, and cell flexibility. Ultrastructural data from an isolate of P. mariagerensis collected from Antarctica demonstrated a uniform layer of elongated rod-shaped epibiotic bacteria on the cell surface (Simpson et al., 1996; Yubuki et al., 2013; Figure 1G). The feeding apparatus in P. mariagerensis and C. aureus is very similar and contains distinctive finger-like projections within the feeding apparatus (Yubuki et al., 2013). There is no confirmed molecular phylogenetic data available for P. mariagerensis, but the morphological traits of this species demonstrate a close affinity with other members of the Symbiontida (Yubuki et al., 2013).
Several studies have demonstrated different lineages of euglenozoans enveloped by epibiotic bacteria. Buck et al. (2000), for instance, reported three different euglenozoans (morphotypes A, B, and C) covered with bacteria from a Monterey Bay cold seep. The authors assigned the type A morphotype to Postgaardi mariagerensis based on the overall cell morphology as observed with SEM; they assigned morphotypes B and C to euglenids insertae sedis based on the presence of pellicle strips. Bernhard et al. (2000) reported Calkinsia aureus, Postgaardi mariagerensis and two other unidentified euglenozoans with epibionts from the Santa Barbara Basin using morphological observations (see also Buck and Bernhard, 2002). So far, only two described euglenid species, namely Dylakosoma pelophilum (Wolowski, 1995) and Euglena helicoideus (Leander and Farmer, 2000) have been shown to have epibionts on the cell surface. The epibionts on euglenozoan cells are rod-shaped, except for the spherical extrusive bacteria on Bihospites baccati (Breglia et al., 2010) and the spherical bacteria on Dylakosoma pelophilum (Wolowski, 1995).
Euglenozoans enveloped with epibiotic bacteria constitute about 68% of the protist community associated with bacterial mats living in the top 1 cm of oxygen-depleted sediment in Monterey Bay, California (Buck et al., 2000). Although the specific taxonomic identities of these euglenozoans remain unknown, many of them are likely members of the Symbiontida. Environmental SSU rDNA sequence surveys suggest that members of the Symbiontida have a cosmopolitan distribution: Northern Europe (Framvaren Fjord, Norway; Mariager Fjord, Denmark), Western North America (Saanich inlet, Canada), Black Sea, Caribbean sea (Cariaco basin) and the Red Sea (Zuendorf et al., 2006; Orsi et al., 2011, 2012; Wang et al., 2014). The previous molecular phylogenetic analyses of eDNA sequences demonstrated three distinct symbiontid clades (A, B, and C) as well as a “putative novel euglenozoan class;” (Orsi et al., 2011). Symbiontid clades A and B only contained sequences from the Cariaco basin (Venezuela), and symbiontid clade C contained sequences from the Framvaren and Mariager Fjords. Environmental DNA sequences closely related to Bihospites baccati were found near a cold seep 850 m deep in the Red Sea (Wang et al., 2014).
We performed a molecular phylogenetic analysis using the most current dataset of eDNA sequences, including Calkinsia aureus and Bihospites baccati (Figure 2). In addition to three symbiontid clades reported previously by Orsi et al. (2011) (i.e., symbiontid clades A, B, and C), we recovered a novel symbiontid clade with robust support (i.e., symbiontid clade D); all of these sequences were collected from an anoxic marine water column at a depth of 200 m in Saanich inlet (Vancouver Island, British Columbia, Canada) (Orsi et al., 2012). Orsi et al. (2011) also discussed a sister lineage to the Symbiontida referred to as a “putative novel euglenozoan class”; however, this molecular phylogenetic analysis did not include subsequently acquired sequences from Bihospites baccati. Our molecular phylogenetic analyses including Bihospites baccati demonstrate that the “putative novel euglenozoan class” clearly falls within the Symbiontida, and has been relabeled as two separate lineages: Symbiontid clades E and F (Figure 2). Our molecular phylogenetic analysis confirmed that (i) Bihospites baccati is the earliest branching member of the Symbiontida; (ii) Calkinsia aureus is not closely related to any current eDNA sequences; (iii) the “putative novel euglenozoan class” described by Orsi et al. (2011) is part of the Symbiontida; (iv) the original “symbiontid clade C” (Orsi et al., 2011) is weakly supported; and (v) there are many symbiontid lineages that have yet to be described at the morphological level. There are at least six major eDNA clades of symbiontids in addition to Calkinsia aureus and Bihospites baccati; one of these eDNA clades almost certainly represents Postgaardi mariagerensis. Although statistical support for each major clade is high, the branching order of the clades (i.e., the symbiontid phylogenetic backbone) remains unclear.
One of the traits shared by members of the Symbiontida is a uniform layer of elongated rod-shaped epibiotic bacteria on the cell surface. SEM, TEM, FISH (fluorescence in situ hybridization) and molecular phylogenetic analyses of the bacteria on the surfaces of Calkinsia aureus and Bihospites bacati show that the epibionts are members of the epsilon proteobacteria and group together with the nitrate-reducing, sulfide-oxidizing autotrophic Arcobacter spp. (Buck et al., 2000; Edgcomb et al., 2011). Diverse prokaryotic communities thrive in microaerobic and anaerobic zones, and hydrogen sulfide production by bacteria is widespread in marine sediments. Hydrogen sulfide serves as an electron donor within these environments, so the epibionts are thought to detoxify the immediate surroundings of the unfavorable environment for the host cells through the consumption of sulfide (Edgcomb et al., 2011).
Calkinsia aureus and Bihospites bacati live in low-oxygen environments but were collected from distant geographical locations, which indicates an ancient and well-established relationship between the rod-shaped epibionts and the symbiontid hosts (Yubuki et al., 2009; Breglia et al., 2010; Edgcomb et al., 2011). Molecular phylogenetic analyses of the rod-shaped epibionts and the hosts showed near identical tree topologies and branch lengths, providing evidence for co-speciation between the hosts and the epibionts (Edgcomb et al., 2011).
In addition to the rod-shaped epibiotic bacteria, Bihospites bacati has distinct longitudinal rows of spherical epibionts, each attached within a concavity in the host plasma membrane. The spherical epibionts have a highly distinctive ultrastructure consisting of an operculum with a pore and a coiled internal thread around an electron dense core of DNA (Breglia et al., 2010). When disturbed, the core and thread are rapidly discharged through the pore in the operculum in a manner reminiscent of cnidarian and dinoflagellate nematocysts (Gavelis et al., 2017). The spherical epibionts on B. bacati are nearly identical to the so-called “epixenosomes” on the aerobic ciliate Euplotidium itoi; the spherical epibionts on both hosts have very similar 16S rDNA sequences and form sister lineages within the Verrucomicrobia (unpublished data; Verni and Rosati, 1990; Giambelluca and Rosati, 1996; Petroni et al., 2000; Rosati, 2001). The presence of these extrusive epibionts on two very distantly related lineages of eukaryotes (a ciliate, Alveolata, and a euglenozoan, Excavata) suggests that lateral symbiont transfer occurred from one host to another.
Our molecular phylogenetic analysis of current eDNA sequences demonstrated that symbiontids are cosmopolitan and consist of at least six major subgroups. The phylogenetic position of the Symbiontida within the Euglenozoa remains unclear; however, ultrastructural evidence, such as S-shaped folds in the plasma membrane and a robust rod-based feeding apparatus, suggests the clade is most closely related to the Euglenida. Symbiontid lineages are components of low oxygen environments (e.g., deep sea sediments), and their SSU rDNA sequences tend to be highly divergent, which suggests they could be largely overlooked in eDNA surveys of biodiversity. Current knowledge of symbiontid diversity likely does not reflect the actual number, abundance and biogeographies of species within the group. More comprehensive environmental DNA surveys using symbiontid-specific primers are necessary to understand the hidden diversity of symbiontids. Improved taxon sampling, single-cell transcriptomics, metagenomics, and multigene phylogenetic analyses of diverse lineages of symbiontids will shed considerable light onto the evolutionary history of the Euglenozoa as a whole and the co-evolutionary history of symbiontids and their epibionts in particular. Currently, genomic and transcriptomic data from symbiontids are not available, but these data will ultimately provide the deepest insights into potential modes of metabolic integration between the epibionts and their hosts.
The eDNA sequence dataset for the Symbiontida was generated from EukRef (http://eukref.org). After removing sequences less than 900 bp and possible chimeric sequences predicted by Bellerophon 6 (Huber et al., 2004), the alignment included 146 sequences using euglenids, diplonemids and kinetoplastids as an outgroup (Accession numbers available in the Supplementary Data, See Supplementary Table 1, Supplementary Figure 1). The sequences were automatically aligned using MAFFT with the L-INS-i option (Katoh et al., 2005; Katoh and Standley, 2013). Ambiguously aligned sites were then trimmed using trimAl (Capella-Gutierrez et al., 2009) creating a final alignment consisting of 1,175 unambiguously aligned sites. The phylogenetic tree was inferred using Maximum Likelihood (ML) with the program Garli (Zwickl, 2006) under a GTR + G model, which was selected by jModeltest 2.1.6 (Darriba et al., 2012). Six substitution rates (AC = 0.9712, AG = 2.343, AT = 1.129, CG = 0.7605, CT = 3.433, GT = 1.000), four discrete gamma distributed rate categories (0.8977) and base frequencies (A = 0.2551 C = 0.2173 G = 0.2796 T = 0.2480) were used in the GTR + G model. ML bootstrap analyses were carried out with 1,000 pseudoreplicates.
NY and BL contributed equally in building the figures and writing the mini-review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Tula Foundation's Center for Microbial Diversity and Evolution at the University of British Columbia.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00100/full#supplementary-material
Supplementary Table 1. GenBank Accession numbers used in the molecular phylogenetic analysis (Figure 2).
Supplementary Figure 1. Phylogenetic tree shown in Figure 1 including all of the GenBank Accession numbers for the 153 SSU rRNA gene sequences analyzed.
Adl, S. M., Simpson, A. G., Lane, C. E., Lukeš, J., Bass, D., Bowser, S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514. doi: 10.1111/j.1550-7408.2012.00644.x
Bauer-Nebelsick, M., Bardele, C. F., and Ott, J. A. (1996). Electron microscopic studies on Zoothamnium niveum (Hemprich&Ehrenberg, 1831) Ehrenberg 1838 (Oligohymenophora, Peritrichida), a ciliate with ectosymbiotic, chemoautotrophic bacteria. Eur. J. Protistol. 32, 202–215. doi: 10.1016/S0932-4739(96)80020-4
Bernhard, J. M., Buck, K. R., Farmer, M. A., and Bowser, S. S. (2000). The Santa Barbara Basin is a symbiosis oasis. Nature 403, 77–80. doi: 10.1038/47476
Breglia, S. A., Yubuki, N., Hoppenrath, M., and Leander, B. S. (2010). Ultrastructure and molecular phylogenetic position of a novel euglenozoan with extrusive episymbiotic bacteria: Bihospites bacati n. gen. et sp. (Symbiontida). BMC Microbiol. 10:145. doi: 10.1186/1471-2180-10-145
Buck, K. R., Barry, J. P., and Simpson, A. G. B. (2000). Monterey Bay cold seep biota: euglenozoa with chemoautotrophic bacterial epibionts. Eur. J. Protistol. 36, 117–126. doi: 10.1016/S0932-4739(00)80029-2
Buck, K. R., and Bernhard, J. M. (2002). “Protistan-prokaryotic symbioses in deep-sea sulfidic sediments,” in Symbiosis: Mechanisms and Model Systems. (Cellular Origin, Life in Extreme Habitats and Astrobiology), Vol. 4, ed J. Seckbach (Dordrecht: Kluwer Academic Publishers), 507–517.
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cavalier-Smith, T. (1998). A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc. 73, 203–266. doi: 10.1017/S0006323198005167
Cavalier-Smith, T. (2016). Higher classification and phylogeny of euglenozoa. Eur. J. Protistol. 56, 250–276. doi: 10.1016/j.ejop.2016.09.003
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Edgcomb, V. P., Breglia, S. A., Yubuki, N., Beaudoin, D., Patterson, D. J., Leander, B. S., et al. (2011). Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 5, 231–243. doi: 10.1038/ismej.2010.121
Fenchel, T., Bernard, C., Esteban, G. F., Finlay, B. J., Hansen, P. J., and Iversen, N. (1995). Microbial diversity and activity in a danish fjord with anoxic deep-water. Ophelia 43, 45–100. doi: 10.1080/00785326.1995.10430576
Fenchel, T., and Finlay, B. J. (1989). Kentrophoros - a mouthless ciliate with a symbiotic kitchen garden. Ophelia 30, 75–93.
Flegontova, O., Flegontov, P., Malviya, S., Audic, S., Wincker, P., de Vargas, C., et al. (2016). Extreme diversity of diplonemid eukaryotes in the ocean. Curr. Biol. 26, 3060–3065. doi: 10.1016/j.cub.2016.09.031
Gavelis, G. S., Wakeman, K. C., Tillman, U., Ripken, C., Mitarai, S., Herranz, M., et al. (2017). Microbial arms race: ballistic “nematocysts” in dinoflagellates represent a new extreme in organelle complexity. Sci. Adv. 3:e1602552. doi: 10.1126/sciadv.1602552
Giambelluca, M., and Rosati, G. (1996). Behavior of epixenosomes and the epixenosomal band during divisional morphogenesis in Euplotidium itoi (Ciliata, Hypotrichida). Eur. J. Protistol. 32, 77–80. doi: 10.1016/S0932-4739(96)80041-1
Guillou, L., Viprey, M., Chambouvet, A., Welsh, R. M., Kirkham, A. R., Massana, R., et al. (2008). Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ. Microbiol. 10, 3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319. doi: 10.1093/bioinformatics/bth226
Jackson, C., Knoll, A. H., Chan, C. X., and Verbruggen, H. (2018). Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids. Sci. Rep. 8:1523. doi: 10.1038/s41598-017-18805-w
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511– 518. doi: 10.1093/nar/gki198
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lackey, J. B. (1960). Calkinsia aureus gen. et sp. nov:, a new marine euglenid. Trans. Am. Microsc. Soc. 79, 105–107. doi: 10.2307/3223980
Lax, G., and Simpson, A. G. (2013). Combining molecular data with classical morphology for uncultured phagotrophic euglenids (Excavata); a single-cell approach. J. Eukaryot. Microbiol. 60, 615–625. doi: 10.1111/jeu.12068
Leander, B. S. (2004). Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 12, 251–258. doi: 10.1016/j.tim.2004.04.001
Leander, B. S., and Farmer, M. A. (2000). Epibiotic bacteria and a novel pattern of strip reduction on the pellicle of Euglena helicoideus (Bernard) Lemmermann. Eur. J. Protistol. 36, 405–413. doi: 10.1016/S0932-4739(00)80046-2
Leander, B. S., Esson, H. J., and Breglia, S. A. (2007). Macroevolution of complex cytoskeletal systems in euglenids. Bioessays 29, 987–1000. doi: 10.1002/bies.20645
Lee, W. J., and Simpson, A. G. (2014a). Morphological and molecular characterisation of Notosolenus urceolatus Larsen and Patterson 1990, a member of an understudied deep-branching euglenid group (Petalomonads). J. Eukaryot. Microbiol. 61, 463–479. doi: 10.1111/jeu.12126
Lee, W. J., and Simpson, A. G. (2014b). Ultrastructure and molecular phylogenetic position of Neometanema parovale sp. nov. (Neometanema gen. nov.), a Marine phagotrophic euglenid with skidding motility. Protist 165, 452–472. doi: 10.1016/j.protis.2014.05.001
Lukeš, J., Skalický, T., Týč, J., Votýpka, J., and Yurchenko, V. (2014). Evolution of parasitism in kinetoplastid flagellates. Mol. Biochem. Parasitol. 195, 115–122. doi: 10.1016/j.molbiopara.2014.05.007
Massana, R., Castresana, J., Balagué, V., Guillou, L., Romari, K., Groisillier, A., et al. (2004). Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70, 3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004
Massana, R., del Campo, J., Sieracki, M. E., Audic, S., and Logares, R. (2014). Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J. 8, 854–866. doi: 10.1038/ismej.2013.204
Orsi, W., Edgcomb, V., Jeon, S., Leslin, C., Bunge, J., Taylor, G. T., et al. (2011). Protistan microbial observatory in the Cariaco Basin, Caribbean. II. Habitat specialization. ISME J. 5, 1357–1373. doi: 10.1038/ismej.2011.7
Orsi, W., Song, Y. C., Hallam, S., and Edgcomb, V. (2012). Effect of oxygen minimum zone formation on communities of marine protists. ISME J. 6, 1586–1601. doi: 10.1038/ismej.2012.7
Petroni, G., Spring, S., Schleifer, K. H., Verni, F., and Rosati, G. (2000). Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. Proc. Natl. Acad. Sci. U.S.A. 97, 1813–1817. doi: 10.1073/pnas.030438197
Rosati, G. (2001). “Ectosymbiosis in ciliated protozoa,” in Symbiosis. Cellular Origin, Life in Extreme Habitats and Astrobiology (Dordrecht: Springer), 475–488.
Simpson, A. G. B. (1997). The identity and composition of the Euglenozoa. Arch. Protistenkd. 148, 318–328. doi: 10.1016/S0003-9365(97)80012-7
Simpson, A. G. B., Van Den Hoff, J., Bernard, C., Burton, H. R., and Patterson, D. J. (1996). The ultrastructure and systematic position of the euglenozoon Postgaardi mariagerensis, Fenchel et al. Arch. Protistenkd. 147, 213–225. doi: 10.1016/S0003-9365(97)80049-8
Verni, F., and Rosati, G. (1990). Peculiar epibionts in Euplotidium itoi (Ciliata, Hypotrichida). J. Protozool. 37, 337–343. doi: 10.1111/j.1550-7408.1990.tb01155.x
Wang, Y., Zhang, W. P., Cao, H. L., Shek, C. S., Tian, R. M., Wong, Y. H., et al. (2014). Diversity and distribution of eukaryotic microbes in and around a brine pool adjacent to the Thuwal cold seeps in the Red Sea. Front. Microbiol. 5:37. doi: 10.3389/fmicb.2014.00037
Wolowski, K. (1995). Dylakosoma pelophilum Skuja, a rare colourless euglenophyte found in Poland. Algol. Stud. 76, 75–78.
Yamaguchi, A., Yubuki, N., and Leander, B. S. (2012). Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: description of Rapaza viridis n. gen. et sp. (Euglenozoa, Euglenida). BMC Evol. Biol. 12:29. doi: 10.1186/1471-2148-12-29
Yubuki, N., Edgcomb, V. P., Bernhard, J. M., and Leander, B. S. (2009). Ultrastructure and molecular phylogeny of Calkinsia aureus: cellular identity of a novel clade of deep-sea euglenozoans with epibiotic bacteria. BMC Microbiol. 9:16. doi: 10.1186/1471-2180-9-16
Yubuki, N., and Leander, B. S. (2012). Reconciling the bizarre inheritance of microtubules in complex (euglenid) microeukaryotes. Protoplasma 249, 859–869. doi: 10.1007/s00709-011-0340-z
Yubuki, N., Simpson, A. G., and Leander, B. S. (2013). Reconstruction of the feeding apparatus in Postgaardi mariagerensis provides evidence for character evolution within the symbiontida (euglenozoa). Eur. J. Protistol. 49, 32–39. doi: 10.1016/j.ejop.2012.07.001
Keywords: biodiversity, eDNA sequences, evolution, Euglenozoa, phylogeny, Symbiontida
Citation: Yubuki N and Leander BS (2018) Diversity and Evolutionary History of the Symbiontida (Euglenozoa). Front. Ecol. Evol. 6:100. doi: 10.3389/fevo.2018.00100
Received: 18 May 2018; Accepted: 26 June 2018;
Published: 18 July 2018.
Edited by:
Eric Wade Linton, Central Michigan University, United StatesReviewed by:
Ugo Cenci, Lille University of Science and Technology, FranceCopyright © 2018 Yubuki and Leander. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian S. Leander, YmxlYW5kZXJAbWFpbC51YmMuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.