- Retired from School of Biological Sciences, The University of Reading, Reading, United Kingdom
Domestication involves changes in various traits of the phenotype in response to human selection. Diversification may accompany or follow domestication, and results in variants within the crop adapted to different uses by humans or different agronomic conditions. Similar domestication and diversification traits may be shared by closely related species (parallel evolution) or by distantly related species (convergent evolution). Many of these traits are produced by complex genetic networks or long biosynthetic pathways that are extensively conserved even in distantly related species. Similar phenotypic changes in different species may be controlled by homologous genes (parallel evolution at the genetic level) or non-homologous genes (convergent evolution at the genetic level). It has been suggested that parallel evolution may be more frequent among closely related species, or among diversification rather than domestication traits, or among traits produced by simple metabolic pathways. Crops domesticated in the Americas span a spectrum of genetic relatedness, have been domesticated for diverse purposes, and have responded to human selection by changes in many different traits, so provide examples of both parallel and convergent evolution at various levels. However, despite the current explosion in relevant information, data are still insufficient to provide quantitative or conclusive assessments of the relative roles of these two processes in domestication and diversification
Introduction
Domestication has been defined in various ways (e.g., Harris, 1989; Harlan, 1992; Clement, 1999; Benz, 2006; Fuller et al., 2010; Abbo et al., 2014; Larson et al., 2014), depending in part on the perspectives of the definer. However, there is a general consensus that domestication occurs in response to selection, predominantly human but also natural. This response involves increase in frequency, often to fixation, of certain traits adaptive to human needs or to the environment created by human activities. These traits constitute the so-called domestication syndrome. They may include increased size (particularly of the harvested organ), loss of dispersal mechanisms, change in plant habit (such as reduction in branching) and, in domesticates from regions with seasonal climates, loss of seed dormancy. Qualitative traits distinguishing domesticates from their wild progenitors are frequently controlled by one or a few genes, most of which have so far proved to be regulatory genes, mainly encoding transcription factors (Doebley et al., 2006; Sang, 2009; Martínez-Ainsworth and Tenaillon, 2016). Quantitative traits are usually controlled polygenically, by quantitative trait loci (QTL), but one or a few of these often have disproportionately large effects, thereby enabling rapid response to selection (Poncet et al., 2004). Genes governing traits of the domestication syndrome have been called domestication genes.
Crop evolution does not cease with domestication, but continues by improvement and diversification. Improvement has progressed by increasingly efficient methods of human-mediated selection, and by gene transfer and gene modification. Improvement often targets the same traits as those involved in domestication. Recurrent cycles of improvement may thus result in favourable alleles accumulating at large-effect QTL that influence the target trait (Poncet et al., 2004). This provides one explanation for apparently contradictory reports that, in a given crop, most domestication traits are under simple genetic control, yet many genes have been affected by domestication and subsequent improvement. An alternative, or additional, explanation is that the “top down” approaches that are often used to identify candidate domestication genes may favour detection of major genes and QTL with large effects (Morrell et al., 2012; Olsen and Wendel, 2013).
Diversification involves development of variants within a crop that are adapted to different uses by humans, or to different agricultural environments. Distinguishing between domestication and diversification is not always easy (Meyer and Purugganan, 2013) and again may depend on definition. Furthermore, the same trait may be part of the domestication syndrome in one crop, but associated with diversification in another. In Capsicum, loss of dispersal results from loss of an abscission zone at the base of the fruit and is a feature of most domesticated chile peppers. Most domesticated tomatoes still have an abscission zone in the fruit stalk, but breeders have recently selected for its loss in tomato cultivars that are harvested mechanically for canning (Mao et al., 2000). Loss of ability to disperse fruits is usually considered part of the domestication syndrome in chile pepper, but is a diversification trait in tomato.
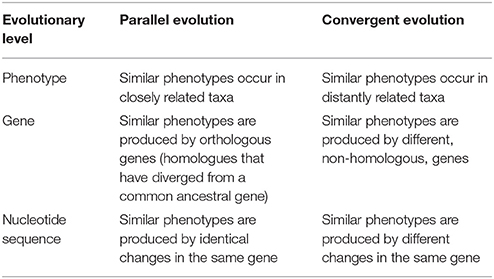
The terms “parallel evolution” and “convergent evolution” are not always used consistently, so may be sources of confusion. They were originally employed for phenotypic characters, but their application has now been extended to the genes controlling these characters and to the nucleotide sequences of these genes. The meanings of parallel and convergent evolution at these different levels, as used in this review, are summarised in Table 1. In the original usage (e.g., Davis and Heywood, 1963), parallel evolution is considered to be independent development of similar phenotypic traits in taxa with a relatively recent common ancestry, while convergent evolution is development of similar traits in distinct phylogenetic lineages, i.e., in taxa that are not closely related. In this paper, genera in the same family are considered to be closely related, whereas genera in different families are considered to be distantly related. Traits associated with both domestication and diversification may be shared by distantly as well as closely related species, hence may have arisen by convergent or by parallel evolution.
At the level of the gene, parallel evolution may be viewed as production of similar phenotypes by orthologous genes, i.e., homologous genes that have diverged from a common ancestor, while convergent evolution occurs when similar phenotypes are produced by genes that are not homologous. At the level of nucleotide sequences, parallel evolution is the occurrence, in different populations, of genotypes with identical changes in DNA sequence in a given gene. This seems to be rare. Different changes in DNA sequences affecting the same gene are more frequent and may produce similar phenotypes. This represents convergence at the level of DNA sequence but parallelism at the level of the gene.
The question of whether, or to what extent, parallel evolution has occurred during domestication and diversification has been much discussed. Glémin and Bataillon (2009) and Martínez-Ainsworth and Tenaillon (2016) concluded that there is little evidence for parallelism at the genetic level and that similar traits of the domestication syndrome in different species are usually controlled by loci that are not homologous. On the other hand, Sang (2009) argued that, since most domestication genes appear to be regulatory genes affecting more than one trait (pleiotropy), the same genes would probably be targeted repeatedly during domestication, because selection would favour consistently those genes for which negative pleiotropic effects are minimal. Poncet et al. (2004) considered that orthologous loci are more likely to be involved when domesticates belong to the same family, while Lenser and Theißen (2013) suggested that traits resulting from alterations in simple metabolic pathways, such as those involved in pigment synthesis, are more likely to be controlled by orthologous genes than complex traits such as seed dormancy. Gross and Olsen (2010) argued that diversification traits are controlled by orthologous genes more often than are domestication traits. Finally, Gaut (2015) concluded that the question of the extent of parallel evolution during domestication of different crops remains open.
The study of domestication and diversification, together with the respective roles of parallel vs. convergent evolution, is relevant to further improvement of existing crops, to possible development of new crops, and to understanding origins and spread of crops in prehistory. When different pathways to increased yield, involving different genes (convergent genetic evolution), have been utilised in related crops, or in independently domesticated lineages within the same crop, plant breeders may be able to combine in one genotype alleles associated with increased yield in different lineages, thereby enabling yield to reach a new plateau. Food production may also be increased by extending agriculture into environments currently considered marginal because of low temperatures, drought or salinity. Cultivated species adapted to such marginal conditions are often undomesticated, hence unsuited to modern mechanised agriculture. It may be possible to identify in the genome of such species homologues of genes known to govern traits of the domestication syndrome in their domesticated relatives, and then to edit these genes to produce a new domesticate by parallel genetic evolution. Finally, techniques for recovering ancient DNA from archaeological specimens are steadily improving, together with the ability to amplify and sequence particular genes in these DNA fragments. This is opening the possibility of being able to study the order in which traits of the domestication syndrome became established in a given crop and hence how human selection was exercised in early stages of domestication of that crop.
The principal regions of crop domestication in the Americas are shown in Table 2, together with the probable regions of domestication of those species mentioned in this text. There are examples of independent domestications of the same species in two continents (e.g., common bean); independent domestications of different species in the same genus for the same purpose (e.g., the New World cottons Gossypium hirsutum and Gossypium barbadense); independent domestications of species in different genera of the same family for the same purpose (e.g., tomato, chile pepper and Physalis spp., all members of the Solanaceae domesticated for their fleshy fruits); and independent domestications of species in different families for the same purpose (e.g., maize in the Poaceae and Amaranthus spp. in the Amaranthaceae, domesticated for their starchy seeds). These crops thus provide an opportunity to evaluate the roles of parallel vs. convergent evolution in domestication and diversification of closely vs. distantly related species.
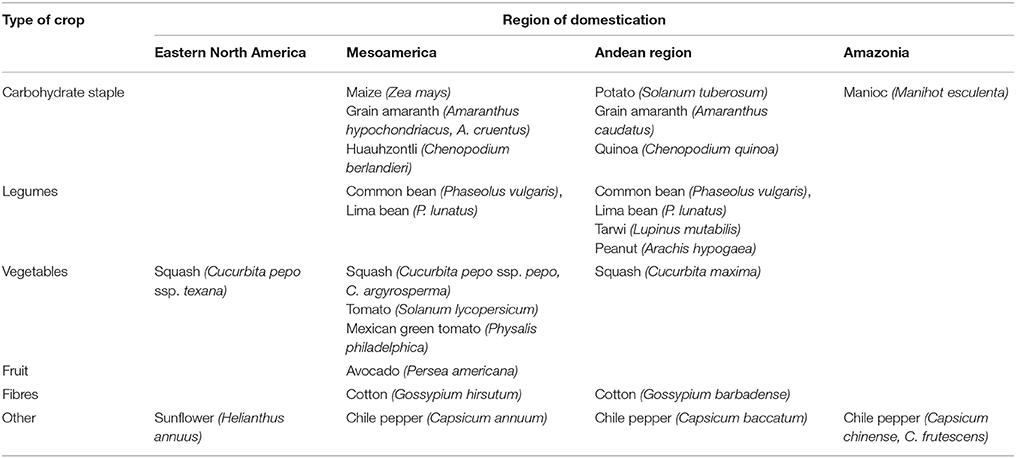
Table 2. Principal regions of crop domestication in the Americas, with probable regions of domestication of those species mentioned in the text.
Parallel vs. Convergent Evolution in Traits of the Domestication Syndrome
Increased size, reduction in natural dispersal mechanisms, loss of dormancy and reduced branching have all been investigated genetically in sufficient detail to be discussed here. Loss of toxic or bitter compounds, considered as part of the domestication syndrome by Abbo et al. (2014), will be treated later, as a diversification trait, since many New World crops are still polymorphic for presence or absence of such compounds.
Increased Size
Increased size results from presence of more cells in a given organ and/or from larger cells. Number of cells in an organ depends on number of cells in the primordium that gave rise to that organ and/or the amount of cell division in the organ after initiation.
Increased Number of Cells in the Primordium
The genetic control of events in the shoot meristem is complex and not yet fully understood (Somssich et al., 2016). Basically, control of cell proliferation vs. differentiation depends on interactions between WUSCHEL (WUS), a gene that encodes a transcription factor promoting cell proliferation, and CLAVATA3 (CLV3), a gene that encodes a product promoting cell differentiation. The CLV3-WUS signalling pathway is in turn regulated by many receptor-like kinases and receptor-like proteins. Somssich et al. (2016) suggested that increased size in various crops is likely to result from changes in genes involved in the CLV3-WUS pathway, since this seems to be significantly conserved between species, but as yet there are very few crops in which candidate genes that affect size have been associated with this pathway. One of these crops is tomato (Solanum lycopersicum), in which an orthologue of WUS (SlWUS) is a candidate gene for LOCULE NUMBER (van der Knaap et al., 2014), a gene that increases the size of the fruit by increasing the number of locules. In Capsicum, selection under domestication has similarly resulted in larger fruits with more locules. Barchi et al. (2009) found a QTL for locule number in Capsicum annuum that they thought might be orthologous to SlWUS, but it has yet to be shown that the gene underlying this QTL functions in the CLV3-WUS pathway, or that orthologous QTL are responsible for parallel evolution of multilocular fruit in different domesticated species of Capsicum. Although the widespread and conserved nature of the CLV3-WUS pathway and associated loci seems to offer opportunities for parallel genetic evolution in crops that have undergone parallel selection for increased size, there is so far little evidence of this.
Increased Cell Division in the Organ Under Selection
Another QTL that influences fruit size in tomato is FW2.2. This encodes a transcription factor that controls the number of cells in the fruit by acting as a negative regulator of cell division (Cong et al., 2002; Tanksley, 2004). FW2.2 is a member of a family of genes known as CELL NUMBER REGULATOR (CNR) genes (van der Knaap et al., 2014), an ancient gene family that occurs also in animals and fungi, but appears to have expanded and radiated more in plants. In maize, the closest orthologue to FW2.2 is ZmCNR1. It seems to act in the same way as FW2.2, in that down-regulation increases organ size, while over-expression reduces size, through changes in cell number, not cell size (Guo et al., 2010). However, ZmCNR1 is not a major domestication gene in maize, whereas human selection for fw2.2 was considered by Blanca et al. (2015) to have been important in the origin of semi-domesticated cherry tomato from wild currant tomato. In avocado, a CNR gene homologous to FW2.2 affects fruit size through similar negative regulation of cell division (Dahan et al., 2010). Thus, convergent changes in fruit size in such distantly related plants as avocado, tomato and maize seem to result from parallel changes involving CNR genes.
However, apparently parallel evolution of similar phenotypes may not extend to structural details, even for genera in the same family. In the Solanaceae, human selection has produced large fruits in both tomato and Capsicum. In C. annuum, at least seven QTL affecting fruit weight are orthologues of corresponding loci in tomato, but their relative contributions differ in the two genera. In tomato, the QTL with greatest effect on fruit size is FW2.2, but its orthologue in Capsicum has only a minor effect (Paran and van der Knaap, 2007). This may be because FW2.2 acts principally on cell division in the placenta and central axis of the fruit, which together make up most of the flesh in large-fruited tomatoes, whereas in the hollow fruit of Capsicum, there is relatively much less placental tissue, so increased size results mainly from increased cell division in the fruit wall (van der Knaap et al., 2014; Wang et al., 2015).
In large-fruited tomatoes, the number of cells in the fruit wall has likewise increased, and is affected by another major QTL, FW3.2. The candidate gene for FW3.2 is an orthologue of KLUH in Arabidopsis, so was designated SlKLUH (Chakrabarti et al., 2013; van der Knaap et al., 2014). A putative orthologue in C. annuum (CaKLUH) is also associated with large fruit (Chakrabarti et al., 2013; Wang et al., 2015). Parallel evolution of large fruits in tomato and chile pepper thus involves both similarities and differences at the levels of fruit structure and its genetic control.
Increased Cell Expansion
An initial phase of cell division in a developing organ is usually followed by a phase of cell expansion. In the Mexican green tomato (Physalis philadelphica), Wang et al. (2014) identified a locus that they named PHYSALIS ORGAN SIZE 1 (POS1). Over-expression of this locus resulted in larger organs, including fruits, whereas virus-induced silencing reduced sizes of these organs. Cell numbers were the same in large or small organs, so changes in POS1 affected cell expansion rather than cell number. Large fruits of domesticated P. philadelphica are therefore phenotypic parallels of large fruits of tomato, but the parallel does not extend to the genetic level.
A more dramatic example of increased cell expansion resulting from human selection during domestication involves lint hairs of cotton. These are single-celled structures borne on the seed coat and provide the commercial fibre. In both of the species domesticated in the Americas, G. hirsutum and G. barbadense, human selection has produced lint hairs that are much longer than those of wild cotton, because they start to elongate earlier, elongate faster and continue elongating for a longer period. Furthermore, formation of secondary cell walls is delayed. Many genes are involved in these differences, but genes governing two sets of processes have been studied in detail. The extended period of cell elongation and delayed formation of secondary cell walls correlates with production of enzymes that break down reactive oxygen species such as hydrogen peroxide. In G. hirsutum, this is achieved by up-regulation of genes encoding catalase, glutathione S-transferase and thioredoxin, while in G. barbadense, genes encoding several peroxidases are up-regulated (Chaudhary et al., 2009). Rapid elongation of lint hairs is attributed to the action of profilins. Five members of the gene family encoding profilins are expressed in lint hairs and all five are up-regulated in both G. hirsutum and G. barbadense (Bao et al., 2011). In the case of the profilins, human selection thus appears to have resulted in parallel genetic as well as phenotypic changes in two independently domesticated, but related, species, whereas in the control of antioxidant activity, independent selection of different sets of genes with similar effects has produced similar phenotypes by convergent genetic routes (Olsen and Wendel, 2013).
Reduction of Dispersal
In crops domesticated for their fruit or seed, or propagated by seed, human selection has frequently resulted in either reduced efficiency or total loss of mechanisms for dispersal. For single-seeded fruits, or multi-seeded fruits that are eaten by animals, this often involves loss of the abscission zone that enables fruit to be removed from the parent plant. For multi-seeded fruits that are not eaten, fruits must open at maturity to release their seeds. This often involves differential lignification of cell layers in the fruit wall so that, as the fruit dries, these layers shrink to different degrees, setting up tensions that are released as the fruit opens along predetermined lines of dehiscence. In peanut, and also in potato, where underground organs are harvested, the structures by which the harvested organs are attached to the parent plant have become shorter, so that harvesting is easier but dispersal occurs over a shorter distance. Parallel reductions in efficiency of dispersal may therefore be achieved by different morphological means, with corresponding differences in genetic control.
To date, genetic control of mechanisms of dispersal has been investigated in detail in a few model species only. Recent reviews (Estornell et al., 2013; Dong and Wang, 2015) have indicated that regions of dehiscence and their associated abscission zones are controlled by regulatory networks involving many transcription factors. Additional networks involve interactions with plant hormones, for example ethylene up-regulates certain genes encoding enzymes involved in breaking down cell walls. Change in any of the genes involved in any of these networks may lead to loss of dispersal. What appear to be similar phenotypes may therefore be produced by changes in quite different genes or sets of genes.
In Phaseolus, five species were domesticated independently, while two of these, common bean (Phaseolus vulgaris) and Lima bean (P. lunatus), were both domesticated at least twice, in Mesoamerica and in the Andean region. Indehiscent pods were selected by humans during each domestication event. In P. vulgaris, a recessive allele of a major gene, stringless (st), causes loss of fibres in the sutures of the pods (Koinange et al., 1996). This results in loss of dispersal, and also in the “stringless” phenotype, favoured in types such as snap beans, in which immature pods are eaten. Additionally St, or a gene tightly linked to St, controls lignification of inner layers of the pod wall, which affects ability of the pods to open explosively. Although St has been mapped, the nature and function of the underlying gene(s) are not yet known. Until a candidate gene is securely identified, it is not possible to determine whether phenotypic parallels in evolution of indehiscent pods in different domesticated species of Phaseolus, and/or in independently domesticated lineages within P. vulgaris and P. lunatus, are paralleled at the genetic or nucleotide levels.
Five different species of Capsicum have been domesticated in different parts of the Americas. Most domesticated peppers in all five species have lost the abscission zone at the base of the fruit that enables the fruit to be removed easily from the parent plant by animal dispersers. Loss of this abscission zone is controlled by a recessive allele, designated s, of a single major gene S, though other QTL must also be involved because the force required to detach the fruit varies. Candidate gene(s) underlying this locus have not been securely identified, though Rao and Paran (2003) suggested that, in C. annuum and C. frutescens, a gene that encodes a polygalacturonase is probably implicated, because this gene maps to the same chromosomal region as S and is active specifically in the fruit. There are no data on whether the same gene is involved in the other domesticated species of Capsicum, hence whether parallel evolution of reduced efficiency of fruit dispersal is caused by parallel evolution at the genetic level.
In tomato, an abscission zone develops in the fruit stalk of wild and most domesticated tomatoes. A mutation of a gene designated JOINTLESS suppresses development of this abscission zone (Mao et al., 2000). JOINTLESS encodes a transcription factor that interacts with transcription factors encoded by two other genes. All three transcription factors seem to regulate the same set of target genes (Estornell et al., 2013; Dong and Wang, 2015). Within the Solanaceae, there has thus been a parallel loss of abscission zones, and hence reduction of fruit dispersal, in most domesticated chile peppers and some domesticated tomatoes, but the sites of loss, the genes involved, and the mechanisms of gene action appear different in the two genera.
Loss of Seed Dormancy
For annual crops grown on a field scale, it is desirable that seeds germinate rapidly and evenly. This helps to suppress competing weeds and produces stands of plants of similar age that will mature at about the same time, so may be harvested in a single operation. Many domesticated crops have therefore lost much of the seed dormancy characteristic of their wild progenitors. However, some degree of dormancy needs to be retained to prevent seeds from germinating before the crop is harvested. Dormancy is therefore a quantitative character (Baskin and Baskin, 2004; Graeber et al., 2012). Dormancy varies over time, with environmental conditions, within species, and even within individuals (Smýkal et al., 2014). It is controlled by nuclear genes, but there may also be maternal effects and epistatic interactions (Baskin and Baskin, 2004). Determining how dormancy is controlled genetically in any given species is therefore challenging.
There are two principal types of seed dormancy among crop species (Baskin and Baskin, 2004). Physical dormancy, known also as hard-seededness, results from impermeability of the seed coat or fruit wall, due to an unbroken cuticle and one or more layers of palisade cells with lignified walls. This impermeability hampers the uptake of water necessary for germination and also, in crops such as grain legumes, the water required if seeds are to become soft when cooked. Physiological dormancy is more widespread than physical dormancy. It may be associated with the seed coat, where compounds such as tannins or pigments may inhibit germination (Smýkal et al., 2014). However, physiological dormancy is more often associated with the seed contents (endosperm and/or embryo). Some species combine both physical and physiological dormancy; for example, sunflower has physiological dormancy, but the fruit wall also provides a physical barrier to germination (Weiss et al., 2013).
Amongst crops domesticated in the Americas, wild species of both Phaseolus and cotton have hard seeds. QTL affecting dormancy have been mapped in common bean (Koinange et al., 1996), but the underlying candidate genes and their mechanisms of action have not been identified. Even less is known about the genetics of hard-seededness in cotton. In sunflower, with its combination of physical and physiological dormancy, Weiss et al. (2013) found that the micropylar end of the two valves of the fruit wall opened earlier in domesticated than in wild sunflowers, facilitating earlier entry of air and water, hence faster germination. However, it is not known what promotes loosening of cells in this region of the fruit wall.
Abscisic acid (ABA) appears to be significant in inducing and maintaining physiological dormancy, while gibberellic acid (GA) releases dormancy and promotes germination (Finch-Savage and Leubner-Metzger, 2006; Nonogaki, 2014). The balance between ABA and GA is therefore important in control of physiological dormancy, while tissues surrounding the embryo frequently appear to play a pivotal role (Graeber et al., 2012). Mechanisms of physiological dormancy have been extensively investigated in Arabidopsis, where numerous QTL are involved, including DELAY OF GERMINATION 1 (DOG1). DOG1 is highly conserved, with homologues in both dicotyledons and monocotyledons, and is required, together with ABA, to induce physiological dormancy (Nonogaki, 2014; Née et al., 2017). Regulation of transcription of DOG1 is complex and is influenced by environmental factors. The light-activated form of the photoreceptor phytochrome B, produced by the gene PHYB, represses transcription of both DOG1 and two further genes that, like DOG1, encode transcriptional regulators (Jiang et al., 2016). The means by which DOG1 acts are not clearly established.
Finch-Savage and Leubner-Metzger (2006) suggested that some at least of the mechanisms involved in control of seed dormancy and germination are widespread and have been conserved during evolution. However, it has not yet been shown that QTL involved in maintenance or loss of dormancy in Arabidopsis have orthologues with similar functions in any of the crop species domesticated in the Americas. The closest approach is the work of Mandel et al. (2014). They searched the literature for genes known to affect domestication-related traits, including germination, then identified homologues of these genes in sunflower, sequenced these from panels of wild, primitive cultivated, and improved sunflowers, and looked for evidence of past selection as shown by reduced nucleotide diversity. They found that one gene that encodes a protein that represses germination in Arabidopsis had apparently been selected during improvement of sunflower. PHYB had also been targeted during sunflower improvement, though this might relate to effects of phytochrome on adaptation to different daylengths, rather than its effects on seed dormancy.
In various plant families, ABA inhibits weakening of the endosperm around the embryo (Graeber et al., 2012). In several members of the Solanaceae, endosperm weakening, facilitating germination, may relate to inhibition by ABA of certain enzymes that degrade callose associated with plasmodesmata and thus break adhesion between adjacent cells in the endosperm (Finch-Savage and Leubner-Metzger, 2006).
It is not possible to reach any conclusion about the respective roles of parallel vs. convergent evolution in genetic control of physiological dormancy until candidate genes underlying QTL affecting this trait are clearly identified, together with their modes of action. Given the complex control of physiological dormancy, mutations in many different genes could produce apparently similar phenotypes in which dormancy has been lost.
Reduced Branching
In species with terminal inflorescences, domestication has often resulted in reduced branching, so that plant resources are channelled into fewer but larger inflorescences, often with larger fruits and seeds. Domesticated maize has fewer basal branches than its closest wild relative, and also has fewer ear-bearing lateral branches on the main stem. In sunflower, modern cultivars have an unbranched stem with a single massive head, unlike the numerous smaller heads, each terminating a lateral branch, of wild sunflowers. In Chenopodium, reduced lateral branching and a much larger terminal inflorescence developed independently in the Andean domesticate quinoa (Chenopodium quinoa) and the Mesoamerican domesticate huauhzontli (Chenopodium berlandieri).
Lateral branches result from outgrowth of buds in axils of leaves on the main stem. This depends on various factors, including genotype, hormonal signals, nutrients, and environmental variables, particularly light. Actions of, and interactions between, these variables are complex (Rameau et al., 2015). Auxin appears to have a key role, with other plant hormones acting both downstream and upstream of auxin. Strigolactones suppress outgrowth of axillary buds, while cytokinins promote this. In Arabidopsis, a protein that interacts with a strigolactone receptor is encoded by MORE AXILLARY GROWTH 2 (MAX2) (Rameau et al., 2015). The recessive mutant max2 has a bushy phenotype. In tomato, a recessive mutant of LATERAL SUPPRESSOR (LS) does not form side shoots, and shoot apices have greatly increased levels of auxin and gibberellin, but decreased levels of cytokinin (Schumacher et al., 1999). A key step in cytokinin biosynthesis is controlled by isopentenyl transferase enzymes, encoded by IPT genes. Transcript levels of IPT genes are in turn modified by auxin levels. Mechanisms by which genetic control of branching is achieved are not yet fully understood, but disruptions at different stages or in different pathways seem likely to produce the same phenotype of reduced branching by different genetic means.
Mandel et al. (2014) identified homologues in sunflower of genes known to affect branching in other species, then looked for evidence of selection acting on these genes during domestication or improvement of sunflower. The homologue of LS showed evidence of selection during domestication, while the homologue of MAX2 was subjected to selection during improvement, and selection at an unknown stage of either domestication or improvement affected IPT5, hence levels of cytokinin. Mandel et al. (2014) have therefore shown that genes affecting auxins, gibberellins, cytokinins, and strigolactones may all be involved in genetic control of branching in sunflower.
In maize, reduced branching seems to involve changes in a pathway that may be unique to maize. The principal gene involved is TEOSINTE BRANCHED 1 (TB1), while a second gene, GRASSY TILLERS 1 (GT1) apparently acts in the same pathway, but downstream of TB1 (Whipple et al., 2011). Both genes encode transcription factors. In rice (Oryza sativa), pea (Pisum sativum) and Arabidopsis, TB1 is a key target of strigolactone signalling, but in maize, both TB1 and GT1 appear to be no longer regulated by strigolactone signalling, suggesting that, even within the cereals, selection under domestication for reduced branching may have been achieved by different routes (Guan et al., 2012).
The limited evidence so far available therefore suggests that parallel evolution of phenotypes with reduced branching in different crops results from convergent evolution at the genetic level.
Parallel vs. Convergent Evolution in Diversification
Diversification in response to human selection affects many different features. These depend on the crop concerned, but include plant habit, colour and/or shape of the harvested organ, flavour and/or palatability, cooking properties, and adaptation to different day-lengths.
Plant Habit
In pumpkins and squashes (Cucurbita spp.), and also in common bean, there are differences in habit within the crop. In Cucurbita, some landraces and cultivars have a trailing vine-like habit, like their wild relatives. This is advantageous in subsistence agriculture, when pumpkins and squashes are often interplanted among maize and beans. Their viny habit then provides complete ground cover, suppressing weeds and minimising soil erosion. Bush types have semi-erect stems with short internodes, can be planted more closely, and are favoured when immature fruits are harvested frequently in order to maintain production. Bush types occur in some cultivars of Cucurbita maxima and in both domesticated subspecies of Cucurbita pepo. Deficiencies in gibberellin synthesis or signalling have been associated with bush phenotypes (Zhang et al., 2015). The bush habit is dominant in the early stages of growth but incompletely dominant later (Denna and Munger, 1963). Zhang et al. (2015) identified three QTL associated with bush habit in C. maxima and suggested that a candidate gene for the QTL with greatest effect is an orthologue of a gene in Arabidopsis that encodes gibberellin 20-oxidase, which catalyses the final stages in gibberellin biosynthesis. A large deletion and two single nucleotide polymorphisms (SNPs) in the promoter region of the bush allele in C. maxima apparently reduce its expression. Zhang et al. (2015) suggested that other genes encoding gibberellin 20-oxidases might become active later in development, partially compensating for the defective gene. It has still to be conclusively demonstrated that the same gene controls bush habit in C. maxima and in the two subspecies of C. pepo, and that the candidate gene in all three cases encodes gibberellin 20-oxidase. It has also yet to be determined whether identical changes in nucleotide sequence independently inactivate this gene.
The bush habit has also become established in some landraces and cultivars of common bean, but here the terminal meristem switches to a reproductive state relatively early in development, so that growth of the main stem is determinate, whereas in bush types of Cucurbita, the main stem remains indeterminate. Bush beans can be grown without support, unlike wild and many domesticated beans, which are climbers. Bush beans flower earlier than climbing beans, so are better adapted to cooler climates. In most bush beans, habit is controlled by a single gene, fin (Koinange et al., 1996). The candidate gene underlying fin is an orthologue of TERMINAL FLOWER 1 (TFL1) in Arabidopsis, and is designated PvTFL1y (Repinski et al., 2012). Common bean was domesticated independently in Mesoamerica and the Andean region and bush types occur in both regions. Kwak et al. (2012) identified eight mutations in the nucleotide sequence of PvTFL1y that were expected to change function of the gene. Different mutations tend to be restricted to one or other region: deletion of the entire PvTFL1y sequence is confined to Andean bush beans, while a large retrotransposon-mediated insertion is likewise virtually restricted to Andean bush beans. A phylogenetic tree of nucleotide sequences from PvTFL1y had two major branches, corresponding to Mesoamerican vs. Andean accessions. This suggested to Kwak et al. (2012) that the bush habit was selected independently in each region.
The bush habit in common bean is therefore an example of parallel evolution at phenotypic and genetic levels, but independent mutations at the nucleotide level, hence multiple origins of the same trait. Convergent phenotypic similarities in bush habit in Phaseolus and Cucurbita are superficial only; morphology and genetic control are fundamentally different.
Flavour, Palatability, and Cooking Qualities
Meyer et al. (2012) found that, in domesticated species in general, the most common changes associated with human selection involve flavour, toxicity, or colour. Loss of bitterness often results in loss of toxicity as well as increased palatability. Abbo et al. (2014) considered that, in those crops in which bitterness has been lost, this is a crucial element of the domestication syndrome. Several crops in the Americas were domesticated from wild progenitors containing toxic or unpalatable compounds, but human selection has not eliminated these compounds entirely. Instead, elaborate methods of processing have been devised. Bitter potatoes, adapted to cold temperatures and high altitudes in the Andes, contain glycoalkaloids in amounts that may be toxic to humans (Johns and Galindo Alonso, 1990). Harvested tubers are repeatedly frozen and thawed, trampled or leached to express the bitter juice, and finally dried. Tubers of sweet landraces of manioc (Manihot esculenta) contain relatively little toxic cyanogenic glycoside, mainly in the outer layers, so the tubers can be cooked and eaten safely after peeling. Bitter tubers, containing more glycoside, distributed throughout the tubers, must be shredded, squeezed, and the pulp heated to drive off any remaining hydrogen cyanide. In both potato and manioc, variation in content of bitter compounds is associated with diversification, rather than domestication.
Many different compounds confer bitterness or unpalatability: cyanogenic glycosides in Lima bean as well as manioc; toxic alkaloids in seeds of tarwi (Lupinus mutabilis); steroidal glycoalkaloids in potato; saponins in grains of quinoa (C. quinoa); cucurbitacins in species of Cucurbitaceae. These different compounds are synthesised by different pathways, so their loss or reduction in different domesticates involves changes in different genes. However, in independently domesticated species of the same genus, containing similar unpalatable compounds, human selection for parallel changes in palatability might lead to parallel decreases in these compounds, controlled by parallel genetic changes.
In Cucurbita, cucurbitacins have been lost independently from fruits of all five domesticated species. In C. pepo and C. maxima, a single gene controls the difference between bitter and non-bitter fruits. Bitterness is dominant, but it is not clear whether the same gene is involved in both species (Paris and Brown, 2005). In C. argyrosperma, non-bitterness is due to recessive alleles at two independent loci, both different from the locus responsible for non-bitterness in C. pepo (Borchers and Taylor, 1988). Presumably the three loci govern different steps in the biosynthesis of cucurbitacins, though the precise biochemical functions of the loci have still to be determined. Parallel evolution of non-bitter fruits in different species of Cucurbita thus does not necessarily involve parallel evolution at the genetic level.
During diversification in chile peppers (Capsicum spp.), people have selected for different levels of pungency. In three independently domesticated species, C. annuum, C. frutescens, and Capsicum chinense, the same gene, Pun1, controls presence vs. absence of pungency. Independent mutations in each species have resulted in loss of function of this gene. In non-pungent C. annuum, the promoter and most of the first exon have been deleted; in non-pungent C. frutescens, part of the second exon has been deleted, causing a frameshift mutation and loss of transcription; while in non-pungent C. chinense, a different small deletion has again caused a frameshift mutation, affecting transcription (Stellari et al., 2010). In these three closely-related species, parallel evolution of non-pungent fruits results from parallel changes in the same gene, but the parallels do not extend to the level of DNA sequences.
In cereals and pseudocereals, starch is the principal reserve stored in the seeds. Starch consists of a mixture of amylopectin and amylose. Starch that lacks amylose is glutinous. In various unrelated species, absence of amylose is associated with mutation in the same gene, waxy. This encodes an enzyme involved in starch synthesis. In grain amaranths, landraces with waxy starch occur in all three domesticated species, but result from different mutations: a SNP in exon 6 in Amaranthus hypochondriacus, a SNP in exon 10 in A. cruentus, and a single nucleotide insertion in exon 8 in Amaranthus caudatus. All three mutations produce a premature stop codon, hence no active enzyme (Park et al., 2010). Waxy starch also occurs in maize. This mutation has not been favoured in Mexico, where maize originated (Whitt et al., 2002), but in China human selection has produced many waxy landraces (Fan et al., 2008). Sequencing studies have shown that these are due to at least two independent mutations, both deletions but affecting different exons. Both differ from the three mutations reported for Amaranthus. Waxy starch therefore represents an example of convergent evolution in dicotyledons and monocotyledons, involving parallel changes in the same gene, though the parallels do not extend to the nucleotide level.
In sweet corn, mutants of the gene sugary 1 (su1) accumulate sucrose rather than starch in the grain. All sweet corns from the United States carry the same amino acid substitution at the same conserved position in the encoded enzyme, presumably making it unable to convert sucrose to starch. This change is not found in sweet corn from Mexico, which carries instead an insertion of a transposable element that disrupts translation of su1 (Whitt et al., 2002). There have therefore been at least two parallel origins of the sweet corn phenotype controlled by su1, but again these parallels do not extend to the level of DNA sequences.
Colour and Shape of the Harvested Organ
Charles Darwin (1868) observed that variation in cultivated plants is greatest in that part of the plant used by man. The most striking variations are in colour and shape. Some may have been selected simply for aesthetic reasons, but different colours and shapes may also act as visible markers for invisible differences in flavour or cooking properties (Boster, 1985). For example, the Aztecs used chile peppers of different colours to flavour different dishes: yellow chile with white fish and also with axolotl, red chile with greyish-brown fish, green chile with frogs (Coe, 1994).
The principal classes of plant pigments are anthocyanins and carotenoids. Biosynthetic pathways for both have been worked out and are common to species in many different families. This affords opportunities for both parallel and convergent evolution. In tomato, the characteristic red fruit colour is due to the carotenoid lycopene. Tomatoes with yellow fruit are homozygous for mutations in the gene PHYTOENE SYNTHASE 1 (PSY1). This encodes an enzyme that catalyses formation of the first carotenoid in the pathway to lycopene. Two different psy1 mutants are known in tomato. One involves insertion of a transposable element, the other involves a short deletion (Jiang et al., 2012). Both result in loss of function of the enzyme, hence absence of red pigment. Parallel yellow phenotypes in different cultivars of tomato therefore result from parallel changes in the same gene, but changes that must have occurred independently. By contrast, red chile pepper fruits owe their colour to two pigments, capsanthin and capsorubin, produced later in the carotenoid pathway than lycopene. Yellow fruit in Capsicum results from mutation in the gene CAPSANTHIN-CAPSORUBIN SYNTHASE (CCS), which encodes the enzyme catalysing the final step in synthesis of these pigments (Lefebvre et al., 1998; Popovsky and Paran, 2000). Parallel evolution of yellow fruit in tomato and Capsicum therefore results from mutations in different genes.
Within Capsicum, at least four independent mutations in CCS are known to produce yellow fruit. In C. annuum, a deletion at the 5′ end of the gene occurs in yellow bell pepper (Lefebvre et al., 1998; Popovsky and Paran, 2000), while a Chinese pungent yellow pepper carries a SNP that produces a premature stop codon (Li et al., 2013). Both changes result in a non-functional enzyme. In C. chinense, one mutation to yellow involves a small deletion that produces a frameshift, hence premature termination of translation, while a second involves a SNP that produces a premature stop codon (Ha et al., 2007), but the SNP in C. chinense is in a different position from that reported by Li et al. (2013) in C. annuum. As in tomato, different changes in the same gene have resulted in repeated parallel evolution of yellow fruit.
PSY1 affects fruit colour in Capsicum as well as in tomato, but psy1 mutants in Capsicum have orange fruits containing reduced amounts of red pigments, rather than yellow fruits with no red pigment. The psy1 mutation in Capsicum is thought to affect a splice site, resulting in premature termination of translation and hence a truncated enzyme. Kim et al. (2010) suggested that the truncated enzyme is unable to anchor properly to the chromoplast membrane, hence has less access to its substrate, but is nevertheless able to produce reduced quantities of red pigments. Orange fruits, visually indistinguishable from those resulting from the psy1 mutation, may also result when red pigments are totally absent, so that colour is provided by carotenoids produced earlier in the carotenoid pathway. In an orange-fruited cultivar of this type, absence of red pigments is caused by deletion of a single nucleotide in the coding sequence of CCS, producing a premature stop codon (Guzman et al., 2010). The two visually indistinguishable classes of orange fruits represent parallel evolution of the same phenotype, but convergent evolution at the genetic level.
In tomatoes grown for canning, elongated fruits are preferred to round. Fruit shape is influenced by the genes fs8.1, SUN, and OVATE (Tanksley, 2004). A candidate gene for fs8.1 is SlSUN22, one of 34 members of the tomato SUN family (Huang et al., 2013). The original sun mutation was caused by a transposon-mediated duplication of a segment of chromosome 10, carrying another member of the SUN family, which was then inserted into chromosome 7 (Xiao et al., 2008). In its original location, this SUN locus is expressed only at a very low level, but in its new location, under control of a different promoter, it is expressed at a much higher level (van der Knaap et al., 2014). A QTL on chromosome 10, previously known to affect fruit shape, may correspond to the SUN locus that was duplicated and transposed. This QTL of tomato is orthologous with fs10.1, associated with round vs. elongate fruits in C. annuum (Ben Chaim et al., 2001), and maps to the same region of the Solanaceae genome as the potato gene Ro, a major QTL affecting tuber shape (van Eck et al., 1994; Borovsky and Paran, 2011). Orthologous QTL may therefore control parallel variations in shape in organs as different as tubers and fruits.
OVATE in tomato is a member of a family of genes called OVATE FAMILY PROTEINS (OFPs). These are widespread in the plant kingdom and act as novel plant growth regulators (Wang et al., 2016). The only mutation to ovate known in tomato results from a SNP that produces a premature stop codon, hence loss of function of the gene product (Tsaballa et al., 2011). In C. annuum, a mutant of the presumed orthologue, CaOvate, does not contain a premature stop codon. Instead, changed expression seems to be responsible for changed fruit shape. In round fruits, CaOvate is expressed most strongly after anthesis, whereas in elongate fruits, it is expressed most strongly before anthesis (Tsaballa et al., 2011).
Much remains to be learned about control of fruit shape in tomato and Capsicum, but evidently there are instances where phenotypic parallels in shape are controlled by parallel changes in orthologous genes, and these parallels may even extend to control of tuber shape in potato.
Adaptation to Long Days
Most crops that originated in the Americas were domesticated in the tropics, many from wild progenitors that require days shorter than a certain critical length to induce flowering. As these crops spread to temperate latitudes, they had to adapt to growing seasons of increasingly longer days. This adaptation usually involved loss of sensitivity to daylength. Closer adaptation to specific environments, in which length of growing season is influenced by factors such as drought or cold, was achieved by selection for earlier or later flowering under any given photoperiod.
Control of time to flowering is complex. More than 90 relevant genes have been identified in Arabidopsis and their homologues in crop species are being isolated and studied. Gibberellin, vernalisation and autonomous pathways act and interact, together with, most importantly, the photoperiodic pathway. Basic features of the photoperiodic pathway appear to be conserved across a wide range of species, though there are differences in detail (Andrés and Coupland, 2012; Blümel et al., 2015). The names used here for genes and gene products are those employed in Arabidopsis thaliana, a model species for these investigations. The flower-inducing signal (FT), also known as florigen, is encoded by the gene FLOWERING LOCUS T (FT), which is activated by the transcription factor CO, produced by the gene CONSTANS. Transcription of CO is repressed by certain factors that are degraded by a light-induced interaction between the product of the gene GIGANTEA (GI) and an enzyme produced by a further gene. Light and dark affect the stability of CO after translation. Interactions with light are mediated by the photoreceptor phytochrome B, encoded by PHYTOCHROME B (PHYB). FT and CO are both expressed in leaves. FT then moves through the phloem to the shoot apical meristem, where it combines with FD, the protein product of FLOWERING LOCUS D. The FT-FD complex activates SUPPRESSION OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which encodes a transcription factor that activates a series of other transcription factors. These in turn activate floral identity genes that irreversibly convert a vegetative to a floral meristem.
Given the complexity of this pathway, it is not surprising that photoperiodic sensitivity has been lost through different genetic changes in different crops. In Sea Island cotton (G. barbadense), a major locus, Gb-Ppd1, is associated with the day-neutral phenotype. The underlying gene has not yet been identified, despite examination of 110 possible candidates (Zhu and Kuraparthy, 2014). In contrast, in upland cotton (G. hirsutum), many genes appear to be involved, including three members of the CONSTANS-LIKE (COL) gene family, all three of which have apparently undergone selection during domestication. Both G. hirsutum and G. barbadense are AADD allotetraploids. Song et al. (2017) observed that the COL2 locus in the A subgenome is hypermethylated, hence repressed, in both species, but in photoperiodically insensitive accessions of both species, the D subgenome homoeologue of COL2 is less methylated, hence more strongly expressed. As Song et al. (2017) noted, there are also other QTL associated with loss of sensitivity to photoperiod in cotton. Nevertheless, the data do suggest that change in a CO-like gene, or in its regulation, is one cause of adaptation to long-day growing seasons, possibly in both species of cotton.
Sunflower (Helianthus annuus) is unusual in that genotypes adapted to short or long days, as well as genotypes insensitive to photoperiod, all occur among wild populations and among domesticated accessions of this single species (Blackman, 2013). In wild sunflowers, insensitivity to photoperiod and adaptation to long days are restricted to different parts of their extensive range, outside the region in which sunflower was probably domesticated (Blackman, 2013). Duplication of the original FT locus has produced four paralogues, HaFT1 to HaFT4, which seem to have diverged in function (Blackman, 2013). HaFT1 is expressed in the shoot apex and is a candidate gene for a major QTL that affects time to flowering. HaFT2 and HaFT4 seem to encode the florigen. They are expressed in leaves under inductive photoperiods, like FT genes in other species. In one wild population and one cultivar, which have independently developed insensitivity to photoperiod, HaFT4 is expressed under both long- and short-days. Blackman (2013) considered that changes in either the promoter of HaFT4 or in other genes that regulate HaFT4 are probably involved in both cases, but could not determine whether the same changes are implicated in both. In contrast, adaptation to long days apparently evolved by different means in wild and domesticated sunflowers. Long days are usually non-inductive, but in one long-day cultivar, both HaFT2 and HaFT4 are expressed in long days, indicating that there has been some change in the regulation of these genes. However, in a long-day wild sunflower, neither HaFT2 nor HaFT4 is expressed, though expression of the homologue of SOC1 has changed (Blackman, 2013). Loss of sensitivity to photoperiod in wild and domesticated sunflowers may therefore represent parallel evolution at the genetic as well as phenotypic level, but parallel evolution of phenotypes adapted to long days in wild and domesticated sunflowers represents convergent evolution at the genetic level.
In common bean, two QTL are consistently implicated in control of flowering time (Kornegay et al., 1993; Koinange et al., 1996; Gu et al., 1998). Ppd has the larger effect and determines sensitivity to photoperiod, while Hr determines degree of response to photoperiod. Kornegay et al. (1993) considered that insensitivity to long days involved the same two QTL in both Andean and Mesoamerican common bean, although these were independently domesticated, but Gu et al. (1998) found different markers associated with earliness in Mesoamerican and Andean beans. Bellucci et al. (2014) presented evidence that a DNA sequence homologous to GI was selected during domestication of Mesoamerican beans, but there is no other information on candidate genes for QTL associated with photoperiodicity or time to flowering in common bean, or their modes of action (Kwak et al., 2008).
Tropical maize has retained the short-day adaptation of its wild relative, teosinte, whereas temperate maize is insensitive to day-length, so flowers sufficiently early in the long-day growing seasons of temperate latitudes to mature its grain before the short days and cold temperatures of autumn. Although a large number of QTL affect flowering time in maize, relatively few affect sensitivity to photoperiod. One of these few is ZmCCT (Hung et al., 2012). This gene represses expression of the probable orthologue of FT, ZCN8 (Lazakis et al., 2011; Yang et al., 2013; Mascheretti et al., 2015). An insertion of a transposable element in the upstream regulatory region of ZmCCT suppresses transcription of ZmCCT under long days, thereby up-regulating expression of ZCN8, resulting in earlier flowering. The insertion is apparently absent in teosinte, but was subjected to a strong selective sweep in photoperiod-insensitive accessions of temperate maize. Yang et al. (2013) therefore concluded that the insertion occurred after domestication and was favoured by selection as maize spread into long-day growing seasons of temperate latitudes. Lazakis et al. (2011) suggested that in temperate maize, the photoperiodic pathway, in which ZCN8 is activated in short days, has been largely superseded by the autonomous pathway, in which ZCN8 is activated by the protein product of the gene indeterminate 1(id1). Although all higher plants contain a large family of id1-related genes, there is no clear orthologue of id1 in dicotyledons.
Convergent evolution of photoperiod-insensitivity has therefore been achieved by different means in different crops, even though the pathways that regulate the transition to flowering, particularly the CO-FT pathway, are extensively conserved. CO-like genes have been major players in cotton, FT genes in sunflower, and suppression of ZmCCT, together with probable increased importance of the autonomous pathway, in maize.
Flowering is not the only process in plants that is controlled by photoperiod. In both potato (Solanum tuberosum) and Jerusalem artichoke (Helianthus tuberosus), tubers are formed in response to short days, and orthologues of FT appear to be involved (Navarro et al., 2011; Blackman, 2013). The process has been most studied in potato, since its success in temperate latitudes depends on ability to form tubers in long days. The orthologue of FT in potato is StSP6A, while the othologue of CO is StCOL1 (Navarro et al., 2011; González-Schain et al., 2012). The product of StCOL1 activates StSP5G and the StSP5G protein in turn represses transcription of StSP6A. StCOL1 is degraded in the dark, so only a low level is present in short days. This releases repression by StSP5G of StSP6A, so tubers are formed. StCOL1 is stabilised in the light and this is dependent on phytochrome B. When PHYB is suppressed, tuber formation is no longer sensitive to photoperiod (Abelenda et al., 2016). Adaptation to long days has apparently evolved at least twice; once in Chile as potatoes spread south from the Andean region, and again in Europe after post-Conquest introduction of Andean short-day potatoes. However, it has yet to be shown what genes are involved in loss of sensitivity to daylength in each case, and whether the same genes are involved in different photoperiod-insensitive potatoes.
Discussion and Conclusions
Parallel vs. convergent evolution are terms that were initially used for discussion of phenotypic similarities between closely vs. distantly related species. Their definition is subjective, in that it does not specify how long ago lineages must have diverged to be considered no longer closely related. For example, waxy starch occurs in some dicotyledonous and some monocotyledonous crops. Given that all flowering plants descend ultimately from a common ancestor, this may be considered as either parallel or convergent evolution, depending on whether dicotyledons and monocotyledons are regarded as closely related, because they are all flowering plants, or distantly related, because they belong to lineages that diverged early in the evolution of flowering plants.
At the genetic level, parallel evolution implies that similar phenotypes are controlled by the same (i.e., homologous) genes, whereas convergent evolution implies that similar phenotypes are controlled by genes that are not homologous. This again raises the problem of subjectivity, this time in the definition of homology. Flowering plants have undergone many cycles of gene duplication, so many genes now occur in multi-gene families. If the original gene controlled multiple functions, these may become partitioned among different members of the family. Alternatively, or additionally, some family members may acquire new functions and/or changes in expression. Different members of the same gene family may then be favoured by selection during domestication in different crops. All this leads to differences in degrees of homology and blurs the distinction between parallel vs. convergent evolution at the genetic level. An example occurs among cereals (Meng et al., 2011). In rice, the gene Hd3a encodes the florigen. In maize, 15 members of the ZCN family are phylogenetically related to Hd3a. ZCN15 maps to the same region of the genome as Hd3a, so in that respect is its closest homologue, but ZCN15 is expressed in maize kernels, not leaves, and is not involved in control of flowering. ZCN8 is expressed in leaves and does encode a florigen, so is functionally homologous to Hd3a although located elsewhere in the genome. The control of flowering in rice and maize thus may, or may not, be considered to involve parallels at the genetic level.
Many phenotypic traits listed as components of the domestication and diversification syndromes are complex characters. When these are broken down into their components, some apparent parallels disappear. For example, increased size of fruit is a frequent response to human selection in species domesticated for their fruit. In tomato and Mexican green tomato (P. philadelphica), development of large fruits in domesticated accessions of both species initially seems to be an example of parallel phenotypic evolution in species belonging to the same family. But on closer examination, large fruits in tomato are found to result from increased numbers of cells in various parts of the fruit, together with more carpels in the fruit, while in Mexican green tomato, large fruits are due primarily to increased cell size (Wang et al., 2014). Carpel number, cell size and cell number are controlled by different genes so, not surprisingly, large fruits in tomato and Mexican green tomato are controlled principally by non-homologous genes. Dissection of this complex character, fruit size, into simpler components thus shows that the phenotypic parallel is more limited than it initially appeared.
In C. annuum, fruit size has likewise increased under human selection. This increase is controlled by numerous QTL, many of which are orthologous to those of tomato. This therefore seems a good example of parallel evolution at levels of both phenotype and gene. However, the QTL with greatest effect on fruit weight in tomato, FW2.2, has a lesser effect in C. annuum, probably because FW2.2 appears to act principally in the placenta and central axis of the fruit, which make up much of the mass of tomato fruits but disproportionately less of the hollow fruits of Capsicum (van der Knaap et al., 2014). Large fruits of both Capsicum and tomato have more cells in the fruit wall, but the QTL primarily responsible has a smaller effect in tomato than its orthologue does in C. annuum. The phenotypic and genetic parallels between large fruits in tomato and C. annuum therefore become less perfect when examined in detail. This suggests that parallelism is a matter of degree, like relationship or homology. Furthermore, in assessing possible cases of parallel evolution, it is important to compare only the comparable.
Difficulties in defining parallel and convergent evolution led Arendt and Reznick (2008) to conclude that applying these terms at the level of molecular genetics leads only to confusion. They advocated use of a single term, and suggested “convergent evolution,” for evolution at both phenotypic and genetic levels. However, this term carries the baggage of past usage. I have therefore continued to distinguish parallel and convergent evolution and have applied these terms at phenotypic, genetic, and DNA sequence levels. This may sometimes seem confusing, and may also seem to be governed by the principle employed by Lewis Carroll's Humpty-Dumpty, who told Alice: “When I use a word, it means exactly what I choose it to mean – neither more nor less.” Nevertheless, clarity in definition and consistency in use of existing terms seems preferable to coining new, hence unfamiliar, ones.
No examples of parallel changes in DNA sequence exist among the cases of parallel genetic evolution reviewed here. I know of only one possible example among crops domesticated in the Americas: occurrence, in semi-domesticated cherry tomato and fully domesticated modern tomatoes, of bushy plants associated with a recessive mutation in the SELF-PRUNING (SP) gene. Pnueli et al. (1998) found that mutants in both types of tomato carry the same amino acid substitution at the same position in the encoded protein. They argued that the two identical mutations arose independently. However, this example seems to be the exception, not the rule. In most cases of parallel genetic evolution, parallel phenotypes have been produced by different (convergent) changes in DNA sequence of the same gene. These changes usually result in loss of function of the gene or its product, less often in changed expression of the gene.
When phenotypic and genetic levels are considered together, any combination of parallel and convergent evolution may occur, as Table 3 shows. There are examples of parallel phenotypes controlled by homologous (parallel) genes in closely related species; convergent phenotypes controlled by homologous genes in distantly related species; parallel phenotypes controlled by non-homologous (convergent) genes in closely related species; and convergent phenotypes controlled by non-homologous genes in distantly related species. Moreover, all these combinations are found in both domestication and diversification traits.
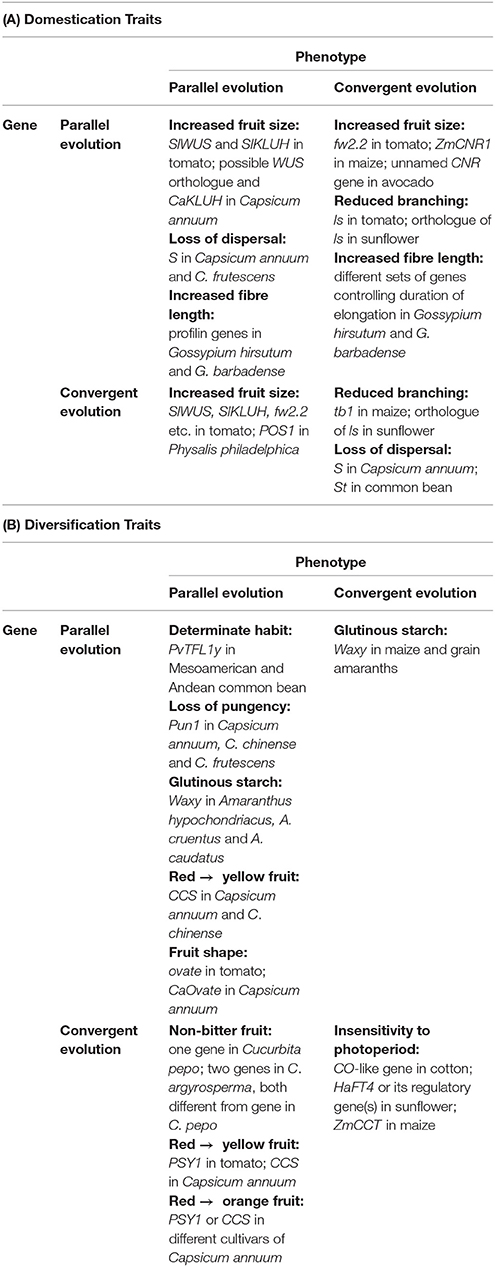
Table 3. Examples of parallel and convergent evolution at levels of phenotype and gene in some domestication and diversification traits.
Glémin and Bataillon (2009) and Martínez-Ainsworth and Tenaillon (2016) considered that evolution of similar phenotypes during domestication generally does not result from parallel genetic changes. Some traits, particularly those that involve complex developmental pathways and/or complex networks of genes, do indeed seem to involve evolution of similar phenotypes by different genetic routes. For example, in loss of sensitivity to daylength, CO-like genes appear to be major players in cotton (Song et al., 2017), whereas genes of the FT family appear more important in sunflower (Blackman, 2013). On the other hand, development of large fruit in two distantly related dicotyledons, tomato and avocado, seems to involve genes of the CELL NUMBER REGULATOR (CNR) family in both genera (Dahan et al., 2010; van der Knaap et al., 2014).
Poncet et al. (2004) considered that parallel evolution at the genetic level, involving orthologous loci, is more likely in crops that belong to the same family. Some of the cases discussed here support this view, but others do not. For example, in the Solanaceae, several orthologous loci are implicated in evolution of large fruit in tomato and C. annuum, but parallel evolution of yellow fruit is controlled by different, non-orthologous, genes in these two crops. Available data are as yet insufficient to assess reliably the number of examples for or against the opinion of Poncet et al. (2004).
Lenser and Theißen (2013) suggested that traits resulting from simple metabolic pathways, rather than from complex developmental networks, are more likely to provide examples of parallel genetic variation. Once more, examples can be cited both for and against this view. Waxy starch results from a change in a relatively simple biosynthetic pathway, controlled by an orthologous gene in both grain amaranths and maize (Whitt et al., 2002; Park et al., 2010). On the other hand, two visually indistinguishable types of orange fruit in C. annuum result from changes in different genes in the pathway of pigment biosynthesis (Guzman et al., 2010; Kim et al., 2010). Again, there are insufficient data to quantify relative frequencies of parallel vs. convergent evolution in traits resulting from simple vs. complex pathways.
Gross and Olsen (2010) argued that diversification traits are more likely to provide examples of parallel genetic evolution than domestication traits. This seemed plausible, because diversification often involves changes in simpler characters than domestication, but the data are again inconclusive and examples cited above include some for and some against this proposition.
Technical advances, such as next-generation sequencing and various methods for studying levels and locations of gene expression, are producing an information explosion regarding details of developmental processes and their control in different plant species. Many traits of the domestication syndrome, and some associated with diversification, are now known to be controlled by complex networks of genes. These networks, or portions of them, are often conserved to considerable extents, though regulation of the genes involved may vary. This widespread conservation suggests that similar phenotypic changes, even in distantly related crop species, could result from parallel genetic changes. On the other hand, the complexity of the pathways suggests that disruption at different points (convergent evolution) could produce parallel phenotypes. Most current investigations aim to elucidate the pathways and genes involved in these networks for a few model species only. Less attention has so far been directed toward establishing how, or at what points, these pathways and networks have been changed or disrupted through natural and/or human selection, and whether, or to what extent, these pathways or networks differ in different species in the same family.
This review is therefore both premature and, given the rate at which new information is accumulating, likely to be out-of-date before it is published. Nevertheless, it may serve to highlight the many gaps in the information available and the many areas where additional data are needed. Both domestication and diversification evidently involve both parallel and convergent evolution, acting at any or all levels of phenotype, gene, or nucleotide sequence, with relative frequencies still to be determined. As Doebley and Stec (1991) concluded many years ago, evolution seems essentially opportunistic, making use of whatever variation is available.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
Frontiers have granted me a full fee waiver.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbo, S., van-Oss, R., Gopher, A., Saranga, Y., Ofner, I., and Peleg, Z. (2014). Plant domestication versus crop evolution: a conceptual framework for cereals and grain legumes. Trends Plant Sci. 19, 351–360. doi: 10.1016/j.tplants.2013.12.002
Abelenda, J. A., Cruz-Oró, E., Franco-Zorrilla, J. M., and Prat, S. (2016). Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26, 872–881. doi: 10.1016/j.cub.2016.01.066
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Arendt, J., and Reznick, D. (2008). Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32. doi: 10.1016/j.tree.2007.09.011
Bao, Y., Hu, G., Flagel, L. E., Salmon, A., Bezanilla, M., Paterson, A. H., et al. (2011). Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc. Natl. Acad. Sci. U.S.A.108, 21152–21157. doi: 10.1073/pnas.1115926109
Barchi, L., Lefebvre, V., Sage-Palloix, A. M., Lanteri, S., and Palloix, A. (2009). QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 118, 1157–1171. doi: 10.1007/s00122-009-0970-0
Baskin, J. M., and Baskin, C. C. (2004). A classification system for seed dormancy. Seed Sci. Res. 14, 1–16. doi: 10.1079/SSR2003150
Bellucci, E., Bitocchi, E., Ferrarini, A., Benazzo, A., Biagetti, E., Klie, S., et al. (2014). Decreased nucleotide and expression diversity and modified coexpression patterns characterize domestication in the common bean. Plant Cell 26, 1901–1912. doi: 10.1105/tpc.114.124040
Ben Chaim, A., Paran, I., Grube, R. C., Jahn, M., Van Wijk, R., and Peleman, J. (2001). QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 102, 1016–1028. doi: 10.1007/s001220000461
Benz, B. F. (2006). “Maize in the Americas,” in Histories of Maize: Multidisciplinary Approaches to the Prehistory, Linguistics, Biogeography, Domestication and Evolution of Maize, eds J. E. Staller, R. H. Tykot, and B. F. Benz (Burlington, MA: Academic Press), 9–20.
Blackman, B. K. (2013). Interacting duplications, fluctuating selection, and convergence: the complex dynamics of flowering time evolution during sunflower domestication. J. Exp. Bot. 64, 421–431. doi: 10.1093/jxb/ers359
Blanca, J., Montero-Pau, J., Sauvage, C., Bauchet, G., Illa, E., Díez, M. J., et al. (2015). Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genomics 16:257. doi: 10.1186/s12864-015-1444-1
Blümel, M., Dally, N., and Jung, C. (2015). Flowering time regulation in crops—what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Borchers, E. A., and Taylor, R. T. (1988). Inheritance of fruit bitterness in a cross of Cucurbita mixta × C. pepo. HortScience 23, 603–604.
Borovsky, Y., and Paran, I. (2011). Characterization of fs10. 1, a major QTL controlling fruit elongation in Capsicum. Theor. Appl. Genet. 123, 657–665. doi: 10.1007/s00122-011-1615-7
Boster, J. S. (1985). Selection for perceptual distinctiveness: evidence from Aguaruna cultivars of Manihot esculenta. Econ. Bot. 39, 310–325. doi: 10.1007/BF02858802
Chakrabarti, M., Zhang, N., Sauvage, C., Muños, S., Blanca, J., Cañizares, J., et al. (2013). A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Nat. Acad. Sci. U.S.A. 110, 17125–17130. doi: 10.1073/pnas.1307313110
Chaudhary, B., Hovav, R., Flagel, L., Mittler, R., and Wendel, J. F. (2009). Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium). BMC Genomics 10:378. doi: 10.1186/1471-2164-10-378
Clement, C. R. (1999). 1492 and the loss of Amazonian crop genetic resources. I. The relation between domestication and human population decline. Econ. Bot. 53, 188–202. doi: 10.1007/BF02866498
Cong, B., Liu, J., and Tanksley, S. D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. U.S.A. 99, 13606–13611. doi: 10.1073/pnas.172520999
Dahan, Y., Rosenfeld, R., Zadiranov, V., and Irihimovitch, V. (2010). A proposed conserved role for an avocado fw2.2-like gene as a negative regulator of fruit cell division. Planta 232, 663–676. doi: 10.1007/s00425-010-1200-3
Davis, P. H., and Heywood, V. H. (1963). Principles of Angiosperm Taxonomy. Edinburgh: Oliver and Boyd.
Denna, D. W., and Munger, H. M. (1963). Morphology of the bush and vine habits and the allelism of the bush genes in Cucurbita maxima and C. pepo squash. Proc. Am. Soc. Horticult. Sci. 82, 370–377.
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Doebley, J., and Stec, A. (1991). Genetic analysis of the morphological differences between maize and teosinte. Genetics 129, 285–295.
Dong, Y., and Wang, Y. Z. (2015). Seed shattering: from models to crops. Front. Plant Sci. 6:476. doi: 10.3389/fpls.2015.00476
Estornell, L. H., Agust,í, J., Merelo, P., Talón, M., and Tadeo, F. R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 199, 48–60. doi: 10.1016/j.plantsci.2012.10.008
Fan, L., Quan, L., Leng, X., Guo, X., Hu, W., Ruan, S., et al. (2008). Molecular evidence for post-domestication selection in the Waxy gene of Chinese waxy maize. Mol. Breed. 22, 329–338. doi: 10.1007/s11032-008-9178-2
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Fuller, D. Q., Allaby, R. G., and Stevens, C. (2010). Domestication as innovation: the entanglement of techniques, technology and chance in the domestication of cereal crops. World Archaeol. 42, 13–28. doi: 10.1080/00438240903429680
Gaut, B. S. (2015). Evolution is an experiment: assessing parallelism in crop domestication and experimental evolution: (Nei Lecture, SMBE 2014, Puerto Rico). Mol. Biol. Evol. 32, 1661–1671. doi: 10.1093/molbev/msv105
Glémin, S., and Bataillon, T. (2009). A comparative view of the evolution of grasses under domestication. New Phytol. 183, 273–290. doi: 10.1111/j.1469-8137.2009.02884.x
González-Schain, N. D., Díaz-Mendoza, M., Zurczak, M., and Suárez-López, P. (2012). Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 70, 678–690. doi: 10.1111/j.1365-313X.2012.04909.x
Graeber, K., Nakabayashi, K., Miatton, E., Leubner-Metzger, G., and Soppe, W. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35, 1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x
Gross, B. L., and Olsen, K. M. (2010). Genetic perspectives on crop domestication. Trends Plant Sci. 15, 529–537. doi: 10.1016/j.tplants.2010.05.008
Gu, W., Zhu, J., Wallace, D. H., Singh, S. P., and Weeden, N. F. (1998). Analysis of genes controlling photoperiod sensitivity in common bean using DNA markers. Euphytica 102, 125–132. doi: 10.1023/A:1018340514388
Guan, J. C., Koch, K. E., Suzuki, M., Wu, S., Latshaw, S., Petruff, T., et al. (2012). Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 160, 1303–1317. doi: 10.1104/pp.112.204503
Guo, M., Rupe, M. A., Dieter, J. A., Zou, J., Spielbauer, D., Duncan, K. E., et al. (2010). Cell Number Regulator1 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. Plant Cell 22, 1057–1073. doi: 10.1105/tpc.109.073676
Guzman, I., Hamby, S., Romero, J., Bosland, P. W., and O'Connell, M. A. (2010). Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant Sci. 179, 49–59. doi: 10.1016/j.plantsci.2010.04.014
Ha, S. H., Kim, J. B., Park, J. S., Lee, S. W., and Cho, K. J. (2007). A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J. Exp. Bot. 58, 3135–3144. doi: 10.1093/jxb/erm132
Harlan, J. R. (1992). Crops and Man. Madison, Wis.: American Society of Agronomy: Crop Science Society of America.
Harris, D. R. (1989). “An evolutionary continuum of people – plant interaction,” in Foraging and Farming: The Evolution of Plant Exploitation, eds D. R. Harris and G. C. Hillman (London: Unwin Hyman), 11–26.
Huang, Z., Van Houten, J., Gonzalez, G., Xiao, H., and van der Knaap, E. (2013). Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genomics 288, 111–129. doi: 10.1007/s00438-013-0733-0
Hung, H. Y., Shannon, L. M., Tian, F., Bradbury, P. J., Chen, C., Flint-Garcia, S. A., et al. (2012). ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. U.S.A. 109, E1913–E1921. doi: 10.1073/pnas.1203189109
Jiang, N., Visa, S., Wu, S., and van der Knaap, E. (2012). “Rider transposon insertion and phenotypic change in tomato,” in Plant Transposable Elements, eds M. A. Grandbastien and J. M. Casacuberta (Heidelberg: Springer). 297–312.
Jiang, Z., Xu, G., Jing, Y., Tang, W., and Lin, R. (2016). Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 7:12377. doi: 10.1038/ncomms12377
Johns, T., and Galindo Alonso, J. (1990). Glycoalkaloid change during the domestication of the potato, Solanum Section Petota. Euphytica 50, 203–210. doi: 10.1007/BF00023646
Kim, O. R., Cho, M. C., Kim, B. D., and Huh, J. H. (2010). A splicing mutation in the gene encoding phytoene synthase causes orange coloration in Habanero pepper fruits. Mol. Cells 30, 569–574. doi: 10.1007/s10059-010-0154-4
Koinange, E. M., Singh, S. P., and Gepts, P. (1996). Genetic control of the domestication syndrome in common bean. Crop Sci. 36, 1037–1045. doi: 10.2135/cropsci1996.0011183X003600040037x
Kornegay, J., White, J. W., Dominguez, J. R., Tejado, G., and Cajiao, C. (1993). Inheritance of photoperiod response in Andean and Mesoamerican common bean. Crop Sci. 33, 977–984. doi: 10.2135/cropsci1993.0011183X003300050021x
Kwak, M., Toro, O., Debouck, D. G., and Gepts, P. (2012). Multiple origins of the determinate growth habit in domesticated common bean (Phaseolus vulgaris). Ann. Bot. 110, 1573–1580. doi: 10.1093/aob/mcs207
Kwak, M., Velasco, D., and Gepts, P. (2008). Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 99, 283–291. doi: 10.1093/jhered/esn005
Larson, G., Piperno, D. R., Allaby, R. G., Purugganan, M. D., Andersson, L., Arroyo-Kalin, M., et al. (2014). Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. U.S.A. 111, 6139–6146. doi: 10.1073/pnas.1323964111
Lazakis, C. M., Coneva, V., and Colasanti, J. (2011). ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Bot. 62, 4833–4842. doi: 10.1093/jxb/err129
Lefebvre, V., Kuntz, M., Camara, B., and Palloix, A. (1998). The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Mol. Biol. 36, 785–789. doi: 10.1023/A:1005966313415
Lenser, T., and Theißen, G. (2013). Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 18, 704–714. doi: 10.1016/j.tplants.2013.08.007
Li, Z., Wang, S., Gui, X.-L., Chang, X.-B., and Gong, Z.-H. (2013). A further analysis of the relationship between yellow ripe-fruit colour and the capsanthin-capsorubin synthase gene in pepper (Capsicum sp.) indicated a new mutant variant in C. annuum and a tandem repeat structure in promoter region. PLoS ONE 8:e61996. doi: 10.1371/journal.pone.0061996
Mandel, J. R., McAssey, E. V., Nambeesan, S., Garcia-Navarro, E., and Burke, J. M. (2014). Molecular evolution of candidate genes for crop-related traits in sunflower (Helianthus annuus L.). PLoS ONE 9:e99620. doi: 10.1371/journal.pone.0099620
Mao, L., Begum, D., Chuang, H. W., Budiman, M. A., Szymkowiak, E., Irish, E. J., et al. (2000). JOINTLESS is a MADS-box gene controlling flower abscission zone development. Nature 406, 910–913. doi: 10.1038/35022611
Martínez-Ainsworth, N. E., and Tenaillon, M. I. (2016). Superheroes and masterminds of plant domestication. C. R. Biol. 339, 268–273. doi: 10.1016/j.crvi.2016.05.005
Mascheretti, I., Turner, K., Roberta, S. B., Hand, A., Colasanti, J., and Rossi, V. (2015). Florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiol. 168, 1351–1363. doi: 10.1104/pp.15.00535
Meng, X., Muszynski, M. G., and Danilevskaya, O. N. (2011). The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23, 942–960. doi: 10.1105/tpc.110.081406
Meyer, R. S., DuVal, A. E., and Jensen, H. R. (2012). Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196, 29–48. doi: 10.1111/j.1469-8137.2012.04253.x
Meyer, R. S., and Purugganan, M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
Morrell, P. L., Buckler, E. S., and Ross-Ibarra, J. (2012). Crop genomics: advances and applications. Nat. Rev. Genet. 13, 85–96. doi: 10.1038/nrg3097
Navarro, C., Abelenda, J. A., Cruz-Oró, E., Cuéllar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. doi: 10.1038/nature10431
Née, G., Kramer, K., Nakabayashi, K., Yuan, B., Xiang, Y., Miatton, E., et al. (2017). DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 8:72. doi: 10.1038/s41467-017-00113-6
Nonogaki, H. (2014). Seed dormancy and germination—emerging mechanisms and new hypotheses. Front. Plant Sci. 5:233. doi: 10.3389/fpls.2014.00233
Olsen, K. M., and Wendel, J. F. (2013). Crop plants as models for understanding plant adaptation and diversification. Front. Plant Sci. 4:290. doi: 10.3389/fpls.2013.00290
Paran, I., and van der Knaap, E. (2007). Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 58, 3841–3852. doi: 10.1093/jxb/erm257
Park, Y.-J., Nemoto, K., Nishikawa, T., Matsushima, K., Minami, M., and Kawase, M. (2010). Waxy strains of three amaranth grains raised by different mutations in the coding region. Mol. Breed. 25, 623–635. doi: 10.1007/s11032-009-9360-1
Pnueli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., et al. (1998). The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989.
Poncet, V., Robert, T., Sarr, A., and Gepts, P. (2004). “Quantitative trait locus analyses of the domestication syndrome and domestication process,” in Encyclopedia of Plant and Crop Science, ed R. M. Goodman (New York, NY: M. Dekker), 1069–1074.
Popovsky, S., and Paran, I. (2000). Molecular genetics of the y locus in pepper: its relation to capsanthin-capsorubin synthase and to fruit colour. Theor. Appl. Genet. 101, 86–89. doi: 10.1007/s001220051453
Rameau, C., Bertheloot, J., Leduc, N., Andrieu, B., Foucher, F., and Sakr, S. (2015). Multiple pathways regulate shoot branching. Front. Plant Sci. 5:741. doi: 10.3389/fpls.2014.00741
Rao, G. U., and Paran, I. (2003). Polygalacturonase: a candidate gene for the soft flesh and deciduous fruit mutation in Capsicum. Plant Mol. Biol. 51, 135–141. doi: 10.1023/A:1020771906524
Repinski, S. L., Kwak, M., and Gepts, P. (2012). The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor. Appl. Genet. 124, 1539–1547. doi: 10.1007/s00122-012-1808-8
Sang, T. (2009). Genes and mutations underlying domestication transitions in grasses. Plant Physiol. 149, 63–70. doi: 10.1104/pp.108.128827
Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G., and Theres, K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. U.S.A. 96, 290–295. doi: 10.1073/pnas.96.1.290
Smýkal, P., Vernoud, V., Blair, M. W., Soukup, A., and Thompson, R. D. (2014). The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 5:351. doi: 10.3389/fpls.2014.00351
Somssich, M., Je, B. I., Simon, R., and Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. doi: 10.1242/dev.133645
Song, Q., Zhang, T., Stelly, D. M., and Chen, Z. J. (2017). Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 18:99. doi: 10.1186/s13059-017-1229-8
Stellari, G. M., Mazourek, M., and Jahn, M. M. (2010). Contrasting modes for loss of pungency between cultivated and wild species of Capsicum. Heredity 104, 460–471. doi: 10.1038/hdy.2009.131
Tanksley, S. D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16, S181–S189. doi: 10.1105/tpc.018119
Tsaballa, A., Pasentsis, K., Darzentas, N., and Tsaftaris, A. S. (2011). Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 11:46. doi: 10.1186/1471-2229-11-46
van der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J. P., Illa-Berenguer, E., Huang, Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5:227. doi: 10.3389/fpls.2014.00227
van Eck, H. J., Jacobs, J. M., Stam, P., Ton, J., Stiekema, W. J., and Jacobsen, E. (1994). Multiple alleles for tuber shape in diploid potato detected by qualitative and quantitative genetic analysis using RFLPs. Genetics 137, 303–309.
Wang, L., He, L., Li, J., Zhao, J., Li, Z., and He, C. (2014). Regulatory change at Physalis Organ Size 1 correlates to natural variation in tomatillo reproductive organ size. Nat Commun. 5:4271. doi: 10.1038/ncomms5271
Wang, L., Li, J., Zhao, J., and He, C. (2015). Evolutionary developmental genetics of fruit morphological variation within the Solanaceae. Front. Plant Sci. 6:248. doi: 10.3389/fpls.2015.00248
Wang, S., Chang, Y., and Ellis, B. (2016). Overview of OVATE FAMILY PROTEINS, a novel class of plant-specific growth regulators. Front. Plant Sci. 7:417. doi: 10.3389/fpls.2016.00417
Weiss, A. N., Primer, S. B., Pace, B. A., and Mercer, K. L. (2013). Maternal effects and embryo genetics: germination and dormancy of crop–wild sunflower hybrids. Seed Sci. Res. 23, 241–255. doi: 10.1017/S0960258513000226
Whipple, C. J., Kebrom, T. H., Weber, A. L., Yang, F., Hall, D., Meeley, R., et al. (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. U.S.A.108, E506–E512. doi: 10.1073/pnas.1102819108
Whitt, S. R., Wilson, L. M., Tenaillon, M. I., Gaut, B. S., and Buckler, E. S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 12959–12962. doi: 10.1073/pnas.202476999
Xiao, H., Jiang, N., Schaffner, E., Stockinger, E. J., and van der Knaap, E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530. doi: 10.1126/science.1153040
Yang, Q., Li, Z., Li, W., Ku, L., Wang, C., Ye, J., et al. (2013). CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. U.S.A. 110, 16969–16974. doi: 10.1073/pnas.1310949110
Zhang, G., Ren, Y., Sun, H., Guo, S., Zhang, F., Zhang, J., et al. (2015). A high-density genetic map for anchoring genome sequences and identifying QTLs associated with dwarf vine in pumpkin (Cucurbita maxima Duch.). BMC Genomics 16:1101. doi: 10.1186/s12864-015-2312-8
Keywords: domestication, diversification, parallel evolution, convergent evolution, American crops
Citation: Pickersgill B (2018) Parallel vs. Convergent Evolution in Domestication and Diversification of Crops in the Americas. Front. Ecol. Evol. 6:56. doi: 10.3389/fevo.2018.00056
Received: 05 September 2017; Accepted: 17 April 2018;
Published: 07 May 2018.
Edited by:
Charles Roland Clement, National Institute of Amazonian Research, BrazilReviewed by:
Vigouroux Yves, Institut de recherche pour le développement (IRD), FranceAlessandro Alves-Pereira, Universidade Estadual de Campinas, Brazil
Copyright © 2018 Pickersgill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Pickersgill, Yi5waWNrZXJzZ2lsbDg4OEBidGludGVybmV0LmNvbQ==
 Barbara Pickersgill
Barbara Pickersgill